Abstract
STIM1 and ORAI1 regulate store-operated Ca2+ entry (SOCE) in most cell types, and mutations in these proteins have deleterious and diverse effects. We established a mouse line expressing the STIM1 R304W gain-of-function mutation causing Stormorken syndrome to explore effects on organ and cell physiology. While STIM1 R304W was lethal in the homozygous state, surviving mice presented with reduced growth, skeletal muscle degeneration, and reduced exercise endurance. Variable STIM1 expression levels between tissues directly impacted cellular SOCE capacity. In contrast to patients with Stormorken syndrome, STIM1 was downregulated in fibroblasts from Stim1R304W/R304W mice, which maintained SOCE despite constitutive protein activity. In studies using foetal liver chimeras, STIM1 protein was undetectable in homozygous megakaryocytes and platelets, resulting in impaired platelet activation and absent SOCE. These data indicate that downregulation of STIM1 R304W effectively opposes the gain-of-function phenotype associated with this mutation, and highlight the importance of STIM1 in skeletal muscle development and integrity.
Keywords: Stim1 R304W, muscle degeneration, Stormorken syndrome, Stim1 expression, Platelet dysfunction
Introduction
Stromal interaction molecule 1 (STIM1) is an endoplasmic reticulum (ER) resident transmembrane protein able to sense Ca2+ levels in the ER, the main Ca2+ repository in most cells. The STIM1 N-terminal region is located in the ER lumen, and is separated from the cytosolic C-terminal region by a transmembrane domain. The STIM1 C-terminal interacts with the plasma membrane (PM) resident calcium release-activated calcium modulator 1, ORAI1. A decrease in ER Ca2+ levels is sensed by the EF-hand domain in the STIM1 N-terminal, resulting in its activation, oligomerisation, and finally lateral translocation in the ER membrane. STIM1 interacts with ORAI1 through a coiled coil domain (CC), forming STIM1-ORAI1 puncta at ER-PM junctions [1–2]. Thus, binding of STIM1 activates ORAI1, the pore forming subunit of calcium release activated Ca2+ (CRAC) channel, allowing Ca2+ influx. This mechanism is known as store operated calcium entry (SOCE) [3–5]. STIM1 and ORAI1 together facilitate SOCE in both excitable and non-excitable cells, providing a key mechanism for cellular Ca2+ influx [6–7].
Mutations in ORAI1 or STIM1 have been shown to cause five Mendelian diseases in humans [5, 8–12]. Both biallelic loss of function (LoF) mutations and heterozygous gain of function (GoF) mutations are pathogenic. Biallelic LoF mutations in STIM1 or ORAI1 result in absence of SOCE and patients present with Immunodeficiency 10 (IMD10, OMIM #612783) or Immunodeficiency 9 (IMD9, OMIM #612782) [7, 11]. In mice, global knockout (KO) of Stim1 or Orai1 results in homozygous lethality. Indeed, sixty to seventy percent of homozygous Stim1 or Orai1 KO pups were observed to die within a few hours of birth, while remaining animals exhibited significant growth retardation and increased mortality within the first 4 weeks of life due to muscle weakness or respiratory failure [7, 13–15].
GoF mutations in ORAI1 or in the STIM1 EF-hand domain result in constitutively active SOCE and cause tubular aggregate myopathy 1 or 2 in patients (TAM1, OMIM #160565, TAM2, OMIM #615883) [8, 10, 16]. One mouse model expressing the GoF STIM1 EF-hand mutation D84G (the Stim1Sax mice) has been described [17]. In contrast to patients carrying the same mutation and presenting with TAM1, the heterozygous Stim1Sax mice showed macro-thrombocytopenia and a bleeding disorder. This haematological dysfunction was more severe in the homozygous Stim1Sax mice, which died in utero [17]. No muscular defect has been described in the Stim1Sax mice [17].
Another STIM1 GoF mutation, R304W, is located in the STIM1 coiled-coil 1 (CC1) domain and has been previously shown to result in constitutively active SOCE [9, 12, 18], causing Stormorken syndrome (STRMK, OMIM #185070). The syndrome manifests with a skeletal muscle phenotype (including tubular aggregate myopathy, muscle spasms and weakness, lack of endurance, and increased serum creatine kinase levels) and a haematological phenotype (including mild anaemia and mild bleeding disorder related to thrombocytopathy and thrombocytopenia). The syndrome also manifests miosis, ichthyosis, and asplenia [9, 12, 18–20]. Although GoF mutations in the CC1 domain typically cause the complex Stormorken syndrome, three patients expressing the STIM1 R304Q mutation presented with myalgia and only a single patient exhibited thrombocytopenia [21]. Thus, TAM1 and Stormorken syndrome may be described as part of a clinical continuum, with overlapping manifestations of STIM1 GoF mutations [19, 21].
Due to the diverse and complex phenotype caused by STIM1 R304W in humans we studied its effect on cellular Ca2+ handling and its phenotypic consequences in a Stim1 R304W knock-in mouse model with focus on skeletal muscle and platelets. We found that the mutation caused constitutive STIM1 activation and cell and species specific regulation of the mutant STIM1 expression resulting in muscle degradation and platelet dysfunction.
Methods
Establishment Stim1 R304W mice
The mouse model carrying the Stim1 (NC_000073.6) chr7:102421471A>T (GRCm38.p4) mutation was established using CompoZr™ custom Zinc Finger Nuclease (ZFN) technology (Sigma Aldrich, Missouri, USA) combined with a repair fragment to stimulate homology directed repair (HDR) at the target loci. The Fok-I based DNA binding heterodimeric ZFN pair was custom designed to bind and cleave ~20 bp upstream of the Stim1 target site; 5’-ACCTTGCCAAGCaggaagcTCAGCGGCTGAAGGAGCT-3’ (the ZFN binding sequence in uppercase and the nuclease Fok1 recognition sequence in lowercase). To introduce the mutation, ~1600 dsDNA fragment containing the A>T substitution at the center was synthesized and sequence verified (Genewix Inc., South Plainfield, NJ, USA). Three to four week old female B6CBAF1/JRj mice (Janvier, France) were superovulated by injecting 5 IU pregnant mares’ serum (PMS, Sigma Aldrich) followed by a second injection 45 hours later with 5 IU of human chorionic gonadotropin (HCG, Sigma Aldrich). Superovulated females were subsequently mated with C57BL/6NTac males (Taconic, Ejby, Denmark) and fertilized embryos were collected from oviducts the next morning. The ZFN pair (1 ng/μl) and the repair template (2 or 3 ng/μl) were diluted in EmbryoMax Injection buffer (Speciality media, Sigma Aldrich) and co-injected into zygotes. Injected zygotes were cultured in KSOM (Merck Millipore, Burlington, MA, USA) at 37°C at 5 % CO2 until implanted later the same day. Pseudo-pregnant CD1 (Envigo, The Netherlands) foster mothers were generated by mating with vasectomized CD1 males. Ear biopsies for identification of founders were taken at weaning.
Maintenance of mice
All animals were housed under standard conditions (12:12-hr light/dark, 21±2°C temperature and 55±5% relative humidity) with ad libitum access to water and standard chow. Mice were weighed weekly to monitor their growth. All animal experimental protocols for generation, breeding and use of transgenic animals in Norway were registered and approved by the Norwegian Food Safety Authority (Mattilsynet, Experimental animal welfare supervision and application system ID 7216 and 6991) and followed the ethical guidelines given in Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. The procedures conducted at the University of North Carolina, USA were approved by The University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee (IACUC).
Identification and genotyping of Stim1 R304W mice
Offspring following ZFN modifications were screened for the genomic insertion of the ZFN cassette using Platinum™ Taq DNA Polymerase High Fidelity (Invitrogen, CA, USA) and primers Stim1-F1 and Stim1-R1, followed by Sanger sequencing (BigDye™ Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems, CA, USA) (primer sequences listed in the Supplementary table 1). Animals positive for Stim1 (NC_000073.6) chr7:102421471A>T (GRCm38.p4) were then selected to determine if the mutation was specifically targeted into the Stim1 gene in the mice. A PCR fragment of 2166 bp covering the complete ZFN cassette was amplified with long range PCR (TaKaRa LA DNA polymerase, Takara Bio USA, Inc., CA, USA) using primers: Stim1-F2 and Stim1-R2 upstream and downstream of the ZFN cassette flanking the gene (Supplementary Table 1). Sanger sequencing of the complete fragment was performed using the primers designed along the length of the fragment; Stim1-F3, Stim1-R3, Stim1-F4, Stim1-R4, Stim1-R6 (Supplementary Table 1). All animals negative for the A>T substitution at the target site and/or with additional mutations detected in the cassette were excluded. The 2166 bp PCR-fragments from the remaining animals were cloned by TOPO TA cloning according to the manufacturer’s instructions (TOPO TA cloning kit for sequencing, Invitrogen). Sanger sequencing of 10 independent clones from the founder animal showed only the Stim1 mutation at Chr7:102421471bp (A>T) and no additional modifications in the same allele. The sequence reads were assembled using Sequencing Analysis software v5.2 (Applied Biosystems) and aligned with the mouse reference genome in the UCSC genome browser (Mouse Dec. 2011 (GRCm38/mm10) Assembly) and MEGA4 software [22]. The mouse line described here is referred to as Stim1R304W and originated from one founder female backcrossed to C57bl/6J male mouse (Envigo) and further intercrossed to establish the Stim1 R304W colony.
Whole genome seauencing of the Stim1R304W female founder mouse
Whole genome sequencing (WGS) of the female founder mouse DNA was performed (Novogene, China). In brief, reads passing quality filters were mapped to the GRCm38.p4 reference genome using the BWA-MEM algorithm [23]. SAMtools was used to remove duplicates and to sort the reads in the resulting bam file [24]. Metrics and statistics of coverage and depth were collected with Picard version 2.12.1 (Broad Institute, Cambridge, MA, USA). For the detection of any structural variation caused by transgene integration, paired-end reads and split-read based variant calling analysis were performed with the software DELLY [25] and Breakdancer [26]. SNP and InDel variations were detected by SAMtools mpileup [27]. The high confidence integration sites reported by variant calling were extracted and further checked as visualized by The Integrative Genomics Viewer (IGV) [28], and the copy number of each transgene was assumed by their effective depth relative to the genome-wide sequencing depth. A total of 779337818 reads were obtained with a 99.28% mapping rate. The whole genome coverage was 91% ≥1x, 90.1% ≥ 5x and 88% ≥ 15x.
