Evolution of a cell specific invertase enzyme creates phenotypic diversity in wild tomato insecticidal metabolite production.
Abstract
Plants produce a myriad of taxonomically restricted specialized metabolites. This diversity—and our ability to correlate genotype with phenotype—makes the evolution of these ecologically and medicinally important compounds interesting and experimentally tractable. Trichomes of tomato and other nightshade family plants produce structurally diverse protective compounds termed acylsugars. While cultivated tomato (Solanum lycopersicum) strictly accumulates acylsucroses, the South American wild relative Solanum pennellii produces copious amounts of acylglucoses. Genetic, transgenic, and biochemical dissection of the S. pennellii acylglucose biosynthetic pathway identified a trichome gland cell–expressed invertase-like enzyme that hydrolyzes acylsucroses (Sopen03g040490). This enzyme acts on the pyranose ring–acylated acylsucroses found in the wild tomato but not on the furanose ring–decorated acylsucroses of cultivated tomato. These results show that modification of the core acylsucrose biosynthetic pathway leading to loss of furanose ring acylation set the stage for co-option of a general metabolic enzyme to produce a new class of protective compounds.
INTRODUCTION
Plants synthesize hundreds of thousands of structurally diverse and lineage-, tissue-, or cell type–specific specialized metabolites (1). These compounds act as antiherbivory, antimicrobial, or allelopathic compounds (2–4). The enzymes that produce these specialized metabolites arise from a variety of proteins through gene duplication and neo- or subfunctionalization. Examples include diversification of existing specialized metabolic enzymes (5) and co-option of core metabolic activities (6). In the latter mechanism, gene duplication can remove stabilizing selective pressure on one of the paralogs associated with core metabolism, enabling evolution of the novel enzymatic activity without disrupting growth and development. This leads to new and modified pathways that produce structurally and functionally diverse specialized metabolites.
Glandular trichome–synthesized acylated sugars (“acylsugars”) are structurally diverse specialized metabolites found throughout the Solanaceae (7–13). These compounds have documented roles in direct and indirect protection against herbivores and microbes (14, 15), as well as allelopathic properties (15, 16). Their low toxicity to vertebrates generates interest in generating plant breeding strategies for deploying acylsugars in crop protection (17, 18). These metabolites consist of a sugar core—typically sucrose—with aliphatic chains of variable length, structure, and number attached by ester linkages. Acylsugars were reported from genera across the Solanaceae family, including Datura, Nicotiana, Petunia, Physalis, Salpiglossis, and Solanum with single species producing at least three dozen chromatographically distinct acylsugars (10, 13, 16, 19–22).
In recent years, several evolutionarily related enzymes were implicated in the core acylsucrose biosynthetic pathways in species across the family, including the cultivated tomato Solanum lycopersicum, Petunia axillaris, and Salpiglossis sinuata (7, 9–12, 23). These biosynthetic pathways consist of trichome-expressed BAHD-family (BEAT, AHCT, HCBT, and DAT) acylsugar acyltransferases (ASATs) (7, 9, 23), which sequentially transfer acyl groups from acyl–coenzyme A (acyl-CoA) substrates to specific hydroxyl groups of sucrose (7, 9, 23).
The cultivated tomato biosynthetic network is well characterized, with four ASATs—SlASAT1 to SlASAT4—catalyzing consecutive reactions to produce tri- and tetra-acylated sucroses. SlASAT1 acts first by transferring an acyl chain to the R4 hydroxyl of the pyranose ring of sucrose, and SlASAT2 transfers an acyl chain to the R3 position of the monoacylated sucrose (23). Next, SlASAT3 acylates the diacylated sucroses at the furanose ring R3′ position (9). SlASAT4 completes the pathway by transferring an acetyl group to the pyranose ring R2 position of a triacylsucrose (7, 22). Enzyme promiscuity and the presence of an array of acyl-CoAs result in the production of a diverse group of acylsucroses in S. lycopersicum (11, 24).
The metabolic diversity in acylsugars is even greater in the broader Solanum genus. The wild relative of tomato, Solanum pennellii LA0716, is a prime example, producing a mixture of abundant acylsucroses that are distinct from those in S. lycopersicum. While S. lycopersicum accumulates acylsucroses with two or three acylations on the pyranose ring and a single acylation at the furanose ring R3′ position (termed F-type acylsucroses), S. pennellii accumulates distinct P-type triacylsucroses acylated only on the pyranose R2, R3, and R4 positions (11). P-type acylsucroses are synthesized by S. pennellii orthologs of the S. lycopersicum ASAT1, ASAT2, and ASAT3 enzymes (Fig. 1A). The different acylation pattern observed in S. pennellii results from altered substrate specificity and acylation position of SpASAT2 and SpASAT3 relative to their S. lycopersicum counterparts (11).
Fig. 1. Three S. pennellii LA0716 regions condition acylglucose accumulation.
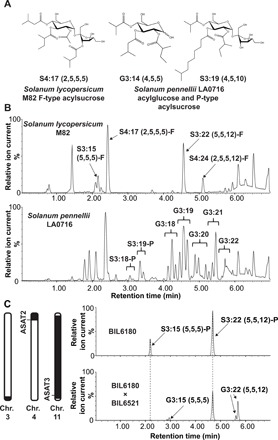
(A) Examples of nuclear magnetic resonance (NMR)–resolved S. lycopersicum and S. pennellii acylsugar structures. Acylsugars from S. lycopersicum are composed of sucrose acylated on both the pyranose and furanose rings (“F-type”). S. pennellii acylsugars are a mixture of sucrose-based (“P-type”) and glucose-based compounds with acylation exclusively on the pyranose ring. (B) Acylsugar electrospray ionization (ESI−)–mode LC-QToF (Quadrupole time-of-flight) MS profiles. Top: S. lycopersicum M82 with acylsucroses S3:15 (5,5,5)-F, S4:17 (2,5,5,5)-F, S3:22 (5,5,12)-F, and S4:24 (2,5,5,12)-F annotated. Bottom: S. pennellii LA0716 acylsucroses and acylglucoses. (C) Left: Representation of S. pennellii chromosomal introgressions in BIL6521 × BIL6180 progeny that contain quantitative trait loci (QTLs) affecting acylglucose biosynthesis (30). The black portions of the chromosomes correspond to S. pennellii introgressions, while the white portions correspond to the chromosomal regions in the M82 background. Right: ESI− mode liquid chromatography–mass spectrometry (LC-MS) analysis of BIL6180 compared with the BIL6180 × BIL6521 F2 progeny reveals acylglucose accumulation in the hybrid, but not in BIL6180. All ESI− mode acylsugars were identified as formate adducts. Chr., chromosome.
S. pennellii LA0716 has other acylsugar characteristics that differentiate it from cultivated tomato (Fig. 1B). First, it produces copious amounts of acylsugars that render the plant sticky, representing up to ~20% of leaf dry weight (19, 25). Second, the vast majority of S. pennellii LA0716 acylsugars are glucose molecules with three acyl chains (termed “acylglucoses”) (Fig. 1, A and B), while only 7 to 16% of total acylsugars are acylsucroses (26). In contrast to the well-characterized S. pennellii acylsucrose biosynthetic enzymes (9, 11), no complete acylglucose metabolic pathway has yet been described. This is despite the fact that acylglucoses were also characterized in several additional Solanaceae species (16, 20). A previously proposed partial S. pennellii pathway invoked two glucosyltransferases capable of creating 1-O-acyl-d-glucose from uridine diphosphate (UDP)–glucose and free fatty acids of differing structures (27). This mechanism proposed a second step in which a serine carboxypeptidase-like (SCPL) acyltransferase catalyzed disproportionation of two 1-O-isobutyryl-D-glucose molecules to yield one 1,2-O-di-isobutyryl-D-glucose (28, 29). However, this pathway is unlikely to function in vivo as the 1,2,-O-diacylglucoses obtained in vitro differ from the 2,3,4-O-tri-acylglucoses observed in S. pennellii both in the number (two instead of three) and in the position of acyl chains: S. pennellii acylglucoses bear chains at the R2, R3, and R4 positions rather than at the R1 position (19).
In contrast to the unsubstantiated published biosynthetic pathway, compelling quantitative trait locus (QTL) and biochemical results implicate multiple genetic loci in acylglucose accumulation in S. pennellii LA0716. The combination of three S. pennellii regions on chromosomes 3, 4, and 11 causes S. lycopersicum breeding line CU071026 to accumulate acylsugars comprising up to 89% acylglucoses (30). The presence of QTLs on both chromosomes 3 and 11 yields detectable acylglucoses, while addition of the chromosome 4 locus leads to elevated accumulation. Notably, chromosome 4 and 11 QTLs respectively include the SpASAT2 and SpASAT3 genes responsible for accumulation of P-type acylsucroses in S. pennellii (9, 23). The agreement of QTL and biochemical data is consistent with the hypothesis that SpASAT2 and SpASAT3 produce P-type acylsucroses that are substrates for a chromosome 3 factor that then synthesizes triacylglucoses.
We report the characterization of the plant specialized metabolic invertase-like enzyme acylsucrose fructofuranosidase 1 (SpASFF1; Sopen03g040490), a chromosome 3 β-fructofuranosidase capable of cleaving the glycosidic bond of P-type acylsucroses. Genetic and transgenic plant approaches demonstrate that S. pennellii LA0716 acylglucose production requires SpASFF1. This work also documents a three-gene epistatic interaction between the SpASAT2, SpASAT3, and SpASFF1 loci that conditions high-level acylglucose accumulation. While yeast invertase and other variants involved in core metabolism have been studied since the 19th century (31–33), this work documents a new type of role for β-fructofuranosidase–type enzymes in specialized metabolism. These results extend our understanding of evolutionary mechanisms leading to trichome specialized metabolic diversity by demonstrating how neofunctionalization led to co-option of invertase from general metabolism into a cell type–specific specialized metabolic network.
