Abstract
Heterochromatin protein 1α (HP1α) undergoes liquid-liquid phase separation (LLPS) and forms liquid droplets and gels in vitro, properties that also appear to be central to its biological function in heterochromatin compaction and regulation. Here we use solid-state NMR spectroscopy to track the conformational dynamics of phosphorylated HP1α during its transformation from the liquid to the gel state. Using experiments designed to probe distinct dynamic modes, we identify regions with varying mobilities within HP1α molecules and show that specific serine residues uniquely contribute to gel formation. The addition of chromatin disturbs the gelation process while preserving the conformational dynamics within individual bulk HP1α molecules. Our study provides a glimpse into the dynamic architecture of dense HP1α phases and showcases the potential of solid-state NMR to detect an elusive biophysical regime of phase separating biomolecules.
Keywords: Biophysics, structural biology, Heterochromatin protein 1α (HP1α), liquid-liquid phase separation (LLPS), solid-state NMR
Graphical Abstract:
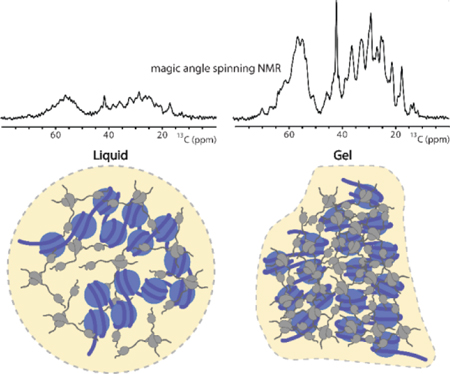
Within the mesh: Magic angle spinning solid-state NMR spectroscopy unveils residue-specific details of the liquid to gel transition of heterochromatin protein 1α (HP1α). The conformational dynamics of the protein are monitored over time and extended to physiological conditions that suggest a defense mechanism against gelation within cells.
Proteins and nucleic acids have evolved collective properties that enable the spatial and temporal regulation of cellular functions without the need of lipid membranes.[1] Such phenomena are often mediated by a process called liquid-liquid phase separation (LLPS) and have been observed in a diverse set of biological events including the stress response, gene regulation, signaling and transport.[2–5] LLPS results in the formation of droplets where molecules form a dense liquid phase but can still diffuse freely. Sometimes, the droplets mature into less mobile gel or solid-like states where a stronger interaction network limits molecular diffusion and reorientation.[4,6–8] For example, immobile condensates have been identified under physiological conditions in heterochromatin, nucleoli and stress granules.[3,4,7,9–11] While reduced condensate dynamics can have profound implications for physiological function, aging, and disease, the molecular basis of these transformations has largely remained elusive.
Here, we focus our attention on the gel transition of HP1α, a highly conserved protein that mediates gene silencing, genome stability and chromatin compaction.[12,13] HP1α is composed of two folded domains, the chromodomain (CD) that binds methylated histone H3K9, and the chromoshadow (CSD) domain that enables formation of the stable 44 kDa HP1α dimer (Fig. 1a).[14,15] These domains interrupt three disordered regions enriched in charged amino acids: the N-terminal segment (NTE), the hinge region, and the C-terminal tail (CTE). For many years, it had been known that HP1α in cells exhibits differential dynamics ranging from fast to immobile, and recent studies propose that this is due to LLPS behavior.[3,7,16,17] For example, HP1α can form liquid droplets and gels in vitro upon N-terminal phosphorylation or the addition of DNA.[7] HP1α foci with properties consistent with LLPS behavior have also been observed at heterochromatin domains in Drosophila and mammalian cells.[3] These studies suggest that HP1α forms distinct dilute, dense liquid and gel-like phases within the nucleus to partition heterochromatin from the transcriptional machinery by either phase incompatibility or a physical sieving mechanism.[18]
Figure 1.
