Abstract
The amputation of a teleost fin rapidly triggers an intricate maze of hierarchically regulated signalling processes which ultimately reconstruct the diverse tissues of the appendage. Whereas the generation of the fin pattern along the proximodistal axis brings with it several well‐known developmental regulators, the mechanisms by which the fin widens along its dorsoventral axis remain poorly understood. Utilizing the zebrafish as an experimental model of fin regeneration and studying more than 1000 actinopterygian species, we hypothesized a connection between specific inter‐ray regulatory mechanisms and the morphological variability of inter‐ray membranes found in nature. To tackle these issues, both cellular and molecular approaches have been adopted and our results suggest the existence of two distinguishable inter‐ray areas in the zebrafish caudal fin, a marginal and a central region. The present work associates the activity of the cell membrane potassium channel kcnk5b, the fibroblast growth factor receptor 1 and the sonic hedgehog pathway to the control of several cell functions involved in inter‐ray wound healing or dorsoventral regeneration of the zebrafish caudal fin. This ray‐dependent regulation controls cell migration, cell‐type patterning and gene expression. The possibility that modifications of these mechanisms are responsible for phenotypic variations found in euteleostean species, is discussed.
Keywords: actinopterygii, euteleostei, evo‐devo, fibroblast growth factor receptor 1, fin regeneration, inter‐ray, knck5b
Introduction
The dermal component of zebrafish fins consists of rays and inter‐rays. Each ray comprises two contralateral, bracket‐like hemi‐rays. Both hemi‐rays are symmetrically segmented, branched and show distal actinotrichia fibrils. These rays are connected by inter‐ray membranes. In the last 15 years, hundreds of articles have reported on the regulatory properties acting during the development and regeneration of the caudal fin, providing a better view of these processes. During development, the caudal fin is ventral, but a dorsal flexion changes its position to the caudal margin of the body. This morphological change transforms the original anteroposterior (AP) axis into the final ventrodorsal (VD), normally named dorsoventral (DV), axis of the fin. Whereas the fin grows along the proximodistal (PD) axis at its distal margin, it widens along the DV axis (see Fig. 1A) and thickens along a third contralateral left‐right (LR) axis (Fig. 1A,B; Marí‐Beffa & Murciano, 2010) at all fin positions. Cell lineage specification, pattern formation and size control along specific axes are general features of the development of the vertebrate dermoskeleton (Durán et al. 2015) and these are also present in zebrafish fins (Marí‐Beffa & Murciano, 2010; Durán et al. 2015; Wehner & Weidinger, 2016).
Figure 1.
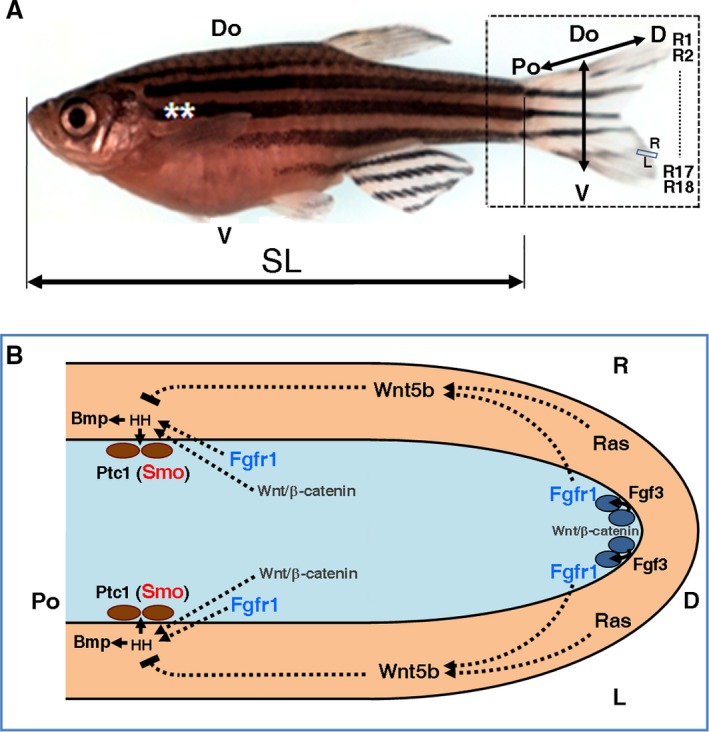
Anatomy and some signalling pathways controlling regeneration of the caudal fin of Danio rerio. (A) Adult zebrafish specimen. Rectangle engulfs caudal fin. Discontinuous line shows rays between R2 and R17. Double arrows show body and fin axes. Double asterisks represent pectoral fins. Do, dorsal; SL, standard length. V, ventral. (B) Model of interactions between three different cell types – epidermis (beige), mesenchyme fibroblast (blue ovals) and osteoblasts (brown ovals) – in several ray blastema domains (Lee et al. 2009; Wehner & Weidinger, 2016). Light blue area is blastema mesenchyme. Arrows and broken lines, respectively, suggest activation and repression. Wnt‐β catenin signalling activates fgf3 transcription and FGF3 activates FGFR1 receptor in distal mesenchyme (Wehner & Weidinger, 2016). Ptc1 is expressed in osteoblasts (Laforest et al. 1998). The activity of proteins in blue and red have been modified experimentally. L and R, left and right; Po and D, proximal and distal.
The caudal fin regenerates after a cut (Broussonet, 1786), forming a very complex (Yoshinari et al. 2009) and increasingly interesting blastema of nine different proliferating cell lineages (Knopf et al. 2011; Sousa et al. 2011; Tu & Johnson, 2011; Stewart & Stankunas, 2012). Whereas caudal fin regeneration has been the subject of classic papers (e.g. Morgan, 1902; Goss & Stagg, 1957; Kemp & Park, 1970), recent molecular studies of this process in zebrafish disclose several controlling molecular mechanisms (e.g. Akimenko et al. 2003; Pfefferli & Jaźwińska, 2015; Wehner & Weidinger, 2016). These mechanisms are normally active along one of the three different axes of the fin (Fig. 1A; Marí‐Beffa & Murciano, 2010). The best examples of these one‐axis studies are those focused on the regulation of regeneration along the PD axis of the fin (e.g. Géraudie et al. 1994; White et al. 1994; Akimenko et al. 1995; Quint et al. 2002; Lee et al. 2005; Stoick‐Cooper et al. 2007; Sims et al. 2009). These studies have revealed cross‐interactions between specific distal epidermal domains (Lee et al. 2009; Wehner & Weidinger, 2016) and the underlying mesenchyme. These interactions are mediated by specific signalling pathways (e.g. Laforest et al. 1998; Poss et al. 2000; Quint et al. 2002; Lee et al. 2005; Whitehead et al. 2005; Smith et al. 2008; Chablais & Jaźwińska, 2010; Blum & Begemann, 2015) and control blastema formation and distal outgrowth (Johnson & Bennett, 1999; Nechiporuk & Keating, 2002; Akimenko et al. 2003; Marí‐Beffa & Murciano, 2010; Wehner & Weidinger, 2016). These signalling processes regulate the hierarchical transcription of genes in cell type‐specific lineages (Wehner et al. 2014; Durán et al. 2015; Wehner & Weidinger, 2016; Fig. 1B). For example, the osteoblast lineage that ultimately up‐regulates osterix (Brown et al. 2009; Knopf et al. 2011) and osteocalcin transcripts (Sousa et al. 2011) are regulated by the epidermal Sonic (Quint et al. 2002) or Indian (Avaron et al. 2006) hedgehog signalling pathway (Armstrong et al. 2017), which are activated by several other regulators from the distal fibroblast/epidermal domain (Laforest et al. 1998; Lee et al. 2009; Wehner & Weidinger, 2016). Recently, bioelectric signalling has also been found to regulate fin growth rate (Monteiro et al. 2014), pattern and size (Perathoner et al. 2014), opening new avenues of research. However, few of these studies have analysed the potential molecular mechanisms controlling fin morphogenesis along the other two axes, or have studied other fish species.
Regarding the DV axis, our group has updated classic experiments (Nabrit, 1929) on caudal fin regeneration (Marí‐Beffa et al. 1996, 1999; Murciano et al. 2001, 2002; Marí‐Beffa & Murciano, 2010). From these studies, ray/inter‐ray interactions have been proposed locally to regulate both the rate of outgrowth (Morgan, 1902) and the morphogenesis of each ray of the regenerating fin (Marí‐Beffa & Murciano, 2010). These local inter‐tissue interactions would control the widening of rays and inter‐rays and the branching of the rays in the strict vicinity during outgrowth of the distal fin blastema (Marí‐Beffa et al. 1999; Murciano et al. 2001, 2002; Marí‐Beffa & Murciano, 2010). The generation of the pattern and size of the complete fin would depend on the iteration of these inter‐tissue interactions along the DV axis of the fin at each ray/inter‐ray boundary (Marí‐Beffa & Murciano, 2010). These ray/inter‐ray boundaries restrict positioning of migrating osteoblast and mesenchyme during development and regeneration (Tu & Johnson, 2011), which is established by retinoic acid‐dependent signals (Blum & Begemann, 2015).
In this paper, we study both the cellular and molecular mechanisms controlling ray‐dependent inter‐ray formation and widening in zebrafish, and the natural variation of inter‐ray membranes among actinopterygian species. Ray/inter‐ray interactions have been studied by partial ablation or ray grafting (Murciano et al. 2002). Proliferation and migration of specific cell lineages have been tracked by bromodeoxyuridine (BrdU; Santamaría et al. 1996), epidermal DiI (Poleo et al. 2001) and endothelial transgenic (Lawson & Weinstein, 2002; Bayliss et al. 2006) labelling. Bioelectric signalling during inter‐ray wound healing and regeneration have been analysed using the another long fin (alf) dty86d gain‐of‐function mutation of the two‐pore domain potassium (K+) channel coded by the zebrafish knck5b gene (Perathoner et al. 2014). The involvement of fibroblast growth factor receptor 1 (FGFR1)‐like tyrosine kinases and the Sonic hedgehog (SHH) signalling pathway (Poss et al. 2000; Quint et al. 2002; Fig. 1B) activities during inter‐ray healing and regeneration have also been studied by chemical inhibition. Finally, inferences about variations observed in a large sample of euteleostean species have also been drawn from zebrafish experiments to propose hypotheses for future Evo‐Devo studies.
Methods
Animal husbandry
Fishes were kept in a circulating system (Aquatic Habitats, USA). We obtained AB wild type (Fig. 1A), Tg(fli1a:EGFP)/AB transgenic and alf dty86d mutant specimens of zebrafish Danio rerio as offspring (Westerfield, 1995) of males and females commercially supplied from ZIRC, Oregon. We also used specimens of goldfish Carassius auratus obtained from a local pet shop. Regeneration experiments with zebrafish took place at 28.5 °C and those with goldfish were carried out at 25 °C. Fishes showing abnormal adult or regenerating fin morphologies were discarded. Operations and handling of fishes were done under the principles approved by National Laws (Directives 98/81/CE and 2000/54/CE, Law 32/2007 BOE 268, and Royal Decree Laws 178/2004 and 367/2010, Spain).
Experimental fin conditions
Fishes were anaesthetized with 0.2 mg mL−1 tricaine methanosulphonate (MS222, Sigma, St. Louis. MO, USA) in Tris buffer (Westerfield, 1995). Each operated fish was then laid on a glass slide inclined over the side of a small Petri dish with the head in anaesthetic solution and the caudal fin dry, out of the solution. The caudal fins were considered symmetrical and thus either the dorsal or the ventral lobe was indiscriminately chosen for operation. The rays were numbered R1 to R18, following their serial position along the dorsoventral axis (Fig. 1A).
The caudal fin of 11 developing zebrafish were first dissected after euthanasia in an overdose of 0.2 mg mL−1 MS222, embedded in paraffin, sectioned and stained for histological studies (Becerra et al. 1983). Six other live adult transgenic fishes and four goldfish were anaesthetized and used for pigment and endothelium studies without any previous operation. Zebrafish spontaneously showing abnormal fin morphologies in tanks were also anaesthetized and studied.
Experiments to study inter‐ray wound healing and regeneration (Fig. 2)
Complete fin cuts. Caudal fins were cut approximately two segments proximal to the first ray branching (Fig. 2A; after Géraudie et al. 1994). To study fin morphometry or cell‐type presence after complete regeneration, 12 wild type and eight Tg(fli1a:EGFP)/AB and alf dty86d zebrafish fins were cut. Caudal fins from five wild type specimens were cut and sampled 4–5 days post‐amputation (dpa) for histological studies. Five additional wild type C. auratus fishes were also cut and BrdU injected at 4 dpa. The caudal fins of 76 specimens were cut and used for inhibitor experiments.
Ray cuts. In general, one or several neighbouring rays (always including the third dorsal or ventral) were transversally cut at proximal positions (Goss & Stagg, 1957). Healing and regenerating fins were used for morphological, cell function and gene expression studies (Fig. 2B; Supporting Information Table S1). Cell migration was studied by DiI‐labelling before cutting. Cell division studies used intraperitoneal injection of BrdU 2 days post operation (dpo). In situ hybridization was carried out to study gene expression.
Ray grafts. A proximal fragment of the first large ray (R1 or R18) was grafted into the proximal region of the inter‐ray between the central rays (R9–R10) of the caudal fin. Before implantation, grafted rays were extracted and rotated 90° clockwise or anti‐clockwise (Fig. 2C,D) to join the internal margin of the ray and the central host inter‐ray (Fig. 2D). The distal part of the graft normally remained outside of the fin after implantation. Leucophores were used for labelling (Murciano et al. 2002). In all, 37 specimens were used in this experiment, five of them for histology and morphometry (years 2000–2002).
Figure 2.
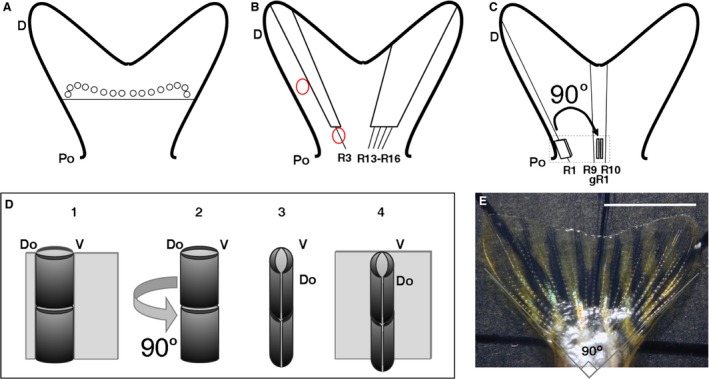
Surgical operations carried out in the experiments. (A) Transversal cut of a caudal fin about two joints proximal to first ray branching positions (circles). (B) One ray and four neighbouring rays cut in the lobes of the same fin. Red circles show positions of DiI injection. (C) Grafting with previous 90° rotation of a proximal fragment of ray 1 (R1) into the inter‐rays between rays 9 and 10 in the fin. (D) 3D drawing showing the operation in the discontinuous square in (C). A proximal R1 fragment (in dark grey) (1) is cut out from the ray, rotated 90° (2 and 3), and grafted (4) in the central inter‐ray. Do and V, dorsal and ventral positions in the original ray fragment. (E) Caudal fin completely opened over a calibrated slide. The angle between lines is 90°. D, distal; gR1, grafted ray 1; Po, proximal; R1–3, R9, R10, R13–R18, rays 1–3, 9, 10, 13–18 in dorsoventral series. Scale bar: 5 mm.
