Abstract
Interleukin-6 (IL-6)-type cytokines share the common receptor glycoprotein 130 (gp130), which activates a signaling cascade involving Janus kinases (JAKs) and signal transducer and activator of transcription (STAT) transcription factors. IL-6 and/or its signaling pathway is often deregulated in diseases, such as chronic liver diseases and cancer. Thus, the identification of compounds inhibiting this pathway is of interest for future targeted therapies. We established novel cellular screening systems based on a STAT-responsive reporter gene (Cypridina luciferase). Of a library containing 538 microRNA (miRNA) mimics, several miRNAs affected hyper-IL-6-induced luciferase activities. When focusing on candidate miRNAs specifically targeting 3′ UTRs of signaling molecules of this pathway, we identified, e.g., miR-3677-5p as a novel miRNA affecting protein expression of both STAT3 and JAK1, whereas miR-16-1-3p, miR-4473, and miR-520f-3p reduced gp130 surface expression. Interestingly, combination treatment with 2 or 3 miRNAs targeting gp130 or different signaling molecules of the pathway did not increase the inhibitory effects on phospho-STAT3 levels and STAT3 target gene expression compared to treatment with single mimics. Taken together, we identified a set of miRNAs of potential therapeutic value for cancer and inflammatory diseases, which directly target the expression of molecules within the IL-6-signaling pathway and can dampen inflammatory signal transduction.
Keywords: microRNAs, signal transduction, interleukin-6, gp130, cytokine, JAK1, STAT3, cancer, inflammation, liver
Graphical Abstract
Introduction
The inflammatory cytokine interleukin-6 (IL-6) signals via a receptor complex consisting of IL6R (IL6Rα, gp80) and the signaling subunit glycoprotein 130 (gp130) (IL6Rβ, IL6ST), shared among all IL-6-type cytokines. gp130-associated Janus kinase 1 (JAK1) phosphorylates different tyrosine residues in the cytoplasmic tail of gp130, to which mainly signal transducer and activator of transcription 3 (STAT3) transcription factors are recruited. Upon phosphorylation, STATs translocate into the nucleus and regulate the expression of various genes, including the one encoding suppressor of cytokine signaling 3 (SOCS3), an important negative regulator of this pathway.1 Next to this classical signaling pathway triggered by membrane IL6R-gp130 complexes, IL-6 can also act on cells, which do not express IL6R (but only gp130) when complexed to a soluble form of IL6R (sIL6R), a process termed trans-signaling.2
In addition to its manifold physiological roles, e.g., in the immune system and in liver regeneration,3, 4 IL-6 has important systemic and local effects in the pathogenesis of cancers, such as multiple myeloma, endometrial cancer, lung cancer, colorectal cancer, renal cell carcinoma, cervical cancer, breast cancer, and ovarian carcinoma.5, 6 IL-6 is important for tumor development and angiogenesis, affecting the proliferation, migration, and invasion of cells, as well as for protecting them against drug treatments and apoptosis.5, 6 Tumor-induced systemic IL-6 can induce systemic metabolic changes, e.g., lipolysis in the white adipose tissue and insulin resistance of the skeletal muscle, which may ultimately lead to cancer inflammation-induced cachexia (reviewed in White7). In addition, IL-6 can act locally in the tumor on immune cells to suppress anti-cancer immunity.8, 9 For example, IL-6 favors the development of M2 macrophages and myeloid-derived suppressor cells (MDSCs), leading to increased levels of immunosuppressive factors such as IL-10 and IL-4, while it attenuates the differentiation of Th1 cells. Tumor-associated macrophages have also been described to produce IL-6, thus promoting the expansion of hepatocellular carcinoma (HCC) stem cells and, therefore, tumorigenesis.10
IL-6 plays an important role in HCC, the focus of this study. IL-6 and sIL6R levels have been shown to be increased in the sera of patients with HCC,11 and high levels of serum IL-6 can serve as a biomarker for an elevated risk to develop HCC.12 Increased activity of STAT3 was observed in liver tumors of mice treated with N-nitrosodiethylamine (DEN);13 later, IL-6 was found to be crucially implicated in this HCC mouse model,14, 15 which, according to gene expression data, best resembles liver cancer with poor prognosis in humans.16 Autocrine production of IL-6 by liver cancer progenitor cells also contributes to their malignant progression.17 While liver-specific knockout of SOCS3 promoted DEN-induced hepatocarcinogenesis,18, 19 deletion of gp130 or STAT3 reduced HCC prevalence and resulted in smaller tumors in DEN-treated mice.15, 20 Similarly, in another chemically induced HCC mouse model (thioacetamide based), the absence of STAT3 in liver parenchymal cells attenuated HCC development.21 Interestingly, IL-6-trans-signaling, rather than classical signaling, seems to be responsible for DEN-dependent tumor development.22, 23 Of note, a whole-genome sequencing study of 88 matched pairs of (hepatitis B virus [HBV]-related) HCC tumor and surrounding tissues indicated that the (IL-6)-JAK-STAT-signaling pathway is a major oncogenic driver in HCC, with mutations in almost half of the cases investigated, including IL6R gene amplifications in 26% of the samples and (in part, gain-of-function) mutations in the JAK1 gene in 9.1%.24
Altogether, these data strongly suggest an important role of IL-6 signaling in the initiation and development of HCC, and they have spawned the interest of targeting molecules involved in the IL-6-JAK-STAT3-signaling pathway as a strategy to block STAT3 activation.25, 26, 27 Compounds targeting IL-6-STAT3 signaling have been reported to suppress the transformed phenotype and tumor progression.5, 6 For example, treatment with antisense oligonucleotides (ASOs) targeting STAT3 leads to an inhibition of proliferation of various hepatoma cell lines, a reduction of tumor size, and an improved survival of mice bearing orthotopically xenografted HCC.28 Interestingly, STAT3 ASO therapy has entered clinical trials, yielding the first promising results particularly for the treatment of lymphoma and lung cancer.29 Other studies on hepatoma describe stronger effects of anti-cancer drugs when this pathway is concomitantly targeted, pointing at the anti-apoptotic effects of JAK-STAT3 signaling and its implication in drug resistance.26, 30
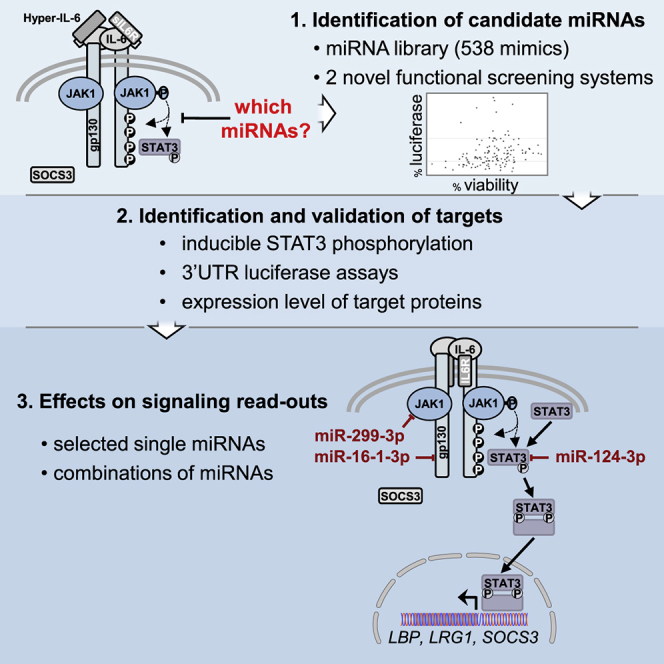
MicroRNAs (miRNAs) have been recognized as promising drug candidates (miRNA replacement therapy) as well as drug targets (e.g., of anti-miRNAs), and nucleic acid-based therapeutic approaches are particularly promising if the target organ is the liver.31, 32, 33 Therefore, we aimed to identify miRNAs that can modulate the IL-6-JAK1-STAT3-signaling pathway in hepatoma cells. To do so, we established 2 novel functional cellular screening systems, which allowed for the selection of miRNAs affecting expression of STAT3-responsive reporter genes. Of a library of 538 mimics, we identified a set of miRNAs targeting 3′ UTR sequences of the main signaling molecules of the pathway. Further characterization revealed their effects on STAT3 or JAK1 protein expression, on gp130 surface availability, as well as on cytokine-mediated signaling events.
Results
Two Novel Cellular High-Throughput Screening Systems Identify Candidate miRNAs as Modulators of the IL-6-JAK-STAT3-Signaling Pathway
To identify miRNAs that regulate the IL-6-JAK-STAT3-signaling pathway, we first performed a pre-screen of a miRNA mimic-based library in HEK293T reporter cells (whose growth is not affected by IL-6 signaling), engineered to express the secreted Cypridina luciferase gene under the control of 6 (synthetic) STAT3-responsive elements (see Figure S1 for a schematic flowchart of this study). Since sIL6R is known to be upregulated in HCC and IL-6 trans-signaling has been implied in HCC,11, 23 we stimulated the cells with hyper-IL-6 (hy-IL-6) to detect miRNAs, which modulate the signaling independently of the membrane-bound IL6R (hy-IL-6 is a designer cytokine composed of sIL6R linked to IL-634).
From 538 molecules tested (see Table S1), 129 miRNA candidates, which either increased or decreased luciferase activity without a major effect on growth (data not shown), were retained for the screen in the hepatoma cell line Hep3B. The latter was engineered to express the secreted Cypridina luciferase gene under the control of a highly IL-6-inducible STAT3-specific rat pancreatitis-associated protein 1 (rPAP1) promoter35, 36 (Figure S2). miRNA candidates were selected based on a reduction or increase in the luciferase activity, normalized to the negative control mimic (NMC) as well as to the viability of the Hep3B reporter cells (Figure S3; it should be noted that hy-IL-6 does not stimulate Hep3B cell proliferation). Based on this analysis, 34 miRNA mimics were selected to be further studied regarding their ability to modulate phosphorylation and, thus, activation of STAT3. These included miR-124-3p and miR-142-3p, previously shown to target molecules involved in the JAK-STAT pathway (STAT3,37 IL6R,38 and gp130,39 respectively), thus serving as positive controls.
