Despite the severity and global burden of Cryptosporidium infection, treatments are less than optimal, and there is no effective vaccine. Egress from host cells is a key process for the completion of the life cycle of apicomplexan parasites.
KEYWORDS: calcium-dependent protein kinase, Cryptosporidium parvum, subtilisin, cryptosporidiosis, egress
ABSTRACT
Despite the severity and global burden of Cryptosporidium infection, treatments are less than optimal, and there is no effective vaccine. Egress from host cells is a key process for the completion of the life cycle of apicomplexan parasites. For Plasmodium species, subtilisin-like serine protease (SUB1) is a key mediator of egress. For Toxoplasma species, calcium-dependent protein kinases (CDPKs) are critical. In this study, we characterized Cryptosporidium SUB1 expression and evaluated its effect using an infection model. We found increased expression between 12 and 20 h after in vitro infection, prior to egress. We induced silencing of SUB1 (ΔSUB1) mRNA using SUB1 single-stranded antisense RNA coupled with human Argonaute 2. Silencing of SUB1 mRNA expression did not affect parasite viability, excystation, or invasion of target cells. However, knockdown led to a 95% decrease in the proportion of released merozoites in vitro (P < 0.0001). In contrast, silencing of CDPK5 had no effect on egress. Overall, our results indicate that SUB1 is a key mediator of Cryptosporidium egress and suggest that interruption of the life cycle at this stage may effectively inhibit the propagation of infection.
INTRODUCTION
Cryptosporidium species are protozoan parasites that infect intestinal epithelial cells (1). While human infection is self-limiting in immunocompetent adults (2), studies in resource-poor countries have identified Cryptosporidium as an important cause of morbidity and mortality in young children, particularly those under the age of 2 years (3, 4). Millions of children in South Asia and sub-Saharan Africa are infected, with an estimate of 60,000 to 200,000 child deaths per year (5, 6). Cryptosporidiosis is also a major contributor to childhood malnutrition (1, 7–11). Recurring and persistent gastrointestinal infections may lead to defects in cognitive and physical development in children (8, 12, 13). Unfortunately, current treatments are not optimal, and there is no effective vaccine (14). Therefore, there is a clear need for further studies on the biology of the parasite.
Cryptosporidium is an obligately intracellular parasite. Egress is required for the reinvasion of cells, leading to parasite proliferation and persistence. Egress appears to be driven by parasite molecules (15–17). Mechanisms of egress have been studied in other apicomplexan parasites. Toxoplasma gondii calcium-dependent protein kinase 3 (CDPK3) has been identified as a crucial moderator of parasite egress (15, 18). In Plasmodium falciparum, egress has been found to be related to the cleavage of proteins found on the parasitophorous vacuolar membrane (PVM), as well as the host cell membrane, by subtilisin-like serine protease (SUB1) (17, 19–21).
While invasion has been studied in Cryptosporidium (11, 14, 22–25), there is little information regarding the molecular mechanisms of egress. We identified homologues of P. falciparum SUB1 and Toxoplasma CDPK3 in Cryptosporidium. We hypothesized that Cryptosporidium parvum SUB1 (CpSUB1) and/or CDPK5 (CpCDPK5; the homologue of Toxoplasma CDPK3) are key mediators of Cryptosporidium egress from host cells. For this study, we first determined the expression patterns of SUB1 and CDPK5 throughout the asexual stage of infection. We then determined the specific roles of SUB1 and CDPK5 by a modified small interfering RNA (siRNA) method (26, 27) and evaluated the effects on parasite viability, infection of host intestines, and the egress of merozoites during infection.
RESULTS
SUB1 expression peaks prior to the peak of parasite release from in vitro culture.
To determine the time of parasite egress from human epithelial cells, supernatants from infected cells were collected at different time points. Parasite RNA appeared at 18 h, and the highest levels were detected at ∼24 h postinoculation (Fig. 1A). We found that SUB1 mRNA expression increased significantly between 12 and 20 h postinfection and then decreased at 22 to 24 h (Fig. 1B). As a control, we determined the expression of a second serine protease and observed a peak at 1 h postinfection, with no significant increase in expression throughout infection (see Fig. S1 in the supplemental material). We also observed the pattern of expression of CDPK5 and found that expression of this target peaked at 24 h postinfection (see Fig. S2 in the supplemental material). All data are representative of at least two independent experiments with consistent results.
FIG 1.
