Abstract
Introduction
Primary progressive aphasia (PPA) is a neurological syndrome, associated with both frontotemporal dementia and Alzheimer’s disease, in which progressive language impairment emerges as the most salient clinical feature during the initial stages of disease.
Methods
We screened the main genes associated with Alzheimer’s disease and frontotemporal dementia for pathogenic and risk variants in a cohort of 403 PPA cases.
Results
In this case series study, 14 (3.5%) cases carried (likely) pathogenic variants: four C9orf72 expansions, nine GRN, and one TARDBP. Rare risk variants, TREM2 R47H and MAPTA152T, were associated with a three- to seven-fold increase in risk for PPA.
Discussion
Our results show that while pathogenic variants within the most common dementia genes were rarely associated with PPA, these were found almost exclusively in GRN and C9orf72, suggesting that PPA is more TDP43- than tau-related in our series. This is consistent with the finding that PPA frequency in dominantly inherited dementias is the highest in kindreds with GRN variants.
Keywords: Primary progressive aphasia, Genetics, C9orf72, GRN, TARDBP
1. Introduction
Primary progressive aphasia (PPA) is a neurodegenerative syndrome characterized by prominent, relatively isolated language impairment that develops gradually, while other cognitive and behavioral domains are relatively preserved [1]. In the early stages of disease, activities of daily living (except those related to language) are maintained; however, some patients go on to develop more widespread symptoms of dementia or rto present behavioral deficits typical of the behavioral variant of frontotemporal dementia (FTD). In recent years, PPA has been divided into three main distinct variants: progressive nonfluent agrammatic, semantic, and logopenic [2].
Nonfluent variant PPA (nfvPPA), also known as progressive nonfluent aphasia or PPA-agrammatic, is mainly characterized by disrupted language production (short, simple phrase structure and omission of grammatical morphemes) and slow, labored speech. Apraxia of speech is common, but single-word comprehension is preserved. Many patients with nfvPPA will eventually progress to a syndrome with generalized motor problems, such as corticobasal syndrome or progressive supranuclear palsy [3,4]. One of the most distinctive features of nfvPPA is atrophy within the left inferior frontal gyrus, where Broca’s area is located [5]. Nonfluent cases with agrammatism or motor speech disturbance will most often show frontotemporal lobar degeneration (FTLD) with tau or, less often, FTLD with TDP-43, usually of type A, pathologies.
Semantic variant PPA (svPPA), also known as semantic dementia, is mainly characterized by profound impairment in object naming and single-word comprehension deficits, especially for low-frequency items. Other features include surface dyslexia (reading that is literally phonetic) and dysgraphia, whereas articulation, grammar, and repetition are spared. These patients often show a very distinct pattern of atrophy in the ventral and lateral portions of the anterior temporal lobes, sometimes predominantly left-sided, but often bilateral [6]. This variant has been associated with FTLD with TDP-43 pathology, usually of type C [7].
In the logopenic variant PPA (lvPPA), patients have halting anomic speech, with single-word retrieval deficits, and often show inability to repeat phrases and sentences, while grammatical processing, word comprehension, and motor speech are relatively preserved. These patients show abnormalities in the posterior (temporoparietal) part of the language network [5], with Alzheimer’s disease (AD) being the most common underlying pathology [8,9].
Although most PPA cases are sporadic, there have been reports of familial PPA cases with disease-causing variants in the three most common FTD genes, MAPT, GRN, and C9orf72, and also in the AD gene PSEN1. However, the overall genetic contribution to each PPA variant, and its underlying processes, is not clear. Based on underlying pathology mostly associated with the distinct PPA variants, it would be expected to observe mostly MAPT and perhaps some GRN variants in nfvPPA cases, GRN/TARDBP in svPPA cases, and PSEN1/PSEN2/APP variants in lvPPA. To test this hypothesis and to determine the relative frequency of pathogenic variants in the various PPA subgroups, we screened the main causative and risk-associated genes for AD and FTD in the largest cohort of PPA cases to date.
2. Methods
2.1. Cohort description
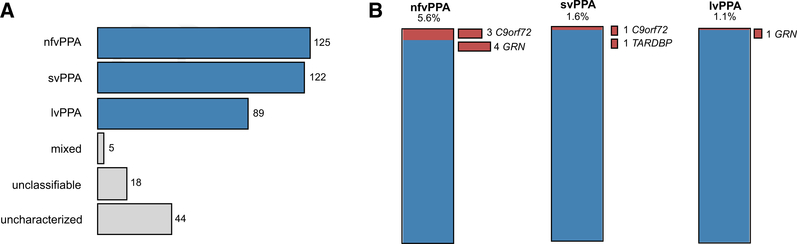
We screened 403 patients diagnosed with PPA recruited from two collaborating centers: University of California San Francisco Memory and Aging Center (UCSF series: 238 individuals) and Northwestern University (Northwestern series: 165 individuals). Application of consensus criteria for PPA (as described in [2,10] for the UCSF and Northwestern series, respectively) categorized patients into 125 nonfluent, 122 semantic, 89 logopenic, and five mixed PPA cases (Fig. 1A). The remaining cases were either unclassifiable (18 with too severe, mild, or other symptoms) or uncharacterized (44) at the time of inclusion. Two hundred nineteen cases (54.3%) were female, and most (at least 91.8%, no available information for 13 cases) were of European descent. All participants signed informed consent for genetic analyses.
