Abstract
Bile salt export pump BSEP (ABCB11) is a liver-specific adenosine triphosphate-cassette binding transporter that mediates canalicular bile salt excretion from hepatocytes. Human mutations in ABCB11 cause progressive familial intrahepatic cholestasis type II (PFIC2). Although over 150 ABCB11 variants have been reported, our understanding of their biological consequences is limited by the lack of experimental model that recapitulates the patient phenotypes. We applied CRISPR/Cas9-based genome editing technology to knockout abcb11b, the ortholog of human ABCB11, in zebrafish and found that these mutants died prematurely. Histological and ultrastructural analyses showed that abcb11b mutant zebrafish exhibited hepatocyte injury similar to that seen in patients with PFIC2. Hepatocytes of the mutant zebrafish failed to excrete the fluorescently tagged bile acid that is a substrate of human BSEP. Multi-drug resistance protein 1 (MDR1), which is thought to play a compensatory role in Abcb11 knockout mice, was mislocalized to the hepatocyte cytoplasm in abcb11b mutant zebrafish and in a patient lacking BSEP protein due to nonsense mutations in ABCB11. We discovered that BSEP deficiency induced autophagy in both human and zebrafish hepatocytes. Treatment with rapamycin restored bile acid excretion, attenuated hepatocyte damage, and extended lifespan of abcb11b mutant zebrafish, correlating with the recovery of canalicular Mdr1 localization. Conclusions: Collectively, these data suggest a model that rapamycin rescues BSEP-deficient phenotypes by prompting alternative transporters to excreting bile salts. MDR1 is a candidate for such alternative transporter.
Keywords: PFIC2, canalicular transporter, Mdr1, rapamycin, BSEP
Bile secretion from hepatocyte is the main route of excretion for exogenous toxic lipophilic substances, cholesterol, and bilirubin (1). It is also required for digestion of dietary fats and fat-soluble vitamins in the intestine. The rate-limiting step in bile secretion is mediated by the bile salt export pump (BSEP) ABCB11 that belongs to the adenosine triphosphate-binding cassette (ABC) transporter superfamily (2). It is selectively enriched at the apical membrane of hepatocyte to transport monovalent bile salts across the canalicular membrane (3). Impairment of BSEP function leads to intrahepatic accumulation of toxic bile acids. Lack of luminal bile acids also causes fat malabsorption, malnutrition and fat-soluble vitamin deficiency.
The most severe form of BSEP deficiency, progressive familial intrahepatic cholestasis type II (PFIC2), is an early-onset childhood disease which progresses rapidly to end-stage liver failure, and predisposes to hepatocellular carcinoma (4). Liver transplantation remains as the only effective treatment (5). Studies to classify ABCB11 mutations based on their impact on protein expression and function were mostly conducted in vitro and yielded controversial results (6). The only in vivo model, Abcb11 knockout mouse, develops mild cholestasis (7–9). This may result, in part, from detoxification of hydrophobic bile acids by hydroxylation in rodents (8). Increased expression of another canalicular transporter multi-drug resistance protein 1 (MDR1) may compensate for the loss of ABCB11 (7). MDR1 transcript expression does not increase in the livers of patients with PFIC2 (10). It is not clear whether MDR1 has a compensatory role in humans with BSEP deficiency.
Since Abcb11 knockout mice do not fully recapitulate PFIC2 pathology, additional animal model is needed. The small teleost zebrafish (Danio rerio) has gained increasing popularity in liver research. The zebrafish liver contains counterparts of mammalian liver cell types and becomes functional by four days post fertilization (11). The cholangiocyte and hepatocyte-specific transgenic zebrafish allows direct observation of the intrahepatic bile ducts and hepatocytes, respectively (12). Bile flow can be tracked in live larvae following feeding with fluorescent lipid analogs (13, 14). Zebrafish has been applied in the study of inherited cholestasis syndromes including Alagille syndrome (15) and biliary atresia (16, 17). Although zebrafish express several canalicular transporters with high homology to the human proteins (18), it has not been used to study bile secretion.
In the present study, we modeled BSEP deficiency using zebrafish. We detected impaired bile excretion, mislocalized Mdr1 expression, and induction of autophagy in the hepatocytes of abcb11b mutant zebrafish and a patient with nonsense mutations in ABCB11. Rapamycin partially restored bile excretion, recovered the canalicular localization of Mdr1 in the zebrafish mutants, and improved their survival, suggesting novel strategies to treat cholestasis caused by BSEP deficiency.
Materials and Methods
Additional information can be found in the Supporting Information.
HUMAN SUBJECTS
Patient A, a Caucasian female, presented at 7 months of age with jaundice, malnutrition, severe pruritus and underwent percutaneous liver biopsy, when the liver tissues for the current study were obtained. At that time, serum aspartate aminotransferase, alanine aminotransferase, conjugated bilirubin, γ-Glutamyl transferase, total bile acid levels were 331 IU/L, 316 IU/L, 4.1 mg/dL, 19 IU/L and >300 mcmol/L, respectively. At most recent follow up at 8.5 year of age, the patient had survived with the native liver, but showed refractory pruritus, malnutrition and severe fat-soluble vitamin deficiency.
Patient B, an African American female, was diagnosed in infancy with PFIC2 because of chronic cholestasis and the genetic diagnosis of BSEP disease in the biological sister whose case was reported before (19). She rapidly progressed to liver failure with a total bilirubin level of 23.3 mg/dL and encephalopathy with hyperammonemia of 120 mcmol/L for which she underwent liver transplantation at 13 months of age. The liver tissues for the study were obtained from the explanted liver.
The control tissue originated from the explant of a 20-month-old male who underwent liver transplantation for urea cycle defect. Human study protocols conformed to the ethical guidelines of the 1975 declaration of Helsinki and were approved by the Cincinnati Children’s Hospital Medical Center (CCHMC) Institutional Review Board.
ZEBRAFISH
Wild type, Tg(EPV.Tp1-Mmu.Hbb:EGFP)um14, Tg(fabp10a:hBSEP-GFP)ci202, Tg(CMV:mCherry-map1lc3b)scf4tg, Tg(CMV:EGFP-map1lc3b)zf155Tg, and abcb11bci200+/− zebrafish were maintained under standard conditions (20) in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 86–23, revised 1985) and approved by the Institutional Animal Care and Use Committee at CCHMC. Animals of both genders were studied. Detailed methods for generating abcb11b mutant and Tg(fabp10a:hBSEP-GFP)ci202 transgenic zebrafish are provided in the Supporting Information.
BODIPY AND CHOLYLGLYCYLAMIDOFLUORESCEIN (CGAMF) FEEDING ASSAYS
For BODIPY assay, 6-day-old larvae were fed with 6.4 μM BODIPY C5.0 (Invitrogen, Waltham, MA) for 7 hours before being scored live for gallbladder fluorescence (13). For CGamF assay, 6-day-old larvae were incubated in egg water containing 10 μM CGamF (gift from Dr. Alan Hofmann, University of California San Diego) (21) in the dark at room temperature with agitation for 7.25 hours. Larvae were imaged live on a Nikon A1Rsi inverted confocal microscope (Nikon Instruments Inc., Melville, NY).
STATISTICAL ANALYSIS
One-way ANOVA and Tukey’s post-hoc test were performed using GraphPad Prism (GraphPad Software, La Jolla, CA). Statistical analyses of the BODIPY and CGamF feeding experiments were performed using Fisher’s exact test and Ridit test, respectively, followed by a pairwise comparison on R software (R-project, Vienna, Austria).
