Abstract
This article contains further data and information from our published manuscript [1]. We aim to identify significant transcriptome alterations of total normal human thyroid vs. histologically normal thyroid adjacent to papillary thyroid carcinoma. We performed a systematic meta-analysis of all the available gene expression profiles for the whole organ also collecting gene expression data for the normal thyroid adjacent to papillary thyroid carcinoma. A differential quantitative transcriptome reference map was generated by using TRAM (Transcriptome Mapper) software able to combine, normalize and integrate a total of 35 datasets from total normal thyroid and 40 datasets from histologically normal thyroid adjacent to papillary thyroid carcinoma from different sources. This analysis identified genes and genome segments that significantly discriminated the two groups of samples. Differentially expressed genes were grouped and enrichment function analyses were performed identifying the main features of the differentially expressed genes between total normal thyroid and histologically normal thyroid adjacent to papillary thyroid carcinoma. The search for housekeeping genes retrieved 414 loci.
Specifications table
| Subject area | Biology |
| More specific subject area | Genomics, bioinformatics |
| Type of data | Table, figure |
| How data was acquired | Microarray data repository: Gene Expression Omnibus (GEO) provided by the National Center for Biotechnology Information (NCBI) and Array Express provided by the European Bioinformatics Institute (EBI) |
| Data format | Raw data |
| Experimental factors | Database search, dataset selection, TRAM (Transcriptome Mapper) analysis |
| Experimental features | Analysis of gene expression data by TRAM software; enrichment function analysis |
| Data source location | Data sources are public database entries and are listed in theSupplementary Table 1Meta-analysis results have been obtained in Bologna, Italy, DIMES Department at University of Bologna |
| Data accessibility | Data are with this article |
| Related research article | [1] |
Value of the data
|
1. Data
1.1. Database searching and database building
The systematic search performed in gene expression data repositories retrieved 35 datasets from 10 microarray experiments on the total normal human thyroid and 40 datasets from 4 microarray experiments on the histologically normal thyroid adjacent to papillary thyroid carcinoma. The 35 datasets of the total thyroid were already used in Ref. [1]. Sample identifiers (GEO and EBI ID numbers) and main sample features are listed in the Supplementary Table 1.
1.2. Total normal thyroid vs. histologically normal thyroid adjacent to papillary thyroid carcinoma transcriptome map
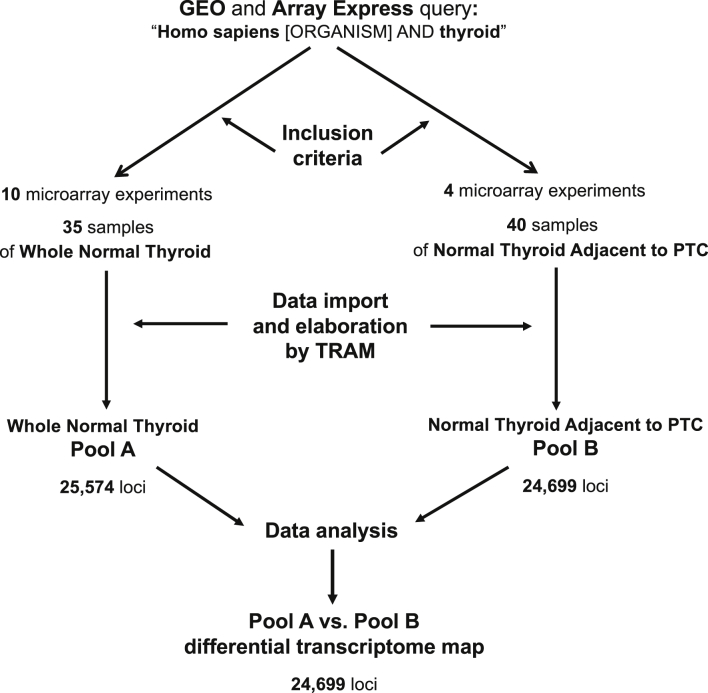
The differential transcriptome map was performed integrating 35 datasets from total normal human thyroid [1] and 40 datasets from histologically normal thyroid adjacent to papillary thyroid carcinoma. The 35 datasets included in the Pool A folder provided reference gene expression values for 25,574 loci coming from 947,816 data points (data from Ref. [1] updated after the analysis with TRAM 1.3 software version), the 40 datasets included in the Pool B folder provided reference gene expression values for 24,699 loci (Supplementary Table 2) coming from 1,917,840 data points, and the differential transcriptome map obtained provided reference gene expression values for 24,699 loci (Supplementary Table 3) common to both pools (Fig. 1).
