Summary
Opioid receptors (ORs) precisely modulate behavior when activated by native peptide ligands but distort behaviors to produce pathology when activated by non-peptide drugs. A fundamental question is how drugs differ from peptides in their actions on target neurons. Here we show that drugs differ in the subcellular location at which they activate ORs. We develop a genetically encoded biosensor that directly detects ligand-induced activation of ORs and uncover a real-time map of the spatiotemporal organization of OR activation in living neurons. Peptide agonists produce a characteristic activation pattern initiated in the plasma membrane and propagating to endosomes after receptor internalization. Drugs produce a different activation pattern by additionally driving OR activation in the somatic Golgi apparatus and Golgi elements extending throughout the dendritic arbor. These results establish an approach to probe the cellular basis of neuromodulation and reveal that drugs distort the spatiotemporal landscape of neuronal OR activation.
Introduction
Opiate alkaloid drugs such as morphine are among the most effective agents known for alleviating pain. However, such drugs produce significant toxicity and have high abuse potential. These factors have contributed to opioid addiction becoming a large and growing public health problem globally, which has now reached epidemic proportions in the US (Volkow and McLellan, 2016). Opiate drugs produce their biological effects by binding to the same subfamily of G proteincoupled receptors (GPCRs), the opioid receptors (ORs), as endogenously produced opioid peptide ligands (Kieffer and Evans, 2009). This supports a long-held view (Bradbury et al., 1976) that opiate drugs mimic the actions of peptide ligands at the level of individual receptor-expressing target neurons. Despite an urgent need to develop improved analgesic therapies, and compounded by the fact that opiate drugs remain mainstays in the pharmacological management of severe pain, it remains unclear to what degree opiate drugs differ in their cellular effects (Thompson et al., 2015). It also remains unknown if opiate drugs, beyond producing cellular effects that are similar to peptide ligands, have the potential to produce discrete or additional receptor-based effect(s) that opioid peptides cannot.
Drugs are well known to differ substantially from peptides in physicochemical properties that affect membrane permeability and drug bioavailability within tissues (Weber et al., 1993). However, such differences have been thought to have little impact at the cellular level because present models assume that receptor activation is restricted to the plasma membrane (PM) (von Zastrow and Williams, 2012). It has become clear recently that a number of other GPCRs are subject to ligand-dependent activation at internal membrane locations as well as the PM (Calebiro et al., 2010; Irannejad et al., 2015; Jong et al., 2017). With this in mind, and in light of the exquisitely high degree of membrane compartmentation that is characteristic of neurons, we wondered if ORs have the ability to undergo ligand-dependent activation at more than one membrane location in neurons. If so, might chemically distinct ligands differ in the subcellular location(s) at which they produce OR activation?
Resolving the precise subcellular location(s) at which wild type ORs are activated by ligands was previously impossible due to a lack of suitable experimental tools. Recent advances in GPCR structural biology have made feasible the development of single-domain antibody (or nanobody) probes that bind to receptors specifically based on a defined activation-associated conformational change (Manglik et al., 2017). Such reagents can be adapted to produce genetically encoded probes, or conformational biosensors, to detect receptor activation in living cells and to resolve ligand-induced activation of catecholamine receptors both in the PM and at internal membrane sites (Irannejad et al., 2017; 2013). No such biosensor has yet been described to specifically detect ligand-dependent activation of ORs and, to our knowledge, no previous study has investigated the ability of conformational biosensor technology to detect activation of any receptor in neurons.
Here we describe a biosensor derived from a conformation-specific nanobody that is capable of detecting ligand-induced activation of mu- and delta-ORs (MORs and DORs) in living neurons. We demonstrate that this conformational biosensor provides precise spatial and temporal resolution of OR activation and deactivation in situ with minimal perturbation of function. Using this tool, we show that neuronal OR activation is not restricted to the cell surface as previously assumed. Instead, activation by peptide ligands begins in the PM and propagates to endosomes after ligand-induced OR internalization. We then demonstrate that both antagonist and agonist drugs distort this pattern. In particular, we show that non-peptide drugs, including the prototypic opiate alkaloid morphine, drive a discrete wave of internal OR activation in the somatic Golgi apparatus and Golgi outposts distributed throughout the dendritic arbor. We provide functional evidence supporting the hypothesis that internal OR activation contributes to the cellular signaling response. These results reveal a characteristic pattern of subcellular OR activation generated by peptides and its profound distortion by drugs. We propose a discrete principle of biased drug action that is imposed at the level of individual target neurons and manifest through the spatiotemporal landscape of receptor activation.
Results
A conformational biosensor for direct and specific detection of OR activation
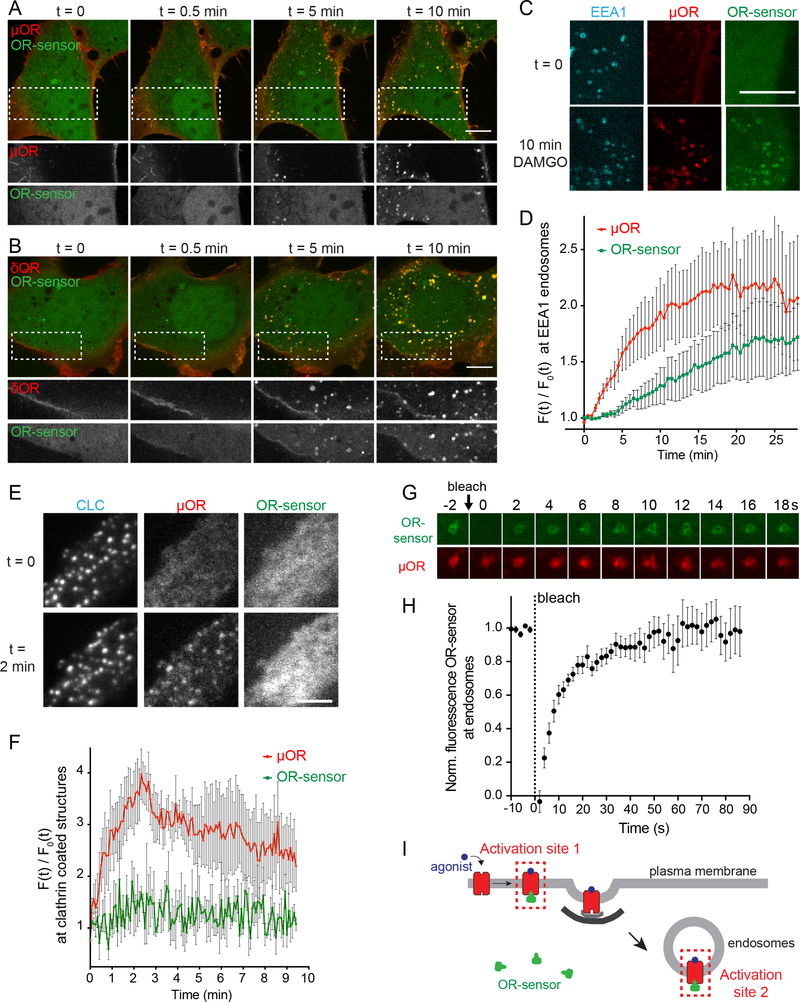
To develop an OR activation biosensor we began with clones selected from a camelid-derived antibody fragment (nanobody) library that bind in vitro to MOR specifically in an active (agonistbound) relative to inactive (antagonist-bound) conformation (Figure 1A) (Huang et al., 2015; Manglik et al., 2012). We selected four nanobody clones in which key residues that engage MOR in the active structure are conserved (Figure S1A), prepared N- and C-terminal fluorescent protein fusions from each nanobody clone, and tested them for cytoplasmic expression. The ability of nanobody fusion proteins to act as sensors of active-conformation ORs was first examined by co-expression with MOR in human embryonal kidney (HEK293) cells and imaging of cells using total internal reflection fluorescence microscopy (TIR-FM). TIR-FM allowed precise examination of nanobody recruitment to the PM because it selectively detects fluorophores near (within ~100 nm from) the cell surface (Figure 1B) (Mattheyses et al., 2010). Several constructs exhibited ligand-dependent recruitment to the PM and N-terminally tagged Nb33 (named OR-sensor) was selected for further study (Figure S1B). In cells not exposed to opioid ligands, little fluorescence of ORsensor was observed in the TIR-FM illumination volume, consistent with localization in the cytoplasm. Activation of co-expressed MOR with a peptide agonist (DAMGO) triggered a pronounced increase of OR-sensor fluorescence in the TIR-FM field (Figures 1C and 1D, Movie S1). PM recruitment of OR-sensor had a rapid onset (t½ <20 s) after DAMGO application and was rapidly reversed (t½ ~20 s) after agonist washout or addition of the competitive antagonist naloxone (Figure 1E).
Figure 1: A conformational biosensor for direct detection of OR activation.
(A) Crystal structures of inactive, β-FNA-bound (PDB: 4DKL) and active, BU27-bound (PDB: 5C1M) MOR (red). The active conformation is stabilized by a nanobody (Nb39, green), named OR-sensor. Ligands are in blue. Arrow: outward movement of TM6 upon activation. See also Figure S1. (B) Schematic of OR-sensor and MOR localization in cells and expected OR-sensor re-localization upon agonist addition. Total internal reflection fluorescence microscopy (TIR-FM) light beam is indicated. PM = plasma membrane. (C) TIR-FM images of a time series of a HEK293 cell, expressing EGFP-OR-sensor (and MOR, not shown). Media exchange to DAMGO (1 μM, agonist) or naloxone (10 μM, antagonist) by perfusion. Scale bar: 10 μm. See also Movie S1. (D) EGFP-OR-sensor intensity during TIR-FM time-lapse series with media exchange to DAMGO and naloxone. 2 s between frames. F0 = average fluorescence intensity before agonist. See also Figure S2. (E & F) Recruitment and detachment kinetics of OR-sensor to MOR or DOR, measured by EGFP-OR-sensor intensity increase using TIR-FM. 2 s between frames. Normalization of EGFP-intensity values (range [0–1]). Data fit with one-phase exponential association or decay formula. (E) HEK293 cells expressing MOR and OR-sensor. Left: DAMGO (1 μM) was applied by media perfusion starting at t = 0. (n=9, average +/− sem). Right: media exchange from DAMGO (1 μM) to naloxone (10 μM) or no agonist (washout) (naloxone: n=4, washout n=8, average +/− sem). (F) HEK293 cells expressing DOR and OR-sensor. Left: DADLE (1 μM) was applied by media perfusion starting at t = 0 (n=5, average +/− sem). Right: Media exchange from DADLE (1 μM) to naloxone (10 μM) (n=5, average +/− sem). (G) EGFP-OR-sensor intensity measured by TIR-FM in a HEK293 cell expressing OR-sensor and DOR (not shown). Repeated media exchange to 1 μM DADLE (1 min), 10 μM naloxone (2 min), and wash out (2 min). (H) Maximal cAMP response measured by pGloSensor cAMP sensor in HEK293 cells expressing DOR and EGFP-control or EGFP-OR-sensor. Cells were stimulated with Forskolin (FSK, 2 μM) and different concentrations of DAMGO. Data normalized to 100% FSK control. (n=3, average +/− SD, n.s. = no significant difference, unpaired t test). (I) EGFP-OR-sensor intensity during TIR-FM time-lapse series of a HEK293 cell, co-expressing EGFP-OR-sensor and MOR, adding increasing concentrations of DAMGO. 5 s between frames. (J) Concentration-dependent recruitment of ORsensor to MOR measured by TIR-FM (n=5, average +/− sem). Normalization of EGFP-intensity values (range [0–1]). Regression curves with Hill slope of 1. EC50: DAMGO: 57 nM, morphine: 20 nM, etorphine: 0.7 nM. (K) Western Blot analysis of EGFP-OR-sensor biotinylation by engineered ascorbate peroxidase (APEX)-fused to MOR captured by streptavidin agarose resin, showing untreated cells or cells treated with DAMGO, naloxone, or DAMGO followed by naloxone.
