Abstract
The anti-hyperglycemic effect of metformin is believed to be caused by its direct action on signaling processes in hepatocytes, leading to lower hepatic gluconeogenesis. Recently, metformin was reported to alter the gut microbiota community in humans, suggesting that the hyperglycemia-lowering action of the drug could be the result of modulating the population of gut microbiota. However, the critical microbial signaling metabolites and the host targets associated with the metabolic benefits of metformin remained elusive. Here, we performed metagenomic and metabolomic analysis of samples from individuals with newly diagnosed type 2 diabetes (T2D) naively treated with metformin for 3 d, which revealed that Bacteroides fragilis was decreased and the bile acid glycoursodeoxycholic acid (GUDCA) was increased in the gut. These changes were accompanied by inhibition of intestinal farnesoid X receptor (FXR) signaling. We further found that high-fat-diet (HFD)-fed mice colonized with B. fragilis were predisposed to more severe glucose intolerance, and the metabolic benefits of metformin treatment on glucose intolerance were abrogated. GUDCA was further identified as an intestinal FXR antagonist that improved various metabolic endpoints in mice with established obesity. Thus, we conclude that metformin acts in part through a B. fragilis–GUDCA–intestinal FXR axis to improve metabolic dysfunction, including hyperglycemia.
Metformin has been the first-line antidiabetic drug for more than 60 years because of its distinct glucose-lowering effect and safety profile. The mechanism by which metformin reduces glucose production has received considerable attention, yet controversy still exists. The prevailing view is that metformin acts directly on hepatocytes to decrease gluconeogenesis in part through modulation of mitochondrial complex I activity, AMP-activated protein kinase (AMPK) activation and AMP concentration1–3. Metformin was also shown to enhance hepatic cytosolic redox state by inhibiting glycerol-3-phosphate dehydrogenase activity to decrease gluconeogenesis4,5. However, metformin is an orally administered drug that reaches high concentrations in the small intestine with much lower serum concentrations6; thus, the possibility cannot be excluded that its metabolic benefits might be due in part to actions in the intestine. In the gut, metformin could act on the gut microbiota population. The gut microbiota influences development of metabolic diseases, including T2D and non-alcoholic fatty liver disease (NAFLD)7,8, and metformin was recently shown to alter the gut microbiome of individuals with T2D9,10. Transfer of fecal samples from metformin-treated donors to germ-free mice phenocopied metformin treatment, and this effect was thought to be partially due to altered metal homeostasis10. Some studies also suggest that metformin treatment improves glucose homeostasis in obese mice and rats by modulating the gut microbiota11–14. However, which species of the gut microbiota were affected by metformin and the underlying mechanism of how changes in the gut microbiota regulates host glucose metabolism is not clear.
The gut microbiota influences host metabolism via the modulation of metabolites, including the endotoxin LPS, bile acids, trimethylamine N-oxide, and short chain fatty acids, in part by mediating the interaction between the gastrointestinal system and other organs15. The primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA) in humans and α/β-muricholic acid (α/βMCA) in mice, are synthesized from cholesterol in the liver and secreted as glycine and taurine conjugates into the intestine, respectively. In the gut, primary bile acids are transformed into secondary bile acids by the metabolic activities of enteric anaerobic bacteria16. Bile acids can alter metabolism by binding to nuclear receptors, including FXR (encoded by FXR, official symbol: NR1H4), pregnane X receptor and vitamin D receptor, as well as to the Takeda G-protein-coupled receptor 5 (TGR5)16,17. Recent reports revealed that gut microbiota-mediated deconjugation of bile acids influences hepatic de novo bile acid synthesis, dependent on the FXR-fibroblast growth factor (FGF)15 axis in mice18–20.
FXR is a ligand-activated nuclear receptor that regulates hepatic bile acid biosynthesis, transport and secretion and is involved in multiple metabolic diseases16. Bile acids such as CA and CDCA are agonists of FXR, and activation of the FXR–small heterodimer partner (SHP, encoded by SHP, official symbol: NR0B2) axis in the liver inhibits the sterol 12 α-hydroxylase (CYP8B1) expression and bile acid synthesis20. In contrast, the conjugated bile acid, tauro-β-muricholic acid (TβMCA), is a natural FXR antagonist in mice21–23. Furthermore, inhibition of intestinal FXR prevented obesity-related metabolic dysfunction in mice23–26. However, human and mouse bile acid profiles are known to differ, most notably due to the synthesis of MCA in mice and the predominance of taurine conjugation in mice and glycine conjugation in humans23. Because humans do not produce TβMCA, it remains unclear whether there is a natural bile acid that has FXR antagonist activity in humans.
The current study revealed that metformin treatment increased the levels of the bile acid glycoursodeoxycholic acid (GUDCA) in the gut by decreasing the abundance of species of B. fragilis and its bile salt hydrolase (BSH) activity in the intestines of individuals with T2D, as revealed by metagenomics sequencing analysis and metabolomics analysis. Furthermore, we identified GUDCA as a new endogenous antagonist of intestinal FXR. These results suggested that the metformin-induced improvement of metabolism is mediated by a B. fragilis–GUDCA–intestinal FXR axis in humans. Moreover, oral GUDCA supplementation or a synthetic FXR antagonist may be of potential translational value in the clinical treatment of T2D.
Results
Oral metformin modulates gut microbiota and bile acid metabolism in individuals with T2D
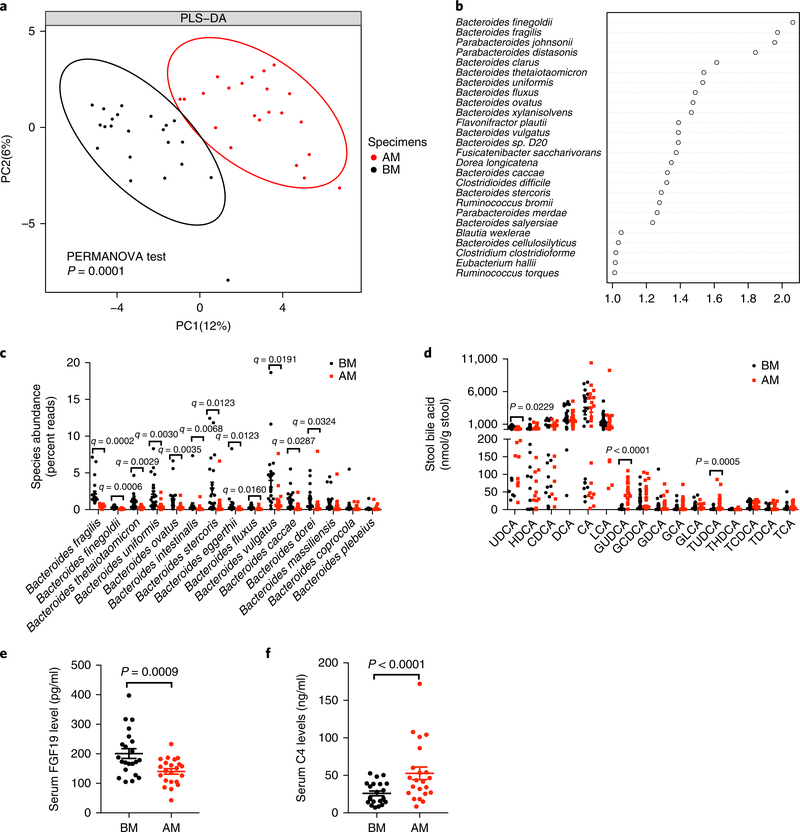
To investigate how oral metformin controls gut microbiota and metabolites in humans, we collected serum and stool samples from 22 individuals with T2D before and after being naively treated with 1,000 mg metformin twice daily (b.i.d) for 3 d. We performed whole-genome shotgun sequencing of stool samples, and association studies of metagenomics profiling and clinical indicators provided in-depth insight into the function of specific gut microbiota on host metabolism. Although whole-gene counts were not changed, the Shannon diversity index (α-diversity) of gut microbiota was slightly decreased after metformin treatment (Supplementary Fig. 1a,b). The plot from the partial least squares discriminant analysis (PLS-DA) showed that the gut microbiota composition was substantially reshaped after metformin treatment (Fig. 1a). The variable importance in projection (VIP) score of the gut microbiota indicated that the species Bacteroides finegoldii and B. fragilis were the top gut microbiota that resulted in the group separation (Fig. 1b). Metagenomic analysis further revealed that the genus Bacteroides showed the largest reduction in abundance (Supplementary Fig. 1c). Among the genus Bacteroides, the abundances of multiple species were reduced after metformin treatment, with the decrease in level of B. fragilis exhibiting the most striking change (Fig. 1c). Clinical metformin treatment was found to be related to the modulation of bile acid profiles in individuals with T2D10. We adopted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to suggest that Blautia obeum and B. fragilis were the top changed bacterial species on the pathway of secondary bile acid metabolism regulated by metformin (Supplementary Fig. 1d). We further used ultra-performance liquid chromatography–coupled time-of-flight mass spectrometry (UPLC-ESI-QTOFMS) metabolite profiling to quantitate bile acid levels in the serum and stool. The levels of GUDCA and tauroursodeoxycholic acid (TUDCA), conjugated secondary bile acids in humans, were predominantly elevated after metformin treatment (Fig. 1d and Supplementary Fig. 1e). Total bile acid levels remained unchanged, whereas noticeable elevation in ratios of conjugated to unconjugated bile acids was observed (Supplementary Fig. 1f–i). There was no difference in the ratio of 12α-OH to non-12α-OH bile acids (Supplementary Fig. 1j,k). These results suggest that oral metformin treatment regulates the gut microbiota and bile acid metabolism in individuals with T2D.