Inhibitor administration
Cyclopamine (alkaloid inhibitor of Sonic/Indian hedgehog, SHH/IHH, pathway, Sigma‐Aldrich) and tomatidine (control alkaloid, Sigma‐Aldrich; Watkins et al. 2003) were dissolved in ethanol 100° to final stock concentrations of respectively 1 and 100 mm and stored at −20 °C. This inhibitor attaches to the protein Smoothened (Smo), a transducer regulated by SHH/IHH receptor Ptc1, and prevents signal transduction of the pathway (Chen et al. 2002). Recently, off‐target effects of cyclopamine on cell proliferation have been documented in zebrafish that preclude any conclusion on cell division control (Armstrong et al. 2017). Moreover, different concentrations of SU5402 (inhibitor of the tyrosine kinase activity of FGFR1 and other FGFRs; Calbiochem, Germany) was dissolved in dimethylsulphoxide (DMSO) to a stock concentration of 34 mm and stored at −20 °C. SU5402 most efficiently inhibits FGFR1 activity by repressing its tyrosine kinase activity (IC50 = 0.03 μm; Sun et al. 1999). DMSO was used as an SU5402 control. Fish treatments were carried out in beakers with 100 mL of water from the circulating system. Different quantities of stock solutions were added to the water to obtain the final experimental concentration of each reagent (Supporting Information Table S2A,B). Fishes were fed with ZM‐fish Ltd food once every experimental day except at 1–2 and 8–9 dpa.
Different concentrations of cyclopamine were administered to 39 fishes to inhibit the Ptc1‐pathway to various degrees (Table S2A). Different concentrations of SU5402 were also added to the water of a total of 23 fishes (Table S2B). As a cyclopamine‐negative control, tomatidine was added. DMSO at 0.01 or 0.004% was indiscriminately used as a control of SU5402 treatments. Regenerating fishes were left in 100 mL of system water in groups of two or three. All fins were sampled and processed for morphometry at 12 dpa (see commentaries on Supporting Information Figures).
Specimens on loan
In all, 49 adult or young fish specimens of 42 actinopterygian species were lent by the Museo Nacional de Ciencias Naturales (MNCN) in Madrid, Spain (years 2001–2002, 2007). These specimens were used to characterize inter‐ray morphologies. Each specimen shown here is identified by its Museum code.
Anatomy and morphometry
Fin anatomies were first visualized under the dissecting microscope (SMZ800, Nikon, Japan). The standard length (SL; length from mouth to caudal fin base) of most fishes (Fig. 1A, Table S2A,B) was measured. To visualize pigment and endothelial cell patterns, adult and regenerating Tg(fli1a:EGFP)/AB transgenic fins were photographed in a light and fluorescence magnifying microscope (Multizoom AZ100, Nikon; Axioskop, Zeiss). For correct morphometry, all fins regenerated under inhibitor treatments were opened before photographing or sampling (Fig. 2E). The complete extensions of these caudal fins were done with a thin brush over a glass cover‐slide where the fins were slowly dried for 2 min (Fig. 2E). As the angle generated by the caudal fin web normally ranges between 80° and 90° (data not shown), two lines at a 90° angle were drawn over the slide to guide the correct fin extension (Fig. 2E). Fins were discarded if the distal margin was partially broken during an extra‐extension or if the mounted fin was incompletely opened, showing creased inter‐rays. Opened fins were cut, fixed in phosphate‐buffered saline (PBS) 4% paraformaldehyde pH 7.4 (12 h at 4 °C), mounted and photographed with a Polaroid DMC camera for morphometry. All actinopterygian specimens lent by MNCN were received in 100° ethanol. Before photographing, each specimen was sequentially immersed in 96°, 75°, 50° ethanol and water for 1 h.
Digital morphometry was done using an imagej 1.43u program (nih.gov, USA). Morphometric variables were measured to analyse inter‐ray widening and distal outgrowth (Fig. S1A–D and below). Pigment cell distribution was measured by counting the number of marginal and central inter‐rays with pigments in digital images. Non‐parametric statistical analysis of morphometric data used the Mann–Whitney U‐test at various levels of significance (spss, version 11.5, SPSS Inc., Chicago, IL, USA). Basic statistics (mean and standard deviation) were obtained from an excel datasheet (Microsoft Office Excel 2007).
Transgenic, BrdU and DiI‐labelling
Endothelial cells were tracked in vivo using a Tg(fli1a:EGFP)/AB zebrafish line specimen as stated above (Lawson & Weinstein, 2002; Bayliss et al. 2006). Fishes with complete fin or ray cuts were injected intraperitoneally at 2 or 4 dpo with BrdU (Sigma) in Hanks solution (Westerfield, 1995) at a dose of 0.25 mg g−1 wet weight. At 24 h after injection, samples were obtained, processed histologically and immuno‐stained (Santamaría et al. 1996). DiI‐labelling was carried out by injections (Poleo et al. 2001) in the stump of cut rays or in neighbouring rays (Fig. 2B).
Histology and in situ hybridization
Histological sections were obtained after fixation in PBS 4% paraformaldehyde pH 7.4 (12 h at 4 °C), paraffin or ‘CryoWax’ (Durán et al. 2011) embedding and staining with haematoxylin‐eosin‐picrosirius (Becerra et al. 1983) or Mallory's trichrome (Pearse, 1985; Kiernan, 2015). Sections were photographed in a Zeiss Axioskop (Zeiss, Germany) or a Multizoom Nikon AZ‐100 microscope under Nomarski optics. RNA antisense probes against msxa, msxc, msxd (Akimenko et al. 1995), shh (Laforest et al. 1998), bmp4 (Murciano et al. 2002) or dlx3 (Akimenko et al. 1994) genes were obtained after Quint et al. (2002). Fins after ray cuts were fixed with 4% paraformaldehyde stained with probes and whole‐mounted (Akimenko et al. 1995; Laforest et al. 1998). Photographs were taken in a Nikon Eclipse E 800 (Nikon, Japan) microscope.
Results
To investigate the mechanisms controlling the widening during outgrowth of the inter‐rays, we studied caudal fin development and regeneration in zebrafish. The rays of the caudal fin of Danio rerio are branched except for the dorsalmost and the ventralmost large rays and all lateral, small procumbent rays. We have studied both inter‐rays that neighbour the rays until the first branching and the new inter‐ray formed between both branches. To avoid compensatory inter‐ray narrowing after formation of ray dichotomies, other inter‐rays distal to these positions or those neighbouring non‐branched rays were not considered here.
Histological features can define two distinct inter‐ray regions in zebrafish
In young or adult zebrafish caudal fins (15–38 mm SL), the inter‐rays are not histologically homogeneous. The marginal region abutting the ray is always thicker than the central regions located more than 150 μm away (Fig. 3A; Table 1). This is also observed in the flag‐like inter‐ray (Fig. 3B) external to the long, lateral non‐branched rays.
Figure 3.
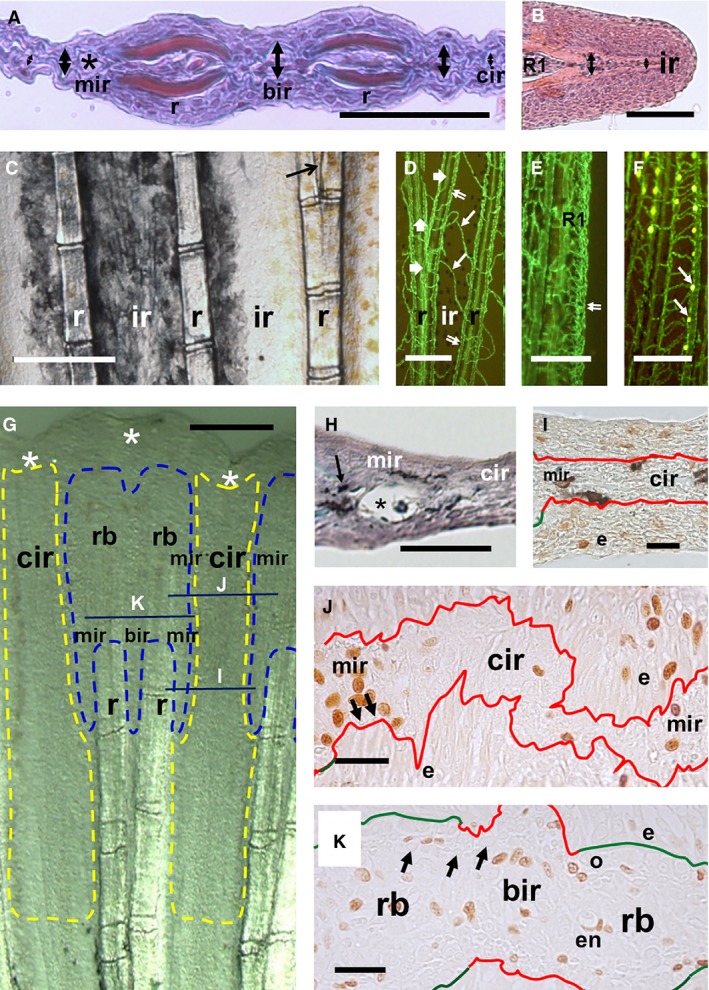
Histological features of inter‐ray regions in adult and regenerating caudal fins. (A) Detail of Mallory's trichrome‐stained cross‐section of an adult zebrafish caudal fin. The marginal and the branching inter‐ray regions are thick and show blood capillaries (asterisk). The central region is thinner. (B) Haematoxylin‐eosin‐stained section of the first long, non‐branched ray (R1). The small flag‐like inter‐ray lateral to R1 shows different thickness. (A, B) Thickness is shown by double‐pointing arrows. (C) Pigment pattern of a live zebrafish caudal fin. Black and orange pigment cells are melanophores and xanthophores. Arrow shows a branching inter‐ray. (D–F) EGFP fluorescence (green) of endothelial cells in live Tg(fli1a:EGP)y1/AB transgenic caudal fins. (D) Rays and inter‐rays at ray branching. Double and thick arrows show veins and arteries at marginal inter‐ray and ray branching, respectively. Thin arrow shows central inter‐ray vessels. (E) R1, non‐branched ray. Double arrow shows capillary plexus. (F) White leucophores (yellow cells and white arrows) on capillaries in long rays. (G–I) Histological domains of inter‐ray blastema. Transversal line in (G) shows approximate section levels in (I–K). Yellow and blue discontinuous lines in (G) show potential boundaries of high and low proliferating blastema regions, respectively. Asterisks show inter‐ray incisions. (H) Mallory's trichrome‐stained section of a regenerated inter‐ray processed with the Cryowax technique. Asterisk shows a large capillary. Arrow indicates a pigment cell. (I–K) BrdU immunostained sections of zebrafish (I) and goldfish (J, K) inter‐rays. Observe the higher frequency of positive cells at the marginal inter‐ray blastema. Green and red lines show ray and inter‐ray basement membranes, respectively. Arrows show actinotrichia sections. e, en, and o, epidermal, endothelial cells, and osteoblast; mir, cir and bir, marginal, central and branching inter‐ray regions; rb, r and ir, ray blastema, ray and inter‐ray. Scale bars: 10 (J, K), 20 (I), 50 (B, H), 200 (G), 250 (A) and 500 (C–F) μm.
Table 1.
Histological features of inter‐ray domains at ray branching in zebrafish
| Histological domains | n | Thickness (μm) | Cell density (x mm–2) | n | Melanophore (1) | Xanthophore (1) | Leucophore (1) | Endothelium (1) |
|---|---|---|---|---|---|---|---|---|
| Adult marginal IR | 46 | 275.8 ± 41.8 | 143.7 ± 31.1 | 207 | 42.5 ± 5.6 | 76.4 ± 6.4 | 68.2 ± 9.9** | 100.0 ± 0.0 |
| Adult Central IR | 40 | 82.4 ± 6.2† , *** | 254.8 ± 71.2*** | 103 | 23.6 ± 10.8† , * | 70.9 ± 4.1 | 29.2 ± 26.8† , ** | 64.0 ± 33.8** |
| Adult Branch IR | 53 | 281.4 ± 52.3 | 130.6 ± 32.9† , * | 164 | 36.7 ± 5.3 | 87 ± 4.9† , * | 86.3 ± 16.1** | 100.0 ± 0.0 |
| Adult Ray | 84 | nd | 138.4 ± 46 | 106 | 36.8 ± 11.4 | 78.2 ± 10.6 | 14.5 ± 16.1 | 100.0 ± 0.0 |
| Reg marginal IR | 60 | 406 ± 147.3† , *** | 99.7 ± 28.2** | 94 | 65.2 ± 47.89 (·) | nd | nd | |
| Reg central IR | 39 | 117.6 ± 24.3† , *** | 160.2 ± 58.1*** | 47 | 46.6 ± 50.4 (·) | nd | nd | |
| Reg branch IR | 11 | 295.1 ± 14.6† , *** | 72.4 ± 14.9 | 56 | 74.1 ± 44.2 (·) | nd | nd | |
| Reg ray | 51 | nd | 86.5 ± 22.4 | 119 | 74.5 ± 18.3 (·) | nd | nd | |
Reg, regenerated. ***P < 0.001, **P < 0.01 and *P < 0.05 when compared with data from any other domain.
†When compared with the other inter‐ray domains, n is number of inter‐rays studied. (1) Percentage of cell‐type presence in Tg(fli1a:EGFP)/AB line. nd, not determined. (·) Sum of variables obtained from regenerated fins.
This marginal region shows mesenchyme fibroblasts and a vein, and is frequently pigmented (Fig. 3A). Irrespective of the pigment band or PD position studied in Tg(fli1a:EGFP)/AB transgenic fins, melanophores, long‐ray leucophores or blood vessels are more often found, and the cell nuclei density is significantly lower in marginal than in central inter‐ray regions (Fig. 3C–F; Table 1). The marginal vein (Huang et al. 2003) runs parallel to the ray, whereas the central vessels are randomly oriented (Fig. 3D). Veins also run parallel to ray branches at the internal inter‐ray margins arising from vessels outside the ray in many different sprouting patterns. The new inter‐rays formed after ray dichotomy and normal marginal regions are similar but show low‐significance differences in cell density and xantophore or leucophore presence (Table 1). Both are equal to rays and do not show significant differences in any histological parameter studied (Table 1). Distal actinotrichia also differentiate in ray and marginal inter‐rays, but not in centralmost regions. During development, all marginal regions show similar features and remain almost constant in width, whereas central regions widen at different rates depending on their positions (not shown). This general pattern is also observed in the caudal fin of goldfish (Supporting Information Fig. S2).
Two specializations are found in zebrafish marginal inter‐rays. The first is found in the inter‐ray external to the lateral long rays (R1 or R18, Fig. 3B). The marginal region of this inter‐ray is highly pigmented, and its parallel vein is transformed into a plexus that does not produce vein sprouts to the rest of this flag‐like inter‐ray (Fig. 3E). A second specialization is found in the distal margin of the longest rays where leucophore differentiate (Fig. 3F). These cells show endogenous fluorescence at excitation wavelengths similar to those of enhanced green fluorescent protein (EGFP), although it emits light at different wavelengths. Every single leucophore can thus be easily located over the longitudinal vein (Fig. 3F), or rarely over central inter‐ray or ray vessels, always in complete co‐distribution. This suggests regulation of leucophore patterning by endothelial cells during fin development.