Material from patients with Stormorken syndrome and healthy controls
Cultured skin derived fibroblast cells and whole blood samples from two patients with Stormorken syndrome (F1 III-1 and F2 II-3 in Misceo. et al 2014) were obtained [9]. Similarly derived, fibroblast cell cultures from two healthy individuals were used as controls for SOCE experiments. Blood samples from two healthy individuals were used as controls for platelet-protein preparation. All samples were obtained following informed consent.
Real time qPCR of Stim1 cDNA
RNA was harvested from whole blood in PAX-solution as described previously [29]. RNA concentration was measured with Qubit nanodrop RNA high sensitivity (Thermo Fischer Scientific, Waltham, MA, USA), and quality assessed with Agilent BioAnalyzer RNA Nano (Agilent Technologies, Waldbronn, Germany). Samples with RIN values below 7 were excluded from experiment. cDNA was synthesized from 0.25–1μg RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
qPCR was run in an QuantStudio 12k instrument, using manufacturers software and instructions (Thermo Fischer). SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma Aldrich) was used for the amplification of cDNA with Stim1 specific primers; Stim1_cDNA_F, Stim1_cDNA_R (Supplementary Table 1). All primer pairs were tested for efficiency with a 5-point dilution series, and melt curve analysis was used to confirm the presence of a single amplicon for each reaction. Data was analysed by normalizing against three reference genes; Rplp0, Ppib and Eef2 (Supplementary Table 1).
Examination of mouse embryos
Mouse embryos collected at 12.5, 13.5, 14.5, 15.5, 16.5 and 17dpc were evaluated macroscopically for gross abnormalities and signs of bleeding. A blood sample was collected for quantification of platelets and tail biopsies were collected for genotyping and gender determination. Embryos at 14.5 and 16.5dpc were fixed, paraffin embedded and sectioned across the midline to obtain whole mount sections of the embryo and stained with Hematoxylin and eosin (HE). Following deparaffinization and antigen retrieval according to the manufacturer’s instructions, sections from 14.5 and 16.5dpc embryos were immuno-stained with an antibody binding the STIM1 N-terminal (LS-B10807–50, LS-Bio Inc., Seattle, WA, USA). The anti-rabbit IgG-Alexa 488 (Thermo Fischer Scientific) was used as secondary antibody to detect and visualise STIM1 and evaluate the expression in embryonic tissues.
Whole cell measurements of Ca2+ concentration and SOCE in cultured mouse neonatal fibroblast cells
Fibroblasts were grown from skin biopsies harvested from 0–2 days old mice in 10% FBS and 10 mM PenStrep supplemented DMEM media (Gibco, Thermo Fischer Scientific). Cells were harvested after two weeks and plated on glass coverslips coated with gelatin-fibronectin and incubated overnight at 37°C supplemented with 5% CO2. The fibroblast-containing coverslips were then loaded with Ca2+ measuring dyes, Indo-1-AM (Molecular probes/Invitrogen, Carlsbad, CA, USA) or Fluo-4-AM (Molecular probes/Invitrogen) for 20 and 15 minutes, respectively, and the experiments were conducted as previously described [9]. Indo-1-AM loaded cells were superfused with Hepes Tyrode (HT) buffer (140 mM NaCl, 0.5 mM MgCl2, 5.0 mM HEPES, 5.5 mM glucose, 0.4 mM NaH2PO4, 5.4 mM KCl, pH 7.4 at 37°C) containing 2 mM Ca2+ and whole cell fluorescence was measured. The corresponding resting [Ca2+]i was calculated as described previously [9]. For assessment of SOCE, Ca2+ stores were first depleted by application of the SERCA inhibitor thapsigargin (2 μM, Sigma Aldrich) in the presence of Ca2+-free HT for 5 min, followed by rapid reintroduction of 10 mM Ca2+. SOCE was calculated as the resulting increase in fluorescence intensity, and calibrated to [Ca2+] as previously described [9]..
Transfection of NIH 3T3 cells
NIH 3T3 mouse embryonic fibroblast cell line (Sigma Aldrich) was grown in DMEM with high glucose (4500 mg/L) (Sigma Aldrich), supplemented with 10% Bovine Calf Serum (BCS, Thermo Fisher Scientific), 2% PenStrep (Thermo Fisher Scientific) and 2mM L-glutamine (Thermo Fisher Scientific). The cells were seeded on glass coverslips in a 6 well plate coated with gelatin-fibronectin and incubated overnight prior to transfection with Polyjet transfection reagent (SignaGen Laboratories, MD, USA). Vectors expressing mouse STIM1_WT-YFP or STIM1_R304W-YFP [12] (kind gift from Dr. Leonidas Tsiokas, University of Oklahoma Health Sciences Centre, USA) were transfected with a vector expressing mouse Orai1 in a 1:1 ratio (kind gift from Dr. Stefan Feske, New York University Medical Centre, USA). Cells were imaged at 24 hours post-transfection using Zeiss LSM 800 with Airyscan (ZEISS microscopy, Oberkochen, Germany) microscope and with ZEN imaging software (ZEISS microscopy). Images were analysed to measure the intensity of YFP signals using Fiji image processing software [30]. Raw images were cropped to remove any debris or secondary cells in the field of view and analysed with a macro which used contrast enhancement, background subtraction, blurring and binarization. Regions of interest identified by elevated fluorescent signals were analysed to determine the number and intensity of STIM1 aggregates.
Western blotting
Whole cell protein lysates were prepared from primary fibroblasts and NIH 3T3 cells using 1x RIPA buffer (Sigma Aldrich). Cells were washed twice with 1x PBS, pH 7.4 (Gibco, Life technologies) and the cell pellet was lysed with 1x RIPA buffer supplemented with Protease and Phosphatase inhibitor cocktail (Thermo Fischer Scientific) for 15 minutes at 4°C followed by centrifugation at 13000 rpm for 15 minutes at 4°C.
Tissue lysates from snap frozen skeletal muscle samples were made with 1x RIPA buffer using gentleMACS™ Octo Dissociator (Miltenyi Biotec, Germany). Lysates were centrifuged at 13000 rpm for 15 minutes at 4°C and supernatant stored at −80°C until use.
Protein concentrations were measured with Pierce BCA Protein Assay kit according to manufacturer’s instructions (Thermo Fischer Scientific). Protein lysates from washed platelets (described in flowcytometry studies on platelets section) were prepared using 2x RIPA buffer. All western blots were performed according to standard protocols using Mini-PROTEAN TGX Stain-Free Precast Gels and apparatus (Bio-rad Laboratories, CA, USA). List of the antibodies used is provided in Supplementary Table 2.
Necropsy
A full necropsy was performed on eight mice per genotype in Stim1+/+ and Stim1R304W/+ and three Stim1R304W/R304W mice euthanized at 5–6 months age. Tissues were harvested and snap frozen and/or fixed in 10% neutral buffered formalin. Blood samples were collected by cardiac puncture to measure blood cell counts, total Ca2+, total protein, haemoglobin and albumin.
Histological staining of formalin-fixed tissue sections was performed according to routine protocols. A standard examination for muscle degeneration parameters were done on HE stained sections of quadriceps muscle and the sections were evaluated blindly for the severity of the phenotype in each genotype. Muscle degeneration was evaluated and scored for variation in fibre size, loss of striation, hypereosinophilia, centralised nuclei and inflammation/inflammatory cells in HE stained sections of quadriceps muscle. A severity score of 0–3 was used with 0 being unaffected, 1-weak/present, 2-medium and 3-severe.
Muscle fibrosis and calcinosis was evaluated in formalin-fixed heart tissue using van Gieson stain (Elastica van Gieson staining kit, VWR, Pennsylvania, USA) and Von Kossa stain [31] respectively. Lung pathology was evaluated and scored for alveolar oedema, congestion and alveolar histiocytosis in HE stained sections of lung tissue. A severity score of 0–3 was used with 0 being unaffected, 1-weak/present, 2-medium and 3-severe.
General locomotor activity and behavioural testing
General locomotor activity and behaviour examinations were carried out on 5–6 month old male Stim1+/+ and Stim1+/R304W animals (n=10 per genotype) in an open field test and endurance test in that order. To reduce bias, the experimental parameters were scored blinded to the genotypes. Mice were video-tracked during experiments (ANY-Maze™ Behavioural tracking software, Dublin, Ireland). All parameters were compared between Stim1+/+ and Stim1+/R304W animals.
In the open field test, mice were introduced to a brightly lit closed maze and were monitored and video tracked for 45 minutes. The distance travelled, time spent in the centre of the maze, time spent mobile and maximum speed were monitored and automatically analysed for each mouse with the tracking system.
The endurance tests were performed in a 5-lane mouse treadmill with an electric grid (Panlab-Harvard apparatus, Barcelona, Spain). Mild electric shock of 0.2 A (adjustable 0 – 2 A) activated at 5 seconds intervals was used as motivation for mice to run. Mice were accustomed to the instrument for two days prior to test. Exercise endurance was tested by running mice till exhaustion which was defined as inability to continue running following mild electric shock, standing on electric grid for >4 seconds or running close to the grid for longer than 5 seconds. The time and distance run before reaching exhaustion were recorded.
Experiments on isolated skeletal muscle fibres
Skinned hind feet were transported overnight to the laboratory at room temperature in oxygenated 10% foetal calf serum (Gibco) supplemented DMEM media (Sigma Aldrich). The flexor digitorum brevis (FDB) muscle was split into individual digits and the central three digits were used in these experiments. Muscle fibres were isolated with a pair of micro-iris scissors and jeweler’s forceps [32].
Fibres were chosen if it contracted and then relaxed upon stimulation with an electrode that delivered 1 ms, 5–8 V pulses. All other fibres were killed and removed to leave a single fibre. Platinum micro-clips were attached to the residual tendinous material and mounted in a perfusion chamber between a force transducer (AE801, Kronex Technologies, CA, USA) and an adjustable holder.
Fibres were superfused with Tyrode solution containing 121 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 0.5 mM MgCl2, 0.4 mM NaH2PO4, 24 mM NaHCO3, 0.1mM EDTA, and 5.5 mM glucose at 31°C which is the temperature that the muscle fibres experience in the living foot. Solution was supplemented with 0.2% FBS and bubbled with 95% O2 – 5% CO2, giving a pH of 7.4. The fibre was stimulated with 15 to 150Hz, 350 ms tetani of supramaximal electrical pulses (0.5 ms in duration) delivered via platinum electrodes placed parallel to the long axis of the fibre. The length of the preparation was adjusted to the length at which tetanic force was maximal. Tetanic force was normalized to cross-sectional area and expressed as specific force (kN/m2).
