Abstract
Salinity stress represents one of the most harmful abiotic stresses for agricultural productivity. Tibetan hulless barley is an important economic crop widely grown in highly stressful conditions in the Qinghai-Tibet Plateau and is often challenged by salinity stress. To investigate the temporal metabolic responses to salinity stress in hulless barley, we performed a widely targeted metabolomic analysis of 72 leaf samples from two contrasting cultivars. We identified 642 compounds 57 % of which were affected by salt stress in the two cultivars, principally amino acids and derivatives, organic acids, nucleotides, and derivatives and flavonoids. A total of 13 stress-related metabolites including piperidine, L-tryptophan, L-glutamic acid, L-saccharopine, L-phenylalanine, 6-methylcoumarin, cinnamic acid, inosine 5′-monophosphate, aminomalonic acid, 6-aminocaproic acid, putrescine, tyramine and abscisic acid (ABA) represent the core metabolome responsive to salinity stress in hulless barley regardless of the tolerance level. In particular, we found that the ABA signalling pathway is essential to salt stress response in hulless barley. The high tolerance of the cultivar 0119 is due to a metabolic reprogramming at key stress times. During the early salt stress stages (0–24 h), 0119 tended to save energy through reduced glycolysis, nucleotide metabolism and amino acid synthesis, while increased antioxidant compounds such as flavonoids. Under prolonged stress (48–72 h), 0119 significantly enhanced energy production and amino acid synthesis. In addition, some important compatible solutes were strongly accumulated. By comparing the two cultivars, nine salt-tolerance biomarkers, mostly unreported salt-tolerance compounds in plants, were uncovered. Our study indicated that the salt tolerant hulless barley cultivar invokes a tolerance strategy which is conserved in other plant species. Overall, we provide for the first time some extensive metabolic data and some important salt-tolerance biomarkers which may assist in efforts to improve hulless barley tolerance to salinity stress.
Keywords: Biomarkers, hulless barley, salinity stress, tolerance mechanisms, widely targeted metabolites
Tibetan hulless barley is an important economic crop widely grown in highly stressful conditions in the Qinghai-Tibet Plateau and is often challenged by salinity stress. We investigated the temporal metabolic responses to salinity stress by employing the widely targeted metabolomic analysis in leaf samples from two contrasting cultivars. We successfully identified 13 stress-related metabolites representing the core metabolome responsive to salinity stress. Moreover, we revealed that the efficient management of glycolysis and energy consumption, especially during the most stressful time is the key strategy for tolerating salt stress. Finally, some unreported salt-tolerance biomarkers were unveiled in hulless barley.
Introduction
Most of the food and feed crops are salt-sensitive; therefore, salinity stress represents one of the most harmful abiotic stresses for agricultural productivity (Roy et al. 2014). Worldwide, over 45 million hectares of irrigated lands have been damaged by salt, and 1.5 million hectares are out of production each year because of the high salinity levels in the soil (Munns and Tester 2008). The vast areas affected and the seriousness of salt affecting agricultural land are predicted to worsen due to an inadequate drainage of the irrigated lands, poor management of the fertilizers applied and global warming (Munns and Gilliham 2015). The primary cause of the salinity stress is an increase in sodium (Na+) and chloride (Cl−) ions, the most abundant salt ions in natural sources (Shabala and Mackay 2011). High salinity induces several disorders in plants: water stress, ion toxicity, nutritional disorders, oxidative stress, alteration of metabolic processes, membrane disorganization, reduction of cell division and expansion, genotoxicity, etc. (Munns 2002; Flowers 2004; Demidchik et al. 2010; Flowers and Colmer 2015). Together, these effects reduce plant growth, productivity and survival. Many plants have evolved several mechanisms for either to exclude salt from the cells or to tolerate its presence within the cells, which include the regulation of ion homeostasis, osmotic adjustment, the regulation of phytohormones, the induction of antioxidant enzyme activity, etc. (Ashraf et al. 2008). Gradually, an increasing number of salt-responsive genes have been identified in plants, including ion transporters, free radical scavengers, aquaporins, heat shock proteins, transcription regulators, osmoprotectants and late embryogenesis abundant proteins (Kawasaki et al. 2001; Wang et al. 2016; Yong et al. 2015).
