Abstract
A series of new benzene-based derivatives was designed, synthesized and comprehensively characterized. All of the tested compounds were evaluated for their in vitro ability to potentially inhibit the acetyl- and butyrylcholinesterase enzymes. The selectivity index of individual molecules to cholinesterases was also determined. Generally, the inhibitory potency was stronger against butyryl- compared to acetylcholinesterase; however, some of the compounds showed a promising inhibition of both enzymes. In fact, two compounds (23, benzyl ethyl(1-oxo-1-phenylpropan-2-yl)carbamate and 28, benzyl (1-(3-chlorophenyl)-1-oxopropan-2-yl) (methyl)carbamate) had a very high selectivity index, while the second one (28) reached the lowest inhibitory concentration IC50 value, which corresponds quite well with galanthamine. Moreover, comparative receptor-independent and receptor-dependent structure–activity studies were conducted to explain the observed variations in inhibiting the potential of the investigated carbamate series. The principal objective of the ligand-based study was to comparatively analyze the molecular surface to gain insight into the electronic and/or steric factors that govern the ability to inhibit enzyme activities. The spatial distribution of potentially important steric and electrostatic factors was determined using the probability-guided pharmacophore mapping procedure, which is based on the iterative variable elimination method. Additionally, planar and spatial maps of the host–target interactions were created for all of the active compounds and compared with the drug molecules using the docking methodology.
Keywords: benzene-based carbamates, in vitro cholinesterase inhibition, CoMSA, IVE-PLS, molecular docking study
1. Introduction
Several clinically implemented drug and pesticide molecules contain amide (–CONH–) and/or carbamate (–OCONH–) groups that can be variously substituted to form privileged structural fragments [1,2,3,4]. These molecular motifs are able to interact with a wide range of receptors/enzymes to induce a biological response [5,6,7]. As a result, a growing interest in amide- and carbamate-based compounds is being observed among medicinal chemists [8,9]. Based on the cholinergic hypothesis that assumes the increase in the acetylcholine (ACh) level, carbamate-like drugs can be regarded as one of the mainstays for the contemporary pharmacotherapy for Alzheimer’s disease (AD) by inhibiting cholinesterases (ChEs) [10]. AD is a progressive central nervous system (CNS) disorder that irreversibly degenerates the memory and cognitive abilities (dementia), which causes the everyday activities of older adults to be severely limited. Even though the etiology of AD has not yet been comprehensively revealed, a few factors are postulated as playing valid roles in the pathogenesis of this disease, including the aggregation and accumulation of amyloid-β peptide deposits in the brain, oxidative stress and a low level of ACh [11]. Basically, two types of the ChE enzymes that belong to the group of serine hydrolases occur in the human nervous system: acetylcholinesterase (AChE) and butyrylcholinesterase (BChE) [12]. In fact, both enzymes are able to catalyze ACh hydrolysis in the cholinergic synapses, but BChE can hydrolyze other esters as well; therefore, its concentration in the brain seems to be important for those suffering from AD. It is reported that AChE has a higher hydrolytic capacity, but its levels decrease as the changes become more pronounced during the course of the disease, while the BChE levels are supposedly increased or unchanged, which can somewhat compensate for the higher AChE activity [13]. Inhibiting both enzymes produces an increment of the ACh concentration in the cholinergic synapses that show a symptomatic efficacy in AD treatment; therefore, competitive cholinesterase inhibitors (ChEIs), e.g., galanthamine or rivastigmine as well as non-competitive ChEIs, e.g., tacrine or donepezil are clinically applied to alleviate the neuromuscular symptoms of AD [14]. In fact, novel BChE selective inhibitors such as anti-Alzheimer agents have recently been proposed, but the potential effectiveness that they offer is frequently accompanied by the occurrence of central and peripheral side effects [15,16,17,18]. In this context, further research for new ChEIs seems to be required to achieve success in the treatment of AD.
Potency modeling of prospective drug molecules seems to be useful as an initial screening test (rank order) and a decisive factor in avoiding “chasing” false positives and/or eradicating “bad actors” at the early stages of drug design or development [19,20]. The art of specifying molecules that have a potential therapeutic value at the “pre-synthesis” stage can be assisted by computer-aided molecular design (CAMD) techniques for the comprehensive mapping of the topology and/or topography of a compound into the property-based chemical space. A key question that confronts computational chemists is how to quantitatively transform a myriad of feature-based descriptors that have been a priori calculated for the compounds into an ADMET-related molecular potency. From among the more optimistic approaches, modeling the multidimensional quantitative structure–activity relationship (mD–QSAR) techniques seem to be pragmatic provided that a congeneric series of compounds is produced—the analogy is crucial in chemistry, especially in cases in which the investigated process is not fully understood or when the variety of observed guest–host interactions makes the optimization of the pharmacological response a cost/resources/knowledge challenging issue [21,22,23]. The “Holy Grail” of in silico modeling is to produce statistically robust models capable of making accurate quantitative predictions that include the binding affinity, metabolic fate and pharmacokinetic or toxicity parameters (ADMET) on the route “towards the prediction paradise” [24]. Hence, huge attempts have been undertaken to implement “direct” receptor-dependent (RD) and “indirect” receptor-independent (RI) in silico protocols, respectively. The working paradigm of fragment-based design, or fragonomics, assumes that well-placed fragments account for the crucial ligand–receptor interactions, while the rest of the molecule primarily serves as a scaffold that holds the “active” fragments together [25]. With regard to the structure of a ligand as a “negative” image of an active target site (pharmacophore), the question about a reliable quantitative measure of its “intrinsic” activity and its correspondence to robust empirical receptor–ligand interaction data naturally appears. In the target-guided QSAR procedures. the complementary (bio)effector-binding mode is retrieved based on the intrinsic dependence of the atomic coordinates of both the receptor and ligand in the binding/active site while the target spatial arrangement of the atoms is also available [26]. The adopted spatial distribution of the ligand property space is mediated by the corresponding mapping of the target steric, electronic or lipophilic patterns. In fact, drugs are generally engineered to have a tightly fitting interface with the receptor/enzyme bonding/active site; however, the “goodness” of a favorable guest–host binding assessment is still questionable. In other words, the Achilles’ heel of docking procedures is the deficiency of “truly” selective scoring functions to rank the receptor–ligand complexes according to their actual binding affinities [27,28]. To overcome these constraints, it is advisable to use both ligand- and structure-based approaches on the path from the relationships to the models.
The current paper can be regarded as a follow-up of the recently reported carbamates as prospective ChEIs [29,30,31]; therefore, the synthesis and biological assessment of a new series of benzene and naphthalene derivatives is provided. Moreover, the electronic/steric/lipophilic determinants, which are potentially valid for characterizing the structure-inhibitory potency for a set of the carbamate analogs, are discussed using RI and RD in silico procedures: comparative molecular surface analysis (CoMSA) and molecular docking [32]. Additionally, a systematic space inspection was performed to investigate the statistical estimators using the stochastic model validation (SMV) method to derive the “average” pharmacophore pattern in the probability-guided pharmacophore approach [33]. The molecular pharmacokinetic profiles were comprehensively screened using several descriptors that revealed the physicochemical and structural features, which might be essential to map the inhibition potency of carbamates.
2. Results and Discussion
2.1. Design and Synthesis
The concept of the synthesis of the designed compounds was based on a modification of the structure of the selected α-aminoketones. Cathinone is biologically active natural product and its derivatives have broad pharmacological properties. All carbamates were formed in a four-step synthesis that begins with the basic reagents (see Scheme 1 and Scheme 2).
Scheme 1.
Synthesis of benzene derivatives 1–34. Reagents and conditions: (a) AlCl3, CH2Cl2; (b) Br2, CH2Cl2; (c) HCl; and (d) CH2Cl2.
Scheme 2.
Synthesis of the naphthalene derivatives 35–41. Reagents and conditions: (a) AlCl3, CH2Cl2; (b) Br2, CH2Cl2; (c) HCl; and (d) CH2Cl2.
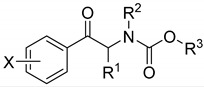
In the first step, the corresponding aromatic ketone was obtained in the Friedel–Crafts reaction. Next, the bromination of the obtained aromatic ketone with elemental bromine led to a 2-bromo derivative, which was subjected to the ammonolysis of the obtained halide without secretion. In the final step, the amine in a free form reacted with the appropriate chloroformate to produce desired carbamate. The final products were purified using column chromatography with a mixture of ethyl acetate and hexane, respectively. The structures of the obtained compounds are presented in Table 1. All of the derivatives were analyzed in order to confirm their chemical structures and were also tested for their biological activity.
Table 1.
Structures of the carbamate derivatives 1–41; in vitro AChE and BChE inhibition (IC50) compared with the standards rivastigmine (RIV) and galanthamine (GLT). ChE inhibitions are expressed as the mean ± SD (n = three experiments).
Benzene derivatives
| |||||||
| No. | X | R1 | R2 | R3 |
AChE
IC50 [µM] |
BChE
IC50 [µM] |
SI * |
| methyl carbamates | |||||||
| 1. | H | CH3 | C2H5 | CH3 | 245.33 ± 0.75 | 227.91 ± 0.90 | 1.08 |
| 2. | H | C2H5 | CH3 | CH3 | 328.86 ± 9.72 | 440.56 ± 7.54 | 0.75 |
| 3. | H | C2H5 | C2H5 | CH3 | 273.74 ± 2.16 | 362.87 ± 7.41 | 0.75 |
| 4. | 3-CH3 | CH3 | CH3 | CH3 | 133.98 ± 2.30 | 115.89 ± 2.16 | 1.16 |
| 5. | 3-Cl | CH3 | CH3 | CH3 | 329.68 ± 42.06 | 113.61 ± 0.29 | 2.90 |
| 6. | 4-OCH3 | CH3 | CH3 | CH3 | 146.91 ± 1.51 | 186.60 ± 1.55 | 0.79 |
| 7. | 4-CH3 | CH3 | CH3 | CH3 | 50.63 ± 0.63 | 180.77 ± 11.99 | 0.28 |
| 8. | 4-Cl | CH3 | CH3 | CH3 | 308.80 ± 6.16 | 131.57 ± 4.55 | 2.35 |
| 9. | 4-Br | CH3 | CH3 | CH3 | 282.74 ± 0.71 | 224.08 ± 2.19 | 1.26 |
| 10. | 3,4-OCH2O | CH3 | CH3 | CH3 | 225.46 ± 2.72 | 194.93 ± 1.93 | 1.16 |
| phenyl carbamates | |||||||
| 11. | H | CH3 | C2H5 | Ph | 166.72 ± 1.75 | 27.64 ± 0.55 | 6.03 |
| 12. | H | C2H5 | CH3 | Ph | 179.02 ± 6.21 | 52.96 ± 1.06 | 3.38 |
| 13. | H | C2H5 | C2H5 | Ph | 95.69 ± 5.37 | 15.62 ± 0.06 | 6.13 |
| 14. | H | C3H7 | CH3 | Ph | 97.97 ± 0.36 | 16.55 ± 0.13 | 5.92 |
| 15. | 3-CH3 | CH3 | CH3 | Ph | 165.62 ± 2.75 | 48.29 ± 0.31 | 3.43 |
| 16. | 3-Cl | CH3 | CH3 | Ph | 108.81 ± 1.10 | 14.81 ± 0.01 | 7.35 |
| 17. | 4-OCH3 | CH3 | CH3 | Ph | 90.21 ± 0.73 | 105.73 ± 1.94 | 0.85 |
| 18. | 4-CH3 | CH3 | CH3 | Ph | 138.64 ± 1.45 | 87.63 ± 2.37 | 1.58 |
| 19. | 4-Cl | CH3 | CH3 | Ph | 44.24 ± 2.35 | 38.96 ± 0.28 | 1.14 |
| 20. | 4-Br | CH3 | CH3 | Ph | 51.46 ± 1.38 | 44.05 ± 1.71 | 1.17 |
| 21. | 3,4-CH3 | CH3 | CH3 | Ph | 47.95 ± 2.09 | 26.41 ± 0.05 | 1.82 |
| 22. | 3,4-OCH2O | CH3 | CH3 | Ph | 84.00 ± 0.43 | 53.56 ± 0.72 | 1.57 |
| benzyl carbamates | |||||||
| 23. | H | CH3 | C2H5 | Bn | 158.41 ± 0.87 | 11.37 ± 0.05 | 13.93 |
| 24. | H | C2H5 | CH3 | Bn | 104.15 ± 2.91 | 21.38 ± 0.26 | 4.87 |
| 25. | H | C2H5 | C2H5 | Bn | 119.47 ± 1.23 | 21.29 ± 0.48 | 5.61 |
| 26. | H | C3H7 | CH3 | Bn | 134.65 ± 0.70 | 16.30 ± 0.17 | 8.26 |
| 27. | 3-CH3 | CH3 | CH3 | Bn | 83.32 ± 2.49 | 12.50 ± 0.24 | 6.67 |
| 28. | 3-Cl | CH3 | CH3 | Bn | 84.37 ± 2.27 | 5.51 ± 0.20 | 15.31 |
| 29. | 4-OCH3 | CH3 | CH3 | Bn | 62.57 ± 0.65 | 57.39 ± 0.02 | 1.09 |
| 30. | 4-CH3 | CH3 | CH3 | Bn | 40.06 ± 0.01 | 26.43 ± 0.40 | 1.52 |
| 31. | 4-Cl | CH3 | CH3 | Bn | 52.83 ± 0.34 | 7.02 ± 0.15 | 7.53 |
| 32. | 4-Br | CH3 | CH3 | Bn | 32.01 ± 0.54 | 11.28 ± 0.15 | 2.84 |
| 33. | 3,4-CH3 | CH3 | CH3 | Bn | 76.78 ± 2.76 | 16.87 ± 0.22 | 4.55 |
| 34. | 3,4-OCH2O | CH3 | CH3 | Bn | 75.57 ± 1.25 | 51.89 ± 1.18 | 1.46 |
Naphthalene derivatives
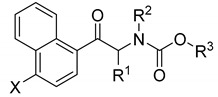
| |||||||
| methyl carbamates | |||||||
| 35. | H | CH3 | CH3 | CH3 | 126.82 ± 1.74 | 150.36 ± 7.13 | 0.84 |
| phenyl carbamates | |||||||
| 36. | H | CH3 | CH3 | Ph | 99.58 ± 5.88 | 12.72 ± 0.19 | 7.83 |
| 37. | Cl | CH3 | CH3 | Ph | 30.64 ± 1.57 | 13.54 ± 0.47 | 2.26 |
| 38. | Br | CH3 | CH3 | Ph | 61.68 ± 1.42 | 12.97 ± 0.28 | 4.76 |
| benzyl carbamates | |||||||
| 39. | H | CH3 | CH3 | Bn | 105.20 ± 0.99 | 18.94 ± 0.18 | 5.55 |
| 40. | Cl | CH3 | CH3 | Bn | 103.28 ± 0.41 | 42.75 ± 0.31 | 2.42 |
| 41. | Br | CH3 | CH3 | Bn | 128.27 ± 0.88 | 235.91 ± 16.3 | 0.54 |
| RIV | 56.1 ± 1.41 | 38.4 ± 1.97 | 1.46 | ||||
| GLT | 1.54 ± 0.02 | 2.77 ± 0.15 | 0.56 | ||||
* SI (index selectivity) = IC50 (AChE)/IC50 (BChE). A higher selectivity to BChE is indicated in bold.
