Summary:
The anti-cancer immune response against mutated peptides of potential immunological relevance (neoantigens) is primarily attributed to MHC-I-restricted cytotoxic CD8+ T-cell responses. MHC-II-restricted CD4+ T-cells also drive anti-tumor responses, but their relation to neoantigen selection and tumor evolution has not been systematically studied. Modeling the potential of an individual’s MHC-II genotype to present 1,018 driver mutations in 5,942 tumors, we demonstrate that the MHC-II genotype constrains the mutational landscape during tumorigenesis in a manner complementary to MHC-I. Mutations poorly bound to MHC-II are positively selected during tumorigenesis, even more than mutations poorly bound to MHC-I. This emphasizes the importance of CD4+ T-cells in anti-tumor immunity. In addition, we observed less inter-patient variation in mutation presentation for MHC-II than for MHC-I. These differences were reflected by age at diagnosis, which was correlated with presentation by MHC-I only. Collectively, our results emphasize the central role of MHC-II presentation in tumor evolution.
In Brief
Inter-individual variation in MHC-II genotype influences the evolution of patient-specific tumor mutational spectra, emphasizing the key role of CD4+ T cells in anti-tumor immunity.
Introduction:
The Major Histocompatibility Complex (MHC) is one of the most polymorphic gene systems found in living organisms. This formidable polymorphism has led to the idea that MHC diversity is maintained so that heterozygous individuals are able to recognize and cope with a wide variety of microbial pathogens (overdominance model) (Carrington et al., 1999; Doherty and Zinkernagel, 1975; Messaoudi et al., 2002). Thus, in the interplay between microbial pathogens and an individual immune system, immune escape by mutation necessitates that the pathogen must accumulate multiple mutations before immunity is lost (immune pressure) (Bonhoeffer and Sniegowski, 2002; Nowak and Bangham, 1996).
Genomic interrogation of cancer has made it possible to catalogue non-synonymous mutations that could create cell surface peptide/MHC complexes potentially recognizable by T cells (e.g., neoantigens). This information is central to efforts to understand the role of the MHC in cancer evolution. To this end, we recently reported that MHC-I molecules play a significant role in shaping the mutational landscape of cancer (Marty et al., 2017). We demonstrated that mutations were more likely to be observed in tumors if the background MHC-I genotype resulted in poor affinity for peptides harboring the mutation, with implications for individual mutation probability and population mutation frequencies. These findings demonstrated that genomic variation in the MHC can play a significant role in shaping tumor genomes.
MHC-II molecules typically present 12-16 amino acid peptides to CD4+ T cells. CD4+ T cells play a more complex role than CD8+ T cells. While possessing cytotoxic effector properties similar to CD8+ T cells, they also exert a wide range of regulatory functions that distinguish them from CD8+ T cells. Classically, CD4+ T cells provide functional help to B cells, CD8+ T cells and CD4+ T cells in the form of cooperation involving cognate interaction with an antigen presenting cell (B cell or dendritic cell) (Cassell and Forman, 1988; Gerloni et al., 2000; Mitchison, 1971; Schoenberger et al., 1998). The role of CD4+ T cells in tumor immunity and protection has been proven in many studies in the mouse ((Haabeth et al., 2014; Hung et al., 1998; Wang, 2001). Indeed, patients responding to immunotherapy show a strong proliferative CD4+ T cell response to tumor-associated antigens Adoptive CD4+ T cell therapy has been associated with durable clinical responses in melanoma (Hunder et al., 2008) and cholangiocarcinoma (Tran et al., 2014) patients.
While these results suggest the essential role of MHC-II molecules in antigen presentation and in immune detection of mature tumors through neoantigen recognition, little is known about how MHC-II impacts emerging tumors. MHC-II, like MHC-I, is highly variable amongst humans with 4,802 documented alleles (Robinson et al., 2015). However, the antigen affinity of each MHC-II molecule is influenced by two genes, producing a combinatorial effect that leads to higher variation than MHC-I (Unanue et al., 2016). Because the average MHC binding affinity for MHC-II-restricted peptides required to activate CD4+ T cells is less stringent than that for MHC-I restricted peptides (Southwood et al., 1998), the MHC-II peptide binding groove structure allows more promiscuous binding of peptides (Consogno et al., 2003; Kobayashi et al., 2000), and CD4+ T cell responses can extend to encompass additional antigens after initial activation (epitope spreading (Hunder et al., 2008)), understanding the impact of MHC-II molecules in cancer immunity and cancer evolution is also more complex than for MHC-I.
Does high inter-individual variation in MHC-II genotype contribute to shaping the prevalence of mutations in cancer and directly influence patient-specific tumor development? To test this hypothesis, we adapted our strategy for analysis of MHC-I presentation to MHC-II. First, we adjusted the MHC-I specific Patient Harmonic-mean Best Rank (PHBR-I) score to account for differences in MHC-II peptide binding. We used multi-allelic mass spectrometry data to validate the effectiveness of the score to represent peptide presentation by an individual’s MHC-II genotype. Next, we determined and analyzed the MHC-II PHBR scores (PHBR-II) of 1,018 recurrent mutations for 5,942 patients in The Cancer Genome Atlas (TCGA). We found that MHC-II genotype has an even stronger influence over mutation probability than the MHC-I genotype.
Results:
Creating an affinity based MHC-II genotype scoring scheme
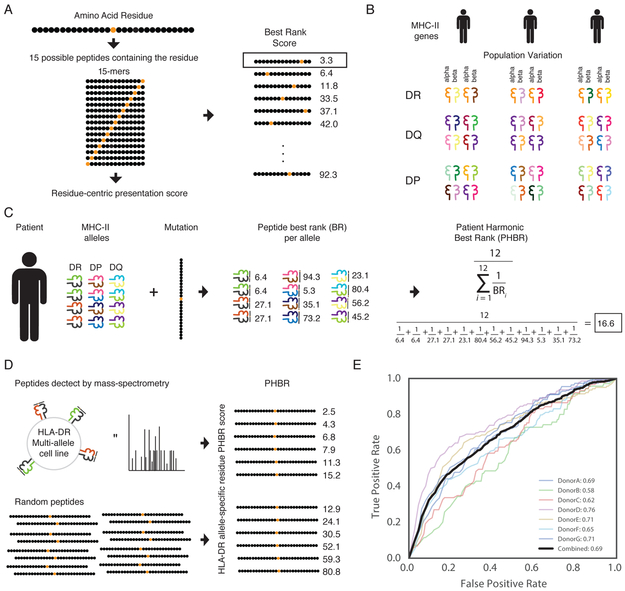
To study the role of MHC-II during tumorigenesis, we needed a score linking MHC-II genotype to presentation of specific mutations. We first constructed a score representing the ability of a single MHC-II molecule to present a residue. We previously established that using the best rank among peptides provided the best performance for predicting MHC-I presentation. We therefore adapted this scoring scheme to reflect the structure and composition of MHC-II. Three molecules (HLA-DR, HLA-DP, and HLA-DQ) make up the MHC-II, all of which are heterodimers formed by an alpha and beta chain (Unanue et al., 2016). Both the alpha and the beta chain influence the binding affinity of a peptide. In contrast to MHC-I, the MHC-II binding groove is open at both ends, allowing longer peptides to bind. To predict binding affinity to each alpha and beta paired MHC-II molecule, we used netMHCIIpan-3.1 which returns a single rank for the pair with each peptide (Karosiene et al., 2013). Unlike netMHCpan-3.0, netMHCIIpan- 3.1 has only been optimized for 15-mers and not for varying lengths. As previously, we assigned the best rank of all 15-mers containing the desired residue for the single MHC-II molecule presentation score (Figure 1A).
Figure 1:
Development of a residue-specific, patient-specific MHC-II presentation score. A graphical representation (A) of the Best Rank (BR) presentation score for a residue, (B) MHC-II genetic diversity in the population, and (C) of the the Patient Harmonic-mean Best Rank Class II (PHBR-II) presentation score. (D) An experimental schematic of the MS-based validation of the PHBR-II score. HLA-DR MS data from 7 donors was used to validate the PHBR-II score. (E) ROC AUC curves showing the accuracy of the PHBR-II for classifying the extracellular presentation of a residue by a patient’s HLA-DR genes for 7 donors (colors) and for all donors combined (black). The aggregated PHBR-II presentation scores for the 7 donors expression HLA-DR alleles was compared to a sent of random residues for the same HLA-DR alleles. See also Figure S1, Figure S2 and Table S1.
Next, we combined single molecule residue-centric presentation scores into an MHC-II genotype score. Previously, MHC-I single allele best rank scores were combined using the harmonic mean resulting in the Patient Best-rank Harmonic Mean (PHBR-I) score, as this outperformed all other tested formulations. To create an analogous score for MHC-II, we modified the PHBR-I score to account for the different composition of MHC-II molecules. The MHC-II genotype comprises 2 copies each of HLA-DR alpha and beta, HLA-DP alpha and beta and HLA-DR alpha and beta. HLA-DRA is the only non-variable gene in the population, resulting in only 2 possible HLA-DR heterodimers. Each individual can form four possible alpha-beta heterodimers from HLA-DP and HLA-DQ. This results in a total of 10 possible unique heterodimeric MHC-II molecules (Figure 1B). To weight each gene equally in the final presentation score, each HLA-DRB1 allele is considered twice, bringing the total number of complexes to twelve. To evaluate the combined effect of these complexes on the presentation of a residue, the best rank score is calculated for all twelve complexes and those twelve values are combined using the harmonic mean to create a PHBR-II score (Figure 1C).
To assess the performance of the PHBR-II score at predicting extracellular presentation, we compared the scores for peptides derived from several multi-allelic HLA-DR expressing cell lines against matched scores for randomly derived peptides (Ciudad et al., 2017) (Figure 1D). The combined AUC across all cell lines was 0.69 (Figure 1E). This formulation of the PHBR-II score out-performed another scoring variation where peptides of varying lengths were considered (Figure S1). Two reasons contribute to the reduced performance relative to MHC-I (ROC AUC 0.75) (Marty et al., 2017). First, predicting single allele MHC-II binding has higher error than predicting single allele MHC-I binding (Andreatta et al., 2018; Paul et al., 2015; Wang et al., 2008). Second, computing an AUC value requires a non-binding negative set of residues. We employ a random set of residues when evaluating PHBR scores for both MHC classes; however, MHC-II has a larger effective binding range than MHC-I. As a result, the negative set should have an order of magnitude more actual binding residues for MHC-II than MHC-I. Thus, lack of an appropriate negative set for MHC-II deflates the calculated AUC value. For this application, namely using predicted MHC class II binding affinities to identify T cell epitopes for which the exact restricting MHC class II molecule is not known, performance measured by AUC values is typically around 0.7 (Alessandro Sette and Peter Bjoern, private communication). Despite these limitations, the PHBR-II score contains significant signal that renders it useful for further analysis.
Finally, we applied the HLA-HD tool (Kawaguchi et al., 2017) to predict HLA-II alleles for patients in TCGA with exome sequencing data (Table S1). To the best of our knowledge, HLA-HD is currently the only tool that can call alpha and beta alleles for HLA-DR, HLA-DP and HLA-DQ with high accuracy. Thus, from a total of 8,333 patients with exome sequencing, we successfully typed 7,929 patients at all three genes. To validate these HLA types, we also applied xHLA (Xie et al., 2017), which calls the beta alleles for HLA-DR, HLA-DP and HLA-DQ. We restricted our patient set to samples where both HLA-HD and xHLA completely agreed, leaving 5,942 patients (Table S1, Figure S2A). Within the typed TCGA patients, HLA-DPA1 revealed the least population variation, with only 14 types represented and the most common allele (HLA-DPA1*0103) at a frequency of 0.76 in the population. HLA-DRB1 had the most variation in the population, with 74 types represented, the most common of which (HLA-DRB1*0701) was observed at only a frequency of 0.20 (Figure S2B-F).
