Abstract
Purpose: Triple-negative breast cancer (TNBC) is a highly heterogeneous disease. It is very important to explore novel biomarkers to better clarify the characteristics of TNBC. It has been reported that polymorphisms in claudin 1 (CLDN1) are associated with risk of several cancers. But till now, there is no report about these polymorphisms and TNBC. Patients and methods: Between January 2004 and December 2013, 267 patients with stage I–III primary TNBC were included in our study. We investigated the association between polymorphisms in CLDN1 gene and clinicopathological characteristics or survival of these patients. We used Haploview 4.2 software to identify Tag single nucleotide polymorphisms (SNPs). MassARRAY MALDI-TOF System was used for genotyping. Results: We found that rs10513846 GA genotype was associated with older age [P=0.013, hazard ratios (HR) = 2.231, 95% confidence interval (CI): 1.186–4.195]. Rs10513846 AA genotype carriers were more likely to develop grade 3 tumors (P=0.005, HR = 2.889, 95% CI: 1.389–6.007). And rs9283658 genotypes were also related to grade, more patients with grade 3 tumors were rs9283658 CC genotype carriers (P=0.023, HR = 0.446, 95% CI: 0.222–0.894). There was no association between polymorphisms in CLDN1 and survival of TNBC patients. After multivariate analysis, tumor size (P=0.021, HR = 3.146, 95% CI: 1.185–8.354) and lymph node status (P<0.001, HR = 10.930, 95% CI: 3.276–36.470) were demonstrated to be independent prognostic factors. Conclusion: We first demonstrated that polymorphisms in CLDN1 gene were associated with age and differentiation of TNBC patients.
Keywords: Claudin, clinicopathological characteristics, polymorphism, TNBC
Introduction
Breast cancer is one of the most common cancers in women around the world [1]. It is a highly heterogeneous disease which for decades has been divided into several subgroups according to immunohistochemical staining (IHC) of three receptors: estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor 2 (HER2) [2]. Triple-negative breast cancer (TNBC) is defined as lacking expression of ER, PR, and HER2. It accounts for 15–20% of all breast cancers and is characterized by enhanced invasiveness and metastatic capacity, young age of onset and poor prognosis [3]. However, even within TNBC patients, distinct response to treatments and prognosis was observed [4]. With the development of molecular profile, four to six distinct subtypes have been defined within TNBC, such as basal-like and claudin-low [5]. Investigators are making more efforts to explore novel biomarkers to clarify the characteristics of TNBC [6].
Claudins (CLDNs) are key cell adhesion molecules, which compose tight junctions (TJs), regulate paracellular permeability, and maintain cell polarity [7]. There are 27 members in CLDNs family, each member is predicted to possess four transmembrane domains with intracellular amino and carboxyl-termini in the cytoplasm and two extracellular loops [8]. It has been reported that CLDN1 expression levels were decreased in breast cancer [9], colorectal carcinoma [10], glioblastoma [11], and melanoma brain metastasis [12]. In contrast, only a few literatures reported about polymorphisms in CLDN genes and their role in cancer development [13,14]. Our study was designed to explore the relationship between genetic variants in CLDN1 and clinicopathological characteristics or survival of TNBC.
Materials and methods
Study population
Between January 2004 and December 2013, 267 patients with stage I–III primary TNBC according to American Join Committee on Cancer 2010 classification [15] were included in our study. ER, PR, and HER2 status were evaluated according to the guidelines issued by the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) in 2010 [16,17]. Tumors negative for ER, PR, and HER2 were defined as TNBCs. Clinical data such as age, tumor sizes, regional lymph node status, histopathologic grading, and vascular invasion were collected. Follow-up visits were performed every 3 months for 2 years, then every 6 months for 3 years, then annually. Patients were followed until December 2017 to collect data on recurrence and death.
This investigation was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Hospital and Jiangxi Cancer Hospital. It was conducted in accordance with the ethical standards of the Declaration of Helsinki and following the national and international guidelines. Written informed consent was obtained from all patients.
