Abstract
The Janus kinases (JAKs) consist of four similar tyrosine kinases and function as key hubs in the signaling pathways that are implicated in both innate and adaptive immunity. Among the four members, JAK3 is probably the more attractive target for treatment of inflammatory diseases because its inhibition demonstrates the greatest immunosuppression and most profound effect in the treatment of such disorders. Although many JAK3 inhibitors are already available, certain shortcomings have been identified, mostly acquired drug resistance or unwanted side effects. To discover and identify new promising lead candidates, in this study, the structure of JAK3 (3LXK) was obtained from the Protein Data Bank and used for simulation modeling and protein-ligand interaction analysis. The ~36,000 Chinese herbal compounds obtained from TCM Database@Taiwan were virtually screened by AutoDock Vina docking program and filtered with Lipinski's Rules and ADME/T virtual predictions. Because of high occurrence of fake hits during docking, we selected 12 phytochemicals which have demonstrated modulating JAKs expressions among the top 50 chemicals from docking results. To validate whether these compounds are able to directly mediate JAK3 kinase, we have investigated the inhibitory activity using enzymatic activity assays, western blot, and HEK 293 cell STAT5 transactivity assays. The molecular analysis included docking and molecular dynamics (MD) simulations in order to investigate structural conformations and to explore the key amino acids in the interaction between JAK3 kinase and its putative ligands. The results demonstrated that Cryptotanshinone, Icaritin, and Indirubin exhibited substantial inhibitory activity against JAK3 kinase in vitro. The results also provide binding models of the protein-ligand interaction, detailing the interacting amino acid residues at the active ATP-binding domains of JAK3 kinase. In conclusion, our work discovered 3 potential natural inhibitors of JAK3 kinase and could provide new possibilities and stimulate new insights for the treatment of JAK3-targeted diseases.
1. Introduction
The Janus kinases (JAKs) are a family of four similar tyrosine kinases (JAK1, JAK2, JAK3, and TYK2) that function as key hubs in the signaling pathways and receptors for cytokines implicated in both innate and adaptive immunity [1, 2]. In recent years, JAKs have been identified as attractive therapeutic targets for inflammatory diseases in the fields of both biotechnology and pharmaceuticals. Among the four JAK family members, JAK3 is probably the more attractive target because it exhibits the greatest immunosuppression and causes the most profound effect in the treatment of inflammatory diseases [1, 3]. However, the current approved JAK3 inhibitors showed undesirable side effects including anaemia and neutropenia [4, 5].
Chinese herbal compounds have been recognized as a tremendous contribution to drug development because they are generally well tolerated and have better biocompatibility and diversity in molecular structure and bioactive substructures [6].
Thus, Chinese herbal chemicals could be developed as promising lead candidates for the inhibitors against JAK3. The structure of JAK3 was obtained from the Protein Data Bank (PDB ID code: 3LXK) and used for simulation modeling, and protein-ligand interaction analysis. The ~36,000 Chinese herbal compounds obtained from TCM Database@Taiwan [7] were virtually screened by AutoDock Vina docking program [8] and filtered with Lipinski's Rules [9, 10] and ADME/T virtual predictions [11].
Although docking simulation has been commonly used in virtual screening for drugs development, the docking results sometimes have been questioned because of high occurrence of fake top potent hits [12, 13]. It has been reported that selecting candidates from validated traditional Chinese medicines could effectively improve the success rate of drug development [14]. Therefore, to discover and identify the most potential candidates of JAK3 inhibitor, we carefully evaluated and selected 12 Chinese herbal chemicals among the top 50 chemicals from docking results. The selected 12 phytochemicals, which modulate the expression of JAKs and have thus been effective in the treatment of inflammatory diseases, have been demonstrated using western blot analysis or immunohistochemistry assays through searches on the Web of Science Core Collection. These phytochemicals were tested for their ability to bind to JAK3 using biological activity assays. Subsequently, based on the results of the bioactivity assays, the 3 compounds with the highest JAK3 affinity (IC50<100μM) were selected and their interaction with JAK3 kinase was investigated by docking computational again and evaluated by molecular dynamics (MD) simulations to provide further insight into the probable JAK3 protein-ligand interactions (the 12 chemicals were listed in Table 1).
Table 1.
Chinese herbal compounds analyzed.
| No. | Phytochemical | Plant Species | Reference |
|---|---|---|---|
| 1 | Andrographolide | Andrographis paniculata L. | [15] |
| 2 | Arctigenin | Fructus Arctii L. | [16] |
| 3 | Baicalein | Scutellaria baicalensis Fisch. | [17] |
| 4 | Berbamine | Berberis amurensis Rupr. | [18] |
| 5 | Cryptotanshinone | Salvia miltiorrhiza Bunge. | [19] |
| 6 | Curcumin | Curcuma longa L. | [20] |
| 7 | Ginkgolide B | Ginkgo biloba L. | [21] |
| 8 | Icaritin | Epimedium grandiflorum C. Morren | [22] |
| 9 | Indirubin | Strobilanthes formosanus Moore | [23] |
| 10 | Quercetin | - | [24] |
| 11 | Salvianolic acid B | Salvia miltiorrhiza Bunge. | [25] |
| 12 | Ursolic acid | - | [26] |
2. Materials and Methods
2.1. Reagents and Materials
Twelve Chinese herbal compounds modulating the JAK kinase were selected from the first-round docking results and the published literature, purchased and tested, as detailed in Table 1.
