Graphical abstract
Abbreviations: HNE, human neutrophil elastase; UM, ultramarine protein; UM10, ultramarine engineered without the last 11th β-strand; E11, 11th β-strand of ultramarine inserted in a eglin c portion with an HNE cleavage sequence; RMSF, root-mean-square fluctuation; pNa, p-nitroaniline; pep11, synthetic peptide equivalent to free 11th β-sheet of ultramarine
Keywords: Human neutrophil elastase (HNE), Split GFP, Chromogenic GFP-like proteins, Ultramarine (UM), Proteases, Molecular dynamics simulations
Highlights
-
•
GFP-like chromoproteins can be used for protease signalling through colour changes.
-
•
Absorbance changes obtained on split UM system upon HNE cleavage were transient.
-
•
Molecular dynamics confirmed the instability registered in vitro studies.
-
•
In silico studies provide further understanding on split and reassembly mechanisms.
-
•
Split-GFP technology for chromoproteins isn’t straightforward as for fluoroproteins.
Abstract
Recent studies have demonstrated that human neutrophil elastase (HNE) can be used as marker for inflammation/infection of chronic wounds since it was found to be present in high concentration in exudate collected from chronic wounds. Biosensors used in wound care benefit from a chromogenic signalling due to the readiness of signal interpretation, but the most common use faint yellow chromogenic molecules such as p-nitroaniline (pNa). In addition, if to be converted into smart dressings, the colour of the detection system should not be masked by the exudate’s colour.
In this work, we designed a chromogenic substrate for HNE aiming to be incorporated in a smart dressing as a colour switch sensor. The substrate was developed using the GFP-like chromoprotein ultramarine (UM), following the split GFP technology. The cleavage sequence for HNE (Ala-Ala-Pro-Val) was embedded into the sensing moiety of the substrate corresponding to the 11th β-sheet. In the presence of HNE, the 11th β-sheet is able to interact to the signalling moiety composed of the β1-β10 incomplete barrel, allowing the re-establishment of the chromophore environment and, hence, the colour production. Structural homology and molecular dynamics simulations were conducted to aid on the disclosure of the structural changes that are the base of the mechanism of action of this HNE switch substrate. Our findings explore the possible application of GFP-like chromogenic sensors in point-of-care devices for the evaluation of the wounds status, representing a major step in the medical field.
1. Introduction
Human neutrophil elastase (HNE) plays a critical role during inflammation. In spite of its protective role, HNE is also implicated in numerous disorders, including delayed chronic wound healing [[1], [2], [3], [4]]. It is difficult, expensive and time consuming to accurately assess the wound status and there is an urgent need for a new diagnostic tool [5]. Early detection of incipient wound infection and chronic inflammation reduces the severity of the lesion and decreases health care expenses [6].
The progress of wound healing and the assessment of the wound status can be achieved through monitoring and detecting HNE in wounds. Even though several approaches have been developed to monitor HNE in the wound environment [2,[7], [8], [9], [10], [11], [12],[13], [14], [15], [16], [17], [18]], alternative strategies are in demand as those present known limitations. Most of these approaches are based on fluorogenic methods and always require an off-line detection system or an imaging analysis. For example, the commercially available Neutrophil Elastase Activity Assay Kit from BioVision presents a detection limit below 1 ng, but relies on other components essential for the visualization, and demands post hoc analysis and interpretation of the results. Other systems based on chromogenic approaches make use on p-nitroaniline (pNa) as a chromophore (yellow) [8,10,19], being difficult the visualization and discrimination of the signal from the wound fluids.
Green fluorescent protein (GFP) is one of the most used tagging and detection molecules in several biological and molecular studies, in some of them due to its inherent ability of self-reassembling of the β-strands [20]. GFP family of proteins folds into a distinctive β-barrel composed of 11 β-sheets surrounding a α-helix core that restrains the chromophore group. Maturation of the chromophore is a spontaneous 3-step autocatalytic process (cyclization, oxidation and dehydration) in the Ser65-Tyr66-Gly67 tripeptide (in GFP from jellyfish Aequorea victoria) though the manifestation of colour or fluorescence does require a network of polar interactions involving the chromophore group and several closely-apposed amino acids [[21], [22], [23], [24], [25]]. According to recent studies, self-reassembly of GFP and its homologous proteins is greatly dependent on C-terminus sheets (β7-11) [25]. Specifically, the 11th β-sheet is known to protect the chromophore from the surrounding environment [26], being significantly involved on the refolding pathways required for final chromophore maturation [27].
Cabantous and colleagues [27] engineered a split GFP system involving two non-fluorescent portions of GFP that when mixed together restored the green fluorescent signal: one portion corresponds to GFP 1–10 fragment (amino acids 1–214, 10 β-sheets) and another portion corresponds to GFP11 (amino acids 215–231, 11th β-sheet). In following works, this split GFP system has been extensively adapted to integrate a protease-sensitive switch-on system for HIV protease, caspase-3, factor Xa, enteropeptidase and thrombin [28] and is commercially available as a kit to monitor protein expression (Interchim).
In our work, we used the same described rationale to develop a HNE switch-on coloured system, for application on a smart wound dressing, using a non-fluorescent GFP-like chromoprotein. The two following criteria had to be met regarding colour change upon HNE proteolytic activity to accomplish the intended application of the system: (1) easy discrimination by the human eye in the wound dressing, and (2) direct correlation between colour intensity/development time vs the amount of HNE in the wound. Considering the reddish to brownish colour of the wound environment (due to blood and pus) [29], we determined that the colour of choice for the sensor should be dark blue, dark green or deep purple [30,31], to avoid misleading interpretations. Taking this into consideration, we selected as subject of our work ultramarine (UM), a chromogenic dark blue coloured protein derived by genetic modifications [23,24] from Rtms5 (a naturally occurring GFP-like fluorescent protein) [32].
We developed our strategy, inspired by the method described in [28], to convert UM into a protease switch for HNE (schematized in Fig. 1). Briefly, we genetically modified UM to delete its last 11th β-sheet, leading to a colourless protein composed of a barrel with 10 β-sheets (UM10 protein). The sequence of the 11th β-sheet was produced hybridized with the stable surface-exposed α-helix from eglin c containing the specific HNE-cleavage sequence, Alanine-Alanine-Proline-Valine (AAPV) (E11 portion). As observed by Callahan and colleagues [28], we expected to restore the chromogenic environment when both polypeptides are mixed together in the presence of HNE. Similarly, to GFP, the released 11th β-strand should interact with the incomplete β-barrel from UM10 leading to the appearance of a blue colour (Fig. 1). To better understand the phenomenon, molecular dynamics studies were conducted to compare the structures of split UM upon HNE cleavage.
