Abstract
Bacteria from the Saccharibacteria phylum (formerly known as TM7) are ubiquitous members of the human oral microbiome and are part of the Candidate Phyla Radiation. Recent studies have revealed remarkable 16S rRNA diversity in environmental and mammalian host-associated members across this phylum, and their association with oral mucosal infectious diseases has been reported. However, due to their recalcitrance to conventional cultivation, TM7’s physiology, lifestyle, and role in health and diseases remain elusive. The recent cultivation and characterization of Nanosynbacter lyticus type strain TM7x (HMT_952)—the first Saccharibacteria strain coisolated as an ultrasmall obligate parasite with its bacterial host from the human oral cavity—provide a rare glimpse into the novel symbiotic lifestyle of these enigmatic human-associated bacteria. TM7x is unique among all bacteria: it has an ultrasmall size and lives on the surface of its host bacterium. With a highly reduced genome, it lacks the ability to synthesize any of its own amino acids, vitamins, or cell wall precursors and must parasitize other oral bacteria. TM7x displays a highly dynamic interaction with its bacterial hosts, as reflected by the reciprocal morphologic and physiologic changes in both partners. Furthermore, depending on environmental conditions, TM7x can exhibit virulent killing of its host bacterium. Thus, Saccharibacteria potentially affect oral microbial ecology by modulating the oral microbiome structure hierarchy and functionality through affecting the bacterial host’s physiology, inhibiting the host’s growth dynamics, or affecting the relative abundance of the host via direct killing. At this time, several other uncharacterized members of this phylum have been detected in various human body sites at high prevalence. In the oral cavity alone, at least 6 distinct groups vary widely in relative abundance across anatomic sites. Here, we review the current knowledge on the diversity and unique biology of this recently uncovered group of ultrasmall bacteria.
Keywords: candidate phyla radiation, ultra-small bacteria, interspecies interaction, epiparasite, symbiont, periodontitis
A monumental milestone in human microbiology within the past few decades was uncovering the vast microbial abundance and diversity associated with the human body (Peterson et al. 2009; Chen et al. 2010; Dewhirst et al. 2010). These microbial communities coevolve with their host and significantly influence health and disease in humans (Nishihara and Koseki 2004; Ley et al. 2006). Due to its easy access and clinical relevance, the human oral cavity has become a model for advanced microbiome analysis to gain an understanding of microbial ecology and functionality (Chen et al. 2010; Edlund et al. 2013; Edlund et al. 2015; Baker et al. 2017; Nowicki et al. 2018). However, a significant portion (≈30%) of oral microbial species remains uncultivable (http://www.homd.org); thus, their metabolic potential, physiologic properties, ecologic function, and influence in human health are unknown. A primary example is the bacterial phylum Saccharibacteria (formerly known as phylum TM7 and referred to as such in this review). Members of the TM7 phylum are ubiquitous in the human oral microbiome, and accumulating evidence links their association with periodontal disease. However, due to the lack of cultivated representatives, knowledge of TM7 was scarce until the recent cultivation of the first oral strain, which provided the first view on its unique cell size and lifestyle and the means to study these bacteria (He et al. 2015). In this review, we provide an update on the current knowledge of this enigmatic bacterial phylum.
TM7: Ubiquitous yet “Enigmatic” Bacterial Phylum Associated with the Human Oral Cavity
General Introduction to TM7
TM7 16S rRNA gene sequences were first detected from a peat bog sample in northern Germany (TM stands for Torf, mittlere Schicht; German for “a middle layer of peat”) in the mid-1990s by using a culture-independent molecular approach (Rheims, Rainey, and Stackebrandt 1996; Rheims, Sproer, et al. 1996) with highly divergent sequences from a number of additional candidate bacteria, such as TM6 (McLean et al. 2013). TM7 was later proposed as a distinct bacterial phylum based on partial 16S rDNA sequences (Hugenholtz et al. 1998; Hugenholtz et al. 2001) and designated “Candidate Division TM7.” Subsequently, its presence has been detected in extremely diverse natural habitats, including soil, seawater, underground water, rhizospheres, deep-sea sediments, a wastewater treatment plant, hot springs, termite guts, and mammals (Hugenholtz et al. 2001; Brinig et al. 2003; Kumar et al. 2003; Nakajima et al. 2005; Dewhirst et al. 2012; Kindaichi et al. 2016; Dudek et al. 2017; Zhang et al. 2017; Starr et al. 2018), with 16S rRNA-based molecular approaches. As of 2011, 255 TM7 phylotypes have been indicated by 16S rRNA gene sequencing, with 160 listed as environmental, 42 as animal associated, and 53 as human associated (Dinis et al. 2011). This list continues to expand as studies unveil new phylotypes (i.e., distinct sequences with no cultured representatives) from increasingly diverse environmental niches (Liu et al. 2012; Lowe et al. 2012; Segata et al. 2012; Winsley et al. 2014; Dudek et al. 2017; Zhang et al. 2017; Starr et al. 2018). Single-cell sequencing approaches then provided the first fragmented partial genomes of the oral TM7a (Marcy et al. 2007) and the soil harboring TM7 bacterium GTL1 (Podar et al. 2007). While valuable, these incomplete genomes provided limited power to predict the functional capacities present in this diverse phylum (Dinis et al. 2011). In 2013, 4 complete TM7 genomes were assembled through differential coverage binning sampled from an activated sludge bioreactor (Albertsen et al. 2013). “Candidatus Saccharibacteria” was proposed as the new phylum name for TM7 based on the genomic analysis, which suggested that these bacteria, with reduced genomes, consume primarily sugar compounds.