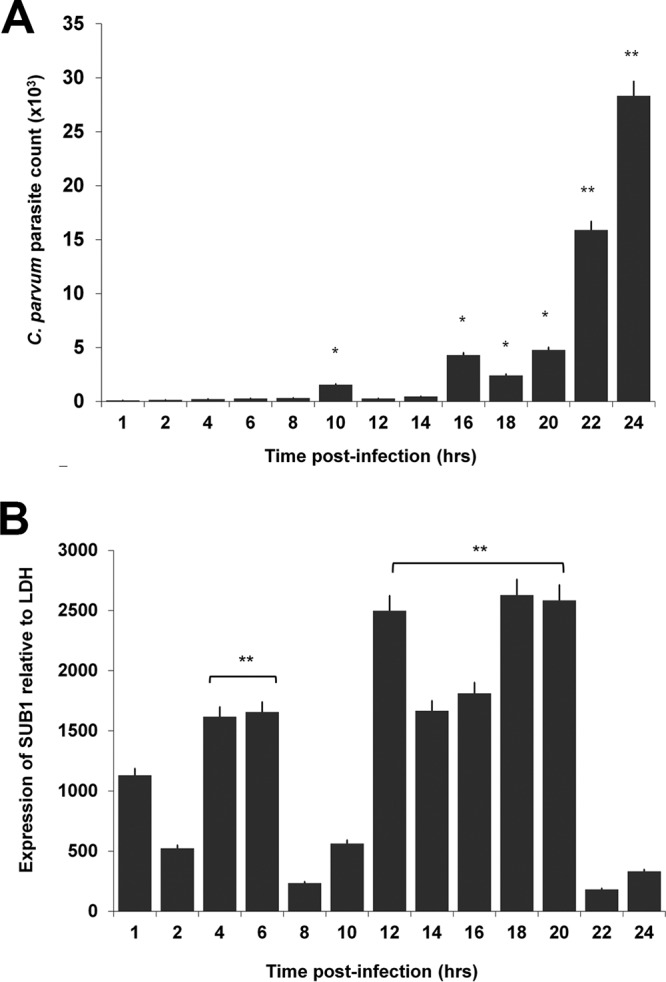
Timing of parasite egress and SUB1 expression. Viable Cryptosporidium parvum sporozoites were used to infect HCT-8 monolayers. (A) Supernatants were collected and assayed for released parasites using qPCR. The peak count of released merozoites was observed at 24 h postinfection. Data are representative of the results of four independent experiments. **, P < 0.0001; *, P < 0.005. (B) Sporozoite-infected HCT-8 monolayers were collected at different times postinoculation. Levels of SUB1 mRNA were analyzed by qRT-PCR and relative quantification. Peak expression of SUB1 was observed at 12 to 20 h postinfection with respect to initial infection at 1 h (P < 0.0001). Data are representative of the results of three independent experiments.
Antisense RNA designed against SUB1 produces potent gene silencing.
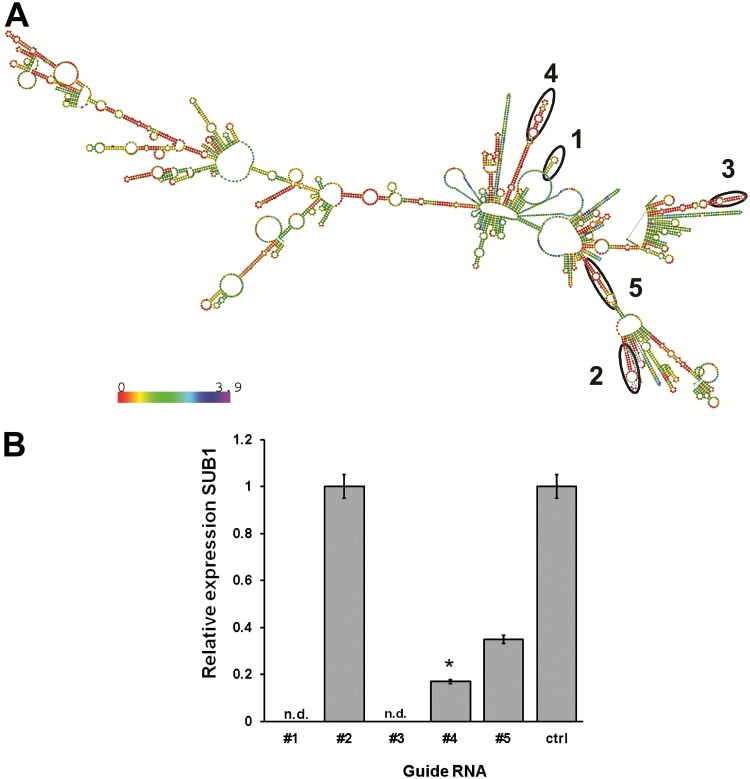
Using base-pairing probability algorithms determined by the predicted secondary structure, we synthesized five guide sequences designed to bind optimally to the mRNA sequence (Fig. 2A). Of the five guide sequences targeting Cryptosporidium SUB1, antisense sequences 4 and 5 resulted in 80% (P < 0.0001) and 60% (P < 0.05) reductions in expression from that for wild-type (WT) parasites. In contrast, guide RNA 2 was not effective, and guide RNAs 1 and 3 were toxic to the parasites, with no observable amplification of the housekeeping gene encoding lactate dehydrogenase (LDH) (Fig. 2B). To confirm that silencing was not detrimental to parasite viability, we stained oocysts with carboxyfluorescein succinimidyl ester (CFSE) after silencing and induced excystation. Using fluorescence microscopy, we observed >95% viability with antisense RNA 4 relative to that of wild-type parasite samples. Thus, using guide RNA 4, we were able to silence SUB1 expression without affecting parasite viability or excystation. Subsequent studies used this guide RNA to create parasites with silenced SUB1 (ΔSUB1).
