Abstract
Using gene modification of hematopoietic stem cells (HSC) to create persistent generation of multilineage immune effectors to target cancer cells directly is proposed. Gene-modified human HSC have been used to introduce genes to correct, prevent, or treat diseases. Concerns regarding malignant transformation, abnormal hematopoiesis, and autoimmunity exist, making the co-delivery of a suicide gene a necessary safety measure. Truncated epidermal growth factor receptor (EGFRt) was tested as a suicide gene system co-delivered with anti-CD19 chimeric antigen receptor (CAR) to human HSC. Third-generation self-inactivating lentiviral vectors were used to co-deliver an anti-CD19 CAR and EGFRt. In vitro, gene-modified HSC were differentiated into myeloid cells to allow transgene expression. An antibody-dependent cell-mediated cytotoxicity (ADCC) assay was used, incubating target cells with leukocytes and monoclonal antibody cetuximab to determine the percentage of surviving cells. In vivo, gene-modified HSC were engrafted into NSG mice with subsequent treatment with intraperitoneal cetuximab. Persistence of gene-modified cells was assessed by flow cytometry, droplet digital polymerase chain reaction (ddPCR), and positron emission tomography (PET) imaging using 89Zr-Cetuximab. Cytotoxicity was significantly increased (p = 0.01) in target cells expressing EGFRt after incubation with leukocytes and cetuximab 1 μg/mL compared to EGFRt+ cells without cetuximab and non-transduced cells with or without cetuximab, at all effector:target ratios. Mice humanized with gene-modified HSC presented significant ablation of gene-modified cells after treatment (p = 0.002). Remaining gene-modified cells were close to background on flow cytometry and within two logs of decrease of vector copy numbers by ddPCR in mouse tissues. PET imaging confirmed ablation with a decrease of an average of 82.5% after cetuximab treatment. These results give proof of principle for CAR-modified HSC regulated by a suicide gene. Further studies are needed to enable clinical translation. Cetuximab ADCC of EGFRt-modified cells caused effective killing. Different ablation approaches, such as inducible caspase 9 or co-delivery of other inert cell markers, should also be evaluated.
Keywords: HSC, CAR, cancer immunotherapy, EGFR, cetuximab, gene therapy
Introduction
In several clinical trials, gene therapy using hematopoietic stem cells (HSC) has shown promise for monogenic disorders of hematopoietic progeny, such as adenosine deaminase-deficient severe combined immunodeficiency (SCID),1 X-linked SCID,2 chronic granulomatous disease,3 Wiskott–Aldrich disease,4,5 and lysosomal disorders.6,7 In addition, trials are underway for beta thalassemia8 and sickle cell disease,9 and gene-modified HSC for use in cancer immunotherapy have also been investigated.10–13
Genotoxicity has been a major concern, as a number of patients in some trials using retroviral vectors developed clonal hematopoietic disorders such as T-cell leukemia,14,15 acute myeloid leukemia,16 and myelodysplastic syndrome,17 all associated with vector integration near proto-oncogenes. To date, the results have been much better with lentiviral vectors with built-in safety mechanisms. However, long-term safety data are not available. In adoptive T-cell cancer immunotherapy, including T-cell receptor (TCR) and chimeric antigen receptor (CAR) therapies, there has been interest in suicide systems. Cytokine release syndromes18 and on-target, off-site toxicities have resulted in several deaths.19,20 If gene-modified allogeneic cells are used, graft versus host disease is also a concern. Therefore, several suicide systems have been under development.12
One of these suicide systems involves the use of a truncated human epidermal growth factor (EGF) receptor polypeptide (EGFRt) with the clinically available chimeric immunoglobulin G1 anti-EGFR monoclonal antibody, cetuximab (Erbitux™).21–24 Truncation of the intracellular domains leads to a lack of intracellular receptor tyrosine kinase activity, and truncation of the extracellular domains leads to the inability to bind EGF and other ligands. EGFRt does, however, retain type I transmembrane cell surface localization and a conformationally intact binding epitope for cetuximab. Lentiviral transduction of human T cells with a CAR and EGFRt resulted in EGFRt successfully providing a cell surface marker for tracking adoptively transferred transduced cells using immunohistochemistry and flow cytometry.21,22 It also provides a target for cetuximab-mediated antibody-dependent cellular cytotoxicity (ADCC), leading to in vivo elimination.21,22 For these considerations, clinical trials have been initiated using T cells modified with this construct co-delivering CAR and EGFRt (named EQ21,22).
In the search for a cancer immunotherapy approach to establish persistent activity with an engineered “off-switch” safety, this study aimed to evaluate EGFRt as a suicide system for human HSC in the setting of gene therapy. It was hypothesized that gene modification with EGFRt would not significantly affect HSC proliferation and differentiation, but in the presence of cetuximab would lead to ablation of the gene-modified HSC and their progeny. In addition, the study aimed to understand the mechanisms of ablation, as patients receiving these therapies might have incomplete immune reconstitution.
Methods
Lentiviral vectors
The construction of pCCL-MNDU3-eGFP has been described previously.9 Vector constructs for both huEGFRt alone and huEGFRt combined with an anti-CD19 second-generation CAR with the CD28 costimulatory molecule and CD3ζ chain (EQ) were developed as described21,22 and generously provided by Stephen Forman (City of Hope, Duarte, CA). Relevant sequences were cloned onto a CCL vector backbone25 with MND LTR U3 as an internal enhance/promoter26 to make lentiviral vectors, which were denominated CCL-MNDU3-EGFRt (EGFRt) and CCL-MNDU3-CD19CARCD28-EGFRt (EQ), respectively. Lentiviral vectors were packaged with a VSV-G pseudotype.
Human cell lines
Cytotoxicity target cells were Jurkat cells (ATCC, Manassas, VA) maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, Damstadt, Germany) containing 10% fetal bovine serum (FBS; R10 medium).
Isolation of primary human cells
Collection of anonymous human cord blood units from delivery rooms at UCLA (Los Angeles, CA) and use of human peripheral blood cells were deemed exempt from need for formal approval by the Institutional Review Board at UCLA. After FiColl Hypaque (Stem Cell Technologies, Vancouver, Canada) isolation of mononuclear cells from fresh umbilical cord blood, human CD34+ cells were isolated using immunomagnetic beads (MACS CD34 MicroBead Cell Separation Kit; Miltenyi, Auburn, CA) at enrichment >70% and stored in liquid nitrogen. Anonymous human peripheral blood samples were obtained from the UCLA CFAR Virology Core Laboratory. Whole leukocytes were isolated using Hetasep (Stem Cell Technologies). Samples were enriched for natural killer (NK) cells by negative selection using the RosetteSep system (Stem Cell Technologies). Enriched samples were immediately used for cytotoxicity experiments.
Vector production and transduction of cell lines and primary human cells
Vector-containing supernatant was harvested from transfected HEK293T cells treated with sodium butyrate, and large-scale preparations (2–5 L) were concentrated by tangential flow filtration, with titers measured by vector copy number (VCN) assessment in transduced HT29 cells, as previously described.27
To generate stably transduced target cells, lentiviral vectors were added to 1 × 105 Jurkat cells at a concentration of >1 × 108 TU/mL and allowed to incubate for 24 h, expanding cells in R10 medium.
For transduction of human CD34+ cells,10–13 thawed cells were pre-stimulated for 14–18 h in X-Vivo15 medium (Lonza, Basel, Switzerland) containing 1 × L-glutamine/penicillin/streptomycin (L-Glut/Pen/Strep; Gemini BioProducts, West Sacramento, CA), enriched with recombinant human (rhu) SCF (50 ng/mL), rhuFlt-3 ligand (50 ng/mL), and rhuThrombopoietin (50 ng/mL; cytokines from R&D Systems, Minneapolis, MN). Transduction was performed for 24 h with the addition of lentiviral vectors at a concentration of 5.5 × 107 TU/mL onto 105 cells in 1 mL transduction medium in RetroNectin-precoated 48-well plates, as previously described.10–13
Proliferation and differentiation cultures of primary human HSC
Myeloid differentiation was performed in culture for 12–14 days in Iscove's modified Dulbecco's medium (IMDM; Mediatech, Inc., Manassas, VA) containing L-Glut/Pen/Strep and 10% FBS, enriched with cytokines rhuSCF (100 ng/mL) and rhuIL-3 (100 ng/mL) from day 1, with the addition of rhuGM-CSF (10 ng/mL) from day 3.10,28 Short-term proliferation of human HSC was performed for 9 days and cultured in basal bone marrow medium (BBMM): IMDM enriched with 20% FCS, 0.5% bovine serum albumin, 5 ng/mL rhuIL-3, 10 ng/mL rhuIL-6, and 25 ng/mL rhuFlt-3 ligand.27 Cell concentrations were maintained at <5 × 105 per well.
