Abstract
Objective
To provide an open source, interoperable, and scalable data quality assessment tool for evaluation and visualization of completeness and conformance in electronic health record (EHR) data repositories.
Materials and Methods
This article describes the tool’s design and architecture and gives an overview of its outputs using a sample dataset of 200 000 randomly selected patient records with an encounter since January 1, 2010, extracted from the Research Patient Data Registry (RPDR) at Partners HealthCare. All the code and instructions to run the tool and interpret its results are provided in the Supplementary Appendix.
Results
DQe-c produces a web-based report that summarizes data completeness and conformance in a given EHR data repository through descriptive graphics and tables. Results from running the tool on the sample RPDR data are organized into 4 sections: load and test details, completeness test, data model conformance test, and test of missingness in key clinical indicators.
Discussion
Open science, interoperability across major clinical informatics platforms, and scalability to large databases are key design considerations for DQe-c. Iterative implementation of the tool across different institutions directed us to improve the scalability and interoperability of the tool and find ways to facilitate local setup.
Conclusion
EHR data quality assessment has been hampered by implementation of ad hoc processes. The architecture and implementation of DQe-c offer valuable insights for developing reproducible and scalable data science tools to assess, manage, and process data in clinical data repositories.
Keywords: data quality, electronic health records, data science, medical informatics applications
INTRODUCTION
The exponential upsurge in the volume of medical records coupled with the advent of electronic data repositories and data-sharing infrastructures is transforming biomedical research into a collaborative and increasingly data-driven area of inquiry. High throughput of clinical data offers unprecedented opportunities to accelerate scientific discoveries that can translate into improved patient care. Electronic health record (EHR) systems provide valuable information about determinants of health and treatment effectiveness. The uptake of EHRs within the past few years1–4 offers huge potential for improved secondary use of EHR data in health care research and decision-making.5–7 Large-scale distributed clinical research data networks, such as the National Patient-Centered Clinical Research Network (PCORnet)8 and the Electronic Medical Records and Genomics (eMERGE) network,9 and regional practice-based research networks, such as Data QUEST,10 are expanding EHR-based data-driven research. However, the massive data constantly produced in EHRs present unique challenges for secondary use,11 and therefore have yet to be maximized to improve the efficiency of the health care system.2
Implementation of ad hoc data-management practices has exacerbated the challenges of integrating diverse clinical data across distributed EHR research data networks.12 Data science contributes invaluable methodologies that can be used to design and implement scalable, reproducible, and interoperable clinical data-management practices. A recent National Institutes of Health initiative (Big Data to Knowledge) fosters application of data science methodologies in health care to improve patient care by supporting creative thinking on generation, management, and analysis of large-scale biomedical data.13–15
Data quality poses important concerns for secondary use of EHR data,16–19 which has not been properly addressed in research data networks using ad hoc solutions. Systematic evaluation of EHR data quality is a priority for reliable secondary use of the data. The absence of standardized data quality assessment measures is a barrier to systematic evaluation of EHR data quality.6,19–23 EHR data quality evaluation requires consistency.21,24 Harmonization across methodologies is key to establishing consistent EHR data quality assessment. Kahn et al.25 introduced a “harmonized framework” for assessing EHR data quality across 3 categories: conformance, completeness, and plausibility. Among these categories, completeness has garnered more attention.6,26 Completeness has often been measured in the EHR data quality assessment research by missing and/or blank data values.21 We build upon the data quality definitions presented in the harmonized framework to develop DQe-c, a tool that operationalizes the completeness category – and a completeness-related dimension of the conformance category – defined in the framework.
DATA QUALITY ASSESSMENT IN CONTEXT
The absence of standardized data quality assessment measures before the harmonized framework has led to subjective, use-oriented21,27,28 approaches to EHR data quality assessment in the clinical research informatics community. Data quality measures have generally been defined based on an intended use case for data, and therefore their definitions are often inconsistent, overlapping (ie, same definition for different terminologies),6,21,29 ambiguous,29 or complementary to each other.30 Nevertheless, the subjective use-oriented approach to data quality assessment is almost a requirement to ensure fitness for use in clinical research using EHR data. In the context of growing clinical research data networks, applying use-oriented approaches to the existing multiplicity of data models presents a challenge for the systematic evaluation of data quality across distributed networks. Scalability and interoperability are integral characteristics of data quality assessment tools. It is important to consider data quality assessment in the context of the data life cycle from patient to researcher (Figure 1).
Figure 1.
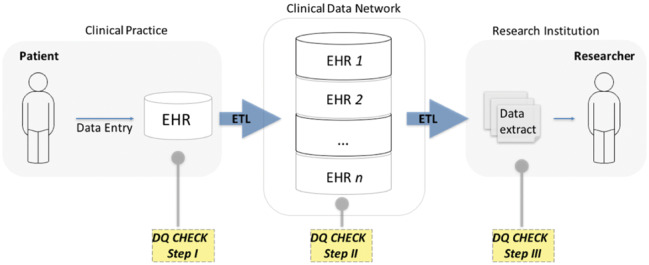
Data life cycle from patient to researcher.
