Abstract
Although relatively small in size, the ZZ-type zinc finger (ZZ) domain is a versatile signaling module that is implicated in a diverse set of cell signaling events. Here, we highlight the most recent studies focused on the ZZ domain function as a histone reader and a sensor of protein degradation signals. We review and compare the molecular and structural mechanisms underlying targeting the amino-terminal sequences of histone H3 and arginylated substrates by the ZZ domain. We also discuss the ZZ domain sensitivity to histone PTMs and summarize biological outcomes associated with the recognition of histone and non-histone ligands by the ZZ domain-containing proteins and complexes.
Keywords: ZZ domain, epigenetics, HAT, acetylation, chromatin, autophagy
Introduction
Recognition of epigenetic marks or post-translational modifications (PTMs) in histones by protein domains, known as readers, is required for a broad range of cellular events. Binding of readers to their cognate histone PTMs helps to define the place and timing for where the host proteins are recruited or stabilized within the genome (Strahl and Allis 2000; Jenuwein and Allis 2001; Kouzarides 2007). In the past two decades, a gigantic effort was put forth to characterize the biological outcomes associated with “reading” the epigenetic marks and identify new histone readers and novel sites and types of PTMs. Among numerous PTMs identified by mass spectrometry and biochemical analyses, acetylation (or acylation in general) of lysine residues, methylation of lysine and arginine residues, and phosphorylation of serine and threonine residues are the most common (Huang et al. 2015). Acetylation and phosphorylation of histones are generally associated with gene activation and cell growth, whereas methylation is linked to either gene activation or repression, depending on which lysine or arginine residue is being modified.
The first reader of acetyllysine PTM, bromodomain (BD), was discovered in 1999 (Dhalluin et al. 1999), followed by the finding of the methyllysine-recognizing modules, chromodomain (CD) (Bannister et al. 2001; Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002) and then a plant homeodomain (PHD) finger (Li et al. 2006; Peña et al. 2006; Shi et al. 2006; Wysocka et al. 2006). The family of epigenetic readers is growing fast and currently contains ~30 members that target unmodified histone tails or histone PTMs either specifically or promiscuously and modulate chromatin function, structure and dynamics (Taverna et al. 2007; Kutateladze 2011; Musselman et al. 2012; Andrews et al. 2016). The most recent addition to the family of epigenetic readers is the ZZ-type zinc finger (ZZ) domain that has been found to recognize unmodified and acetylated histone H3 tail (Mi et al. 2018; Zhang, Xue, et al. 2018), and some associate with nonhistone substrates.
In this work, we review the molecular and structural mechanisms underlying functioning of the ZZ domain as a histone reader and a sensor of protein degradation signals. We discuss the ZZ domain sensitivity to histone PTMs and summarize functional consequences associated with targeting histone and nonhistone ligands by the ZZ domain and its ability to facilitate diverse biological processes.
ZZ domain-containing proteins
Originally identified in dystrophin and its homologs through the sequence alignment (Ponting et al. 1996), the ZZ domain—named after its ability to bind two zinc ions—was found in a set of ~20 eukaryotic proteins. These proteins are implicated in various cellular processes, ranging from transcriptional regulation, chromatin remodeling and ubiquitination to cytoskeletal signaling and autophagy. For example, CBP and p300 are well-established transcriptional co-activators and major histone acetyltransferases (HATs), whereas cytoplasmic polyadenylation element-binding protein 1 (CPEB1) regulates mRNA polyadenylation and translation (Goodman and Smolik 2000; Richter 2007). Other ZZ domain-harboring proteins, ZZ-type zinc finger-containing protein 3 (ZZZ3) and transcriptional adapter 2-alpha/beta (TADA2a/b), are native components of the general control nonderepressible 5 (GCN5)-containing complexes that also acetylate histones (Wang YL et al. 2008; Wang and Dent 2014). The E3 ubiquitin-protein ligases Mib1/2 and HERC2 promote ubiquitination during Notch signaling and in response to DNA damage (Itoh et al. 2003; Danielsen et al. 2012). Dystrophin and related homologs are essential in connecting cytoskeleton with the surrounding extracellular matrix (Constantin 2014). Next to BRCA1 gene 1 protein (NBR1) and sequestosome-1 (SQSTM1/p62) are cargo receptors required for selective autophagy and degradation of protein aggregates (Kirkin et al. 2009; Moscat and Diaz-Meco 2009).
