Abstract
Objectives
The nuclear receptor superfamily is a potential target for the development of new treatments for obesity and metabolic diseases. Increasing evidence has pointed towards the retinoic acid-related orphan receptor-alpha (RORα) as an important nuclear receptor involved in several biological processes. RORα full body knockout mice display improved metabolic phenotypes on both chow and high fat (60% fat, 20% carbohydrate) diets, but also have severe behavioral abnormalities. Here we investigated the effect of hepatic RORα by generating mice with liver-specific RORα deletion to elucidate the role of this nuclear receptor on host metabolism.
Methods
8 week-old mice with liver-specific RORα deletion and littermate controls were fed either chow or western-style diets (40% fat, 40% carbohydrate) for 12 weeks. Metabolic phenotyping was performed at the end of the dietary intervention.
Results
Here, we show that hepatic RORα deletion does not affect the metabolic susceptibility to either chow or western-style diet in terms of glucose metabolism and adiposity.
Conclusions
Our data indicate that liver deletion of RORα does not have a pivotal role in the regulation of hepatic glucose and lipid metabolism on chow or western-style diet.
Keywords: RORα, Obesity, Glucose metabolism, Steatosis
Highlights
-
•
Hepatic deletion of RORα does not affect host metabolism on chow diet.
-
•
Hepatic deletion of RORα does not affect host metabolism on western-style diet.
-
•
Similar phenotypes between male and female mice.
1. Introduction
The incidence of obesity and metabolic associated disorders, such as cardiovascular diseases, type 2 diabetes mellitus and non-alcoholic fatty liver disease (NAFLD), increase worldwide [1], [2], [3]. Furthermore, there are no effective long-term non-surgical treatments for obesity and metabolic associated disorders [4]. However, accumulating evidence points towards nuclear receptors (NRs) as potential molecular treatment targets [5], [6]. The NRs superfamily comprises 48 transcription factors in humans, which are involved in the regulation of a wide range of physiologic and pathophysiologic processes (i.e. development, circadian rhythm, response to steroid hormones, inflammation, obesity, diabetes, NAFLD, and cancer) [7], [8], [9], [10].
The retinoic acid-related orphan receptor α (RORα; NR1F1) is a NR that has been implicated in the regulation of a wide variety of metabolic pathways, including lipid, glucose and steroid metabolism [11], [12], [13]. The initial indications that RORα can affect metabolic processes originate mainly from a natural occurring mutant mouse, the Staggerer mouse, which results in a loss of function [14]. Staggerer mice are protected against diet-induced obesity and associated complications (i.e., adipose tissue inflammation, NAFLD, and impaired glucose metabolism), but also have cerebral phenotypes including impaired locomotion [15], [16]. However, activation of RORα with synthetic ligands also improves NAFLD by reducing hepatic oxidative stress and inflammation suggesting a potential beneficial role of RORα activation on obesity and metabolic diseases [17], [18], [19].
In order to elucidate the role of RORα in obesity and metabolic diseases, tissue specific knockout models have recently been developed. Liver-specific ablation of RORα deteriorates metabolic profile in terms of adiposity, liver steatosis, inflammation and hepatic insulin resistance, when mice were fed a high fat diet (60% of energy from fat and 20% from carbohydrates) [20], [21]. These findings suggest a central role of RORα in the regulation of hepatic lipid and glucose metabolism. To further extend and validate the role of RORα on glucose and lipid metabolism, we generated an independent liver-specific RORα deficient strain that was metabolically phenotyped on both chow and western-style diet (WSD; 40% of energy from fat and 40% from carbohydrate and 20% protein).
2. Materials and methods
2.1. Mice experiments
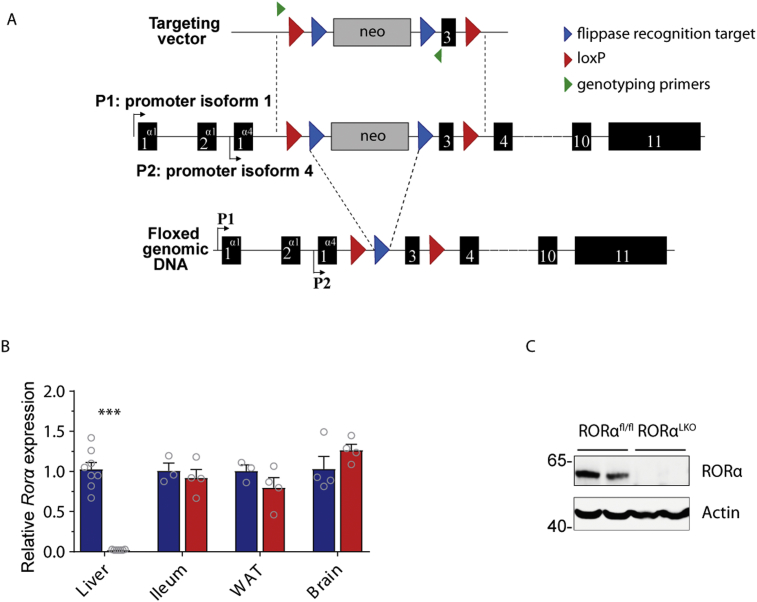
Mice harbouring loxP sequences flanking exon 3, the first common exon coding for both isoform 1 and 4, were generated (Figure 1A). Deletion of the 86 bp exon 3 led to a frame shift resulting in a premature stop codon. Potential translated truncated proteins would be composed of 88 and 32 amino acids instead of 523 and 467 for isoform, 1 and 4 respectively. No functional domain would be present on these putative proteins. Generation of the strain was completed by homologous recombination in C57BL/6J embryonic stem cells followed by microinjection into C57BL/6J blastocysts. The resulting new-borns were crossed with C57BL/6 Flp delete mice to allow the excision of the neomycin selection cassette.
