Cryptococcal meningitis (CM) causes high rates of HIV-related mortality, yet the Cryptococcus factors influencing patient outcome are not well understood. Pathogen-specific traits, such as the strain genotype and degree of antigen shedding, are associated with the clinical outcome, but the underlying biology remains elusive.
KEYWORDS: Cryptococcus, Cryptococcus neoformans, cryptococcosis, human, meningitis, model, mouse, pathogenesis, sequence type, virulence
ABSTRACT
Cryptococcal meningitis (CM) causes high rates of HIV-related mortality, yet the Cryptococcus factors influencing patient outcome are not well understood. Pathogen-specific traits, such as the strain genotype and degree of antigen shedding, are associated with the clinical outcome, but the underlying biology remains elusive. In this study, we examined factors determining disease outcome in HIV-infected cryptococcal meningitis patients infected with Cryptococcus neoformans strains with the same multilocus sequence type (MLST). Both patient mortality and survival were observed during infections with the same sequence type. Disease outcome was not associated with the patient CD4 count. Patient mortality was associated with higher cryptococcal antigen levels, the cerebrospinal fluid (CSF) fungal burden by quantitative culture, and low CSF fungal clearance. The virulence of a subset of clinical strains with the same sequence type was analyzed using a mouse inhalation model of cryptococcosis. We showed a strong association between human and mouse mortality rates, demonstrating that the mouse inhalation model recapitulates human infection. Similar to human infection, the ability to multiply in vivo, demonstrated by a high fungal burden in lung and brain tissues, was associated with mouse mortality. Mouse survival time was not associated with single C. neoformans virulence factors in vitro or in vivo; rather, a trend in survival time correlated with a suite of traits. These observations show that MLST-derived genotype similarities between C. neoformans strains do not necessarily translate into similar virulence either in the mouse model or in human patients. In addition, our results show that in vitro assays do not fully reproduce in vivo conditions that influence C. neoformans virulence.
INTRODUCTION
Cryptococcus neoformans is a fungal pathogen that causes disease mainly in immunocompromised patients, such as individuals living with human immunodeficiency virus (HIV) infection/AIDS or receiving organ transplants. The availability of antiretroviral therapy has reduced HIV-related mortality; however, deaths due to cryptococcal meningitis (CM) have plateaued, with C. neoformans still causing 15% of all HIV-related deaths globally (1, 2). Mortality rates due to Cryptococcus infection vary by region, from 70% of all infected patients in low-income countries to 20% of all infected patients in high-income countries (2). Although differences in mortality rates between high- and low-income countries can be linked to suboptimal antifungal treatments in low-income countries (3), variations in mortality rates between patient groups receiving similar treatments and residing in the same region of the world are observed (1, 4–6). Mortality is a measure influenced by many intrinsic host and pathogen factors (as well as host-pathogen interaction factors). Thus, we need better proxies to understand C. neoformans pathogenesis in patients and identify factors determining clinical outcome.
Although C. neoformans pathogenesis in the mouse model of cryptococcosis has been extensively studied, the correlation between disease characteristics in the human patient and those during clinical isolate infection in mice is unknown. The mouse model has been used to define C. neoformans virulence factors that allow the organism to be pathogenic, such as capsule, melanin, and titan cell formation. The progression of disease in the inhalational mouse model—from initial inhalation into the lungs to disseminated disease, and, ultimately, death due to central nervous system infection—is similar to that in human infection, yet whether the mouse model accurately recapitulates the differences in human disease and whether the model can be used to identify subtle variations in virulence factors that impact the outcome of human disease remain unexplored. Having clinical information from human research participants, we investigated the association between disease parameters in humans and those in mice infected with the same C. neoformans isolate.
Previous studies showed that pathogen-specific characteristics, such as the genotype or the degree of antigen shedding, influence immune responses to C. neoformans and the clinical outcome of patients (5, 7–9). Multilocus sequence typing (MLST) of 7 genetic loci has previously been used to identify genetically similar strains (5–10). For example, studies of clinical isolates in both Uganda and Brazil showed higher patient mortality associated with sequence type 93 (ST93) strains (5, 10). ST93 strains produced increases in type 2 cytokine levels in ex vivo cytokine release assays, suggesting that these strains may shift the Th1/Th2 immune balance (5). Similarly, studies from Vietnam have identified an association between ST5 and infections in non-HIV-infected patients, but the underlying biological differences remain unknown (11). To explore the association between genotype and clinical outcome, we examined individual patient differences in outcome and immune response in patients infected with the same sequence type (ST). We found that human patients infected with C. neoformans strains with the same sequence type can have dramatically different clinical outcomes. This dichotomy in disease outcome was recapitulated in the mouse model of cryptococcosis, suggesting that these differences in clinical outcome were due to strain-specific characteristics.
RESULTS
C. neoformans isolates with the same sequence type were observed in patients who lived and patients who died.
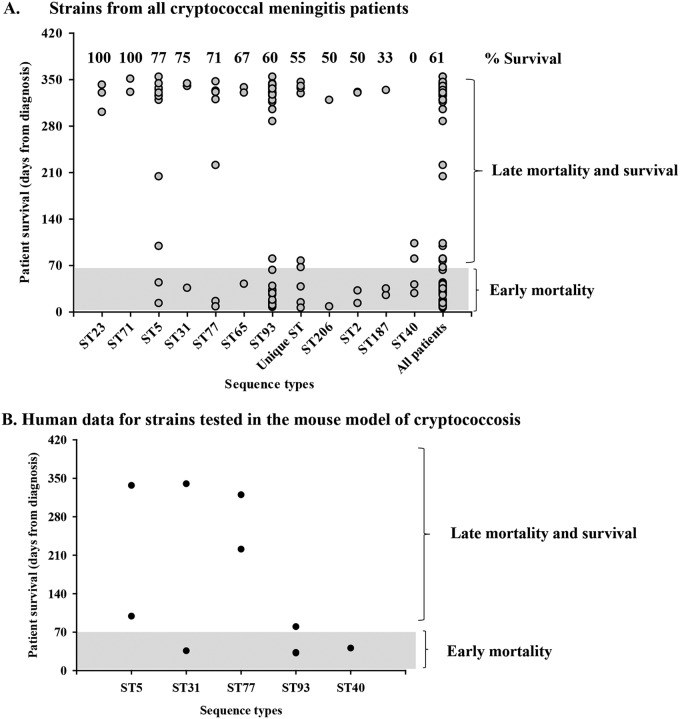
The association between clinical outcome and the C. neoformans MLST-based sequence type (ST) is controversial, with some studies showing differences in patient clinical outcomes associated with specific sequence types and others showing no association (5, 7–9, 12). To explore this phenomenon, we examined individual patient mortality in HIV-infected cryptococcal meningitis participants enrolled in the Cryptococcal Optimal ART Timing (COAT) clinical trial who were infected with C. neoformans strains of the same sequence type (Fig. 1A). Eleven participants had unique sequence types not observed in any of the other patients in this cohort. These singleton sequence types were cumulatively analyzed as unique STs. We observed a combination of patient death within 10 weeks (early mortality) and patient survival for eight of the sequence types (ST5, ST31, ST77, ST65, ST93, ST2, ST206, and ST187). No patients survived infections with ST40, whereas all patients survived ST23 and ST71 infections (Table 1). While the rates of mortality of patients infected with isolates of ST40 and ST23 were examples of the two ends of the spectrum, patient mortality (number of participants with mortality due to cryptococcosis/total number of participants) differed across the sequence types (Fig. 1A and Table 1). For example, infection with ST187 resulted in a high rate of patient mortality, whereas infection with ST5 resulted in a lower rate of patient mortality. A similar dichotomy in patient mortality was observed with the unique sequence types.
FIG 1.
C. neoformans sequence types and human mortality. C. neoformans clinical strains were isolated from the CSF of HIV-infected patients with CM, grown on YPD agar medium at 30°C, and stored at −80°C before their sequence types were identified. (A) Cryptococcus strains from all participants have different sequence types and can be classified as early- versus late-mortality strains. (B) To test the association between human and mouse infections, 11 strains with various sequence types and different degrees of human mortality were chosen to be tested in the mouse model of cryptococcosis. The sequence type and human mortality for this subset of strains are presented. Early mortality indicates that the patient died within 10 weeks from the time of diagnosis, late mortality indicates that the patient died after 10 weeks, and survival indicates that the patient survived the infection. Unique ST, sequence types infecting only one patient.
TABLE 1.