Fig. 1.
Clinical and genetic characteristics of the PPA cohort. (A) Clinical categorization of the 403 PPA patients included in this study, following the consensus criteria for PPA. (B) Carriers of (likely) pathogenic variants in the main AD and FTD genes (red) among the three PPA variants. Abbreviations: PPA, logopenic variant primary progressive aphasia; nfvPPA, nonfluent variant PPA; svPPA, semantic variant PPA.
2.2. Targeted sequencing
Samples were screened using targeted sequencing of a panel of genes previously implicated in neurodegenerative disorders, including the most common causative genes for Mendelian forms of AD and FTD, and eight genes previously implicated in language and reading deficits (Supplementary Material). Exons and flanking intronic regions for these genes were captured using a custom-designed library (SeqCap EZ Choice Library, NimbleGen) and sequenced on an Illumina HiSeq2500 instrument at the UCLA Neuroscience Genomics Core (http://www.semel.ucla.edu/ungc). Sequence reads were mapped to the GRCh37/hg19 reference genome, and variants were joint-called using the GATK software [11]. The joint variant calling file was annotated using ANNOVAR [12].
2.3. Dementia genes screening
The exonic regions of the seven most common AD and FTD genes (APP TARDBP, FUS, GRN, MAPT, PSEN1, and PSEN2) were screened for known (AD&FTD Mutation Database: http://www.molgen.ua.ac.be/ADMutations) or novel pathogenic variants (according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP) published guidelines [13]). The transcripts NM_001136129 (APP), NM_001170634 (FUS), NM_002087 (GRN), NM_001123066 (MAPT), NM_000021 (PSEN1), NM_000447 (PSEN2), and NM_007375 (TARDBP) were used as reference. Known and novel (likely) pathogenic variants were confirmed by Sanger sequencing. The presence of a pathological hexanucleotide repeat expansion in C9orf72 was detected using both fluorescent and repeat-primed PCR, as previously described [14]. Genotypes for MAPT A152T (rs143624519), TREM2 R47H (rs75932628), and APOE (rs429358 and rs7412) risk alleles, and the 17q.21.31 (rs1560310) haplotype, were obtained using TaqMan® SNP assays and/or extracted from the targeted sequencing data. Statistical analysis was performed in R (version 3.1.3, http://www.r-project.org).
3. Results
3.1. Dominant FTD variants in a subset of PPA cases
Overall, we identified a total of 13 PPA cases that carried a pathogenic variant in the known AD- and FTD-associated genes: four with an expanded C9orf72 repeat and nine with GRN loss-of-function variants (Table 1). We also identified one case harboring a likely pathogenic variant in TARDBP, corresponding to a total frequency of 3.5% (14 of 403) carriers of causative variants in this PPA series. Most of these carriers (9 of 14) had a positive family history (i.e., first- or second-degree relatives with a clinical diagnosis of dementia), and their ages at onset ranged from 49 to 69 years. When stratified by PPA variant, we identified seven carriers of pathogenic variants among the 125 nfvPPA cases, corresponding to a frequency of 5.6% (Fig. 1B). Three of the nfvPPA cases carried a C9orf72 repeat expansion, whereas the other four had different pathogenic variants in the GRN gene. By contrast, within the 122 individuals diagnosed with svPPA, we identified one case with a C9orf72 repeat expansion and one with a likely pathogenic TARDBP variant (1.6%, Fig. 1B), whereas among the 89 lvPPA cases, we found only one carrier of a pathogenic GRN variant (1.1%, Fig. 1B).
Table 1.
