Abstract
During both embryonic development and adult tissue regeneration, changes in chromatin structure driven by master transcription factors lead to stimulus-responsive transcriptional programs. A thorough understanding of how stem cells in the skeleton interpret mechanical stimuli and enact regeneration would shed light on how forces are transduced to the nucleus in regenerative processes. Here we develop a genetically dissectible mouse model of mandibular distraction osteogenesis–which is a process that is used in humans to correct an undersized lower jaw that involves surgically separating the jaw bone, which elicits new bone growth in the gap. We use this model to show that regions of newly formed bone are clonally derived from stem cells that reside in the skeleton. Using chromatin and transcriptional profiling, we show that these stem-cell populations gain activity within the focal adhesion kinase (FAK) signalling pathway, and that inhibiting FAK abolishes new bone formation. Mechanotransduction via FAK in skeletal stem cells during distraction activates a gene-regulatory program and retrotransposons that are normally active in primitive neural crest cells, from which skeletal stem cells arise during development. This reversion to a developmental state underlies the robust tissue growth that facilitates stem-cell-based regeneration of adult skeletal tissue.
The facial skeleton exhibits morphological variations that underlie the evolutionary diversification of mammals. The lower jaw comprises mandibular bone, vasculature, dentition, innervation and musculature. Mechanical forces are integral to skeletal homeostasis and skeletal regeneration by defining tissue architecture and driving cell differentiation. In the lower jaw, the mechanical forces applied during distraction osteogenesis promote endogenous bone formation across a mechanically controlled environment, providing functional replacement of tissue1,2. Distraction osteogenesis has revolutionized the treatment of facial malformations that include Pierre–Robin sequence, Treacher Collins syndrome and craniofacial microsomia3–5.
However, little is known about the cell population and molecular signals that drive tissue growth in distraction osteogenesis. Recently, the mouse skeletal stem cell (SSC) lineage has been elucidated and isolated6. Whether this lineage is present in the facial skeleton, which is known to arise from the neural crest, is unknown.
During regenerative processes, adult stem-cell populations change not only in proliferation and location but also in their underlying gene-regulatory programs7,8. Stem cells may reactivate a greater potential for differentiation, while also responding to injury conditions9. Clinical studies comparing acute separation of bone to gradual distraction indicate that the application of constant physical force has a role in driving regeneration at the molecular level1–5. The process of converting mechanical stimuli into a molecular response (mechanotransduction) occurs through multiple pathways, including the FAK pathway, leading to context-dependent transcriptional regulation10. Understanding how SSCs translate mechanical stimuli into productive regeneration will shed light on how force is transduced in other regenerative processes.
Here we use a rigorous model of mandibular distraction osteogenesis in mice and show that new bone is clonally derived from mandibular SSCs. Using the assay for transposase-accessible chromatin (ATAC-seq), as well as RNA sequencing (RNA-seq) to analyse the SSC transcriptome, we show that SSCs have distinct chromatin accessibility and gene expression within the FAK pathway. Activation of FAK through controlled mechanical advancement of the lower jaw in adults is required to induce a primitive neural crest transcriptional network that may allow for the massive tissue regeneration seen in distraction osteogenesis. The cellular mode of regeneration in response to mandibular distraction is of great interest, as this represents a successful strategy to elicit the endogenous potential of postnatal tissue11,12.
Bone regeneration in distraction osteogenesis
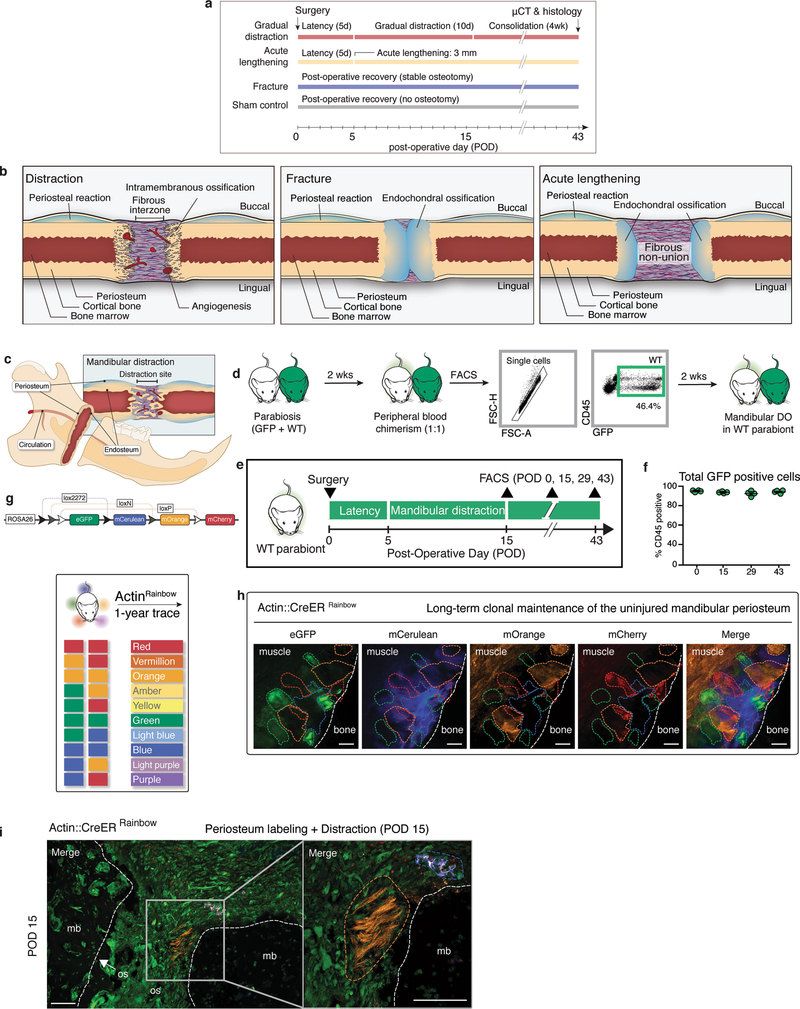
We interrogated the cellular and mechanical mechanisms of adult bone regeneration by developing a mouse model of mandibular distraction osteogenesis, beginning with the design and three-dimensional (3D) printing of distraction devices (Fig. 1a, b). Next, animals were divided into four groups (Extended Data Fig. 1a): sham-operated (in which the mandible was exposed and the distraction device was placed, but there was no surgical cutting of the bone (osteotomy)); fracture (osteotomy without distraction); acutely lengthened (osteotomy with bone segments separated to 3 mm on day 5); and gradually distracted (osteotomy with bone segments separated by 0.15 mm every 12 hours, to a total separation of 3 mm).
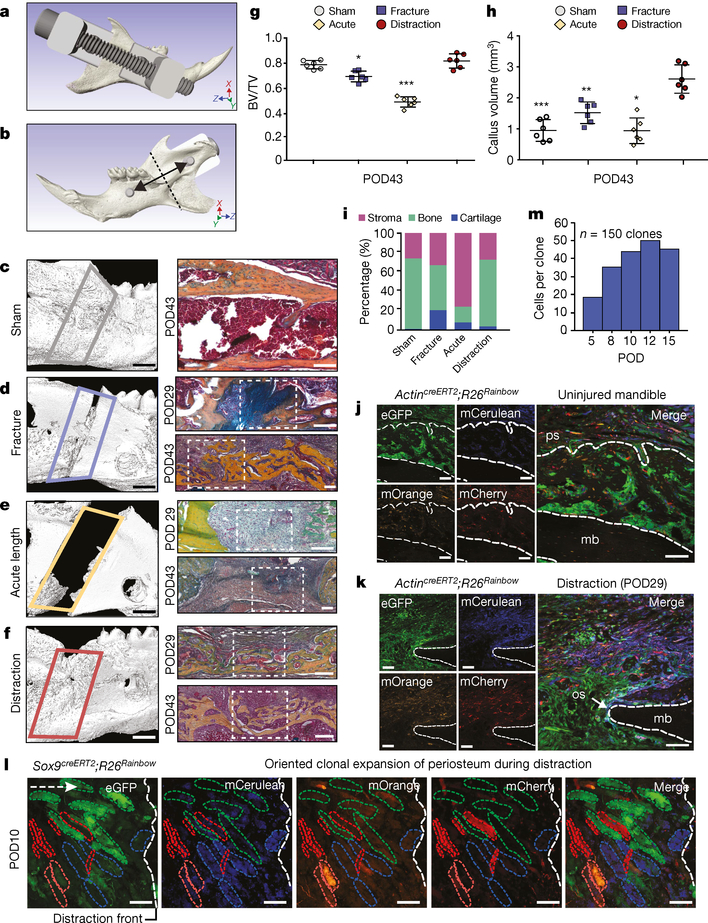
Fig. 1 |. Tissue-resident stem and progenitor cells enact bone regeneration in distraction osteogenesis.
a, Computer-assisted design of a distraction device using 3D μCT of the C57BL/6 mouse hemimandible. b, The lingual aspect illustrates the location of the osteotomy (dotted line), perpendicular to the vector of bidirectional distraction (solid arrow). c, Three-dimensional μCT of a sham-operated mandible (left, lateral view), with pentachrome staining of a transverse section (right) at POD43. The outlined area (left) indicates the volume analysed for new bone formation (n = 5). d, As for c, but for a fractured mandible, and also showing POD29. The white dotted lines indicate the osteotomy area. e, As for d, but for an acutely lengthened mandible. f, As for d, but for a gradually distracted mandible. g, Quantification of bone volume/total tissue volume (BV/TV) analysed at POD43 from μCT (n = 6, *P ≤ 0.05, ***P ≤ 0.001; Tukey’s multiple comparisons). h, Quantification of new bone callus volume formed at POD43 (n = 6, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; Tukey’s multiple comparisons). i, Tissue fraction of bone, cartilage and stroma from pentachrome histology at POD43 (n = 6). j, Confocal micrographs of uninjured transverse mandible sections after one week (n = 4). Uninjured mandibular bone (mb) is between the dotted lines. eGFP, enhanced green fluorescent protein; mCerulean, membrane cerulean (blue fluorescence signal); mCherry, membrane cherry (red fluorescence signal); mOrange, membrane orange (orange fluorescence signal); ps, periosteum. k, As for j, but for POD29 of distraction osteogenesis (n = 4). The dotted outline indicates the mandibular bone at the distraction site. os, osteotomy site. l, Confocal micrographs of whole-mount periosteum at POD10 (clones are indicated by coloured dotted outlines) (n = 5). The vector of distraction is indicated by the dotted white arrow (left). Buccal to lingual view of periosteal callus overlying the distraction sites. m, Quantification of average clone size within the regenerate during distraction (POD5–15; n = 5). Scale bars, 1 mm (c–f left), 200 μm (c–f right, j–l).
Microcomputed tomography (μCT) and pentachrome staining revealed that sham-operated mandibles had normal bone morphology and minimal new bone at postoperative day (POD) 43 (Fig. 1c). Fractured mandibles demonstrated incomplete bone union with persistent cartilage at the middle and end of the consolidation phase (Fig. 1d). Acute lengthening resulted in non-union with primarily fibrous tissue (Fig. 1e). By contrast, gradual distraction resulted in complete union and robust new bone formation at mid- and end-consolidation (Fig. 1f). The callus mineralized volume fraction (bone volume/total tissue volume, or BV/TV) was significantly increased in gradual-distraction specimens compared with acutely lengthened (***P < 0.001) and fracture (*P < 0.05) specimens at POD43 (Fig. 1g). The callus volume (TV) of gradual-distraction specimens was significantly higher than in all other conditions, including acute lengthening (*P < 0.05), fracture (**P < 0.01) and sham (***P < 0.001) (Fig. 1h). Analysis of tissue fraction in distraction and sham mandibles revealed similar proportions of mineralized bone, and confirmed cartilaginous healing in fracture specimens and fibrous healing in acute lengthening specimens (Fig. 1i). Whereas gradual-distraction mandibles exhibited direct intramembranous ossification, fracture mandibles displayed endochondral ossification (Extended Data Fig. 1b).
We next sought to investigate the cellular mechanism responsible for regeneration in distraction osteogenesis. We first ruled out the possibility of a circulating source of regenerated tissue using parabiosis (Extended Data Fig. 1c–f). Then, to test whether the generation of new mandible in distraction osteogenesis involved tissue-resident stem cells (Extended Data Fig. 1c), we performed the procedures on the mandibles of ubiquitous Rainbow mice (ActincreERT2;Rosa26Rainbow; Rosa26 is hereafter referred to R26 and Actin refers to Actb throughout). After recombination of the Rainbow reporter (R26VT2/GK3), cells are genetically marked with one of ten colour combinations, which is passed to all daughter cells (Extended Data Fig. 1g). To determine the location of stem and progenitor cells within the mandible, we traced uninjured tissue over the course of one year, finding large single-coloured clones within the periosteum—demonstrating the presence of a stem and progenitor population (Extended Data Fig. 1h).
We then sought to determine the role of these cells during distraction. ActincreERT2;R26Rainbow mice were distracted, using uninjured mandibles for comparison. At mid-consolidation, we observed expansion of single-coloured clones near the osteotomy site (Fig. 1j, k). To trace the fate of cells within the periosteum during distraction osteogenesis, we developed a strategy of local labelling and found that large clone sizes were enriched (Extended Data Fig. 1i).
