Abstract
Genomic instability is implicated in the etiology of several deleterious health outcomes including megaloblastic anemia, neural tube defects, and neurodegeneration. Uracil misincorporation and its repair are known to cause genomic instability by inducing DNA strand breaks leading to apoptosis, but there is emerging evidence that uracil incorporation may also result in broader modifications of gene expression, including: changes in transcriptional stalling, strand break-mediated transcriptional upregulation, and direct promoter inhibition. The factors that influence uracil levels in DNA are cytosine deamination, de novo thymidylate (dTMP) biosynthesis, salvage dTMP biosynthesis, dUTPase, and DNA repair. There is evidence that the nuclear localization of the enzymes in these pathways in mammalian cells may modify and/or control the levels of uracil accumulation into nuclear DNA. Uracil sequencing technologies demonstrate that uracil in DNA is not distributed stochastically across the genome, but instead shows patterns of enrichment. Nuclear localization of the enzymes that modify uracil in DNA may serve to change these patterns of enrichment in a tissue-specific manner, and thereby signal the genome in response to metabolic and/or nutritional state of the cell.
Keywords: DNA repair, thymidylate, dUTPase, folate deficiency, neural tube defects, uracil in DNA
1. Introduction
Deoxyuracil in DNA can originate from cytosine base deamination or from the misincorporation of dUTP during DNA synthesis by DNA polymerases (1). Genomic uracil is recognized and removed by DNA repair enzymes, and excessive levels of genomic uracil can lead to the accumulation of repair-mediated strand breaks and apoptosis (2). Repair of uracil in DNA is known to induce p53-dependent apoptosis (2–4) and may induce PARP1-dependent necrosis (5, 6). Current literature suggests that strand breaks, apoptosis, and necrosis induced by repair of uracil in DNA likely contribute to the etiology of vitamin-deficiency related megaloblastic anemia, neural tube defects (NTDs), and neurodegenerative disorders. This potential for deleterious effects of uracil in DNA, and the multiple pathways that affect its accumulation and repair, suggest the presence of evolutionally-conserved mechanisms to minimize its accumulation in DNA.
This review will focus on the mechanisms that determine uracil incorporation into DNA and explore the evidence that uracil accumulation in DNA is regulated and may be a genomic signal by influencing transcription. The link between tissue-specific differences in uracil levels in DNA and transcription as now being realized. DNA repair intermediates, which are known to be induced by uracil incorporation, have been shown to induce changes in gene expression (7–11). Uracil levels in DNA, and the expression of enzymes which influence it, have been shown to exhibit tissue-specific expression (12–15). Additionally, DNA sequencing has recently revealed that genomic uracil is not randomly distributed, but shows patterns of enrichment in the bacterial, yeast, and mammalian genomes (16, 17). In mammalian cells, but not in yeast cells, nuclear localization of the enzymes influencing uracil levels in DNA have been shown to be critical in limiting uracil accumulation in DNA (13). Therefore, nuclear localization of these enzymes may modify both patterns of uracil accumulation in DNA and resulting changes in gene expression. Lastly, the impact of uracil-mediated changes in gene expression may underlie disease phenotypes associated with uracil accumulation in nuclear DNA.
2. Overview of factors that influence uracil accumulation in DNA
The presence of uracil in DNA results from spontaneous chemical deamination of cytosine, or enzymatic and metabolic processes governed by the expression, localization, and activity of several enzymes that ether catalyze cytosine deamination or generate biosynthetic intermediates in nucleotide synthesis that control rates of DNA synthesis and repair. Specifically, genomic uracil is generated via spontaneous or enzyme-catalyzed cytosine deamination or by misincorporation of dUTP in to DNA during DNA synthesis. Misincorporation occurs when DNA polymerases incorporate dUTP into DNA, in place of dTTP, and the rate of misincorporation is believed to be determined by the intracellular dUTP:dTTP ratio. This ratio is influenced by rates de novo thymidylate (dTMP) biosynthesis, salvage dTMP biosynthesis, and dUTP degradation by dUTPase. Once uracil has been incorporated into DNA, it can be excised by one of several repair mechanisms. The levels of uracil in DNA vary across species and cell types (Table 1). There are several methods to measure uracil levels in DNA including GC-MS, LC-MS, and DNA blotting-based methods (18–20). Methodological differences introduce technical variation, making it difficult to compare measurements of uracil in DNA across studies (19). The relative contribution of cytosine deamination, uracil misincorporation, and uracil repair to steady state levels of uracil in DNA among tissues remains largely uncharacterized. These mechanisms are described in detail in this section.
Table 1:
Measurements of uracil in DNA across tissue type.
| Cell type | Uracil per genome | Reference |
|---|---|---|
| E. coli | <5 | Lari et al (20) |
| S. cerevisiae | 300 -1200 | Owiti et al (21) |
| Rat colon epithelium | 36,000 | Choi et al (22) |
| Rat fetal liver | 27,000 – 45,000 | Tian et al (12) |
| Rat fetal brain | 87,000 – 153,000 | Tian et al (12) |
| Mouse liver | 1600 – 4000 | Marcfarlane et al (13) Martiniova et al (23) |
| Human whole blood | 90,000 - 400,000 | Blount et al (18) |
| Human bone marrow | 240,000 | Blount et al (18) |
| HeLa | 10,000 - 18,000 | Martiniova et al (23) |
2.1. Nonenzymatic and enzymatic cytosine deamination
Both spontaneous and enzyme-catalyzed cytosine deamination results in U:G pairing in DNA. If not repaired, this can lead to a transition mutation (Figure 1). Cytosine deamination is estimated to occur at a rate of 400 deaminations/day/human genome (1).
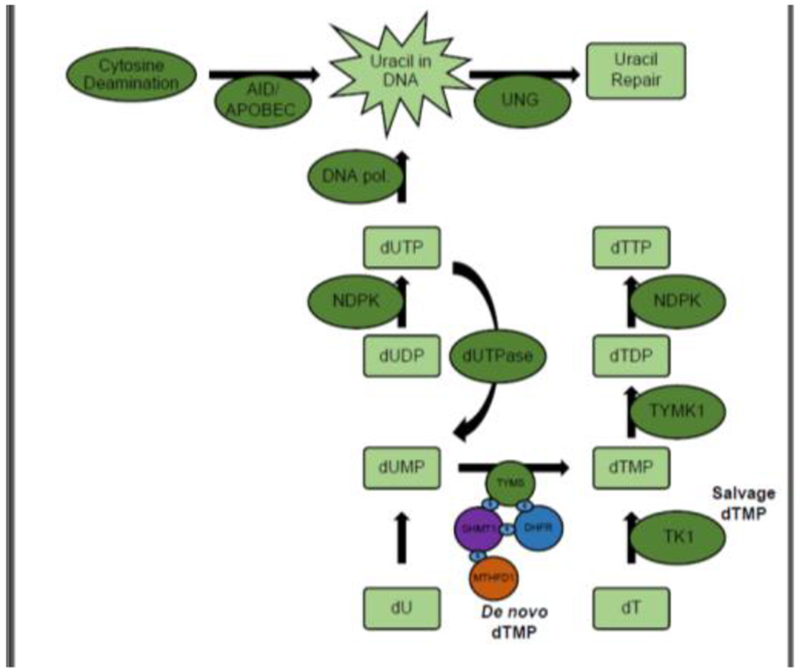
Figure 1. Pathways that influence uracil accumulation in DNA.
In de novo dTMP biosynthesis, TYMS transfers a one-carbon unit from 5,10-methylenetetrahydrofolate, onto dUMP, synthesizing dTMP. SHMT1, DHFR, TYMS, and MTHFD1 are SUMOylated and form a lamin-bound nuclear complex at sites of DNA replication and repair. In the salvage pathway, TK1 generates dTMP via phosphorylation of the nucleoside dT. TYMK phosphorylates dTMP, synthesizing dTDP. NDPK phosphorylates dTDP and dUDP, generating dTTP and dUTP, respectively. DNA polymerases incorporate dUTP into DNA. dUTPase dephosphorylates dUTP into dUMP. Spontaneous and enzymatic cytosine deamination by the enzymes AID or APOBEC can lead to U:G mispairs in DNA. Incorporated uracil is excised primarily by UNG, initiating uracil repair. AID, activation induced cytosine deamination; APOBEC, Apolipoprotein B Editing Complex Catalytic Subunit 1; DHFR, dihydrofolate reductase; dUTPase; dUTP phosphorylase; MTHFD1,methylenetetrahydrofolate dehydrogenase 1; NDPK, nucleoside-diphosphate kinase; S, small ubiquitin-related modifier (SUMO), SHMT1, serine hydroxymethyltransferase 1; TK1, thymidine Kinase 1; TYMK, thymidylate kinase; TYMS, dTMP synthase; UNG, uracil N-glycosylase.
