Abstract
The identification and quantification of proteins lags behind DNA sequencing methods in scale, sensitivity and dynamic range. Here we show that sparse amino acid sequence information can be obtained for individual protein molecules for thousands to millions of molecules in parallel. We demonstrate selective fluorescent labeling of cysteine and lysine residues in peptide samples, immobilization of labeled peptides on a glass surface, and imaging by total internal reflection microscopy to monitor reductions in each molecule’s fluorescence following consecutive rounds of Edman degradation. The obtained sparse fluorescent sequence of each molecule was then assigned to its parent protein in a reference database. We demonstrate the method on synthetic and naturally-derived peptide molecules in zeptomole-scale quantities. We also fluorescently label phosphoserines and demonstrate single-molecule, positional readout of the phosphorylated sites. We measured >93% efficiencies for dye labeling, survival, and cleavage; further improvements should empower studies of increasingly complex proteomic mixtures, with the high sensitivity and digital quantification offered by single molecule sequencing.
Proteins often exist in extremely complex mixtures, with a typical human cell containing >10,000 unique proteins and perhaps ten times as many post-translationally modified proteoforms. Each protein potentially varies in abundance from 1 to 109 copies in a manner often poorly predicted by mRNA transcript levels1. The inability to comprehensively sequence such complex protein samples, and especially to quantify and identify low-abundance proteins and post-translational modifications, is a major road-block in protein biomarker discovery2. Currently, mass spectrometry is the method of choice for large-scale protein identification, but it is limited in its ability to analyze low-abundance samples and map rare amino acid variants3–5. These limitations could be addressed by successful development of highly parallel single-molecule protein sequencing6–12, a concept analogous to nucleic acid technologies that sequence millions to billions of oligonucleotides in complex mixtures in parallel. The approach would offer more than a million-fold improvement in sensitivity over conventional technologies and allow millions of distinct peptide molecules to be sequenced in parallel, identified and digitally quantified (Fig. 1A). Here we describe an implementation of protein fluorosequencing by directly visualizing individual fluorescently labeled peptide or protein molecules as they are subjected to the classic protein sequencing chemistry, Edman degradation13.
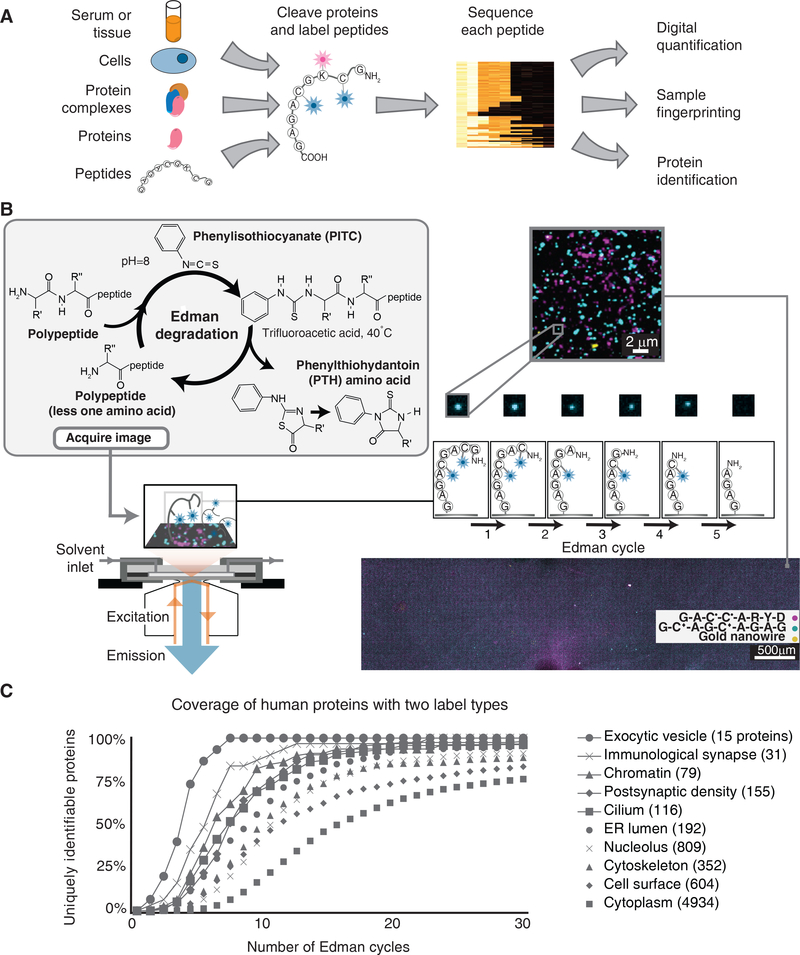
Figure 1: Overview of single-molecule fluorosequencing.
(A) Summary of the approach for protein and peptide analyses. (B) Peptides are covalently labeled with amino-acid specific fluorescent dyes and immobilized in a total internal reflection fluorescence (TIRF) single molecule microscope stage perfusion chamber. Using TIRF, each peptide is imaged, then its amino-terminal (N-terminal) amino acid is chemically removed via Edman degradation, leaving each peptide one amino acid shorter and regenerating its free N-terminus. Repeated cycles of chemistry (each removing one amino acid) and imaging reveal the positions of fluorescent dyes within each molecule. Millions of individual peptide molecules can be analyzed in parallel at reasonable attachment densities, shown for approx. 3 million peptides in a roughly 1.3 × 5 mm area of the cover slip. • indicates TMR conjugated to cysteine, ♦indicates Atto647N conjugated to cysteine; gold nanowires serve as fiducial markers. (C) Even a relatively modest amino acid labeling scheme can be sufficiently information-rich to identify proteins, as illustrated by calculating the proportions of human proteins in specific subcellular compartments (defined by Gene Ontology Cellular Component annotations; numbers indicate protein counts) that are uniquely identifiable with a two-color code. Each curve plots coverage of uniquely identifiable proteins, as a function of Edman cycles performed, considering the scenario of labeling only cysteines and lysines on peptides formed by GluC proteolysis, which cleaves after glutamate or aspartate.
In the protein fluorosequencing concept, one or more amino acid types are selectively labeled with a specific identifier fluorophore14. After immobilizing millions of labeled peptides on a glass cover slip, each molecule’s fluorescence is monitored using TIRF microscopy following consecutive rounds of amino-terminal (N-terminal) amino acid removal by Edman chemistry13 (Fig. 1B). The sequence positions of the labeled amino acids are thus identified for each peptide molecule, providing a partial sequence. These sequences of fluorescent amino acids are compared to a reference proteome for assignment to their proteins of origin. Although only labeled amino acids are visualized, the results can nonetheless be very information-rich because the labeled amino acids’ sequence positions are precisely determined, the terminal amino acids’ identities can be constrained by choice of proteolytic enzyme and surface attachment chemistry, and the intervening amino acids’ identities are partly constrained (because they were not the labeled, cleaved, or attached amino acid types). Fig. 1C illustrates the information-richness by plotting the proportions of human proteins in an assortment of subcellular compartments that can be uniquely identified using only a 2-color code (here, modeling labeling of cysteines and lysines on peptides generated by proteolysis after glutamate or aspartate). Even a 2-color code is sufficiently information-rich to uniquely identify most proteins in mixtures of moderate complexity ranging up to ~1,000 human proteins (Fig. 1C and refs. 6, 7). Monte Carlo simulations have predicted that use of additional labels (e.g. as established for aspartate/glutamate and tryptophan14) should be sufficient to identify a majority of proteins in the human proteome, even when considering the expected effects of experimental errors due to e.g. photo/chemical dye inactivation, incomplete fluorescent labeling, and sporadic failures of Edman reaction cycles6.
Here, we experimentally implement the fluorosequencing concept by labeling and discriminating peptides and simple peptide mixtures, which required developing instrumentation and methods, extensive testing of fluorophores, microfluidic design, chemistry of peptide immobilization and Edman degradation, image processing algorithms for monitoring individual peptide’s fluorescent intensity and classifying and modeling the sources of errors. We analyze samples of increasing complexity, from singly labeled peptide samples to peptides labeled at up to 3 positions, both individually and in simple peptide mixtures, and distinguish specific phosphoserine post-translational modifications.