Recurrent cancer mutations are poorly presented by human MHC-II
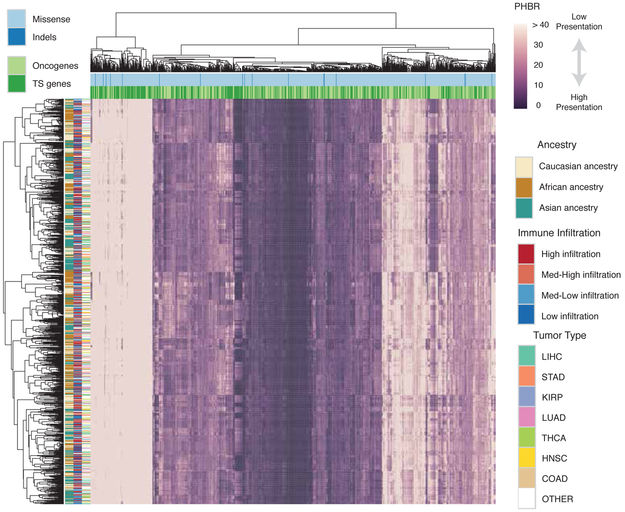
Mutations that drive the early development of tumors should be observed more frequently across tumors (Lawrence et al., 2013). We therefore used recurrence of mutations in established oncogenes and tumor suppressors (Davoli et al., 2013) as criteria to assemble a list of 1,018 cancer driving mutations likely to have occured prior to immune evasion and that could therefore reflect the effects of selection by immunosurveillance. We calculated PHBR-II scores for every mutation-patient combination, resulting in a matrix of 5,942 patients (Table S2, Figure 2; rows) and 1,018 mutations (Figure 2; columns). The matrix provides a high-level overview of the MHC-II presentation landscape across cancer patients and recurrent cancer mutations. Patients and mutations were clustered according to similarity of presentation score profiles. While we observed no obvious clustering of patients by tumor type or infiltration by CD4+ T cells (Newman et al., 2015), we did observe expected clusters of samples with shared ancestry, resulting from population-specific differences in MHC-II allele frequencies. Interestingly, we observed bias toward poor presentation of tumor suppressor mutations by MHC-II across the entire population (Fisher’s Exact Test, PHBR-II ≥ 10, OR (Odds Ratio)=1.43, p=0.006). Notably, this same enrichment was not present for MHC-I presentation (Fisher’s Exact Test, PHBR-I ≥ 2, OR=1.33, p=0.40). Although only a small fraction of the tested mutations were inframe indels, there was no clear difference between the MHC-II presentation of missense mutations and indels. Interestingly, when a similar matrix was generated using the wild type sequences instead of the mutations, the presentation of the sequences across the population were highly concordant (Pearson r = 0.96, Figure S3A-B).
Figure 2:
Pan-cancer overview of patient-mutation MHC-II presentation. A clustered heatmap of patients in TCGA with the 1,018 frequent cancer mutations. Only 1,050 ancestry-distributed, patients are included for spatial reasons. The heatmap is colored by PHBR-II score. Column and row coloring highlight groupings of patients and mutations into different categories. Tumor suppressor is abbreviated as TS. See also Figure S3 and Table S2.
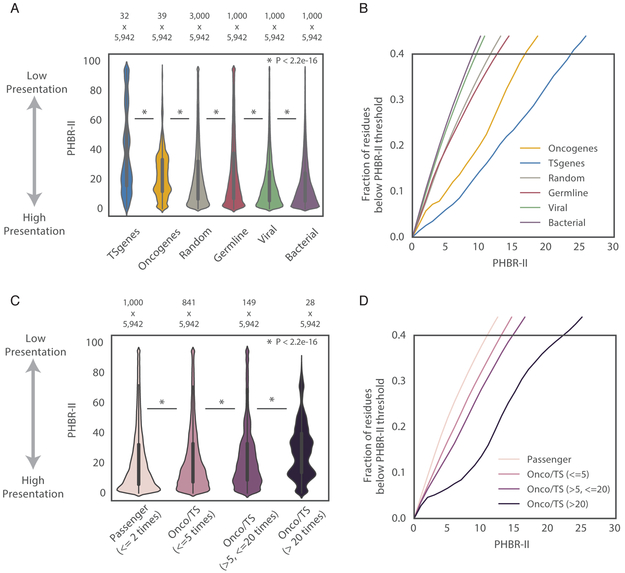
Next, we compared the ability of the 5,942 cancer patients to present different classes of residues by MHC-II. We calculated the PHBR-II scores of every patient for 1,000 viral residues, 1,000 bacterial residues, 1,000 common polymorphisms and 1,000 random mutations (Marty et al., 2017). To compare the behaviors of PHBR-II scores, we visualized raw distribution and the cumulative distribution function (CDF) for each class of residues. Viral and bacterial residues were presented the most effectively out of these classes by the patients in the population (Figure 3A). Assuming that the MHC-II system has primarily evolved to ward off pathogens, it is not surprising that the CDF curves are shifted to the left in comparison with other classes, with more than 27% of viral and 29% of bacterial PHBR-II scores falling below a PHBR-II threshold of 6 (threshold based on 0.2 false positive rate) (Figure 3B, Figure S4A) (Table S3 for confidence intervals (CI)). Common germline polymorphisms and random mutations should in contrast approximate events that are selectively neutral. MHC-II presentation of germline variants should in principle be decoupled by tolerance such that germline variants should not be biased to occur in particularly well or poorly presented peptides. Similarly, randomly selected mutations should represent an unbiased sample of background MHC-II presentation. Consistent with positive selection, pathogen residues are presented significantly better than germline variants or random mutations by MHC-II across the population than, yet still 22% and 23% of PHBR-II scores fall below the 6 PHBR-II threshold, for common germline polymorphisms and random mutations, respectively. In contrast, distributions of PHBR-II scores for recurrent mutations in oncogenes and tumor suppressors (observed >10 times in MHC-II typed population) show a shift upward toward poor presentation relative to random mutations (p < 2.2e-16), with only 12% of scores for mutations in oncogenes falling below the 6 PHBR-II threshold. Strikingly, there was even poorer presentation of mutations in tumor suppressor genes (p < 2.2e-16; relative to random mutations), with only 7% of PHBR-II scores below the 6 PHBR-II threshold. The differences observed in MHC-II presentation for these classes of mutation were robust to to the inclusion of less recurrent (observed >2 times in TCGA) cancer mutations (Figure S4B) and to using different samples of random mutations (Figure S4C, empirical p < 0.05). Interestingly, these trends were not unique to cancer patients but were also observed in alternate human populations, suggesting that MHC-II genotypes do not significantly differ between the two populations (Figure S4D).
Figure 3:
Recurrent cancer mutations have low MHC-II presentation in the TCGA population. (A) A violin plot denoting the distribution of PHBR-II presentation scores across all patients in TCGA for 6 different classes of residue. Tumor Suppressor is abbreviated as TS. Mutations observed >10 times in TCGA are displayed. (B) Cumulative distribution functions (CDF) for the 6 different classes of residue. (C) A violin plot with the distribution of somatic mutations occurring at different frequencies: passenger mutations in non-cancer implicated genes observed ≤2 in TCGA and mutations in cancer implicated genes observed 3-10 times, 11- 40 times and >40 times in TCGA. (D) CDFs for somatic mutations occurring at different frequencies. For both (A, C), the white dots represent the median, the thick dark grey lines denote the interquartile of the data and the thin dark grey lines denote the 1.5 IQR range. See also Figure S4 and Table S4.
We next evaluated whether the recurrence of a mutation was related to its presentation by MHC-II by comparing the PHBR-II score distributions of passenger mutations and varying frequencies of cancer-driving mutations (Figure 3C). Passenger mutations, defined as mutations occurring only 1-2 times across all tumors in non-cancer genes, had a PHBR-II score distribution very similar to that of random mutations with an enrichment for PHBR-II scores near 0, suggesting that many passengers are likely to be effectively presented. This enrichment of presented passenger mutations is consistent with recent reports that HLA loss of heterozygosity is frequent in some tumor types (Chowell et al., 2017; McGranahan et al., 2017; Ryschich et al., 2004) and is associated with the accumulation of mutations that would have been effectively presented by the lost allele (McGranahan et al., 2017). Consequently, 25% percent of the passenger mutation PHBR-II scores fall below the PHBR-II cutoff of 6 (Figure 3D). In comparison, we observed significantly worse presentation with increasing mutation frequency for recurrent mutations (observed > 2 times across typed tumors) in known cancer genes (p < 2.2e-14). The percentage of PHBR-II scores falling below the PHBR-II threshold of 6 falls with each jump in frequency; from 20% for low frequency driver mutations (<=5 times; 841 total) to 16% for medium frequency driver mutations (>5, <=20 times; 149 total) to a dramatic 8% for high frequency driver mutations (>20 times; 28 total) (Figure 3D). Despite the striking shift toward larger PHBR-II scores with increasing recurrence, MHC-II presentation across patients was not quite significantly correlated with mutation frequency (burden) across tumors overall (Spearman rho=0.27, p=0.07, Figure S4E). This is in contrast to the relationship observed for MHC-I (Spearman rho=0.66, p=1.02e-6 within the same patient group). We note that median PHBR-II scores for mutations observed >10 times tend to be elevated equivalently. This may reflect a threshold beyond which presentation no longer occurs, and thus beyond which numeric differences in PHBR-II score should no longer be informative about mutation frequency. Taken together, these results suggest that MHC-II based presentation across the human population constrains the frequency at which mutations arise across tumors.
MHC-II genotype constrains the landscape of cancer mutations in individual tumors
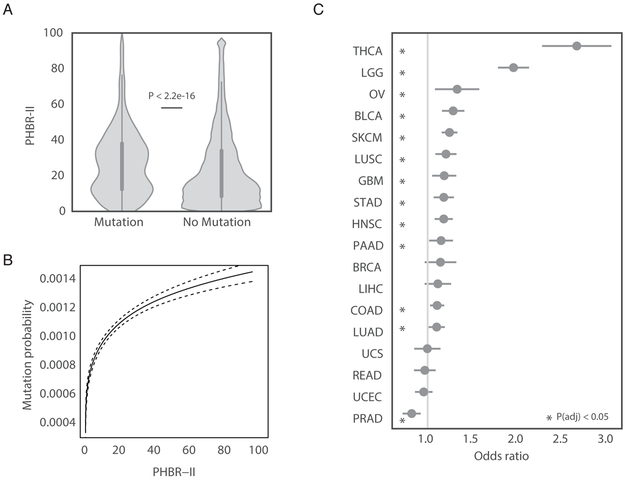
Given observed bias for cancer mutations to be poorly presented by human MHC-II (Figure 3A), we hypothesized that MHC-II genotype could influence patient-specific mutation probability. To explore this hypothesis, we intersected occurrence of mutations with potential of an individual to present those mutations as quantified by their PHBR-II score. PHBR-II scores were separated into two groups: those that corresponded to observed mutations and those that corresponded to unobserved mutations (Figure 4A). Consistent with our hypothesis, we observed a large upward shift in PHBR-II distribution for the observed mutations as opposed to the unobserved mutations. As mutations become less presentable (higher PHBR-II), the probability of mutation increases significantly (Figure 4B), with the most pronounced increase occurring at lower PHBR-II scores.