Fig. 1 |. Oral metformin modulates the composition of gut microbiota and bile acids in individuals with T2D.
a, Partial least-square discriminant analysis (PLS-DA). The BM (before the metformin treatment) group is shown in black, and the AM (after 3-d metformin treatment) group is shown in red. PC1 and PC2 account for 12% and 6%, respectively, of the total variance. PERMANOVA with the Bray–Curtis distance was used to assess the significant difference between the two groups, and the result showed significant separation of the BM and AM groups (P = 0.0001). n = 22 individuals/group. b, VIP scores of PLS-DA. VIP scores were used to rank the ability of different taxa to discriminate between BM and AM groups. A taxon with VIP score >1 was considered important in the discrimination. n = 22 individuals/group. c, Different species abundance (percent reads) of Bacteriodes based on metagenomics data. n = 22 individuals/group. q value (FDR-adjusted P value), P value determined by two-tailed Wilcoxon matched-pairs signed rank test. d, Bile acid levels in the stool. n = 22 individuals/group. P value was determined by two-tailed Wilcoxon matched-pairs signed rank test. e,f, Serum FGF19 (e) and C4 (f) levels. n = 22 individuals/group. P value was determined by two-tailed Wilcoxon matched-pairs signed rank test. All the data are presented as the mean ± s.e.m.
GUDCA is an FXR antagonist
Bile acids and their metabolites are important bioactive ligands that interact with receptors in the gut to regulate metabolic processes19,23. In the distal ileum, FXR is activated by bile acids and induces expression of the FGF19 gene in humans (Fgf15 in mice). FGF19 levels in serum of individuals with T2D were dramatically decreased, and 7α-hydroxy-4-cholesten-3-one (C4) levels were markedly increased after metformin treatment, suggesting that intestinal FXR signaling was suppressed and hepatic CYP7A1 activity was increased after metformin treatment (Fig. 1e, f).
Previously, in silico molecular docking studies of the FXR antagonist glycine-muricholic acid (GMCA) were carried out25. GMCA docked into the FXR ligand binding domain, establishing hydrogen bonds with some of the residues involved in noncovalent interactions with the FXR agonist CDCA25. GW4064, GMCA, TUDCA and GUDCA were docked into the human FXR ligand binding domain (PDB 3DCT, R4). GMCA, TUDCA and GUDCA docked with a much lower score of − 15.68 (GMCA), − 18.29 (TUDCA) and − 13.83 (GUDCA), and with a similar pose surrounded by residues Arg331, Met265, His447 and Tyr369 (Supplementary Fig. 2a,b). GW4064 docked with a score of − 55, reproducing the crystallographic binding pattern (PDB 3DCT, Supplementary Fig. 2c)27. Similar to GW4064, TUDCA and GUDCA establish a salt bridge and a hydrogen bond interaction between the acidic groups and the side chain of Arg331 and backbone NH of Met265 (Supplementary Fig. 2c–e). Additionally, the 7β-OHs of TUDCA and GUDCA do not interact with the side chain of Tyr369.
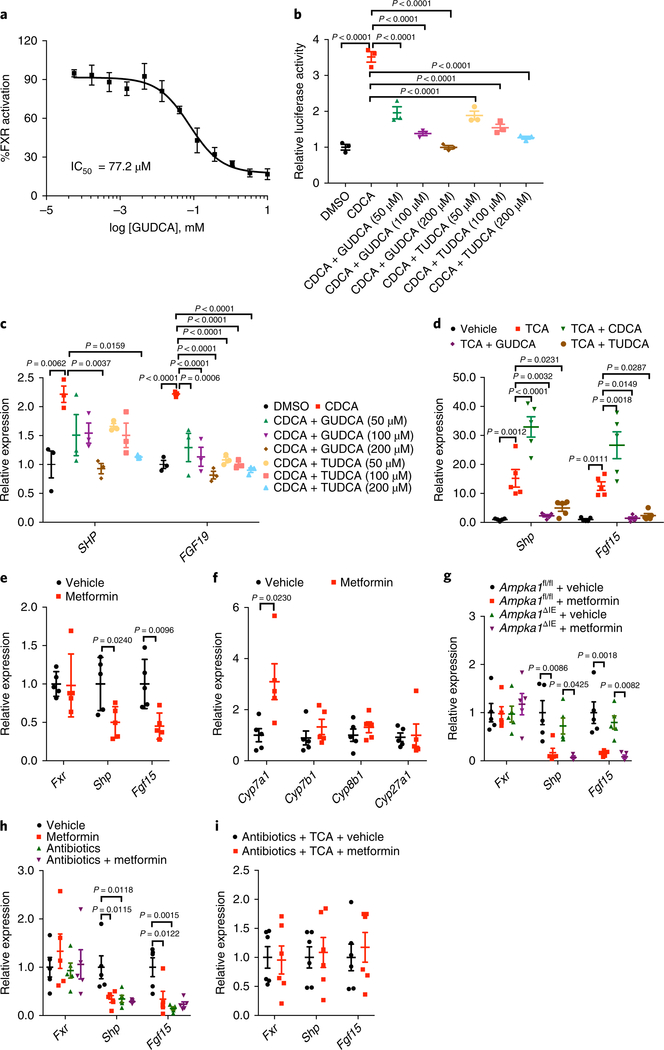
To assess whether GUDCA and TUDCA were direct FXR antagonists, we applied the TR-FRET FXR coactivator assay. Neither GUDCA nor TUDCA showed agonistic action with CDCA as a positive control (EC50 = 11.1 μM, Supplementary Fig. 2f). In the presence of CDCA, we found that GUDCA (IC50 = 77.2 μM) and TUDCA (IC50 = 75.1 μM) were FXR antagonists with comparable IC50s to that of TβMCA (IC50 = 83.3 μM, Fig. 2a and Supplementary Fig. 2g,h). Luciferase reporter gene assays revealed that both GUDCA and TUDCA markedly inhibited CDCA-induced FXR transcriptional activity (Fig. 2b). In the human intestinal Caco-2 cell line, induction of the FXR target genes SHP and FGF19 mRNAs by CDCA were substantially suppressed by GUDCA and TUDCA (Fig. 2c). Furthermore, luciferase reporter gene assays demonstrated that GUDCA had no effects on TGR5 activity (Supplementary Fig. 2i). To examine the inhibition of FXR signaling by GUDCA and TUDCA in vivo, antibiotic-treated, microbiota-depleted mice were given TCA (an FXR agonist) combined with CDCA, GUDCA and TUDCA by gavage. We found that CDCA further elevated TCA-induced intestinal FXR signaling, whereas GUDCA and TUDCA attenuated TCA-activated intestinal FXR signaling (Fig. 2d). These results indicated that GUDCA and TUDCA are potential FXR antagonists in vitro and in vivo.
Fig. 2 |. GuDCA and tuDCA are identified FXR antagonists.
a, TR-FRET FXR coactivator recruitment assay to assess whether GUDCA is a FXR antagonist in the presence of CDCA (20 μM). n = 4 replicates/treatment. b, Luciferase activity of control (DMSO), different concentrations of GUDCA or TUDCA without or with FXR agonist CDCA treatment. n = 3 replicates/treatment. P value was determined by one-way ANOVA with Tukey’s correction (F7,16 = 61.69). c, SHP and FGF19 mRNA expression in differentiated Caco-2 cells after treatment with different concentrations of GUDCA or TUDCA with CDCA. n = 3 replicates/treatment. P value was determined by one-way ANOVA with Tukey’s correction (SHP: F7,16 = 4.871, FGF19: F7,16 = 15.41). d, Relative expression of the target gene mRNAs of intestinal FXR in mice treated with vehicle, TCA or TCA and CDCA, GUDCA or TUDCA. n = 5 mice/group. P value was determined by one-way ANOVA with Tukey’s correction (Shp: F4,20 = 38.47, Fgf15: F4,20 = 25.02). e, Relative expression of intestinal Fxr mRNA and its target gene mRNAs in mice treated with metformin on a HFD for 1 week. n = 5 mice/group. P value was determined by two-tailed Student’s t-test (Shp: t8 = 2.779, Fgf15: t8 = 3.383). f, The relative expression of Cyp7a1, Cyp7b1, Cyp8b1 and Cyp27a1 mRNAs in the livers of metformin-treated mice after 1-week HFD feeding. n = 5 mice/group. P value was determined by two-tailed Student’s t-test (Cyp7a1: t8 = 2.805). g, Relative abundance of intestinal Fxr mRNA and its target gene mRNAs in metformin-treated Ampka1fl/fl and Ampka1ΔIE mice on a HFD for 1 week. n = 5 mice/group. P value was determined by one-way ANOVA with Tukey’s correction (Shp: F3,16 = 8.046, Fgf15: F3,16 = 11.83). h, Relative expression of intestinal Fxr mRNA and its target gene mRNAs in antibiotics-treated, microbiota-depleted mice treated with metformin on a HFD for 1 week. n = 5 mice/group. P value was determined by one-way ANOVA with Tukey’s correction (Shp: F3,16 = 6.859, Fgf15: F3,16 = 8.782). i, In microbiota-depleted mice, metformin had no effect in inhibiting the activation of intestinal FXR on top of TCA. The relative expression of Fxr and its target genes in the ileum. n = 6 mice/group. All the data are presented as the mean ± s.e.m.
