Abstract
Whooping cough also called Pertussis is a highly contagious respiratory infection that affects all age populations. Given recent pertussis outbreaks, there is an urgent need for a point-of-care (POC) device for rapid diagnosis of pertussis. Herein, we report a low-cost microfluidic POC device integrated with loop-mediated isothermal amplification (LAMP) technique for the rapid and accurate diagnosis of pertussis. The 3D-printed bioanalyzer housed not only the biochip but also an in-house-developed portable and fully battery-powered heater for rapid POC detection of pertussis, without the need of external electricity. The fluorescence-based results could be rapidly visualized in about one hour by the naked eye without the need for any additional instrumentation. In addition, a simple centrifuge-free sample preparation process was optimized for the efficient lysis of pertussis samples and successfully used for direct detection of bacteria in nasopharyngeal samples. High sensitivity, with a limit of detection (LOD) of 5 DNA copies per LAMP zone, and high specificity were demonstrated. We envision that the microfluidic POC device can be used in various venues such as medical clinics, schools, and other low-resource settings for the fast detection of pertussis.
Keywords: microfluidic, pertussis diagnosis, point-of-care analysis, instrument-free detection, LAMP (loop-mediated isothermal amplification)
Graphical Abstract
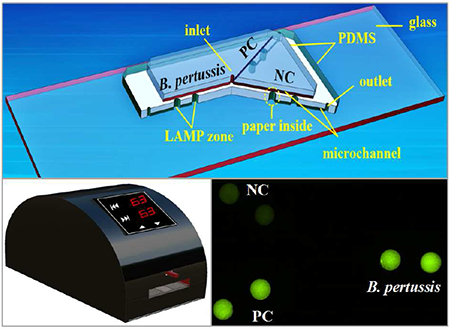
We report a low-cost microfluidic point-of-care (POC) platform integrated with loop-mediated isothermal amplification (LAMP) supported by a portable battery-powered heater for rapid and instrument-free detection of whooping cough in resource-limited settings
1. Introduction
Pertussis, commonly known as whooping cough, is caused by the bacterium Bordetella pertussis (B. pertussis) [1, 2]. This respiratory infection affects people of any age and is potentially life-threatening to young children. In the United States, pertussis has been brought to the forefront of the nation’s attention due to the frequent and recent outbreaks despite the high vaccination coverage. Reported cases in 2012 reached 48,277, the highest in fifty years [3]. In developing countries, pertussis has been considered as a serious health concern. It was estimated that about 24.1 million pertussis cases and 160,700 deaths occurred in children younger than 5-year-old globally in 2014. Most of these cases happened in developing countries [4]. Cases are often underdiagnosed because the symptoms may resemble a common cold at the beginning of infection [5]. Therefore, it is imperative to have a rapid and effective pertussis diagnostic tool to rapidly detect, treat and prevent the spread of the infection.
The current pertussis diagnostic methods include bacterial culture, enzyme-linked immunosorbent assay (ELISA), and real-time polymerase chain reaction (real-time PCR). However, these technologies have their own limitations. For example, bacterial culture takes several days or even weeks for the bacteria to grow and there are many variables that make this method of detection not sensitive [6]. ELISA which is an immunological technique has low specificity because of cross-reactivity with other pathogens and timing of sample collection [7]. Real-time qPCR has high sensitivity and specificity but requires expensive instrumentation [8, 9]. In addition, the IS481 sequence that most real-time qPCR assays target for whooping cough diagnosis also exists in B. holmesii genomes, making it challenging to differentiate B. pertussis with the closely related species B. holmesii [9]. Recently, as a relatively new DNA amplification method, loop-mediated isothermal amplification (LAMP) has received increasing attention for pathogen detection [10, 11]. It can amplify the target DNA at a constant temperature (60-65 °C) by using 4 or 6 different primers and Bst polymerase derived from Bacillus stearothermophilus, which adds a high degree of specificity and has high strand displacement activity [12]. Although LAMP has shown high specificity in disease diagnosis [13–15], the conventional tube-based LAMP method usually requires expensive and/or bulky heating equipment (e.g. thermocyclers) for amplification, and the use of centrifuges and time-consuming procedures for sample preparation, making it unsuitable for point-of-care (POC) pertussis diagnostics for low-resource settings.
The World Health Organization (WHO) provides guidelines for a successful diagnostic technology in the developing world, “ASSURED”, meaning affordable, sensitive, specific, user-friendly, robust and rapid, equipment free, and deliverable [16]. In light of these guidelines, many biomedical researchers and emerging diagnostic companies turn to microfluidic techniques. Microfluidic lab-on-a-chip, also called “micro total analysis systems” (μTAS), is a miniaturized device or system that can perform laboratory analysis functions by integrating one or several functional units and manipulating tiny amounts of fluids (microliters or nanoliters) [17]. The microfluidic technique has demonstrated great potential for the development of POC testing devices that brings rapid and immediate diagnostics nearer to the site of patient care [18–27]. Our previous studies demonstrated the feasibility of the microfluidic biochip-based LAMP method for detection of bacterial meningitis [28–30]. As far as we know, no microfluidic devices have been reported for POC detection of B. pertussis.