Single nucleotide polymorphism selection and genotyping
Peripheral blood samples (5 ml) were collected from each patient upon recruitment and stored in −20°C for DNA extraction. Genotype data from CLDN1 gene regions encompassing 5 kb of upstream and 5 kb of downstream flanking sequences were extracted from the HapMap Chinese Han population. Haploview 4.2 software was used to identify Tag single nucleotide polymorphisms (SNPs). The inclusion criteria were SNPs known in ethnic Han Chinese population and with a minor allele frequency (MAF) >0.05 and r2 > 0.8. A total of five candidate SNPs were selected for genotyping (Table 1). Primers and probes were designed by MassARRAY Typer 4.0 software. MassARRAY MALDI-TOF System (Sequenom Inc., San Diego, CA, U.S.A.) [18,19] was used for genotyping by the method described in the Sequenom Genotyping Protocol.
Table 1. Information for the SNPs genotyped in the present study.
| SNPs | Position | Location | Alleles | MAF |
|---|---|---|---|---|
| rs10513846 | 3:190313200 | Intron variant | A/G | 0.2017 |
| rs1155884 | 3:190323165 | Upstream variant 2KB | A/C | 0.4631 |
| rs8298 | 3:190305763 | UTR variant 3 prime | C/T | 0.2788 |
| rs9842214 | 3:190305586 | Intron variant | C/T | 0.3223 |
| rs9283658 | 3:190306476 | Variant 3 prime | C/T | 0.1749 |
Statistical analyses
Statistical analysis was performed using SPSS version 18.0 (SPSS Inc, Chicago, IL, U.S.A.). The distribution of genotypes in patients with different clinicopathological characteristics was compared by two-sided Pearson’s χ2 tests, odds ratios (ORs) and 95% confidence intervals (CI) were calculated by logistic regression. Disease-free survival (DFS) was calculated from date of diagnosis to date of first locoregional recurrence, first distant metastasis, or death from any cause (whichever came first). Overall survival (OS) was calculated from date of diagnosis to date of death from any reason or last follow-up. Kaplan–Meier curves with the log-rank test were applied to estimate and compare 5-year DFS and OS rates of patients with different genotypes. Hazard ratios (HR) of recurrence/metastasis and death with 95% CI were estimated by Cox-regression model. The multivariate analysis was adjusted for age, histological grade, tumor size, lymph node status, and vascular invasion. All statistical tests were two-sided, and P<0.05 was considered significant.
Results
Clinical characteristics and survival of TNBC patients
A total of 267 TNBC patients were enrolled in our study. The median age at diagnosis is 47 years old (range from 23 to 75). The 5-year OS and DFS rate were 86.6 and 72.1%, respectively. More patients were ≤50 years old and with grade 3 tumors. 81 (30.3%), 135 (50.6%), and 51 (19.1%) subjects were diagnosed at stage I, II, and III. The association between clinicopathological characteristics and survival was listed in Table 2. Patients with grade 3 tumors had a significantly poorer 5-year OS rate than those with grade 1–2 tumors (78.3 vs 93.9%, P=0.029, HR = 2.445, 95% CI: 1.094–5.463). Tumor size and lymph node status were significantly associated with both DFS and OS. There was no significant association between age, vascular invasion, and TNBC survival. After multivariate analysis, tumor size (P=0.021, HR = 3.146, 95% CI: 1.185–8.354) and lymph node status (P<0.001, HR = 10.930, 95% CI: 3.276–36.470) were demonstrated to be independent prognostic factors.
Table 2. Clinicopathological characteristics and survival of TNBC.