Fig. 1.
Meta-analysis study design. A search for thyroid tissue gene expression profiles was performed on the online databases GEO and Array Express. It was followed by the selection of experiments and samples according to the inclusion and exclusion criteria, the import and elaboration of data by TRAM software, and the generation and analysis of the whole normal thyroid, histologically normal thyroid adjacent to papillary thyroid carcinoma as well as of the whole normal thyroid vs. histologically normal thyroid adjacent to papillary thyroid carcinoma transcriptome maps. PTC: papillary thyroid carcinoma.
At single gene level, the known gene HTN3, encoding for histatin 3, has the highest gene expression ratio (360.91) followed by STATH, encoding for statherin (gene expression ratio=324.27), HTN1, encoding for histatin 1 (gene expression ratio=240.67), SMR3B encoding for submaxillary gland androgen regulated protein 3B (gene expression ratio=128.73) and ZG16B encoding for zymogen granule protein 16B (gene expression ratio=116.25) (Table 1). These 5 genes are over-expressed in total normal thyroid and have a very low expression value in the histologically normal thyroid adjacent to papillary thyroid carcinoma (Table 1). Fifty genes have a gene expression ratio between 10 and 100 (Table 1).
Table 1.
List of loci of the differential transcriptome map between whole normal thyroid (Pool A) and histologically normal thyroid adjacent to papillary thyroid carcinoma (Pool B). Loci are sorted in descending order of expression ratio (Ratio A/B). Chr: chromosome. SD: standard deviation. N/A: not available in the “NCBI Gene” database (http://www.ncbi.nlm.nih.gov/gene) when the analysis was performed.
| Gene name | Chr | Location | Value A | Value B | Ratio A/B | Data Points A | Data Points B | SD as % of Expression A | SD as % of Expression B |
|---|---|---|---|---|---|---|---|---|---|
| Gene expression ratio >100 | |||||||||
| HTN3 | chr4 | 4q13.3 | 1,876.98 | 5.20 | 360.91 | 25 | 40 | 130.02 | 10.20 |
| STATH | chr4 | 4q13.3 | 2,224.34 | 6.86 | 324.27 | 33 | 40 | 150.28 | 30.81 |
| HTN1 | chr4 | 4q13.3 | 2,039.62 | 8.47 | 240.67 | 31 | 40 | 155.53 | 48.36 |
| SMR3B | chr4 | 4q13.3 | 1,444.86 | 11.22 | 128.73 | 28 | 40 | 137.51 | 56.34 |
| ZG16B | chr16 | 16p13.3 | 1,779.84 | 15.31 | 116.25 | 16 | 40 | 110.20 | 58.66 |
| Gene expression ratio >10 and <100 | |||||||||
| LINC01521 | chr22 | 22q12.2 | 1,034.91 | 11.42 | 90.65 | 15 | 40 | 260.89 | 77.62 |
| PTH | chr11 | 11p15.3 | 343.99 | 5.10 | 67.45 | 38 | 40 | 126.93 | 24.13 |
| MUC7 | chr4 | 4q13.3 | 495.47 | 8.58 | 57.72 | 53 | 80 | 260.31 | 54.80 |
| NACA2 | chr17 | 17q23.2 | 544.38 | 9.80 | 55.53 | 29 | 40 | 143.02 | 79.12 |
| PRH1 | chr12 | 12p13.2 | 1,346.40 | 26.14 | 51.51 | 40 | 80 | 183.58 | 77.46 |
| HBD | chr11 | 11p15.4 | 1,016.65 | 21.48 | 47.33 | 49 | 40 | 210.97 | 54.06 |
| SMR3A | chr4 | 4q13.3 | 318.57 | 7.86 | 40.53 | 40 | 80 | 220.10 | 33.07 |
| KRT13 | chr17 | 17q21.2 | 332.61 | 9.54 | 34.86 | 30 | 40 | 478.35 | 40.15 |
| LINC01234 | chr12 | 12q24.13 | 214.17 | 6.52 | 32.85 | 24 | 80 | 325.50 | 31.97 |
| CST4 | chr20 | 20p11.21 | 791.16 | 31.20 | 25.35 | 21 | 40 | 191.60 | 29.20 |
| FOXM1 | chr12 | 12p13.33 | 291.00 | 13.33 | 21.82 | 33 | 40 | 395.21 | 29.83 |
