Abstract
Aim:
To investigate the formation of Mycobacterium avium membrane vesicles (MVs) within macrophage phagosomes.
Materials & methods:
A phagosome model was utilized to characterize proteomics and lipidomics of MVs. A click chemistry-based enrichment assay was employed to examine the presence of MV proteins in the cytosol of host cells.
Results:
Exposure to metals at concentrations present in phagosomes triggers formation of bacterial MVs. Proteomics identified several virulence factors, including enzymes involved in the cell wall synthesis, lipid and fatty acid metabolism. Some of MV proteins were also identified in the cytosol of infected macrophages. MVs harbor dsDNA.
Conclusion:
M. avium produces MVs within phagosomes. MVs carry products with potential roles in modulation of host immune defenses and intracellular survival.
Keywords: : AHA, bioorthogonal metabolic labeling, M. avium, macrophages, membrane vesicles, minimal medium, phagosome model
Mycobacterium avium subsp. hominissuis (MAH) belongs to the group of nontuberculous mycobacteria whose worldwide disease incidence and prevalence are on the rise [1]. MAH is one of the leading causes of bacterial infection in patients with HIV/AIDS and in individuals with chronic lung conditions [2,3]. In addition, pulmonary infections in immunocompetent middle aged and elderly individuals without any history of lung diseases have been also documented [4]. MAH has the ability to invade and proliferate within a variety of mammalian cells, including mucosal epithelium cells and macrophages. Following invasion, the pathogen is contained in a cytoplasmic vacuole, and intracellular survival is facilitated by a number of bacterial virulence factors associated with the remodeling of the intracellular compartment and resistance to the host antimicrobial killing mechanisms [5–8].
It has been shown that surface-localized secretion machineries and secreted substrates are important virulence factors for many bacterial pathogens, primarily, because of their roles in the pathogen–host interaction [9–11]. In addition, many bacterial pathogens produce and utilize outer membrane vesicles (OMVs) as a mechanism of exporting the multiple complex factors such as active enzymes, toxins, lipids, polysaccharides, peptidoglycans, lipoproteins, DNA, RNA and quorum sensing molecules [12–14] across the bacterial cell envelope and ultimately delivering them into the host cells [15,16]. While OMVs formation occurs under all physiological conditions, the vesiculation process is accelerated under stress. OMVs exhibit biological activities that play a key role in bacterial communication, defense and resistance to an environmental stress, nutrient acquisition, biofilm production and pathogenesis [12,15,16]. Current evidence suggests that OMVs aid the pathogen in establishing the colonization and survival niche in vivo [17]. Due to the fact that OMVs contain antigens recognized by an innate and acquired immune defenses, components of these secreted vesicles are also plausible candidates for development of effective vaccines [18].
While OMVs have been extensively researched in Gram-negative bacteria [19], researchers have just begun to appreciate the importance of membrane vesicles (MVs) in the physiology and pathogenesis of Gram-positive bacteria, including mycobacteria [14]. It has been shown that Mycobacterium tuberculosis MVs are involved in iron acquisition [20], in TLR2-dependent immune modulation of host cells [21] and inhibition of T-cell activation [22]. Therefore, the characterization of MV cargo that is produced in the biologically relevant environment and is delivered into host cells during bacterial intracellular phase of infection will add to the understanding of pathogenesis mechanisms of mycobacteria.
Our group previously identified the metal content of the phagosomes of pathogenic M. tuberculosis, MAH and nonpathogenic Mycobacterium smegmatis at different time points by using the high energy x-ray microscopy [23], and created an in vitro ‘phagosome’ model mimicking the metal ion content and pH of mycobacterial phagosome. Using this biologically relevant system, we further demonstrated that many mycobacterial virulence-related genes that are expressed inside phagocytic cells are regulated by metals [24], and proteins secreted in this system are also exported in the host macrophage cytosol [25]. The present study is the first report to show that MAH vesiculation is triggered under conditions encountered in the phagosomal environment, and establishes MVs as delivery vehicles of several MAH virulence-associated products within phagocytic cells.
Materials & methods
Bacterial culture & media
The Mycobacterium avium subsp. hominissuis 104 (MAH104) isolate from the blood of an AIDS patient was used in this study. MAH104 was cultured in 7H9 liquid broth supplemented with 10% oleic acid, albumin, dextrose and catalase (OADC, Hardy Diagnostics, CA, USA) at 37°C for 7–8 days. The mid-log phase cultures of MAH104 were centrifuged at 3500 r.p.m. for 20 min, and bacterial pellets were used to prepare inoculum using the McFarland standard #2 (approximately 3 × 108 CFU/ml) for inoculation into the minimal media or 24-h metal mix mimicking the MAH phagosome environment at 24-h postinfection. The minimal media were prepared as described previously [26], which is an established nutrient starvation medium known to stimulate vesiculation in mycobacteria. The metal-mix was made as described previously [24,25]. MAH104 was cultured in the 1 L of minimal media for 2 weeks or in the 1 L of metal mix for 24 h and incubated at 37°C in an orbital shaker rotating at 50 r.p.m. Bacterial viability was tested in terms of CFU per milliliter over the period of 2 weeks in both minimal media and metal mix, and was compared with the growth control of MAH grown in the 7H9 broth with 10% OADC supplement.
Isolation & purification of MAH104 membrane vesicles
MVs were isolated and purified as described previously [26] with few modifications. Briefly, the experimental media was centrifuged at 2000 × g for 20 min at 4°C, and then the supernatant was vacuum filtered through 0.22 μm filter units. The filtrate was concentrated approximately 50-fold using 100 kDa exclusion ultrafiltration concentrators (Thermo Fisher Scientific, MA, USA) and by centrifugation at 2000 × g at 4°C. The concentrator filters were rinsed with the remaining supernatant, and 20 ml of recovered concentrate was further cleared by centrifugation at 15,000 × g for 15 min to remove aggregates. To pellet the MVs, supernatants were transferred into new tubes and ultracentrifuged in the Beckman Optima XPN ultracentrifuge (Beckman Coulter Life Sciences, IN, USA) at 100,000 × g for 1 h at 4°C using fixed rotor. Next, supernatants were discarded and reminder pellets were resuspended in 2 ml of 35% (w/v) Optiprep solution diluted in phosphate-buffered saline (PBS). Samples were overlaid with less dense gradient layers in the range of 30–10%, forming six layers in 12.5 ml clear ultracentrifuge tubes (Beckman). Using the swinging rotor, samples were ultracentrifuged at 140,000 × g for 16 h at 4°C. Next day, 4 ml sample of 20 and 25% fractions (third and fourth layers proven in pre-experiments to contain MAH MVs) were retained, diluted in 6 ml PBS and ultracentrifuged again at 38,400 × g for 2 h at 4°C. MV pellets were resuspended in 600 μl PBS, filtered through 0.22 μm filter units and then processed for downstream experiments. The MV amounts for each experiment were determined based on nanoparticle tracking analysis. Two hundred microliters (out of 600 μl) of MVs with approximately 108-particles/μl concentration were processed for proteomic sequencing, lipidomics and DNA analysis.
Scanning & transmission electron microscopy
MAH104 MVs of the minimal and metal mix were fixed in the Karnovsky fixative (5% glutaraldehyde, 4% formaldehyde, 80 mM cacodylate buffer [pH 7.3] and 5 mM CaCl2). The Scanning Electron Microscopy (SEM) samples were critical point dried using an EMS 850 (Hatfield) and dehydrated at room temperature using graded solutions of ethanol and liquid CO2. Micrographs were taken with the FEI Quanta 600 FEG scanning electron microscope (Thermo Fisher Scientific ) at the Oregon State University Electron Microscopy Facility.
Transmission electron microscopy (TEM) samples were also fixed with the Karnovsky fixative (Thermo Fisher Scientific) overnight at 4°C. After washing samples using the same buffer, MVs were treated with cold 1% osmium tetroxide for 90 min and dehydrated in an acetone gradient made with an ascending order of series of concentrations 10, 30, 50, 70, 90, 100 and 100%, respectively. Next, MVs were fixed in the epoxy resin. Ultrathin sections (80 mm) were cut out of blocks, mounted on grids and stained using uranyl acetate and lead citrate. TEM micrographs were taken using the FEI Titan Chemi STEMT transmission electron microscope (Thermo Fisher Scientific) operating at 200 kV at the Oregon State University Electron Microscopy Facility.
MV size & concentration distribution by nanoparticle tracking analyses
Purified MVs were diluted in PBS (1:1,000) and injected into the chamber of the NanoSight NS300 instrument (Malvern Panalytical, Inc., Malvern, UK) using temperature: 25°C and viscosity of PBS: 0.89 centipoise (cP). The setting of the system was adjusted at camera level: 10 (high shutter speed [600], gain [250]; frames processed: 749; frames per second: 25; blur: auto; detection threshold: 5). Light scattered by the particles and their movement under Brownian motion was captured for 30 s. Three videos for each sample were analyzed to obtain the mean, mode and number of particles per milliliter. The analysis settings were optimized and kept constant between samples.
Freeze-fracture transmission electron microscopy
THP-1 cells infected with MAH for 24 h were fixed in 4% formaldehyde, 1.25% glutaraldehyde and 10 mM CaCl2, washed in PBS, 25% glycerol in PBS and frozen in a liquid ethane cooled by liquid nitrogen. Samples were freeze fractured at -100°C and shadowed by platinum and carbon evaporation. The metal replicas of the fractured samples were cleaned in normal bleach (sodium hypochlorite) and distilled water, mounted on Formvar-coated grids, and observed by TEM.