Effect of Selected miRNAs on Endogenous pSTAT3 and STAT3 Levels following Cytokine Stimulation
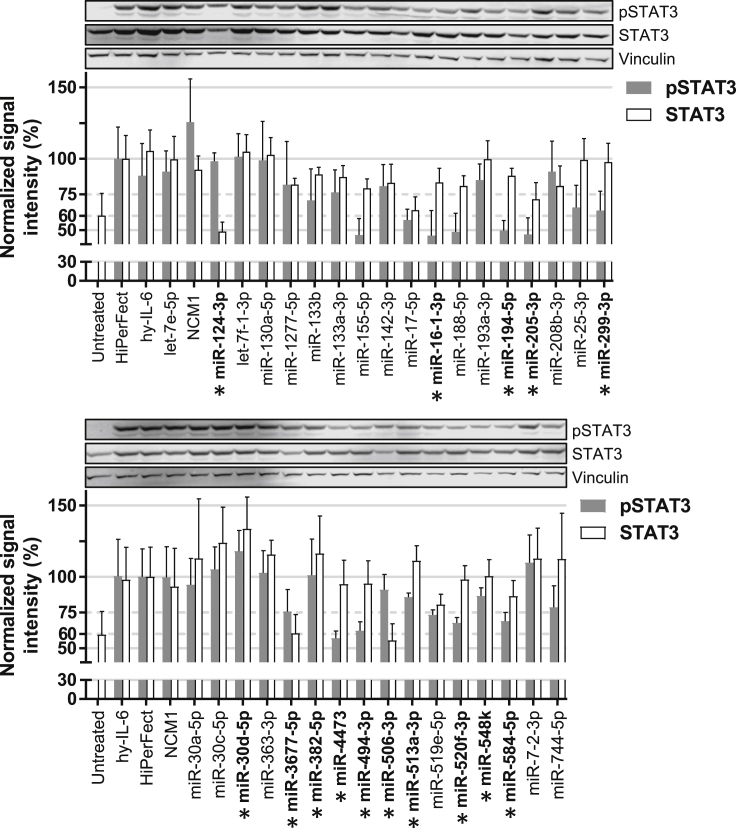
In the previous screening approaches, miRNA candidates were identified, which affected the IL-6-signaling cascade. To further refine this list of interesting miRNAs, the phosphorylation of STAT3 (as functional readout) and total STAT3 (as potential target) were quantified by western blot analysis in Hep3B cells transfected with each of the 34 miRNA mimics and stimulated with hy-IL-6 (Figure 1). miR-142-3p, a miRNA known to target gp130, the common signaling receptor for all IL-6-type cytokines, only slightly decreased phosphorylated (p)STAT3 and STAT3 levels (Figure 1). The other control, miR-124-3p, reduced the expression level of STAT3 by 50%, as previously published,37 but it had no effect on pSTAT3, in line with the finding that only a part of STAT3 proteins generally becomes phosphorylated following cytokine stimulation.40 Based on their potential to reduce pSTAT3 and/or STAT3 levels, 15 candidate miRNAs were selected for the identification of their putative target(s) by luciferase-3′ UTR reporter assays (in bold and marked with a star, Figure 1; some candidates, such as miR-155-5p, were not taken along as they have been extensively studied;41 see also Figure 6).
Figure 1.
Effect of Selected miRNAs on STAT3 and pSTAT3 Protein Levels
Hep3B cells were transfected with 34 selected mimics and 2 days later stimulated for 24 h with hy-IL-6 to activate the JAK-STAT pathway. pSTAT3 and STAT3 protein levels were quantified using the LI-COR device and normalized to total protein staining (data not shown). The normalized signal obtained for each sample was divided by the signal obtained for the transfection control (HiPerFect). Vinculin staining is shown as an additional control. Error bars represent the SD of 3 biological replicates. Mimics selected for luciferase-3′ UTR reporter assays are in bold and marked with a star. A representative western blot result for pSTAT3, STAT3, and Vinculin is shown.
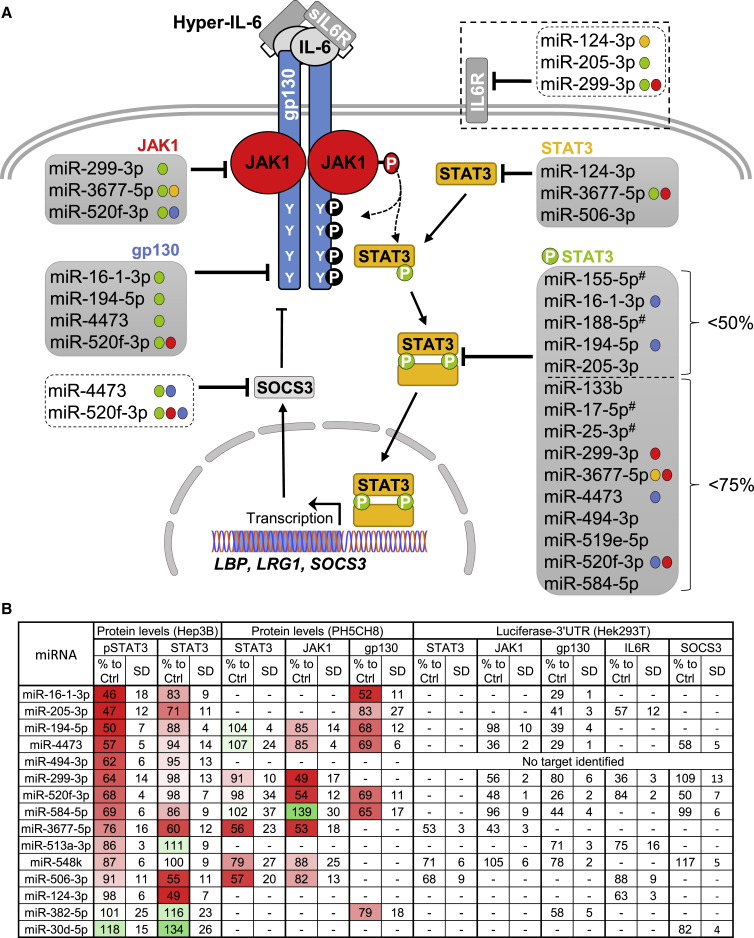
Figure 6.
Summary of the Results Obtained in This Study
(A) Graphical overview of effects of the different miRNA mimics on the IL-6-type cytokine-signaling pathway. For STAT3 phosphorylation, the mimics that reduce the western blot signal to below 75% or below 50% are listed. For JAK1 and STAT3 mimics, those that reduce the western blot signal to below 60% are shown. For gp130, the mimics that reduce surface expression efficiently (below 70%) are listed. The colored dots behind the name of the mimics in the lists indicate additional target genes of the respective miRNA (red, JAK1; blue, gp130; yellow, STAT3; and green, phosphorylation of STAT3). miRNAs labeled with a “#” have already been characterized before. Interestingly, downregulation of STAT3 protein alone was never associated with a reduction of phosphorylated STAT3. (B) Summary of the results obtained upon transfection with each of the 15 selected miRNAs for pSTAT3 (Figure 1, Hep3B), JAK1, and STAT3 (Figure 4, PH5CH8) protein levels, for gp130 surface availability (Figure S5, PH5CH8) and for luciferase-3′ UTR reporter assays (Figures 2, 4, and S7, HEK293T). Results represent the protein and luciferase signals measured for the corresponding target compared to the control (expressed in %). The SD of at least 3 biological replicates is shown. A dash indicates that no data were obtained for this specific miRNA and target. For miR-494-3p, no target could be identified from the ones tested. The list of miRNA effects is colorcoded for the protein levels in Hep3B and PH5CH8 cells (reduction depicted in red and increase in green). In addition, the miRNAs are sorted according to their inhibitory effects on STAT3 activation (from the strongest to the lowest).
Identification of miRNA Mimics Reducing gp130 Surface Availability
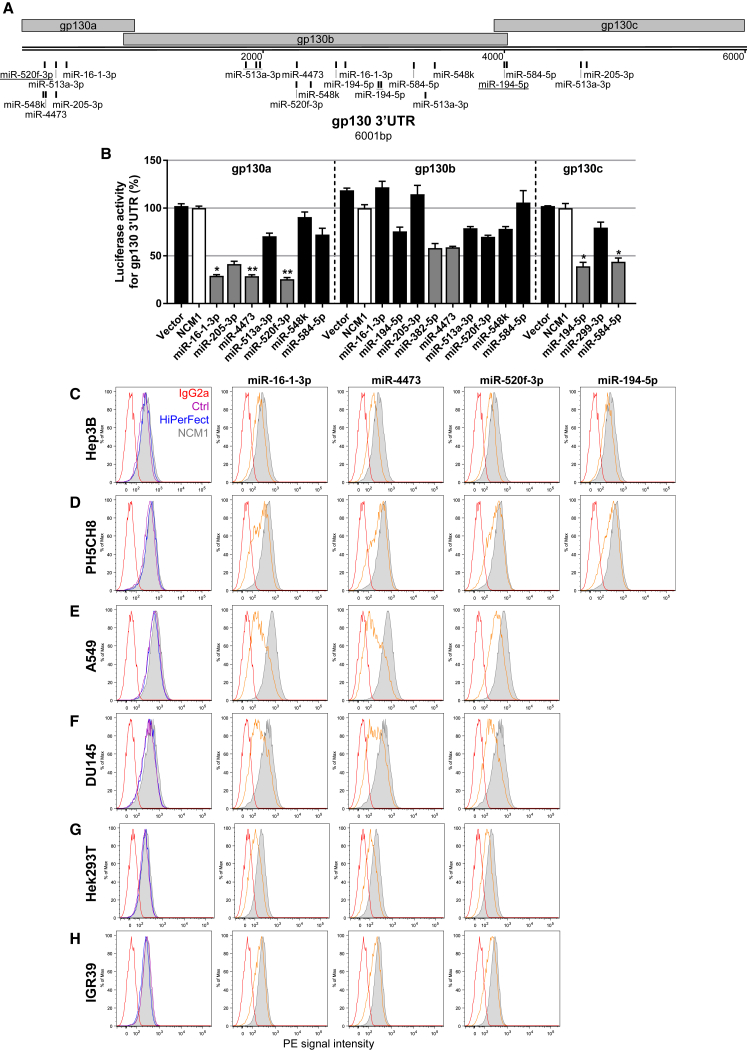
Next, we were interested to screen for miRNAs that specifically and directly target gp130. To do so, luciferase reporter assays were performed using plasmids harboring the cDNA for Gaussia luciferase, a secreted enzyme, followed by 3′ UTR sequences of gp130. As the 3′ UTR of the gene encoding gp130 is very long (6,001 bp), 3 different plasmids with separate, overlapping parts of the gp130 3′ UTR were constructed (Figure 2A). HEK293T cells were used for the co-transfection experiments as they can be better transfected than liver-derived cells, thereby providing a smaller variability between the biological replicates.
Figure 2.
Identification of miRNAs Targeting the Co-receptor gp130 and Affecting Its Surface Availability
(A) Graphic representations of the gp130 3′ UTR as well as the predicted binding sites (based on TargetScan website). Sites conserved among mammals are underlined. (B) HEK293T cells were co-transfected with mimics specifically selected for one of the vectors harboring the Gaussia luciferase cDNA followed by overlapping 3′ UTR sequences of the gene encoding gp130 (gp130a/b/c). Mimics selected for further functional assays are shown in gray, and negative control mimic 1 (NCM1) is in white. Error bars represent the SD of 3 biological replicates. Kruskal-Wallis followed by Dunn’s Post hoc test were performed to assess statistical significance represented with *p < 0.05 and **p < 0.01. (C–H) Selected miRNA candidates affect the surface availability of gp130 in various cell lines. (C) Hep3B hepatoma cells, (D) non-neoplastic PH5CH8 liver cells, (E) A549 lung adenocarcinoma cells, (F) DU145 prostate cancer cells, (G) HEK293T cells, and (H) IGR39 melanoma cells were either left untreated (Ctrl, in purple) or transfected with a negative control (NCM1, in gray) or one of the selected miRNA mimics and analyzed for gp130 surface expression level by flow cytometry. Candidate miRNAs are depicted in orange, the isotype control is in red, and the transfection control is in blue.