Fig. 1.
Schematic representation of the split UM strategy developed as sensor substrate for HNE. The GFP-like chromogenic protein ultramarine (UM) was engineered with the last 11th β-strand removed from the 10 β-sheet-barrel, creating a colourless protein (UM10). The 11th β-strand, preceded by HNE’s cleavage site, was inserted on eglin c moiety (E11). After HNE proteolysis, the 11th β-strand is released and its proximity to the incomplete β-barrel (UM10) should regenerate UM’s blue colour. Structures were created with VMD 1.9.3 software. Incomplete β-barrel is presented in grey, eglin c moiety is coloured golden, 11th β-strand is coloured blue and the cleavage sequence for HNE is represented as a red box (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
The design of a colour switch sensor based on a recombinant chromogenic protein for the monitoring of HNE can revolutionize the field of diagnostics and medical sensors, in particular in the wound care domain, since colour-based switch-on approaches with non-fluorescent GFP-like proteins were never reported before.
2. Materials and methods
2.1. Materials and reagents
All reagents were analytical grade, purchased from commercial suppliers and used as received. Standard molecular biology procedures were used for the gene cloning and protein recombinant expression in Escherichia coli TOP10 (Invitrogen) and E. coli BL21 DE3 (Novagen), respectively. DNA restriction enzymes NotI and NdeI and kanamycin antibiotic were purchased from ThermoFisher. Gold View for DNA staining was from UVAT Bio (Valencia, Spain). Culture media LB Broth and Terrific Broth (TB) (Auto induction medium) were purchased from Grisp (Porto, Portugal). Isopropyl-β-D-thiogalactopyranoside (IPTG) inductor was purchased from Acros Organics (Geel, Belgium). Nickel magnetic beads for His6 tag protein purification and respective magnetic separator were purchased from Bimake (Houston, USA). Protein molecular weight markers, Precision Plus Protein All Blue or Precision Plus Protein Dual Xtra Standards, were purchased from Biorad (Portugal). Chaotropic agent urea was purchased from labkem (Spain). All other chemicals were purchased from Sigma-Aldrich (Spain), including the enzyme elastase from human leukocytes, synthetic elastase substrate N-methoxysuccinyl-Alanine-Alanine-Proline-Valine-p-nitroanilide (MeOSuc-AAPV-pNA), imidazole and N-Laurylsarcosine sodium salt (sarkosyl) detergent. The dialysis tubing cellulose membranes (MWCO, 1–2 kDa and 12–14 kDa) and GenElute Plasmid Miniprep Kit were also obtained from Sigma-Aldrich. The ultramarine’s 11th β-strand peptide (pep11) was synthetized by JPT Peptide Technologies GmbH (Germany).
2.2. Instrumentation
All absorbance and fluorescence measurements were carried out in 96-well microplates (flat bottom, polystyrene and transparent plates, Nunc, ThermoScientific) of 300 μL capacity/well using a Synergy Mx spectrometer with the Gen5 Data Analysis Software (Bio-Tek Instruments, Inc., USA). DNA and total protein were performed quantified in Nanodrop 1000 (ThermoScientific). Bacterial cell lysis was accomplished with an Ultrasonic Processor VCX-500 W (Sonics & Materials, Inc., Connecticut, USA). Purified recombinant proteins were lyophilized in a FreeZone 2.5 Freeze Dry System (Labconco, Missouri, USA).
2.3. Methods
2.3.1. Protein expression and purification
Genes of UM, UM10 (GenBank accession number BankIt2109475, MH346479) and E11 (GenBank accession number BankIt2109475, MH346480) inserted in a pET28a(+) plasmid were ordered from Genscript (New Jersey, USA). Gene size confirmation was performed with GenElute Plasmid Miniprep Kit and double digested with the restriction enzymes NotI and NdeI; the molecular weight of the resultant DNA fragments was confirmed in a 1% agarose gel stained with Gold View. All plasmids were first established in E. coli TOP10 and then expressed in E. coli BL21 (DE3). The TSS method was used for transformations as previously described [33]. Expression and purification conditions were established after optimization protocols performed with UM. Optimized protein expression was accomplished in 24 h using LB medium supplemented with kanamycin (50 μg/mL) and 180 rpm after induction with 0.25 mM IPTG at a culture OD of 0.5. All proteins demanded a culture/flask ratio of 1:10 and aeration via gauze sheets. UM and E11 were incubated at 30 °C, whereas UM10 required a low incubation temperature (16 °C) to increase solubility. After expression, cultures were placed at 4 °C for at least 2 h to enhance chromophore folding. Then, the cells were harvested by centrifugation at 8,000g at 4 °C for 10 min, cell pellets were suspended in lysis buffer (20 mM NaH2PO4, 500 mM NaCl, pH 7.4) and subsequently lysed by ultrasounds. The lysate was centrifuged at 10,000g at 4 °C for 30 min. Proteins were purified using nickel magnetic beads and an imidazole gradient of 10 mM to 500 mM. At every step, the fractions obtained were monitored by SDS-PAGE with Coomassie blue staining. Removal of imidazole from the eluted fractions was performed through dialysis against ultrapure water using a cellulose membrane with a MWCO of 1–2 kDa for E11 protein and 12–14 kDa for the other proteins. The pure protein fractions were then frozen at -80 °C and lyophilized. The molecular weight of all proteins obtained by SDS-PAGE was in accordance with the theoretical size expected from the polypeptide sequences including the amino acids encoded by pET28a(+) vector.
2.3.2. Assessment of the UM-based split protein as HNE protease substrate
HNE activity was first evaluated in the batches used in the experiments with a control assay with the synthetic substrate MeOSuc-AAPV-pNA following the protocol supplied by the manufacturer. Briefly, the enzyme (0.5 U/mL) was mixed with 25–1500 μM of substrate, to a final volume of 300 μL of reaction buffer (100 mM HEPES with 500 mM NaCl, pH 8). A blank consisting only in buffer and substrate was also included. The kinetic constants were determined from the initial rates of hydrolysis by Michaelis-Menten method. All enzymatic assays were conducted at 37 °C for 10 min in triplicate and the absorbance was measured at 405 nm by UV–vis spectrophotometry.