Outside of spontaneous deamination, the enzyme activation induced cytidine deaminase (AID) plays a critical role in antibody diversification via class switch recombination and somatic hypermutation of immunoglobin genes in immune cells. Additionally, the enzyme apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) induces cytosine deamination in mRNA, generating a stop codon; APOBEC1 has also been shown to induce cytosine deamination in DNA, which is thought be critical in antibody diversification (24). However, low level AID expression has been shown in oocytes, embryonic germ cells, and embryonic stem cells (25), suggesting enzymatic cytosine deamination may influence uracil accumulation in DNA outside of the context of antibody diversification.
2.2. Uracil misincorporation into DNA
Rates of uracil misincorporation into DNA is believed to be determined the dUTP:dTTP ratio. In vitro studies indicate that DNA polymerases incorporate both dTTP and dUTP into DNA, but preferentially incorporate dTTP. DNA polymerase from porcine liver incorporated approximately 3 times more dTTP than dUTP when both were present in equimolar concentrations; DNA polymerase from E. coli incorporated approximately 2 times more dTTP than dUTP (26, 27). Mitochondrial polymerases can also incorporate dUTP into mitochondrial DNA (28). Unlike cytostine deamination, uracil misincorporation into DNA is not inherently mutagenic. The pathways which influence the dUTP:dTTP ratio are de novo dTMP biosynthesis, salvage dTMP biosynthesis, and dUTP degradation by dUTPase.
The de novo dTMP synthesis pathway, which is dependent on the cofactor tetrahydrofolate (THF), is composed of four enzymes: serine hydroxymethyltransferase (SHMT1 and SHMT2α), dihydrofolate reductase (DHFR), dTMP synthase (TYMS) and methylenetetrahydrofolate dehydrogenase (MTHFD1) (Figure 1). SHMT1 transfers a one-carbon group from serine to the folate cofactor THF, synthesizing 5,10-methyleneTHF and glycine. TYMS transfers a one-carbon group from 5,10-methyleneTHF onto deoxyuridine monophosphate (dUMP), synthesizing deoxythymidine monophosphate (dTMP) and dihydrofolate. DHFR reduces dihydrofolate to THF, recycling it for another round of dTMP synthesis. Notably, SHMT1 is not the main catalytic contributor of 5,10-methyleneTHF incorporated into dTMP. Instead MTHFD1, which derives its one-carbon from formate instead of serine, produces up to 90% of the 5,10-methyleneTHF for dTMP synthesis in SH-SY5Y cells (29).
Impairment of de novo dTMP biosynthesis and/or reduction of cellular folate pools results in elevated uracil content in DNA (3, 13, 18). Folate-deficient individuals exhibit increased uracil in DNA in blood and bone marrow cells (18). Likewise, mice fed a folate-deficient diet also showed elevated genomic uracil in colon epithelial cells (30). In rats consuming a folate-deficient diet, uracil misincorporation in lymphocytes was elevated relative to incorporation in rats consuming a folate-sufficient diet (31).
A single-allele knockout of Shmt1 in mice increased uracil in liver and colon DNA, which was exacerbated when Shmt1−/+ mice consumed a folate-deficient diet (32). In fact, the two primary phenotypes of Shmt1+/− and Shmt−/− mice were elevated uracil in DNA and development of folate-responsive neural tube defects (NTDs), suggesting that uracil in DNA may be causal in development of folate-responsive NTDs (32, 33). SHMT1 siRNA treatment in A549 lung cancer cells also increased uracil in DNA; this phenotype was rescued by dTMP supplementation in culture medium, indicating that salvage pathway dTMP synthesis can compensate for impaired de novo dTMP synthesis (3). De novo dTMP synthesis is also impaired by anti-cancer drugs known as antifolates, which act to inhibit enzymes in this pathway. These include 5-fluorouracil, methotrexate, pemetrexed, and raltitrexed, all of which have all been shown to increase uracil in DNA in several cell types (34–37). Similarly, MTHFD1-deficient fibroblasts exhibit reduced incorporation of formate into dTMP which results in increased uracil in DNA compared to control fibroblasts (38).
Salvage dTMP biosynthesis is catalyzed by the enzyme thymidine kinase (TK1 and TK2). These enzymes catalyze the addition of a phosphate group to thymidine (dT), forming dTMP. The cytosolic/nuclear isoform is encoded by TK1, while the mitochondrial isoform is encoded by TK2. Knockdown of TK1 via siRNA has been shown to sensitize tumor cells to both 5-fluorouridine and pemetrexed (39). This sensitization is likely the result of increased uracil in DNA caused by inhibition of both de novo dTMP synthesis (targeted by 5-fluorouridine and pemetrexed) and salvage dTMP synthesis (resulting from reduced TK1).
The enzyme dUTPase catalyzes the conversion of dUTP to dUMP. It serves to both degrade dUTP and also to provide the substrate for de novo dTMP biosynthesis, dUMP (Figure 1). Knockdown of dUTPase via siRNA increased intracellular dUTP levels in HeLa and SW620 cells (40), although uracil levels in DNA were not measured.
2.3. Repair of uracil in DNA
The excision of uracil from DNA is necessary to prevent its accumulation. Excision of uracil is primarily carried out by the base excision repair (BER) enzymes, uracil DNA glycosylases. In humans these include uracil N-DNA glycosylase 1/2 (UNG1/2), single-strand selective monofunctional uracil DNA glycosylase (SMUG1), thymine DNA glycosylase 1 (TDG), and methyl-CpG binding domain 4 (MBD4) (41). UNG is considered the primary enzyme that excises uracil in DNA, responsible for >90% of uracil excision activity in human cell extracts (42, 43). SMUG1 has overlapping function with UNG and is also the primary enzyme which excises 5-hydroxymethyluracil from DNA. Both UNG and SMUG1 can excise uracil from U:G or U:A pairs in dsDNA or ssDNA. TDG primarily excises T from T:G mispairs, but also excises U from U:G mispairs to a lesser extent (44). MBD4 excises uracil and thymine primarily from CpG rich regions of DNA (41).
These UDGs catalyze the initial step in base excision repair (BER) (45), excising the uracil base from the sugar-phosphate backbone, resulting in an abasic (AP) site (Figure 2). This site is then cleaved by AP endonuclease I (APE1), which cleaves 5’ to the abasic site creating a single[-strand break, leaving a 3’ OH and a 5’ deoxyribose phosphate (dRP). DNA polymerase β synthesizes a single base at the newly created 3’ OH and excises the 5’ dRP with its AP lyase activity. DNA ligase III seals the remaining nick. This process is known as short patch BER; uracil may also be processed by long patch BER, which involves replacement of several nucleotides (41).
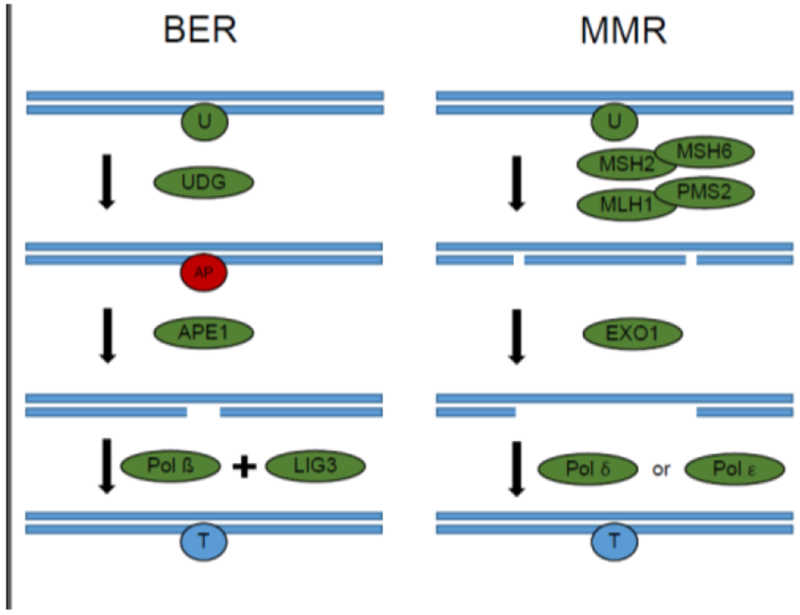
Figure 2. Repair of uracil in DNA. Left: uracil in DNA is excised primarily by base excision repair.