Figure 1 represents a simplified life cycle for research data, where patient data are collected into an EHR (in a clinical practice), extracted, transformed, and loaded into a clinical data repository sponsored by a clinical data network and, after another extract, transform, load process, delivered to a researcher. At least 3 steps of data quality assessment need to happen across the data life cycle from patient to researcher. Each of these steps serves a different purpose and would ideally include data quality measures that build upon one another to form a comprehensive list of data quality assessment measures. Data quality checks in step 1 focus on potential data entry issues. As data in distributed data networks often come from multiple EHRs, step 2 data quality checks focus on a broad range of harmonization issues due to the diversity of data models and ontologies. The final data extract for research is often designed for a specific purpose,31 which requires a subjective use-oriented assessment of data quality in step 3 to satisfy minimum fitness-for-use requirements. Therefore, subjective use-oriented approaches to data quality are well justified for step 3 checks.
The availability of widely used information models such as i2b2,32,33 and the adoption of common data models (CDMs) such as the PCORnet CDM and the Observational Medical Outcomes Partnership (OMOP) CDM in clinical data networks facilitate designing step 2 data quality assessment solutions that are interoperable across distributed networks. Data quality assessment in steps 1 and 2 hold shared utilities that can lead to interoperable solutions. DQe-c is a scalable and interoperable data science–driven tool for EHR data quality evaluation, focused on completeness in clinical data repositories. The iterative design of DQe-c was tailored for use in step 2, but can also be utilized in steps 1 and 3 with some user-defined modifications.
According to Weiskopf et al.,18 completeness in EHR is a contextual concept, meaning that it can be defined differently based on different data needs and purposes. In the harmonized framework, completeness is defined as a measure of “the absence of data at a single moment over time or when measured at multiple moments over time, without reference to its structure or plausibility.”25 We use this definition in the context of a clinical data repository (step 2 data quality assessment), and also add user-defined flexibility to ensure that our operationalization embraces an inclusive definition of “data” as a representation of factual information. That is, we measure completeness as the presence of “sensical” data, regardless of whether a data point is plausible or not. Accordingly, DQe-c differentiates 2 types of missingness: (1) presence of absence (true null) and (2) presence of nonsense, where the latter category can be defined by the user.
Below we describe DQe-c’s design and architecture, provide an overview of its outputs using a sample dataset from real patient EHR data, and discuss our experience implementing the tool across clinical data repositories and our ongoing efforts to improve its interoperability across major EHR-based data networks.
DQe-C DESIGN AND ARCHITECTURE
We designed, architected, and developed DQe-c through an iterative process, which enabled us to incorporate feedback from a diverse group (including clinicians, medical informaticists, data scientists, database administrators, computer scientists, and research analysts) across multiple institutions. The process was guided by 2 questions. The initial question that inspired us to design DQe-c was: How can we measure completeness in a clinical data repository? A priority list of tests was generated from the data completeness category of the harmonized data quality assessment framework through iterations within the Data QUEST Coordinating Center of experts, which included individuals with expertise in biomedical informatics, biostatistics, and primary clinical care. Following the priority list, the architecture and development of DQe-c are the result of our efforts to answer a principal question: How can we operationalize the list of tests in a scalable interoperable tool that would allow database administrators, data network managers, and analysts to measure completeness in clinical data repositories?
Initial DQe-c development efforts focused on designing its outputs. DQe-c output visualizations were designed through iterations of design and evaluation of the tool’s prototype within the University of Washington Data QUEST Coordinating Center. The prototype was then adapted to the Accessible Research Commons for Health (ARCH) Clinical Data Research Network (CDRN) (formerly known as Scalable Collaborative Infrastructure for a Learning Healthcare System34) and transformed into production through iterative implementations across the network’s diverse partner institutions, resulting in major improvements to the architecture of DQe-c.
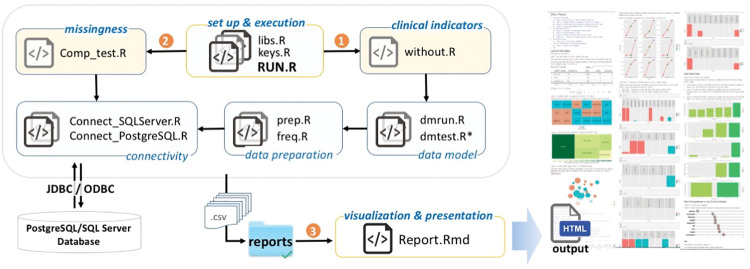
Interoperability, scalability, and open science are 3 key considerations we aimed to instill in the DQe-c architecture. We architected DQe-c to flexibly accommodate site-/user-specific needs as well as future development. DQe-c is modular and was developed in R statistical language. Data preparation, analysis, and visualization are performed in 7 modules, each module consisting of 1 or more R scripts (Figure 2). Modules 1–6 perform data preparation and analysis and store their outputs as comma-separated flat files in the reports directory. The modular design increases the flexibility of the tool for future improvements and facilitates interoperability.
