Abstract
Turnover of cellular proteins is regulated by Ubiquitin Proteasome System (UPS). Components of this pathway, including the proteasome, ubiquitinating enzymes and deubiquitinating enzymes, are highly specialized and tightly regulated. In this mini-review we focus on the de-ubiquitinating enzyme USP7, and summarize latest advances in understanding its structure, substrate specificity and relevance to human cancers. There is increasing interest in UPS components as targets for cancer therapy and here we also overview the recent progress in the development of small molecule inhibitors that target USP7.
1. Introduction
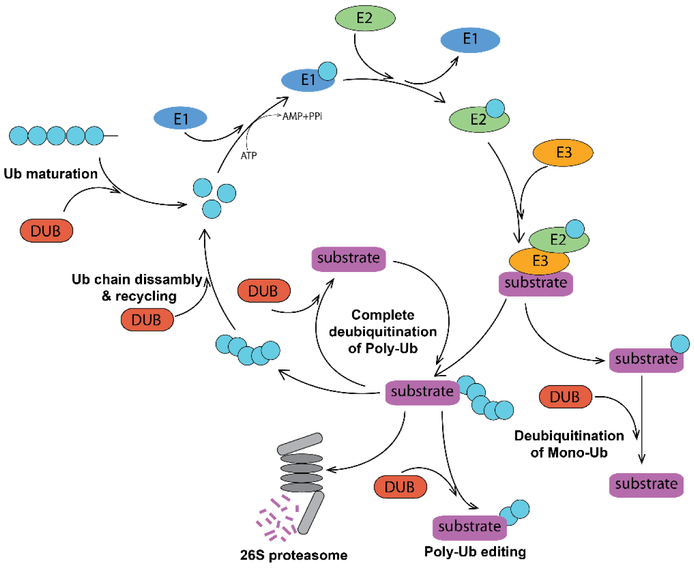
Living cells constantly synthetize proteins, and dispose proteins that are misfolded, aggregated and no longer needed. Maintenance of proper protein homeostasis is essential for cell growth and survival. In eukaryotes, protein degradation is carried out by the ubiquitin-proteasome system (UPS). In this multi–step pathway, a polyubiquitin chain is conjugated to a protein substrate and serves as a signal for the substrate recognition and degradation by the 26S proteasome (1). Protein ubiquitination can be reversed by action of deubiquitinating enzymes (DUBs) (2) that remove ubiquitin moieties from their substrates (Figure 1).
Figure 1. Ubiquitin-Proteasome System and DUBs.
Mono- and polyubiquitination of a protein substrate is catalyzed by consecutive action of E1, E2 and E3 ubiquitinating enzymes. The K48-linked polyubiquitin tag targets the substrate for proteasomal degradation, while monoubiquitination and other ubiquitin linkages result in a different functional outcome. DUBs deubiquitinate both poly- and monoubiquitinated proteins and thus change their fate. DUBs can edit polyubiquitin chains architecture and recycle ubiquitin. They also participate in maturation of the free ubiquitin.
Ub – ubiquitin; E1 – ubiquitin activating enzyme; E2 – ubiquitin conjugating enzyme; E3 – ubiquitin Ligase; DUB – deubiquitinating enzyme.
Human ubiquitin-specific protease 7 (USP7) also known as Herpes virus associated protease (HAUSP) is a cysteine peptidase that belongs to the largest USP family of DUBs (Figure 2) (3). Located primarily in the nucleus, USP7 regulates the stability of multiple proteins involved in diverse cellular processes including DNA damage response, transcription, epigenetic control of gene expression, immune response, and viral infection (Table 1). USP7 has been extensively studied for its ability to regulate the cellular level of tumor suppressor p53 affected in the majority of solid tumors (4-7). USP7 knockout was shown to be lethal in mice (8,9). However, several children have been recently identified carrying USP7 mutations and deletions. The 46 individuals identified so far suffer from neurodevelopmental disorders such as autism spectrum disorder, intellectual disability, and speech/motor impairments (10) (www.usp7.org).
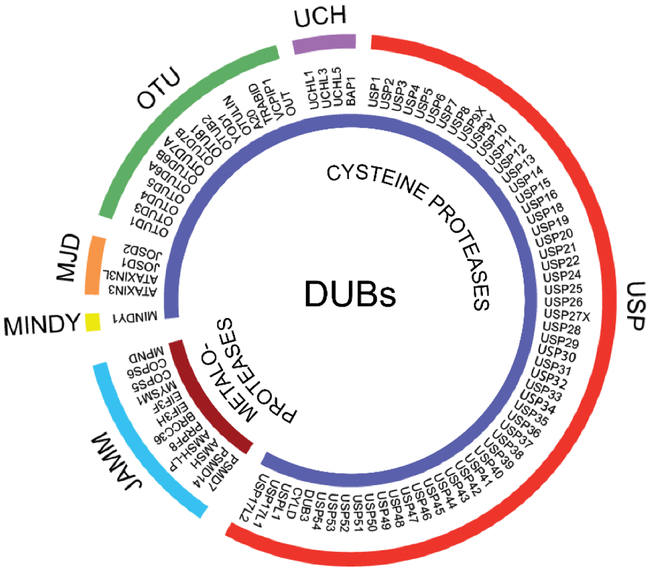
Figure 2. Human DUBs.
Human DUBs consist of two classes of enzymes: cysteine proteases and metalloproteases. Cysteine proteases are further subdivided into five families, including ubiquitin-specific proteases (USPs), ubiquitin carboxyl-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph (Josephin) domain containing (MJD) proteases, and MINDY (motif interacting with Ub-containing novel DUB family). The JAB1/MPN/MOV34 domain containing proteases (JAMMs) family is the only representative of the metalloproteases class of DUBs. Adapted from (2).
Table 1.
USP7 substrates
| Substrate name | Reference | Substrate name | Reference |
|---|---|---|---|
| Transcription factors | Epigenetic control of gene expression | ||
| p53 | (18) | RING1B | (39) |
| HDM2 | (19) | Bmi1 | (40) |
| Rb | (105) | Histone H2B | (106) |
| FOX(O)3 | (46) | DNMT1 | (107) |
| FOX(O)4 | (46) | UHRF1 | (37) |
| PTEN | (12) | RNF168 | (23) |
| β-catenin | (71) | MLL5 | (43) |
| PPARγ | (108) | Tip60 | (42) |
| N-Myc | (109) | UbE2E1 | (75) |
| DNA replication/Cell cycle control | PHF8 | (14) | |
| CHFR | (110) | Telomere proteins | |
| Bub3 | (111) | TPP1 | (112) |
| SUMO | (34) | Immune response | |
| DNA damage response/DNA repair | NFºB | (113) | |
| DAXX | (114) | TRAF6 | (115) |
| Polymerase η | (32) | IKKγ | (115) |
| HLTF | (29) | TRIM27 | (116) |
| Rad 18 | (30) | Viral proteins | |
| Clapsin | (27) | EBNA1 from EBV | (117) |
| Chk1 | (26) | ICP0 from HSV-1 | (49) |
| UVSSA | (28) | vIRF1 from KSHV | (54) |
| Mule/ARF-BP1 | (118) | vIRF4 from KSHV | (53) |
| XPC | (22) | LANA from KSHV | (51) |
| Epigenetic control of gene expression | UL35 from HCMV | (55) | |
| MEL18 | (40) | E1B from Adenovirus | (56) |
In this review, we discuss recent advances in understanding of the USP7 function, structure, regulation and its pharmaceutical relevance.
2. USP7 and Cancer
Given involvement of USP7 in multiple cellular pathways, it is not surprising that its expression is often dysregulated in human malignancies. USP7 overexpression contributes to tumor progression through changes in DNA damage response, apoptosis and cell cycle control. In particular, USP7 is upregulated in chronic lymphocytic leukemia (11) and its overexpression in human prostate cancer correlates with the tumor aggressiveness (12). USP7 expression level was shown to gradually increase with the tumor progression from grade I to grade IV in glioma patients (13). The enzyme is also overexpressed in breast carcinomas (14), as well as in lung squamous cell carcinoma and large cell carcinoma (15). Its dysregulation in non-small cell lung cancers leads to disruption of the HDM2–p53 axis and is associated with induced cell epithelial mesenchymal transition (EMT), (metastasis and overall poor prognosis (15). Similarly, USP7 overexpression in patients with epithelial ovarian cancer was found to induce cell invasion and correlate with poor survival (16,17). Because of the USP7 aberrant expression in many human cancers and its role in important cell signaling pathways, this enzyme has emerged as a promising target for cancer therapy.
