Gram-positive bacteria process and release small peptides, or pheromones, that act as signals for the induction of adaptive traits, including those involved in pathogenesis. One class of small signaling pheromones is the cyclic autoinducing peptides (AIPs), which regulate expression of genes that orchestrate virulence and persistence in a range of microbes, including staphylococci, listeriae, clostridia, and enterococci.
KEYWORDS: Agr, Staphylococcus aureus, accessory gene regulator, peptidase, peptide signaling, quorum sensing, virulence
ABSTRACT
Gram-positive bacteria process and release small peptides, or pheromones, that act as signals for the induction of adaptive traits, including those involved in pathogenesis. One class of small signaling pheromones is the cyclic autoinducing peptides (AIPs), which regulate expression of genes that orchestrate virulence and persistence in a range of microbes, including staphylococci, listeriae, clostridia, and enterococci. In a genetic screen for Staphylococcus aureus secreted virulence factors, we identified an S. aureus mutant containing an insertion in the gene SAUSA300_1984 (mroQ), which encodes a putative membrane-embedded metalloprotease. A ΔmroQ mutant exhibited impaired induction of Toll-like receptor 2-dependent inflammatory responses from macrophages but elicited greater production of the inflammatory cytokine interleukin-1β and was attenuated in a murine skin and soft tissue infection model. The ΔmroQ mutant phenocopies an S. aureus mutant containing a deletion of the accessory gene regulatory system (Agr), wherein both strains have significantly reduced production of secreted toxins and virulence factors but increased surface protein A abundance. The Agr system controls virulence factor gene expression in S. aureus by sensing the accumulation of AIP via the histidine kinase AgrC and the response regulator AgrA. We provide evidence to suggest that MroQ acts within the Agr pathway to facilitate the optimal processing or export of AIP for signal amplification through AgrC/A and induction of virulence factor gene expression. Mutation of MroQ active-site residues significantly reduces AIP signaling and attenuates virulence. Altogether, this work identifies a new component of the Agr quorum-sensing circuit that is critical for the production of S. aureus virulence factors.
INTRODUCTION
Quorum sensing in Gram-positive bacteria occurs via the maturation and release of small signaling oligopeptides (1). These peptides can be linear but may also undergo cyclic ring formation, as is known to occur with the Staphylococcus aureus autoinducing peptide (AIP) (2, 3). In either scenario, a precursor peptide is synthesized and processed prior to or after export from the bacterial cell. While many signaling molecules of Gram-negative bacteria are freely diffusible, the peptides of Gram-positive bacteria generally must transit the membrane via a dedicated transporter (4, 5). After processing and transport, the peptide either is imported back into the bacterial cell or transmits a signal from the extracellular environment by binding to membrane-embedded sensor histidine kinases (6). Peptide signaling culminates in a change in gene expression mediated by transcription factors that respond to the peptide. Many Gram-positive bacterial pathogens use these quorum-sensing peptides to induce gene expression programs that promote virulence adaptations, such as competence, toxin production, biofilm formation, and the establishment of persistence traits. Because of its importance in activating virulence programs in S. aureus, quorum-sensing inhibition has been the focus of many therapeutic initiatives (7–14).
The ability of S. aureus to infect host tissues and cause acute and chronic disease is partially due to its use of complex gene regulatory systems that control virulence factor gene expression (15–17). S. aureus employs 16 two-component systems that contribute to virulence and a range of environmental adaptations (18–25). One of the central two-component systems in S. aureus that regulates virulence factor production is the accessory gene regulatory (Agr) system (15, 26). The components of the Agr system are encoded in an operon containing 4 open reading frames (agrBDCA) (3). These open reading frames encode the histidine kinase AgrC and its cognate response regulator, AgrA, as well as AgrB, a protease that cleaves AgrD, the precursor peptide of AIP. The AgrD precursor peptide is ∼46 amino acids in length with a central stretch of 7 to 9 amino acids that comprise mature AIP. The C-terminal charged tail of AgrD is cleaved by AgrB (27–30), which subsequently mediates peptide cyclization by inducing formation of a thiolactone linkage between the C-terminal carbonyl and the sulfur atom of a conserved cysteine side chain (29). The N-terminal 18 residues form an amphipathic α-helix that anchors the peptide to the cell membrane by lateral association and that is required for processing and transport (31). The remaining processing and transport mechanisms for AIP are not fully defined, but in some S. aureus strains, the type I signal peptidase SpsB is involved in peptide cleavage (32). Clinical isolates of S. aureus can be stratified into one of four Agr groups (I, II, III, and IV) that harbor allelic variants of AgrD, AgrB, and AgrC (3, 30, 33–35). Recognition of AIP from different Agr types is group specific, and AIP variants (except AIP I and IV) inhibit the activity of noncognate AgrCs (36–40). S. aureus strains of all Agr types are known to cause clinical disease; therefore, an understanding of the divergence between Agr types is paramount to our understanding of pathogenesis and population dynamics (33, 41, 42). The final outcome of Agr activation in S. aureus is the production of toxins and other virulence factors that promote disease (3). Coincident with its critical role in regulating the production of virulence factors, strains of S. aureus with defects in Agr function are attenuated in skin and lung infection models, underscoring the significance of this regulatory system to pathogenesis (43–46).
We recently published the results of a screen for S. aureus virulence factors using a transposon mutant library of the methicillin-resistant S. aureus (MRSA) strain JE2 (47). We hypothesized that identification of S. aureus mutants with secreted factors that positively or negatively modulate macrophage activation would yield good virulence factor candidates, given the significant immunopathology associated with S. aureus infection and the major involvement of S. aureus-macrophage interactions in disease. Indeed, the screen successfully uncovered a range of previously unstudied mediators of virulence that our laboratory has been investigating in recent years (47–49). This study relates to an insertion mutant identified in the above-mentioned screen whose supernatant elicited the production of high levels of interleukin-1β (IL-1β) but reduced the secretion of other proinflammatory cytokines, including keratinocyte-derived chemokine (KC) and IL-6, by macrophages (47). The transposon insertion disrupted a single open reading frame, SAUSA300_1984, which encodes a putative type II CAAX metalloprotease.
Type II CAAX proteases are multipass transmembrane proteins with two conserved catalytic motifs, EEXXXR and FXXXH (50). The diglutamate in the EEXXXR motif facilitates the activation of water for nucleophilic attack, whereas the histidine in the FXXXH motif is important for metal binding. Both motifs are thought to be necessary for enzyme activity. In eukaryotes, type II CAAX proteases are involved in prenylation pathways that target proteins to the cell membrane through lipidation of a conserved cysteine within a CAAX motif (50). In bacteria, the function of CAAX proteases is more ambiguous, although their activities have been linked to immunity to bacteriocins in some species (51–54). In S. aureus, there are at least four putative type II CAAX metalloproteases that are also annotated as members of the abortive infectivity (Abi) protein family. The Abi nomenclature for this class of metalloproteases was originally assigned based upon studies of phage resistance mechanisms in Lactococcus lactis (55). Despite the frequent use of this terminology, members of the Abi protein family are not believed to exert functions related to phage resistance (50). Three of the four type II CAAX proteases in S. aureus have roles in surface protein display (SpdA, SpdB, and SpdC [also known as LyrA]) (56, 57). SpdC was recently found to play a role in activation of the WalKR two-component system in S. aureus (58). SpdC directly interacts with the histidine kinase WalK at the division septum, where it appears to have a negative effect on WalK kinase activity. SpdC interacts with WalK through its membrane-spanning domain, and due to sequence divergence from other type II CAAX proteases in S. aureus, its function likely does not require proteolytic activity (56–58). Additional bacterial two-hybrid studies suggest that SpdC interacts with S. aureus histidine kinases SaeS, SrrB, VraS, GraS, and ArlS but not the histidine kinase of the Agr system, AgrC. Whereas SpdA, SpdB, and SpdC orchestrate cell wall homeostasis in a manner that is independent of catalytic activity, the fourth type II CAAX protease, SAUSA300_1984, appears to be functionally distinct and does not modulate cell wall integrity in any known way (56). To date, no additional functions for SAUSA300_1984 have been described. Though it has not yet been investigated in great detail, at least one other study has implicated a type II CAAX protease in the regulation of virulence gene expression. The CAAX protease of group B Streptococcus, Abx1, acts as a regulator of the CovSR two-component system through its histidine kinase CovS, where overexpression of Abx1 promotes activation of CovS (59). Thus, there is mounting evidence for a conserved function of type II CAAX proteases in the regulation of two-component system signaling in bacteria, which in these two instances appears to operate through interaction with a histidine kinase and is independent of catalytic activity.
In this work, we report the identification of SAUSA300_1984, named membrane protease regulator of Agr quorum sensing (MroQ). We determined that MroQ promotes Toll-like receptor 2 (TLR2)-mediated activation of innate immune cells while preventing induction of IL-1β by primary murine macrophages in vitro. This altered inflammatory profile is recapitulated in in vivo infection models, where MroQ promotes inflammatory pathology, cytokine secretion, and bacterial persistence in a model of skin and soft tissue infection. Further investigation into the mode of action of MroQ determined that it acts to regulate the activity of the Agr quorum-sensing system, as evidenced by reduced secreted virulence factors and increased surface protein expression in a ΔmroQ mutant. Our data suggest that MroQ may interface with the Agr system at the level of peptide processing or transport to blunt activation of the AgrC/A two-component system, which remains otherwise responsive in a ΔmroQ mutant background. Altogether, our data support a model whereby MroQ controls gene expression at the level of the Agr system, with significant impacts on the virulence potential of S. aureus.
RESULTS
MroQ is a predicted type II CAAX protease that blunts TLR2-mediated recognition of S. aureus by macrophages.
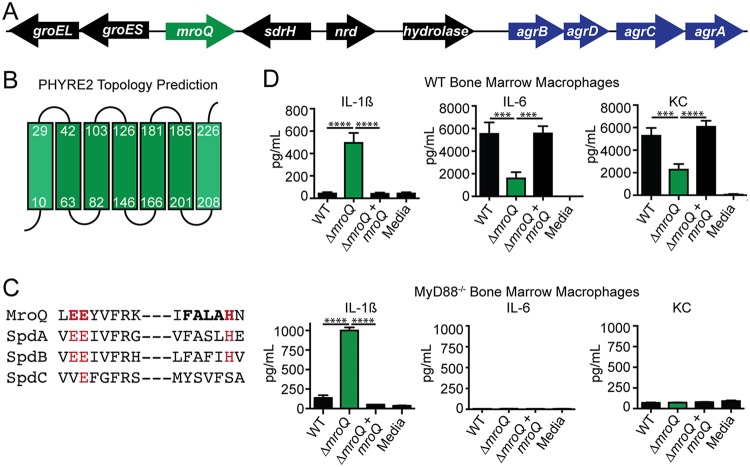
The mroQ open reading frame is located within the core genome of S. aureus and is positioned four open reading frames upstream of the Agr system (Fig. 1A). The gene is flanked by sdrH and groEL groES, which encode the serine aspartate repeat family protein SdrH (60) and the GroEL/ES stress response chaperone system (61), respectively (Fig. 1A). Phyre2 prediction software analysis of the MroQ amino acid sequence suggests a topology containing seven transmembrane helices with the N terminus positioned in the cytosol and the C terminus positioned in the extracellular space, though the weaker predictive strength of the topology analysis at the N and C termini may suggest alternative arrangements in the membrane (Fig. 1B). Consistent with its annotation as a type II CAAX protease, MroQ contains a conserved diglutamate motif, EEXXXR (E141-E142), and the FXXXH (H180) metal coordination motif that shares sequence identity with previously characterized CAAX proteases SpdA and SpdB, with weaker conservation than SpdC (Fig. 1C). We previously determined that supernatant derived from a SAUSA300_1984 transposon insertion mutant compromises macrophage activation (47). To begin interrogating the function of MroQ in greater detail, we first generated an in-frame deletion mutant and complementation strain that expresses mroQ from an ectopic site in the S. aureus chromosome driven by its predicted native promoter (62). The wild-type (WT) strain, the ΔmroQ mutant, and the ΔmroQ mutant complement with mroQ (the ΔmroQ + mroQ strain) replicated identically in tryptic soy broth (TSB) and RPMI medium (see Fig. S1A and B in the supplemental material). We found that, similar to the findings for supernatant derived from the transposon insertion mutant, supernatant derived from a ΔmroQ mutant enhanced the production of IL-1β by murine bone marrow-derived macrophages (BMM) but reduced the production of the inflammatory cytokine IL-6 and the neutrophil chemotactic factor KC (Fig. 1D). The perturbations in IL-6 and KC production were dependent on Toll-like receptor (TLR)-mediated recognition of S. aureus, as evidenced by the elimination of IL-6 and KC production in MyD88−/− and TLR2−/− macrophages but not TLR4−/− macrophages (Fig. 1D and S2A and B). In contrast, the enhanced IL-1β production did not depend on TLRs (Fig. 1D and S2A and B). These data indicate MroQ bears similarities to type II CAAX proteases, is positioned in close proximity to the agr locus in the S. aureus chromosome, and perturbs the activation of innate immune cells in culture through both TLR-dependent and -independent mechanisms.