One of several uracil DNA glycosylases excise uracil, leaving an AP site. APE1 recognizes these sites and creates single strand breaks. POL β fills in these sites, and LIG3 seals the nick. Right: Mismatch repair can excise repair uracil from DNA. Uracil is recognized by the MSH2-MSH6 heterodimer. The MLH1-PMS3 heterodimer creates nick on either side of the uracil moiety. EXO1 degrades the nicked DNA. DNA is filled in by either POL δ or POL ε. BER = base excision repair. U, Uracil; UDG, uracil DNA glycosylase;.AP, abasicsite; APE1, AP endonuclease 1; POL β, DNA pol beta; MSH2, MutS homolog 2; MSH6, MutS homolog 6; MLH1, MutL homolog 1; PMS2, PMS1 homolog 2; EXO1, Exonuclease 1; Pol δ, DNA polymerase δ; Pol ε, DNA polymerase ε.
In addition to BER, uracil can be excised from DNA by mismatch repair (MMR) (46). Mismatch repair is initiated by the binding of MutSα to the mismatch. MutSα is a heterodimer consisting of the Msh2-Msh6. MutLα, a heterodimer consisting of Mlh1 and Pms2, is recruited to MutSα. PCNA activates MutLα incision activity, nicking the strand on either side of the base. Exo1 degrades this double-nicked strand of DNA, and the gap is is filled through DNA synthesis by DNA pol δ or ε (47) (Figure 2).
3. Tissue specific differences in uracil regulation
The levels of uracil in DNA vary among human tissues and cell culture conditions (12, 48). Accordingly, the enzymes that determine uracil levels in DNA also demonstrate tissue-specific expression patterns. For example, SHMT1, an enzyme whose expression affects uracil levels in DNA (3, 32), is present in mouse liver, colon, ileum, and kidney, while undetectable in brain (13). Similar observations have been made for TYMS (14) and UNG across tissue types. Using a uracil excision activity assay, Alsøe et al showed that UNG was the dominant uracil DNA glycosylase in spleen, heart, muscle, and liver tissue, but SMUG1 was the dominant uracil DNA glycosylase in brain tissue (15). The constellation of factors that affect uracil levels in DNA and the differences in their expression among tissues raises the possibility that genomic uracil accumulation in DNA is a dynamic and regulated process, and its misincorporation and excision is not exclusively a housekeeping activity necessitated to manage spontaneous chemistries of cytosine deamination and dUTP misincorporation.
4. Uracil and genome instability
4.1. Repair of uracil induces strand breaks
While uracil misincorporation into DNA is not inherently mutagenic, intermediates generated by repair can lead to genome instability and mutagenesis. The repair of uracil in DNA generates AP sites and subsequent single strand breaks. AP sites have been shown to induce transcriptional stalling, which has been shown to be highly mutagenic (7). Additionally, AP sites require mutagenic translesion DNA synthesis to bypass the lesion (49). Two single-strand breaks on opposing strands can induce a double-strand break (50), which are known to induce DNA mutation when nonhomologous end-joining (NHEJ) is chosen as the route of repair (51) (Figure 3). Additionally double-strand break repair is implicated in the expansion and contraction of tandem repeats in DNA (52). Although DNA damage and repair are normal occurrences in cells, increased rates of uracil incorporation and subsequent generation of repair-mediated strand breaks may surpass a tolerable level, leading to genomic instability and mutagenesis.
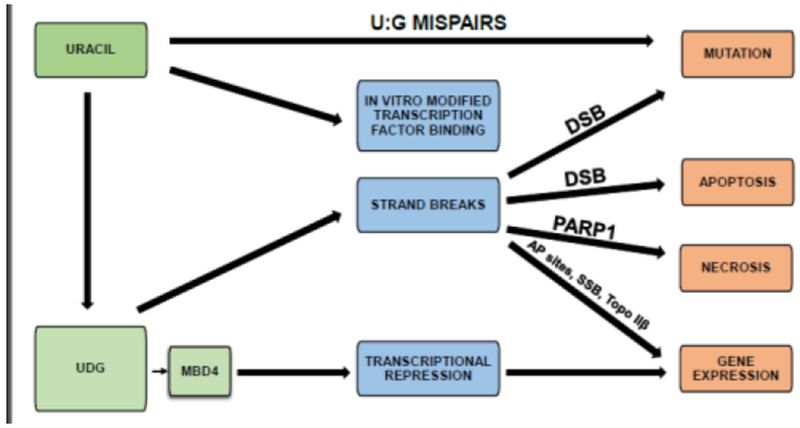
Figure 3. Consequences of uracil in DNA.
Cytosine deamination induces U:G mispairs. In vitro replacement of thymine with uracil modifies transcription factor binding. Repair of uracil induces strand breaks, which can lead to mutation or apoptosis. Potentially, these strand breaks can induce necrosis, and modify gene expression through AP sites, single strand breaks, orTopo IIβ. MBD4 inhibits transcription, modifying gene expression. MBD4, Methyl-CpG binding domain ;protein 4. DSB, double strand breaks; PARP1, Poly (ADP-ribose) polymerase 1; AP, abasic sites; SSB, single strand breaks; Topo IIβ, Topoisomerase IIβ.
Impairment of the enzymes that influence uracil accumulation has been shown to be associated with strand breaks and DNA mutation. Folate deficiency has been shown to lead to double-strand breaks, and increased chromosome breakage (53). In the case of folate fragile sites (discussed in section 4.2), the chromosome breakage induced by folate deficiency occur at regular sites in the genome (54). Genotoxic stress and chromosomal breakage can induce formation of micronuclei, which are extra-nuclear chromosomal fragments bound by nuclear membranes. Folate deficiency has been shown to induce micronuclei formation across several model systems. Human primary lymphocytes cultured in folate-deficient medium (containing 20 nM folic acid) exhibited induction of micronuclei formation, compared to lymphocytes cultured in folic acid-sufficient culture medium (55). Mice fed folic acid-deficient diets exhibited increased micronuclei formation in reticulocytes and red blood cells, compared to mice fed control diets (56). Reduction of dUTPase activity in yeast resulted in a strong mutator phenotype that is rescued by loss of UNG, indicating that the repair of uracil in DNA, not its accumulation, is mutagenic (57).
4.2. Folate fragile sites
Fragile sites are specific loci in the genome which are prone to breakage, when under replication stress. Unlike common fragile sites which are inherent in nearly all humans, folate fragile sites are heritable sites which are prone to breakage as a result of folate deficiency (2, 53, 54, 58) (Table 2). Common fragile site have been shown to be tissue specific; further studies are required to determine if folate fragile sites also occur in a tissue specific manner. Folate fragile sites are characterized by tandem CGG repeat expansion, leading to increased cytosine methylation and gene silencing (54). Loss of interrupting AGG triplets interspersed between CGG repeats is associated with CGG repeat expansion (59). The role of folate deficiency in repeat expansion has not been elucidated; folate deficiency may induce uracil incorporation opposite these AGG triplets leading to strand breaks. Repair of strand breaks has been implicated in the expansion and contract of tandem repeats (52). Hence, folate deficiency induced repair-mediated strand breaks can result in repeat expansion and ultimately gene silencing. Folate fragile sites likely represent predictable genomic loci of uracil incorporation into DNA, but this has not been demonstrated experimentally.
Table 2:
List of folate fragile sites. Adapted from HumCFS: A Database Of Human Chromosomal Fragile Sites. https://webs.iiitd.edu.in/raghava/humcfs/index.html
| Name | Chromosome Location | Name | Chromosome Location |
|---|---|---|---|
| FRA1M | 1p21.3 | FRA11A | 11q13.3 |
| FRA2A | 2q11.2 | FRA11B | 11q23.3 |
| FRA2B | 2q13 | FRA12A | 12q13.1 |
| FRA2K | 2q22.3 | FRA12D | 12q24.13 |
| FRA2L | 2q22.3 | FRA16A | 16p13.11 |
| FRA5G | 5q35 | FRA19B | 19p13 |
| FRA6A | 6p23 | FRA20A | 20p11.23 |
| FRA7A | 7p11.2 | FRA22A | 22q13 |
| FRA8A | 8q22.3 | FRAXA | Xq27.3 |
| FRA9B | 9q32 | FRAXE | Xq28 |
| FRA10A | 10q23.3 | FRAXF | Xq28 |
5. Evidence that uracil in DNA shows patterns of enrichment across the genome
Whereas folate fragile sites may represent hotspots of uracil incorporation into DNA, single-base resolution sequencing of uracil in DNA has not yet been developed in eucaryotes. However lower resolution methods have revealed that uracil in DNA shows specific patterns of enrichment. Notably two methods have been developed to examine the landscape of uracil enrichment throughout the genome. Excision-seq is a method for mapping modified nucleobases in DNA, yielding base-pair level resolution sequencing in E. coli, and read-level resolution sequencing in yeast (16). By replacing uracil with biotinylated dUTP in extracted DNA, Shu et al developed a method to localize uracil in DNA within hundreds of base pairs (17).