While TM7 bacteria can be found in a wide distribution of habitats, their physiology, lifestyle, and ecologic role or pathogenic nature (for host-associated phylotypes) remained elusive due to their apparent recalcitrance to conventional cultivation methods. Attempts were made to isolate TM7 bacteria but failed to achieve sustainable growth. A soil-slurry membrane system, which combined a polycarbonate membrane as growth support and a soil extract as growth substrate, was implemented to cultivate the environmental TM7 phylotypes as tiny microcolonies (Ferrari et al. 2005; Abrams et al. 2012), albeit with no success in further enrichment. Intriguingly, these microcolonies were composed of a mixture of bacterial species, suggesting that TM7 may require cocultivation to propagate (Abrams et al. 2012). These results were consistent with previous studies evaluating uncultivable microorganisms from the environment, which revealed that many recalcitrant bacteria rely on metabolic cooperation with other species for the provision of nutrients (Zengler et al. 2002; Vartoukian et al. 2010; Pham and Kim 2012). Furthermore, using a cultivation-independent metagenomic approach, Banfield and colleagues recovered the complete genome of a TM7 representative (RAAC3) from an acetate-stimulated aquifer sediment (Kantor et al. 2013). Genomic analysis revealed that RAAC3 is missing pathways for biosynthesis of nucleotides, lipids, and amino acids and suggested that TM7 could be auxotrophic and metabolically dependent on other organisms (Kantor et al. 2013).
Human Oral TM7
TM7 bacteria are commonly found constituents of the human microbiome and are detected in various human body sites, including the gastrointestinal tract, skin, and female genital tract (Brinig et al. 2003; Eckburg et al. 2005; Fredricks et al. 2005; Gao et al. 2007). Additionally, 16S rRNA gene sequencing indicates that TM7 has been continuously prevalent in the human oral microbiome from Neanderthals (Weyrich et al. 2017) to modern-day Homo sapiens (Brinig et al. 2003). The association of TM7 with inflammatory mucosal diseases, such as vaginosis and inflammatory bowel disease, has been suggested from studies that found TM7 sequences differentially abundant in the disease state (Brinig et al. 2003; Fredricks et al. 2005; Kuehbacher et al. 2008). TM7 is typically found at low abundance—approximately 1% of the whole oral microbial population—based on culture-independent molecular analyses (Brinig et al. 2003; Podar et al. 2007); however, an increase of TM7 members (as high as 21% of the whole oral bacterial population in some cases) was detected in patients with mucosal infections (Rylev et al. 2011). Even greater abundances of TM7 were associated with gingivitis severity and periodontal disease (Paster et al. 2002; Brinig et al. 2003; Rylev et al. 2011; Liu et al. 2012; Kistler et al. 2013; Camelo-Castillo et al. 2015; Sousa et al. 2017; Nowicki et al. 2018). Certain oral TM7 phylotypes—such as TM7 clones AF125206 (HMT_356, 99.7% identity) and AY134895 (HMT_952, strain: TM7x, 98.9% identity)—can even be detected on or within the host crevicular epithelial cells (Paster et al. 2002). Moreover, the reduction in the abundance of TM7 can be consistently observed in plaque-induced gingivitis after treatment intervention (Huang et al. 2016). This clinical evidence suggests the correlation of TM7 with gingivitis and periodontitis and therefore justifies that several TM7 taxa have been included in the core microbiome associated with periodontitis (Abusleme et al. 2013). Despite these associations, no causative relationship between TM7 and periodontal diseases has been established, which warrants further study.
Oral TM7: Biodiversity and Biogeography
As high-throughput DNA sequencing studies accumulated, many inadvertently grouped these candidate phylum sequences into bins designated as “other” or “unclassified,” primarily as a result of lacking close representative sequences in databases. In addition, although some related sequences were deposited into databases, many were not classified as being phylogenetically related to TM7, which we now hypothesize to be driven by sequence divergence across the phylum. Thorough 16S rRNA gene sequence analyses from publicly available databases have subsequently revealed the diversity of TM7 across many environments. Development of the Human Oral Microbiome Database (Chen et al. 2010)—which contains deposited full-length 16S rRNA gene sequences with group-level designations—allows evaluation of the distribution of these TM7 sequences in data sets that originally did not report group-resolved TM7 hits. One study performed this type of contemporary “reanalysis” with recently developed oligotyping techniques (250-bp sequences grouped at 99% sequence identity), which were applied across the oral samples within the Human Microbiome Project high-throughput 16S rRNA marker gene data set (Eren et al. 2014). The distribution of the oligotypes reported indicated a best “hit” match with TM7 groups G1, G3, and G5, which were found across 9 oral sites in humans with no oral disease (Eren et al. 2014). Consequently, recent studies based on available 16S rRNA gene phylogeny revealed 6 major TM7 phylum groups (G1 to G6), which are detected in the oral cavity alone (Camanocha and Dewhirst 2014). Collectively, the groups vary in prevalence across subjects within the oral cavity, and the enrichment of some groups within specific anatomic sites is evident (Fig. 1). Over 2 decades past the first reported 16S rRNA sequence in 1996 (Rheims, Rainey, and Stackebrandt 1996), only genomes from the G1 group have been uncovered from single cells or metagenomic binning approaches. Very recent studies, however, uncovered additional genomes from the other diverse groups within the phylum from humans and mammals (McLean et al. 2018), including the dolphin oral cavity (Dudek et al. 2017).
Figure 1.
