Abstract
Six new dammarane-type saponins, gypenosides CP1-6 (1–6), along with 19 known compounds 7–25, were isolated and characterized from the aerial parts of Gynostemma pentaphyllum. Among these compounds, eight dammarane-type saponins, 2, 5, 6, 7, 11, 12, 13, and 15, exhibited the greatest antiproliferative effects against two human tumor cell lines (A549 and HepG2).
Keywords: Gynostemma pentaphyllum, Jiaogulan, dammarane-type saponins, gypenosides antiproliferative activity
1. Introduction
Gynostemma pentaphyllum (Thunb.) Makino (family Cucurbitaceae) is an ethnomedicine frequently used in Asian countries as a functional food and tea [1,2,3,4]. Due to its various pharmacological activities, including anti-inflammatory [5,6,7], antioxidative [8,9], anti-hyperlipidemic [3,10], hypoglycemic [3,11,12], and antitumor effects [13,14,15,16,17,18], it is marketed in Asia in dietary supplements, such as Jiaogulan tea and Jiaogulan concentrated juice [4,12,19]. Dammarane-type triterpene saponins or gypenosides are the major components responsible for the plant’s pharmacological activities [1,2,3,4,5,6,7,11,15,17,20,21,22,23,24,25]. To date, more than 210 compounds, including over 180 gypenosides, along with flavonoids [9,15,17] and polysaccharides [8,16] have been isolated from G. pentaphyllum. Moreover, gypenosides are structurally like the ginsenosides, which are well-known pharmacologically active components of ginseng root (Panax ginseng) [26]. Thus, G. pentaphyllum is a unique non-Panax plant rich in gypenosides [26,27].
In our present study, 25 components were isolated from an ethanol extract of the aerial parts of G. pentaphyllum. Their structures were identified from NMR, IR and HRMS spectroscopic data. Among them, six new gypenosides CP1-6 (1–6) (Figure 1) and 10 known dammarane-type saponins 7–16, seven known flavonoid glycosides 17–23, and two known sesquiterpene glycosides 24 and 25, were obtained from the title plant. All isolated compounds were evaluated for antiproliferative activities against human lung cancer (A549) and hepatoma (HepG2) cell lines.
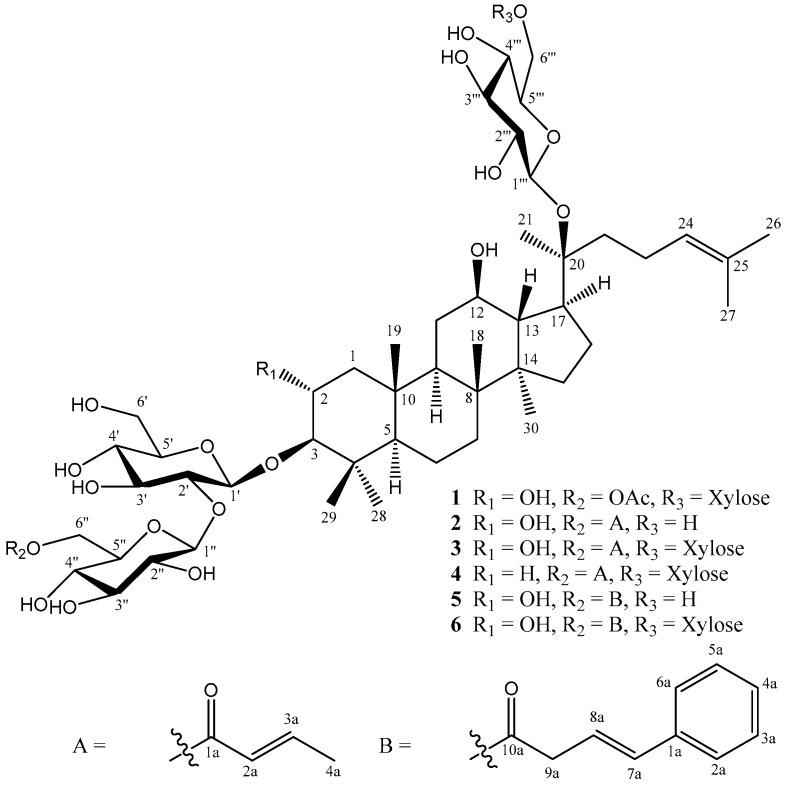
Figure 1.
Chemical structures of compounds 1–6 isolated from G. pentaphyllum.
2. Results and Discussion
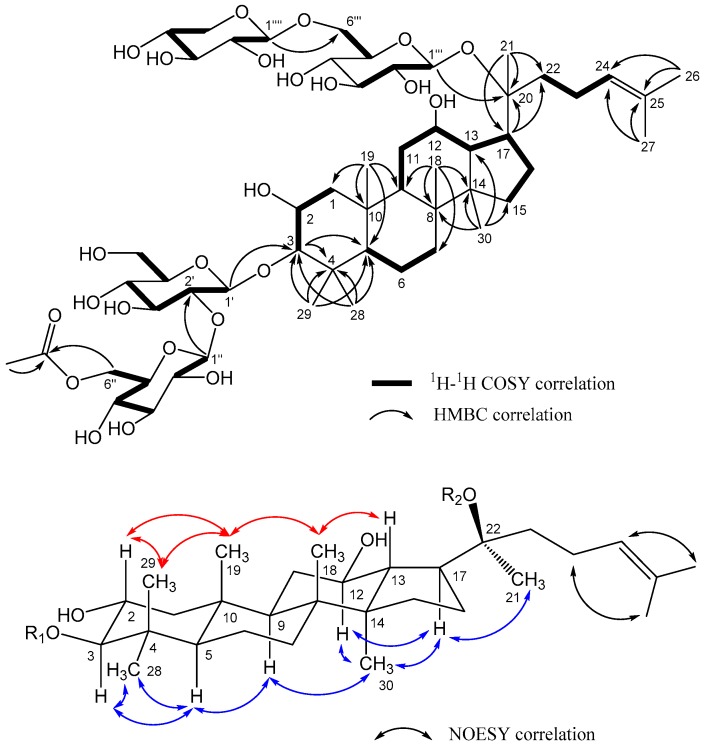
Gypenoside CP1 (1), [α]26D +11.5 (c 0.2, MeOH), has the molecular formula C55H92O24, as established by NMR and HRESIMS (m/z 1159.5874 [M + Na]+, calcd for C55H92O24Na 1159.5876), indicating ten degrees of unsaturation. The IR spectrum showed absorption bands for hydroxyl, carbonyl, and olefinic groups at 3358, 1736, and 1638 cm−1, respectively. In the 1H-NMR spectrum (Table 1 and Table 2), signals were observed for nine tertiary methyl groups [δH 0.86, 0.92, 0.97, 1.00, 1.10, 1.36, 1.62, 1.68, and 2.04 (each 3H, s)] and an olefinic proton [δH 5.13 (1H, bt, J = 7.0 Hz)]. The 13C-NMR (Table 1 and Table 2) and DEPT spectra showed resonances for 55 carbons, among which 30 were aglycone carbons including 8 methyls [δC 16.3 (C-18), 17.4 (C-30), 17.7 (C-29), 17.8 (C-19), 18.0 (C-27), 22.4 (C-21), 25.9 (C-26), 28.6 (C-28)], 4 oxygenated carbons [δC 68.2 (C-2), 71.5 (C-12), 84.9 (C-20), 96.5 (C-3)], and a pair of olefinic carbons [δC 126.1 (C-24), 132.2 (C-25)]. The 1H-1H COSY and HMBC correlations fully established the planar structure of 1 (Figure 2). Furthermore, oxygenations at C-2, C-3, and C-12 were corroborated by 1H-1H COSY correlations between H2-1 (δH 2.09; 0.87)/H-2 (δH 3.72)/H-3 (δH 2.95) and H-9 (δH 1.48)/H2-11 (δH 1.28; 1.82)/H-12 (δH 3.74)/H-13 (δH 1.73)/H-17 (δH 2.28) /H2-16 (δH 1.33; 1.89)/H2-15 (δH 1.03; 1.57) as well as the HMBC long-range correlation of H-3 with carbon signals at δC 41.8 (C-4) and 57.2 (C-5). The molecular formula and 1D and 2D-NMR spectroscopic data of 1 suggested a dammarane-type saponin, a typical constituent of Gynostemma species, with the same aglycone as that of 2α,3β,12β,20(S)-tetrahydroxydammar-24-ene [28,29]. The aglycone accounted for 30 carbon signals, leaving 25 carbon signals assignable to four sugar moieties and one acetyl group [δC 20.9 (CH3) and 172.8 (C=O)] in the 13C NMR spectrum. Four anomeric signals were observed at δH 4.30 (d, J = 7.5 Hz)/δC 105.5, δH 4.42 (d, J = 7.5 Hz)/δC 104.7, δH 4.56 (d, J = 8.0 Hz)/δC 98.1, and δH 4.71 (d, J = 7.5 Hz)/δC 105.0. HMBC cross peaks of H-1′ to C-3, H-1″ to C-2′, H-1‴ to C-20, and H-1″″ to C-6‴ determined the positions of the four sugars. Acid hydrolysis of 1 yielded d-glucose (Glc) and d-xylose (Xyl) in a ratio of 3:1 based on HPLC analysis of the component monosaccharides compared with the standard sugars [30]. A long-range correlation between proton and carbon signals at δH 4.15, 4.31 (H-6″) and δC 172.8 (C=O), respectively, was consistent with acetylation of the 6″-OH. In the NOESY spectrum of 1 (Figure 2), cross peaks were found between H-2 (δH 3.72)/Me-29 (δH 1.10), Me-29 (δH 1.10)/Me-19 (δH 0.97), Me-19 (δH 0.97)/Me-18 (δH 1.00), and Me-18 (δH 1.00)/H-13 (δH 1.73), indicating β-orientations of Me-29, Me-19, Me-18, and H-13. However, the NOESY correlations of H-3 (δH 2.95)/Me-28 (δH 0.86), H-3 (δH 2.95)/H-5 (δH 0.84), H-5 (δH 0.84)/H-9 (δH 1.48), H-9 (δH 1.48)/Me-30 (δH 0.92), and Me-30 (δH 0.92)/H-17 (δH 2.28) suggested α-orientations of H-5, H-9, H-17, Me-28, and Me-30. The configuration of C-20 in 1 was determined to be S based on a comparison of the 13C-NMR spectroscopic data of 1 and gypenoside XLVI [29]. The complete structure of 1 (gypenoside CP1) was elucidated as 2α,3β,12β,20S-tetrahydroxydammar-24-ene-3-O-[(6-O-acetyl-β-d-glucopyranosyl)-(1→2)-β-d-glucopyranosyl]-20-O-[β-d-xylopyranosyl-(1→6)-β-d-glucopyranoside].
