Abstract
Background
The genetic diversity in Australian populations of Xanthomonas species associated with bacterial leaf spot in tomato, capsicum and chilli were compared to worldwide bacterial populations. The aim of this study was to confirm the identities of these Australian Xanthomonas species and classify them in comparison to overseas isolates. Analysis of whole genome sequence allows for the investigation of bacterial population structure, pathogenicity and gene exchange, resulting in better management strategies and biosecurity.
Results
Phylogenetic analysis of the core genome alignments and SNP data grouped strains in distinct clades. Patterns observed in average nucleotide identity, pan genome structure, effector and carbohydrate active enzyme profiles reflected the whole genome phylogeny and highlight taxonomic issues in X. perforans and X. euvesicatoria. Circular sequences with similarity to previously characterised plasmids were identified, and plasmids of similar sizes were isolated. Potential false positive and false negative plasmid assemblies were discussed. Effector patterns that may influence virulence on host plant species were analysed in pathogenic and non-pathogenic xanthomonads.
Conclusions
The phylogeny presented here confirmed X. vesicatoria, X. arboricola, X. euvesicatoria and X. perforans and a clade of an uncharacterised Xanthomonas species shown to be genetically distinct from all other strains of this study. The taxonomic status of X. perforans and X. euvesicatoria as one species is discussed in relation to whole genome phylogeny and phenotypic traits. The patterns evident in enzyme and plasmid profiles indicate worldwide exchange of genetic material with the potential to introduce new virulence elements into local bacterial populations.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5600-x) contains supplementary material, which is available to authorized users.
Keywords: Cell wall degrading enzymes, CAZymes, Secretion system, Pan genome
Background
In recent years, whole genome sequences of a variety of bacterial plant pathogens have been used to investigate the phylogenetic relationships between species, as well as the genetic basis for pathogenicity and potential diagnostic target genes [1]. Next generation sequencing (NGS) and population genomics provides insight into many facets of host-pathogen interactions [2]. The wealth of information generated with NGS technology gives plant pathologists an opportunity to investigate pathogen movement, infection strategies and phenotypic trait association with the ultimate goal of providing targeted management strategies and better biosecurity. For example, genome sequence analysis of pathogenic and non-pathogenic Xanthomonas species on Prunus spp. resulted in a molecular diagnostic assay to differentiate pathogenic and non-pathogenic strains where previous tests did not [3]. Similar studies have examined the genetic diversity of Xanthomonas species that cause bacterial leaf spot (BLS) of tomato, capsicum and chilli worldwide [1], but not yet to Australian Xanthomonas strains associated with this disease.
Xanthomonas species reported to cause BLS in Australian tomato and pepper have been assigned to X. euvesicatoria, X. perforans and X. vesicatoria, with non-pathogenic strains of X. arboricola and Xanthomonas sp. also isolated [4]. A draft genome comparison of BLS-causing X. vesicatoria, X. euvesicatoria, X. gardneri and X. perforans provided the basis for many subsequent studies using genomic data. Insights into the virulence and pathogenicity of Xanthomonas has been provided by genomic studies that have revealed much about plasmids and secretion systems that deliver effectors and host cell wall degrading enzymes [1, 5]. Plasmid transferral via conjugation is a major mechanism of gene transfer throughout bacterial populations, accounting for rapid shifts in pathogen response to chemicals, antibiotics and host resistance genes [6–8]. Plasmids of BLS-causing Xanthomonas species vary in size and carry virulence and resistance genes [9, 10]. Gene cassettes and integrons are also responsible for genome diversity in Xanthomonas [11]. The characteristic structure and content of a number of Xanthomonas species has been described as an open pan genome that readily exchanges mobile elements within a population [12]. Other features of Xanthomonas genomes include products of bacterial secretion systems involved with host interactions such as effectors and carbohydrate active enzymes. Understanding these elements of the bacterial genome are key to understanding how genetics reflects species phylogeny and pathogenicity.
Plant defence responses to bacterial pathogens involve recognition of molecular patterns or proteins associated with bacterial secretion systems [13]. Pathogen associated molecular patterns are recognised by pattern receptors in the host that then triggers immunity. Proteins introduced by bacterial secretion systems, known as effectors may also induce immunity. Effectors of the type III secretion system (T3SS) were shown to be the main source of virulence in X. campestris pv. campestris [14], and integral to pathogenicity in Xanthomonas [13]. The T3SS introduces a complex of proteins to the host cell that target plant cell structures, alter the regulation of host genes or act as chaperones and delivery systems for the secreted effectors [15]. Effectors of plant pathogens are complex and diverse; some of the better studied include the TAL/ TALE (transcription activator-like) classes of effectors [16]. The Xop (Xanthomonas outer protein) effector classes and general effector nomenclature is described by White et al. [13], and are identified in strains of many Xanthomonas species. They note that the complex interactions between secreted proteins and host cells will likely be expanded and refined with additional genomic data. The need to understand the impact of effectors is demonstrated by the X. perforans host range expansion partially correlated with the loss of the effector AvrBsT [17]. Interestingly, AvrBsT has been described as a fitness factor [18], demonstrating that effectors may influence disease severity as well as host range. Other effectors have been linked to pathogenic function, such as AvrHah1 inducing a water soaking effect common in many bacterial diseases [19] by upregulating the intake of water into cells. Even as more genomes are sequenced every year, there is still much to be investigated about effector function [20].
In addition to the T3SS, the type II secretion system (T2SS) has also been described as important for pathogenicity in Xanthomonas species [21]. The type II secretion system is a common feature of many plant and animal pathogens as well as non-pathogenic species, involved in a range of infection and colonisation processes [22]. The T2SS is typically associated with the secretion of carbohydrate active enzymes (CAZymes), families of enzymes involved in carbohydrate processing pathways. Carbohydrate degradation has traditionally been used as a diagnostic trait in bacteriology [23], and have also been discussed in structural biology as therapeutic targets [24]. Determining which CAZyme families are present in bacterial strains may indicate substrate preference and pathogenicity. As they are currently classified, CAZymes are described by protein sequence as numbered families of six classes; glycoside hydrolases (GH), glycosyl transferases (GT), polysaccharide lyases (PL), carbohydrate esterases (CE), carbohydrate binding modules (CBM), and auxiliary activity families (AA) [25, 26]. The variety of secreted proteins in Xanthomonas and their impact on pathogenicity has been reviewed previously [1, 27], highlighting the potential for effector and CAZyme profiles to infer pathogenicity and bacterial growth strategies.
The analyses of genome structure and secretion system products contribute to the understanding of bacterial relatedness and function. By comparing genomes of Australian BLS-associated Xanthomonas strains we aim to improve our understanding of the taxonomic status of these species as well as incorporating Australian BLS-causing strains into the global understanding of this pathogen complex. These analyses will provide a foundation for further identification of targets for resistance breeding or future population genetics studies.
Results
Taxonomy and pathogenicity
Genome statistics of all 50 Australian Xanthomonas draft genomes are reported in Table 1. All draft genomes were approximately 5 Mbp in length, ranging from 4,806,110 bp to 5,379,097 bp and had a GC content ranging from 64.02 to 66.14% (average of 64.74%) which is consistent with reference genomes of the sequenced species [10, 27].
Table 1.
Draft genome statistics of 50 Australian Xanthomonas strains and reported genome statistics of public assemblies from Genbank
| Strain | Genbank ID b | Species c | Host | Region | Collection date | Contig number | Mb | CDS f | N50 | GC% | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRIPa 62410 | QFAP01 | xa | Tomato | Stanthorpe | 2015 | 36 | 4.9 | 4077 | 224,446 | 65.76 | this study |
| BRIP 62412 | QFAO01 | xa | Tomato | Stanthorpe | 2015 | 53 | 5.12 | 4295 | 403,341 | 65.65 | this study |
| BRIP 62414 | QFAN01 | xa | Tomato | Stanthorpe | 2015 | 45 | 5.14 | 4304 | 307,902 | 65.63 | this study |
| BRIP 62416 | QFAM01 | xa | Tomato | Stanthorpe | 2015 | 63 | 5.17 | 4266 | 455,337 | 65.66 | this study |