Participant clinical outcome
| Sequence type | No. of participants with: |
Total rate of survival (%) | |||
|---|---|---|---|---|---|
| Early mortality (within 10 wk) | Late mortality (after 10 wk) | Survival | Total | ||
| ST23 | 0 | 0 | 3 | 3 | 100 |
| ST71 | 0 | 0 | 2 | 2 | 100 |
| ST5 | 2 | 1 | 10 | 13 | 77 |
| ST31 | 1 | 0 | 3 | 4 | 75 |
| ST77 | 2 | 0 | 5 | 7 | 71 |
| ST65 | 1 | 0 | 2 | 3 | 67 |
| ST93 | 19 | 1 | 30 | 50 | 60 |
| Unique ST | 4 | 1 | 6 | 11 | 55 |
| ST206 | 1 | 0 | 1 | 2 | 50 |
| ST2 | 2 | 0 | 2 | 4 | 50 |
| ST187 | 2 | 0 | 1 | 3 | 33 |
| ST40 | 2 | 2 | 0 | 4 | 0 |
| All participants | 36 | 5 | 65 | 106 | 61 |
These data show that for the majority of sequence types, there were participants who succumbed to their cryptococcosis and participants who survived their cryptococcosis. There are a number of parameters, both human and fungal, that could explain this difference in patient survival for the same sequence type. We investigated factors that could impact whether a participant survived or succumbed to cryptococcal infection with the same sequence type.
Association between HIV disease and mortality/survival.
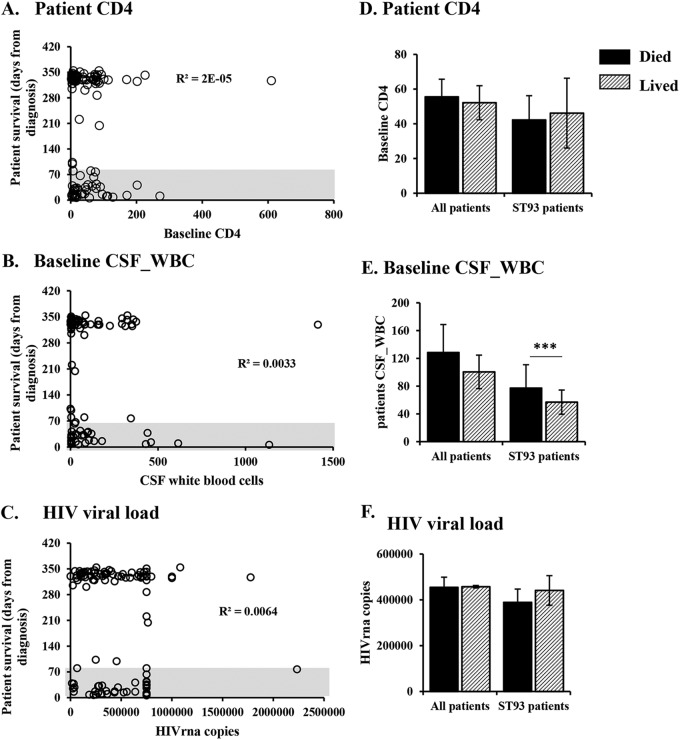
C. neoformans infections are predominantly observed in persons with compromised immune function. To test the hypothesis that differences in the underlying HIV infection account for the observed differences in mortality in the various sequence types, we examined CD4+ counts, the cerebrospinal fluid (CSF) white blood cell count (CSF_WBC), and the plasma HIV viral load in participants who died from cryptococcosis within 10 weeks versus those who survived (Fig. 2A to C). ST93 was the most prevalent sequence type, infecting 60 patients, and the only sequence type that infected enough participants to perform statistical analyses. Therefore, we also analyzed the HIV viral load, the CSF white cell count, and the CD4+ cell count in the cohort of patients infected with ST93 strains and performed analyses for all patients (Fig. 2D to F). The number of CD4+ cells, CSF white cell count, and the HIV viral load were similar in patients that lived and those that died (P > 0.05). Similar results were observed when considering only the strains that were tested in the mouse model of cryptococcosis (see Fig. S1 in the supplemental material).
FIG 2.
Human mortality is not determined by patient parameters. The clinical outcome was compared to baseline CM patient characteristics. (A to C) Data from individual participants (circles); (D to F) patient survival, classified as died (mortality by 10 weeks postdiagnosis) or lived (survival past 10 weeks postdiagnosis). (A, D) CD4+ cell counts (for all participants, 36 died and 70 lived; for ST93-infected patients, 20 died and 30 lived). (B, E) White blood cell counts in the CSF (for all participants, 34 died and 68 lived; for ST93-infected patients, 18 died and 29 lived). (C, F) HIV viral load (for all participants, 35 died and 70 lived; for ST93-infected patients, 20 died and 30 lived). Error bars represent the standard error of the mean. Patients who died and those who survived the infection were compared by the Mann-Whitney test. ***, P < 0.001. CD4 cells, CD4+ T helper cells; CM, cryptococcal meningitis; CSF, cerebrospinal fluid; HIV, human immunodeficiency virus; ST93, sequence type 93; WBC, white blood cells.
Although the CSF white cell count did not differ between patients who died and those who survived the infection in the total cohort, participants who survived the ST93 infection had lower CSF white cell counts than participants who died from the ST93 infection (Fig. 2E) (P < 0.05). These data suggest that observations at the population level do not always represent individual sequence types and that higher CSF white cell counts at the time of presentation are associated with mortality in the ST93-infected patients.
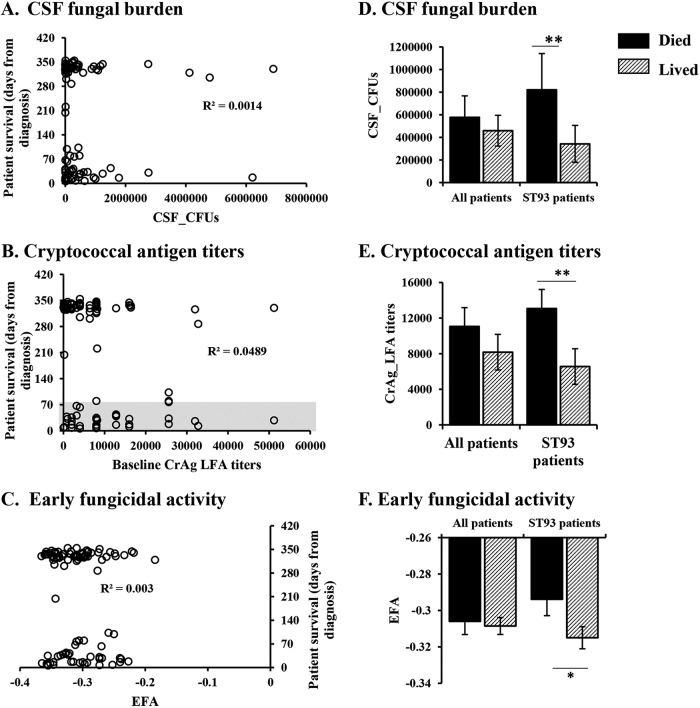
Human mortality is associated with the fungal burden in the CSF.
To explore the hypothesis that fungal factors impact mortality in patients infected with the various sequence types, we analyzed the initial CSF quantitative fungal burden, the amount of CSF cryptococcal antigen, and the rate of CSF fungal clearance (early fungicidal activity [EFA]) in the participants. Our analysis revealed that these fungal parameters partially correlated with patient survival (Fig. 3). Looking at all participants individually, we found that persons with comparable numbers of cryptococcal CFU had different clinical outcomes (Fig. 3A). However, lower numbers of CFU were associated with survival in the ST93-infected patients (Fig. 3D). We observed similar trends for the amount of cryptococcal capsular antigen detected in the participant CSF and the rate of fungal clearance (Fig. 3B and E and Fig. 3C and F, respectively), with higher antigen levels and lower clearance being associated with mortality in patients infected with ST93 genotypes but not in the entire population. These data suggest that fungal parameters, such as the ability to multiply inside the host, resistance to treatment, and the amount of antigen shed by Cryptococcus cells, play a role in infection outcome in a sequence type-dependent manner.
FIG 3.