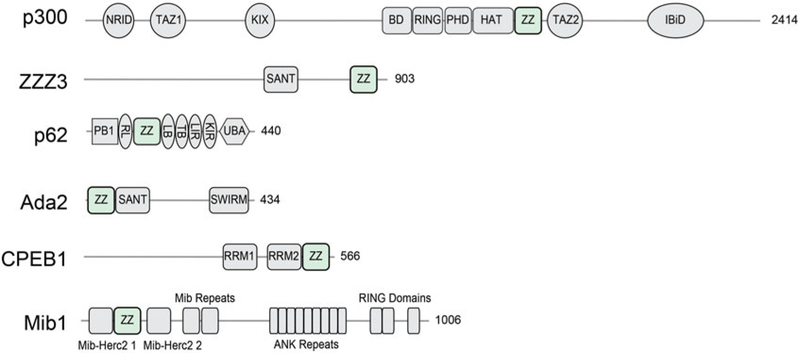
Recent studies have illumined the diverse activities of the ZZ domain itself (Figure 1). Pull-down assays using 16 ZZ domains derived from 15 human proteins reveal that nine ZZ domains are capable of binding to histone H3 tail (Mi et al. 2018). Specifically, the ZZ domain of p300 was shown to recognize unmodified histone H3, whereas the ZZ domain of ZZZ3 selects for the histone H3 tail acetylated on lysine 4 (H3K4ac) (Mi et al. 2018; Zhang, Xue, et al. 2018). The ZZ domain of HERC2 was defined as a SUMO binding module that promotes protein interactions and SUMOylation (Danielsen et al. 2012). While in Mib1 the ZZ domain plays a scaffolding role, the ZZ domain of yeast transcriptional adapter 2 (Ada2) associates with the N-terminal extension of the GCN5 HAT domain, and the ZZ domain of CPEB1 was suggested to interact with proteins and/or RNA (Afroz et al. 2014; McMillan et al. 2015; Sun et al. 2018). In addition to their roles in cellular processes occurred in the nucleus, some ZZ domains are essential in cytoplasmic signaling. In particular, recognition of N-terminally arginylated substrates by the ZZ domain of p62 has been shown to mediate selective autophagy (Cha-Molstad et al. 2015, 2017, 2018; Kwon et al. 2018; Zhang, Mun, et al. 2018).
Figure 1.
ZZ domain containing proteins. Schematic diagram showing domain architecture of representative ZZ-type zinc finger-containing proteins, including human p300 (UniProt Q09472), human ZZZ3 (UniProt Q8IYH5), human SQSTM1/p62 (UniProt Q13501), yeast Ada2 (UniProt Q02336), human CPEB1 (UniProt Q9BZB8) and human Mib1 (UniProt Q86YT6). The drawings are approximately in scale with the length of the protein (see color version of this figure at www.tandfonline.com/ibmg).
The ZZ domain is comprised of ^50 amino acids and topologically belongs to a large family of RING fingers that also includes other zinc fingers, such as the PHD finger and the FYVE domain (Joazeiro and Weissman 2000; Kutateladze 2006; Musselman and Kutateladze 2011). Although all RING fingers contain two clusters of cysteine/histidine residues that coordinate two zinc ions in a cross-braced manner, these modules have distinctly different biological functions and ligands which vary from proteins to RNA to phospholipids. Below, we review and compare the structure-function relationship within a subset of ZZ domains that specifically target the N-terminal sequences of histone H3 tail and arginylated substrates.
The ZZ domain of p300 binds to unmodified H3
P300 is involved in a number of vital signaling pathways that mediate cell proliferation, apoptosis and DNA repair and is associated with many human diseases (Goodman and Smolik 2000; Bedford et al. 2010; Dancy and Cole 2015). p300 has intrinsic HAT activity which markedly impacts chromatin structure and stimulates expression of specific genes. The central region of p300 contains a combination of closely linked bromodomain, a RING finger, a PHD finger and the catalytic HAT domain, followed by the ZZ domain of unclear function.
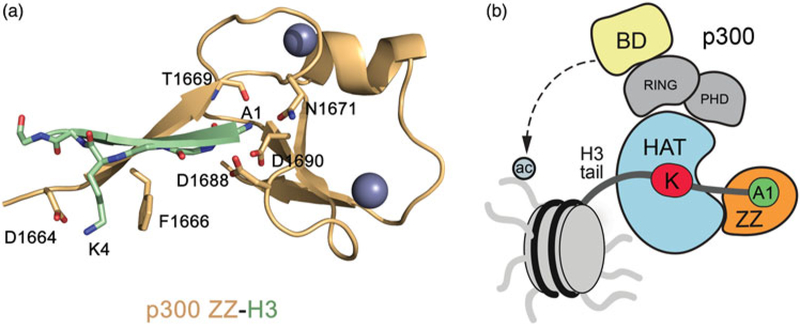
Using histone peptides containing various PTMs in immunoprecipitation and NMR assays, Zhang et al. have recently shown that the ZZ domain of p300 binds to the N-terminus of histone H3, and acetylation or methylation of H3K4 does not affect this interaction (Zhang, Xue, et al. 2018). A 9 µM binding affinity of the p300 ZZ domain to the unmodified H3 peptide, which is in the range of binding affinities of other histone-recognizing modules, further confirms that this domain is a novel epigenetic reader (Taverna et al. 2007; Musselman et al. 2012; Andrews et al. 2016). The crystal structure of the p300 ZZ domain-H3 complex reveals a unique H3-binding mechanism that differs from the binding mechanisms of other known H3-specific readers (Zhang, Xue, et al. 2018). The N-terminal residues of histone H3, Ala1-Gln5, make extensive intermolecular contacts with the ZZ domain, pairing with the β1-stand of the ZZ domain (Figure 2(a)). Ala1 of H3 occupies a highly negatively charged binding site, whereas Lys4 is sandwiched between the aromatic ring of phenylalanine and the negatively charged carboxylic group of aspartate, which forms a salt bridge with the ammonium group of Lys4. Such an arrangement most likely accounts for the indifference of the p300 ZZ domain toward acetylation or methylation of H3K4, which would augment cation-π and hydrophobic interactions with the phenylalanine but reduce the electrostatic contact with the aspartate.