Similar regions can also be found after inter‐ray regeneration (Fig. 3G–I; Tables 1 and 2). The regenerated inter‐ray shows a margin that is thicker and has a lower cell density than the central region, at either ray branching or early post‐branching (Fig. 3G,H; Table 1). Distal actinotrichia are also preferentially observed in the ray and the marginal inter‐ray regions (data not shown). Five days after fin lobe or a one‐ray cut, the epidermis (Fig. 3I,J) or mesenchyme (Fig. 3J) of the blastema in potential marginal inter‐ray regions of zebrafish and goldfish shows a higher density of proliferating (BrdU‐positive) cells compared with the central regions (Table 2). These marginal regions can be recognized by the absence of lepidotrichia bone and the presence of actinotrichia; in contrast, central regions are devoid of both skeletal structures. In goldfish, the inter‐ray formed at ray branching (Fig. 3K; Table 2) also shows a higher density of proliferating cells compared with the central regions, but an equal density to marginal inter‐ray or ray blastema (Table 2). The histological study of adult, developing and regenerating fins thus discloses two different inter‐ray membrane regions, the marginal and the central inter‐rays. We have analysed the cellular mechanisms underlying this regionalization.
Table 2.
Proliferative features of inter‐ray regions in the caudal fin blastema
| Histological regions | Danio rerio | Carassius auratus | |||||
|---|---|---|---|---|---|---|---|
| Epidermis | Mesenchyme | Mesenchyme | |||||
| n | BrdU+ % | n | BrdU+ % | n | Cell density (x mm–2) | BrdU+ % | |
| Inter‐ray marginal | 20 | 28.4 ± 9 | 12 | 32.39 ± 15.4 | 19 | 80.91 ± 21.05 | 53.47 ± 17.63 * |
| Central inter‐ray | 21 | 19 ± 12.76† , * | 9 | 21.94 ± 17.97 | 10 | 45.48 ± 17.7** / † , ** | 16.95 ± 12.13**/† , ** |
| Branching inter‐ray | – | – | – | – | 6 | 99.9 ± 28.2† , * | 46.7 ± 16.1* |
| Ray blastema | 11 | 22.5 ± 12.1 | 6 | 26.15 ± 14.7 | 14 | 83.9 ± 20.86 | 67.21 ± 12.28 |
n, number of inter‐rays studied; x, number of cells counted; %, percentage of BrdU+ cell presence.
**P < 0.01 and *P < 0.05, when compared with data from the ray blastema.
† Statistical significance when compared with inter‐ray data.
Intercalation between ray and central inter‐ray cells may form new inter‐rays
We have investigated the involvement of ray cells in inter‐ray formation and widening. To this end, a proximal fragment of the first long, non‐branched ray (R1) was grafted onto the central inter‐ray of a caudal fin with a previous 90° angle rotation (Figs 2C,D and 4). During the first 24 h, the epidermis heals over the grafted fragment (Fig. 4A) maintaining it in its original surgical position in 34 of 37 cases. Several days after healing, the distal part of the graft remains outside of the fin and begins to grow (28 of 34 cases). In nine of these graft regenerations, the ray re‐rotates to its normal position and regenerates as a graft without a previous 90° rotation (see Murciano et al. 2002). However, this ray normally regenerates from the fin in isolation, maintaining its previous angle (19 of 28 cases; Fig. 4B). This regeneration apparently occurs in two stages: blastema formation and distal outgrowth (Fig. 4A,B). During this process, the inter‐ray may not form initially, showing a strong ‘serrate’ phenotype (Jaźwińska et al. 2007) with a cleft of several segments in size. In some instances, an inter‐ray forms between the ray regeneration and the centralmost inter‐ray (six of 19 grafts; Fig. 4C,D). This ectopic membrane always grows at right angles to the host inter‐ray, showing a characteristic ‘T‐shape’ (Fig. 4D), being narrow proximally (Fig. 4C) and widening medial or distally (Fig. 4D). After 30 dpo, the maximum width of the ectopic inter‐rays (99.1 ± 7.7 μm; n = 12) is about half that of the host inter‐rays (111 ± 18.3 μm; n = 8, P ≫ 0.05) and very different from the half‐width of the inter‐ray near the R1 graft (51.97 ± 17.73 μm; n = 8, P < 0.01).
Figure 4.
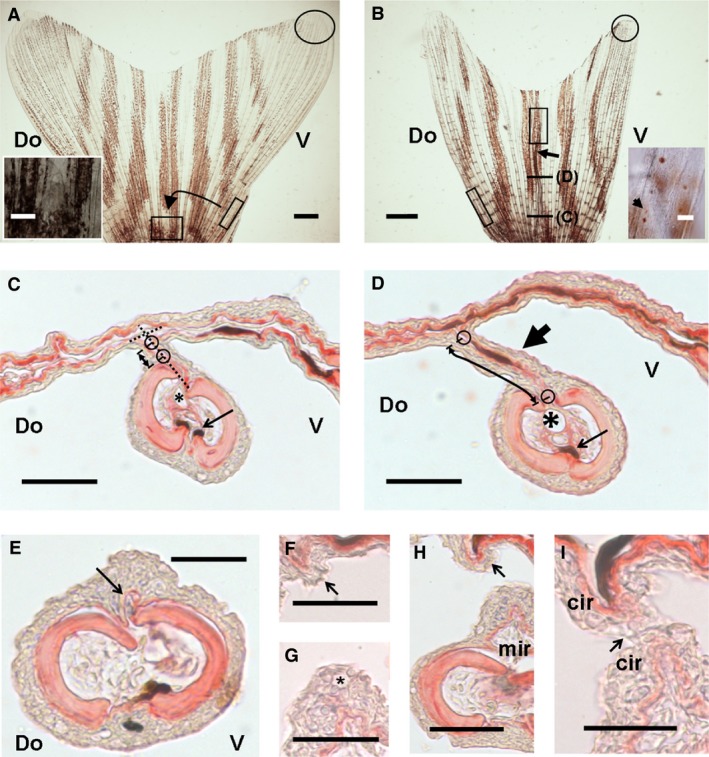
Results after 90° rotated ray graft experiment. (A, B) General views of 90° rotated ray grafted caudal fins. (A) Proximal square is host central inter‐ray. Inset is amplification of the square with ray graft. (B) Fin with graft regenerated for 30 dpo (arrow). Inset is distal region of regenerated graft (rectangle). Arrow in inset is a leucophore. Ovals and oblique rectangles in (A, B) are leucophore regions and graft origins. Fin in (B) has not been opened completely. (B) Section levels in (C) and (D). (C, D) Proximal (C) and medial (D) cross‐sections of a 90° rotated R1 regeneration. (C) Proximal region shows a ray graft regeneration at a right angle (discontinuous lines). (D) Distal region of a 90° rotated regeneration forming a perpendicular, ectopic inter‐ray (large arrow). Small circles in (C, D) show limits of intercalated inter‐ray. Asterisks show ray veins. Small arrows show ray melanophores. Double arrows show ectopic inter‐ray. (E–I) Cross‐sections of the distal margin of an ectopic inter‐ray. A marginal region is distally formed de novo (E, F) and gradually develops (G, H) into a ‘meniscus’‐like leading edge (arrows). Meniscus of ectopic inter‐ray at host inter‐ray (F, H) or grafted ray regeneration (E, G, H). Asterisk shows a mucous cell. (H, I) Distal fusion (arrow) of marginal edges. Do and V, dorsal and ventral; mir and cir, marginal and central inter‐rays. Scale bars: 20 μm (F, G, I), 50 μm (C–E, H; insets in A, B) and 1 mm (A, B).
A ‘meniscus‐like’ leading edge forms at both ends of the ectopic inter‐ray (Fig. 4E–I). The margin near the regenerating ray shows a large vessel and pigment cells, whereas the opposite ectopic inter‐ray region in contact with the host inter‐ray never forms a longitudinal vessel or thickens. In some instances, the vessel and pigment cells form in the margin of the isolated ray instead of the neighbouring marginal inter‐ray region (Fig. 4C,D). This may occur when a strong cleft is formed during isolated ray regeneration. When the vessel is in the ray margin, a marginal region is absent, and the neighbouring inter‐ray region resembles a central‐like region (Fig. 4D). When a marginal inter‐ray with vein is formed, potentially when a slight ‘serration’ occurs, the above‐mentioned histological differences with the central region of the ectopic inter‐ray are statistically significant (Table 3). This suggests that the margins of ray blastema are a default migration fate of vein endothelial cells and that the ectopic membrane is a half inter‐ray. In these ectopic inter‐rays, leucophores are very rare (Fig. 4A,B), precluding any clear conclusion about cell lineage origins.
Table 3.
Histological features of ectopic inter‐ray domains after implantation of 90° rotated ray grafts
| Ectopic inter‐ray domains | n | Cell density (x mm–2) | n | Thickness (μm) | n | Melanophore (%) | n | Endothelium (%) |
|---|---|---|---|---|---|---|---|---|
| Marginal inter‐ray | 6 | 88.1 ± 41.4 | 7 | 10.3 ± 11.4 | 32 | 10.9 ± 15.4 | 24 | 65.9 ± 48.2 |
| Central inter‐ray | 3 | 108.4 ± 30 | 6 | 2.8 ± 0.5† , * | 31 | 53 ± 13.5 | 15 | 45.8 ± 41.7 |
| Marginal ray | 8 | 46.3 ± 19.9* | 8 | 11.1 ± 10.4 | 33 | 38.3 ± 25.8 | 32 | 94.4 ± 7.9 |
*P < 0.05 when compared with data from the other domains.
†When compared with marginal inter‐ray data.
n, number of inter‐rays studied; x, is number of cells counted.
The contact between central inter‐ray and ray cells may induce the formation of an inter‐ray. This intercalation forms a ‘meniscus‐like’ leading edge at both sides of the new inter‐ray. Several arguments suggest that this new membrane is a half ‘host‐type’ inter‐ray. The different positions of interacting tissues in this experiment suggest a wide distribution of competence for these interactions along the fin.
Different cell migration and gene expression patterns occur during wound healing and regeneration of inter‐rays
Additional cellular properties have been inferred from ray‐cut experiments. A single inter‐ray, or groups of one, two, three or four neighbouring long rays were cut and cell migration, cell proliferation and gene expression were studied. After these cuts, a small remnant of the marginal inter‐ray always remained attached to neighbouring non‐operated rays.
When an inter‐ray is cut out, wound healing restores the continuity of the inter‐ray membrane (Fig. 5A) in less than 2 days. This is also observed after a one‐ray cut, although at slightly slower rates (Fig. 5B; Goss & Stagg, 1957; Marí‐Beffa et al. 1996, 1999). After this operation, ray regeneration is delayed. Wound healing is seldom observed after two‐ray cuts and never when three or four neighbouring rays are cut (Fig. 5C). This healing membrane shows variable width depending on the space left by the non‐operated neighbouring rays (Fig. 5D). Wound healing after one‐ or two‐ray cuts shows similar sigmoid growth rates (Fig. 5E). Regeneration is slower when more than two rays are cut (Fig. 5E). In this case, rays and inter‐rays regenerate slightly more quickly, the closer they are to intact flanking rays (see below). Irrespective of the number of clipped rays, a ‘meniscus’‐like leading edge is always observed in the marginal regions (Fig. 5A–C). During wound healing after a one‐ray cut, the size of this ‘meniscus’ (Fig. 5B) in a proximal position reaches 250 μm, gradually reducing to 80–100 μm in more distal positions (Fig. 5F). One month after two‐ray cuts (Supporting Information Fig. S3A), fusion of regenerating rays has been found in four of 15 cases. Fusion of neighbouring rays is very rarely observed after a fin cut. Both ray fusions and ray separations with proximal connecting membranes (Fig. S3B) can in rare cases be observed spontaneously during normal zebrafish development.
Figure 5.
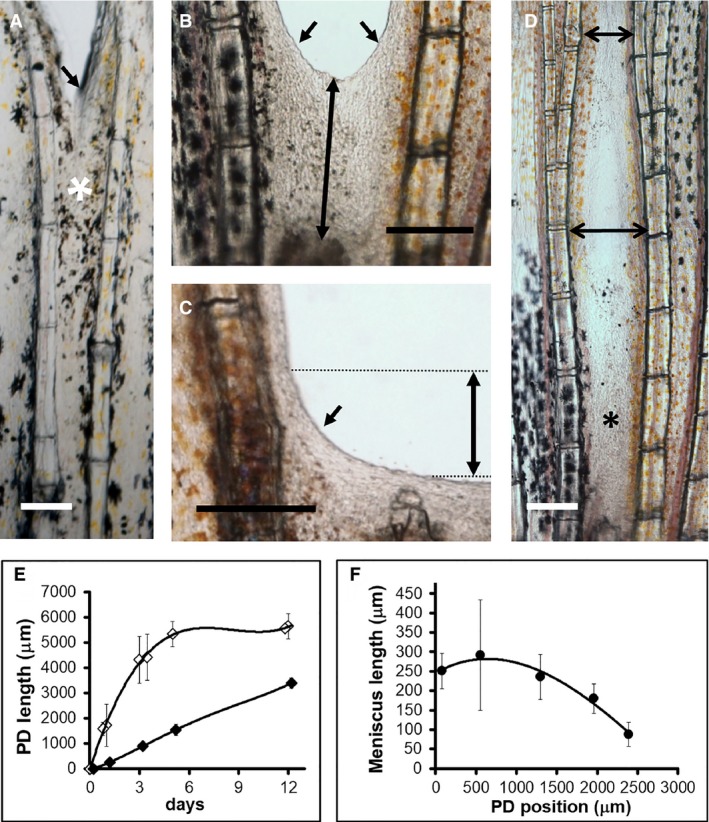
Wound healing after inter‐ray cut experiments. (A) Inter‐ray wound healing at 1 dpo. Observe the many pigment cells in distal regions (white asterisk). (B, C) Inter‐ray wound healing 1 day after one‐ (B) and four‐ray (C) cuts. (D) Varying width (double‐pointing arrows) of healing inter‐ray (asterisk). (E) Inter‐ray lengths (double arrow in B) during inter‐ray (white rhombus, R3 or R3‐R4 cuts) wound healing or ray + inter‐ray (black rhombus, R3–R5 or R3–R6 cuts) regeneration. (F) PD variations of inter‐ray meniscus lengths (double arrow in C) during inter‐ray wound healing. Oblique arrows in (A–C) indicate marginal leading edges (meniscus) of wound healing inter‐rays. Vertical bars in (E, F) are standard deviations. Scale bars: 100 (A) and 250 (B–D) μm.