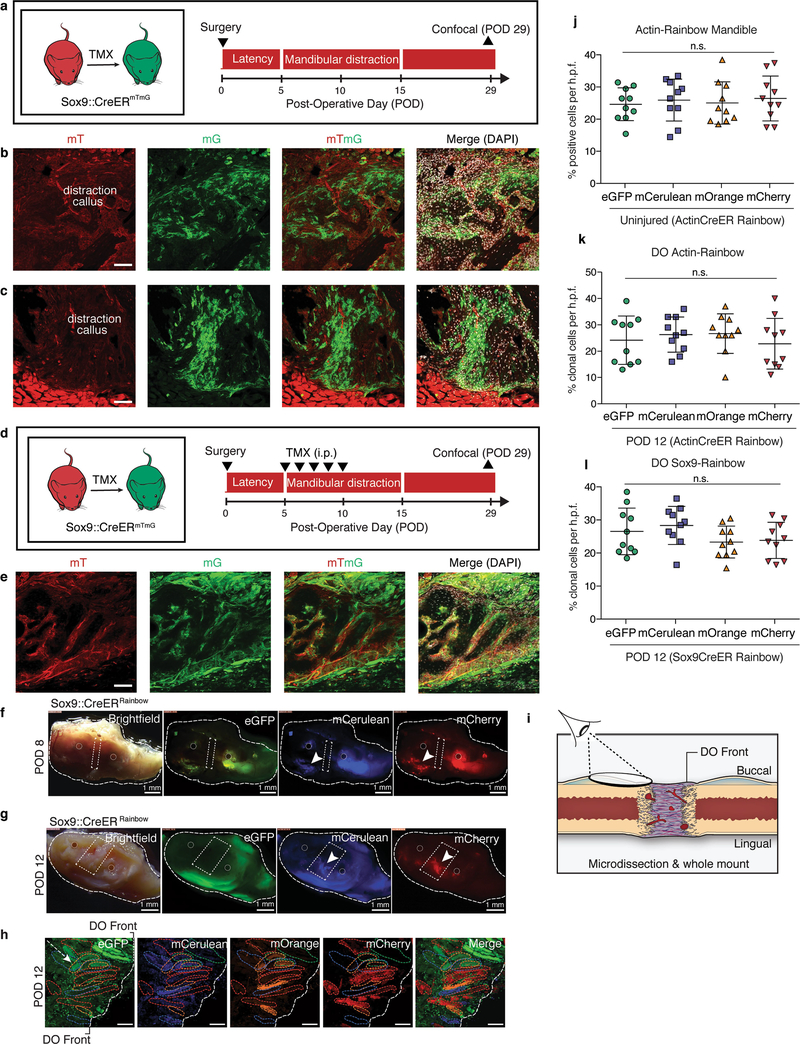
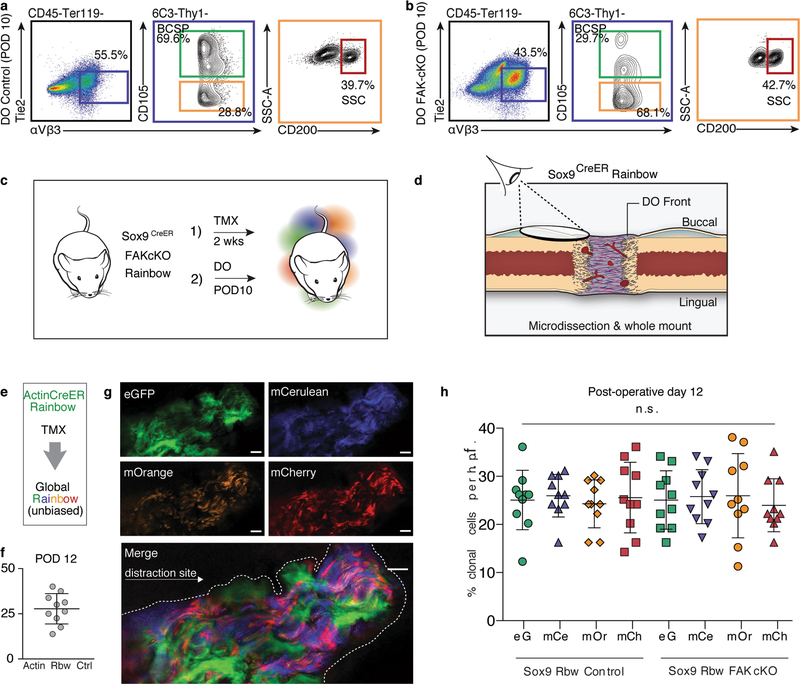
To assess the lineage-specific characteristics of regeneration in distraction, we distracted mandibles from skeleton-specific Sox9creERT2;R26mT/mG mice and found that cells of the Sox9-expressing lineage gave rise to new bone (Extended Data Fig. 2a–e). We then crossed the Sox9creERT2 driver to the Rainbow construct and observed infiltration of multiple single-coloured clones into the distraction gap at early and late distraction (PODs 8 and 12) (Extended Data Fig. 2f, g). Clonal analysis of the regenerate over time revealed that cells within the lateral periosteum expanded into large clones with linear alignment (Fig. 1l and Extended Data Fig. 2h, i). As with ActincreERT2;R26Rainbow mandibles, Sox9creERT2;R26Rainbow mandibles exhibited equivalent recombination frequencies at POD12 for each fluorescent protein (Extended Data Fig. 2j–l). The average size of the Sox9-lineage clone increased over the course of distraction, peaking at POD12 (Fig. 1m). Thus, skeletal-lineage-specific stem or progenitor cells are responsible for new bone formation during distraction osteogenesis.
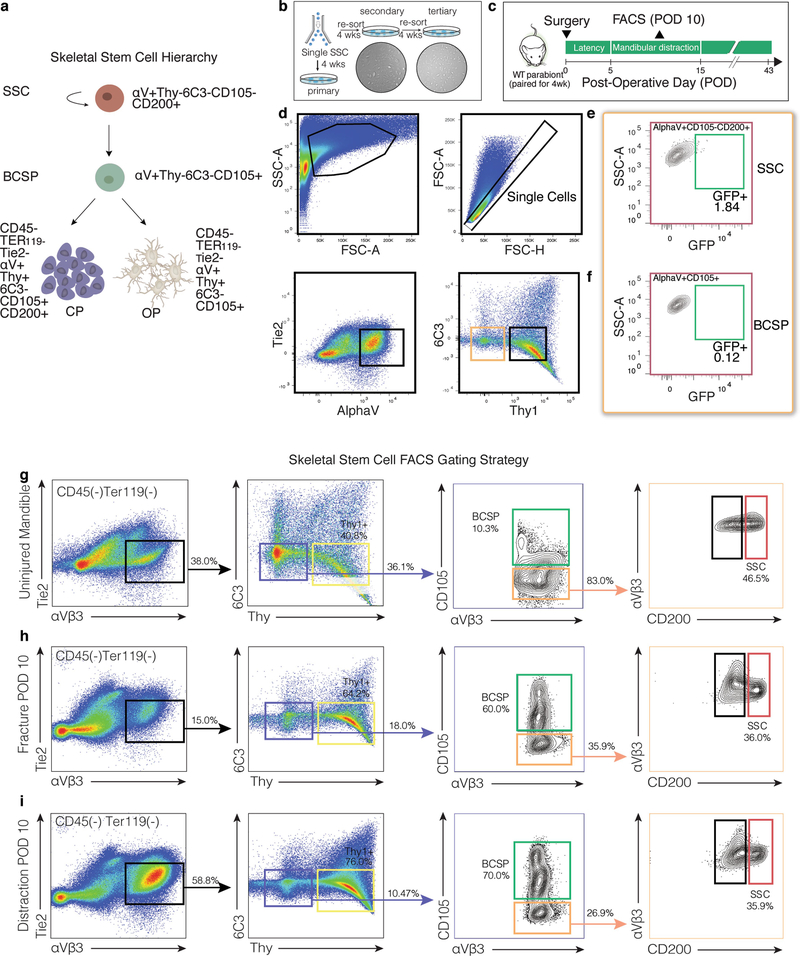
Next, we purified skeletogenic populations from mandibles using a strategy that has been used for long bones6. Lineage tracing of Sox9creERT2;R26Rainbow mice for two weeks revealed clonal expansion within the growth plate of the mandibular condyle (Fig. 2a). We therefore collected tissues from the condylar growth plate to enrich for skeletal stem cells (SSCs, with markers CD45− Ter119− CD202b− Thy1− 6C3− CD51+ CD105− CD200+), their multipotent progenitors (bone, cartilage and skeletal progenitors (BCSPs): CD45− Ter119− CD202b− Thy1− 6C3− CD51+ CD105+) and their unipotent progenitors (osteoprogenitors: CD45− Ter119− CD202b− Thy1+ 6C3− CD51+ CD105+ CD200−; and chondroprogenitors: CD45-Ter119− CD202b− Thy1+ 6C3− CD51+ CD105+ CD200+) (Extended Data Fig. 3a). We sorted single SSCs and demonstrated tertiary colony formation (Extended Data Fig. 3b). There was a complete absence of circulating (green fluorescent protein (GFP)+) SSCs and BCSPs, as assessed by parabiosis (Extended Data Fig. 3c–f).
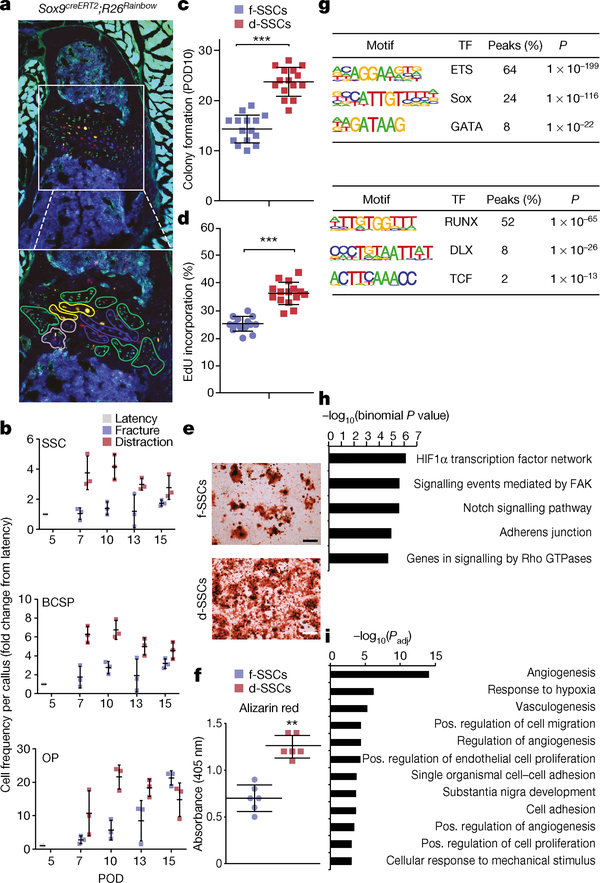
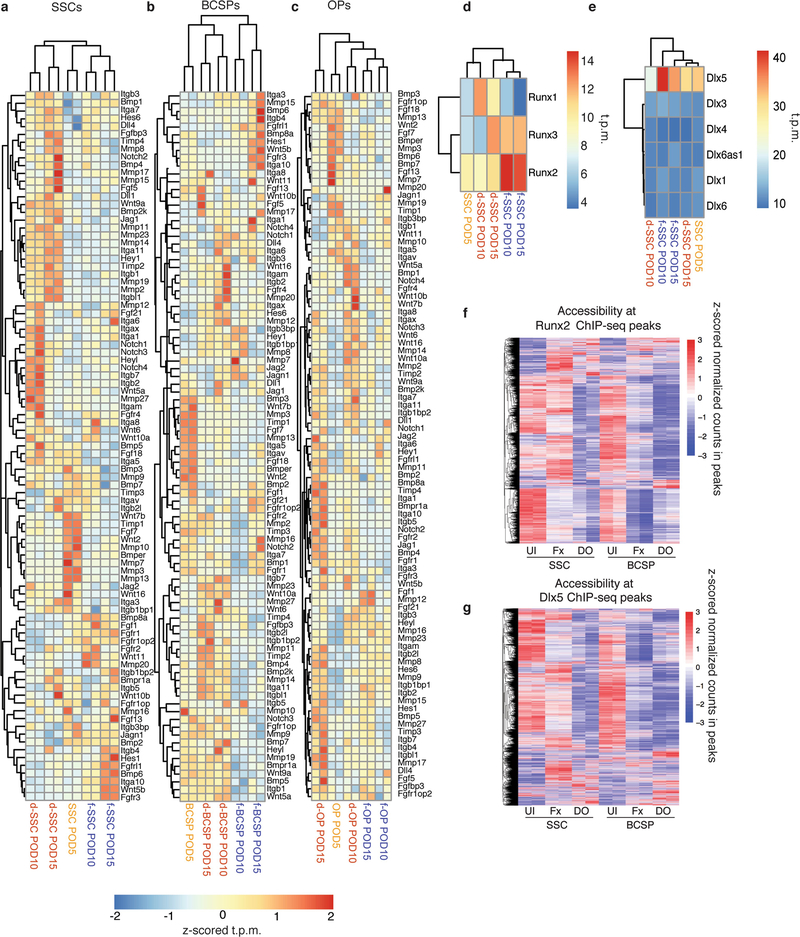
Fig. 2 |. Transcriptional regulation underlying SSC function.
a, Top, confocal micrograph of a transverse section from a Sox9creERT2;R26Rainbow mandibular condyle (n = 6); bottom, expanded image of the area in the white box. Coloured outlines indicate single-colour clones. b, Quantification of cellular frequency within mandibular calluses in fracture and distraction conditions (n = 3 per time point; boundaries indicate standard deviation (s.d.)). c, Quantification of colonies formed from f- or d-SSCs isolated at POD10 (n = 15, ***P < 0.0001; t-test). d- (n = 15, **P < 0.01) and f- (n = 15, *P < 0.05) SSCs demonstrated elevated colony-forming capacity compared with uninjured SSCs (n = 6). d, Quantification of proliferation rate via incorporation of 5-ethynyl-2′-deoxyuridine (EdU) into SSCs at POD10 (n = 15, ***P < 0.0001; t-test). e, f, Alizarin red staining of f- versus d-SSCs (n = 6, **P = 0.001; t-test) at POD10. g, Motif analysis for differential peaks shared between SSCs and BCSPs. Top, chromatin sites that are more accessible in distraction than in fracture (n = 1,617 sites). Bottom, sites that are more accessible in fracture than in distraction (n = 1,506 sites). The top three motifs and the transcription factors (TFs) that bind them, the percentage of sites that contain the motif and the one-sided binomial P-values with Benjamini–Hochberg corrections are shown. h, GO terms, identified by ATAC-seq, enriched for genes near peaks that are more accessible in d- than in f-SSCs and BCSPs (n = 1,617 sites). Selected GO terms (right) with significant P-values (one-sided binomial, with Benjamini–Hochberg corrections; left) are shown. i, GO terms enriched for genes upregulated in d- versus f-SSCs at PODs 10 and 15, identified by RNA-seq. Select GO terms with significant P-values (one-sided binomial, with Benjamini–Hochberg corrections) are shown. Scale bars, 200 μm (a, e).