Pathogenic variants identified within the main AD and FTD genes
| Patient | PPA variant | Gender | Age at onset | Family history* | Gene | Variant | gnomAD frequency (allele count) |
|---|---|---|---|---|---|---|---|
| 1 | svPPA | M | 51 | Yes | TARDBP | c.A1147G:p.Ile383Val | 1.949E-05 (5) |
| 2 | svPPA | M | 49 | No | C9orf72 | Repeat expansion | - |
| 3 | nfvPPA | F | 65 | Yes | C9orf72 | Repeat expansion | - |
| 4 | nfvPPA | M | 62 | N/A | C9orf72 | Repeat expansion | - |
| 5 | nfvPPA | F | 49 | Yes | C9orf72 | Repeat expansion | - |
| 6 | nfvPPA | F | 63 | No | GRN | c.264+2T>C | - |
| 7 | Uncharacterized | M | 57 | Yes | GRN | c.388_391delCAGT:p.Gln130SerfsTer125 | 7.217E-06 (2) |
| 8 | Uncharacterized | F | 56 | No | GRN | c.675_676delCA:p.Ser226TrpfsTer28 | - |
| 9 | Uncharacterized | M | 53 | Yes | GRN | c.709–2A>G | - |
| 10 | nfvPPA | F | 50 | Yes | GRN | c.910_911dupTG:p.Trp304CysfsTer58 | 4.064E-06 (1) |
| 11 | nfvPPA | M | 69 | Yes | GRN | c.1145delC:p.Thr382SerfsTer30 | 4.081E-06 (1) |
| 12 | nfvPPA | F | 64 | Yes | GRN | c.1256_1263dupGAAGCGAG:p.Ile422GlufsTer72 | - |
| 13 | lvPPA | F | 60 | Yes | GRN | c.C1477 T:p.Arg493Ter | 4.065E-06 (1) |
| 14 | Mixed | F | 67 | No | GRN | c.C1477 T:p.Arg493Ter | 4.065E-06 (1) |
Abbreviations: AD, Alzheimer’s disease; FTD, frontotemporal dementia; lvPPA, logopenic variant primary progressive aphasia (PPA); nfvPPA, nonfluent variant PPA; svPPA, semantic variant PPA.
Patients were considered to have family history if any of their first or second degree relatives had a clinical diagnosis of dementia.
Most of the GRN variants identified in this series—frameshift deletions Gln130SerfsTer125, Ser226TrpfsTer28, Trp304CysfsTer58, and Thr382SerfsTer30, splice variants c.264+2T>C and c.709–2A>G, and stop gain variant Arg493Ter—were already reported in the literature in association with FTD cases (first described [15–19]). We also identified one novel GRN loss-of-function variant: frameshift deletion Ile422GlufsTer72 (absent from the population database gnomAD, http://gnomad.broadinstitute.org/). In addition to these pathogenic variants, we also identified a known, likely pathogenic variant in the TARDBP gene, Ile383Val. This variant was first described as pathogenic in familial amyotrophic lateral sclerosis patients [20] and is associated with a substantial increase in TDP-43 truncation products in vitro [21]; however, it has also been reported in 5/128,243 individuals (minor allele frequency, MAF = 1.949E-05) in the gnomAD population database and is predicted to be tolerated by both SIFT and Polyphen, therefore we are considering it likely pathogenic. None of the rare, deleterious variants identified in the eight genes previously associated with language or reading deficits (Supplementary Material) had sufficient evidence to be classified as disease causing or risk associated.
3.2. MAPT A152T and TREM2 R47H rare variants increase risk for PPA
MAPT variant A152T, shown to reduce tau binding to microtubules while increasing tau oligomer formation, was identified as a genetic risk factor for both FTLD-spectrum disorders and AD [22,23]. In our PPA series, we identified a total of seven (1.74%) MAPT A152T carriers (three nonfluent, two logopenic, one semantic, and one mixed PPA cases). In our controls, recruited worldwide across collaborating centers, we identified only 10/4351 (0.23%) A152T carriers [23], consistent with the frequency reported in gnomAD (0.29%, with two homozygous carriers). A combined analysis performed on the 403 patients placed the estimated odds ratio (OR) for PPA in an individual carrying MAPT A152 T at 7.67 (CI: 2.462–22.45, Fisher’s P-value = .00027).
Another rare variant that has been, mostly, associated with AD risk is the R47H variant in the TREM2 gene [24,25]. We identified a total of six (1.49%) carriers of this variant (one logopenic, two nonfluent, and three uncharacterized/unclassifiable cases). In a subset of the National Institute of Mental Health controls, we identified 17/3855 (0.44%) carriers, a frequency similar to that reported in the gnomAD database (0.49%, with two homozygous carriers). A combined analysis placed the estimated OR for PPA in an individual carrying TREM2 R47H at 3.41 (CI: 1.09–9.13, Fisher’s P-value = .01735).
The APOE ε4 allele is the most prevalent genetic risk factor for sporadic AD, with a two- to three-fold increase in risk in people with one APOE ε4 allele and about 12-fold increase in those with two alleles (reviewed in [26]). We identified 114 carriers of at least one ε4 allele, of which 14 were ε4-ε4 (also six e2-ε4, and 94 ε3-ε4).
The 17q.21.31 haplotype, also commonly referred as MAPT/tau haplotype, has been linked to many neurological diseases, with H1 haplotype being consistently associated with progressive supranuclear palsy. In this series, most samples (273, 67.7%) had a H1-H1 tau haplotype, whereas 114 (28.3%) and 16 (4%) had a H1-H2 and H2-H2 haplotype, respectively.