3. Cellular Function of USP7
3.1. USP7 in DNA damage response
USP7 controls cellular levels of several key proteins involved in DNA damage response. The enzyme has been extensively studied due to its ability to regulate the level of tumor suppressor p53, a central regulator of the cell fate during DNA damage response. Interestingly, USP7 can deubiquitinate both p53 and its negative regulator HDM2, an E3 ubiquitin ligase responsible for polyubiquitination and subsequent degradation of p53 (18-20). Under normal conditions, USP7 preferentially interacts with HDM2 and prevents its degradation (4). In turn, HDM2 ubiquitinates p53, which results in low cellular levels of p53 during normal homeostasis. DNA damage causes ATM-dependent dephosphorylation of USP7 by PPM1G that decreases the affinity of the enzyme towards HDM2 (21). This allows USP7 to associate with p53, protecting it from ubiquitination. This interaction results in p53 stabilization and initiation of the p53-dependent DNA damage response.
USP7 is involved in regulation of the nucleotide excision repair (NER) pathway. Specifically, it prevents degradation of the XPC protein that recognizes helix-distorting DNA lesions and initiates the repair (22). USP7 stabilizes both RNF168 and RNF169 that control access of DNA damage response proteins to chromatin at the sites of double stranded DNA breaks (23,24). USP7 also deubiquitinates the cell cycle checkpoint CDC25A upon DNA damage in the BRCA-deficient cells and contributes to cell survival despite the loss of BRCA-mediated genome integrity (25).
USP7 plays a significant role in the ATR-Chk1 branch of DNA damage response where it deubiquitinates Chk1, an important checkpoint kinase (26), and an adapter protein claspin (27). Another important substrate of USP7 is UVSSA, one of initiating factors of the UV-induced transcription-coupled DNA repair (28). Additionally, in response to genotoxic stress, USP7 stabilizes HLTF, which regulates error-free bypass replication over DNA lesions (29).
Furthermore, USP7 is involved in DNA damage tolerance through controlling the stability of the E3 ubiquitin ligase Rad 18 that monoubiquitinates DNA sliding clamp PCNA in response to replication forks stalled at DNA lesions (30). PCNA monoubiquitination facilitates the recruitment of specialized translesion synthesis (TLS) DNA polymerases, which can bypass DNA lesions in an error-prone manner, thereby allowing replication to proceed (31). The stability of at least one TLS DNA polymerase, polη, is also controlled by USP7 (32).
3.2. USP7 in DNA Replication
In addition to DNA damage repair, USP7 plays an important role in bulk DNA replication by stabilizing several key proteins in this process. For example, USP7 was shown to be a part of the minichromosome maintenance (MCM) complex, which is loaded onto chromatin following mitosis and remains on chromatin until the completion of DNA synthesis. USP7 is required for efficient unloading of the MCM complex from chromatin at the end of S-phase (33). Recently, USP7 was shown to be enriched at the replication forks, where it counteracts ubiquitination of SUMO and SUMOylated proteins and, thus, creates SUMO-rich environment at sites of DNA replication, which promotes replication fork progression (34,35). USP7 also stabilizes Geminin, an inhibitor protein involved in prevention of DNA re-replication (36).
3.3. USP7 in Epigenetics
USP7 is known to associate with chromatin and participates in epigenetic control of gene expression. It stabilizes multiple proteins responsible for maintenance of epigenetic modifications of chromatin, including “readers” and “writers” of epigenetic modifications of DNA and histones. For example, tissue-specific DNA methylation patterns are maintained by replisome components UHRF1 and methyltransferase DNMT1. Both proteins are the substrates of USP7 (37,38). Monoubiquitination of histones is another common epigenetic modification that control gene expression. USP7 can modulate ubiquitination status of histones. It associates with and stabilizes the components of the Polycomb Repressive Complexes 2 and 1 (PRC2, PRC1), resulting in monoubiquitination of histone H2A(K119), which serves as a marker for gene repression (39-41).
USP7 stabilizes several other histone-associated proteins including acetyltransferase TIP60 (42), demethylase PHF8 (14), methyltransferase MLL5 (43), lysine-specific demethylase 1 (LSD1/KDM1) (44), and directly deubiquitinates histone H3 (45).
Although USP7 is mostly known for its role in substrate stabilization, it has multiple other cellular functions. For example, deubiquitination of monoubiquitinated transcription factors PTEN and FOX(O)4 (12,46) by USP7 serves to negatively regulate transcription activity of these proteins. Deubiquitination of both PTEN and FOX(O)4 causes their translocation from the nucleus to cytoplasm leading to their inactivation. In addition, USP7 was shown to remove Lys-63-linked polyubiquitin chain from the SIRT7 histone deacetylase, which leads to inhibition of its activity.
3.4. USP7 in viral infections
In addition to numerous cellular processes, USP7 is known to play a role in viral infection. Originally, USP7 was discovered as an enzyme associated with ICP0, an immediate-early protein from Herpes Simplex Virus-1 (47). ICP0 is an E3 ubiquitin ligase required for efficient lytic infection (48). Association with USP7 prevents auto-ubiquitination and proteasomal degradation of ICP0 (49). Later studies revealed that USP7 interacts with proteins from three other viruses of the Herpesviridae family. Thus, binding of USP7 to LANA from Kaposi’s sarcoma-associated herpes virus (KSHV) and its functional homologue EBNA-1 from Epstein-Barr virus (EBV) has regulatory effect on latent viral replication (50,51). Furthermore, EBNA-1–USP7 interaction prevents the binding of USP7 to p53 and thereby diminishes p53 stabilization (52). Similar viral mechanism that disrupts the p53-mediated antiviral response was proposed for USP7 substrates vIRF1 and vIRF4 (viral interferon regulatory factors 1 and 4) from KSHV (53,54), while the significance of interaction between USP7 and UL35 from human cytomegalovirus (HCMV) remains elusive (55). Finally, besides its role in infections mediated by Herpesviruses, USP7 also promotes adenoviral replication via interaction with viral multifunctional protein E1B-55K (56) and enhances HIV viral production by deubiquitinating its Tat protein (57).
4. Regulation of USP7 in the cell
USP7 is an important component of UPS and its activity in the cell is tightly regulated to avoid uncontrolled stabilization of its multiple substrates. There are several levels of USP7 regulation including intramolecular mechanisms, post-transcriptional modifications, and protein–protein interactions. Intramolecular mechanisms include domain reorganization required for the enzyme activation and active site rearrangement (58-64). Post-transcriptional modifications can further tune the activity of USP7. In particular, the enzyme was shown to be phosphorylated at Ser18, Ser963, and ubiquitinated at Lys869 (65). Phosphorylation at Ser18 by the protein kinase CK2 alters affinity of USP7 towards its substrates HDM2 and p53 in a way that the phosphorylated enzyme preferably binds to HDM2, while its dephosphorylation results in the higher affinity to p53 (21). USP7 ubiquitination at Lys869 by E3 ligase TRIM27 promotes the TNF-α-induced apoptosis through deubiquitination of RIPK1 (66) and the role of phosphorylation at Ser963 remains to be determined. In addition, USP7 is aberrantly phosphorylated at Tyr243 by chimeric protein p210 BCR-ABL in chronic myeloid leukemia (CML) cells. This post-translational modification enhances deubiquitinase activity of the enzyme towards the tumor suppressor PTEN whose dysregulation is linked to CML pathogenesis (67). Efficient USP7 regulation is mediated by several proteins that interact with the enzyme and mediate its stability and/or activity. Thus, Trip12 was recently identified as an E3 ligase for USP7 ubiquitination (68). In the absence of DNA damage, the DAXX protein binds to USP7 and HDM2, facilitating the HDM2 deubiquitination (69). DAXX–USP7 complex also regulates stability of the E3 ligase CHFR and Aurora-A kinase involved in mitosis (70). In the Wnt/β-catenin signaling pathway, the USP7 mediated stabilization of β-catenin depends on the E3 ligase RNF220 that forms a ternary complex with both USP7 and β-catenin and promotes deubiquitination of the latter (71). Another protein, MBD4, recruits USP7 to heterochromatin foci where it co-localizes with UHRF1 and DNMT1 promoting their interaction (72). Finally, at least one protein, TRAF4, negatively regulates USP7 activity. It binds to the same region of USP7 as p53 blocking p53 accesses to the enzyme. This inhibition mechanism was shown to play an important role in breast cancers where TRAF4 overexpression prevents USP7-mediated p53 stabilization and diminishes cytotoxic stress response (73).