Figure 4:
Patient-specific MHC-II genotype influences mutation probability. (A) A violin plot denoting the difference in PHBR-II scores when the 5,942 patients are split by mutation occurrence, considering only mutations observed ≥2 times across tumors. (B) Nonparametric estimate of the logit-mutation probability as a function of PHBR-II scores considering mutations observed ≥2 times across tumors (C) The MHC-II ORs (grey circles) and 95% CIs associated with a 1-unit increase in log-PHBR-II score for different cancer types. See also Table S4 and Table S5.
Next, we used a logistic regression with non-linear effects to model the relationship between MHC-II genotype and the probability of observing a recurrent somatic mutation in a pan-cancer setting. We found a substantial increase in odds of acquiring a mutation as PHBR-II scores increased (OR=1.23, p < 9.9e-58, Table 1). Importantly, passenger mutations, established non-driver mutations (Table S4) and germline polymorphisms did not exhibit the same increase (OR=1.00, OR=0.99 and OR=0.99 respectively, Table 1). In addition, the OR decreased when less stringent HLA type calls were used (OR=1.20), suggesting the importance of accurate HLA typing. Since the immune environment can vary considerably across tissue sites, we revisited our analysis for each tumor type separately (Figure 4C, Table S5). Twelve of the eighteen tissues had significant positive ORs (p <0.05) after multiple testing correction. Similar to MHC-I, MHC-II genotype had the strongest effect in thyroid cancer; however, the effects of MHC-II were even greater than MHC-I (OR=2.63 versus OR=2.21, considering only thyroid cancer patients with confident MHC-I and MHC-II typing) (Figure 4C).
Table 1:
The association between PHBR-II score and mutation occurrence. ORs, 95% CIs and p values are shown for logistic regression model relating PHBR-II scores to set of mutations observed ≥2 times in set of tumors. Models relating PHBR-II score to sets of passenger mutations, non-driver mutations and germline variants serve as controls. See also Table S4 and Table S5.
| MHC-II PHBR | |||
|---|---|---|---|
| Odds ratio | 95% CI | P-value | |
| ≥2 mutations | 1.23 | (1.19, 1.26) | 9.9e-58 |
| Passenger mutations | 1.00 | (0.94, 1.06) | 0.99 |
| Non-driver mutations | 0.99 | (0.96, 1.04) | 0.96 |
| Germline variants | 0.99 | (0.99, 0.99) | 5.8e-07 |
MHC-II works together with MHC-I to influence mutation probability in individual tumors
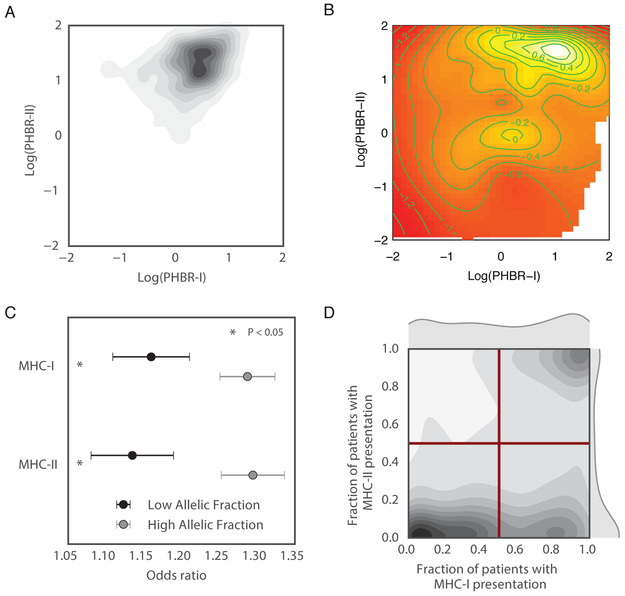
We previously established the influence of germline MHC-I genotype on the probability of observing specific mutations in tumors (Marty et al., 2017). To assess the combined influence of MHC-I and MHC-II on mutation probability, we evaluated the correlation between PHBR-I and -II scores across recurrent cancer mutations. The range and distribution of PHBR-I and -II scores differs substantially (Figure S5A), and while lower PHBR scores are indicative of more effective presentation in both cases, the range of values where most presentation takes place is expected to differ as MHC-II binds peptides with lesser stringency for peptide affinity and more promiscuity than MHC-I. These differences suggest the potential for MHC-I and MHC-II to contribute to presentation and thus constrain mutation probability in complementary ways. Indeed, we observed only a weak positive correlation between PHBR-I and -II score distributions across recurrent cancer mutations (Spearman rho=0.36; Figure 5A, Figure S5B). Consequently, we modeled the relationship between the probability of observing a mutation and both classes of PHBR scores across the 1,018 recurrent mutations (Figure 5B). Mutations with low PHBR scores (effective presentation) for either class had a much lower probability of being observed in tumors than mutations that had high PHBR scores (poor presentation) for both classes.
Figure 5:
MHC-II and MHC-I presentation have different effects. (A) A kernel density plot with the density of PHBR-II and -I scores across cancer driving mutations. (B) A heatmap of mutation probability for all combinations of PHBR-II and -I scores. Dark red represents low probability and white represents high probability. (C) The MHC-I and MHC-II ORs (grey circles) and 95% CIs associated with a 1-unit increase in log-PHBR-II score. Results are shown for mutations with low allelic fraction (dark grey) and high allelic fraction (light grey). (D) A kernel density plot showing the density of mutations according to the fraction of patients who can present it with MHC-I and MHC-II. The red bars denote the four quadrants of the graph. See also Figure S5 and Table S6.
To quantify the influence of MHC-I and MHC-II on probability of mutation, we used an additive logistic regression model with non-linear effects that incorporated both PHBR-I and -II scores in the pan-cancer setting. Since the distributions of PHBR-I and -II are very different, we calculated the odds ratios between the 25th and 75th percentile PHBR, such that the odds ratio represents the increase in odds of observing a mutation amongst individuals with a high PHBR score relative to a low PHBR score for each MHC class. Notably, we found the impact of MHC-II on the probability of a mutation to be larger than the impact of MHC-I (single model incorporating both classes: OR=1.74 with CI [1.67, 1.80] and OR=1.60 with CI [1.54, 1.64], respectively). To better understand the relative effects of presentation by MHC II versus MHC I in a tissue specific setting, we also estimated their individual effects on mutation probability in a joint model. Consistent with our pan-cancer analysis, we found MHC-II to have more extreme effect sizes in most tissues (Figure S5C).
The same driver mutations can occur early or late during tumor development; however, in a model where immune selection is impaired later in tumorigenesis by mechanisms of immune evasion, selection should be stronger on early clonal occurrences. Therefore, we further annotated mutations according to whether they were more likely clonal or subclonal based on relative allelic fraction of the mutations (Methods). Consistent with our assumption, likely subclonal mutations had decreased ORs relative to PHBR II and PHBR I scores (single class model, reference Table 1: PHBR-II OR=1.13 as compared to 1.21 for all mutations, PHBR-I OR=1.16 as compared to 1.20 for all mutations, Figure 5C), confirming that subclonal events are subject to weaker selection. Moreover, when restricting analysis of selection to likely clonal mutations, ORs for both PHBR II and PBHR I scores increased (single class model, reference Table 1: PHBR-II OR=1.29 as compared to 1.21 for all mutations, PHBR-I OR=1.29 as compared to 1.20 for all mutations). Though mutation calls may be less confident for subclonal mutations, these results suggest that true effect sizes may be higher than previously reported.
Differences in MHC-II versus MHC-I presentation specificities
Next, we explored whether practical differences exist in the presentation of particular driver mutations by MHC-II versus MHC-I. We compared the fraction of patients wherein a mutation was presented by MHC-II with the same fraction for MHC-I (Figure 5D, Table S6), and further divided mutations into four categories: rarely presented by either MHC-I or MHC-II, more frequently presented by MHC-I, more frequently presented by MHC-II and frequently presented by both. Interestingly, we observed that MHC-II-based presentation tended to be bimodal, such that a mutation was presented by most patients, or by almost no patients, with a few notable exceptions including KRAS G12 (Figure S5D). In contrast, MHC-I-based presentation spanned the full range, with many mutations presented in varying fractions of patients. Though these trends may be impacted by the higher sensitivity of the PHBR-I score as compared to the PHBR-II score, they were constant across several thresholds (Figure S5E). This suggests that MHC-II-based presentation may be more shared across patients, whereas MHC-I based presentation is more individual specific. We further investigated the mutations frequently presented by both MHC-I and MHC-II because we would expect them to arise with low likelihood in cancer. Indeed, these mutations had lower allelic fractions than mutations presented well by at least MHC-I or MHC-II (Mann-Whitney, p=0.03), suggesting these mutations are subclonal, arising after immune evasion and could be effectively eliminated by the immune system.
Based on this analysis, the relative abundance of class I peptides appears to be higher than that for class II, suggesting better potential for engineering class I anti-tumor responses; however, recents reports suggest a bias for responses to be CD4+ driven in practice (Ott et al., 2017). This could indicate that TCR availability is a major bottleneck for effective CD8+ immune responses.
Evidence for distinct effects of class II versus class I driven immunosurveillance
Differences in the dynamics of peptide presentation and immune response for MHC-I versus MHC-II may have important implications for tumor-immune interactions. Whereas MHC-I binds peptides with high specificity, MHC-II binds a broader array of peptides with a high degree of promiscuity. CD4+ T cells activated by MHC-II-peptide complexes can play either a regulatory or an effector role, whereas CD8+ T cells are strictly (cytotoxic) effectors. The different properties of class I and II based immunity (Unanue et al., 2016; Yewdell and Haeryfar, 2005) are essential for an effective defense against pathogens, but the implications for anti-tumor responses are less clear. We therefore sought to further quantify the potential for these distinct roles to introduce measurable differences between class I and class II mediated immunosurveillance during tumor development.
Because of its established regulatory role in cancer, we reasoned that MHC II driven immunosurveillance could have a larger effect on the immune microenvironment than MHC I. Using CIBERSORT (Newman et al., 2015) to evaluate infiltration by different immune cell types into tumors, we sought to identify a relationship between immune infiltrates, cytotoxicity score (Rooney et al., 2015) and strength of immune selection. We divided patients into groups based on their immune infiltrates and cytotoxicity scores and tested for differences in immune selection (Figure S6A-D), but did not find any significant relationships. This apparent lack could be an artifact of the timing of the MHC-imposed selection relative to when the RNA samples were taken.
