Abstract
Mammalian glutamate transporters are crucial players in neuronal communication as they perform neurotransmitter reuptake from the synaptic cleft. Besides L-glutamate and L-aspartate, they also recognize D-aspartate, which might participate in mammalian neurotransmission and/or neuromodulation. Much of the mechanistic insight in glutamate transport comes from studies of the archeal homologs GltPh from Pyrococcus horikoshii and GltTk from Thermococcus kodakarensis. Here, we show that GltTk transports D-aspartate with identical Na+: substrate coupling stoichiometry as L-aspartate, and that the affinities (Kd and Km) for the two substrates are similar. We determined a crystal structure of GltTk with bound D-aspartate at 2.8 Å resolution. Comparison of the L- and D-aspartate bound GltTk structures revealed that D-aspartate is accommodated with only minor rearrangements in the structure of the binding site. The structure explains how the geometrically different molecules L- and D-aspartate are recognized and transported by the protein in the same way.
Research organism: Other
Introduction
Mammalian excitatory amino acid transporters (EAATs) are responsible for clearing the neurotransmitter glutamate from the synaptic cleft (for review see Grewer et al., 2014; Takahashi et al., 2015; Vandenberg and Ryan, 2013). EAATs are secondary transporters that couple glutamate uptake to co-transport of three sodium ions and one proton and counter-transport of one potassium ion (Levy et al., 1998; Owe et al., 2006; Zerangue and Kavanaugh, 1996). EAATs transport L-glutamate, L- and D-aspartate with similar affinity (Arriza et al., 1994).
D-aspartate is considered as a putative mammalian neurotransmitter and/or neuromodulator (Brown et al., 2007; D'Aniello et al., 2011; Spinelli et al., 2006) (reviewed in D'Aniello, 2007; Genchi, 2017; Ota et al., 2012). Such a role is also proposed for L-aspartate (Cavallero et al., 2009), however this is still a matter of debate (Herring et al., 2015). Both stereoisomers bind to and activate N-methyl-D-aspartate receptors (NMDARs) (Patneau and Mayer, 1990) and might be involved in learning and memory processes (reviewed in Errico et al., 2018; Errico and Usiello, 2017; Katane and Homma, 2011; Ota et al., 2012).
Although it is well established that EAATs take up D-aspartate (Arriza et al., 1994; Gundersen et al., 1993), structural insight in the binding mode of the enantiomer is lacking. The best structurally characterized members of the glutamate transporter family are the archeal homologs GltPh and GltTk (Akyuz et al., 2015; Boudker et al., 2007; Guskov et al., 2016; Jensen et al., 2013; Reyes et al., 2013; Reyes et al., 2009; Scopelliti et al., 2018; Verdon et al., 2014; Verdon and Boudker, 2012; Yernool et al., 2004), which share 32–36% sequence identity with eukaryotic EAATs (Jensen et al., 2013; Slotboom et al., 1999; Yernool et al., 2004). In contrast to EAATs, GltPh and GltTk are highly selective for aspartate over glutamate, and couple uptake only to co-transport of three sodium ions (Boudker et al., 2007; Groeneveld and Slotboom, 2010; Guskov et al., 2016). Despite these differences, the amino acid residues in the substrate-binding sites of mammalian and prokaryotic glutamate transporters are highly conserved (Boudker et al., 2007; Jensen et al., 2013). The first structures of human members of the glutamate transporter family (Canul-Tec et al., 2017; Garaeva et al., 2018), showed that the substrate-binding sites are indeed highly similar among homologs (Figure 2—figure supplement 1).
Here, we present the structure of GltTk with the enantiomeric substrate D-aspartate. The crystal structure was obtained in the outward-facing state with the substrate oriented in a very similar mode as L-aspartate, showing that the two enantiomers bind almost identically regardless of the mirrored spatial arrangement of functional groups around the chiral Cα atom.
Results
Affinity of D-aspartate and stoichiometry of sodium binding to GltTk
Using Isothermal Titration Calorimetry (ITC), we determined the binding affinities of D-aspartate to GltTk in the presence of varying concentrations of sodium ions (Figure 1A, Table 1). The affinity of the transporter for D-aspartate was strongly dependent on the concentration of sodium, similar to what has been reported for L-aspartate binding to GltPh and GltTk (Boudker et al., 2007; Hänelt et al., 2015; Jensen et al., 2013; Reyes et al., 2013). At high sodium concentration (500 mM), the Kd values of GltTk for D- and L-aspartate binding level off to 374 ± 30 nM and 62 ± 3 nM, respectively. The ΔH values for binding of both substrates were favorable, with a more negative value of ~1 kcal mol−1 for L-aspartate, indicating a better binding geometry for L- than for D-aspartate. For both substrates, the ΔS contribution was unfavorable (Table 1). When plotting the observed Kd values for L- and D-aspartate against the sodium concentration (on logarithmic scales), the slopes of both curves in the lower limit of the sodium concentration are close to −3, indicating that binding of both compounds is coupled to the binding of three sodium ions (Boudker et al., 2007; Lolkema and Slotboom, 2015) (Figure 1B).