To measure resting and tetanic [Ca2+]i, muscle fibres were pressure injected with the fluorescent indicator Indo-1 (Molecular Probes/Invitrogen, Carlsbad, CA, USA). Indo-1 was excited at 360 ± 5 nm and the light emitted at 405 ± 5 and 495 ± 5 nm was measured with a pair of photomultiplier tubes (Photon Technology International, Wedel, Germany). Indo-1 fluorescence signals were converted into [Ca2+]i as described previously [33]. Mean resting [Ca2+]i was measured during the 200 ms preceding a tetanus. Tetanic [Ca2+]i was measured as the mean of the final 250 ms of the 350 ms tetanus and tetanic force was measured as the peak force during the tetanus.
The force-frequency relationship was determined for each fibre by producing single tetanic contractions at different frequencies at 1 minute intervals. Fatigue was induced by stimulating fibres with a train of 150 tetani (70 Hz, 500 ms) given at 2 second intervals. Ca2+ entry into the fibre was assessed by a Mn2+-quench method which relies on the fact that Mn2+ reduces the fluorescent signal of many Ca2+ sensitive indicators including Indo-1 and is not taken up by the SR Ca2+-pumps [34]. Rested fibres were exposed to Tyrode solution containing 1 mM Mn2+ and the 405 nm light emission of Indo-1 was followed before, during and after exposure to Mn2+.
Preparation of STIM1 R304W foetal liver cell chimeras
Foetal liver cells from E14.5 mouse embryos derived from Stim1+/R304W mice inter-crosses were used to generate Stim1+/+→WT, Stim1+/R304W→WT, and Stim1R304W/R304W→WT chimeras as previously described [35]. The platelet specific STIM1 KO mice; Stim1fl/flPF4-Cre have been previously described [36].
Flow cytometry studies on platelets
All flow cytometry experiments were performed using washed platelets or diluted whole blood collected into heparinised capillary tubes. Washed platelets were prepared as described previously [37]. In brief, centrifugation of diluted whole blood at 130 g for 4 minutes to obtain platelet rich plasma (PRP) followed by pelleting of platelets in the PRP in the presence of prostaglandin I2 (PGI2, Caymen Chemical, Michigan, USA). The pellet was washed twice, and the platelets re-suspended in modified Tyrode’s buffer at pH 7.4.
Platelet counting was performed in a 5 μl aliquot of heparinised whole blood labelled with the platelet specific antibody detecting GPIX (anti GPIX, clone Xia-B4, Emfret analytics) labelled with AlexaFluor-647 for 10 minutes at room temperature. The samples were diluted with 1x PBS and immediately analysed on an Accuri C6 flow cytometer (BD Bio Sciences, CA, USA). Platelets were gated on GPIX labelling and forward scatter parameters.
In order to measure platelet activation, whole blood was diluted in Tyrode’s buffer containing 2 mM CaCl2 (1 mM final), activated with PAR4p or Cvx for 10 minutes in the presence of antibodies recognizing activated integrin αIIβ3 (JON/A-PE, Emfret analytics, Eibelstadt, Germany) and P-selectin expression (α-P selectin-FITC, BD Biosciences), diluted and analysed on an Accuri C6 flow cytometer. Platelets were gated on GPIX-Alexa 647 labelling and JON/A and P-selectin antibody binding was determined by mean fluorescence intensity (MFI). PS exposure was measured in washed platelets activated with PAR4p (400 μΜ) + Cvx (Low dose or High dose; Appropriate doses were determined by testing activation of wild-type platelets) for 10 mins in the presence of anti-GPIX-647 antibody (2 μg/ml) and AnnexinV-488 (2 μg/ml) in 1mM Ca2+ supplemented Tyrode buffer.
In order to measure Ca2+ fluxes, washed platelets were incubated with 5 μM Fluo-4-AM for 30 minutes in the absence of Ca2+ and then treated with 1 μM thapsigargin followed by re-introduction of 2 mM Ca2+. Fluo-4 intensity was then measured over time.
For the measurement of STIM1 expression in platelets, whole blood was fixed in 4% PFA for 10 mins at RT. The cells were pelleted, and the RBCs were lysed with RBC lysis buffer for 10 min. Samples were then centrifuged and supernatant removed. The platelet/WBC pellet was washed with 1x PBS and cell counts were normalized between samples. The cells were then permeabilized with 0.5% Triton X-100 (Sigma Aldrich) for 5 mins at RT. The cell pellet obtained after centrifugation was resuspended with PBS+1% BSA and stained with STIM1 (5ug/ml final concentration) for 10 mins at RT and co-stained with GPIX-Alexa 647 platelet marker. Secondary antibody, Alexa-488- α-rabbit was used to detect the STIM1 antibody in a 1:1000 dilution. Cells were pelleted, re-suspended in 1x PBS and analysed by flow cytometry.
Statistics
Results are presented as mean ± SEM. Chi-square test was used to calculate the statistical significance in the deviation from Mendelian birth ratio in the mouse line. Differences between Stim1+/+ and Stim1+/R304W or Stim1R304W/R304W were assessed using Student’s t-test. Graph Pad Prism 7 (GraphPad Software Inc., CA, USA) was used for statistical analyses and graph plots.
Statistical analyses of [Ca]i were performed using the SigmaPlot software (Systat Software, Inc., San Jose, CA, USA). Unpaired or paired t-tests were applied to compare the differences between groups. A p-value <0.05 was defined as statistically significant.
Results
Generation of the Stim1R304W mouse model
A knock-in mouse line expressing the Stim1 (NC_000073.6) chr7:102421471A>T (GRCm38.p4) mutation was established using the CompoZr™ ZFN technology [38]. Out of 144 progeny, two heterozygous Stim1+/R30AW founder females were identified. Targeted insertion of the Stim1 mutation was confirmed in the two founders by Sanger sequencing of a PCR fragment obtained with primers flanking the region corresponding to the ZNF modification template (chr7:102420366–102422512 bp, GRCm38/mm10). One founder died spontaneously before breeding, and thus the line described here originated from a single female founder mouse. Whole genome sequencing of DNA from the founder mouse detected no off-target insertions of the repair template in the genome and confirmed the specific Stim1 chr:7102421471A>T substitution with no other modifications in Stim1. Thus, the mouse line generated carried a mutation that resulted in the R304W substitution in STIM1, and is hereafter referred to as Stim1R304W.
Lethality of the Stim1R304W/R304W mice
Crossbreeding of heterozygous animals was performed to generate homozygous mice. Following 25 heterozygous crosses, 130 offspring survived more than three days. The total number of pups obtained and the genotype ratio in these mice, 36:91:3 (Stim1+/+:Stim1+/R304W:Stim1R304W/R304W), deviated significantly from the expected Mendelian ratio (chi-square test: p<0.0001) (Supplementary Table 3), highlighting the lethality of the Stim1R304W/R304W mice.
Examination of embryos following 14 Stim1+/R304W x Stim1+/R304W timed crosses revealed litter sizes ranging from 2 to 11 embryos (average 6.6), with less than 10 residual bodies or resorption sites observed in total. The genotype ratio in embryos extracted at 12.5, 13.5, 14.5, 15.5, 16.5 and 17 dpc was 20:53:19 (Stim1+/+:Stim1+/R304W:Stim1R304W/R304W), which did not deviate from the Mendelian ratio (Supplementary Table 4). The pups found dead or euthanized at P0-P1 stages showed a genotype ratio of 14:27:25 (Stim1+/+:Stim1+/R304W:Stim1R304W/R304W), demonstrating no decrease in the fraction of Stim1R304W/R304W pups at parturition. Taken together these data show survival of Stim1R304W/R304W embryos up to late gestational stages with subsequent perinatal lethality.
On inspection, the Stim1R304W/R304W embryos appeared slightly smaller than their littermates, without apparent dysmorphisms (Figure 1A). As cyanosis was not observed in the dead P0-P1 pups, respiratory distress or defects in blood oxygenation were considered unlikely causes of death. Histological examinations of the diaphragm performed in formalin-fixed, paraffin-embedded sections from three 14.5 dpc Stim1R304W/R304W embryos revealed no obvious abnormalities in morphology or thickness of the diaphragms (Figure 1B).
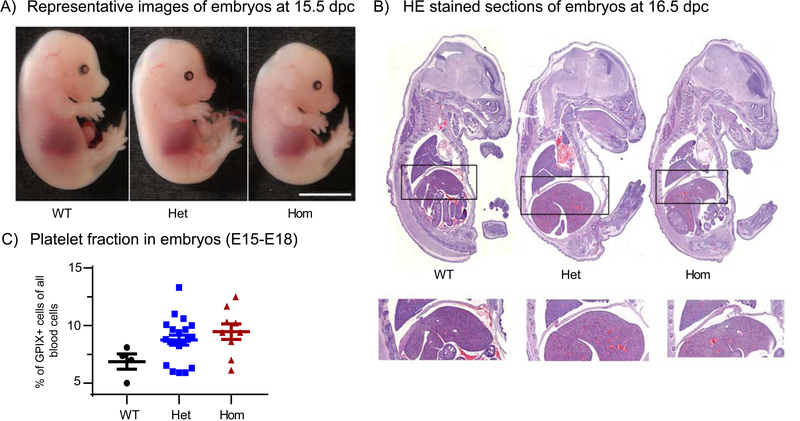
Figure 1. Evaluation of Stim1R304W embryos.
(A) Representative images of embryos at 15.5 dpc showed overall normal morphology and development with slightly smaller body size in Stim1R304W/R304W embryos (Hom) compared to heterozygous Stim1+/R304W (Het) and Stim1+/+ (WT) littermates. Scale bar= 5mm.
(B) No apparent abnormalities in morphology were detected in the HE stained sections of paraffin embedded embryos at 16.5 dpc. The diaphragms (indicated by black boxes) of the analysed sections showed no signs of abnormal thickness or degeneration (bottom).
(C) Whole blood from embryos at E15-E18 stages showed similar fraction of platelets in WT (n=4), Het (n=20) and Hom (n=9) mice. The platelet fractions were measured by flow cytometry as the percentage of cells positive for the platelet marker GPIX.
Guided by the thrombocytopenia and bleeding diathesis in patients with Stormorken syndrome expressing the STIM1 R304W GoF mutation and by the lethal bleeding disorder observed in Stim1Sax GoF mouse model [9, 17], embryos were examined macroscopically for signs of bleeding and blood and lymphatic vascular mixing. No visible signs of bleeding or differences in the fraction of platelets were detected in blood from Stim1R304W/R304W embryos at 15–17 dpc compared to their Stim1+/R304W and Stim1+/+ littermates (Figure 1A and C). Thus, bleeding and thrombocytopenia were unlikely to underlie the lethality documented in the homozygous Stim1R304W mice.
