Abstract
Mutations in the Kelch domain of the K13 gene (PF3D7_1343700) were previously associated with artemisinin resistance in Plasmodium falciparum. This study followed the dynamics of the K13 polymorphisms in P. falciparum parasites from the China-Myanmar border area obtained in 2007–2016, and their in vitro sensitivities to artesunate (AS) and dihydroartemisinin (DHA). The 50% effective concentration (EC5072h) values of 133 culture-adapted field isolates to AS and DHA, measured by the conventional 72 h SYBR Green I-based assay, varied significantly among the parasites from different years; all were significantly higher than that of the reference strain 3D7. Compared with parasites from 2007 to 2008, ring survival rates almost doubled in parasites obtained in later years. Sequencing the full-length K13 genes identified 11 point mutations present in 85 (63.9%) parasite isolates. F446I was the predominant (55/133) variant, and its frequency was increased from 17.6% (3/17) in 2007 to 55.9% (19/34) in 2014–2016. No wild-type (WT) Kelch domain sequences were found in the 34 samples obtained from 2014 to 2016. In the 2014–2016 samples, a new mutation (G533S) appeared and reached 44.1% (15/34). Collectively, parasites with the Kelch domain mutations (after amino acid 440) had significantly higher ring survival rates than the WT parasites. Individually, F446I, G533S and A676D showed significantly higher ring survival rates than the WT. Although the drug sensitivity phenotypes measured by the RSA6h and EC5072h assays may be intrinsically linked to the in vivo clinical efficacy data, the values determined by these two assays were not significantly correlated. This study identified the trend of K13 mutations in parasite populations from the China-Myanmar border area, confirmed an overall correlation of Kelch domain mutations with elevated ring-stage survival rates, and emphasized the importance of monitoring the evolution and spread of parasites with reduced artemisinin sensitivity along the malaria elimination course.
Keywords: Plasmodium falciparum, K13 kelch gene, Polymorphism, Artemisinin resistance, Correlation
Graphical abstract

Highlights
-
•
P. falciparum field isolates were collected from the China-Myanmar border in 2007–16.
-
•
133 isolates showed annually varied in vitro sensitivities to artemisinins.
-
•
Parasites showed temporally increased prevalence in K13 kelch domain mutations.
-
•
F446I was the predominant mutation and a new mutation G533S emerged in later years.
-
•
Correlation between K13 kelch mutations and increased ring survival was confirmed.
1. Introduction
Drug resistance has emerged as one of the greatest challenges facing malaria control. Due to widespread resistance in malaria parasites to multiple antimalarial drugs, WHO recommended artemisinin (ART)-based combination therapies (ACT) to be the frontline treatment of falciparum malaria since 2005. To our dismay, reports of resistance to ART and its partner drugs started emerging in 2006–2007 in the Greater Mekong Subregion (GMS) of Southeast Asia. First reports of reduced in vivo susceptibility to ART, characterized by delayed clearance of parasites, came from northwestern Cambodia after artesunate (AS) monotherapy or ACT (Dondorp et al., 2009; Noedl et al., 2008) followed by numerous reports from other parts of GMS including Cambodia, Thailand, Vietnam, Myanmar, and the China-Myanmar border areas, due to spread, independent emergence, or both (Ashley et al., 2014; Bustos et al., 2013; Cheeseman et al., 2012; Hien et al., 2012; Kyaw et al., 2013; Phyo et al., 2012; Takala-Harrison et al., 2013). Spread of ART resistance from GMS to Africa would be catastrophic and an enormous threat to global malaria control programs. Such resistance creates an urgent need to identify molecular markers for the surveillance of ART-resistant parasites in order to contain the spread of resistance.
The two important phenotypes associated with ART resistance are delayed parasite clearance half-life (>5 h) in vivo (Ariey et al., 2014; Ashley et al., 2014) and increased ring-stage survival rates in vitro after a brief ART exposure (Witkowski et al., 2013a). The ART-resistant mechanism was first identified in parasites cultured under ART drug pressure in vitro for over a period of five years, which involved a mutation (M476I) in the Kelch 13 gene (K13, P. falciparum 3D7_1343700) (Ariey et al., 2014). This finding was further supported by genome-wide association studies, which identified a region of chromosome 13 to be associated with slow parasite clearance (Cheeseman et al., 2012; Takala-Harrison et al., 2013). Multiple reports from the GMS and other areas from Asia, Africa and Latin America ensued and the identification of more than 200 different non-synonymous mutations in the Kelch domain of the K13 protein (codons 441–726) (Menard et al., 2016; Nyunt et al., 2015; Takala-Harrison et al., 2013; Tun et al., 2015; Wang et al., 2015c). Parasites engineered to have some of these mutations conclusively showed increased ring-stage survival after exposure to ART in vitro (Ghorbal et al., 2014; Straimer et al., 2015). Although the molecular mechanism of K13-mediated ART resistance is not completely understood, an upregulated unfolded response pathway was observed in clinically ART-resistant parasites with mutant K13 genes by transcriptome analysis (Mok et al., 2015). In addition, K13 mutations were also implicated to act via phosphatidylinositol-3-kinase signaling (Mbengue et al., 2015). Certain K13 mutations were found to enhance the parasite's stress response involving the ubiquitin/proteasome pathway, as resistant parasites harboring K13 mutations showed reduced ubiquitination (Dogovski et al., 2015).
Given the different drug histories and evolutionary backgrounds (Talundzic et al., 2015), there is substantial geographic heterogeneity within the GMS with regard to the patterns as well as frequencies of K13 mutations (Huang et al., 2015; Tun et al., 2015). C580Y was the most prevalent mutation and approached fixation in western Cambodia, whereas in the Thai–Cambodian/Thai–Myanmar border areas C580Y, Y493H, R539T, and I543T were all present (Ariey et al., 2014; Nyunt et al., 2015; Takala-Harrison et al., 2015; Thriemer et al., 2014). In northern Myanmar and the China–Myanmar border areas, F446I was the most prevalent and showed association with reduced ART susceptibility (Huang et al., 2015; Wang et al., 2015c, 2018). The heterogeneity in K13 mutations over the whole GMS requires a regional approach to study the occurrence and spread of ART resistance in this area. At the China-Myanmar border area, where ART family drugs have been used for over three decades, there have been reports of an increase in the day-3 parasite-positive cases and increased prevalence of K13 mutations (Huang et al., 2015; Wang et al., 2015c, 2018), suggesting possible emergence of ART resistance.
In this study, K13 polymorphisms were investigated in longitudinally archived parasite samples from the China-Myanmar border. In vitro sensitivities of the parasites to AS and dihydroartemisinin (DHA) were evaluated using the 72 h traditional SYBR Green I-based assay (EC5072h) and the ring survival assay (RSA6h), where early (0–3 h) ring-stage parasites were exposed to DHA for 6 h, in order to track the evolution of ART resistance in the parasite populations over a 10-year period.
