Abstract
Background
Mathematical transmission models are increasingly used to guide public health interventions for infectious diseases, particularly in the context of emerging pathogens; however, the contribution of modeling to the growing issue of antimicrobial resistance (AMR) remains unclear. Here, we systematically evaluate publications on population-level transmission models of AMR over a recent period (2006–2016) to gauge the state of research and identify gaps warranting further work.
Methods
We performed a systematic literature search of relevant databases to identify transmission studies of AMR in viral, bacterial, and parasitic disease systems. We analyzed the temporal, geographic, and subject matter trends, described the predominant medical and behavioral interventions studied, and identified central findings relating to key pathogens.
Results
We identified 273 modeling studies; the majority of which (> 70%) focused on 5 infectious diseases (human immunodeficiency virus (HIV), influenza virus, Plasmodium falciparum (malaria), Mycobacterium tuberculosis (TB), and methicillin-resistant Staphylococcus aureus (MRSA)). AMR studies of influenza and nosocomial pathogens were mainly set in industrialized nations, while HIV, TB, and malaria studies were heavily skewed towards developing countries. The majority of articles focused on AMR exclusively in humans (89%), either in community (58%) or healthcare (27%) settings. Model systems were largely compartmental (76%) and deterministic (66%). Only 43% of models were calibrated against epidemiological data, and few were validated against out-of-sample datasets (14%). The interventions considered were primarily the impact of different drug regimens, hygiene and infection control measures, screening, and diagnostics, while few studies addressed de novo resistance, vaccination strategies, economic, or behavioral changes to reduce antibiotic use in humans and animals.
Conclusions
The AMR modeling literature concentrates on disease systems where resistance has been long-established, while few studies pro-actively address recent rise in resistance in new pathogens or explore upstream strategies to reduce overall antibiotic consumption. Notable gaps include research on emerging resistance in Enterobacteriaceae and Neisseria gonorrhoeae; AMR transmission at the animal-human interface, particularly in agricultural and veterinary settings; transmission between hospitals and the community; the role of environmental factors in AMR transmission; and the potential of vaccines to combat AMR.
Electronic supplementary material
The online version of this article (10.1186/s12916-019-1314-9) contains supplementary material, which is available to authorized users.
Keywords: Antimicrobial, Resistance, Mathematical, Computational, Models, Communicable diseases, Epidemiology, Transmission
Background
Antibiotics are commonly regarded as one the greatest discoveries of the twentieth century; however, antibiotic or antimicrobial resistance (AMR) is now a significant threat to global health. According to a World Health Organization (WHO) global report [1], healthcare-acquired infections (HCAI) with AMR pathogens such as methicillin-resistant Staphyloccus aureus are a serious problem in high- and middle-income countries where surveillance is well established. There are also indications that the prevalence of HCAIs in low-income countries may be greater than in higher-income regions, although epidemiological data are scarce [1, 2]. In addition to the threat posed by HCAIs, low-income countries need to contend with the emergence of drug resistance to long-standing pathogens, namely human immunodeficiency virus (HIV), tuberculosis (TB), and Plasmodium parasites (malaria) [1].
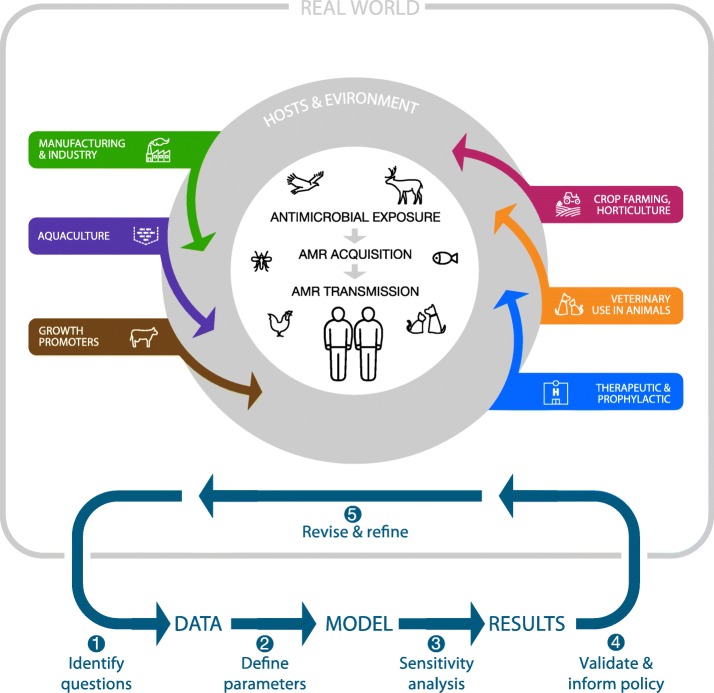
There is an abundance and diversity of sources of drug pressure favoring the emergence of AMR (Fig. 1) [1, 3, 4]. Antimicrobials produced by pharmaceutical manufacturers are distributed widely across a diverse array of industries and applications. Unnecessary or suboptimal use of antimicrobials in humans and animals for medical or prophylactic purposes can promote AMR. Antimicrobial use in animals for growth promotion and intensive crop farming also facilitate evolution of AMR organisms, which can then enter the food chain. Other nonmedical uses of antimicrobials include industrial manufacturing (anti-fouling paint, detergents, ethanol production, food preservations, etc.). Solid or liquid waste contaminated with either AMR organisms or antimicrobials from these many sources may then enter municipal sewer systems or waterways. Thus, antimicrobial release from pharmaceutical manufacturers and non-pharmaceutical industries, combined with human and agricultural use, can lead to contamination of the soil and water [3, 4].
Fig. 1.
Sources of antimicrobial contamination, transmission of AMR, and development of mathematical models. Drivers of AMR as well as resistant pathogens themselves (antimicrobial, biocides, metals) may enter the environment through water (as effluent or through water sanitation systems) or soil (manure application or illegal dumping) from various sources including (i) medical therapeutic and prophylactic use in humans, (ii) veterinary use in companion or food animals, (iii) non-veterinary use in animals (growth promoters), (iv) direct or indirect use in horticulture and crop farming, (v) industrial scale prophylactic use in aquaculture, and (vi) pharmaceutical manufacturers themselves and various industrial applications. Resistant pathogens may then be transmitted to various living organisms through various routes including foodborne, waterborne, airborne, vectorborne, or direct contact. Zoonotic transmission is possible between humans and animals (domestic and wild). Transmission can be further intensified by insect vectors such as mosquitoes and flies, as well as human activity, such as global travel (tourism, migration) and food importation. The goal of mathematical modeling is to synthesize the data collected on AMR and design models to inform public health policy: step 1, identify key questions; step 2, extract or estimate disease parameters based on available data to build a model; step 3, assess model uncertainty/sensitivity; step 4, validate model results with an independent dataset and use to inform policy; and step 5, refine and revise model as needed with new data.
Once primary antimicrobial resistance arises in an organism, it can spread through numerous routes, both within hosts (e.g., via plasmids or mobile elements that are common in bacterial genomes) and between hosts, or via contaminated environment (Fig. 1). There are multiple recognized routes of transmission of AMR pathogens from agricultural farms to humans [5, 6]. Soil and water can also transmit AMR organisms to humans, animals, and plants. Aerosol or airborne transmission is common for respiratory pathogens that may carry resistance such as influenza or tuberculosis, while vectors can facilitate the spread of resistant malaria or bacteria, facilitating rapid diffusion over vast geographic areas [7, 8]. While AMR cannot be realistically eradicated, it may be possible to slow down or reduce its occurrence through antimicrobial stewardship, namely, strategies designed to improve the appropriate use of antimicrobials.
Mathematical models are increasingly used to help understand and control infectious diseases, particularly to identify key parameters driving disease spread, assess the effect of potential interventions, and forecast the trajectory of epidemics [9]. The most impactful modeling studies typically involve close feedback between modelers, public health experts, and clinicians, to identify an actionable research question, design and calibrate a model against empirical data, perform sensitivity analyses, refine the model as more data become available, and eventually issue policy guidance (Fig. 1). Modeling AMR organisms can be particularly challenging compared to modeling sensitive pathogens for several reasons (see Box 1). In addition to crucial data gaps, modelers have to contend with issues of pathogen heterogeneity, fitness costs, co-infections, and competition, which are important features of resistance that remain poorly understood and quantified.
The contribution of mathematical modeling to the control of emerging infections is well established [9], and mathematical modeling can also be a powerful tool to guide policies to control AMR. Here, we undertake a systematic review to assess how population-level mathematical and computational modeling has been applied in the field of AMR over a period of 11 years (2006–2016). Previous reviews of AMR modeling were either completed some time ago [10, 11], only applied to a specific subset of AMR, such as HCAIs [12, 13], or focused on acquired resistance [14]. Our goals in this study were to (1) identify the predominant pathogens, populations, and interventions studied; (2) highlight recent advances in the field; (3) assess the influence of the research; and (4) identify gaps in both modeling of AMR and data availability.
Methods
Search strategy and selection criteria
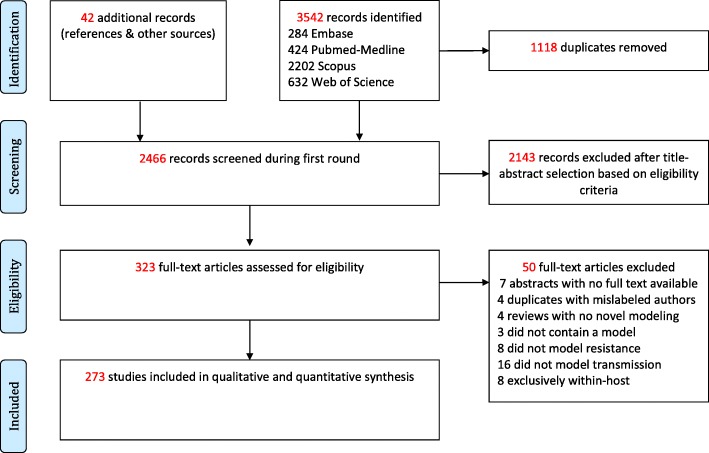
We undertook a systematic search and review of publications relevant to the transmission modeling of AMR. Searches were carried out in PubMed-MEDLINE, Scopus, Web of Science, and Embase. Publications were limited by date (January 1, 2006–December 31, 2016) and journal type (original research and review articles only). Data extraction was initially carried out on November 15, 2016 and updated in January 2018. The search query included terms specific to transmission models, resistance issues, and individual pathogens known to acquire resistance (see Additional file 1 for details of the query). We removed duplicate publications and continued with the selection of relevant publications according to the inclusion/exclusion criteria listed below. A summary of the process is outlined in the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) diagram in Fig. 2 and in Additional file 2.
Fig. 2.
PRISMA flowchart outlining selection of studies included in the review.
Inclusion and exclusion criteria
We included any mathematical or computational models describing AMR in an infectious disease pathogen and considering transmission at the population level (i.e., publications on between-host transmission dynamics). We excluded within pathogen/host models of resistance (e.g., exclusively within-host models based on in vitro data), pharmacokinetic-pharmacodynamic models (i.e., pharmacological models focused on optimizing drug dosage that did not include a transmission component), molecular modeling studies (studies focused on molecular structure of chemical compounds), reviews that did not present original work, non-journal articles or reviews (poster or conference abstracts), and descriptive statistical models not incorporating mechanistic principles (such as models based on probability distributions, e.g., regression, clustering analysis).
Selection and analysis of publications
An initial round of title and abstract screening was performed by AMN. Articles identified as potentially relevant were then reviewed by both AMN and CV, and the publication list for full-text analysis was agreed upon by consensus. Full texts for 313 articles were then retrieved, evaluated by AMN, and relevant data was extracted for further analysis (see below). For details on the number of articles excluded at each step, see Fig. 2.
Data extraction
The following data were retrieved from articles: disease system (type: viral (V), bacterial (B), parasitic (P), fungal (F) or non-specific (NS)); drug type; control measures (pharmaceutical and non-pharmaceutical interventions, vaccines, behavioral); location (year, country, WHO region); host population: type (human, animal, plant) and setting (school/family, hospital, community, farm, etc.); data: data used for parameterization (epidemiological, clinical, behavioral, demographic, geospatial), data availability (public, on request, private); methodology: model class (compartmental or individual-based), inference method, and study type (explicative, predictive, interventions vs. forecasting); and metadata (authors, institutions, funding). Pathogen types were also later compared with the published WHO and center for disease control (CDC) lists of most urgent threats in AMR [1, 15].
Time trend and impact analysis
A goal of our systematic review was to explore trends in the publication output for AMR modeling studies and their impact in the field, as AMR is emerging as a global health threat. Our review focused on the period 2006–2016; to explore publication trends in earlier years, we used a prior review by Temime et al. [11] which covered the period 1993–2006. Further, for comparison with a related area of infectious disease modeling, we compiled trends in publication of individual-based transmission models (defined as a model tracking the characteristics of an individual, including infection and transmission, over time), based on a recent systematic review [16]. In addition to the volume of AMR modeling publications, we assessed the impact of these publications in the field using the metric field-weighted citation impact (FWCI) [17]. The FWCI is the ratio between the number citations for a specific article and the average number of citations received by similar articles in the same field, type, and year of publication, thus making values comparable across these three variables. A FWCI greater than 1.0 indicates that publications have been cited more than would be expected; for example, a score of 1.2 means that an article has been cited 20% more than average. It should be noted that a FWCI score can vary over time and that data in our manuscript is based on a snapshot of the Scopus database taken on November 21, 2018.
Intervention analysis
We used a seminal 2016 Review on Antimicrobial Resistance as a framework to classify interventions [18]. The report identified 10 intervention categories, of which only the first six were relevant to our study: (1) education or awareness campaigns, (2) improved hygiene and infection control, (3) reduction in use of antimicrobials, (4) improved surveillance of resistance, (5) improvement and development of rapid diagnostics, and (6) use of antimicrobial alternatives such as vaccines and alternatives. We also added a seventh category to consider antimicrobial regimen changes, as this is an area of high interest for public health (e.g., antimicrobial switching, cycling, introduction of new drug class).
Further, we identified whether interventions were modeled on a “micro” (institution level) or “macro” level (structural or policy interventions that might affect large populations, communities, countries, or regions). We also assessed whether the aim of the study was to prevent the development/acquisition of AMR (de novo resistance) or direct transmission of a resistant pathogen.
Results
Details of the screening process can be found in the PRISMA diagram in Fig. 2. A total of 2466 articles were identified after removing duplicates. Two rounds of title and abstract screening removed a further 2143 records. A total of 323 articles were earmarked for full-text review. Upon reading these, we found that 50 articles did not meet the inclusion criteria specified above, which resulted in a final tally of 273 records included in our analyses. We describe the characteristics of all studies below and then focus on key findings for the five pathogens or diseases most commonly modeled: methicillin-resistant Staphylococcus aureus (MRSA), tuberculosis (TB), human immunodeficiency virus (HIV), influenza, and malaria.
Trends in the number of published modeling studies
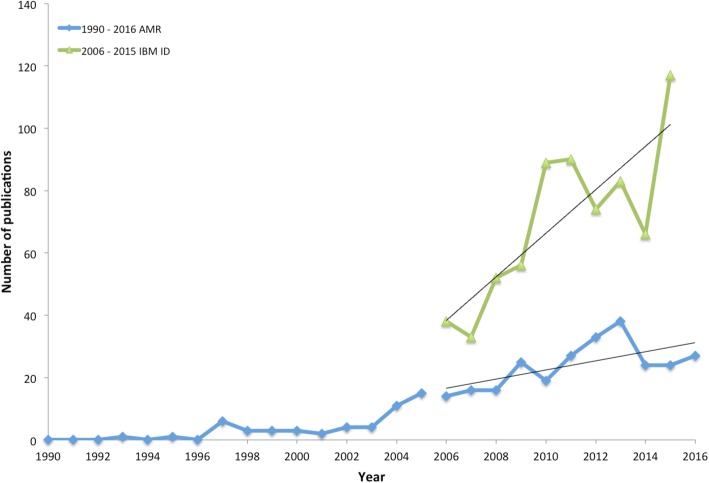
We found an increasing trend (Fig. 3) in the annual number of AMR modeling studies between 2006 and 2016 (linear trend, slope = 1.5, R2 = 0.43), building off the steady increase shown by Temime et al. [11]. Since 2013, the pace of AMR modeling publications has leveled off at around 25 articles/year. In contrast, as described by Willem et al. [16], publications on individual-based models of infectious diseases have experienced a faster increase over the same time period (linear trend, slope = 7, R2 = 0.66), with on average three to four times more articles published on infectious disease related individual-based models than on AMR (Fig. 3). A histogram showing the number of AMR modeling articles published per year since 1990 can be found in Additional file 1: Fig. S1.