Metabolomics has become a robust approach for plant biologists to understand the complex metabolic responses to various abiotic pressures such as salt stress (Nakabayashi and Saito 2015). Systems biology has mainly focused on studying genomics, transcriptomics and proteomics which only represent the potential for a biological outcome. Unlike the above-mentioned ‘omics’ measures, metabolites and their concentrations directly reflect the underlying biochemical activity and state of cells, tissues or organisms (Kosmides et al. 2013). Metabolomics approach has been successfully employed to uncover important metabolites, serving as osmolytes and osmoprotectants, involved in salt stress responses in tomato, Arabidopsis thaliana, barley, peanut, alfalfa and safflower (Cramer et al. 2007; Zuther et al. 2007; Shulaev et al. 2008; Widodo et al. 2009; Gengmao et al. 2015; Song et al. 2017; Cui et al. 2018).
Tibetan hulless barley (Hordeum vulgare var. nudum) has developed a strong tolerance system to cope with environmental stresses as compared to other plant species within the genus of Hordeum (Li et al. 2016). In fact, the crop is widely grown in highly stressful conditions in the Qinghai-Tibet Plateau generally called ‘roof of the world’ because of its extremely high altitude and harsh environment (Dai et al. 2012). Therefore, the crop has to suffer various types of abiotic stresses such as cold, drought and salinity (Deng et al. 2013), which seriously impair its productivity. As the most important staple food and economic crop for the population in Tibet and its vicinity, it is important to enhance hulless barley stress tolerance for a higher yield and productivity. Unfortunately, to date, only limited progress has been made to enhance the stress tolerance in this crop. More importantly, several studies have focused on drought and cold tolerance enhancement in hulless barley (Deng et al. 2013; Zeng et al. 2016; Liang et al. 2017; Xu et al. 2017; Yuan et al. 2017) but research on the salinity tolerance has been largely overlooked. The salinization of cropland soil in the Tibetan Plateau has become an increasingly serious concern for agricultural production (Wang et al. 2018). It is estimated that 4 % of the total crop land soil is salinized land with an increasing annual rate of 0.15 % (Wang et al. 2008). Recently, we launched a breeding programme for hulless barley improvement towards salt tolerance. Over 1700 diverse Tibetan hulless barley accessions were screened for salt-tolerance and some contrasting accessions, including cultivars 0119 and 0226 were identified. The tolerant cultivar 0119 can survive for a week under high salinity stress while the sensitive cultivar 0226 dies just after 4 days. The goal of this study is to get an insight into the dynamic metabolic response to salinity stress and to uncover the major metabolites conferring salinity tolerance, which could be regarded as key biomarkers in hulless barley breeding programmes. To achieve this objective, we extracted, quantified and analysed a large number of metabolites accumulated under progressive salinity stress and we highlighted the divergent metabolic responses of the two contrasting cultivars 0119 and 0226.
Materials and Methods
Plant materials and growth conditions
In the present study, two Tibetan hulless barley cultivars 0119 (tolerant to salinity stress) and 0226 (sensitive to salinity stress) were used. They were identified after an initial screening of 1700 accessions. The seeds were surface-sterilized with 10 % (v/v) H2O2 for 15 min, rinsed thoroughly with distilled water, and then sown in a plastic tray (37 × 35 × 25 cm). The tray was filled with nutritional soil (pH 5.8–6.5, particle size: 0.5–3.0 mm, N: 200 mg L−1, PO3: 2500 mg L−1, K: 200 mg L−1 and Mg: 200 mg L−1; Nippi Engei Baido No. 1, Nihon Hiryo, Tokyo, Japan): vermiculite (1:1) and kept in a plant incubator set at 25 °C, 2000 µmol m−2 s−1. At the two-leaf stage, seedlings were removed from the tray and thoroughly washed with tap water. Well-developed and uniform seedlings were selected and transplanted to a 5-L black plastic bucket filled with 4.5 L ½ strength Hoagland nutrient solution (Hoagland and Arnon 1950). Each container was covered with a polystyrol plate with eight evenly spaced holes (one plant per hole) and kept in a growth chamber set at 16–18 °C (with a photoperiod of 18 h light/6 h dark and a relative humidity of 80 %). After 7 days’ growth, the solution was renewed and the corresponding containers were supplemented with 250 mM NaCl. Control plants were maintained in a ½ strength Hoagland nutrient solution without adding NaCl. The experiment was laid in a split-plot design with treatment as the main plot and cultivar as the subplot, and there were five replicates for each treatment. The solution pH was adjusted to 5.8 with NaOH or HCl as required and continuously aerated with pumps throughout the experiment. Then, from the beginning of the stress application, the youngest fully expanded leaves were harvested at 0, 2, 8, 24, 48 and 72 h from the control treatment (CK) and stress treatment (S). Three biological replicates from three different plants were collected at each time point and from each treatment, with a total of 72 samples. Samples were immediately snap-frozen in liquid nitrogen, ground in liquid nitrogen, stored in 15 mL Falcon tubes at −80 °C and later used for metabolomics investigation.