Figure 2.
DQe-c workflow.
The setup and execution module consists of scripts that govern the tool’s execution, initiating 3 modules sequentially (the order of initiations is identified in Figure 2). Scripts within each module initiate their dependent scripts. Overall, DQe-c works on PCORnet CDM (versions 3 and 3.1) and OMOP CDM (versions 4 and 5), and operates on 2 Relational Database Management Systems (RDBMSs): Microsoft Structured Query Language (SQL) Server and Oracle. It calls SQL queries from within R commands via a Java database connectivity (JDBC)/open database connectivity (ODBC) connection; connection settings need to be set in the connectivity module. This capability increases the scalability of the tool against large-scale repositories.
Two flat files provide the CDM templates for DQe-c to operate in the data preparation module. The first step in expanding the tool’s functionality to other CDMs is to create a new CDM template and modify the data preparation module in order to direct the tool to the correct CDM template flat file.
The visualization and presentation module includes an R Markdown document that generates the Hypertext Markup Language (HTML) report from completeness tests conducted through DQe-c. This module uses the outputs of its preceding modules, as they are stored with specific names as comma-separated flat files in the reports directory. We provide a brief description of the DQe-c report in the next section with example visualizations. Data used for all visualizations in this article are from a sample database of 200 000 randomly selected records of real patients with encounters since January 1, 2010, extracted from the RPDR at Partners HealthCare.35 Use of RPDR data was approved by Partners’ Institutional Review Board.
DQe-C OUTPUTS
Each run of DQe-c generates an HTML report that summarizes outputs from its data preparation and analytics in tables and graphs. The report is organized into 4 sections.
Load and test details
The first section of the report presents a database-level snapshot summary of the latest data loaded in the clinical data repository. The summary begins with a table presenting a list of CDM tables, their availability status, and the gigabyte (GB) size and number of rows for each table. This information is then presented in 3 visualizations (Figure 3): 2 treemaps and an interactive visualization of table-level completeness that will help users obtain a comprehensive understanding of the relative importance of missing tables in relation to the CDM. Data for this section of the report are generated by the data preparation module. For example, information presented in Figure 3 shows that 3 of the PCORnet CDM tables were not loaded into the data repository. The third plot shows that no other table in the CDM has relational dependencies on the 3 unavailable tables; therefore, users may consider investigating the reasons behind the unavailability of the 3 tables as a low-priority task.
Figure 3.
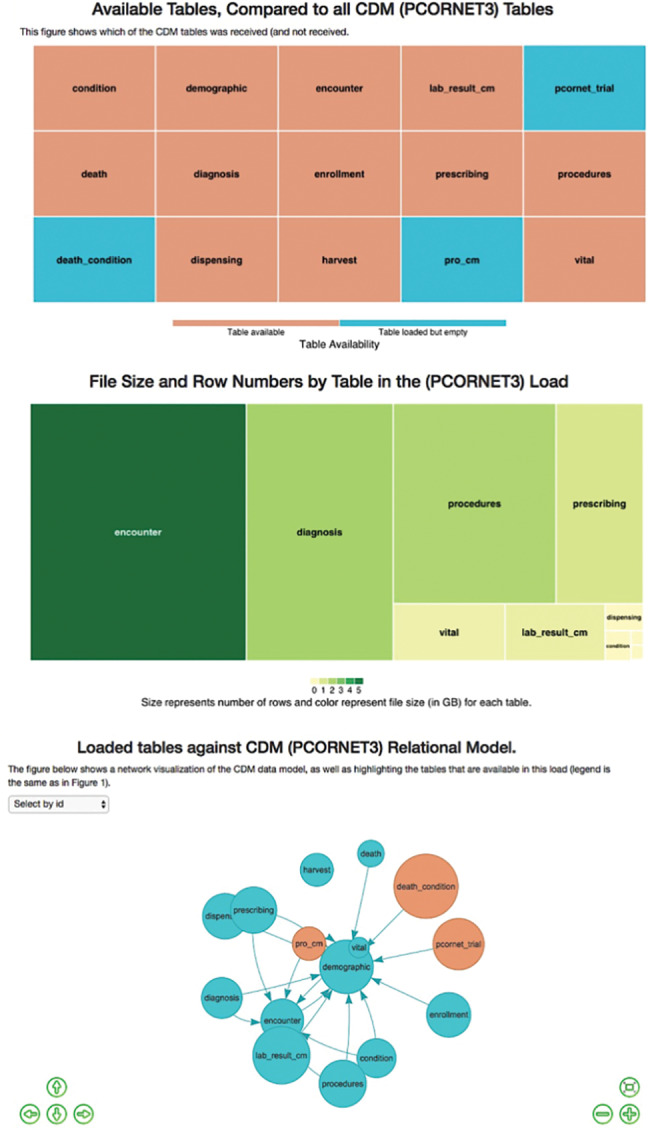
Load details visualizations.