Cell migration during wound healing was studied by injection of a DiI solution (Fig. 2B) or BrdU at 3 dpo, or by transgenic labelling of endothelial cells (see Material and methods). DiI injection in marginal non‐operated rays did not label central regions (Fig. 6A,B), whereas injections proximal to the cut (Fig. 6C) completely labelled the central healing membrane (Fig. 6D). This labelled membrane only comprises two abutting epidermis in cross‐section (data not shown), suggesting distal migration of epidermal cells. Proximal melanophores have also been found to invade the healing non‐pigmented inter‐ray from neighbouring pigmented regions (Fig. 6E). In daily photographs of these healing membranes (Supporting Information Fig. S4A), marginal inter‐ray melanophores were observed to lose their stellate phenotype at 1 dpo (Fig. S4B) and to migrate distally at 2 or 3 dpo (Fig. S4C,D) to reach the distal margin, where they initiate re‐positioning (Fig. 5A). During the first 2 days, some DiI‐labelled cells were also found in proximal inner positions, suggesting proximally invading mesenchyme cells. At 4 dpo, there is proliferation of epidermal cells but not of these mesenchyme cells (Fig. 6F). Small vessel sprouts are also formed from the vein in marginal inter‐rays during these stages (Fig. 6G). Nevertheless, the original endothelial and pigment pattern is only restored once the ray, or rays, regenerate. After 3 dpo, the pigment pattern around the regions originating the migrating melanophore, changes slightly (Fig. S4D). This suggests potential replenishment of absent migrating cells from neighbouring regions.
Figure 6.
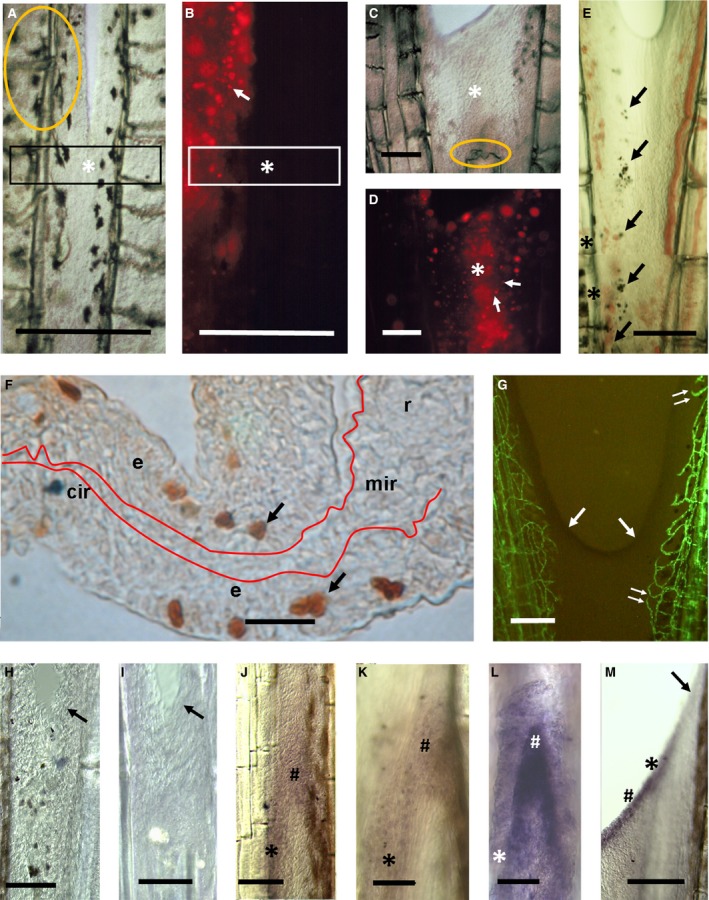
Cell migration, proliferation and gene expression during inter‐ray wound healing and regeneration. (A–D) DiI‐labelling of wound healing inter‐ray (asterisks) after one‐ray cut. The same inter‐rays under light (A, C) and fluorescence (B, D) microscopy. Orange ovals are injection points. White arrows show labelled cells. Rectangles in (A, B) are for comparative reference. (E) Melanophores (arrows) migrating from proximal pigmented inter‐ray margins (asterisk) at 2 dpo. (F) BrdU incorporation (arrows) during late inter‐ray wound healing after one‐ray cut. Red line shows inter‐ray basement membrane. e, epidermis; r, mir and cir, ray, marginal and central inter‐ray regions. (G) Endothelial cells (green) during inter‐ray wound healing. Double arrow shows early endothelial invasion. (H–M) bmp4 (H), msxa (I, J), msxd (K, M) and dlx3 (L) gene expression during inter‐ray wound healing (H, I) and regeneration (J–L) after one‐ray cut or during inter‐ray regeneration after four‐ray cut (M). Oblique arrows in (G–I, M) indicate inter‐ray meniscus. Symbols in J–M are wound epidermis over ray (#) and inter‐ray (asterisk) blastema. Scale bars: 10 (F), 100 (H, I, K, L), 150 (E), 200 (J), 250 (A–D, G) and 300 (M) μm.
During ray regeneration, an endothelial and pigment pattern is completely restored by lateral invasion from neighbouring tissues. Sprouts from ray or marginal inter‐ray vessels reach the regenerating ray to restore the pattern (Supporting Information Fig. S5A; MGC, unpublished results). At 30 dpo, pigments are found in the regenerated inter‐ray neighbouring the pigmented non‐operated rays (Fig. S5B). Both endothelium and pigments show a slightly oblique distribution which is more distal, nearer to the regenerating ray. This suggests a moving chemo‐attractant signalling centre, potentially at the distal ray blastema according to the distribution of blood vessels (Fig. S5A,B). In serial photographs, melanophores and the lateral vein are seen to migrate distally through the new marginal inter‐ray to reach distal positions at a distance from the distalmost blastema. Finally, the inter‐ray spreads (Fig. S5C), reaching a width similar to that of the healing membrane, the new ray and each new marginal region (Fig. S5D).
Gene expression may also reveal inter‐ray regionalization during regeneration. Expression domains of msxa, msxc, msxd (Akimenko et al. 1995), bmp4 (Murciano et al. 2002) and dlx3 (Akimenko et al. 1994) in the distal blastema are wider than shh expression (Laforest et al. 1998) in the proximal ray blastema. Thus, distally expressed genes could be transcribed in presumptive marginal inter‐ray regions. We have compared the expression of these five genes with shh expression in regenerating inter‐rays after one‐ray and several‐ray cuts. The msxa, msxd, bmp4 and dlx3 are not expressed in the healing inter‐ray (see Fig. 6H,I) and only msxc is inconsistently expressed by mesenchyme cells that invade the central inter‐ray distal to the regenerating ray (data not shown; LL, MAA, unpublished results). Nevertheless, during ray regeneration after a one‐ray cut, msxa (Fig. 6J) and msxd (Fig. 6K) are expressed in the epidermis covering the complete neighbouring inter‐rays, whereas bmp4 and dlx3 (Fig. 6L) are expressed in the epidermis covering a region only slightly wider (not shown) than that of shh (Murciano et al. 2002). At this stage, this gene expression pattern resembles that observed after a fin cut. After four‐ray cuts (see Fig. 6L), expression of these genes is similar to that shown after complete fin lobe cuts (not shown) or fin cuts (Akimenko et al. 1994, 1995; Murciano et al. 2002 ). During these events, the leading ‘meniscus’‐like edge shows complete absence of expression of these genes (Fig. 6H,I,M).
We have shown that epidermal, early melanophore and fibroblast‐like cells invade the inter‐ray proximodistally during wound healing, whereas endothelial cells and late melanophores migrate dorsoventrally during ray regeneration. We have studied potential molecular mechanisms controlling these processes.
kcnk5b, FGFR1 and SHH pathway control inter‐ray patterning and size
Among available zebrafish mutants (van Eeden et al. 1996), alf dty86d (Perathoner et al. 2014) was selected to show abnormalities after one‐, two‐, three‐ or four‐ray cuts in a caudal fin screening (data not shown). The fins of alf dty86d mutant show inter‐rays with varying widths. When one ray is cut, a healing membrane forms that may show symmetric or asymmetric ‘meniscus‐like’ leading edges (Fig. 7A). Interestingly, a membrane also forms when three or four rays are cut in this mutant (Fig. 7B). This membrane is about double width (P < 0.01) and shows larger meniscal edges (P < 0.01) (Fig. 7B) when compared with inter‐rays formed after one‐ or two‐ray cuts in wild type fins. As in wild type (Supporting Information Fig. S4), marginal melanophores migrate distally after one‐ray cut (Supporting Information Fig. S6) but are drastically reduced at 3 dpo (Fig. S6A–D). This may be due to the large size of the fin, assuming a signalling centre at distal regions. During ray regeneration, lateral migration of endothelial cells and melanophores also occurs after one‐ray or three‐ray cuts (Supporting Information Fig. S7A–D). The oblique distributions of the enlarged vessels of this mutant (Fig. S7A) and melanophores (Fig. S7B,D) also suggest chemo‐attraction/electrotaxis from the ray blastema. Distal migration along the margins of the regenerating rays (Fig. S7A) or varying widths of the membrane healing the space left by flanking rays (Fig. S7C) also occur, as in wild type. Angiogenesis between two regenerating rays enhances when compared with vessels sprouting from non‐operated rays after two‐ or three‐ray cuts (Fig. S7A). After ray regeneration, the width of the inter‐ray and ray also suggests the formation of new marginal inter‐rays during the process (Fig. S7D). The new longitudinal veins show a sinusoidal/plexus profile suggesting electrotaxis‐dependence of distal angiogenesis at the marginal inter‐ray (Fig. S7D).
Figure 7.
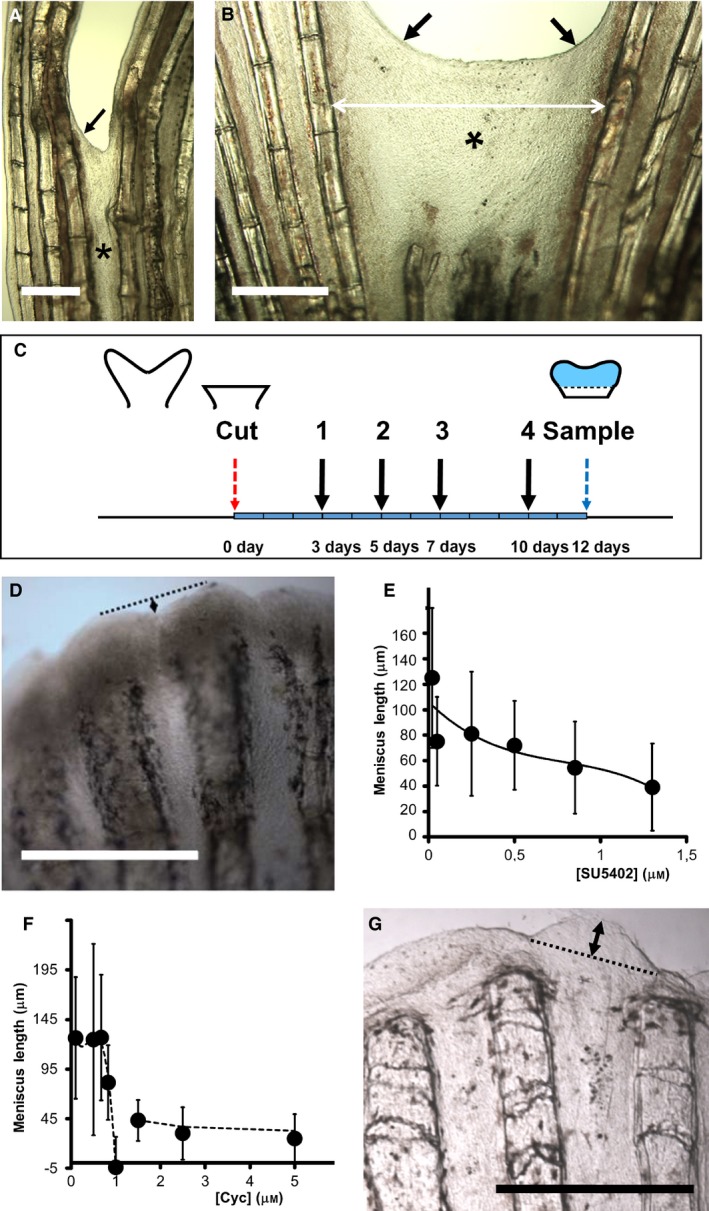
Effects of alf dty86d mutation and inhibitors on regenerating inter‐ray clefts during wound healing and regeneration. (A, B) Inter‐ray wound healing (asterisk) after one‐ray (A) and three‐ray (B) cuts in alf dty86d caudal fins. Double arrows are healing membrane width. Oblique arrows are meniscus. (C) Scheme of inhibitor experiments. 3, 5, 7 and 10 days, days post‐operation (red arrow) of drug administration/replacement (black arrows 1–4). Light blue fin region is regenerated tissue at 12 dpo. Blue arrow is sampling day. (D–G) Incision lengths (double arrows in D and G) between largest rays regenerated under increasing concentrations (μm) of SU5402 (D, E) and cyclopamine (Cyc; F–G). Negative values in (F) show inter‐ray overgrowth (G). Vertical bars (E, F) are standard deviations. (D, G) Fins regenerated under 1.3 μm SU5402 (D) and 1 μm cyclopamine (G). Scale bars: 250 (A–B) and 500 (D, G) μm.
We have further studied the involvement of FGFR1 and SHH signalling in the regulation of inter‐ray size and pigment positioning after fin cuts. Several concentrations of the FGFR1 (and other tyrosine kinases) inhibitor SU5402 and the Smo inhibitor cyclopamine were used after fin blastema formation to study canonical activities on ray branching (Armstrong et al. 2017) or inter‐ray width (Figs 2A and 7C). Inhibitor water changes and fin sampling were done in a 12‐day experiment. (Fig. 7C; Commentary on Supporting Information Figures). Slight SL variations of fishes (Table S2) are unrelated to the observed results.
We first measured the size of these distal incisions in fins regenerated under chemical inhibition (Fig. 7D–G). The size reduced when the concentration of SU5402 (Fig. 7D,E) or cyclopamine (Fig. 7F–G) increased. Nevertheless, whereas this reduction is gradual with the increasing concentration of SU5402 (Fig. 7E), it shows a sudden change at 1 μm cyclopamine (Fig. 7F). At this concentration, the central inter‐ray shows a convex profile overgrowing the marginal regions (Fig. 7G), acquiring negative values (Fig. 7F) and suggesting growth arrest of ray, but not inter‐ray, blastemas. At higher concentrations, both ray and inter‐ray growths are arrested. Growth arrest is an off‐target effect of this inhibitor (Armstrong et al. 2017), precluding any conclusion on SHH/IHH regulation. Nevertheless, these results suggest independent regulation of rays and inter‐rays.
We have also studied the effect of these inhibitors on inter‐ray and ray widths and melanophore presence in marginal or central inter‐rays at and post‐branching (Fig. S1A). Unlike control fins (Figs 8A and S1D, data not shown), cyclopamine‐ (Fig. 8B,C) and SU5402‐treated fins (Fig. 8D) showed modified widths and/or a ray branching pattern as concentration increased. The ray width at dichotomy increases gradually with FGFR1 inhibitor concentration but that of inter‐rays suddenly decreases over 0.85 μm SU5402 (Fig. 8E). At this concentration, melanophores in the marginal inter‐rays are more frequent at branching rays compared with DMSO controls (P < 0.01). This narrowing (Fig. 8E) and modification in melanophore distribution is not observed after cyclopamine treatments (Fig. 8F) (P ≫ 0.05). Nevertheless, the new inter‐ray between branches after ray dichotomy (see Fig. 8B) gradually narrows with increasing SU5402 (Fig. 8G) or cyclopamine (Fig. 8H) concentrations. Melanophore frequency also increases at this inter‐ray over 0.85 μm SU5402 (P < 0.05). Finally, inter‐ray width along PD positions between fin cut and ray dichotomy was also measured (Fig. 8I,J). In control fins, the inter‐rays widen gradually along this axis but fluctuate around a critical width during ray branching. Only the administration of 1.3 μm SU5402 prevents inter‐ray widening, maintaining the width (Fig. 8I), although the ray widens along the PD axis (data not shown). This suggests differential sensitivities of rays and inter‐rays to this inhibitor. Interestingly, although cyclopamine prevents ray dichotomy, inter‐ray width at those distal dichotomies similar to control widths.