| Variables | Patients (%) | 5-year DFS (%) | HR (95% CI) | P-value | 5-year OS (%) | HR (95% CI) | P-value |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| ≤50 | 164 (61.4) | 68.2 | 1 (Ref) | 86.7 | 1 (Ref) | ||
| > 50 | 103 (39.4) | 78.7 | 0.590 (0.342–1.017) | 0.058 | 86.4 | 0.876 (0.403–1.901) | 0.737 |
| Grade | |||||||
| 1–2 | 123 (46.1) | 69.9 | 1 (Ref) | 93.9 | 1 (Ref) | ||
| 3 | 144 (53.9) | 76.0 | 1.112 (0.679–1.821) | 0.674 | 78.3 | 2.445 (1.094–5.463) | 0.029 |
| Vascular invasion | |||||||
| Negative | 250 (93.6) | 72.9 | 1 (Ref) | 86.8 | 1 (Ref) | ||
| Positive | 17 (6.4) | 60.7 | 1.603 (0.691–3.723) | 0.272 | 80.4 | 2.261 (0.781–6.546) | 0.133 |
| Tumor size | |||||||
| ≤2cm | 123 (46.1) | 83.9 | 1 (Ref) | 95.4 | 1 (Ref) | ||
| >2cm | 144 (53.9) | 62.6 | 2.508 (1.440–4.369) | 0.001 | 79.9 | 3.876 (1.473–10.198) | 0.006 |
| Lymph node | |||||||
| Negative | 159 (59.6) | 80.7 | 1 (Ref) | 98.0 | 1 (Ref) | ||
| Positive | 108 (40.4) | 59.5 | 3.074 (1.843–5.126) | <0.001 | 71.3 | 13.252 (4.000–43.901) | <0.001 |
| TNM | |||||||
| I | 81 (30.3) | 86.3 | 1 (Ref) | 100.0 | 1 (Ref) | ||
| II | 135 (50.6) | 74.3 | 2.049 (0.933–4.499) | 0.074 | 92.7 | 5.596 (0.716–43.745) | 0.101 |
| III | 51 (19.1) | 44.7 | 7.780 (3.534–17.127) | <0.001 | 52.0 | 34.557 (4.589–260.235) | 0.001 |
Abbreviations: Ref, reference.
Polymorphisms in CLDN1 and clinicopathological features
The interactions between CLDN1 genotypes and various clinicopathological characteristics were summarized in Supplementary Table 1. The distribution of rs10513846 genotypes was significantly associated with age and grade (Table 3). Rs10513846 GA genotype was associated with older age (P=0.013, HR = 2.231, 95% CI: 1.186–4.195). Rs10513846 AA genotype (P=0.005, HR = 2.889, 95% CI: 1.389–6.007) carriers were more likely to develop grade 3 tumors. And rs9283658 genotypes were also related to grade, more patients with grade 3 tumors were rs9283658 CC genotype carriers (P=0.023, HR = 0.446, 95% CI: 0.222–0.894). There was no significant association between other genetic variants in CLDN1 and clinicopathological features.
Table 3. Relationship between genotypes and clinicopathological features.
| Variables | Age | Grade | ||||||
|---|---|---|---|---|---|---|---|---|
| ≤50 (n, %) | >50 (n, %) | P-value | HR (95% CI) | 1–2 (n, %) | 3 (n, %) | P-value | HR (95% CI) | |
| rs10513846 | ||||||||
| GG | 52 (31.7) | 18 (17.5) | 1 (Ref) | 40 (32.5) | 30 (20.8) | 1 (Ref) | ||
| GA | 79 (48.2) | 61 (59.2) | 0.013 | 2.231 (1.186–4.195) | 65 (52.8) | 75 (52.1) | 0.144 | 1.538 (0.863–2.743) |
| AA | 33 (20.1) | 24 (23.3) | 0.053 | 2.101 (0.992–4.452) | 18 (14.7) | 39 (27.1) | 0.005 | 2.889 (1.389–6.007) |
| rs9283658 | ||||||||
| CC | 40 (24.4) | 31 (30.1) | 1 (Ref) | 26 (21.1) | 45 (31.3) | 1 (Ref) | ||
| CT | 79 (48.2) | 55 (53.4) | 0.718 | 0.898 (0.502–1.607) | 62 (50.4) | 72 (50.0) | 0.185 | 0.671 (0.372–1.211) |
| TT | 45 (27.4) | 17 (16.5) | 0.053 | 0.487 (0.235–1.010) | 35 (28.5) | 27 (18.7) | 0.023 | 0.446 (0.222–0.894) |
Abbreviations: n, number of patient; Ref, reference.
Polymorphisms in CLDN1 and survival of TNBC patients
Tables 4 and 5 listed the 5-year DFS and OS rate for patients with different genotypes. There was no association between polymorphisms in CLDN1 and survival of TNBC patients. Rs9842214 TT genotype carriers had less DFS rate than CC genotype carriers, the 5-year DFS was 74.8 and 39.8%, respectively. However, the difference was not significant, P-values were 0.050 and 0.056 in univariant and multi-variant analyses, respectively. Since only ten patients carried rs9842214 TT genotype, the results need to be verified.
