Abstract
Purpose
Colorectal cancer (CRC) is the third most common cancer in China and poses high morbidity and mortality. In recent years, increasing evidence has indicated that microRNAs played important functions in the occurrence and development of tumors. The purpose of this study was to identify the biological mechanisms of miR-362 in CRC.
Materials and Methods
Quantitative real-time PCR was carried out to assess the expression of miR-362 and SIX1. The Kaplan-Meier method was employed to evaluate the 5-year overall survival of CRC patients. The proliferative and invasive abilities of CRC cells were assessed by MTT and transwell assays.
Results
miR-362 was significantly decreased in CRC tissues and cell lines, compared to the normal tissues and normal cells. A significant connection was confirmed between the overall survival of 53 CRC patients and low expression of miR-362. Downregulation of miR-362 inhibited the proliferation and invasion through binding to the 3′-UTR of SIX1 mRNA in CRC. Additionally, we discovered that SIX1 was a direct target gene of miR-362 and that the expression of miR-362 had a negative connection with SIX1 expression in CRC. SIX1 could reverse partial functions in the proliferation and invasion in CRC cells.
Conclusion
miR-362 may be a prognostic marker in CRC and suppress CRC cell proliferation and invasion in part through targeting the 3′-UTR of SIX1 mRNA. The newly identified miR-362/SIX1 axis provides insight into the progression of CRC.
Keywords: miR-362, SIX, proliferation, invasion, colorectal cancer
INTRODUCTION
With approximately 600000 deaths every year, colorectal cancer (CRC) accounts for 10% of all cancer cases and mortalities, making CRC the third leading cause of cancer deaths.1,2 The clinic outcomes of CRC patients are poor because of high rates of local recurrence and distant metastasis.3,4 Therefore, identifying tumor molecular markers for metastasis and recurrence in patients with CRC is critical.
MicroRNAs (miRNAs), non-coding short RNAs of 19–24 nucleotides in length, cleave mRNA or bind to target mRNA to cause mRNA degradation or translational repression at the post-transcriptional level.5,6 miRNAs have been found to act as tumor suppressors or oncogenes in CRC and to play crucial roles in multiple biological processes.7,8 miR-362 has been reported to act as a tumor suppressor, inhibiting cell growth and the cell cycle and inducing cell apoptosis in cervical adenocarcinoma and chronic myelocytic leukemia.9,10 Xia, et al.11 demonstrated cell growth induced cell apoptosis in gastric cancer. Similarly, Wang, et al.12 indicated that cell proliferation and metastasis are affected by miR-362 in preeclampsia. In CRC, miR-362 expression appears to below and to induce cell cycle arrest.13 However, as research on miR-362 in CRC is limited, we aimed to explore the function of miR-362 in cell metastasis and growth in CRC in this study.
The sineoculis homeobox homolog 1 (SIX1), a member of the SIX family, shares a lysine within the DNA-binding helix in the homeodomain.14,15 SIX1 has been reported to be a conserved transcription factor associated with tumor development.16,17 In hepatocellular carcinoma, Cheng, et al.18 demonstrated that SIX1 induces cell proliferation and inhibits apoptosis. Moreover, SIX1 has been shown to promote proliferation and clone formation in endometrial carcinoma through ERK/AKT signaling.19 Similarly, Lerbs, et al.20 indicated that SIX1 could be a stem cell marker and promote cell metastasis in pancreatic cancer. Additionally, SIX1 was deemed a prognostic factor predictive of poor prognosis in glioma.21 In the present study, we found miR-362 to be downregulated in CRC tissues and cell lines, and discovered that low expression of miR-362 predicted poor overall survival in CRC patients. Additionally, we noted that overexpression of miR-362 suppressed cell proliferation and invasion through targeting SIX1 in CRC cell line SW480.
MATERIALS AND METHODS
Tissue samples
Cancer and matched paracancerous tissues were obtained from CRC patients who were diagnosed in the Department of Anorectal Surgery in the Affiliated Hospital of Qingdao University from January 2016 to June 2018. The clinical information of the patients and the associations between the expression of miR-362 and the clinicopathological features are shown in Table 1. The fresh tissues were instantly frozen in liquid nitrogen and stored at −80℃. The study protocol was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University, and all the patients provided informed consent before specimen collection.