DOR is closely homologous in structure to MOR, and all residues contacting OR-sensor in the activated conformation of MOR are conserved in DOR (Figure S1C). Verifying that OR-sensor also detects ligand-activated DOR, the peptide agonist DADLE triggered rapid OR-sensor recruitment to the PM that was rapidly reversible by antagonist (Figure 1F). Verifying receptordependence of this response, neither DAMGO nor DADLE produced a recruitment signal in cells not expressing ORs (Figure S2). Verifying receptor-specificity, expression of the Gi-coupled M2 muscarinic receptor at similar levels as OR failed to produce detectable recruitment of OR-sensor following application of the muscarinic agonist carbachol (Figure S2).
A potential caveat of conformational biosensors is that, because the core nanobody structure binds directly to the active receptor conformation, they could ‘force’ receptor activation in a ligand-independent manner. This is unlikely because OR-sensor recruitment to the PM was not observed in the absence of agonist, and because recruitment of OR-sensor was rapidly reversible upon agonist removal. Emphasizing reversibility of OR-sensor recruitment, alternating agonist and antagonist application to the same cell produced repeated rounds of OR-sensor recruitment and dissociation from the PM (Figure 1G). Another potential caveat is that OR-sensor could block receptor function. This was also not the case because OR-sensor did not produce any detectable effect on the ability of receptors to mediate ligand-dependent inhibition of adenylyl cyclase, an assay requiring receptor coupling to G protein (Figure 1H). OR-sensor recruitment to the PM was dependent on ligand concentration and allowed us to detect differences in agonist potency and efficacy that parallel known pharmacology (Figure 1I and 1J). APEX proximity labeling provided independent confirmation of reversible recruitment of OR-sensor to activated receptors (Figure 1K) (Lobingier et al., 2017). Together, these results indicate that OR-sensor provides direct and specific detection of OR activation and deactivation in living cells with minimal perturbation of receptor function.
Peptide agonists drive sequential OR activation waves in the PM and endosomes
Peptide-induced activation of both MOR and DOR promotes rapid, clathrin-dependent endocytosis of receptors. Peptide binding to GPCRs can be reduced by acidification and proteolysis (Grady et al., 1995; Gupta et al., 2014), and current models of OR regulation involving endocytosis assume that OR activation is restricted to the PM (von Zastrow and Williams, 2012). The development of OR-sensor provided an opportunity to directly test this assumption. We labeled FLAG-tagged MOR or DOR in the cell surface with a fluorescent monoclonal antibody and used confocal microscopy to simultaneously image receptor and OR-sensor in living cells. MOR remained in the PM in the absence of agonist and accumulated in endosomes within several minutes after application of DAMGO (Figure 2A, Movie S2). OR-sensor was transiently recruited to the PM, consistent with the TIR-FM observations, and then localized to OR-containing endosomes (Figure 2A). Similar results were observed in experiments using DOR rather than MOR (Figure 2B, Movie S3). The kinetics of receptor and OR-sensor recruitment to endosomes were estimated by three-color confocal imaging, using Early Endosome Antigen 1 (EEA1) as a marker of the early endosome population (Figure 2C). We measured the average intensity of MOR and OR-sensor fluorescence within a mask defined by EEA1 as a function of time after peptide agonist addition. Accumulation of MOR in endosomes began ~1 min after DAMGO application and reached a plateau at ~20 min, consistent with previously published rates of MOR endocytosis and approach to steady state through continuous endocytosis and recycling that occurs in the presence of peptide agonist (Henry et al., 2012). Ligand-induced OR-sensor recruitment to endosomes lagged receptor accumulation in this compartment and also reached a plateau within ~20 min (Figure 2D). These results indicate that active-conformation ORs are not restricted to the PM but are also present in endosomes.
Figure 2: An endosome-localized wave of OR activation.
(A & B) Confocal images of time series of HEK293 cells, expressing EGFP-OR-sensor and FLAGMOR (A) or FLAG-DOR (B). Receptors were surface-labeled with anti-FLAG M1-AF555. 10 μM DAMGO (A) or DADLE (B) was added at t=0. Boxed areas are displayed separately for both fluorophores below respective images. Scale bar: 10 μm. See also Movies S2 and S3. (C) Maximum Z-projection of 12 confocal slices of a HEK293 cell, expressing EGFP-OR-sensor, FLAG-MOR (surface-labeled with M1-AF647, red), and an early endosomal marker (DsRed-EEA1-FYVE, blue) before (t=0) and after (t=15 min) adding DAMGO (10 μM). Images are representative examples used for quantification shown in (D). Scale bar 10 μm. (D) Quantification and kinetics of MOR and OR-sensor recruitment to EEA1 endosomes. F0 = average fluorescence signal in EEA1 mask before agonist addition. Movie length 25 min, 30 s between Z-stacks (n=5, average +/− sem). (E) TIR-FM images of a time series of a HEK293 cell, co-expressing clathrinlight-chain (CLC)-DsRed, FLAG-MOR (surface-labeled with M1-AF647) and EGFP-OR-sensor. DAMGO (10 μM) was added at t=0. Scale bar 5 μm. See also Figure S3. (F) Quantification of MOR and OR-sensor intensity in clathrin-coated pits (CCPs) vs. PM over time. CLC-positive spots were used as quantification mask. F0 = average fluorescence signal in CCPs before agonist addition. Movie length 10 min, 5 s between frames (n=3, average +/− sem). (G) Fluorescencerecovery after photobleaching (FRAP) series displaying intensity of EGFP-OR-sensor (photobleached) and MOR (surface-labeled with M1-AF647) at a bleached endosome, 2 s intervals. (H) FRAP of EGFP-OR-sensor at MOR-loaded endosomes in HEK293 cells, stimulated for 15 min with DAMGO (10 μM). Normalized EGFP-OR-sensor florescence in bleached area over time. 2 s interval between frames during acquisition (n=7, average +/− sem) (I) Scheme depicting the two distinct sites of OR-sensor recruitment to peptide ligand activated receptors: activation site 1: plasma membrane, activation site 2: endosomes.
A potential caveat to this interpretation is that OR-sensor could accumulate in endosomes as an artifact of a persistent receptor-biosensor complex that forms at the PM and remains associated during endocytosis. Two lines of evidence argue against this. First, TIR-FM time-lapse analysis showed that ORs robustly cluster in clathrin-coated pits (CCPs) without concomitant recruitment of OR-sensor to the endocytic site (Figures 2E and 2F), and CCPs pinch off from the PM containing ORs but not associated with OR-sensor (Figure S3). Second, when we applied fluorescence recovery after photobleaching (FRAP) analysis to examine exchange kinetics of ORsensor at individual OR-containing endosomes, the OR-sensor signal fully recovered with a halftime of ~15 s (Figures 2G and 2H), much faster than the kinetics of receptor trafficking (Figure 2D). This rules out the possibility that OR-sensor localization to endosomes occurs as an artifact of stable receptor-biosensor complex formation. To the contrary, OR-sensor exchanges remarkably rapidly and therefore provides a continuous readout of the receptor’s activation state. Together, these results reveal spatiotemporal organization of cellular OR activation induced by peptide agonists, with sequential activation occurring in the PM and then in endosomes following the process of ligand-induced internalization of ORs (Figure 2I).
Endosome OR activation is ligand-dependent and sustained
We next tested if OR activation in endosomes is indeed dependent on the presence of agonist ligand. If endosomal activation requires agonist binding to receptors, as it does at the PM, we expected competitive antagonist ligands to reverse endosomal OR-sensor recruitment depending on the ability of the antagonist to access internalized receptors (Figure 3A). Activation of MOR in endosomes remained ligand-dependent because naloxone, a membrane-permeant alkaloid antagonist, fully reversed OR-sensor localization to endosomes. Reversal occurred within 30 s, a time period shorter than required for MOR recycling, as verified by internalized receptors still being present in endosomes after OR-sensor dissociation (Figure 3B). In contrast, a membrane-impermeant peptide antagonist (CTOP) failed to cause such rapid reversal of OR-sensor recruitment to endosomes. Similarly, agonist removal by washout also did not cause rapid reversal of the endosomal OR-sensor signal (Figure 3C), even though the PM-localized ORsensor was rapidly reversed under the same conditions (Figure 1E). These results indicate that activation of MORs in endosomes is ligand-dependent but differs from PM activation in that it persists after agonist removal from the extracellular media.
Figure 3: Endosomal OR activation is ligand-dependent and sustained.