Table 1.
1H- and 13C-NMR spectroscopic data of the aglycone of gypenosides CP1-6 (1–6).
| No. | 1 | 2 | 3 | 4 | 5 | 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δH (mult, J in Hz) | δC | δH (mult, J in Hz) | δC | δH (mult, J in Hz) | δC | δH (mult, J in Hz) | δC | δH (mult, J in Hz) | δC | δH (mult, J in Hz) | δC | |
| 1 | 2.09 (dd, 5.0, 13.0) | 47.8 | 2.07 (m) | 47.8 | 2.07 (dd, 4.8, 12.6) | 47.8 | 1.70 (m) | 40.2 | 2.03 (dd, 4.2, 12.6) | 47.7 | 2.03 (dd, 4.2, 12.6) | 47.7 |
| 0.87 (m) | 0.91 (t, 8.0) | 0.88 (m) | 0.98 (m) | 0.87 (m) | 0.87 (m) | |||||||
| 2 | 3.72 (m) | 68.2 | 3.70 (m) | 68.2 | 3.69 (m) | 68.2 | 1.68 (m); 1.95 (m) | 27.3 | 3.70 (m) | 68.2 | 3.70 (m) | 68.2 |
| 3 | 2.95 (d, 9.0) | 96.5 | 2.93 (d, 9.5) | 96.5 | 2.93(d, 9.6) | 96.5 | 3.11 (dd, 3.0, 9.0) | 91.0 | 2.92 (d, 9.6) | 96.6 | 2.92 (d, 9.6) | 96.6 |
| 4 | - | 41.8 | - | 41.8 | 41.8 | 40.5 | - | 41.8 | - | 41.8 | ||
| 5 | 0.84 (d, 11.5) | 57.2 | 0.84 (d, 10.5) | 57.2 | 0.82 (m) | 57.2 | 0.74 (d, 11.4) | 57.6 | 0.79 (d, 11.4) | 57.2 | 0.79 (d, 11.4) | 57.2 |
| 6 | 1.49 (m); 1.56 (m) | 19.3 | 1.46 (m); 1.54 (m) | 19.4 | 1.46 (m); 1.53 (m) | 19.4 | 1.43 (m); 1.53 (m) | 19.3 | 1.37 (m); 1.50 (m) | 19.4 | 1.37 (m); 1.50 (m) | 19.4 |
| 7 | 1.57 (m); 1.29, (m) | 35.7 | 1.55 (m); 1.30 (m) | 35.7 | 1.57 (m); 1.28 (m) | 35.7 | 1.55 (m); 1.26 (m) | 35.9 | 1.47 (m); 1.19 (m) | 35.6 | 1.47 (m); 1.19 (m) | 35.6 |
| 8 | - | 41.0 | - | 41.0 | - | 41.0 | - | 41.0 | 40.9 | - | 40.9 | |
| 9 | 1.48 (m) | 51.0 | 1.50 (m) | 51.0 | 1.47 (m) | 51.0 | 1.42 (dd, 3.0, 13.2) | 51.1 | 1.45 (m) | 50.9 | 1.45 (m) | 51.0 |
| 10 | - | 38.8 | - | 38.8 | - | 38.8 | 37.9 | - | 38.8 | - | 38.7 | |
| 11 | 1.82 (m);1.28 (m) | 31.0 | 1.86 (m); 1.28 (m) | 31.1 | 1.82 (m); 1.28 (m) | 31.0 | 1.78 (m); 1.22 (m) | 30.8 | 1.81 (m); 1.22 (m) | 31.1 | 1.81 (m); 1.22, (m) | 30.9 |
| 12 | 3.74 (m) | 71.5 | 3.67 (m) | 71.8 | 3.73 (m) | 71.5 | 3.71 (m) | 71.7 | 3.66 (m) | 71.8 | 3.66 (m) | 71.5 |
| 13 | 1.73 (d, 11.5) | 49.7 | 1.74 (d, 10.5) | 49.8 | 1.73 (d, 10.8) | 49.8 | 1.72 (t, 10.8) | 49.7 | 1.70 (t, 10.8) | 49.7 | 1.70 (t, 10.8) | 49.7 |
| 14 | - | 52.4 | 52.5 | - | 52.4 | - | 52.4 | - | 52.5 | - | 52.4 | |
| 15 | 1.57 (m); 1.03 (m) | 31.5 | 1.58 (m); 1.06 (m) | 31.6 | 1.58 (m); 1.04 (m) | 31.5 | 1.57 (m); 1.03 (m) | 31.5 | 1.55 (m); 1.03 (m) | 31.6 | 1.55 (m); 1.03 (m) | 31.5 |
| 16 | 1.89 (m); 1.33 (m) | 27.3 | 1.92 (m); 1.38 (m) | 27.2 | 1.89 (m); 1.33 (m) | 27.3 | 1.95 (m), 1.32 (m) | 27.3 | 1.91 (m); 1.38 (m) | 27.2 | 1.91 (m); 1.38 (m) | 27.2 |
| 17 | 2.28 (m) | 52.9 | 2.27 (m) | 53.1 | 2.28 (m) | 52.9 | 2.28 (m) | 52.9 | 2.28 (m) | 53.1 | 2.28 (m) | 52.9 |
| 18 | 1.00 (s) | 16.3 | 1.00 (s) | 16.2 | 0.99 (s) | 16.3 | 0.99 (s) | 16.3 | 0.89 (s) | 16.2 | 0.89 (s) | 16.2 |
| 19 | 0.97 (s) | 17.8 | 0.94 (s) | 17.8 | 0.93 (s) | 17.8 | 0.88 (s) | 16.8 | 0.84 (s) | 17.9 | 0.84 (s) | 17.9 |
| 20 | - | 84.9 | - | 84.9 | - | 84.9 | - | 85.0 | - | 84.9 | - | 84.9 |
| 21 | 1.36 (s) | 22.4 | 1.34 (s) | 22.8 | 1.35 (s) | 22.4 | 1.35 (s) | 22.4 | 1.33 (s) | 22.9 | 1.33 (s) | 22.4 |
| 22 | 1.79 (m); 1.53(m) | 36.8 | 1.80 (m); 1.60 (m) | 36.7 | 1.79 (m); 1.52 (m) | 36.7 | 1.80 (m); 1.52 (m) | 36.7 | 1.80 (m); 1.61 (m) | 36.7 | 1.80 (m); 1.61 (m) | 36.8 |
| 23 | 2.02 (m); 2.15 (m) | 23.8 | 2.00 (m) | 24.2 | 2.05 (m); 2.16(m) | 23.8 | 2.05 (m); 2.14 (m) | 23.8 | 2.06 (m) | 24.3 | 2.06 (m) | 24.1 |
| 24 | 5.13 (bt, 7.0) | 126.1 | 5.10 (brt, 7.0) | 125.9 | 5.11 (m) | 126.1 | 5.12 (m) | 126.1 | 5.11 (bt, 7.2) | 125.9 | 5.11 (bt, 7.2) | 126.1 |
| 25 | - | 132.2 | - | 132.3 | 132.2 | - | 132.2 | - | 132.3 | - | 132.2 | |
| 26 | 1.68 (s) | 25.9 | 1.68 (s) | 25.9 | 1.68 (s) | 25.9 | 1.68 (s) | 25.9 | 1.69 (s) | 25.9 | 1.69 (s) | 25.9 |
| 27 | 1.62 (s) | 18.0 | 1.62 (s) | 17.9 | 1.62 (s) | 18.0 | 1.62 (s) | 18.0 | 1.62 (s) | 18.0 | 1.62 (s) | 18.0 |
| 28 | 0.86 (s) | 28.6 | 0.82 (s) | 28.5 | 0.82 (s) | 28.5 | 0.77 (s) | 28.4 | 0.83 (s) | 28.6 | 0.83 (s) | 28.6 |
| 29 | 1.10 (s) | 17.7 | 1.08 (s) | 17.8 | 1.07 (s) | 17.79 | 1.02 (s) | 16.7 | 1.07 (s) | 17.9 | 1.07 (s) | 17.9 |
| 30 | 0.92 (s) | 17.4 | 0.92 (s) | 17.2 | 0.91 (s) | 17.3 | 0.91 (s) | 17.4 | 0.89 (s) | 17.1 | 0.89 (s) | 17.3 |
Table 2.