Monoclonal Antiphosphotyrosine–Peroxidase antibody raised in mouse (catalog # A5964), synthetic polypeptide poly(Glu, Tyr), sodium salt, 20-50 kDa (catalog # P0275), globulin bovine serum albumin (BSA) (catalog #A3059), DL-dithiothreitol (catalog #D9779), adenosine-50-triphosphate disodium salt hydrate (ATP) (catalog # A2383), and sodium orthovanadate (Na3VO4) (catalog #S6508) were purchased from Sigma-Aldrich GmbH, Germany. JAK3 kinase domain (amino acids 781 to 1124) was purchased from Millipore, UK (catalog #D8EN006U-H). 2-[4-(2-hydroxyethyl)piperazin-1-yl] ethanesulfonic acid (HEPES) (catalog # L1613) was obtained from Biochrom GmbH, Germany. MgCl2·6H2O (catalog # 105833) was purchased from Merck KGA, Germany. 3,3',5,5'-tetramethylbenzidine (TMB) peroxidase reagent (catalog # 555214) was obtained from BD Biosciences Europe. Nunc MaxiSorp® microtiter plates (catalog # 442404) were obtained from Fisher Scientific GmbH, Germany. Plasmids pGL4.52-luc2P/STAT5RE (catalog # E465A) and pRL-TK (catalog # E2241) and the dual luciferase assay kit (catalog # E1980) were purchased from Promega, USA. Lipofectamine 2000 (catalog # 11668019) was obtained from Invitrogen, USA. HEK 293 cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences. Ultrapure water produced using a Millipore Synergy UV water purification device was used throughout the study. All reagents were of ultrapure grade.
2.2. JAK3 Enzymatic Activity Assay
Kinase activity was determined using an ELISA-based assay according to the procedure reported by Bauer [27].
Kinase buffer (KB), containing 100 mM HEPES, 10 mM MgCl2, 4 mM dithiothreitol, 0.1 mM Na3VO4, and 5 μM ATP, was prepared in ultrapure water, the pH adjusted to 6.8–6.9 and used to prepare JAK3 kinase solution at a concentration of 100 ng/mL and for dilution of the test compounds. The synthetic polypeptide poly (Glu, Tyr) served as the substrate for kinase JAK3. It was dissolved in PBS at a concentration of 10 μg/mL and then each well of a 96-well microplate was coated by adding 100 μL of this working solution. The plate was sealed and incubated overnight at 4°C. Into the wells of the assay plate, 50 μL of either pure KB, KB containing JAK3 kinase (100ng/mL of final concertation), or KB containing JAK3 kinase and a test compound were added followed by incubation at 37°C for one hour. One hundred μL of antiphosphotyrosine–peroxidase conjugated antibody (diluted 1:10 000) were added to each well of the plate, which was incubated at 37°C for an additional hour. Finally, TMB substrate reagent was added to the wells and incubated at room temperature for 5 min. The reaction was blocked by the addition of 25 μL of 1 M sulfuric acid. The optical density (OD) of the wells was measured immediately at 450 nm using a microplate reader. The quantity of kinase substrate that was phosphorylated was proportional to JAK3 kinase activity. CP-690,550 was selected as positive control. The OD of self-stimulation (STIM) without inhibitors indicated the maximum phosphorylation, and the OD of plain KB (NSB) was considered as negative control. The inhibition activity is calculated as the following equation: Inhibition(%)= 100%×[1-(ODSample-ODNSB)/(ODSTIM-ODNSB)].
2.3. Western Blot Analysis
To validate whether the hit compounds are able to mediate JAK3 kinase in cells, we investigated the JAK3 phosphorylation using western blot in cellulo.
HEK 293 cells were seeded in 6-well plates at a density of 5×104 cells/well in 2 ml cell culture per well. Cells were treated with Cryptotanshinone, Icaritin, and Indirubin at 25 and 50 μM concentrations for 1 h prior to stimulation with 20 ng/mL of IL-2 for 3 h and were harvested for phoso-JAK3 signaling determination. Cells without drug treatment were selected and used as control group. The above analyses were performed in three independent experiments.
Cells of all the groups were lysed and total proteins were extracted using RIPA lysis buffer (Beyotime, Shanghai, China) plus the PhosStop (Roche, Indianapolis, IN, USA) phosphatase inhibitors and Complete Ultra protease inhibitors (Roche). Equal amounts of protein were used to perform electrophoresis on a 12% SDS-polyacrylamide gel and subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% BSA in Tris-buffered saline (TBS) containing 0.1% Tween-20 for 1 h at room temperature, the membranes were incubated overnight at 4°C with the primary antibodies (1:1000). The primary antibodies, rabbit monoclonal phospho-JAK3, were purchased from Cell Signaling Technology (BSN, USA). Membranes were washed three times using Tris-buffered saline Tween-20 (TBST) for 5 min and then incubated for 2 h at room temperature with horseradish peroxidase-conjugated secondary antibody (1:1,000; Boster, Wuhan, China). After three TBST washes, the target proteins bands were detected by enhancing Pierce ECL Plus (ThermoFisher, Rockford, IL, USA) reagents and exposed to X-ray film (Eastman Kodak, Rochester, NY, USA) for the visualization. GAPDH was used as the internal loading control for protein normalization.
2.4. Stat5 Transactivation Activity
Activation of JAKs can induce constitutive expression of STAT5 that can be detected during an inflammatory response. STAT5 is one of the principal downstream signaling proteins induced by activated JAK3 and serves a critical role in inflammation and cell survival. To investigate STAT5 regulation by the Chinese herbal compounds, transcriptional activity of STAT5 was quantified by the use of a pSTAT5-Luc plasmid.
HEK 293 cells were transiently cotransfected with STAT5-responsive luciferase reporter pGL4.52-luc2P/STAT5RE/Hygro (firefly luciferase) and pRL-TK (renilla luciferase) plasmids, the latter acting as a transfection control [28, 29]. HEK 293-STAT5-Luc cells that were proliferating exponentially were seeded into the wells of a 24-well plate at a concentration of 5×104 cells/well. Twenty-four h after transfection, the cells were preincubated with an appropriate concentration of each Chinese herbal compound for 1 h. The cells in each well were then stimulated with 20 ng/mL of IL-2 for an additional 3 h, after which passive lysis buffer was added (50 μL/well). The plate was then incubated for 15 min on a shaking platform. A dual luciferase assay kit was used to measure luciferase expression using a GloMAX 20/20 luminometer. Transcriptional activity corresponded to the ratio of firefly luciferase luminescence to that of renilla luciferase.
2.5. Computational Molecular Simulation
The Chinese herbal compounds were employed to explore their interactions with JAK3 protein according to Rashid's methods [30].