Herein, by taking advantages of microfluidics and LAMP, we developed a microfluidic device that integrated with LAMP for instrument-free POC detection of B. pertussis. In this study, the microfluidic device was composed of both the paper substrate for storage and protection of DNA primers and the transparent polydimethylsiloxane (PDMS) substrate for robust liquid manipulation. As our previous study confirmed [21, 28], this type of hybrid microfluidic devices with paper inside enabled a better primer distribution and more stable diagnostic performance over a long period of time than paper-free non-hybrid devices. Because the previous microfluidic devices we developed for meningitis detection still required external AC power with limited capacity for onsite detection or field diagnosis, we devised a 3D-printed integrated bioanalyzer which housed not only the biochip but also an in-house-developed battery-powered heater for rapid POC detection of pertussis. The results could be easily observed by the naked eye under a portable UV light pen within one hour. We further demonstrated the high specificity and sensitivity of our device in pertussis diagnosis with a limit of detection (LOD) as low as 5 DNA copies per LAMP zone. Furthermore, we successfully performed instrument-free detection of B. pertussis directly from nasopharyngeal samples by using an optimized centrifuge-free lysis approach. Thus, our hybrid microfluidic device has great potential for rapid and accurate diagnosis of whooping cough in low-resource settings such as in the field.
2. Experimental Section
2.1. Microfluidic device design
The design of the hybrid microfluidic device is shown in Figure 1. The microfluidic device is composed of two PDMS layers and a glass slide layer. The top PDMS layer is mainly used for reagent delivery, consisting of an inlet reservoir (diameter 1.0 mm, depth 1.5 mm) and microchannels (width 100 μm, depth 100 μm). The middle PDMS layer is mainly used for on-chip LAMP reactions as well as positive control (PC) and negative control (NC), and consists of 6 LAMP zones (diameter 2.0 mm, depth 1.5 mm), 3 outlet reservoirs (diameter 1.0 mm, depth 1.5 mm), and microchannels. The glass slide (75 mm × 25 mm) is used for structural support. A paper disk with a diameter of 2.0 mm was placed inside each LAMP zone for storage of DNA primers specific to pertussis.
Figure 1.
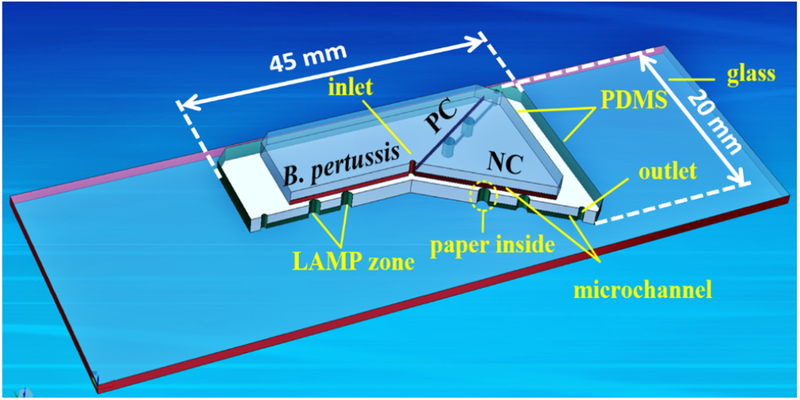
Schematic of the PDMS/paper hybrid microfluidic device integrated with LAMP for B. pertussis detection. The cross-section view illustrates the LAMP zone structure with the paper substrate inside.
2.2. On-chip LAMP process
The LAMP reaction mixture included (1) primers: 1.6 μM each of the FIP/BIP (inner primer), 0.2 μM each of the F3/B3 (outer primer), 0.4 μM each of the LF/BF (loop primer); (2) 8U Bst Polymerase; (3) 1.4 mM dNTPs; (4) salts: 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 8 mM MgSO4, 10 mM (NH4)2SO4, 0.1% Tween 20, 0.8 M Betaine, and 0.5 mM MnCl2; (5) calcein (1 μL/26 μL reaction mixture). LAMP primers are listed in Table 1, which are designed to target the B. pertussis PT promoter region [13].
Table 1.
LAMP primer sequences for PT promoter region of B. pertussis
| Primer | Sequences (5’-3’) | No. of bases |
|---|---|---|
| FIP | TTGGATTGCAGTAGCGGGATGTGCATGCGTGCAGATTCGTC | 41 |
| BIP | CGCAAAGTCGCGCGATGGTAACGGATCACACCATGGCA | 38 |
| F3 | CCGCATACGTGTTGGCA | 17 |
| B3 | TGCGTTTTGATGGTGCCT | 18 |
| FL | ACGGAAGAATCGAGGGTTTTGTAC | 24 |
| BL | GTCACCGTCCGGACCGTG | 18 |
This study used two different negative controls, the omission of primers (NC1) and omission of template DNA (NC2), which demonstrated consistent results as shown in the traditional tube-based LAMP detection method (see Figure S1). So we used NC1 as the main NC for practical application for the convenient delivery of reagent and samples, and used NC2 in this study to investigate and validate the performance of the microfluidic device for specificity and sensitivity and direct detection of B. pertussis.
NC1 and PC1:
B. pertussis LAMP primers (see Table 1) were pre-loaded in the B. pertussis detection LAMP zone, and the PC DNA with its primer mix from the commercial LAMP kit was pre-loaded in the PC LAMP zones (PC1). No primer was pre-loaded in the negative control LAMP zone (NC1). Then, a 26 μL LAMP reaction mixture with the B. pertussis template DNA was introduced from the inlet reservoir to fill the LAMP zones. That is, the NC1 was in the presence of template DNA but not the primers.