Metformin inhibits intestinal FXR signaling via the gut microbiota in an AMPK-independent manner
The clinical therapeutic dose of metformin is 1,000 mg b.i.d (~25 mg/kg/d), similar to 225 mg/kg/d in mice after allometric scaling. Mice were given 1.5 mg/ml metformin in water, which was equivalent to 200 mg/kg/d, the safe and efficacious dose of metformin. Similar to GUDCA in humans, TβMCA and TUDCA levels in the ileum were substantially higher in metformin-treated mice than in vehicle-treated mice, whereas GUDCA was undetectable in mice (Supplementary Fig. 3a). Expression of FXR target gene mRNAs were markedly suppressed in the ileum of metformin-treated mice but remained unchanged in the liver (Fig. 2e and Supplementary Fig. 3b). Metformin treatment upregulated hepatic Cyp7a1 mRNA levels, resulting from downregulation of the intestinal FXR–FGF15 axis (Fig. 2f). As intestinal AMPK is a proposed target of metformin, we administered metformin (in water) to mice and saw no evidence of AMPKα activation in the intestine, whereas when metformin was administered using gavage, we observed a transient activation, but only with a high dose (Supplementary Fig. 3c,d). To further determine whether intestinal AMPK was involved in inhibition of intestinal FXR by metformin treatment, we detected AMPKα1 (encoded by Ampka1, official symbol: Prkaa1) and AMPKα2 (encoded by Ampka2, official symbol: Prkaa2) levels in the intestine. As AMPKα1 is the main form of intestinal AMPKα (Supplementary Fig. 3e,f), we adopted intestine-specific AMPKα1 knockout (Ampka1ΔIE) mice in which AMPKα1 expression is dramatically decreased in the intestine compared with that of the AMPKα1-floxed (Ampka1fl/fl) mice (Supplementary Fig. 3g). We found that in this mouse strain, metformin still attenuated intestinal FXR signaling (Fig. 2g and Supplementary Fig. 3h). To investigate whether the gut microbiota was involved in inhibition of intestinal FXR signaling by metformin, antibiotic-treated, microbiota-depleted mice were fed a HFD and given drinking water containing metformin. Metformin treatment suppressed intestinal FXR activation, but a further inhibition of FXR signaling was not observed in the group treated with metformin plus antibiotics compared with the group treated with antibiotics only (Fig. 2h and Supplementary Fig. 3i). Similar to the intestinal FXR levels, levels of TβMCA and TUDCA were substantially higher after treatment with metformin or antibiotics, but there was no further upregulation in the group treated with metaformin plus antibiotics compared with the group treated with antibiotics only (Supplementary Fig. 3j). Furthermore, in antibiotics-treated, microbiota-depleted mice, metformin did not inhibit TCA-activated intestinal FXR signaling, indicating that the inhibition of intestinal FXR signaling by metformin was dependent on the microbiota (Fig. 2i). These results revealed that inhibition of intestinal FXR signaling by metformin depends on gut microbiota but not intestinal AMPK.
B. fragilis is correlated with changes in bile acid metabolites and FXR signaling in metformin-treated individuals with T2D
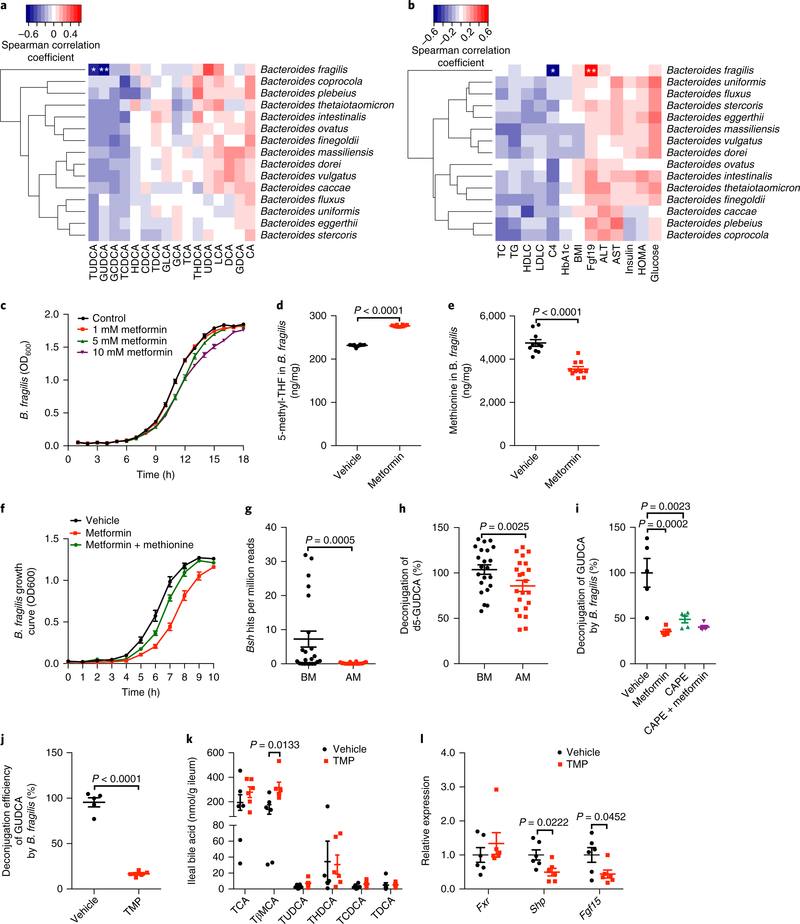
To identify the bacterium responsible for GUDCA and TUDCA metabolism in individuals with T2D, we evaluated the correlation between changes in the gut microbiome and bile acids. GUDCA and TUDCA levels in stool and serum samples were only negatively correlated with the presence of species B. fragilis (Fig. 3a and Supplementary Fig 4a–e). We further analyzed the correlation between changes in the gut microbiota and clinical biochemical indexes. The abundance of B. fragilis was positively correlated with serum FGF19 levels and was negatively correlated with serum C4 levels (Fig. 3b and Supplementary Fig 4f,g). Metformin was reported to influence the growth of bacteria in vitro28 and regulate microbial folate and methionine metabolism29. Metformin directly inhibited the growth of B. fragilis in a dose-dependent manner (Fig. 3c); more interestingly, KEGG pathway analysis of metagenomic data showed that B. fragilis was the most top-changed bacterial species on the pathway of folate synthesis and one-carbon pool production affected by metformin (Supplementary Fig. 4h,i). Metabolomic analysis revealed that the level of 5-methyl-tetrahydrofolate (5-methyl-THF), a methyl donor for homocysteine methylation to methionine, in metformin-treated B. fragilis, was higher than that in vehicle-treated B. fragilis, and the content of methionine in B. fragilis was lower after metformin treatment (Fig. 3d, e and Supplementary Fig. 4j,k). Supplementation of methionine restored metformin inhibition of B. fragilis growth in culture (Fig. 3f). BSH, which deconjugates conjugated bile acids, was purified and characterized from B. fragilis30. Indeed, B. fragilis mediated the deconjugation of GUDCA and TUDCA in vitro (Supplementary Fig. 4l,m). To clarify whether the accumulation of GUDCA and TUDCA was due to suppression of BSH activity by metformin, we performed further analysis of the metagenomic data. The B. fragilis Bsh gene copy number and BSH activity were dramatically reduced after metformin treatment (Fig. 3g, h). Metformin substantially diminished deconjugation of GUDCA in vehicle-treated B. fragilis, but not when treated with caffeic acid phenethyl ester (CAPE), a BSH inhibitor (Fig. 3i). Furthermore, the dihydrofolate reductase inhibitor trimethoprim (TMP) was found to inhibit B. fragilis–mediated hydrolysis of GUDCA (Fig. 3j). In the mice fed a HFD, supplementation with TMP mimicked metformin effects on bile acid metabolism and intestinal FXR signaling (Fig. 3k, l). These results suggested that metformin upregulates GUDCA levels via downregulation of BSH activity in B. fragilis, mainly resulting from disruption of B. fragilis folate and methionine metabolism. Thus, the modulation of B. fragilis levels by metformin could result in a change of bile acid composition and FXR signaling in the gut.
Fig. 3 |. Metformin-induced downregulation of B. fragilis abundance was negatively correlated with the modulation of bile acid profiles.
a, Heat map of the correlation between the species of Bacteroides and the bile acid levels in the stool of individuals with T2D in response to metformin treatment. b, Heat map of the correlation between the species of Bacteroides and biochemical indexes in individuals with T2D in response to metformin treatment. n = 22 individuals/group. Spearman’s rank test. *q < 0.05, **q < 0.01 versus BM, q value (FDR-adjusted P value). c, Growth curve of B. fragilis with or without metformin in a laboratory culture. n = 9 replicates/treatment. d,e, The content of 5-methyl-tetrahydrofolic acid (5-methyl-THF (d) and methionine (e) in B. fragilis treated with and without metformin or in a laboratory culture. n = 10 replicates/treatment. P value was determined by two-tailed Student’s t-test (d, t18 = 43.34; e, t18 = 6.249). f, Reconstitution with methionine restored the growth of B. fragilis in the presence of metformin in culture. n = 5 replicates/treatment. g, The abundance of Bsh gene of B. fragilis in individuals with T2D. n = 22 individuals/group. P value was determined by two-tailed Wilcoxon matched-pairs signed rank test. h, The hydrolysis efficiency of d5-GUDCA by stool samples from individuals with T2D before (BM) and after (AM) metformin treatment. n = 22 individuals/group. P value was determined by two-tailed Wilcoxon matched-pairs signed rank test. i, The hydrolysis efficiency of GUDCA mediated by vehicle- and metformin-treated B. fragilis with or without caffeic acid phenethyl ester (CAPE, BSH activity inhibitor) in culture. n = 5 replicates/treatment. P values were determined by one-way ANOVA with Tukey’s correction (F3,16 = 13). j, The hydrolysis efficiency of GUDCA mediated by B. fragilis with or without TMP in culture. n = 5 replicates/treatment. P value was determined by two-tailed Student’s t-test (t8 = 15.2). k,l, Bile acid levels (k) and the relative expression of Fxr and its target genes (l) in the ileum after 1 week of TMP treatment on a HFD. n = 6 mice/group. P values were determined by two-tailed Student’ t-test (TβMCA: t10 = 3, Shp: t10 = 2.703, Fgf15: t10 = 2.288). All data are presented as the mean ± s.e.m.