Figure 1.
Generation of liver-specific RORα knock out mice. Schematic representation of the Rora gene-targeting strategy (A, see methods section for details). Expression level of Rora in the liver, ileum, white adipose tissue (WAT), and brain in RORαfl/fl and RORαLKO mice (B, 3–8 mice per group). Protein expression level of RORα and actin in liver extract from RORαfl/fl and RORαLKO mice (C). Data are plotted as mean – SEM. ***p < 0.001.
Albumin-Cre mice were purchased from The Jackson Laboratory. Cre efficacy and specificity towards hepatocytes was confirmed by breading the mice with ROSAmT/mG mice (data not shown). 8-weeks old male RORαLKO and RORαfl/fl littermate were fed a chow diet (Labdiet) or WSD (Harland, TD.96132, 40% fat, 40% sugar, 20% proteins) ad libitum for 12 weeks. MRI, insulin, glucose and pyruvate tolerance tests, and measurements of insulin levels were performed as earlier described [22]. Briefly, insulin, glucose and pyruvate tolerance tests were performed by injecting insulin (0.75 U/kg body weight), glucose (2 g/kg body weight) or pyruvate (2 g/kg weight) respectively, intraperitoneally after a 4 (ITT and GTT) or 12 (PTT) h fast. Tail blood samples were collected at 0, 15, 30, 60, 90 and 120 min and blood glucose levels were determined using a glucose meter (Accu-Check Aviva, Roche). Insulin was measured with ELISA (Crystal Chem) according to the manufacturers' protocols. All experiments including tissue harvesting were performed at the same time of the day (12.00 PM). The local ethics committee in Gothenburg approved all animal experiments.
2.2. Analysis of liver lipids content
Lipids were extracted using the BUME method [23]. The cholesteryl esters, triglycerides and free cholesterol were analyzed using straight phase HPLC coupled to evaporative light scattering detection according to previous work [24]. Briefly, the extract was evaporated under nitrogen and reconstituted in 500 μl heptane:isopropanol [9:1]. Cholesteryl esters, triglycerides, and free cholesterol were separated using straight-phase HPLC (Sunfire 2.1 × 100 mm, Waters) coupled to an evaporative light-scattering detector (PL-ELS 1000, Polymer Laboratories), as described previously [24]. Quantification was made using external standards (Larodan Fine Chemicals).
2.3. Primary hepatocytes
Primary hepatocytes were isolated from RORαLKO and RORαfl/fl littermates mice by digesting liver with perfusion of collagenase type IV, as described previously [25]. After perfusion, 16 × 105 cells were plated on collagen-coated 60 mm dishes in Dulbecco's modified Eagle's medium (DMEM)/F12 (Thermo Fisher) supplemented with fetal bovine serum (Thermo Fisher), penicillin/streptomycin, and 100 nM dexamethasone (Sigma Aldrich). After 4 h, the medium was changed to DMEM/F12 containing penicillin/streptomycin and then cells were harvested after 18h.
2.4. Preparation of protein extracts and immunoblotting
RORα deletion in the liver was evaluated by extracting proteins from the snap-frozen liver or primary cells with lysis buffer, as described previously [26] and immunoblotting with anti-RORα (Abcam, ab60134, 1:300 dilution), GADPH (Cell Signaling #2118, 1:1000 dilution), and anti-actin (Cell Signaling #4970, 1:1000 dilution) antibody.
2.5. Quantitative RT-PCR
Total RNA was isolated using RNeasy kit with DNase treatment (Qiagen) and cDNA templates were generated using the high-capacity cDNA reverse transcription kit using random hexamers (Applied Biosystems). qRT-PCR assays were performed in 10 μl reactions with SYBR Green Master Mix buffer (Thermo Scientific). Gene expression data were normalized to the ribosomal protein L32. Primer sequences used are listed in Supplementary Table 1 (Sigma–Aldrich).
2.6. Statistical analysis
Data are presented as mean - SEM. Analyses between groups were performed by Student's t-test using GraphPad Prism 7 software.
3. Results
3.1. Generation of mice with a hepatocyte specific RORα deletion
We generated mice with a hepatocyte specific Rora deletion of by introducing loxP sites flanking exon 3 of the Rora allele and crossing them with mice expressing Cre under the albumin promotor (Figure 1A and Supplementary Fig. 1A).