2.5.1. JAK3 Kinase Protein Structures
The crystal structure of JAK3 kinase (PDB ID: 3LXK) was obtained from the Protein Data Bank (PDB) (http://www.rcsb.org). Energy minimization of the structure was employed using Amber's force field and conjugate gradient algorithms provided in UCSF Chimera 1.5.6 software [31].
2.5.2. Ligand Preparation
In the first-round docking simulation, the Chinese herbal compounds (~40,000) were obtained from the TCM Database@Taiwan provided by Dr. Chen's, and employed using AutoDock 4.2 tools for JAK3 structure-based virtual screening.
In the second-round docking simulation, three Chinese herbal compounds identified as inhibiting JAK3 demonstrating high JAK3 affinity (IC50<100 μM) were selected and compared with CP690,550 (2-cyano-3-(3,4-dihydroxyphenyl)-N-(phenylmethyl)-2-propenamide), a positive control to investigate the ligand-binding interaction of JAK3–PTK domains.
2.5.3. Docking Simulations
Molecular docking simulations of JAK3 kinase were conducted using AutoDock 4.2 and AutoDock Vina software, (Scripps Research Institute) according to published methods [32, 33]. It was anticipated that the docking results should provide the primary binding conformations of JAK3 with its ligands prior to MD simulation, after which optimized binding sites and interactions would be ascertained. Firstly, all hydrogen atoms had to be added to either the protein or the ligands. However, only polar hydrogen coordinates are utilized during docking simulations. Gasteiger-Marsili atomic charges are incorporated into the calculations of electrostatic interactions and desolvation energies by AutoDock. During docking simulations all rotatable bonds of the chemical ligands were allowed free choice of torsional degrees of freedom. Rigid receptor and flexible ligand were assumed in the docking simulations. A grid 60 Å × 60 Å × 60 Å was defined on the structure of JAK3 kinase in order to generate a grid map. Each docking simulation was repeated 100 times and employed empirical free energy and a Lamarckian genetic algorithm with the following parameters: the population was set at 150 randomly selected individuals, a maximum of 27,000 generations, a mutation rate of 0.02, crossover rate of 0.80, and energy evaluation of 2.5 × 106. The docked complex conformation with lowest binding energy for each receptor was selected for further MD analysis. The hydrophobic and electrostatic interactions of docked complexes were mapped using Ligplot+ [34] and Discovery Studio 3.5 visualizer tools (http://accelrys.com/events/webinars/discovery-studio/index.php).
2.5.4. MD Simulations
The MD simulations were performed using Groningen Machine for Chemicals Simulations (GROMACS) 4.6.5 software [35], using a GROMOS96 43A1 force field [36] and SPC/E molecular water model [37, 38]. The topology file and force field parameters of the chemical ligands were generated using PRODRG software [39]. The MD simulation was constructed in a cube with dimensions of 120 Å along each edge. Sodium or chloride ions were added to the MD simulation to neutralize the net charge. The SHAKE algorithm was utilized to fix the lengths of the bonds containing hydrogen atoms [40] and Particle Mesh Ewald (PME) used to calculate long-range electrostatic interactions. Before performing the MD simulations, energy minimization was performed with a maximum force per complex not greater than 1000 kJ/mol Å. Constant temperature and pressure equilibrations of complexes were performed over 1000 ps. Finally, the constructed MD simulations were performed over 20 ns at a temperature of 300 K and 1 atmosphere pressure. Coordinates were recorded every 100 ps.
After conducting the MD simulations, the conformations, trajectories, and behaviors of each simulation were analyzed using VMD [41], PyMol (http://www.pymol.org) [42] and GROMACS software.
2.5.5. Statistical Analysis
Statistical analysis was performed using GraphPad Prism Version 5.01. Data are expressed as mean ± standard error of the mean (SEM). The significance of differences between groups was estimated by one-way ANOVA, adjusting for repeated measures with Dunnett's multiple comparison tests.
3. Results
3.1. JAK3 Enzymatic Activity Assay
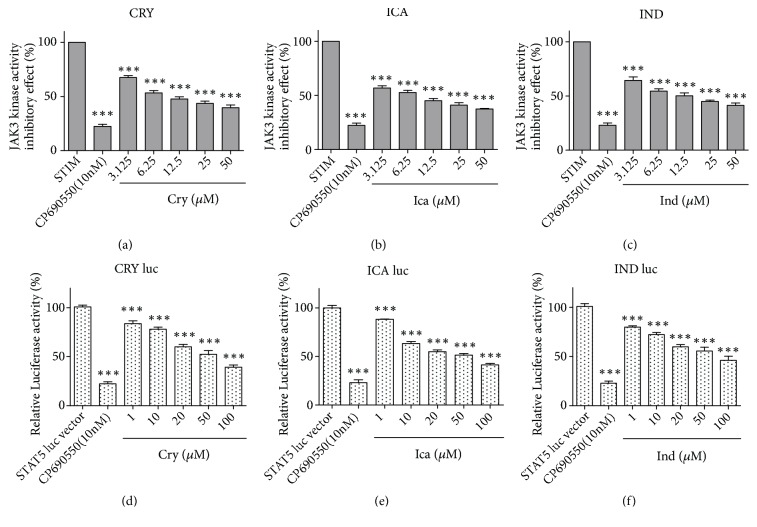
Based on the first-round docking results and the effects of Chinese herbal compounds on the JAKs/STATs pathway published in the literature, we selected and investigated the effect of 12 compounds on JAK3 phosphorylation activity. Among them, 3 compounds, Cryptotanshinone, Icaritin, and Indirubin, demonstrated significant reduction in JAK3 activity (IC50<100 μM). The results of the 3 natural compounds showing highest affinity are shown in Figure 1 and S2.
Figure 1.
