Abstract
Although the number of sequenced insect genomes numbers in the hundreds, little is known about gene regulatory sequences in any species other than the well-studied Drosophila melanogaster. We provide here a detailed protocol for using SCRMshaw, a computational method for predicting cis-regulatory modules (CRMs, also “enhancers”) in sequenced insect genomes. SCRMshaw is effective for CRM discovery throughout the range of holometabolous insects and potentially in even more diverged species, with true-positive prediction rates of 75% or better. Minimal requirements for using SCRMshaw are a genome sequence and training data in the form of known Drosophila CRMs; a comprehensive set of the latter can be obtained from the SCRMshaw download site. For basic applications, a user with only modest computational know-how can run SCRMshaw on a desktop computer. SCRMshaw can be run with a single, narrow set of training data to predict CRMs regulating a specific pattern of gene expression, or with multiple sets of training data covering a broad range of CRM activities to provide an initial rough regulatory annotation of a complete, newly-sequenced genome.
Keywords: Non-model insects, Regulatory genomics, Transcriptional gene regulation, Genome annotation, Enhancer prediction
1. Introduction
Considerable resources have been invested into insect genome sequencing over the last decade. As of May 2017 over 230 individual insect genomes had undergone some degree of genome sequencing and assembly, including orders from each of the major insect groups (Palaeoptera, Polyneoptera, Paraneoptera, Holometabola), in addition to arthropods from the Crustacea, Chelicerata, and Myriapoda. There is particular depth in the Holometabola, especially in the Hymenoptera, Diptera, and Lepidoptera (see http://i5k.github.io/arthropod_genomes_at_ncbi). Community efforts such as the i5k consortium seek to increase the number of sequenced arthropod genomes to as many as 5000 [1]. However, while sequence has become readily available, annotation has lagged. First-pass annotation of the coding genome is typically conducted with an automated pipeline such as MAKER [2], followed by manual curation to refine and/or supplement computational predictions. Subsequent improved annotation can be a labor- and resource-intensive endeavor requiring additional high-quality data from paired-end RNA-seq, directed PCR, CAGE-seq, and the like (for review of genome annotation, see [3, 4]). Nonprotein-coding transcription units are often poorly represented in the initial annotation, and in most cases, there is no concerted effort at all to annotate regulatory sequences. Thus a major functional genomic component—the regulatory genome—does not factor into early rounds of insect genome annotation, and with the exception of the intensively studied model insect Drosophila melanogaster, knowledge of insect gene regulatory sequences remains limited (for review of insect regulatory genomics, see [5]).
Regulation of gene transcription is a fundamental process, and detailed characterization of gene regulatory sequences represents an important step toward understanding the basic biology of any organism. In insects, as in most of the Metazoa, a significant amount of gene regulation is mediated through distal regulatory sequences that can range from immediately upstream of the promoter to hundreds of kilobases away from the transcription start site. We refer to such sequences collectively as cis-regulatory modules (CRMs), a classification that includes, although is not strictly limited to, what are commonly referred to as “enhancers” (for recent reviews see [6–11]). CRMs tend to be organized in a modular fashion, with each controlling a discrete subset of a gene’s overall expression pattern. They are typically a few hundred basepairs in length and can be located 5′ to, 3′ to, within introns of, or in some cases even within exons of, their target genes. Although characterizing CRMs has historically been a laborious and low-throughput activity, the advent of genome-wide approaches coupled with next-generation sequencing technologies has led to a renaissance of sorts in methods for CRM discovery (for review see [8, 12–14]). However, current methods are often problematic for insects, where issues of small organism size, lack of extensive tissue-specific cell lines, and lack of mature transgenic methods for most species make these methods technically challenging and often prohibitively expensive. While the well-established community invested in Drosophila research has led to initiatives such as the NIH-funded modENCODE project [15] and the production of extensive experimental and computational resources, a similar infusion of funding and effort seems unlikely for non-model insects.
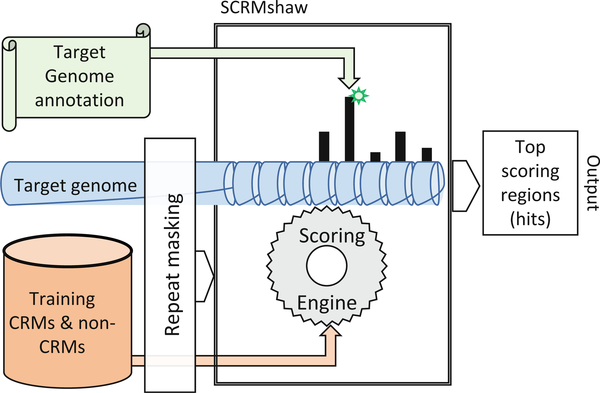
We present here a detailed protocol for using SCRMshaw, a computational method we developed, that leverages the wealth of available data on Drosophila CRMs [16] to predict regulatory sequences in both Drosophila and in non-model, non-drosophilid, insect species [17, 18]. SCRMshaw (for “Supervised Cis-Regulatory Module prediction”) is effective for CRM discovery in species at least as evolutionarily distant from Drosophila as the 345 My diverged Hymenoptera [19]. SCRMshaw is a machine-learning based method that takes as input a sequenced insect genome along with a training set of Drosophila CRMs, defined by a common functional characterization (e.g., midgut expression), and outputs a set of functionally related predicted CRMs [17, 18] (Fig. 1). It does this by building statistical models that compare the short subsequence (k-mer) count distribution in the training CRMs to a set of randomly selected, noncoding sequences and then scoring the target genome using these models. Essentially, SCRMshaw relies on the idea that CRMs with similar function will have similar sequence characteristics, not visible by eye but identifiable using machine-learning methods, and that these regulatory mechanisms will be conserved across insect evolution. Although not universally true, we have found that these assumptions are robust enough to allow effective CRM discovery without requiring knowledge of transcription factor binding sites or of the expression patterns of the genes being regulated, between species with a virtually complete lack of observable sequence alignment at the noncoding level. Both empirical validation and comparison of predictions to regulatory regions identified by open-chromatin profiling methods suggests true-positive prediction rates averaging between 75% and 85% ([19] and MSH and Y. Tomoyasu, unpublished results). SCRMshaw can be used with all available training datasets to provide a first-pass regulatory annotation for newly sequenced insect genomes, or can be used in a targeted fashion to predict CRMs driving specific gene expression patterns of interest (e.g., [20]).