Distraction osteogenesis elicited an expansion of SSCs and BCSPs that is not seen following fracture (Fig. 2b). Expansion after fracture was delayed and restricted to osteoprogenitors. Colony formation and proliferation were greatly elevated in SSCs (Fig. 2c–f and Extended Data Fig. 3g–i) at the time that distraction-specific kinetics were seen. Formation of mineralized tissue was substantially elevated by distraction (d-)SSCs versus fracture (f-)SSCs (Fig. 2e, f).
Distraction-specific gene regulation
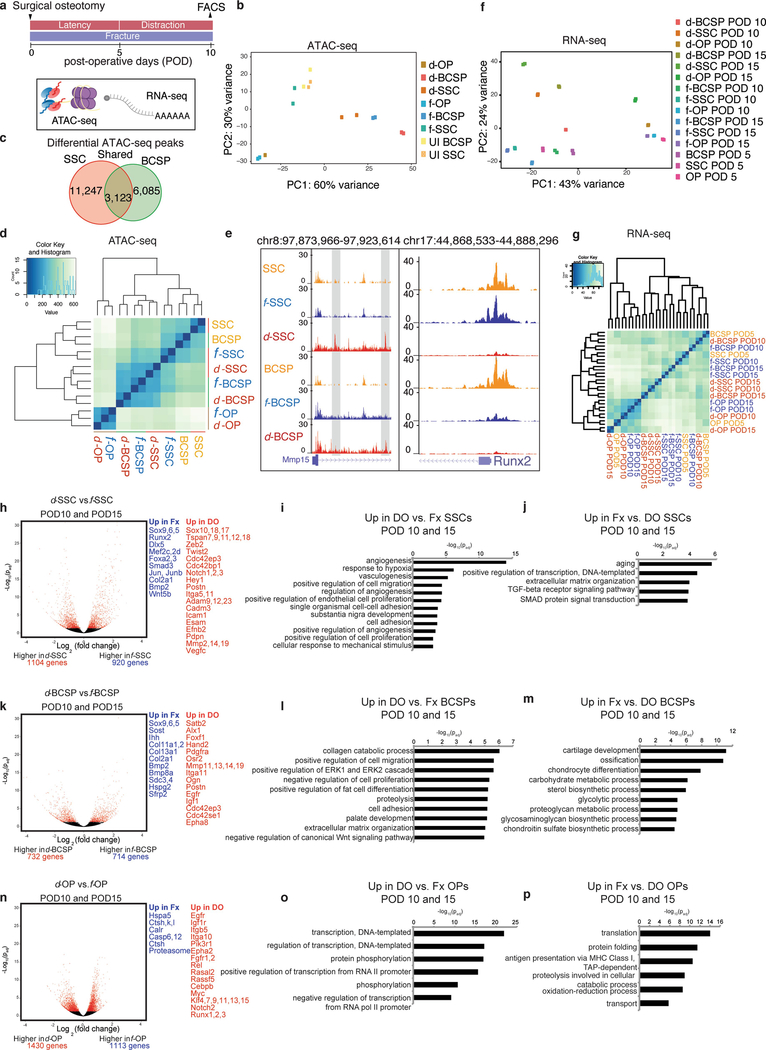
We next investigated the molecular changes that drive the marked differences in regenerative capacity displayed by the SSC lineage in response to mechanotransduction. We performed ATAC-seq on SSCs, BCSPs and osteoprogenitors isolated from uninjured, fractured and distracted mandibles at POD10 to capture transcription-factor-binding sites throughout the genome13 (Extended Data Fig. 4a). Samples clustered into three groups: differentiated osteoprogenitors (d-OPs and f-OPs), quiescent cells (SSCs, BCSPs and f-SSCs), and activated or regenerative cells (d-SSCs, d-BCSPs and f-BCSPs) (Extended Data Fig. 4b, d).
We performed RNA-seq on d-SSCs, f-SSCs, d-BCSPs, f-BCSPs, d-OPs and f-OPs at PODs 5, 10 and 15, and demonstrated major differences between fracture and distraction osteogenesis at the level of SSCs and BCSPs, whereas the osteoprogenitors were highly similar in fracture and distraction conditions (Extended Data Fig. 4f, g). d-SSCs at PODs 10 and 15 were more similar to one another than d-SSCs were to f-SSCs at a single time point, suggesting that their transcriptional programs bifurcated according to the applied mechanical force. The expression of molecules from developmental signalling pathways and of functional molecules was highly variable between fracture and distraction osteogenesis, and only somewhat variable between time points (Extended Data Fig. 5a–c).
We directly compared ATAC-seq from d- and f-SSCs and d- and f-BCSPs. We found many changes in accessible chromatin sites (14,370 changes in SSCs; 9,208 in BCSPs), of which 3,123 were shared (Extended Data Fig. 4c, e). Motif analysis showed that sites that are less accessible in distraction osteogenesis than in fracture are enriched for binding motifs for the core transcription factors that drive skeletal development (RUNX and DLX) (Fig. 2g). Runx2 and Dlx5 were highly and differentially expressed and accessible between f- and d-SSCs, making them strong candidates for binding factors (Extended Data Fig. 4e and Extended Data Fig. 5d, e). Binding sites for these factors in preosteoblasts were less accessible in d-SSCs and d-BCSPs than in uninjured or fracture cells14,15 (Extended Data Fig. 5f, g).
Sites that gain accessibility during distraction osteogenesis are enriched for ETS, SOX and GATA motifs (Fig. 2g). Genes near distraction-specific sites are enriched for Gene Ontology (GO) terms associated with the hypoxia-inducible factor 1α (HIF1α), FAK, Notch and RAS signalling pathways, as well as with adherens junctions (Fig. 2h).
At the RNA level, GO terms associated with vascularization, adherens junctions, cell migration and responses to mechanical stimuli are enriched in d-SSCs (Fig. 2i and Extended Data Fig. 4h–j). Genes upregulated in d-BCSPs are enriched for migration and adhesion terms, whereas genes upregulated in f-BCSPs are enriched for cartilage-formation terms (Extended Data Fig. 4k–m). Osteoprogenitors show fewer changes in expression in fracture versus distraction, indicating that most molecular changes during distraction osteogenesis occur in SSCs and BCSPs (Extended Data Fig. 4n–p).
We next explored the role of FAK signalling in distraction, because this pathway was upregulated in d-SSCs, and because FAK is a known mechanotransducer. FAK propagates information about physical stimuli at integrin-mediated cell–matrix contacts to the nucleus, affecting proliferation, differentiation, migration and more. The downstream targets of FAK are context-dependent and thus we sought to understand its role in our model9,10.
Inhibiting FAK disrupts bone formation
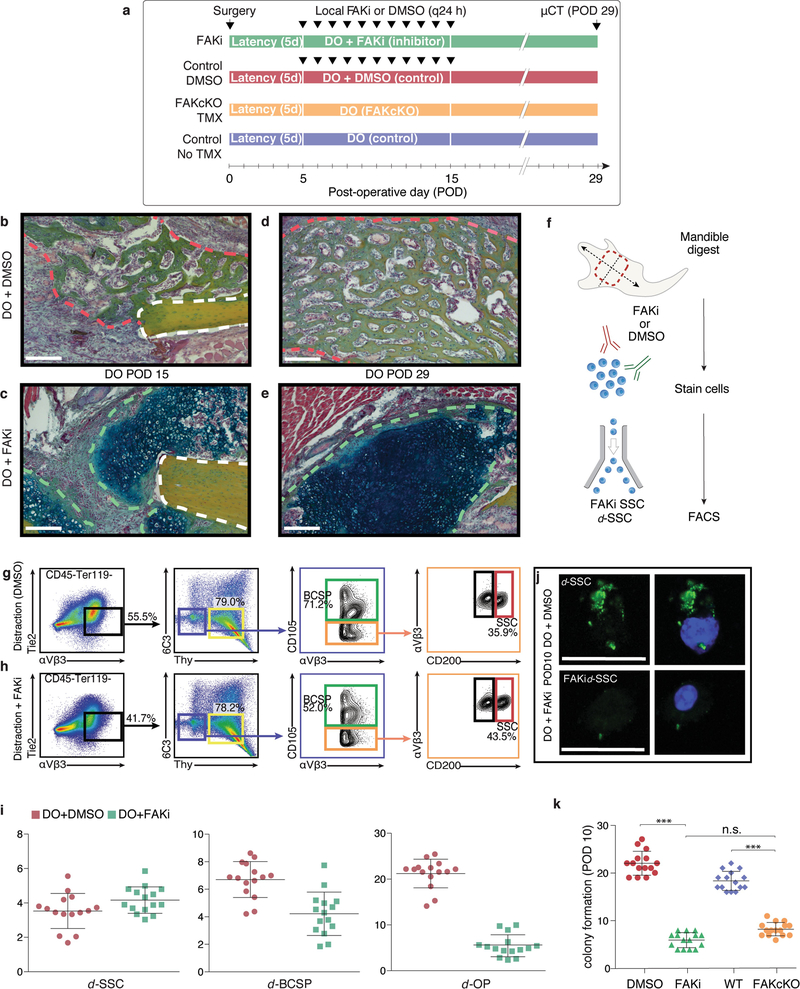
To investigate whether mechanotransduction via FAK is essential in distraction osteogenesis, we inhibited FAK with the small molecule PF-573228 (FAKi condition). In addition, we evaluated a skeleton-specific conditional knockout of FAK (FAK(cKO)) using Sox9creERT2;Ptk2fl/fl animals (Extended Data Fig. 6a). μCT analysis revealed that bone formation was diminished in both FAKi and FAK(cKO) mandibles compared with controls (Fig. 3a–e; ***P < 0.001). Pentachrome staining revealed cartilage formation under FAKi conditions that was not present in controls at PODs 15 and 29 (Extended Data Fig. 6b–e). Disruption of bone formation and induction of cartilage at the distraction site was a notable tissue-level response to FAK inhibition, congruent with disruption of the integrin-mediated cell–matrix interactions that are responsible for intramembranous bone formation in distraction osteogenesis.
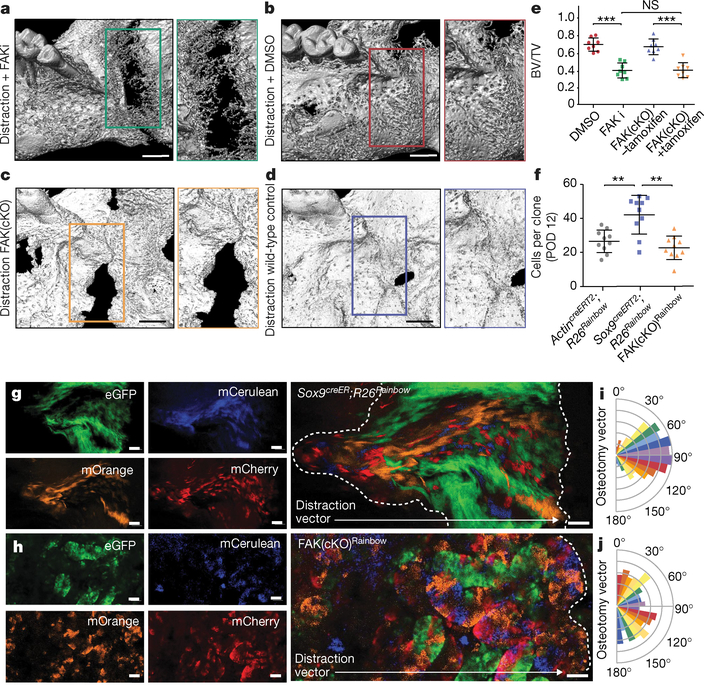
Fig. 3 |. FAK inhibition disrupts bone formation during distraction.
a, Three-dimensional μCT of a distracted mandible treated with FAK inhibitor (FAKi) during distraction osteogenesis, collected at POD29. An absence of bone union is indicated by the green outline (with high magnification shown at the right). The view is a lingual aspect. n = 5. b, As for a, but using dimethylsulfoxide (DMSO) as a control in place of the FAK inhibitor. c, As for a, but using conditional FAK knockout (FAK(cKO)) animals treated with tamoxifen two weeks before distraction. d, As for a, but using wild-type animals and a corn-oil control treatment. e, BV/TV analysed at POD29 (n = 8, ***P ≤ 0.001; NS, not significant; Tukey’s multiple comparisons). f, Quantification of average clone size within the regenerate over the course of distraction (POD12) in ActincreERT2;R26Rainbow, Sox9creERT2;R26Rainbow, and FAK(cKO)Rainbow mice (n = 10 per condition; Tukey’s multiple comparisons). g, Lateral to medial view of the callus overlying the distraction site. Confocal micrographs of whole-mount Sox9creERT2;R26Rainbow periosteum at POD12 (the white dotted outline shows the distraction gap; n = 6). h, As for g, but for FAK(cKO)Rainbow mandible. i, Quantification of angular clonal expansion in Sox9creERT2;R26Rainbow mice at POD12. j, As for i, but for FAK(cKO)Rainbow mice. Scale bars, 1 mm (a–d), 200 μm (g, h).
FAKi stunted skeletal lineage differentiation and progenitor expansion (Extended Data Fig. 6f–i). Ex vivo imaging and intracellular fluorescence-activated cell sorting (FACS) confirmed that FAKi inhibited FAK phosphorylation (Extended Data Fig. 6j). Similarly, induction of FAK(cKO) led to a disruption of the normal skeletal stem and progenitor expansion in distraction osteogenesis (Extended Data Fig. 7a, b). SSCs isolated from FAKi and FAK(cKO) mandibles at POD10 revealed significantly reduced colony-forming potential (Extended Data Fig. 6k; FAKi: ***P < 0.001; FAK(cKO): ***P < 0.001).