Completeness results
The second section of the report illustrates the results of data preparation and missingness modules. Upon completion of each run of DQe-c, a reference table is produced and saved in the reports directory, which includes frequencies of rows, unique values, missingness, and percent missingness for each column and table (Table 1 provides a description of the columns and their contents in the reference table). This table in the DQe-c architecture is called the “Master Completeness Results” table.
Table 1.
Columns and contents of the reference table.
| Column Name | Description |
|---|---|
| TabNam | Table name |
| ColNam | Column name |
| test_date | DQe-c run date |
| FRQ | Number of rows in column |
| UNIQFRQ | Number of unique values in column |
| MS1_FRQ | Frequency of missing values, missingness category 1a in column |
| MS2_FRQ | Frequency of missing values, missingness category 2b in column |
| MSs_PERC | Percent total missingness in column |
| Organization | Organization name |
| CDM | Common data model |
aCategory 1 of missingness measures the presence of absence (true null).
bCategory 2 of missingness measures the presence of “nonsense.”
DQe-c’s approach to missingness/completeness encompasses a broad yet flexible connotation. We measure completeness as the presence of “sensical” data, regardless of whether a data point is plausible or not. That is, if for any reason a data point does not include a value or an attribute that is compatible for analysis – ie, the analyst needs to treat the data point with missing data procedures – we will consider it as missing data. Accordingly, if needed, DQe-c can differentiate 2 categories of missingness: (1) presence of absence (MS1) and (2) presence of nonsense (MS2). Category 1 of missingness (MS1) is the conventional definition for missingness, counting the frequency of NULL/NA values or empty strings in each column. Category 2 of missingness (MS2) can be defined by the user. In implementing DQe-c in ARCH CDRN, we currently consider data points with characters that do not represent meaning (including “+,” “-,” “_,” “#,” “$,” “*,” “\,” “?,” “.,” “&,” “^,” “%,” “!,” “@,” and “NI”) in the MS2 category. This list can be easily modified in the missingness module using a text editor or R, or can be left empty to not be counted as missingness.
Changes in primary keys across loads
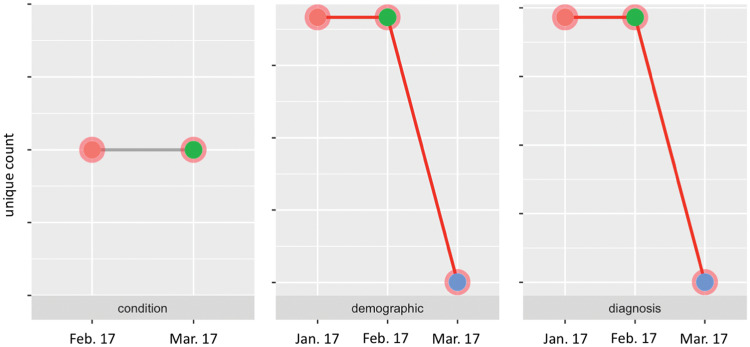
Before presenting the results of missingness for each table, DQe-c first visualizes how completeness in key variables changes over time/data loads. The purpose of this visualization is to compare key quantities of presence over time to track potential significant changes in a clinical data repository. For this purpose, DQe-c profiles changes over time in primary keys for available tables across loads. Figure 4 shows a snapshot of the visualizations produced for a PCORnet CDM database. The tool also utilizes reports generated from the previous runs to automatically compile data for this visualization. For example, Figure 4 shows significant drops in the count of unique patients (demographic table) and diagnoses in the latest data refresh, compared with the previous 2 data refreshes (as documented by the previous 2 DQe-c runs). Such a drastic decline in number of patients would raise a flag for the database administrators and IT staff as a likely sign of issues with the extract, transform, load or CDM transformation. The figure also shows that while the number of unique conditions did not change between the last 2 loads, the condition table was not available in the first data load. This may indicate a potential issue in mapping data onto the condition table.
Figure 4.
A snapshot of changes in primary keys across loads.
The proportion of missing data in loaded tables
The DQe-c report visualizes the missingness percentages by column for each of the tables available from the CDM, differentiating the 2 types of missingness (MS1 and MS2). Figure 5 presents an example of 2 tables (encounter and diagnosis) from the PCORnet CDM. The figure shows that, for instance, there is around 15% missingness in the columns “providerid,” “enc_type,” and “pdx” from the tables “encounter” and “diagnosis,” which in this example case is due to the existence of a nonsensical character.
Figure 5.
