Abstract
Background
Cancer cells possess a common metabolic phenotype, rewiring their metabolic pathways from mitochondrial oxidative phosphorylation to aerobic glycolysis and anabolic circuits, to support the energetic and biosynthetic requirements of continuous proliferation and migration. While, over the past decade, molecular and cellular studies have clearly highlighted the association of oncogenes and tumor suppressors with cancer-associated glycolysis, more recent attention has focused on the role of microRNAs (miRNAs) in mediating this metabolic shift. Accumulating studies have connected aberrant expression of miRNAs with direct and indirect regulation of aerobic glycolysis and associated pathways.
Scope of review
This review discusses the underlying mechanisms of metabolic reprogramming in cancer cells and provides arguments that the earlier paradigm of cancer glycolysis needs to be updated to a broader concept, which involves interconnecting biological pathways that include miRNA-mediated regulation of metabolism. For these reasons and in light of recent knowledge, we illustrate the relationships between metabolic pathways in cancer cells. We further summarize our current understanding of the interplay between miRNAs and these metabolic pathways. This review aims to highlight important metabolism-associated molecular components in the hunt for selective preventive and therapeutic treatments.
Major conclusions
Metabolism in cancer cells is influenced by driver mutations but is also regulated by posttranscriptional gene silencing. Understanding the nuanced regulation of gene expression in these cells and distinguishing rapid cellular responses from chronic adaptive mechanisms provides a basis for rational drug design and novel therapeutic strategies.
Keywords: Metabolism, Warburg effect, microRNA, Aerobic glycolysis, Cancer
Highlights
-
•
Various oncogenes and tumor suppressors are associated with cancer cell metabolism.
-
•
Metabolic reprogramming involves pathways that rely upon microRNA regulation.
-
•
MicroRNAs ensure post-transcriptional buffering of adaptive metabolic responses.
-
•
Dysregulated microRNAs can impact energy flux in cancer cells.
-
•
Cancer metabolism may be disrupted by novel, multi-target, therapeutic interventions.
1. Introduction
In the 1920s, Otto Warburg reported for the first time that while cells under normal conditions utilize glucose to derive 70% of required ATP through mitochondrial oxidative phosphorylation (OXPHOS), cancer cells metabolize glucose by glycolysis even in the presence of adequate oxygen supply [1], [2]. Since then, aerobic glycolysis has been regarded as a hallmark of cancer that provides bioenergetic, biosynthetic and redox balance advantages for cancer cells [3].
Although Warburg's seminal studies resulted in a misinterpretation that irreversible inactivation of mitochondrial respiration is the primary and sole cause of aerobic glycolysis in cancer cells, later it was reported that impaired respiration is inadequate to explain the metabolic shift [4]. The study of cancer cell glycolysis continues to surprise, revealing further associations between a metabolic switch in cancer cells, mutations in mitochondrial metabolic enzymes and altered mitochondrial function [5], [6]. In addition, discoveries that associate oncogene and tumor suppressor gene dysfunction with metabolic reprogramming suggest that both environmental and genetic factors underlie the metabolic heterogeneity of tumors [7], [8]. Moreover, in light of numerous microRNA-related studies, it is now important to consider the roles of these small non-coding RNAs in fine-tuning gene expression at different stages of tumourigenesis. Accumulating evidence supports the involvement of miRNAs in modulating cancer cell metabolism by directly and indirectly regulating genes associated with aerobic glycolysis [9].
microRNAs (miRNAs) are small non-coding RNAs that canonically play a major role in post-transcriptional gene repression. Themselves the products of RNA polymerase II or III dependent transcription, primary (pri)-miRNA transcripts are 5′-7-methylguanosine capped, spliced and 3′-polyadenylated and may give rise to one or more mature miRNAs. Some miRNAs may also derive from processed intronic sequences [10]. In the nucleus, pri-miRNAs are subjected to cleavage by Drosha releasing precursor (pre)-miRNA hairpin structures. Pre-miRNAs are then transported to the cytoplasm where cleavage by Dicer results in a 19–24 nucleotide double-stranded miRNA of which one strand, the mature miRNA, is transferred to the Argonaute (AGO) component of the RNA-induced silencing complex (RISC). AGO acts as a RISC effector protein modulating mRNA stability and translation [11], [12].
This review summarizes recent knowledge of the causes and consequences of the Warburg effect, paying particular attention to the contribution of miRNAs. It also aims to further discuss complex interactions between metabolic pathways and mitochondrial function, as well as oncogenic and tumor suppressor mutations. Finally, in view of recent findings, future approaches that can be exploited for therapeutic benefit are discussed.
2. Metabolic reprogramming
Proliferating cells and, indeed, cancer cells require constant cell division. In order to maintain this, there is an urgent need to provide a consistent energy source, macromolecular biosynthesis, and controlled redox status. Therefore, to optimize proliferation, growth, and survival, cancer cells redirect their metabolic pathways and alter the production and consumption of numerous metabolites [13], [14].
To support cancer cell proliferation, glycolysis provides the precursors for major macromolecules including the carbohydrates, proteins, lipids, and nucleic acids needed to produce a new cell. Therefore, aerobic glycolysis imbues cancer cells with ribose, amino acids and fatty acids [15], [16]. The upregulation of glycolysis is mostly due to the increased expression of enzymes and transporters involved in glucose uptake, lactate production, and lactate secretion. These proteins include glucose transporters (GLUT1-4), hexokinase 2 (HK2), glyceraldehyde-3- phosphate dehydrogenase (GAPDH), 6-phosphofructo-1-kinase (PFK1), aldolase (ALDO), triose-phosphate isomerase (TPI), phosphoglycerate kinase 1 (PGK1), phosphoglycerate mutase (PGM), enolase 1 (ENO1), pyruvate kinase (PKM2), lactate dehydrogenase (LDHA) and monocarboxylate transporters (MCTs).
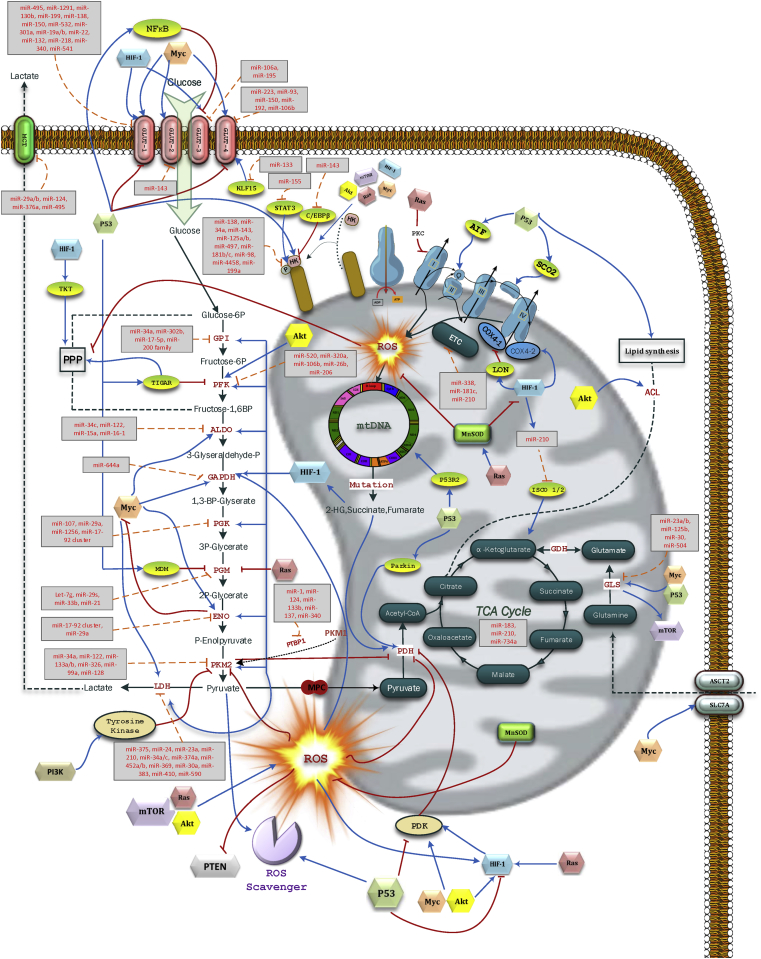
There is substantial evidence regarding the importance of aberrant expression of oncomiRs and tumor suppressor miRNAs targeting key players in aerobic glycolysis to give proliferation, growth, and invasion advantages to cancer cells (Figure 1). Such changes in miRNA activity reflect a mechanism by which cancer cells bypass checkpoints that determine thresholds of biosynthetic enzyme activities.
Figure 1.
miRNAs targeting glycolytic and mitochondrial enzymes.
In addition to miRNAs directly targeting genes involved in cancer cell glycolysis, summarized in Table 1, several indirect mechanisms have been reported for miRNA-mediated regulation of glycolytic genes. Horie et al. [17] showed that forced expression of miR-133 decreases GLUT4 expression by directly targeting Kruppel-like factor 15 (KLF15) in cardiomyocytes. KLF15 is a transcription factor required for GLUT4 transcription. Also, miR-155 was reported to upregulate HK2 through signal transducer and activator of transcription 3 (STAT3) activation, as well as through miR-143 repression by targeting CCAAT-enhancer-binding protein β (C/EBPβ). Moreover, miR-143 was found to target HK2 directly, linking inflammatory miR-155-related signaling with cancer-associated changes in metabolism [18], [19]. PKM is one of the rate limiting enzymes in glycolysis. While PKM1 expression was shown to be active in normal cells, cancer cells switch PKM1 to the tumor-associated PKM2. Also, some miRNAs were reported to regulate polypyrimidine tract-binding protein 1 (PTB-1), which processes PKM transcripts and is involved in PKM1 to PKM2 conversion in tumor cells. These miRNAs, including miR-1, miR-124, miR-133b, miR-137 and miR-340 were shown to directly inhibit cancer cell proliferation and may also explain the repressed PTB-1 expression associated with tumor progression in vivo [20], [21], [22], [23], [24].
Table 1.
Summary of miRNAs directly targeting glycolytic enzymes and transporters.
| Gene | miRNAs | Diseases | References |
|---|---|---|---|
| GLUT1 | miR-495, miR-1291, miR-130b, miR-199a, miR-138, miR-150, miR-532, miR-301a, miR-19a/b, miR-22, miR-132, miR-218, miR-340, miR-541 | Renal Cell Carcinoma, Glioma, Breast Cancer, Prostate Cancer, Bladder Cancer, Oral Squamous Cell Carcinoma, Glioblastoma Multiforme | [25], [26], [27], [28], [29], [30], [31], [32], [33] |
| GLUT2 | miR-143 | – | [34] |
| GLUT3 | miR-195, miR-106a | Bladder Cancer, Glioblastoma | [35], [36] |
| GLUT4 | miR-223, miR-93, miR-150, miR-192, miR-106b | Cardiomyocytes, Polycystic Ovary Syndrome, Diabetes Mellitus | [37], [38], [39], [40] |
| HK1 | miR-138 | Head and Neck Squamous Cell Carcinoma | [41] |
| HK2 | miR-34a, miR-143, miR-125a/b, miR-497, miR-181b/c, miR-98, miR-4458, miR-199a | Colorectal Cancer, Head and Neck Squamous Cell Carcinoma, Breast Cancer, Lung Cancer, Glioblastoma, Hepatocellular Carcinoma, Chronic Lymphocytic Leukemia, Primary keratinocytes, Osteocarcinoma, Prostate Cancer, Gastric Cancer | [18], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53] |
| GPI | miR-34a, miR-302b, miR-17, miR-200 family | Colorectal Cancer, Primordial Germ Cells, Breast Cancer | [42], [54], [55] |
| PFK | miR-520, miR-320a, miR-106b, miR-26b, miR-206 | Hepatocellular Carcinoma, Lung Adenocarcinoma, Renal Cell Carcinoma, Osteosarcoma, Breast Cancer | [56], [57], [58], [59], [60], [61] |
| ALDOA | miR-34c, miR-122, miR-15a, miR-16-1 | Emphysematous Lung, Hepatocellular Carcinoma, Leukemia, Lung Cancer | [62], [63], [64], [65] |
| GAPDH | miR-644a | Prostate Cancer | [66] |
| TPI1 | miR-15a, miR-16-1, miR-107, miR-195 | Leukemia, Renal Cell Carcinoma, Lung Cancer, Bladder Cancer | [63], [65], [67], [68] |
| PGK1 | miR-107, miR-29a, miR-1256, miR-17-92 cluster | Renal Cell Carcinoma, Prostate Cancer, Lung Cancer, Pancreatic Cancer, Squamous Cell Lung Carcinoma | [67], [69], [70], [71] |
| PGM | Let-7g, miR-29a, miR-33b, miR-21 | Primary Human Hepatocytes, Lung Cancer, Renal Cell Carcinoma | [60], [70], [72], [73] |
| ENO1 | miR-17-92 cluster, miR-29a | Lung Cancer | [70], [71] |
| PKM2 | miR-34a, miR-122, miR-133a/b, miR-326, miR-99a, miR-128 | Colorectal Cancer, Hepatocellular Carcinoma, Squamous Cell Carcinoma of Tongue, Glioblastoma, Type 2 Diabetes, Prostate Cancer | [42], [74], [75], [76], [77] |
| LDHA | miR-375, miR-24, miR-23a, miR-210, miR-30a, miR-34a/c, miR-374a, miR-383, miR-4524a/b, miR-369, miR-410, miR-590 | Maxillary Sinus Squamous Cell Carcinoma, Acute Myocardial Ischemia, Breast Cancer, Colorectal Cancer, Hypoxia-Induced Cardiomyocytes Dysfunction, Ovarian Cancer, Cervical Cancer, Gestational Diabetes Mellitus, Type2 Diabetes | [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88] |
| MCTs | miR-29a/b, miR-124, miR-376a-5p, miR-495 | Pancreatic β Cells, Medulloblastomas, Type2 Diabetes | [89], [90], [91] |
Although several decades have passed since the first report on cancer metabolism, with many studies since, a definitive mechanism underpinning the Warburg metabolic shift has remained obscure. Moreover, how individually disrupted metabolic pathways converge to coordinate a global metabolic shift and facilitate the tumor phenotype remains to be fully elucidated.
3. Inter-connection between aerobic glycolysis and mitochondria
Whilst glycolysis accounts for the generation of almost two thirds of the ATP required for tumor cells, in most cancer cells mitochondria are still functional and generate the remaining energy requirements [92]. Mitochondria also contribute to pivotal roles in controlling anaplerotic and cataplerotic pathways within cancer cells. Indeed, several roles for mitochondria in carcinogenesis, other than ATP production for cellular demands, have been established [93]. As a result, functions including hypoxia resistance, apoptosis escape, reactive oxygen species (ROS) control, and bio-synthetic contributions are attributed to mitochondria. Mutations in mitochondrial TCA cycle genes, encoded by nuclear DNA, were found in various types of cancers. Mutational inactivation of these enzymes contributed to a metabolic shift through direct adaptation to decreased OXPHOS or, alternatively, by epigenetic modification caused by cytosolic and mitochondrial accumulation of oncometabolites such as 2-hydroxyglutarate (2HG) [94], [95], [96], [97], [98].
Studies of miRNA localization from nucleus to mitochondria have led to the discovery of mitochondria-related miRNAs (mitomiRs). A considerable body of literature demonstrated the miRNA contributions to every aspect of mitochondrial metabolism, respiration, and dynamics [99]. Additionally, ROS generated within mitochondria were found to be strictly regulated by several miRNAs (reviewed in [100]). miRNAs that regulate tricarboxylic acid (TCA) cycle transcripts include miR-183, miR-210 and miR-734a, which target isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH), and malate dehydrogenase (MDH), respectively [101], [102], [103] (Figure 1). Moreover, several electron transport chain components are reportedly regulated by miRNAs. For instance, miR-338 and miR-181c downregulate cytochrome c oxidase complex COX4 and COX1, respectively. Hypoxically regulated miR-210 represses iron-sulfur cluster scaffold (ISCU) and COX10 translation [104], [105], [106]. Glutaminase (GLS) is a rate-limiting enzyme in glutamine metabolism which converts glutamine to glutamate. An increasing number of reports revealed cooperation of c-Myc and p53 with several miRNAs such as miR-23a/b, miR-125b, miR-30 and miR-504 in modulating GLS activity [107]. Based on these reports, it is clear that miRNAs target both nuclear mRNAs and mitochondrial mRNAs. Moreover, the Crabtree effect, originally identified in fermenting yeast, enables some cancer cells to switch between glycolysis and OXPHOS in spite of functional mitochondria and also challenges the “purely glycolytic cancer cell” paradigm. The Crabtree effect is considered to be a short-term and reversible mechanism and an adaptive response of mitochondria to the heterogeneous microenvironment of cancer cells [108]. Hence, there is still a need to fully determine whether changes in mitochondrial functionality, mediated by several miRNAs, contribute to cellular transformation. Otherwise it may be considered a secondary phenomenon, which arises from changes in cell glycolysis and/or other signaling pathways also regulated by miRNAs.
4. Hypoxia and glycolysis
Hypoxia is a common feature in proliferating solid tumors. In normal cells, hypoxia leads to cellular adaptation, or p53-dependent apoptosis and cell death. However, cancer cells acquire mutations in p53 and other genes, along with changes in their metabolic pathways in order to survive and even proliferate under hypoxic stress. A key mediator of responses to hypoxia is hypoxia-inducible factor-1 (HIF-1), a transcription factor that plays a pivotal role in responding to decreased oxygen levels, initiating hypoxia-related processes such as OXPHOS repression and induced glycolysis [109].
Although prolyl-4-hydroxylase (PHD) and factor inhibiting HIF-1 (FIH-1; also known as HIF1AN) dependent regulation of HIF-1 is primarily thought to be the sole mechanism of HIF-1 regulation [110] it is now clear that hypoxia influences miRNA biogenesis and these miRNAs can regulate HIF-1α and HIF-1β expression [111]. HIF-1α is also regulated at the DNA, RNA, protein and DNA binding levels [112]. Translational regulation of HIF-1α could also be a consequence of activating the mechanistic target of rapamycin (mTOR) signaling pathway in cancer cells. Many miRNAs, such as miR-99a, were shown to repress HIF-1α expression by targeting mTOR [76]. The abnormal activation of HIF-1 under normoxia could alternatively be a result of changes in cancer-associated genes. Such tumourigenic mutations include loss of function in tumor suppressors such as P53, phosphatase and tensin homolog (PTEN) [113], Von Hippel-Lindau (VHL) [114], LKB1 [115], promyelocytic leukemia protein (PML) [116], and tuberous sclerosis proteins (TSC1/TSC2) [117] along with mutational activation of oncogenes such as Ras [118], V-Src [119], phosphoinositide 3-kinase (PI3K) [120], and human epidermal growth factor receptor 2 (Her2/Neu) [121]. PKM2 was also reported to enhance HIF-1 transcription, through binding to its promoter, and promote HIF-1 stabilization by inhibiting PHD interactions [122]. Mitochondria also act as both targets and effectors of HIF-1 activation [100]. To adapt to a hypoxic microenvironment and acquire lethal cancer characteristics, HIF-1 activation leads to a range of physiological responses [123]. At the transcriptional level, HIF-1α activates a variety of genes following translocation into the nucleus, dimerization with HIF-1β and binding to hypoxia response elements (HREs) upstream of target genes. Besides HRE-dependent responses, HIF-1α interacts with other signal transduction pathways including Notch [124], Wnt [125] and c-Myc [126].
Activated HIF-1 is directly and indirectly associated with increased expression of virtually all glycolytic transporters and enzymes [123]. Moreover, HIF-1 affects mitochondria through various mechanisms and stimulates glycolysis indirectly by supressing mitochondrial oxidative metabolism, which enables HIF-1 to function as a switch between glycolysis and OXPHOS [127]. HIF-1 represses mitochondrial pyruvate dehydrogenase (PDH) activity [109], which is a gate-keeping enzyme feeding the TCA cycle by converting pyruvate to acetyl-CoA. HIF-1 suppresses PDH expression by actively upregulating pyruvate dehydrogenase kinase (PDK1), a PDH suppressor [128]. By such regulation, pyruvate is converted to lactate, cytosolic NADH is re-oxidized and glycolysis is continued. As a consequence, PDH suppression by activated HIF-1 protects cells from increased ROS generated within mitochondria [129]. In addition, HIF-1 regulates mitochondrial function in response to oxygen by mediating a subunit switch in COX4. HIF-1 induces COX4I2 subunit expression under hypoxic conditions, while the normoxic COX4I1 subunit is downregulated through HIF-1-mediated activation of LON, a mitochondrial protease. This subunit switch optimizes the efficiency of respiration in response to hypoxia by influencing H2O2 levels in an oxygen-dependant manner [127]. Zhao et al. [130] showed that HIF-1α upregulates TKT and TKTL2, two transketolase enzymes of the pentose phosphate pathway, to elevate the ribose production required for nucleic acid anabolic pathways.
Thus far, no mutations within the HIF-1 genes have been associated with its activation or related regulation of glucose metabolism. However, aberrant HIF1 activity has proved to be important in the initiation and maintenance of some tumors [112].
Hypoxia is a significant mediator of miRNA biosynthesis, at both the transcriptional and post-transcriptional levels [131]. A recently identified subset of miRNAs are known as “hypoxia regulated miRNAs” (also termed hypoxiamiRs or HRMs). Hypoxia regulates hypoxiamiRs in either a HIF-1 dependent or independent manner [132]. First reported by Kulshreshtha et al. [111], hypoxia is capable of upregulating miRNA expression (Table 2 and Figure 2). Among these hypoxia-inducible miRNAs, miR-210 and miR-26 were found to have dynamic recruitment of HIF-1 to their promoters. Upon activation, HIF-1α translocates to the nucleus and targets HREs of downstream genes, including miRNA encoding genes. Interestingly, hypoxia is also associated with miRNA downregulation. In that regard, the miR-17–92 cluster was downregulated by hypoxia in p53 wild type cells [133]. Similarly, Lei et al. [134] reported miR-20b upregulation in HIF1-knockdown cells. Other hypoxia-suppressed miRNAs are listed in Table 2. Nevertheless, contrasting reports, with miRNAs such as miR-26 and miR-19, demonstrate that hypoxia-dependent regulation of miRNAs is cell type and microenvironment dependent [111], [135]. Among downregulated hypoxiamiRs, HIF-1 was shown to downregulate miR-17 and miR-199a [136], [137]. HIF-1 also regulates miRNA expression indirectly by mediating the expression of other transcription factors, examples being activation of miR-10b by HIF-1-dependent TWIST1 expression and regulation of miR-20a/b through vascular endothelial growth factor A (VEGFA) targeting by HIF-1 [138], [139]. Beside miRNAs directly regulated by hypoxia, it is evident that hypoxia is post-transcriptionally involved in the regulation of hypoxiamiR biogenesis, processing and function in both a HIF-dependent and independent manner. It was shown that hypoxia accelerates Ago2 assembly to RISC and its translocation to stress granules by upregulating Ago2 prolyl-hydroxylation and increasing its endonuclease activity [140]. Moreover, HIF-1 regulates expression of the prolyl 4-hydroxylase, alpha polypeptide I (P4HA1) by regulating miR-124 expression [141]. In fact, stress granule formation increased in a hypoxia-dependent manner. Nonetheless, ADP-ribosylaton of Ago2 in response to oxidative stress is another mechanism that eventually leads to relief of miRNA-mediated repression. Interestingly, it was reported that some miRNA maturation that is not dependent on Dicer activity [142], might be processed by the endonuclease activity of Ago2, the levels of which are induced by hypoxia. Accordingly, Dicer was found to be downregulated by hypoxia, while miR-451 was upregulated [143], [144].
Table 2.
