Abstract
The stoichiometry of a ligand binding reaction to a protein is given by a parameter (n). The value of this parameter may indicate the presence of protein monomer or dimers in the binding complex. Members of the lipocalin superfamily show variation in the stoichiometry of binding to ligands. In some cases the stoichiometry parameter (n) has been variously reported for the same protein as mono- and multimerization of the complex. Prime examples include retinol binding protein, β lactoglobulin and tear lipocalin, also called lipocalin-1(LCN1). Recent work demonstrated the stoichiometric ratio for ceramide:tear lipocalin varied (range n = 0.3–0.75) by several different methods. The structure of ceramide raises the intriguing possibility of a lipocalin dimer complex with each lipocalin molecule attached to one of the two alkyl chains of ceramide. The stoichiometry of the ceramide-tear lipocalin binding complex was explored in detail using size exclusion chromatography and time resolved fluorescence anisotropy. Both methods showed consistent results that tear lipocalin remains monomeric when bound to ceramide. Delipidation experiments suggest the most likely explanation is that the low ‘n’ values result from prior occupancy of the binding sites by native ligands. Lipocalins such as tear lipocalin that have numerous binding partners are particularly prone to an underestimated apparent stoichiometry parameter.
Keywords: tear lipocalin, lipocalin-1, LCN1, Lipid binding, ceramide retinol binding protein
1. Introduction
Members of the lipocalin superfamily are linked by structural homology. Eight antiparallel β-strands form a fold or calyx, which accommodate small hydrophobic ligands (Fig. 1). Accurate quantification of binding generally requires calculation of the stoichiometry binding parameter, ‘n’. In the analysis of binding studies for lipocalins, ‘n’ is often not reported [1–4]. In other reports the assumption is made that the association reaction is bimolecular and that the ligand binds to a single binding site, i.e., n = 1 [1,5–7]. This is a reasonable assumption in cases where the binding site has been well studied for the particular ligand. This may not be the case for heterogeneous ligands and binding sites especially in situations where multimeric complexes are formed. For example lipocalins are known to form higher order oligomers [8–10]. For apolipoprotein-D, crustacyanin and β lactoglobulin the oligomeric state has been linked to ligand binding functions [11,12]. Apolipoprotein-D was initially claimed as monomeric. Later dimerization was discovered when the protein binds lipids through conversion of Met 93 to methionine sulfoxide [13,14]. Lipocalins may form transient multimeric complexes induced by ligand binding, changes in pH, as well as increases in concentration [15–17]. The importance of an accurate stoichiometry parameter is evident in controversies over multimerization in the lipocalin family. β lactoglobulin and tear lipocalin (LCN1) have both been variably reported to exist as monomers and dimers in solution and crystal form [18–24]. Several authors noted that tear lipocalin (monomer mass = 17,446) [25] elutes in size exclusion chromatography at the apparent molecular mass for a dimer complex and even tetrameric complexes [21,26]. Gouveia and Tiffany suggested that apo-lipocalin was monomeric but holo-lipocalin was dimeric implying ligand induced dimer formation [22]. Later, multiangle laser light scattering and rotational time constants (measured by both fluorescence anisotropy and electron paramagnetic resonance) indicated that both holo- and apo-tear lipocalin generally exist as monomers with a small percentage of dimer [24]. Most published studies of ligand binding in the lipocalin family assume or show a stoichiometric parameter of one [1,5–7,27–36]. However in the case of tear lipocalin calculations may be lower. Redl et al. calculated 0.3 for the ratio of retinol to tear lipocalin at saturation [21]. A recent study with ceramide revealed stoichiometric ratios of TL:ceramide ligand from n = 0.32–0.67, accompanied by a very low dissociation constant [37]. Several explanations are possible including the formation of a multimeric complex with 2 lipocalin molecules flanking the 2 alkyl chains of the ceramide or a binding site pre-occupied by other ligands. The current study sought an explanation for the low ‘n’ in this case with an overall analysis of the lipocalin family.
Fig. 1.
Cartoon of the backbone structure of tear lipocalin showing the 8 antiparallel β strands that form the lipocalin fold. Orange arrow indicates cavity entrance. Residue 9 from the N terminus was chosen for the cysteine substitution because of its exposed position. (modified from published solution structure of tear lipocalin) [42].
2. Materials and Methods
2.1. Reagents
C18-ceramide (ceramide- N-(octadecanoyl) sphing-4-enine, also known as N-Stearoyl-D-erythro-sphingosine), was obtained from ACROS Organic (Pittsburg, PA). C6-NBD ceramide (N-[6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-sphingosine) and C12-NBD ceramide (N-[12-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino] dodecanoyl]-D-erythro-sphingosine) were obtained from Avanti Polar Lipids (Alabaster, AL). 4-Chloro-7-nitrobenzofurazan (NBD–Cl) and dimethyl sulfoxide (DMSO) were obtained from MilliporeSigma (St.Louis, MO).
2.2. Collection of Human Tears
Stimulated human tears were collected from healthy volunteers in accordance with the tenets of the Declaration of Helsinki and approved by the institutional review board. Informed consent was obtained from donors after explanation of the nature and possible consequences. Collection was performed as previously published [38–40] with polished glass tips and glass transfer pipettes. Tears were pooled in polytetrafluoroethylene-lined glass vials and stored under nitrogen at −80 °C until use.