Results
Instrumentation for single molecule fluorescent peptide imaging and Edman sequencing
Edman sequencing employs harsher reagents than conventional aqueous microscopy experiments—including strong organic acids, bases, solvents, and heat—so we first identified fluorescent dyes that survive the chemistry (Supplementary Fig. 1), adapted a microscope stage perfusion chamber with chemically-resistant tubing, connectors, and perfluoroelastomer gaskets (Supplementary Fig. 2B), and automated chemical manipulations within the chamber using computer-controlled pumps and valves to exchange reagents under nitrogen (Supplementary Fig. 2C). Tests of bulk fluorescent peptides on beads confirmed the dyes did not strongly affect Edman degradation (Supplementary Fig. 3). We next confirmed that fluorescent peptides could be covalently tethered via aminosilane to a glass cover slip and survive extended imaging (Supplementary Fig. 4), exposure to Edman solvents, and heat, without significant loss of fluorescence (Supplementary Fig. 5). Fig. 1B shows ~3 million peptides in an approx. 1.3mm × 5mm area of cover slip; covalently tethered gold nanowires additionally provide unique constellations of fiducial markers in each field of view. Thus, even reasonably sparse peptide densities allow millions of individual peptide molecules to be imaged in the apparatus, and the immobilized peptides and dyes survive the necessary reagents.
Identifying positions of single labels within peptide molecules
To demonstrate that consecutive cycles of Edman chemistry can be performed on peptides with high efficiency in the apparatus, we considered a series of experiments with control peptides of increasing sample and label complexity. In order to interpret these experiments, we developed custom image processing algorithms (Supplementary Figs. 6, 7). These (1) identify individual fluorescent molecules within each micrograph, (2) align fluorescent peaks from the same field of view, imaged across consecutive Edman cycles, using fiducial markers to correct for microscope stage variation, then (3) identify peptides whose fluorescent signals were stable and successfully removed by the final Edman cycle, computationally flagging contaminating fluorescent objects and non-sequenced peptides.
We first compared a uniform population of copies of the peptide GK†AGAG († indicates the fluorophore Atto647N covalently coupled via NHS ester to the lysine side chain) to a second uniform population of that peptide blocked from sequencing by N-terminal acetylation, serving as a negative control (Fig. 2A). We performed several cycles of Edman chemistry with all reagents and incubation steps, but omitting the key reagent, phenylisothiocyanate (PITC). Dyes disappearing during these “mock” Edman cycles allowed us to estimate background dye loss rates at roughly 7% per cycle, from a combination of photobleaching (Supplementary Fig. 4), chemical destruction, and loss of non-covalently bound molecules. Subsequent Edman cycles incorporating PITC confirmed that peptides most frequently lost dyes at the expected second cycle, in contrast to blocked negative control peptides, demonstrating successful identification of dye position for 98,945 individual molecules (out of 238,503 molecules analyzed over 3 replicate experiments, imaging 100 image fields each), in a manner requiring a free peptide amino-terminus.
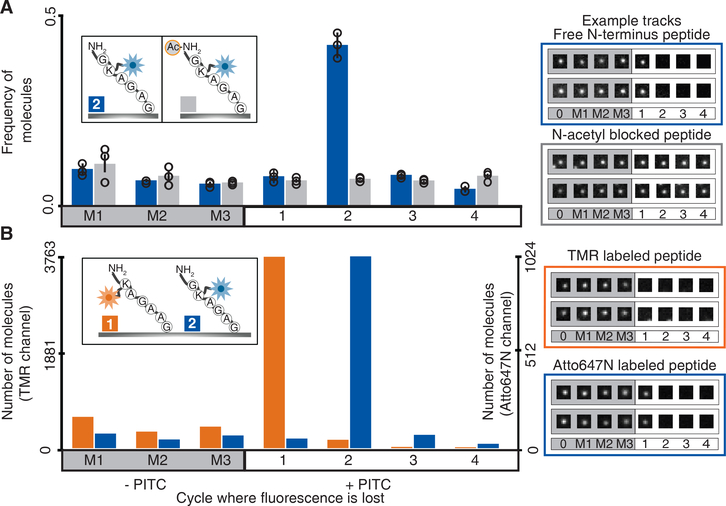
Figure 2: Fluorescent amino acid positions can be determined at single molecule sensitivity.
(A) Sequencing of individual peptide molecules requires a free amino terminus, as shown by comparing fluorescent sequences of the 6-mer GK†AGAG and its non-sequenceable N-terminally acetylated version. The histogram at left plots relative frequencies of peptide molecules exhibiting dye loss at each Edman cycle (mean +/− s.d. of 3 replicates; n=59434, 80541, 98528 molecules measured across 100 image fields each). M1, M2, and M3 denote negative control (“mock”) Edman cycles in which PITC was omitted. Individual traces are illustrated at right with extracted TIRF images for 4 individual molecules (2 blocked and 2 unblocked) across cycles. (B) The sequence positions of dye-labeled amino acids can be accurately determined within individual peptide molecules, shown by deconvoluting a mixture of two control peptides differing both in label position and color. The histogram displays counts of individual molecules of each color, K*AGAAG and GK†AGAG (n=5683 and n=1598, across 20 fields), indicating the cycle numbers at which the dyes are removed, and with example single molecule TIRF images at right. † indicates Atto647 conjugated to lysine, and * indicates TMR conjugated to lysine. Fluorescence intensity measurements are provided for all single molecule image tracks in Supplementary File 1.
We further confirmed the apparatus and chemistry by analyzing a control mixture of many copies of two peptides distinguishable both by fluorophore color and sequence position, with one set labeled by the red-emitting dye tetramethylrhodamine (TMR) at position 1, and the other set labeled by far red Atto647N at position 2. We determined the positions of dye loss for approx. 4,000 individual peptide molecules across 9 cycles of chemistry (3 mock Edman and 6 complete Edman cycles). The predominant patterns observed were PITC-dependent and matched the expected positions for each dye (Fig. 2B). Similar to Fig. 2A, we observed a low background rate of dye loss per cycle, consistent with non-specific, PITC-independent dye destruction. Because each fluorescent channel independently reports on a different dye, the sequence positions of multiple amino acid types on a single peptide can be determined by labeling each type with a different fluorophore, as in Supplementary Fig. 8. Overall, the efficiencies of Edman degradation, dye attachment, detection, and stability, as well as peptide surface attachment chemistry, all appear sufficiently robust to support fluorosequencing.
Determining the precise amino acid positions of dyes within multiply-labeled peptide molecules
Determining the positions of multiple dyes within one peptide requires accurately determining which Edman cycles elicit step-wise intensity decreases in that molecule’s fluorescence; each step corresponds to removing one or more dye molecules. We demonstrated this key requirement by determining the positions of two labeled cysteine amino acids within many identical copies of peptide GC♦AGC♦AGAG (♦indicates Atto647N coupled by iodoacetamide to cysteine). For each copy of GC♦AGC♦AGAG, we expected losses of the fluorescent cysteines after the 2nd and 5th Edman cycles (Fig. 3A).
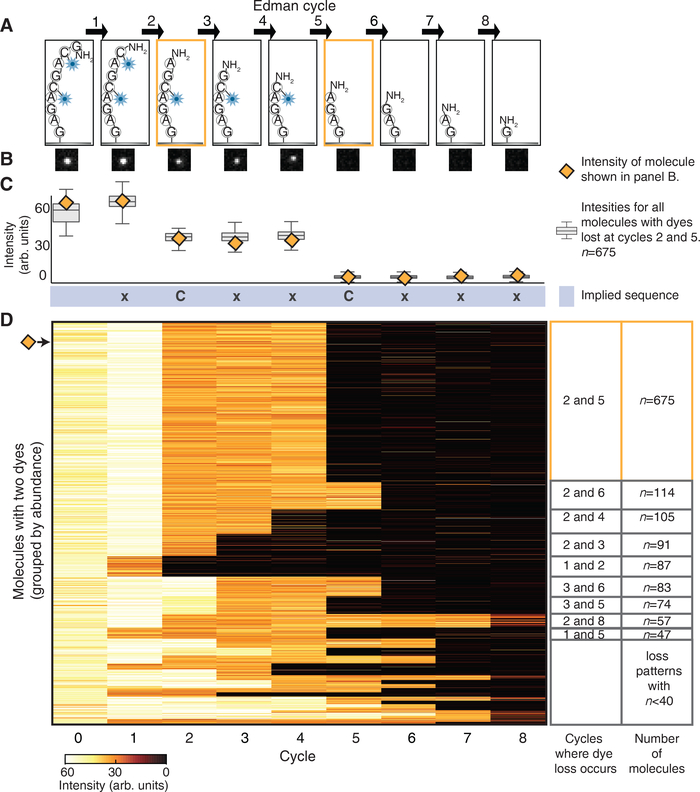
Figure 3: Step-wise decreases in fluorescent intensity occur at the Edman cycles that correspond to the removal of the dye-labeled amino acids.