Summary of associations between miRNAs and hypoxia.
| miRNA | Disease/cell line | Regulation of HIF/mechanism | Regulation of miRNA by Hypoxia | References |
|---|---|---|---|---|
| miR-17-92 cluster | Lung Cancer, Cervical Adenocarcinoma, Inflammation, Colon Cancer, Brest Cancer, Hepatocarcinoma | Downregulation/targeting HIF-1α and HIF-2 | Downregulation | [71], [134], [136], [159], [160], [163], [166] |
| miR-15b | Hemophilia, Nasopharyngeal Carcinoma | Downregulation/targeting HIF-2 | Downregulation | [166], [167] |
| miR-16 | Nasopharyngeal Carcinoma | NA | Downregulation | [166] |
| miR-19a | Oral Squamous Cell Carcinoma, Human Atherosclerotic Lesions | NA | Downregulation/Upregulation | [135], [168] |
| miR-20a/b | Nasopharyngeal Carcinoma, Lung Cancer | Downregulation/targeting HIF-1α and HIF-2α | Downregulation | [71], [166] |
| miR-21 | Breast Cancer, Prostate Cancer | Upregulation | Upregulation | [111], [150], [169] |
| miR-22 | Clear Cell Renal Cell Carcinoma, Colorectal Cancer, Heart muscle, Oral Squamous Cell Carcinoma | Downregulation/targeting HIF-1α | Upregulated/Downregulated | [132], [135], [170], [171] |
| miR-23a/b | Colorectal Cancer, Breast Cancer | NA | Upregulated | [111], [172] |
| miR-24 | Colorectal Cancer, Breast Cancer | NA | Upregulated | [172] |
| miR-26a/b | Nasopharyngeal Carcinoma, Colorectal Cancer, Breast Cancer | NA | Upregulated/Downregulated | [166], [172] |
| miR-27a/b | Heart Muscle, Colorectal Cancer, Breast Cancer | NA | Upregulated | [132], [172] |
| miR-30 b/d/e | Oral Squamous Cell Carcinoma, Colorectal Cancer, Breast Cancer, Nasopharyngeal Carcinoma | NA | Downregulation/Upregulated | [135], [166] |
| miR-29b | Oral Squamous Cell Carcinoma | NA | Downregulated | [135], [172] |
| miR-31 | Colorectal Cancer, Human Corneal Epithelial Keratinocytes, Oral Squamous Cell Carcinoma | Upregulation/targeting FIH1 | Upregulated | [135], [145], [147], [173], [174] |
| miR-33a | Melanoma | Downregulation/targeting HIF-1α | NA | [175] |
| miR-93 | Colorectal Cancer, Breast Cancer | NA | Upregulation | [111] |
| miR-99a | Type 2 Diabetes | Downregulation/targeting HIF-1α | NA | [76] |
| miR-101 | Oral Squamous Cell Carcinoma | NA | Downregulation | [135] |
| miR-103 | Colorectal Cancer, Breast Cancer | NA | Upregulated | [111] |
| miR-106b | Colorectal Cancer, Breast Cancer | NA | Upregulated | [111] |
| miR-107 | Ischemic Heart Disease, Colorectal Cancer, Colorectal Cancer, Breast Cancer | Downregulation/targeting HIF-1β | Upregulation | [111], [151], [152] |
| miR-122a | Oral Squamous Cell Carcinoma | NA | Downregulation | [135] |
| miR-125b | Colorectal Cancer, Breast Cancer | NA | Upregulated | [111] |
| miR-128 | Prostate Cancer | Downregulation/targeting HIF-1α | NA | [77] |
| miR-135b | Prostate Cancer, Breast Cancer | Upregulation/targeting FIH-1 | NA | [142], [176] |
| miR-138 | Clear Cell Renal Cell Carcinoma, Ovarian Cancer | Downregulation/targeting HIF-1α | NA | [177], [178] |
| miR-141 | Oral Squamous Cell Carcinoma | NA | Downregulation | [111] |
| miR-155 | Cervical Adenocarcinoma, Nasopharyngeal Carcinoma | Downregulation/targeting HIF-1α | Upregulation | [159], [166] |
| miR-181a/b/c | Colorectal Cancer, Breast Cancer, Nasopharyngeal Carcinoma, Heart Muscle | NA | Upregulated/Downregulated | [111], [132], [166] |
| miR-184 | Glioma | Upregulation/targeting FIH-1 | NA | [146] |
| miR-186 | Oral Squamous Cell Carcinoma, Gastric Cancer | Downregulation/targeting HIF-1α | Downregulation | [135], [179] |
| miR-192 | Colorectal Cancer, Breast Cancer | NA | Upregulated | [111] |
| miR-195 | Hypoxic Chondrocytes, Colorectal Cancer, Breast Cancer | Downregulation/targeting HIF-1α | Upregulation | [111], [180] |
| miR-197 | Oral Squamous Cell Carcinoma | NA | Downregulation | [135] |
| miR-199a/b | Ovarian Cancer, Sickle Cell Disease, Lung Cancer exposed to arsenic, Heart muscle | Downregulation/targeting HIF-1α and HIF-2α | Downregulation | [132], [137], [181], [182] |
| miR-204 | Pulmonary Arterial Hypertension | Downregulation/targeting HIF-1α | NA | [183] |
| miR-206 | Pulmonary Arterial Hypertension | Downregulation/targeting HIF-1α | NA | [149] |
| miR-210 | Cervical Cancer, Head and Neck Paragangliomas, Hypotriploid Human Kidney Cell Line, Ischemia, Breast Cancer, Nasopharyngeal Carcinoma, Oral Squamous Cell Carcinoma, Heart Muscle, Colorectal Cancer, Breast Cancer | NA | Upregulation | [111], [132], [135], [154], [166], [184], [185], [186] |
| miR-213 | Colorectal Cancer, Breast Cancer | NA | Upregulation | [111] |
| miR-361 | Umbilical Vein Endothelial Cells (HUVEC) | Downregulation/targeting HIF-1α | Downregulation | [187] |
| miR-374 | Oral Squamous Cell Carcinoma, Breast Cancer | Upregulation/targeting TXNIP | Downregulation | [135], [188] |
| miR-422b | Oral Squamous Cell Carcinoma | NA | Downregulation | [135] |
| miR-424 | Ovarian Cancer, Oral Squamous Cell Carcinoma | Upregulation/targeting CUL2 | Downregulation | [134], [135], [156] |
| miR-429 | Human Endothelial Cells | Downregulation/targeting HIF-1α | Upregulation | [165] |
| miR-494 | Lung Cancer | NA | NA | [157] |
| miR-519c | Hepatic Cancer | Downregulation/targeting HIF-1α | NA | [189] |
| miR-565 | Oral Squamous Cell Carcinoma | NA | Downregulation | [135] |
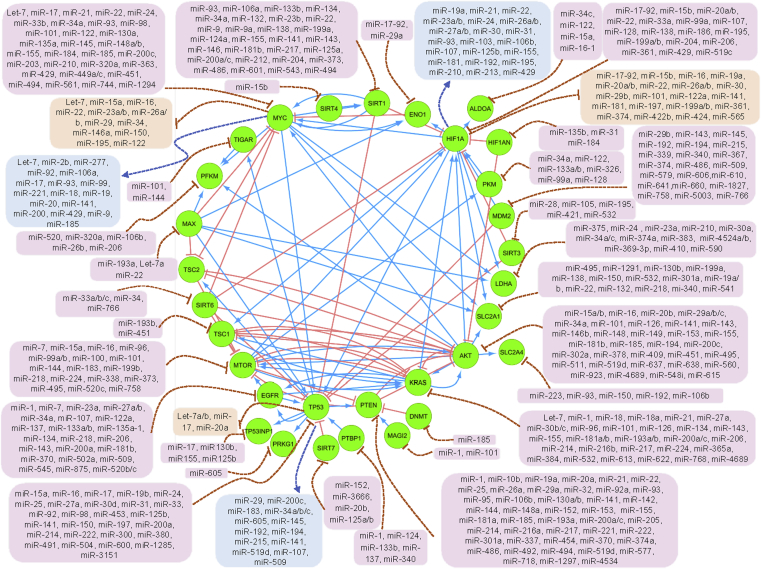
Figure 2.
Interconnections between the drivers and suppressors of glycolysis, and the role of miRNAs in these networks. Protein–protein interactions identified using String V10.0. Solid blue lines indicate protein activation while solid red lines indicate protein inhibition. Dotted blue and red lines represent transcription factor-mediated activation or inhibition of the miRNAs, respectively. miRNAs in pink boxes repress gene expression, while those in orange and blue boxes indicate miRNAs that are inhibited or activated by the transcription factors, respectively. Specific miRNAs present in both the pink boxes and either the orange or blue boxes, may represent feedback loops in particular cellular contexts.
HIF-1α may be directly targeted by miRNAs in various diseases, including cancer (Table 2). Besides direct translational repression, some miRNAs inhibit other factors that modulate HIF-1 expression and stability. As FIH-1 inhibits the transcriptional activation of HIF-1α; miRNAs that suppress FIH-1, such as miR-31, miR-135b, and miR-184, result in HIF-1 activation [142], [145], [146]. FIH-1 was also shown to regulate cell metabolism through reducing glycogen and attenuating AKT signaling [147]. miR-92-1 supresses HIF-1 degradation by targeting pVHL [148]. miR-206 targets the HIF-1/FHL-1 pathway on pulmonary artery smooth muscle cells to promote hypertension [149]. Increased expression of miR-21 was shown to increase HIF-1α and VEGF expression in prostate cancer possibly through a PTEN-dependant pathway [150]. miR-107 downregulates mRNA and protein levels of HIF-1β in endothelial progenitor cells while overexpression of HIF-1β also blocks the effects of miR-107 [151], [152]. miR-185 targets HIF-2a transcripts and, thus, indirectly moderates HIF-1 expression and stability [153].
Feedback loops have been reported in the miRNA regulation of HIF-1. miR-210 forms a positive feedback loop with HIF-1 where hypoxia-induced miR-210 further induces HIF-1α protein stability [154]. Kelly et al. [154] showed that miR-210 targets glycerol-3-phosphate dehydrogenase 1 like (GPD1L), a HIF-1 regulator, and overexpression of miR-210 results in decreased HIF-1 proline hydroxylation and increased accumulation during hypoxia. What's more, HIF-1 directly induces miR-210 expression, which then causes synthesis of cytochrome c oxidase 2/1 (SCO2/1) protein activation and enhanced TCA cycle function [155]. Hypoxia was shown to induce C/EBP levels, which, in turn, increase PU1 activation and binding to the miR-424 promoter to induce its expression. Upregulated miR-424 inhibits cullin 2 (CUL2) and leads to HIF-1 stabilization and nuclear translocation [156]. Overexpression of miR-494 and miR-21 significantly increases Akt phosphorylation and subsequently induces HIF-1 activity [150], [157]. Recent evidence that the activities of non-coding RNAs, including oncogenic miR-21, can be manipulated by small molecules suggests that such processes may be druggable [158].
As a predominant oncomiR, the miR-17–92 cluster has been heavily investigated for its association with hypoxia. Bertozzi et al. [159] showed that miR-17-5p reduced HIF-1α at low camptothecin exposure. miR-17 and miR-20a also target the 3′UTR of HIF-1 and HIF-2 in primary human macrophages [160]. All members of this cluster were shown to directly target HIF-1 in lung cancer [71]. miR-17 and miR-20a were downregulated by HIF-1 through a transcriptional and HIF-1β-independent manner and by downregulating c-Myc expression [136]. miR-20a is a hypoxia-responsive miRNA that targets HIF-1 in breast cancer, lung adenocarcinoma, colorectal cancer, and endometriotic stromal cells [71], [138], [160], [161], [162]. In the paralogous miR-106a∼363 cluster, miR-20b is known to target HIF-1 in hepatocellular carcinoma (HCC) and breast cancer cells [163], [164]. Also, chromatin immunoprecipitation analyses revealed that miR-20b prevents HIF-1 binding to the VEGF promoter and, thus, modulates VEGFA expression [163].
Aberrant expression of miRNAs, which can result from hypoxia encountered during tumor progression, may play a critical role in HIF-1 regulation and altered downstream effects (Figure 2). Interestingly, some miRNAs that target HIF-1 were also reported to be modulated by hypoxia in both a HIF-dependent and independent manner. However, some anomalies regarding hypoxiamiRs and miRNAs that regulate HIF-1 still exist. For instance, Bartoszewska et al. [165] showed that HIF-1 is a direct target of miR-429 in HUVEC cells and is induced during hypoxia. However, Sun et al. [157] showed that overexpression of miR-429 increases HIF-1α expression, under both hypoxia and normoxia, and couldn't find a miR-429 target sequence in the 3′UTR of HIF-1α in liver cells. These inconsistencies likely depend on cellular context and experimental conditions. Moreover, as HIF is mainly post-translationally regulated, miRNA activity may be largely redundant in some systems. Table 2 summarizes the associations between hypoxia and miRNAs in different cancers. It has been proposed [92] that the observed Warburg effect is entirely attributable to the in vivo tumor hypoxia and is, in fact, a manifestation of the Pasteur Effect.
5. Metabolic consequences of miRNA associations with driver mutations and transformation
While oncoproteins and tumor suppressor proteins are well-known for their roles in regulating cellular processes such as cell proliferation, they are also capable of affecting cancer cell metabolism. Activation of certain oncogenic signals is important for stimulating glycolysis. Various mutations in different oncogenes and tumor suppressors show that cancer cells alter metabolism to adapt to their microenvironment [190]. These fundamental genes include oncogenes such as KRAS, MYC, AKT, and MTOR, along with their inhibitors (PTEN and TSC1/2) and activator (EGFR). They also include tumor suppressor genes such as TP53, along with its negative regulator murine double mutant 2 (MDM2) and metabolic effector TP53-induced glycolysis and apoptosis regulator (TIGAR). Sirtuins are further regulatory molecules that can act both as oncogenes and tumor suppressors and will be discussed later. Accumulating evidence highlights the association of miRNAs with oncogenes and tumor suppressors. Some cancer associated genes, such as HIF1, MYC and TP53, regulate both the expression and functions of some miRNAs and are regulated by miRNAs. Table 5 and Figure 2 summarize recent findings on miRNA-mediated regulation of oncogenes and tumor suppressors.
Table 5.
Summary of miRNAs targeting metabolism-related oncogenes and tumor suppressors S indicates references in Supplementary file.
| miRNAs | Gene | Disease | References |
|---|---|---|---|
| Let-7 | KRAS, MYC | Breast Cancer, Colorectal Cancer, Lung Cancer, Glioma, Malignant Mesothelioma, Oropharyngeal Squamous Cell Carcinoma, Pancreatic Ductal Adenocarcinoma, Gastric Cancer, Prostate Cancer, Burkitt Lymphoma, Malignant Bronchial Epithelial Cell, Pulmonary Hypertension | S [1–8], [199], [205], [224], [234], [236] |
| miR-1 | KRAS, EGFR, PTEN | Nasopharyngeal Carcinoma, Head and Neck Squamous Cell Carcinoma, Cardiovascular Disease | S [9–11], [298] |
| miR-100 | mTOR | Esophageal Squamous Cell Carcinoma, Bladder Cancer, Endometrioid Endometrial Carcinoma, Breast Cancer | S [12], [325], [326], [327] |
| miR-101 | KRAS, MYC, AKT, mTOR, TIGAR | Hepatocellular Carcinoma, Osteosarcoma, Clear Cell Renal Cell Carcinoma, Prostate Cancer | S [13–15], [331], [332] |
| miR-105 | SIRT3 | Ovarian Cancer | [449] |
| miR-106a/b | SIRT1, PTEN | Pituitary Tumor, Breast Cancer | S [16–18] |
| miR-107 | EGFR | Non-Small Cell Lung Cancer | S [19] |
| miR-10b | PTEN | Breast Cancer | S [20] |
| miR-122/a | MYC, EGFR | Hepatocellular Carcinoma, Inflammatory Bowel Disease | S [21], [231] |
| miR-124a | SIRT1 | Neuropathic Pain | S [22] |
| miR-125a/b | SIRT1, SIRT7, TP53 | Hepatocellular Carcinoma, Age-Related Cataract, Multiple Myeloma, Non-Small Cell Lung Cancer, Colorectal Cancer, Neuroblastoma, Cataract | S [23–28], [422] |
| miR-126 | KRAS, AKT | Pancreatic Cancer, Squamous Tongue Cell Carcinoma, Glioma, Colorectal Cancer | S [29,30], [204], [263] |
| miR-1285 | TP53 | Neuroblastoma, Hepatoblastoma | S [31] |
| miR-1294 | MYC | Esophageal Squamous Cell Carcinoma | S [32] |
| miR-1297 | PTEN | Breast Cancer | S [33] |
| miR-130a/b | MYC, PTEN | Osteocarcinoma, Bladder Carcinoma, Non-small Cell Lung Cancer | S [34–36], [223] |
| miR-132 | SIRT1 | Glioma, Type2 Diabetes Mellitus, Gastric Cancer, Colitis | S [37–40] |
| miR-133a/b | EGFR, SIRT1 | Pancreatic Cancer, Ovarian Cancer, Hepatocellular Carcinoma | S [41,42] |
| miR-134 | KRAS, EGFR | Rena Cell Carcinoma, Glioblastoma, Non-Small Cell Lung Cancer, Colorectal Cancer | S [43–45], [207] |
| miR-135a/a-1 | MYC, EGFR | Renal Cell Carcinoma, Prostate Cancer | S [46,47] |
| miR-137 | EGFR | Glioblastoma Multiforme, Thyroid Cancer | S [48,49] |
| miR-138 | SIRT1 | Diabetic Vascular Smooth Muscle Cells, Intervertebral Disc Degeneration, Pancreatic Cancer, Osteocarcinoma | S [50–53] |
| miR-141 | PTEN, AKT, TP53, SIRT1 | Esophageal Cancer, Osteosarcoma, Multiple Myeloma, Pluripotent Stem Cells, Glioma, HB infection | S [25,54–57], [272] |
| miR-142 | PTEN | Cutaneous Squamous Cell Carcinoma | S [58] |
| miR-143 | KRAS, EGFR, AKT, MDM2, SIRT1 | Colorectal Cancer, Non-Small Cell Lung Cancer, Bladder Cancer, Head and Neck Squamous Cell Carcinoma, Pancreatic Cancer | S [59–63], [266], [357], [410] |
| miR-144 | PTEN, mTOR, TIGAR | Pancreatic Neuroendocrine Tumor, Preeclampsia, Salivary Adenoid Carcinoma, Renal Cell Carcinoma, Inflammation of Microglia, Lung Cancer | S [64–67], [350] |
| miR-145 | MYC, MDM2, SIRT1 | Non-Small Cell Lung Cancer, Esophageal Squamous Cell Carcinoma, Ovarian Cancer, Oral Squamous Cell Carcinomas, Glioblastoma, Head and Neck Squamous Cell Carcinoma, Pancreatic Cancer | S [68–70], [266], [357], [410] |
| miR-146b | AKT | Osteosarcoma | [272] |
| miR-148a/b | PTEN, AKT, MYC | Osteosarcoma, Renal Cell Carcinoma, Hepatocellular Carcinoma | S [71,72], [2], [229] |
| miR-149 | AKT | Hepatocellular Carcinoma, Neuroblastoma, Glioblastoma Multiforme | S [73–75] |
| miR-150 | TP53 | Lung Cancer | S [76] |
| miR-152 | PTEN, SIRT7 | Hepatic Insulin Resistance, Human Dental Pulp Stem Cells | S [77,78] |
| miR-153 | PTEN, AKT | Prostate Cancer, Lung Cancer | [275], [276] |
| miR-155 | KRAS, MYC, PTEN, AKT, SIRT5, SIRT1 | Gastric Carcinoma, hepatocellular Carcinoma, Waldenström Macroglobulinemia, Leukemia, Colorectal Cancer, Neuropathic Pain | S [22,79–81], [220], [301], [450] |
| miR-15a/b, miR-16 | TP53, mTOR, AKT, SIRT4 | Multiple Myeloma, Glioma, Ischemia, Dermal Fibroblasts | S [25,82–85] |
| miR-17 | MYC, TP53 | Neuroblastoma, Cervical Cancer | S [86], [361] |
| miR-18/a | KRAS | Human Squamous Carcinoma, Colorectal Cancer, Liver Cancer, Ovarian Cancer | S [87,88] |
| miR-181a/b/d | PTEN, KRAS, EGFR, AKT, SIRT1 | Colorectal Cancer, Osteosarcoma, Oral Squamous Cell Carcinoma, Glioma, Cutaneous Squamous Cell Carcinoma, Acute Myeloid Leukemia, Glioblastoma Multiforme, Hepatic Stellate Cells, Non-Alcoholic Fatty Liver Diseases | S [89–97] |
| miR-1827 | MDM2 | Colorectal Cancer | S [98] |
| miR-183 | mTOR | Neuropathic Pain | S [99] |
| miR-184 | MYC | Non-Small Cell Lung Cancer, Nasopharyngeal Carcinoma | S [100,101] |
| miR-185 | MYC, PTEN, AKT | Colorectal Cancer, Breast Cancer, Hepatocellular Carcinoma, Idiopathic Pulmonary Fibrosis, Non-Small Cell Lung Cancer | S [102–104], [228], [297] |
| miR-192 | MDM2 | Colorectal Cancer, Multiple Myeloma | [358], [376] |
| miR-193a/b | PTEN, KRAS, TSC1/2 | Renal Cell Carcinoma, Colon Cancer, Breast Cancer, Pancreatic Ductal Adenocarcinoma, Cutaneous Squamous Cell Carcinoma, Amyotrophic Lateral Sclerosis | S [105–108], [202], [208] |
| miR-194 | AKT, MDM2 | Gall Bladder Cancer, Multiple Myeloma | S [109], [358], [376] |
| miR-195 | SIRT3 | Myocardium | [446] |
| miR-197 | TP53 | Non-Small Cell Lung Cancer | S [110] |
| miR-199a/b | SIRT1, mTOR | Pluripotent Stem Cells, Hyperglycemia-Induced Pancreaticβ-Cell Loss, Endometrioid Endometrial Carcinoma | S [111,112], [325] |
| miR-19a/b | PTEN, TP53 | Bladder Cancer, Osteosarcoma, Myeloma, Liver Cancer, Breast Cancer | S [113–116] |
| miR-200a/c | EGFR, TP53, KRAS, PTEN, SIRT1, MYC, AKT | Bladder Cancer, Breast Cancer, Multiple Myeloma, Nasopharyngeal Carcinoma, Colorectal Cancer, Hepatic Stellate Cell, Pluripotent Stem Cell, Lung Adenocarcinoma, Renal Cell Carcinoma, Ovarian Cancer, Esophageal Cancers | S [25,117–126], [271], [334] |
| miR-203 | MYC | Cutaneous Squamous Cell Carcinoma | S [127] |
| miR-204 | SIRT1 | Osteosarcoma, Spermatogonial Stem Cell, Hepatocellular Carcinoma | S [128,129], [405] |
| miR-205 | PTEN | Ovarian Cancer | S [130] |
| miR-206 | KRAS, EGFR | Gastric Cancer, Pancreatic Ductal Adenocarcinoma, Oral Squamous Cell Carcinoma, Head and Neck Squamous Cell Carcinoma | S [10,131–133] |
| miR-20a/b | PTEN, AKT, SIRT7 | Coronary Artery Disease, Diabetic Retinopathy, Diabetic Nephropathy | S [134–136] |
| miR-21 | KRAS, MYC, PTEN | Lung Cancer, Breast Cancer, Diabetic Kidney Disease, Colorectal Cancer, Hepatocellular Carcinoma, Leukemia, Vestibular Schwannomas, Glioblastoma, Bladder Cancer, Radio-resistance Lung Cancer | S [5,137–142], [296], [305] |
| miR-210 | MYC | Colorectal Cancer, Glioblastoma, Cervical Cancer, Breast Cancer | S [143] |
| miR-212 | SIRT1 | Prostate Cancer | S [144] |
| miR-214 | KRAS, PTEN, TP53 | Non-small Cell Lung Cancer, Ovarian Cancer, Breast Cancer, Ovarian Cancer Stem Cells | S [145–148], [291] |
| miR-215 | MDM2 | Colorectal Cancer, Multiple Myeloma | [358], [376] |
| miR-216a/b | PTEN, KRAS | Acute Pancreatitis, Kidney Disorders, Ovarian Cancer, Nasopharyngeal Carcinoma | S [149–151], [289] |
| miR-217 | KRAS, PTEN, SIRT1 | Pancreatic Ductal Adenocarcinoma, Lung Cancer, Kidney Disorders, Podocyte Injury, Aging | S [152–154], [289], [411] |
| miR-218 | EGFR, mTOR | Non-Small Cell Lung Cancer, Prostate Cancer | S [155,156] |
| miR-22 | MYC, PTEN, SIRT1 | Leukemia, Clear Cell Renal Cell Carcinoma, Glioblastoma, Ischemia-Reperfusion Injury | S [157–161], [225] |
| miR-221 | PTEN | Radiosensitive Cancer Cells, Glioblastoma | S [162], [293] |
| miR-222 | PTEN, TP53 | Radiosensitive Cancer Cells, Oral Squamous Cell Carcinoma | S [162], [346] |
| miR-224 | KRAS, mTOR | Colorectal Cancer, Gastric Cancer | S [163,164] |
| miR-23a/b | EGFR, SIRT5, SIRT1 | Coronary Artery Disease, Colorectal Cancer, Diabetic Retinopathy | S [165,166], [450] |
| miR-24 | MYC, TP53 | Leukemia, Embryonic Stem Cells, Hepatocellular Carcinoma | S [167,168], [227] |
| miR-25 | PTEN, TP53 | Diabetic Nephropathy, Multiple Myeloma, Non-Small Cell Lung Cancer, Colorectal Cancer | S [25,169,170] |
| miR-26a | PTEN | Glioblastoma | [282] |