| BRIP 62432 | QFAQ01 | xa | Tomato | Tenterfield | 2015 | 49 | 4.95 | 4060 | 227,845 | 65.61 | this study |
| NCPPB 1630 | JPHE01 | xa pv ce d | Banana | New Zealand | 1960 | 121 | 4.98 | 4062 | 80,994 | 65.5 | Harrison et al. 2014 (Unpublished) |
| NCPPB 1832 | JPHC01 | xa pv ce | Banana | New Zealand | 1960 | 75 | 4.91 | 4044 | 172,772 | 65.6 | Harrison et al. 2016 |
| NCPPB 1447 | AJTL01 | xa pv j d | na e | na | na | 371 | 5.02 | 4257 | 26,883 | 65.4 | Noh et al. (Unpublished) |
| NCCB 100457 | APMC02 | xa pv co d | Hazel | USA | 2010 | 281 | 5.23 | 4346 | 35,479 | 65.5 | Ibarra Caballero et al. 2013 |
| xa3004 | AZQY01 | xa | Barley | Russia | 1998 | 132 | 4.76 | 3936 | 61,665 | 66 | Ignatov et al. 2015 |
| IVIA 2626.1 | LJGN01 | xa pv p d | Plum (Prunus salicina) | Spain | 2009 | 214 | 5.03 | 4268 | 47,650 | 65.4 | Garita-Cambronero et al. 2016 |
| CITA 44 | LJGM01 | xa | Cherry (Prunus mahaleb) | Spain | 2002 | 71 | 4.76 | 3951 | 120,981 | 65.8 | Garita-Cambronero et al. 2016 |
| J303 | LSGZ01 | xa pv j | Walnut | Chile | 2011 | 161 | 5.07 | 4227 | 53,673 | 65.5 | Santander et al. (Unpublished) |
| CITA 124 | LXKK01 | xa | Peach (Prunus persica) | Spain | 2012 | 128 | 4.75 | 4041 | 60,818 | 65.8 | Garita-Cambronero et al. (Unpublished) |
| CITA 14 | LXIB01 | xa | Peach (Prunus persica) | Spain | 2008 | 72 | 4.86 | 4001 | 100,946 | 65.6 | Garita-Cambronero et al. (Unpublished) |
| CFBP 2528 | JZEF01 | xa pv j | Walnut | New Zealand | 1956 | 19 | 5.08 | 4264 | 431,859 | 65.5 | Cesbron et al. (Unpublished) |
| CFBP 7634 | JZEH01 | xa | Walnut | France | 2002 | 5 | 4.93 | 4102 | 3,063,886 | 65.6 | Cesbron et al. (Unpublished) |
| CFBP 7179 | JZEG01 | xa pv j | Walnut | France | 2002 | 23 | 5.16 | 4358 | 479,113 | 65.4 | Cesbron et al. (Unpublished) |
| CFBP 7651 | JZEI01 | xa | Walnut | France | 2008 | 8 | 5.03 | 4188 | 2,811,770 | 65.5 | Cesbron et al. (Unpublished) |
| CFBP 3894 | LOMI01 | xa pv p | Plum (Prunus salicina) | New Zealand | 1953 | 78 | 5.06 | 4204 | 162,135 | 65.4 | Lopez-Soriano et al. (Unpublished) |
| MAFF301420 | BAVC01 | xa pv p | na | na | na | 396 | 5.002 | 4485 | 21,373 | 65.3 | Fujikawa et al. (Unpublished) |
| MAFF311562 | BAVB01 | xa pv p | na | na | na | 432 | 5.08 | 4630 | 23,374 | 65.3 | Fujikawa et al. (Unpublished) |
| MAFF301427 | BAVD01 | xa pv p | na | na | na | 356 | 4.9 | 4444 | 27,529 | 65.4 | Fujikawa et al. (Unpublished) |
| Xap33 | JHUQ01 | xa pv p | Almond | Spain | 2009 | 474 | 5.1 | 4388 | 34,438 | 65.4 | Garita-Cambronero et al. 2014 |
| BRIP 63464 | QEZX01 | xe | Capsicum | Bundaberg | 2015 | 140 | 5.34 | 4561 | 194,822 | 64.53 | this study |
| DAR 82542 | QEYY01 | xe | Capsicum | Bundaberg | 1981 | 426 | 5.12 | 4482 | 30,590 | 64.66 | this study |
| BRIP 62441 | QFAC01 | xe | Chilli | Bundaberg | 2015 | 135 | 5.34 | 4565 | 159,122 | 64.52 | this study |
| BRIP 62555 | QFAB01 | xe | Chilli | Bundaberg | 2015 | 139 | 5.34 | 4570 | 164,574 | 64.53 | this study |
| BRIP 62757 | QFAA01 | xe | Chilli | Bundaberg | 2015 | 149 | 5.33 | 4558 | 167,015 | 64.53 | this study |
| BRIP 62858 | QEZZ01 | xe | Chilli | Bundaberg | 2015 | 144 | 5.34 | 4554 | 146,014 | 64.53 | this study |
| BRIP 62959 | QEZY01 | xe | Chilli | Bundaberg | 2015 | 129 | 5.34 | 4560 | 169,175 | 64.53 | this study |
| BRIP 38997 | QEZV01 | xe | Chilli | Bundaberg | 1986 | 112 | 5.3 | 4513 | 163,565 | 64.55 | this study |
| BRIP 39016 | QEZW01 | xe | Apple of Peru (Nicandra physalodes) | Christmas creek | 1973 | 96 | 5.06 | 4290 | 171,827 | 64.6 | this study |
| DAR 34895 | QEYX01 | xe | Capsicum | Dural | 1981 | 69 | 5.23 | 4455 | 176,952 | 64.62 | this study |
| BRIP 62390 | QFAL01 | xe | Capsicum | Gatton | 2014 | 137 | 5.34 | 4550 | 153,551 | 64.53 | this study |
| BRIP 62391 | QFAK01 | xe | Capsicum | Gatton | 2014 | 131 | 5.34 | 4559 | 113,982 | 64.53 | this study |
| BRIP 62392 | QFAJ01 | xe | Capsicum | Gatton | 2014 | 183 | 5.34 | 4562 | 169,141 | 64.53 | this study |
| BRIP 62395 | QFAI01 | xe | Capsicum | Gatton | 2014 | 169 | 5.35 | 4574 | 179,351 | 64.53 | this study |
| BRIP 62396 | QFAH01 | xe | Chilli | Gatton | 2014 | 161 | 5.38 | 4604 | 102,920 | 64.5 | this study |
| BRIP 62400 | QFAG01 | xe | Chilli | Gatton | 2014 | 133 | 5.34 | 4548 | 194,822 | 64.53 | this study |
| BRIP 62403 | QFAF01 | xe | Chilli | Gatton | 2014 | 144 | 5.34 | 4557 | 135,453 | 64.53 | this study |
| BRIP 62438 | QFAD01 | xe | Capsicum | Hawkesbury Heights | 2015 | 99 | 5.12 | 4310 | 161,933 | 64.71 | this study |
| BRIP 62425 | QFAE01 | xe | Capsicum | Stanthorpe | 2015 | 163 | 5.34 | 4576 | 164,091 | 64.53 | this study |
| DAR 26930 | QEYW01 | xe | Tomato | Liverpool | 1976 | 101 | 5.05 | 4201 | 103,617 | 64.9 | this study |
| 66b | JSZG01 | xe | Capsicum (C. annuum cv. Kambi) | Bulgaria | 2012 | 384 | 5.31 | 4550 | 28,793 | 64.6 | Vancheva et al. 2015 |
| 83 M | JSZH01 | xe | Capsicum (C. annuum cv. Kurtovska kapija) | Macedonia | 2013 | 317 | 5.1 | 4330 | 31,385 | 64.7 | Vancheva et al. 2015 |
| LMG 27970 | JPYC01 | xe | pepper | Belgium | 1947 | 1491 | 5.04 | 4380 | 9186 | 64.7 | Constantin et al. 2015 |
| Xe329 | JZLQ01 | xe | Pepper | USA | 1997 | 203 | 5.09 | 4242 | 210,541 | 64.7 | Schwartz et al. 2015 |
| Xe206 | JZLN01 | xe | Pepper | USA | 1995 | 133 | 5.44 | 4648 | 133,864 | 64.5 | Schwartz et al. 2015 |
| Xe515 | JZLV01 | xe | Pepper | USA | 1999 | 118 | 5.32 | 4525 | 163,350 | 64.5 | Schwartz et al. 2015 |
| Xe526 | JZLW01 | xe | Pepper | USA | 2000 | 178 | 5.39 | 4598 | 176,627 | 64.5 | Schwartz et al. 2015 |
| Xe586 | JZLX01 | xe | Pepper | USA | 2003 | 120 | 5.31 | 4522 | 140,453 | 64.6 | Schwartz et al. 2015 |
| Xe679 | JZLZ01 | xe | Pepper | USA | 2008 | 109 | 5.14 | 4324 | 144,310 | 64.7 | Schwartz et al. 2015 |
| Xe681 | JZMA01 | xe | Pepper | USA | 2009 | 104 | 5.16 | 4360 | 210,555 | 64.7 | Schwartz et al. 2015 |
| Xe678 | JZLY01 | xe | Pepper | USA | 2008 | 202 | 5.42 | 4643 | 132,312 | 64.4 | Schwartz et al. 2015 |
| Xe259 | JZLO01 | xe | Pepper | USA | 1995 | 139 | 5.25 | 4423 | 187,535 | 64.6 | Schwartz et al. 2015 |
| Xe199 | JZLM01 | xe | Pepper | USA | 1994 | 110 | 5.27 | 4470 | 174,842 | 64.6 | Schwartz et al. 2015 |
| Xe315 | JZLP01 | xe | Pepper | USA | 1996 | 629 | 5.51 | 4638 | 164,442 | 64.3 | Schwartz et al. 2015 |
| Xe354 | JZLR01 | xe | Pepper | USA | 1998 | 287 | 5.43 | 4629 | 176,627 | 64.4 | Schwartz et al. 2015 |
| Xe376 | JZLS01 | xe | Pepper | USA | 1995 | 128 | 5.3 | 4500 | 176,635 | 64.5 | Schwartz et al. 2015 |
| Xe455 | JZLT01 | xe | Pepper | USA | 1998 | 137 | 5.33 | 4549 | 148,783 | 64.5 | Schwartz et al. 2015 |
| Xe490 | JZLU01 | xe | Pepper | USA | 1999 | 132 | 5.37 | 4587 | 187,439 | 64.5 | Schwartz et al. 2015 |
| Xe683 | JZRP01 | xe | Pepper | USA | 2010 | 108 | 5.32 | 4544 | 163,371 | 64.5 | Schwartz et al. (Unpublished) |
| Xe684 | JZRQ01 | xe | Pepper | USA | 2010 | 157 | 5.38 | 4594 | 148,783 | 64.5 | Schwartz et al. (Unpublished) |
| Xe685 | JZRR01 | xe | Pepper | USA | 2010 | 131 | 5.32 | 4536 | 148,770 | 64.5 | Schwartz et al. (Unpublished) |
| F4–2 | JZRU01 | xe | Pepper | USA | 2004 | 123 | 5.35 | 4591 | 159,113 | 64.5 | Schwartz et al. (Unpublished) |
| H3–2 | JZRW01 | xe | Pepper | USA | 2004 | 108 | 5.32 | 4531 | 163,306 | 64.5 | Schwartz et al. (Unpublished) |
| Xe689 | JZRS01 | xe | Pepper | USA | 2011 | 143 | 5.37 | 4588 | 163,336 | 64.5 | Schwartz et al. (Unpublished) |
| Xe695 | JZRT01 | xe | Pepper | USA | 2012 | 110 | 5.36 | 4588 | 163,310 | 64.5 | Schwartz et al. (Unpublished) |
| G4–1 | JZRV01 | xe | Pepper | USA | 2004 | 112 | 5.35 | 4586 | 163,306 | 64.5 | Schwartz et al. (Unpublished) |
| L3–2 | JZRX01 | xe | Pepper | USA | 2004 | 152 | 5.36 | 4584 | 159,114 | 64.5 | Schwartz et al. (Unpublished) |
| Xe181 | JZRY01 | xe | Pepper | USA | 1994 | 107 | 5.31 | 4518 | 163,975 | 64.5 | Schwartz et al. (Unpublished) |
| ATCC 19865 | AEQX01 | xg | Tomato | Yugoslavia | 1961 | 552 | 5.53 | 4786 | 22,686 | 63.7 | Potnis et al. 2011 |
| SM406–11 | JZJW01 | xg | Tomato | USA | 2011 | 461 | 5.26 | 4508 | 18,995 | 63.5 | Schwartz et al. 2015 |
| SM234–10 | JZJV01 | xg | Tomato | USA | 2010 | 250 | 5.33 | 4521 | 42,583 | 63.5 | Schwartz et al. 2015 |
| SM413–11 | JZJX01 | xg | Tomato | USA | 2011 | 313 | 5.31 | 4527 | 29,049 | 63.5 | Schwartz et al. 2015 |
| SM605–11 | JZJY01 | xg | Tomato | USA | 2011 | 242 | 5.34 | 4517 | 45,830 | 63.5 | Schwartz et al. 2015 |
| SM194–10 | JZJR01 | xg | Tomato | USA | 2010 | 309 | 5.39 | 4550 | 41,678 | 63.5 | Schwartz et al. 2015 |
| SM177–10 | JZJS01 | xg | Tomato | USA | 2010 | 245 | 5.31 | 4509 | 41,491 | 63.5 | Schwartz et al. 2015 |
| SM182–10 | JZJT01 | xg | Tomato | USA | 2010 | 251 | 5.3 | 4505 | 36,743 | 63.5 | Schwartz et al. 2015 |
| SM220–10 | JZJU01 | xg | Tomato | USA | 2010 | 260 | 5.29 | 4506 | 38,544 | 63.5 | Schwartz et al. 2015 |
| SM795–12 | JZKA01 | xg | Tomato | USA | 2012 | 293 | 5.13 | 4325 | 37,712 | 63.6 | Schwartz et al. 2015 |
| SM775–12 | JZJZ01 | xg | Tomato | USA | 2012 | 241 | 5.24 | 4420 | 45,190 | 63.6 | Schwartz et al. 2015 |
| BRIP 62383 | QEZU01 | xp | Tomato | Bowen | 2012 | 61 | 5.27 | 4462 | 302,799 | 64.68 | this study |
| BRIP 62384 | QEZT01 | xp | Tomato | Bowen | 2012 | 77 | 5.25 | 4450 | 312,509 | 64.66 | this study |
| BRIP 62398 | QEZP01 | xp | Tomato | Bowen | 2014 | 57 | 5.23 | 4410 | 187,438 | 64.67 | this study |
| BRIP 62404 | QEZO01 | xp | Tomato | Bowen | 2014 | 109 | 5.25 | 4446 | 140,260 | 64.66 | this study |