Figure 2.
The ZZ domain of p300 binds to histone H3 and directs the HAT specificity of p300 toward H3K18 and H3K27. (a) A ribbon diagram of the complex (pdb 6DS6) between the p300 ZZ domain (light orange) and histone H3 tail (residues 1–6) (light green). (b) A schematic diagram for the regulation of the p300 HAT activity by the ZZ domain and bromodomain. Dashed line indicates interaction of bromodomain with acetylated histones (see color version of this figure at www.tandfonline.com/ibmg).
Most of histone readers recruit or stabilize their host proteins at specific chromatin regions, and the ZZ domain is not an exception. The robust association of the p300 fragment containing bromodomain, the RING and PHD fingers, the HAT domain and the ZZ domain (hereafter referred to as BRPHZ) with chromatin is shown to depend on concurrent interactions of the ZZ domain with H3 and of bromodomain with acetylated histones (Zhang, Xue, et al. 2018). Importantly, the H3-binding function of the ZZ domain is found to be vital for p300 to specifically acetylate H3K18 and H3K27 sites in vitro and in vivo. Mutation of the ZZ residues essential for binding to H3 drastically reduces the HAT activity of p300 BRPHZ on H3K27 and H3K18 but does not affect acetylation of other histone sites, such as H3K4 and H3K9. In contrast, mutation that disrupts acetyllysine binding of bromodomain results in a reduction of acetylation at all sites, with acetylation of H4 being most notably impeded. These observations lead to the conclusion that while acetylation of histones, H4 in particular, by p300 necessities the acetyllysine binding function of BD, specific acetylation of H3K18 and H3K27 (Jin et al. 2011; Tang et al. 2013) depends on the H3-binding activity of the ZZ domain (Figure 2(b)) (Zhang, Xue, et al. 2018).
Why does the recognition of H3 by the ZZ domain direct the p300 HAT activity toward H3K18 and H3K27? Considering that the ZZ domain and the catalytic HAT domain in p300 are only about two amino acids apart, Zhang et al. hypothesized that the ZZ domain acts as a ruler that limits the availability of histone H3 lysine residues to be primed in the active site of the HAT domain (Zhang, Xue, et al. 2018). A model of the BRPHZ:H3 complex generated using the simulated annealing method and the crystal structures of p300 BRPH (Delvecchio et al. 2013) and H3-ZZ reveals a ~38 Å distance between the H3A1-binding site of the ZZ domain and the catalytic site in the HAT domain (Zhang, Xue, et al. 2018). This distance is too long for Lys9 or Lys14 of H3 to occupy the active site of the HAT domain and thus be acetylated in cis when the N-terminus of H3 is locked through the interaction with the ZZ domain; however, other lysine residues (K18, K23, K27) in the H3 tail can reach the active site. We note that the ZZ-dependent HAT activity of p300 on the H3K23 site has not been tested, and therefore, it will be interesting to explore this idea. In support, the p300 region containing both the HAT and ZZ domains indeed binds to a long histone H3 peptide (residues 1–31 of H3) substantially stronger than either the isolated HAT domain or the isolated ZZ domain bind to the same peptide, demonstrating a cooperative binding of the natively linked HAT-ZZ domains and an in cis H3-binding mechanism.
Given the critical regulatory role of the ZZ domain in chromatin binding and HAT activity of p300, this reader could be a new target for therapeutic applications, as p300 and its paralog CBP are linked to a number of human diseases. Heterozygous mutations/deletions of CBP/p300 cause Rubinstein-Taybi syndrome, a genetic disorder characterized by mental retardation (Petrij et al. 1995; Solomon et al. 2015). P300/CBP are among the most frequently mutated genes in human cancers and are often mutated in B-cell lymphoma (Morin et al. 2011; Pasqualucci et al. 2011; Haery et al. 2014) and microsatellite instability colon cancer (Gayther et al. 2000; Ionov et al. 2004). High expression of p300/CBP is correlated with poor prognosis in prostate cancer (Debes et al. 2003), hepatocellular carcinomas (Yokomizo et al. 2011) and lung cancers (Hou et al. 2012; Gao et al. 2014). Overall, p300 and CBP have been shown to act as either tumor suppressors or oncogenes in a context-dependent manner.