Figure 1. Binding and transport of D-aspartate by GltTk.
(A) ITC analysis of D-aspartate binding to GltTk in presence of 300 mM NaCl (Kd of 0.47 ± 0.17 µM). Insets show no D-aspartate binding in absence of NaCl. (B) Sodium and aspartate binding stoichiometry. Logarithmic plot of Kd values (nM) for L-aspartate (black squares; slope is −2.8 ± 0.4; taken for reference from Guskov et al., 2016) and D-aspartate (gray circles; slope is −2.9 ± 0.2) against logarithm of NaCl concentration (mM). The negative slope of the double logarithmic plot (red line) in the limit of low sodium concentrations indicates the number of sodium ions that bind together with aspartate. Error bars represent the ±SD from at least three independent measurements. (C) GltTk transport rate of D-aspartate in presence of 100 mM NaCl. The solid line reports the fit of the Michaelis-Menten model to the data revealing a Km value of 1.1 ± 0.11 µM. Error bars represent the ±SD from duplicate experiments. (D) Determination of Na+ : aspartate coupling stoichiometry in GltTk using equilibrium potential measurement. The uptake or efflux of radiolabeled aspartate was determined by comparing the lumenal radioactivity associated with the liposomes after 2 min of incubation with the radioactivity initially present (Δcpm). Gray circles and black squares show the measurements for D- and L-aspartate, respectively. The solid and dashed lines are the best linear regression for the D- and L-aspartate data, respectively. The 95% confidence interval for D-aspartate is displayed by gray curves. Numbers in parentheses are the coupling stoichiometries expected to give zero flux conditions for each membrane voltage. Error bars represent the ± SD obtained in five replicates.
Table 1. Thermodynamic parameters of D- and L-aspartate binding at high (300 mM) and low (75 mM) Na+ concentration.
| Substrate/ Na+ | Kd (µM) | ΔH (cal mol−1) | ΔS (cal mol−1 K−1) |
|---|---|---|---|
| L-aspartate/300 mM NaCl | 0.12 ± 0.04 | −1.61 (±0.08) x 104 | −22.1 ± 2.2 |
| D-aspartate/300 mM NaCl | 0.47 ± 0.17 | −1.48 (±0.11) x 104 | −20.6 ± 3.6 |
| L-aspartate/75 mM NaCl | 1.04 ± 0.39 | −1.22 (±0.13) x 104 | −13.2 ± 5.2 |
| D-aspartate/75 mM NaCl | 5.66 ± 1.59 | −1.14 (±0.41) x 104 | −14.3 ± 14.3* |
*At low Na+ concentrations high errors prevented accurate measuring of ΔS values.
To test whether D-aspartate is a transported substrate, purified GltTk was reconstituted into proteoliposomes and uptake of [3H]-D-aspartate was assayed. GltTk catalyzed transport of the radiolabeled substrate into the proteoliposomes. The Km for transport was 1.1 ± 0.11 μM at a sodium concentration of 100 mM (Figure 1C). This value is comparable to the Km for L-aspartate uptake under the same conditions (0.75 ± 0.17 μM). The stoichiometry Na+: D-aspartate was determined by flux measurements of radiolabeled D-aspartate at different membrane voltages (Fitzgerald et al., 2017). Depending on the concentrations of Na+ and D-aspartate on either side of the membrane, the imposed voltages either lead to flux of radiolabeled D-aspartate across the membrane (accumulation into or depletion from the lumen), or does not cause net flux (when the voltage equals the equilibrium potential) (Fitzgerald et al., 2017). The equilibrium potentials for different possible stoichiometries are calculated by:
where n and m are the stoichiometric coefficients for Na+ and substrate S, respectively. Membrane voltages were chosen that would match the equilibrium potential for stoichiometries of 2:1 (−78 mV), 3:1 (−39 mV) or 4:1 (−26 mV), and flux of radiolabeled D-aspartate was measured (Figure 1D). At −78 mV D-aspartate was taken up into the lumen; at −26 mV it was released from the liposomes; and at −39 mV there was little flux. From these data, we conclude that D-aspartate is most likely symported with three sodium ions. However, the flux was not exactly zero at the calculated equilibrium potential of −39 mV for 3:1 stoichiometry. This small deviation could be caused by systematic experimental errors, or by leakage or slippage (Parker et al., 2014; Shlosman et al., 2018). To exclude that it was caused specifically by D-aspartate, we repeated the experiment using radiolabeled L-aspartate. The equilibrium potentials for the experiments using D- and L-aspartate were identical, showing that the two stereoisomers use the same coupling stoichiometry.