The lyophilized proteins were suspended immediately prior to the assay in 100 mM Tris−HCl at pH 8. The absorbance spectra of the UM-based split proteins prior and after addition of HNE (fixed concentration of 150mU, previously determined by optimization) were recorded in a spectrophotometer for the assessment of colour change. The split UM system was prepared by mixing UM10 (fixed at 1.5 mg/mL or 3.4 mg/mL) and E11 in a final molar ratio of 167:1 or 167:5 (UM10:E11), or by mixing in the same ratios UM10 and pep11 (the synthetic peptide equivalent to free 11th β-sheet). Split UM system was first allowed to stabilize overnight before the addition of HNE, as done by Callahan and colleagues [28]. To prevent any inaccuracy measurements over time due to evaporation, the assay testing the split system was evaluated at RT. In addition, to improve the sensibility of the assay, the same assays were performed in a 96-well microplate firstly blocked with a solution of 0.5% bovine serum albumin (BSA), as described in [34].
2.4. Molecular dynamics simulations
The molecular dynamics simulations were performed with the Gromacs 4.0.7 package applying the GROMOS 54A7 united-atom force-field [[35], [36], [37], [38], [39]], with periodic boundary conditions, at 300 K and 1 atm.
The molecular models for the UM and E11 proteins were obtained through structure homology using the SWISS-MODEL web service (http://swissmodel.expasy.org). The X-ray model of the Rtms5 protein (PDB code: 3vk1) was used as template for UM while the NMR model of eglin c (PDB code: 1 EG L) was used for E11. The UM10, pep11 and cleaved E11 molecular structures were obtained through manual edition of the UM and E11 models. The simulation pictures were rendered with VMD (Visual Molecular Dynamics) software [40].
2.5. Simulation systems and protocol
Nine different systems, composed of water and proteins models, were built in this work. The proteins models were the UM, UM10, pep11, eglin c, E11, cleaved E11, UM10 with pep11, UM10 with E11 and UM10 with cleaved E11. The systems were subjected to energy minimization, three equilibration simulations, and finally to the production run. At least to replicas of each system were built, and 100 ns of total production simulation were calculated. The solvated UM, UM10, pep11, eglin c, E11 and cleaved E11 systems were simulated to equilibrate its molecular structures. The remaining UM10 with pep11, UM10 with E11 and UM10 with cleaved E11 system were simulated to unveil the possible interactions between these molecules, with main focus on the β11 sequence (free or inserted in the eglin c protein) and UM10 interactions. After the production runs visual inspection and several properties such as the temperature, pressure, and potential energy were evaluated to check if the system were well equilibrated. More details about the molecular dynamics simulations is presented in supplementary information.
3. Results
3.1. Production of the split UM substrate for HNE detection
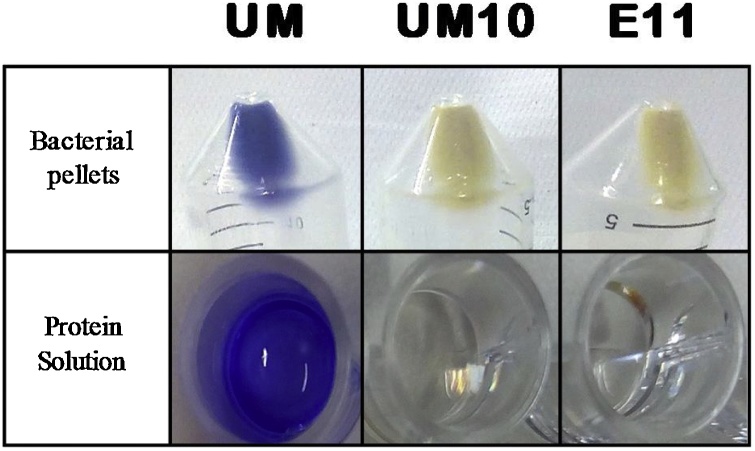
Ultramarine (UM) protein, its incomplete version made of 10 β-sheets (UM10) and the sensing substrate composed by the HNE cleavage sequence AAPV plus the sequence of the 11th β-sheet incorporated in eglin c moiety (E11) were expressed in E. coli with distinct solubility profiles. The original UM presented a dark blue colour and the engineered proteins UM10 and E11 were non-coloured, as expected (Fig. 2). UM and E11 proteins were mainly obtained in the soluble fraction, but the expression of UM10 was challenging. After verifying its low initial protein expression yield, with the same protocol used for the other two proteins, we undertook an optimization of the conditions to control the culture’s growth rate and increase protein proper expression. Culture’s growth rate was slowed-down by reducing the incubation temperature from 30 °C to 16 °C to improve the chances of correct folding of the protein being expressed. In addition, after cell lysis, several ice-vortex cycles were performed to aid its solubilisation, thus enhancing the correct folding of the protein. Under these optimized conditions, UM10 was expressed ≈50% soluble (data not shown).
Fig. 2.
Colour of the UM, UM10 and E11 proteins. Bacterial pellets after expression under optimized laboratorial set-up and in solution after purification at 1.5 mg/mL for UM and UM10, and at 0.5 mg/mL for E11.
3.2. Effect of HNE on split UM system
Prior to the assessment of the split UM substrate in the presence of HNE, we evaluated the protease effect in the original UM as a measure of specificity analysis. Foremost, loss of colour was not observed in ultramarine after incubation with HNE, even over 24 h of incubation, indicating no significant proteolytic effect of elastase on our casting protein. However, a slight and transient increase in absorbance was registered upon HNE addition (≈ 0.18 a.u. corresponding to ≈ 11% increase) (Supplementary Figure S1A). Somehow, in the first moments, the interaction of the protein with HNE led to a gain on the absorbance, but, since it returned to baseline values, this effect was disregarded. However, by SDS-PAGE we detected in ultramarine a small molecular weight decrease of approximately 1 kDa after incubation with HNE (Supplementary Figure S1B). After analysis of the theoretical cleavage map provided by ExPASy Peptide Cutter software (SIB ExPASy Bioformatics Resources Portal, available online tool) [41], we inferred, by the corresponding fragment’s size, that it probably resulted from a small cleavage right after the His6 tag at protein’s N-terminal (Supplementary Figure S1C). Still, this cleavage does not affect the protein conformation nor chromophore’s arrangement, since the colour remained blue even after several days of incubation with the enzyme (Supplementary Figure S1 and data not shown). Hence, any colour change in the split UM system as specific to the action of HNE at the inserted cleavage site located on E11.