The ability of p300/CBP to function in development also relies on targeting of chromatin and the HAT activity. Mechanistically, activation of key development genes depends on these two activities of p300/CBP. Particularly, the genes governing cell identity are associated with large and clustered enhancer elements containing very high levels of p300/CBP and H3K27ac and named super-enhancers (Hnisz et al. 2013; Whyte et al. 2013). Chromatin association of p300/CBP and production of H3K27ac are required for super-enhancers to drive transcription of the cell identity genes, which is crucial to development. In multiple types of cancers, the super-enhancers machinery is hijacked to sustain the high expression of oncogenes, potentiating carcinogenesis (Hnisz et al. 2013; Loven et al. 2013; Niederriter et al. 2015). Consequently, small-molecule inhibitors targeting p300/CBP bromodomain that displace p300/CBP from chromatin and disrupt the super-enhancers functions have been developed. These inhibitors show promising results in impeding the growth of hematopoietic malignancies (Hammitzsch et al. 2015; Conery et al. 2016; Ghosh et al. 2016), and inhibitors targeting the catalytic HAT domain of p300/CBP efficiently repress androgen-sensitive prostate cancer (Lasko et al. 2017; Lee et al. 2017) and acute myeloid leukemia (Giotopoulos et al. 2016). Because the functional ZZ domain is necessary for both recruitment of p300 to chromatin and generation of H3K27ac, inhibition of this reader could provide an efficient way to moderate both p300 activities in cancer.
The ZZ domain of ZZZ3 binds to H3K4ac
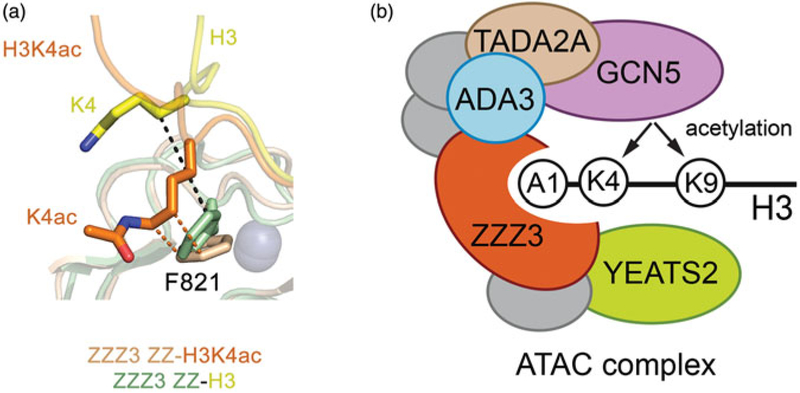
The human protein ZZZ3, a core component of the large multisubunit Ada2A-containing (ATAC) HAT complex (Wang et al. 2008; Wang and Dent 2014), harbors the ZZ domain in the C-terminus. Mi et al. have found that the ZZ domain of ZZZ3 binds to the histone H3 peptide, and while this binding is insensitive to methylation on H3K4 or H3R2, acetylation on H3K4 causes a moderate, ~3-fold enhancement effect (Mi et al. 2018). The selectivity of the ZZZ3 ZZ domain toward H3K4ac has been thoroughly characterized and confirmed by multiple orthogonal approaches, including peptide pulldowns and microarrays, microscale thermophoresis and fluorescence measurements, and NMR titration experiments, which collectively demonstrate that the ZZ domain of ZZZ3 is the first example of readers that select for the H3K4ac mark (Figure 3(a)).
Figure 3.
The ZZ domain of ZZZ3 recognizes histone H3K4ac and modulates ATAC-dependent histone acetylation. (a) Superimposition of the mean position of acetylated and unmodified Lys4 in the ensembles of the structures of the ZZZ3 ZZ-H3K4ac and ZZ-H3K4 complexes (pdb 6E83 and 6E86). Each ensemble contains 20 NMR structures. (b) Schematic diagram of the human ATAC complex and the interpaly between writing of and binding to the indicated acetylated PTMs (see color version of this figure at www.tandfonline.com/ibmg).
The preference of the ZZZ3 ZZ domain for H3K4ac distinguishes it from other nonselective ZZ domains (Mi et al. 2018). Comparison of the two solution structures of the ZZZ3 ZZ domain in complex with histone H3K4ac peptide and in complex with unmodified H3 peptide provides a possible explanation for such selectivity. Acetylation eliminates the positive charge in the lysine side chain and increases hydrophobicity. Although Lys4 lays in the same hydrophobic groove in both ZZZ3 complexes, the acetylated side chain of Lys4 in the ensemble of the NMR structures is positioned closer to the aromatic ring of phenylalanine compared to the position of the unmodified side chain of Lys4.