Fig. 3.
Yearly number of AMR modeling studies (1990–2016). This figure compares the yearly number of AMR modeling studies (based on data from Temime et al. (1990–2006) [11] as well our analysis (2006–2016), with the number of individual-based models used to analyze infectious disease (IBM ID) identified by Willem et al. between 2006 and 2015 [16]
In addition to overall publication output, we assessed the influence of AMR modeling publications in the field using the FWCI score. The three publications with the highest FWCI during this period had a FWCI greater than 10 (two articles on TB [19, 20] and one on pandemic flu [21]). Excluding these three highly cited outliers, we found that the median FWCI for publications ranged between 0.47 and 2.65, with an overall median of 0.96, indicating that AMR modeling publications are being cited at a rate on par with other studies in their field (Additional file 1: Figure S2).
Distribution of modeling studies by pathogen type
Approximately 65% of the AMR studies focused on bacterial diseases, 25% on viral diseases, 13% on parasitic diseases, and 2% on plant fungal pathogens. The top five pathogens most prominently studied were MRSA (25%), TB (16%), Plasmodium falciparum (8%), HIV (13%), and influenza (11%). For a detailed list of pathogens studied in each publication, see Additional file 1: Table S1. There was no significant time trend in the modeling of specific pathogens (Additional file 1: Figure S3).
Host and population settings used in AMR modeling
Of the 273 publications considered in our review, 89% (n = 234) concerned human hosts, 7% (n = 18) focused on animal diseases, and 2% (n = 5) considered plant hosts. Only 2% (n = 6) addressed transmission between humans and animals in the same model. Animal transmission studies were mainly on animals of agricultural importance, although one explored transmission between humans and companion animals [22]. Only one study modeled the interaction of AMR pathogens between their hosts and the environment [23]. The majority of studies were either set exclusively in the community (n = 151, 55%) or in a healthcare facility (n = 74, 27%), with few (n = 11, 4%) exploring the link between these two (Table 1). Only eight studies (3%) modeled the transmission of AMR in long-term care facilities such as nursing homes, which are thought to be major reservoirs of AMR. The model populations were largely homogeneous and did not allow for variable mixing rates. A minority of the studies (n = 48, 18%) included heterogeneity in age, gender, sexual activity, and treatment status for pathogens such as TB, HIV, influenza, or malaria [24, 25]. Details can be found in Additional file 3: Table S4.
Table 1.
Distribution of selected studies according to study characteristics.
| TOTAL | MRSA | TB | HIV | Influenza | Malaria | |
|---|---|---|---|---|---|---|
| Host type | (n = 273) | (n = 65) | (n = 43) | (n = 34) | (n = 30) | (n = 22) |
| Human | 233 (89%) | 62 (95%) | 43 (100%) | 34 (100%) | 28 (93%) | 21 (95%) |
| Animal | 18 (7%) | 2 (3%) | – | – | 2 (7%) | 1 (5%) |
| Human-animal | 7 (2%) | 1 (2%) | – | – | – | – |
| Plant | 5 (2%) | – | – | – | – | – |
| Population type | ||||||
| Community (endemic) | 154 (58%) | 7 (11%) | 40 (93%) | 34 (100%) | 25 (83%) | 21 (95%) |
| Healthcare facility | 71 (27%) | 49 (75%) | 1 (2%) | – | – | – |
| Community-healthcare facility | 11 (4%) | 5 (8%) | 2 (5%) | – | 1 (3.5%) | – |
| Agricultural-farming | 20 (8%) | 1 (1.5%) | – | – | 1 (3.5%) | – |
| Other | 8 (3%) | 3 (4.5%) | – | – | 3(10%) | 1 (5%) |
| Model parameters | ||||||
| Referenced data | 191 (72%) | 34 (52%) | 38 (88%) | 32 (94%) | 29 (97%) | 17 (77%) |
| Primary data | 73 (28%) | 31 (48%) | 5 (12%) | 2 (6%) | 1 (3%) | 5 (23%) |
| Model type | ||||||
| Deterministic | 175 (66%) | 30 (46%) | 36 (84%) | 23 (70%) | 18 (60%) | 18 (82%) |
| Stochastic | 57 (22%) | 30 (46%) | 6 (14%) | 5 (12%) | 3 (10%) | 2 (9%) |
| Both (D and S) | 25 (9%) | 5 (8%) | 1 (2%) | 4 (12%) | 8 (27%) | 1 (5%) |
| Hybrid | 7 (3%) | – | – | 2 (6%) | 1 (3%) | 1 (5%) |
| Model class | ||||||
| Compartmental | 201 (76%) | 34 (52%) | 37 (86%) | 27 (79%) | 26 (87%) | 19 (86%) |
| Individual-based | 33 (12%) | 19 (29%) | 3 (7%) | 4 (12%) | 1 (3%) | 1 (5%) |
| Both | 7 (3%) | 1 (2%) | – | 2 (6%) | 1 (3%) | 1 (5%) |
| Other | 23 (9%) | 11 (17%) | 3 (7%) | 1 (3%) | 2 (7%) | 1 (5%) |
| Model features | ||||||
| Multi-strain model | 190 (72%) | 23 (35%) | 42 (98%) | 32 (94%) | 29 (97%) | 17 (77%) |
| Co-infection of hosts | 22 (8%) | 5 (8%) | 9 (21%) | 5 (15%) | 3 (10%) | 3 (14%) |
| Fitness cost modeled | 132 (50%) | 15 (23%) | 32 (74%) | 20 (59%) | 25 (83%) | 13 (59%) |
| Acquired and transmitted resistance | 89 (34%) | 1 (2%) | 26 (60%) | 28 (82%) | 20 (67%) | 5 (23%) |
| Within-host model | 17 (6%) | 1 (2%) | 3 (7%) | 2 (6%) | 1 (3%) | 4 (18%) |
| Population stratification | 48 (18%) | 4 (6%) | 11 (26) | 18 (53%) | 3 (10%) | 4 (18%) |
| Model rigor | ||||||
| Calibration | 115 (43%) | 33 (51%) | 26 (60%) | 16 (47%) | 4 (13%) | 3 (14%) |
| Validation | 36 (14%) | 10 (15%) | 11 (26%) | 5 (15%) | 0 (0%) | 1 (5%) |
| Sensitivity analyses | 159 (60%) | 36 (55%) | 32 (74%) | 25 (74%) | 16 (53%) | 14 (64%) |
| Economics | ||||||
| Cost/benefit analysis | 23 (9%) | 8 (12%) | 6 (14%) | 6 (18%) | 1 (3%) | 3 (14%) |
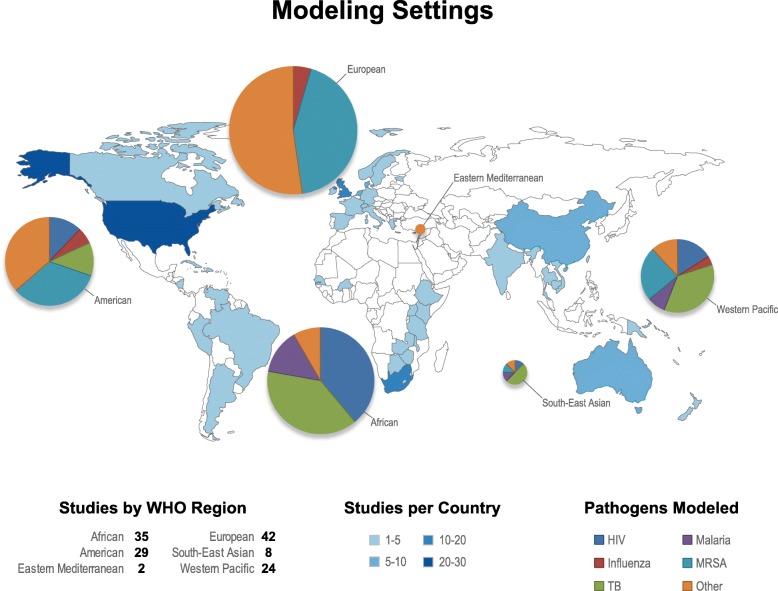
A large fraction of studies (n = 121, 44%) did not focus on a particular geographic area. Those that did were approximately evenly split between four regions: Africa (n = 35, 13%), the Americas (n = 36, 13%), Europe (n = 43, 16%), and Western Pacific (n = 24, 9%) (Fig. 4). Few studies modeled AMR in either the Eastern Mediterranean (n = 2, 1%) or South East Asian (n = 8, 3%) regions. Most models that did specify a geographic location focused on only one country and did not model transmission between countries. Five studies modeled global transmission of the pathogen of interest [26–30]. There was an association between the pathogens modeled and country income status: 91% of studies (74/81) that specified locations and modeled HCAI were restricted to high-income countries (Table 2). On the other hand, the majority of TB and malaria modeling studies were set in low- and middle-income countries (LMIC) (Table 2). HIV was the only disease modeled across all regions (Table 2).
Fig. 4.
Geographic locations of models and pathogens modeled. A visual representation of 146 models that used parameters specific to geographic settings. One hundred seventeen models did not specify a particular geographic location. We also show the percentage of modeling studies by WHO region, categorized by the most highly represented pathogen types (HIV, human immunodeficiency virus; Influenza; Malaria; MRSA, methicillin-resistant Staphylococcus aureus; TB, tuberculosis). The size of the pie charts is proportional to the number of studies
Table 2.
Pathogens modeled by World Bank income level.
| Infectious disease system | Total | High | Upper-middle | Low-middle | Low | ND |
|---|---|---|---|---|---|---|
| MRSA | 65 | 38 | 2 | 1 | 24 | |
| TB | 43 | 3 | 15 | 7 | 25 | |
| HIV | 34 | 4 | 9 | 4 | 2 | 15 |
| Influenza | 30 | 5 | 25 | |||
| Plasmodium falciparum | 21 | 1 | 5 | 1 | 14 | |
| General bacteria | 17 | 4 | 13 | |||
| Streptococcus pneumoniae | 12 | 6 | 1 | 5 | ||
| Enterococci (VRE) | 10 | 7 | 3 | |||
| Escherichia coli | 9 | 6 | 1 | 2 | ||
| Neisseria gonorrhoeae | 6 | 3 | 3 | |||
| Acinetobacter baumannii | 4 | 1 | 1 | 2 | ||
| Zymoseptoria tritici | 4 | 3 | 1 | |||
| Enterobacteriaceae (ESBL-E) | 3 | 2 | 1 | |||
| Klebsiella pneumoniae | 3 | 3 | ||||
| Teladorsagia circumcincta | 3 | 3 | ||||
| Campylobacter | 2 | 2 | ||||
| General protozoan | 2 | 1 | 1 | |||
| Nematodes | 2 | 1 | 1 | |||
| Pseudomonas aeruginosa | 2 | 2 | ||||
| Salmonella spp. | 2 | 1 | 1 | |||
| Schistosoma mansoni | 2 | 2 | ||||
| Blumeria graminis | 1 | 1 | ||||
| Enterobacteriaceae, CRE | 1 | 1 | ||||
| Leishmania donovani | 1 | 1 | ||||
| NS | 1 | 1 | ||||
| Plasmodium chabaudii | 1 | 1 | ||||
| Shigella spp. | 1 | 1 | ||||
| Trichostrongylus colubriformis | 1 | 1 | ||||
| Wucheria bancrofti | 1 | 1 |
A representation of pathogens modeled by the World Bank income level classification: high, upper-middle, low-middle, low, or not described (ND).
Modeling structure, dynamics, and model fitting
Of the 273 studies analyzed, most used deterministic models (n = 175, 66%). Other studies adopted stochastic models (n = 57, 22%), or hybrid deterministic models containing some elements of stochasticity (n = 7, 3%). A few studies compared the results of deterministic and stochastic methods (n = 25, 9.5%). Models were predominantly compartmental (n = 201, 76%) relative to individual-based models (n = 33, 12%). Several studies compared AMR outcomes using both model strategies (n = 7, 3%) (Table 1). A full breakdown of models by class is available in Additional file 1: Table S2.
Most studies considered more than one pathogen strain (n = 190, 72%), but the majority of the studies did not allow for co-infection of hosts, with a few exceptions (n = 22, 8%) (see Additional file 3: Table S4 for details). Half of the studies considered that the resistant strain carried a fitness cost (n = 132, 50%); however, fitness cost was often assumed, and few studies used primary data to infer this parameter (n = 21, 8%). With regard to the type of resistance studied, many models (n = 119, 45%) focused exclusively on transmitted resistance (secondary resistance) and significantly fewer models (n = 36, 14%) explored acquired or de novo resistance. Approximately a third of models (n = 89, n = 34%) accounted for both acquired and transmitted resistance, and some (n = 20, 8%) did not differentiate. Interestingly, a few studies integrated within- and between-host models (n = 17, 6%), allowing for joint exploration of emergence and transmission of AMR.
Model calibration against epidemiological or experimental data is an important feature of mathematical modeling. Some form of calibration (partial or full parameter calibration) was reported in just under half of the studies (n = 115, 43%). In addition to model calibration, sensitivity analysis testing the impact of varying parameter values on model outputs is critical to explore the robustness of conclusions. Out of 273 studies, 159 (60%) reported some level of parameter sensitivity or uncertainty analysis.
The accuracy of model results can also be assessed by out-of-sample validation techniques, in which model predictions are compared to independent observations that have not been used for model calibration. Only 36 studies (14%) reported out-of-sample model validation. From these, 31 used a statistical approach, while 5 simply conducted “face validity” tests by qualitative comparisons to empirical epidemiological datasets. There was no significant time trend in the type of models used, nor in the proportion of studies presenting a calibration or validation step (Additional file 1: Figure S4).
Finally, integration of economic frameworks in mathematical models to project economic costs can help to inform public health decision makers, by translating model results into more tangible cost-benefit analyses. Only 23 studies (n = 23, 9%) included financial components and proposed cost-benefit or savings analyses.
Intervention analysis
Mathematical models can be particularly useful to assess the effectiveness of intervention strategies (Table 3). Studies modeling interventions were approximately evenly split between interventions targeting non-resistant pathogens (n = 99) and those aimed specifically at suppressing resistance (n =100). Several articles (n = 17) explored interventions that could be classified as being aimed at the suppression of both susceptible and resistant pathogens. Of those aimed at reducing resistance (n = 117), few (n = 20) focused on reducing the emergence or acquisition of resistance, while the majority (n = 82) focused on the transmission of resistant pathogens, and some (n = 15) considered both (Table 3). Perhaps unsurprisingly, the majority of models (n = 85) focused on micro-level interventions affecting institutions (such as hospital-level interventions), with fewer (n = 32) focusing on macro-level interventions such as national policy changes or vaccines (Table 3).
Table 3.