B. fragilis administration abrogates improvement of glucose metabolism by metformin
To uncover the effects of metformin-treated microbiota on host glucose metabolism, we transplanted stool obtained from four individuals with T2D before (trans-BM) and after (trans-AM) metformin treatment to antibiotic-treated, microbiota-depleted mice fed a HFD. After stool transplantation, the abundance of B. fragilis in the trans-AM mice was much lower than that in trans-BM mice (Supplementary Fig. 5a). We observed a slight body weight loss in the trans-AM mice compared with the trans-BM mice (Supplementary Fig. 5b). Glucose intolerance and insulin resistance were substantially improved in mice colonized with gut microbiota after metformin treatment (Supplementary Fig. 5c–h). Stool transplantation altered bile acid metabolism in mice, as the levels of intestinal TβMCA and TUDCA were higher, and intestinal FXR signaling was substantially suppressed in trans-AM mice compared with trans-BM mice (Supplementary Fig. 5i–k). These observations showed that the beneficial effects of metformin could be transmitted by stool transplantation.
We found that colonization of B. fragilis resulted in higher body weight gain, impaired glucose tolerance, and lower insulin sensitivity compared with control, heat-killed B. fragilis, on a HFD (Supplementary Fig. 6a–g). TβMCA and TUDCA levels in the ileum were substantially reduced, and intestinal FXR signaling was greatly activated after B. fragilis colonization (Supplementary Fig. 6h–j). It is well established that inhibition of intestinal FXR ameliorates obesity-related metabolic diseases via white adipose tissue browning25. Expression of thermogenic gene mRNAs, including Ucp1, Elovl3, and Pgc1a (official symbol: Ppargc1a), was markedly downregulated in subcutaneous white adipose tissue (sWAT) but not in epididymal white adipose tissue (eWAT) or brown adipose tissue (BAT), after B. fragilis treatment (Supplementary Fig. 6k–m).
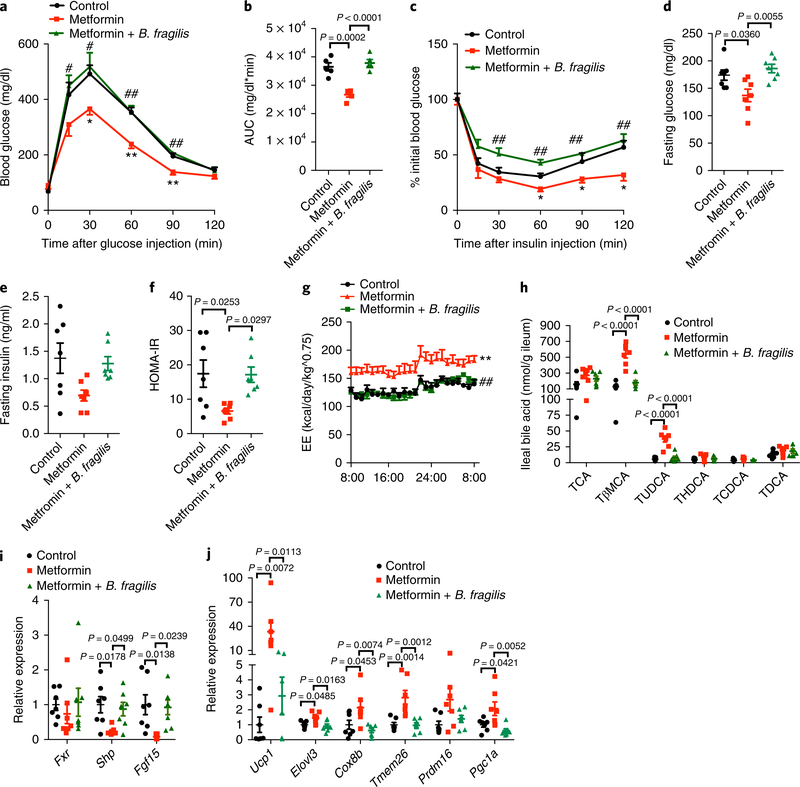
To further clarify whether B. fragilis participated in metformin-induced improvements of metabolic diseases, we gave B. fragilis to metformin-treated mice on a HFD using gavage for 4 weeks. The reduced body weight gain caused by metformin treatment was eliminated after B. fragilis treatment (Supplementary Fig. 7a). B. fragilis treatment impaired improvements in glucose tolerance and insulin sensitivity by metformin (Fig. 4a–f). The enhanced energy expenditure by metformin was substantially diminished by B. fragilis treatment (Fig. 4g); however, food intake was not affected (Supplementary Fig. 7b). B. fragilis administration markedly abrogated up-regulation of Tβ MCA and TUDCA levels and decrease of intestinal FXR signaling in metformin-treated mice (Fig. 4h, i and Supplementary Fig. 7c). The enhanced levels of thermogenic gene mRNAs in sWAT of metformin-treated mice was blunted by B. fragilis (Fig. 4j), but not in eWAT or BAT (Supplementary Fig. 7d,e). To further explore the vital role of B. fragilis BSH in metformin’s action on glucose metabolism, mice fed a HFD were treated with metformin with or without CAPE upon B. fragilis colonization after microbiota elimination by antibiotics. Metformin substantially ameliorated B. fragilis colonization–induced metabolic dysfunction, although no further improvements were seen in mice treated with metformin plus CAPE compared with mice treated with CAPE only (Supplementary Fig. 8a–g). Metformin-treated mice that were colonized with B. fragilis displayed a higher metabolic rate than control; however, CAPE-treated mice were unresponsive to metformin in this regard (Supplementary Fig. 8h,i). Metformin noticeably inhibited intestinal FXR signaling in mice that were colonized with B. fragilis, but the drug had no further effects in CAPE-treated mice (Supplementary Fig. 8j,k). These results revealed that reduction of B. fragilis abundance and its BSH activity contributes to the improvements of glucose intolerance by metformin.
Fig. 4 |. B. fragilis reverses the metabolic improvements of metformin.
Mice were divided to three groups (control, metformin and metformin plus B. fragilis) under HFD treatment for 4 weeks to explore the reversal effects. Mice in the control and metformin groups were given the same dose of heatkilled B. fragilis. a,b, Glucose tolerance test (GTT, a) and area under the curve (AUC, b). n = 5 mice/group. b, F2,12 = 26.93. c, Insulin tolerance test (ITT). n = 5 mice/group. d,e, Fasting glucose (d) and insulin (e) levels. n = 7 mice/group. d, F2,18 = 7.043. f, HOMA-IR. n = 7 mice/group. F2,18 = 5.415. g, Energy expenditure. n = 5 mice/group. h, Bile acid levels in the ileum. n = 7 mice/group. TβMCA: F2,18 = 43.27, TUDCA: F2,18 = 31.76. i, Relative mRNA abundance of intestinal Fxr and its target gene mRNAs. n = 7 mice/group. Shp: F2,18 = 5.37, Fgf15: F2,18 = 6.213. j, Relative expression levels of the indicated thermogenic genes in sWAT; mice were placed at 4 °C for 24 h before killing. n = 7 mice/group. Ucp1: F2,18 = 7.604, Elovl3: F2,18 = 5.488, Cox8b: F2,18 = 6.511, Tmem26: F2,18 = 12.09, Pgc1a: F2,18 = 7.038. All the P value was determined by one-way ANOVA with Tukey’s correction; a,c,g, *P < 0.05, **P < 0.01 versus control; #P < 0.05, ##P < 0.01 versus metformin. All data are presented as the mean ± s.e.m.
Intestinal FXR is essential for metformin-induced metabolic improvements
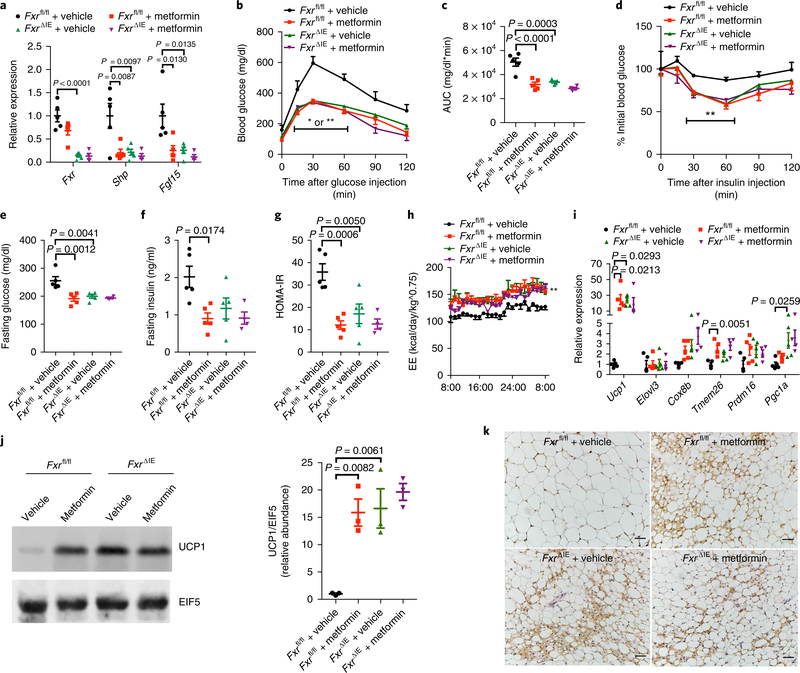
To further address the role of intestinal FXR in the action of metformin and evaluate the long-term effects, we placed intestinal-specific Fxr knockout (FxrΔIE) mice and floxed control (Fxrfl/fl) mice on a HFD and treated them with metformin for 12 weeks. Metformin suppressed intestinal FXR signaling and restricted body weight again and enhanced glucose tolerance and insulin sensitivity in Fxrfl/fl mice, but not in FxrΔIE mice (Fig. 5a–g and Supplementary Fig. 9a,b). Metformin substantially elevated energy expenditure in Fxrfl/fl mice, but had no further effects in FxrΔIE mice (Fig. 5h and Supplementary Fig. 9c). In sWAT, metformin markedly upregulated the expression levels of thermogenic genes in Fxrfl/fl mice; whereas FxrΔIE mice were unresponsive to metformin (Fig. 5i and Supplementary Fig. 9d,e). Immunohistochemical staining and Western blot analysis confirmed the elevated biogenesis of beige adipocytes and the expression level of UCP1 in metformin-treated Fxrfl/fl mice, but no further upregulation was noted in FxrΔIE mice (Fig. 5j, k). These data revealed that metformin has potential long-term effects in improving metabolic diseases dependent on intestinal FXR signaling.