Table 1. miR-362 Expression and Clinicopathological Features in 53 Colorectal Cancer Patients.
| Clinicopathological features | Cases (n=53) | miR-362 expression | p value | |
|---|---|---|---|---|
| High (n=25) | Low (n=28) | |||
| Age (yr) | 0.353 | |||
| >50 | 24 | 13 (54.2) | 11 (45.8) | |
| ≤50 | 29 | 12 (41.4) | 17 (58.6) | |
| Sex | 0.114 | |||
| Male | 30 | 17 (56.7) | 13 (43.3) | |
| Female | 23 | 8 (34.8) | 15 (65.2) | |
| Tumor size (mm) | 0.072 | |||
| >5.0 | 27 | 16 (59.3) | 11 (40.7) | |
| ≤5.0 | 26 | 9 (34.6) | 17 (65.4) | |
| TNM stage | 0.040* | |||
| III–IV | 26 | 16 (61.5) | 10 (38.5) | |
| I–II | 27 | 9 (33.3) | 18 (66.7) | |
| Lymph-node metastasis | 0.033* | |||
| >2 | 30 | 18 (60.0) | 12 (40.0) | |
| 0–2 | 23 | 7 (30.4) | 16 (69.6) | |
| Tumor location | 0.037 | |||
| Left-side | 28 | 17 (60.7) | 11 (39.3) | |
| Right-side | 25 | 8 (32.0) | 17 (68.0) | |
Variables are expressed as a number (percentage).
*p<0.05 (p values are calculated with chi-square test).
Cell lines and cell culture
Human CRC cells LOVO and SW480 and normal colorectal epithelial cells CCD-18Co were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Little Chalfont, UK) in a humidified atmosphere of 5% CO2 at 37℃.
Cell transfection
The miR-362 inhibitor and miR-362 mimic sequences were designed and synthesized from RiboBio (Guangzhou, China); SIX1 overexpressed plasmid (pcDNA-SIX1) was purchased from GenePharma Company (Shanghai, China). SW480 cells were seeded into six-well plates, and transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific) according to the company's instructions. Cells were harvested at 48 h for further analysis.
RNA isolation and quantitative real-time PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was applied to extract total RNA according to the manufacturer's protocol. For SIX1 analysis, the first cDNA chain was synthesized using a PrimeScript™ Reverse Transcription Reagent Kit (TaKaRa Bio, Otsu, Japan). Subsequently, SYBR Premix Ex Taq II (Takara Biotechnology, Dalian, China) was utilized to perform quantitative real-time PCR (RT-qPCR). For miRNA, reverse transcription was performed using the miScript Reverse Transcription Kit, and subsequent RT-qPCR was conducted using miScript SYBR Green PCR kits (Qiagen, New York, NY, USA), according to the manufacturer's protocol. GAPDH and U6 small nuclear RNA were used as internal normalization controls for SIX1 and miR-362, respectively. The primers for RT-qPCR were miR-362, forward 5′-TCGGAATCCTTGGAAC CTAGGTG, reverse 5′-ATCCAGTGCAGGGTCCGAGG; U6, forward 5′-AA CGCTTCACGAATTTGCGT, reverse 5′-CGCTTCA CGAATTT GCGTGTCAT; SIX1, forward 5′-AAGGAGAAGTCG AGGGGT GT-3′, reverse 5′-TGCTTGTTGGAGGAGGAGTT-3′; and GAP DH, forward 5′-GTTTGTGATGGGCGTGAAC, reverse 5′-ATG GACCTGGGTCATGAGT. Cycling parameters were as follows: initial denaturation for 3 min at 95℃, followed by 45 cycles of 5 s at 95℃ and 30 s at 60℃. The relative expression of each gene was calculated using the 2−ΔΔCt method.
MTT assay
The SW480 cells at a density of 2000 cells per well were seeded in 96-well plates and maintained for 24, 48, 72, or 96 h in an atmosphere containing 5% CO2 at 37℃. Subsequently, 100 µL of sterile3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide dye (MTT; Sigma-Aldrich, St. Louis, MO, USA) was added to each well and incubated for 4 h at 37℃. Next, the medium was removed, and 150 µL of dimethyl sulfoxide (DMSO; Sigma-Aldrich) were added to dissolve the formazan. Absorbance was measured at 490 nm using an automatic multi-well spectrophotometer (Bio-Rad, Richmond, CA, USA). All experiments were performed in triplicate.