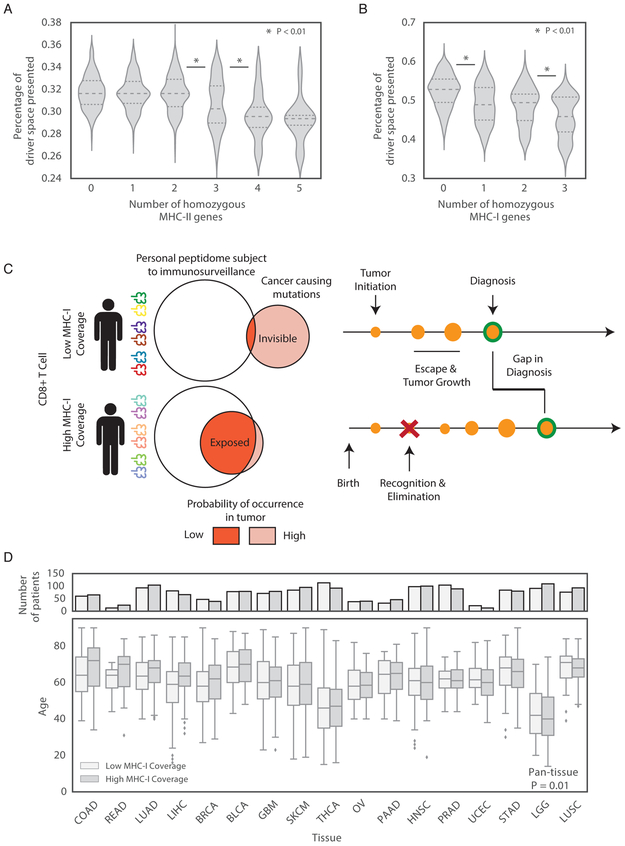
Population level variation in effectiveness of cancer-relevant immunosurveillance could also relate directly to cancer susceptibility. We reasoned that patients whose MHC genotype could present a larger fraction of driver mutations to the immune system would be more resistant to developing cancer. As homozygous genotype at MHC alleles could reduce the diversity of presented peptides, we compared presentation across patients with different levels of homozygosity. We quantified coverage of cancer causing mutations as the fraction of the 1,018 driver mutations that could be presented by the MHC-II genotype of each patient (Methods) and henceforth refer to this fraction as MHC-II coverage. As expected, patients with more homozygous MHC-II alleles were able to present a smaller fraction of the space due to their decreased MHC diversity (Figure 6A). MHC-I (using a PHBR-I cutoff of 2 (Nielsen and Andreatta, 2016)) and showed a similar trend (Figure 6B).
Figure 6:
MHC presentation of driver mutations and cancer susceptibility. (A-B) Violin plots depicting the distributions of the percentage of the 1,018 driver mutations presented by (A) MHC-II and (B) MHC-I for patients with varying numbers of homozygous genes. (C) A schematic showing the effect of MHC coverage on age at diagnosis. (D) A box plot of the distributions of age at diagnosis for patients separated by tumor type and percentage of the driver space presented for MHC-I. See also Figure S6 and Figure S7.
Next, we asked whether higher MHC coverage could delay the development of cancer. We reasoned that if two patients acquired a cancer driving mutation at the same time, the patient with higher MHC coverage would be more likely to expose their mutation to the immune system and stop expansion of the cancer. Thus, high MHC coverage should lead to diagnosis with cancer later in life, and vice-versa (Figure 6C). First, we tested MHC-II, but found no relationship between age at diagnosis and coverage (p = 0.51, Figure S7A). In contrast, patients with higher MHC-I coverage of driver mutations were more often diagnosed with cancer at a later age (p = 0.01, controlling for tumor type and ancestry, Figure 6D). Across tumor types, the 5% of patients with the highest MHC-I coverage were diagnosed with cancer 4 years later than the 5% of patients with the lowest coverage (p=0.004, Figure S7B), versus a 2 year difference when the highest and lowest 10% was used (p=0.02). Across tumor types, hepatocellular carcinoma showed the most significant difference after multiple testing correction and was diagnosed on average 7 years earlier when MHC-I coverage was low. Though coverage of driver and passenger mutations was strongly correlated (MHC-I Pearson R=0.79, MHC-II Pearson R=0.68), the significant association with age at diagnosis with MHC-I coverage was not observed for passengers (p=0.11). Within tumor types, MHC-I coverage did not correlate with overall mutation burden (Figure S7C). These findings suggest that the effect on age is specific to MHC-I coverage of driver mutations rather than to effects of coverage on mutagenesis in general. Using the number of homozygous MHC-I genes in place of coverage showed the same association with age at diagnosis but was more granular since patients fall into discrete bins of homozygous genes counts (p=0.024).
The observation that MHC-I but not MHC-II coverage is correlated with age at diagnosis supports a protective role for CD8+ driven cytotoxicity. The lack of association with MHC-II suggests that MHC-II driven CD4+ effector responses against key driver mutations are weaker than CD8+ responses. In addition, either the regulatory role of CD4+ driven immune responses does not depend on coverage of driver mutations or, as indicated in Figure 2, low variance in inter-patient coverage by MHC-II causes this effect to be undetectable.
Discussion:
The role of MHC-II in immunity to cancer is of clear relevance (Haabeth et al., 2014; Hung et al., 1998; Wang, 2001). MHC-II represents a tumor-autonomous phenotype that predicts therapeutic response to immune checkpoint inhibitors (Johnson et al., 2016). CD4+ T cells system-wide play a central role in the initiation of an anti-tumor response in the context of immune checkpoint inhibition (Spitzer et al., 2017) and the response to neoantigens (Zanetti, 2015). Surprisingly, melanoma patients immunized against autologous tumor neopeptides predicted to bind MHC-I, mounted preferentially MHC-II-restricted CD4+ T cell responses (Ott et al., 2017). Besides effector functions (Haabeth et al., 2014), CD4+ T cells exert important regulatory function contributing in a fundamental way to the activation and maintenance of CD8+ and CD4+ T cell responses (Cassell and Forman, 1988; Gerloni et al., 2000; Janssen et al., 2003; Langlade-Demoyen et al., 2003; Shedlock and Shen, 2003; Sun and Bevan, 2003; Sun et al., 2004). Thus, it appears that MHC-II and MHC-II restricted T cell responses play a pivotal role in the anti-tumor response independent of the role of MHC-I and MHC-I-restricted T cell responses.
To assess how MHC-II genotype impacts emerging tumors, we systematically interrogated the role of MHC-II genotype in shaping tumor genomes using a quantitative PHBR-II score representing the ability of an individual’s MHC-II genotype to present specific residues. We investigated the relationship between MHC-II genotype, somatic mutation probability and tumor susceptibility across thousands of tumors.
Recurrent cancer mutations were presented more poorly by MHC-II genotypes across the population than either pathogenic residues or passenger mutations. The strong MHC-II presentation of pathogenic residues suggests an evolutionary advantage to MHC-II genotypes able to present and evoke elimination of infection (Hedrick, 2002). On the other hand, the difference in MHC-II presentation between recurrent driver and passenger mutations suggests that tumors develop under changing selective pressure from tumor surveillance by CD4+ T cells, suggestive of immunoediting. As the tumor grows and activates mechanisms to evade the immune system, MHC-II presentation becomes less important and passenger mutations are acquired regardless of their affinity to MHC-II.
In individual tumors, we found that MHC-II presentation impacts the probability of a patient acquiring a driver mutation, using passenger mutations and germline variants as negative controls. Because the most frequently observed mutations in the TCGA cohort are those that are not presented by most HLA genotypes, shuffling HLA alleles does not result in a significant change to the effect sizes estimated by our models. We anticipate that carefully designed experiments that provide serial monitoring of mutation profiles in the presence of an active immune system may be required to understand the true strength of MHC-driven selection. Differences in the effect of MHC-II presentation across tumor types further suggests that this selection is greater in some tissue settings than others. The differences could also reflect some of the distinct molecular alterations of each tumor type. For example, the strongest effects were observed in thyroid cancer, which is characterized by frequent BRAF mutations and RAS mutations (NRAS Q61R, HRAS Q61R and NRAS Q61K) (Cancer Genome Atlas Research Network, 2014) that are all poorly presented by MHC-II. The weakest effects occurred in prostate cancer, which has a somatic landscape dominated by gene fusion and copy number alterations but fewer somatic point mutations (Cancer Genome Atlas Research Network, 2015). Developing strategies to measure the effect of MHC-II selection on gene fusions and copy number alterations could provide further insight into the apparent tumor-type specific differences and role of MHC-II in specific tumor types.
Although gene expression was shown to be a determinant of MHC-driven selection across all somatic mutations (Yang et al., 2017), our analysis focused on a small number of driver mutations that should be enriched for early events in tumor development. Since gene expression in the TCGA was measured from samples taken at diagnosis, it may not accurately reflect the levels of expression at earlier times during tumor development; therefore, we omitted expression levels from our model. We also found no relationship between MHC-II based mutation and expression-derived estimates of tumor infiltrating immune cells. Indeed, while high levels of T cell infiltration at the time of diagnosis is associated with longer disease-free survival (Galon et al., 2013), T cell densities decrease along with tumor progression (Bindea et al., 2013). Thus, our data may reflect the evolving nature of immune infiltration in tumors.
Notably, MHC-II had stronger effects than MHC-I in shaping the driver mutations of a tumor. Interestingly, these effects appear to be less patient-specific than MHC-I, perhaps due to the promiscuous nature of MHC-II peptide binding. Furthermore, these effects could be driven by a faster evasion of MHC-I presentation than MHC-II presentation due to mechanisms like HLA mutation or HLA loss of heterozygosity that would occur within the tumor but are unlikely to affect the MHC-II on professional APCs (McGranahan et al., 2017; Shukla et al., 2015). Another possibility is that MHC-II presentation and CD4+ T cell recognition may be a necessary prerequisite to CD8+ T cell cytotoxicity and tumor elimination, in agreement with the regulatory role of CD4+ T cells. We reason that the stronger effect of MHC-II on the odds of acquiring a mutation is consistent with a dual regulatory and effector CD4+ role. If the role of CD4+ T cells was purely regulatory, MHC-I specificity would be expected to drive mutation probability. Therefore, the role of the MHC-II genotype and MHC-II presentation needs to be properly weighted to understand the role of the interplay between mutational burden and tumor evolution. This understanding will be essential in the development of immunotherapies, likely being a critical component of their future success.
Early detection, diagnosis and treatment of tumors is a major determinant of patient morbidity and mortality. Accurate predictions of when, where and how tumors are likely to arise would have enormous implications for cancer screening and could improve survival rates. While the main contributor to the development of most adulthood tumors is sporadic somatic mutation, germline variants have been implicated as a determinant of tumor characteristics (Carter et al., 2017). Here, we propose that the MHC-II genotype is an additional such germline influence.
We also noted a direct association between patient MHC-I diversity and age at diagnosis. Interestingly, we do not see the same age at diagnosis associations with MHC-II. This observation along with the increased effect of MHC-II as compared to MHC-I over mutation selection suggests different roles of the two MHC classes. MHC-II appears to exert a stronger selective pressure, leading to a stronger effect on somatic mutation probability. This role aligns with the understanding of CD4+ T cells as a necessary component of the activation and regulation of CD8+ T cells. On the other hand, the diversity of an individual’s MHC-I may play a role in tumor susceptibility, but MHC-I appears to have weaker effects on mutation selection. Importantly, our estimate of the protective effect of MHC-I is based on an incomplete list of early cancer driving mutations, and may therefore underestimate the magnitude of the effect.
These findings suggest that the combination of MHC genotype with other relevant information such as germline risk factors or mutagenic exposures could improve prediction of cancer susceptibility. In addition, future improvement of the algorithms that predict peptide binding to MHC-II molecules based on affinity, which currently lag behind such algorithms for MHC-I, could lead to better accuracy in predicting presentation. Improvements in predictive algorithms will increase our ability to predict cancer risk and predispositions. Importantly, while MHC-restricted presentation is the initial event in antigen-specific T cell activation, it may not be the only factor required for subsequent elimination by T cell. Thus, accurate prediction of presentation and T cell activation may be required to predict the occurrence of specific mutations. Finally, in addition to the well-recognized roles of MHC and T cells, one needs to factor in additional mechanisms of immune escape and intratumor heterogeneity such as polarization of myeloid cells (Zelenay et al., 2015), the local effects of ER stress response (Cubillos-Ruiz et al., 2015; Rodvold et al., 2017), and NK cells (O’Sullivan et al., 2012).
In conclusion, we found that predicted MHC-II presentation of cancer-related somatic mutations shape tumor development through variation in antigen presentation in complementary fashion to MHC-I, highlighting the need to consider the independent yet complementary roles of CD4+ and CD8+ T cells in the selection and elimination of tumors.
