Abstract
Background:
Although inhaled corticosteroid (ICS) medication is considered the cornerstone treatment for patients with persistent asthma, few ICS pharmacogenomic studies have involved non-white populations.
Objective:
To identify genetic predictors of ICS response in multiple population groups with asthma.
Methods:
The discovery group comprised African American participants from the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE) who underwent 6 weeks of monitored ICS therapy (n=244). A genome-wide scan was performed to identify single nucleotide polymorphism (SNP) variants jointly associated (i.e., the combined effect of the SNP and SNP x ICS treatment interaction) with changes in asthma control. Top associations were validated by assessing the joint association with asthma exacerbations in three additional groups – African Americans (n=803 and n=563) and Latinos (n=1,461). RNA-seq data from 408 asthma cases and 405 controls were used to examine whether genotype was associated with gene expression.
Results:
One variant, rs3827907, was significantly associated with ICS-mediated changes in asthma control in the discovery set (P=7.79×10−8) and was jointly associated with asthma exacerbations in three validation cohorts (P=0.023, P=0.029, and P=0.041). RNA-seq analysis found the rs3827907 C-allele to be associated with lower RNASE2 expression (P=6.10×10−4). RNASE2 encodes eosinophil-derived neurotoxin (EDN), and the rs3827907 C-allele appeared to particularly influence ICS treatment response in the presence of eosinophilic inflammation (i.e., high pre-treatment EDN levels or blood eosinophil counts).
Conclusion:
We identified a variant, rs3827907, which appears to influence response to ICS treatment in multiple population groups, and likely mediates its effect through eosinophils.
Clinical Implications:
African Americans and Latinos are disproportionately affected by asthma and its complications. Here we identify a pharmacogenomic variant that may assist in identifying individuals from these groups who will respond to ICS treatment.
Capsule Summary:
This is the first study to use a non-white study population for the discovery of pharmacogenomic predictors of asthma controller response. Variant rs3827907 genotype appeared to influenced ICS response especially in the setting of eosinophilic inflammation.
Keywords: pharmacogenetics, EDDM3B, RNASE2, eosinophil-derived neurotoxin, transcriptome, eosinophils
INTRODUCTION
African American individuals appear to be disproportionately affected by asthma and its complications with rates of asthma-related emergency room visits, hospitalizations, and deaths that are approximately 2–3 times higher when compared with white individuals.1 Inhaled corticosteroid (ICS) medication is one of the most effective treatments for persistent asthma and its use has been associated with lower rates of severe exacerbations including death.2, 3 However, to date there has been little to no African American representation in pharmacogenomic studies assessing genetic predictors of ICS response.4–6
Despite previous work by us suggesting that an individual’s overall proportion of African ancestry may not be a strong determinant of ICS response,7, 8 population-specific risk variants influencing asthma medication response may still exist. The possibility of the latter is supported by genome-wide association studies demonstrating both shared and group-specific risk variants for asthma9, 10 and pharmacogenomic studies for other disease conditions showing populationspecific variants associated with medication response.11
We and others have also shown that ICS medication adherence varies widely among individuals and is on average quite poor among patients being treated for asthma.12, 13 Not surprisingly, this variation in patient use is a major predictor of treatment response and asthma-related exacerbations.14, 15 As a result, studies examining predictors of ICS response should account for patient medication adherence, along with dose and preparation, as all of these factors influence the level of anti-inflammatory exposure.
The Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE) is an ongoing prospective cohort study to understand the genetic predictors of asthma medication treatment response in a large, diverse patient population from southeast Michigan. A subgroup of SAPPHIRE participants who met strict criteria received 6 weeks of ICS treatment to directly quantify corticosteroid response; our discovery set consisted of African American participants in this treatment group (n=244). The validation groups included African American individuals with prospective clinical information on ICS exposure and asthma exacerbations from the SAPPHIRE cohort; Latino individuals from the Genes-environments & Admixture in Latino Americans Study (GALA II); and African American individuals from the Study of African Americans, Asthma, Genes and Environments (SAGE II). The latter 2 cohorts had retrospective information on ICS use and asthma exacerbations. We also reassessed our top association in a set of European American individuals from the SAPPHIRE cohort who underwent the same 6-week ICS treatment course as the discovery group.
METHODS
Study Overview and Patient Population
The SAPPHIRE cohort participants were recruited within a large health system serving southeast Michigan and the Detroit metropolitan area. Individuals were eligible to participate if they were 12–56 years of age, had a previously documented physician diagnosis of asthma, and no prior diagnosis of congestive heart failure or chronic obstructive pulmonary disease. Study participants underwent a clinical evaluation which included a detailed questionnaire and lung function testing. Further details about the SAPPHIRE patient evaluation can be found in the online supplement.
The GALA II and SAGE II cohorts are described below and in the methods section of the online supplement. All studies had institutional review board approval at their respective institutions, and all studies required informed written consent prior to study participation.
Discovery Group
The discovery group consisted of a subset of SAPPHIRE participants who completed 6 weeks of monitored ICS treatment, had existing genome-wide genotype data, and were African American by self-report (n=244). These individuals completed the Asthma Control Test (ACT), a validated 5-question instrument used for assessing asthma control in individuals ≥12 years of age (Optum Inc., Eden Prairie, MN),16 both before and after ICS treatment (320μg of inhaled beclomethosone dipropionate hydrofluoroalkane [beclomethasone HFA] daily x 6 weeks). The phenotype for ICS response in the discovery set was the change in ACT score over the course of the treatment trial. We have previously demonstrated a “dose-response” relationship between ICS use and change in ACT score.8 Since the primary purpose of this study was to identify genetic factors that modify ICS treatment response, we reasoned that the strength and direction of an association would be contingent on the level of ICS exposure/adherence. Adherence was measured using an electronic counting device (DOSER-CT, Meditrack, Easton, MA) attached to the ICS metered dose inhaler.
African American SAPPHIRE participants in the discovery and validation groups were genotyped using the Axiom AFR array (Affymetrix Inc., Santa Clara, CA).17 After quality control, 574,370 of the genotyped single nucleotide polymorphisms (SNPs) were available for analysis.
Validation Groups
Validation was carried out in 4 groups. These groups included a separate set of African American individuals from the SAPPHIRE cohort, Latino individuals from GALA II, African American participants from SAGE II, and European American individuals from the SAPPHIRE cohort.
The primary validation analysis was performed in African American individuals from SAPPHIRE (n=803) who met the following criteria: available prospective clinical data on asthma exacerbations, recorded ICS fills through pharmacy claims records, existing genome-wide genotype data, and African American race-ethnicity by participant self-report. Our method of calculating ICS exposure using pharmacy data can be found in the online supplement and elsewhere.18 Additional validation was performed among Latino participants in the GALA II study (n=1,461) and African American participants in the SAGE II study (n=563). The latter two cohorts had retrospective information on asthma exacerbations. Genotyping in these cohorts was performed using the Affymetrix Axiom LAT1 array. Lastly, we had a group of European American individuals from the SAPPHIRE cohort (n=98) who underwent the same ICS treatment protocol as the discovery set; we used the change in asthma control (i.e., the change in the ACT composite score) as the outcome in this group.
RNA-seq Data
At the time of enrollment, SAPPHIRE participants had whole blood RNA collected, preserved, and stored in PAXgene Blood RNA tubes (BD Biosciences, San Jose, CA) until the time of analysis. Sequencing libraries were constructed using TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Globin (Illumina, San Diego, CA) on 408 African American individuals with asthma and 405 healthy African American controls. Illumina HiSeq RNA-seq reads were mapped to human genome (build GRCh38.p5) using the software STAR and quantified at the gene level using the program RSEM.19, 20 We normalized raw read counts for human protein-coding genes using the variance stabilizing transformation implemented in the program DESeq2.21 As done by others,22 we only analyzed genes with ≥1 read count in ≥50% of individuals.
Blood Eosinophil Counts and Eosinophil-Derived Neurotoxin (EDN) Protein
Measurements
Blood and serum samples were collected from SAPPHIRE participants at the time of study enrollment. A complete blood count with 5-part white blood cell differential was obtained immediately for individuals with available samples. Serum specimens were placed into long-term storage at −80°C until the time of analysis. We used a commercial enzyme-linked immunosorbent assay (ELISA) kit (MBL International Corp., Woburn, MA) and followed the manufacturer’s instructions to measure EDN levels in the thawed serum samples.