2.2. In Vitro Assessment of the AChE- and BChE-Inhibitory Profiles
The AChE- and BChE-inhibiting activity for all of the tested carbamates was evaluated and compared with the internal standards rivastigmine (RIV, Exelon®) and galantamine (GLT, Reminyl®). These standards were selected because of their different structures since rivastigmine is a classical acylating pseudo-reversible carbamate ChEI that inhibits both AChE and BChE, whereas galanthamine is a non-acylating competitive reversible ChEI as well as an allosteric ligand at the nicotinic ACh receptors. The choice of these reference drugs, which have different mechanisms of action, can provide relevant results. The findings are summarized in Table 1 and expressed as the 50% inhibitory concentration (IC50 [μM]) or the concentration of the inhibitor that was required for the 50% inhibition of the mentioned enzymes.
All of the considered derivatives exhibited a very good to moderate inhibitory activity. The IC50 values for AChE ranged from 32.01 ± 0.54 to 329.68 ± 42.06 µM and for BChE from 5.51 ± 0.20 to 440.56 ± 7.54 µM. It was observed that some of them had an even better activity than a known drug—rivastigmine. Surprisingly, more of the compounds exhibited a better inhibitory potency towards BChE than to AChE. Some of the compounds proved to be highly selective for BChE with respect to AChE (11, 13, 14, 16, 23–28, 31, and 33). Specifically, two compounds, 23 and 28, had a very high selectivity index (SI = 13.93 and 15.31). In fact, compound 28 had the lowest IC50 value and had an approximately seven-fold higher inhibitory activity against BChE than RIV (IC50 = 5.51 vs. 38.40 µM), which corresponds quite well with GLT (IC50 = 2.77 µM). Similarly, a five-fold higher inhibitor activity against BChE relative to RIV was also observed for compound 31 (IC50 = 7.02 µM) and was more than three-fold higher for compounds 23 and 32 as well. It should be emphasized that compound 32 exhibited a promising inhibitory activity against AChE with an IC50 value of 32.01 µM—approximately two times better compared to RIV (IC50 = 56.1 µM). Among the naphthalene derivatives, phenyl carbamates 36–38 revealed a three-fold higher inhibitor activity towards BChE relative to RIV, whereas only one benzyl carbamate, 39, exhibited a two-fold higher anti-BChE activity to RIV. In fact, compounds 36 and 39 might serve as selective inhibition agents towards BChE with respect to AChE (SI = 7.83 vs. 5.55). Moreover, compounds 37 and 38 also exhibited a high activity against AChE. Not surprisingly, the substitution of the aryl ring had a direct impact on the variations in the potency of the investigated compounds. It was observed that the methyl-substituted carbamates generally exhibited a lower inhibitory ability compared to their phenyl or benzyl counterparts. Interestingly, compounds 17 and 29, which have the methoxy group in the same position were weaker inhibitors—the presence of a hydrophilic electron-donating -OCH3 substituent of the phenyl ring at the para-position decreased the potency of the compounds, especially against the BChE enzyme (see Table 1). On the other hand, the presence of methyl group(s) in the meta/para-position(s) of the phenyl ring (compounds 15, 18, 21, 27, 30, and 33) resulted in an improvement in the IC50 value for the BChE enzyme, thus suggesting the significance of the hydrophobic interactions with the enzyme. The placement of an electron-withdrawing chlorine substituent in the meta-position of the phenyl ring appeared to be strongly preferable, especially for the BChE inhibition activity (compounds 16 or 28).
2.3. In Silico Evaluation of AChE- and BChE-Inhibitory Profile
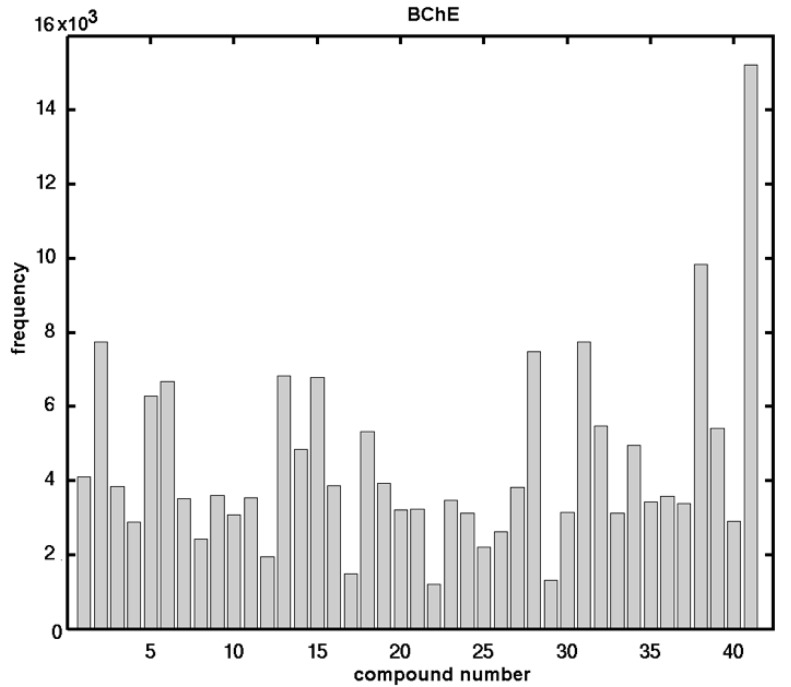
The principal objective of the ligand-based study was to comparatively analyze the molecular surface (CoMSA) to gain insight into the electronic and/or steric factors that potentially determine the inhibitory AChE/BChE activities of the investigated compounds. The findings for the surface descriptors were compared with their force field counterparts (CoMFA), when modeling thinhibiting potency for the multiple training/test subsets. First, the performance of the AChE and BChE profiles for the entire benzene and naphthalene carbamate dataset 1–41 was investigated as the training set using the CoMFA and CoMSA approach. In this case, both methods performed comparably for the AChE/BChE activity (expressed in a logarithmic scale as pAChE and pBChE); however, in general, the modeling of pBChE data produced superior outcomes of the statistical metrics (CoMFA = 0.61 vs. CoMSA = 0.61), irrespective of the map size (20 × 20 to 50 × 50) or the probe atom that was used (CH3+ or H+). It was clear that relying exclusively on data fitting with cross-validated leave-one-out (CV-LOO) procedure is not satisfactory, and therefore the external validation by splitting the molecules into training/test subsets was conducted to assess the predictive power of a model using the SDEP, MAE and statistics. The naphthalene-based carbamates were excluded from the training set in order to form the test set. We observed an improvement in the performance (CoMFA = 0.72 vs. CoMSA = 0.76); however, the poor predictive abilities of the models ( < 0.5) confirmed the dichotomic nature of the / parameters for which a high value of did not automatically imply a high model predictability [34]. From a philosophical point of view, it is not possible to determine an “absolute” measure of predictivity, as it significantly depends on the choice of datasets and the statistical approach that is used, but the great advantage of the QSAR paradigm lies not in the extrapolation, as was stated by Hansch [35]. In other words, the separation of objects into training/test subgroups is not a trivial issue, and therefore, an additional evaluation, namely the Stochastic Model Validation (SMV), was performed as a kind of “perturbation” procedure to examine the data structure [36]. Hence, the fluctuations of the statistical estimators during the CoMSA BChE modeling were examined since the original dataset of 41 molecules was repeatedly sampled into 30/11 training/test subseries (ratio 3:1). Unfortunately, it was not technically feasible to scrutinize the entire pool of systematically generated training/test populations (≈ 3 × 109), and, therefore, the overall number of samplings was restricted to a relatively small fraction of approximately 3 × 106 systematically generated populations (1 out of 1000). Not surprisingly, the generated vs. fluctuation pattern revealed the areas with a greater modeling ability that were depicted for the BChE potency ( ≥ 0.75) and were accompanied by areas that had a higher ( > 0.5) or lower ( < 0.5) predictive power. Obviously, the preferential selection of objects into the training set resulted in a decrease in the predictive performance of the test set. Figure 1 illustrates the frequency distribution of the compounds in the test subgroup when sampling the models that had the best statistical parameters ( ≥ 0.7 and > 0.5). A relatively smooth compound frequency distribution within the test set population was disrupted by the larger numbers of compounds 2, 5, 6, 13, 15, 28, 31, 38, and 41, respectively.
Figure 1.
Histogram indicating number of individual compounds that accured in test set within the regions of > 0.7 and 0.5 for the training set against L–7–O CV for modeling the BChE potency of the carbamate derivatives (1–41) using CoMSA approach.
In general, the specified molecules covered the entire structural space as well as the BChE activity range (5.51–440.65 µM) of the investigated compounds evenly, which partially explains the good ability and predictability of the model.
Additionally, the similarity was analyzed using the PCA procedure on the pool of descriptors that was derived from Dragon 6.0 software, in which the final dataset of 2777 variables was arranged in matrix X41×2777 with the rows representing molecules (objects) and the columns preserving the numerical values of the variables (parameters) [37,38]. The experiment was carried out for the centered and standardized data to illustrate any meaningful variations in the modeling performance of the examined set of carbamate derivatives with respect to their structure and inhibitory profiles. The PCA model with the first four PCs described 80.66% of the total data variance, whereas the first two PCs accounted for 67.37%.
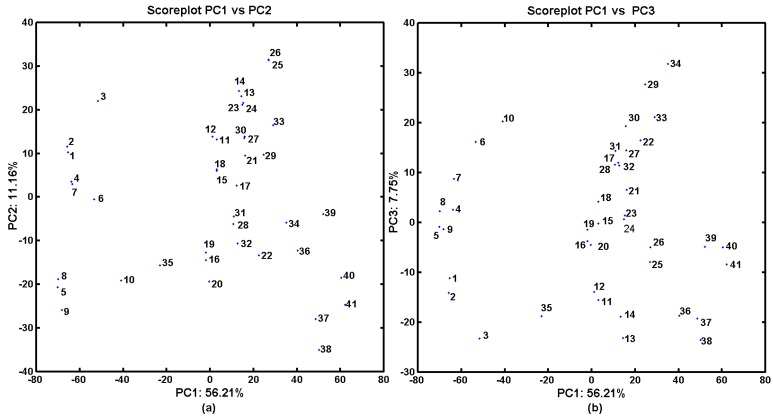
Figure 2 presents the respective score plots of PC1 vs. PC2 (Figure 2a) and of PC1 vs. PC3 (Figure 2b), which indicated that the methyl-based carbamate derivatives (1–10 and 35) can be classified into one structurally related group along the first principal component (PC1 < −20). Moreover, the naphthalene-based derivatives were generally grouped together (PC1 > 40), which indicated major structural variations from all of the remaining ones.
Figure 2.
Projection of carbamates 1–41 on a plane defined by two principal components for the Dragon descriptors: the first vs. second (a); and the first vs. third (b).
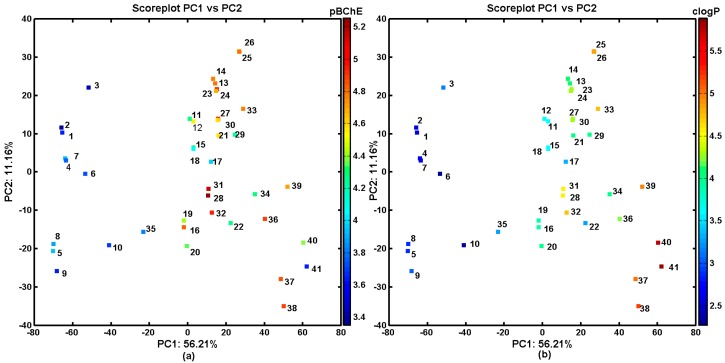
A detailed inspection of the experimental BChE inhibitory profile and compound lipophilicity, which were color-coded according to the values of logP (calculated in Sybyl-X) for the objects that were projected on the plane specified by the two first components, revealed some correlation (R = 0.67). This was especially visible for the methyl-based carbamates (1–10 and 35), for which the was the lower activity (pBChE < 4) the lower was the lipophilicity (clogP < 3). On the other hand, some of the more active naphthalene derivatives were accompanied by fairly high values of the calculated lipophilicity (clogP > 5) as observed in Figure 3.
Figure 3.
Projection of carbamates 1–41 on the plane defined by the first vs. second principal components with: BChE activity in the logarithmic scale (a); and the calculated lipophilicity profile (b). Colors indicate the numerical values of the pBChE activity and clogP.
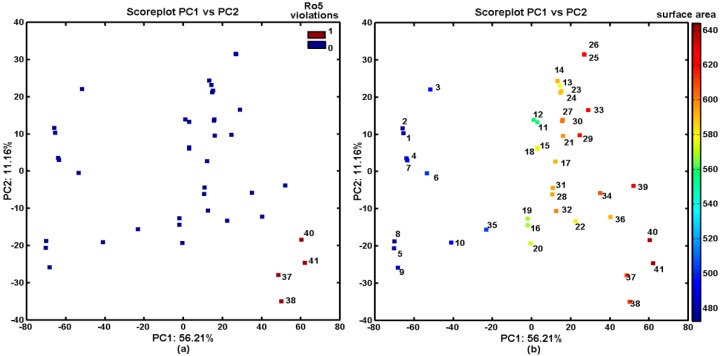
One can conclude that the lipophilic profile for compounds should be directly related with their chemical structure, and, therefore, structurally similar molecules (chemotypes) might have similar properties. This tendency can be confirmed for halogenated naphthalene derivatives (37, 38, 40, and 41), which violated the Lipinski’s Rule of Five (Ro5), as can be observed in Figure 4a. Obviously, the above chlorine- and bromine-based molecules were characterized by the greatest values of the surface area descriptors, which were obtained by the Sybyl-X software, as visualized in Figure 4b.
Figure 4.
Projection of carbamates 1–41 on the plane defined by the first vs. second principal components with: Ro5 rule violations (a); and the calculated surface area (b). Colors indicate the number of Ro5 violations and the numerical values of surface area.