Mass spectrometric sequencing
Isolated MV samples were incubated with four volumes of acetone at 20°C for overnight and then at -20°C for 4 h. The precipitate was centrifuged at 25,000 r.p.m. at 4°C for 10 min and then treated with 50 mM ammonium bicarbonate with 0.1% RapiGest SF surfactant (Waters, M, USA). Subsequently, samples were incubated with 5 mM DTT solution at 56°C for 1 h to reduce disulfide bonds, and then with 5 mM iodoacetamide again for 1 h in dark condition to prevent the formation of disulfide bonds. Samples were digested with trypsin at 37°C overnight and sequenced in the Mass Spectrometry Center at Oregon State University using a Thermo Orbitrap Fusion Lumos MS (Thermo Fisher Scientific) coupled with a Waters nano-Acquity UPLC system (Waters). All raw files were analyzed by Proteome Discoverer 2.2 software. Mycobacterium avium proteins were classified into distinct groups based on their function. The functional classification was conducted by blasting the amino acid sequences of identified M. avium proteins against the protein sequence database of well-characterized M. tuberculosis H37Rv strain using the Institute Pasteur's TubercuList web server (http://genolist.pasteur.fr/TubercuList). Mycobacterium avium proteins that did not align with M. tuberculosis H37Rv genome were classified based on their domains/motifs using Conserved Domain search tool at the National Center for Biotechnology Information or assigned to the conserved hypothetical functional group.
Lipidomics
MVs were disrupted by sonication, and lipids were extracted with dichloromethane: isopropyl alcohol: methanol ratio of 20:10:65 by volume at room temperature. Samples were vortexed and the organic dichloromethane layer was collected for the lipid analysis. Chromatographic separation was carried out using the Waters HSS T3 column and the lipid extracts were measured using the Applied Biosystems 5600 Triple TOF mass spectrometer (Applied Biosystems, CA, USA) coupled to a Shimadzu Nexera UHPLC (Shimadzu, Kyoto Prefecture, Japan) at the Oregon State University Mass Spectrometry Center.
Host cell culture & growth media
Human THP-1 cells were purchased from the American Type Culture Collection (ATCC, VA, USA) and maintained in Roswell Park Memorial Institute medium-1640 (RPMI 1640, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gemini, CA, USA) at 37°C with 5% CO2. THP-1 cells were differentiated using 10 ng/ml of phorbol-12-myristate-13-acetate (Sigma–Aldrich, MO, USA) for 24 h at 37°C with 5% CO2 and seeded at 90–100% confluency in T-200 tissue culture flasks. Following 24-h phorbol-12-myristate-13-acetate treatment, the monolayers were replenished with fresh RPMI media and were allowed to rest for additional 48 h at 37°C with 5% CO2. Differentiated monolayers were used for MAH infection experiments.
Bioorthogonal noncanonical amino acid tagging of MAH104 & macrophage infection
MAH104 was grown in 7H9 broth supplemented with methionine or 2 mM azide-bearing methionine surrogate azidohomoalanine (AHA) for 7 days at 37°C. Bacterial inoculums were prepared as described above, and THP-1 macrophages were infected with multiplicity of infection (MOI) 10 bacteria to 1 cell. After 2 h, extracellular bacteria were removed by washing three-times with the Hank's Balanced Salt Solution (Life Technologies, CA, USA). Infected THP-1 macrophages were incubated for additional 24 h at 37°C with 5% CO2.
Protein enrichment using the click chemistry
Following 24-h incubation, the cell culture media was removed and the infected monolayers were resuspended into 3 ml of 0.1% triton X-100 in PBS. Cells were mechanically lysed by passing the concentrated cell suspension through a 27-gauge needle several times and then filtered via 0.22 μm filter units to remove cell debris and whole bacteria. Cleared samples were processed for the enrichment of MAH104 AHA-labeled proteins by using Click-iT® Protein Enrichment kit (Molecular Probes, OR, USA) as per manufacturer's protocol. Briefly, the click reaction was set up by adding lysate and catalyst solution over the agarose resin slurry, and allowed to take place for overnight at room temperature on a rotator. Next day, resin bound proteins were reduced and alkylated with Dithiothreitol (DTT) and then, iodoacetamide solutions in the Sodium Dodecyl Sulfate (SDS) wash buffer. Several washing steps were performed initially with the SDS wash buffer, and then with 8 M urea/100 mM Tris (pH 8.00) and with 20% acetonitrile. The resin-bound proteins were overnight digested with trypsin at 37°C and processed for protein sequencing at the Oregon State University Mass Spectrometry Facility.
Quantification of external & internal MV-associated DNA
PicoGreen dsDNA assay (Molecular Probes) was used to quantify MV-associated DNA as described previously [27], with few modifications. Similar concentrations of purified MVs of minimal media and metal mix determined using nanoparticle tracking analysis were incubated with or without DNase I (Promega, WI, USA) for 2 h to remove the externally associated DNA. The DNase was inactivated using DNA-free™ DNA removal kit (Thermo Fisher Scientific). MVs were either lysed by sonicating samples (OMNI International, Inc., GA, USA) to release internally associated DNA or kept intact. Subsequently, the samples were assayed using the PicoGreen dsDNA kit following the manufacturer's instructions. Fluorescence was measured using infinite 200q fluorescent plate reader (Tecan, Switzerland) with fluorescein excitation/emission at 480/520 nm.
Visualization of MV-associated DNA using laser confocal microscope
Purified MV samples were kept intact or treated with DNase I as described above. Samples were stained with a lipophilic nucleic acid stain SYTO®-61 (Molecular Probes, Eugene, OR, USA) at a ratio of 1:1000, and visualized using Zeiss LSM 780 NLO confocal microscope system (Carl Zeiss, Germany) in the Center for Genome Research and Biocomputing microscopy facility, Oregon State University. Stained MVs were excited at 633 nm Helium–Neon laser (of power of 5%) and emitted at 636–758 nm that was collected with a Zeiss plan Apochromat 40×/1.4 oil DIC M27 objective. Zen 2012 software was used to capture and merge images. As a control experiment, to confirm MVs presence on the transmitted light images, we sonicated samples and then visualized for its presence in the bright field. We did not observe any MVs structures in sonicated samples (data not shown).
Construction of MAH104 FLAG-tag overexpression clones
Eleven MAH unique proteins of metal mix were selected and primers listed in the Table 1 were designed to FLAG tag and clone into mycobacterium pMV261 overexpression plasmid under Hps60 promoter. The DNA sequence of GATTACAAGGATGACGACGATAAG encoding the FLAG peptide (DYKDDDDK) was incorporated into the primer design. Either two forward (F) or two reverse (R) primers were designed for each of the protein depending on whether the FLAG-tag sequence was inserted at the N- or C-terminus of the gene. Initially PCR amplification was carried with pair of the first primer (either F1 or R1) coupled respective R or F primer. For this round of amplification, MAH104 genomic DNA was used as a template. The DNA was obtained as previously described [28]. The second round of PCR amplification was done using the pair of second primer (either F2 or R2) coupled respective R or F primer, in which the template used was the PCR product of the first round of amplification. All amplicons were tested by using electrophoresis on 1% agarose gel. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Germany). All genes were cloned in the restriction sites EcoRI and HpaI (except MAV_2964 cloned in PstI and HpaI) of pMV261 vector harboring kanamycin resistance cassette. Positive vectors were transformed into MAH104 as described previously [25] and plated on 7H10 Middlebrook agar plates containing 400 μg/ml kanamycin. After 2-weeks incubation, resulting colonies were tested for the presence of kanamycin and positive clones were used for THP-1 macrophage infection experiments.
Table 1. . Primers used in the study.
| Gene name | Primer | Sequence (5′ to 3′) |
|---|---|---|
| MAV_5153 | F1 F2 R |
AGTGAGTTCACAATTCCG TTTTTGAATTCGATTACAAGGATGACGACGATAAGAGTGAGTTCA TTTTTGTTAACTCAGGATTTGCCG |
| MAV_1082 | F R1 R2 |
TTTTTGAATTCCGCGAGATTACGA ACCGGTGGTGACGGTGGC TTTTTGTTAACGATTACAAGGATGACGACGATAAGACCGGTGGTG |
| MAV_2909 | F1 F2 R |
TTCTATGGGGCCTTTCCG TTTTTGAATTCGATTACAAGGATGACGACGATAAGTTCTATGGGG TTTTTGTTAACTCAACCCAACTGG |
| MAV_2345 | F1 F2 R |
GCGTTCAGTAAGCCACCG TTTTTGAATTCGATTACAAGGATGACGACGATAAGGCGTTCAGTA TTTTTGTTAACCTAGTTGTTGCCC |
| MAV_4365 | F1 F2 R |
AGCAAGATCATTGAGTAC TTTTTGAATTCGATTACAAGGATGACGACGATAAGAGCAAGATCA TTTTTGTTAACTCAGTGATGGTGA |
| MAV_2833 | F1 F2 R |
CAAGGGGATCCGGAAGTT TTTTTGAATTCGATTACAAGGATGACGACGATAAGCAAGGGGATC TTTTTGTTAACTCAGCTCGGTGGC |
| MAV_2964 | F R1 R2 |
TTTTTCTGCAGTCGCGGGGGTGGC CTAGCCGCCCAGCCCCTT TTTTTGTTAACGATTACAAGGATGACGACGATAAGGCCGCCCAGC |
| MAV_3813 | F1 F2 R |
GCCGACAACCCCAAACGT TTTTTGAATTCGATTACAAGGATGACGACGATAAGGCCGACAACC TTTTTGTTAACTCACATGGCGGCC |
| MAV_3310 | F R1 R2 |
TTTTTGAATTCTCTGCCAGCTGTC TCATCCGCCGTGGCCGGG TTTTTGTTAACGATTACAAGGATGACGACGATAAGTCCGCCGTGG |
| MAV_0740 | F1 F2 R |
AACAACCTCTACCGCGAT TTTTTGAATTCGATTACAAGGATGACGACGATAAGAACAACCTCTA TTTTTGTTAACTCACGAAGTGAGC |
| MAV_2054 | F1 F2 R |
ACGTCGGCTCAAAATGAG TTTTTGAATTCGATTACAAGGATGACGACGATAAGACGTCGGCTC TTTTTGTTAACTCACTTGTACTCA |
Western-blot analysis of MAH-secreted proteins
THP-1 macrophages were differentiated into three 125 cm2 tissue culture flasks for each clone, and infected with MAH clones expressing selected 11 MX proteins for 24 h. The cytosolic FLAG_RFP protein expressing MAH clone was used as a control for bacterial cell lysis if any within 24-h infection of THP-1 cells. Cell cultures were lysed with 0.1% Triton X-100, centrifuged at 3500 r.p.m. for 20 min and filtered via 0.22 μm filter units to remove cell debris and whole bacteria. Precleared cell lysates were incubated with FLAG primary agarose conjugate antibody overnight at 4°C to immunoprecipitate FLAG-tagged MAH proteins. Samples were separated by electrophoresis on 12.5% SDS-PAGE gels, transferred to nitrocellulose membranes and blocked for overnight with 5% blocking reagent. Membranes were probed with a FLAG primary antibody (1:200) for 2 h, washed three-times and incubated with the corresponding IRDye secondary antibody (Li-Cor Biosciences, Inc., NE, USA) at a dilution of 1:5000 for 30 min. The reactivity and the photon emission means for protein bands were assessed using the Odyssey Imager (Li-Cor).