Table 4. CLDN1 genotypes and DFS.
| Variables | Patients (%) | 5-year DFS(%) | Crude | Adjusted | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |||
| rs10513846 | ||||||
| GG | 70 (26.2) | 69.6 | 1 (Ref) | 1 (Ref) | ||
| GA | 140 (52.4) | 69.7 | 0.961 (0.544–1.698) | 0.891 | 0.933 (0.520–1.675) | 0.817 |
| AA | 57 (21.4) | 82.3 | 0.763 (0.360–1.617) | 0.480 | 0.757 (0.349–1.640) | 0.480 |
| rs1155884 | ||||||
| AA | 144 (54.0) | 78.0 | 1 (Ref) | 1 (Ref) | ||
| AC | 98 (36.7) | 62.2 | 1.470 (0.882–2.449) | 0.139 | 1.278 (0.756–2.161) | 0.359 |
| CC | 25 (9.3) | 78.2 | 1.032 (0.399–2.667) | 0.949 | 1.009 (0.382–2.669) | 0.986 |
| rs8298 | ||||||
| CC | 157 (58.8) | 74.3 | 1 (Ref) | 1 (Ref) | ||
| CT | 83 (31.1) | 69.8 | 1.006 (0.577–1.754) | 0.983 | 1.068 (0.608–1.876) | 0.819 |
| TT | 27 (10.1) | 63.2 | 1.570 (0.756–3.263) | 0.227 | 1.656 (0.783–3.500) | 0.187 |
| rs9842214 | ||||||
| CC | 167 (62.5) | 74.8 | 1 (Ref) | 1 (Ref) | ||
| CT | 90 (33.7) | 68.2 | 1.003 (0.585–1.721) | 0.991 | 1.166 (0.674–2.017) | 0.583 |
| TT | 10 (3.8) | 38.9 | 2.542 (0.999–6.470) | 0.050 | 2.527 (0.976–6.543) | 0.056 |
| rs9283658 | ||||||
| CC | 71 (26.6) | 83.7 | 1 (Ref) | 1 (Ref) | ||
| CT | 134 (50.2) | 68.0 | 1.445 (0.750–2.787) | 0.272 | 1.436 (0.742–2.781) | 0.283 |
| TT | 62 (23.2) | 71.7 | 1.571 (0.749–3.295) | 0.232 | 1.575 (0.736–3.369) | 0.242 |
Abbreviation: Ref, reference.
Table 5. CLDN1 genotypes and OS.
| Variables | 5-year OS (%) | Crude | Adjusted | ||
|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| rs10513846 | |||||
| GG | 91.4 | 1 (Ref) | 1 (Ref) | ||
| GA | 86.7 | 2.222 (0.747–6.606) | 0.151 | 1.514 (0.491–4.673) | 0.471 |
| AA | 81.1 | 2.561 (0.747–8.779) | 0.135 | 1.803 (0.492–6.615) | 0.374 |
| rs1155884 | |||||
| AA | 88.8 | 1 (Ref) | 1 (Ref) | ||
| AC | 82.9 | 1.984 (0.911–4.321) | 0.085 | 1.856 (0.833–4.135) | 0.130 |
| CC | 89.7 | 1.144 (0.253–5.171) | 0.861 | 1.121 (0.235–5.339) | 0.886 |
| rs8298 | |||||
| CC | 89.2 | 1 (Ref) | 1 (Ref) | ||
| CT | 82.9 | 1.256 (0.564–2.798) | 0.577 | 1.169 (0.504–2.716) | 0.716 |
| TT | 86.0 | 0.995 (0.287–3.445) | 0.994 | 1.266 (0.347–4.616) | 0.721 |
| rs9842214 | |||||
| CC | 87.9 | 1 (Ref) | 1 (Ref) | ||
| CT | 86.3 | 1.058 (0.471–2.380) | 0.891 | 1.419 (0.613–3.285) | 0.413 |
| TT | 75.0 | 1.587 (0.364–6.922) | 0.538 | 2.510 (0.549–11.473) | 0.235 |
| rs9283658 | |||||
| CC | 81.6 | 1 (Ref) | 1 (Ref) | ||
| CT | 88.4 | 0.769 (0.318–1.860) | 0.560 | 0.782 (0.330–1.855) | 0.577 |
| TT | 87.9 | 0.807 (0.246–2.653) | 0.724 | 0.573 (0.187–1.760) | 0.331 |
Abbreviation: Ref, reference.