2.4. Probability-Guided Pharmacophore Mapping
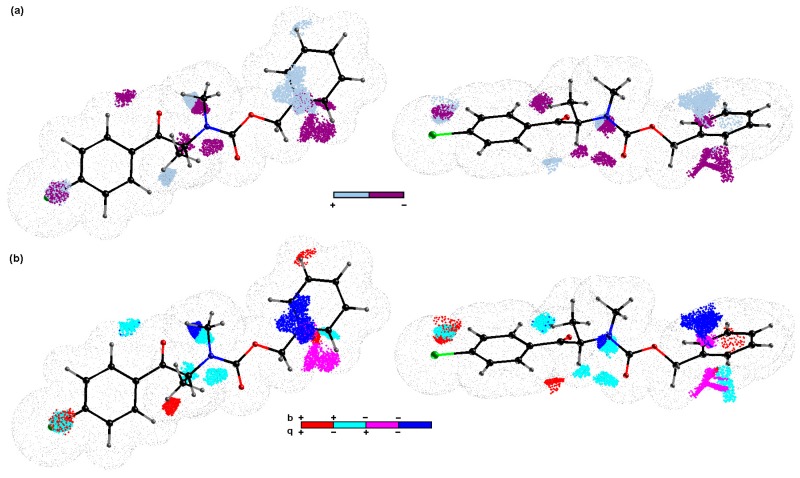
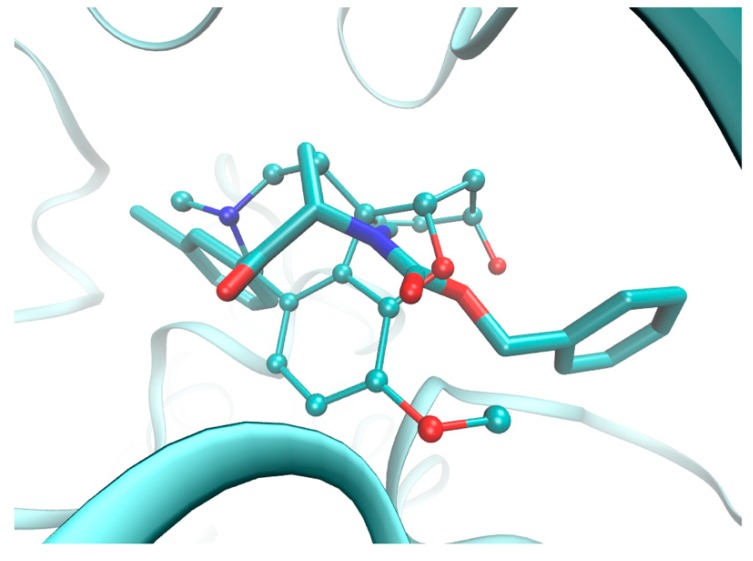
It is characteristic that a vast set of highly inter-correlated topologic/topographic descriptors is generated for mD-QSAR studies, and, therefore, the uninformative variable elimination procedure can be regarded as a pre-processing stage to prune the input data ensemble. As a result, the iterative variable elimination procedure (IVE-PLS) was used as a filter to specify the descriptors with the highest individual weightings towards the inhibitory activity [39]. Thus, 10% of the 30/11 training/test samples was randomly selected from regions that had a fairly high model ability ( > 0.70) and predictability ( > 0.5) to produce “consensus-based” pharmacophore maps. Taking the number of molecules into account, the maximum number of PLS components to generate the model was truncated to seven. The variables with the highest stability abs(mean(b)/std(b)) for each of the randomly selected models were specified using the IVE-PLS algorithm. In general, the predictive power of a model that was monitored by was stable for a considerable range of the variables that were eliminated, whereas there was a slight improvement of the performance when the columns with the lowest values of stability were extracted. The backward column elimination was iteratively looped until the optimal number of variables to be included within the model was accomplished—the moment that the deterioration determined the number of relevant columns. The cumulative sum of the common columns for all of the investigated 1661 BChE models was calculated and normalized to a range of <0,1>. The set of columns with a value above the pre-selected cut-off of 0.4 was selected; however, the spatial pattern, as depicted in Figure 5, was produced by filtering a further 80% of the CoMSA descriptors that had a relatively small statistical significance for the BChE inhibitory activity. To simplify the visual inspection of the key pharmacophore patterns, colors were used to indicate the impact of the descriptors on the inhibitory potency to show the areas with a positive and/or negative activity contribution (Figure 5a) as well as the four possible combinations of the mean regression coefficients and charge values (Figure 5b), respectively.
Figure 5.
The spatial sectors with the greatest contribution into the BChE inhibitory potency specified as CoMSA IVE-PLS for the selected 30/11 training/test set samplings. Colors indicate their effect (a) and the four possible combination of the mean charge q and correlation coefficient b values (b). Reference compound 32 is plotted in two different orientations.
The spatial areas with a potentially detrimental impact on the BChE inhibitory potency, primarily due to steric or electrostatic factors, are indicated by the dark spheres in Figure 5a, whereas the bright polyhedrals depict the 3D pattern that was predicted to be positioned by an atom or substituent to reinforce the molecular inhibitory profile. It appeared that the elongation of the side chains R1 and R2 bonded to the peptide-bond-like motif in the scaffold contributed favorably to the BChE inhibition activity of the analyzed carbamates, as was suggested by the bright areas close to the nitrogen attached directly to the carbonyl group. This confirms the tendency that was observed among the carbamate homologs in which an additional methylene group improved the BChE inhibition affinity slightly (see Table 1). The obtained findings demonstrate the significance of the side chain R3, where the mixed (positive/negative) steric contribution to the inhibitory potency was observed, as shown in Figure 5a. The increase in the bulkiness in this area appears to be a favorable structural variation that partially elucidates the increase in the BChE potency for phenyl and benzyl carbamates compared to the methyl counterparts, as presented in Table 1. The presence of negatively charged motifs (delocalized π electrons) probably contributed favorably (negative regression coefficients) to the inhibition profile of the analyzed series, as depicted in Figure 5b. Basically, the spatially disallowed areas that were attributed by the negative mean charge values of the IVE-PLS CoMSA models were specified for the para position of the phenyl ring (X substituent). In other words, the indicated area might have been occupied by negatively charged atoms/groups, but an increase in the volume/surface may be detrimental to the inhibition potential, which corresponds quite well with the depletion of the BChE inhibitory abilities that were observed for the bromine- vs. the chlorine-based carbamates (see Table 1).
The visualization of the pharmacophore maps, which were generated using the consensus 3D-QSAR methodology, provided valuable knowledge about the interaction mode that was required to enhance the inhibitory potential of the analyzed carbamates as prospective ChEIs.
2.5. Molecular Docking
Site-directed computer-assisted docking algorithms optimally position a (bio)effector molecule in a binding/active site, which is based on the intrinsic relationship of the target–host atomic coordinates, while the spatial geometry, or at least the homology models of macromolecule, are accessible [40]. In fact, target-guided docking algorithms mainly employ descriptor-matching approaches in which the ligand property space is adapted to the corresponding map of the target electronic, steric and lipophilic features. How to in silico estimate the “natural” forces that combine the building blocks together in structure-based drug-design (SBDD) is still unclear, since the method does not always produce a reliable quantitative correlation between the experimental and predicted activity assays [41]. In fact, the computational procedures that attempt to reconstruct a ligand–receptor complex using the molecular docking approaches involve an efficient search procedure and effective scoring function, respectively [42]. Rapidly covering the relevant conformational space at the search stage as well as the effective discrimination between native or non-native docked conformation at the scoring stage are basically the two critical elements of the docking procedure; however, the SBDD methodology is typically used for hit virtual screening (flash docking) and lead to optimization [43,44].
The geometries of human acetyl (AChE) and butyrylcholinesterase (BChE) enzymes that are co-crystallized with pharmacologically used drugs (e.g., galanthamine or rivastigmine) are still under intense scrutiny, which enabled us to compare the findings of the ligand- and structure-based protocols. On the other hand, a comprehensive examination of the guest–target interactions using molecular dynamic simulations (MDs) was beyond the scope of this study.
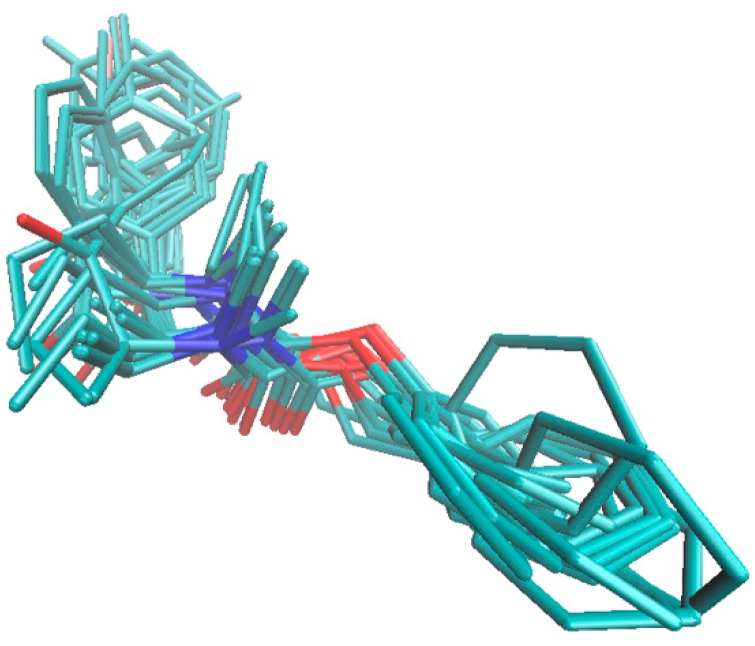
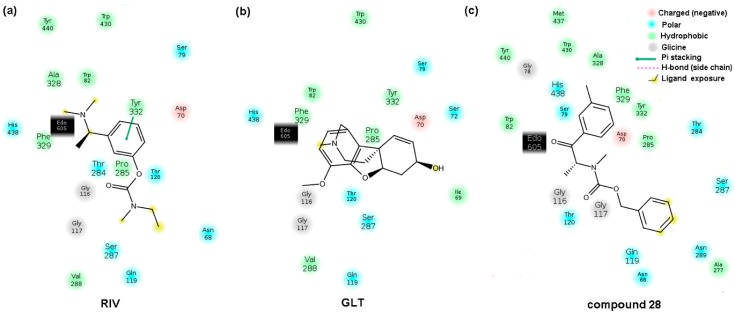
The crystallographic data of butyrylcholinesterase with a catalytic core that is specified at a higher resolution of 2.6 Å in the liganded state (holo) with the rivastigmine (RIV) analog ([3-[(1~{R})-1-(dimethylamino)ethyl]-4-oxidanyl-phenyl]~{N}-ethyl-~{N}-methyl-carbamate) were retrieved from the Protein Data Bank repository (PDB entry: 6eul). The drug analog was subsequently (re)docked in the active site AC2 of the enzyme chain A, which is composed of six amino acid residues (Asn68, Ile69, Asp70, Trp82, Thr120, and Pro285) and a 1,2-ethanediol molecule (EDO605), using the AutoDock Vina program [45]. Moreover, the population of the BChE inhibitors (IC50 < 30 µM) that are comparatively active to rivastigmine was investigated to compare the interacting mode of the carbamate derivatives with the drug–enzyme spatial interaction pattern. In fact, the specific conformations and orientations (poses) of the most active compounds (11, 13, 14, 16, 23–28, 30–33, and 36–39), as illustrated in Figure 6, confirmed our intuitive selection of the trial alignment in the ligand-based study.
Figure 6.
Comparison of the poses for the most active BChE inhibitors in the enzyme active site AC2. As an additional experiment galanthamine (GLT), which is a very active AChE/BChE inhibitor (IC50 < 3 µM) was docked in the enzyme active site AC2. Although the structural variations between GLT and RIV and the most active molecules are easily noticeable, similar spatial patterns of the host–BChE complex within the catalytic core was observed for the negatively charged atoms (nitrogen and oxygen), as presented in Figure 7.
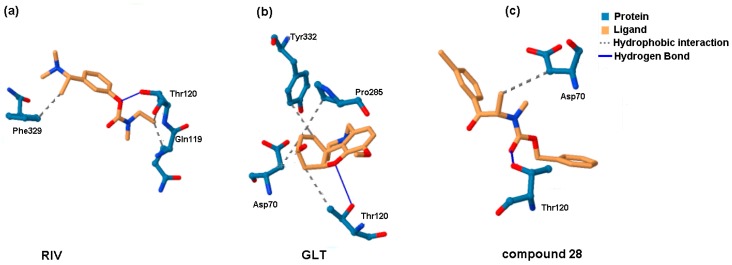
Moreover, a run-through was performed to investigate the interacting mode for the most active compounds, which basically revealed two types of non-binding interactions: hydrophobic and hydrogen bond formation. Planar (2D) and spatial (3D) maps of the host–target interactions were created for all of the active compounds using Schrödinger Maestro software and the Protein-Ligand Interaction Profiler (PLIP) and were then compared with the drug (GLT and RIV) molecules [46]. Overall, the hydrophobic interactions were primarily generated with Asp70 (94%), Trp82 (28%), Pro285 (22%) and Tyr332 (22%), while Thr120 (83%) was basically specified as the hydrogen bond donor (HBD) that interacted with one of the oxygen atoms in the molecule carboxylic group. The 2D- and 3D-binding patterns for GLT, RIV and the most active BChE inhibitor (28) that was tested are illustrated in Figure 8 and Figure 9, respectively. The hydroxyl substituent of Thr120 appeared to be crucial to form the hydrogen bond with the ether (Figure 9a,b) or carbonyl oxygen (Figure 9c). For the examined BChE inhibitors, the regions close to the nitrogen atom (R1 substituent) seemed to be valid for the hydrophobic interactions with the Asp70 amino acid residue, which is in line with our previous receptor-independent findings (Figure 5).
Figure 8.
Planar (2D) comparision of the binding mode for: rivastigmine (a); galanthamine (b); and the most active BChE inhibitor 28 that was tested (c), as generated by the Maestro software.
Figure 9.
Spatial (3D) comparison of the binding mode for: rivastigmine (a); galanthamine (b); and the the most active BChE inhibitor 28 that was tested (c), as generated by the PLIP software.
Unfortunately, the obtained docking findings did not provide a clear explanation of the variations that the meta/para-positioned carbamate derivatives exerted on the enzyme reaction site; however, the close proximity of a positively charged nitrogen atom of His438 (Figure 8c) might have been potentially beneficial to the inhibition potential, especially the negatively charged chlorine- or bromine-based carbamates, which corresponds relatively well with the IVE-PLS CoMSA results. The electrostatic repulsion between the negatively charged atoms and the oxygen of Ser79 can partially explain the detrimental impact of the –OCH3 or –OCH2O- groups attached to the phenyl ring. On the other hand, the postulated hydrophobic interactions with Phe329 might favorably contribute to the inhibitory potential, as was observed for (di)methyl derivatives.
Obviously, we should beware of q2 [47] and beware of docking [48]; therefore, a combination of the consensus pharmacophore mapping with a systematic screening of multifaceted guest–host interactions with target-tailored procedures is highly advisable in rational drug design.
3. Materials and Methods
3.1. Chemistry
All of the reagents and solvents were purchased from ACROS Organics, Asta-Tech, Maybridge, Santa Cruz Biotechnology and Sigma-Aldrich. The melting points were measured in an OptiMelt—Automated Melting Point System (MPA 100, Stanford Research Systems). The 1H and 13C NMR spectra were recorded on a Bruker Ascend 500 MHz spectrometer at frequencies of 500 MHz and 126 MHz and a Bruker Avance III 400 MHz FT-NMR spectrometer at frequencies of 400 MHz and 101 MHz using CDCl3-d6 or DMSO-d6 as the solvent and TMS as the internal standard. The NMR solvents were purchased from ACROS Organics. The chemical shifts (δ) are given in ppm and the values of the coupling constants (J) are reported in hertz (Hz). The purity of all of the compounds was tested using the HPLC/MS method. The HPLC-MS analyses were performed on a Varian model 920 liquid chromatograph equipped with a Varian 900-LC model autosampler, a gradient pump, a Varian Pro Star 510 model column oven and a Varian 380-LC model evaporative light scattering detection (ELSD) detector. This was coupled with a Varian 500-MS IT. The HRMS were determined using a Waters LCT Premier XE high resolution mass spectrometer with electrospray ionization (ESI).