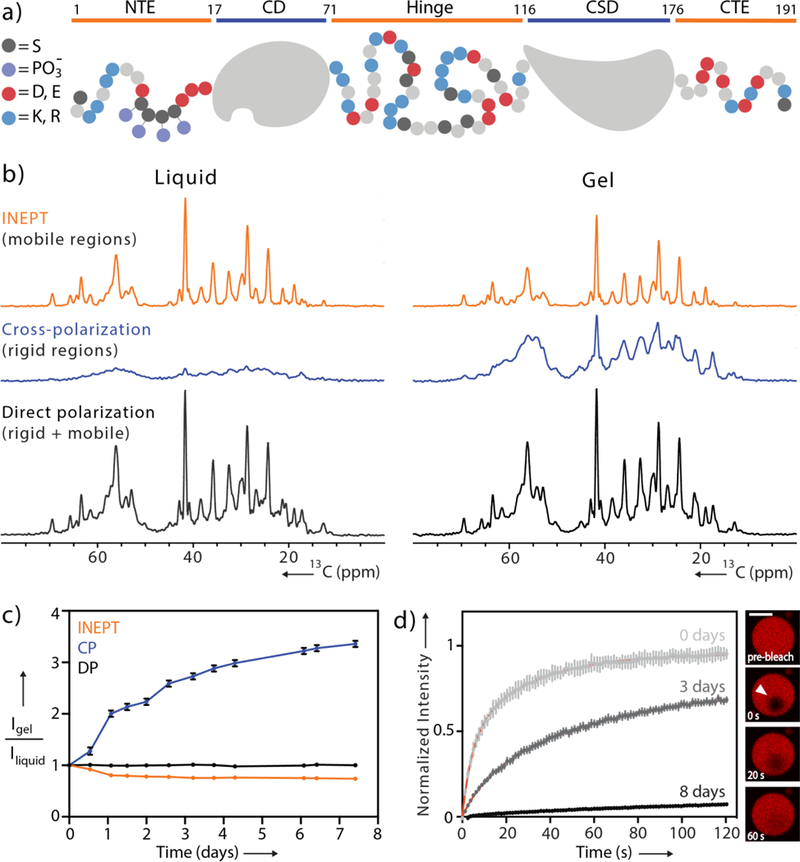
pHP1α dynamics monitored during gelation. (a) Domain map of monomeric pHP1α. Negatively charged residues are colored in red, positively charged residues are in blue, serine residues are in black and phosphorylation sites are in purple. (b) 1D 13C INEPT, CP and DP spectra acquired during the initial dense liquid state and the final gel state. (c) Integrated signal of the aliphatic regions of the spectra collected every 12 hours. Data are normalized to the starting point of each respective experiment. For error analysis, see SI. (d) FRAP recovery curves for pHP1α collected immediately upon droplet formation, 3 days and 8 days later. Red lines represent exponential fits. Inset: FRAP of a single pHP1α droplet at day 3, scale bar 20 μm.
Biophysical treatments of LLPS proteins such as HP1α suggest that the compaction state, interaction strength and multivalency of the biomolecule can shape the collective material property.[19–21] Within this framework, however, specific molecular interactions and dynamics have been challenging to describe experimentally due to the dense and amorphous nature of biological condensates.[22] Some details have emerged from solution nuclear magnetic resonance (NMR) experiments of the dense liquid phase that point to the importance of intrinsically disordered protein regions and their transient interactions with other LLPS components.[22–24] Solution NMR spectroscopy, however, cannot capture the molecular picture of biomolecules in the gel state where the increased viscosity and cross-linking interactions reduce molecular tumbling and cause severe line-broadening in the NMR spectra. To circumvent this problem, here we monitor the gelation of HP1α by magic angle spinning (MAS) solid-state NMR spectroscopy. With this methodology, line-broadening interactions such as the chemical shift anisotropy are averaged out by mechanical rotation at the magic angle, allowing the acquisition of resolved spectroscopic data independent of molecular size. Information rich interactions such as the dipolar and scalar nuclear couplings can then be reintroduced by selective pulse sequences to allow the characterization of secondary and tertiary protein structure, protein-protein interactions and the description of dynamic and rigid regions of the molecule. Already successfully applied to biological hydrogels and fibrils,[25–29] MAS NMR can be a powerful technique for studying the molecular driving forces of biomolecular LLPS and gelation. In the context of HP1α condensates, this approach allowed us to observe the mobile and rigid regions of HP1α molecules at residue specific resolution and to identify specific sites that participate in the transformation from a dense liquid to a gel state.
