Abstract
Background:
Allergic and autoimmune diseases comprise a group of inflammatory disorders caused by aberrant immune responses in which CD25+ Forkhead box P3-positive (FOXP3+) T regulatory (Treg) cells that normally suppress inflammatory events are often poorly functioning. This has stimulated an intensive investigative effort to find ways of increasing Tregs as a method of therapy for these conditions. One such line of investigation includes the study of how ligation of Toll-like receptors (TLRs) by CpG oligonucleotides (ODN) results in an immunostimulatory cascade that leads to induction of T-helper (Th) type 1 and Treg-type immune responses.
Objective:
The present study investigated the mechanisms by which calf thymus mammalian double-stranded DNA (CT-DNA) and a synthetic methylated DNA CpG ODN sequence suppress in vitro lymphoproliferative responses to antigens, mitogens, and alloantigens when measured by [3H]-thymidine incorporation and promote FoxP3 expression in human CD4+ T cells in the presence of transforming growth factor (TGF) beta and interleukin-2 (IL-2).
Methods:
Lymphoproliferative responses of peripheral blood mononuclear cells from four healthy subjects or nine subjects with systemic lupus erythematosus to CT-DNA or phytohemagglutinin (PHA) was measured by tritiated thymidine ([3H]-TdR) incorporation expressed as a stimulation index. Mechanisms of immunosuppressive effects of CT-DNA were evaluated by measurement of the degree of inhibition to lymphoproliferative responses to streptokinase-streptodornase, phytohemagglutinin (PHA), concanavalin A (Con A), pokeweed mitogen (PWM), or alloantigens by a Con A suppressor assay. The effects of CpG methylation on induction of FoxP3 expression in human T cells were measured by comparing inhibitory responses of synthetic methylated and nonmethylated 8-mer CpG ODN sequences by using cell sorting, in vitro stimulation, and suppressor assay.
Results:
Here, we showed that CT-DNA and a synthetic methylated DNA 8-mer sequence could suppress antigen-, mitogen-, and alloantigen-induced lymphoproliferation in vitro when measured by [3H]-thymidine. The synthetic methylated DNA CpG ODN but not an unmethylated CpG ODN sequence was shown to promote FoxP3 expression in human CD4+ T cells in the presence of TGF beta and IL-2. The induction of FoxP3+ suppressor cells is dose dependent and offers a potential clinical therapeutic application in allergic and autoimmune and inflammatory diseases.
Conclusion:
The use of this methylated CpG ODN offers a broad clinical application as a novel therapeutic method for Treg induction and, because of its low cost and small size, should facilitate delivery via nasal, respiratory, gastrointestinal routes, and/or by injection, routes of administration important for vaccine delivery to target sites responsible for respiratory, gastrointestinal, and systemic forms of allergic and autoimmune disease.
Keywords: Allergic, autoimmune, CpG oligonucleotides, DNA sequence, immune response, inflammatory disorders, Treg
Atopic allergic and autoimmune disorders are now considered the result of a breakdown of mechanism(s) responsible for maintaining immunologic tolerance, a fundamental biologic process that functions to avoid immune reactivity against normally innocuous environmental or self-antigens.1 This immunologic dysfunction leads to proinflammatory events mediated by cells and cellular components of the innate and adaptive (i.e., T and B cell) immune systems that promote tissue injury in affected target organs. Defective function or deficient numbers of regulatory T cells (Tregs), which normally function to suppress or regulate immune responses, have been suggested to substantially contribute to the loss of peripheral tolerance associated with the inflammatory clinical sequelae of both the allergic diseases and the autoimmune disorders.2,3
A major therapeutic strategy for the treatment of the autoimmune and allergic diseases involves a search for modalities that can increase the diminished quantities or functions of Treg cells in these disorders. Fortuitously, for the field of allergy/immunology, allergen immunotherapy is the singular modality in all of medicine that provides a clinically acceptable treatment regimen for allergic disease whose mechanism of action has been shown, in major part, to be mediated by the induction of tolerance as defined by a decrease of an interleukin-4 (IL-4) secreting type 2 helper (Th2) cell population to an IL-10–, transforming growth factor beta (TGF-β)–secreting inducible Treg cell population.4 Currently, subcutaneous immunotherapy and sublingual immunotherapy are the two major forms of allergen immunotherapy for allergic rhinitis (AR) and asthma whose clinical usage has been correlated with the improvement of allergic symptomatology.5
Over the years, several studies have highlighted the possible important role of immunostimulatory function of DNA as an immunotherapeutic agent in disease processes.3–5 After the 1893 discovery by Coley6 that a toxin, i.e., “Coley toxin,” derived from a mixture of bacterial cell lysates possessed immunostimulatory properties that could reduce the progression of some carcinomas, it was not until 1984 that Tokunaga et al.7 specifically identified bacterial DNA as the underlying component of the lysate that elicited the response. It was later demonstrated by Krieg et al.8 that CpG motifs commonly found within bacterial DNA were responsible for the immunostimulatory effects. In addition, because the immunostimulatory CpGs of microbial DNA were found to be unmethylated, in contrast to mammalian methylated CpG moieties found in healthy humans that were both noninflammatory and lacked immunogenicity, it indicated that DNA methylation could be a key determinant molecular switch that controls their regulatory properties.9–11
Recent studies of epigenetics, i.e., changes in gene expression that do not involve alterations in the underlying DNA sequence, such as DNA methylation, have been shown to play an important role in the pathogenesis of autoimmune diseases, e.g., systemic lupus erythematosus (SLE)12 and cancer13 and, more recently, in allergic diseases.14,15 In mammalian cells, the term DNA methylation refers to the addition of a methyl (CH3) group to the fifth carbon atom of a cytosine ring to form 5-methylcytosine found in juxtaposition to the guanine residues that, together with a phosphate group, formulate the CpG motifs. These CpGs are usually found clustered in the DNA molecule in regions referred to as CpG islands that, when hypermethylated, promote gene silencing, and when hypomethylated facilitate transcriptional activation.12