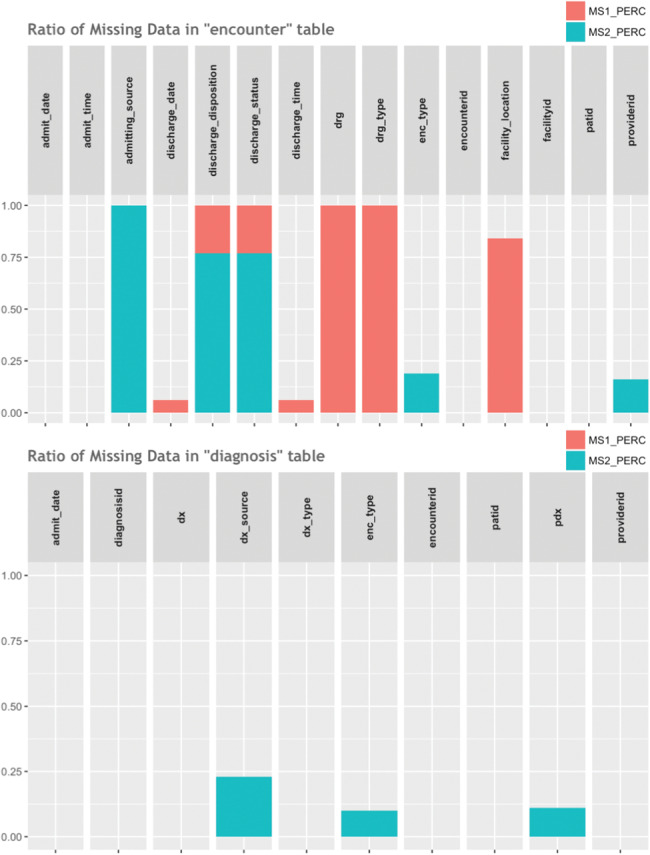
A snapshot of missingness percentages by table and column.
As Figure 5 illustrates, some columns have missingness of category 1 (NULL/NA/empty string), and some have a combination of cells with both categories of missingness. The bar charts distinguish between the two and allow users to see the overall missingness percentage.
Data model conformance test
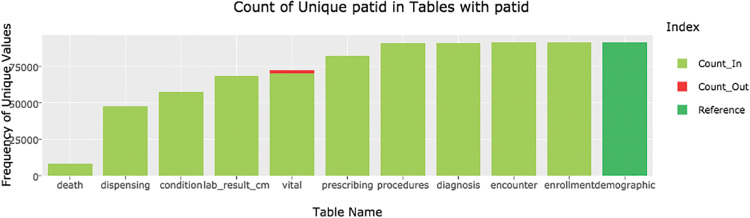
The data model module in DQe-c enables the tool to perform data model tests that are related to completeness. The tool performs a test that looks for orphan records among common key variables based on the CDM constraints. Results are visualized in a series of interactive bar charts. Figure 6 presents an example of the “patid” variable in PCORnet CDM.
Figure 6.
Count of unique patids in tables with patid in PCORnet v3 CDM. There were no Count_Out patid records in the vital table from the sample RPDR database. For purposes of visualization, we manually added some values to the table that were used to generate this graphic.
The procedure to identify and visualize orphan records begins with identifying common variables among tables of the CDM. DQe-c uses the CDM template to generate a reference table for each common variable in the data model module. For example, in the PCORnet v3 CDM, the demographic table is the reference table for patid. That is, all other tables’ patids should be included in the demographic table, otherwise they are identified as orphan rows. The data model module has functions to categorize each unique value in common keys under “Count_In” or “Count_Out” (orphan rows), based on the unique values available in the reference table. Figure 6, for instance, shows that there is a small proportion of orphan patids in the “vital” table – ie, a small number of patids in the “vital” table are not available in the “demographic” table.
Test of missingness in key clinical indicators
All of the completeness tests that DQe-c performs up to here are agnostic to clinical meaning – ie, they treat data without reference to clinical meaning. The clinical indicators module in DQe-c enables users to evaluate data completeness from a subjective viewpoint. The tool calculates the proportion of patient records that are missing data on key clinical indicators, such as height, weight, blood pressure, medication, diagnosis, and encounter records, and demographic data including gender, race, and ethnicity (Figure 7). The selection of clinical indicators in this module is flexible and can be customized based on local needs. To calculate these percentages, DQe-c first calculates a denominator from unique patients in the “demographic” table in PCORnet CDM and the “person” table in OMOP CDM with a valid date of birth; a minimum date of birth can also be set. The missingness percentage is the product of dividing the total number of unique patient records that are missing any data whatsoever for a given clinical indicator (eg, no medication record) by the denominator.
Figure 7.