FIG 2.
Design of ssRNA guides and inhibition of SUB1. (A) SUB1 ssRNA and SUB1 mRNA secondary structure. (B) Silencing was carried out in oocysts, and levels of silencing were determined by RT-PCR. With guide sequences 1 and 3, parasites were not detectable (n.d.). Sequence 2 failed to silence the expression of SUB1. Guide sequence 5 demonstrated moderate silencing, while sequence 4 showed significant silencing (P < 0.0001). Data are representative of the results of three independent experiments.
Silencing SUB1 blocks parasite egress from host cells.
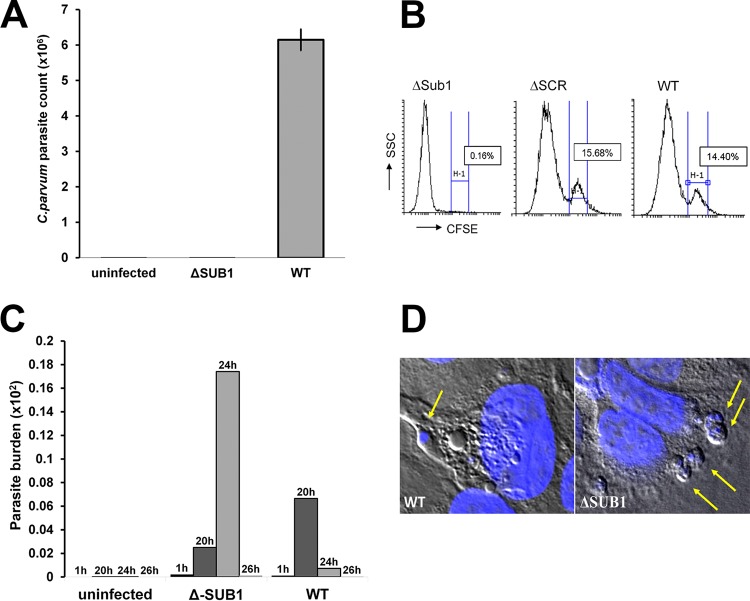
ΔSUB1 parasites were used to infect cultured HCT-8 cells. We collected sample supernatants at 24 h postinfection. With ΔSUB1 sporozoites, there was a 95% decrease (P < 0.0001) in parasite RNA in supernatants compared to supernantants from wild-type parasites (Fig. 3A). In contrast, there was no change in egress when CDPK5 was silenced (see Fig. S3 in the supplemental material). Merozoites that had egressed were also quantified using flow cytometry. Three independent experiments demonstrated a 99.5% lower proportion of merozoites in the supernatant for ΔSUB1 parasites compared to wild-type parasite controls (Fig. 3B) (P, <0.0001 by analysis of variance [ANOVA]).
FIG 3.
(A) SUB1-silenced samples had reduced levels of egress. We observed significantly lower levels of parasites that had egressed in supernatants collected from SUB1-silenced samples than in supernatants from controls at 24 h postinfection (P < 0.05). Data are representative of the results of three independent experiments. (B) Quantification of free merozoites after silencing. Supernatants of infected cells were collected and quantified with a fluorescence-activated cell sorter. The proportion of stained free merozoites is highlighted in the H-1 subpopulation and was found to be significantly lower in ΔSUB1 (SUB1 silenced) parasites than in wild-type parasite controls (P < 0.05). Data are representative of the results of two independent experiments. ΔSCR, scrambled antisense RNA. (C) The parasite burden in the HCT-8 layer was quantitated by qPCR for parasite LDH at various time points after infection. There was no significant difference between wild-type and ΔSUB1 parasites at 1 h postinfection. However, at the 24-h time point, the parasite burden was markedly higher for ΔSUB1 parasites than for the wild type (P < 0.001). (D) C. parvum-infected HCT-8 cells stained with DAPI and visualized using confocal microscopy. ΔSUB1 parasites demonstrated a higher number of intracellular parasites than wild-type parasite controls.