RESULTS
An S. pennellii chromosome 3 locus is necessary for acylglucose production from P-type acylsucroses
Published QTL mapping studies indicate that introgression of S. pennellii LA0716 loci on chromosomes 3, 4, and 11 leads to accumulation of acylglucoses in a cultivated tomato S. lycopersicum background (30, 34). Three introgression lines harboring individual acylglucose QTLs in the S. lycopersicum background were screened, but none of the single introgressions in lines IL3-5, IL4-1, or IL11-3 (35) yielded detectable leaf acylglucoses (fig. S1). These observations are consistent with the hypothesis that multiple S. pennellii loci are needed for S. lycopersicum acylglucose accumulation. There are low but detectable levels of acylglucoses (87% of total acylsugars; fig. S2) in backcross inbred line BIL6521 (36), which contains S. pennellii LA0716 introgressions from chromosomes 1, 3, and 11. This BIL accumulates four acylglucoses (table S1), with the major one, G3:22 (5,5,12) (figs. S3 and S4), resembling the pyranose ring of the P-type acylsucrose S3:22 (5,5,12)-P detected at low levels in trichomes of the single chromosome 11 introgression line, IL11-3 (9). BIL6521 accumulates a P-type acylsucrose, S3:22 (figs. S3 and S4). These results are consistent with the hypothesis that the chromosome 3 region is necessary for acylglucose production but only when P-type acylsucroses are produced. Note that, in our nomenclature, “S” and “G” refer to a sucrose or glucose core, respectively, and 3:22 (5,5,12) indicates that there are three ester-linked acyl chains of 5, 5, and 12 carbons, for a total of 22 chain carbons (9). When nuclear magnetic resonance (NMR)–derived structural information is available, superscripts indicate acyl chain positions with R representing the pyranose ring and R′ representing the furanose ring (Fig. 1A).
Because the acylsugar levels in BIL6521, which lack a chromosome 4 introgression, were much lower than most other lines, we tested the impact of adding a chromosome 4 introgression carrying the SpASAT2 locus. A cross was made between BIL6521 and BIL6180, a recombinant line harboring introgressions on chromosomes 4, 5, and 11, which includes both the SpASAT2 and SpASAT3 loci (Fig. 1C). BIL6180 was previously found to produce only P-type acylsucroses as a result of the chromosome 4 and 11 introgressions (Fig. 1C); however, it accumulated significantly higher overall levels of acylsucroses compared to BIL6521 (fig. S2) and other short chain–containing P-type acylsucroses not present in BIL6521. If all P-type acylsucroses are substrates for an S. pennellii LA0716 factor on chromosome 3, we predict that both of the corresponding acylglucoses G3:15 (5,5,5) and G3:22 (5,5,12) would accumulate in a line harboring the chromosome 3, 4, and 11 introgressions. The F2 progeny of BIL6521 × BIL6180, genotyped as heterozygous for the S. pennellii chromosome 3 and 4 introgressions and homozygous for the S. pennellii chromosome 11 region, produced these two predicted acylglucoses (Fig. 1C and figs. S3 and S4). These findings, in combination with the published QTL results, indicate that the S. pennellii chromosome 3 introgression is necessary for acylglucose biosynthesis and suggests that P-type acylsucroses are acylglucose biosynthetic precursors.
The chromosome 3 locus encodes a glandular trichome–expressed β-fructofuranosidase
We sought candidate glycoside hydrolase (GH) genes in the 1.7-Mb QTL AG3.2, the acylglucose-associated region from S. pennellii LA0716 previously mapped to the bottom of chromosome 3 (Fig. 2A) (30). Three of the 238 genes in this region of the S. lycopersicum Heinz 1706 genome assembly SL2.50 annotation (37) are predicted as encoding GHs (members of the GH32, GH35, and GH47 families; table S2). We focused on the GH32 family Sopen03g040490 gene because all previously characterized members of the family have β-fructofuranosidase or fructosyltransferase activity (38). As acylsucroses are β-fructofuranosides, we hypothesized that the GH32 enzyme cleaves the glycosidic bond of P-type acylsucroses to generate acylglucoses. On the basis of the full results of this study, we designate this gene ACYLSUCROSE FRUCTOFURANOSIDASE 1 (ASFF1).
Fig. 2. GH 32 family gene SpASFF1 from QTL AG3.2 shows trichome-specific expression.
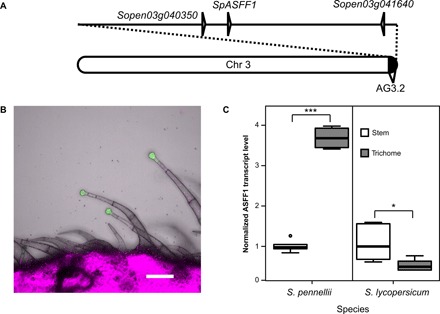
(A) Chromosome 3 with the AG3.2 QTL (30). Positions of three GH genes (Sopen03g040350, Sopen03g041640, and SpASFF1) are indicated. (B) Expression of GFP-GUS under control of the native ASFF1 promoter from S. pennellii LA0716 yields a GFP signal in S. lycopersicum M82 type I/IV trichome tip cells, but not in stalk cells or stem tissue. The green channel indicates the GFP signal; the magenta channel shows chlorophyll fluorescence. Scale bar, 100 μm. (C) qRT-PCR analysis of transcript abundance indicates that ASFF1 transcripts are higher in S. pennellii LA0716 trichomes than in underlying stem tissue but lower in S. lycopersicum M82 trichomes than in underlying stem tissue. Whiskers represent minimum and maximum values less than 1.5 times the interquartile range from the first and third quartiles, respectively. Values outside this range are represented as circles. *P < 0.05 and ***P < 0.001, Welch two-sample t test; n = 6 for all species and tissue types.
S. lycopersicum acylsugars accumulate in type I/IV glandular trichome tip cells (39), and trichome tip cell–specific gene expression is a hallmark of all characterized acylsugar biosynthetic genes (e.g., ASAT1/2/3/4, IPMS3) (7, 9, 23, 24). We used a reporter gene approach to ask whether SpASFF1 exhibits trichome-specific expression. The 1.8-kb region immediately upstream of the SpASFF1 open reading frame (ORF) in the S. pennellii LA0716 genome drove expression of a green fluorescent protein–β-glucuronidase (GFP-GUS) fusion protein in S. lycopersicum M82 plants. A GFP signal in transformed plants was observed in the tip cells of type I/IV trichomes but not in the trichome stalk cells or underlying stem epidermis (Fig. 2B). This result is consistent with a role of SpASFF1 enzyme in type I/IV trichome metabolism.
We cross-validated the trichome-enriched expression pattern of ASFF1 in S. pennellii LA0716 using quantitative reverse transcription PCR (qRT-PCR). ASFF1 transcript levels were 3.7-fold higher in trichomes of S. pennellii LA0716 stems than in underlying shaved stem tissue (P < 0.001, Welch two-sample t test) (Fig. 2C). The observed enrichment of transcripts in trichome samples is similar to analysis of previously identified acylsugar biosynthetic genes from tomato, petunia, and tobacco (7, 12, 15, 23, 24). Together, transcript enrichment in trichomes and restriction of gene expression to trichome tip cells support the hypothesis that SpASFF1 acts in acylsugar biosynthesis.
Acylglucoses accumulate in S. pennellii LA0716 but not in S. lycopersicum M82 (8, 19). However, ASFF1 is predicted to encode a full ORF in both the S. lycopersicum Heinz 1706 and the S. pennellii LA0716 genomes (37, 40). In contrast to enrichment of ASFF1 transcripts in trichomes of S. pennellii LA0716, ASFF1 transcripts are half as abundant in S. lycopersicum M82 trichomes as in underlying stem tissue (P < 0.05, Welch two-sample t test) (Fig. 2C). Together, the tissue- and species-level specificity of ASFF1 expression is consistent with a role for the gene in acylglucose biosynthesis.
Gene editing reveals that SpASFF1 is necessary for S. pennellii LA0716 acylglucose accumulation
We used CRISPR-Cas9–mediated gene editing in S. pennellii LA0716 to test whether SpASFF1 is necessary for acylglucose accumulation. Two small guide RNAs (sgRNAs) targeting the third SpASFF1 exon were used to promote site-specific DNA cleavage by hCas9 in the stably transformed plants (Fig. 3A and fig. S5). Three homozygous T1 mutants were obtained with different site-specific mutations, each of which is predicted to cause complete loss of function through translational frameshifts and premature protein termination. Two of them (spasff1-1-1 and spasff1-1-2), which carry 228–base pair (bp) and 276-bp indels, respectively, are derived from segregation of one heteroallelic T0 plant. The third mutant (spasff1-2) with a 1-bp insertion is the descendant of a homozygous T0 mutant.
Fig. 3. CRISPR-Cas9–mediated S. pennellii LA0716 spasff1 knockouts eliminate detectable acylglucoses.
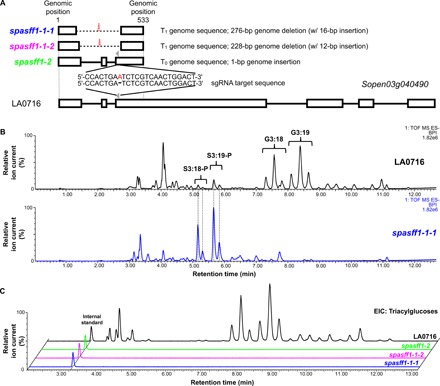
(A) Schematic representation of mutagenesis strategy with two sgRNAs (gray arrowheads; only one sgRNA is shown) targeting the SpASFF1 ORF that result in three homozygous knockout lines. White boxes indicate exons, horizontal bars indicate introns, dashed lines indicate deletions, and the red letter indicates insertion. Mutant allele DNA sequences are found in fig. S5. (B) Mutant line spasff1-1-1 accumulates abundant acylsucroses but no detectable acylglucoses. ESI− base-peak intensity (BPI) LC-MS chromatograms are shown for spasff1-1-1 and LA0716. (C) ESI− mode analysis of formate adducts of triacylglucose extracted ion chromatograms of trichome extracts from S. pennellii LA0716 and three spasff1 mutant plants show that homozygous asff1 lines produce undetectable levels of triacylglucose. Extracted ion chromatogram values displayed the following: G3:12 [mass/charge ratio (m/z) 435.19], G3:13 (m/z 449.2), G3:14 (m/z 463.22), G3:15 (m/z 477.23), G3:16 (m/z 491.28), G3:17 (m/z 505.26), G3:18 (m/z 519.28), G3:19 (m/z 533.30), G3:20 (m/z 547.31), G3:21 (m/z 561.33), G3:22 (m/z 575.34), and telmisartan (internal standard) (m/z 513.23). ESI− mode BPI LC-MS chromatograms for all lines are shown in fig. S6. For (B) and (C), spasff1-1-1/1-1-2 are homozygous T2 lines, while spasff1-2 are homozygous T1 lines that were all grown together. spasff1 lines were diluted 100-fold before LC-MS analysis to avoid saturation of the LC-MS detector. This is due to differences in ionization between acylsucroses and acylglucoses in the ESI− mode.