Fig. 1.
Overview of the SCRMshaw cis-regulatory module discovery pipeline
2. Materials
2.1. Genomes and Annotation Files
Genome sequence: Table 1 lists suggested websites for availability of insect genomes, although genomes can be downloaded from any available source (see Note 1). Minimally required is a genome FASTA file. Depending on the state of genome assembly, this may be listed simply as “genome” or as “chromosomes,” “scaffolds,” or “contigs.” We recommend using the most complete assembly available (see Note 2).
Genome Annotation: Genome annotation, minimally consisting of a set of genes and more preferably a set of genes and exons, is recommended but optional. Annotation is typically found in GFF3 format [21]. For evaluating results, it is often useful to know the Drosophila orthologs of the genes in your genome (see Subheading 3.6.4).
Drosophila genome and annotation: For creating customized training datasets (see Subheading 3.2), the Drosophila melanogaster genome and its annotation is required, along with (optionally) the genomes for related drosophilids. These can most easily be obtained from FlyBase [22] (ftp://ftp.flybase.net/genomes/). Genomes can be found in the respective “fasta/” directory for each species, and annotation in the “gff/” directory.
Table 1.
Resources for insect genomes and genome annotation (nonexclusive)
| Resource | URL | Represented species |
|---|---|---|
| BIPAA: Bioinformatics platform for agroecosystem arthropods | http://bipaa.genouest.org/is/ | |
| -aphidbase | Aphids | |
| -lepidoBD | Lepidoptera | |
| -ParWaspDB | Parasitoid wasps | |
| Diamondback Moth genomic database | http://dbm.dna.affrc.go.jp/px/ | Plutella xylostella |
| EnsemblMetazoa | http://metazoa.ensembl.org/index.html | Broad collection including dipterans, hymenopterans, lepidopterans, coleopterans, hemipterans, pthirapterans |
| FlyBase | http://flybase.org | Drosophila melanogaster and other drosophilids |
| Fourmidable | http://www.antgenomes.org | Ants |
| Hymenoptera Genome Database | http://hymenopteragenome.org | Bees, wasps, ants |
| i5k Workspace@NAL | https://i5k.nal.usda.gov | Broad collection spanning many arthropod orders |
| InsectBase | http://www.insect-genome.com | Broad collection spanning many arthropod orders |
| KAIKObase | http://sgp.dna.affrc.go.jp/KAIKObase/ | Bombyx mori |
| lepbase | http://lepbase.org | Lepidoptera |
| SilkDB | http://silkworm.genomics.org.cn | Bombyx mori |
| SpodoBase | http://bioweb.ensam.inra.fr/spodobase/ | Spodoptera frugiperda |
| Spotted Wing Flybase | http://spottedwingflybase.org | Drosophila suzukii |
| UCSC Genome Database | https://genome.ucsc.edu/ | Various Diptera; many non-insects |
| VectorBase | https://www.vectorbase.org/ | Mosquitoes and other vector species |
2.2. SCRMshaw Software
Operating system and software dependencies: SCRMshaw software is tested under Linux-based operating systems such as Ubuntu and CentOS. Other operating systems including Mac and Windows OS will require additional steps for installation. The core scoring methods are written in the C++ programming language and the utility scripts are implemented in Perl. The BioPerl modules “DB” and “SeqIO” are required for reading the input FASTA files. Preinstallation requirements are Perl (v5 or higher), BioPerl, and the gcc compiler (v4.8 or higher).
SCRMshaw and related programs: The SCRMshaw software and associated utility programs can be downloaded from http://veda.cs.uiuc.edu/SCRMshaw/ (Sinha lab) or http://halfonlab.ccr.buffalo.edu/scrmshaw.html (Halfon lab).
2.3. Training Data
SCRMshaw requires “training data” in the form of known Drosophila CRMs with common function (see Note 3). Precompiled sets of training data can be downloaded from the SCRMshaw website at http://veda.cs.uiuc.edu/SCRMshaw/ (Sinha lab) or http://halfonlab.ccr.buffalo.edu/scrmshaw.html (Halfon lab). These sets include, for each constituent CRM from D. melanogaster, orthologous sequences from 10 related Drosophila species; inclusion of these sequences improves SCRMshaw’s predictive power [18]. Custom training sets can be developed by downloading selected sets of CRMs from the REDfly database (http://redfly.ccr.buffalo.edu; [16]) and following the steps in Subheading 3.2 (see Subheading 3.2 below).
2.4. Computational Requirements
User expertise: Although extensive computational know-how is not required for using SCRMshaw, a basic familiarity with the Linux command line is recommended. This includes being able to launch the terminal shell, navigate between directories, and upload/download/view files. There are many online tutorials for learning basic Linux commands (e.g., https://diyhacking.com/linux-commands-for-beginners/). Programming skills are not necessary, but the source codes (in C++ and Perl) are provided for advanced users who may want to adapt the code based on their project.
Time and memory considerations: The computational time required for the scoring, and the required storage space, scales with the genome size. A coarse computational time estimate required per training set per million basepairs of the target genome for all three scoring methods is about 1 min on a 2.4 GHz processor. The storage space requirement for the same calculation (with the default window size = 500 bp, shift size 250 bp) is about 75 MB. Thus for a 350 MB genome, for example, computational time is ~6 h with a required 26 GB of storage. Decreasing the shift size will provide higher resolution scoring profiles; however, it will require more storage space. After the scoring, most of the storage space can be released (see Note 4). When having multiple training datasets, the computational time can be distributed among multiple processors by running an instance of SCRMshaw for each training dataset (see Note 5).