Similar mode of enantiomers binding
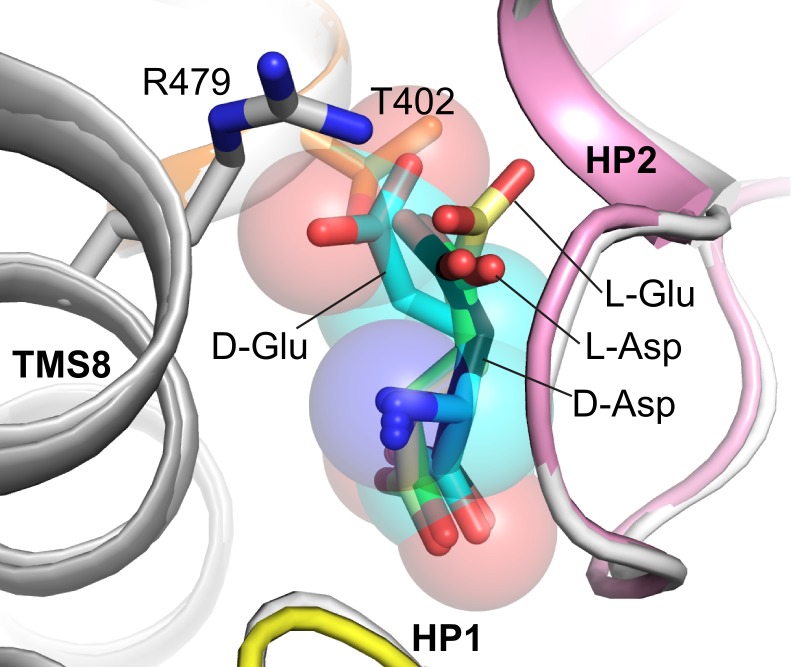
We determined a crystal structure of GltTk in complex with D-aspartate at 2.8 Å resolution (Figure 2A,B). The obtained structure is highly similar to the previously described GltTk and GltPh structures with the transport domains in the outward-oriented occluded state. Comparison of the GltTk structures in complex with L- and D-aspartate revealed a highly similar binding mode of the substrates with analogous orientation of amino and carboxyl groups. Despite the impossibility to superimpose two enantiomers, D- and L-aspartate are capable of forming almost identical hydrogen bonding networks with conserved amino acid residues of the substrate-binding site (Figure 2C). There are only small changes in the positions of the Cα atoms and Cβ carboxyl groups due to the constitutional differences. However, this divergence leads to only minor changes in the interaction network, consistent with the comparable Kd and ΔH values determined by ITC (Table 1).
Figure 2. The crystal structure of GltTk with D-aspartate.
The model contains one protein molecule in the asymmetric unit with the substrate present in each protomer of the homotrimer. (A) Cartoon representation of the homotrimer viewed from the extracellular side of the membrane. Lines separate protomers. Each protomer consists of the scaffold domain (pale green) and the transport domain. In the transport domain HP1 (yellow), HP2 (red), TMS7 (orange) are shown. D-aspartate is shown as black sticks and Na+ ions as purple spheres. Like in most GltPh structures a part of the long flexible loop 3–4 between the transport and scaffold domain is not visible. It is indicated by a dashed connection. (B) A single protomer is shown in the membrane plane. (C) Comparison of the substrate-binding site of GltTk in complex with L-aspartate (gray; PDB code 5E9S) and D-aspartate (black). Cartoon representation; substrates and contacting amino acid residues are shown as sticks; hydrogen bonds are shown as dashed lines. The GltTk structures with D- and L-aspartate can be aligned with Cα-RMSD = 0.38 Å for the three transport domains. (D) Composite omit map (cyan mesh) for D-aspartate (contoured at 1σ) and sodium ions (2σ) calculated using simulated annealing protocol in Phenix (Terwilliger et al., 2008). Color coding in all panels is the same.
Figure 2—figure supplement 1. Superposition of substrate-binding sites of L-aspartate bound GltTk and thermostabilized human EAAT1.
Figure 2—figure supplement 2. Superposition of substrate and sodium binding sites in L-aspartate and D-aspartate bound GltTk.
Figure 2—figure supplement 3. Model of glutamate binding in EAAT1.
Three peaks of electron density (Figure 2D; Figure 2—figure supplement 2) located at the same positions as three sodium ions in the GltTk complex with L-aspartate (Guskov et al., 2016) most probably correspond to sodium ions, consistent with a 3:1 Na+: D-aspartate coupling stoichiometry (Figure 1B,D).
Discussion
Most proteins selectively bind a single stereoisomer of their substrates (for a review see Nguyen et al., 2006). On the other hand, some proteins are able to bind different stereoisomers of a ligand, which is believed to be possible due to different binding modes, because enantiomers cannot be superimposed in the three-dimensional space and thus cannot interact with the binding site identically.
Based on three- and four-point attachment models (Easson and Stedman, 1933; Mesecar and Koshland, 2000; Ogston, 1948) it has been suggested that stereoisomers can bind in the same site but with significant differences. This hypothesis was supported by crystal structures of enzymes with different enantiomeric substrates (Brem et al., 2016; Sabini et al., 2008), including enantiomeric amino acids (Aghaiypour et al., 2001; Bharath et al., 2012; Driggers et al., 2016; Temperini et al., 2006). In contrast, the binding poses of enantiomers in some other enzymes are remarkably similar, for instance in aspartate/glutamate racemase EcL-DER, where active site forms pseudo-mirror symmetry (Liu et al., 2016).