Discussion
The principle functions of TJs include preventing the mixing of membrane proteins between the apical and basolateral membranes; and controlling the paracellular passage of ions and solutes in between cells [20]. TJs play important roles in tumor progression and metastasis [21]. A disruption of TJs during tumorigenesis generally leads to invasiveness, loss of cohesion, and lack of differentiation in cancer cells [21]. CLDNs are the first components identified to be involved in sealing in TJs [22]. To date, 27 CLDN family members have been identified; each CLDN has a different tissue-expression pattern and function [22,23]. Thus, CLDNs may serve both as biomarkers in detecting cancer and as possible targets in cancer therapeutics [24]. CLDN1 is one of the most commonly investigated CLDNs, but the association between polymorphisms in CLDN1 and TNBC has never been reported.
CLDN1 is a 17 kb gene that codes for a 3.4 kb transcript which translates to an important protein CLDN1 [25]. It has been reported that polymorphisms in CLDN1 are associated with the risk of cancer [14], small vessel vascular dementia [26], leukoaraiosis [27], and hepatitis C virus infection [28,29]. In Hahn-Strömberg V’s study, they found that CLDN1 rs9869263 genotype was related to risk of colon cancer and polymorphisms in CLDN7 were associated with differentiation and age of colon cancer [14]. Chen et al. reported that CLDN1 rs17501976 polymorphism was significantly associated with a decreased susceptibility to colorectal cancer in a Chinese population [30]. Polymorphisms investigated in our study have never been reported in cancer patients. We first demonstrated that rs10513846 and rs9283658 genotypes were significantly associated with age and grade in TNBC patients. As age and differentiation have been proved to be prognostic factors for breast cancer [31,32], our results indicate the potential role of polymorphisms in CLDN1 as biomarkers for tumor invasion or prognosis.
Though researches about polymorphisms in CLDN1 are rare, protein CLDN1 has been widely investigated in cancers. CLDN1 can promote or suppress tumor proliferation in different cancers or even in different histological subtypes of the same cancer. The over expression of CLDN1 has been reported to increase cell invasion in colorectal cancer [33] and oral squamous cell carcinoma (OSCC) [34]. CLDN1 has long been considered as a tumor suppressor in breast cancer. But recently, some studies showed that the expression level of CLDN1 was low in luminal-like and claudin-low breast cancers, while the expression level of CLDN1 was high in basal-like, most ER negative, BRCA1, medullary breast cancers [35]. Whether CLDN1 plays tumor-facilitating role in basal-like breast cancer or TNBC still needs to be proved.
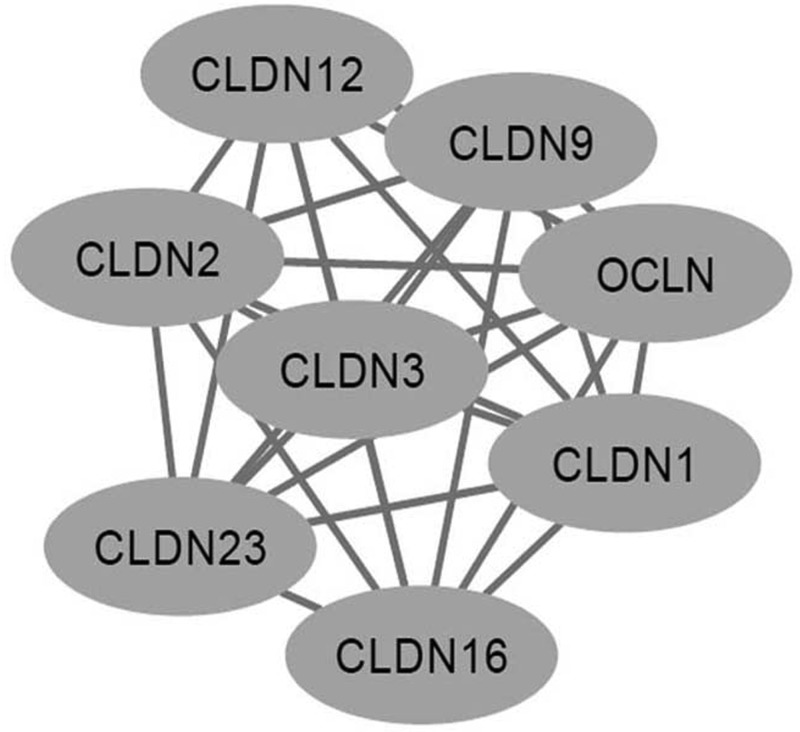
Down-regulation of CLDN1 was associated with shorter DFS of breast cancer patients [9]. In TNBC, CLDN1-negative phenotype also predicted poor prognosis [36,37]. In our study, no association between polymorphisms in CLDN1 and survival of TNBC patients was observed. There are some explanations. First, the sample size was relatively small and we did not do subgroup analysis. Second, though polymorphisms in CLDN1 might influence the expression level of CLDN1 protein, the complex interactions between CLDN family members could also affect the results. By using STRING, we found that CLDN1 interacts with some other proteins in CLDN family, such as CLDN2, CLDN3, CLDN6, CLDN12 and so on (Figure 1). CLDN 12 expression could be clinically useful for predicting the survival of the ER-negative subgroup of patients with breast cancer [38]. High expression of CLDN6 confers chemoresistance on breast cancer [39]. So, combination analysis of CLDN1 and other related proteins might help us to better understand the role of CLDN1 in TNBC.