Ability of extracellular MVs to cross the plasma membrane
Purified MV samples (108 particles/μl) were incubated in the PBS containing 1 mM Texas Red hydrazide (Molecular Probes) for 2 h at room temperature, and then washed two-times to remove unbound dye using ultracentrifugation at 40,000 × g for 4 h. Labeled MVs were either directly added to THP-1 macrophages or cells were first pretreated for 2 h with 3 μM of cytochalasin B (Sigma–Aldrich) in DMSO; 250 μM of monodansylcadaverine (Sigma–Aldrich) in DMSO; 80 μM dynasore (Sigma–Aldrich) in DMSO; 2.5 mM methyl-β-cyclodextrin (Sigma–Aldrich) in DMSO; or 0.05% DMSO as a control and then incubated with host cells. Extracellular MVs were removed by washing cells with PBS four-times. Fluorescent readings were recorded up to 48 h using Tecan Infinity 200 cytofluorometer with corresponding filter sets. Treatments were made in duplicates and the experiment was repeated three times. In addition, fluorescent micrographs were taken at 4-h time point using confocal microscopy. Cells were fixed with 2% paraformaldehyde in PBS for 20 min and permeabilized with 0.1% Triton X-100. THP-1 macrophages were incubated with DAPI and phalloidin in PBS at a 1:1000 dilution for 30 min followed by three washing steps with PBS.
Data analysis
Proteomic and lipidomic studies for both conditions were performed three times and only common constituents present within each experimental group are reported as results. Venn diagrams were created using the Bioinformatics & Evolutionary Genomics website of the Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/). MAH104 survival, DNA quantification and MV endocytosis assays were performed three times and GraphPad PRISM version 7.04 was used for plotting. Statistical analysis results are presented as means ± standard deviations of results from three biological experiments unless otherwise indicated. The significance level was determined by using Student's t-test. A p-value of < 0.05 was considered statistically significant.
Results
Exposure to minimal media & ‘phagosome-mimicking’ model induces a robust vesiculation of MAH
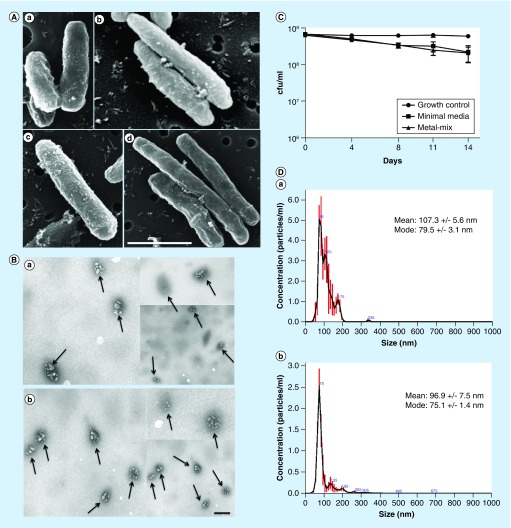
Previous studies have established that under starvation condition of a minimal media, deprived of most of the essential nutrients, M. tuberculosis efficiently forms MVs [26]. MVs were demonstrated to play important role in mycobacterial pathogenesis [20–22]. With the exception of the starvation aspect, the minimal media do not represent the intraphagosomal environment of a macrophage [8,29]. To address this limitation and investigate MV formation under a biologically relevant environment, we utilized recently developed ‘phagosome-mimicking’ model of metal mix, proven to induce the expression of an intracellular phenotype of MAH [24]. By culturing MAH in minimal media for 2 weeks and in metal mix for 24 h and then performing SEM, we could demonstrate that exposure to both conditions resulted in vigorous vesiculation of MAH104 (Figure 1A). The SEM micrographs clearly show membrane bulges budding off from the MAH104 cell surface. To substantiate the finding, MVs were isolated and purified, and observed under TEM. Micrographs showed intact MAH104 MVs in their native form (Figure 1B), and validated our experimental procedure of MV sample preparation.
Figure 1. . Characterization of Mycobacterium avium subsp. hominissuis membrane vesicles.
(A) Scanned electronic microscopy micrographs of MAH104 exposed to minimal media for 2 weeks (a & b) and metal-mix mimicking 24 h phagosome environment (c & d). Scanned electronic microscopy micrographs demonstrate bulges on the bacterial membranes that are about to bud off from the membrane. In addition, bacteria of phagosome model are much elongated when compared with Mycobacterium avium subsp. hominissuis exposed to environmental stress, a phenotype typically seen in intracellular mycobacteria isolated from host cells. Scale bar: 1 μm. (B) Transmission electron microscopy micrographs of purified MV samples with closed membrane structure. MVs are indicated by arrows. (a) Purified membrane vesicle (MV) sample from MAH104 exposed to the minimal media for 2 weeks. (b) Purified MV sample from MAH104 exposed to the metal-mix for 24 h. Scale bar: 300 nm. (C) MAH104 viability assay tested in both minimal media and metal mix for up to 2 weeks. CFUs of bacteria exposed to both minimal media and metal mix show slight decrease over the 2-week course when compared with the 7H9 Middlebrook broth grown control supplemented with 10% oleic acid, albumin, dextrose and catalase. Data were plotted as mean of two independent trials and standard deviation. (D) The size and concentration of membrane vesicles isolated from the minimal media (a) and metal mix (b) were determined by nanoparticle tracking analysis on a Nanosight NS300.
To rule out the possibility of MAH death under stress environments of minimal media and metal mix, bacterial viability was established based on the colony forming units per milliliter of both experimental media for up to 2 weeks. Results indicate that MAH104 survival is slightly impacted in the minimal media and in metal mix when compared with control bacteria grown in 7H9 nutrient-rich medium supplemented with 10% OADC. Nonetheless, 97% of bacteria survived and were completely viable after 14 days exposure to both stress conditions (Figure 1C).
Furthermore, using the nanoparticle tracking analysis, which utilizes a light scatter technology, we measured MV size and concentration in 1 l of minimal media and 1 l of metal mix (Figure 1D). The measured mode of vesicle size ranged from 62.4 to 152.6 nm with concentration of 8.24e+08 +/- 1.41e+08 particles/μl. The study demonstrates that MAH produces comparable concentrations of MVs in both tested models.
Proteomic profiling of MAH104 MVs formed under minimal media & metal-mix condition
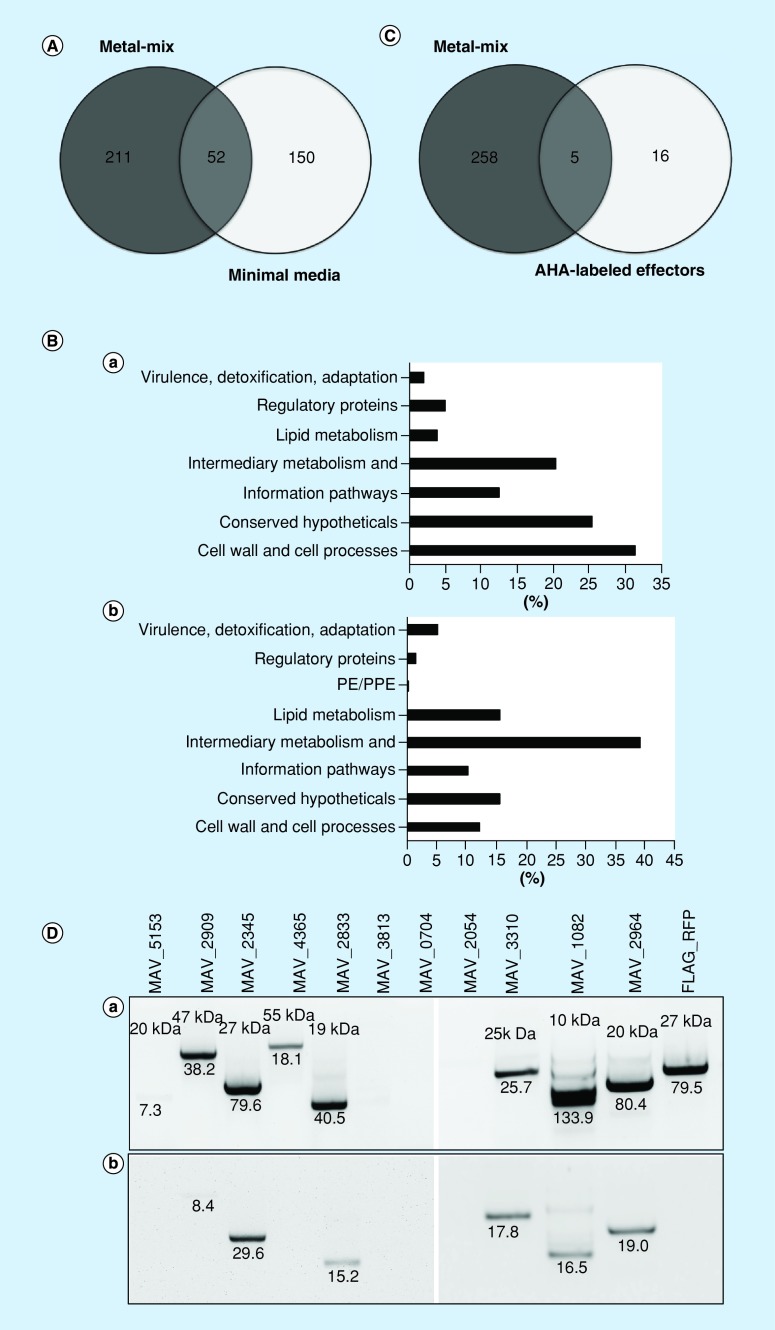
Numerous studies have shown that bacteria export proteins via MVs, including virulence effectors [19]. Therefore, we characterized proteomics of MVs under starvation and intracellular stress settings and identified differences between two conditions. Mass spectrometric analysis of MVs isolated from the minimal media identified a total of 202 cargo proteins (hereafter referred to as MM proteins; Supplementary Table 1 ), and 263 proteins from the metal mix (hereafter referred to as MX proteins; Supplementary Table 2 ). Between MM and MX proteins, 52 proteins were found to be common, whereas 211 proteins were unique to metal mix and 150 to minimal media (Figure 2A). Proteins were categorized based on their functional groups as described in the methods. Among the most represented category of MM proteins was the cell wall and cell processes (63 proteins), 41 proteins involved in intermediate metabolism and respiration, and 25 proteins associated with the information pathways. The 25% of MM proteins belonged to the conserved hypothetical proteins of unknown function. Among the least represented categories, we found proteins to belong to regulatory group (4.95%), lipid metabolism (3.96%) and related to virulence, detoxification and adaptation (1.98%) (Figure 2Ba). MX proteomic analysis identified the most represented categories of the intermediate metabolism and respiration (103 proteins), lipid metabolism (41 proteins) and proteins involved in cell wall and cell processes (32 proteins). The 15% of MX proteins belonged to the conserved hypothetical proteins of unknown function. The least represented categories identified in MX proteomics were the virulence, detoxification and adaptation (5.32%), regulatory proteins (1.52%) and PE/PPE family protein (0.38%) (Figure 2Bb). Comparative genomic analysis between M. avium and M. tuberculosis was performed as previously described [30].