Results from a liquid chromatography–mass spectrometry (LC-MS) analysis of leaf surface metabolites from these lines were consistent with the hypothesis that SpASFF1 is necessary for acylglucose biosynthesis. All spasff1 lines failed to accumulate detectable acylglucoses (Fig. 3, B and C, and fig. S6) but produced acylsucroses at levels comparable to those of total acylsugars in wild-type S. pennellii plants (Table 1).
Table 1. Quantitative analysis of acylsugars from S. pennellii LA0716 and three spasff1 lines.
Acyl chains were saponified from the acylsugars, and the resulting sugar cores were analyzed by UPLC-ESI-Multiple Reaction Monitoring. Data are shown from individual T1 homozygous plants grown together but independently from those in Fig. 3. These extracts include other glycosylated compounds such as flavonoids, which could be responsible for the nonzero values for glucose measurements in plants lacking detectable acylglucoses (61). DW, dry weight.
| Line |
Plant number |
Sucrose (%) |
Glucose (%) |
Total sugar core (nmol/ mg DW) |
| LA0716 | 1 | 1 | 99 | 41.93 |
| 2 | 3 | 97 | 15.78 | |
| 3 | 1 | 99 | 33.70 | |
| 4 | 2 | 98 | 55.89 | |
| 5 | 1 | 99 | 138.80 | |
| 6 | 1 | 99 | 29.34 | |
| spasff1-1-1 | 1 | 98 | 2 | 25.82 |
| 2 | 99 | 1 | 23.23 | |
| 3 | 98 | 2 | 26.79 | |
| 4 | 99 | 1 | 21.49 | |
| spasff1-1-2 | 1 | 99 | 1 | 18.74 |
| 2 | 99 | 1 | 24.55 | |
| 3 | 99 | 1 | 28.72 | |
| 4 | 99 | 1 | 18.35 | |
| spasff1-2 | 1 | 97 | 3 | 80.01 |
| 2 | 98 | 2 | 37.51 | |
| 3 | 95 | 5 | 84.44 |
SpASFF1 converts pyranose ring–acylated P-type acylsucroses to acylglucoses both in vivo and in vitro
The results described above strongly suggest that SpASFF1 converts pyranose ring–acylated P-type acylsucroses to acylglucoses. In addition, IL3-5 does not accumulate detectable acylglucoses despite having S. pennellii ASFF1, suggesting that F-type acylsucroses are not substrates for SpASFF1 (Fig. 4A and fig. S7). We took a transgenic approach to ask whether SpASFF1 alone is sufficient to confer acylglucose accumulation in a P-type acylsucrose-accumulating background. We transformed the P-type acylsucrose-producing S. lycopersicum double-introgression BIL6180 (Fig. 4B) with a transferred DNA (T-DNA) containing the ORF of SpASFF1 and the 1.8 kb immediately upstream of its start codon (Fig. 4C). In addition to the P-type acylsucroses in the parental BIL6180, the SpASFF1 transgenics accumulated major hexose acylsugars with MS characteristics consistent with G3:15 (5,5,5) and G3:22 (5,5,12) (Fig. 4C and fig. S8). The acyl chain composition of these acylglucoses matches the S3:15 (5R2,5R3,5R4) and S3:22 (5R2,5R4,12R3) P-type acylsucroses detected in BIL6180 (fig. S4). Acylglucoses in the transgenic lines are also identical to those seen in BIL6521 × BIL6180 based on LC retention time and MS fragmentation. This confirms that SpASFF1 converts S. pennellii P-type acylsucroses produced by SpASAT2 and SpASAT3 to acylglucoses. Together, these in vivo results indicate that the presence of SpASFF1 is sufficient to yield acylglucoses in vivo when P-type acylsucroses are present, but not in plants accumulating only F-type acylsucroses.
Fig. 4. Expression of SpASFF1 in P-type acylsucrose producing BIL6180 trichomes results in accumulation of acylglucoses in surface extracts.
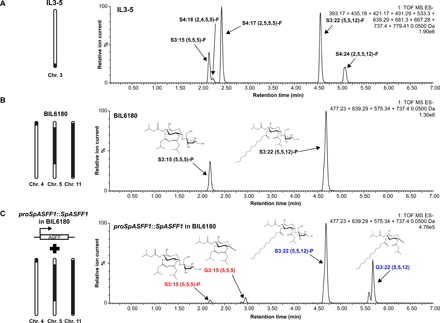
(A) IL3-5 accumulates F-type acylsucroses without detectable acylglucoses. ESI− mode LC-MS analysis of trichome extracts of IL3-5 are shown. Extracted ion chromatograms of S3:15 (m/z 639.29), S4:16 (m/z 667.28), S4:17 (m/z 681.30), S3:22 (m/z 737.40), and S4:24 (m/z 779.41) in addition to their glucose cognates (missing a C5 chain present on the furanose ring), G2:10 (m/z 393.17), G3:11 (m/z 421.17), G3:12 (m/z 435.18), G2:17 (m/z 491.29), and G3:19 (m/z 533.30) are shown. (B) BIL6180 accumulates P-type acylsucroses with no detectable acylglucoses. ESI− mode LC-MS analysis of trichome extracts of BIL6180 are shown. Extracted ion chromatograms of S3:15 (m/z 639.29), S3:22 (m/z 737.40), G3:15 (m/z 477.23), and G3:22 (m/z 575.34) are shown. (C) Introduction of SpASFF1 driven by its endogenous promoter in BIL6180 is sufficient to cause accumulation of detectable G3:15 and G3:22 acylglucoses. ESI− mode LC-MS analysis of trichome extracts of a proSpASFF1::SpASFF1 in a BIL6180 T2 line is shown. Extracted ion chromatograms of S3:15 (m/z 639.29), S3:22 (m/z 737.40), G3:15 (m/z 477.23), and G3:22 (m/z 575.34) are shown. All m/z values correspond to the formate adducts of those acylsugars. The mass window is 0.05 Da in all experiments. Acylglucose structure is inferred from collision-induced dissociation-mediated fragmentation (fig. S8). All ESI− mode acylsugars were identified as formate adducts.
In vitro assays supported the hypothesis that SpASFF1 accepts P-type but not F-type acylsucroses as substrates. Initial attempts to express SpASFF1 fusion proteins in Escherichia coli did not produce soluble protein. For this reason, recombinant His-tagged SpASFF1 was expressed using the Nicotiana benthamiana transient expression system (41). The enzyme was tested with both P-type and F-type acylsucrose substrates purified from S. pennellii asff1 and S. lycopersicum M82, respectively. Consistent with in vivo observations, SpASFF1 demonstrated hydrolytic activity with purified P-type S3:19 (4R4,5R2,10R3) (42), yielding a compound with a mass/charge ratio (m/z) consistent with a G3:19 (4,5,10) structure (Fig. 5A and fig. S9). In contrast, SpASFF1 demonstrated no hydrolytic activity with F-type S3:22 (5R4,5R3′,12R3) (8) (Fig. 5B), suggesting that the presence of an acyl chain on the sucrose furanose ring prevents enzymatic hydrolysis. We further observed that SpASFF1 activity was undetectable with unmodified sucrose, while a commercially available yeast invertase hydrolyzed sucrose but not S3:19 (fig. S10). This SpASFF1 in vitro substrate specificity corroborates the in vivo results showing that acylglucoses only accumulate in lines containing P-type acylsucroses.
Fig. 5. SpASFF1 cleaves a P-type S3:19 acylsucrose but not an F-type S3:22 acylsucrose.
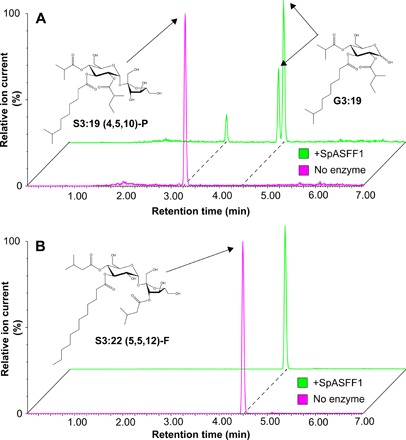
(A) LC-MS analysis of in vitro enzyme assay products indicates that SpASFF1 hydrolyzed P-type S3:19 (4R4,5R2,10R3) acylsucrose, yielding two compounds with m/z 533.3. This m/z is consistent with an acylglucose product with a G3:19 (4,5,10) configuration; the two peaks represent the α and β anomers of the acylglucose. (B) LC-MS analysis of in vitro assays with F-type S3:22 (5R4,5R3′,12R3) acylsucrose indicates no hydrolysis products with SpASFF1. Acylglucose structure is inferred from collision-induced dissociation-mediated fragmentation (fig. S9). All ESI− mode acylsugars were identified as formate adducts.
DISCUSSION
The results described above show that S. pennellii LA0716 synthesizes acylglucoses from P-type acylsucroses via the action of a previously uncharacterized trichome invertase (Fig. 5A), a homolog of the most venerable enzyme in the history of biochemistry, yeast invertase. The canonical GH32 enzyme was first characterized in the 1840s through studies of “optical inversion” of cane sugar (sucrose) into a mixture of glucose and fructose. The enzyme was assayed two decades later, and its study by Maud Leonora Menten and Leonor Michaelis led to the theory of enzyme kinetics early in the 20th century (31). Since that time, other general metabolic activities have been identified for diverse GH32 β-fructofuranosidases, including plant glycan biosynthesis, cell wall modification, and hormone metabolism (43, 44).
Our results stand in contrast to the previously proposed direct synthesis of acylglucoses from UDP-glucose and free fatty acids (27–29, 45). Steffens and co-workers (27–29, 45) identified two glucosyltransferases and an SCPL acyltransferase from S. pennellii capable of generating 1-O-monos-acylglucoses and 1,2-O-di-acylglucoses in vitro. Multiple lines of evidence indicate that these enzymes are not involved in S. pennellii acylglucose biosynthesis. First, acylglucoses generated in vitro by these enzymes are structurally distinct from the 2,3,4-O-tri-acylglucoses detected from S. pennellii (19); the in vitro products had two acyl chains instead of three and were acylated at the R1 position. Next, comparative transcriptomic data suggest that the SCPL acyltransferase shows similar expression levels in S. lycopersicum M82 and S. pennellii LA0716, yet there are no acylglucoses detected in M82 (46). In addition, the SCPL acyltransferase described by Li and Steffens (29) is encoded on chromosome 10 (Solyc10g049210), in a region not implicated in acylglucose accumulation in QTL mapping studies (30). In contrast, QTLs linked to acylglucose accumulation in S. pennellii on chromosomes 4 and 11 include SpASAT2 and SpASAT3, suggesting a connection between acylsucrose and acylglucose biosynthesis (23, 30).