3.2. General Procedure Used to Synthesize Carbamates 1–41
A mixture of a suitable acid chloride (1 moL) and an anhydrous aluminum chloride (1.05 moL) was stirred until the aluminum chloride was completely dissolved, and then cooled in a water bath and diluted in 250 mL of methylene chloride. Next, a suitable aromatic hydrocarbon (1.10 moL) was added dropwise to a magnetically stirred solution. The reaction mixture was poured into ice-cooled water, the dish was washed with distilled water three times and the organic layer was separated. Methylene chloride was evaporated and the reaction product was crystallized or purified by distillation. In the next step, 1.1 moL of bromine was added dropwise to the solution of the suitable aromatic ketone (1 moL) dissolved in 300 mL of methylene chloride. After the bromine was dropped into the magnetically stirred mixture, 300 mL of distilled water was added and mixed until the color of the mixture changed to white or cream. Then, the organic layer was separated and used in that form in the next step. A bromine derivative (1 moL) was dissolved in methylene chloride, a suitable amine (2 moL) was added to the obtained aromatic ketone and then sodium hydroxide was added dropwise. After 12 h of stirring, the organic layer was separated and poured into a beaker filled with ice and hydrochloric acid. An aqueous sodium hydroxide solution was then added to the separated aqueous phase. The extracted amine was washed three times with water, dried with anhydrous magnesium sulfate, diluted with diethyl ether and treated with gaseous hydrogen chloride until an acidic pH solution was obtained. The isolated hydrochloride of the 2-amino-1-phenylpropane-1-one derivative in the form of a white solid was washed with acetone and dried. Then, 0.01 mol of 2-amino-1-phenylpropane-1-one derivative dissolved in 50 mL of methylene chloride and 0.01 mol of triethyloamine was added to the cooled and magnetically stirred mixture, after which approximately 9 mmoL of suitable chloroformate was added dropwise. After dropping, the mixture was washed with diluted hydrochloric acid to remove any excess unreacted 1-phenylpropylamine derivative. The organic layer was separated and dried with anhydrous magnesium sulfate. The solvent was evaporated under reduced pressure and the product was purified on a column chromatography using hexane: ethyl acetate 9:1 as the eluent.
Methyl ethyl(1-oxo-1-phenylpropan-2-yl)carbamate (1). Yield 88%;1H-NMR (CDCl3, 400 MHz, ppm): δ 7.99 (t, H, aromat, J = 20.6 Hz); 7.83 (d, H, aromat, J1 = 51.2 Hz); 7.57 (ddd, H, aromat, J1 = 8.5 Hz, J2 = 2.4 Hz, J3 = 1.2 Hz); 7.46 (dd, H, aromat, J1 = 10.5 Hz, J2 = 4.7 Hz); 7.28 (s, H, aromat); 5.75 (d, H, CH, J = 6.6 Hz); 3.75 (s, 3H, CH3); 3.30 (s, H, CH2); 3.16 (m, H, CH2); 1.71–1.24 (m, 3H, CH3); 1.10–0.78 (m, 3H, CH3); 13C-NMR (CDCl3, 100 MHz, ppm): δ 199.2; 155.6; 135.6; 133.3; 128.6; 128.6; 128.4; 128.4; 55.2; 52.8; 38.5; 14.9; 14.3; HR-MS (ESI): for C13H17NO3Na [M+Na]+ calculated: 258.1106. m/z, found: 258.1102 m/z.
Methyl methyl(1-oxo-1-phenylbutan-2-yl)carbamate (2). Yield 83%; 1H-NMR (DMSO, 400 MHz, ppm): δ 7.90 (dd, 2H, aromat, J1 = 21.8 Hz, J2 = 7.5); 7.71–7.41 (m, 3H, aromat), 5.35 (ddd, 1H, CH, J1 = 52.6 Hz, J2 = 9.6 Hz, J3 = 5.1 Hz), 3.47 (d, 3H, CH3, J = 111.4 Hz), 2.79–2.44 (m, 2H, CH2), 1.92-1.62 (m, 3H, CH3), 1.05-0.72 (m, 3H, CH3); 13C-NMR (DMSO, 100 MHz, ppm): δ 204.0; 161.8; 140.7; 138.4; 133.9; 133.9; 133.1; 133.1; 65.9; 57.8; 34.9; 25.6; 15.5; HR-MS (ESI): for C13H17NO3Na [M+Na]+ calculated: 258.1106 m/z, found: 258.1103 m/z.
Methyl ethyl(1-oxo-1-phenylbutan-2-yl)carbamate (3). Yield 85%; 1H-NMR (CDCl3, 400 MHz, ppm): δ 8.01 (dd, 2H, aromat, J1 = 48.8 Hz, J2 = 6.8); 7.62–7.25 (m, 3H, aromat), 5.54 (dd, 1H, CH, J1 = 65.9 Hz, J2 = 58.5 Hz), 3.79 (d, 3H, CH3, J = 17.3 Hz), 3.14 (dd, 2H, CH2, J1 = 22,1 Hz, J2 = 7,1 Hz), 2.10–1.92 (m, 2H, CH2), 1.08–0.81 (m, 3H, CH3), 1.08-0.81 (m, 3H, CH3); 13C-NMR (CDCl3, 100 MHz, ppm): δ 198.8; 157.3; 135.9; 133.3; 128.5; 128.5; 127.9; 127.9; 60.1; 52.9; 38.1; 21.8; 15.0; 10.4; HR-MS (ESI): for C14H19NO3 Na [M+Na]+ calculated: 272.1263 m/z, found: 272.1263 m/z.
Methyl methyl(1-oxo-1-(m-tolyl)propan-2-yl)carbamate (4). Yield 86%; 1H-NMR (CDCl3, 400 MHz, ppm): 7.75 (dd, 2H, aromat, J1 = 42.0 Hz, J2 = 8,6); 7.44–7.28 (m, 2H, aromat); 5.79–5.25 (m, 1H, CH), 3.95–3.45 (m, 3H, CH3), 2.77 (d, 3H, CH3, J = 34,6 Hz), 2.30 (d, 3H, CH3, J = 91,9); 1.69–1.22 (m, 3H, CH3); 13C-NMR (CDCl3, 101 MHz, ppm): 199.5; 156.9; 138.5; 135.4; 134.1; 128.8; 128.5; 125.6; 55.0; 52.9; 29.3; 21.3; 13.5; HR-MS (ESI): for C13H17NO3 Na [M+Na]+ calculated: 258.1106 m/z, found: 258.1105 m/z.
Methyl (1-(3-chlorophenyl)-1-oxopropan-2-yl)(methyl)carbamate (5). Yield 89%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.15–7.69 (m, 2H, aromat); 7.64–6.99 (m, 2H, aromat); 5.77–5.28 (m, 1H, CH), 3.76 (d, 3H, CH3 J = 16.0 Hz), 2.76 (d, 3H, CH3, J = 42.1 Hz), 1.46–1.19 (m, 3H, CH3); 13C-NMR (CDCl3, 126 MHz, ppm): 198.1; 156.8; 136.9; 135.0; 133.2; 130.0; 128.4; 126.5; 55.3; 53.1; 29.4; 13.3; HR-MS (ESI): for C12H14ClNO3 [M+H]+ calculated: 256.0740 m/z, found: 256.0741 m/z.
Methyl (1-(4-methoxyphenyl)-1-oxopropan-2-yl)(methyl)carbamate (6). Yield 79%; 1H-NMR (CDCl3, 400 MHz, ppm): 7.97 (dd, 2H, aromat, J1 = 42.0 Hz, J2 = 8.4); 7.01–6.87 (m, 2H, aromat); 5.84–5.14 (m, 1H, CH), 3.88 (s, 3H, CH3); 3.76 (d, 3H, CH3, J = 11.1 Hz), 2.93-2.58 (m, 3H, CH3), 1.37 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 101 MHz, ppm): 197.3; 163.7; 156.8; 130.8; 130.5; 128.2; 113.9; 113.9; 55.4; 54.4; 53.0; 29.1; 13.4; HR-MS (ESI): for C13H17NO4 Na [M+Na]+ calculated: 274.1055 m/z, found: 274.1043 m/z.
Methyl methyl(1-oxo-1-(p-tolyl)propan-2-yl)carbamate (7). Yield 87%; m.p.: 36-37.5 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 7.87 (dd, 2H, aromat, J1 = 53.2 Hz, J2 = 7.8); 7.28 (dd, 2H, aromat, J1 = 5.2 Hz, J2 = 4.6); 5.65 (dt, 1H, CH, J1 = 156.3 Hz, J2 = 6.8), 3.76 (d, 3H, CH3 J = 8.6 Hz), 2.76 (d, 3H, CH3, J = 49.2 Hz), 2.44 (d, 3H, CH3, J = 15.6); 1.81–0.70 (m, 3H, CH3); 13C-NMR (CDCl3, 126 MHz, ppm): 198.7; 156.8; 144.2; 132.8; 129.4; 129.4; 128.6; 128.6; 54.8; 53.0; 29.2; 21.7; 13.4; HR-MS (ESI): for C13H17NO3 Na [M+Na]+calculated: 258.1106, m/z, found: 258.1108 m/z.
Methyl (1-(4-chlorophenyl)-1-oxopropan-2-yl)(methyl)carbamate (8). Yield 87%; 1H-NMR (CDCl3, 500 MHz, ppm): 7.91 (dd, 2H, aromat, J1 = 55.8 Hz, J2 = 8.1); 7.50–7.16 (m, 2H, aromat); 5.72–5.31 (m, 1H, CH), 3.75 (s, 3H, CH3), 2.75 (d, 3H, CH3, J = 53.5 Hz), 1.49–1.23 (m, 3H, CH3); 13C-NMR (CDCl3, 126 MHz, ppm): 197.9; 156.8; 139.8; 133.6; 129.9; 129.9; 129.0; 129.0; 54.0; 53.1; 29.2; 13.2; HR-MS (ESI): for C12H14ClNO3 [M+H]+ calculated: 256.0740 m/z, found: 256.0743 m/z.
Methyl (1-(4-bromophenyl)-1-oxopropan-2-yl)(methyl)carbamate (9). Yield 87%; 1H-NMR (DMSO, 400 MHz, ppm): 7.77 (dd, 4H, aromat, J1 = 23.0 Hz, J2 = 8.6); 5,35 (dt, 1H, CH, J1 = 21.8 Hz, J2 = 6.7 Hz), 3.58–3.26 (m, 3H, CH3), 2.85–2.44 (m, 3H, CH3), 1.35–1.24 (m, 3H, CH3); 13C-NMR (DMSO, 100 MHz, ppm): 203.3; 160.8; 139.8; 136.9; 136.9 135.1; 135.1; 132.3; 61.9; 57.9; 35.8; 18.2; HR-MS (ESI): for C12H14BrNO3 [M+H]+ calculated: 300.0235 m/z, found: 300.0222 m/z.
Methyl (1-(benzo[d][1,3]dioxol-5-yl)-1-oxopropan-2-yl)(methyl)carbamate (10). Yield 76%; 1H-NMR (CDCl3, 500 MHz, ppm): 7.54 (ddd, 1H, aromat, J1 = 57.3 Hz, J2 = 45.8, J3 = 16.5); 7.28 (s, 1H, aromat); 6.87 (d, 1H, aromat, J = 8.2); 6.06 (s, 2H, CH2); 5.77–5.26 (m, 1H, CH), 3.77 (d, 3H, CH3, J = 15.2); 2.94–2.48 (m, 3H, CH3); 1.61 (s, 3H, CH3); 13C-NMR (CDCl3, 126 MHz, ppm): 197.0; 156.8; 152.4 148.2; 129.9; 125.0; 108.2; 101.8; 100,4; 54.5; 53.0; 29.1; 13.5; HR-MS (ESI): for C13H15NO5Na [M+Na]+ calculated: 288.0848 m/z, found: 288.0842 m/z.
Phenyl ethyl(1-oxo-1-phenylpropan-2-yl)carbamate (11). Yield 87%; 1H-NMR (CDCl3, 400 MHz, ppm): 8.03 (dd, 2H, aromat, J1 = 27.1 Hz, J2 = 7.5); 7.68–6.71 (m, 8H, aromat); 5.69 (dq, 1H, CH, J1 = 92.3, J2 = 6,8); 3.56-3.15 (m, 2H, CH2), 1.56 (dd, 3H, CH3, J1 = 36.3 Hz, J2 = 6.6); 1.20 (dt, 3H, CH3, J1 = 22.3 Hz, J2 = 7.1); 13C-NMR (CDCl3, 101 MHz, ppm): 199.1; 154.9; 151.4 135.5; 133.4; 129.4; 129.4; 128.8; 128.8; 128.5; 128.5; 125.5; 121.6; 121.6; 55.5; 39.1; 15.6; 14.6; HR-MS (ESI): for C18H19NO3 Na [M+Na]+ calculated: 320.1263m/z, found: 320.1258 m/z.
Phenyl methyl(1-oxo-1-phenylbutan-2-yl)carbamate (12). Yield 89%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.07 (ddd, 2H, aromat, J1 = 21.3 Hz, J2 = 7.4, J3 = 1.2); 7.65–7.59 (m, 1H, aromat); 7.51 (td, 2H, aromat, J1 = 7.4 Hz, J2 = 3.7); 7.43-7.35 (m, 2H, aromat); 7.29–7.20 (m, 1H, aromat); 7.14–7.09 (m, 2H, aromat); 5.62 (ddd, 1H, CH, J1 = 34.9, J2 = 9.5, J3 = 5.6); 2.91 (d, 3H, CH3, J = 32.3); 2.09–1.81 (m, 2H, CH2); 1.08 (dt, 3H, CH3, J1 = 35.9 Hz, J2 = 7.4); 13C-NMR (CDCl3, 126 MHz, ppm): 197.8; 155.5; 151.3 135.7; 133.5; 129.3; 129.3; 128.8; 128.8; 128.6; 128.6; 125.5; 121.7; 121.7; 60.4; 29.8; 20.9; 10.4; HR-MS (ESI): for C18H19NO3Na [M+Na]+ calculated: 320.1263 m/z, found: 320.1260 m/z.
Phenyl ethyl(1-oxo-1-phenylbutan-2-yl)carbamate (13). Yield 88%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.21–7.90 (m, 2H, aromat); 7.62 (dd, 1H, aromat, J1 = 14.2 Hz, J2 = 6.9); 7.54–7.48 (m, 2H, aromat); 7.46–7.37 (m, 2H, aromat); 7.33–7.21 (m, 1H, aromat); 7.18–7.09 (m, 2H, aromat); 5.61 (ddd, 1H, CH, J1 = 44.6, J2 = 8.7, J3 = 6.1); 3.63–3.12 (m, 2H, CH2); 2.12-1.82 (m, 2H, CH2); 1.13 (td, 3H, CH3, J1 = 7.2 Hz, J2 = 3.4); 1.06 (dt, 3H, CH3, J1 = 14.8 Hz, J2 = 7.2); 13C-NMR (CDCl3, 126 MHz, ppm): 198.6; 155.4; 151.4 135.8; 133.5; 129.4; 129.4; 128.8; 128.8; 128.6; 128.6; 125.4; 121.6; 121.6; 60.4; 38.8; 21.8; 15.3; 10.5; HR-MS (ESI): for C19H21NO3Na [M+Na]+ calculated: 334.1419 m/z, found: 334.1411 m/z.
