Abstract
Genome-wide association studies identified a single nucleotide polymorphism (SNP) in the MSRB3 gene encoding Methionine Sulfoxide Reductase-B3 (MsrB3) to be associated with the risk for low hippocampal volume and late onset Alzheimer’s disease (AD). Subsequently, we identified AD-associated abnormal patterns of neuronal and vascular MsrB3 expression in postmortem hippocampi. The present study investigated the relationship between the MSRB3 SNP rs61921502, G (minor/risk allele) and MRI measures of brain injury including total brain volume, hippocampal volume, and white matter hyperintensities using linear regression models; the presence of brain infarcts using logistic regression models; and the incidence of stroke, dementia, and AD using Cox proportional hazards models in 2,038 Framingham Heart Study Offspring participants with MRI administered close to examination cycle 7 (1998–2001). Participants with neurological conditions that impede evaluation of vascular pathology by MRI, i.e., brain tumors, multiple sclerosis, and major head trauma, were excluded from the study. When adjusted for age and age squared at MRI exam, sex, and presence of Apolipoprotein ɛ4 allele (APOE4), individuals with MSRB3 rs61921502 minor allele had increased odds for brain infarcts on MRI compared to those with no minor allele. However, in stratified analyses, MSRB3 rs61921502 minor allele was significantly associated with increased odds for MRI brain infarcts only in the absence of APOE4.
Keywords: Framingham Heart Study, hippocampus, MSRB3, vascular pathology
INTRODUCTION
The oxidation of the sulfur atom within free methionine, as well as within the methionine residues of proteins, generates methionine sulfoxide [1]. The former reaction has no biological function but depletes the pool of methionine [2]—an essential amino acid, while the latter, being oxidation product, is generally considered damaging to proteins [1]. The regeneration of the native amino acid is catalyzed by methionine sulfoxide reductases (MSRBs) [3]. MSRBA catalyzes the conversion of both the free- and protein methionine [3], while MSRB1, 2, and 3 are thought to reduce primarily methionine sulfoxide residues within polypeptide chains. The MSRB3 gene is polymorphic. Its intronic SNP rs61921502 harbors minor (G) allele associated with low hippocampal volume (HV) and late onset Alzheimer’s disease (AD) [4]. Our recent study on postmortem hippocampi of 23 individuals uncovered previously unknown locations of MsrB3 protein: synaptic vesicles and arteriolar walls, providing clues for possible mechanisms whereby the rs61921502 SNP may affect HV and AD risks by influencing neurotransmission and arteriolar homeostasis [5, 6]. We discovered MsrB3 immunoreactivity (MsrB3-IR) in arteriolar smooth muscle layer as well as endothelial/pericyte cell layer and observed variation in MsrB3-IR intensity in the hippocampal arteriolar walls depending on the Braak stage and Clinical Dementia Rating (CDR) score [5]. MsrB3 signal intensity in arteriolar walls of hippocampal white matter was significantly decreased in AD patients compared to controls and subjects with early AD-associated pathology [5]. However, among 23 studied subjects, those with a history of stroke or transitory ischemic attack (TIA) had significantly increased MsrB3-IR signal in hippocampal white matter arteriolar walls compared to subjects without stroke or TIA history [5].
To elucidate the significance of changes in MsrB3-IR in the arteriolar walls in the hippocampal white matter of patients with evidence of AD and cerebrovascular diseases, we set to determine the associations between the genomic presence of minor allele (G) in MSRB3 rs61921502 SNP and specific pathologies on magnetic resonance imaging (MRI) by investigating subjects enrolled in Framingham Heart Study (FHS), the multi-generational epidemiological cohort study with cognitive, imaging, clinical, and risk factor data available antemortem for the participants, in addition to detailed postmortem neuropathological assessment [6, 7].
MATERIALS AND METHODS
Study cohort
The FHS is a community-based, prospective cohort study initiated in 1948 to identify risk factors of cardiovascular disease. Since 1975, ancillary studies have focused on risk factors of neurological outcomes [8]. The present analysis focuses on the Offspring Cohort, which enrolled 5,124 offspring of the Original Cohort participants between 1971 and 1975, and since that time has undergone comprehensive examinations approximately every four years [9, 10]. The seventh clinical examination was held from 1998 to 2001 [10]. Participants self-reported their medical history and lifestyle habits (physical activity, alcohol and nicotine consumption, occupation, and socio-demographic circumstances), and underwent anthropometry (height, weight, girth), blood pressure, and chemistry (cholesterols, triglycerides, glucose, etc.) assessments, as well as urinalysis and a physical exam including pulmonary function measurements. In addition, participants submitted themselves to echocardiography, carotid Doppler, electrocardiogram, and Holter monitoring. Their psychosocial and cognitive status were also evaluated [by self-report and Center for Epidemiologic Studies-Depression Scale (CES-D), and by Mini-Mental Status Examination (MMSE), respectively]. Participants who attended the seventh clinical examination (1998–2001) were invited to take part in a brain MRI study and those who did were included in incident dementia analyses. Of the 3,539 participants who attended the exam, 2,265 underwent brain MRI and, among those, 2,069 also had information on MSRB3 SNP rs61921502 and APOE gene polymorphism status. We excluded 31 participants with neurological disorders that might affect the accuracy of MRI to diagnose vascular pathology, i.e., brain tumors, multiple sclerosis, and major head trauma. Thus, our final sample included 2,038 participants. The study protocol was approved by the Institutional Review Board of Boston University Medical Center. All participants provided written, informed consent.