As the acylsucroses and acylglucoses in S. pennellii differ only by the presence or absence of a furanose ring (Fig. 1A), we hypothesized that a GH converts the S. pennellii acylsucroses (42) to acylglucoses. Three GH genes were identified in the third acylglucose-linked QTL on chromosome 3. These genes represent members of GH families 32, 35, and 47 (table S2). Most characterized plant GH35 enzymes act as β-galactosidases, while GH47 enzymes function as α-mannosidases in posttranslational protein modification (47, 48). Thus, these were not compelling candidates for cleavage of acylated sucrose substrates. Conversely, GH32 enzymes act on a variety of β-fructofuranoside in plants, including sucrose and fructans (38, 43). Our results indicate that SpASFF1 is a “derived” β-fructofuranosidase, with an active site that can accommodate pyranose, but not furanose-acylated sucrose esters. Understanding the structural features that allow SpASFF1 to hydrolyze P-type acylsucroses could inform engineering of novel specialized metabolites in plants and microbes.
We identified the GH32 SpASFF1 β-fructofuranosidase as being necessary and sufficient for conversion of P-type acylsucroses into acylglucoses. The most direct evidence is that ablation of the SpASFF1 gene using CRISPR-Cas9 gene editing led to acylsucrose-accumulating wild tomato S. pennellii LA0716 mutants with undetectable acylglucoses, showing that the enzyme is necessary for production of acylglucoses (Fig. 3). Multiple lines of genetic and biochemical evidence support the hypothesis that SpASFF1 uses P-type acylsucrose substrates. For example, no acylglucoses were detected in the F-type acylsucrose-producing introgression line IL3-5, despite the presence of SpASFF1 in the introgressed region (Fig. 4A and fig. S1). In contrast, transgenic trichome expression of the SpASFF1 invertase in the P-type acylsucrose-producing SpASAT2 and SpASAT3 double introgression line S. lycopersicum BIL6180 resulted in acylglucose accumulation (Fig. 4). Our in vitro assay results support the in vivo evidence that P-type acylsucroses are SpASFF1 substrates. In vitro assays with recombinant SpASFF1 demonstrated conversion of the purified P-type S3:19 (4R4,5R2,10R3) to the cognate acylglucose G3:19 (4,5,10) (Fig. 5A). In contrast, the enzyme was inactive against F-type S3:22 (5R4,5R3′,12R3) (Fig. 5B) and did not hydrolyze unacylated sucrose (fig. S10).
Together, these data indicate that S. pennellii acylglucose metabolism results from evolution of a three-gene epistatic system, where the innovation of P-type acylsucrose synthesis by modification of the core BAHD acyltransferases potentiated evolution of SpASFF1 to produce acylglucoses. Our results reveal that a member of the GH32 β-fructofuranosidase enzyme family acquired expression in the trichome glandular tip cell (Fig. 2) and the ability to cleave acylated sucrose (Fig. 5), which led to an increase in the diversity of Solanaceae trichome specialized metabolites. This is a remarkable evolutionary innovation, where a member of an enzyme family long recognized as important in general metabolism was co-opted into specialized metabolism by the “blind watchmaker” of evolution.
Acylsugar accumulation is widespread throughout the Solanaceae with occurrences in genera as distantly related as Salpiglossis and Solanum, sharing a last common ancestor >30 million years (Ma) ago (8, 10, 49). While acylsugars show wide structural variation in the number and length of acyl chains throughout the family, sucrose is the most prominent sugar core. Acylsucroses not only accumulate in genera whose lineages diverged <20 Ma ago, such as Solanum and Physalis (8, 21), but also accumulate in species representing earlier diverging lineages, including Salpiglossis and Petunia (10, 12). In addition, acyl chains are present on the furanose ring in at least some members of each of these genera, suggesting that accumulation of F-type acylsucroses evolved long ago.
Although apparently limited in distribution relative to acylsucroses, acylglucoses occur in diverse genera including Solanum, Datura, and Nicotiana (16, 19, 20). Despite the fact that acylglucose accumulation is common to species in both Solanum and Nicotiana, which diverged approximately 24 Ma ago, the differences in SpASFF1 gene expression and SpASAT substrate specificity that facilitated acylglucose accumulation in S. pennellii arose in the ~7 Ma since divergence from the last ancestor in common with S. lycopersicum (11, 49, 50). This supports independent evolutionary origins of acylglucoses in distinct lineages. In the Solanum genus, P-type acylsucroses are a prerequisite for acylglucose accumulation. The predominance of F-type acylsucroses within the Solanaceae may explain the relative rarity of acylglucoses in the family. However, characterization of the ASAT enzymes responsible for acylsucrose biosynthesis in Salpiglossis, Petunia, and Solanum demonstrates multiple changes in enzyme substrate specificity throughout the evolutionary history of the acylsucrose pathway (10–12). Plasticity of the acylsugar pathway may have given rise to P-type acylsucroses multiple times throughout evolutionary history. If so, this would provide independent opportunities for co-option of acylglucose-producing GHs into the acylsugar pathway. Are the enzymes responsible for hydrolyzing acylsucroses to yield acylglucoses restricted to the GH32 family or have other enzyme families evolved in different acylglucose-accumulating lineages? Whether and to what extent multiple origins of acylglucose biosynthesis share common features remains to be explored.
Over the past decade, discovery of pathways and enzymes of plant specialized metabolism has improved at an increasing rate. Before this, the taxonomic restriction of specialized metabolism biased deep analysis toward pathways found in model organisms: for instance, glucosinolates in Arabidopsis, cyclic hydroxamic acids in maize and other well-studied grasses, and isoflavonoids in Medicago and soybean. Marked improvements in sensitivity and selectivity of MS- and NMR-based analytical chemistry helped broaden the scope of well-studied metabolic networks (51, 52). In parallel, development of species-agnostic DNA sequencing and functional genomics screening tools (notably, virus-induced gene silencing and genetic transformation) permitted rigorous correlation of in vitro activities and in vivo phenotypes. The rapid advancement of gene editing techniques using CRISPR-Cas on agriculturally important and undomesticated species markedly expands the specialized metabolism functional genomics toolkit. These methods allow not only direct tests of in vivo function but also elimination of the T-DNA by simple genetic crossing. The removal of the T-DNA permits growing edited mutants in agricultural fields or common gardens with lower regulatory barriers. For example, the spasff1 mutant lines can be used to understand the impacts of acylsucroses versus acylglucoses on the fitness of S. pennellii both in the greenhouse and in the field. These studies could lead to crops with engineered natural pesticides, broaden our understanding of the roles of specialized metabolites in mediating environmental interactions, and inform our understanding of the mechanisms underpinning specialized metabolic evolution.
MATERIALS AND METHODS
Plant material
Seeds of S. lycopersicum M82 were obtained from the C.M. Rick Tomato Genetics Resource Center (TGRC; University of California, Davis, CA); seeds of IL3-5, BIL6180, and BIL6521 were obtained from D. Zamir (Hebrew University of Jerusalem, Rehovot, Israel) (36); seeds of S. pennellii LA0716 were provided by M. Mutschler (Cornell University, Ithaca, NY). Seeds were treated with half-strength bleach for 30 min and rinsed three times in deionized water for 5 min before sowing on moist filter paper in petri dishes. Upon germination, seedlings were transferred to soil. Young plants were grown in 9-cm pots in a peat-based propagation mix (Sun Gro Horticulture, Agawam, MA). S. lycopersicum and introgression lines were watered four times weekly with deionized water and supplemented once weekly with half-strength Hoagland’s solution; S. pennellii was watered once weekly with deionized water and supplemented once weekly with half-strength Hoagland’s solution. Plants used for analysis were grown in a growth chamber under a 16-hour photoperiod [190 μmol m−2 s−1 photosynthetic photon flux density (PPFD)] with 28°C day and 22°C night temperatures set to 50% relative humidity. BIL lines used for crosses were grown in a soil mix consisting of four parts SUREMIX (Michigan Grower Products Inc., Galesburg, MI) to one part sand in a greenhouse with a daytime maximum temperature of 30°C and a nighttime minimum temperature of 16°C; sunlight was supplemented with high-pressure sodium bulbs on a 16-hour light/8-hour dark cycle. For seed production, S. pennellii asff1 T0 plants were grown in soil containing one part Canadian sphagnum (Mosser Lee Co., Millston, WI), one part coarse sand (Quikrete, Atlanta, GA), one part white pumice (Everwood Farm, Brooks, OR), and one part redwood bark (Sequoia Bark Sales, Reedley, CA) supplemented with 1.8 kg of crushed oyster shell (Down to Earth Distributors Inc., Eugene, OR), 1.8 kg of hydrated lime (Bonide Products Inc., Oriskany, NY), and 0.6 kg of triple super phosphate (T & N Inc., Foristell, MO) per cubic meter.
Acylsugar analysis
The acylsugar extraction interactive protocol is available in Protocols.io at http://dx.doi.org/10.17504/protocols.io.xj2fkqe. Leaf surface acylsugars were extracted from single leaflets with 1 ml of a mixture of isopropanol (J.T.Baker, Phillipsburg, NJ)/acetonitrile (Sigma-Aldrich, St. Louis, MO)/water (3:3:2) with 0.1% formic acid and 1 μM telmisartan (Sigma-Aldrich, St. Louis, MO) as a high-performance liquid chromatography (HPLC) standard. The leaf tissue was gently agitated on a rocker in this extraction solvent for 2 min. The extraction solvent was collected and stored in 2-ml LC-MS vials at −80°C.
LC-MS samples (both enzyme assays and plant samples) were run on a Waters ACQUITY UPLC coupled to a Waters Xevo G2-XS QToF mass spectrometer. Ten microliters of the acylsugar extracts were injected into an Ascentis Express C18 HPLC column (10 cm by 2.1 mm, 2.7 μm) (Sigma-Aldrich, St. Louis, MO), which was maintained at 40°C. The LC-MS methods used the following solvents: 10 mM ammonium formate (pH 2.8) as solvent A and 100% acetonitrile as solvent B. Compounds were eluted using one of two gradients.
A 7-min linear elution gradient consisted of 5% B at 0 min, 60% B at 1 min, 100% B at 5 min, held at 100% B until 6 min, 5% B at 6.01 min, and held at 5% B until 7 min. A 21-min linear elution gradient consisted of 5% B at 0 min, 60% B at 3 min, 100% B at 15 min, held at 100% B until 18 min, 5% B at 18.01 min, and held at 5% B until 21 min.