Inhibition of JAK3 phosphorylation activity (a–c). Inhibition of JAK3 (%) by 3 Chinese herbal compounds: (a) Cryptotanshinone (CRY), (b) Icaritin (ICA), and (c) Indirubin (IND), at various concentrations. JAK3 phosphorylation was measured using monoclonal antiphosphotyrosine–peroxidase conjugated antibody. Data represent means ± SEM of three independent experiments. The self-simulation (STIM) without inhibitors indicated the maximum phosphorylation. Compound CP690,550 was selected as positive control. Inhibition of cellular JAK3-mediated STAT5 activity (d–f). Inhibitory effect of treatment with 3 Chinese herbal compounds on luciferase activity of STAT5 at various concentrations. HEK 293 cells were transfected with a STAT5-dependent luciferase reporter then treated with herbal compounds for 1 h prior to conducting the luciferase reporter assay. Data represent means ± SEM of three independent experiments. ∗∗∗p <0.001 versus control group; significance was determined using one-way ANOVA.
3.2. JAK3 Enzymatic Activity Assay
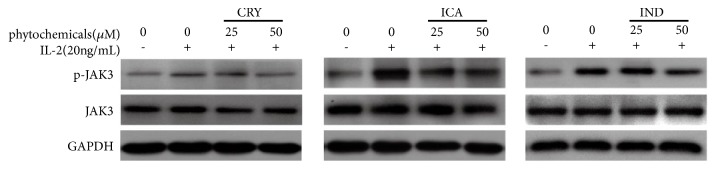
The 3 hit compounds, Cryptotanshinone, Icaritin, and Indirubin, demonstrated directly inhibitory effects on the enzyme activity. Therefore, in this work we measured the changes of phoso-JAK3 content in HEK 293 cells by western blot. The result showed that 3 Chinses herbal compounds, Cryptotanshinone, Icaritin, and Indirubin, attenuated IL-2-induced JAK3 phosphorylation in HEK 293 cell line in Figure 2.
Figure 2.
Effects of hit Chinses herbal chemicals on JAK3 phosphorylation. IL-2 induced phosphorylated JAK3 protein in HEK-293 cell line was determined by western blot after treatment with phytochemicals at the indicated concentrations for 2 h prior to stimulation with IL-2 for 1 h. GAPDH was used as the internal control for protein normalization.
3.3. SATA5 Transactivation Activity
3.3.1. Luciferase Experiments
Inhibition of JAK3 phosphorylation activity would be expected to lead to a decrease in downstream STAT5 phosphorylation levels. Thus, we sought to investigate the ability of the hit compounds to inhibit JAK3 signaling in human cells using a luciferase reporter assay. HEK 293 cells transfected with the luciferase reporter gene driven by a promoter containing multiple copies of the STAT5 response element were used in this study. The transcriptional activity of STAT5 was quantified by measuring the luciferase activity of cell lysates using a luminometer. We performed a dose response analysis of the 3 Chinese compounds in their ability to attenuate IL-2-induced STAT5 signaling (Figure 1).
The compounds inhibited IL-2 induced luciferase activity in a dose-dependent manner. Although the other three compounds also inhibited IL-2-induced luciferase activity, the potency of their inhibition was approximately 1% that of CP- 690,550.
3.4. Docking Simulation
The workflow of the docking simulation is outlined in Scheme 1. The molecular model of JAK3 for docking analysis was constructed using the reported X-ray cocrystal structure of JAK3 using the reference inhibitor CP-690,550. The binding site of JAK3 was defined to be within 3Å of the bound inhibitor, situated at the ATP-binding pocket of the JAK3 phosphorylated tyrosine kinase (PTK) domain.
Scheme 1.
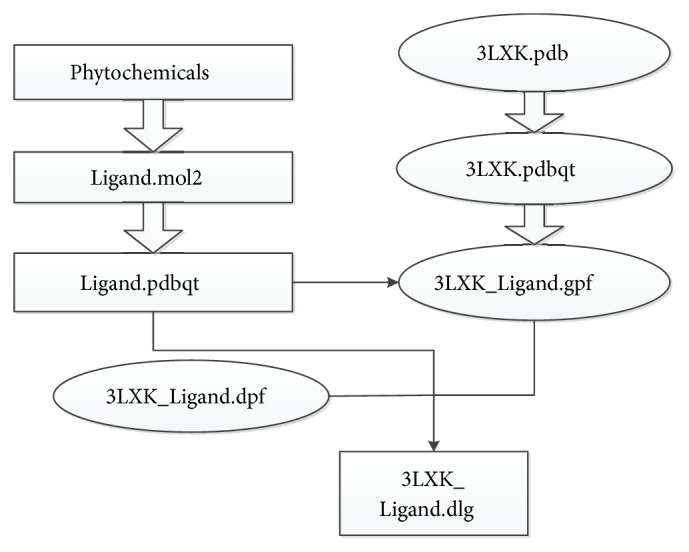
The workflow of the docking simulation.
On the first round of virtual screen, the phytochemicals of high docking scores were filtered using Lipinski's Rules of five and ADME/T virtual predictions. As phytochemicals are derived from natural plants, one or two violations of Lipinski's Rules were permitted [43]. The chemicals with toxicity under grade 2 of ADME/T virtual predictions were permitted [11]. The top 50 chemicals were listed in S1.
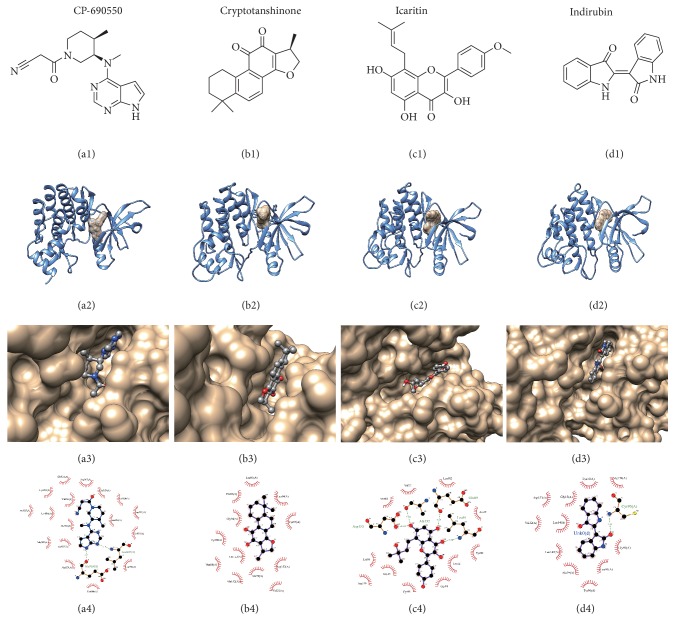
Based on the results of the bioassays, the 3 compounds, Cryptotanshinone (CRY), Icaritin (ICA), and Indirubin (IND), with the highest bioactivity, were investigated by docking computation using AutoDock 4.2 again. Contact between compounds and the JAK3 binding pocket was analyzed using LigPlus 2.1 and Discovery Studio Viewer 3.5. Docking analysis results indicated that the interaction of the 3 herbal compounds could be situated in the binding-pocket region of JAK3. In the analysis, Icaritin formed hydrogen bonds with the same kinase domain (KD) residues as reference compound CP-690,550, including Glu903 and Leu 905, whereas Indirubin formed hydrogen bonds with KD residue Cys909. Cryptotanshinone could not form hydrogen bonds. Critical residues and interactions within the ATP-binding domain of JAK3-ligand complex are listed in Table 2. The lowest energy binding pose and the protein-ligand interactions are shown in Figure 3.