To further characterize the effect of FAK(cKO), we carried out in vivo clonal analysis using Sox9creERT2;Ptk2fl/fl;R26Rainbow mice (hereafter FAK(cKO)Rainbow) for distraction osteogenesis (Extended Data Fig. 7c, d). FAK(cKO)Rainbow mice exhibited clones of significantly smaller size in the distraction site versus controls (Fig. 3f–h; **P < 0.01). Whereas a normal clonal recombination frequency was observed in Sox9creERT2;R26Rainbowand FAK(cKO) mandibles during distraction (Extended Data Fig. 7e–h), FAK(cKO) disrupted the oriented clonal expansion observed in Sox9creERT2;R26Rainbow mandibles (Fig. 3i, j), highlighting that integrin-based cell–matrix interactions are required for vector-aligned expansion of progenitors.
To understand the molecular role of FAK during distraction osteogenesis, we performed ATAC-seq on SSCs and BCSPs treated with FAKi, finding that many distraction-specific accessible chromatin sites were no longer accessible in d-SSCs + FAKi. In FAKi mandibles, d-SSCs and d-BCSPs have chromatin accessibility resembling that of f- SSCs and BCSPs at a large subset of sites that differ between the fracture and distraction osteogenesis conditions (Fig. 4a; BCSP peaks not shown). k-means clustering divided all differential peaks between SSCs, f-SSCs and d-SSCs into eight clusters. We assigned GO terms to nearest genes. Injury-related terms were more accessible in fracture, distraction and distraction + FAKi samples (clusters 1, 3 and 6), whereas Wnt and fibroblast growth factor (FGF) signalling were enriched in clusters that were more accessible in the uninjured state (clusters 4, 7 and 8). We focused on distraction-specific sites the gain in accessibility of which was blocked with FAK inhibition, representing mechanotransduction-sensitive sites (clusters 2 and 5). These sites were enriched for adhesion and migration terms, along with terms for vascular/endothelial growth factor (VEGF), HIF1, and Cdc42 signalling (Fig. 4a).
Fig. 4 |. Changes in chromatin state during distraction osteogenesis with FAK inhibition.
a, Heat map showing differential peaks (log2(normalized reads in peaks)) between SSCs, f-SSCs and d-SSCs. k-means clusters are indicated by coloured bars at the left. Enriched GO terms for each cluster are shown at the right. b, Graphical demonstration of FAK-R sites. c, Pie charts showing the proportion of accessible sites that are FAK-R in SSCs and BCSPs. For SSCs, 4,432/9,937 sites (44.6%) that are more accessible in the distraction condition than in the fracture condition are FAK-R, versus 167/4,523 sites (3.7%) in the fracture condition (P < 1 × 10−16; one-sided Fisher’s exact test). For BCSPs, 1,738/4,883 sites (35.6%) that are more accessible in distraction are FAK-R, versus 495/4,415 sites (11.2%) that are more accessible in the fracture condition (P < 2.2 × 10−16; one-sided Fisher’s exact test). d, Genomic enrichment for peaks in five categories of SSC: R, FAK-R; NR, FAK-NR; DO, distraction osteogenesis; Fx, fracture; and FAKi. e, Expression of Sox9, Sox10 and Sox18 as determined by RNA-seq in d- and f-SSCs from PODs 5, 10 and 15. The y-axis shows the average t.p.m. (transcripts per million) from two biological replicates. All genes are significantly differentially expressed (P < 0.05, from DESEq2; see Methods) between the fracture and distraction conditions at at least one time point. f, Confocal micrographs of a distraction regenerate in a Sox10creERT2;R26mT/mG mandible collected at POD29 (the area of new bone is outlined with the dotted white lines). mTomato, background; mGFP, Sox10-expressing lineage; white, 4′,6-diamidino-2-phenylindole (DAPI) staining. Representative of three independent experiments. Scale bar, 200 μm.
We separated sites that were differentially accessible between the fracture and distraction conditions into FAK-responsive (FAK-R; more or less accessible in distraction than in fracture and FAKi) or FAK-non-responsive (FAK-NR; more or less accessible in distraction and distraction plus FAKi than in fracture; Fig. 4b and Supplementary Table 1). The majority of FAK-R sites in SSCs and BCSPs increased in accessibility (44.6% and 35.6%, respectively) rather than decreased (3.7 and 11.2% respectively) compared with fracture (Fig. 4c), suggesting that FAK is required for activation of a large part of the distraction-specific regenerative program. Genomic annotation revealed that FAK-R sites were more enriched for transposable long interspersed nuclear elements (LINEs) and long terminal repeats (LTRs) than were FAK-NR sites or all sites (Fig. 4d). Transposable elements constitute around 50% of the genome and contain transcription-factor-binding sites. Interestingly, LINEs are the largest class that contains active enhancers in the neural crest, from which the mandible is derived16.
To understand the mechanosensitive program further, we performed motif enrichment in FAK-R and FAK-NR sites. CCAAT/enhancer-binding protein (C/EBP) and activating transcription factor (ATF) motifs were enriched in FAK-NR sites in SSCs and BCSPs, consistent with the roles of C/EBP, ATF4 and RUNX2 in bone differentiation in vitro17. FAK-R sites in SSCs were enriched for Sox and ETS motifs, whereas responsive sites in BCSPs were enriched for Sox but not ETS motifs (Extended Data Fig. 8a). Sox9 is a core transcription factor in osteogenesis, and other Sox proteins are involved in mandibular development and other regenerative processes. RNA-seq revealed that Sox9 was downregulated in d-SSCs. Sox18 and Sox10 were upregulated, indicating that they may direct the FAK response (Fig. 4e and Extended Data Fig. 8b, d). The ETS factor Elk3 showed similar upregulation (Extended Data Fig. 8c). We were surprised to find Sox10, Sox18 and Elk3 upregulated in distraction osteogenesis, as they are not involved in postnatal bone repair, but are critical for neural crest development18.
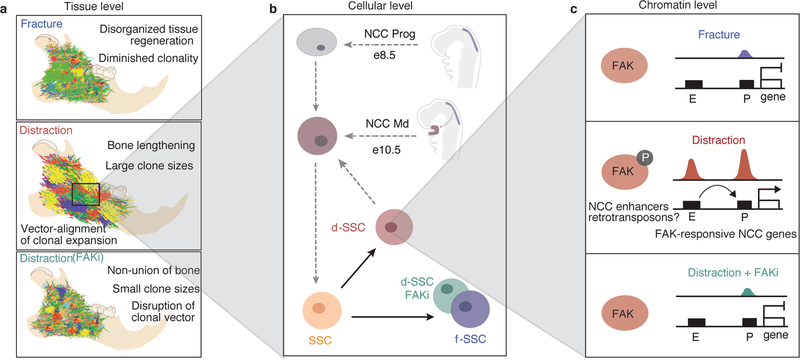
Lineage tracing with Sox10creERT2;R26mT/mG mice confirmed Sox10 expression within newly forming bone (Fig. 4f and Extended Data Fig. 8e–g). Given the expression of neural crest transcription factors by d-SSCs and the migratory properties of these cells (Fig. 1m, n and Fig. 3g, h), we hypothesized that d-SSCs access a primitive, neural crest cell (NCC)-like state to achieve productive lengthening.
SSCs adopt an NCC-like state during distraction
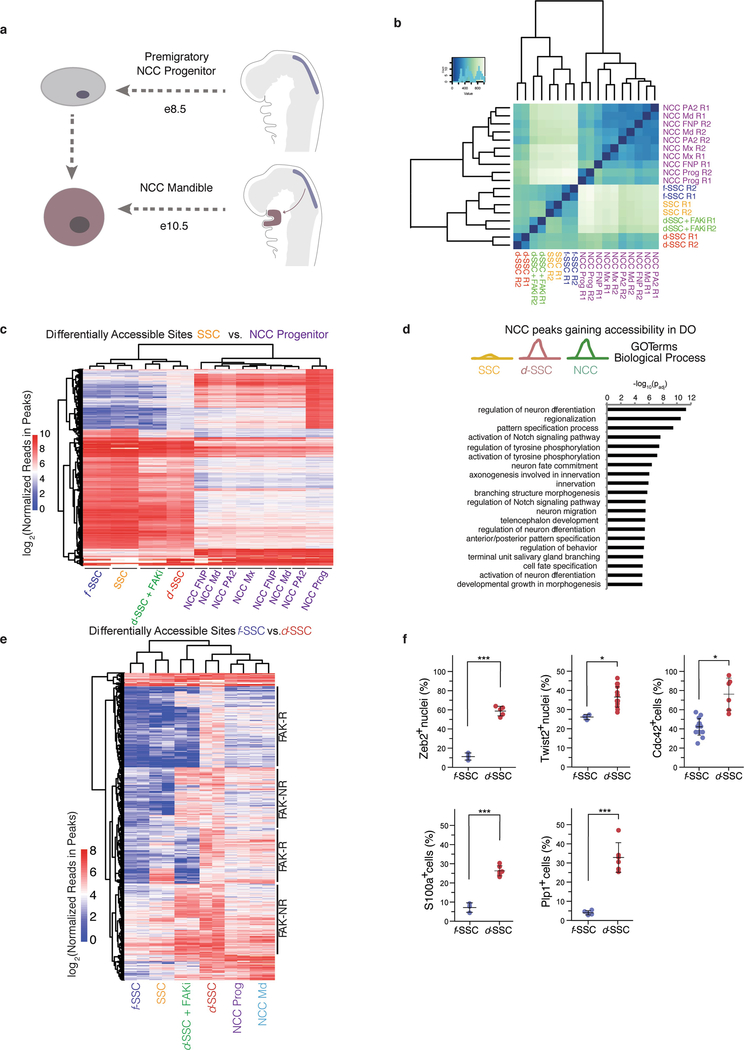
NCCs are highly plastic embryonic cells that give rise to diverse tissues including the Schwann cells, teeth and bones of the mandible19–22. To assess whether d-SSCs take on a more NCC-like identity, we compared their chromatin accessibility to that of premigratory mandibular NCCs (NCC Prog) and postmigratory populations (NCC Md (mandibular), Mx (maxillary), PA2 (pharyngeal arch 2) and FNP (frontonasal projection))23 (Extended Data Fig. 9a). Principal component analysis (PCA) showed that SSCs, f-SSCs, and d-SSCs + FAKi cluster close together, whereas d-SSCs fall closer to NCCs (Fig. 5a). Clustering on all open chromatin sites, d-SSCs are more similar to NCC populations than are SSCs, f-SSCs or d-SSCs + FAKi (Extended Data Fig. 9b). We identified regulatory elements that are inaccessible in homeostatic SSCs but accessible in d-SSCs and NCCs, and found that they are enriched for embryonic, developmental and migratory terms (Extended Data Fig. 9c, d). Inaccessibility of these DNA elements in f-SSCs and d-SSCs + FAKi indicated that this activation is FAK-dependent. RNA-seq confirmed that key neural crest genes are expressed specifically in d-SSCs (Fig. 5b). d-SSCs gained accessibility at bona fide developmental neural crest enhancers (FaceBase24,25) in a FAK-dependent manner (Fig. 5c). Profiling of SSC and NCC populations revealed a subset of FAK-R sites that are active in neural crest populations, further indicating FAK dependency (Extended Data Fig. 9e). The association of FAK-R sites with LINEs (Fig. 4d) led us to consider the relationship between this NCC program and transposable-element insertions. Transposable elements that were accessible in SSCs were different from those accessible in NCCs, and the d-SSCs clustered closely with NCCs (Fig. 5d).
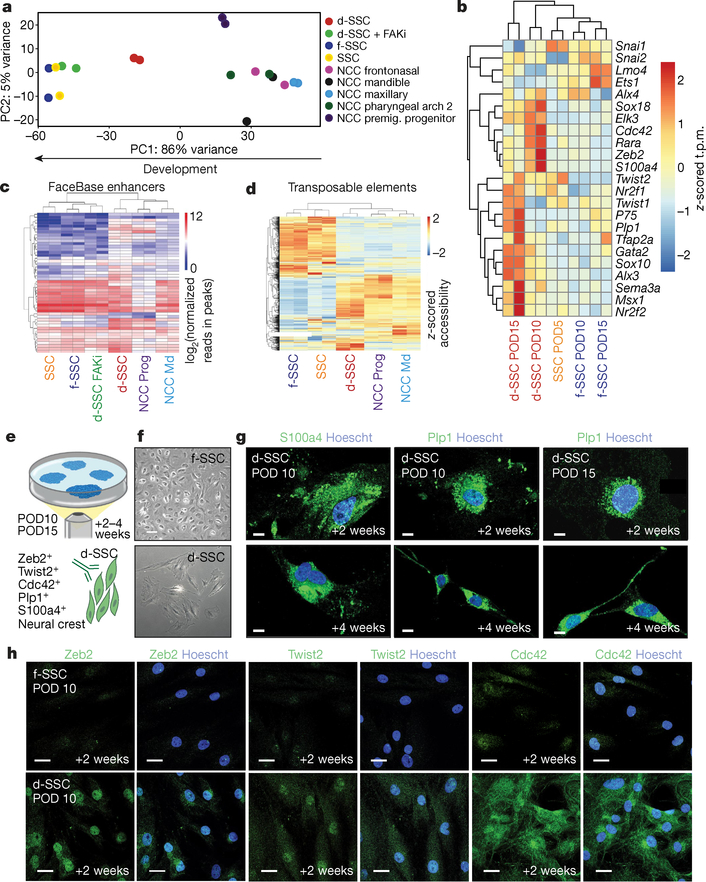
Fig. 5 |. SSCs adopt an NCC-like state during distraction.
a, Principal component analysis (for principal components (PC) 1 and 2) of ATAC-seq data from SSCs and NCCs (n = 269,900 peaks). The arrow at the bottom shows the direction of cellular differentiation from NCC to SSC. b, Heat map of neural crest gene expression in d- and f-SSCs at PODs 5,10 and 15. The z-scored t.p.m. refers to the number of standard deviations from the mean for each t.p.m. value in a sample. P75 is also known as Ngfr. c, Accessibility at mouse neural crest enhancers (from FaceBase24,25) at neural crest genes. Shown are log2(reads in peaks) for ATAC-seq peaks that overlap the annotated enhancers. d, Heat map of z-scored accessibility at all families of transposable elements from ATAC-seq data. e, Schematic showing f- or d-SSC culture and staining from postoperative mandibles. FACS-isolated f-SSCs and d-SSCs at PODs 10 and 15 form colonies at two weeks and four weeks in culture (top). Then d- and f-SSCs (bottom right, green cells) are stained with the known NCC markers indicated at the bottom left (indicated by the green antigen). f, Representative images of d- or f-SSCs plated and cultured for two weeks. g, Immunofluorescent staining of POD10 and POD15 d-SSCs for NCC markers (S100a4 and Plp1; green) at two and four weeks in culture. Images are representative of three independent experiments. Blue (Hoechst) staining identifies the nucleus. h, Immunofluorescent staining of f-SSCs versus d-SSCs at POD10 for the markers Zeb2, Twist2 or Cdc42 (green) and nuclear staining (blue) (representative of three independent experiments). Scale bars, 200 μm (f), 100 μm (g, h).