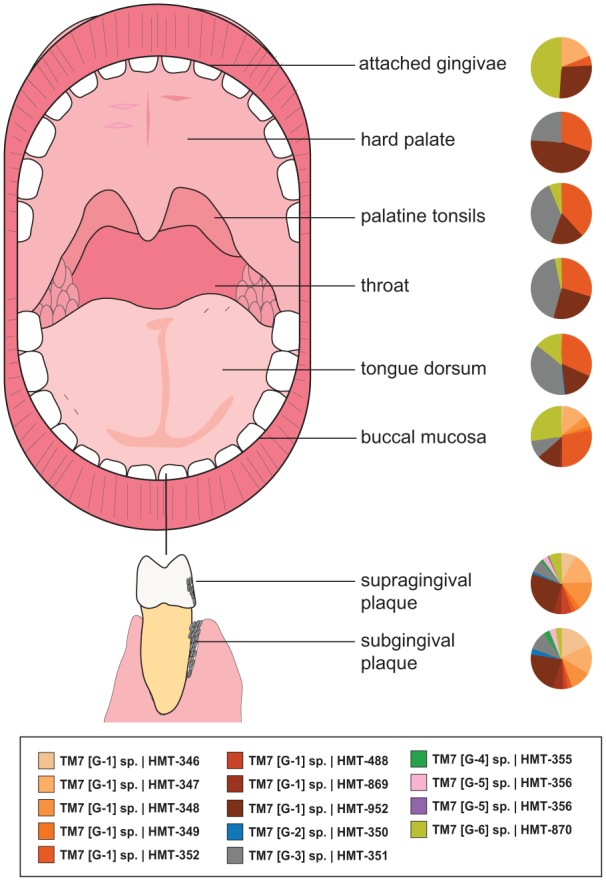
Saccharibacteria (TM7) biogeography in the oral cavity. With the accumulated availability of 16S rRNA gene sequences in databases, the phylogenetic diversity of the phylum results in at least 6 major taxonomic groups, which are detected at various levels in the oral cavity. Pie charts displaying the percentage distribution of the different groups within the phylum across oral cavity sites were derived from a reanalysis of the Human Microbiome Project data (https://hmpdacc.org/) with recently established reference sequences from the different groups.
TM7x: The First Cultivated TM7 Strain
TM7x: An Obligate Parasite That Infects Oral Bacteria
He et al. (2015; McLean et al. 2016) combined targeted antibiotic enrichment with a newly developed culturing medium that supports the growth of multiple TM7 phylotypes within an in vitro multispecies oral microbial community (Tian et al. 2010; Edlund et al. 2013). In doing so, the authors were able to achieve the successful coisolation of the first cultivable representative of TM7, Nanosynbacter lyticus type strain TM7x (HMT_952), with its host bacterium, Actinomyces odontolyticus actinosynbacter strain XH001.
Initial observations showed for the first time that TM7x has an ultrasmall cell size (200 to 300 nm) and was attached to a bacterial host. The TM7x genome is also reduced (about 700 genes) and lacks the ability to produce membrane lipids or nucleotides. Additionally, genes required for de novo biosynthesis of essential amino acids, nucleotides, and cofactors are lacking (He et al. 2015), which could explain the previous failed attempts in isolating TM7 species as a pure monoculture. TM7x has a high coding density genome and ranks among the smallest bacteria found in the human body in both genome size and gene count to date. TM7x falls within the G1 phylum group, which has several environmental G1 relatives; this group contains members from mixed sources, from mammals to deep groundwater (McLean et al. 2018). In comparison to the environmental genomes available, the oral TM7x genome is smaller and has therefore likely become reduced within mammalian hosts over time (He et al. 2015). The gene loss, however, is clustered mostly within a single genomic region. The rest of the genome is highly syntenic when compared with the metagenomically derived and closed environmental genomes isolated from an aquifer (RAAC3) as well as a sludge bioreactor (“Ca. Saccharibacteria aalborgensis”; He et al. 2015). The genome of TM7x bears some similarities to known bacterial symbionts in insects (McCutcheon and Moran 2011)—specifically, the highly reduced gene content with diminished functional capacity and auxotrophy for various vitamins as well as amino acids. Unlike TM7x, insect symbionts are generally reliant on a eukaryotic host instead of a free-living bacterium.
Further studies demonstrated a highly dynamic interaction between TM7x and its bacterial host in response to physical association and environmental conditions, such as oxygen level and nutritional availability. This is reflected by their reciprocal morphologic and physiologic changes during episymbiotic growth (Bor et al. 2016). Specifically, XH001 cells grow as short rods when alone but exhibit elongated cell morphology when physically associated with TM7x under nutrient-replete conditions. In contrast, upon starvation, TM7x-associated host cells manifest a variety of cell morphologies, including swollen cell body, clubbed ends, and even cell lysis. In addition, a large portion of the TM7x cells transform from ultrasmall cocci into more elongated cells under starvation condition (Bor et al. 2016). Transcriptomic gene profiling of the roughly 2,000 genes in XH001 revealed roughly 340 differentially regulated genes. A total of 70 genes were upregulated (>3-fold) when XH001 was physically associated with TM7x. Most interesting, of the 35 most upregulated genes, 8 (23%) encoded functions related to the stress response, and 8 more (23%) were involved in carbohydrate metabolism (He et al. 2015). The gene expression data aligned with the physical observation that the association of TM7x resulted in a slightly reduced growth in XH001, a characteristic feature of a parasitic relationship (Bor et al. 2016). Overall, the combined physiologic, genomic, and transcriptional characteristics led to the classification of TM7x as an obligate epibiont that is parasitic (an epiparasite; He et al. 2015).
Although epiparasitism is widespread between microorganisms and the domain Eukarya (Bidartondo et al. 2002), TM7x represents the first epiparasite discovered within the domain Bacteria that requires other bacteria as a host (Bor et al. 2016). Interestingly, similar symbiotic relationships have been reported in the domain Archaea. The first evidence was reported in 2002, when a nanosized hyperthermophilic archaeon, Nanoarchaeum equantum, was found to proliferate while attached to a specific archaeal host, a member of the genus Ignicoccus (Huber et al. 2002). Like TM7x, N. equitans is an obligate parasite with reduced genome capacity that cannot sustain host-free independent cellular growth. In high cell density, N. equitans is inhibitory and prevents Ignicoccus host growth (Jahn et al. 2008). Further studies indicated that these ultrasmall archaeal “parasites” can be found in various environmental settings, particularly in extreme environments, including deep-sea hydrothermal vents (Huber et al. 2002), acid mine drainage (Baker et al. 2006), and hot springs (Wurch et al. 2016). These nanoarchaeum species are members of a larger lineage termed DPANN Archaea, which are inferred to be mostly symbionts, based on genomic information similar to Candidate Phyla Radiation (CPR), and it was even shown that they share similar metabolic traits (Castelle and Banfield 2018).