Clonogenic assays
Erythro-myeloid colony formation was assayed by culturing human HSC immediately after transduction in duplicate 35 mm gridded cell culture dishes, using complete methylcellulose (MethoCult™ H4435 Enriched, Stem Cell Technologies) for 14 days. Colony-forming units (CFU) on plates were then counted under microscopy, and colony type was scored based on morphology. Plating efficiency was calculated as 100% × (total number CFU counted/total number cells plated).29 Colonies were then collected for genomic DNA extraction and quantitative droplet digital PCR (ddPCR) analysis.
Cytotoxicity assays
Direct antibody cytotoxicity was assessed by culturing EGFRt-transduced and non-transduced Jurkat cells with different concentrations of cetuximab antibody—0.3, 1, and 10 μg/mL—or vehicle alone. Complement-dependent cytotoxicity was assessed by culturing EGFRt-transduced and non-transduced Jurkat cells with different dilutions of rabbit complement, with 1 μg/mL of cetuximab or vehicle (phosphate-buffered saline [PBS]) alone. Target cell cytotoxicity was measured by incubating cells with 4′,6-diamidino-2-phenylindole (DAPI; BD Biosciences, San Jose, CA) at room temperature for 15 min. Cells positive for DAPI by flow cytometry were counted as nonviable.
Assays for ADCC were performed using the flow-cytometry-based Live/Dead Cell Mediated Cytotoxicity Kit (Invitrogen),10 with 2 × 104 EGFRt-transduced or non-transduced Jurkat target cells/well pre-labeled with 3,3′-dioctadecyloxacarbocyanine (DiOC). Effector cells were added at different effector:target (E:T) ratios, with or without cetuximab at a concentration of 1 μg/mL. To determine maximal lysis, 20% ethanol30 was added to target cells. Cell mixtures were incubated in propidium iodide (PI) containing R10 for 4–6 h. DiOC+ cells were counted as target cells, and cells double positive for DiOC and PI were counted as nonviable target cells. All samples were loaded in triplicate, and all assays had controls with isolated effectors and targets.
Determination of VCN
Genomic DNA of cell samples was extracted using NucleoSpin Tissue XS (Macherey-Nagel, Düren, Germany) or a PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA). VCN was determined by multiplex ddPCR of the HIV-1 packaging signal sequence in the provirus and normalized to the cellular autosomal gene SDC4 to calculate the average VCN.9,31
Flow cytometry
To evaluate the cell expression of huEGFRt, transduced cells were washed with blocking solution and incubated with biotinylated cetuximab antibody. Pharmaceutical-grade cetuximab (Erbitux, Eli Lilly and Company, Indianapolis, IN) was biotinylated by using the One-Step Antibody Biotinylation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Primary human cells were incubated with 1 × Human TruStain FcX Fc Receptor Blocking Solution (BioLegend, San Diego, CA) before incubation with biotinylated cetuximab, followed by incubation with streptavidin-R–phycoerythrin conjugate (BD Biosciences) after washing with blocking solution. Transduced Jurkat cells with >95% EGFRt expression served as positive controls for EGFRt detection.
NK cell markers were assessed with fluorescent-labeled murine monoclonal antibodies to human CD16-PE, CD56-APC, and CD3-PerCP. After washing, cells were analyzed on an LSR II flow cytometer (BD Biosciences).
Flow cytometry was used to analyze the cell target lysis percentage based upon the following formula: % specific lysis = 100% × [(# of nonviable target cells in co-culture with antibody, complement, or effector cells/# of target cells in co-culture) – (# of nonviable targets cultured alone/# of target cells cultured alone)]/[(# of nonviable target cells cultured in maximal lysis conditions/# of target cells cultured in maximal lysis conditions) – (# of nonviable targets cultured alone/# of target cells cultured alone)].10 DIVA analysis software (BD Biosciences) and FlowJo software (FlowJo, LLC, Ashland, OR) were used for data evaluation.
In vivo studies
NOD/SCID/γ-chainnull (NSG) mice (NOD.Cg.Prkdcscid Il2rgtm1Wjl/SzJ; stock no. 005557; Jackson Laboratory, Bar Harbor, ME) were housed in accordance with approved protocol. Newborn pups aged 3–7 days were injected with 3 × 105 cells/pup via intrahepatic injection of transduced or non-transduced human CD34+ cells 1 day after conditioning with 125 cGy of sub-lethal body irradiation from a 137Ce source with attenuator and allowed to engraft over 12 weeks.10 Engraftment was assessed by flow cytometry and VCN assessment from retro-orbital venous plexus blood sampling and harvested bone marrow and spleen.
Humanized mice received tumor challenges with subcutaneous injection of 1 × 106 CD19+ human non-Hodgkin lymphoma Raji cells on week 13 after injection of human HSC, followed by tumor size measurements every other day, as previously published.10,13 Euthanasia was performed when tumor interfered with ambulation or nutrition, obstructed any orifice opening, developed skin ulceration, or otherwise achieved a size that interfered with normal activity or body functions.
Cohorts of mice were treated with cetuximab at 1 mg/mouse intraperitoneally daily for 12 days before euthanasia, blood sampling, and spleen and bone marrow harvest, as described.21
Conjugation of cetuximab
As previously published,32,33 the pH of 75 μL (150 μg) of cetuximab was adjusted with Na2CO3, and molar excess of p-isothiocyanatobenzyl-desferrioxamine (SCN-DFO) was added for incubation at 37°C for 30 min. During incubation, a Bio-Rad Micro Bio-Spin Chromatography column was prepped per manufacturer instructions. After incubation, the sample was added to the column and placed in the centrifuge at 1,000 g for 4 min. This sample was then measured in Nanodrop to determine concentration and recovery, and run on a sodium dodecyl sulfate polyacrylamide gel with the unconjugated cetuximab in non-reducing and reducing conditions to confirm conjugation.
Cetuximab radiolabeling with 89zirconium
A previously published protocol was followed.32,33 The activity and volume of [89Zr]Zr-oxalic were measured, and 40% of 2 M Na2CO3 was added, with pH adjustment with HEPES. The desired amount of Zr-89 was added to obtain approximately 500 μCi per 100 μg of cetuximab, and then incubated for 1 h at room temperature. To determine radiolabeling efficiency, instant thin-layer chromatography (ITLC) was done using chromatography strips and citric acid 20 mM, and samples were run through a gamma counter. Radiolabeling efficiency was determined by calculating the cpm in section #1 on the strip, representing the conjugated antibody, divided by the cpm of section #1 plus the cpm of section #2. To determine radiochemical purity, a BioRad Micro Bio-Spin Chromatography column was prepped as described above. ITLC was repeated with this sample, and the purity was determined by calculating the cpm of section #1 divided by the cpm of section #1 plus cpm of section #2.
Positron emission tomography/computed tomography imaging of gene-modified NSG mice
All mice for imaging were injected via the tail vein with 10 μg 89Zr-cetuximab and imaged on Inveon positron emission tomography (PET) and CrumpCAT computed tomography (CT) at the California NanoSystems Institute at 24 and 120 h after injection. The images were processed and analyzed using Amide's Medical Imaging Data Examiner. Immediately after animal imaging, mouse tissues were dissected and imaged for biodistribution calculations, defined as percentage of injected dose per gram of tissue (% ID/g).
Statistical analysis
Descriptive statistics such as the mean and standard error of the mean (SEM) are reported graphically for continuous variables. Normal distribution of the data was confirmed. For continuous dependent variables, one-way analysis of variance (ANOVA) was used to compare groups by one factor, and two-way ANOVA was used to compare groups by more than one factor. Repeated-measures analysis was used with ANOVA to account for day-to-day experiment variation. Statistical analysis was performed using R v3.1.0 (http://cran.R-project.org/), with the Ggplot2 and easyGgplot2 packages. Two-sided p-values <0.05 were considered statistically significant.