(A) Probing OR activation in endosomes using OR-sensor. (B) Confocal images of a HEK293 cell expressing MOR (surface-labeled with M1-AF555) and EGFP-OR-sensor. Top: 15 min after adding DAMGO (1 μM). Bottom: 30 s after adding naloxone (10 μM). Scale bar: 10 μm. (C) Quantification of EGFP-OR-sensor intensity at MOR-containing endosomes (treated for 15 min with 1 μM DAMGO) after adding 10 μM of non-peptide (naloxone) or peptide (CTOP) antagonist or upon agonist washout. EEA1-endosomes were used as quantification mask and intensity normalized to cytoplasmic OR-sensor signal at each time point. Naloxone: 2 s between frames, n=6, CTOP: 30 s between frames, n=4, washout: 30 s between frames, n=3, average +/− sem. (D) Quantification of EGFP-OR-sensor intensity at DOR-containing endosomes (15 min 1 μM DADLE) after adding 10 μM of non-peptide (naltrindole) or peptide (TIPPpsi) antagonist or upon agonist washout. DOR-positive endosomes were used as quantification mask and intensity normalized to cytoplasmic OR-sensor signal at each time point. Naltrindole: 2 s between frames, n=5, TIPPpsi: 30 s between frames n=5, washout: 30 s between frames, n=5, average +/− sem. (E) Experimental setup to test endosomal OR signaling. (F) cAMP response in living HEK293 cells expressing DOR and the luciferase cAMP reporter GloSensor-20F. Forskolin (FSK, 2 μM)-induced cAMP response (black curve) and effect of persistent presence of DOR agonist (1 μM DADLE, grey curve) and agonist removal (1 μM DADLE, followed by washout, blue curve) (n=5, average +/− sem). (G) Same experiments as in (F) in the presence of the endocytosis inhibitor Dyngo-4a (30 μM) (n=4, average +/− sem).
Sustained endosomal activation was even more striking for the DOR. While the membranepermeant antagonist naltrindole fully reversed endosomal OR-sensor recruitment within <1 min, little reversal was observed for >30min in the presence of a membrane-impermeant antagonist (TIPPpsi) or after agonist washout (Figure 3D). This provided an ideal experimental condition to test the potential functional significance of endosomal OR activation (Figure 3E). If DOR signaling occurs exclusively from the PM, the cellular response to peptide agonist should fully reverse upon washout. However, if endosome-localized DORs also signal, we anticipated that a fraction of the cellular response would persist after agonist peptide washout. We tested this by monitoring DORmediated inhibition of cAMP accumulation in living cells, a readout of G protein-linked OR signaling. We applied DADLE for 12 min, a time sufficient to produce significant DOR accumulation in endosomes, and then compared inhibition observed in the continuous presence of agonist peptide to that observed after peptide washout. As expected, continuous exposure of cells to DADLE strongly inhibited the forskolin (FSK)-induced cAMP response (Figure 3F, compare black and gray curves). Remarkably, significant inhibition persisted after peptide agonist washout (compare black and blue curves). Further, the persistent DOR response was lost when the same experiment was conducted after blockade of DOR internalization using Dyngo, a chemical inhibitor of the dynamin GTPase that is essential for OR endocytosis (Murray et al., 1998). Together, these results support the hypothesis that endosome-localized, ligand-activated DORs contribute to the overall G protein-mediated signaling response in intact cells.
Sequential waves of OR activation in neurons
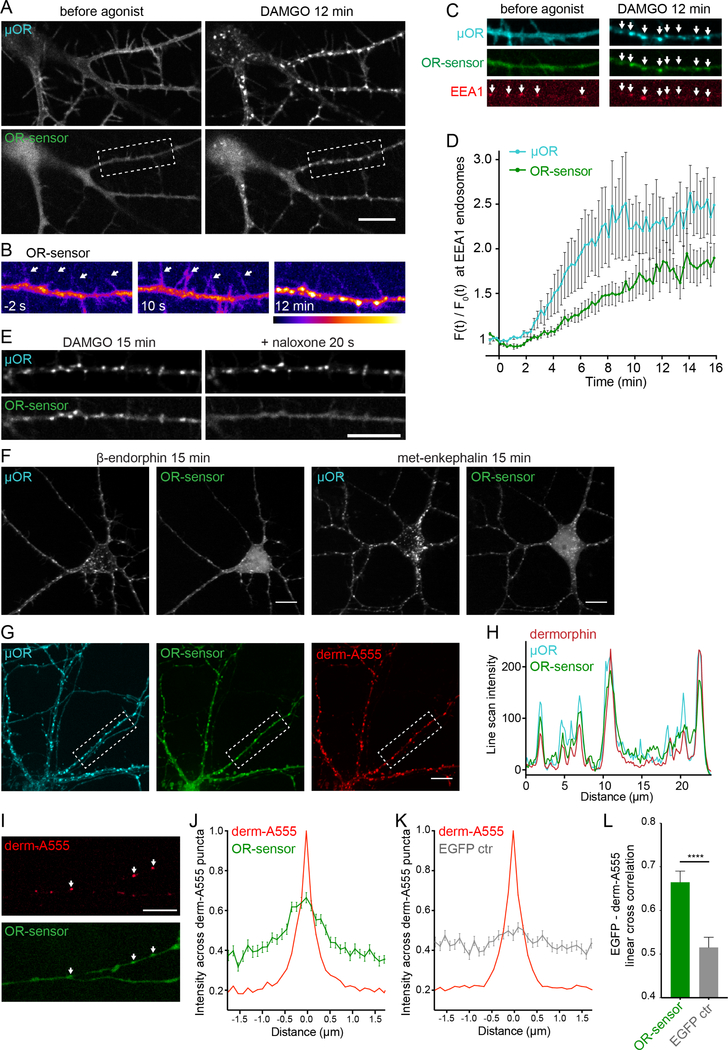
We next investigated whether OR-sensor can detect receptor activation in neurons, and chose primary cultures of striatal medium spiny neurons that are known to express MOR and DOR endogenously in a fraction of neurons. To start, we expressed OR-sensor together with epitopetagged OR to allow reliable detection of both proteins in the same cells, and carried out simultaneous imaging of both using time-lapse confocal microscopy. Prior to agonist addition, OR-sensor showed a diffusive cytoplasmic distribution in soma and neurites (Figure 4A). The majority of these structures in medium spiny neurons are dendrites and we did not specifically label for axons. DAMGO addition caused a rapid accumulation of OR-sensor at the PM, which was particularly evident in dendritic processes (Figure 4B, left and middle panels). Within minutes thereafter, DAMGO addition induced MOR redistribution from the PM to endosomes that appeared as punctate structures in soma and dendrites (Figures 4A and 4B, Movie S4). Strikingly, the MOR-loaded endosomes also recruited OR-sensor. We verified that these structures indeed represent endosomes by co-labeling with EEA1 (Figure 4C) and then used EEA1 as marker for quantifying MOR and OR-sensor recruitment to endosomes over time. As in non-neural cells, ORsensor recruitment to the endosome compartment was strong and appeared to lag MOR accumulation (Figure 4D). Further, the biosensor signal at endosomes was reversed within seconds after addition of naloxone, a time point clearly preceding MOR exit from endosomes (Figure 4E). The endogenous peptide ligands β-endorphin and met-enkephalin also produced OR-sensor recruitment to MOR-loaded endosomes (Figure 3F). In similar experiments, we also detected activation of internalized DORs in response to the agonists DADLE, DPDPE, and deltophin II in neurons (Figure S4).
Figure 4: OR activation in somatodendritic endosomes.
(A) Striatal neuron (12 DIV), expressing FLAG-MOR (surface-labeled with M1-AF555) and EGFPOR-sensor. Images of a time series, before and 12 min after adding DAMGO (10 μM). See also Movie S4. (B) Pseudocolor images (low to high intensity) of OR sensor, corresponding to boxed area in (A). Dendrite before (−2 s), immediately after (10 s) and 12 min after adding DAMGO. Arrows depict dendritic processes. (C) Dendrite of a striatal neuron (12 DVI), expressing FLAGMOR (surface-labeled with M1-AF647), OR-sensor, and EEA1 before agonist and 12 min after adding DAMGO (10 μM). Arrows depict EEA1-positive endosomes that contain MOR and recruit OR-sensor after agonist addition. (D) Quantification and kinetics of MOR and OR-sensor recruitment to EEA1 endosomes in dendrites. F0 = average fluorescence signal before agonist addition. Movie length 16 min, 5 s between frames (n=5, average +/− sem). (E) Localization of MOR and OR-sensor 15 min after adding DAMGO, and 20 s after adding naloxone (10 μM). (F) Confocal images of striatal neurons (12–14 DIV) expressing FLAG-MOR (surface-labeled with M1-AF647) and EGFP-OR-sensor, 15 min after adding β-endorphin (1 μM) or met-enkephalin (10 μM). See also Figure S4. (G) Confocal images showing localization of FLAG-MOR (surfacelabeled with M1-AF647, blue), EGFP-OR-sensor, and fluorescent dermorphin (derm-A555, red) in striatal neurons 15 min after derm-A555 addition. (H) Fluorescence intensity profile (line scan) of dermorphin (red), MOR (blue), and OR-sensor along dendritic process, boxed in (F). (I) Dendrite of a striatal neuron (14 DIV) expressing OR-sensor, incubated with derm-A555 (1 μM) for 15 min. Arrows depict dermorphin-loaded endosomes that co-localize with OR-sensor. (J,K) Average fluorescence intensity profile (line scans) of EGFP-OR-sensor (J) or EGFP control (K) along dendrite across derm-A555-labeled endosomes in striatal neurons. (OR-sensor: n=110 endosomes from 15 cells, EGFP-control: n=102 endosomes from 18 cells, average +/− sem.) (L) Peak cross correlation at derm-A555 puncta (average +/− sem). EGFP-OR-sensor = 0.6645 ± 0.0257, EGFP control = 0.5152 ± 0.0233, ****p <0.0001, unpaired t test. All scale bars: 10 μm.
Our antagonist and washout experiments had shown that the active conformation of ORs at the PM depended on the presence of agonist. To verify this, we asked if MOR-containing endosomes that recruit OR-sensor also contain agonist peptide. We used the opioid peptide dermorphin conjugated to AlexaFluor555 (derm-A555), shown previously to retain biological activity (Arttamangkul et al., 2000). Verifying this, dermorphin application to neurons drove the rapid internalization of MOR as well as OR-sensor recruitment to MOR-containing endosomes (Figure 4F, left and middle panel). Importantly, all of the endosomes containing activated MOR colocalized with dermorphin (Figures 4F and 4G), demonstrating that opioid peptide is indeed present at sites of endosomal MOR activation.
Because fluorescent dermorphin is sufficiently sensitive to label endosomes containing endogenous MORs (Arttamangkul et al., 2006), we next asked if endosomal activation also occurs in neurons expressing only endogenous receptors. Indeed, dermorphin-containing endosomes were detected throughout dendrites in a subset of neurons and, at many of these endosomes, OR-sensor recruitment was visually evident (Figure 4H). We used cross-correlation analysis to assess endosomal OR-sensor recruitment mediated by endogenous ORs in an unbiased manner and across hundreds of endosomes in multiple experiments. This analysis revealed a clear cross-correlation signal between OR-sensor fluorescence intensities and endosomes marked by derm-A555 (Figure 4I). In contrast, no cross-correlation signal was detected in control experiments using soluble EGFP in place of EGFP-OR-sensor (Figures 4J and 4K). These results indicate that OR-sensor can detect activation of ORs expressed at endogenous levels in dissociated neuronal cultures and that opioid peptides can drive endosomal activation of endogenous ORs.