The ATAC complex is known to acetylate histone H3K9, H3K14 and H3K4 sites (Wang et al. 2008; Wang and Dent 2014). Mi et al. have proposed that recognition of H3 by the ZZZ3 ZZ domain could facilitate the association of the ATAC complex with substrates and thus promote H3 acetylation (Mi et al. 2018). Indeed, in vitro HAT assays show the robust acetylation activity of the ATAC complex on H3K9 but relatively weak activity on H3K4, and this activity requires functional ZZ domain of ZZZ3. When binding of the ZZZ3 ZZ domain to histone H3 is eliminated, this leads to a great reduction of the HAT activity of the ATAC complex on either histone peptide substrates or reconstituted nucleosomes (Figure 3(b)). Furthermore, Mi et al. demonstrate that the H3K4ac peptide is a better substrate than the unmodified H3 peptide for the ATAC complex. Consistent with the in vitro data, chromatin immunoprecipitation sequencing (ChIP-seq) and qPCR analysis using ZZZ3 knockdown cells shows a global reduction of H3K9ac and H3K4ac levels, which is specific to the ATAC target genes.
What is the role of ZZZ3 in the ATAC complex in regulation of gene expression? The ATAC complex has emerged later during evolution from the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex and is exclusive to multicellular eukaryotes (Spedale et al. 2012). Both the ATAC and SAGA complexes contain the conserved catalytic HAT subunit GCN5, or highly related p300/CBP-associated factor (PCAF), that mainly acetylates K9, K14 and K4 of histone H3, and the second acetyltransferase ATAC2 in the ATAC complex has been reported to modify H4K16 (Guelman et al. 2006; Suganuma et al. 2008; Riss et al. 2015). Although the SAGA complex has been extensively studied, biological activities of subunits within the ATAC complex, with the exception of GCN5, remain largely unknown.
Mi et al. (2018) show that ZZZ3 occupies primarily gene promoters and co-localizes with YEATS2 (YEATS domain containing 2), another component of the ATAC complex, and with H3K4ac, H3K9ac, and H3K4me3 in ChIP-seq experiments. Further analysis of the RNA-seq and ChIP-seq data reveals a set of downregulated and upregulated genes that are likely direct targets of ZZZ3. Although the up-regulated direct target genes are not significantly enriched in any pathway, the down-regulated genes are strongly enriched in the cell growth-related pathways, including ribosome biogenesis, DNA replication and the cell cycle. Rescue experiments using ectopically expressed wild type ZZZ3 or histone-binding deficient mutants in the ZZZ3-depleted cells provide an evidence that the ZZ domain of ZZZ3 is critical for the function of ATAC at chromatin. The finding that wild-type ZZZ3 but not loss-of-function ZZ-mutants restores ZZZ3 occupancy at the ATAC target gene promoters points to the essential role of histone binding by the ZZ domain for the association with chromatin. Furthermore, wild type ZZZ3 but not loss-of-function ZZ-mutants reinstates the ATAC-mediated H3K9 acetylation and global transcriptional changes in the ZZZ3-depleted cells, underscoring the importance of the functional ZZ domain for the enzymatic activity of the ATAC complex.
Besides the ZZ domain in ZZZ3, the ATAC complex contains several other epigenetic readers. These include bromodomain of GCN5 or PCAF that targets acetylated histones (Hudson et al. 2000; Zeng et al. 2008; Filippakopoulos et al. 2012), the H3K4me3-recognizing double Tudor domain of SGF29 (Vermeulen et al. 2010; Bian et al. 2011) and the YEATS domain of YEATS2 that binds to H3K27ac (Mi et al. 2017). Interestingly, both SAGA and ATAC are known to occupy gene promoter regions enriched in histone acetylation and H3K4me3; therefore, it is likely that the ZZ domain of ZZZ3 cooperates with other readers in the ATAC complex to facilitate its chromatin recruitment and catalytic activity.
The ZZ domain of p62 recognizes arginylated substrates
p62 functions as a major autophagic receptor and a positive mediator of the mTORC1 (mechanistic target of rapamycin complex 1) signaling pathway. It has multiple diverse binding partners and is known to be dysregulated in cancer and neurodegenerative disorders (Ramesh Babu et al. 2008; Mathew et al. 2009; Moscat and Diaz-Meco 2009; Duran et al. 2016; Todoric et al. 2017). Recent studies have shown that the ZZ domain of p62 recognizes a degradation signal in proteins – the N-terminal arginine residue (Nt-R) (Cha-Molstad et al. 2015, 2017, 2018). The Nt-R signal is typically generated either through proteolytic cleavage of the protein or enzymatically added by Arg-tRNA transferases.