We analyzed the levels of residual parasites in the infected monolayers. At 1 h postinfection, there were similar quantities of parasite DNA for wild-type parasites and ΔSUB1 parasites (Fig. 3C). However, at 24 h postinfection, the levels of parasites in wild-type samples decreased drastically, due to merozoite egress. The ΔSUB1 parasite burden at 24 h postinfection was significantly higher than the wild-type parasite burden (P < 0.001) (Fig. 3C). Additionally, we used confocal microscopy to visualize retained intracellular meronts in host cells. C. parvum ΔSUB1-infected cells demonstrated higher numbers of intracellular parasites than wild-type controls (Fig. 3D).
DISCUSSION
Egress is a key step in the Cryptosporidium life cycle. Motile merozoites are released, facilitating the spread of infection from the infected cell throughout the epithelium. Cryptosporidium SUB1 and CDPKs have been implicated in the invasion of host cells (26, 28). Here we studied the effects of SUB1 silencing on merozoite egress into the cell supernatant. Using quantitative PCR (qPCR) and flow cytometry, we demonstrated that silencing of CpSUB1 led to a 95% reduction in the proportion of parasites egressing from infected cells. This was associated with an increase in “trapped” parasites within the host cell monolayer. In contrast, silencing of CpCDPK5 did not significantly affect parasite egress.
The release of asexual apicomplexan parasites from the host cell is driven by parasite molecules (17). Two main mechanisms have been described. P. falciparum subtilisin-like serine protease 1 (PfSUB1) has been found to proteolytically activate serine repeat antigen (SERA), which subsequently cleaves the parasitophorous vacuolar membrane (PVM) and host membrane to release parasites (17, 29, 30). In contrast, Toxoplasma CDPKs involved in parasite motility have been found to play a crucial role in the egress of the parasite (15, 31). PfSUB1 is synthesized and packaged into exonemes (17, 29, 30). The exonemes discharge PfSUB1 into the parasitophorous vacuolar space immediately prior to egress (17, 29, 30). SUB1 then proteolytically activates SERA, which cleaves the PVM and host membrane to release parasites (17, 29). Since the active site of apicomplexan SUB1 homologues is highly conserved, we reasoned that CpSUB1 would play a similar role. Interestingly, we noted that mRNA expression for CpSUB1 was increased between 12 and 20 h postinfection, corresponding to the timing of merozoite development. This is consistent with the synthesis and export of preformed SUB1 into excretory vesicles, as was noted for P. falciparum. However, we were unable to study this directly, due to the lack of availability of antibodies to CpSUB1. However, silencing of CpSUB1 clearly blocked egress.
CDPKs have shown to be important factors in the egress of related apicomplexans and to be crucial drivers of development and motility (31–33). A previous study observed the expression patterns of CDPKs in Cryptosporidium parvum, demonstrating that a few had cyclical patterns of expression and that expression was upregulated prior to egress. In this study, CDPK5, a homologue of T. gondii CDPK3, had peak expression at 24 h postinfection (Fig. S2 in the supplemental material). However, after the silencing of CDPK5, the quantities of merozoites found in the supernatant were not significantly different from the quantities of wild-type parasite controls. Based on these observations, we conclude that CDPK5 is not crucial for egress in C. parvum. However, other calcium-dependent protein kinases present in the genome of C. parvum have increased expression prior to egress as well (34). Further studies will have to be conducted in order to investigate whether CDPKs play a role in C. parvum egress.
In summary, our data demonstrate that CpSUB1 is essential for merozoite egress from infected epithelial cells. In contrast, silencing of CDPK5 did not affect egress. Thus, Cryptosporidium may use a mechanism resembling that of P. falciparum more closely than that of Toxoplasma. Further studies are needed to confirm the localization of CpSUB1 and, if it is localized to exonemes, to determine the signals involved in its release. For now, our results show that silencing of SUB1 reduces parasite egress from infected cells, and thus, SUB1 may be a target for future vaccine and drug development.
MATERIALS AND METHODS
Parasites, infection model, and excystation.