Collectively, a congeries of studies that evaluated immunogenicity of DNA from prokaryotic and eukaryotic systems support the potential for use of an eukaryotic methylated DNA sequence as a potential vaccine for induction of Tregs in humans.16–19 In 2004, Moseman et al.20 reported that human plasmacytoid dendritic cells activated by B-type CpG oligodeoxynucleotides with 23 base pairs on a phosphodiester backbone could induce the generation of CD4+CD25+ Treg cells. None of these reports, however, identified methylated DNA as the molecular inducer of suppressor and/or Treg cells. These findings, however, indicated that DNA molecules in prokaryotes and eukaryotes not only differed in their immunogenic potential but also that the presence of unique methylated sequences or structures within the DNA played a determinative role. This was recently confirmed by the studies of Notley et al.,21 which showed that the methylation status of cell DNA governs the regulation of inflammation as seen in healthy subjects compared with DNA from patients with rheumatoid arthritis and SLE. Apoptotic and mammalian double-stranded (ds) DNAs are methylated and were found to be immunosuppressive by activating Tregs either directly by surface activation or through dendritic cells as intermediary antigen-presenting cells. In contrast, dsDNA from patients with rheumatoid arthritis or SLE was found to be demethylated and to promote inflammation.21 These findings supported the rationale of the present study for the use of the methylated CpG octamer as a potential therapeutic agent. The discovery of these critical epigenetic mechanisms of gene function is now finding clinical application in ways of increasing Tregs as a method of therapy for allergic and autoimmune disorders. One such line of investigation includes the study of how ligation of the CpG oligonucleotides (ODN) to Toll-like receptor 9 (TLR-9), the principal CpG binding site, results in an immunostimulatory cascade that leads to induction of Th1 and Treg-type immune responses.22,23
The present study investigated the mechanisms by which calf thymus derived methylated dsDNA (CT-DNA) and a synthetic methylated DNA CpG ODN octamer sequence suppressed in vitro lymphoproliferative responses to antigens, mitogens, and methylated CpG ODN when measured by [3H]-TdR incorporation, and examined the possibility that CpG ODN may be an operative inducer of Treg cells as measured by increased Forkhead box P3 (FOXP3) expression in human CD4+ T cells in the presence of TGF-β and IL-2.
METHODS
Study Subjects
Healthy subjects and patients with SLE were recruited during the period of 1978 to 1980 from the rheumatology/immunology laboratory and clinic of Walter Reed Army Medical Center, Washington, D.C., and Georgetown University Medical Center, Washington, D.C., during the 2012–2016 period. All protocols that involved human subjects were approved by the institutional review boards (IRBs) at Walter Reed Army Medical Center and Georgetown University. The nine patients with SLE were all adults (mean age, 27 years), predominantly women (8/9) and African-American (5/9). The four control subjects were all adults (mean age, 27 years), predominantly men (3/4) and half African-American (2/4) and half white (2/4).
Lymphoproliferative Assays
Heparinized whole blood, 10 mls, was obtained from each subject, after which purified suspensions of peripheral blood mononuclear cells (PBMC) were prepared by using Ficoll-Hypaque density centrifugation, and lymphoproliferative assays were performed in a microculture system by methods previously described.24 Briefly, tritiated thymidine [3H]-TdR incorporation, which resulted from stimulation of the PBMCs by general mitogens phytohemagglutinin (PHA) and concanavalin A (Con A), CT-DNA (obtained from Sigma-Aldrich Corp., St. Louis, MO), the global antigen streptokinase-streptodornase (SK-SD), or allogeneic leukocytes in a one way mixed lymphocyte culture (MLC) was measured, and the resultant data were expressed in two different ways: (1) as mean counts per minute (cpm) of tritiated thymidine incorporation, or (2) as a stimulation index (experimental cpm/control cpm).
Immunosuppressive Assay
A modification of the immunosuppressive assay of Shou et al.25,26 was used to measure the presence of suppressor cells in the peripheral blood of normal subjects and of patients with SLE. Briefly, the technique consists of a binary set of interactive cellular responses: (1) a first phase, in which PBMCs were preincubated with Con A or CT-DNA for 48 hours, after which the cells were washed and treated with mitomycin C (“stimulatory cells”); and (2) a second phase, in which equal numbers of freshly isolated autologous or homologous normal PBMCs (“responder cells”) were cocultured with the mitomycin C–inactivated PBMCs from the first phase and were allowed to respond to SK-SD; to the mitogens PHA, Con A, pokeweed mitogen (PWM); or to alloantigens; 2 μCi of [3H]-TdR were added to each well during the last 24 hours of the seventh day of incubation, after which the cells were harvested and radioactivity was measured as described in the section Lymphoproliferative Assays. The lack of [3H] thymidine [3H]-TdR incorporation into the “responder cells” due to a lack of stimulation by the mitomycin C cells in the second phase was defined as a measure of suppression and was expressed as the percentage reduction of experimental from medium control values.
Timing of Performance of Assays
The studies described in the present report were performed at two time periods. The blood samples for the lymphoproliferative studies were drawn during the earlier 1978–1980 period. Blood samples for performance of the Treg studies were drawn more recently, from 2012 to 2016. Although the Systemic Lupus Erythematosus Disease Activity Index criteria for patients with SLE were not used,27 fluorescent antinuclear antibody (FANA) testing, DNA binding; percentage capacity; and avidity, interferon, circulating immune complexes, C3, CH50 were measured, all of which were elevated above normal values (data not shown) and consistent with significant active immune activation seen in patients with SLE.
ODN Sequences
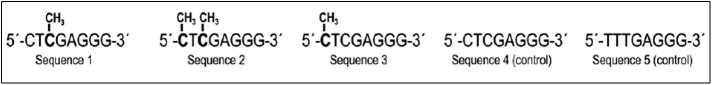
Five separate synthetic nucleotide sequences were obtained from Integrated DNA Technologies, Coralville, Iowa, as dry powder products. These differed in content and positioning of methyl groups at cytosine residues (Fig. 1). Each of the five methylated DNA sequences contained eight backbone nucleotide residues. Sequences 1–4 were composed of an identical backbone nucleotide structure; sequence 5 had a different backbone nucleotide structure. Sequences 1–3 were methylated with the location of the CH3 moieties placed on different cytosine residues; sequence 2 had two CH3 moieties placed on two different cytosine residues. Sequences 4 and 5 were unmethylated and served as methodologic controls.