| CST1 | chr20 | 20p11.21 | 206.18 | 9.48 | 21.74 | 31 | 40 | 208.13 | 30.56 |
| MGC16025 | chr2 | 2q37.3 | 149.46 | 7.63 | 19.59 | 15 | 40 | 245.74 | 35.23 |
| GBP4 | chr1 | 1p22.2 | 542.60 | 28.00 | 19.38 | 39 | 80 | 408.89 | 59.18 |
| TAS2R1 | chr5 | 5p15.31 | 350.21 | 18.07 | 19.38 | 31 | 40 | 408.60 | 21.54 |
| HCAR3 | chr12 | 12q24.31 | 285.79 | 15.54 | 18.39 | 22 | 40 | 309.92 | 55.06 |
| GABRD | chr1 | 1p36.33 | 278.26 | 15.72 | 17.70 | 47 | 80 | 475.42 | 54.59 |
| PPIAL4A | chr1 | 1p11.2 | 104.49 | 6.03 | 17.34 | 21 | 40 | 231.22 | 44.18 |
| LINC02078 | chr17 | 17q25.3 | 170.77 | 10.00 | 17.08 | 13 | 40 | 227.57 | 31.84 |
| BPIFB2 | chr20 | 20q11.21 | 290.54 | 17.27 | 16.82 | 26 | 40 | 275.56 | 35.69 |
| Hs.649237 | chr16 | N/A | 393.84 | 24.42 | 16.13 | 11 | 40 | 41.67 | 64.68 |
| PIP | chr7 | 7q34 | 360.81 | 22.48 | 16.05 | 35 | 40 | 187.82 | 49.66 |
| AKAP6 | chr14 | 14q12 | 191.95 | 12.63 | 15.20 | 51 | 80 | 650.30 | 48.25 |
| RHOC | chr1 | 1p13.2 | 180.95 | 13.47 | 13.44 | 24 | 40 | 103.28 | 35.67 |
| CCDC103 | chr17 | 17q21.31 | 463.69 | 34.98 | 13.26 | 13 | 40 | 231.41 | 20.32 |
| CRCT1 | chr1 | 1q21.3 | 92.27 | 7.04 | 13.10 | 31 | 40 | 493.70 | 29.20 |
| SBSN | chr19 | 19q13.12 | 187.27 | 14.41 | 13.00 | 22 | 40 | 414.04 | 53.84 |
| NPHS2 | chr1 | 1q25.2 | 275.83 | 21.32 | 12.93 | 31 | 40 | 382.71 | 63.61 |
| KRT6A | chr12 | 12q13.13 | 105.18 | 8.35 | 12.60 | 50 | 80 | 576.30 | 55.43 |
| BPIFA2 | chr20 | 20q11.21 | 236.54 | 18.92 | 12.50 | 26 | 40 | 133.55 | 48.56 |
| AGXT | chr2 | 2q37.3 | 132.65 | 10.75 | 12.34 | 73 | 120 | 548.67 | 44.40 |
| CNFN | chr19 | 19q13.2 | 165.81 | 13.79 | 12.02 | 26 | 40 | 355.33 | 18.49 |
| CRISP3 | chr6 | 6p12.3 | 63.55 | 5.30 | 11.98 | 33 | 40 | 158.98 | 20.82 |
| CEP19 | chr3 | 3q29 | 277.20 | 23.20 | 11.95 | 45 | 80 | 348.49 | 62.90 |
| KLF8 | chrX | Xp11.21 | 290.06 | 24.35 | 11.91 | 47 | 80 | 473.99 | 83.04 |
| PLA2G1B | chr12 | 12q24.31 | 93.86 | 7.99 | 11.74 | 35 | 40 | 153.96 | 38.29 |
| KIR3DX1 | chr19 | 19q13.42 | 190.23 | 16.53 | 11.51 | 53 | 80 | 446.28 | 61.41 |
| PSORS1C1 | chr6 | 6p21.33 | 154.59 | 13.49 | 11.46 | 23 | 40 | 301.22 | 22.86 |
| KRT4 | chr12 | 12q13.13 | 191.37 | 16.96 | 11.28 | 53 | 80 | 621.34 | 64.17 |
| ALPP | chr2 | 2q37.1 | 209.50 | 18.93 | 11.07 | 51 | 80 | 467.86 | 70.41 |
| SPRR3 | chr1 | 1q21.3 | 155.41 | 14.05 | 11.06 | 52 | 80 | 455.13 | 43.64 |
| PRR4 | chr12 | 12p13.2 | 233.81 | 21.42 | 10.92 | 33 | 40 | 169.40 | 26.73 |
| FOXL2 | chr3 | 3q22.3 | 99.01 | 9.25 | 10.70 | 33 | 40 | 211.90 | 53.22 |
| DMRTC2 | chr19 | 19q13.2 | 57.74 | 5.47 | 10.56 | 26 | 40 | 174.35 | 16.37 |
| DNMT3L | chr21 | 21q22.3 | 101.54 | 9.68 | 10.49 | 31 | 40 | 357.89 | 28.81 |
| POLE | chr12 | 12q24.33 | 328.71 | 31.44 | 10.46 | 49 | 40 | 417.34 | 25.33 |
| GALNT4 | chr12 | 12q21.33 | 376.31 | 36.31 | 10.36 | 36 | 80 | 411.01 | 74.50 |
| MTPN | chr7 | 7q33 | 256.91 | 24.88 | 10.33 | 26 | 40 | 127.05 | 38.70 |
| C9 | chr5 | 5p13.1 | 49.48 | 4.82 | 10.26 | 33 | 40 | 307.49 | 14.30 |
| GPR88 | chr1 | 1p21.2 | 53.33 | 5.25 | 10.17 | 33 | 40 | 249.83 | 13.38 |
The genome segment that has the highest statistically significant expression value is on chromosome 4 (4q13.3) (Table 2) including the over-expressed known genes (STATH, HTN1, HTN3, SMR3A, SMR3B, MUC7). There are no significantly under-expressed segments.