Transwell assays
Transwell chambers (8-µm; Millipore, Billerica, MA, USA) covered with Matrigel (BD Biosciences, San Jose, CA, USA) were utilized to carry out the invasion assay. SW480 cells re-suspended in serum-free medium were placed into the upper chamber transwell insert. Meanwhile, the lower chamber was filled with normal medium containing 20% FBS as the chemoattractant. Subsequently, the non-invaded cells were wiped off by a cotton swab, while the invasive cells were fixed and stained with 100% methanol and 0.5% crystal violet solution, in that order. Cells were then counted using a microscope (CX31; Olympus, Tokyo, Japan). Each experiment was repeated at least three times.
Dual-luciferase reporter assay
The wild type 3′-UTR fragment of the SIX1 mRNA that contained the complementary sequences of miR-362 or the mutant sequences were inserted into pmirGlo vector (Promega, Madison, WI, USA), which were confirmed by sequencing. The miR-362 mimic or negative control and the pmirGlo constructs were co-transfected into SW480 cells using the Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer's instructions. The cells were lysed after calculated at 48 h, and the Dual-Luciferase Reporter Assay kit (Promega) was used to calculate the luciferase activity, according to the manufacturer's instructions. The firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
All data were analyzed by SPSS 19.0 software (IBM Corp., Armonk, NY, USA) and expressed as the mean±standard error. Two-sided Student's t-tests or one-way analysis of variance was employed to compare with differences between two groups or more than two groups. p values <0.05 were considered to indicate a statistically significant difference.
RESULTS
miR-362 expression reduced in CRC and downregulation of miR-362 predicts poor prognosis
The miR-362 mRNA levels in 53 pairs of CRC and adjacent normal tissues were assessed by RT-qPCR in an attempt to determine the importance of miR-362. As expected, we discovered that the expression of miR-362 was higher in CRC than in adjacent normal tissues (p<0.001) (Fig. 1A). Furthermore, the overall survival of all 53 patients was assessed, and Kaplan-Meier analysis indicated that miR-362 downregulation elicited shorter overall survival than miR-362 upregulation in CRC patients (p=0.048) (Fig. 1B).
Fig. 1. miR-362 expression was low in CRC, and downregulation of miR-362 predicted poor prognosis. (A) The expression of miR-362 was higher in CRC than paracancerous tissues (PT). (B) Kaplan-Meier analysis indicated that miR-362 downregulation predicted poor prognosis in CRC. (C) The expression of miR-362 was lower in LOVO and SW480 CRC cells than in normal CCD-18Co cells. (D) The efficiency of transfected miR-362 mimic or inhibitor was evaluated by qRT-PCR. **p<0.01, ***p<0.001. CRC, colorectal cancer.
The expressions of miR-362 in two human CRC cell lines (LOVO and SW480) and normal colon cells (CCD-18Co) were also evaluated using RT-qPCR. In comparison with the normal CCD-18Co colon cells, the expression of miR-362 was lower in LOVO (p=0.002) and SW480 CRC cells (p<0.001) (Fig. 1C). To investigate the biological mechanisms of miR-362 in CRC, miR-362 mimic or miR-362 inhibitor was injected into SW480 cells, and the transfection efficiency was evaluated by qRT-PCR. As shown in Fig. 1D, the mRNA levels of miR-362 increased in miR-362 mimic-transfected cells (p<0.001) and decreased in miR-362 inhibitor-transfected SW480 cells (p=0.006).
We also assessed associations between miR-362 expression and clinicopathological features in CRC. As shown in Table 1, the expression of miR-362 held significant correlations with TNM stage (p=0.040), lymph node metastasis (p=0.033), and tumor location (p=0.037). Meanwhile, miR-362 tended to exhibit a negative correlation with tumor size (p=0.072). However, there were no significant correlations between the expression of miR-362 and age (p=0.353) or sex (p=0.114) (Table 1).