Figure 1.
Five candidate synthetic CpG sequences.
Rationale for the Design of the CpG ODN Sequences
The methylated DNA sequences were based on size and compositional studies of the CT-DNA and it derivatives. CT-DNA was purchased from Sigma-Aldrich and was sized by French press shearing into fragments that ranged in size from >100,000 to 75,000, and 50,000 by Dr. Doctor Doctor, director of biochemistry at Walter Reed Army Institute of Research. Because none of these preparations showed any reactivity by [3H]-TdR uptake in vitro, it was determined that molecular size disparity could be eliminated as a potential negative influence on the assays. Subsequently, the design of the sequences was based on the compositional structure of CpGs known to have Treg suppressor effects.16
The molecular weights of the DNA fragments were determined by agarose gel electrophoresis by using markers of known molecular weight (DNA ladders; ThermoFisher, Waltham, MA). Because the use of the French press results in DNA fragments of differing sizes, the removal of the sheared DNA fragments was achieved by hydroxyapatite chromatography to fractionate ssDNA, dsDNA, and RNA nucleic acids from the mixture.28
Measurement of Treg Cells
The measurement of Treg cells was performed by measurement of FoxP3 expression levels by using flow cytometry techniques previously described.29,30 Briefly, purified suspensions of PBMCs from healthy subjects and from patients with SLE were prepared by Ficoll-Hypaque density centrifugation as described above (under Lymphoproliferative Assays) and were then sorted to obtain naive CD4+ T cells by using a human naive CD4+ T cells isolation kit (Miltenyi Biotec Co., Auburn, CA). The cells were then cultured with antihuman CD3/CD28 beads (beads-to-cells ratio is 1:10) in the presence of TGF-β and IL-2 for either 3 or 5 days. Methylated sequence 1 (DNA-1) and unmethylated sequence 4 (DNA-4) with 0, 10, 50, and 100 μg doses were added at day 1 and renewed at day 3, and FoxP3 expression was measured in both the 3- and 5-day cultures. Because FoxP3 expression levels are closely associated with Treg function, when FoxP3 is upregulated, the Treg functional activity is known to be concomitantly upregulated. Accordingly, Treg cells were measured by FoxP3 expression in CD4+ T cells, and the results were expressed as percentage suppression.
Statistics and Methods of Analysis
All analyses were done by using R software (version 3.3), (R Foundation for Statistical Computing, Vienna, Austria). Data analyses were performed by using the paired or unpaired Student's t-test. Normality of the data was not formally tested. A one-way analysis of variance (ANOVA) was used when more than two group means were compared; p values <0.05 were deemed statistically significant.
RESULTS
CT-Methylated dsDNA Is Itself Nonimmunogenic
In a set of preliminary in vitro experiments performed in four healthy subjects and nine patients with SLE, lymphoproliferative responses of PBMCs to CT-DNA or PHA were measured by [3H]-TdR incorporation expressed as a stimulation index to examine whether this type of mammalian DNA was immunogenic (Table 1). As can be seen, there was significantly little stimulation of PBMCs by CT-DNA in comparison with a more-robust stimulation index seen with PHA, which confirmed the nonimmunogenicity of a known methylated form of mammalian DNA.
Table 1.
Proliferative responses of PBMCs from healthy subjects or subjects with SLE to CT-DNA or PHA as measured by [3H]-TdR incorporation expressed as SI
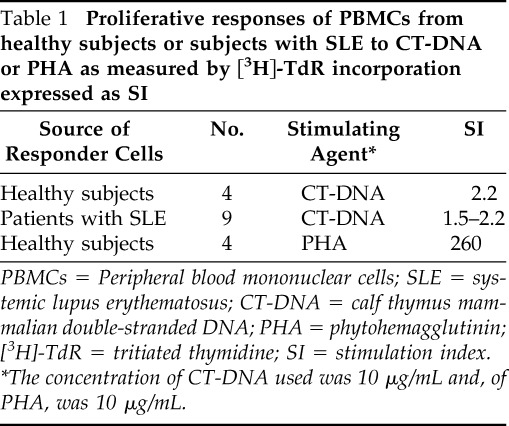
PBMCs = Peripheral blood mononuclear cells; SLE = systemic lupus erythematosus; CT-DNA = calf thymus mammalian double-stranded DNA; PHA = phytohemagglutinin; [3H]-TdR = tritiated thymidine; SI = stimulation index.
*The concentration of CT-DNA used was 10 μg/mL and, of PHA, was 10 μg/mL.
CT-DNA Inhibited T- and B-cell Stimulation Induced by Antigens, Mitogens, and Alloantigens In Vitro
To determine whether the immunosuppressive effect of CT-DNA was due to preferential activation of a suppressor cell component of the PBMC population, a series of experiments were conducted by using a modification of the protocol of Shou et al.25,26 An initial set of experiments was performed to evaluate whether the immunosuppressive effects of CT-DNA was inhibitory for stronger stimuli elicited by T- and B-cell mitogenic stimuli, including PHA, Con A, and PWM as well as by alloantigens. Shown in Table 2 are the responder responses of PHA, Con A, and PWM to Con A and CT-DNA induced stimulatory responses.
Table 2.