To our knowledge GltTk is the first amino acid transporter for which the binding of enantiomeric substrates has been characterized. The only other transporter for which structures have been determined in the presence of D- and L-substrates is the sodium-alanine symporter AgcS. However, in that case, limited resolution prevented determination of the absolute orientation of bound enantiomers (Ma et al., 2019). In the substrate-binding site of GltTk, L- and D-aspartate take similar poses leading to almost identical networks of contacts. Since mirror imaged substrates inevitably have differences in angles between donors and acceptors of hydrogen bonds, the binding affinities are not identical, with 4–6 times higher Kd of the GltTk-D-aspartate complex in comparison with L-aspartate (Table 1). Similar differences in binding affinities between these enantiomers were also found for the GltPh homologue (Boudker et al., 2007). The higher Kd values for the D-aspartate enantiomer might be explained by a higher dissociation rate (koff) in comparison with L-aspartate, that was shown in kinetic studies of sodium and aspartate binding on GltPh (Ewers et al., 2013; Hänelt et al., 2015). GltTk couples binding and transport of three sodium ions to one D-aspartate molecule (Figure 1B,D), the same number as for L-aspartate. Although the affinity for D-aspartate is lower than for L-aspartate, the binding of D-aspartate is not accompanied by a loss of sodium binding sites, which is in line with the observation that none of the sodium binding sites are directly coordinated by the substrate L-aspartate. In the crystal structure of GltTk with D-aspartate peaks of density were resolved at positions corresponding to the three sodium ions in the L-aspartate bound GltTk structure (Figure 2D) (Guskov et al., 2016). Altogether our data suggest that the mechanism of D- and L-aspartate transport in GltTk is most probably identical.
Mammalian glutamate transporters take up D-aspartate, L-glutamate and L-aspartate with similar micromolar affinity, but have significantly lower affinity (millimolar) for D-glutamate (Arriza et al., 1997; Arriza et al., 1994). In the absence of the structures of human SLC1A transporters with different stereoisomeric substrates, one can only speculate why EAATs can readily bind and transport both L- and D-aspartate, but only L-glutamate. It seems that the extra methylene group in D-glutamate compared to D-aspartate could cause sterical clashes within the binding site (Figure 2— Figure 2—figure supplement 3), which might affect affinity of binding.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene | TK0986 | UniProt database | Q5JID0 | |
| Strain, strain background (E. coli) | MC1061 | Casadaban and Cohen, 1980 | ||
| Biological sample (Thermococcus kodakarensis KOD1) |
ATCC BAA-918/JCM 12380/KOD1 | |||
| Recombinant DNA reagent | pBAD24-GltTk-His8 | Jensen et al., 2013 | Expression plasmid for C-terminally His8-tagged GltTk. | |
| Chemical compound | D-Asp | Sigma-Aldrich | 219096–25G | ReagentPlus99% |
| Software | Origin 8 | OriginLab | ||
| Other | GltTk-D-aspartate coordinate file and structural factors | This paper | accession number PDB ID code 6R7R |
Crystal structure of the glutamate transporter homologue GltTk in complex with D-aspartate |
Protein purification and crystallization
GltTk was expressed and purified as described previously (Guskov et al., 2016). It was shown that L-aspartate binds to GltTk only if sodium ions are present, and the protein purified in absence of sodium ions is in the apo state (Jensen et al., 2013). For crystallization with D-aspartate the apo protein was purified by size exclusion chromatography (SEC) on a Superdex 200 10/300 GL (GE Healthcare) column equilibrated with buffer containing 10 mM Hepes KOH, pH 8.0, 100 mM KCl, 0.15% DM. Crystals of GltTk with D-aspartate were obtained in presence of 300 mM NaCl, 300 µM D-aspartate (Sigma-Aldrich, 99%) by the vapour diffusion technique (hanging drop) at 5°C by mixing equal volumes of protein (7 mg ml−1) and reservoir solution (20% glycerol, 10% PEG 4000, 100 mM Tris/bicine, pH 8.0, 60 mM CaCl2, 60 mM MgCl2, 0.75% n-octyl-b-D-glucopyranoside (OG)).
Data collection and structure determination
Crystals were flash-frozen without any additional cryo protection and data sets were collected at 100K at the beamline ID23-1 (ESRF, Grenoble). The data were indexed, integrated and scaled in XDS (Kabsch, 2010) and the structure was solved by Molecular Replacement with Phaser (McCoy et al., 2007) using structure of GltTk (PDB ID 5E9S) as a search model. Manual model rebuilding and refinement were carried out in COOT (Emsley et al., 2010) and Phenix refine (Afonine et al., 2012). Data collection and refinement statistics are summarized in Table 2. Coordinates and structure factors for GltTk have been deposited in the Protein Data Bank under accession codes PDB 6R7R. All structural figures were produced with an open-source version of PyMol.
