Abstract
Genome-wide association studies on alcohol dependence, by themselves, have yet to account for the estimated heritability of the disorder and provide incomplete mechanistic understanding of this complex trait. Integrating brain ethanol-responsive gene expression networks from model organisms with human genetic data on alcohol dependence could aid in identifying dependence-associated genes and functional networks in which they are involved. This study used a modification of the Edge-Weighted Dense Module Searching for genome-wide association studies (EW-dmGWAS) approach to co-analyze whole-genome gene expression data from ethanol-exposed mouse brain tissue, human protein-protein interaction databases and alcohol dependence-related genome-wide association studies. Results revealed novel ethanol-responsive and alcohol dependence-associated gene networks in prefrontal cortex, nucleus accumbens, and ventral tegmental area. Three of these networks were overrepresented with genome-wide association signals from an independent dataset. These networks were significantly overrepresented for gene ontology categories involving several mechanisms, including actin filament-based activity, transcript regulation, Wnt and Syndecan-mediated signaling, and ubiquitination. Together, these studies provide novel insight for brain mechanisms contributing to alcohol dependence.
Introduction
Alcohol Use Disorder [1], which spans the spectrum from abusive drinking to full alcohol dependence (AD), has a lifetime prevalence of 29.1% among adults in the United States [2]. Alcohol misuse ranks third in preventable causes of death in the U.S. [3] and fifth in risk factors for premature death and disability, globally [4]. Although pharmacological therapy for AUD exists [5], the effectiveness is limited and the relapse rate is high. Improvement in AUD treatment requires research on the underlying genetic and biological mechanisms of the progression from initial exposure to misuse, and finally to dependence.
Twin studies estimate that AUD is roughly 50% heritable [6, 7]. Multiple rodent model studies have used selective breeding to enrich for ethanol behavioral phenotypes or have identified ethanol-related behavioral quantitative trait loci [8–10], further confirming the large genetic contribution to alcohol behaviors. Recent studies have also documented genetic factors influencing the effectiveness of existing pharmacological treatments for AD, further substantiating genetic contributions to the mechanisms and treatment of AUD [11]. Genome-wide association studies (GWAS) in humans have identified several genetic variants associated with alcohol use and dependence [12–15]. However, they have yet to account for a large portion of the heritability estimated by twin studies. Lack of power, due to a large number of variants with small effects, is believed to the source of this “missing heritability”” [16]. Although recent large-scale studies have shown promise in identifying novel genetic contributions to alcohol consumption, these studies do not contain the deep phenotypic information necessary for identifying variants associated with dependence [14, 15]. Further, such GWAS results still generally lack information about how detected single gene variants are mechanistically related to the disease phenotype.
Genome-wide gene expression studies are capable of improving the power of GWAS by providing information about the gene networks and biological mechanisms in which GWAS variants function [17–20]. Although gene expression in brain tissue has been studied in AD humans [17, 18], these studies are often difficult to conduct and interpret, due to lack of control over experimental variables and small sample sizes. However, studies have found high conservation in gene expression correlation patterns between mice and humans, particularly in brain tissue [21]. Furthermore, extensive studies in rodent models have successfully identified ethanol-associated gene expression differences and gene networks in brain tissue [22–26]. Multiple ethanol-behavioral rodent models exist to measure different aspects of the developmental trajectory from initial exposure to compulsive consumption [27]. Acute administration to naïve mice models the response of initial alcohol exposure in humans, which is an important predictor of risk for AD [28, 29]. Wolen et al. used microarray analysis across a mouse genetic panel to identify expression correlation-based networks of acute ethanol-responsive genes (genes whose expression levels change after ethanol consumption or treatment), along with significantly associated expression quantitative trait loci in brain regions subserving the mesolimbocortical dopamine reward pathway—the prefrontal cortex (PFC), nucleus accumbens (NAc), and ventral tegmental area (VTA) [25]. Furthermore, specific networks also correlated with ethanol behavioral data derived from the same mouse genetic panel (BXD recombinant inbred lines) [10]. Importantly, these gene expression responses to acute ethanol in BXD mice were later shown by our laboratory to have highly significant overlap with expression responses in a chronic ethanol exposure model known to mimic aspects of alcohol dependence in humans [30], and also contained a gene expression network associated with alcohol dependence thatwe recently identified Gsk3b as a potential candidate gene for treatment of alcoholism [31]. Together, these results support our premise that acute ethanol-exposed rodent brain gene expression could provide insight into relevant mechanistic frameworks and pathways underlying ethanol behaviors and risk for dependence in humans.
Several studies have integrated GWAS and gene expression or gene network data to cross-validate behavioral genetic findings [17]. For instance, the Psychiatric Genomics Consortium [32] tested for enrichment of nominally significant genes from human GWAS in previously identified functional pathways, and found shared functional enrichment of signals for schizophrenia, major depression disorder, and bipolar disorder in several categories. These pathways included histone methylation, neural signaling, and immune pathways [32]. Mamdani et al. reversed this type of analysis by testing for significant enrichment of previously identified GWAS signals in gene networks from their study. They found that expression quantitative trait loci for AD-associated gene expression networks in human nucleus accumbens tissue had significant enrichment with AD diagnosis and symptom count GWAS signals from the Collaborative Study on the Genetics of Alcoholism dataset [17]. Additional approaches have taken human GWAS significant (or suggestive) results for AD and provided additional confirmation by showing that expression levels for such genes showed correlations with ethanol behaviors in rodent models [33]. Such methods are informative with respect to analyzing the function of genes that have already reached some association significance threshold. However, they do not provide information about genes not reaching such statistical thresholds, but possibly still having important contributions to the genetic risk and mechanisms of AUD.
Dense module searching for GWAS (dmGWAS) is an algorithm for directly integrating GWAS data and other biological network information so as to identify gene networks contributing to a genetic disorder, even if few of the individual network genes exceed genome-wide statistical association thresholds [34]. The initial description of this approach utilized Protein-Protein Interaction (PPI) network data to identify networks associated with a GWAS phenotype. Modules derived from protein-protein interactions were scored from node-weights based on gene-level GWAS p-values. This approach was used to identify AD-associated PPI networks that replicated across ethnicities and showed significant aggregate AD-association in independent GWAS datasets [35], thus demonstrating the potential utility of the method. A more recent iteration of the dmGWAS algorithm, termed Edge-Weighted dense module searching for GWAS (EW-dmGWAS), allows integration of gene expression data to provide a direct co-analysis of gene expression, PPI, and GWAS data [36].
Utilization of the EW-dmGWAS algorithm would allow for identification of gene networks coordinately weighted for GWAS significance for AD in humans and ethanol-responsiveness in model organism brain gene expression data. We hypothesized that such an approach could provide novel information about candidate gene networks likely contributing to the genetic risk for AUD, while also adding mechanistic information about the role of such networks in ethanol behaviors. We show here the first use of such an approach for the integration of human PPI connectivity with mouse brain expression responses to acute ethanol and human GWAS results on AD. Our design incorporated the genome-wide microarray expression dataset derived from the acute ethanol-exposed mouse brain tissue used in Wolen et al. [10, 25], human protein-protein interaction data from the Protein Interaction Network database, and AD GWAS summary statistics from the Irish Affected Sib-Pair Study of Alcohol Dependence [33]. Importantly, we validated the identified ethanol-responsive and AD-associated networks by co-analysis with an additional, independent AD GWAS study on the Avon Longitudinal Study of Parents and Children dataset. Our results, although requiring further detailed investigation, could provide important methodological and biological function insight for future studies on the mechanisms and treatment of AUD.
Materials and methods
Samples
Mouse gene expression data
In order to maximize the utility of the mouse model by minimizing effects of behavioral and environmental variation, and to afford the identification of dependence-contributing genes involved in initial ethanol response, this study utilized gene expression data from ethanol-naïve mice treated with a single dose of acute ethanol. All mouse brain microarray data (Affymetrix GeneChip Mouse Genome 430 2.0) are from Wolen et al., 2012 [25] and can be downloaded from the GeneNetwork resource (www.genenetwork.org), via accession numbers GN135-137, GN154-156 and GN228-230, respectively for PFC, NAc and VTA data. Treatment and control groups each contained one sample (pooled RNA from 3 mice) from each strain and were given IP injections of saline or 1.8 g/kg of ethanol, respectively. Euthanasia and brain tissue collection took place 4 hours later. Data used for edge weighting in EW-dmGWAS analysis included Robust Multi-array Average (RMA) values, background-corrected and normalized measures of probe-wise expression, from the PFC, VTA, and NAc of male mice in 27–35 BXD recombinant inbred strains and two progenitor strains (DBA/2J and C57BL/6J).
Ethanol-responsive genes are predicted to be involved in pathways of neural adaptations that lead to dependence [25]. We predicted they would also be involved in mechanistic pathways from which GWAS signals are being detected. We therefore filtered for ethanol-responsive gene expression as done in Wolen et al.[25] prior to EW-dmGWAS so as to ensure edge weighting focused on ethanol responsiveness. Probe-level expression differences between control and ethanol-treated groups using the S-score algorithm which performs a probe-level analysis of expression between two groups [23, 37, 38] were obtained from the Wolen study [25] (S1 Table). Fisher’s Combined Test determined S-score significance values for ethanol responsiveness of each probeset across the entire BXD panel, and empirical p-values were calculated by 1,000 random permutations. Finally, q-values were calculated from empirical p-values to correct for multiple testing[25]. We defined an ethanol responsive gene set using a S-score probeset-level threshold of qFDR<0.1 for differential expression, in any one of the three brain regions. Genes associated with these probesets were carried forward in our analysis (Fig 1). Multiple probesets from single genes were reduced to single gene-wise expression levels within a particular brain region by selecting the maximum brain region-specific RMA value for each gene. After removing genes that were absent from the human datasets, 6,050 genes remained with expression values across all three brain regions (Fig 1).
Fig 1. Data pipeline for determining ethanol-responsiveness and merging datasets.
Pipeline used to prepare the data for the present analysis. The first cell contains the starting number of genes in the BXD mouse PFC, NAc, and VTA gene expression dataset.