Figure 8.
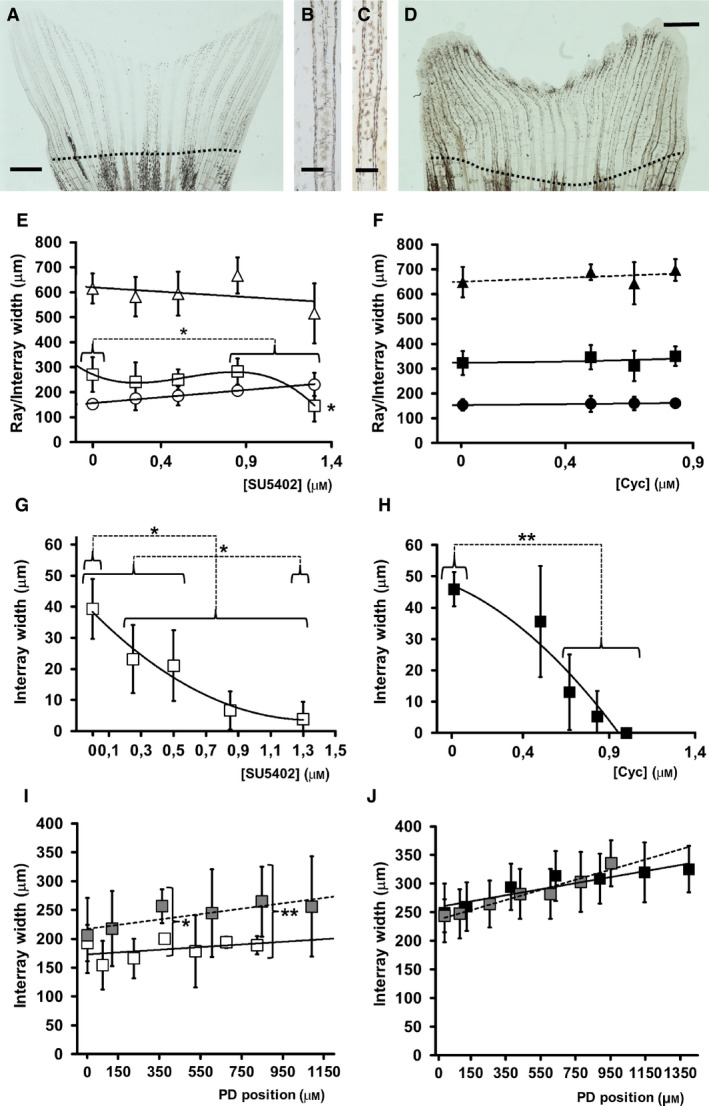
Morphometric data from 12 dpo caudal fin R3 after zebrafish inhibitor experiments. (A) Fin regenerated in water. (B, C) Detail of branched (B) and non‐branched (C) rays after 0.83 μm cyclopamine treatment. (D) Fin regenerated in 1.3 μm SU5402. Discontinuous lines in (A, D) show cut planes. (E, F) Widths of ray (circles), neighbouring inter‐rays (squares) and addition of both, pinnamere, (triangles) at ray branching. (G, H) Internal inter‐ray widths two‐joints distal to ray branch. (I, J). Inter‐ray width variations in the distance to cut plane (0). (E, G, I) Data from SU5402‐treated fins (white symbols). (F, H, J) Data from cyclopamine‐treated fins (black symbols). Concentration units are μm. Vertical bars represent standard deviations. Horizontal brackets show data groups with similar statistical properties (see text). Vertical bracket in (I) compares experimental and control (grey) data from similar positions. *P < 0.05 and and **P < 0.01. Connecting discontinuous lines join compared groups. Upper and lower asterisks in (E) compare rays and inter‐rays, respectively. Scale bars: 150 (B, C) and 1000 (A, D) μm.
Inter‐ray membranes of euteleostean fins vary from incised to overgrown
Inter‐ray membranes connect neighbouring rays in the fins of most actinopterygian species (i.e. Whitehead et al. 1986; Nelson, 1994). Each of these membranes shows a specific width, length and pigment pattern depending on the species, fin and position within the fin. These morphologies and sizes are sometimes important in fish systematics (Nelson, 1994). We have studied, in the literature, fin inter‐ray morphologies of 1103 euteleostean species of 193 families of 44 orders (Whitehead et al. 1986; Anam & Mostarda, 2012 and references therein). Fixed loaned specimens from the Museo Nacional de Ciencias Naturales (Madrid, Spain) were also described anatomically.
Most inter‐ray membranes in actinopterygian fishes connect neighbouring rays from the most proximal (Fig. 9A; Lepidorhombus whiffiagonis dorsal fin, MNCN‐72077) to the most distal positions where they show slight incisions (Fig. 9B; Taurulus bubalis dorsal fin, MNCN‐044260). The width of these inter‐rays gradually increases along the proximodistal axis up to the level of ray branches or the distal end of the non‐branched rays (Fig. 9A,B). In the former case, the inner inter‐ray between branches widens at expense of a gradual narrowing of the inter‐rays between the rays (see Whitehead et al. 1986; Anam & Mostarda, 2012). Nevertheless, actinopterygian inter‐rays can significantly differ from this basic anatomy. We selected 410 species from 11 euteleostean orders that show these fin morphological variations in a significant number of genera. During this selection, those orders with very few species showing very common morphological variants were eliminated. The selected species were classified into six groups according to the presence of these morphologies in at least one of their fins (Supporting Information Table S3). In these species, rays are normally unperturbed or rarely change. These modifications will be studied elsewhere (SCV, JG, JAH, CM, TDF, MMB, unpublished data).
Figure 9.
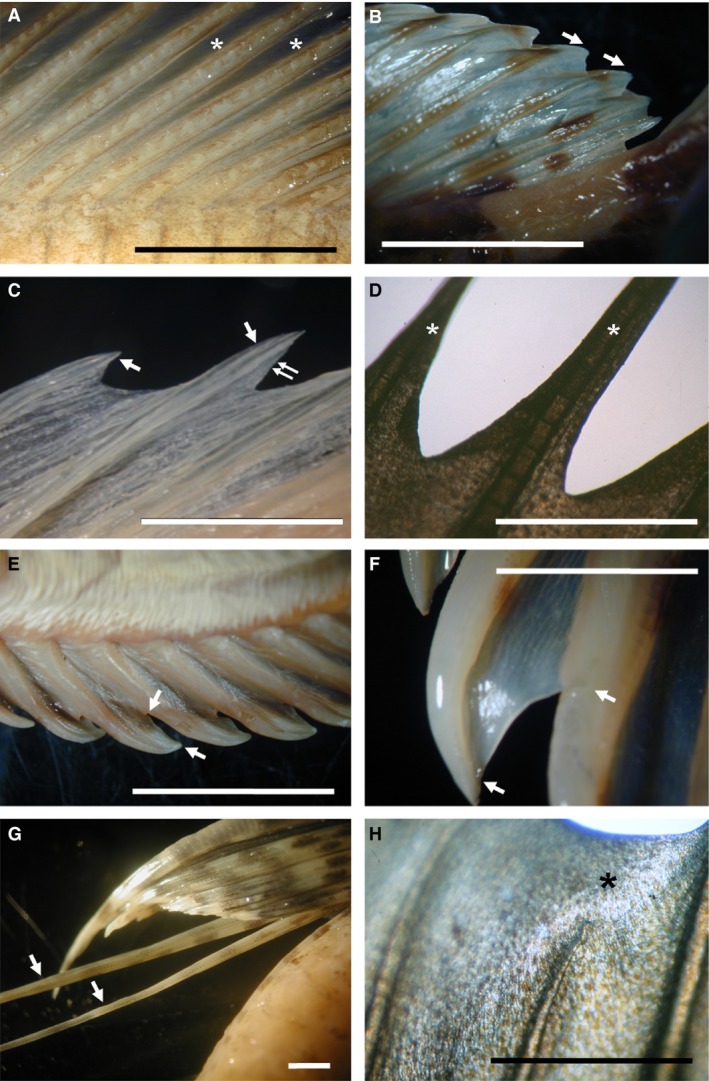
Morphological variants of actinopterygian inter‐ray membranes. (A) Lepidorhombus whiffiagonis dorsal fin, MNCN‐72077. Asterisks show inter‐rays. (B) Taurulus bubalis dorsal fin, MNCN‐044260. (C) Aspitriglas cuculus dorsal fin, MNCN‐107555. Double arrow shows distal inter‐ray incision. Arrows in (B, C) show distal ray margins. (D) Parablennius incognitus second dorsal fin, MNCN‐72911. Asterisks show distal ray regions. (E) Lipophrys pholis anal fin, MNCN‐010863. (F) Pectoral fin of T. bubalis, MNCN‐044260. Arrows in (E, F) show margins of asymmetric inter‐rays. (G) Pantodon buchholzi pelvic fins, MNCN‐235355. Arrows show long rays without a neighbouring inter‐ray. (H) P. incognitus first‐second dorsal fin boundary, MNCN‐72911. Asterisk shows inter‐ray overgrowth. Scale bars: 2(G) and 5 (A–F, H) mm.
The first phenotypic group is characterized by an incision of the distal margin of the inter‐ray membrane (Fig. 9C,D). This phenotype ranges from a slight incision the size of a single ray segment (Fig. 9C; Aspitriglas cuculus dorsal fin, MNCN‐107555) to larger incisions the size of many ray segments (Fig. 9D; Parablennius incognitus second dorsal fin, MNCN‐72911). In these instances, inter‐ray length gradually increases in the neighbourhood of the rays showing a symmetric ‘meniscus‐like’ morphology (Fig. 9B,D). The second group shows asymmetrically incised inter‐rays (i.e. Kanayama, 1991) that connect neighbouring rays at very different positions. This morphology can be observed in dorsal and ventral median fins of Lipophrys pholis (MNCN‐010863; Fig. 9E) or in the pectoral fin of Taurulus bubalis (MNCN‐044260; Fig. 9F). A third phenotypic group is characterized by the complete, or almost complete, absence of the inter‐ray membrane. This separates the flanking rays at both sides (Fig. 9G; Pantodon buchholzi pelvic fins, MNCN‐235355). Our fourth group comprises species with fin inter‐rays only connected to one neighbouring ray as a flag (i.e. Pterois volitans; Anam & Mostarda, 2012). This phenotype is similar to the inter‐ray external to the lateral‐most long rays of zebrafish caudal fin. The fifth group is formed by species with fins in which the inter‐ray membranes overgrow the rays. These inter‐rays may differentiate distal to a single ray (Fig. 9H; i.e. P. incognitus first‐second dorsal fin boundary, MNCN‐72911, a non‐selected river species) or may show a convex distal profile with the central region distal to the margins (i.e. the second dorsal fin of Epinephelus coioides; Anam & Mostarda, 2012). In the most extreme phenotypes, rudiment rays may differentiate into an otherwise ‘rayless’ caudal fin in species of the Anguilliformes order (Whitehead et al. 1986; Nelson, 1994 and references therein; data not shown). Although rare, a sixth phenotypic group shows absent or reduced inter‐ray membranes leading to fused, or nearly fused, rays (i.e. the pelvic fin of Dactylopterus volitans, Fischer et al. 1981, or Dactiloptena orientalis; Anam & Mostarda, 2012).
Many species with these phenotypic variants have been found in the order Perciformes, Gadiformes, Scorpaeniformes and Stomiformes. These phenotypes are mostly found at the dorsal, anal and pelvic fins. The caudal and the pectoral fins, used mostly during swimming, show the smallest number of inter‐ray abnormalities (Table S3). Most of these phenotypes involve changes along the proximodistal axis of the inter‐ray (incised, asymmetric, absent and overgrown inter‐ray groups – A–C and E in Table S3). In this group, phenotypic variants with incised inter‐rays (groups A–D), also called fin clefts (Kanayama, 1991), are much more abundant than those with convex‐shaped inter‐rays (group E) or rayless fin webs (Nelson, 1994). In fewer instances, morphological variations along the dorsoventral axis (asymmetric, flag‐like inter‐ray and fused ray groups – B, D and F in Table S3) occur.
Discussion
During fin development and regeneration, the widths of the global fin web and of each inter‐ray membrane gradually increase (see Marí‐Beffa & Murciano, 2010). As morphology influences the swimming efficiency of fishes (see Lauder et al. 2002; Alben et al. 2007), fin widening and patterning might be adaptive traits, expected to be both finely regulated by specific cellular and genetic mechanisms and under strict evolutionary selective pressure. The experimental and comparative evidence shown in this article supports this view.
Qualitative and quantitative DV variations of cell features characterize the inter‐rays of zebrafish and goldfish caudal fin
Each caudal fin inter‐ray comprises one central and two marginal regions that join the flanking rays. The differences between these two regions are both qualitative and quantitative in adult, developing and regenerating zebrafish, and in goldfish fins. The marginal regions show an almost constant width, a vein that runs along the PD axis (Huang et al. 2003) and the absence of lateral line neuromasts (Wada et al. 2008). Central regions may present variable widths, randomly oriented blood vessels that connect marginal veins or ray arteries (Xu et al. 2014), and neuromasts (Wada et al. 2008). In addition, the marginal and central regions show quantitative differences, the former being thicker and showing a higher abundance of melanophores and leucophores, lower cell density, and greater and more distal proliferative activity during regeneration compared with the latter. These quantitative features show gradual transitions between both regions. In agreement with previous data (Tu & Johnson, 2011), these two regions do not show proliferation restriction boundaries. Moreover, the marginal inter‐ray region and the ray show qualitative and quantitative similarities not shared with central regions. After a 90° rotated ray graft, a large vessel will only form in the marginal region of the regenerating ray in the absence of the marginal vein in the ectopic inter‐ray. Moreover, a similar cell density and frequency of pigment, endothelium or proliferating cells have been observed in rays and marginal inter‐rays, even in those formed internally, early after ray dichotomy. This suggests a somehow similar developmental signature of rays and marginal inter‐rays.
These variations and similarities along DV axis of the caudal fin also involve gene expression domains in the fin blastema (Fig. 10A,B). Distal wound epidermis‐expressed genes, such as msxa and msxd, are transcribed over both ray and inter‐ray blastema (Akimenko et al. 1995), Nevertheless, most of the genes expressed in the distal ray blastema and the wound epidermis over it, e.g. msxc (Akimenko et al. 1995), dlx5 (Akimenko et al. 1994) and several wnt genes (Stoick‐Cooper et al. 2007), are apparently also expressed in marginal inter‐ray regions or in inter‐rays formed early after dichotomies (e.g. Quint et al. 2002), but are not expressed in central inter‐rays (e.g. Murciano et al. 2002). More proximally expressed genes are exclusively transcribed either in the ray blastema, e.g. shh pathway (Laforest et al. 1998), or in inter‐rays, e.g. keratin 8 (Martorana et al. 2001), id1 (Thorimbert et al. 2015) in zebrafish or the tmsβ‐like gene in Xiphophorus (Offen et al. 2009) (Fig. 10A). Interestingly, two of these inter‐ray‐expressed transcripts, those coded by zebrafish keratin 8 (Martorana et al. 2001) and the Xiphophorus tmsβ‐like genes (Offen et al. 2009), show quantitative variations along the DV axis of the inter‐ray epidermis.