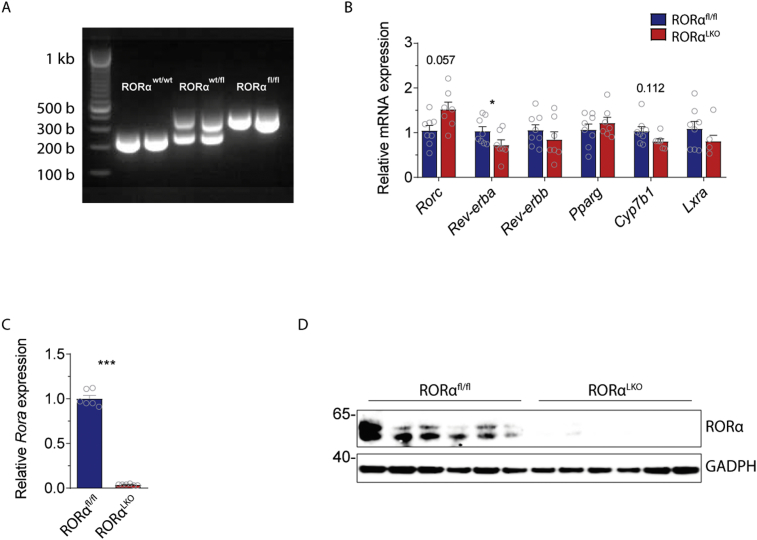
Analyses of Rora expression by qRT-PCR in the liver, ileum, white adipose tissue (WAT), and brain, as well as RORα protein levels by Western blotting, confirmed a selective and specific liver deletion in RORαLKO mice compared to littermate RORαfl/fl mice (Figure 1B–C). Liver RT-qPCR analysis showed that there was a trend (p = 0.057) towards a compensatory increased expression of Rorc by the selective and specific liver RORα deletion (Supplementary Fig. 1B). Rorb was not detectable in the livers (data not shown). Moreover, the expression of Rev-erba, one of the known RORs target genes, was reduced by the selective and specific liver RORα deletion (Supplementary Fig. 1B). The expression of other RORs target genes (i.e. Rev-erbb, Pparg, Cyp7b1, and Lxra) was not affected by the selective and specific liver RORα deletion (Supplementary Fig. 1B).
To further confirm a specific hepatocyte RORα deletion, we isolated primary hepatocytes from RORαLKO mice and littermate RORαfl/fl mice. Analyses of Rora expression by qRT-PCR as well as RORα protein levels by Western blotting, confirmed a specific RORα hepatocyte deletion in RORαLKO mice compared to littermate RORαfl/fl mice (Supplementary Figs. 1C–D). Mice with a specific liver RORα deletion displayed a normal fertility and fecundity generating an equal number of male and female pups during RORαLKO and RORαfl/fl cross-breading.
3.2. RORα hepatic deletion does not affect glucose and lipid metabolism in mice fed chow diet
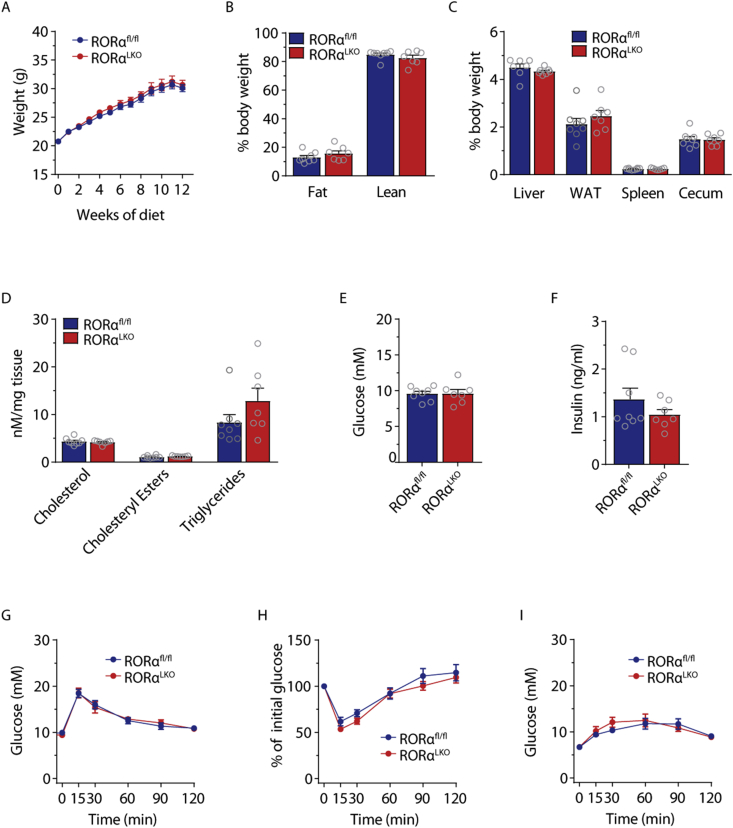
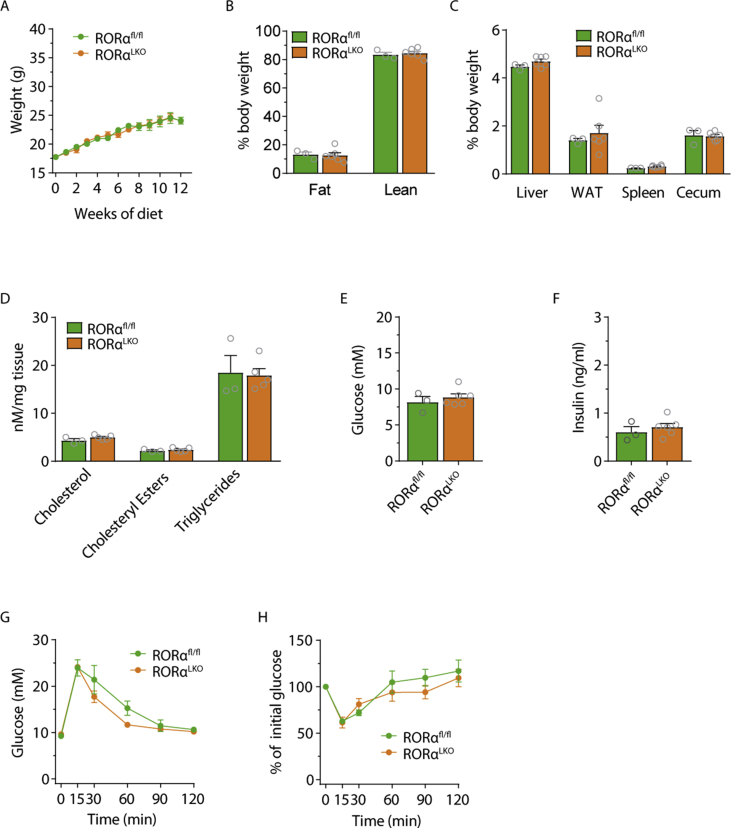
Weight gain was monitored of 8-week-old male RORαLKO and littermate RORαfl/fl mice on chow diet for 12 weeks. We did not observe any significant differences in weight gain (Figure 2A), whole body adiposity determined by MRI (Figure 2B) or liver, WAT, spleen and cecum weights (Figure 2C). Liver RT-qPCR analysis as well as liver lipid measurements did not reveal any differences in lipid metabolism genes nor in cholesterol, cholesteryl ester or triglyceride levels between RORαLKO and RORαfl/fl littermates (Figure 2D and Supplementary Fig. 2).