4. Discussion
We report the largest genetic study of PPA to date, where we identified a total of 14 out of 403 cases with causative variants. We found mostly GRN pathogenic variant carriers, as well as four C9orf72 expansion and one likely pathogenic TARDBP variant carriers, while no pathogenic variants were identified in either MAPT or in the AD-associated genes, suggesting that PPA is more TDP43- than tau-related in our series. The frequency of disease-causing variants was higher within the nfvPPA cases (7 of 125, 5.6%), when compared to the semantic (2 of 122, 1.6%) and logopenic cases (1 of 89, 1.1%). Importantly, nfvPPA cases only carried GRN/C9orf72 variants and not MAPT mutations, as it would be expected based on the multiple reports of predominant tau pathology in nfvPPA.
Although disease-associated variants in the three most common FTD genes, MAPT, GRN, and C9orf72, and also a few in the AD genes, PSEN1 and TREM2, have been reported in the literature, these were mostly single PPA case reports (summarized in Table 2). Within this group, most nfvPPA cases were reported to carry either GRN or MAPT pathogenic variants, while PSEN1 variants and C9orf72 expansions were also reported. The number of case reports with disease-causing variants in svPPA is far scarcer, with MAPT and GRN causative variants associated with familial cases, while a protein-truncating variant in TREM2 has been reported in two sporadic svPPA cases, and a C9orf72 expansion in a few sporadic and familial cases. On the other hand, the few logopenic cases reported in the literature with causative variants have been primarily associated with GRN variants, and—in one case—with a C9orf72 expansion (Table 2).
Table 2.
Summary of pathogenic variants in PPA cases reported in the literature
| PPA variant | Gene | Variant | Number of cases | References |
|---|---|---|---|---|
| nfvPPA | GRN | p.Cys31LeufsTer34 | 1 familial | [27] |
| p.Gly35GlufsTer19 | 1 familial | [28] | ||
| IVS3 –2delA | 1 familial | [29] | ||
| p.Gln130SerfsTer124 | 1 familial | [27] | ||
| p.Cys139Arg | 1 sporadic | [30] | ||
| p.Cys157LysfsTer97 | 1 familial and 1 sporadic | [31–33] | ||
| p.Arg161GlyfsTer36 | 1 sporadic | [34] | ||
| IVS7+ 1delTGAG | 1 familial | [35] | ||
| IVS7–1G > A | 5 familial | [36] | ||
| p.Gln257ProfsTer27 | 1 familial | [37] | ||
| p.Thr272SerfsTer10 | 6 familial and 1 sporadic | [38,39] | ||
| p.Gln300Ter | 1 familial | [37] | ||
| p.Cys366fsTer1 | 1 familial | [30] | ||
| p.Gln415Ter | 1 sporadic | [27] | ||
| p.Val452TrpfsTer38 | 1 familial | [27] | ||
| p.Arg493Ter | 1 familial | [27] | ||
| p.Cys521Tyr | 1 familial | [40] | ||
| MAPT | p.Gly639Ser | 1 familial | [41] | |
| p.Val698Ile | 1 familial | [42] | ||
| p.Gly724Arg | 1 sporadic | [43] | ||
| C9orf72 | repeat expansion | 3 familial | [30,44] | |
| 1 sporadic | ||||
| PSEN1 | p.Thr147Ile | 1 familial | [45] | |
| p.Pro264Leu | 1 familial | [46] | ||
| svPPA | GRN | p.Thr409Met | 1 familial | [47] |
| MAPT | p.Pro636Leu | 1 familial | [48] | |
| IVS10+ 16C>T | 13 familial | [27] | ||
| C9orf72 | Repeat expansion | 1 familial and 2 sporadic | [44,49–51] | |
| 3 not specified | ||||
| TREM2 | p.Gln33Ter | 2 sporadic | [52] | |
| lvPPA | GRN | p.M1? | 1 familial | [53] |
| Not specified | 5 familial | [49,54] | ||
| 1 sporadic | ||||
| C9orf72 | Repeat expansion | 1 not specified | [55] |
Abbreviations: lvPPA, logopenic variant primary progressive aphasia (PPA); nfvPPA, nonfluent variant PPA; svPPA, semantic variant PPA.
Genetic screens of the three causative FTD genes in smaller PPA cohorts (32 and 100 cases) identified a few causative variants in GRN, as well as C9orf72 expansion carriers (Table 2) [30,49]. Although in one study (including 32 patients) all causative variants were identified in nfvPPA cases (two in GRN and two C9orf72 repeat expansions), in the larger study (including 100 patients), three logopenic cases carried GRN variants, while one semantic and one unclassified PPA had C9orf72 expansions. Although in these smaller genetic studies FTD causative genes by themselves accounted for about 5%–12.5%, in our large cohort, the frequency of PPA cases with causative variants in the eight most common dementia genes was only 3.5%. Low frequencies of familial PPA have been previously reported [56], and disease-causing variants in MAPT, GRN, and C9orf72 are mostly associated with familial cases. This could provide a possible explanation for such low frequency of carriers, as well as the fact that most FTD cases with these causative genes seem to be associated with the behavioral variant of FTD, not language variants.