After a first, preliminary experiment with all 15 miRNA candidates (data not shown), 10 were further evaluated for their binding to the gp130 3′ UTR sequence: miR-16-1-3p, miR-205-3p, miR-4473, and miR-520f-3p reproducibly decreased the luciferase activity of the respective gp130 3′ UTR plasmid by more than 50% (gp130a, Figure 2B). Similarly, miR-194-5p and miR-584-5p as well as miR-382-5p and miR-4473 reduced the luciferase activity of the plasmids harboring the part b or c, respectively, by at least 40% (Figure 2B). No off-target effects of these miRNA mimics were found in control experiments using only the backbone vector (Figure S4). In total, 7 different miRNA mimics (miR-16-1-3p, miR-194-5p, miR-205-3p, miR-382-5p, miR-4473, miR-520f-3p, and miR-584-5p, depicted in gray in Figure 2B) had a strong effect on gp130 3′ UTR.
As IL-6 (classical or trans-) signaling is initiated from gp130 receptors present at the plasma membrane, we evaluated the effects of selected miRNAs on the surface expression of endogenous gp130 by flow cytometry in different cell lines from various tissues. In comparison to the cells transfected with NCM1 (in gray), miR-16-1-3p, miR-194-5p, miR-4473, and miR-520f-3p decreased gp130 surface expression in both Hep3B hepatoma cells (Figure 2C) and non-transformed PH5CH8 liver cells (Figure 2D). Two miRNAs had cell-specific effects: miR-382-5p reduced gp130 surface expression only in Hep3B cells, while miR-584-5p affected gp130 surface expression (relative to the NCM1 control) only in PH5CH8 cells (Figures S5A and S5B). Finally, miR-205-3p had almost no effects on the gp130 levels in both tested liver cell lines (Figures S5A and S5B), although it had clear effects in the reporter gene assays.
miR-16-1-3p, miR-4473, and miR-520f-3p were further tested in a variety of other cells types, such as lung adenocarcinoma (A549), prostate carcinoma (DU145), HEK293T, and melanoma cells (IGR39). Upon transfection of each miRNA tested (orange), histograms were strongly shifted to the left in comparison to control cells (Figures 2E–2H), indicating that these 3 miRNAs have a robust effect on the surface expression of gp130 in a variety of cells.
miR-16-1-3p, miR-4473, and miR-520f-3p Inhibit hy-IL-6-Inducible Phosphorylation of STAT3 and Differentially Affect the Expression of a Target Gene
Surface availability of gp130 has been shown to regulate the cell responsiveness to IL-6-type cytokines. For example, gp130 expression is downregulated during granulocyte differentiation42 and upon activation of T cells.43 Moreover, pre-stimulation with IL-1β reduces gp130 availability at the surface of human hepatocytes44 or monocytes45 in a p38-dependent manner. Therefore, we investigated if the effects of miR-16-1-3p, miR-4473, and miR-520f-3p on gp130 were sufficient to modulate the activation of STAT3 upon hy-IL-6 stimulation. First of all, we confirmed that the corresponding miRNA expression level was indeed increased upon mimic transfection (Figure S6). An only slightly suppressive effect of the 3 miRNA mimics on gp130 mRNA expression was observed (Figure 3A), suggesting that translational repression, rather than mRNA decay, may substantially contribute to gp130 downregulation (for a review, see Iwakawa and Tomari46). Interestingly, all 3 miRNA mimics largely reduced inducible STAT3 phosphorylation in comparison to the NCM1, whereas STAT3 protein levels remained unchanged (single mimics, Figure 3B). The basal level of STAT3 phosphorylation (i.e., without stimulation) was not affected by any of the 3 mimics (or combinations thereof), indicating that this “tonic” STAT3 phosphorylation may be mediated by signaling events independent of gp130 (e.g., elicited by growth factors or non-IL-6-type cytokines).
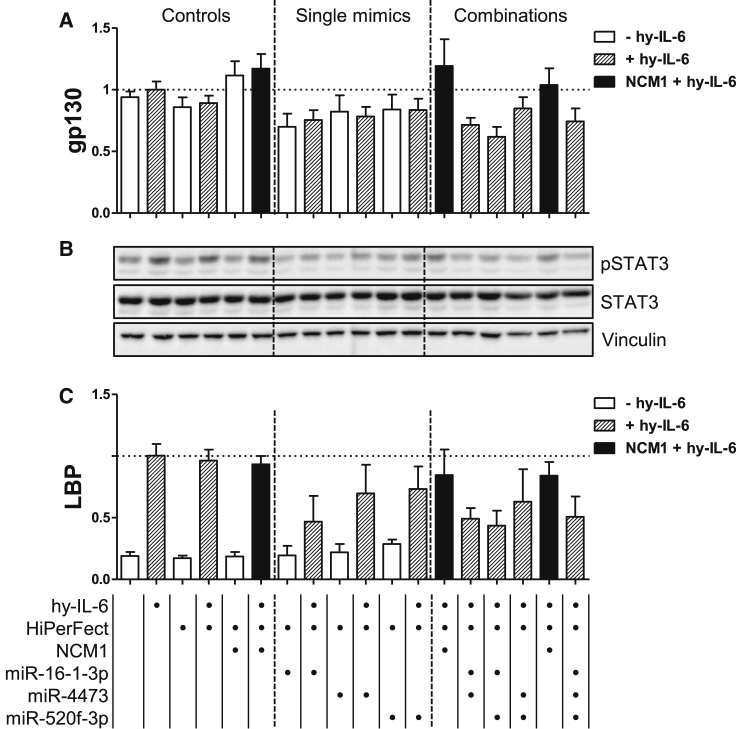
Figure 3.
miR-16-1-3p, miR-4473, and miR-520f-3p Partially Prevent the Activation of STAT3 upon hy-IL-6 Stimulation by Targeting gp130 mRNA, Reducing the Expression of a hy-IL-6 Target Gene in PH5CH8 Cells
Cells were left untreated, reverse transfected with NCM1 or miR-16-1-3p, miR-4473, and miR-520f-3p mimics (20 nM), and were stimulated with hy-IL-6 (20 ng/mL). Single mimics as well as combinations were tested and compared to the respective NCM1 control (in black). For the double or triple combinations, 40 or 60 nM NCM1 was used as the respective control. (A) Normalized mRNA expression level of gp130. (B) Representative western blot analysis of both the activated and total forms of STAT3 and of the Vinculin as loading control. (C) Normalized mRNA expression level of Lipopolysaccharide-binding protein (LBP). The mRNA levels were monitored upon mimic transfection and hy-IL-6 stimulation (depicted with dashed lines). Error bars represent the SD of at least 3 biological replicates. Bottom indicates the applied treatments per lane.
To further address the impact of these miRNAs on a signaling readout, we monitored their effect on the inducibility of Lipopolysaccharide-binding protein (LBP), a target gene of IL-6 signaling.47 All 3 miRNAs reduced inducible LBP mRNA expression in PH5CH8 cells; the effect was strongest for miR-16-1-3p, with a reduction by more than 50% in comparison to the cytokine-stimulated control cells (Figure 3C). Combination of 2 or all 3 miRNAs had no additional effect than the one measured for miR-16-1-3p alone. Of note, miR-4473 and miR-520f-3p, which were less effective in dampening inducible LBP mRNA expression compared to miR-16-1-3p, also target the 3′ UTR of SOCS3, the major negative regulator of the pathway (Figure S7A).
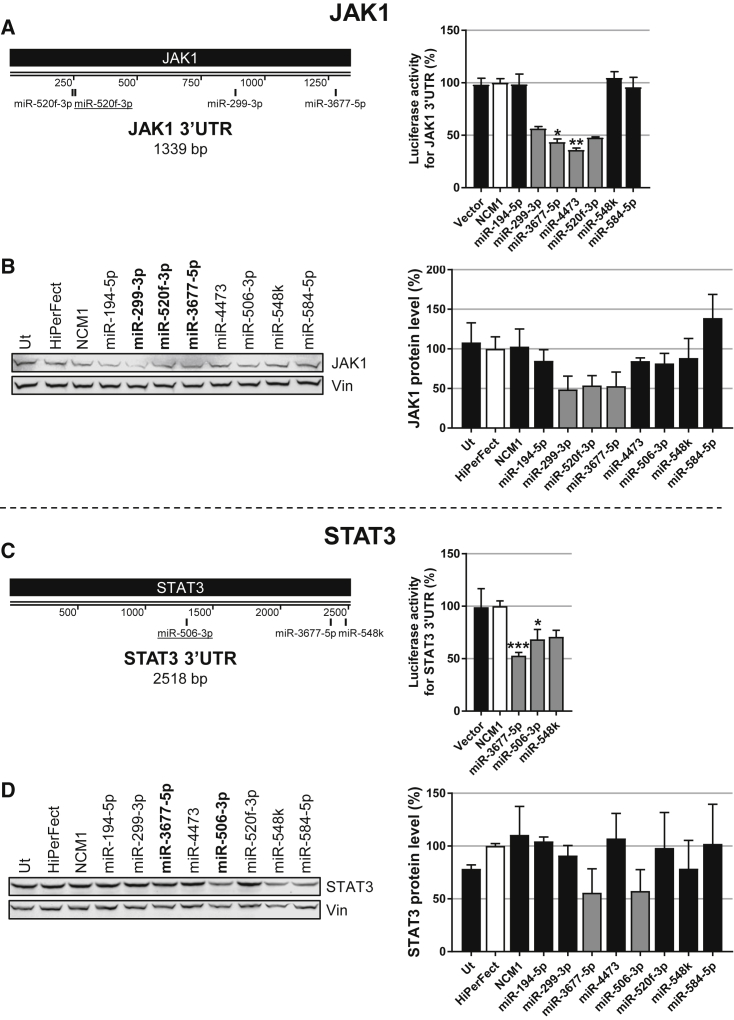
Identification of miRNA Mimics Directly Targeting JAK1 and STAT3
In similar approaches, we also screened for miRNAs targeting JAK1 and STAT3, 2 other major players of the pathway. In luciferase-3′ UTR reporter assays, we identified 8 miRNA candidates (in gray, Figures 4A and 4C) to be further analyzed for their effects on endogenous JAK1 and/or STAT3 protein levels. To do so, PH5CH8 cells were transfected with either NCM1 or one of the selected mimics (miR-194-3p, miR-299-3p, miR-3677-5p, miR-4473, miR-506-3p, miR-520f-3p, miR-548k, and miR-584-5p).
Figure 4.
Identification of miRNAs Targeting JAK1 and STAT3 and Affecting Their Endogenous Protein Levels
(A and C) Graphic representations of (A) JAK1 and (C) STAT3 3′ UTRs as well as predicted binding sites (TargetScan). Sites conserved among mammals are underlined. HEK293T cells were co-transfected with the mimics specifically selected for each construct and one of the vectors harboring the Gaussia luciferase cDNA followed by the 3′ UTR of the gene encoding (A) JAK1 or (C) STAT3. Mimics selected for further functional assays are shown in gray, and NCM1 is in white. Kruskal-Wallis followed by Dunn’s post hoc test were performed to assess statistical significance represented with *p < 0.05, **p < 0.01, and ***p < 0.001. (B and D) Selected miRNAs downregulate endogenous JAK1 and STAT3 protein levels in PH5CH8 cells. (B) miR-299-3p, miR-3677-5p, and miR-520f-3p reduce JAK1 protein levels. (D) miR-3677-5p and miR-506-3p reduce STAT3 protein levels. A representative western blot as well as quantifications are depicted. Error bars represent the SD of at least 3 biological replicates.