2.5. Additional Software
1. Tandem Repeat Finder
Tandem Repeat Finder [23] is used for masking tandem repeat sequences and is strongly recommended for both the genome and training sequences (see Subheading 3.3). The correct version of Tandem Repeat Finder for your system can be down-loaded from https://tandem.bu.edu/trf/trf.download.html.
2. BEDTools
BEDTools [24] is a convenient and easy-to-use software suite for working with genome data. While not required for using SCRMshaw, it is highly recommended for use in associated protocols such as working with SCRMshaw output data (see Subheading 3.6) or creating customized training data (see Subheading 3.2). BEDTools can be obtained from https://github.com/arq5x/bedtools2/releases and is easily installed on a UNIX/Linux or Mac OS X computer. Documentation can be found at http://bedtools.readthedocs.io/en/latest/.
3. Methods
3.1. Installing SCRMshaw
Download the SCRMshaw software (see Subheading 2.2 above).
- Decompress the tar archive. This will create a single “SCRMshaw” directory with the following subdirectories:
id="P17">bin/ src/ code/ include/ example/
Navigate to the SCRMshaw directory and enter the “make” command.
Detailed information on how to install and run the software is provided in the “README.txt” file located within the software package.
3.2. Constructing Custom Training Sets
For ease of use, we recommend using precompiled training sets available on the SCRMshaw download pages (see Subheading 2.3). We will continue to provide updated and improved training data at these sites. However, customized training sets can also be constructed (see Note 6).
3.2.1. Obtaining CRMs from REDfly
Go to the REDfly database (http://redfly.ccr.buffalo.edu).
Use the menu bar to navigate to the “search” page and click on “Advanced Search.”
Under “Advanced Search>Data Type” check the “CRM” button.
If desired, set a maximum CRM length using the “Max Size” box. We typically restrict our training CRMs to ≤2000 bp.
Click in the “Ontology/Expression Term” box and begin typing the name of the expression pattern to which you want to restrict the activity of the returned CRMs. An auto-complete drop-down list will enable you to select the desired term. Alternatively, you can enter a Drosophila Anatomy Ontology ID number (FBbt:xxxxxxxx). See the REDfly online “Help” for additional information on expression pattern searching (also see Note 7).
Click “Search.”
A list of CRMs meeting the search criteria will appear in the “Search Results” area. Click the box on the left of the header bar to select all CRMs, then click “Download Selected” at the bottom of the page.
In the dialogue box, select “File Type: FASTA,” “Coord Version: R6,” and “FASTA Options: Sequence Only.” Click download.
The downloaded file contains a multi-FASTA file with the Drosophila melanogaster members of the training set. This can be used as is or checked for overlapping sequences and refined (see Note 8) before moving on to obtain orthologous sequences (see Subheading 3.2.2) and generating the negative training data (see Subheading 3.2.3). To check for overlapping sequences and/or to obtain orthologous CRMs, it is convenient to also download the same selected set of CRMs in BED format. This is accomplished by following the same steps as in step 8, above, substituting “File Type: BED” and “BED File Type: BED simple.”
3.2.2. Obtaining Orthologous Regions from Other Drosophila Species
Many of the sequenced Drosophila species are evolutionarily close enough to D. melanogaster (within up to 40 My of divergence) that there is a high level of conservation of their noncoding genomes, and as such extensive alignments of noncoding regions can be constructed. We have shown that augmenting the training set CRMs with the orthologous sequences from additional species improves SCRMshaw performance [18]. (Presumably this is because the most important k-mers within the CRMs are under evolutionary constraint and therefore more conserved than background k-mers, boosting the weight of the most relevant k-mers within the training data.) Orthologous CRMs can be obtained using the liftOver tool at the UCSC Genome Browser [25]:
Obtain the FASTA and BED files for the training CRMs from steps 8 and 9 (see Subheading 3.2.1 above) and rename them “crm.fa” and “crm.bed,” respectively.
Navigate to the liftOver tool at https://genome.ucsc.edu/cgi-bin/hgLiftOver (see Note 9).
For “Original Genome” select D. melanogaster. Choose an appropriate Original Assembly, “New Genome,” and New Assembly from the drop-down lists (see Note 10).
[Optional] To increase the stringency of mapping between genomes, change the “Minimum ratio of bases that must remap” from 0.1 (default) to 0.25.
Upload the “crm.bed” file obtained in step 9 (see Subheading 3.2.1 above) using “Choose File,” followed by “Submit.”
Download and save the successful conversions from the link “View Conversions” as “species1_crms.bed.”
Download the correct genome assembly for the “New genome” (“species1_genome.fa”; see Subheading 2.1, item 3).
- Use “bedtools getfasta” from the BEDTools software suite [24] to obtain the FASTA formatted sequences for the CRMs from the orthologous species:
$bedtools getfasta -fi species1_genome.fa -bed species1_crms.bed -fo species1_crms.fa
Repeat steps 2–6 for all other orthologous species of interest.
- Combine the FASTA files to create one training CRM file, e.g.:
$cat species*_crms.fa crm.fa > project1/dataset1/crms. Fasta
3.2.3. Creating the Negative Training Sets (Non-CRMs, or Background Sequences)
Two of our scoring methods (HexMCD and IMM) maximize the likelihood ratio between the similarity of a target sequence to the training CRMs versus its similarity to a “background set” of non-CRMs, in terms of their k-mer profiles. The ideal negative training set should be depleted of the k-mers that distinguish CRMs from non-CRMs, while retaining all the other k-mers (see Note 11).