Besides AD and FTD causative variants, we also screened our series for rare neurodegeneration risk variants: MAPT A152 T, associated with FTD and progressive supranuclear palsy [23], and TREM2 R47H, associated with AD [24,25]. We identified seven (1.74%) MAPT A152T carriers among the 403 PPA patients compared to 10/4351 (0.23%) in our controls, corresponding to a 7.7-fold increase in risk for PPA. The risk for PPA is not as high as that reported for progressive supranuclear palsy (OR = 8.13) but is considerably higher than the risk for FTD (OR = 3.35) [23]. Four of the seven MAPT A152T carriers in our series fall under the general heading of FTD (nfvPPA and svPPA), which could reflect a contribution of PPA cases to the effect observed in FTD [23]. On the other hand, many patients with nfvPPA eventually progress to progressive supranuclear palsy [3,4], suggesting that the MAPT A152T effect we observed in this series could also be driven by progressive supranuclear palsy cases initially manifesting as PPA.
We also observed a higher frequency of the rare TREM2 R47H variant (1.49%) in PPA patients compared to a subset of our controls (0.44%), suggesting that this variant also increase the risk for PPA (3.4-fold increase). This might be explained by the fact that (1) we included in our series a smaller subset of logopenic cases, for which AD is reported to be the most common underlying pathology [8,9]; (2) while we identified one logopenic case with the TREM2 R47H variant, the other carriers were mostly uncharacterized/unclassifiable PPA cases, so it remains possible that the TREM2 effect we observed in our series is driven by the lvPPA cases.
The APOE ε4 allele is by far the strongest genetic risk factor for AD, as over 40% of cases are ε4 carriers. In our series, 28.3% of PPA cases carried at least one APOE ε4 allele, while 3.5% carried two e4 alleles. These frequencies are quite similar to those reported in over 74,000 individuals from a prospective study in the Danish general population (30% ε4 and 2.8% ε4-ε4 carriers) [57]. Previous studies have also looked at the frequency of e4 carriers in PPA subjects, where frequencies ranged from 20% (in the FTLD-tau group) to 30% (in the AD group), similar to what was observed in their set of 190 control subjects (26% ε4 carriers) [7,58]. However, the rate of APOE ε4 positives in our series was higher, and closer to reported AD frequencies, among lvPPA patients (36.0% ε4 and 7.9% ε4-ε4 carriers vs. 21.6% and 1.6% in nfvPPA vs. 26.2% and 1.6% in svPPA), consistent with the reported association between lvPPA and AD pathology [8,9].
The H1 MAPT/tau haplotype has been consistently associated with progressive supranuclear palsy [59]. Most of our PPA cases were carriers of the H1-H1 haplotype, 67.8%, a frequency slightly higher than that reported in population databases, such as the 1000 Genomes and Wellderly projects (58%–64% for European populations) [60,61]. When categorized by each subset of PPA variants, the frequency of H1-H1 carriers in our series was higher among nfvPPA cases (72.0% vs. 64.0% in lvPPAvs. 65.6% in svPPA), which is consistent with the fact that nfvPPA cases are mostly associated with FTLD-tau pathology.
The present series suggested that there was no genetic feature that can reliably distinguish among the different PPA variants. For example, based on literature reports, we had hypothesized that nfvPPA cases would be mostly associated with MAPT mutations; however, we only observed C9orf72 and GRN mutations in this PPA subgroup. In addition, mutations in the same gene, for example, GRN, were associated with both nfvPPA and lvPPA syndromes, suggesting that the causal mechanism may be complex, with other risk alleles in different genes, possibly interacting with nongenetic factors and producing variation in clinical presentation. We studied four such candidates, rare variants in MAPT and TREM2, and common variants at the APOE and 17q21.31 loci, and found them to be overrepresented in PPA, sometimes in PPA subgroups, raising the possibility that rare and common variants may contribute to clinical presentation across PPA subgroups.
Like most genetic screens in clinically diagnosed syndromes, our study has a number of limitations: first, cases were recruited at two different US tertiary referral centers, which could lead to some ascertainment bias; second, although we applied uniform methods of disease phenotyping and genetic analysis, slight differences in the application of diagnostic criteria across these centers could also lead to measurement errors. Third, symptoms observed in PPA cases do not always clearly allow classification in one of the three variants, as shown by the 18 unclassifiable and 44 uncharacterized cases in our cohort.
In conclusion, our genetic screen of AD- and FTD-associated genes in the largest PPA cohort reported to date showed that both causative and risk-associated variants are rarely associated with PPA. Distinct clinical syndromes are not specifically associated with one pathology or one type of mutation, pointing to a complex etiology, possibly including rare and common risk-associated variants, and nongenetic factors. Further studies with even larger sample sizes will be needed to clarify this complex etiology and the genetic basis of the various clinical syndromes in PPA. Specifically, (1) pathological examination in all cases will reduce the inevitable measurement error associated with clinical diagnosis across multiple clinical centers and will provide valuable information about the relationship between asymmetry of degeneration, clinical presentation, and risk-associated genetic variants; (2) genome-wide sequence analysis will identify additional rare and common risk-associated variants; (3) calculation of polygenic risk scores [62] will contribute to estimating the genome-wide contribution of common variation to these phenotypes.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: Disease-causing variants in the frontotemporal dementia genes MAPT, GRN, and C9orf72, and also a few in the Alzheimer’s disease genes PSEN1 and TREM2, have been reported in the literature for patients diagnosed with primary progressive aphasia (PPA). However, these were mostly single case reports while large cohorts have not been studied in depth.