Cryptosporidium parvum oocysts, Iowa strain II, were purchased from the Animal and Comparative Biomedical Sciences Department at the University of Arizona. Excystation of C. parvum was induced by incubating the parasites with acidic water (60 μl; pH ∼2.5), followed by 250 μl of 0.8% taurocholate in serum-free RPMI medium as described previously (26). Sporozoites were separated from unexcysted and residual oocysts by filtration using a 3-μm-pore-size membrane (Millipore Sigma, Burlington, MA). In vitro infection was carried out with human ileocecal adenocarcinoma epithelial cells (HCT-8; ATCC, Manassas, VA) as described previously (35–37). Briefly, approximately 5 × 105 HCT-8 cells were plated overnight on 24-well plates (Corning, Tewksbury, MA) to approximately 80% confluence, after which the medium was removed and C. parvum sporozoites (∼2 × 106) in excystation medium were added to establish a basal infection (2 h at 37°C under 5% CO2). For RNA extraction, cells and 250 μl of supernatant were collected at the following time points postinfection: 1, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, and 26 h. We also collected the residual monolayers at 1, 20, 24, and 26 h postinfection. For RNA extraction and silencing experiments, supernatants from infected monolayers were removed and were passed through a 3-μm-pore-size filter to separate the parasite from the host cells. The supernatant was then added to 350 μl of Qiagen lysis buffer and was stored at –20°C for later RNA extraction and analysis by reverse transcription-quantitative PCR (qPCR). In addition, infected monolayers were lysed directly from culture plates by adding 350 μl of lysis buffer from the Qiagen (Valencia, CA) RNeasy kit. Infection and silencing were each confirmed in three independent experiments with biological duplicates and technical triplicates. Student’s t test was used in time-of-egress experiments to determine statistical significance. For experiments detailing the expression profile of SUB1, statistical significance was determined by ANOVA.
Antisense ssRNA design.
Antisense single-stranded RNA (ssRNA) sequences of 19 to 29 nucleotides, targeting different regions of CpSUB1 and CpCDPK5, were designed using RNAfold software (Institute for Theoretical Chemistry, University of Vienna). All antisense RNAs contained the modifications described previously (26). Targets were selected by including hairpins and loops in the secondary structure, low GC counts, a lack of internal repeats, and the distance from the start codon in order to design optimal antisense RNAs. Single-stranded antisense sequences are listed in Table S2 in the supplemental material. The target sequences were CDPK5 (ApiDB_CryptoDB: cgd2_1300; NCBI Protein database accession no. XP_626355.1) and SUB1 (ApiDB_CryptoDB: cgd6_4840; NCBI Protein database accession no. XP_627811.1) and contained the modifications described previously (26).
Silencing of RNA expression in oocysts.
To carry out silencing experiments, we assembled a silencing complex and transfected it into parasites using a lipid-based transfection system. Purified recombinant human Argonaute 2 (hAGO2) (250 ng; Sino-Biological, Beijing, China) was coupled with 1 μM single-stranded RNA with assembly buffer (30 mM HEPES [pH 7.4], 150 mM potassium acetate, and 2 mM magnesium chloride [MgCl2]) (26, 38) and incubated at room temperature for 1 h. To encapsulate the complex, 10 μl of protein transfection reagent (PTR) (Thermo Fisher Scientific, Waltham, MA) was added to the sample, which was then incubated at room temperature for 30 min. For transfection experiments, 5 × 105 Cryptosporidium oocysts were added to the sample containing the complexes, and the mixture was incubated at room temperature for 2 h. While oocysts are relatively impermeable, we have demonstrated previously by use of fluorescent immunoglobulin or Argonaute that protein transfection reagent facilitates the transfer of large proteins into the oocyst (26). While oocysts are relatively metabolically inactive, mRNA is continuously produced and is further increased by incubation in vitro at 37°C. Thus, to activate silencing, the transfected parasites were incubated at 37°C for 2 h. To confirm silencing, ∼20 ng of total RNA was extracted from the oocysts using the Qiagen (Valencia, CA) RNeasy kit and was stored at –20°C until further analysis by real-time PCR. Silencing potency was determined in three independent experiments, each with biological duplicates and technical triplicates. Student’s t test was used in these experiments to determine statistical significance.
Quantification of egress by flow cytometry.