We assessed neural crest potential in these cells by sorting SSCs from distraction versus fracture mandibles (Fig. 5e, f). d-SSCs stained positive for the NCC-associated markers S100a4 and Plp1 at two and four weeks of culture (Fig. 5g). Additionally, canonical neural crest markers were specifically expressed in the d-SSC condition, further supporting the idea that an NCC-like program is activated (Fig. 5h and Extended Data Fig. 9f).
Discussion
Here we have comprehensively modelled the process of bone regeneration in the jaw from the tissue to the chromatin level, to understand how controlled mechanical separation of bones leads to lengthening of the mandible (Extended Data Fig. 10). We have shown that the skeletal stem cell lineage gives rise to new bone in this paradigm, and can be isolated using the same markers as for long bone. The differences in chromatin accessibility and gene expression between the fracture and distraction conditions suggest that, while stem-cell proliferation and differentiation to produce restricted progenitors are important for this massive tissue regeneration, they are not the only factors at play. The downregulation of the canonical skeletal program during distraction osteogenesis suggested that the homeostatic program for tissue renewal is not sufficient for regeneration in distraction. We have shown that an alternative regenerative program activated in distraction osteogenesis is dependent on the FAK pathway, which has a known role in mechanotransduction. This FAK-dependent program is an embryonic NCC-like program that reverts d-SSCs to a more plastic, developmental state.
Postnatal regeneration is highly restricted in vertebrates. Newts use both dedifferentiation and activation of tissue-resident stem cells and progenitors, whereas the axolotl uses dedifferentiation followed by redifferentiation in the limb26–31. Studies in mouse digits have demonstrated the formation of new bone with lineage-restricted origins11,12. We have shown, however, that adult stem cells revert to a developmentally plastic state during regeneration of the jaw. Reversion to an embryonic-like NCC has also been seen during the initiation of melanoma tumours32. In the context of the massive tissue regeneration seen in distraction osteogenesis and the cellular hyperproliferation that occurs in cancer, a more potent stem-cell-like program may be required, whereas the more restricted cell (that is, the postnatal SSC in uninjured tissue) is sufficient during homeostatic renewal.
Retrotransposon sequences are co-opted as transcriptional enhancers during development, and related sequences make up large transcriptional networks33–37. The accessibility of differing retrotransposon families between uninjured and fractured SSCs versus distraction SSCs suggests that a novel transcriptional network is active in this regenerative context, and may have a role during neural crest development. Understanding the relationship between retrotransposon activation and developmental mechanisms may unveil key processes underlying regenerative paradigms.
Methods
Animals
Rainbow mice38 containing reporter gene R26VT2/GK3 were crossed with ActincreERT2 and Sox9creERT2 mice (Jackson Laboratories) to obtain ActincreERT2;R26Rainbow and Sox9creERT2;R26Rainbow mice. Additionally, mice containing R26mT/mG (Jackson Laboratories) were crossed with Sox9creERT2 and Sox10creERT2 animals to obtain Sox9creERT2;R26mT/mG and Sox10creERT2;R26mT/mG mice (Jackson Laboratories). Tamoxifen (Sigma-Aldrich) was dissolved in 90% corn oil/10% ethanol (vol./vol.), and filtered through a 0.2-μm membrane. Ten-week-old male mice were given intraperitoneal injections of 200 mg per kg body weight of tamoxifen daily for five consecutive days. After one week, distraction osteogenesis was surgically applied to hemimandibles as specified. For all conditions, age- and sex-matched littermates served as control animals and were given corn-oil injections (no tamoxifen). Ten-week old male C57BL/6J mice (Jackson Laboratories) were used for isolation of mouse SSCs and skeletal progenitors for gene-expression and ATAC-sequencing experiments as indicated. Ten-week-old male C57BL/6J-Tg(CAG-EGFP)10sb/J mice (Jackson Laboratories) were used for parabiosis experiments.
All experiments were performed in accordance with Stanford University Animal Care and Use Committee guidelines. Animals were housed in a light- and temperature-controlled environment and given food and water ad libitum. Sample size was no less than three animals for all experiments. Specific numbers are stated in the figure legends. Sample size was determined by the number of viable animals of the right age and genotype at the time of experiment. All data analysis was conducted in a blinded manner for experiments in which the investigator could affect the outcome, such as any μCT analyses, cell counting in immunofluorescence-based assays, assessment of molecular treatments, clonal analyses related to lineage tracing, and so on. Animals with the appropriate genotype were randomly allocated to experimental conditions.
Device design and manufacturing
Mandibular distraction devices were manufactured via computer-aided design (CAD) in SolidWorks (SolidWorks) for 3D printing (ProJet 3510 HD Plus, 3DSYSTEMS) at 16-μm resolution.
Mandibular distraction surgery
Animals were divided into four groups in this study: sham-operated, fractured, acutely lengthened and gradually distracted. In brief, animals were anaesthetized (with 20 mg kg−1 Ketaset, 1.5 mg kg−1 xylazine and 0.2 mg kg−1 acepromazine maleate), given a preoperative dose of antibiotics (10 mg kg−1 cefazolin) and prepped with Betadine, and their incisors were clipped. An incision was made over the right hemimandible, the masseter muscle divided, and the mandible exposed. One 0.6-mm hole was drilled 3 mm anterior, and one 3 mm posterior, to a line dividing the mandibular ramus just posterior to the third molar. An osteotomy was then performed posterior to the third molar using a diamond disc saw under constant saline irrigation (Brasseler). Distraction plates were secured with insertion of tight-fit 0.65-mm screws (McMaster-Carr). The muscle and skin were then closed in layers. All animals tolerated the procedure well and received appropriate postoperative analgesia. Postoperative mortality was 5%, and all deaths were replaced with new animals to obtain the final numbers.
Mandibular distraction protocol
The gradual-distraction protocol consisted of a 5-day latency period after the initial osteotomy and fixation of the distraction device, followed by 10 days of distraction at a rate of 0.15 mm every 12 h (for a total of 3.0 mm) and then 28 days of consolidation. For our acute-lengthening protocol, lengthening was performed equal to the total distraction amount (3.0 mm) following a 5-day latency period, with a consolidation period ending at 43 days postoperation. All specimens were collected at either 29 days or 43 days postoperation, the latter being the end of bone consolidation.
Micro-computed tomography scanning
Devices were removed carefully before fixation. Specimens were manually palpated as a screening test for complete bone union before overnight fixation (in 2% paraformaldehyde (PFA) at 4 °C), then were processed to 70% ethanol and scanned using a MicroXCT-200 (Carl Zeiss Microscopy) at 40 kV and 160 lA. We took 1,200 projection images at a total integration time of 6 s, with linear magnification of ×2 and a pixel size of 10 μm. The morphological data were reconstructed in a 3D solid volume in the 3D image data visualization program Avizo Fire (FEI). Following μCT scanning, tissues from POD43 were processed for histomorphometric and histologic evaluation. A total of six ten-week old C57BL/6J male mice were included per treatment group (n = 6).
Histological analysis
Dissected specimens were fixed in 2% PFA at 4 °C overnight. Specimens were decalcified in 400 mM EDTA in phosphate-buffered saline (PBS; pH 7.2) at 4 °C for four weeks, with a change of EDTA every 48 h. The specimens were then dehydrated in 30% sucrose at 4 °C overnight. Specimens were then embedded in optimal cutting temperature (OCT) compound and sectioned at 8 μm. Representative sections were stained with Movat’s modified pentachrome or with alizarin red depending on the individual experiment.
Histomorphometric analysis
Histomorphometry was performed using micrographs of Movat’s pentachrome staining obtained for subsequent quantitative analysis of relative bone (yellow) and cartilage (blue) formation in the distracted callus. A region of interest (ROI) was assigned in Adobe Photoshop for pixel-based quantification (bone pixels/total ROI pixels; cartilage pixels/total ROI pixels; stromal tissue pixels/total ROI pixels) using ten slides (one every 50 μm) per specimen across five specimens.
Immunofluorescence
Immunofluorescence on cryopreserved ectopic bone specimens were performed using a mouse-on-mouse immunodetection kit from Vector Laboratories according to the manufacturer’s instructions. In brief, specimens were treated with a blocking reagent, then probed with primary antibody at 4 °C overnight. Specimens were next washed with PBS, probed with Alexa-dye-conjugated antibodies, washed, cover-slipped and imaged with a Leica DMI6000B inverted microscope system.
Tamoxifen induction of the Rainbow reporter system
We used Rainbow mice for clonal analysis of bone regeneration during mandibular distraction. Rainbow mice were crossed with the ubiquitous ActincreERT2 or Sox9creERT2 driver so as to mark all cells or skeletal-lineage-derived cells after systemic tamoxifen induction by intraperitoneal injection. The Rainbow reporter (R26VT2/GK3) is a multicolour Cre-dependent marker system with a four-colour reporter construct in the ROSA locus. Once recombination occurs, cells are randomly and genetically marked with one of ten possible colour combinations and daughter cells will be marked with the same colour as the parent cell, creating a fluorescent mosaic pattern upon analysis. Nine-week old male ActincreERT2;R26Rainbow or Sox9creERT2;R26Rainbow mice were administered intraperitoneal injections with tamoxifen at 200 mg kg−1 daily for five consecutive days. At ten weeks of age, distraction osteogenesis was surgically applied to hemimandibles in systemically induced Rainbow mice. Hemimandibles were collected 15 and 29 days after initial surgery and immediately fixed in 2% paraformaldehyde (wt/vol.) overnight at 4 °C in the dark. Tissue samples were prepared for cryo-embedding by soaking in 30% (vol./vol.) sucrose in PBS at 4 °C for 24 h. Samples were removed from the sucrose solution and tissue blocks prepared by embedding in Tissue Tek OCT (Fisher). Cryosections were obtained and counterstained for nuclei with Hoechst 33342 dye for subsequent confocal microscopy (Leica TCS Sp8).
Imaging analysis
Laser scanning confocal microscopy was performed with a Leica TCS SP8 X confocal microscope (Leica Microsystems) with an objective lens (×10 HC PL APO, air, numerical aperture 0.40; ×20 HC PL APO IMM CORR CS2, H2O/glycerol/oil, numerical aperture 0.75), located in the Cell Sciences Imaging Facility (Stanford University). Raw image stacks were imported into Fiji (NIH) or Imaris (Bitplane/Perkin Elmer) software for further analysis. Imaris software was used to analyse cells and obtain xyz coordinates of individual clones. All clones were examined individually to confirm that they reported a single colour. Only clones that could be visually determined to consist of five cells or more were included in the analysis of clone size. The theoretical recombination frequency resulting in a cell being marked by one colour in Rainbow mice is 1/10 possible combinations. The probability of obtaining by chance single-colour clones containing five or more cells would then be approximately greater than or equal to 1 in 2.1 million. Therefore, we included only clones of five cells or more in our analysis. Imaris (Bitplane) software was used to render 3D volumes of z-stacks and to perform image analysis. Cell-spotting analysis of Rainbow fluorescent clones was performed using the volume surface and spot creation tools, thresholding by volume and signal quality. For Rainbow clonal analysis, adjacency was determined by thresholding maximum distance to the nearest cell using the spot-to-spot distance tool. Quantification of images was performed on Imaris using the spots rendering statistics tool as per the manufacturer’s protocol, within a 1–3-mm-wide ROI containing the distraction gap and callus. For the osteotomy site, a surface was manually defined in Imaris by the edge of dissection within the callus area. The Imaris distance transformation module from their XTensions library was then used to determine the vector of each clone for a determination of angles, and the axis of each clone was measured with respect to the osteotomy edge. For determination of angular expansion, clones of at least ten cells were evaluated throughout five distracted mandibles for each group. Measured clonal vectors were plotted within each 10° segment. Provided images are typically presented as a maximal projection of either 8–12-μm optical sections or 30–50-μm whole-mount renderings, unless otherwise specified. For visualization of individual labelled cells expressing the Rainbow reporter, the brightness and contrast were adjusted accordingly for the green (eGFP), blue (mCerulean), orange (mOrange) and red (mCherry) channels, and composite serial image sequences were assembled. Tiled images were stitched by a grid and collection stitching plugin in Fiji.