Interpretation: Our findings show that causative variants within the main dementia genes are rarely associated with PPA, accounting for only 3.5% of cases in our series, while known rare risk variants for Alzheimer’s disease and frontotemporal dementia also increase the risk for PPA.
Future directions: This study suggests that these language variants have a complex etiology and genetic basis, possibly including rare and common risk-associated variants, and nongenetic factors. Future studies with even larger sample sizes, including genome-wide sequence analysis and calculation of polygenic risk scores, will be needed to elucidate the genetic architecture of PPA.
Acknowledgments
The authors thank contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible.
Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials are being collected by the “Molecular Genetics of Schizophrenia II” (MGS-2) collaboration. The investigators and coinvestigators are as follows: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. (PI).
This work was supported by the NIH (RC1 AG035610 andR01 AG26938 to G.C., P50 AG023501 and P01 AG019724 to B.L.M., NS050915, AG052943, and DC015544 to M.L.G. -T., DC008552 and AG13854 to M.-M.M.), the John Douglas French Alzheimer’s Foundation, and the Tau Consortium. Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
There are no conflicts of interest to declare.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jalz.2018.10.009.
References
- [1].Mesulam MM. Slowly progressive aphasia without generalized dementia. Ann Neurol 1982;11:592–8. [DOI] [PubMed] [Google Scholar]
- [2].Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, et al. Classification of primary progressive aphasia and its variants. Neurology 2011;76:1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Josephs KA, Duffy JR, Strand EA, Whitwell JL, Layton KF, Parisi JE, et al. Clinicopathological and imaging correlates of progressive aphasia and apraxia of speech. Brain 2006;129:1385–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Knibb JA, Xuereb JH, Patterson K, Hodges JR. Clinical and pathological characterization of progressive aphasia. Ann Neurol 2006; 59:156–65. [DOI] [PubMed] [Google Scholar]
- [5].Gorno-Tempini ML, Dronkers NF, Rankin KP, Ogar JM, Phengrasamy L, Rosen HJ, et al. Cognition and anatomy in three variants of primary progressive aphasia. Ann Neurol 2004;55:335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mummery CJ, Patterson K, Price CJ, Ashburner J, Frackowiak RS, Hodges JR. A voxel-based morphometry study of semantic dementia: relationship between temporal lobe atrophy and semantic memory. Ann Neurol 2000;47:36–45. [PubMed] [Google Scholar]
- [7].Mesulam MM, Weintraub S, Rogalski EJ, Wieneke C, Geula C, Bigio EH. Asymmetry and heterogeneity of Alzheimer’s and frontotemporal pathology in primary progressive aphasia. Brain 2014; 137:1176–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mesulam M, Wicklund A, Johnson N, Rogalski E, Leger GC, Rademaker A, et al. Alzheimer and frontotemporal pathology in subsets of primary progressive aphasia. Ann Neurol 2008;63:709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rabinovici GD, Jagust WJ, Furst AJ, Ogar JM, Racine CA, Mormino EC, et al. Abeta amyloid and glucose metabolism in three variants of primary progressive aphasia. Ann Neurol 2008;64:388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mesulam MM, Rogalski EJ, Wieneke C, Hurley RS, Geula C, Bigio EH, et al. Primary progressive aphasia and the evolving neurology of the language network. Nat Rev Neurol 2014;10:554–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011;72:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006;442:916–9. [DOI] [PubMed] [Google Scholar]
- [16].Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet 2006;15:2988–3001. [DOI] [PubMed] [Google Scholar]
- [17].Gijselinck I, Van Broeckhoven C, Cruts M. Granulin mutations associated with frontotemporal lobar degeneration and related disorders: an update. Hum Mutat 2008;29:1373–86. [DOI] [PubMed] [Google Scholar]
- [18].Huey ED, Grafman J, Wassermann EM, Pietrini P, Tierney MC, Ghetti B, et al. Characteristics of frontotemporal dementia patients with a Progranulin mutation. Ann Neurol 2006;60:374–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spina S, Murrell JR, Huey ED, Wassermann EM, Pietrini P, Baraibar MA, et al. Clinicopathologic features of frontotemporal dementia with progranulin sequence variation. Neurology 2007; 68:820–7. [DOI] [PubMed] [Google Scholar]