1H- and 13C-NMR spectroscopic data of the sugar moieties of gypenosides CP1-6 (1–6).
| No. | 1 | 2 | 3 | 4 | 5 | 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | δC | |
| 1′ | 4.42 (d, 7.5) | 104.7 | 4.42 (d, 7.5) | 104.6 | 4.41 (d, 7.8) | 104.6 | 4.40 (d, 7.8) | 105.2 | 4.42 (d, 7.8) | 104.6 | 4.41 (d, 7.8) | 104.6 |
| 2′ | 3.56 (m) | 82.4 | 3.55 (m) | 82.7 | 3.56 (m) | 82.7 | 3.45 (m) | 83.4 | 3.54 (dd, 7.8, 9.0) | 83.2 | 3.55 (dd, 7.8, 9.0) | 83.2 |
| 3′ | 3.58 (m) | 78.7 | 3.58 (m) | 78.7 | 3.58 (m) | 78.7 | 3.54 (t, 9.0) | 78.6 | 3.59 (m) | 78.8 | 3.58 (m) | 78.8 |
| 4′ | 3.37 (m) | 70.9 | 3.36 (m) | 70.9 | 3.36 (m) | 70.9 | 3.32 (m) | 71.2 | 3.36 (m) | 70.9 | 3.36 (m) | 70.9 |
| 5′ | 3.34 (m) | 78.0 | 3.19 (m) | 77.9 | 3.34 (m) | 78.0 | 3.23 (m) | 77.5 | 3.20 (m) | 77.9 | 3.35 (m) | 78.0 |
| 6′ | 3.85 (dd, 5.0, 12.0) | 62.3 | 3.85 (dd, 5.0, 12.0) | 62.3 | 3.86 (dd, 1.8, 12.0) | 62.3 | 3.83 (dd, 1.8, 12.0) | 62.7 | 3.86 (dd, 1.8, 12.0) | 62.3 | 3.86 (dd, 1.8, 12.0) | 62.3 |
| 3.66 (dd, 5.0, 12.0) | 3.66 (dd, 5.0, 12.0) | 3.66 (dd, 5.4, 12.0) | 3.65 (dd, 5.4, 12.0) | 3.66 (m) | 3.65 (dd, 5.4, 12.0) | |||||||
| 1″ | 4.71 (d, 7.5) | 105.0 | 4.71 (d, 7.5) | 105.2 | 4.70 (d, 7.8) | 105.2 | 4.62 (d, 7.8) | 105.5 | 4.71 (d, 7.8) | 105.4 | 4.71 (d, 7.8) | 105.4 |
| 2″ | 3.23 (m) | 76.1 | 3.24 (dd, 7.5, 9.0) | 76.1 | 3.24 (t, 8.4) | 76.1 | 3.23 (t, 8.4) | 76.4 | 3.24 (t, 8.4) | 76.1 | 3.23 (t, 8.4) | 76.1 |
| 3″ | 3.35 (m) | 77.9 | 3.35 (m) | 77.9 | 3.35 (m) | 77.9 | 3.35 (m) | 77.7 | 3.35 (m) | 78.0 | 3.34 (m) | 78.1 |
| 4″ | 3.29 (m) | 71.4 | 3.32 (m) | 71.4 | 3.32 (m) | 71.4 | 3.32 (m) | 71.3 | 3.33 (m) | 71.4 | 3.33 (m) | 71.3 |
| 5″ | 3.43 (m) | 75.3 | 3.45 (m) | 75.3 | 3.45 (m) | 75.3 | 3.44 (m) | 75.5 | 3.46 (dd, 1.8, 5.4) | 75.2 | 3.46 (m) | 75.3 |
| 6″ | 4.15 (dd, 5.0, 12.0) | 65.0 | 4.18 (dd, 5.0, 12.0) | 64.7 | 4.18 (dd, 4.8, 12.0) | 64.7 | 4.18 (dd, 4.8, 12.0) | 64.7 | 4.20 (dd, 5.4, 12.0) | 65.2 | 4.20 (dd, 4.8, 12.0) | 65.1 |
| 4.31 (dd, 5.0, 12.0) | 4.37 (dd, 5.0, 12.0) | 4.36 (dd, 1.8, 12.0) | 4.37 (dd, 1.8, 12.0) | 4.38 (dd, 1.8, 12.0) | 4.38 (dd, 1.8, 12.0) | |||||||
| 1a | 172.8 | 168.1 | 168.1 | 168.2 | 138.4 | 138.4 | ||||||
| 2a | 2.04 (s) | 20.9 | 5.88 (dq, 1.5, 15.5) | 123.6 | 5.88 (dq, 1.5, 15.6) | 123.5 | 5.89 (dq, 1.8, 15.6) | 123.5 | 7.38 (d, 7.2) | 127.4 | 7.38 (d, 7.2) | 127.3 |
| 3a | 7.00 (dq, 7.0, 15.5) | 146.6 | 7.00 (dq, 7.2, 15.5) | 146.6 | 7.00 (dq, 7.2, 15.6) | 146.6 | 7.29 (t, 7.2) | 129.6 | 7.29 (t, 7.2) | 129.7 | ||
| 4a | 1.89 (dd, 1.5, 7.0) | 18.1 | 1.89 (dd, 1.8, 7.2) | 18.1 | 1.89 (dd, 1.8, 7.2) | 18.1 | 7.20 (t, 7.2) | 128.6 | 7.21 (t, 7.2) | 128.6 | ||
| 5a | 7.29 (t, 7.2) | 129.6 | 7.29 (t, 7.2) | 129.7 | ||||||||
| 6a | 7.38 (d, 7.2) | 127.4 | 7.38 (d, 7.2) | 127.3 | ||||||||
| 7a | 6.52 (d, 16.2) | 134.6 | 6.52 (d, 16.2) | 134.6 | ||||||||
| 8a | 6.33 (dd, 7.2, 16.2) | 122.7 | 6.33 (dd, 7.2, 16.2) | 122.7 | ||||||||
| 9a | 3.30 (m) | 38.7 | 3.30 (m) | 38.7 | ||||||||
| 10a | 173.4 | 173.4 | ||||||||||
| 1’’’ | 4.56 (d, 8.0) | 98.1 | 4.60 (d, 8.0) | 98.3 | 4.56 (d, 7.8) | 98.1 | 4.56 (d, 8.4) | 98.1 | 4.59 (d, 7.8) | 98.3 | 4.56 (d, 7.8) | 98.1 |
| 2’’’ | 3.12 (m) | 75.3 | 3.08 (m) | 75.4 | 3.11 (m) | 75.3 | 3.11 (m) | 75.3 | 3.07 (m) | 75.4 | 3.12 (m) | 75.3 |
| 3’’’ | 3.33 (m) | 78.6 | 3.34 (m) | 78.3 | 3.32 (m) | 78.6 | 3.32 (m) | 78.6 | 3.35 (m) | 78.2 | 3.33 (m) | 78.6 |
| 4’’’ | 3.31 (m) | 71.4 | 3.32 (m) | 71.2 | 3.32 (m) | 71.4 | 3.32 (m) | 71.4 | 3.33 (m) | 71.2 | 3.32 (m) | 71.4 |
| 5’’’ | 3.32 (m) | 76.7 | 3.18 (m) | 78.0 | 3.39 (m) | 76.7 | 3.39 (m) | 76.7 | 3.20 (m) | 77.9 | 3.38 (m) | 76.7 |
| 6’’’ | 3.73 (dd, 5.5, 11.5) | 70.1 | 3.64 (dd, 5.5, 11.5) | 62.5 | 3.72 (dd, 5.4, 11.4) | 70.1 | 3.73 (m) | 70.1 | 3.64 (m) | 62.5 | 3.73 (dd, 5.4, 11.4) | 70.1 |
| 4.00 (dd, 2.0, 11.5) | 3.77 (dd, 2.0, 11.5) | 4.00 (dd, 2.4, 11.4) | 4.00 (dd, 1.8, 11.4) | 3.77 (dd, 2.4, 12.0) | 4.00 (dd, 1.8, 11.4) | |||||||
| 1’’’’ | 4.30 (d, 7.5) | 105.5 | 4.29 (d, 7.2) | 105.6 | 4.29 (d, 7.8) | 105.6 | 4.29 (d, 7.8) | 105.6 | ||||
| 2’’’’ | 3.20 (m) | 74.8 | 3.20 (m) | 74.8 | 3.19 (d, 7.2) | 74.8 | 3.20 (d, 7.2) | 74.8 | ||||
| 3’’’’ | 3.30 (m) | 77.5 | 3.30 (m) | 77.5 | 3.29 (m) | 77.5 | 3.30 (m) | 77.5 | ||||
| 4’’’’ | 3.47 (m) | 71.2 | 3.46 (m) | 71.2 | 3.47 (m) | 71.2 | 3.46 (m) | 71.2 | ||||
| 5’’’’ | 3.18 (m) | 66.8 | 3.18 (m) | 66.8 | 3.18 (m) | 66.8 | 3.18 (m) | 66.8 | ||||
| 4.00 (dd, 2.0, 11.5) | 3.84 (dd, 5.4, 11.4) | 3.84 (dd, 5.4, 11.4) | 3.84 (dd, 5.4, 11.4) | |||||||||
Figure 2.