Figure 2.
RORα hepatic deletion does not affect glucose and lipid metabolism in mice fed chow diet. Body weight (A), body composition (B), relative organ weights (C), liver cholesterol, cholesteryl esters, triglycerides levels (D), fasting glucose (E) and insulin levels (F), intraperitoneal glucose (G), insulin (H), and pyruvate (I) tolerance test in RORαfl/fl and RORαLKO male mice fed 12 weeks chow diet (n = 7–8 mice per group). Data are plotted as mean – SEM.
Fasting glucose and insulin levels as well as glucose levels during glucose-, insulin-, and pyruvate tolerance tests did not demonstrate any differences between RORαLKO and RORαfl/fl littermates (Figure 2E–I). Similar findings where obtained for female mice (see Supplementary Fig. 3).
Taken together, our findings suggest that hepatocyte RORα deletion does not affect glucose and lipid metabolism on chow diet neither in males nor in females.
3.3. RORα hepatic deletion does not affect glucose and lipid metabolism in mice fed Western style diet
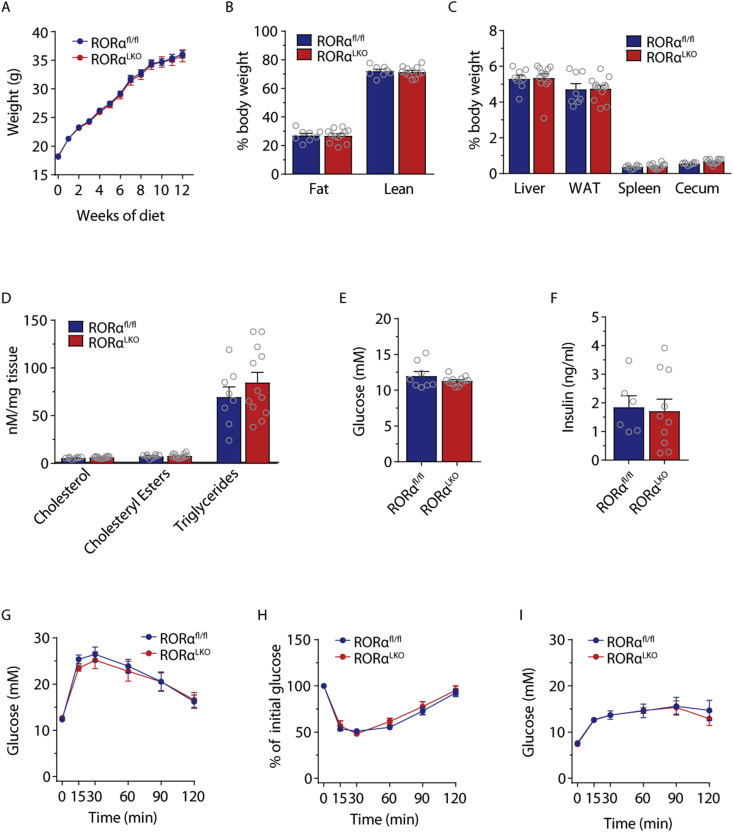
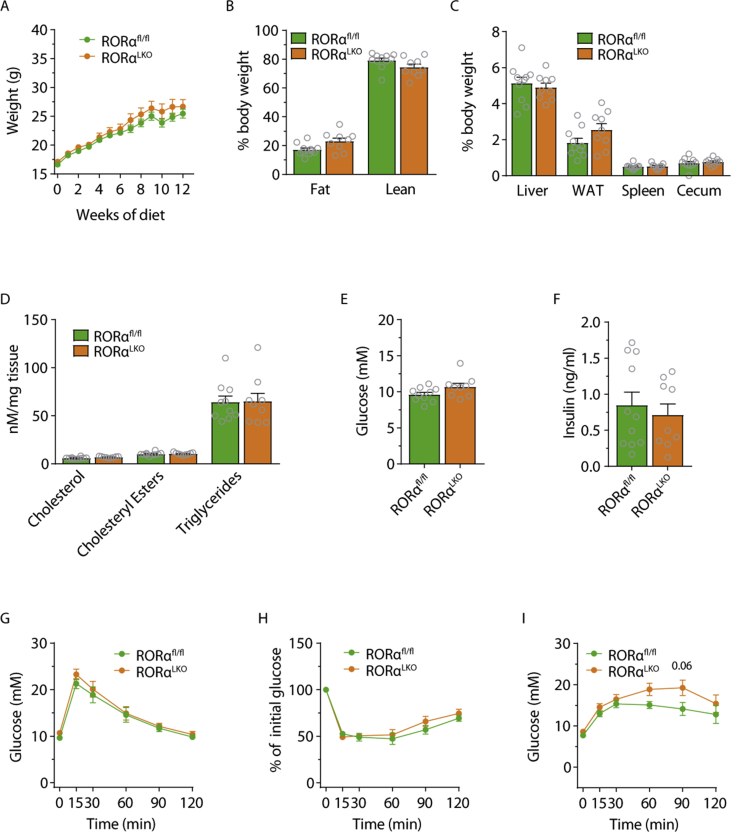
To further study the effect of RORα on metabolic associated disorders, we fed 8-week-old RORαLKO and littermate RORαfl/fl male mice with a WSD for 12 weeks. We did not observe any significant differences in weight gain (Figure 3A), whole body adiposity measured with MRI (Figure 3B) or liver, WAT, spleen or cecum weights (Figure 3C) suggesting that RORα does not affect diet-induced obesity. Liver RT-qPCR analysis as well as liver lipid measurements did not reveal any differences in in lipid metabolism genes nor in cholesterol, cholesteryl esters or triglyceride levels between RORαLKO and RORαfl/fl littermates (Figure 3D and Supplementary Fig. 4).
Figure 3.
RORα hepatic deletion does not affect glucose and lipid metabolism in mice fed western-style diet (WSD). Body weight (A), body composition (B), relative organ weights (C), liver cholesterol, cholesteryl esters, triglycerides levels (D), fasting glucose (E) and insulin levels (F) intraperitoneal glucose (G) insulin (H) and pyruvate (I) tolerance test in RORαfl/fl and RORαLKO male mice fed 12 weeks WSD (n = 8–12 mice per group). Data are plotted as mean – SEM.
Next, we assessed whether RORα contributes to glucose metabolism during WSD by analyzing fasting glucose and insulin levels as well as performing glucose-, insulin- and pyruvate tolerance tests. We did not observe any differences between RORαLKO and RORαfl/fl littermates (Figure 2E–I). Similar findings were obtained when we used female mice (see Supplementary Fig. 5).
Taken together, our findings suggest that a hepatocyte RORα deletion does not affect glucose and lipids metabolism on WSD in both males and females.
4. Discussion