Table 2. Data collection and refinement statistics.
| GltTk D-Asp | |
|---|---|
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 116.55, 116.55, 314.77 |
| α, β, γ (°) | 90.00, 90.00 120.00 |
| Resolution (Å) | 48.06-2.80 (2.87-2.80)* |
| Rmeas | 0.11 (>1) |
| CC1/2 | 99.9 (11.7) |
| I / σI | 8.40 (0.98) |
| Completeness (%) | 99.3 (98.9) |
| Redundancy | 5 (4) |
| Refinement | |
| Resolution (Å) | 2.80 |
| No. reflections | 301,077 |
| Rwork/Rfree (%)s | 23.4/27.2 |
| No. of atom | |
| Protein | 9262 |
| PEG/detergent | 181/33 |
| Ligand/ion | 27/9 |
| Water | - |
| B-factors | |
| Protein | 127 |
| PEG/detergent | 147/174 |
| Ligand/ion | 114/117 |
| Water | - |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.162 |
*Values in parentheses are for the highest-resolution shell.
Isothermal titration calorimetry
ITC experiments were performed at a constant temperature of 25°C using an ITC200 calorimeter (MicroCal). Varying concentrations of the indicated substrates (in 10 mM Hepes KOH, pH 8.0, 100 mM KCl, 0.15% DM and indicated sodium concentrations) were titrated into a thermally equilibrated ITC cell filled with 250 μl of 3–20 μM GltTk supplemented with 0 to 1000 mM NaCl. Data were analyzed using the ORIGIN-based software provided by MicroCal.
Reconstitution into proteoliposomes
A solution of E. coli total lipid extract (20 mg ml−1 in 50 mM KPi, pH 7.0) was extruded with a 400-nm-diameter polycarbonate filter (Avestin, 11 passages) and diluted with the same buffer to a final concentration of 4 mg ml−1. The lipid mixture was destabilized with 10% Triton-X100. Purified GltTk and the destabilized lipids were mixed in a ratio of 1:1600 or 1:250 (protein: lipid) and incubated at room temperature for 30 min. Bio-beads were added four times (25 mg ml−1, 15 mg ml−1, 19 mg ml−1, 4 mg ml−1 lipid solution) after 0.5 hr, 1 hr, overnight and 2 hr incubation, respectively, on a rocking platform at 4°C. The Bio-beads were removed by passage over an empty Poly-Prep column (Bio-Rad). The proteoliposomes were collected by centrifugation (20 min, 298,906 g, 4°C), subsequently resuspended in 50 mM KPi, pH 7.0 to the concentration of the protein 33.4 µg ml−1 and freeze-thawed for four cycles. The proteoliposomes were stored in liquid nitrogen until subsequent experiments.
Uptake assay
Stored proteoliposomes with reconstitution ratio of 1:1600 were thawed and collected by centrifugation (20 min, 298,906 g, 4°C), the supernatant was discarded and the proteoliposomes were resuspended in buffer containing 10 mM KPi, pH 7.5, 300 mM KCl. The internal buffer was exchanged by three cycles of freezing in liquid nitrogen and thawing, and finally extruded through a polycarbonate filter with 400 nm pore size (Avestin, 11 passages). The proteoliposomes were finally pelleted by centrifugation (20 min, 298,906 g, 4°C) and resuspended to the concentration of the protein 625 ng µl−1. 2 µl of proteoliposomes were diluted 100 times in reaction buffer containing 10 mM KPi, pH 7.5, 100 mM NaCl, 200 mM Choline-Cl, 3 µM valinomycin and 0.2–15 µM D-aspartate (each concentration point contained 0.2 µM [3H]-D-aspartate). After 15 s the reaction was quenched by adding 2 ml of ice-cold buffer (10 mM KPi, pH 7.5, 300 mM KCl) and immediately filtered on nitrocellulose filter (Protran BA 85-Whatman filter), finally the filter was washed with 2 ml of quenching buffer. The filters were dissolved in scintillation cocktail and the radioactivity was measured with a PerkinElmer Tri-Carb 2800RT liquid scintillation counter.
Measuring transporter equilibrium potentials
Stored proteoliposomes with reconstitution ratio of 1:250 were thawed and collected by centrifugation (20 min, 298,906 g, 4°C), the supernatant was discarded and the proteoliposomes were resuspended to a concentration of 10 mg ml−1 of lipids in buffer containing 20 mM Hepes/Tris, pH 7.5, 200 mM NaCl, 50 mM KCl, 10 μM D-aspartate (containing 1 μM [3H]-D-aspartate). The internal buffer was exchanged by freeze-thawing and extrusion as described above. The experiment was started by diluting the proteoliposomes 20 times into a buffer containing 20 mM Hepes/Tris, pH 7.5, 200 mM NaCl, 3 μM valinomycin, varying concentrations of KCl and Choline Cl were added in order to obtain the desired membrane potential as shown in (Figure 1—source data 1).