MRI outcomes
The Framingham Heart Study’s MRI methods have been previously described [11]. MRIs were performed with a 1 or 1.5 Tesla Siemens Magnetom scanner. Three-dimensional T1, double echo proton density, and T2 images were acquired in 4 mm contiguous slices. Images were transferred to University of California Davis for centralized reading (QUANTA 6.2, Sun Microsystems Ultra 5 workstation). MRI images were analyzed by a neurologist, who was blinded as to participants’ demographic and clinical characteristics. Segmentation and quantification of brain volumes were performed using automated procedures, which have been described in further detail elsewhere [11–13]. Brain infarcts (BI) were defined as an area of abnormal signal density in a vascular distribution. Contrast in size, location, shape, and tissue characteristics were used to distinguish BIs from dilated perivascular spaces.
Assessment of incident stroke and dementia
FHS monitors incident stroke and dementia via continuous surveillance. For this study, follow-up for incident stroke and dementia began at the seventh examination and continued until the time of event or last known event-free contact with the participant. For incident dementia, participants undergo cognitive screening at each examination cycle with the MMSE [14]. Participants are flagged for suspected cognitive impairment based on MMSE performance or concerns from the participant, family members, or physicians. Flagged participants undergo annual neuropsychological and neurological evaluations to track progression to dementia. Dementia status and diagnosis dates are determined by a dementia review committee, including a neuropsychologist and neurologist and defined in accordance with the Diagnostic and Statistical Manual of Mental Disorders, Text Revision (DSM-IV-TR, Edition 4, 2000). AD dementia was defined in accordance with the National Institute of Neurological and Communicative Disorders and Stroke and the AD and Related Disorders Association for definite, probable, or possible AD [15]. Stroke was defined as the acute onset of focal neurological symptoms of presumed vascular origin lasting 24 hours or more [16].
MSRB3 rs61921502 minor allele and covariates
The SNP rs61921502 is located in position 65438688 on chromosome 12. As the genotyping arrays do not have probes for rs61921502, participants were genotyped with Affymetrix 500K (250K Nsp and 250K Sty) MIPS 50K genotyping platforms. MSRB3 rs61921502 data were imputed with validated and freely available MaCH software. Data were imputed to the 1000 Genomes Version I Phase III reference panel (European ancestry 1000G set). The estimated genotype score of MSRB3 rs61921502 is a continuous variable and used as such in the analysis. The estimated genotype score is a weighted average of possible genotype scores with weights being posterior probability of each score value estimated from the imputation.
The imputation quality of rs61921502 is 0.72, which is the estimated R-squared of observed genotypes with true genotypes. We define the presence of the risk allele in terms of unit increases in the continuous imputed genotype score, which can be interpreted as numbers of alleles. APOE genotype was determined by TaqMan assay. We considered the presence of APOE4 as having at least one APOE4 allele.
MsrB3 immunohistochemistry analysis
FFPE blocks were sectioned at 5μm thickness, dried at room temperature for 24 h, and heated at 80°C for 24 h before immunohistochemistry (IHC) experiments. Deparaffinization, antigen retrieval, and subsequent staining were performed with Ventana Benchmark Ultra automated IHC instrument using commercially available primary antibodies, Horseradish Peroxidase-conjugated secondary antibody with diaminobenzidine chromogen, and hematoxylin counterstain. Primary antibodies included rabbit anti-MsrB3 polyclonal antibody, a pan-MsrB3 antibody [17] (1:40; HPA014432 Atlas Antibodies, Stockholm, Sweden, and NBP1-84259, Novus Biologicals, Littleton, CO), mouse anti-human beta-amyloid [6F/3D] monoclonal antibody (1:25, Dako, Glostrup, Denmark), rabbit anti-human tau [A0024] polyclonal antibody (1:3200, Dako, Glostrup, Denmark), and mouse anti-human phospho-Paired Helical Filament-tau [AT8] monoclonal antibody (1:2000, Pierce, Rockford, IL) for analyses. IHC was performed in independent, triplicate experiments, conducted on the Ventana Benchmark Ultra to remove human error and diminish variability between independent experiments. Representative cases with established immunoreactivity (IR) patterns were stained together with subsequently added samples in order to verify consistency in IHC experiments.
We previously discovered MrsB3-IR signal in the arteriolar walls of vasculature of the hippocampal sections and evaluated the changes in the signal intensity (see Fig. 1 and 8 in [5]) as described [5]. Briefly, images of the low power hippocampal sections, needed to capture the arterioles on a given section, included other MsrB3-immunoreactive structures that would have contaminated our analysis of the arteriolar walls alone, if processed through a simple threshold-based determination of the percent area of signal. For that reason, we performed semi-quantitative analysis of DAB staining commonly utilized in traditional histological scoring. Before starting our scoring process by at least two blinded, independent observers, we reviewed the staining range of arteriolar walls in all the sections and established a pre-determined scoring scale. The intensity of arteriolar staining was assessed across all cases, and a range of weakest to strongest signal was established for semi-quantification using a scale of 0–3; 0 = no MsrB3-IR, 3 = strongest MsrB3-IR [5]. Using this scale, semi-quantitative scoring of MsrB3-IR in the arteriolar walls of hippocampal white matter was conducted using triplicate IHC sections in all subjects (n = 10) as previously described [5]. De-identified slides were viewed at 40x magnification and scored by at least two independent, blinded observers. Semi-quantitative scores of triplicate IHC experiments were averaged to find the mean MsrB3-IR in the hippocampal blood vessel walls for each subject [5].