Patient survival is partly associated with fungal parameters. The clinical outcome was compared to fungal parameters. (A to C) Data from individual participants (circles); (D to F) patient survival classified as died (mortality by 10 weeks postdiagnosis) or lived (survival past 10 weeks postdiagnosis). (A, D) CSF fungal burden determined by enumeration of CFU (for all participants, 36 died and 70 lived; for ST93-infected patients, 20 died and 30 lived). (B, E) Cryptococcal antigen titers in the CSF (for all participants, 31 died and 55 lived; for ST93-infected patients, 16 died and 23 lived). (C, F) Rate of fungal clearance determined by the early fungicidal activity (for all participants, 32 died and 69 lived; for ST93-infected patients, 18 died and 29 lived). Patients who died and those who survived the infection were compared by Student’s t test or the Mann-Whitney test. *, P < 0.05; **, P < 0.01. Error bars represent the standard error of the mean. CSF, cerebrospinal fluid; CrAg LFA, cryptococcal antigen lateral flow assay; EFA, early fungicidal activity; ST93, sequence type 93.
When only the participants infected with strains that were tested in the mouse model of cryptococcosis were considered, only the CSF fungal burden was associated with virulence (Fig. S2A and D). The cryptococcal antigen detected in the participant CSF as well as the rate of fungal clearance were similar between the participants infected with high-, intermediate-, and low-virulence strains (Fig. S2B and E and Fig. S2C and F).
C. neoformans virulence in humans correlates with virulence in a mouse model of cryptococcosis.
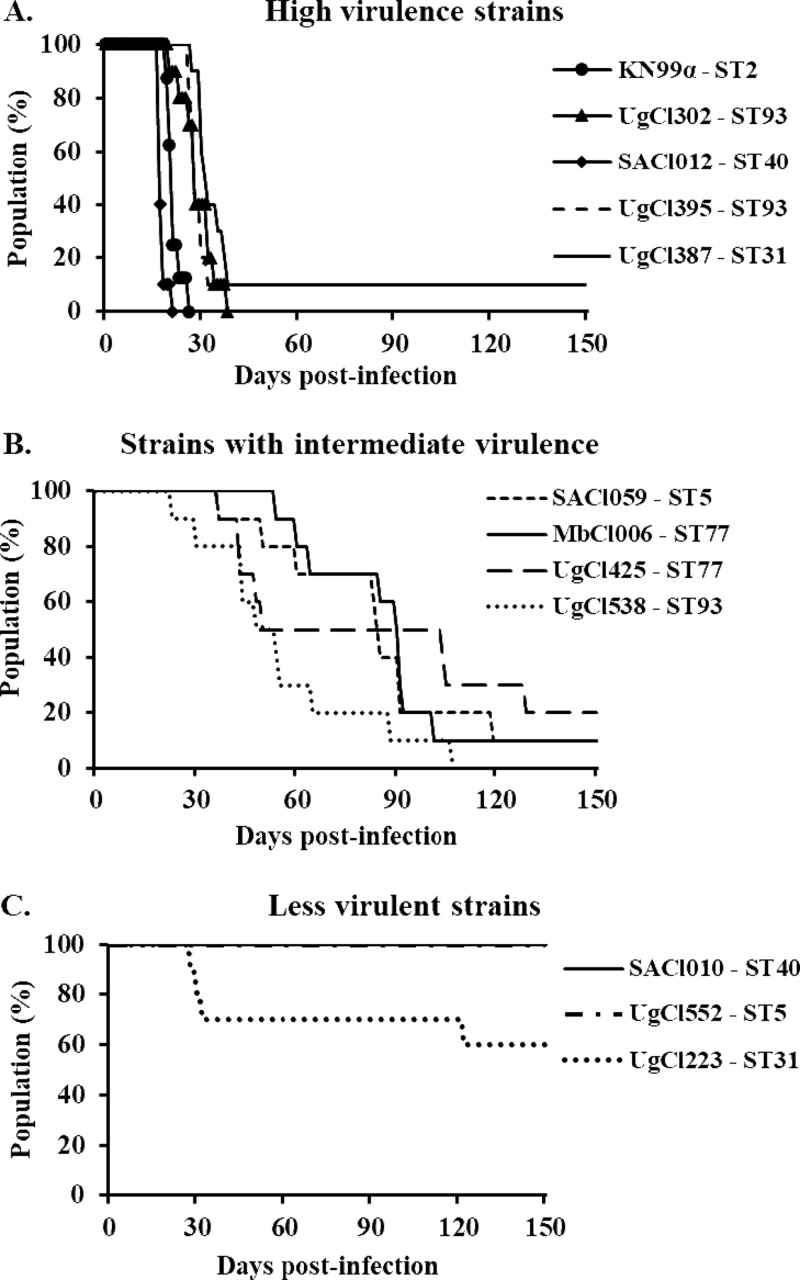
After observing that both human and fungal parameters were partially associated with patient survival, we determined whether differences in Cryptococcus virulence in humans could be recapitulated in the mouse inhalation model of cryptococcosis. We selected paired strains—one from a participant who died and one from a participant who lived—of ST5, ST31, and ST77 for analysis in the mouse model (Fig. 1B). In addition, we analyzed two ST40 strains causing high rates of mortality, two ST93 strains causing high rates of mortality, and one ST93 strain causing late mortality (Fig. 1B). Mice infected with the clinical strains showed variation in disease progression in the mouse inhalation model (Fig. 4). Based on mouse survival, the clinical strains were divided into three groups: high-, intermediate-, and low-virulence strains (Fig. 4A to C). The high-virulence strains were defined as strains that caused 80% mortality within 37 days postinfection (Fig. 4A). The intermediate-virulence strains caused mortality at between 69 and 129 days postinfection (Fig. 4B). The low-virulence group consisted of three strains: two strains, UgCl552 and SACl010, produced no overt signs of the disease in mice at 150 days postinfection, and the third strain, UgCl223, had reduced virulence, with 60% of mice surviving through 150 days postinfection (Fig. 4C). By histologic examination of the lungs, no cryptococci were identified in UgCl552-infected mice. In SACl010-infected mice, a few cryptococci were scattered in pulmonary alveoli. Of the two mice infected with UgCl223, both had a low to moderate number of organisms within the lungs, and one mouse had the infiltration of small cryptococcal organisms into the brain with necrosis in the affected areas.
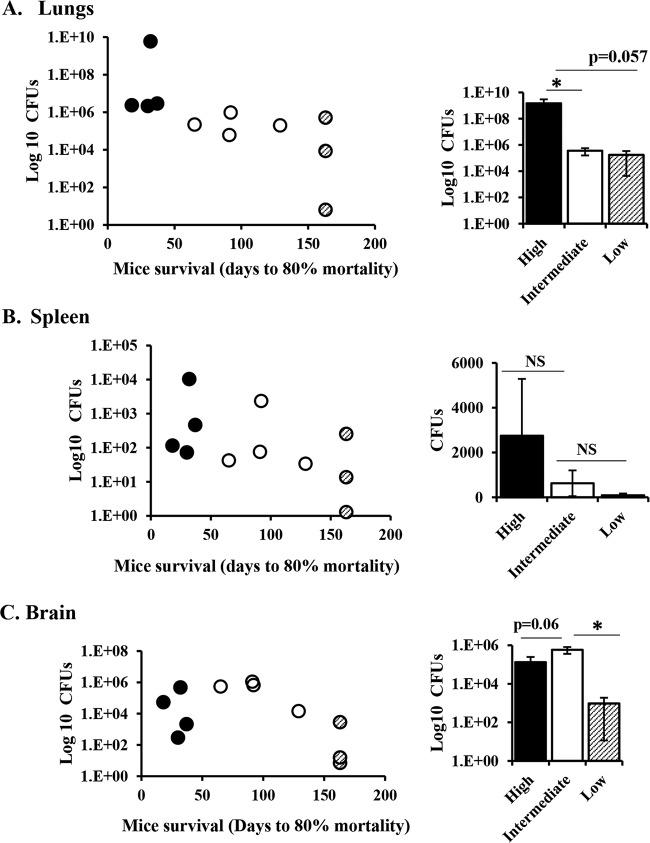
FIG 4.
C. neoformans clinical strains show differential virulence in mice. Groups of 10 6- to 8-week-old A/J mice were infected intranasally with 5 × 104 cells from C. neoformans clinical strains isolated from the CSF of HIV-infected patients with CM. Progression to severe morbidity was monitored for 150 days, and mice were sacrificed when the endpoint criteria were reached. (A) High-virulence strains; (B) strains with intermediate virulence; (C) low-virulence strains.