Inhibited indicator responses of PHA, Con A and PWM to Con A and CT-DNA–induced stimulatory responses*
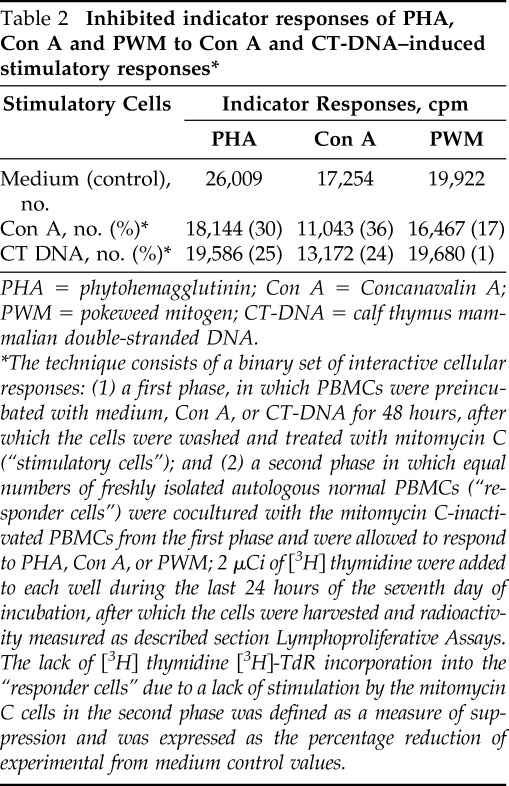
PHA = phytohemagglutinin; Con A = Concanavalin A; PWM = pokeweed mitogen; CT-DNA = calf thymus mammalian double-stranded DNA.
The technique consists of a binary set of interactive cellular responses: (1) a first phase, in which PBMCs were preincubated with medium, Con A, or CT-DNA for 48 hours, after which the cells were washed and treated with mitomycin C (“stimulatory cells”); and (2) a second phase in which equal numbers of freshly isolated autologous normal PBMCs (“responder cells”) were cocultured with the mitomycin C-inactivated PBMCs from the first phase and were allowed to respond to PHA, Con A, or PWM; 2 μCi of [3H] thymidine were added to each well during the last 24 hours of the seventh day of incubation, after which the cells were harvested and radioactivity measured as described section Lymphoproliferative Assays. The lack of [3H] thymidine [3H]-TdR incorporation into the “responder cells” due to a lack of stimulation by the mitomycin C cells in the second phase was defined as a measure of suppression and was expressed as the percentage reduction of experimental from medium control values.
As can be seen, the degree of inhibition of indicator responses to all three mitogens to mitomycin-inactivated Con A and CT-DNA stimulation were similar, and ranged from 17 to 36% for Con A– and 1 to 25% for CT-DNA–induced cellular responses. More specifically, there was a significant decrease of the inhibited indicator responses of PHA to either Con A (p = 0.06) or CT-DNA (p = 0.02). Either Con A– or dsDNA–stimulated suppressor cells inhibited stimulation of the indicator cells to a similar degree of 45% when compared with controls, which indicated that CT-DNA–induced inhibitory effects included both T- and B-cellular responses (data not shown). In a subsequent set of experiments, the immunosuppressive effects of CT-DNA or Con A on allogeneic responses was evaluated next. Similar inhibition of allogeneic indicator responses was found in a one-way mixed lymphocyte culture (MLC) assay to Con A– or CT-DNA–induced stimulation (Table 3).
Table 3.
Inhibited indicator allogeneic responses to Con A and CT-DNA–induced stimulatory responses*
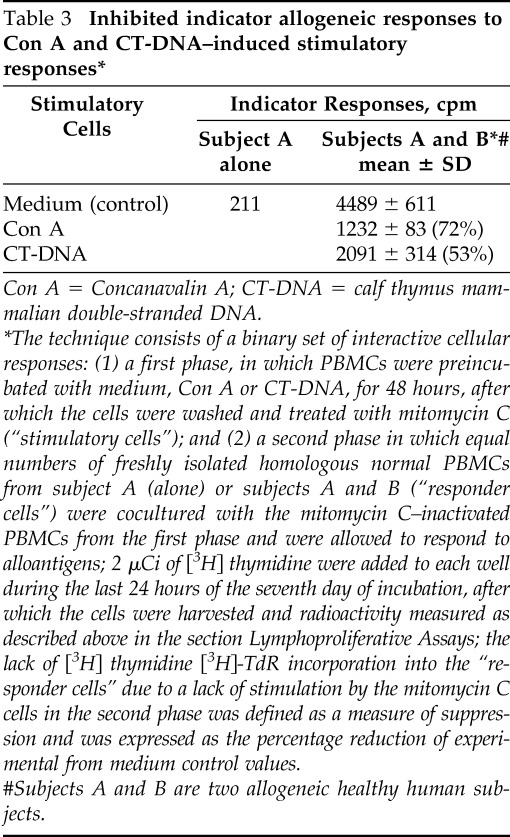
Con A = Concanavalin A; CT-DNA = calf thymus mammalian double-stranded DNA.
The technique consists of a binary set of interactive cellular responses: (1) a first phase, in which PBMCs were preincubated with medium, Con A or CT-DNA, for 48 hours, after which the cells were washed and treated with mitomycin C (“stimulatory cells”); and (2) a second phase in which equal numbers of freshly isolated homologous normal PBMCs from subject A (alone) or subjects A and B (“responder cells”) were cocultured with the mitomycin C–inactivated PBMCs from the first phase and were allowed to respond to alloantigens; 2 μCi of [3H] thymidine were added to each well during the last 24 hours of the seventh day of incubation, after which the cells were harvested and radioactivity measured as described above in the section Lymphoproliferative Assays; the lack of [3H] thymidine [3H]-TdR incorporation into the “responder cells” due to a lack of stimulation by the mitomycin C cells in the second phase was defined as a measure of suppression and was expressed as the percentage reduction of experimental from medium control values.
#Subjects A and B are two allogeneic healthy human subjects.