Figure 1. Interactions between CLDN1 and related proteins.
The expression level of CLDN1 was found to be associated with tumor differentiation and age in several kinds of cancers. In OSCCs, the highest expression of CLDN1 was observed in well-differentiated OSCCs, whereas poorly differentiated OSCCs exhibited mostly negative staining for CLDN1 [40]. In hepatocellular cancer, down-regulation of CLDN1 was associated with poor differentiation [41]. In basal-like breast cancer patients, the highest level of CLDN1 protein expression was observed in patients who were older than 55 years of age [42]. So, we assume that polymorphisms in CLDN1 might influence the expression level of CLDN1 protein and then influence the tumor differentiation. However, the underlying mechanisms need to be further investigated. Our results also suggested that polymorphisms in CLDN1 might help us to identify subtypes of TNBC, as TNBC patients with different age and grade were proved to have unique molecular features [32,43].
Conclusion
In conclusion, we first demonstrated that polymorphisms in CLDN1 gene were associated with age and differentiation of TNBC patients. Since most polymorphisms have never been reported and the underlying mechanisms are still unknown, more researches are needed to verify our results.
Supporting information
Supplemental Table S1. Relationship between genotypes and clinicopathological features.
Abbreviations
- BRCA
breast cancer susceptibility gene
- CI
confidence interval
- CLDN 1
Claudin 1
- DFS
disease-free survival
- ER
estrogen receptor
- HER2
epidermal growth factor receptor 2
- HR
hazard ratio
- OS
overall survival
- OSCC
oral squamous cell carcinoma
- PR
progesterone receptor
- SNP
single nucleotide polymorphism
- TJ
tight junction
- TNBC
triple-negative breast cancer
Funding
The present study was funded by Natural Science Foundation of Jiangxi Province of China [grant number 20151BAB205043].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
All authors contributed significantly to this work. Y.F. conceived and designed the present study. A.H., J.L., S.R., and Y.L. collected the samples and performed the research study. Y.L. and A.H. wrote the paper. Y.F. reviewed and revised the paper.
References
- 1.Siegel R.L., Miller K.D. and Jemal A. (2017) Cancer Statistics, 2017. CA Cancer J. Clin. 67, 7–30 [DOI] [PubMed] [Google Scholar]
- 2.Blows F.M., Driver K.E., Schmidt M.K.. et al. (2010) Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 7, e1000279 10.1371/journal.pmed.1000279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haffty B.G., Yang Q., Reiss M.. et al. (2006) Locoregional relapse and distant metastasis in conservatively managed triple negative early stage breast cancer. J. Clin. Oncol. 24, 5652–5657 10.1200/JCO.2006.06.5664 [DOI] [PubMed] [Google Scholar]
- 4.Saraiva D.P., Guadalupe Cabral M., Jacinto A. and Braga S. (2017) How many diseases is triple negative breast cancer: the protagonism of the immune microenvironment. ESMO Open 2, e000208 10.1136/esmoopen-2017-000208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson T.R., Udyavar A.R., Chang C.W., Spoerke J.M.. et al. (2019) Genomic alterations associated with recurrence and TNBC subtype in high-risk early breast cancers. Mol. Cancer. Res. 17, 97–108 10.1158/1541-7786.MCR-18-0619 [DOI] [PubMed] [Google Scholar]
- 6.Burstein M.D., Tsimelzon A., Poage G.M.. et al. (2015) Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 21, 1688–1698 10.1158/1078-0432.CCR-14-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piontek J., Winkler L., Wolburg H.. et al. (2008) Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 22, 146–158 10.1096/fj.07-8319com [DOI] [PubMed] [Google Scholar]