Statistical Analysis
A full description of the models used for each analysis can be found in the online supplement. Both the discovery and validation analyses assessed the joint association of SNP and SNP x ICS exposure interactions on asthma outcomes related to disease control. Our primary outcome for the discovery analysis was the change in asthma control (i.e., ACT score) from before to immediately following 6 weeks of ICS treatment. The conservative Bonferroni correction for this analysis resulted in significance threshold of P-value <8.71×10−8 (0.05/574,370). In the validation analyses, we used time-to-severe exacerbation as a prospective measure of asthma control in African American SAPPHIRE participants. For validating in GALA II and SAGE II, we used a dichotomous outcome of exacerbation in the 12-month period preceding study enrollment. We defined a severe asthma exacerbation as one requiring burst oral corticosteroids, an emergency department visit, or hospitalization. For European American SAPPHIRE participants who completed 6 weeks of observed ICS treatment, we used the same model as the discovery analysis with change in ACT score as the outcome.
To assess for expression quantitative trait loci (eQTL), we used linear regression to model the relationship between rs3827907 genotype and gene expression. A Bonferroni corrected P-value <1.56×10−3 (0.05/32 genes) was considered statistically significant in the eQTL analyses.
To assess the potential effect of pre-treatment EDN levels and eosinophil counts on ICS treatment response, we assessed the relationship between rs3827907 genotype (i.e., C-allele carrier status) on the 6-week change in ACT score after stratifying by median levels of the aforementioned variables in the discovery group. To assess the impact of ICS usage, we also stratified by ICS adherence (i.e., ≤75% and >75%) based on our earlier work demonstrating that this was a meaningful threshold for asthma outcomes.15 A P-value<0.05 was considered statistically significant.
As a post hoc analysis, we imputed missing genotypes among SAPPHIRE African Americans using the program Minimac3;23 the 1000 Genomes Project (1KGP; version 5) cosmopolitan panel was used as the reference for imputation. The models employed in the discovery and eQTL analyses were used to assess imputed variants surrounding our lead candidate, rs3827907.
RESULTS
Study Populations
Our discovery set comprised 244 African American individuals whose characteristics are shown in Table 1. Individuals in the discovery set received 6 weeks of ICS treatment, during which time we assessed for changes in asthma control. The validation samples used in this analysis are also shown. The validation groups included African American SAPPHIRE participants (n=803), Latino patients from GALA II patients (n=1,461), African American patients from SAGE II (n=563), and European American SAPPHIRE participants (n=98). As can be seen in Table 1, SAPPHIRE participants were on average considerably older when compared with GALA II and SAGE II participants. In addition, there were no active smokers in GALA II and SAGE II as this was an exclusion criterion for participation in both of these studies.
Table 1.
Characteristics of subjects with asthma in both the discovery and validation sets
| Discovery Set | Validation Sets |
||||
|---|---|---|---|---|---|
| SAPPHIRE (n=244) | SAPPHIRE – African Americans (n=803) | GALA II (n=1,461) | SAGE II (n=563) | SAPPHIRE –European Americans (n=98) | |
| Age in years – mean ± SD | 32.2 ± 13.0 | 31.9 ± 15.1 | 12.2 ± 3.0 | 13.5 ± 3.4 | 36 ± 14.9 |
| Female – no. (%) | 146 (59.8) | 509 (63.4) | 623 (42.6) | 262 (46.5) | 53 (54.1) |
| Race-ethnicity – no. (%) | |||||
| African American | 244 (100.0) | 803 (100.0) | -- | 563 (100.0) | -- |
| European American | -- | -- | -- | -- | 98 (100.0) |
| Latino | -- | -- | 1,461 (100.0) | -- | -- |
| Genetic ancestry percentage – mean ± SD* | |||||
| African | 79.8 ± 10.4 | 79.4 ± 10.4 | 15.6 ± 13.8 | 79.0 ± 12.7 | NA |
| European | 20.2 ± 10.4 | 20.6 ± 10.4 | 53.7 ± 19.4 | 21.0 ± 12.7 | NA |
| Native American | -- | -- | 30.7 ± 25.5 | -- | NA |
| Body mass index in kg/m2 – mean ± SD | 32.4 ± 9.5 | 30.8 ± 8.8 | -- | -- | 29.6 ± 9.1 |
| Body mass index percentile – mean ± SD† | -- | -- | 73.3 ± 29.8 | 77.1 ± 24.3 | -- |
| Smoking status – no. (%)‡ | |||||
| Never smoker | 217 (88.9) | 658 (81.9) | 1,432 (98.0) | 535 (95.0) | 91 (92.9) |
| Past smoker | 24 (9.9) | 68 (8.5) | 29 (2.0) | 28 (5.0) | 7 (7.1) |
| Current smoker | 3 (1.2) | 77 (9.6) | -- | -- | 0 (0.0) |
| Secondhand smoke exposure – no. (%)§ | 76 (31.1) | 219 (27.3) | 306 (20.9) | 163 (29.0) | 18 (18.4) |
| Initial ACT score – mean ± SD|| | 18.0 ± 5.2 | 20.6 ± 3.9 | NA | NA | 20.2 ± 4.5 |
| Percent of predicted FEV1 in liters – mean ± SD | 73.3 ± 12.9 | 92.5 ± 17.7 | 90.8 ± 16.3 | 99.2 ± 15.0 | 72.0 ± 13.6 |
SAPPHIRE denotes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; GALA II, the Genes-environments and Admixture in Latino Americans study; SAGE II, the Study of African Americans, Asthma, Genes, and Environments; ICS, inhaled corticosteroid; SD, standard deviation; NA, not available; kg/m2, weight in kilograms per height in meters squared; ACT, asthma control test; and FEV1, forced expiratory volume in one second.
The total proportion of continental group ancestry (i.e., global ancestry) was estimated using genome-wide genotype data and the methods described in the online supplement. Genome-wide genotype data was not available for European American individuals in the SAPPHIRE cohort.
Body mass index percentiles were based on growth charts (http://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/sas.htm) and represented where each individual ranked in comparison to the age- and sex-matched population distribution.
Current smoking was an exclusion criterion for enrollment into GALA II and SAGE II.
Based on participant-reported exposure to secondhand tobacco smoke at home.
Asthma Control Test scores range from 5–25 with higher values representing better control. An ACT score ≤19 represents poor control, whereas scores >19 are considered good asthma control.
Evaluation for ICS Pharmacogenomic Interactions in the Discovery Set
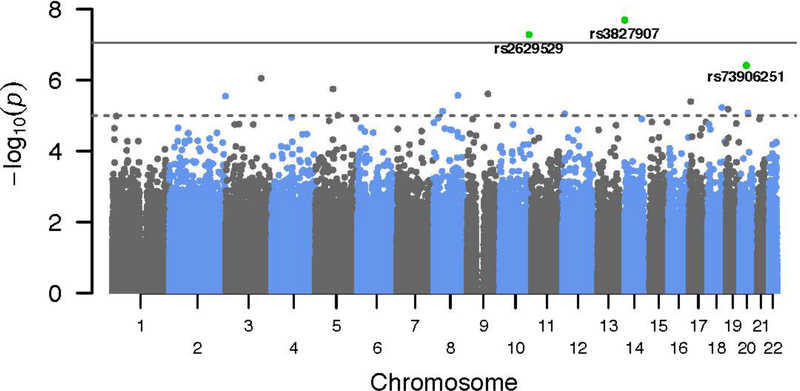
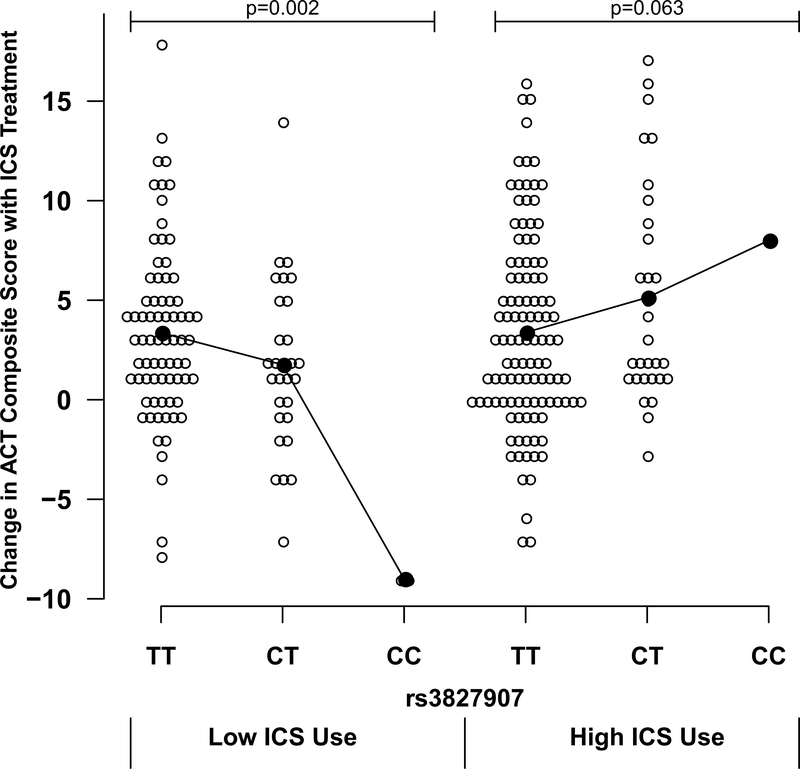
We assessed for the genetic predictors of ICS response (defined in the discovery analysis as a change in ACT score over 6 weeks of treatment). To account for differences in ICS use among patients in the discovery set, we incorporated measures of ICS adherence which had been gathered using electronic monitoring devices. Average ICS adherence was 0.76 (± 0.22 standard deviation [SD]), representing the proportion of the prescribed amount taken. Since all patients in the discovery set were prescribed the same type and amount of ICS medication, the adherence estimate was used to account for relative differences in ICS exposure between individuals. In our tests of association with drug response, we assessed the joint effect of genotype and a genotype x ICS exposure multiplicative interaction, while simultaneously adjusting for potential confounders. The quantile-quantile plot for this association analysis is shown in Figure E1 (online supplement). As the plot showed mild genomic inflation (λ=1.09), we adjusted our Pvalues accordingly using genomic control. Three SNPs – rs3827907 (chromosome [chr] 14), rs2629529 (chr 10), and rs7390625 (chr 20) – were found to have joint association P-values <5.0×10−7 prior to genomic control (Table 2). However, only SNP rs3827907, a variant located in the 3’UTR of the transcribed sequence for EDDM3B, had a joint test P-value, which still met the conservative Bonferroni threshold after applying genomic control – P=7.79×10−8 (Table 2, Figure 1, and Figure E2 [locus zoom plot]). For this variant, both the main effect for the SNP and the interaction term were statistically significant, suggesting that the effect of rs3827907 on asthma control was simultaneously dependent upon the degree of ICS exposure. Specifically, a seemingly detrimental effect of the rs3827907 C-allele (i.e. worsened asthma control) was reversed at prescribed levels of ICS. This relationship was also observed (Figure 2) when plotting the actual values for change in ACT score versus rs3827907 genotype stratified by ICS adherence (i.e., ≤75% adherence [low use] and >75% [high use]). A similar pattern was observed when restricting the analytic set to individuals whose asthma was not controlled (ACT composite score ≤19) prior to initiating ICS treatment (Figure E3). Lastly, adjusting for local ancestry at rs3827907 (in addition to adjusting for global ancestry) did not diminish the overall statistical significance of this variant (P=8.57×10−8 after genomic control).
