Abstract
Non-membrane bound organelles such as nucleoli, processing bodies, cajal bodies, and germ granules form via spontaneous self-assembly of specific proteins and RNAs. How these biomolecular condensates form and interact are poorly understood. Here we identify two proteins, ZNFX-1 and WAGO-4, that localize to C. elegans germ granules (P granules) in early germline blastomeres. Later in germline development, ZNFX-1/WAGO-4 separate from P granules to define an independent liquid-like condensate that we term the Z granule. In adult germ cells, Z granules assemble into ordered tri-condensate assemblages with P granules and Mutator foci, which we term the PZM granule. Finally, we show that one biological function of ZNFX-1 and WAGO-4 is to interact with silencing RNAs in the C. elegans germline to direct transgenerational epigenetic inheritance (TEI). We speculate that the temporal and spatial ordering of liquid droplet organelles may help cells organize and coordinate the complex RNA processing pathways underlying gene regulatory systems, such as RNA-directed TEI.
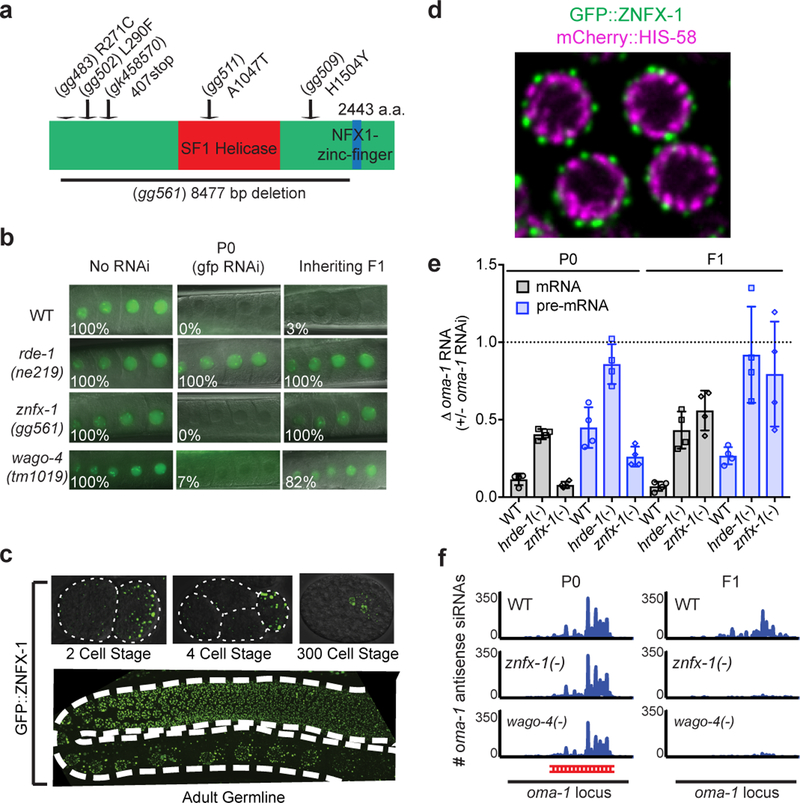
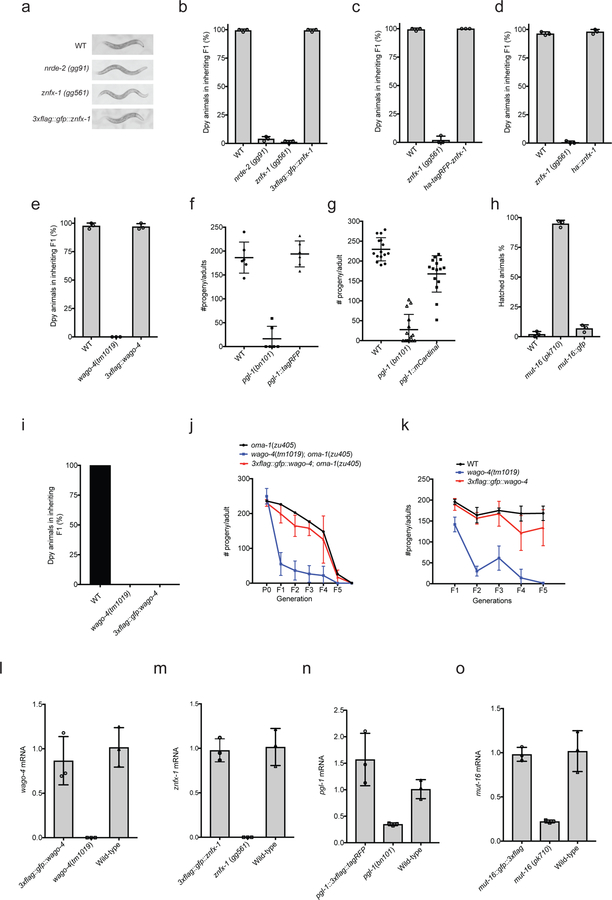
Epigenetic information can be inherited for multiple generations (TEI)1,2. Non-coding RNAs have emerged as important mediators of TEI (RNA-directed TEI), although the mechanism(s) by which RNA mediates TEI remains poorly understood. In many eukaryotes, dsRNAs silence other cellular RNAs that exhibited sequence complementarity to trigger dsRNAs; a process termed RNA interference (RNAi)3. In C. elegans, RNAi is heritable: distant progeny of animals exposed to dsRNAs continue to silence complementary RNAs in the absence of further dsRNA exposure (RNAi inheritance)4–6. To further our understanding of RNA-directed TEI, we conducted a genetic screen to identify factors required for RNA inheritance (Extended Data figure 1). Our screen identified 37 mutations that disrupted RNAi inheritance. We subjected DNA from these 37 mutant strains to whole-genome sequencing and identified four independent mutations in the gene zk1067.2 (Fig. 1a). To confirm that zk1067.2 is required for RNAi inheritance, we tested two additional alleles of zk1067.2 (gk458570 and gg561) for RNAi inheritance defects. gk458570 and gg561 animals responded normally to dsRNA treatment, however, the progeny of these mutant animals were largely unable to inherit gene silencing (Fig. 1b, and Extended Data figure 2). We conclude that zk1067.2 is required for RNAi inheritance.
Figure 1. ZNFX-1 is a conserved RNA helicase required for RNAi inheritance in C. elegans.
a, znfx-1 alleles are indicated. b, Animals expressing a pie-1::gfp::h2b transgene 8 were exposed to gfp dsRNA. F1 progeny were grown in the absence of dsRNA, and GFP expression in oocytes was visualized by fluorescence microscopy. % of animals expressing pie-1::gfp::h2b is shown (n=6 biologically independent samples for WT, n=3 for rde-1, znfx-1 and wago-4, each n=at least 30 animals). P0, parental generation; WT, wild-type. c, Fluorescence micrograph of gfp::znfx-1 animals. Images are representative of >3 animals visualized at each lifestage. d, Pachytene germ cells of animals expressing GFP::ZNFX-1 and a chromatin marker mCherry::HIS-58. Image representative of 3 animals. e, WT, hrde-1(tm1200), and znfx-1(gg561) animals were exposed to oma-1 dsRNA. Total RNA from RNAi (P0) and inheriting (F1) generations was isolated. RNA was quantified using qRT-PCR using primers 5’ to the site of RNAi and data were normalized to eft-3 pre-mRNA. n= 4 biologically independent samples. +/− standard deviation of the mean (SD). Note, independent mRNA and pre-mRNA primer sets gave similar results (data not shown). f, siRNA libraries (material and methods) were prepared from WT, znfx-1(gg561), and wago-4(tm1019) animals exposed to oma-1 dsRNA (P0) and progeny (F1). Antisense reads mapping to oma-1 locus are shown. Red line indicates region of oma-1 locus targeted by dsRNA. Reads counts were normalized to total number of sequenced reads (n=2 biologically independent samples ).
ZNFX-1 is required for TEI
Sequence analysis showed that ZK1067.2 encodes a 2443 amino acid protein that contains a superfamily one (SF1) RNA helicase domain and a Zn finger domain (Fig. 1a). A single putative ortholog of ZK1067.2 was found in most eukaryotic genomes. Fungal orthologs have been linked to RNAi pathways in S. pombe and N crassa9–10. Homology between ZK1067.2 and its mammalian ortholog ZNFX1 extend to a Zn finger domain not present in fungal orthologues. We conclude that ZK1067.2 is a conserved protein involved in RNAi-mediated gene silencing in many eukaryotes. Henceforth, we refer to ZK1067.2 as ZNFX-1.
To begin to understand the function of ZNFX-1 during RNAi inheritance, we used CRISPR/Cas9 to insert a 3xflag::gfp epitope immediately upstream of the znfx-1 start codon. Note, CRISPR-mediated gene conversion was used throughout this work. Tagged loci were expressed near wild-type levels and resultant fusion proteins were functional unless otherwise indicated (Extended Data figure 3). We observed GFP::ZNFX-1 expression in the adult germline as well as in developing germ cells during all stages of embryonic and larval development (Fig. 1c). No GFP::ZNFX-1 expression was observed in somatic tissues. Post fertilization, C. elegans zygotes undergo a series of asymmetric cell divisions in which germline determinants segregate with germline blastomeres. During embryonic development, ZNFX-1 foci were concentrated in, and segregated with, the germline blastomeres (Fig 1c and see below). In adult germ cells, GFP::ZNFX-1 was concentrated in foci that were distributed in a perinuclear pattern around nuclei (Fig 1d). We conclude that znfx-1 encodes a germline-expressed protein that segregates with the germline and localizes to perinuclear foci in adult germ cells.
Treatment of animals with oma-1 dsRNA silences the oma-1 gene for multiple generations5,6. To address when ZNFX-1 acts to promote RNAi inheritance, we used qRT-PCR to measure oma-1 mRNA and pre-mRNA levels in znfx-1(−) animals exposed to oma-1 RNAi, as well as in the progeny of these animals. znfx-1(−) animals responded normally to oma-1 RNAi; however, their progeny failed to inherit silencing, suggesting that ZNFX-1 acts during the inheritance phase of RNAi (Fig. 1e). During RNAi inheritance, siRNAs targeting genes undergoing RNAi silencing are expressed for multiple generations6. In znfx-1(−) animals exposed directly to oma-1 dsRNA, oma-1 siRNAs were produced at wild-type levels; however, progeny of these mutant animals failed to express oma-1 siRNAs (Fig. 1f). Three additional genetic and biochemical analyses supported the idea that ZNFX-1 acts during the inheritance phase of RNAi (Extended Data figure 4). The data establish that ZNFX-1 is a dedicated RNAi inheritance factor.