Excision-seq revealed that uracil content in DNA correlated with replication timing in both E. coli and budding yeast. In ungΔ E. coli with a hypomorphic dUTPase, uracil content in DNA was found to negatively correlate with origins of replication. In ung1Δ budding yeast with a hypomorphic dUTPase, total genome uracil content was found to be depleted in uracil at early-replicating regions, and at several origins of replication compared to the rest of the genome. Very late replicating regions also showed a more modest depletion of uracil in DNA in yeast. The authors suggest that the depletion of uracil in DNA in early replicating regions of DNA reflects higher dTTP availability at the start of replication (16). It is important to note that the pattern of uracil incorporation into DNA is the result of misincorporation, as opposed to the equilibrium between incorporation and excision in E. coli and yeast void of UNG. Overall these data suggest that uracil incorporation into DNA is depleted at origins of replication in E. coli and yeast due to increased dTTP availability at the start of replication.
In three human cell lines, (K562, WPMY-1, and HEK293T), Shu et. al. found that uracil in DNA was enriched at the centromere, at centromere protein A (CENP-A) binding regions, and at intergenic regions. CENP-A is a centromeric histone variant critical in kinetochore assembly (17). In Xenopus egg extracts, inhibition of UNG2 has been shown to block CENP-A assembly (60). Therefore, it is hypothesized that the uracil in DNA may serve as a platform for CENP-A assembly (17), though this has not been tested in mammalian systems. Enrichment of uracil in DNA at intergenic regions may indicate regulated excision of uracil from DNA within genes and suggests one mechanism whereby excision and repair of uracil could lead to changes in gene expression. Thus, in the E.coli, yeast, and human cell lines, current uracil sequencing technology has revealed distinct patterns of uracil enrichment in the genome. Further studies are required to elucidate the effects of such site-specific uracil incorporation on both genome stability and gene expression.
6. Nuclear localization of components may modify uracil incorporation into DNA
Nuclear localization of key enzymes associated with genomic uracil accumulation has been shown for every pathway in mammalian cells, whereas in yeast they are localized exclusively to the cytosol (61). The de novo dTMP biosynthesis enzymes SHMT1, TYMS, DHFR, and MTHFD1, have been shown to be SUMOylated during S phase, leading to their nuclear translocation and to the formation of a lamin-bound multi-enzyme complex at sites of DNA replication and repair (29, 62). SHMT1 acts as a scaffold for this complex and is essential for proper localization of the other components (29). The salvage enzyme TK1 is found in the cytoplasm and nucleus, with increasing expression and nuclear localization during S phase (63). In the case of DNA repair, the gene Ung produces the mitochondrial isoform UNG1 containing a mitochondrial leader sequence, and the nuclear isoform UNG2 containing a nuclear localization signal, resulting from alternative splicing (64). Similarly, the mitochondrial and nuclear isoforms of dUTPase are generated from alternative splicing of a single gene (65). Expression of mitochondrial dUTPase is constitutive, whereas expression of nuclear dUTPase coincides with DNA replication (66).
Nuclear localization appears to play an important role in reducing uracil accumulation in DNA. Whereas de novo dTMP biosynthesis occurs in both the nucleus and cytosol, there is evidence show that nuclear synthesis of dTMP is required to meet the needs of replication. Loss of nuclear localization of SHMT1 and TYMS was associated with increased uracil in liver DNA (13) in mice. Overexpression of SHMT1-DN2, an SHMT1 mutant that was catalytically inactive but could still act as a scaffold, was associated with increased de novo dTMP biosynthesis activity in SH-SY5Y cells, indicating that formation of the nuclear metabolic multienzyme complex, and not SHMT1 enzyme activity, is critical for de novo dTMP synthesis (29). Higher nuclear expression of TYMS was associated with poorer response to 5-FU in colorectal carcinoma (67), suggesting that nuclear dTTP synthesis prevents 5-FU incorporation into DNA. Similarly, increased nuclear expression of dUTPase was associated with tumor resistance to 5-fluorouracil (68), suggesting reduced dUTP availability may have enhanced dTTP incorporation into cells. Localization of these enzymes at sites of replication may create local microenviroments with altered dUTP:dTTP ratios and affect probability of dUTP incorporation into DNA.
There is also evidence that nuclear localization of enzymes that influence uracil accumulation is responsive to challenges to uracil accumulation. Following folate deficiency, MTHFD1 is enriched in mouse liver, protecting liver from increased uracil incorporation into DNA (69).
7. Uracil and Gene Expression
Although mutation and apoptosis are well established consequences of excision and repair of uracil in DNA, transcriptomic analysis have revealed that folate deficiency is also associated with changes in gene expression. Maternal folate deficiency was associated with transcriptomic changes in liver tissue from male adult offspring; folic acid supplementation prevented changes in genes associated with reactive oxygen species response, and steroid hormone response (70). Both folate and vitamin B12 status are known to affect chromatin methylation by influencing the concentrations of S-adenosylmethionine, the primary methyl donor in the cell (71–73). It is unclear the degree to which uracil misincorporation in DNA is also responsible for changes to gene expression in folate deficiency. However, the repair intermediates triggered by uracil in DNA are known to affect gene expression at the transcriptional level by several mechanisms.
7.1. AP sites and strand breaks modify gene expression
AP sites and single-strand breaks generated by repair of uracil can induce transcriptional stalling (7, 8). Studies with uracil-substituted reporter plasmids show data consistent with these findings. Single uracil substitutions in the coding region of an EGFP reporter plasmid in HeLa cells results in decreased EGFP expression compared to the uracil-free control plasmid, in a strand-independent manner. EGFP expression was rescued by UNG1/2 shRNA. Together, these data suggest repair of uracil in DNA, but not stalling of RNA polymerase II at uracil bases, impairs gene expression (9) (Figure 3).
Although strand breaks are known to impair gene expression, emerging literature shows double-strand breaks can be gene activating. Madabhushi et. al. showed that strand breaks mediated by Topoisomerase IIβ are critical in gene activation in dissociated cortical neurons, and that induction of double-strand breaks is sufficient to stimulate gene expression (10). Ju et. al. showed that Topoisomerase IIβ-mediated strand breaks at the promoter were required for activation of the PS2 gene in MCF7 cells. It is thought that strand breaks alter DNA topology at promoters, enhancing transcriptional initiation (11). Together, these data show induction of strand breaks can increase gene expression. Therefore uracil-induced AP sites and strand breaks may modify gene expression either positively or negatively, throughout the genome (Figure 3).
There is evidence that strand breaks generated by uracil incorporation occur at specific loci. Rats fed folate deficient diets exhibited strand breaks in exons 5-8 of p53 gene, but not in other exons of p53, the Ape gene, or the β-actin gene in colon tissue (74). Sohn et al found that folate deficiency reduced steady state levels of p53 mRNA in rat colon (75). These data suggest that strand breaks induced by folate deficiency occur in a nonrandom fashion, and also lead to changes in gene expression. Loci-specific incorporation of uracil would suggest a role and mechanism for uracil regulating gene expression.
7.2. Secondary function of MBD4 may affect global gene expression
Uracil DNA glycosylases also exhibit secondary functions in regulating gene expression, though it is not clear whether this linked to their role in uracil excision. For example, methyl-CpG binding domain protein 4 MBD4) binds the Glucocorticoid-Induced TNFR-Related Protein (Gitr) promoter in T regulatory cells, directly inhibiting its transcription (76). MBD4 has also been shown to bind the hypermethylated promoters of p16INK4A and MLH1, repressing transcription from these promoters (77). Therefore, MBD4 is a uracil DNA glycosylase capable of regulating gene expression in a site-specific manner; it is unclear if the glycosylase activity of MBD4 has any role in recruiting MBD4 to specific loci or in repressing transcription of GITR. Proteins in the MBD family all have a methyl binding domain, and have been shown to repress transcription in other genes (78). However, conditions which modify MBD4 activity have the potential lead to transcriptional repression (Figure 3). Because repair of uracil is DNA is both tissue-specific and responsive to challenges to uracil incorporation into DNA (15, 36), MBD4 expression may vary among tissue types or in response to challenges to uracil, modifying the level of transcriptional repression, though this has not been rigorously assessed in mammalian cells.