Table 2.
Details of interactions between compounds and the JAK3 ATP-binding site.
| JAK3 binding region residues | Nature of interaction | |||
|---|---|---|---|---|
| CP690550 | Cry | Ica | Ind | |
| Leu828 | Van der Waals | VDW | Electrostatic | Electrostatic |
| Gly829 | Electrostatic | - | VDW | VDW |
| Lys830 | Electrostatic | - | VDW | VDW |
| Gly831 | Electrostatic | - | - | - |
| Gly834 | Electrostatic | - | - | - |
| Ser835 | Electrostatic | - | - | - |
| Val836 | Electrostatic | VDW | VDW | VDW |
| Ala853 | VDW | VDW | VDW | VDW |
| Lys855 | VDW | - | - | - |
| Val884 | VDW | - | VDW | - |
| Met902 | VDW | VDW | VDW | - |
| Glu903 | H-Bond | Electrostatic | H-Bond | - |
| Tyr904 | VDW | VDW | VDW | VDW |
| Leu905 | H-Bond | Electrostatic | H-Bond | VDW |
| Pro906 | - | VDW | - | - |
| Gly908 | - | VDW | Electrostatic | Electrostatic |
| Cys909 | VDW | VDW | VDW | H-Bond |
| Arg953 | VDW | - | Electrostatic | VDW |
| Asn954 | VDW | - | Electrostatic | - |
| Ile955 | VDW | - | - | - |
| Leu956 | VDW | VDW | VDW | VDW |
| Ala966 | VDW | VDW | H-Bond | - |
| Asp967 | VDW | VDW | H-Bond | Electrostatic |
Figure 3.
Analysis of Chinese herbal compound binding to JAK3 kinase. (a) CP-690550; (b) Cryptotanshinone; (c) Icaritin; (d) Indirubin. JAK3 kinase is depicted in marine-blue ribbon. Chemicals are shown as sticks & balls and transparent solids. JAK3 protein-ligand interactions were mapped using Ligplot+. CP-690,550 was the positive control.
3.5. Molecular Dynamic Simulation
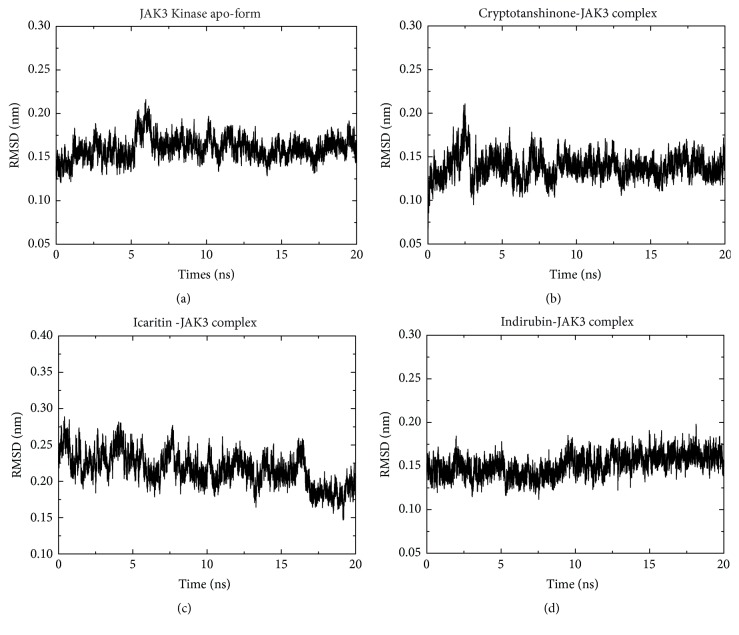
To investigate the stability of each JAK3-ligand complex, binding conformations of optimally docked complexes were studied through MD simulation assays. A least squares fit of the binding complexes was used in each case in the calculation of the root-mean-square deviations (RMSD). The RMSD for each complex was calculated over 5 ns. The output plots displayed RMSDs for each JAK3-ligand structure in its minimized, equilibrated state. MD simulation analysis indicated that the backbone RMSD profile for JAK3-compound complexes was quite stable (1.5–3 Å) throughout the MD trajectories, demonstrating stability of complexes during the MD simulations (Figure 4). The converging behavior indicated by these results provides credibility to the docking results.
Figure 4.
Plots demonstrating JAK3 kinase-ligand stability. RMSD versus time (20ns) graph through each MD trajectory file. (a) JAK3 Kinase apo-form; (b) Cryptotanshinone-JAK3 complex; (c) Icaritin -JAK3 complex; (d) Indirubin-JAK3 complex.
4. Discussion
JAK kinases perform pivotal roles in inflammatory and oncological disorders [44]. Several JAK inhibitors have been developed for the treatment of inflammatory disease such as rheumatoid arthritis (RA) and autoimmune disorders such as inflammatory bowel disease and psoriasis [1, 3].