WAGO-4 acts with ZNFX-1 to direct TEI
The C. elegans genome encodes ~27 Argonaute (AGO) proteins. The molecular function of many of these AGOs remains a mystery. Two of the mutant strains identified by our genetic screen harbored mutations in the AGO-encoding gene wago-4. To confirm WAGO-4 is required for RNAi inheritance, we tested two additional wago-4 deletion alleles (tm1019 and tm2401) for RNAi inheritance defects. Both alleles exhibited RNAi inheritance defects (Fig. 1b and Extended Data Figure 5). Thus, like ZNFX-1, WAGO-4 is required for RNAi inheritance. Additionally, when we appended a gfp tag to the wago-4 locus, we observed that, like ZNFX-1, WAGO-4 is a germline-expressed protein that segregates with the P lineage blastomeres and localizes to perinuclear foci (Extended Data Figure 5). For unknown reasons, our GFP::WAGO-4 fusion protein was fully functional for RNAi inheritance in some RNAi inheritance assays but only partially functional in other assays (Extended Data Figure 3). TagRFP::ZNFX-1 and GFP::WAGO-4 colocalized in germ cells, hinting that WAGO-4 and ZNFX-1 may act together to promote RNAi inheritance (Fig. 2a). Three additional lines of evidence support this idea. First, 3xFLAG::WAGO-4 co-precipitates with HA::ZNFX-1, but not a HA tagged negative control protein HA::HRDE-1, suggesting a physical interaction between the two proteins (Fig. 2b). Second, wago-4 mutant animals behaved like znfx-1 mutant animals in molecular assays of RNAi inheritance (Fig. 1f). Third, znfx-1 and wago-4 animals share a pleiotropic phenotype: Both mutant animals exhibited a temperature-sensitive mortal germline (Mrt) phenotype, whereby mutant animals became sterile several generations after populations were shifted to growth at a higher temperature (25C) (Fig. 2c). Taken together these data show that WAGO-4 functions with ZNFX-1 to transmit RNA-based epigenetic information across generations.
Figure 2. ZNFX-1 and WAGO-4 act cooperatively to drive RNAi inheritance.
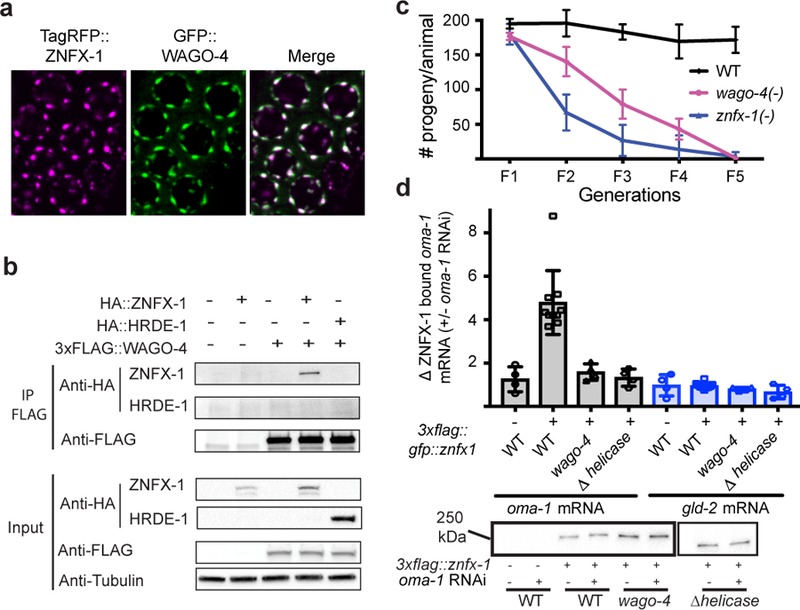
a, Fluorescent micrographs of pachytene germ cells expressing GFP::WAGO-4 and TagRFP::ZNFX-1. Image representative of >3 animals. b, CoIP analysis HA::ZNFX-1 and FLAG::WAGO-4 (n=1 for WT and HA::HRDE-1, n=3 for others. n=independent experiments). HA::HRDE-1 is a negative control. c, Animals of indicated genotypes were shifted to growth at 25℃ and progeny were counted for five generations (the mean of n=3 biologically independent samples. +/− SD is displayed). d, Top panel: FLAG::ZNFX-1 was IP’ed in RNAi generation from animals treated +/− with oma-1 dsRNA. Co-precipitating RNA was subjected to qRT-PCR to quantify oma-1 mRNA co-precipitating with ZNFX-1 in wild-type, or wago-4(tm1019) animals. Note, ZNFX-1 also binds RNAi-targeted RNAs in inheriting generation (data not shown). ZNFX-1 Δ helicase harbors a 1487 bp in frame deletion of ZNFX-1 helicase domain. gld-2 is a control mRNA. The mean of n=10 for WT, n=4 for others, n=biologically independent samples, +/− SD is displayed. Bottom panel, Western blot of IP’ed ZNFX-1 from one RNA IP replicate shown in top panel. Two unrelated lanes were removed from this image (see Supplemental Figure 1).
How do WAGO-4 and ZNFX-1 promote RNAi inheritance? The closest homolog of ZNFX-1 is SMG-2/UPF1, which marks mRNAs containing premature termination codons9. We wondered if, by analogy, ZNFX-1 might bind and mark mRNAs encoded by genes undergoing heritable gene silencing. To test this idea, we subjected 3XFLAG::ZNFX-1 expressing animals to oma-1 RNAi, immunoprecipitated 3XFLAG::ZNFX-1, and used qRT-PCR to ask if oma-1 RNAi caused ZNFX-1 to interact with oma-1 mRNA. Indeed, oma-1 RNAi caused ZNFX-1 to co-precipitate with oma-1 mRNA (Fig. 2d). The following three lines of evidence show that the interaction of ZNFX-1 with TEI-related RNAs is a sequence-specific event directed by the RNAi machinery. First, RNAi targeting the lin-15b gene caused ZNFX-1 to interact with the lin-15b mRNA, but not the oma-1 mRNA (and vice versa), indicating that ZNFX-1/mRNA interactions are sequence-specific (data not shown). Second, most RNA helicases bind RNA via their helicase domains. Deletion of the ZNFX-1 helicase domain did not affect ZNFX-1 expression but did prevent ZNFX-1 from interacting with oma-1 mRNA (Fig. 2d). Third, in wago-4 mutant animals, ZNFX-1 failed to interact with the oma-1 mRNA in response to oma-1 RNAi (Fig. 2d). We conclude that RNAi directs ZNFX-1 to interact with mRNAs undergoing heritable silencing and that WAGO-4 is required for this property of ZNFX-1.
ZNFX-1/WAGO-4 separate from P granules
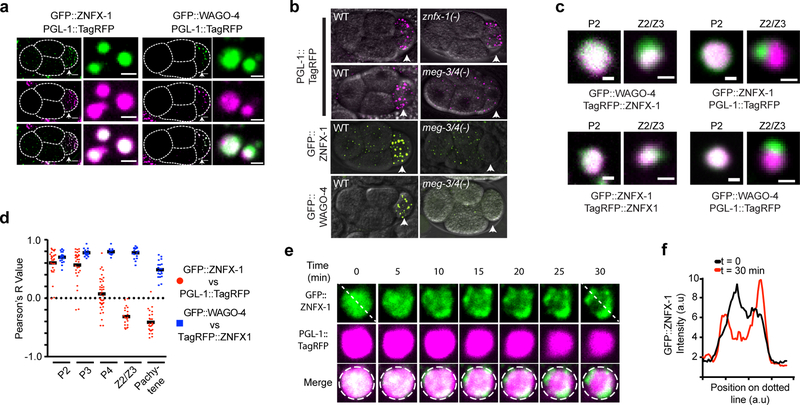
P granules are biomolecular condensates that, like ZNFX-1/WAGO-4 foci, segregate with the germline blastomeres (P0-P4) during embryonic development10,11. The low-complexity protein PGL-1 marks P granules12. GFP::ZNFX-1 and GFP::WAGO-4 colocalized with PGL-1::TagRFP in P1-P3 germline blastomeres, suggesting that ZNFX-1 and WAGO-4 are P granule factors (Fig. 3A). MEG-3/MEG-4 are low-complexity domain proteins redundantly required for P granule formation in the P lineage (Fig. 3b)13. In meg-3/4(−) embryos, ZNFX-1/WAGO-4 foci failed to segregate with the P lineage (Fig. 3b). Thus, in early P1-P3 germline blastomeres, ZNFX-1 and WAGO-4 localize to P granules.
Figure 3. ZNFX-1/WAGO-4 appear to separate from P granules to form new foci during germline development.
a-b, Micrographs of 4 cell embryos expressing indicated proteins are shown. P2 blastomeres are indicated by arrowheads. a, Magnifications of P granules are shown to right. Images are representative of >3 animals. b, Genotypes are znfx-1(gg561), or meg-3(tm4259);meg-4(ax2026). Images are representative of >3 animals. c, Micrographs of, and d, quantification of colocalization between (see methods), indicated fluorescent proteins at indicated stages of germline development. c, Images are representative >3 animals. d, Each data points represent quantification of an individual granule (n>15 granules imaged in >3 animals for each lifestage). Mean is indicated by solid black line. e, Time-lapse micrographs in early (~300 cell embryos) Z2/Z3 cells. Image representative of 3 animals. f, Fluorescent intensity along dotted lines shown in e. Scale bars: a, whole embryo 6 μm, individual granules 1 μm, c, 0.5 μm.