Table 2.
The genomic segments significantly over-expressed in the total normal thyroid (Pool A) vs. histologically normal thyroid adjacent to papillary thyroid carcinoma (Pool B) differential transcriptome map. Over-expressed genes are in bold, under-expressed genes are with an asterisk and in bold. Under-expressed genomic segments were not found.
| Chr and location | Segment start | Segment end | Value A/B | q-value | Genes in the segment |
|---|---|---|---|---|---|
| chr4 (4q13.3) | 70,000,001 | 70,500,000 | 105.52 | 0.00000017 | STATH HTN3 HTN1 MR3A SMR3B MUC7 |
| chr4 (4q13.3) | 70,250,001 | 70,750,000 | 21.34 | 0.00201816 | SMR3A SMR3B MUC7 UTP3* |
| chr12 (12p13.2) | 11,000,001 | 11,500,000 | 9.31 | 0.00214630 | PRH1 PRB3 PRB4 |
| chr20 (20p11.21) | 23,500,001 | 24,000,000 | 6.99 | 0.00000413 | CST3 CST4 CST1 CST2 CST5 |
| chr12 (12p13.2) | 10,750,001 | 11,250,000 | 6.05 | 0.00335694 | TAS2R10 PRR4 PRH1 |
| chr16 (16q23.1-q23.2) | 78,500,001 | 79,000,000 | 4.50 | 0.00024953 | Hs.649237 Hs.648714 Hs.649874 |
| chr17 (17q21.2) | 41,250,001 | 41,750,000 | 3.61 | 0.00021980 | KRT31 KRT35 KRT13 KRT15 KRT14 |
| chr1 (1q21.3) | 152,250,001 | 152,750,000 | 3.44 | 0.00113717 | CRNN CRCT1 LCE2B |
| chr1 (1q21.3) | 153,000,001 | 153,500,000 | 3.15 | 0.00004193 | SPRR3 SPRR1B PGLYRP3 S100A9 S100A12 |
| chrX (Xq13.2) | 74,000,001 | 74,500,000 | 2.88 | 0.00011566 | Hs.720466 FTX Hs.607917 Hs.625698 |
| chr17 (17q21.2) | 41,000,001 | 41,500,000 | 2.51 | 0.00003900 | KRTAP1-5 KRTAP4-6 KRTAP4-4 KRTAP4-1 KRTAP9-3 KRT31 KRT35 |
| chr17 (17q12.2) | 40,750,001 | 41,250,000 | 2.51 | 0.00016288 | KRTAP3-2 KRTAP1-5 KRTAP4-6 KRTAP4-4 KRTAP4-1 KRTAP9-3 |
| chr11 (11q14.1) | 85,750,001 | 86,250,000 | 2.15 | 0.00045054 | Hs.658368 Hs.658335 Hs.656225 |
| chr19 (19q13.33) | 49,750,001 | 50,250,000 | 1.98 | 0.01225630 | AKT1S1 TBC1D17 ATF5 |
1.3. Functional enrichment analysis
The results of functional enrichment analysis, performed by “ToppFun” from the “ToppGene Suite” Gene Ontology tool, of over- and under-expressed genes (with expression ratios between 1.30 and 10.00 and 0.69 and 0, respectively) in the total normal thyroid vs. histologically normal thyroid adjacent to papillary thyroid carcinoma differential transcriptome map, are shown in Table 3 and Table 4. Input gene lists included 5,012 out of 6,686 over-expressed and 4,258 out of 4,854 under-expressed genes resulted following exclusion of all the EST clusters (Supplementary Table 3).