| miR-27a/b | KRAS, TP53, EGFR, SIRT5 | Esophageal Squamous Cells Carcinoma, Colorectal Cancer, Renal Cell Carcinoma, Non-Small Cell Lung Cancer | S [47,171–173], [450] |
| miR-28 | SIRT3 | Primary human tenocytes | [448] |
| miR-29a/b/c | PTEN, AKT, MDM2 | Colorectal Cancer, Gastric Cancer, Prostate Cancer, Breast Cancer, Non-Small Cell Lung Cancer | S [174–177], [265], [288], [356] |
| miR-300 | TP53 | Lung Cancer, Colorectal Cancer | S [178,179] |
| miR-301a | PTEN | Pancreatic Cancer, Malignant Melanoma | S [180,181] |
| miR-302a | AKT | Prostate Cancer | [264] |
| miR-30b/c/d | KRAS, TP53 | Colorectal Cancer, Breast Cancer, Non-Small Cell Lung Cancer, Multiple Myeloma, Cardiac Disease | S [170,182–184], [347] |
| miR-31 | TP53 | Breast Cancer | S [185] |
| miR-3151 | TP53 | Malignant Melanoma | S [186] |
| miR-32 | PTEN | Hepatocellular Carcinoma | [287] |
| miR-320a | MYC | Hepatocellular Carcinoma | S [187] |
| miR-33 | TP53 | Hematopoietic Stem Cells | S [188] |
| miR-337 | PTEN | Endometrial Carcinoma | S [189] |
| miR-338 | mTOR | Colon Cancer | S [190] |
| miR-339 | MDM2, SIRT2 | Breast Cancer, Neuroblastoma | S [191], [431] |
| miR-33a/b/c | SIRT6, MYC | Liver Cancer, Osteosarcoma | [219], [418] |
| miR-340 | MDM2 | Prostate Cancer | S [192] |
| miR-34a | MYC, EGFR, AKT, SIRT6, SIRT1 | Hepatocellular Carcinoma, Prostate Cancer, Renal Cell Carcinoma, Non-Small Cell Lung Cancer, Glioma, Colorectal Cancer, Non-Alcoholic Fatty Liver Diseases, Pancreatic Cancer | S [193–197], [222], [240], [268], [410] |
| miR-363 | MYC | Prostate Cancer, Hepatocellular Carcinoma | S [198], [2] |
| miR-365a | KRAS | Cutaneous Squamous Cell Carcinoma | S [106] |
| miR-3666 | SIRT7 | Breast Cancer | S [199] |
| miR-367 | MDM2 | Hepatocellular Carcinoma | S [200] |
| miR-370 | EGFR, PTEN | Gastric Cancer, Colorectal Cancer, Gastric Cancer | S [45,201,202] |
| miR-373 | mTOR, SIRT1 | Fibrosarcoma | [333] |
| miR-374/a | MDM2, PTEN | Bladder Cancer, Breast Cancer | S [203,204] |
| miR-377 | SIRT1 | Obesity | S [205] |
| miR-378 | AKT | Breast Cancer | S [206] |
| miR-380 | TP53 | Neuroblastoma | S [207] |
| miR-384 | KRAS | Colorectal Cancer | S [208] |
| miR-409 | AKT | Breast Cancer | S [209] |
| miR-421 | SIRT3 | Non-Alcoholic Fatty Liver Disease | S [210] |
| miR-429 | MYC | Gastric Cancer, Breast Cancer | S [119,211] |
| miR-449a/c | MYC | Glioblastoma, Gastric Carcinoma, Osteosarcoma, Prostate Cancer | S [212–214] |
| miR-451 | MYC, AKT | Docetaxel-Resistant Lung Adenocarcinoma, Dilated Cardiomyopathy, Bladder Cancer, Non-Small Cell Lung Cancer, Glioblastoma | S [215–220] |
| miR-451 | TSC1/2 | Multiple Myeloma, Hypertrophic Cardiomyopathy | S [221], [335] |
| miR-453 | TP53 | Lung Cancer | S [222] |
| miR-4534 | PTEN | Prostate Cancer | S [223] |
| miR-454 | PTEN | Non-Small Cell Lung Cancer | S [224] |
| miR-4689 | KRAS, AKT | Colorectal Cancer, | [203] |
| miR-486 | PTEN, MDM2, SIRT1 | Cardiac Myocytes, Lung Cancer, Erythroleukemia | S [225,226], [283] |
| miR-491 | TP53 | Pancreatic Cancer | S [227] |
| miR-492 | PTEN | Hepatic Cancer | S [228] |
| miR-494 | MYC, PTEN, SIRT1 | Epithelial Ovarian Cancer, Cardiac Disease, Myeloid-Derived Suppressor Cells, Cervical Cancer, Gastric Carcinoma, Pancreatic Cancer | S [229–233], [294], [304] |
| miR-495 | AKT, mTOR | Prostate Cancer | S [234] |
| miR-496 | mTOR | Aging | S [235] |
| miR-5003 | MDM2 | Breast Cancer | S [236] |
| miR-502a | EGFR | Colorectal Cancer | S [237] |
| miR-504 | TP53 | Non-Small Cell Lung Cancer, Colorectal Cancer, Multiple Myeloma, Abdominal Aortic Aneurysm | S [170,238], [360] |
| miR-509 | EGFR, MDM2 | Tongue Squamous Cell Carcinoma, Cervical Cancer, Hepatocellular Carcinoma, Prostate Cancer | S [239,240], [355] |
| miR-511 | AKT | Prostate Cancer | S [241] |
| miR-519d | PTENAKT | Hepatocellular Carcinoma | [286] |
| miR-520 b/c/e | EGFR, mTOR, SIRT1 | Gastric Cancer, Fibrosarcoma | S [242], [333] |
| miR-532 | KRAS, SIRT3 | Lung Adenocarcinoma, Ovarian Cancer | S [243], [449] |
| miR-543 | SIRT1 | Hypertension, Gastric Cancer | S [244,245] |
| miR-545 | EGFR | Colorectal Cancer | S [246] |
| miR-548i | AKT | Non-Small Cell Lung Cancer | S [247] |
| miR-561 | MYC | Gastric Cancer | S [248] |
| miR-577 | PTEN | Glioblastoma | [283] |
| miR-579 | MDM2 | Melanoma | S [249] |
| miR-600 | TP53 | Colorectal Cancer | S [250] |
| miR-601 | SIRT1 | Pancreatic Cancer | S [251] |
| miR-606 | MDM2 | Breast Cancer, Lung Cancer, Colorectal Cancer | [353] |
| miR-610 | MDM2 | Glioma | S [252] |
| miR-613 | KRAS | Ovarian Cancer | S [253] |
| miR-615 | AKT | Pancreatic Ductal Adenocarcinoma | S [254] |
| miR-622 | KRAS | Lung Cancer, Colorectal Cancer | S [255,256] |
| miR-637 | AKT | Glioma | [267] |
| miR-638 | AKT | Lung Cancer | S [257] |
| miR-641 | MDM2 | Lung Cancer | S [258] |
| miR-650 | AKT | Rheumatoid Arthritis | S [259] |
| miR-660 | MDM2 | Lung Cancer | [354] |
| miR-7 | EGFR, mTOR | Ovarian Cancer, Glioblastoma, Lung Cancer, Breast Cancer, Hepatocellular Carcinoma, Gastric Cancer | S [260,261], [277], [278], [279], [329], [330] |
| miR-718 | PTEN | Kaposi's Sarcoma, Inflammation | S [262], [292] |
| miR-744 | MYC | Hepatocellular Carcinoma | [221] |
| miR-758 | mTOR, MDM2 | Hepatocellular Carcinoma | S [263] |
| miR-766 | MDM2, SIRT6 | Breast Cancer, Dermal Fibroblast | S [264], [421] |
| miR-768 | KRAS | Lung Cancer | S [265] |
| miR-875 | EGFR | Prostate Cancer | S [266] |
| miR-9/a | SIRT1PTEN | Hepatic Stellate Cells, Non-Alcoholic Fatty Liver Disease, Acute Myeloid Leukemia, Colorectal Cancer, Nasopharyngeal Carcinoma, Non-Small Cell Lung Cancer | S [267–272] |
| miR-92 | TP53 | Multiple Myeloma, Pluripotent Stem Cells | S [25,55] |
| miR-923 | AKT | Lung Cancer | S [257] |
| miR-93 | MYC, PTEN, SIRT1 | Colon Cancer, Ovarian Cancer, Myocardial Ischemia/Reperfusion(I/R) Injury, Breast Cancer, Aging | S [18,273,274], [290], [412] |
| miR-95 | PTEN | Radioresistance Lung Cancer | S [138] |
| miR-96 | KRAS, mTOR | Pancreatic Ductal Adenocarcinoma, Pancreatic Cancer, Colorectal Cancer, Myocardial Hypertrophy | S [184,275–277], [206] |
| miR-98 | MYC, TP53 | Breast Cancer, Lung Cancer | S [5,222] |
| miR-99a/b | mTOR | Breast Cancer, Esophageal Squamous Cell Carcinoma, Cervical Cancer, Endometrioid Endometrial Carcinoma | [324], [325], [327] |
5.1. KRAS proto-oncogene
The KRAS oncogene features as an early mutation in up to 45% of colorectal tumors, notable because it can drive many hallmarks of cancer [191]. KRAS-mediated transformation is linked with mitochondrial respiratory dysfunction and elevated NADPH oxidase (NOX)-mediated ROS generation [192], [193]. Wang et al. [194] postulated that oncogenic KRAS influences complex I activity in the electron transport chain, most likely by downregulating complex I assembly factor protein (NDUFAF1) and, as a consequence, induces mitochondrial dysfunction. However, additional oncogenic signals and/or loss of tumor suppressors, including dysregulated miRNAs, are required for tumourigenesis.
Unsurprisingly, KRAS is a target of multiple miRNAs, including let-7, miR-96, miR-134 and miR-143 (Summarized in Table 5). These miRNAs affect cancer cell metabolism, cell cycle arrest, apoptosis, cell migration and invasion, especially by modulating RAS/MAPK signaling (Figure 2)
KRAS is frequently mutated in human neoplasia including pancreatic, colorectal and lung cancer. The oncogenic KRASG12V variant, which leads to higher KRAS activity, was reported to be the most frequent mutation. However, despite low KRAS mutation frequency in glioblastoma and breast cancer cells, activation of the wild-type KRAS pathway is common in these cancers. Also, sequence variants in the KRAS 3′UTR (rs712) were found in gastric cancer, colorectal cancer, papillary thyroid cancer, breast cancer, and non-small cell lung cancer, which disrupt let-7 binding site and subsequent miRNA-mediated downregulation [195], [196], [197], [198], [199].
The expression of some miRNAs such as let-7, miR-126, miR-200c, miR-193b, and miR-4689 was found to be lower in KRAS mutant cells, as compared to tumors expressing wild-type KRAS [199], [200], [201], [202], [203], confirming the context dependent activity of miRNAs, even in regulating KRAS itself. Kopp et al. [200] reported that in breast cancer cells harboring the KRASG13D mutation, miR-200c targets KRAS transcripts and inhibits proliferation and cell cycle progression, while in KRAS wild type cells miR-200c affects proliferation through other targets. Despite different miR-126 expression levels in KRAS mutant and wild type colon cancer cells, Hara et al. [201] showed that over-expression of miR-126 does not alter KRAS expression and function. In contrast, Jiao et al. [204] showed KRAS regulation by miR-126 in pancreatic cancer. Such variations suggest that the activity of some miRNAs is subjected to changes through both transcriptional and post-transcriptional processes during tumourigenesis. Examples are erythropoietin-producing hepatoma receptor A1 (EphA1) upregulating let-7 in multiple myeloma [205], EVI1 suppressing miR-96 in pancreatic ductal adenocarcinoma [206], KLF4 downregulating miR-134 in glioblastoma [207] and MYC associated factor X (MAX) inhibiting miR-193a in breast cancer [208]. Therefore, coordinated suppression of miRNAs, as is found in various cancers, would not only influence oncogenic KRAS activity but may also influence other genes involved in KRAS-related signaling to cooperatively initiate tumourigenesis, including genes in metabolic pathways.
5.2. MYC proto-oncogene
Overexpression of the c-MYC proto-oncoprotein plays pivotal roles in sustaining the transformed phenotype of most cancer cells [209]. The discovery that LDHA is among 20 putative targets of c-MYC provided evidence that c-MYC directly regulates glycolysis. Since then, other glycolytic genes including GLUTs, GAPDH, PGK, HK2, ENO1, PGM, PKM2, and MCTs are also reportedly induced by c-MYC [210].
Along with its role in glycolysis, c-MYC was found to regulate mitochondrial biogenesis, respiration, and function [211]. Upregulation of some nuclear genes that encode proteins for mitochondrial function, mitochondrial DNA replication and transcription of mitochondrial DNA are known to be direct consequences of c-MYC overexpression [212]. c-MYC also contributes to mitochondrial biogenesis and gives rise to the synthesis of acetyl-CoA and fatty acid biosynthesis required for cancer cell proliferation. In parallel, c-MYC upregulates the glutamine catabolism required for biosynthetic processes by inducing GLS and the glutamine transporters, ASCT2 and SLC7A25 [213], [214]. Overall, while c-MYC enhances glycolysis and consequently depletes pyruvate required for mitochondrial OXPHOS, it also confers the ability for cancer cells to utilize non-glucose substrates and maintain mitochondrial respiration to support cancer cell proliferation and progression.
c-MYC cooperates with HIF-1, or acts independently, to regulate glycolysis and OXPHOS [215]. In normal cells, MYC enhances glycolytic flux to OXPHOS. However, in cancer cells, c-MYC cooperates with HIF-1 and PKM2 to upregulate glycolysis and provide adequate metabolic intermediates for biomass synthesis [216]. While upregulation of HIF-1-mediated glycolysis was observed under hypoxic conditions, c-MYC regulates glycolytic genes independently under normal oxygen tension. In addition, while HIF-1 upregulates PDK1 under hypoxia, c-MYC cooperates with HIF-1 to further upregulate PDK1 and, thus, amplifies the hypoxic response. Therefore, under normoxia, c-MYC enhances glycolysis, but it cooperates with HIF-1 to upregulate PDK1 and reduce mitochondrial respiration under hypoxic conditions [217]. Intriguingly, elevated ENO1 was shown to form a negative feedback loop with activated c-MYC. c-MYC-induced ENO1 increases the expression of MBP1, a transcription factor, and suppresses c-MYC expression [218].
c-MYC both regulates miRNA expression and is, in turn, controlled by them (Table 3, Table 5). Several miRNAs have been shown to modulate c-MYC expression by different mechanisms (Figure 2). Let-7a, miR-22, miR-33b, miR-34a, miR-130a, miR-145, and miR-155 were found to suppress c-MYC after binding with canonical target sequences in the c-MYC 3′UTR [219], [220], [221], [222], [223], [224], [225], [226]. miR-24 binds to a seedless, but highly complementary, sequence while miR-18-5p and miR-774 bind to the protein coding region of c-MYC mRNA [221], [227], [228]. Some other miRNAs, such as miR-363-3p act more indirectly. In HCC, miR-363-3p destabilizes c-MYC through targeting USP28, a ubiquitin protease of MYC, and promoting the degradation of pre-existing c-MYC protein [229]. Several reports indicate a coordinated and reciprocal relationship between c-MYC and miRNA expression levels. For instance, Liao et al. [228] showed a negative feedback and auto-regulatory role for c-MYC levels, as monitored by miR-185-3p. They confirmed that miR-185-3p is a genuine transcriptional target of c-MYC but also that miR-185-3p inhibits c-MYC translation by targeting the coding region of c-MYC transcripts.
Table 3.
Summary of miRNAs regulated by the transcription factor MYC.
| miRNA | Regulation | Disease/cells | References |
|---|---|---|---|
| miR-2b | Upregulation | Drosophila S2 Cells | [242] |
| miR-277 | Upregulation | Drosophila S2 Cells | [242] |
| miR-92 | Upregulation | Neuroblastoma, Burkitt Lymphoma | [233], [243] |
| miR-106a | Upregulation | Neuroblastoma, Burkitt Lymphoma | [233], [243] |
| Let-7a/c | Up/Downregulation | Neuroblastoma, Burkitt Lymphoma, Breast Cancer, Prostate cancer | [234], [235], [243], [244] |
| miR-17 | Upregulation | Neuroblastoma, Burkitt Lymphoma | [233], [243] |
| miR-93 | Upregulation | Neuroblastoma | [243] |
| miR-99 | Upregulation | Neuroblastoma | [243] |
| miR-221 | Upregulation | Neuroblastoma | [243] |
| miR-18 | Upregulation | Burkitt Lymphoma | [233] |
| miR-19 | Upregulation | Burkitt Lymphoma | [233] |
| miR-20 | Upregulation | Burkitt Lymphoma | [233] |
| miR-15a | Downregulation | Lymphoma | [245] |
| miR-16 | Downregulation | Lymphoma | [245] |
| miR-22 | Downregulation | Lymphoma | [245] |
| miR-23a/b | Downregulation | Lymphoma, Prostate Cancer | [107] |
| miR-26a/b | Downregulation | Lymphoma, Burkitt Lymphoma, Prostate Cancer | [245], [246], [247] |
| miR-29 | Downregulation | Lymphoma, lung Adenocarcinoma | [245], [248] |
| miR-34 | Downregulation | Lymphoma | [245] |
| miR-146a | Downregulation | Lymphoma | [245] |
| miR-150 | Downregulation | Lymphoma | [245] |
| miR-195 | Downregulation | lymphoma | [245] |
| miR-141 | Upregulation | Embryonic Stem Cells, Nasopharyngeal Carcinoma | [249], [250] |
| miR-200 | Upregulation | Embryonic Stem Cells | [249] |
| miR-429 | Upregulation | Embryonic Stem Cells | [249] |
| miR-9 | Upregulation | Breast Cancer | [251], [252] |
| miR-185 | Upregulation | Non-small Cell Lung Cancer | [228] |
| miR-122 | Downregulation | Hepatocellular Carcinoma | [230], [231], [232] |
c-MYC activates or represses a variety of genes, including miRNA genes, mainly through interactions with different complexes and proteins. c-MYC supresses MIR122 gene transcription in liver tumors through association with a conserved promoter region upstream of the MIR122 gene. It also downregulates hepatocyte nuclear factor 3-beta (HNF3β), which normally activates miR-122 and enhances its stability [230], [231].
miR-122 was reported to supress c-MYC expression indirectly by targeting E2F1 and TFDP2 (E2F dimerization partner 2) mRNA [232]. In addition, feedback regulation was reported for miR-17-5p/c-MYC/E2F in some cancers, including breast and prostate [233]. Nadiminty et al. [234] reported a LIN28/let-7/c-MYC loop that plays an important role in some cancers. Relief of c-MYC repression occurs when LIN28, a highly conserved RNA-binding factor, binds let-7 precursors and inhibits miRNA maturation [235]. There is a direct relationship between c-MYC, its dimerization partner, MAX, and the expression of some miRNAs such as let-7a and miR-22 [225], [236]. c-MYC can also transcriptionally activate some miRNAs, including the miR-17-92 cluster, through interaction with MAX protein at the polycistronic promoter region [233], [237]. Ting et al. [225] showed that increased miR-22 limits the amount of MAX protein available for c-MYC binding by directly targeting it and, therefore, affects the expression of downstream targets of the c-MYC/MAX complex. In contrast, interaction of c-MYC with MIZ-1 represses expression of some c-MYC target genes through displacement of p300 co-activator protein [238]. There is also a miRNA/c-MYC negative feedback loop in HCC with miR-148a-5p directly targeting c-MYC and, as previously mentioned, miR-363-3p indirectly destabilizing c-MYC by targeting ubiquitin specific peptidase 28 (USP28) [229]. Other miRNAs that are repressed transcriptionally and post-transcriptionally by c-MYC are summarized in Table 3.
The activation of c-MYC alone is unable to transform cells. Therefore, there is cooperation between oncogenic partners, such as RAS, and inactivation of tumor suppressors such as p53 in c-MYC dependant tumor development [239], [240], [241]. Hence, along with passive adaptation of tumor cells, oncogenic mutations and transcriptional controls, such as the reciprocal association of c-MYC with several miRNAs, enhance the ability of cancer cells to consume non-glucose substrates and fuel mitochondria. This may explain the inefficiency of drugs which only target glycolysis and add another layer of complexity to therapeutic strategies.
5.3. PI3K/AKT pathway
The PI3K intracellular signaling pathway plays a critical role in cell apoptosis, proliferation, and protein synthesis. Its role in regulation of glucose uptake and metabolism is equally definitive. PI3K dysregulation was reported in several human cancers and several drugs targeting this pathway are currently in clinical trials [253]. Activation of PI3K leads to an upregulation of downstream effectors such as AKT and mTOR.
The evolutionarily conserved serine/threonine kinase, AKT, was reported to be one of the most prevalent and constitutively activated onco-proteins in malignant cells [254], [255]. AKT is an important activity-dependent stimulus for cancer cell metabolism, influencing glycolysis by both direct and indirect mechanisms. AKT plays a central role in the regulation of cellular energy metabolism and glucose homeostasis. It stimulates ATP generation by accelerating both glycolytic and oxidative metabolism with a concomitant increase in oxygen consumption to preserve energy. AKT activation results in ROS generation and, therefore, contributes to tumourigenesis by inducing mutations and facilitating tumor-promoting signaling pathways and inducing mutations [256]. Elevated, AKT-mediated, glycolysis plays a major role in proliferation and survival of transformed cells. AKT increases glucose uptake, directly, by increasing the expression and plasma membrane translocation of glucose transporters (GLUT1, GLUT2, and GLUT4) [257]. It also maintains MMP and promotes the association of HK2 with the mitochondrial outer membrane by mediating HK2 phosphorylation and inhibiting glucose-6 phosphate dissociation from the mitochondrial membrane [258]. This may enhance enzymatic efficiency of the kinase, promote metabolic coupling between glycolysis and OXPHOS, increase ATP synthesis through OXPHOS and decrease susceptibility to apoptosis [256]. Indirectly, AKT activates PFK1 phosphorylation and activation by inducing PFK2 and releases forkhead box O1 (FOXO)-mediated repression of glycolysis. AKT also activates mTORC1 indirectly through phosphorylating and, thus, inactivating TSC2, an mTOR inhibitor [259], [260], [261]. The ability of AKT to increase glucose uptake and glycolysis in tumor cells may also require cooperation from other cancer-associated proteins, such as c-MYC and HIF-1. Although AKT-transformed cells show elevated levels of amino acid and lipid transporters that are linked to cell growth, constitutive activation of AKT renders cells dependent on an extracellular glucose supply for survival [256]. Together these findings demonstrate the coordinated regulation of glycolysis and OXPHOS by oncogenic AKT.