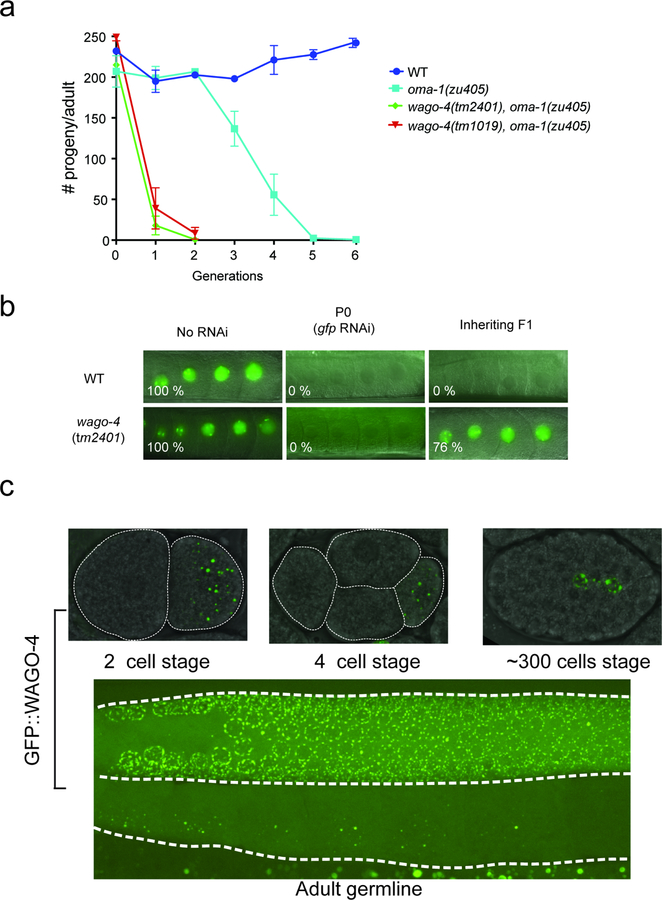
At the ~100 cell stage of embryonic development the P4 blastomere divides to give rise to Z2 and Z3, which are the primordial germ cells of C. elegans. Surprisingly, we found that GFP::ZNFX-1 no longer colocalized with PGL-1::TagRFP in Z2/Z3 (Fig. 3c). Rather, GFP::ZNFX-1 appeared in foci that were adjacent to (see below), yet distinct from, PGL-1::TagRFP foci (Fig. 3c). Similar results were seen when antibodies were used to visualize PGL and ZNFX-1, indicating that failure to colocalize was not an artifact of GFP/TagRFP epitopes (Extended Data figure 6). Quantitative analyses showed that the degree to which ZNFX-1 and PGL-1 colocalized changed during development, with a transition from colocalized to non-colocalized occurring between the P3 and Z2/Z3 (Fig. 3d). The ZNFX-1/WAGO-4 foci seen in Z2/Z3 could form de novo or by the separation of ZNFX-1/WAGO-4 and PGL-1 from within pre-existing foci. We favor the later model as time-lapse imaging in Z2/Z3 cells captured what appeared to be ZNFX-1/PGL-1 separation events (Fig. 3e/f). Separation of ZNFX-1/WAGO-4 into discrete foci could be triggered by phase separation or by segregation of pre-existing sub-structures into discrete areas14. We conclude that, late in germline development, ZNFX-1 and WAGO-4 are concentrated in foci adjacent to P granules.
Z granules are liquid-like condensates
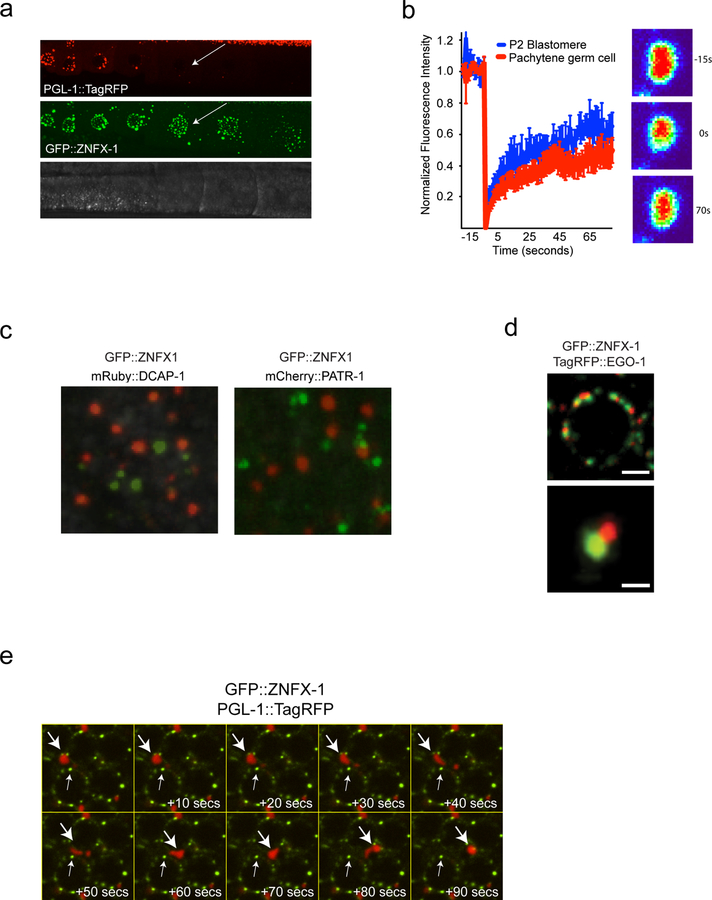
Liquid-like condensates are self-assembling cellular structures that form when specific proteins and RNAs undergo liquid-liquid phase transitions from surrounding cytoplasm. The ability of ZNFX-1 and WAGO-4 to separate from P granules hints that ZNFX-1/WAGO-4 foci may also be liquid-like condensates. Liquid-like condensates are typically spherical in shape and their internal constituents undergo rapid internal rearrangements15,16. Consistent with the idea that ZNFX-1 foci are liquid-like condensates, we observed that during oocyte maturation, ZNFX-1 foci detached from nuclei and assumed spherical shapes (Extended Data figure 7). Additionally, fluorescence recovery after photobleaching (FRAP) experiments showed that within ZNFX-1 foci, GFP::ZNFX-1 fluorescence recovered rapidly from bleaching (t=~8 seconds), which is a rate similar to that reported for PGL-1 FRAP in P granules (Extended Data figure 7)10. Thus, ZNFX-1/WAGO-4 foci (post Z2/Z3) exhibit properties reminiscent of liquid-like condensates and, therefore, we refer to these foci as Z granules.
Z granules assemble into tri-droplet structures
C. elegans germ cells possess at least two other foci (processing bodies and Mutator foci) with properties akin to liquid-like condensates17,18. TagRFP::ZNFX-1 did not co-localize with MUT-16::GFP, which marks Mutator foci, nor did GFP::ZNFX-1 colocalize with mCherry::PATR-1 or mRuby::DCAP-1, which mark processing bodies (Fig. 4a and Extended Data Figure 7). Interestingly, although Z granules did not colocalize with Mutator foci the relative positions of these two foci were not random. Z granules were usually [89% of the time, (n=35)] found closely apposed to (no empty space between fluorescence signals) a Mutator foci (Fig. 4a). Similarly, Z granules were usually [91% of the time, (n=35)] found closely apposed to a P granule (Fig. 4a). Quantification of distances between surfaces and centers of fluorescence for the three foci supported the idea that Z granules localize adjacent to P granules and Mutator foci in adult germ cells (Fig. 4b). This analysis also showed that the distance between the surfaces of Z granules and P granules/Mutator foci (but not P granules and Mutator foci) lies within the diffraction limit of light, indicating that Z granules exist in very close proximity to, and may be in direct physical contact with P granules and Mutator foci (Fig. 4b). Note, although Z granules are intimately associated with P granules/Mutator foci in adult germ cells (and throughout most of germline development), they can exist independently. For instance, in the adult germline, Z granules remained visible at developmental time points when P granules were no longer present (Extended Data figure 7). Similarly, Z granules are present in developing germ cells at time points (i.e. P blastomeres) when Mutator foci are not thought to be present17. Additionally, shearing force causes P granules in pachytene stage germ cells to disengage from nuclei and flow through the germline syncytium10. After applying shearing force, we found that P granules flowed through the cytoplasm, however, Z granules remained largely static (Extended Data figure 7). Thus, Z granules can be separated from P granules and Mutator foci both developmentally and physically. We conclude that Z granules represent an independent form of liquid-like condensate, which closely mirror P granules and Mutator foci in adult germ cells.
Figure 4. Z granules assemble into tri-condensate (PZM) structures with P granules and Mutator foci.
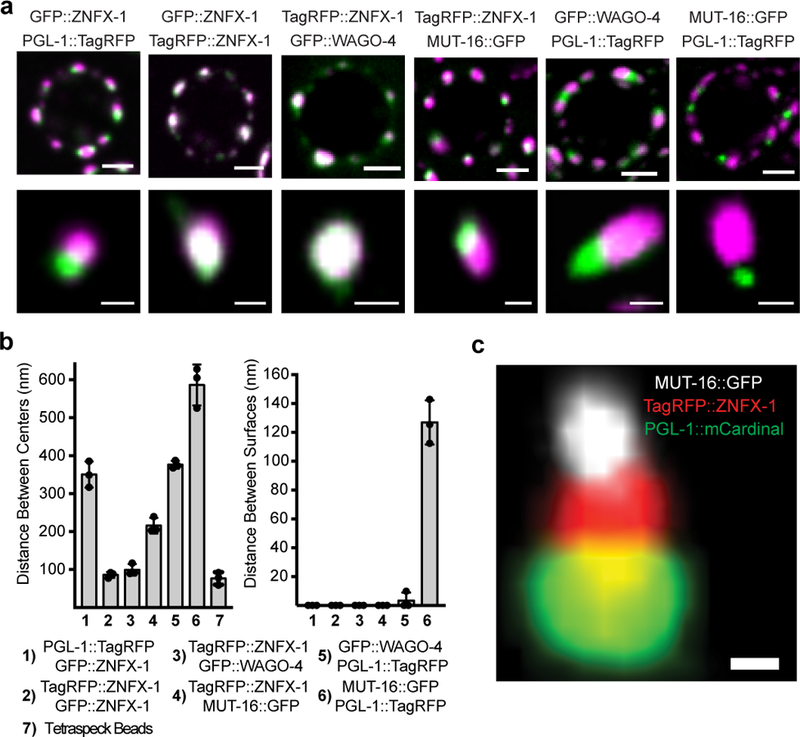
a, Top, Fluorescent micrographs of a pachytene germ cell nucleus from animals expressing the indicated fluorescent proteins. Bottom, 3D renders of representative foci. Images are representative of >3 animals. b, Distance between the centers (left) and surfaces (right) of the spaces occupied by the indicated fluorescent proteins was calculated as described in Methods. Means of 10 granules measured in 3 animals (30 total) +/− SD is shown. Column 7 shows chromatic shift associated with tetraspeck beads. Data in right panel have been corrected for this shift. c, Fluorescent micrograph of indicated fluorescent proteins from pachytene germ cell. Image is representative of >3 animals (see Extended Data Figure 8 for quantifications). Scale bars: a, germ cell, 2 μm, single granule, 0.5 μm. c, 0.25 μm. Position of nuclear membrane and nuclear pores and PZM segments is not yet known.
Our data suggest that Z granules may localize between (bridge) P granules and Mutator foci. To test this idea, we imaged the three foci simultaneously using PGL-1::mCardinal, TagRFP::ZNFX-1, and MUT-16::GFP expressing animals19. This analysis confirmed the idea that Z granules bridge P granules and Mutator foci (Fig. 4c): In 60% (52/86) of cases, we observed a Z granule in close apposition to both a P granule and a Mutator foci, while in 92% (48/52) of these cases the Z granule lay between the other two foci. In no case (0/52) did a P granule or a Mutator foci bridge the other two types of foci, respectively. Quantification of the distances between the centers and surfaces of Z granules, P granules, and Mutator foci from triple-marked images support the idea that Z granules act as a bridge between P granules and Mutator foci in adult germ cells (Extended Data figure 8). We conclude that P granules, Z granules, and Mutator foci form tri-condensate assemblages (henceforth PZMs) in adult germ cells and that the relative position of the three liquid-like condensates constituting the PZM is ordered.