Previous studies have indicated that the LLPS behavior of HP1α depends on the phosphorylation of several serine residues in the N-terminal region of the protein (Fig. 1a).[7] Following published protocols,[7,30] we obtained high yields (50 – 80 mg) of pure pHP1α per liter of 13C, 15N enriched culture (Fig. S1). Upon phase separation of pHP1α, we coalesced the dense pHP1α condensates into a single phase to be used in NMR experiments (Fig. S2). As expected, the solution NMR spectra of pHP1α condensates lost all resolved information leaving only a congested region in the middle of the spectrum presumably corresponding to the disordered segments of the protein (Fig. S3).
Proceeding with MAS NMR spectroscopy, we performed three simple one-dimensional (1D) experiments that nevertheless provide rich information about the overall dynamics in the condensate (Fig. 1b).[31] First, the scalar-based 1D INEPT transfer experiment selects for highly mobile sites that have long transverse relaxation times similar to those detected under solution NMR conditions. Second, rigid regions that exhibit strong dipole-dipole couplings can be selected with 1D cross-polarization (CP) experiments. And finally, the 13C direct polarization (DP) experiment can in principle be used to detect all the 13C nuclei in the sample independent of dynamics. At the start of our experiments, the 1H-13C CP signal was remarkably lower than the 1H-13C INEPT signal (Fig. 1b). Comparing the intensities of these spectra with the DP experiment, it also became clear that CP and INEPT together do not account for all the 13C signals in the sample. Thus, in the dense liquid droplet state there are pHP1α molecules or regions of each individual protein that exhibit motion in an intermediate dynamic range rendering them invisible in both scalar-based and dipolar-based MAS NMR experiments, similar to the behavior of other complex biological assemblies.[31]
Expecting that pHP1α condensates will equilibrate into gels given sufficient time,[7] we continuously performed MAS and monitored the intensities of the CP and INEPT spectra at 12 hour intervals over the course of a week (Fig. 1c, S4). Reaching a gel state after 7 days (Fig. S2), the total CP signal increased 3.3 fold, the INEPT signal decreased 1.35 fold and the 13C DP signal remained unchanged. We interpret the dramatic change in total CP intensity to arise from an expanding cross-linking network during gelation that restricts the motion of individual pHP1α dimers and shifts their overall dynamics to a slower timescale where dipolar-based polarization transfer mechanisms are more efficient. As an independent measure of this process, we also performed concurrent fluorescence recovery after photobleaching (FRAP) experiments using Cy3-labeled HP1α (Fig. 1d). In the initial liquid droplet state, the recovery of fluorescence intensity occurs on a timescale of several seconds, consistent with observations in other dense liquid systems.[16,22] Over the course of a week, the fluorescence recovery slowed down, eventually reaching a flat line. Reminiscent of the observations made at heterochromatin foci, these results align well with our NMR time course and provide further evidence that the motion of individual molecules within the gel state is severely limited.
Next, we turned our attention to the INEPT signals, which remained strong even after gelation was complete. The superior resolution in these spectra allowed a more careful analysis of the intensity changes for individual peaks. For example, in the Ser/Thr Cα/Cβ region certain peaks (e.g. at 65.8 ppm) decreased steadily, while other peaks (e.g. at 64.4 ppm) lost intensity much more slowly (Fig. 2a,b, S5). To obtain further insight, we proceeded to record two-dimensional (2D) 13C-13C correlation experiments (Fig. 2c, S6). In particular, INEPT can be coupled with TOBSY[32] (total through-bond correlation spectroscopy) mixing that relies on the isotropic scalar couplings to transfer polarization between adjacent nuclei and to report on highly mobile residues. A comparison of the INEPT-TOBSY spectra of the dense liquid and gel state revealed several Ser Cα/Cβ cross-peaks that remained strong and one Ser cross-peak that lost intensity over time. The downfield shift of the Cβ atom of the latter Ser compared to random coil values is consistent with phosphorylation. Thus, it appears likely that phosphorylated residues in the N-terminal tail experience changes in dynamics during gelation distinct from all other residues (Fig. S5).