After 1, 2 and 3 min the reaction was quenched with ice-cold quenching buffer containing 20 mM Hepes/Tris, pH 7.5, 250 mM Choline Cl and immediately filtered on nitrocellulose filter (Protran BA 85-Whatman filter), finally the filter was washed with 2 ml of quenching buffer. The initial amount of radiolabeled aspartate was measured by filtering the proteoliposomes immediately after diluting them in quenching buffer. The filters were dissolved in scintillation cocktail and the radioactivity was measured with a PerkinElmer Tri-Carb 2800RT liquid scintillation counter. The equilibrium, or reversal, potential, Erev, for each condition was calculated as described in Fitzgerald et al. (2017).
Acknowledgements
This work is funded by the Netherlands Organisation for Scientific Research (Vici grant 865.11.001 to DJS and Vidi grant 723.014.002 to AG) and European Research Council starting grant 282083 to DJS. We thank A Garaeva and M Ejby for synchrotron data collection. The European Synchrotron Radiation Facility beamlines ID23-1 and ID29 (Grenoble, France) and EMBL beamlines P13 and P14 (Hamburg, Germany) are acknowledged for beamline facilities. This work has been supported by iNEXT, grant number 653706, funded by the Horizon 2020 programme of the European Commission.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Dirk J Slotboom, Email: d.j.slotboom@rug.nl.
Albert Guskov, Email: a.guskov@rug.nl.
José D Faraldo-Gómez, National Heart, Lung and Blood Institute, National Institutes of Health, United States.
Richard Aldrich, The University of Texas at Austin, United States.
Funding Information
This paper was supported by the following grants:
Nederlandse Organisatie voor Wetenschappelijk Onderzoek 865.11.001 to Dirk J Slotboom.
European Research Council 282083 to Dirk J Slotboom.
Nederlandse Organisatie voor Wetenschappelijk Onderzoek 723.014.002 to Albert Guskov.
Additional information
Competing interests
No competing interests declared.
Author contributions
Formal analysis, Supervision, Validation, Investigation, Visualization, Writing—original draft, Project administration, Writing—review and editing.
Formal analysis, Investigation, Methodology, Writing—review and editing.
Formal analysis, Investigation.
Resources, Investigation.
Conceptualization, Resources, Data curation, Formal analysis, Supervision, Funding acquisition, Methodology, Project administration, Writing—review and editing.
Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Methodology, Writing—review and editing.
Additional files
Data availability
Diffraction data and the derived model have been deposited in PDB under accession number 6R7R.
The following dataset was generated:
Arkhipova V, Dirk Slotboom. 2019. Diffraction data and the derived model. Protein Data Bank. 6R7R
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghaiypour K, Wlodawer A, Lubkowski J. Structural basis for the activity and substrate specificity of erwinia chrysanthemi L-asparaginase. Biochemistry. 2001;40:5655–5664. doi: 10.1021/bi0029595. [DOI] [PubMed] [Google Scholar]
- Akyuz N, Georgieva ER, Zhou Z, Stolzenberg S, Cuendet MA, Khelashvili G, Altman RB, Terry DS, Freed JH, Weinstein H, Boudker O, Blanchard SC. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 2015;518:68–73. doi: 10.1038/nature14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriza JL, Fairman WA, Wadiche JI, Murdoch GH, Kavanaugh MP, Amara SG. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. The Journal of Neuroscience. 1994;14:5559–5569. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriza JL, Eliasof S, Kavanaugh MP, Amara SG. Excitatory amino acid transporter 5, a retinal glutamate transporter coupled to a chloride conductance. PNAS. 1997;94:4155–4160. doi: 10.1073/pnas.94.8.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharath SR, Bisht S, Harijan RK, Savithri HS, Murthy MR. Structural and mutational studies on substrate specificity and catalysis of salmonella typhimurium D-cysteine desulfhydrase. PLOS ONE. 2012;7:e36267. doi: 10.1371/journal.pone.0036267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- Brem J, van Berkel SS, Zollman D, Lee SY, Gileadi O, McHugh PJ, Walsh TR, McDonough MA, Schofield CJ. Structural basis of Metallo-β-Lactamase inhibition by captopril stereoisomers. Antimicrobial Agents and Chemotherapy. 2016;60:142–150. doi: 10.1128/AAC.01335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ER, Piscopo S, Chun JT, Francone M, Mirabile I, D'Aniello A. Modulation of an AMPA-like glutamate receptor (SqGluR) gating by L- and D-aspartic acids. Amino Acids. 2007;32:53–57. doi: 10.1007/s00726-006-0349-3. [DOI] [PubMed] [Google Scholar]
- Canul-Tec JC, Assal R, Cirri E, Legrand P, Brier S, Chamot-Rooke J, Reyes N. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature. 2017;544:446–451. doi: 10.1038/nature22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. Journal of Molecular Biology. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Cavallero A, Marte A, Fedele E. L-aspartate as an amino acid neurotransmitter: mechanisms of the depolarization-induced release from cerebrocortical synaptosomes. Journal of Neurochemistry. 2009;110:924–934. doi: 10.1111/j.1471-4159.2009.06187.x. [DOI] [PubMed] [Google Scholar]