Key COSY, HMBC, and NOESY correlations of 1.
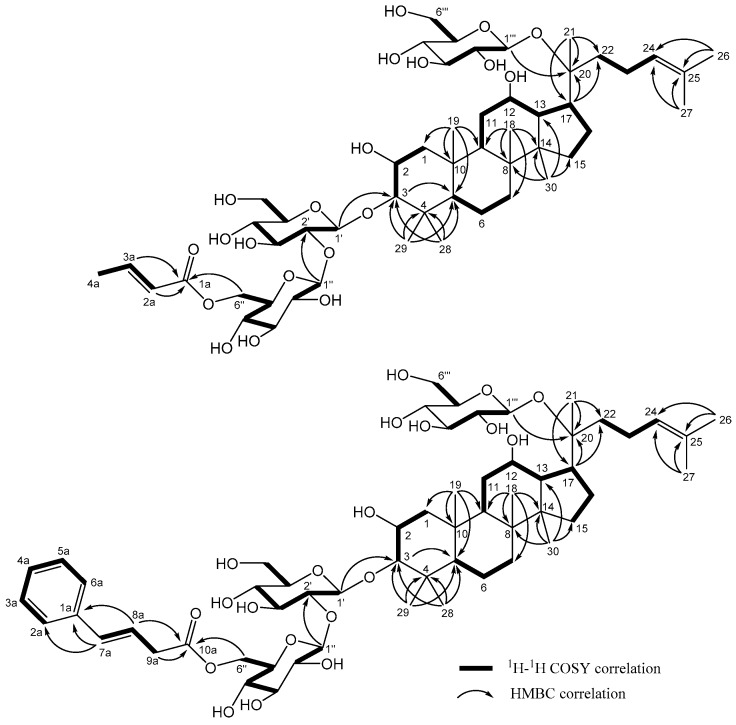
Gypenoside CP2 (2) was isolated as a white powder. Its molecular formula was determined to be C52H86O20 from HRESIMS and 13C-NMR spectroscopic analysis. Comparison of the 1H and 13C-NMR spectroscopic data of 1 and 2 (Table 1 and Table 2) indicated that both compounds have the same aglycone; however, compound 2 contains a but-2-enoyl unit (δH 7.00, 5.88, and 1.89/δC 168.1, 146.6, 123.6, and 18.1) but lacks the xylose and acetyl group found in 1. The location of the but-2-enoyl group at Glc-C-6″ was confirmed by the correlations observed in the HMBC between δH 4.18, 4.37 (H-6″) and δC 168.1 (C-1a, C=O) (Figure 3). The NMR spectra showed the presence of three β-glucopyranosyl signals [δH 4.42 (d, J = 7.5 Hz)/δC 104.6, δH 4.71 (d, J = 7.5 Hz)/δC 105.2, δH 4.60 (d, J = 8.0 Hz)/δC 98.3], which were confirmed to be from d-glucose via acid hydrolysis. Long-range correlations were also observed between δH 4.42 (H-1′) and δC 96.5 (C-3), δH 4.71 (H-1″) and δC 82.7 (C-2′), and δH 4.60 (H-1‴) and δC 84.9 (C-20) indicating that the three sugars were attached to C-3, C-2′, and C-20, respectively. Thus, the structure of gypenoside CP2 (2) was elucidated as 2α,3β,12β,20S-tetra-hydroxydammar-24-ene-3-O-{[6-O-(E)-but-2-enoyl]-β-d-glucopyranosyl-(1→2)-β-d-glucoyranosyl}-20-O-β-d-glucopyranoside.
Figure 3.
Key COSY and HMBC correlations of 2 and 5.
Gypenoside CP3 (3) was isolated as a white powder. Its molecular formula was determined as C57H94O24 from HRESIMS and 13C-NMR spectroscopic analysis. The 1H- and 13C-NMR spectra (Table 1 and Table 2) of 3 showed signals assignable to a 3-O-{[6-O-(E)-but-2-enoyl]-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosyl} moiety and a 20-O-[β-d-xylopyranosyl-(1→6)-β-d-glucopyranosyl] moiety, which were virtually superimposable onto those of 2; however, carbon signals (δC 66.8, 71.2, 74.8, 77.5, and 105.6) consistent with an additional sugar moiety were also present. Four anomeric signals were found at δH 4.29 (d, J = 7.2 Hz)/δC 105.6, δH 4.41 (d, J = 7.8 Hz)/δC 104.6, δH 4.56 (d, J = 7.8 Hz)/δC 98.1, and δH 4.70 (d, J = 7.8 Hz)/δC 105.2. Acid hydrolysis of 3 yielded d-glucose and d-xylose (3:1). The signals for CH2-6‴ (δH 3.72, 4.00/δC 70.1) in 3 were shifted downfield compared to those in 2 (δH 3.64, 3.77/δC 62.5), indicative of the attachment of a d-xylose at CH2-6‴ in 3 [29]. Moreover, long-range correlations (HMBC) between δH 4.41 (H-1′) and δC 96.5 (C-3), δH 4.70 (H-1″) and δC 82.7 (C-2′), δH 4.56 (H-1‴) and δC 84.9 (C-20), and δH 4.29 (H-1″″) and δC 70.1 (C-6‴) indicated the following sugar locations, d-glucose at C-3, C-2′, and C-20, respectively and d-xylose at C-6‴. Accordingly, compound 3 (gypenoside CP3) was determined as 2α,3β,12β,20S-tetrahydroxydammar-24-ene-3-O-{[6-O-(E)-but-2-enoyl]-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosyl}-20-O-[β-d-xylopyranosyl-(1→6)-β-d-glucopyranoside].
Gypenoside CP4 (4) was isolated as a white powder. The HRESIMS and 13C-NMR spectroscopic data of 4 suggested its molecular formula to be C57H94O23. Analysis of the 1H- and 13C-NMR spectra (Table 1 and Table 2) gave 57 signals, of which 30 were assigned to the triterpene skeleton. The further comparison of the 1D and 2D NMR data of 3 and 4 indicated the structural similarity in a 3β,12β,20S-trihydroxydammar-24-ene with four sugar moieties, except for the replacement of an oxymethine (δC 68.2, C-2) by a methylene (δC 27.3, C-2) at aglycone in 4. Detailed checking the NMR data together with the analysis of acid hydrolysis, the glycone moiety of 4 were composed 3 units of of d-glucose and one d-xylose. Thus, gypenoside CP4 was determined as 3β,12β,20S -trihydroxydammar-24-ene-3-O-{[6-O-(E)-but-2-enoyl]-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosyl}-20-O-[β-d-xylopyranosyl-(1→6)-β-d-glucopyranoside].