We next compared the mortality of humans and mice infected with the same Cryptococcus strain (Fig. 5 and Table 2). All but two of the isolates associated with early mortality in humans showed high virulence in mice (Fig. 5A). Strains SACl010 and UgCl552 had low virulence in mice but produced early mortality in humans. Histological examination of the lungs from mice infected with SACl010 revealed low numbers of cryptococci within alveoli, which were eliciting an inflammatory response. Interestingly, while the human participant infected with SACl010 died 41 days after diagnosis, the clinical records indicated that the participant died from immune reconstitution inflammatory syndrome (IRIS) and not directly from cryptococcosis. IRIS is defined as detrimental inflammatory responses despite fungal clearance (e.g., negative cultures) (13), consistent with the inflammation observed in the mouse model. The participant infected with strain UgCl552 cleared the initial infection but then died at 99 days postdiagnosis of an unknown cause at home, which was, again, consistent with the mouse model showing the low virulence of the Cryptococcus strain.
FIG 5.
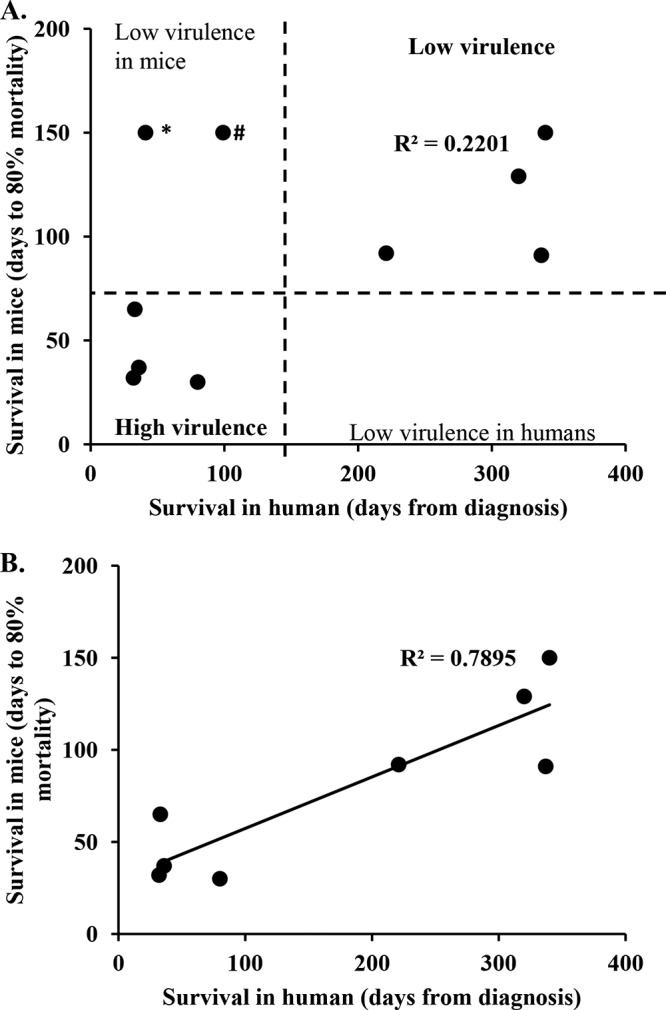
Disease outcome in humans is associated with disease outcome in mice. The clinical outcome in human participants was compared to the infection outcome in mice infected with the same C. neoformans strain. (A) Correlation between human and mouse survival for all 10 clinical strains. (B) Correlation between human and mouse survival when outliers (the SACl010 and UgCl552 strains) were removed. *, the SACl010 strain was associated with low virulence in mice, but the human participant died of IRIS at 41 days postdiagnosis; #, the participant infected with UgCl552 died at home after clearing the initial C. neoformans infection. The cause of death was not determined.
TABLE 2.
Association between clinical strain virulence in humans and mice
| Virulence | Strain name | Sequence type | Human infectionsa |
Mouse infections |
||
|---|---|---|---|---|---|---|
| Day of mortality | Outcome | Day of 80% mortality | Outcome | |||
| High | SACl012 | ST40 | NA | NA | 18 | All died |
| KN99α | ST2 | RS | RS | 21 | All died | |
| UgCl395 | ST93 | 80 | Died | 30 | Died (9/10)b | |
| UgCl302 | ST93 | 32 | Died | 32 | Died (9/10) | |
| UgCl387 | ST31 | 36 | Died | 37 | Died (9/10)c | |
| Intermediate | UgCl538 | ST93 | 33 | Died | 65 | All died |
| SACl059 | ST5 | Survived | 91 | Died (9/10)d | ||
| MbCl006 | ST77 | 221 | Died | 92 | Died (9/10)e | |
| UgCl425 | ST77 | Survived | 129 | Died (8/10)f | ||
| Low | UgCl223 | ST31 | Survived | NA | Survived | |
| UgCl552 | ST5 | 99 | IRIS | NA | Survived | |
| SACl010 | ST40 | 41 | NI | NA | Survived | |
NA, not available; RS, laboratory reference strain; NI, not identified (participant died at home); IRIS, participant died of immune reconstitution inflammatory syndrome.
Nine out of 10 mice died, with the remaining mouse being sacrificed after 150 days; the terminal numbers of CFU in the surviving mouse were 6.4 × 105 C. neoformans cells in the lungs and 4 × 102 cells in the brain.
Nine out of 10 mice died, with the remaining mouse being sacrificed after 150 days; the terminal number of CFU in the surviving mouse was 7 × 105 C. neoformans cells in the lungs; no cells were detected in the brain.
Nine out of 10 mice died, with the remaining mouse being sacrificed after 150 days; the terminal number of CFU was 6 × 105 C. neoformans cells in the lungs; no cells were detected in the brain.
Nine out of 10 mice died, with the remaining mouse being sacrificed after 150 days; the terminal numbers of CFU were 6 × 106 C. neoformans cells in the lungs and 6 × 103 cells in the brain.
Eight out of 10 mice died, with the remaining mice being sacrificed after 150 days; the terminal numbers of CFU were 4 × 105 C. neoformans cells in the lungs of each mouse, 4 × 104 cells in the brain of one mouse, and 3.6 × 104 cells in the brain of the second mouse.
All strains associated with late mortality in humans showed reduced virulence in mice (Fig. 5 and Table 2). No strain had high virulence in mice but low virulence in humans, providing further support for the association between human and mouse virulence. When all 10 strains (including the outliers SACl010 and UgCl552) were considered, a trend between human and mouse survival was observed (R2 = 0.2201; Fig. 5A). However, when both outliers were removed, the association between human and mouse mortality was robust (R2 = 0.7895; Fig. 5B). These data show that human and mouse mortality are similar when humans and mice are infected with the same C. neoformans strain, although unrelated human causes of mortality can influence the human clinical outcome.
Mouse virulence is associated with fungal burden.
To further investigate the factors that influence the infection outcome in mice, we compared mouse survival with tissue fungal burden at the time of sacrifice. High-virulence strains were associated with high fungal burdens, while intermediate- and low-virulence strains correlated with low numbers of CFU in the lungs of infected mice (Fig. 6A). Similar trends were observed for fungal burdens in the spleen, but the differences were not statistically significant (Fig. 6B). We observed similar terminal fungal burdens in the brains of mice infected with high- and intermediate-virulence strains and lower fungal burdens in the brains of mice infected with low-virulence strains sacrificed at 150 days postinfection (Fig. 6C). These data suggest that both high- and intermediate-virulence strains can disseminate to the brain, with the only difference being the time that it took to reach the brain (Fig. 6C and Table 2). Histologic examination of lung and brain from two intermediate-virulence strains, UgCl425 and SACl059, revealed moderate to large numbers of cryptococci and cryptococcoma formation with a moderate to strong inflammatory response (Tables S1 and S2).
FIG 6.
Mouse survival correlates with organ fungal burden. Groups of 10 6- to 8-week-old A/J mice were infected intranasally with 11 C. neoformans clinical strains. Progression to severe morbidity was monitored for 150 days, and mice were sacrificed at 150 days or when the endpoint criteria were reached. Lungs (A), spleen (B), and brain (C) were harvested and homogenized, and serial dilutions were plated on agar plates to determine tissue fungal burdens. Strains were classified into three groups as high-, intermediate-, and low-virulence strains, as determined in the mouse survival experiment. (Left) Individual C. neoformans strains plotted as the tissue fungal burden (number of CFU) against time to 80% mortality of infected mice. Each dot represents the average for mice infected with the same strain (n = 3 to 8 mice). (Right) Average number of CFU of high-virulence (n = 4 strains), intermediate-virulence (n = 4 strains), and low-virulence (n = 3 strains) strains. Error bars represent the standard error of the mean. The difference between high-, intermediate-, and low-virulence strains was compared by Student’s t test or the Mann-Whitney U test. *, P < 0.05; NS, not statistically significant.