5. Structure of USP7
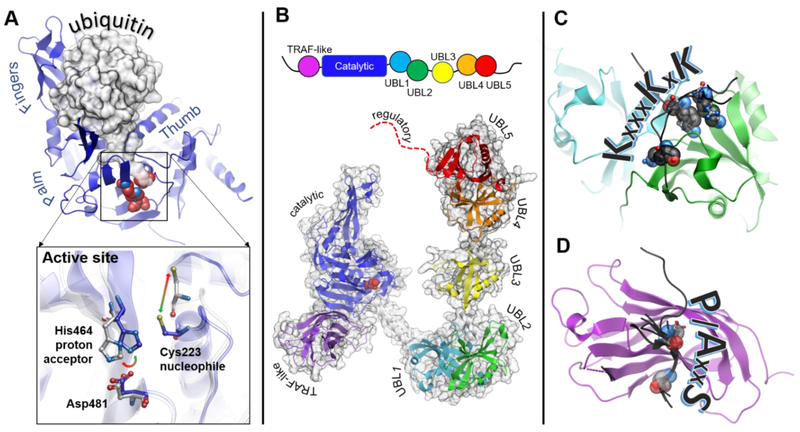
Schematic representation of the USP7 domain architecture is shown in Figure 3B. USP7 is a 135 kDa protein that consists of seven domains, including the N-terminal TRAF-like (Tumor necrosis factor Receptor–Associated Factor) domain, followed by the catalytic core domain and the five C-terminal ubiquitin-like domains, UBL1-5, (Figure 3B) (50,59). The TRAF-like and the UBL domains recognize various USP7 substrates (4,7,33,52-54,74,75), and the regulatory C-terminus of the protein is thought to be crucial for enhancement of the USP7 catalytic activity (5,59,64). The domains of this dynamic DUB are connected by linkers that allow for flexibility of interdomain arrangement (Figure 3B and Supplementary Movie 1). Such flexibility is likely important for regulation of the enzyme’s activity (59,63,64,76).
Figure 3. Structural features of USP7.
A. Structure of the catalytic core of USP7 (blue) has a overall right-hand shape formed by Palm, Thumb and Fingers regions. The Fingers grasp ubiquitin molecule, shown as a gray surface. The C-terminal tail of ubiquitin enters the narrow catalytic cleft and extends towards the catalytic triad (shown as red spheres). The inset shows conformational rearrangements of the active site switching between “active” (blue, PDB 1NBF) and “inactive” (grey PDB 1NB8) conformations. B. Schematic representation of the protein domain organization is shown on top. A structural model of the full length USP7 assembled based on overlapping structures of USP7 fragments including PDB IDs 2F1Z, 5FWI and 2YLM. The individual domains are labeled and color-coded: TRAF-like (purple), catalytic (blue), UBL1 (cyan), UBL2 (green), UBL3 (yellow), UBL4 (orange), UBL5 (red), the disordered extreme C-terminus is shown as red dotted line. C. Structure of UBL12 domains (cyan and green, respectively) in complex with its known substrates (black) including DNMT1 (PDB 4YOC), ICP0 (PDB 4WPH) and UHRF1 (PDB 5C6D), The lysine residues of the shared KxxxKxK motif are shown as spheres. The ICP0 peptide has a reversed RxKxxxK motif and binds in the opposite direction. D. Structure of the TRAF-like domain in complex with its known substrates including p53 (PBD 2F1X), HDM2 (PDB 2FOP), HDMX (PDB 3MQR), UbE2E1 (PDB 4JJQ), MCM-BP (PDB 4KG9), vIRF1 (PDB 4YSI), vIRF4 (PDB 2XXN) and EBNA1 (PDB 1YY6). The TRAF-like domain is shown as purple ribbon, and the substrate peptides are black. The P/AxxS motif shows the direction of the peptides. The conserved P/A and S substrate residues are shown as spheres.
5.1. Substrate recognition by the TRAF-like domain
The N-terminal TRAF-like domain spans residues 53-205 and has a characteristic eight-stranded, antiparallel β-sandwich fold (52) (purple in Figure 3B, D). The function of TRAF-like domain is to bind USP7 substrates. Tumor suppressor p53 is the first characterized substrate of the TRAF-like domain (58) and later studies revealed many others (Table 2) (4,7,33,52-54,74,75). All these substrates share common P/AxxS motif that is recognized by the same shallow grove on the surface of the TRAF-like domain, in which the USP7 residues D164 and W165 anchor the conserved motif of a substrate (Figure 3D) (4,7,33,75). A novel ExxS motif that binds to the same site was recently reported (54). This common mode of interaction suggests that the substrates compete for USP7 binding. Such competition between the tumor suppressor p53 and its E3 ubiquitin ligase HDM2, for example, serves as a switch to fine-tune the level of p53 in a cell (4).
Table 2.
Structures of USP7
| pdb ID | Ligand | Reference | pdb ID | Ligand | Reference |
|---|---|---|---|---|---|
| TRAF-like domain | Catalytic domain | ||||
| 2F1W | no | (4) | 2F1Z | no | (4) |
| 2F1Y | no | (4) | 5J7T | no | (64) |
| 1YZE | no | (52) | 1NBF | Ubiquitin-aldehyde | (58) |
| 2F1X | p53 | (4) | 5JTV | Ubiquitin-bromoethylamine | (64) |
| 2FOO | p53 | (7) | 5JTJ | Ubiquitin-bromoethylamine | (64) |
| 2FOJ | p53 | (7) | USP7 UBL domains | ||
| 2FOP | HDM2 | (7) | 5J7T | no | (64) |
| 3MQS | HDM2 | (74) | 5FWI | no | (62) |
| 3MQR | HDMX | (74) | 2YLM | no | (59) |
| 4KG9 | MCM-BP | (33) | 4WPI | no | (63) |
| 4JJQ | UbE2E1 | (75) | 4PYZ | no | unpublished |
| 1YY6 | EBNA1 | (52) | 4YOC | DNMT1 | (76) |
| 4YSI | vIRF1 | (54) | 4Z96 | DNMT1 | unpublished |
| 2XXN | vIRF4 | (53) | 5C56 | ICP0 | (86) |
| Catalytic domain | 4Z97 | DNMT1(K11 15Q) | unpublished | ||
| 1NB8 | no | (58) | 4WPH | ICP0 | (63) |
| 4M5W | no | (77) | 5GG4 | RNF169 | (24) |
| 4M5X | no | (77) | 5C6D | UHRF1 | (91) |
5.2. Unique structure of the USP7 catalytic core
The function of the DUB catalytic core domain is to bind ubiquitin and cleave the isopeptide bond between ubiquitin and a substrate. The USP7 catalytic core is the largest domain within the protein (residues 208-560) centrally located between the TRAF-like and UBL domains. It adopts characteristic for all USP enzymes fold resembling the right hand. Residues C223, H464 and D481 form the catalytic triad of the enzyme that are located in a cleft between the two protein regions referred to as Thumb and Palm. The third region, Fingers, is responsible for binding to ubiquitin (58,77) (Figures 3A, 3B).
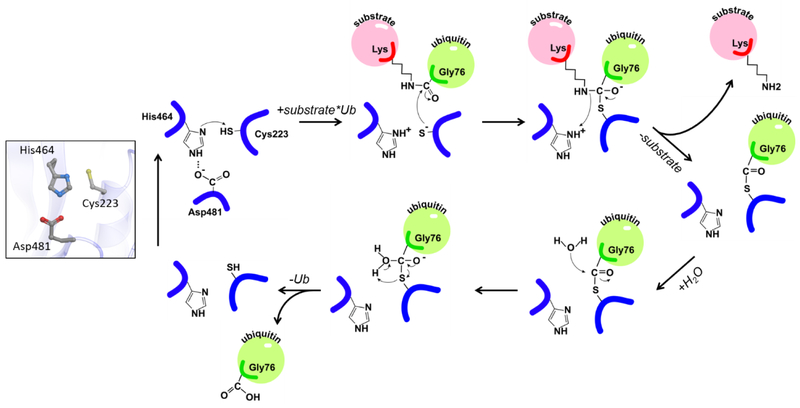
The catalytic mechanism of USP7 and other cysteine proteases is similar to that of well-studied plant enzymes papains (78) (Figure 4). The role of the catalytic histidine is to deprotonate the thiol group of the cysteine and initiate its nucleophilic attack on the isopeptide bond, while the aspartate restricts the side-chain rotation of the histidine leading to its polarization. Remarkably, active site conformation of the isolated USP7 catalytic domain differs from those observed in the available structures of other USPs. While in USP4, USP8, USP14 and CYLD the catalytic residues are properly positioned for catalysis (79-82), the USP7 catalytic triad is found in an unproductive conformation (Figure 3A inset and Supplementary Movie 1). The catalytically competent active site conformation was captured in the crystal structure of USP7 in complex with ubiquitin covalently attached to the catalytic cysteine (58,64). Based on the conformational changes seen in crystal structures, it was postulated that ubiquitin binding triggers activation of the enzyme (Figure 3A, inset).