- 8.Tabaries S. and Siegel P.M. (2017) The role of claudins in cancer metastasis. Oncogene 36, 1176–1190 10.1038/onc.2016.289 [DOI] [PubMed] [Google Scholar]
- 9.Morohashi S., Kusumi T., Sato F.. et al. (2007) Decreased expression of claudin-1 correlates with recurrence status in breast cancer. Int. J. Mol. Med. 20, 139–143 [PubMed] [Google Scholar]
- 10.Suren D., Yildirim M., Kaya V.. et al. (2014) Loss of tight junction proteins (Claudin 1, 4, and 7) correlates with aggressive behavior in colorectal carcinoma. Med. Sci. Monit. 20, 1255–1262 10.12659/MSM.890598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liebner S., Fischmann A., Rascher G.. et al. (2000) Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol. (Berl) 100, 323–331 10.1007/s004010000180 [DOI] [PubMed] [Google Scholar]
- 12.Izraely S., Sagi-Assif O., Klein A.. et al. (2015) The metastatic microenvironment: claudin-1 suppresses the malignant phenotype of melanoma brain metastasis. Int. J. Cancer 136, 1296–1307 10.1002/ijc.29090 [DOI] [PubMed] [Google Scholar]
- 13.Victoria H.S., Henrik E., Lennart B. and Lennart F. (2010) Claudin 1 and claudin 7 gene polymorphisms and protein derangement are unrelated to the growth pattern and tumor volume of colon carcinoma. Int. J. Biomed. Sci. 6, 96–102 [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn-Stromberg V., Askari S., Befekadu R., Matthiessen P., Karlsson S. and Nilsson T.K. (2014) Polymorphisms in the CLDN1 and CLDN7 genes are related to differentiation and tumor stage in colon carcinoma. APMIS 122, 636–642 10.1111/apm.12211 [DOI] [PubMed] [Google Scholar]
- 15.Edge S.B. and Compton C.C. (2010) The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 17, 1471–1474 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 16.Wolff A.C., Hammond M.E., Hicks D.G.. et al. (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 31, 3997–4013 10.1200/JCO.2013.50.9984 [DOI] [PubMed] [Google Scholar]
- 17.Hammond M.E., Hayes D.F., Dowsett M.. et al. (2010) American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 28, 2784–2795 10.1200/JCO.2009.25.6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiu L., Zhang C., Wu Z. and Peng J. (2017) Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front. Microbiol. 8, 1510 10.3389/fmicb.2017.01510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung K.W., Peng Q., He L.. et al. (2016) Rapid and simultaneous detection of major drug resistance mutations in reverse transcriptase gene for HIV-1 CRF01_AE, CRF07_BC and subtype B in China using Sequenom MassARRAY(R) system. PLoS ONE 11, e0153641 10.1371/journal.pone.0153641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartsock A. and Nelson W.J. (2008) Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta 1778, 660–669 10.1016/j.bbamem.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leech A.O., Cruz R.G., Hill A.D. and Hopkins A.M. (2015) Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Ann. Transl. Med. 3, 184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunzel D. and Fromm M. (2012) Claudins and other tight junction proteins. Compr. Physiol. 2, 1819–1852 [DOI] [PubMed] [Google Scholar]
- 23.Zihni C., Mills C., Matter K. and Balda M.S. (2016) Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 17, 564–580 10.1038/nrm.2016.80 [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto Y., Fukasawa M., Kuniyasu H., Yagi K. and Kondoh M. (2017) Claudin-targeted drug development using anti-claudin monoclonal antibodies to treat hepatitis and cancer. Ann. N. Y. Acad. Sci. 1397, 5–16 10.1111/nyas.13337 [DOI] [PubMed] [Google Scholar]
- 25.Furuse M. and Tsukita S. (2006) Claudins in occluding junctions of humans and flies. Trends Cell Biol. 16, 181–188 10.1016/j.tcb.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan V. and Braidy N. (2017) Association of genetic polymorphisms of claudin-1 with small vessel vascular dementia 44, 623–630 [DOI] [PubMed] [Google Scholar]