2. Materials and methods
2.1. Sample collection
P. falciparum clinical isolates were collected in 2007–2016 from the China-Myanmar border. Most isolates were convenience samples collected at local health facilities. Giemsa-stained thick smears were made on febrile patients with uncomplicated malaria to identify P. falciparum infections. Written informed consent was signed by the patients or by the guardians of children before enrollment into this study. Prior to drug treatment, up to 5 ml of venous blood were drawn from patients, cryopreserved and stored in liquid nitrogen. Patients were then treated with DHA-piperaquine per local malaria treatment guidelines. Treatment was generally not supervised and the outcome was not followed. Monoclonal infections were determined by analysis of the merozoite surface protein 1 (msp1) and msp2) and glutamate-rich protein (glurp) genes as described earlier (Meng et al., 2010; Yuan et al., 2013). Only isolates with monoclonal infections were used for drug assays. The study was approved by the Institutional Review Boards of Pennsylvania State University, Kunming Medical University, China Medical University and Department of Health of Kachin.
2.2. In vitro culture of parasites
Samples of frozen strains were thawed in solutions of NaCl and glucose. P. falciparum parasite isolates were cultured in RPMI 1640 culture medium with 6% human AB serum, supplemented with 0.5% Albumax II. Cultures were maintained in 90% N2, 5% O2 and 5% CO2. In general, about three weeks of continuous culture were performed before the parasites were used for drug assays.
2.3. Drug susceptibility assay
Sensitivities of the parasite isolates to AS and DHA were assessed using a standard SYBR green I-based assay with a 72 h duration and using 3D7 as the reference strain (Smilkstein et al., 2004; Wang et al., 2016). Parasites in the ring stage were tightly synchronized by 5% D-sorbitol, and diluted to 2% hematocrit and 0.5% parasitemia with complete medium. Drug assay was conducted in 96-well culture plates with different concentrations of drugs. Results were expressed as the 50% effective concentration (EC50) calculated from the variable-slope sigmoidal function for each measurement. Each isolate was measured in three biological experiments, each with two technical replicates.
2.4. Ring survival assay
RSA was performed as previously described (Siddiqui et al., 2018; Witkowski et al., 2013a; Zhang et al., 2017). Briefly, highly synchronous parasite cultures at the young ring stage (0–3 h) were exposed to 700 nM DHA or 0.1% dimethyl sulfoxide (DMSO) as the control for 6 h. The drugs were then washed off with RPMI 1640, and parasites cultivated further for 66 h under the standard in vitro culture conditions. At 72 h after the assay initiation, survival rates were calculated in Giemsa-stained thin smears by counting the viable parasites surviving in DHA-treated versus DMSO-treated cultures. Parasite isolates demonstrating >1% survival in the RSA were considered to display reduced susceptibility to ART (Amaratunga et al., 2014). The RSA value of each parasite isolate was determined in three biological replicates.
2.5. Amplification and sequencing of K13 gene
DNA was extracted from the blood samples by using the High Pure PCR Template Preparation Kit (Roche, Germany) following the manufacturer's instructions. The full-length K13 gene was amplified by three nested PCR reactions and sequenced by using the Sanger sequencing method. Specific primers and cycling conditions are listed in Table S1. Assembly and alignment of DNA sequences were performed using the DNASTAR software.
2.6. Statistical analysis
All data were analyzed with Microsoft Excel and GraphPad Prism 6.0. Normally distributed data were expressed as means with standard deviations, while data that were not normally distributed were expressed as medians with interquartile ranges (IQR). Mann-Whitney U test was used to compare between two groups. The EC5072h value for each drug was estimated using GraphPad Prism 6.0. Estimates of EC50 and RSA survival rates were analyzed with one-way ANOVA. Fisher's exact test was used to compare mutation frequency data between two groups. For data involving multiple comparisons, P values were corrected using the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995).
3. Results
3.1. In vitro sensitivities of adapted field isolates to AS and DHA
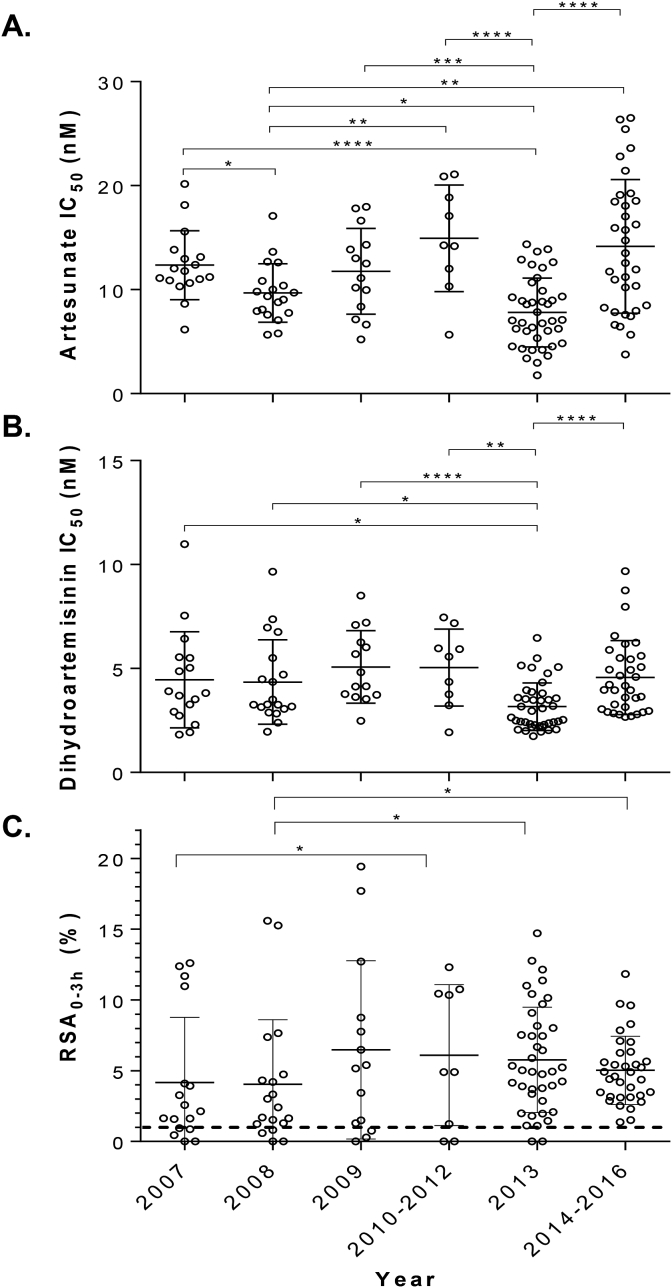
A total of 133 field parasite isolates collected between 2007 and 2016 from the China-Myanmar border area were selected for this study. Parasites were culture-adapted and confirmed to be monoclonal after genotyping three polymorphic genes, msp1, msp2 and glurp (Meng et al., 2010). In vitro susceptibilities to AS and DHA of 133 parasite isolates and the laboratory clone 3D7 were tested using the 72 h SYBR Green I assay and RSA (Fig. 1A and B and Table 1). The mean EC5072h values for the 133 field parasite isolates were 11.2 nM (range 1.8–26.5 nM) to AS and 4.2 nM (range 1.7–11.0 nM) to DHA, which were ∼2 fold higher than those for 3D7 (P < 0.05, Mann Whitney U test). EC5072h values for both AS and DHA showed significant differences among the years (P < 0.001, one-way ANOVA), but they did not follow a clear trend over the years. RSA6h values of the 133 clinical isolates (4.3%) were significantly higher than that for 3D7 (Table 1). The range of the RSA6h values for the field isolates was relatively wide (0–19.4%). If the 1% RSA value is used as the cutoff value for ART resistance, 117 (88.0%) parasite isolates had RSA values greater than 1%. It is noteworthy that RSA6h values rose consistently from 2007 to 2016 with significant difference in samples obtained after 2013 when compared to samples collected before 2009 (P < 0.05, Mann-Whitney U test) (Fig. 1C & Table 1). Pair-wise comparison between the RSA6h and EC5072h values for both AS and DHA showed no significant correlation (P > 0.05, Spearman's correlation test) (Fig. S1).