NC2 and PC2:
All LAMP zones were pre-loaded with B. pertussis primers. Then, each of a 13 μL LAMP reaction mixture including a nasopharyngeal sample and lysis buffer, or positive control consisting of purified B. pertussis DNA (PC2), or negative control which had equal volumes of water (NC2), was added separately to each outlet reservoir and their corresponding LAMP zones. That is, the NC2 was in the presence of LAMP primers but not the template DNA. The PC2 used the purified B. pertussis template DNA and its primer mix to evaluate the performance for direct detection of B. pertussis in nasopharyngeal samples.
The reaction mixture was introduced into the microfluidic device using a pipette and the flow is driven by capillary action. After that, the inlet and outlets reservoirs were sealed with Epoxy glue to prevent the reagent evaporation during on-chip LAMP reactions. Then the microfluidic device was heated at 63 °C for 45 min by using a 3D-printed in-house-developed portable and fully battery-powered heater (see Figure 2), followed by increasing the temperature to 95 °C for 2 min for the termination of on-chip LAMP reactions.
Figure 2.
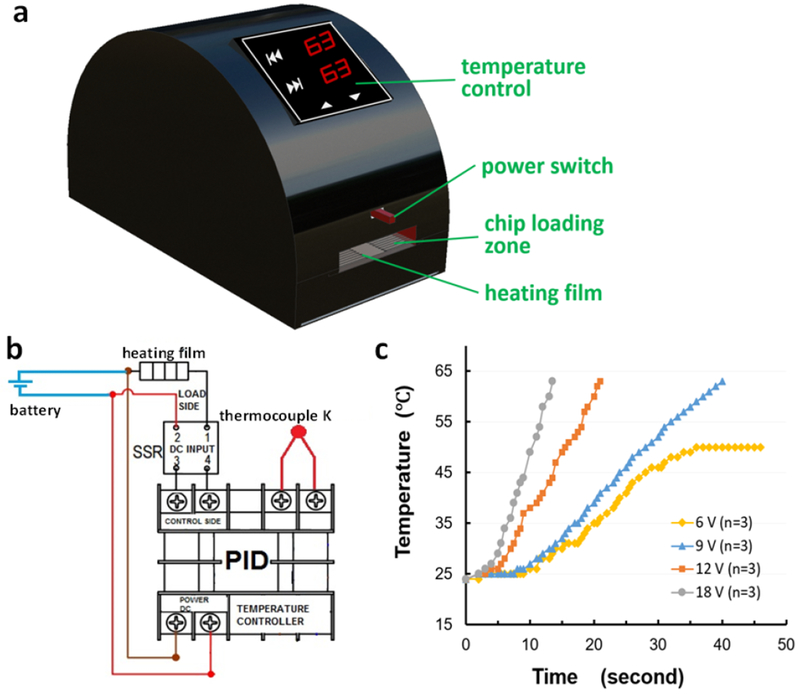
Design of the portable battery-powered heater. (a) 3D schematic of the heater holder fabricated by using a 3D printer; (b) Schematic of the PID-based temperature controller; (c) Diagram of the battery heating speeds to achieve the desired temperature at four different battery voltages: 6, 9, 12, and 18 volts.
2.3. Detection and confirmatory tests
Results of LAMP reactions were directly detected by the generated fluorescence in the LAMP zones using a portable UV light pen. An image of the LAMP zones was captured by using a cellular phone camera (e.g. iPhone 5) and the NIH software ImageJ was used to measure the brightness of selected areas (i.e. LAMP zones) to obtain the corresponding gray values for further analysis.
Fluorescence detection was also carried out by using a Nikon Ti-E fluorescence microscope (Melville, NY) equipped with a motorized stage and a cooled charge-coupled device (CCD) camera under a FITC optical filter (Ex = 495 nm; Em = 520 nm) to confirm results. DNA amplification was also confirmed by Gel electrophoresis (Sub-Cell GT, Bio-Rad, CA) analyzing the extracted LAMP products from the microfluidic device. During the gel electrophoresis process, LAMP products were resolved by applying 90 V for 1 h in 1.5% agarose gel.
2.4. Optimization of the centrifuge-free lysis protocol
In order to verify if our lysis buffer would be efficient for the preparation of a clinical sample, nasopharyngeal samples were collected from healthy individuals using a nasopharyngeal swab. The swab was then rinsed in 1 mL sterile saline and B. pertussis was added afterward. The suspension was adjusted to a cell density of 1.5 × 108 CFU/mL using a McFarland standard (Key Scientific Products, TX) [31].
Optimization of lysis process: 3 μL of the prepared nasopharyngeal sample was mixed with different volumes of the lysis buffer (50 mM Tris buffer at pH 7.5, 4 M Urea and 0.1% Triton) to generate a serial of sample/lysis buffer mixtures with ratios ranging from 1:0.5 to 1:5 (v/v). Mixtures were incubated at room temperature for 10 min. DNA was purified from each mixture using the Qiagen kit (Valencia, CA) and the DNA was quantitated by Nanodrop (Nanodrop 1000, Thermo Scientific, MA). DNA amounts in each of those lysed mixtures were obtained to reflect the lysis performance at different ratios between the samples and the lysis buffers.
2.5. Direct detection of nasopharyngeal samples
Based on the optimized lysis procedure, direct detection of B. pertussis from nasopharyngeal swab was performed. Specifically, 3 μL of the nasopharyngeal sample (B. pertussis bacteria: ~1.5 × 108 CFU/mL) was mixed with a 3 μL lysis buffer and incubated at room temperature for 10 min. Then, 1 μL of the lysate was directly used for the subsequent on-chip LAMP reactions.
3. Results and Discussion
3.1. Design of the portable battery-powered heater
The conventional LAMP technique for pathogen detection is often limited for low-resource settings due to the requirement of bulky heating instruments such as thermal cyclers or water baths and external AC power. Herein, we developed a cost-effective, portable, and fully battery-powered heater to support on-chip LAMP reactions.