Opioid drugs produce a distinct and additional Golgi-localized OR activation wave
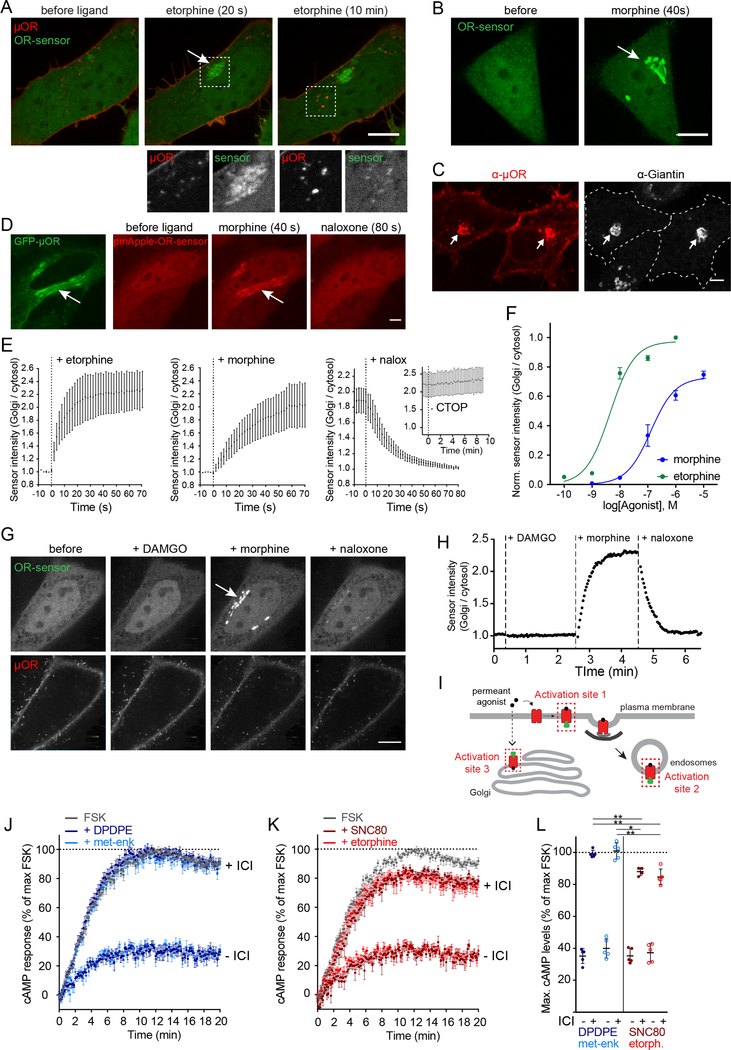
Another unexpected result was obtained when we examined the effect of agonist drugs. The alkaloid agonist etorphine drove OR-sensor recruitment to a perinuclear membrane compartment within seconds after application, before detectable internalization and almost simultaneous with MOR activation in the PM (Figure 5A, Movie S5). Morphine produced a similar effect (Figure 5B). The compartment to which drugs drove OR-sensor recruitment did not originate from the endocytic pathway because it failed to label with receptors internalized from the PM as visualized using our surface labeling protocol (Figure 5A). Neurons are known to contain an internal membrane pool of ORs that originates from the biosynthetic rather than endocytic pathway and localizes at steady state to ER and Golgi membranes (Erbs et al., 2014; Scherrer et al., 2006; Shiwarski et al., 2017). Consistent with this, an internal pool of MORs and DORs was detected by immunostaining of fixed cells after permeabilization, and this pool colocalized with the Golgi marker Giantin (Figure 5C, Figure S5A). We used GFP-fused OR constructs to visualize this internal membrane pool in living cells and observed that alkaloid agonists drove rapid OR-sensor recruitment precisely to this compartment (Figure 5D, Movie 6). Verifying specificity of the activation signal, OR-sensor recruitment to Golgi membranes was rapidly reversed by opiate antagonist (Figure 5D).
Figure 5: Non-peptide drugs activate Golgi-localized ORs.
(A) Confocal images of a time series of a HEK293 cell, expressing EGFP-OR-sensor and FLAGMOR (surface-labeled with M1-AF555). Etorphine (1 μM) was added at t=0. Boxed areas are displayed separately for both fluorophores below respective images. See also Movie S5. (B) Confocal images of time series of a HeLa cells, expressing EGFP-OR-sensor and FLAG-MOR (not depicted). OR-sensor localization is shown before and 20 s after adding morphine (1 μM). (C) Confocal images of HeLa cells, expressing FLAG-MOR. Cells were fixed, permeabilized, and immunolabelled with anti-FLAG (red) and anti-Giantin (grey) antibodies. The internal OR pool colocalizes with Golgi marker (arrows). See also Figure S5A. (D) Rapid activation of Golgi-localized MORs by morphine. Confocal images of a time series of a HeLa cell, expressing MOR-EGFP and mCherry-OR-sensor. Cell was treated with morphine (1 μM), followed by naloxone (10 μM). See also Movie S6. (E) Quantification and kinetics of EGFP-OR-sensor intensity at Golgi apparatus upon agonist or antagonist addition in HeLa cells, expressing OR-sensor, GalT-DsRed, and MOR. GalT-marked Golgi apparatus was used as quantification mask and intensity normalized to cytoplasmic OR-sensor signal. Morphine n=4, etorphine n=3, 1 μM etorphine followed by 10 μM naloxone n=6, or by 10 μM CTOP n=5. 2 s intervals for non-peptide ligands, 15 s intervals for peptide ligand (CTOP), average +/− sem. See also Figure S5B–D. (F) Ligand concentrationdependent recruitment of OR-sensor to MOR in GalT-marked Golgi apparatus (n=5, average +/− sem). Normalization of EGFP-intensity values (range [0–1]). Regression curves with Hill slope of 1. EC50: morphine: 130 nM, etorphine: 4.5 nM. See also Figure S5E. (G) Confocal images of a time series of a HeLa cell, expressing EGFP-OR-sensor, FLAG-MOR (surface labeled with M1–647) and GalT-DsRed (Golgi-marker, not shown). Cell was treated with DAMGO (10 μM, t = 20 s), morphine (1 μM, t = 150 s), and naloxone (10 μM, t = 270 s). See also Figure S5F. (H) Quantification of EGFP-OR-sensor intensity at GalT-marked Golgi apparatus, normalized to cytoplasmic OR-sensor signal. 10 s between frames. (I) Scheme depicting three distinct sites of OR-sensor recruitment to activated receptors following etorphine addition: activation site 1: plasma membrane, activation site 2: endosomes, activation site 3: Golgi apparatus. (J) cAMP response in living HEK293 cells expressing DOR and the luciferase cAMP reporter GloSensor-20F. Forskolin (FSK, 2 μM)-induced cAMP response (grey curve) and effect of 100 nM DADLE or met-enkephalin in the absence (-ICI) or presence (+ICI) of 100 μM ICI-174,864 (n=5, average +/− sem). (K) Forskolin (FSK, 2 μM)-induced cAMP response (grey curve) and effect of 100 nM SNC80 or etorphine in the absence (-ICI) or presence (+ICI) of 100 μM ICI-174,864 (n=5, average +/− sem). (L) Maximum cAMP levels of experiments presented in (J) and (K) (average +/− SD). Unpaired t test between peptide or non-peptide agonists in the presence of ICI: DPDPE vs. SNC80 (p<0.05), or vs. etorphine (p<0.01), and met-enkephalin vs. SNC80 (p<0.01) or vs. etorphine (p<0.01). All scale bars: 10 μm.
To quantify the kinetics of the Golgi-localized activation signal, we labeled the Golgi apparatus in living cells with GalT-DsRed and quantified OR-sensor recruitment throughout confocal time series after application of agonist drug. Both morphine and etorphine drove robust recruitment of OR-sensor to Golgi-localized MORs with t½ < 20s (Figure 5E). Etorphine, as well as the non-peptide DOR agonists ARM390 and SNC80, produced a similarly rapid and robust Golgi activation response in cells expressing DOR (Figure S5B–D). All of these non-peptide drugs are thought to be relatively membrane-permeant when compared to peptides, and the remarkably fast kinetics of the Golgi-localized OR activation response suggests that ligand access from the cell surface is not mediated by membrane trafficking because this process typically has slower kinetics. Instead, it suggests that non-peptide drugs selectively produce Golgi-localized OR activation because they diffuse across membranes. Supporting this hypothesis, Golgi-localized activation of MOR and DOR was reversed rapidly after application of naloxone and naltrindole, membrane-permeant alkaloid antagonists, but it persisted after application of CTOP, TIPPpsi, or ICI-174,864 (ICI) that are peptide-based antagonists thought to penetrate membranes slowly (Figure 5E, Figure S5C). Activation of Golgi-localized ORs detected by OR-sensor was concentration-dependent and exhibited similar pharmacology as activation at the PM (Figure 5F and Figure S5E).
To directly verify that Golgi-localized OR activation is specific to drugs, we challenged individual MOR-expressing cells sequentially – first with the opioid peptide agonist DAMGO and then with the alkaloid agonist morphine – while recording time-lapse series of OR-sensor localization (Figure 5F). Strikingly, DAMGO failed to produce detectable recruitment of OR-sensor to Golgi membranes but subsequent application of morphine produced a robust activation signal that was receptor-dependent and reversible by naloxone (Figure 5F and 5G). This difference persisted even after prolonged exposure because DAMGO failed to produce detectable OR-sensor recruitment to the Golgi apparatus after 90 min of continuous application at saturating concentration, whereas subsequent drug application produced rapid OR-sensor recruitment to the Golgi apparatus (Figure S5F). Together these results identify Golgi membranes as a discrete internal site of OR activation that is rapidly accessible to alkaloid (and other non-peptide) drugs but is inaccessible to opioid peptides (Figure 5H).