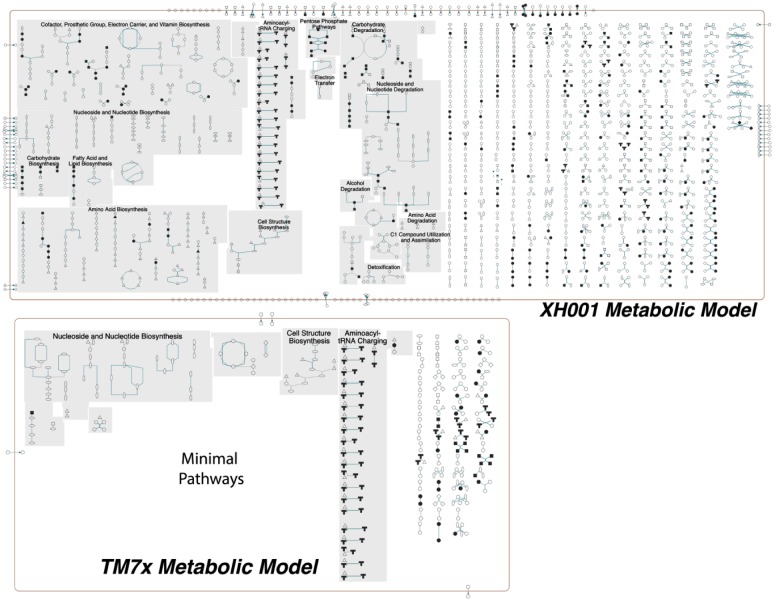
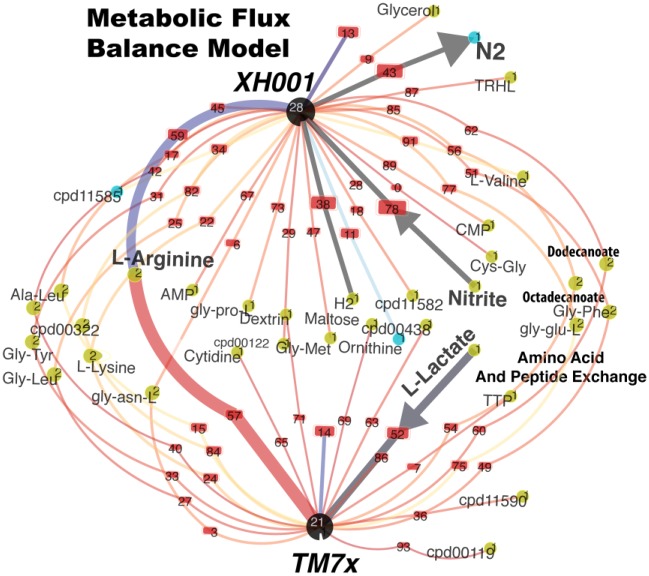
One of the key features of parasites is their metabolic dependency on their hosts, which is evident from the minimal metabolic pathways predicted in TM7x, as shown in Figure 2. These annotated genes in pathways are derived from our collaboration with BioCyc (Pathway/Genome Database Collection) and displayed with Pathway Tools (Caspi et al. 2016; Karp et al. 2016). XH001 genome is teeming with a multitude of pathways that can satisfy the cellular requirements of TM7x (Fig. 2). At the genome scale, the flux balance model (Orth et al. 2010)—which predicts the metabolic interactions of TM7x and XH001 in minimal media—indicates that lactate is likely the main carbohydrate source for TM7x derived from XH001 (Fig. 3). This was generated with the Seed Models (Henry et al. 2010), combined with a community metabolic model (Henry et al. 2016), and subsequently visualized in Kbase (Arkin et al. 2018). Additionally, there is a predicted flux of L-arginine as well as other amino acids, peptides (Gly-Glu-L, Gly-Phe), and fatty acid metabolites (Fig. 3). Validation of these predictions and the mechanisms by which TM7x transports these components can now be investigated with the strains available in stable coculture (Bor et al. 2018; He et al. 2015).
Figure 2.
Genome scale metabolic maps of the XH001 and TM7x interaction. Curated, genome scale metabolic models of XH001 and TM7x derived from collaboration with the Biocyc team, which is publically available (https://biocyc.org/). There is a major contrast in the predicted functions for each of these bacteria. Actinomyces odontolyticus XH001 has 123 pathways and 774 reactions and 501 predicted enzymes. The reduced genome parasite TM7x is auxotrophic for all essential amino acids and in comparison has only 20 pathways and 151 reactions with 101 predicted enzymes. XH001: Actinomyces odontolyticus strain XH00; TM7x: Nanosynbacter lyticus type strain TM7x.
Figure 3.
Flux balance analysis of the XH001 and TM7x interaction. Flux balance analysis using the Seed Model (http://modelseed.org/) enables a cursory prediction of the possible metabolic interaction between the two species in minimal media with glucose as the carbon source. Lactate under these conditions is the predicted substrate for TM7x metabolism. Colored nodes represent compounds and lines with numbers represent the predicted flux of metabolites. XH001: Actinomyces odontolyticus strain XH001; TM7x: Nanosynbacter lyticus type strain TM7x.
TM7x: A Bacterial Parasite That Enjoys Long-term Stable Relationship with Its Host
Once the epiparasitic relationship between TM7x and its bacterial host XH001 is established, it remains stable and easily maintained under nutrient-replete in vitro laboratory conditions (He et al. 2015; Bor et al. 2016). This long-term, stable relationship is possible because the TM7x/XH001 coculture appears to maintain a subpopulation of uninfected XH001 bacterial cells, which accounts for ~28% to 52% of the population on average (Bor et al. 2018). While the TM7x-infected host cell population in the coculture displays reduced growth and severely inhibited cell division, the uninfected XH001 population exhibits normal cell division and could serve as a reservoir to facilitate another infection cycle by TM7x via horizontal transmission (Bor et al. 2018).