Is PZM assembly required for RNA-directed TEI? Factors concentrated in Mutator foci8,17,20,21 and Z granules (this work) contribute to RNA-directed TEI. Additionally, we find that several factors, known to be required for P granule assembly, are also needed for efficient RNAi inheritance (Extended Data figure 9). Thus, factors associated with all three segments of the PZM have now been linked to TEI. Additionally, we find that in mutant animals with defective P granules, Z granules become malformed and ZNFX-1 fails to bind TEI-related RNAs, hinting that the segments of the PZM may communicate with each other during TEI (Extended Data figure 9). The results are consistent with the idea that PZM assembly is important for RNA-directed TEI.
Here we show that the inheritance factors ZNFX-1 and WAGO-4 localize to a liquid-like condensate that we name the Z granule. Given that Z granules segregate with the germline, we speculate that one function of the Z granule is to concentrate and segregate silencing factors into the germline to promote RNA-based TEI. ZNFX-1 is a conserved RNA helicase, which marks RNAs produced from genes undergoing heritable silencing. The S. pombe ortholog of ZNFX-1 is Hrr1, which forms a nuclear complex (termed the RDRC) with Argonaute and RdRP to amplify siRNA populations directing pericentromeric heterochromatin22. We speculate that C. elegans RDRC acts in the cytoplasm where it promotes RNAi inheritance by: 1) binding inherited siRNAs (via WAGO-4), 2) marking mRNAs complementary to inherited siRNAs (via ZNFX-1), 3) using marked mRNAs as templates for RdRP-based siRNA amplification, and 4) repeating this cycle each generation (Extended Data figure 10). Note, a related study suggests that the function of ZNFX-1 in RNA marking may involve positioning RdRP enzymes to prevent 5’ drift of AGOs targeting mRNAs (Craig Mello, personal communication).
ZNFX-1 and WAGO-4 separate from components of the P granule during early embryogenesis to form an independent liquid-like condensate. Separation occurs at a developmental time that correlates roughly with the first association of P granules with nuclear pores and the advent of germline transcription11,23–26. We speculate that condensate separation might be triggered when newly synthesized mRNAs transit P granules and interact with RNA binding proteins to alter local protein concentration and initiate separation. In addition to temporal ordering, we find that Z granules are spatially ordered relative to P granules and Mutator foci, with Z granules forming the centerpiece of PZM tri-condensate assemblages in adult germ cells. These results show that mechanism(s) exist to organize and arrange liquid-like condensates in space as well as time. Additional work is needed to understand how PZM segments assemble in the correct order and if/how PZM assembly contributes to RNA-based TEI. Small RNA-based pathways in animals are complex with many thousands of small regulatory RNAs regulating thousands of mRNAs at virtually all levels of gene expression. We speculate that the ordering of liquid-like condensates in space and time helps organize and coordinate these small RNA pathways, including RNA-directed TEI (Extended Data figure 10). Similar strategies may be used by cells to organize and coordinate other gene regulatory or biochemical pathways.
Methods
Strain list:
N2 (WT); (NL1870) mut-16(pk710); (YY009) eri-1(mg366), (YY193) eri-1(mg366); nrde-2(gg91), (YY502) nrde-2(gg91), (YY503) nrde-2(gg90), (YY538) hrde-1(tm1200), (YY562) hrde-1(tm1200); oma-1(zu405), (YY913) nrde-2(gg518[nrde-2::3xflag::ha]), (YY916) znfx-1(gg544[3xflag::gfp::znfx-1]), (YY947) hrde-1(tm1200); nrde-2(gg518), (YY967) pgl-1(gg547[ pgl-1::3xflag::tagrfp]), (YY968) znfx-1(gg544); pgl-1(gg547), (YY996) znfx-1(gg561), (TX20) oma-1(zu405), (YY998) znfx-1(gg544); ego-1(gg644[ha::tagrfp::ego-1]), (YY1020) znfx-1(gg561); oma-1(zu405), (SX461) mjIS31(pie-1::gfp::h2b), (SS579) pgl-1(bn101), (JH3225) meg-3(tm4259); meg-4(ax2026), (DG3226) deps-1(bn124), (YY1006) eri-1(mg366); znfx-1(gg561), (YY1003) eri-1(mg366); znfx-1(gk458570), (YY1021) znfx-1(gg561); nrde-2(gg518), (YY1062) znfx-1 (gk458570), (YY1081) deps-1(bn124); mjIS31, (YY1083) wago-4(tm1019), (YY1084) wago-4(tm2401), (YY1093) wago-4(tm1019); mjIS31, (YY1094) wago-4(tm2401); mjIS31, (YY1108) znfx-1(gg561); mjlS31, (YY1109) mjIS31; dpy-10(cn64), (YY1110) eri-1(mg366); wago-4(tm1019), (YY1111) eri-1(mg366); wago-4(tm2401), (YY1153) wago-4(tm1019), znfx-1(gg544),mjlS31, (YY1175) hrde-1(gg594[ha::hrde-1]) (YY1287) znfx-1(gg611[ha::znfx-1]), (YY1305) meg-3(tm4259); meg-4(ax2026), znfx-1(gg544), (YY1308) meg-3(tm4259); meg-4(ax2026), pgl-1(gg547), (YY1325) wago-4(gg620[3xflag::gfp::wago-4]), (YY1326) wago-4(gg620); znfx-1(gg561), (YY1327) pgl-1(gg547); wago-4(gg620), (YY1346) pgl-1(gg547); znfx-1(gg561), (YY1364) meg-3(tm4259); meg-4(ax2026), wago-4(gg620), (YY1388) wago-4(gg627[3xflag::wago-4]), (YY1393) znfx-1(gg611); wago-4(gg627), (YY1408) znfx-1(gg561); mjlS31; dpy-10(cn64), (YY1416) znfx1(gg544); axIs1488, (YY1419) znfx-1(gg561); wago-4(tm1019); oma-1(zu405), (YY1442) znfx1(gg544); hjSi397, (YY1446) znfx-1(gg634[ha::tagrfp::znfx-1]), (cmp3) mut-16::gfp::flag+loxP, (YY1444) znfx-1(gg634); mut-16[mut-16::gfp::flag+loxP], (YY1452) znfx-1(gg544); Itls37(pie-1::mcherry::his58), (YY1453) znfx-1(gg634); wago-4(gg620), (YY1460) mut-16(mut-16::gfp::flag); znfx-1(gg561); (YY1461) mut-16(mut-16::gfp::flag); wago-4(tm1019); (YY1486) znfx1(gg631[3xflag::gfp::znfx-1 ∆ helicase]), (YY1491) wago-4(gg620); oma-1(zu405) (YY1492)pgl-1(gg640[pgl-1::3xflag::mcardinal]); mut-16[mut-16::gfp::flag+loxP]; znfx-1(gg634), (YY1494) wago-4(tm2401); pgl-1(gg547) (YY1503) pgl-1(gg547); mut-16[mut-16::gfp::flag+loxP], (YY1556) wago-4(gg627); hrde-1(gg594).
CRISPR/Cas9:
gRNAs were chosen using Ape according to following standards: first, PAM sites are in the context of GGNGG 27 or GNGG; second, GC content of 20 bp spacer sequence was 40% to 60%; third, high specificity according to crispr.mit.edu. All CRISPR was done using co-CRISPR strategy 28. Plasmids were purified with PureLink™ HiPure Plasmid Kits (Thermofisher). For deletions: two gRNAs (20ng/μl) were co-injected into gonads with pDD162(50ng/μl), unc-58 gRNA(20ng/μl), AF-JA-76 (20ng/μl) and 1X taq buffer. For 3xFLAG or HA epitope tagging, single strand oligos (4nM ultramer from IDT, purified by isopropanol precipitation) with 50bp homology regions were used as repair templates. gRNA (20ng/μl) and repair template (20ng/μl) were co-injected into gonads with pDD162(50ng/μl), unc-58 gRNA(20ng/μl), AF-JA-76 (20ng/μl) and 1X taq buffer. For GFP, tagRFP or mCardinal tagging, repair templates contained homologous arms of 500bp to 1000bp and were cloned into pGEM-7zf(+). Sequences were confirmed by Sanger sequencing. Repair templates were amplified with PCR, gel purified and isopropanol precipitated. PCR product was heated at 95 ℃ for 5min and then immediately put on ice for at least 2min. Injection mix was prepared: pDD162(50ng/μl), unc-58 gRNA (20ng/μl), AF-JA-76(20ng/μl), gRNAs close to N terminal or C terminal of the genes (20ng/μl), heated and cooled repair template (50ng/μl) and 1X standard taq buffer. Injected animals were maintained at 25°C, Unc animals were isolated 4 days later and grown at 20℃. Animals were screened for deletion or tagging by PCR.
RNA IP:
animals were flash frozen in liquid nitrogen and stored at −80℃. Animals were resuspended in sonication buffer (20 mM Tris.HCl PH 7.5, 200mM NaCl, 2.5mM MgCl2, 10% glycerol, 0.5% NP-40, 80U/ml RNaseOUT, 1mM DTT and protease inhibitor cocktail without EDTA) and sonicated (30s on, 30s off, 20%−30% output for 2min on a Qsonica Q880R sonicator, repeat once). Lysates were clarified by centrifuging at 14000 rpm for 15min. Supernatants were precleared with protein A agarose beads and incubated with FLAG-M2 agarose beads for 2–3 hours at 4℃. Beads were washed with RIP buffer (20 mM Tris.HCl PH 7.5, 200mM NaCl, 2.5mM MgCl2, 10% glycerol, 0.5% NP-40) 6x. Protein and associated RNAs were eluted with 100ug/ml 3xFLAG peptide. RNAs were treated with Turbo DNase I for 20 min at 37°C and then extracted with TRIzol reagent followed by precipitation with isopropanol.
RT-qPCR:
mRNA isolated from total RNA or from RNA IP experiments, was converted to cDNA using the iScript cDNA synthesis kit according to vendor’s instructions. The following primer sequence were used to quantify mRNA levels. oma-1 mRNA:GCTTGAAGATATTGCATTCAACC (Forward primer); AACTGTTGAAATGGAGGTGC (Reverse primer). oma-1 pre-mRNA: GTGCGTTGGCTAATTTCCTG (Forward primer); CTGAATCGCGCGAACTTG (Reverse primer). gld-2 mRNA: ACGTGTAGAAAGGGCTGCAC (Forward primer); GTCGATGCAGATGATGATGG (Reverse primer). gld-2 pre-mRNA: CCTTATTAATTTCAGAGCTGCTGTC (Forward primer); AAGACTAGCACACGCAATCG (Reverse primer). eft-3 pre-mRNA: CCTGCAAGTTCAACGAGCTTA (Forward primer); TGAAAAACAAATTGGTACATAAAC (Reverse primer).