Widely targeted metabolomics
The sample preparation, extract analysis, metabolite identification and quantification were performed at Wuhan MetWare Biotechnology Co., Ltd (www.metware.cn) following their standard procedures.
Sample preparation
The freeze-dried leaf samples were crushed using a mixer mill (MM 400, Retsch) with a zirconia bead for 1.5 min at 30 Hz. One hundred (100) mg powder was weighed and aliquots were extracted overnight at 4 °C with 1 mL 70 % aqueous methanol. Following centrifugation at 10 000 g for 10 min, the extracts were absorbed (CNWBOND Carbon-GCB SPE Cartridge, 250 mg, 3 mL; ANPEL, Shanghai, China, www.anpel.com.cn/cnw) and filtrated (SCAA-104, 0.22 µm pore size; ANPEL, Shanghai, China, www.anpel.com.cn/cnw) before the LC-MS analysis (Chen et al. 2013).
HPLC conditions
The sample extracts were analysed using an LC-ESI-MS/MS system (HPLC, Shim-pack UFLC SHIMADZU CBM30A system, www.shimadzu.com.cn; MS, Applied Biosystems 4500 Q TRAP, www.appliedbiosystems.com.cn). The analytical conditions were as follows, HPLC: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 µm, 2.1 mm × 100 mm); solvent system, water (0.04 % acetic acid):acetonitrile (0.04 % acetic acid); gradient programme, 95:5 V/V at 0 min, 5:95 V/V at 11 min, 5:95 V/V at 12 min, 95:5 V/V at 12.1 min, 95:5 V/V at 15 min; flow rate, 0.40 mL min−1; temperature, 40 °C; injection volume: 5 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS.
ESI-Q TRAP-MS/MS
Linear ion trap (LIT) and triple quadrupole (QQQ) scans were acquired on a Q TRAP-MS, API 4500 Q TRAP LC/MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in a positive ion mode and controlled by the Analyst 1.6 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 550 °C; ion spray voltage 5500 V; ion source gas I, gas II, curtain gas were set at 55, 60 and 25 psi, respectively; the collision gas was high. Instrument tuning and mass calibration were performed with 10 and 100 μmol L−1 polypropylene glycol solutions in QQQ and LIT modes, respectively. Based on the self-built database MetWare Database (http://www.metware.cn/) and metabolite information in public database, the materials were qualitatively analysed according to the secondary spectrum information and the isotope signal was removed during the analysis. QQQ scans were acquired as multiple reaction monitoring (MRM) experiments with collision gas (nitrogen) set to 5 psi (Fraga et al. 2010). De-clustering potential (DP) and collision energy (CE) for individual MRM transitions were done with further DP and CE optimization (Chen et al. 2013). A specific set of MRM transitions were monitored for each period according to the metabolites eluted within this period.
Metabolomics data analysis
Data matrices with the intensity of metabolite features at the six time points under both salt and control conditions for the two cultivars were uploaded to the Analyst 1.6.1 software (AB SCIEX, Ontario, Canada). For statistical analysis, missing values were assumed to be below the limits of detection, and these values were imputed with a minimum compound value (Chen et al. 2013). The relative abundance of each metabolite was log transformed before analysis to meet normality. A Dunnett’s test was used to compare the abundance of each metabolite between different time points. False discovery rate was used for controlling multiple testing. The supervised multivariate method, partial least squares-discriminant analysis (PLS-DA), was used to maximize the metabolome difference between the control and salt-treated samples, as well as the difference between the two cultivars. The relative importance of each metabolite to the PLS-DA model was checked using a parameter called the variable importance in projection (VIP). Metabolites with VIP > 1.0 were considered as differential metabolites for group discrimination. Principal component analysis (PCA), hierarchical cluster analysis (HCA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed in the R software (www.r-project.org). In addition, the central pathway analysis was performed using the Vanted software v.2.2.1 (Junker et al. 2006) for the identified important metabolites using A. thaliana pathway libraries.