The efficiency of the system was assayed by adding HNE to purified UM10 (fixed at 3.4 mg/mL) previously mixed with E11 in the molar ratios 167:1 and 167:5 of UM10:E11. The synthetic peptide equivalent to the 11th β-strand (pep11) was also included in this assay and mixed with UM10 at the same ratios as E11. Evaluation of the interaction of the 11th β-strand and the incomplete β-barrel was made by measurement of the absorbance over time (Fig. 3).
Fig. 3.
Efficiency of (A) the split UM system in detecting HNE (150 mU) and comparison with (B) the mixture of UM10 with the synthetic peptide pep11, in molar ratios of 167:1 or 167:5. Absorbance was measured at 585 nm for 24 or 44 h, respectively, and normalized against the absorbance of UM10. All assays were performed at room temperature. Bars representing E11 (control) and pep11 (control) are correspondent to 4.09 nmol (equivalent amount used in 167:5 ratio). Registered absorbances of UM10 (control) presented as dashedlines in (A) and black dots in (B) are points referred to the secondary vertical axis.
For split UM system, a measurable increase in absorbance was observed for a period of 6 h after the addition of HNE (≈ 0.2 a.u of absorbance in UM10 + E11+HNE mixture, corresponding to ≈75% increase) (Fig. 3A and Supplementary Figure S2A). However, we could not detect by naked-eye a gain of colour of the solution. After this time, the absorbance steeply decreased and then stabilized at the 7th hour of incubation, continuing with a steady and very slow decrease in the absorbance verified until the end of the assay.
We let the solution to dry in the wells and then we observed some purplish-blue (Supplementary Figure S3) similar to those that formed after evaporation of UM which evidence the formation of the UM10 + E11 complex.
To rule out a poor enzymatic cleavage by HNE on E11, the interaction of UM10 with a free and readily available 11th β-sheet was evaluated using pep11 peptide instead of E11 protein (Fig. 3B and Supplementary Figure S2B). This system followed almost the same trend as the described for UM10 + E11+HNE, although the interaction of UM10+pep11 differed in each tested ratio, contrarily to the observed with E11. When mixing UM10+pep11 in 167:1 ratio, the absorbance of the complex slowly increased for 42 h. Then, in 30 min, it rapidly recovered the initial values and continued to drop. However, increasing the number of pep11 molecules, the decrease in absorbance occurred in only 18 h, then it continued to drop for 24 h to much lower values than those initially recorded, and finally it stabilized in the last hours of the assay.
3.3. Molecular dynamics studies for comprehension of the split UM system
Structural homology analysis was done between UM and UM10 proteins to compare both β-barrels (Supplementary Figure S4A). Despite missing the last β-sheet, UM10 showed a closely superposing structure with the original UM, without relevant relaxation or constriction of the barrel’s diameter (top and bottom views in Supplementary Figure S4). Still, as expected, the alignment of both structures showed a significant displacement of the 10th β-strand in UM10 which occupies the gap left by the absence of the 11th β-strand.
As a quantifying measure, we used the root-mean-square fluctuation (RMSF) values of UM and UM10 since it can provide physical information regarding the residues mobility through the in silico simulations (Supplementary Figure S4B). Terminal residues have free movement due to the absence of a constraining force at one end. For this reason, residues 1–7 and 217–220 in UM and 1–7 and 199–201 in UM10 were not considered in the analysis as their RMSF values are biased by the inherited freedom. Removal of the last 11th β-strand significantly affected the fluctuation of some residues, namely the ones located in the internal loop (49–80) that holds the chromophore group, but also in β5 (107–112) and in β6 (121–124) (Supplementary Figure S4B). In particular, chromophore interacting residues Pro59, Tyr78 and Lys80 and conserved residues Gly122 and Thr123 showed fluctuation values increase by 2-fold in comparison to UM.
To better understand the interaction between UM10 and E11, cleaved E11 was placed together with UM10 in the simulation box which was run 4 times (Fig. 4). Two of the four replicates showed an interaction of the sequence of β11 hanging from E11 with UM10 (Fig. 4C and D), although not at the original position of β11.
Fig. 4.
Simulation of the interactions between UM10 signalling moiety (in grey) and cleaved E11 substrate moiety, with eglin c in golden, sequence of the 11th β-strand in blue and cleavage site in red. Conformations of the final stage resulting from four simulations are presented separately (A–D), showing front, back and side views. Ultramarine original conformation is shown in E. All structures were generated with VMD 1.9.3 software. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Additionally, six replicas of the interaction of pep11 with UM10 were run (Fig. 5). Four out of six showed interaction of pep11 with UM10, however only in replica 3 pep11 was located at a given time in the β11 gap of the UM10 barrel. During this simulation, pep11 interacted in other faces of UM10, interspersing with searches for other sites of the barrel.
Fig. 5.
Simulation of the interaction between UM10 (in grey) and pep11, with each of the 6 replicas presented in different colours, showing back and side views. Replicas were combined in the same figure, fixing the UM10 barrel. Side box distinguishes the only system conformation where pep11 interacted in the β11 gap. All structures were generated with VMD 1.9.3 software.
4. Discussion
4.1. HNE signalling occurs though transiently
The rationale published years ago by [28] showing the adaptation of the split-GFP technology [27] into a protease switch together with the need of a naked-eye detection system led us to engineer a switch-on system for HNE based on ultramarine (UM), a chromogenic GFP-like protein. To produce the sensing moiety, we inserted the sequence of the 11th β-strand from UM into the stable surface-exposed α-helix loop of eglin c and added AAPV peptide, the HNE specific cleavage sequence, upstream the β11 sequence. The signalling moiety corresponds to the incomplete UM10 β-barrel which bears the chromophore group, at this stage, unable to produce all the required interactions for colour generation.
The difficulties encountered on the production of soluble UM10 should be mainly related to the absence of the last β-sheet as it is known that hysteresis of GFP-like proteins is directly related to the C-terminal β-sheets (β7-β11) [25]. Our total extracts had a good amount of the desired protein, but unable to fold correctly. The modifications made to the production protocol allowed the recovery 50% of the protein in the soluble fraction.