Mrt assay:
Each generation, 3–6 L4 animals were picked to a single plate and grown at 25°C, average brood sizes were calculated by counting the total number of progeny per plate.
RNAi inheritance assays:
dpy-11 and gfp RNAi inheritance: Embryos were collected via hypochlorite treatment and placed onto HT115 bacteria expressing dsRNA against dpy-11 or gfp. F1 embryos were collected by hypochlorite treatment from RNAi or control treated adults and placed onto non-RNAi plates. Worms were scored at late L4 (dpy-11) or early young adult (gfp) stages. oma-1 RNAi inheritance: Experiments were done at 20°C. Embryos were collected via hypochlorite treatment and placed onto HT115 bacteria expressing dsRNA against oma-1. 6 F1 embryos were picked onto a single OP50 plate. From F2 to F6, 6 L4 animals were picked onto a single OP50 plate. tm1019 is a 571 bp deletion which removes part of the PIWI domain. tm1019 also introduces a frameshift that would be expected to prevent translation of the rest of the PIWI domain. znfx-1 (gg561) is a 8476 bp deletion that deletes most (2300/2400 a.a.) of ZNFX-1, including the helicase domain. Both alleles were presumably null.
Co-immunoprecipitation:
Young adults were flash frozen in liquid nitrogen. Animals were ground into powder in liquid nitrogen and resuspended in 1ml 1X lysis buffer (20mM Hepes pH7.5, 100mM NaCl, 5mM MgCl2, 1mM EDTA, 10% Glycerol, 0.25% Triton, 1mM fresh made PMSF, 1X complete protease inhibitor from Roche without EDTA) and rotated for 45min at 4℃. Lysate was cleared by spinning at 5000 rpm for 15min, 30μl protein G beads were added to preclear lysate for 30min. 3xFLAG::WAGO-4 proteins were pulled down by 30μl agarose beads conjugated to α-FLAG antibody (A2220, Sigma-Aldrich). Input and IP proteins were separated by SDS-PAGE and detected by FLAG M2 antibody and HA antibody (Roche, 3F10).
Small RNA sequencing:
Total RNA was extracted using TRIzol. 20ug total RNA were separated by 15% Urea gel. Small RNA from about 18 nt to 35 nt were cut from gel. Small RNAs were cloned using a 5’ monophosphate independent small RNA protocol as previously described 29. Libraries were multiplexed with a 4 nt 5’ barcode and a 6 nt 3’ barcode and pooled for next generation sequencing on a NextSeq 500. FastX 0.0.13 was used to separate reads that contained the 3’ adapter and filter low quality reads for further analysis. Reads >14 nt were mapped to the C. elegans genome (WS220) using Bowtie. Read counts were normalized to the total number of reads matching the genome. Two independent libraries were prepared and the two replicates were combined for Figure 1.
Microscopy and Analysis:
To image larval and adult stages, animals were immobilized in M9 with 0.1% Sodium Azide, and mounted on glass slides. To image embryos, gravid adults were dissected on a coverslip containing 10 μl of 1X egg buffer, and then mounted on freshly made 3% agarose pads. Animals were imaged immediately with a Nikon Eclipse Ti microscope equipped with a W1 Yokogawa Spinning disk with 50 um pinhole disk and an Andor Zyla 4.2 Plus sCMOS monochrome camera. A 60X/1.4 Plan Apo Oil objective was used unless otherwise stated.
Colocalization:
The degree of colocalization between different fluorescently labeled proteins across development was calculated using the Coloc2 plugin from ImageJ. Animals were imaged as described above with the exception of using a 100X/1.45 Plan Apo Oil objective. 3–5 granules were selected from at least 3 different animals across each stage of development specified. Region of interest (ROI) masks were generated using the 3D ROI Manager plugin in ImageJ to eliminate black regions surrounding granules 30. Coloc2 was used to generate a Pearson’s R Value for degree of colocalization between two channels in the region defined by the ROI mask.
FRAP:
Fluorescence recovery after photobleaching (FRAP) experiments were conducted using a Zeiss LSM 780 point scanning confocal equipped with a Quasar PMT x2 + GAaSP 32 Channel Spectral Detector using a 63X/1.4 Plan Apo Oil objective. Adult animals (for pachytene germ cells) or embryos (for P2 blastomere) were suspended in a mixture of 0.5% sodium azide and 50% 0.1 μm polystyrene beads (Polysciences) to inhibit movement. The mixture was added to a coverslip and placed on a fresh 3% agarose pad. Slides were sealed with nail polish. The bleaching plugin within the Zeiss Black software was used to specify the ROI to be bleached. One ROI was used for all data points. Single z slice images were acquired at 1 second intervals for 15 seconds, followed by bleaching, then continued at 1 second intervals for 85 seconds. Images were aligned using neighboring granules in ImageJ to account for subtle shifts in movement. An ROI was generated around the bleached region and continuously measured across all time points using the plot profile function within ImageJ. Data was normalized to an unbleached control granule to account for background bleaching throughout the 100 second period. Normalized data points were averaged across all 7 granules and plotted using Prism. The heat map of a representative granule was generated using the thermal LUT within ImageJ.
Quantification of distances between foci centers and surfaces.
We imaged pachytene germ cell nuclei in 3 animals. ~10 granules were selected from each animal. Confocal z stacks were opened with the 3D objects counter plugin from ImageJ to generate x, y, and z coordinates for the center of each object 31. To account for chromatic shift between channels, 0.1 μm Tetraspek beads were imaged and granule distances were corrected accordingly. Distances between foci surfaces was calculated with 3D ROI manager in ImageJ30. Thresholding function within 3D ROI manager was used to eliminate background signal.
Immunofluorescence:
~30 animals were sliced open in 8 ul of 1X egg buffer (25 mM HEPEs, pH 7.3, 118 mM NaCl2, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2) to isolate gonads and embryos. A coverslip was added and slides were placed on a metal block (chilled on dry ice) for 10 minutes. Coverslips were popped off and slides were submerged in methanol at −20°C for 10 minutes, followed by acetone at −20°C for 5 minutes. Samples were allowed to dry at room temperature for 3 minutes. 500 ul of 1XPBST was added to each sample and incubated for 5 minutes at room temperature followed by 500 ul of 1XPBST + 1% BSA for 30 minutes at room temperature. 50 ul of antibody solution (1XPBST, 1% BSA, 1:20 dilution of anti PGL-1 antibody (K76 from DSHB), and 1:250 dilution of anti HA antibody (abcam ab9110)) was added to each sample. Samples were covered with parafilm and incubated overnight at room temperature inside a humid chamber. Samples were washed 3 times in 1XPBST at room temperature for 10 minutes. Secondary antibodies (Alexa Fluor 555 goat anti-rabbit (Life technologies A21429) and Alexa Fluor 488 goat anti-mouse (Life technologies A10667) were diluted 1:50 in 1XPBST. 50 ul of secondary solution was added to each sample, covered with parafilm, and incubated for 90 minutes in the dark at room temperature. Samples were washed 3 times in 1XPBST at room temperature for 10 minutes. 15 ul of Vectashield antifade + DAPI was added to each sample. Slides were sealed with nail polish.
Extended Data
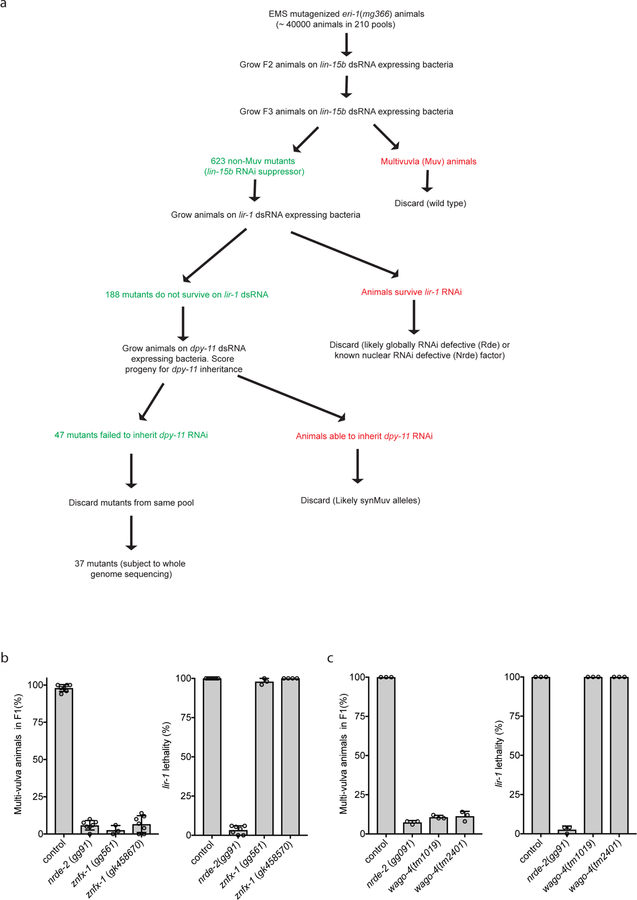
Extended Data figure 1. Genetic screen to identify novel RNAi inheritance mutants.