Results
IDENTIFICATION OF THE ZEBRAFISH ORTHOLOG OF ABCB11
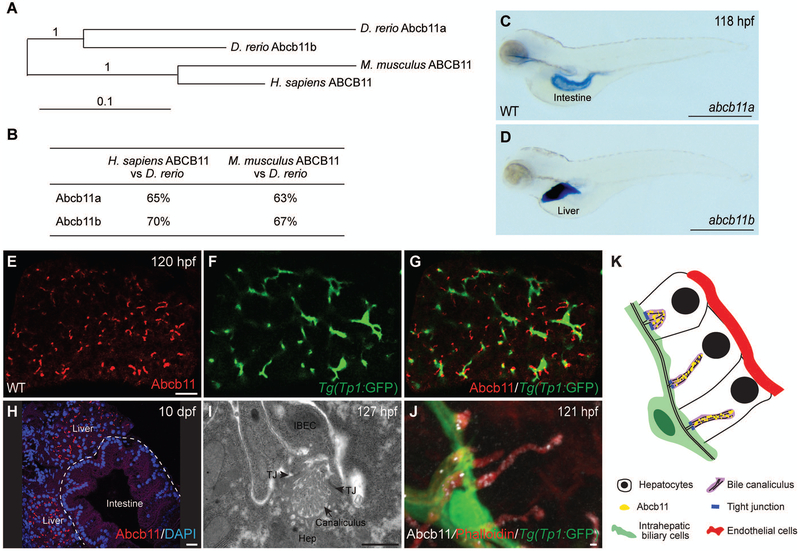
ENSEMBL genome assembly predicts that zebrafish possess two co-orthologs of the human ABCB11 gene (Fig. 1A). Zebrafish Abcb11a and Abcb11b proteins show 65% and 70% identity to human ABCB11, and are 63% and 67% identical to their mouse counterpart, respectively (Fig. 1B and Supporting Fig. S1). Whole-mount in situ hybridization of wild-type larvae showed that abcb11a transcript was enriched in the intestine (Fig. 1C), whereas abcb11b was specifically expressed in the liver (Fig. 1D). Zebrafish genome contains 11 members of the Abcb gene family. By quantitative real-time PCR (qPCR), we found that in addition to abcb11b, 6 other Abcb genes were expressed at modest levels in the livers of 5 day-old wild-type larvae (Supporting Fig. S2A). Both mammalian Abcb11 and zebrafish abcb11b genes encode full transporters with two sets of transmembrane motifs and nucleotide binding folds (18). Among the other Abcb genes that are expressed in the larval livers, abcb4 also encodes a full transporter and is shown to be the ortholog of mammalian Abcb1 (22). abcb6b, abcb7, abcb8, abcb9, and abcb10 encode half transporters with only one transmembrane motif and one nucleotide binding fold (18). The similar gene expression pattern and protein sequence implicate that zebrafish abcb11b is the ortholog of mammalian Abcb11.
FIG. 1. The zebrafish ortholog of human ABCB11 gene, abcb11b, is expressed in bile canaliculi.
(A) Phylogenetic tree of the human, mouse, and zebrafish ABCB11 proteins. (B) ABCB11 protein sequence identity for zebrafish, mouse, and human. (C,D) Expression of the abcb11a and abcb11b transcripts in wild-type (WT) larvae at 118 hours post fertilization (hpf) by whole mount in situ hybridization. Lateral views, anterior to the left. (E-G) Confocal single-plane images showing expression of Abcb11 protein (E) and Tg(EPV.Tp1-Mmu.Hbb:EGFP)/Tg(Tp1:GFP) transgene that labels the intrahepatic biliary cells (F) in WT larva. (G) is a merged image of (E) and (F). Ventral views, anterior to the top. (H) Confocal image showing sagittal section of a WT larva at 10 days post fertilization (dpf): Abcb11 protein (red) is detected in the liver, but not in the intestine (marked by the dashed line). (I) TEM image of a bile canaliculus connecting a hepatocyte (Hep) and an intrahepatic biliary epithelial cell (IBEC) in a WT larva. TJ, tight junction. (J) Confocal three-dimensional projection image showing punctate expression of Abcb11 protein (white) along the bile canaliculi in a WT larva. The canaliculi can be recognized by bright phalloidin staining (red) due to its enrichment of F-actin cytoskeleton. Tg(Tp1:GFP) transgene expression (green) marks the intrahepatic biliary cells. (K) Diagram showing the organization of liver cells in zebrafish. Scale bars: (C,D) 1 mm; (E-H) 20 μm; (I,J) 1 μm.
In mammalian liver, the bile canaliculus is located at the apical surface of adjacent hepatocytes, with its lumen sealed by tight junctions to prevent diffusion of bile acids (23). Transmission electron microscopy showed that the zebrafish bile canaliculus is formed on the apical side of hepatocytes and sealed by tight junctions between the intrahepatic biliary cell and hepatocyte (Fig. 1I,K) (15, 24, 25). It was enriched with actin microvilli, similar to mammals. Using a polyclonal antibody raised against mouse ABCB11 (26), we detected Abcb11 protein in the zebrafish hepatocytes at 120 hours post fertilization (hpf) (Fig. 1E-H). High-magnification confocal imaging revealed punctate Abcb11 protein expression along the canaliculi (Fig. 1J), similar to the expression of mammalian ABCB11 (27).
GENERATION OF ABCB11B MUTANT ZEBRAFISH
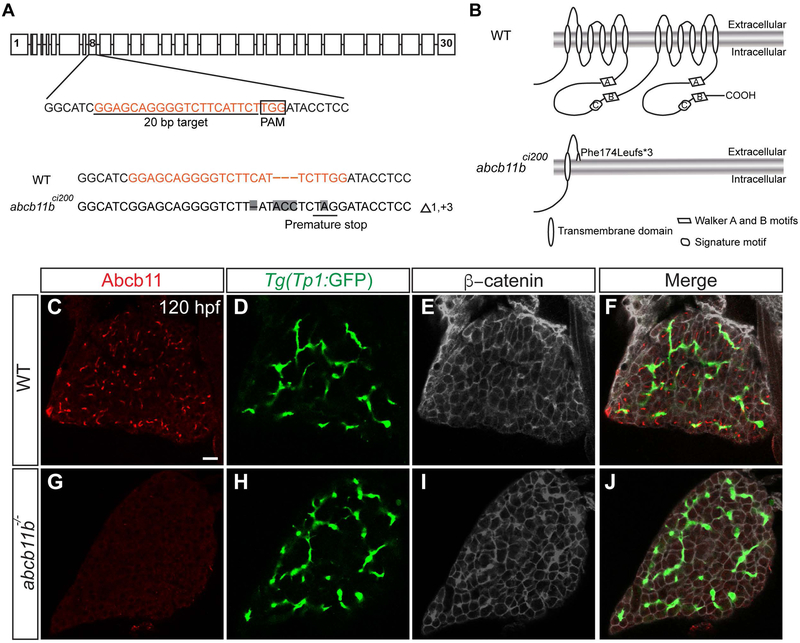
To determine whether zebrafish Abcb11b has similar function as mammalian ABCB11, we knocked out abcb11b in zebrafish by using CRISPR/Cas9-mediated genome editing technology (28). We targeted the eighth exon of the abcb11b gene and obtained the abcb11bci200 allele with a 1 bp deletion, a 3 bp insertion, and a T to A nucleotide change. The mutation resulted in frameshift and premature stop codon (Fig. 2A). The mutant protein is predicted to retain only the first transmembrane helix and the first extracellular loop, but lack the remaining 11 transmembrane helices, and both nucleotide binding folds (Fig. 2B). The mutation caused reduced abcb11b transcript expression and an absence of Abcb11b protein (Fig. 2C-J; Supporting Fig. S2B-C), and thus was likely nonfunctional. We did not observe an obvious increase in abcb11a transcript expression in the abcb11b mutant liver by in situ hybridization (Supporting Fig. S2D-E).
FIG. 2. Generation of abcb11b mutant zebrafish.