The absence of colour of UM10 (Fig. 2) is due to the inexistence of the 11th β-sheet. The absence of the two chromophore interacting residues Gln209 and Glu211 from the 11th β-sheet prevents the establishment of the complete network of interactions paramount for chromophore maturation [27]. The 11th β-sheet itself is significantly involved in the refolding pathways required for the complete chromophore maturation and directly protects the chromophore from the surrounding environment [26]. Being absent, β10 acquires a conformation slightly distorted. Nevertheless, the overall barrel structure of UM10 is very similar to the original structure of UM (Supplementary Figure S4A), as observed in the crystallography of protein GFP 1–9 (GFP lacking β10 and β11) [42].
Changes in the dynamic behaviour of UM10 were better analysed by the calculation of the root-mean-square fluctuation (RMSF) of residues in comparison to the original UM (Supplementary Figure S4B). A higher RMSF value is related to more flexible movement of a given residue during the simulation time in relation to its average position, whereas a low RMSF value indicates limited movement. The 2-fold increase in the fluctuation of some chromophore-interacting residues (Pro59, Tyr78 and Lys80) and conserved residues (Gly122 and Thr123) in comparison to the original UM is also indicative of alterations on the conditions for the establishment of chromophore environment.
The close neighbouring of Pro59, Tyr78 and Lys80 and the element of the chromophore triad Tyr63 (Tyr66 in GFP), which allows the establishment of the strong interaction network that compose the chromophore environment is, hence, modified [[43], [44], [45], [46], [47]]. Particularly, Pro59 (Thr63 in GFP) is reported to be restricted within the GFP for the establishment of carbon-proton bounds with the vicinity, instead in UM10 this residue possesses higher fluctuation and it is freely rotating [45]. Furthermore, the absent residues Gln209 and Glu211 from the 11th β-sheet are known to promote the intramolecular complex formation of the chromophore group, being instrumental for stabilization of the key reaction intermediates from precyclized tripeptide to the dehydrated intermediate [43,46]. In particular, Gln209 (Glu222 in GFP) is required to interact to Tyr63 for the completion of this process.
We expected that chromophore environment, and hence colour production, could be restored by the proximity of the 11th β-strand to the incomplete β-barrel, following the same mechanism of split GPF technology nowadays broadly implemented [27,28,34,42,[48], [49], [50], [51], [52]]. HNE cleavage on AAPV at E11 should release the N-terminal of the 11th β-strand, being available for interaction with the incomplete β-barrel from UM10.
Our switch-on system was evaluated in vitro by measurements of the absorbance that was expected to increase upon HNE addition to the mixture of UM10 + E11. Indeed, a ≈75% increase in the absorbance was observed in the first 6 h, but the signal was lost afterwards (Fig. 3A and Supplementary Figure S2A). Our interpretation is that the interaction of UM10 with the 11th β-strand hanging from C-terminal fraction of E11 was not stable and the chromogenic state of the chromophore was not maintained for long in solution. The registered decrease in absorbance in the last part of the assay results from the instability of the established interaction between the 11th β-strand and UM10; the 11th β-strand might have drifted to the solution and no longer interacted with UM10 protein. The dynamics of this interaction was not dependent on number of E11 molecules, since the same absorbance changes were observed for both tested ratios of UM10:E11 (167:1 and 167:5). The same was observed in the work of Callahan et al., as the fluorescence changes were mainly influenced by the GFP1–10 molecules rather than GPF11 [28]. We observed the formation of purplish-blue crystals at the bottom of the well after complete solvent evaporation (Supplementary Figure S3). The concentration effect occurring during evaporation might have promoted re-interaction of the system which crystalized in correct conformation. This effect can favour the application here proposed, as the coloured signal of this split substrate should be kept stable even after drying of the exudate on the dressing.
Molecular dynamics simulations helped us to better understand the possible interactions occurring in the system UM10 + E11+HNE. Indeed, the interaction of the 11th β-strand in cleaved-E11 with UM10 did not present a specific tendency to the β11 gap β-barrel (Fig. 4). Instead, the interaction seemed to be quite misguided. In addition, the structure of cleaved-E11 suffered no distortion compared to E11 (Supplementary Figure S4). This led us to infer that the high stability of this loop from eglin c restrains the mobility of the 11th β-strand, which is unable to escape and find β11 gap at UM10.
To exclude as cause for the transient increase in absorbance the eventual entropy caused by the presence of HNE or the inefficacy of the enzyme in releasing the 11th β-strand, we made readily available the peptide corresponding to the sequence of the 11th β-strand by adding pep11 to UM10. Like in UM10 + E11+HNE system, the phenomenon was transitory, but on the opposite, this system’s behaviour seemed to be dependent on the number of molecules of pep11 available (Fig. 3B and Supplementary Figure S2B). The interaction was faster and steeper when more molecules of pep11 were added to UM10. In addition, we observed protein precipitation after 42 h for 167:1 ratio or 18 h for 167:5 ratio (data not shown) which coincided with the decrease in the absorbance; the rate of precipitation was higher and faster when more pep11 molecules were involved. The same happened with the pep11-only control assay (grey bar in Supplementary Figure S2B), with precipitation occurring after 18 h of incubation which correlates with increase in absorbance of pep11 from 18 to 42 h. The precipitation of pep11 impends its interaction with the barrel, as less soluble molecules are available in the surrounding milieu. In silico studies confirm the instability and randomness of the interaction of pep11 with UM10, with only one out of six replicas displaying a correct approach and placement of pep11 in the β11 gap of UM10 (Fig. 5).
At the available amount of protein UM10 (limiting factor at this moment), the gain of colour (corresponding to an increase of 0.2 a.u) was not detectable by naked-eye in solution. Only after complete evaporation, the resulting crystals had a slight gain of purplish-blue colour, however we were not able to resolubilize them and verify the colour once again (Supplementary Figure S3). The observed results at this stage, though promising, were still inconclusive since the mechanism of interaction here presented is not yet fully understood.
The commercially available Neutrophil Elastase Activity Assay Kit (BioVision) is said to have a sensibility of 1 ng of HNE by off-line fluorometric detection. Making a direct correlation with our assays, the 150 mU correspond to 7.5 ng of HNE. Considering this as the limit of detection of our system, the split UM assay is 7.5 times less sensible than the already existing kit. However, in the practice, it is sensible enough for the detection of HNE in chronic wounds since the enzyme’s activity in infected wounds reaches ∼ 20 U/mL, corresponding to ∼ 40,000 mU while non-infected wounds is ∼ 2 U/mL (∼ 4000 mU) [2]. Furthermore, our system here under development is destined for naked-eye read-out in real-time and in-situ (on the dressing), which is not possible using the BioVision fluorometric kit.