Table 3.
Results of functional enrichment analysis, performed by “ToppFun” from the “ToppGene Suite” Gene Ontology tool, of over-expressed genes (with expression ratios between 1.30 and 10.00) in the total normal thyroid vs. histologically normal thyroid adjacent to papillary thyroid carcinoma differential transcriptome map. The first 10 results of each Gene Ontology categories are listed. Complete results are provided in Supplementary Table 4.
| Gene expression ratio 1.30–10.00 | |||
|---|---|---|---|
| Genes from input | GO: molecular function | Name | p-Value |
| 148 | GO:0022838 | substrate-specific channel activity | 5.76E-09 |
| 143 | GO:0005216 | ion channel activity | 1.19E-08 |
| 155 | GO:0022803 | passive transmembrane transporter activity | 1.91E-08 |
| 154 | GO:0015267 | channel activity | 2.92E-08 |
| 294 | GO:0022857 | transmembrane transporter activity | 4.18E-08 |
| 114 | GO:0022836 | gated channel activity | 1.02E-07 |
| 256 | GO:0015075 | ion transmembrane transporter activity | 1.15E-07 |
| 14 | GO:0005132 | type I interferon receptor binding | 2.04E-07 |
| 358 | GO:0005215 | transporter activity | 3.17E-07 |
| 58 | GO:0022834 | ligand-gated channel activity | 1.52E-06 |
| Genes from input | GO: Biological process | Name | p-Value |
| 15 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 1.82E-06 |
| 432 | GO:0006811 | ion transport | 2.85E-06 |
| 15 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 4.55E-06 |
| 19 | GO:0002323 | natural killer cell activation involved in immune response | 9.71E-06 |
| 38 | GO:0060349 | bone morphogenesis | 2.38E-05 |
| 22 | GO:0003009 | skeletal muscle contraction | 2.51E-05 |
| 16 | GO:0042501 | serine phosphorylation of STAT protein | 3.03E-05 |
| 21 | GO:0033275 | actin-myosin filament sliding | 3.74E-05 |
| Genes from input | GO: Cellular component | Name | p-Value |
| 422 | GO:0005615 | extracellular space | 1.14E-11 |
| 472 | GO:0031226 | intrinsic component of plasma membrane | 4.23E-09 |
| 453 | GO:0005887 | integral component of plasma membrane | 1.49E-08 |
| 297 | GO:0098590 | plasma membrane region | 1.36E-07 |
| 79 | GO:0045211 | postsynaptic membrane | 3.42E-06 |
| 97 | GO:0097060 | synaptic membrane | 1.19E-05 |
| 339 | GO:0098589 | membrane region | 1.26E-05 |
| 91 | GO:0034702 | ion channel complex | 1.15E-04 |
| 99 | GO:1902495 | transmembrane transporter complex | 2.26E-04 |
| 128 | GO:0031012 | extracellular matrix | 3.14E-04 |
Table 4.
Results of functional enrichment analysis, performed by “ToppFun” from the “ToppGene Suite” Gene Ontology tool, of under-expressed genes (with expression ratios between 0 and 0.69) in the total normal thyroid vs. histologically normal thyroid adjacent to papillary thyroid carcinoma differential transcriptome map.