FIG 1.
MroQ is a putative membrane-embedded protease that affects macrophage activation by S. aureus secreted factors. (A) Illustration of the gene arrangement surrounding mroQ and its proximity to the agr operon. nrd, nitroreductase family protein; sdrH, serine aspartate repeat family protein; hydrolase, carbon-nitrogen family hydrolase. (B) Phyre2 topology prediction analysis of MroQ within the membrane. Green boxes, predicted α-helices. (C) Amino acid sequence alignment of the EEXXXR and FXXXH motifs that comprise the type II CAAX protease active site among four predicted family members in S. aureus. (D) IL-1β, IL-6, and KC production by BMM after addition of supernatant from the WT, ΔmroQ, or ΔmroQ + mroQ strain grown in TSB. (Top) WT BMM; (bottom) MyD88−/− BMM. The data shown are from one of at least three experiments conducted in triplicate. Means ± SD are shown (n = 3). P values were determined by one-way ANOVA with Tukey’s posttest. ***, P < 0.001; ****, P < 0.0001.
MroQ is required for S. aureus virulence.
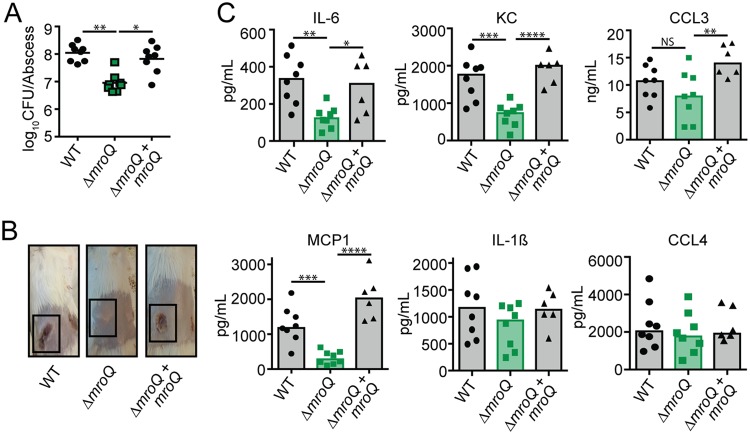
Given the dramatic effect of the ΔmroQ mutant on macrophage activation and the predicted effects that this might have on the inflammatory milieu during infection, we reasoned that the mutant would be compromised for virulence. To test this possibility, we infected mice intradermally with the WT, ΔmroQ, and ΔmroQ + mroQ strains. There were no differences in the number of bacterial CFU between strains at 72 h postinfection (Fig. S3A). However, significant dermonecrosis was evident in mice infected with the WT and ΔmroQ + mroQ strains but not those infected with the ΔmroQ mutant (Fig. S3B). Consistent with reduced inflammatory tissue damage, mice infected with a ΔmroQ mutant showed significantly reduced levels of the inflammatory chemokine monocyte chemotactic protein 1 (MCP-1) and trends toward reduced levels of IL-6, KC, CCL3, and CCL4 at 72 h (Fig. S3C). At 120 h postinfection, 10-fold fewer CFU were recovered from the abscesses of mice infected with a ΔmroQ mutant (Fig. 2A). Gross pathological signs of disease, including large, ruptured, dermonecrotic abscesses, were not observed in ΔmroQ mutant-infected mice, and the abscesses remained closed with little evidence of inflammation (Fig. 2B). There were significant reductions in IL-6, KC, MCP-1, and CCL3 at the abscess site, consistent with the findings of in vitro studies using BMM (Fig. 1D and 2C). IL-1β levels were equally elevated under all infection conditions compared to those at 72 h (Fig. 2C and S3C). These data indicate that MroQ is important for S. aureus skin and soft tissue infection.
FIG 2.
MroQ is important for S. aureus skin and soft tissue infection. (A) Bacterial burden in skin abscesses of mice at 120 h postinfection with the WT (n = 8), ΔmroQ mutant (n = 8), or ΔmroQ + mroQ mutant (n = 8). P values were determined by a nonparametric one-way ANOVA (Kruskal-Wallis test) with Dunn’s posttest. *, P < 0.05; **, P < 0.01. (B) Representative images of skin abscesses at 120 h postinfection with the WT, ΔmroQ, and ΔmroQ + mroQ strains. (C) IL-6, KC, MCP-1, IL-1β, CCL3, and CCL4 levels in abscess homogenates of mice infected with the WT (n = 8), ΔmroQ mutant (n = 8), or ΔmroQ + mroQ mutant (n = 6). P values were determined by a nonparametric one-way ANOVA (Kruskal-Wallis test) with Dunn’s posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant.
A ΔmroQ mutant phenocopies a Δagr mutant.
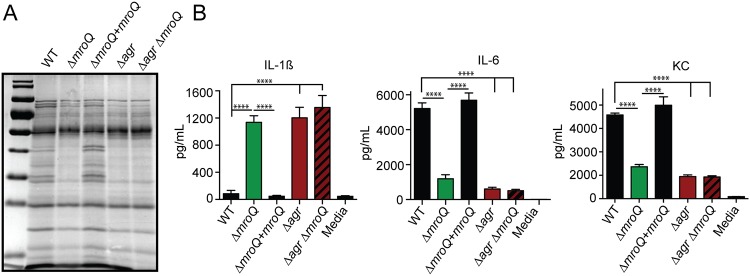
mroQ was identified in a screen that used secreted factors to probe innate immune cell activation (47). Therefore, we sought to test if a ΔmroQ mutant had notable differences in its secretome that might explain the BMM activation defects and reduced virulence. Exoprotein profile analysis indicated that the levels of secreted proteins were reduced in ΔmroQ mutant supernatants (Fig. 3A). This defect in protein secretion closely resembled the profiles of an S. aureus strain with a deletion of the agr locus, the Δagr mutant (Fig. 3A). Exoprotein analysis of a ΔmroQ Δagr double mutant revealed profiles identical to those of the ΔmroQ and Δagr mutants (Fig. 3A). Furthermore, ΔmroQ, Δagr, and ΔmroQ Δagr mutant supernatants induced BMM production of IL-1β but led to reduced levels of IL-6 and KC with the same observed dependency on TLR2 (Fig. 3B and S4A to C). Incidentally, both mroQ and agrB transposon insertion mutants were identified to be inducers of IL-1β secretion in our prior transposon mutant library screen (47).
FIG 3.
A ΔmroQ mutant resembles a Δagr mutant for protein secretion and perturbations in macrophage activation. (A) TCA-precipitated exoproteins from the WT, ΔmroQ, ΔmroQ + mroQ, Δagr, and Δagr ΔmroQ strains collected after growth in TSB. (B) IL-1β, IL-6, and KC production by BMM after addition of supernatant from the WT, ΔmroQ, ΔmroQ + mroQ, Δagr, and Δagr ΔmroQ strains grown in TSB. The data shown are from one of at least three experiments conducted in triplicate. Means ± SD are shown (n = 3). ****, P < 0.0001 by one-way ANOVA with Tukey’s posttest.
Given the similarities between the ΔmroQ and Δagr mutants for defects in protein secretion and secreted factor-mediated perturbations in macrophage activation, we sought to determine whether or not a ΔmroQ mutant exhibits other common phenotypes associated with a Δagr mutant. The Agr system regulates toxin secretion and surface protein production (3). Therefore, we compared the levels of lytic toxins and the Agr-regulated surface protein, protein A, between the ΔmroQ and Δagr mutants (3, 63). We found that the toxins leukocidin AB (LukA), Panton-Valentine (PV) leukocidin (LukS-PV), and alpha-hemolysin (Hla) were reduced in abundance in a ΔmroQ and Δagr mutant, while surface protein A levels were increased (Fig. 4A). Indeed, a prior transposon mutagenesis screen for deficiencies in hemolytic activity, conducted by Fey et al., also identified SAUSA300_1984 (mroQ) to be a gene important for Hla production (64). Consistent with these observations, the ΔmroQ and Δagr mutants exhibited significantly reduced hemolytic activity compared to the WT and ΔmroQ + mroQ strains (Fig. 4B), and both strains were similarly attenuated in a murine intradermal infection model (Fig. 4C). In summary, MroQ appears to be required for Agr function in some unknown way.
FIG 4.
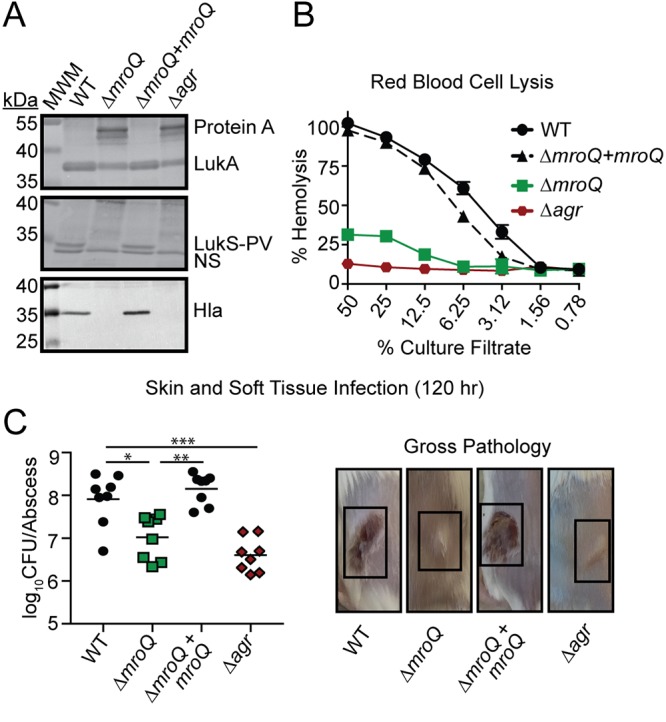
MroQ contributes to Agr function. (A) LukA, LukS-PV, and Hla immunoblots of TCA-precipitated exoproteins from WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains. Protein A levels were detected based upon binding of anti-LukA antibody to protein A. MWM, molecular weight marker; NS, nonspecific band. (B) Rabbit red blood cell lysis of cell-free culture filtrates derived from WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains. (C) (Left) Bacterial burden in skin abscesses of mice at 120 h postinfection with the WT (n = 8), ΔmroQ (n = 8), ΔmroQ + mroQ (n = 8), and Δagr (n = 8) strains. P values were determined by a nonparametric one-way ANOVA (Kruskal-Wallis test) with Dunn’s posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (Right) Representative images of skin abscesses at 120 h postinfection with the WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains are shown. Hemolysis assay data are from one of at least three experiments conducted in triplicate. Means ± SD are shown (n = 3).