The 3′ UTR of the mRNA encoding JAK1 was found to be strongly targeted by miR-299-3p, miR-3677-5p, miR-4473, and miR-520f-3p (Figure 4A). Importantly, of these 4, 3 mimics (miR-299-3p, miR-520f-3p, and miR-3677-5p) also strongly decreased JAK1 protein level by approximately 50% (depicted in gray, Figure 4B), while miR-4473 only had a modest effect.
Regarding STAT3, miR-3677-5p, miR-506-3p, and miR-548k were identified as mimics targeting its 3′ UTR (Figure 4C). STAT3 protein levels in non-neoplastic PH5CH8 cells were reduced by ∼50% upon transfection with miR-3677-5p and miR-506-3p (Figure 4D, depicted in gray). However, miR-548k had less impact on STAT3 protein expression (Figure 4D). Similar findings were observed previously in Hep3B hepatoma cells (Figure 1C).
Single Mimics Targeting Various Proteins of the IL-6-Signaling Pathway Attenuate IL-6-STAT3-Induced Gene Readouts as Effectively as Combination Treatments
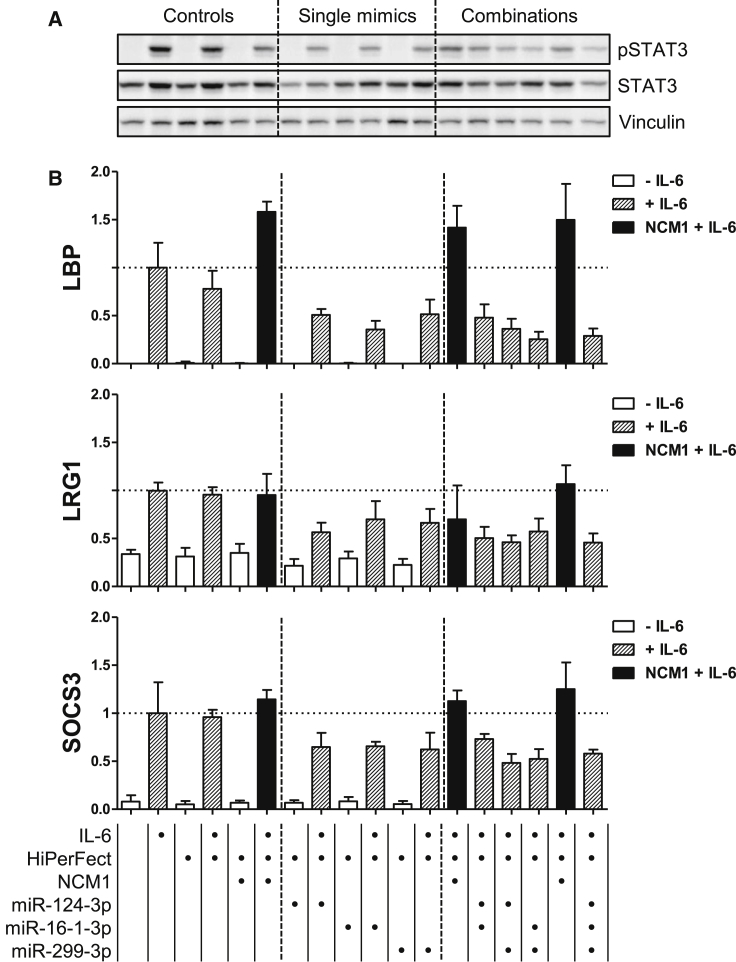
As a next step, we tested whether a combination of miRNAs with different targets in the IL-6-signaling pathway would have stronger inhibitory effects on cytokine signaling compared to single mimics. For these combinatory experiments, we selected miR-16-1-3p, miR-299-3p, and miR-124-3p: miR-16-1-3p targets gp130, miR-299-3p targets JAK1 (and likely IL6R, see Figure S7B), and miR-124-3p targets STAT3 (and likely IL6R, see Figure S7B) (see Figure 6 for a summary). We now stimulated with IL-6 (and not with hy-IL-6) so that the potential effects of a lower IL6R expression would also be reflected in the results. In this context, we utilized Hep3B cells as they respond better to IL-6 stimulation than PH5CH8 cells (data not shown).
Figure S8 shows the controls for the efficient transfection of the mimics as well as their effects on the different target mRNAs. JAK1 mRNA expression was only affected by miR-299-3p; basal and IL-6-inducible expression of STAT3 mRNA was attenuated only by miR-124-3p. In contrast, expression of gp130 was affected not only by miR-16-1-3p (in line with the previous experiments) but also by miR-299-3p. Interestingly, IL6R mRNA expression was reduced by all 3 mimics, which was expected for miR-124-3p and miR-299-3p (see Figure S7B), but not for miR-16-1-3p (miR-299-3p and miR-16-1-3p had little effect in the preliminary experiment with reporter plasmids comprising 3′ UTR sequences of gp130 or IL6R, respectively; data not shown).
Clear inhibitory effects of all 3 miRNAs were observed for the activation of STAT3 (Figure 5A) as well as for the IL-6-inducible expression of the STAT3 target genes LBP, Leucine-rich alpha-2 glycoprotein (LRG1), and SOCS3 (Figure 5B). Interestingly, combination treatments were not more effective than single treatments (Figure 5, combinations).
Figure 5.
miR-124-3p, miR-16-1-3p, and miR-299-3p Partially Prevent the Activation of STAT3 upon IL-6 Stimulation by Targeting STAT3, IL6R, gp130, and JAK1 mRNA, and They Reduce the Induction of the Expression of IL-6 Target Genes in Hep3B Cells
Cells were left untreated, reverse transfected with NCM1 or miR-124-3p, miR-16-1-3p, and miR-299-3p mimics (20 nM), and stimulated with IL-6 (20 ng/mL). Single mimics as well as combinations were tested and compared to the respective NCM1 control (in black). For the double or triple combinations, 40 or 60 nM NCM1 was used as the respective control. (A) Western blot analysis for both the activated and total forms of STAT3 and of Vinculin as loading control. (B) Normalized mRNA expression levels of LBP, Leucine-rich alpha-2 glycoprotein (LRG1), and SOCS3 were monitored upon mimic transfection and IL-6 stimulation (depicted with dashed lines). Error bars represent the SD of at least 3 biological replicates. Bottom indicates the applied treatments per lane.
Discussion
In this study, (1) we show that a screen of 538 miRNA mimics designed to identify miRNAs influencing IL-6 (trans-)signaling yielded a panel of interesting candidate miRNAs affecting the (hyper-)IL-6-gp130-JAK1-STAT3 pathway. (2) We validated several selected miRNAs by showing that they specifically interact with the 3′ UTR of the gp130, JAK1, and/or STAT3 mRNAs. (3) We confirmed some of those miRNAs to specifically affect the expression of different proteins in this pathway, notably, gp130, JAK1, and STAT3. (4) We found that treatment with single mimics was as effective in downregulating cytokine signaling as combinations of multiple miRNA mimics targeting either gp130 or various molecules in the IL-6-signaling pathway.
As described in the Introduction, IL-6 has important systemic and local effects in the pathogenesis of cancer. It influences tumor development, angiogenesis, cell proliferation, migration, and invasion; it has anti-apoptotic and immunomodulatory effects.5, 6 Previous studies have demonstrated miRNAs as regulators of the JAK-STAT-signaling pathway. For instance, miR-124 has been extensively studied and shown to target both STAT337 and IL6R.38, 48 Another example is miR-142-3p, which was shown to directly target the co-receptor gp130 in cardiac myocytes,39 macrophages,49 and endometrial stromal cells.50
The aim of this study was to identify miRNAs with the potential for inhibiting the IL-6-JAK-STAT3-signaling pathway and that, therefore, might be of therapeutic relevance. Although there are still issues with miRNA-based therapies (e.g., delivery, poor penetration in tissues, stability, off-target effects; reviewed in Chen),51 the potential of nucleic acids as versatile drug candidates is more and more recognized.32, 33 For example, miRNA-based drugs have been successfully used in pre-clinical studies (e.g., miR-2652 and anti-miR-1753, 54) as well as in first clinical trials, such as for miR-16-based mimics.55 Here we describe and characterize the effects of several novel miRNAs and of some previously described ones (e.g., miR-124-3p and miR-506-3p) on the IL-6-signaling pathway (see Figure 6). We focused on miRNAs that did not affect cell viability, but it would also be very interesting to further investigate those miRNAs that alter cellular viability, as previously done for cholangiocarcinoma.56 For example, the miR-374 family, from which 3 of 6 members were included in the large library and led to an enhanced cell proliferation (data not shown), has already been associated with an increased proliferation and survival, e.g., in gastrointestinal stromal tumors,57 gastric cancer,58 and hepatoma.59
miRNA-Mediated Effects on STAT3
Interestingly, we noticed that among those miRNAs that most strongly reduced STAT3 protein levels (miR-124-3p, miR-3677-5p, and miR-506-3p), only miR-3677-5p reduced pSTAT3 signals (Figure 1). miR-3677-5p was found to also target JAK1 (see below), which may explain that this miRNA is more effective on STAT3 phosphorylation than the others. Our results indicate that a reduction of STAT3 levels by 50% does not translate into decreased pSTAT3 levels. Indeed, only a fraction of STAT3 is phosphorylated upon IL-6 stimulation.40 A certain reduction in the expression can thereby be tolerated without a necessary effect on the absolute amounts of pSTAT3.
Another interesting aspect of STAT3 silencing is that, in the absence of STAT3, IL-6 leads to a sustained STAT1 activation60 and switches to an interferon (IFN)γ-like response in hepatoma cells.61 IFNγ is one of those cytokines long known to be involved in anti-tumor defense, since interferon gamma receptor (IFNγR) and STAT1 deficiency has been described to increase cancer susceptibility upon treatment with chemical pathogens in mice.62, 63 Of note, the observed reduction of STAT3 expression (by 50%) is not only insufficient to reduce the phosphorylation of STAT3 but also not able to induce a compensatory phosphorylation of STAT1 (data not shown).
In contrast to our first findings (Figure 1), several studies have observed a decrease of pSTAT3 signals by miR-124-3p.37, 38, 48 However, it needs to be taken into consideration that this miRNA also targets IL6R (seen also in our hands in the luciferase-3′ UTR assay [Figure S7B] and confirmed at the mRNA levels [Figure S8]), which can translate into a reduced signaling input when analyzing signals elicited by IL-6 (as observed in our final experiment utilizing IL-6 as a stimulating cytokine; Figure 5). Since we used the designer cytokine hy-IL-6 for the first experiments, the observed signaling effects were independent of the surface level of IL6R.
miRNA-Mediated Effects on JAK1
miRNA-mediated downregulation of JAK1 in cancer has been shown to suppress metastasis and invasion (e.g., miR-214 in lung carcinoma,64 miR-340 in HCC,65 and miR-488 in pancreatic ductal adenocarcinoma66). We found the 3 miRNAs miR-299-3p, miR-3677-5p, and miR-520f-3p to diminish JAK1 protein levels by at least 40%. While JAK1 knockdown only partially inhibits IL-6 signaling, expression of kinase-deficient mutants of JAK1 totally abolishes IL-6 signaling (unpublished data), indicating that the partial signaling reduction in the knockdown scenario is due to compensatory binding of the other JAKs (JAK2 or Tyk2) to gp130. Thus, even a very efficient downregulation of JAK1 by a miRNA is unlikely to have drastic effects on IL-6 signaling.