- 27.Yadav B.K. and Shin B.S. (2015) Single-nucleotide polymorphisms of tight junction component claudin-1 associated with leukoaraiosis. J. Stroke Cerebrovasc. Dis. 24, 1662–1670 10.1016/j.jstrokecerebrovasdis.2015.03.038 [DOI] [PubMed] [Google Scholar]
- 28.Bekker V., Chanock S.J., Yeager M.. et al. (2010) Genetic variation in CLDN1 and susceptibility to hepatitis C virus infection. J. Viral Hepat. 17, 192–200 10.1111/j.1365-2893.2009.01166.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun S., Jin G. and Kang H. (2015) CD81 and CLDN1 polymorphisms and hepatitis C virus infection susceptibility: a case control study. Gene 567, 87–91 10.1016/j.gene.2015.04.072 [DOI] [PubMed] [Google Scholar]
- 30.Chen J.J., Zhong M., Dou T.H., Wu Z.Y. and Tang W.J. (2015) rs17501976 polymorphism of CLDN1 gene is associated with decreased risk of colorectal cancer in a Chinese population. Int. J. Clin. Exp. Med. 8, 1247–1252 [PMC free article] [PubMed] [Google Scholar]
- 31.Anders C.K., Hsu D.S., Broadwater G.. et al. (2008) Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J. Clin. Oncol. 26, 3324–3330 10.1200/JCO.2007.14.2471 [DOI] [PubMed] [Google Scholar]
- 32.Geyer F.C., Pareja F., Weigelt B.. et al. (2017) The spectrum of triple-negative breast disease: high- and low-grade lesions. Am. J. Pathol. 187, 2139–2151 10.1016/j.ajpath.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinugasa T., Huo Q., Higashi D.. et al. (2007) Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res. 27, 3729–3734 [PubMed] [Google Scholar]
- 34.JC D.E.V., Fernandez-Valle A., Vivanco-Allende B.. et al. (2015) The prognostic role of claudins -1 and -4 in oral squamous cell carcinoma. Anticancer Res. 35, 2949–2959 [PubMed] [Google Scholar]
- 35.Zhou B., Moodie A., Blanchard A.A., Leygue E. and Myal Y. (2015) Claudin 1 in breast cancer: new insights. J. Clin. Med. 4, 1960–1976 10.3390/jcm4121952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma F., Ding X., Fan Y.. et al. (2014) A CLDN1-negative phenotype predicts poor prognosis in triple-negative breast cancer. PLoS ONE 9, e112765 10.1371/journal.pone.0112765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katayama A., Handa T., Komatsu K.. et al. (2017) Expression patterns of claudins in patients with triple-negative breast cancer are associated with nodal metastasis and worse outcome. Pathol. Int. 67, 404–413 10.1111/pin.12560 [DOI] [PubMed] [Google Scholar]
- 38.Iravani O., Yip G.W., Thike A.A.. et al. (2016) Prognostic significance of Claudin 12 in estrogen receptor-negative breast cancer. J. Clin. Pathol. 69, 878–883 10.1136/jclinpath-2015-203265 [DOI] [PubMed] [Google Scholar]
- 39.Yang M., Li Y., Shen X.. et al. (2017) CLDN6 promotes chemoresistance through GSTP1 in human breast cancer. J. Exp. Clin. Cancer Res. 36, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouban A. and Ahmed A. (2015) Analysis of the distribution and expression of claudin-1 tight junction protein in the oral cavity. Appl. Immunohistochem. Mol. Morphol. 23, 444–448 [DOI] [PubMed] [Google Scholar]
- 41.Higashi Y., Suzuki S., Sakaguchi T.. et al. (2007) Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J. Surg. Res. 139, 68–76 10.1016/j.jss.2006.08.038 [DOI] [PubMed] [Google Scholar]
- 42.Blanchard A.A., Ma X., Dueck K.J.. et al. (2013) Claudin 1 expression in basal-like breast cancer is related to patient age. BMC Cancer 13, 268 10.1186/1471-2407-13-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez-Flores-Ramos L., Castro-Sanchez A., Pena-Curiel O. and Mohar-Betancourt A. (2017) Molecular biology in young women with breast cancer: from tumor gene expression to DNA mutations. Rev. Invest. Clin. 69, 181–192 [DOI] [PubMed] [Google Scholar]