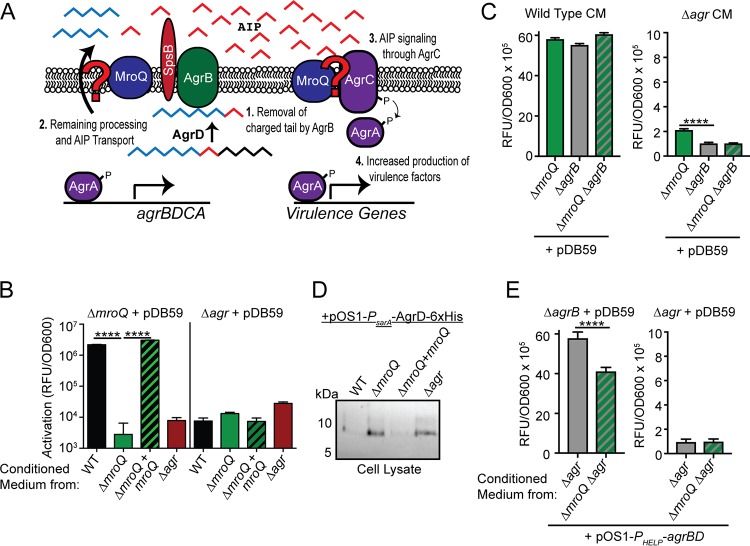
A ΔmroQ mutant is compromised for AIP processing or export.
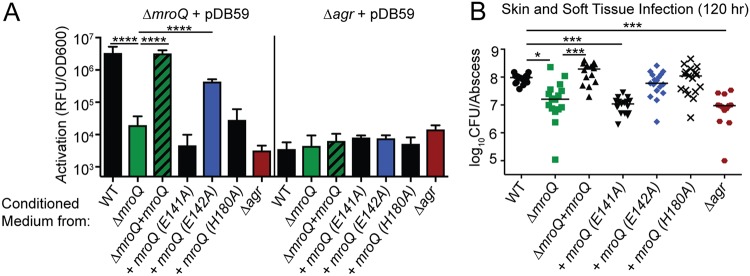
MroQ is a hypothetical type II CAAX metalloprotease. Despite the clear connections between Agr and S. aureus pathobiology, there exist key gaps in our knowledge of the mechanics of Agr system activation (Fig. 5A) (5, 65). Most notably, (i) the complete range of proteins needed for maturation of AgrD has not been fully elucidated, (ii) a dedicated transporter for AIP or its leader peptide has not been conclusively identified, and (iii) differential processing of AIP variants I to IV has not been investigated. We wondered if the function of MroQ within the Agr system might be to facilitate processing and/or export of the AgrD precursor peptide to generate AIP or if it promotes the function of the histidine kinase AgrC in a manner similar to that of SpdC and WalK (Fig. 5A). To test these possibilities, we first introduced a reporter plasmid that drives the expression of green fluorescent protein (GFP) via an AgrA-regulated promoter known as P3 (pDB59-P3-gfp) into ΔmroQ and Δagr mutant strains (66). We collected conditioned medium (CM) from early-stationary-phase cultures (in which AIP production is high) of the WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains and applied them to the reporter strains, followed by measurement of GFP fluorescence. Conditioned medium derived from all strains tested on the Δagr/pDB59-P3-gfp reporter strain did not induce promoter activation (Fig. 5B). This was expected because the Δagr mutant lacks all components of the Agr system, including those that transmit the AIP signal (AgrC and AgrA). In contrast, when conditioned medium from the WT or ΔmroQ + mroQ strains was added to the ΔmroQ/pDB59-P3-gfp reporter strain, we observed a significant increase in fluorescence intensity, indicating that the AgrC-AgrA signaling axis is intact and able to recognize AIP. However, conditioned medium from the ΔmroQ and Δagr mutants was unable to induce P3 promoter activity in this reporter strain background (Fig. 5B). Similarly, no difference in the ability to transmit the signal through AgrC and AgrA was seen by introduction of the pDB59-P3-gfp reporter into the ΔagrB and ΔagrB ΔmroQ strains, followed by the addition of CM derived from WT S. aureus (Fig. 5C). These data suggest that the Agr system defect in the ΔmroQ mutant does not occur at the level of AgrC or affect its ability to propagate a signal to AgrA.
FIG 5.
Interrogating the effects of MroQ on Agr peptide processing or signaling. (A) Model depicting the potential locations where MroQ might facilitate Agr function. We propose that MroQ functions at the level of the peptide-processing module (AgrBD) or at the level of the membrane-embedded histidine kinase (AgrC). (B) P3-gfp (pDB59) promoter activation (relative fluorescence units [RFU]/OD600) in the ΔmroQ and Δagr strains upon addition of conditioned medium from the WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains. (C) P3-gfp/pDB59 promoter activation (RFU/OD600) in the ΔmroQ, ΔagrB, and ΔmroQ ΔagrB strains upon addition of conditioned medium (CM) from the WT and Δagr strains. (D) Whole-cell lysates of the WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains constitutively expressing AgrD-6×His (pOS1-PsarA-agrD-6×His) followed by immunoblotting with anti-His monoclonal antibody to detect full-length (unprocessed) AgrD. (E) P3-gfp/pDB59 promoter activation (RFU/OD600) in ΔagrB and Δagr strains upon addition of conditioned medium from the Δagr and Δagr ΔmroQ strains constitutively expressing agrBD/pOS1-PHELP-agrBD. ****, P < 0.0001 by one-way ANOVA with Tukey’s posttest (C) and a two-tailed t test (E). The data shown are from one of at least three experiments conducted in triplicate. Means ± SD are shown (n = 3).
The absence of a clear effect of MroQ on AgrC signaling led us to interrogate whether or not MroQ affects AgrD protein processing/export. We generated a plasmid that expresses AgrD-6×His under the control of a constitutive promoter (pOS1-PsarA-agrD-6×His) (67, 68). The plasmid was introduced into the WT, ΔmroQ, ΔmroQ + mroQ, and Δagr strains. AgrD accumulates when AgrB peptidase activity is compromised, indicating that processing of AgrD by AgrB precedes export (3, 27–30). We hypothesized that if MroQ is involved in promoting an AgrD processing event similar to that of AgrB, then we should detect full-length AgrD-6×His in the ΔmroQ mutant. Indeed, full length AgrD-6×His was detectable when it was expressed in the ΔmroQ and Δagr strains (Fig. 5D). To further determine if MroQ is involved in regulating AgrBD functions related to AIP processing/export, we generated a plasmid that constitutively expresses agrBD driven by the synthetic promoter PHELP and introduced this plasmid (pOS1-PHELP-agrBD) into Δagr or Δagr ΔmroQ strains followed by determination of whether or not conditioned medium derived from these strains was able to activate the pDB59-P3-gfp reporter in a ΔagrB or Δagr mutant background. We found that conditioned medium derived from the Δagr ΔmroQ/pOS1-PHELP-agrBD strain exhibited a 30% reduction in P3 promoter activity compared to that of conditioned medium derived from the Δagr/pOS1-PHELP-agrBD strain (Fig. 5E). These data support the hypothesis that MroQ plays a role in promoting the functionality or stability of the AgrB-AgrD peptide processing/export axis of the Agr system.
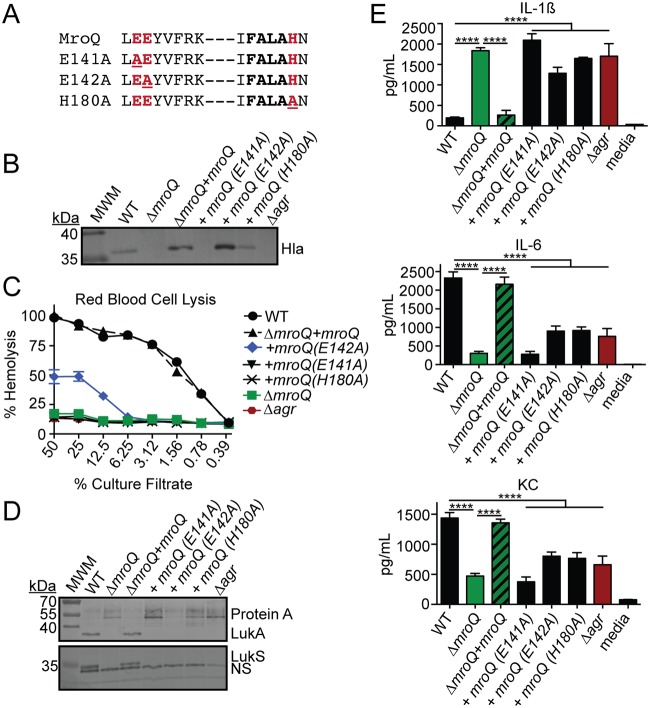
Conserved active-site amino acids are required for MroQ regulation of the Agr system.
The type II CAAX protease family of enzymes is conserved in both prokaryotes and eukaryotes. In eukaryotes, proteolytic activity occurs at a CAAX motif on the C-terminal end of target proteins, ultimately leading to lipidation. In bacteria, the functions of the CAAX protease family members are not as clear, but some proteins in this family use proteolytic activity to confer immunity against bacteriocins (69), while others engage in intramembrane proteolysis events to promote adaptive traits (52, 70). Of the three additional characterized CAAX protease enzymes in S. aureus (SpdA, SpdB, and SpaC), it is known that the conserved active-site glutamate of SpdC does not appear to be required for function (57). This is also true for the Abx1 enzyme of group B Streptococcus (59). Thus, the CAAX proteases of bacteria appear to have diverse functions, some of which depend on proteolytic activity and others of which do not. To test if the active site of MroQ is required to promote AgrBD activity, we introduced mutations into the conserved CAAX protease amino acid residues within the EEXXXR and FXXXH motifs previously determined to be required for enzymatic activity in a range of type II CAAX proteases (50, 52, 69, 71, 72). Alanine substitutions were introduced at E141, E142, and H180, followed by expression of point mutations in a single copy under the control of the mroQ native promoter in S. aureus ΔmroQ (Fig. 6A). As evidenced by immunoblot analysis, the expression of MroQ(E141A) was unable to complement alpha toxin secretion defects, whereas the MroQ(E142A) and MroQ(H180A) mutants exhibited some restoration in alpha toxin production by immunoblot analysis, with the MroQ(E142A) strain supernatant containing larger amounts of alpha toxin than the MroQ(H180A) strain supernatant (Fig. 6B). The observed differences in alpha toxin levels by immunoblot analysis were recapitulated in hemolytic activity assays where the MroQ(E142A) strain exhibited a partial restoration of hemolytic activity on rabbit red blood cells (RBCs) but the MroQ(E141A) and MroQ(H180A) strains did not (Fig. 6C). Consistent with the observed effects on alpha toxin secretion and hemolytic activity, MroQ(E142A) partially complemented protein A production defects (Fig. 6D). In contrast, none of the three mutations restored LukS or LukA production to an observable degree by immunoblot analysis, and the MroQ point mutations failed to complement changes in macrophage activation caused by supernatant derived from a ΔmroQ mutant (Fig. 6D and E). Together, these data suggest that conserved amino acids implicated in type II CAAX protease activity are important for the effects of MroQ on AgrBD function. Residue E141 is required for MroQ function, whereas residues E142 and H180 promote optimal activity.