The HRESIMS of gypenoside CP5 (5) showed a quasimolecular ion at m/z 1129.5928 [M + Na]+ (calcd. for C58H90O20Na 1129.5923), corresponding to the molecular formula C58H90O20. Like previous isolates, compound 5 has a 2α,3β,12β,20 (S)-tetrahydroxydammar-24-ene skeleton, due to the similarity of the 1H- and 13C-NMR spectroscopic data (Table 1 and Table 2). Detailed analysis of the NMR and HRESIMS data of 5 and 2, suggested that both compounds possess the same aglycone and d-glucopyranosyl moieties, while compound 5 contains a phenyl moiety not found in 2. The cross peaks between δH 4.20, 4.38 (H-6″) and δH 173.4 (C-10a) in the HMBC spectrum of 5 (Figure 3) indicated that the (E)-but-2-enoyl ester at Glc C-6″ in 2 was replaced by a (E)-4-phenylbut-3-enoyl unit in 5. Accordingly, the structure of 5 (gypenoside CP5) was confirmed as 2α,3β,12β,20S-tetrahydroxydammar-24-ene-3-O-{[6-O-(E)-4-phenyl-but-3-enoyl]-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosyl}-20-O-β-d-glucopyranoside.
The positive HRESIMS of compound 6 showed a molecular ion peak at m/z 1261.6340 [M + Na]+ (calcd for C63H98O24Na, 1261.6346), which was 132 amu more than the molecular ion of 5, presumably corresponding to a xylose group. The 1D and 2D NMR spectroscopic data of 6 showed similar signals as those of 5 except for an additional unit in 6 characterized by a xylose signals (δC 105.6, 77.5, 74.8, 71.2, and 66.8). Acidic hydrolysis of 6 also furnished d-glucose and d-xylose. Based on the above corroborations, the structure of 6 (gypenoside CP6) was determined as 2α,3β,12β,20S-tetrahydroxydammar-24-ene-3-O-{[6-O-(E)-4-phenyl-but-3-enoyl]-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosyl}-20-O-[β-d-xylopyranosyl-(1→6)-β-d-glucopyranoside].
After the detailed spectroscopic analysis and chemical hydrolysis mentioned above, compounds 1-6 were proved as novel chemical structures and named as gypenosides CP1-CP6, respectively. The remaining nineteen isolates were identified as 2α,3β,12β,20S-tetrahydroxydammar-24-ene-3-O-β-d-glucopyranosyl-20-O-[β-d-6-O-acetylglucopyranosyl-(1→2)-β-d-glucopyranoside (7) [31], gypenoside XLVI (8) [29], gypenoside LVI (9) [29], gypenoside LVII (10) [32], gypenoside LXXVII (11) [33], gypenoside L (12) [29], 2α,3β,12β,20S-tetrahydroxydammar-24-ene-3-O-β-d-glucopyranosyl-20-O-β-d-glucopyranoside (13) [34], gypenoside XLII (14) [35], gypenoside Rd (15) [36] and 2α,3β,20S-trihydroxydammar-24-ene-3-O-[β-d-glucopyranosyl-(1→2)-β-d-glucopyranosyl]-20-O-[β-d-xylo-pyranosyl-(1→6)-β-d-glucopyranoside] (16) [37], together with seven flavonoids, quercetin-3-O-α-l-rhamnopyranosyl(1→2)-β-d-galactopyranoside (17) [38], quercetin-3-neohesperidoside (18) [39], kaempferol-3-O-α-l-rhamnopyranosyl-(1→2)-β-d-galactopyranoside (19) [40], kaempferol-3-O-α-l-rhamnopyranosyl-(1→2)-β-d-glucopyranoside (20) [41], quercetin-7-O-β-d-glucoside (21) [42], kaempferol-7-O-β-d-galactopyranoside (22) [43], and isorhamnetin-7-O-β-d-glucopyranoside (23) [44], and two sesquiterpene glucosides, (6R,7E,9R)-9-hydroxy-megastigman-4,7-dien-3-one-9-O-β-d-glucopyranoside (24) [45], and (E)-4-[3′-(β-d-glucopyranosyloxy)butylidene]-3,5,5-trimethyl-2-cyclohexen-1-one (25) [46]. The structures of the known compounds were identified by comparing their NMR data with published literature.
All isolates 1–25 were evaluated for antiproliferative activities against two human tumor cell lines, adenocarcinoma (A549) and human liver carcinoma (HepG2) and the results are shown in Table 3. Although none of the isolates showed significant cytotoxcity against the two human cell lines, certain dammarane-type triterpene saponins (2, 5, 6, 7, 11, 12, 13, and 15) were more potent than the remaining compounds. The EC50 values of these eight compounds against the HepG2 cell line ranged from 29.3 to 100.6 μM, while only four compounds (2, 11, 13 and 15) exhibited EC50 values of less than 100 μM (EC50 59.4~87.3 μM) against A549 cells. Among the six new gypenosides, compound 2 was the most potent against A549 cells and compound 5 was among the most potent against HepG2 cells.
Table 3.
Antiproliferative data for compounds 1–25 against cancer cell lines.
| Cmpd. | A549 Cell Line | HepG2 Cell Line | Number of Sugars | ||
|---|---|---|---|---|---|
| Inhibition (%) a | EC50 (μM) | Inhibition (%) | EC50 (μM) | ||
| 1 | 16.4 ± 3.97 | (-) b | 38.9 ± 3.82 | (-) | 4 |
| 2 | 84.1 ± 7.97 | 59.4 ± 2.51 | 83.0 ± 3.91 | 60.4 ± 0.63 | 3 |
| 3 | 23.3 ± 6.20 | (-) | 44.0 ± 2.28 | (-) | 4 |
| 4 | 17.0 ± 2.00 | (-) | 46.4 ± 2.60 | (-) | 4 |
| 5 | 42.7 ± 1.41 | (-) | 71.1 ± 0.60 | 29.3 ± 0.26 | 3 |
| 6 | 29.7 ± 5.34 | (-) | 55.5 ± 6.45 | 54.2 ± 2.07 | 4 |
| 7 | 37.1 ± 0.78 | (-) | 53.8 ± 3.43 | 89.1 ± 3.75 | 3 |
| 8 | 6.5 ± 5.16 | (-) | 30.7 ± 5.32 | (-) | 3 |
| 9 | 14.5 ± 6.04 | (-) | 31.1 ± 7.76 | (-) | 4 |
| 10 | 20.9 ± 2.62 | (-) | 44.1 ± 1.96 | (-) | 3 |
| 11 | 94.4 ± 0.28 | 70.1 ± 2.34 | 93.1 ± 0.99 | 76.2 ± 2.10 | 2 |
| 12 | 27.8 ± 11.35 | (-) | 60.4 ± 6.34 | 100.7 ± 1.36 | 2 |
| 13 | 65.1 ± 7.29 | 87.3 ± 3.39 | 73.3 ± 1.81 | 68.4 ± 0.57 | 2 |
| 14 | 17.9 ± 2.72 | (-) | 24.5 ± 2.79 | (-) | 4 |
| 15 | 43.2 ± 3.67 | 73.8 ± 2.86 | 56.5 ± 1.55 | 75.4 ± 1.30 | 3 |
| 16 | 25.2 ± 1.35 | (-) | 37.3 ± 0.53 | (-) | 4 |
| 17 | 13.1 ± 2.88 | (-) | 11.5 ± 1.17 | (-) | - |
| 18 | 16.1 ± 5.32 | (-) | 9.4 ± 3.02 | (-) | - |
| 19 | 19.3 ± 1.40 | (-) | 32.5 ± 3.83 | (-) | - |
| 20 | 13.0 ± 3.40 | (-) | 18.0 ± 2.99 | (-) | - |
| 21 | 15.5 ± 2.73 | (-) | 29.2 ± 2.45 | (-) | - |
| 22 | 22.9 ± 12.78 | (-) | 26.6 ± 6.30 | (-) | - |
| 23 | 21.7 ± 9.46 | (-) | 31.2 ± 3.45 | (-) | - |
| 24 | 10.9 ± 6.06 | (-) | 25.2 ± 3.65 | (-) | - |
| 25 | 5.6 ± 3.12 | (-) | 6.5 ± 3.17 | (-) | - |
a Inhibition (%) of pure compounds against cell lines at 100 μg/mL; b (-): ED50 > 100 g/mL.