The Responder Responses of SK-SD to CT-DNA–Induced Stimulation Were Reduced in Patients with SLE
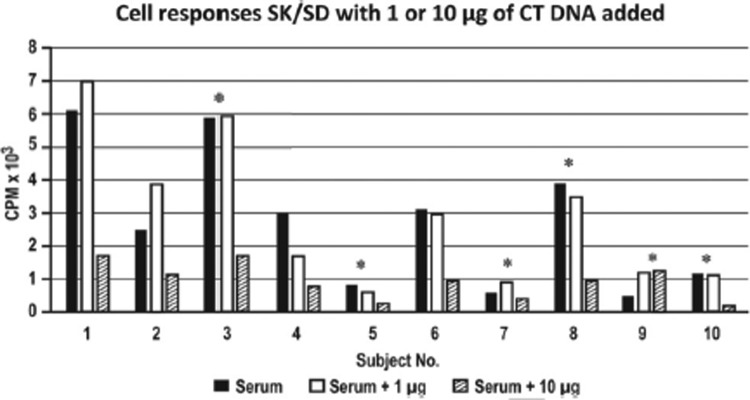
To explore the clinical translational validities of these findings, we next validated these results by using PBMCs isolated from patients with SLE, a prototype of autoimmune diseases. The responder responses of SK-SD to CT-DNA–induced stimulation was next examined in nine patients with SLE and in one healthy subject by using CT-DNA responder doses of 1 μg and 10 μg for each. As shown in Fig. 2, little inhibitory effect was seen at a dose of 1 μg. However, at a dose of 10 μg, there was a severalfold inhibitory effect in both the healthy subjects as well as in eight of the nine patients with SLE tested (subjects 2–8 and subject 10), which indicated that the inhibitory effect was dose dependent.
Figure 2.
Responder responses of streptokinase-streptodornase (SK-SD) in cell cultures from nine patients with systemic lupus erythematosus (SLE) (subjects 2–10) and one healthy subject (subject 1) when 1 or 10 μg of calf thymus mammalian double-stranded DNA (CT-DNA) was added to cell cultures supplemented with pooled normal serum. *Patients with SLE and with active disease.
Dose-Response Studies of the Inhibitory Effects of CT-DNA
To further characterize the dose-response inhibitory properties of CT-DNA, a separate set of experiments was conducted in which PBMCs from healthy subjects were pretreated for 48 hours with “stimulatory cell” doses of 1, 10, 50, and 100 μg of CT-DNA, after which freshly isolated autologous PBMCs were cocultured with Con A–stimulated “responder cells.” As shown in Table 4, there was a dose-dependent inhibitory response seen, beginning at a dose of 1 μg and increasing to a maximum inhibition observed at 100 μg. An overall inverse dose-response relationship was observed (p < 0.01).
Table 4.
Dose-response inhibitory effects of varying doses of CT-DNA on Con A-induced responses
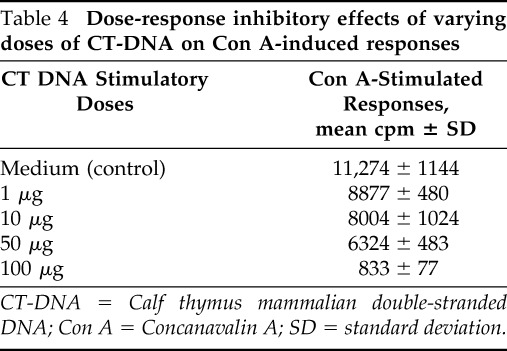
CT-DNA = Calf thymus mammalian double-stranded DNA; Con A = Concanavalin A; SD = standard deviation.
Results of In Vitro Suppression Assays for Measurement of Treg Cells with Methylated and Nonmethylated ODN Sequences
Because CT-DNA is a large molecule, it was possible that the observed inhibitory effects could have resulted from steric inhibition related to molecular size. To examine this possibility, a set of pilot studies was performed in which smaller ODN sequences with molecular weights that ranged from 10, 50, to 100,000 were prepared by French press cleavage, and all were found to be nonstimulatory in the indicator cell system at any of the doses tested (data not shown). Based on these initial observations and the knowledge of preferential methylation sites within CpG, a more-detailed study of specific synthetic methylated ODN sequences was conducted, which compared the capacity of methylated with nonmethylated sequences to stimulate Treg responses.
Methylated Sequence 1 Induced Greater FoxP3 Expression Than Unmethylated Sequence 4 in Human Naive CD4+ T Cells
The methodologies described in publications from the Zheng group29–33 were used to evaluate the role of CpG methylation on induction of FoxP3 expression in human T cells by comparing inhibitory responses of sequence 1 with sequence 4 (Lu et al.,29 Zheng et al.,31 Lu et al.,32). Purified suspensions of PBMCs were isolated from healthy subjects and from patients with SLE, and then PBMCs was sorted to obtain naive CD4+ T cells by using a human naive CD4+ T cells isolation kit. The cells were then cultured with antihuman CD3-CD28 beads in the presence of TGF-β and IL-2 for either 3 or 5 days.