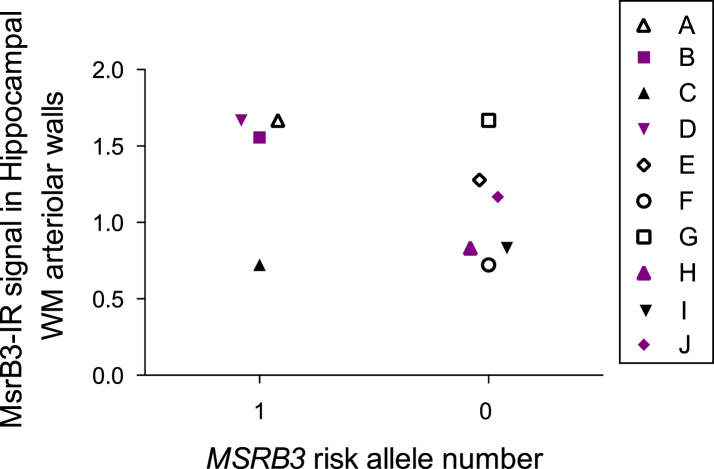
Fig.1.
MSRB3 rs61921502 risk allele and MsrB3-IR in hippocampal white matter (WM) arteriolar walls in 10 FHS participants- brain donors. Solid symbols represent AD patients (CDR1-3, BB IV-VI), empty symbols are either cognitively intact (CDR 0) or have mild cognitive deficit (CDR 0.5) and early AD pathology (Braak I-III). Colored symbols represent subjects with APOE4 subjects, a trait that appears to exhibit an organized pattern according to the presence of MSRB3 rs61921502 risk allele.
Statistical analyses
We analyzed total brain, hippocampal, and white matter hyperintensity volumes as percentages of total cranial volume to account for differences in head size. White matter hyperintensity volume was natural log-transformed due to its skewed distribution. We fit multivariable linear regression models for total brain volume, HV, and white matter hyperintensity volume, and multivariable logistic regression models for the presence of BIs, each adjusted for age at MRI, age at MRI squared, sex and presence of APOE4. We fit multivariable Cox proportional hazards models for the incidence of stroke and dementia outcomes, adjusted for age, sex, and presence of APOE4. We assessed the proportional hazard assumptions via martingale residuals. We also assessed interaction between presence of APOE4 and MSRB3 rs61921502 minor allele by including interaction terms, and report estimates stratified by APOE4 presence. We employed a false discovery rate (FDR) correction to account for multiple testing within primary multivariable analyses and within analyses stratified by APOE4 presence. For these analyses, we report FDR adjusted p-values and consider adjusted p-values less than 0.05 as significant. We did not apply FDR correction when assessing interaction terms, and consider nominal p-values less than 0.10 as evidence of interaction. Further, in the FHS brain bank we identified the 10 donors that all had not only brain tissue available accompanied by neuropathology reports, CDR score, and APOE status, but also had known MSRB3 genotype. We analyzed MsrB3-IR in the hippocampal white matter arteriolar walls [5] and plotted the MsrB3-IR signal intensity against the rs61921502 minor allele presence. All analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, North Carolina).
RESULTS
Descriptive statistics
All descriptive statistics are available in Table 1. Our sample consisted of 2,038 participants, with 53.0% women and a mean age at examination 7 of 62.0±9.4 years. Imputed genotype scores were rounded to generate pseudo numbers of alleles: 587 (28.8%) participants had one MSRB3 rs61921502 minor (risk) allele, and 44 (2.2%) participants had two.
Table 1.
Descriptive Statistics
| Variable | Overall (n = 2038) |
| Covariates | |
| Age in years at examination 7 MRI, mean±SD | 62.0±9.4 |
| Male, n (%) | 958 (47.0%) |
| APOE4 presence, n (%) | 469 (23.0%) |
| Number of MSRB3 risk alleles | |
| 0 alleles, n (%) | 1407 (69.0%) |
| 1 allele, n (%) | 587 (28.8%) |
| 2 alleles, n (%) | 44 (2.2%) |
| Outcomes | |
| Total brain volume %, mean±SD | 79.3±3.5 |
| Hippocampal volume %, mean±SD | 0.5±0.05 |
| White matter hyperintensities %, median [Q1, Q3] | 0.05 [0.03, 0.1] |
| Presence of brain infarcts, n (%) | 232 (11.4%) |
| Incident dementia, n (%) | 116 (11.2%) |
| Incident Alzheimer’s disease, n (%) | 91 (8.8%) |
| Incident stroke, n (%) | 96 (4.9%) |
Association of MSRB3 rs61921502 risk allele and MRI outcomes
The association between the risk (minor) allele G on rs61921502 with low HV and with AD has been demonstrated by Hibar et al. [4] who reported that the (major) allele T was associated with greater HV (Z = 9.02, p = 1.94×10–19). Therefore, we designated the risk allele to be G. MSRB3 rs61921502 G allele was significantly associated with increased odds of BIs after false discovery rate correction. With each allele increase, the odds of BI increased by 56% (OR [95% CI]: 1.56 [1.16, 2.08]). MSRB3 rs61921502 minor allele was not significantly associated with incident dementia, incident stroke, or other MRI measures, including HV ( [95% CI]: –0.004 [–0.01, 0.001], adjusted p-value: 0.304). However, the direction of the hazard ratio for incident stroke (HR [95% CI]: 1.44 [0.95, 2.19]) is consistent with the odds ratio for BI, despite not reaching statistical significance. We did not find evidence of violation of the proportional hazards assumptions in analyses of incident dementia or stroke. All results are available in Table 2.