Characteristics of AMR-specific interventions reviewed
| Resistance target | |
| Acquired | 20 (17%) |
| Transmitted | 82 (70%) |
| Both | 15 (13%) |
| Intervention scale | |
| Micro | 85 (73%) |
| Macro | 32 (27%) |
| Both | 0 |
| Recommended strategies to combat AMR | |
| 1. Education or awareness campaigns | 3 (3%) |
| 2. Improved hygiene and Infection Control | 59 (50%) |
| 3. Reduction in use of antimicrobials | 16 (14%) |
| 4. Improved surveillance of resistance | 32 (27%) |
| 5. Improved and rapid diagnostics | 10 (9%) |
| 6. Vaccines and alternatives | 11 (9%) |
| 7. Changes to drug regimens | 46 (39%) |
We categorized the interventions from 117 studies that were specifically aimed at blocking AMR based on whether the interventions targeted acquired or transmitted resistance, the scale of interventions, and the type of intervention strategy modeled, motivated by the categories identified in a seminal report [18]. It should be noted that several models investigated the effects of more than one intervention; therefore, the sum of total strategies evaluated (n = 177) exceeds the total number of studies evaluated (n = 117).
We analyzed interventions based on the categories identified in a seminal report on AMR [18] (Table 3).The interventions studied were primarily improved hygiene or infection control measures (n = 59, 50%) such as hand hygiene, isolation, and decolonization. The impact of different drug regimens was often explored (n = 46, 39%) and included techniques such as mixing, switching, and cycling of drugs as well as changes to drug dosage and frequency. Surveillance of resistance (n = 32, 27%), rapid diagnostic techniques (n = 10, 9%), and a reduction in exposure to antimicrobials (n = 16, 14%) were also modeled. Relatively few studies included alternative treatment strategies or vaccines (n = 11, 9%). Only three studies modeled behavioral interventions (n = 3, 3%). Generally, many interventions modeled were organism specific, and further details can be found in Additional file 1: Table S3 and Additional file 3: Table S4.
The five most common resistant pathogens modeled
We provide a short summary of the main findings of AMR modeling efforts for each of the top five diseases included in our review: MRSA, TB, HIV, influenza, and malaria.
Methicillin-resistant Staphylococcus aureus (MRSA)
Almost all of the 58 MRSA transmission studies focused exclusively on humans, except for three that explored MRSA in animals or the associations between animals and humans [22, 31, 32] (Table 1). The studies were mainly set in healthcare facilities (n = 49, 75%), with a few modeling transmission between hospitals and other settings (n = 5, 8%). Only one model was set in low-middle-income country. Key findings of these studies include: (1) reaffirming the importance of hand hygiene compliance; (2) the prediction of coexistence of community-acquired and hospital-acquired MRSA [33–35], rather than the dominance of one over the other (although Webb et al. predict that community-acquired MRSA will dominate [36]); (3) the importance of effectively implementing appropriate screening, followed by isolation and/or decolonization; (4) the importance of hygiene and infectious disease control measures; and finally (5) two studies that proposed the intriguing concept of vaccines as a new weapon against MRSA [37, 38].
Tuberculosis
We identified a total of 43 models studying the dynamics of TB resistance in humans, mainly in community settings (n = 40, 93%). The studies modeled general transmission dynamics of multidrug-resistant (MDR) or extensively drug-resistant (XDR) TB and considered multiple interventions, most commonly intermittent preventative therapy (IPT); directly observed treatment, short-course (DOTS); and surveillance and drug susceptibility testing (Additional file 1: Table S3). Major conclusions include the following: (1) the vast majority of MDR-TB incidence is due to transmitted resistance rather than de novo treatment-related acquisition [30, 39, 40]; (2) to combat resistance, drug susceptibility testing and TB surveillance should be emphasized [41–44]; (3) treatment and drug susceptibility testing should be expanded in community settings in Africa and the private sector in India [42, 43, 45–47]; (4) controlling HIV would help decrease the transmission rates of resistant -TB [48, 49]; (5) isolation or quarantine strategies would help prevent transmission and decrease the number of patients lost to follow-up [50, 51]; and (6) while community-wide intermittent preventative therapy may increase the incidence of drug resistance, the benefits in reducing primary TB infections outweigh the risks. However, such therapy should be coupled with appropriate diagnostic and treatment policies [48, 52–54].
Human immunodeficiency virus
HIV studies represented 13% of our data (n = 34). Topics modeled included the dynamics of HIV resistance in the context of the introduction of new pharmaceutical interventions (e.g., antiretroviral therapy, pre-exposure prophylaxis, vaginal microbicides, or structural interventions such as changes in diagnostics or treatment policy (Additional file 1: Table S3)). Seven additional papers modeled HIV-TB co-infection. Several manuscripts reached similar conclusions, most notably the following: (1) while oral pre-exposure prophylaxis is expected to reduce new HIV infections, a rise in de novo resistance is projected if prophylaxis is administered to those unknowingly infected with HIV [55–62]; (2) similar findings apply to vaginal microbicides [63–65]; and (3) modeling stresses the likelihood of accumulation of resistance over time as a response to various therapies and the importance of regular viral load testing and early diagnosis [66–69]. Various changes in HIV treatment policy or diagnostics were also modeled [66, 68–75].
Influenza
Influenza resistance modeling studies (n = 30) mostly focused on humans, with few exceptions (one transmission model in chickens and one between ferrets) [76, 77]. Interventions modeled included use of antivirals (matrix ion channel or neuraminidase inhibitors), vaccines, antibiotics for treatment of secondary infections, and non-pharmaceutical interventions (isolation and social distancing) (Additional file 1: Table S3). Three repeating themes emerged: (1) there is support for the use of prophylactic drugs despite the risk of developing resistance during pandemic situations, but conditions varied [21, 78–85]; (2) timing, dosage, and coverage levels of drugs are important when it comes to determining treatment effectiveness [82–91]; and (3) there is a need for monitoring the transmissibility and/or fitness of the resistant virus [28, 77, 78, 92–94].
Malaria
A total of 22 studies described mathematical models for transmission of Plasmodium species in the context of AMR. All studies modeled Plasmodium falciparum in humans with the exception of one study of Plasmodium chabaudi in mice [95]. Geographically defined studies were restricted to Sub-Saharan Africa and the Thai-Cambodian region. Pharmaceutical interventions included the following drugs: artemisinin or artemisinin combination therapy (ACT), chloroquine, sulphadoxine, and pyrimethamine. Various non-pharmaceutical interventions were also modeled (Additional file 1: Table S3). Major conclusions include (1) the importance of using artemisinin as part of combination therapy regime (rather than monotherapy) [25, 96–99] and (2) intermittent preventive therapy should be used carefully in areas where resistance is not already established [24, 100].
Discussion
Our systematic review of transmission modeling of AMR over a decade highlights a continuous increase in publications during 1996–2012, a peak in 2013 (n = 38), and a plateau in the following 3 years (average annual publications = 25). Modeling of AMR overall experiences a slower progression than a related field such as individual-based infectious disease models. Five infectious diseases have dominated mathematical models of AMR during 2006–2016: MRSA, TB, HIV, influenza, and malaria. The majority of AMR articles focused exclusively on humans, either in community or healthcare settings, rather than modeled interactions between hosts or multiple settings. Over the study period, a majority of models remained data-free and few were validated against independent datasets. Many models assumed a fitness cost for resistant organisms; however, this was often not derived from primary experimental or epidemiological data. Few models integrated within-host dynamics or economic factors into their transmission framework. Most of the interventions aimed at combating AMR were primarily focused on transmitted rather than acquired resistance and were implemented on a micro-level scale. The interventions considered the impact of different drug regimens, hygiene and infection control measures, or screening and diagnostics, while less than 5% addressed alternative therapeutic strategies or behavioral changes.
The predominance of five pathogens in AMR transmission modeling is likely driven by a long history of disease modeling for at least four of these pathogens (TB, HIV, influenza, and malaria) and an early recognition of MRSA as an important drug-resistant pathogen, combined with availability of epidemiological and surveillance data. Historically, these diseases have taken a large toll on global morbidity and mortality rates; however, it has been predicted that the consequences of AMR in other pathogens may rapidly outpace them by 2050 [18]. More research should be undertaken before resistance in other disease systems becomes a major crisis.
The observed skew towards these 5 diseases was in stark contrast in comparison to the WHO’s priority list of 12 antibiotic resistant bacteria [1] and the CDC’s list of 18 drug-resistant threats in the USA [15]. Only a handful of studies modeled the diseases categorized as the most urgent by the WHO and CDC: Neisseria gonorrhoeae (n = 6), Acinetobacter baumannii (n = 4), ESBL-producing enterobactriaceae (n = 3), Pseudomonas aeruginosa (n = 2), carbapenem-resistant enterobactriaceae (n = 1), and Clostridium difficile (n = 0) (Table 4). The lack of Clostridium difficile resistance studies is puzzling as several mathematical models exist for sensitive strains of this pathogen (e.g., [101]). In contrast, the top two bacteria represented in our AMR review, MRSA and TB, were the focus of 65 and 43 studies respectively. And while modeling of an intermediate-level threat like vancomycin-resistant enterococci is gaining momentum (n = 10), much remains to be done to understand transmission in the community and environmental settings.
Table 4.
The number of modeling studies compared to the WHO and CDC lists of important AMR threats.
| Pathogen | WHO category | CDC category | AMR models | References |
|---|---|---|---|---|
| Enterobacteriaceae, carbapenem-resistant | C1 | C1 | 1 | [156] |
| Enterobacteriaceae, ESBL-producing | C1 | C2 | 3 | [141, 157, 158] |
| Acinetobacter baumannii, carbapenem- or multidrug-resistant | C1 | C2 | 4 | [158–161] |
| Pseudomonas aeruginosa, carbapenem-or multidrug-resistant | C1 | C2 | 2 | [139, 158] |
| Neisseria gonorrhoeae, cephalosporin- or fluoroquinolone-resistant | C2 | C1 | 6 | [162–167] |
| Staphylococcus aureus, methicillin-resistant | C2 | C2 | 65 | [22, 31–38, 151, 158, 168–221] |
| Enterococcus faecium, vancomycin-resistant | C2 | C2 | 10 | [158, 202, 222–229] |
| Campylobacter spp., fluoroquinolone-resistant | C2 | C2 | 2 | [229, 230] |
| Salmonellae, (Typhi and non-typhoidal), fluoroquinolone-resistant | C2 | C2 | 2 | [231, 232] |
| Staphylococcus aureus, vancomycin-intermediate and resistant | C2 | C3 | – | – |
| Helicobacter pylori | C2 | – | – | – |
| Haemophilus influenzae, ampicillin-resistant | C3 | – | – | – |
| Streptococcus pneumoniae, penicillin-non-susceptible | C3 | C2 | 12 | [37, 233–240] |
| Shigella spp., fluoroquinolone-resistant | C3 | C2 | 1 | [241] |
| Clostridium difficile | – | C1 | – | – |
| Mycobacterium tuberculosis, MDR, XDR | – | C2 | 43 | [19, 20, 30, 39–54, 242–265] |
| Fluconazole-resistant Candida | – | C2 | – | – |
| Streptococcus agalactiae, clindamycin-resistant group B MDR, XDR | – | C3 | – | – |
| Streptococcus pyogenes, erythromycin-resistant group A MDR, XDR | – | C3 | – | – |
The pathogens that pose the greatest threat to human health according to the WHO and the top drug-resistant threats in the USA according to the CDC. Category 1 (C1) threats are described as “Critical” (WHO) or “Urgent” (CDC); category 2 (C2) as “High” (WHO) or “Serious” (CDC); and category 3 (C3) as “Medium” (WHO) or “Concerning” (CDC).
Other serious threats based on WHO or CDC criteria that are rarely modeled include Campylobacter (n = 2), Salmonellae spp. (n = 2), Neisseria gonorrhoeae, and Shigella spp. (n = 1). Importantly, we were unable to find any published AMR models for the following serious threats: Helicobacter pylori, Haemophilus influenzae, fluconazole-resistant Candida, clindamycin-resistant group B strep, and erythromycin-resistant group A strep. While mathematical transmission models do exist for wild-type H. pylori [102], H. influenzae [103], and Candida parapsilosis [104], we are not aware of any models for resistant strains, which may have different transmission parameters than susceptible strains.
Most models did not consider pathogen heterogeneity, such as multiple viral or bacterial strains, parasite species, or multiple resistance mechanisms (e.g., membrane permeability, enzymatic degradation, mutation of antimicrobial targets), which might affect transmission potential. As a case in point, most malaria modeling has dealt with the Plasmodium falciparum species in Africa or East Asia. This is presumably based on the long-held assumption that the majority of malaria burden is caused by P. falciparum rather than other plasmodium species. However, there is growing evidence that Plasmodium vivax, which is endemic in South and South-East Asia as well as Central and South America, is associated with a significant burden of morbidity and associated mortality [105, 106]. P. vivax is already largely resistant to chloroquine [107], though resistance to artemisinin has not yet been reported. A similar issue exists in regard to mathematical modeling studies of HIV, where no distinction was made between HIV-1 and HIV-2, which are known to have markedly different resistance profiles to the various antiretroviral drugs used [108, 109]. This is likely because HIV-2 has historically infected a much smaller, but significant, proportion of the population. It was estimated in 2006 that one to two million people [110] in several West African countries were infected with HIV-2, though we could not find more recent estimates.
While there has been increasing effort to design models with explicit interactions between community and hospital populations, few include long-term care facilities, which often lack effective antimicrobial stewardship programs [111–113]. Most worrisome perhaps, almost all models were set in humans and there were few attempts to tackle the hypothesized connection between veterinary/agricultural use of antibiotics and AMR. No studies modeled AMR transmission in aquaculture, despite the growing body of evidence that AMR resistance could enter the food chain through these means [114, 115]. Similarly, there were few ecological studies on the transmission of AMR from the environment (water, soil, etc.) to potential hosts, despite the increasing evidence for a link between antimicrobial contamination of the environment, and the development and transfer of resistance to human pathogens [116–118]. This is particularly concerning given the large quantity of antibiotics used in agricultural facilities, the lack of regulation on their waste disposal and the inability of many sanitation systems to filter out antimicrobials and AMR elements. Another environmental factor that was not modeled was the effect of climate change on the rates of AMR. Recent research has shown that increasing temperatures are associated with increased levels of resistance [119, 120], but there is no projection of AMR patterns under climate change scenarios.
We found that the vast majority of HCAI and influenza models were set in high-income countries, although this is an increasingly recognized threat in LMIC [1]. The lack of studies in developing countries is particularly concerning because of unregulated or poorly regulated antimicrobial manufacturing and usage [121, 122]. This is likely due to lack of appropriate diagnostics and surveillance in low-resource settings [1, 122].
A major reason for the lack of modeling studies on particular pathogens or certain settings is likely to be a deficiency in available data needed for model calibration and design. There is a need for more precise data on antibiotic consumption rates in both humans and animals [18], which is often not made publicly available [123–125]. In addition, improved surveillance of AMR incidence is required in humans, animals, and the environment (soil and water) [126]. There have been several examples of zoonotic transmission of AMR in both domestic [127, 128] and wild animals [129, 130] as well as evidence of transmission of genetic determinants of AMR into the environment [3, 116], which in turn may facilitate further dissemination of resistance.
In terms of AMR-specific model dynamics, half of the reviewed studies factored in a fitness cost for the resistant strain; however, this was often assumed and rarely estimated from primary data. Additionally, many models did not distinguish between acquired (de novo) or transmitted resistance. This is important for accurately defining model parameters such as reversion [131] or transmission rates [78, 132], which ultimately affect model outcomes. Most studies modeled homogeneous infections with a single pathogen strain and therefore did not investigate host co-infection and strain competition. Host populations were also largely assumed to be mixing homogeneously with no stratification by age, susceptibility, or contact patterns. Integration of within- and between-host models was also rare; multi-scale modeling is an important frontier for AMR and more broadly for the field of infectious disease modeling [133].