2.3. Size-exclusion chromatography.
Fractionations of proteins, including tears, synthetic dimers and ligand binding complexes, were performed by gel filtration liquid chromatography using the AKTA purifier Versatile FPLC with a HiLoad Superdex 75 column (GE Healthcare, Piscataway, NJ). In general, one ml column fractions were collected with elution in 50 mM Tris-HCl, 100 mM NaCl pH 8.4 run at 1 ml/min as previously described [37]. For purification of tear lipocalin monomers peak fractions from gel filtration of 1 ml of tears were pooled and applied to an anion exchange column using DEAE-Sephadex A-25. Purity was confirmed by tricine polyacrylamide gel electrophoresis as published [40,41]. The appropriate absorbance wavelengths for NBD (480 nm) and protein (280 nm) were monitored simultaneously for analyzing NBD-derivatized proteins and NBD ligand complexes as necessary.
2.3. Expression of tear lipocalin and construction of tear lipocalin dimer
Tear lipocalin, wild type, was expressed and purified as previously described [23,25]. The free cysteine at residue position 101 in native tear lipocalin is buried in the G strand and is relatively inaccessible to chemicals for derivatization [25,42]. Tear lipocalin was altered by site directed mutagenesis to create a single free cysteine in the exposed portion (amino acid position 9) of the A strand (Fig. 1). The cysteine at position 101, was replaced with leucine as previously published [25]. A dimer of tear lipocalin was constructed by crosslinking the cysteine substituted monomers with 1,11-bismaleimido-triethyleneglycol, BM (PEG)3, (Thermo Scientific™ Pierce) according to the manufacturer’s protocol. The product was a cysteine linked three unit polyethylene glycol spacer arm joining two tear lipocalin molecules. The reaction in brief included TL, 0.3 mM final concentration, incubated for 16 h 37 °C with a three fold molar excess of BM(PEG)3, 0.9 mM, final concentration in 10 mM Na–P, 130 mM NaCl, 5 mM EDTA pH 7.0. Formation of the dimer was confirmed with tricine gel electrophoresis stained with Coomassie blue followed by silver nitrate as previously described [40]. The crosslinker added 352 Da of net mass to the two monomers. Fractions, containing dimer were combined, concentrated on 10,000 MW cutoff centrifugal membrane filter by centrifugation at 14,000 g for 30 min.
2.4. Tear lipocalin labeling with NBD
Recent studies have indicate that at pH 7.0 NBD-Cl preferentially labels the N terminus of proteins [43]. In an attempt to verify specific labeling, NBD-Cl was reacted with both monomeric and dimeric tear lipocalin. A 900 ul reaction mixture comprised of tear lipocalin (10 μM) and NBD-Cl (100 μM) was incubated in 10 mM Na–P buffer at pH 8.0 overnite at 37 °C as previously described for NBD labeling of proteins [44]. In addition, identical reactions were run at pH 6.2 in sodium citrate buffer, pH 7.0 in sodium phosphate buffer. The reaction mixture was concentrated with a centrifugal membrane filter (Amicon 10 m 10,000 MW cutoff) and washed with 5 × 500 μl of the respective buffers. The final retentate was adjusted to 800 μl in their respective buffers. Labeling efficiency was estimated from absorbance spectrophotometry using the molar absorptivity, ε, for NBD-lysyl residues and TL at λ = 480 and λ = 280 nm as 26,000 and 13,760 M-1 cm-1 respectively [37,45,46]. The molar ratio of NBD label/tear lipocalin was multiplied by 100 to determine percent efficiency. The dimer was considered to have 2 fold the moles of tear lipocalin..
2.5. Delipidation of tear lipocalin
Lipids were extracted from purified native tear lipocalin (0.8 ml, 5 mg/ml) using chloroform/methanol extraction as previous published [41]. C6-NBD labeled ceramide bound to tear lipocalin bound was extracted in parallel and served as a control. Delipidation was confirmed by the disappearance of fluorescence at 523 nm in the control sample (see Section 2.7.1). Two extractions in series were required..
2.6. Spectroscopy
2.6.1. Absorption spectrophotometry
Absorption spectrophotometric measurements were obtained with a Shimadzu UV-2401PC instrument, (Kyoto, Japan). The concentrations of proteins and fluorescent lipids were calculated from molar extinction coefficients that have been published [37].
2.6.2. Steady State Fluorescence
Fluorescence measurements were made at 20 °C in a Jobin Yvon-SPEX Fluorolog-3 spectrofluorimeter (Jobin Yvon, Edison, NJ); band-widths for excitation and emission were 2 and 4 nm, respectively. For NBD measurements, the excitation wavelength was 465 nm. Raman and background scattering by the solvent were corrected where necessary using appropriate blank solutions. Delipidated tear lipocalin, (0.5 ml) 1 μM in 10 mM sodium phosphate, pH 7.0, was titrated with 0.4 μl increments of 1 mM C6-NBD ceramide in DMSO to a final concentration 2.4 μM. The final concentration of DMSO did not exceed 1%. The binding was assessed by fluorescence of the binding complex. The accumulated means at each point from at least 5 measurements were used to construct the emission spectra. Fluorescence intensities at 523 nm were fit to the hyperbola curve and Hill equation (Microcal Origin, Northhampton, MA, USA) as previously described.11
2.6.3. Fluorescence anisotropy experiments
In general, for anisotropy experiments, an excess of the unlabeled reagent was used to avoid the unbound labeled component in fitting curves. NBD labeled tear lipocalin was incubated with a 10 M excess of C18-ceramide ligand in DMSO. Alternatively, NBD labeled lipid was incubated with an approximate 3 fold excess of tear lipocalin. Fluorescence decays were measured with a time-resolved fluorescence spectrometer, FluoTime 100 (PicoQuant, Berlin, Germany), using a time-correlated single-photon counting scheme. Samples were excited at 470 nm by a subnanosecond pulsed diode laser, LDH D-C 470 (PicoQuant, Berlin, Germany), with a repetition rate of 10 MHz. The excitation path included a 475 nm short pass filter (Tech Spec Edmund Optics) and rotatable linear polarizer. An emission OG-530 nm longpass filter and second rotatable linear polarizer and detector were oriented 90 degrees from the excitation path. The fluorescence intensity decay was analyzed using FluoFit iterative-fitting software based on the Marquardt algorithm (PicoQuant, Berlin, Germany). Anisotropic decay (r(t)) was measured using the formula:
where VV(t) and VH (t) are the parallel and perpendicular polarized time decays respectively. The G factor, is the instrument responsive function/wavelength correction factor and was measured to compensate for polarization bias with the excitation polarizer set in the horizontal position. The emission polarizer was set at the magic angle to measure lifetime intensity decays. The rotational correlation time (ϕ) was derived from the fluorescence anisotropic decay r(t) of a sphere by:
r(t) = roe−t/ϕ where ro is the anisotropy at time (t) 0. Fitting of anisotropic decays was made using both a reconvolution fit and modified tailfit. The modified tailfit involved adjusting range to avoid the short lifetimes and the instrument response functions. The tailfit was simpler, involved fitting to fewer parameters, and is discussed in more detail elsewhere (MethodsX, submitted). Fluofit software adds parameters for channel shifts both parallel and perpendicular as well as scatter if necessary.