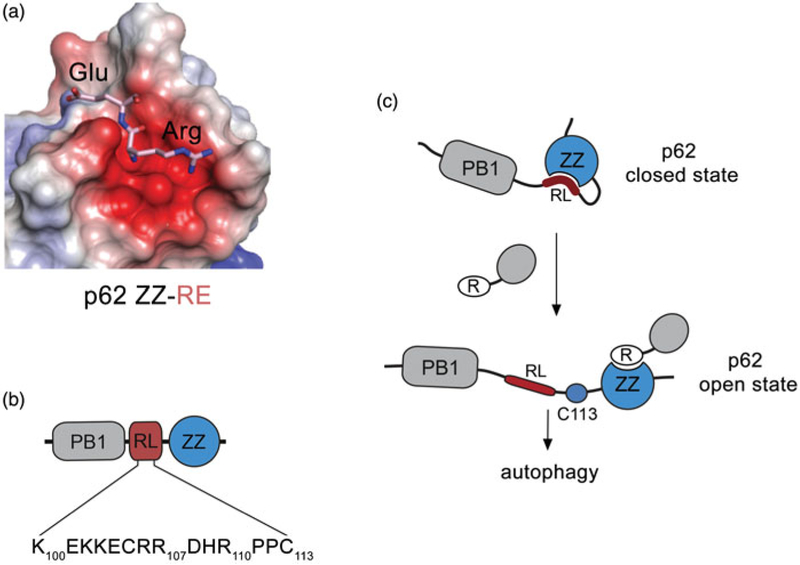
Two studies have elucidated the structural basis for the selective recognition of Nt-R by p62. Zhang et al. report the crystal structure of the p62 ZZ domain in complex with the amino acid sequence commonly found in arginylated substrates, Arg1-Glu2 (Zhang, Mun, et al. 2018), and Kwon et al. report the crystal structures of the p62 ZZ domain complexes with various type-1 and type-2 degradation signals, including the arginylated Arg1-Glu2 sequence (Kwon et al. 2018). In the complex, Arg1 is bound in the negatively charged pocket of the p62 ZZ domain (Figure 4(a)). The NH3+ group of Arg1 is coordinated almost identical to that of how the NH3+ group of Ala1 of H3 is coordinated in the p300 and ZZZ3 complexes; however, each complex is characterized by a unique set of intermolecular and intramolecular contacts that distinctively restrain each individual ligand, thus accounting for the diverse selectivity of these ZZ domains.
Figure 4.
The ZZ domain of p62 targets arginylated substrates and mediates selective autophagy. (a) Electrostatic surface potential of the p62 ZZ domain (pdb 6MIU) colored blue and red for the positive and negative charges, respectively. The bound Nt-R substrate (residues RE) is shown in stick. (b) Schematic representation of the N-terminal region of p62 containing the PB1 domain, RL, and the ZZ domain. (c) A model for potential p62 autoregulation. The ZZ domain binds to the RL region, locking p62 in a closed state. Upon binding of the ZZ domain to arginylated substrates, the autoinhibition is released. Cys113 in RL which is implicated in the formation of disulfide-linked conjugates is shown (see color version of this figure at www.tandfonline.com/ibmg.
In addition to its well-known function as an ubiquitinated cargo adaptor, new studies have implicated p62 in promoting selective autophagy through its ZZ domain (Zhang, Mun, et al. 2018). Particularly, functional p62 ZZ domain is required to induce p62 puncta formation and enhance macroautophagy, autophagy stimulated by proteasome inhibitor, and autophagy under starved conditions. Additionally, it plays a role in p62 aggregation, a process which also depends on the adjacent Phox and Bem1p (PB1) domain of p62 and the formation of disulfide conjugates.
The PB1 and ZZ domains in p62 are separated by a short, 20-residue linker, which contains two positively charged motifs that can mimic arginylated substrates as suggested by modeling of the peptide mimetics (Figure 4(b)). Indeed, Zhang et al. show that the p62 ZZ domain is capable of binding to the peptide corresponding to the residues 100–110 of p62, which is named a regulatory linker (RL). Interestingly, a similar set of NMR resonances of the p62 ZZ domain is perturbed by either arginylated substrate or RL, implying that the same binding site of the ZZ domain accommodates both ligands. The binding affinity of the p62 ZZ domain to the RL peptide is ~5-fold tighter than to a free arginine amino acid but ~60-fold weaker than to the arginylated substrate peptide REEE; however, the interaction of the ZZ domain with RL should be stronger in the physiologically relevant condition, because RL and ZZ are naturally connected in p62. Nevertheless, the difference in binding affinities of the p62 ZZ domain to Nt-R, RL, and arginine could direct p62 toward distinct autophagic and mTORC1 signaling pathways, and recognition of the internal RL sequence could further modulate p62 activities (Figure 4(c)) (Zhang, Mun, et al. 2018). Change in pH may provide an additional layer of regulation because the interaction between the p62 ZZ domain and Nt-R is pH sensitive, being the most robust around pH 5.5–6.0 (Kwon et al. 2018). Although the ideas of pH-dependent regulation and autoregulation involving RL need to be explored in cells, these findings provide a new exciting avenue for future studies aimed at understanding how p62 senses various stimuli and mediates different signaling pathways.
Concluding remarks
Altogether, recent reports have illuminated the ZZ domain as a versatile signaling module with diverse modes of interaction. Here, we summarized new findings regarding the ZZ domains that target the amino-terminal sequences of histone H3 and arginylated substrates, predominantly focusing on the three ZZ-containing proteins, p300, ZZZ3 and p62. In future studies, it will be important to characterize the structure-function relationships of other histone-binding ZZ domains and determine the precise roles the ZZ-histone interactions play in biological activities of their host proteins. Furthermore, it will be interesting to identify functions of seven ZZ domains insensitive to histone H3 (Mi et al. 2018), such as the ZZ domains of TADA2A and TADA2B that, intriguingly, are also components of the ATAC and SAGA complexes.