(A) A schematic of the peptide molecule, GC♦AGC♦AGAG, losing dye labeled amino acids at the 2nd and 5th Edman degradation cycles. ♦indicates Atto647N conjugated to cysteine. (B) The decrease in fluorescence intensity accompanying dye loss is illustrated in a representative set of TIRF images for a single peptide molecule. (C) Intensities for the representative molecule shown in panel B (orange diamond) and a box plot of intensities (Center line, median; limits, 75% and 25%; whiskers, +/− 1.5 IQR) for all 675 molecules collected across all 49 images correctly identified as having drops at amino acid positions 2 and 5. By noting the Edman cycle corresponding to the step-wise intensity decrease, the partial sequence of the peptide (xCxxCxxx) can be inferred. (D) The heatmap of fluorescent intensity values for each of the 1,695 peptides with two dyes, observed after every Edman cycle, shows that the predominant pattern corresponds to dye losses after the 2nd and 5th cycles (n=675 peptide molecules).
Indeed, monitoring an individual peptide molecule (Fig. 3B) and measuring its fluorescence after every Edman cycle revealed clear step-wise decreases in its intensity after the 2nd and 5th cycles (Fig. 3C, orange diamonds). We collated such intensity patterns for all 1,695 individual double labeled peptide molecules (Fig. 3D) and observed that the largest proportion of the peptide tracks (675 molecules) had distinct intensity drops after the 2nd and 5th Edman cycles (Fig. 3C, box plots). Thus, by noting which Edman cycle elicited a step-wise intensity decrease for a peptide molecule, we could correctly localize the two cysteine-coupled dyes within each individual molecule, sufficient to infer the sequence xCxxCxxx (C=cysteine, x=any amino acid except C).
To better interpret data for other dye positions and counts, we empirically determined single peptide molecule fluorescence intensity distributions, then used these empirical distributions as the basis for a maximum likelihood statistical model for assigning the most probable dye positions to an observed peptide intensity track (Supplementary Figs. 9, Methods). We found it useful to summarize these sequence assignments across a population of molecules by representing them as a heatmap of counts of peptides with given dye positions. Such heatmaps allowed us to quickly determine the most prevalent sequences and to assess systematic errors. Fig. 4A (right panel) plots the histogram corresponding to the GC♦AGC♦AGAG experiment described above. Notably, 675 molecules (also presented in Fig. 3D) were correctly determined to have dyes at the expected 2 and 5 positions, corresponding to the peak of the doubly labeled sequences (illustrated schematically in the left panel).
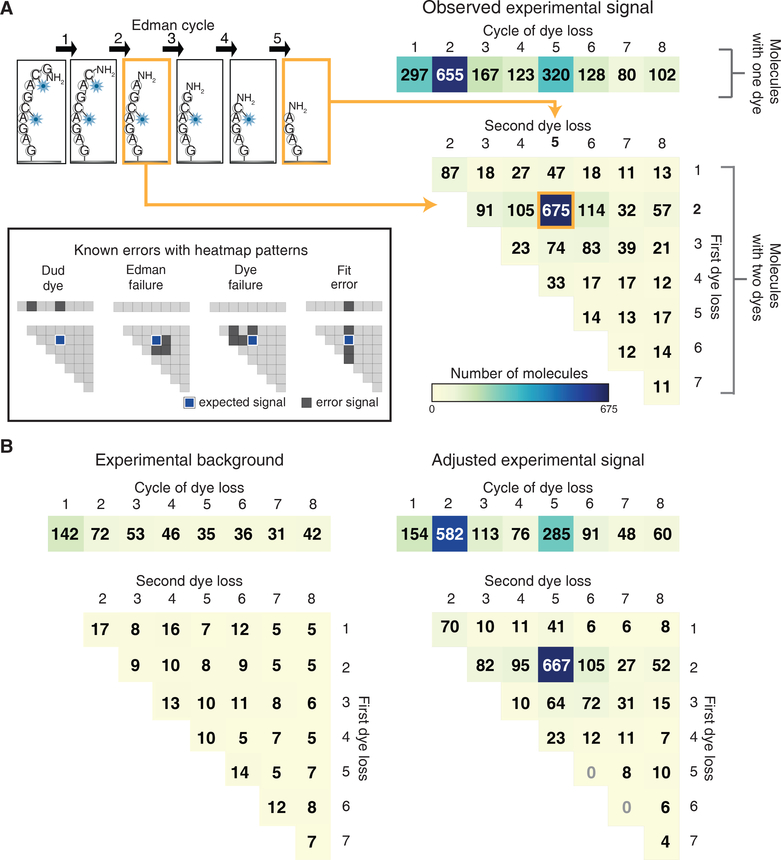
Figure 4: Fluorescent sequences can be interpreted computationally to identify dye positions and quantify errors.
A maximum likelihood statistical model allows for correct sequencing of multi-labeled peptides, evident in (A) histograms of the fit fluorescent sequences obtained for GC♦AGC♦AGAG (right panels, based on 49 image fields;♦indicates Atto647N conjugated to cysteine). We summarized dye loss positions for peptides with only one detectable dye as a 1D histogram (top right panel) and dye loss positions for doubly labeled peptides as a 2D histogram (bottom right histogram). As an aid for interpreting the 2D histogram, the example at top left shows a schematic of a peptide exhibiting dye losses at the 2nd and 5th cycles, which correspond to the 2nd row and 5th column of the 2D histogram. 675 peptide molecules exhibited this pattern. In this experiment, all other patterns correspond to specific sequencing errors, as illustrated graphically in the inset at left. (B) By sequencing an N-terminally acetylated population over 49 image fields of the same sequence, background observations expected from non-Edman events were determined (left panel) and the foreground counts adjusted to determine the signal above background (right panel, see Methods for calculation).
In parallel, to isolate and quantify specific sources of sequencing error, we tested N-terminally acetylated versions of the peptide. The resulting histogram (Fig. 4B, left panel), arising from background dye/molecule loss rates, established an empirical baseline for correcting observed sequence frequencies for losses by chance, and allowed us to calculate the signal relative to expected noise (Fig. 4B, right panel).
Characterizing and modeling errors
Although we observed the correct sequence in about 40% of the examples of Figs. 3 and 4, these were accompanied by certain previously expected systematic errors6. These errors arose from defective dyes or failed dye attachment (collectively referred to as “dud dyes”), molecule-by-molecule failures of Edman chemistry resulting in missed cleavage events, position-independent background rates of dye or molecule loss in each cycle (directly quantified by N-acetylated peptide controls), and assignment errors in computing dye positions from observed fluorescent sequences. Each type of error introduces a distinct bias into the sequencing histogram (Fig. 4A, inset), thus allowing us to estimate error rates by comparing our observed signal to that obtained from Monte Carlo simulations of sequencing with errors. Simulations of the GC♦AGC♦AGAG sequencing experiment (Supplementary Fig. 10) agreed well with observed sequences with low residuals (35%), confirming high rates of Edman cleavage (94% efficiency per cycle), 95% of dyes surviving per cycle, and molecular surface retention of 95%, with the largest error arising from dud dyes (7%). Because we chromatographically purified doubly-labeled peptides and verified their labels by mass spectrometry before analysis (Supplementary Fig. 11), this effect was attributable to correctly coupled dyes that failed to fluoresce. A survey of multiple dyes and manufacturer batches revealed this to be a feature of several commercial dyes, identifying a clear need for future improvement. Lastly, to confirm these experimental conditions allowed for high rates of Edman cleavage independent of amino acid composition, we studied peptides containing proline, characterized historically by lower Edman cleavage rates15. We observed only a modest reduction in cleavage efficiency to 91%, as compared to 95% for alanine and 97% for repetitive glycine/alanine residues (Supplementary Fig. 12).
Deconvolution of peptide mixtures into groups of individual molecules
We next performed experiments on peptides in simple mixtures and from naturally occurring proteins, and demonstrated identification of a protein from a database. In addition, since all acetylated control experiments exhibited similar sequencing patterns (because they simply lost dyes at background rates due to chemical destruction and other factors), we collected all acetylated experiments with each given dye count to obtain general background distributions that we could subtract from any experimental histogram to better estimate sequenced label positions (see Methods).