- [20].Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet 2008;4:e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gendron TF, Rademakers R, Petrucelli L. TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J Alzheimer’s Dis 2013;33:S35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Coppola G, Chinnathambi S, Lee JJ, Dombroski BA, Baker MC, Soto-Ortolaza AI, et al. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer’s diseases. Hum Mol Genet 2012;21:3500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lopez A, Lee SE, Wojta K, Ramos EM, Klein E, Chen J, et al. A152T tau allele causes neurodegeneration that can be ameliorated in a zebra-fish model by autophagy induction. Brain 2017;140:1128–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med 2013;368:117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 2013;368:107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Michaelson DM. APOE epsilon4: the most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimers Dement 2014; 10:861–8. [DOI] [PubMed] [Google Scholar]
- [27].Pickering-Brown SM, Rollinson S, Du Plessis D, Morrison KE, Varma A, Richardson AM, et al. Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: comparison with patients with MAPT and no known mutations. Brain 2008;131:721–31. [DOI] [PubMed] [Google Scholar]
- [28].Skoglund L, Brundin R, Olofsson T, Kalimo H, Ingvast S, Blom ES, et al. Frontotemporal dementia in a large Swedish family is caused by a progranulin null mutation. Neurogenetics 2009;10:27–34. [DOI] [PubMed] [Google Scholar]
- [29].Cioffi SM, Galimberti D, Barocco F, Spallazzi M, Fenoglio C, Serpente M,et al. Non Fluent Variant of Primary Progressive Aphasia Due to the Novel GRN g.9543delA(IVS3–2delA) Mutation. J Alzheimers Dis 2016;54:717–21. [DOI] [PubMed] [Google Scholar]
- [30].Gil-Navarro S, Llado A, Rami L, Castellvi M, Bosch B, Bargallo N, et al. Neuroimaging and biochemical markers in the three variants of primary progressive aphasia. Demen Geriatr Cogn Disord 2013; 35:106–17. [DOI] [PubMed] [Google Scholar]
- [31].Caso F, Agosta F, Magnani G, Galantucci S, Spinelli EG, Galimberti D, et al. Clinical and MRI correlates of disease progression in a case of nonfluent/agrammatic variant of primary progressive aphasia due to progranulin (GRN) Cys157LysfsX97 mutation. J Neurol Sci 2014;342:167–72. [DOI] [PubMed] [Google Scholar]
- [32].Caso F, Villa C, Fenoglio C, Santangelo R, Agosta F, Coppi E, et al. The progranulin (GRN) Cys157LysfsX97 mutation is associated with nonfluent variant of primary progressive aphasia clinical phenotype. J Alzheimers Dis 2012;28:759–63. [DOI] [PubMed] [Google Scholar]
- [33].Milan G, Napoletano S, Pappata S, Gentile MT, Colucci-D’Amato L, Della Rocca G, et al. GRN deletion in familial frontotemporal dementia showing association with clinical variability in 3 familial cases. Neurobiol Aging 2017;53:193.e9–193.e16. [DOI] [PubMed] [Google Scholar]
- [34].Gazzina S, Archetti S, Alberici A, Bonomi E, Cosseddu M, Di Lorenzo D, et al. Frontotemporal Dementia due to the Novel GRN Arg161GlyfsX36 Mutation. J Alzheimers Dis 2017;57:1185–9. [DOI] [PubMed] [Google Scholar]
- [35].Galimberti D, Cioffi SM, Fenoglio C, Serpente M, Oblak AL, Rodriguez-Porcel F, et al. Rapidly progressive primary progressive aphasia and parkinsonism with novel GRN mutation. Mov Disord 2017; 32:476–8. [DOI] [PubMed] [Google Scholar]
- [36].Moreno F, Indakoetxea B, Barandiaran M, Alzualde A, Gabilondo A, Estanga A, et al. “Frontotemporoparietal” dementia: clinical phenotype associated with the c.709–1G>A PGRN mutation. Neurology 2009;73:1367–74. [DOI] [PubMed] [Google Scholar]
- [37].Takada LT, Bahia VS, Guimaraes HC, Costa TV, Vale TC, Rodriguez RD, et al. GRN and MAPT Mutations in 2 Frontotemporal Dementia Research Centers in Brazil. Alzheimer Dis Assoc Disord 2016;30:310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bonvicini C, Milanesi E, Pilotto A, Cattane N, Premi E, Archetti S, et al. Understanding phenotype variability in frontotemporal lobar degeneration due to granulin mutation. Neurobiol Aging 2014; 35:1206–11. [DOI] [PubMed] [Google Scholar]
- [39].Cerami C, Marcone A, Galimberti D, Villa C, Scarpini E, Cappa SF. From genotype to phenotype: two cases of genetic frontotemporal lobar degeneration with premorbid bipolar disorder. J Alzheimers Dis 2011;27:791–7. [DOI] [PubMed] [Google Scholar]
- [40].Cruchaga C, Fernandez-Seara MA, Seijo-Martinez M, Samaranch L, Lorenzo E, Hinrichs A, et al. Cortical atrophy and language network reorganization associated with a novel progranulin mutation. Cereb Cortex 2009;19:1751–60. [DOI] [PubMed] [Google Scholar]