| BRIP 62397 | QEZQ01 | xp | Tomato | Brisbane | 2014 | 45 | 4.97 | 4178 | 288,612 | 64.99 | this study |
| BRIP 62386 | QEZS01 | xp | Tomato | Bundaberg | 2012 | 80 | 5.29 | 4484 | 187,311 | 64.67 | this study |
| BRIP 62405 | QEZN01 | xp | Tomato | Bundaberg | 2014 | 77 | 5.28 | 4478 | 293,980 | 64.67 | this study |
| BRIP 63262 | QEZM01 | xp | Tomato | Bundaberg | 2015 | 100 | 5.33 | 4545 | 372,915 | 64.65 | this study |
| BRIP 63565 | QEZL01 | xp | Tomato | Bundaberg | 2015 | 73 | 5.23 | 4421 | 312,487 | 64.68 | this study |
| BRIP 63666 | QEZK01 | xp | Tomato | Bundaberg | 2015 | 59 | 5.28 | 4473 | 312,512 | 64.68 | this study |
| BRIP 62389 | QEZR01 | xp | Tomato | South Turramurra | 2013 | 70 | 5.27 | 4466 | 225,394 | 64.68 | this study |
| Xp91–118 | CP019725.1 | xp | Tomato | USA | 2004 | 1 | 4.89 | 4450 | 4,898,349 | 65 | Potnis et al. 2011 |
| Xp5–6 | JZVA01 | xp | Tomato | USA | 2006 | 70 | 5.3 | 4501 | 202,262 | 64.6 | Schwartz et al. 2015 |
| Xp7–12 | JZVB01 | xp | Tomato | USA | 2006 | 65 | 5.12 | 4343 | 207,802 | 64.9 | Schwartz et al. 2015 |
| Xp9–5 | JZVD01 | xp | Tomato | USA | 2006 | 113 | 5.38 | 4624 | 193,597 | 64.6 | Schwartz et al. 2015 |
| Xp10–13 | JZVE01 | xp | Tomato | USA | 2006 | 78 | 5.17 | 4410 | 158,830 | 64.8 | Schwartz et al. 2015 |
| Xp11–2 | JZVF01 | xp | Tomato | USA | 2006 | 50 | 5.35 | 4568 | 302,422 | 64.6 | Schwartz et al. 2015 |
| Xp15–11 | JZVG01 | xp | Tomato | USA | 2006 | 63 | 5.31 | 4520 | 236,137 | 64.6 | Schwartz et al. 2015 |
| GEV839 | JZVK01 | xp | Tomato | USA | 2012 | 109 | 5.44 | 4669 | 107,802 | 64.4 | Schwartz et al. 2015 |
| Xp3–15 | JZUY01 | xp | Tomato | USA | 2006 | 93 | 5.46 | 4692 | 226,886 | 64.4 | Schwartz et al. 2015 |
| Xp8–16 | JZVC01 | xp | Tomato | USA | 2006 | 62 | 5.26 | 4469 | 241,328 | 64.6 | Schwartz et al. 2015 |
| Xp4–20 | JZUZ01 | xp | Tomato | USA | 2006 | 71 | 5.29 | 4515 | 250,796 | 64.7 | Schwartz et al. 2015 |
| Xp17–12 | JZVH01 | xp | Tomato | USA | 2006 | 84 | 5.2 | 4387 | 201,083 | 64.7 | Schwartz et al. 2015 |
| Xp18–15 | JZVI01 | xp | Tomato | USA | 2006 | 78 | 5.35 | 4568 | 178,519 | 64.6 | Schwartz et al. 2015 |
| GEV909 | JZVO01 | xp | Tomato | USA | 2012 | 71 | 5.14 | 4388 | 128,792 | 64.9 | Schwartz et al. 2015 |
| GEV940 | JZVS01 | xp | Tomato | USA | 2012 | 98 | 5.14 | 4384 | 118,066 | 64.9 | Schwartz et al. 2015 |
| GEV968 | JZVT01 | xp | Tomato | USA | 2012 | 96 | 5.14 | 4392 | 114,913 | 64.9 | Schwartz et al. 2015 |
| GEV1001 | JZVV01 | xp | Tomato | USA | 2012 | 78 | 5.18 | 4402 | 123,606 | 64.8 | Schwartz et al. 2015 |
| GEV872 | JZVL01 | xp | Tomato | USA | 2012 | 80 | 5.14 | 4401 | 143,709 | 64.9 | Schwartz et al. 2015 |
| GEV893 | JZVM01 | xp | Tomato | USA | 2012 | 116 | 5.13 | 4387 | 105,783 | 64.9 | Schwartz et al. 2015 |
| Xp4B | JZUX01 | xp | Tomato | USA | 1998 | 214 | 5.22 | 4469 | 73,796 | 64.6 | Schwartz et al. 2015 |
| GEV904 | JZVN01 | xp | Tomato | USA | 2012 | 134 | 5.14 | 4405 | 77,145 | 64.9 | Schwartz et al. 2015 |
| GEV936 | JZVR01 | xp | Tomato | USA | 2012 | 104 | 5.14 | 4390 | 111,234 | 64.9 | Schwartz et al. 2015 |
| GEV915 | JZVP01 | xp | Tomato | USA | 2012 | 70 | 5.12 | 4356 | 180,795 | 64.9 | Schwartz et al. 2015 |
| GEV917 | JZVQ01 | xp | Tomato | USA | 2012 | 122 | 5.14 | 4390 | 85,814 | 64.9 | Schwartz et al. 2015 |
| GEV1054 | JZVY01 | xp | Tomato | USA | 2012 | 85 | 5.23 | 4450 | 128,850 | 64.7 | Schwartz et al. 2015 |
| GEV1044 | JZVX01 | xp | Tomato | USA | 2012 | 82 | 5.24 | 4456 | 128,850 | 64.7 | Schwartz et al. 2015 |
| TB6 | JZWA01 | xp | Tomato | USA | 2013 | 148 | 5.27 | 4473 | 249,903 | 64.6 | Schwartz et al. 2015 |
| TB9 | JZWB01 | xp | Tomato | USA | 2013 | 268 | 5.32 | 4483 | 236,007 | 64.6 | Schwartz et al. 2015 |
| GEV993 | JZVU01 | xp | Tomato | USA | 2012 | 69 | 5.15 | 4385 | 176,938 | 64.9 | Schwartz et al. 2015 |
| GEV1026 | JZVW01 | xp | Tomato | USA | 2012 | 70 | 5.15 | 4373 | 158,062 | 64.9 | Schwartz et al. 2015 |
| GEV1063 | JZVZ01 | xp | Tomato | USA | 2012 | 93 | 5.16 | 4384 | 128,850 | 64.8 | Schwartz et al. 2015 |
| TB15 | JZWC01 | xp | Tomato | USA | 2013 | 261 | 5.33 | 4505 | 233,353 | 64.6 | Schwartz et al. 2015 |
| Xp2010 | JZVJ01 | xp | Tomato | USA | 2010 | 13 | 5.25 | 4575 | 1,113,511 | 64.6 | Schwartz et al. 2015 |
| 4P1S2 | JRWW01 | xp | Tomato | Sicily | 2011 | 442 | 5.51 | 5216 | 41,480 | 64.7 | Torelli et al. (Unpublished) |
| DAR 33341 | QEYV01 | x sp | Capsicum | Peats Ridge | 1979 | 57 | 5.18 | 4387 | 199,700 | 64.66 | this study |
| BRIP 62409 | QEZJ01 | x sp | Tomato | Stanthorpe | 2015 | 37 | 4.92 | 4033 | 210,840 | 65.92 | this study |
| BRIP 62411 | QEZI01 | x sp | Tomato | Stanthorpe | 2015 | 46 | 4.81 | 4002 | 334,519 | 66.14 | this study |
| BRIP 62415 | QEZH01 | x sp | Tomato | Stanthorpe | 2015 | 49 | 4.85 | 3983 | 325,857 | 66.06 | this study |
| BRIP 62418 | QEZG01 | x sp | Tomato | Stanthorpe | 2015 | 67 | 5.09 | 4274 | 193,024 | 65.8 | this study |
| BRIP 38864 | QEZA01 | xv | Wild tomato | Bowen | 1982 | 69 | 5.22 | 4439 | 377,679 | 64.23 | this study |
| DAR 26931 | QEYU01 | xv | Capsicum | Burrundulla | 1977 | 46 | 5.18 | 4386 | 243,089 | 64.25 | this study |
| DAR 26933 | QEYT01 | xv | Capsicum | Redland bay | 1977 | 1851 | 4.93 | 4899 | 4596 | 64.09 | this study |
| BRIP 62388 | QEZD01 | xv | Tomato | South Turramurra | 2012 | 85 | 5.31 | 4488 | 142,449 | 64.02 | this study |
| BRIP 62413 | QEZF01 | xv | Tomato | Stanthorpe | 2015 | 56 | 5.37 | 4574 | 377,588 | 64.12 | this study |
| BRIP 62423 | QEZE01 | xv | Tomato | Stanthorpe | 2015 | 69 | 5.37 | 4582 | 467,775 | 64.12 | this study |
| BRIP 62428 | QEZC01 | xv | Tomato | Tenterfield | 2015 | 66 | 5.36 | 4575 | 298,833 | 64.12 | this study |
| BRIP 62429 | QEZB01 | xv | Tomato | Tenterfield | 2015 | 60 | 5.36 | 4567 | 198,567 | 64.13 | this study |
| BRIP 38861 | QEYZ01 | xv | Tomato | Victoria Point | 1981 | 46 | 5.23 | 4442 | 377,697 | 64.22 | this study |
| ATCC 35937 | AEQV01 | xv | Tomato | New Zealand | 1955 | 296 | 5.53 | 4768 | 41,380 | 64.1 | Potnis et al. 2011 |
| 53 M | JSYJ01 | xv | Capsicum annuum cv. Kurtovska kapija | Macedonia | 2005 | 344 | 5.29 | 4526 | 29,340 | 64 | Vancheva et al. 2015 |
| 15b | JSXZ01 | xv | Capsicum annum | Bulgaria | 2005 | 338 | 5.29 | 4517 | 31,449 | 64 | Vancheva et al. 2015 |
| LMG 919 | JTEF01 | xv | Tomato | Zimbabwe | na | 420 | 5.12 | 4391 | 21,731 | 64.3 | Lefeuvre et al. (Unpublished) |
| LMG 920 | JTEG01 | xv | Tomato | Italy | 1956 | 326 | 5.16 | 4430 | 28,850 | 64.2 | Lefeuvre et al. (Unpublished) |
aBrisbane pathology herbarium ID
bWGS project number or Genbank ID where complete genome is available
cxa X. arboricola, xe X. euvesicatoria, xp X. perforans, xv X. vesicatoria, x sp Xanthomonas sp.
dxa pv ce: X. arboricola pv. celebensis, xa pv j: X. arboricola pv. juglandis, xa pv co: X. arboricola pv. corylina, xa pv p: X. arboricola pv. pruni
eData not available
fCDS as determined by the annotation of this study for consistency
Included statistics are contig number, length in Mbp, CDS, N50 and GC content. All reported CDS numbers were generated in this study. The metadata for Australian strains was sourced from Roach et al. 2017
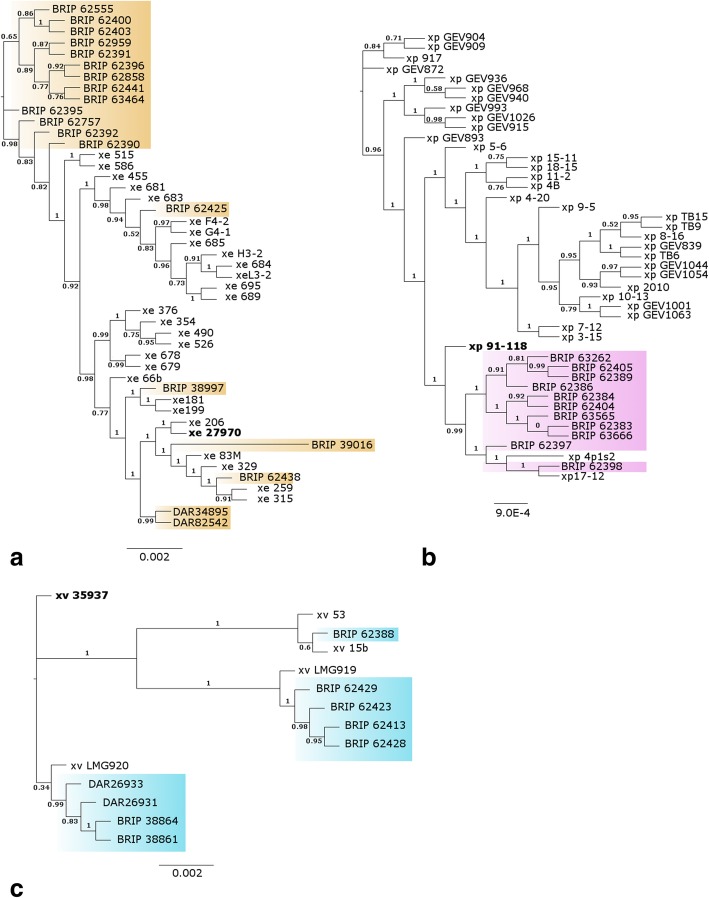
The SNP-based phylogenetic tree arranges most strains in this study into distinct clades (Additional file 1: Figure S1). The X. euvesicatoria and X. perforans clades grouped distinctly from the X. vesicatoria, X. gardneri and X. arboricola clades. The X. vesicatoria and X. arboricola clades contain three and four distinct subclades respectively (branch support values of 1). Four strains from tomato in Stanthorpe (BRIP 62409, 62411, 62415, and 62418, designated the uncharacterised Xanthomonas sp. clade) resolved in a clade distinct from its closest relative X. arboricola. The core genome phylogenies of individual BLS-causing species (excluding X. gardneri, X. arboricola and the uncharacterised Xanthomonas clade) clustered Australian strains in clades with multiple strains from other countries (Fig. 1). Australian strains of X. perforans cluster with overseas isolates xp 91–118, xp 4p1s2 and xp 17–12. Nine Australian X. euvesicatoria strains cluster in a clade by themselves, with the other ten distributed across clades with overseas strains. Australian X. vesicatoria strains cluster in three clades with overseas strains, distant from the type strain ATCC 35937.
Fig. 1.
Phylogeny of Australian and Genbank genomes of a) X. euvesicatoria, b) X. perforans and c) X. vesicatoria based on core genome alignments generated by the Roary program. Australian strains are indicated by BRIP and DAR collection prefixes and highlighted; all others are public genomes of related species. Type strains are indicated in bold and branch support values are displayed to clade level (measured with the Shimodaira-Hasegawa test within FastTree). Branch length is indicated by the scale bar
All X. euvesicatoria strains apart from BRIP 39016 tested were pathogenic on both capsicum [4] and tomato, where BRIP 39016 was determined to be non-pathogenic on both hosts. Strains of X. vesicatoria and X. perforans were pathogenic on tomato as determined previously [4], and non-pathogenic on capsicum. Strains of the uncharacterised Xanthomonas sp. were non-pathogenic on both hosts, and strains of X. arboricola were designated non-pathogenic on tomato [4] and capsicum. Pathogenicity of X. euvesicatoria on tomato was observed as small, dark lesions with yellow halo that displayed bacterial streaming. Isolations resulted in yellow, gram negative colonies.