Phenyl methyl(1-oxo-1-phenylpentan-2-yl)carbamate (14). Yield 76%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.11–8.02 (m, 2H, aromat); 7.62 (dd, 1H, aromat, J1 = 14.4 Hz, J2 = 7.2 Hz); 7.54–7.47 (m, 2H, aromat); 7.47–7.35 (m, 2H, aromat); 7.32–7.20 (m, 1H, aromat); 7.13–7.08 (m, 2H, aromat); 5.73 (ddd, 1H, CH, J1 = 28.4, J2 = 9.0, J3 = 5.8); 2.92 (d, 3H, CH3, J = 30.6); 1.98–1.81 (m, 2H, CH2); 1.49–1.40 (m, 2H, CH2); 1.04 (t, 3H, CH3, J = 7.4 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 199.0; 155.4; 151.4; 135.7; 133.5; 129.5; 129.5; 128.8; 128.8; 128.6; 128.6; 125.4; 121.7; 121.7; 58.6; 30.2; 29.8; 19.2; 13.9; HR-MS (ESI): C19H21NO3 [M+H]+ calculated for: 312.1600 m/z, found: 312.1605 m/z.
Phenyl methyl(1-oxo-1-(m-tolyl)propan-2-yl)carbamate (15). Yield 84%; m.p.: 52.5-53.5 °C; 1H-NMR (CDCl3, 400 MHz, ppm): 7.94–7.73 (m, 2H, aromat); 7.40 (dt, 2H, aromat, J1 = 15.7 Hz, J2 = 7.5); 7.28 (s, 2H, aromat); 7.23 (t, 1H, aromat, J = 7.4 Hz); 7.09 (t, 2H, aromat, J = 8.3 Hz); 5.74 (dq, 1H, CH, J1 = 50.8, J2 = 6.9); 2.94 (d, 3H, CH3, J = 10.2 Hz); 2.44 (s, 3H, CH3); 1.48 (d, 3H, CH3, J = 7,0 Hz); 13C-NMR (CDCl3, 101 MHz, ppm): 199.3; 154.9; 151.4; 138.6; 135.4; 134.3; 129.3; 129.3 128.9; 128.7; 125.7; 125.4; 121.6; 121.6; 55.4; 30.0; 21.3; 13.6; HR-MS (ESI): for C18H19NO3Na [M+Na]+ calculated: 320.1263 m/z, found: 320.1277 m/z.
Phenyl (1-(3-chlorophenyl)-1-oxopropan-2-yl)(methyl)carbamate (16). Yield 92%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.01 (dd, 1H, aromat, J1 = 7.5 Hz, J2 = 5.7 Hz); 7.91 (dd, 1H, aromat, J1 = 27.4Hz, J2 = 7.8 Hz); 7.63–7.56 (m, 1H, aromat); 7.49–7.42 (m, 1H, aromat); 7.42–7.35 (m, 2H, aromat); 7.27–7.19 (m, 1H, aromat); 7.13–7.06 (m, 2H, aromat); 5.68 (dq, 1H, CH, J1 = 61.2, J2 = 6.9); 2.93 (d, 3H, CH3, J = 17.1 Hz); 1.49 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 198.0; 155.0; 151.3; 137.0; 135.1; 133.5; 130.2; 129.4; 129.4; 128.5; 126.5; 125.6; 121.6; 121.6; 55.6; 30.0; 13.4; HR-MS (ESI): for C17H16ClNO3 Na [M+Na]+ calculated: 340.0717 m/z, found: 340.0714 m/z.
Phenyl (1-(4-methoxyphenyl)-1-oxopropan-2-yl)(methyl)carbamate (17). Yield 78%; m.p.: 53–56 °C; 1H-NMR (CDCl3, 400 MHz, ppm): 8.04 (dd, 2H, aromat, J1 = 17.9 Hz, J2 = 8.8); 7.39 (dd, 2H, aromat, J1 = 10.7 Hz, J2 = 5.1); 7.30–7.19 (m, 1H, aromat); 7.11 (t, 2H, aromat, J = 8.8 Hz); 6.97 (d, 2H, aromat, J = 8.9 Hz); 5.74 (dq, 1H, CH, J1 = 53.9, J2 = 6.9); 3.90 (s, 3H, CH3); 2.93 (d, 3H, CH3, J = 8,2 Hz); 1.47 (d, 3H, CH3, J = 6.7 Hz); 13C-NMR (CDCl3, 101 MHz, ppm): 199.3; 154.9; 151.4; 138.6; 135.4; 134.3; 129.3; 129.3 128.9; 128.7; 125.7; 125.4; 121.6; 121.6; 55.4; 30.0; 21.3; 13.6; HR-MS (ESI): for C18H19NO4Na [M+Na]+ calculated: 336.1212 m/z, found: 336.1222 m/z.
Phenyl methyl(1-oxo-1-(p-tolyl)propan-2-yl)carbamate (18). Yield 82%; m.p.: 42.5–44 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 7.94 (dd, 2H, aromat, J1 = 22.3 Hz, J2 = 8.2); 7.39 (t, 2H, aromat, J = 7.9 Hz); 7.30 (dd, 2H, aromat, J1 = 11.7 Hz, J2 = 3.7); 7.23 (t, 1H, aromat, J = 7.4 Hz); 7.14–7.07 (m, 2H, aromat); 5.74 (dq, 1H, CH, J1 = 72.3, J2 = 6.9); 2.93 (d, 3H, CH3, J = 9.3 Hz); 2.44 (s, 3H, CH3); 1.48 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 198.5; 154.9; 151.4; 144.4; 132.8; 129.5; 129.5; 129.3; 129.3; 128.7; 128.7; 125.5; 121.7; 121.7; 55.0; 29.9; 21.7; 13.6; HR-MS (ESI): for C18H19NO3Na [M+Na]+ calculated: 320.1263 m/z, found: 320.1262 m/z.
Phenyl (1-(4-chlorophenyl)-1-oxopropan-2-yl)(methyl)carbamate (19). Yield 91%; m.p.: 52–54 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 7.98 (dd, 2H, aromat, J1 = 27.4 Hz, J2 = 8.5 Hz); 7.48 (dd, 2H, aromat, J1 = 11.8 Hz, J2 = 4.9Hz); 7.43–7.36 (m, 2H, aromat); 7.24 (t, 1H, aromat, J = 7.4 Hz); 7.08 (dd, 2H, aromat, J1 = 22.8 Hz, J2 = 7.8 Hz); 5.68 (dq, 1H, CH, J1 = 82.3, J2 = 6.9); 2.93 (s, 3H, CH3); 1.48 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 197.7; 154.9; 151.3; 140.0; 133.7; 130.0; 130.0; 129.4; 129.4; 129.1; 129.1; 125.6; 121.6; 121.6; 55.2; 29.9; 13.4; HR-MS (ESI): for C17H16ClNO3Na [M+Na]+ calculated: 340.0717 m/z, found: 340.0712 m/z.
Phenyl (1-(4-bromophenyl)-1-oxopropan-2-yl)(methyl)carbamate (20). Yield 88%; m.p.: 54–55 °C; 1H-NMR (CDCl3, 400 MHz, ppm): 7.90 (dd, 2H, aromat, J1 = 21.9 Hz, J2 = 8.5 Hz); 7.65 (dd, 2H, aromat, J1 = 8.3 Hz, J2 = 6.5 Hz); 7.40 (dd, 2H, aromat, J1 = 11.0 Hz, J2 = 4.8 Hz); 7.25 (dd, 1H, aromat, J1 = 15.9 Hz, J2 = 8.5 Hz); 7.08 (dd, 2H, aromat, J1 = 18.7 Hz, J2 = 7.8 Hz); 5.67 (dq, 1H, CH, J1 = 66.0, J2 = 6.9); 2.93 (s, 3H, CH3); 1.48 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 101 MHz, ppm): 198.0; 155.0; 151.3; 134.1; 132.1; 132.1; 130.0; 130.0; 129.4; 129.4; 128.8; 125.6; 121.6; 121.6; 55.2; 29.9; 13.4; HR-MS (ESI): for C17H16BrNO3Na [M+Na]+ calculated: 384.0211 m/z, found: 384.0226 m/z.
Phenyl (1-(3,4-dimethylphenyl)-1-oxopropan-2-yl)(methyl)carbamate (21). Yield 74%; m.p.: 94.5–95.5 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 7.84–7.74 (m, 2H, aromat); 7.42–7.36 (m, 2H, aromat); 7.29–7.19 (m, 2H, aromat); 7.11 (dd, 2H, aromat, J1 = 11.6, J2 = 4.1); 5.75 (dq, 1H, CH, J1 = 57.6, J2 = 6.9); 2.92 (d, 3H, CH3, J = 14.5); 2.41–2.25 (m, 6H, CH3); 1.47 (d, 3H, CH3, J = 6.9); 13C-NMR (CDCl3, 126 MHz, ppm): 198.8; 154.9; 151.4; 143.2; 137.2; 133.2; 130.0; 129.6; 129.3; 129.3; 126.3; 125.4; 121.7; 121.7; 55.1; 29.9; 20.1; 19.8; 13.7; HR-MS (ESI): for C19H21NO3Na [M+Na]+ calculated: 334.1419 m/z, found: 334.1418 m/z.
Phenyl (1-(benzo[d][1,3]dioxol-5-yl)-1-oxopropan-2-yl)(methyl)carbamate (22). Yield 76%; m.p.: 94–95 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 7.75–7.63 (m, 1H, aromat); 7.54–7.48 (m, 1H, aromat); 7.39 (dd, 2H, aromat, J1 = 10.8 Hz, J2 = 5.1); 7.24 (dt, 1H, aromat, J1 = 13.8 Hz, J2 = 5.9Hz); 7.12 (dd, 2H, aromat, J1 = 12.5 Hz, J2 = 4.8 Hz); 6.89 (dd, 1H, aromat, J1 = 8.2 Hz, J2 = 4.9 Hz); 6.08 (d, 2H, CH2, J = 7.7 Hz); 5.69 (dq, 1H, CH, J1 = 58.8, J2 = 6.9); 2.93 (d, 3H, CH3, J = 19.9 Hz); 1.46 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 196.8; 154.9; 152.2; 151.4; 148.4; 129.1; 129.4; 129.4; 125.5; 125.0; 121.7; 121.7; 108.2; 108,0; 102.0; 54.7; 29.8; 13.7; HR-MS (ESI): for C18H17NO5Na [M+Na]+ calculated: 350.1004 m/z, found: 350.0998 m/z.
Benzyl ethyl(1-oxo-1-phenylpropan-2-yl)carbamate (23). Yield 82%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.01 (d, 2H, aromat, J = 7.6); 7.81 (d, 1H, aromat, J = 7.0 Hz); 7.60–7.26 (m, 7H, aromat); 5.76 (q, 1H, CH, J = 6.8 Hz); 5.31–5.05 (m, 2H, CH2); 3.32–3.10 (m, 2H, CH2), 1.43 (t, 3H, CH3, J = 8.0); 1.05–0.98 (m, 3H, CH3); 13C-NMR (CDCl3, 101 MHz, ppm): 199.3; 156.2; 136.7 135.5; 133.3; 128.7; 128.7; 128.5; 128.5; 128.5; 128.5; 128.0; 127.6; 127.6; 67.4; 55.4; 38.7; 15.5; 14.6; HR-MS (ESI): for C19H21NO3 [M+H]+ calculated: 312.1600 m/z, found: 312.1645 m/z.
Benzyl methyl(1-oxo-1-phenylbutan-2-yl)carbamate (24). Yield 87%; 1H-NMR (d6-aceton, 500 MHz, ppm): 8.03-7.87 (m, 2H, aromat); 7.62 (dt, 1H, aromat, J1 = 22.0, J2 = 7.4); 7.52–7.29 (m, 7H, aromat); 5.52 (ddd, 1H, CH, J1 = 46.7, J2 = 9.6, J3 = 5.3); 5.29–5.10 (m, 2H, CH2); 2.76 (s, 3H, CH3); 1.99–1.73 (m, 2H, CH2); 0.98-0.92 (m, 3H, CH3); 13C-NMR (d6-aceton, 126 MHz, ppm): 198.4; 156.4; 137.3 136.1; 133.1; 128.6; 128.6; 128.4; 128.4; 128.2; 128.2; 127.8; 127.5; 127.5; 66.8; 60.7; 29.6; 20.5; 9.8; HR-MS (ESI): for C19H21NO3Na [M+Na]+ calculated: 334.1419 m/z, found: 334.1418 m/z.
Benzyl ethyl(1-oxo-1-phenylbutan-2-yl)carbamate (25). Yield 83%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.10-7.83 (m, 2H, aromat); 7.60-7.26 (m, 8H, aromat); 5.62 (dd, 1H, CH, J1 = 8.3, J2 = 6.6); 5.22 (q, 2H, CH2, J = 12.5 Hz) 3.14 (dd, 2H, CH2, J1 = 14.1, J2 = 7.0); 2.07–1.72 (m, 2H, CH2); 0.99 (dq, 6H, 2CH3, J1 = 13.5 Hz, J2 = 6.9); 13C-NMR (CDCl3, 126 MHz, ppm): 198.8; 156.7; 136.7; 135.9; 133.3; 128.7; 128.7; 128.5; 128.5; 128.5; 128.5; 128.0; 127.6; 127.6; 67.5; 60.3; 38.3; 21.7; 15.1; 10.5; HR-MS (ESI): for C20H23NO3 [M+H]+ calculated: 326.1756 m/z, found: 326.1763 m/z.
Benzyl methyl(1-oxo-1-phenylpentan-2-yl)carbamate (26). Yield 80%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.06-7.82 (m, 2H, aromat); 7.56 (dt, 1H, aromat, J1 = 23.9 Hz, J2 = 7.4 Hz); 7.48–7.27 (m, 7H, aromat); 5.60 (ddd, 1H, CH, J1 = 115.5, J2 = 9.2, J3 = 5.6); 5.33-5.12 (m, 2H, CH2); 2.78 (d, 3H, CH3, J = 15.1); 1.93-1.72 (m, 2H, CH2); 1.43-1.30 (m, 2H, CH2); 0.99 (dt, 3H, CH3, J1 = 15.0 Hz, J2 = 7.4 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 199.0; 156.7; 136.7; 135.7; 133.3; 128.7; 128.7; 128.5; 128.5; 128.3; 128.3; 128.0; 127.6; 127.6; 67.5; 58.5; 29.7; 29.3; 19.1; 13.9; HR-MS (ESI): for C20H23NO3Na [M+Na]+ calculated: 348.1576 m/z, found: 348.1574 m/z.
Benzyl methyl(1-oxo-1-(m-tolyl)propan-2-yl)carbamate (27). Yield 81%; 1H-NMR (CDCl3, 400 MHz, ppm): 7.83–7.58 (m, 2H, aromat); 7.41–7.18 (m, 7H, aromat); 5.63 (dq, 1H, CH, J1 = 104.2, J2 = 6.8); 5.29-5.10 (m, 2H, CH2); 2.80 (d, 3H, CH3, J = 22.4 Hz); 2.36 (d, 3H, CH3, J = 25.2 Hz); 1.41 (d, 3H, CH3, J = 6.9); 13C-NMR (CDCl3, 101 MHz, ppm): 199.4; 156.2; 138.5; 136.7; 135.4; 134.1; 128.9; 128.5; 128.5; 128.5; 128.5; 128.0; 127.6; 125.6; 67.4; 55.2; 29.4; 21.3; 13.5; HR-MS (ESI): for C19H21NO3Na [M+Na]+ calculated: 334.1419 m/z, found: 334.1419 m/z.