3. Materials and Methods
3.1. General Experimental Procedures
The optical rotations were determined using a JASCO P-2000 polarimeter (Jasco Co., Tokyo, Japan). The infrared (IR) spectra were measured on a Mattson Genesis II spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Electrospray ionization mass spectrometry (ESIMS) data were obtained on an LCQ mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). High-resolution electronic ionization mass spectrometry (HREIMS) data were measured on a Finnigan MAT-95XL mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Nuclear magnetic resonance (NMR) spectra were recorded using by Bruker AC-400 FT-NMR (Bruker BioSpin, Rheinstetten, Germany), Varian unit Inova 500 MHz, and Varian VNMRS 600 MHz spectrometers (Aglient Technologies, Santa Clara, CA, USA). Diaion HP-20 (Mitsubishi Chemical Co., Tokyo, Japan), Sephadex LH-20 (GH Healthcare, Uppsala, Sweden), and silica gel 60 (Merck 70–230 and 230–400 mesh, Merck, Darmstadt, Germany) were used for column chromatography, and precoated silica gel (Merck 60 F-254) plates were used for TLC. The spots on TLC were detected by spraying with an anisaldehyde-sulfuric acid solution and heating at 100 °C. HPLC separations were performed on a Shimadzu LC-6AD series instrument (Shimadzu Inc., Kyoto, Japan) with an SPD-10A UV detector and a 380-LC ELSD detector (Aglient Technologies, Santa Clara, CA, USA), which was equipped with a Cosmosil 5C18 AR-II column (Nacalai Tesque, Inc., Kyoto, Japan).
3.2. Plant Material
The aerial parts of Gynostemma pentaphyllum (8.4 kg) were purchased from Zheng Yuen Tang Biotech Co. Ltd. in Kaohsiung, Taiwan in August 2013. A voucher specimen (NRICM, No. 20130101) has been deposited in the National Research Institute of Chinese Medicine, Taipei, Taiwan.
3.3. Extraction and Isolation
The dried aerial parts of G. pentaphyllum (8.4 kg) were extracted three times at 60 °C with 95% ethanol (EtOH). The EtOH soluble portion was concentrated to give a crude extract (8 L). The concentrated EtOH extract was partitioned with n-hexane and H2O (1:1, v/v) to give a n-hexane portion (847.2 g). The aqueous layer was further partitioned with EtOAc to give an EtOAc portion (279.5 g). Then, the H2O portion was loaded onto a Diaion HP-20 column (11 × 72 cm), and successively eluted with H2O, 25% MeOH, 50% MeOH, 75% MeOH, 100% MeOH, and 100% EtOAc to obtain five fractions (Fr-1 to 6). Fr-4 (298.7 g) was further chromatographed on a Sephadex LH-20 column with 60% MeOH as the eluent to give four fractions (Fr-4-1 to Fr-4-4). Fr-4-3 was further purified by semi-preparative HPLC using 35% CH3CN in H2O as the solvent system at a flow rate of 2.0 mL/min to give compounds 1 (71.5 mg), 7 (30.3 mg), 8 (54.2 mg), 9 (32.1 mg), and 16 (30.4 mg). Fr-3 was chromatographed on a LH-20 column with 60% MeOH as the eluent to yield eight fractions (Fr-3-1 to Fr-3-8). Fr-3-2 and Fr-3-3 were further separated by semi-preparative HPLC with 35%, and 18% CH3CN in H2O as the solvent system, respectively. Compounds 24 (17.2 mg) and 25 (5.4 mg) were obtained from Fr-3-3, and compound 14 (15.4 mg) was separated from Fr-3-2. Fr. 5 was further fractioned with a step gradient elution of H2O-MeOH (from 30:70 to 0:100, v/v) on a C18-gel flash column to afford six fractions (Fr-5-1 to Fr-5-6). Fr-5-2, and Fr-5-3 were further purified by semi-preparative HPLC using 38% CH3CN in H2O as the solvent system at a flow rate of 2.0 mL/min to give compounds 2 (5.2 mg), 3 (3.1 mg), 4 (2.8 mg), and 10 (4.5 mg). Fr-5-4, was further purified by semi-preparative HPLC using 48% CH3CN in H2O as the solvent system at a flow rate of 2.0 mL/min to give compounds 5 (2.6 mg), 6 (2.1 mg), 12 (11.2 mg), and 13 (7.9 mg). The EtOAc portion was loaded onto a LH-20 column eluting with CH2Cl2/MeOH (1:1, v/v) to afford 10 fractions (Fr-E-1 to Fr-E-10). Fr-E-2 was further purified by semi-preparative HPLC using 80% CH3CN in H2O as the solvent system at a flow rate of 2.0 mL/min to give compound 11 (8.7 mg). Fr-E-8 was further purified by semi-preparative HPLC using 38% CH3CN in H2O as the solvent system at a flow rate of 2.0 mL/min to give compounds 17 (13.7 mg), 18 (42.7 mg), 19 (30.2 mg), 20 (48.0 mg), 21 (7.0 mg), 22 (4.2 mg), and 23 (16.2 mg).
3.4. Spectroscopic Data (1H- and 13C-NMR spectra of 1-6 were also provided by the supplementary materials)
Gypenoside CP1 (1), White amorphous powder; [α]26D +11.5 (c 0.2, MeOH); IR (KBr) νmax 3358, 2945, 2876, 1736, 1638, 1082 cm−1; 1H- (500 MHz, methanol-d4) and 13C- (125 MHz, methanol-d4) NMR data, see Table 1 and Table 2, respectively; HRESIMS m/z 1159.5874 [M + Na]+ (calcd for C55H92O24Na, 1159.5876).
Gypenoside CP2 (2), White amorphous powder; [α]26D +15.7 (c 0.2, MeOH); IR (KBr) νmax 3362, 2941, 2872, 1715, 1650, 1074 cm−1; 1H- (500 MHz, methanol-d4) and 13C- (125 MHz, methanol-d4) NMR data, see Table 1 and Table 2, respectively; HRESIMS m/z 1053.5607 [M + Na]+ (calcd for C52H86O20Na, 1053.5610).
Gypenoside CP3 (3), White amorphous powder; [α]26D +12.9 (c 0.2, MeOH); IR (KBr) νmax 3370, 2928, 2876, 1715, 1650, 1078 cm−1; 1H- (600 MHz, methanol-d4) and 13C- (150 MHz, methanol-d4) NMR data, see Table 1 and Table 2, respectively; HRESIMS m/z 1185.6030 [M + Na]+ (calcd for C57H94O24Na, 1185.6033).
Gypenoside CP4 (4), White amorphous powder; [α]26D +6.6 (c 0.2, MeOH); IR (KBr) νmax 3350, 2937, 2868, 1728, 1650, 1078 cm−1; 1H- (600 MHz, methanol-d4) and 13C- (150 MHz, methanol-d4) NMR data, see Table 1 and Table 2, respectively; HRESIMS m/z 1169.6084 [M + Na]+ (calcd for C57H94O23Na, 1169.6084).
Gypenoside CP5 (5), White amorphous powder; [α]26D +7.7 (c 0.2, MeOH); IR (KBr) νmax 3370, 2921, 2872, 1679, 1074 cm−1; 1H- (600 MHz, methanol-d4) and 13C- (150 MHz, methanol-d4) NMR data, see Table 1 and Table 2, respectively; HRESIMS m/z 11129.5928 [M + Na]+ (calcd for C58H90O20Na, 1129.5923).
Gypenoside CP6 (6), White amorphous powder; [α]26D +12.9 (c 0.2, MeOH); IR (KBr) νmax 3370, 2921, 2864, 1687, 1070 cm−1; 1H- (600 MHz, methanol-d4) and 13C- (150 MHz, methanol-d4) NMR data, see Table 1 and Table 2, respectively; HRESIMS m/z 1261.6340 [M + Na]+ (calcd for C63H98O24Na, 1261.6346).
3.5. Acid Hydrolysis of Dammarane-Type Glycosides
Each isolated compound (1.0 mg) was treated with 2 N methanolic HCl (2 mL) under conditions of reflux at 90 °C for 1 h. Each mixture was extracted with CH2Cl2 to afford the aglycone portion, and the aqueous layer was neutralized with Na2CO3 and filtered. To the evaporated filtrate was added 1-(trimethylsilyl)imidazole and pyridine (0.2 mL), and the mixture was stirred at 60 °C for 5 min. After the reaction mixture was dried under a stream of N2, each residue was partitioned between CHCl3 and H2O. Each CH2Cl2 fraction was subjected to gas chromatography (GC, column: Varian capillary column CP-chirasil-L-val for optical isomers, 25 m × 0.25 mm, 0.12 μm; column temperature, 50–150 °C, 30 °C/min, 150–180 °C, 0.8 °C /min; injector temperature, 200 °C; He carrier gas, 2.0 kg/cm3; mass detector, Thermo, DSQ2; electron energy, 70 eV). Under these conditions, the sugars of each reactant were identified by comparison with authentic standards (d-glucose and d-xlyose).