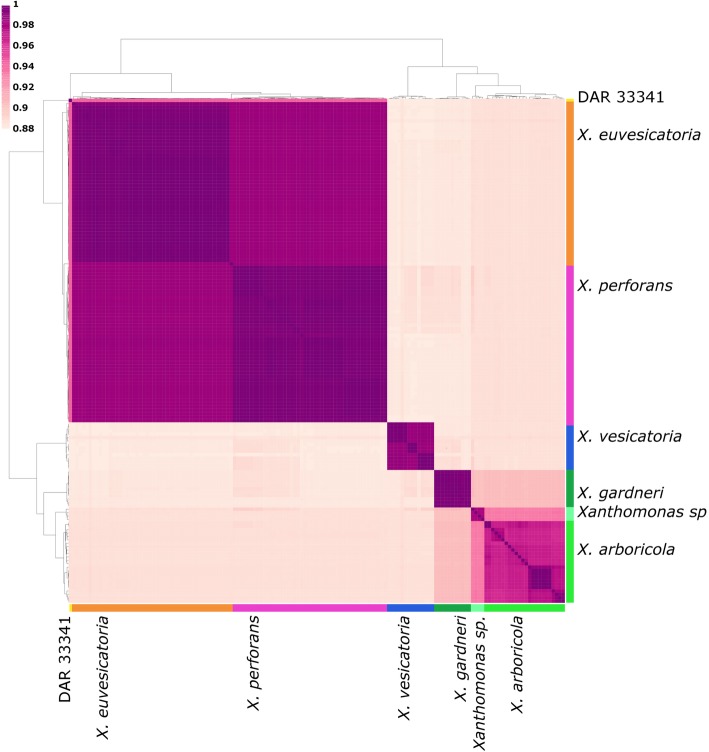
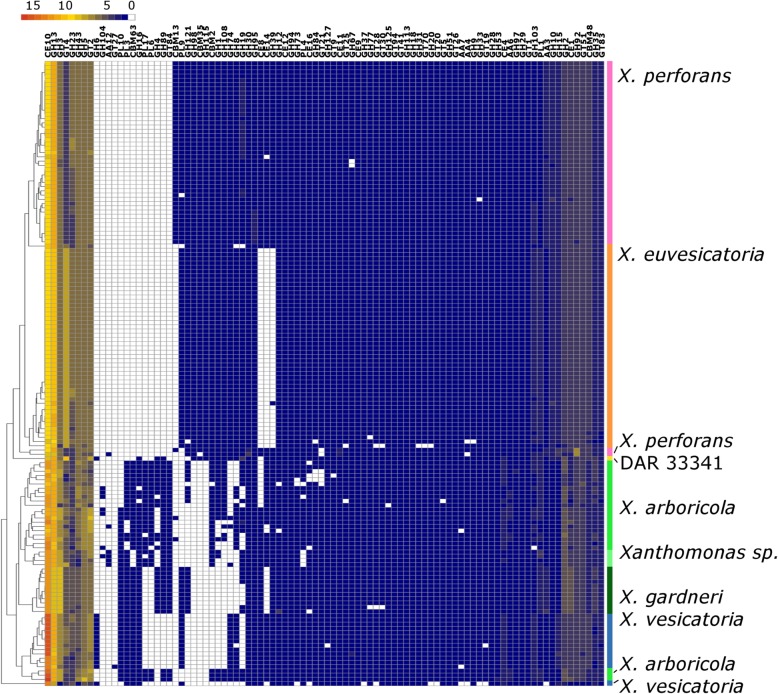
Average nucleotide identity (ANI) of core genome sequence analysed in this study are presented in Fig. 2. An ANI of 93% supports the separation of X. arboricola and the four uncharacterised Xanthomonas sp. strains into two separate species. ANI values of > 98% showed the genetic similarity of X. perforans and X. euvesicatoria while also displaying conserved separation. ANI analysis indicates that BRIP 39016 and DAR 26930 are also strains of X. euvesicatoria (ANI > 98%). Strain DAR 33341 has an ANI < 95% to all other strains in the analysis and 94% to X. euvesicatoria and was therefore excluded from X. euvesicatoria.
Fig. 2.
A heat map and dendrogram of average nucleotide identity (ANI) between 147 Xanthomonas genomes. The coloured bars represent the species as indicated in the SNP phylogeny and supported ANI values shown here. Xanthomonas perforans strains of X. euvesicatoria are indicated separately to highlight ANI differences. ANI is depicted as the colour gradient indicated by the legend: darker = 1 (100% ANI), lighter = 0.88 (88% ANI)
Pan genome composition
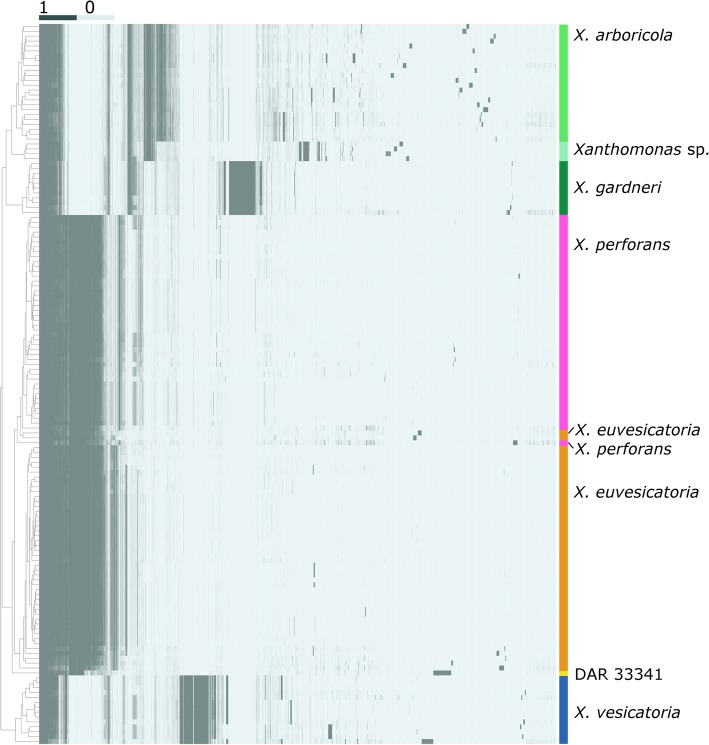
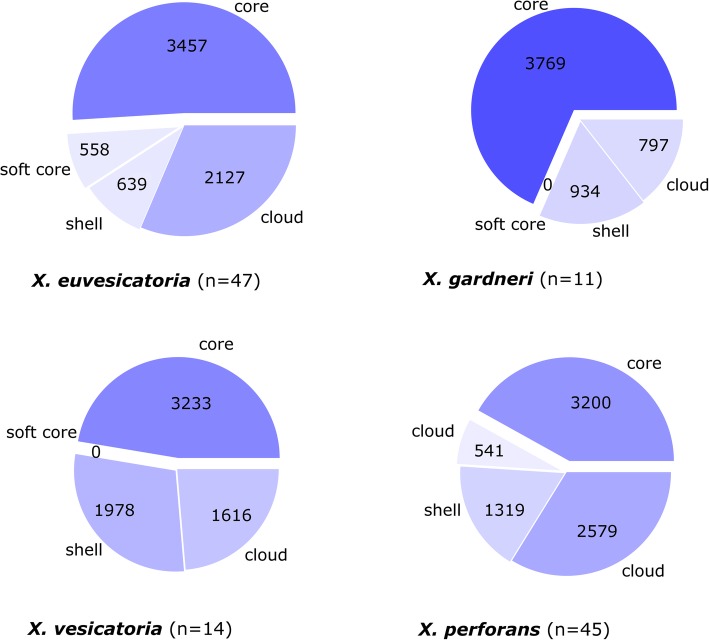
The nucleotide homologue cluster matrix grouped all strains (Fig. 3) in a generally similar topology to the phylogeny while also highlighting distinct differences between species. All species contained unique homologues (280 in X. arboricola, 70 in X. euvesicatoria, 69 in X. perforans, 416 in the combined X. euvesicatoria and X. perforans clades, 1639 in X. gardneri, 1646 in X. vesicatoria, and 609 in the uncharacterised Xanthomonas sp. clade). Pan genome pie charts (Fig. 4) based on the homologue matrix (Fig. 3) describe the core, soft core, shell and cloud genome content of each species. Gene discovery plots for each species (Additional file 2: Figure S2) showed that the number of new genes approached zero as genome number increased.
Fig. 3.
Cluster matrix of 147 Xanthomonas genomes with dendrogram based on homologue presence (dark) and absence (light). Species groupings are indicated with coloured bars as determined by phylogeny and ANI. Xanthomonas perforans strains of X. euvesicatoria are indicated separately to highlight homologue differences. The four Australian strains most closely related to X. arboricola are designated in the text as an uncharacterised Xanthomonas species
Fig. 4.
Pie plots of gene content in core, soft core, shell and cloud genomes describing the pan genome for X. euvesicatoria, X. perforans, X. vesicatoria and X. gardneri. The core genome is defined as genes present in 99–100% of strains; soft core, shell and cloud genomes are defined as 95–99%, 15–95% and 0–15% respectively. Number of genomes in each pan genome is indicated as ‘n’
Predicted and isolated plasmids and predicted effector content
Contigs originating from plasmids were assembled for 48 of 50 sequenced Australian strains, resulting in a total of 61 plasmids reconstructed for 41 strains (Table 2). A 31 kb plasmid (13 plasmids of 31,328 bp and two of 31,318 bp) was present in 15 of the X. euvesicatoria strains studied, although it was not found in any X. perforans strains. A 150 kb plasmid (six plasmids of 159,114 bp and one of 159,115 bp) was present in seven X. euvesicatoria strains and none of the X. perforans strains. An 88 kb plasmid (3 plasmids of 88,047 bp, 88,057 bp, 88,063 bp) was present in three of the X. vesicatoria strains. A 47 kb plasmid (three plasmids of 47,218 bp, one of 47,214 bp) was present in four strains of X. vesicatoria. The remaining plasmids were not unique to individual species. A 17 kb plasmid (two of 17,360 kb, one of 17,359 kb, 17,382 kb, and 17,377 kb) was present in four X. euvesicatoria strains and one X. arboricola strain. Plasmids of 40–41 kb were present in two X. euvesicatoria strains, one X. perforans strain and two X. vesicatoria strains. A single X. euvesicatoria strain contained two plasmids of 28,462 kb and 32,900 kb, while a strain of the uncharacterised Xanthomonas sp. contained a plasmid of 28,836 kb. The 19 plasmids less than 10 kb in size were found in five X. euvesicatoria strains, nine strains of X. perforans, one strain of the uncharacterised Xanthomonas clade, and DAR 33341. Protein sequence with homology to established effectors were detected in reconstructed plasmid sequence. Homologues of AvrBsT were detected in each of seven strains of X. vesicatoria (four 47 kb plasmids and three 88 kb plasmids); AvrBs3 in two strains of X. euvesicatoria on 40 kb plasmids and XopH in an X. perforans strain on a 41 kb plasmid.
Table 2.
Reconstructed circular sequence assembled for Australian Xanthomonas draft genomes from whole genome sequence data
| Strain | Species a | Plasmid number b | Plasmid .0 c | Plasmid .1 | Plasmid .2 | Plasmid .3 |
|---|---|---|---|---|---|---|
| BRIP 62410 | xa | 0 | ||||
| BRIP 62412 | xa | 0 | ||||
| BRIP 62414 | xa | 0 | ||||
| BRIP 62416 | xa | 1 | 17,377 | |||
| BRIP 62432 | xe | 1 | 17,382 | |||
| BRIP 62441 | xe | 1 | 31,328 | |||
| BRIP 62555 | xe | 1 | 31,328 | |||
| BRIP 62757 | xe | 2 | 159,114 | 31,328 | ||
| BRIP 62858 | xe | 3 | 8247 | 159,114 | 31,328 | |
| BRIP 62959 | xe | 1 | 159,115 | |||
| BRIP 38997 | xe | 2 | 17,360 | ***40,439 | ||
| BRIP 63464 | xe | 2 | 159,114 | 31,328 | ||
| BRIP 39016 | xe | 2 | 32,900 | 28,462 | ||
| BRIP 62396 | xe | 2 | 31,328 | 4316 | ||
| BRIP 62400 | xe | 1 | 31,328 | |||
| BRIP 62403 | xe | 2 | 31,328 | 159,114 | ||
| BRIP 62390 | xe | 2 | 159,114 | 31,328 | ||
| BRIP 62391 | xe | 2 | 8247 | 31,328 | ||
| BRIP 62392 | xe | 1 | 31,328 | |||
| BRIP 62395 | xe | 1 | 31,328 | |||
| BRIP 62438 | xe | 0 | ||||
| BRIP 62425 | xe | 3 | 31,328 | 159,114 | 17,360 | |
| DAR 34895 | xe | 1 | 31,318 | |||
| DAR 82542 | xe | 4 | 2587 | 17,350 | ***40,429 | 31,318 |
| DAR 26930 | xe | 1 | 8201 | |||
| BRIP 62383 | xp | 1 | 1418 | |||
| BRIP 62384 | xp | 1 | 2808 | |||
| BRIP 62398 | xp | 1 | 7812 | |||
| BRIP 62404 | xp | 1 | 2808 | |||
| BRIP 62397 | xp | 1 | **41,842 | |||
| BRIP 62386 | xp | 1 | 2808 | |||
| BRIP 62405 | xp | 2 | 2589 | 5031 | ||
| BRIP 63262 | xp | 2 | 2589 | 5031 | ||
| BRIP 63565 | xp | 1 | 2808 | |||
| BRIP 63666 | xp | 1 | 1418 | |||
| BRIP 62389 | xp | 0 | ||||
| BRIP 62409 | x sp | 0 | ||||
| BRIP 62411 | x sp | 1 | 28,836 | |||
| BRIP 62415 | x sp | 0 | ||||
| BRIP 62418 | x sp | 1 | 1418 | |||
| DAR 33341 | x sp | 2 | 7154 | 4921 | ||
| BRIP 38864 | xv | 2 | *88057 | 41,641 | ||
| BRIP 62388 | xv | 0 | ||||
| BRIP 62413 | xv | 1 | *47218 | |||
| BRIP 62423 | xv | 1 | *47218 | |||
| BRIP 62428 | xv | 1 | *47218 | |||
| BRIP 62429 | xv | 1 | *47214 | |||
| BRIP 38861 | xv | 2 | *88063 | 41,641 | ||
| DAR 26931 | xv | 1 | *88047 | |||
| DAR 26933 | xv | 0 |
aspecies as determined by whole genome SNP phylogeny and ANI
bcircular sequences assembled by recycler for each strain
ccircular sequence ID is indicated by decimal value after strain ID
Length is described in base pairs. * indicates the predicted plasmid contains AvrBsT, ** XopH, ***AvrBs3
Predicted plasmids were investigated by isolating and visualising plasmid DNA of strains BRIP 38864, BRIP 62858, BRIP 62416, BRIP 62423, BRIP 62388, BRIP 62397, BRIP 63464, BRIP 38997 and the extraction control DC 3000. (Additional file 3: Figure S3). Bands approximately of the size predicted were observed in strains BRIP 38864, BRIP 62858, BRIP 62423, BRIP 62397, BRIP 63464 and DC 3000. Where no plasmids were predicted for strain BRIP 62388, bands similar to those of other extractions were observed. For strains BRIP 62416, no plasmids were recovered despite the prediction of 17 kbp circular sequence. BRIP 62423, BRIP 62397, BRIP 38997, and BRIP 62858 may have additional large bands that could not be separated effectively. Sizing is only approximate due to the possibility of multiple plasmid structures (nicked circular sequence, linear sequence, supercoiled plasmids) migrating through the gel at different rates.