Results
Temporal metabolic profiling in leaves of 0119 and 0226 under salt stress
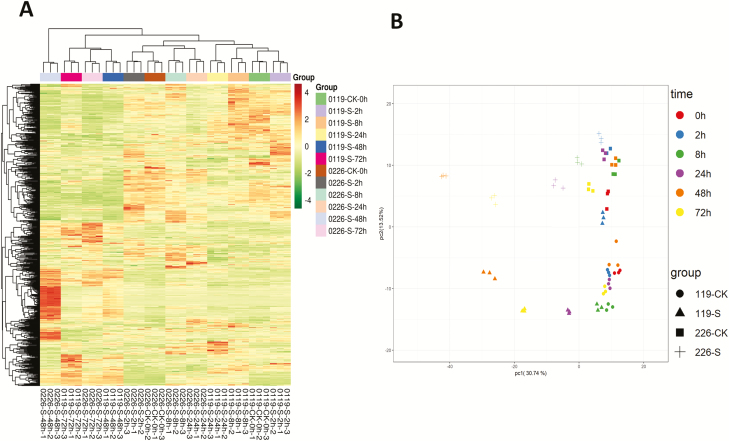
In the present study, we profiled the metabolic changes in two hulless barley cultivars displaying contrasting responses to salinity stress. In total, we detected 642 metabolites grouped into 32 major classes using the widely targeted metabolomics approach [seeSupporting Information—Table S1]. The majority of the identified metabolites belong to the classes of organic acids, amino acid and derivatives, and nucleotide and its derivative. Conversely, very few metabolites from the classes of catechin derivatives, pyridine derivatives and isoflavone were identified in hulless barley leaves (Table 1). Clustering analysis of the metabolic profiles from the two cultivars showed that all the biological replicates were grouped together, indicating a good correlation between replicates and the high reliability of our data (Cui et al. 2018). Moreover, we observed that metabolic data from the early stages of salinity stress (0–24 h) was clearly separated from those from the late stages (48–72 h) (Fig. 1A). Consistently, based on the PCA, a clear separation between the time points could be observed by the PC1. Furthermore, the PC2 distinctly distinguished the two cultivar (Fig. 1B). These results suggest that distinct metabolic programmes are engaged at the early and late stages of salt stress and the differential metabolic responses between 0119 and 0226 could be the basis of their contrasting tolerance to salinity stress.
Table 1.
Classification of the 642 detected metabolites in hulless barley accessions into major classes.
| Class | Number of compounds | Class | Number of compounds |
|---|---|---|---|
| Organic acids | 59 | Phytohormones | 15 |
| Amino acid derivatives | 58 | Vitamins | 14 |
| Nucleotide and its derivates | 56 | Carbohydrates | 13 |
| Flavone | 54 | Benzoic acid derivatives | 11 |
| Flavone C-glycosides | 45 | Tryptamine derivatives | 10 |
| Hydroxycinnamoyl derivatives | 32 | Flavonolignan | 10 |
| Lipids_glycerophospholipids | 31 | Indole derivatives | 9 |
| Amino acids | 27 | Cholines | 7 |
| Others | 26 | Alkaloids | 7 |
| Phenolamides | 26 | Anthocyanins | 5 |
| Quinate and its derivatives | 20 | Nicotinic acid derivatives | 4 |
| Lipids_fatty acids | 19 | Alcohols and polyols | 4 |
| Lipids_glycerolipids | 18 | Terpenoids | 3 |
| Flavanone | 18 | Catechin derivatives | 3 |
| Coumarins | 17 | Pyridine derivatives | 2 |
| Flavonol | 17 | Isoflavone | 2 |
Figure 1.
(A) Heatmap hierarchical clustering of detected metabolites. Hierarchical trees were drawn based on detected metabolites in leaves of 0119 and 0226 at 0, 2, 8, 24, 48 and 72 h in control (CK) and salt stress treatment (S). The columns correspond to cultivar at different time points, while the rows represent different metabolites; (B) PCA loading plot of the two first principal components based on the metabolic data from leaves of 0119 and 0226 at 0, 2, 8, 24, 48 and 72 h in control (CK) and salt stress treatment (S).