Previous reviews predicted that technological advances in computational tools could allow for more complex models and calibration to larger datasets [9, 13]. Consistent with this prediction, a sharp increase was reported in the field of individual-based models of infectious diseases, but this increase has not percolated to the field of AMR [16]. The majority of AMR transmission models reviewed here remain theoretical, with little attempt to compare model predictions to epidemiological data, and calibration with independent data is scarce. It should also be noted that improvements could also be made in terms of documenting modeling methods. Only 47% of the studies assessed cited the modeling software or computational tools used and few described modeling techniques in a way that might be able to be reproduced by researchers who are not already experienced modelers. Even fewer manuscripts provided the computational code used: two manuscripts provided a link (both were expired at the time of this writing), and three were willing to share the code upon request. Some attempts have been made to standardize the terminology, methodology, and reporting structure for infectious disease transmission models [134–136], but better documentation of modeling methods is needed for reproducibility. Furthermore, it would also be useful to make the underlying AMR epidemiological datasets publicly available to aid reproducibility.
With regard to interventions aimed at combating AMR, many models incorporated elements of improved hygiene or infection control in order to combat the spread of AMR. No model focused on “macro” scale interventions such as improved access to water and sanitation facilities that can curb the transmission and development of resistance. Improved water, sanitation, and hygiene can lead to a decrease in respiratory and diarrheal disease, both of which are often unnecessarily treated with antibiotics although the causative agents may be viral [137, 138]. Numerous interventions examined improved surveillance or diagnostic methods, particularly for HIV and TB, but were lacking for many bacterial diseases outside of healthcare settings. Many diagnostic methods for antimicrobial resistance are culture based, and confirmation of resistance, let alone specific genotyping, may take several days. There is an urgent need for rapid molecular diagnostics in order to improve antimicrobial stewardship; more modeling work in this area could highlight the transmission and cost-effectiveness benefits of such technologies.
Surprisingly, few studies modeled reduction in the use of antimicrobials as an intervention, particularly when supplied to food animals either as a growth supplement or prophylaxis. Several models studied the effects of reducing antimicrobial exposure levels in healthcare settings [139–142], but there were fewer for animals [143–145]. No models for AMR or AMR-related interventions in aquaculture settings exist.
Many infectious disease models increasingly incorporate features of human behavior [123–125, 146]; however, this is not common in the field of AMR modeling outside of healthcare facilities. In addition, most models did not consider how social, cultural, or behavioral differences might affect resistance development or transmission. Those that did were mainly focused on sexually transmitted infections such as HIV or N. gonorrhoeae. Similarly, few models included vaccination despite increasing appreciation for the role they could play in reducing antimicrobial consumption [147, 148]. Vaccines can also have indirect effects on antimicrobial consumption [147, 148] by reducing the number of pharmaceuticals erroneously prescribed for viral infections. Several vaccine candidates are under development for C. difficile, S. aureus, group B Streptococcus, E. coli, and respiratory syncytial virus [149]; mathematical models could be used to evaluate their potential effects at a population level and inform cost-effectiveness analyses.
The increasing availability of multiple epidemiological and pathogen genetic data streams offers exciting new possibilities to improve and expand modeling capabilities. Enhanced access to, and integration of, digital disease surveillance data [150] into epidemiological analyses could help further strengthen model validation. Pathogen genomic sequences (together with relevant metadata such as date, location) can also inform many aspects of transmission dynamics. And although some have started integrating genomic data [151] into modeling studies, this is the exception rather than the norm in the field of AMR. An integrative approach will be required to synthesize large amounts of data together, which will ideally help to develop more realistic AMR models tailored to specific populations. It is noteworthy that few publications addressed the spatial diffusion of AMR; a lack of spatially resolved AMR datasets may explain this gap.
This review has some limitations. We have only searched four databases most relevant to biomedical sciences. Furthermore, in an effort to keep the amount of search results to a manageable number, we use certain keywords specific to population dynamic studies of AMR organisms. Therefore, we may have inadvertently excluded some publications (without these keywords) relevant to this review. However, we are confident that this review provides an accurate overview of overall trends in the field.
Conclusions
The field of AMR modeling is growing but is limited by both the quantity and quality of available data. Success stories include accurate predictions of the emergence of resistance in malaria [152], MDR-TB [153], and influenza [154], and modeling is also frequently used to inform AMR stewardship programs in healthcare facilities [155]. Our review suggests a need for more applied, data-driven models, better tuned to and diversified to reflect the public health concerns highlighted by the WHO and the CDC. Although the overall increase in AMR transmission modeling in the last decade is encouraging, the recent plateau in published work and scarcity of studies on high-concern pathogens should be addressed. Most importantly perhaps, more forward-thinking models should be developed to predict the emergence of resistance in pathogens where the issue is not yet rampant and evaluate how policy and behavioral changes can curb drug pressure and mitigate AMR. Research programs in support of AMR modeling, increased data collection efforts, and stronger links between modelers and public health experts are warranted to stimulate this field.
Box 1: Challenges to mathematical modeling of AMR.
Data gaps:
Lack of sufficient data on antimicrobial use in humans and animals, antimicrobial environmental contamination, and resistance rates in unmonitored industries and low-income countries.
Lack of standardization in data definitions or collection methods.
Complexity of model dynamics:
Lack of understanding of disease ecological dynamics or model too complex.
Pathogen heterogeneity: resistance governed by multiple genetic and epigenetic factors, so that a diversity of strains can exhibit the same resistance phenotype (single nucleotide polymorphisms, acquisition or deletion of genes or plasmids, up- or downregulation of genes).
Dynamic fitness landscapes: resistance carries fitness costs that are poorly understood and can decrease transmission potential, while compensatory mutations can restore transmission.
Co-infection dynamics between sensitive and resistant strains: strain coexistence, competition, conversion, or replacement are possible depending on the disease studied.
Model assessment:
Validation cannot take place without proper surveillance data.
Inability to accurately evaluate AMR interventions in the field for ethical, practical, or political reasons.
Inability to validate model parameters in a changing environment (changes in transmission rates, fitness costs, and growth potential under antibiotic treatment, as resistance evolves).
Additional files
Figure S1. Trend in the number of AMR models per year. Figure S2. (a) Field-weighted citation impact (FWCI) of AMR publications by year (excludes 3 publications with FWCI greater than 10) and (b) FWCI by pathogen category: viral (V), bacterial (B), parasitic (P), and fungal (F). Figure S3. Pathogen trends over time. The proportion of articles on selected infectious diseases relative to the total publications per year published between 2006 and 2016 (articles/year): healthcare-acquired infections (HCAI), methicillin-resistant staphylococcus aureus (MRSA), tuberculosis (TB), human immunodeficiency virus (HIV), influenza and plasmodium falciparum (malaria). Figure S4. Model characteristic trends over time: The proportion of articles with a particular model approach (deterministic or stochastic, compartmental or individual-based models) or rigor (calibrated or validated) by the total publications per year published between 2006 and 2016. Table S1. References for publications divided by diseases they model. Table S2. References for publications divided by model class. Table S3. References for publications divided by disease specific interventions. (DOCX 2019 kb)
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2009 checklist. (DOC 62 kb)
Table S4. Complete table of data extracted from each publication included in this review. (XLSX 203 kb)
Acknowledgements
We thank Alicia A. Livinski, National Institutes of Health Library for her assistance with search strategy development and the overall process. We sincerely thank the reviewers and editors, whose comments helped improve and clarify this manuscript. This study does not represent the views of the NIH or the US government.
Funding
This project was funded by the in-house research division of the Fogarty International Center, NIH.
Availability of data and materials
Not applicable.
Abbreviations
- ACT
Artemisinin combination therapy
- AMR
Antimicrobial resistance
- CDC
Center for Disease Control and Prevention
- ESBL
Extended spectrum beta-lactamases
- FWCI
Field-weighted citation impact
- HCAI
Healthcare-acquired infections
- HIV
Human immunodeficiency virus
- IBM
Individual-based model
- LMIC
Low- and middle-income countries
- MDR or XDR TB
Multidrug- or extremely drug-resistant tuberculosis
- MRSA
Methicillin-resistant Staphylococcus aureus
- ND
Not described
- NS
Non-specific
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-analyses
- TB
Tuberculosis
- WHO
World Health Organization
Authors’ contributions
AMN, BJ, and CV designed the study. AMN extracted and analyzed the data and wrote the first draft of the report. AMN, LW, BJ, JCS, DS, BG, and CV reviewed the data and revised the final report. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anna Maria Niewiadomska, Email: amniewiadomska@gmail.com.
Bamini Jayabalasingham, Email: b.jayabalasingham@elsevier.com.
Jessica C. Seidman, Email: jessica.seidman@gmail.com
Lander Willem, Email: lander.willem@uantwerpen.be.
Bryan Grenfell, Email: grenfell@princeton.edu.
David Spiro, Email: david.spiro@nih.gov.
Cecile Viboud, Email: viboudc@mail.nih.gov.
References
- 1.WHO . Antimicrobial resistance: global report on surveillance 2014. 2014. p. 257. [Google Scholar]
- 2.Allegranzi B, Bagheri Nejad S, Combescure C, Graafmans W, Attar H, Donaldson L, Pittet D. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet. 2011;377(9761):228–241. doi: 10.1016/S0140-6736(10)61458-4. [DOI] [PubMed] [Google Scholar]
- 3.Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014;22(1):36–41. doi: 10.1016/j.tim.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Agga GE, Arthur TM, Durso LM, Harhay DM, Schmidt JW. Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS One. 2015;10(7):e0132586. doi: 10.1371/journal.pone.0132586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silbergeld EK, Graham J, Price LB. Industrial food animal production, antimicrobial resistance, and human health. Annu Rev Public Health. 2008;29:151–169. doi: 10.1146/annurev.publhealth.29.020907.090904. [DOI] [PubMed] [Google Scholar]
- 6.Hao H, Sander P, Iqbal Z, Wang Y, Cheng G, Yuan Z. The risk of some veterinary antimicrobial agents on public health associated with antimicrobial resistance and their molecular basis. Front Microbiol. 2016;7:1626. doi: 10.3389/fmicb.2016.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaumburg F, Onwugamba FC, Akulenko R, Peters G, Mellmann A, Kock R, Becker K. A geospatial analysis of flies and the spread of antimicrobial resistant bacteria. Int J Med Microbiol. 2016;306(7):566–571. doi: 10.1016/j.ijmm.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Onwugamba FC, Fitzgerald JR, Rochon K, Guardabassi L, Alabi A, Kuhne S, Grobusch MP, Schaumburg F. The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Med Infect Dis. 2018;22:8–17. doi: 10.1016/j.tmaid.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, Eames KT, Edmunds WJ, Frost SD, Funk S, et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347(6227):aaa4339. doi: 10.1126/science.aaa4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundmann H, Hellriegel B. Mathematical modelling: a tool for hospital infection control. Lancet Infect Dis. 2006;6(1):39–45. doi: 10.1016/S1473-3099(05)70325-X. [DOI] [PubMed] [Google Scholar]
- 11.Temime L, Hejblum G, Setbon M, Valleron AJ. The rising impact of mathematical modelling in epidemiology: antibiotic resistance research as a case study. Epidemiol Infect. 2008;136(3):289–298. doi: 10.1017/S0950268807009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opatowski L, Guillemot D, Boelle PY, Temime L. Contribution of mathematical modeling to the fight against bacterial antibiotic resistance. Curr Opin Infect Dis. 2011;24(3):279–287. doi: 10.1097/QCO.0b013e3283462362. [DOI] [PubMed] [Google Scholar]
- 13.van Kleef E, Robotham JV, Jit M, Deeny SR, Edmunds WJ. Modelling the transmission of healthcare associated infections: a systematic review. BMC Infect Dis. 2013;13:294. doi: 10.1186/1471-2334-13-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birkegard AC, Halasa T, Toft N, Folkesson A, Graesboll K. Send more data: a systematic review of mathematical models of antimicrobial resistance. Antimicrob Resist Infect Control. 2018;7:117. doi: 10.1186/s13756-018-0406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention OoIDArtitUS . Antibiotic Resistance Threats in the United States, 2013. 2013. [Google Scholar]
- 16.Willem L, Verelst F, Bilcke J, Hens N, Beutels P. Lessons from a decade of individual-based models for infectious disease transmission: a systematic review (2006-2015) BMC Infect Dis. 2017;17(1):612. doi: 10.1186/s12879-017-2699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colledge L, Verlinde R. SciVal Metrics Guidebook. 2014. [Google Scholar]
- 18.O’Neill J, RESISTANCE" TROA . Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. [Google Scholar]
- 19.Abu-Raddad LJ, Sabatelli L, Achterberg JT, Sugimoto JD, Longini IM, Jr, Dye C, Halloran ME. Epidemiological benefits of more-effective tuberculosis vaccines, drugs, and diagnostics. Proc Natl Acad Sci U S A. 2009;106(33):13980–13985. doi: 10.1073/pnas.0901720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu S, Andrews JR, Poolman EM, Gandhi NR, Shah NS, Moll A, Moodley P, Galvani AP, Friedland GH. Prevention of nosocomial transmission of extensively drug-resistant tuberculosis in rural South African district hospitals: an epidemiological modelling study. Lancet. 2007;370(9597):1500–1507. doi: 10.1016/S0140-6736(07)61636-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipsitch M, Cohen T, Murray M, Levin BR. Antiviral resistance and the control of pandemic influenza. PLoS Med. 2007;4(1):0111–0121. doi: 10.1371/journal.pmed.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heller J, Innocent GT, Denwood M, Reid SWJ, Kelly L, Mellor DJ. Assessing the probability of acquisition of meticillin-resistant Staphylococcus aureus (MRSA) in a dog using a nested stochastic simulation model and logistic regression sensitivity analysis. Prev Vet Med. 2011;99(2–4):211–224. doi: 10.1016/j.prevetmed.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Call DR, Matthews L, Subbiah M, Liu J. Do antibiotic residues in soils play a role in amplification and transmission of antibiotic resistant bacteria in cattle populations? Front Microbiol. 2013;4:193. doi: 10.3389/fmicb.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Meara WP, Smith DL, McKenzie FE. Potential impact of intermittent preventive treatment (IPT) on spread of drug-resistant malaria. PLoS Med. 2006;3(5):633–642. doi: 10.1371/journal.pmed.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pongtavornpinyo W, Yeung S, Hastings IM, Dondorp AM, Day NP, White NJ. Spread of anti-malarial drug resistance: mathematical model with implications for ACT drug policies. Malar J. 2008;7:229. doi: 10.1186/1475-2875-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JT, Leung GM, Lipsitch M, Cooper BS, Riley S. Hedging against antiviral resistance during the next influenza pandemic using small stockpiles of an alternative chemotherapy. PLoS Med. 2009;6(5):e1000085. doi: 10.1371/journal.pmed.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tchuenche JM, Chiyaka C, Chan D, Matthews A, Mayer G. A mathematical model for antimalarial drug resistance. Math Med Biol. 2011;28(4):335–355. doi: 10.1093/imammb/dqq017. [DOI] [PubMed] [Google Scholar]