FIG 6.
MroQ active-site residues are required for regulation of Agr by MroQ. (A) Amino acid sequences of the EEXXXR and FXXXH motifs that comprise the type II CAAX protease active site. The locations of site-directed amino acid substitutions are underlined. (B) Hla immunoblots of TCA-precipitated exoproteins from the WT, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains. (C) Rabbit red blood cell lysis of cell-free culture filtrates derived from the WT, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains. (D) LukA and LukS-PV immunoblots of TCA-precipitated exoproteins from the WT, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains. Protein A levels were detected based upon the binding of anti-LukA antibody to protein A. MWM, molecular weight marker; NS, nonspecific band. (E) IL-1β, IL-6, and KC production by BMM after addition of supernatant from the WT, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains grown in TSB. ****, P < 0.0001 by one-way ANOVA with Tukey’s posttest. The data shown are from one of at least three experiments conducted in triplicate. Means ± SD are shown (n = 3).
Investigation of the role of MroQ amino acid residues E141, E142, and H180 in Agr-mediated gene expression and virulence.
We next sought to determine the extent to which ΔmroQ mutant strains expressing MroQ(E141A) [ΔmroQ + MroQ(E141A)], MroQ(E142A) [ΔmroQ + MroQ(E142A)], and MroQ(H180A) [ΔmroQ + MroQ(E180A)] alter gene expression and pathogenesis. We first conducted P3 promoter activation analyses to evaluate whether or not the generation and release of functional AIP occurs in strains expressing each point mutation. As anticipated, conditioned medium derived from the ΔmroQ + MroQ(E141A) and ΔmroQ + MroQ(H180A) strains was unable to appreciably activate the P3-gfp promoter fusion above background levels when applied to the ΔmroQ/pDB59 reporter strain (Fig. 7A). In contrast, conditioned medium derived from the ΔmroQ + MroQ(E142A) strain led to partial activation with a greater than 10-fold increase in fluorescence intensity compared to background levels (Fig. 7A). When the ΔmroQ + MroQ(E141A), ΔmroQ + MroQ(E142A), and ΔmroQ + MroQ(H180A) strains were used in an intradermal infection model, we found that the ΔmroQ + MroQ(E141A) strain exhibited a 10-fold reduction in the number of CFU, similar to a ΔmroQ and Δagr mutant. In contrast, the ΔmroQ + MroQ(E142A) and ΔmroQ + MroQ(H180A) strains restored infection to nearly WT levels (Fig. 7B). Thus, MroQ(E141A) appears to be the most crucial active-site mutation, as it is necessary for MroQ function in vitro and in vivo, whereas MroQ(E142A) and MroQ(H180A) confer nearly normal virulence characteristics during skin and soft tissue infection, an outcome consistent with their partial restoration of in vitro defects.
FIG 7.
MroQ amino acid substitution E141A is required for Agr activation and full virulence. (A) P3-gfp (pDB59) promoter activation (relative fluorescence units [RFU]/OD600) in ΔmroQ and Δagr strains upon addition of conditioned medium from the WT, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains. The data shown are from one of at least three experiments conducted in triplicate. Means ± SD are shown (n = 3). ****, P < 0.0001 by one-way ANOVA with Tukey’s posttest. (B) Bacterial burden in skin abscesses of mice at 5 days postinfection with the WT (n = 16), ΔmroQ (n = 16), ΔmroQ + mroQ (n = 14), ΔmroQ + mroQ(E141A) (n = 17) ΔmroQ + mroQ(E142A) (n = 17), ΔmroQ + mroQ(H180A) (n = 17), and Δagr (n = 16) strains. P values were determined by a nonparametric one-way ANOVA (Kruskal-Wallis Test) with Dunn’s posttest. *, P < 0.05; ***, P < 0.001.
DISCUSSION
Many Gram-positive pathogens use small-peptide signaling pathways to control the expression of virulence genes. In this work, we present the discovery of a membrane-embedded protease that plays a major role in promoting the function of the Agr system of S. aureus. Our data suggest that the putative type II CAAX peptidase SAUSA300_1984 (MroQ) directly interfaces with the Agr system to promote virulence factor gene expression and pathogenesis in a murine skin and soft tissue infection model. Unlike other type II CAAX proteases of S. aureus and group B Streptococcus (57, 59), MroQ requires its conserved active-site amino acids to confer normal functionality to the Agr system. Our studies to probe the location in the Agr pathway where MroQ acts indicate that the AgrC/A signaling axis remains functional and that the AgrBD axis, involved in peptide maturation, is likely compromised. Together, these findings expand the view of peptide-based signaling in S. aureus and highlight a newly uncovered regulatory input into virulence factor gene expression that dramatically affects virulence outcomes. Gram-positive pathogens of urgent clinical importance, including Clostridioides difficile and Enterococcus faecalis, as well as significant threats to food safety, such as Listeria monocytogenes, each harbor Agr systems that contribute to disease (73–79). Thus, this work provides insights into alternative pathways of regulation of the Agr system that may extend to other Gram-positive microbes of clinical importance.
Our studies have determined that the secreted factors derived from either a ΔmroQ mutant or a Δagr mutant cause the significant release of IL-1β but the release of low levels of other inflammatory cytokines and chemokines from primary murine macrophages in culture (Fig. 1). The production of the inflammatory cytokine IL-1β in superficial tissues is known to be critical for controlling skin and soft tissue infection caused by S. aureus (80–84). In contrast, the production of IL-1β during systemic infection is associated with increased immunopathology (85, 86). Agr-defective isolates of S. aureus are routinely isolated from patients with bacteremia (65). The reason that these Agr-defective isolates arise and their association with morbidity in bacteremia are not well understood; however, the observation is recapitulated in murine infection models, wherein agr mutants persist in deep tissues during bacteremia (87). Some potential mechanisms behind these observations have been proposed, including a role for agr-defective isolates in the development of a persister state, as well as disruption of the optimal immune environment by agr-defective strains (65). Although a ΔmroQ mutant did not elicit the increased production of IL-1β compared to the WT at the time points tested in vivo, we postulate that the reduced levels of proinflammatory cytokines and maintenance of IL-1β may be linked to improved disease outcomes (Fig. 2). The possibility that the inflammatory shifts imparted by a loss of Agr functionality might dictate clearance or survival in superficial versus systemic infection is currently being interrogated by our laboratory.
The mechanics of cyclic AIP maturation by S. aureus are not fully understood (5). Generation of AIP absolutely requires C-terminal cleavage and formation of a thiolactone ring by the Agr locus enzyme AgrB (27–30). The events that occur after thiolactone ring formation remain ambiguous but at a minimum must involve additional proteins for proteolytic cleavage and transport (5, 32). Our data demonstrate that the putative metalloprotease MroQ does not seem to influence the ability of the histidine kinase AgrC to transmit a signal to AgrA (Fig. 5C). Thus, it does not appear that MroQ directly regulates the activity of AgrC. This observation is different from that for Abx1 from group B Streptococcus or SpdC of S. aureus, which are implicated in the regulation of CovS and WalK histidine kinases, respectively (58, 59). These studies used bacterial two-hybrid assays to determine that Abx1 and SpdC interact with WalK and CovS and that deletion or overexpression changes kinase activity. Aside from these physical associations, no additional evidence was provided to describe how each protein mediates its function, but the studies do note that SpdC negatively regulates WalK and that overexpression of Abx1 promotes CovS activity (58, 59). Uncoupling agrBD gene expression from its autoregulatory circuit has allowed us to identify a defect in AIP peptide production when MroQ is absent (Fig. 5E). We found that conditioned medium derived from the Δagr/pOS1-PHELP-agrBD strain was better able to activate a P3-gfp reporter than conditioned medium derived from the Δagr ΔmroQ/pOS1-PHELP-agrBD strain. Importantly, the absence of MroQ did not eliminate reporter activation, suggesting either that overexpression of agrBD can compensate or that MroQ activity on AgrBD is not essential for peptide maturation/release; rather, it improves its efficiency in some way. Thus, MroQ appears to behave differently from Abx1 and SpdC, where it acts to promote the functionality of the peptide maturation module of Agr, and is important for the optimal generation or export of AIP in the type I Agr MRSA strain LAC. We do not yet know precisely how MroQ mediates its effects on the AgrBD module; however, a ΔmroQ mutant does not release appreciable amounts of active AIP (Fig. 5 and 7) and harbors increased amounts of unprocessed cytosolic AIP (Fig. 5D). It is notable that although a ΔmroQ mutant closely resembles a Δagr mutant, there appears to be some low-level intrinsic activation of the Agr system in a ΔmroQ mutant, as evidenced by P3 promoter activity that rises above the levels in a Δagr mutant (Fig. 5C and 7A). This observation indicates that mature AIP is at least capable of being produced by a ΔmroQ mutant, though at low levels. Taking these findings together, we propose that MroQ promotes efficient AgrD proteolytic processing, transport, or both. This may occur through direct effects on AgrB function or on the AgrD peptide itself. We are currently exploring these possibilities.
S. aureus strains encode one of four allelic variants of AIP, all of which are capable of causing clinical disease in humans (36–40). Our data indicate that MroQ is critical for the Agr activity of a type I strain. AgrB, the protease that cleaves the C terminus of AgrD, has considerable sequence variability among Agr types, rendering C-terminal cleavage specific to the four Agr groups (3, 30, 33). There is negligible sequence variation at the amino acid level for MroQ in strains that harbor each of the four Agr alleles (data not shown). Therefore, we suspect either that the activity of MroQ on the Agr system is broad and not influenced by Agr type or that alternative enzymes, perhaps other CAAX proteases, modulate the functions of different Agr types. Given the strict dependency on the length and amino acid composition of the N-terminal tails of AIP for signaling and given that precise peptide cleavage at the N terminus is a requisite for activity, we favor the possibility that additional peptidases may promote the full maturation or export of these AIP sequence variants (39).
One important difference in the function of MroQ compared to that of other type II CAAX proteases noted in S. aureus is that conserved active-site residues appear to be important for the activity of MroQ but not for that of the CAAX protease SpdC or group B Streptococcus Abx1 (57, 59). This represents an additional departure from the models put forth for these CAAX proteases and suggests that MroQ may alternatively function in a way that is similar to that of proteases whose activities involve peptide hydrolysis or intramembrane proteolysis (50, 52, 69, 71, 72). Thus far, we know that the glutamate at position 141 is critical for MroQ function, whereas mutations of glutamate at position 142 and the histidine at position 180 do not confer complete null phenotypes. In other type II CAAX protease family members, the second glutamic acid in the EEXXXR motif is usually essential for activity, an observation that is contrary to what was observed for MroQ (50). While the E142A and H180A mutations conferred a partial restoration of Agr activity, only the mutant with the E141A point mutation was attenuated in vivo to the same level as a ΔmroQ or Δagr mutant during skin and soft tissue infection. This outcome suggests that the partial Agr activity conferred by expression of MroQ(E142A) and MroQ(H180A), but not that conferred by expression of MroQ(E141A), is sufficient to confer normal infection kinetics.