To quantify the egress of merozoites from infected cells, sporozoites were stained with the vital dye carboxyfluorescein succinimidyl ester (CFSE) at 0.5 μM prior to infection of the HCT-8 cells (Thermo Fisher Scientific, Waltham, MA). After 24 h of infection, the supernatants were analyzed by flow cytometry. Sporozoites from approximately 5 × 105 oocysts were separated from unexcysted oocysts by filtration. HCT-8 cells were infected with sporozoites for 2 h at 37°C to establish a basal infection; then the medium was removed and the monolayer washed once with ∼250 μl phosphate-buffered saline (PBS). RPMI medium supplemented with 10% fetal bovine serum (FBS) was added to cells and incubated for 26 h. To evaluate the effect of silencing on egress, we quantified the labeled parasites in supernatants from infected cells. For these experiments, 300 μl of supernatant was collected at 1 h postinfection and then every 2 h starting at 2 h and continuing to 26 h. To quantify parasites in the supernatant, we used flow cytometry to observe stained viable merozoites. We separated parasites from sloughed-off HCT-8 cells that may have been collected by use of a 3-μm-pore-size filter. Samples were then centrifuged at 8,000 × g for 3 min and were resuspended with 10 μl of 4% paraformaldehyde in water. To add volume, 500 μl of PBS was added to all samples. Samples containing stained parasites were analyzed by flow cytometry using the Stratedigm SE500 analyzer (Stratedigm, San Jose, CA). Viable (CFSE-stained) merozoites were quantified using side-scatter (SSC) and forward-scatter size exclusion to identify positively stained parasites. Samples from two independent experiments with biological duplicates and technical triplicates were analyzed.
Quantification of merozoites by reverse transcriptase qPCR.
The total number of parasites released into the supernatant was determined by reverse transcriptase qPCR. The controls in these experiments were wild-type (untreated) and scrambled (treated with nonsense ssRNA) parasites. We collected supernatants and extracted RNA at 1, 22, 24, and 26 h postinfection. To evaluate silencing efficiency, we compared the gene expression of SUB1-silenced (ΔSUB1), wild-type (untreated), and scrambled-ssRNA-treated parasite samples. One hundred nanograms of total RNA was used as a template with a SuperScript III Platinum SYBR green one-step reverse transcription-qPCR kit (Life Technologies, Grand Island, NY), The following conditions were used: 55°C for 20 min, 95°C for 5 min, and 40 cycles at 95°C for 15 s and 65°C for 1 min (AB7500 Fast RT-PCR system for 98-well plates). Primers are shown in Table S1 in the supplemental material. The expression of SUB1 RNA relative to the expression of parasite lactate dehydrogenase (LDH) (ApiDB_CryptoDB: cgd7_480; NCBI Protein database accession no. XP_628238.1) RNA in ΔSUB1, scrambled-ssRNA-treated, and wild-type parasites was evaluated using the comparative cycle threshold method. Furthermore, we also analyzed a second serine protease (ApiDB_CryptoDB: cgd2_3660; XP_001388237.1 and XM_001388200.1) normalized to parasite LDH. The parasite burden was determined through quantification of LDH as a parasite housekeeping gene and was compared to a standard curve prepared from defined numbers of oocysts. All samples were analyzed in triplicate in at least two independent experiments with biological duplicates. The Student t test was used in these experiments to determine statistical significance.
Observation of intracellular parasites by confocal microscopy.
To evaluate the effect of silencing SUB1 on the blockage of egress, we determined the number of intracellular organisms at 24 h postinfection by confocal microscopy. For these experiments, HCT-8 cells were grown on 22- by 22- by 1.5-mm coverslips until confluent. Sporozoites from 5 × 105 C. parvum oocysts of SUB1-silenced (ΔSUB1) or wild-type (untreated) parasites were used to infect host cells for 24 h at 37°C under 5% CO2. At the end of the infection period, the supernatant was removed, and infected monolayers were washed with PBS, fixed with 10 μl 4% paraformaldehyde (at room temperature for 25 min), and then washed again with PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min, washed three times with PBS, and stained with 4',6-diamidino-2-phenylindole (DAPI) (Vectashield Antifade Mounting Medium with DAPI; Vector Labs, Burlingame, CA). Samples were then visualized using the Zeiss LSM 880 confocal microscope with Airyscan (Zeiss, Oberkochen, Germany).
Supplementary Material
ACKNOWLEDGMENTS
S.N. was supported by a fellowship from the Sealy Center for Vaccine Development at the University of Texas Medical Branch at Galveston. This project was supported by the Bill & Melinda Gates Foundation (grant OPP1161026) and by grant 5R21AI12627502 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00784-18.
