Abstract
Antibiotic resistance has become increasingly prevalent over the past few decades, and this combined with a dearth in the development of new classes of antibiotics to treat multidrug resistant Gram-negative infections has led to a significant global health problem and the increased usage of colistin as the last resort antibiotic. Colistin, however, presents dose dependent toxicity in the clinic. One potential approach to combatting this problem is the use of an antibiotic adjuvant, a compound that is non-toxic to the bacteria, that enhances the potency of colistin and ultimately allows for reducing dosing. Herein, we present a new urea-containing class of 2-aminoimidazole based adjuvants that potentiates colistin activity against colistin-sensitive Acinetobacter baumannii. Lead compounds enabled 1000-fold reduction in the minimum inhibitory concentration of colistin in vitro, and showed efficacy in a Galleria mellonella infection model, representing the first step towards validating the potential of employing these adjuvants to lower colistin dosage.
Keywords: Antibiotic resistance, Acinetobacter baumannii, antibiotic adjuvant, colistin
Graphical Abstract

The discovery of antibiotics has enabled significant advances in medicine and agricultural practices.1-2 Unfortunately, bacterial resistance has evolved against every antibiotic placed into the clinic. For example, resistance to linezolid and daptomycin, two Gram-positive selective antibiotics, was observed within a few years of deployment.3 The Centers for Disease Control and Prevention (CDC) estimates that approximately 2 million people per year acquire an antibiotic-resistant bacterial infection and from those 2 million, about 23,000 deaths result.4 A significant number of these deaths stem from Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp., dubbed the ESKAPE pathogens as they are known to be capable of “escaping” the effects of antibiotics due to the existence of multidrug resistant (MDR) strains.2 The Gram-negative members of this class are of particular concern given the well documented difficulties associated with the discovery of new antibiotics.5
A. baumannii accounts for 10% of hospital-acquired infections in the United States and has a >50% mortality rate when the infected patient presents with sepsis and/or pneumonia.6 A. baumannii is an evolving, opportunistic pathogen that is found worldwide, especially in hospital settings, predominantly in intensive care units.7 This species is exceptionally problematic for wounded soldiers as it has been observed in injured warfighters returning from conflicts in the Middle East.8 Recently isolated strains have exhibited resistance to most classes of antibiotics.9
Lately, there has been an increase in use of the polymyxin antibiotic colistin due to the rising levels of MDR Gram-negative infections and the lack of new viable Gram-negative antibiotics. Currently, colistin is viewed as the antibiotic of last resort for the treatment of these infections. Colistin became clinically available in 1959, however its use rapidly faded in the 1970s due to renal and neurological toxicity.10 Following its reintroduction for the treatment of MDR Gram-negative infections, it has been shown that over the last 10 years, on average, 30% of patients receiving a full regimen of colistin experienced at least mild nephrotoxicity with some patients’ treatment being stopped due to toxicity.10 Colistin toxicity needs to be addressed if clinical use is to be continued; one approach is the usage of combination therapies with the aim of lowering the required colistin dosage. The use of antibiotic/antibiotic combinations have been investigated, for example pairing tigecycline with colistin resulted in bactericidal and synergistic effects in vitro at most concentrations and time intervals against eight carbapenemase-producing K. pneumoniae strains.11
As an alternative approach to the development of new antibiotics or antibiotic combinations, our group and others have pursued the use of antibiotic adjuvants – compounds that themselves exhibit little or no toxicity against bacteria, but enhance the effects of antibiotics typically by blocking resistance mechanisms.12-13 Within our lab, we have focused on analogs of nitrogen dense, sponge-derived natural products that possess innate properties of inhibiting and dispersing bacterial biofilms.14 Compounds containing a 2-aminoimidazole (2-AI) moiety are one such class of these marine-derived analogs and have been further reported to potentiate the activity of several classes of antibiotics against a broad spectrum of drug-resistant bacteria.15-17 For example, in A. baumannii, resistance to colistin is underpinned by phosphoethanolamine modification of the lipid A anchor of lipopolysaccharides, which is controlled by the PmrAB two-component signaling system. We have demonstrated that certain aryl-derived 2-AI derivatives (Figure 1) downregulate the PmrAB system, reverse lipid A modification and in the process, break colistin resistance in colistin-resistant strains of A. baumannii, as well as K. pneumoniae and P. aeruginosa.13 Recently, we have also demonstrated that these 2-AIs are able to reverse colistin resistance in bacterial strains harboring the recently reported mobile colistin resistance (mcr-1) gene, a plasmid-borne gene that encodes a lipid A phosphoethanolamine transferase.17 The mobile nature of plasmids encoding mcr-1 and related genes provides the potential to rapidly disseminate colistin resistance in the human pathogen pool, rendering colistin therapy ineffective. During these studies, we also noted that these adjuvants also had the ability to enhance colistin efficacy 16- to >2048-fold in colistin-susceptible strains of bacteria, thus raising the possibility that adjuvant pairing could allow lower dosage of colistin in vivo against colistin-sensitive, MDR Gram-negative pathogens and potentially reduce/mitigate colistin’s dose-dependent toxicity. Herein, we detail our efforts to identify an alternative 2-AI scaffold that enhances colistin efficacy in vitro against colistin-sensitive A. baumannii along with an assessment of the activity of lead compounds in a Galleria mellonella model of infection.
Figure 1.

Aryl-derived 2-AI adjuvants that potentiate colistin activity against colistin-resistant and colistin-sensitive MDR bacteria.
Results & Discussion
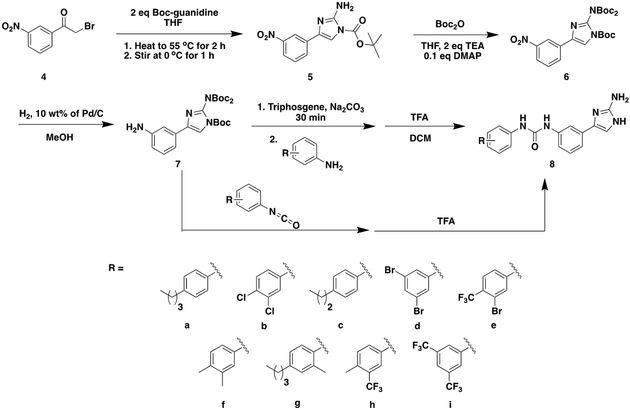
From a screen of our in-house library focusing on enhancement of colistin activity, we discovered that meta-urea derived 2-AIs (Scheme 1) of general structure 8 had modest potentiation activity. To explore this activity, we elected to first synthesize a pilot library that varied the distal aromatic ring (Scheme 1) and study the ability of each analog to enhance colistin efficacy against the colistin-sensitive A. baumannii strains 19606 and 17978 as well as the hypervirulent, MDR A. baumannii strain 5075. Library synthesis commenced by reacting commercially available 2-bromo-3’-nitroacetophenone with Boc-guanidine (Scheme 1). After 2 hours stirring at 55 ºC and an additional hour stirring at 0 ºC, the resulting mono-Boc-protected 2-AI, compound 5, precipitated out of solution allowing for purification by filtration. The resulting white solid was immediately dissolved in tetrahydrofuran and added a solution of Boc anhydride affording intermediate 6 that served as the key building block for accessing each member of the library. To complete the synthesis, the nitro group was reduced to afford compound 7 and then either coupled with a commercially available isocyanate or converted to the isocyanate in situ then subsequently coupled with an aniline derivative to access the desired urea. Trifluoroacetic acid mediated deprotection followed by counter ion exchange delivered 2-AI HCl salts 8a-i for testing.