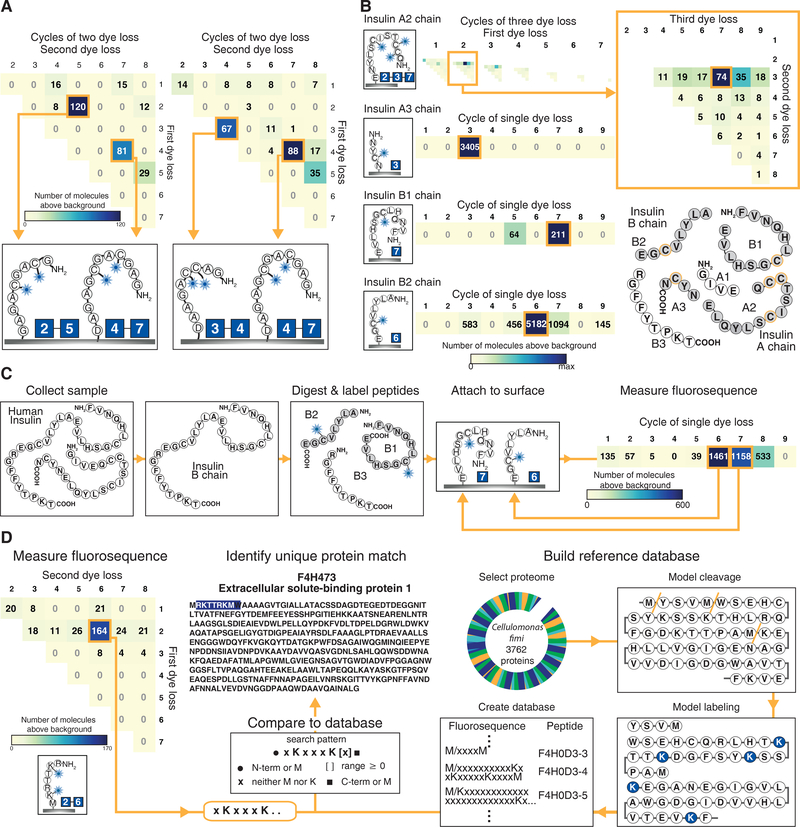
Fig. 5A and Supplementary Fig. 13A show zeptomole-scale experiments on two mixtures of peptide pairs, clearly distinguishing several hundred peptide molecules labeled on cysteines at the (2,5), (4,7), and (3,4) positions. To show identification of peptides derived from a natural human protein, we also synthesized peptides corresponding to GluC protease-digested insulin fragments fluorescently labeled on cysteine residues. The major peak in each histogram corresponded to the correct dye-labeled amino acid positions, thus correctly detecting the specific labeling patterns of two singly labeled insulin B chain fragments, one singly labeled - A chain fragment, and one triply labeled A chain fragment (Fig. 5B and Supplementary Fig. 14). Notably, fluorescent sequences of these four peptides, when considered together, are sufficient to uniquely identify insulin in the human proteome. We obtained equivalent results for biologically derived insulin, as shown in Fig 5C, using a peptide mixture obtained following GluC protease digestion of recombinant insulin B chain.
Figure 5: Fluorosequencing can discriminate individual peptide molecules in zeptomole-scale mixtures and uniquely identify their parent proteins.
(A) Histograms tallying counts of molecules sequenced from a mixture of GC♦AGC♦AGAG with GAGC♦GAC♦GAGAD (left panel, 98 image fields) and GAC♦C♦AGAAD with GAGC♦GAC♦GAGAD (right panel, 49 image fields) highlight the ability to distinguish individual peptides within mixtures. (B) Data on 4 individual insulin peptides that, in combination, uniquely identify insulin in the human proteome. (Top panel) adjusted three dye histogram for insulin A2 chain (QC♦C♦TSIC♦SLYNE) showing the expected signal at amino acid positions 2, 3, and 7 (magnified in inset). The remaining panels plot adjusted single dye histograms for insulin A3 (NYC♦N), B1 (FVNQHLC♦GSHLVE), and B2 chains (ALYLVC♦GE), respectively; each histogram represents 100 image fields. (C) Data on recombinant human insulin B chain after purification, GluC proteolysis and cysteine labeling shows the expected peaks at cycles 6 and 7 (100 image fields) as expected for the mixture of B2 and B1 peptides, respectively. (D) The fluorescent sequence of peptide RK†TTRK†M is sufficient to uniquely identify its parent protein F4H473 from the Cellulomonas fimi protein. The adjusted two dye sequencing histogram (left panel, 49 image fields) reveals the sequence as xKxxxK[x]≥0 which can be compared to a reference database (center panel) created by modeling fluorescent sequences for all possible peptides in the proteome assuming predefined protease cleavage and dye labeling specificities (right panel), here modeling cyanogen bromide cleavage after M and labeling K. ♦ indicates Atto647N conjugated to cysteine and † indicates Atto647N coupled to lysine residues. Supplementary Figs. 13–14 provide full single, double, and triple dye histograms, as appropriate.
Protein identification
To illustrate how a single experimentally determined partial sequence might be used to identify a parent protein from a reference proteome, we studied peptide RK†TTRK†M (†indicates Atto647N coupled to lysine residues) from the bacterium Cellulomonas fimi, modeling a scenario in which peptides are generated by cyanogen bromide proteolysis, which cleaves proteins after methionines, followed by fluorescent labeling of lysine residues (Fig. 5D. and Supplementary Fig. 13B). The observed partial sequence XKXXXK[X]≥0 (K=lysine, X=any amino acid except lysine or methionine), when constrained by knowledge of the proteolysis cleavage specificity (i.e., adjacent to a methionine or protein terminus), occurs only once in a database of all 3,762 proteins from the bacterium Cellulomonas fimi (strain ATCC 484), uniquely identifying the protein “Extracellular solute-binding protein family 1” (Uniprot database identifier F4H473_CELFA). Thus, even for simple labeling schemes, there exist peptides for which partial sequencing suffices to uniquely identify their parent protein from a reference proteome. In practice, the identification of proteins in a reference database will be limited by sequencing errors. A computational model that incorporates our experimentally determined error rates (Supplementary Fig. 15) suggests that the technique is currently sufficiently empowered to discriminate proteins in samples of tens to hundreds. Incorporating additional labels or information-rich constraints from proteolysis or attachment specificity should serve to increase the power of this approach.
Single molecule sequencing of serine phosphorylation sites
We demonstrated identification of the specific amino acid positions of phosphoserine residues at single-molecule sensitivity. We considered the peptide YSPTSPSK, found in high-copy tandem repeats within the C-terminal domain of RNA polymerase II and whose phosphorylation patterns on Ser2 and Ser5 are implicated in transcriptional regulation16. Fig 6A indicates the scheme used to selectively label serine or threonine phosphorylation sites, consisting of beta-elimination followed by conjugate addition via thiols17 in order to substitute thiol-linked fluorophores in place of phosphates. Analysis of the peptides YpS°PTSPSK and YSPTpS°PSK (° indicates Atto647N coupled at phosphoserine residues) clearly discriminated serine phosphorylation sites within 3 amino acids of each other at single molecule sensitivity (Fig. 6B).
Figure 6: Direct single molecule sequencing of phosphoserine positions within RNA polymerase II C-terminal domain (CTD) repeat peptides.
(A) Phosphorylated serines or threonines can be specifically labeled with fluorescent dyes by beta-elimination and conjugate addition17, then sequenced to determine the amino acid positions of the phosphorylated residues within each molecule, as demonstrated in (B) for CTD repeat peptides phosphorylated at either Serine 2 (top panel) or Serine 5 (bottom panel). Histograms in panels A and B each report observations from 49 image fields.
Discussion
Single-molecule protein sequencing combines aspects from DNA sequencing, mass spectrometry proteomics, and classic Edman sequencing, and as such, it is useful to compare it to these technologies to get a better sense of its likely scalability, limits of dynamic range, applications, and other properties. Broadly speaking, the approach shares upstream protein isolation and proteolysis with shotgun mass spectrometry, as well as computationally matching peptide sequence-dependent patterns (fluorescent sequences vs. spectra) to a reference proteome database, and combining evidence from peptide identifications into protein identifications. Thus, it should be able to take advantage of established protocols for these aspects. However, as the sensitivity of the approach is inherently single molecule, as opposed to the attomoles to femtomoles (106-109 molecules) typically analyzed by a conventional Orbitrap mass spectrometer3, 4, there are reasonable prospects for reducing sample volumes and protein abundance requirements. Provided that the challenge of fluorescently labeling low abundance proteins can be met, this could open up the potential for, e.g. single cell proteomics experiments18.
In other respects, the approach resembles DNA and RNA sequencing pipelines, whose basis is the acquisition of large numbers of (often short) reads in parallel. Parallels include the fact that the data are intrinsically amenable to digital quantification simply by counting reads and that longer reads tend to be more information-rich. In principle, the method will work for both peptides (short reads) and full proteins (long reads). Currently, the partial sequence information gained by knowing protease specificity and the observed dye destruction rates make application to peptides more practical. Parallel efforts are underway to develop long-read single-molecule protein sequencing based on nanopores7–10, 19–21
The error spectrum of the method strongly resembles that of nucleic acid sequencing, as it is characterized by indels (insertions/deletions) and substitutions, rather than the attribution errors that predominate in mass spectrometry due to isobaric amino acids or peptides. It also shares many of the same concerns as N-terminal Edman sequencing for optimization of PITC attachment and cleavage of PTH amino acids (Fig. 1C), requiring similar optimizations to temperature and reagent incubation time for efficient cleavage15. However, unlike Edman sequencers, it does not rely on detecting PTH amino acids and thus is not affected by many challenges to the traditional method, including inefficient extraction and detection of PTH molecules and amino acid modification effecting PTH retention times13, 22. Also, while Edman suffers from lag and reductions to repetitive yield caused by loss of population synchrony, our approach differs in that a missed cleavage on one molecule has no effect on a different molecule, and cleavage efficiencies (91–97%) are simply modeled into database lookup probabilities.