We next investigated the possibility that Golgi-localized OR activation may contribute to the cellular signaling response. If so, we anticipated that non-peptide agonists would selectively drive a response that cannot be blocked by a membrane-impermeant antagonist. To test this, we focused on DOR and used ICI because it selectively blocks DORs present in the PM but not Golgi apparatus (Figure S5C). ICI fully reversed the ability of two peptide agonists, DPDPE and metenkephalin, to inhibit FSK-induced cAMP accumulation in intact cells (Figure 5J, note the +ICI curves overlap FSK alone). However, ICI did not completely reverse inhibition by two non-peptide agonist drugs, SNC80 and etorphine, even when ICI was pre-applied in 1000-fold molar excess (Figure 5J, note the +ICI curves fall below FSK alone). This residual, ICI-resistant component of adenylyl cyclase inhibition was significant for both non-peptide drugs (Figure 5L). These results are consistent with Golgi-localized DORs making a partial contribution to the cellular signaling response produced by permeant agonists.
OR activation in somatic Golgi and Golgi outposts of neurons
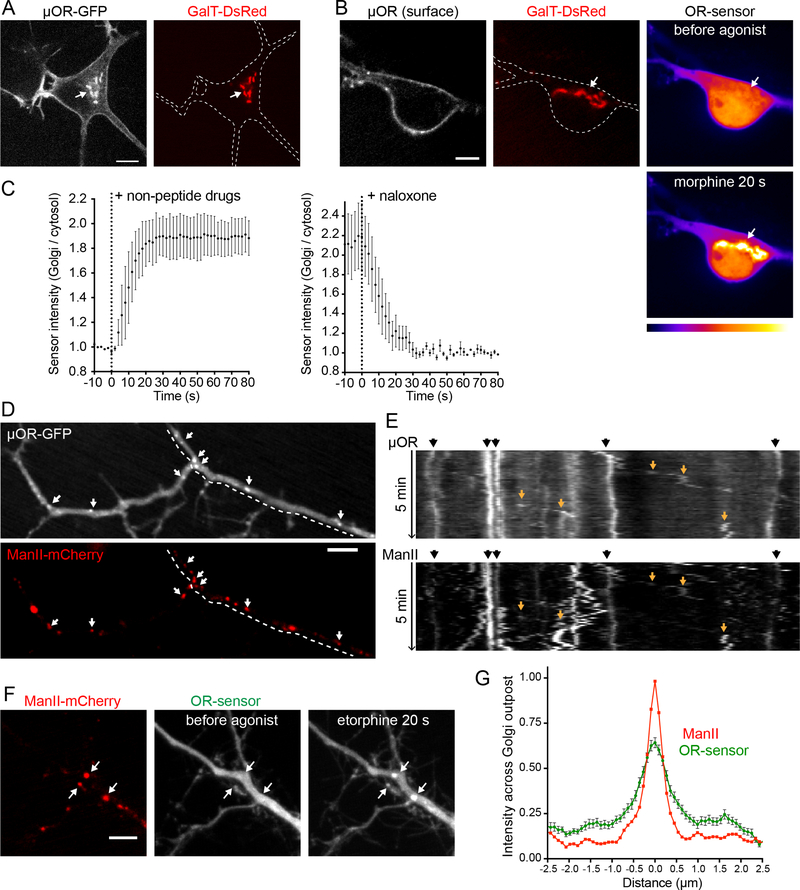
Striatal neurons also contained a Golgi-localized pool of ORs, as detected in the soma by colocalization with GalT (Figure 6A). Non-peptide agonists including etorphine (Figure S6A) and morphine (Figure 6B, Movie S7) drove rapid recruitment of OR-sensor to this internal membrane pool. We verified that the Golgi pool is not accessible to ORs internalized from the PM (Figure S6B). Moreover, we detected rapid kinetics of drug-induced OR activation in Golgi membranes of neurons and its reversibility by membrane-permeant antagonist (Figure 6C). Therefore, nonpeptide drugs rapidly and specifically promote OR activation in the somatic Golgi apparatus of neurons.
Figure 6: OR activation in somatic Golgi and Golgi outposts of neurons.
(A) Soma of a striatal neuron (12 DIV), expressing MOR-EGFP and GalT-DsRed. Internal MORs co-localize with GalT-marked Golgi apparatus (arrow). (B) Soma of striatal neuron (14 DIV), expressing FLAG-MOR (surface-labeled with M1-AF647), GalT-DsRed, and OR-sensor (pseudocolored low to high intensity). OR-sensor distribution is depicted before agonist and 20 s after adding morphine (1 μM). See also Movie S7 and Figure S6. (C) Quantification and kinetics of EGFP-OR-sensor intensity at somatic Golgi upon non-peptide agonist or antagonist addition in striatal neurons, expressing OR-sensor, FLAG-MOR, and GalT-DsRed. Time series with 5 s intervals. Left: Averaged data using non-peptide drugs, from n=2 morphine (1 μM) and n=2 etorphine (1 μM). Left: 1 μM etorphine followed by naloxone (10 μM), n=3. Average +/− sem. (D) Dendrite of a striatal neuron (13 DIV), expressing MOR-EGFP and ManII-mCherry (Golgi-outpost (GOP) marker). Puncta of MOR co-localize with GOP (arrows). (E) 5 min kymograph along the dendritic branch highlighted by dashed line in (D). Black arrows (top): stable dendritic GOPs appear as straight, vertical lines. Orange arrows (in kymograph): sloped traces of mobile secretory carriers. (F) Dendrite of a striatal neuron (12 DIV), expressing ManII-mCherry, EGFP-OR-sensor, and FLAG-MOR (not depicted). OR-sensor distribution before agonist and 20 s after adding etorphine (1 μM). GOPs recruit OR-sensor upon receptor activation (arrows). (G) Average fluorescence intensity profile (line scan) of EGFP-OR-sensor across ManII-labeled GOPs after adding etorphine. n=47 GOPs from 14 cells, average +/− sem. All scale bars: 10 μm.
In addition to the classical Golgi apparatus localized in the soma, many neurons contain functional Golgi-related membrane clusters in dendrites that are called Golgi outposts (Horton and Ehlers, 2003; Ori-McKenney et al., 2012; Pierce et al., 2001). We verified the presence of such membrane structures, marked by Alpha-mannosidase 2 (ManII), throughout dendrites of spiny neurons and established that they contain ORs (Figure 6D). Consistent with the characteristic properties of Golgi outposts (Horton and Ehlers, 2003), these structures were frequently observed at dendritic branch points and were immobile in live image series (Figure 6E, black arrows). We also observed mobile OR-containing membrane structures that appeared to emerge from static ManII-marked clusters (Figure 6E, orange arrows), consistent with transport carriers derived from Golgi outposts. Together, these observations raised the question of whether non-peptide opiate drugs, in addition to producing a discrete component of OR activation in the somatic Golgi apparatus, might have the ability to drive additional internal membrane activation more broadly by accessing Golgi outposts. This was indeed the case because alkaloid agonists produced robust OR-sensor recruitment to ManII-marked membrane structures throughout dendrites (Figure 6F and 6G). Like Golgi activation in the soma, OR-sensor recruitment to dendritic Golgi outposts occurred in <20 s, distinguishing this component of OR activation temporally from OR activation in endosomes that develops over several minutes (Figure S6). Taken together, these results indicate the ability of opiate alkaloids to drive a rapid and discrete component of OR activation in Golgi membranes is not restricted to the soma. Rather, OR activation by drugs extends throughout the dendritic arbor via Golgi outposts.
Discussion
The present study establishes a genetically encoded conformational biosensor that is capable of directly resolving the subcellular location of OR activation in living neurons and in real time. To our knowledge, the present results are the first to describe a nanobody-based conformational biosensor for detecting activation of any peptidergic GPCR, and the first to apply conformational biosensor technology to assess any receptor type in neurons. Our results delineate a characteristic spatiotemporal pattern of neuronal OR activation that is produced by opioid peptides and reveal distinct activation effects of opioid drugs, thereby suggesting a cellular framework for understanding specificity and diversity of opioid drug action at the level of individual target neurons.
The traditional understanding of OR activation in neurons holds that ligand-dependent activation of receptors is restricted to the cell surface. While neuropeptides, including opioids, are clearly influenced by endosome acidification and proteases (Grady et al., 1995; Gupta et al., 2014), the idea that ORs are necessarily inactive in endosomes is based on assumption. The development of OR-sensor provided an opportunity to test this assumption directly. Unexpectedly, we found endosomes to represent a discrete site of ligand-dependent OR activation. Using OR-sensor, we delineated a characteristic spatiotemporal pattern of OR activation that is elicited by peptide ligands in living neurons, beginning in the PM and propagating to endosomes after receptors undergo ligand-induced internalization.
Endosomal OR activation remains dynamically ligand-dependent but differs from activation in the PM due to ligand trapping. Accordingly, endosomal OR activation is longer-lasting than activation in the PM, and we directly demonstrate slow reversal after peptide removal from the extracellular milieu. This suggests that OR activation in endosomes provides a form of cellular ‘memory’ of previous opioid exposure. The effect of membrane-permeant antagonists such as naloxone is different from agonist washout because naloxone rapidly reverses endosomal OR activation. This suggests that administration of an opiate antagonist drug does not simply mimic the effect of agonist removal but, instead, it fundamentally distorts the neuronal OR activation landscape by blocking or reversing a discrete component that would normally persist. Alkaloid agonists distort the activation landscape even more profoundly by driving conformational activation of ORs at discrete and additional internal membrane locations, including the somatic Golgi apparatus and Golgi outposts throughout the dendritic arbor, which are inaccessible to peptides and do not require receptor trafficking for activation.
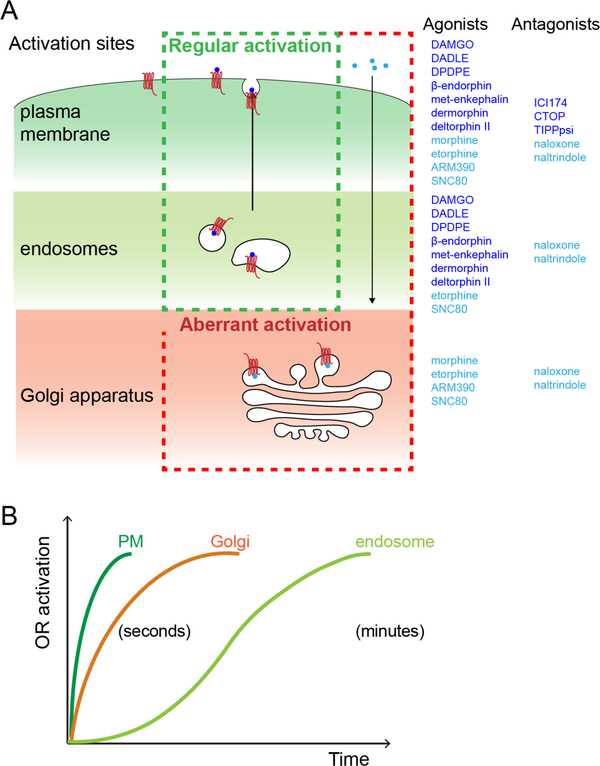
Together, these results support a model in which OR activation occurs in living neurons through spatially (Figure 7A) and temporally (Figure 7B) resolved ‘waves’. Opioid peptides drive a ‘regular’ activation pattern that is comprised of sequential waves of OR activation in the PM and propagating to endosomes. Agonist and antagonist drugs distort this pattern, with clinically relevant alkaloid agonists, such as morphine, uniquely driving a third wave of Golgi-localized OR activation in the soma and dendrites.