miR-362 suppresses cell proliferation and invasion
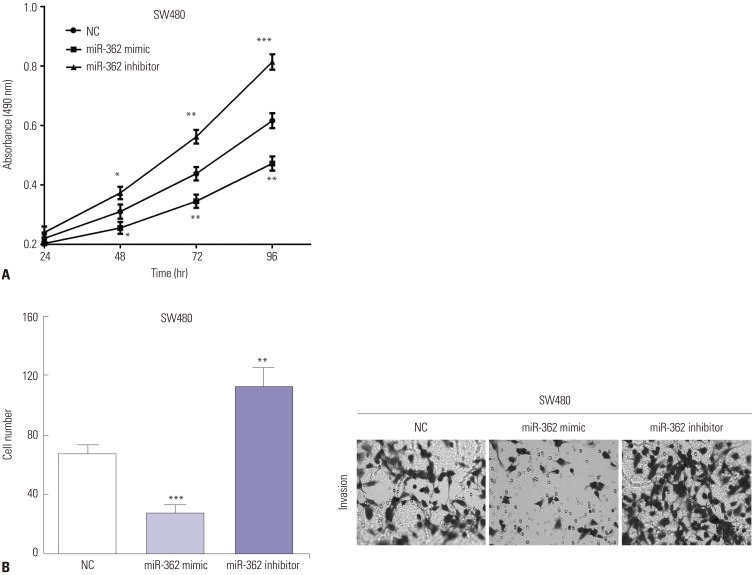
To elucidate the roles of miR-362 in CRC, MTT and transwell assays were performed to evaluate proliferative and invasive abilities. MTT data indicated that cell viability decreased (p=0.038, p=0.007, and p=0.002 of 48 h, 72 h, and 96 h) in miR-362 mimic-transfected SW480 cells, while the opposite was observed in miR-590 inhibitor-transfected cells (p=0.027, p=0.003, and p<0.001 of 48 h, 72 h, and 96 h) (Fig. 2A). Similar with the MTT results, transwell assay revealed that miR-362 mimic reduced (p<0.001) cell invasion, while miR-362 inhibitor promoted (p=0.004) invasive ability in SW480 cells (Fig. 2B).
Fig. 2. miR-362 suppressed cell proliferation and invasion. (A) The cell viability was regulated by exogenous changing miR-362 in SW480 cells. (B) Transwell assay revealed miR-362 mimic inhibits cell invasion, while miR-362 inhibitor promotes invasive ability in SW480 cells (0.5% crystal violet, ×200). *p<0.05, **p<0.01, ***p<0.001.
SIX1 upregulated in CRC and overexpression of SIX1 predicts poor prognosis
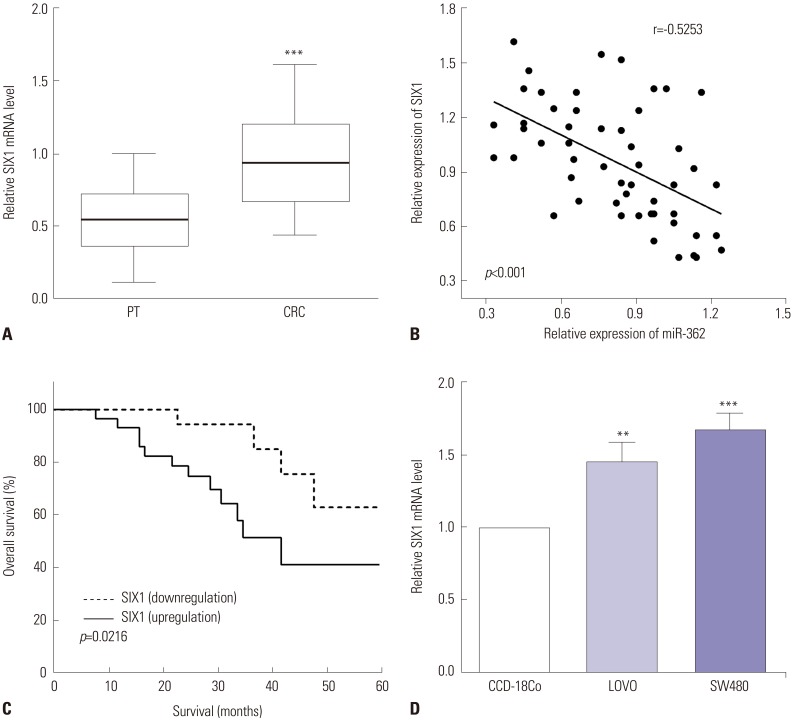
The expression of SIX1 was calculated by RT-qPCR in CRC tissues and cell lines. As expected, the expression of SIX1 showed significant increases in CRC tissues, compared to the paracancerous tissues (p<0.001) (Fig. 3A). Therefore, the associations between the expression of miR-362 and SIX1 were assessed, and we discovered that miR-362 had a negative relationship with the expression of SIX1 in CRC tissues (r=−0.525, p<0.001) (Fig. 3B). Moreover, Kaplan-Meier analysis revealed that upregulation of SIX1 predicts poor 5-year overall survival in CRC patients (p=0.021) (Fig. 3C). In addition, the expression of SIX1 in CRC cells LOVO and SW480 and normal colon cell CCD-18Co was evaluated by RT-qPCR. We discovered that the expression of SIX1 was higher in LOVO and SW480 cells than in CCD-18Co cells (p=0.004 and p<0.001) (Fig. 3D).