Fig. 1.
In vitro susceptibilities of 133 P. falciparum parasite isolates from the China-Myanmar border to two artemisinin derivatives stratified by years. (A) EC5072h values (nM) to artesunate; (B) EC5072h values (nM) to dihydroartemisinin; (C) Ring-stage survival rates after exposure of ring-stage parasites to 700 nM of dihydroartemisinin for 6 h *, **, ***, and **** denote significance at P < 0.05, 0.01, 0.001, and 0.0001, respectively (Mann-Whitney U test).
Table 1.
Susceptibilities of P. falciparum parasites collected in different years to artesunate and dihydroartemisinin.
| Year | N | AS EC50 (nM)* | P | DHA EC50 (nM)* | P | RSA (%)# | P |
|---|---|---|---|---|---|---|---|
| 2007 | 17 | 12.4 ± 3.3 | <0.0001a | 4.5 ± 2.3 | 0.0002a | 2.1 (0.9–7.5) | 0.1353a |
| 2008 | 19 | 9.7 ± 2.8 | 4.3 ± 2.0 | 2.4 (1.2–4.7) | |||
| 2009 | 14 | 11.8 ± 4.1 | 5.1 ± 1.7 | 5.3 (1.2–9.8) | |||
| 2010–2012 | 9 | 14.9 ± 5.1 | 5.0 ± 1.9 | 4.9 (0.6–10.6) | |||
| 2013 | 40 | 7.8 ± 3.3 | 3.2 ± 1.1 | 5.0 (3.0–8.1) | |||
| 2014–2016 |
34 |
14.2 ± 6.4 |
4.6 ± 1.8 |
4.8 (3.2–6.3) |
|||
| Total |
133 |
11.2 ± 5.1 |
<0.0001b |
4.2 ± 1.8 |
<0.0001b |
4.3 (1.9–7.5) |
<0.0001b |
| 3D7 | 5.0 ± 0.1 | 2.4 ± 0.2 | 0 |
*The EC50 values for artesunate (AS) and dihydroartemisinin (DHA) are expressed as geometric mean ± standard deviation (SD), while the ring survival assay (RSA) values (#) are shown as median (interquartile range).
Comparison among all the years (Kruskal-Wallis test).
Comparison between all field isolates and 3D7 (Wilcoxon signed rank test).
3.2. K13 gene polymorphisms
Full-length K13 genes were amplified from the 133 culture-adapted field isolates and sequenced. A total of 11 single nucleotide polymorphisms (SNPs) were detected, all being nonsynonymous mutations. Of the 11 amino acid (aa) mutations, two (K189T and E252Q) are located before the Kelch propeller domain (<440 aa), while nine (P441L, F446I, N458Y, G533S, R539T, P574L, C580Y, A676D, and H719N) are within the propeller domain (>440 aa). In addition, an NN insertion at 136–137 aa (this is outside of the propeller and the BTZ domains) was observed in 103 (77.4%) parasite isolates (Table 2). Compared with the 3D7 K13 sequence, 20 (15%) were wild-type (WT) K13 gene, whereas 113 (85%) carried the NN insertion and/or SNPs. Of the 85 (63.9%) parasites that carried nonsynonymous SNPs, only one parasite from 2008 carried two nonsynonymous SNPs (E252Q and F446I) in the same gene. Interestingly, the NN insertion gradually increased its frequency and approached fixation after 2014. Among the SNPs, seven were observed in earlier samples collected in 2007–2009. F446I was the most predominant mutation, and its frequency increased from 17.6% (3/17) in 2007 to 55.9% (19/34) in 2014–2016. A new mutation, G533S, never reported earlier in China-Myanmar border, appeared in the latest samples obtained in 2014–2016 and was the second most prevalent mutation (44.1%) in these samples (Table 2). The rest of the mutations were relatively rare (<2.3%). Specifically, the C580Y and R539T mutations associated with delayed parasite clearance and common in other areas of GMS were only found in one sample each. For mutations outside the Kelch domain, two samples from 2008 carried the E252Q mutation, while two samples from 2013 carried the K189T mutation.
Table 2.
Prevalence (%) of mutations in the K13 gene in clinical isolates from the China-Myanmar border.
| Year | n | NNa | K189T | E252Q | P441L | F446I | N458Y | G533S | R539T | P574L | C580Y | A676D | H719N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2007 | 17 | 47.1 | – | – | 5.9 | 17.6 | – | – | – | 11.8 | – | – | – |
| 2008 | 19 | 47.4 | – | 10.5 | – | 21.1 | – | – | 5.3 | 5.3 | 5.3 | 5.3 | – |
| 2009 | 14 | 64.3 | – | – | – | 7.1 | – | – | – | – | – | 7.1 | 21.4 |
| 2010–2012 | 9 | 77.8 | – | – | – | 33.3 | – | – | – | – | – | – | – |
| 2013 | 40 | 90.0 | 5.0 | – | – | 62.5 | 2.5 | – | – | – | – | – | – |
| 2014–2016 | 34 | 100.0 | – | – | – | 55.9 | – | 44.1 | – | – | – | – | – |
| Total | 133 | 77.4 | 1.5 | 1.5 | 0.8 | 41.4 | 0.8 | 11.3 | 0.8 | 2.3 | 0.8 | 1.5 | 2.3 |
NN insertion at amino acid 136–137 of K13 gene.