Figure 2.
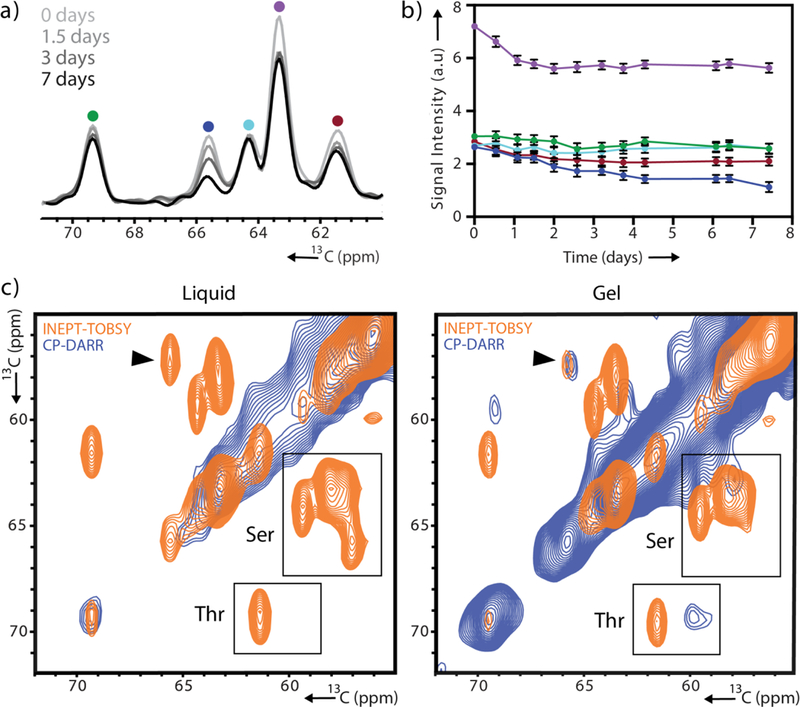
Site-specific variation in the gelation dynamics of pHP1α. (a) 1D INEPT spectra collected 0, 1.5, 3 and 7 days after initiation of phase separation. (b) Signal intensity of the five peaks shown in (a) tracked over time with colors corresponding to the color scheme in (a). For error analysis, see SI. (c) Serine/threonine region of 2D 13C-13C CP-DARR and INEPT-TOBSY spectra recorded 3 hours after droplet formation and 7 days later. A black arrow denotes the serine cross-peak with greatest signal loss.
To complement the INEPT-TOBSY 2D correlation experiment, we also recorded a 2D CP-DARR[33] (dipolar assisted rotational resonance) correlation spectrum which enables transfer between dipolar coupled and hence more rigid 13C spin systems (Fig. 2c, 3a). A visual inspection of the complete DARR and INEPT-TOBSY spectra illustrates the dramatic difference in content and linewidths for the two sets of targeted residues (Fig. 3a). The INEPT-TOBSY spectrum shows narrow cross-peaks located at typical random coil chemical shifts that could easily be assigned to specific amino acid types. We leveraged the distinct sequence composition of the structured and unstructured regions of HP1α to match the assigned amino types with those enriched in the disordered regions (Fig. 3b). We note the cross-peak for an Ile residue, potentially originating from the hinge, the Ala and Thr correlations consistent with the N- and C-terminal tails and the multiple Ser cross-peaks consistent with all three disordered regions. On the other hand, the DARR spectrum shows a greater diversity of chemical shifts, indicative of the sequence variety in the CD and CSD. The relatively broad Cα-Cβ region may also indicate the presence of intermediate motions that interfere with decoupling during acquisition and that contribute to the linewidth.[34] Nevertheless, many individually resolved peaks are present in the spectrum suggesting that the addition of other spectral dimensions (e.g. 15N) will enable sequence assignments and the interrogation of HP1α gel interactions in more detail. Note that these regions are completely intractable in solution NMR experiments of the dense liquid or gel state.