Results
Lentiviral transduction of human HSC
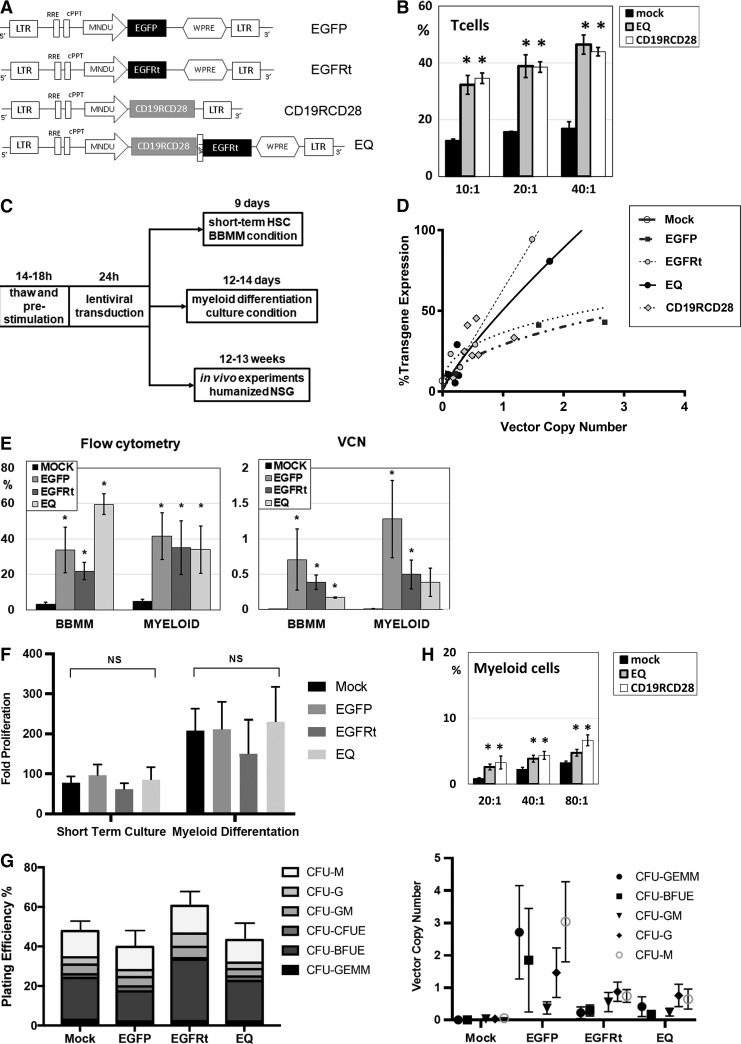
Four different lentiviral constructs were generated (Fig. 1A), and the pCCL-MNDU-eGFP (eGFP) construct to express eGFP was used as a transduction control. Isolated EGFRt was delivered by using the pCCL-MNDU-EGFRt (EGFRt) construct. CD19-specific second-generation CAR was delivered by the vector construct pCCL-MNDU-CD19RCD28 (CD19RCD28).10 For the co-delivery of CD19-specific CAR and EGFRt, pCCL-MNDU-CD19CARCD28-EGFRt (EQ) was generated by cloning the previously described EQ construct into the MNDU promoter lentiviral vector backbone.21,22
Figure 1.
Lentiviral transduction of human HSC with CAR. (A) Lentiviral vector constructs driven by promoter MNDU-3. (B) CD19-specific cytotoxicity by non-transduced (mock) or CAR-transduced (EQ or CD19RCD28) human T cells against the Raji cell line. (C) Experimental schema of lentiviral transduction of human HSC. (D) Transgene expression as a function of VCN in transduced human HSC for all the lentiviral vectors studied. (E) Flow cytometry transgene expression and VCN of human HSC transduced with eGFP, EGFRt, and EQ compared to non-transduced (mock) cells. (F) Proliferation of mock and gene-modified HSC in vitro, in BBMM short-term culture, or myeloid differentiation culture. (G) CFU assay of mock and gene-modified HSC: plating efficiency (left panel) and VCN determination by ddPCR of different colonies (right panel). (H) CD19-specific cytotoxicity by non-transduced (mock) or CAR-transduced (EQ or CD19RCD28) human myeloid cells against the Raji cell line. These myeloid cells were differentiated from non-transduced or CAR-transduced HSC. Representative of 4 (B and H), 3 (F and G), and over 10 experiments (D and E), respectively. Data are presented as the mean ± SEM; *p < 0.05 by Student's t test, and NS indicates statistically not significant. BBMM, basal bone marrow medium; CAR, chimeric antigen receptor; CFU, colony-forming unit; ddPCR, droplet digital polymerase chain reaction; eGFP, enhanced green fluorescent protein; EGFRt, truncated epidermal growth factor receptor; HSC, hematopoietic stem cells; SEM, standard error of the mean; VCN, vector copy number; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
As previously published, these second-generation CD19-specific CAR engineer antigen-specific cytotoxicity in T, NK, and myeloid cells.10,21,22 CAR-engineered T cells had two to three times higher in vitro cytotoxicity against CD19+ targets in comparison to non-transduced (mock) T cells (Fig. 1B), with no difference between CD19RCD28 and EQ constructs.
After transduction with 5.5 × 107 TU/mL of each vector, cord blood–derived CD34+ cells were cultured in BBMM short-term culture for 9 days (which preserves HSC in the short term) or myeloid differentiation culture for 14 days (Fig. 1C), and then cells were analyzed by flow cytometry and VCN assays. The expression curve of VCN analysis was plotted (Fig. 1D), which is consistent with previous experiments of lentiviral vector transductions of human HSC.10 In the short-term BBMM culture, the mean transgene expression percentages of eGFP- (n = 4), EGFRt- (n = 5), and EQ-transduced (n = 4) cells detected by flow cytometry were 33.8%, 21.8%, and 59.6% compared to 3.2% for the mock-transduced cells (n = 6; Fig. 1E, left panel). In the myeloid differentiated transduced cells, the mean transgene expression percentages of eGFP- (n = 4), EGFRt- (n = 5), and EQ-transduced (n = 6) cells were 46.1%, 35.0%, and 34.0% compared to 6.8% for the mock-transduced cells (n = 7; Fig. 1E, left panel). Copy number analysis of transduced cells in both BBMM and myeloid cultures resulted mean copies/cell of 0.70 for eGFP-BBMM and 1.2 for eGFP-myeloid cells, 0.38 for EGFRt-BBMM and 0.49 for EGFRt-myeloid cells, and 0.16 for EQ-BBMM and 0.38 for EQ-myeloid cells (Fig. 1E, right panel).
Gene modification of human HSC did not impair proliferation or differentiation
Transduced HSC and their progeny were evaluated for proliferation adjusted for the starting cell population. There was no significant difference in proliferation of eGFP-, EGFRt-, or EQ-transduced cells using either the short-term BBMM culture or myeloid differentiation cultures (Fig. 1F). The mean (SEM) fold proliferation after 9 days in short-term BBMM culture was 95.9 (27.7) for eGFP (n = 5), 61.1 (15.2) for EGFRt (n = 8), and 85.5 (31.7) for EQ (n = 6) compared to 77.6 (16.3) for mock-transduced cells (n = 11, p = 0.93). The mean (SEM) fold proliferation after 14 days in myeloid differentiation culture was 210.8 (68.9) for eGFP (n = 4), 149.5 (85.9) for EGFRt (n = 5), and 230.1 (87.2) for EQ (n = 4) compared to 207.8 (54.9) for mock-transduced cells (n = 7, p = 0.66; Fig. 1F). These results are similar to previously reported findings on human HSC transduced with CD19-specific CAR.10
Similar to published evaluations of clonogenic assays of CAR-modified HSC,10 gene-modified HSC were plated at 100 cells/plate in methylcellulose for clonogenic assays, and then evaluated by optic microscopy 14 days later. Once again, there was no significant difference in the plating efficiency between the different transduced human HSC (Fig. 1G, left panel). The percentages (SEM) of plating efficiency in eGFP- (n = 2), EGFRt- (n = 2), and EQ-transduced (n = 2) cells were 37.7% (4.9), 61% (8), and 43.3% (9.3), respectively, compared to the mock-transduced cells (47.7 [4.8]; p = 0.28; Fig. 1G, left panel). There was a similar proportion of CFU-monocyte, CFU-granulocyte, CFU-granulocyte/monocyte (CFU-GM), CFU-erythroid, burst forming unit-erythroid (BFU-E), and CFU-granulocyte/erythroid/monocyte/megakaryocyte in non-transduced and transduced cells (Fig. 1G, left panel). The plating efficiency of BFU-E in EGFRt-transduced cells was 1.5 × higher than in mock-transduced cells. However, statistical analysis showed no significant difference (p = 0.146; Fig. 1G, left panel). To demonstrate that transduced cells contributed to overall plating efficiency counted on clonogenic assay, colonies were individually selected and evaluated for VCN analysis, and the results confirm that gene-modified cells are well represented in the CFU counted on the clonogenic assay, with a similar pattern to the copy numbers found on cells cultured in BBMM and myeloid differentiating conditions (Fig. 1G, right panel). The overall mean VCN of colonies arising from EGFRt-transduced HSC was 0.59, and the mean VCN of colonies from EQ-transduced HSC was 0.49 (Fig. 1G, right panel).