STAR Methods
Contact for Reagent and Resource Sharing
“Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Hannah Carter (hkcarter@ucsd.edu).”
Method Details
Data Acquisition
Data were obtained from publicly available sources including The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov/), The Allele Frequency Net Database (González-Galarza et al., 2015), Ensembl, Exome Variant Server, UniProt (The UniProt Consortium, 2014), or cited literature (Ciudad et al., 2017). TCGA normal exome sequences and TCGA clinical data were also downloaded from the GDC on September 21-24th, 2017 and April 25th, 2017 respectively. Furthermore, TCGA somatic mutations were accessed from the NCI Genomic Data Commons (https://portal.gdc.cancer.gov/) on May 14th, 2017. Population level HLA frequencies were obtained from the Allele Frequency Net Database on October 9th, 2015. Common germline variants were downloaded from the Exome Variant Server NHLBI GO Exome Sequencing Project (ESP), Seattle, WA on August 13, 2015. Finally, viral and bacterial peptides were obtained from UniProt on October 13th, 2015.
Single Allele Presentation Score Construction
To create a residue-centric presentation score, we evaluated allele-based ranks for peptides containing the residue of interest. Each allele-based rank was predicted using the NetMHCIIPan-3.1 tool, downloaded from the Center for Biological Sequence Analysis on August 15, 2016 (Karosiene et al., 2013). NetMHCIIPan-3.1 takes a peptide and an MHC-II protein (HLA-DRB1, HLA-DPA1/DPB1 or HLA-DQA1/DQBP) and returns binding affinity IC50 scores and corresponding allele-based ranks. Peptides with rank < 10 and < 2 are considered to be weak and strong binders respectively. Allele-based ranks were used to represent peptide binding affinity. Based on previous analysis (Marty et al., 2017), we previously established the best rank of possible peptides containing the residue as an effective estimator of extracellular presentation. Here we evaluated two approaches to selecting the set of peptides containing the residue to consider:
All 15-mers: Every peptide of length 15 containing the residue of interest, totaling 15 peptides.
13-mers through 25-mers: Every peptide of length 13 through length 25 containing the peptide, totaling in 247 peptides (Wieczorek et al., 2017).
Insertion and deletion mutations were modeled by the resulting peptides that differed from the native sequence and tested with the same peptide-set parameters. These two peptide selection models were compared based on performance in a multi-allelic setting and all 15-mers model was selected (see below).
Multi-allele Presentation Score Construction
We defined a patient presentation score to represent a particular patient’s ability to present a residue given their distinct set of 12 HLA-encoded MHC-II molecules (4 combinations of HLA-DPA1/DPB1 and HLA-DQA1/DQB1; 2 alleles of HLA-DRB1 considered twice each -- since HLA-DRA1 is invariant -- for consistency between resulting molecules). The Patient Harmonic-mean Best Rank (PHBR) score was assigned as the harmonic mean of the best residue presentation scores for each of the 12 MHC-II molecules A lower patient presentation score indicates that the patient’s MHC-II molecules are more likely to present a residue on the cell surface.
Mass Spectrometry-based Presentation Score Validation
In order to test the performance of the different peptide sets that could compose the multi-allelic PHBR score to predict presentation, we used published MS data for 7 cell lines expressing 2-3 HLA-DRB1 alleles typed to the fourth digit (Ciudad et al., 2017). Ciudad et al. catalogs peptides observed in complex with MHC-II (HLA-DR) on the cell surface for 7 different combinations of 2-3 HLA-DRB1 alleles, with 70 to 240 mappable peptides each. These data were combined with a set of random peptides to construct a benchmark for evaluating the performance of scoring schemes for identifying residues presented on the cell surface as follows:
Converting MS peptide data to residues: The Ciudad et al. MS data provides peptides observed in complex with the MHC-II, whereas our presentation score is residue-centric. For each peptide in the MS data, we selected the residue at the center (or one residue before the center in the case of peptides of even length) as the residue for calculating the residue-centric presentation score.
Selection of background peptides: We selected 3000 residues at random from the Ensembl human protein database (Release 89) (Aken et al., 2017) to ensure balanced representation of MS-bound and random residues. The randomly selected residues represent an approximation of a true negative set of residues that would likely not be presented on the cell surface. If this assumption is flawed, the resulting AUC will underestimate the true accuracy.
Scoring benchmark set residues: We calculated PHBR presentation scores with each peptide set for all of the selected residues from the Ciudad et al. data and the 3000 random residues against each of the 7 cell lines.
Evaluating scoring scheme performance using the benchmark: For each scoring scheme, scores were calculated for each cell line and pooled across the 7 cell lines. We plotted and compared ROC curves for each score formulation by calculating the True Positive Rate (% of observed MS residues predicted to bind at a given threshold) and the False Positive Rate (% of random residues predicted to bind at a given threshold) from 0 to 100 with steps of 0.5. Finally, we assessed overall score performance using the area under the curve (AUC) statistic. Based on this analysis, the 15-mer peptide set was used to construct the PHBR presentation score for all subsequent analyses.
HLA-II Typing
HLA genotyping was performed for genes HLA-DRB1, HLA-DPA1, HLA-DPB1, HLA-DQA1 and HLA-DQB1, which encode three protein determinants of MHC-I peptide binding specificity, HLA-DR, HLA-DP, and HLA-DQ. TCGA samples (Table S1) were typed with HLA-HD (Kawaguchi et al., 2017), using default parameters. HLA-HD requires germline (whole blood or tissue matched) whole exome sequenced samples. The tool reports 100% 4-digit validation accuracy across 90 low-coverage exomes. Samples with very low of coverage on specific genes are left untyped by HLA-HD. Patients were assigned an HLA-DR type if they were successfully typed for HLA-DRB1. Patients were assigned HLA-DP and -DQ types if they had successful typing for HLA-DPA1/HLA-DPB1 and HLA-DQA1/HLA-DQB1, respectively. Samples were validated by xHLA (Xie et al., 2017), run with default parameters, and only patients were all alleles agreed were included in the analysis (Table S1, Figure S2A). Allele frequencies were visualized with horizontal bar graphs (Figure S2B-F).
Selection of Recurrent Oncogenic Mutations, Passenger-like and Non-driver Mutations
Somatic mutations were considered to be recurrent and oncogenic if they occurred in one of the 100 most highly ranked oncogenes or tumor suppressors described by Davoli et al. (Davoli et al., 2013) and were observed in at least 3 TCGA samples. Among these, we retained only mutations that would result in predictable protein sequence changes that could generate neoantigens, including missense mutations and inframe indels. A total 1,018 mutations (512 missense mutations from oncogenes, 488 missense mutations from tumor suppressors, 11 indels from oncogenes and 7 indels from tumor suppressors) were obtained (Marty et al., 2017). All mutations observed in TCGA patients that did not fall into the 200 most highly ranked cancer genes were designated passenger-like mutations. Furthermore, we created an additional set of established non-cancer mutations. To do so, we selected a set of genes from Lawrence et al. that were known non-cancer genes and and selected mutations in these genes regardless of their recurrence in TCGA (Table S4) (Lawrence et al., 2013).
Selection of Other Classes of Residues
Peptides from pathogens, common germline human variants and randomly mutated human peptides were assembled for comparison with recurrent oncogenic mutations (Marty et al., 2017). The proteomes of 10 virus species and 10 bacterial species were downloaded from UniProt (The UniProt Consortium, 2014). One thousand residues were selected at random from both the viral and the bacterial set. A random set of mutations was generated by sampling 3,000 possible amino acid substitutions across human proteins from Ensembl (release 90; GRCh38) (Aken et al., 2017). A set of 1,000 common germline variants was sampled from the Exome Variant Server.
Generating Mutant Peptide Sequences
To allow determination of peptide sequences incorporating missense mutations, protein sequences were obtained from Ensembl (release 90; GRCh38) (Aken et al., 2017) and updated with the new amino acid. For indels, we modified the corresponding mature messenger RNA transcript sequences (CDS) by inserting or deleting nucleotides then translated the modified mRNA to protein sequence.
Patient Presentation Score-based Clustering
A matrix of PHBR scores was constructed with 5,942 TCGA samples as rows, 1,018 recurrent oncogenic mutations as columns, and PHBR score in each cell. The matrix was clustered using hierarchical agglomerative clustering on rows and columns. For convenience of visualization, a partial matrix is displayed in Figure 2. In order to use the dynamic range in heatmap color to display variation in patient presentation scores relevant to MHC-II based presentation, the PHBR color scheme only varies from 0 to 40. Colorbars provide additional information about patients and mutations, including ancestry, tumor type and T-cell infiltration levels (patients) and mutation type and gene category (mutations). CD4 T-cell infiltration was determined using CIBERSORT (Newman et al., 2015), an mRNA-based immune infiltration prediction algorithm. Patients were mapped to high, medium-high, medium-low and low CD4+ T-cell infiltration categories if their CIBERSORT scores fell into upper to lower quartiles respectively.
Comparison of Presentation Scores for Different Classes of Residue
PHBR presentation scores were calculated for 5,942 TCGA patients across different classes of residue including 71 highly-recurrent (≥10) oncogenic missense mutations, 1000 random amino acid substitution, 1000 germline variants, 1000 viral residues and 1000 bacterial residues (see Selection of Other Classes of Residues). Across categories, this resulted in 24,189,882 PHBR scores (oncogenes: 231,738, tumor suppressor genes: 190,144, random: 5,942,000, common: 5,942,000, viral: 5,942,000, bacterial: 5,942,000). The distributions of PHBR scores in each category were compared with Mann-whitney U tests and visualized with violin plots (Figure 3A). Furthermore, we plotted cumulative distributions to demonstrate the practical presentation of each class across several thresholds and calculated the confidence intervals of each curve with bootstrapping (Figure 3B, Table S4). Finally, we tested 20 independent sets of 1,000 random mutations to evaluate the confidence of the cumulative distributions (Figure S4C).
Generation of non-cancer population
As a control population, we used dbGaP samples (dbGaP accession numbers: Phs000398, Phs000254,Phs000632, Phs000209, Phs000290, Phs000179, Phs000422, Phs000291, Phs000631 and Phs000518) typed at MHC-II using HLA-HD (Kawaguchi et al., 2017), with default parameters and typed at MHC-I using Optitype (Szolek et al., 2014), with default parameters. Both tools require germline (whole blood or tissue matched) whole exome sequenced samples. We successfully typed the HLA-I genes for 1,386 patients and the HLA-II genes for 1,219 patients who had alleles in the netMHCpan-3.0 and the netMHCIIpan-3.1 database. This control population was used to look at the MHC-II population of different classes of peptides by a non-cancer specific population (Figure S4D). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession numbers: Phs000398, Phs000254, Phs000632, Phs000209, Phs000290, Phs000179, Phs000422, Phs000291, Phs000631 and Phs000518. We would like to acknowledge the following dbGaP studies and all of their contributors:
Phs000398.v1.p1: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C,HHSN268201100006C,HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). The authors thank the staff and participants of the ARIC study for their important contributions. This study is part of the NHLBI Grand Opportunity Exome Sequencing Project (GO-ESP). Funding for GO-ESP was provided by NHLBI grants RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO) and RC2 HL102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO). HeartGO gratefully acknowledges the following groups and individuals who provided biological samples or data for this study. DNA samples and phenotypic data were obtained from the following studies supported by the NHLBI: the Atherosclerosis Risk in Communities (ARIC) study, the Coronary Artery Risk Development in Young Adults (CARDIA) study, Cardiovascular Health Study (CHS), the Framingham Heart Study (FHS), the Jackson Heart Study (JHS) and the Multi-Ethnic Study of Atherosclerosis (MESA).