AKT, which is described as “Warburg's kinase”, provides selective advantages to tumor cells by increasing both glycolysis and OXPHOS [262]. Several miRNAs were reported to modulate AKT expression directly by targeting AKT mRNA, and protein phosphorylation and/or indirectly regulating its upstream stimuli, such as EGFR and its upstream repressors, such as PTEN (Figure 2).
While some miRNAs, such as miR-637 in glioma, miR-302a and miR-29b in prostate cancer and miR-143 in bladder cancer, directly bind the AKT 3′UTR and inhibit its translation, some other miRNAs, reduce AKT phosphorylation without affecting total AKT levels. For instance, miR-126 reduces AKT phosphorylation by inhibiting by phosphatidylinositol 3-kinase regulatory subunit beta (p85β) [263], [264], [265], [266], [267] (Table 5).
Other proteins and regulatory factors also contribute to regulating AKT activation in different cell types and conditions. For instance, the over-expression of Rictor, a target of miR-34a and mTORC2 component, causes activation of AKT in glioma stem cells [268]. Rictor activation results in mTORC2 activation and consequently, AKT is further activated by mTORC2 mediated phosphorylation [269]. In breast cancer cells miR-205, which is often downregulated in cancer, targets HER3 receptor transcripts and supresses the activation of AKT [270]. Protein phosphatase 2 scaffold subunit Abeta (PPP2R1B) is another intermediate in AKT signal transduction, directly interacting with AKT, and is a target of miR-200c in esophageal cancer cells [271]. Al-Khalaf and Aboussekhra [272] showed that miR-141 and miR-146b-5p target an RNA binding protein, AUF1, which has an important role in PI3K/AKT/mTOR pathway regulation. AUF1 binds to and stabilizes PDK1 mRNA and promotes AKT phosphorylation and activation. AUF1 was also reported to negatively regulate PTEN phosphatase and activate PI3K [273], [274]. Additionally, some AKT-targeting miRNAs were shown to regulate drug sensitivity in cancer cells, such as miR-29b and miR-200c that influence chemotherapy responses in prostate and esophageal cancers, respectively [265], [271].
However, the miRNAs that regulate AKT signaling do not act to fully repress AKT and its mediators. Rather, they fine tune expression in a context-specific manner. Therefore, it is likely that AKT is not exclusively regulated by specific miRNAs and further, it is not surprising that some miRNAs, such as miR-153 which targets both PTEN and AKT [275], [276], play complex pleiotropic roles in regulating PI3K/AKT signaling.
Although a number of studies have reported EGFR gene amplification in some cancers, post-transcriptional modulation remains a significant cause of EGFR overexpression in cancer cells (Table 5). For instance, miR-7 was found to regulate expression of multiple effectors of the EGFR signaling pathway, as well as directly targeting EGFR mRNA. Zhou and Hu et al. [277] showed that miR-7 overexpression in epithelial ovarian carcinoma (EOC) cells results in reduced expression of EGFR without any changes in EGFR phosphorylation. A feedback loop between miR-7 and EGFR was reported [277], [278], as increased EGFR activity results in extracellular-signal-regulated kinase (ERK)-mediated degradation of YAN, which is a miR-7 repressor. Further, miR-7 binds to the YAN 3′UTR and represses its expression [279].
PTEN has a central role in cell cycle progression. Although mutational loss of PTEN was reported in some cancers, epigenetic factors, including miRNAs, also regulate PTEN expression [280] (Table 5). Due to the unusually long 3′UTR of PTEN, it contains binding sites for many miRNAs, which can reduce its mRNA levels (including miR-32, miR-29, miR-26a/b, miR-217, miR-486, miR-193a, miR-519d) [281], [282], [283], [284], [285], [286], [287], [288], [289] or PTEN translation without affecting its mRNA levels (miR-93, miR-214, miR-221, miR-494, miR-21) [290], [291], [292], [293], [294], [295], [296].
Furthermore, miR-185 in HCC and miR-26a in low-grade glioma alter PTEN promoter methylation and play a subordinate role in PTEN gene regulation by targeting DNA (cytosine-5)-methyltransferase 1 (DNMT1) and enhancer of zeste homolog 2 histone methyltransferase (EZH2) [282], [297]. Therefore, along with direct regulation of PTEN by the aforementioned miRNAs, several miRNAs regulate PTEN through indirect mechanisms. Examples include PTEN repression via miR-101 and miR-1 both targeting the PTEN activator, membrane-associated guanylate kinase inverted 2 (MAGI-2); as well as PTEN induction following the miR-185 targeting of PTEN silencer, DNMT1 [297], [298], [299].
High glucose was shown to affect some PTEN targeting miRNAs, such as stimulating miR-21 levels in renal cancer or lowering miR-32 levels in HCC, depending on the physiological status of the cells, which results in AKT activation or suppression, respectively [300], [301].
PTEN dephosphorylates PIP3, generated by PI3K, to inhibit AKT activation. Suppression of PTEN, through miRNA-mediated mechanisms, enhances AKT phosphorylation and signaling and supports cell proliferation and survival [302]. PTEN inhibition also results in cystic vestibular schwannoma development and cancer cell invasion via induced metalloproteinase-2 (MMP-2) [303]. Transforming growth factor beta 1 (TGF-β) mediated AKT activation is another consequence of reduced PTEN activity [289], [304]. Decreased PTEN expression was also shown to impair p53-dependant responses in cancer cells [286]. Moreover, some miRNAs were shown to induce drug- and radio-resistance by inhibiting PTEN. For instance, miR-21 induces daunorubicin resistance in leukemia, miR-214 induces cisplatin resistance in ovarian cancer cells and miR-221 induces TRAIL- and radio-resistance in glioma cells by inhibiting PTEN [288], [293], [305]. Breast cancer metastases in the brain also display increased aggression due to suppression of PTEN by astrocyte exosomal miRNAs [306].
5.4. Mechanistic target of rapamycin kinase (mTOR)
Mechanistic target of rapamycin (mTOR), also known as mammalian target of rapamycin, consists of two divergent complexes: complex 1 (mTORC1) and (mTORC2). mTORC1 acts as a metabolic hub, integrating extracellular stimuli with nutrient availability and cellular energy to coordinate responses. mTORC1 is mainly involved in cellular proliferation, translation and metabolic programming while mTORC2 regulates cell survival, cytoskeletal organization, and degradation of newly synthesized polypeptides [307], [308].
mTOR is stimulated by loss of function of some inhibitors including LKB1, PML, PTEN, and TSC1/2 or activation of some oncogenes such as AKT and RAS [115], [116], [262], [309]. Activated mTOR, in turn, dramatically enhances the translational machinery and ribosome biogenesis, increases cell growth in response to mitogens, growth factors and hormones, and upregulates some transcription factors [310]. It also activates several glycolytic enzymes such as GLUT1, LDHA, PKM2, and HK2 [311], [312], [313]. The connection between hypoxia and mTOR is of particular interest. Although it has been shown that mTOR is able to induce HIF-1 translation, mTOR activity is reduced in hypoxia, likely through negative feedback [314], [315]. Hypoxia-mediated inhibition of mTOR could be through activation of tuberous sclerosis protein (TSC1/2) via AMPK, REDD1 or BNIP3 activation [117], [309], [316], [317]. However, there is also evidence that hypoxia-mediated inhibition of mTOR is more prevalent in normal cells compared with cancer cells [318]. Therefore, it may be concluded that mutations in the mTOR signaling pathway account for the reduced hypoxia-mediated mTOR inhibition. It was discovered that mTOR, along with p53, spares the available serine for glutathione synthesis by stimulation of PKM2 protein synthesis, which links glycolysis to anabolic pathways [319]. Moreover, mTOR suppresses autophagy and mitophagy and, therefore, produces ROS. AMP-activated protein kinase (AMPK), an mTOR inhibitor, plays a vital role in metabolic flux and regulates GLUT4 expression, mitochondrial biogenesis and fatty acid oxidation. Complex interaction between mTOR, AKT, and AMPK to regulate GLUT4 translation has also been shown [320]. Activated AKT phosphorylates and inhibits AS160 Rab GTPase activating protein in the cytoplasm leading to increased translocation of the insulin-responsive glucose transporter, GLUT4 to the membrane [321]. Also, ADP and ATP play a critical role in the stability of AKT phosphorylation at residues T308 and S473 and, therefore, act as on/off switches as ATP binds to these phosphorylated sites and protects them against phosphatases. Consequently, AMPK regulates AKT phosphorylation by responding to the equilibrium of the adenylate pool [320], [322]. On the other hand, Kumar et al. [323], reported that FRic−/− murine fat cells, with ablated Rictor, showed impaired insulin-stimulated GLUT4 translocation to the plasma membrane and decreased glucose transport.
Given the integral role that mTOR plays in oxygen and nutrient sensing, it is notable that several miRNAs may directly or indirectly influence mTOR activity. Increased expression of MTOR coexists with downregulation of several miRNAs in various types of cancer (Table 5 and Figure 2). Examples include miR-99a/b, miR-100, and miR-199b in cancers, including endometrial cancer, esophageal squamous cell carcinoma, and bladder cancer [324], [325], [326], [327]. miR-99 and miR-100 were also reported to be endogenous inhibitors of mTOR protein abundance [328]. miR-7 was found to target MTOR directly and form a negative feedback loop by also directly repressing EGFR and thus results in pleiotropic inhibition of protein translation [329], [330]. Chen et al. and Lin and Shao et al. [331], [332] reported a significant inhibition of mTOR expression, at both RNA and protein levels, by miR-101. Also, miR-373 and miR-520c were reported to reduce MTOR mRNA and protein levels and increase MMP9, which consequently results in the increased migration and invasion capability of cancer cells [333]. A negative regulator of mTOR is TSC1/2 complex. miR-451 was found to target TSC1 and stimulate the stemness phenotype of myeloma cells through activation of the PI3K/AKT/mTOR pathway [334], [335]. These findings further highlight the role of mTOR, situated at the crossroads of cancer-related signaling pathways. They show the interplay between components of signaling cascades and miRNAs, with practical implications for cancer therapy.
5.5. Tumor protein p53 (TP53)
p53 is a transcription factor and tumor suppressor that plays critical roles in controlling cell cycle progression through DNA damage response and apoptosis, which has been shown to regulate both glycolysis and OXPHOS [190]. In general, p53 inhibits glycolysis transcriptionally by supressing GLUT1, GLUT3, and PGM expression. Therefore, loss of p53 function in many cancers contributes to either glycolysis or the pentose phosphate pathway (PPP) [155], [336]. Mutated p53 was shown to reduce oxygen consumption and mitochondrial respiration. First, diminished p53 activity reduces OXPHOS by eliminating its suppression of SCO2, a protein essential for COX assembly and mitochondrial respiration [337]. Moreover, p53 may affect mtDNA by regulating the expression of ribonucleotide reductase subunit p53R2 and, ultimately, regulating mitochondrial oxidative respiration [338]. P53R2 plays important roles in both the biogenesis of mitochondria and mtDNA maintenance [339]. Although p53 induces oxidative stress by its pro-apoptotic function, it can also adversely impact redox maintenance [340]. Anti-oxidant roles of p53 include upregulation of GLS2 and subsequent increase in glutathione as well as enhanced stability of NRF2, an important antioxidant transcription factor, under oxidative stress [341], [342]. Other p53 functions that regulate metabolism include induced PTEN expression, which inhibits the PI3K pathway and glycolysis, cooperation with the OCT1 transcription factor to modulate the balance between glycolysis and OXPHOS and reduced fatty acid oxidation in response to metabolic flux [343], [344], [345].
The identification of several miRNAs that target p53 implies complex regulation and may explain the development of malignancies in cells with wild-type p53, where miRNA-mediated repression of TP53 and its transactivational genes, such as CDKN1A, BBC3, DNM1L, and BAX, is sufficient to cause tumourigenesis [346], [347]. p53 both regulates, and is regulated by, miRNAs. Many of these miRNAs were shown to directly target TP53 in different systems (summarized in Table 5). It is becoming clear that most of these miRNAs represent conservative regulation of p53 activity, targeting multiple components of the p53 pathway. Also, the functional overlap between these miRNAs indicates the potential for cumulative miRNA dysregulation influencing the p53 network during tumourigenesis. p53 suppresses glucose transporters and glycolytic enzymes by enhancing TIGAR [348]. TIGAR is best characterized by its negative regulation of fructose-2, 6-bisphosphatase. Eventually, TIGAR directs glucose to PPP and enhances NADPH production [349]. miR-144 targets TIGAR and modulates autophagy, apoptosis and metastasis in lung cancer cells [350]. In order to survive, cancer cells can also render p53 inactive by point mutation or through degradation induced by the E3 ubiquitin ligase, (MDM2) [351], [352]. Aside from gene mutations, promoter (de)methylation and proteolytic degradation, MDM2 is regulated by miRNAs. miRNAs such as miR-605 and miR-660 directly target MDM and modulate MDM:p53 interaction, aiding rapid stabilization and accumulation of p53. On the other hand, p53 trans-activates the expression of the miR-605 host gene PRKG1 through binding to its promoter region, which results in a positive feedback loop and increased p53 activity [353], [354]. Other miRNAs that suppress MDM2 include miR-509-5p in HCC and cervical cancer, miR-29b in non-small cell lung cancer (NSCLC), miR-143/145 in head and neck squamous cell carcinoma (HNSCC), miR-192, miR-215, miR-194, and miR-339-5p in renal cell adenocarcinoma, breast cancer, and colorectal cancer [355], [356], [357], [358] (Figure 2).
In addition to the aforementioned functions of p53 in regulating cell metabolism, miRNA biosynthesis also involves p53-signaling components. p53 interacts with the Drosha complex and accelerates the processing of targeted primary miRNA sequences to precursor miRNA fragments [359]. Specific miRNAs are also transcriptionally regulated by p53 [355], [360], [361] (Table 4). Most of these p53-responsive miRNAs are involved in both positive and negative feedback loops. For instance, members of the miR-34 family are induced through p53 binding to their promoter in response to stress and, in turn, TP53 mRNA has been validated as a direct target of miR-34 [362], [363]. miR-605 and miR-509-5p/MDM2/p53 are examples of positive feedback loops where p53 induces miRNA synthesis and miR-509-5p and miR-605 target MDM2 to increase p53 protein levels [353], [355]. miR-17-5p/TP53INP1/p53 is another regulatory feedback loop. miR-17-5p targets TP53INP1 mRNA transcript which encodes a p53-induced nuclear protein and also is a direct target of p53; so, miR-17-5p functions as a mediator in a regulatory loop in colon and cervical cancer [361]. Other miRNAs that target tumor protein P53 inducible nuclear protein 1 (TP53INP1) include miR-130b in hepatocarcinoma, miR-155 in pancreatic cells, and miR-125b in endometrial carcinoma [364], [365], [366]. Therefore, both regulation of the p53 network by miRNAs, and p53 induction of miRNA levels, are tightly coordinated to enable response to stimuli.
Table 4.
Summary of miRNAs regulated by the transcription factor p53.
| miRNA | Regulation | Disease/cells | References |
|---|---|---|---|
| Let-7a/b | Downregulation | Colorectal Cancer | [367] |
| miR-17 | Downregulation | Colorectal Cancer | [133] |
| miR-20a | Downregulation | Colorectal Cancer | [133] |
| miR-29 | Upregulation | Colorectal Cancer | [368] |
| miR-200c | Upregulation | Mammary Gland, Colorectal Cancer | [369], [370] |
| miR-183 | Upregulation | Mammary Gland | [369] |
| miR-34a/b/c | Upregulation | Colorectal Cancer, Non-Small Cell Lung Cancers, Ovary Clear Cell Carcinoma, Osteosarcoma, Pancreatic Cancer, Prostate Cancer, Ovarian Carcinoma | [362], [371], [372], [373], [374], [375] |
| miR-605 | Upregulation | Breast Cancer, Lung Carcinoma | [353] |
| miR-145 | Upregulation | Breas Cancer, Colorectal Cancer | [241] |
| miR-192 | Upregulation | Colorectal Cancer, Multiple Myeloma, Ovary Clear Cell Carcinoma, Osteosarcoma | [358], [370], [372], [376] |
| miR-194 | Upregulation | Multiple Myeloma, Colorectal Cancer | [358], [376] |
| miR-215 | Upregulation | Colorectal Cancer, Multiple Myeloma, Ovary Clear Cell Carcinoma, Osteosarcoma | [358], [370], [372], [376] |
| miR-141 | Upregulation | Colorectal Cancer | [370] |
| miR-519d | Upregulation | Hepatocellular Carcinoma | [286] |
| miR-107 | Upregulation | Colorectal Cancer | [151], [377] |
| miR-509 | Upregulation | Cervical Cancer, Hepatocellular Carcinoma | [355] |
These findings show that the p53 network is more complex than previously envisioned and suggest that additional regulatory layers, incorporating miRNAs, provide derepression of TP53 enabling it to accumulate rapidly in response to cell stress. The aforementioned functions establish a new driver of the Warburg effect and demonstrate that p53 may act as a “brake” on glycolysis and neoplastic cell proliferation.
5.6. Sirtuins
Sirtuins are a conserved family of NAD+-dependent deacetylases. Advances in sirtuin biology have identified multiple targets for the seven mammalian sirtuins (SIRT1-7) and, recently, their participation in tumourigenesis and regulation of cancer cell metabolism [378].
SIRT1 is a nuclear protein that shuttles between the nucleus and cytoplasm, especially when insulin signaling is inhibited [379]. SIRT1 modulates several cellular pathways by deacetylating a subset of nuclear and cytosolic targets. AMPK and SIRT1 cooperate in the induction of gluconeogenesis, glycolysis and lipid catabolism, mitochondrial biogenesis and respiration by phosphorylation and then deacetylation of PPARgamma coactivator 1alpha (PGC1α) and FOXO transcription factors [380], [381], [382], [383]. A homeostatic and negative feedback loop has been reported among SIRT1, p53, FOXO3A, and FOXO1. During energy stress FOXO3A binds to p53 promoter, repressing SIRT1 expression, and in turn SIRT1 inhibits p53 activity by excessive deacetylation and also through FOXO3A activation [384], [385], [386]. In addition, SIRT1 is involved in the oxidative response, working together with HIF-1, p53, and Myc [387].
SIRT6 is another member of the sirtuin nuclear histone deacetylase (HDAC) family, which exerts both nuclear ADP-ribosyltransferase activity and deacetyltransferase activity with roles in epigenetic regulation of genomic stability, cellular metabolism, stress response, aging, and cancer [388], [389], [390], [391]. Yin and Gao et al. [392] showed that neuronal SIRT6 overexpression significantly suppresses insulin-like factor 2 (IGF2) activity and other proteins such as AKT and mTOR at the chromatin level. SIRT6 activation results in inhibition of HIF-1, glycolysis, and respiration, as well as induction of homologous and non-homologous DNA repair. The latter function of SIRT6 occurs through ADP-ribosylation of poly(ADP-Ribose) polymerase 1 (PARP-1) [393], [394]. SIRT6 regulates HIF-1 and c-Myc expression, at the transcriptional level, through chromatin deacetylation and also regulates HIF-1 stability through an unknown mechanism [395]. Mostoslavsky et al. [396] reported a novel role for SIRT6 in glucose homeostasis in mice. Accordingly, subsequent studies confirmed its vital role in direct and indirect regulation of glucose uptake and metabolism. Nevertheless, SIRT6 was found to transcriptionally regulate some c-Myc targets involved in ribosomal biogenesis and glutamine utilization, rather than those involved in regulating cancer cell glycolysis [397]. In contrast to SIRT6 that acts independently, SIRT1 cooperates with c-Myc to suppress p53 activity and increase c-Myc-induced LDHA expression [395]. SIRT7 is a nucleolar sirtuin member that activates transcription by binding to RNA polymerase [398], [399]. Vakhrusheva et al. [400] reported a p53 hyperacetylation state in SIR7 knockout mice, which results in increased apoptosis and decreased resistance to oxidative stress.
The miRNA-mediated regulation of nuclear sirtuins, with an emphasis on SIRT1 and SIRT6, has highlighted their roles in glycolysis. SIRT1 has been the most extensively studied member in this context. miR-34a was the first discovered SIRT1 targeting miRNA. miR-34 is a p53-related miRNA that most importantly regulates cell cycle. miR-34 downregulates SIRT1 expression by directly binding to its 3′UTR and indirectly through targeting nicotinamide phosphoribosyltransferase (NAMPT), the rate limiting enzyme in NAD+ biosynthesis [401], [402], [403]. Xu et al. [404] reported SIRT1 targeting by miR-22, which modulates the retinoblastoma signaling pathway. miR-204 targets SIRT1 in osteocarcinoma cells and inhibits epithelial–mesenchymal transition (EMT) of the cancer cells [405]. Similarly, miR-200c has been reported to form a negative feedback loop with SIRT1, attenuating epithelial to mesenchymal transition (EMT) in breast cancer cells [406]. Likewise, miR-181a and miR-9 regulate SIRT1 and impact insulin signaling, glucose homeostasis and cell apoptosis [407], [408], [409]. miR-143, miR-93 and miR-217 lead to decreased glucose uptake, downregulated microsomal glutathione S-transferase 1 and inhibited angiogenesis, respectively, by targeting SIRT1 [410], [411], [412]. Several other miRNAs that modulate SIRT1 expression and activation include miR-9, miR-34c, miR-132, miR-135, miR-146, miR-181b, miR-195, miR-199, and miR-499 [413], [414], [415], [416], [417] (Figure 2).
Post-transcriptional regulation of SIRT6 by miR-33a/b plays a vital role in regulation of cholesterol and lipid metabolism via acetylation of its targets [392], [418], [419], [420]. Sharma et al. [421] reported a negative feedback loop between SIRT6 and miR-766 in dermal fibroblasts. SIRT7 expression is elevated in highly metabolic and proliferative cells and was reported to be a target of miR-125a/b inducing G1 cell cycle arrest [422].
SIRT2 is predominantly cytosolic but it also shuttles to the nucleus and is mainly enriched in the brain [423]. SIRT2 was reported to deacetylate histone H3, p300, FOXO1, FOXO3A, adenomatous polyposis coli (APC), cell division cycle 20 (CDC20), p65, PGM, phosphoenolpyruvate carboxykinase 2 (PEPCK), and receptor-interacting protein 1 (RIP1) and, therefore, regulates cell cycle, genome integrity, energy homeostasis, gluconeogenesis, glycolysis, oxidative stress modulation, cell growth, and death [424], [425], [426], [427], [428], [429], [430]. miR-339 was shown to target SIRT2, increasing NF-KB and FOXO1 acetylation in neuroblastoma cells [431]. Moreover, in silico analysis revealed a longevity associated SNP of SIRT2 within the binding site of three miRNAs (called miRSNPs). Therefore, miR-3170, miR-92a-1-5p and, more importantly, miR-615-5p were predicted to target SIRT2 resulting in reduction in SIRT2 expression [432]. Li and Dai et al. [284] showed that SIRT2 is downregulated in glioma. SIRT2 acts as a tumor suppressor and inhibits glioma growth by targeting miR-21 expression through deacetylating p65 and blocking p65 binding to the miR-21 promoter. Regulation of miR-21 activity is particularly important as this miRNA displays significant oncogenic activity [433].