Altogether, this work has expanded our understanding of the regulatory inputs that dictate cyclic peptide quorum sensing in S. aureus. Our studies represent an expansion of the Agr regulatory circuit that may have broad impacts on therapeutic design for noncomplicated infection (12, 14, 88–90). Further, the work highlights the complexities of peptide-based signaling and the adaptive traits that promote virulence in S. aureus. Our knowledge of the activities of proteins from the type II CAAX protease family in Gram-positive pathogens remains in its infancy. This work adds to the growing compendium of work on proteins in this protein class and their diverse range of functions, some of which are crucial for Gram-positive bacterial virulence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. aureus LAC (AH-1263) was used as the wild-type (WT) strain for the experiments in this study. LAC (AH-1263) is an S. aureus USA300 clinical isolate cured of its plasmids (91). All other bacterial strains used in this work are described in Table 1. Most recombinant plasmids were passaged through Escherichia coli DH5α before propagation in S. aureus. When constructing complementation plasmids harboring the mroQ gene, we found that mroQ exhibited signs of toxicity in E. coli, resulting in a high frequency of secondary mutations in the coding sequence. As an alternative, E. coli BH10C was used as a host strain; BH10C reduces the copy number of plasmids to nearly single-copy levels to limit the abundance of potentially toxic genes in E. coli and reduces the likelihood of mutations (92). After passage through E. coli, S. aureus RN4220 and RN9011 (RN4220/pRN7023) (62) were used as intermediate strains, followed by electroporation or transduction into AH-1263 or its isogenic mutant derivatives. All E. coli strains were grown in lysogeny broth (LB; Amresco), while S. aureus strains were grown in either tryptic soy broth (TSB; Criterion) or RPMI medium (Corning) supplemented with 1% Casamino Acids (Amresco) and 2.4 mM sodium bicarbonate (Amresco). When necessary, the media were supplemented with the following antibiotics at the indicated concentrations: ampicillin (Amp) (GoldBio), 100 μg/ml; erythromycin (Erm) (Amresco), 5 μg/ml; chloramphenicol (Cm) (Amresco), 10 μg/ml; anhydrous tetracycline (AnTet) (Acros Organics), 1 μg/ml; and tetracycline (Tet) (Amresco and Acros Organics), 10 μg/ml. Cadmium chloride (Alfa Aesar) was used at 0.1 to 0.3 mM to select for pJC1111 transductants. Bacterial growth was monitored using a Genesys 10S UV-visible spectrophotometer by measuring the culture optical density at 600 nm (OD600; Thermo).
TABLE 1.
Strains used in this study
| Strain | Description | Designation | Source or reference |
|---|---|---|---|
| AH-1263 | S. aureus USA300 strain LAC, plasmid cured | LAC (WT) | 91 |
| DH5α | E. coli strain for recombinant plasmids | DH5α | |
| BH10C | E. coli strain that restricts the plasmid copy number for complementation of mroQ | BH10C | 92 |
| RN4220 | Restriction-deficient S. aureus | RN4220 | 96 |
| RN9011 | RN4220/pRN7023 expressing SaPI1 integrase | RN9011 | 62 |
| FA-S922 | AH-1263 with an in-frame deletion of mroQ | ΔmroQ mutant | This work |
| FA-S982 | FA-S922 complemented with pJC1112-mroQ | ΔmroQ + mroQ mutant | This work |
| FA-S1008 | AH-1263 containing a gene replacement of agrBDCA with tetracycline resistance cassette | Δagr::tet mutant | This work |
| FA-S995 | FA-S922 transduced with Δagr::tet mutation | ΔmroQ Δagr::tet mutant | This work |
| FA-S1732 | AH-1263 with an in-frame deletion of agrB | ΔagrB mutant | This work |
| FA-S1736 | FA-S922 containing an in-frame deletion of agrB | ΔagrB ΔmroQ mutant | This work |
| AH-E462 | E. coli containing reporter plasmid pDB59 with gfp expression driven by the P3 promoter | 66, 97 | |
| FA-S945 | FA-S922 with GFP reporter plasmid pDB59 | ΔmroQ/pDB59 mutant | This work |
| FA-S984 | LAC Δagr with GFP reporter plasmid pDB59 | Δagr/pDB59 mutant | This work |
| FA-S1972 | FA-S1732 with GFP reporter plasmid pDB59 | ΔagrB/pDB59 mutant | This work |
| FA-S1973 | FA-S1736 with GFP reporter plasmid pDB59 | ΔagrB ΔmroQ/pDB59 mutant | This work |
| FA-S1145 | AH-1263 containing pOS1-PsarA-sodRBS-agrD-6×His | WT/pOS1-PsarA-agrD-6×His | This work |
| FA-S1146 | FA-S922 containing pOS1-PsarA-sodRBS-agrD-6×His | ΔmroQ/pOS1-PsarA-agrD-6×His mutant | This work |
| FA-S1147 | FA-S1008 containing pOS1-PsarA-sodRBS-agrD-6×His | Δagr/pOS1-PsarA-agrD-6×His mutant | This work |
| FA-S1265 | FA-S982 containing pOS1-PsarA-sodRBS-agrD-6×His | ΔmroQ + mroQ/pOS1-PsarA-agrD-6×His mutant | This work |
| FA-S1841 | FA-S922 containing pJC1111-mroQ(E141A) | ΔmroQ + mroQ(E141A) mutant | This work |
| FA-S1898 | FA-S922 containing pJC1111-mroQ(E142A) | ΔmroQ + mroQ(E142A) mutant | This work |
| FA-S1837 | FA-S922 containing pJC1111-mroQ(H180A) | ΔmroQ + mroQ(H180A) mutant | This work |
| FA-S2012 | FA-S1008 containing pOS1-PHELP-agrBD | Δagr/pOS1-PHELP-agrBD mutant | This work |
| FA-S2013 | FA-S995 containing pOS1-PHELP-agrBD | Δagr ΔmroQ/pOS1-PHELP-agrBD mutant | This work |
Molecular genetic techniques.
To isolate genomic DNA from S. aureus, cultures were grown overnight in 5 ml TSB at 37°C with shaking at 200 rpm. Bacteria (1.5 ml) were pelleted by centrifugation and resuspended in 200 μl of TSM buffer (50 mM Tris, 0.5 M sucrose, 10 mM MgCl2 [pH 7.5]) and 2.5 μl of a lysostaphin (Ambi Products) stock (2 mg/ml) to achieve a final concentration of 25 μg/ml. Samples were incubated at 37°C for 15 min to digest the cell wall of S. aureus, followed by centrifugation at 14,000 rpm for 5 min to pellet the bacteria. The supernatants were discarded, and the genomic DNA was isolated using a Wizard Genomic DNA purification kit (Promega). PCR was performed in a FlexID Mastercycler (Eppendorf) using Phusion High-Fidelity DNA polymerase (New England Biolabs) or GoTaq DNA polymerase (Promega) and deoxynucleoside triphosphates (Quanta BioSciences). For all PCRs, oligonucleotides were ordered from Eurofins (Table 2). Electrophoresis of DNA samples was performed in 0.8% agarose (Amresco) gels. DNA digestions were performed using the restriction endonucleases XhoI, BamHI, EcoRI, KpnI, XbaI, SacI, and PstI (New England Biolabs) following the manufacturer’s suggested protocols, and all digested plasmids were subsequently treated with shrimp alkaline phosphatase (Amresco). Ligations were performed using T4 DNA ligase (New England Biolabs), and the ligation mixtures were incubated in an Eppendorf ThermoMixer at 16°C overnight. When necessary, PCR purification and DNA gel extraction were carried out using Qiagen QIAquick kits.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| MroQ-1 | CCC-GGTACC(KpnI)-CCATAAATGATAAACCTCCAT |
| MroQ-2 | GTGTGATTCGTTTTTTTTATTA-GGCGCC(KasI)-CATAATTTTCCTCCAAATATT |
| MroQ-3 | AATATTTGGAGGAAAATTATG-GGCGCC(KasI)-TAATAAAAAAAACGAATCACAC |
| MroQ-4 | CCC-GAGCTC(SacI)-ATTTTTAGCCTTGGCAAATG |
| 37F | GGCGGATCCAGCACTATCAGTTAAAACAAT |
| 37R | GGCGAATTCACAAGAAGATAATAAGAAAAG |
| agrB-1 | ATAT-GGTACC(KpnI)-TAACTCTACTAGCAAATGTTACTC |
| agrB-2 | GTTAAATAATGTATTCATTTTTACACCACTCTCCTCA |
| agrB-3 | TGAGGAGAGTGGTGTAAAAATGAATACATTATTTAAC |
| agrB-4 | ATAT-GAGCTC(SacI)-TATTACAAATATACTATCAGAAATTTTGGTG |
| MroQPM-CompF | GGC-CTGCAG(PstI)-AGCACTATCAGTTAAAACAAT |
| MroQPM-CompR | GGC-TCTAGA(XbaI)-ACAAGAAGATAATAAGAAAA |
| MroQ(H180A)-2 | AAATTTGAAATCATTGGCTGCTAATGCAAATATTAATG |
| MroQ(H180A)-3 | CATTAATATTTGCATTAGCAGCCAATGATTTCAAATTT |
| MroQ(E141A)-2 | TTTTCTGAATACATATTCTGCTAATAAAGGACCAATAA |
| MroQ(E141A)-3 | TTATTGGTCCTTTATTAGCAGAATATGTATTCAGAAA |
| MroQ(E142A)-2 | TTTTCTGAATACATATGCTTCTAATAAAGGACCAATAA |
| MroQ(E142A)-3 | TTATTGGTCCTTTATTAGAAGCATATGTATTCAGAAAA |
| phelp-agrBD-1 | ATAT-GTCGAC(SalI)-ATCCCATTATGCTTTGGCA |
| phelp-agrBD-2 | CAATTTTATTATCAAAATAATTCAAGGGTTTCACTCTCCTTCTA |
| phelp-agrBD-3 | TAGAAGGAGAGTGAAACCCTTGAATTATTTTGATAATAAAATTG |
| phelp-agrBD-4 | ATAT-GAATTC(EcoRI)-TTATTCGTGTAATTGTGTTAAT |
| PsarA-AgrD6×His-1 | ATAT-CTGCAG(PstI)-CTGATATTTTTGAC |
| PsarA-AgrD6×His-2 | AAATAAGTTAAATAATGTATTCAT-AAATAATCATCCTCCTAAGG |
| PsarA-AgrD6×His-3 | CCTTAGGAGGATGATTATTT-ATGAATACATTATTTAACTTATTT |
| PsarA-AgrD6×His-4 | ATAT-GAATTC(EcoRI)-TTAGTGATGGTGAATGGTGA |
aUnderlining and boldface indicate locations of nucleotide substitutions.
To isolate plasmids from S. aureus, 5-ml overnight cultures of bacteria were grown in TSB at 37°C with shaking at 200 rpm. On the next day, the bacteria were pelleted by centrifugation at 4,200 rpm for 15 min and resuspended in 400 μl of TSM buffer containing 20 μl of lysostaphin stock solution (2 mg/ml) to achieve a final concentration of 0.1 mg/ml. Samples were incubated for 10 min at 37°C to digest the cell wall, followed by centrifugation at 13,000 rpm for 2 min to pellet the bacterial cells. Thereafter, a Qiagen miniprep kit was used to isolate plasmid DNA. DNA was eluted with sterile water in all cases. DNA sequencing was performed by GenScript or Genewiz.