As seen with human T cells transduced with CD19RCD28 and EQ (Fig. 1B), myeloid cells differentiated from gene-modified HSC had engineered antigen-specific cytotoxicity (Fig. 1H), confirming previously published results.10,21,22 In the experiments, the myeloid cells had lower cytotoxicity compared to T cells due to lower transduction efficiency and because unsorted cells were used on the cytotoxicity assays against the CD19+ Raji cell line. CAR expression in myeloid cells was 5.3% in EQ-transduced cells and 16.9% in CD19RCD28-transduced cells, but these populations determined statistically significant antigen-specific killing compared to non-transduced (mock) cells (Fig. 1H),
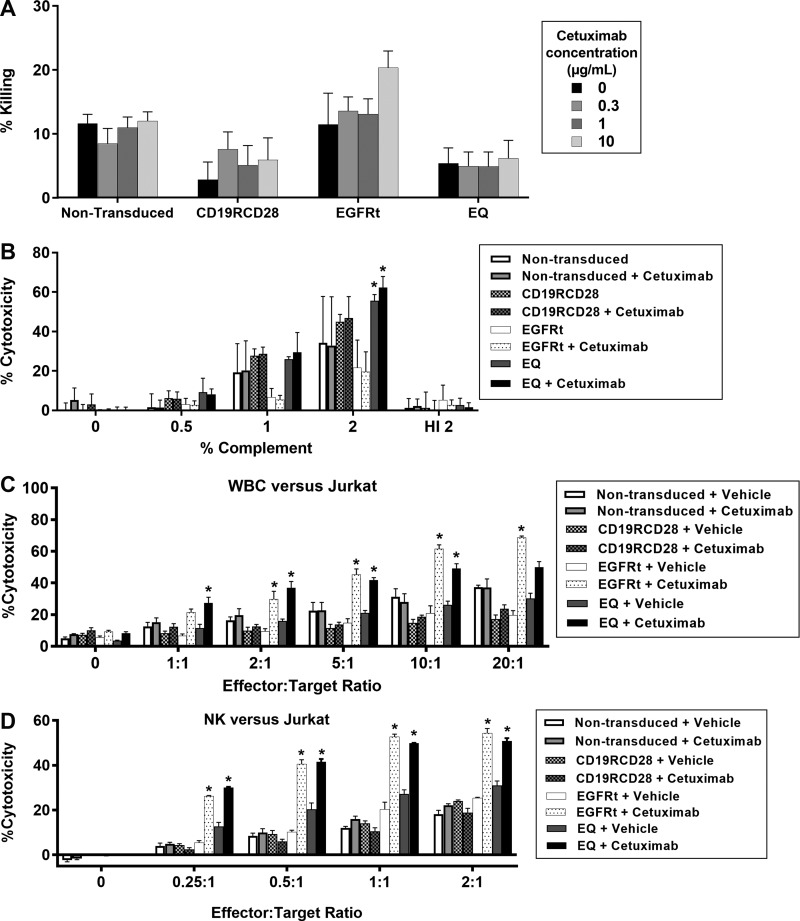
Direct antibody cytotoxicity of cetuximab on EGFRt-transduced cells
To evaluate direct antibody toxicity of the commercially available monoclonal antibody cetuximab, mock-transduced Jurkat cells and Jurkat cells transduced with CD19RCD28, EGFRt, and EQ with >95% of transgene expression were used as targets for in vitro cytotoxicity analysis (Fig. 2A). There was no significant increase in cytotoxicity with different concentrations (0, 0.3, 1, and 10 μg/mL) of cetuximab in any of the Jurkat cells (p = 0.38). There was an isolated significant increase in cytotoxicity in Jurkat-EGFRt cells with the supra-therapeutic cetuximab concentration (10 μg/mL) compared to CD19RCD28 (p = 0.0235) or EQ (p = 0.0165), but not with any of the other cetuximab concentrations (Fig. 2A). These findings support the conclusion that cetuximab is not expected to exert direct cytotoxicity on targets expressing the truncated EGFRt antigen (solely expressed on the cell membrane without EGFR signaling transduction in the cells transduced with EQ and EGFRt constructs used in this study) binding the antibody at concentrations up to 10 μg/mL.21,22,34
Figure 2.
In vitro ablation of gene-modified cells by cetuximab. (A) Jurkat cell line non-modified or transduced with CD19RCD28, EGFRt, or EQ, treated in culture with cetuximab for assessment of direct antibody-mediated cytotoxicity at different concentrations. Statistical analysis: two-way ANOVA for antibody concentration (p = 0.38 comparing the different cetuximab concentrations) and among groups. There was isolated significant cytotoxicity increase in Jurkat-EGFRt cells with the supra-therapeutic cetuximab concentration (10 μg/mL) when compared to CD19RCD28 (p = 0.0235) or EQ (p = 0.0165), but not on any of the other cetuximab concentrations. (B) Complement-dependent cytotoxicity on non-modified Jurkat cells or transduced with CD19RCD28, EGFRt, or EQ in the presence or absence of cetuximab. Statistical analysis: two-way ANOVA for the different complement doses and among groups. (C) ADCC assay of non-transduced and EGFRt-transduced Jurkat cells using primary human leukocytes in the presence or absence of cetuximab. Statistical analysis: two-way ANOVA for the different vector constructs (p < 0.0001) and the presence of cetuximab (p < 0.0001). (D) ADCC assay of non-transduced and EGFRt-transduced Jurkat cells using primary human NK cells in the presence or absence of cetuximab. Statistical analysis: two-way ANOVA for the different vector constructs (p < 0.0001) and the presence of cetuximab (p < 0.0001). Representative of four experiments. Data are presented as the mean ± SEM; *p < 0.03, and NS indicates statistically not significant. ADCC, antibody-dependent cell-mediated cytotoxicity; ANOVA, analysis of variance; HI, heat inactivated; NK, natural killer.
Complement-dependent cytotoxicity of cetuximab on EGFRt-transduced cells
To evaluate direct complement toxicity on target cells, non-transduced Jurkat cells and Jurkat cells transduced with CD19RCD28, EGFRt, and EQ with >95% of transgene expression were incubated in medium with different concentrations of rabbit complement. All cells incubated at 2% dilution of complement revealed a significant increase of cytotoxicity compared to complement doses of 0.5% or 1% and to medium only (p < 0.0001). However, there was no difference in cytotoxicity between transduced Jurkat cells versus non-transduced Jurkat, or between cells treated with 1 μg/mL cetuximab versus non-treated cells, except for the EQ-transduced cells in both non-treated (p = 0.0025) and cetuximab-treated (p < 0.0001) conditions at 2% complement (Dunnett's multiple comparisons test against non-transduced, non-treated target cells). These findings are consistent with a direct dose-dependent cytotoxicity of complement on all target cells, which is not specific to the presence of cetuximab or EGFRt expression on cells transduced with EQ or EGFRt constructs (Fig. 2B).
To confirm whether the addition of complement determined direct cytotoxicity, the same Jurkat target cells, with or without cetuximab treatment at a dose of 1 μg/mL, were exposed to 2% complement inactivated at 56°C for 60 min. With heat-inactivated (HI) complement, cytotoxicity decreased to 0–16% across all groups, showing that complement is required for cytotoxicity and supporting the assertion the direct cytotoxicity by complement is not antigen-specific nor antibody-mediated at the cetuximab dose of 1 μg/mL (Fig. 2B).