Figure 2. . Membrane vesicle proteomics and conformational studies for secretion of MX proteins.
(A) Venn diagram showing the number of overlapping and unique sets of membrane vesicle cargo proteins found in MAH104 membrane vesicles (MV) released in response to minimal media and metal mix at chosen time points. (B) Functional classification of MV cargo proteins into categories was done by finding the protein homologs of M. tuberculosis H37Rv strain and using the functional categorization available on TubercuList webserver of Institute Pasteur or based on predicted or known function for those Mycobacterium avium proteins that do not match to any proteins of H37Rv strain. Bars show percentage representation of each category in MV cargo proteins of minimal media (a) and metal mix (b). (C) Venn diagram showing common and unique proteins found in MAH104 MVs released in response to metal mix and azidohomoalanine-labeled M. avium subsp. hominissuis presynthesized secreted effectors found in the host cell cytosol at 24 h postinfection of THP-1 macrophages. (D) Analysis of FLAG-tagged MX protein secretion in the cytoplasm of infected macrophages. MAH104 MAV_5152, MAV_2909, MAV_2345, MAV_4365, MAV_2833, MAV_3813, MAV_0740, MAV_2054, MAV_3310, MAV_1082 and MAV_2964 protein overexpression clones were constructed using the pMV261:FLAG vector. (a) Mycobacterium avium subsp. hominissuis clones were mechanically disturbed using bead beater and cleared samples were processed for western-blot analysis to confirm the protein expression within mycobacterial cells. (b) THP-1 cells were infected with MAH104 clones at a multiplicity of infection 1: 10. Precleared and concentrated bacterial and cell lysate-free supernatants were subjected to immunoprecipitation and then western blotting using FLAG-tag antibody. The photon emission means for each protein band are recorded to quantify the signal intensity on the Odyssey Imager (Li-Cor).
Among metal mix-specific virulence factors, we discovered enhanced intracellular survival enhanced intracellular survival (Eis) protein of the family of N-acyltransferase, trehalose synthase involved in mycobacterium cell wall modification, five heat shock proteins of DnaK, HtpG, 10 kDa chaperonin, 60 kDa chaperonin 1 and 2 employed by mycobacteria to survive under different environmental stress including the host cell. It was also found that PE/PPE family protein and four antioxidant enzymes (superoxide dismutase [SOD], catalase [katE], catalase peroxidase [katG] and alkyl-hydroperoxide reductase [ahpC] were produced in the cargo of MVs of the metal mix. The glyoxylate cycle enzyme isocitrate lyase was only observed in the metal mix-induced MV proteome. This fully characterized enzyme has been selected as a potential antituberculosis drug target due to its function associated with the fatty acid catabolism and important role in M. tuberculosis virulence and persistence of the murine model [31,32]. Unlike above-described virulence factors that were present only in the metal mix MV cargo, we identified 12 lipoproteins in the cargo of minimal media-induced MVs, consistent with the reported minimal media-induced MVs from M. tuberculosis (H37Rv) and Mycobacterium bovis Bacillus Calmette–Guérin (BCG) [21]. The physiological function of bacterial lipoproteins is to acquire ions and nutrients, facilitating survival within hostile host environment [33]. LprG, a cell envelope lipoprotein, was found in both minimal media and metal mix-induced MVs. It plays a crucial role in the expression of surface lipoarabinomannan and interaction with mannose receptors on macrophages has been suggested to be necessary for cell entry, inhibition of phagolysosome fusion and intracellular survival [34].
Bioorthogonal noncanonical amino acid tagging labeling & subsequent protein enrichment revealed some MAH104 effectors common with MX proteins secreted within the cytosol of 24-h MAH-infected THP-1 macrophages
To substantiate the relevance of discovered MX proteins and MV-mediated export within the host cells, we used a novel metabolic labeling approach termed BONCAT (bioorthogonal noncanonical amino acid tagging). Secreted effectors of Yersinia enterocolitica [35] Salmonella typhimurium [36], M. tuberculosis [37] and a protozoal pathogen Toxoplasma gondii [38] have been successfully investigated using this strategy. In order to avoid the incorporation of AHA during the host protein synthesis, MAH proteins were metabolically labeled with 2 mM methionine surrogate AHA in the culture medium, and then used for infection assays. Using this approach, we identified only MAH presynthesized proteins within infected THP-1 macrophages that were secreted by the pathogen during early phase of infection. AHA-containing proteins carry azide groups, which can be tagged with a copper-catalyzed ‘click’ reaction. Affinity-tagged proteins were separated from the pre-existing proteome by affinity purification and identified by tandem mass spectrometry. Twenty-one MAH104 proteins were found within the cytosol of 24-h postinfected human macrophages (Table 2). Out of 21 proteins found, 9 proteins belonged to the cell wall and cell processes, 6 proteins were involved in intermediary metabolism and respiration, 3 proteins belonged to virulence, detoxification and adaptation, and 3 conserved hypotheticals. Interestingly, five proteins were found to be common between the MX proteome and MAH104 effectors identified within host cells (Figure 2C), suggesting MVs as one of delivery mechanisms to the host cell cytosol.
Table 2. . MAH104 presynthesized secreted proteins found in the lysate of 24-h postinfected THP-1 macrophages through azidohomoalanine metabolic labeling and subsequent click-reaction-mediated enrichment.