Table 2.
Associations of MSRB3 risk allele and MRI outcomes at Exam 7
| Outcome | n | Measure of Association | Estimate [95% CI] | Adjusted p-value |
| Total brain volume | 1,987 | β | 0.14 [–0.15, 0.42] | 0.518 |
| Hippocampal volume | 1,977 | β | –0.004 [–0.01, 0.001] | 0.304 |
| White matter hyperintensities | 1,987 | β | 0.04 [–0.05, 0.13] | 0.518 |
| Presence of brain infarcts | 2,038 | OR | 1.56 [1.16, 2.08] | 0.022* |
| Incident dementia | 1,032 | HR | 1.06 [0.70, 1.60] | 0.931 |
| Incident Alzheimer’s disease | 1,032 | HR | 1.00 [0.62, 1.61] | 0.997 |
| Incident stroke | 1,952 | HR | 1.44 [0.95, 2.19] | 0.304 |
*FDR < 0.05. OR, odds ratio; HR, hazard ratio. Adjusted for age, age squared (MRI outcomes), sex, and APOE4 presence.
MSRB3 rs61921502 risk allele and APOE4 interaction
We observed a significant interaction between presence of APOE4 and MSRB3 rs61921502 minor (risk) allele in their effects on the presence of BIs. In the absence of APOE4 allele, the odds of BI increased by 84% with each allele increase in MSRB3 rs61921502 (OR [95% CI]: 1.84 [1.30, 2.60]). However, the effect of MSRB3 rs61921502 on the odds of BI was not significant in the presence of APOE4 (OR [95% CI]: 0.88 [0.45, 1.72]). No other stratified results or interactions reached statistical significance. Despite not reaching statistical significance, the stratified results of incident stroke appear consistent with BI results, with increased risk of stroke in the absence of APOE4 (HR [95% CI: 1.53 0.94, 2.50]) compared to those in the presence of APOE4 (HR [95% CI: 1.16 0.52, 2.62]). All results are available in Table 3.
Table 3.
APOE4 and MSRB3 risk allele interaction
| Outcome | Measure of Association | Interaction nominal p-value | APOE4 Present (n = 469) | APOE4 Absent (n = 1,569) | ||||
| n | Estimate [95% CI] | Adjusted p-value | n | Estimate [95% CI] | Adjusted p-value | |||
| Total brain volume | β | 0.170 | 455 | –0.20 [–0.82, 0.42] | 0.715 | 1,532 | 0.25 [–0.07, 0.57] | 0.548 |
| Hippocampal volume | β | 0.519 | 455 | –0.01 [–0.02, 0.004] | 0.548 | 1,522 | 0.00 [–0.01, 0.003] | 0.548 |
| White matter hyperintensities | β | 0.552 | 455 | 0.09 [–0.08, 0.27] | 0.548 | 1,532 | 0.03 [–0.08, 0.13] | 0.715 |
| Presence of brain infarcts | OR | 0.044 | 469 | 0.88 [0.45, 1.72] | 0.715 | 1,569 | 1.84 [1.30, 2.60] | 0.008* |
| Incident dementia | HR | 0.387 | 234 | 1.34 [0.69, 2.58] | 0.598 | 798 | 0.90 [0.52, 1.55] | 0.715 |
| Incident Alzheimer’s disease | HR | 0.115 | 234 | 1.55 [0.78, 3.07] | 0.548 | 798 | 0.66 [0.33, 1.33] | 0.548 |
| Incident stroke | HR | 0.593 | 449 | 1.16 [0.52, 2.62] | 0.715 | 1,503 | 1.53 [0.94, 2.50] | 0.548 |
*FDR < 0.05. OR, odds ratio; HR, hazard ratio. Adjusted for age, age squared (MRI outcomes), and sex.
MSRB3 rs61921502 risk allele and MSRB3-IR in hippocampal white matter arteriolar walls in the context of APOE4
We analyzed MsrB3-IR signal in white matter arteriolar walls in postmortem hippocampi available at the FHS brain depository as described before [5] from participants with known MSRB3 rs61921502 SNP and APOE status (Table 4, Supplementary Table 1). We plotted the MsrB3 signal intensity in hippocampal white matter arteriolar walls against the presence of MSRB3 risk allele (Fig. 1) in the context of AD and the presence of APOE4. All four APOE4 subjects were AD patients. Two of those four subjects that had no MSRB3 risk allele yielded a lower MsrB3 signal in hippocampal arteriolar walls in comparison to the two subjects with MSRB3 risk allele.
Table 4.