2.7. Stoichiometric analysis of lipocalin binding curves
The literature was reviewed to analyze the stoichiometry of lipocalins and ligands. The stoichiometric binding coefficient ‘n’ was categorized as: reported, not reported, not available and assumed. For cases where ‘n’ was not reported, determination was possible from simple analysis of the protein and ligand concentrations reported at saturation or by digitizing the given binding curves and/or Scatchard plots. Scatchard plots were fit by linear regression using least squares analysis. Digitized binding curves were analyzed as described for binding curves in Section 2.7.2.
3. Results and Discussion
3.1. Construction of covalent dimer of tear lipocalin
A covalent dimer of tear lipocalin served as a positive control in size exclusion chromatography, gel electrophoresis and anisotropy experiments to evaluate potential multimer formation of tear lipocalin upon binding ceramides. The amino acid residue 9, glutamic acid was chosen for substitution with cysteine for 2 reasons. First, residue 9 was shown to be accessible to solvent in site directed tryptophan fluorescence studies. Solvent exposure facilitates chemical reactivity for dimerization. Second, residue 9 is flexible and situated away from the primary amine at the N terminus. NBD labeling would not be hindered by crosslinking at residue 9 to form a dimer (Fig. 1). After crosslinking the C9 mutant of tear lipocalin with bis, the FPLC profile showed a new peak at 55 min ahead of the peak of the monomer, at 67 min (Fig. 2). Pooled fractions from the presumed oligomeric elution peak were reapplied to the size exclusion FPLC column and eluted at the same time. The later eluting peak was much smaller peak indicating relative dimer enrichment (Fig. 2). Silver nitrate stained tricine electrophoresis gels of the fractions showed adequate separation of monomeric and dimeric fractions. A faint monomeric band was present in the dimer fractions (Fig. 2 inset) but the enrichment of dimer was adequate to serve as a control. Circular dichroism and binding studies were also performed to demonstrate that the dimeric construct was functionally similar to monomeric tear lipocalin (see MethodsX, submitted).
Fig. 2.
Size exclusion chromatography showing absorbance at λ =280 nm after loading the sample from tear lipocalin dimer synthesis. Two peaks appear over fractions 52–57 (dimer) and fractions 61–72 (monomer). Both the dimer (small peak) and the monomer (large peak) are evident in the samples after cross linking (black solid line). The fractions containing enriched dimer from pooling the peaks at 53–59 show both a large peak of dimer and small monomer peak. Inset: Silver nitrate stained SDS tricine gel after electrophoresis of fractions in the FPLC. Samples in lanes: 1, sample from pooled fractions 62–70; 2, pooled fractions 52–57; 3, human tears 1 μl; 4, molecular weight markers.
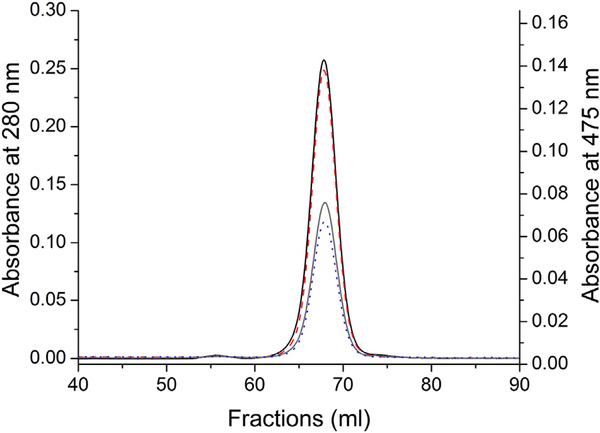
3.2. Size Exclusion Gel Chromatography (FPLC) of C6 and C12 Ceramidetear lipocalin complexes
Mixtures of native tear lipocalin and C6 and C12 ceramides showed identical elution times to that of known monomeric lipocalin in size exclusion chromatography (Fig. 3). Prior size exclusion chromatography data from 2 groups reported that native tear lipocalin elutes at an apparent mass of 35–36kDA and 45 kDa, respectively [21,26]. In the current study, the covalent dimeric construct of tear lipocalin elutes prior to the native form (Fig. 2). Prior studies combining size exclusion chromatography and multiangle laser light scattering interrogated dimer formation for native and recombinant tear lipocalin [23]. Not-withstanding the small shoulders of dimer formation most evident in the recombinant form, Debye plots verified that the central chromatographic peaks reflected monomers.23 In the present study the elution times of native tear lipocalin and the TL-ceramide binding complex are identical and appear at the same elution time of prior studies. A significant ligand induced non-covalent aggregation is effectively excluded because the monomer is discretely separated from the synthetic dimer (Figs. 2 and 3). Column matrix interactions and/or chromatographic conditions may play a role in the aforementioned apparent altered elution time in prior publications [21,26]. Tear lipocalin remains a monomer when bound to ceramide.