| Accession no. | Gene name | Description | Functional category | MW (kDa) | Peptides | H37Rv homolog |
|---|---|---|---|---|---|---|
| A0A0H2ZST3 | MAV_1432 | OppC ABC transporter, permease protein | Cell wall and cell processes | 33 | 2 | Rv1282 |
| A0A0H2ZUA4 | MAV_3419 | ABC transporter permease protein | Virulence, detoxification, adaptation | 28 | 2 | – |
| A0A0H2ZWB3 | MAV_3084 | amt ammonium transporter | Cell wall and cell processes | 44 | 2 | – |
| A0A0H3A0D0 | MAV_2087 | Amino acid transporter | Cell wall and cell processes | 48 | 6 | Rv2320c |
| A0A0H3A0X6 | MAV_4750 | GluB bacterial extracellular solute-binding protein | Cell wall and cell processes | 36 | 2 | Rv0411c |
| A0A0H3A326 | MAV_4059 | Monoxygenase | Intermediary metabolism and respiration | 95 | 2 | – |
| A0A0H3A3W0 | MAV_0467 | dppD ABC transporter, ATP-binding protein | Cell wall and cell processes | 60 | 3 | Rv3663c |
| A0QFE1 | MAV_2434 | SsuB aliphatic sulfonates import ATP-binding protein | Cell wall and cell processes | 28 | 3 | – |
| A0QHM5 | MAV_3231 | lspA lipoprotein signal peptidase | Cell wall and cell processes | 20 | 2 | Rv1539 |
| A0QK47 | MAV_4137 | UPF0182 protein | Cell wall and cell processes | 108 | 3 | Rv3193c |
| A0A0H2ZYT4 | MAV_1694 | Single-stranded DNA-binding protein | Conserved hypotheticals | 17 | 2 | Rv2478c |
| A0A0H2ZVH6 | MAV_2217 | TraSA:integrase fusion protein | Conserved hypotheticals | 43 | 4 | – |
| A0A0H2ZTJ2 | MAV_2056 | Putative sulfate exporter family transporter | Cell wall and cell processes | 40 | 7 | – |
| A0A0H2ZVV6 | MAV_2267 | glnA glutamine synthetase | Intermediary metabolism and respiration | 54 | 3 | Rv2220 |
| A0A0H2ZZP6 | MAV_0406 | Glycerol kinase | Intermediary metabolism and respiration | 55 | 2 | Rv3696c |
| A0A0H2ZZZ9 | MAV_2296 | Cytochrome b6 | Intermediary metabolism and respiration | 63 | 2 | Rv2196 |
| A0A0H3A2U3 | MAV_4248 | BPL_C biotin-[acetyl-CoA-carboxylase] ligase | Intermediary metabolism and respiration | 27 | 2 | Rv3279c |
| A0QCX7 | MAV_1526 | ATP synthase γ chain | Intermediary metabolism and respiration | 34 | 2 | Rv1309 |
| A0A0H2ZQZ9 | MAV_0558 | Uncharacterized protein | Conserved hypotheticals | 27 | 3 | – |
| A0A0H2ZVL5 | MAV_0059 | ABC transporter, permease protein, His/Glu/Gln/Arg/opine family protein | Virulence, detoxification, adaptation | 63 | 2 | – |
| A0A0H2ZXB8 | MAV_2433 | SsuC putative aliphatic sulfonates transport permease protein | Virulence, detoxification, adaptation | 30 | 2 | – |
FLAG-tagging of MX proteins identified additional MAH-secreted effectors within host macrophages
In order to confirm relevance of MX protein to macrophage infection and their export within the host cells, 11 proteins were selected from the metal mix MV proteome, FLAG tagged and overexpressed in MAH. Unlike BONCAT metabolic labeling, where we explored secretion of presynthesized effector proteins at 24-h postinfection and then compared results with MV proteome of 24-h metal mix, in case of FLAG-tag approach, we directly studied MX protein secretion within THP-1 macrophages. The western-blot analysis confirmed overexpression of 9 MAH proteins out of 11 tested (Figure 2Da). Using immunoprecipitation approach with FLAG agarose-conjugate antibody, we could demonstrate that out of nine MAH protein-overexpressed clones six MX proteins were also secreted within the cytosolic fraction of THP-1-infected cells (Figure 2Db). The Red Fluorescent Protein (RFP) construct served as a control for bacterial cell lysis. While cytosolic RFP protein was highly expressed within MAH104, we did not detect presence of this protein in the cytoplasm of THP-1 cells.
Lipidomics of MAH104 MVs found no differences between minimal media & metal mix
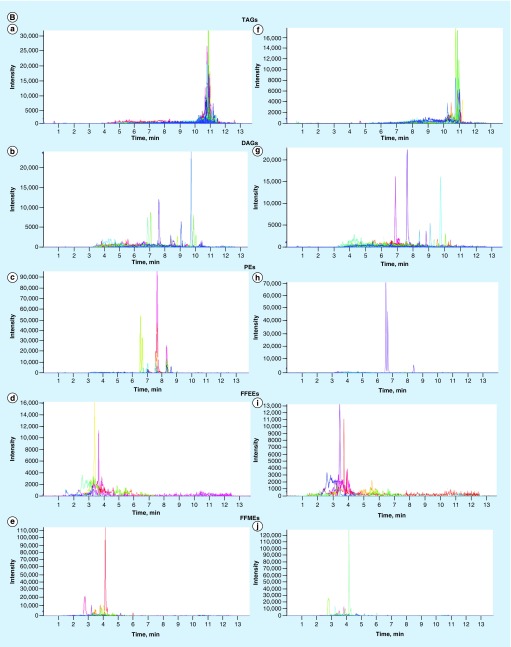
Being armed with highly complex and extremely lipid-rich cell wall, mycobacterial lipids play direct roles in host–pathogen interactions, virulence and host immunomodulation [39]. For example, T-cell receptor signaling is inhibited by M. tuberculosis lipoglycans, lipoarabinomannan and lipomannan, and it was demonstrated that trafficking of these lipoglycans from intracellular tubercle bacilli to T cells occurred though MVs that ultimately inhibited activation of CD4+ T cells [22]. Hence, we decided to conduct lipidomic analysis of MAH104 MVs and found numerous polar and nonpolar lipids in association with the MVs released in response to both minimal media and metal mix. More specifically, triacylglycerols, diacylglycerols, phosphatidylethanolamines, free fatty acid ethyl esters and free fatty acid methyl esters representing the uppermost layers of the MAH104 cell wall were identified in both samples (Figure 3A & B). We did not detect any differences in the MV lipid composition between these two experimental groups.
Figure 3. . Membrane vesicle lipidomics.
(A) Total/positive-ion chromatograms of membrane vesicles of minimal media and metal mix. (B) Extracted ion chromatograms for individual lipid classes of membrane vesicles of minimal media (a, b, c, d & e) and metal mix (f, g, h, i & j).
DAGs: Diacyl gycerols; FFEE: Free fatty acid ethyl esters; FFME: Free fatty acid methyl esters; PEs: Phosphatidylethanolamines; TAGs: Triacylglycerols.
MAH104 MVs carry DNA as part of the internal cargo & DNA is also externally associated with the vesicle membrane
Several groups have demonstrated in Gram-negative bacterial pathogens dsDNA in association with OMVs [27,40–43]. We investigated whether MAH104 also could carry DNA within its vesicles. Using dsDNA PicoGreen assay, we established DNA presence in MVs released in response to both minimal media and metal-mix condition (Figure 4A). MVs released in response to minimal media carried significantly higher amount of total DNA when compared with MVs released in response to metal mix. Furthermore, higher amount of external DNA was present on MVs surface in both conditions when compared with the internally associated DNA as part of the MV-packaged cargo. However, internal DNA of MX condition was significantly lower making impossible to sequence DNA at this stage. Results were substantiated by laser confocal microscope, where MV-associated total and internal DNAs were visualized by staining samples with the lipophilic nucleic acid stain SYTO-61 confirming our observation (Figure 4B).
Figure 4. . Quantification of membrane vesicle-associated DNAs of minimal media and metal mix.
(A) Total, external and internal Membrane vesicle (MV)-associated DNAs were quantified using Quant-iT PicoGreen dsDNA assay (n = 3). Internal DNA was measured by treating intact MV sample with DNase before lysing MVs, whereas a total DNA was measured without treating samples with DNase. *p < 0.05 of internal and external DNA concentrations between the minimal media and metal mix. (B) MV samples were stained with a lipophilic nucleic acid stain SYTO-61, which emits red fluorescence upon DNA binding. Laser confocal images of the DNase treated MVs visualizing the internally associated DNA of minimal media (a) and metal-mix (b). Intact MVs are also seen in the transmitted light images (right panels). Scale bar: 10 μm.
MAH forms membrane vesicles within the host macrophages
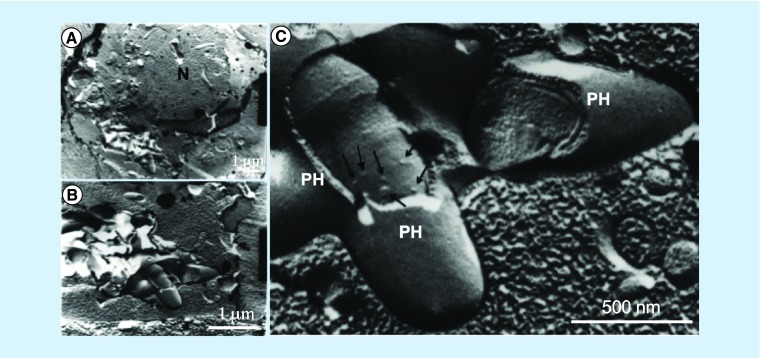
In attempts to identify MV formation on the MAH104 cell surface within infected THP-1 cells, we performed freeze-fracture electron microscopy (Figure 5). Freeze-fracture TEM is a unique method among electron microscopic techniques because the deep imprint of rapidly frozen samples helps visualization of the detailed surface structures on the cell and organelles, including precise structure of bacterial cell wall [44]. In the Figure 5, MAH-containing phagosome is clearly visible and opened, exposing bacterial cell surface. MVs budding from the bacterial surface can be observed on intracellular bacteria (Figure 5C).
Figure 5. . Freeze-fracture transmission electron microscopy.
Freeze-fracture transmission electron microscopy of Mycobacterium avium subsp. hominissuis-infected THP-1 macrophages reveals numerous membrane vesicles on the cell wall of intracellular bacteria shown by arrows. Several M. avium subsp. hominissuis-containing phagosomes are seen in the cytoplasm of THP-1 cells.
N: Nucleus of macrophage; PH: Phagosomes.
Mycobacterial MVs are translocated into the cytosol of macrophages via clathrin-mediated endocytosis
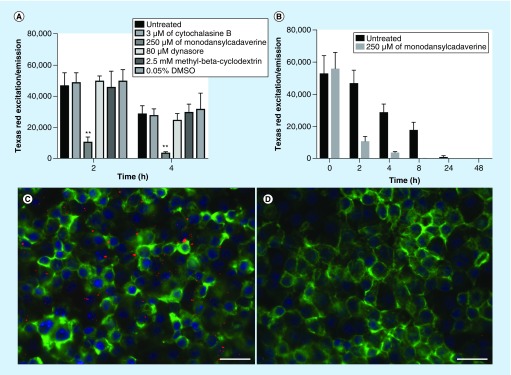
Due to the fact that it is very challenging to study MV formation within the bacterial phagosomes and to track MV trafficking via phagosome membrane into the cytosol of infected macrophages, we investigated ability of MVs to cross the plasma membrane of THP-1 cells. During bacterial invasion of host cells, the membrane of the phagosome is formed by the fusion of the plasma membrane, and the basic composition of the phospholipid bilayer of the phagosome remains the same as it is seen in the plasma membrane. To examine whether extracellular MVs can cross the plasma membrane of THP-1 cells and to reveal the possible uptake mechanism of mycobacterial MVs, we used several inhibitors of the host-cell endocytic pathway and lipid raft. More specifically, cytochalasin B inhibits actin filament polymerization; dynasore interferes with mobilization of the cellular membrane by inhibiting dynamin protein, reducing the plasma membrane cholesterol and lipid raft organization; methyl-cyclodextrin extracts cholesterol from the plasma membrane and, thus, disrupts lipid raft causing inhibition of endocytosis process; and monodansylcadaverine diminishes function of clathrin and clathrin-coated vesicles.