3.6. Antiproliferation Assay
The isolates were tested for antiproliferative effects against HepG2 (human hepatocellular carcinoma), A549 (human lung adenocarcinoma) tumor cell lines and the M10 (human mammary epithelial) cell line in vitro using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] colorimetric method based on previously published procedures [47]. Two cell lines were maintained optimal medium (Life Technologies) supplemented with 2 mM l-glutamine and 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies) under standard culture conditions. After treatment with serial dilutions of tested compounds for 48 h, the alamar blue assay (Biosource International, Nivelles, Belgium) was used to obtain the half maximal inhibitory concentration (IC50). Doxorubicin was used as a positive control. Plates were incubated at 37 °C for 6 h prior to measure the absorbance at 570 nm and at 600 nm wavelengths using a spectrophotometric plate reader (DYNEX Technologies, Chantilly, VA, USA). Experimental data were normalized to control values. Mitomycin c was used as a positive control (A549 cell line: 0.1 ± 0.01 μg/mL; HepG2 cell line: 0.1 ± 0.01 μg/mL).
4. Conclusions
In this study, we isolated and characterized six new and ten known dammarane-type triterpene saponins as well as eight known flavonoids and two known sesquiterpene glucosides from a 95% EtOH extract of dried aerial parts of G. pentaphyllum. All the new chemical structures of compounds 1–6 were elucidated completely and tentatively named gypenosides CP1–CP6. These new isolates were obtained from the titled plant for the first time, and not yet found in other natural resources or synthesized molecules before. For the derived chemical structures of 5 and 6, (E)-4-phenylbut-3-enyl unit is first time to be found in nature. This study adds to the present phytochemical and properties information on this plant species, together with the studies performed and compiled by others [1], could assist in future modification of dammarane-type compounds as anticancer or other therapeutic agents.
Supplementary Materials
The 1H- and 13C-NMR spectra of compounds 1–6 are available online.
Author Contributions
Y.-H.K. and K.-H.L. supervised the study. P.-Y.C., N.-L.N. and T.-H.V. performed the experiments. L.-J.Z., C.-C.L., and Y.-C.L. analyzed the data. C.-C.C., H.-C.H., C.-C.L., Y.-C.L., Y.-Y.C. and S.L.M.-N. wrote the paper.
Funding
This work was supported by grants from the National Science Council (NSC098-2811-B-077-002 and NSC98-2320-B-077-005-MY3) and Ministry of Science and Technology (MOST104-2320-B-077 -006 -MY3), Taiwan, as well as from the Ministry of Health and Welfare (MM10601-0160), Taiwan.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 1–6 are available from the authors.
References
- 1.Razmovski-Naumovski V., Huang T.H.-W., Tran V.H., Li G.Q., Duke C.C., Roufogalsi B.D. Chemistry and Pharmacology of Gynostemma pentaphyllum. Phytochem. Rev. 2005;4:197–219. doi: 10.1007/s11101-005-3754-4. [DOI] [Google Scholar]
- 2.Shi G., Wang X., Zhang H., Zhang X., Zhao Y. New dammarane-type triterpene saponins from Gynostemma pentaphyllum and their anti-hepatic fibrosis activities in vitro. J. Funct. Foods. 2018;45:10–14. doi: 10.1016/j.jff.2018.03.016. [DOI] [Google Scholar]
- 3.Nguyen P.H., Gauhar R., Hwang S.L., Dao T.T., Park D.C., Kim J.E., Song H., Huh T.L., Oh W.K. New dammarane-type glucosides as potential activators of AMP-activated protein kinase (AMPK) from Gynostemma pentaphyllum. Bioorgan. Med. Chem. 2011;19:6254–6260. doi: 10.1016/j.bmc.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Hung T.M., Hoang D.M., Kim J.C., Jang H.-S., Ahn J.S., Min B.-S. Protein tyrosine phosphatase 1B inhibitory by dammaranes from Vietnamese Giao-Co-Lam tea. J. Ethnopharmacol. 2009;124:240–245. doi: 10.1016/j.jep.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Quan Y., Qian M.Z. Effect and mechanism of gypenoside on the inflammatory molecular expression in high-fat induced atherosclerosis rats. Chin. J. Integr. Tradit.l Western Med. 2010;30:403–406. [PubMed] [Google Scholar]
- 6.Cai H., Liang Q., Ge G. Gypenoside attenuates β-amyloid-induced inflammation in N9 microglial cells via SOCS1 signaling. Neural Plast. 2016;2016:1–10. doi: 10.1155/2016/6362707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang F., Shi H., Zhang X., Yang H., Zhou Q., Yu L.L. Two new saponins from tetraploid jiaogulan (Gynostemma pentaphyllum), and their anti-inflammatory and α-glucosidase inhibitory activities. Food Chem. 2013;141:3606–3613. doi: 10.1016/j.foodchem.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Li B., Zhang X., Wang M., Jiao L. Characterization and antioxidant activities of acidic polysaccharides from Gynostemma pentaphyllum (Thunb.) Markino. Carbohyd. Polym. 2015;127:209–214. doi: 10.1016/j.carbpol.2015.03.069. [DOI] [PubMed] [Google Scholar]
- 9.Jang H., Lee J.W., Lee C., Jin Q., Lee M.K., Lee C.K., Lee M.K., Hwang B.Y. Flavonol glycosides from the aerial parts of Gynostemma pentaphyllum and their antioxidant activity. Arch. Pharm. Res. 2016;39:1232. doi: 10.1007/s12272-016-0793-x. [DOI] [PubMed] [Google Scholar]
- 10.Cour B.L., Mølgaard P., Yi Z. Traditional Chinese medicine in treatment of hyperlipidaemia. J Ethnopharmacol. 1995;46:125–129. doi: 10.1016/0378-8741(95)01234-5. [DOI] [PubMed] [Google Scholar]
- 11.Gao D., Zhao M., Qi X., Liu Y., Li N., Liu Z., Bian Y. Hypoglycemic effect of Gynostemma pentaphyllum saponins by enhancing the Nrf2 signaling pathway in STZ-inducing diabetic rats. Arch. Pharm. Res. 2016;39:221–230. doi: 10.1007/s12272-014-0441-2. [DOI] [PubMed] [Google Scholar]
- 12.Huyen V.T.T., Phan D.V., Thang P., Hoa N.K., Östenson C.G. Antidiabetic effect of Gynostemma pentaphyllum tea in randomly assigned type 2 diabetic patients. Horm. Metab. Res. 2010;42:353–357. doi: 10.1055/s-0030-1248298. [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Lin W., Huang J., Xie Y., Ma W. Anti-cancer effects of Gynostemma pentaphyllum (Thunb.) Makino (Jiaogulan) Chinese Med. 2016;11:43. doi: 10.1186/s13020-016-0114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan G., Wei J., Zhou J., Guo X., Yang M. Apoptosis of human hepatoma cells induced by Gynostemma pentaphyllum Makino. Chin.-Ger. J. Clin. Oncol. 2006;5:173–177. doi: 10.1007/s10330-005-0436-z. [DOI] [Google Scholar]
- 15.Tsai Y., Lin C., Chen B. Preparative chromatography of flavonoids and saponins in Gynostemma pentaphyllum and their antiproliferation effect on hepatoma cell. Phytomedicine. 2010;18:2–10. doi: 10.1016/j.phymed.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Zhang L., Ren Y., Gao Y., Kang L., Qiao Q. Anticancer and immunoregulatory activity of Gynostemma pentaphyllum polysaccharides in H22 tumor-bearing mice. Int. J. Biol. Macromol. 2014;69:1–4. doi: 10.1016/j.ijbiomac.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Cheng T.-C., Lu J.-F., Wang J.-S., Lin L.-J., Kuo H.-I., Chen B.-H. Antiproliferation effect and apoptosis mechanism of prostate cancer cell PC-3 by flavonoids and saponins prepared from Gynostemma pentaphyllum. J. Agr. Food Chem. 2011;59:11319–11329. doi: 10.1021/jf2018758. [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Huang J., Lin W., Yuan Z., Feng S., Xie Y., Ma W. In vitro anticancer activity of a nonpolar fraction from Gynostemma pentaphyllum (Thunb.) Makino. Evid-Based Compl. Alt. 2016;2016:1–11. doi: 10.1155/2016/6308649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P.K., Tai W.C., Choi R.C., Tsim K.W., Zhou H., Liu X., Jiang Z.-H., Hsiao W.W. Chemical and DNA authentication of taste variants of Gynostemma pentaphyllum herbal tea. Food Chem. 2011;128:70–80. doi: 10.1016/j.foodchem.2011.02.078. [DOI] [PubMed] [Google Scholar]