Further knowledge was gained by infecting naïve host cells with isolated TM7x in the laboratory—specifically, an independent Actinomyces odontolyticus isolate from the human oral cavity (designated XH001n) that had not been exposed to TM7x. Bor et al. (2018) observed that XH001n suffered a dramatic cell death, which was a result of an overwhelmingly high number of TM7x cells (on average >50) infecting individual host cells. This virulent killing is transient, however, where the XH001n cells can quickly evolve a reduced susceptibility to TM7x infection and enter a stable, long-term epiparasitic relationship akin to XH001. This reduced susceptibility is hypothesized to be driven by rapid host evolution via genetic adaptation, where genome sequencing revealed that these stable host cells gained multiple mutations in the transporter and regulatory genes, potentially allowing partial protection from infection (Bor et al. 2018).
The rapidly developed long-term association between TM7x with XH001n was also observed through infection of a number of closely related Actinomyces species with TM7x (Bor et al. 2018). The ability of TM7x to quickly adapt to multiple hosts postinfection and the subsequent establishment of the stable epiparasitic relationship could allow TM7 species to associate with many bacterial lineages and persist within the oral cavity or other natural environmental niches. This is an intriguing evolutionary question that warrants further investigation.
TM7x: A Parasite with Benefits
As an obligate epiparasite, TM7x represents a burden to its host bacteria and exerts various negative impacts, including increased stress responses (Bor et al. 2016), reduced cellular growth rate and division (Bor et al. 2016), and even cell lysis under nutrient starvation conditions in batch culture (He et al. 2015). However, recent studies indicated that the association with TM7x is potentially beneficial to the host bacteria—for example, by promoting biofilm formation (Bedree et al. 2018).
In most ecologic habitats, including the human oral cavity, microbes form complex and highly structured multispecies communities known as biofilms. This structure is facilitated through an intricate extracellular matrix consisting of polysaccharides, proteins, DNA, and lipids, and it is an essential part of the many microbial lifecycles in the oral cavity (Bowen et al. 2018). It protects oral bacteria from saliva flow, daily oral hygiene interventions, and clearance by the human immune system, thereby allowing persistence within the oral cavity (Sanz et al. 2017). Bedree et al. (2018) recently demonstrated that the association of TM7x with XH001 promotes biofilm formation partially via autoinducer-2 (AI-2) quorum sensing (QS). This was validated by chromosomal deletions of key AI-2 QS genes, luxS and lsrB, encoding the AI-2 QS molecule synthase and binding receptor, respectively, in XH001, which resulted in biofilm formation deficiencies of TM7x-associated XH001 (Bedree et al. 2018). In addition to TM7x and its host withstanding physical disturbances within oral cavity, a plausible advantage could be that of evading the immune system in vivo by directly reducing inflammation via biofilm formation. While this recent study did not directly evaluate this possibility in the oral cavity, existing literature reports that biofilm-inducing bacteria impede activation of the immune system (Donlan and Costerton 2002; Domenech et al. 2013) and diminish the proinflammatory response by reducing TNF-α production (Thurlow et al. 2011). This is consistent with the observation that TM7x association repressed XH001-induced TNF-α expression in macrophages, which suggests that TM7x may prevent detection of XH001 by macrophages or possibly directly suppress TNF-α expression in macrophages (He et al. 2015). Altogether, these data are suggestive of ultrasmall bacteria potentially having the capacity to modulate the normal host immune response (He et al. 2015). The full molecular mechanism behind this QS-regulated, enhanced biofilm formation phenotype in TM7x-associated XH001 remains to be elucidated. Furthermore, determining if biofilm formation could provide evolutionary fitness in the survival of TM7x’s bacterial hosts merits further investigation.
Conventionally, a parasitic relationship is commonly defined as one partner gaining benefit at the expense of its counterpart. While recent evaluation of the epiparasitic relationship between TM7x and XH001 corroborates the increasing lines of evidence showing that while parasites negatively affect the growth of their hosts, they could offer evolutionary advantages by enabling the persistence of their host species (Dunn et al. 2008). Thus, in assessing the benefits and disadvantages of parasitism, the outcome of the relationship should not solely be studied in isolation but rather in a natural context.
Beyond TM7x: Into the Ultrasmall World
TM7 and Candidate Phyla Radiation
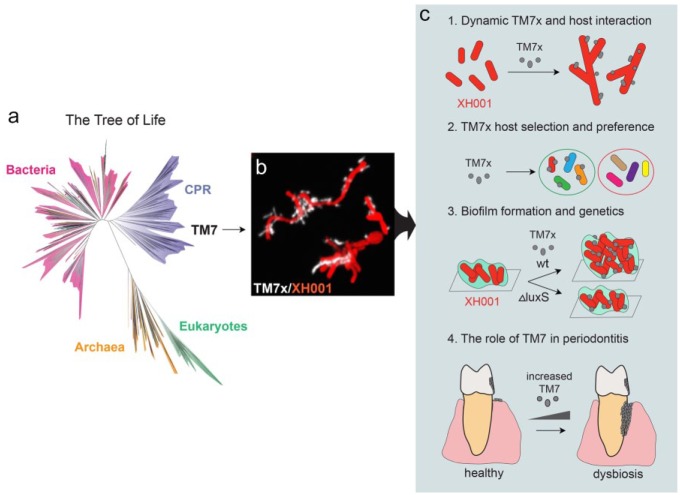
CPR is a newly described bacterial major lineage (superphylum) containing >70 phyla, potentially accounting for >25% of bacterial diversity, and until recently, no cultured representative had been reported (Brown et al. 2015; Hug et al. 2016; Castelle and Banfield 2018). This discovery has greatly expanded the view of microbial diversity but simultaneously engendered fundamental questions that remain unanswered regarding their biology and their potential influence on microbial ecology. TM7, in addition to the SR1 and GN02 phyla, is among the only CPR that is detected within the human microbiome and specifically enriched in the oral cavity (Camanocha and Dewhirst 2014). Other members of the TM7 and CPR share characteristics with TM7x, including highly reduced genomes, restricted metabolic capacities, and an inferred ultrasmall cell size (Kantor et al. 2013; Brown et al. 2015; Luef et al. 2015; McLean et al. 2018). Strikingly, since most available genomes reveal numerous absent biosynthetic pathways across the CPR lineage, they are predicated to have a symbiotic lifestyle similar to TM7x (Kantor et al. 2013; McLean et al. 2013; Brown et al. 2015; Castelle and Banfield 2018). TM7x represents the first cultivated member of CPR and has thus far enabled detailed studies of these ultrasmall epiparasites, such as their dynamic interaction with their hosts, their host preference and range, their biofilm formation, and their pathogenic potential (Fig. 4). As the impetus for studying the CPR increases (Brown et al. 2015; Attar 2016; Burstein et al. 2016; Castelle et al. 2017; Danczak et al. 2017; Castelle and Banfield 2018), TM7x/XH001 may continue to provide fundamental knowledge on the underlying mechanisms governing the lifestyle of bacteria with reduced genomes, such as those belonging to the CPR group.