The findings described in Fig. 2B reflect the published literature suggesting that complement is not a relevant mediator for cetuximab cytotoxicity on cancer cells, as complement-dependent cytotoxicity by cetuximab has only been described on cell lines with very high expression of EGFR.34
ADCC of cetuximab on EGFRt-transduced cells
Having evaluated the direct cytotoxicities of cetuximab and complement on cells expressing EGFRt, the next experiments addressed ADCC. Total human leukocytes were isolated from whole donor blood, with a yield of 2–5 × 105 cells/mL of blood for in vitro cell-mediated cytotoxicity assays. By flow cytometry analysis, a typical composition of leukocyte phenotype subsets was 7.5% CD8+, 7.9% CD4+, 5.3% CD14+, 14.6% CD3+, 6.2% CD33+, 5.1% CD19+, and 16.2% CD56+ (data not shown).
After 4 h of incubation with target cells and cetuximab, increased cytotoxicity was significant (p < 0.0001) only in leukocytes incubated with EGFRt-expressing or EQ-expressing Jurkat cells with cetuximab at 1 μg/mL. This was true for E:T ratios of 2:1, 5:1, and 10:1 at p < 0.01, and only for EQ-transduced cells at 1:1 (p = 0.0210), and only for EGFRt-transduced cells at 20:1 (p < 0.0001; Fig. 2C).
Similarly, human NK cells were isolated from whole donor blood, with a yield of 0.4–0.5 × 106 cells/mL of blood. A typical flow cytometry analysis of a NK cell suspension was 0.1% CD14+, 1.4% CD33+, 95.5% CD56+, 0.2% CD19+, and 0.3% CD3+ (data not shown). Cytotoxicity assays of NK cells incubated for 4 h at different E:T ratios with target cells and cetuximab (1 μg/mL) showed significantly higher NK-mediated cytotoxicity (p < 0.0001) against EGFRt-expressing or EQ-expressing Jurkat cells compared to non-transduced cells (Fig. 2D). This was true for all E:T ratios at p < 0.0001.
The results described on Fig. 2C and D provide evidence that cytotoxicity by cetuximab is dependent on ADCC mediated by neutrophils and NK cells, in accordance with previous publications.34
CAR-modified HSC successfully engraft in vivo, establish survival advantage to tumor challenge, and are amenable to ablation
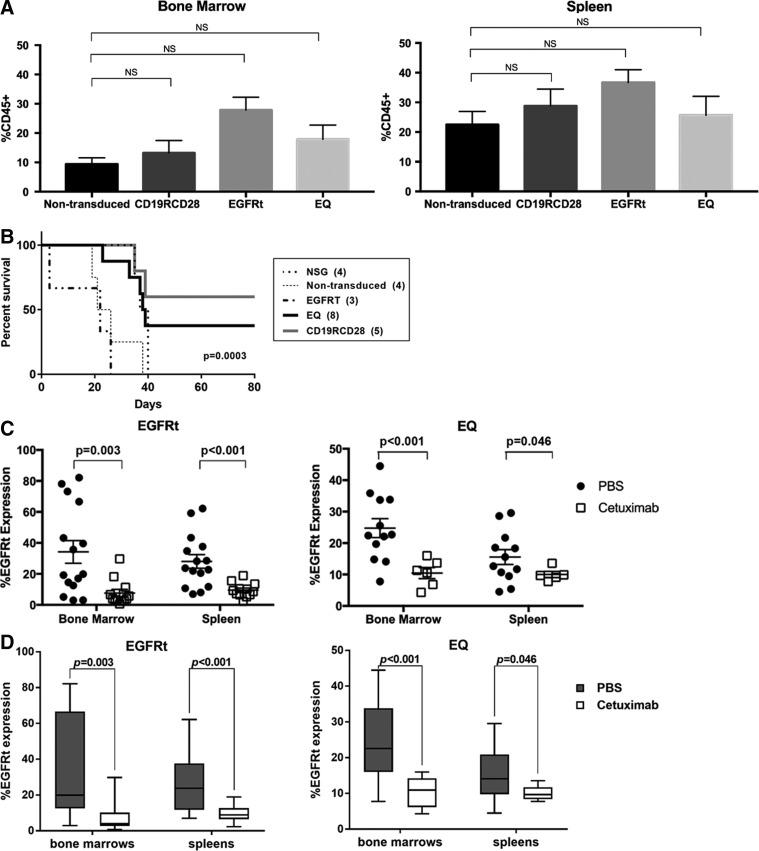
In vivo assessment of the engraftment, proliferation, and differentiation of modified human HSC was performed by the intrahepatic injection of mock-transduced, EGFRt-transduced, and EQ-transduced human HSC into sub-lethally irradiated NSG pups. Human hematopoiesis in transplanted mice was analyzed through harvest of bone marrows and spleens at 16 weeks post transplant. Human hematopoietic engraftment (as detected by the percentage of human CD45+ cells by flow cytometry) for both compartments was not significantly different between the EQ-, EGFRt-, and mock-transduced arms (Fig. 3A), confirming that gene modification did not impair HSC proliferation.
Figure 3.
In vivo engraftment, anti-CD19 activity and ablation of gene-modified human HSC. (A) Humanization of NSG mice injected with non-transduced and gene-modified HSC in the bone marrow (left panel) and spleen (right panel), defined by the detection of the human CD45 marker as a percentage of all viable cells. (B) Kaplan–Meier curve of humanized mice challenged with subcutaneous injection of CD19+ Raji cell line and followed over 80 days; Log-rank test with p = 0.0003 for the comparison among the different mouse populations. (C) Comparison of flow cytometry expression of EGFRt after treatment with vehicle (PBS; black circles) versus cetuximab (white squares) in the bone marrow and spleen of humanized NSG. (D) Box and whiskers graphical representation of the data presented in (C). Each box extends from the 25th to 75th percentiles, and the whiskers depict minimum to maximum values. (E) Comparison of lentiviral VCN after treatment with vehicle (PBS; black circles) versus cetuximab (white squares) in the bone marrow and spleen of humanized NSG. (F) Box and whiskers graphical representation of the data presented on (E). Each box extends from the 25th to 75th percentiles, and the whiskers depict minimum to maximum values. Representative of four experiments. Data are presented as the mean ± SEM; *p < 0.05 by Student's t-test (A and C–F), and NS indicates statistically not significant. In (C) and (E), each symbol represents an individual mouse for each tissue. PBS, phosphate-buffered saline.
Humanized mice were challenged with a subcutaneous injection of CD19-expressing Raji human cell line to evaluate engineered antigen-specific cytotoxicity using previously published methods.10,13 All wild-type non-humanized NSG mice injected with Raji cells did not survive the tumor challenge, similar to the NSG humanized with non-transduced HSC or EGFRt-transduced HSC (Fig. 3B). Similar to previously published data,10 NSG mice humanized with CAR-modified HSC had a survival advantage facing the tumor challenge, with 60% of mice engrafted with HSC transduced with CD19RCD28 and 28.5% of mice engrafted with HSC transduced with EQ surviving (p = 0.0003; Fig. 3B).
To evaluate the ablation in vivo of gene-modified cells, additional cohorts of mice humanized with EQ-, EGFRt-, or mock-transduced human HSC were then given either an intraperitoneal injection of cetuximab 1 mg/mouse or PBS for 12 consecutive days. On day 13, cells harvested from the bone marrow and spleen were evaluated by flow cytometry detection of EGFRt expression and with VCN assessment by ddPCR.