Figure 4. Chemical reaction mechanism of catalysis by USP7.
The catalytic triad of USP7 is shown on the left. Cleavage of ubiquitinated substrate is shown on the right. The substrate and ubiquitin are shown in pink and green, respectively and labeled. The nucleophilic attack of the isopeptide bond is carried out by C223 of the catalytic triad. H464 serves as a proton acceptor and D481 restricts the side-chain rotation of the histidine.
Remarkably, recent NMR studies revealed that structural realignment into productive conformation is nit induced by interaction with free ubiquitin in solution. Although ubiquitin binds to the Fingers region of USP7 (KD of 106 ± 16 μM), its C-terminal tail does not enter the catalytic cleft and does not affect the active site conformation (83). These results suggest that the substrate-conjugated rather than free ubiquitin is necessary to activate USP7. This is not surprising considering that the C-terminus of free ubiquitin is negatively charged and unlikely to enter the hydrophobic catalytic cleft, while the conjugated ubiquitin lacks such negative charge. Upon the cleavage of ubiquitin moiety from the substrate the negative charge is restored, thus, causing free ubiquitin to leave the active site of the enzyme. Furthermore, interaction with other domains (64) and a substrate (59,60) may also be necessary to achieve the full activation of the enzyme.
5.3. USP7 specificity for polyubiquitin chain type:
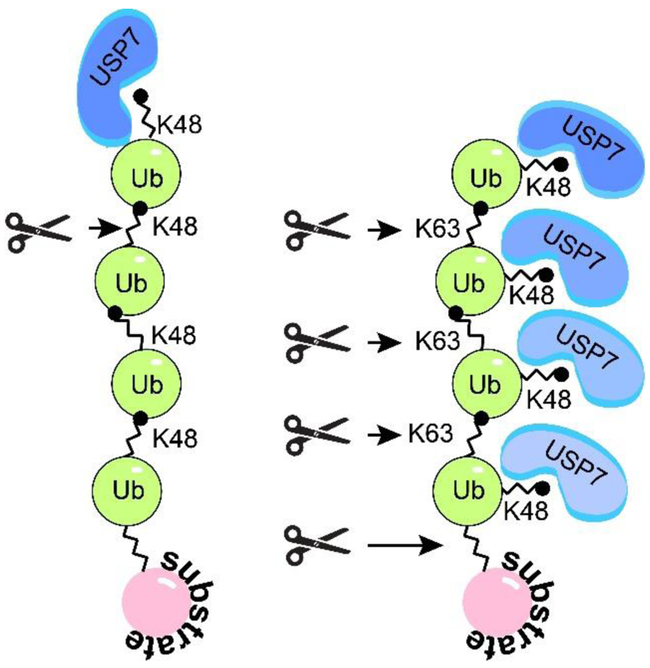
Ubiquitin contains seven lysine residues, including K6, K11, K27, K29, K33, K48, and K63, and all of them can be utilized during polyubiquitin chain formation, thus, determining the type of a polyubiquitin chain formed. USP7 deubiquitinates both mono- and poly-ubiquitinated substrates. Similar to other members of the USP family, USP7 is fairly promiscuous towards the type of poly-ubiquitin chains. While it cannot cleave the linear M1-linked poly-ubiquitin, USP7 cleaves K6, K11, K33, K48, and K63-linked chains connected by iso-peptide bonds, and less efficiently K27 and K29 chains (84). However, a recent study (85) revealed a marked difference between the way USP7 cleaves the two most common linkage types, a degrative K48 and a non-degrative K63 (Figure 5). The study suggests that USP7 requires free K48 ubiquitin side chain for the efficient binding, which means that the K48-linked chains can only be de-polymerized by one ubiquitin moiety at a time starting from the most distal ubiquitin in a chain (exo-cleavage), while the K63-linked chains can be cleaved between any two consecutive ubiquitin moieties (exo- and endo-cleavage) as well as between the substrate and the proximal ubiquitin (base-cleavage). This preference results in significantly faster depolymerization of K63- compared to K48-linked ubiquitin chains.
Figure 5. USP7 polyubiquitin cleavage specificity.
USP7 preferentialy binds free K48 side chains, which directs USP7 to the distal ubiquitin subunit of K48-polyubiquitin and promotes sequential exo-cleavage of K48-linked poly-ubiquitin. Whereas USP7 can bind all subunits of K63-polyubiquitin and promotes exo-, endo-, and base-cleavage. Adapted from (85).
5.4. Five UBL domains act as an additional substrate-binding platform
The C-terminal region of USP7 (C-USP7) spans residues 564-1102. Its five UBL domains are separated from the catalytic core by a 39 Å long α-helix. The extended nature of this rigid “connector’ helix is important for the USP7 activity (62). The UBL domains are arranged in the following manner: UBL1 closely interacts with UBL2, UBL4 shares the extensive interface with UBL5, and UBL3 is separated from UBL2 and UBL4 by flexible linkers (Figure 3B) (59,76,86). UBL domains are not unique to USP7; 18 other DUBs of the USP family also contain UBLs (87). Interestingly, the number of the domains varies from one in USP9X/Y, USP19, USP31 and several others to five in USP7, USP40 and USP47. Position of the UBLs within different USPs also varies, with some domains located at either N- or C-terminus, and others embedded in the catalytic core. Although such divergence within the USP protein family is surprising, the UBL domains appear to share a common regulatory function. They regulate catalytic activity of USPs by either inhibiting, or enhancing the enzyme’s activity, or localizing it to proteasome (79,88-90).
UBLs of USP7 can serve as a substrate-binding platform. Interestingly, despite the overall structural similarity, sequences of USP7 UBLs significantly vary from that of ubiquitin and each other resulting in unique surface charge distributions for each domain. The presence of differently charged surfaces implies that several distinct protein interaction sites may be harbored within the C-USP7. Mounting biochemical evidence suggests that different USP7 substrates are recognized by distinct UBL domains in its C-terminal region. Thus, UBL12 binds human XPC, RNF168, RNF169, DNMT1, UHRF1 and viral ICP0 (22-24,50,63,76,86,91), UBL3 contains secondary binding site for HDM2 (5), and UBL45 was shown to recognize p53 (5) and FOX(O)4 transcription factors (46). The distinct substrate-specific binding sites in the C-terminus on USP7 provide attractive targets for potential therapeutic intervention by manipulating USP7 activity toward a specific substrate or a group of substrates.
Recent structural studies of the USP7 UBL domains uncovered a mechanism of substrate recognition by UBL12 (24,63,76,86,91,92) (Table 2). A viral E3 ubiquitin ligase ICP0 from Herpes Simplex Virus 1 was identified as the first USP7 substrate that binds exclusively to its UBL domains. Several structures of UBLs in complex with ICP0 has now been reported (63,86,92) and revealed that the positively charged region of ICP0 interacts with the negatively charged patch on UBL2 surface. Furthermore, solution NMR studies showed that the ICP0-binding stabilizes the interface between UBL1 and UBL2 domains (92). In addition, crystal structures of the UBL domains in complexes with UHRF1, DNMT1 and RNF169 were recently published (24,76,91). Interestingly, structural analysis showed that all three proteins recognize UBL12 in a manner similar to ICP0, but in reverse orientation (Figure 3C). Specifically, these proteins share a positively charged KxxxKxK motif in their sequence that binds to the same negatively charged surface of UBL2 as ICP0, which has a RxKxxxK motif (61,63). The presence of such motif in a USP7 substrate can now be used to predict its interaction with the UBL12 tandem in a way P/A/ExxS motif has been used to predict binding to the N-terminal TRAF-like domain.