The design of the portable and fully battery-powered heater is shown in Figure 2. The heater holder was simply fabricated by using a 3D printer. The 3D schematic of the holder in Figure 2a illustrates the location of different components inside, where the microfluidic device rested above the heating film. Figure 2b shows the schematic of the battery-powered heater with a proportional-integral-derivative (PID)-based temperature controller. The PID controller was used to control the heater under precise temperature ranges. A thermocouple K was included in this system in order to monitor the temperature in real time during the operating process, providing feedback to the PID controller. The output signal was provided by a solid state relay to increase or decrease the temperature of the heater.
To meet the desired detection speed without sacrificing battery life or overweighing the bioanalyzer, we conducted the heating speed test at four different voltages of 6, 9, 12 and 18 volts to achieve the desired temperature of 63 °C for LAMP reactions. From these tests, as can be seen in Figure 2c, the battery at 9, 12, and 18 volts could reach the desired temperature of 63 °C within 40, 22, and 13 sec respectively. However, the desired temperature of 63 °C at 6 volts could not be reached regardless of the time increase. Therefore, a 9-volt battery was used as the power supply since it could accelerate the temperature to 63 °C within 2 min for on-chip LAMP reactions without adding excessive weight to the heating device. In addition, according to the following formula: Battery Life = Watt Hours / (Voltage × Current in Ampere), the life of the 9-volt battery was estimated to be 2.3 h, which met the required time for two LAMP reactions (45 min/LAMP).
The total material cost of this inexpensive heater was about $60. This heater weighs only 45 g with small dimensions of 15 cm in length, 8 cm in width and 10 cm in depth. More importantly, the heater was fully battery-powered without relying on external electricity. Those characteristics make the inexpensive, portable and fully battery-powered heater very suitable for POC detection in poor areas where there is no stable electricity.
3.2. On-chip LAMP detection of B. pertussis using purified DNA
We first tested the feasibility of the paper/polymer hybrid microfluidic device for B. pertussis detection by using purified bacterial DNA. The LAMP primers specific to B. pertussis, and the PC DNA with its primer mix from the commercial LAMP kit (PC1) were separately pre-loaded in the corresponding LAMP zones with paper substrates inside. Paper has a high surface-to-volume ratio which makes it an ideal 3D porous substrate for storage of DNA primers. After LAMP reactions, the results can be easily visually observed by the naked eye or imaged by a smartphone camera based on the restored fluorescence of calcein in the LAMP zones by applying a portable UV light pen to shine LAMP products near the chip. The detection principle is based on the fluorescence recovery of pre-quenched calcein [10, 32], as illustrated in the cross-section view of the LAMP zone in Figure S2. This endpoint detection step is easy and flexible, and doesn’t require the use of any specialized laboratory instrument, leveraging POC detection of pertussis in low-resource settings.
A fluorescence image captured by a cellphone camera (Figure 3a) shows that both the B. pertussis and the PC DNA sample exhibited bright green fluorescence while the NC1 showed a weak background. The image was processed by using the software ImageJ to obtain the gray values for further analysis. As shown in Figure 3c, a > 3-fold difference between the B. pertussis or PC1 and the NC1 was observed. The results were further confirmed by high-sensitivity fluorescent microscopy. Similarly, strong fluorescence was observed in LAMP zones for B. pertussis and PC1, but not for NC1 (Figure 3b). The fluorescence intensity of the B. pertussis LAMP products was about 4.5-fold higher than that of the NC1 (Figure 3d). Subsequently, we confirmed the results by gel electrophoresis using the extracted LAMP products from the outlet, as shown in Figure 3e. The multiple ladder-pattern DNA bands from Lane 2 for B. pertussis confirmed the success of the on-chip LAMP reaction.
Figure 3.
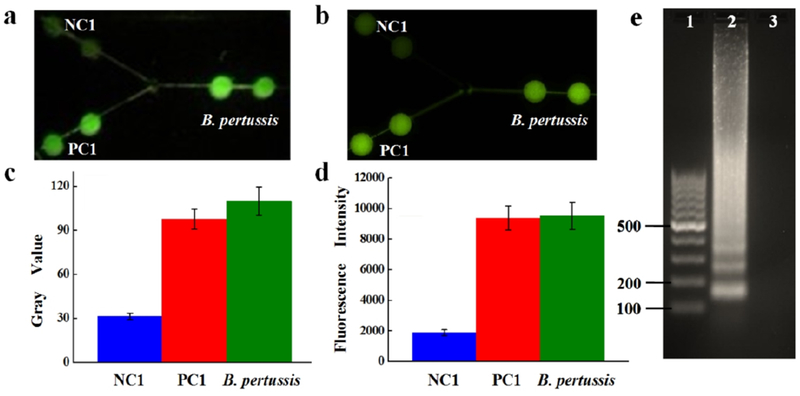
On-chip LAMP detection of B. pertussis using purified DNA by a cellphone camera (a) and fluorescence microscopy (b). (c) Gray values measured by ImageJ; (d) Fluorescent intensities measured by a fluorescence microscope. (e) Gel electrophoresis analysis. Lanes 1-3: 100 bp ladder, B. pertussis LAMP products, NC1. The template DNA used was 5 × 106 copies per LAMP zone.