Benzyl (1-(3-chlorophenyl)-1-oxopropan-2-yl)(methyl)carbamate (28). Yield 91%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.00–7.86 (m, 1H, aromat); 7.84–7.63 (m, 1H, aromat); 7.56–7.49 (m, 1H, aromat); 7.47–7.44 (m, 1H, aromat); 7.42–7.23 (m, 5H, aromat); 5.55 (dq, 1H, CH, J1 = 141.1, J2 = 6,8); 5.27–5.11 (m, 2H, CH2); 2.79 (d, 3H, CH3, J = 30.2 Hz); 1.41 (d, 3H, CH3, J = 6.9 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 198.0; 156.2; 136.9; 136.5; 136.1; 135.0; 133.2; 130.0; 128.4; 128.4; 128.7; 127.7; 127.7; 126.5; 67.6; 55.5; 29.5; 13.3; HR-MS (ESI): for C18H18ClNO3Na [M+Na]+ calculated: 354.0873 m/z, found: 354.0863 m/z.
Benzyl (1-(4-methoxyphenyl)-1-oxopropan-2-yl)(methyl)carbamate (29). Yield 79%; m.p.: 48–50 °C; 1H-NMR (CDCl3, 400 MHz, ppm): 7.91 (dd, 2H, aromat, J1 = 74.8 Hz, J2 = 8.6); 7.43–7.24 (m, 5H, aromat); 6.85 (dd, 2H, aromat, J1 = 55.0 Hz, J2 = 8.6); 5.67 (dt, 1H, CH, J1 = 93.9, J2 = 6.8); 5.36-5.08 (m, 2H, CH2); 3.87 (d, 3H, CH3, J = 9.7 Hz); 2.77 (d, 3H, CH3, J = 14.7 Hz); 1.42-1.35 (m, 3H, CH3); 13C-NMR (CDCl3, 101 MHz, ppm): 197.3; 163.7; 156.2; 136.7; 130.9; 130.9; 128.5; 128.5; 128.2; 128.0 127.7; 127.7; 113.9; 113.9; 67.4; 55.4; 54.6; 29.2; 13.4; HR-MS (ESI): for C19H21NO4 [M+H]+ calculated: 328.1549 m/z, found: 328.1537 m/z.
Benzyl methyl(1-oxo-1-(p-tolyl)propan-2-yl)carbamate (30). Yield 84%; m.p.: 62–63 °C; 1H-NMR (d6-aceton, 500 MHz, ppm): 7.89-7.74 (m, 2H, aromat); 7.42-7.21 (m, 7H, aromat); 5.60 (dq, 1H, CH, J1 = 44.4, J2 = 6.9); 5.24-5.04 (m, 2H, CH2); 2.86–2.76 (m, 3H, CH3); 2.07 (tt, 3H, CH3, J1 = 4.4, J2 = 2.2); 1.43-1.33 (m, 3H, CH3); 13C-NMR (d6-aceton, 126 MHz, ppm): 198.1; 155.7; 143.8; 137.3; 133.2; 129.2; 129.2; 128.4; 128.4; 128.3; 128.3; 127.8; 127.5; 127.5; 66.8; 55.6; 30.0; 20.7; 12.8; HR-MS (ESI): for C19H21NO3Na [M+Na]+ calculated: 334.1419 m/z, found: 334.1419 m/z.
Benzyl (1-(4-chlroophenyl)-1-oxopropan-2-yl)(methyl)carbamate (31). Yield 90%; m.p.: 72–73 °C; 1H-NMR (d6-aceton, 500 MHz, ppm): 7.90 (dd, 2H, aromat, J1 = 50.6 Hz, J2 = 8.4 Hz); 7.47 (dd, 2H, aromat, J1 = 38.9 Hz, J2 = 48.4 Hz); 7.40–7.27 (m, 5H, aromat); 5.53 (dq, 1H, CH, J1 = 26.0, J2 = 6.7); 5.22–5.02 (m, 2H, CH2); 2.87–2.76 (m, 3H, CH3); 1.39 (t, 3H, CH3, J = 8.3 Hz); 13C-NMR (d6-aceton, 126 MHz, ppm): 197.6; 155.7; 138.5; 137.2; 134.6; 129.9; 129.9; 128.7; 128.7; 128.4 128.4; 127.9; 127.9; 127.5; 66.8; 56.3; 29.7; 12.5; HR MS (ESI): for C18H18 ClNO3Na [M+Na]+ calculated: 354.0873 m/z, found: 354.0864 m/z.
Benzyl (1-(4-bromophenyl)-1-oxopropan-2-yl)(methyl)carbamate (32). Yield 92%; m.p.: 63–64 °C; 1H-NMR (d6-aceton, 500 MHz, ppm): 7.90–7.74 (m, 2H, aromat); 7.63 (dd, 2H, aromat, J1 = 38.6 Hz, J2 = 8.3 Hz); 7.40–7.27 (m, 5H, aromat); 5.57–5.45 (m, 1H, CH); 5.22–5.02 (m, 2H, CH2); 2.82 (dd, 3H, CH3, J1 = 12.5 Hz, J2 = 4.2 Hz); 1.39 (t, 3H, CH3, J = 8.2 Hz); 13C-NMR (d6-aceton, 126 MHz, ppm): 197.8; 155.7; 137.2; 134.9; 131.7; 131.7; 129.9; 129.9; 128.4; 128.4; 127.8; 127.5; 127.5; 127.2; 66.8; 56.3; 29.8; 12.5; HR-MS (ESI): for C18H18BrNO3 Na [M+Na]+ calculated: 398.0368 m/z, found: 398.0367 m/z.
Benzyl (1-(3,4-dimethylphenyl)-1-oxopropan-2-yl)(methyl)carbamate (33). Yield 79%, 1H-NMR (d6-aceton, 500 MHz, ppm): 7.78–7.59 (m, 2H, aromat); 7.41–7.28 (m, 5H, aromat); 7.21 (dd, 1H, aromat, J1 = 30.0 Hz, J2 = 7.8 Hz); 5.61 (dq, 1H, CH, J1 = 45.9, J2 = 6.8); 5.15 (ddd, 2H, CH2, J1 = 31.2 Hz, J2 = 15.7 Hz, J3 = 8.9 Hz) 2.88-2.74 (m, 3H, CH3); 2.29 (dd, 3H, CH3, J1 = 24.4 Hz, J2 = 15.7 Hz); 2.07 (ddt, 3H, CH3, J1 = 6.6 Hz, J2 = 4.4 Hz, J3 = 2.2 Hz) 1.41-1.33 (m, 3H, CH3); 13C-NMR (d6-aceton, 126 MHz, ppm): 198.2; 155.7; 142.5; 137.3; 136.9; 133.6; 129.7; 129.7; 129.3; 128.4; 128.4; 127.8; 127.5; 125.9; 66.7; 55.5; 30.0; 19.1; 18.9; 12.9; HR-MS (ESI): for C20H23NO3Na [M+Na]+ calculated: 348.1576 m/z, found: 348.1575 m/z.
Benzyl (1-(benzo[d][1,3]dioxol-5-yl)-1-oxopropan-2-yl)(methyl)carbamate (34). Yield 79%; m.p.: 60–61.5 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 7.67 (dd, 1H, aromat, J1 = 8.2 Hz, J2 = 1.4 Hz); 7.51-7.26 (m, 6H, aromat); 6.85-6.64 (m, 1H, aromat); 6.06 (s, 2H, CH2); 5.58 (dq, 1H, CH, J1 = 130.5, J2 = 6.8); 5.33-5.12 (m, 2H, CH2) 2.78 (d, 3H, CH3, J = 20.4 Hz); 1.44-1.33 (m, 3H, CH3); 13C-NMR (CDCl3, 126 MHz, ppm): 197.0; 156.2; 152.0; 148.2; 136.6; 130.0; 128.5; 128.5; 128.0; 127.7; 127.7; 125.0; 108.3; 108.1; 101.8; 67.5; 54.6; 29.2; 13.6; HR MS (ESI): for C19H19NO5 [M+H]+ calculated: 342.1342 m/z, found: 342.1354 m/z.
Methyl methyl(1-(naphthalen-2-yl)-1-oxopropan-2-yl)carbamate (35). Yield 69%; 1H-NMR (CDCl3, 400 MHz, ppm): 8.41 (dd, 1H, aromat, J1 = 29.5, Hz, J2 = 8.3); 8.00 (d, 2H, aromat, J = 8.3 Hz); 7.93–7.87 (m, 1H, aromat); 7.63–7.46 (m, 3H, aromat); 5.80-5.29 (m, 1H, CH), 3.68 (d, 3H, CH3, J = 13.5); 2.87 (d, 3H, CH3, J = 42.2 Hz); 1.53-1.42 (m, 3H, CH3); 13C-NMR (CDCl3, 101 MHz, ppm): 203.7; 157.1; 134.2 134.0; 132.5; 130.5; 128.5; 127.8; 127.2; 126.4; 125.3; 124.5; 58.0; 52.9; 30.0; 13.6; HR-MS (ESI): for C16H17NO3Na [M+Na]+ calculated: 294.1106 m/z, found: 294.1107 m/z.
Phenyl (1naphthalen-1-yl)-1-oxopropan-2-yl)(methyl)carbamate (36). Yield 68%; 1H-NMR (CDCl3, 400 MHz, ppm): 8.38 (dd, 1H, aromat, J1 = 56.8, Hz, J2 = 8.6); 7.94 (ddd, 3H, aromat, J1 = 28.4 Hz, J2 = 14.9 Hz, J3 = 7.9 Hz); 7.65-7.44 (m, 3H, aromat); 7.41-7.22 (m, 5H, aromat); 5.63 (gq, 1H, CH; J1 = 103.9, Hz, J2 = 7.0); 2.95–2.82 (m, 3H, CH3); 1.50–1.45 (m, 3H, CH3); 13C-NMR (CDCl3, 101 MHz, ppm): 203.5; 156.4; 136.6; 134.2; 134.0; 132.5; 130.6; 128.5; 128.5; 128.5; 128.5; 127.9; 127.8; 127.6; 127.2; 126.4; 125.3; 124.5; 58.2; 30.2; 13.6; HR-MS (ESI): for C21H19NO3 [M+H]+ calculated: 334.1443 m/z, found: 334.1450 m/z.
Phenyl (1-(4-chloronaphthalen-1-yl)-1-oxopropan-2-yl)(methyl)carbamate (37). Yield 71%; m.p.: 85-87 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 8.50-8.35 (m, 2H, aromat); 7.87 (dd, 1H, aromat, J1 = 73.5 Hz, J2 = 7.8 Hz); 7.72–7.58 (m, 3H, aromat); 7.39–7.18 (m, 3H, aromat); 7.06–6.92 (m, 2H, aromat) 5.66 (dq, 1H, CH, J1 = 63.0 Hz, J2 = 7.0 Hz); 3.03 (d, 3H, CH3, J = 15.8); 1.54 (d, 3H, CH3, J = 7.1 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 202.8; 155.2; 151.3; 136.7; 133.4; 131.7; 131.3; 129.3; 129.3; 128.6; 127.6; 127.0; 125.7; 125.5; 125.0; 124.8; 121.6; 121.6; 58.6; 30.9; 13.6; HR-MS (ESI): for C21H18ClNO3 [M+H]+ calculated: 368.1053 m/z, found: 368.1067 m/z.
Phenyl (1-(4-bromonaphthalen-1-yl)-1-oxopropan-2-yl)(methyl)carbamate (38). Yield 72%; m.p.: 103–104 °C; 1H-NMR (CDCl3, 500 MHz, ppm): 8.46–8.30 (m, 2H, aromat); 7.88-7.83 (m, 1H, aromat); 7.72–7.56 (m, 3H, aromat); 7.39–7.19 (m, 3H, aromat); 7.06–6.91 (m, 2H, aromat); 5.64 (dq, 1H, CH, J1 = 61.2 Hz, J2 = 7.1 Hz); 3.03 (d, 3H, CH3, J = 16.6); 1.54 (d, 3H, CH3, J = 7.1 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 203.0; 155.2; 151.2; 134.3; 132.5; 131.6; 129.3; 129.3; 128.8; 128.6; 127.9; 127.8; 127.1; 126.2; 125.7; 125.5; 121.6; 121.6; 58.7; 30.9; 13.6; HR MS (ESI): for C21H18BrNO3Na [M+Na]+ calculated: 434.0368 m/z, found: 434.0352 m/z.
Benzyl methyl(1-(naphthalen-1-yl)-1-oxopropan-2-yl)carbamate (39). Yield 68%; 1H-NMR (CDCl3, 500 MHz, ppm): 8.48–8.27 (m, 1H, aromat); 7,.(ddd, 3H, aromat; J1 = 35.0 Hz, J2 = 16.7 Hz, J3 = 9.3); 7.65–7.45 (m, 3H, aromat); 7.41–7.22 (m, 5H, aromat); 5.63 (dq, 1H, CH, J1 = 130.5 Hz, J2 = 7.0 Hz); 5.24–5.02 (m, 2H, CH2); 2.89 (d, 3H, CH3, J = 33.7 Hz); 1.48 (dd, 3H, CH3, J1 = 7.0 Hz, J2 = 3.9 Hz); 13C-NMR (CDCl3, 126 MHz, ppm): 203.6; 156.4; 136.6; 134.2; 134.0; 132.6; 130.5; 128.5; 128.5; 127.9; 127.8; 127.8; 127.6; 127.6; 127.2; 126.4; 125.3; 124.5; 67.4; 58.2; 30.2; 13.6; HR-MS (ESI): for C22H21NO3 [M+H]+ calculated: 348.1600 m/z, found: 348.1608 m/z.
Benzyl (1-(4-chloronaphthalen-1-yl)-1-oxopropan-2-yl)(methyl)carbamate (40). Yield 72%; m.p.: 65–66 °C; 1H-NMR (CDCl3, 400 MHz, ppm): 8.50–8.28 (m, 2H, aromat); 7.87 (d, 1H, aromat; J = 7.8 Hz,); 7.70–7.46 (m, 3H, aromat); 7.43-7.19 (m, 5H, aromat); 5.55 (dq, 1H, CH, J1 = 91.4 Hz, J2 = 7.0 Hz); 5.26-4.98 (m, 2H, CH2); 2.88 (d, 3H, CH3, J = 30.1 Hz); 1.48 (t, 3H, CH3, J = 5.8 Hz); 13C-NMR (CDCl3, 101 MHz, ppm): 202.9; 156.3; 136.5; 136.4; 133.3; 131.7; 131.3; 128.7; 128.5; 128.5; 128.5; 128.5; 128.0; 127.6; 126.9; 126.2; 125.8; 124.9; 67.4; 58.3; 30.3; 13.4; HR-MS (ESI): for C22H20ClNO3Na [M+Na]+ calculated: 404.1029 m/z, found: 404.1014 m/z.
Benzyl (1-(4-bromonaphthalen-1-yl)-1-oxopropan-2-yl)(methyl)carbamate (41). Yield 73%; m.p.: 59–60 °C; 1H-NMR (CDCl3, 400 MHz, ppm): 8.47–8.22 (m, 2H, aromat); 7.78 (d, 1H, aromat; J = 7.9 Hz,); 7.70–7.51 (m, 3H, aromat); 7.44–7.16 (m, 5H, aromat); 5.54 (dq, 1H, CH, J1 = 89.1 Hz, J2 = 7,0 Hz); 5.26-4.96 (m, 2H, CH2); 2.88 (d, 3H, CH3, J = 29.8 Hz); 1.48 (t, 3H, CH3, J = 6.1 Hz); 13C-NMR (CDCl3, 101 MHz, ppm): 203.0; 156.3; 136.5; 134.1; 132.5; 131.6; 128.7; 128.7; 128.5; 128.5; 128.5; 128.5; 128.0; 127.8; 127.7; 127.6; 127.0; 125.8; 67.4; 58.4; 30.4; 13.4; HR-MS (ESI): for C22H20BrNO3 Na [M+Na]+ calculated: 448.0524 m/z, found: 448.0512 m/z.