In addition, the downregulation of JAK1 is likely to impact on a variety of other cytokine signals, as JAK1 is the most promiscuous Janus kinase from the 4 family members. It is used for the signaling of the majority of the ∼40 cytokine receptors. For instance, IFNα/β, IFNγ, and IL-27 signaling, all associated with anti-tumor functions, are concomitantly reduced when JAK1 is downregulated.61 Interferon-mediated antiviral responses have already been shown to be impaired by miRNAs targeting JAK1 (e.g., miR-30c67 and miR-37368). Therefore, it needs to be evaluated whether an inhibition of JAK1 would be useful in a tumor situation, as IFNs essentially have anti-tumor effects.69 On the other hand, these miRNAs may be useful to diminish inflammatory processes involving both IFNs and IL-6-type cytokines. Similarly, due to their strong anti-inflammatory effects, JAK inhibitors are regarded as highly promising in inflammatory diseases.70
miR-299-3p was shown to target the 3′ UTR of JAK1 and to efficiently suppress JAK1 mRNA and protein expression (Figures S8C and 4B). Interestingly, it also affected the 3′ UTR of the IL6R (Figure S7B). Known targets of miR-299-3p include vascular endothelial growth factor A (VEGFA),71, 72 androgen receptor,73 and ATP Binding Cassette Subfamily E Member 1 (ABCE1),74 which have been linked to tumor-suppressive functions (less angiogenesis, viability, or drug resistance). Moreover, miR-299-3p has been shown to be downregulated in The Cancer Genome Atlas (TCGA) HCC tumor samples and to inhibit growth and invasion of SK-HEP-1 hepatoma cells.75
We also identified miR-3677-5p as a novel miRNA involved in the regulation of the JAK-STAT pathway. Interestingly, it targets the expression of both STAT3 and JAK1 with poorly conserved binding sites present in their 3′ UTRs (Figures 4A and 4C). Overexpression of miR-3677-5p reduces mRNA and protein levels of STAT3 and JAK1 in Hep3B and Huh-7 hepatoma cells as well as in non-neoplastic PH5CH8 cells (Figures 1 and 4; data not shown). Very little is known about this miRNA. In contrast to most other miRNAs discussed here, which are associated with tumor-suppressive functions, miR-3677-5p is upregulated in HCC and an increased expression correlates with poor survival,76, 77 indicating a potential tumor-promoting role. Of note, however, this is one of the very few miRNAs showing divergent effects in the 2 reporter cell lines: while it reduced luciferase activity in the Hep3B reporter cells, it increased reporter gene activity in the first screen in Hek-293T reporter cells (see Table S2). Clearly, this miRNA warrants further analysis.
miRNA-Mediated Effects on gp130
Finally, we identified miRNAs efficiently decreasing gp130 surface expression. miR-16-1-3p, miR-4477, miR-520f-3p, and miR-194-5p showed a reduction of at least 25% of gp130 surface expression. As gp130 is the signaling receptor shared by the IL-6-type cytokines, these miRNAs may be particularly relevant in situations requiring the inhibition of various cytokines of the IL-6 family. Indeed, monotherapies with monoclonal antibodies targeting IL-6 or IL6R have failed in clinical trials against multiple myeloma,78, 79, 80 metastatic renal cell carcinoma,81 and prostate cancer,82, 83 probably due to the compensation of the IL-6-signaling pathway by other gp130 ligands.84, 85
One of the miRNAs identified here to target gp130 was also reducing JAK1 protein levels (miR-520f-3p). Interestingly, 2 of the miRNAs efficiently reducing gp130 surface availability (miR-16-1-3p and miR-194-5p) caused a strong reduction of phosphorylated STAT3 (suppression of greater than 50%; Figure 1). The 2 other miRNAs (miR-4473 and miR-520f-3p) only reduced pSTAT3 levels at least 25% compared to the control. miR-4473 and miR-520f-3p were also less potent than miR-16-1-3p when monitoring the effects on LBP expression, a STAT3-inducible gene (Figure 3C). Interestingly, those 2 miRNAs also bound to SOCS3 3′ UTR (Figure S7A), the major inhibitor of IL-6 signaling, which might explain their overall weaker effect. A careful comparison with the effects of small interfering RNAs (siRNAs) specifically downregulating their target mRNAs would allow for a further dissection of the impact of “other” miRNA targets on the signaling readouts. However, in order to facilitate this comparison, doses of siRNAs would have to be adjusted to achieve only partially suppressed levels similar to the ones obtained with miRNAs.
miR-16-1-3p is part of a well-characterized tumor suppressor cluster (mir-15a∼16-1, reviewed in Huang),86 located in the chromosomal region 13q14.2, which is often deleted in cancer.87, 88, 89, 90 Interestingly, min-icells loaded with miR-16-based mimics (TargomiRs) have entered first clinical trials for patients with malignant mesothelioma.55 While many targets have been identified for miR-16-5p, its passenger strand is less studied. Recently, it was shown to target TWIST1 in gastric91 and non-small-cell lung92 cancer cells. To our knowledge, no link between miR-16-1-3p and the IL-6-JAK-STAT3-signaling pathway had been established so far.
miRNAs affecting the receptor availability may be particularly promising candidates for further investigation (see Figure 6B), and they will have more specific effects than miRNAs targeting signaling molecules shared by many cytokines. Notably, several of the miRNAs identified to affect IL-6 signal transduction have actually multiple targets within the pathway (see Figure 6). Combinations of miRNAs, either sharing the same target (gp130; Figure 3) or regulating different signaling molecules of the pathway (gp130, JAK1, STAT3, and/or IL6R; Figure 5) did not increase effects compared to single treatments, indicating that drug candidates based on a single miRNA might be able to achieve optimal signal attenuation. However, possible saturation effects need to be addressed by detailed dose-response analyses (we used up to 60-nM mimics for the combination treatments shown in Figures 3 and 5; for comparison, Liang et al.93 described that transfection of 10-fold less siRNA already led to considerable AGO2 competition of endogenous miRNAs). A more detailed analysis of miRNA cooperation may also be warranted, considering that the spacing between two miRNAs on the 3′ UTR of a common target mRNA may contribute to the efficiency of target mRNA downregulation. Saetrom et al.94 have determined the optimal spacing for downregulation to be between 17 and 35 nt between the seed sites.
Subsequent studies will have to evaluate the therapeutic relevance of selected miRNAs on cellular behavior (proliferation, migration, and invasion) as well as in the context of IL-6-dependent disease models, in particular for inflammatory diseases (e.g., rheumatoid arthritis) and cancer (e.g., HCC).
Materials and Methods
Materials and Cell Culture
All cells were grown at 37°C in a water-saturated atmosphere at 5%C CO2. HEK293T, Hep3B, PH5CH8, and DU145 cells were maintained in DMEM (Lonza, Basel, Switzerland) and A549 cells in DMEM-F12 medium (Lonza, Basel, Switzerland), both supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, Erembodegem, Belgium), 25 mM HEPES (Lonza, Basel, Switzerland), and 100 μg/mL normocin (InvivoGen, Toulouse, France). IGR39 cells were cultured in RPMI 1640 medium (Lonza, Basel, Switzerland) supplemented with 10% FBS and 100 μg/mL normocin.
All miRIDIAN miRNA mimics were obtained from Dharmacon (GE Healthcare, Diegem, Belgium). Vectors containing the 3′ UTR sequence of the genes coding for IL6R (GenBank: NM_181359.1), gp130 (GenBank: NM_001190981.1), JAK1 (GenBank: NM_002227.2), SOCS3 (GenBank: NM_003955.4), and STAT3 (GenBank: NM_213662.1) were purchased from GeneCopoeia (Tebu-Bio, Boechout, Belgium), as well as the backbone vector (pEZX-MT05) is used as a control to monitor the off-target effects. Mimic transfections were performed using HiPerFect Transfection Reagent (QIAGEN, Venlo, the Netherlands) and 20 nM miRNA mimic (or control), according to the manufacturer’s instructions. Co-transfections of 3′ UTR reporter vectors and miRNA mimics were performed using DharmaFect Duo (Dharmacon, GE Healthcare, Diegem, Belgium), 100 ng 3′ UTR reporter vector, and 20 nM miRNA mimic (or control), according to the manufacturer’s protocol.
2 days after mimic transfection, cells were stimulated for 24 h with 20 ng/mL hy-IL-6, a designer cytokine comprising IL-6 bound to the extracellular domain of IL6R (sIL6R) through a flexible polypeptide linker.34 For Figure 5, Hep3B cells were stimulated with 20 ng/mL IL-6 (PeproTech, Tebu-Bio, Boechout, Belgium) instead of hy-IL-6.
Engineering of Stable Cell Lines Allowing for High-Throughput Screening of miRNAs Affecting the JAK-STAT(3)-Signaling Pathway
HEK293T cells were plated in 6-well plates in complete medium, and, 24 h later, they were transduced with lentiviral particles containing 6 STAT3-responsive elements (STAT3-REs, sequence: 5-TTCTGGGAA-3′), followed by a minimal cytomegalovirus (CMV) promoter in front of the secreted Cypridina luciferase gene and linked to a puromycin resistance gene. 48 h after transduction, selection of transduced cells was initiated with 3 μg/mL puromycin (InvivoGen, Toulouse, France). After several rounds of selection, hy-IL-6 responsiveness of the construct was assessed by measuring the Cypridina luciferase activity in the supernatant of stimulated HEK293T-STAT3-RE-Cypridina luciferase (Cluc) cells (20 ng/mL hy-IL-6). The transfected clone showing the highest fold induction of luciferase activity upon cytokine treatment was selected and amplified.
Hep3B cells (hepatoma cells with a ploidy status of n = 3.2, see Wilkens et al.95; for mutations in cancer genes see Table S1 in Ewald)96 were plated in 6-well plates in complete medium and transfected with a linearized plasmid containing the STAT3-inducible promoter rPAP135, 36 in front of the secreted Cypridina luciferase gene, using the PromoFectin-Hepatocyte reagent (PromoCell, Bio-Connect, Huissen, the Netherlands), following the manufacturer’s protocol. 4 h later, serum-reduced Optim-MEM medium (Life Technologies, Merelbeke, Belgium) was replaced by normal DMEM. After 4 days, selection of transfected cells was initiated with 500 μg/mL G418 (Sigma-Aldrich, Diegem, Belgium). After several rounds of selection, a pool of Hep3B-rPAP1-Cluc cells was tested for successful inducible luciferase production and secretion upon stimulation with 20 ng/mL hy-IL-6 (Figure S2B) and amplified.
Selection of miRNAs for the Pre-screen
We designed a library consisting of 538 miRNAs, which were selected from miRBase version (v.)21 by the following criteria: (1) presence in the high-confidence list from miRBase,97 (2) predicted by Diana web server v.5.098, 99 to target molecule(s) related to IL-6 signaling, (3) presence in the top 100 list of miRNAs regulated upon hy-IL-6 stimulation in primary hepatocytes (microarray analysis, ArrayExpress: E-MTAB-6572),100 and (4) miRNAs previously published as regulated by STAT3 (for a complete list, see Table S1).