Phs000254.v2.p1: This study is part of the NHLBI Grand Opportunity Exome Sequencing Project (GO-ESP). Funding for GO-ESP was provided by NHLBI grants RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO) and RC2 HL102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO). Collection of the cystic fibrosis data and specimens was supported by Awards GIBSON07K0, KNOWLE00A0, OBSERV04K0, and RDP R026 from the Cystic Fibrosis Foundation; NHLBI grants R01 HL068890 and R01 HL095396; NCRR grant UL1RR025014 and NHGRI grant R00 HG004316.
Phs000632.v1.p1: This study is part of the NHLBI Grand Opportunity Exome Sequencing Project (GO-ESP). Funding for GO-ESP was provided by NHLBI grants RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO) and RC2 HL102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO). The Hematological Cancer specimens and data were collected in the laboratory of Dr. Benjamin L. Ebert, Brigham & Womens Hospital/Broad Institute, Boston, USA.
Phs000209.v13.p3: MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-RR-025005, and UL1-TR-000040.
Phs000290.v1.p1: Exome data provided by ARRA – NHLBI Lung Cohorts Sequencing Project 1RC2HL102923-01. The authors wish to thank the supported effort of the faculty and staff members of the Johns Hopkins University Bayview Genetics Research Facility and the Johns Hopkins University ‘Genomics and Genetics of Pulmonary Arterial Hypertension’ program (NIH P50 HL084946, P.M. Hassoun, NIH K23 AR52742–01, L.K. Hummers, and NHLBI F32 HL083714-01 S. C. Mathai).
Phs000179.v5.p2: This research used data generated by the COPDGene study, which was supported by NIH grants U01HL089856 and U01HL089897. The COPDGene project is also supported by the COPD Foundation through contributions made by an Industry Advisory Board comprised of Pfizer, AstraZeneca, Boehringer Ingelheim, Novartis, and Sunovion.
Phs000422.v1.p1: This study is part of the NHLBI Grand Opportunity Exome Sequencing Project (GO-ESP). Funding for GO-ESP was provided by NHLBI grants RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO) and RC2 HL102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO). The following NHLBI Severe Asthma Research Program (SARP) sites have contributed parent study data and DNA samples for exome sequencing in this project: Wake Forest School of Medicine (R01 HL069167), University of Wisconsin (R01 HL069116), University of Virginia, Cleveland Clinic (R01 HL069170), National Jewish Health, University of Pittsburgh (R01 HL069174), Washington University (R01 HL069149), Brigham and Women’s Hospital (R01 HL069349) and genotyping was supported by NHLBI HL87665 and 1RC2 HL101487).
Phs000291.v2.p1: This study is part of the NHLBI Grand Opportunity Exome Sequencing Project (GOESP). Funding for GO-ESP was provided by NHLBI grants RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO) and RC2 HL102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO). The authors wish to thank the supported effort of the faculty and staff members of the Johns Hopkins University Bayview Genetics Research Facility, NHLBI grant HL066583 (Garcia/Barnes, PI) and NHGRI grant HG004738 (Barnes/Hansel, PI). The Lung Health Study was supported by U.S. Government contract No. N01-HR-46002 from the Division of Lung Diseases of the National Heart, Lung and Blood Institute. The principal investigators and senior staff of the clinical and coordinating centers, the NHLBI, and members of the Safety and Data Monitoring Board of the Lung Health Study can be found at http://www.biostat.umn.edu/lhs/ and as follows: Case Western Reserve University, Cleveland, OH: M.D. Altose, M.D. (Principal Investigator), C.D. Deitz, Ph.D. (Project Coordinator); Henry Ford Hospital, Detroit, MI: M.S. Eichenhorn, M.D. (Principal Investigator), K.J. Braden, A.A.S. (Project Coordinator), R.L. Jentons, M.A.L.L.P. (Project Coordinator); Johns Hopkins University School of Medicine, Baltimore, MD: R.A. Wise, M.D. (Principal Investigator), C.S. Rand, Ph.D. (Co-Principal Investigator), K.A. Schiller (Project Coordinator); Mayo Clinic, Rochester, MN: P.D. Scanlon, M.D. (Principal Investigator), G.M. Caron (Project Coordinator), K.S. Mieras, L.C. Walters; Oregon Health Sciences University, Portland: A.S. Buist, M.D. (Principal Investigator), L.R. Johnson, Ph.D. (LHS Pulmonary Function Coordinator), V.J. Bortz (Project Coordinator); University of Alabama at Birmingham: W.C. Bailey, M.D. (Principal Investigator), L.B. Gerald, Ph.D., M.S.P.H. (Project Coordinator); University of California, Los Angeles: D.P. Tashkin, M.D. (Principal Investigator), I.P. Zuniga (Project Coordinator); University of Manitoba, Winnipeg: N.R. Anthonisen, M.D. (Principal Investigator, Steering Committee Chair), J. Manfreda, M.D. (Co-Principal Investigator), R.P. Murray, Ph.D. (Co-Principal Investigator), S.C. Rempel-Rossum (Project Coordinator); University of Minnesota Coordinating Center, Minneapolis: J.E. Connett, Ph.D. (Principal Investigator), P.L. Enright, M.D., P.G. Genomics & Genetics of the Lung Health Study June 10, 2011 version Page 6 of 8 Lindgren, M.S., P. O'Hara, Ph.D., (LHS Intervention Coordinator), M.A. Skeans, M.S., H.T. Voelker; University of Pittsburgh, Pittsburgh, PA: R.M. Rogers, M.D. (Principal Investigator), M.E. Pusateri (Project Coordinator); University of Utah, Salt Lake City: R.E. Kanner, M.D. (Principal Investigator), G.M. Villegas (Project Coordinator); Safety and Data Monitoring Board: M. Becklake, M.D., B. Burrows, M.D. (deceased), P. Cleary, Ph.D., P. Kimbel, M.D. (Chairperson; deceased), L. Nett, R.N., R.R.T. (former member), J.K. Ockene, Ph.D., R.M. Senior, M.D. (Chairperson), G.L. Snider, M.D., W. Spitzer, M.D. (former member), O.D. Williams, Ph.D.; Morbidity and Mortality Review Board: T.E. Cuddy, M.D., R.S. Fontana, M.D., R.E. Hyatt, M.D., C.T. Lambrew, M.D., B.A. Mason, M.D., D.M. Mintzer, M.D., R.B. Wray, M.D.; National Heart, Lung, and Blood Institute staff, Bethesda, MD: S.S. Hurd, Ph.D. (Former Director, Division of Lung Diseases), J.P. Kiley, Ph.D. (Former Project Officer and Director, Division of Lung Diseases), G. Weinmann, M.D. (Former Project Officer and Director, Airway Biology and Disease Program, DLD), M.C. Wu, Ph.D. (Division of Cardiovascular Sciences).
Phs000631.v1.p1: The datasets were obtained as part of the identification of SNPs Predisposing to Altered ALI Risk (iSPAAR) study funded by the NHLBI (RC2 HL101779).
Phs000518.v1.p1: The authors wish to acknowledge the support of the National Heart, Lung and Blood Institute (NHLBI) and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. This work was supported in part by grants R01 HL071798 from the NHLBI and U54 HL096458 from the NHLBI (previously supported by the NCRR), the components of NIH. This study is part of the NHLBI Grand Opportunity Exome Sequencing Project (GO-ESP). Funding for GO-ESP was provided by NHLBI grants RC2 HL103010 (HeartGO), RC2 HL102923 (LungGO) and RC2 HL102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL102925 (BroadGO) and RC2 HL102926 (SeattleGO).
Analysis of Presentation versus Mutation Frequency Among Tumors
The PHBR scores of 5,942 patients in TCGA were calculated for 1000 passenger mutations (observed 1 or 2 times in the 5,942 patients; not occurring in 200 cancer-implicated genes). PHBR scores were calculated for 1,018 recurrent driver mutations (from 200 cancer-implicated genes) in the 7137 patients. The distribution of passenger PHBR scores was compared to 841 low frequency (<=5 times), 149 medium frequency (>5, <=20 times) and 28 high frequency oncogenic mutations (>20 times). The distributions of PHBR scores in each category were compared with Mann-whitney U tests and visualized with violin plots (Figure 3C). Furthermore, we plotted cumulative distributions to demonstrate the practical presentation of each frequency grouping across several thresholds (Figure 3D).
Modeling the effect of PHBR-II on mutation probability
To assess the role of MHC-II in regards to mutation probability, we further restricted the recurrent oncogenic mutations to those occurring at least two times in the set of patients, resulting in 787 mutations and 5,942 patients. To first visualize the difference in PHBR-II distributions for mutations observed versus absent from tumors, PHBR-II scores from the 1,018 mutations × 5,942 patient matrix were grouped according to mutation status and plotted in side-by-side violin plots. Next, we built a 5,942 × 787 binary mutation matrix yij ∋ {0, 1} indicating whether patient i has a specific mutation j. We evaluated the relationship between this binary matrix and the matched 5,942 × 787 matrix with PHBR-II scores xij of patient i and for mutation j. We fitted a generalized additive model for the PHBR-II score and mutation probability with the GAM function in the MGCV R package (Wood, 2010). To estimate the effect of xij on yij we considered the following random effects model:
where ηi ~ N(0, θη) are random effects capturing different mutation propensities among patients.
In these models, γ measures the effect of the log-PHBR-II. We fitted this model using the glmer function from the lme4 R package (Bates et al., 2015) and tested the null hypothesis that γ = 0. To analyze the PHBR-mutation relationship in different tumor types, we fit separate models for each tumor type where there were at least 50 total number of driver mutations in the cohort. Furthermore, we used this same method to evaluate the difference in selection between mutations high allelic fraction and low allelic fraction (see ‘Clonality of mutations’ section).
Modeling the interaction between MHC-I and MHC-II effects
To assess the interaction between MHC-I and MHC-II in regards to mutation probability, we reduced the set of patients to those successfully typed for both MHC-I and MHC-II (Marty et al., 2017). We further restricted the recurrent oncogenic mutations to those occurring at least twice in the set of patients, resulting in 787 mutations and 5,942 patients. Then, we checked the correlation between MHC-I and MHC-II presentation using a Spearman Rank Test between MHC-I and MHC-II scores for each patient across all 1,018 mutations. These correlations were displayed as a histogram (Figure S4B). After finding low correlation scores, we built a model of the interaction.
We built a 5,942 × 787 binary mutation matrix yij ∋ {0, 1} indicating whether patient i has a specific mutation j. We evaluated the relationship between this binary matrix and two matched 5,942 × 787 matrices with MHC-I PHBR scores wij of patient i and for mutation j and MHC-II PHBR scores xij of patient i and for mutation j. To visualize the relationship between wij and xij with yij, we fit an generalized additive model for the PHBR scores of both classes using the GAM function in the mgcv R package (Wood, 2010).
Finally, to estimate the effect of xij and wij on yij we considered the following random effects model:
A within-patient model relating xij and wij to yij for a given patient
where αis the intercept term and ηi ~ N(0, θη) are random effects capturing different mutation propensities among patients.
In these models, γ measures the effect of the log-PHBR-I and ß measures the effect of the log-PHBR-II on the probability of a mutation being observed. We fitted this model using the glmer function from the lme4 R package (Bates et al., 2015) and tested the null hypothesis that γ = 0 and ß = 0. To analyze the PHBR-mutation relationship in different tumor types, we fit separate models for each tumor type where there were at least 50 total number of driver mutations in the cohort.