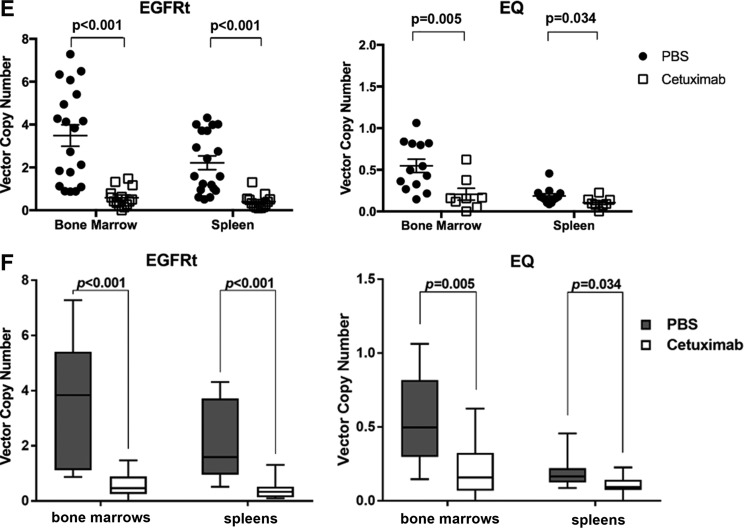
When evaluated for EGFRt expression by flow cytometry, human cells from spleens of EGFRt-HSC-transplanted mice were on average 28.02% positive for EGFRt if treated with vehicle, and 9.51% positive for EGFRt if treated with cetuximab (p < 0.001). When comparing human cells from cetuximab-treated versus vehicle-treated mice in bone marrow, there was 34.18% EGFRt positivity in the vehicle group versus 7.60% in the cetuximab group (p = 0.003; Fig. 3C). For EQ-HSC-transplanted mice, evaluation of EGFRt expression by flow cytometry from spleens was 15.54% if treated with vehicle compared to 10% if treated with cetuximab (p = 0.046). For bone marrow, there was 24.75% EGFRt positivity in the mice in the vehicle group compared to 10.43% in mice treated with cetuximab (p < 0.001). The significant decrease in EGFRt-expressing cells in the spleen and bone marrow after cetuximab treatment demonstrate the elimination of gene-modified cell progeny in mice humanized with EGFRt- or EQ-transduced HSC. Mice harvested up to 5 weeks after cetuximab treatment showed similar levels of expression of EGFRt, suggesting long-lasting effect of the ablation. Figure 3D presents the same data in Fig. 3C in a box and whiskers graphic representation, allowing clear observation of the significant drop in EGFRt expression in the bone marrow and spleen of humanized mice from both EGFRt- and EQ-transduced HSC groups.
When evaluated for lentiviral transduction by VCN, cells from the spleen of EGFRt-HSC-transplanted mice had an average VCN of 2.21 if treated with vehicle compared to 0.40 if treated with cetuximab (p < 0.001). In the bone marrow, the average VCN was 3.48 in the vehicle-treated group, which decreased to 0.59 in the cetuximab-treated group (p < 0.001; Fig. 3E). Cells from the spleen of EQ-HSC-transplanted mice had an average VCN of 0.18 if treated with vehicle compared to 0.10 if treated with cetuximab (p = 0.034). In the bone marrow, the average VCN was 0.54 in the vehicle group and 0.20 in the cetuximab-treated group (p = 0.005). Consistent with the flow cytometry analysis, in the spleen and bone marrow, there were significant differences in VCN between the vehicle group and the cetuximab-treated group (Fig. 3E and F). Mice harvested up to 5 weeks after cetuximab treatment showed similar levels of VCN, suggesting a long-lasting effect of the ablation.
ImmunoPET imaging of in vivo ablation
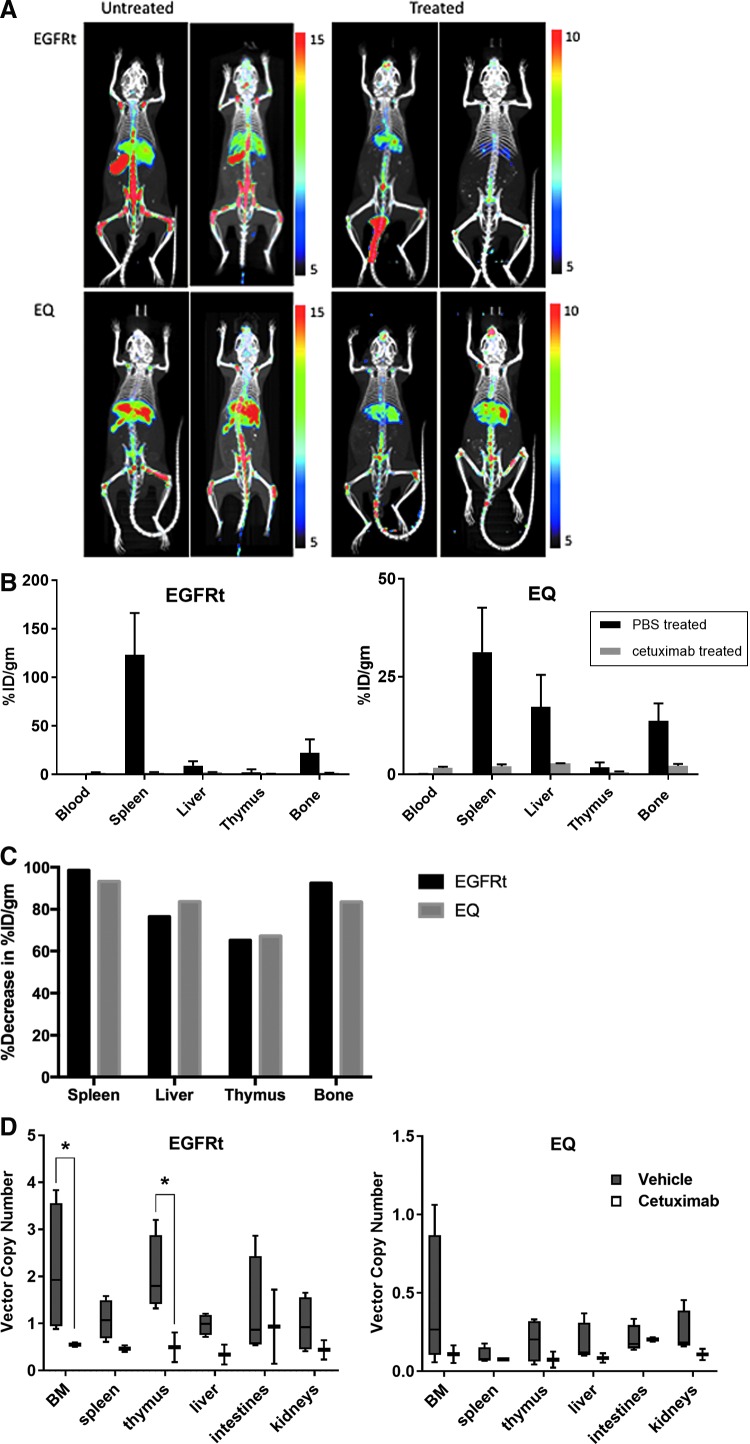
Additional cohorts of humanized mice were used for ImmunoPET imaging of gene-modified cells employing radiolabeled cetuximab. The concentration of conjugated cetuximab was 1 μg/μL with 57% recovery. The radiolabeling efficiency was 78%, and the radiochemical purity was 97%. Each mouse was injected with 9.2 μg 89Zr-cetuximab, with radioactivity between 28 and 23 μCi. Screening of transgene expression prior to imaging using flow cytometry showed higher expression in the mice engrafted with EGFRt-modified HSC (65 ± 10.5%) compared to those engrafted with EQ-HSC (34.6 ± 1.8%). The PBS-treated mice versus those treated intraperitoneally with 1 mg cetuximab over 12 days were imaged at 24 and 120 h after injection of the 89Zr-labeled cetuximab (Fig. 4A). PBS-treated EQ-engrafted mice had a % ID/g of 31.3 ± 5.6% in the spleen and 13.7 ± 2.2% in the bone compared to 2.1 ± 0.3% in the spleen and 2.3 ± 0.3% in the bone in the cetuximab-treated mice. The EGFRt-engrafted mice had a % ID/g in the spleen of 123.2 ± 21.5% and 22.2 ± 6.9% in the bone in the PBS-treated group compared to 1.8 ± 0.6% in the spleen and 1.67 ± 0.1% in the bone of the cetuximab-treated group (Fig. 4B). An overall decrease in % ID/g of 65–98% was detected in the cetuximab-treated mice (Fig. 4C).
Figure 4.
PET imaging of humanized mice with gene-modified HSC. (A) Imaging of humanized mice engrafted with gene-modified HSC at 20 weeks post engraftment untreated or treated with cetuximab for 12 days; mice imaged at 24 and 120 h after injection of the 89Zr-labeled cetuximab. (B) Distribution of PET intensity per gram of tissue of different organs from humanized mice injected with radiolabeled cetuximab. (C) Percentage of decrease of PET intensity in the spleen, liver, thymus, and bone marrow of humanized mice after cetuximab treatment. (D) Box and whiskers graphical representation of the viral VCN detected in the different organs removed from mice in the PET imaging experiment. Each box extends from the 25th to 75th percentiles, and the whiskers depict minimum to maximum values. Representative of three experiments. Data are presented as the mean ± SEM; *p < 0.02 by two-way ANOVA by tissue and treatment, with Tukey's multiple comparisons test. PET, positron emission tomography.