Luciferase Reporter Gene Assays
For the miRNA library screens, 104 HEK293T-STAT3-RE-Cluc or Hep3B-rPAP1-Cluc engineered cells were reverse transfected on a 96-well plate with 20 nM of the respective miRIDIAN miRNA mimic using HiPerFect Transfection Reagent (QIAGEN, Venlo, the Netherlands). 48 h after transfection, cells were stimulated for 24 h, as described in the figure legends, after which supernatants were collected and kept at −20°C until measurement. Briefly, 10 μL supernatant was used to measure the secreted Cypridina luciferase activity using the BioLux Cypridina Luciferase assay kit (New England Biolabs, Bioke, Leiden, the Netherlands), following the manufacturer’s instructions. Automatic injection of substrate and measurement were performed with the CLARIOstar plate reader (BMG Labtech, ISOGEN Life Science, De Meern, the Netherlands).
For the identification of IL6R, gp130, JAK1, SOCS3, and STAT3 as potential target(s) of selected miRNA mimics, 104 HEK293T cells were plated on a 96-well plate and co-transfected the next day with a mimic and either the backbone vector or a vector containing the Gaussia luciferase gene followed by (a portion of) the 3′ UTR sequence of IL6R (IL6Ra or b), gp130 (gp130a, b, or c), JAK1, SOCS3, or STAT3. After 2 days of transfection, cell supernatants were collected to assess the activities of both the secreted Gaussia luciferase and the secreted Alkaline phosphatase, used for normalization purposes, using the Secrete-pair dual luminescence assay kit (GeneCopoeia, Tebu-Bio, Boechout, Belgium), according to the manufacturer’s protocol. Both activities were determined with the CLARIOstar plate reader. For all luciferase experiments, at least 3 independent biological replicates, each with 3 technical replicates, were performed.
Cell Viability Assessment
To monitor the effects of the miRNA mimics on cell growth (and thereby reduce false-positive results), cell viability was assessed for the same wells as the luciferase measurements using the PrestoBlue reagent (Invitrogen, Thermo Fisher Scientific, Erembodegem, Belgium), according to the manufacturer’s instructions. The fluorescence of each well was then measured with the CLARIOstar plate reader and normalized to the fluorescence emitted by the cells treated with HiPerFect transfection reagent only, present on each plate.
Flow Cytometry
The effects of miRNA mimics on gp130 receptors were assessed by flow cytometry. Briefly, transfected cells were resuspended in cold PBS supplemented with 5% FBS and 0.1% sodium azide (Sigma-Aldrich, Diegem, Belgium) and incubated with 0.5 μg antibody against gp130 (555757, BD Biosciences, Erembodegem, Belgium) or the corresponding isotype control antibody, immunoglobulin G (IgG)2α (21225021, Immunotools, Friesoythe, Germany), for 1 h at 4°C. Cells were then washed with cold PBS-azide and incubated with secondary labeled antibodies (715-116-150, Jackson ImmunoResearch Laboratories, Bio-Connect, Huissen, the Netherlands). After 1 h of incubation at 4°C, cells were again washed with cold PBS-azide and then analyzed on a FACSCanto II flow cytometer using the FACSDiva software (BD Biosciences, Erembodegem, Belgium). Overlay plots were created using the FlowJo software (FlowJo, Ashland, OR).
Cell Lysis and Western Blot Analysis
Cultured cells were lysed on the plate with ice-cold Laemmli 1× buffer and kept at −20°C. Before separation by SDS-PAGE and blotting either onto a polyvinylidene fluoride (PVDF)-PSQ (for LI-COR) or a PVDF-FL (for enhanced chemiluminescence [ECL]) membrane (Millipore, Overijse, Belgium), protein extracts were heated for 10 min at 96°C. Blots were incubated overnight with antibodies against human JAK1 (610232, BD Biosciences, Erembodegem, Belgium), STAT3 (610189, BD Biosciences, Erembodegem, Belgium), pSTAT3 (9145, Cell Signaling Technology, Bioke, Leiden, the Netherlands), or Vinculin (13901, Cell Signaling Technology, Bioke, Leiden, the Netherlands). After washing, membranes were incubated for 1 h at room temperature with either fluorescent (LI-COR Biosciences, Westburg, Leusden, the Netherlands) or horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology, Bioke, Leiden, the Netherlands) for LI-COR or ECL detection, respectively. Prior to ECL detection, membranes were incubated in an ECL solution containing 2.5 mM luminol, 2.6 mM hydrogen peroxide, 100 mM Tris/HCl (pH 8.8), and 0.2 mM para-coumaric acid.101 Signals were detected with the Odyssey classic (LI-COR Biosciences, Westburg, Leusden, the Netherlands) or Fusion SL device (Vilber, Analis, Suarlee, Belgium). Protein levels were quantified by using the Image Studio Lite software (version 5.2, LI-COR Biosciences, Lincoln, NE) and normalized to the total protein content (REVERT Total Protein Stain, LI-COR Biosciences, Westburg, Leusden, the Netherlands), following the manufacturer’s instructions.
Total RNA Isolation and qPCR
Total RNA was extracted using the Quick-RNA MiniPrep Kit (Zymo Research, Laborimpex, Brussels, Belgium) and reverse transcribed with the miScript II RT kit (QIAGEN, Venlo, the Netherlands) in a volume of 10 μL, according to the respective manufacturer’s instructions. Real-time qPCR was carried out on a CFX384 Detection System (Bio-Rad, Temse, Belgium), using 5- or 50-ng (miRNA or mRNA detection, respectively) RNA input in a 10-μL reaction volume, 2× iTaq SYBR Green Supermix (Bio-Rad, Temse, Belgium), and 1 μL 10× miRNA-specific primers (QIAGEN, Venlo, the Netherlands) or either 2.5 pmol gene-specific primer pairs or 1 μL 10× QuantiTech primers (QIAGEN, Venlo, Netherlands). miRNAs and mRNAs of interest, as well as small RNAs used as miRNA normalizers (RNU1A, SCARNA17, and SNORD95) and reference genes used for RNA normalization (HRPT, PPIA, and TBP), were assessed in parallel for each sample and run in triplicates. Calculations were carried out by using the CFX Manager software (Bio-Rad, Temse, Belgium) provided with the machine.
Primers were purchased from Eurogentec (Liège, Belgium) and the sequences were as follows: IL6R forward, 5′-GTATTCCCAGGAGTCCCAGA-3′; IL6R reverse, 5′-GCAAGATTCCACAACCCTG-3′; gp130 forward, 5′-TGAAACTGCTGTGAATGTGG-3′; gp130 reverse, 5′-CATCCTTCCCACCTTCATCT-3′; HPRT forward, 5′-TGGACAGGACTGAACGTCTT-3′; HPRT reverse, 5′-GAGCACACAGAGGGCTACAA-3′; JAK1 forward, 5′-TCTTGGAATCCAGTGGAGGCATAAA-3′; JAK1 reverse, 5′-CACTCTTCCCGGATCTTGTTTTTCT-3′; LBP forward, 5′-AGGTGATGTTTAAGGGTGAAAT-3′; LBP reverse, 5′-ATAATCCGAGATGGCAAAGTA-3′; PPIA forward, 5′-CAGACAAGGTCCCAAAGACA-3′; PPIA reverse, 5′-CCATTATGGCGTGTGAAGTC-3′; SOCS3 forward, 5′-ATGAGAACTGCCAGGGAATC-3′; SOCS3 reverse, 5′-CCCAGGCTCCACAACCA-3′; STAT3 forward, 5′-ACACAGATAAACTTGGTCTTCAGGTA-3′; STAT3 reverse, 5′-GCCAGAGAGCCAGGAGCA-3′; TBP forward, 5′-ACCCAGCAGCATCACTGTT-3′; and TBP reverse, 5′-CGCTGGAACTCGTCTCACTA-3′.
Statistical Analysis
Statistical significance between control (NCM1) and the candidate miRNA for western blot analysis (Figures 1 and 4) and luciferase assays (Figures 2, 4, S4, and S7) was assessed by non-parametric one-way ANOVA (Kruskal-Wallis test), followed by Dunn’s multiple comparisons test performed with the statistical program Prism 7 (GraphPad, La Jolla, CA, USA).
Author Contributions
Conceptualization, C.H., S.K., and I.B.; Methodology, F.A.S., M.K., and I.B.; Investigation, F.A.S., M.K., N.W.E.M., and M.H.; Resources, S.R.-J.; Writing – Original Draft, F.A.S., C.H., and I.B.; Writing – Review & Editing, F.A.S., C.H., S.K., and I.B.; Supervision, S.K., and I.B.; Funding Acquisition, C.H., S.K., and I.B.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was funded by the Luxembourg National Research Fund (FNR) and the Deutsche Forschungsgemeinschaft (C12/BM/3975937, Inter project “HepmiRSTAT”), by the FNR funding scheme PRIDE (Doctoral Training Unit CANBIO, project number 10675146), and by the Internal Research Project «IL6LongLiv» of the University of Luxembourg. We are grateful to Professor Nobuyuki Kato, Okayama University, Japan, for providing the PH5CH8 cells. We thank Demetra Philippidou, Catherine Rolvering, Sébastien Plançon, Odile Lecha, Aurélien Ginolhac, and Eric Koncina for helpful technical advice. We thank Michèle Gaetti for the work performed during her master thesis in our research group.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.03.007.