(A) Schematic representation of the zebrafish abcb11b locus along with the CRISPR target site, PAM motif, and WT and indel sequences. Nucleotide changes are highlighted in gray. (B) Putative topological models of WT (top) and ci200 mutant (bottom) Abcb11b protein. (C-J) Confocal single-plane images of the livers in WT and abcb11b mutant at 120 hpf. (C,G) Abcb11 protein expression; (D,H) intrahepatic biliary cells expressing Tg(Tp1:GFP) transgene; (E,I) β-catenin expression that marks the hepatocyte periphery; (F,J) merged images. All 10 WT and 10 mutant larvae examined showed the representative results. Ventral views, anterior to the top. Scale bar, 20 μm.
ABCB11B MUTANT ZEBRAFISH EXHIBITED HEPATOCELLULAR INJURY SIMILAR TO THOSE OBSERVED IN PATIENTS WITH PFIC2
We raised the embryos from the abcb11b+/− incrosses and started to observe mutant death at 11 dpf (Supporting Fig. S3A). Of the 54 fish that survived to adult, there were 19 wild types and 33 heterozygotes, but only two mutants (2/54 = 3.7%), confirming that the abcb11b mutation was lethal. At 11 dpf, the mutant fish were shorter than the wild-type and heterozygous siblings and had enlarged livers (Supporting Fig. S3B-C). H&E staining revealed hepatocyte swelling and necrosis (Fig. 3B, red box and arrow, respectively), and disorganized intestinal epithelium with increased apoptosis (Supporting Fig. S3D-F). qPCR analyses showed that the expression levels of hepatic function genes were drastically decreased in the abcb11b mutant livers at 10 dpf, right before they started dying (Supporting Fig. S3G). This result indicates that liver function is severely compromised in abcb11b mutants.
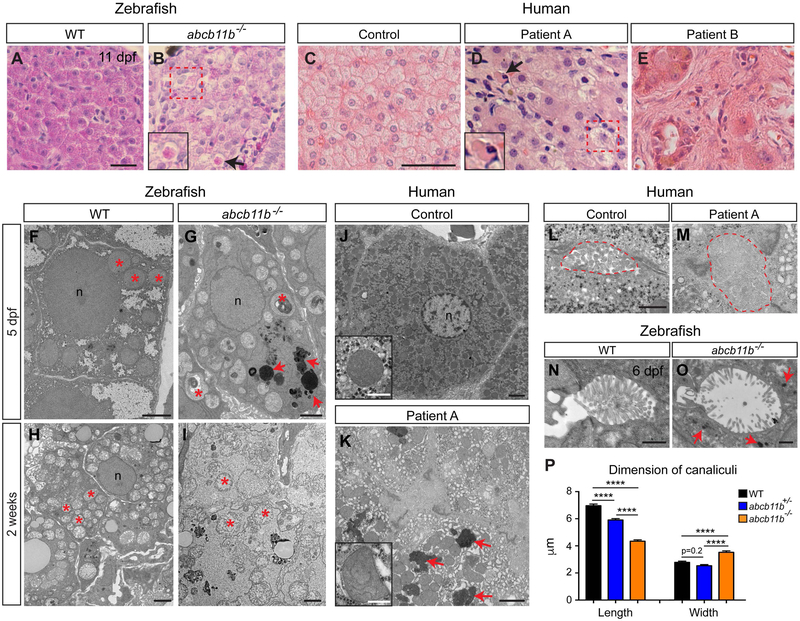
FIG. 3. abcb11b mutant developed hepatocellular injury similar to that observed in patients with PFIC2.
(A,B) H&E staining of the liver sections in WT (n=5) and abcb11b mutants (n=4) at 11 dpf. (C-E) H&E staining of the liver sections from control and two patients with PFIC2. (B,D) arrows point to necrotic hepatocytes with condensed eosinophilic cytoplasm (enlarged in inserts). Dashed red squares label the swollen hepatocytes. (F-I) TEM images of the hepatocytes in WT and abcb11b mutant zebrafish at 5 dpf and 2 weeks of age. 4 WT and 4 mutants were examined at both stages. Red asterisks mark mitochondria. (J,K) TEM images of the hepatocytes in control and patient A. Inserts show mitochondria. Red arrows in (G,K) point to electron-dense amorphous bile-like granular deposits. (F-K) n, nucleus. (L,M) TEM images of the bile canaliculi (marked by dashed lines) in control and patient A. (N,O) TEM images of the bile canaliculi in WT and abcb11b mutant zebrafish at 6 dpf. Red arrows in O point to amorphous bile-like deposits near the bile canaliculus. (P) Length and width (mean±s.e.m.) of the bile canaliculi in 5 dpf zebrafish. Measurements were performed on the confocal three-dimensional projections of 6 WT, 6 heterozygous, and 6 mutant larvae stained with phalloidin to mark the canaliculi. Statistical significance was calculated by one-way ANOVA and Tukey’s post-hoc test. ****, p<0.0001. Scale bars: (A,B) 20 μm; (C-E) 50 μm; (F-K) 2 μm, inserts: 500 nm; (L-O) 1 μm.
To compare the liver phenotypes in abcb11b mutant zebrafish with human PFIC2, we performed histological analyses on liver tissues from control and two patients with PFIC2. Patient A carries two heterozygous pathogenic variants, c.1408C>T(p.R470*) and c.3945delC(p.T1316Lfs*64), in the ABCB11 gene. The c.1408C>T variant is a previously reported nonsense mutation that abolishes ABCB11 protein expression (29, 30). The c.3945delC variant is predicted to cause frameshift that eliminates the stop codon and extends the open reading frame of the mutant allele for 57 extra amino acids. The resulting mutant protein is predicted to preserve all the transmembrane and nucleotide binding domains. H&E revealed normal liver architecture, minimal inflammation, patchy giant cell transformation (Fig. 3D), zone 3 pericellular fibrosis, and mild periportal fibrosis (data not shown). Patient B carries two heterozygous pathogenic variants, c.2782C>T(p.R928*) and c.3268C>T(p.R1090*), in the ABCB11 gene. Both variants were nonsense mutations causing an absence of ABCB11 protein (19). H&E revealed severely distorted lobular structure with prominent giant cells surrounded by fibrous tissue (Fig. 3E) (19). Similar to that seen in the zebrafish mutant, human PFIC2 exhibited hepatocyte swelling and necrosis (red box and insert in Fig. 3D, respectively). At 11 dpf, the zebrafish mutant hepatocytes had less defined boundaries compared to wild type, but no giant cells (Fig. 3B). Immunofluorescence showed increased laminin deposition in the mutant liver, suggestive of fibrogenic responses (Supporting Fig. S4A-F). However, the amount of extracellular matrix protein could not be readily detected by Sirius Red stain (data not shown). Premature death of the mutants prohibited evaluation of advanced fibrosis.
TEM showed that at 5 dpf, the hepatocytes in the zebrafish mutants had reduced glycogen content and accumulation of amorphous bile-like granular deposits (Fig. 3G, red arrows). The mitochondria were pleomorphic with irregularities in arrangement of cristae and often contained electron-dense whorled membranes (comparing Fig. 3F and G, red asterisks). The hepatocyte phenotypes progressively worsened over time. At 2 weeks, the mutant hepatocytes no longer had defined cell membrane (Fig. 3I). The mitochondria were swollen with pale matrix and disintegrating cristae (Fig. 3I, red asterisks). Similarly, the hepatocytes in patient A exhibited mitochondrial pleomorphism (Fig. 3K, insert), glycogen depletion, and accumulation of amorphous bile-like granular deposits (Fig. 3K, red arrows). There was modest dilatation of smooth endoplasmic reticulum (ER) and patchy increase in rough ER (Fig. 3K). ER stress was also induced in the zebrafish mutant livers as revealed by elevated expression of unfolded protein response genes (Supporting Fig. S4G). We observed dilatation of the bile canaliculi and effacement of canalicular microvilli in both the zebrafish mutants and human PFIC2 (Fig. 3M,O,P) (19). In the zebrafish mutants, bile-like granular deposits were detected in close proximity to the bile canaliculi (Fig. 3O, arrows), consistent with a failure of bile salt excretion across the canalicular membrane.