Figure 4.
TM7x represents the first CPR bacteria cocultivated with its host. (a) Current view of the tree of life highlights TM7 and CPR (tree inferred from concatenated ribosomal gene data set provided by Hug et al. 2016). (b) Fluorescence in situ hybridization image shows the epiparasitic relationship between TM7x and its bacterial host XH001. (c) TM7x/XH001 provides a better understanding of bacterial epiparasitic interaction: (1) a detailed mechanistic understanding of the dynamic parasitic interaction between TM7x and its host bacterium XH001; (2) the host selection and host range of TM7x; (3) the impact of interaction on bacterial physiology; and (4) pathogenic potential. CPR, Candidate Phyla Radiation. XH001: Actinomyces odontolyticus strain XH00; TM7x: Nanosynbacter lyticus type strain TM7x.
Novel Approach for Isolating and Cultivating TM7 Strains
Cultivation of the first TM7x strain with its bacterial host XH001 has provided invaluable knowledge. The requirement of TM7x to grow only with a host bacterium and the discovery of the ability of TM7x to horizontally infect new host bacteria (Bor et al. 2018) have recently led to a novel host “baiting” methodology (http://www.homd.org/doc/Saccharibacteria Isolation and Cultivation.pdf). The efficacy of this isolation methodology in part prompted organization of the inaugural Forsyth Institute symposium “The Uncultivable Bacteria” (October 2018). The conclusion of the symposium consisted of a hands-on workshop demonstrating TM7 bacteria isolation and cultivation from the human oral cavity. This symposium enabled exposure and dissemination of critical cultivation knowledge to the global research community. This cultivation methodology has resulted in stable cultures of several new TM7 strains of different species (unpublished data), greatly expanding the current isolated TM7 bacterial representatives. The addition and sequencing of new cultivated TM7 strains are pivotal to elucidating the molecular underpinnings of this novel parasitic mechanism as well as the universal TM7 bacterial ecology. It is anticipated that the attendees at this symposium and abroad will implement the validated method to isolate even more TM7 bacteria from diverse environments. This method removed a major roadblock and paved the way for cultivating and studying TM7 strains, and it may enable isolation of other oral candidate phyla that are otherwise difficult to cultivate, including “yet to be cultivated” species from the GN02 and SR1 phyla in humans.
Future Perspectives
The obligate epiparasitic relationship between TM7x and XH001 represents a novel yet presumably common and long-established interspecies interaction in the oral microbiota, as suggested by the presence of CPR from ancient to modern humans (Camanocha and Dewhirst 2014; Weyrich et al. 2017; Baker et al. 2018). There is strong evidence that CPR organisms, particularly TM7, interact with the core members of the microbiome, such as Actinomyces, and play a critical yet poorly understood function in the development of the human community composition in health and disease (Abusleme et al. 2013; Costalonga and Herzberg 2014; Baker et al. 2017). The distinct interactions between TM7 and their bacterial hosts may have a considerable impact on oral microbial ecology at various levels. Depending on local microenvironments, TM7 species could modulate the oral microbiome structure hierarchy and functionality through affecting their bacterial host’s physiology, inhibiting host growth dynamics, or influencing the relative abundance of the host via direct killing. It is also possible that bacterial species carrying TM7 parasites could invade other hosts or host niches via parasite-mediated competition, effectively using the parasites as biological weapons, similar to how human intestinal bacteria, such as certain Enterococcus faecalis strains, use phage as a bioweapon to kill off competitors (Duerkop et al. 2012). Furthermore, the observed TM7x repression of the XH001-induced TNF-α mRNA expression in macrophages implies that interactions between TM7 species and their bacteria are likely to be more complex than a simple metabolic dependency and may affect the human host responses (He et al. 2015), thus indirectly modulating human oral microbiome. While accumulating evidence has established enrichment and subsequent association of these ultrasmall parasites of bacteria with disease states, the next critical step is to investigate the causation between TM7 and oral infectious diseases. As interest in the study of ultrasmall bacteria increases, the isolation and investigation of additional TM7 strains and other CPR bacteria will provide much-needed knowledge on the biology of this distinct bacterial lineage and will confirm whether any of the findings obtained from studies on the human associated TM7x-XH001 pair are generalizable.