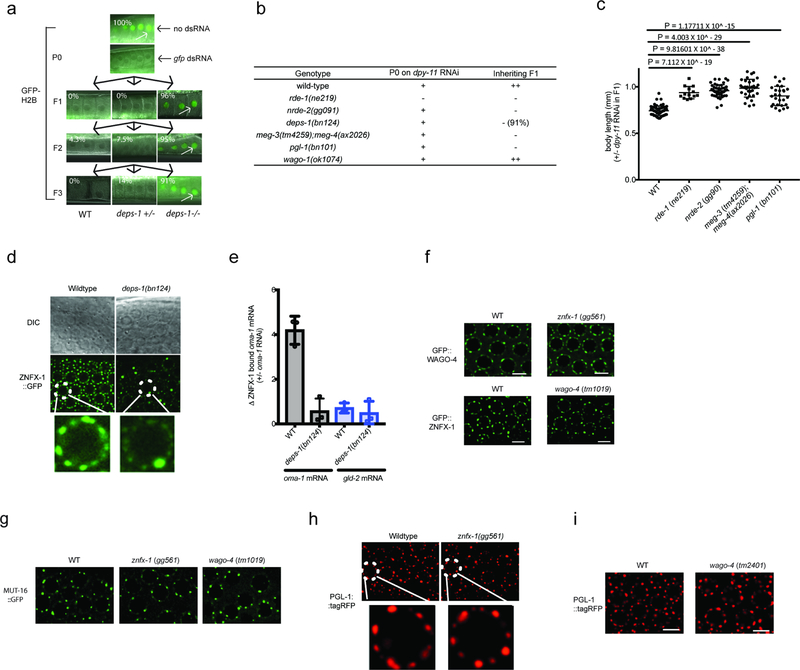
a, We conducted a genetic screen to identify components of the C. elegans RNAi inheritance machinery. The screen contained several filters (see below) to remove known RNAi inheritance factor and, therefore, to allow us to focus on new RNAi inheritance factors. We know that factors defective for RNAi inheritance are also defective for nuclear RNAi 6. Therefore, our screen began with selections for mutant alleles that disrupt nuclear RNAi. We have developed two selections for nuclear RNAi mutants. First, The lin-15b and lin-15a genes are transcribed as a polycistronic message that is spliced within the nucleus into lin-15b and lin-15a mRNAs 32. Animals harboring mutations in both lin-15b and lin-15a exhibit a multivulva (Muv) phenotype 33,34. RNAi targeting lin-15b (in eri-1(−) animals) silences lin-15b and lin-15a co-transcriptionally, thus inducing a Muv phenotype 35. The previously identified nuclear RNAi factors are required for lin-15b RNAi-induced co-transcriptional silencing of lin-15a and, therefore, for lin-15b RNAi induced Muv. A second assay for nuclear RNAi is lir-1 RNAi. lir-1 RNAi is lethal because lir-1 is in an operon with lin-26, and co-transcriptional silencing of lin-26 by lir-1 RNAi causes lethality 35. Nuclear RNAi defective (NRDE) animals do not die in response to lir-1 RNAi because they fail to silence lin-26 35. Previous genetic screens in the lab have used suppression of lir-1 RNAi to find factors required for nuclear RNAi. These screens have reached saturations: we have identified multiple alleles in all the nrde genes using this approach. Unpublished work from the lab shows, however, that hypomorphic alleles of the nrde genes will often block lin-15b RNAi-induced Muv and yet still die in response to lir-1 RNAi. We interpret these data to mean that survival from lir-1 RNAi is a much stronger selection for nuclear RNAi mutants than a failure to form Muv in response to lin-15b RNAi. In other words, factors contributing to nuclear RNAi, but not being 100% required for nuclear RNAi, would not be identified by lir-1 RNAi suppression screens. For these reasons, our screen looked for suppressors of lin-15b RNAi, which did not suppress lir-1 RNAi, because we felt this screen might identify genes missed in our previous genetic screens. Step #1. Identify factors required for nuclear RNAi. eri-1(mg366) animals were mutagenized with EMS. F2 progeny were exposed to bacteria expressing lin-15b dsRNA. Non-Muv animals were kept as candidate novel nuclear RNAi factors. Step #2. Discard known nuclear RNAi factors. We know all non-essential genes that can mutate to suppress lir-1 RNAi. Therefore, we discarded mutants that suppressed lir-1 RNAi as these alleles are likely known nuclear RNAi factors. Mutants that did not suppress lir-1 may harbor mutations in factors important, but not essential, for nuclear RNAi. Step #3. Identify mutations that suppress RNAi inheritance. The last filter in our screen was to identify mutant alleles that disrupted RNAi inheritance. To do this, we subjected remaining mutant animals to dpy-11 RNAi. dpy-11 RNAi causes animals exposed to dpy-11 dsRNA to become Dumpy (Dpy). Progeny of animals exposed to dpy-11 dsRNA inherit dpy-11 silencing and are Dpy 36. RNAi inheritance mutants become Dpy in response to dpy-11 RNAi, however, the progeny of these animals fail to inherit dpy-11 silencing, and, therefore, are not Dpy. Thus, any of our mutant animals that became Dpy in response to dpy-11 RNAi, but whose progeny were not Dpy, were kept for further analysis. Finally, only one mutant was kept from each pool (pools were maintained as independent populations throughout the screen). b-c, The data in panels B and C show that the alleles of znfx-1 and wago-4 identified by our screen are (as expected) defective for lin-15b RNAi and not defective for lir-1 RNAi (the mean of n>3 biologically independent samples, +/− SD is shown).
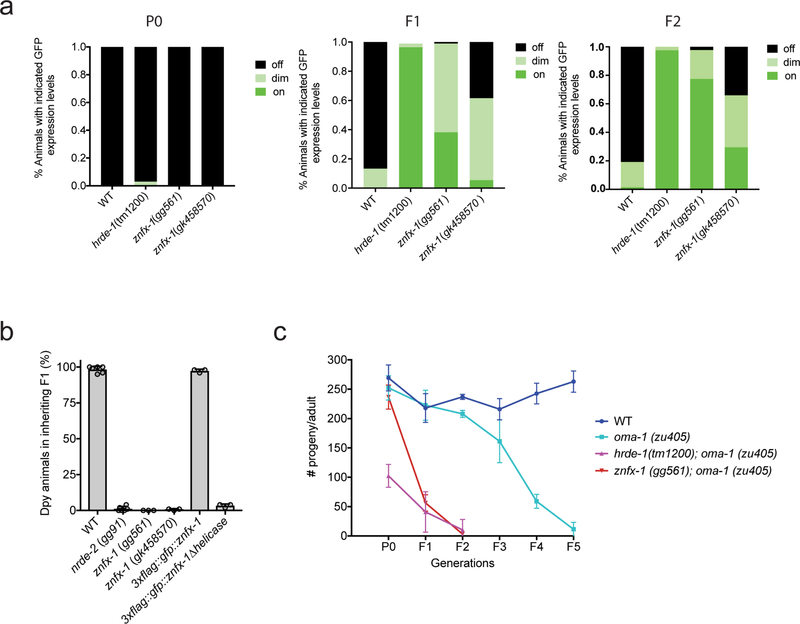
Extended Data figure 2. ZNFX-1 is required for RNAi inheritance.
a, Animals expressing a pie-1::gfp::h2b transgene were exposed to gfp dsRNA 4. The % of P0, F1 and F2 progeny, of the indicated genotypes, expressing GFP was quantified. Data represent scoring of at least 80 animals in each generation and for each genotype. Note, the gfp reporter transgene used in this study is a multi-copy version of the single copy version used in Figure. 1b of the main text. Also note, that some RNAi inheritance can be seen in znfx-1 mutant animals using this reporter transgene. Thus, in some cases, some RNAi inheritance can occur in the absence of ZNFX-1. b, Animals of indicated genotypes were exposed to dpy-11 dsRNA. The F1 progeny of these animals, were grown in the absence of dpy-11 dsRNA, and were scored for Dpy phenotypes. (the mean of n>3 biologically independent samples, bars, S.D is shown). Consistent with the idea that ZNFX-1 (and NRDE-2) is required specifically for inheritance, znfx-1 mutant animals exposed directly to dpy-11 dsRNA are Dpy (data not shown). c, zu405ts is a temperature sensitive (ts) lethal (embryonic arrest at 20ºC) allele of oma-1 37. oma-1 RNAi suppresses oma-1(zu405ts) lethality, and this effect is heritable 5,6. Animals of the indicated genotypes were exposed to oma-1 dsRNA and the fertility of the progeny of these animals was scored over generations. (the mean of n=3 biologically independent samples, +/− SD is shown). Data show that znfx-1 mutant animals are defective for oma-1 RNAi inheritance.
Extended Data figure 3. CRISPR/Cas9-epitope tagged genes used in this study, with one exception b, produce functional proteins and are expressed at or near wild-type levels.
The data in a panels a-h show that the addition of epitope tags by CRISPR/Cas9-mediated gene conversion of znfx-1 or wago-4 did not affect function of tagged proteins in these RNAi inheritance. (a-e) show dpy-11 RNAi inheritance assays in which the progeny of animals exposed to dpy-11 dsRNA are visually scored for the inheritance of Dpy phenotypes. These data show that the indicated epitope tagged proteins are functional in this RNAi inheritance assay (n=3 biologically independent samples, bars, S.D). (f-g) pgl-1 mutant animals show a temperature sensitive (25C) sterile phenotype. Addition of epitope tags by CRISPR/ Cas9-mediated gene conversion to pgl-1 locus did not affect PGL-1 function as these animals were fertile. L4 animals were singled from 20℃ to 25℃ and brood sizes were scored. (h) pgl-1-tagrfp (n=6 animals) and pgl-1::mcardinal; tagrfp::znfx-1; mut-16::gfp (n=15 animals). mut-16(−) animals are defective for pos-1 RNAi. Embryos of indicated genotype were grown on pos-1 dsRNA expressing bacteria. 6 L4 animals were picked to pos-1 dsRNA expressing bacteria and laid egg overnight. Unhatched embryos and hatch animals were scored. The data shows that addition of gfp to mut-16 locus did not affect MUT-16 function. (the mean of n=3 biologically independent samples, bars, S.D is shown). The data in (i-k) show that in some case, FLAG::GFP::WAGO-4 expressing animals are defective in RNAi inheritance, indicating that FLAG::GFP::WAGO-4 is not fully functional. (i) Animals of indicated genotypes were exposed to dpy-11 dsRNA and F1 progeny were grown in the absence of dpy-11 dsRNA. % of Dpy animals is shown. At least 150 animals of each genotype were scored. These data show that 3xFLAG::GFP::WAGO-4 is not functional for dpy-11 inheritance. (n=3 biologically independent samples). (j) This panel shows that 3xFLAG::GFP::WAGO-4 is functional during oma-1 RNAi inheritance. See Extended Data figure 2c for details of oma-1 RNAi inheritance assay. (n=3 biologically independent samples, bars, S.D). In figure 2d of the main text, we show that both wago-4 and znfx-1 exhibit mortal germline (Mrt) phenotypes at 25C. The data in this panel show that 3xflag::gfp::wago-4 animals are not Mrt, indicating that 3xFLAG::GFP:: WAGO-4 is capable of promoting germline immortality. (n=3 biologically independent samples, bars, S.D). The data in (l-m) show that CRISPR tags did not seem to affect gene expression. To address the possibility that epitope tagging of the genes used in this study changed gene expression levels we isolated total RNA from animals of the indicated genotypes and used qRT-PCR to quantify indicated mRNA levels. Primers target exon-intron junctions. Early stop or deletion alleles for each of these loci were used as controls. wago-4(tm1019) and znfx-1(gg561) are deletions and primers were located within deleted regions. pgl-1(bn101) and mut-16 (pk710) are nonsense alleles. Decrease of mRNA levels in these mutants is likely due to nonsense mediated decay (n=3 biologically independent samples, +/−SD).