Bacteriophage-mediated generalized transduction.
Transduction was used to transfer stably integrated complementation plasmids between strains, as well as to mobilize marked mutations within the S. aureus chromosome (Δagr::tet). S. aureus-specific bacteriophage ϕ11 or 80α was used in this study. Donor strains were grown overnight in 3 ml of TSB-LB (1:1) supplemented with 5 mM calcium chloride (CaCl2; Amresco) and 5 mM magnesium sulfate (MgSO4; Amresco) at 37°C with shaking at 200 rpm. On the next day, overnight cultures were diluted 1:100 in 10 ml fresh TSB-LB (1:1) supplemented with 5 mM CaCl2 and 5 mM MgSO4. Samples were grown for ∼3 h at 37°C with shaking at 200 rpm until reaching an OD600 of 0.3 to 0.9. To package donor DNA into transducing phage, 100 μl of 10-fold serial dilutions (10−1 to 10−10) of bacteriophage stock in TMG (10 mM Tris, pH 7.5, 5 mM MgCl2, 0.01% [vol/vol] gelatin) was added to 15-ml conical tubes and incubated with 500 μl of the donor S. aureus culture. The phage-bacterium mixture was gently vortexed and incubated at room temperature for 30 min. Melted and cooled CY top agar (2.5 ml; Casamino Acids, 5 g/liter; yeast extract, 5 g/liter; glucose, 5 g/liter; NaCl, 6 g/liter; agar, 7.5 g/liter) supplemented with 5 mM CaCl2 and 5 mM MgSO4 was added to the bacterium-phage mixture and immediately poured onto tryptic soy agar (TSA) plates, followed by incubation overnight at 30°C. On the following day, the phages were harvested from two to three plates with confluent plaques. Bacteriophage stocks were stored at 4°C.
To transduce plasmids and marked mutations, recipient strains were first grown overnight in 20 ml of TSB-LB (1:1) supplemented with 5 mM CaCl2 at 37°C. Samples were then centrifuged for 15 min at maximum speed, the supernatants were discarded, and the bacterial pellets were suspended in 3 ml of CY medium (Casamino Acids, 5 g/liter; yeast extract, 5 g/liter; glucose, 5 g/liter; NaCl, 6 g/liter) supplemented with 5 mM CaCl2. Four infection conditions were set up with either bacteria alone (mock) or 10-fold serial dilutions of bacteria infected with 100 μl of a bacteriophage stock containing donor DNA. The tubes were mixed by inverting every 10 min for 30 min at room temperature, followed by the addition 40 mM sodium citrate to each tube and incubation for 30 min at room temperature. The tubes were centrifuged at maximum speed for 5 min and washed twice in 500 μl of CY medium supplemented with 40 mM sodium citrate, followed by resuspension in 200 μl of CY medium supplemented with 40 mM sodium citrate and spreading onto CY agar or TSA plates containing 10 mM sodium citrate and any antibiotics necessary for selection. The plates were placed at 37°C overnight. Potential transductants were screened for antibiotic resistance and acquisition of the desired mutations by PCR and DNA sequencing.
Generation of mroQ and agrB in-frame deletion mutants.
mroQ and agrB in-frame deletion mutants were generated using the temperature-sensitive mutagenesis plasmid pIMAY (93). For mroQ, two fragments that correspond to ∼500-bp regions of sequence homology immediately upstream and downstream of mroQ were amplified. Oligonucleotides MroQ-1 and MroQ-2 were used to amplify the region upstream of mroQ, while oligonucleotides MroQ-3 and MroQ-4 were used to amplify the region downstream of mroQ (Table 2). The fragments were joined by splicing by overlap extension (SOEing) PCR using oligonucleotides MroQ-1 and MroQ-4 and subcloned into the multicloning site of pIMAY using restriction endonucleases KpnI and SacI. For agrB, two fragments that correspond to ∼500-bp regions of sequence homology immediately upstream and downstream of agrB were amplified. Oligonucleotides agrB-1 and agrB-2 were used to amplify the region upstream of agrB, while oligonucleotides agrB-3 and agrB-4 were used to amplify the region downstream of agrB (Table 2). The fragments were joined by splicing by overlap extension (SOEing) PCR using oligonucleotides agrB-1 and agrB-4 and subcloned into the multicloning site of pIMAY using restriction endonucleases KpnI and SacI. Mutagenesis was carried out according to our previously published protocols (48, 49). PCR and DNA sequencing analysis were used to determine the successful generation of mutants.
Generation of ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), and ΔmroQ + mroQ(H180A) complementation strains.
A single-copy chromosomal complementation strain driving mroQ expression under its native promoter was constructed using the integrative plasmid pJC1112 (62). The pJC1112-mroQ plasmid was generated by PCR using oligonucleotides 37F and 37R (Table 2). The resulting amplicon was then subcloned into the pJC1112 vector using the EcoRI and BamHI restriction endonucleases, followed by transformation into E. coli BH10C and electroporation into the RN9011 strain, where the plasmid integrates at the SapI1 site. The integrated complementation vector was then transduced into a ΔmroQ mutant as described above. Successful introduction of the complementation construct was determined by PCR and sequencing. To generate complementation strains harboring amino acid substitutions in MroQ catalytic residues, primer MroQPM-CompF was paired with primer MroQ(E141A)-2, MroQ(E142A)-2, or MroQ(H180A)-2 and primer MroQPM-CompR was paired with MroQ(E141A)-3, MroQ(E142A)-3, or MroQ(H180A)-3. The resulting amplicons from these two PCRs were used in a SOEing PCR with primers MroQPM-CompF and MroQPM-CompR, followed by subcloning the product into pJC1111 using the XbaI and PstI restriction endonucleases and then transformation into E. coli BH10C and electroporation into the RN9011 strain, where the plasmid integrates at the SapI1 site. The integrated complementation vector was then transduced into a ΔmroQ mutant as described above.
Construction of Δagr::tet mutants.
A marked deletion mutation of agrBDCA (Δagr::tet) was transduced into the AH-1263 (WT), ΔmroQ, and ΔagrB strains by bacteriophage-mediated transduction as described above.
Construction of P3-gfp reporter strains.
The pDB59 reporter plasmid contains the P3 promoter of the Agr system driving the expression of gfp (56). The plasmid was isolated from E. coli and passaged through RN4220, followed by electroporation into the WT, ΔmroQ, ΔagrB, ΔagrB ΔmroQ, and Δagr::tet strains.
Construction of pOS1-PsarA-agrD-6×His expression plasmid.
An AgrD-6×His expression plasmid was generated by fusing the PsarA promoter linked to the S. aureus superoxide dismutase (sod) ribosomal binding site (sodRBS) (67, 68) and to the coding sequence of AgrD harboring a 6×His tag on its C terminus. PsarA-sodRBS was amplified using primers PsarA-AgrD6×His-1 and PsarA-AgrD6×His-2, and agrD-6×His was amplified using oligonucleotides PsarA-AgrD6×His-3 and PsarA-AgrD6×His-4. A SOEing PCR was performed to splice these amplicons together using primers PsarA-AgrD6×His-1 and PsarA-AgrD6×His-4. The resulting amplicon was cloned into pOS1 using the PstI and EcoRI restriction endonucleases, and the product was transformed into DH5α, passaged through RN4220, and electroporated into the wild-type, ΔmroQ, ΔmroQ + mroQ, and Δagr isogenic strains.
Construction of pOS1-PHELP-agrBD plasmid.
An agrBD expression plasmid was generated by fusing the PHELP promoter (48, 49, 94) to agrBD. PHELP was amplified using primers phelp-agrBD-1 and phelp-agrBD-2, and agrBD was amplified using oligonucleotides phelp-agrBD-3 and phelp-agrBD-4. A SOEing PCR was performed to splice the resulting amplicons together using primers phelp-agrBD-1 and phelp-agrBD-4. The final amplicon was cloned into pOS1 using the SalI and EcoRI restriction endonucleases, and the resulting plasmid was transformed into E. coli BH10C, passaged through RN4220, and electroporated into the wild-type, Δagr, and Δagr ΔmroQ isogenic strains.
Whole-cell lysate preparation.
Five milliliters of S. aureus strains was grown overnight at 37°C with shaking at 200 rpm. Overnight cultures were diluted 1:100 in 20 ml of fresh medium and allowed to replicate for an additional 8 h at 37°C with shaking at 200 rpm. After 8 h, the OD600 was measured and samples were centrifuged at 4,200 rpm for 15 min to pellet the bacteria. The supernatants were removed, and the bacterial cell pellets were stored at −80°C until use. The pellets were thawed on ice and resuspended in 250 μl of phosphate-buffered saline (PBS) supplemented with 1 M NaCl and 6 M urea (VWR). The resuspended bacteria were then added to screw-cap microcentrifuge lysing tubes (Fisher Scientific) containing 250 μl 0.1-mm glass cell disruption beads (Scientific Industries, Inc.). Cells were lysed using a Fast Prep-24 5G homogenizer (MP Biomedicals) at speed 5.0 for 20 s with subsequent incubation on ice for 5 min and were then disrupted a second time at speed 4.5 for 20 s. Fifty microliters of 6× sodium dodecyl sulfate (SDS) sample buffer (0.375 M Tris, pH 6.8, 12% SDS, 60% glycerol, 0.6 M dithiothreitol, 0.06% bromophenol blue) was added directly to the lysed cells, and the mixture was boiled for 10 min, followed by centrifugation at maximum speed for 15 min at 4°C. Samples were stored at −20°C for no longer than a few days.
Exoprotein preparations.
S. aureus strains were grown at 37°C with shaking at 200 rpm overnight. Overnight cultures were diluted 1:100 in fresh medium and allowed to grow to stationary phase at 37°C with shaking at 200 rpm (∼8 h). The OD600 was measured, and samples were centrifuged at 4,200 rpm for 15 min. The supernatants (1.3 ml) were collected in 1.5-ml microcentrifuge tubes and cooled on ice for approximately 30 min, followed by addition of 150 μl of 100% trichloroacetic acid (TCA) and overnight incubation at 4°C. On the next day, samples were centrifuged at maximum speed for 15 min at 4°C and the supernatants were removed and discarded. Precipitated proteins were washed with 1 ml of 100% ethanol (Decon Laboratories Inc.) and incubated at 4°C for 30 min, followed by a 15-min spin at maximum speed at 4°C. The protein pellets were air dried at room temperature for about 1 h, followed by dissolving the pellet in TCA-SDS sample buffer (0.5 M Tris-HCl buffer, 4% SDS mixed 1:1 with 2× SDS sample buffer) and boiling for 10 min before storage at −20°C.
Immunoblot analysis for secreted virulence factors.