Fig. 3.
Size exclusion chromatography after ligand binding of purified tear lipocalin and NBD-C6 and –C12 ceramides. Traces: solid lines black and grey are absorbance at λ = 280 nm; dashed orange and grey are absorbance at λ = 480nm.
3.3. NBD labeling of tear lipocalin
NBD-Cl is a robust reagent for labeling proteins because several amino acids may be labeled and result in a potentially greater fluorescent signal. A possible method for a high yield specific and rapid N terminal labeling of proteins has recently emerged [43]. N terminal labeling accrues potential advantages over a more specific sulhydryl adduct for tear lipocalin. The latter require a double mutation whereas no mutations are required for the native protein. For sulfhydryl labeling in tear lipocalin the buried cysteine at position 101 must be replaced and a new cysteine must be positioned in a solvent exposed site in order to facilitate chemical reactivity [25]. Attempts at NBD labeling of tear lipocalin at all pH’s revealed a lower than expected labeling efficiency ranging from 20 to 25%. The low labeling efficiency may in part be due to the number of buried residues. However this value is only approximate since only the molar absorption coefficient for NBD lysyl adducts are available. The ε for adducts of tyrosine, cysteine and the N terminal amine are unknown. The UV–Vis absorbance spectra of NBD labeled tear lipocalin at various pH values, 6.2, 7.0 and 8.0 showed a broad peaks of absorbance with maxima at about 450, 453, and 460 nm respectively. The constructed tear lipocalin dimer labeled at pH 8 showed a broad peak at 470 nm spectral (MethodsX, submitted). This latter peak has been described for other proteins labeled with NBD [47,48]. Primary and secondary amines have absorption maxima at 465 and 485 nm. Houk demonstrated that cysteine of a protein may be labeled at acidic pH while lysines are preferentially labeled above neutrality. [49] NBD adducts of cysteine and tyrosine produce absorption maxima at 425 nm. NBD lysine adducts show a peak at about 480 nm. Native tear lipocalin sports eleven lysines, five tyrosines and one free cysteine residues. Our absorbance maxima are broad and intermediate between that of lysine and other residues suggesting a mixture of amino acid labeling. However, the increase in absorption maxima at higher pH is consistent with some preference for lysine labels at pH 8.0. All 5 tyrosine residues and the native cysteine are buried in tear lipocalin and relatively inaccessible for labeling [42]. There is no free cysteine in the constructed dimer. Lysines are all located in solvent exposed sites available to react with NBD–Cl. Although we cannot exclude concomitant N terminal labeling, the labeling process resulted in broad spectra pH 6.2, 7.0 and 8.0. and with the dimer that do not match the expected absorbance of selective N terminal labeling under any condition in tear lipocalin. These results contrast with those of Bernal et al. in which N terminal amines of the Z-domain, a small protein derived from staphylococcal protein A, were selectively reacted at pH 7.0 whereas lysines, tyrosines and cysteines were not reactive [43]. The results from time resolved fluorescence are consistent with multiple NBD labeled adducts in tear lipocalin under all conditions tested (Section 3.4 and MethodsX, submitted).
3.4. Time resolved fluorescence spectroscopy
Time resolved fluorescence lifetimes reveal information about the molecular environment of the fluorophore (Table 1, Fig. 4a). The fluorescence lifetime decay for NBD labeled TL (Table 1) fits two exponentials of about 10.6 and 4.3 ns with fractional intensities of 60 and 40% respectively. This suggests that the NBD labels reside in 2 distinct environments on tear lipocalin. Similarly fit experimental lifetimes were obtained at all conditions of NBD labeling including native tear lipocalin at various pH’s and synthetic dimer labeling at pH 8.0 (MethodsX, submitted). Several explanations are possible for 2 lifetimes including 2 different labeling sites, rotameric configurations, differential interactions with solvent and other residues. The uniformity of the data is somewhat surprising considering the number of available labeling sites. The rotational correlation time is related to the average angular displacement of the NBD labeled molecule that occurs during the time of fluorescence emission of a photon or the time resolved fluorescence anisotropy. The rate of rotational diffusion depends on the size and shape of the molecule so that a dimer is expected to have about a 2 fold increase in the correlation time. NBD labeled tear lipocalin shows a single correlation time of about 10 ns, reassuring for a monomer (Table 1). The experiment interrogating the molecular interactions of C18 ceramide, a lipid found in tears, complexed with NBD labeled tear lipocalin is shown in Fig. 4. Inspection of Fig. 4 reveals that tail matching was possible as the anisotropic decay was faster than the intensity decay. As a result there was no residual anisotropy and the longer rotational correlation time of the entire molecule is captured in the time of the decay. The fluorescence lifetimes and rotational correlation times are comparable to that for NBD-tear lipocalin without a ligand (Table 1, Fig. 4b). The time dependent anisotropy decay Fig. 4b fit reasonably well to a single rotational correlation time of about 9.5 ns. This correlation time matches that found for monomeric tear lipocalin in prior experiments where a dansyl label was placed at position 99, a specific and buried site [23]. In a separate experiment, anisotropic decays of NBD-tear lipocalin were carefully measured at the same acquisition times before and after C18 ceramide was added (Fig. 5). No difference was seen in vertical and horizontal decays, both of which had a correlation time of about 9.5 ns. The NBD labeled control cysteine dimer of tear lipocalin showed a correlation time of 19 ns (Table 1 and MethodsX, submitted). The rotational correlation time of a molecule is linearly related to the mass of the protein. The correlation time of the control dimer is twice that of the known monomer and the NBD-tear lipocalin C18 ceramide complex providing further evidence that tear lipocalin remain monomeric.