Figure 10.
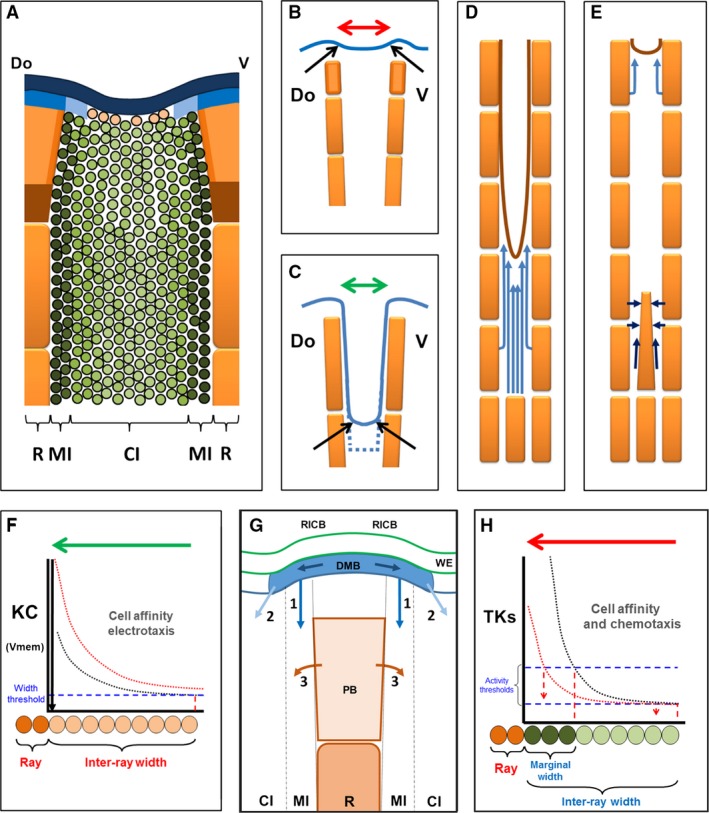
Patterning gradients controlling inter‐ray wound healing and regeneration. (A) Patterned gene expression during inter‐ray regeneration. Blue and dark blue areas are dlx3 and msxa/msxd domains, respectively. Blue plus light blue areas are bmp4/fgfr1 domains. Orange and brown areas are ray blastema and shh/ihh/ptc1 domain. Green area is id1 inter‐ray domain and green intensity is the opposite of zfk8 expression in zebrafish. Darkest green areas are tmsβ‐like expression in Xiphophorus. References are in text. (B, C) Marginal leading edges (arrows) during fin (B) and inter‐ray (C) regeneration. Upper rectangles (B) are ray blastema. Discontinuous line is cut plane. (D, E) Wound healing (D) and regeneration (E) patterns of cell migration. Light and dark blue arrows are early and late migratory directions. (F–H) Bioelectricity‐dependent (F) and signalling‐dependent (G, H) gradients controlling inter‐ray formation, cell patterning and widening. (F) Cell affinity and/or positioning during distal migration quantitatively depend on the K+ channel activity (KC) or an opposite variable, the resting potential membrane (Vmem). Below a discontinuous blue line, the healing inter‐ray is not formed. Red line is gradient activity increase occurring in alf dty86d mutant. (G) RICB and dotted lines are ray/inter‐ray compartment boundaries, DMB is distalmost blastema, WE is wound epidermis, PB is proximal blastema, MI and CI are marginal and central inter‐ray regions, R is ray. 1 (marginal Fgfs, BMPs) and 2 (central, Fgfs) are inter‐ray signals from distalmost blastema controlling cell migration and tissue size, 3 is signalling from shh‐expressing cells controlling short‐range tissue size, upper dark arrows are potential signals from wnt5b‐expressing cells. Discontinuous lines are the boundary between marginal and central inter‐ray regions. (H) Fgfr SU5402‐sensitive tyrosine kinases (TKs) pathway‐dependent inter‐ray‐patterning gradient. Cell affinity/distal migration and/or lateral migration are quantitatively regulated. Upper discontinuous line is marginal region formation threshold. Lower line is central inter‐ray widening threshold. Green or red gradients show gradual experimental inhibition and discontinuous vertical arrows are interpretations of observed phenotypes. Green and red arrows (F–G) are activity domains of KC and specific gradients in half inter‐rays, respectively. Orange and dark and light green ovals are ray, marginal and central inter‐ray cells, respectively. Pink circles in (A, F) are potential bioelectricity‐dependent cells. Do and V, dorsal and ventral.
Our gene expression results after a one‐ray cut, agree with this transition of distal overlapping to proximal ray vs. inter‐ray regionalization (Fig. 10A). This gene expression evidence would account for both the similar signature inferred for rays and marginal inter‐rays, and the gradual variation of cell features observed within inter‐rays.
During wound healing, width is established by a bioelectricity‐dependent gradient
After a fin cut, wound healing occurs by epidermal cell migration. This is followed by epidermal cell proliferation (Santos‐Ruiz et al. 2002) and migration (Poleo et al. 2001) of several cell types (Knopf et al. 2011; Sousa et al. 2011; Tu & Johnson, 2011; Stewart & Stankunas, 2012) to form a blastema beneath wound epidermis. As in inter‐ray cuts (Fig. 10A), cuts of one ray lead to wound healing spatially and temporally distinct from blastema formation (Nabrit, 1929; Marí‐Beffa et al. 1999). This distinction has facilitated wound‐healing studies. Besides epidermal cells, melanophores (Fig. 10D) and fibroblasts can also be traced to migrate distally during wound healing, whereas they migrate laterally during regeneration (Fig. 10E). Potential migration attractants during healing are Igf (Chablais & Jaźwińska, 2010) and ROS (Sehring et al. 2016), which regulate wound closure and are expressed at wound epidermis (Gauron et al. 2013). actβa (Jaźwińska et al. 2007) or retinoic acid (Blum & Begemann, 2015) signalling is also involved in maintenance and/or de‐differentiation of distally migrating fibroblasts or osteoblasts.
In the gain‐of‐function kcnk5b mutant alf dty86d, the healing membrane after one or several ray cuts widens and forms an enlarged meniscus at the margins. Zebrafish knck5b gene codifies for a two‐pore domain K+ channel (Perathoner et al. 2014) homologous to the gene ‘Tandem of P domains in a weak inwardly rectifying K+ channel’ (TASK‐2) in vertebrates (Cid et al. 2013). TASK‐2 (K2P5.1) regulates cancer proliferation, modulating the resting membrane potential spatiotemporally and being regulated by direct G‐protein interaction (Cid et al. 2013; Inanobe & Kurachi, 2014; McCudden et al. 2005 and references within). TASK‐2 activates instructive signals controlling proliferation, differentiation, cell shape, gene expression (Levin, 2014), and persistent direction and invasiveness by electrotaxis (Özkucur et al. 2011) of several cell types in culture and pattern formation during planaria, tadpole or zebrafish regeneration (Sundelacruz et al. 2009; Levin, 2014; Perathoner et al. 2014). This strongly suggests an underlying bioelectricity‐dependent regulation of the healing membrane width in zebrafish.
A histochemical estimation of cell membrane polarization in adult wild type and alf dty86d mutants (Perathoner et al. 2014) further suggests variations in the resting membrane potential. Cell membranes appear to be hyperpolarized at central inter‐ray regions (low channel activity) and gradually depolarized (high activity) at marginal regions of the wild type caudal fin inter‐rays. This activity gradient increases (depolarizes) in the mutant (Perathoner et al. 2014; Fig 10F). This gradient could directly control epidermal cell migration to form the ‘meniscus’ profile and healing tissue size by mechanisms such as electrotaxis. Below a threshold channel activity, migration would be prevented and the inter‐ray would not form. The increase of the bioelectric gradient in the mutant would enhance cell migration, leading potentially to wider inter‐rays (Fig. 10F), depending on the space left between flanking rays. Contact‐inhibition between migrating cells would explain this behaviour. Gene expression associated to wound‐healing/regeneration defects, the control of tissue size or cell interactions are modulated in developing frog, axolotl and humans under experimental Vmem depolarization (Pai et al. 2015). As previously proposed, initial hyperpolarization/depolarization would regulate voltage‐sensitive mechanisms that would ultimately regulate signalling pathways, such as FGFR1 or SHH in this study, regulating cellular functions (Sundelacruz et al. 2009; Urrego et al. 2014). These positional and size control functions agree with previous suggestions (Marí‐Beffa & Murciano, 2010; Levin, 2014; Perathoner et al. 2014) and would account for the instructive control of widening direction of inter‐rays exerted by cells at ray margins after 90° ray graft rotation. How the distribution of this gradient is regulated is unknown, but the spatiotemporal pattern of Vmem of a tissue has been shown in experiments to depend on Vmem in neighbouring cells (Levin, 2014).
During regeneration, FGFR1 and SHH pathway‐dependent gradients modulate widening and cell migration
As ray regeneration is delayed with respect to inter‐ray formation, a one‐ray cut may be a good model to study regeneration of single rays or interactions between ray and inter‐rays. Expression of genes during one‐ray regeneration suggests a PD ordering of domains equal to that of fin blastema (Yoshinari et al. 2009). In the one‐ray blastema, genes are expressed in similar domains in wound epidermis, msxa or msxd, distal mesenchyme, dlx3 or bmp4 (Murciano et al. 2002), or proximal fin blastema, shh (Murciano et al. 2002). This indeed supports the regulation by the ray blastema during this process. Differentiating inter‐rays after wound healing are only able to activate msxa or msxd in epidermis, or dlx3 in marginal mesenchyme, when in the neighbourhood of the regenerating ray blastema.
Cells from ray blastema or ray margins also regulate cell activities at neighbouring inter‐ray compartments (Fig. 10G). The gradual decrease of the meniscus size at the marginal inter‐ray when FGFR1 is gradually inhibited (Fig. 10H) or the ectopic ‘T‐shaped’ inter‐ray obtained after 90° rotation of the ray graft supports this regulation. Migration of several cell types also occurs next to the regenerating ray after one‐ray cuts. Melanophores and endothelial cells migrate laterally across inter‐rays and distally along the new marginal inter‐rays towards the ray blastema (Fig. 10E). The lateral migration of pigments may have been impaired after high SU5402 concentration treatments during fin regeneration. This suggests the involvement of ray blastema FGFR1‐dependent chemo‐attractant signals (Fig. 10G,H). Pigment migration through the inter‐ray has also been suggested after grafting sword rays in Xiphophorus (Eibner et al. 2008) and a migration‐driving function of FGFR1 has been found during early pectoral fin bud formation (Mao et al. 2015). At least two tentative genes involved in endothelium and keratinocyte migration have been found to be dependent on FGFR1 activity and are expressed in distal mesenchyme (dusp1) and basal epidermal layers (tmsβ‐like gene) in rays and inter‐rays in regenerating Xiphophorus fins (Offen et al. 2009). The expression patterns of these genes as well as previous functional studies of similar genes in other vertebrates (Malinda et al. 1997, 1999; Kinney et al. 2008) further support this view (as cited in Offen et al. 2009). The distal migration of the longitudinal vein or pigments may also occur inside the ray when the inter‐ray is absent, as observed after 90° ray graft rotation. This agrees with the similar gene expression patterns and histological features observed in rays and the marginal inter‐ray. Bmps (Thorimbert et al. 2015) are candidate signals that regulate distal migration; they have been proposed to regulate the formation of the longitudinal vein (Fig. 10G). A distal wnt/β‐catenin signalling centre has also been involved in regulation of inter‐ray‐expressing genes (Fig. 10G; Wehner et al. 2014). These veins in distal positions of long rays would ultimately regulate leucophore position and differentiation. This emerging network of interactions would also explain the high density of proliferating cells or actinotrichia differentiation in the marginal inter‐ray blastema during normal regeneration in zebrafish and goldfish.
The results obtained after 90° graft rotation further suggest that the margin of a ray, or ray blastema, regulates the formation of a half inter‐ray. This agrees with previous proposals (Marí‐Beffa et al. 1999; Murciano et al. 2001, 2002; Marí‐Beffa & Murciano, 2010). All zebrafish (Murciano et al. 2001, 2007) and swordtail fish (Eibner et al. 2008) caudal fin ray margins are capable of inducing inter‐ray formation and all inter‐rays, or half inter‐rays, are able to respond to these inductions. This would be mediated by SHH and FGFR1 pathways. SHH has been shown to control inter‐ray formation during ray branching (Quint et al. 2002; Armstrong et al. 2017). Although we have used an inhibitor with off‐target effects on cell proliferation (Armstrong et al. 2017), our results also support a regulation of initial inter‐ray widening by this pathway, as they agree with narrow inter‐ray phenotypes observed after laser ablation of shh‐expressing (co‐expressing fgfr1, Lee et al. 2009) cells (Zhang et al. 2012) or after administration of a canonical SHH pathway inhibitor (Armstrong et al. 2017). The reduction of inter‐ray width observed after high‐dose SU5402 inhibition also supports the involvement of FGFR1‐signalling in inter‐ray widening. FGFR1 is expressed in ray and potentially in marginal inter‐ray blastema, but not in central inter‐ray (Poss et al. 2000). High levels of these FGFR1 pathway‐dependent gradients would control marginal inter‐ray formation and meniscus size, controlling cell proliferation (Shibata et al. 2016) and/or cell affinity properties. Low levels of these FGFR‐dependent signals would control central inter‐ray widening (Fig. 10H). The different widths shown by inter‐rays along the DV axis of the caudal fin could be the result of autonomous responding properties and not of a differential regulation by signals from rays. The final host‐like size of inter‐rays after 90° graft rotation supports this autonomous response. Although conclusions about SHH activity‐dependent gradients on size or outgrowth control cannot be drawn from our study (Armstrong et al. 2017), the convex inter‐ray phenotype observed at 1 μm cyclopamine further suggests an autonomous control of inter‐ray outgrowth. Phenotypes shown after fin regeneration by alf dty86d (Murciano et al. 2007; Sims et al. 2009; Perathoner et al. 2014) or H+ pump activity inhibitors (Monteiro et al. 2014) also support a bioelectric control of regeneration, but this has not been definitely assigned to any of these compartments. As stated above, the absence of gene expression in the meniscus and wound‐healing membrane, and its large size in alf dty86d mutant fins, suggests these structures depend on bioelectric signals. The increased area of the marginal region and the longer size of rays nearer to flanking non‐operated rays after four ray cuts, where the meniscus forms, would also suggest a regulation of ray regeneration by bioelectric control during wound healing.