The wild-type, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains were grown in 5 ml of TSB for 8 h at 37°C with shaking at 200 rpm, and the supernatants were collected for TCA precipitation as described above. Prior to loading, protein samples were normalized based on the OD600 and subsequently separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 12% polyacrylamide gels at 120 V in a Quadra Mini-Vertical PAGE/blotting system (CBS Scientific). The resolved proteins were then transferred from the polyacrylamide gels to 0.45-μm-pore-size polyvinylidene difluoride membranes (Immobilon; Roche) at 200 V for 1 h. After transfer, the membranes were stored overnight in Tris-buffered saline (TBS)–Tween 20 (TBST; 0.1% Tween 20 [Amresco] in TBS [Corning]) at 4°C. Membranes were then blocked in TBST–5% bovine serum albumin (BSA) (Amresco) for 1 h while rocking at room temperature. Rabbit anti-LukA (1:50,000), rabbit anti-LukS-PV (1:50,000), and rabbit anti-Hla (1:50,000) antibodies (63) were incubated with the membranes in TBST for 1 h. Although these antibodies recognize LukA, LukS-PV, and Hla, they also permit the detection of protein A due to its binding of the Fc region of antibodies (95). Following incubation with primary antibodies, the membranes were washed three times in TBST for 15 min and goat anti-rabbit IgG (H+L) conjugated to alkaline phosphatase (Thermo) was added at a 1:5,000 dilution in TBST with 5% BSA for 1 h. The membranes were washed 3 times in TBST for 15 min each time, and the blots were developed with 5-bromo-4-chloro-3-indoyl-phosphate–nitroblue tetrazolium (BCIP/NBT) color development substrate (VWR).
AIP reporter assays.
Reporter strains containing pDB59 (described above) and test strains (from which conditioned medium was collected) were cultured overnight in TSB supplemented with Cm at 37°C with shaking at 200 rpm. On the next day, samples were diluted 1:100 in fresh TSB and incubated for 5 h at 37°C with shaking at 200 rpm. Samples were centrifuged at 4,200 rpm for 10 min. The supernatants from the test strains were collected and filter sterilized through a 0.22-nm-pore-size syringe filter and mixed 2:1 with fresh TSB. The reporter strain pellets were then resuspended in 5 ml of the 2:1 conditioned medium-fresh medium mix and incubated for an additional 3 h. The bacteria were washed twice in 3.5 ml PBS and resuspended in 1 ml of PBS. Samples were then diluted 1:1 in PBS and aliquoted in triplicate into a clear-bottom 96-well black plate. The bacterial optical density (OD600) and GFP fluorescence (excitation wavelength, 495 nm; emission wavelength, 509 nm) were measured using a BioTek Synergy H1 microplate reader. The data were normalized and displayed as GFP fluorescence/OD.
Generation of murine BMM.
Murine bone marrow-derived macrophages (BMM) were differentiated from the bone marrow of 6- to 8-week-old female or male C57BL/6J (WT), TLR2−/−, TLR4−/−, and MyD88−/− mice as previously described (47).
Macrophage cytokine profiles.
Bacteria were grown overnight in triplicate in a 96-well round-bottom plate (Corning) in 150 μl of TSB at 37°C with shaking at 200 rpm. Samples were diluted 1:50 in 147 μl of fresh TSB in a 96-well round-bottom plate and incubated for 8 h at 37°C with shaking at 200 rpm. Bacterial growth was assessed by reading the OD600, followed by centrifugation at 3,700 rpm for 15 min to pellet the cells. The supernatants were collected and stored at −80°C. Primary bone marrow-derived macrophages (BMM) from WT, TLR2−/−, TLR4−/−, and MyD88−/− mice were thawed and incubated at 37°C with 5% CO2 for 48 h before seeding in a 96-well plate at 65,000 cells per well. The BMM were allowed to rest overnight before being treated with 10 μl of thawed bacterial supernatant for 24 h at 37°C with 5% CO2. After 24 h, 50 μl of the supernatants was collected and stored at −80°C. To assess cytokine levels, a BD cytometric bead array mouse soluble protein set and a soluble protein master buffer kit were used. The assay was conducted using the manufacturer’s instructions.
Rabbit RBC lysis assay.
For relative quantitation of the hemolytic activity of WT S. aureus and the mutants, strains were grown overnight in a round-bottom 96-well plate in TSB with shaking at 37°C. On the next day, overnight cultures were diluted (1:50) in 150 μl TSB in a 96-well round-bottom plate and incubated at 37°C with shaking for 6 h. A 2% suspension of defibrinated rabbit red blood cells (RBCs; Hemostat) was prepared by washing the cells twice in 1× phosphate-buffered saline (PBS) and diluting cells from the stock RBC packed cell volume in PBS to a final concentration of 2% RBCs. This preparation was kept on ice until use. After 6 h, the bacterial culture was centrifuged for 5 min at 3,900 rpm. One hundred microliters of OD600-normalized cell-free supernatants was added to the top row of round-bottom 96-well plates, and a 2-fold serial dilution series was set up with these supernatants. To each supernatant dilution, 2% RBCs were added (1:1), followed by incubation for 1 h at 37°C. The plates were then centrifuged for 5 min at 1,500 rpm to pellet the nonlysed RBCs, the supernatants were transferred to a flat-bottom 96-well plate, and red blood cell lysis was measured at OD450 on a spectrophotometer.
Murine skin and soft tissue infection model.
Cultures of the wild-type, ΔmroQ, ΔmroQ + mroQ, ΔmroQ + mroQ(E141A), ΔmroQ + mroQ(E142A), ΔmroQ + mroQ(H180A), and Δagr strains were inoculated from freshly isolated single colonies and grown with shaking at 200 rpm at 37°C overnight. On the next day, a 1:100 subculture in 15 ml of TSB was incubated at 37°C for 3 h with shaking. Cultures were then centrifuged for 5 min at maximum speed, and cell pellets were washed twice in 5 ml PBS. Bacterial suspensions were diluted 2 ml into 8 ml of PBS, normalized with PBS to an OD600 of approximately 0.32 to 0.33 (1 × 108 CFU/ml), and mixed 1:1 with sterile Cytodex beads (Sigma). The mice were deeply anesthetized with 2,2,2-tribromoethanol (Avertin; Sigma) via intraperitoneal injection, followed by removal of fur and infection with 100 μl PBS containing 1 × 107 CFU of bacteria and Cytodex beads on the right and left sides by intradermal injection. Mice were monitored daily; and at 72 or 120 h postinfection, they were euthanized. Skin abscesses were collected, homogenized, spread onto TSA plates, and incubated overnight at 37°C in order to enumerate the CFU. Skin homogenates were also used for cytometric bead array analysis of local cytokine levels as described above.
Ethics statement.
All experiments were performed following the ethical standards of the institutional Biosafety Committee and the Institutional Animal Care and Use Committee (IACUC) at Loyola University Chicago, Health Sciences Division. The institution is approved by the Public Health Service (PHS; A3117-01 through 28 February 2022), is fully accredited by AAALAC International (000180, certification dated 17 November 2016), and is registered/licensed by USDA (33-R-0024 through 24 August 2020). All animal experiments were performed in animal biosafety level 2 facilities with IACUC-approved protocols (IACUC protocol 2017028) under the guidance of the Office of Laboratory Animal Welfare (OLAW) following USDA and PHS policy and guidelines on the humane care and use of laboratory animals.
Statistical analyses.
All experiments were repeated at least three independent times. For in vitro macrophage and AIP reporter data, statistical significance was analyzed with data from representative experiments that were conducted in triplicate and that were repeated a minimum of three independent times. All statistical significance was analyzed using GraphPad Prism (version 7.0) software by the statistical tests specified in the figure legends. Prior to conducting statistical analyses on data derived from animal studies, a D’Agostino and Pearson or Shapiro-Wilk normality test was conducted. Based on normality testing, either analysis of variance (ANOVA) or a nonparametric test was performed. Post hoc statistical significance was calculated using Dunn’s post hoc test. The number of animals per treatment group is indicated by the n values in the figure legends. For all other data, statistical significance (P < 0.05) was determined by one-way ANOVA with Tukey’s post hoc test.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the F. Alonzo laboratory for helpful discussions and Alex Argianas for assistance with preliminary studies that helped guide project directions.
This work was supported by grants NIH R01 AI120994 (to F.A.), AHA 17PRE33660173 (to J.P.G.), and AHA 19POST34380259 (to W.P.T.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00019-19.
For a companion article on this topic, see https://doi.org/10.1128/IAI.00002-19.
REFERENCES
- 1.Monnet V, Juillard V, Gardan R. 2016. Peptide conversations in Gram-positive bacteria. Crit Rev Microbiol 42:339–351. doi: 10.3109/1040841X.2014.948804. [DOI] [PubMed] [Google Scholar]
- 2.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus, and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem Rev 111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang B, Muir TW. 2016. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol 23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyon GJ, Novick RP. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 7.LaSarre B, Federle MJ. 2013. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev 77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A 110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scutera S, Zucca M, Savoia D. 2014. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin Drug Discov 9:353–366. doi: 10.1517/17460441.2014.894974. [DOI] [PubMed] [Google Scholar]
- 10.Remy B, Mion S, Plener L, Elias M, Chabriere E, Daude D. 2018. Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front Pharmacol 9:203. doi: 10.3389/fphar.2018.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salam AM, Quave CL. 2018. Targeting virulence in Staphylococcus aureus by chemical inhibition of the accessory gene regulator system in vivo. mSphere 3:e00500-17. doi: 10.1128/mSphere.00500-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brackman G, Coenye T. 2015. Inhibition of quorum sensing in Staphylococcus spp. Curr Pharm Des 21:2101–2108. doi: 10.2174/1381612821666150310101014. [DOI] [PubMed] [Google Scholar]
- 13.Parashar V, Aggarwal C, Federle MJ, Neiditch MB. 2015. Rgg protein structure-function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci U S A 112:5177–5182. doi: 10.1073/pnas.1500357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quave CL, Horswill AR. 2018. Identification of staphylococcal quorum sensing inhibitors by quantification of õ-hemolysin with high performance liquid chromatography. Methods Mol Biol 1673:363–370. doi: 10.1007/978-1-4939-7309-5_27. [DOI] [PubMed] [Google Scholar]
- 15.Le KY, Otto M. 2015. Quorum-sensing regulation in staphylococci—an overview. Front Microbiol 6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Yeo WS, Bae T. 2016. The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7:E81.27706107 [Google Scholar]
- 17.Cheung AL, Zhang G. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front Biosci 7:d1825–d1842. doi: 10.2741/A882. [DOI] [PubMed] [Google Scholar]
- 18.Burgui S, Gil C, Solano C, Lasa I, Valle J. 2018. A systematic evaluation of the two-component systems network reveals that ArlRS is a key regulator of catheter colonization by Staphylococcus aureus. Front Microbiol 9:342. doi: 10.3389/fmicb.2018.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker JN, Crosby HA, Spaulding AR, Salgado-Pabon W, Malone CL, Rosenthal CB, Schlievert PM, Boyd JM, Horswill AR. 2013. The Staphylococcus aureus ArlRS two-component system is a novel regulator of agglutination and pathogenesis. PLoS Pathog 9:e1003819. doi: 10.1371/journal.ppat.1003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4:e00696-13. doi: 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall JW, Yang J, Guo H, Ji Y. 2015. The AirSR two-component system contributes to Staphylococcus aureus survival in human blood and transcriptionally regulates sspABC operon. Front Microbiol 6:682. doi: 10.3389/fmicb.2015.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue T, You Y, Hong D, Sun H, Sun B. 2011. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect Immun 79:2154–2167. doi: 10.1128/IAI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins RL, Zurek OW, Pallister KB, Voyich JM. 2013. The SaeR/S two-component system induces interferon-gamma production in neutrophils during invasive Staphylococcus aureus infection. Microbes Infect 15:749–754. doi: 10.1016/j.micinf.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Yang SJ, Bayer AS, Mishra NN, Meehl M, Ledala N, Yeaman MR, Xiong YQ, Cheung AL. 2012. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun 80:74–81. doi: 10.1128/IAI.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang SJ, Xiong YQ, Yeaman MR, Bayles KW, Abdelhady W, Bayer AS. 2013. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob Agents Chemother 57:3875–3882. doi: 10.1128/AAC.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet 248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 27.Thoendel M, Horswill AR. 2009. Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem 284:21828–21838. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Gray L, Novick RP, Ji G. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J Biol Chem 277:34736–34742. doi: 10.1074/jbc.M205367200. [DOI] [PubMed] [Google Scholar]