Taken together, zebrafish abcb11b mutants recapitulate several common histological and ultrastructural features of human PFIC2 (summarized in Table 1).
Table 1.
Liver histological and ultrastructural features in the spectrum of PFIC2 and zebrafish abcb11b mutant.
| Histological features | |||
|---|---|---|---|
| Patient A | Patient B | Zebrafish abcb11b mutant |
|
| Lobular structure | Largely preserved | Distorted | Largely preserved |
| Hepatocyte swelling | Yes | Yes | Yes |
| Hepatocyte necrosis | Yes | Yes | Yes |
| Giant cells | Yes | Yes | Not evident |
| Fibrosis | Moderate | Severe | Mild |
| Inflammation | Mild | Mild | Not evident |
| Bile duct paucity or ductular reaction | No | No | No |
| Ultrastructural changes | |||
| Amorphous bile-like granular deposits | Yes | Yes | Yes |
| Bile canaliculi dilatation, effacement of microvilli | Yes | Yes | Yes |
| Glycogen depletion | Yes | Yes | Yes |
| Mitochondria pleomorphism | Yes | No | Yes |
| Swelling of ER | Yes | Yes | Not evident |
BILE EXCRETION WAS IMPAIRED IN ABCB11B MUTANT ZEBRAFISH
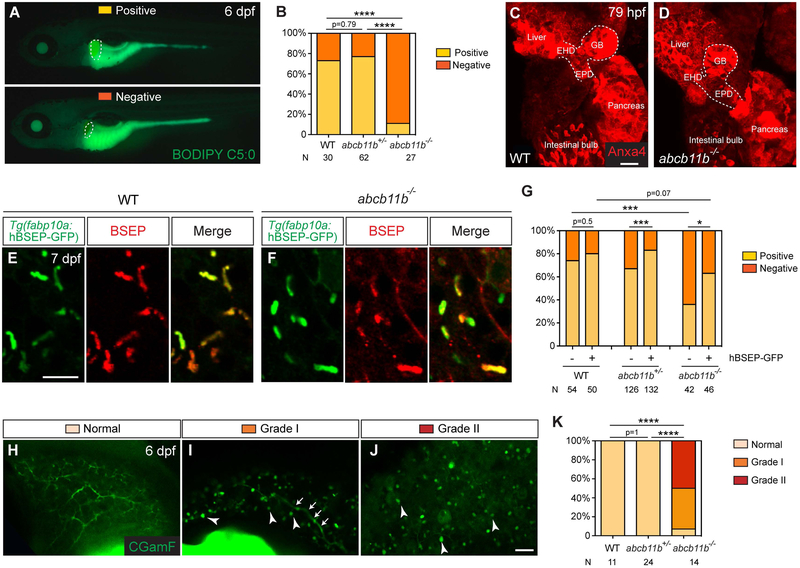
To evaluate hepatobiliary function of the abcb11b mutant larvae, we administered fluorescent lipid analog BODIPY C5:0 that consists of saturated acyl chain of five-carbons tagged with the BODIPY fluorophore (13). After being ingested, BODIPY C5:0 is metabolized in the intestine, delivered to the liver, and secreted into bile by hepatocytes. The intrahepatic bile ducts and gallbladder exhibit green fluorescence signal under normal bile flow (31). At 6 dpf, 73% of the wild-type and 77% of the abcb11b heterozygous larvae, but only 11% of the abcb11b mutants showed green fluorescence signal in the gallbladder (Fig. 4A,B). Whole-mount immunostaining with the anti-Annexin A4 (Anxa4) antibody revealed an intact hepatopancreatic ductal system in the mutants (Fig. 4C,D) (32), suggesting that the lack of BODIPY fluorescence in the mutant gallbladder was not due to malformation of the extrahepatic duct or gallbladder.
FIG. 4. Bile excretion was impaired in abcb11b mutant larvae.
(A) Fluorescent micrographs of 6 dpf live larvae after BODIPY C5:0 feeding. The larva at the top showed filling of the gallbladder (Positive), and the one at the bottom had no fluorescence signal in the gallbladder (Negative). Lateral views, anterior to the left. Dashed lines outline the gallbladder. (B) Percentages of larvae with either positive or negative BODIPY filling of the gallbladder. (C,D) Confocal three-dimensional projections of larvae stained with the Anxa4 antibody at 79 hpf. The dashed lines outline the hepatopancreatic ductal system. EHD, extrahepatic duct; GB, gallbladder; EPD, extrapancreatic duct. 6 WT and 6 abcb11b mutant larvae were examined. Ventral views, anterior to the top. (E,F) Confocal single-plane images of 7 dpf larvae expressing the Tg(fabp10a:human BSEP-GFP) transgene (green). The GFP expression is largely colocalized with BSEP antibody staining (red). (G) Percentages of transgenic and control larvae with either positive or negative BODIPY filling of the gallbladder. (H-J) Confocal single plane images of the livers in 6 dpf live larvae after CGamF feeding. Arrowheads in (I,J) mark the CGamF signal close to the bile canaliculi. Arrows in (I) label the large bile duct connecting to the extrahepatic duct. Lateral views, anterior to the top. (K) Percentages of larvae showing different patterns of CGamF labeling. Statistical significance in (B, G) was calculated by Fisher’s exact test, in (K) was by Ridit test. *, p<0.05, ***, p<0.001, ****, p<0.0001. The numbers of animals analyzed for each genotype are indicated. Scale bar, (C,D, H-J) 20 μm; (E,F) 10 μm.
To confirm that zebrafish Abcb11b and human BSEP have conserved function, we generated transgenic construct that expresses human BSEP-GFP fusion protein under the control of the hepatocyte-specific promoter fabp10a (33). We injected the construct into wild-type and abcb11b mutant zebrafish at the one-cell stage and detected mosaic expression of the fusion protein in the hepatocytes (data not shown). The fusion protein was detected in the canaliculus, colocalized with BSEP antibody staining (Fig. 4E,F). We raised the injected fish to adult. Whereas in the uninjected sibling group, only 1 out of 39 adult fish was abcb11b mutant (2.6%), in the injected group, 5 out of 44 fish were the mutants (11.4%). Next we established stable transgenic fish and performed BODIPY assay on the larvae. 63% of the transgenic mutants showed positive BODIPY filling in the gallbladder (Fig. 4G). Thus expression of human BSEP in the hepatocytes of the mutant zebrafish restored bile flow and improved their survival.
To directly examine whether zebrafish Abcb11b is capable of exporting bile acid substrate of human BSEP, we administered fluorescent bile acid derivative cholylglycylamidofluorescein (CGamF) that has been used as surrogate for natural cholylglycine to monitor bile acid transport in mammalian hepatocytes (21). We examined the subcellular distribution of CGamF in the 6-dpf larval livers by confocal live imaging and observed three different patterns (Fig. 4H-J). In wild-type and abcb11b heterozygous larvae, CGamF fluorescence was detected throughout the intrahepatic biliary network, the “normal” pattern (Fig. 4H). The abcb11b mutant larvae had no obvious defect in bile duct morphology (Fig. 2H), but only few exhibited normal CGamF distribution (Fig. 4K). 43% of the mutants showed the “Grade I” pattern as CGamF signal was detected only in the large bile duct connecting to the extrahepatic duct (Fig. 4I). 50% of the mutants showed the “Grade II” pattern with no CGamF signal in the intrahepatic biliary network (Fig. 4J). In the mutant livers, we detected bright CGamF fluorescence signal close to the bile canaliculi (Fig. 4I,J, arrowheads), consistent with the TEM observation (Fig. 3O). These results confirm the requirement of zebrafish Abcb11b for bile excretion from hepatocytes.