Acknowledgments
Funding
Research in the Kutateladze lab is supported by grants from NIH GM106416, GM125195 and GM100907 and research in the Shi lab is supported by grants from NIH CA204020 and Leukemia & Lymphoma Society (1339–17).
Footnotes
Disclosure statement
X.S. is a Scientific Advisory Board member of EpiCypher. All other authors declare no competing interests.
References
- Afroz T, Skrisovska L, Belloc E, Guillen-Boixet J, Mendez R, Allain FH. 2014. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev. 28: 1498–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews FH, Strahl BD, Kutateladze TG. 2016. Insights into newly discovered marks and readers of epigenetic information. Nat Chem Biol. 12:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 410:120–124. [DOI] [PubMed] [Google Scholar]
- Bedford DC, Kasper LH, Fukuyama T, Brindle PK. 2010. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 5:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.]. EMBO J. 30:2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Lee SH, Kim JG, Sung KW, Hwang J, Shim SM, Ganipisetti S, McGuire T, Mook-Jung I, Ciechanover A, et al. 2018. Regulation of autophagic proteolysis by the N-recognin SQSTM1/p62 of the N-end rule pathway. Autophagy. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Sung KS, Hwang J, Kim KA, Yu JE, Yoo YD, Jang JM, Han DH, Molstad M, Kim JG, et al. 2015. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 17:917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Yu JE, Feng Z, Lee SH, Kim JG, Yang P, Han B, Sung KW, Yoo YD, Hwang J, et al. 2017. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat Commun. 8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conery AR, Centore RC, Neiss A, Keller PJ, Joshi S, Spillane KL, Sandy P, Hatton C, Pardo E, Zawadzke L, et al. 2016. Bromodomain inhibition of the transcriptional coactivators CBP/EP300 as a therapeutic strategy to target the IRF4 network in multiple myeloma. eLife. 5:e10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin B 2014. Dystrophin complex functions as a scaffold for signalling proteins. Biochim Biophys Acta. 1838: 635–642. [DOI] [PubMed] [Google Scholar]
- Dancy BM, Cole PA. 2015. Protein lysine acetylation by p300/CBP. Chem Rev. 115:2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen JR, Povlsen LK, Villumsen BH, Streicher W, Nilsson J, Wikstrom M, Bekker-Jensen S, Mailand N. 2012. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 197:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes JD, Sebo TJ, Lohse CM, Murphy LM, Haugen DA, Tindall DJ. 2003. p300 in prostate cancer proliferation and progression. Cancer Res. 63:7638–7640. [PubMed] [Google Scholar]
- Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. 2013. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 20:1040–1046. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 399:491–496. [DOI] [PubMed] [Google Scholar]
- Duran A, Hernandez ED, Reina-Campos M, Castilla EA, Subramaniam S, Raghunandan S, Roberts LR, Kisseleva T, Karin M, Diaz-Meco MT, et al. 2016. p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell. 30:595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. 2012. Histone recognition and large-scale structural analysis of the human bromodomain family [Research Support, Non-U.S. Gov’t]. Cell. 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Geng J, Hong X, Qi J, Teng Y, Yang Y, Qu D, Chen G. 2014. Expression of p300 and CBP is associated with poor prognosis in small cell lung cancer. Int J Clin Exp Pathol. 7:760–767. [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, et al. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 24:300–303. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Taylor A, Chin M, Huang HR, Conery AR, Mertz JA, Salmeron A, Dakle PJ, Mele D, Cote A, et al. 2016. Regulatory T cell modulation by CBP/EP300 bromodomain inhibition. J Biol Chem. 291:13014–13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giotopoulos G, Chan WI, Horton SJ, Ruau D, Gallipoli P, Fowler A, Crawley C, Papaemmanuil E, Campbell PJ, Gottgens B, et al. 2016. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene. 35:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. 2000. CBP/p300 in cell growth, transformation, and development . Genes Dev. 14:1553–1577. [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, Washburn MP, Abmayr SM, et al. 2006. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 26:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haery L, Lugo-Pico JG, Henry RA, Andrews AJ, Gilmore TD. 2014. Histone acetyltransferase-deficient p300 mutants in diffuse large B cell lymphoma have altered transcriptional regulatory activities and are required for optimal cell growth. Mol Cancer. 13:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammitzsch A, Tallant C, Fedorov O, O’Mahony A, Brennan PE, Hay DA, Martinez FO, Al-Mossawi MH, de Wit J, Vecellio M, et al. 2015. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc Natl Acad Sci USA. 112:10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell. 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Li Y, Luo RZ, Fu JH, He JH, Zhang LJ, Yang HX. 2012. High expression of the transcriptional co-activator p300 predicts poor survival in resectable non-small cell lung cancers. Eur J Surg Oncol. 38:523–530. [DOI] [PubMed] [Google Scholar]