REFERENCES
- 1.Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabada MM, White AC. 2010. Treatment of cryptosporidiosis: do we know what we think we know? Curr Opin Infect Dis 23:494–499. doi: 10.1097/QCO.0b013e32833de052. [DOI] [PubMed] [Google Scholar]
- 3.Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH, van Eijk A, Adegbola RA, Alonso PL, Breiman RF, Faruque AS, Saha D, Sow SO, Sur D, Zaidi AK, Biswas K, Panchalingam S, Clemens JD, Cohen D, Glass RI, Mintz ED, Sommerfelt H, Levine MM. 2012. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis 55(Suppl 4):S232–S245. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 5.Sow SO, Muhsen K, Nasrin D, Blackwelder WC, Wu Y, Farag TH, Panchalingam S, Sur D, Zaidi AK, Faruque AS, Saha D, Adegbola R, Alonso PL, Breiman RF, Bassat Q, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ahmed S, Qureshi S, Quadri F, Hossain A, Das SK, Antonio M, Hossain MJ, Mandomando I, Nhampossa T, Acácio S, Omore R, Oundo JO, Ochieng JB, Mintz ED, O'Reilly CE, Berkeley LY, Livio S, Tennant SM, Sommerfelt H, Nataro JP, Ziv-Baran T, Robins-Browne RM, Mishcherkin V, Zhang J, Liu J, Houpt ER, Kotloff KL, Levine MM. 2016. The burden of Cryptosporidium diarrheal disease among children <24 months of age in moderate/high mortality regions of sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis 10:e0004729. doi: 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korpe PS, Haque R, Gilchrist C, Valencia C, Niu F, Lu M, Ma JZ, Petri SE, Reichman D, Kabir M, Duggal P, Petri WA Jr. 2016. Natural history of cryptosporidiosis in a longitudinal study of slum-dwelling Bangladeshi children: association with severe malnutrition. PLoS Negl Trop Dis 10:e0004564. doi: 10.1371/journal.pntd.0004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Checkley W, Epstein LD, Gilman RH, Black RE, Cabrera L, Sterling CR. 1998. Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am J Epidemiol 148:497–506. doi: 10.1093/oxfordjournals.aje.a009675. [DOI] [PubMed] [Google Scholar]
- 8.Mondal D, Haque R, Sack RB, Kirkpatrick BD, Petri WA. 2009. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am J Trop Med Hyg 80:824–826. doi: 10.4269/ajtmh.2009.80.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agnew DG, Lima AA, Newman RD, Wuhib T, Moore RD, Guerrant RL, Sears CL. 1998. Cryptosporidiosis in northeastern Brazilian children: association with increased diarrhea morbidity. J Infect Dis 177:754–760. doi: 10.1086/514247. [DOI] [PubMed] [Google Scholar]
- 10.Mølbak K, Andersen M, Aaby P, Højlyng N, Jakobsen M, Sodemann M, da Silva AP. 1997. Cryptosporidium infection in infancy as a cause of malnutrition: a community study from Guinea-Bissau, West Africa. Am J Clin Nutr 65:149–152. doi: 10.1093/ajcn/65.1.149. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Bolick DT, Kolling GL, Fu Z, Guerrant RL. 2016. Protein malnutrition impairs intestinal epithelial cell turnover, a potential mechanism of increased cryptosporidiosis in a murine model. Infect Immun 84:3542–3549. doi: 10.1128/IAI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrant RL, Oriá RB, Moore SR, Oriá MO, Lima AA. 2008. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev 66:487–505. doi: 10.1111/j.1753-4887.2008.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrant RL, Deboer MD, Moore SR, Scharf RJ, Lima AA. 2013. The impoverished gut—a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 10:220–229. doi: 10.1038/nrgastro.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sparks H, Nair G, Castellanos-Gonzalez A, White AC. 2015. Treatment of Cryptosporidium: what we know, gaps, and the way forward. Curr Trop Med Rep 2:181–187. doi: 10.1007/s40475-015-0056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy JM, Whitehead L, van Dooren GG, Tonkin CJ. 2012. TgCDPK3 regulates calcium-dependent egress of Toxoplasma gondii from host cells. PLoS Pathog 8:e1003066. doi: 10.1371/journal.ppat.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roiko MS, Carruthers VB. 2009. New roles for perforins and proteases in apicomplexan egress. Cell Microbiol 11:1444–1452. doi: 10.1111/j.1462-5822.2009.01357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackman MJ, Carruthers VB. 2013. Recent insights into apicomplexan parasite egress provide new views to a kill. Curr Opin Microbiol 16:459–464. doi: 10.1016/j.mib.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lourido S, Tang K, Sibley LD. 2012. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J 31:4524–4534. doi: 10.1038/emboj.