LOSS OF ABCB11 CAUSED MISLOCALIZATION OF MDR1 IN BOTH HUMAN AND ZEBRAFISH
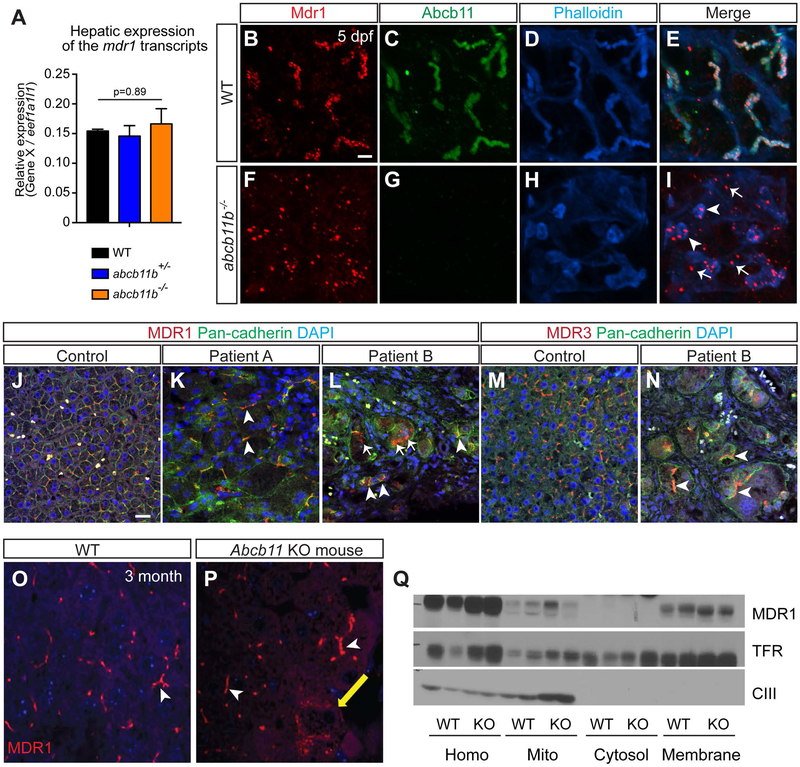
In Abcb11 knockout mice, increased expression of MDR1 is thought to compensate for the loss of BSEP, resulting in mild cholestasis (7). To explore the mechanisms underlying the severe cholestasis seen in abcb11b mutant zebrafish, we examined the expression of abcb4, which is the zebrafish ortholog of human MDR1/ABCB1 (22).Whereas Mdr1 transcript is upregulated in Abcb11 knockout mice (7), its expression was not significantly increased in the zebrafish abcb11b mutant livers compared to wild type (Fig. 5A). Mdr1 protein was distributed evenly along the canaliculi in the wild-type larvae (Fig. 5B-E). In abcb11b mutants, Mdr1 protein formed aggregates in the canaliculi (Fig. 5I, arrowheads). Abundant Mdr1 aggregates were also seen in the cytoplasm of the hepatocytes (Fig. 5I, arrows).
FIG. 5. MDR1 protein was mislocalized in abcb11b mutant zebrafish and a patient with nonsense mutations in ABCB11.
(A) qPCR analysis comparing mdr1 transcript expression in the livers of WT, heterozygous, and homozygous mutant larvae at 5 dpf. Triplicates were performed. The results are represented as relative expression levels that were normalized to the house keeping gene eef1a1l1 (mean±s.e.m.). Statistical significance was calculated by one-way ANOVA and Tukey’s post-hoc test. (B-I) Confocal three-dimensional projections of WT (n=10) and abcb11b mutant (n=10) livers at 5 dpf. (B,F) Mdr1 protein expression; (C,G) Abcb11 protein expression. (D,H) phalloidin staining; (E,I) merged images. Ventral views, anterior to the top. (J-L) Confocal single-plane images of liver sections from control and two PFIC2 patients that were stained with MDR1 antibody (red), Pan-cadherin antibody staining cell membranes (green), and DAPI (blue). Arrowheads in (I,K,L) mark MDR1 expression at the membrane. Arrows in (I,L) mark MDR1 expression in the hepatocyte cytoplasm. (M,N) Confocal single-plane images of liver sections from control and patient B stained with MDR3 antibody (red), Pan-cadherin antibody (green), and DAPI (blue). Arrowheads show that MDR3 was localized at the hepatocyte cell membrane. (B-N) Scale bars, 20 μm. (O,P) Confocal images showing MDR1 protein expression in the livers of WT and Abcb11 knockout mice at 3 month of age. White arrowheads mark MDR1 expression in the canaliculi and yellow arrow marks the expression in the cytoplasm. (Q) Subcellular fractionation assay showing that MDR1 protein is mainly expressed at the cell membrane in both WT and Abcb11 knockout mice. TfR is the transferrin receptor that marks the plasma membrane and CIII represents mitochondrial complex.
In human, all the hepatocytes from the control liver expressed MDR1 at the cell membrane (Fig. 5J, 636/636 cells) (34). In patient A who likely retains some BSEP function based on the mutations, 98% of the hepatocytes, including giant cells, expressed MDR1 at the cell membrane (Fig. 5K, arrowheads; 145/148 cells). In patient B with a complete loss of BSEP protein, however, 40% of the hepatocytes expressed MDR1 in the cytoplasm (Fig. 5L, arrows; 105/264 cells). MDR3, another canalicular transporter in the same protein family as BSEP and MDR1, is localized at the cell membrane in 99% of the hepatocytes in the control liver (Fig. 5M, 2769/2771 cells). 85% of the hepatocytes in patient B still showed membrane localization of MDR3 (Fig. 5N, arrowheads; 305/355 cells) (10), indicating that mislocalization of MDR1 seen in patient B is not caused by a universal change in canalicular transporter localization.
Our zebrafish and human data suggest a model that MDR1 is mislocalized in the absence of BSEP, thus is incapable of playing a compensatory role in bile excretion. This is in contrast to what is proposed for the Abcb11 knockout mice in which MDR1 may compensate for BSEP deficiency (7). We examined the localization of MDR1 in the knockout mice at 3 month by immunofluorescence and found that MDR1 was localized at the canalicular membrane in majority of the hepatocytes (Fig. 5O,P). Subcellular fractionation experiment further confirmed that MDR1 was mainly located on the cell membrane in the knockout mice (Fig. 5Q). Therefore, loss of BSEP has different impact on MDR1 localization in mouse hepatocytes versus zebrafish and human hepatocytes.