Scheme 1.
General Synthetic Route for 1st Library of Meta-Urea 2-AIs
Initially, we measured the MICs of 2-AI analogs 8a-i against three strains of colistin-sensitive A. baumannii and observed stand alone MIC values of 5 to >200 μM (Table 1). Next measured was the MIC of colistin in the presence of the 2-AI at 30% of its MIC or 30 μM (whichever concentration was lower). A majority of these compounds showed no activity (zero to two-fold enhancement), with only three compounds (8c, 8e, and 8i) showing a modest four-fold potentiation of colistin activity (lowering the MIC from 1.0 μg/mL to 0.25 μg/mL). As varying the distal aromatic ring appeared unproductive for augmenting activity, we next elected to introduce substitutions on the 2-AI ring. Specifically, we elected to synthesize 1,4-disubstituted derivatives with N-1 bearing an isopropyl substituent that was previously shown in other 2-AI adjuvants to enhance colistin potentiation activity.15
Table 1.
2-AI Potentiation of Colistin Against Three Colistin-Sensitive A. baumannii Strains
| Compd | A. baumannii 5075 | A. baumannii 19606 | A. baumannii 17978 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (μg/mL) |
MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (μg/mL) |
MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (μg/mL) |
|
| Colistin | 1 | 1 | 1 | ||||||
| 8a | >200 | 30 | 1 | >200 | 30 | 0.5 | >200 | 30 | 0.5 |
| 8b | 50 | 15 | 0.5 | 50 | 15 | 0.25 | 25 | 7.5 | 0.25 |
| 8c | 200 | 30 | 0.25 | >200 | 30 | 0.5 | >200 | 30 | 0.5 |
| 8d | 100 | 30 | 0.5 | 50 | 15 | 0.25 | 50 | 15 | 0.5 |
| 8e | 25 | 7.5 | 0.25 | 5 | 2 | 1 | 12.5 | 3.75 | 0.5 |
| 8f | >200 | 30 | 1 | >200 | 30 | 0.5 | 200 | 30 | 0.5 |
| 8g | >200 | 30 | 1 | 100 | 30 | 1 | 100 | 30 | 1 |
| 8h | >200 | 30 | 1 | 200 | 30 | 1 | >200 | 30 | 0.5 |
| 8i | 25 | 7.5 | 0.25 | 5 | 2 | 1 | 12.5 | 3.75 | 0.25 |
This second-generation library was synthesized using the strategy outlined in Scheme 2. Reaction of 2-bromo-3’-nitroacetophenone with isopropylamine followed by cyclization with cyanamide afforded the 1,4-disubstituted 2-AI, compound 9. This intermediate was di-Boc-protected with Boc anhydride and purified to deliver compound 10. All analogs were then synthesized using the same strategy as the mono-substituted 2-AI library to afford HCl salts 12a-k.
Scheme 2.
General Synthetic Route for Meta-Urea Derived 1,4-Disubstituted 2-AIs
The second-generation library (Table 2) exhibited a substantial increase in colistin potentiation activity compared to the first-generation library, and as we found previously for other 1,4-disubstituted 2-AIs, exhibited significantly reduced inherent toxicity when dosed alone, with most compounds having a MIC of ≥200 μM.15 This follows a general trend we have empirically noted in which N-substitution significantly reduces inherent toxicity of these adjuvants, yet increases adjuvant activity. The five most active compounds from this series were 12b, 12d, 12e, 12i, and 12k, which effected a reduction in the colistin MIC between 128 and >1024-fold, with compounds 12b, 12d, and 12e reducing the MIC from 1 μg/mL to ≤0.98 ng/mL against A. baumannii 5075 at an adjuvant concentration of 30 μM. A dose response study with these five compounds was then performed against all three strains at concentrations of 20, 15, and 7.5 μM (Table 3). Most of these select compounds exhibited upwards of 16-fold reductions in the MIC of colistin at only 7.5 μM, with compound 12i exhibiting the highest activity, reducing the colistin MIC from 1 μg/mL to 62.5, 31.3, and 31.3 ng/mL against 5075, 19606, and 17978, respectively.
Table 2.
1,4-Disubstituted 2-AI Meta-Ureas Potentiation of Colistin Against Three A. baumannii Strains
| Compd | A. baumannii 5075 | A. baumannii 19606 | A. baumannii 17978 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (ng/mL) |
MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (ng/mL) |
MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (ng/mL) |
|
| Colistin | 1000 | 1000 | 1000 | ||||||
| 12a | 50 | 15 | ≤ 3.9 | 15 | 7.5 | 62.5 | 50 | 15 | ≤ 15.6 |
| 12b | 100 | 30 | ≤ 0.98 | 50 | 15 | ≤ 3.9 | 100 | 30 | ≤ 0.98 |
| 12c | >200 | 30 | 15.6 | 200 | 30 | 3.9 | 50 | 15 | 125 |
| 12d | 200 | 30 | ≤ 0.98 | >200 | 30 | ≤ 0.98 | >200 | 30 | ≤ 7.8 |
| 12e | >200 | 30 | ≤ 0.98 | >200 | 30 | ≤ 0.98 | >200 | 30 | ≤ 7.8 |
| 12f | >200 | 30 | 500 | >200 | 30 | 125 | 200 | 30 | 250 |
| 12g | >200 | 30 | 500 | >200 | 30 | 500 | >200 | 30 | 500 |
| 12h | 200 | 30 | 1.95 | 200 | 30 | ≤ 1.95 | 100 | 30 | 15.6 |
| 12i | >200 | 30 | 3.9 | 200 | 30 | ≤ 0.98 | >200 | 30 | 7.8 |
| 12j | >200 | 30 | 250 | >200 | 30 | 250 | >200 | 30 | 250 |
| 12k | 200 | 30 | ≤ 3.9 | >200 | 30 | ≤ 0.98 | >200 | 30 | ≤ 15.6 |
Table 3.