Author Contributions
B. Bor, contributed to acquisition, analysis, and interpretation, drafted and critically revised the manuscript; J.K. Bedree, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; W. Shi, contributed to design, data analysis, and interpretation, critically revised the manuscript; J.S. McLean, X. He, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
Financial support is acknowledged from the National Institute of Dental and Craniofacial Research of the National Institutes of Health under awards 1R01DE023810, 1R01DE020102, 1R01D E026186 (to X.H., J.S.M., and W.S.); and 5T90DE022734, F32DE025548, and 1K99DE027719 (to B.B.) and F31DE026057 (J.K.B.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Abrams M, Barton DE, Vandaei E, Romero D, Caldwell A, Ouverney CC. 2012. Genomic characteristics of an environmental microbial community harboring a novel human uncultivated TM7 bacterium associated with oral diseases. Open Access Sci Rep. 1:276. [Google Scholar]
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 31(6):533–538. [DOI] [PubMed] [Google Scholar]
- Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F, Canon S, et al. 2018. KBase: the United States Department of Energy systems biology knowledgebase. Nat Biotechnol. 36(7):566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar N. 2016. Bacterial evolution: CPR breathes new air into the tree of life. Nat Rev Microbiol. 14(6):332–333. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, Banfield JF. 2006. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 314(5807):1933–1935. [DOI] [PubMed] [Google Scholar]
- Baker JL, Bor B, Agnello M, Shi W, He X. 2017. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 25(5):362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Lindsay EL, Faustoferri RC, To TT, Hendrickson EL, He X, Shi W, McLean JS, Quivey RG., Jr. 2018. Characterization of the trehalose utilization operon in Streptococcus mutans reveals that the TreR transcriptional regulator is involved in stress response pathways and toxin production. J Bacteriol. 200(12):e00057-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedree JK, Bor B, Cen L, Edlund A, Lux R, McLean JS, Shi W, He X. 2018. Quorum sensing modulates the epibiotic-parasitic relationship between Actinomyces odontolyticus and its Saccharibacteria epibiont, a Nanosynbacter lyticus strain, TM7x. Front Microbiol. 9:2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Redecker D, Hijri I, Wiemken A, Bruns TD, Dominguez L, Sersic A, Leake JR, Read DJ. 2002. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature. 419(6905):389–392. [DOI] [PubMed] [Google Scholar]
- Bor B, McLean JS, Foster KR, Cen L, To TT, Serrato-Guillen A, Dewhirst FE, Shi W, He X. 2018. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc Natl Acad Sci U S A. 115(48):12277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, Poweleit N, Bois JS, Cen L, Bedree JK, Zhou ZH, Gunsalus RP, Lux R, McLean JS, He X, et al. 2016. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont Actinomyces. Microb Ecol. 71(1):243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Burne RA, Wu H, Koo H. 2018. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26(3):229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. 2003. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 69(3):1687–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain bacteria. Nature. 523(7559):208–211. [DOI] [PubMed] [Google Scholar]
- Burstein D, Sun CL, Brown CT, Sharon I, Anantharaman K, Probst AJ, Thomas BC, Banfield JF. 2016. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat Commun. 7:10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camanocha A, Dewhirst FE. 2014. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J Oral Microbiol. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, Tomas I. 2015. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol. 6:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44(D1):D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, Banfield JF. 2018. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 172(6):1181–1197. [DOI] [PubMed] [Google Scholar]
- Castelle CJ, Brown CT, Thomas BC, Williams KH, Banfield JF. 2017. Unusual respiratory capacity and nitrogen metabolism in a parcubacterium (OD1) of the Candidate Phyla Radiation. Sci Rep. 7:40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 162:22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danczak RE, Johnston MD, Kenah C, Slattery M, Wrighton KC, Wilkins MJ. 2017. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome. 5(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, Buckley CM, Davis IJ, Bennett ML, Marshall-Jones ZV. 2012. The canine oral microbiome. PLoS One. 7(4):e36067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinis JM, Barton DE, Ghadiri J, Surendar D, Reddy K, Velasquez F, Chaffee CL, Lee MC, Gavrilova H, Ozuna H, et al. 2011. In search of an uncultured human-associated TM7 bacterium in the environment. PLoS One. 6(6):e21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech M, Ramos-Sevillano E, Garcia E, Moscoso M, Yuste J. 2013. Biofilm formation avoids complement immunity and phagocytosis of Streptococcus pneumoniae. Infect Immunity. 81(7):2606–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 15(2):167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek NK, Sun CL, Burstein D, Kantor RS, Aliaga Goltsman DS, Bik EM, Thomas BC, Banfield JF, Relman DA. 2017. Novel microbial diversity and functional potential in the marine mammal oral microbiome. Curr Biol. 27(24):3752–3762.e3756. [DOI] [PubMed] [Google Scholar]
- Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A. 109(43):17621–17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DW, Segar ST, Ridley J, Chan R, Crozier RH, Yu DW, Cook JM. 2008. A role for parasites in stabilising the fig-pollinator mutualism. PLoS Biol. 6(3):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science. 308(5728):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Yang Y, Hall AP, Guo L, Lux R, He X, Nelson KE, Nealson KH, Yooseph S, Shi W, et al. 2013. An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome. 1(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Yang Y, Yooseph S, Hall AP, Nguyen DD, Dorrestein PC, Nelson KE, He X, Lux R, Shi W, et al. 2015. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J. 9(12):2605–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A. 111(28):E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari BC, Binnerup SJ, Gillings M. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol. 71(12):8714–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN, Fiedler TL, Marrazzo JM. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 353(18):1899–1911. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 104(8):2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 112(1):244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CS, Bernstein HC, Weisenhorn P, Taylor RC, Lee JY, Zucker J, Song HS. 2016. Microbial community metabolic modeling: a community data-driven network reconstruction. J Cell Physiol. 231(11):2339–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 28(9):977–982. [DOI] [PubMed] [Google Scholar]