NRs have been shown to play a crucial role in the regulation of glucose and lipid metabolism. Evidences from the RORα whole body knockout (Staggerer) mice have pointed towards RORα as a potential molecular target for the treatment of metabolic diseases [14], [15], [16]. However, the improved metabolic phenotype of Staggerer mice is highly affected by the staggering phenotype induced by the cerebellum RORα deletion [14], [15], thus it is difficult to evaluate the role of RORα in metabolic active tissues, such as the liver. Here, by generating a liver specific RORα knockout mouse model, we did not detect any effects of RORα liver deletion on glucose or lipids metabolism on chow diet or after a WSD exposure.
Our data are in contrast with recent published data that showed that liver-specific ablation of RORα dramatically worsens metabolic profiles in mice eg. increased adiposity, liver fat accumulation, inflammation, and hepatic insulin resistance, when mice are fed a high fat diet [20], [21]. Several factors should be taken into account when comparing the discrepancy between the distinct phenotype reported by other groups and our present study. First, we targeted RORα exon 3 while the Cre-lox system used by others targeted exon 4 and 4–5, leaving the exon 3 that encodes the beginning of the DNA binding domain. Although the stability of potentially produced recombinant proteins, which in our experience usually is low, we cannot exclude that the generation of different truncated proteins containing a portion of the DNA binding domain may explain some of the divergent results between the studies. Importantly, we demonstrated an almost complete elimination of RORα in hepatocytes similar to other groups and also a significant effect on some of the known RORα target genes [20], [21], [27], [28]. Second, we challenged our mice with WSD while other reports used HFD. Importantly, it has been well-documented that, despite containing fewer calories, WSD induces more pronounced steatosis and liver inflammation compared with HFD [29], [30]. Thus, it may be plausible that in our model, the effect of WSD on steatosis and inflammation overwhelms the effect of RORα liver deletion.
In support to our findings Zhang et al. recently reported that the single knockout of RORα or RORγ in hepatocyte has almost no effect on liver gene expression while the double deletion of both RORα and RORγ in hepatocytes has a substantial impact on gene expression profiles, increases lipogenesis and therefore predisposes to HFD-induced steatosis [31]. RORα and RORγ bind to the same response element and are both highly expressed in liver. Thus, deletion of only one of them may not be sufficient to induce a metabolic phenotype due to their functional redundancy. In our experimental model upon deletion of RORα, we observed a trend towards increased Rorc expression, which may be a compensation for the lack of RORα activity. Taken together, it is thus likely that double targeting of RORα and RORγ may be required for achieving maximum effect.