The MS settings were as follows for negative ion-mode electrospray ionization (ESI−): capillary voltage, 2.00 kV; source temperature, 100°C; desolvation temperature, 350°C; desolvation nitrogen gas flow rate, 600 liters/hour; cone voltage, 35 V; and mass range, m/z 50 to 1000 (with spectra accumulated at 0.1 s per function). Three separate acquisition functions were set up to test different collision energies (0, 15, and 35 V).
The MS settings for positive ion-mode ESI were as follows: capillary voltage, 3.00 kV; source temperature, 100°C; desolvation temperature, 350°C; desolvation nitrogen gas flow rate, 600 liters/hour; cone voltage, 35 V; and mass range, m/z 50 to 1000 (with spectra accumulated at 0.1 s per function). Three separate acquisition functions were set up to test different collision energies (0, 15, and 45 V). Lock mass correction was performed using leucine enkephalin as the reference compound for data acquired in both negative and positive ion mode.
Acylsugar quantification
To accurately quantify total acylsugars, samples were saponified before LC-MS analysis and sugar cores were quantified with authentic isotopically labeled standards. A leaflet was immersed in 2 ml of dichloromethane (VWR International, Radnor, PA) and 500 μl of water with 30-s vortexing. After phase separation, 1 ml of the dichloromethane layer was removed to a borosilicate glass vial and evaporated to dryness under flowing air. Dried samples were dissolved in 1 ml of acetonitrile with 0.1% formic acid for storage. Twenty-microliter aliquots of acylsugar extracts were dried in a 1.7-ml microcentrifuge tube using SpeedVac and dissolved in 100 μl of methanol. An equal volume of 3 N aqueous ammonia solution (Sigma-Aldrich, St. Louis, MO) was added, and the reaction was incubated in a sealed 1.5-ml microcentrifuge tube for 48 hours in a fume hood. Before LC-MS analysis, samples were dissolved in 200 μl of ammonium bicarbonate (pH 7 to 8) in 90% acetonitrile containing 0.5 μM 13C12-sucrose and 0.5 μM 13C6-glucose as internal standards and transferred to 2-ml LC-MS vials. Compounds were analyzed on a Waters ACQUITY TQD Tandem Quadrupole UPLC/MS/MS system (Waters, Milford, MA). Ten microliters of the acylsugar extracts were injected into a Waters ACQUITY UPLC BEH amide column (2.1 mm by 100 mm, 1.7 μM) in a column oven with a temperature of 40°C and with a flow rate of 0.5 ml/min. The LC-MS methods used 10 mM ammonium bicarbonate (pH 8) in 50% acetonitrile as solvent A and 10 mM ammonium bicarbonate (pH 8) in 90% acetonitrile as solvent B. The chromatography gradient was as follows: 100% B at 0 min, 0% B at 5 min, 100% B at 5.01 min, and held at 100% B until 10 min. Multiple-reaction monitoring mode was operated to detect each sugar. For glucose: precursor ion, m/z 179; product ion, m/z 89; cone voltage, 16 V; collision energy, 10 V. For 13C6-glucose: precursor ion, m/z 185; product ion, m/z 92; cone voltage, 16 V; collision energy, 10 V. For sucrose: precursor ion, m/z 341; product ion, m/z 89; cone voltage, 40 V; collision energy, 22 V. For 13C12-sucrose: precursor ion, m/z 353; product ion, m/z 92; cone voltage, 40 V; collision energy, 22 V. Quantification of glucose and sucrose was conducted by standard curves with authentic glucose and sucrose standards (Sigma-Aldrich, St. Louis, MO). Glucose and sucrose standard solutions of 31.25, 62.5, 125, 250, and 500 μM in water were prepared and processed using the same protocol for acylsugars, as described above.
Acylsucrose purification
All purifications were performed using a Waters 2795 Separations Module (Waters, Milford, MA) and an Acclaim 120 C18 HPLC column (4.6 mm by 150 mm, 5 μm; Thermo Fisher Scientific, Waltham, MA) with a column oven temperature of 30°C and a flow rate of 1 ml/min. The mobile phase consisted of water (solvent A) and acetonitrile (solvent B). Fractions were collected using a 2211 Superrac fraction collector (LKB Bromma, Stockholm, Sweden).
For purification of acylsucroses from S. pennellii LA0716, approximately 75 g of fresh aboveground tissue of mature S. pennellii asff1-1 was harvested into a 1-liter glass beaker to which 500 ml of 100% methanol was added. Tissue was stirred for 2 min and filtered through Miracloth (EMD Millipore, Billerica, MA) prewetted with methanol into a 1-liter round-bottom flask. The solvent was removed with a rotary evaporator in a water bath held between 35° and 40°C, and the residue was dissolved in 5 ml of acetonitrile. A 5-μl aliquot of this solution was diluted 1000-fold in 9:1 water/acetonitrile with 0.1% formic acid for chromatographic purification. The S3:19 compound was purified from 20 injections of 100 μl each using a linear elution gradient of 1% B at 0 min, 63% B at 10 min, 65% B at 30 min, 100% B at 35 min, brought back to 1% B at 35.01 min, and held at 1% B until 40 min. Eluted compounds were collected in 10-s fractions. Fraction collection tubes contained 333 μl of 0.1% formic acid in water, and the S3:19 product was eluted at 18 to 19 min.
For purification of the S3:22 acylsucrose from S. lycopersicum M82, approximately 75 g of fresh aboveground tissue was harvested from mature plants into a 500-ml glass beaker to which 250 ml of 100% methanol was added. The tissue was stirred for 2 min and filtered through Miracloth into a 1-liter round-bottom flask. Methanol was removed with a rotary evaporator in a water bath held between 35° and 40°C, and the residue was dissolved in 5 ml of acetonitrile. This solution was diluted 50-fold in 9:1 water/acetonitrile with 0.1% formic acid for further processing. The S3:22 compound was purified from 10 injections of 100 μl using a linear elution gradient of 1% B at 0 min, 50% B at 5 min, 70% B at 30 min, 100% B at 32 min, held at 100% until 35 min, brought back to 1% B at 35.01 min, and held at 1% B until 40 min. Eluted compounds were collected in 1-min fractions. Fraction collection tubes contained 333 μl of 0.1% formic acid in water, and the S3:22 product was eluted at 7 to 8 min.
qPCR analysis
Tissue of 10-week-old S. pennellii LA0716 and S. lycopersicum M82 were harvested as follows: stems were flash-frozen in liquid nitrogen, and trichomes were shaved into 1.5-ml microcentrifuge tubes with a clean razor blade. Trichomes and denuded stems were kept in liquid nitrogen and ground with plastic micropestles in 1.5-ml microcentrifuge tubes. RNA was extracted from ground trichomes and stems (six biological replicates for each species and tissue type) using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. For each sample, 250 ng of RNA as quantified using a NanoDrop 2000c (Thermo Fisher Scientific, Waltham, MA) was used to synthesize cDNA using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). qRT-PCR was carried out using SYBR Green PCR Master Mix on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Warrington, UK) using the following cycling conditions: 48°C for 30 min, 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by melt curve analysis. RT_ASFF_F and RT_ASFF_R primers were used to detect the ASFF1 transcript; RT_EF-1a_F/R, RT_actin_F/R, and RT_ ubiquitin_F/R primers were used to detect transcripts of the EF-1α, actin, and ubiquitin genes, respectively (table S3). For each biological replicate, relative levels of the ASFF1 transcript were determined using the ΔΔCt method (53) and normalized to the geometric mean of EF-1α, actin, and ubiquitin transcript levels.
Genotyping of progeny of BIL6521 × BIL6180
DNA from the progeny of the cross between BIL6521 and BIL6180 was extracted from leaf material that were archived on FTA (Flinders Technology Associates) PlantSaver cards (GE Healthcare, Uppsala, Sweden) and purified according to the manufacturer’s specifications. Extracted DNA from the FTA cards were used for PCR amplification with GoTaq Green Mastermix to genotype the sample using 04g011460_Marker_Indel-F/R and ASFF_Chr3_Indel_002_F/R (table S3).
DNA construct assembly
All Sanger DNA sequencing confirmation in this study was performed with the indicated sequencing primers at the Research Technology Support Facility Genomics Core, Michigan State University, East Lansing, MI.
For proSpASFF1::SpASFF1 ORF (pK7WG), a 1.8-kb region of the upstream region and ORF of SpASFF1 was split into four amplicons using four sets of primers: ASFF_001_F/R, ASFF 002_F/R, ASFF_003_F/R, and ASFF_004_F/R (table S3). The first and fourth amplicon contained adapters for assembly into pENTR-D-TOPO that has been digested with NotI/AscI, respectively. The construct was assembled using New England Biolabs (NEB) Gibson Assembly according to manufacturer specifications (NEB, Ipswich, MA). The construct was verified by Sanger sequencing using M13 Forward, T7 promoter primers, and cloning primers. The insert was subcloned into pK7WG (54) using the LR Clonase II Enzyme mix (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Presence of the insert was determined by colony PCR using ASFF_001F/R. Completed vectors were transformed into Agrobacterium strain AGL0. Leaf material from recovered plants were archived on FTA PlantSaver cards (GE Healthcare, Uppsala, Sweden) and genotyped by PCR amplification with GoTaq Green Mastermix and pK7WG-Kan-F/R primers (table S3).
For proSpASFF1::GFP/GUS (pKGWFS7), a 1.8-kb region of the upstream region of ASFF1 was amplified from S. pennellii LA0716 genomic DNA using ASFF_promoter_F1/R1 primers (table S3). pENTR-D-TOPO was digested with Not I/Asc I to linearize the vector and create overhangs compatible for Gibson Assembly. The amplicon also contained adapters for insertion into pENTR-D-TOPO digested with Not I/Asc I. Constructs were Sanger-sequenced using M13F/R primers in addition to the ASFF_promoter_F1/R1 primers. LR Clonase II Enzyme mix was used to subclone the fragment into pKGWFS7 (54). The construct was transformed into AGLO for plant transformation using the described protocol.