Fig. 5 |. Intestinal FXR signaling is essential for the metformin-induced long-term improvements in metabolic diseases.
HFD-fed Fxrfl/fl and FxrΔIE mice were treated with metformin for 12 weeks. a, Relative mRNA abundance of intestinal Fxr and its target gene mRNAs. n = 4 or 5 (5, 5, 5, 4) mice/group. Fxr: F3,15 = 23.16, Shp: F3,15 = 7.384, Fgf15: F3,15 = 7.121. b,c, GTT (b) and AUC (c). n = 4 or 5 (5, 5, 5, 4) mice/group. c, F3,15 = 20.58. d, ITT. n = 4 or 5 (5, 5, 5, 4) mice/group. e,f, Fasting glucose (e) and insulin (f) levels. n = 4 or 5 (5, 5, 5, 4) mice/group. e, F3,15 = 10.23; f, F3,15 = 5.074. g, HOMA-IR. n = 4 or 5 (5, 5, 5, 4) mice/group. F3,15 = 11.49. h, Energy expenditure. n = 4 or 5 (5, 5, 5, 4) mice/group. i, Relative mRNA abundance of the indicated thermogenic genes in sWAT; mice were placed at 4 °C for 24 h before killing. n = 4 or 5 (5, 5, 5, 4)/group. Ucp1: F3,15 = 4.722, Tmem26: F3,15 = 8.348, Pgc1a: F3,15 = 5.669. j, Western blot analysis (cropping of blot images) of UCP1 protein expression in sWAT and the statistical graph, mice were placed at 4 °C for 24 h before killing. n = 3 mice/group. F3,8 = 13. k, UCP1 immunohistologic staining of sWAT sections; mice were placed at 4 °C for 24 h before killing. n = 3 mice/group, three images per mouse. Scale bars, 50 μm. All P values were determined by one-way ANOVA with Tukey’s correction, *P < 0.05, **P < 0.01 versus Fxrfl/fl + vehicle. All data are presented as the mean ± s.e.m.
GUDCA has therapeutic effects on glucose intolerance and insulin resistance
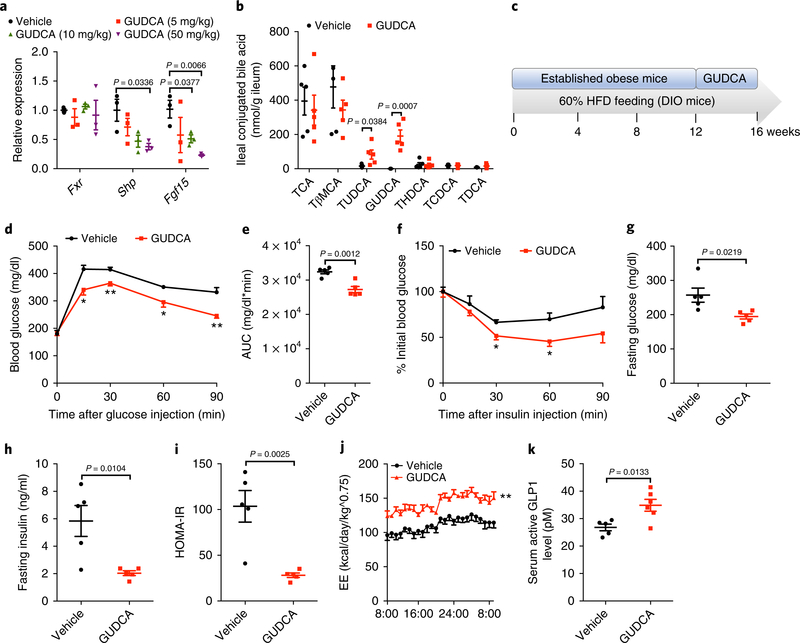
Because GUDCA was identified as a novel FXR antagonist, we further investigated whether GUDCA produced metabolic benefits in vivo. Oral administration of GUDCA suppressed FXR signaling in the gut, but did not affect FXR signaling in the liver (Fig. 6a and Supplementary Fig. 10a,b). Analysis of bile acid profiles revealed that GUDCA and TUDCA levels were markedly higher, whereas other bile acid metabolites were not changed (Fig. 6b). GUDCA treatment substantially restored glucose intolerance and insulin resistance and attenuated body weight gain in Fxrfl/fl mice, but not in FxrΔIE mice on a HFD (Supplementary Fig. 10c–i). The metabolic rates were markedly enhanced in GUDCA-treated Fxrfl/fl mice; however, FxrΔIE mice were unresponsive to GUDCA treatment (Supplementary Fig. 10j,k). Mechanistically, GUDCA treatment significantly induced the expression of thermogenesis-related genes in Fxrfl/fl mice and had no further induction in FxrΔIE mice (Supplementary Fig. 10l). These results demonstrated that GUDCA selectively antagonizes intestinal FXR signaling and thus ameliorates insulin resistance.
Fig. 6 |. GuDCA supplementation had therapeutic effects in improving glucose tolerance dependent on intestinal FXR.
a, Intestinal Fxr and its target gene mRNAs relative abundance after 1-week GUDCA treatment by gavage (5 mg/kg/d, 10 mg/kg/d and 50 mg/kg/d) on a HFD. n = 3 mice/group. Shp: t4 = 3.178, Fgf15: t4 (10 mg/kg) = 3.06, t4 (50 mg/kg) = 5.171. b, Bile acid profiles in the ileum after 1-week GUDCA treatment by gavage (50 mg/kg/d). n = 5 mice/group. TUDCA: t8 = 2.475, GUDCA: t8 = 5.369. c, The schematic diagram of animal model applied in GUDCA therapeutic experiments; after a 12-week HFD treatment, mice were given vehicle or GUDCA (50 mg/kg/d) for 4 weeks. DIO, diet-induced obesity. d,e GTT (d) and AUC (e). n = 5 mice/ group. e: t8 = 4.891. (f) ITT. n = 5 mice/group. g,h Fasting glucose (g) and insulin (h) levels. n = 5 mice/group. (g): t8 = 2.839, (h): t8 = 3.332. i, HOMA-IR. n = 5 mice/group. t8 = 4.321. j, Energy expenditure. n = 5 mice/group. k, Serum active GLP1 levels. Vehicle or GUDCA-treated (50 mg/kg/d) mice on a HFD for 1 week. n = 5 or 6 mice/group. t9 = 3.072. All P values were determined by two-tailed Student’s t-test, *P < 0.05, **P < 0.01 versus vehicle. All data are presented as the mean ± s.e.m.
Furthermore, GUDCA treatment had therapeutic effects in reversing metabolic disorders in established obese mice (Fig. 6c–i and Supplementary Fig. 11a–c). The levels of serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were lower after GUDCA supplementation, indicating that there was no liver injury (Supplementary Fig. 11d,e). These metabolic improvements were consistent with an elevated metabolic rate (Fig. 6j and Supplementary Fig. 11f). In addition, GUDCA substantially elevated active glucagon-like peptide 1 (GLP1) production, possibly contributing to the improvement of glucose homeostasis (Fig. 6k).
Discussion
Although AMPK is believed to be one of the principal targets of metformin1,3, some studies revealed that metformin effects may be independent of AMPK2, as verified in the present study. Notably, metformin still suppressed intestinal FXR signaling in Ampka1ΔIE mice. In microbiota-depleted mice, metformin did not affect intestinal FXR signaling on either a HFD or after FXR agonist (TCA) treatment. These results revealed that metformin inhibits intestinal FXR signaling via modulation of the gut microbiota, and not through intestinal AMPK signaling. While there are several studies associating the gut microbiota with metformin treatment in mouse models or individuals with T2D, the major changes in the gut microbiota (especially at the species level) are not consistent9–13,31. The gut microbiota varies widely among populations and life style and can influence the effectiveness of drugs9,32. Thus, it is of great interest to find the important microbial-derived metabolites and determine their host targets rather than focusing only on changes in specific gut microbiota. The current study aimed to investigate the impact of the gut microbiota, bile acid metabolism and related host targets on the regulation of glucose metabolism in individuals with T2D and obese mice.