3.3. Evaluating the In Vitro AChE- and BChE-Inhibition Potency
The ability of all the prepared compounds to inhibit AChE from electric eel (Electrophorus electricus) and BChE from equine serum (both purchased from Sigma, St. Louis, MO, USA) was determined in vitro using a modified Ellman’s method. The effectiveness of the inhibitors, which are expressed as the IC50 values, represent the concentration of an inhibitor that was required for reduction of enzyme activity (or reaction rate) to 50% (sometimes referred to as the negative logarithm of the molar concentration required to inhibit the enzyme activity by 50%, pIC50 = log 1/IC50). The Ellman’s method is widely used for measuring cholinesterase activity and the effectivity of ChEIs [49]. It is a simple, quick and direct method to determine the content of the SH and –S–S– groups in proteins [50]. Cholinesterase activity is measured indirectly by quantifying the concentration of the 5-thio-2-nitrobenzoic acid (TNB) ion that is formed in the reaction between disulfide reagent 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) and thiocholine, which is a product of substrate (i.e., acetylthiocholine, ATCh) hydrolysis that is catalyzed by cholinesterase [51].
All of the examined compounds were dissolved in DMSO (concentration 0.01 M) and diluted in demineralized water (concentrations 0.001 M and 0.0001 M). The ability of the tested compounds to inhibit AChE (from electric eel) and BChE (from equine serum) was determined using a modified Ellman’s method at 25 °C in the presence of phosphate buffered saline (PBS, 0.1 M, pH 7.4) in a glass cuvette with a 1 cm optical path. The enzyme activity in the total reaction mixture (2 mL) was 0.2 U/mL, the concentration of the substrate ATCh 40 μM and the concentration of DTNB 0.1 mM for all of the reactions. The IC50 values were obtained from the dependence of v0/vi on the concentration of the tested compound (inhibitor), where v0 is the reaction rate of an uninhibited reaction and vi is the reaction rate of an inhibited reaction (for a given concentration of the inhibitor). First, v0 was determined. PBS (0.1 M, pH 7.4), DTNB and ATCh were put into the cuvette. The enzymatic reaction was started by adding the enzyme. The dependence of absorbance (λ = 412 nm) on time was observed for 70 s (the reference solution contained PBS, DTNB and ATCh), and then the reaction rate (v0) was calculated (v = ΔA/Δt). The measurement was performed at least in triplicate, and average v0 was determined. Then, vi (for a given concentration of the inhibitor) was determined. DTNB, ATCh and the selected volume of a suitably diluted inhibitor (to achieve the required concentration of the inhibitor in the total reaction mixture) and a certain volume of PBS (to achieve the total volume of the reaction mixture 2 mL after adding the enzyme) were put into the cuvette. The enzymatic reaction was started by adding the enzyme. The dependence of absorbance (λ = 412 nm) on time was observed for 70 s (the reference solution was the same as for uninhibited reaction), and then the reaction rate (vi) was calculated. To determine the IC50 values, twelve different concentrations of the inhibitor were used and each measurement was performed at least in triplicate. Finally, the dependence of v0/vi on the concentration of the inhibitor was determined, and the IC50 values were calculated from the obtained equation of the regression curve for y = 2 (resulting from the definition of the IC50 value) [52]. The obtained results are presented in Table 1.
3.4. Comparative Molecular Surface Analysis using Iterative PLS-Based Variable Elimination
Self-organizing neural mapping (SOM) is regarded as being a nonlinear projection tool that decreases the dimensionality of the input object, e.g., converts 3D objects to 2D, while preserving the topological relationships between the input and output data. Moreover, a trained network can be used to project the specified molecular property (expressed as a vector) by generating a 2D color-coded clustering pattern that is called a feature map. Hence, the SOM algorithm was used to generate an electrostatic potential map in the form of a 2D topographic pattern produced from input signals (points) that were sampled randomly at the molecular surface [53]. In this case, specifying the closest neighbor and then projecting the signals into this particular neuron is based on a comparison of each 3D input vector that consists of x, y and z coordinates with a three-element weight vector that describes each neuron. The shape of a specific molecular surface (template) that is encoded in the weights of the trained Kohonen network can be used to process the signals coming from the surface of the other molecule(s) (counter-template), thereby producing a series of comparative SOM maps can be used to compare/contrast the superimposed molecular geometry.
Applying SOMs to compress/visualize/classify the structural data has been widely reported, e.g., for the 2D mapping of the electrostatic potential on 3D molecular surfaces or partial atomic charges for atomic molecular representation [54].
The set of CoMSA shape/electronic descriptors is subsequently processed using the PLS method and expresses the relationship between variable y and the set of predictors X in a form that is represented by the following equation:
| y = X × b + e | (1) |
where b is the vector of the regression coefficients and e is the vector of the errors. Generally, PLS models are constructed for centered/autoscaled data, and their complexity is estimated using, e.g., the leave-one-out cross-validation (LOO-CV) procedure. The cross-validated is calculated as:
| (2) |
where obs is the observed value; pred is the predicted value; mean is the mean value of obs; and i refers to the object index, which ranges from 1 to m.
The quality of the external predictions was measured using the standard deviation of error of the prediction (SDEP), and the mean absolute error (MAE) parameters, which are defined, respectively, as:
| (3) |
| (4) |
| (5) |
where n is the number of objects in a test set.
Redundant variables can impede the interpretation of a model by increasing its complexity; therefore, reducing the number of variables is advisable. The iterative variable elimination (IVE-PLS) procedure, which is a modification of the UVE-PLS algorithm (developed by Centner et al.) has been proposed, to analyze the stability of the regression coefficients, which are expressed as the mean(b)/s(b) ratio, where s(b) represents the standard deviation of the regression coefficient b [55]. Generally, the entire procedure is composed of the following steps: (i) a standard PLS analysis with LOO-CV to assess the performance of the PLS model (); (ii) elimination of the matrix column with the lowest stability value; (iii) a standard PLS analysis of the new matrix without the column that was eliminated in Step (ii); and (iv) the recurrent repetition of Steps (i)–(iii) to maximize the LOO parameter.
A molecule might be encoded by an ensemble of structural (S) and physicochemical (P) properties that are organized in a vector, which represents an object in the chemical space (CS). The distribution of the empirically (FCS) and virtually (VCS) produced compounds can be visually scrutinized using, e.g., a linear projection procedure called Principal Component Analysis (PCA). PCA is regarded as being a classical method to explore data that permits the data dimensionality to be reduced, visualized and the relationships between the objects (molecules) and parameters (descriptors) to be interpreted. On the condition that the reduction of the data dimensionality is efficient, it is possible to capture interesting information about the data structure (uncover groups of objects and atypical objects) to indicate the importance of the original data variables that contribute to the observed structure, and finally, to illustrate and interpret the relationships between the objects and the parameters in the X matrix (using the first few principal components PCs) [56,57].
3.5. Building the Model and Molecular Modeling
The same laboratory was employed to specify all of the pharmacological data to eliminate any potential data noise that might have been introduced by pooling the datasets from various sources. The in vitro AChE and BChE inhibition values (IC50) for the set of carbamate derivatives are listed in Table 1. The CACTVS/csed molecular editor was used to build the set of the respective compound models. The spatial geometry of the molecules was specified using a CORINA 3D generator. The (inter)change file format converter OpenBabel was used to convert the chemical data.
The majority of the modeling studies were performed with a Sybyl-X 2.0/Certara software package running on a HP workstation with a Debian 6.0 operating system. The standard Tripos force field (POWELL conjugate gradient algorithm) with a 0.01 kcal/mol energy gradient convergence criterion and a distant dependent dielectric constant was used to optimize the initial geometry of each compound (MAXMIN2 module). For the electrostatic potential calculations, the Gasteiger–Hückel method (implemented in Sybyl-X) was initially used to produce the partial atomic charges. One 13-ordered atom trial alignment on 32 was selected to cover the entire bonding topology in the maximal common structure (MCS) using the atom FIT method, which is based on matching the positions of the atoms between the corresponding atom pairs.
The SONNIA software was used in the CoMSA analysis to simulate 20 × 20 to 50 × 50 SOMs with a winning distance that varied within a range of 0.2–2.0. The Cartesian coordinates of the molecular surfaces for the superimposed molecules were produced by a SOM network to form a 2D map of the electrostatic potential. Pretty active against AChE and BChE 32 was selected to form the template molecules. The output maps were subsequently transformed into a 400–2500-element vector, which was used by the PLS method implemented in the MATLAB programming environment.
The crystallographic structure of BChE, which contained one amino acid chain and rivastigmine analog molecule, was retrieved from the PDB repository (code 6eul). Only the 1,2-ethanediol molecule was retained, because it was specified as being valid in the enzyme active site AC2. The rest of the heteroatoms, including the crystallographic waters, were extracted prior to the calculations. The ligand/protein structures for the docking study were prepared in the pdbqt file format with the Gasteiger charges calculated. During the AutoDock simulation, various poses (default nine) were generated progressively from a single conformer (an energy-optimized molecule) by applying a collection of the preferred torsion angles to the rotatable bonds and were evaluated using the united-atom scoring function. All of the predicted binding 2D/3D modes, including the positions of the flexible side chains, were visualized using PyMol, Maestro and VMD molecular graphics viewers and the Protein-Ligand Interaction Profiler (PLIP).
4. Conclusions
To summarize, a series of novel benzene-based derivatives was designed, synthesized and characterized using 1H NMR spectroscopy and HRMS. All 41 of the tested compounds were evaluated for their in vitro ability to potentially inhibit AChE and BChE, respectively. The selectivity index of the individual molecules to cholinesterases was also specified. Roughly speaking, a rather moderate inhibitory effect against AChE was revealed; however, some of the compounds (11, 13, 14, 16, 23–28, 31, and 33) proved to be very selective for the BChE enzyme. In fact, two compounds (23 and 28) had a very high selectivity index (SI = 13.93 and 15.31). Specifically, compound 28 had the lowest IC50 value revealing an approximately seven-fold higher inhibitory activity against BChE than RIV (IC50 = 5.51 vs. 38.40 μM), which corresponds quite well with GLT (IC50 = 2.77 μM). It was observed that the methyl-substituted carbamates generally exhibited a lower inhibitor ability compared to their phenyl or benzyl counterparts. Interestingly, compounds 17 and 29, which had a methoxy group in the same position, were weaker inhibitors—the presence of the hydrophilic electron-donating -OCH3 substituent of the phenyl ring at the para-position decreased the potency of the compounds, especially against the BChE enzyme. On the other hand, the presence of methyl group(s) in the meta/para-position(s) of the phenyl ring (compounds 15, 18, 21, 27, 30, and 33) resulted in an improvement of the IC50 value for the BChE enzyme, thus suggesting the significance of a hydrophobic character for the interactions with the enzyme. Placing the electron-withdrawing chlorine substituent in the meta-position of the phenyl ring appeared to be strongly preferable, especially for the BChE inhibition activity (compounds 16 or 28). To partially explain the observed variations in the anti-AChE/BChE potential of the investigated carbamate series, comparative receptor-independent (RI) and receptor-dependent (RD) structure–activity studies were conducted, respectively. The principal purpose of the ligand-based study was to comparatively analyze the molecular surface (CoMSA) to gain insight into the electronic and/or steric factors that govern the ability of the tested compounds to inhibit the AChE/BChE activities. The findings for the surface descriptors were compared with their force field counterparts (CoMFA) when modeling the inhibiting potency for multiple training/test subsets using the stochastic model validation (SMV) procedure. Moreover, a similarity analysis was performed using the PCA approach on the pool of Dragon descriptors. The spatial distribution of the potentially important steric and electrostatic factors on the BChE inhibitory potency was specified using the probability-guided pharmacophore mapping procedure based on the iterative variable elimination (IVE-PLS) method. Finally, a comprehensive scrutiny of the guest–target interactions for the inhibitors that were comparatively active to rivastigmine BChE (IC50 < 30 μM) was conducted using the site-directed computer-assisted docking methodology. Planar (2D) and spatial (3D) maps of the host–target interactions were created for all of the active compounds and compared with the marketed drug (GLT and RIV) molecules, which generally revealed two types of non-binding interactions—hydrophobic and hydrogen bond formation, respectively. The hydroxyl substituent of Thr120 appeared to be crucial in forming the hydrogen bond with the ether or carbonyl oxygen (Figure 9c). For the examined BChE inhibitors, the regions in close proximity to the nitrogen atom (R1 substituent) seemed to be valid for hydrophobic interactions with the Asp70 amino acid residue, which is in line with ligand-based findings. Regrettably, a clear explanation of the variations that the meta/para-positioned carbamate derivatives exerted on the BChE reaction site was not revealed; however, some regularities were observed in the ligand–receptor interaction pattern. It appeared that the close proximity of the positively charged nitrogen atom of His438 may potentially be beneficial to the inhibition potential, especially the negatively charged chlorine- or bromine-based carbamates, which corresponds relatively well with the IVE-PLS CoMSA results. The electrostatic repulsion between the negatively charged atoms and the oxygen of Ser79 can partially explain the detrimental impact of the –OCH3 or –OCH2O- groups that were attached to the phenyl ring. On the other hand, the postulated hydrophobic interactions with Phe329 can favorably contribute to the inhibitory potential, as was observed for (di)methyl derivatives.
The combination of consensus pharmacophore mapping with a systematic screening of the multifaceted guest–host interactions using target-tailored approaches seems to be the path towards an intelligent drug design system.
Figure 7.
Structure of the BChE enzyme (displayed as ribbons) bound with GLT (depicted as balls and sticks), RIV (presented as thin sticks) and the most active molecule 28 (illustrated as thick sticks).
Acknowledgments
We would like to acknowledge the Schrödinger, OpenBabel and OpenEye for the free academic software licenses. The authors wish to acknowledge the financial support to the Faculty of Chemical Technology, University of Pardubice as well as to the project of Grant Agency of Czech Republic—Project No.: GA 18-03847S.
Abbreviations
| AChE | Acetylcholinesterase |
| BChE | Butyrylcholinesterase |
| CAMD | Computer Asisted Molecular Design |
| ADMET | Absorption Distribution Metabolism Excretion Toxicity |
| CoMSA | Comparative Molecular Surface Analysis |
| SMV | Stochastic Model Validation |
| RIV | Rivastigmine |
| GLT | Galanthamine |
| PCA | Principal Component Analysis |
| IVE-PLS | Iterative Variable Elimination Partial Least Squares |
| PLIP | Protein Ligand Interaction Profiler |
Author Contributions
A.B., V.K.; CoMSA, CoMFA, and SMV: docking analysis, and writing, editing, reviewing of the paper. D.J.S.: HRMS analyses. D.K., K.G., and J.P.: design, synthesis and compound characterisations. A.S. (Aleksandra Swietlicka); descriptor calculations. and editing of the paper. A.S. (Adam Smolinski), J.J.: editing and writing of the paper. S.S. and A.I.: AChE/BChE evaluation.