Unlike other JAK family members, the expression of JAK3 is mostly in hematopoietic cells where it exclusively associates with cytokine receptors bearing the common gamma-chain (γc) subunit, enabling JAK3 to specifically operate in immune cells [45]. Consequently, JAK3 represents an attractive target for the treatment of inflammatory immunosuppression [46]. The JAK3 inhibitor Tofacitinib has been approved in US for treating RA patients [47].
Although many JAK3 inhibitors are already available, certain shortcomings have been identified, mostly acquired drug resistance or unwanted side effects [48, 49]. Physiological environments are more complicated than in vitro experimental conditions and unpredicted toxic effects can arise from the use of a potent inhibitor [50]. For example, European regulatory agencies did not approve Tofacitinib because of concerns over efficacy and safety [49]. Animal studies indicated the occurrence of carcinogenesis, mutagenesis, and impairment of fertility (XELJANZ prescribing information, Labeling.Pfizer.com). Thus, the development of novel JAK3 inhibitors or the application of medicines that inhibit JAK3 with limit side effects is required.
In this study, we performed an inhibition assay to determine the inhibitory activity of 12 phytochemicals from Chinese herbs against JAK3, using a cell-free enzyme assay and an HEK 293 cell pSTAT5 transactivity assay.
The kinase and transactivity assays revealed that the majority of 12 test compounds did not exhibit kinase activity (IC50<100μM) that qualified them as a “hit”. Some exhibited only limited activity while others showed no direct activity at all. Thus, these nonhit compounds probably modulate the JAK/STAT pathway through other mechanisms or factors. However, 3 compounds showed direct inhibition of JAK3 kinase in vitro. These results indicate that the IC50 of Cryptotanshinone was ca. 25 μM, Icaritin ca. 20 μM, and Indirubin ca. 50 μM.
These compounds also demonstrated inhibition at low concentration in cell-based assays. There might be two reasons for this. Firstly, in cell culture conditions, there is a dynamic balance between kinase and phosphatase, whereas the kinase catalytic reaction persists in vitro until the substrates are saturated. The compounds in low concentration could disrupt this balance. Secondly, the compounds might affect STAT3 phosphorylation through other mechanisms. However, the nonhit compounds did not demonstrate activity either in the kinase assay or cell-based assays.
There is a tradition in Chinese medicine of use of the herbs containing the hit phytochemicals since ancient times, demonstrating a record of safety and efficacy [6, 51]. Thus, our study indicated that the hit compounds and their original Chinese herbs provide efficacy in the treatment of inflammatory diseases and immune disorders via direct interaction with JAK3 kinase.
To investigate the structural conformations and to explore the key amino acids involved in the interaction between JAK3 kinase and the hit ligands, we performed computational calculations using docking and molecular dynamics simulations.
In previous reports, structure-based computational molecular simulation has been used to identify candidate compounds able to inhibit JAK3 phosphorylation [52, 53]. The JAK3 ATP-binding pocket domain structure was used for computational chemical assays of small molecules. To explore models of compound binding to the JAK3 binding-pocket domain, computational modeling was conducted.
ATP-induced phosphorylation of Tyr on the JH1 domain of the JAK3 protein is critical for the function and modulation of JAK3 kinase [54]. The ATP-binding site of JAK3 kinase consists of a narrow and hydrophobic cleft located between the N- and C-lobes of the kinase domain (KD), the two lobes linked together by a binding-pocket region consisting of a hydrogen bond donor and acceptor residues from the protein backbone. The key factor in determining kinase inhibitory activity is the identity of residues in the glycine, hinge, and activation loops which control access of the inhibitor to the hydrophobic pocket. For this reason, we evaluated the magnitude of JAK3 inhibition of the 3 compounds targeting the ATP-binding pocket domain using computational assays.
Docking strategies generate binding or affinity scores for different sites and poses on the target protein. Although docking has been popularly applied in drug discovery, some docking results show high dock scores that are not well correlated with actual bioactivity, also failing in MD simulation [13]. To enhance the credibility of our docking results, we also performed MD simulations, the results of which demonstrated stable and converged behavior of complexes throughout the MD trajectories.
The docking simulation results of the 3 compounds using AutoDock 4.2, which are slightly lower than that using AutoDock Vina, showed a high dock score (the binding energy of Cryptotanshinone to JAK3 kinase was 9.2 kcal/mol, Icaritin 9.3 kal/mol, Indirubin 9.1 kcal/mol and CP-690,550 8.8 kcal/mol), indicating that the 3 phytochemicals are indeed excellent potential inhibitors. The poses found in this modeling approach indicated that the 3 Chinese herbal compounds could possibly bind to the JH1 domain of JAK3. The refined model predicted that these compounds bind at the specific site at which the binding-pocket residues are located within the JAK3 JH1 domain (Table 2). The model also predicted that Icaritin can form a number of hydrogen bonds with nearby amino acid residues, including Glu903, Leu905, Ala966, and Asp967, and that Indirubin can form a hydrogen bond with Cys909 (Figure 3). The results also indicate that the amino acid residues Leu828, Gly829, Lys830, Val836, Ala853, Met902, Glu903, Tyr904, Leu905, Gly908, Cys909, Leu956, Ala966, and Asp967 are those mostly involved in the interaction between JAK3 kinase and the compounds and are possibly the most important residues in the binding-pocket for protein-ligand interaction.
Thus, by combining bioactivity assays and computational research, the present study identified 3 Chinese herbal compounds that could serve as potential candidates for JAK3 kinase inhibition and JAK3-targeted diseases.
5. Conclusions
In conclusion, we have discovered 3 potential natural inhibitors of JAK3, Cryptotanshinone, Icaritin, and Indirubin. Our work could provide new possibilities and stimulate new insights for the treatment of JAK3-targeted diseases.
Acknowledgments
This work was supported by “Natural Science Foundation of Guangdong Province of China” (Grant number 2016A030313366;2018A030313419), “Administration of Traditional Chinese Medicine of Guangdong Province of China” (Grant number 20182169), “Science and Technology Planning Project of Zhongshan City of China” (Grant number 2018B1037;201811110), and Cultivation of Guangdong College Students' Scientific and Technological Innovation Project & Climbing Program Special Funds (Grant number pdjh2017b0916).