Table 1.
Fluorescence lifetimes and correlation times for tear lipocalin-NBD and ceramides.
| Binding components | τ1(ns)/fi | τ2(ns)/fi | ϕ(ns)/Ro |
|---|---|---|---|
| NBD-TL + C18 ceramide | 9.6/0.55 | 3.3/0.45 | 9.5/100 |
| NBD-TL | 10.6/0.6 | 4.3/0.50 | 9.0/100 |
| NBD-TL dimer (synthetic) | 10.4 | 4.3 | 19.3(100) |
fi = fractional intensity of positive decay component.
ϕ = rotational correlation time.
Ro= Percent of positive anisotropy component.
Fig. 4.
Time resolved decays and fitting curves for NBD-tear lipocalin (14 μm) and C18 ceramide (28 μM) in 10 mM sodium phosphate buffer pH 7. Dark green- vertically polarized emission decay curve (VV). Light green= horizontally polarized emission decay curve (VH). Inset shows anisotropic decay with fitting curve and residuals (lower panel), χ2 = 0.999.
Fig. 5.
Time resolved decays for NBD-tear lipocalin, 21 μM before (colored curves) and after (black dashed curves) addition of concentrated C18 ceramide, final concentration of 50 μM. The decays overlap precisely. The vertically polarized emission decays are green and large black dashes above in the figure. Horizontally polarized emissions are yellow and fine black dots, respectively, in the lower part of the figure.
4. Stoichiometry of other lipocalin ligand binding curves
The lack of oligomeric complex formation for lipocalin and ceramide despite stoichiometric parameters < 0.5 prompted investigation of published works of other lipocalins. Of the 60 lipocalin-ligand binding reactions surveyed less than half directly measured the stoichiometric binding parameter [1,21,28,29,31,32,34–36,51–53]. In 24 cases n was assumed to be one and the mathematical analysis of Kd was made under that assumption [5–7]. In 8 instances, the stoichiometry was not reported.1–4 In deference to the authors, where n = 1 was assumed, there were sound biochemical reasons for the assumption. Some other studies preceded readily available software for curve fitting by least squares analysis. Our concern focuses on instances when the n was calculated. For lipocalins in which the stochiometric parameter was measured, ‘n’ was generally only slightly less than one. α2μ-globulin, retinal binding protein, and tear lipocalin are clearly exceptions [21,37,52,54]. The ligand to protein ratio at saturation for 2,4,4-trimethyl-2-pentanol:α2μ-globulin was reported to be 0.01. Binding stoichiometries close to 0.5 can be computed from graphs provided for several ligands of retinol binding protein [1,3]. An example is shown in Fig. 6. In this case the original tryptophan quenching curve was replotted in a form amenable to fitting [1]. The Scatchard plot could be fit by linear least squares to determine that n = 0.5, originally not reported. One can speculate the binding site of retinol binding protein could be pre-occupied with its native ligand. However, the binding study was not preceded by delipidation. Redl reported a binding ratio of retinol to tear lipocalin of 0.3 at saturation (n = 0.3) [21]. These results are similar to ours with ceramides and tear lipocalin (Table 2) [37]. However, the size exclusion chromatography gel filtration and fluorescence anisotropy confirm a monomeric complex of tear lipocalin and ceramide. It is evident that the calculated ‘n’ may not reflect the true stoichiometric ratio of protein and ligand for some lipocalins.
Fig. 6.
Fitting of binding curve from replotted data for human apo-retinol binding protein and retinoic acid [1]. The original data (black solid circles) replotted from quenching of tryptophan fluorescence but now in the form of a binding curve. The published Kd =0.21 μM, n not reported. Curves: Black solid, best fit by non linear least squares analysis (Kd =0.92 μM, n = 0.51); Blue dashed, is the fitting done using published Kd =0.21 μM as a fixed parameter, then n =0.34; Red dashed, uses n =0.55 as a fixed parameter which was calculated from linear least squares plot of published data (inset), then Kd = 1.08 μM. Inset: Linear least square plot of published titration data, Po = concentration of protein, Ro =concentration of total ligand, α = fraction of free binding sites of protein molecule.
Table 2.
Stoichiometric Parameters for binding of tear lipocalin with C6-NBD labeled ceramide.
5. Influence of delipidation on assessment of the stoichiometric binding parameters
The source of the disparities may be that ligands tenaciously occupy the calyx. Pre-occupation of the binding sites with native ligands may reduce capacity for additional ligand and lower the apparent ‘n’. Complete removal of these ligands may be extremely difficult or impossible. Among lipocalins, tear lipocalin is known to be particularly promiscuous i.e. having multiple ligand binding partners. To determine if pre-occupation of the binding site by other ligands was responsible for the apparent low stoichiometric parameter ‘n’, tear lipocalin was delipidated. Two rounds of extraction with chloroform and methanol were necessary to remove NBD labeled ceramide from the control tear lipocalin complex. The resulting delipidated tear lipocalin was then reused to study binding of NBD-6 ceramide. The ‘n’ increased 2 fold compared that of native purified tear lipocalin and the Kd was reduced (Fig. 7, Table 2). Recombinant tear lipocalin has been taken as a surrogate for the apo-form without exposure to solvents for delipidation [19,55]. During expression tear lipocalin binds some lipids, but fewer than native tear lipocalin [56,57]. The fluorescence binding curve demonstrates that the n is intermediate between the native and delipidated native form (Fig. 7 and Table 2). These data provide further support for the notion that pre-occupation by a native ligand accounts for the disparity of the ‘n’ to the actual stoichiometry. Further, the data exclude solvents as the cause of the change in the stoichiometric parameter.
Fig. 7.