Given the distinct PHBR score ranges for MHC-I and MHC-II, we constructed an odds ratio analysis to compare the relative effects in the population. Instead of reporting the odds ratio for a single unit increase, we reported the odds of observing a mutation in the 25th PHBR percentile relative to the 75th PHBR percentile.
Fraction of patients with presentation
For each mutation in our set of 1,018 driver mutations, we calculated the fraction of patients that could present the mutation based on their MHC-I and MHC-II genotype respectively. We used the standard weak binding cutoffs of 2 for MHC-I and 10 for MHC-II. These results were visualized with a density plot (Figure 5D) and a scatter plot of the high frequency mutations (Figure S5D). Furthermore, we compared the distributions for fraction of MHC-I and MHC-II presentation across several thresholds (0.25, 0.5, 1, and 2 for MHC-I and 1, 2, 5, and 10 for MHC-II) to ensure robustness (Figure S5E).
Clonality of mutations
The occurences of mutations within the set of 1,018 driver mutations were designated as likely clonal or likely subclonal based on the allelic fraction annotation provided by TCGA. Mutations that were among the lowest 30th percentile were designated likely subclonal and all the remaining were considered likely clonal. We modeled the independent effect of PHBR-II and PHBR-I on mutation probability separately for subclonal and clonal occurrences as described above in the section ‘Modeling the effect of PHBR-II on mutation probability’.
MHC-based selection with different immune infiltration phenotypes
Immune infiltration levels were quantified from expression using CIBERSORT (Newman et al., 2015) and patient-specific cytotoxicity scores were derived (Rooney et al., 2015). Tumors were divided into “high” and “low” groups for each of the following categories using the tumor-type specific 30th and 70th percentile: APC infiltration (B cells, dendritic cells and macrophages), cytolytic activity, CD8+ T cell infiltration and CD4+ T cell infiltration. We modeled the independent effect of PHBR-II and PHBR-I on mutation probability in the high and low groups as described above in the section ‘Modeling the effect of PHBR-II on mutation probability’.
MHC coverage
MHC-I and MHC-II coverage of driver mutations was determined by calculating the fraction of the 1,018 driver mutation PHBR scores for each patient that fell below the binding thresholds, 2 and 10 for MHC-I and MHC-II respectively. This analysis resulted in each patient being assigned two MHC coverage values (MHC-I and MHC-II). Furthermore, two more values were calculated for each patient using 1,000 passenger mutations. The number of homozygous genes was determined for each patient by adding the number of identical alleles for MHC-I (-A, -B, -C) and MHC-II (-DRB, -DPA, -DPB, -DQA, -DQB) separately. The MHC coverage values were calculated for these patients as well and compared to the TCGA MHC coverage values with a Mann Whitney U test.
Age at diagnosis analysis
To visualize the association between MHC coverage and age at diagnosis, the patients with MHC coverage values in the lowest quartile and the patients with MHC coverage values in the highest quartile were compared. To determine statistical significance, a linear model in R was applied with age as the independent variable and MHC coverage, ancestry and tumor type as the dependent variables. Statistical significance was also determined for MHC-I and MHC-II coverage of passenger mutations and MHC homozygosity count as a replacement for MHC coverage. To assess the practical effect size of the extreme cases of MHC coverage, we compared the ages at diagnosis of the 5% of patients with the lowest MHC-I coverage with the ages at diagnosis for the 5% of patients with the highest MHC-I coverage with a two sample T-test. We also performed the same analysis for the patients with the highest and lowest 10% of MHC-I coverage. A pearson correlation test was used to determine the correlation between MHC coverage of driver mutations and MHC coverage of passenger mutations for both MHC-I and MHC-II.
Quantification and Statistical Analysis
Additional Statistical Considerations
For all individual tests, a p-value of less than 0.05 was considered significant. When multiple comparisons were made, p-values were adjusted using the Benjamini-Hochberg method unless otherwise specified. For all box plots, whiskers indicate the 1.5 IQR range.
Data and Software Availability
The python (2.7) and R code used to perform the analyses described in this manuscript and generate all main and supplemental figures is available in Data File S1 and at https://github.com/Rachelmarty20/MHC_II.
Supplementary Material
Figure S1: Development of a residue-specific, patient-specific MHC-II presentation score, related to Figure 1. (A) ROC AUC curves showing the accuracy of the PHBR-II including peptides of length 13-25 for classifying the extracellular presentation of a residue by a patient’s HLA-DR genes for 7 donors (colors) and for all donors combined (black). The aggregated PHBR-II presentation scores for the 7 donors expression HLA-DR alleles was compared to a set of random residues for the same HLA-DR alleles.
Figure S2: HLA-II typing of patients in TCGA. (A) The agreement of hla types for patients typed with HLA-HD and xHLA. (B-F) Frequencies of MHC-II alleles occuring in TCGA - (B) HLA-DPA, (C) HLA-DPB, (D) HLA-DQA, (E) HLA-DQB and (F) HLA-DRB. Related to Figure 1.
Figure S3: Native germline presentation, related to Figure 2. (A) A clustered heatmap of patients in TCGA with the native germline sequence 1,018 frequent cancer mutations. The same 1,050 patients are represented as in Figure 2. The heatmap is colored by PHBR-II score. Column and row coloring highlight groupings of patients and mutations into different categories. (B) A scatter plot showing the median population PHBR-II score for each of the 1,018 mutations and their native germline sequence.
Figure S4: Population presentation with MHC-II, related to Figure 3. (A) Cumulative distribution functions denoting the fraction of true positive and false positive residues detected for each PHBR-II score in the mass spectrometry validation. (B) A violin plot denoting the distribution of PHBR-II presentation scores across all TCGA patients for 6 different classes of residue. Cancer mutations observed > 2 times in TCGA are displayed. White dots represent the median. (C) The cumulative distribution of 20 sets of random 1,000 mutations. Shown alongside the cumulative distribution from oncogenes and tumor suppressor genes. (D) A violin plot denoting the distribution of PHBR-II presentation scores across non-cancer dbGaP patients for 6 different classes of residue. White dots represent the median. (E) Two dot plots showing the median PHBR-II and -I presentation scores for all 5,942 patients of the 1,018 recurrent cancer mutations grouped by their mutation count in TCGA and displayed as a median. The number of times the mutation group is observed in TCGA is plotted below the plots. The light grey line highlights the mutations observed 10 times.
Figure S5: Patient-specific MHC presentation prediction of mutation probability, related to Figure 5. (A) The distribution of PHBR-II and PHBR-I scores. (B) The distribution of spearman rho correlations for PHBR-II and PHBR-I scores across all driver mutations for every patient in TCGA. (C) A scatter plot showing the relationship between tissue specific ORs for MHC-II and MHC-I with a joint model for tumor types with at least 100 patients. (D) A scatter plot showing mutations observed at least 20 times in TCGA. Each point is placed according to the fraction of patients who can present it with MHC-I and MHC-II. (E) Histograms showing the variation in the number of mutations with different fractions of presentation by both MHC-I and MHC-II across several presentation thresholds.
Figure S6: MHC-based mutation selection for differing levels of immune activity, related to Figure 6. The MHC-I and MHC-II ORs (circles) and 95% CIs associated with a 1-unit increase in log-PHBR-II score. The results are shown for patients with low and high (A) APC infiltration, (B) cytolytic activity, (C) CD8+ T cell infiltration and (D) CD4+ T cell infiltration.
Figure S7: MHC presentation of driver mutations and age at diagnosis, related to Figure 6. (A) A box plot denoting the distributions of age at diagnosis for patients separated by tumor type and percentage of the driver space presented for MHC-II. The number of patients in each category is visualized above with a bar plot. (B) Box plots showing the age at diagnosis for patients with extreme 5% of patients for MHC-I and MHC-II coverage. (C) A histogram representing the spearman rho correlations for each tumor type between MHC-I coverage and mutation burden.
Table S1: HLA typing of patients in TCGA, related to Figure 1. Table shows patients with attempted MHC-II typing, patients with successful MHC-II typing, patients with successful MHC-I typing and patients with validated MHC-II typing.
Table S2: PHBR-II scores for driver mutations, related to Figure 2. The matrix of PHBR-II scores for all 1,018 driver mutations and 5,942 patients (represented visually in Figure 2).
Table S5: Tumor-type specific association, related to Figure 4. Table shows tumor-type specific ORs, 95% CIs and p values for logistic regression model with random effects relating PHBR-II scores to mutation occurrence.
Table S6: Fraction of patient presentation, related to Figure 5. Table shows the fraction of patients with MHC-I presentation and the fraction of patients with MHC-II presentation for all 1,018 mutations..
Highlights.
Patient- and residue-specific PHBR-II score estimates mutation presentation by MHC-II
Tumors are less likely to harbor driver mutations that bind well to MHC-II
Frequent driver mutations are universally poorly presented by MHC-II
MHC-II shows less inter-patient variability but stronger selective effects than MHC-I
Acknowledgements:
We would like to thank Alessandro Sette, Peter Bjoern, Julien Racle, David Gfeller and Trey Ideker for scientific discussion and the TCGA research network for providing data used in the analyses. Please see the methods for acknowledgements of other dbGaP data. Furthermore, we would like to acknowledge the reviewers of this manuscript for their wonderful suggestions that vastly improved our work. This work was supported by an NSF graduate fellowship (2015205295 to R.M.), a CIFAR fellowship (to H.C.), and NIH (K99/R00CA191152 to J.F.-B., DP5-OD017937 to H.C., R01-GM104400 to W.K.T., R01-CA220009 to M.Z. and H.C., and P41-GM103504 for the computing resources).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental data and legends
Data S1: Code used to produce results and figures, related to Figure 1. This file includes Python and R code used to perform the analyses described in this manuscript and generate all main and supplemental figures. The code is also available at https://github.com/Rachelmarty20/MHC_II.