Mice infected with low-virulence strains showing no overt disease were sacrificed at 150 days postinfection. These mice had moderate numbers of CFU in the lungs but low numbers of CFU in the spleen and brain, suggesting that the low virulence of these strains was due to reduced fungal dissemination (Fig. 6B and C). In addition, we observed differences in disease progression/pathogenesis in mice infected with the three low-virulence strains. One of the low-virulence strains (UgCl223) had high numbers of terminal lung CFU (1 × 107) and very few CFU in the spleen (10 colonies) and brain (10 colonies). Histologic examination of lung and brain from 2 additional animals revealed low numbers of cryptococci in the lung with moderate inflammation, and one mouse had small cryptococci infiltrating the meninges, scattered in small areas of necrosis within the neuropil (Tables S1 and S2). These data suggest that UgCl223 can multiply in the lungs and may disseminate to the brain but may not remain viable in the brain. Most mice infected with the other two low-virulence strains (SACl010 and UgCl552) cleared the infection. Seven out of nine mice infected with SACl010 had no CFU in the lung, spleen, or brain tissues at 150 days postinfection. Five out of nine mice infected with UgCl552 had no CFU in the lungs, and only one mouse with lung CFU had detectable numbers of CFU in the spleen or brain.
C. neoformans virulence did not correlate with known virulence factors under in vitro conditions.
After observing that the infection outcome was strain specific in both human and mouse infection, we tested whether the production of virulence factors previously shown to be associated with C. neoformans pathogenesis (14–19) was associated with survival in our study. The virulence factors tested included the ability to growth at a high temperature (37°C), capsule formation, and the ability to form titan cells both in vitro and in vivo. Virulence did not correlate with the ability to grow in vitro either under nutrient-rich or nutrient-limited nutrient conditions or at low and high temperatures (30°C and 37°C) (Fig. S3). Similarly, no significant difference in the doubling time of the high-, intermediate-, and low-virulence strains was observed at either 30°C or 37°C (Table S3).
Using two previously described methods (20, 21), we found that virulence was also not associated with the ability to form titan cells in vitro (Table S4). However, we observed a trend where low-virulence strains formed more titan cells than high- and intermediate-virulence strains when grown in Dulbecco modified Eagle medium (DMEM) supplemented with serum (Table 3). Finally, we also analyzed in vitro capsule formation under capsule-inducing conditions (Table 3). While the capsules varied across the strains, there was no difference in the size of the capsule between low- and high-virulence strains (Fig. S4).
TABLE 3.
Cellular phenotypes and growth in the presence of cell wall stressorsa
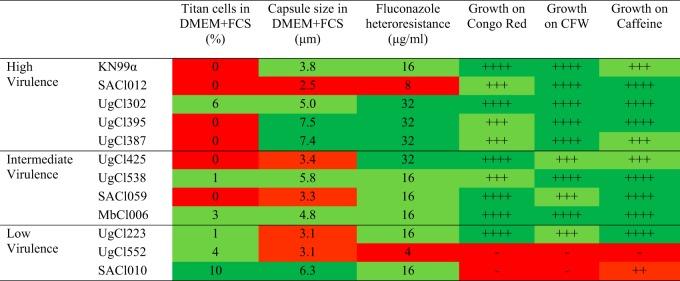 |
Red color denotes a weak or no phenotype, pale green denotes a moderate phenotype, and dark green denotes a strong phenotype. CFW, calcofluor white.
We next compared the susceptibility of the clinical strains to the antifungal drugs fluconazole and amphotericin B, which were used to treat the CM patients in our cohort. The in vitro susceptibility to the antifungal drugs fluconazole and amphotericin B was not associated with strain virulence (Table S5). Next, we analyzed the levels of heteroresistance to fluconazole, a factor that was previously associated with C. neoformans virulence (22), as well as the ability to grow in the presence of cell wall stressors calcofluor white, Congo red, and caffeine (Table 3). We observed a trend where high-virulence strains manifested heteroresistance at high levels of fluconazole and low-virulence strains developed heteroresistance at low levels of fluconazole (Table 3). However, not all high-virulence strains had high levels of fluconazole heteroresistance. Two highly virulent strains, SACl012 and KN99α, manifested fluconazole heteroresistance at 8 and 16 μg/ml respectively, while the other three high-virulence strains showed heteroresistance at 32 μg/ml (Table 3). In addition, high-virulence strains grew better in the presence of cell wall stressors than two of the three low-virulence strains, SACl010 and UgCl552 (Table 3). However, the low-virulence strain UgCl223 did not show any growth defect under any of the tested conditions (Table 3). Our in vitro assays did not identify a single condition or a virulence-determining factor that exclusively distinguished our high- and low-virulence strains, but there was a trend where most of the low-virulence strains grew slower or did not grow in the presence of various stresses.
DISCUSSION
In this study, we showed that humans infected with C. neoformans strains of the same sequence type could have different clinical outcomes. Using a mouse inhalation model of cryptococcosis, we showed that these differences in virulence were strain specific, with similar mortality rates being observed in both humans and mice infected with the same strain. In both humans and mice, we identified an association between an increased fungal burden and early mortality, but no single in vitro virulence factor could explain the observed in vivo differences between closely related strains.
We sought to identify factors associated with infection outcome, both in humans and in infected mice, which could explain the differences in mortality observed between C. neoformans strains with the same sequence type. We found that C. neoformans growth in vivo was associated with mortality in mice and humans. Our observations are in accord with those of previous clinical trials showing that the CSF fungal burden and the rate of fungal clearance were associated with patient mortality (23–26). However, underlying human immune deficiencies did not correlate with patient outcome. Some patients died and others survived the infection, while they had similar CD4+ cell counts, HIV viral loads, and white blood cell numbers in their CSF. These data are also consistent with previous observations that baseline CD4+ cell counts, CSF white cell counts, and HIV viral loads do not influence mortality in HIV-infected cryptococcal meningitis patients (24, 27, 28). Taken together, these data show that differences in patient outcome are due, in part, to differences in the strains of C. neoformans with which the patients are infected. A previous study from our group showed that differences in the C. neoformans MLST genotype influence the immune response and clinical outcome in HIV-infected cryptococcal meningitis patients (5). To further explore this phenomenon, we examined the impact of the sequence type on patient outcome. We found that patients infected with C. neoformans isolates of the same sequence type had differential clinical outcomes; some patients survived, while others died from the infection. Thus, whether the sequence type is associated with human disease parameters is dependent upon the composition of the cohort being studied. This includes not only the ratio of high- and low-virulence strains within the sequence type but also the number of patients infected with each sequence type analyzed. Analysis of large numbers of patients infected with strains of the same sequence type, such as ST93 in the study of Wiesner et al. (5), likely has the power to detect associations not readily apparent in patient cohorts with a more diverse representation of sequence types. Similarly, different regions of the world may have sequence types with different ratios of high- and low-virulence strains. For example, high-virulence ST5 strains may have been more prevalent among the Vietnamese strains analyzed by Day et al. (11).
To determine whether these differences in clinical outcome were inherent to the individual C. neoformans strains, we tested the virulence of a subset of the clinical isolates in a mouse inhalational model of cryptococcosis. We showed a robust association between human and mouse mortality for the majority of C. neoformans strains tested. These data not only show that the mouse model accurately recapitulates human disease but also strongly suggest that a large proportion of patient mortality is due to inherent differences between the infecting C. neoformans strains.