| Gene expression ratio 0.69–0 | |||
|---|---|---|---|
| Genes from input | GO: molecular function | Name | p-Value |
| 463 | GO:0003723 | RNA binding | 8.64E-16 |
| 499 | GO:0019899 | enzyme binding | 8.25E-10 |
| 467 | GO:0035639 | purine ribonucleoside triphosphate binding | 6.77E-09 |
| 472 | GO:0001882 | nucleoside binding | 7.30E-09 |
| 480 | GO:0017076 | purine nucleotide binding | 1.13E-08 |
| 469 | GO:0001883 | purine nucleoside binding | 1.17E-08 |
| 469 | GO:0032549 | ribonucleoside binding | 1.17E-08 |
| 468 | GO:0032550 | purine ribonucleoside binding | 1.31E-08 |
| 475 | GO:0032555 | purine ribonucleotide binding | 1.91E-08 |
| 478 | GO:0032553 | ribonucleotide binding | 2.33E-08 |
| Genes from input | GO: Biological process | Name | p-Value |
|---|---|---|---|
| 274 | GO:0032446 | protein modification by small protein conjugation | 6.27E-13 |
| 294 | GO:0070647 | protein modification by small protein conjugation or removal | 1.24E-10 |
| 233 | GO:0016567 | protein ubiquitination | 2.16E-10 |
| 478 | GO:0065003 | protein-containing complex assembly | 5.47E-10 |
| 38 | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 2.40E-09 |
| 264 | GO:1903047 | mitotic cell cycle process | 3.45E-09 |
| 284 | GO:0000278 | mitotic cell cycle | 4.09E-09 |
| 39 | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 4.13E-09 |
| 259 | GO:0006396 | RNA processing | 4.70E-09 |
| 163 | GO:0044772 | mitotic cell cycle phase transition | 1.06E-08 |
| Genes from input | GO: cellular component | Name | p-Value |
|---|---|---|---|
| 483 | GO:0005739 | mitochondrion | 4.47E-14 |
| 270 | GO:0005730 | nucleolus | 2.18E-13 |
| 225 | GO:1990904 | ribonucleoprotein complex | 3.42E-11 |
| 116 | GO:0016604 | nuclear body | 9.51E-08 |
| 261 | GO:0044429 | mitochondrial part | 8.96E-07 |
| 16 | GO:0022624 | proteasome accessory complex | 1.08E-06 |
| 50 | GO:0000784 | nuclear chromosome, telomeric region | 2.84E-06 |
| 220 | GO:0005768 | endosome | 3.85E-06 |
| 310 | GO:0005773 | vacuole | 4.78E-06 |
| 10 | GO:0008540 | proteasome regulatory particle, base subcomplex | 5.08E-06 |
1.4. Housekeeping gene search
In the histologically normal thyroid adjacent to papillary thyroid carcinoma transcriptome map, the search for housekeeping genes with the described criteria (Methods section) retrieved 414 loci, including the known genes RPL41, and TG, encoding for ribosomal protein L41 and thyroglobulin, respectively, having low standard deviation (SD), the highest expression values and a high number of data points (n=40) (Table 5). This search did not give the same results of total normal thyroid transcriptome map (see Table 4 [1]). The two transcriptome maps have in common only seven genes: ACTG1, BLOC1S2, DIABLO, OCIAD2, GTPBP6, EIF2B2, AKR1B1.
Table 5.
List of the first 20 (out of 414) predicted housekeeping genes for the histologically normal thyroid adjacent to papillary thyroid carcinoma transcriptome map.
| Gene name | Chromosome | Location | Expression value B | Data points B | SD as % of expression B |
|---|---|---|---|---|---|
| RPL41 | chr12 | 12q13.2 | 7,559.25 | 40 | 7.67 |
| RPL39 | chrX | Xq24 | 4,195.49 | 40 | 8.47 |
| RPL9 | chr4 | 4p14 | 4,586.08 | 40 | 10.91 |
| TG | chr8 | 8q24.22 | 7,619.57 | 40 | 11.06 |
| ACTG1 | chr17 | 17q25.3 | 3,836.40 | 80 | 11.55 |
| RPL27 | chr17 | 17q21.31 | 2,928.50 | 40 | 13.95 |
| TOMM6 | chr6 | 6p21.1 | 368.69 | 40 | 14.09 |
| WASHC5 | chr8 | 8q24.13 | 118.48 | 40 | 14.34 |
| RRAGA | chr9 | 9p22.1 | 594.55 | 40 | 14.51 |
| NDUFA4 | chr7 | 7p21.3 | 1,563.68 | 40 | 14.70 |
| PTDSS1 | chr8 | 8q22.1 | 493.86 | 40 | 14.98 |
| RPS13 | chr11 | 11p15.1 | 2,900.71 | 40 | 15.08 |
| KIAA1191 | chr5 | 5q35.2 | 496.47 | 40 | 15.33 |
| CTR9 | chr11 | 11p15.4 | 254.23 | 40 | 15.58 |
| DUSP11 | chr2 | 2p13.1 | 180.36 | 40 | 15.78 |
| RPS4X | chrX | Xq13.1 | 4,512.67 | 80 | 16.15 |
| SLTM | chr15 | 15q22.1 | 558.06 | 40 | 16.18 |
| MRPS35 | chr12 | 12p11.22 | 275.88 | 40 | 16.37 |
| UNC50 | chr2 | 2q11.2 | 435.55 | 40 | 16.45 |
| FAM96A | chr15 | 15q22.31 | 499.93 | 40 | 16.80 |
Genes are sorted in ascending order of SD as percentage of the mean value. In bold, the two best genes at behaving like housekeeping genes due to a combination of a low SD, a high expression value and a high number of data points. Following checking in the “Values B” TRAM table, the 40 (TG), and the 40 (RPLP41) data points are derived from all the 40 samples of the histologically normal thyroid adjacent to papillary thyroid carcinoma dataset analyzed.