- 28.Chao DL, Bloom JD, Kochin BF, Antia R, Longini IM., Jr The global spread of drug-resistant influenza. J R Soc Interface. 2012;9(69):648–656. doi: 10.1098/rsif.2011.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao DL. Modeling the global transmission of antiviral-resistant influenza viruses. Influenza Other Respir Viruses. 2013;7(Suppl 1):58–62. doi: 10.1111/irv.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kendall EA, Fofana MO, Dowdy DW. Burden of transmitted multidrug resistance in epidemics of tuberculosis: a transmission modelling analysis. Lancet Respir Med. 2015;3(12):963–972. doi: 10.1016/S2213-2600(15)00458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciccolini M, Dahl J, Chase-Topping ME, Woolhouse ME. Disease transmission on fragmented contact networks: livestock-associated methicillin-resistant Staphylococcus aureus in the Danish pig-industry. Epidemics. 2012;4(4):171–178. doi: 10.1016/j.epidem.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Hetem DJ, Bootsma MC, Troelstra A, Bonten MJ. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2013;19(11):1797–1802. doi: 10.3201/eid1911.121085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Agata EM, Webb GF, Pressley J. Rapid emergence of co-colonization with community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus strains in the hospital setting. Math Model Nat Phenom. 2010;5(3):76–73. doi: 10.1051/mmnp/20105306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pressley J, D'Agata EMC, Webb GF. The effect of co-colonization with community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus strains on competitive exclusion. J Theor Biol. 2010;264(3):645–656. doi: 10.1016/j.jtbi.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouyos R, Klein E, Grenfell B. Hospital-community interactions foster coexistence between methicillin-resistant strains of Staphylococcus aureus. PLoS Pathog. 2013;9(2):e1003134. doi: 10.1371/journal.ppat.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb GF, Horn MA, D'Agata EM, Moellering RC, Jr, Ruan S. Competition of hospital-acquired and community-acquired methicillin-resistant Staphylococcus aureus strains in hospitals. J Biol Dyn. 2010;4(1):115–129. doi: 10.1080/17513750903026411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joice R, Lipsitch M. Targeting imperfect vaccines against drug-resistance determinants: a strategy for countering the rise of drug resistance. PLoS One. 2013;8(7):e68940. doi: 10.1371/journal.pone.0068940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hogea C, van Effelterre T, Acosta CJ. A basic dynamic transmission model of Staphylococcus aureus in the US population. Epidemiol Infect. 2014;142(3):468–478. doi: 10.1017/S0950268813001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luciani F, Sisson SA, Jiang H, Francis AR, Tanaka MM. The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2009;106(34):14711–14715. doi: 10.1073/pnas.0902437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trauer JM, Denholm JT, McBryde ES. Construction of a mathematical model for tuberculosis transmission in highly endemic regions of the Asia-Pacific. J Theor Biol. 2014;358:74–84. doi: 10.1016/j.jtbi.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Dowdy DW, Chaisson RE, Maartens G, Corbett EL, Dorman SE. Impact of enhanced tuberculosis diagnosis in South Africa: a mathematical model of expanded culture and drug susceptibility testing. Proc Natl Acad Sci U S A. 2008;105(32):11293–11298. doi: 10.1073/pnas.0800965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basu S, Frledland GH, Medlock J, Andrews JR, Shah NS, Gandhi NR, Moll A, Moodley P, Sturm AW, Galvani AP. Averting epidemics of extensively drug-resistant tuberculosis. Proc Natl Acad Sci U S A. 2009;106(18):7672–7677. doi: 10.1073/pnas.0812472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uys PW, Warren R, van Helden PD, Murray M, Victor TC. Potential of rapid diagnosis for controlling drug-susceptible and drug-resistant tuberculosis in communities where Mycobacterium tuberculosis infections are highly prevalent. J Clin Microbiol. 2009;47(5):1484–1490. doi: 10.1128/JCM.02289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shrestha S, Knight GM, Fofana M, Cohen T, White RG, Cobelens F, Dowdy DW. Drivers and trajectories of resistance to new first-line drug regimens for tuberculosis. Open Forum Infect Dis. 2014;1(2):ofu073. doi: 10.1093/ofid/ofu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu S, Galvani AP. The transmission and control of XDR TB in South Africa: an operations research and mathematical modelling approach. Epidemiol Infect. 2008;136(12):1585–1598. doi: 10.1017/S0950268808000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen T, Hedt BL, Pagano M. Estimating the magnitude and direction of bias in tuberculosis drug resistance surveys conducted only in the public sector: a simulation study. BMC Public Health. 2010;10:355. doi: 10.1186/1471-2458-10-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suen SC, Bendavid E, Goldhaber-Fiebert JD. Cost-effectiveness of improvements in diagnosis and treatment accessibility for tuberculosis control in India. Int J Tuberc Lung Dis. 2015;19(9):1115–1124. doi: 10.5588/ijtld.15.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen T, Lipsitch M, Walensky RP, Murray M. Beneficial and perverse effects of isoniazid preventive therapy for latent tuberculosis infection in HIV-tuberculosis coinfected populations. Proc Natl Acad Sci U S A. 2006;103(18):7042–7047. doi: 10.1073/pnas.0600349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sergeev R, Colijn C, Murray M, Cohen T. Modeling the dynamic relationship between HIV and the risk of drug-resistant tuberculosis. Sci Transl Med. 2012;4(135):135ra67. doi: 10.1126/scitranslmed.3003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhunu CP. Mathematical analysis of a three-strain tuberculosis transmission model. Appl Math Model. 2011;35(9):4647–4660. doi: 10.1016/j.apm.2011.03.037. [DOI] [Google Scholar]
- 51.Agusto F. B., Cook J., Shelton P. D., Wickers M. G. Mathematical Model of MDR-TB and XDR-TB with Isolation and Lost to Follow-Up. Abstract and Applied Analysis. 2015;2015:1–21. doi: 10.1155/2015/828461. [DOI] [Google Scholar]
- 52.Basu S, Maru D, Poolman E, Galvani A. Primary and secondary tuberculosis preventive treatment in HIV clinics: simulating alternative strategies. Int J Tuberc Lung Dis. 2009;13(5):652–658. [PubMed] [Google Scholar]
- 53.Mills HL, Cohen T, Colijn C. Community-wide isoniazid preventive therapy drives drug-resistant tuberculosis: a model-based analysis. Sci Transl Med. 2013;5(180):80ra49. doi: 10.1126/scitranslmed.3005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunkel A, Crawford FW, Shepherd J, Cohen T. Benefits of continuous isoniazid preventive therapy may outweigh resistance risks in a declining tuberculosis/HIV coepidemic. Aids. 2016;30(17):2715–2723. doi: 10.1097/QAD.0000000000001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Supervie V, Garcia-Lerma JG, Heneine W, Blower S. HIV, transmitted drug resistance, and the paradox of preexposure prophylaxis. Proc Natl Acad Sci U S A. 2010;107(27):12381–12386. doi: 10.1073/pnas.1006061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abbas UL, Hood G, Wetzel AW, Mellors JW. Factors influencing the emergence and spread of HIV drug resistance arising from rollout of antiretroviral pre-exposure prophylaxis (PrEP) PLoS One. 2011;6(4):e18165. doi: 10.1371/journal.pone.0018165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Supervie V, Barrett M, Kahn JS, Musuka G, Moeti TL, Busang L, Blower S. Modeling dynamic interactions between pre-exposure prophylaxis interventions & treatment programs: predicting HIV transmission & resistance. Sci Rep. 2011;1:185. doi: 10.1038/srep00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abbas UL, Glaubius R, Mubayi A, Hood G, Mellors JW. Antiretroviral therapy and pre-exposure prophylaxis: combined impact on HIV transmission and drug resistance in South Africa. J Infect Dis. 2013;208(2):224–234. doi: 10.1093/infdis/jit150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichols BE, Boucher CA, van Dijk JH, Thuma PE, Nouwen JL, Baltussen R, van de Wijgert J, Sloot PM, van de Vijver DA. Cost-effectiveness of pre-exposure prophylaxis (PrEP) in preventing HIV-1 infections in rural Zambia: a modeling study. PLoS One. 2013;8(3):e59549. doi: 10.1371/journal.pone.0059549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vijver DAMCVD, Nichols BE, Abbas UL, Boucher CAB, Cambiano V, Eaton JW, Glaubius R, Lythgoe K, Mellors J, Phillips A, et al. Preexposure prophylaxis will have a limited impact on HIV-1 drug resistance in sub-Saharan Africa: a comparison of mathematical models. AIDS. 2013;27(18):2943–2951. doi: 10.1097/01.aids.0000433237.63560.20. [DOI] [PubMed] [Google Scholar]
- 61.Dimitrov DT, Boily MC, Hallett TB, Albert J, Boucher C, Mellors JW, Pillay D, van de Vijver DA. How much do we know about drug resistance due to PrEP use? Analysis of experts’ opinion and its influence on the projected public health impact. PLoS One. 2016;11(7):e0158620. doi: 10.1371/journal.pone.0158620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glaubius RL, Parikh UM, Hood G, Penrose KJ, Bendavid E, Mellors JW, Abbas UL. Deciphering the effects of injectable pre-exposure prophylaxis for combination human immunodeficiency virus prevention. Open Forum Infect Dis. 2016;3(3):ofw125. doi: 10.1093/ofid/ofw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson DP, Coplan PM, Wainberg MA, Blower SM. The paradoxical effects of using antiretroviral-based microbicides to control HIV epidemics. Proc Natl Acad Sci U S A. 2008;105(28):9835–9840. doi: 10.1073/pnas.0711813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dimitrov DT, Masse B, Boily MC. Who will benefit from a wide-scale introduction of vaginal microbicides in developing countries? Stat Commun Infect Dis. 2010;2(1):1012. doi: 10.2202/1948-4690.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dimitrov DT, Boily MC, Baggaley RF, Masse B. Modeling the gender-specific impact of vaginal microbicides on HIV transmission. J Theor Biol. 2011;288:9–20. doi: 10.1016/j.jtbi.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vardavas R, Blower S. The emergence of HIV transmitted resistance in Botswana: “when will the WHO detection threshold be exceeded?”. PLoS One. 2007;2(1):e152. doi: 10.1371/journal.pone.0000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoare A, Kerr SJ, Ruxrungtham K, Ananworanich J, Law MG, Cooper DA, Phanuphak P, Wilson DP. Hidden drug resistant HIV to emerge in the era of universal treatment access in Southeast Asia. PLoS One. 2010;5(6):e10981. doi: 10.1371/journal.pone.0010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Phillips AN, Pillay D, Garnett G, Bennett D, Vitoria M, Cambiano V, Lundgren J. Effect on transmission of HIV-1 resistance of timing of implementation of viral load monitoring to determine switches from first to second-line antiretroviral regimens in resource-limited settings. AIDS. 2011;25(6):843–850. doi: 10.1097/QAD.0b013e328344037a. [DOI] [PubMed] [Google Scholar]
- 69.Pham QD, Wilson DP, Nguyen TV, Do NT, Truong LX, Nguyen LT, Zhang L. Projecting the epidemiological effect, cost-effectiveness and transmission of HIV drug resistance in Vietnam associated with viral load monitoring strategies. J Antimicrob Chemother. 2016;71(5):1367–1379. doi: 10.1093/jac/dkv473. [DOI] [PubMed] [Google Scholar]
- 70.Lima VD, Johnston K, Hogg RS, Levy AR, Harrigan PR, Anema A, Montaner JS. Expanded access to highly active antiretroviral therapy: a potentially powerful strategy to curb the growth of the HIV epidemic. J Infect Dis. 2008;198(1):59–67. doi: 10.1086/588673. [DOI] [PubMed] [Google Scholar]
- 71.Lou J, Bu L, Han E, Ruan Y, Xing H, Shao Y. Modeling primary and secondary drug resistances under China’s “four-free-one-care policy”. Int J Biomath. 2012;5(5):1–19.
- 72.Sood N, Wagner Z, Jaycocks A, Drabo E, Vardavas R. Test-and-treat in Los Angeles: a mathematical model of the effects of test-and-treat for the population of men who have sex with men in Los Angeles County. Clin Infect Dis. 2013;56(12):1789–1796. doi: 10.1093/cid/cit158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cambiano V, Bertagnolio S, Jordan MR, Pillay D, Perriëns JH, Venter F, Lundgren J, Phillips A. Predicted levels of HIV drug resistance: potential impact of expanding diagnosis, retention, and eligibility criteria for antiretroviral therapy initiation. AIDS. 2014;28(SUPPL. 1):S15–S23. doi: 10.1097/QAD.0000000000000082. [DOI] [PubMed] [Google Scholar]
- 74.Nichols BE, Sigaloff KC, Kityo C, Hamers RL, Baltussen R, Bertagnolio S, Jordan MR, Hallett TB, Boucher CA, de Wit TF, et al. Increasing the use of second-line therapy is a cost-effective approach to prevent the spread of drug-resistant HIV: a mathematical modelling study. J Int AIDS Soc. 2014;17:19164. doi: 10.7448/IAS.17.1.19164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nichols BE, Sigaloff KC, Kityo C, Mandaliya K, Hamers RL, Bertagnolio S, Jordan MR, Boucher CA, Rinke de Wit TF, van de Vijver DA. Averted HIV infections due to expanded antiretroviral treatment eligibility offsets risk of transmitted drug resistance: a modeling study. Aids. 2014;28(1):73–83. doi: 10.1097/01.aids.0000433239.01611.52. [DOI] [PubMed] [Google Scholar]
- 76.Iwani S, Suzuki T, Takeuchi Y. Paradox of vaccination: is vaccination really effective against avian flu epidemics? PLoS One. 2009;4(3):e4915. doi: 10.1371/journal.pone.0004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCaw JM, Arinaminpathy N, Hurt AC, McVernon J, AR ML. A mathematical framework for estimating pathogen transmission fitness and inoculum size using data from a competitive mixtures animal model. PLoS Comput Biol. 2011;7(4):e1002026. doi: 10.1371/journal.pcbi.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Regoes RR, Bonhoeffer S. Emergence of drug-resistant influenza virus: population dynamical considerations. Science. 2006;312(5772):389–391. doi: 10.1126/science.1122947. [DOI] [PubMed] [Google Scholar]