Binding curves of C6 NBD ceramide added to native tear lipocalin before (black solid circles) and after (open circles) delipidation as well as to recombinant tear lipocalin (open triangles). Red dashed lines- fit to Hill equation), grey lines- fit to hyperbola. Binding parameters are given in Table 2.
6. Conclusion
The stochiometric parameter ‘n’ must be interpreted with caution for promiscuous binding proteins and especially for lipocalins. Underestimation of the parameter may be related to pre-occupation of the binding site with other ligands. Delipidation can improve the analysis but other methods may be necessary to confirm or exclude the formation of a multimeric binding complex.
Acknowledgements
Funding: This work was supported by the National Eye Institute [EY11224] BG and the Edith and Lew Wasserman Professorship (BG).
References
- [1].Cogan U, Kopelman M, Mokady S, Shinitzky M, Binding affinities of retinol and related compounds to retinol binding proteins, Eur. J. Biochem 65 (1976) 71–78. [DOI] [PubMed] [Google Scholar]
- [2].Zanotti G, Malpeli G, Berni R, The interaction of N-ethyl retinamide with plasma retinol-binding protein (RBP) and the crystal structure of the retinoid-RBP complex at 1.9-A resolution, J. Biol. Chem. 268 (1993) 24873–24879. [DOI] [PubMed] [Google Scholar]
- [3].Noy N, Blaner WS, Interactions of retinol with binding proteins: studies with rat cellular retinol-binding protein and with rat retinol-binding protein, Biochemistry 30 (1991) 6380–6386. [DOI] [PubMed] [Google Scholar]
- [4].Müller HN, Skerra A, Functional Expression of the Uncomplexed Serum Retinol-binding Protein in Escherichia coli, J. Mol. Biol. 230 (1993) 725–732, 10.1006/jmbi.1993.1194. [DOI] [PubMed] [Google Scholar]
- [5].Breustedt DA, Schönfeld DL, Skerra A, Comparative ligand-binding analysis of ten human lipocalins, Biochim. Biophys. Acta 1764 (2006) 161–173, 10.1016/j.bbapap.2005.12.006. [DOI] [PubMed] [Google Scholar]
- [6].Vogt M, Skerra A, Bacterially produced apolipoprotein D binds progesterone and arachidonic acid, but not bilirubin or E-3M2H, J. Mol. Recognit 14 (2001) 79–86, . [DOI] [PubMed] [Google Scholar]
- [7].Narayan M, Berliner LJ, Mapping fatty acid binding to beta-lactoglobulin: Ligand binding is restricted by modification of Cys 121, Protein Sci. 7 (1998) 150–157, 10.1002/pro.5560070116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huber R, Schneider M, Mayr I, Müller R, Deutzmann R, Suter F, Zuber H, Falk H, Kayser H, Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution, J. Mol. Biol 198 (1987) 499–513. [DOI] [PubMed] [Google Scholar]
- [9].Sanchez D, Ortega-Cubero S, Åkerström B, Herrera M, Bastiani MJ, Ganfornina MD, Molecular interactions of the neuronal GPI-anchored lipocalin Lazarillo, J. Mol. Recognit 21 (2008) 313–323, 10.1002/jmr.902. [DOI] [PubMed] [Google Scholar]
- [10].Ghosh S, Yu C-L, Ferraro DJ, Sudha S, Pal SK, Schaefer WF, Gibson DT, Ramaswamy S, Blue protein with red fluorescence, Proc. Natl. Acad. Sci. U. S. A 113 (2016) 11513–11518, 10.1073/pnas.1525622113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gamiz-Hernandez AP, Angelova IN, Send R, Sundholm D, Kaila VRI, Protein-Induced Color Shift of Carotenoids in β-Crustacyanin, Angew. Chemie Int. Ed 54 (2015) 11564–11566, 10.1002/anie.201501609. [DOI] [PubMed] [Google Scholar]
- [12].Gutiérrez-Magdaleno G, Bello M, Portillo-Téllez MC, Rodríguez-Romero A, García-Hernández E, Ligand binding and self-association cooperativity of β-lacto-globulin, J. Mol. Recognit 26 (2013) 67–75, 10.1002/jmr.2249. [DOI] [PubMed] [Google Scholar]
- [13].Bhatia S, Knoch B, Wong J, Kim WS, Else PL, Oakley AJ, Garner B, Selective reduction of hydroperoxyeicosatetraenoic acids to their hydroxy derivatives by apolipoprotein D: implications for lipid antioxidant activity and Alzheimer’s disease, Biochem. J 442 (2012) 713–721, 10.1042/BJ20111166. [DOI] [PubMed] [Google Scholar]
- [14].Oakley AJ, Bhatia S, Ecroyd H, Garner B, Molecular dynamics analysis of apo-lipoprotein-D - lipid hydroperoxide interactions: Mechanism for selective oxidation of Met-93, PLoS One 7 (2012) e34057,, 10.1371/journal.pone.0034057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qin BY, Jameson GB, Bewley MC, Baker EN, Creamer LK, Functional implications of structural differences between variants A and B of bovine β-lacto-globulin, Protein Sci. 8 (2008) 75–83, 10.1110/ps.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hallin EI, Hasan M, Guo K, Åkerlund H-E, Molecular studies on structural changes and oligomerisation of violaxanthin de-epoxidase associated with the pH-dependent activation, Photosynth. Res 129 (2016) 29–41, 10.1007/s11120-016-0261-y. [DOI] [PubMed] [Google Scholar]