Human GWAS data
Although many GWAS datasets now exist for AD, alcohol consumption and other ethanol responses, we chose two AD-related datasets for our analysis because of the phenotypic and methodological similarity between the studies and their availability at the time this work was initiated. The Irish Affected Sib-Pair Study of Alcohol Dependence (IASPSAD) AD GWAS dataset was used for the EW-dmGWAS analysis. It contains information from 1,748 unscreened controls (43.2% male) and 706 probands and affected siblings (65.7% male) from a native Irish population, after quality control [33]. Samples were genotyped on Affymetrix v6.0 SNP arrays. Diagnostic criteria for AD were based on the DSM-IV, and probands were ascertained from in- and out-patient alcoholism treatment facilities. Association of each Single Nucleotide Polymorphisms (SNP) with AD diagnosis status was tested by the Modified Quasi-Likelihood Score method [39], which accounts for participant relatedness. SNPs were imputed using IMPUTE2 [40] to hg19/1000 Genomes, and gene-wise p-values were calculated using Knowledge-Based mining system for Genome-wide Genetic studies (KGG2.5) [41]. This dataset was chosen because of its deep phenotyping and its theoretical consistency with findings from mouse experiments. The expression of the top-scoring genes in IASPSAD (COL6A3, RYR3, and KLF12) in mouse brain correlates with handling-induced convulsions, anxiety-like behavior, and acute functional tolerance to ethanol, respectively[33].
The Avon Longitudinal Study of Parents and Children (ALSPAC) GWAS gene-wise p-values were used to examine the ability of EW-dmGWAS to validate the EW-dmGWAS networks. This GWAS tested SNP association with a factor score calculated from 10 Alcohol Use Disorder Identification Test items for 4,304 (42.9% male) participants from Avon, UK. Samples were genotyped by the Illumina HumanHap550 quad genome-wide SNP platform [42]. This dataset was chosen because of its overall similarity to IASPSAD. Although the analyzed phenotypes were not identical between these two datasets, they were similar in that they both studied dependence symptoms, as opposed to non-diagnostic drinking measures. Additionally like IASPSAD, ALSPAC possessed the following important qualities: 100% of the sample was from the United Kingdom; the male to female ratio was roughly 1:1; SNPs were imputed to hg19/1000 Genomes; and gene-wise p-values were calculated by KGG2.5. No other GWAS dataset is as similar to IASPSAD to our knowledge, with respect to ancestral origin, genotyping, and phenotyping.
Protein network data
The Protein-Protein Interaction (PPI) network was obtained from the Protein Interaction Network Analysis (PINA 2.0) Platform (http://omics.bjcancer.org/pina/interactome.pina4ms.do). This platform was chosen because it includes PPI data from a wide array of databases, including: Intact, MINT, BioGRID, DIP, HPRD, and MIPS/Mpact. The Homo sapiens dataset was used for this analysis, due to it having much more content (166,776 binary interactions) than the mouse repository (only 13,865 binary interactions) [43, 44]. Uniprot IDs were used to match protein symbols to their corresponding gene symbols [45].
Statistical methods
EW-dmGWAS
The edge-weighted dense module searching for GWAS (dmGWAS_3.0) R package was used to identify treatment-dependent edge-weighted modules (small, constituent networks) nested within the background network(s) of non-weighted, binary interactions (https://bioinfo.uth.edu/dmGWAS/). We used the PPI framework for the background network, IASPSAD GWAS gene-wise p-values [33] for the node-weights, and RMA values from acute ethanol- and saline-exposed mouse PFC, VTA, and NAc genomic data for edge-weights [25]. For the remainder of this manuscript, we will use the term “network” to refer to the background PPI framework, and “module” to refer to the resulting groups of interrelated genes nested within this larger network. By the EW-dmGWAS algorithm, higher node-weights represent lower (i.e. more significant) GWAS p-values, whereas higher edge-weights represent a greater difference in the correlation of two genes between ethanol and control groups. This is calculated by taking the difference of correlations in RMA expression values of the two genes in control vs. ethanol treated BXD lines. The module score algorithm incorporated edge- and node-weights, which were each weighted to prevent bias towards representation of nodes or edges in module score calculations. Such bias could cause some modules to be identified based almost solely on edge-weights or node-weights, as opposed to the two combined, which would defeat the purpose of integration. The respective weighting depends upon a parameter (λ) which is calculated prior to module searching, based on the entire set of node- and edge-weights and used across all module score calculations, as part of the EW-dmGWAS algorithm. Higher module scores thus represent higher edge- and node-weights. Genes were kept in a module if they increased the standardized module score (Sn) by 0.5%. Sn corresponding to a permutation-based, empirical qFDR<0.05 were considered significant. A significant Sn (i.e. more significant qFDR values) indicates that a module’s constituent genes are more highly associated with AD in humans, and their interactions with each other are more strongly perturbed by acute ethanol exposure in mice than randomly constructed modules of the same size.
Due to the redundancy of genes between modules, we modified the EW-dmGWAS output by iteratively merging significant modules that overlapped >80% until no modules had >80% overlap, for each brain region. Percent overlap represented the number of genes contained in both modules (for every possible pair) divided by the number of genes in the smaller module. We call the final resulting modules “mega-modules”. Standardized mega-module scores (MM-Sn) were calculated using the algorithms employed by EW-dmGWAS. MM-Sn corresponding to qFDR<0.05 were considered significant (S1 Fig). Finally, connectivity (k) and Eigen-centrality (EC) were calculated using the igraph R package for each gene in each module to identify hub genes. Nodes with EC>0.2 and in the top quartile for connectivity for a module were considered to be hub genes.
Overlap with ALSPAC
Genes with an ALSPAC GWAS gene-wise p<0.001 were considered nominally significant, and will be referred to as “ALSPAC-nominal genes”. We used linear regression to test MM-Sn’s prediction of mean ALSPAC GWAS gene-wise p-value of each mega-module. Given our hypothesis that EW-dmGWAS would identify alcohol-associated gene networks and prioritize them by association, we predicted that higher MM-Sn’s would predict lower (i.e. more significant) mean GWAS p-values. Empirical p-values<0.017, reflecting Bonferroni correction for 3 independent tests (one per brain region): α = 0.05/3, were considered to represent significant association.
Overrepresentation of ALSPAC-nominal genes within each mega-module was analyzed for those modules containing >1 such gene. For each of these mega-modules, 10,000 modules containing the same number of genes were permuted to determine significance. Empirical p-values < 0.05/n (where n = total number of mega-modules tested) were considered significant.
Functional enrichment analysis
To determine if mega-modules with significant overrepresentation of ALSPAC-nominal genes represented an aggregation of functionally related genes, ToppGene (https://toppgene.cchmc.org/) was used to analyze functional enrichment. Categories of biological function, molecular function, cellular component, mouse phenotype, human phenotype, pathways, and drug interaction were tested for over-representation. All genes in the human genome were included in the reference gene set. This set was not limited to the ethanol-responsive genes included in this analysis, in order to preclude functional bias. Significant over-representation results were defined as p<0.01 (uncorrected), n≥3 genes overlap and n≤1000 genes per functional group. Given the number of categories and gene sets tested, our discussion below was narrowed to the most relevant categories, defined as Bonferroni-corrected p<0.1.
Results
Of the initial 45,037 probesets for the mouse gene expression arrays, 16,131 were associated with human-mouse homologues and had qFDR<0.1 for ethanol responsiveness (S-score) in at least one of the three brain regions (Fig 1). These probesets corresponded to a total of 7,730 genes and were trimmed to a single probeset per gene by filtering for the most abundant probeset as described in Methods. After removing genes that were absent from either the PPI network or the IASPSAD dataset, the final background PPI network for EW-dmGWAS analysis contained 6,050 genes (nodes) and 30,497 interactions (edges). The nodes contained 25 of the 78 IASPSAD-nominal genes and 24 of the 100 ALSPAC-nominal genes. There was no overlap between the IASPSAD and ALSPAC nominal gene sets.
Prefrontal cortex
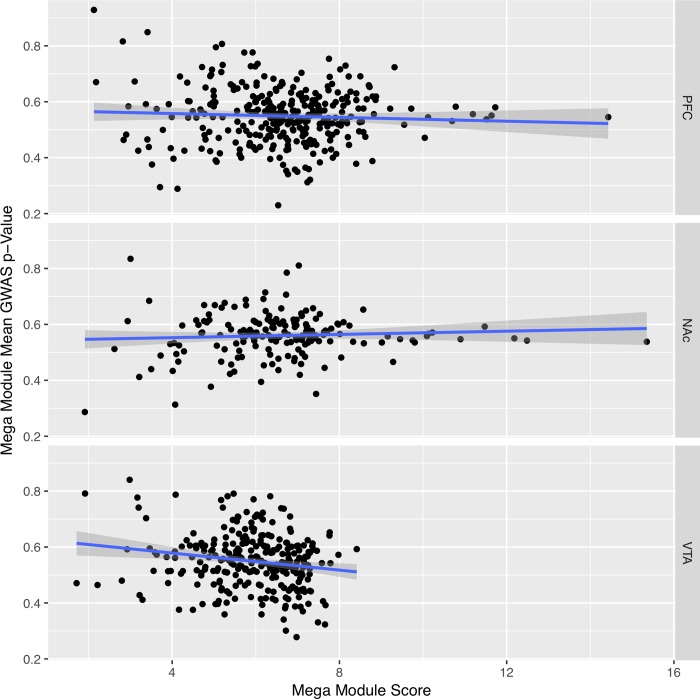
For analysis using PFC expression data for edge-weights, results revealed 3,545 significant modules (qFDR<0.05) containing a total of 4,300 genes, with 14 ALSPAC-nominal genes and 18 IASPSAD-nominal genes. These modules were merged to form 314 mega-modules, all with significant MM-Sn. Twelve mega-modules contained at least one ALSPAC-nominal gene, and 160 contained at least one IASPSAD-nominal gene. However, MM-Sn did not significantly predict mean ALSPAC GWAS gene-wise p-value (β = -0.003, p = 0.327, Fig 2).
Fig 2. Mega module score v. module average ALSPAC GWAS p-Value.
Correlation between each Mega Module’s score and average ALSPAC gene-wise GWAS p-value, for the Prefrontal Cortex (PFC) (β = -0.003, p = 0.327), Nucleus Accumbens (Nac) (β = 0.003, p = 0.390), and Ventral Tegmental Area (VTA) (β = -0.02, p = 0.003). Blue lines represent the line of best fit, estimated by linear regression, surrounded by their 95% confidence intervals (shaded gray).