- 79.Debarre F, Bonhoeffer S, Regoes RR. The effect of population structure on the emergence of drug resistance during influenza pandemics. J R Soc Interface. 2007;4(16):893–906. doi: 10.1098/rsif.2007.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y, Allen LJ, Perelson AS. Stochastic model of an influenza epidemic with drug resistance. J Theor Biol. 2007;248(1):179–193. doi: 10.1016/j.jtbi.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCaw JM, Wood JG, McCaw CT, McVernon J. Impact of emerging antiviral drug resistance on influenza containment and spread: influence of subclinical infection and strategic use of a stockpile containing one or two drugs. PLoS One. 2008;3(6):e2362. doi: 10.1371/journal.pone.0002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Handel A, Longini IM, Jr, Antia R. Intervention strategies for an influenza pandemic taking into account secondary bacterial infections. Epidemics. 2009;1(3):185–195. doi: 10.1016/j.epidem.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moghadas SM, Bowman CS, Rost G, Fisman DN, Wu J. Post-exposure prophylaxis during pandemic outbreaks. BMC Med. 2009;7:73. doi: 10.1186/1741-7015-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Den Dool C, Hak E, Bonten MJM, Wallinga J. A model-based assessment of oseltamivir prophylaxis strategies to prevent influenza in nursing homes. Emerg Infect Dis. 2009;15(10):1547–1555. doi: 10.3201/eid1510.081129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dafilis MP, Moss R, McVernon J, McCaw J. Drivers and consequences of influenza antiviral resistant-strain emergence in a capacity-constrained pandemic response. Epidemics. 2012;4(4):219–226. doi: 10.1016/j.epidem.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 86.Moghadas SM. Management of drug resistance in the population: influenza as a case study. Proc Biol Sci. 2008;275(1639):1163–1169. doi: 10.1098/rspb.2008.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moghadas SM, Bowman CS, Rost G, Wu J. Population-wide emergence of antiviral resistance during pandemic influenza. PLoS One. 2008;3(3):e1839. doi: 10.1371/journal.pone.0001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alexander ME, Dietrich SM, Hua Y, Moghadas SM. A comparative evaluation of modelling strategies for the effect of treatment and host interactions on the spread of drug resistance. J Theor Biol. 2009;259(2):253–263. doi: 10.1016/j.jtbi.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arino J, Bowman CS, Moghadas SM. Antiviral resistance during pandemic influenza: implications for stockpiling and drug use. BMC Infect Dis. 2009;9:8. doi: 10.1186/1471-2334-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shim E, Chapman GB, Galvani AP. Decision making with regard to antiviral intervention during an influenza pandemic. Med Decis Mak. 2010;30(4):E64–E81. doi: 10.1177/0272989X10374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patterson-Lomba O, Althouse BM, Goerg GM, Hebert-Dufresne L. Optimizing treatment regimes to hinder antiviral resistance in influenza across time scales. PLoS One. 2013;8(3):e59529. doi: 10.1371/journal.pone.0059529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alexander ME, Bowman CS, Feng Z, Gardam M, Moghadas SM, Rost G, Wu J, Yan P. Emergence of drug resistance: implications for antiviral control of pandemic influenza. Proc Biol Sci. 2007;274(1619):1675–1684. doi: 10.1098/rspb.2007.0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaberi-Douraki M, Heffernan JM, Wu J, Moghadas SM. Optimal treatment profile during an influenza epidemic. Differential Equations and Dynamical Systems. 2013;21(3):237–252. doi: 10.1007/s12591-012-0149-z. [DOI] [Google Scholar]
- 94.Jaberi-Douraki M, Moghadas SM. Optimality of a time-dependent treatment profile during an epidemic. J Biol Dyn. 2013;7:133–147. doi: 10.1080/17513758.2013.816377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hansen J, Day T. Coinfection and the evolution of drug resistance. J Evol Biol. 2014;27(12):2595–2604. doi: 10.1111/jeb.12518. [DOI] [PubMed] [Google Scholar]
- 96.Laxminarayan R, Over M, Smith DL. Will a global subsidy of new antimalarials delay the emergence of resistance and save lives? Health Aff. 2006;25(2):325–336. doi: 10.1377/hlthaff.25.2.325. [DOI] [PubMed] [Google Scholar]
- 97.Maude RJ, Pontavornpinyo W, Saralamba S, Aguas R, Yeung S, Dondorp AM, Day NP, White NJ, White LJ. The last man standing is the most resistant: eliminating artemisinin-resistant malaria in Cambodia. Malar J. 2009;8:31. doi: 10.1186/1475-2875-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maude RJ, Socheat D, Nguon C, Saroth P, Dara P, Li G, Song J, Yeung S, Dondorp AM, Day NP, et al. Optimising strategies for Plasmodium falciparum malaria elimination in Cambodia: primaquine, mass drug administration and artemisinin resistance. PLoS One. 2012;7(5):e37166. doi: 10.1371/journal.pone.0037166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maude RJ, Nguon C, Dondorp AM, White LJ, White NJ. The diminishing returns of atovaquone-proguanil for elimination of Plasmodium falciparum malaria: modelling mass drug administration and treatment. Malar J. 2014;13:380. doi: 10.1186/1475-2875-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alexander N, Sutherland C, Roper C, Cissé B, Schellenberg D. Modelling the impact of intermittent preventive treatment for malaria on selection pressure for drug resistance. Malar J. 2007;6:9. doi: 10.1186/1475-2875-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gingras G, Guertin MH, Laprise JF, Drolet M, Brisson M. Mathematical modeling of the transmission dynamics of Clostridium difficile infection and colonization in healthcare settings: a systematic review. PLoS One. 2016;11(9):e0163880. doi: 10.1371/journal.pone.0163880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rupnow MF, Shachter RD, Owens DK, Parsonnet J. A dynamic transmission model for predicting trends in Helicobacter pylori and associated diseases in the United States. Emerg Infect Dis. 2000;6(3):228–237. doi: 10.3201/eid0603.000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coen PG, Heath PT, Barbour ML, Garnett GP. Mathematical models of Haemophilus influenzae type b. Epidemiol Infect. 1998;120(3):281–295. doi: 10.1017/S0950268898008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Panackal AA. Optimizing containment and control of Candida parapsilosis Fungemia among neonates in the outbreak setting using a mathematical modeling approach. J Mycol. 2013;2013:11. [Google Scholar]
- 105.Anstey NM, Russell B, Yeo TW, Price RN. The pathophysiology of vivax malaria. Trends Parasitol. 2009;25(5):220–227. doi: 10.1016/j.pt.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 106.Baird JK. Evidence and implications of mortality associated with acute Plasmodium vivax malaria. Clin Microbiol Rev. 2013;26(1):36–57. doi: 10.1128/CMR.00074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14(10):982–991. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther. 2004;9(1):57–65. [PubMed] [Google Scholar]
- 109.Boyer PL, Clark PK, Hughes SH. HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J Virol. 2012;86(10):5885–5894. doi: 10.1128/JVI.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Visseaux B, Damond F, Matheron S, Descamps D, Charpentier C. Hiv-2 molecular epidemiology. Infect Genet Evol. 2016;46:233–240. doi: 10.1016/j.meegid.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 111.Morrill HJ, Caffrey AR, Jump RL, Dosa D, LaPlante KL. Antimicrobial stewardship in long-term care facilities: a call to action. J Am Med Dir Assoc. 2016;17(2):183 e181–183 e116. doi: 10.1016/j.jamda.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Augustine S, Bonomo RA. Taking stock of infections and antibiotic resistance in the elderly and long-term care facilities: a survey of existing and upcoming challenges. Eur J Microbiol Immunol (Bp) 2011;1(3):190–197. doi: 10.1556/EuJMI.1.2011.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Buul LW, van der Steen JT, Veenhuizen RB, Achterberg WP, Schellevis FG, Essink RT, van Benthem BH, Natsch S, Hertogh CM. Antibiotic use and resistance in long term care facilities. J Am Med Dir Assoc. 2012;13(6):568 e561–568 e513. doi: 10.1016/j.jamda.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 114.Cabello FC, Godfrey HP, Buschmann AH, Dolz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis. 2016;16(7):e127–e133. doi: 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- 115.Watts Joy, Schreier Harold, Lanska Lauma, Hale Michelle. The Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Marine Drugs. 2017;15(6):158. doi: 10.3390/md15060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegard B, Soderstrom H, Larsson DG. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One. 2011;6(2):e17038. doi: 10.1371/journal.pone.0017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol. 2012;46(21):11541–11549. doi: 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 118.Hocquet D, Muller A, Bertrand X. What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect. 2016;93(4):395–402. doi: 10.1016/j.jhin.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 119.Caminade Cyril, McIntyre K. Marie, Jones Anne E. Impact of recent and future climate change on vector-borne diseases. Annals of the New York Academy of Sciences. 2018;1436(1):157–173. doi: 10.1111/nyas.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.MacFadden DR, McGough SF, Fisman D, Santillana M, Brownstein JS. Antibiotic resistance increases with local temperature. Nat Clim Chang. 2018;8(6):510–514. doi: 10.1038/s41558-018-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Holloway K, Mathai E, Gray A, Community-Based Surveillance of Antimicrobial U, Resistance in Resource-Constrained Settings Project G Surveillance of antimicrobial resistance in resource-constrained settings - experience from five pilot projects. Tropical Med Int Health. 2011;16(3):368–374. doi: 10.1111/j.1365-3156.2010.02696.x. [DOI] [PubMed] [Google Scholar]
- 122.Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6:47. doi: 10.1186/s13756-017-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Funk S, Salathe M, Jansen VA. Modelling the influence of human behaviour on the spread of infectious diseases: a review. J R Soc Interface. 2010;7(50):1247–1256. doi: 10.1098/rsif.2010.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oraby T, Thampi V, Bauch CT. The influence of social norms on the dynamics of vaccinating behaviour for paediatric infectious diseases. Proc Biol Sci. 2014;281(1780):20133172. doi: 10.1098/rspb.2013.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Funk S, Bansal S, Bauch CT, Eames KT, Edmunds WJ, Galvani AP, Klepac P. Nine challenges in incorporating the dynamics of behaviour in infectious diseases models. Epidemics. 2015;10:21–25. doi: 10.1016/j.epidem.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 126.Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Burgmann H, Sorum H, Norstrom M, Pons MN, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13(5):310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 127.Fernández Javier, Guerra Beatriz, Rodicio M. Resistance to Carbapenems in Non-Typhoidal Salmonella enterica Serovars from Humans, Animals and Food. Veterinary Sciences. 2018;5(2):40. doi: 10.3390/vetsci5020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vossenkuhl B, Brandt J, Fetsch A, Kasbohrer A, Kraushaar B, Alt K, Tenhagen BA. Comparison of spa types, SCCmec types and antimicrobial resistance profiles of MRSA isolated from turkeys at farm, slaughter and from retail meat indicates transmission along the production chain. PLoS One. 2014;9(5):e96308. doi: 10.1371/journal.pone.0096308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jobbins SE, Alexander KA. From whence they came--antibiotic-resistant Escherichia Coli in African wildlife. J Wildl Dis. 2015;51(4):811–820. doi: 10.7589/2014-11-257. [DOI] [PubMed] [Google Scholar]
- 130.Bonnedahl J, Jarhult JD. Antibiotic resistance in wild birds. Ups J Med Sci. 2014;119(2):113–116. doi: 10.3109/03009734.2014.905663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baggaley RF, Powers KA, Boily MC. What do mathematical models tell us about the emergence and spread of drug-resistant HIV? Curr Opin HIV AIDS. 2011;6(2):131–140. doi: 10.1097/COH.0b013e328343ad03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426(6967):658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gog JR, Pellis L, Wood JL, McLean AR, Arinaminpathy N, Lloyd-Smith JO. Seven challenges in modeling pathogen dynamics within-host and across scales. Epidemics. 2015;10:45–48. doi: 10.1016/j.epidem.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 134.Patyk K, Caraguel C, Kristensen C, Forde-folle K. Lexicon of disease spread modelling terms. Rev Sci Tech. 2011;30(2):547–554. doi: 10.20506/rst.30.2.2049. [DOI] [PubMed] [Google Scholar]
- 135.Mishra S, Fisman DN, Boily MC. The ABC of terms used in mathematical models of infectious diseases. J Epidemiol Community Health. 2011;65(1):87–94. doi: 10.1136/jech.2009.097113. [DOI] [PubMed] [Google Scholar]
- 136.Grimm V, Berger U, DeAngelis DL, Polhill JG, Giske J, Railsback SF. The ODD protocol: a review and first update. Ecol Model. 2010;221:2760–2768. doi: 10.1016/j.ecolmodel.2010.08.019. [DOI] [Google Scholar]
- 137.Lim YW, Steinhoff M, Girosi F, Holtzman D, Campbell H, Boer R, Black R, Mulholland K. Reducing the global burden of acute lower respiratory infections in children: the contribution of new diagnostics. Nature. 2006;444(Suppl 1):9–18. doi: 10.1038/nature05442. [DOI] [PubMed] [Google Scholar]
- 138.Bruzzese E, Giannattasio A, Guarino A. Antibiotic treatment of acute gastroenteritis in children. F1000Res. 2018;7:193. doi: 10.12688/f1000research.12328.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hurford A, Morris AM, Fisman DN, Wu J. Linking antimicrobial prescribing to antimicrobial resistance in the ICU: before and after an antimicrobial stewardship program. Epidemics. 2012;4(4):203–210. doi: 10.1016/j.epidem.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 140.Talaminos A, Lopez-Cerero L, Calvillo J, Pascual A, Roa LM, Rodriguez-Bano J. Modelling the epidemiology of Escherichia coli ST131 and the impact of interventions on the community and healthcare centres. Epidemiol Infect. 2016;144(9):1974–1982. doi: 10.1017/S0950268816000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pelat C, Kardas-Sloma L, Birgand G, Ruppe E, Schwarzinger M, Andremont A, Lucet JC, Yazdanpanah Y. Hand hygiene, cohorting, or antibiotic restriction to control outbreaks of multidrug-resistant Enterobacteriaceae. Infect Control Hosp Epidemiol. 2016;37(3):272–280. doi: 10.1017/ice.2015.284. [DOI] [PubMed] [Google Scholar]
- 142.D'Agata EMC, Horn MA, Ruan S, Webb GF, Wares JR. Efficacy of infection control interventions in reducing the spread of multidrug-resistant organisms in the hospital setting. PLoS One. 2012;7(2):e30170. doi: 10.1371/journal.pone.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park Andrew W., Haven James, Kaplan Ray, Gandon Sylvain. Refugia and the evolutionary epidemiology of drug resistance. Biology Letters. 2015;11(11):20150783. doi: 10.1098/rsbl.2015.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Laurenson YCSM, Kahn LP, Bishop SC, Kyriazakis I. Which is the best phenotypic trait for use in a targeted selective treatment strategy for growing lambs in temperate climates? Vet Parasitol. 2016;226:174–188. doi: 10.1016/j.vetpar.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 145.Leathwick DM, Waghorn TS, Miller CM, Candy PM, Oliver AM. Managing anthelmintic resistance--use of a combination anthelmintic and leaving some lambs untreated to slow the development of resistance to ivermectin. Vet Parasitol. 2012;187(1–2):285–294. doi: 10.1016/j.vetpar.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 146.Verelst Frederik, Willem Lander, Beutels Philippe. Behavioural change models for infectious disease transmission: a systematic review (2010–2015) Journal of The Royal Society Interface. 2016;13(125):20160820. doi: 10.1098/rsif.2016.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lipsitch M, Siber GR. How can vaccines contribute to solving the antimicrobial resistance problem? MBio. 2016;7(3):e00428–16. [DOI] [PMC free article] [PubMed]
- 148.Ginsburg AS, Klugman KP. Vaccination to reduce antimicrobial resistance. Lancet Glob Health. 2017;5(12):e1176–e1177. doi: 10.1016/S2214-109X(17)30364-9. [DOI] [PubMed] [Google Scholar]
- 149.Jansen KU, Knirsch C, Anderson AS. The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med. 2018;24(1):10–19. doi: 10.1038/nm.4465. [DOI] [PubMed] [Google Scholar]
- 150.Bansal S, Chowell G, Simonsen L, Vespignani A, Viboud C. Big data for infectious disease surveillance and modeling. J Infect Dis. 2016;214(suppl_4):S375–S379. doi: 10.1093/infdis/jiw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Worby CJ, Chang HH, Hanage WP, Lipsitch M. The distribution of pairwise genetic distances: a tool for investigating disease transmission. Genetics. 2014;198(4):1395–1404. doi: 10.1534/genetics.114.171538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brock Aleisha, Gibbs Carole, Ross Joshua, Esterman Adrian. The Impact of Antimalarial Use on the Emergence and Transmission of Plasmodium falciparum Resistance: A Scoping Review of Mathematical Models. Tropical Medicine and Infectious Disease. 2017;2(4):54. doi: 10.3390/tropicalmed2040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cohen T, Dye C, Colijn C, Williams B, Murray M. Mathematical models of the epidemiology and control of drug-resistant TB. Expert Rev Respir Med. 2009;3(1):67–79. doi: 10.1586/17476348.3.1.67. [DOI] [PubMed] [Google Scholar]