Several factors, such as cellular phenotypes and stress response mechanisms, have been associated with C. neoformans virulence and pathogenesis in the mouse model of cryptococcosis (14, 17, 18). In addition, a few studies showed that pleomorphism and the production of some virulence factors under in vitro conditions correlated with infection outcome in human patients (7, 19, 29). Therefore, we tested whether the ability to produce known virulence factors, as well as other phenotypes previously associated with pathogenesis, correlates with the virulence level of the subset of C. neoformans clinical isolates used in our study. The dominant virulence factors in C. neoformans are high-temperature growth and capsule formation (30, 31). High- and low-virulence strains grew similarly at a high temperature under both nutrient-rich and low-nutrient conditions. We observed similar patterns in capsule formation, where high- and low-virulence strains produced capsules of a similar size. However, the capsule formed during infection was significantly larger than the capsule induced in vitro, suggesting differences in capsule structure, composition, and/or function (32–34). This might explain our observations and those from other studies (35–37) that capsule size in vitro does not correlate with C. neoformans virulence. Next, we tested whether increased virulence was associated with increased resistance to two antifungal drugs, fluconazole and amphotericin B, used to treat patients in our cohort. In vitro susceptibility to fluconazole and amphotericin B also did not correlate with the virulence of our clinical strains. The lack of an association between infection outcome and the susceptibility to antifungal drugs in in vitro assays can be explained by the physiological differences between in vitro conditions and the in vivo environments in which C. neoformans cells have to survive (38). We also tested the ability to form titan cells, a phenotype that has been associated with C. neoformans virulence/pathogenesis (14, 15, 39). Using recently described techniques to induce titan cells in vitro (20, 21), we found that the ability to form titan cells in vitro did not correlate with the degree of virulence of our clinical strains. Similar results were observed with in vivo titan cell formation in the mouse model.
Under in vivo conditions, C. neoformans cells have to withstand multiple host defense mechanisms. Our data show that, individually, the in vitro analyses that we performed were unable to distinguish high- versus low-virulence strains. Thus, no single in vitro assay/condition can be used as a proxy for in vivo C. neoformans virulence. However, we did observe a trend when comparing the various in vitro assays (Table 3). Increased virulence was associated with heteroresistance at high levels of fluconazole, the ability to grow in the presence of cell wall stressors, and the inability to form titan cells in DMEM and serum. In contrast, strains with low virulence were unable to grow in the presence of cell wall stressors, had low fluconazole heteroresistance, and formed more titan cells in DMEM and serum. Most mice infected with strains that were unable to grow in the presence of multiple stresses in vitro cleared the infection, and the strains did not disseminate to the brain, showing that growth defects in vitro in response to multiple stresses might predict an inability to multiply in vivo during infection. These observations suggest that a panel of in vitro stresses could be developed to differentiate between high- and low-virulence C. neoformans strains. Future studies that incorporate a large number of clinical isolates are necessary to determine whether an appropriate panel of in vitro stresses can be identified to accurately predict in vivo virulence. If confirmed, this could be a valuable tool in clinical laboratories to help in the early identification and follow-up of patients who are infected with high-virulence strains and who are at a high mortality risk.
An alternative explanation for differences in in vivo virulence that do not correlate with the findings of single in vitro assays is that the observed differences are due to novel virulence factors. In vivo studies with evolutionarily closely related strains, such as those with strains with identical STs presented here, need to be combined with genomic analyses to identify novel genes that are critical in vivo. Our observation that strains with identical sequence types can have dramatically different virulence in both humans and the mouse model suggests that the multilocus sequence type does not have a sufficient resolution to identify the underlying differences between strains. Whole-genome sequencing of closely related strains and at the population level may be required to identify these in vivo virulence factors.
In summary, our study had two major findings. First, the mouse inhalation model of cryptococcosis accurately recapitulates human infection and human outcomes. Thus, this model can be used to explore how differences between C. neoformans clinical isolates impact human disease outcome. Second, C. neoformans isolates of the same sequence type were associated with different clinical outcomes. The association between sequence type and clinical outcome has been contentious, with some patient cohorts showing no association and some showing robust associations (5, 7, 40, 41). Our data show that belonging to the same lineage or sequence type does not necessarily mean that C. neoformans strains will have a comparable degree of virulence. The virulence-determining factor(s) is not the lineage/sequence type of the C. neoformans strain but, instead, is other genotypic or phenotypic characteristics specific to individual isolates within the sequence type. Future studies investigating the relationship between individual C. neoformans genotypes and virulence are needed to fully understand the pathogen-associated factors that influence in vivo virulence and, ultimately, clinical outcome.
MATERIALS AND METHODS
Ethical statement.
Animal experiments were done in accordance with the Animal Welfare Act, United States federal law, and NIH guidelines. Mice were handled in accordance with guidelines defined by the University of Minnesota Animal Care and Use Committee (IACUC) under protocol 1308-30852A.
The study population consisted of human immunodeficiency virus (HIV)-infected, antiretroviral therapy (ART)-naive individuals with a first episode of cryptococcal meningitis screened for the Cryptococcal Optimal ART Timing (COAT) trial (ClinicalTrials.gov registration number NCT01075152) (42). Participants were enrolled from Uganda (Mulago Hospital in Kampala, Uganda, and Mbarara Hospital in Mbarara, Uganda) and South Africa (GF Jooste Hospital in Cape Town, South Africa) between November 2010 and April 2012. Written informed consent was obtained from all subjects or their proxy, and all data were deidentified. Institutional review board approvals were obtained from each participating site. A total of 106 participants that had cerebrospinal fluid (CSF) culture positive for Cryptococcus and from which isolates were sequenced for genotypic analyses (5, 8) were included in this study. As with the parent COAT trial, survival was decreased with early ART initiation (42), and all clinical isolates used for mouse infections were selected from the standard-of-care (deferred ART treatment) arm of the clinical trial.
Strains and media.
Cryptococcus clinical isolates were colony purified from CSF specimens from participants enrolled in the COAT trial, and the multilocus sequence type (MLST) was determined previously (5, 8). The strains used in this study are listed in Table 4. Strains were stored in glycerol stocks at −80°C. Strains from glycerol stocks were grown on yeast-peptone-dextrose (YPD) agar for 24 to 48 h. C. neoformans cells from agar plates were transferred to YPD broth and grown overnight at 30°C with shaking before being used in experiments.
TABLE 4.
Strains used in this study
| Strain | Sequence type | Source (reference) |
|---|---|---|
| KN99α | ST2 | United States (44) |
| UgCl223 | ST31 | Kampala, Uganda (8) |
| UgCl302 | ST93 | Kampala, Uganda (8) |
| UgCl387 | ST31 | Kampala, Uganda (8) |
| UgCl395 | ST93 | Kampala, Uganda (8) |
| UgCl425 | ST77 | Kampala, Uganda (8) |
| UgCl538 | ST93 | Kampala, Uganda (8) |
| UgCl552 | ST5 | Kampala, Uganda (8) |
| MbCl006 | ST77 | Mbarara, Uganda (8) |
| SACl010 | ST40 | Cape Town, South Africa (8) |
| SACl012 | ST40 | Cape Town, South Africa (8) |
| SACl059 | ST5 | Cape Town, South Africa (8) |
To analyze in vitro growth curves, 5 × 103 cells were inoculated in triplicate into 96-well plates containing 200 μl YPD medium, RPMI medium, or DMEM. The plates were incubated at 30°C or 37°C in a Synergy H1 incubating plate reader (BioTek, Inc., Winooski, VT), and the optical density at 600 nm was measured every 15 min for 72 h. To determine the doubling time, YPD medium was inoculated with approximately 5 × 105 cells/ml and the cells were incubated at 30°C or 37°C. At 4 and 6 h of incubation during log-phase growth, the cell number was enumerated by use of a hemocytometer count, and the cells were diluted and placed onto YPD plates to determine the number of CFU. The doubling time was calculated using the following equation: Δt [log(2)/log(number of CFU at 6 h/number of CFU at 4 h)], where Δt is the change in time.
Mouse infection.
C. neoformans strains were cultured overnight in YPD broth at 30°C. After incubation, the C. neoformans cells were washed 3 times in sterile phosphate-buffered saline (PBS), enumerated by use of a hemocytometer, and resuspended in sterile PBS at a concentration of 1 × 106 yeast cells/ml. Groups of 6- to 8-week-old female A/J mice (Jackson Laboratory, Bar Harbor, ME) were anesthetized by intraperitoneal pentobarbital injection. Ten mice per strain were infected intranasally with 5 × 104 yeast cells in 50 μl PBS. The animals were monitored daily for morbidity and sacrificed when endpoint criteria were reached. Endpoint criteria were defined as a loss of 20% of total body weight, a loss of 2 g of body weight in 2 consecutive days, or signs of neurological disease. Mice that survived to 150 days postinfection without exhibiting signs of disease were sacrificed, and their tissues were processed to determine lung, spleen, and brain fungal burdens.
Tissue burden analysis.
Terminal lung, spleen, and brain tissues were collected at the time of mouse sacrifice to determine the organ fungal burden. The collected tissues were homogenized in 2 ml PBS. Serial dilutions of tissue homogenates were plated on YPD medium supplemented with 0.04 mg/ml chloramphenicol and incubated at 30°C. C. neoformans colonies were counted after 48 h of incubation. The data presented are representative of those for 2 to 8 mice per strain.