Dose Response of Five Active 1,4-Disubstituted 2-AI Meta-Ureas
| Colistin MIC (ng/mL) | ||||
|---|---|---|---|---|
| Compd | Concentration Tested (μM) |
A. baumannii 5075 |
A. baumannii 19606 |
A. baumannii 17978 |
| Colistin | 1000 | 1000 | 1000 | |
| 12b | 30 | ≤ 0.98 | Toxic | ≤ 0.98 |
| 20 | 7.8 | ≤ 0.98 | 3.9 | |
| 15 | 7.8 | ≤ 3.9 | 3.9 | |
| 7.5 | 125 | 250 | 125 | |
| 12d | 30 | ≤ 0.98 | ≤ 0.98 | ≤ 7.8 |
| 20 | 15.6 | ≤ 3.9 | 7.8 | |
| 15 | 15.6 | 15.6 | 15.6 | |
| 7.5 | 62.5 | 125 | 62.5 | |
| 12e | 30 | ≤ 0.98 | ≤ 0.98 | ≤ 7.8 |
| 20 | 15.6 | ≤ 3.9 | 7.8 | |
| 15 | 31.3 | 15.6 | 31.3 | |
| 7.5 | 62.5 | 125 | 125 | |
| 12i | 30 | 3.9 | ≤ 0.98 | 7.8 |
| 20 | 15.6 | 2.0 | 7.8 | |
| 15 | 15.6 | 7.8 | 15.6 | |
| 7.5 | 62.5 | 31.3 | 31.3 | |
| 12k | 30 | ≤ 3.9 | ≤ 0.98 | ≤ 15.6 |
| 20 | 15.6 | ≤ 3.9 | 15.6 | |
| 15 | 31.3 | 15.6 | 31.3 | |
| 7.5 | 15.6 | 62.5 | 62.5 | |
Next, we selected two of the five lead compounds, 12d and 12i, for additional colistin potentiation screening. First, they were tested at 20 μM against 30 additional colistin-sensitive A. baumannii strains (MIC 1-2 μg/mL) isolated by Walter Reed Army Institute of Research (supporting information).18 This diverse set of A. baumannii isolates represents every major clade and many of the minor clades found clinically in A. baumannii infections. Both compounds showed potentiation activity against all strains; compound 12d effected reductions in the MIC of colistin between eight-fold and 512-fold, while compound 12i exhibited between 64-fold and ≥512-fold reductions. The fold reduction of MIC50 and MIC90 values were then calculated. Compound 12d had exhibited a 64-fold reduction and 32-fold reduction, respectively; while compound 12i had exhibited a 128-fold and 64-fold reduction, respectively. We further tested activity of compounds 12d and 12i against colistin-sensitive strains of K. pneumoniae and P. aeruginosa along with colistin-resistant A. baumannii harboring the mcr-1 plasmid. As shown in Table 4, we observed moderate activity for KP2146 (32-fold reduction), some activity for PAO1 (4-fold), and high activity against A. baumannii 17978+mcr-1. Activity against A. baumannii 17978+mcr-1 rivaled the 1024-fold reduction of colistin’s MIC observed in the parent strain A. baumannii 17978 that did not harbor the mcr-1 plasmid. Overall, we observed that these two compounds are effective colistin potentiating adjuvants in vitro across a range of clinically isolated strains.
Table 4.
Colistin Potentiation Against Various Bacterial Strains at
| Compd | K. pneumoniae 2146 | P. aeruginosa PAO1 | A. baumannii 17978+mcr-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (ng/mL) |
MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (ng/mL) |
MIC (μM) |
Concentration Tested (μM) |
Colistin MIC (ng/mL) |
|
| Colistin | 1000 | 1000 | 16000 | ||||||
| 12d | 200 | 30 | 31.3 | >200 | 30 | 250 | >200 | 30 | ≤ 7.8 |
| 12i | >200 | 30 | 31.3 | >200 | 30 | 250 | >200 | 30 | ≤ 7.8 |
The activity of lead compound 12i was then evaluated in the presence of Tween 80 to rule out activity due to non-specific compound aggregation effects (i.e. a PAINS mechanism).19 The adjuvant suppressed the MIC of colistin against A. baumannii 5075 to the same degree in the absence or presence of the detergent (supporting information). Thus, activity is not driven by non-specific compound aggregation.
Finally, we wanted to explore the potential in vivo relevance of this adjuvant approach for enhancing colistin efficacy by examining activity in a G. mellonella model of infection, using A. baumannii 5075.6 We tested the five lead compounds (12b, d, e, i, and k), while rifampin was used as a positive control. For comparison we also tested the original isopropyl adjuvant compound 3 (Figure 1).15 We first determined the maximum tolerated dose for the adjuvants by injecting compound 12d in uninfected worms, and found that single doses up to 400 mg/kg were well tolerated with no worm mortality noted after 6 days (supporting information). For comparison, toxicity of colistin was tested at 10 mg/kg and 100 mg/kg; colistin displayed no toxicity at 10 mg/kg, but at 100 mg/kg, worm survival was 73% at day 6 (supporting information). Next, we determined that a dose of 1 mg/kg colistin provided minimal protection from infection and was subsequently employed for all combination dosing. As a positive control, a single dose of 30 mg/kg rifampin afforded 90% survival of infected worms (vs. 3% survival for untreated worms). Treatment of infected worms with only compound 12d at 50 mg/kg provided no improvement in survival in comparison to untreated controls (supporting information). Treatment, however, with a combination of each urea derived adjuvant at 50 mg/kg in combination with colistin at 1 mg/kg provided an increase in survival, showing 36-50% worm survival from a single dose (Figure 2). This level of survival compares to previous studies in infected G. mellonella treated with penicillin/clavulanic acid, one of the few clinically employed antibiotic/adjuvant pairs.20 The original adjuvant, compound 3, provided only a moderate increase in survival when paired with colistin (10% increase vs. colistin alone). Finally, we tested the hemolytic activity of compounds 12d, 12i, and inactive compound 12g. At 200 μM, all three compounds showed <10% red blood hemolysis.
Figure 2.
Percent survival of G. mellonella after antibiotic-adjuvant, antibiotic, or DMSO (control, i.e. bacteria only) dosage followed with an inoculation of A. baumannii 5075.
In conclusion, we report a series of new urea containing 2-AIs that potentiate the activity of colistin against colistin-sensitive A. baumannii both in vivo and in vitro. Compounds 12d and 12i effected ≥512-fold reductions in the MIC of colistin when tested at 20 μM against some colistin-sensitive A. baumannii strains and exhibited activity against a diverse panel of A. baumannii strains in vitro, exhibiting MIC50 reduction values of 64- and 128-fold, respectively. Clavulanic acid, the prototypical adjuvant that is paired with β-lactam antibiotics to overcome β-lactam resistance in the clinic, is also employed at 20 μM, thus establishing that these urea adjuvants are active at relevant concentrations. Examination in a G. mellonella model indicated that these compounds have in vivo activity and provides the first step towards validating the approach of using these small molecule adjuvants to lower colistin dosage. As the Galleria model has inherent limitations, namely that it is typically limited to a single dose, it is anticipated that moving to a more clinically relevant model that employs a multiple dosing regimen will lead to a significant increase in survival. Further studies are currently underway to validate this approach in vivo for other colistin-sensitive Gram-negative pathogens as well as those that are colistin-resistant.
Experimental
All reagents used for chemical synthesis were purchased from commercially available sources (Sigma Aldrich U.S. or Fisher Scientific U.S.) and used without further purification. Flash chromatography was performed using 60 Å mesh standard grade silica gel from Sorbtech. NMR solvents were obtained from Cambridge Isotope Laboratories and used as is. All 1H NMR (400 MHz) and 13C NMR (400 MHz) spectra were recorded at 25 °C on a Varian Mercury spectrometer. Chemical shifts (δ) are given in parts per million relative to the respective NMR solvent; coupling constants (J) are in hertz (Hz). Abbreviations used are s, singlet; bs, broad singlet; d, doublet; dd, doublet of doublets; t, triplet; dt, doublet of triplets; m, multiplet. Mass spectra were obtained at the NCSU Department of Chemistry Mass Spectrometry Facility. The purities of the tested compounds were all verified to be >95% by LC-MS analysis on a Shimadzu LC-MS 2020 with Kinetex, 2.6 mm, C18 50 × 2.10 mm.