Extended Data figure 4. ZNFX-1 acts specifically during the inheriting phase of RNAi.
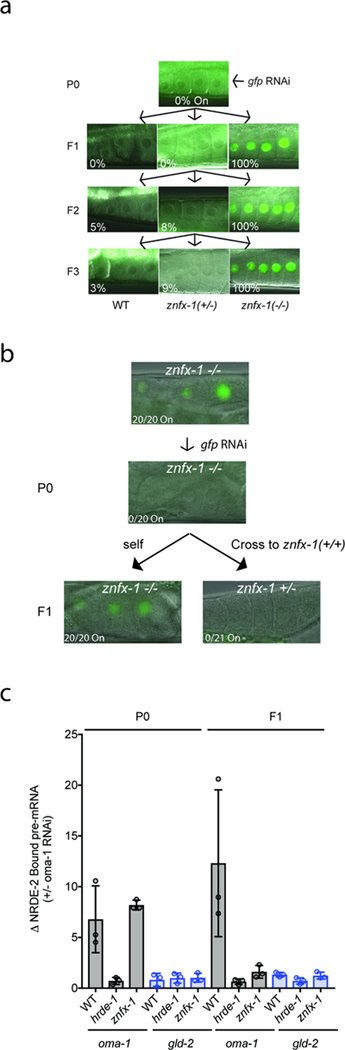
a, The data in (a )show that znfx-1 is required in inheriting generations for RNAi inheritance to occur. Briefly, we initiated gene silencing in znfx-1/+ heterozygous animals and scored the +/+ and −/− progeny for their ability to inherit gene silencing. Progeny harboring at least one wild-type copy of znfx-1 were capable of inheriting gene silencing while −/− progeny were not. More specifically, znfx-1(gg561) +/− animals expressing pie-1::gfp::h2b transgene 38 were exposed to gfp dsRNA, progeny from F1 to F3 generations were scored. Micrographs of GFP fluorescence in oocytes are shown. In order to identify cross progeny the following strategy was employed via CRISPR. pie-1::gfp::h2b was marked by dpy-10 (cn64) (dpy-10 is ~0.77 cM from pie-1::gfp::h2b). dpy-10 (cn64) /+ animals are Dpy Rol and dpy-10 (cn64) homozygous animals are Dpy. znfx-1 genotypes was inferred based upon WT, Dpy, and Rol phenotypes, n>30 animals. b, The data in (b) show that znfx-1 is sufficient in inheriting generations for RNAi inheritance to occur. We initiated gene silencing in znfx-1(−/−) animals, introduced a wild-type copy of znfx-1 to progeny (via mating), and scored znfx-1/+ cross-progeny for inheritance. The data show that znfx-1(+/−) progeny, from parents that lack a wild-type copy of znfx-1, were able to inherit silencing. znfx-1(gg561) was marked by dpy-10 (cn64) (dpy-10 is ~1.09 cM from znfx-1). dpy-10 (cn64) /+ animals are Dpy Rol and dpy-10 (cn64) homozygous animals are Dpy. znfx-1 genotypes was inferred based upon WT, Dpy, and Rol phenotypes. n>20 animals. c, The data in (c) show provide additional biochemical evidence that ZNFX-1 acts in inheriting generations to promote inheritance. The nuclear RNAi factor NRDE-2 binds to pre-mRNA of genes undergoing heritable silencing 6. When znfx-1(−) animals were exposed directly to oma-1 dsRNA, NRDE-2 bound to the oma-1 pre-mRNA at wild-type levels, however, in progeny of znfx-1(−) mutant animals NRDE-2 failed to bind oma-1 pre-mRNA. Animals expressing NRDE-2::3xFLAG were treated with +/− oma-1 RNAi. Extracts were generated from these animals as well as the progeny of these animals (which were not treated directly with oma-1 RNAi). NRDE-2::3xFLAG was IP’ed with an anti-FLAG antibody and NRDE-2 co-precipitating oma-1 pre-mRNA was quantified by qRT-PCR with exon/intron primer sets designed to detect unspliced RNAs (pre-mRNAs) of the oma-1 gene as well as a control germline expressed pre-mRNA gld-2. Data are expressed as ratio of signals ± oma-1 RNAi. (the mean of n=3, bars, S.D is shown).
Extended Data Figure 5. WAGO-4 is an Argonaute that localizes to the peri-nucleus and is required for RNAi inheritance.
a, oma-1(zu405) is a temperature sensitive lethal (embryonic arrest at 20C) allele of oma-1. oma-1 RNAi suppresses oma-1(zu405) lethality and this effect is heritable 5. Animals of indicated genotypes were exposed to oma-1 dsRNA and F1 to F5 progeny were grown in the absence of oma-1 dsRNA. Number of viable progeny of P0 (directly exposed to oma-1 RNAi) and inheriting generations (F1 to F6, grown in the absence of oma-1 RNAi) were scored (20℃). (the mean of n=3 biologically independent samples, bars, S.D is shown). b, Animals of the indicated genotypes and expressing a pie-1::gfp::h2b transgene were exposed to gfp dsRNA 38. Micrographs of animals +/− gfp RNAi as well as the F1 progeny of these animals are shown. % of animals expressing GFP is indicated. Percentages represent scoring of at least 90 animals in each generation and for each genotype. c, We used CRISPR/Cas9 to append a gfp tag upstream of the predicted wago-4 atg start codon. Top panels, fluorescent micrographs of gfp::wago-4 in 2 cell, 4 cell, and ~300 cell embryos. Bottom pane, fluorescent micrograph of the germline of an adult gfp::wago-4 animal. Images are representative of >3 animals at each lifestage.
Extended Data figure 6. Visualization of Z granule formation with antibodies targeting PGL-1 (P granule) and HA::ZNFX-1 (Z granule).
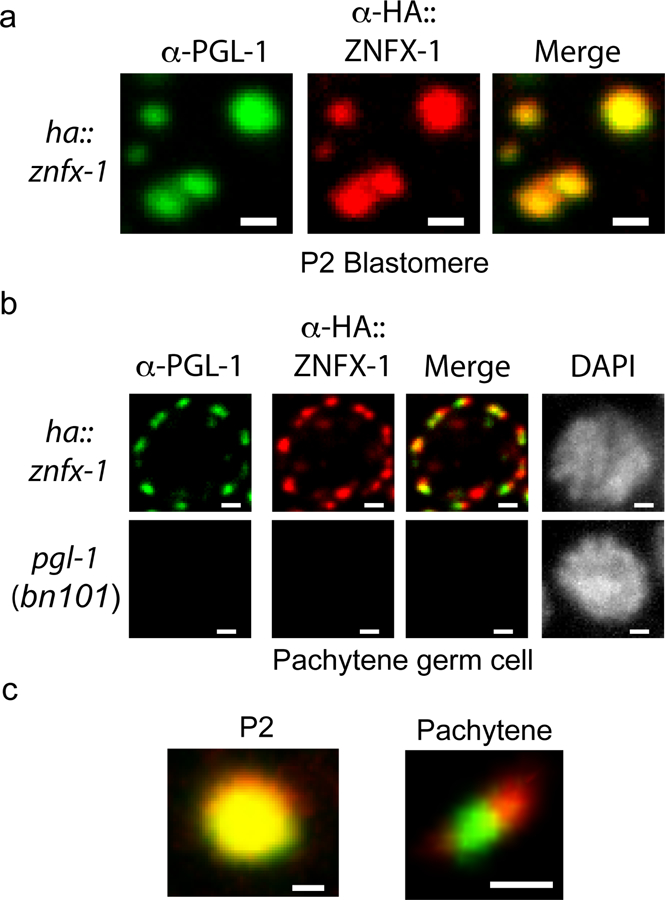
To control for possible artifacts caused by fluorescent epitopes we conducted immunofluorescence on HA::ZNFX-1 expressing animals using α-PGL-1 (K76 from Developmental Studies Hybridoma Bank) and α-HA (Abcam ab9110) antibodies. a, α-PGL-1 and α-HA signals colocalized in the P2 blastomeres of 4 cell embryos. b, α-PGL-1 and α-HA signals were adjacent, yet distinct in, in pachytene germ cells. No PGL-1 or HA::ZNFX-1 signal was detected in pgl-1(bn101) animals, which do not express PGL-1 or HA::ZNFX-1, establishing that IF signals were specific. c, Magnification of foci from (a-b). Images in (a-c) are representative of 3 independent animals at each life stage. Scale bars for (a) = 1 um, (b) = 1 um, (c) = 0.5 um.
Extended Data Figure 7. Five experiments lumped together so that we don’t have more than ten total Extended Data files.
a, The data in (a) show that during oocyte maturation ZNFX-1 foci detach from nuclei, assume spherical shapes, and move away from the nucleus; behavior consistent with Z foci being liquid-like condensates. Additionally, the data show that Z foci can exist at developmental stages during which P granules are no longer visible, indicating that Z foci can be temporally separated from P granules. Image is of maturing oocytes of animals expressing the indicated fluorescent proteins. Long arrows indicates oocyte that contains Z granules, but not P granules. −1 indicates −1 oocyte. Image representative of >3 animals. b, The data in (b) show that Z foci exhibit properties reminiscent of liquid droplets. Left, GFP::ZNFX-1 expressing animals were subjected to FRAP (see methods) and fluorescence was monitored in bleached area over indicated time. Data is normalized to a non-bleached control granule from same sample. Mean of n=7 individual granules from 7 animals +/− SEM is shown for both life stages. Right, heat maps showing recovery of ZNFX-1 fluorescence in a representative bleached Z granule. c, The data in (c) show that Z foci do not colocalize with other known liquid droplets. GFP::ZNFX-1 does not colocalize with markers of processing bodies. PATR-1 and DCAP-1 localize to processing bodies 18. Fluorescent micrographs of somatic blastomeres of embryos expressing the indicated fluorescent proteins. The data show that ZNFX-1 does not colocalize with markers of processing bodies in these cells. Images are representative of >3 independent animals. d, The data in (d) show that Z foci in adult pachytene germ cells do not colocalize with another marker of P granules (EGO-1). ZNFX-1 foci form adjacent to EGO-1 foci. Fluorescent micrographs of a single pachytene germ cell nucleus from animals expressing GFP::ZNFX-1 and TagRFP::EGO-1. 3D renders of a representative foci is shown below. Images are representative of 3 independent animals. Scale Bar = 0.5 um. e, The data in (e) show that Z foci can be physically separated from P granules. Gonads were isolated from animals expressing GFP::ZNFX-1 and PGL-1::TagRFP and subjected to shearing force as described10. Time lapse imaging at 10 second intervals is shown. A PGL-1-labeled P granule detaching from the nucleus and flowing throughout the cytoplasm is shown(large arrow). ZNFX-1 labeled Z granules remain immobile (small arrow). Physical shearing was induced as previously described10. Briefly, GFP::ZNFX-1 and PGL-1::TAGRFP adults were dissected to extrude gonads. Isolated gonads were squeezed between two coverslips to generate shearing force. Coverslips were then mounted on a slide and imaged immediately with a spinning disc confocal. Z stacks were acquired every 10 seconds. Image is representative of 4 independent animals.