Tissue-specific Rainbow clonal analysis
To further explore the periosteum as a cellular source of progenitors during mandibular distraction, we devised a strategy of local genetic labelling of the mandibular periosteum for tissue-specific clonal analysis. To induce site-specific recombination in ActincreERT2;R26Rainbow mice, we delivered activated 4-hydroxytamoxifen (10 μg) in a liposomal formulation (20 μg μl−1) to the periosteum on the buccal surface of the mandible at a 1-mm distance posterior to the third molar. After one week, tissues were collected to evaluate the local labelling strategy in intact mandibles, at which time distraction osteogenesis was surgically applied to hemimandibles in locally induced Rainbow mice and tissues were harvested at POD15. Periosteum-specific clonal analysis was performed with four animals (including controls receiving liposomal formulation without 4-hydroxytamoxifen) across two independent experiments (n = 8) using ActincreERT2;R26Rainbow mice.
Whole-mount Rainbow clonal analysis
Nine-week-old male ActincreERT2; R26Rainbow, Sox9creERT2;R26Rainbow and Sox9creERT2;R26Rainbow;Ptk2fl/fl mice were intraperitoneally induced with tamoxifen as described. Intact periosteum (one-year lineage trace) or distraction calluses (PODs 5, 8, 10, 12 and 15) were collected using fine-precision surgical techniques to expose the mandibular periosteum. Upon exposure, mandibular periosteum was preserved in Fluoromount-G medium (SouthernBiotech) and mounted onto microscope slides (Fisher Scientific). Preserved periosteum specimens were imaged immediately using confocal microscopy techniques as described above. Whole-mount Rainbow clonal analysis was performed using a minimum of ten biological replicates for ActincreERT2;R26Rainbow, and ten biological replicates for each of Sox9creERT2;R26Rainbow and Sox9creERT2;R26Rainbow;Ptk2fl/fl animals.
Lineage tracing of Sox-expressing progenitors in vivo
To evaluate Sox-factor-expressing cells in mandibular distraction, we used Sox10creERT2;R26mT/mG mice to trace progenitors. Nine-week-old male Sox10creERT2;R26mT/mG mice were administered intraperitoneal injections with tamoxifen as described above. The hemimandibles of Sox10creERT2;R26mT/mG mice were isolated and transverse sections prepared for confocal microscopy as described above. Lineage tracing of Sox-expressing progenitors was performed using a minimum of six separate animals as biological replicates (n = 6).
Parabiosis for circulating cell fate in distraction
To determine the contribution of circulating cells to newly formed bone, we carried out parabiosis as previously described39. In brief, sex- and age-matched GFP-labelled mice (C57BL/6 J-Tg(CAG-EGFP)10sb/J; Jackson Laboratories) and non-GFP littermates (C57BL/6 wild-type; Jackson Laboratories) were used. Animals were anaesthetized via inhalational anaesthesia. An incision in the skin was made from the base of the right foreleg to the base of the right hind leg of one parabiont, and from the left foreleg to the base of the left hindleg of the other parabiont. The skins were sutured together at the foreleg and hind-leg joints. The remaining dorsal and ventral flaps were stapled together. Analgesia was administered postoperatively. After one month of parabiosis, peripheral samples were collected from the tail, and FACS was used to assess parabiont blood chimaerism. After peripheral blood chimaerism reached a 1/1 ratio—representing a complete fusion of the circulatory systems of both parabionts—distraction osteogenesis was performed on non-GFP mice as described above. Hemimandibles were collected to analyse the distracted callus for the presence of GFP cells after two weeks of consolidation (POD29), when bone union was complete. Tissue processing and histology was performed on hemimandibles as previously described. Parabiosis experiments were performed using four biological replicates (n = 4) with data represented as means ± s.d.
Sample preparation and FACS isolation
Bones were dissected and serially digested in collagenase digestion buffer supplemented with DNase at 37 °C for 40 min under constant agitation; total dissociated cells were filtered through 40-mm nylon mesh, pelleted at 200g at 4 °C, resuspended in staining medium (2% fetal calf serum (FCS) in PBS), and stained with fluorochrome-conjugated antibodies against CD45, Ter119, CD202b, Thy1.1, Thy 1.2, CD105, CD51 and 6C3, and with a streptavidin-conjugated antibody for CD200. Propidium iodide staining was performed to exclude dead cells. FACS analysis was performed on a FACS Aria II Instrument (BD Biosciences) using a 70-mm nozzle. Gating schemes were established with fluorescence-minus-one controls and propidium iodide was used for viability staining. All cell populations were double-sorted for purification and subsequently evaluated for their functional responses as outlined below. Flow-cytometry plots are representative of a minimum of three independent experiments.
To calculate cell population frequencies (Figs. 3e, 5l), we assessed five post- operative calluses (n = 5) by FACS and represent data as an average across three independent experiments. Single uninjured mandibles (dissected anterior of condyle but posterior to third molar) contain approximately 1 × 103 to 1.2 × 103 SSCs, 2.8 × 103 to 3.5 × 103 BCSPs, and 4.5 × 103 to 5 × 103 osteoprogenitors. All molecular and flow-cytometric analyses of FACS-purified cell populations in the SSC hierarchy throughout this study were performed using double-sorted cells to ensure purity of each population. The double-sorting technique reduces the yield of purified populations by roughly one half, necessitating surgical operations on approximately twice the number of animals expected to achieve the desired cell number for a given experiment (see the Methods sections below on ATAC-seq and RNA-seq). To double-sort SSCs and BCSPs, we first performed FACS isolation of the ‘double-negative’ population (CD45− Ter119− CD202b− Thy1− 6C3− CD51+) on the basis of yield, and then re-sorted on the basis of purity for SSCs (CD45− Ter119− CD202b− Thy1− 6C3− CD51+ CD105− CD200+) and BCSPs (CD45− Ter119− CD202b− Thy1− 6C3− CD51+ CD105+). For double-sorting of terminally differentiated Thy1+ populations, we first performed FACS isolation of the ‘Thy’ population (CD45− Ter119− CD202b− Thy1+ 6C3− CD51+) on the basis of yield, and then re-sorted cells on the basis of purity for osteoprogenitors (CD45− Ter119− CD202b− Thy1+ 6C3− CD51+ CD105+ CD200−) and chondrocytic progenitors (CD45− Ter119− CD202b− Thy1+ 6C3− CD51+ CD105+ CD200+).
EdU analysis in vivo
Mice were injected intraperitoneally with 100 mg kg−1 EdU (Life Technologies) 12 h before euthanasia. Cells were double-sorted for purity through FACS isolation as described above before being fixed and subject to EdU staining using the Click-iT Plus EdU Alexa-488 flow-cytometry assay kit (Life Technologies). Propidium iodide was used to stain for total DNA viability content and the percentage of GFP-negative and GFP-positive cells showing EdU incorporation was analysed using a BD FACSAria II. SSCs were isolated from ten-week-old male mice according to strain, and colony-formation assays were performed in triplicate, using a minimum of three biological replicates across three independent experiments. Graphs depict means ± s.d. across the biological replicates.
Colony formation in vitro
Isolated SSCs from uninjured wild-type or FAK(cKO) (uninduced, no tamoxifen) mice were directly plated onto precoated (0.1% gelatin) culture plates (100 cells per well in a 10 cm2 well plate) in MEMa medium with 20% FCS under low O2 (2% atmospheric oxygen, 7.5% CO2) conditions. After four days of attachment, SSCs from wild-type mandibles were treated with the FAK inhibitor PF-573228 (FAKi) or with DMSO (control), while SSCs from the FAK(cKO) mice were treated with activated tamoxifen (4-OHT) or control medium every other day for one week. SSCs isolated from uninjured wild-type mandibles for serial colony-forming units, and plated in the same conditions described above, were not treated with any substance. Colony-forming units were identified using an inverted microscope under ×40 magnification. Specimens were examined under phase microscopy and a cloning ring was used for quantification. Colonies were assessed for size and cell morphology as previously described6. The cells were subsequently lifted for staining and analysis by FACS, or plated for tertiary colonies. Similarly, isolated SSCs from uninjured wild-type mice were plated onto precoated dishes (1 cell per well in a 4-cm2 well plate), cultured and analysed in identical conditions to those described above. SSCs were isolated from ten-week-old male mice according to strain, and colony-formation assays were performed in triplicate using a minimum of three biological replicates across three independent experiments (n = 9); graphs depict means ± s.d. across the biological replicates.
Osteogenic differentiation assay
Upon isolation of cells by FACS, colonies were grown over two weeks as described above. Each cell-type condition (f-SSC and d-SSC) was incubated with osteogenic medium for two weeks with the medium changed every other day. After undergoing two weeks of osteogenic differentiation, cells were washed with PBS followed by ultrapure water. The monolayer of cells was fixed with 100% ethanol for 15 min and stained with alizarin red solution for 1 h at room temperature. The cells were washed several times with ultrapure water and imaged for osteogenic potential immediately under a bright-field microscope. Following imaging, the cells were treated with a methanol/acetic acid mixture for 15 min and absorbance was detected with an Ultraspec 2100 UV/Visible Spectrophotometer (Biochrom, Harvard Bioscience) at 450 nm to measure alizarin red protein concentration across each cell type.
ATAC-seq
ATAC-seq was performed as previously described, using the omni-ATAC protocol40. In brief, 10,000 cells per replicate were pelleted and lysed in 50 μl lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2) with 0.1% NP40, 0.1% Tween-20 and 0.01% digitonin for 3 min on ice. We then added 1 ml of cold lysis buffer plus 0.1% Tween-20, and centrifuged cells at 500g for 10 min at 4 °C. Following centrifugation, pelleted nuclei were resuspended in 50 μl transposition mix (25 μl 2× tagmentation DNA (TD) buffer, 2.5 μl transposase (final concentration 100 nM), 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, 5 μl H2O) and incubated for 30 min at 37 °C with shaking at 1,000 r.p.m. Transposition mix was cleaned up using Qiagen MinElute columns. Library preparation was performed exactly as previously described13. To achieve adequate cell numbers (minimum 10,000 double-sorted cells per replicate), we required 18–22 total ‘distracted’ mandibles (one per animal) per replicate in a single experiment, and 26–30 total ‘fracture’ mandibles per replicate in a single experiment. Comparisons between surgical conditions were strictly performed using littermates randomly assigned and mandibles operated upon at the same date. Cells were FACS-isolated within the same session for parallel library preparation (for example, f- and d- SSC, BCSP and OP populations isolated on the same date for a direct comparison). A minimum of two biological replicates (as two independent operations in separate animal cohorts and separate FACS isolation experiments) were used for each condition in our ATAC-seq analyses.
ATAC-seq analysis
ATAC-seq libraries were sequenced on an Illumina NextSeq. Adaptor sequences were trimmed using CutAdapt software and reads were then mapped to the genome (mm9 build) using Bowtie2. Mapped reads were filtered for quality, and then reads mapping to segmental duplications and non-unique sequences were removed. Duplicates were further filtered out using PICARD. Peaks were called on each individual sample using MACS2 with a q-value cut-off of 0.01 and with no shifting model. Peaks from all samples were merged and bedtools was used to calculate the read depth at each peak for each sample. DESEq2 was used for pairwise differential peak calling with a cut-off of fold change >2 and padj <0.05 for differential sites. To calculate P values in DESEq2, for each gene the counts were modelled using a generalized linear model (GLM) of negative binomial distribution among samples. The two-sided Wald statistics test was processed for significance of the GLM coefficients. The Benjamini–Hochberg correction was applied to all P values to account for the multiple tests performed. This gives the final adjusted P values used for assessing significance. For motif-enrichment analysis, we used HOMER v.4.8.3 with default settings. The de novo motif search results are shown. For all motif enrichment, background sets of all peaks were used. For genomic annotations, HOMER’s Annotate peaks tool was used.
Analysis of GO terms from ATAC-seq data
We carried out GO-term analysis of ATAC-seq sites using GREAT v.3.0.0 (ref.41; http://great.stanford.edu/public/html/), with a background set of all peaks for a given cell type.
Analysis of retroviral elements in ATAC-seq data
To analyse retroviral-element accessibility in ATAC-seq data, we mapped all ATAC-seq reads to a bowtie2 index containing the sequences of all retroviral insertions from the RepeatMasker database (http://www.repeatmasker.org/). Reads mapping to each retroviral-element family were summed for each sample. Transfer RNAs and simple repeats were removed. The read counts were then normalized for each repeat type using DESEq2.
ATAC-seq data from neural crest cells
ATAC-seq data from NCC populations were downloaded from the Gene Expression Omnibus (accession number GSE89436). Data were downloaded in raw form (fastq) and processed using the same pipeline as above.
Small-molecule inhibition of FAK
Mice undergoing distraction were treated with local subcutaneous injections of the FAK inhibitor PF-573228 (Tocris) at a concentration of 50 μM. PF-573228 specifically inhibits the phosphorylation of FAK at tyrosine 397—the active site required for canonical activation of the FAK pathway in the context of integrin-based signalling events. We determined the minimum effective dose as the minimum required to inhibit FAK phosphorylation in vitro and in vivo, in both SSCs and BCSPs, without disrupting cell viability according to increased propidium iodide staining via FACS. We evaluated both SSCs and BCSPs after in vitro treatment using FAKi at 10 μM, 20 μM, 40 μM, 50 μM, 70 μM, 80 μM or 100 μM, compared with DMSO treatment alone. FAKi was added at each 24-h medium change over the course of one week. Using intracellular FACS and immunocytochemical staining for phosphorylated FAK, we determined that the 50 μM treatment substantially inhibited phosphorylation without disrupting cell viability when compared with DMSO treatment alone. Ex vivo intracellular FACS and immunocytochemical staining confirmed inhibition of FAK phosphorylation (Extended Data Fig. 9e) without disrupting cell viability when FAKi was administered subcutaneously every 24 h at a concentration of 50 μM in a 100-μl volume in vivo. We determined the minimum toxic concentration to be 70 μM in vitro and 80 μM in vivo, according to propidium iodide staining on FACS. To determine the effect of inhibiting the mechanotransduction pathway on the rate of bone healing during distraction osteogenesis, we administered wild-type C57BL/6 mice with daily local injections (100 μl total per day) of the FAK small-molecule inhibitor or control vector (DMSO) throughout the distraction period.