7.3. Uracil alters transcription factor binding in vitro
Substitution of thymine for uracil in transcription factor binding sites has been shown to modify transcription factor binding in vitro (79, 80) (Table 3). Substitution of a T:A pair with a U:A pair in the cAMP response element (CRE) decreases binding of cAMP response element binding protein (CBP) in vitro, while substitution of C:G with a U:G pair increases binding affinity for CBP (80). DNA uracilation has been also shown to alter the secondary structure of DNA through several mechanisms (81) (Table 3). Computational modelling suggests that uracil substitution in A-tracts of DNA widens the minor groove of DNA (82) (Table 3). Additionally, uracilation alters the cleavage pattern of DNA using both DNase I and micrococcal nuclease (83) (Table 3). Whether uracilation of DNA modifies interaction of DNA with proteins in vivo has not been assessed.; further RNA-seq and ChIP-seq studies will reveal if uracil in DNA leads to these in vivo changes.
Table 3:
Effect of uracil substitution on DNA structure and protein interaction.
8. Hotspots of uracil repair may indicate targeted regulation of gene expression
There is evidence that excision and repair of uracil in DNA does not occur stochastically across the genome, but instead is enriched at certain loci. Using ChIP-seq for γ-H2AX (a marker of DNA damage), Weeks et. al. showed that UNG−/− DLD1 human colon cancer cells have differential distribution of γ-H2AX hotspots compared to UNG+/+ cells in response to the antifolate pemetrexed. Compared to UNG+/+ cells, UNG−/− cells showed increased peak enrichment (representing increased γ-H2AX binding) at transcription factor binding sites, origins of replication, transcription factor CTCF binding sites, and CpG islands, but decreased peak enrichment at lamin-associated domains (84). These data suggest uracil is present in DNA in distinct patterns in the presence and absence of repair; excision and repair of uracil may contribute to targeted changes in gene expression. AP sites and strand breaks generated at these regular sites may generate predictable changes in gene expression, or uracil in DNA may modify transcription factor binding (as described in the section 7.3).
9. Uracil, cell fate and human disease
9.1. Uracil repair triggers cell death
A major consequence of high levels of uracil incorporation into DNA is apoptosis (Figure 3). Uracil in DNA is excised and induces repair-mediated strand breaks. In conditions of increased uracil incorporation, “futile cycles” of uracil excision and uracil re-incorporation into DNA occur resulting in continuous formation of strand breaks. This excessive generation of strand breaks can also signal p53, leading to p53-mediated apoptosis (85).
Challenges to the pathways influencing uracil incorporation have been shown to induce apoptosis. Paone et al showed knockdown of SHMT1 in A549 lung cancer cells increases uracil in DNA and leads to p53-mediated apoptosis, and that dTMP supplementation in media rescued both uracil in DNA and apoptosis in these cells (3). Folate depletion has been repeatedly shown to lead to apoptosis across several cell culture and mouse models (4). Arsenic trioxide treatment was associated with proteolytic degradation of SHMT1 and MTHFD1, increased uracil incorporation into DNA and increased DNA damage in HeLa cells (86). Arsenic trioxide has also been shown to induce apoptosis in HeLa cells (87). Additionally loss of dUTPase is known to be lethal in both E. coli and budding yeast (57, 88). Thus, apoptosis has been established as a consequence of uracil incorporation into DNA.
9.2. Necrosis
In addition to apoptosis, induction of strand breaks can lead to PARP1-dependent necrosis (Figure 3). Poly(ADP-ribose) polymerases (PARP) covalently modify proteins, by adding ADP-ribose (generated from NAD+) onto target proteins, a process known as PARylation. PARP1 and PARylation of proteins are critical in homologous recombination HR, NHEJ, and BER (89). Excessive activation PARP1 can lead to NAD depletion, which then leads to ATP depletion and subsequent necrosis (90). DNA damaging agents have shown to induce this PARP1-dependent necrosis (91, 92).
Treatment with antifolates, which are known to increase uracil incorporation into DNA, have been shown to induce necrosis. 5-FU treatment induced necrosis HepG2 and Hep3B cells (5). Treatment with the methotrexate (an inhibitor of DHFR) showed increased PARP1 expression and increased cell death as measured by TUNEL in rat kidneys, while co-treatment with the PARP inhibitor ISO significantly reduced cell death (6). Considering the rescue effect of ISO, and that TUNEL is not apoptosis-specific (93), it may be that methotrexate is inducing PARP1-dependent necrosis. Therefore, these studies indicate that activation of PARP1 in the repair of uracil leads to necrosis.
9.3. Megaloblastic anemia
In megaloblastic anemia, impaired DNA synthesis prevents cell division, leading to larger but fewer red blood cells (94). Folate deficiency has been established as a cause of megaloblastic anemia; both folate deficiency and megaloblastic anemia are associated with apoptosis. Folate deficient erythroblasts cultured in folate deficient medium demonstrated increased uracil misincorporation into DNA and increased p53 and p21 accumulation compared to control cells (95). Patients with megaloblastic anemia often demonstrate primary folate deficiency or a secondary folate deficiency due to vitamin B12 deficiency, as well as increased p53 accumulation in bone marrow, compared to non-megaloblastic anemia controls (96). Additional challenges to de novo dTMP biosynthesis, including deleterious mutations in DHFR and antifolate treatment, are associated with megaloblastic anemia (97, 98). Furthermore, mutation of dUTPase also causes apoptosis and megaloblastic anemia in humans (99). Lastly, deoxyuridine incorporation into DNA correlated with megaloblastic scoring in the bone marrow of anemic patients (100, 101). Therefore, apoptosis induced by impaired de novo dTMP biosynthesis and uracil misincorporation is the primary cause of megaloblastic anemia.
9.4. Neural tube defects
Increased apoptosis in the neural epithelium results in failure of the neural tube to close during embryonic development in animal models of neural tube defects (NTDs). Increased p53 accumulation, increased caspase cleavage (a hallmark of apoptosis), and decreased Pax3 expression, have been associated with NTD affected births. Pax3, a homeobox transcription factor, enhances MDM2-mediated ubiquitination and degradation of p53 and its loss-of-function has been associated with NTD risk. (102). In mice, Inhibition of p53 was also shown to rescue NTDs in embryos. Apoptosis was associated with the occurrence of NTDs in Pax3 deficient embryos; loss of p53 rescued both apoptosis and the occurrence of NTDs (103). Central nervous tissue from NTD terminated human pregnancies exhibits increased apoptosis compared to nervous tissue from unaffected terminated pregnancies. Spinal cord from anencephalic human samples demonstrated increased expression of p53, cleaved caspase 3 and cleaved caspase 8, and decreased expression of Pax3, all biomarkers of elevated apoptosis (104). Therefore, p53-mediated apoptosis is associated with the occurrence of NTDs.
Folate deficiency is established as a risk factor of NTDs. Folic acid supplementation has been shown to reduce the risk of NTD affected births (105). Mouse models of folic-acid responsive NTDs indicate that impaired de novo dTMP biosynthesis is the mechanism by which folate deficiency causes NTDs. Treatment with the DHFR inhibitor methotrexate increased occurrence of NTDs in mice (106). Shmt1−/+ mice have folate responsive NTDs and increased uracil in DNA in liver and colon tissue (32, 33), linking DNA uracil-induced apoptosis to folic-acid responsive NTDs.
9.5. Neurodegeneration
Recently, BER has been shown to be critical in mediating neurodegeneration. Hoch et al showed that single strand break repair was required for neurodegeneration to occur. Mutations in the BER protein Xrcc1 were associated with ocular motor apraxia, axonal neuropathy, and cerebellar ataxia. Loss of Parp1 partially rescued the occurrence of neuronal cell death and ataxia in Xrcc1 deficient mice (107). This suggests apoptosis or PARP1-mediated necrosis is the mechanism by which neurodegeneration occurs. Given that uracil in DNA is removed by BER, apoptosis or necrosis induced by uracil incorporation is linked to neurodegeneration.