Moreover, it is known that RORα is a clock gene involved in circadian rhythm regulation [31], [32]. In an attempt to reduce experimental variation all metabolic phenotyping was performed at the same time of the day. Thus we cannot exclude that the discrepancy between our data and previous published data may be due to differences in experimental set up such as time of analyses.
5. Conclusions
In conclusion, our data show that the deletion of RORα in the liver does not affect glucose or lipid metabolism during WSD in mice. Further studies testing the role of liver RORα or the double RORα and RORγ deletion in different dietary settings are needed to further elucidate the role of RORα on obesity and metabolic associated disorders.
Author contributions
AM, BS, and FB: conceived the project and designed the experiments. AM, RC, AK, LL, and MS: performed and analyzed experiments. AM and FB wrote the paper. All authors commented and approved the paper.
Acknowledgements
The authors thank Zacharias Gulic for superb technical assistance and prof. Martin Bergö for kind donation of ROSAmT/mG mice. This study was supported by the Novo Nordisk Foundation, the Swedish Research Council, Swedish Diabetes Foundation, Swedish Heart-Lung Foundation, Göran Gustafsson’s Foundation, Inga-Britt och Arne Lundberg’s Foundation, Knut and Alice Wallenberg Foundation, the regional agreement on medical training and clinical research (ALF) between Region Västra Götaland and Sahlgrenska University Hospital. FB is Torsten Söderberg Professor in Medicine and recipient of an ERC Consolidator Grant (European Research Council, Consolidator grant 615362 – METABASE). BS holds an ERC Advanced Grant 694717.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.02.010.
Conflicts of interests
The authors report no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1. Generation of liver-specific RORα knock out mice. PCR analyses (A) of genomic DNA extracted from tails of wild type (RORα−/-), heterozygote (RORα−/fl), homozygote (RORαfl/fl) mice. (B) Relative mRNA expression for Rorc (Retinoic acid-related orphan receptor gamma), Rev-erba (Nuclear Receptor Subfamily 1 Group D Member 1), Rev-erbb (Nuclear Receptor Subfamily 1 Group D Member 2), Pparg (Peroxisome proliferator-activated receptor gamma), Cyp7b1 (25-hydroxycholesterol 7-alpha-hydroxylase), and Lxra (Liver X receptor alpha) in RORαfl/fl and RORαLKO male mice (n = 7–8 mice per group). (C) Relative mRNA expression for Rora and proteins levels for RORα (D) in primary hepatocytes isolated from RORαfl/fl and RORαLKO mice (n = 3 mice per group). Data are plotted as mean – SEM. *p < 0.05, ***p < 0.001.
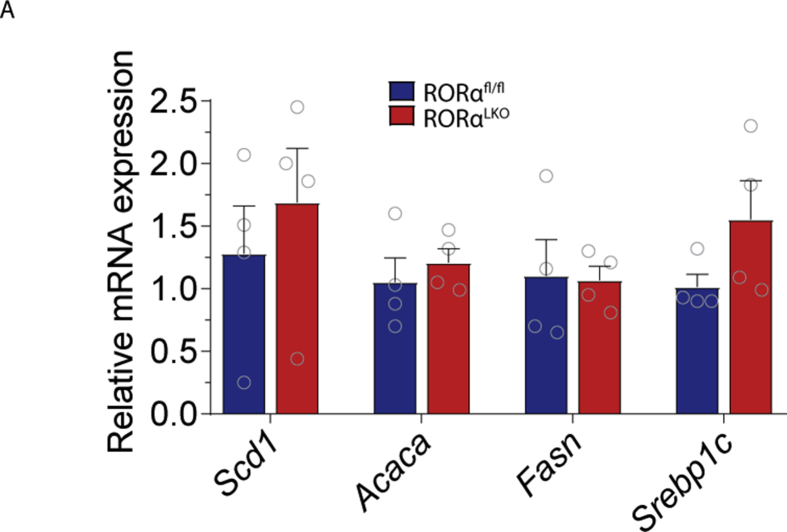
Supplementary figure 2. RORα hepatic deletion does not affect lipogenic gene expression in the liver on chow diet. Relative mRNA expression for Scd1 (Stearoyl-CoA desaturase), Acaca (Acetyl-CoA carboxylase), Fasn (Fatty acid synthase), Srebp1c (sterol regulatory element-binding protein 1c) in RORαfl/fl and RORαLKO male mice on chow diet (n = 4 mice per group). Data are plotted as mean – SEM.
Supplementary figure 3. RORα hepatic deletion does not affect glucose or lipid metabolism in female mice fed chow diet. Body weight (A), body composition (B), relative organ weights (C), liver cholesterol, cholesteryl esters, triglycerides levels (D), fasting glucose (E) and insulin levels (F), intraperitoneal glucose (G), and insulin (H), tolerance test in RORαfl/fl and RORαLKO female mice fed 12 weeks chow diet (n = 3–6 mice per group). Data are plotted as mean – SEM.