Effector and CAZyme content
The effector profiles of the dataset (Fig. 5) grouped species in the same general topology as the whole genome SNP phylogeny (Additional file 1: Figure S1). Effectors that were core to each species and the entire dataset, as well as effectors discussed in other studies, are listed in supplementary material (Additional file 4: Table S1). The occurrence of important effectors identified in previous studies [1, 17] are also listed here. Retained as core to BLS-causing species (X. euvesicatoria, X. perforans, X. vesicatoria and X. gardneri) as listed previously (AvrBs2, XopR, XopX, XopZ1, XopAD, XopN, XopF1, XopK, XopL, XopQ, XopD) [1] are AvrBs2, XopR, XopX, XopZ1 and XopAD. Several of these effectors previously considered core were detected in all but a few strains of certain species; XopN was absent in one X. vesicatoria and one X. gardneri strain, XopF1 was absent in one X. gardneri strain, XopK was absent in one X. euvesicatoria strain, and XopL was absent in four X. gardneri strains. XopQ was absent in all X. vesicatoria strains and XopD was absent in eight X. vesicatoria strains.
Fig. 5.
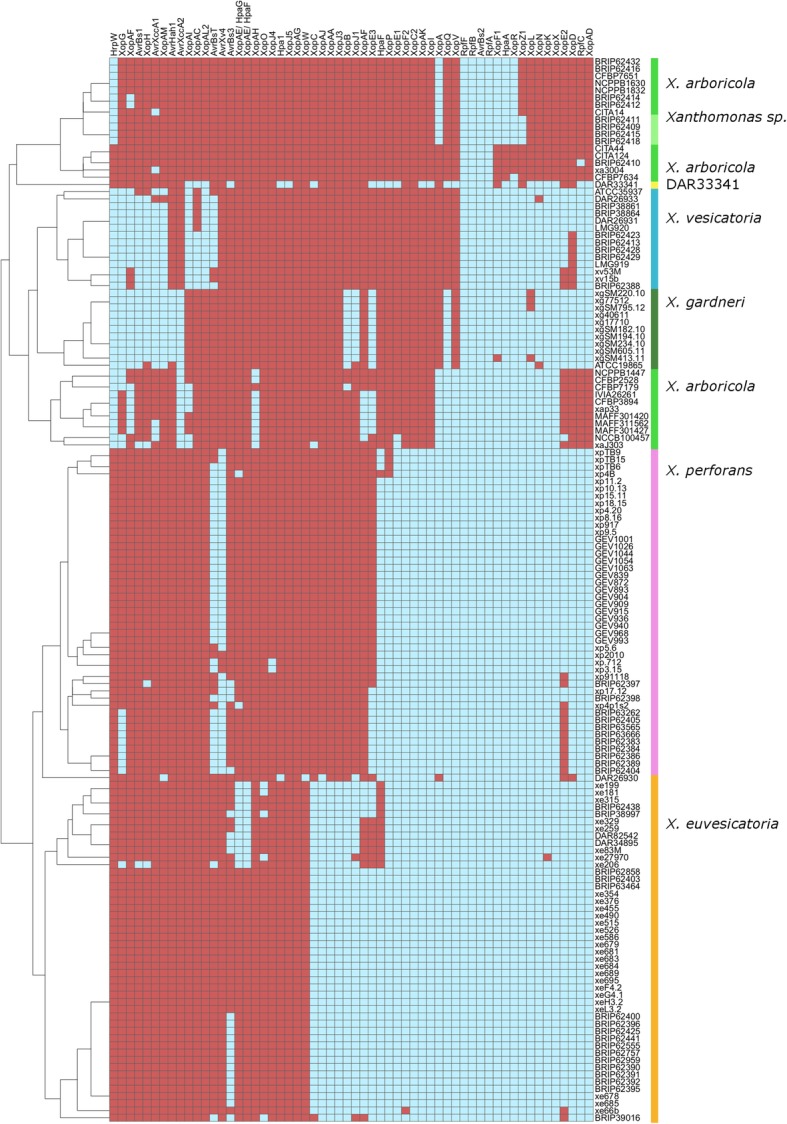
Presence/ absence matrix with dendrogram of effectors identified in 147 Xanthomonas genomes. Effector presence is indicated by colour as described in the legend (presence = blue, absence = red). Names and Genbank numbers of identified effectors are listed vertically. Species groupings as determined by phylogeny and ANI are indicated by the horizontal coloured bar as follows: X. euvesicatoria; orange, X. perforans; pink, X. vesicatoria; blue, X. gardneri; dark green, X. arboricola; green, Xanthomonas sp.; light green
The dendrogram based on CAZyme family data (Fig. 6) grouped species distinctly in the same general topology as seen in the genome SNP phylogeny (Additional file 1: Figure S1). In contrast to the SNP phylogeny, the CAZyme dendrogram clusters some X. arboricola and X. perforans strains outside of their group. A total of 92 carbohydrate active and facilitator enzyme families were identified, revealing groups present or absent in certain species and clades (Fig. 6, Additional file 5: Table S2). CAZyme families of cell wall degrading enzyme genes identified in BLS-causing Xanthomonas by Potnis et al. [27] were also identified in this dataset (Additional file 5: Table S2).
Fig. 6.
Cluster matrix and dendrogram based on number of CAZyme families identified in 147 Xanthomonas genomes. Number of CAZyme families present in each strain is indicated by the red-blue scale of the figure legend (17 families = red, 1 = blue, 0 = white). CAZyme families are listed vertically. Horizontal coloured bars represent species as indicated by SNP phylogeny and ANI. Xanthomonas perforans strains of X. euvesicatoria are indicated separately to highlight differences in cazyme profile
Discussion
Genomic analysis of BLS-associated Xanthomonas strains revealed diverse groups with distinguishing features that will have implications for future pathogenicity and taxonomic studies. Phylogenetic analysis (SNP, core genome, ANI) supports the close relationship between X. euvesicatoria and X. perforans. An uncharacterised Xanthomonas species (BRIP 62409, BRIP 62411, BRIP 62415 and BRIP 62418) was demonstrated to be distinct from closely related strains of X. arboricola according to the SNP phylogeny and ANI. The effector and CAZyme profiles of species that differ in pathogenicity displayed clear differences that may reflect differences in epidemiology and niche survival.
Taxonomic status of BLS-causing Xanthomonas species
The phylogenies and homologue matrix generated in this study support the current taxonomic status of X. euvesicatoria, X. perforans and X. vesicatoria [28], also confirming previous findings [4] that X. gardneri was not detected in Australian strains of this study. A recent study based on whole genome ANI determined that X. euvesicatoria and X. perforans were not genetically distinct enough to be considered separate species [29]. Our study found an ANI of 98.6% between strains of X. euvesicatoria and X. perforans, supporting these findings. While genetically very similar, strains of X. perforans were still clearly distinguished from X. euvesicatoria and other species in the phylogenetic analyses and the analyses of secretion systems. This may reflect differences in phenotype and pathogenicity, as Australian strains of X. euvesicatoria and X. perforans are generally isolated from capsicum and tomato respectively [4]. This example of genetically similar species being reliably differentiated by other measures is important to consider in the ongoing debate of how to classify bacteria, as a name could reflect phylogenetic groupings or phenotypic (and pathogenic) differences.
The core genome phylogenies of the individual BLS-causing species X. euvesicatoria, X. perforans and X. vesicatoria cluster Australian strains in multiple clades with strains from overseas (Fig. 1). Australian X. vesicatoria strains are similar to strains from Italy (LMG 920), Zimbabwe (LMG 919), Macedonia (53 M) and Bulgaria (15b). Australian strains of X. perforans generally resolved in a clade of their own, closely related to the type strain. A subclade clustered BRIP 62398 and BRIP 62397 with xp 4p1s2 and xp 17–12, two strains from Sicily and the USA respectively (Table 1). Australian strains of X. euvesicatoria generally clustered in their own clade, with strains BRIP 62425, BRIP 38997, BRIP 39016, BRIP 62438, DAR34895 and DAR82542 dispersed throughout the phylogeny with overseas strains, generally from the USA. The presence of Australian and overseas strains together in different clades may represent direct introductions of pathogens or the general distribution of the species across the world over time.
Pan genome of collected Xanthomonas species
The homologue matrix of strains in this study reflected the whole genome SNP phylogeny, while also highlighting blocks of unique and shared regions containing hundreds of genes that may be relevant to host specificity, virulence, other phenotypic traits and niche adaptations. This matrix shows that the genomes of X. euvesicatoria strains BRIP 39016 and DAR 26930 were included in the X. perforans group, indicating there may be some recombination events present in these strains/ species. The Australian strains were not significantly differentiated from overseas strains by this matrix, indicating a certain level of species homogeneity distributed across the world.
The core genomes of species in this dataset represent conserved functionally essential genes, while the larger accessory and cloud genomes contained genes that may be specific to growth or pathogenicity, particularly as species with different pathogenic capabilities are present in the homologue matrix. Most species analysed individually reflected this trend of large accessory genomes, with the exception of X. gardneri, which was influenced by the small sample size of highly similar strains. The gene discovery plots for the pan genomes of these species showed that the genomes of X. euvesicatoria and X. perforans can be considered closed. Plotting gene discovery indicates there is some potential that additional genomes would result in detection of new genes for X. vesicatoria and X. gardneri. The large accessory genomes observed in many of these species reflects the genetic diversity seen in other studies of Xanthomonas species [3], and also suggests that genome plasticity could result in new genes being added to the population.
Predicted plasmids reconstructed from collected Xanthomonas strains
The seven 150 kb plasmids from X. euvesicatoria had high homology to the X. euvesicatoria plasmid pLMG930.2 of similar size (167,496 bp). Similarly, the 31 kb group of Australian plasmids found in X. euvesicatoria shared homology with X. euvesicatoria and X. citri plasmids pLMG930.4 (GenBank, unpublished) and CFBP6167 plasmid pG [7]. This homology, and their presence in X. euvesicatoria strains (notably absent in X. perforans strains) indicates these 150 kb and 31 kb sequences are previously characterised plasmids. This is likely also the case for plasmids of 47 kb found in X. vesicatoria, as they are similar in size and homology to the X. vesicatoria plasmid pLMG911.2 (CP018727.1, 47 kb). By contrast, three 88 kb plasmids in X. vesicatoria, although similar by sequence homology, are much smaller than the X. vesicatoria plasmid pLMG911.1 (192,558 bp).
Plasmids of 40 kb found in X. vesicatoria, X. euvesicatoria and X. perforans were most similar to previously reported plasmids of X. campestris pv. campestris strains CN14 (GenBank: CP017318.1) and CN15 (CP017325.1), and X. perforans pLH3.3 (NZCP018474.1) and pLH3.2 (NZCP018476.1), all of varying sizes. Three plasmids of approximately 28–32 kb were slightly different in size and homology to the 31 kb plasmids, their presence in older and uncharacterised Xanthomonas strains indicating they may be more distantly related. Interestingly, five 17 kb plasmids of X. euvesicatoria did not significantly match any plasmid sequence and were not recovered in the plasmid isolations of BRIP 62416 and BRIP 38997. Evidence for large (80–150 kbp) plasmids was observed in the plasmid extractions, as well as bands that are likely 30–40 kbp in size (Additional file 3: Figure S3). No definitive bands at 8 kbp or 17 kbp were observed in strains BRIP 62858, BRIP 62416 and BRIP 38997, indicating some smaller plasmids may be a result of computational reconstruction. There also appeared to be some plasmids present that were not detected by the selected programs, as in BRIP 62388. False positives and negatives may be a result of integrative conjugative elements or repeat regions that may require further sequencing to fully resolve.