The mutant allele is expressed in the Stim1R304W mice
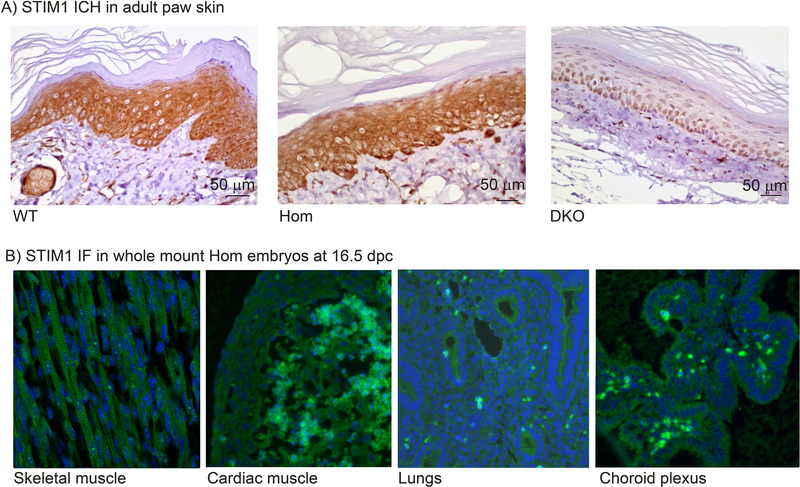
To verify the expression of the Stim1 mutant allele, immunohistochemistry (IHC) staining was performed on skin sections from homozygous mice. Specific STIM1 staining was detected in Stim1+/+ and Stim1R304W/R304W mice (Figure 2A, left and center), but not in cells from an epithelial cell specific Stim1/Stim2 double KO mouse (DKO) [39], which served as a negative control (Figure 2A, right). Furthermore, STIM1 expression was detected by immunofluorescence (IF) microscopy in tissues of endodermal, mesodermal, and ectodermal origin including skeletal muscle, heart, lungs and choroid plexus in the Stim1R304W/R304W mice (Figure 2B).
Figure 2. Stim1 expression in tissues from Stim1R304W mice.
(A) STIM1 IHC in adult paw skin demonstrates strong cytoplasmic STIM1 staining (brown) in epidermal keratinocytes in Stim1+/+ (WT, left), and Stim1R304W/R304W mice (Hom, middle). No staining was detected in the epithelial cell specific Stim1/2 double KO mouse (DKO, right) [39].
(B) STIM1 immunofluorescence (IF) in whole mount homozygous Stim1R304W embryos at 16.5 dpc demonstrates STIM1 staining (green) in cells of mesodermal origin (skeletal muscle cells and cardiac muscle cells), in cells of endodermal origin (lung cells), and in cells of ectodermal origin (cells in choroid plexus), as indicated.
Constitutive STIM1 activation in Stim1R304W mice
To explore the functional effect of the mutation in the Stim1R304W mice, we performed whole cell Ca2+ flux measurements in cultured skin-derived fibroblasts from P0-P2 day old mice. Representative traces are illustrated in Figure 3A (top panels). At baseline, intracellular cytosolic Ca2+ levels ([Ca2+]i) were significantly lower in fibroblasts from Stim1+/R304W mice than in wildtype mice (p<0.02), and also tended to be lower in Stim1R304W/R304W fibroblasts (Figure 3A, lower-left panel). SOCE was assessed by depleting Ca2+ stores with thapsigargin in Ca2+-free conditions, followed by subsequent reintroduction of Ca2+. Similar magnitude SOCE was observed in wildtype, Stim1+/R304W and Stim1R304W/R304W cells (Figure 3A, lower-right panel). In contrast, skin-derived fibroblasts from patients carrying the same mutation showed significantly elevated resting [Ca2+]i under basal conditions (p<0.02) and increased SOCE compared to healthy controls (p<0.05) (Figure 3B).
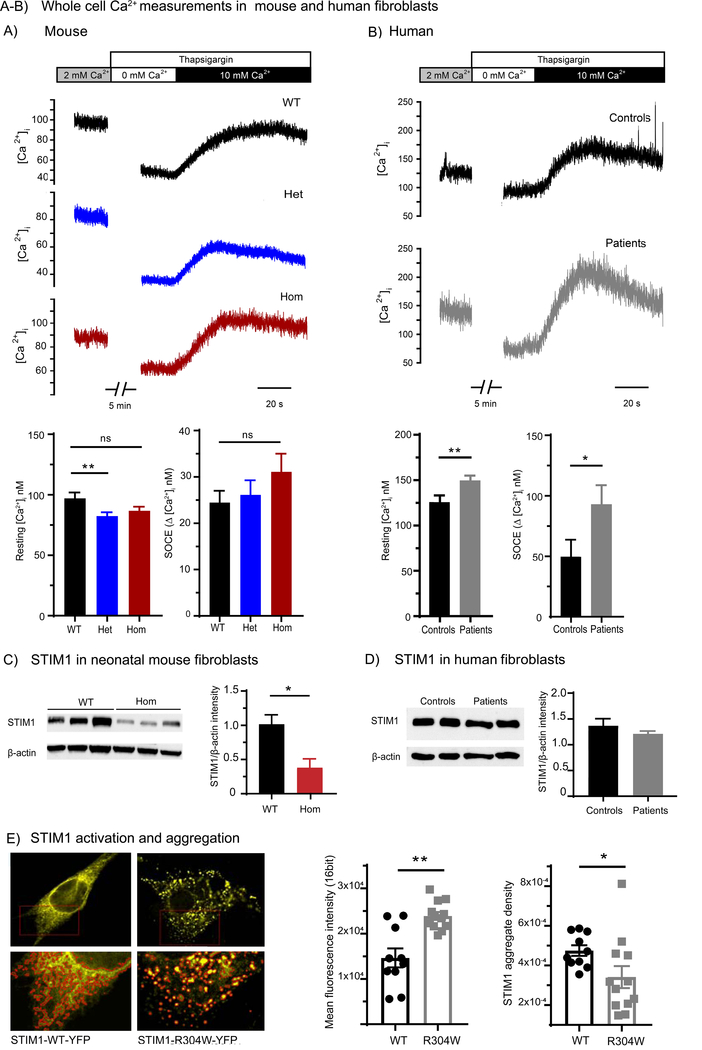
Figure 3. Calcium handling in human and neonatal mouse fibroblast cells.
(A) Whole cell Ca2+ measurements were performed in cultured skin-derived fibroblast cells from neonatal mice. Representative recordings and mean data (top and lower panels, respectively) show that under baseline conditions, resting [Ca2+]i was significantly lower in fibroblasts from heterozygous Stim1+/R304W mice (Het, n=93 cells) compared to wildtype Stim1+/+ (WT, n=76 cells, p<0.02), and tended to be lower in homozygous Stim1R304W/R304W (Hom, n=95 cells). Following Ca2+ store depletion with 2 μΜ thapsigargin and Ca2+-free conditions, reintroduction of 10 mM Ca2+ revealed similar magnitude SOCE in WT, Het, and Hom cells (n=66, 86, 78 cells, respectively).
(B) A similar experiment performed in human fibroblast cells showed significantly elevated resting Ca2+ levels in cells from patients with Stormorken syndrome (n=39 cells) compared to healthy controls (n=39 cells) (p<0.02). Fibroblasts from the two patients (n=26 cells) also exhibited augmented SOCE compared to control cells (n=14 cells) (p<0.05).
(C) Western blot results showed significant reduction of STIM1 expression in fibroblasts from homozygous animals (Hom) compared to Stim1+/+ (WT) (p<0.05); β-actin was used as a loading control.
(D) No difference in STIM1 expression was detected in fibroblast cells of two patients with Stormorken syndrome [9] compared to healthy controls, as indicated.
(E) Representative images of NIH 3T3 murine cells transfected with mouse STIM1-WT-YFP or STIM1-R304W-YFP plasmids. Cells transfected with mouse STIM1-R304W-YFP showed increased clustering of YFP signal at resting conditions indicating constitutive STIM1 aggregation and activation (left). Measurement of fluorescence intensity of STIM1-YFP signal in regions of interest (red outlines in lower panel) in STIM1-R304W-YFP (n=12) transfected cells showed a 28% decrease in aggregate density (p<0.05) compared to STIM1-WT-YFP (n=10) and a 63% increase in mean intensity (p<0.02) (right).
In all panels, p-values for significance are indicated as: ns=not significant, *=p<0.05, **=p<0.02, ****=p<0.0001.
Measurements of STIM1 protein levels in homozygous skin-derived fibroblasts showed an approximate 3-fold reduction in STIM1 expression in Stim1R304W/R304W mice compared to Stim1+/+ littermates (p<0.05) (Figure 3C), while STIM1 protein level was comparable in fibroblasts from patients and controls (Figure 3D). Thus, in the fibroblasts from patients, the expression of the constitutively active R304W mutation in STIM1 at normal protein levels resulted in increased resting [Ca2+]i, and SOCE. In contrast, over-activity of STIM1 was counterbalanced in Stim1R304W/R304W mice by reduced protein expression.
To further characterise the effect of the Stim1 R304W mutation in murine cells, YFP tagged mouse STIM1-WT and mouse STIM1-R304W expression vectors were co-transfected with untagged wildtype mouse Orai1 expression vector into NIH 3T3 cells. Greater aggregation of the YFP-signal was detected under resting conditions in cells transfected with the STIM1-R304W-YFP vector than in those transfected with STIM1-WT-YFP (Figure 3E). This was manifested as distinct fluorescent puncta, found at lower density than in those in WT-transfected cells (p<0.05), with increased fluorescence intensity (p<0.02) (Figure 3E graphs). These findings show that the R304W mutation results in constitutive STIM1 activation and oligomerization.