Table 2.
Interactions between genetic variants and inhaled corticosteroid use on changes in asthma control among SAPPHIRE individuals in the discovery set (n=244)*
| SNP | Chr | Position | Allele† | MAF† | HWE P-value‡ | Gene | Parameter estimate for ICS§ | P-value for ICS§ | Parameter estimate for SNP§ | P-value for SNP term§ | Parameter estimate for SNP x ICS interaction§ | P-value for interactio n term§ |
P-value for joint test║ | Adjusted P-value for joint test¶ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3827907 | 14 | 21238798 | C/T | 0.13 | 0.59 | EDDM3B | −0.51 | 0.641 | −9.67 | 3.06×10−9 | 12.35 | 5.98×10−9 | 2.04×10−8 | 7.79×10−8 |
| rs2629529 | 10 | 126534415 | C/A | 0.06 | 1.00 | -- | 3.43 | 0.001 | 2.07 | 0.357 | −7.74 | 7.40×10−3 | 5.25×10−8 | 1.86×10−7 |
| rs73906251 | 20 | 29991274 | T/C | 0.05 | 0.48 | -- | 1.89 | 0.065 | −14.51 | 1.13×10−5 | 14.60 | 2.77×10−4 | 3.85×10−7 | 1.16×10−6 |
SAPPHIRE, denotes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; and ICS, inhaled corticosteroid.
The phenotype was the change in Asthma Control Test (ACT) score from before to immediately following 6 weeks of inhaled corticosteroid (ICS) treatment.
The effect allele (minor allele) is shown first, and the referent allele follows. The allele frequency of the “effect” allele is provided, and in the current table, this estimate is based on the observed frequency in the discovery set.
P-values are from the exact test assessing deviation in the discovery group from the expected the Hardy-Weinberg distribution. Low P-values signify more significant deviation.
The parameter estimates demonstrate the direction and magnitude for a change in the ACT score over the course of 6 weeks of ICS treatment. A positive parameter estimate indicates that the variable was associated with an improvement in asthma control over the course of treatment. The parameter estimate for ICS represents the effect of increasing ICS use (i.e., going from zero to complete adherence to the prescribed dose [a continuous variable ranging from 0 to 1]). The parameter estimate for the SNP can be interpreted as the effect on asthma control for each additional “effect” allele (i.e., none [=0], one [=1], or two alleles [=2]). The parameter estimate for the interaction term can be interpreted as the combined effect of increasing ICS use (i.e., going from zero to complete adherence to the prescribed dose [a continuous variable ranging from 0 to 1]) and the number of “effect” alleles on the change in asthma control over 6 weeks of ICS treatment. The parameter estimates and P-values for the ICS, SNP, and SNP × ICS interaction variables are shown for the model simultaneously including these variables, as well as adjusting for patient age, sex, ACT score at 660 baseline (i.e., time of study enrollment), smoking status (coded as never, past, and present), the first three principal components, and 661 ICS adherence.
The F-test assessed the significance of the difference in model fit between the full model (i.e., all of the aforementioned variables included) and the reduced model (i.e., the full model minus both the SNP and SNP x ICS interaction terms). In other words, a joint test was used to simultaneously assess the combined significance of the SNP and SNP x ICS interaction terms.
Adjusted using genomic control for an inflation factor of λ=1.09.
Figure 1.
Manhattan plot of the p-values from the genome wide association analysis of inhaled corticosteroid (ICS) response in the discovery set (n=244). Each p-value represents the statistical significance of jointly testing the contribution of both the SNP and SNP x ICS interaction terms. The Bonferroni-adjusted significance level (p=8.72×10−8) is shown as a solid gray line, and a pvalue threshold of 10−5 is shown as a dashed gray line.
Figure 2.
Relationship between rs3827907 genotype and absolute change in Asthma Control Test (ACT) composite score among African American participants in the SAPPHIRE cohort following 6 weeks of observed inhaled corticosteroid (ICS) treatment (n=244). Results are stratified by levels of patient ICS use (≤75% adherence was considered low use and >75% adherence was considered high use). Black lines connect the means for each group.
Based on the regression models (Table 2), a similar pattern of association was seen for SNP rs73906251 on chromosome 20 (P=3.85×10−7, joint test), where an observed detrimental association of the T-allele at this locus also appeared to be mitigated by ICS treatment. In contrast, variant rs2629529 on chromosome 10 (P=5.25×10−8, joint test) showed a salutary association between the C-allele and asthma control, but this effect appeared to diminish with increasing ICS exposure. These two variants were no longer statistically significant after applying genomic control (P=1.16×10−6 for rs73906251 and P=1.86×10−7 for rs26295290).
To exclude the possibility that minor allele homozygotes unduly influenced our results, we repeated our analysis using a dominant genetic model (i.e., collapsing the homozygous and heterozygous carriers into a single group). The joint test under a dominant genetic model identified the same top three SNPs as the discovery analysis (data not shown). The main effect and interaction term P-values for rs3827907 were 1.12×10−7 and 1.45×10−7, respectively (P=6.51×10−7, joint test). This relationship was also observed when plotting the actual values for change in ACT score versus rs3827907 genotype categorized dichotomously as TT homozygotes and C-allele carriers (Figure E4).
Validation Analysis
Due to the absence of African American replication populations with prospectively measured ICS treatment response on asthma control, we elected to support and validate our discovery findings using diverse cohorts with information on both ICS exposure and severe asthma exacerbation outcomes. For this analysis we used a separate group of 803 SAPPHIRE participants who had both prospectively-collected clinical information and genome-wide genotype data. We used nested models to assess the joint effect of the SNP and the SNP x ICS exposure interaction on time-to-severe asthma exacerbation. The joint test P-values for rs3827907, rs2629529, and rs7390625 were 0.023, 5.65×10−6, and 0.043, respectively (Table 3). Overall, rs3827907 was the only one of the three SNPs evaluated to show a direction of effect that was consistent with the discovery analysis. The regression model suggested that the rs3827907 C-allele was associated with an increased risk of severe exacerbation and was mitigated by increasing ICS exposure (although the interaction term alone was not statistically significant). Therefore, rs3827907 appeared to affect response to ICS treatment in terms of both asthma control and exacerbations.
Table 3.