- Huang S, Li Z, He T, Bo C, Chang J, Li L, He Y, Liu J, Charbonneau D, Li R, et al. 2016. Microbiota-based signature of gingivitis treatments: a randomized study. Sci Rep. 6:24705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. 2002. A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature. 417(6884):63–67. [DOI] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. 2016. A new view of the tree of life. Nat Microbiol. 1:16048. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 180(18):4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl Environ Microbiol. 67(1):411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn U, Gallenberger M, Paper W, Junglas B, Eisenreich W, Stetter KO, Rachel R, Huber H. 2008. Nanoarchaeum equitans and Ignicoccus hospitalis: new insights into a unique, intimate association of two archaea. J Bacteriol. 190(5):1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. 2013. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 4(5):e00708–00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Latendresse M, Paley SM, Krummenacker M, Ong QD, Billington R, Kothari A, Weaver D, Lee T, Subhraveti P, et al. 2016. Pathway tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform. 17(5):877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindaichi T, Yamaoka S, Uehara R, Ozaki N, Ohashi A, Albertsen M, Nielsen PH, Nielsen JL. 2016. Phylogenetic diversity and ecophysiology of candidate phylum Saccharibacteria in activated sludge. FEMS Microbiol Ecol. 92(6):fiw078. [DOI] [PubMed] [Google Scholar]
- Kistler JO, Booth V, Bradshaw DJ, Wade WG. 2013. Bacterial community development in experimental gingivitis. PLoS One. 8(8):e71227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher T, Rehman A, Lepage P, Hellmig S, Folsch UR, Schreiber S, Ott SJ. 2008. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 57:1569–1576. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J Dent Res. 82(5):338–344. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 124(4):837–848. [DOI] [PubMed] [Google Scholar]
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, et al. 2012. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 7(6):e37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe BA, Marsh TL, Isaacs-Cosgrove N, Kirkwood RN, Kiupel M, Mulks MH. 2012. Defining the “core microbiome” of the microbial communities in the tonsils of healthy pigs. BMC Microbiol. 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luef B, Frischkorn KR, Wrighton KC, Holman HY, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, et al. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 6:6372. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, et al. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 104(29):11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- McLean JS, Bor B, To TT, Liu Q, Kearns KA, Solden LM, Wrighton KC, He X, Shi W. 2018. Evidence of independent acquisition and adaption of ultra-small bacteria to human hosts across the highly diverse yet reduced genomes of the phylum Saccharibacteria. BioRxiv. doi: 10.1101/258137 [DOI] [Google Scholar]
- McLean JS, Liu Q, Bor B, Bedree JK, Cen L, Watling M, To TT, Bumgarner RE, He X, Shi W. 2016. Draft genome sequence of Actinomyces odontolyticus subsp. actinosynbacter strain xh001, the basibiont of an oral TM7 epibiont. Genome Announc. 4(1):e01685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, et al. 2013. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci U S A. 110(26):E2390–E2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Hongoh Y, Usami R, Kudo T, Ohkuma M. 2005. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol Ecol. 54(2):247–255. [DOI] [PubMed] [Google Scholar]
- Nishihara T, Koseki T. 2004. Microbial etiology of periodontitis. Periodontol 2000. 36:14–26. [DOI] [PubMed] [Google Scholar]
- Nowicki EM, Shroff R, Singleton JA, Renaud DE, Wallace D, Drury J, Zirnheld J, Colleti B, Ellington AD, Lamont RJ, et al. 2018. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. MBio. 9(2):e00575-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BO. 2010. What is flux balance analysis? Nat Biotechnol. 28(3):245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Russell MK, Alpagot T, Lee AM, Boches SK, Galvin JL, Dewhirst FE. 2002. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Ann Periodontol. 7(1):8–16. [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, et al. 2009. The NIH Human Microbiome Project. Genome Res. 19(12):2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VH, Kim J. 2012. Cultivation of unculturable soil bacteria. Trends Biotechnol. 30(9):475–484. [DOI] [PubMed] [Google Scholar]
- Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA, Holland T, Cotton D, Hauser L, Keller M. 2007. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 73(10):3205–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheims H, Rainey FA, Stackebrandt E. 1996. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 17(3–4):159–169. [Google Scholar]
- Rheims H, Sproer C, Rainey FA, Stackebrandt E. 1996. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 142:2863–2870. [DOI] [PubMed] [Google Scholar]
- Rylev M, Bek-Thomsen M, Reinholdt J, Ennibi OK, Kilian M. 2011. Microbiological and immunological characteristics of young Moroccan patients with aggressive periodontitis with and without detectable Aggregatibacter actinomycetemcomitans JP2 infection. Mol Oral Microbiol. 26(1):35–51. [DOI] [PubMed] [Google Scholar]
- Sanz M, Beighton D, Curtis MA, Cury JA, Dige I, Dommisch H, Ellwood R, Giacaman RA, Herrera D, Herzberg MC, et al. 2017. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases: consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J Clin Periodontol. 44 Suppl 18:S5–S11. [DOI] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. 2012. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13(6):R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa V, Nibali L, Spratt D, Dopico J, Mardas N, Petrie A, Donos N. 2017. Peri-implant and periodontal microbiome diversity in aggressive periodontitis patients: a pilot study. Clin Oral Implants Res. 28(5):558–570. [DOI] [PubMed] [Google Scholar]
- Starr EP, Shi S, Blazewicz SJ, Probst AJ, Herman DJ, Firestone MK, Banfield JF. 2018. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome. 6(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 186(11):6585–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, He X, Torralba M, Yooseph S, Nelson KE, Lux R, McLean JS, Yu G, Shi W. 2010. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol. 25(5):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. 2010. Strategies for culture of “unculturable” bacteria. FEMS Microbiol Lett. 309(1):1–7. [DOI] [PubMed] [Google Scholar]
- Weyrich LS, Duchene S, Soubrier J, Arriola L, Llamas B, Breen J, Morris AG, Alt KW, Caramelli D, Dresely V, et al. 2017. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 544(7650):357–361. [DOI] [PubMed] [Google Scholar]
- Winsley TJ, Snape I, McKinlay J, Stark J, van Dorst JM, Ji M, Ferrari BC, Siciliano SD. 2014. The ecological controls on the prevalence of candidate division TM7 in polar regions. Front Microbiol. 5:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurch L, Giannone RJ, Belisle BS, Swift C, Utturkar S, Hettich RL, Reysenbach AL, Podar M. 2016. Genomics-informed isolation and characterization of a symbiotic nanoarchaeota system from a terrestrial geothermal environment. Nat Commun. 7:12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. 2002. Cultivating the uncultured. Proc Natl Acad Sci U S A. 99(24):15681–15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Bai L, Goel N, Bailey A, Jang CJ, Bushman FD, Meerlo P, Dinges DF, Sehgal A. 2017. Human and rat gut microbiome composition is maintained following sleep restriction. Proc Natl Acad Sci U S A. 114(8):E1564–E1571. [DOI] [PMC free article] [PubMed] [Google Scholar]