Fig. 3. SIX1 was upregulated in CRC, and overexpression of SIX1 predicted poor prognosis. (A) The expression of SIX1 reflected significant increases in CRC tissues, compared to paracancerous tissues (PT). (B) miR-362 exhibited negative correlations with the expression of SIX1 in CRC tissues. (C) Upregulation of SIX1 predicted poor 5-year overall survival in CRC patients. (D) The expression of SIX1 was higher in LOVO and SW480 cells, compared with CCD-18Co cells. **p<0.01, ***p<0.001. CRC, colorectal cancer.
SIX1 is a target of miR-362 and expression of SIX1 is mediated by miR-362
TargetScan was employed to predict the potential target genes of miR-362, and SIX1 was discovered to be a direct target of miR-362, showing putative binding sequences. To investigate whether miR-362 directly binds to the 3′-UTR of SIX1 mRNA, the putative binding sequences on SIX1 mRNA were mutated, as shown in Fig. 4A. Luciferase reporter assay was applied to identify the prediction, and the luciferase ability of the wild type of SIX1 3′-UTR was notably decreased (p=0.002) by miR-362 mimic, but had no effect (p=0.800) on the mutant type of SIX1 3′-UTR (Fig. 4B). Moreover, after transfecting SW480 cells with miR-362 mimic or miR-362 inhibitor, we evaluated the expression of SIX1, the results of which indicated that the mRNA levels of SIX1 were reduced in miR-362 mimic-transfected cells (p=0.005). On the contrary, SIX1 mRNA levels were increased when miR-362 was knocked down in SW480 cells (p=0.002) (Fig. 4C).
Fig. 4. SIX1 was a target of miR-362, and the expression of SIX1 was mediated by miR-362. (A) TargetScan was employed to predict SIX1 as a potential target gene of miR-362. (B) Luciferase reporter assay was applied to determine whether miR-362 targets SIX1. (C) miR-362 regulated the expression of SIX1. #p>0.05, **p<0.01, ***p<0.001.
Knockdown of SIX1 inhibits the proliferation and invasion of SW480 cells
To explore the functions of SIX1 in CRC, the siRNA-SIX1 was utilized to knockdown SIX1 in SW480 cells detected by RT-qPCR (p<0.01) (Fig. 5A). MTT and transwell assays were performed to calculate the proliferative and invasive abilities after knockdown of SIX1. In comparison with siRNA-NC, MTT data indicated that cell viability was decreased by transfecting siRNA-SIX1 in SW480 cells (p<0.01) (Fig. 5B). Similarly, transwell assay revealed that siRNA-SIX1 reduces (p<0.01) the invasive ability of SW480 cells (Fig. 5C).
Fig. 5. Knockdown of SIX1 inhibited the proliferation and invasion of SW480 cells. (A) The siRNA-SIX1 was conducted to knockdown SIX1 in SW480 cells. (B) In comparison with siRNA-NC, the cell viability was decreased by transfecting siRNA-SIX1 in SW480 cells. (C) Transwell assay revealed that siRNA-SIX1 reduces the invasive ability in SW480 cells (0.5% crystal violet, ×200). **p<0.01.