Figure 7: Spatiotemporal landscape of OR activation in the cell.
(A) Summary of findings: Activation of OR occurs at distinct cellular membrane compartments in a ligand-dependent manner. Peptides agonists tested (dark blue) drive a ‘regular’ activation pattern, with two sequential waves of OR activation, first in plasma membrane and then in endosomes following internalization of the receptor. Non-peptide agonist (light blue) distort this pattern by activating a Golgi-localized internal OR pool (‘aberrant’ activation). The distinct OR activation sites are differentially affected by peptide (dark blue) and non-peptide (light blue) antagonists. (B) Kinetics of OR activation at distinct cellular membrane compartments. Activation of ORs in PM and the Golgi apparatus occurs within seconds of agonist application. The endosome activation wave lags several minutes.
An important future direction is to determine the functional significance of location-specific OR activation. The present results suggest that intracellular OR activation can contribute to the overall cellular response at the level of cellular cAMP accumulation. We provide evidence that endosome-localized ORs mediate a sustained component of adenylyl cyclase inhibition that is lost after endocytic blockade. We also provide evidence suggesting that Golgi-localized ORs contribute to the cellular cAMP response but, here, the evidence is limited to differential antagonist access. Additional study is therefore needed to more fully delineate the signaling consequences of internal OR activation from both intracellular membrane locations, to extend the analysis beyond the cAMP cascade, and to extend analyses into neurons and neuronal tissues that natively respond to opioids. A number of GPCRs are now recognized to initiate signaling from internal membranes, including endosomes and the Golgi apparatus (Calebiro et al., 2009; Irannejad et al., 2017; 2013; Vilardaga et al., 2014), and location-specific activation has been demonstrated to produce distinct downstream effects by affecting signal duration as well as pathway selectivity through proximity to downstream effectors (Godbole et al., 2017; Jong et al., 2014; Tsvetanova and von Zastrow, 2014).
The present results have interesting implications for opioid biology and therapeutics. Differences in the cellular actions of opioid drugs are presently understood to arise through ligand-selective bias imposed on the conformational landscape of individual receptors (Kenakin, 2011; Raehal et al., 2011; Staus et al., 2016). The present results identify an additional layer of selectivity that is imposed through ligand-specific bias of the spatiotemporal landscape of OR activation in individual neurons. Location-specific activation of ORs therefore suggests an expanded framework for understanding diversity and specificity of cellular opioid actions. In this regard, the Golgi-localized activation wave is of particular interest because it is robustly induced by opiate alkaloid drugs but apparently not at all by peptide ligands.
The present findings also have more general implications for neural cell biology and neuropharmacology. Most neuromodulator receptors belong to the GPCR superfamily, and neurons represent some of the most structurally elaborate cells in the body. Thus, the spatiotemporal activation landscape that we delineate here for ORs may be widely applicable to neuromodulator receptors, particularly those whose natural ligands are hydrophilic. We also note that internal membrane pools of various GPCRs (including ORs) have been previously identified in neurons, but were generally thought to represent inactive reserves for later mobilization to the PM. That such internal pools are subject to ligand-dependent activation could be broadly relevant to GPCR-directed drug action because most CNS-accessible drugs in current use are relatively hydrophobic, and thus are likely to be sufficiently membrane-permeant to effectively access internal membrane pools at the level of individual target neurons.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for resources, reagents, or questions about methods should be directed to Lead Contact Mark von Zastrow (mark@vzlab.org).
EXPERIMETNAL MODEL AND SUBJECT DETAILS
Mammalian Cell Culture Conditions
HEK293 (CRL-1573, ATCC, female) and HeLa cells (CRM-CCL-2, ATCC, female) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO), supplemented with 10% fetal bovine serum (UCSF Cell Culture Facility). Stably transfected HEK293 cells expressing N-terminally FLAG-tagged MOR or DOR were cultured in the presence of 250 μg/ml Geneticin (Gibco). For transient DNA expression, Lipofectamine 2000 (Invitrogen) was used according to manufacturer’s instructions. For live cell imaging, cells were plated on poly-L-lysine-coated 35-mm glassbottomed culture dishes (MatTek Corporation) 48 h before the experiments. Cells were transfected 24h prior to imaging. Per 35-mm culture dish, 200 ng cDNA was used for OR-sensor or organelle markers (EEA1, GalT, CLC), and 1.5 μg DNA was used for receptor constructs.
Primary rat striatal neurons
Striatal neurons were prepared from embryonic day 18 Sprague-Dawley rats (the embryo sex was not determined). The striatum, including the caudate-putamen and nucleus accumbens, was identified as described (Ventimiglia and Lindsay, 1998). Striata were dissected in ice cold Hank’s Buffered Saline solution, Calcium/magnesium/phenol red-free (UCSF Cell Culture Facility). Striata were dissociated in 1x trypsin/EDTA for 15 min at 37°C. Cells were washed in Gibco DMEM (Invitrogen) supplemented with 10% fetal bovine serum (UCSF Cell Culture Facility) and 30 mM Hepes, then mechanically separated with a flame-polished Pasteur pipette. Striatal neurons were transfected using electroporation (Rat Neuron Nucleofector Kit; Lonza) before plating or using Lipofectamine 2000 on DIV 8. Cells were plated on poly-D-lysine-coated 35mm glass-bottom dishes (MatTek) in DMEM (Gibco) supplemented with 10% FBS. Media was exchanged 1–4 days after plating for Neurobasal (Invitrogen) supplemented with Glutamax 1x (Invitrogen) and B27 1x (Thermo Fischer). Half of the media was exchanged every week, Cytosine arabinosine 2μM was added at 8 DIV. Transfection using Lipofectamine 2000 was performed using 1μl of Lipofectamine and 1.5 μg DNA in 1ml of media per 35mm imaging dish, media was exchanged 6 hours later. Cells were maintained in a humidified incubator with 5% CO2 at 37°C and imaged at 10–15 DIV. All animal procedures have been reviewed and approved by the UCSF Institutional Animal Care and Use Committee (IACUC).
METHOD DETAILS
cDNA constructs
EGFP- and pmApple-OR-sensor was created by amplifying Nb33 DNA using 5’−3’ TCGAAGCTTCCGGTAGCGGCAGCGGTATGGCACAGGTGCAGCTGG and TGCGGATCCTTATGCGGCCCCGTGATGGTG primers, adding the linker sequence GSGSG, and inserted into EGFP-C1 or pmApple-C1 vectors using HindIII and BamHI sites. FLAG-MORAPEX2 was cloned into pcDNA3.1 by PCR-amplifying FLAG-tagged murine MOR and inserting into pcDNA3.1 using the restriction enzymes NheI and EcoRI. APEX2 was amplified by PCR and inserted in-frame to FLAG-MOR using EcoRI and NotI restriction enzymes. For neuronal expression, signal sequence FLAG ssf-MOR, ssf-DOR, EGFP-OR-sensor, ssf-MOR-EGFP, ManII(1–112)-pmApple were PCR-amplified and cloned into pCAGGS/SE expression vector with CAG promoter, using EcoR1/KpnI restriction sites.
Total internal reflection fluorescence microscopy
Live cell image series testing OR-sensor recruitment to the PM were acquired on a Nikon TE-2000 inverted microscope equipped with 60× 1.45 NA Plan Apo TIRF objective, a Bioptechs objective warmer and an Andor iXon EM+ EMCCD camera. The illumination from an argon laser source was focused on the periphery of the back focal plane of the objective with a micrometer guided illuminator (Nikon) to achieve total internal reflection. For perfusion, an insert was 3D printed and placed inside the imaging dish where it left a dead volume of about 300 μl. It was used to perfuse solution (Hepes buffered saline (HBS) with 135 mM NaCl, 5 mM KCl, 0.4 mM MgCl2,1.8 mM CaCl2, 20 mM Hepes, 5 mM d-glucose adjusted to pH 7.4 and 300–315 mOsmol/l) containing agonists or antagonists at concentrations indicated in the figure legends with a flow rate of 1.5ml/min.
Other TIR-FM imaging experiments were performed at 37 °C using a Nikon Ti-E microscope equipped for through-the-objective TIR-FM with a temperature-, humidity- and CO2-controlled chamber (Okolab) and an Andor DU897 EMCCD camera. Images were obtained with a 100× 1.49 NA Apo TIRF objective (Nikon) with solid-state lasers of 488, 561 and 647 nm (Keysight Technologies). Cells were imaged in DMEM without phenol red supplemented with 30 mM HEPES, pH 7.4 (UCSF Cell Culture Facility) and agonists or antagonists were added by bath application at concentrations indicated in the figure legends.
Spinning disc confocal imaging
Live-cell spinning disc confocal imaging was carried out using Yokagawa CSU-22 confocal Nikon Ti microscope with a 60x or 100× 1.4 NA Plan Apo VC oil objective and a temperature-, humidity- and CO2-controlled Okolab enclosure Bold Line incubator. 488-nm, 568-nm and 640-nm Coherent OBIS lasers were used as light sources. Time-lapse images were acquired with an Evolve Delta EMCCD camera (Photometrics) driven by Micro-Manager 1.4 or Nikon Elements 4.5. Cells were imaged every 2–30 s for up to 30 min in DMEM without phenol red supplemented with 30 mM HEPES, pH 7.4 (UCSF Cell Culture Facility). Agonists or antagonists were added by bath application during acquisition as indicated in the figure legends. For immunofluorescence imaging, cells were cells grown on coverslips and fixed using 4% formaldehyde in PBS. Cells were permeabilized with 0.05% saponin and 1% BSA in PBS and incubated with primary (1:1000) and secondary (1:1000) antibodies. Specimens were mounted using ProLong Gold (Thermo Fisher).