5.5. Auto-regulation of USP7 activity by its C-terminus
A conserved disordered peptide (residues 1080-1102) at the extreme C-terminus of USP7 is required for the enzyme’s activation. Its deletion significantly decreases the deubiquitinase activity of the DUB (5,59). The molecular mechanism of activation, however, remains unknown and raised controversy in the literature. According to the crystal structure of the UBL1-5 tandem, UBL domains are found in the extended conformation (Figure 3B), which positions the extreme C-terminus about 80 Å away from the enzyme’s active site. Furthermore, the isolated USP7 C-terminal peptide fails to directly interact with the catalytic core and only partially restores the enzymatic activity in trans (59). On the other hand, the catalytic domain of USP7 fused to the C-terminal peptide via a 10 amino residue linker has deubiquitinase activity comparable to that of the full-length protein (64). A crystal structure of the fusion complex engaged with ubiquitin offered a potential molecular mechanism for such stabilization. It shows that ubiquitin trapped in the active site of USP7 causes rearrangements of the catalytic triad and the adjacent “switching” loop of USP7. The conformational change in the “switching” loop, in turn, creates a binding site for the regulatory peptide, resulting in stabilization of catalytically competent enzyme (64).
The apparent discrepancy between the current structural models of the full-length USP7, which places the C-terminus 30-80 Å away from the catalytic site, and evidence of direct interaction between the catalytic and C-terminal domains suggests that a major conformational change is taking place during protein activation (Supplementary Movie 2). Flexible inter-domain linkers of USP7 may provide the means for domain rearrangement (62,63,86). Therefore, structural characterization of the USP7 conformations and dynamic exchange between them is necessary to fully understand the mechanism of its action.
6. Inhibition of USP7
Manipulating stability of proteins that are mutated, overexpressed or downregulated in human malignancies represents a promising therapeutic strategy for cancer treatment. Between the two protein degradation pathways, lysosomal proteolysis and UPS, the latter is highly selective and, therefore, its inhibition provides a strategy for the development of highly specific targeted therapies (93). E3 ligases and DUBs are of special interest since they determine the selectivity of UPS.
USP7 is a promising pharmaceutical target because of (i) its role in cellular pathways involving regulators of DNA damage, oncogenes and tumor suppressors and (ii) growing evidence of its aberrant expression in various cancer cell lines. Despite several USP7 inhibitors have been reported in the literature (94,95), the lack of co-crystal structures of USP7 with small-molecule compounds until recently has been a limiting factor in the development of potent and selective USP7 inhibitors. In the past year, several groups reported structures of USP7 in complexes with small molecule inhibitors (Figure 6, Supplementary Table 1) (83,85,96-98) , which triggered a rapid structure-based design of a number of potent and highly specific analogues (96-100).
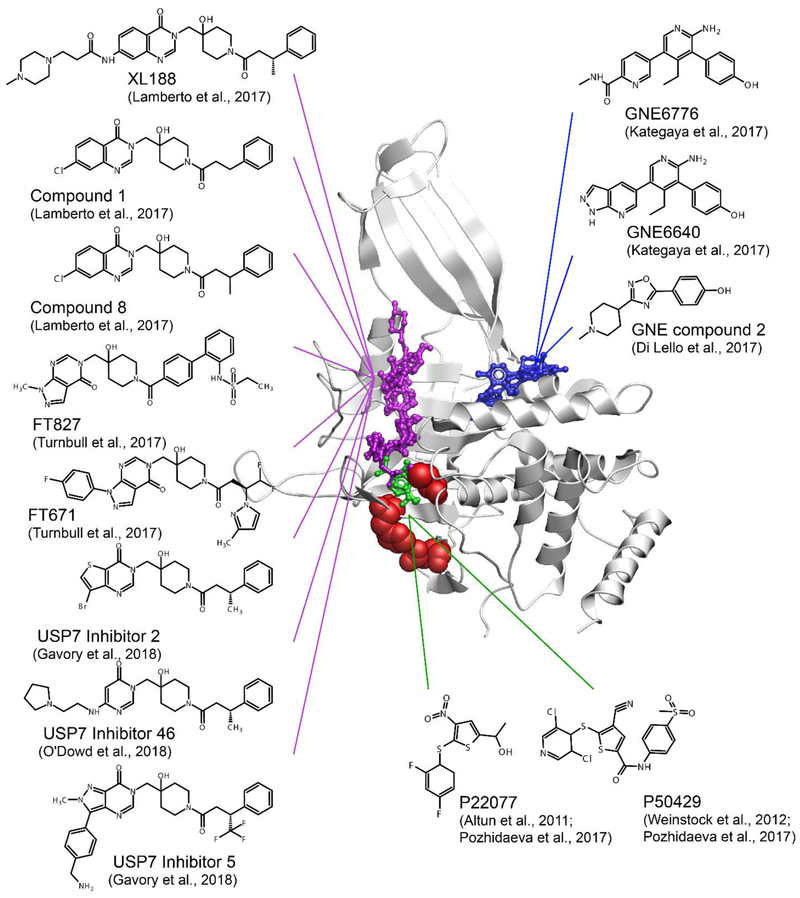
Figure 6. Small molecule inhibitors of USP7.
USP7 inhibitor complexes are overlayed and colored according to the region of the catalytic domain they target. Residues of the catalytic triad is shown as red spheres. The compounds that bind to the catalytic cleft are shown in purple, alosteric inhibitors that bind outside of the catalytic cleft are shown in blue, and those directly targeting the catalytic triad are shown in green. FT827 targers the catalytic cleft and extends to the catalytic triad. P22077, P50429 and FT827 covalently modify catalytic cystein C223. PDB 5UQX was used to display USP7 catalytic domain.
Solution NMR and mass spectrometry studies of interactions between the USP7 catalytic domain and P22077 (101) and P50429 (102) inhibitors uncovered the molecular mechanism of action of these thiophene-based compounds (Figure 6, green, and Supplementary Table 1). Both inhibitors bind to the active site of USP7 and covalently and irreversibly modify the catalytic cysteine C223 with remarkable specificity (83).
Co-crystal structures of several pyrimidinone based USP7 inhibitors were reported by several independent groups (96-99) and shown to bind the same narrow and long grove of the catalytic site normally occupied by the C-terminal tail of ubiquitin (Figure 6, purple). One of the inhibitors, FT827, carries vinylsulfonamide moiety, which reaches the catalytic triad and covalently modifies the catalytic cysteine. All other reported USP7 inhibitors are non-covalent.
Another class of allosteric inhibitors, GNE-6640, GNE-6776 (85) and USP7 inhibitor 2 (103), were shown to bind 12 Å away from the catalytic cysteine and impede ubiquitin binding (85) (Figure 6, blue). Using a different approach, Zhang et al. reported ubiquitin variants that selectively interact with USP7 blocking its interaction with ubiquitin (104). All these USP7 inhibitors exhibit cytotoxic activity in several cancer cell lines making them promising leads for further development and optimization that through rational structure-based design.
7. Concluding Remarks and Perspectives
USP7 plays an essential role in regulation of p53 pathway and, therefore, is a promising target for development of novel therapies for cancer treatment. Research efforts of recent years brought significant breakthroughs in our understanding of USP7 structure, regulation and inhibition. However, many questions remain to be answered. What is the structure of the full length enzyme and what conformations can it sample? What role does conformational dynamics play in USP7 activation and substrate recognition? How does USP7 achieve specificity to its many diverse substrates, and can it be pharmaceutically manipulated to change affinity to a particular substrate or a group of substrates?