3.3. Sensitivity and specificity test
We further investigated the sensitivity of our approach by testing a serial of 10-fold diluted B. pertussis DNA samples (i.e. 5 × 105, 5 × 104, 5 × 103, 5 × 102, 5 × 101, 5 × 100, and 5 × 10−1 copies per LAMP zone) on this microfluidic device. As shown in Figure 4a, strong fluorescence was generated in the LAMP products when the initial template DNA was equal to or greater than 5 copies per LAMP zone, while no obvious fluorescence was observed when the initial template DNA was less than 5 copies. The corresponding fluorescence intensities show the same results. On the basis of 3-fold standard deviations of the mean fluorescence intensity of NC2, the cutoff LOD value was calculated to be 2245 a.u. (arbitrary unit) as shown by the dashed line in Figure 4b. The fluorescence intensities of the LAMP products from 5 DNA copies was much higher than the LOD cutoff value. The results were further confirmed by gel electrophoresis (see Figure 4c). The ladder-pattern bands of the LAMP products from 5 copies of the template DNA indicated the success of the on-chip LAMP reactions. However, when the initial template DNA was less than 5 copies, no DNA bands were observed. Therefore, the LOD of the on-chip LAMP approach for detection of B. pertussis was determined as low as 5 DNA copies per LAMP zone, indicating the high detection sensitivity of our approach for pertussis diagnosis.
Figure 4.
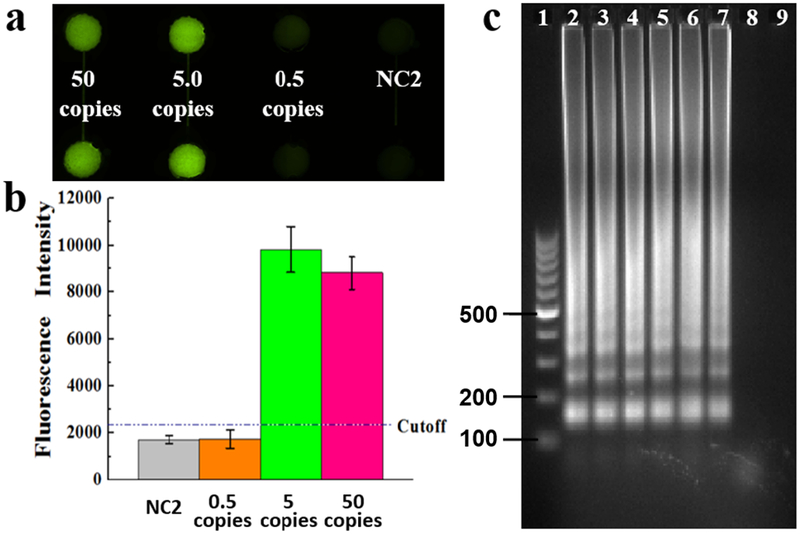
Sensitivity investigation, (a) Fluorescence microscopy image of LAMP products using a series of 10-fold diluted B. pertussis template DNA ranging from 50, 5 and 0.5 copies per LAMP zone, as well as the NC. (b) Gray values of LAMP products, (c) Gel electrophoresis analysis for LOD test. Lanes 1-9: 100 bp marker, LAMP products from 5 × 105, 5 × 104, … , 5 × 100, 5 × 10−1 copies of the template DNA per LAMP zone, and NC2, respectively.
To investigate the specificity of our approach for pertussis diagnosis, we simultaneously tested B. pertussis with its similar Bordetella species, B. parapertussis and B. holmesii, which are also associated with respiratory infections in humans [33]. We introduced DNA samples of these three bacteria separately into different LAMP zones that were all preloaded with B. pertussis LAMP primers for on-chip LAMP reactions. As indicated by their gray values of fluorescence images captured by a smartphone camera in Figure 5a, we found only the LAMP zones with B. pertussis DNA samples exhibited strong fluorescence, while non-specific DNA samples showed weak signals similar to negative control NC2. The corresponding LAMP products from different bacterial DNA samples were separately extracted for gel electrophoresis analysis to confirm the results. As shown in Figure 5b, only the LAMP products extracted from the LAMP zones with B. pertussis DNA sample exhibited ladder-pattern bands, indicating the high specificity of our approach for B. pertussis detection. These results indicated that our approach can obtain both high sensitivity and high specificity for pertussis diagnosis, which is challenging for the current diagnostic methods including advanced real-time qPCR [7, 13].
Figure 5.
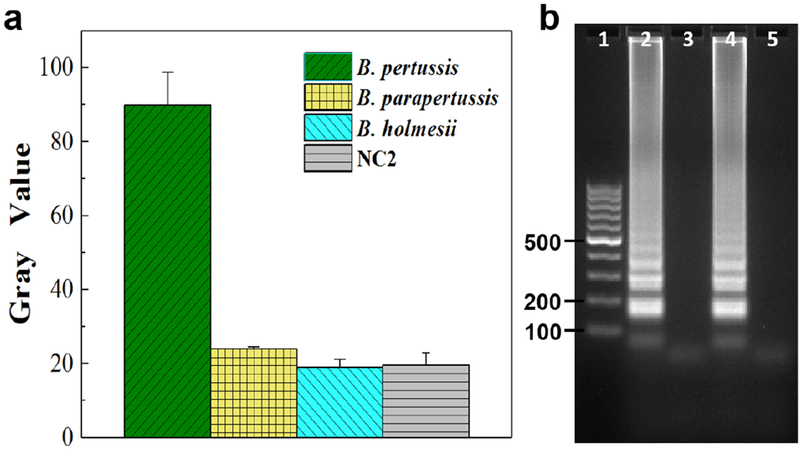
Specificity investigation, (a) Gray values of the specificity testing results using a smartphone camera by simultaneously identifying B. pertussis from B. parapertussis and B. holmesii. (b) Gel electrophoresis analysis. Lane 1: 100 bp marker; Lanes 2-3: LAMP products from B. pertussis and B. parapertussis DNA samples; Lanes 4-5: LAMP products from B. pertussis and B. holmesii DNA samples. All LAMP zones were preloaded with B. pertussis LAMP primers. Different DNA samples of B. pertussis, B. parapertussis, and B. holmesii (5 × 106 copies per LAMP zone), as well as the NC2 (without template DNA), were individually introduced into their corresponding LAMP zones.