The CRISPR-ASFF1 vector was constructed as follows. CRISPR sgRNAs were designed using the site finder toolset in Geneious v10 (www.geneious.com). Two target sequences located on the exon were selected for their high on-target activity scores, based on a published algorithm (55), and low off-target scores against published S. pennellii genome database (56). Each sgRNA was obtained as a gBlock synthesized in vitro by IDT (Integrated DNA technologies) (www.idtdna.com) (table S3) and subsequently assembled with pICH47742::2x35S-5′UTR-hCas9(STOP)-NOST [Addgene plasmid no. 49771, provided by S. Kamoun (Sainsbury Lab, Norwich, UK)] (57), pICH41780 (Addgene plasmid no. 48019) and pAGM4723 (Addgene plasmid no. 48015, both gifts from S. Marillonnet) (58), and pICSL11024 [Addgene plasmid no. 51144, a gift from J. D. Jones (Sainsbury Lab, Norwich, UK)] using Golden Gate Assembly. In short, the restriction-ligation reactions (20 μl) were set up by mixing 15 ng of synthesized sgRNAs with 1.5 μl of T4 ligase buffer (NEB), 320 U of T4 DNA ligase (NEB), 1.5 μl of bovine serum albumin (BSA; 0.1 mg/ml; NEB), 8 U of BpiI (Thermo Fisher Scientific), and 100 to 200 ng of the intact plasmids. The reactions were incubated at 37°C for 30 s, followed by 26 cycles (37°C, 3 min; 16°C, 4 min), and then incubated at 50°C for 5 min and 65°C for 5 min. The ligated products were directly used to transform E. coli competent cells. Positive clones were chosen based on colony PCR and sequenced at the Michigan State University Research Technology Support Facility (MSU RTSF) using the pAGM4723_SeqF1, pAGM4723_SeqR1, pICSL11024_SeqF1, pICH47742CAS9_SeqF2, pICH47742_SeqF1, pICH41780_SeqR1, and ASFF_SeqR primers (table S3). The construct was transformed into S. pennellii LA0716 using the plant transformation protocol described below. Leaf material from recovered plants were archived on FTA PlantSaver cards (GE Healthcare, Uppsala, Sweden) and genotyped by PCR amplification with ASFF_F/R, followed by Sanger sequencing with ASFF_SeqR (table S3).
For spasff1 line transcript analysis, RNA was extracted from spasff1-1-1/1-1-2 lines using RNeasy Plant Mini Kit according to the kit specifications (Qiagen, Venlo, The Netherlands). RNA was quantified using a NanoDrop 2000c (Thermo Fisher Scientific, Waltham, MA). One microgram of RNA was used for cDNA synthesis using Superscript II Reverse Transcriptase according to the manufacturer’s specifications. The ASFF1_transcript_amp_01F/R primers (final concentration, 0.5 μM) were used to amplify the region within the ASFF1 CDS (coding sequence), which was cloned into pMINI-T 2.0 (NEB, Ipswich, MA). T7 and SP6 promoter primers were used for Sanger sequence confirmation of the inserts, and ClustalW was used for alignment of the transcripts (www.ebi.ac.uk/Tools/msa/clustalo/).
Competent cell preparation and transformation of constructs into Agrobacterium
A single colony of AGL0 or LBA4404 Agrobacterium was inoculated into two 5-ml cultures of YEP media [10 g of yeast extract, 10 g of Bacto peptone, and 5 g of NaCl per liter (pH 7)] with rifampicin (50 μg/ml). Cultures were incubated in borosilicate glass test tubes (18 mm by 150 mm) with foam plugs overnight at 30°C, shaken at 200 rpm. One hundred ninety milliliters of LB was inoculated with 10 ml of the overnight cultures in an autoclaved 500-ml Erlenmeyer flask. Cultures were grown in a shaking incubator (200 rpm) at 30°C to OD600 (optical density at 600 nm) = 1.0, incubated in ice for 10 min, and centrifuged at 4°C in 50-ml conical tubes at 3200g for 5 min. Pellets were resuspended in 1 ml of sterile 20 mM CaCl2 and 100 -μl aliquots; dispensed into sterile, prechilled 1.7-ml microcentrifuge tubes; snap-frozen using liquid nitrogen; and stored at −80°C.
For Agrobacterium transformation, 1 μg of construct DNA purified using an Omega EZNA Plasmid DNA Mini Kit I (Omega Bio-Tek, Norcross, GA) was added to the frozen Agrobacterium aliquots on ice. Cells were thawed in a 37°C water bath for 5 min, mixed well by flicking, and snap-frozen in liquid nitrogen. Cells were thawed, and 1 ml of YEP was added to the tube. The transformations were incubated at 28°C and shaken at 200 rpm for 4 hours. Cells were centrifuged at 17,000g for 30 s, the supernatant was decanted, and the cell pellet was resuspended in 100 μl of fresh YEP. The cell pellet was resuspended, and the entire suspension was plated onto an LB plate with spectinomycin (100 μg/ml).
Presence of the insert containing vector was verified by colony PCR. Colonies were collected with a pipette tip and resuspended in 20 μl of sterile water. Two microliters of the cell suspension was added to a PCR tube with a reaction with a final volume of 25 μl. GoTaq Green Mastermix (2×) was used for the colony PCR according to the manufacturer’s specifications (Promega, Madison, WI). Primers (final concentration, 0.4 μM) pertaining to the insert were used for amplification.
Plant transformation
In all cases, petri plates containing plant tissue were sealed with a single layer of micropore paper tape (3M, Maplewood, MN). Transformation of S. lycopersicum and S. pennellii LA0716 was performed using AGL0 using a modification of published protocols (59, 60). Fifty to sixty seeds were incubated in 40% bleach and agitated for 5 min. Seeds were rinsed six times, each with 40 ml of sterile double-distilled H2O with 5 min of rocking and decanting of wash solution. A flame-sterilized spatula was used to distribute the seeds onto the surface of 1/2× MSO medium (Murashige and Skoog media with no sucrose) (59) in a Phytatray II (Sigma-Aldrich, St. Louis, MO). Containers were incubated at 25°C on a 16-hour light/8-hour dark cycle with a light intensity of 70 μmol m−2 s−2 PPFD.
At day 8 for S. lycopersicum or day 11 for S. pennellii LA0716, the seedlings were removed from the 1/2× MSO medium jar. The hypocotyl and radicle were excised and discarded. The cotyledon explant was placed on a sterile petri dish. One to two millimeters was removed from the base and tip of the cotyledon. An autoclaved piece of Whatman #1 filter paper (GE Healthcare, Uppsala, Sweden) was placed on the surface of a sterile D1 media plate (59) on which the cotyledons were placed adaxial side up. Approximately 100 explants were added per plate. The plates were placed under the same conditions for 2 days until day 10.
For cocultivation, the Agrobacterium containing the construct was streaked out onto an LB plate containing the appropriate antibiotic. A single colony was inoculated into a 25-ml LB culture with the same antibiotic plus rifampicin (50 mg/liter) in a 250-ml Erlenmeyer flask. The culture was incubated at 30°C in a shaking incubator (225 rpm) for 2 days. The culture was transferred to a sterile 50-ml conical tube and centrifuged at 3200g for 10 min at 20°C. The supernatant fluid was decanted, and 10 ml of MSO media (59) was added to the tube (with no pellet resuspension). The cell pellet was centrifuged at 2000g for 5 min, and this washing step was then repeated. The cell pellet was resuspended in 10 to 20 ml of MSO liquid media. Absorbance of the culture was measured at 600 nm. The suspension was diluted with MSO to OD600 = 0.5. Acetosyringone dissolved in dimethyl sulfoxide (DMSO) was added at a final concentration of 375 μM, and 5 ml of the Agrobacterium suspension was pipetted onto the cotyledons on the plate and incubated with swirling at room temperature for 10 min, at which point the excess culture was pipetted off. Using a scalpel, cotyledons were transferred to a fresh D1 medium plate containing autoclaved Whatman paper. Approximately 50 cotyledons per plate were placed abaxial side up. Plates were incubated at 24°C for 2 days with a 16-hour light/8-hour dark cycle at 70 μmol m−2 s−2 PPFD.
For transgenic callus selection, 2 days after cocultivation, the cotyledons were transferred directly onto sterile 2Z media plates (60) containing kanamycin (100 μg/ml) and timentin (200 μg/ml; no filter paper). Explants were placed abaxial side up with 20 to 30 cotyledons per plate. Plates were incubated under the same growth conditions for 10 days. Cotyledons were then transferred to a sterile petri dish and, using a scalpel, calluses were cut and then placed onto fresh 2Z media plates with the same selection. Subsequently, explants were transferred to new 2Z plates every 2 weeks. Throughout the process, dying tissue was removed, and growing tissue was placed on the media. Five to eight weeks after cocultivation, shoots were harvested from the explants and placed into Phytatray II (Sigma-Aldrich, St. Louis, MO) containing 100 ml of Murashige and Skoog salts, 3% sucrose, and Nitsch Vitamins, 0.8% bactoagar, pH 6.0 (MSSV) media (Murashige and Skoog salts, 3% sucrose, and Nitsch Vitamins, 0.8% bactoagar, pH 6.0) (60) supplemented with timentin (100 μg/ml), kanamycin (50 μg/ml), and indole-3 butyric acid (1 μg/ml). MSSV-containing Phytatrays were incubated under the same growth conditions (16-hour light/8-hour dark cycle at 70 μmol m−2 s−2 PPFD). Shoots were monitored for leaf and root production, and shoots with roots and leaves were placed into pots containing Redi-Earth soil. Flats were covered with a plastic dome under the same growth conditions. Domes were removed from flats after 3 to 4 days.