Fluorescence recovery after photobleaching
FRAP experiments of EGFP-OR-sensor at endosomes were performed at 37 °C on a Andor Borealis CSW-W1 spinning disk confocal Nikon Ti Microscope with Andor 4-line laser launch and a temperature-, humidity- and CO2-controlled chamber (Okolab). HEK293 cells transiently expressing EGFP-OR-sensor and stably expressing ssf-MOR (labeled with anti-FLAG M1–647) were imaged using a 100× 1.49 NA Apo TIRF objective (Nikon) and an Andor Zyla 4.2 sCMOS camera controlled by MicroManager software. Cells were incubated with 10 μM DAMGO for 15 min prior to imaging. EGFP-OR-sensor was bleached at 2–3 individual receptor-loaded endosomes per cell using a Rapp Optoelectronic UGA-40 photobleaching system with 473 laser (Vortran). EGFP fluorescence recovery was monitored by acquiring images every 2 s for 5 min. The receptor signal was also acquired throughout the image series to correct for possible movement of endosomes. The mean EGFP-OR-sensor intensity at bleached endosomes at each time point was corrected for background signal and photobleaching of the cell. Fluorescence intensity before bleaching was normalized to 1 and directly after bleaching to 0.
Luminescence-based live cell cAMP accumulation assay
Cellular cAMP was detected in living cells as described previously (Irannejad et al., 2013). To measure the effect of OR-sensor expression on G-protein signaling (Fig. 1) HEK293 cells, stably expressing ssf-MOR were transiently transfected with GloSensor-20F cAMP reporter (Promega), and EGFP-OR-sensor or empty EGFP-C1 vector and plated in 24-well dishes. 24 hours post transfection, cells were treated with Forskolin (2 μM) and varying concentrations of DAMGO, immediately followed by imaging. To probe endosomal signaling (Fig. 3), HEK293 cells stably expressing ssf-DOR were transiently transfected with GloSensor-20F and plated in poly-L-Lysinecoated 24-well dishes. 24 hours post transfection, cells were treated with DADLE (1 μM) for 12 min. Wells were washed 5x with 1 mL media. Subsequently DADLE was re-added (continuous presence of agonist) or not (washout condition), and cells treated with Forskolin (2 μM), immediately followed by imaging. In experiments with endocytic block, cells were pre-incubated for 15 min in serum-free media containing 30 μM Dyngo-4a before adding DADLE and the assay performed in the continuous presence of Dyngo-4a. To probe Golgi signaling (Fig. 5), HEK293 cells stably expressing ssf-DOR were transiently transfected with GloSensor-20F and plated in 24-well dishes. 24 hours post transfection, cells were treated or not for 5 min with ICI-174,864 (100 μM), followed by addition of peptide or non-peptide agonists (100 nM) and Forskolin (2 μM), immediately followed by imaging. In all experiments, images were collected every 10 s for 30 min using a CCD camera (Hamamatsu C9100–13) in a light-proof cabinet at 37 °C. During the assay, cells were in DMEM without FBS and phenol red containing 250 μg/ml luciferin. Luminescence signals generated were measured using ImageJ and normalized to the maximum luminescence signal measured in the presence of 2 μM Forskolin (or 2 μM Forskolin + 100 μM ICI).
APEX proximity labeling
HEK293 cells stably expressing FLAG-MOR-APEX2 were transfected with EGFP-OR-sensor for 24 hrs. For the ‘DAMGO+nalox’ condition, 10 μM DAMGO was incubated with cells for 30 min and then cells were washed once. Next, 500 μM biotin-phenol was pre-incubated for all conditions for 30 min at 37 °C. During this time-window, cells were either untreated, incubated with 10 μM DAMGO for 30 min (‘DAMGO’ condition), or incubated with 10 μM naloxone (both ‘DAMGO+nalox’ and ‘nalox’ conditions). APEX labeling was initiated by 1:1 mixing of room temperature media supplemented H202 (cf, 1 mM) and the reaction was allowed to proceed for 30 s. Cells were washed three times in ice cold quenching buffer (TBS supplemented with 1 mM CaCl2, 10 mM sodium ascorbate, 1 mM sodium azide, and 1 mM Trolox). Cells were left in the third wash for 20 min on ice. Cells were lysed in RIPA (50 mM Tris, 150 mM NaCl, 1% TritonX-100, 0.5% sodium deoxycholate, 0.1 % SDS, pH 7.4) supplemented with 10 mM sodium ascorbate and 1 mM Trolox. Samples were briefly sonicated and spun down at 10,000 x g for 10 min. Supernatant was applied to streptavidin agarose resin and incubated overnight at 4 °C. Resins were washed five times, and biotinylated proteins eluted by heating the resin to 85 °C for 7 min in NuPAGE LDS sample buffer supplemented with 143 mM B-mercaptoethanol. Samples were separated on 4%−12% SDS-PAGE gels, transferred to nitrocellulose membranes, and blocked in Li-COR Odyssy Blocking Buffer (TBS). To probe eluate blot, anti-GFP (1:750) was incubated for 2 h at room-temperature. To probe lysate blot, anti-GFP (1:2000) and anti-GAPH (1:200) were incubated for 2 h at room-temperature. Membranes were washed in TBS-Tween (0.1% v/v), incubated with secondary fluorescent antibodies (1:3000 for 680-donkey-anti-rabbit; 1:3000 for 800-donkey-anti-mouse) for 1 h at room-temperature. Membranes were washed in TBS-Tween and imaged using an Odyssey Infrared Imaging System (Li-COR).
Concentration dependence of OR-sensor recruitment
For probing OR-sensor recruitment to the PM, HEK293 cell co-expressing EGFP-OR-sensor and MOR were imaged using TIR-FM. Cells were treated with increasing concentrations of agonist (bath application) and imaged at a frame rate of 0.2/s (total movie length 4.5 min). OR-sensor intensity during time lapse series was measured using ImageJ. For DAMGO and etorphine, values were normalized between 0 (before agonist) and 1 (10 μM agonist). For morphine, values were normalized between 0 (before agonist) and 1 (10 μM full agonist DAMGO). Regression curves with Hill slope of 1 were fit using Prism 7.0c. For probing OR-sensor recruitment to the Golgi, HEK293 cell co-expressing EGFP-OR-sensor, GalT-DsRed and MOR or DOR were imaged using spinning disc confocal. Cells were treated with increasing concentrations of agonist (bath application) and imaged at a frame rate of 0.05/s (total movie length 8–10 min). OR-sensor intensity at GalT-labeled Golgi was measured for each frame as described under quantitative image analysis. For etorphine and SNC80, values were normalized between 0 (before agonist) and 1 (10 μM agonist), for morphine, values were normalized between 0 (before agonist) and 1 (10 μM full agonist etorphine).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
Quantification of data are presented as mean ± standard error of the mean (SEM) based on at least 3 biologically independent experiments with the precise number indicated in the figure legends. Statistical analysis was performed using Prism (7.0c, GraphPad) and using unpaired or paired two tailed Student’s t test.
Quantitative image analysis
All quantitative image analysis was performed on unprocessed images using MATLAB (MathWorks, R2014b) or ImageJ (2.0.0). For quantifying enrichment of OR and OR-sensor at endosomes, Golgi apparatus, or clathrin-coated pits, time-lapse movies were divided in subregions containing the fluorescent marker proteins used for labeling (EEA1 (aa 1262–1411), GalT (aa 1–82), or CLC). Movies were thresholded to encompass signal from marker proteins throughout the movie and to generate a mask of the structures. Filtering to exclude regions smaller than 5 pixels was applied, remaining objects were smoothened with a dilatation of 1 pixel before an erosion of 1 pixel. The generated mask was applied to the OR or OR-sensor channel to measure the fluorescence at the location of marker objects. The average fluorescence in the mask was background subtracted and normalized to baseline (before agonist addition) or to the cellular fluorescence outside of the thresholded structures. To determine the linear correlation coefficient between agonist-loaded endosomes (marked by dermorphin) or Golgi outposts (marked by ManII) and OR-sensor or soluble EGFP, a 5-pixel line was drawn along the dendrite on individual endosomes identified in the marker channel. The fluorescence profile obtained in both channels for each endosome was normalized with the maximal value set to 1 and the minimal value set to 0. Linescans were aligned to the maximal value in the red channel and the linear correlation coefficient calculated for every endosome as the value of the GFP channel for this linescan point.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-FLAG (M1) | Sigma | Cat# F-3040; RRID: AB_439712 |
| Rabbit anti-Giantin | Covance | Cat# PRB-114C-200, RRID: AB_291560 |
| Goat anti-Mouse IgG Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A32723, RRID: AB_2633275 |
| Goat anti-Rabbit IgG Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A32733, RRID: AB_2633282 |
| Mouse anti-GFP | Roche | Cat# 11814460001; RRID: AB_390913 |
| Mouse anti-GAPDH | EMD Millipore | Cat# MAB374; RRID: AB_2107445 |
| Donkey anti-mouse (800 conjugate) | LI-COR | Cat# MAB374; RRID: AB_2107445 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DAMGO, [D-Ala2, N-Me-Phe4, Gly5-ol]-Enkephalin acetate salt | Sigma-Aldrich | E7384 |
| DADLE, [D-Ala2, N-Me-Phe4, Gly5-ol]-Enkephalin acetate salt | Sigma-Aldrich | E7131 |
| DPDPE, [D-Pen2,5]-Enkephalin hydrate | Sigma-Aldrich | E3888 |
| Carbamoylcholine chloride, Charbachol) | Sigma-Aldrich | C4382 |
| β-Endorphin, human | Sigma-Aldrich | E6261 |
| [Met5]-Enkephalin acetate salt hydrate | Sigma-Aldrich | M6638 |
| [D-Ala2]-Deltorphin II | Sigma-Aldrich | T0675 |
| Morphine sulfate | Sigma-Aldrich | 1448005 |
| Forskolin | Sigma-Aldrich | F6886 |
| Etorphine-HCl | NIDA | N/A |
| AR-M 1000390 hydrochloride (ARM390) | Tocris | 4335 |
| SNC80 | Tocris | 0764 |
| ICI 174,864 | Tocris | 0820 |
| CTOP | Tocris | 1578 |
| Naltrindole hydrochloride | Tocris | 0740 |
| dermorphin-A555 | (Arttamangkul et al., 2000) | N/A |
| TIPPpsi | (Schiller et al., 1993) | N/A |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher Scientific | Cat# A20173 |
| Alexa Fluor 555 Protein Labeling Kit | Thermo Fisher Scientific | Cat# A20173 |
| ProLong Gold Antifade Mountant | Thermo Fisher Scientific | P36930 |
| Experimental Models: Cell Lines | ||
| Human HEK293 cells | ATCC | CRL-1573 |
| Human HeLa cells | ATCC | CRM-CCL-2 |
| Experimental Models: Organisms/Strains | ||