RAPAMYCIN TREATMENT PARTIALLY RESTORED BILE EXCRETION IN ABCB11B MUTANT ZEBRAFISH
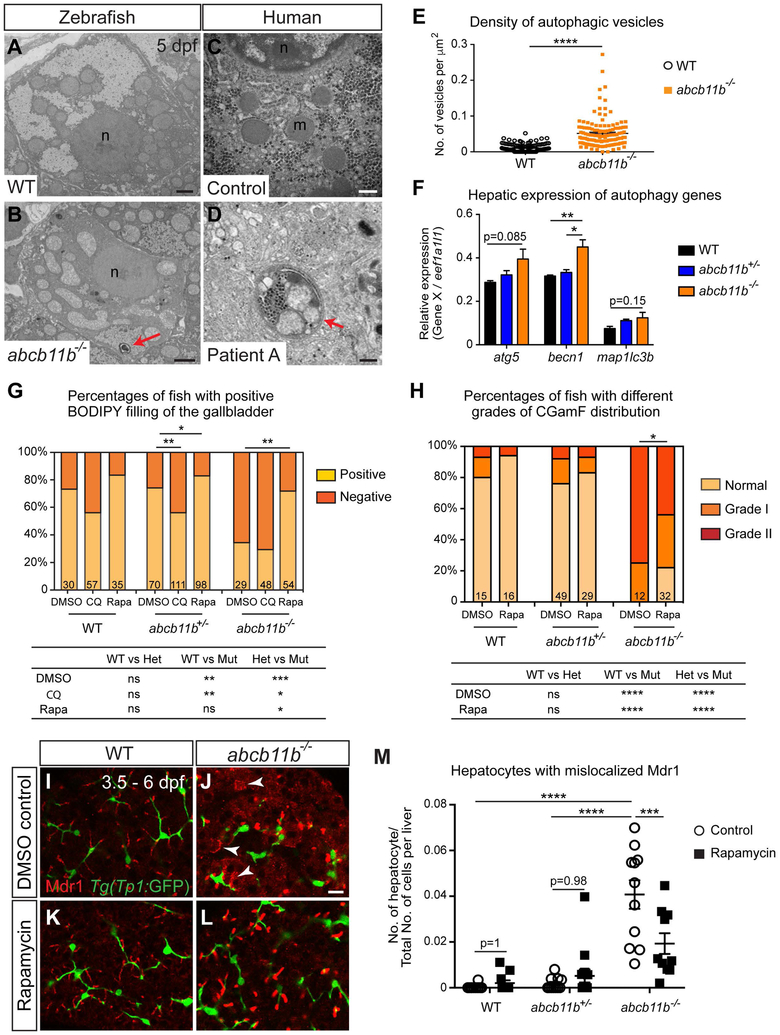
Our TEM analyses of the zebrafish abcb11b mutant hepatocytes detected double-membrane microbodies with cytoplasmic materials in the center, resembling autophagic vesicles (Fig. 6B, arrowhead). The density of these vesicles was significantly higher in the mutants than the wild types (Fig. 6E). qPCR revealed that expression of autophagic genes was elevated in the mutants compared to the wild types (Fig. 6F), further demonstrating activation of autophagy in the zebrafish abcb11b mutant livers. Autophagic vesicles were also seen in the hepatocytes from patient with PFIC2 (Fig. 6D).
FIG. 6. Rapamycin treatment partially restored bile excretion in abcb11b mutants.
(A-D) TEM images of the hepatocytes in WT larva, abcb11b mutant, human control, and patient A. Red arrowheads in (B,D) point to autophagic vesicles with double-layered membrane and cytosolic contents. n, nucleus; m, mitochondria. (E) Density (mean±s.e.m.) of the autophagic vesicles in 7 WT and 7 abcb11b mutants measured by the number of vesicles divided by the area of each hepatocyte. Each dot represents a hepatocyte. (F) qPCR analyses showing the expression of autophagy genes in zebrafish livers at 5 dpf. Triplicates were performed. The results are represented as relative expression levels that are normalized to the house keeping gene eef1a1l1 (mean±s.e.m.). (G) Percentages (mean±s.e.m.) of the larvae with positive or negative BODIPY filling in the gallbladder after being treated with DMSO, 50 μM CQ, or 5 μM rapamycin from 3.5 to 6 dpf. (H) Percentages of the fish with different grades of CGamF distribution after DMSO or rapamycin treatment. (G,H) The comparisons among different genotype groups for each experimental condition are shown at the bottom. (I-L) Confocal single-plane images of WT and abcb11b mutant livers after DMSO or rapamycin treatment. Mdr1 protein is in red, and the intrahepatic biliary cells are in green. Arrowheads mark Mdr1 expression outside of the apical membrane. Ventral views, anterior to the top. (M) The numbers of hepatocytes with mislocalized Mdr1 divided by the total numbers of cells within the liver, including both parenchymal and non-parenchymal cells (mean±s.e.m.). Each dot represents the result from individual liver. Scale bars: (A,B) 2 μm; (C,D) 500 nm; (I-L) 20 μm. Statistical significance in (E,F,M) was calculated by one-way ANOVA and Tukey’s post-hoc test, in (G) was by Fisher’s exact test, and in (H) was by Ridit test. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, not significant.
To investigate the relevance between autophagy and bile excretion defects in abcb11b mutant zebrafish, we treated the fish with autophagy inhibitor chloroquine (CQ) (35). We started the treatment at 3.5 dpf when we first detected Abcb11 protein in the bile canaliculi (36), and stopped at 6 dpf when the mutants showed obvious defects in bile excretion. CQ treatment caused a significant decrease in the percentage of abcb11b heterozygous larvae with positive filling of BODIPY (Fig. 6G). Thus inhibition of autophagy perturbed bile flow in the larvae with reduced Abcb11 protein expression. When we treated the larvae with rapamycin, an mTOR inhibitor known to induce autophagy, there was a significant increase in the percentage of abcb11b mutants with positive gallbladder filling (Fig. 6G). To further support that activation of autophagy restores bile flow, treatment with 2% trehalose, which induces autophagy by blocking glucose transport and is independent of mTOR (37), also improved BODIPY gallbladder filling in the mutants (Supporting Fig. S5). To confirm that rapamycin restored canalicular bile salt excretion, we administered CGamF to these animals. In the control group, 75% of the DMSO-treated mutants showed Grade II pattern with no CGamF signal in the intrahepatic bile ducts and none of them had normal pattern. In contrast, only 44% of the rapamycin-treated mutants showed Grade II pattern and 21% of them even exhibited normal CGamF distribution (Fig. 6H and Supporting Fig. S6A,B). TEM showed that rapamycin treatment rescued the ultrastructural phenotypes of the mutant hepatocytes: the bile canaliculi were less dilated, the microvilli were more densely packed, and bile deposits were no longer evident near the canaliculi (Supporting Fig. S6C-F). Rapamycin treatment did not cause significant changes in the body length and liver volume (Supporting Fig. S6G,H). Consistent with the improved liver phenotypes, more mutants survived at 2 weeks of age after continuous rapamycin treatment compared to egg water and DMSO-treated controls (Supporting Table S1). We examined the localization of Mdr1 and found that its expression was restored in the canaliculi in abcb11b mutants upon rapamycin treatment (Fig. 6L,M). These results suggest that rapamycin may prompt other transporters to excrete bile in abcb11b mutants.
Discussion
In this study, we show that in both human and zebrafish, loss of Abcb11 is associated with activation of autophagy, impaired bile salt excretion, and retention of amorphous bile-like deposits in hepatocytes. Ultimately, diminished canalicular transport results in severe liver damage. Treating zebrafish abcb11b mutants with rapamycin partially restores bile excretion and prolongs lifespan. It also correlates with recovery of Mdr1 localization from the cytoplasm to the canaliculi in the mutant hepatocytes. Our study establishes a novel animal model for studying BSEP deficiency that complements the existing in vitro systems and knockout mouse model. It also suggests that rapamycin may have therapeutic potential by directing alternative transporters to the canaliculus to compensate for BSEP deficiency.
Compensation from MDR1 may account for the mild cholestasis seen in BSEP knockout mice (7). We show that MDR1 protein is located on the canalicular membrane in these mice. In abcb11b mutant zebrafish, however, Mdr1 is mislocalized to the hepatocyte cytoplasm. Restoration of bile excretion by rapamycin treatment correlates with recovery of canalicular Mdr1 localization in the zebrafish mutant, raising the possibility that Mdr1 may play an evolutionarily conserved role as alternative bile acid transporter when positioned in the canaliculus. Conducting rapamycin treatment on abcb11b;mdr1 compound mutants will be informative to test this hypothesis. Our data also implicate that MDR1 localization may contribute to the phenotypic heterogeneity in patients with PFIC2: patient A with one truncating and one non-truncating ABCB11 mutation and preserved canalicular MDR1 expression has survived with the native liver beyond 8 years of age, whereas patient B with two truncating mutations and mislocalized MDR1 required liver transplantation in infancy to prolong survival. The correlation between MDR1 localization and disease severity is currently under investigation.