Transient expression and purification of SpASFF1 protein
The ASFF1 CDS was amplified from S. pennellii LA0716 trichome cDNA using ASFF_F and ASFF_R primers (table S3) and cloned into the pGEM backbone using the pGEM-T Easy cloning kit (Promega, Madison, WI). The ASFF1 CDS was subsequently reamplified with the (pEAQ-HT)-ASFF-His_F and (pEAQ-HT)-ASFF-His_R primers (table S3) to add adapters for Gibson Assembly. The resulting PCR product was transferred to pEAQ-HT vector (41) previously digested with NruI–HF and SmaI restriction enzymes (NEB, Ipswich, MA) using 2× Gibson Assembly master mix (NEB, Ipswich, MA) according to the manufacturer’s instructions to create an expression clone coding for the full-length protein with a C-terminal 6× His tag (ASFF1-HT-pEAQ). The completed vector was subsequently transformed into LBA4404 cells, as described above. For transient expression, an Agrobacterium tumefaciens LBA4404 strain carrying the ASFF1-HT-pEAQ construct was streaked onto LB agar containing rifampicin (50 μg/ml) and kanamycin (50 μg/ml) and incubated for 3 days at 28°C. Single colonies were used to inoculate 250-ml Erlenmeyer flasks containing 50 ml of YEP medium with rifampicin (50 μg/ml) and kanamycin (50 μg/ml); cultures were incubated at 28°C and shaken at 300 rpm overnight. Cultures were harvested by centrifugation at 800g at 20°C for 20 min. The supernatant was discarded, and the resulting loose pellet was resuspended in 50 ml of buffer A [10 mM 2-ethanesulfonic acid (MES; Sigma-Aldrich, St. Louis, MO) at pH 5.6 and 10 mM MgCl2]. This cell suspension was centrifuged at 800g at 20°C for 20 min, and the resulting pellet was resuspended to a final OD600 = 1.0 with buffer A. A 200 mM solution of acetosyringone (Sigma-Aldrich, St. Louis, MO) dissolved in DMSO was added to the suspension at a final concentration of 200 μM, and the suspension was incubated at room temperature with gentle rocking for 4 hours. This suspension was infiltrated into fully expanded leaves of 6-week-old N. benthamiana plants using a needleless 1-ml tuberculin syringe. Plants were grown under a 16-hour photoperiod (70 μmol m−2 s−1 PPFD) and at a constant 22°C set to 70% relative humidity. At 8 days after infiltration, 28 g of infiltrated leaves was harvested, deveined, and flash-frozen in liquid nitrogen. Tissue was powdered under liquid nitrogen with a mortar and pestle and added to 140 ml of ice-cold buffer B [25 mM 3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid (EPPS) at pH 8.0, 1.5 M NaCl, 1 mM EDTA with 2 mM dithiothreitol (DTT), 1 mM benzamidine, 0.1 mM phenylmethansesulfonyl fluoride, 10 μM trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E-64), and 5% (w/v) polyvinylpolypyrrolidone (PVPP); all reagents were obtained from Sigma-Aldrich (St. Louis, MO) except DTT, which was obtained from Roche Diagnostics (Risch-Rotkreuz, Switzerland)]. The mixture was stirred for 4 hours at 4°C, filtered through six layers of Miracloth, and centrifuged at 27,000g at 4°C for 30 min. The supernatant was decanted and passed through a 0.22-μm polyethersulfone filter (EMD Millipore, Billerica, MA) before being loaded onto a HisTrap HP 1-ml affinity column and eluted using a gradient of 10 to 500 mM imidazole in buffer B using an ÄKTA start FPLC module (GE Healthcare, Uppsala, Sweden). Fractions were analyzed by SDS–polyacrylamide gel electrophoresis, and the presence of ASFF1-HT was confirmed by immunoblot using the BMG–His-1 monoclonal antibody (Roche, Mannheim, Germany) to detect His-tagged proteins. Purified ASFF1-HT was subsequently transferred to 100 mM sodium acetate (pH 4.5) with 50% glycerol using a 10DG desalting column (Bio-Rad, Hercules, CA). Protein was quantified against a standard curve of BSA (Thermo Fisher Scientific, Waltham, MA) using a modified Bradford reagent (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions.
Enzyme assays
For activity assays, 100 ng of ASFF1-HT or 1 μg of Saccharomyces cerevisiae invertase (catalog no. I4504, Grade VII, Sigma-Aldrich, St. Louis, MO) and 0.1 nmol F- or P-type acylsucrose or 10 nmol sucrose (Sigma-Aldrich, St. Louis, MO) were added to 30 μl of 50 mM sodium acetate (pH 4.5) in 250-μl thin-wall PCR tubes. Reactions were incubated for 1 hour at 30°C and stopped by the addition of 60 μl of 1:1 acetonitrile/isopropanol containing 1.5 μM telmisartan as internal standard and centrifuged for 10 min at 16,000g to remove precipitated protein. The supernatant was transferred to 2-ml autosampler vials with 250-μl glass inserts and analyzed by LC-MS as described above.
Statistical analysis
All statistical analyses were performed using the “stats” R package (R Core Team, 2017). One-way analysis of variance (ANOVA) was executed on acylsugar data using the “aov” command. Between- and within-group variances were determined using the sum-of-squares values obtained from ANOVA; these values were subsequently used to determine the power of the ANOVA using the “power.anova.test” function. Analysis by Tukey’s post hoc mean-separation test was executed using the “TukeyHSD” command, with the results of one-way ANOVA as input. Welch two-sample t tests were executed on transcript abundance data using the “t.test” command. The power of these analyses was determined using the “power.t.test” function.
Supplementary Material
Acknowledgments
We thank M. Mutschler, D. Zamir, and the C. M. Rick TGRC (University of California, Davis, CA) for providing germplasm essential to this study. Plant transformation was performed by K. Imre. We are also grateful to R. Chetelat and X. Qin for advice about S. pennellii transformation and D. Jones, L. Chen, and the MSU RTSF Mass Spectrometry and Metabolomics facility for analytical chemistry advice. We thank C. Schenck for editorial suggestions and E. Pichersky and members of the MSU Solanaceae Specialized Metabolism Evolution group for advice. Funding: This work was funded by National Science Foundation grant IOS-PGRP-1546617 (to R.L.L.) and National Institute of General Medical Sciences of the National Institutes of Health graduate training grant no. T32-GM110523 (to B.J.L. and D.B.L.). Author contributions: Design: B.J.L., D.B.L., Y.-R.L., P.F., A.L.S., and R.L.L. Experiments and data analysis: B.J.L., D.B.L., and Y.-L.R. Interpretation and preparation of figures and table: B.J.L., D.B.L., Y.-R.L., P.F., A.L.S., and R.L.L. Writing: B.J.L., D.B.L., Y.-R.L., P.F., A.L.S., and R.L.L. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Sequence data for SpASFF1 are available in GenBank: SpASFF1 (MK297330). The following materials require a material transfer agreement: pEAQ-HT, pK7WG, pKGWFS7, pICH47742::2x35S-5′UTR-hCas9(STOP)-NOST, pICH41780, pAGM4723, and pICSL11024. Requests for biological materials or data should be submitted to R.L.L. at lastr@msu.edu.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaaw3754/DC1
Fig. S1. Acylglucoses are not detected in trichome extracts of IL3-5, IL4-1, or IL11-3.
Fig. S2. Quantification of acylsugars in S. lycopersicum M82 and breeding lines containing S. pennellii LA0716 introgressions.
Fig. S3. Comparison of major acylsugars in BIL6521 and BIL6521 × BIL6180 F2 progeny using LC-MS.
Fig. S4. Mass spectra of major acylsugars S3:15, S3:22, G3:15, and G3:22 from BIL6521 × BIL6180—F2 lines, BIL6180, and BIL6521.
Fig. S5. Mutated genomic sequence of three homozygous spasff1 CRISPR-Cas9 lines.
Fig. S6. Acylsugars in BPI chromatograms of spasff1 and LA0716 plants.
Fig. S7. Comparison of acylsugars from IL3-5 and parental M82 using LC-MS.
Fig. S8. LC-MS analysis of P-type acylsucrose-producing S. lycopersicum BIL6180 stably transformed with proSpASFF1::SpASFF1.
Fig. S9. Mass spectra of G3:19-derived from SpASFF1 in vitro assay.
Fig. S10. SpASFF1 cleaves a purified P-type triacylsucrose but not unmodified sucrose while yeast invertase cleaves unmodified sucrose but not triacylsucrose.
Table S1. Annotation of acylsugars identified in BIL6521 and BIL6521 × BIL6180 F2 using LC-MS and collision-induced dissociation.
Table S2. Annotation of three GH candidates for SpASFF1 identified in the AG3.2.
Table S3. Primers/gBlocks/sgRNAs used in this study.
REFERENCES AND NOTES
- 1.Pichersky E., Lewinsohn E., Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Niculaes C., Abramov A., Hannemann L., Frey M., Plant protection by benzoxazinoids—Recent insights into biosynthesis and function. Agronomy 8, 143 (2018). [Google Scholar]
- 3.Tsunoda T., van Dam N. M., Root chemical traits and their roles in belowground biotic interactions. Pedobiologia 65, 58–67 (2017). [Google Scholar]
- 4.Block A. K., Vaughan M. M., Schmelz E. A., Christensen S. A., Biosynthesis and function of terpenoid defense compounds in maize (Zea mays). Planta 249, 21–30 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Leong B. J., Last R. L., Promiscuity, impersonation and accommodation: Evolution of plant specialized metabolism. Curr. Opin. Struct. Biol. 47, 105–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moghe G. D., Last R. L., Something old, something new: Conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol. 169, 1512–1523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilmiller A. L., Charbonneau A. L., Last R. L., Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl. Acad. Sci. U.S.A. 109, 16377–16382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh B., Westbrook T. C., Jones A. D., Comparative structural profiling of trichome specialized metabolites in tomato (Solanum lycopersicum) and S. habrochaites: Acylsugar profiles revealed by UHPLC/MS and NMR. Metabolomics 10, 496–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schilmiller A. L., Moghe G. D., Fan P., Ghosh B., Ning J., Jones A. D., Last R. L., Functionally divergent alleles and duplicated loci encoding an acyltransferase contribute to acylsugar metabolite diversity in Solanum trichomes. Plant Cell 27, 1002–1017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moghe G. D., Leong B. J., Hurney S. M., Jones A. D., Last R. L., Evolutionary routes to biochemical innovation revealed by integrative analysis of a plant-defense related specialized metabolic pathway. eLife 6, e28468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan P., Miller A. M., Liu X., Jones A. D., Last R. L., Evolution of a flipped pathway creates metabolic innovation in tomato trichomes through BAHD enzyme promiscuity. Nat. Commun. 8, 2080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadakuduti S. S., Uebler J. B., Liu X., Jones A. D., Barry C. S., Characterization of trichome-expressed BAHD acyltransferases in Petunia axillaris reveals distinct acylsugar assembly mechanisms within the Solanaceae. Plant Physiol. 175, 36–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Enright M., Barry C. S., Jones A. D., Profiling, isolation and structure elucidation of specialized acylsucrose metabolites accumulating in trichomes of Petunia species. Metabolomics 13, 85 (2017). [Google Scholar]
- 14.Weinhold A., Baldwin I. T., Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc. Natl. Acad. Sci. U.S.A. 108, 7855–7859 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luu V. T., Weinhold A., Ullah C., Dressel S., Schoettner M., Gase K., Gaquerel E., Xu S., Baldwin I. T., O-acyl sugars protect a wild tobacco from both native fungal pathogens and a specialist herbivore. Plant Physiol. 174, 370–386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzaki T., Shinozaki Y., Suhara S., Shigematsu H., Koiwai A., Isolation and characterization of tetra- and triacylglucose from the surface lipids of Nicotiana miersii. Agric. Biol. Chem. 53, 3343–3345 (1989). [Google Scholar]