Alteration of the metabolite levels induced by salt stress
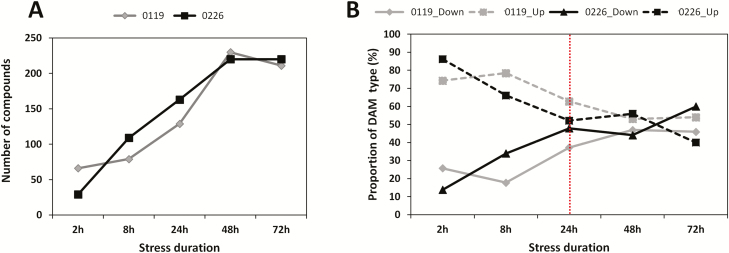
In order to identify the differentially accumulated metabolites (DAMs) under salinity stress, we compared the metabolite levels in the control condition to their levels under stress at each time point. We identified from 66 to 230 DAMs (364 DAMs in total) in 0119 and from 29 to 220 DAMs (364 DAMs in total) in 0229 at the different time points under stress (VIP > 1) [seeSupporting Information—Table S2]. These salt-responsive compounds are mainly primary metabolites including organic acids, nucleotide and its derivates, and amino acid derivatives. We observed an increasing DAM number over the stress duration in both cultivars, indicating an active metabolic readjustment to respond to the stress (Fig. 2A). However, after 2 h of salt stress, the number of DAMs affected in the sensitive cultivar 0226 was significantly higher than 0119, which denote the strong metabolic perturbation experienced by 0226. By scrutinizing the types of DAM (up- and down-accumulated) in both cultivars during the time points, it could be noticed that the tolerant cultivar responded quickly to the stress by strongly decreasing key metabolites from the class of nucleotides and its derivatives (adenosine 3′-monophosphate, adenosine, inosine 5′-monophosphate, deoxyadenosine) to control the energy homeostasis, while strikingly increased flavonoids (>2300-fold increase of chrysoeriol O-glucuronic acid), and phytohormone compounds (>173-fold increase of methyl jasmonate) (Fig. 2A; seeSupporting Information—Table S3). However, as the stress duration was elapsing, the metabolic processes were impaired leading to the decrease of a large number of metabolites in both cultivars (Fig. 2B; seeSupporting Information Tables S3andS4). Nonetheless, the tolerant cultivar (0119) could still accumulate the salt stress-responsive metabolites and better maintain the metabolic processes than cultivar 0226, especially after 24 h under stress, a time point which appears to be critical for the salinity stress endurance. After 24 h under stress, as the energy is critically needed, the metabolites from the nucleotide metabolism were strongly accumulated as well as the important stress-related compounds such as vitamin (L-ascorbate), phenolamide (tyramine), coumarin (esculetin), amino acid derivatives (L-tryptophan), phytohormone (abscisic acid (ABA)), polyamine (putrescine), etc. At 72 h under stress, the sensitive cultivar (0226) was overwhelmed and failed to accumulate in optimal levels the majority of the metabolites, which might lead to the death of the seedlings (Fig. 2B).
Figure 2.
Temporal changes of metabolic reprogramming in 0119 and 0226 under salt stress. (A) DAMs at 2, 8, 24, 48 and 72 h under salt stress; (B) proportion of DAM types (down- and up-accumulated) at 2, 8, 24, 48 and 72 h under salt stress. The red dashed line depicts the critical time point for salt stress endurance in hulless barley.