Supplemental Information
References
- 1.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Müller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int. J. Biol. Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taub R. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y., Xu F., Lu T., Duan Z., Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat. Rev. 2012;38:904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Johnson D.E., O’Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White J.P. IL-6, cancer and cachexia: metabolic dysfunction creates the perfect storm. Transl. Cancer Res. 2017;6(Suppl 2):S280–S285. doi: 10.21037/tcr.2017.03.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkholder B., Huang R.-Y., Burgess R., Luo S., Jones V.S., Zhang W., Lv Z.Q., Gao C.Y., Wang B.L., Zhang Y.M., Huang R.P. Tumor-induced perturbations of cytokines and immune cell networks. Biochim. Biophys. Acta. 2014;1845:182–201. doi: 10.1016/j.bbcan.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Tsukamoto H., Fujieda K., Senju S., Ikeda T., Oshiumi H., Nishimura Y. Immune-suppressive effects of interleukin-6 on T-cell-mediated anti-tumor immunity. Cancer Sci. 2018;109:523–530. doi: 10.1111/cas.13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan S., Zhao E., Kryczek I., Vatan L., Sadovskaya A., Ludema G., Simeone D.M., Zou W., Welling T.H. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147:1393–1404. doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soresi M., Giannitrapani L., D’Antona F., Florena A.M., La Spada E., Terranova A., Cervello M., D’Alessandro N., Montalto G. Interleukin-6 and its soluble receptor in patients with liver cirrhosis and hepatocellular carcinoma. World J. Gastroenterol. 2006;12:2563–2568. doi: 10.3748/wjg.v12.i16.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aleksandrova K., Boeing H., Nöthlings U., Jenab M., Fedirko V., Kaaks R., Lukanova A., Trichopoulou A., Trichopoulos D., Boffetta P. Inflammatory and metabolic biomarkers and risk of liver and biliary tract cancer. Hepatology. 2014;60:858–871. doi: 10.1002/hep.27016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sánchez A., Nagy P., Thorgeirsson S.S. STAT-3 activity in chemically-induced hepatocellular carcinoma. Eur. J. Cancer. 2003;39:2093–2098. doi: 10.1016/s0959-8049(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 14.Naugler W.E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A.M., Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 15.He G., Yu G.Y., Temkin V., Ogata H., Kuntzen C., Sakurai T., Sieghart W., Peck-Radosavljevic M., Leffert H.L., Karin M. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J.S., Grisham J.W., Thorgeirsson S.S. Comparative functional genomics for identifying models of human cancer. Carcinogenesis. 2005;26:1013–1020. doi: 10.1093/carcin/bgi030. [DOI] [PubMed] [Google Scholar]
- 17.He G., Dhar D., Nakagawa H., Font-Burgada J., Ogata H., Jiang Y., Shalapour S., Seki E., Yost S.E., Jepsen K. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–396. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogata H., Kobayashi T., Chinen T., Takaki H., Sanada T., Minoda Y., Koga K., Takaesu G., Maehara Y., Iida M., Yoshimura A. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology. 2006;131:179–193. doi: 10.1053/j.gastro.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 19.Riehle K.J., Campbell J.S., McMahan R.S., Johnson M.M., Beyer R.P., Bammler T.K., Fausto N. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J. Exp. Med. 2008;205:91–103. doi: 10.1084/jem.20070820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatting M., Spannbauer M., Peng J., Al Masaoudi M., Sellge G., Nevzorova Y.A., Gassler N., Liedtke C., Cubero F.J., Trautwein C. Lack of gp130 expression in hepatocytes attenuates tumor progression in the DEN model. Cell Death Dis. 2015;6:e1667. doi: 10.1038/cddis.2014.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe M., Yoshida T., Akiba J., Ikezono Y., Wada F., Masuda A., Sakaue T., Tanaka T., Iwamoto H., Nakamura T. STAT3 deficiency prevents hepatocarcinogenesis and promotes biliary proliferation in thioacetamide-induced liver injury. World J. Gastroenterol. 2017;23:6833–6844. doi: 10.3748/wjg.v23.i37.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong J., Wang H., Shen G., Lin D., Lin Y., Ye N., Guo Y., Li Q., Ye N., Deng C., Meng C. Recombinant soluble gp130 protein reduces DEN-induced primary hepatocellular carcinoma in mice. Sci. Rep. 2016;6:24397. doi: 10.1038/srep24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmann J., Müller M., Baumann N., Reichert M., Heneweer C., Bolik J., Lücke K., Gruber S., Carambia A., Boretius S. IL-6 trans-signaling is essential for the development of hepatocellular carcinoma in mice. Hepatology. 2017;65:89–103. doi: 10.1002/hep.28874. [DOI] [PubMed] [Google Scholar]
- 24.Kan Z., Zheng H., Liu X., Li S., Barber T.D., Gong Z., Gao H., Hao K., Willard M.D., Xu J. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schust J., Sperl B., Hollis A., Mayer T.U., Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Li P.K., Li C., Lin J. Inhibition of STAT3 signaling blocks the anti-apoptotic activity of IL-6 in human liver cancer cells. J. Biol. Chem. 2010;285:27429–27439. doi: 10.1074/jbc.M110.142752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moser C., Lang S.A., Mori A., Hellerbrand C., Schlitt H.J., Geissler E.K., Fogler W.E., Stoeltzing O. ENMD-1198, a novel tubulin-binding agent reduces HIF-1alpha and STAT3 activity in human hepatocellular carcinoma(HCC) cells, and inhibits growth and vascularization in vivo. BMC Cancer. 2008;8:206. doi: 10.1186/1471-2407-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W.C., Ye S.L., Sun R.X., Liu Y.K., Tang Z.Y., Kim Y., Karras J.G., Zhang H. Inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting signal transducer and activator of transcription 3. Clin. Cancer Res. 2006;12:7140–7148. doi: 10.1158/1078-0432.CCR-06-0484. [DOI] [PubMed] [Google Scholar]
- 29.Hong D., Kurzrock R., Kim Y., Woessner R., Younes A., Nemunaitis J., Fowler N., Zhou T., Schmidt J., Jo M. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015;7:314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie L., Zeng Y., Dai Z., He W., Ke H., Lin Q., Chen Y., Bu J., Lin D., Zheng M. Chemical and genetic inhibition of STAT3 sensitizes hepatocellular carcinoma cells to sorafenib induced cell death. Int. J. Biol. Sci. 2018;14:577–585. doi: 10.7150/ijbs.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenzer C., Dirin M., Winkler A.M., Baumann V., Winkler J. Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J. Control. Release. 2015;203:1–15. doi: 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Hosseinahli N., Aghapour M., Duijf P.H.G., Baradaran B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell. Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 33.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer M., Goldschmitt J., Peschel C., Brakenhoff J.P., Kallen K.J., Wollmer A., Grötzinger J., Rose-John S. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 1997;15:142–145. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- 35.Dusetti N.J., Ortiz E.M., Mallo G.V., Dagorn J.C., Iovanna J.L. Pancreatitis-associated protein I (PAP I), an acute phase protein induced by cytokines. Identification of two functional interleukin-6 response elements in the rat PAP I promoter region. J. Biol. Chem. 1995;270:22417–22421. doi: 10.1074/jbc.270.38.22417. [DOI] [PubMed] [Google Scholar]
- 36.Eyckerman S., Broekaert D., Verhee A., Vandekerckhove J., Tavernier J. Identification of the Y985 and Y1077 motifs as SOCS3 recruitment sites in the murine leptin receptor. FEBS Lett. 2000;486:33–37. doi: 10.1016/s0014-5793(00)02205-5. [DOI] [PubMed] [Google Scholar]
- 37.Koukos G., Polytarchou C., Kaplan J.L., Morley-Fletcher A., Gras-Miralles B., Kokkotou E., Baril-Dore M., Pothoulakis C., Winter H.S., Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–852.e2. doi: 10.1053/j.gastro.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatziapostolou M., Polytarchou C., Aggelidou E., Drakaki A., Poultsides G.A., Jaeger S.A., Ogata H., Karin M., Struhl K., Hadzopoulou-Cladaras M., Iliopoulos D. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S., Liu J., Wei J., Yuan H., Zhang T., Bishopric N.H. Repression of miR-142 by p300 and MAPK is required for survival signalling via gp130 during adaptive hypertrophy. EMBO Mol. Med. 2012;4:617–632. doi: 10.1002/emmm.201200234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobotta S., Raue A., Huang X., Vanlier J., Jünger A., Bohl S., Albrecht U., Hahnel M.J., Wolf S., Mueller N.S. Model Based Targeting of IL-6-Induced Inflammatory Responses in Cultured Primary Hepatocytes to Improve Application of the JAK Inhibitor Ruxolitinib. Front. Physiol. 2017;8:775. doi: 10.3389/fphys.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lind E.F., Ohashi P.S. Mir-155, a central modulator of T-cell responses. Eur. J. Immunol. 2014;44:11–15. doi: 10.1002/eji.201343962. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson A.N., Gartlan K.H., Kelly G., Samson L.D., Olver S.D., Avery J., Zomerdijk N., Tey S.K., Lee J.S., Vuckovic S., Hill G.R. Granulocytes Are Unresponsive to IL-6 Due to an Absence of gp130. J. Immunol. 2018;200:3547–3555. doi: 10.4049/jimmunol.1701191. [DOI] [PubMed] [Google Scholar]
- 43.Betz U.A., Müller W. Regulated expression of gp130 and IL-6 receptor alpha chain in T cell maturation and activation. Int. Immunol. 1998;10:1175–1184. doi: 10.1093/intimm/10.8.1175. [DOI] [PubMed] [Google Scholar]
- 44.Radtke S., Wüller S., Yang X.P., Lippok B.E., Mütze B., Mais C., de Leur H.S., Bode J.G., Gaestel M., Heinrich P.C. Cross-regulation of cytokine signalling: pro-inflammatory cytokines restrict IL-6 signalling through receptor internalisation and degradation. J. Cell Sci. 2010;123:947–959. doi: 10.1242/jcs.065326. [DOI] [PubMed] [Google Scholar]
- 45.Honke N., Ohl K., Wiener A., Bierwagen J., Peitz J., Di Fiore S., Fischer R., Wagner N., Wüller S., Tenbrock K. The p38-mediated rapid down-regulation of cell surface gp130 expression impairs interleukin-6 signaling in the synovial fluid of juvenile idiopathic arthritis patients. Arthritis Rheumatol. 2014;66:470–478. doi: 10.1002/art.38245. [DOI] [PubMed] [Google Scholar]
- 46.Iwakawa H.O., Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Schumann R.R., Kirschning C.J., Unbehaun A., Aberle H.P., Knope H.P., Lamping N., Ulevitch R.J., Herrmann F. The lipopolysaccharide-binding protein is a secretory class 1 acute-phase protein whose gene is transcriptionally activated by APRF/STAT/3 and other cytokine-inducible nuclear proteins. Mol. Cell. Biol. 1996;16:3490–3503. doi: 10.1128/mcb.16.7.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y., Wang J., Yan W., Zhou Y., Chen Y., Zhou K., Wen J., Wang Y., Cai W. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting IL-6/STAT3 signalling. J. Hepatol. 2015;62:889–896. doi: 10.1016/j.jhep.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 49.Sonda N., Simonato F., Peranzoni E., Calì B., Bortoluzzi S., Bisognin A., Wang E., Marincola F.M., Naldini L., Gentner B. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity. 2013;38:1236–1249. doi: 10.1016/j.immuni.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Kästingschäfer C.S., Schäfer S.D., Kiesel L., Götte M. miR-142-3p is a novel regulator of cell viability and proinflammatory signalling in endometrial stroma cells. Reprod. Biomed. Online. 2015;30:553–556. doi: 10.1016/j.rbmo.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Gao D.Y., Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhanasekaran R., Gabay-Ryan M., Baylot V., Lai I., Mosley A., Huang X., Zabludoff S., Li J., Kaimal V., Karmali P., Felsher D.W. Anti-miR-17 therapy delays tumorigenesis in MYC-driven hepatocellular carcinoma (HCC) Oncotarget. 2017;9:5517–5528. doi: 10.18632/oncotarget.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X., Magnus J., Kaimal V., Karmali P., Li J., Walls M., Prudente R., Sung E., Sorourian M., Lee R. Lipid Nanoparticle-Mediated Delivery of Anti-miR-17 Family Oligonucleotide Suppresses Hepatocellular Carcinoma Growth. Mol. Cancer Ther. 2017;16:905–913. doi: 10.1158/1535-7163.MCT-16-0613. [DOI] [PubMed] [Google Scholar]