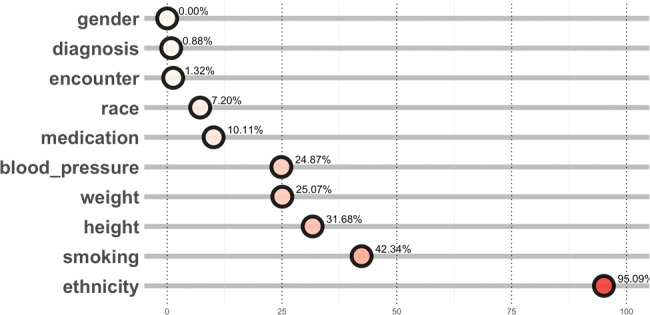
Test of missingness in key clinical indicators – percentage of missingness.
For example, Figure 7 shows that 24.87% and 95.09% of the patient records in the sample data did not have any blood pressure or ethnicity data, respectively. Information from this visualization would guide the database management team to investigate what proportion of the missingness percentages are due to lack of records (eg, patient ID not found in vital table) or to missingness in the respective table (eg, blood pressure record missing for patient in vital table). We found that a high percentage of missingness in ethnicity was due to counting “NI” as a missingness character, under category 2 missingness, that was not of concern to the ARCH network.
DISCUSSION
Open science is a key consideration in the design and architecture of DQe-c. Making computer code available is a requirement for open science.36 We use GitHub (https://github.com/hestiri/DQe-c) as the main platform to distribute DQe-c under an i2b2 open source license, as well as to involve users in development of the tool – as we strive to add new analytics to the tool, users can also recommend new features. With the advent of large-scale health data research networks, interoperability across major clinical research informatics platforms is another key consideration in DQe-c’s architecture. The tool works on 2 commonly used CDMs, PCORnet and OMOP. DQe-c has been successfully implemented in the University of Washington WWAMI region Practice and Research Network’s Data QUEST37 network and within the DARTNet Institute.38 As part of a data quality testing standard operating procedure, the University of Washington team has developed a procedure to run DQe-c that posts the HTML output onto a protected web page as data are refreshed in the Data QUEST OMOP warehouse. The University of Washington Data QUEST Coordinating Center additionally supports the DARTNet Institute’s use of the tool to test completeness across national OMOP data repositories used to conduct clinical research.
DQe-c has also been successfully implemented in 8 of the ARCH partner institutions. Installing and running the tool on the ARCH network revealed issues that do not directly fit into specific categories of data quality assessment. For example, we found spelling mismatches in table names compared with the PCORnet CDM and an instance of very slow query performance, which has spurred improvements in the network. We provided individual assistance to the sites for installation and first run of DQe-c. Due to institutional and sociotechnical limitations across ARCH partner institutions (eg, staff availability and expertise, server and database environment), implementation on this CDRN has directed us to find ways to facilitate local setup. Some of the main issues for the first installation and execution of DQe-c on ARCH sites involved installing Java on secured servers and the JDBC connection setup, which is related to a lack of local experience with using R to analyze relational data. Installing R and DQe-c’s required packages might be facilitated by wrapping the tool and all its software and systems settings into a standardized container for software development, such as Docker. We are considering this approach for a future release. We also made changes to DQe-c to address concerns expressed during the implementation phase, such as enhanced password security. We believe that the time to help sites install the software requirements (ie, R, R Studio, Java) was time well spent, as it will augment the network’s capacity to develop and apply harmonized data science methodologies for processing and analysis of EHR data.
Scalability to large databases has been an active area of exploration in DQe-c’s development. The first prototype of the tool used in-memory processing – ie, it needed to load the entire dataset into memory for data processing. Although we were able to successfully test the prototype on an OMOP database with >1 million patients (which contained >140 million observation records), we learned that scaling up to very large databases would be a challenge. We applied an ad hoc solution to this issue by developing an add-on that uses the flat file reports (that are small in size) from individual DQe-c runs to generate an aggregated report. Using this add-on will allow large organizations to run the tool in chunks (eg, per hospital or clinical unit) and to generate an aggregated report for the entire federated network. One of the major improvements in DQe-c from prototype to production is the adoption of an out-of-memory solution. DQe-c operates on 2 RDBMSs, MS SQL Server and Oracle, calling SQL queries from within R to create the flat file reports and generate the final HTML report. This upgrade has significantly reduced the memory reliance. For instance, running DQe-c prototype on an OMOP database with 1 million unique patients required a server with >30 GB memory, whereas a DQe-c run on a PCORnet database with 2 million unique patients runs on any laptop, with virtually no significant memory implications, because all the queries are handled on the database server’s RDBMS. The upgrade also can accelerate the data-processing time, as RDBMSs are faster at querying data than R.