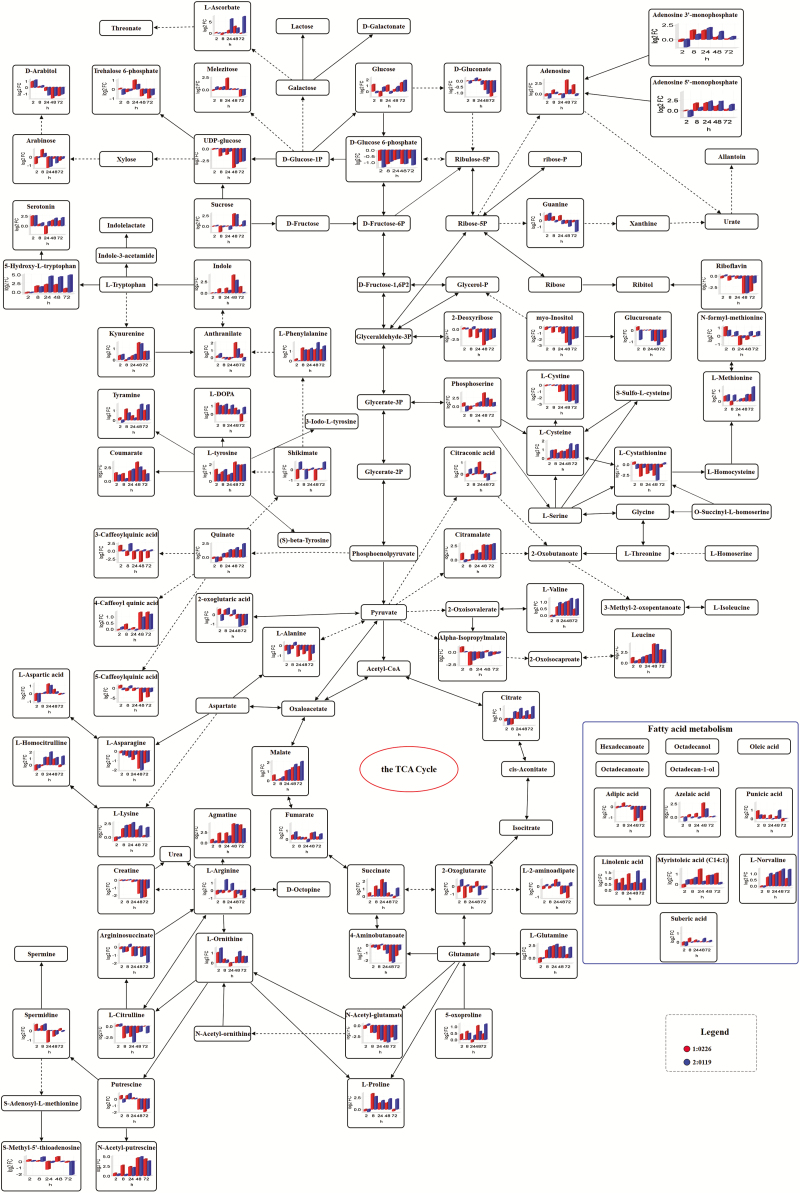
Through the general metabolic pathway mapping, the DAMs in response to salt stress were found to be mainly involved in amino acid metabolism, glycolysis, tricarboxylic acid (TCA) cycle and urea cycle, suggesting that these metabolic pathways play important roles in adaptive response to salt stress in hulless barley (Fig. 3). It also revealed that most of these metabolites, particularly the amino acids, organic acids and osmolytes were highly accumulated mainly during the late stages of salt stress (from 48 to 72 h) in the tolerant cultivar 0119 and a reduced glycolysis activity was maintained until 48 h as compared to the sensitive cultivar 0226. For example, malate, succinate, fumarate, citrate, proline, lysine, linoleic acid, ascorbate, phenylalanine, quinate, etc. were lower in the tolerant cultivar than in the sensitive one at the early stage of salt exposure but, an opposite scenario was observed during the late stage (Fig. 3).
Figure 3.
Metabolic pathways of the salt-responsive metabolites in hulless barley. Log2 fold change (FC) of significantly changed metabolites mapped to the central metabolic pathways in the leaves of 0119 and 0226 at 2, 8, 24, 48 and 72 h under salt treatment as compared to control treatment. Metabolic pathways were constructed according to the KEGG (http://www.genome.jp/kegg/) metabolic database.