Three mitochondrial sirtuins are SIRT3, SIRT4, and SIRT5. SIRT3 is the major mitochondrial sirtuin, which promotes ATP production by regulating TCA cycle enzymes such as acetyl COA synthetase, IDH2, glutamate dehydrogenase 1 (GDH) and SDH during energy stress. GDH upregulation leads to an induction in glutamine metabolism which consequently produces more ATP and releases insulin [434], [435], [436], [437]. It also upregulates Mn superoxide dismutase (MnSOD), downregulates ROS, HIF-1, and p53 and activates FOXO3A to modulate redox homeostasis and maintain mitochondrial membrane potential [438], [439], [440], [441], [442]. SIRT3-mediated regulation of OXPHOS components has been shown. The targets include components of complex I, II, III and V such as NDUFA11/S8, SDHA/B and ATP5A1/B1/F1 [438], [443], [444]. Altogether, SIRT3 is capable of reversing the Warburg effect toward mitochondrial predominance and ATP synthesis. SIRT4 is another mitochondrial sirtuin which seems to function as a tumor suppressor by downregulating GDH through ADP-ribosylation activity and consequently suppressing glutamine utilization and the flow of amino acids into the TCA cycle [434]. Nasrin et al. [445] showed that reduced SIRT4 results in increased fatty acid oxidation and mitochondrial metabolism. They also demonstrated an increase in SIRT1 expression levels. miR-193 mediated suppression of SIRT3 leads to impaired energy metabolism and ATP synthesis in myocardium [446]. miR-23a was also shown to target PGC1 and thereby indirectly modulate SIRT3 expression [447]. Moreover, upregulation of miR-28-5p, resulting from oxidative stress, directly targets SIRT3 [448]. Liang et al. [449] reported SNPs in the miR-105 and miR-532 binding sites in the SIRT3 3′UTR that are associated with ovarian cancer treatment responses. In addition, Slaby et al. [450] reported three miRNAs that are regulated by natural agents called isothiocyanates in colorectal cancer (CRC) cells. In silico analysis also revealed CRC-related SNPs within the 3′UTR of genes, including SIRT5, may influence binding of these isothiocyanate-regulated miRNAs.
Collectively, SIRTs play important roles in a wide range of metabolic pathways and interact with many transcriptional regulators. miRNAs targeting SIRTs (summarized in Table 5) may modulate SIRT-related signaling transduction and downstream effectors, providing insight into novel therapeutic strategies.
6. Towards future applications for disrupting cancer cell glycolysis
Metabolomics provides a new exciting platform to explore potential anti-cancer drugs. A universally observed phenotype of malignant cells is their propensity to import glucose and secrete lactate, even in the presence of oxygen. The characterization of aerobic glycolysis has led to dramatic advances in tumor imaging. Positron emission tomography (PET) scans, widely used for cancer diagnosis, exploit the ability of cancer cells to sequester excessive glucose from the blood stream. Ever since aerobic glycolysis was found to be a characteristic of tumor cells and was accepted as a hallmark of cancer, it has been proposed that suppressing aerobic glycolysis would be a promising strategy to treat cancer. As a consequence, several studies have reported the use of glycolytic enzyme inhibitors. For instance, lonidamine as a HK2 inhibitor, PEP analogues as PKM inhibitors, as well as FX-11 and panepoxydone as LDHA inhibitors, have been considered potential therapeutic agents (reviewed in [451], [452]). However, as glycolysis is also a vital metabolic pathway in normal cells, inhibition of aerobic glycolysis remains challenging when identifying potential cancer-specific targets. Although a definitive explanation for Warburg's observations is overdue, the control of this process by oncogenes and tumor suppressors, coupled with epigenetic factors including microRNAs, provides additional insight. So far, ample evidence supports associations between the metabolic shift in cancer cells and oncogene activation or inactivation of tumor suppressors. The elusive nature of metabolic rewiring and branching in cancer cells, along with influences upon other signaling pathways, raise concerns as to whether targeting a single component of this complex circuit will be sufficient to eradicate cancer cells with minimal side effects. Despite several reports of the involvement of miRNA-mediated gene regulation, there is still much to learn about how miRNAs contribute to the Warburg effect. Development of new miRNA-mediated strategies, that target metabolic pathways rather than single components, have the potential to enhance future cancer treatment. Systems biology approaches that iteratively couple massively parallel gene expression analytical technologies with high throughput functional screens, may identify additional miRNAs or miRNA-targets with promise for cancer diagnosis, prognosis and drug development. Polymorphisms in the miRNA binding sites of oncogenes are known to influence cancer predisposition and therapeutic response, which may further inform target selection [198], [453]. Conversely, acquired somatic mutations in miRNA-binding sites may also lead to the reduced efficacy of miRNA-based therapies. Similarly, the demands of other tissues, such as the highly glucose-dependent nature of brain and retina, will necessitate tissue-specific delivery of anti-glycolysis miRNAs in a therapeutic context where administration already presents challenges. Regardless, multi-faceted solutions are required to provide hope for cancer patients who currently have limited options.
7. Conclusion
Aerobic glycolysis, a hallmark of cancer, is the consequence of specific driver mutations and re-equilibrated homeostasis in tumor cells. While cellular responses to the environment continue to involve the existing signaling pathways, longer adaptive responses invoke post-transcriptional and epigenetic control of gene expression. By regulating multiple cellular pathways and multiple components of individual pathways, microRNAs fine-tune expression to ensure high level buffering of adaptive responses. Thorough understanding of these regulatory processes should provide the capacity to suppress metabolism and inhibit cancer cell survival under stress. With the advent of RNA-based therapies and the development of drugs that modulate the activity of microRNA targets, or even microRNAs themselves, this review has highlighted metabolic processes that may be disrupted by novel therapeutic interventions
Authors' contributions
AVO structured and drafted the article, she also designed and produced the tables and figures. MZM contributed to the structure, design of tables, writing and revision of the article. JP and RAM participated in revising the article critically for important content.
Funding
AVO is supported by a Research Training Program Scholarship through Flinders University. AVO and MZM were supported by research grants from Tour de Cure, Australia and Flinders Foundation. JP was supported by a research grant from Flinders Foundation. RAM is supported by the Beat Cancer Project, funded by Cancer Council SA and the South Australian Government. The funding bodies did not influence the content of this manuscript.
Conflict of interest
None declared.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.01.014.
Contributor Information
Ayla V. Orang, Email: ayla.orang@flinders.edu.au.
Janni Petersen, Email: janni.petersen@flinders.edu.au.
Ross A. McKinnon, Email: ross.mckinnon@flinders.edu.au.
Michael Z. Michael, Email: michael.michael@flinders.edu.au.
Abbreviations
- 2HG
2-hydroxyglutarate
- AGO
argonaute
- ALDO
aldolase
- AMPK
AMP-activated protein kinase
- APC
adenomatous polyposis coli
- C/EBPβ
CCAAT-enhancer-binding protein β
- CDC20
cell division cycle 20
- COX
cytochrome c oxidase complex
- CUL2
cullin 2
- DNMT1
DNA (cytosine-5)-methyltransferase 1
- EMT
epithelial to mesenchymal transition
- ENO1
enolase 1
- EphA1
erythropoietin-producing hepatoma receptor A1
- ERK
extracellular-signal-regulated kinase
- EZH2
enhancer of zeste homolog 2
- FIH-1
factor inhibiting HIF-1
- FOXO
forkhead box O
- GAPDH
glyceraldehyde-3- phosphate dehydrogenase
- GDH
glutamate dehydrogenase 1
- GLS
glutaminase
- GLS
hypoxia-inducible factor-1
- GLUTS:
glucose transporters
- GPD1L:
glycerol-3-phosphate dehydrogenase 1 like
- HDAC
histone deacetylase
- Her2/Neu
human epidermal growth factor receptor 2
- HK2
hexokinase 2
- HNF3β
hepatocyte nuclear factor 3-beta
- HRE
hypoxia response element
- IDH
isocitrate dehydrogenase
- IGF2
insulin-like factor 2
- ISCU
iron-sulfur cluster scaffold
- KLF15
Kruppel-like factor 15
- LDHA
lactate dehydrogenase A
- MAGI2
membrane-associated guanylate kinase inverted 2
- MAX
MYC associated factor X
- MCT
monocarboxylate transporters
- MDH
malate dehydrogenase
- miRNA
microRNA
- mitomiRs
mitochondria-related microRNAs
- MMP-2
metalloproteinase-2
- MnSOD
Mn Superoxide Dismutase
- mTOR
mechanistic target of rapamycin
- NOX
NADPH oxidase
- OXPHOS
oxidative phosphorylation
- PARP-1
poly(ADP-ribose) polymerase 1
- PDH
pyruvate dehydrogenase
- PDK1
pyruvate dehydrogenase kinase
- PEPCK
phosphoenolpyruvate carboxykinase 2
- PET
positron emission tomography
- PFK1
6-phosphofructo-1-kinase
- PGC1α
PPARgamma coactivator 1alpha
- PGK1
phosphoglycerate kinase 1
- PGM
phosphoglycerate mutase
- PHD
prolyl-4-hydroxylase
- PKM1/2
pyruvate kinase isozymes M1/M2
- PPP2R1B
protein phosphatase 2 scaffold subunit Abeta
- pre-miRNA
precursor microRNA
- pri-miRNA
primary microRNA
- PTEN
phosphatase and tensin homolog
- RIP1
receptor-interacting protein 1
- RISC
RNA-induced silencing complex
- ROS
reactive oxygen species
- SCO2/1
synthesis of cytochrome c oxidase 2/1
- SDH
succinate dehydrogenase
- SIRT
sirtuin
- STAT3
signal transducer and activator of transcription 3
- TCA
tricarboxylic acid
- TGF-β:
transforming growth factor beta 1
- TIGAR
TP53-induced glycolysis and apoptosis regulator
- TLK
transketolase
- TP53INP1
tumor protein P53 inducible nuclear protein 1
- TPI
triose-phosphate isomerase
- TSC
tuberous sclerosis proteins
- USP28
ubiquitin specific peptidase 28
- VEGFA
vascular endothelial growth factor A
- VHL:
Von Hippel-Lindau
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 2.Zhang H., Wang Y., Xu T., Li C., Wu J., He Q. Increased expression of microRNA-148a in osteosarcoma promotes cancer cell growth by targeting PTEN. Oncology Letters. 2016;12(5):3208–3214. doi: 10.3892/ol.2016.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., Capaldi R.A. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Research. 2004;64(3):985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L., Cardaci S., Jerby L., MacKenzie E.D., Sciacovelli M., Johnson T.I. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nature Communications. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardella C., Pollard P.J., Tomlinson I. SDH mutations in cancer. Biochimica et Biophysica Acta. 2011;1807(11):1432–1443. doi: 10.1016/j.bbabio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nature Reviews Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 8.Varmus H., Pao W., Politi K., Podsypanina K., Du Y.C. Oncogenes come of age. Cold Spring Harbor Symposia on Quantitative Biology. 2005;70:1–9. doi: 10.1101/sqb.2005.70.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P.K., Mehla K., Hollingsworth M.A., Johnson K.R. Regulation of aerobic glycolysis by microRNAs in cancer. Molecular and Cellular Pharmacology. 2011;3(3):125–134. [PMC free article] [PubMed] [Google Scholar]
- 10.Ruby J.G., Jan C.H., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 12.Valinezhad Orang A., Safaralizadeh R., Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. International Journal of Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer D.E., Harris M.H., Plas D.R., Lum J.J., Hammerman P.S., Rathmell J.C. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. The FASEB Journal. 2004;18(11):1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramanathan A., Wang C., Schreiber S.L. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(17):5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong X., Zhao F., Thompson C.B. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Current Opinion in Genetics & Development. 2009;19(1):32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature Reviews Cancer. 2007;7(10):763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 17.Horie T., Ono K., Nishi H., Iwanaga Y., Nagao K., Kinoshita M. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochemical and Biophysical Research Communications. 2009;389(2):315–320. doi: 10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- 18.Jiang S., Zhang L.F., Zhang H.W., Hu S., Lu M.H., Liang S. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. The EMBO Journal. 2012;31(8):1985–1998. doi: 10.1038/emboj.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao M., Wang X., Tang Y., Zhang W., Cui B., Liu Q. Dicer mediating the expression of miR-143 and miR-155 regulates hexokinase II associated cellular response to hypoxia. American Journal of Physiology – Lung Cellular and Molecular Physiology. 2014;307(11):L829–L837. doi: 10.1152/ajplung.00081.2014. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi K., Ito Y., Sugito N., Kumazaki M., Shinohara H., Yamada N. Organ-specific PTB1-associated microRNAs determine expression of pyruvate kinase isoforms. Scientific Reports. 2015;5:8647. doi: 10.1038/srep08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y., Zhao X., Zhou Y., Hu Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncology Reports. 2012;28(4):1346–1352. doi: 10.3892/or.2012.1958. [DOI] [PubMed] [Google Scholar]
- 22.Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Letters. 2015;363(1):17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 23.Sugiyama T., Taniguchi K., Matsuhashi N., Tajirika T., Futamura M., Takai T. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Science. 2016;107(12):1767–1775. doi: 10.1111/cas.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi K., Sakai M., Sugito N., Kumazaki M., Shinohara H., Yamada N. PTBP1-associated microRNA-1 and -133b suppress the Warburg effect in colorectal tumors. Oncotarget. 2016;7(14):18940–18952. doi: 10.18632/oncotarget.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamasaki T., Seki N., Yoshino H., Itesako T., Yamada Y., Tatarano S. Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Science. 2013;104(11):1411–1419. doi: 10.1111/cas.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow T.F., Mankaruos M., Scorilas A., Youssef Y., Girgis A., Mossad S. The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. The Journal of Urology. 2010;183(2):743–751. doi: 10.1016/j.juro.2009.09.086. [DOI] [PubMed] [Google Scholar]
- 27.Nie S., Li K., Huang Y., Hu Q., Gao X., Jie S. miR-495 mediates metabolic shift in glioma cells via targeting Glut1. Journal of Craniofacial Surgery. 2015;26(2):e155–e158. doi: 10.1097/SCS.0000000000001385. [DOI] [PubMed] [Google Scholar]
- 28.Chen B., Tang H., Liu X., Liu P., Yang L., Xie X. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Letters. 2015;356(2 Pt B):410–417. doi: 10.1016/j.canlet.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 29.Qu W., Ding S.M., Cao G., Wang S.J., Zheng X.H., Li G.H. miR-132 mediates a metabolic shift in prostate cancer cells by targeting Glut1. FEBS Open Bio. 2016;6(7):735–741. doi: 10.1002/2211-5463.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King B.C., Esguerra J.L., Golec E., Eliasson L., Kemper C., Blom A.M. CD46 activation regulates mir-150-mediated control of GLUT1 expression and cytokine secretion in human CD4+ T cells. The Journal of Immunology. 2016;196(4):1636–1645. doi: 10.4049/jimmunol.1500516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P., Yang X., Cheng Y., Zhang X., Yang C., Deng X. MicroRNA-218 increases the sensitivity of bladder cancer to cisplatin by targeting Glut1. Cellular Physiology and Biochemistry. 2017;41(3):921–932. doi: 10.1159/000460505. [DOI] [PubMed] [Google Scholar]
- 32.Xu P., Li Y., Zhang H., Li M., Zhu H. MicroRNA-340 mediates metabolic shift in oral squamous cell carcinoma by targeting glucose transporter-1. Journal of Oral and Maxillofacial Surgery. 2016;74(4):844–850. doi: 10.1016/j.joms.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 33.Guo H., Nan Y., Zhen Y., Zhang Y., Guo L., Yu K. miRNA-451 inhibits glioma cell proliferation and invasion by downregulating glucose transporter 1. Tumour Biology. 2016;37(10):13751–13761. doi: 10.1007/s13277-016-5219-3. [DOI] [PubMed] [Google Scholar]
- 34.Trakooljul N., Hicks J.A., Liu H.C. Identification of target genes and pathways associated with chicken microRNA miR-143. Animal Genetics. 2010;41(4):357–364. doi: 10.1111/j.1365-2052.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 35.Fei X., Qi M., Wu B., Song Y., Wang Y., Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Letters. 2012;586(4):392–397. doi: 10.1016/j.febslet.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Dai D.W., Lu Q., Wang L.X., Zhao W.Y., Cao Y.Q., Li Y.N. Decreased miR-106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer. 2013;13:478. doi: 10.1186/1471-2407-13-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu H., Buchan R.J., Cook S.A. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovascular Research. 2010;86(3):410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 38.Karolina D.S., Armugam A., Tavintharan S., Wong M.T., Lim S.C., Sum C.F. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0022839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y.H., Heneidi S., Lee J.M., Layman L.C., Stepp D.W., Gamboa G.M. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62(7):2278–2286. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou T., Meng X., Che H., Shen N., Xiao D., Song X. Regulation of insulin resistance by multiple MiRNAs via targeting the GLUT4 signalling pathway. Cellular Physiology and Biochemistry. 2016;38(5):2063–2078. doi: 10.1159/000445565. [DOI] [PubMed] [Google Scholar]
- 41.Peschiaroli A., Giacobbe A., Formosa A., Markert E.K., Bongiorno-Borbone L., Levine A.J. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32(6):797–802. doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 42.Kim H.R., Roe J.S., Lee J.E., Cho E.J., Youn H.D. p53 regulates glucose metabolism by miR-34a. Biochemical and Biophysical Research Communications. 2013;437(2):225–231. doi: 10.1016/j.bbrc.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 43.Zhao S., Liu H., Liu Y., Wu J., Wang C., Hou X. miR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Letters. 2013;333(2):253–260. doi: 10.1016/j.canlet.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 44.Jiang J.X., Gao S., Pan Y.Z., Yu C., Sun C.Y. Overexpression of microRNA-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Molecular Medicine Reports. 2014;10(2):995–1002. doi: 10.3892/mmr.2014.2271. [DOI] [PubMed] [Google Scholar]
- 45.Song J., Wu X., Liu F., Li M., Sun Y., Wang Y. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochemical and Biophysical Research Communications. 2017;490(2):217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 46.Li W., Hao J., Zhang L., Cheng Z., Deng X., Shu G. Astragalin reduces hexokinase 2 through increasing mir-125b to inhibit the proliferation of hepatocellular carcinoma cells in vitro and in vivo. Journal of Agricultural and Food Chemistry. 2017;65(29):5961–5972. doi: 10.1021/acs.jafc.7b02120. [DOI] [PubMed] [Google Scholar]
- 47.Tao T., Chen M., Jiang R., Guan H., Huang Y., Su H. Involvement of EZH2 in aerobic glycolysis of prostate cancer through miR-181b/HK2 axis. Oncology Reports. 2017;37(3):1430–1436. doi: 10.3892/or.2017.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu W., Huang Y., Pan Q., Xiang P., Xie N., Yu H. MicroRNA-98 suppress Warburg effect by targeting HK2 in colon cancer cells. Digestive Diseases and Sciences. 2017;62(3):660–668. doi: 10.1007/s10620-016-4418-5. [DOI] [PubMed] [Google Scholar]
- 49.Li L.Q., Yang Y., Chen H., Zhang L., Pan D., Xie W.J. MicroRNA-181b inhibits glycolysis in gastric cancer cells via targeting hexokinase 2 gene. Cancer Biomarkers. 2016;17(1):75–81. doi: 10.3233/CBM-160619. [DOI] [PubMed] [Google Scholar]
- 50.Qin Y., Cheng C., Lu H., Wang Y. miR-4458 suppresses glycolysis and lactate production by directly targeting hexokinase2 in colon cancer cells. Biochemical and Biophysical Research Communications. 2016;469(1):37–43. doi: 10.1016/j.bbrc.2015.11.066. [DOI] [PubMed] [Google Scholar]
- 51.Lan H., Luo L., Qi X., Gong Y., Chen Y. [miR-181c inhibits glycolysis by targeting hexokinase 2 in cancer-associated fibroblasts] Nan Fang Yi Ke Da Xue Xue Bao. 2015;35(11):1619–1623. [PubMed] [Google Scholar]
- 52.Zhou P., Chen W.G., Li X.W. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. American Journal of Cancer Research. 2015;5(6):2056–2063. [PMC free article] [PubMed] [Google Scholar]
- 53.Guo W., Qiu Z., Wang Z., Wang Q., Tan N., Chen T. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62(4):1132–1144. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]
- 54.Rengaraj D., Park T.S., Lee S.I., Lee B.R., Han B.K., Song G. Regulation of glucose phosphate isomerase by the 3'UTR-specific miRNAs miR-302b and miR-17-5p in chicken primordial germ cells. Biology of Reproduction. 2013;89(2):33. doi: 10.1095/biolreprod.112.105692. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad A., Aboukameel A., Kong D., Wang Z., Sethi S., Chen W. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Research. 2011;71(9):3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park Y.Y., Kim S.B., Han H.D., Sohn B.H., Kim J.H., Liang J. Tat-activating regulatory DNA-binding protein regulates glycolysis in hepatocellular carcinoma by regulating the platelet isoform of phosphofructokinase through microRNA 520. Hepatology. 2013;58(1):182–191. doi: 10.1002/hep.26310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang H., Lee M., Sharpe O., Salamone L., Noonan E.J., Hoang C.D. Oxidative stress-responsive microRNA-320 regulates glycolysis in diverse biological systems. The FASEB Journal. 2012;26(11):4710–4721. doi: 10.1096/fj.11-197467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White N.M., Bui A., Mejia-Guerrero S., Chao J., Soosaipillai A., Youssef Y. Dysregulation of kallikrein-related peptidases in renal cell carcinoma: potential targets of miRNAs. Biological Chemistry. 2010;391(4):411–423. doi: 10.1515/BC.2010.041. [DOI] [PubMed] [Google Scholar]
- 59.Du J.Y., Wang L.F., Wang Q., Yu L.D. miR-26b inhibits proliferation, migration, invasion and apoptosis induction via the downregulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 driven glycolysis in osteosarcoma cells. Oncology Reports. 2015;33(4):1890–1898. doi: 10.3892/or.2015.3797. [DOI] [PubMed] [Google Scholar]
- 60.Seliger B., Jasinski S., Dressler S.P., Marincola F.M., Recktenwald C.V., Wang E. Linkage of microRNA and proteome-based profiling data sets: a perspective for the priorization of candidate biomarkers in renal cell carcinoma? Journal of Proteome Research. 2011;10(1):191–199. doi: 10.1021/pr1011137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ge X., Lyu P., Cao Z., Li J., Guo G., Xia W. Overexpression of miR-206 suppresses glycolysis, proliferation and migration in breast cancer cells via PFKFB3 targeting. Biochemical and Biophysical Research Communications. 2015;463(4):1115–1121. doi: 10.1016/j.bbrc.2015.06.068. [DOI] [PubMed] [Google Scholar]
- 62.Savarimuthu Francis S.M., Davidson M.R., Tan M.E., Wright C.M., Clarke B.E., Duhig E.E. MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genomics. 2014;15:88. doi: 10.1186/1471-2164-15-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calin G.A., Cimmino A., Fabbri M., Ferracin M., Wojcik S.E., Shimizu M. MiR-15a and miR-16-1 cluster functions in human leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(13):5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boesch-Saadatmandi C., Wagner A.E., Wolffram S., Rimbach G. Effect of quercetin on inflammatory gene expression in mice liver in vivo – role of redox factor 1, miRNA-122 and miRNA-125b. Pharmacological Research. 2012;65(5):523–530. doi: 10.1016/j.phrs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Nana-Sinkam S.P., Croce C.M. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biology. 2014;15(9):445. doi: 10.1186/s13059-014-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sikand K., Singh J., Ebron J.S., Shukla G.C. Housekeeping gene selection advisory: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-actin are targets of miR-644a. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White N.M., Newsted D.W., Masui O., Romaschin A.D., Siu K.W., Yousef G.M. Identification and validation of dysregulated metabolic pathways in metastatic renal cell carcinoma. Tumour Biology. 2014;35(3):1833–1846. doi: 10.1007/s13277-013-1245-6. [DOI] [PubMed] [Google Scholar]
- 68.Ichimi T., Enokida H., Okuno Y., Kunimoto R., Chiyomaru T., Kawamoto K. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. International Journal of Cancer. 2009;125(2):345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 69.Li Y., Kong D., Ahmad A., Bao B., Dyson G., Sarkar F.H. Epigenetic deregulation of miR-29a and miR-1256 by isoflavone contributes to the inhibition of prostate cancer cell growth and invasion. Epigenetics. 2012;7(8):940–949. doi: 10.4161/epi.21236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muniyappa M.K., Dowling P., Henry M., Meleady P., Doolan P., Gammell P. MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. European Journal of Cancer. 2009;45(17):3104–3118. doi: 10.1016/j.ejca.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 71.Taguchi A., Yanagisawa K., Tanaka M., Cao K., Matsuyama Y., Goto H. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Research. 2008;68(14):5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 72.Leder A., Raschzok N., Schmidt C., Arabacioglu D., Butter A., Kolano S. Micron-sized iron oxide-containing particles for microRNA-targeted manipulation and MRI-based tracking of transplanted cells. Biomaterials. 2015;51:129–137. [Google Scholar]