- 55.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 56.Palumbo T., Poultsides G.A., Kouraklis G., Liakakos T., Drakaki A., Peros G., Hatziapostolou M., Iliopoulos D. A functional microRNA library screen reveals miR-410 as a novel anti-apoptotic regulator of cholangiocarcinoma. BMC Cancer. 2016;16:353. doi: 10.1186/s12885-016-2384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long Z.W., Wu J.H., Hong C., Wang Y.N., Zhou Y. MiR-374b Promotes Proliferation and Inhibits Apoptosis of Human GIST Cells by Inhibiting PTEN through Activation of the PI3K/Akt Pathway. Mol. Cells. 2018;41:532–544. doi: 10.14348/molcells.2018.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X., Wang W., Su N., Zhu X., Yao J., Gao W., Hu Z., Sun Y. miR-374a promotes cell proliferation, migration and invasion by targeting SRCIN1 in gastric cancer. FEBS Lett. 2015;589:407–413. doi: 10.1016/j.febslet.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 59.Li H., Chen H., Wang H., Dong Y., Yin M., Zhang L., Wei J. MicroRNA-374a Promotes Hepatocellular Carcinoma Cell Proliferation by Targeting Mitogen-Inducible Gene-6 (MIG-6) Oncol. Res. 2018;26:557–563. doi: 10.3727/096504017X15000784459799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Costa-Pereira A.P., Tininini S., Strobl B., Alonzi T., Schlaak J.F., Is’harc H., Gesualdo I., Newman S.J., Kerr I.M., Poli V. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc. Natl. Acad. Sci. USA. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rolvering C., Zimmer A.D., Kozar I., Hermanns H.M., Letellier E., Vallar L., Nazarov P.V., Nicot N., Ginolhac A., Haan S. Crosstalk between different family members: IL27 recapitulates IFNγ responses in HCC cells, but is inhibited by IL6-type cytokines. Biochim Biophys Acta Mol Cell Res. 2017;1864:516–526. doi: 10.1016/j.bbamcr.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Kaplan D.H., Shankaran V., Dighe A.S., Stockert E., Aguet M., Old L.J., Schreiber R.D. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin Z., Kim H.J., Hemme J., Blankenstein T. Inhibition of methylcholanthrene-induced carcinogenesis by an interferon gamma receptor-dependent foreign body reaction. J. Exp. Med. 2002;195:1479–1490. doi: 10.1084/jem.20011887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X., Du J., Jiang R., Li L. MicroRNA-214 inhibits the proliferation and invasion of lung carcinoma cells by targeting JAK1. Am. J. Transl. Res. 2018;10:1164–1171. [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan J., Ji H., Xiao F., Lin Z., Zhao X., Wang Z., Zhao J., Lu J. MicroRNA-340 inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting JAK1. Biochem. Biophys. Res. Commun. 2017;483:578–584. doi: 10.1016/j.bbrc.2016.12.102. [DOI] [PubMed] [Google Scholar]
- 66.Yu D.L., Zhang T., Wu K., Li Y., Wang J., Chen J., Li X.Q., Peng X.G., Wang J.N., Tan L.G. MicroRNA-448 suppresses metastasis of pancreatic ductal adenocarcinoma through targeting JAK1/STAT3 pathway. Oncol. Rep. 2017;38:1075–1082. doi: 10.3892/or.2017.5781. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Q., Huang C., Yang Q., Gao L., Liu H.C., Tang J., Feng W.H. MicroRNA-30c Modulates Type I IFN Responses To Facilitate Porcine Reproductive and Respiratory Syndrome Virus Infection by Targeting JAK1. J. Immunol. 2016;196:2272–2282. doi: 10.4049/jimmunol.1502006. [DOI] [PubMed] [Google Scholar]
- 68.Mukherjee A., Di Bisceglie A.M., Ray R.B. Hepatitis C virus-mediated enhancement of microRNA miR-373 impairs the JAK/STAT signaling pathway. J. Virol. 2015;89:3356–3365. doi: 10.1128/JVI.03085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 70.Vainchenker W., Leroy E., Gilles L., Marty C., Plo I., Constantinescu S.N. JAK inhibitors for the treatment of myeloproliferative neoplasms and other disorders. F1000Res. 2018;7:82. doi: 10.12688/f1000research.13167.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai H., Liu X., Zheng J., Xue Y., Ma J., Li Z., Xi Z., Li Z., Bao M., Liu Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 72.Liu L., Chen X., Zhang Y., Hu Y., Shen X., Zhu W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget. 2017;8:31386–31394. doi: 10.18632/oncotarget.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Östling P., Leivonen S.K., Aakula A., Kohonen P., Mäkelä R., Hagman Z., Edsjö A., Kangaspeska S., Edgren H., Nicorici D. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 74.Zheng D., Dai Y., Wang S., Xing X. MicroRNA-299-3p promotes the sensibility of lung cancer to doxorubicin through directly targeting ABCE1. Int. J. Clin. Exp. Pathol. 2015;8:10072–10081. [PMC free article] [PubMed] [Google Scholar]
- 75.Wang F., Dai M., Chen H., Li Y., Zhang J., Zou Z., Yang H. Prognostic value of hsa-mir-299 and hsa-mir-7706 in hepatocellular carcinoma. Oncol. Lett. 2018;16:815–820. doi: 10.3892/ol.2018.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Chong C.C., Chen G.G., Lai P.B. A Seven-microRNA Expression Signature Predicts Survival in Hepatocellular Carcinoma. PLoS ONE. 2015;10:e0128628. doi: 10.1371/journal.pone.0128628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu M., Kong X., Wang H., Huang G., Ye C., He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775–8784. doi: 10.18632/oncotarget.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Voorhees P.M., Manges R.F., Sonneveld P., Jagannath S., Somlo G., Krishnan A., Lentzsch S., Frank R.C., Zweegman S., Wijermans P.W. A phase 2 multicentre study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with relapsed or refractory multiple myeloma. Br. J. Haematol. 2013;161:357–366. doi: 10.1111/bjh.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.San-Miguel J., Bladé J., Shpilberg O., Grosicki S., Maloisel F., Min C.K., Polo Zarzuela M., Robak T., Prasad S.V., Tee Goh Y. Phase 2 randomized study of bortezomib-melphalan-prednisone with or without siltuximab (anti-IL-6) in multiple myeloma. Blood. 2014;123:4136–4142. doi: 10.1182/blood-2013-12-546374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orlowski R.Z., Gercheva L., Williams C., Sutherland H., Robak T., Masszi T., Goranova-Marinova V., Dimopoulos M.A., Cavenagh J.D., Špička I. A phase 2, randomized, double-blind, placebo-controlled study of siltuximab (anti-IL-6 mAb) and bortezomib versus bortezomib alone in patients with relapsed or refractory multiple myeloma. Am. J. Hematol. 2015;90:42–49. doi: 10.1002/ajh.23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rossi J.F., Négrier S., James N.D., Kocak I., Hawkins R., Davis H., Prabhakar U., Qin X., Mulders P., Berns B. A phase I/II study of siltuximab (CNTO 328), an anti-interleukin-6 monoclonal antibody, in metastatic renal cell cancer. Br. J. Cancer. 2010;103:1154–1162. doi: 10.1038/sj.bjc.6605872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dorff T.B., Goldman B., Pinski J.K., Mack P.C., Lara P.N., Jr., Van Veldhuizen P.J., Jr., Quinn D.I., Vogelzang N.J., Thompson I.M., Jr., Hussain M.H. Clinical and Correlative Results of SWOG S0354: a Phase II Trial of CNTO328 (siltuximab), a Monoclonal Antibody against Interleukin-6, in Chemotherapy-Pretreated Patients with Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2010;16:3028–3034. doi: 10.1158/1078-0432.CCR-09-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fizazi K., De Bono J.S., Flechon A., Heidenreich A., Voog E., Davis N.B., Qi M., Bandekar R., Vermeulen J.T., Cornfeld M., Hudes G.R. Randomised phase II study of siltuximab (CNTO 328), an anti-IL-6 monoclonal antibody, in combination with mitoxantrone/prednisone versus mitoxantrone/prednisone alone in metastatic castration-resistant prostate cancer. Eur. J. Cancer. 2012;48:85–93. doi: 10.1016/j.ejca.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 84.Rossi J.F., Lu Z.Y., Jourdan M., Klein B. Interleukin-6 as a therapeutic target. Clin. Cancer Res. 2015;21:1248–1257. doi: 10.1158/1078-0432.CCR-14-2291. [DOI] [PubMed] [Google Scholar]
- 85.Burger R., Günther A., Klausz K., Staudinger M., Peipp M., Penas E.M., Rose-John S., Wijdenes J., Gramatzki M. Due to interleukin-6 type cytokine redundancy only glycoprotein 130 receptor blockade efficiently inhibits myeloma growth. Haematologica. 2017;102:381–390. doi: 10.3324/haematol.2016.145060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang E., Liu R., Chu Y. miRNA-15a/16: as tumor suppressors and more. Future Oncol. 2015;11:2351–2363. doi: 10.2217/fon.15.101. [DOI] [PubMed] [Google Scholar]
- 87.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klein U., Lia M., Crespo M., Siegel R., Shen Q., Mo T., Ambesi-Impiombato A., Califano A., Migliazza A., Bhagat G., Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 89.Porkka K.P., Ogg E.L., Saramäki O.R., Vessella R.L., Pukkila H., Lähdesmäki H., van Weerden W.M., Wolf M., Kallioniemi O.P., Jenster G., Visakorpi T. The miR-15a-miR-16-1 locus is homozygously deleted in a subset of prostate cancers. Genes Chromosomes Cancer. 2011;50:499–509. doi: 10.1002/gcc.20873. [DOI] [PubMed] [Google Scholar]
- 90.Lovat F., Fassan M., Gasparini P., Rizzotto L., Cascione L., Pizzi M., Vicentini C., Balatti V., Palmieri D., Costinean S., Croce C.M. miR-15b/16-2 deletion promotes B-cell malignancies. Proc. Natl. Acad. Sci. USA. 2015;112:11636–11641. doi: 10.1073/pnas.1514954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang T., Hou J., Li Z., Zheng Z., Wei J., Song D., Hu T., Wu Q., Yang J.Y., Cai J.C. miR-15a-3p and miR-16-1-3p Negatively Regulate Twist1 to Repress Gastric Cancer Cell Invasion and Metastasis. Int. J. Biol. Sci. 2017;13:122–134. doi: 10.7150/ijbs.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feng Q.Q., Dong Z.Q., Zhou Y., Zhang H., Long C. miR-16-1-3p targets TWIST1 to inhibit cell proliferation and invasion in NSCLC. Bratisl. Lek Listy. 2018;119:60–65. doi: 10.4149/BLL_2018_012. [DOI] [PubMed] [Google Scholar]
- 93.Liang X.H., Hart C.E., Crooke S.T. Transfection of siRNAs can alter miRNA levels and trigger non-specific protein degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:455–468. doi: 10.1016/j.bbagrm.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 94.Saetrom P., Heale B.S., Snøve O., Jr., Aagaard L., Alluin J., Rossi J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilkens L., Hammer C., Glombitza S., Müller D.E. Hepatocellular and cholangiolar carcinoma-derived cell lines reveal distinct sets of chromosomal imbalances. Pathobiology. 2012;79:115–126. doi: 10.1159/000334100. [DOI] [PubMed] [Google Scholar]
- 96.Ewald F., Nörz D., Grottke A., Bach J., Herzberger C., Hofmann B.T., Nashan B., Jücker M. Vertical Targeting of AKT and mTOR as Well as Dual Targeting of AKT and MEK Signaling Is Synergistic in Hepatocellular Carcinoma. J. Cancer. 2015;6:1195–1205. doi: 10.7150/jca.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reczko M., Maragkakis M., Alexiou P., Grosse I., Hatzigeorgiou A.G. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- 99.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kirchmeyer M., Servais F.A., Hamdorf M., Nazarov P.V., Ginolhac A., Halder R., Vallar L., Glanemann M., Rubie C., Lammert F., Kreis S., Behrmann I. Cytokine-mediated modulation of the hepatic miRNome: miR-146b-5p is an IL-6-inducible miRNA with multiple targets. J. Leukocyte Biol. 2018;104:987–1002. doi: 10.1002/JLB.MA1217-499RR. [DOI] [PubMed] [Google Scholar]
- 101.Haan C., Behrmann I. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J. Immunol. Methods. 2007;318:11–19. doi: 10.1016/j.jim.2006.07.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.