SIX1 could reverse the effects of miR-362 on cell proliferation and invasion
In an attempt to further explore the biological mechanisms of miR-362 in cell proliferation and invasion, rescue experiment was performed to verify the results above. pcDNA3.1-SIX1 was utilized to re-express SIX1 in miR-362 mimic-transfected SW480 cells, and the transfection efficiency was detected by RT-qPCR, as shown in Fig. 6A (p=0.011). The proliferative and invasive capacities were assessed by MTT or transwell assays. As expected, the proliferative ability was increased by re-expressing SIX1 in miR-362 mimic transfected SW480 cells (p=0.045) (Fig. 6B). Similar with the results of MTT, the transwell assay results demonstrated that the invasive ability was enhanced by transfection of pcDNA3.1-SIX1 in SW480 cells in which miR-362 was overexpressed (p=0.030) (Fig. 6C).
Fig. 6. SIX1 could reverse partial roles of miR-362 on cell proliferation and invasion. (A) RT-qPCR detected the transfection efficiency after re-expressed SIX1 in miR-362 mimic-transfected SW480 cells. Alterations of the proliferative (B) and invasive (C) abilities after re-expressing SIX1 in miR-362 mimic transfected SW480 cells (0.5% crystal violet, ×200). *p<0.05, **p<0.01, ***p<0.001.
DISCUSSION
CRC is a frequent malignant tumor with high metastasis and recurrence, and rates thereof are increasing every year.22,23 Therefore, identifying tumor molecular markers for metastasis and recurrence is essential in the diagnosis of CRC patients. miRNAs have been reported to act as a tumor promoter or suppressor in various cancers.24,25 Zou, et al.26 revealed that miR-362 functions as a tumor suppressor by reducing cell proliferation, migration, invasion, and cell cycle and by enhancing cell apoptosis in renal cancer. Similarly, Wu, et al.27 indicated that miR-362 suppresses the proliferation and migration of neuroblastoma cells. Consistent with these findings, we discovered that miR-362 was downregulated in CRC tissues and cell lines. Moreover, the CRC patients in the low miR-362 group showed poorer overall survival than patients in the high miR-362 group, consistent with the findings of Kheirollahi, et al.28 in glioma. Ni, et al.29 revealed that miR-362 repressed proliferation and metastasis in breast cancer. Consistent with the findings mentioned above, we deemed that overexpression of miR-362 suppressed proliferation and invasion in the CRC cell line SW480. In reverse, the cell proliferative and invasive capacities were enhanced by miR-362 inhibitor.
SIX1 was reported to enhance the proliferation and migration of osteosarcoma cells.30 Similarly, SIX1 was found to enhance cell proliferation and tumorigenesis through PI3K/AKT pathway in osteosarcoma.31 Moreover, Sun, et al.32 indicated that SIX1 regulated cell motility, tumor metastasis, and EMT in cervical cancer. Consistent with the findings above, we revealed that SIX1 was upregulated in CRC tissues and cell lines. Also, upregulation of SIX1 predicted shorter overall survival than SIX1 downregulation, which was consistent with previously reported findings in pancreatic ductal adenocarcinoma, gastric adenocarcinoma, hepatocellular carcinoma, and esophageal squamous cell carcinoma.33,34,35,36 In addition, SIX1 was a target gene of several miRNAs, including miR-204, miR-30a, miR-188 and miR-27a.37,38,39,40 We proposed that miR-362 targets and regulates the expression of SIX1. Moreover, knockdown of SIX1 had the same effect on the proliferation and migration as transfection of miR-362 mimic. SIX1 reversed the functions of miR-362 on cell proliferation and invasion in CRC.
In conclusion, miR-362 was significantly decreased in CRC tissues and cell lines, compared to normal tissues and cells. We confirmed a significant association between the overall survival of 53 CRC patients and low expression of miR-362. Downregulation of miR-362 inhibited cell proliferation and invasion through SIX1 in CRC. Additionally, we also discovered that SIX1 was a direct target gene of miR-362 and that the expression of miR-362 had negative correlations with SIX1 in CRC. SIX1 could partially reverse the functions of miR-362 on proliferation and invasion in CRC cells.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Gang Zhao.
- Data curation: Jin'e Wan and Jian Yang.
- Formal analysis: Jian Yang and Cuixia Qiao.
- Funding acquisition: Gang Zhao.
- Investigation: Xiaomei Sun.