The work here presented proves the difficult process on re-forming a stable chromophore after cleavage in UM, which might be transversal to all chromoproteins. Our findings suggest that we must first overcome the low heterologeous expression yield of UM10 to further characterize the UM split system. Nowadays, methods using fluorescence-based switch-on approaches with GFP-like proteins are widely used [27,28,34,53,46,49,54,55], however GFP-like colour-based approaches are still a long way to be achieved. Even though we could not obtain a stable colour change on our split UM system, we were able to register an interaction between the incomplete β-barrel and the 11th β-strand upon HNE cleavage translated into absorbance changes and precipitation. Hence, this work serves as a proof-of-concept that a chromogenic protein engineered in a split system may be used as a switch sensor. Combination of molecular dynamics studies supported the in vitro observations and helped us to understand the physicochemical phenomenon behind the split technology. It became clear the challenging implications for the production of colour in our system. To our knowledge, this is the first attempt in the development of a split system based on a chromogenic GFP-like protein. Our findings can guide the following research in this chromogenic sensing systems using GFP-like proteins.
4.2. Field of application
The focus of this work was the detection of elastase, having in mind a biomedical application in medical sensor devices for wound care. The embedding of a chromogenic HNE-specific substrate into a wound dressing material could allow in-situ and real-time assessment of the status of a wound at an early stage of inflammation and/or infection. HNE enzyme present in the exudate of a chronic wound would act on the sensor leading to a colour change on the dressing. By simple visualization of the coloured signal, the medical staff could monitor the activity levels of HNE in the wound, and consequently acknowledge whether the current treatment is being effective for the wound healing. Monitoring of the wound status can be further explored through a combination of other markers of inflammation/infection aside from HNE enzyme, for example matrix metalloproteinases (MMPs), proteinase 3, collagenase, reactive oxygen species (ROS) or even pH [[56], [57], [58], [59]]. This technology can be explored for other uses, not only adapted for different proteases, but also for alternative applications. Here, we studied the use of the chromogenic protein UM specifically chosen due to its blue colour which is easily discriminated in a dressing covering a non-healing wound. However, the existing variety in colours of GFP-like chromoproteins along with other properties [46,[60], [61], [62]] further broads the usage of this technology. Split GFP-like chromoproteins offer a promising and compelling research focus with a length of possibilities, that once fully understood can revolutionize naked-eye visual sensing.
Funding
This study was funded by the European project InFact – Functional materials for fas diagnosis of wound infection (FP7-NMP-2013-SME-7 - Grant agreement no. 604,278). The work done at Centre of Biological Engineering (CEB) was also supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UID/BIO/04469/2013 unit, COMPETE 2020 (POCI-01-0145-FEDER-006684) and BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by European Regional Development Fund under the scope of Norte2020 - Programa Operacional Regional do Norte.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We greatly acknowledge the European project InFact – Functional materials for fast diagnosis of wound infection (FP7-NMP-2013-SME-7 - Grant agreement no. 604,278) for funding the work and also the grant for Joana Cunha and José Egipto Antunes. Ana Ferreira, Artur Ribeiro and Teresa Matamá would also like to acknowledge the Portuguese Foundation for Science and Technology (FCT) for funding their scholarships with the references SFRH/BD/113247/2015, SFRH/BPD/98388/2013 and SFRH/102153/2014, respectively. The work done at Centre of Biological Engineering (CEB) was supported by FCT under the scope of the strategic funding of UID/BIO/04469/2013 unit, COMPETE 2020 (POCI-01-0145-FEDER-006684) and BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by European Regional Development Fund under the scope of Norte2020 - Programa Operacional Regional do Norte.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2019.e00324.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Barros S.C., Louro R.O., Micaêlo N.M., Martins J.A., Marcos J.C., Cavaco-Paulo A. NMR and molecular modelling studies on elastase inhibitor-peptides for wound management. React. Funct. Polym. 2013;73:1357–1365. [Google Scholar]
- 2.Hasmann A., Gewessler U., Hulla E., Schneider K.P., Binder B., Francesko A., Tzanov T., Schintler M., Van der Palen J., Guebitz G.M., Wehrschuetz-Sigl E. Sensor materials for the detection of human neutrophil elastase and cathepsin G activity in wound fluid. Exp. Dermatol. 2011;20:508–513. doi: 10.1111/j.1600-0625.2011.01256.x. [DOI] [PubMed] [Google Scholar]
- 3.Vasconcelos A., Azoia N.G., Carvalho A.C., Gomes A.C., Güebitz G., Cavaco-Paulo A. Tailoring elastase inhibition with synthetic peptides. Eur. J. Pharmacol. 2011;666:53–60. doi: 10.1016/j.ejphar.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 4.Vasconcelos A., Pêgo A.P., Henriques L., Lamghari M., Cavaco-Paulo A. Protein matrices for improved wound healing: elastase inhibition by a synthetic peptide model. Biomacromolecules. 2010;11:2213–2220. doi: 10.1021/bm100537b. [DOI] [PubMed] [Google Scholar]