Extended Data Figure 8. Quantification of centers and surfaces of fluorescence for Z granules, P granules, and Mutator foci.
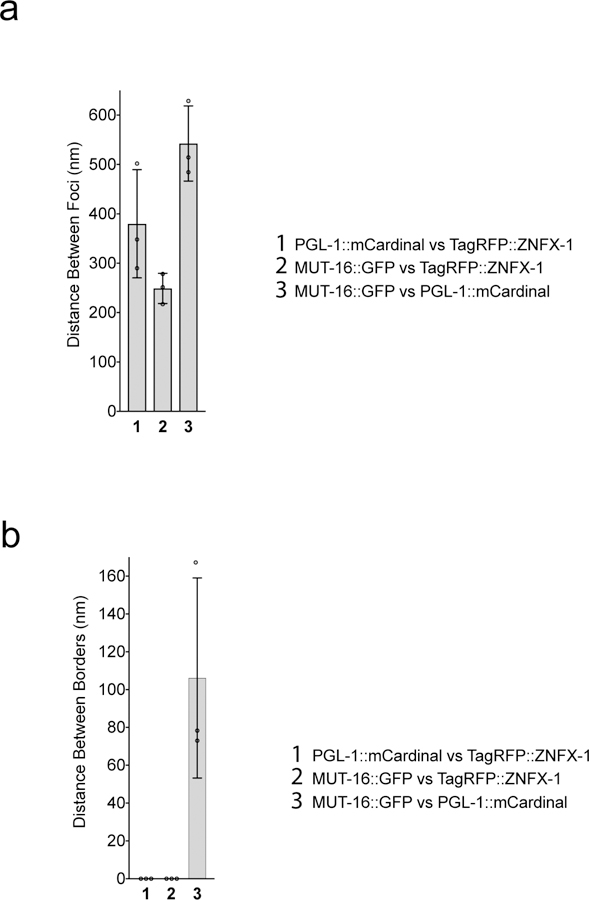
Distance between centers (a) and surfaces (b) of the spaces occupied by PGL::mCardinal, tagRFP::ZNFX-1, and MUT-16::GFP were calculated as described in Methods. The mean of 10 granule measurements across 3 independent animals +/− SD is plotted. Distances in (a-b) have been corrected for chromatic shift.
Extended Data Figure 9. P granule assembly factors contribute to RNAi inheritance, normal Z granule morphology, and the ability of ZNFX-1 to bind mRNAs.
a, DEPS-1 is required for P granule formation in adult germ cells (13,39,40. deps-1 (bn124)/+) animals expressing pie-1::gfp::h2b transgene 38 were exposed to gfp dsRNA. Progeny were grown in absence of gfp dsRNA for three generations. Fluorescent micrographs show GFP expression in oocytes. % of animals expressing pie-1::gfp::h2b is shown. These data show that DEPS-1 activity is required in inheriting generations to allow for gfp RNAi inheritance. (n=32 animals for P0, n>100 for F1 to F3). b, MEG-3/4 and PGL-1 also contribute to P granule formation 13,39,40. dpy-11 RNAi causes animals exposed to dpy-11 dsRNA to become Dumpy (Dpy). Progeny of animals exposed to dpy-11 dsRNA inherit dpy-11 silencing and are Dpy 36. RNAi inheritance mutants become Dpy in response to dpy-11 RNAi however, progeny fail to inherit dpy-11 silencing, and, therefore, are not Dpy. Animals of indicated genotypes were exposed to dpy-11 dsRNA. F1 progeny were grown in the absence of dpy-11 dsRNA. (−) indicates Non-Dpy, (+) indicates mild Dpy phenotype, (++) indicates strong Dpy (pgl-1, deps-1, and meg-3/4) are defective for dpy-11 RNA inheritance. n=3 biologically independent samples for each condition. c, Animals of indicated genotypes were exposed to dpy-11 dsRNA and F1 progeny were grown in the absence of dpy-11 dsRNA. Body length of F1 animals were measured by Image J. Data are expressed as body length from progeny of dpy-11 RNAi treated animals divided by average body length from control animals. The mean of n>12 animals with P values calculated by student’s two tail t test is shown. d, In deps-1(bn124) animals, most Z granules are smaller than normal while one Z granule/nucleus becomes enlarged. Images are from pachytene region of germline. Images are representative of >3 animals. e, In deps-1(bn124) animals, ZNFX-1 doesn’t bind RNA. Wild-type or 3xFLAG::ZNFX-1 expressing animals were treated with oma-1 dsRNA. ZNFX-1 was IP’ed in RNAi generation with α-FLAG antibodies and co-precipitating RNA was subjected to qRT-PCR to quantify oma-1 mRNA co-precipitating with ZNFX-1 in wild-type or deps-1(bn124) animals. gld-2 is a germline expressed control mRNA. The mean of n=3 biologically independent samples. +/−SD is shown. Panels (f–i) show that loss of ZNFX-1 or WAGO-4 does not seem to affect the formation of (f) Z granules marked by GFP::WAGO-4 or GFP::ZNFX-1, (g) Mutator foci marked by MUT-16::GFP, or (h,i) P granules marked by PGL-1::RFP. Note, in late embryonic germline development, PGL-tagRFP foci may not be efficiently concentrated into Z2/Z3 in wago-4 mutant (data not shown). (f-i) are representative images from n>3 animals.
Extended Data Figure 10. Working model for role of PZM assemblages in RNAi inheritance.
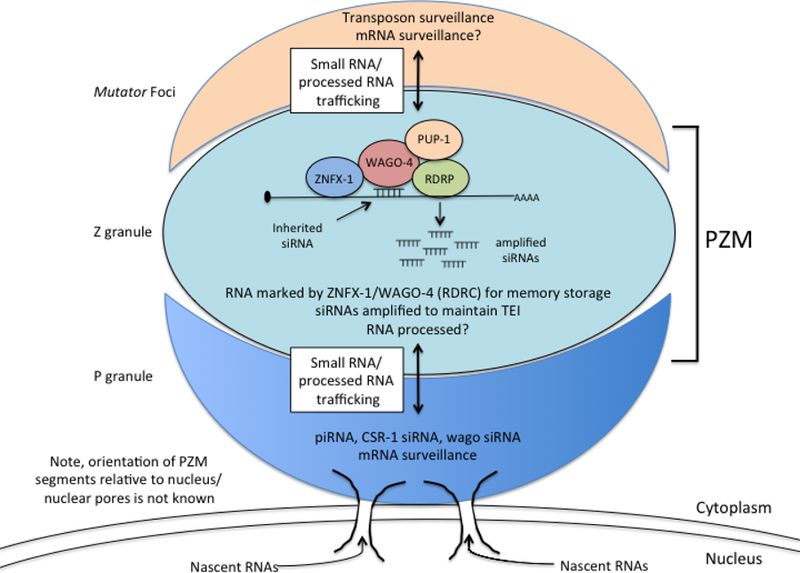
P granules make contacts with nuclear pores 24,26. The relationship between the Z and M segments of the PZM granule and nuclear pores are not yet known.
Supplementary Material
Acknowledgements:
We thank members of the Kennedy lab for discussions. We thank Takao Ishidate and Craig Mello for sharing unpublished data. We thank Ho Yi Mak for sharing strains. Some strains were provided by the CGC (P40 OD010440). Some strains were provided by the MITANI Lab. This work was supported by the National Institutes of Health, RO1 GM088289 (S.K.). B.D.F and A.S. were supported by NSF graduate research fellowships.
Footnotes
The authors declare no competing financial interests.
Code Availability. Custom python scripts used to generate small RNA plots in Figure 1f are available upon request.
Data Availability. Small RNA sequencing data that support the findings of this study have been deposited in the Expression Omnibus Database (GEO) with the accession code GSE112109. Source data for Figure 2b,2d is located in Supplemental File 1. The remaining data that support the findings of this study are available from the corresponding author upon reasonable request.
Bibliography:
- 1.Heard E & Martienssen RA Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim JP & Brunet A Bridging the transgenerational gap with epigenetic memory. Trends Genet 29, 176–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Vastenhouw NL et al. Gene expression: long-term gene silencing by RNAi. Nature 442, 882 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Alcazar RM, Lin R & Fire AZ Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 180, 1275–1288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley BA et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spracklin G et al. The RNAi Inheritance Machinery of Caenorhabditis elegans. Genetics 206, 1403–1416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashe A et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brogna S, McLeod T & Petric M The Meaning of NMD: Translate or Perish. Trends Genet 32, 395–407 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Brangwynne CP et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Strome S & Wood WB Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35, 15–25 (1983). [DOI] [PubMed] [Google Scholar]
- 12.Strome S & Wood WB Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 79, 1558–1562 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JT et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife 3, e04591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JT et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife 3, e04591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toretsky JA & Wright PE Assemblages: functional units formed by cellular phase separation. J. Cell Biol 206, 579–588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber SC & Brangwynne CP Getting RNA and protein in phase. Cell 149, 1188–1191 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Phillips CM, Montgomery TA, Breen PC & Ruvkun G MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev 26, 1433–1444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo CM, Munro E, Rasoloson D, Merritt C & Seydoux G Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol 323, 76–87 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Chu J, Haynes RD, Corbel SY & Li P Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nature (2014). [DOI] [PMC free article] [PubMed]
- 20.Grishok A, Tabara H & Mello CC Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494–2497 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Shirayama M et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motamedi MR et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Seydoux G & Dunn MA Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124, 2191–2201 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Pitt JN, Schisa JA & Priess JR P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol 219, 315–333 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Furuhashi H et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin 3, 15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheth U, Pitt J, Dennis S & Priess JR Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137, 1305–1314 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farboud B & Meyer BJ Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199, 959–971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arribere JA et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gent JI et al. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics 183, 1297–1314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ollion J, Cochennec J, Loll F, Escudé C & Boudier T TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolte S & Cordelières FP A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Blumenthal T et al. A global analysis of Caenorhabditis elegans operons. Nature 417, 851–854 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Clark SG, Lu X & Horvitz HR The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137, 987–997 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang LS, Tzou P & Sternberg PW The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5, 395–411 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guang S et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321, 537–541 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burton NO, Burkhart KB & Kennedy S Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 108, 19683–19688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin R A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol 258, 226–239 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Ashe A et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawasaki I et al. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Spike CA, Bader J, Reinke V & Strome S DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development 135, 983–993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.