Mislocalization of MDR1 in human PFIC2 is not due to global impairment of hepatocyte polarity or bile canaliculus integrity because canalicular transporters MDR3 and MRP2 are located at the hepatocyte cell membrane in patients with PFIC2 (10). Newly synthesized canalicular transporters are targeted directly from the Golgi to the apical membrane of the hepatocytes (38). Both MDR3 and MRP2 contain a PDZ domain-binding motif at the C terminus and their apical localization relies on interactions with other PDZ domain proteins (39–41). In contrast, ABCB11 and MDR1 do not contain obvious PDZ domains. Differential regulation of intracellular trafficking may explain why MDR1, but not MDR3 and MRP2, is mislocalized in the absence of ABCB11. In rat hepatocytes ABCB11 and MDR1 are in the same complex at the canalicular membrane (42, 43). Both ABCB11 and MDR1 recycle between the apical cell membrane and the intracellular vesicles (38, 44). Thus it is possible that interaction between ABCB11 and MDR1 is required for their trafficking to the canaliculi.
We discover that the hepatocytes in abcb11b mutant zebrafish activate autophagy and that treatment with autophagy inducers rapamycin and trehalose restore their bile excretion. We also detect autophagic vesicles in two patients with PFIC2, and more patients should be examined to confirm this observation. Several studies suggest that autophagy could be a treatment target for cholestasis. Augmenting autophagy ameliorates liver injury in mice after bile duct ligation by eliminating accumulation of reactive oxygen species (45). Autophagic flux is impaired in livers of Fxr knockout mice and in primary mouse hepatocytes treated with bile acids (46). Increasing bile acid synthesis induces autophagy via CYP7A1-AKT-mTOR pathway in mice (47). However, to our knowledge, no previous report has specifically connected autophagy with canalicular bile salt excretion or PFIC2. Autophagy is required for polarization of the hepatocytes cultured in a collagen sandwich system (48). It is plausible that rapamycin increases autophagy and provides more recycled materials to enhance the synthesis of polarized plasma membrane domains, cytoskeleton, and intracellular organelles. This in turn promotes trafficking of other transporters, possibly Mdr1, to the canaliculi to excrete bile. Alternatively, rapamycin may also restore bile excretion through other autophagy-independent mechanisms.
Our work illustrates that abcb11b mutant zebrafish have the potential to reveal novel pathogenic mechanisms and therapeutic strategies for PFIC2. Different from the in vitro systems, abcb11b mutant zebrafish permit studying of BSEP deficiency in living organism. In human, PFIC2 is early onset and can rapidly progress to end-stage liver disease with cirrhosis, liver failure, and death commonly occurring in the first year of life (30). In contrast to Abcb11 knockout mice that survive to adult and only develop mild cholestasis (8), abcb11b mutant zebrafish show severe cholestasis and are premature lethal, similar to that seen in patients. Zebrafish bile is predominantly composed of bile alcohols, differing from human (49). However, the substrate spectrum of zebrafish Abcb11b and human ABCB11 may partially overlap as they can both transport CGamF. Furthermore, we show that expressing human BSEP in the zebrafish mutant hepatocytes rescues the liver phenotypes and improves mutant survival. These findings signify the relevance of the zebrafish mutants to human PFIC2. The small size of the zebrafish larvae and the ease of tracking liver cells and bile transport in live animals make it an ideal model for whole-organism chemical screens. abcb11b mutants can be applied in high-throughput chemical screens for compounds that restore bile acid excretion and ameliorate hepatobiliary damage. Given the high sequence similarity between human and zebrafish ABCB11 proteins, it will also be interesting to express human ABCB11 mutations in zebrafish to study their biological consequences.
Supplementary Material
Acknowledgments:
We would like to thank Drs Jorge Bezerra, William Balistreri, Kimberley Evason, Stacey Huppert, Shuji Kishi, and Akihiro Asai for critical comments and reagents, and Alan Hofmann for generous gift of CGamF. We acknowledge Dr. Matt Kofron at CCHMC Confocal Imaging Core for assistance with confocal imaging and post analyses, Dr. Lin Fei for statistical analyses, Cynthia Cline and Kari Huppert for immunohistochemistry, Drs Jordan Shavit and Megan Rost for coagulation analyses, CCHMC veterinary service for fish care, and Zenab Saeed for research assistance. This project was supported in part by NIDDK P30 DK078392 Integrative morphology core of the Digestive Disease Research Core Center in Cincinnati.
Financial support: This work was supported by National Institutes of Health Grant R00AA020514 (C.Y.), R01DK096001 (A.M.), HL114066, GM60904, and GM034496 (J.D.S.), ALSAC and Cancer Center support grant P30 CA021765, and funding from Center for Pediatric Genomics at Cincinnati Children’s Hospital Medical Center.
List of abbreviations:
- Anxa4
annexin A4
- ANOVA
analysis of variance
- ABC
Adenosine triphosphate-binding cassette
- BODIPY
4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Pentanoic Acid
- bp
base pairs
- BSEP
Bile salt export pump
- CCHMC
Cincinnati Children’s Hospital Medical Center
- cDNA
complementary DNA
- CGamF
cholylglycylamidofluorescein
- CQ
chloroquine diphosphate salt
- CRISPR
clustered regularly interspaced short palindromic repeats
- DAPI
4’,6-Diamidino-2-Phenylindole, Dihydrochloride
- DMSO
dimethyl sulfoxide
- dpf
days post fertilization
- EHD
extrahepatic duct
- EPD
extrapancreatic duct
- ER
endoplasmic reticulum
- gb
gallbladder
- Hep
hepatocyte
- Het
heterozygote
- hpf
hours post fertilization
- m
mitochondria
- Mdr1
multi-drug resistance protein 1
- mRNA
message RNA
- Mut
mutant
- n
nucleus
- ns
not significant
- PAM
protospacer adjacent motif
- PCR
polymerase chain reaction
- PFIC2
progressive familial intrahepatic cholestasis type 2
- qPCR
quantitative real-time PCR
- Rapa
rapamycin
- sgRNA
single-guide RNA
- TEM
transmission electron microscopy
- TfR
transferrin receptor
- TJ
tight junction
- Tp1:GFP
EPV.Tp1-Mmu.Hbb:EGFP
- Treh
trehalose
- UPR
unfolded protein response
- WT
wild type
REFERENCES
- 1.Boyer JL. Bile formation and secretion. Compr Physiol 2013;3:1035–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Childs S, Yeh RL, Georges E, Ling V. Identification of a sister gene to P-glycoprotein. Cancer Res 1995;55:2029–2034. [PubMed] [Google Scholar]
- 3.Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, et al. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem 1998;273:10046–10050. [DOI] [PubMed] [Google Scholar]
- 4.Knisely AS, Strautnieks SS, Meier Y, Stieger B, Byrne JA, Portmann BC, Bull LN, et al. Hepatocellular carcinoma in ten children under five years of age with bile salt export pump deficiency. Hepatology 2006;44:478–486. [DOI] [PubMed] [Google Scholar]
- 5.Jacquemin E Progressive familial intrahepatic cholestasis. Clin Res Hepatol Gastroenterol 2012;36 Suppl 1:S26–35. [DOI] [PubMed] [Google Scholar]
- 6.Telbisz A, Homolya L. Recent advances in the exploration of the bile salt export pump (BSEP/ABCB11) function. Expert Opin Ther Targets 2016;20:501–514. [DOI] [PubMed] [Google Scholar]
- 7.Lam P, Wang R, Ling V. Bile acid transport in sister of P-glycoprotein (ABCB11) knockout mice. Biochemistry 2005;44:12598–12605. [DOI] [PubMed] [Google Scholar]