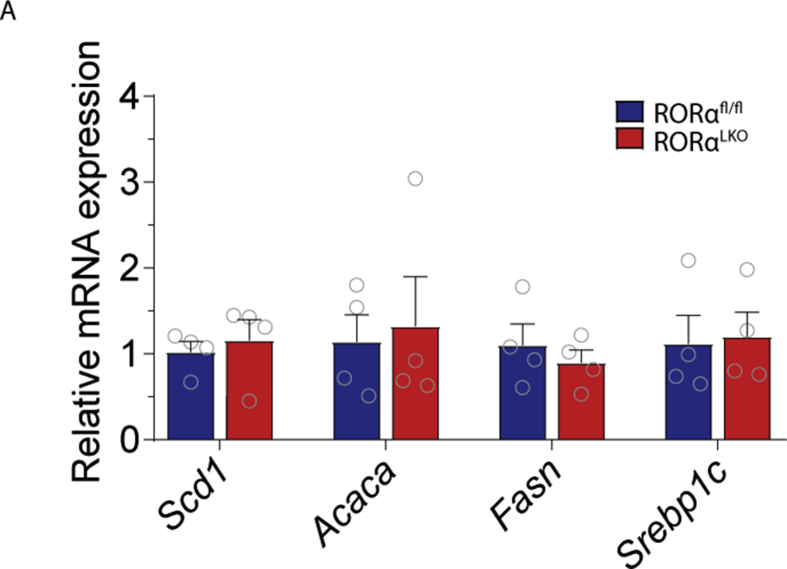
Supplementary figure 4. RORα hepatic deletion does not affect lipogenic gene expression in the liver on western style diet. Relative mRNA expression for Sdc1 (Stearoyl-CoA desaturase), Acaca (Acetyl-CoA carboxylase), Fasn (Fatty acid synthase), Srebp1c (sterol regulatory element-binding protein 1c) in RORαfl/fl and RORαLKO male mice on western style diet (n = 4 mice per group). Data are plotted as mean – SEM.
Supplementary Figure 5. RORα hepatic deletion does not affect glucose and lipid metabolism in female mice fed western-style diet (WSD). Body weight (A), body composition (B), relative organ weights (C), liver cholesterol, cholesteryl esters, triglycerides levels (D), fasting glucose (E) and insulin levels (F) intraperitoneal glucose (G) insulin (H) and pyruvate (I) tolerance test in RORαfl/fl and RORαLKO female mice fed 12 weeks WSD (n = 9–11 mice per group). Data are plotted as mean – SEM.
References
- 1.Afshin A., Forouzanfar M.H., Reitsma M.B., Sur P., Estep K., Lee A. Health effects of overweight and obesity in 195 countries over 25 years. New England Journal of Medicine. 2017;377(1):13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkelstein E.A., Khavjou O.A., Thompson H., Trogdon J.G., Pan L., Sherry B. Obesity and severe obesity forecasts through 2030. American Journal of Preventive Medicine. 2012;42(6):563–570. doi: 10.1016/j.amepre.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Boyle J.P., Honeycutt A.A., Narayan K.M., Hoerger T.J., Geiss L.S., Chen H. Projection of diabetes burden through 2050: impact of changing demography and disease prevalence in the U.S. Diabetes Care. 2001;24(11):1936–1940. doi: 10.2337/diacare.24.11.1936. [DOI] [PubMed] [Google Scholar]
- 4.Sjöström L. Review of the key results from the Swedish Obese Subjects (SOS) trial - a prospective controlled intervention study of bariatric surgery. Journal of Internal Medicine. 2013;273(3):219–234. doi: 10.1111/joim.12012. [DOI] [PubMed] [Google Scholar]
- 5.Pearen M.A., Muscat G.E. Orphan nuclear receptors and the regulation of nutrient metabolism: understanding obesity. Physiology (Bethesda) 2012;27(3):156–166. doi: 10.1152/physiol.00007.2012. [DOI] [PubMed] [Google Scholar]
- 6.Schulman I.G. Nuclear receptors as drug targets for metabolic disease. Advanced Drug Delivery Reviews. 2010;62(13):1307–1315. doi: 10.1016/j.addr.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhiman V.K., Bolt M.J., White K.P. Nuclear receptors in cancer – uncovering new and evolving roles through genomic analysis. Nature Reviews Genetics. 2018;19(3):160–174. doi: 10.1038/nrg.2017.102. [DOI] [PubMed] [Google Scholar]
- 8.Germain P., Staels B., Dacquet C., Spedding M., Laudet V. Overview of nomenclature of nuclear receptors. Pharmacological Reviews. 2006;58(4):685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka N., Aoyama T., Kimura S., Gonzalez F.J. Targeting nuclear receptors for the treatment of fatty liver disease. Pharmacology & Therapeutics. 2017;179:142–157. doi: 10.1016/j.pharmthera.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanson H.I., Wada T., Xie W., Renga B., Zampella A., Distrutti E. Role of nuclear receptors in lipid dysfunction and obesity-related diseases. Drug Metabolism & Disposition. 2013;41(1):1–11. doi: 10.1124/dmd.112.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jetten A.M. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nuclear Receptor Signaling. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzsimmons R.L., Lau P., Muscat G.E. Retinoid-related orphan receptor alpha and the regulation of lipid homeostasis. The Journal of Steroid Biochemistry and Molecular Biology. 2012;130(3–5):159–168. doi: 10.1016/j.jsbmb.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Marciano D.P., Chang M.R., Corzo C.A., Goswami D., Lam V.Q., Pascal B.D. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARγ, RORs, and Rev-erbs. Cell Metabolism. 2014;19(2):193–208. doi: 10.1016/j.cmet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Dussault I., Fawcett D., Matthyssen A., Bader J.A., Giguère V. Orphan nuclear receptor ROR alpha-deficient mice display the cerebellar defects of staggerer. Mechanisms of Development. 1998;70(1–2):147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 15.Lau P., Fitzsimmons R.L., Raichur S., Wang S.C., Lechtken A., Muscat G.E. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. Journal of Biological Chemistry. 2008;283(26):18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 16.Kadiri S., Monnier C., Ganbold M., Ledent T., Capeau J., Antoine B. The nuclear retinoid-related orphan receptor-α regulates adipose tissue glyceroneogenesis in addition to hepatic gluconeogenesis. American Journal of Physiology Endocrinology and Metabolism. 2015;309(2):E105–E114. doi: 10.1152/ajpendo.00518.2014. [DOI] [PubMed] [Google Scholar]