Stim1R304W/R304W mice show reduced muscle mass and reduced endurance
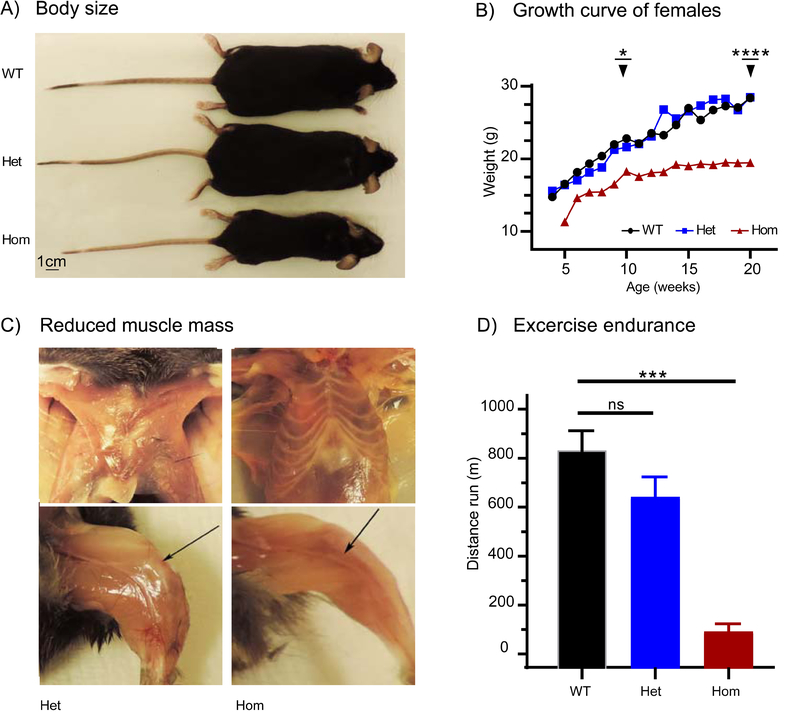
Three homozygous Stim1R304W female animals survived until they were euthanized and necropsied at 5–6 months of age. They showed reduced body size (Figure 4A) and significantly reduced body weight compared to Stim1+/+ and Stim1+/R304W littermates (p<0.05 at 10 weeks age and p<0.0001 at 20 weeks age) (Figure 4B). They also exhibited reduced hind limb muscle mass and a near absence of pectoral muscles in comparison with their Stim1+/+ and Stim1+/R304W littermates (Figure 4C).
Figure 4. Reduced growth and endurance in Stim1R304W mice.
(A) Homozygous Stim1R304W/R304W (Hom) mice showed reduced overall body size compared to their heterozygous Stim1+/R304W (Het) and wild type Stim1+/+ (WT) littermates. Mice were 5 months old at the time of imaging. Scale bar= 1 cm.
(B) Female wildtype and heterozygous mice followed similar growth curves, while female homozygous mice showed reduced linear growth. Each curve represents an average weight of ≥3 animals with significant differences at age 10 weeks (p<0.05) and 20 weeks (p<0.0001) between wildtype and homozygous mice.
(C) Homozygous Stim1R304W/R304W (Hom) mice showed reduced pectoral muscle mass and hindlimb muscle mass (arrow, bottom) compared to their heterozygous littermates (Het).
(D) Test of endurance in wildtype, heterozygous and homozygous mice measured with distance run on treadmill showed significantly reduced endurance in homozygous mice (Hom, n=3) compared to age matched wildtype mice (WT, n=21) (p<0.005). Endurance in heterozygous mice (Het, n=21) was similar to the wildtype controls.
In all panels, p-values for significance are indicated as: ns=not significant, *=p<0.05, ***=p<0.005, ****=p<0.0001.
There were no significant differences in general locomotor activity such as distance travelled, average speed, maximum speed and total time mobile in an open-field test in the Stim1+/R304W compared to Stim1+/+ mice (n=10 per genotype, data not shown). However, in endurance examination, performance was severely reduced in the three surviving Stim1R304W/R304W mice compared to Stim1+/R304W and wildtype mice (p<0.002, Figure 4D).
Stim1R304W mice show skeletal muscle degeneration
Histological evaluation was performed in order to assess if heart or lung pathology may play a role in the reduced endurance observed in the Stim1R304W/R304W mice. No connective tissue ingrowth or Ca2+ salt deposits were detected by van Gieson stain and Von Kossa stain in the ventricular walls or in the atrium. Hematoxylin and eosin (HE) staining of the lung sections in the homozygous Stim1R304W mice showed no alveolar oedema, congestion or alveolar histiocytes, indicating an absence of cardiac pulmonary pathologies (data not shown). These results suggest that the reduced endurance is not related to heart and lung defects in these animals.
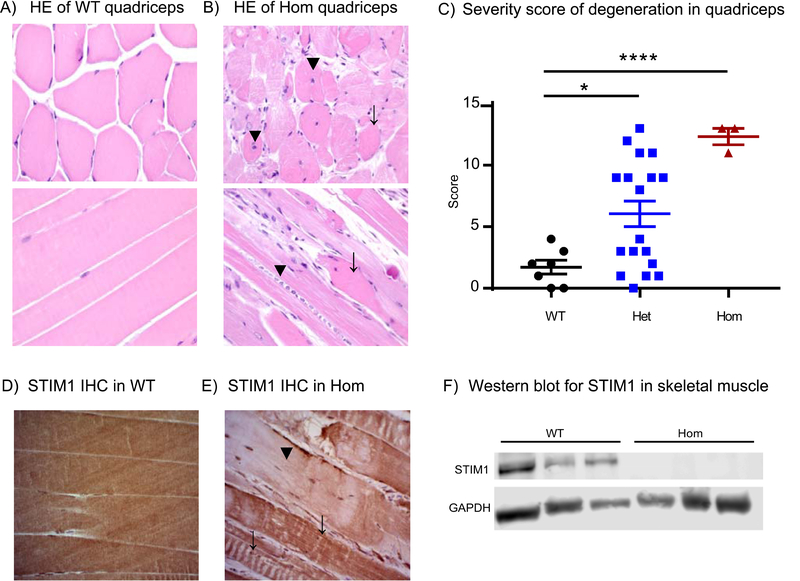
HE staining of quadriceps muscles from 5–6 months old animals showed muscle degeneration in Stim1+/R304W mice of both genders and in the three surviving Stim1R304W/R304W mice. Staining in quadriceps muscle cross sections in Stim1R304W/R304W mice showed marked variation in fibre size and infiltration of mononuclear cells indicating inflammation compared to wildtype mice (Figure 5A-B, top). The number of muscle fibres with centralised nuclei were increased in the Stim1+/R304W (data not shown) and Stim1R304W/R304W mice compared to wildtypes (Figure 5B, top), indicating both muscle degeneration and regeneration. Longitudinal sections of muscles from Stim1R304W/R304W mice showed an increased number of hypereosinophilic cells and loss of the characteristic striation pattern (Figure 5B, bottom). Blinded severity scoring of muscle fibre size, loss of striation, hypereosinophilia, centralised nuclei, and inflammation showed a significantly increased severity score of degeneration in Stim1R304W/R304W (p<0.0001) mice compared to the wildtype mice (Figure 5C). Severity scoring in quadriceps from Stim1+/R304W mice also showed a significantly increased mean score (p<0.05) compared to the wildtypes. No signs of muscle degeneration were observed in the age-matched Stim1+/+ animals. HE stains of diaphragm in adult Stim1+/R304W showed a slight increase in the mean score of degeneration of the diaphragm compared to Stim1+/+. However, the scoring showed a variation between the heterozygous animals, while the one Stim1R304W/R304W diaphragm analysed showed an elevated score (Supplementary Figure 1).
Figure 5. Skeletal muscle degeneration in Stim1R304W mice.
(A) Cross-section (top) and longitudinal section (bottom) of HE stained quadriceps muscles showed regular fibre structure and diameter and peripheral nuclei with presence of characteristic striation in Stim1+/+ wildtype mice (WT).
(B) Stim1R304W/R304W homozygous (Hom) mice showed basophilic fibres containing centralized nuclei (top, arrowheads), irregular fibre diameter and staining intensity (top, arrow), aggregates of inflammatory cells (bottom, arrowhead) and degenerated muscle fibres (bottom, arrow).
(C) Severity scoring of skeletal muscle degeneration parameters of mice with all three genotypes. Quadriceps muscle showed significant degeneration in Stim1+/R304W heterozygous (Het, n=18) (p<0.05) and homozygous (Hom, n=3) (p<0.0001) compared to wildtypes (WT, n=7). Sections were scored for degeneration on variation in fibre size, loss of striation, hypereosinophilia, centralised nuclei and inflammation/inflammatory cells with a score of 0–3 (0-unaffected, 1-weak/present, 2-medium and 3-severe) with a potential maximum total score of 15.
(D) IHC show homogenous STIM1 staining in quadriceps muscle fibres from Stim1+/+ mice.
(E) Some STIM1 staining was detected (arrow) but most was lost in degenerate fibres in quadriceps from homozygous mice (arrowhead).
(F) Western blot on proteins extracted from frozen skeletal muscle samples shows STIM1 expression in lysates from Stim1+/+ (WT) but not in lysates from homozygous animals (Hom); GAPDH was used as a loading control.
p-values for significance are indicated as: *=p<0.05, ****=p<0.0001.
STIM1 expression was detected in skeletal muscle in the wildtype and most fibres from three surviving Stim1R304W/R304W mice on IHC stained sections of the quadriceps (Figure 5D-E). However, the STIM1 level was too low to be detected on western blots indicating a marked reduction in STIM1 expression relative to wildtype (Figure 5F).