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackman MJ. 2008. Malarial proteases and host cell egress: an ‘emerging’ cascade. Cell Microbiol 10:1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmon BL, Oksman A, Goldberg DE. 2001. Malaria parasite exit from the host erythrocyte: a two-step process requiring extraerythrocytic proteolysis. Proc Natl Acad Sci U S A 98:271–276. doi: 10.1073/pnas.011413198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickham ME, Culvenor JG, Cowman AF. 2003. Selective inhibition of a two-step egress of malaria parasites from the host erythrocyte. J Biol Chem 278:37658–37663. doi: 10.1074/jbc.M305252200. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Roellig DM, Guo Y, Guo Y, Li N, Frace MA, Tang K, Zhang L, Feng Y, Xiao L. 2016. Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genomics 17:1006. doi: 10.1186/s12864-016-3343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith HV, Nichols RA, Grimason AM. 2005. Cryptosporidium excystation and invasion: getting to the guts of the matter. Trends Parasitol 21:133–142. doi: 10.1016/j.pt.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Singh P, Mirdha BR, Srinivasan A, Rukmangadachar LA, Singh S, Sharma P, Hariprasad G, Gururao H, Gururao H, Luthra K. 2015. Identification of invasion proteins of Cryptosporidium parvum. World J Microbiol Biotechnol 31:1923–1934. doi: 10.1007/s11274-015-1936-9. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P, Chitnis CE. 2013. Key molecular events during host cell invasion by apicomplexan pathogens. Curr Opin Microbiol 16:432–437. doi: 10.1016/j.mib.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Castellanos-Gonzalez A, Perry N, Nava S, White AC. 2016. Preassembled single-stranded RNA-Argonaute complexes: a novel method to silence genes in Cryptosporidium. J Infect Dis 213:1307–1314. doi: 10.1093/infdis/jiv588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 28.Wanyiri JW, Techasintana P, O’Connor RM, Blackman MJ, Kim K, Ward HD. 2009. Role of CpSUB1, a subtilisin-like protease, in Cryptosporidium parvum infection in vitro. Eukaryot Cell 8:470–477. doi: 10.1128/EC.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeoh S, O'Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, Hackett F, Withers-Martinez C, Mitchell GH, Bannister LH, Bryans JS, Kettleborough CA, Blackman MJ. 2007. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell 131:1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal S, Singh MK, Garg S, Chitnis CE, Singh S. 2013. Ca2+-mediated exocytosis of subtilisin-like protease 1: a key step in egress of Plasmodium falciparum merozoites. Cell Microbiol 15:910–921. doi: 10.1111/cmi.12086. [DOI] [PubMed] [Google Scholar]
- 31.Billker O, Lourido S, Sibley LD. 2009. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe 5:612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagamune K, Sibley LD. 2006. Comparative genomic and phylogenetic analyses of calcium ATPases and calcium-regulated proteins in the Apicomplexa. Mol Biol Evol 23:1613–1627. doi: 10.1093/molbev/msl026. [DOI] [PubMed] [Google Scholar]
- 33.Lourido S, Moreno SN. 2015. The calcium signaling toolkit of the apicomplexan parasites Toxoplasma gondii and Plasmodium spp. Cell Calcium 57:186–193. doi: 10.1016/j.ceca.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Etzold M, Lendner M, Daugschies A, Dyachenko V. 2014. CDPKs of Cryptosporidium parvum—stage-specific expression in vitro. Parasitol Res 113:2525–2533. doi: 10.1007/s00436-014-3902-0. [DOI] [PubMed] [Google Scholar]
- 35.Castellanos-Gonzalez A, Yancey L, Wang H, Pantenburg B, Liscum KR, Lewis DE, White AC Jr. 2008. Cryptosporidium infection of human intestinal epithelial cells increases expression of osteoprotegerin: a novel mechanism for evasion of host defenses. J Infect Dis 197:916–923. doi: 10.1086/528374. [DOI] [PubMed] [Google Scholar]
- 36.Mele R, Gomez Morales MA, Tosini F, Pozio E. 2004. Cryptosporidium parvum at different developmental stages modulates host cell apoptosis in vitro. Infect Immun 72:6061–6067. doi: 10.1128/IAI.72.10.6061-6067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sifuentes LY, Di Giovanni GD. 2007. Aged HCT-8 cell monolayers support Cryptosporidium parvum infection. Appl Environ Microbiol 73:7548–7551. doi: 10.1128/AEM.01579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi K, Uejima H, Nagami-Okada T, Siomi H, Siomi MC. 2008. In vitro RNA cleavage assay for Argonaute-family proteins. Methods Mol Biol 442:29–43. doi: 10.1007/978-1-59745-191-8_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.