- Methodology: Jin'e Wan and Aiting Di.
- Project administration: Gang Zhao.
- Resources: Gang Zhao.
- Software: Lize Zhang.
- Supervision: Cuixia Qiao and Dandan Wang.
- Validation: Jian Yang.
- Visualization: Gang Zhao.
- Writing—original draft: Jin'e Wan.
- Writing—review & editing: Gang Zhao and Cuixia Qiao.
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Shimada H, Tanaka K, Endou I, Ichikawa Y. Treatment for colorectal liver metastases: a review. Langenbecks Arch Surg. 2009;394:973–983. doi: 10.1007/s00423-009-0530-8. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju J. Implications of miRNAs in colorectal cancer chemoresistance. Int Drug Discov. 2011;2011:2063. [PMC free article] [PubMed] [Google Scholar]
- 7.Ke SB, Qiu H, Chen JM, Shi W, Chen YS. MicroRNA-202-5p functions as a tumor suppressor in colorectal carcinoma by directly targeting SMARCC1. Gene. 2018;676:329–335. doi: 10.1016/j.gene.2018.08.064. [DOI] [PubMed] [Google Scholar]
- 8.Chen E, Li Q, Wang H, Zhang P, Zhao X, Yang F, et al. MiR-32 promotes tumorigenesis of colorectal cancer by targeting BMP5. Biomed Pharmacother. 2018;106:1046–1051. doi: 10.1016/j.biopha.2018.07.050. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Wang H, Li Y, Li Q. MiR-362-3p functions as a tumor suppressor through targeting MCM5 in cervical adenocarcinoma. Biosci Rep. 2018;38:BSR20180668. doi: 10.1042/BSR20180668. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Yang P, Ni F, Deng RQ, Qiang G, Zhao H, Yang MZ, et al. MiR-362-5p promotes the malignancy of chronic myelocytic leukaemia via down-regulation of GADD45α. Mol Cancer. 2015;14:190. doi: 10.1186/s12943-015-0465-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, et al. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-κB signaling. J Transl Med. 2014;12:33. doi: 10.1186/1479-5876-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N, Feng Y, Xu J, Zou J, Chen M, He Y, et al. miR-362-3p regulates cell proliferation, migration and invasion of trophoblastic cells under hypoxia through targeting Pax3. Biomed Pharmacother. 2018;99:462–468. doi: 10.1016/j.biopha.2018.01.089. [DOI] [PubMed] [Google Scholar]
- 13.Christensen LL, Tobiasen H, Holm A, Schepeler T, Ostenfeld MS, Thorsen K, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer. 2013;133:67–78. doi: 10.1002/ijc.28010. [DOI] [PubMed] [Google Scholar]
- 14.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 15.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu W, Ren Z, Liu H, Wang L, Huang R, Chen J, et al. Core promoter analysis of porcine Six1 gene and its regulation of the promoter activity by CpG methylation. Gene. 2013;529:238–244. doi: 10.1016/j.gene.2013.07.102. [DOI] [PubMed] [Google Scholar]
- 17.Micalizzi DS, Wang CA, Farabaugh SM, Schiemann WP, Ford HL. Homeoprotein Six1 increases TGF-beta type I receptor and converts TGF-beta signaling from suppressive to supportive for tumor growth. Cancer Res. 2010;70:10371–10380. doi: 10.1158/0008-5472.CAN-10-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Q, Ning D, Chen J, Li X, Chen XP, Jiang L. SIX1 and DACH1 influence the proliferation and apoptosis of hepatocellular carcinoma through regulating p53. Cancer Biol Ther. 2018;19:381–390. doi: 10.1080/15384047.2018.1423920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin X, Li Y, Yang X. SIX1 is overexpressed in endometrial carcinoma and promotes the malignant behavior of cancer cells through ERK and AKT signaling. Oncol Lett. 2016;12:3435–3440. doi: 10.3892/ol.2016.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerbs T, Bisht S, Schölch S, Pecqueux M, Kristiansen G, Schneider M, et al. Inhibition of Six1 affects tumour invasion and the expression of cancer stem cell markers in pancreatic cancer. BMC Cancer. 2017;17:249. doi: 10.1186/s12885-017-3225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Xu R. Six1 expression is associated with a poor prognosis in patients with glioma. Oncol Lett. 2017;13:1293–1298. doi: 10.3892/ol.2017.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou X, Zhong J, Li J, Su Z, Chen Y, Deng W, et al. miR-362-3p targets nemo-like kinase and functions as a tumor suppressor in renal cancer cells. Mol Med Rep. 2016;13:994–1002. doi: 10.3892/mmr.2015.4632. [DOI] [PubMed] [Google Scholar]