- 29.Wang B, Zhao A, Novick RP, Muir TW. 2015. Key driving forces in the biosynthesis of autoinducing peptides required for staphylococcal virulence. Proc Natl Acad Sci U S A 112:10679–10684. doi: 10.1073/pnas.1506030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Ji G. 2004. Identification of a staphylococcal AgrB segment(s) responsible for group-specific processing of AgrD by gene swapping. J Bacteriol 186:6706–6713. doi: 10.1128/JB.186.20.6706-6713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Lin J, Ji G. 2004. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum-sensing pheromone in Staphylococcus aureus. J Biol Chem 279:19448–19456. doi: 10.1074/jbc.M311349200. [DOI] [PubMed] [Google Scholar]
- 32.Kavanaugh JS, Thoendel M, Horswill AR. 2007. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol 65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 33.Geisinger E, Chen J, Novick RP. 2012. Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus. J Bacteriol 194:2854–2864. doi: 10.1128/JB.06685-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, Zhao A, Xie Q, Olinares PD, Chait BT, Novick RP, Muir TW. 2017. Functional plasticity of the AgrC receptor histidine kinase required for staphylococcal virulence. Cell Chem Biol 24:76–86. doi: 10.1016/j.chembiol.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tal-Gan Y, Ivancic M, Cornilescu G, Blackwell HE. 2016. Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus. Org Biomol Chem 14:113–121. doi: 10.1039/c5ob01735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B, Zhao A, Novick RP, Muir TW. 2014. Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol Cell 53:929–940. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geisinger E, Muir TW, Novick RP. 2009. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci U S A 106:1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geisinger E, George EA, Chen J, Muir TW, Novick RP. 2008. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem 283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyon GJ, Wright JS, Muir TW, Novick RP. 2002. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- 40.Ji G, Beavis R, Novick RP. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 41.Wright JS III, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. 2005. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol 187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun 70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, Ramos M, Bayer AS. 1994. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Invest 94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heyer G, Saba S, Adamo R, Rush W, Soong G, Cheung A, Prince A. 2002. Staphylococcus aureus agr and sarA functions are required for invasive infection but not inflammatory responses in the lung. Infect Immun 70:127–133. doi: 10.1128/IAI.70.1.127-133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Bubeck Wardenburg J, Schneewind O, Otto M, Deleo FR. 2011. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis 204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grayczyk JP, Harvey CJ, Laczkovich I, Alonzo F III.. 2017. A lipoylated metabolic protein released by Staphylococcus aureus suppresses macrophage activation. Cell Host Microbe 22:678–687.e9. doi: 10.1016/j.chom.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laczkovich I, Teoh WP, Flury S, Grayczyk JP, Zorzoli A, Alonzo F III.. 16 April 2018. Increased flexibility in the use of exogenous lipoic acid by Staphylococcus aureus. Mol Microbiol doi: 10.1111/mmi.13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zorzoli A, Grayczyk JP, Alonzo F III.. 2016. Staphylococcus aureus tissue infection during sepsis is supported by differential use of bacterial or host-derived lipoic acid. PLoS Pathog 12:e1005933. doi: 10.1371/journal.ppat.1005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pei J, Mitchell DA, Dixon JE, Grishin NV. 2011. Expansion of type II CAAX proteases reveals evolutionary origin of gamma-secretase subunit APH-1. J Mol Biol 410:18–26. doi: 10.1016/j.jmb.2011.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kjos M, Borrero J, Opsata M, Birri DJ, Holo H, Cintas LM, Snipen L, Hernandez PE, Nes IF, Diep DB. 2011. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157:3256–3267. doi: 10.1099/mic.0.052571-0. [DOI] [PubMed] [Google Scholar]
- 52.Ellermeier CD, Losick R. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev 20:1911–1922. doi: 10.1101/gad.1440606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biswas S, Biswas I. 2014. A conserved streptococcal membrane protein, LsrS, exhibits a receptor-like function for lantibiotics. J Bacteriol 196:1578–1587. doi: 10.1128/JB.00028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Draper LA, Cotter PD, Hill C, Ross RP. 2015. Lantibiotic resistance. Microbiol Mol Biol Rev 79:171–191. doi: 10.1128/MMBR.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Connor L, Coffey A, Daly C, Fitzgerald GF. 1996. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl Environ Microbiol 62:3075–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. 2010. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol Microbiol 78:238–252. doi: 10.1111/j.1365-2958.2010.07334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grundling A, Missiakas DM, Schneewind O. 2006. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol 188:6286–6297. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poupel O, Proux C, Jagla B, Msadek T, Dubrac S. 2018. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog 14:e1006917. doi: 10.1371/journal.ppat.1006917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Firon A, Tazi A, Da Cunha V, Brinster S, Sauvage E, Dramsi S, Golenbock DT, Glaser P, Poyart C, Trieu-Cuot P. 2013. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog 9:e1003179. doi: 10.1371/journal.ppat.1003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qoronfleh MW, Streips UN, Wilkinson BJ. 1990. Basic features of the staphylococcal heat shock response. Antonie Van Leeuwenhoek 58:79–86. doi: 10.1007/BF00422721. [DOI] [PubMed] [Google Scholar]
- 62.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. 2014. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76:1–7. doi: 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alonzo F III, Benson MA, Chen J, Novick RP, Shopsin B, Torres VJ. 2012. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol Microbiol 83:423–435. doi: 10.1111/j.1365-2958.2011.07942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. 2014. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol 22:676–685. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dumont AL, Yoong P, Day CJ, Alonzo F III, McDonald WH, Jennings MP, Torres VJ. 2013. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc Natl Acad Sci U S A 110:10794–10799. doi: 10.1073/pnas.1305121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DuMont AL, Yoong P, Surewaard BG, Benson MA, Nijland R, van Strijp JA, Torres VJ. 2013. Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect Immun 81:1830–1841. doi: 10.1128/IAI.00095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol 192:2068–2076. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heinrich J, Wiegert T. 2006. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol Microbiol 62:566–579. doi: 10.1111/j.1365-2958.2006.05391.x. [DOI] [PubMed] [Google Scholar]
- 71.Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. 2000. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry 39:4096–4104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 72.Plummer LJ, Hildebrandt ER, Porter SB, Rogers VA, McCracken J, Schmidt WK. 2006. Mutational analysis of the Ras converting enzyme reveals a requirement for glutamate and histidine residues. J Biol Chem 281:4596–4605. doi: 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Autret N, Raynaud C, Dubail I, Berche P, Charbit A. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect Immun 71:4463–4471. doi: 10.1128/IAI.71.8.4463-4471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riedel CU, Monk IR, Casey PG, Waidmann MS, Gahan CG, Hill C. 2009. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol Microbiol 71:1177–1189. doi: 10.1111/j.1365-2958.2008.06589.x. [DOI] [PubMed] [Google Scholar]
- 75.Zetzmann M, Sanchez-Kopper A, Waidmann MS, Blombach B, Riedel CU. 2016. Identification of the Agr peptide of Listeria monocytogenes. Front Microbiol 7:989. doi: 10.3389/fmicb.2016.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paspaliari DK, Mollerup MS, Kallipolitis BH, Ingmer H, Larsen MH. 2014. Chitinase expression in Listeria monocytogenes is positively regulated by the Agr system. PLoS One 9:e95385. doi: 10.1371/journal.pone.0095385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, Pettit L, Dougan G, Lawley TD, Wren BW. 2013. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol 195:3672–3681. doi: 10.1128/JB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin X, Singh KV, Weinstock GM, Murray BE. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun 68:2579–2586. doi: 10.1128/IAI.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Engelbert M, Mylonakis E, Ausubel FM, Calderwood SB, Gilmore MS. 2004. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect Immun 72:3628–3633. doi: 10.1128/IAI.72.6.3628-3633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho JS, Zussman J, Donegan NP, Ramos RI, Garcia NC, Uslan DZ, Iwakura Y, Simon SI, Cheung AL, Modlin RL, Kim J, Miller LS. 2011. Noninvasive in vivo imaging to evaluate immune responses and antimicrobial therapy against Staphylococcus aureus and USA300 MRSA skin infections. J Invest Dermatol 131:907–915. doi: 10.1038/jid.2010.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melehani JH, Duncan JA. 2016. Inflammasome activation can mediate tissue-specific pathogenesis or protection in Staphylococcus aureus infection. Curr Top Microbiol Immunol 397:257–282. doi: 10.1007/978-3-319-41171-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller LS, Cho JS. 2011. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol 11:505–518. doi: 10.1038/nri3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller LS, O'Connell RM, Gutierrez MA, Pietras EM, Shahangian A, Gross CE, Thirumala A, Cheung AL, Cheng G, Modlin RL. 2006. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 24:79–91. doi: 10.1016/j.immuni.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 84.Miller LS, Pietras EM, Uricchio LH, Hirano K, Rao S, Lin H, O'Connell RM, Iwakura Y, Cheung AL, Cheng G, Modlin RL. 2007. Inflammasome-mediated production of IL-1beta is required for neutrophil recruitment against Staphylococcus aureus in vivo. J Immunol 179:6933–6942. doi: 10.4049/jimmunol.179.10.6933. [DOI] [PubMed] [Google Scholar]
- 85.Kitur K, Wachtel S, Brown A, Wickersham M, Paulino F, Penaloza HF, Soong G, Bueno S, Parker D, Prince A. 2016. Necroptosis promotes Staphylococcus aureus clearance by inhibiting excessive inflammatory signaling. Cell Rep 16:2219–2230. doi: 10.1016/j.celrep.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Powers ME, Becker RE, Sailer A, Turner JR, Bubeck Wardenburg J. 2015. Synergistic action of Staphylococcus aureus alpha-toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe 17:775–787. doi: 10.1016/j.chom.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benson MA, Lilo S, Wasserman GA, Thoendel M, Smith A, Horswill AR, Fraser J, Novick RP, Shopsin B, Torres VJ. 2011. Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol Microbiol 81:659–675. doi: 10.1111/j.1365-2958.2011.07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bebbington C, Yarranton G. 2008. Antibodies for the treatment of bacterial infections: current experience and future prospects. Curr Opin Biotechnol 19:613–619. doi: 10.1016/j.copbio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 89.Tan L, Li SR, Jiang B, Hu XM, Li S. 2018. Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front Microbiol 9:55. doi: 10.3389/fmicb.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Howell-Adams B, Seifert HS. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol Microbiol 37:1146–1158. doi: 10.1046/j.1365-2958.2000.02067.x. [DOI] [PubMed] [Google Scholar]
- 93.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riedel CU, Monk IR, Casey PG, Morrissey D, O'Sullivan GC, Tangney M, Hill C, Gahan CG. 2007. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Appl Environ Microbiol 73:3091–3094. doi: 10.1128/AEM.02940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Forsgren A, Sjoquist J. 1966. Protein A from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J Immunol 97:822–827. [PubMed] [Google Scholar]
- 96.Fairweather N, Kennedy S, Foster TJ, Kehoe M, Dougan G. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect Immun 41:1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Benson MA, Lilo S, Nygaard T, Voyich JM, Torres VJ. 2012. Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J Bacteriol 194:4355–4365. doi: 10.1128/JB.00706-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.