FHS subjects, grouped according to CDR score, with analyzed MsrB3 immunoreactivity in the arteriolar walls of the hippocampal WM and the information on the presence of MSRB3 risk allele, APOE4, and vascular disease
| Subject | Number of alleles | Arteriolosclerosis in hippocampus | Brain infarcts | History pathology | WM | |||
| MSRB3 rs61921502 risk allele (G) | APOE4 | Cortex | WM | Stroke or TIA | HTN | |||
| CDR 0–0.5 | ||||||||
| E | 0 | 0 | 0 | 0 | BG &WM | N | Y | WM gliosis |
| F | 0 | 0 | 0 | 2+ | Striatum, neocortex | N | Y | VWMD |
| A | 1 | 0 | 0 | 2+ | N | N | N | WM gliosis |
| G | 0 | 0 | 0 | 1–2 | OCC, BG, WM | N | Y | Some gliosis |
| CDR 1–3 | ||||||||
| B | 1 | 1 | 0 | 1+ | BG, WM | N | Y | WM gliosis; corpora amylacea |
| H | 0 | 1 | 0 | 0 | N | N | Y | Minimal gliosis |
| C | 1 | 0 | 0 | 0 | N | N | Y | VWMD in STG, PFC, OCC |
| I | 0 | 0 | 0 | 1+ | Temporal WM | N | N | VWMD |
| J | 0 | 1 | 0 | 2+ | Right motor cortex and lateral angle of the lateral ventricle | Y | Y | Large acute infarct in the right MCA territory |
| D | 1 | 1 | 0 | 2+ | N | N | Y | WM gliosis and macrophages in the hippocampus |
*0 = none; 1+ = mild; 2+ = moderate; 3+ = severe. HTN, hypertension; VWMD, vascular white matter disease; BG, basal ganglia; STG, superior temporal gyrus; PFC, prefrontal cortex; OCC, occipital pole; CRBL, cerebellum; TIA, transient ischemic attack; MCA, middle cerebral artery. Analyzed hippocampi of subjects, divided according to Clinical Dementia Rating (CDR) score; subjects with CDR 1–3 have clinical dementia. Arteriolosclerosis of hippocampal cortex and white matter was graded by severity of disease on H&E sections from hippocampus, from 0 (none) to 2 + . Arteriolosclerosis of neocortex was also graded on H&E in some of the subjects. Infarcts and other vascular/ vascular- related pathology were recorded according to the neuropathology reports. History of stroke and/or hypertension was recorded according to the clinical histories available through brain bank or de-identified autopsy reports.
DISCUSSION
The elucidation of the pathophysiologic mechanisms of brain disease has been approached by both unbiased- and hypothesis-driven experimental designs. Unbiased approaches, such as GWAS, have generated a wealth of information by pointing to specific genes or genomic regions that are associated with risk of disease. Indeed, the association of the MSRB3 rs61921502 SNP with low HV and increased risk of AD was discovered by GWAS [4]. However, once such risk alleles are identified by an unbiased approach, the next challenge is to define their expression and function of their protein products in healthy brain and in neurological diseases. As most of such newly-discovered disease-associated genes tend to be understudied, it is not surprising that this information is almost universally lacking. In our previous study, we described high expression of the MsrB3 protein in arteriolar walls of the hippocampal white matter [5], prompting the hypothesis that cerebral blood vessels constitute possible sites of vulnerability to genetic polymorphisms in MSRB3. In the current study, we tested this idea. By analyzing 2,038 FHS Offspring participants we provide evidence that the minor allele in MSRB3 rs61921502 is significantly associated with increased odds for BIs in the absence of APOE4 (Tables 1–3). Thus, APOE4 attenuates the vulnerability to BI conferred by the MSRB3 rs61921502 minor allele. A prior meta-analysis and systematic review of data from three different populations, including FHS, concluded that APOE4 allele has no effect on BI [18]. It would be interesting to re-examine the reported data by including the MSRB3 rs61921502 SNP in the analysis. Given that APOE4 is the ancestral allele from which APOE3 and APOE2 evolved [19], it would be expected that in some biological contexts APOE4 may provide adaptive advantages. Indeed, studies of age-related macular degeneration have previously implicated APOE4 as a protective allele in this disease as compared to the other two [20–22], APOE4 is reportedly associated with better cognition in a non-industrial Amazonian population characterized by high parasite and pathogen load [23], and there is evidence that APOE4 may confer survival and neurodevelopmental advantage in infants and children [19].
In our sample MSRB3 rs61921502 minor allele was not significantly associated with either AD or HV as previously reported by Hibar et al. [4]. The complexity of the relationship between AD and HV is underlined by large longitudinal studies in which the hallmarks of AD pathology, beta-amyloid and tau deposition, did not associate with rates of hippocampal atrophy in MRI-histological exams [24, 25] while hippocampal TDP43 presence did [26]. Additionally, in Hibar et al. [4] the association between the rs61921502 SNP and low HV appeared lateralized as the larger effect was reported for the right hippocampal formation. In contrast, our study analyzed HV data as the sums of right and left hippocampi. Similarly, total brain volume, white matter hyperintensities, and incident stroke were not significantly associated with MSRB3 rs61921502 minor allele in our analysis.