3.4. Optimization of the centrifuge-free lysis process
To avoid complicated and time-consuming sample preparation processes such as DNA extraction, we developed and optimized a simple centrifuge-free lysis approach for direct detection of B. pertussis bacteria. Considering the complex matrix in clinical samples, we collected nasopharyngeal swabs specimens from healthy volunteers and added B. pertussis bacteria to prepare nasopharyngeal samples to investigate the bacterial lysis performance. Figure 6a shows the epithelial cells and traces of mucus from the collected nasopharyngeal swab.
Figure 6.
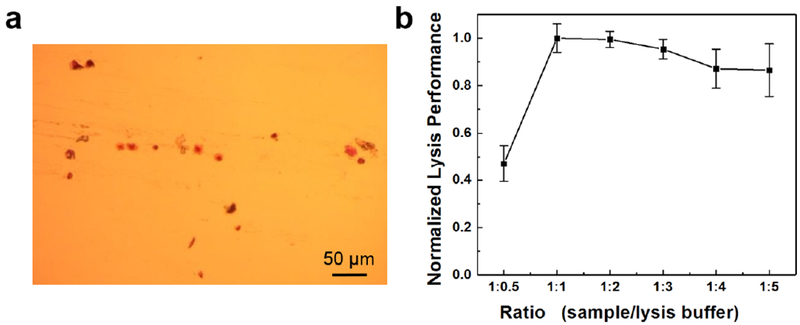
Nasopharyngeal swab specimen and optimization of the lysis performance. (a) A Gram-stained smear of nasopharyngeal swabs showing epithelial cells and traces of mucus. (b) Optimization of the ratio between the sample and the lysis buffer (v/v) based on the normalized lysis performance of different mixtures.
A serial of the nasopharyngeal sample and lysis buffer mixtures were prepared at different ratios. The corresponding lysis performance was evaluated based on the obtained DNA amounts from each of the mixtures, as shown in Figure 6b. It demonstrated that high lysis performance could be achieved when the ratio between the sample and the lysis buffer was below 1.0. The lysis performance in this ratio range (1:1 to 1:5) was 1.8-fold higher than that at the ratio of 1:0.5. However, smaller ratios meant more lysis buffer needed, which could dilute the concentration of released DNA from bacteria in samples. Therefore, the ratio of 1.0 between the sample and the lysis buffer was chosen as the optimal ratio for the following test.
3.5. Direct and instrument-free detection of B. pertussis bacteria in nasopharyngeal samples
We finally performed the direct detection of B. pertussis bacteria in nasopharyngeal samples based on the optimized lysis procedure. To eliminate the false positives or false negatives, NC2 and PC2 (using purified B. pertussis template DNA, 1 μL ~7.5 × 104 copies/μL) were employed. All LAMP zones were pre-loaded with B. pertussis primers, then the purified B. pertussis DNA sample as the positive control, the lysate (i.e. nasopharyngeal sample/lysis buffer mixture after incubation), and the NC2 with their LAMP reaction mixture were introduced from each outlet to their corresponding LAMP zones.
Figure 7 shows the results of the direct detection of the nasopharyngeal sample. It can be seen that strong fluorescence could be produced from both the nasopharyngeal sample and the purified DNA sample (Figure 7a and 7b), which was much higher than that of the NC2. No obvious differences were observed by comparing the gray values and fluorescence intensities of the LAMP products from both the nasopharyngeal sample and the purified DNA sample (Figure 7a and 7b). The results were further confirmed by the gel electrophoresis analysis. As shown in Figure 7c, the LAMP products from both the nasopharyngeal sample and the extracted B. pertussis DNA sample showed clear ladder-pattern DNA ladders. The successful detection indicated that our optimized lysis approach was very efficient and totally compatible with on-chip LAMP reactions for direct detection of nasopharyngeal swab samples, in which the human cells (Figure 6a) and cellular matrix didn’t cause any inhibitory problems. Therefore, it demonstrated that our approach can achieve direct detection of clinical samples without relying on any equipment (e.g. centrifuge and water bath) or any complicated and time-consuming sample preparation process (i.e. DNA extraction).
Figure 7.
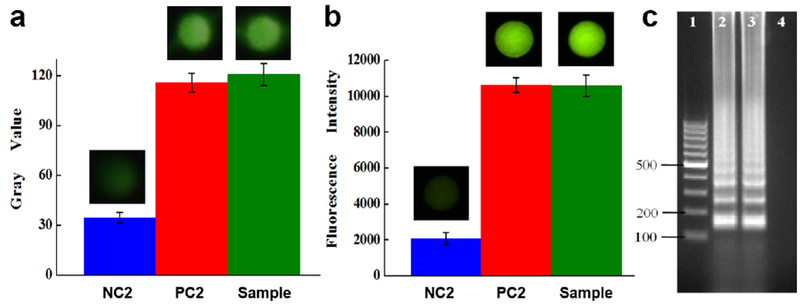
Direct detection of B. pertussis bacteria in nasopharyngeal samples. Gray values (a) and fluorescent intensities (b) of the LAMP products. Insets are fluorescence images, (c) Gel electrophoresis analysis. Lanes 1-4: 100 bp marker; LAMP products from the nasopharyngeal sample and purified B. pertussis DNA; NC2.