To obtain an appropriate set of randomly selected noncoding sequences to use as the background set, we have provided a script called “randomWithSameGC.pl” in the SCRMshaw package that extracts random regions in the genome with the same GC content as the CRMs (±1%). This script can be run from within the SCRMshaw directory as:
$perl code/randomWithSameGC.pl --crm crms.fasta --output neg. fasta --size 10 --genomedir rootDIR --gene gene.gff3
The genome root directory (“rootDIR”) is the path to the genome FASTA file (i.e., the D. melanogaster genome, see Subheading 2.1, item 3 above). For example, if the genome is located at “project1/genome.fa” then the root directory is “project1/.” The size option defines the number of background CRMs matched to each training CRM (see Note 12). The gene annotation file is used to filter out regions that overlap coding sequences.
3.3. Masking Tandem Repeats
Tandem repeats, as the name indicates, consist of a repeated pattern of one or more nucleotides that occur directly after each other. An example would be ACACACACAC, where the dinucleotide “AC” is repeated five times. The occurrence of a long tandem repeat in the training dataset or the region to be scored significantly skews the distribution of k-mer counts toward the repeated pattern, which would result in assignment of a “false” high score to regions with one or more occurrences of the repeated pattern. To avoid this potential issue, tandem repeats in both the training datasets and the genome to be searched are masked prior to running SCRMshaw using Tandem Repeat Finder [23]. To mask tandem repeats:
Navigate to “https://tandem.bu.edu/trf/trf.download.html” and download the current version of TRF for your system type (e.g., trf409.linux64).
- Change the downloaded TRF file to an executable form:
$chmod +x trf409.linux64
- Run TRF on your FASTA files (e.g., genome.fa, crms.fa, neg.fa) using parameters as shown:
$trf409.linux64 genome.fa 2 7 7 80 10 50 500 –m –h $trf409.linux64 crms.fasta 2 7 7 80 10 50 500 –m –h $trf409.linux64 neg.fasta 2 7 7 80 10 50 500 –m –h
- Move the TRF output to your training set directory and rename:
$mv genome.fa.2.7.7.80.10.50.500.mask project1/genome.fa $mv crms.fasta.2.7.7.80.10.50.500.mask project1/dataset1/crms.fasta $mv neg.fasta.2.7.7.80.10.50.500.mask project1/dataset1/neg.fasta
TRF substitutes the repeated nucleotides with “N” characters that can be directly fed into SCRMshaw. If using a different tool for masking repeats, be sure that the repeated nucleotides are replaced by the same number of “N” characters.
3.4. Running SCRMshaw
SCRMshaw requires the following files and directories:
A genome file (e.g., “genome.fa”), a multi-FASTA file including all of the genomic sequences needing to be scored. This is typically a downloaded genome file (see Subheading 2.1, item 1) that has been masked for tandem repeats (see Subheading 3.3 above), and is passed to the program using the “--genome” option.
-
One or more dataset directories each containing two files, “crms.fasta” and “neg.fasta.” These files are respectively the positive and negative training sequences, both in FASTA file format, as described above (see Subheadings 2.3 and 3.2).
We highly recommend tandem repeat masking of genome.fa, crms.fasta, and neg.fasta (see Subheading 3.3 above).
A text file containing a list of the dataset directories (e.g., “trainingSet.lst”; see below step 4). This is specified using the “--traindirlst” option.
To run SCRMshaw:
- Create a project directory (e.g., project1/):
$mkdir project1/
- Under the project directory, create one or more dataset directories (here, dataset1/, dataset2/, etc.) (see Note 13):
$mkdir project1/dataset1/
- Move each pair of positive and negative training datasets to the appropriate dataset directory:
$mv crms.fasta project1/dataset1/ $mv neg.fasta project1/dataset1/
-
Under theproject directory, create a list file “trainingSet.lst” that contains the path to the datasets (e.g., project1/dataset1/), one line per dataset. You can create this file using a simple text editor or using the following “echo” commands:
$echo “project1/dataset1/” >> project1/trainingSet.lst $echo “project1/dataset2/” >> project1/trainingSet.lst $echo “project1/dataset3/” >> project1/trainingSet.lst
etc.
- Move your gene annotation file to the project directory.
$mv gene.gff3 project1/
-
For simplicity, move the genome FASTA file to your project directory:
$mv genome.fa project1/
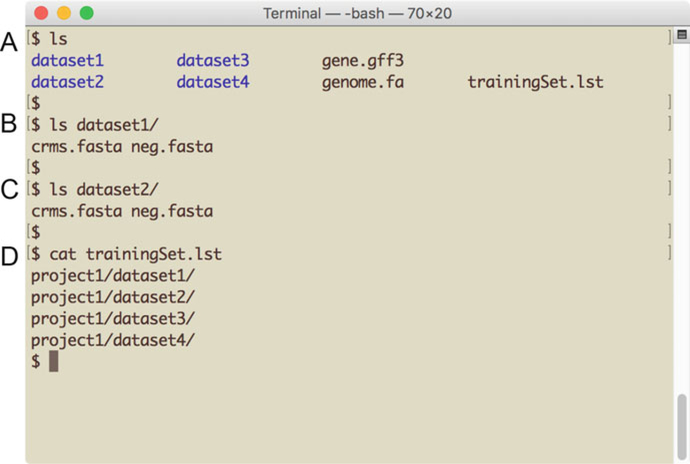
At the conclusion of these steps, your directory will resemble Fig. 2.
- Run SCRMshaw, e.g.:
$perl code/scrm.pl --thitw 2000 --gff project1/gene.gff3 --imm --hexmcd --pac --genome project1/genome.fa --traindirlst project1/trainingSet.lst --outdir project1/results/--step 123
Fig. 2.
Preparing to run SCRMshaw. The screenshot illustrates the directory setup and required user-added files for using SCRMshaw. (a) Directory structure showing the genome file, annotation file, training set list file, and training set subdirectories. (b, c) Each training set subdirectory contains a positive (crms.fasta) and negative (neg.fasta) training data file in the form of a pair of multi-FASTA sequence files. (d) The “trainingSet. lst” file is a text file containing a list of paths to the training set subdirectories
The SCRMshaw results will be placed in the directory specified by “--outdir,” which will be generated automatically if it doesn’t exist (here, “project1/results/”).