Avirulence genes have been found in many described plasmids of most genera, including Xanthomonas pathogens [8]. Sequence with homology to three effectors (AvrBs3, AvrBsT and XopH) were detected in assembled plasmids of ca. 40–47 kb. AvrBsT was detected in most X. vesicatoria plasmids (seven of nine), and has been known to exist on plasmids since its characterisation [9, 30]. As in other studies of plasmid-borne effectors [31], the presence of effectors here demonstrates these circular elements have the potential to influence pathogenicity. XopH, detected in one plasmid of X. perforans, has been suggested as a potential determinant of pathogenicity in X. arboricola pv. corylina [32]. It has also been found in X. campestris pv. campestris [33], and here was found in the majority of X. gardneri and X. vesicatoria chromosomes. Other genes, for example copper tolerance genes, have been found on Xanthomonas plasmids [8], suggesting other significant adaptive genes in addition to effectors may be investigated in future studies.
Effector profiles of Xanthomonas
Many studies have presented core effector lists for Xanthomonas pathogens and found that these effectors are integral to certain strains/species and play key roles in pathogenicity [1, 13, 17, 29, 34]. The effector profiles determined by this study show distinct patterns specific to species and clades. We have revised the core and specific effector list for Xanthomonas species causing BLS and contrast them with species displaying different pathogenic abilities.
Core and specific effectors
Few effectors were found to be shared between closely related phylogenetic groups, a finding consistent with a previous study on the type strains of four BLS-causing Xanthomonas species [27]. Subsequent studies have noted that strains may display different effector profiles to that of the type strain of their species [17], a pattern also observed in this study. The core effectors previously identified in the type strains of the BLS pathogens X. gardneri, X. perforans, X. vesicatoria and X. campestris pv. vesicatoria (X. euvesicatoria) [27] were expanded [17] with the addition of XopE2 and a member of the YopJ family (AvrBsT and XopJ1). Barak et al. [29] further refined the list of core effectors, finding all effectors previously identified [27], with the exception of XopAD that displayed internal stop codons in some X. euvesicatoria strains. The analysis of effectors in species that do not cause BLS provides an opportunity to compare and contrast effector profiles with X. vesicatoria, X. euvesicatoria and X. perforans. The Xanthomonas sp. clade has few effectors, most of which are shared with some X. arboricola and X. vesicatoria strains. Reduced T3SS effector repertoires do not necessarily indicate a lack of pathogenic capability [35], however it is likely these effectors (Additional file 4: Table S1) are not directly involved in pathogenicity on tomato or pepper due to their presence in non-pathogenic strains. The Australian X. arboricola strains in this study have varied effector profiles with few common effectors. As there are relatively few sequenced strains of X. arboricola from different hosts, it is difficult to draw meaningful conclusions about effector profiles in relation to their pathogenicity. The variation observed in these profiles is likely a result of wider host range, presenting a point of contrast to the other groups.
In this study, the core effectors AvrBs2, XopAD, XopR, XopX and XopZ1 were found in the majority of strains of X. euvesicatoria, X. gardneri, X. perforans and X. vesicatoria. All of these strains together with those of X. arboricola and the uncharacterised Xanthomonas sp. clade contained rpfA, rpfB and rpfF, members of the rpf gene family that regulate pathogenicity factors and biofilm production [36]. Homologues of AvrBs2, involved in the modulation of effector delivery [37] were also found in all strains of this study. Other effectors previously listed as core to BLS-causing Xanthomonas species were detected in most strains of these species with some exceptions as listed above. Interestingly, many of these effectors are also present in X. arboricola strains and the uncharacterised Xanthomonas clade. For example, XopF1 was only absent in one strain of X. gardneri, but was detected in BLS-causing species as well as most X. arboricola and all strains of the uncharacterised clade. The core effectors XopK, XopL and XopN, were also found in strains not isolated from tomato or pepper, which indicates these proteins may not be associated with specificity to these hosts. No single effector in this study appeared to be consistently associated with pathogenicity on tomato based on comparison with the X. arboricola and Xanthomonas spp. clades. This was also the case for pepper pathogenic strains, though XopAA and XopJ3 were present only in the pepper pathogenic X. euvesicatoria and the non- pepper pathogenic BRIP 39016. The profiles presented here represent homologues in predicted protein sequence, so it is possible inactivation in effector gene sequences play a role in pathogenicity as well.
As demonstrated by Barak et al. [29], the core effectors listed above do not determine pathogenicity on tomato due to their presence in an X. euvesicatoria strain isolated from rose. One particular clade of X. arboricola (containing the MAFF strains) shared many effectors with BLS-causing species, further emphasising the need for comprehensive pathogenicity studies to tie effector profile to functional traits.
Effectors and host range of the X. euvesicatoria and X. perforans clades
While X. euvesicatoria is commonly reported as a pathogen of tomato and pepper, all but two X. euvesicatoria strains (BRIP 39016 and DAR 26930) from Australian crops were found in capsicum and chilli [4]. Recent reports indicate it is more common to observe X. perforans (and X. gardneri) in tomato and X. euvesicatoria in peppers [17, 38, 39]. Prior to 1991, X. euvesicatoria was the main BLS pathogen on tomato in Florida [1]. This indicates it was once more common to find X. euvesicatoria in tomato than it is today. As the only Australian X. euvesicatoria strains isolated from tomato were from 1973 and 1976, Australian X. euvesicatoria populations reflect this host shift observed overseas. One X. perforans strain (Xp2010) from Florida displayed dual infecting ability in pepper and tomato [27]. An Australian strain, BRIP 62398, phylogenetically related to Xp2010 did not share this trait, as all tested Australian X. perforans strains were pathogenic only on tomato. While this indicates pathogenicity traits are not necessarily reflected in phylogenies, variation in virulence on pepper of certain phylogenetic groups has been noted [17].
Strains of X. euvesicatoria and X. perforans are genetically similar and share a similar effector profile, while still displaying notable differences. The core effectors XopF1, XopL, XopN, XopQ, XopR, XopX, XopAK, were conserved in the X. euvesicatoria and X. perforans strains in this study as well as in a previous study (that did not include X. perforans strains) [29]. While core effectors may indicate evolutionary history, several studies note that functionality of effector genes must be investigated in addition to presence or absence [17, 29]. Australian strains of X. euvesicatoria displayed almost identical profiles to those of overseas strains, apart from a group of 11 that contain an AvrBs3 homologue along with xe678 and xe685 that likely reflect pathogenicity differences. Australian X. perforans effector profiles were also similar to other X. perforans strains, though they (as well as xp91–118 and xp4p1s2) lack XopE2. Australian X. perforans (excluding BRIP 62397) appear to have XopE3 where all other X. perforans strains do not. These presences and absences may have pathogenicity implications according to the description of the XopE family [40], though this pattern does also reflect their clade groupings in the core genome phylogeny. XopE3 has also been implicated in citrus pathogenicity [41]. Further investigation into the function of these effectors may reveal the significance of these patterns.
An effector that has been used to track population changes is the 600 amino acid protein XopAE, which is a fusion of the HpaF and hpaG effectors [27]. The majority of Australian X. euvesicatoria and X. perforans strains contained a XopAE homologue, while four X. euvesicatoria strains (BRIP 62438, BRIP 38997, DAR 34895 and DAR 82542) had hpaG and hpaF as separate effectors. These strains were collected from locations and/ or time points different from the rest of the collection, possibly reflecting different introductions or outbreaks. Barak et al. [29] suggested the presence of the translational mutation and the single alleles represented separate introductions, as they observed in strain LMG918. The difference in effector profiles between strains separated by time has also been noted previously [17] and is reflected in historical strains of X. euvesicatoria in this study, in particular BRIP 39016 and DAR29630 that are also separated geographically.
Effectors of X. vesicatoria and X. gardneri
Strains of X. vesicatoria have a distinct effector profile similar to X. gardneri, which reflects their position in the whole genome SNP phylogeny. The variation of effector profile within X. vesicatoria reflects the phylogenetic clades identified, rather than specific differences in Australian and overseas strains. Homologues of XopAG and XopAI have previously been identified as specific to X. vesicatoria [27]. However, we have shown that homologues of XopAG exist in DAR 33341 (Xanthomonas sp.) and an X. arboricola strain (NCPPB 100457). XopAI was also detected in these and an additional five strains of X. arboricola. Previous studies of the X. gardneri effector profile found differences between the type and other strains, which is also evident in this study [17].
Key CAZymes
Similar to the effector profiles, the cazyme profiles grouped strains mostly into species, highlighting regions of difference. No differences between Australian and overseas strains within species were detected. CAZyme genes and families have been identified previously in the type strains of BLS-causing Xanthomonas species [27] and reflect the profiles seen in this study. Cellulases are known to be common to the gammaproteobacteria [42], and the abundance of GH families was expected. The xylanase families GH10 and GH30 [25, 43] were present in all strains, indicating these groups share similar strategies for degrading plant cells. Identifying core carbohydrate active enzymes needs further investigation into the proteins and genes of these enzyme families [27]. Carbohydrate utilisation reactions have been used as a diagnostic tool for many years to differentiate bacterial species based on substrate usage [23]. It has also been suggested that secreted cell wall degrading enzymes play a role in host adaptation, with several studies linking these enzymes and pathogenicity [35].
CAZyme differences between species
The CAZyme family profiles of the X. euvesicatoria and X. perforans strains were nearly identical, apart from the absence of three families (CE8, CE14, and GH39) in the X. euvesicatoria strains. The absence of these families of plant polysaccharides, acetylases and pectinases highlights a distinct difference between these two groups. The absence of CE8 (a pectin methylesterase family) in X. euvesicatoria strains correlates with their lack of ability to degrade pectin [25, 43].
The X. euvesicatoria strains had mostly glycosyl hydrolase (GH) and polysaccharide lyase (PL) families (PL10, PL3, CBM63, GH16, PL17, PL6, GH4, GH89, GT84) and lacked many CAZyme families present in other species. The CAZyme families PL3, CBM63 and GH16 were found in all X. vesicatoria and X. gardneri strains and many X. arboricola strains. The conserved presence of some CAZyme families that are present or absent in some species may indicate different substrate utilisation capabilities. Groups of cellulose degrading enzymes also display different profiles, which indicates that species have different modes of action on this substrate. The families GH5, GH9 and GH12 were found in most or all of the strains in this study, GH8 and GH6 had a more restricted distribution. In particular, GH6 was identified only in strains of X. vesicatoria and DAR 33341, which may reflect different strategies or evolutionary pathways for degrading cellulose.
Conclusions
This study has provided an overview of the genome structure and content of several Xanthomonas species and expanded the original identification of Australian species associated with BLS. We support the taxonomic status of X. euvesicatoria and X. perforans as one species, though it is clear these strains also have conserved differences that complicate taxonomy. Our analysis of effector proteins and carbohydrate active enzymes links pathogenic data with proteins detected in the genomic analysis, demonstrating that while these profiles differ between species no single pathogenicity factor was identified. It is clear that some differences may also exist in Australian populations regarding effector content. The limitations of bioinformatic reconstruction of plasmids was also highlighted. This study has furthered the understanding of species that cause BLS and provided several points of future study to improve the understanding of Australian bacterial populations.
Methods
Isolate collection, pathogenicity testing and sequencing
Strains of Xanthomonas spp. associated with BLS in Australia were collected as described in Roach et al. [4]. To determine pathogenicity on tomato and capsicum, all isolates (excluding DAR strains as only genomic data was available) were inoculated on susceptible Capsicum annuum var. Yolo Wonder and susceptible Solanum lycopersicum var. Grosse Lisse. Overnight cultures of bacteria were diluted in distilled H20 to concentrations of 1 × 108 cfu/ml and sprayed onto plants until run-off. Pathogenicity was recorded after approximately 7 days. Pathogenicity on host of isolation has been reported [4]. Pathogenicity on the alternative host was observed as small, dark lesions with yellow halo that displayed bacterial streaming.
The dataset of 50 Australian strains was comprised of 44 strains held in the Queensland Plant Pathology Herbarium (BRIP) culture collection and six sequenced Australian strains provided by the NSW Plant Pathology Herbarium (DAR) (Table 1). Selected strains of each identified species represented a range of taxa, host and geographical distribution. Strains were grown overnight in lysogeny broth (Luria-Bertani) [44] and the DNA was extracted using a Qiagen DNeasy Blood and Tissue kit (Qiagen; Hilden, Germany). Genomic libraries were prepared using an Illumina Nextera XT Library Preparation Kit according to the manufacturer instructions (Illumina; San Diego, USA). Sequencing was conducted using a Miseq v3 reagent kit on an Illumina Miseq®.