Prospective relationship between genetic variants and inhaled corticosteroid use on asthma exacerbations within the SAPPHIRE validation population (n=803)*
| SNP | Chr | Position | Allele† | MAF† | Gene | Parameter estimate for ICS‡ | P-value for ICS‡ | Parameter estimate for SNP‡ | P-value for SNP‡ | Parameter estimate for SNP × ICS interaction‡ | P-value for interaction‡ | P-value for joint test§ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3827907 | 14 | 21238798 | C/T | 0.13 | EDDM3B | −1.75 | 7.13×10−5 | 0.35 | 0.007 | −0.07 | 0.679 | 0.023 |
| rs2629529 | 10 | 126534415 | C/A | 0.06 | -- | −1.75 | 2.06×10−5 | 0.39 | 0.042 | −1.27 | 1.58×10−4 | 5.65×10−6 |
| rs73906251 | 20 | 29991274 | T/C | 0.05 | -- | −1.92 | 5.06×10−6 | 0.22 | 0.173 | 0.28 | 0.182 | 0.043 |
SAPPHIRE, denotes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; and ICS, inhaled corticosteroid.
The validation analyses used Cox proportional hazards models to assess the time-to-severe asthma exacerbation (i.e., burst oral corticosteroid use, asthma-related emergency department visits, and hospitalizations for asthma).
The effect allele (minor allele) is shown first, and the referent allele follows. The allele frequency of the “effect” allele is provided, and in the current table, this estimate is based on the observed frequency in the validation set.
The parameter estimates represent the risk (i.e., hazard) of having a severe asthma exacerbation following the baseline assessment (i.e., the assessment at study enrollment). A negative parameter estimate indicates that the variable was associated with a lower risk of experiencing a severe asthma exacerbation. The parameter estimate for ICS can be interpreted as the effect of increasing ICS use (i.e., average number of ICS doses taken per day over the preceding 6 months).15, 18 The parameter estimate for the SNP can be interpreted as the effect on exacerbation risk for each additional “effect” allele (i.e., none [=0], one [=1], or two alleles [=2]). The parameter estimate for the interaction term can be interpreted as the combined effect of increasing ICS use and the number of “effect” alleles on the risk of a severe asthma exacerbation. The parameter estimates and P-values for the ICS, SNP, and SNP x ICS interaction variables are shown for the model simultaneously including variables, as well as adjusting for patient age, sex, BMI, smoking status, ACT score at baseline (i.e., time of study enrollment), baseline asthma severity score, LABA use (an indicator variable denoting use of an ICSLABA combination inhaler), time-updated measures of SABA MDI and nebulizer use, and the first 3 principal components.
The likelihood ratio test assessed the significance of the difference in model fit between the full model (i.e., all of the aforementioned variables included) and the reduced model (i.e., the full model minus both the SNP and SNP x ICS interaction terms). In other words, a joint test was used to simultaneously assess the combined significance of the SNP and SNP x ICS interaction terms.
We assessed the promoted SNPs in the GALA II and SAGE II cohorts. For these crosssectional studies, the dichotomous outcome was an asthma exacerbation in the year prior to enrollment. The respective joint test P-values for rs3827907, rs2629529, and rs7390625 were 0.029, 0.281, and 0.413 for GALA II and 0.041, 0.876, and 0.004 for SAGE II (Table 4). While the joint test was statistically significant for rs3827907 in both GALA II and SAGE II, the direction of the SNP main effect for C-allele was most consistent between African Americans in SAPPHIRE (parameter estimate 0.35, Table 3) and Latinos from GALA II (parameter estimate 0.21, Table 4), but differed for African Americans in SAGE II (parameter estimate −1.04, Table 4).
Table 4.
Validation of the relationship between genetic variants and inhaled corticosteroid use on asthma exacerbations within Latinos (n=1,461) from GALA II and African Americans (n=563) from SAGE II*
| Study | SNP | Chr | Position | Allele† | MAF† | Gene | Parameter estimate for ICS‡ | P-value for ICS‡ | Parameter estimate for SNP‡ | P-value for SNP‡ | Parameter estimate for SNP x ICS interaction‡ | P-value for interaction‡ | P-value for joint test§ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GALA II | rs3827907 | 14 | 21238798 | C/T | 0.21 | EDDM3B | −0.16 | 0.521 | 0.21 | 0.194 | 0.15 | 0.503 | 0.029 |
| rs2629529 | 10 | 126534415 | C/A | 0.12 | -- | −0.12 | 0.634 | 0.17 | 0.405 | 0.08 | 0.766 | 0.281 | |
| rs73906251 | 20 | 29991274 | T/C | 0.02 | -- | −0.12 | 0.611 | −0.45 | 0.312 | 1.08 | 0.224 | 0.413 | |
| SAGE II | rs3827907 | 14 | 21238798 | C/T | 0.16 | EDDM3B | −1.01 | 0.183 | −1.04 | 0.021 | 0.96 | 0.053 | 0.041 |
| rs2629529 | 10 | 126534415 | C/A | 0.06 | -- | −0.93 | 0.204 | 0.15 | 0.794 | −0.01 | 0.995 | 0.876 | |
| rs73906251 | 20 | 29991274 | T/C | 0.08 | -- | −0.89 | 0.204 | −0.69 | 0.219 | −0.12 | 0.851 | 0.004 |
GALA II denotes the Genes-environments and Admixture in Latino Americans study; SAGE II, the Study of African Americans, Asthma, Genes, and Environments; SNP, single nucleotide polymorphism; Chr, chromosome; MAF, minor allele frequency; and ICS, inhaled corticosteroid.
The validation analyses used logistic regression to model the retrospective relationship between severe asthma exacerbation in the year prior to study enrollment (i.e., burst oral corticosteroid use, asthma-related emergency department or unscheduled physician visits, and hospitalizations for asthma) and both SNPs and SNP x ICS interactions.
The effect allele (minor allele) is shown first, and the referent allele follows. The allele frequency of the “effect” allele is provided, and in the current table, this estimate is based on the observed frequency among individuals with asthma in each study.
The parameter estimates represent the risk of having a severe asthma exacerbation in the year prior to baseline assessment (i.e., the assessment at study enrollment). A negative parameter estimate indicates that the variable was associated with a lower risk of experiencing a severe asthma exacerbation. The parameter estimate for ICS represents the effect of ICS, a dichotomous indicator variable for participant-reported ICS use at the time of study enrollment. The parameter estimate for the SNP can be interpreted as the effect on exacerbation risk for each additional “effect” allele (i.e., none [=0], one [=1], or two alleles [=2]). The parameter estimate for the interaction term can be interpreted as the combined effect of ICS use at enrollment and the number of “effect” alleles on the risk of a severe asthma exacerbation. In both GALA II and SAGE II, ICS use was represented as a single dichotomous variable based patient reported use at the time of enrollment. The parameter estimates and P-values for the ICS, SNP, and SNP x ICS interaction variables are shown for the model simultaneously including these variables, as well as adjusting for patient age, sex, BMI percentile, secondhand smoke exposure, medication step at enrollment (a proxy for asthma severity based on Expert Panel Report-3 guidelines [National Asthma Education and Prevention Program. Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 120, 2007, S94-S138.]), and the first 3 principal components.
The likelihood ratio test assessed the significance of the difference in model fit between the full model (i.e., all of the aforementioned variables included) and the reduced model (i.e., the full model minus both the SNP and SNP x ICS interaction terms). In other words, a joint test was used to simultaneously assess the combined significance of the SNP and SNP x ICS interaction terms.
The relationship between rs3827907 genotype and change in ACT score stratified by ICS adherence among 98 European American individuals in SAPPHIRE who received 6 weeks of ICS treatment is shown in Figure E5 (online supplement). The rs3827907 C-allele frequency in this group was 40.3%. While not statistically significant, the direction of the allelic effect was similar to that observed in the discovery set.
Assessment of Previously Described Pharmacogenetic Variants
We also attempted to validate 32 such variants that have previously been identified in the literature.4–6, 24–31 The results for change in ACT score are presented in Supplementary Table E1, and those for time-to-severe asthma exacerbation are presented in Supplementary Table E2. While none of the variants were significantly associated with a change in ACT score (Table E1), 12 (38%) of the 32 variants had a joint test P<0.05 for the association with time-to-severe exacerbation (Table E2).