The SCRMshaw options include:
the name of scoring method(s) to be used for prediction (“--imm” for IMM, “--hexmcd” for HexMCD, and “--pac” for PAC): required. Any individual or combination of methods can be selected to run (e.g., “--imm --pac” to run IMM and PAC). For details about the individual scoring methods see [17, 18].
the gene annotation file of the genomic regions to be scored in GFF3 format (e.g., “--gff gene.gff3”): optional but strongly recommended. Alternatively, separate “gene” and “exon” files can be used (see Note 14). It is also possible to use SCRMshaw without providing any annotation, but this will not allow for exclusion of coding sequences or calculation of “local rank” (see below).
the number of top scoring regions to be reported as CRM predictions (--thitw N): optional, default = 2000 (see Note 15).
which steps of the SCRMshaw pipeline to run (--step 123): optional, default = 123. Step 1 processes the genome and genome annotation, step 2 processes the training data, and step 3 scores the genome (see Note 5).
Detailed information on how to run SCRMshaw from the command line and additional available options can be obtained by the following command: “perl code/scrm.pl” or from the README document accompanying the SCRMshaw distribution. An example benchmark data set along with instructions is provided in the example directory within the software package.
3.5. The SCRMshaw Output
SCRMshaw provides several pieces of information about each predicted CRM, or “hit.” These include:
Chromosome_ID:start_coordinate. This is the name of the chromosome/scaffold/contig and the nucleotide coordinate of the start of the scored window. The end coordinate can be determined by simply adding the length of the window to the start coordinate.
Score. The score for the window (see Note 16).
Gene_ID:Gene_symbol:distance:location. The Gene_ID and Gene_symbol are drawn from the genome annotation (these will sometimes be identical, depending on how the annotation was constructed). distance provides the distance in base pairs between the start of the window and the annotated gene, while location takes the value “upstream,” “downstream,” or “inside” depending on whether the window is 5′, 3′, or within an intron of the gene (for windows within introns, distance is given as 0).
local_rank: The local rank refers to the rank score of the window considering only a 50 kb range to either side of the annotated gene [19]. (This distance can be set to a different value at run time using the “--distance” option.) Local ranks are only provided when an annotation file is used (see Note 16).
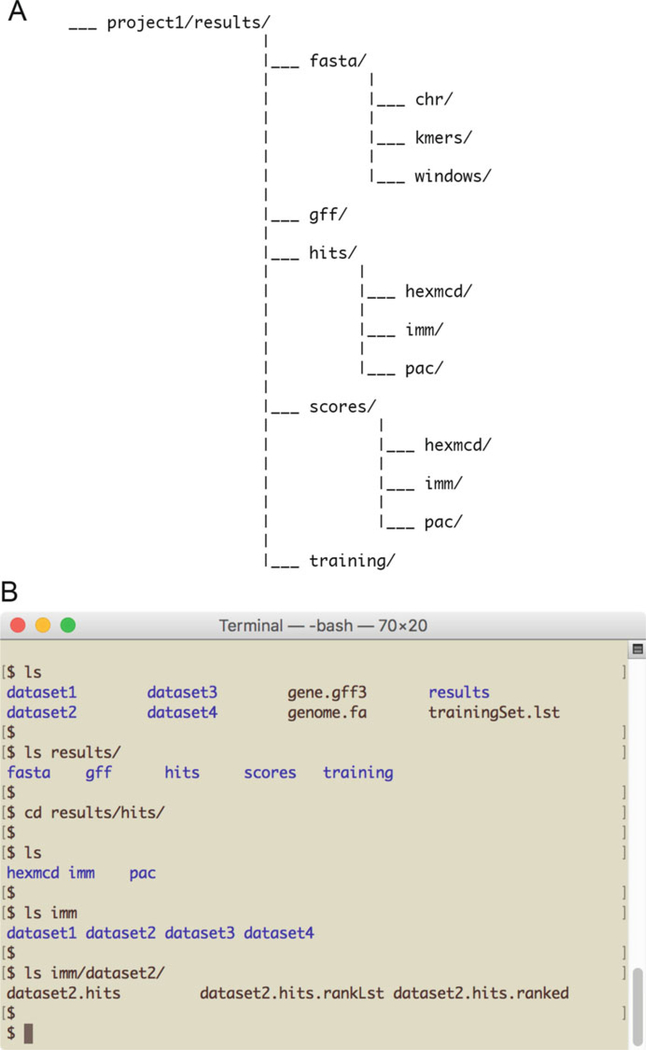
The SCRMshaw output is placed in the directory specified by the “--outdir” option and has the following subdirectories (Fig. 3a):
-
hits/. This is the main subdirectory of interest and contains the top hits (i.e., predicted CRMs) for each of the methods for each training dataset. A separate subdirectory is created for each method (hexmcd, imm, pac). Each contains a subdirectory for each training set, with three “hits” files (Fig. 3b):
dataset1.hits. This file contains the top-scored windows in rank order in FASTA format:>chromosome_ID:start_coordinate score gene_ID:gene_symbol:distance: location ATGCCCAGAGAATGGGCAACAAGTAGCGGCGAATTAGCAATCCTATCATGCTTTTATGGCCGGCCAACTCTTGCC
dataset1.hits.ranked. This file contains the top-scored windows in rank order in FASTA format, but with local ranks also included:>chromosome_ID: start_coordinate score gene_ID: gene_symbol:distance: location:local_rank AATGCCCAGAGAATGGGCAACAAGTAGCGGCGAATTAGCAATCCTATCATGCTTTTATGGCCGGCCAACTCTTGCC
dataset1.hits.rankLst. This file contains the top-scored windows without sequence in tabular format:
Gene_ID, chromosome_ID: start_coordinate, score, local_rank
If no GFF file or gene/exon annotation was provided, only the “dataset1.hits” file will be generated.