- Huang H, Lin S, Garcia BA, Zhao Y. 2015. Quantitative proteomic analysis of histone modifications. Chem Rev. 115: 2376–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol. 304:355–370. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Matsui S, Cowell JK. 2004. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc Natl Acad Sci USA. 101:1273–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. 2003. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 4:67–82. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 295(5562):2080–2083. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science. 293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 102:549–552. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 33:505–516. [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell. 128:693–705. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG. 2006. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta. 1761:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG. 2011. SnapShot: histone readers. Cell. 146: 842–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DH, Park OH, Kim L, Jung YO, Park Y, Jeong H, Hyun J, Kim YK, Song HK. 2018. Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat Commun. 9:3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 550:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yang B, Lindahl AJ, Damaschke N, Boersma MD, Huang W, Corey E, Jarrard DF, Denu JM. 2017. Identifying dysregulated epigenetic enzyme activity in castrate-resistant prostate cancer development. ACS Chem Biol. 12: 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 442:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 137:1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan BJ, Schnute B, Ohlenhard N, Zimmerman B, Miles L, Beglova N, Klein T, Blacklow SC. 2015. A tail of two sites: a bipartite mechanism for recognition of notch ligands by mind bomb E3 ligases. Mol Cell. 57:912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, Andrews FH, Wang X, Gagea M, Wen H, et al. 2017. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat Commun. 8:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Zhang Y, Lyu J, Wang X, Tong Q, Peng D, Xue Y, Tencer AH, Wen H, Li W, et al. 2018. The ZZ-type zinc finger of ZZZ3 modulates the ATAC complex-mediated histone acetylation and gene activation. Nat Commun. 9: 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 476:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 137:1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG. 2011. Handpicking epigenetic marks with PHD fingers [Research Support, N.I.H., Extramural Review]. Nucleic Acids Res. 39:9061–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederriter AR, Varshney A, Parker SC, Martin DM. 2015. Super enhancers in cancers, complex disease, and developmental disorders. Genes. 6:1183–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 416:103–107. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 471:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 442:100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ. 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 376:348–351. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ. 1996. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci. 21:11–13. [PubMed] [Google Scholar]
- Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, et al. 2008. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 106:107–120. [DOI] [PubMed] [Google Scholar]
- Richter JD. 2007. CPEB: a life in translation. Trends Biochem Sci. 32:279–285. [DOI] [PubMed] [Google Scholar]
- Riss A, Scheer E, Joint M, Trowitzsch S, Berger I, Tora L. 2015. Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) coactivator complexes enhance the acetyltransferase activity of GCN5. J Biol Chem. 290:28997–29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 442:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Bodian DL, Khromykh A, Mora GG, Lanpher BC, Iyer RK, Baveja R, Vockley JG, Niederhuber JE. 2015. Expanding the phenotypic spectrum in EP300-related Rubinstein-Taybi syndrome. Am J Med Genet. 167: 1111–1116. [DOI] [PubMed] [Google Scholar]
- Spedale G, Timmers HT, Pijnappel WW. 2012. ATAC-king the complexity of SAGA during evolution. Genes Dev. 26: 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature. 403:41–45. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. 2008. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 15:364–372. [DOI] [PubMed] [Google Scholar]
- Sun J, Paduch M, Kim SA, Kramer RM, Barrios AF, Lu V, Luke J, Usatyuk S, Kossiakoff AA, Tan S. 2018. Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2. Proc Natl Acad Sci USA. 115: 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, Sengoku T, McGinty RK, Fernandez JP, Muir TW, et al. 2013. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 154:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 14:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todoric J, Antonucci L, Di Caro G, Li N, Wu X, Lytle NK, Dhar D, Banerjee S, Fagman JB, Browne CD, et al. 2017. Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell. 32:824–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers [Research Support, Non-U.S. Gov’t]. Cell. 142: 967–980. [DOI] [PubMed] [Google Scholar]
- Wang L, Dent SY. 2014. Functions of SAGA in development and disease. Epigenomics. 6:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E. 2008. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 283:33808–33815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 442:86–90. [DOI] [PubMed] [Google Scholar]
- Yokomizo C, Yamaguchi K, Itoh Y, Nishimura T, Umemura A, Minami M, Yasui K, Mitsuyoshi H, Fujii H, Tochiki N, et al. 2011. High expression of p300 in HCC predicts shortened overall survival in association with enhanced epithelial mesenchymal transition of HCC cells. Cancer Lett. 310: 140–147. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. 2008. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 16:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mun SR, Linares JF, Ahn J, Towers CG, Ji CH, Fitzwalter BE, Holden MR, Mi W, Shi X, et al. 2018. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat Commun. 9:4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue Y, Shi J, Ahn J, Mi W, Ali M, Wang X, Klein BJ, Wen H, Li W, et al. 2018. The ZZ domain of p300 mediates specificity of the adjacent HAT domain for histone H3. Nat Struct Mol Biol. 25:841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]