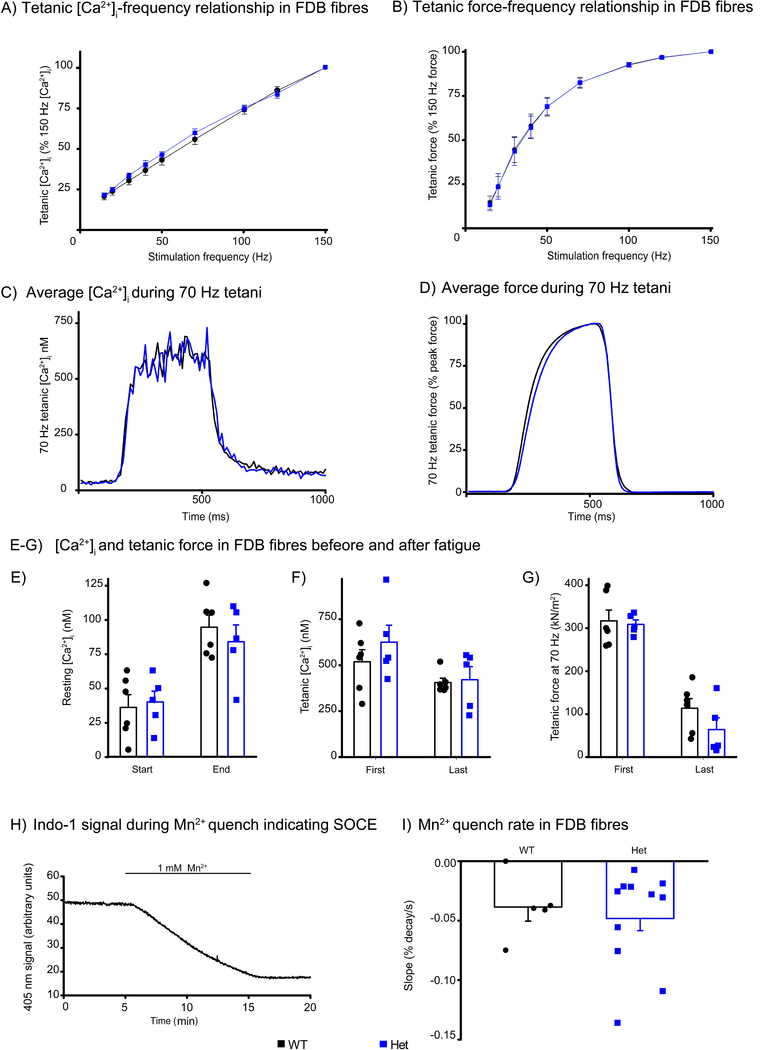
Cellular muscle function was studied in intact single flexor digitorum brevis (FDB) fibres of adult Stim1+/R304W and Stim1+/+ mice [32]. Resting [Ca2+]i did not differ between Stim1+/+ and Stim1+/R304W fibres. The [Ca2+]i-frequency and force-frequency relationships were studied by stimulating fibres with brief (350 ms) trains of electrical pulses at 1 minute intervals. Tetanic [Ca2+]i and force were similar in Stim1+/R304W and Stim1+/+ fibres, and the [Ca2+]i-frequency and force-frequency relationships were virtually identical in the two groups (Figure 6A-B). Average [Ca2+]i and force records from 70 Hz tetanic stimulations show clearly that there were no differences in the time course of changes in [Ca2+]i and force at the onset of contraction or during the relaxation after the end of stimulation (Figure 6C-D). As a result of fatigue induced by 150 repeated 70 Hz tetanic contractions, resting [Ca2+]¡ increased and tetanic [Ca2+]i and force decreased to a similar degree in Stim1+/R304W and Stim1+/+ FDB fibres (Figure 6E-G).
Figure 6. Ca2+ handling in mouse FDB fibres.
(A-B) Tetanic [Ca2+]i-frequency (A) and force-frequency relationships (B) in FDB fibres from wildtype (WT, n=9 fibres) and heterozygous Stim1+/R304W (Het, n=7 fibres) mice showed no difference between the two groups. Tetani were evoked at 1 minute intervals.
(C-D) Comparable averaged tetanic [Ca2+]i records (C) and force records (D) were observed from 70 Hz tetanic contractions in wildtype (WT, n=9) and Stim1+/R304W heterozygous (Het, n=7) FDB fibres from at least three mice in each group.
(E-G) Resting [Ca2+]i (E), tetanic [Ca2+]i (F), and tetanic force (G) were similar in wildtype (n=6) and Stim1+/R304W (n=5) FDB fibres both at the start and end of fatigue induced by 150 repeated tetani. Symbols represent the individual values obtained in fibres.
(H) A typical record of the Indo-1 fluorescent signal at 405 nm during a Mn2+ quenching experiment performed to assess SOCE in a single FDB fibre. Mn2+ present during the experiment indicated.
(I)Mn2+ quenched the Indo-1 signal at the same rate in wildtype (WT, n=5) and heterozygous Stim1+/R304W (Het, n=11) fibres.
Mn2+ ions traverse most Ca2+ permeable channels and can substitute for Ca2+ in SOCE but are not taken up by the SR Ca2+-pumps. Once inside the cell, Mn2+ quenches the fluorescence signal of fluorescent indicators, including Indo-1. Figure 6H shows the general procedure for carrying out the Mn2+ quench experiments and the mean data in Figure 6I show that the influx of Mn2+ was not significantly different in Stim1+/+ and Stim1+/R304W fibres. Together these results indicate no differences in [Ca2+]i handling and force production in isolated muscle fibres between Stim1+/R304W and wildtype mice.
Platelets from stim1R304W/R304W mice showed normal numbers, but lacked STIM1 expression
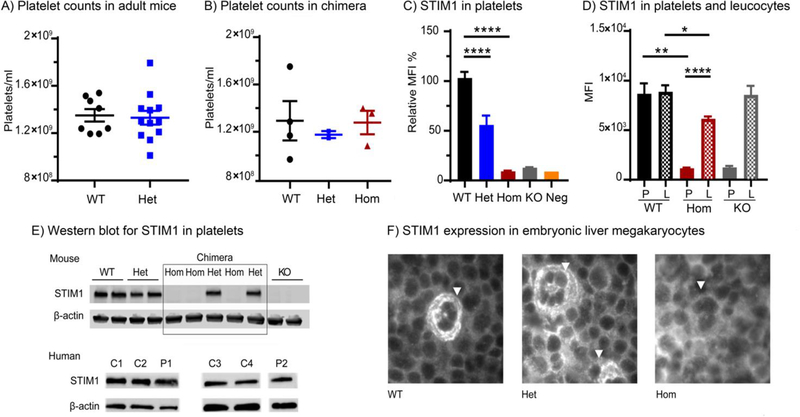
In the Stim1+/R304W mice and in two surviving stim1R304W/R304W mice, red and white blood cell counts (total and differential), haemoglobin levels, mean cell volume, haematocrit and calculated free serum Ca2+ levels showed counts within reference ranges and comparable to Stim1+/+ animals (data not shown). In addition, flow cytometry on blood samples from Stim1+/R304W mice showed normal platelet numbers (Figure 7A) and size (data not shown).
Figure 7. Platelet counts and STIM1 expression in Stim1R304W mice.
(A) Platelet counts in adult wildtype Stim1+/+ (WT, n=8) and heterozygous Stim1+/R304W (Het, n=12) mice were within reference ranges (reference mean platelet count= 1–1.3×109/ml).
(B) Foetal liver cell chimeric mice from all three Stim1 genotypes showed platelet counts within reference ranges (reference mean platelet count= 1–1. 3×109/ml, WT: n=4, Het: n=2, Hom’ n=3).
(C-D) STIM1 expression in platelets (P) and leucocytes (L) measured by flow cytometry in chimeric mice. (C) In platelets, the expression was significantly reduced in heterozygous Stim1+/R304W →WT (Het, n=6) (p<0.0001) and homozygous Stim1R304W/R304W→WT mice (Hom, n=3) (p<0.0001) compared to WT (n=5). (D) Similarly, in leucocytes the STIM1 expression was significantly reduced in homozygous Stim1R304W/R304W→WT mice (n=3) (p<0.05), but not in platelet-specific Stim1 knockout mice (KO).
(E) Western blot analysis documents reduced STIM1 expression in platelets from Stim1R304W mice (top) while platelets from two patients with Stormorken syndrome (P1 and P2) show similar levels compared to healthy controls (C1–4) (bottom). Expression of β-actin was used as a loading control.
(F) Immunofluorescence staining of STIM1 in 14.5 dpc embryonic liver from wildtype (WT), heterozygous (Het) and homozygous Stim1R304W (Hom) embryos detected STIM1 expression in WT and heterozygous, but not in homozygous megakaryocytes (arrowheads).
In panels C and D, p-values for significance are indicated as: *=p<0.05, **=p<0.02, ****=p<0.0001.
Due to the lethality of stim1R304W/R304W mice, the consequences of the R304W mutation in the haematological lineage were studied in foetal liver cell chimeric mice. Three Stim1R304W/R304W→WT, two Stim1+/R304W→WT and four Stim1+/+→WT chimeras were created. Following a 4–6 week recovery period post transplantation, Sanger sequencing detected DNA only from the donor cells in peripheral blood samples from the chimeric mice, documenting an efficient reconstitution of the haematopoietic cell lineage in these animals (Supplementary Figure 2). Platelet counts of Stim1+/R304W→WT and Stim1R304W/R304W→WT chimeric animals were within reference ranges and comparable to Stim1+/+→WT controls (Figure 7B). Intracellular staining demonstrated significantly reduced expression of STIM1 protein in platelets isolated from Stim1+/R304W→WT and Stim1R304W/R304W→WT chimeric animals when compared to controls (Figure 7C). In fact, STIM1 staining in Stim1R304W/R304W→WT platelets was as low as that in platelets isolated from platelet specific Stim1 knockout mice. In contrast, STIM1 protein expression was detectable in leukocytes from Stim1R304W/R304W→WT chimeras (Figure 7D). Absence of STIM1 expression was confirmed in platelets and megakaryocytes from Stim1R304W/R304W→WT chimeras by western blotting (Figure 7E) and in fetal liver in Stim1R304W/R304W embryos by immunofluorescence staining (Figure 7F). Quantification of the transcript level by real-time quantitative PCR (qPCR) of cDNA derived from whole blood RNA demonstrated a significant reduction in the Stim1 levels in Stim1+/R304W mice compared to wildtype mice (p<0.05) (Supplementary Figure 3). In platelets from patients with Stormorken syndrome expressing the STIM1 R304W mutation and healthy controls, however, similar STIM1 expression was observed by western blot (Figure 7E, bottom). Together these results indicate a significant reduction of STIM1 expression in the megakaryocytic lineage of the STIM1 R304W mutant mice, which differs from the patients with Stormorken syndrome.
No SOCE and impaired activation of homozygous Stim1 R304W platelets
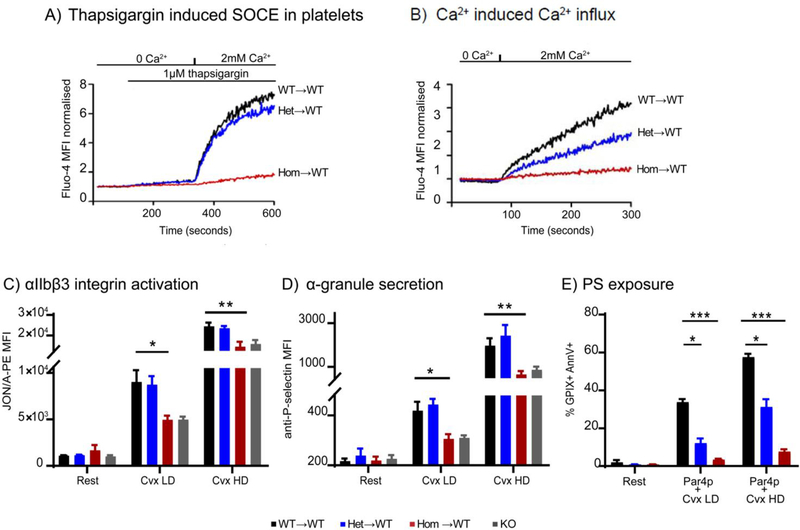
In line with reduced STIM1 expression, platelets from Stim1+/R304W→WT mice showed slightly reduced SOCE following Ca2+ depletion with thapsigargin, while no SOCE induction was detected in platelets from Stim1R304W/R304W→WT chimeras (Figure 8A). Platelets from Stim1+/R304W→WT chimeras showed markedly reduced Ca2+ induced Ca2+ entry compared to Stim1+/+ and platelets from Stim1R304W/R304W→WT chimeras completely lacked Ca2+ influx following re-introduction of Ca2+ in extracellular media (Figure 8B).