Although we do not evaluate quantification of peptides here, the intrinsically digital nature of the data offers both advantages and disadvantages over mass spectrometry. Unlike in mass spectrometry, in which assay dynamic ranges are largely set by mass detector dynamic ranges of 103-104 (as for Orbitrap detectors3, 4) or counts of mass spectra collected (typically no more than 105), dynamic ranges for single-molecule protein identification should scale in a manner similar to imaging-based nucleic acid sequencing methods, set fundamentally by the surface area of the flow cell, density of attached molecules, and imaging times. Current generation Illumina sequencers routinely collect hundreds of millions of reads per run, and one previously developed single molecule DNA sequencing instrument using a similar TIRF microscope setup to those in this study has reported scaling to > 1 billion molecules sequenced23. In principle, single-molecule protein sequencing should scale similarly, offering multiple order of magnitude increases in dynamic range over current generation proteomics platforms. However, a potential confounding issue distinct from DNA and RNA sequencing is the substantially larger dynamic range exhibited by some natural proteomes, e.g. plasma protein concentrations can vary over 12 orders of magnitude24. In such cases, approaches will be needed to simplify the samples, such as affinity-based subtraction of highly abundant proteins or biochemical fractionation prior to sequencing. Finally, directions for future development include methods for multiplexing samples, low-abundance protein/peptide preparation, and expanding the palettes and stabilities of dyes and labelable amino acids or their modifications.
Methods
All methods are described in full in the Online Methods section.
Online Methods
Fluorophore selection
We observed that many commonly available fluorophores underwent significant spectral shifts (>100 nm) or irreversible fluorescence loss following exposure to the Edman reagents, primarily the trifluoroacetic acid (TFA) and phenylisothiocyanate (PITC) / pyridine mixture. We screened 26 fluorophores (Supplementary Table 1) in order to identify those most resistant to the Edman solvents by covalently attaching the dyes to Tentagel beads (Chem-Impex International Inc; Cat # 04773) and measuring their fluorescence following 24 incubation with TFA or pyridine/PITC in 9:1 at 40 °C (Supplementary Fig. 1). Non-specifically bound fluorophores were removed by repeated washing with dimethylformamide (DMF), dichloromethane (DCM), and methanol. Atto647N, Alexa555, and rhodamine variants including tetramethylrhodamine (TMR) showed minimal (< 5%) change in fluorescence and had quantum yields high enough for effective sequencing. We used Atto647N (quantum yield = 0.65) and rhodamine variants, including the improved TMR analog JF594 (quantum yield = 0.88), for all subsequent experiments.
Widefield microscopy for bead-based assays
Beads labeled with fluorescent dyes or peptides were suspended in 20 μL of phosphate-buffered saline (PBS, pH 7.2) and added to a glass cover slip. The samples were imaged using a Apo 60×/NA 0.95 objective mounted on an Eclipse TE2000-E inverted microscope (Nikon) equipped with a Cascade II 512 camera (Photometrics), Lambda LS Xenon light source and Lambda 10–3 filter wheel control (Sutter Instrument), and a motorized stage (Prior Scientific), all operated via Nikon NIS Elements Imaging Software. Images were acquired at 1 frame per second through a 89000ET filter set (Chroma Technology) with channels “DAPI” (Ex 350/50, Em 455/50), “FITC” (Ex 490/20, Em 525/36 ) “TRITC” (Ex 555/25, Em 605/52) “Cy5” (Ex 645/30 Em 705/72), and bead fluorescence quantified from the images.
Peptide synthesis, purification, and labeling
All peptides were synthesized using a standard automated solid-phase peptide synthesizer (Liberty Blue microwave peptide synthesizer; CEM Corporation) and purified by analytical high-performance liquid chromatography (HPLC) (Shimadzu Inc.) with an Agilent Zorbax column (4.6×250 mm) operating at 10 mL/min flow rate and eluting with a gradient of 5–95% acetonitrile (0.1% TFA) over 90 minutes. Solvents used were HPLC grade. Peptides were labeled with flourophores using standard coupling schemes14 by reaction with Atto647N-NHS, Atto647N-iodoacetamide, TMR-NHS, or JF549-NHS, as appropriate, to label lysines (via NHS) or cysteines (via iodoacetamide) (Supplementary Table 1). Purities, including presence and count of fluorescent labels, were confirmed by mass spectrometry (6530 Accurate Mass QTofLC/MS, Agilent Technologies). N-terminal amines of synthetic peptides were typically blocked with a tert-butyloxycarbonyl (boc) or a fluorenylmethyloxycarbonyl (fmoc) protecting group prior to immobilizing peptides, preventing peptide concatenation of the activated C-termini with free peptide amino termini.
Labeling phosphoserines
Phosphorylated serines were fluorescently labeled (Fig. 6) by mixing solubilized phosphopeptide with a saturated solution of barium hydroxide and sodium hydroxide for 3 hours at room temperature for beta-elimination of the phosphate17. Atto647N-NHS was reacted with cystamine to produce Atto647N-S-S-Atto647N, which was subsequently incubated overnight with the peptide solution and Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in DMF in order to fluorescently label the relevant serines. Peptides were purified by HPLC and labeling verified by mass spectrometry. Note that this chemistry is known to additionally label phosphothreonines17 and also has the potential to eliminate O-glycans or to eliminate water from hydroxy amino acids25.
Fluidics
We adapted an FCS2 temperature-controlled perfusion chamber (Bioptechs), substituting the gaskets with custom gaskets die-cut from 0.05 mm thick Kalrez-0040 rubber (Dupont), based upon its compressibility and inertness to the Edman reagents (Supplementary Fig. 2B). We used a USB-controlled piston syringe (Cavro) and 10-port valve (Valco) to dispense reagents through polytetrafluoroethylene tubing into the perfusion chamber, which was affixed on the microscope stage (Supplementary Fig. 2C).
Tentagel bead-based confirmation of Edman sequencing through fluorescent amino acids
As prior literature was unclear as to the applicability of Edman chemistry to fluorescent dye-modified amino acid residues, we used the bead-based assay to test if Edman sequencing could be observed in bulk studies of fluorescent peptides. Synthetic peptides with known positions of TMR labeled lysine residues were covalently coupled to Tentagel beads via EDC/NHS chemistry (described below). We measured the reduction in peripheral bead fluorescence (attributable to covalent binding) after consecutive Edman cycles adapting established protocols26, observing ~80% efficiency per amino acid residue without optimization, thus confirming the general capacity of the Edman degradation chemistry to sequence peptides with bulky and hydrophobic fluorophore-tagged residues (Supplementary Fig. 3). We did not attempt to further optimize Edman chemistry on bulk peptides or beads.
Reducing photobleaching
We took advantage of the perfusion chamber’s compatibility with diverse solvents to optimize the solution conditions for single molecule imaging, testing imaging quality (dye brightness and half-life) in a range of organic and aqueous solvents. We observed optimal performance from methanol with 1 mM trolox (Sigma; Cat # 238813–1G), purged 30 min with nitrogen gas. The methanol/trolox imaging solution increased the half-life of the TMR and Atto647N fluorophores to 105 and 110 seconds, respectively, corresponding to >100 Edman cycles, assuming a 1 sec exposure per cycle (Supplementary Fig. 4).
Peptide surface immobilization
For single molecule Edman sequencing, a #1 (1.7mm) glass cover slip surface was first cleaned by UV/ozone (Jelight Company) and functionalized by amino-silanization with aminopropyltriethoxysilane (APTES) (Gelest, Cat # SIA0610.1) using the vendor-supplied protocol (http://www.gelest.com/wp-content/uploads/09Apply.pdf). Slide surfaces were further passivated (for experiments in Figs. 3 - 6 and Supplementary Figs. 10, 13 and 14) by overnight incubation with polyethylene glycol (PEG)-NHS solution, prepared by dissolving a mixture of 80 mg mPEG-SVA and 4 mg tboc-PEG-SVA (Laysan Bio Inc; Cat # MPEG-SVA-2000 and Cat # tBOC-NH-PEG-SVA-5K, respectively) in sodium bicarbonate solution (pH 8.2). Functionalized slides were stored in a vacuum desiccator until use. The t-butyloxycarbonyl protecting groups were removed by incubating a slide with 90% TFA (v/v in water) for 5 h before use, exposing free amine groups for peptide immobilization. Additionally, to aid in surface passivation, PEG sides were optionally treated with a 2% solution of Tween 20 (Biorad; Cat #170–6531) in TRIS for 30 min (as for experiments in Fig 5C). In control experiments, we confirmed that an amino-silanized glass surface was stable to multiple cycles of Edman degradation and after washes with wash buffer (1% sodium dodecyl sulfate (SDS) and 0.1% Triton in PBS), determined by assaying the retention of N-hydroxysuccinimide (NHS)-derivatized Atto647N covalently attached to free amines on the surface (Supplementary Fig. 5).