- 73.Ramirez C.M., Goedeke L., Rotllan N., Yoon J.H., Cirera-Salinas D., Mattison J.A. MicroRNA 33 regulates glucose metabolism. Molecular and Cellular Biology. 2013;33(15):2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong T.S., Liu X.B., Chung-Wai Ho A., Po-Wing Yuen A., Wai-Man Ng R., Ignace Wei W. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. International Journal of Cancer. 2008;123(2):251–257. doi: 10.1002/ijc.23583. [DOI] [PubMed] [Google Scholar]
- 75.Kefas B., Comeau L., Erdle N., Montgomery E., Amos S., Purow B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro-Oncology. 2010;12(11):1102–1112. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W., Wang J., Chen Q.D., Qian X., Li Q., Yin Y. Insulin promotes glucose consumption via regulation of miR-99a/mTOR/PKM2 pathway. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0064924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tao T., Li G., Dong Q., Liu D., Liu C., Han D. Loss of SNAIL inhibits cellular growth and metabolism through the miR-128-mediated RPS6KB1/HIF-1alpha/PKM2 signaling pathway in prostate cancer cells. Tumour Biology. 2014;35(9):8543–8550. doi: 10.1007/s13277-014-2057-z. [DOI] [PubMed] [Google Scholar]
- 78.Kinoshita T., Nohata N., Yoshino H., Hanazawa T., Kikkawa N., Fujimura L. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. International Journal of Oncology. 2012;40(1):185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- 79.Li D.F., Tian J., Guo X., Huang L.M., Xu Y., Wang C.C. Induction of microRNA-24 by HIF-1 protects against ischemic injury in rat cardiomyocytes. Physiological Research. 2012;61(6):555–565. doi: 10.33549/physiolres.932270. [DOI] [PubMed] [Google Scholar]
- 80.Saumet A., Vetter G., Bouttier M., Antoine E., Roubert C., Orsetti B. Estrogen and retinoic acid antagonistically regulate several microRNA genes to control aerobic glycolysis in breast cancer cells. Molecular BioSystems. 2012;8(12):3242–3253. doi: 10.1039/c2mb25298h. [DOI] [PubMed] [Google Scholar]
- 81.Kaller M., Liffers S.T., Oeljeklaus S., Kuhlmann K., Roh S., Hoffmann R. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Molecular & Cellular Proteomics. 2011;10(8) doi: 10.1074/mcp.M111.010462. M111 010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J., Wang H., Liu A., Fang C., Hao J., Wang Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget. 2015;6(23):19456–19468. doi: 10.18632/oncotarget.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Liu G., Gao X. Attenuation of miR-34a protects cardiomyocytes against hypoxic stress through maintenance of glycolysis. Bioscience Reports. 2017;37(6) doi: 10.1042/BSR20170925. BSR20170925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li L., Kang L., Zhao W., Feng Y., Liu W., Wang T. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Letters. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 85.Han R.L., Wang F.P., Zhang P.A., Zhou X.Y., Li Y. miR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma. 2017;64(2):244–252. doi: 10.4149/neo_2017_211. [DOI] [PubMed] [Google Scholar]
- 86.Zhang R., Su J., Xue S.L., Yang H., Ju L.L., Ji Y. HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. American Journal of Cancer Research. 2016;6(2):312–320. [PMC free article] [PubMed] [Google Scholar]
- 87.Mi Y., Guo N., He T., Ji J., Li Z., Huang P. miR-410 enhanced hESC-derived pancreatic endoderm transplant to alleviate gestational diabetes mellitus. Journal of Molecular Endocrinology. 2015;55(3):219–229. doi: 10.1530/JME-15-0100. [DOI] [PubMed] [Google Scholar]
- 88.Chen Y., Wang X., Shao X. A combination of human embryonic stem cell-derived pancreatic endoderm transplant with LDHA-repressing miRNA can attenuate high-fat diet induced type II diabetes in mice. Journal of Diabetes Research. 2015;2015:796912. doi: 10.1155/2015/796912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pullen T.J., da Silva Xavier G., Kelsey G., Rutter G.A. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1) Molecular and Cellular Biology. 2011;31(15):3182–3194. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li K.K., Pang J.C., Ching A.K., Wong C.K., Kong X., Wang Y. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Human Pathology. 2009;40(9):1234–1243. doi: 10.1016/j.humpath.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 91.Liang D., Zhang Y., Han J., Wang W., Liu Y., Li J. Embryonic stem cell-derived pancreatic endoderm transplant with MCT1-suppressing miR-495 attenuates type II diabetes in mice. Endocrine Journal. 2015;62(10):907–920. doi: 10.1507/endocrj.EJ15-0186. [DOI] [PubMed] [Google Scholar]
- 92.Zu X.L., Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochemical and Biophysical Research Communications. 2004;313(3):459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 93.Mathupala S.P., Ko Y.H., Pedersen P.L. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochimica et Biophysica Acta. 2010;1797(6–7):1225–1230. doi: 10.1016/j.bbabio.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ward P.S., Patel J., Wise D.R., Abdel-Wahab O., Bennett B.D., Coller H.A. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Janeway K.A., Kim S.Y., Lodish M., Nose V., Rustin P., Gaal J. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):314–318. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Linehan W.M., Rouault T.A. Molecular pathways: fumarate hydratase-deficient kidney cancer – targeting the Warburg effect in cancer. Clinical Cancer Research. 2013;19(13):3345–3352. doi: 10.1158/1078-0432.CCR-13-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moreadith R.W., Lehninger A.L. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. Journal of Biological Chemistry. 1984;259(10):6215–6221. [PubMed] [Google Scholar]
- 98.Chowdhury R., Yeoh K.K., Tian Y.M., Hillringhaus L., Bagg E.A., Rose N.R. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Reports. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li P., Jiao J., Gao G., Prabhakar B.S. Control of mitochondrial activity by miRNAs. Journal of Cellular Biochemistry. 2012;113(4):1104–1110. doi: 10.1002/jcb.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ilaria D., Marco C., Elisa D.P., Giulia B., Massimo D., Marta P. Antioxidant mechanisms and ROS-related MicroRNAs in cancer stem cells. Oxidative Medicine and Cellular Longevity. 2015;2015:425708. doi: 10.1155/2015/425708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tanaka H., Sasayama T., Tanaka K., Nakamizo S., Nishihara M., Mizukawa K. MicroRNA-183 upregulates HIF-1alpha by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. Journal of Neuro-Oncology. 2013;111(3):273–283. doi: 10.1007/s11060-012-1027-9. [DOI] [PubMed] [Google Scholar]
- 102.Puissegur M.P., Mazure N.M., Bertero T., Pradelli L., Grosso S., Robbe-Sermesant K. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death and Differentiation. 2011;18(3):465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi Q., Gibson G.E. Up-regulation of the mitochondrial malate dehydrogenase by oxidative stress is mediated by miR-743a. Journal of Neurochemistry. 2011;118(3):440–448. doi: 10.1111/j.1471-4159.2011.07333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aschrafi A., Schwechter A.D., Mameza M.G., Natera-Naranjo O., Gioio A.E., Kaplan B.B. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(47):12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jung K.A., Lee S., Kwak M.K. NFE2L2/NRF2 activity is linked to mitochondria and AMP-activated protein kinase signaling in cancers through mir-181c/mitochondria-encoded cytochrome c oxidase regulation. Antioxidants and Redox Signaling. 2017;27(13):945–961. doi: 10.1089/ars.2016.6797. [DOI] [PubMed] [Google Scholar]
- 106.Saenz-de-Santa-Maria I., Bernardo-Castineira C., Secades P., Bernaldo-de-Quiros S., Rodrigo J.P., Astudillo A. Clinically relevant HIF-1alpha-dependent metabolic reprogramming in oropharyngeal squamous cell carcinomas includes coordinated activation of CAIX and the miR-210/ISCU signaling axis, but not MCT1 and MCT4 upregulation. Oncotarget. 2017;8(8):13730–13746. doi: 10.18632/oncotarget.14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rodriguez-Enriquez S., Juarez O., Rodriguez-Zavala J.S., Moreno-Sanchez R. Multisite control of the Crabtree effect in ascites hepatoma cells. European Journal of Biochemistry. 2001;268(8):2512–2519. doi: 10.1046/j.1432-1327.2001.02140.x. [DOI] [PubMed] [Google Scholar]
- 109.Semenza G.L. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends in Pharmacological Sciences. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mahon P.C., Hirota K., Semenza G.L. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes & Development. 2001;15(20):2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulshreshtha R., Ferracin M., Wojcik S.E., Garzon R., Alder H., Agosto-Perez F.J. A microRNA signature of hypoxia. Molecular and Cellular Biology. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Keith B., Johnson R.S., Simon M.C. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature Reviews Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zundel W., Schindler C., Haas-Kogan D., Koong A., Kaper F., Chen E. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes & Development. 2000;14(4):391–396. [PMC free article] [PubMed] [Google Scholar]
- 114.Wiesener M.S., Munchenhagen P.M., Berger I., Morgan N.V., Roigas J., Schwiertz A. Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1alpha in clear cell renal carcinomas. Cancer Research. 2001;61(13):5215–5222. [PubMed] [Google Scholar]
- 115.Shackelford D.B., Vasquez D.S., Corbeil J., Wu S., Leblanc M., Wu C.L. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(27):11137–11142. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442(7104):779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 117.Brugarolas J., Lei K., Hurley R.L., Manning B.D., Reiling J.H., Hafen E. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & Development. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lim J.H., Lee E.S., You H.J., Lee J.W., Park J.W., Chun Y.S. Ras-dependent induction of HIF-1alpha785 via the Raf/MEK/ERK pathway: a novel mechanism of Ras-mediated tumor promotion. Oncogene. 2004;23(58):9427–9431. doi: 10.1038/sj.onc.1208003. [DOI] [PubMed] [Google Scholar]
- 119.Jiang B.H., Agani F., Passaniti A., Semenza G.L. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Research. 1997;57(23):5328–5335. [PubMed] [Google Scholar]
- 120.Courtnay R., Ngo D.C., Malik N., Ververis K., Tortorella S.M., Karagiannis T.C. Cancer metabolism and the Warburg effect: the role of HIF-1 and PI3K. Molecular Biology Reports. 2015;42(4):841–851. doi: 10.1007/s11033-015-3858-x. [DOI] [PubMed] [Google Scholar]
- 121.Laughner E., Taghavi P., Chiles K., Mahon P.C., Semenza G.L. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Molecular and Cellular Biology. 2001;21(12):3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaelin W.G., Jr., Ratcliffe P.J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Molecular Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 124.Gustafsson M.V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Developmental Cell. 2005;9(5):617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 125.Kaidi A., Williams A.C., Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nature Cell Biology. 2007;9(2):210–217. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- 126.Yeung S.J., Pan J., Lee M.H. Roles of p53, MYC and HIF-1 in regulating glycolysis – the seventh hallmark of cancer. Cellular and Molecular Life Sciences. 2008;65(24):3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fukuda R., Zhang H., Kim J.W., Shimoda L., Dang C.V., Semenza G.L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 128.Sutendra G., Dromparis P., Kinnaird A., Stenson T.H., Haromy A., Parker J.M. Mitochondrial activation by inhibition of PDKII suppresses HIF1a signaling and angiogenesis in cancer. Oncogene. 2013;32(13):1638–1650. doi: 10.1038/onc.2012.198. [DOI] [PubMed] [Google Scholar]
- 129.Majmundar A.J., Wong W.J., Simon M.C. Hypoxia-inducible factors and the response to hypoxic stress. Molecular Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao F., Mancuso A., Bui T.V., Tong X., Gruber J.J., Swider C.R. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene. 2010;29(20):2962–2972. doi: 10.1038/onc.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nallamshetty S., Chan S.Y., Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radical Biology & Medicine. 2013;64:20–30. doi: 10.1016/j.freeradbiomed.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Azzouzi H.E., Leptidis S., Doevendans P.A., De Windt L.J. HypoxamiRs: regulators of cardiac hypoxia and energy metabolism. Trends in Endocrinology and Metabolism. 2015;26(9):502–508. doi: 10.1016/j.tem.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 133.Yan H.L., Xue G., Mei Q., Wang Y.Z., Ding F.X., Liu M.F. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. The EMBO Journal. 2009;28(18):2719–2732. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lei Z., Li B., Yang Z., Fang H., Zhang G.M., Feng Z.H. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hebert C., Norris K., Scheper M.A., Nikitakis N., Sauk J.J. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Molecular Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.He M., Wang Q.Y., Yin Q.Q., Tang J., Lu Y., Zhou C.X. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death & Differentiation. 2013;20(3):408–418. doi: 10.1038/cdd.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li C., Mpollo M.S., Gonsalves C.S., Tahara S.M., Malik P., Kalra V.K. Peroxisome proliferator-activated receptor-alpha-mediated transcription of miR-199a2 attenuates endothelin-1 expression via hypoxia-inducible factor-1alpha. Journal of Biological Chemistry. 2014;289(52):36031–36047. doi: 10.1074/jbc.M114.600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li J.Y., Zhang Y., Zhang W.H., Jia S., Kang Y., Zhu X.Y. Differential distribution of miR-20a and miR-20b may underly metastatic heterogeneity of breast cancers. Asian Pacific Journal of Cancer Prevention. 2012;13(5):1901–1906. doi: 10.7314/apjcp.2012.13.5.1901. [DOI] [PubMed] [Google Scholar]
- 139.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 140.Wu C., So J., Davis-Dusenbery B.N., Qi H.H., Bloch D.B., Shi Y. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Molecular and Cellular Biology. 2011;31(23):4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chakravarthi B.V., Pathi S.S., Goswami M.T., Cieslik M., Zheng H., Nallasivam S. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5(16):6654–6669. doi: 10.18632/oncotarget.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aakula A., Leivonen S.K., Hintsanen P., Aittokallio T., Ceder Y., Borresen-Dale A.L. MicroRNA-135b regulates ERalpha, AR and HIF1AN and affects breast and prostate cancer cell growth. Molecular Oncology. 2015;9(7):1287–1300. doi: 10.1016/j.molonc.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leung A.K., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular Cell. 2011;42(4):489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu Y.P., Karg M., Harwig A., Herrera-Carrillo E., Jongejan A., van Kampen A. Mechanistic insights on the dicer-independent AGO2-mediated processing of AgoshRNAs. RNA Biology. 2015;12(1):92–100. doi: 10.1080/15476286.2015.1017204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen T., Yao L.Q., Shi Q., Ren Z., Ye L.C., Xu J.M. MicroRNA-31 contributes to colorectal cancer development by targeting factor inhibiting HIF-1alpha (FIH-1) Cancer Biology & Therapy. 2014;15(5):516–523. doi: 10.4161/cbt.28017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yuan Q., Gao W., Liu B., Ye W. Upregulation of miR-184 enhances the malignant biological behavior of human glioma cell line A172 by targeting FIH-1. Cellular Physiology and Biochemistry. 2014;34(4):1125–1136. doi: 10.1159/000366326. [DOI] [PubMed] [Google Scholar]
- 147.Peng H., Hamanaka R.B., Katsnelson J., Hao L.L., Yang W., Chandel N.S. MicroRNA-31 targets FIH-1 to positively regulate corneal epithelial glycogen metabolism. The FASEB Journal. 2012;26(8):3140–3147. doi: 10.1096/fj.11-198515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ghosh A.K., Shanafelt T.D., Cimmino A., Taccioli C., Volinia S., Liu C.G. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113(22):5568–5574. doi: 10.1182/blood-2008-10-185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yue J., Guan J., Wang X., Zhang L., Yang Z., Ao Q. MicroRNA-206 is involved in hypoxia-induced pulmonary hypertension through targeting of the HIF-1alpha/Fhl-1 pathway. Laboratory Investigation. 2013;93(7):748–759. doi: 10.1038/labinvest.2013.63. [DOI] [PubMed] [Google Scholar]
- 150.Liu L.Z., Li C., Chen Q., Jing Y., Carpenter R., Jiang Y. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamakuchi M., Lotterman C.D., Bao C., Hruban R.H., Karim B., Mendell J.T. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(14):6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meng S., Cao J., Wang L., Zhou Q., Li Y., Shen C. MicroRNA 107 partly inhibits endothelial progenitor cells differentiation via HIF-1beta. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0040323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ho J.J., Metcalf J.L., Yan M.S., Turgeon P.J., Wang J.J., Chalsev M. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. Journal of Biological Chemistry. 2012;287(34):29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kelly T.J., Souza A.L., Clish C.B., Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Molecular and Cellular Biology. 2011;31(13):2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Johnson R.F., Perkins N.D. Nuclear factor-kappaB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends in Biochemical Sciences. 2012;37(8):317–324. doi: 10.1016/j.tibs.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 156.Ghosh G., Subramanian I.V., Adhikari N., Zhang X., Joshi H.P., Basi D. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. Journal of Clinical Investigation. 2010;120(11):4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sun G., Zhou Y., Li H., Guo Y., Shan J., Xia M. Over-expression of microRNA-494 up-regulates hypoxia-inducible factor-1 alpha expression via PI3K/Akt pathway and protects against hypoxia-induced apoptosis. Journal of Biomedical Sciences. 2013;20:100. doi: 10.1186/1423-0127-20-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Velagapudi S.P., Costales M.G., Vummidi B.R., Nakai Y., Angelbello A.J., Tran T. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chemical Biology. 2018;25(9):1086–1094. doi: 10.1016/j.chembiol.2018.05.015. e1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bertozzi D., Marinello J., Manzo S.G., Fornari F., Gramantieri L., Capranico G. The natural inhibitor of DNA topoisomerase I, camptothecin, modulates HIF-1alpha activity by changing miR expression patterns in human cancer cells. Molecular Cancer Therapeutics. 2014;13(1):239–248. doi: 10.1158/1535-7163.MCT-13-0729. [DOI] [PubMed] [Google Scholar]
- 160.Poitz D.M., Augstein A., Gradehand C., Ende G., Schmeisser A., Strasser R.H. Regulation of the Hif-system by micro-RNA 17 and 20a – role during monocyte-to-macrophage differentiation. Molecular Immunology. 2013;56(4):442–451. doi: 10.1016/j.molimm.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 161.Lin S.C., Wang C.C., Wu M.H., Yang S.H., Li Y.H., Tsai S.J. Hypoxia-induced microRNA-20a expression increases ERK phosphorylation and angiogenic gene expression in endometriotic stromal cells. The Journal of Clinical Endocrinology and Metabolism. 2012;97(8):E1515–E1523. doi: 10.1210/jc.2012-1450. [DOI] [PubMed] [Google Scholar]
- 162.Kang S.G., Lee W.H., Lee Y.H., Lee Y.S., Kim S.G. Hypoxia-inducible factor-1alpha inhibition by a pyrrolopyrazine metabolite of oltipraz as a consequence of microRNAs 199a-5p and 20a induction. Carcinogenesis. 2012;33(3):661–669. doi: 10.1093/carcin/bgr320. [DOI] [PubMed] [Google Scholar]
- 163.Cascio S., D'Andrea A., Ferla R., Surmacz E., Gulotta E., Amodeo V. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. Journal of Cellular Physiology. 2010;224(1):242–249. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 164.Xue T.M., Tao L.D., Zhang M., Zhang J., Liu X., Chen G.F. Clinicopathological significance of MicroRNA-20b expression in hepatocellular carcinoma and regulation of HIF-1alpha and VEGF effect on cell biological behaviour. Disease Markers. 2015;2015:325176. doi: 10.1155/2015/325176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bartoszewska S., Kochan K., Piotrowski A., Kamysz W., Ochocka R.J., Collawn J.F. The hypoxia-inducible miR-429 regulates hypoxia-inducible factor-1alpha expression in human endothelial cells through a negative feedback loop. The FASEB Journal. 2015;29(4):1467–1479. doi: 10.1096/fj.14-267054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hua Z., Lv Q., Ye W., Wong C.K., Cai G., Gu D. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sen D., Jayandharan G.R. MicroRNA-15b modulates molecular mediators of blood induced arthropathy in hemophilia mice. International Journal of Molecular Sciences. 2016;17(4):492. doi: 10.3390/ijms17040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Akhtar S., Hartmann P., Karshovska E., Rinderknecht F.A., Subramanian P., Gremse F. Endothelial hypoxia-inducible factor-1alpha promotes atherosclerosis and monocyte recruitment by upregulating MicroRNA-19a. Hypertension. 2015;66(6):1220–1226. doi: 10.1161/HYPERTENSIONAHA.115.05886. [DOI] [PubMed] [Google Scholar]
- 169.Han M., Wang Y., Liu M., Bi X., Bao J., Zeng N. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1alpha expression in third-sphere forming breast cancer stem cell-like cells. Cancer Science. 2012;103(6):1058–1064. doi: 10.1111/j.1349-7006.2012.02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.White N.M., Masui O., Newsted D., Scorilas A., Romaschin A.D., Bjarnason G.A. Galectin-1 has potential prognostic significance and is implicated in clear cell renal cell carcinoma progression through the HIF/mTOR signaling axis. British Journal of Cancer. 2014;110(5):1250–1259. doi: 10.1038/bjc.2013.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yamakuchi M., Yagi S., Ito T., Lowenstein C.J. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS One. 2011;6(5) doi: 10.1371/journal.pone.0020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 173.Peng H., Kaplan N., Hamanaka R.B., Katsnelson J., Blatt H., Yang W. microRNA-31/factor-inhibiting hypoxia-inducible factor 1 nexus regulates keratinocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(35):14030–14034. doi: 10.1073/pnas.1111292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Peng H., Katsnelson J., Yang W., Brown M.A., Lavker R.M. FIH-1/c-kit signaling: a novel contributor to corneal epithelial glycogen metabolism. Investigative Ophthalmology & Visual Science. 2013;54(4):2781–2786. doi: 10.1167/iovs.12-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou J., Xu D., Xie H., Tang J., Liu R., Li J. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1alpha. Cancer Biology & Therapy. 2015;16(6):846–855. doi: 10.1080/15384047.2015.1030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Umezu T., Tadokoro H., Azuma K., Yoshizawa S., Ohyashiki K., Ohyashiki J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124(25):3748–3757. doi: 10.1182/blood-2014-05-576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Song T., Zhang X., Wang C., Wu Y., Cai W., Gao J. MiR-138 suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in clear cell renal cell carcinoma 786-O cells. Asian Pacific Journal of Cancer Prevention. 2011;12(5):1307–1311. [PubMed] [Google Scholar]
- 178.Yeh Y.M., Chuang C.M., Chao K.C., Wang L.H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. International Journal of Cancer. 2013;133(4):867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 179.Liu L., Wang Y., Bai R., Yang K., Tian Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1alpha regulation. Oncogenesis. 2017;6(4):e318. doi: 10.1038/oncsis.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bai R., Zhao A.Q., Zhao Z.Q., Liu W.L., Jian D.M. MicroRNA-195 induced apoptosis in hypoxic chondrocytes by targeting hypoxia-inducible factor 1 alpha. European Review for Medical and Pharmacological Sciences. 2015;19(4):545–551. [PubMed] [Google Scholar]
- 181.Joshi H.P., Subramanian I.V., Schnettler E.K., Ghosh G., Rupaimoole R., Evans C. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(14):5331–5336. doi: 10.1073/pnas.1317242111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.He J., Wang M., Jiang Y., Chen Q., Xu S., Xu Q. Chronic arsenic exposure and angiogenesis in human bronchial epithelial cells via the ROS/miR-199a-5p/HIF-1alpha/COX-2 pathway. Environmental Health Perspectives. 2014;122(3):255–261. doi: 10.1289/ehp.1307545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guillaume M., Marjorie B., François P., Jolyane M., Audrey C., Christian C. miR204/RUNX2 axis regulates HIF-1α activation in pulmonary arterial hypertension. The FASEB Journal. 2013;31:650–659. [Google Scholar]
- 184.Merlo A., de Quiros S.B., Secades P., Zambrano I., Balbin M., Astudillo A. Identification of a signaling axis HIF-1alpha/microRNA-210/ISCU independent of SDH mutation that defines a subgroup of head and neck paragangliomas. The Journal of Cinical Endocrinology and Metabolism. 2012;97(11):E2194–E2200. doi: 10.1210/jc.2012-2410. [DOI] [PubMed] [Google Scholar]
- 185.Biswas S., Roy S., Banerjee J., Hussain S.R., Khanna S., Meenakshisundaram G. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(15):6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Camps C., Buffa F.M., Colella S., Moore J., Sotiriou C., Sheldon H. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical Cancer Research. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 187.Dal Monte M., Landi D., Martini D., Bagnoli P. Antiangiogenic role of miR-361 in human umbilical vein endothelial cells: functional interaction with the peptide somatostatin. Naunyn-Schmiedeberg's Archives of Pharmacology. 2013;386(1):15–27. doi: 10.1007/s00210-012-0808-1. [DOI] [PubMed] [Google Scholar]