Histopathology.
Terminal lungs, spleen, and brain were harvested from infected mice, fixed in 10% buffered formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E). Tissue sections were examined for cell size and morphology by microscopy. The data presented are representative of those for 1 to 4 mice per strain.
In vivo titan cell analysis.
For a subset of infected mice, lungs were lavaged three times with 1.5 ml sterile PBS using a 20-gauge needle placed in the trachea. Cells in the lavage fluid were pelleted at 15,000 × g and washed three times with PBS. Yeast cells were fixed with 3.7% formaldehyde at room temperature for 40 min, washed 3 times with sterile PBS, and then resuspended in 200 μl sterile PBS. Cell body and capsule sizes were analyzed by microscopy. To visualize the yeast cells, a drop of India ink was added to the cell suspension on the slide and observed using a Zeiss Axioplan microscope (AxioImager; Carl Zeiss, Inc.). Cell body size was determined as the diameter of the yeast cell body and did not include the capsule. The data presented are representative of those for 1 to 3 mice per strain, and a total of 200 to 400 C. neoformans cells per mouse were analyzed.
In vitro titan cell analysis.
To induce the formation of titan cells in vitro, two previously described methods were used (20, 21). For the first method (20), C. neoformans cells from a YPD agar plate were transferred to 10 ml YPD broth medium in a T25cm3 flask (TPP, Switzerland), grown at 30°C for 22 h with shaking (150 rpm), and washed with sterile double-distilled water, and the cell number was enumerated with a hemocytometer. Aliquots of 1 × 106 cells in 1 ml of minimal medium (15 mM glucose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine, 3 μM vitamin B1, pH 5.5) were transferred into 1.5-ml Eppendorf tubes and incubated in a thermomixer at 30°C with shaking (800 rpm) for 48 h. For the second method (21), yeast cells were grown overnight at 30°C and 150 rpm in 2 ml yeast nitrogen base without amino acids supplemented with 2% glucose. Yeast cells from the overnight culture were washed twice with sterile PBS, the cell number was enumerated with a hemocytometer, and the cells were resuspended in 0.5 ml PBS supplemented with fetal calf serum (FCS; Thermo Fisher Scientific) to a final concentration of 1 × 103 cells/ml in each well of a 24-well plate. The plates were then incubated at 37°C in 5% CO2 for 48 h. After incubation, cell morphology was analyzed by microscopy (AxioImager; Carl Zeiss, Inc.). Cell body diameter was measured as described above. For in vitro titan cell analysis, the data presented are from 2 biological replicates per strain, with 300 cells being counted for each replicate.
Capsule formation.
Capsule formation was analyzed from C. neoformans cells generated from (i) in vitro titan cell formation as described above and (ii) C. neoformans cells grown in DMEM supplemented with fetal calf serum (FCS). For capsule induction in DMEM, C. neoformans cells were grown overnight in YPD at 30°C with shaking (250 rpm), washed with sterile PBS, and counted with a hemocytometer. Yeast cells were then inoculated at a final concentration of 1 × 106 cells/ml into DMEM supplemented with 10% FCS and incubated at 37°C in 5% CO2 for 5 days. After incubation, the yeast cells were fixed with 3.7% formaldehyde and analyzed by microscopy (AxioImager, Carl Zeiss, Inc.). The capsule width was defined as the difference between the diameter of the whole yeast cell (cell body and capsule) and the cell body diameter (no capsule) divided by two. The data presented are averages for a total of 100 C. neoformans cells per strain.
Antifungal drug susceptibility assays.
Antifungal drug susceptibility assays were performed and the results were assessed as described previously (43). Briefly, a microdilution assay was performed according to CLSI guidelines using 2.5 × 103 CFU/ml. Fluconazole and amphotericin B were tested as 2-fold dilutions ranging from 512 μg/ml to 0.0625 μg/ml and from 8 μg/ml to 0.0315 μg/ml, respectively, in a final volume of 200 μl per well. Spectrophotometric analysis of the well turbidity at 600 nm was used to determine the MIC for each strain. The plates were scanned in a BioTek Synergy H1 hybrid reader (Winooski, VT) prior to and after 72 h of incubation at 37°C. The amphotericin B MIC was defined as the drug concentration at which no growth was observed at 72 h (100% inhibition of growth). The fluconazole MIC was defined as a 50% reduction in growth (turbidity) compared to the growth of the no-drug control.
Statistical analysis.
An unpaired t test was used to compare the clinical parameters between patients who lived and those who died. The Mann-Whitney U test determined differences between mouse survival. Fisher’s exact test was used to analyze the association between survival in mice and survival in human patients. A P value of <0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Aleeza Gerstein, Sophie Altamirano, and Marina Yoder for helpful discussions and the University of North Carolina Lineberger Comprehensive Cancer Center Animal Histopathology Core Facility.
The University of North Carolina Lineberger Comprehensive Cancer Center Animal Histopathology Core Facility is supported in part by an NCI Center core support grant (CA016086). This research was supported by NIH grants R01AI134636 (to K.N.), R21AI122352 (to K.N.), and U01AI089244 (to D.R.B.). L.M. received support from the NIH Fogarty International Center (R25TW009345) and the University of Minnesota Center for Translational Science Institute (UL1TR000114). D.B.M. is a DELTAS/THRiVE fellow under grant DEL-15-001/07742//z/15/Z.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00046-19.
REFERENCES
- 1.Kambugu A, Meya DB, Rhein J, O’Brien M, Janoff EN, Ronald AR, Kamya MR, Mayanja-Kizza H, Sande MA, Bohjanen PR, Boulware DR. 2008. Outcome of cryptococcal meningitis in Uganda before and after the availability of HAART. Clin Infect Dis 46:1694–1701. doi: 10.1086/587667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perfect JR, Bicanic T. 2015. Cryptococcosis diagnosis and treatment: what do we know now. Fungal Genet Biol 78:49–54. doi: 10.1016/j.fgb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bicanic T, Meintjes G, Wood R, Hayes M, Rebe K, Bekker LG, Harrison T. 2007. Fungal burden, early fungicidal activity, and outcome in cryptococcal meningitis in antiretroviral-naive or antiretroviral-experienced patients treated with amphotericin B or fluconazole. Clin Infect Dis 45:76–80. doi: 10.1086/518607. [DOI] [PubMed] [Google Scholar]
- 5.Wiesner DL, Moskalenko O, Corcoran JM, McDonald T, Rolfes MA, Meya DB, Kajumbula H, Kambugu A, Bohjanen PR, Knight JF, Boulware DR, Nielsen K. 2012. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 3:e00196-12. doi: 10.1128/mBio.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguiar P, Pedroso RDS, Borges AS, Moreira TA, Araujo LB, Roder D. 2017. The epidemiology of cryptococcosis and the characterization of Cryptococcus neoformans isolated in a Brazilian university hospital. Rev Inst Med Trop Sao Paulo 59:e13. doi: 10.1590/S1678-9946201759013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beale MA, Sabiiti W, Robertson EJ, Fuentes-Cabrejo KM, O’Hanlon SJ, Jarvis JN, Loyse A, Meintjes G, Harrison TS, May RC, Fisher MC, Bicanic T. 2015. Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across southern Africa. PLoS Negl Trop Dis 9:e0003847. doi: 10.1371/journal.pntd.0003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulware DR, von Hohenberg M, Rolfes MA, Bahr NC, Rhein J, Akampurira A, Williams DA, Taseera K, Schutz C, McDonald T, Muzoora C, Meintjes G, Meya DB, Nielsen K, Huppler Hullsiek K, Cryptococcal Optimal ART Timing Trial Team. 2016. Human immune response varies by the degree of relative cryptococcal antigen shedding. Open Forum Infect Dis 3:ofv194. doi: 10.1093/ofid/ofv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desnos-Ollivier M, Patel S, Raoux-Barbot D, Heitman J, Dromer F. 2015. Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. mBio 6:e00311-16. doi: 10.1128/mBio.00311-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira-Paim K, Andrade-Silva L, Fonseca FM, Ferreira TB, Mora DJ, Andrade-Silva J, Khan A, Dao A, Reis EC, Almeida MTG, Maltos A, Junior VR, Trilles L, Rickerts V, Chindamporn A, Sykes JE, Cogliati M, Nielsen K, Boekhout T, Fisher M, Kwon-Chung J, Engelthaler DM, Lazéra M, Meyer W, Silva-Vergara ML. 2017. MLST-based population genetic analysis in a global context reveals clonality amongst Cryptococcus neoformans var. grubii VNI isolates from HIV patients in southeastern Brazil. PLoS Negl Trop Dis 11:e0005223. doi: 10.1371/journal.pntd.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day JN, Qihui S, Thanh LT, Trieu PH, Van AD, Thu NH, Chau TTH, Lan NPH, Chau NVV, Ashton PM, Thwaites GE, Boni MF, Wolbers M, Nagarajan N, Tan PBO, Baker S. 2017. Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLoS Negl Trop Dis 11:e0005628. doi: 10.1371/journal.pntd.0005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyazika TK, Hagen F, Machiridza T, Kutepa M, Masanganise F, Hendrickx M, Boekhout T, Magombei-Majinjiwa T, Siziba N, Chin A, Ombe N, Mateveke K, Meis JF, Robertson VJ. 2016. Cryptococcus neoformans population diversity and clinical outcomes of HIV-associated cryptococcal meningitis patients in Zimbabwe. J Med Microbiol 65:1281–1288. doi: 10.1099/jmm.0.000354. [DOI] [PubMed] [Google Scholar]