Expression Quantitative Trait Locus (eQTL) Analysis for rs3827907
To investigate possible functionality, we evaluated whether rs3827907 was an eQTL for genes located within 1Mb upstream and downstream of the variant. RNA-seq data derived from whole blood (i.e., whole blood transcriptome) were available for 813 African American SAPPHIRE participants (408 with asthma and 405 without asthma). Expression of EDDM3B in whole blood was low and did not meet our criterion for evaluation (≥50% of individuals with ≥1 read count). Therefore, if rs3827907 had a substantial regulatory effect on expression in whole blood, it was likely for genes other than EDDM3B. The correlation in expression between the 32 genes analyzed is shown in Figure E6 of the online supplement. In the combined analysis of all 813 individuals (Table 5), rs3827907 was significantly associated with both RNASE2 expression (P=6.10×10−4) and RNASE1 expression (P=7.92×10−4) using a conservative Bonferroni threshold (P<0.05/32=1.56×10−3). The association with RNASE3 was of borderline significance (P=0.008). In addition to being the most significant association in the combined sample, the parameter estimates (PE) and ranks for the eQTL association between rs3827907 and RNASE2 expression were consistent among individuals with and without asthma (PE=−0.243, rank 3 and PE=−0.238, rank 2, respectively). The consistently negative estimates across all groups suggested that the main effect of the rs3827907 C-allele was to decrease expression of RNASE2.
Table 5.
Expression quantitative trait locus analysis for rs3827909 and its association with nearby gene expression on chromosome 14 among African American SAPPHIRE participants with (n=408) and without asthma (n=405)*
| Gene | Start | End | Distance to rs3827907 (bp)† | Rank | Cases (n=408) Parameter estimate‡ | P-value | Rank | Controls (n=405) Parameter estimate‡ | P-Value | Rank | Total (n=81 Parameter estimate‡ | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR11H4 | 20242739 | 20243820 | +526,819 | 27 | −0.046 | 0.666 | 8 | −0.088 | 0.339 | 18 | −0.066 | 0.346 |
| TTC5 | 20256558 | 20305994 | +464,645 | 6 | −0.195 | 0.058 | 24 | −0.018 | 0.851 | 8 | −0.109 | 0.121 |
| CCNB1IP1 | 20311368 | 20333312 | +437,327 | 25 | −0.055 | 0.575 | 31 | 0.003 | 0.975 | 23 | −0.027 | 0.703 |
| PARP2 | 20343582 | 20357905 | +412,734 | 4 | 0.211 | 0.025 | 4 | 0.134 | 0.202 | 5 | 0.173 | 0.014 |
| TEP1 | 20365667 | 20413429 | +357,210 | 10 | −0.117 | 0.230 | 21 | −0.028 | 0.779 | 17 | −0.074 | 0.292 |
| KLHL33 | 20428811 | 20435642 | +334,997 | 24 | 0.054 | 0.571 | 12 | −0.080 | 0.437 | 27 | −0.011 | 0.874 |
| OSGEP | 20446411 | 20455105 | +315,534 | 2 | −0.246 | 0.015 | 5 | −0.107 | 0.273 | 4 | −0.179 | 0.110 |
| APEX1 | 20455191 | 20457772 | +312,867 | 28 | −0.036 | 0.715 | 20 | 0.038 | 0.701 | 32 | 0.000 | 0.998 |
| TMEM55B | 20457719 | 20461612 | +309,027 | 22 | 0.062 | 0.526 | 16 | 0.070 | 0.488 | 19 | 0.066 | 0.348 |
| PNP | 20468954 | 20477094 | +293,545 | 20 | −0.067 | 0.501 | 7 | 0.098 | 0.323 | 26 | 0.013 | 0.850 |
| RNASE10 | 20505537 | 20511169 | +259,470 | 9 | 0.142 | 0.178 | 15 | 0.065 | 0.485 | 9 | 0.104 | 0.138 |
| RNASE4 | 20684100 | 20701215 | +69,424 | 14 | 0.101 | 0.321 | 3 | 0.183 | 0.060 | 6 | 0.141 | 0.045 |
| ANG | 20684177 | 20698971 | +71,668 | 21 | −0.069 | 0.503 | 11 | −0.087 | 0.360 | 14 | −0.078 | 0.267 |
| RP11–903H12.5 | 20684587 | 20700576 | +70,063 | 7 | −0.187 | 0.062 | 17 | −0.062 | 0.531 | 7 | −0.126 | 0.072 |
| RNASE6 | 20781051 | 20782467 | −10,412 | 13 | −0.102 | 0.319 | 9 | −0.091 | 0.346 | 10 | −0.097 | 0.170 |
| RNASE1 | 20801228 | 20803278 | −30,589 | 5 | −0.210 | 0.027 | 1 | −0.262 | 0.012 | 2 | −0.235 | 7.92×10−4 |
| RNASE3 | 20891399 | 20892348 | −120,760 | 1 | −0.261 | 0.007 | 6 | −0.109 | 0.288 | 3 | −0.187 | 0.008 |
| RNASE2 | 20955452 | 20956436 | −184,813 | 3 | −0.243 | 0.018 | 2 | −0.238 | 0.013 | 1 | −0.240 | 6.10×10−4 |
| METTL17 | 20989770 | 20997035 | −219,131 | 23 | −0.059 | 0.533 | 10 | −0.095 | 0.360 | 15 | −0.077 | 0.275 |
| NDRG2 | 21016763 | 21070872 | −246,124 | 15 | −0.090 | 0.355 | 18 | −0.061 | 0.549 | 16 | −0.076 | 0.280 |
| ARHGEF40 | 21070270 | 21090240 | −299,631 | 11 | −0.114 | 0.250 | 14 | −0.072 | 0.472 | 12 | −0.094 | 0.184 |
| ZNF219 | 21090046 | 21104722 | −319,407 | 12 | 0.109 | 0.251 | 13 | 0.078 | 0.458 | 11 | 0.094 | 0.183 |
| TMEM253 | 21098937 | 21103724 | −328,298 | 29 | −0.036 | −0.725 | 23 | 0.021 | 0.830 | 28 | −0.008 | 0.910 |
| HNRNPC | 21209136 | 21269494 | −438,497 | 17 | 0.081 | 0.420 | 19 | −0.044 | 0.655 | 24 | 0.020 | 0.771 |
| RPGRIP1 | 21287939 | 21351301 | −517,300 | 19 | 0.076 | 0.449 | 27 | 0.006 | 0.954 | 22 | 0.042 | 0.552 |
| SUPT16H | 21351472 | 21384266 | −580,833 | 30 | 0.002 | 0.982 | 28 | 0.004 | 0.963 | 30 | 0.003 | 0.962 |
| CHD8 | 21385194 | 21456126 | −614,555 | 32 | 0.001 | 0.990 | 26 | 0.007 | 0.942 | 29 | 0.004 | 0.954 |
| RAB2B | 21459020 | 21476973 | −688,381 | 16 | 0.085 | 0.368 | 30 | 0.005 | 0.965 | 21 | 0.046 | 0.514 |
| TOX4 | 21476597 | 21499175 | −705,958 | 18 | 0.076 | 0.441 | 22 | 0.028 | 0.783 | 20 | 0.052 | 0.457 |
| METTL3 | 21498133 | 21511375 | −727,494 | 8 | 0.154 | 0.116 | 29 | 0.005 | 0.963 | 13 | 0.081 | 0.248 |
| SALL2 | 21521081 | 21537216 | −750,442 | 31 | 0.002 | 0.986 | 32 | −0.001 | 0.994 | 31 | 0.001 | 0.993 |
| OR10G2 | 21633836 | 21634940 | −863,197 | 26 | −0.046 | 0.641 | 25 | 0.011 | 0.916 | 25 | −0.019 | 0.792 |
SAPPHIRE denotes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity, and bp, nucleotide base pair.
Gene expression was measured using RNA-seq data derived from whole blood RNA. To assess whether rs3827907 was a cis- expression quantitative trait locus for nearby genes, we restricted the analysis to genes located within 1 mega-base upstream and downstream of the variant.
Positive (+) distance indicates that the start codon for the gene is located 5’to rs3827907 on sense strand, whereas negative (−) distance indicates that the start codon for the gene is located 3’ to the variant on the sense strand.
Linear regression was used to examine the relationship between rs3827907 genotype (assuming an additive genetic model where genotypes were coded as TT=0, CT=1, CC=2) and gene expression (dependent variable). The model adjusted for patient age, sex, BMI, the absolute cell counts for each of the 5 white blood cell types measured (i.e., neutrophils, monocytes, lymphocytes, eosinophils, and basophils from complete blood counts obtained at the time that the RNA was collected), sequence batch, and the first 30 probabilistic estimation of expression residuals (PEERs). The parameter estimates are the adjusted genotype main effect estimates for rs3827907 from the model. A positive parameter estimate suggests that an increasing dose of the C-allele is associated with increased expression of a given gene, whereas a negative parameter estimate suggests that the C-allele is associated with decreased gene expression. Rank represents the P-value order for the association between rs3827907 genotype and gene expression; the most statistically significant P-value is ranked first.