4. Conclusion
We developed a low-cost hybrid microfluidic platform for rapid and instrument-free POC detection of whooping cough, with high sensitivity and specificity. The 3D-printed battery-powered bioanalyzer and the optimized centrifuge-free sample lysis procedure significantly enhanced the portability and the field detection capacity of the hybrid biochip for the POC diagnosis of pertussis. This microfluidic approach for pertussis diagnosis has the following features. (1) It is low-cost. The cost of the ready-to-use paper/polymer hybrid microfluidic device was only ~30 cents, and the cost per assay was ~3 dollars. (2) It is rapid. The whole assay process took less than one hour. (3) It is instrument-free. The testing results can be easily detected by the naked eye under a portable UV light pen without relying on any instruments. The detection system is portable and fully battery-powered without relying on external AC power supply. (4) It is highly sensitive and specific. The LOD was found as low as 5 DNA copies per LAMP zone. Specific identification for B. pertussis among other similar Bordetella species has been achieved. (5) Samples can be directly detected without any time-consuming and complicated sample preparation procedures or the use of centrifuges. All these significant features make this microfluidic approach suitable for POC detection of B. pertussis, which can be potentially applied for fast and accurate pertussis diagnosis after clinical validation in low-resource settings such as developing nations, physician’s offices, and schools. In addition, it should have broad applications in POC testing of a variety of other pathogens (e.g. foodborne pathogens).
Supplementary Material
Highlights:
We for the first time report a low-cost hybrid microfluidic platform integrated with LAMP for rapid and instrument-free detection of whooping cough (pertussis). This is also the first point-of-care (POC) device for pertussis diagnosis.
We developed a portable and fully battery-powered heater to support on-chip LAMP reactions, which significantly enhanced the capacity for point-of-care detection of B. pertussis without the requirement of the external AC power supply. This allows rapid pertussis diagnosis on site or in the field.
The microfluidic approach demonstrated high specificity by detecting B. pertussis and other Bordetella species. The limit of detection of 5 DNA copies per LAMP zone was achieved, demonstrating high detection sensitivity.
By using an optimized centrifuge-free lysis protocol, direct detection of B. pertussis bacteria was achieved without laborious and time-consuming DNA extraction procedures.
Our novel microfluidic approach has great potential for point-of-care detection of many other pathogens such as foodborne pathogens, especially for low-resource settings.
Acknowledgments
We would like to acknowledge the financial support from the National Institute of Allergy and Infectious Disease of the NIH (R21AI107415), the Philadelphia Foundation, the Medical Center of the Americas Foundation, and the U.S. NSF-PREM program (DMR 1205302 and DMR 1827745). Financial support from the National Institute of General Medical Sciences of the NIH (SC2GM105584), the NIH RCMI Pilot grant, the NIH BUILDing Scholar Summer Sabbatical Award (NIGMS Award Numbers RL5GM118969, TL4GM118971, and UL1GM11897), the University of Texas at El Paso (UTEP) for the IDR Program, and University of Texas (UT) System for the STARS award is also greatly acknowledged. We would like to express special thanks to Dr. Delfina Dominguez for her contribution in microbiology and the staff of the Genomic Analysis Core Facility of UTEP This core is supported by Grant G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors XL and MD submitted a USA patent (pending) and have financial interest on the work. Other authors have no financial interest on the work.
References:
- [1].Kilgore PE, Salim AM, Zervos MJ, Schmitt H-J, Pertussis: microbiology, disease, treatment, and prevention, Clinical Microbiology Reviews, 29 (2016) 449–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].WHO, WHO vaccine-preventable diseases: monitoring system, 2009 global summary, http://apps.who.int/iris/bitstream/10665/70149/1/WHO_IVB_2009_eng.pdf, 2009.
- [3].CDC, Centers for Disease Control and Prevention, About pertussis outbreaks, 2017.
- [4].CDC, Centers for Disease Control and Prevention, Pertussis in other countries, 2017.