Although an elevation of Escherichia and a decrease of Intestinibacter were found in metformin-treated Danish and Swedish patients, there were slight or no changes in a Chinese cohort9, consistent with the present findings. A recent study of human stool samples collected after metformin treatment for 2, 4 and 6 months revealed that the abundance of genus Bacteroides was reduced after metformin treatment10. To exclude the possibility that the metabolic phenotype improvements after long-term metformin treatment modified the gut microbiota, we collected samples from individuals with T2D naively metformin treated for only 3 d. The results revealed that the consistent decrease of Bacteroides abundance was due to a direct effect of metformin treatment, and not derived from the subsequent metabolic improvements. Another report also demonstrated that there was a remarkable decrease of Bacteroides levels in Asian individuals with T2D after acarbose treatment33. Surprisingly, the improvement effects of acarbose were more remarkable in cluster B (rich in Bacteroides) than cluster P (rich in Prevotella), indicating that the decrease of Bacteroides abundance was beneficial in T2D treatment33. Among Bacteroides, we found that only the species B. fragilis was strongly correlated with GUDCA and TUDCA levels and intestinal FXR signaling. Furthermore, according to the KEGG pathway, B. fragilis showed the most striking change on the pathway of folate biosynthesis and one-carbon pool under metformin treatment. Folate provides one-carbon units for various biosynthetic processes, including methionine, purine and thymine, which are required for bacterial growth and survival34. The current study found that metformin inhibited the growth of B. fragilis through modification of folate and methionine metabolism. The exact mechanism by which metformin regulates folate and methionine metabolism of B. fragilis needs further investigation. We further found that B. fragilis colonization exacerbated metabolic disorders induced by a HFD regimen. Consistent with this, B. fragilis decreased the levels of Tβ MCA and TUDCA, resulting in activated intestinal FXR signaling in mice. Moreover, improvements of glucose metabolism in metformin-treated mice were reversed by B. fragilis. Through the application of the BSH inhibitor CAPE, we further verified that the beneficial effects of metformin were dependent on inhibition of BSH activity in B. fragilis.
The gut microbiota is known to influence host intestinal homeostasis, in part via metabolic pathways such as the short chain fatty acid (SCFA) and bile acid metabolism. We found that the levels of GUDCA and TUDCA, which were identified in this study as novel endogenous FXR antagonists, were upregulated after metformin treatment in T2D individuals, but downregulated in B. fragilis–colonized mice. CDCA is converted to UDCA by gut microbial 7α/β-dehydrogenation and UDCA is then transported from the intestine to the liver in humans through the enterohepatic circulation. In liver, the majority of UDCA is conjugated with glycine and taurine to produce GUDCA and TUDCA, which are transported into the gut and then reabsorbed into the ileum epithelial cells19. In mice, CDCA is oxidized to βMCA by hepatic Cyp2c70 and conjugated with taurine to produce the mouse-specific TβMCA35, an intestinal FXR antagonist21,22. The conjugated bile acids also go through deconjugation mediated by gut microbiota–produced BSH, and thus they are markedly elevated in germ-free and antibiotic-treated mice21,22. In the current study, we observed that deconjugation of GUDCA and TUDCA in humans was suppressed due to the decrease of B. fragilis BSH. UDCA treatment was reported to have protective effects on hepatic metabolic diseases36, and oral administration of UDCA resulted in lower serum FGF19 levels, indicating that intestinal FXR signaling might be suppressed37. Because UDCA was found not to be a direct FXR antagonist38, the underlying mechanism remains unclear. Surprisingly, the present study revealed that GUDCA and TUDCA were bona fide FXR antagonists. Thus, the metabolic benefits of UDCA may in part be due to GUDCA. We confirmed that GUDCA supplementation suppressed intestinal FXR signaling and alleviated obesity-related metabolic diseases in vivo. In the current study, we found that oral GUDCA supplementation did not result in disorders in bile acid metabolism or liver injury, thus indicating the potential safety of this compound. The observation of similar metabolic effects of GUDCA in mice and humans suggest that the gut microbiota–intestinal FXR signaling pathway and associated mechanism as detailed in mouse studies21,23,24 translates to humans and suggests that this pathway can be exploited for drug development.
FXR, a ligand-activated member of the nuclear receptor super-family, is mainly expressed in the liver and ileum and plays a critical role in the development of metabolic diseases16. In contrast to the role of hepatic FXR in metabolism, inhibition of intestinal FXR signaling was demonstrated to improve metabolic diseases21,23–25,39–41. A recent study showed that metformin improved the SGLT1-dependent glucose-sensing glucoregulatory pathway by elevating the upper small intestinal levels of genus Lactobacillus in a HFD-treated rat model14. In agreement with this study, we also observed that the abundance of species Lactobacillus sanfranciscensis was increased in T2D individuals after a brief (3-day) clinical metformin treatment, and Sglt1 mRNA was elevated in the upper small intestine of HFD-treated mice after metformin treatment (L. Sun, Q. Wu, X. Wang, and C. Jiang, unpublished observations). Furthermore, the genus Lactobacillus was found to affect FXR signaling in the upper small intestine42. Inhibition of intestinal FXR could be a common pathway that connects gut microbiota remodeling with the anti-diabetic effects of metformin. The selective intestinal FXR antagonist GMCA had preventive and therapeutic effects on metabolic dysfunctions, thus indicating that this pathway can be pharmacologically targeted25,43. The FXR and TGR5 dual agonist INT-767 was observed to stimulate GLP-1 secretion, which suggested that FXR and TGR5 may coordinately stimulate secretion of GLP-1 (ref. 44). Supporting this view, a recent study showed that the metabolic benefits of fexeramine, an intestine-specific FXR agonist, are in part through modulating the gut microbiota resulting in higher levels of the bile acid lithocholic (generated through bacterial 7β-dehydroxylation of UDCA) to activate TGR5/GLP-1 signaling45. Interestingly, FXR deficiency and FXR inhibition in intestinal L cells promoted GLP-1 production and secretion in a glycolysis-dependent manner40. The current study revealed that inhibition of intestinal FXR by GUDCA treatment substantially elevated serum active GLP1 levels. Additionally, previous results revealed that activation of intestinal FXR signaling enhanced the synthesis of ceramide in the gut and elevated serum ceramide levels; ceramide treatment was shown to suppress the expression of thermogenesis-related genes in beige adipocytes25. In the current study, sWAT thermogenesis-related genes were elevated in metformin-treated Fxrfl/fl mice and FxrΔIE mice, but there was no further effect in metformin-treated FxrΔIE mice. Further studies are needed to explore the role of intestinal ceramides in the metabolic improvements by metformin.
The present study partially explains how metformin improves obesity-induced glucose intolerance and insulin resistance via an AMPK-independent mechanism (Supplementary Fig. 11g). We conclude that metformin can act through the gut microbiota to improve metabolic dysfunction and does so by a B. fragilis–GUDCA–intestinal FXR axis. As the target of metformin in the gut, GUDCA signaling shows promise as a potential target for the treatment of metabolic disease in humans.
Methods
Human subjects
Human serum and feces samples were collected from 22 newly diagnosed individuals with T2D who received oral metformin hydrochloride treatment (1000 mg b.i.d, Merck Serono) for 3 d. Venous blood samples were collected after 10 h of overnight fasting. All blood samples were set at room temperature for 30 min and were then centrifuged at 3,000g for 20 min to obtain the serum. Feces samples were collected on the same day, snap frozen in dry ice, and stored at − 80 °C until analysis. All of the subjects enrolled satisfied the following criteria: newly diagnosed with T2D and has never been treated with antidiabetic drugs before this study. The exclusion criteria were: type 1 diabetes; pregnancy; severe diabetic complications (diabetic retinopathy, diabetic neuropathy, diabetic nephropathy and diabetic foot); severe hepatic diseases including chronic persistent hepatitis, liver cirrhosis or the co-occurrence of positive hepatitis and abnormal hepatic transaminase; continuous antibiotic use for over 3 d within 3 months prior to enrollment; continuous use of weight-loss drug for over 1 month; gastrointestinal surgery (except for appendicitis or hernia surgery); severe organic diseases, including cancer, coronary heart disease, myocardial infarction or cerebral apoplexy; infectious diseases, including pulmonary tuberculosis and AIDS; and alcoholism. Clinical parameters were determined at Beijing Chao-Yang Hospital. The demographic characteristics, insulin resistance relative indicators, liver enzymes and lipid analyses of patients involved in the study are listed in Supplementary Table 1. The study protocol was approved by the Conjoint Health Research Ethics Board of Beijing Chao-Yang Hospital, and the study was conducted in accordance with the principles of the Declaration of Helsinki. All participants provided written informed consent.
DNA extraction and preparation
Genomic DNA from human stool samples clinically collected was extracted by Stool Genomic DNA Kit (CWBIO, Cat# CW20925). Degradation and contamination of DNA were monitored on 1% agarose gels. DNA purity was checked by the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). The Qubit® DNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA) measured the concentration of DNA.
Metagenomics sequencing
DNA, 700 ng per sample, was used for sample preparation. Sequencing libraries were established by NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, USA); the manufacturer’s recommendations and index codes were adopted to attribute sequences to each sample. The fragmented DNA ends were repaired, polyA-tailed, and ligated with a sequencing adaptor for Illumina sequencing. PCR amplification and purification (AMPure XP system) were performed. The concentration of DNA was measured by the Qubit® DNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA) and diluted to 2 ng/μl. The insert size of library was assessed by using the Agilent Bioanalyzer 2100 system. qPCR ensured accurate concentration (>3 nM) of the library. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq 4000 PE Cluster Kit (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 4000 platform and 150-bp paired-end reads were generated.