Figure 8. SOCE and platelet activation assessed in Stim1R304W chimeric mice by flow cytometry.
(A) Platelets from chimeric mice were washed and loaded with Fluo-4-AM, and Fluo-4 intensity was measured in real time by flow cytometry. Store operated Ca2+ entry (SOCE) in platelets was assessed by incubation with thapsigargin in Ca2+-free buffer, followed by the reintroduction of 2 mM Ca2+. Homozygous Stim1R304W/R304W→WT platelets (Hom→WT) did not show thapsigargin induced SOCE, which was detected in heterozygous Stim1+/R304W→WT (Het→WT) and WT (WT→ WT) platelets.
(B) Heterozygous Stim1+/R304W→WT platelets showed reduced Ca2+ induced Ca2+ entry compared to WT (WT→ WT) platelets, while homozygous Stim1R304W/R304W→WT (Hom→WT) platelets showed lack of Ca2+ induced Ca2+ entry.
(C-D) Platelets stimulated with low doses (LD) and high doses (HD) of Cvx showed impaired activation of homozygous platelets measured by Integrin activation with JON/A-PE antibody (C) and reduced granule secretion with anti-P-selectin antibody (D). Significantly impaired activation was detected at both LD (p<0.05) and HD (p<0.02) Cvx in homozygous platelets. (WT→ WT: n=6, Het→WT: n=4, Hom→WT: n=5)
(E) Dual agonist stimulation (Par4p + Cvx) in washed platelets to induce phosphatidylserine (PS) exposure showed significant impairment of PS exposure measured with AnnexinV-488 in heterozygous (Het→WT, n=2) (p<0.05) and homozygous (Hom→WT, n=3) (p<0.0001) compared to WT (WT→WT, n=2) platelets.
In panels C, D and E, p-values for significance are indicated as: *=p<0.05, **=p<0.02, ***=p<0.005.
Platelet activation studies were performed on Stim1R304W/R304W→WT and Stim1+/R304W→WT chimeric mice using the immunoreceptor tyrosine-based activation motif (ITAM) receptor agonist, convulxin, (Cvx) and the G-protein coupled receptor (GPCR) agonist, Par4 activating peptide (Par4p). Platelets from Stim1R304W/R304W→WT mice and mice lacking STIM1 in the megakaryocyte lineage (KO) showed impaired activation of integrin αIIbβ3 (Figure 8C) and impaired granule secretion (Figure 8D) in response to low dose (LD) and high dose (HD) Cvx. Similar results were obtained with platelets activated with PAR4p (data not shown). Stim1+/R304W→WT platelets showed a normal integrin activation and granule secretion response. We previously demonstrated that SOCE is particularly important for the platelet pro-coagulant response, i.e. their ability to externalize phosphatidylserine (PS) and to assemble plasma clotting factors on their cell surface [36]. Consistent with the reduced STIM1 protein levels, platelets from both Stim1+/R304W→WT and Stim1R304W/R304W→WT chimeras showed a significant defect in AnnexinV binding (a measure of surface PS exposure) in response to stimulation with Par4p and Cvx agonists (Figure 8E).
Discussion
We generated a mouse line expressing the STIM1 R304W mutant protein. The specific integration of the mutation was verified by on-target PCR and by whole genome sequencing of the founder mouse. The mutant protein showed expression in most tissues assessed in heterozygous and homozygous embryos and adult mice. However, we detected a reduced level of STIM1 in homozygous Stim1R304W/R304W haematopoietic cells, fibroblasts and skeletal muscle, which appears to effectively compensate for constitutive activity of the protein.
Although STIM1 GoF mutations are pathogenic in humans and mice, our results contribute to a growing consensus that the specific consequences of these mutations may be species dependent. The Stim1 EF-hand GoF, D84G, mutation caused a severe bleeding disorder resulting in embryonic lethality in the Stim1Sax mice [17], but it causes only TAM in humans [8]. In humans, the R304W mutation results in constitutively active STIM1 causing Stormorken syndrome, a multi-organ disease which includes TAM [9, 12, 18–19]. The results presented here suggest that the Stim1 R304W encodes a constitutively active protein also in mice, with the protein being expressed at reduced levels in skin fibroblasts, skeletal muscle and in haematopoietic cells. The reduced expression of a constitutively active STIM1 R304W protein explains the similar resting [Ca2+]i and SOCE amplitude detected in the fibroblasts from Stim1+/+ and Stim1R304W/R304W mice, despite the 3 fold reduction in STIM1 levels in the latter.
All but three homozygous Stim1R304W mice died perinatally. The requirement of a well-regulated SOCE during embryonic and neonatal stages is demonstrated by the lethality of the Stim1 KO mouse models [40–41]. Both the Stim1R304W and the Stim1Sax GoF mouse models with constitutive STIM1 activation manifested homozygous lethality [17], but stage and aetiology of the lethality were profoundly different. The homozygous Stim1R304W embryos did not show the bleeding and the blood lymphatic vascular mixing seen in Stim1Sax mice which caused death as early as <14.5 dpc [17]. Conceivably the lack of STIM1 in platelets and megakaryocytes explains the lack of bleeding in the Stim1R304W embryos and survival until birth. Platelet specific Stim1 KO was not lethal in mice [36], therefore the perinatal lethality in the homozygous Stim1R304W mice was likely not caused by the lack of STIM1 in platelets. Common causes of perinatal lethality involve problems at parturition, such as abnormalities in breathing, suckling and homeostasis [42]. However, we detected no cyanosis or morphological abnormalities in the diaphragm in the homozygous Stim1R304W pups that were found dead at day P0, which would have indicated breathing dysfunction. Lethality of the global Stim1 KO was ascribed to dysfunction in respiratory muscle [41]. However, lethality was also observed in skeletal muscle specific Stim1 KO and smooth muscle specific Stim1 KO mice, highlighting the importance of STIM1-mediated SOCE in both types of muscle at early developmental stages [40, 43].
A large body of evidence underscores the importance of STIM1 and ORAI1 mediated regulation of Ca2+ signalling during skeletal muscle development (reviewed in [44] [45]). The STIM1 expression is developmentally regulated and it peaks postnatally in skeletal muscle in mice [46]. Furthermore, patients that lack SOCE due to STIM1 mutations present with congenital skeletal muscle myopathy [11, 47] and mice lacking functional STIM1 in skeletal muscles died in perinatal stages from a skeletal muscle myopathy [14, 43]. Growth, differentiation and regeneration of muscle after birth are also dependent on STIM1 regulated Ca2+ signalling; for instance, a skeletal-muscle-specific STIM1 transgenic mouse model showed signs of muscle degeneration (35). In contrast, humans with GoF STIM1 mutations manifest myopathy and TAM, but no muscle degeneration [6, 9, 16]. No muscular phenotype was reported in heterozygous GoF Stim1Sax mice [17]. We presently show markedly decreased muscle mass and reduced endurance exercise performance in the three surviving Stim1R304W/R304W mice, and our histological investigation of the skeletal muscle showed muscle degeneration and regeneration. On the other hand, we did not detect any significant differences between Stim1+/R304W and wildtype mice regarding either in vivo endurance exercise performance or [Ca2+]i handling and force production in isolated muscle fibres in the rested state or during fatigue. Major changes in STIM1 function are accompanied by marked alterations in skeletal muscle, or even an absence of skeletal muscle phenotype. Conversely, contractile activity, including fatiguing exercise, in adult skeletal muscle fibres mainly depends on bi-directional Ca2+ fluxes between the sarcoplasmic reticulum and the cytosol. SOCE may therefore be of limited importance for contractile function in the short-term [48], which can explain the apparently functional skeletal muscles in Stim1+/R304W mice. However, further studies are required to reveal the exact mechanisms involved in the role of SOCE in long-term muscle maintenance at adult stages.
Previous work showed elevated resting [Ca2+]i, constitutive SOCE and impaired platelet function leading to a bleeding diathesis in patients with Stormorken syndrome, as well as variable severity of thrombocytopenia [9, 12]. Impaired platelet activation and function in Stim1Sax and Stim1 KO mice, also highlight the importance of STIM1 for platelet functions [17, 41]. Furthermore, Stim1 GoF mutations resulted in embryonal lethality due to bleeding in the Stim1Sax mice [17]. When we assessed the STIM1 expression in a range of tissues in the Stim1R304W mice and in chimeric mice, we detected reduced expression in platelets, megakaryocytes and leucocytes. Interestingly, the data indicated that the level of the Stim1 mutant transcript was reduced compared to the wildtype transcript which resulted in reduced protein levels in heterozygotes and especially in homozygotes compared to cells from the wildtype mice. In fact, no STIM1 protein expression was detected in megakaryocytes and platelets in Stim1R304W/R304W mice and Stim1R304W/R304W→WT chimeras, resulting in a platelet specific STIM1 KO. In line with the observations previously described in the platelet specific Stim1 KO mice [36], the homozygous Stim1R304W mice and the chimeras with undetectable STIM1 levels, showed impaired platelet activation, no SOCE and presented with normal platelet counts.
Conclusion
We describe a Stim1 knock-in mouse model expressing the constitutive STIM1 R304W mutation. The perinatal lethality and the severe muscle degeneration documented in the Stim1R304W line highlight the critical role of a tightly controlled SOCE for skeletal muscle development and long-term integrity. We showed a specific reduction of the mutant protein in particular in the haematopoietic lineage, where it possibly compensated constitutive STIM1 activation and prevented cytosolic Ca2+ overload. Understanding what promotes downregulation of STIM1 in this mouse model but not in the patients with Stormorken syndrome carrying the same R304W mutation is the obvious next step to study.