- [5].Pierce VM, Elkan M, Leet M, McGowan KL, Hodinka RL, Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children, Journal of Clinical Microbiology, (2011) JCM-05996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dragsted DM, Dohn B, Madsen J, Jensen JS, Comparison of culture and PCR for detection of Bordetella pertussis and Bordetella parapertussis under routine laboratory conditions, Journal of Medical Microbiology, 53 (2004) 749–754. [DOI] [PubMed] [Google Scholar]
- [7].Orenstein WA, Pertussis in adults: epidemiology, signs, symptoms, and implications for vaccination, Clinical Infectious Diseases, 28 (1999) S147–S150. [DOI] [PubMed] [Google Scholar]
- [8].Martini H, Detemmerman L, Soetens O, Yusuf E, Pierard D, Improving specificity of Bordetella pertussis detection using a four target real-time PCR, PLoS One, 12 (2017) e0175587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Williams MM, Taylor TH, Warshauer DM, Martin MD, Valley AM, Tondella ML, Harmonization of Bordetella pertussis real-time PCR diagnostics in the US, 2012, Journal of Clinical Microbiology, 53 (2015) 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tomita N, Mori Y, Kanda H, Notomi T, Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products, Nature Protocols, 3 (2008) 877. [DOI] [PubMed] [Google Scholar]
- [11].Notomi T, Mori Y, Tomita N, Kanda H, Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects, Journal of Microbiology, 53 (2015) 1–5. [DOI] [PubMed] [Google Scholar]
- [12].Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, Loop-mediated isothermal amplification of DNA, Nucleic Acids Research, 28 (2000) e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kamachi K, Toyoizumi-Ajisaka H, Toda K, Soeung SC, Sarath S, Nareth Y, Horiuchi Y, Kojima K, Takahashi M, Arakawa Y, Development and evaluation of a loop-mediated isothermal amplification method for rapid diagnosis of Bordetella pertussis infection, Journal of Clinical Microbiology, 44 (2006) 1899–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brotons P, de Paz HD, Esteva C, Latorre I, Munoz-Almagro C, Validation of a loop-mediated isothermal amplification assay for rapid diagnosis of pertussis infection in nasopharyngeal samples, Expert Review of Molecular Diagnostics, 16 (2016) 125–130. [DOI] [PubMed] [Google Scholar]
- [15].Fujino M, Suzuki E, Watanabe M, Nakayama T, Loop-mediated isothermal amplification (LAMP) aids the clinical diagnosis of pertussis, Japanese Journal of Infectious Diseases, 68 (2015) 532–533. [DOI] [PubMed] [Google Scholar]
- [16].Urdea M, Penny LA, Olmsted SS, Giovanni MY, Kaspar P, Shepherd A, Wilson P, Dahl CA, Buchsbaum S, Moeller G, Requirements for high impact diagnostics in the developing world, Nature, 444 (2006) 73–79. [DOI] [PubMed] [Google Scholar]
- [17].Volpatti LR, Yetisen AK, Commercialization of microfluidic devices, Trends in Biotechnology, 32 (2014) 347–350. [DOI] [PubMed] [Google Scholar]
- [18].Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE, Point of care diagnostics: status and future, Analytical Chemistry, 84 (2011) 487–515. [DOI] [PubMed] [Google Scholar]
- [19].Xu X, Akay A, Wei H, Wang S, Pingguan-Murphy B, Erlandsson B-E, Li X, Lee W, Hu J, Wang L, Advances in smartphone-based point-of-care diagnostics, Proceedings of the IEEE, 103 (2015) 236–247. [Google Scholar]
- [20].Sanjay ST, Fu G, Dou M, Xu F, Liu R, Qi H, Li X, Biomarker detection for disease diagnosis using cost-effective microfluidic platforms, Analyst, 140 (2015) 7062–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dou M, Sanjay ST, Benhabib M, Xu F, Li X, Low-cost bioanalysis on paper-based and its hybrid microfluidic platforms, Talanta, 145 (2015) 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dou M, Lopez J, Rios M, Garcia O, Xiao C, Eastman M, Li X, A fully battery-powered inexpensive spectrophotometric system for high-sensitivity point-of-care analysis on a microfluidic chip, Analyst, 141 (2016) 3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sanjay ST, Dou M, Sun J, Li X, A paper/polymer hybrid microfluidic microplate for rapid quantitative detection of multiple disease biomarkers, Scientific Reports, 6 (2016) 30474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sanjay ST, Dou M, Fu G, Xu F, Li X, Controlled drug delivery using microdevices, Current Pharmaceutical Biotechnology, 17 (2016) 772–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sanjay ST, Zhou W, Dou M, Tavakoli H, Ma L, Xu F, Li X, Recent advances of controlled drug delivery using microfluidic platforms, Advanced Drug Delivery Reviews, 128 (2018) 3–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dou M, Garcia JM, Zhan S, Li X, Interfacial nano-biosensing in microfluidic droplets for high-sensitivity detection of low-solubility molecules, Chemical Communications, 52 (2016) 3470–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pang B, Fu K, Liu Y, Ding X, Hu J, Wu W, Xu K, Song X, Wang J, Mu Y, Development of a self-priming PDMS/paper hybrid microfluidic chip using mixed-dye-loaded loop-mediated isothermal amplification assay for multiplex foodborne pathogens detection, Analytica Chimica Acta, 1040 (2018) 81–89. [DOI] [PubMed] [Google Scholar]
- [28].Dou M, Dominguez DC, Li X, Sanchez J, Scott G, A versatile PDMS/paper hybrid microfluidic platform for sensitive infectious disease diagnosis, Analytical Chemistry, 86 (2014) 7978–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dou M, Sanjay ST, Dominguez DC, Liu P, Xu F, Li X, Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip, Biosensors and Bioelectronics, 87 (2017) 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dou M, Sanjay ST, Dominguez DC, Zhan S, Li X, A paper/polymer hybrid CD-like microfluidic SpinChip integrated with DNA-functionalized graphene oxide nanosensors for multiplex qLAMP detection, Chemical Communications, 53 (2017) 10886–10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].de Almeida Gomes BPF, Vianna ME, Sena NT, Zaia AA, Ferraz CCR, de Souza Filho FJ, In vitro evaluation of the antimicrobial activity of calcium hydroxide combined with chlorhexidine gel used as intracanal medicament, Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology, 102 (2006) 544–550. [DOI] [PubMed] [Google Scholar]
- [32].Mori Y, Notomi T, Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases, Journal of Infection and Chemotherapy, 15 (2009) 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Templeton KE, Scheltinga SA, van der Zee A, Diederen BMW, Kruijssen AM, Goossens H, Kuijper E, Claas ECJ, Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis, Journal of Clinical Microbiology, 41 (2003) 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