Pattern and size along the DV axis (Marí‐Beffa & Murciano, 2010) has been claimed to be dependent on interactions at the ray/inter‐ray compartment boundary under the control of a positional identity (PI) gradient along the PD axis. Ray/inter‐ray boundaries at each side of the ray may not only control inter‐ray formation or ray branching (Murciano et al. 2002) but also regulate formation of the marginal region, direction of the central region widening, and cell type patterning of inter‐rays and marginal rays. The final inter‐ray size would be an autonomous property of each compartment in response to conserved FGFR1 and SHH signals in all rays. The evidence shown here further suggests that FGFR1 and knck5b, previously involved in PD patterning (Lee et al. 2005; Murciano et al. 2007; Sims et al. 2009; Perathoner et al. 2014), are also involved in inter‐ray DV patterning and size.
An underlying patterning gradient can also explain the natural homoplastic variability of inter‐ray membranes in euteleostean fins
Actinopterygian actinotrichia/lepidotrichia and sarcopterygian dermotrichia/camptotrichia have been considered homologous structures in the dermal skeleton of osteichthyan fins (Géraudie & Meunier, 1980, 1984; Géraudie, 1988; Johanson et al. 2005). Moreover, no relationship has been established between chondrichthyes or agnatha fin skeleton and these osteichthyan structures (Cole & Currie, 2007; Ota et al. 2013). Due to loss of preservation of soft tissues in fossil records, the ancestry of complete inter‐rays between rays in osteichthyan fins cannot be definitely established. Living basal actinopterygian groups, such as bichirs and reedfishes in the order Polypteriformes, sturgeons and paddlefishes in the order Acipenseriformes, and bowfins in the order Amiiformes, or living sarcopterygians, coelacanths (Friedman et al. 2007) and dipnoans (Géraudie & Meunier, 1984), show complete inter‐rays (Whitehead et al. 1986; Nelson, 1994). This character can thus be considered synapomorphic for crown‐osteichthyan fishes and plesiomorphic for crown‐actinopterygian fishes (Hennig, 1950; Sallan, 2014). Only sturgeons display a tendency to reduced inter‐ray widths as a potential mild phenotype of ray fusion (group E; Nelson, 1994).
The developmental origin of the complete inter‐rays in zebrafish is also related to ray formation. The fin fold of zebrafish does not show an actinotrichia‐devoid region (Grandel & Schulte‐Merker, 1998; Durán et al. 2011), such as the central inter‐ray, but a continuous palisade. When the rays form (Grandel & Schulte‐Merker, 1998), inter‐rays appear, maintaining their presence in the fin web until adulthood. This has also been described during fin development in many actinopterygian species (Bone et al. 1995; Mabee et al. 2002) and dipnoans (Géraudie, 1984). In this sense, changes in the mechanisms found in zebrafish that generate a complete inter‐ray may be candidates for generating morphological novelties (groups A–F). As these new morphologies are found in many unrelated ray‐finned orders, they must be considered convergent, or homoplastic, characters. These types of characters are not useful for taxonomic studies but are potentially interesting in Evo‐Devo approaches (e.g. Metscher & Ahlberg, 1999; Zauner et al. 2003). In this article, we assume a close similarity between fin development and regeneration regulatory mechanisms (Marí‐Beffa & Murciano, 2010), so that the results of regeneration studies could also be considered of interest when proposing candidate genes potentially causing euteleostean inter‐ray variants.
In principle, inter‐rays may be considered largely independent of rays in bony fish fins. The gene expression pattern in regenerating inter‐rays (Martorana et al. 2001; Jaźwińska et al. 2007; Thorimbert et al. 2015) is different to that shown by ray blastema (Yoshinari et al. 2009; Marí‐Beffa & Murciano, 2010; Whener et al. 2014). Moreover, the osteoblast and fibroblast lineage restriction found between rays and inter‐rays during fin regeneration and development (Tu & Johnson, 2011) supports the view of two compartments controlled by different cellular and molecular mechanisms. Finally, the morphological variations of euteleostean inter‐rays found in this study also suggest independent regulatory mechanisms of inter‐rays and rays (SCV, JG, JAH, CM, TDF, MMB, unpublished data).
The incised distal margins of groups A, B and C actinopterygian species always show two ‘menisci’ at both sides of the inter‐ray clefts. In these menisci, marginal inter‐ray regions are located more distally than the central regions, suggesting differences along the DV axis (Fig. 9B–D). Spontaneous clefts in developing fins and the healing membrane after fin and one‐, two‐, three‐ or four‐ray cuts in zebrafish also show a ‘meniscus‐like’ marginal edge. This is enhanced in alf dty86d mutant during the early stages after a three‐ray cut, and is modified when SU5402 and cyclopamine are administrated after fin cuts. These epigenetic and genetic regulators of fin regeneration and development are the first candidate mechanisms to be involved in the evolutionary innovation of these morphologies. Moreover, group B (asymmetric inter‐rays) and group D (flag‐like inter‐rays) phenotypes suggest differential ray/inter‐ray interactions at both ray sides. Local regulation has been inferred from the half‐sized ectopic inter‐ray induced during regeneration after 90° rotated ray grafts. A genetic regionalization has also been described in developing and regenerating zebrafish pectoral fins (Nachtrab et al. 2013), suggesting mechanisms potentially involved in ray/inter‐ray polarization. The asymmetric healing membrane found in developing or healing after one‐ray cut alf dty86d fins also suggests a candidate bioelectricity mechanism potentially involved in the evolutionary generation of these morphologies. Finally, group C (absent inter‐rays) and F (ray fusion) phenotypes also suggest specific inter‐ray perturbations along the DV axis. The fusion of ray branches after inhibition with SU5402 or cyclopamine (see above) or ray fusions after retinoic acid administration (Santamaría et al. 1993; Géraudie et al. 1994; White et al. 1994; Blum & Begemann, 2015) during fin regeneration further provide candidate mechanisms responsible for these evolutionary variations. The overall explanation also agrees with previous morphogenetic proposals of a ray/inter‐ray boundary organizer (Marí‐Beffa & Murciano, 2010) and extends the importance of the results to evolutionary innovations in euteleostean species.
The gradual variations shown by the phenotypes of incised, asymmetric and absent inter‐ray groups (A, B and C in Table S3, Fig. 9C–G), similar to a phenotypic series previously described in an euteleostean family (Kanayama, 1991), and the overgrown inter‐rays of group E (Fig. 9H) suggest modifications of regulatory mechanisms acting along PD axis. The spontaneous clefts of developing fins and the overgrown inter‐rays after the administration of 1 μm cyclopamine to zebrafish regenerating fins (Fig. 7G) are similar to these natural variants. Due to the off‐target effect described by Armstrong et al. (2017) no candidate mechanism can be suggested. Finally, this A–C group series shows variations along both DV and PD axes. This could be due to cross‐regulations between mechanisms acting along both axes. This agrees with a previous suggestion of a cross‐regulation between a gradient controlling PD positional identity (Lee et al. 2005) and the ray/inter‐ray boundary organizers (Marí‐Beffa & Murciano, 2010).
Evolutionary novelties, such as inter‐ray clefting or overgrowth, or even ray fusion, could be the result of genetic or epigenetic changes of specific inter‐ray regulatory mechanisms. Moreover, specific adaptive conditions may have selected these evolutionary innovations in euteleostean species to support a convergent homoplastic origin. In principle, these morphological variants are not observed to the same extent in all fins, being rare in caudal and pectoral fins (Table S3). Caudal fin has been associated to thrust generation (Mwaffo et al. 2017), whereas pectoral fins have been related to fish manoeuvring or hovering (Lauder et al. 2002; Alben et al. 2007). Inter‐ray widths and the rigidity of rays have been involved in vortex‐generation, a hydrodynamic structure crucial for motion control during swimming (Lauder et al. 2002). Under a simple Evo‐Devo hypothesis, specific changes in inter‐ray patterning mechanisms could have occurred, affecting the fins of many euteleostean families. Fins under a high hydrodynamic selection pressure would show less frequent inter‐ray morphological variants, as they would not have been maintained in natural populations, being eliminated by natural selection. Some of these morphological variants have already been associated to specific derived uses only partially related to motion control (Fish, 1990; Baldauf et al. 2010). This could be a third stage in the generation of these fin inter‐ray innovations. Some testable molecular hypotheses from this model are being refuted following experiments with appropriate species (JG, MMB, unpublished data).
Supporting information
Fig. S1. Morphometry of inter‐rays of the regenerating caudal fin of Danio rerio.
Fig. S2. Anatomy and pigment distribution of the caudal fin of Carassius auratus.
Fig. S3. Melanophores from marginal inter‐rays migrate distally during inter‐ray wound healing.
Fig. S4. Endothelial cell and melanophore pattern and inter‐ray size during ray regeneration.
Fig. S5. Special inter‐ray phenotypes in zebrafish.
Fig. S6. Melanophore distribution during wound healing after one‐ray cut in an alf dty86d caudal fin.
Fig. S7. Pigment and blood vessel distribution and inter‐ray size during ray regeneration after three‐ray cuts in alf dty86d/+ caudal fins.
Table S1. Ray‐cut experiments.
Table S2. (A) Number and standard length statistics of cyclopamine‐treated fishes. (B) Number and standard length statistics of SU5402‐treated fishes.
Table S3. Number of species and fins showing morphological variations of inter‐rays.
Acknowledgements
All specimens of actinopterygian species were kindly loaned by the Museo Nacional de Ciencias Naturales of Madrid, Spain. We thank Jesús Dorda Dorda and Laura Hernández Javier for technical assistance on specimen loaning. We thank J. Becerra, Marie Andrée Akimenko (2000), Axel Meyer and Gerrit Begemann (2002–2004) and colleagues at their labs for discussion about results and the ray/inter‐ray boundary organizer hypothesis in this paper. The authors are indebted to M.‐A. Akimenko and Lynda Laforest (Ottawa, Canada) for sharing results and assistance with the in situ hybridization study during a sabbatical leave of M.M.B. supported by a PIEE fellowship from the Spanish MCT. The authors are also indebted to M. García‐Caballero for sharing unpublished results, to M. L. Altozano for manuscript typewriting and to L. Cerón for the photograph in Figure 2E. This work has been supported by the grant PB98‐1409 and BOS2002‐00347 from the DGESC of the Spanish MEC and MCT. The authors state no conflict of interest.
References
- Akimenko MA, Ekker M, Wegner J, et al. (1994) Combinatorial expression of three zebrafish genes related to distal‐less: part of a homeobox gene code for the head. J Neurosci 14, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimenko M‐A, Johnson SL, Westerfield M, et al. (1995) Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development 121, 347–357. [DOI] [PubMed] [Google Scholar]
- Akimenko M‐A, Marí‐Beffa M, Becerra J, et al. (2003) Old questions, new tools and some answers to the mistery of fin regeneration. Dev Dyn 226, 190–201. [DOI] [PubMed] [Google Scholar]
- Alben S, Madden PG, Lauder GV (2007) The mechanics of active fin‐shape control in ray‐finned fishes. J R Soc Interface 4, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anam R, Mostarda E (2012) Field Identification Guide to the Living Marine Resources of Kenya. FAP Species Identification Guide for Fishery Purposes. Rome: FAO. [Google Scholar]
- Armstrong BE, Henner A, Stewart S, et al. (2017) SHH promotes direct interactions between epidermal cells and osteoblast progenitors to shape regenerated zebrafish bone. Development 144, 1165–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avaron F, Hoffman L, Guay D, et al. (2006) Characterization of two new zebrafish members of the hedgehog family, atypical expression of a zebrafish Indian Hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev Dyn 235, 478–489. [DOI] [PubMed] [Google Scholar]
- Baldauf SA, Bakker TCM, Herder F, et al. (2010) Male mate choice scales female ornament allometry in a cichlid fish. BMC Evol Biol 10, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss PE, Bellavance KL, Whitehead GG, et al. (2006) Chemical modulation of receptor signaling inhibits regenerative angiogenesis in adult zebrafish. Nat Chem Biol 2, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra J, Montes GS, Bexiga SR, et al. (1983) Structure of the tail fin in teleosts. Cell Tissue Res 230, 127–137. [DOI] [PubMed] [Google Scholar]
- Blum N, Begemann G (2015) Retinoic acid signaling spatially restricts osteoblasts and controls ray‐inter‐ray organization during zebrafish fin regeneration. Development 142, 2888–2893. [DOI] [PubMed] [Google Scholar]
- Bone Q, Marshall NB, Blaxter JHS (1995) Biology of Fishes. London: Chapman and Hall. [Google Scholar]
- Broussonet PMA (1786) Observations sur la regenerations de quelques parties du corps de poisons [Observations of regeneration of some parts of the body of fish]. In: Histoire de l´Académie royale Des Sciences (ed. Boudot J.), pp. 684–688, Paris: Imprimerie Roy. [Google Scholar]
- Brown AM, Fischer S, Iovine MK (2009) Osteoblast maturation occurs in overlapping proximo‐distal compartments during fin regeneration in zebrafish. Dev Dyn 238, 2922–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F, Jaźwińska A (2010) IGF signaling between blastema and wound epidermis is required for fin regeneration. Development 137, 871–879. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, et al. (2002) Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 16, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid PL, Roa‐Rojas HA, Niemeyer MI, et al. (2013) TASK‐2, a K2P K+ channel with complex regulation and diverse physiological functions. Front Physiol 4, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NJ, Currie PD (2007) Insights from sharks: evolutionary and developmental models of fin development. Dev Dyn 236, 2421–2431. [DOI] [PubMed] [Google Scholar]
- Durán I, Marí‐Beffa M, Santamaría JA, et al. (2011) Freeze substitution followed by low melting point was embedding preserves histomorphology and allows protein and mRNA localization techniques. Microsc Res Tech 74, 440–448. [DOI] [PubMed] [Google Scholar]
- Durán I, Ruiz‐Sánchez J, Santamaría M, et al. (2015) Holmgren's principle of delamination and fin morphogenesis. Mec Dev 35, 16–30. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, et al. (1996) Genetic analysis of fin formation in the zebrafish, Danio rerio . Development 123, 255–262. [DOI] [PubMed] [Google Scholar]
- Eibner C, Pittlik S, Meyer A, et al. (2008) An organizer controls the development of the ‘sword’, a sexually selected trait in swordtail fish. Evol Dev 10, 403–412. [DOI] [PubMed] [Google Scholar]
- Fischer W, Bianchi G, Scott WB (1981) FAO Species Identification Sheets for Fishery Purposes. Eastern Central Atlantic. Fishing Areas 34 and 47 (in part), vol. II Ottawa: Canada Funds‐in‐Trust, FAO. [Google Scholar]
- Fish FE (1990) Wing design and scaling of flying fish with regard to flight performance. J Zool 221, 391–403. [Google Scholar]