| Rat: Sprague Dawley | Charles River | |
| Oligonucleotides | ||
| OR-sensor fwd (5’-TCGAAGCTTCCGGTAGCGGCAGCGGTATGGC ACAGGTGCAGCTGG-3’) | This study | N/A |
| OR-sensor rvs (5’-TGCGGATCCTTATGCGGCCCCGTGATGGTG-3) | This study | N/A |
| Recombinant DNA | ||
| EGFP-C1: EGFP-OR-sensor | This study | N/A |
| pCAGGS/SE: EGFP-OR-sensor | This study | N/A |
| pmApple-C1: pmApple-OR-sensor | This study | N/A |
| pCAGGS/SE: pmApple-OR-sensor | This study | N/A |
| pCAGGS/SE: ManII(1–112)-pmApple | This study | N/A |
| pCAGGS/SE: signal sequence FLAG (ssf)-MOR, murine | This study | N/A |
| pCAGGS/SE: ssf-MOR-EGFP | This study | N/A |
| pCAGGS/SE: signal sequence FLAG (ssf)-DOR, murine | This study | N/A |
| FLAG-MOR-APEX2 | This study | N/A |
| pcDNA3: signal sequence FLAG (ssf)-MOR, murine | (Tanowitz and von Zastrow, 2003) | N/A |
| pcDNA3: signal sequence FLAG (ssf)-DOR, murine | (Tanowitz and von Zastrow, 2002) | N/A |
| GalT-DsRed2 (GalT aa 1–82) | (Irannejad et al., 2017) | N/A |
| EEA1-DsRed2 (EEA1 aa 1262–1411) | (Irannejad et al., 2013) | N/A |
| CLC-DsRed2 | (Irannejad et al., 2013) | N/A |
| pGLO-20F | Promega | Cat# E1171 |
| Software and Algorithms | ||
| Prism | GraphPad | 7.0c |
| ImageJ | Imagej.net/contributors | 2.0.0-rc-54/1.51g |
| MATLAB | MathWorks | R2014b |
| Excel | Microsoft | 16.11.1 |
| PyMOL | Schrödinger | v1.7.4.5 |
| Illustrator CC | Adobe | 21.0.2 |
Acknowledgements
We thank Seksiri Arttamangkul, Brian Kobilka, and John Williams for valuable discussion and providing critical reagents. We thank members of the Kobilka and von Zastrow laboratories for valuable discussion and advice. Some of the imaging experiments were carried out in the UCSF Nikon Imaging Center directed by DeLaine Larsen. We thank Simon Braun for advice and critical reading of the manuscript. This study was supported by research grants from the NIH (DA010711 and DA012864 to M.vZ., DA004443 to P.S., and DP5 OD 02304801 to A.M.) and the Canadian Institutes of Health Research (MOP-89716 to P.S.). M.S. was supported by the Swiss National Science Foundation (P2EZP3_152173 and P300PA_164712).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Arttamangkul S, Alvarez-Maubecin V, Thomas G, Williams JT, and Grandy DK (2000). Binding and internalization of fluorescent opioid peptide conjugates in living cells. Mol. Pharmacol 58, 1570–1580. [DOI] [PubMed] [Google Scholar]
- Arttamangkul S, Torrecilla M, Kobayashi K, Okano H, and Williams JT (2006). Separation of mu-opioid receptor desensitization and internalization: endogenous receptors in primary neuronal cultures. J. Neurosci 26, 4118–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury AF, Smyth DG, and Snell CR (1976). Biosynthetic origin and receptor conformation of methionine enkephalin. Nature 260, 165–166. [DOI] [PubMed] [Google Scholar]
- Calebiro D, Nikolaev VO, Gagliani MC, de Filippis T, Dees C, Tacchetti C, Persani L, and Lohse MJ (2009). Persistent cAMP-Signals Triggered by Internalized G-Protein-Coupled Receptors. PLoS Biol. 7, e1000172–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D, Nikolaev VO, Persani L, and Lohse MJ (2010). Signaling by internalized Gprotein-coupled receptors. Trends Pharmacol. Sci 31, 221–228. [DOI] [PubMed] [Google Scholar]
- Erbs E, Faget L, Scherrer G, Matifas A, Filliol D, Vonesch J-L, Koch M, Kessler P, Hentsch D, Birling M-C, et al. (2014). A mu–delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct. Funct 220, 677–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbole A, Lyga S, Lohse MJ, and Calebiro D (2017). Internalized TSH receptors en route to the TGN induce local Gs-protein signaling and gene transcription. Nat. Commun 8, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady EF, Garland AM, Gamp PD, Lovett M, Payan DG, and Bunnett NW (1995). Delineation of the endocytic pathway of substance P and its seven-transmembrane domain NK1 receptor. Mol. Biol. Cell 6, 509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Gomes I, Wardman J, and Devi LA (2014). Opioid receptor function is regulated by post-endocytic peptide processing. J. Biol. Chem 289, 19613–19626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry AG, Hislop JN, Grove J, Thorn K, Marsh M, and von Zastrow M (2012). Regulation of endocytic clathrin dynamics by cargo ubiquitination. Dev. Cell 23, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC and Ehlers MD (2003). Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J. Neurosci 23, 6188–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Manglik A, Venkatakrishnan AJ, Laeremans T, Feinberg EN, Sanborn AL, Kato HE, Livingston KE, Thorsen TS, Kling RC, et al. (2015). Structural insights into μ-opioid receptor activation. Nature 524, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Pessino V, Mika D, Huang B, Wedegaertner PB, Conti M, and von Zastrow M (2017). Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol 13, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SGF, Sunahara RK, El-Samad H, Huang B, et al. (2013). Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Tsvetanova NG, Lobingier BT, and von Zastrow M (2015). Effects of endocytosis on receptor-mediated signaling. Curr. Opin. Cell Biol 35, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong Y-JI, Harmon SK, and O’Malley KL (2017). GPCR signalling from within the cell. Br. J. Pharmacol doi: 10.1111/bph.14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong Y-JI, Sergin I, Purgert CA, and O’Malley KL (2014). Location-dependent signaling of the group 1 metabotropic glutamate receptor mGlu5. Mol. Pharmacol 86, 774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T (2011). Functional selectivity and biased receptor signaling. J. Pharmacol. Exp. Ther 336, 296–302. [DOI] [PubMed] [Google Scholar]
- Kieffer BL and Evans CJ (2009). Opioid receptors: from binding sites to visible molecules in vivo. Neuropharmacology 56, 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, Hüttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, and Krogan NJ (2017). An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 169, 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kobilka BK, and Steyaert J (2017). Nanobodies to Study G Protein-Coupled Receptor Structure and Function. Annu. Rev. Pharmacol. Toxicol 57, 19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, and Granier S (2012). Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheyses AL, Simon SM, and Rappoport JZ (2010). Imaging with total internal reflection fluorescence microscopy for the cell biologist. J. Cell. Sci 123, 3621–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SR, Evans CJ, and Zastrow, von M (1998). Phosphorylation is not required for dynamin-dependent endocytosis of a truncated mutant opioid receptor. J. Biol. Chem 273, 24987–24991. [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, and Jan Y-N (2012). Golgi Outposts Shape Dendrite Morphology by Functioning as Sites of Acentrosomal Microtubule Nucleation in Neurons. Neuron 76, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JP, Mayer T, and McCarthy JB (2001). Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr. Biol 11, 351–355. [DOI] [PubMed] [Google Scholar]
- Raehal KM, Schmid CL, Groer CE, and Bohn LM (2011). Functional Selectivity at the mu-Opioid Receptor: Implications for Understanding Opioid Analgesia and Tolerance. Pharmacol. Rev 63, 1001–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer G, Tryoen-Tóth P, Filliol D, Matifas A, Laustriat D, Cao YQ, Basbaum AI, Dierich A, Vonesh J-L, Gavériaux-Ruff C, et al. (2006). Knockin mice expressing fluorescent delta-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc. Natl. Acad. Sci. USA 103, 9691–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller PW, Weltrowska G, Nguyen TMD, Wilkes BC, Chung NN, and Lemieux C (1993). TIPP(psi): a highly potent and stable pseudopeptide delta opioid receptor antagonist with extraordinary delta selectivity. J. Med. Chem 36, 3182–3187. [DOI] [PubMed] [Google Scholar]
- Shiwarski DJ, Tipton A, Giraldo MD, Schmidt BF, Gold MS, Pradhan AA, and Puthenveedu MA (2017). A PTEN-Regulated Checkpoint Controls Surface Delivery of δ Opioid Receptors. J. Neurosci 37, 3741–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staus DP, Strachan RT, Manglik A, Pani B, Kahsai AW, Kim TH, Wingler LM, Ahn S, Chatterjee A, Masoudi A, et al. (2016). Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature 535, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanowitz M and von Zastrow M (2003). A Novel Endocytic Recycling Signal That Distinguishes the Membrane Trafficking of Naturally Occurring Opioid Receptors. J. Biol. Chem 278, 45978–45986. [DOI] [PubMed] [Google Scholar]
- Tanowitz M and von Zastrow M (2002). Ubiquitination-independent trafficking of G proteincoupled receptors to lysosomes. J. Biol. Chem 277, 50219–50222. [DOI] [PubMed] [Google Scholar]
- Thompson GL, Lane JR, Coudrat T, Sexton PM, Christopoulos A, and Canals M (2015). Biased Agonism of Endogenous Opioid Peptides at the μ-Opioid Receptor. Mol. Pharmacol 88, 335–346. [DOI] [PubMed] [Google Scholar]
- Tsvetanova NG and von Zastrow M (2014). Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol 10, 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia R and Lindsay RM (1998). Rat Striatal Neurons in Low-Density, Serum-Free Culture In Culturing Nerve Cells, Banker G and Goslin K, eds. (The MIT Press; ), pp. 371–393. [Google Scholar]
- Vilardaga J-P, Jean-Alphonse FG, and Gardella TJ (2014). Endosomal generation of cAMP in GPCR signaling. Nat. Chem. Biol 10, 700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND and McLellan AT (2016). Opioid Abuse in Chronic Pain - Misconceptions and Mitigation Strategies. N. Engl. J. Med 374, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Weber SJ, Abbruscato TJ, Brownson EA, Lipkowski AW, Polt R, Misicka A, Haaseth RC, Bartosz H, Hruby VJ, and Davis TP (1993). Assessment of an in vitro blood-brain barrier model using several [Met5]enkephalin opioid analogs. J. Pharmacol. Exp. Ther 266, 1649–1655. [PubMed] [Google Scholar]
- von Zastrow M and Williams JT (2012). Modulating Neuromodulation by Receptor Membrane Traffic in the Endocytic Pathway. Neuron 76, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.