Conditional knockout of FAK
To determine whether FAK-pathway inhibition affects the functional response of skeletal stem and progenitor cells during mandibular distraction, we administered FAK(cKO) mice (Sox9creERT2;Ptk2fl/fl) with injections of either tamoxifen (200 mg kg−1 in corn oil) or corn oil for five consecutive days. Distraction osteogenesis was then applied to hemimandibles of FAK(cKO) (tamoxifen) and control (no tamoxifen) littermates.
RNA-seq library preparation
RNA was extracted from FACS-sorted SSCs, BCSPs and osteoprogenitors using the Qiagen RNEasy Micro kit. For library preparation we modified the SMART-seq protocol as follows: following extraction, 1.5 ng of total RNA was reverse transcribed using an oligo-dT primer to make complementary DNA from polyadenylated transcripts. Reverse-transcribed cDNA was then pre-amplified for 11 PCR cycles. Next, 1 ng of cDNA was transposed with 5 μl of Nextera amplicon tagment mix for 5 min at 55 °C. Transposed libraries were then amplified for 11 cycles using Nextera polymerase chain reaction (PCR) master mix and adapters. Libraries were sequenced (single-end 75-base-pair reads) on an Illumina NextSeq 500 instrument.
RNA-seq analysis
Reads in fastq format were mapped and assigned to a transcript GTF file from UCSC and RefSeq using Tophat 2 (version 2.1.1). Reads in each transcript were then counted using FeatureCounts (featureCounts.v4.R) in R. For global clustering and PCA, read counts per transcript were normalized using DESeq2 and plotted using heatmap.2 and plotPCA(DESEq2) all in R. For differential-expression analysis, DESeq2 was used to make pairwise comparisons using default normalization parameters. Significantly differentially expressed genes are those showing a fold change of 1.5 or greater and an adjusted P value of less than 0.05. David V.6.8 was used for GO-term analysis. To achieve adequate cell numbers (minimum 5,000 cells per replicate), 18 total distracted mandibles (one per animal) were required to generate two technical replicates at each time point. Comparisons between postoperative time points were strictly performed using littermates randomly assigned and mandibles operated at successive time points leading up to the same collection or FACS isolation date. Cells were FACS-isolated within the same session for parallel library preparation. A minimum of two biological replicates (as two independent operations in separate animal cohorts and separate FACS-isolation experiments) were used for each condition in our RNA-seq analyses.
Immunofluorescence staining in vitro
SSCs were sorted through FACS isolation and culture for colonies from two to four weeks as described above. Cells were washed with PBS and fixed in 2% PFA then permeablized with Triton X 100. Specimens were treated with a blocking reagent, then probed with primary antibody at 4 °C overnight. Specimens were washed with PBS, probed with Alexa-dye-conjugated antibodies, and again washed in PBS. Cells were stained with Hoechst dye and mounted on cover slides to be imaged with a Leica DMI6000B inverted microscope system.
Antibodies used were as follows: rabbit anti-mouse-FAK, Abcam ab81298, clone EP2160Y, lot GR237911–21; rabbit anti-mouse-S100A4, Abcam ab41532, lot GR322940–2; rabbit anti-mouse myelin proteolipid protein, Abcam ab28486, lot GR268116–9; rabbit anti-mouse-CDC42, Abcam ab155940, lot GR3177223–5; rabbit anti-mouse-Twist2, Biologicals, Lot R87976; rabbit anti-mouse-ZEB2, ThermoFisher PA5–20980, lot QL2127782; goat anti-rabbit immunoglobulin G with conjugated Alexa Fluor 488, Thermo Fisher A-11034, lot 1885241.
Statistical analysis
Data are expressed either as absolute numbers or as percentages ± s.d. Statistical significance was assigned for P ≤ 0.05. Data analysis was performed using Student’s t-test assuming two-tailed distribution, and/or one-way analysis of variance (ANOVA) and post hoc Tukey correction. Note that *P < 0.05 to ****P < 0.0001 indicate a significant difference. Statistical calculations were performed using the Prism software package (GraphPad), except where stated otherwise. No statistical method was used to predetermine sample size. Figure panels showing representative images are representative of at least two independent experiments and up to five, as indicated in the figure legends. Flow-cytometry plots are representative of at least eight independent experiments and up to twelve, as indicated in the figure legends.
Statistics and reproducibility
For all figures, n indicates the number of animals per independent experiment unless otherwise indicated. All experiments were performed three times unless otherwise indicated. For all graphs, values plotted are the means with errors bars representing ± the s.d.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data to support the conclusions in this manuscript can be found in the figures. All source data for graphs are available in the online version of the paper. Any other data can be requested from the corresponding authors. All ATAC-seq and RNA-seq data can be accessed from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE104473.
Extended Data
Extended Data Fig. 1 |. Analysis of tissue sources of regeneration in distraction.
a, Experimental timeline of distraction model. b, Illustration of the tissue response (at POD15) in the mouse model of mandibular distraction (left), fracture (middle) and acute lengthening (right). c, Putative cellular sources of bone regeneration in mandibular distraction, including periosteum, endosteum and circulating progenitors. d, Experimental scheme for detecting circulating progenitor cells in mandibular distraction. GFP mice are surgically fused to their wild-type (WT) littermates through parabiosis. Peripheral blood chimaerism is confirmed via flow cytometry. After GFP-positive cells are confirmed in the WT through FACS, mandibular distraction is performed on the WT parabiont. FSC-A, forward scatter area; FSC-H, forward scatter height. e, Upon detection of 1/1 blood chimaerism, WT parabionts undergo mandibular distraction according to the timeline outlined. f, Quantification of the haematopoietic fraction of GFP-positive cells obtained from distraction calluses (n = 4 biological replicates per time point). g, Representation of the Rainbow reporter construct at the R26 locus and the colours produced by random recombination. h, One-year tracing of mandibles under normal homeostasis (uninjured), with confocal micrographs of whole-mount periosteum one year after recombination. Clones are shown with coloured dotted outlines. The white dotted line demarcates skeletal muscle (upper left quadrant) from the periosteum. The view is a buccal-to-lingual view of the posterior periosteum overlying the body of the mandible. n = 50 clones, with 16–151 cells per clone. i, Confocal micrograph of the transverse mandible section from a Rainbow mouse at POD15 after targeted labelling of the periosteum for subsequent mandibular distraction (n = 8). Coloured outlines indicate single clones; the white dotted outline indicates mandibular bone (mb) at the distraction site. n refers to the number of animals in each independent experiment.
Extended Data Fig. 2 |. Skeletal lineage tracing demonstrates labelling of bone regenerate in mandibular distraction.
a, Experimental scheme for using tamoxifen (TMX; administered at 9 weeks of age, before surgery at 10 weeks) to induce recombination in Sox9creERT2;R26mT/mG mice to carry out skeletal lineage tracing, with isolation of mandibles at POD29 (n = 6). b, Confocal micrograph (lingual mandible) of the distraction callus in Sox9creERT2;R26mT/mG mice after TMX induction. Filters are shown in the following order from left to right: mT (mTomato, background), mG (mGFP, Sox9 lineage), mTmG (merged) and merged (mTmG with DAPI) (n = 6). c, As for b, but for the buccal mandible. d, Experimental scheme for tamoxifen induction of Sox9creERT2;R26mT/mG mice during the early phase of distraction (POD5–10) for lineage tracing of Sox9+ cells during distraction. Mandibles were isolated at POD29 for confocal microscopy (n = 4). i.p., intraperitoneal. e, Confocal micrographs showing the contribution of the skeletal lineage (mGFP, Sox9 lineage) to new bone formed in mandibular distraction. f, Low-magnification images of the Sox9creERT2;R26Rainbow mandible during distraction at POD8, including bright-field, eGFP, mCerulean and mCherry filters under a dissection microscope. The induced Rainbow-coloured clones (white arrowheads) are anterior and posterior to the site of distraction (dotted white box) between the pin holes (dotted white circles) (n = 8). g, As for f, but for POD12. h, As for Fig. 1l, but for POD12 (n = 5). i, Schematic showing the vantage point for clonal analysis of the skeletal-lineage (Sox9creERT2;R26Rainbow) contribution to regeneration during distraction, through the use of whole-mount specimens from mandibular callus microdissection. DO, distraction osteogenesis. j–l, Quantification of clones (whole-mount imaging) that are positive for each fluorophore per high-pass filter (h.p.f.; ×40 magnification) two weeks after tamoxifen induction (one injection per day, five days total) plus distraction until POD12 using ActincreERT2;R26Rainbow mice (j, k) and Sox9creERT2;R26Rainbow mice (l). Means ± s.d. are shown Scale bars, 200 μm (b, c, e), 1 mm (f). n refers to the number of animals in each independent experiment.
Extended Data Fig. 3 |. Dynamics of SSCs and progenitor cells in parabiosis and distraction.
a, The skeletal stem cell lineage, showing the immunological phenotype of each cell. CP, chondroprogenitor; OP, osteoprogenitor. b, FACS isolation of single SSCs for evaluation of serial colony-forming potential in vitro. Secondary (left) and tertiary (right) colonies are shown. c, Experimental scheme for detecting circulating cells in mandibular distraction at POD10. d, FACS gating of Thy1−6C3− cells (light brown box) for detection of circulating SSCs (green box in e) and progenitor cells (green box in f) in mandibular distraction. e, FACS gating shows an absence of circulating (GFP+) SSCs in mandibular distraction (n = 8; mean = 2.02 ± 0.78). f, As for e, but for BCSPs (n = 8; mean = 1.83 ± 0.91). g–i, FACS isolation from uninjured (g; 38.0% αVβ3-positive events within the gate) versus fracture (h; 15.0%) and distraction (i; 58.8%) conditions reveal expansion of the SSC hierarchy (left column) in response to distraction by POD10. Representative of three independent experiments. n refers to the number of animals in each independent experiment.
Extended Data Fig. 4 |. Changes in gene regulation in response to distraction.
a, Schematic showing FACS isolation of stem and progenitor cells from uninjured, fractured and distracted mandibles for ATAC-seq (assay for accessibility of chromatin, which is shown with purple spheres and yellow lines, to depict nucleosomes) and RNA-seq (shown with a polyA strand). Samples were collected in duplicate for ATAC-seq at POD10 and in duplicate for RNA-seq at PODs 5, 10 and 15. b, PCA showing PC1 and PC2 for all ATAC-seq data (n = 164,266 peaks). UI, uninjured. c, Venn diagram showing the number of differential peaks between the fracture and diffraction conditions in both SSCs and BCSPs. Overlap between these (shared peaks; twofold change with P < 0.05 in both SSCs and BCSPs) is shown in the centre. d, Cluster dendrogram for ATAC-seq, showing clustering of samples based on all ATAC-seq peaks from SSCs, BCSPs and OPs. e, Example loci (Mmp15 and Runx2, with their locations on chromosomes 8 and 17, respectively, shown at the top), revealing accessibility that is distraction-specific (Mmp15) and fracture-specific (Runx2), respectively. The height of the genome browser tracks shows the number of reads normalized by read depth and overall peak enrichment in the library. f, PCA showing PC1 and PC2 for all RNA-seq data at PODs 5,10 and 15 (n = 17,491 genes). g, Cluster dendrogram for RNA-seq, showing clustering of samples based on all genes from SSCs, BCSPs and OPs on PODs 5, 10 and 15. h, Volcano plot showing differential gene expression between d-SSCs at PODs 10 and 15 and f-SSCs at PODs 10 and 15. Red dots represent genes that are significantly differentially expressed with an adjusted P-value cut-off of 0.05 (DESeq2). Differentially expressed genes that are upregulated in f-SSCs (Fx) are shown in blue (fold change greater than 1.5; Padj less than 0.05), and differentially expressed genes that are upregulated in d-SSCs (DO) are shown in red. i, Significantly enriched GO terms for genes upregulated in d-SSCs at PODs 10 and 15, from GREAT version 3.0.0, with P values (one-sided binomial) corrected using the Benjamini–Hochberg correction. j, Significantly enriched GO terms for genes upregulated in f-SSCs at PODs 10 and 15 from GREAT, with P values (one-sided binomial) corrected using the Benjamini–Hochberg correction. k, As for h, but for BCSPs. l, As for i, but for BCSPs. m, As for j, but for BCSPs. n, As for h, but for OPs. o, As for i, but for OPs. p, As for j, but for OPs.
Extended Data Fig. 5 |. Expression of core signalling genes during distraction.
a, Heat map showing the expression of Wnt, Notch, bone morphogenetic protein (BMP) and FGF family members, as well as integrins, matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs), in SSCs. Values from RNA-seq in transcripts per million (t.p.m.) are z-scored for each gene to reflect differences across conditions and time. Only genes that are expressed (t.p.m. > 3) in at least one sample are shown. b, As for a, but for BCSPs. c, As for a, but for OPs. d, Heat map showing expression (in t.p.m.) of Runx-family transcription factors in POD5 SSCs, and in POD10 and POD15 f-SSCs and d-SSCs. e, As for d, but for Dlx-family transcription factors. f, Heat map showing accessibility from ATAC-seq in SSCs and BCSPs at Runx2 chromatin immunoprecipitation (ChIP)-seq peaks from MC3T3 preosteoblasts;12,673 binding sites from ChIP-seq are shown41. Plotted are the row z-scored normalized counts from SSC and BCSP ATAC-seq data. g, As for f, but for Dlx5 ChIP-seq data38. ChIP-seq binding sites (24,365) from MC3T3 preosteoblasts are shown.