For a typical single-molecule peptide sequencing experiment, peptides were covalently coupled to the cover slip surface via amide bonds between the carboxylic acid of the C-terminal amino acid residue and the glass surface amines. Fresh solutions of 4 mM of 1-ethyl-3-(3-dimethylamino) propyl carbodiimide, hydrochloride (EDC, Sigma; Cat # 03449–1G) and 10 mM of N-hydroxysuccinimide (NHS, Sigma; Cat # 130672–5G) or N-hydroxysulfosuccinimide (NHS, Thermo; Cat # PG82071) was made in 0.1 M MES buffer in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES, Pierce; Cat # 28390) just before use (notably, use of fresh EDC was critical). A solution of fluorescently labeled peptide (typically 200 μM) was diluted with EDC-NHS solution (a 1:1 mixture by volume) to a final concentration of 20 μM peptide, 1.6 mM EDC, and 4 mM NHS. This was mixed for 4 h at room temperature before preparing an initial dilution series in 0.1 M MES. We titrated peptides from a secondary dilution series to between 20 pM and 2 nM peptide in 0.1M M NaHCO3 to provide an attachment density on the slide of approximately 10 molecules per square nanometer (Fig. 1B). Peptides were typically incubated on the slide for 20 mins before washing with water and methanol to remove unbound peptide. Additionally, 1 μm long 12mercaptododecanoic acid NHS ester functionalized gold nanorods (Nanopartz; Cat # B14–1000-12CNHS-0.25-DMF) were covalently attached via the amines to serve as fiducial markers for focusing and image registration. After attaching peptides and nanorods, the slide was incubated in 90% TFA (v/v in water) for 5 h and then rinsed with methanol to remove boc groups and expose the peptides’ free amino termini. Alternatively, to remove fmoc protecting groups, peptides were incubated for 1 h in 20% piperidine solution (in dimethylformamide (DMF)), then washed with DMF and methanol to remove residual piperidine. An optional 1 hour incubation of 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU, Sigma; Cat# 33682) was used to remove peptides non-specifically bound to the surface experiments in Figs. 5C, 6, and Supplementary Fig. 8. To assist with focus stability, the chamber and microscope were allowed to equilibrate at 40 °C during de-blocking and up to an additional 12 h.
Total internal reflection (TIRF) microscopy
Single molecule TIRF microscopy experiments were performed with two similar systems, each with a Nikon Ti-E inverted microscope equipped with CFI Apo 60X/1.49NA oil immersion objective lens, motorized stage with 100 nm resolution linear encoder (ProScan II; Prior Scientific), an iXon3 DU-897E 512×512 EMCCD detector (Andor) operated at −70 °C, and a MLC400B (Keysight) laser combiner with 561 nm (N1245AL34) and 647 nm (N1245AL44 and N1245BL56, systems A and B respectively) lasers as diagrammed in Supplementary Fig. 2C. Fluorescence from Atto647N was excited using 6.0 mW (50%, system A) or 2.8 mW (12.5%, system B) of 647 nm laser power via 647LP dichroic and collected through 665LP and 705/72BP emission filters. Fluorescence for tetramethylrhodamine (TMR) was excited using 2.7 mW (35%) of 561 nm laser power via 561LP dichroic and collected through 575LP and 600/50BP emission filters. Gold nanorod reflection was excited by using <0.01 mW (3%) of 561 nm laser light using a 95/5 reflectance cube. To increase the number of pixels in an individual diffraction limited spot and to maximize the flat-field portion of the image collected, an additional 1.5X tube lens was inserted into the beam path. Laser powers were measured prior to the objective. All data presented in figures, except those in Fig. 5B and Supplementary Figs. 8, 12, and 14, were collected using system A. All peptide sequencing results were independently confirmed on both systems.
Automated Edman degradation
For single-molecule Edman sequencing experiments, the sample temperature was maintained at 40 °C by heating both the perfusion chamber and microscope objective. Edman reagents were bubbled with dry nitrogen gas for 10 min, and then kept under nitrogen gas throughout the experiment. Solvent exchanges in the fluidic device were controlled using in-house Python scripts and coordinated with image acquisition via custom macros in the Nikon Elements software package. Reagents (highest purity available from Sigma) were introduced to the perfusion chamber as follows:
| Protocol step | Reagents | Incubation time (min) System A | Incubation time (min) System B | |
|---|---|---|---|---|
| Step 1 | Pump wash | Water | wash | wash |
| Step 2 | Methanol wash | Methanol | wash | wash |
| Step 3 | Free basing solution | 1. Acetonitrile, pyridine, triethylamine, water (10:3:2:1 v/v) 2. Acetonitrile, triethylamine (2:1 v/v) |
wash | 5 |
| Step 4 | Mock or Edman solution | 100% acetonitrile or acetonitrile, phenylisothiocyanate (9:1 v/v) | 30 | 20 |
| Step 5 | Free basing solution | 1. Acetonitrile, pyridine, triethylamine, water (10:3:2:1 v/v) 2. Acetonitrile, triethylamine (2:1 v/v) |
wash | NA |
| Step 6 | Ethylacetate/ACN wash | Ethylacetate (A) or acetonitrile (B) | wash | wash |
| Step 7 | Cleavage solution | 100% trifluoroacetic acid | 30 | 15 |
| Step 8 | Ethylacetate wash | Ethylacetate | wash | wash |
| Step 9 | Pump wash | Water | wash | wash |
| Step 10 | Methanol wash | Methanol | wash | wash |
| Step 11 | Oxygen scavenging solution | Trolox in methanol (1mM) | 10 | 5.5 |
“Wash” denotes exchanging the solvents in the flow chamber (approx. 3 minutes). On system A, free base solution 1 was used for experiments in Figs. 1–4, 5A and 5D and Supplementary Figs. 10 and 13, and free base solution 2 for experiments in Figs. 5C and 6. On system B, free base solution 1 was used for Fig. 5B and Supplementary Fig. 12B and 14; free base solution 2 was used for Supplementary Figs. 8 and 12A. Typically, to distinguish signal loss due specifically to Edman chemistry, as many as four mock Edman cycles using all reagents except PITC were performed prior to Edman cycles. In total, steps 1–11 take approx. 1 to 1.5 hours.
Image processing and photometry
Images of each field of view taken after each consecutive Edman cycle were stored as PNG files, with sets of images from each Edman cycle (henceforth, “frames”) sequentially collated into fields of view by filename.
To identify individual peptide molecules in frames, we applied a median filter to locate candidate fluorescent point-sources in images (Supplementary Fig. 6). Candidate point-sources were then fit with a two-dimensional Gaussian as an approximation to their Airy disc, as implemented in AGPY (authored by Adam Ginsburg; downloaded April 7th 2015 from https://github.com/keflavich), and an R2 quality of fit was assessed, retaining point-sources with R2 > 0.7. Further criteria were applied as described below to remove potential contaminants from analysis.
To track individual fluorescent point-sources through an experiment, each field of view’s frames were aligned pairwise across cycles using fast-Fourier transform cross-correlation27 (implemented in Python by scikit-image, http://scikit-image.org) of the gold nanorod reflection channel images, if present, or one of the fluorescence channels otherwise. We collated instances of each fluorescent point-source across aligned frames by matching their coordinates using the alignment offsets, within error tolerance. If a fluorescent point source was absent in one or more frames, its position was extrapolated to those frames using the alignment offsets. Point sources that mapped outside of a frame in any imaging cycle were discarded.
We quantified the fluorescence intensity of each point-source across frames using Mexican hat photometry. In each frame, we summed the innermost 7×7 pixels centered about the point-source to obtain its raw photometry, then subtracted the median of the enclosing 19×19 pixel area (excluding the 7×7 center) to adjust for background. Any point-source whose Mexican hat was not contained entirely in all frames was discarded.