The discovered association of MSRB3 rs61921502 minor allele with BIs is intriguing in the light of our initial observation of MsrB3-IR in arteriolar smooth muscle and endothelial/pericyte cell layers [5]. We had found that MsrB3-IR signal in arteriolar walls varied in intensity in the hippocampal white matter depending on vascular and AD pathology [5]. In the current analysis of MsrB3-IR in hippocampi of FHS brain donors with known MSRB3 rs61921502 status we sought to explore the relationship between the MsrB3-IR signal intensity and the presence of MSRB3 risk allele in the context of AD (Fig. 1, Table 4, Supplementary Table 1). Out of 6 subjects with no MSRB3 risk allele, 3 had AD and 3 were either cognitively intact or had minimal cognitive dysfunction (Table 4, Supplementary Table 1). Out of 4 subjects with MSRB3 risk allele, 3 had AD (Table 4, Supplementary Table 1). Non-AD subjects tended to have, as expected, no APOE4 and also no MSRB3 risk allele (Table 4, Supplementary Table 1). These available cases do not constitute a large enough sample to determine if MsrB3-IR arteriolar signal stratifies according to the presence of MSRB3 risk allele (Fig. 1). However, in that small sample, APOE4 appeared to promote dependency between MsrB3-IR signal intensity and the presence of MSRB3 risk allele in APOE4 carriers (Fig. 1). The implication of this observation is unclear. MsrB3 deficiency has been shown to stimulate the expression of heme oxygenase-1 (HO-1) [27] which is a target of cognitive-enhancing and neuroprotective drug Neotrofin [28]. HO-1 pathway activation prevents hippocampal neuronal damage and cognitive function deficits in vascular dementia [29].
Our study has limitations. First, as in Hibar et al. [4], the studied population, FHS Offspring cohort, is of European descent, and thus our results may not be generalizable to individuals with other ancestries. However, we note that the rs61921502 minor G allele tends to be rare in people of non-European heritage. In particular, our findings are not applicable to Africans and East Asians, not because we did not study them, but because the rs61921502 G allele is very rare in those groups (its frequency is less than 1% in the 1000 Genomes Project; see https://ldlink.nci.nih.gov/?tab=snpclip). It would be interesting to determine if other MSRB3 gene variants result in low hippocampal volume, BI, and AD-related risk phenotypes in other populations not studied here. Second, the observational design prohibits us from establishing a causal link, as there is a possibility for residual confounding. Third, our analysis of MsrB3-IR signal was performed in a sample of brains too small for statistical inference. Instead, we focus on description information and plots. Finally, we were unable to replicate the significant association between MSRB3 rs61921502 minor allele and HV, previously demonstrated in a meta-analysis of two large consortia [4]. However, the direction of the observed effect is consistent with their findings, which indicates an inverse association between MSRB3 rs61921502 minor allele and HV.
Our study contributed further evidence in support of the role of MSRB3 in brain health. Our data from MRIs of 2,038 FHS subjects revealed MSRB3 rs61921502 minor allele to be associated with presence of BIs. Our MsrB3 expression analysis in the white matter arteriolar walls of the hippocampus affected early in AD suggests a multifaceted interplay between APOE4 and MSRB3 rs61921502 minor allele, influencing the decline of hippocampal vascular health and cognitive functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kerry Cormier of Framingham Heart Study Brain Bank for specimen procurement.
This work is supported by the National Institutes of Health, National Institute on Aging grants R01 AG033193 (SS, ID, JJH), AG049505 (SS, JJH), AG008122 (SS, JJH), AG054076 (SS, JJH) AG057768 (JKB), AG045031 (JKB) and the National Institute of General Medical Sciences Interdisciplinary Training Grant for Biostatisticians T32GM74905-14 (SCC). The Framingham Heart Study is supported by the National Heart, Lung, and Blood Institute (contract no. N01-HC-25195 and no. HHSN268201500001I).
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/18-0977r2).
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-180977.
REFERENCES