Folate deficiency has been shown to be associated with neurodegeneration. Embryonic cortical neurons and SH-SY-5Y human neuroblastoma cells cultured in folate deficient media exhibited Alzheimer’s-like changes including increased ROS, phosphorylation of the protein Tau, and increased cell death (108). In this study, phosphatidylserine externalization and DAPI staining of pyknotic nuclei were used to measure cell death; neither of these methods distinguish between apoptosis and necrosis. Therefore, folate deficiency may be inducing apoptosis or necrosis via uracil incorporation, although direct measures of uracil in DNA in neurodegeneration are lacking.
Conversely, loss of the repair enzyme UNG has also been shown to induce neurodegeneration in mice. Kronenberg et al showed that folate deficiency induced CA3 hippocampal neurodegeneration, impaired cognitive function, and alterations in mood, in UNG−/− but not UNG +/+ mice. Uracil in DNA from brain tissue was increased in UNG-deficient mice, compared to control; this phenotype was further exacerbated by folate deficiency. Therefore, increased uracil incorporation, and not DNA repair following uracil incorporation, was associated with increased neurodegeneration. The authors suggest increased mitochondrial mutation induced by loss of the mitochondrial UNG1 may lead to impaired mitochondrial function, leading to neurodegeneration (109).
10. Uracil in DNA as a biochemical signal
More evidence is required to determine if uracil in DNA acts as more than a source of DNA damage. The relative contribution of cytosine deamination, uracil misincorporation, and uracil repair in maintaining steady state levels of uracil is still unclear. Studies looking at all these aspects simultaneously in a single model system are required. Understanding how these pathways are affected across different nutritional conditions and among tissues would help determine if uracil incorporation in DNA is simply “tolerated” or is being regulated at certain levels.
It is still unclear whether or not uracil in DNA, or its excision, induce meaningful changes in gene expression. In vitro transcription factor binding studies have revealed changes in binding capacity following uracil substitution (79, 80), but in vivo studies are required. Uracil in DNA may signal changes in gene expression through modified transcription factor binding and/or recruitment, or repair-mediated transcriptional stalling or gene activation. Low resolution uracil sequencing technologies have revealed that genomic distribution of uracil in DNA is nonrandom (16, 17). Concurrent RNA sequencing and high resolution uracil sequencing, when this technology becomes available, will help reveal if uracil in DNA regulates gene expression and contributes to disease etiology.
12. Acknowledgements
This work was supported by DK58144 and HD059120 to PJS from the US Public Health Service.
Abbreviations
- dTMP
deoxythymidine triphosphate
- dUTP
deoxyuridine triphosphate
- AID
activation induced cytidine deaminase
- APOBEC
Apolipoprotein B Editing Complex Catalytic Subunit 1
- DHFR
dihydrofolate reductase
- dUTPase
dUTP phosphorylase
- MTHFD1
methylenetetrahydrofolate dehydrogenase 1
- S
small ubiquitin-related modifier (SUMO)
- SHMT1
serine hydroxymethyltransferase 1
- TK1
thymidine Kinase 1
- TYMS
dTMP synthase
- UNG
uracil N-glycosylase
- SMUG1
single-strand selective monofunctional uracil DNA glycosylase
- TDG
thymine DNA glycosylase 1
- MBD4
methyl-CpG binding domain 4
- APE1
AP endonuclease 1
- dRP
deoxyribose phosphate
- BER
base excision repair
- POL β,
DNA pol beta
- MSH2
MutS homolog 2
- MSH6
MutS homolog 6
- MLH1
MutL homolog 1
- PMS2
PMS1 homolog 2
- EXO1
Exonuclease 1
- Pol δ
DNA polymerase δ
- Pol ε
DNA polymerase ε
- NHEJ
nonhomologous end-joining
- PARP1
Poly (ADP-ribose) polymerase 1
- GITR
Glucocorticoid-Induced TNFR-Related Protein
- CRE
cAMP response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflicts of interest.
13. References
- 1.Kunkel TA, Diaz M (2002) Enzymatic Cytosine Deamination: Friend AND Foe. Mol Cell 10(5): 962–963. [DOI] [PubMed] [Google Scholar]
- 2.Fenech M (2001) The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutat Res Mol Mech Mutagen 475(1 ):57–67. [DOI] [PubMed] [Google Scholar]
- 3.Paone A, et al. (2014) SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis 5:e1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li GM, Presnell SR, Gu L (2003) Folate deficiency, mismatch repair-dependent apoptosis, and human disease. J Nutr Biochem 14(10):568–575. [DOI] [PubMed] [Google Scholar]
- 5.Brenes O, Arce F, Gatjens-Boniche O, Diaz C (2007) Characterization of cell death events induced by anti-neoplastic drugs cisplatin, paclitaxel and 5-fluorouracil on human hepatoma cell lines: Possible mechanisms of cell resistance. Biomed Pharmacother 61(6):347–355. [DOI] [PubMed] [Google Scholar]
- 6.Dalaklioglu S, Sahin P, Ordueri EG, Celik-Ozenci C, Tasatargil A (2012) Potential Role of Poly(ADP-Ribose) Polymerase (PARP) Activation in Methotrexate-Induced Nephrotoxicity and Tubular Apoptosis. Int J Toxicol 31(5):430–440. [DOI] [PubMed] [Google Scholar]
- 7.Yu S-L, Lee S-K, Johnson RE, Prakash L, Prakash S (2003) The Stalling of Transcription at Abasic Sites Is Highly Mutagenic. Mol Cell Biol 23(1):382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kathe SD, Shen G-P, Wallace SS (2004) Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J Biol Chem 279(18):18511–18520. [DOI] [PubMed] [Google Scholar]
- 9.Lühnsdorf B, Epe B, Khobta A (2014) Excision of Uracil from Transcribed DNA Negatively Affects Gene Expression. J Biol Chem 289(32):22008–22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madabhushi R, et al. (2015) Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 161 (7): 1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ju B-G, et al. (2006) A Topoisomerase IIβ-Mediated dsDNA Break Required for Regulated Transcription. Science (80- ) 312(5781):1798 LP-1802. [DOI] [PubMed] [Google Scholar]
- 12.Tian Y-J, et al. (2014) Maternal nicotinamide supplementation causes global DNA hypomethylation, uracil hypo-incorporation and gene expression changes in fetal rats. Br J Nutr 111 (9): 1594–1601. [DOI] [PubMed] [Google Scholar]
- 13.MacFarlane AJ, et al. (2011) Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J Biol Chem 286(51 ):44015–44022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Boyer C, Lee JY, Shepard HM (2001) A Novel Approach to Thymidylate Synthase as a Target for Cancer Chemotherapy. Mol Pharmacol 59(3):446–452. [DOI] [PubMed] [Google Scholar]
- 15.Alsøe L, et al. (2017) Uracil Accumulation and Mutagenesis Dominated by Cytosine Deamination in CpG Dinucleotides in Mice Lacking UNG and SMUG1. Sci Rep 7(1):7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan DS, Ransom M, Adane B, York K, Hesselberth JR (2014) High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res 24(9): 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu X, et al. (2018) Genome-wide mapping reveals that deoxyuridine is enriched in the human centromeric DNA. Nat Chem Biol 14(7):680–687. [DOI] [PubMed] [Google Scholar]
- 18.Blount BC, et al. (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A 94(7): 3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galashevskaya A, et al. (2013) A robust, sensitive assay for genomic uracil determination by LC/MS/MS reveals lower levels than previously reported. DNA Repair (Amst) 12(9):699–706. [DOI] [PubMed] [Google Scholar]
- 20.Lari S-U, Chen C-Y, Vertessy BG, Morré J, Bennett SE (2006) Quantitative determination of uracil residues in Escherichia coli DNA: Contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst) 5(12):1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owiti N, Wei S, Bhagwat AS, Kim N (2018) Unscheduled DNA synthesis leads to elevated uracil residues at highly transcribed genomic loci in Saccharomyces cerevisiae. PLOS Genet 14(7):e1007516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S-W, et al. (2004) Vitamin B-12 Deficiency Induces Anomalies of Base Substitution and Methylation in the DNA of Rat Colonic Epithelium. J Nutr 134(4):750–755. [DOI] [PubMed] [Google Scholar]
- 23.Martiniova L, Field MS, Finkelstein JL, Perry CA, Stover PJ (2015) Maternal dietary uridine causes, and deoxyuridine prevents, neural tube closure defects in a mouse model of folate-responsive neural tube defects. Am J Clin Nutr 101 (4):860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen-Mahrt SK, Neuberger MS (2003) In Vitro Deamination of Cytosine to Uracil in Single-stranded DNA by Apolipoprotein B Editing Complex Catalytic Subunit 1 (APOBEC1). J Biol Chem 278(22): 19583–19586. [DOI] [PubMed] [Google Scholar]