- 5.Sen C.K., Gordillo G.M., Roy S., Kirsner R., Lambert L., Hunt T.K., Gottrup F., Gurtner G.C., Longaker M.T. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2010;17:763–771. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macgregor L., Calne S., Day K., Jones J., Pugh A. Wound infection in clinical practice. An international consensus. Int. Wound J. 2008;5(Suppl 3) doi: 10.1111/j.1742-481X.2008.00488.x. iii-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards J., Fontenot K., Prevost N., Pircher N., Liebner F., Condon B. Preparation, characterization and activity of a peptide-cellulosic aerogel protease sensor from cotton. Sensors. 2016;16:1789. doi: 10.3390/s16111789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards J., Caston-Pierre S., Bopp A.F., Goynes W. Detection of human neutrophil elastase with ethoxylate acrylate resin analogs. J. Pept. Res. 2005;66:160–168. doi: 10.1111/j.1399-3011.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 9.Edwards J.V., Prevost N.T., French A.D., Concha M., Condon B.D. Kinetic and structural analysis of fluorescent peptides on cotton cellulose nanocrystals as elastase sensors. Carbohydr. Polym. 2015;116:278–285. doi: 10.1016/j.carbpol.2014.04.067. [DOI] [PubMed] [Google Scholar]
- 10.Edwards J.V., Prevost N., Sethumadhavan K., Ullah A., Condon B. Peptide conjugated cellulose nanocrystals with sensitive human neutrophil elastase sensor activity. Cellulose. 2013;20:1223–1235. [Google Scholar]
- 11.Faccio G., Salentinig S. Enzyme-triggered dissociation of a FRET-Based protein biosensor monitored by synchrotron SAXS. Biophys. J. 2017;113:1731–1737. doi: 10.1016/j.bpj.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontenot K.R., Edwards J.V., Haldane D., Pircher N., Liebner F., Condon B.D., Qureshi H., Yager D. Designing cellulosic and nanocellulosic sensors for interface with a protease sequestrant wound-dressing prototype: implications of material selection for dressing and protease sensor design. J. Biomater. Appl. 2017;32:622–637. doi: 10.1177/0885328217735049. [DOI] [PubMed] [Google Scholar]
- 13.Fontenot K.R., Edwards J.V., Haldane D., Graves E., Citron M.S., Prevost N.T., French A.D., Condon B.D. Human neutrophil elastase detection with fluorescent peptide sensors conjugated to cellulosic and nanocellulosic materials: part II, structure/function analysis. Cellulose. 2016;23:1297–1309. [Google Scholar]
- 14.Hasmann A., Wehrschuetz-Sigl E., Kanzler G., Gewessler U., Hulla E., Schneider K.P., Binder B., Schintler M., Guebitz G.M. Novel peptidoglycan-based diagnostic devices for detection of wound infection. Diagn. Microbiol. Infect. Dis. 2011;71:12–23. doi: 10.1016/j.diagmicrobio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Henares T.G., Takaishi M., Yoshida N., Terabe S., Mizutani F., Sekizawa R., Hisamoto H. Integration of multianalyte sensing functions on a capillary-assembled microchip: simultaneous determination of ion concentrations and enzymatic activities by a “drop-and-sip” technique. Anal. Chem. 2007;79:908–915. doi: 10.1021/ac061245i. [DOI] [PubMed] [Google Scholar]
- 16.Schulenburg C., Faccio G., Jankowska D., Maniura-Weber K., Richter M. A FRET-based biosensor for the detection of neutrophil elastase. Analyst. 2016;141:1645–1648. doi: 10.1039/c5an01747e. [DOI] [PubMed] [Google Scholar]
- 17.Stair J.L., Watkinson M., Krause S. Sensor materials for the detection of proteases. Biosens. Bioelectron. 2009;24:2113–2118. doi: 10.1016/j.bios.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Zagorevski D.V., Stenken J.A. In situ and multisubstrate detection of elastase enzymatic activity external to microdialysis sampling probes using LC-ESI-MS. Anal. Chem. 2008;80:2050–2057. doi: 10.1021/ac702047w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards J.V. Protease biosensors based on peptide-nanocellulose conjugates: from molecular design to dressing interface. Int. J. Med. Nano Res. 2016;3 [Google Scholar]
- 20.Stepanenko O.V., Stepanenko O.V., Kuznetsova I.M., Verkhusha V.V., Turoverov K.K. Beta-barrel scaffold of fluorescent proteins. Int. Rev. Cell Mol. Biol. 2013:221–278. doi: 10.1016/B978-0-12-407699-0.00004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craggs T.D. Green fluorescent protein: structure, folding and chromophore maturation. Chem. Soc. Rev. 2009;38:2865. doi: 10.1039/b903641p. [DOI] [PubMed] [Google Scholar]
- 22.Ong W.J.-H., Alvarez S., Leroux I.E., Shahid R.S., Samma Aa, Peshkepija P., Morgan A.L., Mulcahy S., Zimmer M. Function and structure of GFP-like proteins in the protein data bank. Mol. Biosyst. 2011;7:984. doi: 10.1039/c1mb05012e. [DOI] [PubMed] [Google Scholar]
- 23.Pettikiriarachchi A., Gong L., Perugini M.A., Devenish R.J., Prescott M. Ultramarine, a chromoprotein acceptor for förster resonance energy transfer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott M., Ling M., Beddoe T., Oakley A.J., Dove S., Hoegh-Guldberg O., Devenish R.J., Rossjohn J. The 2.2 Å crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation. Structure. 2003;11:275–284. doi: 10.1016/s0969-2126(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 25.Zimmer M.M.H.M., Li B., Shahid R., Peshkepija P., Zimmer M.M.H.M. Structural consequences of chromophore formation and exploration of conserved lid residues amongst naturally occurring fluorescent proteins. Chem. Phys. 2014;429:5–11. doi: 10.1016/j.chemphys.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B., Shahid R., Peshkepija P., Zimmer M. Water diffusion in and out of the β-barrel of GFP and the fast maturing fluorescent protein. TurboGFP Chem. Phys. 2012;392:143–148. doi: 10.1016/j.chemphys.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabantous S., Terwilliger T.C., Waldo G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 28.Callahan B.P., Stanger M.J., Belfort M. Protease activation of split green fluorescent protein. ChemBioChem. 2010;11:2259–2263. doi: 10.1002/cbic.201000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen G.L., Sparrow E.M., Kokate J.Y., Leland K.J., Iaizzo P.A. Wound status evaluation using color image processing. IEEE Trans. Med. Imaging. 1997;16:78–86. doi: 10.1109/42.552057. [DOI] [PubMed] [Google Scholar]
- 30.Schiffer D., Tegl G., Vielnascher R., Weber H., Herrero-Rollett A., Sigl E., Heinzle A., Guebitz G.M. Myeloperoxidase-responsive materials for infection detection based on immobilized aminomethoxyphenol. Biotechnol. Bioeng. 2016;113:2553–2560. doi: 10.1002/bit.26025. [DOI] [PubMed] [Google Scholar]
- 31.Tegl G., Rollett A., Dopplinger J., Gamerith C., Guebitz G.M. Chitosan based substrates for wound infection detection based on increased lysozyme activity. Carbohydr. Polym. 2016;151:260–267. doi: 10.1016/j.carbpol.2016.05.069. [DOI] [PubMed] [Google Scholar]