Genomic DNA was extracted from the organs harvested from the mice after the PET imaging experiments to confirm the elimination of gene-modified cells by cetuximab treatment through VCN analysis (Fig. 4D). Although the analyses were limited by the numbers of mice, there was a significant decrease in the vector copies in the bone marrow (p = 0.0115) and thymus (p = 0.0156) of mice humanized with EGFRt-transduced cells treated with cetuximab. A clear decrease in copy numbers can be noted in these tissues, similar to the findings described in Fig. 3E and F, even in the cells from mice humanized with EQ-transduced cells, which have lower VCN (Fig. 4D).
Discussion
Gene therapy using modified HSC has shown promise for the treatment of certain monogenic diseases and is under exploration in cancer immunotherapy.10–13,35–37 Nevertheless, there has been concern about insertional oncogenesis leading to hematopoietic malignancy in several patients receiving gene-modified HSC.14–17 In addition, on-target, off-site effects of adoptive T-cell immunotherapy have also dampened enthusiasm.18–20 These concerns highlight the need for the control of gene-modified cells.12,21,22,24 Safety elements have been incorporated into vector design, including insulators and internal promoters with self-inactivating long terminal repeats.25 In addition, there is considerable interest in controlling gene-modified cells using suicide systems consisting of a targetable suicide gene included in the vector construct along with the disease-modifying gene.12
Inspired by the results obtained by the research group at City of Hope,21,22 the EQ construct was used for gene modification of human HSC for preclinical evaluation in a humanized mouse model. The surface marker EGFRt expressed on gene-modified cells presents a unique opportunity as a dual-purpose reagent for the detection and ablation of such cells. The use of the available clinical-grade monoclonal antibody cetuximab is an added advantage for clinical translation consideration. In fact, the EGFRt construct has been incorporated into multiple clinical trials of gene-modified T cells, many of them still currently open, due to its proven efficacy at eliminating gene-modified cells and its safety profile. The use of additional labeling of cetuximab allowed flow cytometry and PET imaging deployed in this study, disclosing other translational opportunities.
Besides EGFRt, the subject of this study, other suicide systems that have been studied include herpes simplex virus-thymidine kinase (HSV-TK) paired with ganciclovir, inducible caspase 9 (iCas9) paired with the chemical inducer of dimerization AP1903 (rimiducid), and CD20 paired with rituximab.12,38,39 The HSV-TK system has the most clinical experience in terms of both suicide gene therapy against cancer and also in the setting of gene therapy.40 So far, it has the most convincing data in HSC.41 It also allows for in vivo tracking of gene-modified cells through PET imaging.42 The iCas9 system has also been studied in limited clinical trials.43 It rapidly responds to AP1903, with the elimination of 85–95% of circulating iCas9-transduced cells within 30 min, when used against gene-modified T cells. In HSC, however, iCas9 was unable to reproduce the complete ablation in rhesus macaques seen in HSV-TK. This was thought to be linked to lower iCas9 expression in residual cells.44
EGFRt is promising as a suicide system for several reasons. Unlike the CD20 system, the EGFRt system uses cetuximab, which has not been shown to cause myelosuppression or hematopoietic cell aplasia in clinical trials,45 and it would allow the concurrent use of rituximab in the treatment of B-lineage lymphomas. A similar anti-EGFR drug M225 did not have effects on proliferation of marrow granulocyte/macrophage progenitors (CFU-GM) or marrow stromal cells.46 In contrast to the HSV-TK system, cetuximab-mediated ablation of EGFRt-expressing cells is not cell cycle dependent, and in clinical care, it would allow ganciclovir to be given for treatment of CMV infection without concern of losing the graft. Furthermore, since EGFRt is based on human EGFR, the potential for immunogenicity is lower than for the virally derived HSV-TK, decreasing the chances of undesired clearance by the host immune system. In contrast to the iCas9 system, the EGFRt system utilizes cetuximab, a Food and Drug Administration–approved monoclonal antibody with significant clinical experience in the treatment of various solid tumors.
In addition, EGFRt is encoded by a relatively small cDNA, facilitating its incorporation into most vector types along with other transgenes. It is specifically engineered to avoid binding with EGF and other EGFR ligands, and it can also be tracked in vitro and in vivo using biotinylated cetuximab staining.21
Here, it is demonstrated that HSC can be modified to express EGFRt as part of a cetuximab-mediated suicide system. EGFRt modification did not alter the ability of HSC to proliferate and differentiate in vitro or in vivo. When exposed to cetuximab, cells expressing EGFRt are killed through ADCC, rather than direct antibody toxicity or complement-dependent cytotoxicity. In addition, monocytes and NK cells contribute to ADCC, allowing ablation of gene-modified cells even in the absence of full immune reconstitution after HSC transplantation.
In vivo, this suicide system worked to eliminate most of the EGFRt-transduced cells when cetuximab was administered to mice after EGFRt-HSC transplant and engraftment, documented by flow cytometry and ddPCR. These data are similar to the published results of using the same system to ablate gene-modified T cells.21,22,24 One possible reason for no complete ablation in the humanized mice is that cetuximab might be more reliant on human effector cells rather than murine effector cells for ADCC, especially when the human cell engraftment of each mouse averaged 10–15%. Other possible reasons for the presence of the remaining gene-modified cells could be insufficient dose or duration of cetuximab treatment. Cetuximab may also have had poor penetration beyond the peritoneum, where it had been administered in the study mice. Preclinical trials of cetuximab, however, were performed in nude mice against human tumor xenografts and therefore showed cetuximab is not reliant on human effectors.47 In the same study, the effective dose for each mouse was 0.5–1 mg/mouse, twice weekly for 5 weeks, with significant differences by treatment group by day 10–14, in which cetuximab was also given intraperitoneally. Some of the residual flow cytometry staining in ablated mice (Fig. 3C and D) might be explained by nonspecific staining with biotinylated cetuximab on hematopoietic cells. While EGFR itself has low levels of protein expression in hematopoietic cells,48,49 it is well known that hematopoietic cells are rich in Fc receptors and may thus bind the Fc portion of biotinylated cetuximab, despite the use of Fc blockers in all the flow cytometry staining protocols, leading to background staining of 2–4%. The results of VCN in ablated mice confirm the ablation of gene-modified cells (Fig. 3E and F).
The development of a radiolabeled cetuximab may lead to a clinically relevant EGFR-specific reagent for PET imaging. Besides its use as suicide system as in this project, a novel ImmunoPET reagent would have possible applications in all cancers bearing EGFR overexpression, such as colorectal, lung, breast, ovarian, and cervical carcinomas. The development of an antibody fragment such as a cys-diabody would decrease nonspecific binding and increase tissue penetration.
There are currently about 20 clinical trials using gene-modified T cells with EGFRt to allow their ablation as a safety feature. Similar to other studies, this study demonstrated that co-delivery of EGFRt and CD19-specific CAR did not impair cell function or engineered antigen-specific cytotoxicity. The findings support the use of a cetuximab-mediated suicide system for gene-modified human HSC, which did not affect HSC proliferation or engraftment in the humanized mouse model, with the added advantage of its use for imaging purposes by ImmunoPET.
Acknowledgments
Support was provided by the UCLA Department of Pediatrics 3K12HD034610 and the NIH/NHLBI 2T32HL086345-06 Training Grant in Developmental Hematology, NCI 1K23CA222659, American Society of Hematology Scholar Award, Hyundai Hope on Wheels Scholar Award, Miranda D. Beck Pediatric Cancer Research Foundation, Gwynne Hazen Cherry Memorial Fund, the Pediatric Cancer Research Fund, UCLA Children's Discovery and Innovation Institute, UCLA Jonsson Comprehensive Cancer Center, UCLA Cancer Research Coordinating Committee, UCLA Clinical and Translational Science Institute Grant UL1TR000124, and the St. Baldrick's Foundation. All the authors acknowledge the collaboration with Drs. Stephen Forman and Christine Brown, from City of Hope, who provided the EGFRt and EQ CAR constructs, and the training and technical support of the Flow Cytometry Core of the UCLA Broad Stem Cell Research Center (BSCRC), where all flow cytometry experiments were performed. The authors also acknowledge the valuable scientific advice and technical support from Dr. Gay Crooks, MBBS, and Donald B. Kohn, MD, and the help on data acquisition and analyses from Felix Bergara Salazar, Andy Tu, Tulika Tyagi, Therese Dinoso, and Nezia Rahman.