Metagenomics analysis
Adaptor contamination and low-quality reads were discarded from the raw sequencing reads, and the remaining reads were filtered to eliminate human host DNA based on the human genome reference (hg18). We acquired 340 Gb high-quality pair-end reads for the 44 samples with an average of 7.7 Gb per sample after removing human DNA reads. All high-quality reads from each sample was assembled to contigs by using metaSPAdes from SPAdes v3.10.0 package with default settings, and a nonredundant gene catalog was constructed by MetaGeneMark and CD-HIT. Gene profile was calculated according to the high-speed quantification tool Kallisto46 and normalized by gene length. Genes were annotated by blasting against the NCBI database using Diamond. The taxonomic composition was acquired by MEGAN and KO composition obtained using the KEGG Orthology-Based Annotation System (KOBAS) tool. The sum of relative abundance of all genes involved in a KEGG pathway was defined to be pathway abundance. For the analysis of bile salt hydrolase (BSH) genes, a local gene database containing all available nonredundant bsh gene sequences was constructed by searching the UniProt (Universal Protein Resource) protein database and further used for targeted reads screening. Gene counts were calculated by counting the number of genes in each sample. Alpha diversity was performed on the basis of the gene profile of each sample according to the Shannon index. Wilcoxon matched-pair signed-rank tests (two-tailed) with adjustments according to Benjamini–Hochberg correction (FDR) were conducted to analyze differences in two groups using R 3.4.0. Spearman’s Rankorder correlation analysis was performed on the genus and species associated with clinical indices. Partial least square discriminant analysis (PLS-DA) was used to determine taxonomic changes in two groups, and VIP (variable importance) scores were adopted to rank the ability of different taxa to discriminate between different groups. The significant difference between the two groups was assessed by PERMANOVA (permutation multivariate analysis of variance) with the Bray–Curtis distance. The number of permutations was 9999 (performed using the vegan package in R 3.4.0).
Bile acid analysis
Serum and ileum samples were prepared by precipitation. Then, chlorpropamide (Sigma-Aldrich, Cat# C1290) was added as an internal standard to quantify bile acid levels. The bile acid concentrations in the supernatants were measured by a UPLC/Synapt G2-Si QTOF MS system (Waters Corp., Milford, MA) with an ESI source. Chromatographic separation was operated on an Acquity BEH C18 column (100 mm × 2.1 mm i.d., 1.7 μm, Waters Corp.). Column temperature was 45 °C, and the flow rate was 0.4 ml/min. The mobile phase included a mixture of 0.1% formic acid in water and 0.1% formic acid in acetonitrile. The gradient elution was applied and MS detection proceeded in negative mode. A mass range of m/z 50 to 850 was acquired. Standards for all bile acids were used to identify the different bile acid metabolites detected by LC-MS.
Serum C4 analysis
An equal volume of serum was mixed with acetonitrile containing 1 μM chlorpropamide (internal standard). The samples were vortexed and then spun at 10 min at 4 °C to precipitate particulates. The supernatant was transferred to an auto sampler vial and 20 μl was separated by reverse phase on a Prominence 20 UFLCXR system (Shimadzu, Columbia, MD) with a Waters BEH C18 column (100 mm × 2.1 mm, 1.7 um particle size) and the eluate delivered into a 5600 TripleTOF using a Duospray ion source (SCIEX, Framingham, MA) operating in enhanced mode and monitoring the transition m/z 401.3–177.1.
TR-FRET FXR coactivator recruitment assay
CDCA, GUDCA, TUDCA and TβMCA were tested for direct FXR activity using the commercial TR-FRET FXR coactivator recruitment assay kit (Thermo Fisher, Cat# A15140).
Cell culture
Caco-2 (ATCC, Cat# HTB-37) cells and HEK293T (ATCC, Cat# CRL-3216) cells were cultured in DMEM with 10% fetal bovine serum (FBS). The cells were incubated for 4–6 h in DMEM media with 1% FBS and then exposed to the indicated concentrations of bile acids for 16 h. HEK293T cells were used for the luciferase report assays.
Luciferase reporter gene assays
G. L. Guo (Rutgers University) supplied the PGL4-Shp-TK firefly luciferase construct and human FXR expression plasmid. P. A. Dawson (Emory University) provided the human ASBT expression plasmid, and M. Miyazaki (University of Colorado) and K. Schoonjans (École Polytechnique Fédérale de Lausanne) gifted the pCMVSPORT6/hTGR5 and cAMP response element-driven luciferase reporter plasmids. HEK293 cells (ATCC, Cat# CRL-1573) were co-transfected with (1) human FXR expression vector, human ASBT expression vector for conjugated bile acid transport, a firefly luciferase reporter vector and a Renilla luciferase control vector (pRL-luciferase; Promega, Madison, WI) or (2) pCMVSPORT6/hTGR5 and cAMP response element-driven luciferase reporter plasmids using Lipofectamine 3000 transfection reagent (ThermoFisher Scientific). After 24 h post-transfection, the cells were exposed to GUDCA (50–200 μM, Sigma-Aldrich, Cat# 06863), or TUDCA (50–200 μM, Sigma-Aldrich, Cat# T0266) in the presence of CDCA (20 μM, Targetmol, Cat# T0847) for 16 h. Luciferase assays were performed using the dual-luciferase assay system (Promega). Firefly and Renilla luciferase activities were measured by Veritas microplate illuminometer (Turner Biosystems).
B. fragilis culture, verification and growth curve
B. fragilis was streaked onto a BHIS plate in a 37 °C anaerobic incubator for 2 d. A single colony was used to inoculate 5 ml PPS (supplemented Proteose Peptone media), and seed cultures were shaken overnight at 37 °C. A 1:500 dilution of each seed culture was subcultured into PPS at 37 °C, until the culture reached OD = 0.7. The culture was split into 1.5 ml EP tubes (1,000 μL/each), and aliquots were flash frozen using liquid nitrogen and stored at − 80 °C. B. fragilis were recovered by shaking at 37 °C for 30 min, and the concentration was made 5 × 108 CFU/ml PBS. Mice were given 108 CFU B. fragilis or the same dose of heat-killed B. fragilis by gavage twice per week. B. fragilis was cultured in anaerobic tubes in the presence or absence of metformin (1 mM, 5 mM and 10 mM) and OD600 value was measured hourly.
Hydrolysis efficiency of GUDCA and TUDCA
B. fragilis proteins were prepared in pH 7.4 PBS using sonication. Incubations were carried out in 3 mM sodium acetate buffer, pH 5.2, containing 0.1 mg/ml protein and bile acids (GUDCA, TUDCA, or d5-GUDCA (Santa Cruz Biotechnology, Cat# sc-280757)) in a final volume of 200 μl. After an hour incubation at 37 °C, the reactions were stopped in ice. Acetonitrile (200 μl) was directly added to the reaction mix. After centrifuging at 14,000g for 20 min, 5 μl of the supernatant was transferred to an auto sampler vial and subjected to analysis.
Folic acid and methionine analysis in B. fragilis
Five milliliters B. fragilis in PPS was centrifuged at 7,000 r.p.m., 3 min. The supernatant was removed, 500 μl methanol with 1 mg/ml butylated hydroxytoluene and 1 mg/ml vitamin C was added, and the mixture was shaken for 30 min. Next, 500 μl of methanol with 1 mg/ml butylated hydroxytoluene and 1 mg/ml vitamin C was added, and the samples shaken for 30 min. Centrifugation was carried out at 14,000g for 10 min and the supernatants measured using the AB SCIEX QTRAP® 5500 LC/MS/MS System. An ultra-high-pressure LC system (Shimadzu) was interfaced to a Q-TRAP 5500 mass spectrometer (SCIEX) for metabolite separation and detection. An HSS T3 C18 column (2.1 × 100 mm i.d., 1.7 μm; Waters) was employed for compound separation at 40 °C. Mobile phase A was 0.1% formic acid; mobile phase B was acetonitrile. The linear gradient used was as follows: 1 min, 0% B; 2 min, 100% B, 5.5 min, 100% B; 5.6 min, 0% B, and end at 8 min, 0% B. The flow rate was 0.3 ml/ min. The injection volume was 1 μl.
Mice
All animal protocols were approved by the Animal Care and Use Committee of Peking University. All the mice were housed in a standard specific-pathogen-free environment. For the two-group experiment, 6- to 8-week-old male C57BL/6 J mice were fed a HFD (Research diets, Cat# D12492) and drinking water with vehicle or 1.5 mg/ml metformin (Sigma-Aldrich, Cat# D150959) for 1 week. 1.5 mg/ml metformin in water was similar to 200 mg/kg/d in mice. For the gut microbiota experiment, after 3 d of antibiotics cocktail (neomycin 1 mg/ml, streptomycin 1 mg/ml and bacitracin 1 mg/ml) treatment in drinking water, 6- to 8-week-old male C57BL/6 J mice were treated with metformin (200 mg/kg/d) under HFD treatment for 1 week. To validate the inhibitory effects of GUDCA and TUDCA (Sigma-Aldrich, Cat# T0266) on intestinal FXR signaling, 6- to 8-weekold male C57BL/6J mice were treated with an antibiotic cocktail in drinking water for 3 d. Vehicle, TCA (Sigma-Aldrich, Cat# T4009), TCA plus CDCA, TCA plus TUDCA, and TCA plus GUDCA were orally administered to mice at doses of 50 mg/kg/d for 1 week. Under antibiotic treatment, mice were given vehicle or metformin in water (200 mg/kg/d) in the presence of TCA (50 mg/kg/d) for 1 week to verify that metformin did not suppress the TCA-induced activation of intestinal FXR signaling in microbiota-depleted mice. To explore the activation of AMPKa in the ileum under oral metformin treatment, HFD-fed mice received 200 mg/kg metformin in water for 1 week or 500 mg/kg by gavage for 3 h.
To determine the role of metformin and TMP (Sigma-Aldrich, Cat# T7883) in regulating bile acid metabolism and intestinal FXR signaling, metformin in water (200 mg/kg/d) and TMP by gavage (160 mg/kg/d) were given to HFD-treated mice for 1 week.
AMPK, Ampka1fl/fl mice47 were purchased from Jackson Laboratory (Farmington, CT, USA). For intestine-specific AMPK disruption, Ampka1fl/fl female mice were crossed with male mice expressing Cre recombinase under the control of the Villin promoter. Six- to eight-week-old male Ampka1fl/fl and Ampka1ΔIE mice were fed a HFD and drinking water with vehicle or 200 mg/kg/d metformin for 1 week.