2. Experimental design, materials and methods
2.1. Database search and selection
Gene expression data repositories were systematically searched for any single human thyroid sample available from subjects explicitly stated as “healthy” or “normal” as previously described [1]. The criteria for inclusion or exclusion in the analysis of each retrieved dataset were as previously described [2]. In addition, datasets from histologically normal thyroid adjacent to papillary thyroid carcinoma were collected when available in the experiments retrieved as described.
2.2. TRAM analysis
TRAM software [3] allows the import, decoding of probe set identifiers to gene symbols via UniGene data parsing [4], integration and normalization of gene expression data recorded in the GEO and ArrayExpress databases or in a custom source in tab-delimited text format for the generation and analysis of transcriptome maps [1], [5]. Furthermore, it creates a graphical representation of gene expression profiles along the chromosomes and determines the statistical significance of differential expression of chromosomal segments through hypergeometric distribution [3], [6].
The most current version of TRAM has been used (TRAM 1.3, set up on November 11, 2017) [5]. Pool A was composed of whole normal thyroid tissue datasets, while Pool B included histologically normal thyroid adjacent to papillary thyroid carcinoma datasets (Supplementary Table 1), thus allowing the creation of a differential expression map between the two biological conditions along with the maps for each separate condition. Thresholding of sample expression values equal to or lower than “0” (≤0) [2], calculation of the mean expression value for each locus and determination of percentiles of expression for each gene have been previously described [2], [3].
The parameters for the “Map” mode graphical representation were chosen based on the gene distribution in human genome [7], [8] (window size of 500,000 base pairs or bp and a shift of 250,000 bp). For each segment, its expression value, the over-/under-expression and the statistical significance have been calculated by TRAM as described [3], [5].
Apart from gene expression analyses, these data might be used in metabolic network models [9], [10], [11], [12] for the validation of hypotheses about the relationships among mRNA levels, corresponding enzymatic proteins and the quantities of their substrates or products obtained by metabolome experiments [13], [14].
The data related to the human normal whole thyroid have already been experimentally validated by “Real-Time” reverse transcription polymerase chain reaction obtaining an excellent correlation coefficient (r=0.93) between in vitro and in silico data as previously described [1].
It was not possible to validate the data related to histologically normal thyroid gland adjacent to papillary thyroid carcinoma because commercial RNA of this particular type of tissue is not available, however experimental validations of the results obtained in several previous studies [1], [2] show that the results provided by the TRAM tool were highly reliable [15].
2.3. Functional enrichment analysis
An enrichment function analysis was performed for three arbitrarily chosen intervals of ratio of the mean gene expression values: expression ratio close to one (0.70–1.29) implying that the genes are not differentially expressed between histologically normal thyroid adjacent to papillary thyroid carcinoma and total thyroid, expression ratio ≥1.30 (1.30–10.00) and expression ratio <0.7 (0.69–0). The first interval includes 13,104 loci, the second 6,686, the third 4,854, respectively (Supplementary Table 3). The analysis was performed using “ToppFun” from the “ToppGene Suite” Gene Ontology tool [16]. We submitted the list of genes with expression ratio ≥1.30 and a list of genes of all the chromosomes with expression ratio <0.7, excluding EST clusters. The selected genes were categorized according to GO classification based on their hypothetical molecular functions and biological processes. The analysis was assessed for Molecular Function and Biological Process and Cellular Component categories.
2.4. Housekeeping gene search
A search of housekeeping genes best suitable for the study of histologically normal thyroid adjacent to papillary thyroid carcinoma (Pool B) has been performed using an optimal combination of parameters [15], [17]: in this case, expression value >100, number of data points ≥20 and SD, expressed as a percentage of the mean value, ≤30.
Acknowledgements
This work was supported by RFO grants to MCP, PS and LV and by FFABR grants to MCP and LV.
Footnotes
Transparency document associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2019.103835.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.103835.
Contributor Information
Lorenza Vitale, Email: lorenza.vitale@unibo.it.
Allison Piovesan, Email: allison.piovesan2@unibo.it.
Francesca Antonaros, Email: francesca.antonaros2@unibo.it.