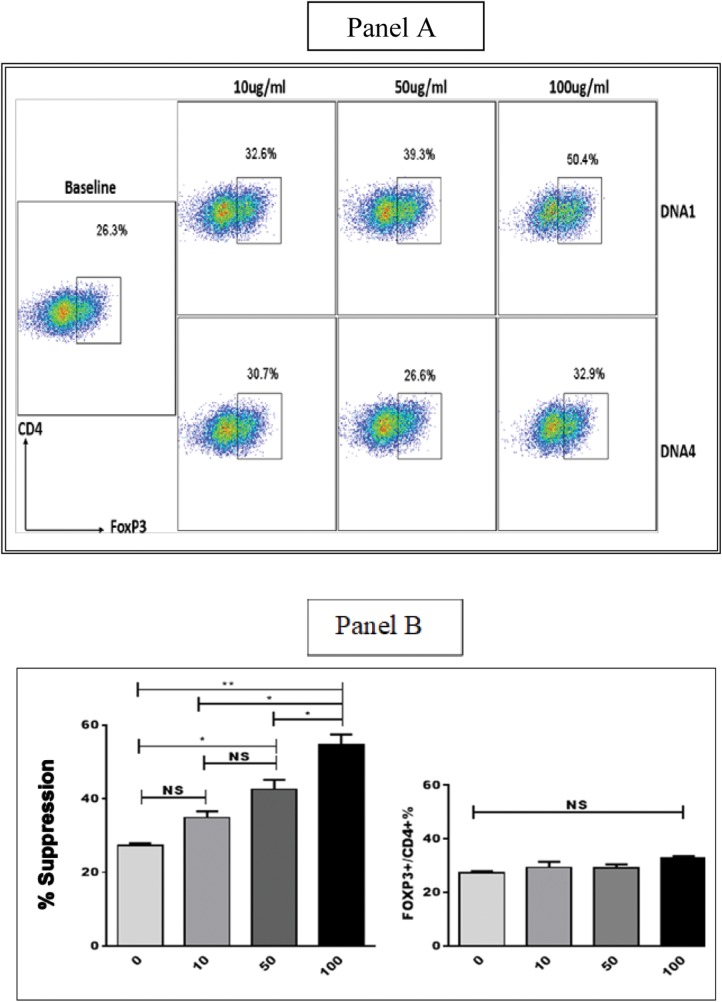
Sequence 1 and sequence 4 with 0, 10, 50, and 100 μg doses were renewed at day 3, and FoxP3 expression was measured in both the 3- and 5-day cultures. Although no differences in FoxP3–CD4 expression were observed between the sequence 1 and sequence 4 cultures at 3 days in different doses (data not shown), there were statistically significant differences of greater FoxP3 expression observed in sequence 1 than in sequence 4 cultures in the 5-day cultures (Fig. 3). It can be seen that CD4+ FoxP3+ T cells increased from a baseline of 26.3 to 32.6% for 10 μg of sequence 1 and to 39.3% for 50 μg, and further increase to 50.4% when 100 μg of sequence 1 was tested. Sequence 1 could also increase FoxP3 expression in naive CD4+ T cells in patients with SLE in a dose-dependent manner (data not shown). Conversely, sequence 4 did not increase FoxP3 induction even in the presence of its high doses (Fig. 3).
Figure 3.
Panel A. Fluorescence-activated cell sorting (FACS) data. Panel B. Comparative studies of FoxP3 expression by methylated sequence 1 (DNA-1) and unmethylated sequence 4 (DNA-4). Sequence 1 shows greater Forkhead box P3 (FOXP3) expression in CD4+ T cells than sequence 4 in 5-day cultures, and it also increases FoxP3 in a dose-dependent responsive manner. NS = Nonsignificance. *p < 0.05; **p < 0.01).
DISCUSSION
The results of the present study demonstrated that CT-DNA and a synthetic methylated CpG ODN octamer sequence suppressed in vitro lymphoproliferative responses to antigens, mitogens, and alloantigens when measured by 3H-TdR incorporation. Although there may be a number of explanations for this induced unresponsiveness, the study results strongly supported the role of Treg cell generation as the most likely as demonstrated by the greater FoxP3 expression induced by the methylated CpG ODN (sequence 1) than by the unmethylated CpG ODN moiety (sequence 4). To our knowledge, this was the first report that demonstrated in vitro induction of FoxP3+ Treg cells by a methylated CpG ODN.
The relevance of methylation status and immunogenicity of DNA has had an interesting historical trajectory. After the 1893 discovery by Coley6 that a toxin, i.e., “Coley toxin,” derived from a mixture of bacterial cell lysates, possessed immunostimulatory properties that could reduce the progression of some carcinomas, it was not until 1984 that Tokunaga et al.7 specifically identified bacterial DNA as the underlying component of the lysate that elicited the response. Subsequently, in a series of seminal studies, Krieg et al.8 and Krieg9,10 demonstrated that CpG motifs commonly found within bacterial DNA were responsible for the immunostimulatory effects.
In addition, because the immunostimulatory CpGs of microbial DNA were found to be unmethylated in contrast to mammalian methylated CpG moieties found in healthy humans that were both noninflammatory and lacked immunogenicity, it indicated that DNA methylation could be a key determinant molecular switch that controls their regulatory properties.9–11 Since then, synthetic nonmethylated CpG ODNs have been the focus of intense research not only due to their proinflammatory responses but also because of their successful use as vaccine adjuvants. The study by Cooper et al.34 provided the first evidence of an unmethylated CpG ODN as a vaccine adjuvant for the hepatitis B virus (HBV) vaccine in humans.
More relevant to the field of allergy is the use of CpG ODNs conjugated to specific allergens35,36 as adjuvants for vaccines in allergen immunotherapy, which have been shown to provide enhanced immunogenicity and reduced adverse allergic responses. By using this approach, Creticos et al.35 showed that immunotherapy by using conjugated CpG ODN and Amb a1 in adults with AR reduced the acute allergic responses, increased Amb a1 specific immunoglobulin G and reduced the Amb-specific immunoglobulin E. In another study, by Asai et al.,36 immunotherapy with Amb a1 in subjects with AR after seasonal ragweed-pollen exposure was found to increase CD4+CD25+ Treg cells in the nasal mucosa. Unlike these studies, which used the immune stimulatory properties of unmethylated CpG, the present study focused on the gene-silencing proprieties of methylated CpG. In fact, in a study by Feltquate and Robinson,37 in which the effect of CpG methylation of an influenza hemagglutinin–expressing plasmid vaccine on isotype and magnitude of antibody responses was studied, methylation abrogated the immunostimulatory activity of the plasmid DNA and significantly reduced the magnitude of the immune response after immunization.
The findings of Treg cell induction with plant mitogens and with methylated CpG ODN in the present study were supported, in part, by the results of several previous investigations that involved the use of Con A in studies of immunoregulation of T cells.38 In these studies, although distinctive classes of T-suppressor cells were identified, the precise mechanism for suppression was unidentified. The discovery by Moseman et al.20 offered an explanation by demonstrating that human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides could induce the generation of CD4+CD25+ Treg cells. Although the methylation composition of the CpG ODN used in this investigation was not determined, the findings of a selective induction of CD4+ FoxP3+ Treg cells by a methylated CpG ODN moiety support the results of the present study.