- 13.Haddow LJ, Colebunders R, Meintjes G, Lawn SD, Elliott JH, Manabe YC, Bohjanen PR, Sungkanuparph S, Easterbrook PJ, French MA, Boulware DR. 2010. Cryptococcal immune reconstitution inflammatory syndrome in HIV-1-infected individuals: proposed clinical case definitions. Lancet Infect Dis 10:791–802. doi: 10.1016/S1473-3099(10)70170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog 6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols CB, Perfect ZH, Alspaugh JA. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- 17.Kozel TR. 1995. Virulence factors of Cryptococcus neoformans. Trends Microbiol 3:295–299. doi: 10.1016/S0966-842X(00)88957-X. [DOI] [PubMed] [Google Scholar]
- 18.Alspaugh JA. 2015. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol 78:55–58. doi: 10.1016/j.fgb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes KE, Brockway A, Haverkamp M, Cuomo CA, van Ogtrop F, Perfect JR, Carter DA. 2018. Phenotypic variability correlates with clinical outcome in Cryptococcus isolates obtained from Botswanan HIV/AIDS patients. mBio 9:e02016-18. doi: 10.1128/mBio.02016-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, Sturny-Leclère A, Janbon G, Perfect JR, Fraser JA, Casadevall A, Cuomo CA, Dromer F, Nielsen K, Alanio A. 2018. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog 14:e1006982. doi: 10.1371/journal.ppat.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, Taylor-Smith L, LeGrave N, Rasmussen T, Fisher MC, Bicanic T, Harrison TS, Jaspars M, May RC, Brown GD, Yuecel R, MacCallum DM, Ballou ER. 2018. The Cryptococcus neoformans titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog 14:e1006978. doi: 10.1371/journal.ppat.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. 2009. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Chemother 53:2804–2815. doi: 10.1128/AAC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson PA, Bauer M, Leal MAE, Evans SG, Holtom PD, Diamond DM, Leedom JM, Larsen RA. 1999. Early mycological treatment failure in AIDS-associated cryptococcal meningitis. Clin Infect Dis 28:82–92. doi: 10.1086/515074. [DOI] [PubMed] [Google Scholar]
- 24.Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, Taseera K, Rebe K, Loyse A, Jarvis J, Bekker LG, Wood R, Limmathurotsakul D, Chierakul W, Stepniewska K, White NJ, Jaffar S, Harrison TS. 2009. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis 49:702–709. doi: 10.1086/604716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson AT, Nussbaum JC, Phulusa J, Namarika D, Chikasema M, Kanyemba C, Jarvis JN, Jaffar S, Hosseinipour MC, van der Horst C, Harrison TS. 2012. A phase II randomised controlled trial adding oral flucytosine to high dose fluconazole, with short-course amphotericin B, for cryptococcal meningitis. AIDS 26:1363–1370. doi: 10.1097/QAD.0b013e328354b419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvis JN, Bicanic T, Loyse A, Namarika D, Jackson A, Nussbaum JC, Longley N, Muzoora C, Phulusa J, Taseera K, Kanyembe C, Wilson D, Hosseinipour MC, Brouwer AE, Limmathurotsakul D, White N, van der Horst C, Wood R, Meintjes G, Bradley J, Jaffar S, Harrison T. 2014. Determinants of mortality in a combined cohort of 501 patients with HIV-associated cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis 58:736–745. doi: 10.1093/cid/cit794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis JN, Meintjes G, Rebe K, Williams GN, Bicanic T, Williams A, Schutz C, Bekker LG, Wood R, Harrison TS. 2012. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS 26:1105–1113. doi: 10.1097/QAD.0b013e3283536a93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chottanapund S, Singhasivanon P, Kaewkungwal J, Chamroonswasdi K, Manosuthi W. 2007. Survival time of HIV-infected patients with cryptococcal meningitis. J Med Assoc Thai 90:2104–2111. [PubMed] [Google Scholar]
- 29.Sabiiti W, Robertson E, Beale MA, Johnston SA, Brouwer AE, Loyse A, Jarvis JN, Gilbert AS, Fisher MC, Harrison TS, May RC, Bicanic T. 2014. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J Clin Invest 124:2000–2008. doi: 10.1172/JCI72950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaragoza O, Chrisman CJ, Castelli MV, Frases S, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2008. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cell Microbiol 10:2043–2057. doi: 10.1111/j.1462-5822.2008.01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yauch LE, Lam JS, Levitz SM. 2006. Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathog 2:e120. doi: 10.1371/journal.ppat.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera J, Feldmesser M, Cammer M, Casadevall A. 1998. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect Immun 66:5027–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaragoza O, Casadevall A. 2004. Experimental modulation of capsule size in Cryptococcus neoformans. Biol Proced Online 6:10–15. doi: 10.1251/bpo68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clancy CJ, Nguyen MH, Alandoerffer R, Cheng S, Iczkowski K, Richardson M, Graybill JR. 2006. Cryptococcus neoformans var. grubii isolates recovered from persons with AIDS demonstrate a wide range of virulence during murine meningoencephalitis that correlates with the expression of certain virulence factors. Microbiology 152:2247–2255. doi: 10.1099/mic.0.28798-0. [DOI] [PubMed] [Google Scholar]
- 36.Dykstra MA, Friedman L, Murphy JW. 1977. Capsule size of Cryptococcus neoformans: control and relationship to virulence. Infect Immun 16:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Littman ML, Tsubura E. 1959. Effect of degree of encapsulation upon virulence of Cryptococcus neoformans. Proc Soc Exp Biol Med 101:773–777. doi: 10.3181/00379727-101-25090. [DOI] [PubMed] [Google Scholar]
- 38.Grossman NT, Casadevall A. 2017. Physiological differences in Cryptococcus neoformans strains in vitro versus in vivo and their effects on antifungal susceptibility. Antimicrob Agents Chemother 61:e02108-16. doi: 10.1128/AAC.02108-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okagaki LH, Nielsen K. 2012. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell 11:820–826. doi: 10.1128/EC.00121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litvintseva AP, Mitchell TG. 2009. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect Immun 77:3188–3195. doi: 10.1128/IAI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desnos-Ollivier M, Patel S, Spaulding AR, Charlier C, Garcia-Hermoso D, Nielsen K, Dromer F. 2010. Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. mBio 1:e00091-10. doi: 10.1128/mBio.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulware DR, Meya DB, Muzoora C, Rolfes MA, Huppler Hullsiek K, Musubire A, Taseera K, Nabeta HW, Schutz C, Williams DA, Rajasingham R, Rhein J, Thienemann F, Lo MW, Nielsen K, Bergemann TL, Kambugu A, Manabe YC, Janoff EN, Bohjanen PR, Meintjes G. 2014. Timing of antiretroviral therapy after diagnosis of cryptococcal meningitis. N Engl J Med 370:2487–2498. doi: 10.1056/NEJMoa1312884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith KD, Achan B, Hullsiek KH, McDonald TR, Okagaki LH, Alhadab AA, Akampurira A, Rhein JR, Meya DB, Boulware DR, Nielsen K. 2015. Increased antifungal drug resistance in clinical isolates of Cryptococcus neoformans in Uganda. Antimicrob Agents Chemother 59:7197–7204. doi: 10.1128/AAC.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.