Future studies of structure and dynamics of USP7 will undoubtedly provide detailed understanding of regulation of its activity and specificity, leading to development of new approaches to cancer treatment.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C, and Ciechanover A (2016) The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. The international journal of biochemistry & cell biology 79, 403–418 [DOI] [PubMed] [Google Scholar]
- 2.Hanpude P, Bhattacharya S, Dey AK, and Maiti TK (2015) Deubiquitinating enzymes in cellular signaling and disease regulation. IUBMB life 67, 544–555 [DOI] [PubMed] [Google Scholar]
- 3.Kessler BM, Fortunati E, Melis M, Pals CE, Clevers H, and Maurice MM (2007) Proteome changes induced by knock-down of the deubiquitylating enzyme HAUSP/USP7. Journal of proteome research 6, 4163–4172 [DOI] [PubMed] [Google Scholar]
- 4.Hu M, Gu L, Li M, Jeffrey PD, Gu W, and Shi Y (2006) Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS biology 4, e27 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J, Martin JD, Xue Y, Lor LA, Kennedy-Wilson KM, Sinnamon RH, Ho TF, Zhang G, Schwartz B, Tummino PJ, and Lai Z (2010) C-terminal region of USP7/HAUSP is critical for deubiquitination activity and contains a second mdm2/p53 binding site. Archives of biochemistry and biophysics 503, 207–212 [DOI] [PubMed] [Google Scholar]
- 6.Nicholson B, and Kumar KGS (2011) The multifaceted roles of USP7: new therapeutic opportunities. Cell biochemistry and biophysics 60, 61–68 [DOI] [PubMed] [Google Scholar]
- 7.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, and Frappier L (2006) Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nature structural & molecular biology 13, 285–291 [DOI] [PubMed] [Google Scholar]
- 8.Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, and Gu W (2010) Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29, 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kon N, Zhong J, Kobayashi Y, Li M, Szabolcs M, Ludwig T, Canoll PD, and Gu W (2011) Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell death and differentiation 18, 1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fountain MD, Oleson DS, Rech ME, Segebrecht L, Hunter JV, McCarthy JM, Lupo PJ, Holtgrewe M, Moran R, Rosenfeld JA, Isidor B, Le Caignec C, Saenz MS, Pedersen RC, Morgan TM, Pfotenhauer JP, Xia F, Bi W, Kang SL, Patel A, Krantz ID, Raible SE, Smith W, Cristian I, Torti E, Juusola J, Millan F, Wentzensen IM, Person RE, Kury S, Bezieau S, Uguen K, Ferec C, Munnich A, van Haelst M, Lichtenbelt KD, van Gassen K, Hagelstrom T, Chawla A, Perry DL, Taft RJ, Jones M, Masser-Frye D, Dyment D, Venkateswaran S, Li C, Escobar LF, Horn D, Spillmann RC, Pena L, Wierzba J, Strom TM, Parenti I, Kaiser FJ, Ehmke N, and Schaaf CP (2019) Pathogenic variants in USP7 cause a neurodevelopmental disorder with speech delays, altered behavior, and neurologic anomalies. Genetics in medicine : official journal of the American College of Medical Genetics 10.1038/s41436-019-0433-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carra G, Panuzzo C, Torti D, Parvis G, Crivellaro S, Familiari U, Volante M, Morena D, Lingua MF, Brancaccio M, Guerrasio A, Pandolfi PP, Saglio G, Taulli R, and Morotti A (2017) Therapeutic inhibition of USP7-PTEN network in chronic lymphocytic leukemia: a strategy to overcome TP53 mutated/deleted clones. Oncotarget 8, 35508–35522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, and Pandolfi PP (2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng C, Niu C, Yang Y, Wang Y, and Lu M (2013) Expression of HAUSP in gliomas correlates with disease progression and survival of patients. Oncology reports 29, 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Ma S, Song N, Li X, Liu L, Yang S, Ding X, Shan L, Zhou X, Su D, Wang Y, Zhang Q, Liu X, Yu N, Zhang K, Shang Y, Yao Z, and Shi L (2016) Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. The Journal of clinical investigation 126, 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao GY, Lin ZW, Lu CL, Gu J, Yuan YF, Xu FK, Liu RH, Ge D, and Ding JY (2015) USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour biology : the journal of the international Society for Oncodevelopmental Biology and Medicine 36, 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma M, and Yu N (2016) Ubiquitin-specific protease 7 expression is a prognostic factor in epithelial ovarian cancer and correlates with lymph node metastasis. OncoTargets and therapy 9, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Wang H, Tian L, and Li H (2016) Expression of USP7 and MARCH7 Is Correlated with Poor Prognosis in Epithelial Ovarian Cancer. The Tohoku journal of experimental medicine 239, 165–175 [DOI] [PubMed] [Google Scholar]
- 18.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, and Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 [DOI] [PubMed] [Google Scholar]
- 19.Li M, Brooks CL, Kon N, and Gu W (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Molecular cell 13, 879–886 [DOI] [PubMed] [Google Scholar]
- 20.Brooks CL, Li M, Hu M, Shi Y, and Gu W (2007) The p53--Mdm2--HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 26, 7262–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, and Dianov GL (2012) ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Molecular cell 45, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, and Wani AA (2014) Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. The Journal of biological chemistry 289, 27278–27289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Q, Sharma N, He J, Wani G, and Wani AA (2015) USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168. Cell cycle 14, 1413–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An L, Jiang Y, Ng HH, Man EP, Chen J, Khoo US, Gong Q, and Huen MS (2017) Dual-utility NLS drives RNF169-dependent DNA damage responses. Proceedings of the National Academy of Sciences of the United States of America 114, E2872–E2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas K, Philip S, Yadav A, Martin BK, Burkett S, Singh V, Babbar A, North SL, Chang S, and Sharan SK (2018) BRE/BRCC45 regulates CDC25A stability by recruiting USP7 in response to DNA damage. Nature communications 9, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso-de Vega I, Martin Y, and Smits VA (2014) USP7 controls Chk1 protein stability by direct deubiquitination. Cell cycle, 3921–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, and Mailand N (2009) USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. The Journal of cell biology 184, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, and Marteijn JA (2012) UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nature genetics 44, 598–602 [DOI] [PubMed] [Google Scholar]
- 29.Qing P, Han L, Bin L, Yan L, and Ping WX (2011) USP7 regulates the stability and function of HLTF through deubiquitination. Journal of cellular biochemistry 112, 3856–3862 [DOI] [PubMed] [Google Scholar]
- 30.Zlatanou A, Sabbioneda S, Miller ES, Greenwalt A, Aggathanggelou A, Maurice MM, Lehmann AR, Stankovic T, Reverdy C, Colland F, Vaziri C, and Stewart GS (2016) USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene 35, 965–976 [DOI] [PubMed] [Google Scholar]
- 31.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, and Walker GC (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and molecular biology reviews : MMBR 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian J, Pentz K, Zhu Q, Wang Q, He J, Srivastava AK, and Wani AA (2015) USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene 34, 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagannathan M, Nguyen T, Gallo D, Luthra N, Brown GW, Saridakis V, and Frappier L (2014) A role for USP7 in DNA replication. Molecular and cellular biology 34, 132–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lecona E, Rodriguez-Acebes S, Specks J, Lopez-Contreras AJ, Ruppen I, Murga M, Munoz J, Mendez J, and Fernandez-Capetillo O (2016) USP7 is a SUMO deubiquitinase essential for DNA replication. Nature structural & molecular biology 23, 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smits VA, and Freire R (2016) USP7/HAUSP: A SUMO deubiquitinase at the heart of DNA replication. BioEssays : news and reviews in molecular, cellular and developmental biology 38, 863–868 [DOI] [PubMed] [Google Scholar]
- 36.Hernandez-Perez S, Cabrera E, Salido E, Lim M, Reid L, Lakhani SR, Khanna KK, Saunus JM, and Freire R (2017) DUB3 and USP7 de-ubiquitinating enzymes control replication inhibitor Geminin: molecular characterization and associations with breast cancer. Oncogene 36, 4802–4809 [DOI] [PubMed] [Google Scholar]
- 37.Felle M, Joppien S, Nemeth A, Diermeier S, Thalhammer V, Dobner T, Kremmer E, Kappler R, and Langst G (2011) The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic acids research 39, 8355–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin W, Leonhardt H, and Spada F (2011) Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. Journal of cellular biochemistry 112, 439–444 [DOI] [PubMed] [Google Scholar]
- 39.de Bie P, Zaaroor-Regev D, and Ciechanover A (2010) Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochemical and biophysical research communications 400, 389–395 [DOI] [PubMed] [Google Scholar]