Fluorescent analyses of Texas red-labeled MVs endocytosis by THP-1 macrophages demonstrate that monodansylcadaverine pretreated cells had significantly lower amounts of MVs endocytosized when compared with untreated, DMSO control and to other experimental (inhibitor) groups after 2- and 4-h incubation of MVs with cells (Figure 6A). Time-dependent dynamics of MV endocytosis assay show that while untreated cells initially endocytosized significantly greater amount of MVs than monodansylcadaverine pretreated cells, the fluorescent readings gradually decreased over time with no fluorescent readings detected at 48 h (Figure 6B). These findings suggest the disintegration of MV cargo within the host cells. Fluorescent micrographs of untreated and monodansylcadaverine pretreated THP-1 cells were taken after 4-h incubation with Texas red-labeled MVs (Figure 6A & D).
Figure 6. . Translocation of membrane vesicles into the cytosol of macrophages.
(A) Approximately 108 Texas Red-labeled MV particles were either directly added to THP-1 macrophages or initially host cells were pretreated with cytochalasin B, monodansylcadaverine, dynasore, methyl-β-cyclodextrin or just DMSO and then added to macrophages. (B) Time-dependent dynamics of intact MV presence within the cytosol of THP-1 cells. Fluorescent readings were recorded with Texas red corresponding filter sets on the Tecan Infinity 200 cytofluorometer. Treatments were made in duplicates and the experiment was repeated three-times. **p < 0.01 between monodansylcadaverine and untreated, DMSO control and all other inhibitor groups. Fluorescent micrographs of THP-1 macrophages exposed with Texas red-labeled MVs in (C) untreated and (D) monodansylcadaverine-pretreated cells after 4-h incubation. Actin filaments are visualized using phalloidin (in green) and nuclei (in blue) with DAPI.
MV: Membrane vesicle.
Discussion
Following uptake by macrophages, MAH resides in the phagosomal environment where it efficiently interferes with the host immune defense. Bacterial virulence determinants that are exported within the host cells play an essential role during the infectious process, and recent studies in Gram-positive pathogens, including mycobacteria, have highlighted the importance of MVs in protein export [14]. Our study provides detailed characterization of M. avium MVs produced in minimal media, representing environmental stress, and in metal mix, representing an in vitro system mimicking phagosome environment. MAH is an opportunistic pathogen, ubiquitously present as free-living organisms, within biofilms, amoeba and in environments such as house dust, showerheads, water boilers, ice machines and garden soil [1]. Considering the potential involvement of MVs in bacterial communication, synergistic and antagonistic interactions, biofilm formation, nutrient acquisition and in adaptation to stress conditions [45], it is highly likely that MAH104 would produce MVs to adapt lifestyles and persist in diverse environments. Starvation would be a common stress that MAH would face in majority of environments. Therefore, the minimal media formulated by Prados-Rosales et al. [21] allow studying the vesiculation phenomenon in MAH. The fact is that prolonged exposure of mycobacteria to minimal media represents more generalized vesiculation response under nutrient deprivation condition with little relevance to its virulence. Consequently, in this study, we used recently developed in vitro phagosome-mimicking model, termed as the metal mix, to isolate and characterize MV cargo of MAH expressing an intracellular phenotype.
Comparative proteomic analysis of MVs of the minimal media and metal mix unveiled a number of MAH104 virulence-associated proteins, which were uniquely present in the cargo of MVs released in response to 24-h phagosome-mimicking condition. Among metal mix-associated virulence factors, we identified the Eis protein belonging to the N-acyltransferase family, which was shown to enhance the survival of a nonpathogenic Mycobacterium smegmatis within macrophages [46]. Eis has been demonstrated to be a secreted effector of M. tuberculosis capable of interacting with several host targets modulating innate immune responses. In particular, Eis inhibits autophagy, suppresses inflammation and reactive oxygen species production preventing programmed cell death by infected macrophages [47]. The Eis protein has been reported to be present in M. tuberculosis containing phagosome, cytosol of the infected macrophages, and even in the sera from pulmonary tuberculosis patients (using anti-Eis antibody) [48], clearly underscoring its significance in mycobacterial infection. Our findings suggest that in MAH-infected macrophages Eis is exported via MVs during the initial 24-h infection.
Trehalose synthase of MAH104 is another notable virulence protein differentially expressed in the MV cargo of metal mix versus MV obtained from the minimal media. Trehalose is a disaccharide, serving as a reserved carbohydrate and a stress protectant. Findings of numerous studies support the notion that lack of complete trehalose pathway is associated with reduced pathogenic potential [49]. One of the most abundant mycobacterial glycolipids, 6,6-dimycolate (TDM) or cord factor, is formed when trehalose combines with the mycolic acid. TDM has been implied in preventing phagosomal maturation, immune evasion, tissue damage and necrosis through association with the host lipids [49]. TDM is also a well-studied immunostimulatory molecule in M. tuberculosis. It binds to a host receptor named macrophage-inducible Ca2+-dependent lectin (Mincle), triggering the production of proinflammatory cytokines and nitric oxide [50]. In vivo administration of TDM caused elevated proinflammatory cytokine production and subsequent severe lung inflammation and granuloma formation. Considering these findings, we can hypothesize that trehalose synthase delivered via MVs could catalyze trehalose synthesis, and in combination with mycolic acids may facilitate an inflammatory response, including granuloma formation usually seen in M. avium infection [51].
Five chaperones, DnaK, HtpG, 10 kDa chaperonin, 60 kDa chaperonin 1 and 2, encountered in metal mix-induced MVs, were absent in the minimal media. Major function of chaperones is to facilitate folding or prevent misfolding of proteins [52]. Chaperones are also important for bacterial survival upon sudden increase in the temperature of their surroundings, for example, upon entry in the host. We hypothesized that MAH104-derived chaperones, upon MV-mediated secretion, may trigger host proinflammatory immune responses, which actually benefit the colonization and spread of MAH104. This possibility is supported by the finding that M. tuberculosis chaperonins are highly immunogenic and potent proinflammatory cytokine stimulators [53].
Antioxidant enzymes, while found in all aerobically respiring organisms, are important virulence determinants of a number of pathogens that neutralize oxygen and nitrogen radicals produced by macrophages and other immune cells [54]. We identified four antioxidant enzymes (SOD, catalase [katE], catalase peroxidase [katG] and alkyl-hydroperoxide reductase [ahpC] in the cargo of MAH104 MVs. Except for SOD, the remaining three proteins were unique for the metal mix-induced MVs. Secreting antioxidant enzymes in MVs during first 24-h infection may be associated with inhibitory function of phagocytic cells, by enabling MAH104 to efficiently counteract on oxidative attack upon infection with macrophage. In an analogous manner, release of antioxidant enzymes of MVs may divert and ultimately overwhelm NADPH oxidase-induced oxidative burst, helping MAH104 to evade the innate immune mechanism.
The PE/PPE family proteins were also exclusive part of the MV cargo obtained from the phagosome-like environment. PE/PPE proteins have been shown to be involved in macrophage uptake, inhibition of phagosome maturation and acidification, cytokine secretion, stimulation of host cell apoptosis (through interaction with TLR2) and necrosis, dendritic cell activation and antigen processing, reviewed in detail in [55]. It is very tempting to hypothesize that MV-mediated export of specific PE/PPE proteins of MAH104 may stimulate macrophage apoptosis, for two solid reasons: mycobacterial MVs have been shown to induce proinflammatory immune responses in a TLR2-dependent manner [21]; and MAH104 is known to use host cell apoptosis to spread from initially infected primary macrophage to neighboring uninfected secondary macrophages [24,56]. A recent study suggested a role for a specific M. tuberculosis PE protein in the response to environmental stress and intracellular survival [57].
Our study supports the use of metal mix as a valuable in vitro model to study MAH104 pathogenesis-relevant MVs, and vesicle-bound virulence proteins. Likewise, our findings also highlight the significance of vesiculation during the early onset of MAH104 macrophage infection identified by freeze-fracture electron microscopy. This finding is consistent with several studies in both Gram-positive and -negative bacteria, and numerous review articles that discuss the importance of vesiculation in bacterial virulence and pathogenesis [13–15,19,58].
Using BONCAT metabolic labeling of MAH presynthesized proteins, we were able to confirm the secretion of five effector proteins of metal mix into the host cell cytosol at 24 h MAH infection of human macrophages. Among these proteins, we identified a virulence factor – glutamine synthetase (GS). The GS knockout mutant of M. tuberculosis failed to grow in human THP-1 macrophages and was attenuated in the guinea pig model of pulmonary tuberculosis [59]. This enzyme has been well characterized in M. tuberculosis and it was found in large concentrations in the phagosome of M. tuberculosis-infected human monocytes [60]. While GS has also been characterized in MAH [61], there are no reports on its involvement in pathogenesis. Our observation presents a strong evidence for the potential role of GS in MAH104 pathogenesis and suggests MV-mediated export as a possible mechanism. The remaining 16 AHA-labeled MAH104 effector proteins encountered in the host cell cytosol were absent in the metal mix-induced MVs, and seemingly were not secreted as an MV cargo, but most likely through one of the cell-bound secretion systems. The AHA labeling may have missed many effectors due to the limitation of tagging only presynthesized proteins, whereas many virulence factors are synthesized de novo in response to the intracellular stimuli. However, we also employed a complementary approach of FLAG tagging of few metal-mix proteins and further confirmed that 6 out of 11 proteins were secreted in the cytosol of host macrophages. It is also important to stress that MVs that are formed during the first 24 h of host macrophage infection most likely will not be released just at 24 h (the time point of BONCAT experiment), but it will be released at different time points as well. It is possible that some of these bacterial effectors will be processed/cleared by the host defense mechanism and, consequently, we could have missed them with BONCAT. To enrich our BONCAT results, protein secretion at early time points than 24 h should be investigated. Despite the fact that we need to experimentally validate if these effectors are exported across bacterial cell wall via release of MVs, there is a strong evidence suggesting that MVs released by M. tuberculosis within infected macrophages can escape phagosomes [22] and they play important role during host–pathogen interaction [62]. By examining the ability of extracellular MVs to cross the macrophage plasma membrane and by using inhibitors blocking several different uptake mechanisms, this study demonstrates that MVs are translocated via plasma membrane by clathrin-mediated endocytosis and over 48 h vesicles disintegrate in the cytosol. Several studies in Gram-negative bacteria have shown that OMVs interact with the host cell endocytic pathways and lipid rafts. For example, outer MVs from enterotoxigenic Escherichia coli, Pseudomonas aeruginosa and Neisseria gonorrhoeae utilize host cell lipid rafts for internalization into the host cell [40,63,64], in contrast with OMVs from enterohemorrhagic E. coli and Helicobacter pylori that are internalized into the host cell via clathrin-mediated endocytosis [65,66].