- 20.Yan H., Wang X., Wang Y., Wang P., Xiao Y. Antiproliferation and anti-migration induced by gypenosides in human colon cancer SW620 and esophageal cancer Eca-109 cells. Hum. Exp. Toxicol. 2013;33:522–533. doi: 10.1177/0960327113497771. [DOI] [PubMed] [Google Scholar]
- 21.Chew Y.L., Wong H.C. Gypenosides, the cancer buster from Gynostemma pentaphyllum (Thunb.) Makino and the apoptotic pathways: A review. Orient. Pharm. Exp. Med. 2016;16:153–154. doi: 10.1007/s13596-016-0231-0. [DOI] [Google Scholar]
- 22.Wu Q., Jang M., Piao X.-L. Determination by UPLC-MS of four dammarane-type saponins from heat-processed Gynostemma pentaphyllum. Biosci. Biotech. Biochem. 2014;78:311–316. doi: 10.1080/09168451.2014.882751. [DOI] [PubMed] [Google Scholar]
- 23.Piao X.-L., Xing S.-F., Lou C.-X., Chen D.-J. Novel dammarane saponins from Gynostemma pentaphyllum and their cytotoxic activities against HepG2 cells. Bioorg. Med. Chem. Lett. 2014;24:4831–4833. doi: 10.1016/j.bmcl.2014.08.059. [DOI] [PubMed] [Google Scholar]
- 24.Ky P.T., Huong P.T., My T.K., Anh P.T., Kiem P.V., Minh C.V., Cuong N.X., Thao N.P., Nhiem N.X., Hyun J.-H., et al. Dammarane-type saponins from Gynostemma pentaphyllum. Phytochemistry. 2010;71:994–1001. doi: 10.1016/j.phytochem.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Shi L., Lu F., Zhao H., Zhao Y.-Q. Two new triterpene saponins from Gynostemma pentaphyllum. J. Asian. Nat. Prod. Res. 2012;14:856–861. doi: 10.1080/10286020.2012.700925. [DOI] [PubMed] [Google Scholar]
- 26.Cui J.-F., Eneroth P., Bruhn J. Gynostemma pentaphyllum: Identification of major sapogenins and differentiation from Panax species. Eur. J. Pharm. Sci. 1999;8:187–191. doi: 10.1016/S0928-0987(99)00013-5. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y.-G., Zhang H.-G., Zhang G.-Y., Fan J.-S., Li X.-H., Liu Y.-H., Li S.-H., Lian X.-M., Tang Z. Panax notoginseng saponins attenuate atherosclerosis in rats by regulating the blood lipid profile and an anti-inflammatory action. Clin. Exp. Pharmacol. P. 2008;35:1238–1244. doi: 10.1111/j.1440-1681.2008.04997.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuo Y.-H., Huang H.-C., Kuo L.-M.Y., Hsu Y.-W., Lee K.-H., Chang F.-R., Wu Y.-C. New Dammarane-Type Saponins from the Galls of Sapindus mukorossi. J. Agr. Food Chem. 2005;53:4722–4727. doi: 10.1021/jf047963s. [DOI] [PubMed] [Google Scholar]
- 29.Chen D.-J., Liu H.-M., Xing S.-F., Piao X.-L. Cytotoxic activity of gypenosides and gynogenin against non-small cell lung carcinoma A549 cells. Bioorg. Med. Chem. Lett. 2014;24:186–191. doi: 10.1016/j.bmcl.2013.11.043. [DOI] [PubMed] [Google Scholar]
- 30.Huang H.-C., Wu M.-D., Tsai W.-J., Liao S.-C., Liaw C.-C., Hsu L.-C., Wu Y.-C., Kuo Y.-H. Triterpenoid saponins from the fruits and galls of Sapindus mukorossi. Phytochemistry. 2008;69:1609–1616. doi: 10.1016/j.phytochem.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 31.Hung T.M., Thu C.V., Cuong T.D., Hung N.P., Kwack S.J., Huh J.-I., Min B.S., Choi J.S., Lee H.K., Bae K. Dammarane-type glycosides from Gynostemma pentaphyllumand their effects on IL-4-induced eotaxin expression in human bronchial epithelial cells. J. Nat. Prod. 2010;73:192–196. doi: 10.1021/np9006712. [DOI] [PubMed] [Google Scholar]
- 32.Takemoto T., Arihara S., Yoshikawa K. Studies on the constituents of cucurbitaceae plants. XIV. Yakugaku Zasshi. 1986;106:664–670. doi: 10.1248/yakushi1947.106.8_664. [DOI] [PubMed] [Google Scholar]
- 33.Yoshikawa K., Takemoto T., Arihara S. Studies on the constituents of cucurbitaceae plants. XVI. on the saponin constituents of Gynostemma pentaphyllum MAKINO. (11) Yakugaku Zasshi. 1987;107:262–267. doi: 10.1248/yakushi1947.107.4_262. [DOI] [PubMed] [Google Scholar]
- 34.Liu X., Ye W.C., Hsiao H.W.W., Che C.T., Zhao S.X. Studies on chemical constituents of Gynostemma pentaphyllum. J. China Pharm. Unive. 2003;34:21–23. [Google Scholar]
- 35.Takemoto T., Arihara S., Yoshikawa K., Kawasaki J., Nakajima T., Okuhira M. Studies on the constituents of cucurbitaceae plants. XI. on the saponin constituents of Gynostemma pentaphyllum MAKINO (7) Yakugaku Zasshi. 1984;104:1043–1049. doi: 10.1248/yakushi1947.104.10_1043. [DOI] [PubMed] [Google Scholar]
- 36.Lin M.-C., Wang K.-C., Lee S.-S. Transformation of ginsenosides Rg1 and Rb1, and crude sanchi saponins by human intestinal microflora. J. Chin. Chem. Soc. 2001;48:113–120. doi: 10.1002/jccs.200100021. [DOI] [Google Scholar]
- 37.Xiang W.-J., Guo C.-Y., Ma L., Hu L.-H. Dammarane-type glycosides and long chain sesquiterpene glycosides from Gynostemma yixingense. Fitoterapia. 2010;81:248–252. doi: 10.1016/j.fitote.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Tomás-Lorente F., Garcia-Grau M.M., Nieto J.L., Tomás-Barberán F.A. Flavonoids from Cistus ladanifer bee pollen. Phytochemistry. 1992;31:2027–2029. doi: 10.1016/0031-9422(92)80355-I. [DOI] [Google Scholar]
- 39.Li S.-S., Wu J., Chen L.-G., Du H., Xu Y.-J., Wang L.-J., Zhang H.-J., Zheng X.-C., Wang L.-S. Biogenesis of C-glycosyl flavones and profiling of flavonoid glycosides in lotus (Nelumbo nucifera) PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0108860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leite J.P.V., Rastrelli L., Romussi G., Oliveira A.B., Vilegas J.H.Y., Vilegas W., Pizza C. Isolation and HPLC quantitative analysis of flavonoid glycosides from Brazilian beverages (Maytenus ilicifolia and M.aquifolium) J. Agr. Food Chem. 2001;49:3796–3801. doi: 10.1021/jf010294n. [DOI] [PubMed] [Google Scholar]
- 41.Sekine T., Arai Y., Ikegami F., Fujii Y., Shindo S., Yanagisawa T., Ishida Y., Okonogi S., Murakoshi I. Isolation of camelliaside C from “Tea Seed Cake” and inhibitory effects of its derivatives on arachidonate 5-lipoxygenase. Chem. Pharm. Bull. 1993;41:1185–1187. doi: 10.1248/cpb.41.1185. [DOI] [PubMed] [Google Scholar]
- 42.Kwon D.-J., Bae Y.-S. Flavonols from the stem bark of Acer komarovii. Chem. Nat. Compd. 2013;49:131–132. doi: 10.1007/s10600-013-0531-2. [DOI] [Google Scholar]
- 43.Zhang H., Li X., Wu K., Wang M., Liu P., Wang X., Deng R. Antioxidant activities and chemical constituents of flavonoids from the flower of Paeonia ostii. Molecules. 2016;22:5. doi: 10.3390/molecules22010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Yin Z.Q., Liang J.Y. Flavonoids from Trachelospermum jasminoides. Chem. Nat. Compd. 2013;49:507–508. doi: 10.1007/s10600-013-0652-7. [DOI] [Google Scholar]
- 45.Wang Y.-S., Liao Z., Zhu H.-K., Feng X.-F., Jiang K.-M., Huang R., Zhu N., Yang J.-H. Megastigmane O-glucopyranosides from Litsea glutinosa. Chem. Nat. Compd. 2012;48:346–349. doi: 10.1007/s10600-012-0247-8. [DOI] [Google Scholar]
- 46.Khan S.H., Mosihuzzaman M., Nahar N., Rashid M.A., Rokeya B., Ali L., Khan A.K.A. Three megastigmane glycosides from the leaves of Pterospermum semisagittatum. Pharm. Biol. 2003;41:512–515. doi: 10.1080/13880200308951345. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L.-J., Chiou C.-T., Cheng J.-J., Huang H.-C., Kuo L.-M.Y., Liao C.-C., Bastow K.F., Lee K.-H., Kuo Y.-H. Cytotoxic polyisoprenyl benzophenonoids from Garcinia subelliptica. J. Nat. Prod. 2010;73:557–562. doi: 10.1021/np900620y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.