Figure 3.

2D 13C-13C correlation spectra of pHP1α condensates. (a) Overlay of CP-DARR and INEPT-TOBSY spectra, probing the rigid and highly mobile sites of pHP1α 4 days after droplet formation. Amino acid types with characteristic and identifiable chemical shifts are marked in boxes (INEPT-TOBSY) and circles (CP-DARR). (b) Complete sequence of HP1α showing structured regions (gray), and disordered regions with identified amino acid types (orange).
Finally, we wondered how the addition of relevant components such as chromatin will affect the dynamics of the gelation process. We therefore prepared chromatin arrays containing 12 nucleosomes with a lysine methylation mimic at position 9 of histone H3 (Fig. S7). These arrays reproduce the physiologically relevant state of the nuclear heterochromatin environment where HP1α binds and undergoes LLPS.[3,7,16,17] We added the equivalent of 200 μM nucleosomes to freshly prepared dense liquid droplets and repeated the experiments described above. To our surprise, the addition of chromatin significantly slowed down the rigidification process as evidenced by the smaller increase in overall CP signal (1.5X) and the retention of mobility in FRAP experiments (Fig. S8). On the other hand, the content of INEPT experiments was preserved albeit with a smaller change in the dynamics of the phosphorylated Ser peak.
Previous models of HP1α condensation suggest that the pHP1α dimer adopts a more open state that facilitates interactions with neighboring dimers resulting in a dense liquid phase.[7,18] These models, however, do not identify specific interactions and stop short of describing the maturation process from the dense liquid to the gel state. Our study provides the first molecular glimpse of this transformation. In our hands, in the absence of other components, pHP1α dense liquid droplets mature into gels over the course of a week. As the interaction network within the condensate strengthens, the molecular orientation of individual pHP1α molecules becomes restricted, a change reflected in the increase of dipolar based signals in our MAS NMR experiments and in the incomplete recovery of our FRAP data. Yet, different regions of the protein exhibit distinct dynamic states. While the disordered tails and hinge region remain dynamic, other sites such as the folded CD and CSD domains experience much more restricted motions potentially reflecting increased participation in interactions within the gel network. The loss of signal in the INEPT spectra suggests that the cross-linking interactions within the gel state may also involve the phosphorylated Ser residues, a significant observation considering their important role in promoting pHP1α phase separation.[7] The addition of physiologically relevant components such as chromatin disrupts the cross-linking interaction network of pHP1α condensates and prevents their complete maturation into a more rigid gel state. In addition to chromatin, “solubilization” of pHP1α condensates in the nucleus may also be achieved by the presence of non-phosphorylated HP1α and other HP1 isoforms that do not appear to phase separate. We note that due to the overwhelming concentration of pHP1α in our samples, our current study reports primarily on the behavior of individual molecules in the bulk condensed phase and not on the chromatin binding dynamics of pHP1α dimers where complex kinetics and states have been reported.[35] Nevertheless, the preservation of mobility in chromatin-containing pHP1α dense phases has important implications for heterochromatin compaction and regulation in the cell where access to genetic information is preserved and can be granted on demand. We expect that implementing the full suite of solid-state NMR experiments will help illuminate the specific intermolecular interactions and structural ensembles of biomolecules in condensed environments.
Supplementary Material
Acknowledgements
We would like to thank Ratan Rai and Anna de Angelis for assistance with the NMR spectrometers. B.A. was supported by NIH Molecular Biophysics Training Grant T32 GM008326. This work utilized the Biotechnology Research Center for NMR Molecular Imaging of Proteins at UCSD, supported by NIH grant P41 EB002031, and the UCSD Microscopy Core supported by NINDS NS047101.