-
fasta/. This directory has three subdirectories containing intermediate files used by SCRMshaw during the run. These are:
chr/: This directory stores the target sequence FASTA files, one sequence per file. Exon regions will be masked if GFF or exon files were provided.
kmers/: This directory stores k-mer frequency files for each target sequence.
windows/: This directory contains the target sequence with each chromosome/scaffold/contig stored as a multi-FASTA file broken into window-length fragments.
gff/. This directory contains gene and exon locations extracted from the provided GFF annotation file.
scores/. This directory contains the raw scores of a sliding window across the target genome, with one score file per chromosome/scaffold/contig. Each score file contains two columns, the location of the window and its corresponding score. Scores are contained in a separate subdirectory for each method (hexmcd, imm, pac).
training/. This directory contains intermediate files with the word statistics of the training sets that are used for building the statistical models and scoring the target genome.
Fig. 3.
SCRMshaw output. (a) Outline of the subdirectory structure established by SCRMshaw in the assigned “results” directory. (b) Screenshot showing the various results subdirectories and their component files
Once the results have been obtained, storage space can be freed up by keeping the “hits” files but discarding the various intermediate files (see Note 4).
3.6. Working with the SCRMshaw Output
In this section, we provide some protocols and tips for making productive use of the SCRMshaw output.
3.6.1. Getting Top Hits in BED Form
For many applications, it will be useful to generate a list of the top SCRMshaw results of interest as a BED-compatible file. We have developed a simple Perl utility script, “Generate_top_N_SCRM-hits,” to facilitate this task. The script takes a user-defined number “N” of desired results and outputs the top-ranked N predicted CRMs for each training set and each method (hexmcd, imm, pac) found in the /hits directory from a given set of SCRMshaw runs. Output is in the form of a 16-column tab-delimited file as follows:
chromosome
start
end
SCRMshaw score
flanking gene
flanking gene (due to a quirk in the way SCRMshaw results arereported, this will often be a repeat of field 5)
distance of hit from flanking gene (basepairs)
location of hit relative to flanking gene
local rank
next closest flanking gene
next closest flanking gene (due to a quirk in the way SCRMshaw results are reported, this will often be a repeat of field 10)
distance of hit from flanking gene (basepairs)
location of hit relative to flanking gene
local rank
training set
method (hexmcd, imm, pac)
Run the script from the command line with the following options:
d directory, path to directory containing SCRMshaw results (i.e., directory name provided as “--outdir” when running SCRMshaw): required
o outfile, name of file for results of script: required
n number of hits desired: defaults to 250 (for each training set and each method)
h help, displays usage information: overrides other options
v version, displays program version: overrides other options
3.6.2. Sorting and Merging Hits
It may be useful, once having generated the list of top predicted CRMs, to merge together overlapping regions and/or duplicate predictions (i.e., made from more than one method). This is most easily accomplished using BEDTools [24] as follows:
$bedtools sort –i Top_N_Hits_file | bedtools merge –c 4,5,10,15,16 –o max, distinct, distinct, distinct, distinct
or
$sort –k1,1 –k2,2n Top_N_Hits_file | bedtools merge –c 4,5,10,15,16 –o max, distinct, distinct, distinct, distinct
The resulting file will be an 8-column tab-delimited file with columns:
chromosome
start
end
SCRMshaw score (if predicted CRMs were merged, the highest score from the merged set)
flanking gene
next closest flanking gene
training set (if predicted CRMs were merged from multiple training sets, a comma-separated list of the training sets)
method (if predicted CRMs were merged from multiple methods, a comma-separated list of the methods)
3.6.3. Comparing SCRMshaw Predictions to Other Genomic Data Sets
It may often be of interest to compare SCRMshaw results with sets of known CRMs, other sets of predicted CRMs, or other genomic data such as histone modifications, transcription factor binding, or open-chromatin regions. This can be done quite simply using the merged top-hits file generated from the above protocol (see Subheadings 3.6.1 and 3.6.2) and the BEDTools program [24]. We recommend setting a minimum overlap of 10% (−f 0.10) for the predicted CRMs. An unmerged predicted CRM would thus need to overlap another genomic feature by 50 bp, while a longer merged CRM would require 75 bp of overlap (in our experience it is rare for merged features to exceed 750 bp). A typical command for comparing SCRMshaw results to another set of genomic features would be:
$bedtools intersect –loj –f 0.10 –a merged_Top_N_hits_file –b other_genomic_features_file
3.6.4. Enrichment Test
One difficulty when using SCRMshaw to predict CRMs for a newly sequenced, non-model insect genome, where there may be few or no known CRMs to validate against, is determining whether SCRMshaw worked effectively, i.e., with a low false-positive rate. One way to attempt a rough assessment of this is to find out whether the predictions in the target species fall near the orthologs of the training set genes, or other genes whose expression is consistent with that of the training set genes (e.g., for a “salivary gland” training set, a set of genes all expressed in the salivary glands). Although orthologous genes do not always maintain a similar expression pattern, such an analysis can at least provide an indirect confirmation of the results. This sort of “enrichment test” analysis is described in [19], and the necessary software can be obtained from the SCRMshaw download page (click on “pipeline for enrichment test”).
Orthologous gene mappings can be obtained from a variety of sources including OrthoDB (http://www.orthodb.org; [26]); InParanoid (http://inparanoid.sbc.su.se/; [27]); and EggNOG (http://eggnogdb.embl.de/; [28]). Precompiled lists of orthologous genes between Drosophila and many other species can be downloaded from InParanoid (http://inparanoid.sbc.su.se/download/current/Orthologs_other_formats/D.melanogaster/ ) or accessed via FlyBase.
Acknowledgments
We thank Kushal Suryamohan for comments on the manuscript. This work was supported by USDA grant 2012-67013-19361 (MSH) and NIH grant 5K22HL125593-02 (MK).