For each point-source’s progression through frames, we constructed a Boolean logic sequence consisting of two possible states: “ON” and “OFF” (Supplementary Fig. 7). A point-source was considered ON in the frames in which a two-dimensional Gaussian fit with R2 above 0.7 was found, OFF otherwise. For example, a point-source that was well-fitted with a two-dimensional Gaussian in frames 1–3, was not detected in frames 4–6, and was again fittable in frames 7–10 would be assigned a sequence [ON, ON, ON, OFF, OFF, OFF, ON, ON, ON, ON]. Only point sources that turned off monotonically were considered validly sequenced peptides: i.e. they started in the ON state, and if they turned OFF in any frame, they then remained OFF for the rest of the experiment. Fluorescent point-sources that turned ON after being OFF at any point were discarded from further consideration. For each point source, the sequence of its Boolean states and its Mexican hat photometries was collated. This collated sequence is termed the point source’s track.
Before further analysis, dye photometries were adjusted to account for frame-to-frame focus variations. Tracks with ON state across all frames (“remainders”) were collated for each field. The percent deviation in fluorescence intensity was determined at each cycle for each remainder track. The average remainder deviation for each cycle was then applied to all tracks within that field. Fields with fewer than 5 remainders were removed from further analysis.
Overview of maximum likelihood assignment of dye positions
For each peptide track, we sought to infer the number of dyes remaining on the peptide in each sequencing frame. For example, a track’s dye count might be written as [3, 3, 2, 2, 2, 1, 0], representing a peptide that started with three dyes, decreased to two dyes after two Edman cycles, then decreased to one dye after another three cycles, and finally to zero dyes after one more cycle.
Dye counts are not directly observable, but rather must be inferred from the measured photometries and their step-wise intensity losses28, 29. Our general strategy to infer a sequence of dye counts di from a sequence of photometries φi across frames 1, 2, …, i was as follows:
A peptide had a dye count of 0 in a frame if and only if it was in the OFF state as defined above.
We considered all possible monotonically decreasing dye count sequences as competing explanations for the observed sequence of photometries. We considered a maximum of 5 dyes, allowing multiple simultaneous dye losses per cycle.
The probability for the observed intensities to be generated from each dye count sequence was calculated as a quality of fit score S(d|φ) = ∏iS(di|φi), with i indexing the track’s frames. The per-frame scoring function S(di|φi) is the probability density function ρ(φi|di) of a point source with di dyes yielding photometry φi. This probability density function is lognormal, as described below. The dye count sequence d maximizing this score was taken as the best explanation for observed photometries φ.
To guard against poorly behaved fluorescent point sources, if the best fitting dye count sequence d had any frame for which S(di|φi) was below a threshold (defined below), we considered the track uninterpretable and discarded it from further consideration.
Single molecule dye fluorescence intensities are log-normally distributed
Tracks from each experiment represented a population of fluorescent point sources that could be characterized in bulk. Here and in subsequent analysis, the distribution of photometries was binned using the optimal histogram binning algorithm from Shimazaki et al.30.
We first characterized the intensities of peptides with only one dye remaining. Since each track contains a sequence of ON/OFF states, we can assume that the last ON state of each track before an OFF is, for the majority of cases, caused by loss of a single dye regardless of how many dyes the peptide began with. This assumption is valid on a population basis because the probability of two or more dyes turning off in a single cycle is small compared to that of one dye. We defined φfinal as the set of photometries of the last ON frames that are followed by an OFF frame across all tracks. To maximize ON/OFF transitions caused by a single dye loss and not whole molecule loss, tracks with OFF transitions in the first three frames (typically “mock” cycles) were excluded from this definition. We found the distribution of φfinal to be lognormal, matching observations by Mutch et al.31 (Supplementary Fig. 9A). A lognormal distribution for one fluorophore can be written as probability density function ρ of intensity φ:
where the scale parameter μ and shape parameter σ completely characterize the distribution.
For simplicity, we henceforth considered the logarithmic space φ* = ln φ, with the corresponding transformed probability density function and parameters:
Following Mutch et al., the lognormal distribution can be expanded to cases of multiple (c) dyes by increasing the scale parameter μ* to μ* + ln c − Qc, where Qc is a dye-dye interaction factor; the shape parameter σ* is held constant, as per 31. Thus, the probability density function for a point source with multiple dyes can be written as:
In this context, step #4 of the general fitting strategy (thresholding) was based on the deviations of a track’s observed photometries from the lognormal model, . If this deviation was above three in any frame, the track was discarded.
Inference of lognormal fluorescence parameters via simulation
Parameter μ* in ρ*(φ*|c) can be obtained directly by setting . Parameters σ* and Qc are more challenging to extract by applying a straightforward function to datapoints . Instead, we used forward simulation to find a combination of parameters under which our fitted model best matched our data. Specifically, we started with fluorosequencing data from a doubly-labeled peptide GC♦AGC♦AGAG ( ♦ indicates Atto647N conjugated to cysteine) experiment and calculated its μ*. We then computationally generated each possible monotonically decreasing dye count that dropped to 0 within the experiment’s number of cycles. Iterating over 225 parameter combinations of σ* and Qc, we generated 105 tracks for each of the possible dye counts as follows: the intensity of each frame in a track was randomly drawn from the distribution ρ*(φ*|c), with c determined by the corresponding dye count di in that frame. We applied the general fitting strategy to both these simulated tracks and experimental tracks using σ* and Qc. To gauge whether a particular pair of parameters σ* and Qc recapitulated the distribution of photometries well, we collated the dye sequences fitted to the experimental data with their simulated counterparts, and compared the distribution of photometries in each frame. Supplementary Fig. 9B shows the distribution of photometries in each frame for dye sequence [2, 2, 2, 2, 2, 1, 1, 1, 0, 0, 0, 0] (mocks included; frames with 0 dyes are OFF and are omitted) under an overestimated shape parameter σ* and an underestimated dye-dye interaction factor Q2. Supplementary Fig. 9C shows the corresponding distributions for σ* = 0.20 and Q2 = 0.30, which fit with an average R2 of 0.87 across all eight frames. Repeating this parameter sweep for multiple experiments showed these values for σ* and Qc to be generally valid for Atto647N, and we used them for all subsequent analyses.
Using these parameters, for any given track φ we could thus infer the underlying dye count sequence d by maximizing the fit score
Estimation of sequencing errors via Monte Carlo simulations
Peptide fluorescent sequences are subject to experimental error. We took advantage of our previously developed computational model of the likeliest sources of experimental error (Edman failure, photobleaching, and dud dyes)6, for which we had developed an extensible Monte Carlo model of fluorosequencing. We added to our previous model an additional source of error – whole molecule loss – to reflect our observations that the reagent flushes through the perfusion chamber could remove labeled but non-specifically bound peptides from the slide surface, especially during the first few experimental cycles. Using this error model, we could simulate the molecular state of a peptide after each experimental cycle, providing a simulated dye count sequence for a given peptide as it undergoes sequencing. By chaining together this existing framework with our lognormal model of fluorescence, we could thus simulate the complete experimental observations (tracks) that we would expect for any given peptide sequence. Applying our dye count inference fitter to the simulated data, we could thus obtain a set of modeled fluorescent sequences for comparison to an actual experiment.
The primary sources of error are modeled as follows (c.f. 6 for more detailed discussion):
Edman failure is modeled as a Bernoulli variable. The probability of an amino acid being removed after every Edman cycle is p, and is independent of all other events.
Photobleaching is modeled as an exponential decay. The probability of a dye photobleaching after any experimental cycle is e−b.
The probability of a dye being a non-fluorescing dud before the experiment begins is a Bernoulli variable, with dud probability u.
Whole molecule loss is modeled as a bimodal Bernoulli variable, with probability dinitial of a whole molecule being removed after every cycle during the initial c cycles, and probability dsubsequent per cycle thereafter. Following experimental observations, dinitial ≥ dsubsequent.
We were able to recapitulate experiments with simulations (e.g. Supplementary Fig. 10), and found that the error parameters were broadly conserved across multiple experiments.
Adjustment of sequencing histograms for expected background rates of dye destruction and whole molecule loss
We compiled data across multiple acetylated peptide sequencing experiments to establish a background rate of non-specific dye destruction and whole molecule loss, and adjusted sequencing histograms (where indicated) to account for this background. Importantly, acetylated control experiments exhibited fluorescent sequencing patterns in a sequence-independent manner, depending only on the per-molecule dye count, and hence we could pool all acetylated experiments with a given dye count to obtain a general background distribution, which could be used to adjust histograms from a sequencing experiment for observations expected by chance.
First, we standardized each acetylated control experiment by converting the counts at each histogram position to relative frequencies, by dividing each count by the total number of observations in the experiment. We considered all step drop patterns that dropped to a dye count of 0 by the end of the experiment, including those that had a total of four or five drops; step drop patterns that remained above 0 in the last frame (i.e. remainders) were omitted. Multiple standardized acetylated experiments were then averaged together on a per-histogram position basis to obtain the average background rate – i.e. the normalized count of each step drop pattern expected by chance due to non-sequencing-related experimental losses. Likewise, we obtained the variance in the background rate on a per-histogram position basis. We assumed the background rate at each histogram position to follow a normal distribution, defined by the average and variance obtained from multiple acetylated control experiments.