Pre-treatment Serum Eosinophil-Derived Neurotoxin (EDN) Levels and Blood Eosinophil Counts on ICS Treatment Response
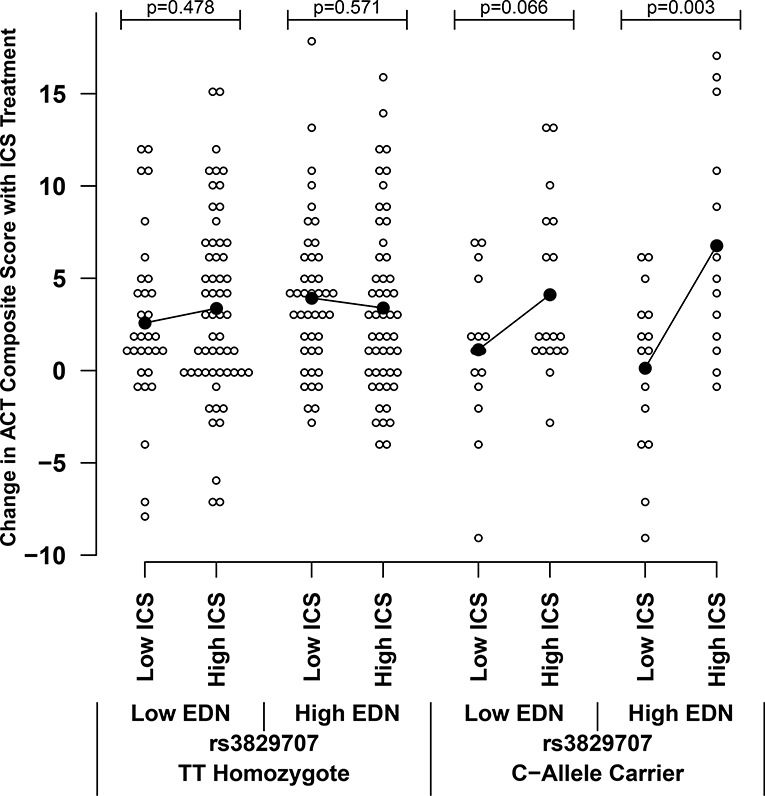
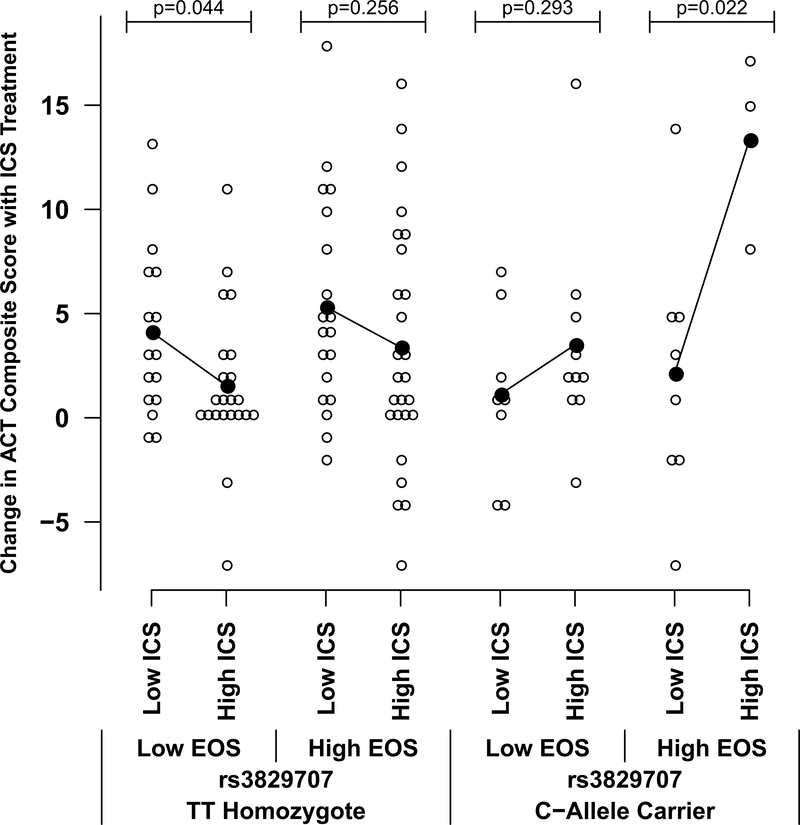
Two of the 3 genes whose expression was most strongly associated with rs3827907 genotype (i.e., RNASE2 and RNASE3) in the eQTL analysis encode for proteins found in eosinophil cytoplasmic granules – namely, EDN and Eosinophil Cationic Protein, respectively. This suggested that the effect of rs3827907 on ICS treatment response may be most apparent in the setting of asthma with ongoing eosinophilic inflammation. Using banked serum from SAPPHIRE participants in the discovery set, we measured EDN levels in serum collected prior to treatment; these data were available for 243 African American individuals. We also evaluated blood eosinophil counts derived from blood cell differentials obtained immediately prior to initiating treatment. As can be seen in Figures 3 and 4, the difference in response to increasing ICS use was statistically significant and prominent in rs3827907 C-allele carriers with high (i.e., greater than the median) baseline serum levels of EDN (>2.34 ng/mL) and blood eosinophil counts (>222 cells/μL). The regression models which support these analyses are shown in Table E3. Replication of these findings in the 98 European American SAPPHIRE participants who also underwent ICS treatment is shown in Figure E7 (EDN stratification) and Figure E8 (blood eosinophil count stratification). While the not statistically significant, the same patterns were observed among the European American participants when compared with the African American participants, particularly for the analyses stratifying by blood eosinophil counts (Figure E8).
Figure 3.
Effect of baseline measures of eosinophil-derived neurotoxin (EDN) on relationship between rs3827907 C-allele carrier status and absolute change in Asthma Control Test (ACT) composite score among African American participants in the SAPPHIRE cohort following 6 weeks of observed inhaled corticosteroid (ICS) treatment (n=243). Results are stratified by levels of patient ICS use (≤75% adherence was considered low use and >75% adherence was considered high use) and baseline serum levels of EDN. Low and high EDN levels were defined as those ≤2.34 ng/mL and >2.34 ng/mL (the median level), respectively. Black lines connect the means for each group.
Figure 4.
Effect of baseline blood eosinophil counts on relationship between rs3827907 C-allele carrier status and absolute change in Asthma Control Test (ACT) composite score among African American participants in the SAPPHIRE cohort following 6 weeks of observed inhaled corticosteroid (ICS) treatment (n=113). Results are stratified by levels of patient ICS use (≤75% adherence was considered low use and >75% adherence was considered high use) and blood eosinophil counts measured before initiating ICS treatment. Low and high eosinophil counts were defined as those ≤222 cells/μL and >222 cells/ μL (the median level), respectively. Black lines connect the means for each group.
Assessment of Imputed Genotypes
To address the possibility that rs3827907 was not the causal variant, we imputed missing genotypes among African Americans in the SAPPHIRE cohort using the cosmopolitan panel from the 1KGP as reference. The locus zoom plot for the joint association with ICS response including imputed genotypes is shown in Figure E9. Only one other SNP in the region, rs12891518, had a joint association P-value <10−5 (joint test P-value without adjustment for genomic control = 3.22×10−7). Evaluation of this variant in 597 African American individuals from SAPPHIRE who had both RNA-seq data and genotype information found that it was less significantly associated with RNASE2 expression when compared with rs3827907 (Table E4). Lastly, rs12891518 was not identified as a cis-eQTL for any gene in publicly available expression data sets using peripheral blood obtained from 5,311 European individuals or in any tissue included in the Genotype-Tissue Expression (GTEx) project (data not shown).32, 33 Taken together, these results support rs3827907 as the causal variant for ICS response in this region.
DISCUSSION
Clinical trials and epidemiologic studies have consistently found ICS therapy to be the single most effective treatment for improving asthma control and mitigating severe complications, such as life-threatening exacerbations.2, 34 Here we identify a novel pharmacogenomic variant, rs3827907, which appears to mediate the relationship between ICS treatment and improved asthma control among African American individuals. This variant located in the 3’ UTR of EDDM3B on chromosome 14 also appeared to influence the relationship between ICS use and severe exacerbations among both African American and Latino individuals. It is unlikely that rs3827907 influenced EDDM3B expression in blood, as we found very few transcripts in our whole blood RNA-seq. We also found almost no expression of EDDM3B in lung (i.e., the other relevant tissue for asthma) from the GTEx project – data not shown.33
In contrast, the eQTL analysis of rs3827907 in 813 African American SAPPHIRE participants with and without asthma suggested that the C-allele was associated with lower expression of RNASE2 and possibly RNASE3. The respective protein products of these genes, EDN and ECP, are major constituents of eosinophil cytoplasmic granules and are released systemically upon cell activation.35 Hence, these proteins have been considered potential biomarkers of eosinophilic inflammation.36–38 Of clinical relevance, we demonstrated that in the setting of eosinophilic inflammation (as evinced by a pre-treatment serum EDN level >2.34 ng/mL or a blood eosinophil count >222 cells/μL), the rs3827907 C-allele distinguished African American individuals with the greatest ICS-related improvement in asthma control. Conversely, African American individuals who were homozygous for the rs3827907 T-allele did not demonstrate a clear dose-response relationship between ICS therapy and asthma control in either the presence or absence of pre-treatment eosinophilic inflammation. In short, our findings suggest that relatively easily measured biomarkers in blood (i.e., rs3827907 genotype along with blood eosinophil counts or serum EDN levels) may help identify the degree to which African American individuals will respond to ICS therapy
Because gene expression was assessed at one time-point, we cannot comment on how rs3827907 genotype and ICS use interact to influence RNASE2 expression over time. Similarly, without additional mechanistic studies, we cannot conclude that RNASE2 or RNASE3 (or their protein products EDN and ECP, respectively) were the mechanism through which the rs3827907 variant influenced ICS-associated improvements in asthma control. However, these limitations do not diminish the potential import of these biomarkers for predicting ICS response.