Funding
This study was supported by the Slovak Research and Development Agency (projects APVV-17-0373 and APVV-14-0547), and by the Ministry of Education of the Czech Republic (LO1305).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lemke T.L., Williams D.A. Foye’s Principles of Medicinal Chemistry. 7th ed. Lippincott Williams & Wilkins and Wolters Kluwer; Baltimore, MD, USA: 2013. [Google Scholar]
- 2.Ghosh A.K., Brindisi M. Organic carbamates in drug design and medicinal chemistry. J. Med. Chem. 2015;58:2895–2940. doi: 10.1021/jm501371s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jampilek J., Brychtova K. Azone analogues: Classification, design, and transdermal penetration principles. Med. Res. Rev. 2012;32:907–947. doi: 10.1002/med.20227. [DOI] [PubMed] [Google Scholar]
- 4.Vettorazzi M., Angelina E., Lima S., Gonec T., Otevrel J., Marvanova P., Padrtova T., Mokry P., Bobal P., Acosta L.M., et al. Search of new structural scaffolds for sphingosine kinase 1 inhibitors. Eur. J. Med. Chem. 2017;139:461–481. doi: 10.1016/j.ejmech.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imramovsky A., Pesko M., Monreal-Ferriz J., Kralova K., Vinsova J., Jampilek J. Photosynthesis-inhibiting efficiency of 4-chloro-2-(chlorophenylcarbamoyl)phenyl alkylcarbamates. Bioorg. Med. Chem. Lett. 2011;21:4564–4567. doi: 10.1016/j.bmcl.2011.05.118. [DOI] [PubMed] [Google Scholar]
- 6.Zadrazilova I., Pospisilova S., Masarikova M., Imramovsky A., Monreal-Ferriz J., Vinsova J., Cizek A., Jampilek J. Salicylanilide Carbamates: Promising antibacterial agents with high in vitro activity against methicillin-resistant Staphylococcus aureus (MRSA) Eur. J. Pharm. Sci. 2015;77:197–207. doi: 10.1016/j.ejps.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Kos J., Nevin E., Soral M., Kushkevych I., Gonec T., Bobal P., Kollar P., Coffey A., O’Mahony J., Liptaj T., et al. Synthesis and antimycobacterial properties of ring-substituted 6-hydroxynaphthalene-2-carboxanilides. Bioorg. Med. Chem. 2015;23:2035–2043. doi: 10.1016/j.bmc.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Kauerova T., Kos J., Gonec T., Jampilek J., Kollar P. Antiproliferative and pro-apoptotic effect of novel nitro-substituted hydroxynaphthanilides on human cancer cell lines. Int. J. Mol. Sci. 2016;17:1219. doi: 10.3390/ijms17081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pospisilova S., Kos J., Michnova H., Kapustikova I., Strharsky T., Oravec M., Moricz A.M., Bakonyi J., Kauerova T., Kollar P., et al. Synthesis and spectrum of biological activities of novel N-arylcinnamamides. Int. J. Mol. Sci. 2018;19:2318. doi: 10.3390/ijms19082318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pizova H., Havelkova M., Stepankova S., Bak A., Kauerova T., Kozik V., Oravec M., Imramovsky A., Kollar P., Bobal P., et al. Proline-based carbamates as cholinesterase inhibitors. Molecules. 2017;14:1969. doi: 10.3390/molecules22111969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss D.E., Perez R.G., Kobayashi H. Cholinesterase inhibitor therapy in Alzheimer’s disease: The limits and tolerability of irreversible CNS-selective acetylcholinesterase inhibition in primates. J. Alzheimers Dis. 2017;55:1285–1294. doi: 10.3233/JAD-160733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajic V., Milovanovic E.S., Spremo-Potparevic B., Zivkovic L., Miliccivc Z., Stanimirovic J., Bogdanovic N., Isenovic E.R. Treatment of Alzheimer’s Disease: Classical therapeutic approach. Curr. Pharm. Anal. 2016;12:82–90. doi: 10.2174/1573412911666150611184740. [DOI] [Google Scholar]
- 13.Skrzypek A., Matysiak J., Niewiadomy A., Bajda M., Szymański P. Synthesis and biological evaluation of 1,3,4-thiadiazole analogues as novel AChE and BuChE inhibitors. Eur. J. Med. Chem. 2013;62:311–319. doi: 10.1016/j.ejmech.2012.12.060. [DOI] [PubMed] [Google Scholar]
- 14.Kumar J., Meena P., Singh A., Jameel E., Maqbool M., Mobashir M., Shandilya A., Tiwari M., Hoda N., Jayaram B. Synthesis and screening of triazolopyrimidine scaffold as multi-functional agents for Alzheimer’s disease therapies. Eur. J. Med. Chem. 2016;119:260–277. doi: 10.1016/j.ejmech.2016.04.053. [DOI] [PubMed] [Google Scholar]
- 15.Mohamed T., Rao P.P. 2,4-Disubstituted quinazolines as amyloid-β aggregation inhibitors with dual cholinesterase inhibition and antioxidant properties: Development and structure-activity relationship (SAR) studies. Eur. J. Med. Chem. 2017;126:823–843. doi: 10.1016/j.ejmech.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Knez D., Brus B., Coquelle N., Sosic I., Sink R., Brazzolotto X., Mravljak J., Colletier J.P., Gobec S. Structure-based development of nitroxoline derivatives as potential multifunctional anti-Alzheimer agents. Bioorg. Med. Chem. 2015;23:4442–4452. doi: 10.1016/j.bmc.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez Y.A., Gutierrez M., Ramirez D., Alzate-Morales J., Bernal C.C., Guiza F.M., Romero Bohorquez A.R. Novel N-allyl/propargyl tetrahydroquinolines: Synthesis via three-component cationic imino Diels-Alder reaction, binding prediction, and evaluation as cholinesterase inhibitors. Chem. Biol. Drug Des. 2016;88:498–510. doi: 10.1111/cbdd.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozakiewicz D., Polanski J., Jampilek J., Imramovsky A., Stepankova S. New Carbamate Derivatives and Their Application. 420626. U.S. Patent. 2017 Feb 23;
- 19.Davis B.J., Erlanson D.A. Learning from our mistakes: The ‘unknowns’ in fragment screening. Bioorg. Med. Chem. Lett. 2013;23:2844–2852. doi: 10.1016/j.bmcl.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 20.Kenny P.W. Comment on the ecstasy and agony of assay interference compounds. J. Chem. Inf. Model. 2017;57:2640–2645. doi: 10.1021/acs.jcim.7b00313. [DOI] [PubMed] [Google Scholar]
- 21.Ghose A.K., Herbertz T., Salvino J.M., Mallamo J.P. Knowledge-based chemoinformatic approaches to drug discovery. Drug Discov. Today. 2006;11:1107–1114. doi: 10.1016/j.drudis.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Stanton D.T. QSAR and QSPR model interpretation using partial least squares (PLS) analysis. Curr. Comput. Aided Drug Des. 2012;8:107–127. doi: 10.2174/157340912800492357. [DOI] [PubMed] [Google Scholar]
- 23.Bak A., Kozik V., Smolinski A., Jampilek J. Multidimensional (3D/4D-QSAR) probability-guided pharmacophore mapping: Investigation of activity profile for a series of drug absorption promoters. RSC Adv. 2016;6:76183–76205. doi: 10.1039/C6RA15820J. [DOI] [Google Scholar]
- 24.Van de Waterbeemd H., Gifford E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 25.Zentgraf M., Steuber H., Koch C., La Motta C., Sartini S., Sotriffer C.A., Klebe G. How reliable are current docking approaches for structure-based drug design? Lessons from aldose reductase. Angew. Chem. Int. Ed. Engl. 2007;46:3575–3580. doi: 10.1002/anie.200603625. [DOI] [PubMed] [Google Scholar]
- 26.Mazur P., Magdziarz T., Bak A., Chilmonczyk Z., Kasprzycka-Guttman T., Misiewicz-Krzeminska I., Skupinska K., Polanski J. Does molecular docking reveal alternative chemopreventive mechanism of activation of oxidoreductase by sulforaphane isothiocyanates? J. Mol. Model. 2010;16:1205–1212. doi: 10.1007/s00894-009-0628-5. [DOI] [PubMed] [Google Scholar]
- 27.Fan H., Irwin J.J., Webb B.M., Klebe G., Shoichet B.K., Sali A. Molecular docking screens using comparative models of proteins. J. Chem. Inf. Model. 2009;49:2512–2527. doi: 10.1021/ci9003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soler M.A., de Marco A., Fortunac S. Molecular dynamics simulations and docking enable to explore the biophysical factors controlling the yields of engineered nanobodies. Sci Rep. 2016;6:34869. doi: 10.1038/srep34869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pejchal V., Stepankova S., Padelkova Z., Imramovsky A., Jampilek J. 1,3-Substituted Imidazolidine-2,4,5-triones: Synthesis and inhibition of cholinergic enzymes. Molecules. 2011;16:7565–7582. doi: 10.3390/molecules16097565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imramovsky A., Stepankova S., Vanco J., Pauk K., Monreal-Ferriz J., Vinsova J., Jampilek J. Acetylcholinesterase-inhibiting activity of salicylanilide N-alkylcarbamates and their molecular docking. Molecules. 2012;17:10142–10158. doi: 10.3390/molecules170910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imramovsky A., Pejchal V., Stepankova S., Vorcakova K., Jampilek J., Vanco J., Simunek P., Kralovec K., Bruckova L., Mandikova J., et al. Synthesis and in vitro evaluation of new derivatives of 2-substituted-6-fluorobenzo[d]thiazoles as cholinesterase inhibitors. Bioorg. Med. Chem. 2013;21:1735–1748. doi: 10.1016/j.bmc.2013.01.052. [DOI] [PubMed] [Google Scholar]
- 32.Bak A., Polanski J. Modeling robust QSAR 3: SOM-4D-QSAR with iterative variable elimination IVE-PLS: Application to steroid, azo dye, and benzoic acid series. J. Chem. Inf. Model. 2007;47:1469–1480. doi: 10.1021/ci700025m. [DOI] [PubMed] [Google Scholar]
- 33.Bak A., Kozik V., Walczak M., Fraczyk J., Kaminski Z., Kolesinska B., Smolinski A., Jampilek J. Towards intelligent drug design system: Application of artificial dipeptide receptor library in QSAR-oriented studies. Molecules. 2018;23:1964. doi: 10.3390/molecules23081964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hann M., Oprea T. Pursuing the leadlikeness concept in pharmaceutical research. Curr. Opin. Chem. Biol. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Kubinyi H. Hansch Analysis and Related Approaches. Wiley-VCH Verlag GmbH; Weinheim, Germany: 1993. [Google Scholar]
- 36.Bak A., Kozik V., Malik I., Jampilek J., Smolinski A. Probability-driven 3D pharmacophore mapping of antimycobacterial potential of hybrid molecules combining phenylcarbamoyloxy and N-arylpiperazine fragments. SAR QSAR Environ. Res. 2018;29:801–821. doi: 10.1080/1062936X.2018.1517278. [DOI] [PubMed] [Google Scholar]
- 37.Todeschini R., Consonni V. Molecular Descriptors for Chemoinformatics. Wiley-VCH Verlag GmbH & Co. KgaA; Weinheim, Germany: 2010. [Google Scholar]
- 38.Todeschini R., Consonni V. Handbook of Molecular Descriptors. Wiley-VCH Verlag GmbH; Weinheim, Germany: 2000. [Google Scholar]
- 39.Polanski J., Bak A., Gieleciak R., Magdziarz T. Modeling robust QSAR. J. Chem. Inf. Model. 2003;46:2310–2318. doi: 10.1021/ci050314b. [DOI] [PubMed] [Google Scholar]
- 40.Kolb P., Irwin J.J. Docking screens: Right for the right reasons? Curr. Top. Med. Chem. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colquhoun D. The quantitative analysis of drug–receptor interactions: A short history. Trends Pharmacol. Sci. 2006;27:149–157. doi: 10.1016/j.tips.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Levoin N., Calmels T., Poupardin-Olivier O., Labeeuw O., Danvy D., Robert P., Berrebi-Bertrand I., Ganellin C.R., Schunack W., Stark H., et al. Refined docking as a valuable tool for lead optimization: Application to histamine H3 receptor antagonists. Arch. Pharm. Chem. Life Sci. 2008;341:610–623. doi: 10.1002/ardp.200800042. [DOI] [PubMed] [Google Scholar]
- 43.Devillers J. Methods for building QSARs. Methods Mol. Biol. 2013;930:3–27. doi: 10.1007/978-1-62703-059-5_1. [DOI] [PubMed] [Google Scholar]
- 44.Bak A., Wyszomirski M., Magdziarz T., Smolinski A., Polanski J. Structure-based modeling of dye-fiber affinity with SOM-4D-QSAR paradigm: Application to set of anthraquinone derivatives. Comb. Chem. High Throughput Screen. 2014;17:485–502. doi: 10.2174/1386207317666140205195252. [DOI] [PubMed] [Google Scholar]
- 45.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:443–447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golbraikh A., Tropsha A. Beware of q2! J. Mol. Graph. Mod. 2002;20:269–276. doi: 10.1016/S1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y.C. Beware of docking. Trends Pharmacol. Sci. 2015;36:78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Ellman G.L., Courtney K.D., Andres V., Featherstone R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 50.Ou S., Kwok K.C., Wang Y., Bao H. An improved method to determine SH and –S–S– group content in soymilk protein. Food Chem. 2004;88:317–320. doi: 10.1016/j.foodchem.2004.05.001. [DOI] [Google Scholar]
- 51.Sinko G., Calic M., Bosak A., Kovarik Z. Limitation of the Ellman method: Cholinesterase activity measurement in the presence of oximes. Anal. Biochem. 2007;370:223–227. doi: 10.1016/j.ab.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 52.Zdrazilova P., Stepankova S., Komers K., Ventura K., Cegan A. Half-inhibition concentrations of new cholinesterase inhibitors. Zeitschrift für Naturforschung C. 2004;59:293–296. doi: 10.1515/znc-2004-3-430. [DOI] [PubMed] [Google Scholar]
- 53.Zupan J., Gasteiger J. Neural Networks and Drug Design for Chemists. 2nd ed. Wiley-VCH; Weinheim, Germany: 1999. [Google Scholar]
- 54.Bak A., Kozik V., Smolinski A., Jampilek J. In silico estimation of basic activity-relevant parameters for a set of drug absorption promoters. SAR QSAR Environ. Res. 2017;28:427–449. doi: 10.1080/1062936X.2017.1327459. [DOI] [PubMed] [Google Scholar]
- 55.Centner V., Massart D.L., de Noord O.E., de Jong S., Vandeginste B.M.V., Sterna C. Elimination of uninformative variables for multivariate calibration. Anal. Chem. 1996;68:3851–3858. doi: 10.1021/ac960321m. [DOI] [PubMed] [Google Scholar]
- 56.Smolinski A., Drobek L., Dombek V., Bak A. Modeling of experimental data on trace elements and organic compounds content in industrial waste dumps. Chemosphere. 2016;162:189–198. doi: 10.1016/j.chemosphere.2016.07.086. [DOI] [PubMed] [Google Scholar]
- 57.Likus-Cieslik J., Smolinski A., Pietrzykowski M., Bak A. Sulphur contamination impact on seasonal and surface water chemistry on a reforested area of a former sulphur mine. Land Degrad. Dev. 2019;30:212–225. doi: 10.1002/ldr.3216. [DOI] [Google Scholar]