- 17.Escobar-Bravo R., Alba J. M., Pons C., Granell A., Kant M. R., Moriones E., Fernández-Muñoz R., A jasmonate-inducible defense trait transferred from wild into cultivated tomato establishes increased whitefly resistance and reduced viral disease incidence. Front. Plant Sci. 7, 1732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leckie B. M., D’Ambrosio D. A., Chappell T. M., Halitschke R., De Jong D. M., Kessler A., Kennedy G. G., Mutschler M. A., Differential and synergistic functionality of acylsugars in suppressing oviposition by insect herbivores. PLOS ONE. 11, e0153345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burke B. A., Goldsby G., Brian Mudd J., Polar epicuticular lipids of Lycopersicon pennellii. Phytochemistry 26, 2567–2571 (1987). [Google Scholar]
- 20.King R. R., Calhoun L. A., 2,3-Di-O- and 1,2,3-tri-O-acylated glucose esters from the glandular trichomes of Datura metel. Phytochemistry 27, 3761–3763 (1988). [Google Scholar]
- 21.Maldonado E., Torres F. R., Martínez M., Pérez-Castorena A. L., Sucrose esters from the fruits of Physalis nicandroides var. Attenuata. J. Nat. Prod. 69, 1511–1513 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Kim J., Kang K., Gonzales-Vigil E., Shi F., Jones A. D., Barry C. S., Last R. L., Striking natural diversity in glandular trichome acylsugar composition is shaped by variation at the Acyltransferase2 locus in the wild tomato Solanum habrochaites. Plant Physiol. 160, 1854–1870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan P., Miller A. M., Schilmiller A. L., Liu X., Ofner I., Jones A. D., Zamir D., Last R. L., In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network. Proc. Natl. Acad. Sci. U.S.A. 113, E239–E248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ning J., Moghe G. D., Leong B., Kim J., Ofner I., Wang Z., Adams C., Jones A. D., Zamir D., Last R. L., A feedback-insensitive isopropylmalate synthase affects acylsugar composition in cultivated and wild tomato. Plant Physiol. 169, 1821–1835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fobes J. F., Mudd J. B., Marsden M. P. F., Epicuticular lipid accumulation on the leaves of Lycopersicon pennellii (Corr.) D’Arcy and Lycopersicon esculentum Mill. Plant Physiol. 77, 567–570 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiro J. A., Steffens J. C., Mutschler M. A., Acylsugars of the wild tomato Lycopersicon pennellii in relation to geographic distribution of the species. Biochem. Syst. Ecol. 22, 545–561 (1994). [Google Scholar]
- 27.Kuai J. P., Ghangas C. S., Steffens J. C., Regulation of triacylglucose fatty acid composition (uridine diphosphate glucose: Fatty acid glucosyltransferases with overlapping chain-length specificity). Plant Physiol. 115, 1581–1587 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li A. X., Eannetta N., Ghangas G. S., Steffens J. C., Glucose polyester biosynthesis. Purification and characterization of a glucose acyltransferase. Plant Physiol. 121, 453–460 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A. X., Steffens J. C., An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc. Natl. Acad. Sci. U.S.A. 97, 6902–6907 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leckie B. M., De Jong D. M., Mutschler M. A., Quantitative trait loci regulating sugar moiety of acylsugars in tomato. Mol. Breed. 31, 957–970 (2013). [Google Scholar]
- 31.Michaelis L., Menten M. L., Johnson K. A., Goody R. S., The original Michaelis constant: Translation of the 1913 Michaelis-Menten paper. Biochemistry 50, 8264–8269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sainz-Polo M. A., Ramírez-Escudero M., Lafraya A., González B., Marín-Navarro J., Polaina J., Sanz-Aparicio J., Three-dimensional structure of Saccharomyces invertase: Role of a non-catalytic domain in oligomerization and substrate specificity. J. Biol. Chem. 288, 9755–9766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan H., Wu L., Yang Y., Zhou G., Ruan Y.-L., Evolution of sucrose metabolism: The dichotomy of invertases and beyond. Trends Plant Sci. 23, 163–177 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Smeda J. R., Schilmiller A. L., Anderson T., Ben-Mahmoud S., Ullman D. E., Chappell T. M., Kessler A., Mutschler M. A., Combination of acylglucose QTL reveals additive and epistatic genetic interactions and impacts insect oviposition and virus infection. Mol. Breed. 38, 3 (2018). [Google Scholar]
- 35.Eshed Y., Zamir D., An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ofner I., Lashbrooke J., Pleban T., Aharoni A., Zamir D., Solanum pennellii backcross inbred lines (BILs) link small genomic bins with tomato traits. Plant J. 87, 151–160 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Tomato Genome Consortium , The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Ende W., Lammens W., Van Laere A., Schroeven L., Le Roy K., Donor and acceptor substrate selectivity among plant glycoside hydrolase family 32 enzymes. FEBS J. 276, 5788–5798 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Nakashima T., Wada H., Morita S., Erra-Balsells R., Hiraoka K., Nonami H., Single-cell metabolite profiling of stalk and glandular cells of intact trichomes with internal electrode capillary pressure probe electrospray ionization mass spectrometry. Anal. Chem. 88, 3049–3057 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Bolger A., Scossa F., Bolger M. E., Lanz C., Maumus F., Tohge T., Quesneville H., Alseekh S., Sørensen I., Lichtenstein G., Fich E. A., Conte M., Keller H., Schneeberger K., Schwacke R., Ofner I., Vrebalov J., Xu Y., Osorio S., Aflitos S. A., Schijlen E., Jiménez-Goméz J. M., Ryngajllo M., Kimura S., Kumar R., Koenig D., Headland L. R., Maloof J. N., Sinha N., van Ham R. C. H. J., Lankhorst R. K., Mao L., Vogel A., Arsova B., Panstruga R., Fei Z., Rose J. K. C., Zamir D., Carrari F., Giovannoni J. J., Weigel D., Usadel B., Fernie A. R., The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46, 1034–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peyret H., Lomonossoff G. P., The pEAQ vector series: The easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 83, 51–58 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Schilmiller A. L., Gilgallon K., Ghosh B., Jones A. D., Last R. L., Acylsugar acylhydrolases: Carboxylesterase-catalyzed hydrolysis of acylsugars in tomato trichomes. Plant Physiol. 170, 1331–1344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Coninck B., Le Roy K., Francis I., Clerens S., Vergauwen R., Halliday A. M., Smith S. M., Van Laere A., Van Den Ende W., Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant Cell Environ. 28, 432–443 (2005). [Google Scholar]
- 44.Minic Z., Physiological roles of plant glycoside hydrolases. Planta 227, 723–740 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Ghangas G. S., Steffens J. C., UDPglucose: Fatty acid transglucosylation and transacylation in triacylglucose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 90, 9911–9915 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koenig D., Jiménez-Gómez J. M., Kimura S., Fulop D., Chitwood D. H., Headland L. R., Kumar R., Covington M. F., Devisetty U. K., Tat A. V., Tohge T., Bolger A., Schneeberger K., Ossowski S., Lanz C., Xiong G., Taylor-Teeples M., Brady S. M., Pauly M., Weigel D., Usadel B., Fernie A. R., Peng J., Sinha N. R., Maloof J. N., Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. U.S.A. 110, E2655–E2662 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanthanuch W., Chantarangsee M., Maneesan J., Ketudat-Cairns J., Genomic and expression analysis of glycosyl hydrolase family 35 genes from rice (Oryza sativa L.). BMC Plant Biol. 8, 84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herscovics A., Structure and function of Class I α-1,2-mannosidases involved in glycoprotein synthesis and endoplasmic reticulum quality control. Biochimie 83, 757–762 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Särkinen T., Bohs L., Olmstead R. G., Knapp S., A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): A dated 1000-tip tree. BMC Evol. Biol. 13, 214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nesbitt T. C., Tanksley S. D., Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162, 365–379 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathew A. K., Padmanaban V. C., Metabolomics: The apogee of the omics trilogy. Int. J. Pharm. Pharm. Sci. 5, 45–48 (2013). [Google Scholar]
- 52.Nagana Gowda G. A., Raftery D., Recent advances in NMR-based metabolomics. Anal. Chem. 89, 490–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfaffl M. W., A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karimi M., Inzé D., Depicker A., GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., Smith I., Tothova Z., Wilen C., Orchard R., Virgin H. W., Listgarten J., Root D. E., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O., Cradick T. J., Marraffini L. A., Bao G., Zhang F., DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belhaj K., Chaparro-Garcia A., Kamoun S., Nekrasov V., Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S., A modular cloning system for standardized assembly of multigene constructs. PLOS ONE 6, e16765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. McCormick, in Plant Tissue Culture Manual, K. Lindsey, Ed. (Springer Netherlands, 1997), vol. 13, pp. 311–319. [Google Scholar]
- 60.Fillatti J. J., Kiser J., Rose R., Comai L., Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730 (1987). [Google Scholar]
- 61.Schilmiller A., Shi F., Kim J., Charbonneau A. L., Holmes D., Daniel Jones A., Last R. L., Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 62, 391–403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaaw3754/DC1
Fig. S1. Acylglucoses are not detected in trichome extracts of IL3-5, IL4-1, or IL11-3.
Fig. S2. Quantification of acylsugars in S. lycopersicum M82 and breeding lines containing S. pennellii LA0716 introgressions.
Fig. S3. Comparison of major acylsugars in BIL6521 and BIL6521 × BIL6180 F2 progeny using LC-MS.
Fig. S4. Mass spectra of major acylsugars S3:15, S3:22, G3:15, and G3:22 from BIL6521 × BIL6180—F2 lines, BIL6180, and BIL6521.
Fig. S5. Mutated genomic sequence of three homozygous spasff1 CRISPR-Cas9 lines.
Fig. S6. Acylsugars in BPI chromatograms of spasff1 and LA0716 plants.
Fig. S7. Comparison of acylsugars from IL3-5 and parental M82 using LC-MS.
Fig. S8. LC-MS analysis of P-type acylsucrose-producing S. lycopersicum BIL6180 stably transformed with proSpASFF1::SpASFF1.
Fig. S9. Mass spectra of G3:19-derived from SpASFF1 in vitro assay.
Fig. S10. SpASFF1 cleaves a purified P-type triacylsucrose but not unmodified sucrose while yeast invertase cleaves unmodified sucrose but not triacylsucrose.
Table S1. Annotation of acylsugars identified in BIL6521 and BIL6521 × BIL6180 F2 using LC-MS and collision-induced dissociation.
Table S2. Annotation of three GH candidates for SpASFF1 identified in the AG3.2.
Table S3. Primers/gBlocks/sgRNAs used in this study.