Footnotes
SCRMshaw can be run with any sequenced insect genome; however, to date we have only used genomes from within the Holometabola. Although empirical evaluations have not been extensive enough to make a definitive determination, initial results suggest that true-positive CRM prediction rates are similar for holometabolous insects regardless of whether they are evolutionarily closer to (e.g., mosquitoes) or further from (e.g., bees, wasps) Drosophila [19, 20].
That is, we recommend favoring chromosome > scaffold > contig. However, the effect of genome assembly quality on SCRMshaw performance has not yet been determined, and we cannot, at this time, suggest a minimal acceptable scaffold N50 or gap percentage (we are currently developing simulations to address this question). Because by default SCRMshaw scores the genome using 500 bp windows, we do not expect the extent of genome assembly to have a major role in determining the effectiveness of CRM prediction.
The “common function” of a set of CRMs can be defined in various ways, and broadly or narrowly. Ideal characteristics are an ongoing area of investigation. As a general rule, the most successful training datasets consist of CRMs with similar (although not necessarily identical) patterns based on well-defined anatomical classifications such as “somatic muscle,” “wing disc,” and “ventral nerve cord.” We expect that the more broadly defined the class, the wider the functional range of the resulting CRM predictions will be. This allows the user to search for a more-or-less specific set of results. We have found that even very broadly constituted datasets give a higher percentage of true-positive results than training data composed of randomly selected noncoding sequences, suggesting that as a whole CRMs are enriched for informative subsequences [17, 29].
SCRMshaw creates a large number of files when processing the genome and training data, which contain breakdowns of individual chromosome/scaffold FASTA sequences (/fasta/chr), details on k-mer frequencies (/fasta/kmers), and FASTA sequences of each 500 bp window used in scoring (/fasta/windows). For a large genome, these files can require significant storage space, but they are not required for routine applications once the predicted CRM results have been obtained. We provide a simple utility shell script on the SCRMshaw website, “cleanup_fastadirectory.sh,” that will delete the contents of the /fasta directory to free up storage space.
For a given genome, we recommend first running SCRMshaw with a limited number of training datasets and the “--step” option set to “--step 123” (see Subheading 3.4, step 7). This will process the genome and annotation, and ensure that all of the proper subdirectories are created. Parallel instances of SCRMshaw can then be run with additional training sets and “--step 23.” To run an additional scoring method for a previously run training set (e.g., if only ran “--imm” initially and now want to add “--hexmcd”), only “--step 3” is necessary.
Training set quality has a significant impact on CRM prediction. Presently, we have no method in place for evaluating training set sensitivity (how many known CRMs are recovered), while the specificity of existing training sets (how many of the predicted CRMs are likely to have the expected regulatory activity) can be inferred from the cross-validation and empirical testing data reported in [17, 18]. New training sets can be evaluated using the “enrichment test” protocol (see Subheading 3.6.4). However, it can be difficult to distinguish by this test if poor performance is due to the training set or to the insect genome being studied. We are developing a comprehensive suite of training set evaluation tests that will allow for assessment of both sensitivity and specificity of new and existing training sets. These tools will be made available on the SCRMshaw website when completed.
REDfly presently allows only a single expression term to be searched. If multiple search terms are desired, search each term individually using the listed procedure, then combine the download results into a single file, manually removing any duplicate CRMs.
It is desirable to remove any sequence overlaps, as including the same sequence twice will artificially weight the k-mers belonging to that sequence. This can be achieved by first sorting the BED file of REDfly CRMs (e.g., using BEDTools sort) and then running the utility Python script “SelectSmallestFeature. py,” available for download at the SCRMshaw and REDfly websites.
LiftOver can also be downloaded and installed locally, following instructions at https://genome-store.ucsc.edu. All options can be used as default with the exception of “minMatch,” which we recommend changing to 0.25.
The current version of the D. melanogaster genome is release 6, listed at the UCSC Genome Browser site as August 2014/dm6. However, few other genomes are mappable using liftOver for this release; most of the relevant other drosophilids are only available through the April 2006/dm3 genome version (release 5). REDfly CRMs through at least REDfly v5.2.1 can be downloaded as release 5/dm3 coordinates and in that form used directly to get orthologous regions via liftOver. Alternatively, a list of REDfly CRM coordinates can be converted from release 6/dm6 to release 5/dm3 using either liftOver or the conversion tool at FlyBase (http://flybase.org/static_pages/downloads/COORD_625.html). While these conversions can be used to obtain FASTA sequences for compiling training data, we recommend using the most up-to-date genome release and annotation when running SCRMshaw.
We used the following ten genomes in our previous work [18]: D. ananassae, D. erecta, D. grimshawi, D. mojavensis, D. persimilis, D. pseudoobscura, D. sechellia, D. simulans, D. virilis, and D. yakuba.
Choosing the “right” negative training set is important for the scoring. One strategy to obtain negative sequences would be to randomly shuffle the nucleotides within CRMs. The negative sequences from this strategy would have the same GC content as the CRM set, but would likely be depleted in the higher order k-mer features shared by the CRMs. However, due to random shuffling they might also inadvertently lack k-mers that are common to the non-CRMs. We prefer to select random noncoding regions in the source (D. melanogaster) genome having the same GC content as the CRMs (±1%). This alleviates the inadvertent effect of the above-mentioned strategy. On the other hand, it is likely that these sequences will contain other, unknown CRMs. Nevertheless, our previous results suggest that this method provides effective negative training data. The script “randomWithSameGC.pl,” included in the SCRMshaw package, can select appropriate sequences.
Note, however, that we have not fully explored the effect of various negative training sets on SCRMshaw performance. Other possibilities include obtaining the negative training sequences from the target genome (i.e., the genome that will be scored), or using the known Drosophila CRMs that do not belong to the positive training set. A version of this latter is used in the “IMMBoost” method recently reported by Yang and Sinha [30]. IMMBoost appear to increase specificity of CRM discovery, but effects on sensitivity and use in a crossspecies (i.e., scoring non-Drosophila genomes) have not yet been evaluated.