The precise mechanism by which a methylated CpG ODN induces FoxP3 expression is not known. One possibility is an effect of the methylated CpG ODN on the methylation status of the Treg gene. Because Treg-specific DNA hypomethylation is required for Treg development,39 a methylated CpG ODN can result (presumably) in the induction of FoxP3 expression by hypomethylation of the FoxP3-related genes. A second possibility is that methylated CpG ODN can result in Treg proliferation (and not FoxP3 intensified expression). In a previous study, by Chen et al.,40 mammalian methylated DNA was shown to be inhibitory through its inhibition effects of NF-kappa B and AP-1.
After T cell receptor (TCR) stimulation by cognate antigens, numerous factors and epigenetic mechanisms of the signaling cascade are known to be required to bring about FoxP3 activation, which includes methylation, acetylation, and ubiquitination status. In previous studies29,37 induced peripheral Treg cells were shown to be antigen driven and dependent on the cytokines IL-2 and TGF-β. In the present study, the induction of FoxP3 expression by a methylated CpG sequence required the presence of TGF-β and IL-2 in a dose-dependent manner, which indicated that ancillary signaling components were necessary to complete the signaling pathway required for Treg cell induction by methylated CpG ODN motifs.41
The most recent studies by Notley et al.21 provide perhaps the most convincing support for the findings of the present study. These investigators showed that the methylation status of cell DNA governs the regulation of inflammation as seen in normal subjects compared with DNA from patients with rheumatoid arthritis and SLE. Apoptotic and mammalian dsDNAs that are methylated were found to be immunosuppressive by activating Tregs either directly by surface activation or through dendritic cells as intermediary antigen-presenting cells. Clearly, further studies will be required to definitively clarify the underlying mechanism(s) responsible for FoxP3 induction by methylated CpG.
CONCLUSION
The findings of the present study have wide potential clinical relevance that may offer innovative therapeutic applications for the management of allergic and autoimmune diseases. In these disorders, CD4+CD25+ FoxP3+ Treg cells that normally suppress inflammatory events are hypofunctioning. If the in vitro Treg inductive properties of a CpG ODN can be replicated in vivo, then initially in animal models and ultimately in the human, this could provide a basis for novel therapeutic development of methylated CpG ODN vaccines for the treatment of these disorders. Because of its small molecular size, low cost, ease of synthesis, nonimmunogenicity, and wide applicability for use by a variety of systemic and mucosal delivery routes, a methylated CpG ODN offers many attractive features for an ideal vaccine candidate.
ACKNOWLEDGMENTS
We thank Jaeil Ahn, Ph.D., for his statistical assistance, and Partha P. Banerjee, Ph.D., for his biochemical expertise.
Footnotes
Supported, in part, by a grant from the MedStar Health Research Institute-Georgetown University partnership (O.J.L., J.A.B., J.G.U., M.L.B.), by UL1TR001409 (K.B., J.G.U.) and by the Martyn A. Vickers Sr, MD Endowment Fund (J.A.B.)
U.S. Patent 7,884,196 B2 “Vaccine Composition Comprising Methylated DNA and Immunomodulatory Motifs” was awarded to O.J. Lawless on February 8, 2011. The remaining authors have no conflicts to report
REFERENCES
- 1. Bellanti JA, editor. Immunology IV: Clinical Applications in Health and Disease. Bethesda, MD: I Care Press; 2012. [DOI] [PubMed] [Google Scholar]
- 2. Lan Q, Fan H, Quesniaux V, Ryffel B, Liu Z, Zheng SG. Induced Foxp3(+) regulatory T cells: a potential new weapon to treat autoimmune and inflammatory diseases? J Mol Cell Biol. 2012; 4:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008; 133:775–787. [DOI] [PubMed] [Google Scholar]
- 4. Palomares O, Akdis M, Martín-Fontecha M, Akdis CA. Mechanisms of immune regulation in allergic diseases. The role of regulatory T and B cells. Immunol Rev. 2017; 278:219–236. [DOI] [PubMed] [Google Scholar]
- 5. Nurmatov U, Dhami S, Arasi S, et al. Allergen immunotherapy for allergic rhinoconjunctivitis: a systematic overview of systematic reviews. Clin Transl Allergy. 2017; 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coley WB. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. Am J Med Sci. 1893; 105:487–511. [PubMed] [Google Scholar]
- 7. Tokunaga T, Yamamoto H, Shimada S, et al. Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J Natl Cancer Inst. 1984; 72:955–962. [PubMed] [Google Scholar]
- 8. Krieg AM, Yi AK, Matson S, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995; 374:546–549. [DOI] [PubMed] [Google Scholar]
- 9. Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002; 20:709–760. [DOI] [PubMed] [Google Scholar]
- 10. Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006; 5:471–484. [DOI] [PubMed] [Google Scholar]
- 11. Vollmer J, Krieg AM. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv Drug Deliv Rev. 2009; 61:195–204. [DOI] [PubMed] [Google Scholar]
- 12. Sekigawa I, Okada M, Ogasawara H, Kaneko H, Hishikawa T, Hashimoto H. DNA methylation in systemic lupus erythematosus. Lupus. 2003; 12:79–85. [DOI] [PubMed] [Google Scholar]
- 13. Pan Y, Liu G, Zhou F, et al. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. July 2017. (Epub ahead of print July 27, 2017). [DOI] [PubMed] [Google Scholar]
- 14. Prescott S, Saffery R. The role of epigenetic dysregulation in the epidemic of allergic disease. Clin Epigenetics. 2011; 2:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li JY, Zhang Y, Lin XP, et al. Association between DNA hypomethylation at IL13 gene and allergic rhinitis in house dust mite-sensitized subjects. Clin Exp Allergy. 2016; 46:298–307. [DOI] [PubMed] [Google Scholar]