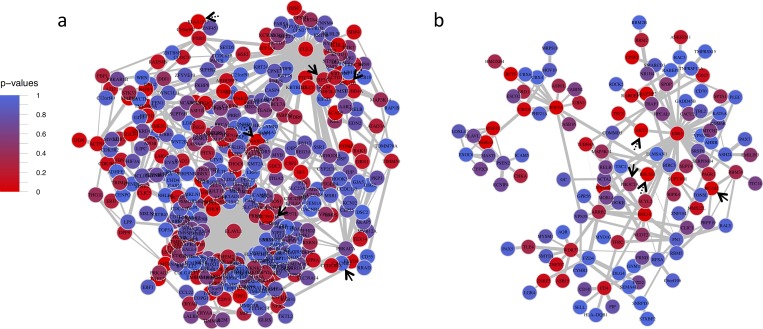
Two mega-modules, Aliceblue and Cadetblue, contained multiple ALSPAC-nominal genes (Table 1). Because overrepresentation was tested for 2 mega-modules, p<0.025 (α = 0.05/2) was considered significant. Cadetblue, was significantly overrepresented with ALSPAC-nominal genes (Table 1). Each of Cadetblue’s ALSPAC- and IASPSAD-nominal genes was connected to one of its most highly connected hub genes, ESR1 (estrogen receptor 1; connectivity (k) = 31, Eigen-centrality (EC) = 1) and ARRB2 (beta-arrestin-2; k = 13, EC = 0.25) (Fig 3). Although the ALSPAC-nominal gene overrepresentation was not significant for Aliceblue, it approached significance (Table 1). Further, Aliceblue had the second-highest MM-Sn in the PFC and contained 3 ALSPAC-nominal genes and 3 IASPSAD-nominal genes (Table 1). For these reasons, Aliceblue was carried through to functional enrichment analysis. Aliceblue’s two hub genes were ELAVL1 ((embryonic lethal, abnormal vision)-like 1; k = 165, EC = 1) and CUL3 (cullin 3; k = 75, EC = 0.21), which were connected to two of the three ALSPAC-nominal genes. Of these, CPM’s (carboxypeptidase M’s) only edge was with ELAVL1, and EIF5A2’s (eukaryotic translation initiation factor 5A2’s) only edge was with CUL3 (Fig 3).
Table 1. ALSPAC nominal gene overrepresentation.
| Brain Region | Mega-modules | kg | MM-Sn | MM-Sn qFDR | Overrep. p | Gene | IASPSAD GWAS p | ALSPAC GWAS p |
|---|---|---|---|---|---|---|---|---|
| PFC | aliceblue | 392 | 11.19 | <1E-16* | 0.063 | CPM | 0.493 | 6.48E-05* |
| CACNB2 | 0.978 | 4.97E-04* | ||||||
| EIF5A2 | 0.163 | 8.06E-04* | ||||||
| RSL1D1 | 3.48E-04* | 0.217 | ||||||
| SMARCA2 | 4.91E-04* | 0.877 | ||||||
| KIAA1217 | 8.84E-04* | 0.904 | ||||||
| cadetblue | 125 | 6.30 | 1.08E-06* | 0.013* | BCAS2 | 0.029 | 4.65E-04* | |
| PIK3C2A | 0.432 | 9.52E-04* | ||||||
| RSL1D1 | 3.48E-04* | 0.217 | ||||||
| AKT2 | 3.90E-05* | 0.980 | ||||||
| NAc | cadetblue2 | 195 | 8.04 | 8.06E-16* | 0.042 | CPM | 0.493 | 6.48E-05* |
| MGST3 | 0.358 | 4.62E-04* | ||||||
| gray26 | 12 | 6.39 | 9.95E-11* | <0.001* | PCDH7 | 0.007 | 2.10E-04* | |
| BCAS2 | 0.029 | 4.65E-04* | ||||||
| VTA | coral | 399 | 4.78 | 1.00E-06* | 0.068 | CPM | 0.493 | 6.48E-05* |
| DENND2C | 0.018 | 4.33E-04* | ||||||
| BIRC7 | 0.930 | 4.37E-04* | ||||||
| MGST3 | 0.358 | 4.62E-04* | ||||||
| PIK3CA | 7.06E-05* | 0.007 | ||||||
| TNN | 3.00E-04* | 0.018 | ||||||
| ANO6 | 6.32E-04* | 0.780 | ||||||
| SMARCA2 | 4.91E-04* | 0.877 | ||||||
| SIMC1 | 2.04E-04* | 0.977 | ||||||
| limegreen | 220 | 5.22 | 1.19E-07* | 0.054 | DENND2C | 0.018 | 4.33E-04* | |
| EIF5A2 | 0.163 | 8.06E-04* | ||||||
| RSL1D1 | 3.48E-04* | 0.217 | ||||||
| CCND2 | 1.94E-04* | 0.603 | ||||||
| AKT2 | 3.90E-05* | 0.980 | ||||||
| bisque | 89 | 6.22 | 7.57E-10* | 0.006* | ACLY | 0.701 | 2.21E-04* | |
| PRKG1 | 0.647 | 8.26E-04* | ||||||
| AKT2 | 3.90E-05* | 0.980 |
The following characteristics are displayed for each mega-module that contained >1 ALSPAC-nominal gene: affiliated brain region; total number of constituent genes (kg); constituent ALSPAC- and IASPSAD-nominal genes; empirical p-values for ALSPAC-nominal overrepresentation (Overrep. p); MM-Sn,and the associated False Discovery Rate (MM-Sn qFDR).
* Significant p-values: p<0.05 for MM Sn; p<0.05/n for ALSPAC overrepresentation, where n = number of tests per brain region; p<0.001 for IASPSAD and ALPSAC GWAS.
Fig 3. Prefrontal cortex mega modules aliceblue and cadetblue.
Prefrontal Cortex Mega Modules Cadetblue (a) and Aliceblue (b). Solid black arrows point to ALSPAC GWAS nominal genes, and dotted black arrows represent IASPSAD nominal genes. Edge-width is proportional to the difference in correlation strength between treatment and control mice, and node color represents IASPSAD GWAS p-values.
Both Cadetblue and Aliceblue showed significant enrichment in several functional categories (S3 Table). In sum, top functional enrichment categories for Aliceblue were related to actin-based movement, cardiac muscle signaling and action, increased triglyceride levels in mice, cell-cell and cell-extracellular matrix adhesion, and syndecan-2-mediated signaling. In contrast, Cadetblue’s top enrichment categories involved transcription-regulatory processes, specifically: RNA splicing, chromatin remodeling, protein alkylation and methylation, DNA replication regulation, several immune-related pathways, NF-κβ and Wnt signaling pathways, and reductase activity (Tables 2 and 3; S3 Table).
Table 2. Top gene ontology enrichment results for pfc mega module cadetblue.
| Category | Name | p-value | q-value Bonferroni | Hit Count in Query List | Hit Count in Genome | Hit in Query List |
|---|---|---|---|---|---|---|
| GO: Biological Process | chromatin organization | 1.50E-09 | 4.12E-06 | 23 | 776 | SMYD1, ESR1, KAT6A, ASH1L, PAGR1, CBX4, KDM6B, ASH2L, MYSM1, PHF21A, BPTF, UBN1, CBX6, SUPT16H, SMARCD3, H3F3B, PAX5, PAX7, BRD1, CABIN1, MGEA5, NR1H4, CBX8 |
| histone modification | 1.97E-06 | 5.40E-03 | 14 | 453 | SMYD1, KAT6A, ASH1L, PAGR1, KDM6B, ASH2L, MYSM1, PHF21A, PAX5, PAX7, BRD1, MGEA5, NR1H4, CBX8 | |
| covalent chromatin modification | 2.87E-06 | 7.89E-03 | 14 | 468 | SMYD1, KAT6A, ASH1L, PAGR1, KDM6B, ASH2L, MYSM1, PHF21A, PAX5, PAX7, BRD1, MGEA5, NR1H4, CBX8 | |
| chromatin remodeling | 1.47E-05 | 4.04E-02 | 8 | 165 | SMYD1, ESR1, ASH2L, MYSM1, BPTF, SMARCD3, H3F3B, PAX7 | |
| RNA splicing | 1.60E-05 | 4.40E-02 | 12 | 403 | SRSF6, NUDT21, BCAS2, RBM39, RALY, RBM5, PRPF19, AKT2, CPSF2, SNRPD3, WDR77, AQR | |
| protein alkylation | 2.44E-05 | 6.71E-02 | 8 | 177 | SMYD1, ASH1L, ASH2L, PAX5, PAX7, SNRPD3, WDR77, NR1H4 | |
| protein methylation | 2.44E-05 | 6.71E-02 | 8 | 177 | SMYD1, ASH1L, ASH2L, PAX5, PAX7, SNRPD3, WDR77, NR1H4 | |
| GO: Cellular Component | nucleoplasm part | 2.23E-05 | 7.49E-03 | 16 | 738 | MMS22L, SRSF6, NUDT21, KAT6A, PAGR1, CBX4, ELMSAN1, ASH2L, RBM39, PHF21A, UBN1, TONSL, PRPF19, SPOP, CPSF2, BRD1 |
| chromosome | 1.21E-04 | 4.07E-02 | 17 | 943 | MMS22L, PSEN2, BCAS2, ESR1, KAT6A, ASH1L, ZNF207, ASH2L, ESCO2, CBX6, TONSL, SUPT16H, PRPF19, SMARCD3, H3F3B, NR1H4, CBX8 | |
| ribonucleoside-diphosphate reductase complex | 1.24E-04 | 4.17E-02 | 2 | 3 | RRM2B, RRM2 | |
| DNA replication factor A complex | 1.39E-04 | 4.67E-02 | 3 | 16 | BCAS2, TONSL, PRPF19 | |
| nuclear replication fork | 1.40E-04 | 4.71E-02 | 4 | 41 | MMS22L, BCAS2, TONSL, PRPF19 | |
| catalytic step 2 spliceosome | 2.96E-04 | 9.94E-02 | 5 | 90 | BCAS2, RALY, PRPF19, SNRPD3, AQR | |
| GO: Molecular Function | oxidoreductase activity, acting on CH or CH2 groups | 3.32E-05 | 1.62E-02 | 3 | 10 | CYP2C8, RRM2B, RRM2 |
| oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor | 1.31E-04 | 6.38E-02 | 2 | 3 | RRM2B, RRM2 | |
| ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor | 1.31E-04 | 6.38E-02 | 2 | 3 | RRM2B, RRM2 | |
| ribonucleoside-diphosphate reductase activity | 1.31E-04 | 6.38E-02 | 2 | 3 | RRM2B, RRM2 | |
| chromatin binding | 1.69E-04 | 8.24E-02 | 12 | 516 | ESR1, KAT6A, ASH1L, RELB, CBX4, KDM6B, ASH2L, PHF21A, TLE4, SMARCD3, H3F3B, CABIN1 | |
| Mouse Phenotype | increased immunoglobulin level | 1.16E-06 | 2.92E-03 | 14 | 307 | TRAF3IP2, GADD45B, SEMA4B, PSEN2, ESR1, SPTA1, ASH1L, BIRC3, RELB, MYSM1, CD4, PIK3C2A, RABGEF1, CABIN1 |
| abnormal humoral immune response | 5.52E-06 | 1.39E-02 | 18 | 566 | TRAF3IP2, GADD45B, SEMA4B, PSEN2, ESR1, SPTA1, MAP3K14, ASH1L, BIRC3, RELB, TNFRSF11A, MYSM1, CD4, PIK3C2A, CD38, RABGEF1, PAX5, CABIN1 | |
| abnormal immunoglobulin level | 7.68E-06 | 1.93E-02 | 17 | 522 | TRAF3IP2, GADD45B, SEMA4B, PSEN2, ESR1, SPTA1, MAP3K14, ASH1L, BIRC3, RELB, TNFRSF11A, MYSM1, CD4, PIK3C2A, RABGEF1, PAX5, CABIN1 | |
| increased IgG level | 9.35E-06 | 2.35E-02 | 11 | 225 | TRAF3IP2, GADD45B, SEMA4B, ESR1, SPTA1, ASH1L, BIRC3, MYSM1, CD4, PIK3C2A, CABIN1 | |
| cortical renal glomerulopathies | 1.18E-05 | 2.96E-02 | 10 | 188 | TRAF3IP2, GADD45B, PSEN2, MYO1E, ESR1, SPTA1, RRM2B, ASH1L, RELB, PIK3C2A | |
| abnormal lymph node morphology | 1.85E-05 | 4.66E-02 | 14 | 390 | SELL, TRAF3IP2, TRAF1, PSEN2, ESR1, SPTA1, RRM2B, MAP3K14, BIRC3, RELB, TNFRSF11A, CD4, PIK3C2A, PIP | |
| glomerulonephritis | 1.95E-05 | 4.91E-02 | 8 | 121 | TRAF3IP2, GADD45B, PSEN2, ESR1, SPTA1, ASH1L, RELB, PIK3C2A | |
| abnormal B cell physiology | 3.21E-05 | 8.07E-02 | 18 | 644 | MYO1G, TRAF3IP2, GADD45B, SEMA4B, PSEN2, ESR1, SPTA1, MAP3K14, ASH1L, BIRC3, RELB, TNFRSF11A, MYSM1, CD4, PIK3C2A, RABGEF1, PAX5, CABIN1 | |
| Pathway | Signaling by Wnt | 2.78E-06 | 2.47E-03 | 13 | 340 | LGR4, ASH2L, FZD4, ARRB2, ZNRF3, TLE4, VPS35, H3F3B, AKT2, GNAO1, FZD2, MOV10, RAC3 |
| NF-kappa B signaling pathway | 1.07E-04 | 9.44E-02 | 6 | 95 | GADD45B, TRAF1, MAP3K14, BIRC3, RELB, TNFRSF11A | |
| Apoptosis | 1.13E-04 | 9.97E-02 | 7 | 138 | GADD45B, TRAF1, SEPT4, SPTA1, MAP3K14, BIRC3, AKT2 |
Functional enrichment results from ToppFun for Prefrontal Cortex Mega Module Cadetblue, where Bonferroni-corrected p<0.1.