Contributor Information
Yu-Qiao Gao, Email: aqiaosky@163.com.
Quan-Xi Mei, Email: meiquanxi@163.com.
Data Availability
The authors declare that the data supporting the findings of this study are available within the article and the supplementary information file.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Table S1: top 50 docking results.
References
- 1.Clark J. D., Flanagan M. E., Telliez J.-B. Discovery and development of Janus kinase (JAK) inhibitors for inflammatory diseases. Journal of Medicinal Chemistry. 2014;57(12):5023–5038. doi: 10.1021/jm401490p. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee S., Biehl A., Gadina M., Hasni S., Schwartz D. M. JAK–STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs. 2017;77(5):521–546. doi: 10.1007/s40265-017-0701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Shea J. J., Kontzias A., Yamaoka K., Tanaka Y., Laurence A. Janus kinase inhibitors in autoimmune diseases. Annals of the Rheumatic Diseases. 2013;72(2):ii111–ii115. doi: 10.1136/annrheumdis-2012-202576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vyas D., O'Dell K. M., Bandy J. L., Boyce E. G. Tofacitinib: the first Janus Kinase (JAK) inhibitor for the treatment of rheumatoid arthritis. Annals of Pharmacotherapy. 2013;47(11):1524–1531. doi: 10.1177/1060028013512790. [DOI] [PubMed] [Google Scholar]
- 5.Winthrop K. L. The emerging safety profile of JAK inhibitors in rheumatic disease. Nature Reviews Rheumatology. 2017;13(4):234–243. doi: 10.1038/nrrheum.2017.23. [DOI] [PubMed] [Google Scholar]
- 6.Harvey A. L. Natural products in drug discovery. Drug Discovery Therapy. 2008;13(19-20):894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Chen C. Y.-C. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0015939.e15939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trott O., Olson A. J. Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipinski C.A. Drug-like properties and the causes of poor solubility and poor permeability. Journal of Pharmacological and Toxicological Methods. 2000;44:235–249. doi: 10.1016/S1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 10.Pollastri M. P. Current Protocols in Pharmacology. 2010. Overview on the rule of five. (chapter 9, unit 9.12). [DOI] [PubMed] [Google Scholar]
- 11.Cheng F., Li W., Zhou Y., et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. Journal of Chemical Information and Modeling. 2012;52(11):3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 12.Hou X., Du J., Zhang J., Du L., Fang H., Li M. How to improve docking accuracy of AutoDock4.2: A case study using different electrostatic potentials. Journal of Chemical Information and Modeling. 2013;53(1):188–200. doi: 10.1021/ci300417y. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y.-C. Beware of docking! Trends in Pharmacological Sciences. 2015;36(2):78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Fang X., Shao L., Zhang H., Wang S. CHMIS-C: A comprehensive herbal medicine information system for cancer. Journal of Medicinal Chemistry. 2005;48(5):1481–1488. doi: 10.1021/jm049838d. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J., Ong C.-N., Hur G.-M., Shen H.-M. Inhibition of the JAK-STAT3 pathway by andrographolide enhances chemosensitivity of cancer cells to doxorubicin. Biochemical Pharmacology. 2010;79(9):1242–1250. doi: 10.1016/j.bcp.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Kou X., Qi S., Dai W., Luo L., Yin Z. Arctigenin inhibits lipopolysaccharide-induced iNOS expression in RAW264.7 cells through suppressing JAK-STAT signal pathway. International Immunopharmacology. 2011;11(8):1095–1102. doi: 10.1016/j.intimp.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Qi Z., Yin F., Lu L., et al. Baicalein reduces lipopolysaccharide-induced inflammation via suppressing JAK/STATs activation and ROS production. Inflammation Research. 2013;62(9):845–855. doi: 10.1007/s00011-013-0639-7. [DOI] [PubMed] [Google Scholar]
- 18.Nam S., Xie J., Perkins A., et al. Novel synthetic derivatives of the natural product berbamine inhibit Jak2/Stat3 signaling and induce apoptosis of human melanoma cells. Molecular Oncology. 2012;6(5):484–493. doi: 10.1016/j.molonc.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin D.-S., Kim H.-N., Shin K. D., et al. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Research. 2009;69(1):193–202. doi: 10.1158/0008-5472.CAN-08-2575. [DOI] [PubMed] [Google Scholar]
- 20.Kim H. Y., Park E. J., Joe E.-H., Jou I. Curcumin Suppresses Janus Kinase-STAT Inflammatory Signaling through Activation of Src Homology 2 Domain-Containing Tyrosine Phosphatase 2 in Brain Microglia. The Journal of Immunology. 2003;171(11):6072–6079. doi: 10.4049/jimmunol.171.11.6072. [DOI] [PubMed] [Google Scholar]
- 21.Song Y., Zeng Z., Jin C., Zhang J., Ding B., Zhang F. Protective effect of ginkgolide B against acute spinal cord injury in rats and its correlation with the JAK/STAT signaling pathway. Neurochemical Research. 2013;38(3):610–619. doi: 10.1007/s11064-012-0959-y. [DOI] [PubMed] [Google Scholar]
- 22.Li S., Priceman S. J., Xin H., et al. Icaritin Inhibits JAK/STAT3 Signaling and Growth of Renal Cell Carcinoma. PLoS ONE. 2013;8(12):p. e81657. doi: 10.1371/journal.pone.0081657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Song Y., Wu Y., et al. Indirubin inhibits tumor growth by antitumor angiogenesis via blocking VEGFR2-mediated JAK/STAT3 signaling in endothelial cell. International Journal of Cancer. 2011;129(10):2502–2511. doi: 10.1002/ijc.25909. [DOI] [PubMed] [Google Scholar]
- 24.Boly R., Gras T., Lamkami T., et al. Quercetin inhibits a large panel of kinases implicated in cancer cell biology. International Journal of Oncology. 2011;38(3):833–842. doi: 10.3892/ijo.2010.890. [DOI] [PubMed] [Google Scholar]
- 25.Chen S. C., Lin Y. L., Huang B., Wang D. L., Cheng J. J. Salvianolic acid B suppresses IFN-γ-induced JAK/STAT1 activation in endothelial cells. Thrombosis Research. 2011;128(6):560–564. doi: 10.1016/j.thromres.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 26.Pathak A. K., Bhutani M., Nair A. S., et al. Ursolic acid inhibits STAT3 activation pathway leading to suppression of proliferation and chemosensitization of human multiple myeloma cells. Molecular Cancer Research. 2007;5(9):943–955. doi: 10.1158/1541-7786.MCR-06-0348. [DOI] [PubMed] [Google Scholar]