Lipids detected in our study belong to the outermost layers of the mycobacterial cell wall. Considering the proposition made by previous study that cell membrane is the likely origin of MVs [21], further studies are needed to determine the exact lipid composition of MAH104 MVs, released in response to both prolonged starvation (i.e., minimal media) and the macrophage phagosomal elemental mixture of 24-h postinfection (i.e., metal mix).
The finding in the current study presents evidence for MV-mediated export of dsDNA in response to both minimal media and metal mix. Consistent with previous studies, increased amounts of DNA seem to be associated with the external surface of MVs, compared with the internal MV-lumen/cargo. Comparable observation was made for DNA-carrying OMVs of H. pylori, P. aeruginosa, Salmonella typhimurium, uropathogenic E. coli and Porphyromonas gingivalis [27]. MV-associated DNA plays a direct role in biofilm formation. Streptococcus mutans is one of the oral pathogens, responsible for dental caries by forming biofilms. It was found that extracellular DNA, released through bacterial MVs, plays a direct role in S. mutans biofilm formation, since DNase I-treated S. mutans cell culture showed significantly reduced biofilm formation. A similar phenomenon may be used by MAH for biofilm formation in patients with underlying conditions such as cystic fibrosis [67]. Nevertheless, the question remains as to how bacteria manage to have their cellular DNA associated with the external surfaces of released MVs? Considering strong association between biofilm formation and quorum sensing documented in several Gram-negative and -positive bacteria [68], we can hypothesize that probable explosive cell lysis of a subpopulation, during biofilm formation [69] in MAH104 could be regulated by quorum sensing. Though quorum sensing in mycobacteria is almost unknown, there is some indirect evidence [70]. Likewise, DNA carried in MVs could also play a role in bacterial cell-to-cell communication through horizontal gene transfer as suggested previously [27].
DNA finding within MVs of metal mix suggests that MV-associated DNA also has certain important functions in MAH104 virulence and/or pathogenesis. DNA carried by P. aeruginosa OMVs was detected in the nuclear fraction of nonphagocytic lung epithelial cells, supporting the notion that vesicle-mediated carriage of bacterial DNA may be involved in bacterial pathogenesis [27]. During macrophage infection with M. tuberculosis and Legionella pneumophila, it has been shown that bacterial DNA enters host cell cytosol and binds to cytosolic dsDNA sensor cyclic GMP–AMP synthase (cGAS), activating type I interferon production via the endoplasmic reticulum-associated stimulator of interferon genes (STING), downstream serine threonine protein kinase (TBK1) and interferon regulatory factor 3 (IRF3) pathway [71]. IRF3 deletion mutant mice, which are unable to respond to cytosolic DNA turned out to be resistant to M. tuberculosis infection, highlighting the importance of bacterial DNA in M. tuberculosis pathogenesis [72]. In M. tuberculosis, cytosolic contact of mycobacterial DNA has been considered to be followed by ESX-1-secreted EsxA-mediated rupture of phagosome [73]. MAH lacks ESX-1 secretion system [10] and, based on our data, we propose that MAH104 MVs, released during the first 24 h of infection, carry bacterial DNA to host cell cytosol where it interacts with one or more cytosolic DNA sensors, activating numerous proinflammatory immune responses, including apoptosis, through which MAH104 has been shown to spread to uninfected cells [24,56]. Further studies are needed to confirm this hypothesis.
Conclusion
MAH vesiculation (i.e., the act of releasing MVs) occurs under all physiological and growth conditions. Nevertheless, this response is considered as a part of bacterial stress response during starvation as well as during infection of host cells, facilitating pathogen's survival and persistence in diverse environments, in nutrient acquisition, biofilm formation, pathogenesis and host immunomodulation. The present study is the first report to demonstrate MAH vesiculation in response to phagosome environment of the macrophage. This research comprehensively characterize MV cargos of the ‘phagosome model’ and starvation stress, and establishes MVs as delivery vehicles of several MAH virulence-associated products such as free lipids, DNA and unique sets of effector proteins. Some virulence-associated proteins found in the phagosomal microenvironment mimicking in vitro system were also released within the cytosol of human macrophages. In addition, this study identifies the clathrin-mediated endocytosis as one of the mechanisms of MV delivery within the macrophage cytosol. Though many questions are yet to be answered, there is a lot of hope and future implications in understanding this highly conserved, pathogenesis-relevant, yet underappreciated aspect of microbial life.
Summary points.
The vesiculation is an indispensable physiological response of an opportunistic mycobacterial pathogen to both prolonged starvation and intracellular environment of macrophage phagosomes.
Phagosomal environment mimicking in vitro system (i.e., metal mix) is a useful model to produce infection-relevant membrane vesicles (MVs) of MAH104.
Molecular characterization implies numerous functions for MVs in MAH104 virulence, host invasion and subsequent colonization.
The vesiculation in response to phagosome mimicking system appears to represent a more host cell invasion- and colonization-focused stress response with vesicles enriched in several known virulence factors and enzymes involved in lipid and fatty acid metabolism, lipids and DNA.
In contrast, the vesiculation in response to prolonged starvation (i.e., minimal media) appears to represent more generalized stress response of MAH104, aimed at acquiring nutrients, maintaining membrane integrity and biofilm formation with vesicles enriched in metabolic enzymes, lipoproteins, other cell wall and cell processes-associated proteins, and higher amounts of internally and externally associated DNA than the one produced under intracellular stress.
MVs released by mycobacteria within infected macrophages escape phagosomes, and this study identifies clathrin-mediated endocytosis as one of the translocation mechanism of MVs into the cytosol of host cells.
Our findings have numerous implications in understanding Mycobacterium avium subsp. hominissuis (MAH) pathogenesis and in the development of vaccine and novel anti-MAH therapies while MAH diseases are on constant rise.
Supplementary Material
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/full/10.2217/fmb-2018-0249
Footnotes
Financial & competing interests disclosure
This work was supported by the Oregon State University Foundation FS062E-VF01 (L Danelishvili) and by the Oregon State University Incentive Programs VBS330-001100 (L Danelishvili). The proteomic sequencing was conducted by the Oregon State University (OSU) Mass Spectrometry Center supported in part by OSU's Research Office and institutional funds. The procurement of the Orbitrap Fusion Lumos was made possible by National Institutes of Health grant S10 OD020111. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Nishiuchi Y, Iwamoto T, Maruyama F. Infection sources of a common non-tuberculous mycobacterial pathogen, Mycobacterium avium complex. Front. Med. (Lausanne) 2017;4:27. doi: 10.3389/fmed.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong D, Gold JW, Dryjanski J, et al. Treatment of infections in patients with the acquired immunodeficiency syndrome. Ann. Intern. Med. 1985;103(5):738–743. doi: 10.7326/0003-4819-103-5-738. [DOI] [PubMed] [Google Scholar]
- 3.Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 2007;175(4):367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 4.Field SK, Fisher D, Cowie RL. Mycobacterium avium complex pulmonary disease in patients without HIV infection. Chest. 2004;126(2):566–581. doi: 10.1378/chest.126.2.566. [DOI] [PubMed] [Google Scholar]
- 5.Mcnamara M, Danelishvili L, Bermudez LE. The Mycobacterium avium ESX-5 PPE protein, PPE25-MAV, interacts with an ESAT-6 family protein, MAV_2921, and localizes to the bacterial surface. Microb. Pathog. 2012;52(4):227–238. doi: 10.1016/j.micpath.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Miltner E, Wu M, Petrofsky M, Bermudez LE. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell Microbiol. 2005;7(4):539–548. doi: 10.1111/j.1462-5822.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- 7.Danelishvili L, Bermudez LE. Mycobacterium avium MAV_2941 mimics phosphoinositol-3-kinase to interfere with macrophage phagosome maturation. Microbes Infect. 2015;17(9):628–637. doi: 10.1016/j.micinf.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danelishvili L, Wu M, Stang B, et al. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc. Natl Acad. Sci. USA. 2007;104(26):11038–11043. doi: 10.1073/pnas.0610746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danelishvili L, Bermudez LE. Mycobacterium avium MAV_2941 mimics phosphoinositol-3-kinase to interfere with macrophage phagosome maturation. Microbes Infect. 2015;17(9):628–637. doi: 10.1016/j.micinf.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdallah AM, Gey Van Pittius NC, Champion PA, et al. Type VII secretion – mycobacteria show the way. Nat. Rev. Microbiol. 2007;5(11):883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 11.Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis . Cell. 2014;159(7):1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74(1):81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13(10):620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host–pathogen interaction. Genes Dev. 2005;19(22):2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 16.Schertzer JW, Whiteley M. Bacterial outer membrane vesicles in trafficking, communication and the host–pathogen interaction. J. Mol. Microbiol. Biotechnol. 2013;23(1–2):118–130. doi: 10.1159/000346770. [DOI] [PubMed] [Google Scholar]
- 17.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74(1):81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012;80(6):1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 2015;15(6):375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 20.Prados-Rosales R, Weinrick BC, Pique DG, Jacobs WR, Jr., Casadevall A, Rodriguez GM. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 2014;196(6):1250–1256. doi: 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prados-Rosales R, Baena A, Martinez LR, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 2011;121(4):1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athman JJ, Sande OJ, Groft SG, et al. Mycobacterium tuberculosis membrane vesicles inhibit T cell activation. J. Immunol. 2017;198(5):2028–2037. doi: 10.4049/jimmunol.1601199. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Membrane vesicles released by intracellular bacteria escape phagosome and play important role in the host–pathogen interaction in the Mycobacterium tuberculosis pathogenicity.