Supplementary Material
Acknowledgements
We are grateful to the patients who participated in this study and provided biological samples. We thank Ms. Guro Lien, Ms. Turid Pedersen from Oslo University Hospital, Oslo, for their valuable input to the project, technical assistance in embryo’s extractions, and flowcytometry studies, respectively. Histological services for embryo characterization were provided by the Histology Research Core Facility at the Department of Cell Biology and Physiology, University of North Carolina, Chapel Hill NC. We also thank Ms. Claire Trincot at Kathleen Caron lab, University of North Carolina, Chapel Hill NC for their assistance and input in immune fluorescence based staining of mouse embryo sections. T.G was supported financially by the Quota scheme of Norwegian State Educational Loan Fund, “Mobility grant for PhD students by the Institute for Clinical medicine, University of Oslo and by UNIFOR. W.B was supported through a grant from National Institute of Health, USA (R01 HL130404), and R.H.L was funded by T32 HL007149. H.W was supported by the Swedish Research Council (K2014–52X-10842–21-5).
Footnotes
Competing Interests
There are no conflicts of interests to declare.
References
- [1].Luik RM, Wang B, Prakriya M, Wu MM, and Lewis RS, Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation, Nature 454 (2008) 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wu MM, Luik RM, and Lewis RS, Some assembly required: constructing the elementary units of store-operated Ca2+ entry, Cell Calcium 42 (2007) 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Putney JW Jr., A model for receptor-regulated calcium entry, Cell Calcium 7 (1986) 1–12. [DOI] [PubMed] [Google Scholar]
- [4].Putney JW Jr., New molecular players in capacitative Ca2+ entry, J. Cell. Sci 120 (2007) 19591965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function, Nature 441 (2006) 179–185. [DOI] [PubMed] [Google Scholar]
- [6].Lacruz RS, and Feske S, Diseases caused by mutations in ORAI1 and STIM1, Ann. N. Y. Acad. Sci 1356 (2015) 45–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Feske S, CRAC channelopathies, Pflugers Archiv : European journal of physiology 460 (2010) 417435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bohm J, Chevessier F, Maues De Paula A, Koch C, Attarian S, Feger C, Constitutive activation of the calcium sensor STIM1 causes tubular-aggregate myopathy, Am. J. Hum. Genet 92 (2013) 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Misceo D, Holmgren A, Louch WE, Holme PA, Mizobuchi M, Morales RJ, A dominant STIM1 mutation causes Stormorken syndrome, Hum. Mutat 35 (2014) 556–564. [DOI] [PubMed] [Google Scholar]
- [10].Endo Y, Noguchi S, Hara Y, Hayashi YK, Motomura K, Miyatake S, Dominant mutations in ORAI1 cause tubular aggregate myopathy with hypocalcemia via constitutive activation of storeoperated Ca(2)(+) channels, Hum. Mol. Genet 24 (2015) 637–648. [DOI] [PubMed] [Google Scholar]
- [11].Picard C, McCarl CA, Papolos A, Khalil S, Luthy K, Hivroz C, STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity, N. Engl. J. Med 360 (2009) 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nesin V, Wiley G, Kousi M, Ong EC, Lehmann T, Nicholl DJ, Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis, Proc. Natl. Acad. Sci. U S A 111 (2014) 4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Feske S, ORAI1 and STIM1 deficiency in human and mice: roles of store-operated Ca2+ entry in the immune system and beyond, Immunol. Rev 231 (2009) 189–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stiber J, Hawkins A, Zhang ZS, Wang S, Burch J, Graham V, STIM1 signalling controls storeoperated calcium entry required for development and contractile function in skeletal muscle, Nat. Cell. Biol 10 (2008) 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baba Y, Nishida K, Fujii Y, Hirano T, Hikida M, and Kurosaki T, Essential function for the calcium sensor STIM1 in mast cell activation and anaphylactic responses, Nat. Immunol 9 (2008) 81–88. [DOI] [PubMed] [Google Scholar]
- [16].Bohm J, Chevessier F, Koch C, Peche GA, Mora M, Morandi L, Clinical, histological and genetic characterisation of patients with tubular aggregate myopathy caused by mutations in STIM1, J. Med. Genet 51 (2014) 824–833. [DOI] [PubMed] [Google Scholar]
- [17].Grosse J, Braun A, Varga-Szabo D, Beyersdorf N, Schneider B, Zeitlmann L, An EF hand mutation in Stim1 causes premature platelet activation and bleeding in mice, J. Clin. Invest 117 (2007) 3540–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Morin G, Bruechle NO, Singh AR, Knopp C, Jedraszak G, Elbracht M, Gain-of-Function Mutation in STIM1 (P.R304W) Is Associated with Stormorken Syndrome, Hum. Mutat 35 (2014) 1221–1232. [DOI] [PubMed] [Google Scholar]
- [19].Markello T, Chen D, Kwan JY, Horkayne-Szakaly I, Morrison A, Simakova O, York platelet syndrome is a CRAC channelopathy due to gain-of-function mutations in STIM1, Mol. Genet. Metab 114 (2015) 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stormorken H, Sjaastad O, Langslet A, Sulg I, Egge K, and Diderichsen J, A new syndrome: thrombocytopathia, muscle fatigue, asplenia, miosis, migraine, dyslexia and ichthyosis, Clin. Genet 28 (1985) 367–374. [DOI] [PubMed] [Google Scholar]
- [21].Harris E, Burki U, Marini-Bettolo C, Neri M, Scotton C, Hudson J, Complex phenotypes associated with STIM1 mutations in both coiled coil and EF-hand domains, Neuromuscul. Disord (2017). [DOI] [PubMed] [Google Scholar]
- [22].Tamura K, Dudley J, Nei M, and Kumar S, MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0, Mol. Biol. Evol 24 (2007) 1596–1599. [DOI] [PubMed] [Google Scholar]
- [23].Li H, and Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics 25 (2009) 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, The Sequence Alignment/Map format and SAMtools, Bioinformatics 25 (2009) 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, and Korbel JO, DELLY: structural variant discovery by integrated paired-end and split-read analysis, Bioinformatics 28 (2012) i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, BreakDancer: an algorithm for high-resolution mapping of genomic structural variation, Nat. Methods. 6 (2009) 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li H, A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data, Bioinformatics 27 (2011) 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Thorvaldsdottir H, Robinson JT, and Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration, Brief Bioinform 14 (2013) 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Krawiec JA, Chen H, Alom-Ruiz S, and Jaye M, Modified PAXgene method allows for isolation of high-integrity total RNA from microlitre volumes of mouse whole blood, Lab. Anim 43 (2009) 394–398. [DOI] [PubMed] [Google Scholar]
- [30].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Fiji: an open-source platform for biological-image analysis, Nat. Methods 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Culling CFA, Allison RT, and Barr WT, (1985) 21 - Hard tissue, In Cellular Pathology Technique (Fourth Edition). (Butterworth-Heinemann; ), pp 408–430. [Google Scholar]
- [32].Cheng AJ, and Westerblad H, Mechanical isolation, and measurement of force and myoplasmic free [Ca(2+)] in fully intact single skeletal muscle fibers, Nat. Protoc 12 (2017) 1763–1776. [DOI] [PubMed] [Google Scholar]
- [33].Andrade FH, Reid MB, Allen DG, and Westerblad H, Effect of hydrogen peroxide and dithiothreitol on contractile function of single skeletal muscle fibres from the mouse, J. Physiol 509 ( Pt 2) (1998) 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hallam TJ, and Rink TJ, Agonists stimulate divalent cation channels in the plasma membrane of human platelets, FEBS Lett 186 (1985) 175–179. [DOI] [PubMed] [Google Scholar]
- [35].Bergmeier W, Oh-Hora M, McCarl CA, Roden RC, Bray PF, and Feske S, R93W mutation in Orai1 causes impaired calcium influx in platelets, Blood 113 (2009) 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ahmad F, Boulaftali Y, Greene TK, Ouellette TD, Poncz M, Feske S, Relative contributions of stromal interaction molecule 1 and CalDAG-GEFI to calcium-dependent platelet activation and thrombosis, J. Thromb. Haemost 9 (2011) 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee RH, Piatt R, Conley PB, and Bergmeier W, Effects of ibrutinib treatment on murine platelet function during inflammation and in primary hemostasis, Haematologica 102 (2017) e89–e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Targeted genome modification in mice using zinc-finger nucleases, Genetics 186 (2010) 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Concepcion AR, Vaeth M, Wagner LE 2nd, Eckstein M, Hecht L, Yang J, Store-operated Ca2+ entry regulates Ca2+-activated chloride channels and eccrine sweat gland function, J. Clin. Invest 126 (2016) 4303–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mancarella S, Potireddy S, Wang Y, Gao H, Gandhirajan RK, Autieri M, Targeted STIM deletion impairs calcium homeostasis, NFAT activation, and growth of smooth muscle, FASEB J 27 (2013) 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Varga-Szabo D, Braun A, Kleinschnitz C, Bender M, Pleines I, Pham M, The calcium sensor STIM1 is an essential mediator of arterial thrombosis and ischemic brain infarction, J. Exp. Med 205 (2008) 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Turgeon B, and Meloche S, Interpreting neonatal lethal phenotypes in mouse mutants: insights into gene function and human diseases, Physiol. Rev 89 (2009) 1–26. [DOI] [PubMed] [Google Scholar]
- [43].Li T, Finch EA, Graham V, Zhang ZS, Ding JD, Burch J, STIM1-Ca(2+) signaling is required for the hypertrophic growth of skeletal muscle in mice, Mol. Cell. Biol 32 (2012) 3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cho CH, Woo JS, Perez CF, and Lee EH, A focus on extracellular Ca(2+) entry into skeletal muscle, Exp. Mol. Med 49 (2017) e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tu MK, Levin JB, Hamilton AM, and Borodinsky LN, Calcium signaling in skeletal muscle development, maintenance and regeneration, Cell Calcium 59 (2016) 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Seth M, Li T, Graham V, Burch J, Finch E, Stiber JA, Dynamic regulation of sarcoplasmic reticulum Ca(2+) stores by stromal interaction molecule 1 and sarcolipin during muscle differentiation, Dev. Dyn 241 (2012) 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McCarl CA, Picard C, Khalil S, Kawasaki T, Rother J, Papolos A, ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia, J. Allergy Clin. Immunol 124 (2009) 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cheng AJ, Place N, and Westerblad H, Molecular Basis for Exercise-Induced Fatigue: The Importance of Strictly Controlled Cellular Ca(2+) Handling, Cold Spring Harb. Perspect. Med 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.