- 27.Wu K, Yang L, Chen J, Zhao H, Wang J, Xu S, et al. miR-362-5p inhibits proliferation and migration of neuroblastoma cells by targeting phosphatidylinositol 3-kinase-C2β. FEBS Lett. 2015;589:1911–1919. doi: 10.1016/j.febslet.2015.05.056. [DOI] [PubMed] [Google Scholar]
- 28.Kheirollahi M, Moodi M, Ashouri S, Nikpour P, Kazemi M. Evaluation of miR-362 expression in astrocytoma of human brain tumors. Adv Biomed Res. 2017;6:129. doi: 10.4103/2277-9175.216782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni F, Gui Z, Guo Q, Hu Z, Wang X, Chen D, et al. Downregulation of miR-362-5p inhibits proliferation, migration and invasion of human breast cancer MCF7 cells. Oncol Lett. 2016;11:1155–1160. doi: 10.3892/ol.2015.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua L, Fan L, Aichun W, Yongjin Z, Qingqing C, Xiaojian W. Inhibition of Six1 promotes apoptosis, suppresses proliferation, and migration of osteosarcoma cells. Tumour Biol. 2014;35:1925–1931. doi: 10.1007/s13277-013-1258-1. [DOI] [PubMed] [Google Scholar]
- 31.Yu C, Zhang B, Li YL, Yu XR. SIX1 reduces the expression of PTEN via activating PI3K/AKT signal to promote cell proliferation and tumorigenesis in osteosarcoma. Biomed Pharmacother. 2018;105:10–17. doi: 10.1016/j.biopha.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 32.Sun SH, Liu D, Deng YT, Zhang XX, Wan DY, Xi BX, et al. SIX1 coordinates with TGFβ signals to induce epithelial-mesenchymal transition in cervical cancer. Oncol Lett. 2016;12:1271–1278. doi: 10.3892/ol.2016.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin A, Xu Y, Liu S, Jin T, Li Z, Jin H, et al. Sineoculis homeobox homolog 1 protein overexpression as an independent biomarker for pancreatic ductal adenocarcinoma. Exp Mol Pathol. 2014;96:54–60. doi: 10.1016/j.yexmp.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Lv H, Cui A, Sun F, Zhang Y, Li Y, Li L, et al. Sineoculis homeobox homolog 1 protein as an independent biomarker for gastric adenocarcinoma. Exp Mol Pathol. 2014;97:74–80. doi: 10.1016/j.yexmp.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Kong J, Zhou X, Liu S, Jin T, Piao Y, Liu C, et al. Overexpression of sineoculis homeobox homolog 1 predicts poor prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:3018–3027. [PMC free article] [PubMed] [Google Scholar]
- 36.He Z, Li G, Tang L, Li Y. SIX1 overexpression predicts poor prognosis and induces radioresistance through AKT signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:1071–1079. doi: 10.2147/OTT.S125330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Y, Zhu Y, Ma T, Pan C, Wang J, He Z, et al. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588:3703–3712. doi: 10.1016/j.febslet.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien JH, Hernandez-Lagunas L, Artinger KB, Ford HL. MicroRNA-30a regulates zebrafish myogenesis through targeting the transcription factor Six1. J Cell Sci. 2014;127(Pt 10):2291–2301. doi: 10.1242/jcs.143677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Liu H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumour Biol. 2016;37:4105–4113. doi: 10.1007/s13277-015-4246-9. [DOI] [PubMed] [Google Scholar]
- 40.Towers CG, Guarnieri AL, Micalizzi DS, Harrell JC, Gillen AE, Kim J, et al. The Six1 oncoprotein downregulates p53 via concomitant regulation of RPL26 and microRNA-27a-3p. Nat Commun. 2015;6:10077. doi: 10.1038/ncomms10077. [DOI] [PMC free article] [PubMed] [Google Scholar]