- 25.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK (2004) Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem 279(50):52353–52360. [DOI] [PubMed] [Google Scholar]
- 26.Mosbaugh DW (1988) Purification and characterization of porcine liver DNA polymerase gamma: utilization of dUTP and dTTP during in vitro DNA synthesis. Nucleic Acids Res 16(12): 5645–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bessman MJ, et al. (1958) ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID.III. THE INCORPORATION OF PYRIMIDINE AND PURINE ANALOGUES INTO DEOXYRIBONUCLEIC ACID. Proc Natl Acad Sci U S A 44(7):633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alonzo JR, Venkataraman C, Field MS, Stover PJ (2018) The mitochondrial inner membrane protein MPV17 prevents uracil accumulation in mitochondrial DNA. J Biol Chem . doi: 10.1074/jbc.RA118.004788 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson DD, Woeller CF, Chiang E-P, Shane B, Stover PJ (2012) Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem 287(10):7051–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linhart HG, et al. (2009) Folate deficiency induces genomic uracil misincorporation and hypomethylation but does not increase DNA point mutations. Gastroenterology 136(1):227–235.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duthie SJ, Grant G, Pirie LP, Watson AJ, Margison GP (2010) Folate deficiency alters hepatic and colon MGMT and OGG-1 DNA repair protein expression in rats but has no effect on genome-wide DNA methylation. Cancer Prev Res (Phila) 3(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macfarlane AJ, Perry CA, McEntee MF, Lin DM, Stover PJ (2011) Shmt1 heterozygosity impairs folate-dependent thymidylate synthesis capacity and modifies risk of Apc(min)-mediated intestinal cancer risk. Cancer Res 71(6):2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaudin AE, et al. (2011) Shmt1 and de novo thymidylate biosynthesis underlie folate-responsive neural tube defects in mice. Am J Clin Nutr 93(4):789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingraham HA, Tseng BY, Goulian M (1982) Nucleotide levels and incorporation of 5-fluorouracil and uracil into DNA of cells treated with 5-fluorodeoxyuridine. Mol Pharmacol 21 (1 ):211–216. [PubMed] [Google Scholar]
- 35.Goulian M, Bleile B, Tseng BY (1980) Methotrexate-induced misincorporation of uracil into DNA. Proc Natl Acad Sci U S A 77(4): 1956–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weeks LD, Fu P, Gerson SL (2013) Uracil–DNA Glycosylase Expression Determines Human Lung Cancer Cell Sensitivity to Pemetrexed. Mol Cancer Ther 12(10):2248 LP-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Connor EE, Berger SH, Wyatt MD (2005) Determination of apoptosis, uracil incorporation, DNA strand breaks, and sister chromatid exchanges under conditions of thymidylate deprivation in a model of BER deficiency. Biochem Pharmacol 70(10): 1458–1468. [DOI] [PubMed] [Google Scholar]
- 38.Field MS, Kamynina E, Watkins D, Rosenblatt DS, Stover PJ (2015) Human mutations in methylenetetrahydrofolate dehydrogenase 1 impair nuclear de novo thymidylate biosynthesis. Proc Natl Acad Sci 112(2):400 LP-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Cresce C, Figueredo R, Ferguson PJ, Vincent MD, Koropatnick J (2011) Combining small interfering RNAs targeting thymidylate synthase and thymidine kinase 1 or 2 sensitizes human tumor cells to 5-fluorodeoxyuridine and pemetrexed. J Pharmacol Exp Ther 338(3): 952–963. [DOI] [PubMed] [Google Scholar]
- 40.Studebaker AW, Lafuse WP, Kloesel R, Williams MV (2005) Modulation of human dUTPase using small interfering RNA. Biochem Biophys Res Commun 327(1):306–310. [DOI] [PubMed] [Google Scholar]
- 41.Jacobs AL, Schär P (2012) DNA glycosylases: in DNA repair and beyond. Chromosoma 121(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kavli B, et al. (2002) hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem 277(42):39926–39936. [DOI] [PubMed] [Google Scholar]
- 43.Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE (2008) The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair (Amst) 7(11):1869–1881. [DOI] [PubMed] [Google Scholar]
- 44.Schormann N, Ricciardi R, Chattopadhyay D (2014) Uracil-DNA glycosylases—Structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci 23(12):1667–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krokan HE, Bjoras M (2013) Base excision repair. Cold Spring Harb Perspect Biol 5(4):a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larson ED, Bednarski DW, Maizels N (2008) High-fidelity correction of genomic uracil by human mismatch repair activities. BMC Mol Biol 9:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunkel TA, Erie DA (2015) Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu Rev Genet 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersen S, et al. (2005) Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis 26(3):547–555. [DOI] [PubMed] [Google Scholar]
- 49.Zhao B, Xie Z, Shen H, Wang Z (2004) Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res 32(13):3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrader CE, Guikema JEJ, Wu X, Stavnezer J (2009) The roles of APE1, APE2, DNA polymerase beta and mismatch repair in creating S region DNA breaks during antibody class switch. Philos Trans R Soc Lond B Biol Sci 364(1517):645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis AJ, Chen DJ (2013) DNA double strand break repair via non-homologous endjoining. Transl Cancer Res 2(3): 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paques F, Leung WY, Haber JE (1998) Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol 18(4):2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reidy JA, Zhou X, Chen AT (1983) Folic acid and chromosome breakage. I. Implications for genotoxicity studies. Mutat Res 122(2):217–221. [DOI] [PubMed] [Google Scholar]
- 54.Debacker K, Kooy RF (2007) Fragile sites and human disease. Hum Mol Genet 16(R2): R150–R158. [DOI] [PubMed] [Google Scholar]
- 55.Fenech MF (2010) Dietary reference values of individual micronutrients and nutriomes for genome damage prevention: current status and a road map to the future. Am J Clin Nutr 91 (5): 1438S–1454S. [DOI] [PubMed] [Google Scholar]
- 56.MacFarlane AJ, et al. (2015) Dietary folic acid protects against genotoxicity in the red blood cells of mice. Mutat Res Mol Mech Mutagen 779:105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guillet M, Van Der Kemp PA, Boiteux S (2006) dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res 34(7):2056–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutherland GR (1979) Heritable fragile sites on human chromosomes I. Factors affecting expression in lymphocyte culture. Am J Hum Genet 31 (2): 125–135. [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz M, Zlotorynski E, Kerem B (2006) The molecular basis of common and rare fragile sites. Cancer Lett 232(1): 13–26. [DOI] [PubMed] [Google Scholar]
- 60.Zeitlin SG, et al. (2011) Uracil DNA N-glycosylase promotes assembly of human centromere protein A. PLoS One 6(3):e17151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poon PP, Storms RK (1994) Thymidylate synthase is localized to the nuclear periphery in the yeast Saccharomyces cerevisiae. J Biol Chem 269(11):8341–8347. [PubMed] [Google Scholar]
- 62.Anderson DD, Woeller CF, Stover PJ (2007) Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin Chem Lab Med 45(12): 1760–1763. [DOI] [PubMed] [Google Scholar]
- 63.Chen Y-L, Eriksson S, Chang Z-F (2010) Regulation and functional contribution of thymidine kinase 1 in repair of DNA damage. J Biol Chem 285(35):27327–27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nilsen H, et al. (1997) Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res 25(4):750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ladner RD, McNulty DE, Carr SA, Roberts GD, Caradonna SJ (1996) Characterization of distinct nuclear and mitochondrial forms of human deoxyuridine triphosphate nucleotidohydrolase. J Biol Chem 271(13):7745–7751. [DOI] [PubMed] [Google Scholar]
- 66.Ladner RD, Caradonna SJ (1997) The human dUTPase gene encodes both nuclear and mitochondrial isoforms. Differential expression of the isoforms and characterization of a cDNA encoding the mitochondrial species. J Biol Chem 272(30): 19072–19080. [DOI] [PubMed] [Google Scholar]