- 154.Wu JT, Cowling BJ. The use of mathematical models to inform influenza pandemic preparedness and response. Exp Biol Med (Maywood) 2011;236(8):955–961. doi: 10.1258/ebm.2010.010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Caudill L, Wares JR. The role of mathematical modeling in designing and evaluating antimicrobial stewardship programs. Curr Treat Options Inf Dis. 2016;8(2):124–138. doi: 10.1007/s40506-016-0074-8. [DOI] [Google Scholar]
- 156.DalBen MF, Teixeira Mendes E, Moura ML, Abdel Rahman D, Peixoto D, Alves Dos Santos S, Barcelos de Figueiredo W, Vitale Mendes P, Utino Taniguchi L, Bezerra Coutinho FA, et al. A model-based strategy to control the spread of Carbapenem-resistant Enterobacteriaceae: simulate and implement. Infect Control Hosp Epidemiol. 2016;37(11):1315–1322. doi: 10.1017/ice.2016.168. [DOI] [PubMed] [Google Scholar]
- 157.Domenech de Cellès M, Zahar JR, Abadie V, Guillemot D. Limits of patient isolation measures to control extended-spectrum beta-lactamase-producing Enterobacteriaceae: model-based analysis of clinical data in a pediatric ward. BMC Infect Dis. 2013;13:187. doi: 10.1186/1471-2334-13-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ballarin A, Posteraro B, Demartis G, Gervasi S, Panzarella F, Torelli R, Paroni Sterbini F, Morandotti G, Posteraro P, Ricciardi W, et al. Forecasting ESKAPE infections through a time-varying auto-adaptive algorithm using laboratory-based surveillance data. BMC Infect Dis. 2014;14:634. doi: 10.1186/s12879-014-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang X, Chen Y, Zhao W, Wang Y, Song Q, Liu H, Zhao J, Han X, Hu X, Grundmann H, et al. A data-driven mathematical model of multi-drug resistant Acinetobacter baumannii transmission in an intensive care unit. Sci Rep. 2015;5:9478. doi: 10.1038/srep09478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Doan TN, Kong DCM, Marshall C, Kirkpatrick CMJ, McBryde ES. Modeling the impact of interventions against Acinetobacter baumannii transmission in intensive care units. Virulence. 2016;7(2):141–152. doi: 10.1080/21505594.2015.1076615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fresnadillo-Martinez MJ, Garcia-Merino E, Garcia-Sanchez E, Martin-del Rey A, Rodriguez-Encinas A, Rodriguez-Sanchez G, Garcia-Sanchez JE. Prevention of an outbreak of Acinetobacter baumannii in intensive care units: study of the efficacy of different mathematical methods. Revista Espanola de Quimioterapia. 2015;28(1):10–20. [PubMed] [Google Scholar]
- 162.Handel A, Regoes RR, Antia R. The role of compensatory mutations in the emergence of drug resistance. PLoS Comput Biol. 2006;2(10):e137. doi: 10.1371/journal.pcbi.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chan CH, McCabe CJ, Fisman DN. Core groups, antimicrobial resistance and rebound in gonorrhoea in North America. Sex Transm Infect. 2012;88(3):200–204. doi: 10.1136/sextrans-2011-050049. [DOI] [PubMed] [Google Scholar]
- 164.Hui BB, Ryder N, Su JY, Ward J, Chen MY, Donovan B, Fairley CK, Guy RJ, Lahra MM, Law MG, et al. Exploring the benefits of molecular testing for gonorrhoea antibiotic resistance surveillance in remote settings. PLoS One. 2015;10(7):e0133202. doi: 10.1371/journal.pone.0133202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Trecker MA, Hogan DJ, Waldner CL, Dillon JA, Osgood ND. Revised simulation model does not predict rebound in gonorrhoea prevalence where core groups are treated in the presence of antimicrobial resistance. Sex Transm Infect. 2015;91(4):300–302. doi: 10.1136/sextrans-2014-051792. [DOI] [PubMed] [Google Scholar]
- 166.Xiridou M, Soetens LC, Koedijk FD, MA VANDERS, Wallinga J. Public health measures to control the spread of antimicrobial resistance in Neisseria gonorrhoeae in men who have sex with men. Epidemiol Infect. 2015;143(8):1575–1584. doi: 10.1017/S0950268814002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fingerhuth SM, Bonhoeffer S, Low N, Althaus CL. Antibiotic-resistant Neisseria gonorrhoeae spread faster with more treatment, not more sexual partners. PLoS Pathog. 2016;12(5):e1005611. doi: 10.1371/journal.ppat.1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bootsma MCJ, Diekmann O, Bonten MJM. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proc Natl Acad Sci U S A. 2006;103(14):5620–5625. doi: 10.1073/pnas.0510077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McBryde ES, Pettitt AN, McElwain DL. A stochastic mathematical model of methicillin resistant Staphylococcus aureus transmission in an intensive care unit: predicting the impact of interventions. J Theor Biol. 2007;245(3):470–481. doi: 10.1016/j.jtbi.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 170.Pettitt AN, Forrester ML, Gibson GJ. Bayesian inference of hospital-acquired infectious diseases and control measures given imperfect surveillance data. Biostatistics. 2007;8(2):383–401. doi: 10.1093/biostatistics/kxl017. [DOI] [PubMed] [Google Scholar]
- 171.Robotham JV, Jenkins DR, Medley GF. Screening strategies in surveillance and control of methicillin-resistant Staphylococcus aureus (MRSA) Epidemiol Infect. 2007;135(2):328–342. doi: 10.1017/S095026880600687X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Drovandi CC, Pettitt AN. Multivariate Markov process models for the transmission of methicillin-resistant Staphylococcus aureus in a hospital ward. Biometrics. 2008;64(3):851–859. doi: 10.1111/j.1541-0420.2007.00933.x. [DOI] [PubMed] [Google Scholar]
- 173.Allen BD, Perla RJ. A long-term forecast of MRSA daily burden using logistic modeling. Clin Lab Sci. 2009;22(1):26–29. [PubMed] [Google Scholar]
- 174.Beggs CB, Shepherd SJ, Kerr KG. How does healthcare worker hand hygiene behaviour impact upon the transmission of MRSA between patients?: an analysis using a Monte Carlo model. BMC Infect Dis. 2009;9:64. doi: 10.1186/1471-2334-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.D'Agata EM, Webb GF, Horn MA, Moellering RC, Jr, Ruan S. Modeling the invasion of community-acquired methicillin-resistant Staphylococcus aureus into hospitals. Clin Infect Dis. 2009;48(3):274–284. doi: 10.1086/595844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Skov RL, Jensen KS. Community-associated meticillin-resistant Staphylococcus aureus as a cause of hospital-acquired infections. J Hosp Infect. 2009;73(4):364–370. doi: 10.1016/j.jhin.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 177.Barnes S, Golden B, Wasil E. MRSA transmission reduction using agent-based modeling and simulation. INFORMS J Comput. 2010;22(4):635–646. doi: 10.1287/ijoc.1100.0386. [DOI] [Google Scholar]
- 178.Donker T, Wallinga J, Grundmann H. Patient referral patterns and the spread of hospital-acquired infections through national health care networks. PLoS Comput Biol. 2010;6(3):e1000715. doi: 10.1371/journal.pcbi.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Friedman A, Ziyadi N, Boushaba K. A model of drug resistance with infection by health care workers. Math Biosci Eng. 2010;7(4):779–792. doi: 10.3934/mbe.2010.7.779. [DOI] [PubMed] [Google Scholar]
- 180.Lee BY, Bailey RR, Smith KJ, Muder RR, Strotmeyer ES, Lewis GJ, Ufberg PJ, Song Y, Harrison LH. Universal methicillin-resistant Staphylococcus aureus (MRSA) surveillance for adults at hospital admission: an economic model and analysis. Infect Control Hosp Epidemiol. 2010;31(6):598–606. doi: 10.1086/652524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Barnes SL, Harris AD, Golden BL, Wasil EA, Furuno JP. Contribution of interfacility patient movement to overall methicillin-resistant Staphylococcus aureus prevalence levels. Infect Control Hosp Epidemiol. 2011;32(11):1073–1078. doi: 10.1086/662375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bootsma MC, Wassenberg MW, Trapman P, Bonten MJ. The nosocomial transmission rate of animal-associated ST398 meticillin-resistant Staphylococcus aureus. J R Soc Interface. 2011;8(57):578–584. doi: 10.1098/rsif.2010.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Christopher S, Verghis RM, Antonisamy B, Sowmyanarayanan TV, Brahmadathan KN, Kang G, Cooper BS. Transmission dynamics of methicillin-resistant Staphylococcus aureus in a medical intensive care unit in India. PLoS One. 2011;6(7):e20604. doi: 10.1371/journal.pone.0020604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kouyos RD, Abelzur Wiesch P, Bonhoeffer S. Informed switching strongly decreases the prevalence of antibiotic resistance in hospital wards. PLoS Comput Biol. 2011;7(3):e1001094. doi: 10.1371/journal.pcbi.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee BY, McGlone SM, Wong KF, Yilmaz SL, Avery TR, Song Y, Christie R, Eubank S, Brown ST, Epstein JM, et al. Modeling the spread of methicillin-resistant Staphylococcus aureus (MRSA) outbreaks throughout the hospitals in Orange County, California. Infect Control Hosp Epidemiol. 2011;32(6):562–572. doi: 10.1086/660014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee BY, Song Y, McGlone SM, Bailey RR, Feura JM, Tai JH, Lewis GJ, Wiringa AE, Smith KJ, Muder RR, et al. The economic value of screening haemodialysis patients for methicillin-resistant Staphylococcus aureus in the USA. Clin Microbiol Infect. 2011;17(11):1717–1726. doi: 10.1111/j.1469-0691.2011.03525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Milazzo L, Bown JL, Eberst A, Phillips G, Crawford JW. Modelling of healthcare associated infections: a study on the dynamics of pathogen transmission by using an individual-based approach. Comput Methods Prog Biomed. 2011;104(2):260–265. doi: 10.1016/j.cmpb.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chamchod F, Ruan S. Modeling methicillin-resistant Staphylococcus aureus in hospitals: transmission dynamics, antibiotic usage and its history. Theor Biol Med Model. 2012;9:25. doi: 10.1186/1742-4682-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chamchod F, Ruan S. Modeling the spread of methicillin-resistant Staphylococcus aureus in nursing homes for elderly. PLoS One. 2012;7(1):e29757. doi: 10.1371/journal.pone.0029757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gurieva T, Bootsma MC, Bonten MJ. Successful Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections revisited. Clin Infect Dis. 2012;54(11):1618–1620. doi: 10.1093/cid/cis272. [DOI] [PubMed] [Google Scholar]
- 191.Gurieva TV, Bootsma MC, Bonten MJ. Decolonization of patients and health care workers to control nosocomial spread of methicillin-resistant Staphylococcus aureus: a simulation study. BMC Infect Dis. 2012;12:302. doi: 10.1186/1471-2334-12-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hall IM, Barrass I, Leach S, Pittet D, Hugonnet S. Transmission dynamics of methicillin-resistant Staphylococcus aureus in a medical intensive care unit. J R Soc Interface. 2012;9(75):2639–2652. doi: 10.1098/rsif.2012.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nielsen KL, Pedersen TM, Udekwu KI, Petersen A, Skov RL, Hansen LH, Hughes D, Frimodt-Moller N. Fitness cost: a bacteriological explanation for the demise of the first international methicillin-resistant Staphylococcus aureus epidemic. J Antimicrob Chemother. 2012;67(6):1325–1332. doi: 10.1093/jac/dks051. [DOI] [PubMed] [Google Scholar]
- 194.Tekle YI, Nielsen KM, Liu J, Pettigrew MM, Meyers LA, Galvani AP, Townsend JP. Controlling antimicrobial resistance through targeted, vaccine-induced replacement of strains. PLoS One. 2012;7(12):e50688. doi: 10.1371/journal.pone.0050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang X, Xiao Y, Wang J, Lu X. A mathematical model of effects of environmental contamination and presence of volunteers on hospital infections in China. J Theor Biol. 2012;293:161–173. doi: 10.1016/j.jtbi.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 196.Deeny SR, Cooper BS, Cookson B, Hopkins S, Robotham JV. Targeted versus universal screening and decolonization to reduce healthcare-associated meticillin-resistant Staphylococcus aureus infection. J Hosp Infect. 2013;85(1):33–44. doi: 10.1016/j.jhin.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 197.Gurieva T, Bootsma MC, Bonten MJ. Cost and effects of different admission screening strategies to control the spread of methicillin-resistant Staphylococcus aureus. PLoS Comput Biol. 2013;9(2):e1002874. doi: 10.1371/journal.pcbi.1002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kardas-Sloma L, Boelle PY, Opatowski L, Guillemot D, Temime L. Antibiotic reduction campaigns do not necessarily decrease bacterial resistance: the example of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57(9):4410–4416. doi: 10.1128/AAC.00711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kong F, Paterson DL, Whitby M, Coory M, AC C. A hierarchical spatial modelling approach to investigate MRSA transmission in a tertiary hospital. BMC Infect Dis. 2013;13:449. doi: 10.1186/1471-2334-13-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lee BY, Bartsch SM, Wong KF, Singh A, Avery TR, Kim DS, Brown ST, Murphy CR, Yilmaz SL, Potter MA, et al. The importance of nursing homes in the spread of methicillin-resistant Staphylococcus aureus (MRSA) among hospitals. Med Care. 2013;51(3):205–215. doi: 10.1097/MLR.0b013e3182836dc2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lee BY, Singh A, Bartsch SM, Wong KF, Kim DS, Avery TR, Brown ST, Murphy CR, Yilmaz SL, Huang SS. The potential regional impact of contact precaution use in nursing homes to control methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol. 2013;34(2):151–160. doi: 10.1086/669091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lee BY, Yilmaz SL, Wong KF, Bartsch SM, Eubank S, Song Y, Avery TR, Christie R, Brown ST, Epstein JM, et al. Modeling the regional spread and control of vancomycin-resistant enterococci. Am J Infect Control. 2013;41(8):668–673. doi: 10.1016/j.ajic.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Moxnes JF, de Blasio BF, Leegaard TM, AEF M. Methicillin-resistant Staphylococcus aureus (MRSA) is increasing in Norway: a time series analysis of reported MRSA and methicillin-sensitive S. aureus cases, 1997-2010. PLoS One. 2013;8(8):e70499. doi: 10.1371/journal.pone.0070499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Plipat N, Spicknall IH, Koopman JS, Eisenberg JNS. The dynamics of methicillin-resistant Staphylococcus aureus exposure in a hospital model and the potential for environmental intervention. BMC Infect Dis. 2013;13:11. doi: 10.1186/1471-2334-13-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang X, Panchanathan S, Chowell G. A data-driven mathematical model of CA-MRSA transmission among age groups: evaluating the effect of control interventions. PLoS Comput Biol. 2013;9(11):e1003328. doi: 10.1371/journal.pcbi.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Worby CJ, Jeyaratnam D, Robotham JV, Kypraios T, O'Neill PD, De Angelis D, French G, Cooper BS. Estimating the effectiveness of isolation and decolonization measures in reducing transmission of methicillin-resistant Staphylococcus aureus in hospital general wards. Am J Epidemiol. 2013;177(11):1306–1313. doi: 10.1093/aje/kws380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Deeny SR, Worby CJ, Tosas Auguet O, Cooper BS, Edgeworth J, Cookson B, Robotham JV. Impact of mupirocin resistance on the transmission and control of healthcare-associated MRSA. J Antimicrob Chemother. 2015;70(12):3366–3378. doi: 10.1093/jac/dkv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gidengil CA, Gay C, Huang SS, Platt R, Yokoe D, Lee GM. Cost-effectiveness of strategies to prevent methicillin-resistant Staphylococcus aureus transmission and infection in an intensive care unit. Infect Control Hosp Epidemiol. 2015;36(1):17–27. doi: 10.1017/ice.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ziakas PD, Zacharioudakis IM, Zervou FN, Mylonakis E. Methicillin-resistant staphylococcus aureus prevention strategies in the ICU: a clinical decision analysis. Crit Care Med. 2015;43(2):382–393. doi: 10.1097/CCM.0000000000000711. [DOI] [PubMed] [Google Scholar]