3.3. Correlation of K13 mutations with RSA phenotypes
We next performed statistical analysis to determine whether certain K13 genotypes were associated with altered drug susceptibilities. Since the mutations in the propeller domain of K13 protein (>440 aa) were associated with clinical ART resistance, we grouped the 133 parasites into two groups based on the presence or absence of mutations in the propeller domain. Parasites with mutations in the propeller domain showed a median RSA6h value of 5.1%, significantly higher than 2.9% in the K13 WT group (P = 0.0021, Mann Whitney U test) (Fig. 2A). One parasite isolate with R539T and two isolates with A676D showed RSA values of >15%. The two most common mutations, F446I (55 isolates) and G533S (15 isolates), also showed significantly higher median RSA6h values of 5.2% (P = 0.002) and 4.6% (P = 0.0193), respectively, than the WT. Some parasites with the K189T (2 isolates) and E252Q (2 isolates) mutations before the propeller domain also displayed RSA6h values > 1% and median values higher than that of the WT parasites, whereas a single parasite isolate carrying the C580Y mutation had a comparable RSA value to that of the 3D7 parasite (Fig. 2B, Table 3). The parasites with the K13 NN insertion showed a slightly higher mean RSA value (4.4%) than that of the parasites without the NN insertion (4.2%), although the difference was not significant (P > 0.05, Mann Whitney U test). Interestingly, parasites carrying the mutations in the K13 propeller domain also showed significantly higher EC5072h values to AS than WT parasites (Fig. S2), although the EC5072h values to DHA were not significantly different between the two groups. In addition, WT parasites and parasites with the NN insertion had similar EC5072h values to either AS or DHA.
Fig. 2.
Correlation of parasite survival rates (RSA0-3h) and K13 polymorphisms. (A) RSA values between parasites with mutations in the kelch propeller domain (>440 mutations) and parasites without kelch mutations (>440 WT). Different K13 mutations were color-coded. P = 0.0021 (Mann-Whitney U test). (B) Comparison of RSA values between parasites with different K13 mutations and the wild type (WT). P values were from Mann-Whitney U test with Benjamini-Hochberg correction. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Ring survival rates (RSA0–3h) of parasite isolates stratified by the K13 alleles.
| Mutations | n | Median RSA (%) | IQR | P* |
|---|---|---|---|---|
| WT# | 48 | 2.5 | 1.0–4.9 | – |
| K189T | 2 | 8.9 | 7.5–10.4 | 0.1403a |
| E252Q | 2 | 5.2 | 3.0–7.4 | 0.451a |
| P441L | 1 | 0.5 | – | 0.0003b |
| F446I | 55 | 5.2 | 3.1–8.2 | 0.002a |
| N458Y | 1 | 7.5 | – | 0.0003b |
| G533S | 15 | 4.6 | 3.3–7.1 | 0.0193a |
| R539T | 1 | 15.3 | – | 0.0003b |
| P574L | 3 | 1.6 | 1.3–11.7 | 0.921a |
| C580Y | 1 | 0.0 | – | 0.0003b |
| A676D | 2 | 16.7 | 15.6–17.7 | 0.0061a |
| H719N | 3 | 5.4 | 1.3–8.8 | 0.4571a |
*Comparison between the isolates carrying individual K13 mutations with the WT. P values shown here are after Benjamini-Hochberg corrections.
#Parasite isolates without any point mutations (including 28 parasites with the NN insertion).
Mann-Whitney U test.
Wilcoxon matched-pairs signed-rank test.
4. Discussion
Through analysis of 133 longitudinally collected clinical samples spanning a decade from the China-Myanmar border area, we detected reduced in vitro sensitivities against two ART drugs, as well as elevated RSA6h values indicative of clinical ART resistance. This was further supported by escalating prevalence of K13 mutant alleles that are correlated with increased ring survival rates. There has been a substantial decline of malaria burden in the GMS, where multidrug-resistant parasites have been rampant. Different drug policies in neighboring countries in the GMS affect the course of evolution of drug resistance (Zeng et al., 2017), and therefore constant surveillance of drug efficacy and resistance is required to ensure timely adjustment of frontline drugs. Notably, international borders in the GMS are extremely porous with high malaria transmission intensity (Cui et al., 2012; Parker et al., 2015), which make the two sides of the border very prone to influx of drug-resistant parasites. Hence, in light of the recently emerged ART resistance in P. falciparum in the GMS, the current study aimed to follow the trend of ART-resistant parasites in the China-Myanmar border area. Together with previous reports documenting delayed parasite clearance, the presence of day-3 positive parasites after ART/ACT treatments, and the high prevalence of K13 polymorphisms in the same border area (Kyaw et al., 2013; Nyunt et al., 2015; Wang et al., 2015c), this study revealed an increasing trend of reduced ART susceptibility in the P. falciparum population at the China-Myanmar border.
Taking advantage of longitudinally archived parasites collected between 2007 and 2016, we evaluated their sensitivities to ART drugs using both EC5072h estimates quantifying parasite proliferation and RSA6h estimating parasite survival at early ring stage. Our data showed significant differences in drug sensitivity over the ten-year period. Parasites banked in different years showed fluctuation of the EC5072h values to both AS and DHA without a clear trend, but the RSA6h data distinctly showed an evident gradual increase over the period of ten years. Consistent with our earlier finding (Wang et al., 2015c), we did not detect significant correlation between the values of the two drug assays, although they both may be intrinsically linked to clinical resistance. It is noteworthy that in the earlier years of emergence of ART resistance in the GMS, a west-to-east increasing trend of EC5072h values estimated using ex vivo 72 h drug assay was reported (Noedl et al., 2009). Given that RSA6h is considered to be able to capture the delayed parasite clearance phenotype attributed to prolonged and dormant ring stages (Hott et al., 2015; Wang et al., 2015c; Witkowski et al., 2013b), the escalating RSA6h values in parasites collected after 2008 suggest a possible emergence of ART resistance in these latter parasites, which are concomitant with the widespread use of ACT in the study area.
A number of mutations in the Kelch propeller domain of K13 were associated with delayed clearance in clinical studies (Ashley et al., 2014). Genotyping analysis for our 133 parasite samples in this study revealed 11 distinct nonsynonymous mutations, of which 9 occurred in the propeller domain. All these mutations have been described previously in Myanmar or the China-Myanmar border area except G533S (Ariey et al., 2014; Ashley et al., 2014; Feng et al., 2015; Nyunt et al., 2015; Tun et al., 2015; Wang et al., 2015b). The parasite population at the China-Myanmar border bears more resemblance to parasite population collected from northern Myanmar, where F446I was the predominant mutation (Ariey et al., 2014; Nyunt et al., 2015; Tun et al., 2015; Wang et al., 2015b). However, it is drastically different from parasite populations from most parts of the GMS. Mutations prevalent in other GMS countries or regions were either completely absent (e.g., I543T and Y493H) or occurred at very low frequencies (C580Y and R539T) in our study population (Ariey et al., 2014; Ashley et al., 2014; Ye et al., 2016). Mutations associated with high RSA6h values but present sparsely in these samples include R539T, A676D and N458Y. A676D has been reported earlier in various studies from the China-Myanmar area but it does not seem to confer ART resistance (Amambua-Ngwa et al., 2012; Feng et al., 2015; Wang et al., 2015c; Win et al., 2016; Ye et al., 2016), whereas N458Y showed significant association with prolonged parasite clearance half-lives in clinical studies (Ariey et al., 2014; Ashley et al., 2014; Tun et al., 2015), and also showed very high ring-stage survival rate (60%) in our previous study (Wang et al., 2015c). While the reasons underlying these geographical differences in K13 mutations are not clear, differences in drug histories and genetic backgrounds of these parasites are likely among the answers (Miotto et al., 2015). Our temporal data revealed that the genetic diversity of K13 was much higher in earlier years (2007–2009), whereas in samples after 2010, the K13 mutations were predominated by F446I and G533S, with the latter only showing up in the 2014–2016 samples and also attaining high prevalence (44.1%). This implies that these two mutations might confer fitness advantages to the parasites under the current ACT regimen in this region.