- 16. Borel Y, Young MC. Nucleic acid-specific suppressor T cells. Proc Natl Acad Sci. 1980; 77:1593–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cowdery JS, Chace JH, Yi AK, Krieg AM. Bacterial DNA induces NK cells to produce IFN-gamma in vivo and increases the toxicity of lipopolysaccharides. J Immunol. 1996; 156:4570–4575. [PubMed] [Google Scholar]
- 18. Messina JP, Gilkeson GS, Pisetsky DS. The influence of DNA structure on the in vitro stimulation of murine lymphocytes by natural and synthetic polynucleotide antigens. Cell Immunol. 1993; 147:148–157. [DOI] [PubMed] [Google Scholar]
- 19. Pisetsky DS, Reich C, Crowley SD, Halpern MD. Immunological properties of bacterial DNA. Ann N Y Acad Sci. 1995; 772:152–163. [DOI] [PubMed] [Google Scholar]
- 20. Moseman EA, Liang X, Dawson AJ, et al. Human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides induce the generation of CD4+CD25+ regulatory T cells. J Immunol. 2004; 173:4433–4442. [DOI] [PubMed] [Google Scholar]
- 21. Notley CA, Jordan CK, McGovern JL, Brown MA, Ehrenstein MR. DNA methylation governs the dynamic regulation of inflammation by apoptotic cells during efferocytosis. Sci Rep. 2017; 7:42204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farrokhi S, Abbasirad N, Movahed A, Khazaei HA, Pishjoo M, Rezaei N. TLR9-based immunotherapy for the treatment of allergic diseases. Immunotherapy. 2017; 9:339–346. [DOI] [PubMed] [Google Scholar]
- 23. Gupta GK, Agrawal DK. CpG oligodeoxynucleotides as TLR9 agonists. therapeutic application in allergy and asthma. BioDrugs. 2010; 24:225–235. [DOI] [PubMed] [Google Scholar]
- 24. Cunningham-Rundles S, Michelis MA, Masur H. Serum suppression of lymphocyte activation in vitro in acquired immunodeficiency disease. J Clin Immunol. 1983; 3:156–165. [DOI] [PubMed] [Google Scholar]
- 25. Shou L, Schwartz SA, Good RA. Suppressor cell activity after concanavalin A treatment of lymphocytes from normal donors. J Exp Med. 1976; 143:1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shou L, Schwartz SA, Good RA, Peng R, Chen CL. A human soluble suppressor factor affecting lymphocyte responses in vitro. Proc Natl Acad Sci U S A. 1980; 77:6096–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Urowitz MB, Gladman DD. Measures of disease activity and damage in SLE. Baillieres Clin Rheumatol. 1998; 12:405–413. [DOI] [PubMed] [Google Scholar]
- 28. Andrews-Pfannkoch C, Fadrosh DW, Thorpe J, Williamson SJ. Hydroxyapatite-mediated separation of double-stranded DNA, single-stranded DNA, and RNA genomes from natural viral assemblages. Appl Environ Microbiol. 2010; 76:5039–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu L, Ma J, Li Z, et al. All-trans retinoic acid promotes TGF-β-induced Tregs via histone modification but not DNA demethylation on Foxp3 gene locus. PLoS One. 2011; 6:e24590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng SG, Wang JH, Gray JD, Soucier H, Horwitz DA. Natural and induced CD4+CD25+ cells educate CD4+CD25- cells to develop suppressive activity: the role of IL-2. TGF-beta, and IL-10. J Immunol. 2004; 172:5213–5221. [DOI] [PubMed] [Google Scholar]
- 31. Zheng SG, Gray JD, Ohtsuka K, Yamagiwa S, Horwitz DA. Generation ex vivo of TGF-beta-producing regulatory T cells from CD4+CD25- precursors. J Immunol. 2002; 169:4183–4189. [DOI] [PubMed] [Google Scholar]
- 32. Lu L, Lan Q, Li Z, et al. Critical role of all-trans retinoic acid in stabilizing human natural regulatory T cells under inflammatory conditions. Proc Natl Acad Sci U S A. 2014; 111:E3432–E3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA. IL-2 is essential for TGF-beta to convert naive CD4+CD25- cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J Immunol. 2007; 178:2018–2027. [DOI] [PubMed] [Google Scholar]
- 34. Cooper CL, Davis HL, Morris ML, et al. CPG 7909, an immunostimulatory TLR9 agonist oligodeoxynucleotide, as adjuvant to Engerix-B HBV vaccine in healthy adults: a double-blind phase I/II study. J Clin Immunol. 2004; 24:693–701. [DOI] [PubMed] [Google Scholar]
- 35. Creticos PS, Schroeder JT, Hamilton RG, et al. Immunotherapy with a ragweed-toll-like receptor 9 agonist vaccine for allergic rhinitis. N Engl J Med. 2006; 355:1445–1455. [DOI] [PubMed] [Google Scholar]
- 36. Asai K, Foley SC, Sumi Y, et al. Amb a 1-immunostimulatory oligodeoxynucleotide conjugate immunotherapy increases CD4+ CD25+ T cells in the nasal mucosa of subjects with allergic rhinitis. Allergol Int. 2008; 57:377–381. [DOI] [PubMed] [Google Scholar]
- 37. Feltquate DM, Robinson HL. Effect of CpG methylation on isotype and magnitude of antibody responses to influenza hemagglutinin-expressing plasmid. DNA Cell Biol. 1999; 18:663–670. [DOI] [PubMed] [Google Scholar]
- 38. Dwyer JM, Johnson C. The use of concanavalin A to study the immunoregulation of human T cells. Clin Exp Immunol. 1981; 46:237–249. [PMC free article] [PubMed] [Google Scholar]
- 39. Morikawa H, Sakaguchi S. Genetic and epigenetic basis of Treg cell development and function: from a FoxP3-centered view to an epigenome-defined view of natural Treg cells. Immunol Rev. 2014; 259:192–205. [DOI] [PubMed] [Google Scholar]
- 40. Chen Y, Lenert P, Weeratna R, et al. Identification of methylated CpG motifs as inhibitors of the immune stimulatory CpG motifs. Gene Ther. 2001; 8:1024–1032. [DOI] [PubMed] [Google Scholar]
- 41. Gray JD, Hirokawa M, Horwitz DA. The role of transforming growth factor beta in the generation of suppression: an interaction between CD8+ T and NK cells. J Exp Med. 1994; 180:1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]