- Friedman M, Coates MI, Anderson P (2007) First discovery of a primitive coelacanth fin fills a major gap in the evolution of lobed fins and limbs. Evol Dev 9, 329–337. [DOI] [PubMed] [Google Scholar]
- Gauron C, Rampon C, Bouzaffour M, et al. (2013) Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci Rep 3, 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géraudie J (1984) Fine structural comparative peculiarities of the developing dipnoan dermal skeleton in the fins of Neoceratodus larvae. Anat Rec 209, 115–123. [DOI] [PubMed] [Google Scholar]
- Géraudie J (1988) Fine structural peculiarities of the pectoral fin dermoskeleton of two brachiopterygii, Polypterus senegalus and Calamoichthys calabaricus (Pisces, Osteichthyes). Anat Rec 221, 455–468. [DOI] [PubMed] [Google Scholar]
- Géraudie J, Meunier FJ (1980) Elastoidin actinotrichia in Coelacanth fins: a comparison with teleosts. Tissue Cell 12, 637–645. [DOI] [PubMed] [Google Scholar]
- Géraudie J, Meunier FJ (1984) Structure and comparative morphology of camptotrichia of lungfish fins. Tissue Cell 16, 217–236. [DOI] [PubMed] [Google Scholar]
- Géraudie J, Brulfert A, Monnot MJ, et al. (1994) Teratogenic and morphogenetic effects of retinoic acid on the regenerating pectoral fin in zebrafish. J Exp Zool 268, 12–22. [Google Scholar]
- Goss RJ, Stagg MW (1957) The regeneration of fins and fin rays in Fundulus heteroclitus . J Exp Zool 136, 487–508. [DOI] [PubMed] [Google Scholar]
- Grandel H, Schulte‐Merker S (1998) The development of the paired fins in the Zebrafish (Danio rerio). Mech Dev 79, 99–120. [DOI] [PubMed] [Google Scholar]
- Hennig W (1950) Grundzüge einer Theorie der phylogenetischen Systematik. Berlín: Deutscher Zentralverlag. [Google Scholar]
- Huang CC, Lawson ND, Weinstein MB, et al. (2003) reg 6 is required for branching morphogenesis during blood vessel regeneration in zebrafish caudal fins. Dev Biol 264, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanobe A, Kurachi Y (2014) Membrane channels as integrators of G‐protein‐mediated signaling. Biochim Biophys Acta 1838, 521–531. [DOI] [PubMed] [Google Scholar]
- Jaźwińska A, Badakov R, Keating MT (2007) Activin‐βA signaling is required for zebrafish fin regeneration. Curr Biol 17, 1390–1395. [DOI] [PubMed] [Google Scholar]
- Johanson Z, Burrow C, Warren A, et al. (2005) Homology of fin lepidotrichia in osteichthyan fishes. Lethaia 38, 27–36. [Google Scholar]
- Johnson SL, Bennett P (1999) Growth control in the ontogenetic and regenerating zebrafish fin. Methods Cell Biol 59, 301–311. [DOI] [PubMed] [Google Scholar]
- Kanayama T (1991) Taxonomy and phylogeny of the family Agonidae (Pisces, Scorpaeniformes). Mem Faculty Fish Hokkaido Univ 38, 1–199. [Google Scholar]
- Kemp NE, Park JH (1970) Regeneration of lepidotrichia and actinotrichia in the tail fin of the teleosts, Tilapia mossambica . Dev Biol 22, 321–342. [DOI] [PubMed] [Google Scholar]
- Kiernan JA (2015) Histological and Histochemical Methods, Theory and Practice. 4th edn Banbury: Scion Publishing Ltd.. [Google Scholar]
- Kinney CM, Chandrasekharan UM, Mavrakis L, et al. (2008) VEGF and thrombin induce MKP‐1 through distinct signalling pathways, role for MKP‐1 in endothelial cell migration. Am J Physiol Cell Physiol 294, C241–C250. [DOI] [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, et al. (2011) Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 20, 713–724. [DOI] [PubMed] [Google Scholar]
- Laforest L, Brown CW, Poleo G, et al. (1998) Involvement of the Sonic hedgehog, patched‐1 and bmp‐2 genes in patterning of the zebrafish dermal fin rays. Development 125, 4175–4184. [DOI] [PubMed] [Google Scholar]
- Lauder GV, Nauen JC, Drucker EG (2002) Experimental hydrodynamics and evolution, functions of median fins in ray‐finned fishes. Integr Comp Biol 42, 1009–1017. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM (2002) In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248, 307–318. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, et al. (2005) Fgf signaling instructs position‐dependent growth rate during zebrafish fin regeneration. Development 132, 5173–5183. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hami D, De Val S, et al. (2009) Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol 331, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M (2014) Molecular bioelectricity, how endogenous voltage potentials control cell behavior and instruct pattern regulation in vivo. Mol Biol Cell 25, 3835–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabee PM, Crotwell PL, Bird NC, et al. (2002) Evolution of median fin modules in the axial skeleton of fishes. J Exp Zool 15, 77–90. [DOI] [PubMed] [Google Scholar]
- Malinda KM, Goldstein AL, Kleinman HK (1997) Thymosin beta 4 stimulates directional migration of human humbilical vein endothelial cells. FASEB J 11, 474–481. [DOI] [PubMed] [Google Scholar]
- Malinda KM, Sidhu GS, Mani H, et al. (1999) Thymosin beta 4 accelerates wound healing. J Invest Dermatol 113, 364–368. [DOI] [PubMed] [Google Scholar]
- Mao O, Stinnett HK, Ho RK (2015) Asymmetric cell convergence‐driven zebrafish fin bud initiation and prepattern requires Tbx5a control of a mesenchymal Fgf signal. Development 142, 4329–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí‐Beffa M, Murciano C (2010) Dermoskeleton morphogenesis in zebrafish fins. Dev Dyn 239, 2779–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí‐Beffa M, Mateos I, Palmqvist P, et al. (1996) Cell to cell interactions during teleosts fin regeneration. Int J Dev Biol Supp 1, 179S–180S. [PubMed] [Google Scholar]
- Marí‐Beffa M, Palmqvist P, Marín‐Girón F, et al. (1999) Morphometrical study of the regeneration of individual rays in teleost tail fins. J Anat 195, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorana ML, Tawk M, Lapointe T, et al. (2001) Zebrafish keratin 8 is expressed at high levels in the epidermis of regenerating caudal fin. Int J Dev Biol 45, 449–452. [PubMed] [Google Scholar]
- McCudden CR, Hains MD, Kimple RJ, et al. (2005) G‐protein signaling, back to the future. CMLS Cell Mol Life Sci 62, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD, Ahlberg P (1999) Zebrafish in context: uses of a laboratory model in comparative studies. Dev Biol 210, 1–14. [DOI] [PubMed] [Google Scholar]
- Monteiro J, Aires R, Becker JD, et al. (2014) V‐ATPase proton pumping activity is required for adult zebrafish appendage regeneration. PLoS ONE 9, e92594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH (1902) Further experiments on the regeneration of the tail fins of fishes. Arch Entw Mech 14, 539–561. [Google Scholar]
- Murciano C, Ruiz J, Maseda D, et al. (2001) Ray and inter‐ray blastemas interact to control bifurcations of Danio rerio fin rays. Int J Dev Biol 45, S129–S130. [Google Scholar]
- Murciano C, Fernández TD, Durán I, et al. (2002) Ray‐inter‐ray interactions during fin regeneration of Danio rerio . Dev Biol 252, 214–224. [DOI] [PubMed] [Google Scholar]
- Murciano C, Pérez‐Claros J, Smith A, et al. (2007) Position dependence of hemiray morphogenesis during tail fin regeneration in Danio rerio . Dev Biol 312, 272–283. [DOI] [PubMed] [Google Scholar]
- Mwaffo V, Zhang P, Romero Cruz S, et al. (2017) Zebrafish swimming in the flow: a particle image velocimetry study. PeerJ 5, e4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabrit SM (1929) The rôle of the fin rays in the regeneration in the caudal fins of fishes in Fundulus and Goldfish. Biol Bull 4, 235–266. [Google Scholar]
- Nachtrab G, Kikuchi K, Tornini VA, et al. (2013) Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140, 3754–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT (2002) A proliferation gradient between proximal and msxb‐expressing distal blastema directs zebrafish fin regeneration. Development 129, 2607–2617. [DOI] [PubMed] [Google Scholar]
- Nelson JS (1994) Fishes of the World. 3rd edn New York: John Wiley & Sons. [Google Scholar]
- Offen N, Meyer A, Begemann G (2009) Identification of novel genes involved in the development of the sword and gonopodium in swordtail fish. Dev Dyn 238, 1674–1687. [DOI] [PubMed] [Google Scholar]
- Ota K, Fujimoto S, Oisi Y, et al. (2013) Late development of hagfish vertebral elements. J Exp Zool B Mol Dev Evol 320B, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkucur N, Perike S, Sharma P, et al. (2011) Persistent directional cell migration requires ion transport proteins as direction sensors and membrane potential differences in order to maintain directedness. BMC Cell Biol 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai VP, Martyniuk CJ, Echeverri K, et al. (2015) Genome‐wide analysis reveals conserved transcriptional responses downstream of resting potential change in Xenopus embryos, axolotl regeneration, and human mesenchymal cell differentiation. Regeneration 3, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse AGE (1985) Histochemistry, Theoretical and Applied. 4th edn, vol. 2 Edinburgh: Churchill Livingstone. [Google Scholar]
- Perathoner S, Daane JM, Henrion U, et al. (2014) Bioelectric signaling regulates size in zebrafish fins. PLoS Genet 10, e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferli C, Jaźwińska A (2015) The art of fin regeneration in zebrafish. Regeneration 2, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleo G, Brown CW, Laforest L, et al. (2001) Cell proliferation and movement during early fin regeneration in zebrafish. Dev Dyn 221, 380–390. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, et al. (2000) Roles of fgf signalling during zebrafish fin regeneration. Dev Biol 222, 347–358. [DOI] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, et al. (2002) Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A 99, 8713–8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallan LC (2014) Major issues in the origins of ray‐finned fish (Actinopterygii) biodiversity. Biol Rev 89, 950–971. [DOI] [PubMed] [Google Scholar]
- Santamaría JA, Marí‐Beffa M, Becerra J (1993) Regeneración de aletas de teleósteos, pp. 281–285. Interacción células‐matriz extracelular. Progresos en Biología Celular. Málaga: Universidad de Málaga. [Google Scholar]
- Santamaría JA, Marí‐Beffa M, Santos‐Ruiz L, et al. (1996) Incorporation of bromodeoxyuridine in regenerating fin tissue of the goldfish Carassius auratus . J Exp Zool 275, 300–307. [DOI] [PubMed] [Google Scholar]
- Santos‐Ruiz L, Santamaría JA, Ruiz‐Sánchez J, et al. (2002) Cell proliferation during blastema formation in the regenerating teleost fin. Dev Dyn 223, 262–272. [DOI] [PubMed] [Google Scholar]
- Sehring IM, Jahn C, Weidinger G (2016) Zebrafish fin and heart, what's special about regeneration? Curr Opin Genet Dev 40, 48–56. [DOI] [PubMed] [Google Scholar]
- Shibata E, Yokota Y, Horita N, et al. (2016) Fgf signalling controls diverse aspects of fin regeneration. Development 143, 2920–2929. [DOI] [PubMed] [Google Scholar]
- Sims KD Jr, Eble M, Iovine MK (2009) Connexin43 regulates joint location in zebrafish fins. Dev Biol 327, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Zhang J, Guay D, et al. (2008) Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole‐mount in situ hybridization method. Dev Dyn 237, 417–425. [DOI] [PubMed] [Google Scholar]
- Sousa S, Afonso N, Bensimon‐Brito A, et al. (2011) Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138, 3897–3905. [DOI] [PubMed] [Google Scholar]
- Stewart S, Stankunas K (2012) Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol 365, 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick‐Cooper CL, Weidinger G, Riehle KJ, et al. (2007) Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489. [DOI] [PubMed] [Google Scholar]
- Sun L, Tran N, Liang C, et al. (1999) Design, synthesis, and evaluations of substituted 3‐[(3‐ or 4‐Carboxyethylpyrrol‐2‐yl)methylidenyl]indolin‐2‐ones as inhibitors of VEGF, FGF, and PDGF receptor tyrosine kinases. J Med Chem 42, 5120–5130. [DOI] [PubMed] [Google Scholar]
- Sundelacruz S, Levin M, Kaplan DL (2009) Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev Rep 5, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorimbert V, König D, Marro J, et al. (2015) Bone morphogenetic protein signaling promotes morphogenesis of blood vessels, wound epidermis, and actinotrichia during fin regeneration in zebrafish. FASEB J 29, 4299–4312. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL (2011) Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 20, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrego D, Tomczak AP, Zahed F, et al. (2014) Potassium channels in cell cycle and cell proliferation. Philos Trans R Soc B 369, 20130094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Hamaguchi S, Sakaizumi M (2008) Development of diverse lateral line patterns on the teleost caudal fin. Dev Dyn 237, 2889–2902. [DOI] [PubMed] [Google Scholar]
- Watkins DN, Berman DM, Burkholder SG, et al. (2003) Hedgehog signalling within airway epithelial progenitors and in small‐cell lung cancer. Nature 422, 313–317. [DOI] [PubMed] [Google Scholar]
- Wehner D, Weidinger G (2016) Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet 31, 336–343. [DOI] [PubMed] [Google Scholar]
- Wehner D, Cizelsky W, Vasudevaro MD, et al. (2014) Wnt/β‐catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep 6, 467–481. [DOI] [PubMed] [Google Scholar]
- Westerfield M (1995) The Zebrafish Book. 3rd edn Eugene: University of Oregon Press. [Google Scholar]
- White JA, Boffa MB, Jones B, et al. (1994) A zebrafish retinoic acid receptor expressed in the regenerating caudal fin. Development 120, 1861–1872. [DOI] [PubMed] [Google Scholar]
- Whitehead PJP, Bauchot M‐L, Hureau J‐C, et al. (1986) Fishes of the North‐eastern Atlantic and the Mediterranean, vols. 1–3 Paris: Unesco. [Google Scholar]
- Whitehead GG, Makino S, Lien CL, et al. (2005) fgf20 is essential for initiating zebrafish fin regeneration. Science 310, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Xu C, Hasan SS, Schmidt I, et al. (2014) Arteries are formed by vein‐derived endothelial tip cells. Nat Commun 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari N, Ishida T, Kudo A, et al. (2009) Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev Biol 325, 71–81. [DOI] [PubMed] [Google Scholar]
- Zauner H, Begemann G, Marí‐Beffa M, et al. (2003) Differential regulation of msx genes in the development of the gonopodium, an intromittent organ, and of the ‘sword’, a sexually selected trait of swordtail fishes (Xiphophorus). Evol Dev 5, 466–477. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jeradi S, Strähle U, et al. (2012) Laser ablation of the sonic hedgehog‐a‐expressing cells during fin regeneration affects ray branching morphogenesis. Dev Biol 365, 424–433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Morphometry of inter‐rays of the regenerating caudal fin of Danio rerio.
Fig. S2. Anatomy and pigment distribution of the caudal fin of Carassius auratus.
Fig. S3. Melanophores from marginal inter‐rays migrate distally during inter‐ray wound healing.
Fig. S4. Endothelial cell and melanophore pattern and inter‐ray size during ray regeneration.
Fig. S5. Special inter‐ray phenotypes in zebrafish.
Fig. S6. Melanophore distribution during wound healing after one‐ray cut in an alf dty86d caudal fin.
Fig. S7. Pigment and blood vessel distribution and inter‐ray size during ray regeneration after three‐ray cuts in alf dty86d/+ caudal fins.
Table S1. Ray‐cut experiments.
Table S2. (A) Number and standard length statistics of cyclopamine‐treated fishes. (B) Number and standard length statistics of SU5402‐treated fishes.
Table S3. Number of species and fins showing morphological variations of inter‐rays.