- 8.Wang R, Salem M, Yousef IM, Tuchweber B, Lam P, Childs SJ, Helgason CD, et al. Targeted inactivation of sister of P-glycoprotein gene (spgp) in mice results in nonprogressive but persistent intrahepatic cholestasis. Proc Natl Acad Sci U S A 2001;98:2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Li F, Patterson AD, Wang Y, Krausz KW, Neale G, Thomas S, et al. Abcb11 deficiency induces cholestasis coupled to impaired beta-fatty acid oxidation in mice. J Biol Chem 2012;287:24784–24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keitel V, Burdelski M, Warskulat U, Kuhlkamp T, Keppler D, Haussinger D, Kubitz R. Expression and localization of hepatobiliary transport proteins in progressive familial intrahepatic cholestasis. Hepatology 2005;41:1160–1172. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Miller SR, Ober EA, Sadler KC. Making It New Again: Insight Into Liver Development, Regeneration, and Disease From Zebrafish Research. Curr Top Dev Biol 2017;124:161–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goessling W, Sadler KC. Zebrafish: an important tool for liver disease research. Gastroenterology 2015;149:1361–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev Biol 2011;360:276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, et al. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science 2001;292:1385–1388. [DOI] [PubMed] [Google Scholar]
- 15.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development 2004;131:5753–5766. [DOI] [PubMed] [Google Scholar]
- 16.Cui S, Leyva-Vega M, Tsai EA, EauClaire SF, Glessner JT, Hakonarson H, Devoto M, et al. Evidence from human and zebrafish that GPC1 is a biliary atresia susceptibility gene. Gastroenterology 2013;144:1107–1115 e1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorent K, Gong W, Koo KA, Waisbourd-Zinman O, Karjoo S, Zhao X, Sealy I, et al. Identification of a plant isoflavonoid that causes biliary atresia. Sci Transl Med 2015;7:286ra267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annilo T, Chen ZQ, Shulenin S, Costantino J, Thomas L, Lou H, Stefanov S, et al. Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics 2006;88:1–11. [DOI] [PubMed] [Google Scholar]
- 19.Evason K, Bove KE, Finegold MJ, Knisely AS, Rhee S, Rosenthal P, Miethke AG, et al. Morphologic findings in progressive familial intrahepatic cholestasis 2 (PFIC2): correlation with genetic and immunohistochemical studies. Am J Surg Pathol 2011;35:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westerfield M The zebrafish book A guide for the laboratory use of zebrafish (Danio rerio). 4th ed. ed. Eugene: Univ. of Oregon Press, 2000. [Google Scholar]
- 21.Holzinger F, Schteingart CD, Ton-Nu HT, Cerre C, Steinbach JH, Yeh HZ, Hofmann AF. Transport of fluorescent bile acids by the isolated perfused rat liver: kinetics, sequestration, and mobilization. Hepatology 1998;28:510–520. [DOI] [PubMed] [Google Scholar]
- 22.Fischer S, Kluver N, Burkhardt-Medicke K, Pietsch M, Schmidt AM, Wellner P, Schirmer K, et al. Abcb4 acts as multixenobiotic transporter and active barrier against chemical uptake in zebrafish (Danio rerio) embryos. BMC Biol 2013;11:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gissen P, Arias IM. Structural and functional hepatocyte polarity and liver disease. J Hepatol 2015;63:1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinton DE, Couch JA. Architectural pattern, tissue and cellular morphology in livers of fishes: relationship to experimentally-induced neoplastic responses. EXS 1998;86:141–164. [DOI] [PubMed] [Google Scholar]
- 25.Langer M [Histological study of the teleost liver. III. The system of biliary pathways]. Z Mikrosk Anat Forsch 1979;93:1105–1136. [PubMed] [Google Scholar]
- 26.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol 2008;18:1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childs S, Yeh RL, Hui D, Ling V. Taxol resistance mediated by transfection of the liver-specific sister gene of P-glycoprotein. Cancer Res 1998;58:4160–4167. [PubMed] [Google Scholar]
- 28.Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, Montague TG, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 2014;9:e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davit-Spraul A, Beinat M, Debray D, Rotig A, Slama A, Jacquemin E. Secondary Mitochondrial Respiratory Chain Defect Can Delay Accurate PFIC2 Diagnosis. JIMD Rep 2014;14:17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davit-Spraul A, Fabre M, Branchereau S, Baussan C, Gonzales E, Stieger B, Bernard O, et al. ATP8B1 and ABCB11 analysis in 62 children with normal gamma-glutamyl transferase progressive familial intrahepatic cholestasis (PFIC): phenotypic differences between PFIC1 and PFIC2 and natural history. Hepatology 2010;51:1645–1655. [DOI] [PubMed] [Google Scholar]
- 31.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, et al. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet 2012;8:e1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D, Golubkov VS, Han W, Correa RG, Zhou Y, Lee S, Strongin AY, et al. Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Dev Biol 2014;395:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Her GM, Chiang CC, Chen WY, Wu JL. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett 2003;538:125–133. [DOI] [PubMed] [Google Scholar]
- 34.Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci U S A 1987;84:7735–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol 2009;625:220–233. [DOI] [PubMed] [Google Scholar]
- 36.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn 2010;239:855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mardones P, Rubinsztein DC, Hetz C. Mystery solved: Trehalose kickstarts autophagy by blocking glucose transport. Sci Signal 2016;9:fs2. [DOI] [PubMed] [Google Scholar]
- 38.Kipp H, Pichetshote N, Arias IM. Transporters on demand: intrahepatic pools of canalicular ATP binding cassette transporters in rat liver. J Biol Chem 2001;276:7218–7224. [DOI] [PubMed] [Google Scholar]
- 39.Harris MJ, Kuwano M, Webb M, Board PG. Identification of the apical membrane-targeting signal of the multidrug resistance-associated protein 2 (MRP2/MOAT). J Biol Chem 2001;276:20876–20881. [DOI] [PubMed] [Google Scholar]
- 40.Kocher O, Comella N, Gilchrist A, Pal R, Tognazzi K, Brown LF, Knoll JH. PDZK1, a novel PDZ domain-containing protein up-regulated in carcinomas and mapped to chromosome 1q21, interacts with cMOAT (MRP2), the multidrug resistance-associated protein. Lab Invest 1999;79:1161–1170. [PubMed] [Google Scholar]
- 41.Venot Q, Delaunay JL, Fouassier L, Delautier D, Falguieres T, Housset C, Maurice M, et al. A PDZ-Like Motif in the Biliary Transporter ABCB4 Interacts with the Scaffold Protein EBP50 and Regulates ABCB4 Cell Surface Expression. PLoS One 2016;11:e0146962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan W, Calderon G, Swift AL, Moseley J, Li S, Hosoya H, Arias IM, et al. Myosin II regulatory light chain is required for trafficking of bile salt export protein to the apical membrane in Madin-Darby canine kidney cells. J Biol Chem 2005;280:23741–23747. [DOI] [PubMed] [Google Scholar]
- 43.Ortiz DF, Moseley J, Calderon G, Swift AL, Li S, Arias IM. Identification of HAX-1 as a protein that binds bile salt export protein and regulates its abundance in the apical membrane of Madin-Darby canine kidney cells. J Biol Chem 2004;279:32761–32770. [DOI] [PubMed] [Google Scholar]
- 44.Fu D Where is it and How Does it Get There - Intracellular Localization and Traffic of P-glycoprotein. Front Oncol 2013;3:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao L, Lv G, Guo X, Jing Y, Han Z, Zhang S, Sun K, et al. Activation of autophagy protects against cholestasis-induced hepatic injury. Cell Biosci 2014;4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manley S, Ni HM, Kong B, Apte U, Guo G, Ding WX. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci 2014;137:478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Ding Y, Li J, Chavan H, Matye D, Ni HM, Chiang JY, et al. Targeting the Enterohepatic Bile Acid Signaling Induces Hepatic Autophagy via a CYP7A1-AKT-mTOR Axis in Mice. Cell Mol Gastroenterol Hepatol 2017;3:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu D, Mitra K, Sengupta P, Jarnik M, Lippincott-Schwartz J, Arias IM. Coordinated elevation of mitochondrial oxidative phosphorylation and autophagy help drive hepatocyte polarization. Proc Natl Acad Sci U S A 2013;110:7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res 2008;49:1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.