To our knowledge, a few other data tools exist that perform similar tasks to DQe-c. Best known in clinical research informatics is the Automated Characterization of Health Information at Large-scale Longitudinal Evidence Systems (ACHILLES), a browser-based tool that visualizes patient demographics and the prevalence of all conditions, drugs, procedures, and observations stored in an OMOP CDM dataset.39 ACHILLES can be used to make interpretations about completeness in the repository. DQe-c provides multiple improvements to ACHILLES, namely in design, architecture, and interoperability. First, the design of DQe-c is the result of a deductive scientific process to operationalize the conception of completeness in clinical data repositories. Second, the development of DQe-c is an outcome of collaboration with actual users. The development and addition of the clinical indicators module was the result of a collaboration with the DARTNet Institute38 while implementing DQe-c on its clinical data repositories. Thus, DQe-c provides a focused snapshot of the clinical data repository that include actionable information – ie, information that can be used by database administrators to initiate exclusive checks. Second, the modular architecture of DQe-c enables it to adapt to different CDMs and easily incorporate new analytics into its HTML-based report. The modular architecture is particularly useful in distributed networks, where DQe-c can be utilized as a foundation for applying more sophisticated data analytics. For example, we are planning to include an extension script into DQe-c runs to extract the data needed to evaluate variability in EHR data across the ARCH network.
LIMITATIONS AND FUTURE DIRECTIONS
Although we continue to improve the tool in collaboration with our growing user base, we acknowledge that a limitation of the DQe-c design is that it has not been systematically user tested. Nevertheless, since DQe-c provides a programmatic foundation that is easy to tweak and customize to incorporate richer data-quality tests and better user interfaces, improving its usability is always possible. In addition, the tool is currently data model–driven, meaning that it will have to be constantly updated once a new version of a CDM is released. We are exploring transitioning the DQe-c architecture from data model–driven to information model–driven. An information model is a flexible model that, unlike a data model, does not require a concrete physical structure for data representation.40 Such a transition would allow the tool to be used more comprehensively (ie, across steps 1–3 data-quality checks). Once the transition is complete, DQe-c will be able to evaluate the completeness and conformance of any input data, given a user-defined flat file or Extensible Markup Language that describes the names of tables, columns, and entity relationships. More data model–related checks will be added to future versions of DQe-c through the data model module. For the short to medium term, we plan to maintain the tool centrally. As the user community grows, we expect more user involvement in maintaining and upgrading DQe-c through GitHub.
CONCLUSION
Data science offers methodologies and guidelines to promote the secondary use of EHR data to improve patient care. DQe-c is an interoperable and scalable data science–driven tool that examines and profiles completeness in clinical data repositories. The tool produces a web-based report that summarizes data completeness and conformance in a given EHR data repository through descriptive graphics and tables. DQe-c has been designed through an iterative, multi-institutional collaborative design process and works on PCORnet and OMOP CDMs. The tool, which is publicly available on GitHub, incorporates data visualization, interoperability with multiple data models, and scalability to high volumes of data, filling a gap for large distributed clinical data networks.
CONTRIBUTORS
HE conceived of the work and designed the tool. HE and KS initiated the design, and JK and SN helped with improvement and implementation. JK performed data extraction. HE conducted data analysis and visualization. HE drafted the article. All authors contributed to critical revision of the article and approved the final manuscript.
FUNDING
This work was partially funded through a Patient-Centered Outcomes Research Institute (PCORI) Award (CDRN-1306-04608) for development of the National Patient-Centered Clinical Research Network, known as PCORnet, NIH NCATS award UL1TR000423, CDC (200-2015-87699), NIH R01-HG009174, and NLM training grant T15LM007092.
Disclaimer
The statements presented in this publication are solely the responsibility of the author(s) and do not necessarily represent the views of the Patient-Centered Outcomes Research Institute (PCORI), its Board of Governors or Methodology Committee or other participants in PCORnet.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
REFERENCES
- 1. Hsiao C-J, Hing E, Socey TC, Cai B. Electronic health record systems and intent to apply for meaningful use incentives among office-based physician practices: United States, 2001-2011. NCHS Data Brief. 2011;(79):1–8. [PubMed] [Google Scholar]
- 2. Murdoch T, Detsky A. The inevitable application of big data to health care. J Am Med Inform Assoc. 2013;30913:1351–52. [DOI] [PubMed] [Google Scholar]
- 3. Liaw ST, Rahimi A, Ray P, et al. . Towards an ontology for data quality in integrated chronic disease management: a realist review of the literature. Int J Med Inform. 2013;82:10–24. [DOI] [PubMed] [Google Scholar]
- 4. Ross MK, Wei W, Ohno-Machado L. “Big data” and the electronic health record. Yearb Med Inform. 2014;9:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hersh WR. Adding value to the electronic health record through secondary use of data for quality assurance, research, and surveillance. Am J Manag Care. 2007;13:277–78. [PubMed] [Google Scholar]