- 40.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, and Peters G (2010) Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. The EMBO journal 29, 2553–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lecona E, Narendra V, and Reinberg D (2015) USP7 cooperates with SCML2 to regulate the activity of PRC1. Molecular and cellular biology 35, 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dar A, Shibata E, and Dutta A (2013) Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Molecular and cellular biology 33, 3309–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding X, Jiang W, Zhou P, Liu L, Wan X, Yuan X, Wang X, Chen M, Chen J, Yang J, Kong C, Li B, Peng C, Wong CC, Hou F, and Zhang Y (2015) Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PloS one 10, e0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi L, Cui Y, Xu Q, and Jiang Y (2016) Stabilization of LSD1 by deubiquitinating enzyme USP7 promotes glioblastoma cell tumorigenesis and metastasis through suppression of the p53 signaling pathway. Oncology reports 36, 2935–2945 [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi L, Nishiyama A, Misaki T, Johmura Y, Ueda J, Arita K, Nagao K, Obuse C, and Nakanishi M (2017) Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation. Scientific reports 7, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, and Burgering BM (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature cell biology 8, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 47.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, and Parkinson J (1997) A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. The EMBO journal 16, 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boutell C, and Everett RD (2013) Regulation of alphaherpesvirus infections by the ICP0 family of proteins. The Journal of general virology 94, 465–481 [DOI] [PubMed] [Google Scholar]
- 49.Canning M, Boutell C, Parkinson J, and Everett RD (2004) A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. The Journal of biological chemistry 279, 38160–38168 [DOI] [PubMed] [Google Scholar]
- 50.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, and Frappier L (2003) Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. The Journal of biological chemistry 278, 47753–47761 [DOI] [PubMed] [Google Scholar]
- 51.Jager W, Santag S, Weidner-Glunde M, Gellermann E, Kati S, Pietrek M, Viejo-Borbolla A, and Schulz TF (2012) The ubiquitin-specific protease USP7 modulates the replication of Kaposi's sarcoma-associated herpesvirus latent episomal DNA. Journal of virology 86, 6745–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, Arrowsmith CH, and Frappier L (2005) Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Molecular cell 18, 25–36 [DOI] [PubMed] [Google Scholar]
- 53.Lee HR, Choi WC, Lee S, Hwang J, Hwang E, Guchhait K, Haas J, Toth Z, Jeon YH, Oh TK, Kim MH, and Jung JU (2011) Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nature structural & molecular biology 18, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chavoshi S, Egorova O, Lacdao IK, Farhadi S, Sheng Y, and Saridakis V (2016) Identification of Kaposi Sarcoma Herpesvirus (KSHV) vIRF1 Protein as a Novel Interaction Partner of Human Deubiquitinase USP7. The Journal of biological chemistry 291, 6281–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, and Frappier L (2012) Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. Journal of virology 86, 806–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ching W, Koyuncu E, Singh S, Arbelo-Roman C, Hartl B, Kremmer E, Speiseder T, Meier C, and Dobner T (2013) A ubiquitin-specific protease possesses a decisive role for adenovirus replication and oncogene-mediated transformation. PLoS pathogens 9, e1003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ali A, Raja R, Farooqui SR, Ahmad S, and Banerjea AC (2017) USP7 deubiquitinase controls HIV-1 production by stabilizing Tat protein. The Biochemical journal 474, 1653–1668 [DOI] [PubMed] [Google Scholar]
- 58.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, and Shi Y (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054 [DOI] [PubMed] [Google Scholar]
- 59.Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, and Sixma TK (2011) Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Molecular cell 44, 147–159 [DOI] [PubMed] [Google Scholar]
- 60.Kim RQ, Geurink PP, Mulder MPC, Fish A, Ekkebus R, El Oualid F, van Dijk WJ, van Dalen D, Ovaa H, van Ingen H, and Sixma TK (2019) Kinetic analysis of multistep USP7 mechanism shows critical role for target protein in activity. Nature communications 10, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim RQ, and Sixma TK (2017) Regulation of USP7: A High Incidence of E3 Complexes. Journal of molecular biology 429, 3395–3408 [DOI] [PubMed] [Google Scholar]
- 62.Kim RQ, van Dijk WJ, and Sixma TK (2016) Structure of USP7 catalytic domain and three Ubl-domains reveals a connector alpha-helix with regulatory role. Journal of structural biology 195, 11–18 [DOI] [PubMed] [Google Scholar]
- 63.Pfoh R, Lacdao IK, Georges AA, Capar A, Zheng H, Frappier L, and Saridakis V (2015) Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7. PLoS pathogens 11, e1004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rouge L, Bainbridge TW, Kwok M, Tong R, Di Lello P, Wertz IE, Maurer T, Ernst JA, and Murray J (2016) Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation. Structure 24, 1335–1345 [DOI] [PubMed] [Google Scholar]
- 65.Fernandez-Montalvan A, Bouwmeester T, Joberty G, Mader R, Mahnke M, Pierrat B, Schlaeppi JM, Worpenberg S, and Gerhartz B (2007) Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. The FEBS journal 274, 4256–4270 [DOI] [PubMed] [Google Scholar]
- 66.Zaman MM, Nomura T, Takagi T, Okamura T, Jin W, Shinagawa T, Tanaka Y, and Ishii S (2013) Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Molecular and cellular biology 33, 4971–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morotti A, Panuzzo C, Crivellaro S, Pergolizzi B, Familiari U, Berger AH, Saglio G, and Pandolfi PP (2014) BCR-ABL disrupts PTEN nuclear-cytoplasmic shuttling through phosphorylation-dependent activation of HAUSP. Leukemia 28, 1326–1333 [DOI] [PubMed] [Google Scholar]
- 68.Liu X, Yang X, Li Y, Zhao S, Li C, Ma P, and Mao B (2016) Trip12 is an E3 ubiquitin ligase for USP7/HAUSP involved in the DNA damage response. FEBS letters 590, 4213–4222 [DOI] [PubMed] [Google Scholar]
- 69.Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, and Yang X (2006) Critical role for Daxx in regulating Mdm2. Nature cell biology 8, 855–862 [DOI] [PubMed] [Google Scholar]
- 70.Giovinazzi S, Morozov VM, Summers MK, Reinhold WC, and Ishov AM (2013) USP7 and Daxx regulate mitosis progression and taxane sensitivity by affecting stability of Aurora-A kinase. Cell death and differentiation 20, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma P, Yang X, Kong Q, Li C, Yang S, Li Y, and Mao B (2014) The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of beta-catenin. Molecular and cellular biology 34, 4355–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meng H, Harrison DJ, and Meehan RR (2015) MBD4 interacts with and recruits USP7 to heterochromatic foci. Journal of cellular biochemistry 116, 476–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi P, Xia W, Wu RC, Lonard DM, Hung MC, and O'Malley BW (2013) SRC-3 coactivator regulates cell resistance to cytotoxic stress via TRAF4-mediated p53 destabilization. Genes & development 27, 274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarkari F, La Delfa A, Arrowsmith CH, Frappier L, Sheng Y, and Saridakis V (2010) Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. Journal of molecular biology 402, 825–837 [DOI] [PubMed] [Google Scholar]
- 75.Sarkari F, Wheaton K, La Delfa A, Mohamed M, Shaikh F, Khatun R, Arrowsmith CH, Frappier L, Saridakis V, and Sheng Y (2013) Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1. The Journal of biological chemistry 288, 16975–16985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, Li Z, and Xu Y (2015) Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nature communications 6, 7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Molland K, Zhou Q, and Mesecar AD (2014) A 2.2 A resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding. Acta crystallographica. Section F, Structural biology communications 70, 283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Storer AC, and Menard R (1994) Catalytic mechanism in papain family of cysteine peptidases. Methods in enzymology 244, 486–500 [DOI] [PubMed] [Google Scholar]
- 79.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, and Shi Y (2005) Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. The EMBO journal 24, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Avvakumov GV, Walker JR, Xue S, Finerty PJ Jr., Mackenzie F, Newman EM, and Dhe-Paganon S (2006) Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). The Journal of biological chemistry 281, 38061–38070 [DOI] [PubMed] [Google Scholar]
- 81.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, and Barford D (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Molecular cell 29, 451–464 [DOI] [PubMed] [Google Scholar]
- 82.Clerici M, Luna-Vargas MP, Faesen AC, and Sixma TK (2014) The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nature communications 5, 5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pozhidaeva A, Valles G, Wang F, Wu J, Sterner DE, Nguyen P, Weinstock J, Kumar KGS, Kanyo J, Wright D, and Bezsonova I (2017) USP7-Specific Inhibitors Target and Modify the Enzyme's Active Site via Distinct Chemical Mechanisms. Cell Chem Biol 24, 1501–1512 e1505 [DOI] [PubMed] [Google Scholar]