- 27.Bauer S. M., Gehringer M., Laufer S. A. A direct enzyme-linked immunosorbent assay (ELISA) for the quantitative evaluation of Janus Kinase 3 (JAK3) inhibitors. Analytical Methods. 2014;6(21):8817–8822. doi: 10.1039/C4AY01589D. [DOI] [Google Scholar]
- 28.Losdyck E., Hornakova T., Springuel L., et al. Distinct acute lymphoblastic leukemia (ALL)-associated Janus Kinase 3 (JAK3) mutants exhibit different cytokine-receptor requirements and JAK inhibitor specificities. The Journal of Biological Chemistry. 2015;290(48):29022–29034. doi: 10.1074/jbc.M115.670224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin C.-H., Bach E. A., Baeg G.-H. Development of a high-throughput cell-based reporter assay for screening of JAK3 inhibitors. Journal of Biomolecular Screening. 2011;16(4):443–449. doi: 10.1177/1087057111400190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashid S., Bibi N., Parveen Z., Shafique S. Inhibition of Janus kinases by tyrosine phosphorylation inhibitor, Tyrphostin AG-490. Journal of Biomolecular Structure and Dynamics. 2015;33(11):2368–2379. doi: 10.1080/07391102.2015.1050696. [DOI] [PubMed] [Google Scholar]
- 31.Meng E. C., Pettersen E. F., Couch G. S., Huang C. C., Ferrin T. E. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forli S., Piche M. E., Sanner M. F. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nature Protocols. 2016;11(5):905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris G. M., Ruth H., Lindstrom W., et al. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. Journal of Computational Chemistry. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski R. A., Swindells M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 35.Pronk S., Páll S., Schulz R., et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerrigan J. E. Molecular dynamics simulations in drug design. Methods in Molecular Biology. 2013;993:95–113. doi: 10.1007/978-1-62703-342-8_7. [DOI] [PubMed] [Google Scholar]
- 37.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. The Journal of Physical Chemistry A. 2001;105(43):9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 38.Berendsen H. J. C., Grigera J. R., Straatsma T. P. The missing term in effective pair potentials. The Journal of Physical Chemistry C. 1987;91(24):6269–6271. doi: 10.1021/j100308a038. [DOI] [Google Scholar]
- 39.Schüttelkopf A. W., van Aalten D. M. F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica Section D: Biological Crystallography. 2004;60(8):1355–1363. doi: 10.1107/s0907444904011679. [DOI] [PubMed] [Google Scholar]
- 40.Li P., Roberts B. P., Chakravorty D. K., Merz K. M. Rational design of particle mesh ewald compatible lennard-jones parameters for +2 metal cations in explicit solvent. Journal of Chemical Theory and Computation. 2013;9(6):2733–2748. doi: 10.1021/ct400146w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 42.DeLano W. L. The PyMOL molecular graphics system. 2002, https://pymol.org/ and http://pymol.sourceforge.net/overview/sld001.htm.
- 43.Beutler J. A. Natural products as a foundation for drug discovery. Current Protocols in Pharmacology. 2009;46 doi: 10.1002/0471141755.ph0911s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark G. R., Darnell J. E. The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilks A. F. The JAK kinases: Not just another kinase drug discovery target. Seminars in Cell & Developmental Biology. 2008;19(4):319–328. doi: 10.1016/j.semcdb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 46.Borie D. C., Si M.-S., Morris R. E., Reitz B. A., Changelian P. S. JAK3 inhibition as a new concept for immune suppression. Current Opinion in Infectious Diseases. 2003;4(11):1297–1303. [PubMed] [Google Scholar]
- 47.Flanagan M. E., Blumenkopf T. A., Brissette W. H., et al. Discovery of CP-690,550: A potent and selective janus kinase (JAK) inhibitor for the treatment of autoimmune diseases and organ transplant rejection. Journal of Medicinal Chemistry. 2010;53(24):8468–8484. doi: 10.1021/jm1004286. [DOI] [PubMed] [Google Scholar]
- 48.Ghoreschi K., Laurence A., O'Shea J. J. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nature Immunology. 2009;10(4):356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cutolo M., Meroni M. Clinical utility of the oral JAK inhibitor tofacitinib in the treatment of rheumatoid arthritis. Journal of Inflammation Research. 2013;6(1):129–136. doi: 10.2147/JIR.S35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoma G., Drückes P., Zerwes H.-G. Selective inhibitors of the Janus kinase Jak3 - Are they effective? Bioorganic & Medicinal Chemistry Letters. 2014;24(19):4617–4621. doi: 10.1016/j.bmcl.2014.08.046. [DOI] [PubMed] [Google Scholar]
- 51.Balunas M. J., Kinghorn A. D. Drug discovery from medicinal plants. Life Sciences. 2005;78(5):431–441. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Wang J. L., Cheng L. P., Wang T. C., Deng W., Wu F. H. Molecular modeling study of CP-690550 derivatives as JAK3 kinase inhibitors through combined 3D-QSAR, molecular docking, and dynamics simulation techniques. Journal of Molecular Graphics and Modelling. 2017;72:178–186. doi: 10.1016/j.jmgm.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 53.Pei H., He L., Shao M., et al. Discovery of a highly selective JAK3 inhibitor for the treatment of rheumatoid arthritis. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-018-23569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chrencik J. E., Patny A., Leung I. K., et al. Structural and Thermodynamic Characterization of the TYK2 and JAK3 Kinase Domains in Complex with CP-690550 and CMP-6. Journal of Molecular Biology. 2010;400(3):413–433. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: top 50 docking results.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and the supplementary information file.