Genome construction
Sequence read adaptors were trimmed with Cutadapt version 1.8.1 and quality trimmed using Trim Galore (q = 25 with ‘paired’ and ‘nextera’ flags) version 0.4.0 [45]. Contigs were assembled with SPAdes version 3.5.0 [46] (with kmers of 127, 117, 107, 97, 87, 77, 67 using the ‘careful’ flag), and annotated with Prokka version 1.11 [47] using the packaged database (using the ‘genus’ and ‘force’ flags). In addition to the 44 sequenced strains, sequence data for 6 Australian strains from DAR were processed with SPAdes and Prokka as above. Genome statistics including GC content, contig number, N50 and genome length were calculated with QUAST version 4.5 [48].
An additional 97 genomes of Xanthomonas strains available in GenBank were downloaded and re-annotated with Prokka (as described above) for standardisation and included in analyses (Table 1). These public genomes represent the majority of sequenced X. arboricola, X. euvesicatoria (X. campestris pv. vesicatoria), X. gardneri, X. perforans, and X. vesicatoria strains in GenBank. Average nucleotide identity of scaffolds was calculated for the entire dataset of 147 genomes with pyani version 0.2.4 [49] using the default settings. Strains were determined to belong to the same species if ANI values were above the 95–96% zone as set in Konstantinidis et al. [50] and utilised in Barak [29].
Plasmid prediction and isolation
Plasmid prediction from the draft genomes of Australian strains was achieved using the plasmidSPAdes option (‘plasmid’ flag) of SPAdes version 3.8.0 [46]. Circular sequences from these assemblies were finished using recycler version 0.62 [51]. Bandage version 0.8.1 [52] was used to view the Recycler paths. The Blast+ algorithm version 2.6.0 [53] was used to compare the plasmid sequences to a custom database of complete Xanthomonas plasmids obtained from GenBank [54]. Plasmid isolation was carried out on a subset of strains (BRIP 38864, BRIP 62858, BRIP 62416, BRIP 62423, BRIP 62388, BRIP 62397, BRIP 63464, BRIP 38997) using the alkaline lysis method described in Chakrabarty [55]. The Pseudomonas strain DC 3000 [56] was used as an extraction control. Strains were grown in LB broth and processed with the described buffers, resuspending the pelleted DNA in distilled H20. Plasmid DNA was visualised on 0.7% agarose gels using standard electrophoresis at 40 V for 4–12 h.
Analysis of genome content
A phylogeny that displays general species relationships was generated using the RedDog pipeline version V1beta.10.3 [57]. Briefly, the RedDog pipeline assembles and aligns raw reads against a reference genome (X. campestris pv. vesicatoria strain 85–10, Table 1), then creates a phylogeny using SNP data generated within the pipeline. Simulated reads were generated for public genomes using WgSim v. 0.3.1-r13 for inclusion in analysis [58]. Support values of the resulting phylogeny calculated by FastTree version 2.1.8 [59] are displayed as a range of 0 to 1. The final tree was annotated in FigTree ver. 1.4.2 [60] and GIMP version 2.8.14 [61].
Roary version 3.8.2 [62] was used to cluster homologues, generate a core alignment (−e and -n flags), and generate the pan-genus homologue matrix. The core alignment was then used to create individual phylogenies of BLS-causing species X. euvesicatoria, X. perforans, and X. vesicatoria with FastTree and annotated with FigTree and GIMP. The homologue matrix was then used to generate pan-genome pie plots using scripts available within the Roary package. Core genes were defined as present in 99–100% strains, soft core genes in 95–99% strains, shell genes in 15–95% strains and cloud genes in 0–15% strains. Gene discovery graphs were plotted using Roary scripts and R version 1.0.136 [62] to determine if the pan-genome was open or closed. These analyses were done for each species as defined in Table 1 (DAR 33341 was not included). The homologue matrix was filtered in R to identify unique genes of each species.
Carbohydrate-active and associated enzyme (CAZyme) coding sequences within each genome were identified using HMMER version 3.1b2 [63] and the DbCAN database [64]. The CAZyme hits were then clustered manually and presented as a heat map and dendrogram in R version 3.3.0 [65] using the pheatmap package [66] and annotated in GIMP. T3SS enzyme protein (effector) sequence and select regulatory protein sequence (rpf) were sourced from GenBank as listed in the Xanthomonas resource ‘effector’ page [67] and compared to the genome sequence and reconstructed plasmid sequence using the Blast+ algorithm. Hits were filtered (e-value 0.00001) and presented as a heat map using R and GIMP as described above.
Additional files
Figure S1. Phylogeny of Australian and Genbank genomes based on whole genome SNP data. Australian strains are indicated by BRIP and DAR collection prefixes; all others are public genomes of related species. Type strains are indicated in bold and branch support values are displayed to clade level (measured with the Shimodaira-Hasegawa test). Branch length is indicated by the scale bar. Clade colouring is based on phylogeny and ANI values to assign strains to species. The four Australian strains most closely related to X. arboricola are designated in the text as an uncharacterised Xanthomonas species. (PNG 278 kb)
Figure S2. Gene discovery graphs for X. euvesicatoria, X. perforans, X. vesicatoria and X. gardneri plot number of new genes in the species pan-genome as genome number increases. The graph curves demonstrate how many new genes will be added with the addition of more sequenced genomes to estimate pan-genome completeness. X axis: genome number; Y axis: number of new genes. (PNG 45 kb)
Figure S3. Standard electrophoresis of plasmid isolations with predicted circular sequence in base pairs below each lane. Ladder = Generuler™ DNA Ladder Mix, ThermoFisher Scientific, Waltham, Massachusetts. The 10 kbp label marks the largest ladder fragment, and the 60–70 kbp label marks the band present in DC 3000 (plasmid extraction control). A) gel was run for approx. 12 h at 40 V B) gel was run for approx. 4 h at 40 V. A and B represent two different extraction experiments (PNG 79 kb)
Table S1. Homologues of effector protein families present in all strains of each Xanthomonas species and unique to each species. Effectors listed include all alleles displayed in the effector matrix for each effector family. a present in all strains of a species and possibly present in other strains/ species. b uncharacterised Xanthomonas sp. of four strains. c core to species in Schwartz et al. 2015. d core to species in Potnis et al. 2011. e Xp4B and Xp4p1S2 have truncated HpaG protein annotations, Xp4p1s2 has truncated XopAE protein annotation. f BRIP 39016 has a larger (650 aa) XopAE protein than all others (546 aa). g present in all Australian strains except BRIP 62397 (DOCX 13 kb)
Table S2. Cazyme families present in all strains of each Xanthomonas species and families unique to each species. Function is described according to the CAZy database.a all three present in DAR 26930, GH39 present in BRIP 39016, CE14 absent in GEV915. b genes of CAZyme family present in all strains of species. c genes of CAZyme family absent in all strains of species. d genes of CAZyme family present in some strains of species. e absent in 1–3 strains. (DOCX 13 kb)
Acknowledgements
The authors would like to thank QLD and NSW herbariums for assistance in maintaining and providing cultures. This project has been funded by Hort Innovation, using the vegetable research and development levy and contributions from the Australian Government. Hort Innovation is the grower owned, not-for-profit research and development corporation for Australian horticulture. This project was also funded through a scholarship at La Trobe University, Bundoora, Australia.
Funding
This project has been funded by Hort Innovation, using the vegetable research and development levy and contributions from the Australian Government. Hort Innovation is the grower owned, not-for-profit research and development corporation for Australian horticulture. The PhD scholarship was provided by La Trobe University.
Availability of data and materials
Contig files for genomic data generated in this study were processed by GenBank and assigned the BioProject ID PRJNA454505. Contigs less than 200 bp were removed and files were submitted as nucleotide fasta files for each reported strain. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QEYT00000000-QFAQ00000000. The version described in this paper is version QEYT01-QFAQ01. Live cultures are available from the Brisbane Pathology (BRIP) Herbarium.
Abbreviations
- AA
Auxiliary activity families
- ANI
Average nucleotide identity
- BLS
Bacterial leaf spot
- BRIP
Queensland Plant Pathology Herbarium
- CAZymes
Carbohydrate active enzymes
- CBM
Carbohydrate binding modules
- CE
Carbohydrate esterases
- DAR
New South Wales Plant Pathology Herbarium
- GH
Glycoside hydrolases
- GT
Glycosyl transferases
- NGS
Next generation sequencing
- NSW
New South Wales
- PL
Polysaccharide lyases
- SNP
Single nucleotide polymorphism
- T2SS
Type two secretion system
- T3SS
Type three secretion system
- TAL/ TALE
Transcription activator-like effectors
- Xop
Xanthomonas outer protein
Authors’ contributions
RR collected and analysed the presented data and wrote the manuscript. RM assisted with genome sequencing and analysis. TC provided raw genome sequence of DAR strains. CG, RS, and BR assisted with experimental planning, analysis and manuscript review. All authors read and approved the final manuscript.
Ethics approval and consent to participate
NA
Consent for publication
NA
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
R. Roach, Email: roach.r@students.latrobe.edu.au
R. Mann, Email: rachel.mann@ecodev.vic.gov.au
C. G. Gambley, Email: cherie.gambley@daf.qld.gov.au
T. Chapman, Email: toni.chapman@dpi.nsw.gov.au
R. G. Shivas, Email: roger.shivas@daf.qld.gov.au
B. Rodoni, Email: brendan.rodoni@ecodev.vic.gov.au
References
- 1.Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol Plant Pathol. 2015;16(9):907–920. doi: 10.1111/mpp.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinatzer BA, Monteil CL, Clarke CR. Harnessing population genomics to understand how bacterial pathogens emerge, adapt to crop hosts, and disseminate. Annu Rev Phytopathol. 2014;52:19–43. doi: 10.1146/annurev-phyto-102313-045907. [DOI] [PubMed] [Google Scholar]
- 3.Garita-Cambronero J, Palacio-Bielsa A, Lopez MM, Cubero J. Pan-genomic analysis permits differentiation of virulent and non-virulent strains of Xanthomonas arboricola that cohabit Prunus spp and elucidate bacterial virulence factors. Front Microbiol. 2017;8:573. doi: 10.3389/fmicb.2017.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roach R, Mann R, Gambley CG, Shivas R, Rodoni B. Identification of Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli crops in eastern Australia. Eur J Plant Pathol. 2017;150(3):595–608. doi: 10.1007/s10658-017-1303-9. [DOI] [Google Scholar]
- 5.Ryan RP, Vorhölter FJ, Potnis N, Jones JB, Van Sluys MA, Bogdanove AJ, Dow JM. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 6.Richard D, Ravigné V, Rieux A, Facon B, Boyer C, Boyer K, Grygiel P, Javegny S, Terville M, Canteros BI, Robene I, Vernière C, Chabirand A, Pruvost O, Lefeuvre P. Adaptation of genetically monomorphic bacteria: copper resistance evolution through multiple horizontal gene transfers of complex and versatile mobile genetic elements. Mol Ecol. 2017;27:2131–2149. doi: 10.1111/mec.14007. [DOI] [PubMed] [Google Scholar]
- 7.Ruh M, Briand M, Bonneau S, Jacques MA, Chen NW. Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics. 2017;18:670. doi: 10.1186/s12864-017-4087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stall RE, Loschke DC, Jones JB. Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonas campestris pv. Vesicatoria. Phytopathology. 1986;76(2):240–243. doi: 10.1094/Phyto-76-240. [DOI] [Google Scholar]
- 9.Minsavage GV, Dahlbeck D, Whalen MC, Kearney B, Bonas U, Staskawicz BJ, Stall RE. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. Vesicatoria - pepper interactions. Mol Plant Microbe In. 1990;3(1):41–47. doi: 10.1094/MPMI-3-041. [DOI] [Google Scholar]