We then adjusted sequencing experiment histograms for expected background using the following iterative algorithm:
Standardize the sequencing histogram as for individual acetylated histograms above.
For each position in the standardized sequencing histogram with standardized frequency S, compute its z-score against the background distribution’s mean μ and standard deviation .
We define a smoothing operation for sequencing histogram position H = (i, j, k, …) as replacing its raw counts with the average of counts at all positions within a Hamming distance of 1. For example, smoothing at position 6 would entail averaging counts at positions 5 and 7, and smoothing at position (3, 4) would entail averaging counts at all eight positions satisfying (3±1, 4±1). Note that after a smoothing operation, a sequencing histogram must be re-standardized using the updated counts in order to compute its z-scores.
- Score all peaks in the sequencing histogram for the largest decrease in Z-score that would result from background correction, using smoothing from adjacent histogram positions to compensate for outliers, calculated as follows:
where: SH, μH, and σH are the histogram value, background mean, and background standard deviation at position H, and SH′ is the corresponding value for the histogram smoothed at position H. Update the histogram by smoothing at the position yielding the best improvement in step 4.
Repeat from step 1 until the highest z-score in step 2 is below a specified threshold (e.g., z = 1), or no further interpolation can be made that lowers a z-score.
This procedure generates a smoothed estimate of the expected background counts for a given sequencing experiment; we simply subtracted these counts from raw foreground sequencing counts to obtain the adjusted foreground counts, setting any negative entries to 0. Note that the z-score threshold applied in step 6 effectively considers any peaks whose z-score is below it as background, and thus removes them from the final results.
Effect of experimental errors on protein identification
To assess the potential impact of our observed experimental error rates on protein identification, we re-simulated the cellular compartments considered under ideal conditions in Fig. 1C with error rates, calculated using the Monte Carlo simulation algorithm as described above and in ref. 6. We considered the case for rates measured for the experimental samples in Figs. 3, 4, and Supplementary Fig. 10 of 94% Edman efficiency, 5% dye destruction, 5% surface degradation, and 7% “dud” dyes. For each set of proteins, we simulated 10,000 copies of each of protein in a Monte Carlo fashion for 30 Edman cycles and tabulated their resulting fluorescent sequences. We defined a protein as being uniquely identified if it yielded a fluorescent sequence at least 10 times (out of 10,000) for which no more than 10% of counts of that fluorescent sequence were emitted by other proteins. The resulting protein coverage curves are plotted in Supplementary Fig. 15.
Statistics and reproducibility
Replicate data is summarized for all figures in Supplementary File 1 and the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgements
We thank Brian Cannon and Rick Russell for early assistance with single molecule imaging, Michelle Gadush for assistance with peptide synthesis, Ian Riddington, Jordan Dinser, and Kristin Suhr for assistance in mass spectrometry analysis of fluorescently labeled peptides, Zack Simpson and Jim Rybarski for assistance with image analysis, Andrew Ellington for many fruitful discussions, and the Texas Advanced Computing Center for high performance computing. This work was supported by fellowships from the HHMI (to J.S.) and NSF (DGE-1610403, to A.A.B.), and grants from DARPA (N66001–14-2–4051, to E.V.A. and E.M.M.), NIH (DP1 GM106408, R01 GM076536, R35 GM122480; to E.M.M.), CPRIT (to E.M.M.), and the Welch foundation (F-1515, to E.M.M., and F-0046, to E.V.A.).
Footnotes
Competing financial interest statement
The authors JS, AB, EMM and EVA are cofounders and shareholders of Erisyon Inc. JS, EMM and EVA are coinventors on granted patent PCT/US2012/043769. JS, AAB, ETH, JLB, AMJ, EVA and EMM are coinventors on pending patent PCT/US2015/050099.
Availability of data and code
Computer code is freely downloadable from GitHub, including image processing algorithms and Monte Carlo simulations, at https://github.com/marcottelab/FluorosequencingImageAnalysis or in Supplementary File 2. All single molecule imaging datasets and an image processing tutorial are available via Zenodo (10.5281/zenodo.782860).
References
- 1.Liu Y, Beyer A & Aebersold R On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 2.da Costa JP, Santos PSM, Vitorino R, Rocha-Santos T & Duarte AC How low can you go? A current perspective on low-abundance proteomics. Trends in Analytical Chemistry 93, 171–182 (2017). [Google Scholar]
- 3.Makarov A et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 78, 2113–2120 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Makarov A, Denisov E, Lange O & Horning S Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J Am Soc Mass Spectrom 17, 977–982 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Hawkridge AM in Quantitative Proteomics (The Royal Society of Chemistry, 2014). [Google Scholar]
- 6.Swaminathan J, Boulgakov AA & Marcotte EM A theoretical justification for single molecule peptide sequencing. PLoS Comput Biol 11, e1004080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Y, Docter M, van Ginkel J, de Ridder D & Joo C Single-molecule protein sequencing through fingerprinting: computational assessment. Phys Biol 12, 055003 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y et al. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat Nanotechnol 9, 466–473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson J, Sloman L, He Z & Aksimentiev A Graphene Nanopores for Protein Sequencing. Adv Funct Mater 26, 4830–4838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy E, Dong Z, Tennant C & Timp G Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat Nanotechnol 11, 968–976 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Sampath G Amino acid discrimination in a nanopore and the feasibility of sequencing peptides with a tandem cell and exopeptidase. RSC Advances 5, 30694–30700 (2015). [Google Scholar]
- 12.Kolmogorov M, Kennedy E, Dong Z, Timp G & Pevzner PA Single-molecule protein identification by sub-nanopore sensors. PLoS Comput Biol 13, e1005356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edman P Method for determination of the amino acid sequence in peptides. Acta Chem. Scand 4, 283–293 (1950). [Google Scholar]
- 14.Hernandez ET, Swaminathan J, Marcotte EM & Anslyn EV Solution-phase and solid-phase sequential, selective modification of side chains in KDYWEC and KDYWE as models for usage in single-molecule protein sequencing. New Journal of Chemistry 41, 462–469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermodson MA, Ericsson LH, Titani K, Neurath H & Walsh KA Application of sequenator analyses to the study of proteins. Biochemistry 11, 4493–4502 (1972). [DOI] [PubMed] [Google Scholar]
- 16.Phatnani HP & Greenleaf AL Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20, 2922–2936 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Stevens SM Jr. et al. Enhancement of phosphoprotein analysis using a fluorescent affinity tag and mass spectrometry. Rapid Commun Mass Spectrom 19, 2157–2162 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Shapiro E, Biezuner T & Linnarsson S Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14, 618–630 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Ohshiro T et al. Detection of post-translational modifications in single peptides using electron tunnelling currents. Nat Nanotechnol 9, 835–840 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Nivala J, Marks DB & Akeson M Unfoldase-mediated protein translocation through an alpha-hemolysin nanopore. Nat Biotechnol 31, 247–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen CB, Rodriguez-Larrea D & Bayley H Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat Biotechnol 32, 179–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wettenhall RE, Aebersold RH & Hood LE Solid-phase sequencing of 32P-labeled phosphopeptides at picomole and subpicomole levels. Methods Enzymol 201, 186–199 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Pushkarev D, Neff NF & Quake SR Single-molecule sequencing of an individual human genome. Nat Biotechnol 27, 847–850 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson NL & Anderson NG The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1, 845–867 (2002). [DOI] [PubMed] [Google Scholar]
Methods only references
- 25.McLachlin DT & Chait BT Improved beta-elimination-based affinity purification strategy for enrichment of phosphopeptides. Anal Chem 75, 6826–6836 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Laursen RA Solid-phase Edman degradation. An automatic peptide sequencer. Eur J Biochem 20, 89–102 (1971). [DOI] [PubMed] [Google Scholar]
- 27.Guizar-Sicairos M, Thurman S & Fienup J Efficient subpixel image registration algorithms. Opt Lett 33, 156–158 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Cannon B, Pan C, Chen L, Hadd AG & Russell R A dual-mode single-molecule fluorescence assay for the detection of expanded CGG repeats in Fragile X syndrome. Mol Biotechnol 53, 19–28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das SK, Darshi M, Cheley S, Wallace MI & Bayley H Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. Chembiochem 8, 994–999 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Shimazaki H & Shinomoto S A method for selecting the bin size of a time histogram. Neural Comput 19, 1503–1527 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Mutch SA et al. Deconvolving single-molecule intensity distributions for quantitative microscopy measurements. Biophys J 92, 2926–2943 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.