Table 3. Top gene ontology enrichment results for pfc mega module aliceblue.
| Category | Name | p-value | q-value Bonferroni | Hit Count in Query List | Hit Count in Genome | Hit in Query List |
|---|---|---|---|---|---|---|
| GO: Biological Process | regulation of actin filament-based movement | 4.76E-08 | 2.07E-04 | 9 | 37 | FXYD1, ATP1A2, DBN1, GJA5, JUP, KCNJ2, DSC2, DSG2, DSP |
| cardiac muscle cell-cardiac muscle cell adhesion | 7.53E-08 | 3.27E-04 | 5 | 7 | CXADR, JUP, DSC2, DSG2, DSP | |
| regulation of cardiac muscle cell contraction | 1.64E-07 | 7.11E-04 | 8 | 31 | FXYD1, ATP1A2, GJA5, JUP, KCNJ2, DSC2, DSG2, DSP | |
| actin filament-based process | 3.57E-07 | 1.55E-03 | 36 | 688 | CDC42EP4, ACTN1, MYOZ1, MKLN1, FXYD1, RHOF, SDC4, CUL3, PRR5, CRYAA, ARHGDIA, ATP2C1, CCDC88A, STAU2, DYNLL1, DIXDC1, ATP1A2, CXADR, DBN1, PTGER4, GJA5, JUP, CDK5R1, NF1, KCNJ2, CACNB2, DSC2, DSG2, DSP, ARHGEF5, CASP4, LCP1, CSRP3, LIMK1, LDB3, LRP1 | |
| cell communication involved in cardiac conduction | 4.34E-07 | 1.89E-03 | 9 | 47 | PRKACA, ATP1A2, CXADR, GJA5, JUP, CACNB2, DSC2, DSG2, DSP | |
| desmosome organization | 8.59E-07 | 3.73E-03 | 5 | 10 | SNAI2, JUP, DSG2, DSP, PKP3 | |
| cardiac muscle cell action potential | 1.07E-06 | 4.65E-03 | 9 | 52 | ATP1A2, CXADR, GJA5, JUP, KCNJ2, CACNB2, DSC2, DSG2, DSP | |
| cardiac muscle cell contraction | 1.07E-06 | 4.65E-03 | 9 | 52 | FXYD1, ATP1A2, GJA5, JUP, KCNJ2, CACNB2, DSC2, DSG2, DSP | |
| bundle of His cell to Purkinje myocyte communication | 1.55E-06 | 6.72E-03 | 5 | 11 | GJA5, JUP, DSC2, DSG2, DSP | |
| regulation of cardiac muscle cell action potential | 2.30E-06 | 9.99E-03 | 6 | 20 | CXADR, GJA5, JUP, DSC2, DSG2, DSP | |
| bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 2.63E-06 | 1.14E-02 | 4 | 6 | JUP, DSC2, DSG2, DSP | |
| regulation of heart rate by cardiac conduction | 2.65E-06 | 1.15E-02 | 7 | 31 | GJA5, JUP, KCNJ2, CACNB2, DSC2, DSG2, DSP | |
| cardiac conduction | 3.37E-06 | 1.46E-02 | 13 | 131 | FXYD1, PRKACA, ATP1A2, ATP1A4, CXADR, GJA5, JUP, KCNJ2, CACNB2, CACNB4, DSC2, DSG2, DSP | |
| cardiac muscle cell action potential involved in contraction | 7.69E-06 | 3.34E-02 | 7 | 36 | GJA5, JUP, KCNJ2, CACNB2, DSC2, DSG2, DSP | |
| regulation of actin filament-based process | 1.05E-05 | 4.58E-02 | 21 | 343 | CDC42EP4, FXYD1, SDC4, ARHGDIA, CCDC88A, STAU2, DIXDC1, ATP1A2, DBN1, PTGER4, GJA5, JUP, CDK5R1, KCNJ2, DSC2, DSG2, DSP, ARHGEF5, CSRP3, LIMK1, LRP1 | |
| lipoprotein localization | 1.34E-05 | 5.83E-02 | 5 | 16 | APOB, APOC2, MSR1, CUBN, LRP1 | |
| lipoprotein transport | 1.34E-05 | 5.83E-02 | 5 | 16 | APOB, APOC2, MSR1, CUBN, LRP1 | |
| regulation of cardiac muscle contraction | 1.36E-05 | 5.91E-02 | 9 | 70 | FXYD1, PRKACA, ATP1A2, GJA5, JUP, KCNJ2, DSC2, DSG2, DSP | |
| GO: Cellular Component | intercalated disc | 2.90E-06 | 1.53E-03 | 9 | 59 | ACTN1, ATP1A2, CXADR, GJA5, JUP, KCNJ2, DSC2, DSG2, DSP |
| cell-cell contact zone | 1.56E-05 | 8.21E-03 | 9 | 72 | ACTN1, ATP1A2, CXADR, GJA5, JUP, KCNJ2, DSC2, DSG2, DSP | |
| desmosome | 1.61E-04 | 8.49E-02 | 5 | 26 | JUP, DSC2, DSG2, DSP, PKP3 | |
| GO: Molecular Function | protein binding involved in heterotypic cell-cell adhesion | 8.62E-07 | 7.88E-04 | 5 | 10 | CXADR, JUP, DSC2, DSG2, DSP |
| protein binding involved in cell adhesion | 1.15E-06 | 1.05E-03 | 6 | 18 | CXADR, ITGA2, JUP, DSC2, DSG2, DSP | |
| protein binding involved in cell-cell adhesion | 2.62E-06 | 2.39E-03 | 5 | 12 | CXADR, JUP, DSC2, DSG2, DSP | |
| cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication | 2.64E-06 | 2.41E-03 | 4 | 6 | JUP, DSC2, DSG2, DSP | |
| Human Phenotype | Dilated cardiomyopathy | 4.35E-05 | 3.89E-02 | 9 | 87 | ACAD9, CRYAB, UBR1, JUP, DSG2, DSP, LAMA4, CSRP3, LDB3 |
| Right ventricular cardiomyopathy | 8.82E-05 | 7.90E-02 | 4 | 13 | JUP, DSC2, DSG2, DSP | |
| Mouse Phenotype | increased circulating triglyceride level | 1.27E-05 | 4.77E-02 | 16 | 179 | ALPI, COL1A1, VLDLR, AGPAT2, WRN, APOB, APOC2, TXNIP, RSBN1, CSF2, PRKACA, BGLAP, MED13, LEPR, LIPC, LRP1 |
| Pathway | Non-integrin membrane-ECM interactions | 3.41E-05 | 4.72E-02 | 7 | 46 | ACTN1, SDC2, SDC4, ITGA2, LAMA3, LAMA4, LAMB3 |
| Syndecan-2-mediated signaling events | 4.44E-05 | 6.14E-02 | 6 | 33 | SDC2, CSF2, PRKACA, ITGA2, NF1, LAMA3 |
Functional enrichment results from ToppFun for Prefrontal Cortex Mega Module Aliceblue, where Bonferroni-corrected p<0.1.
Nucleus accumbens
Using NAc acute ethanol expression data for edge-weights yielded 3,460 significant modules containing a total of 4,213 genes, 15 of which were ALSPAC-nominal and 16 of which were IASPSAD-nominal. After merging by content similarity, there were 171 significant mega-modules. Nineteen MM contained at least one ALSPAC-nominal gene, and 73 MM contained at least one IASPSAD-nominal gene. However, MM Sn did not significantly predict MM mean ALSPAC GWAS gene-wise p-value (β = 0.003, p = 0.390). Two MMs, Cadetblue2 and Gray26, each contained two ALSPAC-nominal genes (Table 1). Because there were 2 tests for overrepresentation, p<0.025 (α = 0.05/2) was considered significant. Gray26, was significantly overrepresented with ALSPAC-nominal genes, and Cadetblue2 showed a trend towards overrepresentation with significance before correcting for multiple testing (Table 1).