- [17].Niemi MH, Rytkönen-Nissinen M, Miettinen I, Jänis J, Virtanen T, Rouvinen J, Dimerization of lipocalin allergens, Nat. Publ. Gr (2015), 10.1038/srep13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qin BY, Bewley MC, Creamer LK, Baker HM, Baker EN, Jameson GB, Structural basis of the Tanford transition of bovine β-lactoglobulin, Biochemistry 34(1998) 14014–23. 10.1021/BI981016T. [DOI] [PubMed] [Google Scholar]
- [19].Breustedt DA, Korndörfer IP, Redl B, Skerra A, The 1.8-A crystal structure of human tear lipocalin reveals an extended branched cavity with capacity for multiple ligands, J. Biol. Chem. 280 (2005) 484–493, 10.1074/jbc.M410466200. [DOI] [PubMed] [Google Scholar]
- [20].Breustedt DA, Chatwell L, Skerra A, A new crystal form of human tear lipocalin reveals high flexibility in the loop region and induced fit in the ligand cavity, Acta Crystallogr. D. Biol. Crystallogr 65 (2009) 1118–1125, 10.1107/S0907444909031011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Redl B, Holzfeind P, Lottspeich F, cDNA cloning and sequencing reveals human tear prealbumin to be a member of the lipophilic-ligand carrier protein superfamily, J. Biol. Chem 267 (1992) 20282–20287. [PubMed] [Google Scholar]
- [22].Gouveia SM, Tiffany JM, Human tear viscosity: An interactive role for proteins and lipids, Biochim. Biophys. Acta - Proteins Proteomics. 1753 (2005) 155–163, 10.1016/J.BBAPAP.2005.08.023. [DOI] [PubMed] [Google Scholar]
- [23].Gasymov OK, Abduragimov AR, Merschak P, Redl B, Glasgow BJ, Oligomeric state of lipocalin-1 (LCN1) by multiangle laser light scattering and fluorescence anisotropy decay, Biochim. Biophys. Acta 1774 (2007) 1307–1315, 10.1016/j.bbapap.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Glasgow BJ, Gasymov OK, Abduragimov AR, Yusifov TN, Altenbach C, Hubbell WL, Side chain mobility and ligand interactions of the G strand of tear lipocalins by site-directed spin labeling, Biochemistry 38 (1999) 13707–13716. [DOI] [PubMed] [Google Scholar]
- [25].Glasgow BJ, Abduragimov AR, Yusifov TN, Gasymov OK, Horwitz J, Hubbell WL, Faull KF, A conserved disulfide motif in human tear lipocalins influences ligand binding, Biochemistry 37 (1998) 2215–2225, 10.1021/bi9720888. [DOI] [PubMed] [Google Scholar]
- [26].Glasgow BJ, Marshall G, Gasymov OK, Abduragimov AR, Yusifov TN, Knobler CM, Tear lipocalins: potential lipid scavengers for the corneal surface., Invest. Ophthalmol. Vis. Sci 40 (1999) 3100–7. [PubMed] [Google Scholar]
- [27].Goessling W, Zucker SD, Role of apolipoprotein D in the transport of bilirubin in plasma, Am. J. Physiol. Liver Physiol 279 (2000) G356–G365, 10.1152/ajpgi.2000.279.2.G356. [DOI] [PubMed] [Google Scholar]
- [28].Briand L, Eloit C, Nespoulous C, Bézirard V, Huet J-C, Henry C, Blon F, Trotier D, Pernollet J-C, Evidence of an odorant-binding protein in the human olfactory lucus: Location, structural characterization, and odorant-binding properties, Biochemistry 41 (2002) 7241–7252, 10.1021/bi015916c. [DOI] [PubMed] [Google Scholar]
- [29].Whitson KWS, Human odorant binding protein 2a has two affinity states and is capable of binding some uremic toxins, Biochem. Anal. Biochem 3 (2014), 10.4172/2161-1009.1000156. [DOI] [Google Scholar]
- [30].Larsson J, Allhorn M, Åkerström B, The lipocalin α1-microglobulin binds heme in different species, Arch. Biochem. Biophys 432 (2004) 196–204, 10.1016/j.abb.2004.09.021. [DOI] [PubMed] [Google Scholar]
- [31].Tanaka T, Urade Y, Kimura H, Eguchi N, Nishikawa A, Hayaishi O, Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter, J. Biol. Chem 272 (1997) 15789–15795. [DOI] [PubMed] [Google Scholar]
- [32].Beuckmann CT, Masaaki Aoyagi AM, Okazaki I, Hiroike T, Toh H, and Hayaishi O, Y. Urade, Binding of biliverdin, bilirubin, and thyroid hormones to lipocalin-type prostaglandin D synthase, Biochemistry 38 (1999) 8006–13 10.1021/BI990261P. [DOI] [PubMed] [Google Scholar]
- [33].Saville JT, Zhao Z, Willcox MDP, Blanksby SJ, Mitchell TW, Detection and quantification of tear phospholipids and cholesterol in contact lens deposits: the effect of contact lens material and lens care solution, Invest. Ophthalmol. Vis. Sci 51 (2010) 2843–2851, 10.1167/iovs.09-4609. [DOI] [PubMed] [Google Scholar]
- [34].Cho Y, Batt CA, Sawyer L, Probing the retinol-binding site of bovine beta-lactoglobulin, J. Biol. Chem 269 (1994) 11102–7. [PubMed] [Google Scholar]
- [35].Dufour E, Bertrand-Harb C, Haertlé T, Reversible effects of medium dielectric constant on structural transformation of β-lactoglobulin and its retinol binding, Biopolymers 33 (1993) 589–598, 10.1002/bip.360330408. [DOI] [PubMed] [Google Scholar]
- [36].Sangamnatdej S, Paesen GC, Slovak M, Nuttall PA, A high affinity serotoninand histamine-binding lipocalin from tick saliva, Insect Mol. Biol 11 (2002) 79–86. [DOI] [PubMed] [Google Scholar]