- 188.Chen D., Dang B.L., Huang J.Z., Chen M., Wu D., Xu M.L. MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1alpha-TWIST signaling axis in breast cancer. Oncotarget. 2015;6(32):32701–32712. doi: 10.18632/oncotarget.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cha S.T., Chen P.S., Johansson G., Chu C.Y., Wang M.Y., Jeng Y.M. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Research. 2010;70(7):2675–2685. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 190.Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Science. 2013;104(3):275–281. doi: 10.1111/cas.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Baldus S.E., Schaefer K.L., Engers R., Hartleb D., Stoecklein N.H., Gabbert H.E. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clinical Cancer Research. 2010;16(3):790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 192.Neuzil J., Rohlena J., Dong L.F. K-Ras and mitochondria: dangerous liaisons. Cell Research. 2012;22(2):285–287. doi: 10.1038/cr.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lu W., Hu Y., Chen G., Chen Z., Zhang H., Wang F. Novel role of NOX in supporting aerobic glycolysis in cancer cells with mitochondrial dysfunction and as a potential target for cancer therapy. PLoS Biology. 2012;10(5) doi: 10.1371/journal.pbio.1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang P., Song M., Zeng Z.L., Zhu C.F., Lu W.H., Yang J. Identification of NDUFAF1 in mediating K-Ras induced mitochondrial dysfunction by a proteomic screening approach. Oncotarget. 2015;6(6):3947–3962. doi: 10.18632/oncotarget.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li Z.H., Pan X.M., Han B.W., Guo X.M., Zhang Z., Jia J. A let-7 binding site polymorphism rs712 in the KRAS 3' UTR is associated with an increased risk of gastric cancer. Tumour Biology. 2013;34(5):3159–3163. doi: 10.1007/s13277-013-0885-x. [DOI] [PubMed] [Google Scholar]
- 196.Pan X.M., Sun R.F., Li Z.H., Guo X.M., Zhang Z., Qin H.J. A let-7 KRAS rs712 polymorphism increases colorectal cancer risk. Tumour Biology. 2014;35(1):831–835. doi: 10.1007/s13277-013-1114-3. [DOI] [PubMed] [Google Scholar]
- 197.Jin H., Liang Y., Wang X., Zhu J., Sun R., Chen P. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Medical Oncology (Northwood, London, England) 2014;31(10):221. doi: 10.1007/s12032-014-0221-3. [DOI] [PubMed] [Google Scholar]
- 198.Chin L.J., Ratner E., Leng S., Zhai R., Nallur S., Babar I. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Research. 2008;68(20):8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kim R.K., Suh Y., Yoo K.C., Cui Y.H., Kim H., Kim M.J. Activation of KRAS promotes the mesenchymal features of basal-type breast cancer. Experimental & Molecular Medicine. 2015;47:e137. doi: 10.1038/emm.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kopp F., Wagner E., Roidl A. The proto-oncogene KRAS is targeted by miR-200c. Oncotarget. 2014;5(1):185–195. doi: 10.18632/oncotarget.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hara T., Jones M.F., Subramanian M., Li X.L., Ou O., Zhu Y. Selective targeting of KRAS-mutant cells by miR-126 through repression of multiple genes essential for the survival of KRAS-mutant cells. Oncotarget. 2014;5(17):7635–7650. doi: 10.18632/oncotarget.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jin X., Sun Y., Yang H., Li J., Yu S., Chang X. Deregulation of the MiR-193b-KRAS axis contributes to impaired cell growth in pancreatic cancer. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0125515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hiraki M., Nishimura J., Takahashi H., Wu X., Takahashi Y., Miyo M. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Molecular Therapy Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jiao L.R., Frampton A.E., Jacob J., Pellegrino L., Krell J., Giamas G. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0032068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Khodayari N., Mohammed K.A., Goldberg E.P., Nasreen N. EphrinA1 inhibits malignant mesothelioma tumor growth via let-7 microRNA-mediated repression of the RAS oncogene. Cancer Gene Therapy. 2011;18(11):806–816. doi: 10.1038/cgt.2011.50. [DOI] [PubMed] [Google Scholar]
- 206.Tanaka M., Suzuki H.I., Shibahara J., Kunita A., Isagawa T., Yoshimi A. EVI1 oncogene promotes KRAS pathway through suppression of microRNA-96 in pancreatic carcinogenesis. Oncogene. 2014;33(19):2454–2463. doi: 10.1038/onc.2013.204. [DOI] [PubMed] [Google Scholar]
- 207.Zhang Y., Kim J., Mueller A.C., Dey B., Yang Y., Lee D.H. Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death & Differentiation. 2014;21(5):720–734. doi: 10.1038/cdd.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Iliopoulos D., Rotem A., Struhl K. Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Research. 2011;71(15):5144–5153. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chung H.J., Levens D. c-myc expression: keep the noise down! Molecular Cells. 2005;20(2):157–166. [PubMed] [Google Scholar]
- 210.Shim H., Dolde C., Lewis B.C., Wu C.S., Dang G., Jungmann R.A. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(13):6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kim J., Lee J.H., Iyer V.R. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3(3) doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O'Donnell K.A. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Molecular and Cellular Biology. 2005;25(14):6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Miller D.M., Thomas S.D., Islam A., Muench D., Sedoris K. c-Myc and cancer metabolism. Clinical Cancer Research. 2012;18(20):5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fan Y., Dickman K.G., Zong W.X. Akt and c-Myc differentially activate cellular metabolic programs and prime cells to bioenergetic inhibition. Journal of Biological Chemistry. 2010;285(10):7324–7333. doi: 10.1074/jbc.M109.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen X., Qian Y., Wu S. The Warburg effect: evolving interpretations of an established concept. Free Radical Biology and Medicine. 2015;79C:253–263. doi: 10.1016/j.freeradbiomed.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Anastasiou D., Poulogiannis G., Asara J.M., Boxer M.B., Jiang J.K., Shen M. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kim J.W., Gao P., Liu Y.C., Semenza G.L., Dang C.V. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Molecular and Cellular Biology. 2007;27(21):7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Subramanian A., Miller D.M. Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. Journal of Biological Chemistry. 2000;275(8):5958–5965. doi: 10.1074/jbc.275.8.5958. [DOI] [PubMed] [Google Scholar]
- 219.Xu N., Li Z., Yu Z., Yan F., Liu Y., Lu X. MicroRNA-33b suppresses migration and invasion by targeting c-Myc in osteosarcoma cells. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0115300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 220.Sun S., Sun P., Wang C., Sun T. Downregulation of microRNA-155 accelerates cell growth and invasion by targeting c-myc in human gastric carcinoma cells. Oncology Reports. 2014;32(3):951–956. doi: 10.3892/or.2014.3288. [DOI] [PubMed] [Google Scholar]
- 221.Lin F., Ding R., Zheng S., Xing D., Hong W., Zhou Z. Decrease expression of microRNA-744 promotes cell proliferation by targeting c-Myc in human hepatocellular carcinoma. Cancer Cell International. 2014;14:58. doi: 10.1186/1475-2867-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yamamura S., Saini S., Majid S., Hirata H., Ueno K., Deng G. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PLoS One. 2012;7(1) doi: 10.1371/journal.pone.0029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Li Y., Challagundla K.B., Sun X.X., Zhang Q., Dai M.S. MicroRNA-130a associates with ribosomal protein L11 to suppress c-Myc expression in response to UV irradiation. Oncotarget. 2015;6(2):1101–1114. doi: 10.18632/oncotarget.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yang X., Cai H., Liang Y., Chen L., Wang X., Si R. Inhibition of c-Myc by let-7b mimic reverses mutidrug resistance in gastric cancer cells. Oncology Reports. 2015;33(4):1723–1730. doi: 10.3892/or.2015.3757. [DOI] [PubMed] [Google Scholar]
- 225.Ting Y., Medina D.J., Strair R.K., Schaar D.G. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochemical and Biophysical Research Communications. 2010;394(3):606–611. doi: 10.1016/j.bbrc.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 226.Wang F., Xia J., Wang N., Zong H. miR-145 inhibits proliferation and invasion of esophageal squamous cell carcinoma in part by targeting c-Myc. Onkologie. 2013;36(12):754–758. doi: 10.1159/000356978. [DOI] [PubMed] [Google Scholar]
- 227.Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O'Day E. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'UTR microRNA recognition elements. Molecular Cell. 2009;35(5):610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liao J.M., Lu H. Autoregulatory suppression of c-Myc by miR-185-3p. Journal of Biological Chemistry. 2011;286(39):33901–33909. doi: 10.1074/jbc.M111.262030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Han H., Sun D., Li W., Shen H., Zhu Y., Li C. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57(6):2378–2389. doi: 10.1002/hep.26302. [DOI] [PubMed] [Google Scholar]
- 230.Xu H., He J.H., Xiao Z.D., Zhang Q.Q., Chen Y.Q., Zhou H. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology (Baltimore, Md) 2010;52(4):1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 231.Coulouarn C., Factor V.M., Andersen J.B., Durkin M.E., Thorgeirsson S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang B., Hsu S.H., Wang X., Kutay H., Bid H.K., Yu J. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59(2):555–566. doi: 10.1002/hep.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 234.Nadiminty N., Tummala R., Lou W., Zhu Y., Zhang J., Chen X. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. Journal of Biological Chemistry. 2012;287(2):1527–1537. doi: 10.1074/jbc.M111.278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chang T.C., Zeitels L.R., Hwang H.W., Chivukula R.R., Wentzel E.A., Dews M. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sampson V.B., Rong N.H., Han J., Yang Q., Aris V., Soteropoulos P. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Research. 2007;67(20):9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 237.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bui T.V., Mendell J.T. Myc: maestro of microRNAs. Genes & Cancer. 2010;1(6):568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Land H., Parada L.F., Weinberg R.A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 240.Xu X., Chen W., Miao R., Zhou Y., Wang Z., Zhang L. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget. 2015;6(6):3988–4004. doi: 10.18632/oncotarget.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Qian J., Zhang Z., Liang J., Ge Q., Duan X., Ma F. The full-length transcripts and promoter analysis of intergenic microRNAs in Drosophila melanogaster. Genomics. 2011;97(5):294–303. doi: 10.1016/j.ygeno.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 243.Schulte J.H., Horn S., Otto T., Samans B., Heukamp L.C., Eilers U.C. MYCN regulates oncogenic MicroRNAs in neuroblastoma. International Journal of Cancer. 2008;122(3):699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 244.Aceto N., Sausgruber N., Brinkhaus H., Gaidatzis D., Martiny-Baron G., Mazzarol G. Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nature Medicine. 2012;18(4):529–537. doi: 10.1038/nm.2645. [DOI] [PubMed] [Google Scholar]
- 245.Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genetics. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sander S., Bullinger L., Klapproth K., Fiedler K., Kestler H.A., Barth T.F. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112(10):4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 247.Koh C.M., Iwata T., Zheng Q., Bethel C., Yegnasubramanian S., De Marzo A.M. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2(9):669–683. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rothschild S.I., Tschan M.P., Federzoni E.A., Jaggi R., Fey M.F., Gugger M. MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene. 2012;31(38):4221–4232. doi: 10.1038/onc.2011.578. [DOI] [PubMed] [Google Scholar]
- 249.Lin C.H., Jackson A.L., Guo J., Linsley P.S., Eisenman R.N. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. The EMBO Journal. 2009;28(20):3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang L., Deng T., Li X., Liu H., Zhou H., Ma J. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31(4):559–566. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 251.Khew-Goodall Y., Goodall G.J. Myc-modulated miR-9 makes more metastases. Nature Cell Biology. 2010;12(3):209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- 252.Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biology. 2010;12(3):247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Greenwell I.B., Flowers C.R., Blum K.A., Cohen J.B. Clinical use of PI3K inhibitors in B-cell lymphoid malignancies: today and tomorrow. Expert Review of Anticancer Therapy. 2017;17(3):271–279. doi: 10.1080/14737140.2017.1285702. [DOI] [PubMed] [Google Scholar]
- 254.Staal S.P. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(14):5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bellacosa A., de Feo D., Godwin A.K., Bell D.W., Cheng J.Q., Altomare D.A. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. International Journal of Cancer. 1995;64(4):280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 256.Elstrom R.L., Bauer D.E., Buzzai M., Karnauskas R., Harris M.H., Plas D.R. Akt stimulates aerobic glycolysis in cancer cells. Cancer Research. 2004;64(11):3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 257.Rathmell J.C., Fox C.J., Plas D.R., Hammerman P.S., Cinalli R.M., Thompson C.B. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Molecular and Cellular Biology. 2003;23(20):7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Roberts D.J., Tan-Sah V.P., Smith J.M., Miyamoto S. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. Journal of Biological Chemistry. 2013;288(33):23798–23806. doi: 10.1074/jbc.M113.482026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Deprez J., Vertommen D., Alessi D.R., Hue L., Rider M.H. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. Journal of Biological Chemistry. 1997;272(28):17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 260.Zhang W., Patil S., Chauhan B., Guo S., Powell D.R., Le J. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. Journal of Biological Chemistry. 2006;281(15):10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 261.Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature Cell Biology. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 262.Robey R.B., Hay N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Seminars in Cancer Biology. 2009;19(1):25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Guo C., Sah J.F., Beard L., Willson J.K., Markowitz S.D., Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes, Chromosomes and Cancer. 2008;47(11):939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhang G.M., Bao C.Y., Wan F.N., Cao D.L., Qin X.J., Zhang H.L. MicroRNA-302a suppresses tumor cell proliferation by inhibiting AKT in prostate cancer. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0124410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Yan B., Guo Q., Nan X.X., Wang Z., Yin Z., Yi L. Micro-ribonucleic acid 29b inhibits cell proliferation and invasion and enhances cell apoptosis and chemotherapy effects of cisplatin via targeting of DNMT3b and AKT3 in prostate cancer. OncoTargets and Therapy. 2015;8:557–565. doi: 10.2147/OTT.S76484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Noguchi S., Mori T., Hoshino Y., Maruo K., Yamada N., Kitade Y. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Letters. 2011;307(2):211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 267.Que T., Song Y., Liu Z., Zheng S., Long H., Li Z. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34(38):4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 268.Rathod S.S., Rani S.B., Khan M., Muzumdar D., Shiras A. Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio. 2014;4:485–495. doi: 10.1016/j.fob.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 270.Iorio M.V., Casalini P., Piovan C., Di Leva G., Merlo A., Triulzi T. microRNA-205 regulates HER3 in human breast cancer. Cancer Research. 2009;69(6):2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 271.Hamano R., Miyata H., Yamasaki M., Kurokawa Y., Hara J., Moon J.H. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clinical Cancer Research. 2011;17(9):3029–3038. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 272.Al-Khalaf H.H., Aboussekhra A. MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. Journal of Biological Chemistry. 2014;289(45):31433–31447. doi: 10.1074/jbc.M114.593004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Alessi D.R., James S.R., Downes C.P., Holmes A.B., Gaffney P.R., Reese C.B. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Current Biology. 1997;7(4):261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 274.Liao Y., Hung M.C. Physiological regulation of Akt activity and stability. American Journal of Translational Research. 2010;2(1):19–42. [PMC free article] [PubMed] [Google Scholar]
- 275.Yuan Y., Du W., Wang Y., Xu C., Wang J., Zhang Y. Suppression of AKT expression by miR-153 produced anti-tumor activity in lung cancer. International Journal of Cancer. 2015;136(6):1333–1340. doi: 10.1002/ijc.29103. [DOI] [PubMed] [Google Scholar]
- 276.Wu Z., He B., He J., Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. The Prostate. 2013;73(6):596–604. doi: 10.1002/pros.22600. [DOI] [PubMed] [Google Scholar]
- 277.Zhou X., Hu Y., Dai L., Wang Y., Zhou J., Wang W. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Research. 2008;68(10):3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 279.Giles K.M., Barker A., Zhang P.M., Epis M.R., Leedman P.J. MicroRNA regulation of growth factor receptor signaling in human cancer cells. Methods in Molecular Biology. 2011;676:147–163. doi: 10.1007/978-1-60761-863-8_11. [DOI] [PubMed] [Google Scholar]
- 280.Cully M., You H., Levine A.J., Mak T.W. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nature Reviews Cancer. 2006;6(3):184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 281.Tumaneng K., Schlegelmilch K., Russell R.C., Yimlamai D., Basnet H., Mahadevan N. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nature Cell Biology. 2012;14(12):1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Huse J.T., Brennan C., Hambardzumyan D., Wee B., Pena J., Rouhanifard S.H. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & Development. 2009;23(11):1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Small E.M., O'Rourke J.R., Moresi V., Sutherland L.B., McAnally J., Gerard R.D. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Li Y.n., Dai D., Lu Q., Fei M., Li M., Wu X. Sirt2 suppresses glioma cell growth through targeting NF-κB–miR-21 axis. Biochemical and Biophysical Research Communications. 2013;441(3):661–667. doi: 10.1016/j.bbrc.2013.10.077. [DOI] [PubMed] [Google Scholar]
- 285.Palumbo T., Faucz F.R., Azevedo M., Xekouki P., Iliopoulos D., Stratakis C.A. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN-AKT pathway. Oncogene. 2013;32(13):1651–1659. doi: 10.1038/onc.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Fornari F., Milazzo M., Chieco P., Negrini M., Marasco E., Capranico G. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. The Journal of Pathology. 2012;227(3):275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 287.Yan S.Y., Chen M.M., Li G.M., Wang Y.Q., Fan J.G. MiR-32 induces cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting PTEN. Tumour Biology. 2015;36(6):4747–4755. doi: 10.1007/s13277-015-3124-9. [DOI] [PubMed] [Google Scholar]
- 288.Zou H., Ding Y., Wang K., Xiong E., Peng W., Du F. MicroRNA-29A/PTEN pathway modulates neurite outgrowth in PC12 cells. Neuroscience. 2015;291:289–300. doi: 10.1016/j.neuroscience.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 289.Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nature Cell Biology. 2009;11(7):881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Fu X., Tian J., Zhang L., Chen Y., Hao Q. Involvement of microRNA-93, a new regulator of PTEN/Akt signaling pathway, in regulation of chemotherapeutic drug cisplatin chemosensitivity in ovarian cancer cells. FEBS Letters. 2012;586(9):1279–1286. doi: 10.1016/j.febslet.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 291.Yang H., Kong W., He L., Zhao J.J., O'Donnell J.D., Wang J. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Research. 2008;68(2):425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 292.Xue M., Yao S., Hu M., Li W., Hao T., Zhou F. HIV-1 Nef and KSHV oncogene K1 synergistically promote angiogenesis by inducing cellular miR-718 to regulate the PTEN/AKT/mTOR signaling pathway. Nucleic Acids Research. 2014;42(15):9862–9879. doi: 10.1093/nar/gku583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Xie Q., Yan Y., Huang Z., Zhong X., Huang L. MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology. 2014;34(5):455–464. doi: 10.1111/neup.12129. [DOI] [PubMed] [Google Scholar]
- 294.Wang X., Zhang X., Ren X.P., Chen J., Liu H., Yang J. MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation. 2010;122(13):1308–1318. doi: 10.1161/CIRCULATIONAHA.110.964684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Liu L., Jiang Y., Zhang H., Greenlee A.R., Han Z. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life Sciences. 2010;86(5–6):192–198. doi: 10.1016/j.lfs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 296.Cioffi J.A., Yue W.Y., Mendolia-Loffredo S., Hansen K.R., Wackym P.A., Hansen M.R. MicroRNA-21 overexpression contributes to vestibular schwannoma cell proliferation and survival. Otology & Neurotology. 2010;31(9):1455–1462. doi: 10.1097/MAO.0b013e3181f20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Qadir X.V., Han C., Lu D., Zhang J., Wu T. miR-185 inhibits hepatocellular carcinoma growth by targeting the DNMT1/PTEN/Akt pathway. American Journal of Pathology. 2014;184(8):2355–2364. doi: 10.1016/j.ajpath.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Glass C., Singla D.K. ES cells overexpressing microRNA-1 attenuate apoptosis in the injured myocardium. Molecular and Cellular Biochemistry. 2011;357(1–2):135–141. doi: 10.1007/s11010-011-0883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Sachdeva M., Wu H., Ru P., Hwang L., Trieu V., Mo Y.Y. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene. 2011;30(7):822–831. doi: 10.1038/onc.2010.463. [DOI] [PubMed] [Google Scholar]
- 300.Zhu G., Chai J., Ma L., Duan H., Zhang H. Downregulated microRNA-32 expression induced by high glucose inhibits cell cycle progression via PTEN upregulation and Akt inactivation in bone marrow-derived mesenchymal stem cells. Biochemical and Biophysical Research Communications. 2013;433(4):526–531. doi: 10.1016/j.bbrc.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 301.Roccaro A.M., Sacco A., Chen C., Runnels J., Leleu X., Azab F. microRNA expression in the biology, prognosis, and therapy of Waldenstrom macroglobulinemia. Blood. 2009;113(18):4391–4402. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kaper F., Dornhoefer N., Giaccia A.J. Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions. Cancer Research. 2006;66(3):1561–1569. doi: 10.1158/0008-5472.CAN-05-3375. [DOI] [PubMed] [Google Scholar]
- 303.Moon K.S., Jung S., Seo S.K., Jung T.Y., Kim I.Y., Ryu H.H. Cystic vestibular schwannomas: a possible role of matrix metalloproteinase-2 in cyst development and unfavorable surgical outcome. Journal of Neurosurgery. 2007;106(5):866–871. doi: 10.3171/jns.2007.106.5.866. [DOI] [PubMed] [Google Scholar]
- 304.Liu Y., Lai L., Chen Q., Song Y., Xu S., Ma F. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. The Journal of Immunology. 2012;188(11):5500–5510. doi: 10.4049/jimmunol.1103505. [DOI] [PubMed] [Google Scholar]
- 305.Bai H., Xu R., Cao Z., Wei D., Wang C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Letters. 2011;585(2):402–408. doi: 10.1016/j.febslet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 306.Zhang L., Zhang S., Yao J., Lowery F.J., Zhang Q., Huang W.C. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 308.Yecies J.L., Manning B.D. mTOR links oncogenic signaling to tumor cell metabolism. Journal of Molecular Medicine (Berlin) 2011;89(3):221–228. doi: 10.1007/s00109-011-0726-6. [DOI] [PubMed] [Google Scholar]
- 309.Brugarolas J.B., Vazquez F., Reddy A., Sellers W.R., Kaelin W.G., Jr. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4(2):147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 310.Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes & Development. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 311.Sun Q., Chen X., Ma J., Peng H., Wang F., Zha X. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(10):4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zha X., Wang F., Wang Y., He S., Jing Y., Wu X. Lactate dehydrogenase B is critical for hyperactive mTOR-mediated tumorigenesis. Cancer Research. 2011;71(1):13–18. doi: 10.1158/0008-5472.CAN-10-1668. [DOI] [PubMed] [Google Scholar]