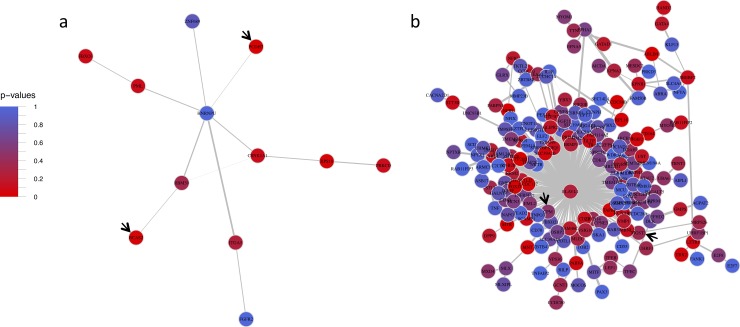
Gray26’s most central hub gene was HNRNPU (heterogeneous nuclear ribonucleoprotein U; connectivity = 6, Eigen-centrality = 1), followed by RBM39 (RNA binding motif protein 39; k = 3, EC = 0.46) and CSNK1A1 (k = 3, EC = 0.37). The two ALSPAC-nominal genes BCAS2 (breast carcinoma amplified sequence 2) and PCDH7 (protocadherin 7), shared their only edges with RBM39 and HNRPNPU, respectively (Fig 4A). As seen in the PFC’s Aliceblue, EAVL1 was a hub gene of Cadetblue2. ELAVL1 (k = 136, EC = 1) was connected to both of the ALSPAC-nominal genes, and served as the only connection for CPM and one of two connections for MGST3 (microsomal glutathione S-transferase 3) (Fig 4B). Strikingly, PFC Aliceblue and NAc Cadetblue 2 showed a highly significant overlap in their gene content, with 72 overlapping genes (S2 Table; p = 2.2 x 10−16).
Fig 4. Nucleus accumbens mega modules Gray26 and Cadetblue2.
Nucleus Accumbens Mega Modules Gray26 (a) and Cadetblue2 (b). Solid black arrows point to ALSPAC GWAS nominal genes. These modules did not contain IASPSAD nominal genes. Edge-width is proportional to the difference in correlation strength treatment and control mice, and node color represents IASPSAD GWAS p-values.
Both Cadetblue2 and Gray26 were significantly enriched with several functional categories (S3 Table). Like PFC Cadetblue, NAc Cadetblue2 was functionally enriched for gene groups related to nuclear function with transcription regulation pathways, particularly those involving RNA polymerase activity. Gray26 was most significantly enriched with genes related to functions involving: telomere maintenance, organelle organization, ribonucleoprotein complexes, and syndecan-mediated signaling (Tables 4 and 5; S3 Table).
Table 4. Top gene ontology enrichment results for nucleus accumbens mega module cadetblue2.
| Category | Name | p-value | q-value Bonferroni | Hit Count in Query List | Hit Count in Genome | Hit in Query List |
|---|---|---|---|---|---|---|
| GO: Biological Process | negative regulation of transcription from RNA polymerase II promoter | 9.38E-06 | 2.93E-02 | 23 | 810 | TGIF2, ZBTB20, SREBF2, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, MNT, TBX2, MLX, YBX3, TFAP2C, MXD4, E2F8, ZBTB14, MLXIPL, UHRF1, TNF, ELK4, PAX3, LEF1 |
| GO: Molecular Function | RNA polymerase II transcription factor activity, sequence-specific DNA binding | 1.80E-09 | 1.20E-06 | 27 | 678 | ZBTB20, SREBF2, GATA4, E2F7, CSRNP1, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, TEAD2, MLX, YBX3, FOXJ3, TFAP2C, E2F8, MLXIPL, KLF13, ELF2, ELK4, PAX3, LEF1 |
| transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding | 3.04E-06 | 2.03E-03 | 11 | 182 | ZBTB20, SREBF2, E2F7, MITF, MNT, TBX2, MLX, YBX3, TFAP2C, E2F8, MLXIPL | |
| transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding | 6.11E-06 | 4.08E-03 | 15 | 365 | ZBTB20, SREBF2, FOXL2, NFIB, NFIC, MITF, NFYA, HAND2, TBX2, TFEB, TFAP2C, E2F8, MLXIPL, KLF13, LEF1 | |
| RNA polymerase II regulatory region sequence-specific DNA binding | 8.95E-06 | 5.98E-03 | 20 | 632 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, MLXIPL, KLF13, LEF1 | |
| transcription regulatory region DNA binding | 9.52E-06 | 6.36E-03 | 24 | 862 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, ZBTB14, MLXIPL, KLF13, UHRF1, TNF, ELK4, LEF1 | |
| regulatory region DNA binding | 1.01E-05 | 6.74E-03 | 24 | 865 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, ZBTB14, MLXIPL, KLF13, UHRF1, TNF, ELK4, LEF1 | |
| RNA polymerase II regulatory region DNA binding | 1.03E-05 | 6.87E-03 | 20 | 638 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, MLXIPL, KLF13, LEF1 | |
| regulatory region nucleic acid binding | 1.07E-05 | 7.14E-03 | 24 | 868 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, ZBTB14, MLXIPL, KLF13, UHRF1, TNF, ELK4, LEF1 | |
| transcription regulatory region sequence-specific DNA binding | 1.32E-05 | 8.82E-03 | 21 | 705 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, MLXIPL, KLF13, UHRF1, LEF1 | |
| sequence-specific double-stranded DNA binding | 2.50E-05 | 1.67E-02 | 21 | 736 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, MLXIPL, KLF13, UHRF1, LEF1 | |
| core promoter proximal region sequence-specific DNA binding | 7.08E-05 | 4.73E-02 | 14 | 399 | SREBF2, GATA4, FOXL2, NFIB, NFIC, MITF, NFYA, TBX2, TFEB, E2F8, MLXIPL, KLF13, UHRF1, LEF1 | |
| core promoter proximal region DNA binding | 7.47E-05 | 4.99E-02 | 14 | 401 | SREBF2, GATA4, FOXL2, NFIB, NFIC, MITF, NFYA, TBX2, TFEB, E2F8, MLXIPL, KLF13, UHRF1, LEF1 | |
| transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | 9.15E-05 | 6.11E-02 | 13 | 358 | GATA4, CSRNP1, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, HAND2, TFEB, TFAP2C, KLF13, LEF1 | |
| double-stranded DNA binding | 1.25E-04 | 8.37E-02 | 21 | 824 | SREBF2, GATA4, E2F7, FOXL2, NFIB, NFIC, NFIX, MITF, NFYA, MNT, HAND2, TBX2, TFEB, MLX, YBX3, TFAP2C, E2F8, MLXIPL, KLF13, UHRF1, LEF1 | |
| Human Phenotype | Synophrys | 3.61E-05 | 2.06E-02 | 5 | 48 | ZBTB20, NFIX, MITF, KLF13, PAX3 |
| Mouse Phenotype | absent coat pigmentation | 2.38E-05 | 6.28E-02 | 4 | 15 | MITF, TFEB, TFEC, PAX3 |
Functional enrichment results from ToppFun for Nucleus Accumbens Mega Module Cadetblue2, where Bonferroni-corrected p<0.1.
Table 5. Top gene ontology enrichment results for nucleus accumbens mega module Gray26.
| Category | Name | p-value | q-value Bonferroni | Hit Count in Query List | Hit Count in Genome | Hit in Query List |
|---|---|---|---|---|---|---|
| GO: Biological Process | negative regulation of telomere maintenance via telomerase | 2.46E-05 | 2.92E-02 | 2 | 12 | HNRNPU, PML |
| negative regulation of organelle organization | 4.65E-05 | 5.52E-02 | 4 | 340 | PRKCD, FGFR2, HNRNPU, PML | |
| negative regulation of telomere maintenance via telomere lengthening | 5.06E-05 | 6.00E-02 | 2 | 17 | HNRNPU, PML | |
| GO: Cellular Component | ribonucleoprotein complex | 8.99E-04 | 8.99E-02 | 4 | 751 | CSNK1A1, RPS18, BCAS2, HNRNPU |
| intracellular ribonucleoprotein complex | 8.99E-04 | 8.99E-02 | 4 | 751 | CSNK1A1, RPS18, BCAS2, HNRNPU | |
| Pathway | Syndecan-4-mediated signaling events | 2.67E-04 | 7.44E-02 | 2 | 31 | PRKCD, ITGA5 |
| Syndecan-2-mediated signaling events | 3.03E-04 | 8.44E-02 | 2 | 33 | PRKCD, ITGA5 |
Functional enrichment results from ToppFun for Nucleus Accumbens Mega Module Gray26, where Bonferroni-corrected p<0.1.
Ventral tegmental area
Use of VTA control/ethanol gene expression responses for edge weighting initially resulted in 3,519 significant modules containing a total of 4,188 genes in EW-dmGWAS analysis. Merging by content similarity, resulted in 276 MMs, each with a significant MM Sn. Seventeen ALSPAC-nominal genes and 19 IASPSAD-nominal genes were spread across 25 and 156 mega-modules, respectively. Furthermore, MM-Sn significantly predicted mean ALSPAC GWAS gene-wise p-value (β = -0.02, p = 0.003).