Author Disclosure
No conflicts of interest to disclose.
References
- 1. Candotti F, Shaw KL, Muul L, et al. Gene therapy for adenosine deaminase-deficient severe combined immune deficiency: clinical comparison of retroviral vectors and treatment plans. Blood 2012;120:3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hacein-Bey-Abina S, Pai S-Y, Gaspar HB, et al. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med 2014;371:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianchi M, Hakkim A, Brinkmann V, et al. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood 2009;114:2619–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castiello MC, Scaramuzza S, Pala F, et al. B-cell reconstitution after lentiviral vector–mediated gene therapy in patients with Wiskott–Aldrich syndrome. J Allergy Clin Immunol 2015;136:692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol 2012;507:187–198 [DOI] [PubMed] [Google Scholar]
- 7. Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:1233158. [DOI] [PubMed] [Google Scholar]
- 8. Boulad F, Wang X, Qu J, et al. Safe mobilization of CD34+ cells in adults with β-thalassemia and validation of effective globin gene transfer for clinical investigation. Blood 2014;123:1483–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romero Z, Urbinati F, Geiger S, et al. β-globin gene transfer to human bone marrow for sickle cell disease. J Clin Invest 2013;123:3317–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Oliveira SN, Ryan C, Giannoni F, et al. Modification of hematopoietic stem/progenitor cells with CD19-specific chimeric antigen receptors as a novel approach for cancer immunotherapy. Hum Gene Ther 2013;24:824–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gschweng E, De Oliveira S, Kohn DB. Hematopoietic stem cells for cancer immunotherapy. Immunol Rev 2014;257:237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larson S, De Oliveira SN. Gene-modified hematopoietic stem cells for cancer immunotherapy. Hum Vaccin Immunother 2014;10:982–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larson SM, Truscott LC, Chiou T-T, et al. Pre-clinical development of gene modification of hematopoietic stem cells with chimeric antigen receptors for cancer immunotherapy. Hum Vaccin Immunother 2017;13:1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 2008;118:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest 2008;118:3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Braun CJ, Boztug K, Paruzynski A, et al. Gene therapy for Wiskott–Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 2014;6:227ra33. [DOI] [PubMed] [Google Scholar]
- 17. Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med 2010;16:198–204 [DOI] [PubMed] [Google Scholar]
- 18. Maude SL, Teachey DT, Porter DL, et al. CD19-targeted chimeric antigen receptor T cell therapy for acute lymphoblastic leukemia. Blood 2015;125:4017–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morgan RA, Yang JC, Kitano M, et al. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther 2010;18:843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morgan RA, Chinnasamy N, Abate-Daga D, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother 2013;36:133–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Chang W-C, Wong CW, et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 2011;118:1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jonnalagadda M, Mardiros A, Urak R, et al. Chimeric antigen receptors with mutated IgG4 Fc spacer avoid Fc receptor binding and improve T cell persistence and anti-tumor efficacy. Mol Ther 2015;23:757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Wong CW, Urak R, et al. CMVpp65 vaccine enhances the antitumor efficacy of adoptively transferred CD19-redirected CMV-specific T cells. Clin Cancer Res 2015;21:2993–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paszkiewicz PJ, Fräßle SP, Srivastava S, et al. Targeted antibody-mediated depletion of CD19 CAR-T cells permanently reverses B cell aplasia. J Clin Invest 2016;126:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zufferey R, Dull T, Mandel RJ, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 1998;72:9873–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robbins PB, Yu XJ, Skelton DM, et al. Increased probability of expression from modified retroviral vectors in embryonal stem cells and embryonal carcinoma cells. J Virol 1997;71:9466–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cooper AR, Patel S, Senadheera S, et al. Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. J Virol Methods 2011;177:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaines P, Berliner N. Differentiation and characterization of myeloid cells. Curr Protoc Immunol 2005;Chapter 22:Unit 22F.5 [DOI] [PubMed] [Google Scholar]
- 29. Munshi A, Hobbs M, Meyn RE. Clonogenic cell survival assay. Methods Mol Med 2005;110:21–28 [DOI] [PubMed] [Google Scholar]
- 30. Tapani E, Taavitsainen M, Lindros K, et al. Toxicity of ethanol in low concentrations. Experimental evaluation in cell culture. Acta Radiol 1996;37:923–926 [DOI] [PubMed] [Google Scholar]
- 31. Hindson BJ, Ness KD, Masquelier DA., et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vosjan M, Perk L, Visser G, et al. Conjugation and radiolabeling of monoclonal antibodies with zirconium-89 for PET imaging using the bifunctional chelate p-isothiocyanatobenzyl-desferrioxamine. Nat Protoc 2010;5:739–743 [DOI] [PubMed] [Google Scholar]
- 33. Zeglis BM, Lewis JS. The bioconjugation and radiosynthesis of Zr-DFO-labeled antibodies video link. J Vis Exp 2015;(96):52521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhuang H, Xue Z, Wang L, et al. Efficacy and immune mechanisms of cetuximab for the treatment of metastatic colorectal cancer. Clin Oncol Cancer Res 2011;8:207–214 [Google Scholar]
- 35. Adair JE, Beard BC, Trobridge GD, et al. Extended survival of glioblastoma patients after chemoprotective HSC gene therapy. Sci Transl Med 2012;4:133ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kramer B, Singh R, Wischusen J, et al. Clinical trial of MGMT(P140K) gene therapy in the treatment of paediatric patients with brain tumours. Hum Gene Ther 2018;29:874–885 [DOI] [PubMed] [Google Scholar]
- 37. Adair JE, Waters T, Haworth KG, et al. Semi-automated closed system manufacturing of lentivirus gene-modified haematopoietic stem cells for gene therapy. Nat Commun 2016;7:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tey S-K. Adoptive T-cell therapy: adverse events and safety switches. Clin Transl Immunol 2014;3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karjoo Z, Chen X, Hatefi A. Progress and problems with the use of suicide genes for targeted cancer therapy. Adv Drug Deliv Rev 2016;99:113–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oliveira G, Greco R, Lupo-Stanghellini MT, et al. Use of TK-cells in haploidentical hematopoietic stem cell transplantation. Curr Opin Hematol 2012;19:427–433 [DOI] [PubMed] [Google Scholar]
- 41. Barese CN, Krouse AE, Metzger ME, et al. Thymidine kinase suicide gene-mediated ganciclovir ablation of autologous gene-modified rhesus hematopoiesis. Mol Ther 2012;20:1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gschweng EH, McCracken MN, Kaufman ML, et al. HSV-sr39TK positron emission tomography and suicide gene elimination of human hematopoietic stem cells and their progeny in humanized mice. Cancer Res 2014;74:5173–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou X, Dotti G, Krance RA, et al. Inducible caspase-9 suicide gene controls adverse effects from alloreplete T cells after haploidentical stem cell transplantation. Blood 2015;125:4103–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barese CN, Felizardo TC, Sellers SE, et al. Regulated apoptosis of genetically modified hematopoietic stem and progenitor cells via an inducible caspase-9 suicide gene in rhesus macaques. Stem Cells 2015;33:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Galizia G, Lieto E, De Vita F, et al. Cetuximab, a chimeric human mouse anti-epidermal growth factor receptor monoclonal antibody, in the treatment of human colorectal cancer. Oncogene 2007;26:3654–3660 [DOI] [PubMed] [Google Scholar]
- 46. Taetle R, Honeysett JM, Houston LL. Effects of anti-epidermal growth factor (EGF) receptor antibodies and an anti-EGF receptor recombinant-ricin A chain immunoconjugate on growth of human cells. J Natl Cancer Inst 1988;80:1053–1059 [DOI] [PubMed] [Google Scholar]
- 47. Goldstein NI, Prewett M, Zuklys K, et al. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res 1995;1:1311–1318 [PubMed] [Google Scholar]
- 48. Kolker E, Higdon R, Haynes W, et al. MOPED: model organism protein expression database. Nucleic Acids Res 2012;40:1093–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilhelm M, Schlegl J, Hahne H, et al. Mass-spectrometry-based draft of the human proteome. Nature 2014;509:582–587 [DOI] [PubMed] [Google Scholar]