- 67.Wong NA, et al. (2001) Nuclear thymidylate synthase expression, p53 expression and 5FU response in colorectal carcinoma. Br J Cancer 85(12): 1937–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladner RD, et al. (2000) dUTP nucleotidohydrolase isoform expression in normal and neoplastic tissues: association with survival and response to 5-fluorouracil in colorectal cancer. Cancer Res 60(13):3493–3503. [PubMed] [Google Scholar]
- 69.Field MS, et al. (2014) Nuclear enrichment of folate cofactors and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) protect de novo thymidylate biosynthesis during folate deficiency. J Biol Chem 289(43):29642–29650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lillycrop KA, et al. (2010) Maternal protein restriction with or without folic acid supplementation during pregnancy alters the hepatic transcriptome in adult male rats. Br J Nutr 103(12): 1711–1719. [DOI] [PubMed] [Google Scholar]
- 71.Brunaud L, et al. (2003) Effects of vitamin B12 and folate deficiencies on DNA methylation and carcinogenesis in rat liver. Clin Chem Lab Med 41 (8): 1012–1019. [DOI] [PubMed] [Google Scholar]
- 72.Garcia BA, Luka Z, Loukachevitch LV, Bhanu NV, Wagner C (2016) Folate deficiency affects histone methylation. Med Hypotheses 88:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berry RJ, Crider KS, Yang TP, Bailey LB (2012) Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate’s Role. Adv Nutr 3(1):21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.YI Kim, et al. (2000) Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology 119(1): 151–161. [DOI] [PubMed] [Google Scholar]
- 75.Sohn K-J, et al. (2003) The effect of dietary folate on genomic and p53-specific DNA methylation in rat colon. Carcinogenesis 24(1):81–90. [DOI] [PubMed] [Google Scholar]
- 76.Shuaiwei W, et al. (2017) DNMT1 cooperates with MBD4 to inhibit the expression of Glucocorticoid-induced TNFR-related protein in human T cells. FEBS Lett 591 (13): 1929–1939. [DOI] [PubMed] [Google Scholar]
- 77.Kondo E, Gu Z, Horii A, Fukushige S (2005) The Thymine DNA Glycosylase MBD4 Represses Transcription and Is Associated with Methylated p16INK4a and hMLH1 Genes. Mol Cell Biol 25(11):4388–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fatemi M, Wade PA (2006) MBD family proteins: reading the epigenetic code. J Cell Sci 119(15):3033–3037. [DOI] [PubMed] [Google Scholar]
- 79.McAlister VJ, Christie GE (2009) Analysis of DNA binding by a eubacterial zinc finger transcription factor. J Bacteriol 191(14):4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verri A, Mazzarello P, Biamonti G, Spadari S, Focher F (1990) The specific binding of nuclear protein(s) to the cAMP responsive element (CRE) sequence (TGACGTCA) is reduced by the misincorporation of U and increased by the deamination of C. Nucleic Acids Res 18(19):5775–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yan N, O’Day E, Wheeler LA, Engelman A, Lieberman J (2011) HIV DNA is heavily uracilated, which protects it from autointegration. Proc Natl Acad Sci USA 108(22): 9244–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marathe A, Bansal M (2010) The 5-Methyl Group in Thymine Dynamically Influences the Structure of A-Tracts in DNA at the Local and Global Level. J Phys Chem B 114(16):5534–5546. [DOI] [PubMed] [Google Scholar]
- 83.Bailly C, Crow S, Minnock A, Waring MJ (1999) Demethylation of thymine residues affects DNA cleavage by endonucleases but not sequence recognition by drugs. J Mol Biol 291 (3):561–573. [DOI] [PubMed] [Google Scholar]
- 84.Weeks LD, Zentner GE, Scacheri PC, Gerson SL (2014) Uracil DNA glycosylase (UNG) loss enhances DNA double strand break formation in human cancer cells exposed to pemetrexed. Cell Death Dis 5(2):e1045–e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaina B (2003) DNA damage-triggered apoptosis: critical role of DNA repair, doublestrand breaks, cell proliferation and signaling. Biochem Pharmacol 66(8): 1547–1554. [DOI] [PubMed] [Google Scholar]
- 86.Kamynina E, et al. (2017) Arsenic trioxide targets MTHFD1 and SUMO-dependent nuclear de novo thymidylate biosynthesis. Proc Natl Acad Sci 114(12):E2319–E2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deng Y, et al. (1999) Mechanisms of arsenic trioxide induced apoptosis of human cervical cancer HeLa cells and protection by Bcl-2. Sci China Ser C, Life Sci 42(6):635–643. [DOI] [PubMed] [Google Scholar]
- 88.Warner HR, Duncan BK, Garrett C, Neuhard J (1981) Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol 145(2):687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wei H, Yu X (2016) Functions of PARylation in DNA Damage Repair Pathways. Genomics Proteomics Bioinformatics 14(3): 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Devalaraja-Narashimha K, Singaravelu K, Padanilam BJ (2005) Poly(ADP-ribose) polymerase-mediated cell injury in acute renal failure. Pharmacol Res 52(1):44–59. [DOI] [PubMed] [Google Scholar]
- 91.Shin H-J, et al. (2015) Doxorubicin-induced necrosis is mediated by poly-(ADP-ribose) polymerase 1 (PARP1) but is independent of p53. Sci Rep 5:15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Douglas DL, Baines CP (2014) PARP1-mediated necrosis is dependent on parallel JNK and Ca(2+)/calpain pathways. J Cell Sci 127(19):4134–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grasl-Kraupp B, et al. (1995) In situ detection of fragmented DNA (tunel assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: A cautionary note. Hepatology 21 (5): 1465–1468. [DOI] [PubMed] [Google Scholar]
- 94.Green R, Datta Mitra A (2017) Megaloblastic Anemias: Nutritional and Other Causes. Med Clin North Am 101 (2):297–317. [DOI] [PubMed] [Google Scholar]
- 95.Koury MJ, et al. (1997) Apoptosis of late-stage erythroblasts in megaloblastic anemia: association with DNA damage and macrocyte production. Blood 89(12):4617–4623. [PubMed] [Google Scholar]
- 96.Yadav MK, Manoli NM, Madhunapantula SV (2016) Comparative Assessment of Vitamin-B12, Folic Acid and Homocysteine Levels in Relation to p53 Expression in Megaloblastic Anemia. PLoS One 11(10):e0164559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cario H, et al. (2011) Dihydrofolate reductase deficiency due to a homozygous DHFR mutation causes megaloblastic anemia and cerebral folate deficiency leading to severe neurologic disease. Am J Hum Genet 88(2):226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scott JM, Weir DG (1980) Drug-induced megaloblastic change. Clin Haematol 9(3):587–606. [PubMed] [Google Scholar]
- 99.Dos Santos RS, et al. (2017) dUTPase (DUT) Is Mutated in a Novel Monogenic Syndrome With Diabetes and Bone Marrow Failure. Diabetes 66(4): 1086–1096. [DOI] [PubMed] [Google Scholar]
- 100.Luzzatto L, Falusi AO, Joju EA (1981) Uracil in DNA in megaloblastic anemia. N Engl J Med 305(19): 1156–1157. [DOI] [PubMed] [Google Scholar]
- 101.Wickramasinghe SN, Fida S (1994) Bone marrow cells from vitamin B12- and folate-deficient patients misincorporate uracil into DNA. Blood 83(6): 1656 LP-1661. [PubMed] [Google Scholar]
- 102.Wang XD, Morgan SC, Loeken MR (2011) Pax3 stimulates p53 ubiquitination and degradation independent of transcription. PLoS One 6(12):e29379–e29379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pani L, Horal M, Loeken MR (2002) Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3-dependent development and tumorigenesis. Genes Dev 16(6):676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, et al. (2017) Apoptosis, Expression of PAX3 and P53, and Caspase Signal in Fetuses with Neural Tube Defects. Birth defects Res 109(19): 1596–1604. [DOI] [PubMed] [Google Scholar]
- 105.Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. MRC Vitamin Study Research Group. (1991) Lancet (London, England) 338(8760):131–137. [PubMed] [Google Scholar]
- 106.Wang X, et al. (2015) Genomic DNA hypomethylation is associated with neural tube defects induced by methotrexate inhibition of folate metabolism. PLoS One 10(3):e0121869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hoch NC, et al. (2016) XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ho PI, et al. (2003) Folate deprivation induces neurodegeneration: roles of oxidative stress and increased homocysteine. Neurobiol Dis 14(1):32–42. [DOI] [PubMed] [Google Scholar]
- 109.Kronenberg G, et al. (2008) Folate deficiency induces neurodegeneration and brain dysfunction in mice lacking uracil DNA glycosylase. J Neurosci 28(28):7219–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]