- [1]. Manta B, Gladyshev VN (2017) Regulated methionine oxidation by monooxygenases. Free Radic Biol Med 109, 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Zhao H, Kim G, Levine RL (2012) Methionine sulfoxide reductase contributes to meeting dietary methionine requirements. Arch Biochem Biophys 522, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Kaya A, Lee BC, Gladyshev VN (2015) Regulation of protein function by reversible methionine oxidation and the role of selenoprotein MsrB1. Antioxid Redox Signal 23, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Hibar DP, Adams HH, Jahanshad N, Chauhan G, Stein JL, Hofer E, Renteria ME, Bis JC, Arias-Vasquez A, Ikram MK, Desrivieres S, Vernooij MW, Abramovic L, Alhusaini S, Amin N, Andersson M, Arfanakis K, Aribisala BS, Armstrong NJ, Athanasiu L, Axelsson T, Beecham AH, Beiser A, Bernard M, Blanton SH, Bohlken MM, Boks MP, Bralten J, Brickman AM, Carmichael O, Chakravarty MM, Chen Q, Ching CR, Chouraki V, Cuellar-Partida G, Crivello F, Den Braber A, Doan NT, Ehrlich S, Giddaluru S, Goldman AL, Gottesman RF, Grimm O, Griswold ME, Guadalupe T, Gutman BA, Hass J, Haukvik UK, Hoehn D, Holmes AJ, Hoogman M, Janowitz D, Jia T, Jorgensen KN, Karbalai N, Kasperaviciute D, Kim S, Klein M, Kraemer B, Lee PH, Liewald DC, Lopez LM, Luciano M, Macare C, Marquand AF, Matarin M, Mather KA, Mattheisen M, McKay DR, Milaneschi Y, Munoz Maniega S, Nho K, Nugent AC, Nyquist P, Loohuis LM, Oosterlaan J, Papmeyer M, Pirpamer L, Putz B, Ramasamy A, Richards JS, Risacher SL, Roiz-Santianez R, Rommelse N, Ropele S, Rose EJ, Royle NA, Rundek T, Samann PG, Saremi A, Satizabal CL, Schmaal L, Schork AJ, Shen L, Shin J, Shumskaya E, Smith AV, Sprooten E, Strike LT, Teumer A, Tordesillas-Gutierrez D, Toro R, Trabzuni D, Trompet S, Vaidya D, Van der Grond J, Van der Lee SJ, Van der Meer D, Van Donkelaar MM, Van Eijk KR, Van Erp TG, Van Rooij D, Walton E, Westlye LT, Whelan CD, Windham BG, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Wolfers T, Yanek LR, Yang J, Zijdenbos A, Zwiers MP, Agartz I, Almasy L, Ames D, Amouyel P, Andreassen OA, Arepalli S, Assareh AA, Barral S, Bastin ME, Becker DM, Becker JT, Bennett DA, Blangero J, van Bokhoven H, Boomsma DI, Brodaty H, Brouwer RM, Brunner HG, Buckner RL, Buitelaar JK, Bulayeva KB, Cahn W, Calhoun VD, Cannon DM, Cavalleri GL, Cheng CY, Cichon S, Cookson MR, Corvin A, Crespo-Facorro B, Curran JE, Czisch M, Dale AM, Davies GE, De Craen AJ, De Geus EJ, De Jager PL, De Zubicaray GI, Deary IJ, Debette S, DeCarli C, Delanty N, Depondt C, DeStefano A, Dillman A, Djurovic S, Donohoe G, Drevets WC, Duggirala R, Dyer TD, Enzinger C, Erk S, Espeseth T, Fedko IO, Fernandez G, Ferrucci L, Fisher SE, Fleischman DA, Ford I, Fornage M, Foroud TM, Fox PT, Francks C, Fukunaga M, Gibbs JR, Glahn DC, Gollub RL, Goring HH, Green RC, Gruber O, Gudnason V, Guelfi S, Haberg AK, Hansell NK, Hardy J, Hartman CA, Hashimoto R, Hegenscheid K, Heinz A, Le Hellard S, Hernandez DG, Heslenfeld DJ, Ho BC, Hoekstra PJ, Hoffmann W, Hofman A, Holsboer F, Homuth G, Hosten N, Hottenga JJ, Huentelman M, Pol HE, Ikeda M, Jack CR Jr., Jenkinson M, Johnson R, Jonsson EG, Jukema JW, Kahn RS, Kanai R, Kloszewska I, Knopman DS, Kochunov P, Kwok JB, Lawrie SM, Lemaitre H, Liu X, Longo DL, Lopez OL, Lovestone S, Martinez O, Martinot JL, Mattay VS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Mecocci P, Melle I, Meyer-Lindenberg A, Mohnke S, Montgomery GW, Morris DW, Mosley TH, Muhleisen TW, Muller-Myhsok B, Nalls MA, Nauck M, Nichols TE, Niessen WJ, Nothen MM, Nyberg L, Ohi K, Olvera RL, Ophoff RA, Pandolfo M, Paus T, Pausova Z, Penninx BW, Pike GB, Potkin SG, Psaty BM, Reppermund S, Rietschel M, Roffman JL, Romanczuk-Seiferth N, Rotter JI, Ryten M, Sacco RL, Sachdev PS, Saykin AJ, Schmidt R, Schmidt H, Schofield PR, Sigursson S, Simmons A, Singleton A, Sisodiya SM, Smith C, Smoller JW, Soininen H, Steen VM, Stott DJ, Sussmann JE, Thalamuthu A, Toga AW, Traynor BJ, Troncoso J, Tsolaki M, Tzourio C, Uitterlinden AG, Hernandez MC, Van der Brug M, van der Lugt A, van der Wee NJ, Van Haren NE, van ‘t Ent D, Van Tol MJ, Vardarajan BN, Vellas B, Veltman DJ, Volzke H, Walter H, Wardlaw JM, Wassink TH, Weale ME, Weinberger DR, Weiner MW, Wen W, Westman E, White T, Wong TY, Wright CB, Zielke RH, Zonderman AB, Martin NG, Van Duijn CM, Wright MJ, Longstreth WT, Schumann G, Grabe HJ, Franke B, Launer LJ, Medland SE, Seshadri S, Thompson PM, Ikram MA (2017) Novel genetic loci associated with hippocampal volume. Nat Commun 8, 13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Adams SL, Benayoun L, Tilton K, Chavez OR, Himali JJ, Blusztajn JK, Seshadri S, Delalle I (2017) Methionine sulfoxide reductase B3 (MsrB3) protein associates with synaptic vesicles and its expression changes in the hippocampi of Alzheimer’s disease patients. J Alzheimers Dis 60, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Satizabal CL, Beiser AS, Chouraki V, Chene G, Dufouil C, Seshadri S (2016) Incidence of dementia over three decades in the Framingham Heart Study. N Engl J Med 374, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Seshadri S, Beiser A, Au R, Wolf PA, Evans DA, Wilson RS, Petersen RC, Knopman DS, Rocca WA, Kawas CH, Corrada MM, Plassman BL, Langa KM, Chui HC (2011) Operationalizing diagnostic criteria for Alzheimer’s disease and other age-related cognitive impairment-Part 2. Alzheimers Dement 7, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Wolf PA (2012) Contributions of the Framingham Heart Study to stroke and dementia epidemiologic research at 60 years. Arch Neurol 69, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP (1975) The Framingham Offspring Study. Design and preliminary data. Prev Med 4, 518–525. [DOI] [PubMed] [Google Scholar]