- 6. Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20:144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stewart WF, Shah NR, Selna MJ, Paulus RA, Walker JM. Bridging the inferential gap: the electronic health record and clinical evidence. Health Aff 2007;262:w181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCarty CA, Chisholm RL, Chute CG, et al. . The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stephens KA, Lin C-P, Baldwin L-M, et al. . LC Data QUEST: a technical architecture for community federated clinical data sharing. AMIA Summits Transl Sci Proc. 2012;2012:57. [PMC free article] [PubMed] [Google Scholar]
- 11. Ohno-Machado L. Data science and informatics: when it comes to biomedical data, is there a real distinction? J Am Med Inform Assoc. 2013;206:1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toga AW, Foster I, Kesselman C, et al. . Big biomedical data as the key resource for discovery science. J Am Med Inform Assoc. 2015;226:1126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bui AAT, Van Horn JD. Envisioning the future of “big data” biomedicine. J Biomed Inform. 2017;69:115–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Margolis R, Derr L, Dunn M, et al. . The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J Am Med Inform Assoc. 2014;216:957–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bourne PE, Bonazzi V, Dunn M, et al. . The NIH Big Data to Knowledge (BD2K) initiative. J Am Med Inform Assoc. 2015;226:1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown JS, Kahn M, Toh S. Data quality assessment for comparative effectiveness research in distributed data networks. Med Care. 2013;51:S22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kahn MG, Batson D, Schilling LM. Data model considerations for clinical effectiveness researchers. Med Care. 2012;50(Suppl):S60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weiskopf NG, Hripcsak G, Swaminathan S, Weng C. Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform. 2013;46:830–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gregori D, Berchialla P. Quality of electronic medical records. In: Faltin FW, Kenett RS, Ruggeri F, eds. Statistical Methods in Healthcare. West Sussex, UK: Wiley; 2012:456–76. [Google Scholar]
- 20. Kahn MG, Raebel MA, Glanz JM, Riedlinger K, Steiner JF. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care. 2012;50:S21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen H, Hailey D, Wang N, Yu P. A review of data quality assessment methods for public health information systems. Int J Environ Res Public Health. 2014;11:5170–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roth CP, Lim Y-W, Pevnick JM, Asch SM, McGlynn EA. The challenge of measuring quality of care from the electronic health record. Am J Med Qual. 2009;24:385–94. [DOI] [PubMed] [Google Scholar]
- 23. Majeed A, Car J, Sheikh A. Accuracy and completeness of electronic patient records in primary care. Fam Pract. 2008;25:213–14. [DOI] [PubMed] [Google Scholar]
- 24. Li R, Abela L, Moore J, et al. . Control of data quality for population-based cancer survival analysis. Cancer Epidemiol. 2014;38:314–20. [DOI] [PubMed] [Google Scholar]
- 25. Kahn MG, Callahan TJ, Barnard J, et al. . A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS (Wash DC). 2016;41:1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan KS, Fowles JB, Weiner JP. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev. 2010;67:503–27. [DOI] [PubMed] [Google Scholar]
- 27. Kerr KA, Norris T, Stockdale R. The strategic management of data quality in healthcare. Health Informatics J. 2008;14:259–66. [DOI] [PubMed] [Google Scholar]
- 28. Hartzema AG, Reich CG, Ryan PB, et al. . Managing data quality for a drug safety surveillance system. Drug Saf. 2013;36(Suppl 1):49–58. [DOI] [PubMed] [Google Scholar]
- 29. Arts DGT, De Keizer NF, Scheffer G-J. Defining and improving data quality in medical registries: a literature review, case study, and generic framework. J Am Med Inform Assoc. 2002;9:600–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hogan WR, Wagner MM. Accuracy of data in computer-based patient records. J Am Med Inform Assoc. 1997;4:342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cole AM, Stephens KA, Keppel GA, Estiri H, Baldwin L-M. Extracting electronic health record data in a practice-based research network: processes to support translational research across diverse practice organizations. EGEMS (Wash DC). 2016;42:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy SN, Mendis M, Hackett K, et al. . Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc. 2007:548–52. [PMC free article] [PubMed] [Google Scholar]
- 33. Murphy SN, Weber G, Mendis M, et al. . Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc. 2010;172:124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mandl KD, Kohane IS, McFadden D, et al. . Scalable collaborative infrastructure for a learning healthcare system (SCILHS): architecture. J Am Med Inform Assoc. 2014;214:615–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nalichowski R, Keogh D, Chueh HC, Murphy SN. Calculating the benefits of a research patient data repository. AMIA Annu Symp Proc 2006:1044. [PMC free article] [PubMed] [Google Scholar]
- 36. Easterbrook SM. Open code for open science? Nat Geosci. 2014;711:779–81. [Google Scholar]
- 37. Stephens KA, Anderson N, Lin C-P, Estiri H. Implementing partnership-driven clinical federated electronic health record data sharing networks. Int J Med Inform. 2016;93:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pace WD, Fox CH, White T, Graham D, Schilling LM, West DR. The DARTNet Institute: seeking a sustainable support mechanism for electronic data enabled research networks. EGEMS. 2014;22:1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hripcsak G, Duke JD, Shah NH, et al. . Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–78. [PMC free article] [PubMed] [Google Scholar]
- 40. Klann JG, Abend A, Raghavan VA, Mandl KD, Murphy SN. Data interchange using i2b2. J Am Med Inform Assoc. 2016;235:909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.