- 210.Agusto FB. Optimal control of methicillin-resistant Staphylococcus aureus transmission in hospital settings. App Math Modell. 2016;40(7–8):4822–4843. doi: 10.1016/j.apm.2015.12.006. [DOI] [Google Scholar]
- 211.Berk Z, Laurenson Y, Forbes AB, Kyriazakis I. Modelling the consequences of targeted selective treatment strategies on performance and emergence of anthelmintic resistance amongst grazing calves. Int J Parasitol Drugs Drug Resist. 2016;6(3):258–271. doi: 10.1016/j.ijpddr.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ding W, Webb GF. Optimal control applied to community-acquired methicillin-resistant Staphylococcus aureus in hospitals. J Biol Dyn. 2016;12:1–14. [DOI] [PubMed]
- 213.Hetem DJ, Bootsma MCJ, Bonten MJM, Weinstein RA. Prevention of surgical site infections: decontamination with mupirocin based on preoperative screening for Staphylococcus aureus carriers or universal decontamination? Clin Infect Dis. 2016;62(5):631–636. doi: 10.1093/cid/civ990. [DOI] [PubMed] [Google Scholar]
- 214.López-García M. Stochastic descriptors in an SIR epidemic model for heterogeneous individuals in small networks. Math Biosci. 2016;271:42–61. doi: 10.1016/j.mbs.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 215.Robotham JV, Deeny SR, Fuller C, Hopkins S, Cookson B, Stone S. Cost-effectiveness of national mandatory screening of all admissions to English National Health Service hospitals for meticillin-resistant Staphylococcus aureus: a mathematical modelling study. Lancet Infect Dis. 2016;16(3):348–356. doi: 10.1016/S1473-3099(15)00417-X. [DOI] [PubMed] [Google Scholar]
- 216.Hetem DJ, Westh H, Boye K, Jarlov JO, Bonten MJ, Bootsma MC. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish hospitals. J Antimicrob Chemother. 2012;67(7):1775–1780. doi: 10.1093/jac/dks125. [DOI] [PubMed] [Google Scholar]
- 217.Kajita E, Okano JT, Bodine EN, Layne SP, Blower S. Modelling an outbreak of an emerging pathogen. Nat Rev Microbiol. 2007;5(9):700–709. doi: 10.1038/nrmicro1660. [DOI] [PubMed] [Google Scholar]
- 218.Wang X, Xiao Y, Wang J, Lu X. Stochastic disease dynamics of a hospital infection model. Math Biosci. 2013;241(1):115–124. doi: 10.1016/j.mbs.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 219.Kardas-Sloma L, Boelle PY, Opatowski L, Brun-Buisson C, Guillemot D, Temime L. Impact of antibiotic exposure patterns on selection of community-associated methicillin-resistant Staphylococcus aureus in hospital settings. Antimicrob Agents Chemother. 2011;55(10):4888–4895. doi: 10.1128/AAC.01626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cooper BS, Kypraios T, Batra R, Wyncoll D, Tosas O, Edgeworth JD. Quantifying type-specific reproduction numbers for nosocomial pathogens: evidence for heightened transmission of an Asian sequence type 239 MRSA clone. PLoS Comput Biol. 2012;8(4):e1002454. doi: 10.1371/journal.pcbi.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Batina NG, Crnich CJ, Anderson DF, Döpfer D. Identifying conditions for elimination and epidemic potential of methicillin-resistant Staphylococcus aureus in nursing homes. Antimicrob Resist Infect Control. 2016;5:32. [DOI] [PMC free article] [PubMed]
- 222.McBryde ES, McElwain DLS. A mathematical model investigating the impact of an environmental reservoir on the prevalence and control of vancomycin-resistant enterococci. J Infect Dis. 2006;193(10):1473–1474. doi: 10.1086/503439. [DOI] [PubMed] [Google Scholar]
- 223.Cooper BS, Medley GF, Bradley SJ, Scott GM. An augmented data method for the analysis of nosocomial infection data. Am J Epidemiol. 2008;168(5):548–557. doi: 10.1093/aje/kwn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wolkewitz M, Dettenkofer M, Bertz H, Schumacher M, Huebner J. Statistical epidemic modeling with hospital outbreak data. Stat Med. 2008;27(30):6522–6531. doi: 10.1002/sim.3419. [DOI] [PubMed] [Google Scholar]
- 225.Ortiz AR, Banks HT, Castillo-Chavez C, Chowell G, Wang X. A deterministic methodology for estimation of parameters in dynamic Markov chain models. J Biol Syst. 2011;19(1):71–100. doi: 10.1142/S0218339011003798. [DOI] [Google Scholar]
- 226.Yahdi M, Abdelmageed S, Lowden J, Tannenbaum L. Vancomycin-resistant enterococci colonization-infection model: parameter impacts and outbreak risks. J Biol Dyn. 2012;6:645–662. doi: 10.1080/17513758.2012.670733. [DOI] [PubMed] [Google Scholar]
- 227.Lowden J, Miller Neilan R, Yahdi M. Optimal control of vancomycin-resistant enterococci using preventive care and treatment of infections. Math Biosci. 2014;249(1):8–17. doi: 10.1016/j.mbs.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 228.Grima DT, Webb GF, D'Agata EM. Mathematical model of the impact of a nonantibiotic treatment for Clostridium difficile on the endemic prevalence of vancomycin-resistant enterococci in a hospital setting. Comput Math Methods Med. 2012;2012:605861. doi: 10.1155/2012/605861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.van Bunnik BA, Ssematimba A, Hagenaars TJ, Nodelijk G, Haverkate MR, Bonten MJ, Hayden MK, Weinstein RA, Bootsma MC, De Jong MC. Small distances can keep bacteria at bay for days. Proc Natl Acad Sci U S A. 2014;111(9):3556–3560. doi: 10.1073/pnas.1310043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Singer RS, Cox LA, Jr, Dickson JS, Hurd HS, Phillips I, Miller GY. Modeling the relationship between food animal health and human foodborne illness. Prev Vet Med. 2007;79(2–4):186–203. doi: 10.1016/j.prevetmed.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 231.Hald T, Lo Fo Wong DM, Aarestrup FM. The attribution of human infections with antimicrobial resistant Salmonella bacteria in Denmark to sources of animal origin. Foodborne Pathog Dis. 2007;4(3):313–326. doi: 10.1089/fpd.2007.0002. [DOI] [PubMed] [Google Scholar]
- 232.Pitzer VE, Feasey NA, Msefula C, Mallewa J, Kennedy N, Dube Q, Denis B, Gordon MA, Heyderman RS. Mathematical modeling to assess the drivers of the recent emergence of typhoid fever in Blantyre, Malawi. Clin Infect Dis. 2015;61(Suppl 4):S251–S258. doi: 10.1093/cid/civ710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Maher MC, Alemayehu W, Lakew T, Gaynor BD, Haug S, Cevallos V, Keenan JD, Lietman TM, Porco TC. The fitness cost of antibiotic resistance in Streptococcus pneumoniae: insight from the field. PLoS One. 2012;7(1):e29407. doi: 10.1371/journal.pone.0029407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Geli P, Rolfhamre P, Almeida J, Ekdahl K. Modeling pneumococcal resistance to penicillin in southern Sweden using artificial neural networks. Microbial Drug Resist. 2006;12(3):149–157. doi: 10.1089/mdr.2006.12.149. [DOI] [PubMed] [Google Scholar]
- 235.Wang YC, Lipsitch M. Upgrading antibiotic use within a class: tradeoff between resistance and treatment success. Proc Natl Acad Sci U S A. 2006;103(25):9655–9660. doi: 10.1073/pnas.0600636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Opatowski L, Mandel J, Varon E, Boelle PY, Temime L, Guillemot D. Antibiotic dose impact on resistance selection in the community: a mathematical model of beta-lactams and Streptococcus pneumoniae dynamics. Antimicrob Agents Chemother. 2010;54(6):2330–2337. doi: 10.1128/AAC.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Domenech de Celles M, Opatowski L, Salomon J, Varon E, Carbon C, Boelle PY, Guillemot D. Intrinsic epidemicity of Streptococcus pneumoniae depends on strain serotype and antibiotic susceptibility pattern. Antimicrob Agents Chemother. 2011;55(11):5255–5261. doi: 10.1128/AAC.00249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Opatowski L, Varon E, Dupont C, Temime L, van der Werf S, Gutmann L, Boelle PY, Watier L, Guillemot D. Assessing pneumococcal meningitis association with viral respiratory infections and antibiotics: insights from statistical and mathematical models. Proc Biol Sci. 2013;280(1764):20130519. doi: 10.1098/rspb.2013.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Boëlle PY, Thomas G. Resistance to antibiotics: limit theorems for a stochastic SIS model structured by level of resistance. J Math Biol. 2016;73(6–7):1353–1378. doi: 10.1007/s00285-016-0996-2. [DOI] [PubMed] [Google Scholar]
- 240.Gao D, Lietman TM, Porco TC. Antibiotic resistance as collateral damage: the tragedy of the commons in a two-disease setting. Math Biosci. 2015;263:121–132. doi: 10.1016/j.mbs.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Stelling J, Yih WK, Galas M, Kulldorff M, Pichel M, Terragno R, Tuduri E, Espetxe S, Binsztein N, O'Brien TF, et al. Automated use of WHONET and SaTScan to detect outbreaks of Shigella spp. using antimicrobial resistance phenotypes. Epidemiol Infect. 2010;138(6):873–883. doi: 10.1017/S0950268809990884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Resch SC, Salomon JA, Murray M, Weinstein MC. Cost-effectiveness of treating multidrug-resistant tuberculosis. PLoS Med. 2006;3(7):1048–1057. doi: 10.1371/journal.pmed.0030241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rodrigues P, Gomes MG, Rebelo C. Drug resistance in tuberculosis--a reinfection model. Theor Popul Biol. 2007;71(2):196–212. doi: 10.1016/j.tpb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 244.Basu S, Orenstein E, Galvani AP. The theoretical influence of immunity between strain groups on the progression of drug-resistant tuberculosis epidemics. J Infect Dis. 2008;198(10):1502–1513. doi: 10.1086/592508. [DOI] [PubMed] [Google Scholar]
- 245.Cohen T, Colijn C, Finklea B, Wright A, Zignol M, Pym A, Murray M. Are survey-based estimates of the burden of drug resistant TB too low? Insight from a simulation study. PLoS One. 2008;3(6):e2363. doi: 10.1371/journal.pone.0002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Gumel AB, Song B. Existence of multiple-stable equilibria for a multi-drug-resistant model of mycobacterium tuberculosis. Math Biosci Eng. 2008;5(3):437–455. doi: 10.3934/mbe.2008.5.437. [DOI] [PubMed] [Google Scholar]
- 247.Colijn C, Cohen T, Murray M. Latent coinfection and the maintenance of strain diversity. Bull Math Biol. 2009;71(1):247–263. doi: 10.1007/s11538-008-9361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jacob BG, Krapp F, Ponce M, Gotuzzo E, Griffith DA, Novak RJ. Accounting for autocorrelation in multi-drug resistant tuberculosis predictors using a set of parsimonious orthogonal eigenvectors aggregated in geographic space. Geospat Health. 2010;4(2):201–217. doi: 10.4081/gh.2010.201. [DOI] [PubMed] [Google Scholar]
- 249.Oxlade O, Schwartzman K, Pai M, Heymann J, Benedetti A, Royce S, Menzies D. Predicting outcomes and drug resistance with standardised treatment of active tuberculosis. Eur Respir J. 2010;36(4):870–877. doi: 10.1183/09031936.00151709. [DOI] [PubMed] [Google Scholar]
- 250.De Espíndola AL, Bauch CT, Troca Cabella BC, Martinez AS. An agent-based computational model of the spread of tuberculosis. J Stat Mech. 2011;(2011):P05003.
- 251.Liu Yongqi, Sun Zhendong, Sun Guiquan, Zhong Qiu, Jiang Li, Zhou Lin, Qiao Yupeng, Jia Zhongwei. Modeling Transmission of Tuberculosis with MDR and Undetected Cases. Discrete Dynamics in Nature and Society. 2011;2011:1–12. [Google Scholar]
- 252.Sergeev R, Colijn C, Cohen T. Models to understand the population-level impact of mixed strain M. tuberculosis infections. J Theor Biol. 2011;280(1):88–100. doi: 10.1016/j.jtbi.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Thomas EG, Barrington HE, Lokuge KM, Mercer GN. Modelling the spread of tuberculosis, including drug resistance and HIV: a case study in Papua New Guinea’s western province. ANZIAM J. 2011;52(1):26–45. doi: 10.1017/S1446181111000587. [DOI] [Google Scholar]
- 254.Liao CM, Lin YJ. Assessing the transmission risk of multidrug-resistant Mycobacterium tuberculosis epidemics in regions of Taiwan. Int J Infect Dis. 2012;16(10):e739–e747. doi: 10.1016/j.ijid.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 255.Agusto FB, Adekunle AI. Optimal control of a two-strain tuberculosis-HIV/AIDS co-infection model. Bio Systems. 2014;119:20–44. doi: 10.1016/j.biosystems.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 256.Ahmadin, Fatmawati Mathematical modeling of drug resistance in tuberculosis transmission and optimal control treatment. Applied Mathematical Sciences. 2014;8:4547–4559. doi: 10.12988/ams.2014.46492. [DOI] [Google Scholar]
- 257.Denholm JT, McBryde ES. Can Australia eliminate TB? Modelling immigration strategies for reaching MDG targets in a low-transmission setting. Aust N Z J Public Health. 2014;38(1):78–82. doi: 10.1111/1753-6405.12161. [DOI] [PubMed] [Google Scholar]
- 258.Lin YJ, Liao CM. Seasonal dynamics of tuberculosis epidemics and implications for multidrug-resistant infection risk assessment. Epidemiol Infection. 2014;142(2):358–370. doi: 10.1017/S0950268813001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Raimundo SM, Yang HM, Venturino E. Theoretical assessment of the relative incidences of sensitive and resistant tuberculosis epidemic in presence of drug treatment. Math Biosci Eng. 2014;11(4):971–993. doi: 10.3934/mbe.2014.11.971. [DOI] [Google Scholar]
- 260.Knight GM, Colijn C, Shrestha S, Fofana M, Cobelens F, White RG, Dowdy DW, Cohen T. The distribution of fitness costs of resistance-conferring mutations is a key determinant for the future burden of drug-resistant tuberculosis: a model-based analysis. Clin Infect Dis. 2015;61:S147–S154. doi: 10.1093/cid/civ579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lin HH, Wang L, Zhang H, Ruan Y, Chin DP, Dye C. Tuberculosis control in China: use of modelling to develop targets and policies. Bull World Health Org. 2015;93(11):790–798. doi: 10.2471/BLT.15.154492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sachdeva KS, Raizada N, Gupta RS, Nair SA, Denkinger C, Paramasivan CN, Kulsange S, Thakur R, Dewan P, Boehme C, et al. The potential impact of up-front drug sensitivity testing on India’s epidemic of multi-drug resistant tuberculosis. PLoS One. 2015;10(7):e0131438. doi: 10.1371/journal.pone.0131438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Trauer JM, Denholm JT, Waseem S, Ragonnet R, McBryde ES. Scenario analysis for programmatic tuberculosis control in Western Province, Papua New Guinea. Am J Epidemiol. 2016;183(12):1138–1148. doi: 10.1093/aje/kwv323. [DOI] [PubMed] [Google Scholar]
- 264.Gilbert JA, Shenoi SV, Moll AP, Friedland GH, Paltiel AD, Galvani AP. Cost-effectiveness of community-based TB/HIV screening and linkage to care in rural South Africa. PLoS One. 2016;11(12):e0165614. doi: 10.1371/journal.pone.0165614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Gilbert JA, Long EF, Brooks RP, Friedland GH, Moll AP, Townsend JP, Galvani AP, Shenoi SV. Integrating community-based interventions to reverse the convergent TB/HIV epidemics in rural South Africa. PLoS One. 2015;10(5):e0126267. doi: 10.1371/journal.pone.0126267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Trend in the number of AMR models per year. Figure S2. (a) Field-weighted citation impact (FWCI) of AMR publications by year (excludes 3 publications with FWCI greater than 10) and (b) FWCI by pathogen category: viral (V), bacterial (B), parasitic (P), and fungal (F). Figure S3. Pathogen trends over time. The proportion of articles on selected infectious diseases relative to the total publications per year published between 2006 and 2016 (articles/year): healthcare-acquired infections (HCAI), methicillin-resistant staphylococcus aureus (MRSA), tuberculosis (TB), human immunodeficiency virus (HIV), influenza and plasmodium falciparum (malaria). Figure S4. Model characteristic trends over time: The proportion of articles with a particular model approach (deterministic or stochastic, compartmental or individual-based models) or rigor (calibrated or validated) by the total publications per year published between 2006 and 2016. Table S1. References for publications divided by diseases they model. Table S2. References for publications divided by model class. Table S3. References for publications divided by disease specific interventions. (DOCX 2019 kb)
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2009 checklist. (DOC 62 kb)
Table S4. Complete table of data extracted from each publication included in this review. (XLSX 203 kb)
Data Availability Statement
Not applicable.