References:
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, et al. (2017). Ensembl 2017. Nucleic Acids Res. 45, D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M, Trolle T, Yan Z, Greenbaum JA, Peters B, and Nielsen M (2018). An automated benchmarking platform for MHC class II binding prediction methods. Bioinformatics 34, 1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, and Walker S (2015). Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw 67, 1–48. [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795. [DOI] [PubMed] [Google Scholar]
- Bonhoeffer S, and Sniegowski P (2002). The importance of being erroneous. Nature 420, 367–369. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014). Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2015). The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, and O’Brien SJ (1999). HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283, 1748–1752. [DOI] [PubMed] [Google Scholar]
- Carter H, Marty R, Hofree M, Gross AM, Jensen J, Fisch KM, Wu X, DeBoever C, Van Nostrand EL, Song Y, et al. (2017). Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer. Cancer Discov. 7, 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassell D, and Forman J (1988). Linked recognition of helper and cytotoxic antigenic determinants for the generation of cytotoxic T lymphocytes. Ann. N. Y. Acad. Sci 532, 51–60. [DOI] [PubMed] [Google Scholar]
- Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, Kuo F, Kendall SM, Requena D, Riaz N, et al. (2017). Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science eaao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciudad MT, Sorvillo N, van Alphen FP, Catalan D, Meijer AB, Voorberg J, and Jaraquemada D (2017). Analysis of the HLA-DR peptidome from human dendritic cells reveals high affinity repertoires and nonconventional pathways of peptide generation. J. Leukoc. Biol 101, 15–27. [DOI] [PubMed] [Google Scholar]
- Consogno G, Manici S, Facchinetti V, Bachi A, Hammer J, Conti-Fine BM, Rugarli C, Traversari C, and Protti MP (2003). Identification of immunodominant regions among promiscuous HLA-DR-restricted CD4+ T-cell epitopes on the tumor antigen MAGE-3. Blood 101, 1038–1044. [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, Zhang S, Bettigole SE, Gupta D, Holcomb K, et al. (2015). ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell 161, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, and Elledge SJ (2013). Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PC, and Zinkernagel RM (1975). Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256, 50–52. [DOI] [PubMed] [Google Scholar]
- Galon J, Angell HK, Bedognetti D, and Marincola FM (2013). The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 39, 11–26. [DOI] [PubMed] [Google Scholar]
- Gerloni M, Xiong S, Mukerjee S, Schoenberger SP, Croft M, and Zanetti M (2000). Functional cooperation between T helper cell determinants. Proc. Natl. Acad. Sci. U. S. A 97, 13269–13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Galarza FF, Takeshita LYC, Santos EJM, Kempson F, Maia MHT, da Silva ALS, Teles e Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, et al. (2015). Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43, D784–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haabeth OAW, Tveita AA, Fauskanger M, Schjesvold F, Lorvik KB, Hofgaard PO, Omholt H, Munthe LA, Dembic Z, Corthay A, et al. (2014). How Do CD4 T Cells Detect and Eliminate Tumor Cells That Either Lack or Express MHC Class II Molecules? Front. Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW (2002). PATHOGEN RESISTANCE AND GENETIC VARIATION AT MHC LOCI. Evolution 56, 1902. [DOI] [PubMed] [Google Scholar]
- Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, and Yee C (2008). Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N. Engl. J. Med 358, 2698–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, and Levitsky H (1998). The central role of CD4(+) T cells in the antitumor immune response. J. Exp. Med 188, 2357–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, and Schoenberger SP (2003). CD4 T cells are required for secondary expansion and memory in CD8 T lymphocytes. Nature 421, 852–856. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, Vilgelm AE, Feld E, Johnson AS, Greenplate AR, et al. (2016). Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD- L1 therapy. Nat. Commun 7, 10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, and Nielsen M (2013). NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 65, 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S, Higasa K, Shimizu M, Yamada R, and Matsuda F (2017). HLA-HD: An accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat 38, 788–797. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Wood M, Song Y, Appella E, and Celis E (2000). Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res. 60, 5228–5236. [PubMed] [Google Scholar]
- Langlade-Demoyen P, Garcia-Pons F, Castiglioni P, Garcia Z, Cardinaud S, Xiong S, Gerloni M, and Zanetti M (2003). Role of T cell help and endoplasmic reticulum targeting in protective CTL response against influenza virus. Eur. J. Immunol 33, 720–728. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. (2013). Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty R, Kaabinejadian S, Rossell D, Slifker MJ, van de Haar J, Engin HB, de Prisco N, Ideker T, Hildebrand WH, Font-Burgada J, et al. (2017). MHC-I Genotype Restricts the Oncogenic Mutational Landscape. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, Birkbak NJ, Veeriah S, Van Loo P, Herrero J, et al. (2017). Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 171, 1259–1271.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, and Nikolich-Zugich J (2002). Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science 298, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Mitchison NA (1971). The carrier effect in the secondary response to hapten-protein conjugates. I. Measurement of the effect with transferred cells and objections to the local environment hypothesis. Eur. J. Immunol 1, 10–17. [DOI] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, and Andreatta M (2016). NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, and Bangham CR (1996). Population dynamics of immune responses to persistent viruses. Science 272, 74–79. [DOI] [PubMed] [Google Scholar]
- O’Sullivan T, Saddawi-Konefka R, Vermi W, Koebel CM, Arthur C, White JM, Uppaluri R, Andrews DM, Ngiow SF, Teng MWL, et al. (2012). Cancer immunoediting by the innate immune system in the absence of adaptive immunity. J. Exp. Med 209, 1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. (2017). An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Lindestam Arlehamn CS, Scriba TJ, Dillon MBC, Oseroff C, Hinz D, McKinney DM, Carrasco Pro S, Sidney J, Peters B, et al. (2015). Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J. Immunol. Methods 422, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, and Marsh SGE (2015). The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43, D423–D431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodvold JJ, Chiu KT, Hiramatsu N, Nussbacher JK, Galimberti V, Mahadevan NR, Willert K, Lin JH, and Zanetti M (2017). Intercellular transmission of the unfolded protein response promotes survival and drug resistance in cancer cells. Sci. Signal 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, and Hacohen N (2015). Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryschich E, Cebotari O, Fabian OV, Autschbach F, Kleeff J, Friess H, Bierhaus A, Buchler MW, and Schmidt J (2004). Loss of heterozygosity in the HLA class I region in human pancreatic cancer. Tissue Antigens 64, 696–702. [DOI] [PubMed] [Google Scholar]
- Schoenberger SP, Toes REM, van der Voort EIH, Offringa R, and Melief CJM (1998). T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature 393, 480–483. [DOI] [PubMed] [Google Scholar]
- Shedlock DJ, and Shen H (2003). Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300, 337–339. [DOI] [PubMed] [Google Scholar]
- Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, Stevens J, Lane WJ, Dellagatta JL, Steelman S, et al. (2015). Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol 33, 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, and Sette A (1998). Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol 160, 3363–3373. [PubMed] [Google Scholar]
- Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. (2017). Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, and Bevan MJ (2003). Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Williams MA, and Bevan MJ (2004). CD4 T cells are required for the maintenance, not programming, of memory CD8 T cells after acute infection. Nat. Immunol 5, 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, and Kohlbacher O (2014). OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2014). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. (2014). Cancer Immunotherapy Based on Mutation-Specific CD4 T Cells in a Patient with Epithelial Cancer. Science 344, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unanue ER, Turk V, and Neefjes J (2016). Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu. Rev. Immunol 34, 265–297. [DOI] [PubMed] [Google Scholar]
- Wang RF (2001). The role of MHC class II-restricted tumor antigens and CD4+ T cells in antitumor immunity. Trends Immunol. 22, 269–276. [DOI] [PubMed] [Google Scholar]
- Wang P, Sidney J, Dow C, Mothé B, Sette A, and Peters B (2008). A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol 4, e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Abualrous ET, Sticht J, Álvaro-Benito M, Stolzenberg S, Noé F, and Freund C (2017). Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SN (2010). Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Series B Stat. Methodol 73, 3–36. [Google Scholar]
- Xie C, Yeo ZX, Wong M, Piper J, Long T, Kirkness EF, Biggs WH, Bloom K, Spellman S, Vierra-Green C, et al. (2017). Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc. Natl. Acad. Sci. U. S. A 114, 8059–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Kim D-K, Nakagawa H, Hayashi S, Imoto S, Stein L, and Roth F (2017). Quantifying Immune-Based Counterselection of Somatic Mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, and Haeryfar SMM (2005). Understanding presentation of viral antigens to CD8+ T cells in vivo: the key to rational vaccine design. Annu. Rev. Immunol 23, 651–682. [DOI] [PubMed] [Google Scholar]
- Zanetti M (2015). Tapping CD4 T cells for cancer immunotherapy: the choice of personalized genomics. J. Immunol 194, 2049–2056. [DOI] [PubMed] [Google Scholar]
- Zelenay S, van der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, et al. (2015). Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 162, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Development of a residue-specific, patient-specific MHC-II presentation score, related to Figure 1. (A) ROC AUC curves showing the accuracy of the PHBR-II including peptides of length 13-25 for classifying the extracellular presentation of a residue by a patient’s HLA-DR genes for 7 donors (colors) and for all donors combined (black). The aggregated PHBR-II presentation scores for the 7 donors expression HLA-DR alleles was compared to a set of random residues for the same HLA-DR alleles.
Figure S2: HLA-II typing of patients in TCGA. (A) The agreement of hla types for patients typed with HLA-HD and xHLA. (B-F) Frequencies of MHC-II alleles occuring in TCGA - (B) HLA-DPA, (C) HLA-DPB, (D) HLA-DQA, (E) HLA-DQB and (F) HLA-DRB. Related to Figure 1.
Figure S3: Native germline presentation, related to Figure 2. (A) A clustered heatmap of patients in TCGA with the native germline sequence 1,018 frequent cancer mutations. The same 1,050 patients are represented as in Figure 2. The heatmap is colored by PHBR-II score. Column and row coloring highlight groupings of patients and mutations into different categories. (B) A scatter plot showing the median population PHBR-II score for each of the 1,018 mutations and their native germline sequence.
Figure S4: Population presentation with MHC-II, related to Figure 3. (A) Cumulative distribution functions denoting the fraction of true positive and false positive residues detected for each PHBR-II score in the mass spectrometry validation. (B) A violin plot denoting the distribution of PHBR-II presentation scores across all TCGA patients for 6 different classes of residue. Cancer mutations observed > 2 times in TCGA are displayed. White dots represent the median. (C) The cumulative distribution of 20 sets of random 1,000 mutations. Shown alongside the cumulative distribution from oncogenes and tumor suppressor genes. (D) A violin plot denoting the distribution of PHBR-II presentation scores across non-cancer dbGaP patients for 6 different classes of residue. White dots represent the median. (E) Two dot plots showing the median PHBR-II and -I presentation scores for all 5,942 patients of the 1,018 recurrent cancer mutations grouped by their mutation count in TCGA and displayed as a median. The number of times the mutation group is observed in TCGA is plotted below the plots. The light grey line highlights the mutations observed 10 times.
Figure S5: Patient-specific MHC presentation prediction of mutation probability, related to Figure 5. (A) The distribution of PHBR-II and PHBR-I scores. (B) The distribution of spearman rho correlations for PHBR-II and PHBR-I scores across all driver mutations for every patient in TCGA. (C) A scatter plot showing the relationship between tissue specific ORs for MHC-II and MHC-I with a joint model for tumor types with at least 100 patients. (D) A scatter plot showing mutations observed at least 20 times in TCGA. Each point is placed according to the fraction of patients who can present it with MHC-I and MHC-II. (E) Histograms showing the variation in the number of mutations with different fractions of presentation by both MHC-I and MHC-II across several presentation thresholds.
Figure S6: MHC-based mutation selection for differing levels of immune activity, related to Figure 6. The MHC-I and MHC-II ORs (circles) and 95% CIs associated with a 1-unit increase in log-PHBR-II score. The results are shown for patients with low and high (A) APC infiltration, (B) cytolytic activity, (C) CD8+ T cell infiltration and (D) CD4+ T cell infiltration.
Figure S7: MHC presentation of driver mutations and age at diagnosis, related to Figure 6. (A) A box plot denoting the distributions of age at diagnosis for patients separated by tumor type and percentage of the driver space presented for MHC-II. The number of patients in each category is visualized above with a bar plot. (B) Box plots showing the age at diagnosis for patients with extreme 5% of patients for MHC-I and MHC-II coverage. (C) A histogram representing the spearman rho correlations for each tumor type between MHC-I coverage and mutation burden.
Table S1: HLA typing of patients in TCGA, related to Figure 1. Table shows patients with attempted MHC-II typing, patients with successful MHC-II typing, patients with successful MHC-I typing and patients with validated MHC-II typing.
Table S2: PHBR-II scores for driver mutations, related to Figure 2. The matrix of PHBR-II scores for all 1,018 driver mutations and 5,942 patients (represented visually in Figure 2).
Table S5: Tumor-type specific association, related to Figure 4. Table shows tumor-type specific ORs, 95% CIs and p values for logistic regression model with random effects relating PHBR-II scores to mutation occurrence.
Table S6: Fraction of patient presentation, related to Figure 5. Table shows the fraction of patients with MHC-I presentation and the fraction of patients with MHC-II presentation for all 1,018 mutations..
Data Availability Statement
The python (2.7) and R code used to perform the analyses described in this manuscript and generate all main and supplemental figures is available in Data File S1 and at https://github.com/Rachelmarty20/MHC_II.