- [37].Glasgow BJ, Abduragimov AR, Interaction of ceramides and tear lipocalin, Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1863 (2018) 399–408, 10.1016/J.BBALIP.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dean AW, Glasgow BJ, Mass spectrometric identification of phospholipids in human tears and tear lipocalin, Invest. Ophthalmol. Vis. Sci 53 (2012) 1773–1782, 10.1167/iovs.11-9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Glasgow BJ, Gasymov OK, Abduragimov AR, Engle JJ, Casey RC, Tear lipocalin captures exogenous lipid from abnormal corneal surfaces, Invest. Ophthalmol. Vis. Sci 51 (2010) 1981–1987, 10.1167/iovs.09-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Glasgow BJ, Tissue expression of lipocalins in human lacrimal and von Ebner’s glands: colocalization with lysozyme, Graefe’s Arch, Clin. Exp. Ophthalmol. = Albr. von Graefes Arch. Für Klin. Und Exp. Ophthalmol 233 (1995) 513–522. [DOI] [PubMed] [Google Scholar]
- [41].Glasgow BJ, Abduragimov AR, Farahbakhsh ZT, Faull KF, Hubbell WL, Tear lipocalins bind a broad array of lipid ligands, Curr. Eye Res 14 (1995) 363–372. [DOI] [PubMed] [Google Scholar]
- [42].Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ, Site-directed tryptophan fluorescence reveals the solution structure of tear lipocalin: evidence for features that confer promiscuity in ligand binding, Biochemistry 40 (2001) 14754–14762. [DOI] [PubMed] [Google Scholar]
- [43].Bernal-Perez LF, Prokai L, Ryu Y, Selective N-terminal fluorescent labeling of proteins using 4-chloro-7-nitrobenzofurazan: A method to distinguish protein N-terminal acetylation, Anal. Biochem 428 (2012) 13–15, 10.1016/j.ab.2012.05.026. [DOI] [PubMed] [Google Scholar]
- [44].Poole LB, Measurement of Protein Sulfenic Acid Content, in: Curr. Protoc. Toxicol., John Wiley & Sons, Inc., Hoboken, NJ, USA, 2008: p. 17.2.1–17.2.27. doi: doi.org/10.1002/0471140856.tx1702s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Aboderin AA, Boedefeld E, Luisi PL, Reaction of chicken egg white lysozyme with 7-chloro-4-nitrobenz-2-oxa-1,3-diazole, Biochim. Biophys. Acta - Protein Struct 328 (1973) 20–30, 10.1016/0005-2795(73)90325-5. [DOI] [PubMed] [Google Scholar]
- [46].Allegrini PR, Sigrist H, Schaller J, Zahler P, Site-Directed Fluorogenic Modification of Bacteriophodopsin by 7-Chloro-4-nitrobenz-2-oxa-1,3-diazole, Eur. J. Biochem 132 (1983) 603–608, 10.1111/j.1432-1033.1983.tb07406.x. [DOI] [PubMed] [Google Scholar]
- [47].Gonçalves MST, Fluorescent Labeling of Biomolecules with Organic Probes, Chem. Rev 109 (2009) 190–212, 10.1021/cr0783840. [DOI] [PubMed] [Google Scholar]
- [48].Doi Y, Hashimoto T, Yamaguchi H, Vertut-Doï A, Modification of gelsolin with 4-fluoro-7-nitrobenz-2-oxa-1,3-diazole, Eur. J. Biochem 199 (1991) 277–83. [DOI] [PubMed] [Google Scholar]
- [49].Houk TW, Ovnic M, Karipides S, pH and polymerization dependence of the site of labeling of actin by 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole, J. Biol. Chem 258 (1983) 5419–23. [PubMed] [Google Scholar]
- [51].Morais Cabral JH, Atkins GL, Sánchez LM, López-Boado YS, López-Otin C, Sawyer L, Arachidonic acid binds to apolipoprotein D: implications for the protein’s function, FEBS Lett. 366 (1995) 53–56. [DOI] [PubMed] [Google Scholar]
- [52].Borghoff SJ, Miller AB, Bowen JP, Swenberg JA, Characteristics of chemical binding to alpha 2u-globulin in vitro–evaluating structure-activity relationships, Toxicol. Appl. Pharmacol 107 (1991) 228–238. [DOI] [PubMed] [Google Scholar]
- [53].Wang Q, Allen JC, Swaisgood HE, Binding of Vitamin D and cholesterol to β-lactoglobulin, J. Dairy Sci 80 (1997) 1054–1059, 10.3168/jds.S0022-0302(97)76030-2. [DOI] [PubMed] [Google Scholar]
- [54].Prescott-Mathews JS, Poet TS, Borghoff SJ, Evaluation of the in Vivo Interaction of Methyltert-Butyl Ether with α2u-Globulin in Male F-344 Rats, Toxicol. Appl. Pharmacol 157 (1999) 60–67, 10.1006/taap.1999.8661. [DOI] [PubMed] [Google Scholar]
- [55].Gasymov OK, Abduragimov AR, T.N., Glasgow BJ, Excited protein states of human tear lipocalin for low- and high-affinity ligand binding revealed by functional AB loop motion, Biophys. Chem. 149 (2010) 47–57. doi: 10.1016/j.bpc.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ, Binding studies of tear lipocalin: the role of the conserved tryptophan in maintaining structure, stability, and ligand affinity, Biochim. Biophys. Acta - Protein Structure and Molecuar Enzymology. 1433 (1999) 307–320, 10.1016/S0167-4838(99)00133-8. [DOI] [PubMed] [Google Scholar]
- [57].Tsukamoto S, Fujiwara K, Ikeguchi M, Fatty acids bound to recombinant tear lipocalin and their role in structural stabilization, J. Biochem 146 (2009) 343–350, 10.1093/jb/mvp076. [DOI] [PubMed] [Google Scholar]