- 23.Wagner D, Maser J, Lai B, et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 2005;174(3):1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]; • Through the x-ray microprobe technology, the presence and concentrations of single elements including iron, calcium, chlorine, potassium, manganese, copper and zinc were identified in the phagosomal microenvironments of 1- and 24-h Mycobacterium avium-infected macrophages.
- 24.Early J, Bermudez LE. Mimicry of the pathogenic mycobacterium vacuole in vitro elicits the bacterial intracellular phenotype, including early-onset macrophage death. Infect. Immun. 2011;79(6):2412–2422. doi: 10.1128/IAI.01120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The metal-mix medium mimmicking the M. avium phagosome inviroment was developed as an in vitro ‘phagosome model’ to study M. avium pathogenesis, and it was confirmed that several bacterial virulence factors known to be upregulated within host macrophages were also highly expressed in the ‘phagosome model’.
- 25.Chinison JJ, Danelishvili L, Gupta R, Rose SJ, Babrak LM, Bermudez LE. Identification of Mycobacterium avium subsp. hominissuis secreted proteins using an in vitro system mimicking the phagosomal environment. BMC Microbiol. 2016;16(1):270. doi: 10.1186/s12866-016-0889-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Mycobacterium avium effector proteins exported in to the cytosol of host macrophages are also secreted in the ‘phagosome model’ upon exposure to the metal mix.
- 26.Prados-Rosales R, Brown L, Casadevall A, Montalvo-Quiros S, Luque-Garcia JL. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX. 2014;1:124–129. doi: 10.1016/j.mex.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The membrane vesicle formation by mycobacteria under different stress conditions has been demonstrated, and membrane vesicle isolation and purification procedures have been developed and optimized.
- 27.Bitto NJ, Chapman R, Pidot S, et al. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 2017;7(1):7072. doi: 10.1038/s41598-017-07288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Soolingen D, De Haas PE, Hermans PW, Van Embden JD. DNA fingerprinting of Mycobacterium tuberculosis . Methods Enzymol. 1994;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 29.Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 2003;198(5):693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffrey B, Rose SJ, Gilbert K, Lewis M, Bermudez LE. Comparative analysis of the genomes of clinical isolates of Mycobacterium avium subsp. hominissuis regarding virulence-related genes. J. Med. Microbiol. 2017;66(7):1063–1075. doi: 10.1099/jmm.0.000507. [DOI] [PubMed] [Google Scholar]
- 31.Mckinney JD, Honer Zu Bentrup K, Munoz-Elias EJ, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406(6797):735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 32.Munoz-Elias EJ, Mckinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005;11(6):638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen MT, Gotz F. Lipoproteins of Gram-positive bacteria: key players in the immune response and virulence. Microbiol. Mol. Biol. Rev. 2016;80(3):891–903. doi: 10.1128/MMBR.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaur RL, Ren K, Blumenthal A, et al. LprG-mediated surface expression of lipoarabinomannan is essential for virulence of Mycobacterium tuberculosis . PLoS Pathog. 2014;10(9):e1004376. doi: 10.1371/journal.ppat.1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahdavi A, Szychowski J, Ngo JT, et al. Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc. Natl Acad. Sci. USA. 2014;111(1):433–438. doi: 10.1073/pnas.1301740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grammel M, Zhang MM, Hang HC. Orthogonal alkynyl amino acid reporter for selective labeling of bacterial proteomes during infection. Angew. Chem. Int. Ed. Engl. 2010;49(34):5970–5974. doi: 10.1002/anie.201002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chande AG, Siddiqui Z, Midha MK, Sirohi V, Ravichandran S, Rao KV. Selective enrichment of mycobacterial proteins from infected host macrophages. Sci. Rep. 2015;5:13430. doi: 10.1038/srep13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wier GM, Mcgreevy EM, Brown MJ, Boyle JP. New method for the orthogonal labeling and purification of Toxoplasma gondii proteins while inside the host cell. MBio. 2015;6(2):e01628. doi: 10.1128/mBio.01628-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barry CE., 3rd Interpreting cell wall ‘virulence factors’ of Mycobacterium tuberculosis . Trends Microbiol. 2001;9(5):237–241. doi: 10.1016/s0966-842x(01)02018-2. [DOI] [PubMed] [Google Scholar]
- 40.Kaparakis M, Turnbull L, Carneiro L, et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12(3):372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 41.Renelli M, Matias V, Lo RY, Beveridge TJ. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology. 2004;150(Pt 7):2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 42.Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl. Environ. Microbiol. 1990;56(6):1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae . J. Bacteriol. 1989;171(5):2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 2008;190(16):5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JH, Lee J, Park J, Gho YS. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015;40:97–104. doi: 10.1016/j.semcdb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Wei J, Dahl JL, Moulder JW, et al. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 2000;182(2):377–384. doi: 10.1128/jb.182.2.377-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin DM, Jeon BY, Lee HM, et al. Mycobacterium tuberculosis Eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010;6(12):e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The enhanced intracellular survival effector secreted within M. tuberculosis-infected macrophages, also constituent of M. avium membrane vesicle, has been well characterized and was found to interact with several host targets interfering innate immune responses, autophagy, reactive oxygen species production and programmed cell death.
- 48.Dahl JL, Wei J, Moulder JW, Laal S, Friedman RL. Subcellular localization of the Iitracellular survival-enhancing Eis protein of Mycobacterium tuberculosis . Infect. Immun. 2001;69(7):4295–4302. doi: 10.1128/IAI.69.7.4295-4302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalscheuer R, Weinrick B, Veeraraghavan U, Besra GS, Jacobs WR., Jr Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis . Proc. Natl Acad. Sci. USA. 2010;107(50):21761–21766. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishikawa E, Ishikawa T, Morita YS, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med. 2009;206(13):2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehlers S, Kutsch S, Benini J, et al. NOS2-derived nitric oxide regulates the size, quantity and quality of granuloma formation in Mycobacterium avium-infected mice without affecting bacterial loads. Immunology. 1999;98(3):313–323. doi: 10.1046/j.1365-2567.1999.00875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart GR, Wernisch L, Stabler R, et al. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148(Pt 10):3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 53.Lewthwaite JC, Coates AR, Tormay P, et al. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect. Immun. 2001;69(12):7349–7355. doi: 10.1128/IAI.69.12.7349-7355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 55.Sampson SL. Mycobacterial PE/PPE proteins at the host–pathogen interface. Clin. Dev. Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bermudez LE, Danelishvili L, Babrack L, Pham T. Evidence for genes associated with the ability of Mycobacterium avium subsp. hominissuis to escape apoptotic macrophages. Front. Cell Infect. Microbiol. 2015;5:63. doi: 10.3389/fcimb.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh P, Rao RN, Reddy JR, et al. PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci. Rep. 2016;6:21624. doi: 10.1038/srep21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pathirana RD, Kaparakis-Liaskos M. Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiol. 2016;18(11):1518–1524. doi: 10.1111/cmi.12658. [DOI] [PubMed] [Google Scholar]
- 59.Tullius MV, Harth G, Horwitz MA. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 2003;71(7):3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harth G, Clemens DL, Horwitz MA. Glutamine synthetase of Mycobacterium tuberculosis: extracellular release and characterization of its enzymatic activity. Proc. Natl Acad. Sci. USA. 1994;91(20):9342–9346. doi: 10.1073/pnas.91.20.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez ME, Mccarthy CM. Glutamine synthetase from Mycobacterium avium . Can. J. Microbiol. 1984;30(3):353–359. doi: 10.1139/m84-052. [DOI] [PubMed] [Google Scholar]
- 62.Gupta S, Rodriguez GM. Mycobacterial extracellular vesicles and host pathogen interactions. Pathog. Dis. 2018;76(4) doi: 10.1093/femspd/fty031. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Highlights the current understanding of the pathogenic mycobacterial membrane vesicle biology with implications in tuberculosis pathogenesis and in therapeutic applications.
- 63.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5(4):e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23(23):4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanaja SK, Russo AJ, Behl B, et al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. 2016;165(5):1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect. Immun. 2010;78(12):5054–5061. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Catherinot E, Roux AL, Vibet MA, et al. Mycobacterium avium and Mycobacterium abscessus complex target distinct cystic fibrosis patient subpopulations. J. Cyst. Fibros. 2013;12(1):74–80. doi: 10.1016/j.jcf.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 68.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13(1):27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 69.Turnbull L, Toyofuku M, Hynen AL, et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016;7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polkade AV, Mantri SS, Patwekar UJ, Jangid K. Quorum sensing: an under-explored phenomenon in the phylum actinobacteria. Front. Microbiol. 2016;7:131. doi: 10.3389/fmicb.2016.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watson RO, Bell SL, Macduff DA, et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 2015;17(6):811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides direct evidence of DNA binding with the cyclic GMP–AMP synthase during M. tuberculosis infection in vivo and highlights importance of bacterial DNA in the host–pathogen interaction.
- 72.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11(5):469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Groschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 2016;14(11):677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.