- 10.Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Buttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, Kirchner O, Lanz C, Linke B, AC MH, Meyer F, Mittenhuber G, Nies DH, Niesbach-Klosgen U, Patschkowski T, Ruckert C, Rupp O, Schneiker S, Schuster SC, Vorholter FJ, Weber E, Puhler A, Bonas U, Bartels D, Kaiser O. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol. 2005;187(21):7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillings MR, Holley MP, Stokes HW, Holmes AJ. Integrons in Xanthomonas: a source of species genome diversity. P Natl Acad Sci USA. 2005;102(12):4419–4424. doi: 10.1073/pnas.0406620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bansal K, Midha S, Kumar S, Patil PB. Ecological and evolutionary insights into Xanthomonas citri pathovar diversity. Appl Environ Microb. 2017;83(9):e02993–e02916. doi: 10.1128/AEM.02993-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White FF, Potnis N, Jones JB, Koebnik R. The type III effectors of Xanthomonas. Mol Plant Pathol. 2009;10(6):749–766. doi: 10.1111/j.1364-3703.2009.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun W, Liu L, Bent AF. Type III Secretion-dependent host defence elicitation and type III secretion-independent growth within leaves by Xanthomonas campestris pv. Campestris. Mol Plant Pathol. 2011;12(8):731–745. doi: 10.1111/j.1364-3703.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesce C, Jacobs JM, Berthelot E, Perret M, Vancheva T, Bragard C, Koebnik R. Comparative genomics identifies a novel conserved protein, HpaT, in proteobacterial type III secretion systems that do not possess the putative translocon protein HrpF. Front Microbiol. 2017;8:1177. doi: 10.3389/fmicb.2017.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Yin Z, White F. TAL effectors and the executor R genes. Front Plant Sci. 2015;6:641. doi: 10.3389/fpls.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz AR, Potnis N, Milsina S, Wilson M, Patane J, Martins J, Minsavage GV, Dahlbeck D, Akhunova A, Almeida N, Vallad GE, Barak JD, White F, Miller SA, Ritchie D, Goss E, Bart RS, Setubal JC, Jones JB, Staskawicz BJ. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front Microbiol. 2015;6:535. doi: 10.3389/fmicb.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrahamian P, Timilsina S, Minsavage GV, Kc S, Goss EM, Jones JB, Vallad GE. The type III effector AvrBsT enhances Xanthomonas perforans fitness in field-grown tomato. Phytopathology. 2018;108(12):1355–1362. doi: 10.1094/PHYTO-02-18-0052-R. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. P Natl Acad Sci USA. 2017;114(5):897–903. doi: 10.1073/pnas.1620407114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Rinaldi FC, Singh P, Doyle EL, Dubrow ZE, Tran TT, Perez-Quintero A, Szurek B, Bogdanove AJ. TAL effectors drive transcription bidirectionally in plants. Mol Plant. 2017;10(2):285–296. doi: 10.1016/j.molp.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Solé M, Scheibner F, Hoffmeister AK, Hartmann N, Hause G, Rother A, Jordan M, Lautier M, Arlat M, Buttner D, Christie PJ. Xanthomonas campestris pv. Vesicatoria secretes proteases and xylanases via the xps type II secretion system and outer membrane vesicles. J Bacteriol. 2015;197(17):2879–2893. doi: 10.1128/JB.00322-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cianciotto NP, White RC. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun. 2017;85(5):e00014–e00017. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward AC. Occurrence of glycoside hydrolases in plant pathogenic and related bacteria. J Appl Bacteriol. 1977;43(3):407–411. doi: 10.1111/j.1365-2672.1977.tb00767.x. [DOI] [Google Scholar]
- 24.Davies GJ, Gloster TM, Henrissat B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr Opin Struc Biol. 2005;15(6):637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233-8. [DOI] [PMC free article] [PubMed]
- 26.Henrissat B. A classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potnis N, Krasileva K, Chow V, Almeida NF, Patil PB, Ryan RP, Sharlach M, Behlau F, Dow JM, Momol MT, White F, Preston JF, Vinatzer BA, Koebnik R, Setubal JC, Norman DJ, Staskawicz BJ, Jones JB. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics. 2011;12:146. doi: 10.1186/1471-2164-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones JB, Lacy GH, Bouzar H, Stall RE, Schaad NW. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol. 2004;27(6):755–762. doi: 10.1078/0723202042369884. [DOI] [PubMed] [Google Scholar]
- 29.Barak JD, Vancheva T, Lefeuvre P, Jones JB, Timilsina S, Minsavage GV, Vallad GE, Keobnik R. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front Plant Sci. 2016;7:1805. doi: 10.3389/fpls.2016.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonas U, Stall RE, Staskawicz B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. Vesicatoria. Mol Gen Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 31.Pothier JF, Vorhölter FJ, Blom J, Goesmann A, Pühler A, Smits THM, Duffy B. The ubiquitous plasmid pXap41 in the invasive phytopathogen Xanthomonas arboricola pv. Pruni: complete sequence and comparative genomic analysis. FEMS Microbiol Lett. 2011;323(1):52–60. doi: 10.1111/j.1574-6968.2011.02352.x. [DOI] [PubMed] [Google Scholar]
- 32.Hajri A, Pothier JF, Fischer-Le Saux M, Bonneau S, Poussier S, Boureau T, Duffy B, Manceau C. Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola. Appl Environ Microb. 2012;78(2):371–384. doi: 10.1128/AEM.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux B, Bolot S, Guy E, Denance N, Lautier M, Jardinaud MF, Fischer-Le-Saux M, Portier P, Jacques MA, Gagnevin L, Pruvost O, Lauber E, Arlat M, Carrere S, Koebnik R, Noel LD. Genomics and transcriptomics of Xanthomonas campestris species challenge the concept of core type III effectome. BMC Genomics. 2015;16:975. doi: 10.1186/s12864-015-2190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timilsina S, Abrahamian P, Potnis N, Minsavage G, White F, Staskawicz BJ, Jones JB, Vallad GE, Goss EM. Analysis of sequenced genomes of Xanthomonas perforans identifies candidate targets for resistance breeding in tomato. Phytopathology. 2016;106(10):1097–1104. doi: 10.1094/PHYTO-03-16-0119-FI. [DOI] [PubMed] [Google Scholar]
- 35.Jacques MA, Arlat M, Boulanger A, Boureau T, Carrère S, Cesbron S, Chen NWG, Cociancich S, Darrasse A, Denance N, Fischer-Le-Saux M, Gagnevin L, Keobnik R, Lauber E, Noel L, Pieretti I, Portier P, Pruvost O, Rieux A, Robene I, Royer M, Szurek B, Verdier V, Verniere C. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas: French network on xanthomonads (FNX) Annu Rev Phytopathol. 2016;54(1):163–187. doi: 10.1146/annurev-phyto-080615-100147. [DOI] [PubMed] [Google Scholar]
- 36.Dow M. Diversification of the function of cell-to-cell signaling in regulation of virulence within plant pathogenic xanthomonads. Sci Signal. 2008;1(21):23. doi: 10.1126/stke.121pe23. [DOI] [PubMed] [Google Scholar]
- 37.Zhao YJ, Zhang YH, Cao Y, Qi JX, Mao LW, Xue YF, Gao F, Peng H, Wang X, Gao GF, Ma Y. Structural analysis of alkaline beta-mannanase from alkaliphilic Bacillus sp N16-5: implications for adaptation to alkaline conditions. PLoS One. 2011;6:e14608. doi: 10.1371/journal.pone.0014608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivey ML, Strayer A, Sidhu JK, Minsavage GV. Bacterial leaf spot of bell pepper (Capsicum annuum) in Louisiana is caused by Xanthomonas euvesicatoria pepper races 1 and 3. Plant Dis Notes. 2016;100(4):853. doi: 10.1094/PDIS-10-15-1150-PDN. [DOI] [Google Scholar]
- 39.Ivey ML, Strayer A, Sidhu JK, Minsavage GV. Bacterial leaf spot of tomato (Solanum lycopersicum) in Louisiana is caused by Xanthomonas perforans, tomato race 4. Plant Dis. 2016;100(6):1233. doi: 10.1094/PDIS-12-15-1451-PDN. [DOI] [Google Scholar]
- 40.Thieme F, Szczesny R, Urban A, Kirchner O, Hause G, Bonas U. New type III effectors from Xanthomonas campestris pv. Vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol Plant Microbe In. 2007;20(10):1250–1261. doi: 10.1094/MPMI-20-10-1250. [DOI] [PubMed] [Google Scholar]
- 41.Moreira LM, Almeida NF, Potnis N, Digiampietri LA, Adi SS, Bortolossi JC, da Silva AC, da Silva AM, de Moraes FE, de Oliveira JC, de Souza RF, Facincani AP, Ferraz AL, Ferro MI, Furlan LR, Gimenez DF, Jones JB, Kitajima EW, Laia ML, Leite RP, Nishiyama MY, Rodrigues Neto J, Nociti LA, Norman DJ, Ostroski EH, Pereira HA, Staskawicz BJ, Tezza RI, Ferro JA, Vinatzer BA, Setubal JC. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics. 2010;11:238. doi: 10.1186/1471-2164-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medie FM, Davies GJ, Drancourt M, Henrissat B. Genome analyses highlight the different biological roles of cellulases. Nat Rev Microbiol. 2012;10(3):227–234. doi: 10.1038/nrmicro2729. [DOI] [PubMed] [Google Scholar]
- 43.Berlemont R, Martiny AC. Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microb. 2015;81(4):1513–1519. doi: 10.1128/AEM.03718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaad NW. Laboratory Guide for Identification of Plant Pathogenic Bacteria (3rd ed.); APS Press. St. Paul, Minnesota USA 2001.
- 45.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 48.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods-UK. 2016;8(1):12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 50.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102(7):2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozov R, Brown Kav A, Bogumil D, Halperin E, Mizrahi I, Shamir R. Recycler: an algorithm for detecting plasmids from de novo assembly graphs. bioRxiv. 2016;33(4):475–482. doi: 10.1093/bioinformatics/btw651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madden T. The BLAST sequence analysis tool. In: McEntyre J, Ostell J, editors. The NCBI Handbook. Bethesda: National Centre for Biotechnology Information; 2003. [Google Scholar]
- 54.NCBI Resource Coordinators Database resources of the National Center for biotechnology information. Nucleic Acids Res. 2017;45(Database issue):D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakrabarty PK, Chavhan RL, Ghosh A, Gabriel DW. Rapid and efficient protocols for throughput extraction of high quality plasmid DNA from strains of Xanthomonas axonopodis pv. Malvacearum and Escherichia coli. J Plant Biochem Biot. 2010;19(1):99–102. doi: 10.1007/BF03323444. [DOI] [Google Scholar]
- 56.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou L, Liu J, Yuan Q, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng WL, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang X, Bender CL, White O, Fraser CM, Collmer A. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003;100(18):10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards DJ, Pope BJ, Holt KE. In prep. 2015. RedDog: comparative analysis pipeline for large numbers of bacterial isolates using high-throughput sequences. [Google Scholar]
- 58.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut A. FigTree. http://tree.bio.ed.ac.uk/software/figtree. Accessed 2016.
- 61.The GIMP team. GNU Image Manipulation program. (2014) www.gimp.org. Accessed 2016.
- 62.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11(1):431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R core team . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 66.Kolde R. pheatmap: Pretty Heatmaps. (2015) https://CRAN.R-project.org/package=pheatmap. Accessed 2017.
- 67.Koebnik R. The Xanthomonas Resource. (2017) http://www.Xanthomonas.org/t3e.html. Accessed 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogeny of Australian and Genbank genomes based on whole genome SNP data. Australian strains are indicated by BRIP and DAR collection prefixes; all others are public genomes of related species. Type strains are indicated in bold and branch support values are displayed to clade level (measured with the Shimodaira-Hasegawa test). Branch length is indicated by the scale bar. Clade colouring is based on phylogeny and ANI values to assign strains to species. The four Australian strains most closely related to X. arboricola are designated in the text as an uncharacterised Xanthomonas species. (PNG 278 kb)
Figure S2. Gene discovery graphs for X. euvesicatoria, X. perforans, X. vesicatoria and X. gardneri plot number of new genes in the species pan-genome as genome number increases. The graph curves demonstrate how many new genes will be added with the addition of more sequenced genomes to estimate pan-genome completeness. X axis: genome number; Y axis: number of new genes. (PNG 45 kb)
Figure S3. Standard electrophoresis of plasmid isolations with predicted circular sequence in base pairs below each lane. Ladder = Generuler™ DNA Ladder Mix, ThermoFisher Scientific, Waltham, Massachusetts. The 10 kbp label marks the largest ladder fragment, and the 60–70 kbp label marks the band present in DC 3000 (plasmid extraction control). A) gel was run for approx. 12 h at 40 V B) gel was run for approx. 4 h at 40 V. A and B represent two different extraction experiments (PNG 79 kb)
Table S1. Homologues of effector protein families present in all strains of each Xanthomonas species and unique to each species. Effectors listed include all alleles displayed in the effector matrix for each effector family. a present in all strains of a species and possibly present in other strains/ species. b uncharacterised Xanthomonas sp. of four strains. c core to species in Schwartz et al. 2015. d core to species in Potnis et al. 2011. e Xp4B and Xp4p1S2 have truncated HpaG protein annotations, Xp4p1s2 has truncated XopAE protein annotation. f BRIP 39016 has a larger (650 aa) XopAE protein than all others (546 aa). g present in all Australian strains except BRIP 62397 (DOCX 13 kb)
Table S2. Cazyme families present in all strains of each Xanthomonas species and families unique to each species. Function is described according to the CAZy database.a all three present in DAR 26930, GH39 present in BRIP 39016, CE14 absent in GEV915. b genes of CAZyme family present in all strains of species. c genes of CAZyme family absent in all strains of species. d genes of CAZyme family present in some strains of species. e absent in 1–3 strains. (DOCX 13 kb)
Data Availability Statement
Contig files for genomic data generated in this study were processed by GenBank and assigned the BioProject ID PRJNA454505. Contigs less than 200 bp were removed and files were submitted as nucleotide fasta files for each reported strain. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QEYT00000000-QFAQ00000000. The version described in this paper is version QEYT01-QFAQ01. Live cultures are available from the Brisbane Pathology (BRIP) Herbarium.