- 313.Buller C.L., Loberg R.D., Fan M.H., Zhu Q., Park J.L., Vesely E. A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. American Journal of Physiology Cell Physiology. 2008;295(3):C836–C843. doi: 10.1152/ajpcell.00554.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Thomas G.V., Tran C., Mellinghoff I.K., Welsbie D.S., Chan E., Fueger B. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nature Medicine. 2006;12(1):122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 315.Land S.C., Tee A.R. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. Journal of Biological Chemistry. 2007;282(28):20534–20543. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 316.Sofer A., Lei K., Johannessen C.M., Ellisen L.W. Regulation of mTOR and cell growth in response to energy stress by REDD1. Molecular and Cellular Biology. 2005;25(14):5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Li Y., Wang Y., Kim E., Beemiller P., Wang C.Y., Swanson J. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. Journal of Biological Chemistry. 2007;282(49):35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 318.Connolly E., Braunstein S., Formenti S., Schneider R.J. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Molecular and Cellular Biology. 2006;26(10):3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Obre E., Rossignol R. Emerging concepts in bioenergetics and cancer research: metabolic flexibility, coupling, symbiosis, switch, oxidative tumors, metabolic remodeling, signaling and bioenergetic therapy. The International Journal of Biochemistry & Cell Biology. 2015;59:167–181. doi: 10.1016/j.biocel.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 320.Vadlakonda L., Dash A., Pasupuleti M., Anil Kumar K., Reddanna P. The paradox of akt-mTOR interactions. Frontiers in Oncology. 2013;3:165. doi: 10.3389/fonc.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sano H., Eguez L., Teruel M.N., Fukuda M., Chuang T.D., Chavez J.A. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metabolism. 2007;5(4):293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 322.Humphrey S.J., James D.E. Uncaging akt. Science Signaling. 2012;5(223):pe20. doi: 10.1126/scisignal.2003085. [DOI] [PubMed] [Google Scholar]
- 323.Kumar A., Lawrence J.C., Jr., Jung D.Y., Ko H.J., Keller S.R., Kim J.K. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59(6):1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wang L., Chang L., Li Z., Gao Q., Cai D., Tian Y. miR-99a and -99b inhibit cervical cancer cell proliferation and invasion by targeting mTOR signaling pathway. Medical Oncology. 2014;31(5):934. doi: 10.1007/s12032-014-0934-3. [DOI] [PubMed] [Google Scholar]
- 325.Torres A., Torres K., Pesci A., Ceccaroni M., Paszkowski T., Cassandrini P. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer. 2012;12:369. doi: 10.1186/1471-2407-12-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Xu C., Zeng Q., Xu W., Jiao L., Chen Y., Zhang Z. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Molecular Cancer Therapeutics. 2013;12(2):207–219. doi: 10.1158/1535-7163.MCT-12-0273. [DOI] [PubMed] [Google Scholar]
- 327.Sun J., Chen Z., Tan X., Zhou F., Tan F., Gao Y. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Medical Oncology. 2013;30(1):411. doi: 10.1007/s12032-012-0411-9. [DOI] [PubMed] [Google Scholar]
- 328.Grundmann S., Hans F.P., Kinniry S., Heinke J., Helbing T., Bluhm F. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123(9):999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 329.Fang Y., Xue J.L., Shen Q., Chen J., Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55(6):1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 330.Webster R.J., Giles K.M., Price K.J., Zhang P.M., Mattick J.S., Leedman P.J. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. Journal of Biological Chemistry. 2009;284(9):5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 331.Lin S., Shao N.N., Fan L., Ma X.C., Pu F.F., Shao Z.W. Effect of microRNA-101 on proliferation and apoptosis of human osteosarcoma cells by targeting mTOR. Journal of Huazhong University of Science and Technology – Medical Sciences. 2014;34(6):889–895. doi: 10.1007/s11596-014-1369-y. [DOI] [PubMed] [Google Scholar]
- 332.Chen K., Fan W., Wang X., Ke X., Wu G., Hu C. MicroRNA-101 mediates the suppressive effect of laminar shear stress on mTOR expression in vascular endothelial cells. Biochemical and Biophysical Research Communications. 2012;427(1):138–142. doi: 10.1016/j.bbrc.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 333.Liu P., Wilson M.J. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-kappaB factor in human fibrosarcoma cells. Journal of Cellular Physiology. 2012;227(2):867–876. doi: 10.1002/jcp.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Vaksman O., Stavnes H.T., Kaern J., Trope C.G., Davidson B., Reich R. miRNA profiling along tumour progression in ovarian carcinoma. Journal of Cellular and Molecular Medicine. 2011;15(7):1593–1602. doi: 10.1111/j.1582-4934.2010.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Du J., Liu S., He J., Liu X., Qu Y., Yan W. MicroRNA-451 regulates stemness of side population cells via PI3K/Akt/mTOR signaling pathway in multiple myeloma. Oncotarget. 2015 doi: 10.18632/oncotarget.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Feng Z., Levine A.J. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends in Cell Biology. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 338.Kulawiec M., Ayyasamy V., Singh K.K. p53 regulates mtDNA copy number and mitocheckpoint pathway. Journal of Carcinogenesis. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Bourdon A., Minai L., Serre V., Jais J.P., Sarzi E., Aubert S. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nature Genetics. 2007;39(6):776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 340.Liu Y., Borchert G.L., Surazynski A., Phang J.M. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene. 2008;27(53):6729–6737. doi: 10.1038/onc.2008.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Suzuki S., Tanaka T., Poyurovsky M.V., Nagano H., Mayama T., Ohkubo S. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(16):7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Molecular Cell. 2009;34(6):663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Ma W., Sung H.J., Park J.Y., Matoba S., Hwang P.M. A pivotal role for p53: balancing aerobic respiration and glycolysis. Journal of Bioenergetics and Biomembranes. 2007;39(3):243–246. doi: 10.1007/s10863-007-9083-0. [DOI] [PubMed] [Google Scholar]
- 344.Stambolic V., MacPherson D., Sas D., Lin Y., Snow B., Jang Y. Regulation of PTEN transcription by p53. Molecular Cell. 2001;8(2):317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 345.Almeida R., Almeida J., Shoshkes M., Mendes N., Mesquita P., Silva E. OCT-1 is over-expressed in intestinal metaplasia and intestinal gastric carcinomas and binds to, but does not transactivate, CDX2 in gastric cells. The Journal of Pathology. 2005;207(4):396–401. doi: 10.1002/path.1861. [DOI] [PubMed] [Google Scholar]
- 346.Jiang F., Zhao W., Zhou L., Zhang L., Liu Z., Yu D. miR-222 regulates the cell biological behavior of oral squamous cell carcinoma by targeting PUMA. Oncology Reports. 2014;31(3):1255–1262. doi: 10.3892/or.2014.2985. [DOI] [PubMed] [Google Scholar]
- 347.Li J., Donath S., Li Y., Qin D., Prabhakar B.S., Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genetics. 2010;6(1) doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 349.Green D.R., Chipuk J.E. p53 and metabolism: inside the TIGAR. Cell. 2006;126(1):30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 350.Chen S., Li P., Li J., Wang Y., Du Y., Chen X. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cellular Physiology and Biochemistry. 2015;35(3):997–1007. doi: 10.1159/000369755. [DOI] [PubMed] [Google Scholar]
- 351.Chen L., Marechal V., Moreau J., Levine A.J., Chen J. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. Journal of Biological Chemistry. 1997;272(36):22966–22973. doi: 10.1074/jbc.272.36.22966. [DOI] [PubMed] [Google Scholar]
- 352.Ishimura A., Terashima M., Kimura H., Akagi K., Suzuki Y., Sugano S. Jmjd2c histone demethylase enhances the expression of Mdm2 oncogene. Biochemical and Biophysical Research Communications. 2009;389(2):366–371. doi: 10.1016/j.bbrc.2009.08.155. [DOI] [PubMed] [Google Scholar]
- 353.Xiao J., Lin H., Luo X., Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. The EMBO Journal. 2011;30(3):524–532. doi: 10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Fortunato O., Boeri M., Moro M., Verri C., Mensah M., Conte D. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death & Disease. 2014;5:e1564. doi: 10.1038/cddis.2014.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ren Z.J., Nong X.Y., Lv Y.R., Sun H.H., An P.P., Wang F. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death & Disease. 2014;5:e1387. doi: 10.1038/cddis.2014.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Avasarala S., Van Scoyk M., Wang J., Sechler M., Vandervest K., Brzezinski C. hsa-miR29b, a critical downstream target of non-canonical Wnt signaling, plays an anti-proliferative role in non-small cell lung cancer cells via targeting MDM2 expression. Biology Open. 2013;2(7):675–685. doi: 10.1242/bio.20134507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Zhang J., Sun Q., Zhang Z., Ge S., Han Z.G., Chen W.T. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32(1):61–69. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 358.Braun C.J., Zhang X., Savelyeva I., Wolff S., Moll U.M., Schepeler T. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Research. 2008;68(24):10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 360.Hu W., Chan C.S., Wu R., Zhang C., Sun Y., Song J.S. Negative regulation of tumor suppressor p53 by microRNA miR-504. Molecular Cell. 2010;38(5):689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wei Q., Li Y.X., Liu M., Li X., Tang H. MiR-17-5p targets TP53INP1 and regulates cell proliferation and apoptosis of cervical cancer cells. IUBMB Life. 2012;64(8):697–704. doi: 10.1002/iub.1051. [DOI] [PubMed] [Google Scholar]
- 362.Bommer G.T., Gerin I., Feng Y., Kaczorowski A.J., Kuick R., Love R.E. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Current Biology. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 363.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Molecular Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 364.Ma S., Tang K.H., Chan Y.P., Lee T.K., Kwan P.S., Castilho A. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7(6):694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 365.Gironella M., Seux M., Xie M.J., Cano C., Tomasini R., Gommeaux J. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Jiang F., Liu T., He Y., Yan Q., Chen X., Wang H. MiR-125b promotes proliferation and migration of type II endometrial carcinoma cells through targeting TP53INP1 tumor suppressor in vitro and in vivo. BMC Cancer. 2011;11:425. doi: 10.1186/1471-2407-11-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Saleh A.D., Savage J.E., Cao L., Soule B.P., Ly D., DeGraff W. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0024429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Ugalde A.P., Ramsay A.J., de la Rosa J., Varela I., Marino G., Cadinanos J. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. The EMBO Journal. 2011;30(11):2219–2232. doi: 10.1038/emboj.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Chang C.J., Chao C.H., Xia W., Yang J.Y., Xiong Y., Li C.W. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nature Cell Biology. 2011;13(3):317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Kim T., Veronese A., Pichiorri F., Lee T.J., Jeon Y.J., Volinia S. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. Journal of Experimental Medicine. 2011;208(5):875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Georges S.A., Biery M.C., Kim S.Y., Schelter J.M., Guo J., Chang A.N. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Research. 2008;68(24):10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 373.Nalls D., Tang S.N., Rodova M., Srivastava R.K., Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Fujita Y., Kojima K., Hamada N., Ohhashi R., Akao Y., Nozawa Y. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochemical and Biophysical Research Communications. 2008;377(1):114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 375.Corney D.C., Flesken-Nikitin A., Godwin A.K., Wang W., Nikitin A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Research. 2007;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 376.Pichiorri F., Suh S.S., Rocci A., De Luca L., Taccioli C., Santhanam R. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18(4):367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 377.Bohlig L., Friedrich M., Engeland K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Research. 2011;39(2):440–453. doi: 10.1093/nar/gkq796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Lavu S., Boss O., Elliott P.J., Lambert P.D. Sirtuins-novel therapeutic targets to treat age-associated diseases. Nature Reviews Drug Discovery. 2008;7(10):841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 379.Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. Journal of Biological Chemistry. 2007;282(9):6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 380.Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 382.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 383.Jager S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Nemoto S., Fergusson M.M., Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306(5704):2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 385.Xiong S., Salazar G., Patrushev N., Alexander R.W. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. Journal of Biological Chemistry. 2011;286(7):5289–5299. doi: 10.1074/jbc.M110.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Chen W.Y., Wang D.H., Yen R.C., Luo J., Gu W., Baylin S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 387.Ferber E.C., Peck B., Delpuech O., Bell G.P., East P., Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death & Differentiation. 2012;19(6):968–979. doi: 10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Zhang P., Tu B., Wang H., Cao Z., Tang M., Zhang C. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(29):10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Van Meter M., Kashyap M., Rezazadeh S., Geneva A.J., Morello T.D., Seluanov A. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nature Communications. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Masri S., Rigor P., Cervantes M., Ceglia N., Sebastian C., Xiao C. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158(3):659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Lerrer B., Cohen H.Y. The guardian: metabolic and tumour-suppressive effects of SIRT6. The EMBO Journal. 2013;32(1):7–8. doi: 10.1038/emboj.2012.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Yin X., Gao Y., Shi H.S., Song L., Wang J.C., Shao J. Overexpression of SIRT6 in the hippocampal CA1 impairs the formation of long-term contextual fear memory. Scientific Reports. 2016;6:18982. doi: 10.1038/srep18982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R.E., Vadysirisack D.D. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Zwaans B.M., Lombard D.B. Interplay between sirtuins, MYC and hypoxia-inducible factor in cancer-associated metabolic reprogramming. Disease Models & Mechanisms. 2014;7(9):1023–1032. doi: 10.1242/dmm.016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 397.Mikawa T., ME L.L., Takaori-Kondo A., Inagaki N., Yokode M., Kondoh H. Dysregulated glycolysis as an oncogenic event. Cellular and Molecular Life Sciences. 2015;72(10):1881–1892. doi: 10.1007/s00018-015-1840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Barber M.F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes & Development. 2006;20(9):1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circulation Research. 2008;102(6):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 401.Lee J., Padhye A., Sharma A., Song G., Miao J., Mo Y.Y. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. Journal of Biological Chemistry. 2010;285(17):12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Choi S.E., Fu T., Seok S., Kim D.H., Yu E., Lee K.W. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12(6):1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Xu D., Takeshita F., Hino Y., Fukunaga S., Kudo Y., Tamaki A. miR-22 represses cancer progression by inducing cellular senescence. The Journal of Cell Biology. 2011;193(2):409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Shi Y., Huang J., Zhou J., Liu Y., Fu X., Li Y. MicroRNA-204 inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in osteosarcoma cells via targeting Sirtuin 1. Oncology Reports. 2015;34(1):399–406. doi: 10.3892/or.2015.3986. [DOI] [PubMed] [Google Scholar]
- 406.Eades G., Yao Y., Yang M., Zhang Y., Chumsri S., Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. Journal of Biological Chemistry. 2011;286(29):25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Zhou B., Li C., Qi W., Zhang Y., Zhang F., Wu J.X. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia. 2012;55(7):2032–2043. doi: 10.1007/s00125-012-2539-8. [DOI] [PubMed] [Google Scholar]
- 408.Lovis P., Roggli E., Laybutt D.R., Gattesco S., Yang J.Y., Widmann C. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57(10):2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Ramachandran D., Roy U., Garg S., Ghosh S., Pathak S., Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. FEBS Journal. 2011;278(7):1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 410.Pramanik D., Campbell N.R., Karikari C., Chivukula R., Kent O.A., Mendell J.T. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Molecular Cancer Therapeutics. 2011;10(8):1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Menghini R., Casagrande V., Cardellini M., Martelli E., Terrinoni A., Amati F. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120(15):1524–1532. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 412.Li N., Muthusamy S., Liang R., Sarojini H., Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mechanism of Ageing and Development. 2011;132(3):75–85. doi: 10.1016/j.mad.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 413.Saunders L.R., Sharma A.D., Tawney J., Nakagawa M., Okita K., Yamanaka S. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2010;2(7):415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Zovoilis A., Agbemenyah H.Y., Agis-Balboa R.C., Stilling R.M., Edbauer D., Rao P. microRNA-34c is a novel target to treat dementias. The EMBO Journal. 2011;30(20):4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Strum J.C., Johnson J.H., Ward J., Xie H., Feild J., Hester A. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Molecular Endocrinology. 2009;23(11):1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Zhu H., Yang Y., Wang Y., Li J., Schiller P.W., Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovascular Research. 2011;92(1):75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 417.Bou Kheir T., Futoma-Kazmierczak E., Jacobsen A., Krogh A., Bardram L., Hother C. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Molecular Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Davalos A., Goedeke L., Smibert P., Ramirez C.M., Warrier N.P., Andreo U. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(22):9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Najafi-Shoushtari S.H., Kristo F., Li Y., Shioda T., Cohen D.E., Gerszten R.E. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Rayner K.J., Suarez Y., Davalos A., Parathath S., Fitzgerald M.L., Tamehiro N. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Sharma A., Diecke S., Zhang W.Y., Lan F., He C., Mordwinkin N.M. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. Journal of Biological Chemistry. 2013;288(25):18439–18447. doi: 10.1074/jbc.M112.405928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Kim J.K., Noh J.H., Jung K.H., Eun J.W., Bae H.J., Kim M.G. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 423.North B.J., Rosenberg M.A., Jeganathan K.B., Hafner A.V., Michan S., Dai J. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. The EMBO Journal. 2014;33(13):1438–1453. doi: 10.15252/embj.201386907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Wang F., Nguyen M., Qin F.X., Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6(4):505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 425.Wang F., Chan C.H., Chen K., Guan X., Lin H.K., Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31(12):1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 426.Black J.C., Mosley A., Kitada T., Washburn M., Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Molecular Cell. 2008;32(3):449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Rothgiesser K.M., Erener S., Waibel S., Luscher B., Hottiger M.O. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. Journal of Cell Science. 2010;123(Pt 24):4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 428.Kim H.S., Vassilopoulos A., Wang R.H., Lahusen T., Xiao Z., Xu X. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Molecular Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Xu Y., Li F., Lv L., Li T., Zhou X., Deng C.X. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Research. 2014;74(13):3630–3642. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Wang J.Y., Li H., Ma C.M., Wang J.L., Lai X.S., Zhou S.F. Acupuncture may exert its therapeutic effect through microRNA-339/Sirt2/NFkappaB/FOXO1 axis. BioMed Research International. 2015;2015:249013. doi: 10.1155/2015/249013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Crocco P., Montesanto A., Passarino G., Rose G. Polymorphisms falling within putative miRNA target sites in the 3'UTR region of SIRT2 and DRD2 genes are correlated with human longevity. Journal of Gerontology Series A, Biological Sciences and Medical Sciences. 2016;71(5):586–592. doi: 10.1093/gerona/glv058. [DOI] [PubMed] [Google Scholar]
- 433.Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 434.Vatrinet R., Iommarini L., Kurelac I., De Luise M., Gasparre G., Porcelli A.M. Targeting respiratory complex I to prevent the Warburg effect. The International Journal of Biochemistry & Cell Biology. 2015;63:41–45. doi: 10.1016/j.biocel.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 435.He W., Newman J.C., Wang M.Z., Ho L., Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends in Endocrinology and Metabolism. 2012;23(9):467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 436.Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C.F., Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. Journal of Molecular Biology. 2008;382(3):790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 437.Someya S., Yu W., Hallows W.C., Xu J., Vann J.M., Leeuwenburgh C. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Jing E., Emanuelli B., Hirschey M.D., Boucher J., Lee K.Y., Lombard D. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(35):14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Finley L.W., Carracedo A., Lee J., Souza A., Egia A., Zhang J. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Pellegrini L., Pucci B., Villanova L., Marino M.L., Marfe G., Sansone L. SIRT3 protects from hypoxia and staurosporine-mediated cell death by maintaining mitochondrial membrane potential and intracellular pH. Cell Death & Differentiation. 2012;19(11):1815–1825. doi: 10.1038/cdd.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Sundaresan N.R., Gupta M., Kim G., Rajamohan S.B., Isbatan A., Gupta M.P. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. Journal of Clinical Investigation. 2009;119(9):2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Chen Y., Zhang J., Lin Y., Lei Q., Guan K.L., Zhao S. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Reports. 2011;12(6):534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Finley L.W., Haas W., Desquiret-Dumas V., Wallace D.C., Procaccio V., Gygi S.P. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Ahn B.H., Kim H.S., Song S., Lee I.H., Liu J., Vassilopoulos A. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Nasrin N., Wu X., Fortier E., Feng Y., Bare O.C., Chen S. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. Journal of Biological Chemistry. 2010;285(42):31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Zhang X., Ji R., Liao X., Brunjes D., Castillero E., George I. miR-195 regulates myocardial SIRT3 expression and mitochondrial enzyme acetylation. American Heart Association. 2015 [Google Scholar]
- 447.Hodzic M., Naaldijk Y., Stolzing A. Regulating aging in adult stem cells with microRNA. Zeitschrift für Gerontologie und Geriatrie. 2013;46(7):629–634. doi: 10.1007/s00391-013-0531-7. [DOI] [PubMed] [Google Scholar]
- 448.Poulsen R.C., Knowles H.J., Carr A.J., Hulley P.A. Cell differentiation versus cell death: extracellular glucose is a key determinant of cell fate following oxidative stress exposure. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Liang D., Meyer L., Chang D.W., Lin J., Pu X., Ye Y. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Research. 2010;70(23):9765–9776. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Slaby O., Sachlova M., Brezkova V., Hezova R., Kovarikova A., Bischofova S. Identification of microRNAs regulated by isothiocyanates and association of polymorphisms inside their target sites with risk of sporadic colorectal cancer. Nutrition and Cancer. 2013;65(2):247–254. doi: 10.1080/01635581.2013.756530. [DOI] [PubMed] [Google Scholar]
- 451.Chen Z., Lu W., Garcia-Prieto C., Huang P. The Warburg effect and its cancer therapeutic implications. Journal of Bioenergetics and Biomembranes. 2007;39(3):267–274. doi: 10.1007/s10863-007-9086-x. [DOI] [PubMed] [Google Scholar]
- 452.Jang M., Kim S.S., Lee J. Cancer cell metabolism: implications for therapeutic targets. Experimental & Molecular Medicine. 2013;45:e45. doi: 10.1038/emm.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Saridaki Z., Weidhaas J.B., Lenz H.J., Laurent-Puig P., Jacobs B., De Schutter J. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clinical Cancer Research. 2014;20(17):4499–4510. doi: 10.1158/1078-0432.CCR-14-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.