General procedure for reduction of nitro group (7 and 11).
Nitro-Boc-protected 2-AI 6 or 10 (0.2 mmol, 1 eq) and 10% by weight of Pd/C (approximately 0.01 g) were dissolved in MeOH (10 mL) and purged with H2 for 30 min. while stirred. The mixture was then allowed to stir under H2 atmosphere for 18 h. before the catalyst was removed by filtration with Celite 545. The solvent was removed under reduced pressure to afford compound 7 or 11 and immediately used for urea formation.
General procedure for urea formation and Boc-deprotection via an isocyanate (8 and 12).
A solution of isocyanate (0.4 mmol, 2 eq) dissolved in THF (10 mL) was added to a solution of amine-Boc-protected 2-AI 7 or 11 (0.2 mmol, 1 eq) in THF (10 mL). The reaction was stirred for 3 h. and then concentrated under reduced pressure. The resulting solid was then dissolved in dichloromethane (5 mL) and trifluoroacetic acid (2 mL) was added dropwise. After stirring for 3 h., the crude product was concentrated and then purified via flash chromatography (0-10% MeOH sat. with NH3/DCM). Purified product was dissolved in 6 M HCl in MeOH and solvent was removed to yield product 8 or 12 as the HCl salt.
General procedure for urea formation and Boc-deprotection via an aniline (8 and 12).
Amine-Boc-protected 2-AI 7 or 11 (0.2 mmol, 1 eq) dissolved in DCM (12 mL) was added to a sodium carbonate (0.03 g, 0.39 mmol, 1.6 eq) solution in H2O (12 mL). After stirring for 5 min., triphosgene (0.03 g, 0.12 mmol, 0.33 eq) was added to the flask. After stirring for 30 min., the aniline (0.4 mmol, 2 eq) was added dropwise and allowed to stir for an additional 2 h. The mixture was then separated and the aqueous layer was washed twice with DCM. The organics were combined, dried with MgSO4, and concentrated under reduced pressure. The resulting solid was then dissolved in dichloromethane (5 mL) and trifluoroacetic acid (2 mL) was added dropwise. After stirring for 3 h., the crude product was concentrated and then purified via flash chromatography (0-10% MeOH sat. with NH3/DCM). Purified product was dissolved in 6 M HCl in MeOH and solvent was removed to yield product 8 or 12 as the HCl salt.
Tri-Boc protected tert-butyl 2-amino-4-(3-nitrophenyl)-1H-imidazole-1-carboxylate, (6).
2-Bromo-3’-nitroacetophenone (50 g, 204 mmol) and Boc-guanidine (65 g, 408 mmol) were dissolved in tetrahydrofuran (300 mL) and the reaction was heated to 56 ºC for 2 h. The reaction was then cooled to 0 ºC, stirred for 1 h., and then the resulting solid was collected by vacuum filtration and washed twice with diethyl ether. After concentration under reduced pressure, compound 5 was dissolved in tetrahydrofuran (200 mL) and a solution of di-tert-butyl dicarbonate (4.3 g, 19.7 mmol) and 4-dimethylaminopyridine (0.04 g, 0.3 mmol) dissolved in tetrahydrofuran (10 mL) was added. The mixture was stirred for 24 h., followed by a dilution with DI H2O and the product was extracted with EtOAc three times. The combined organic layers were then washed three times with 1 N HCl, three times with NaHCO3, brine, dried with MgSO4, and then concentrated under reduced pressure to yield white solid 6 (40%). 1H NMR (400 MHz, DMSO-d6) δ 8.62 (bs, 1H), 8.44 (s, 1H), 8.32-8.28 (m, 1H), 8.15 – 8.11 (m, 1H), 7.69 (t, J = 8.0 Hz, 1H), 1.56 (s, 6H), 1.36 (s, 12H). 13C NMR (100 MHz, DMSO-d6) δ 149.4, 148.8, 146.3, 138.8, 136.3, 134.3, 131.5, 130.8, 122.7, 119.5, 116.7, 87.0, 84.1, 27.9, 27.7. HRMS (ESI) calculated for [M+H]+: 505.2293. Found: 505.2278.
Compound 8a.
Using the general procedure for urea formation via an isocyanate with 4-butylphenyl isocyanate, 8a was obtained as a white solid in 30% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.79 (t, J = 1.9 Hz, 1H), 7.36 – 7.32 (m, 3H), 7.27-7.25 (m, 1H), 7.20 – 7.18 (dt, J = 7.1, 1.7 Hz, 1H), 7.11-7.09 (m, 3H), 2.56 (t, J = 7.7 Hz, 2H), 1.63 – 1.51 (m, 2H), 1.34 (m, 2H), 0.93 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, Methanol-d4) δ 154.1, 140.1, 137.4, 136.4, 129.3, 128.3, 128.1, 127.7, 119.4, 118.8, 118.3, 114.7, 108.6, 34.6, 33.6, 21.9, 12.9. HRMS (ESI) calculated for [M+H]+: 350.1975. Found: 350.1970.
Compound 8b.
Using general procedure for urea formation via an isocyanate with 3,5-dichlorophenyl isocyanate, 8b was obtained as a while solid in 27% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.81 (d, J = 2.5 Hz, 1H), 7.74 (td, J = 1.9, 0.7 Hz, 1H), 7.42 – 7.22 (m, 4H), 7.19 (dt, J = 7.1, 1.7 Hz, 1H), 7.10 (s, 1H). 13C NMR (100 MHz, Methanol-d4) δ 153.3, 139.7, 139.3, 131.9, 130.0, 129.3, 128.1, 127.5, 124.8, 120.0, 118.9, 118.5, 118.2, 114.8, 108.6. HRMS (ESI) calculated for [M+H]+: 362.0570. Found: 362.0564.
Compound 8c.
Using general procedure for urea formation via an aniline with 4-propylaniline, 8c was obtained as a white solid in 35% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.78 (t, J = 1.9 Hz, 1H), 7.36-7.32 (m, 3H), 7.28-7.25 (m,1H), 7.21-7.18 (m, 1H), 7.10 (dd, J = 5.9, 2.4 Hz, 3H), 2.59 – 2.49 (t, 8.4 Hz, 2H), 1.68 – 1.54 (m, 4H), 0.92 (t, J = 7.4 Hz, 2H). 13C NMR (100 MHz, Methanol-d4) δ 154.1, 148.0, 140.1, 137.2, 136.5, 129.3, 128.4, 128.1, 127.7, 119.3, 118.8, 118.3, 114.7, 108.6, 37.0, 24.4, 12.6. HRMS (ESI) calculated for [M+H]+: 336.1819. Found: 336.1813.
Compound 8d.