The most prevalent mutation in our sample set, F446I, is associated with significantly higher ring survival rates (RSA6h) than the WT parasites. Interestingly, its prevalence showed a continuous increase from 2007 to 2016. This mutation has been the most common in the northern part of GMS including Myanmar and the China-Myanmar border area (Feng et al., 2015; Huang et al., 2015; Tun et al., 2015; Wang et al., 2015b, 2015c), but it is lacking in other areas of SE Asia (Ye et al., 2016). Clinical studies showed that F446I was significantly associated with delayed parasite clearance and day-3 positive parasites in the China–Myanmar border region (Huang et al., 2015; Wang et al., 2015c). Recently the role of F446I in mediating ART resistance was confirmed genetically by introduction of this mutation into ART-sensitive laboratory strains (Wang et al., 2018).
The K13 propeller domain consists of six blades: P441L, F446I, and N458Y are located in Blade I, G533S and R539T in Blade III, P574L and C580Y in Blade IV, and A676D and H719N in Blade VI (Ariey et al., 2014). One of the newly found mutations with high prevalence in this geographical location is G533S. This mutation has been reported earlier in P. falciparum samples from Cambodia and Gambia (Amambua-Ngwa et al., 2012; Ariey et al., 2014), whereas an alternative form of this mutation G533A has also been reported from India (Mishra et al., 2015). In our field samples, this mutation appeared for the first time in the most recent samples, and parasites with this mutation showed significantly higher ring survival rates than the WT parasites. Glycine (G), being the smallest amino acid, has the highest conformational flexibility and its substitution by either the polar uncharged serine (S) or the hydrophobic alanine (A) may lead to some structural and functional alterations. Although both substitutions are predicted to be neutral/tolerated for K13 using Provean and SIFT analysis (Mbenda et al., 2018; Siddiqui et al., 2018), G533S may influence the disulfide bond between C532 and C580 in the protein. G to S mutations have also been reported to affect protein stability, resulting in adverse phenotypes in other systems (Nuytinck et al., 1996; Zhao et al., 2000), whereas G to A mutation is generally considered to be well tolerated (Loo and Clarke, 1994). It would be interesting to test the prediction of the structural effects of these mutations on ART resistance by genetic manipulation.
Whereas mutations present in the kelch domain are associated with ART resistance, this study also revealed two point mutations, E252Q and K189T in 2008 and 2013, respectively, outside the Kelch domain. Although these parasites show relatively high RSA values but due to their low frequencies in the sample set and absence of any gene manipulation data, no association can be concluded. Interestingly, E252Q was also the most common mutation before 2011 on the Thai-Myanmar border, and has been recently associated with slow parasite clearance and elevated treatment failure rates (Anderson et al., 2017; Phyo et al., 2016). This mutation was found only in two field isolates from 2008. It is argued that these parasites might be progressively taken over by the ones with K13 propeller mutations, leading to even slower clearance rates (Anderson et al., 2017). The potential role of the E252Q in mediating ART resistance remains to be determined genetically. Another interesting finding from this analysis was the increasing prevalence of parasites with NN insertion at position 136–137, which reached 100% in the most recent parasite samples. Our earlier analysis of a small sample set showed that this insertion was present only in day-3 parasite-positive isolates showing association with increased ring stage survival (Wang et al., 2015c). However, we did not detect a significant association of the NN insertion with altered sensitivity to ART drugs, suggesting that our earlier observation could be due to concurrent Kelch mutations in the NN insertion samples. Since parasites with minisatellite variations in the nhe1 gene show altered sensitivities to quinine (Meng et al., 2010), it would be interesting to test the NN insertion in K13 gene with regard to ART sensitivity.
Besides confirming the contribution of Kelch mutations to increased ring survival rates, we also found a wide range of RSA6h values (0–19.4%) for parasites without the Kelch mutations. Field isolates lacking K13 mutations but showing ART-resistant phenotype were also identified previously in Cambodia (Mukherjee et al., 2017). This indicates involvement of secondary and/or additional mechanisms in ART resistance. We recently reported that falcipain 2a (FP2a) may be an additional contributor to ART resistance as FP2a mutations are associated with reduced enzyme activity, decreased hemoglobin digestion and elevated ring-stage survival after DHA treatment (Klonis et al., 2011; Siddiqui et al., 2018). Genome-wide association studies and gene manipulation experiments have also identified other genetic loci (e.g., ATG18, coronin, pfap2mu) possibly associated with ART resistance (Breglio et al., 2018; Demas et al., 2018; Henriques et al., 2015; Miotto et al., 2015; Wang et al., 2016).
This study has a few limitations. Although efficacy studies in recent years showed excellent clinical efficacies of ACT for treating uncomplicated falciparum malaria in this region (Liu et al., 2015; Wang et al., 2015a), the majority of the clinical samples in this study was convenience samples without a link to the clinical outcomes of the treatment. In addition, although we strived to increase the accuracy of the RSA by performing three biological replicates of the assay, the use of flow cytometry deploying a mitochondrial marker along with SYBR Green staining will further enhance the precision of the results compared to microscopic examination as the read-out for RSA (Henrici et al., 2018). Furthermore, it is noteworthy that there is substantial variability in the RSA results among different laboratories, and the establishment of standardized control isolates will allow better comparison of results from different studies. The small sample numbers collected in certain years make it difficult to draw firm conclusions for association of certain K13 genotypes with reduced ART susceptibility.
Emergence and spread of ART-resistant parasites are a major concern for malaria control and elimination, which justifies continuous monitoring of K13 polymorphisms. In this regard, in vivo observation of clinical efficacy, in vitro (or ex vivo) RSA and K13 genotyping all serve as complementary predictive markers in epidemiological surveillance of ART resistance (Ariey et al., 2014; Ashley et al., 2014; Wang et al., 2015c). Our data further confirm the emergence and/or spread of P. falciparum parasites with reduced ART sensitivity at the China-Myanmar border. Parasites showing increased ring survival rates but lacking the K13 mutations indicate additional mechanisms of ART resistance. Taken together, these findings imply an urgent need for increased regional and phenotypic surveillance and population studies to determine new ART resistance mechanism and monitor the spread of ART resistance in the GMS for designing better containment strategies.
Abbreviations
- ART
Artemisinin
- ACT
Artemisinin-based combination therapy
- AS
Artesunate
- DHA
Dihydroartemisinin
- DMSO
Dimethyl sulfoxide
- GMS
Greater Mekong Subregion
- IQR
Interquartile range
- RSA
Ring survival assay
- SNP
Single nucleotide polymorphism
- WT
Wild-type
Competing interests
The authors have no competing interests to declare.