The association between eosinophilia and asthma severity is well described,39 and it may also identify an asthma phenotype (i.e., eosinophilic asthma vs. non-eosinophilic asthma) that is more responsive to inhaled corticosteroids.40, 41 Similarly, proteins released as a result of eosinophil degranulation may also be useful indicators of ongoing eosinophilic inflammation. For example, Kim et al. showed that EDN blood levels were associated with asthma severity in children,42 and Gon et al. found that EDN levels fell as lung function improved among adults treated with 8 weeks of omalizumab.36 However, it is not clear that these protein biomarkers outperform simple counts of eosinophils in blood or sputum. Meijer et al. showed that ECP levels did not improve upon the use of eosinophil counts to predict corticosteroid-associated improvements in lung function and patient-reported quality of life.43 One of the unique and important findings of our analysis is that we found that markers of pre-treatment eosinophilic inflammation (i.e., either EDN levels or blood eosinophil counts) were only useful in categorizing ICS responsive groups when combined with an additional genetic susceptibility marker. While our findings should be considered preliminary, they do provide a model for refining existing clinical tests by overlaying additional genetic (or “-omic”) data.
Another important feature of our study is its primary focus on U.S. minority populations, particularly African American individuals, who in general disproportionately suffer from asthma complications yet are underrepresented in genetic studies.44, 45 Our ability to replicate the rs3827907 association among a cohort of Latinos with asthma suggests that this variant may be influential in other populations. However, we were unable to replicate our findings in European American individuals, which may have been due in part to the much smaller sample size of this group. Additional testing in these population groups will be necessary to confirm our findings. Westra et al. also showed that the rs3827907 variant was an eQTL for RNASE2; however, in contrast to our findings, this study of the blood transcriptome in Europeans found that the rs3827907 C-allele was associated with increased RNASE2 expression.32 It is not uncommon to find that risk alleles, as well as their magnitude and direction of association, vary between population groups.9, 46
As an ancillary analysis, we analyzed variants previously associated with ICS response in predominantly European American individuals and found that approximately 40% were associated with treatment response in African Americans with asthma (Table E2). Because the studies identifying these variants had different designs and outcomes, it is not possible to confidently comment on whether the direction of effect was consistent. Nevertheless, we believe that there are still important take away points from this analysis. First, our ability to verify previously identify ICS response variants suggests that our method was suitable for validating our discovery findings. Second, at minimum it appears that a substantial proportion of variants identified in individuals of European descent are relevant in African Americans.
There are additional study limitations to consider beyond those already mentioned. First, we did not have a replication set of African American individuals who underwent 6 weeks of observed ICS treatment analogous to the discovery set. This is due in part to the general lack of treatment trial data for minority populations. Fortunately, we were able to reassess potential pharmacogenomic interactions with respect to severe asthma exacerbations in two separate sets of African American individuals with asthma (SAPPHIRE and SAGE II) and one cohort of Latinos with asthma (GALA II). Despite finding a consistent and significant signal in our validation cohorts, characteristic differences between the discovery and validation groups (e.g., baseline differences in lung function) may have also affected the magnitude and direction of the effect estimates observed. Our primary outcome for the discovery set was a change in asthma control, as measured using the ACT, not a change in lung function, as has been used elsewhere.4, 5 However, both the ACT and FEV1 have been shown to be predictive of asthma exacerbations,47, 48 and at least one study has suggested that the ACT is a better predictor of exacerbations.49 We have also recently demonstrated a “dose-response” relationship between ICS use and change in ACT score,8 supporting its use as measure of controller response. For these reasons, we feel that change in ACT score and asthma exacerbations were reasonable phenotypes to use in the discovery and validation of pharmacogenomic variants. While we had a similar replication phenotype for European American SAPPHIRE participants when compared to the discovery group, the sample size of the former was small. As a result, we could not discern whether the lack of statistical significance for rs3827907 in European Americans (despite similar patterns of association) was due to lack of power or population-specific allelic effects. This sample was also too small to be used for discovery. Lastly, we only had gene expression information from whole blood in SAPPHIRE participants. It is possible that variant rs3827907 may have different associations with gene expression in other tissues and cell types. However, the eQTL association with RNASE2, a nearby gene which is highly expressed in eosinophils, suggests that our use of whole blood RNA-seq was both appropriate and fortuitous.
In summary, we identified a novel genetic variant, rs3827907, associated with ICS treatment response. The variant appeared to augment knowledge of pre-treatment eosinophilic inflammation in distinguishing individuals with the greatest ICS response. Future studies are needed to reassess our findings in multiple population groups, to establish the utility of this variant for predicting treatment response in additional independent samples, and to further dissect the mechanism of this pharmacogenetic interaction.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the staff of the University of California San Francisco Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core for their assistance in generating the RNA-seq data for this paper. We would also like to thank all of the past and current investigators and staff members whose hard work and dedication helped create the SAPPHIRE, GALA II, and SAGE II cohorts.
Sources of Funding:
This work was supported by the Fund for Henry Ford Hospital (AML, DEL, and LKW); a fellowship (FI16/00136) from the Instituto de Salud Carlos III (ISCIII) and co-funded by the European Social Funds from the European Union (ESF) – “ESF invests in your future” (NH-P); the Ramón y Cajal Program (RYC-2015–17205) by the Spanish Ministry of Economy, Industry, and Competitiveness (MP-Y); an award (AC15/00015) by the ISCIII through Strategic Action for Health Research (AES) and European Community (EC) within the Active and Assisted Living (AAL) Programme framework (MP-Y); a SysPharmPedia grant from the ERACoSysMed 1st Joint Transnational Call from the European Union under the Horizon 2020 (MP-Y); the American Asthma Foundation (LKW, EGB); the Sandler Family Foundation (EGB); the RWJF Amos Medical Faculty Development Program (EGB); Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II (EGB); and the following institutes of the National Institutes of Health: National Institute of Allergy and Infectious Diseases (U19AI077439 to DJE, and R01AI079139 and R01AI061774 to LKW), the National Heart Lung and Blood Institute (K99HL135403 to WLE; R01HL117004 and X01HL134589 to EGB; and R01HL079055, R01HL118267, and X01HL134589 to LKW), the National Institute of Diabetes and Digestive and Kidney diseases (R01DK064695 and R01DK113003 to LKW), the National Institute of Health and Environmental (R01ES015794 and R21ES024844 to EGB), and the National Institute on Minority Health and Health Disparities (P60MD006902 and R01MD010443 to EGB).