Using general procedure for urea formation via an aniline with 3,5-dibromoaniline, 8d was obtained as a white solid in 40% yield. 1H NMR (400 MHz, DMSO-d6) δ 9.28 (s, 2H), 7.73 – 7.61 (m, 4H), 7.38 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 152.4, 142.5, 127.1, 122.8, 120.3. HRMS (ESI) calculated for [M+H]+: 449.9560. Found: 449.9552.
Compound 8e.
Using general procedure for urea formation via an aniline with 3-bromo-4-(trifluoromethyl)aniline 8e was obtained as a white solid in 39% yield. 1H NMR (400 MHz, Methanol-d4) δ 8.10 (d, J = 2.2 Hz, 1H), 7.81 (s, 1H), 7.63 (d, J = 8.6 Hz, 1H), 7.49 (dd, J = 8.7, 2.5 Hz, 1H), 7.38 – 7.30 (m, 2H), 7.24 – 7.22 (m, 1H), 7.13 (s, 1H). 13C NMR (100 MHz, Methanol-d4) δ 153.0, 143.9, 139.6, 129.4, 128.2, 127.9, 127.6, 123.6, 119.0, 118.8, 116.3, 114.9, 108.7. HRMS (ESI) calculated for [M+H]+: 440.0328. Found: 440.0325.
Compound 8f.
Using general procedure for urea formation via an isocyanate with 3,4-dimethylphenyl isocyanate, 8f was obtained as a white solid in 43% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.79 (t, J = 1.9 Hz, 1H), 7.34 (t, J = 7.9 Hz, 1H), 7.26 – 7.23 (m, 1H), 7.20-7.13 (m, 3H), 7.10 (s, 1H), 7.03 (d, J = 8.1 Hz, 1H), 2.23 (s, 3H), 2.20 (s, 3H). 13C NMR (100 MHz, Methanol-d4) δ 154.1, 140.1, 136.6, 136.4, 130.9, 129.4, 129.3, 128.1, 127.7, 120.6, 118.8, 118.23, 116.9, 114.7, 108.6, 18.6, 17.7. HRMS (ESI) calculated for [M+H]+: 322.1662. Found: 322.1656.
Compound 8g.
Using general procedure for urea formation via an aniline with 4-butyl-2-methylaniline, 8g was obtained white solid in 35% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.86 (m, 1H), 7.44 (d, J = 8.1 Hz, 1H), 7.35 (t, J = 8.0 Hz, 1H), 7.26 – 7.16 (m, 2H), 7.11 (d, J = 0.9 Hz, 1H), 7.07 – 6.95 (m, 2H), 2.55 (t, J = 7.6 Hz, 2H), 2.27 (s, 3H), 1.57 (p, J = 7.4 Hz, 2H), 1.35 (m, 2H), 0.93 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6) δ 153.3, 148.4, 141.1, 137.4, 135.2, 130.5, 129.9, 128.7, 128.6, 127.1, 126.3, 122.2, 118.4, 118.2, 114.2, 109.9, 34.6, 33.7, 22.2, 18.3, 14.2. HRMS (ESI) calculated for [M+H]+: 364.2132. Found: 364.2125.
Compound 8h.
Using general procedure for urea formation via an aniline with 4-methyl-3-(trifluoromethyl)aniline 8h was obtained as a white solid in 46% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.89 (d, J = 2.5 Hz, 1H), 7.81 (t, J = 1.9 Hz, 1H), 7.48 (dd, J = 8.3, 2.5 Hz, 1H), 7.35 (t, J = 7.9, 1H), 7.28-7.26 (m, 2H), 7.23-7.20 (m, 1H), 7.13 (s, 1H), 2.40 (s, 3H). 13C NMR (100 MHz, Methanol-d4) δ 153.7, 139.9, 137.4, 132.2, 130.0, 129.3, 128.2, 127.6, 121.9, 118.9, 118.5, 116.0, 116.0, 114.8, 108.7, 17.3. HRMS (ESI) calculated for [M+H]+: 376.1380. Found: 376.1375.
Compound 8i.
Using general procedure for urea formation via an aniline with 3,5-bis(trifluoromethyl)aniline, 8i was obtained as a white solid in 37% yield. 1H NMR (400 MHz, Methanol-d4) δ 8.11 (s, 1H), 7.84 (t, J = 1.9 Hz, 1H), 7.53 (s, 1H), 7.41 – 7.28 (m, 2H), 7.23 (dt, J = 7.4, 1.5 Hz, 2H), 7.14 (s, 1H). 13C NMR (100 MHz, Methanol-d4) δ 153.2, 141.5, 139.6, 132.3, 131.9, 131.6, 129.4, 128.2, 127.5, 124.7, 119.0, 118.8, 114.9, 108.7. HRMS (ESI) calculated for [M+H]+: 430.1097. Found: 430.1094.
1-Isopropyl-4-(3-nitrophenyl)-1H-imidazol-2-amine (9).
To a solution of 2-bromo-1-(3-nitrophenyl)ethan-1-one (5 g, 20 mmol, 1 eq) in EtOH (125 mL) was added isopropylamine (19 mmol, 0.95 eq) dropwise. The reaction was allowed to stir at room temperature for 10 min. before cyanamide (8.6 g, 200 mmol, 10 eq) was added and the pH adjusted to 4.3 using 1 M HCl. The reaction was heated to 95 °C and stirred for 18 h., before being allowed to cool to room temperature. The solvent was then removed under reduced pressure, and the crude mixture was purified via flash chromatography (0-5% MeOH sat. with NH3/DCM) to yield orange solid 9 (15%). 1H NMR (400 MHz, Methanol-d4) δ 8.45 (s, 1H), 7.97 (m, 2H), 7.51 (t, J = 8.0 Hz, 1H), 7.30 (s, 1H), 4.28 (p, J = 6.6 Hz, 1H), 1.43 (d, J = 6.6 Hz, 6H). 13C NMR (100 MHz, Methanol-d4) δ 136.4, 133.9, 129.5, 129.2, 119.9, 118.0, 116.3, 116.3, 108.5, 46.4, 21.4. HRMS (ESI) calculated for [M+H]+: 247.1190. Found: 247.1185.
Compound 10.
Di-tert-butyl dicarbonate (6 eq) and 4-dimethylaminopyridine (0.1 eq) were added to a solution of 1-isopropyl-4-(3-nitrophenyl)-1H-imidazol-2-amine (1 eq) dissolved in tetrahydrofuran and triethylamine (3 eq). The mixture was stirred at ambient temperature for 24 h. and then diluted with DI H2O. The crude product was extracted with EtOAc three times. The combined organic layers were then washed three times with 1 N HCl, three times with NaHCO3, brine, dried with MgSO4, and then concentrated under reduced pressure. The crude mixture was purified via flash chromatography (20% EtOAc/Hexanes) to yield white solid 10 (20%). 1H NMR (400 MHz, Chloroform-d) δ 8.50 (bs, 1H), 8.08 – 8.01 (m, 2H), 7.49 (t, J = 8.0 Hz, 1H), 7.39 (s, 1H), 4.25-4.18 (m, 1HH), 1.47-1.41 (m, 24H). 13C NMR (100 MHz, Chloroform-d) δ 149.9, 148.5, 138.0, 137.7, 135.9, 130.5, 129.5, 121.2, 119.3, 111.9, 84.2, 47.6, 27.8, 23.1. HRMS (ESI) calculated for [M+H]+: 447.2238. Found: 447.2231.