- D'Aniello A. D-Aspartic acid: an endogenous amino acid with an important neuroendocrine role. Brain Research Reviews. 2007;53:215–234. doi: 10.1016/j.brainresrev.2006.08.005. [DOI] [PubMed] [Google Scholar]
- D'Aniello S, Somorjai I, Garcia-Fernàndez J, Topo E, D'Aniello A. D-Aspartic acid is a novel endogenous neurotransmitter. The FASEB Journal. 2011;25:1014–1027. doi: 10.1096/fj.10-168492. [DOI] [PubMed] [Google Scholar]
- Driggers CM, Kean KM, Hirschberger LL, Cooley RB, Stipanuk MH, Karplus PA. Structure-Based insights into the role of the Cys-Tyr crosslink and inhibitor recognition by mammalian cysteine dioxygenase. Journal of Molecular Biology. 2016;428:3999–4012. doi: 10.1016/j.jmb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easson LH, Stedman E. Studies on the relationship between chemical constitution and physiological action. Biochemical Journal. 1933;27:1257–1266. doi: 10.1042/bj0271257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico F, Usiello A. D-Aspartate, an Atypical Amino Acid with NMDA Receptor Agonist Features: Involvement in Schizophrenia. In: Hashimoto K, editor. The NMDA Receptors. Humana Press, Cham; 2017. pp. 83–101. [DOI] [Google Scholar]
- Errico F, Nuzzo T, Carella M, Bertolino A, Usiello A. The emerging role of altered d-Aspartate metabolism in schizophrenia: new insights from preclinical models and human studies. Frontiers in Psychiatry. 2018;9:559. doi: 10.3389/fpsyt.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers D, Becher T, Machtens JP, Weyand I, Fahlke C. Induced fit substrate binding to an archeal glutamate transporter homologue. PNAS. 2013;110:12486–12491. doi: 10.1073/pnas.1300772110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald GA, Mulligan C, Mindell JA. A general method for determining secondary active transporter substrate stoichiometry. eLife. 2017;6:e21016. doi: 10.7554/eLife.21016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaeva AA, Oostergetel GT, Gati C, Guskov A, Paulino C, Slotboom DJ. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nature Structural & Molecular Biology. 2018;25:515–521. doi: 10.1038/s41594-018-0076-y. [DOI] [PubMed] [Google Scholar]
- Genchi G. An overview on d-amino acids. Amino Acids. 2017;49:1521–1533. doi: 10.1007/s00726-017-2459-5. [DOI] [PubMed] [Google Scholar]
- Grewer C, Gameiro A, Rauen T. SLC1 glutamate transporters. Pflügers Archiv - European Journal of Physiology. 2014;466:3–24. doi: 10.1007/s00424-013-1397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld M, Slotboom DJ. Na(+):aspartate coupling stoichiometry in the glutamate transporter homologue glt(Ph) Biochemistry. 2010;49:3511–3513. doi: 10.1021/bi100430s. [DOI] [PubMed] [Google Scholar]
- Gundersen V, Danbolt NC, Ottersen OP, Storm-Mathisen J. Demonstration of glutamate/aspartate uptake activity in nerve endings by use of antibodies recognizing exogenous D-aspartate. Neuroscience. 1993;57:97–111. doi: 10.1016/0306-4522(93)90114-U. [DOI] [PubMed] [Google Scholar]
- Guskov A, Jensen S, Faustino I, Marrink SJ, Slotboom DJ. Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nature Communications. 2016;7:1–6. doi: 10.1038/ncomms13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänelt I, Jensen S, Wunnicke D, Slotboom DJ. Low affinity and slow na+ binding precedes high affinity aspartate binding in the Secondary-active transporter GltPh. The Journal of Biological Chemistry. 2015;290:15962–15972. doi: 10.1074/jbc.M115.656876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Silm K, Edwards RH, Nicoll RA. Is aspartate an excitatory neurotransmitter? Journal of Neuroscience. 2015;35:10168–10171. doi: 10.1523/JNEUROSCI.0524-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Guskov A, Rempel S, Hänelt I, Slotboom DJ. Crystal structure of a substrate-free aspartate transporter. Nature Structural & Molecular Biology. 2013;20:1224–1226. doi: 10.1038/nsmb.2663. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallographica Section D Biological Crystallography. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katane M, Homma H. d-Aspartate—An important bioactive substance in mammals: A review from an analytical and biological point of view. Journal of Chromatography B. 2011;879:3108–3121. doi: 10.1016/j.jchromb.2011.03.062. [DOI] [PubMed] [Google Scholar]
- Levy LM, Warr O, Attwell D. Stoichiometry of the glial glutamate transporter GLT-1 expressed inducibly in a chinese hamster ovary cell line selected for low endogenous na+-dependent glutamate uptake. The Journal of Neuroscience. 1998;18:9620–9628. doi: 10.1523/JNEUROSCI.18-23-09620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao F, Ma Y, Liu S, Cui Y, Yuan Z, Kang X. Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157. FEBS Letters. 2016;590:1262–1269. doi: 10.1002/1873-3468.12148. [DOI] [PubMed] [Google Scholar]