- 32.Petersen J., Wilmann P.G., Beddoe T., Oakley A.J., Devenish R.J., Prescott M., Rossjohn J. The 2.0-Å crystal structure of eqFP611, a far red fluorescent protein from the sea Anemone entacmaea quadricolor. J. Biol. Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 33.Chung C.T., Niemela S.L., Miller R.H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabantous S., Waldo G.S. In vivo and in vitro protein solubility assays using split GFP. Nat. Methods. 2006;3:845–854. doi: 10.1038/nmeth932. [DOI] [PubMed] [Google Scholar]
- 35.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 36.Huang W., Lin Z., van Gunsteren W.F. Validation of the GROMOS 54A7 force field with respect to β-Peptide folding. J. Chem. Theory Comput. 2011;7:1237–1243. doi: 10.1021/ct100747y. [DOI] [PubMed] [Google Scholar]
- 37.Lindahl E., Hess B., van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001:306–317. [Google Scholar]
- 38.Schmid N., Eichenberger A.P., Choutko A., Riniker S., Winger M., Mark A.E., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 39.Scott W.R.P., Hünenberger P.H., Tironi I.G., Mark A.E., Billeter S.R., Fennen J., Torda A.E., Huber T., Krüger P., van Gunsteren W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A. 1999;103:3596–3607. [Google Scholar]
- 40.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–8) doi: 10.1016/0263-7855(96)00018-5. 27–8. [DOI] [PubMed] [Google Scholar]
- 41.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., Grosdidier A., Hernandez C., Ioannidis V., Kuznetsov D., Liechti R., Moretti S., Mostaguir K., Redaschi N., Rossier G., Xenarios I., Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen H.B., Hung L.W., Yeates T.O., Terwilliger T.C., Waldo G.S. Split green fluorescent protein as a modular binding partner for protein crystallization. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:2513–2523. doi: 10.1107/S0907444913024608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigorenko B.L., Krylov A.I., Nemukhin A.V. Molecular modeling clarifies the mechanism of chromophore maturation in the green fluorescent protein. J. Am. Chem. Soc. 2017;139:10239–10249. doi: 10.1021/jacs.7b00676. [DOI] [PubMed] [Google Scholar]
- 44.Orm M., Cubitt A.B., Kallio K., Gross L.A., Tsien R.Y., Remington S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273(80-):1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 45.Shinobu A., Palm G.J., Schierbeek A.J., Agmon N. Visualizing proton antenna in a high-resolution green fluorescent protein structure. J. Am. Chem. Soc. 2010;132:11093–11102. doi: 10.1021/ja1010652. [DOI] [PubMed] [Google Scholar]
- 46.Stepanenko O.V., Verkhusha V.V., Kuznetsova I.M., Uversky V.N., Turoverov K.K. Fluorescent proteins as biomarkers and biosensors: throwing color lights on molecular and cellular processes. Curr. Protein Pept. Sci. 2008;9:338–369. doi: 10.2174/138920308785132668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vinkenborg J.L., Nicolson T.J., Bellomo E.A., Koay M.S., Rutter G.A., Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrara F., Listwan P., Waldo G.S., Bradbury A.R.M. Fluorescent labeling of antibody fragments using split GFP. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foglieni C., Papin S., Salvadè A., Afroz T., Pinton S., Pedrioli G., Ulrich G., Polymenidou M., Paganetti P. Split GFP technologies to structurally characterize and quantify functional biomolecular interactions of FTD-related proteins. Sci. Rep. 2017;7:14013. doi: 10.1038/s41598-017-14459-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent K.P., Boxer S.G. Light-activated reassembly of split green fluorescent protein. J. Am. Chem. Soc. 2011;133:4046–4052. doi: 10.1021/ja110256c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Listwan P., Terwilliger T.C., Waldo G.S. Automated, high-throughput platform for protein solubility screening using a split-GFP system. J. Struct. Funct. Genomics. 2009;10:47–55. doi: 10.1007/s10969-008-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tansila N., Tantimongcolwat T., Isarankura-Na-Ayudhya C., Nantasenamat C., Prachayasittikul V. Rational design of analyte channels of the green fluorescent protein for biosensor applications. Int. J. Biol. Sci. 2007;3:463–470. doi: 10.7150/ijbs.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang E.D., Y.-M. J. D, Schenkelberg C., Fraser K., Macari S., Bystroff C. GFP-based biosensors. State Art Biosens. Gen. Asp. 2013 [Google Scholar]
- 54.Hense A., Nienhaus K., Nienhaus G.U. Exploring color tuning strategies in red fluorescent proteins. Photochem. Photobiol. Sci. 2015;14:200–212. doi: 10.1039/c4pp00212a. [DOI] [PubMed] [Google Scholar]
- 55.Verkhusha V.V., Lukyanov K.A. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 56.Eming S.A., Krieg T., Davidson J.M. Inflammation in wound repair: molecular and cellular mechanisms. J. Invest. Dermatol. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 57.Power G., Moore Z., O’Connor T. Measurement of pH, exudate composition and temperature in wound healing: a systematic review. J. Wound Care. 2017;26:381–397. doi: 10.12968/jowc.2017.26.7.381. [DOI] [PubMed] [Google Scholar]
- 58.Snyder R.J., Driver V., Fife C.E., Lantis J., Peirce B., Serena T., Weir D. Using a diagnostic tool to identify elevated protease activity levels in chronic and stalled wounds: a consensus panel discussion. Ostomy. Manage. 2011;57:36–46. [PubMed] [Google Scholar]
- 59.Witko-Sarsat V., Rieu P., Descamps-Latscha B., Lesavre P., Halbwachs-Mecarelli L. Neutrophils: molecules, functions and pathophysiological aspects. Lab. Invest. 2000;80:617–653. doi: 10.1038/labinvest.3780067. [DOI] [PubMed] [Google Scholar]
- 60.Labas Y.A., Gurskaya N.G., Yanushevich Y.G., Fradkov A.F., Lukyanov K.A., Lukyanov S.A., Matz M.V. Diversity and evolution of the green fluorescent protein family. Proc. Natl. Acad. Sci. 2002;99:4256–4261. doi: 10.1073/pnas.062552299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pakhomov A.A., Martynov V.I. GFP family: structural insights into spectral tuning. Chem. Biol. 2008;15:755–764. doi: 10.1016/j.chembiol.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Shkrob M.A., Mishin A.S., Chudakov D.M., Labas Y.A., Lukyanov K.A. Chromoproteins of the green fluorescent protein family: properties and applications. Russ. J. Bioorganic Chem. 2008;34:517–525. doi: 10.1134/s1068162008050014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.