Compound 12a.
Using general procedure for urea formation via an isocyanate with 4-butylphenyl isocyanate, 12a was obtained as a white solid in 55% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.76 (s, 1H), 7.37 (s, 1H), 7.30-7.24 (m, 4H), 7.19-7.17 (m, 1H), 7.05 (d, J = 7.5 Hz, 2H), 4.42 – 4.35 (m, 1H), 2.51 (t, J = 7.6 Hz, 2H), 1.56 – 1.47 (m, 2H), 1.43 (d, J = 6.6 Hz H), 1.34-1.23 (m, 2H), 0.88 (t, J = 7.1 Hz, 3H). 13C NMR (100 MHz, Methanol-d4) δ 145.9, 140.1, 137.3, 136.4, 129.3, 128.3, 127.9, 127.4, 119.2, 118.8, 118.3, 114.6, 108.1, 47.8, 34.5, 33.6, 21.8, 20.5, 12.9. HRMS (ESI) calculated for [M+H]+: 392.2449. Found: 392.2436.
Compound 12b.
Using general procedure for urea formation via an isocyanate with 3,4-dichlorophenyl isocyanate, 12b was obtained as a white solid in 55% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.77 (d, J = 2.4 Hz, 1H), 7.70-7.69 (m, 1H), 7.37 (s, 1H), 7.33-7.30 (m, 2H), 7.28 (s, 1H), 7.24-7.22 (m, 1H), 7.19-7.16 (m, 1H), 4.41 (p, J = 6.6 Hz, 1H), 1.46 (d, J = 6.6 Hz, 6H). 13C NMR (100 MHz, Methanol-d4) δ 153.1, 145.9, 139.7, 139.2, 131.8, 130.0, 129.3, 127.8, 127.2, 124.7, 119.8, 118.8, 118.4, 118.0, 114.5, 108.0, 47.7, 20.5. HRMS (ESI) calculated for [M+H]+: 404.1039. Found: 404.1043.
Compound 12c.
Using general procedure for urea formation via an aniline with 4-propylaniline, 12c was obtained as a white solid in 39% yield. H NMR (400 MHz, Methanol-d4) δ 7.67-7.65 (m, 1H), 7.32 – 7.28 (m, 3H), 7.25 – 7.21 (m, 2H), 7.09 – 7.07 (m, 3H), 4.25 (p, J = 6.6 Hz, 1H), 2.52 (t, J = 8.4, 2H), 1.65 – 1.55 (m, 2H), 1.39 (d, J = 6.7 Hz, 6H), 0.91 (t, J = 7.4 Hz, 3H). 13C NMR (100 MHz, Methanol-d4) δ 154.2, 148.5, 139.2, 137.0, 136.6, 135.5, 134.8, 128.6, 128.4, 119.2, 118.5, 116.9, 114.8, 106.7, 46.2, 37.0, 24.4, 21.4, 12.7. HRMS (ESI) calculated for [M+H]+: 378.2288. Found: 378.2285.
Compound 12d.
Using general procedure for urea formation via an aniline with 3,5-dibromoaniline, 12d was obtained as a white solid in 38% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.84 (s, 1H), 7.69 (s, 2H), 7.43 (s, 1H), 7.37 – 7.20 (m, 4H), 4.42 (p, J = 6.6 Hz, 1H), 1.49 (d, J = 6.5 Hz, 6H). 13C NMR (100 MHz, Methanol-d4) δ 153.1, 146.2, 142.0, 139.6, 129.3, 128.1, 127.6, 126.9, 122.3, 119.9, 119.1, 118.5, 114.7, 107.8, 47.6, 20.5. HRMS (ESI) calculated for [M+H]+: 492.0029. Found: 492.0026.
Compound 12e.
Using general procedure for urea formation via an aniline with 3-bromo-4-(trifluoromethyl)aniline, 12e was obtained as a white solid in 23% yield. 1H NMR (400 MHz, Methanol-d4) δ 8.09 (d, J = 2.1 Hz, 1H), 7.80 (d, J = 1.8 Hz, 1H), 7.60 (d, J = 8.7 Hz, 1H), 7.50 – 7.47 (m, 1H), 7.43 (s, 1H), 7.34 (m, 2H), 7.25-7.22 (m, 1H), 4.43 (p, J = 6.5 Hz, 1H), 1.49 (d, J = 6.6 Hz, 6H). 13C NMR (100 MHz, Methanol-d4) δ 152.9, 146.0, 143.8, 139.5, 129.4, 127.9, 127.2, 123.4, 119.0, 118.7, 116.2, 114.7, 108.1, 47.7, 20.5. HRMS (ESI) calculated for [M+H]+: 482.0798. Found: 482.0792.
Compound 12f.
Using general procedure for urea formation via an isocyanate with 3,4-dimethylphenyl isocyanate, 12f was obtained as a white solid in 48% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.82 (bs, 1H), 7.42 (s, 1H), 7.38 – 7.34 (m, 1H), 7.31 (d, J = 8.2 Hz, 1H), 7.24 (m, 2H), 7.21 – 7.17 (m, 1H), 7.04 (d, J = 8.1 Hz, 1H), 4.45 (m, J = 6.7 Hz, 1H), 2.24 (s, 3H), 2.21 (s, 3H), (d, J = 17.3 Hz, 6H), 1.50 50 (d, J = 6.6 Hz, 6H). 13C NMR (100 MHz, Methanol-d4) δ 154.1, 146.0, 140.2, 136.6, 136.5, 130.9, 129.4 (m), 127.9, 127.4, 120.6, 118.9, 118.3, 116.8, 114.7, 108.0, 107.9, 47.7, 20.5, 18.6, 17.8. HRMS (ESI) calculated for [M+H]+: 364.2132. Found: 364.2127.
Compound 12g.
Using general procedure for urea formation via an aniline with 4-butyl-2-methylaniline, 12g was obtained as a white solid in 57% yield. 1H NMR (400 MHz, Methanol-d4) δ 7.81 (m, 1H), 7.45 – 7.37 (m, 2H), 7.32 – 7.30 (m, 2H), 7.22 – 7.19 (m, 1H), 6.97 – 6.91 (m, 1H), 4.40 (h, J = 6.5 Hz, 1H), 2.51 (t, J = 7.7 Hz, 4H), 2.25 (s, 3H), 1.60 – 1.48 (m, 2H), 1.45 (d, J = 6.5 Hz , 6H), 1.41 – 1.23 (m, 2H), 0.92 (t, J = 7.3 Hz, 3H). 13C NMR (100 MHz, Methanol-d4) δ 154.7, 145.9, 140.3, 139.2, 133.7, 130.6, 130.1, 129.3, 127.9, 127.4, 125.9, 123.7, 118.8, 118.2, 114.6, 107.9, 47.2, 34.7, 33.6, 21.9, 20.5, 16.9, 12.9. HRMS (ESI) calculated for [M+H]+: 406.2601. Found: 406.2595.