Pierluigi Strippoli, Email: pierluigi.strippoli@unibo.it.
Maria Chiara Pelleri, Email: mariachiara.pelleri2@unibo.it.
Maria Caracausi, Email: maria.caracausi2@unibo.it.
Transparency document
The following is the transparency document related to this article:
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Table 4
Supplementary Table 5
References
- 1.Vitale L., Piovesan A., Antonaros F., Strippoli P., Pelleri M.C., Caracausi M. A molecular view of the normal human thyroid structure and function reconstructed from its reference transcriptome map. BMC Genomics. 2017;18:739. doi: 10.1186/s12864-017-4049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caracausi M., Piovesan A., Vitale L., Pelleri M.C. Integrated transcriptome map highlights structural and functional aspects of the normal human heart. J. Cell. Physiol. 2017;232:759–770. doi: 10.1002/jcp.25471. [DOI] [PubMed] [Google Scholar]
- 3.Lenzi L., Facchin F., Piva F., Giulietti M., Pelleri M.C., Frabetti F., Vitale L., Casadei R., Canaider S., Bortoluzzi S. TRAM (Transcriptome Mapper): database-driven creation and analysis of transcriptome maps from multiple sources. BMC Genomics. 2011;12:121. doi: 10.1186/1471-2164-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenzi L., Frabetti F., Facchin F., Casadei R., Vitale L., Canaider S., Carinci P., Zannotti M., Strippoli P. UniGene Tabulator: a full parser for the UniGene format. Bioinformatics. 2006;22:2570–2571. doi: 10.1093/bioinformatics/btl425. [DOI] [PubMed] [Google Scholar]
- 5.Pelleri M.C., Cattani C., Vitale L., Antonaros F., Strippoli P., Locatelli C., Cocchi G., Piovesan A., Caracausi M. Integrated quantitative transcriptome maps of human trisomy 21 tissues and cells. Front. Genet. 2018;9:125. doi: 10.3389/fgene.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppe A., Danieli G.A., Bortoluzzi S. REEF: searching REgionally Enriched Features in genomes. BMC Bioinf. 2006;7:453. doi: 10.1186/1471-2105-7-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piovesan A., Caracausi M., Antonaros F., Pelleri M.C., Vitale L. GeneBase 1.1: a tool to summarize data from NCBI Gene datasets and its application to an update of human gene statistics. Database (Oxford) 2016;2016 doi: 10.1093/database/baw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piovesan A., Caracausi M., Ricci M., Strippoli P., Vitale L., Pelleri M.C. Identification of minimal eukaryotic introns through GeneBase, a user-friendly tool for parsing the NCBI Gene databank. DNA Res. 2015;22:495–503. doi: 10.1093/dnares/dsv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schellenberger J., Que R., Fleming R.M., Thiele I., Orth J.D., Feist A.M., Zielinski D.C., Bordbar A., Lewis N.E., Rahmanian S. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: constraints-based reconstruction and analysis for Python. BMC Syst. Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frainay C., Aros S., Chazalviel M., Garcia T., Vinson F., Weiss N., Colsch B., Sedel F., Thabut D., Junot C. MetaboRank: network-based recommendation system to interpret and enrich metabolomics results. Bioinformatics. 2019;35:274–283. doi: 10.1093/bioinformatics/bty577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cicek A.E., Qi X., Cakmak A., Johnson S.R., Han X., Alshalwi S., Ozsoyoglu Z.M., Ozsoyoglu G. An online system for metabolic network analysis. Database (Oxford) 2014;2014 doi: 10.1093/database/bau091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caracausi M., Ghini V., Locatelli C., Mericio M., Piovesan A., Antonaros F., Pelleri M.C., Vitale L., Vacca R.A., Bedetti F. Plasma and urinary metabolomic profiles of Down syndrome correlate with alteration of mitochondrial metabolism. Sci. Rep. 2018;8:2977. doi: 10.1038/s41598-018-20834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrokhi Yekta R., Rezaie Tavirani M., Arefi Oskouie A., Mohajeri-Tehrani M.R., Soroush A.R. The metabolomics and lipidomics window into thyroid cancer research. Biomarkers. 2017;22:595–603. doi: 10.1080/1354750x.2016.1256429. [DOI] [PubMed] [Google Scholar]
- 15.Caracausi M., Piovesan A., Antonaros F., Strippoli P., Vitale L., Pelleri M.C. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol. Med. Rep. 2017;16:2397–2410. doi: 10.3892/mmr.2017.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Z., Wang L., Xu M., Zhou X., Chen T., Sun F. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 4
Supplementary Table 5