- Lolkema JS, Slotboom DJ. The hill analysis and co-ion-driven transporter kinetics. The Journal of General Physiology. 2015;145:565–574. doi: 10.1085/jgp.201411332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lei HT, Reyes FE, Sanchez-Martinez S, Sarhan MF, Hattne J, Gonen T. Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS. PNAS. 2019;116:2086–2090. doi: 10.1073/pnas.1806206116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecar AD, Koshland DE. A new model for protein stereospecificity. Nature. 2000;403:614–615. doi: 10.1038/35001144. [DOI] [PubMed] [Google Scholar]
- Nguyen LA, He H, Pham-Huy C. Chiral drugs: an overview. International Journal of Biomedical Science : IJBS. 2006;2:85–100. [PMC free article] [PubMed] [Google Scholar]
- Ogston AG. Interpretation of experiments on metabolic processes, using isotopic tracer elements. Nature. 1948;162:963. doi: 10.1038/162963b0. [DOI] [PubMed] [Google Scholar]
- Ota N, Shi T, Sweedler JV. D-Aspartate acts as a signaling molecule in nervous and neuroendocrine systems. Amino Acids. 2012;43:1873–1886. doi: 10.1007/s00726-012-1364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owe SG, Marcaggi P, Attwell D. The ionic stoichiometry of the GLAST glutamate transporter in salamander retinal glia. The Journal of Physiology. 2006;577:591–599. doi: 10.1113/jphysiol.2006.116830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Mindell JA, Newstead S. Thermodynamic evidence for a dual transport mechanism in a POT peptide transporter. eLife. 2014;3:e04273. doi: 10.7554/eLife.04273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patneau DK, Mayer ML. Structure-activity relationships for amino acid transmitter candidates acting at N-methyl-D-aspartate and quisqualate receptors. The Journal of Neuroscience. 1990;10:2385–2399. doi: 10.1523/JNEUROSCI.10-07-02385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes N, Oh S, Boudker O. Binding thermodynamics of a glutamate transporter homolog. Nature Structural & Molecular Biology. 2013;20:634–640. doi: 10.1038/nsmb.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabini E, Hazra S, Ort S, Konrad M, Lavie A. Structural basis for substrate promiscuity of dCK. Journal of Molecular Biology. 2008;378:607–621. doi: 10.1016/j.jmb.2008.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopelliti AJ, Font J, Vandenberg RJ, Boudker O, Ryan RM. Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5. Nature Communications. 2018;9:38. doi: 10.1038/s41467-017-02444-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlosman I, Marinelli F, Faraldo-Gómez JD, Mindell JA. The prokaryotic na+/Ca2+ exchanger NCX_Mj transports Na+ and Ca2+ in a 3:1 stoichiometry. The Journal of General Physiology. 2018;150:51–65. doi: 10.1085/jgp.201711897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotboom DJ, Konings WN, Lolkema JS. Structural features of the glutamate transporter family. Microbiology and Molecular Biology Reviews : MMBR. 1999;63:293–307. doi: 10.1128/mmbr.63.2.293-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli P, Brown ER, Ferrandino G, Branno M, Montarolo PG, D'Aniello E, Rastogi RK, D'Aniello B, Baccari GC, Fisher G, D'Aniello A. D-aspartic acid in the nervous system of aplysia limacina: possible role in neurotransmission. Journal of Cellular Physiology. 2006;206:672–681. doi: 10.1002/jcp.20513. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Foster JB, Lin CL. Glutamate transporter EAAT2: regulation, function, and potential as a therapeutic target for neurological and psychiatric disease. Cellular and Molecular Life Sciences. 2015;72:3489–3506. doi: 10.1007/s00018-015-1937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperini C, Scozzafava A, Vullo D, Supuran CT. Carbonic anhydrase activators. activation of isoforms I, II, IV, VA, VII, and XIV with L- and D-phenylalanine and crystallographic analysis of their adducts with isozyme II: stereospecific recognition within the active site of an enzyme and its consequences for the drug design. Journal of Medicinal Chemistry. 2006;49:3019–3027. doi: 10.1021/jm0603320. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Adams PD, Read RJ, Zwart PH, Hung LW. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallographica Section D Biological Crystallography. 2008;64:515–524. doi: 10.1107/S0907444908004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg RJ, Ryan RM. Mechanisms of glutamate transport. Physiological Reviews. 2013;93:1621–1657. doi: 10.1152/physrev.00007.2013. [DOI] [PubMed] [Google Scholar]
- Verdon G, Oh S, Serio RN, Boudker O. Coupled ion binding and structural transitions along the transport cycle of glutamate transporters. eLife. 2014;3:e02283. doi: 10.7554/eLife.02283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdon G, Boudker O. Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nature Structural & Molecular Biology. 2012;19:355–357. doi: 10.1038/nsmb.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- Zerangue N, Kavanaugh MP. Flux coupling in a neuronal glutamate transporter. Nature. 1996;383:634–637. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]