Compound 12h.
Using general procedure for urea formation via an aniline with 4-methyl-3-(trifluoromethyl)aniline, 12h was obtained as a white solid in 56% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.87 (s, 1H), 9.88 (bs, 1H), 9.51 (bs, 1H), 7.95 (bs, 1H), 7.78 (bs, 1H), 7.66 – 7.59 (m, 3H), 7.48 (d, J = 8.2 Hz, 1H), 7.36-7.23 (m, 4H), 4.53 – 4.44 (m, 1H), 2.34 (s, 3H), 1.37 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, DMSO) δ 154.0, 147.2, 141.5, 139.4, 134.0, 130.7, 129.8, 129.4, 128.9, 128.7, 127.6, 126.6, 125.0, 122.6, 119.6, 119.5, 116.0 (q), 115.3, 110.3, 48.2, 22.7, 19.4. HRMS (ESI) calculated for [M+H]+: 418.1849. Found: 418.1844.
Compound 12i.
Using general procedure for urea formation via an aniline with 3,5-bis(trifluoromethyl)aniline, 12i was obtained as a white-yellow solid in 41% yield. 1H NMR (400 MHz, Methanol-d4) δ 8.08 (s, 2H), 7.72 (d, J = 1.9 Hz, 1H), 7.52 (s, 1H), 7.34 – 7.23 (m, 3H), 7.11 (d, J = 1.6 Hz, 1H), 4.34 – 4.21 (m, 1H), 1.42 (dt, J = 6.6, 3.1 Hz, 8H). 13C NMR (100 MHz, Methanol-d4) δ 153.23, 148.6, 141.7, 138.6, 135.5, 135.0, 131.7 (q), 128.6, 119.0, 117.9, 117.1, 115.0, 114.4, 106.8, 46.2, 21.4. HRMS (ESI) calculated for [M+H]+: 472.1567. Found: 472.1560.
Compound 12j.
Using general procedure for urea formation via an aniline with 2-methyl-4-(trifluoromethyl)aniline, 12j was obtained as a white-yellow solid in 45% yield. 1H NMR (400 MHz, Methanol-d4) δ 8.07 (d, J = 8.5 Hz, 1H), 7.81 (t, J = 1.9 Hz, 1H), 7.45 – 7.30 (m, 5H), 7.22 (dt, J = 6.9, 1.7 Hz, 1H), 4.42 (p, J = 6.6 Hz, 1H), 2.36 (s, 3H), 1.47 (d, J = 6.6 Hz, 6H). 13C NMR (100 MHz, Methanol-d4) δ 153.5, 145.9, 140.4, 139.9, 129.3, 128.3, 127.9, 127.2, 126.7 (q), 123.1, 123.0, 122.9, 120.6, 118.8, 118.5, 114.5, 108.0, 48.3, 48.1, 47.8, 47.7, 47.6, 47.4, 47.2, 47.0, 20.5, 17.0. HRMS (ESI) calculated for [M+H]+: 418.1849. Found: 418.1833.
Compound 12k.
Using general procedure for urea formation via an aniline with 4-bromo-3,5-dichloroaniline, 12k was obtained as a purple solid in 62% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.85 (s, 1H), 10.24 (s, 1H), 9.57 (s, 1H), 7.78-7.76 (m, 1H), 7.72 (m, 2H), 7.65 (bs, 1H), 7.61 (bs, 2H), 7.37– 7.26 (m, 3H), 4.48 (p, J = 6.3 Hz, 1H), 1.38 (d, J = 6.3 Hz, 6H). 13C NMR (100 MHz, DMSO-d6) δ 152.8, 146.4, 141.2, 140.2, 135.5, 129.9, 128.6, 126.7, 119.0, 118.9, 118.0, 114.6, 113.5, 109.5, 47.3, 21.8. HRMS (ESI) calculated for [M+H]+: 482.0145. Found: 482.0142.
Broth microdilution method for MIC determination.
Day cultures (6 h) of each bacterial strain in cation adjusted Mueller Hinton II broth (CAMHB, Fisher Scientific U.S.) were subcultured to 5×105 CFU/mL in CAMHB. Aliquots (1 mL) were placed in culture tubes and compound was added from 100 or 10 mM stock samples in DMSO, such that compound concentration equaled highest concentration tested (200 μM). Samples were then aliquoted (200 μL) into the first wells of a 96-well plate, with all remaining wells being filled with 100 μL of initial bacterial subculture. Row one wells were mixed five times, before 100 μL was transferred to row two. Row two was then mixed five times, and 100 μL was transferred to row three. This process was repeated until the final row had been mixed, this served to serially dilute the compound. Plates were then covered with GLAD Press n’ Seal and incubated under stationary conditions at 37 °C for 16 h. MIC values were then recorded as the lowest concentration at which no bacterial growth was observed.
Broth microdilution method for antibiotic potentiation.
Day cultures (6 h) of bacteria in CAMHB were subcultured to 5×105 CFU/mL in CAMHB. Aliquots (4 mL) were placed in culture tubes, and dosed with compound from 100 or 10 mM stock samples to give the desired concentration of the compound to be tested against the particular bacterial strain; this insured non-toxic DMSO concentrations ≤0.3% in each well. 1 mL of the resulting solution was placed in a separate culture, and dosed with antibiotic at the highest concentration to be tested. Bacteria treated with antibiotic alone served as the control. Row one of a 96-well plate was filled with 200 μL of the antibiotic/2-AI solution, and rows 2-12 were filled with 100 μL each of the remaining 4 mL of bacterial subculture containing adjuvant at the desired concentration, except for the control lane which contained only bacterial subculture. Row one was then mixed five times, and 100 μL was transferred to row two, which was then mixed five times before being transferred to row three. This process was repeated until all rows had been mixed, except for row twelve, which would have only 2-AI, to serve as a control. The 96-well plate was then covered in Glad Press n’ Seal and incubated under stationary conditions at 37 °C for 16 h. MIC values were determined as the lowest concentration at which no bacterial growth was observed, fold reductions were determined by comparison to control lane.
Broth microdilution method for antibiotic potentiation with A. baumannii 17978+mcr-1.
Day cultures (6 h) of A. baumannii 17978+mcr-1 in CAMHB dosed with 30 μg/mL gentamicin sulfate were subcultured to 5×105 CFU/mL in CAMHB. Then procedure for ‘broth microdilution method for antibiotic potentiation’ was followed starting with making 4 mL aliquots.
Broth microdilution method for antibiotic potentiation with detergent.
Day cultures (6 h) of bacteria in CAMHB were subcultured to 5×105 CFU/mL in CAMHB and then Tween™ 80 Surfact-Amps™ Detergent Solution (Fisher Scientific, U.S.) was added to make a 0.01% containing bacterial solution. Then procedure for ‘broth microdilution method for antibiotic potentiation’ was followed starting with making 4 mL aliquots from Tween™ 80 bacterial solution.
Galleria mellonella Assay.