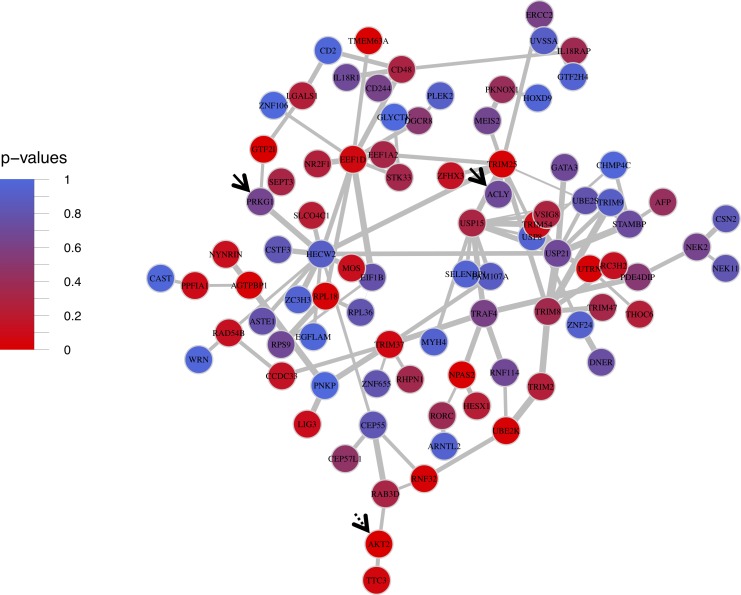
Mega-modules with the highest representation of ALSPAC-nominal genes included Coral, Limegreen, and Bisque (Table 1). Because there were 3 tests for overrepresentation, p<0.017 (α = 0.05/3) was considered significant. Although overrepresentation of ALSPAC-nominal genes was not significant in Coral and Limegreen, it was significant in Bisque, which has the highest MM-Sn of the three (Table 1; Fig 5). Bisque contained four highly interconnected genes: USP21 (ubiquitin specific peptidase 21; k = 10, EC = 1), USP15 (ubiquitin specific peptidase 15; k = 10, EC = 0.65), TRIM25 (tripartite motif-containing 25; k = 10, EC = 0.49), and HECW2 (HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2; k = 12, EC = 0.48). HECW2 and TRIM25 shared edges with this MM’s IASPSAD-nominal genes PRKG1 (protein kinase, cGMP-dependent, type I) and ACLY (ATP citrate lyase), respectively. However, none of the hub genes shared an edge with Bisque’s ALSPAC nominal gene, AKT2 (AKT serine/threonine kinase 2). Finally, Bisque had significant enrichment in several functional categories (S3 Table). It was most significantly enriched with genes associated with ubiquitination, ligase and helicase activity, and eukaryotic translation elongation (Table 6; S3 Table).
Fig 5. Ventral tegmental area mega module bisque.
Ventral Tegmental Area Mega Modules Bisque. Solid black arrows point to ALSPAC GWAS nominal genes, and dotted black arrows represent IASPSAD nominal genes. Edge-width is proportional to the difference in correlation strength between treatment and control mice, and node color represents IASPSAD GWAS p-values.
Table 6. Top gene ontology enrichment results for ventral tegmental area mega module bisque.
| Category | Name | p-value | q-value Bonferroni | Hit Count in Query List | Hit Count in Genome | Hit in Query List |
|---|---|---|---|---|---|---|
| GO: Cellular Component | nucleolus | 6.41E-07 | 1.24E-04 | 17 | 894 | ZNF106, NEK2, EEF1D, RPL36, PNKP, SELENBP1, ZNF655, RPS9, WRN, GATA3, ZFHX3, RORC, DGCR8, TTC3, ARNTL2, NEK11, RPL18 |
| eukaryotic translation elongation factor 1 complex | 1.27E-04 | 2.47E-02 | 2 | 4 | EEF1D, EEF1A2 | |
| GO: Molecular Function | ubiquitin-protein transferase activity | 4.98E-07 | 1.33E-04 | 12 | 414 | RC3H2, TRAF4, UBE2K, TRIM2, TRIM25, TRIM9, HECW2, TRIM8, UBE2S, RNF114, TTC3, TRIM37 |
| ubiquitin-like protein transferase activity | 9.70E-07 | 2.59E-04 | 12 | 441 | RC3H2, TRAF4, UBE2K, TRIM2, TRIM25, TRIM9, HECW2, TRIM8, UBE2S, RNF114, TTC3, TRIM37 | |
| acid-amino acid ligase activity | 3.42E-06 | 9.12E-04 | 9 | 259 | RC3H2, TRIM2, TRIM25, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| ligase activity, forming carbon-nitrogen bonds | 9.78E-06 | 2.61E-03 | 9 | 295 | RC3H2, TRIM2, TRIM25, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| tubulin-glycine ligase activity | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| protein-glycine ligase activity | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| protein-glycine ligase activity, initiating | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| coenzyme F420-0 gamma-glutamyl ligase activity | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| ribosomal S6-glutamic acid ligase activity | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| coenzyme F420-2 alpha-glutamyl ligase activity | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| protein-glycine ligase activity, elongating | 1.87E-05 | 5.00E-03 | 8 | 244 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| tubulin-glutamic acid ligase activity | 2.05E-05 | 5.46E-03 | 8 | 247 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| protein-glutamic acid ligase activity | 2.17E-05 | 5.79E-03 | 8 | 249 | RC3H2, TRIM2, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| ligase activity | 2.38E-05 | 6.35E-03 | 10 | 415 | LIG3, RC3H2, TRIM2, TRIM25, TRIM9, HECW2, TRIM8, RNF114, TTC3, TRIM37 | |
| DNA helicase activity | 2.43E-04 | 6.49E-02 | 4 | 65 | ERCC2, GTF2H4, RAD54B, WRN | |
| Pathway | Eukaryotic Translation Elongation | 1.67E-04 | 8.37E-02 | 5 | 98 | EEF1D, RPL36, RPS9, EEF1A2, RPL18 |
Functional enrichment results from ToppFun for Ventral Tegmental Area Mega Module Bisque, where Bonferroni-corrected p<0.1.
Discussion
To our knowledge, this is the first study to directly co-analyze human GWAS with mouse brain ethanol-responsive gene expression data to identify ethanol-related gene networks relevant to AD. Unlike previous studies that have employed cross-species validation methods for specific genes or gene sets, this study analyzed human and mouse data in tandem to identify gene networks across the entire genome, using the EW-dmGWAS algorithm. This approach successfully identified significantly ethanol-responsive and AD-associated gene networks, or modules. We further improved the existing EW-dmGWAS algorithm by merging highly redundant modules to create more parsimonious mega-modules, thus decreasing complexity without sacrificing significance. Additionally, we validated these results by testing for overrepresentation with, and mega-module score prediction by, signals from an independent GWAS dataset. Overall, our findings suggest that such direct integration of model organism expression data with human protein interaction and GWAS data can productively leverage these data sources. Furthermore, we present initial evidence for novel, cross-validated gene networks warranting further study for mechanisms underlying AUD.
Identification of network-level associations across GWAS datasets
One major concern with existing GWAS studies on AD had been the relative lack of replication across studies. Although some very large GWAS studies on alcohol consumption have shown replicable results [13–15], those do not account for all previously identified associations. We reasoned that our integrative gene network-querying approach might identify networks that shared signals from different GWASs on AD, even if the signals were not from the same genes across GWASs. Concordant with this hypothesis, VTA mega-module scores significantly predicted average gene-wise p-values from an independent GWAS dataset, ALSPAC (Fig 2). This suggests that ethanol-responsive gene expression networks in this brain region may be particularly sensitive to genetic variance and thus are highly relevant to mechanisms contributing to genetic risk for AD. This is possibly attributable to the involvement of VTA dopaminergic reward pathways in the development of AD [46]. Further investigation of dopaminergic neuronal response to acute ethanol administration, and the association between this response and proclivity for developing dependence is needed.
Although scores did not prioritize mega-modules with respect to ALSPAC results in PFC and NAc, individual mega-modules were overrepresented with ALSPAC signals (Table 1). The ALSPAC-overrepresented VTA and PFC mega-modules also contained nominally significant genes from the GWAS dataset used for the network analysis, IASPSAD. These results suggest that the integration of acute ethanol-related expression data from mice and human PPI can identify functional networks that associate signals from different GWAS datasets.
Composition and structure of mega-modules
Functional composition of mega-modules varied between brain regions for the most part. For example, although Aliceblue (PFC) and Cadetblue2 (NAc) shared the hub gene ELAVL1, ALSPAC-nominal gene CPM, and had a significant overlap in their gene content, their functional enrichment results were very different (Tables 3 and 4). These results suggest that brain regional ethanol-responsive gene expression results likely had an important impact on composition of networks, thus leveraging protein-protein interaction network information and GWAS results.
Despite such differences, the mega-modules presented in Table 1 shared certain structural similarities. Most of the IAPSAD- and ALSPAC-nominal genes in these modules shared edges with hub genes (Figs 3–5). These hub genes included: CUL3 and ELAVL1 from PFC Aliceblue; ESR1 from PFC Cadetblue; ELAVL1rom NAc Cadetblue2; TRIM25 and HECW2 from VTA Bisque. Further, GWAS nominally significant genes (IASPAD or ALSPAC) generally were not hub genes in the derived networks (see Figs 3–5; S2 Table). This may be consistent with the general tenet that genetic variation in complex traits does not produce major alterations in cellular function, but rather modulation of cellular mechanisms for maintaining homeostasis. Hub genes may be more functionally more closely related to a given trait, but likely have such widespread influence so as to be evolutionarily resistant to genetic variation in complex traits. This is also consistent with the hypothesis that omnigenic influences are an important feature of complex traits such as AUD [47].
One hub gene was found to influence network structure in both PFC and NAc. ELAVL1 is a broadly expressed gene that acts as a RNA-binding protein in AU-rich domains, generally localized within 3’-UTRs of mRNA. As such, ELAVL1 has been shown to alter mRNA stability by altering binding of miRNA or other factors influencing mRNA degradation [48] and has been implicated in activity-dependent regulation of gene expression in the brain with drug abuse [49]. The large interaction space for ELAVL1 in PFC Alice Blue and NAc Cadetblue 2 and the multiple nominal GWAS hits within these genes suggest that ELAVL1 could have an important modulatory function on the network of genes susceptible to genetic variation in AUD.
Functional aspects of mega-modules
This theory regarding network structure is further supported by our functional enrichment analysis, which revealed several small groups of functionally related genes within each mega-module. All of the mega-modules discussed above (Table 1) contained at least one GWAS-nominal gene in the top enrichment groups, except Cadetblue2, which still had GWAS-nominal genes in its significant enrichment groups (S3 Table).
Another unifying feature across these mega-modules, except Aliceblue, was significant functional enrichment for pathways that regulate gene expression. Specifically, these pathways were related to chromatin organization, RNA splicing, and translation- and transcription-related processes (S3 Table). This is not surprising, as alterations in gene expression have long been proposed as a mechanism underlying long-term neuroplasticity resulting in ethanol-dependent behavioral changes, and eventually dependence [50].
In contrast, the largest functional enrichment groups unique to Aliceblue were related to actin-based filaments and cardiac function (Tables 2 and 3). Actin not only provides cytoskeletal structure to neurons, but also functions in dendritic remodeling in neuronal plasticity, which likely contributes to AD development [51, 52]. Aliceblue was also significantly enriched for the syndecan-2 signaling pathway, and contained the SDC2 gene itself, which functions in dendritic structural changes together with F-actin [53]. Additionally, the most significant enrichment group unique to Cadetblue was the Wnt signaling pathway, which also regulates actin function [54, 55]. Of note, a prior study has shown that ARRB2 (a Cadetblue hub gene and member of Wnt signaling pathway) knockout rats display significantly decreased levels of voluntary ethanol consumption and psychomotor stimulation in response to ethanol [56]. These findings highlight the potential importance of postsynaptic actin-related signaling and dendritic plasticity in PFC gene networks responding to acute ethanol and contributing to genetic risk for AD. Future studies may aim to confirm this association by investigating changes in actin and dendritic processes in response to acute ethanol exposure, and whether or not the degree of these changes is associated with development of dependence.