- [41].Villa C, Ghezzi L, Pietroboni AM, Fenoglio C, Cortini F, Serpente M, et al. A novel MAPT mutation associated with the clinical phenotype of progressive nonfluent aphasia. J Alzheimers Dis 2011;26:19–26. [DOI] [PubMed] [Google Scholar]
- [42].Munoz DG, Ros R, Fatas M, Bermejo F, de Yebenes JG. Progressive nonfluent aphasia associated with a new mutation V363I in tau gene. Am J Alzheimers Dis Other Demen 2007;22:294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tacik P, DeTure MA, Carlomagno Y, Lin WL, Murray ME, Baker MC, et al. FTDP-17 with Pick body-like inclusions associated with a novel tau mutation, p.E372G. Brain Pathol 2017;27:612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AM, et al. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain 2012;135:693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Denvir J, Neitch S, Fan J, Niles RM, Boskovic G, Schreurs BG, et al. Identification of the PS1 Thr147Ile variant in a family with very early onset dementia and expressive aphasia. J Alzheimers Dis 2015; 46:483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mahoney CJ, Downey LE, Beck J, Liang Y, Mead S, Perry RJ, et al. The presenilin 1 P264L mutation presenting as non-fluent/agrammatic primary progressive aphasia. J Alzheimers Dis 2013; 36:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cerami C, Marcone A, Galimberti D, Villa C, Fenoglio C, Scarpini E, et al. Novel missense progranulin gene mutation associated with the semantic variant of primary progressive aphasia. J Alzheimers Dis 2013;36:415–20. [DOI] [PubMed] [Google Scholar]
- [48].Tacik P, Sanchez-Contreras M, DeTure M, Murray ME, Rademakers R, Ross OA, et al. Clinicopathologic heterogeneity in frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) due to microtubule-associated protein tau (MAPT) p.P301L mutation, including a patient with globular glial tauopathy. Neuropathol Appl Neurobiol 2017;43:200–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Flanagan EP, Baker MC, Perkerson RB, Duffy JR, Strand EA, Whitwell JL, et al. Dominant frontotemporal dementia mutations in 140 cases of primary progressive aphasia and speech apraxia. Dement Geriatr Cogn Disord 2015;39:281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cerami C, Marcone A, Galimberti D, Zamboni M, Fenoglio C, Serpente M, et al. Novel evidence of phenotypical variability in the hexanucleotide repeat expansion in chromosome 9. J Alzheimers Dis 2013;35:455–62. [DOI] [PubMed] [Google Scholar]
- [51].Galimberti D, Fenoglio C, Serpente M, Villa C, Bonsi R, Arighi A, et al. Autosomal dominant frontotemporal lobar degeneration due to the C9ORF72 hexanucleotide repeat expansion: late-onset psychotic clinical presentation. Biol Psychiatry 2013; 74:384–91. [DOI] [PubMed] [Google Scholar]
- [52].Borroni B, Ferrari F, Galimberti D, Nacmias B, Barone C, Bagnoli S, et al. Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol Aging 2014;35:934.e7–934.e10. [DOI] [PubMed] [Google Scholar]
- [53].Perry DC, Lehmann M, Yokoyama JS, Karydas A, Lee JJ, Coppola G, et al. Progranulin mutations as risk factors for Alzheimer disease. JAMA Neurol 2013;70:774–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Josephs KA, Duffy JR, Strand EA, Machulda MM, Vemuri P, Senjem ML, et al. Progranulin-associated PiB-negative logopenic primary progressive aphasia. J Neurol 2014;261:604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Saint-Aubert L, Sagot C, Wallon D, Hannequin D, Payoux P, Nemmi F, et al. A case of logopenic primary progressive aphasia with C9ORF72 expansion and cortical florbetapir binding. J Alzheimers Dis 2014; 42:413–20. [DOI] [PubMed] [Google Scholar]
- [56].Goldman JS, Farmer JM, Wood EM, Johnson JK, Boxer A, Neuhaus J, et al. Comparison of family histories in FTLD subtypes and related tauopathies. Neurology 2005;65:1817–9. [DOI] [PubMed] [Google Scholar]
- [57].Rasmussen KL, Tybjaerg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. Plasma levels of apolipoprotein E and risk of dementia in the general population. Ann Neurol 2015;77:301–11. [DOI] [PubMed] [Google Scholar]
- [58].Rogalski E, Sridhar J, Rader B, Martersteck A, Chen K, Cobia D, et al. Aphasic variant of Alzheimer disease: Clinical, anatomic, and genetic features. Neurology 2016;87:1337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 1999;8:711–5. [DOI] [PubMed] [Google Scholar]
- [60].Erikson GA, Bodian DL, Rueda M, Molparia B, Scott ER, Scott-Van Zeeland AA, et al. Whole-Genome Sequencing of a Healthy Aging Cohort. Cell 2016;165:1002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.