- 17.Han Y.H., Kim H.J., Na H., Nam M.W., Kim J.Y., Kim J.S. RORα induces KLF4-mediated M2 polarization in the liver macrophages that protect against nonalcoholic steatohepatitis. Cell Reports. 2017;20(1):124–135. doi: 10.1016/j.celrep.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Kim E.J., Yoon Y.S., Hong S., Son H.Y., Na T.Y., Lee M.H. Retinoic acid receptor-related orphan receptor α-induced activation of adenosine monophosphate-activated protein kinase results in attenuation of hepatic steatosis. Hepatology. 2012;55(5):1379–1388. doi: 10.1002/hep.25529. [DOI] [PubMed] [Google Scholar]
- 19.Han Y.H., Kim H.J., Kim E.J., Kim K.S., Hong S., Park H.G. RORα decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxidants and Redox Signaling. 2014;21(15):2083–2094. doi: 10.1089/ars.2013.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H.J., Han Y.H., Na H., Kim J.Y., Kim T., Shin C. Liver-specific deletion of RORα aggravates diet-induced nonalcoholic steatohepatitis by inducing mitochondrial dysfunction. Scientific Reports. 2017;7(1):16041. doi: 10.1038/s41598-017-16077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K., Boo K., Yu Y.S., Oh S.K., Kim H., Jeon Y. RORα controls hepatic lipid homeostasis via negative regulation of PPARγ transcriptional network. Nature Communications. 2017;8(1):162. doi: 10.1038/s41467-017-00215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molinaro A., Caesar R., Holm L.M., Tremaroli V., Cani P.D., Bäckhed F. Host-microbiota interaction induces bi-phasic inflammation and glucose intolerance in mice. Molecular Metabolism. 2017;6(11):1371–1380. doi: 10.1016/j.molmet.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homan R., Anderson M.K. Rapid separation and quantitation of combined neutral and polar lipid classes by high-performance liquid chromatography and evaporative light-scattering mass detection. Journal of Chromatography B Biomedical Sciences and Applications. 1998;708(1–2):21–26. doi: 10.1016/s0378-4347(97)00651-8. [DOI] [PubMed] [Google Scholar]
- 24.Löfgren L., Forsberg G.B., Ståhlman M. The BUME method: a new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Scientific Reports. 2016;6:27688. doi: 10.1038/srep27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W., Sargis R.M., Volden P.A., Carmean C.M., Sun X.J., Brady M.J. PCB 126 and other dioxin-like PCBs specifically suppress hepatic PEPCK expression via the aryl hydrocarbon receptor. PLoS One. 2012;7(5):e37103. doi: 10.1371/journal.pone.0037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh A., Lee M.N., Yang Y.R., Jeong H., Ghim J., Noh J. C1-Ten is a protein tyrosine phosphatase of insulin receptor substrate 1 (IRS-1), regulating IRS-1 stability and muscle atrophy. Molecular and Cellular Biology. 2013;33(8):1608–1620. doi: 10.1128/MCB.01447-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wada T., Kang H.S., Angers M., Gong H., Bhatia S., Khadem S. Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor alpha (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3) Molecular Pharmacology. 2008;73(3):891–899. doi: 10.1124/mol.107.040741. [DOI] [PubMed] [Google Scholar]
- 28.Delerive P., Chin W.W., Suen C.S. Identification of Reverb(alpha) as a novel ROR(alpha) target gene. Journal of Biological Chemistry. 2002;277(38):35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 29.Bortolin R.C., Vargas A.R., Gasparotto J., Chaves P.R., Schnorr C.E., Martinello K.B. A new animal diet based on human Western diet is a robust diet-induced obesity model: comparison to high-fat and cafeteria diets in term of metabolic and gut microbiota disruption. International Journal of Obesity (Lond) 2017;42(3):525–534. doi: 10.1038/ijo.2017.225. [DOI] [PubMed] [Google Scholar]
- 30.Fleissner C.K., Huebel N., Abd El-Bary M.M., Loh G., Klaus S., Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. British Journal of Nutrition. 2010;104(6):919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Papazyan R., Damle M., Fang B., Jager J., Feng D. The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORα/γ. Genes & Development. 2017;31(12):1202–1211. doi: 10.1101/gad.302323.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X., Cho H., Yu R.T., Atkins A.R., Downes M., Evans R.M. Nuclear receptors rock around the clock. EMBO Reports. 2014;15(5):518–528. doi: 10.1002/embr.201338271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.