- [10]. Tsao CW, Vasan RS (2015) The Framingham Heart Study: Past, present and future. Int J Epidemiol 44, 1763–1766. [DOI] [PubMed] [Google Scholar]
- [11]. DeCarli C, Massaro J, Harvey D, Hald J, Tullberg M, Au R, Beiser A, D’Agostino R, Wolf PA (2005) Measures of brain morphology and infarction in the Framingham Heart Study: Establishing what is normal. Neurobiol Aging 26, 491–510. [DOI] [PubMed] [Google Scholar]
- [12]. Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D (2009) Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage 46, 726–738. [DOI] [PubMed] [Google Scholar]
- [13]. DeCarli C, Fletcher E, Ramey V, Harvey D, Jagust WJ (2005) Anatomical mapping of white matter hyperintensities (WMH): Exploring the relationships between periventricular WMH, deep WMH, and total WMH burden. Stroke 36, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Folstein MF, Folstein SE, McHugh PR (1975) “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12, 189–198. [DOI] [PubMed] [Google Scholar]
- [15]. McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34, 939–944. [DOI] [PubMed] [Google Scholar]
- [16]. Wolf PA, D’Agostino RB, Belanger AJ, Kannel WB (1991) Probability of stroke: A risk profile from the Framingham Study. Stroke 22, 312–318. [DOI] [PubMed] [Google Scholar]
- [17]. Ahmed ZM, Yousaf R, Lee BC, Khan SN, Lee S, Lee K, Husnain T, Rehman AU, Bonneux S, Ansar M, Ahmad W, Leal SM, Gladyshev VN, Belyantseva IA, Van Camp G, Riazuddin S, Friedman TB, Riazuddin S (2011) Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am J Hum Genet 88, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Schilling S, DeStefano AL, Sachdev PS, Choi SH, Mather KA, DeCarli CD, Wen W, Hogh P, Raz N, Au R, Beiser A, Wolf PA, Romero JR, Zhu YC, Lunetta KL, Farrer L, Dufouil C, Kuller LH, Mazoyer B, Seshadri S, Tzourio C, Debette S (2013) APOE genotype and MRI markers of cerebrovascular disease: Systematic review and meta-analysis. Neurology 81, 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Huebbe P, Rimbach G (2017) Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res Rev 37, 146–161. [DOI] [PubMed] [Google Scholar]
- [20]. Toops KA, Tan LX, Lakkaraju A (2016) Apolipoprotein E isoforms and AMD. Adv Exp Med Biol 854, 3–9. [DOI] [PubMed] [Google Scholar]
- [21]. Xiying M, Wenbo W, Wangyi F, Qinghuai L (2017) Association of apolipoprotein E polymorphisms with age-related macular degeneration subtypes: An updated systematic review and meta-analysis. Arch Med Res 48, 370–377. [DOI] [PubMed] [Google Scholar]
- [22]. Liutkeviciene R, Vilkeviciute A, Smalinskiene A, Tamosiunas A, Petkeviciene J, Zaliuniene D, Lesauskaite V (2018) The role of apolipoprotein E (rs7412 and rs429358) in age-related macular degeneration. Ophthalmic Genet 39, 457–462. [DOI] [PubMed] [Google Scholar]
- [23]. Trumble BC, Stieglitz J, Blackwell AD, Allayee H, Beheim B, Finch CE, Gurven M, Kaplan H (2017) Apolipoprotein E4 is associated with improved cognitive function in Amazonian forager-horticulturalists with a high parasite burden. FASEB J 31, 1508–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Erten-Lyons D, Dodge HH, Woltjer R, Silbert LC, Howieson DB, Kramer P, Kaye JA (2013) Neuropathologic basis of age-associated brain atrophy. JAMA Neurol 70, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Silbert LC, Quinn JF, Moore MM, Corbridge E, Ball MJ, Murdoch G, Sexton G, Kaye JA (2003) Changes in premorbid brain volume predict Alzheimer’s disease pathology. Neurology 61, 487–492. [DOI] [PubMed] [Google Scholar]
- [26]. Josephs KA, Dickson DW, Tosakulwong N, Weigand SD, Murray ME, Petrucelli L, Liesinger AM, Senjem ML, Spychalla AJ, Knopman DS, Parisi JE, Petersen RC, Jack CR Jr, Whitwell JL (2017) Rates of hippocampal atrophy and presence of post-mortem TDP-43 in patients with Alzheimer’s disease: A longitudinal retrospective study. Lancet Neurol 16, 917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Kwak GH, Kim KY, Kim HY (2016) Methionine sulfoxide reductase B3 deficiency stimulates heme oxygenase-1 expression via ROS-dependent and Nrf2 activation pathways. Biochem Biophys Res Commun 473, 1033–1038. [DOI] [PubMed] [Google Scholar]
- [28]. Wang X, Hauptmann N, Taylor E, Foreman M, Khawli LA, Maines MD (2003) Neotrofin increases heme oxygenase-1 selectively in neurons. Brain Res 962, 1–14. [DOI] [PubMed] [Google Scholar]
- [29]. Xu X, Zhang B, Lu K, Deng J, Zhao F, Zhao BQ, Zhao Y (2016) Prevention of hippocampal neuronal damage and cognitive function deficits in vascular dementia by dextromethorphan. Mol Neurobiol 53, 3494–3502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.