References:
- [1].Mitrea DM, Kriwacki RW, Cell Comm. Signal. 2016, 14, DOI 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S, eLife 2015, 4, DOI 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH, Nature 2017, 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. , Cell 2015, 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- [5].Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD, Science 2016, 352, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Alberti S, Hyman AA, BioEssays 2016, 38, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ, Nature 2017, 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC-H, Eckmann CR, Myong S, Brangwynne CP, Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP, Cell 2016, 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA, Cell 2017, 168, 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S, Cell Rep. 2018, 23, 3327–3339. [DOI] [PubMed] [Google Scholar]
- [12].Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC, Proc. Natl. Acad. Sci. USA 1990, 87, 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Allshire RC, Madhani HD, Nature Rev. Mol. Cell Biol. 2017, 19, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kwon SH, Workman JL, Mol. Cell. 2008, 26, 217–227. [PubMed] [Google Scholar]
- [15].Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ, Current Biology 2000, 10, 517–525. [DOI] [PubMed] [Google Scholar]
- [16].Cheutin T, Science 2003, 299, 721–725. [DOI] [PubMed] [Google Scholar]
- [17].Dialynas GK, Terjung S, Brown JP, Aucott RL, Baron-Luhr B, Singh PB, Georgatos SD, J. Cell Sci 2007, 120, 3415–3424. [DOI] [PubMed] [Google Scholar]
- [18].Larson AG, Narlikar GJ, Biochemistry 2018, 57, 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].T. S. Harmon, 2017, DOI 10.7554/elife.30294.001. [DOI]
- [20].Lin Y-H, Forman-Kay JD, Chan HS, Biochemistry 2018, 57, 2499–2508. [DOI] [PubMed] [Google Scholar]
- [21].Leibler L, Rubinstein M, Colby RH, Macromolecules 1991, 24, 4701–4707. [Google Scholar]
- [22].Mitrea DM, Chandra B, Ferrolino MC, Gibbs EB, Tolbert M, White MR, Kriwacki RW, J. Mol. Biol. 2018, DOI 10.1016/j.jmb.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Burke KA, Janke AM, Rhine CL, Fawzi NL, Molecular Cell 2015, 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brady JP, Farber PJ, Sekhar A, Lin Y-H, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, et al. , Proc. Natl. Acad. Sci. USA 2017, 114, E8194–E8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kennedy SB, deAzevedo ER, Petka WA, Russell TP, Tirrell DA, Hong M, Macromolecules 2001, 34, 8675–8685. [Google Scholar]
- [26].Ader C, Frey S, Maas W, Schmidt HB, Gorlich D, Baldus M, Proc. Natl. Acad. Sci. USA 2010, 107, 6281–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lin H-K, Boatz JC, Krabbendam IE, Kodali R, Hou Z, Wetzel R, Dolga AM, Poirier MA, van der Wel PCA, Nat. Comm. 2017, 8, 15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tycko R, Annu. Rev. Phys. Chem. 2011, 62, 279–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R, Cell 2017, 171, 615–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Munari F, Gajda MJ, Hiragami-Hamada K, Fischle W, Zweckstetter M, FEBS Lett. 2014, 588, 1094–1099. [DOI] [PubMed] [Google Scholar]
- [31].Matlahov I, van der Wel PCA, Methods 2018, 148, 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M, J. Am. Chem. Soc. 2005, 127, 12965–12974. [DOI] [PubMed] [Google Scholar]
- [33].Takegoshi K, Nakamura S, Terao T, Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar]
- [34].Bajaj VS, van der Wel PCA, Griffin RG, J. Am. Chem. Soc. 2009, 131, 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bryan LC, Weilandt DR, Bachmann AL, Kilic S, Lechner CC, Odermatt PD, Fantner GE, Georgeon S, Hantschel O, Hatzimanikatis V, et al. , Nucl. Ac. Res. 2017, 45, 10504–10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