ABBREVIATIONS:
- ACT
Asthma Control Test
- CHR
chromosome
- ECP
eosinophil cationic protein
- EDN
eosinophil derived neurotoxin
- eQTL
expression quantitative trait locus (or loci)
- FEV1
forced expiratory volume at one second
- GALA II
Genes-environments & Admixture in Latino Americans Study
- ICS
inhaled corticosteroid
- LABA
long-acting beta-agonist
- MDI
metered dose inhaler
- SABA
short-acting beta-agonist
- SAGE II
Study of African Americans, Asthma, Genes and Environments
- SAPPHIRE
Study of Asthma Phenotypes and Pharmacogenomic Interactions by Raceethnicity
- SNP
single nucleotide polymorphism
Footnotes
Conflicts of Interest: NH-P reports grant funding through a fellowship from the Instituto de Salud Carlos III. WLE reports grant funding from the NIH. MP-Y reports grant funding from the Spanish Ministry of Economy, Industry and Competitiveness and Instituto de Salud Carlos III. DJE reports grant funding from the NIAID, NIH; an honorarium and travel expenses from Boehringer Ingelheim; and consulting and travel expenses from Coherus. LKW reports grant funding from the NIDDK, NHBLI and NIAID, NIH and the American Asthma Foundation. The remaining authors have no disclosures to reports.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Akinbami LJ, Moorman JE, Bailey C, Zahran HS, King M, Johnson CA, et al. Trends in asthma prevalence, health care use, and mortality in the United States, 2001–2010. NCHS Data Brief 2012:1–8. [PubMed] [Google Scholar]
- 2.Sin DD, Man J, Sharpe H, Gan WQ, Man SF. Pharmacological management to reduce exacerbations in adults with asthma: a systematic review and meta-analysis. JAMA 2004; 292:367–76. [DOI] [PubMed] [Google Scholar]
- 3.Suissa S, Ernst P, Benayoun S, Baltzan M, Cai B. Low-dose inhaled corticosteroids and the prevention of death from asthma. N Engl J Med 2000; 343:332–6. [DOI] [PubMed] [Google Scholar]
- 4.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med 2011; 365:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawkins GA, Lazarus R, Smith RS, Tantisira KG, Meyers DA, Peters SP, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol 2009; 123:1376–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet 2004; 13:1353–9. [DOI] [PubMed] [Google Scholar]
- 7.Gould W, Peterson EL, Karungi G, Zoratti A, Gaggin J, Toma G, et al. Factors predicting inhaled corticosteroid responsiveness in African American patients with asthma. J Allergy Clin Immunol 2010; 126:1131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells KE, Cajigal S, Peterson EL, Ahmedani BK, Kumar R, Lanfear DE, et al. Assessing differences in inhaled corticosteroid response by self-reported race-ethnicity and genetic ancestry among asthmatic subjects. J Allergy Clin Immunol 2016; 137:1364–9 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 2011; 43:887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galanter JM, Torgerson D, Gignoux CR, Sen S, Roth LA, Via M, et al. Cosmopolitan and ethnic-specific replication of genetic risk factors for asthma in 2 Latino populations. J Allergy Clin Immunol 2011; 128:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther 2011; 89:408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams LK, Joseph CL, Peterson EL, Wells K, Wang M, Chowdhry VK, et al. Patients with asthma who do not fill their inhaled corticosteroids: a study of primary nonadherence. J Allergy Clin Immunol 2007; 120:1153–9. [DOI] [PubMed] [Google Scholar]
- 13.Apter AJ, Boston RC, George M, Norfleet AL, Tenhave T, Coyne JC, et al. Modifiable barriers to adherence to inhaled steroids among adults with asthma: it’s not just black and white. J Allergy Clin Immunol 2003; 111:1219–26. [DOI] [PubMed] [Google Scholar]
- 14.Williams LK, Pladevall M, Xi H, Peterson EL, Joseph C, Lafata JE, et al. Relationship between adherence to inhaled corticosteroids and poor outcomes among adults with asthma. J Allergy Clin Immunol 2004; 114:1288–93. [DOI] [PubMed] [Google Scholar]
- 15.Williams LK, Peterson EL, Wells K, Ahmedani BK, Kumar R, Burchard EG, et al. Quantifying the proportion of severe asthma exacerbations attributable to inhaled corticosteroid nonadherence. J Allergy Clin Immunol 2011; 128:1185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatz M, Sorkness CA, Li JT, Marcus P, Murray JJ, Nathan RA, et al. Asthma Control Test: reliability, validity, and responsiveness in patients not previously followed by asthma specialists. J Allergy Clin Immunol 2006; 117:549–56. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann TJ, Zhan Y, Kvale MN, Hesselson SE, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics 2011; 98:422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells KE, Peterson EL, Ahmedani BK, Severson RK, Gleason-Comstock J, Williams LK. The relationship between combination inhaled corticosteroid and long-acting betaagonist use and severe asthma exacerbations in a diverse population. J Allergy Clin Immunol 2012; 129:1274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013; 501:506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet 2016; 48:1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlin A, Denny J, Roden DM, Brilliant MH, Ingram C, Kitchner TE, et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun Inflamm Dis 2015; 3:350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijverberg SJ, Koster ES, Tavendale R, Leusink M, Koenderman L, Raaijmakers JA, et al. ST13 polymorphisms and their effect on exacerbations in steroid-treated asthmatic children and young adults. Clin Exp Allergy 2015; 45:1051–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Tong C, Wang Z, Wang Z, Mauger D, Tantisira KG, et al. Pharmacodynamic genome-wide association study identifies new responsive loci for glucocorticoid intervention in asthma. Pharmacogenomics J 2015; 15:422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tantisira KG, Hwang ES, Raby BA, Silverman ES, Lake SL, Richter BG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A 2004; 101:18099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tantisira KG, Damask A, Szefler SJ, Schuemann B, Markezich A, Su J, et al. Genomewide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med 2012; 185:1286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park HW, Dahlin A, Tse S, Duan QL, Schuemann B, Martinez FD, et al. Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. J Allergy Clin Immunol 2014; 133:664–9 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KC, et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet 2011; 7:e1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu W, Rogers AJ, Damask A, Raby BA, Klanderman BJ, Duan QL, et al. Pharmacogenomics: novel loci identification via integrating gene differential analysis and eQTL analysis. Hum Mol Genet 2014; 23:5017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45:1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suissa S, Ernst P. Inhaled corticosteroids: impact on asthma morbidity and mortality. J Allergy Clin Immunol 2001; 107:937–44. [DOI] [PubMed] [Google Scholar]
- 35.Walsh GM. Eosinophil granule proteins and their role in disease. Curr Opin Hematol 2001; 8:28–33. [DOI] [PubMed] [Google Scholar]
- 36.Gon Y, Ito R, Hattori T, Hiranuma H, Kumasawa F, Kozu Y, et al. Serum eosinophilderived neurotoxin: correlation with persistent airflow limitation in adults with housedust mite allergic asthma. Allergy Asthma Proc 2015; 36:e113–20. [DOI] [PubMed] [Google Scholar]
- 37.Niimi A, Amitani R, Suzuki K, Tanaka E, Murayama T, Kuze F. Serum eosinophil cationic protein as a marker of eosinophilic inflammation in asthma. Clin Exp Allergy 1998; 28:233–40. [DOI] [PubMed] [Google Scholar]
- 38.Tomassini M, Magrini L, De Petrillo G, Adriani E, Bonini S, Balsano F, et al. Serum levels of eosinophil cationic protein in allergic diseases and natural allergen exposure. J Allergy Clin Immunol 1996; 97:1350–5. [DOI] [PubMed] [Google Scholar]
- 39.Price DB, Rigazio A, Campbell JD, Bleecker ER, Corrigan CJ, Thomas M, et al. Blood eosinophil count and prospective annual asthma disease burden: a UK cohort study. Lancet Respir Med 2015; 3:849–58. [DOI] [PubMed] [Google Scholar]
- 40.Pavord ID, Brightling CE, Woltmann G, Wardlaw AJ. Non-eosinophilic corticosteroid unresponsive asthma. Lancet 1999; 353:2213–4. [DOI] [PubMed] [Google Scholar]
- 41.Berry M, Morgan A, Shaw DE, Parker D, Green R, Brightling C, et al. Pathological features and inhaled corticosteroid response of eosinophilic and non-eosinophilic asthma. Thorax 2007; 62:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim CK, Callaway Z, Fletcher R, Koh YY. Eosinophil-derived neurotoxin in childhood asthma: correlation with disease severity. J Asthma 2010; 47:568–73. [DOI] [PubMed] [Google Scholar]
- 43.Meijer RJ, Postma DS, Kauffman HF, Arends LR, Koeter GH, Kerstjens HA. Accuracy of eosinophils and eosinophil cationic protein to predict steroid improvement in asthma. Clin Exp Allergy 2002; 32:1096–103. [DOI] [PubMed] [Google Scholar]
- 44.Asthma prevalence and control characteristics by race/ethnicity--United States, 2002. MMWR Morb Mortal Wkly Rep 2004; 53:145–8. [PubMed] [Google Scholar]
- 45.Bustamante CD, Burchard EG, De L,V Genomics for the world. Nature 2011; 475:163–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B Associated with Asthma in Children. N.Engl.J Med 2009. [DOI] [PubMed] [Google Scholar]
- 47.Kitch BT, Paltiel AD, Kuntz KM, Dockery DW, Schouten JP, Weiss ST, et al. A single measure of FEV1 is associated with risk of asthma attacks in long-term follow-up. Chest 2004; 126:1875–82. [DOI] [PubMed] [Google Scholar]
- 48.Sato R, Tomita K, Sano H, Ichihashi H, Yamagata S, Sano A, et al. The strategy for predicting future exacerbation of asthma using a combination of the Asthma Control Test and lung function test. J Asthma 2009; 46:677–82. [DOI] [PubMed] [Google Scholar]
- 49.Ko FW, Hui DS, Leung TF, Chu HY, Wong GW, Tung AH, et al. Evaluation of the asthma control test: a reliable determinant of disease stability and a predictor of future exacerbations. Respirology 2012; 17:370–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.