We typically use a tenfold excess of negative training sequences (i.e., --size 10). Although we have not explored the effects of varying this parameter, our assumption is that the larger the negative set, the more accurate the statistics of the k-mer distribution.
If using SCRMshaw in a multi-user setting, other directory structures might be more practical. For example, all of the training datasets can be placed in a single “datasets” directory outside of the “project” directory where they will be accessible for multiple projects. Paths in the “trainingSet.lst” file (step 4) should then be adjusted accordingly (e.g., “../datasets/dataset1”).
Individual “genes” and “exons” files can be used in lieu of an annotation GFF3 file; one or both files can be provided. These should be tab-delimited lists of gene or exon coordinates, respectively, in the form chromosome (or scaffold), start coordinate, end coordinate, strand (+ or −), gene ID/exon ID (e.g., gene_name:1). In the absence of an annotation GFF3 file, running SCRMshaw without a “genes” file will preclude calculation of local rank and provision of results indicating closest flanking genes. Omitting an “exons” file will prevent masking of exons and allow coding regions to be scored as potential CRMs.
The number of predictions to be reported can be adjusted depending on expectations of how many CRMs might be recovered, for instance based on how functionally broad or specific the training CRMs are. At the moment, we do not have a systematic way of learning this parameter from the input training sets. We recommend using a generous value, and then if desired working with a smaller subset of results obtained using the “Generate_top_N_SCRMhits” script (see Subheading 3.6.1).
Scores are used in determining the “global rank” of each hit [19] using the rule of thumb that higher scores are stronger predictions than lower scores. However, we do not at present have a clear guideline for “good” versus “poor” absolute scores. In [19], an msIMM score threshold of four provided good discrimination between pattern-specific (expression matching the training data) and nonspecific predicted CRMs, but our overall experience suggests that this number does not generalize. Negative-scoring windows from IMM and HexMCD should be avoided, as the negative score indicates they are more similar to the background than the training CRMs.
“Local rank” refers to the rank of a window score with respect only to the 50 kb region to either side of an annotated gene (more strictly, with respect to the region specified by the “--distance” option) [19]. In general, we would expect a true CRM that activates a nearby gene in a manner consistent with the training data to fall within one of the highest-scoring windows in the locus. (As genes may have multiple similar CRMs [31], a true-positive prediction does not necessarily need to be the top-ranked local window.)
References
- 1.i5k Consortium (2013) The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered 104:595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekblom R, Wolf JB (2014) A field guide to whole-genome sequencing, assembly and annotation. Evol Appl 7:1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yandell M, Ence D (2012) A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet 13:329–342 [DOI] [PubMed] [Google Scholar]
- 5.Suryamohan K, Halfon M (2015) Insect regulatory genomics In: Raman C et al. (eds) Short views on insect genomics and proteomics. Springer International Publishing, pp 119–155 [Google Scholar]
- 6.Cho KW (2012) Enhancers. Wiley interdisciplinary reviews developmental biology, vol. 1, pp 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long HK et al. (2016) Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167:1170–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlyueva D et al. (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15:272–286 [DOI] [PubMed] [Google Scholar]
- 9.Smith E, Shilatifard A (2014) Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21:210–219 [DOI] [PubMed] [Google Scholar]
- 10.Vernimmen D, Bickmore WA (2015) The hierarchy of transcriptional activation: from enhancer to promoter. Trends Genet 31:696–708 [DOI] [PubMed] [Google Scholar]
- 11.Buffry AD et al. (2016) The functionality and evolution of eukaryotic transcriptional enhancers. Adv Genet 96:143–206 [DOI] [PubMed] [Google Scholar]
- 12.Suryamohan K, Halfon MS (2015) Identifying transcriptional cis-regulatory modules in animal genomes. Wiley Interdiscip Rev Dev Biol 4:59–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y et al. (2015) The identification of cis-regulatory elements: a review from a machine learning perspective. Biosystems 138:6–17 [DOI] [PubMed] [Google Scholar]
- 14.Murakawa Y et al. (2016) Enhanced identification of transcriptional enhancers provides mechanistic insights into diseases. Trends Genet 32:76–88 [DOI] [PubMed] [Google Scholar]
- 15.modENCODE Consortium et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330:1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo SM et al. (2011) REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res 39:D118–D123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantorovitz MR et al. (2009) Motif-blind, genome-wide discovery of cis-regulatory modules in Drosophila and mouse. Dev Cell 17:568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazemian M et al. (2011) Improved accuracy of supervised CRM discovery with interpolated Markov models and cross-species comparison. Nucleic Acids Res 39:9463–9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazemian M et al. (2014) Evidence for deep regulatory similarities in early developmental programs across highly diverged insects. Genome Biol Evol 6:2301–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suryamohan K et al. (2016) Redeployment of a conserved gene regulatory network during Aedes aegypti development. Dev Biol 416:402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein L (2013) Generic Feature Format Version 3 (GFF3). https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md
- 22.Gramates LS et al. (2017) FlyBase at 25: looking to the future. Nucleic Acids Res 45: D663–D671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ et al. (2002) The human genome browser at UCSC. Genome Res 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zdobnov EM et al. (2017) OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res 45:D744–D749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnhammer EL, Ostlund G (2015) InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43:D234–D239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huerta-Cepas J et al. (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suryamohan K (2016) PhD Thesis: Regulatory networks in development: understanding the role of cis-regulatory modules in Gene Regulatory Network evolution. Department of Biochemistry, University at Buffalo-State University of New York [Google Scholar]
- 30.Yang W, Sinha S (2017) A novel method for predicting activity of cis-regulatory modules, based on a diverse training set. Bioinformatics 33:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barolo S (2012) Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]