G. mellonella larvae (Speedy Worm, Alexandria, MN) were used within 10 days of shipment from the vendor. After reception of worms, larvae were kept in the dark at room temperature for at least 24 h. before infection. Larvae weighing between 200 to 300 mg were used in the survival assay. Using a 10 μL glass syringe (Hamilton, Reno, NV) fitted with a 30 G needle (Exel International, St. Petersburg, Fl), a 5 μL solution of the desired compound(s) and concentration(s) were injected into the last left proleg. Colistin was dosed at 1 mg/kg dissolved in DI water, 2-AIs at 50 mg/kg dissolved in DMSO, rifampin at 30 mg/kg dissolved in DMSO, and DMSO was injected for “bacteria only” as a control. For bacterial injections, 50 μL from an overnight culture of A. baumannii 5075 in Miller LB broth (Fisher Scientific U.S.) was subcultured into 5 mL Miller LB broth and incubated for an additional 3 h before use. Then, 2.5 h. after the first injection, a second 5 μL injection containing 105 CFU of A. baumannii 5075 in phosphate buffer solution (Fisher Scientific U.S.) was injected into the second to last left proleg. Injected worms were left at room temperature in the dark while being assessed at 24 h. intervals over 6 days. Larvae were considered dead if they did not respond to physical stimuli. Experiment was repeated 3 times using 10 larvae per experimental group. No ethics approval was needed for G. mellonella.
Supplementary Material
Acknowledgements
The authors would like to thank the National Institutes of Health (GM05579) for funding.
Abbreviations
- 2-AI
2-aminoimidazole
- MDR
multidrug-resistant
Footnotes
Supporting Information
Colistin potentiation in presence of compounds 12d and 12i against colistin-sensitive A. baumannii from Walter Reed Army Institute of Research; Colistin potentiation of A. baumannii 5075 in presence of compound 12i with Tween 80 detergent; G. mellonella day-by-day results, including toxicity of compound 12d at 400 mg/kg and testing of antibiotic activity of compound 12d at 50 mg/kg; representative 1H and 13C Nuclear Magnetic Resonance spectra.
References
- 1.Rice LB, Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 2008, 197 (8), 1079–1081. DOI 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 2.Pendleton JN; Gorman SP; Gilmore BF, Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 2013, 11 (3), 297–308. DOI 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 3.Clatworthy AE; Pierson E; Hung DT, Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol 2007, 3 (9), 541–548. DOI 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 4.CDC, Antibiotic resistance threats in the U. S 2013. [Google Scholar]
- 5.Hay M; Thomas DW; Craighead JL; Economides C; Rosenthal J, Clinical development success rates for investigational drugs. Nat Biotechnol 2014, 32 (1), 40–51. DOI 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs AC; Thompson MG; Black CC; Kessler JL; Clark LP; McQueary CN; Gancz HY; Corey BW; Moon JK; Si Y; Owen MT; Hallock JD; Kwak YI; Summers A; Li CZ; Rasko DA; Penwell WF; Honnold CL; Wise MC; Waterman PE; Lesho EP; Stewart RL; Actis LA; Palys TJ; Craft DW; Zurawski DV, AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio 2014, 5 (3), e01076–14. DOI 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray CK; Hospenthal DR, Acinetobacter infection in the ICU. Crit Care Clin 2008, 24 (2), 237–248, vii. DOI 10.1016/j.ccc.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Davis KA; Moran KA; McAllister CK; Gray PJ, Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg Infect Dis 2005, 11 (8), 1218–1224. DOI 10.3201/eid1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dizbay M; Tozlu DK; Cirak MY; Isik Y; Ozdemir K; Arman D, In vitro synergistic activity of tigecycline and colistin against XDR-Acinetobacter baumannii. J Antibiot (Tokyo) 2010, 63 (2), 51–53. DOI 10.1038/ja.2009.117. [DOI] [PubMed] [Google Scholar]
- 10.Spapen H; Jacobs R; Van Gorp V; Troubleyn J; Honore PM, Renal and neurological side effects of colistin in critically ill patients. Ann Intensive Care 2011, 1 (1), 14 DOI 10.1186/2110-5820-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pournaras S; Vrioni G; Neou E; Dendrinos J; Dimitroulia E; Poulou A; Tsakris A , Activity of tigecycline alone and in combination with colistin and meropenem against Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae strains by time-kill assay. Int J Antimicrob Agents 2011, 37 (3), 244–247. DOI 10.1016/j.ijantimicag.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 12.Ejim L; Farha MA; Falconer SB; Wildenhain J; Coombes BK; Tyers M; Brown ED; Wright GD, Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 2011, 7 (6), 348–350. DOI 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 13.Harris TL; Worthington RJ; Hittle LE; Zurawski DV; Ernst RK; Melander C, Small molecule downregulation of PmrAB reverses lipid A modification and breaks colistin resistance. ACS Chem Biol 2014, 9 (1), 122–127. DOI 10.1021/cb400490k. [DOI] [PubMed] [Google Scholar]
- 14.Richards JJ; Reed CS; Melander C, Effects of N-pyrrole substitution on the anti-biofilm activities of oroidin derivatives against Acinetobacter baumannii. Bioorg Med Chem Lett 2008, 18 (15), 4325–4327. DOI 10.1016/j.bmcl.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 15.Brackett CM; Furlani RE; Anderson RG; Krishnamurthy A; Melander RJ; Moskowitz SM; Ernst RK; Melander C, Second generation modifiers of colistin resistance show enhanced activity and lower inherent toxicity. Tetrahedron 2015, 72 (25), 3549–3553. DOI 10.1016/j.tet.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens MD; Hubble VB; Ernst RK; van Hoek ML; Melander RJ; Cavanagh J; Melander C, Potentiation of Francisella resistance to conventional antibiotics through small molecule adjuvants. Medchemcomm 2016, 7 (1), 128–131. DOI 10.1039/C5MD00353A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker WT; Martin SE; Chandler CE; Nguyen TV; Harris TL; Goodell C; Melander RJ; Doi Y; Ernst RK; Melander C, Small molecule adjuvants that suppress both chromosomal and mcr-1 encoded colistin-resistance and amplify colistin efficacy in polymyxin-susceptible bacteria. Bioorg Med Chem 2017, 25 (20), 5749–5753. DOI 10.1016/j.bmc.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taitt CR; Leski TA; Stockelman MG; Craft DW; Zurawski DV; Kirkup BC; Vora GJ, Antimicrobial resistance determinants in Acinetobacter baumannii isolates taken from military treatment facilities. Antimicrob Agents Chemother 2014, 58 (2), 767–781. DOI 10.1128/AAC.01897-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baell JB; Holloway GA, New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 2010, 53 (7), 2719–2740. DOI 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 20.Ba X; Harrison EM; Lovering AL; Gleadall N; Zadoks R; Parkhill J; Peacock SJ; Holden MT; Paterson GK; Holmes MA, Old Drugs To Treat Resistant Bugs: Methicillin-Resistant Staphylococcus aureus Isolates with mecC Are Susceptible to a Combination of Penicillin and Clavulanic Acid. Antimicrob Agents Chemother 2015, 59 (12), 7396–404. DOI 10.1128/AAC.01469-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.