Finally, although the NAc Cadetblue2 mega-module was highly enriched for functions related to transcriptional regulation, it also contained the gene FGF21 within its interaction space (S2 Table and Fig 4B). FGF21 is a member of the fibroblast growth factor gene family and is a macronutrient responsive gene largely expressed in liver. Importantly FGF21 has been shown to be released from the liver by ethanol consumption and negatively regulates ethanol consumption by interaction with brain FGF-receptor/beta-Klotho complexes. Beta-Klotho, a product of the KLB gene, is an obligate partner of the FGF receptor and has recently been shown to have a highly significant association with alcohol consumption in recent very large GWAS studies [14, 15]. Although the role of FGF21 and KLB in AD are not currently known, the association of FGF21 with the Cadetblue2 mega-module, containing nominally associated genes from AD GWAS studies, is a possible additional validation of the utility of our studies integrating protein-protein interaction information (tissue non-specific), AD GWAS (tissue non-specific) and brain ethanol-responsive gene expression.
Potential weaknesses and future studies
The studies presented here provide evidence for the utility of integrating genomic expression data with protein-protein interaction networks and GWAS data in order to gain a better understanding of the genetic architecture of complex traits, such as AD. Our analysis also generated several testable hypotheses regarding gene networks and signaling mechanisms related to ethanol action and genetic burden for AD. However, these studies utilized acute ethanol-related expression data in attempting to identify mechanisms of AD, a chronic ethanol exposure disease. Use of a chronic exposure model could provide for a more robust integration of the expression data and GWAS signals. However, we feel the current study is valid, since acute responses to ethanol have been repeatedly shown to be a heritable risk factor for AD [57–59]. Further, large GWAS studies have recently shown significant genetic correlation and overlapping significant genes between alcohol consumption and alcohol dependence phenotypes [60, 61]. We have also recently demonstrated a very high degree of overlap in mouse brain expression changes between acute ethanol exposure and a chronic ethanol vapor exposure model thought to mimic aspects of alcohol dependence [30]. Our laboratory has also recently reported that an acute ethanol-responsive gene network from the same microarray data used for studies in this manuscript showed significant association, at a network level, with AD in data from the COGA GWAS analysis of AD [31]. Finally, the cross-species analysis of acute ethanol responses and AD allowed us to explore networks involved in specific brain regional initial response to ethanol that are also related to dependence. Therefore, our findings may have implications for mechanistic activations or changes occurring upon initial ethanol exposure, and ultimately contributing to the development of dependence.
A potential shortcoming for this work regards the limited size of the GWAS studies utilized and differences in phenotypic assessment. The IASPSAD study was based on AD diagnosis, whereas ALSPAC was based on a symptom factor score. Had we used larger GWAS studies based on the same assessment criteria, it is possible that greater overlap of GWAS signals within mega-modules would have been observed. Recent large GWAS studies on ethanol have, to date, generally concerned measures of ethanol consumption, rather than a diagnosis of alcohol dependence per se [14, 15]. For this reason, we focused this initial effort on GWAS studies concerned with alcohol dependence. However, using the IASPAD and ALSPAC studies allowed us to identify gene networks that are robust across both the severe end of the phenotypic spectrum (i.e. diagnosable AD), and for symptoms at the sub-diagnostic level.
Overall, this analysis successfully identified novel ethanol-responsive, AD-associated, functionally enriched gene expression networks in the brain that may play a causal role in the developmental pathway from first ethanol exposure to AD. This is the first analysis to identify such networks by directly co-analyzing brain regional gene expression data, protein-protein interaction data, and GWAS summary statistics. The identified modules provided insight into common pathways between differing signals from independent, largely underpowered, yet deeply phenotyped GWAS datasets. This supports the conjecture that the integration of different GWAS results at a gene network level, rather than simply looking for replication of individual gene signals, could make use of previously underpowered datasets and identify common genetic mechanisms relevant to AD. Future expansion of such approaches to integrate additional model organism chronic ethanol-responsive gene sets with the rapidly evolving GWAS literature on alcohol consumption and dependence, together with validation of key targets by gene targeting in animals models, may provide both novel insight for the neurobiology of AD and the development of improved therapeutic approaches.
Supporting information
Empirical p-values were calculated from standardized module scores based on a Z-distribution. The original EW-dmGWAS module score, permutation, and score standardization algorithms were used to calculate the respective Mega Modules parameters. Modules were considered to have >80% overlap if >80% of the genes in the smaller module was contained in the larger module. False Discovery Rates were calculated based on the Benjamini-Hochberg algorithm, using the “stats” package in R. Intramodular connectivity was defined as the number of edges (i.e. connections) attached to that node (i.e. gene). Eigen-Centrality was calculated using the “igraph” package in R.
(PDF)
One table per brain region, containing each of the following values: RMA values and S-scores from the maximally expressed probeset per gene, for each BXD strain; the associated probeset IDs, human gene symbols, and mouse gene symbols; and the Fisher’s combined False Discovery Rate (q-value) for each probeset.
(XLSX)
One table per brain region, containing each of the following characteristics, for all significant Mega Modules: name; constituent genes; ALPSAC and IASPSAD p-values for each gene; Mega Module score (Sn), p-value (Sn_p), and False Discovery Rate (Sn_qFDR); and intramodular eigencentrality and connectivity. Significance values < 10−16 are rounded to 0.
(XLSX)
One table for each ALSPAC-overrepresented Mega Module, containing ToppFun output for gene ontology enrichment groups with p<0.01 and minimum group size of 3 genes and maximum size of 1,000 genes, for the following categories: Biological Process, Cellular Component, Molecular Function, Human Phenotype, Mouse Phenotype, and Pathways.
(XLSX)
Acknowledgments
The authors wish to thank members of the Miles laboratory and the Virginia Institute for Psychiatric and Behavioral Genetics for their helpful comments and suggestions during the course of these studies. Additionally, we thank Dr. Zhongming Zhou at the University of Texas Health Sciences Center for suggestions regarding the utilization of EW-dmGWAS.
Data Availability
Most relevant data are contained within the paper and Supporting Information. Additional raw microarray data are found on GeneNetwork (www.genenetwork.org) under accession numbers GN135-137, GN154-156 and GN228-230 and in the Gene Expression Omnibus (GEO) database (accession number GSE28515). Primary human GWAS data is available from cited publications.
Funding Statement
This work was supported by NIAAA grant P50AA022537 (BR, SB, MFM) and by intramural funds of the VCU Alcohol Research Center (https://arc.vcu.edu). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Takahashi T, Lapham G, Chavez LJ, Lee AK, Williams EC, Richards JE, et al. Comparison of DSM-IV and DSM-5 criteria for alcohol use disorders in VA primary care patients with frequent heavy drinking enrolled in a trial. Addiction science & clinical practice. 2017;12(1):17 Epub 2017/07/19. 10.1186/s13722-017-0082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, et al. Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA psychiatry. 2015;72(8):757–66. Epub 2015/06/04. 10.1001/jamapsychiatry.2015.0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahre M, Roeber J, Kanny D, Brewer RD, Zhang X. Contribution of excessive alcohol consumption to deaths and years of potential life lost in the United States. Preventing chronic disease. 2014;11:E109 Epub 2014/06/27. 10.5888/pcd11.130293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. The Lancet. 2012;380(9859):2224–60. 10.1016/s0140-6736(12)61766-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reus VI, Fochtmann LJ, Bukstein O, Eyler AE, Hilty DM, Horvitz-Lennon M, et al. The American Psychiatric Association Practice Guideline for the Pharmacological Treatment of Patients With Alcohol Use Disorder. The American journal of psychiatry. 2018;175(1):86–90. Epub 2018/01/06. 10.1176/appi.ajp.2017.1750101 . [DOI] [PubMed] [Google Scholar]
- 6.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. The American journal of psychiatry. 1999;156(1):34–40. Epub 1999/01/19. 10.1176/ajp.156.1.34 . [DOI] [PubMed] [Google Scholar]
- 7.Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychological medicine. 2015;45(5):1061–72. Epub 2014/08/30. 10.1017/S0033291714002165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1997;17(10):3946–55. Epub 1997/05/15. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saba LM, Bennett B, Hoffman PL, Barcomb K, Ishii T, Kechris K, et al. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology. 2011;60(7–8):1269–80. Epub 2010/12/28. 10.1016/j.neuropharm.2010.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putman AH, Wolen AR, Harenza JL, Yordanova RK, Webb BT, Chesler EJ, et al. Identification of quantitative trait loci and candidate genes for an anxiolytic-like response to ethanol in BXD recombinant inbred strains. Genes, brain, and behavior. 2016;15(4):367–81. Epub 2016/03/08. 10.1111/gbb.12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Zhang Y, Wang F, Sun Y, Shi J, Lu L. From genetic studies to precision medicine in alcohol dependence. Behavioural pharmacology. 2016;27(2–3 Spec Issue):87–99. Epub 2015/11/19. 10.1097/FBP.0000000000000202 . [DOI] [PubMed] [Google Scholar]