- 84.Faesen AC, Luna-Vargas MP, Geurink PP, Clerici M, Merkx R, van Dijk WJ, Hameed DS, El Oualid F, Ovaa H, and Sixma TK (2011) The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chemistry & biology 18, 1550–1561 [DOI] [PubMed] [Google Scholar]
- 85.Kategaya L, Di Lello P, Rouge L, Pastor R, Clark KR, Drummond J, Kleinheinz T, Lin E, Upton JP, Prakash S, Heideker J, McCleland M, Ritorto MS, Alessi DR, Trost M, Bainbridge TW, Kwok MCM, Ma TP, Stiffler Z, Brasher B, Tang Y, Jaishankar P, Hearn BR, Renslo AR, Arkin MR, Cohen F, Yu K, Peale F, Gnad F, Chang MT, Klijn C, Blackwood E, Martin SE, Forrest WF, Ernst JA, Ndubaku C, Wang X, Beresini MH, Tsui V, Schwerdtfeger C, Blake RA, Murray J, Maurer T, and Wertz IE (2017) USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550, 534–538 [DOI] [PubMed] [Google Scholar]
- 86.Cheng J, Li Z, Gong R, Fang J, Yang Y, Sun C, Yang H, and Xu Y (2015) Molecular mechanism for the substrate recognition of USP7. Protein & cell 6, 849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, and Urbe S (2013) Deubiquitylases from genes to organism. Physiological reviews 93, 1289–1315 [DOI] [PubMed] [Google Scholar]
- 88.Faesen AC, Luna-Vargas MP, and Sixma TK (2012) The role of UBL domains in ubiquitin-specific proteases. Biochemical Society transactions 40, 539–545 [DOI] [PubMed] [Google Scholar]
- 89.Zhao B, Velasco K, Sompallae R, Pfirrmann T, Masucci MG, and Lindsten K (2012) The ubiquitin specific protease-4 (USP4) interacts with the S9/Rpn6 subunit of the proteasome. Biochemical and biophysical research communications 427, 490–496 [DOI] [PubMed] [Google Scholar]
- 90.Zhang Q, Harding R, Hou F, Dong A, Walker JR, Bteich J, and Tong Y (2016) Structural basis of the recruitment of ubiquitin-specific protease USP15 by spliceosome recycling factor SART3. The Journal of biological chemistry 291, 17283–17292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang ZM, Rothbart SB, Allison DF, Cai Q, Harrison JS, Li L, Wang Y, Strahl BD, Wang GG, and Song J (2015) An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell reports 12, 1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pozhidaeva AK, Mohni KN, Dhe-Paganon S, Arrowsmith CH, Weller SK, Korzhnev DM, and Bezsonova I (2015) Structural Characterization of Interaction between Human Ubiquitin-specific Protease 7 and Immediate-Early Protein ICP0 of Herpes Simplex Virus-1. The Journal of biological chemistry 290, 22907–22918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buckley DL, and Crews CM (2014) Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system. Angewandte Chemie 53, 2312–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, Calabrese A, Borg-Capra C, Sippl W, Collura V, Boissy G, Rain JC, Guedat P, Delansorne R, and Daviet L (2009) Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular cancer therapeutics 8, 2286–2295 [DOI] [PubMed] [Google Scholar]
- 95.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, and Colland F (2012) Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry & biology 19, 467–477 [DOI] [PubMed] [Google Scholar]
- 96.Gavory G, O'Dowd CR, Helm MD, Flasz J, Arkoudis E, Dossang A, Hughes C, Cassidy E, McClelland K, Odrzywol E, Page N, Barker O, Miel H, and Harrison T (2018) Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nature chemical biology 14, 118–125 [DOI] [PubMed] [Google Scholar]
- 97.Lamberto I, Liu X, Seo HS, Schauer NJ, Iacob RE, Hu W, Das D, Mikhailova T, Weisberg EL, Engen JR, Anderson KC, Chauhan D, Dhe-Paganon S, and Buhrlage SJ (2017) Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7. Cell Chem Biol 24, 1490–1500 e1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, Tonkin LM, Townsend EC, Buker SM, Lancia DR, Caravella JA, Toms AV, Charlton TM, Lahdenranta J, Wilker E, Follows BC, Evans NJ, Stead L, Alli C, Zarayskiy VV, Talbot AC, Buckmelter AJ, Wang M, McKinnon CL, Saab F, McGouran JF, Century H, Gersch M, Pittman MS, Marshall CG, Raynham TM, Simcox M, Stewart LMD, McLoughlin SB, Escobedo JA, Bair KW, Dinsmore CJ, Hammonds TR, Kim S, Urbe S, Clague MJ, Kessler BM, and Komander D (2017) Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Dowd CR, Helm MD, Rountree JSS, Flasz JT, Arkoudis E, Miel H, Hewitt PR, Jordan L, Barker O, Hughes C, Rozycka E, Cassidy E, McClelland K, Odrzywol E, Page N, Feutren-Burton S, Dvorkin S, Gavory G, and Harrison T (2018) Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors. ACS medicinal chemistry letters 9, 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang F, Wang L, Wu J, Sokirniy I, Nguyen P, Bregnard T, Weinstock J, Mattern M, Bezsonova I, Hancock WW, and Kumar S (2017) Active site-targeted covalent irreversible inhibitors of USP7 impair the functions of Foxp3+ T-regulatory cells by promoting ubiquitination of Tip60. PloS one 12, e0189744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KGS, Konietzny R, Fischer R, Kogan E, Mackeen MM, McGouran J, Khoronenkova SV, Parsons JL, Dianov GL, Nicholson B, and Kessler BM (2011) Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chemistry & biology 18, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 102.Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Kumar KGS, Goldenberg SJ, Mattern MR, and Nicholson B (2012) Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS medicinal chemistry letters 3, 789–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Di Lello P, Pastor R, Murray JM, Blake RA, Cohen F, Crawford TD, Drobnick J, Drummond J, Kategaya L, Kleinheinz T, Maurer T, Rouge L, Zhao X, Wertz I, Ndubaku C, and Tsui V (2017) Discovery of Small-Molecule Inhibitors of Ubiquitin Specific Protease 7 (USP7) Using Integrated NMR and in Silico Techniques. Journal of medicinal chemistry 60, 10056–10070 [DOI] [PubMed] [Google Scholar]
- 104.Zhang W, Sartori MA, Makhnevych T, Federowicz KE, Dong X, Liu L, Nim S, Dong A, Yang J, Li Y, Haddad D, Ernst A, Heerding D, Tong Y, Moffat J, and Sidhu SS (2017) Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10. Journal of molecular biology 429, 3546–3560 [DOI] [PubMed] [Google Scholar]
- 105.Bhattacharya S, and Ghosh MK (2014) HAUSP, a novel deubiquitinase for Rb - MDM2 the critical regulator. The FEBS journal 281, 3061–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, and Verrijzer CP (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Molecular cell 17, 695–707 [DOI] [PubMed] [Google Scholar]
- 107.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, Sedwick D, Ewing RM, and Wang Z (2010) DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science signaling 3, ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee KW, Cho JG, Kim CM, Kang AY, Kim M, Ahn BY, Chung SS, Lim KH, Baek KH, Sung JH, Park KS, and Park SG (2013) Herpesvirus-associated ubiquitin-specific protease (HAUSP) modulates peroxisome proliferator-activated receptor gamma (PPARgamma) stability through its deubiquitinating activity. The Journal of biological chemistry 288, 32886–32896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tavana O, Li D, Dai C, Lopez G, Banerjee D, Kon N, Chen C, Califano A, Yamashiro DJ, Sun H, and Gu W (2016) HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nature medicine 22, 1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oh YM, Yoo SJ, and Seol JH (2007) Deubiquitination of Chfr, a checkpoint protein, by USP7/HAUSP regulates its stability and activity. Biochemical and biophysical research communications 357, 615–619 [DOI] [PubMed] [Google Scholar]
- 111.Giovinazzi S, Sirleto P, Aksenova V, Morozov VM, Zori R, Reinhold WC, and Ishov AM (2014) Usp7 protects genomic stability by regulating Bub3. Oncotarget 5, 3728–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zemp I, and Lingner J (2014) The shelterin component TPP1 is a binding partner and substrate for the deubiquitinating enzyme USP7. The Journal of biological chemistry 289, 28595–28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Colleran A, Collins PE, O'Carroll C, Ahmed A, Mao X, McManus B, Kiely PA, Burstein E, and Carmody RJ (2013) Deubiquitination of NF-kappaB by Ubiquitin-Specific Protease-7 promotes transcription. Proceedings of the National Academy of Sciences of the United States of America 110, 618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang J, Qu L, Pang M, and Yang X (2010) Daxx is reciprocally regulated by Mdm2 and Hausp. Biochemical and biophysical research communications 393, 542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Daubeuf S, Singh D, Tan Y, Liu H, Federoff HJ, Bowers WJ, and Tolba K (2009) HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood 113, 3264–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hao YH, Fountain MD Jr., Fon Tacer K, Xia F, Bi W, Kang SH, Patel A, Rosenfeld JA, Le Caignec C, Isidor B, Krantz ID, Noon SE, Pfotenhauer JP, Morgan TM, Moran R, Pedersen RC, Saenz MS, Schaaf CP, and Potts PR (2015) USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Molecular cell 59, 956–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, and Frappier L (2003) Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. The Journal of biological chemistry 278, 29987–29994 [DOI] [PubMed] [Google Scholar]
- 118.Khoronenkova SV, and Dianov GL (2013) USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic acids research 41, 1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.