Acknowledgements
We thank Dr. Chengqi Wang for assistance with statistical analysis. This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health USA (U19AI089672 and R01AI128940), Yunnan Provincial Project (2013HA026), National Science Foundation of China (31860604 and U1802286), Major science and technology projects of Yunnan Province (2018ZF0081), Yunnan Applied Basic Research Projects-Union Foundation, China (No. 2015FB034, 2017FE468-185, 2018FE001-190). YW was funded by the Hundred-Talent Program of Kunming Medical University (No. 60117190439) and Foundation of the Education Department of Yunnan Province (No. 2018JS151).
Contributor Information
Liwang Cui, Email: lcui@health.usf.edu.
Zhaoqing Yang, Email: zhaoqingy92@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Amambua-Ngwa A., Tetteh K.K., Manske M., Gomez-Escobar N., Stewart L.B., Deerhake M.E., Cheeseman I.H., Newbold C.I., Holder A.A., Knuepfer E., Janha O., Jallow M., Campino S., Macinnis B., Kwiatkowski D.P., Conway D.J. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga C., Witkowski B., Dek D., Try V., Khim N., Miotto O., Menard D., Fairhurst R.M. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob. Agents Chemother. 2014;58:4935–4937. doi: 10.1128/AAC.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T.J., Nair S., McDew-White M., Cheeseman I.H., Nkhoma S., Bilgic F., McGready R., Ashley E., Pyae Phyo A., White N.J., Nosten F. Population parameters underlying an ongoing soft sweep in Southeast Asian malaria parasites. Mol. Biol. Evol. 2017;34:131–144. doi: 10.1093/molbev/msw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Menard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., Sopha C., Chuor C.M., Nguon C., Sovannaroth S., Pukrittayakamee S., Jittamala P., Chotivanich K., Chutasmit K., Suchatsoonthorn C., Runcharoen R., Hien T.T., Thuy-Nhien N.T., Thanh N.V., Phu N.H., Htut Y., Han K.T., Aye K.H., Mokuolu O.A., Olaosebikan R.R., Folaranmi O.O., Mayxay M., Khanthavong M., Hongvanthong B., Newton P.N., Onyamboko M.A., Fanello C.I., Tshefu A.K., Mishra N., Valecha N., Phyo A.P., Nosten F., Yi P., Tripura R., Borrmann S., Bashraheil M., Peshu J., Faiz M.A., Ghose A., Hossain M.A., Samad R., Rahman M.R., Hasan M.M., Islam A., Miotto O., Amato R., MacInnis B., Stalker J., Kwiatkowski D.P., Bozdech Z., Jeeyapant A., Cheah P.Y., Sakulthaew T., Chalk J., Intharabut B., Silamut K., Lee S.J., Vihokhern B., Kunasol C., Imwong M., Tarning J., Taylor W.J., Yeung S., Woodrow C.J., Flegg J.A., Das D., Smith J., Venkatesan M., Plowe C.V., Stepniewska K., Guerin P.J., Dondorp A.M., Day N.P., White N.J. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Breglio K.F., Amato R., Eastman R., Lim P., Sa J.M., Guha R., Ganesan S., Dorward D.W., Klumpp-Thomas C., McKnight C., Fairhurst R.M., Roberts D., Thomas C., Simon A.K. A single nucleotide polymorphism in the Plasmodium falciparum atg18 gene associates with artemisinin resistance and confers enhanced parasite survival under nutrient deprivation. Malar. J. 2018;17:391. doi: 10.1186/s12936-018-2532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos M.D., Wongsrichanalai C., Delacollette C., Burkholder B. Monitoring antimalarial drug efficacy in the Greater Mekong Subregion: an overview of in vivo results from 2008 to 2010. Southeast Asian J. Trop. Med. Publ. Health. 2013;44(Suppl. 1):201–230. [PubMed] [Google Scholar]
- Cheeseman I.H., Miller B.A., Nair S., Nkhoma S., Tan A., Tan J.C., Al Saai S., Phyo A.P., Moo C.L., Lwin K.M., McGready R., Ashley E., Imwong M., Stepniewska K., Yi P., Dondorp A.M., Mayxay M., Newton P.N., White N.J., Nosten F., Ferdig M.T., Anderson T.J. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Yan G., Sattabongkot J., Chen B., Cao Y., Fan Q., Parker D., Sirichaisinthop J., Su X.Z., Yang H., Yang Z., Wang B., Zhou G. Challenges and prospects for malaria elimination in the greater Mekong subregion. Acta Trop. 2012;121:240–245. doi: 10.1016/j.actatropica.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demas A.R., Sharma A.I., Wong W., Early A.M., Redmond S., Bopp S., Neafsey D.E., Volkman S.K., Hartl D.L., Wirth D.F. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogovski C., Xie S.C., Burgio G., Bridgford J., Mok S., McCaw J.M., Chotivanich K., Kenny S., Gnadig N., Straimer J., Bozdech Z., Fidock D.A., Simpson J.A., Dondorp A.M., Foote S., Klonis N., Tilley L. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhou D., Lin Y., Xiao H., Yan H., Xia Z. Amplification of pfmdr1, pfcrt, pvmdr1, and K13 propeller polymorphisms associated with Plasmodium falciparum and Plasmodium vivax isolates from the China-Myanmar border. Antimicrob. Agents Chemother. 2015;59:2554–2559. doi: 10.1128/AAC.04843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbal M., Gorman M., Macpherson C.R., Martins R.M., Scherf A., Lopez-Rubio J.J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- Henrici R.C., van Schalkwyk D.A., Sutherland C.J. Transient temperature fluctuations severely decrease P. falciparum susceptibility to artemisinin in vitro. Int. J. Parasitol. Drugs Drug Resist. 2018;9:23–26. doi: 10.1016/j.ijpddr.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques G., van Schalkwyk D.A., Burrow R., Warhurst D.C., Thompson E., Baker D.A., Fidock D.A., Hallett R., Flueck C., Sutherland C.J. The Mu subunit of Plasmodium falciparum clathrin-associated adaptor protein 2 modulates in vitro parasite response to artemisinin and quinine. Antimicrob. Agents Chemother. 2015;59:2540–2547. doi: 10.1128/AAC.04067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien T.T., Thuy-Nhien N.T., Phu N.H., Boni M.F., Thanh N.V., Nha-Ca N.T., Thai le H., Thai C.Q., Toi P.V., Thuan P.D., Long le T., Dong le T., Merson L., Dolecek C., Stepniewska K., Ringwald P., White N.J., Farrar J., Wolbers M. In vivo susceptibility of Plasmodium falciparum to artesunate in Binh phuoc Province, Vietnam. Malar. J. 2012;11:355. doi: 10.1186/1475-2875-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hott A., Casandra D., Sparks K.N., Morton L.C., Castanares G.G., Rutter A., Kyle D.E. Artemisinin-resistant Plasmodium falciparum parasites exhibit altered patterns of development in infected erythrocytes. Antimicrob. Agents Chemother. 2015;59:3156–3167. doi: 10.1128/AAC.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Takala-Harrison S., Jacob C.G., Liu H., Sun X., Yang H., Nyunt M.M., Adams M., Zhou S., Xia Z., Ringwald P., Bustos M.D., Tang L., Plowe C.V. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J. Infect. Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonis N., Crespo-Ortiz M.P., Bottova I., Abu-Bakar N., Kenny S., Rosenthal P.J., Tilley L. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyaw M.P., Nyunt M.H., Chit K., Aye M.M., Aye K.H., Aye M.M., Lindegardh N., Tarning J., Imwong M., Jacob C.G., Rasmussen C., Perin J., Ringwald P., Nyunt M.M. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yang H.L., Tang L.H., Li X.L., Huang F., Wang J.Z., Li C.F., Wang H.Y., Nie R.H., Guo X.R., Lin Y.X., Li M., Wang J., Xu J.W. In vivo monitoring of dihydroartemisinin-piperaquine sensitivity in Plasmodium falciparum along the China-Myanmar border of Yunnan Province, China from 2007 to 2013. Malar. J. 2015;14:47. doi: 10.1186/s12936-015-0584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo T.W., Clarke D.M. Functional consequences of glycine mutations in the predicted cytoplasmic loops of P-glycoprotein. J. Biol. Chem. 1994;269:7243–7248. [PubMed] [Google Scholar]
- Mbenda H.G.N., Zeng W., Bai Y., Siddiqui F.A., Yang Z., Cui L. Genetic diversity of the Plasmodium vivax phosphatidylinositol 3-kinase gene in two regions of the China-Myanmar border. Infect. Genet. Evol. 2018;61:45–52. doi: 10.1016/j.meegid.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R.V., Rizk S.S., Njimoh D.L., Ryan Y., Chotivanich K., Nguon C., Ghorbal M., Lopez-Rubio J.J., Pfrender M., Emrich S., Mohandas N., Dondorp A.M., Wiest O., Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D., Khim N., Beghain J., Adegnika A.A., Shafiul-Alam M., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., Bustos M.D., Cao J., Chen J.H., Collet L., Cui L., Thakur G.D., Dieye A., Djalle D., Dorkenoo M.A., Eboumbou-Moukoko C.E., Espino F.E., Fandeur T., Ferreira-da-Cruz M.F., Fola A.A., Fuehrer H.P., Hassan A.M., Herrera S., Hongvanthong B., Houze S., Ibrahim M.L., Jahirul-Karim M., Jiang L., Kano S., Ali-Khan W., Khanthavong M., Kremsner P.G., Lacerda M., Leang R., Leelawong M., Li M., Lin K., Mazarati J.B., Menard S., Morlais I., Muhindo-Mavoko H., Musset L., Na-Bangchang K., Nambozi M., Niare K., Noedl H., Ouedraogo J.B., Pillai D.R., Pradines B., Quang-Phuc B., Ramharter M., Randrianarivelojosia M., Sattabongkot J., Sheikh-Omar A., Silue K.D., Sirima S.B., Sutherland C., Syafruddin D., Tahar R., Tang L.H., Toure O.A., Tshibangu-wa-Tshibangu P., Vigan-Womas I., Warsame M., Wini L., Zakeri S., Kim S., Eam R., Berne L., Khean C., Chy S., Ken M., Loch K., Canier L., Duru V., Legrand E., Barale J.C., Stokes B., Straimer J., Witkowski B., Fidock D.A., Rogier C., Ringwald P., Ariey F., Mercereau-Puijalon O., Consortium K. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H., Zhang R., Yang H., Fan Q., Su X., Miao J., Cui L., Yang Z. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob. Agents Chemother. 2010;54:4306–4313. doi: 10.1128/AAC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M., Imwong M., Woodrow C., Manske M., Stalker J., Drury E., Campino S., Amenga-Etego L., Thanh T.N., Tran H.T., Ringwald P., Bethell D., Nosten F., Phyo A.P., Pukrittayakamee S., Chotivanich K., Chuor C.M., Nguon C., Suon S., Sreng S., Newton P.N., Mayxay M., Khanthavong M., Hongvanthong B., Htut Y., Han K.T., Kyaw M.P., Faiz M.A., Fanello C.I., Onyamboko M., Mokuolu O.A., Jacob C.G., Takala-Harrison S., Plowe C.V., Day N.P., Dondorp A.M., Spencer C.C., McVean G., Fairhurst R.M., White N.J., Kwiatkowski D.P. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Prajapati S.K., Kaitholia K., Bharti R.S., Srivastava B., Phookan S., Anvikar A.R., Dev V., Sonal G.S., Dhariwal A.C., White N.J., Valecha N. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob. Agents Chemother. 2015;59:2548–2553. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S., Ashley E.A., Ferreira P.E., Zhu L., Lin Z., Yeo T., Chotivanich K., Imwong M., Pukrittayakamee S., Dhorda M., Nguon C., Lim P., Amaratunga C., Suon S., Hien T.T., Htut Y., Faiz M.A., Onyamboko M.A., Mayxay M., Newton P.N., Tripura R., Woodrow C.J., Miotto O., Kwiatkowski D.P., Nosten F., Day N.P., Preiser P.R., White N.J., Dondorp A.M., Fairhurst R.M., Bozdech Z. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Bopp S., Magistrado P., Wong W., Daniels R., Demas A., Schaffner S., Amaratunga C., Lim P., Dhorda M., Miotto O., Woodrow C., Ashley E.A., Dondorp A.M., White N.J., Wirth D., Fairhurst R., Volkman S.K. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar. J. 2017;16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M., Artemisinin Resistance in Cambodia 1 Study, C Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Noedl H., Socheat D., Satimai W. Artemisinin-resistant malaria in Asia. N. Engl. J. Med. 2009;361:540–541. doi: 10.1056/NEJMc0900231. [DOI] [PubMed] [Google Scholar]
- Nuytinck L., Dalgleish R., Spotila L., Renard J.P., Van Regemorter N., De Paepe A. Substitution of glycine-661 by serine in the alpha1(I) and alpha2(I) chains of type I collagen results in different clinical and biochemical phenotypes. Hum. Genet. 1996;97:324–329. doi: 10.1007/BF02185764. [DOI] [PubMed] [Google Scholar]
- Nyunt M.H., Hlaing T., Oo H.W., Tin-Oo L.L., Phway H.P., Wang B., Zaw N.N., Han S.S., Tun T., San K.K., Kyaw M.P., Han E.T. Molecular assessment of artemisinin resistance markers, polymorphisms in the k13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin. Infect. Dis. 2015;60:1208–1215. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- Parker D.M., Carrara V.I., Pukrittayakamee S., McGready R., Nosten F.H. Malaria ecology along the Thailand-Myanmar border. Malar. J. 2015;14:388. doi: 10.1186/s12936-015-0921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo A.P., Ashley E.A., Anderson T.J.C., Bozdech Z., Carrara V.I., Sriprawat K., Nair S., White M.M., Dziekan J., Ling C., Proux S., Konghahong K., Jeeyapant A., Woodrow C.J., Imwong M., McGready R., Lwin K.M., Day N.P.J., White N.J., Nosten F. Declining efficacy of artemisinin combination therapy against P. Falciparum malaria on the Thai-Myanmar border (2003-2013): the role of parasite genetic factors. Clin. Infect. Dis. 2016;63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo A.P., Nkhoma S., Stepniewska K., Ashley E.A., Nair S., McGready R., ler Moo C., Al-Saai S., Dondorp A.M., Lwin K.M., Singhasivanon P., Day N.P., White N.J., Anderson T.J., Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui F.A., Cabrera M., Wang M., Brashear A., Kemirembe K., Wang Z., Miao J., Chookajorn T., Yang Z., Cao Y., Dong G., Rosenthal P.J., Cui L. Plasmodium falciparum falcipain-2a polymorphisms in Southeast Asia and their association with artemisinin resistance. J. Infect. Dis. 2018;218:434–442. doi: 10.1093/infdis/jiy188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilkstein M., Sriwilaijaroen N., Kelly J.X., Wilairat P., Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J., Gnadig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P.D., Urnov F.D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R.M., Menard D., Fidock D.A. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Clark T.G., Jacob C.G., Cummings M.P., Miotto O., Dondorp A.M., Fukuda M.M., Nosten F., Noedl H., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Socheat D., Ariey F., Phyo A.P., Starzengruber P., Fuehrer H.P., Swoboda P., Stepniewska K., Flegg J., Arze C., Cerqueira G.C., Silva J.C., Ricklefs S.M., Porcella S.F., Stephens R.M., Adams M., Kenefic L.J., Campino S., Auburn S., MacInnis B., Kwiatkowski D.P., Su X.Z., White N.J., Ringwald P., Plowe C.V. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl. Acad. Sci. U. S. A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Jacob C.G., Arze C., Cummings M.P., Silva J.C., Dondorp A.M., Fukuda M.M., Hien T.T., Mayxay M., Noedl H., Nosten F., Kyaw M.P., Nhien N.T., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Ariey F., Mercereau-Puijalon O., Menard D., Newton P.N., Khanthavong M., Hongvanthong B., Starzengruber P., Fuehrer H.P., Swoboda P., Khan W.A., Phyo A.P., Nyunt M.M., Nyunt M.H., Brown T.S., Adams M., Pepin C.S., Bailey J., Tan J.C., Ferdig M.T., Clark T.G., Miotto O., MacInnis B., Kwiatkowski D.P., White N.J., Ringwald P., Plowe C.V. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talundzic E., Okoth S.A., Congpuong K., Plucinski M.M., Morton L., Goldman I.F., Kachur P.S., Wongsrichanalai C., Satimai W., Barnwell J.W., Udhayakumar V. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thriemer K., Hong N.V., Rosanas-Urgell A., Phuc B.Q., Ha do M., Pockele E., Guetens P., Van N.V., Duong T.T., Amambua-Ngwa A., D'Alessandro U., Erhart A. Delayed parasite clearance after treatment with dihydroartemisinin-piperaquine in Plasmodium falciparum malaria patients in central Vietnam. Antimicrob. Agents Chemother. 2014;58:7049–7055. doi: 10.1128/AAC.02746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun K.M., Imwong M., Lwin K.M., Win A.A., Hlaing T.M., Hlaing T., Lin K., Kyaw M.P., Plewes K., Faiz M.A., Dhorda M., Cheah P.Y., Pukrittayakamee S., Ashley E.A., Anderson T.J., Nair S., McDew-White M., Flegg J.A., Grist E.P., Guerin P., Maude R.J., Smithuis F., Dondorp A.M., Day N.P., Nosten F., White N.J., Woodrow C.J. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Huang Y., Zhao Y., Ye R., Zhang D., Pan W. Introduction of F446I mutation in the K13 propeller gene leads to increased ring survival rates in Plasmodium falciparum isolates. Malar. J. 2018;17:248. doi: 10.1186/s12936-018-2396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang Z., Yuan L., Zhou G., Parker D., Lee M.C., Yan G., Fan Q., Xiao Y., Cao Y., Cui L. Clinical efficacy of dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria at the China-Myanmar border. Am. J. Trop. Med. Hyg. 2015;93:577–583. doi: 10.4269/ajtmh.15-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cabrera M., Yang J., Yuan L., Gupta B., Liang X., Kemirembe K., Shrestha S., Brashear A., Li X., Porcella S.F., Miao J., Yang Z., Su X.Z., Cui L. Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border. Sci. Rep. 2016;6:33891. doi: 10.1038/srep33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Shrestha S., Li X., Miao J., Yuan L., Cabrera M., Grube C., Yang Z., Cui L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007-2012. Malar. J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang Y., Cabrera M., Zhang Y., Gupta B., Wu Y., Kemirembe K., Hu Y., Liang X., Brashear A., Shrestha S., Li X., Miao J., Sun X., Yang Z., Cui L. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob. Agents Chemother. 2015;59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win A.A., Imwong M., Kyaw M.P., Woodrow C.J., Chotivanich K., Hanboonkunupakarn B., Pukrittayakamee S. K13 mutations and pfmdr1 copy number variation in Plasmodium falciparum malaria in Myanmar. Malar. J. 2016;15:110. doi: 10.1186/s12936-016-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., Lim P., Mao S., Sopha C., Sam B., Anderson J.M., Duong S., Chuor C.M., Taylor W.R., Suon S., Mercereau-Puijalon O., Fairhurst R.M., Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Khim N., Chim P., Kim S., Ke S., Kloeung N., Chy S., Duong S., Leang R., Ringwald P., Dondorp A.M., Tripura R., Benoit-Vical F., Berry A., Gorgette O., Ariey F., Barale J.C., Mercereau-Puijalon O., Menard D. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Hu D., Zhang Y., Huang Y., Sun X., Wang J., Chen X., Zhou H., Zhang D., Mungthin M., Pan W. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci. Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Zhao H., Wu L., Li X., Parker D., Xu S., Zhao Y., Feng G., Wang Y., Yan G., Fan Q., Yang Z., Cui L. Plasmodium falciparum populations from northeastern Myanmar display high levels of genetic diversity at multiple antigenic loci. Acta Trop. 2013;125:53–59. doi: 10.1016/j.actatropica.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Bai Y., Wang M., Wang Z., Deng S., Ruan Y., Feng S., Yang Z., Cui L. Significant divergence in sensitivity to antimalarial drugs between neighboring Plasmodium falciparum populations along the eastern border of Myanmar. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01689-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Feng G.H., Zou C.Y., Su P.C., Liu H.E., Yang Z.Q. Overview of the improvement of the ring-stage survival assay-a novel phenotypic assay for the detection of artemisinin-resistant Plasmodium falciparum. Zool. Res. 2017;38:317–320. doi: 10.24272/j.issn.2095-8137.2017.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Hutton H.M., Meyer T.E., Walker F.A., MacKenzie N.E., Cusanovich M.A. Structure and stability effects of the mutation of glycine 34 to serine in Rhodobacter capsulatus cytochrome c(2) Biochemistry. 2000;39:4053–4061. doi: 10.1021/bi992979a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.