For stool transplantation experiments, stool samples were collected from four individuals with T2D before and after metformin treatment. Stool (20 mg) was dissolved in 1 ml saline, shaken for 3 min, centrifuged for 3 min at 4 °C, and 200 μl supernatant collected. Mice were fed a HFD and administered 100 μl of the above supernatant by gavage after 3 d of antibiotics treatment.
To transplant B. fragilis (ATCC, Cat# ATCC25285), mice were fed a HFD and heat-killed B. fragilis (control, 108CFU/mouse) or B. fragilis (108CFU/mouse) by gavage for 4 weeks48. Additionally, the mice were given heat-killed B. fragilis (control, 108CFU/mouse), metformin (200 mg/kg/d) plus heat-killed B. fragilis (108CFU/mouse), or metformin (200 mg/kg/d) plus B. fragilis (108CFU/mouse) by gavage on a HFD for 4 weeks to explore the reversal effects. After 3-d antibiotic treatment, microbiota-depleted mice were fed a HFD and given metformin (200 mg/kg/d) with or without CAPE (75 mg/kg/d) under B. fragilis treatment for 4 weeks.
Six- to eight-week-old male Fxrfl/fl and FxrΔIE mice49 were fed a HFD and drinking water with vehicle or 200 mg/kg/d metformin for 12 weeks. Six- to eight-week-old male Fxrfl/fl and FxrΔIE mice were fed a HFD and given 50 mg/kg/d GUDCA (Sigma-Aldrich, Cat# 06863) by gavage for 4 weeks. The diet-induced obese (DIO) mice model used in our experiment: after a 12-week HFD treatment, mice were given vehicle or GUDCA (50 mg/kg/d) for 4 weeks. Mice had free access to water, and all of the sample collections were performed at consistent times after 4 h fasting.
Metabolic assays
Glucose tolerance tests were performed after 16–18 h fasting. Blood glucose concentrations were measured with a glucometer, and blood samples were taken from the tail tip at 0, 15, 30, 60 and 120 min after intraperitoneal injection of glucose (1.5 g/kg body weight). For the insulin tolerance test, insulin (1 U/kg body weight) was administered via intraperitoneal injection after 4 h fasting. All of the ITT and GTT tests were performed at indicated times.
Serum active GLP1 detection
Vehicle- or GUDCA- (50 mg/kg/d) treated mice on a HFD for 1 week received sitagliptin (25 mg/kg, DPP4 inhibitor) by gavage 1 h before serum collection and oral glucose (2 g/kg) challenge 15 min before serum collection. Serum active GLP1 levels were measured by GLP1 (active) assay kit (Millipore, Cat# EGLP-35K).
Histological analysis
Adipose tissues were fixed in 4% paraformaldehyde, and paraffin sections were cut and stained by immunostained with UCP1 (Abcam, Cat# ab10983) antibody. At least three discontinuous sections were evaluated for each sample in over three samples per group.
Real-time PCR analysis
Mouse inguinal adipose tissue and intestinal mucosa scraped from the ileum and liver were frozen in liquid nitrogen and stored at − 80 °C. A standard phenol-chloroform extraction was performed to isolate total RNA from frozen tissues with Trizol reagent. cDNA was synthesized from 2 μg of total RNA with a Reverse Transcription Kit (ABM, Cat# G490). The real-time PCR primer sequences are summarized in Supplementary Table 2. The relative count of genes was calculated by normalizing to 18S mRNA.
Western blot analysis
Adipose tissues were homogenized in RIPA buffer with protease and phosphatase inhibitors; the protein extracts were separated by SDSPAGE electrophoresis and transferred to a PVDF membrane. The membrane was incubated with antibodies against UCP1 (Abcam, Cat# ab10983), AMPKa1 (Abcam, Cat# ab32047), AMPKa2 (Abcam, Cat# ab3760), GAPDH (Cell Signaling, Cat# 2118), EIF5 (Santa Cruz, Cat# 135854) overnight at 4 °C. Full western blot gel panels are shown in Supplementary Fig. 12.
Data analysis
GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA) was used for statistical treatment. The sample sizes were determined by power analysis using StatMate version 2.0 (GraphPad Software, San Diego, CA). Experimental data were shown as the mean ± SEM. The investigators involved in the current study were not completely blinded in experiments during sample collection and analysis. No data were excluded from data analysis. Two-tailed unpaired Student’s t-test and one-way ANOVA with Tukey’s correction were used for all comparisons of mice-related experiments, and a Wilcoxon matched-pairs signed rank test was used for clinical indicators in individuals with T2D. P values < 0.05 were considered significant. The sample distribution was determined using a Kolmogorov–Smirnov normality test. Correlation analysis of gut microbiome and host metabolome were investigated using nonparametric Spearman’s test.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Sequence data are available for download from the Sequence Read Archive with accession number PRJNA486795.
Supplementary Material
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFC0903100 and 2016YFC0903102) to C.J., the National Natural Science Foundation of China (91439206 and 91739303 to X.W., and 81522007, 81470554 and 31401011 to C.J.), the National Program for Support of Top-notch Young Professionals (82008Y0005) to C.J., the Fundamental Research Funds for the Central Universities: Clinical Medicine Plus X—Young Scholars Project of Peking University (PKU2018LCXQ013) to C.J., and the National Cancer Institute Intramural Research Program to F.J.G.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-018-0222-4.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41591-018-0222-4.
References
- 1.Zhou G et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest 108, 1167–1174 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rena G, Hardie DG & Pearson ER The mechanisms of action of metformin. Diabetologia 60, 1577–1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lien F et al. Metformin interferes with bile acid homeostasis through AMPK-FXR crosstalk. J. Clin. Invest 124, 1037–1051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madiraju AK et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510, 542–546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madiraju AK et al. Metformin inhibits gluconeogenesis via a redox-dependent mechanism in vivo. Nat Med 24, 1384–1394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey CJ, Wilcock C & Scarpello JH Metformin and the intestine. Diabetologia 51, 1552–1553 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Geach T Gut microbiota: mucin-munching bacteria modulate glucose metabolism. Nat Rev Endocrinol 13, 66 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Friedman SL, Neuschwander-Tetri BA, Rinella M & Sanyal AJ Mechanisms of NAFLD development and therapeutic strategies. Nat. Med 24, 908–922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forslund K et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med 23, 850–858 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Lee H & Ko G Effect of metformin on metabolic improvement and gut microbiota. Appl. Environ. Microbiol 80, 5935–5943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep 5, 14405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin NR et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Bauer PV et al. Metformin alters upper small intestinal microbiota that impact a glucose-SGLT1-sensing glucoregulatory pathway. Cell Metab. 27, 101–117 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Schroeder BO & Backhed F Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med 22, 1079–1089 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Matsubara T, Li F & Gonzalez FJ FXR signaling in the enterohepatic system. Mol. Cell. Endocrinol 368, 17–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Aguiar Vallim TQ, Tarling EJ & Edwards PA Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S & Moschetta A Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 7, 12–18 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Wahlstrom A, Sayin SI, Marschall HU & Backhed F Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Kong B et al. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 56, 1034–1043 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun 4, 2384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayin SI et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez FJ, Jiang C & Patterson AD An intestinal microbiota-farnesoid X receptor axis modulates metabolic disease. Gastroenterology 151, 845–859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang C et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest 125, 386–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang C et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat. Commun 6, 10166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie C et al. An intestinal farnesoid X receptor-ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes 66, 613–626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akwabi-Ameyaw A et al. Conformationally constrained farnesoid X receptor (FXR) agonists: naphthoic acid-based analogs of GW 4064. Bioorg. Med. Chem. Lett 18, 4339–4343 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Singhal A et al. Metformin as adjunct antituberculosis therapy. Sci. Transl. Med 6, 263ra159 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Cabreiro F et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stellwag EJ & Hylemon PB Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. Biophys. Acta 452, 165–176 (1976). [DOI] [PubMed] [Google Scholar]
- 31.Napolitano A et al. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS ONE 9, e100778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deschasaux M et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med 24, 1526–1531 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Gu Y et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat. Commun 8, 1785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olgun A “Metformin-resistant” folic acid producing probiotics or folic acid against metformin’s adverse effects like diarrhea. Med. Hypotheses 106, 33–34 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Takahashi S et al. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res 57, 2130–2137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuchida T, Shiraishi M, Ohta T, Sakai K & Ishii S Ursodeoxycholic acid improves insulin sensitivity and hepatic steatosis by inducing the excretion of hepatic lipids in high-fat diet-fed KK-Ay mice. Metabolism 61, 944–953 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Mueller M et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J. Hepatol 62, 1398–1404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita K, Iguchi Y, Une M & Watanabe S Ursodeoxycholic acid suppresses lipogenesis in mouse liver: Possible role of the decrease in beta-muricholic acid, a farnesoid X receptor antagonist. Lipids 52, 335–344 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Parseus A et al. Microbiota-induced obesity requires farnesoid X receptor. Gut 66, 429–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trabelsi MS et al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat. Commun 6, 7629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziętak M et al. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab. 23, 1216–1223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer PV et al. Lactobacillus gasseri in the upper small intestine impacts an ACSL3-dependent fatty acid-sensing pathway regulating whole-body glucose homeostasis. Cell Metab. 27, 572–587e576 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Zhang L et al. Farnesoid X receptor signaling shapes the gut microbiota and controls hepatic lipid metabolism. mSystems 1, e00070–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pathak P et al. Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism. J. Biol. Chem 292, 11055–11069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pathak P et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 68, 1574–1588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaeffer L, Pimentel H, Bray N, Melsted P & Pachter L Pseudoalignment for metagenomic read assignment. Bioinformatics 33, 2082–2088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakada D, Saunders TL & Morrison SJ Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653–658 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vetizou M et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim I et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res 48, 2664–2672 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Sequence data are available for download from the Sequence Read Archive with accession number PRJNA486795.