- 12.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry. 2014;19(1):41–9. Epub 2013/10/30. 10.1038/mp.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart AB, Kranzler HR. Alcohol Dependence Genetics: Lessons Learned From Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcoholism, clinical and experimental research. 2015;39(8):1312–27. Epub 2015/06/26. 10.1111/acer.12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumann G, Liu C, O'Reilly P, Gao H, Song P, Xu B, et al. KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(50):14372–7. Epub 2016/12/03. 10.1073/pnas.1611243113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N = 112 117). Molecular psychiatry. 2017;22(10):1376–84. Epub 2017/09/25. 10.1038/mp.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. Epub 2009/10/09. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, et al. Integrating mRNA and miRNA Weighted Gene Co-Expression Networks with eQTLs in the Nucleus Accumbens of Subjects with Alcohol Dependence. PloS one. 2015;10(9):e0137671 Epub 2015/09/19. 10.1371/journal.pone.0137671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Molecular psychiatry. 2015;20(11):1438–47. Epub 2014/12/03. 10.1038/mp.2014.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. American Journal of Human Genetics. 2010;86(4):581–91. 10.1016/j.ajhg.2010.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98(1):1–8. Epub 2011/05/14. 10.1016/j.ygeno.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowell RD. The similarity of gene expression between human and mouse tissues. Genome Biol. 2011;12(1):101 Epub 2011/01/19. 10.1186/gb-2011-12-1-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farris SP, Miles MF. Fyn-dependent gene networks in acute ethanol sensitivity. PloS one. 2013;8(11):e82435 Epub 2013/12/07. 10.1371/journal.pone.0082435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25(9):2255–66. Epub 2005/03/05. 10.1523/JNEUROSCI.4372-04.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith ML, Lopez MF, Archer KJ, Wolen AR, Becker HC, Miles MF. Time-Course Analysis of Brain Regional Expression Network Responses to Chronic Intermittent Ethanol and Withdrawal: Implications for Mechanisms Underlying Excessive Ethanol Consumption. PloS one. 2016;11(1):e0146257 Epub 2016/01/06. 10.1371/journal.pone.0146257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, et al. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PloS one. 2012;7(4):e33575 10.1371/journal.pone.0033575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osterndorff-Kahanek EA, Tiwari GR, Lopez MF, Becker HC, Harris RA, Mayfield RD. Long-term ethanol exposure: Temporal pattern of microRNA expression and associated mRNA gene networks in mouse brain. PloS one. 2018;13(1):e0190841 Epub 2018/01/10. 10.1371/journal.pone.0190841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crabbe JC. Use of animal models of alcohol-related behavior. Handbook of clinical neurology. 2014;125:71–86. Epub 2014/10/14. 10.1016/B978-0-444-62619-6.00005-7 . [DOI] [PubMed] [Google Scholar]
- 28.Trim RS, Schuckit MA, Smith TL. The relationships of the level of response to alcohol and additional characteristics to alcohol use disorders across adulthood: a discrete-time survival analysis. Alcoholism, clinical and experimental research. 2009;33(9):1562–70. Epub 2009/06/03. 10.1111/j.1530-0277.2009.00984.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Wit H, Phillips TJ. Do initial responses to drugs predict future use or abuse? Neuroscience and biobehavioral reviews. 2012;36(6):1565–76. Epub 2012/05/01. 10.1016/j.neubiorev.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Vaart AD, Wolstenholme JT, Smith ML, Harris GM, Lopez MF, Wolen AR, et al. The allostatic impact of chronic ethanol on gene expression: A genetic analysis of chronic intermittent ethanol treatment in the BXD cohort. Alcohol. 2017;58:93–106. Epub 2016/11/14. 10.1016/j.alcohol.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Vaart A, Meng X, Bowers MS, Batman AM, Aliev F, Farris SP, et al. Glycogen synthase kinase 3 beta regulates ethanol consumption and is a risk factor for alcohol dependence. Neuropsychopharmacology. 2018;43(13):2521–31. Epub 2018/09/07. 10.1038/s41386-018-0202-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Network, Pathway Analysis Subgroup of Psychiatric Genomics C. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nature neuroscience. 2015;18(2):199–209. Epub 2015/01/20. 10.1038/nn.3922 PubMed Central PMCID: PMCPMC4378867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adkins AE, Hack LM, Bigdeli TB, Williamson VS, McMichael GO, Mamdani M, et al. Genomewide Association Study of Alcohol Dependence Identifies Risk Loci Altering Ethanol-Response Behaviors in Model Organisms. Alcoholism, clinical and experimental research. 2017;41(5):911–28. Epub 2017/02/23. 10.1111/acer.13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics (Oxford, England). 2011;27(1):95–102. Epub 2010/11/04. 10.1093/bioinformatics/btq615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han S, Yang BZ, Kranzler HR, Liu X, Zhao H, Farrer LA, et al. Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. American Journal of Human Genetics. 2013;93(6):1027–34. Epub 2013/11/26. 10.1016/j.ajhg.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Yu H, Zhao Z, Jia P. EW_dmGWAS: edge-weighted dense module search for genome-wide association studies and gene expression profiles. Bioinformatics (Oxford, England). 2015;31(15):2591–4. Epub 2015/03/26. 10.1093/bioinformatics/btv150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Wang L, Ravindranathan A, Miles MF. A new algorithm for analysis of oligonucleotide arrays: application to expression profiling in mouse brain regions. Journal of molecular biology. 2002;317(2):225–35. Epub 2002/03/21. 10.1006/jmbi.2001.5350 . [DOI] [PubMed] [Google Scholar]
- 38.Kennedy RE, Archer KJ, Miles MF. Empirical validation of the S-Score algorithm in the analysis of gene expression data. BMC Bioinformatics. 2006;7:154 Epub 2006/03/21. 10.1186/1471-2105-7-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. American Journal of Human Genetics. 2007;81(2):321–37. Epub 2007/08/02. 10.1086/519497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature genetics. 2012;44(8):955–9. Epub 2012/07/24. 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li MX, Sham PC, Cherny SS, Song YQ. A knowledge-based weighting framework to boost the power of genome-wide association studies. PloS one. 2010;5(12):e14480 Epub 2011/01/11. 10.1371/journal.pone.0014480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards AC, Aliev F, Wolen AR, Salvatore JE, Gardner CO, McMahon G, et al. Genomic influences on alcohol problems in a population-based sample of young adults. Addiction (Abingdon, England). 2015;110(3):461–70. Epub 2014/12/03. 10.1111/add.12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6(1):75–7. 10.1038/nmeth.1282 . [DOI] [PubMed] [Google Scholar]
- 44.Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, et al. PINA v2.0: mining interactome modules. Nucleic acids research. 2012;40(Database issue):D862–5. Epub 2011/11/10. 10.1093/nar/gkr967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods in molecular biology (Clifton, NJ). 2007;406:89–112. Epub 2008/02/22. . [DOI] [PubMed] [Google Scholar]
- 46.Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3(8):760–73. 10.1016/S2215-0366(16)00104-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169(7):1177–86. Epub 2017/06/18. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279(13):12974–9. Epub 2004/01/09. 10.1074/jbc.M310433200 . [DOI] [PubMed] [Google Scholar]
- 49.Tiruchinapalli DM, Caron MG, Keene JD. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J Neurochem. 2008;107(6):1529–43. Epub 2008/11/19. 10.1111/j.1471-4159.2008.05718.x . [DOI] [PubMed] [Google Scholar]
- 50.Wilke N, Sganga M, Barhite S, Miles MF. Effects of alcohol on gene expression in neural cells. Exs. 1994;71:49–59. Epub 1994/01/01. . [DOI] [PubMed] [Google Scholar]
- 51.Seo D, Sinha R. Neuroplasticity and Predictors of Alcohol Recovery. Alcohol research: current reviews. 2015;37(1):143–52. Epub 2015/08/11. [PMC free article] [PubMed] [Google Scholar]
- 52.Kyzar EJ, Pandey SC. Molecular mechanisms of synaptic remodeling in alcoholism. Neuroscience letters. 2015;601:11–9. Epub 2015/01/28. 10.1016/j.neulet.2015.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu HT, Hsueh YP. Calcium influx and postsynaptic proteins coordinate the dendritic filopodium-spine transition. Developmental neurobiology. 2014;74(10):1011–29. Epub 2014/04/23. 10.1002/dneu.22181 . [DOI] [PubMed] [Google Scholar]
- 54.Stamatakou E, Hoyos-Flight M, Salinas PC. Wnt Signalling Promotes Actin Dynamics during Axon Remodelling through the Actin-Binding Protein Eps8. PloS one. 2015;10(8):e0134976 Epub 2015/08/08. 10.1371/journal.pone.0134976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen CH, He CW, Liao CP, Pan CL. A Wnt-planar polarity pathway instructs neurite branching by restricting F-actin assembly through endosomal signaling. PLoS genetics. 2017;13(4):e1006720 Epub 2017/04/07. 10.1371/journal.pgen.1006720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bjork K, Rimondini R, Hansson AC, Terasmaa A, Hyytia P, Heilig M, et al. Modulation of voluntary ethanol consumption by beta-arrestin 2. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22(7):2552–60. Epub 2008/03/28. 10.1096/fj.07-102442 . [DOI] [PubMed] [Google Scholar]
- 57.Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychological medicine. 1999;29(5):1069–81. Epub 1999/11/27. . [DOI] [PubMed] [Google Scholar]
- 58.Morean ME, Corbin WR. Subjective response to alcohol: a critical review of the literature. Alcoholism, clinical and experimental research. 2010;34(3):385–95. Epub 2009/12/24. 10.1111/j.1530-0277.2009.01103.x . [DOI] [PubMed] [Google Scholar]
- 59.Ray LA, Mackillop J, Monti PM. Subjective responses to alcohol consumption as endophenotypes: advancing behavioral genetics in etiological and treatment models of alcoholism. Substance use & misuse. 2010;45(11):1742–65. Epub 2010/07/02. 10.3109/10826084.2010.482427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature genetics. 2019;51(2):237–44. Epub 2019/01/16. 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, and Me Research Team tSUDWGotPGC, Adams MJ, et al. Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. The American journal of psychiatry. 2019;176(2):107–18. Epub 2018/10/20. 10.1176/appi.ajp.2018.18040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Empirical p-values were calculated from standardized module scores based on a Z-distribution. The original EW-dmGWAS module score, permutation, and score standardization algorithms were used to calculate the respective Mega Modules parameters. Modules were considered to have >80% overlap if >80% of the genes in the smaller module was contained in the larger module. False Discovery Rates were calculated based on the Benjamini-Hochberg algorithm, using the “stats” package in R. Intramodular connectivity was defined as the number of edges (i.e. connections) attached to that node (i.e. gene). Eigen-Centrality was calculated using the “igraph” package in R.
(PDF)
One table per brain region, containing each of the following values: RMA values and S-scores from the maximally expressed probeset per gene, for each BXD strain; the associated probeset IDs, human gene symbols, and mouse gene symbols; and the Fisher’s combined False Discovery Rate (q-value) for each probeset.
(XLSX)
One table per brain region, containing each of the following characteristics, for all significant Mega Modules: name; constituent genes; ALPSAC and IASPSAD p-values for each gene; Mega Module score (Sn), p-value (Sn_p), and False Discovery Rate (Sn_qFDR); and intramodular eigencentrality and connectivity. Significance values < 10−16 are rounded to 0.
(XLSX)
One table for each ALSPAC-overrepresented Mega Module, containing ToppFun output for gene ontology enrichment groups with p<0.01 and minimum group size of 3 genes and maximum size of 1,000 genes, for the following categories: Biological Process, Cellular Component, Molecular Function, Human Phenotype, Mouse Phenotype, and Pathways.
(XLSX)
Data Availability Statement
Most relevant data are contained within the paper and Supporting Information. Additional raw microarray data are found on GeneNetwork (www.genenetwork.org) under accession numbers GN135-137, GN154-156 and GN228-230 and in the Gene Expression Omnibus (GEO) database (accession number GSE28515). Primary human GWAS data is available from cited publications.