Extended Data Fig. 6 |. Expansion of SSCs and progenitor cells depends on FAK.
a, Timeline for FAK inhibition and conditional genetic knockout during distraction, with respective control conditions, for μCT analysis (Fig. 3a–d) at mid-consolidation (POD29). b, Pentachrome staining of transverse section from mandibles treated with DMSO during distraction osteogenesis and collected at POD15. The white dotted line indicates cortical bone at the osteotomy site; the red dotted line indicates bone that newly formed in response to distraction. Representative of three independent replicates. c, As for b, but for FAKi treatment. The green dotted line indicates cartilage that newly formed in response to FAKi during distraction osteogenesis. d, As for b, but for POD29. e, As for c, but for POD29. f, Schematic showing FACS isolation of SSCs and BCSPs from mandible calluses for subsequent in vitro analyses. g, h, FACS isolation of SSCs and BCSPs from distraction control mandibles receiving DMSO injections (g; 55.5%) versus FAK-inhibitor injections (FAKi) (h; 41.7%), revealing diminished expansion of the SCC hierarchy (first column, black gate) in response to FAK inhibition by POD10. The ratio of SSCs (red gate) to their committed bone progenitors (BCSPs, green gate) during distraction was substantially disrupted by FAK inhibition, such that the proportion of SSCs was higher. Representative of three independent replicates. i, Quantification of the frequency of d-SSCs, d-BCSPs and d-OPs within mandibular calluses collected at POD10 after FAKi or control injections (n = 6; error bars indicate s.d. from the mean). j, Representative fluorescence micrographs showing phosphorylated-FAK activity (left column, green) in d-SSCs after treatment with DMSO (top) or with FAK inhibitor (bottom). Right column, phospho-FAK fluorescence merged with DAPI fluorescence. Representative of three independent replicates. k, Quantification of d-SSC colony formation in vitro in response to FAKi and in a FAK(cKO), compared with their respective control conditions (n = 15 per condition; ***P < 0.001, Tukey’s multiple comparisons; means ± s.d.). Direct comparison of colony formation in FAKi and FAK(cKO) conditions was not significantly different. n refers to the number of animals in each independent experiment.
Extended Data Fig. 7 |. Cellular dynamics during distraction osteogenesis in FAK(cKO) mice.
a, b, FACS isolation of cells from control mice (a, Sox9creERT2;Ptk2fl/fl, no TMX) and FAK(cKO) mice (b, Sox9creERT2;Ptk2fl/fl, TMX treatment) shows that the SSC hierarchy (first column, black gate) is disrupted similarly in FAK(cKO) mice and in FAKi (Extended Data Fig. 6h). In the FAK(cKO) mice, the proportion of downstream multipotent BCSPs (green gate) compared with SSCs (orange and red gates) is lower than in controls. Representative of three independent replicates. c, Experimental strategy for clonal analysis of FAK(cKO) Rainbow contribution to regeneration in response to distraction, using whole-mount specimens from callus microdissection of mandibles in Sox9creERT2;R26Rainbow mice. d, Vantage point for the acquisition of the confocal images of whole-mount specimens shown in Fig. 3g, h. e, Experimental strategy for clonal analysis in ActincreERT2;R26Rainbow mice. f, Quantification of average clone size at POD12 during distraction osteogenesis in ActincreERT2;R26Rainbow mice (n = 10; mean ± s.d.). g, Whole-mount imaging at the site of distraction at POD12 in ActincreERT2;R26Rainbow mice. The view is a lateral-to-medial view of callus overlying the distraction site (indicated by the white dotted outline overlying the distraction gap). The distraction gap contains large clones with a migratory spreading phenotype (n = 4 for each of three independent replicates). h, Quantification of clones (whole-mount imaging) that are positive for each fluorophore per h.p.f. (×40 magnification) two weeks after tamoxifen induction (one injection per day, five days total) plus distraction until POD12, using Sox9creERT2;R26Rainbow mice (left) and FAK(cKO)Rainbow mice (right) (n = 10; mean ± s.d.). n refers to the number of animals in each independent experiment.
Extended Data Fig. 8 |. Involvement of neural crest transcription factors in SSCs during distraction.
a, Motifs enriched in FAK-R and FAK-NR sites in SSCs and BCSPs. The size of each diamond represents the negative P value for enrichment (one-sided binomial, Benjamini–Hochberg correction); colours represent the percentage of sites in target/percentage of sites in background. b, Heat map showing expression of Sox-family transcription factors. Colours represent z-scored t.p.m. values from RNA-seq at PODs 5, 10 and 15 for d-SSCs and f-SSCs. Only factors with t.p.m. greater than or equal to 3 in at least one sample are shown. c, As for b, but for Ets-family transcription factors. d, The Sox10 locus (with its location on chromosome 15 shown at the top), showing accessibility at the promoter that is distraction-specific (red), congruent with the expression data in b and the RNA-seq track below. The signal for tracks is normalized by read depth and overall peak enrichment in the library. Tracks are representative of two biological replicates. e, Experimental scheme for tamoxifen induction of Sox10creERT2;R26mT/mG mice during the early and mid-distraction phase (POD5–10) to trace the Sox10+ lineage in a distraction-specific context. Mandibles were collected for confocal microscopy at POD29 (n = 4). f, Confocal micrograph of Sox10creERT2;R26mT/mG mandible at POD29, demonstrating the capability of Sox10+ cells to give rise to the distraction regenerate. Filters are, from left to right, mT (mTomato, background), mG (mGFP, Sox10+ lineage), mTmG (merged) and merged (mTmG with DAPI). g, Confocal micrograph of Sox10creERT2;R26mT/mG mandible at POD29, showing the presence of Sox10-lineage cells within surrounding callus and periosteum to give rise to the regenerate. Scale bars, 200 μm (b, c). n refers to the number of animals in each independent experiment.
Extended Data Fig. 9 |. NCC transcriptional networks in d-SSCs.
a, Diagram showing the developmental origin of NCC-derived SSCs. e, embryonic day of development. b, Clustering of SSC and NCC samples using all accessible sites in SSCs and NCCs merged. c, Heat map showing all accessible sites that are significantly differentially accessible between uninjured SSCs and NCC progenitors. Colour represents the log2-transformed normalized read counts within peak regions. d, GO terms enriched in sets of genes near peaks that are more accessible in NCC and d-SSCs than in uninjured SSCs. GO terms shown are those that are significantly enriched after false discovery rate correction in GREAT (two-sided binomial P value shown). e, Heat map showing all accessible sites that are differentially accessible between f-SSCs and d-SSCs. The sites that are FAK-R or FAK-NR are highlighted along the right. Colour represents the log2-transformed normalized read counts within peak regions. f, Quantification of immunofluorescence staining for each of the NCC markers observed on RNA-seq analysis and evaluated in this figure, including nuclear Zeb2 (n = 15, ***P < 0.001), nuclear Twist2 (n = 15, *P < 0.05), Cdc42 (n = 6, P < 0.05), S100a (n = 6, ***P < 0.001) and Plp1 (n = 6, ***P < 0.001) (Student’s t-test; shown are means ± s.d.). n refers to the number of animals in each independent experiment.
Extended Data Fig. 10 |. Controlled mechanical advancement of the lower jaw unlocks neural crest potential for regeneration of the mandible.
a, At the tissue level, clonality within the mandible during distraction (middle) is observed to occur in a highly linear and directional manner in parallel to the vector of distraction. By contrast, clonality observed in fracture (top) and distraction plus FAKi (bottom) was highly mesenchymal with less apparent organization in its morphology, indicating a nondirectional clonal proliferation. b, The cellular level: the developmental origin of the NCC-derived SSCs of the mandible (top), and the postnatal SSCs of the mandible (bottom) that are present in our experiments. During distraction the d-SSC (shown in pink) demonstrates plasticity and takes on an NCC- derived signature, whereas the f-SSC (purple) retains its postnatal SSC characteristics with no NCC signature. In the absence of FAK signalling, the d-SSC FAKi (green) reverts functionally and epigenomically to the fracture state without emergence of the NCC signature. ‘NCC Prog’ indicates the premigratory (e8.5) NCC progenitor population; ‘NCC Md’ indicates the postmigratory (e10.5) NCC population arriving within the mandible. c, At the chromatin level, distraction induces a gain in accessibility at promoters (P) of FAK-responsive NCC craniofacial genes through the activation of their enhancers (E), with a parallel gain in accessibility of retroviral elements near NCC-specific craniofacial enhancers. Thus mechanotransduction during mandible distraction unlocks FAK-responsive craniofacial enhancers, potentially through retrotransposons, enacting a developmental NCC program in d-SSCs similar to that of the e10.5 NCC Md population (b). This does not occur under fracture conditions (top) or during distraction plus FAKi (bottom). These differential epigenomic responses correlate with the degree of clonality and patterning seen in Rainbow mice (a) that occurs in response to distraction. Circled P represents phosphorylation.
Supplementary Material
Acknowledgements
We thank J. Wysocka for her review of the manuscript and helpful suggestions. We thank the Stanford Functional Genomics Facility, Stanford Cell Sciences Imaging Facility, Lorry Lokey Imaging Facility, and Stanford Shared FACS Facility Cores. We thank D. J. Hunter and D. Atashroo for their respective contributions to the design of the distraction device. This work was supported by the National Institutes of Health (NIH) grants R01DE026730 (to M.T.L. and R.C.R.), U24DE026914 (to M.T.L.) and K08DE024269 (to D.C.W.); the Child Health Research Institute (CHRI) at Stanford University (D.C.W.); The Hagey Laboratory for Pediatric Regenerative Medicine (M.T.L.); the Steinhart/Reed Award (M.T.L); the Gunn–Oliver Fund (M.T.L.); and NIH grant P50-HG007735 and the Scleroderma Research Foundation (H.Y.C.). H.Y.C. is an Investigator of the Howard Hughes Medical Institute.
Reviewer information Nature thanks C. Tabin, L. Gerstenfeld and P. Scacheri for their contribution to the peer review of this work.
Footnotes
Competing interests The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41586-018-0650-9.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-018-0650-9.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41586-018-0650-9.
References
- 1.Ilizarov GA The tension-stress effect on the genesis and growth of tissues: part II. The influence of the rate and frequency of distraction. Clin. Orthop. Relat. Res 239, 263–285 (1989). [PubMed] [Google Scholar]
- 2.Ilizarov GA The tension-stress effect on the genesis and growth of tissues. Part I. The influence of stability of fixation and soft-tissue preservation. Clin. Orthop. Relat. Res 238, 249–281 (1989). [PubMed] [Google Scholar]
- 3.Tahiri Y, Viezel-Mathieu A, Aldekhayel S, Lee J & Gilardino M The effectiveness of mandibular distraction in improving airway obstruction in the pediatric population. Plast. Reconstr. Surg 133, 352e–359e (2014). [DOI] [PubMed] [Google Scholar]
- 4.McCarthy JG, Schreiber J, Karp N, Thorne CH & Grayson BH Lengthening the human mandible by gradual distraction. Plast. Reconstr. Surg 89, 1–8 (1992). [PubMed] [Google Scholar]
- 5.Khansa I et al. Airway and feeding outcomes of mandibular distraction, tongue-lip adhesion, and conservative management in Pierre Robin sequence: a prospective study. Plast. Reconstr. Surg 139, 975e–983e (2017). [DOI] [PubMed] [Google Scholar]
- 6.Chan CK et al. Identification and specification of the mouse skeletal stem cell. Cell 160, 285–298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adam RC et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge Y et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frechin M et al. Cell-intrinsic adaptation of lipid composition to local crowding drives social behaviour. Nature 523, 88–91 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Bell S & Terentjev EM Focal adhesion kinase: the reversible molecular mechanosensor. Biophys. J 112, 2439–2450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinkevich Y, Lindau P, Ueno H, Longaker MT & Weissman IL Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476, 409–413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehoczky JA, Robert B & Tabin CJ Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl Acad. Sci. USA 108, 20609–20614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hojo H, Ohba S, He X, Lai LP & McMahon AP Sp7/Osterix is restricted to bone-forming vertebrates where it acts as a Dlx co-factor in osteoblast specification. Dev. Cell 37, 238–253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer MB, Benkusky NA & Pike JW The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J. Biol. Chem 289, 16016–16031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prescott SL et al. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163, 68–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tominaga H et al. CCAAT/enhancer-binding protein β promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol. Biol. Cell 19, 5373–5386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers CD, Phillips JL & Bronner ME Elk3 is essential for the progression from progenitor to definitive neural crest cell. Dev. Biol 374, 255–263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santagati F & Rijli FM Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci 4, 806–818 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Kaukua N et al. Glial origin of mesenchymal stem cells in a tooth model system. Nature 513, 551–554 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Kaucka M et al. Analysis of neural crest-derived clones reveals novel aspects of facial development. Sci. Adv 2, e1600060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaucka M et al. Oriented clonal cell dynamics enables accurate growth and shaping of vertebrate cartilage. eLife 6, e25902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minoux M et al. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science 355, eaal2913 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Brinkley JF et al. The FaceBase Consortium: a comprehensive resource for craniofacial researchers. Development 143, 2677–2688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attanasio C et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342, 1241006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka HV et al. A developmentally regulated switch from stem cells to dedifferentiation for limb muscle regeneration in newts. Nat. Commun 7, 11069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez Alvarado A Developmental biology: a cellular view of regeneration. Nature 460, 39–40 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Kragl M et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Purnell BA Regrow like an axolotl. Science 355, 592 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Nacu E, Gromberg E, Oliveira CR, Drechsel D & Tanaka EM FGF8 and SHH substitute for anterior–posterior tissue interactions to induce limb regeneration. Nature 533, 407–410 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Roensch K, Tazaki A, Chara O & Tanaka EM Progressive specification rather than intercalation of segments during limb regeneration. Science 342, 1375–1379 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Kaufman CK et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coufal NG et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivancevic AM, Kortschak RD, Bertozzi T & Adelson DL LINEs between species: evolutionary dynamics of LINE-1 retrotransposons across the eukaryotic tree of life. Genome Biol. Evol 8, 3301–3322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bridier-Nahmias A et al. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348, 585–588 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Mashanov VS, Zueva OR & García-Arrarás JE Retrotransposons in animal regeneration: overlooked components of the regenerative machinery? Mob. Genet. Elements 2, 244–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baillie JK et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueno H & Weissman IL Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev. Cell 11, 519–533 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Wright DE, Wagers AJ, Gulati AP, Johnson FL & Weissman IL Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933–1936 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to support the conclusions in this manuscript can be found in the figures. All source data for graphs are available in the online version of the paper. Any other data can be requested from the corresponding authors. All ATAC-seq and RNA-seq data can be accessed from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE104473.