The core metabolites involved in salt stress response in hulless barley
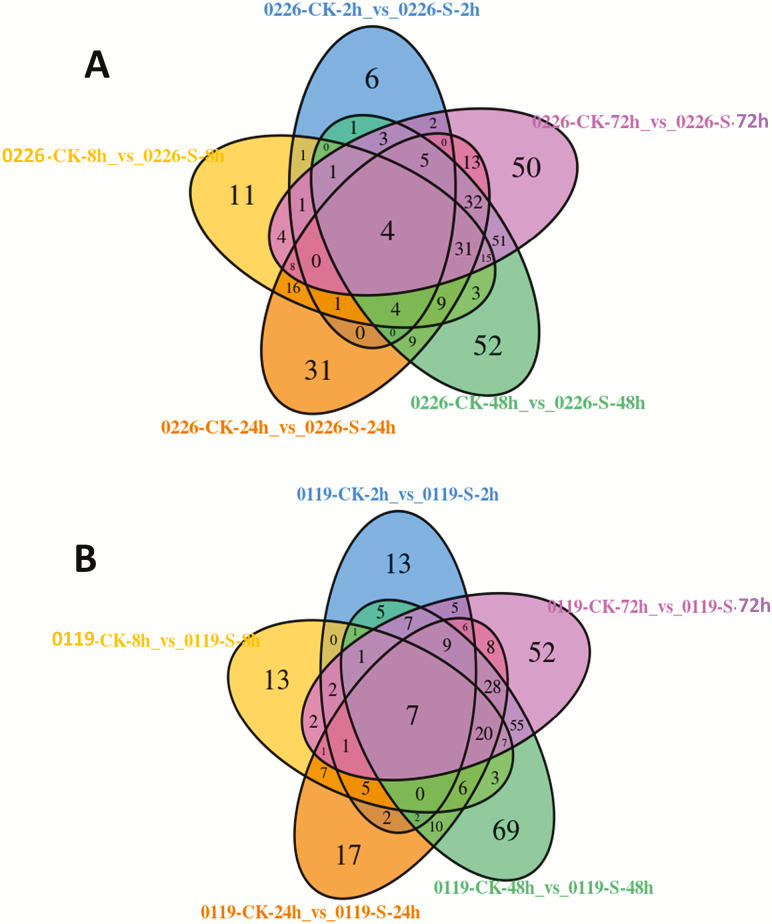
Figure 4A and B present the shared and specific DAMs at the different time points under salinity stresses. It further confirms that specific metabolic adjustments occur at the different time points under salinity stress in hulless barley, though similar metabolites were regulated at the late stages of salinity stress (48–72 h) in both cultivars. We identified 4 and 41 metabolites whose levels significantly changed during all the time points or at four time points, respectively, under salt stress in 0226 [seeSupporting Information—Table S5]. Likewise, the levels of 7 and 31 metabolites were constitutively and significantly changed at five or four time points under stress in 0119, respectively [seeSupporting Information—Table S6]. Among these major salt-responsive metabolites, 13 compounds (piperidine, 5-hydroxy-L-tryptophan, N-acetyl-L-glutamic acid, L-saccharopine, L-phenylalanine, 6-methylcoumarin, cinnamic acid, inosine 5′-monophosphate, aminomalonic acid, 6-aminocaproic acid, N′,N-p-coumaroyl-feruloyl putrescine, N-feruloyl tyramine and ABA) were shared by both cultivars representing the core metabolites associated with salt stress responses in hulless barley, regardless of the tolerance levels. In particular, we identified ABA as the only metabolite commonly and constantly changed (increased) during all the time points under salt stress in both cultivars.
Figure 4.
(A) Venn diagram depicting the shared and common DAMs between the time point series under salt stress in 0226; (B) Venn diagram depicting the shared and common DAMs between time point series under salt stress in 0119.
Detecting important salt-tolerance biomarkers in hulless barley
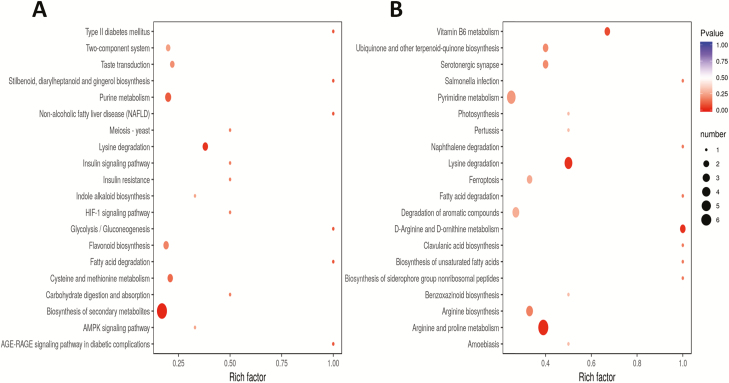
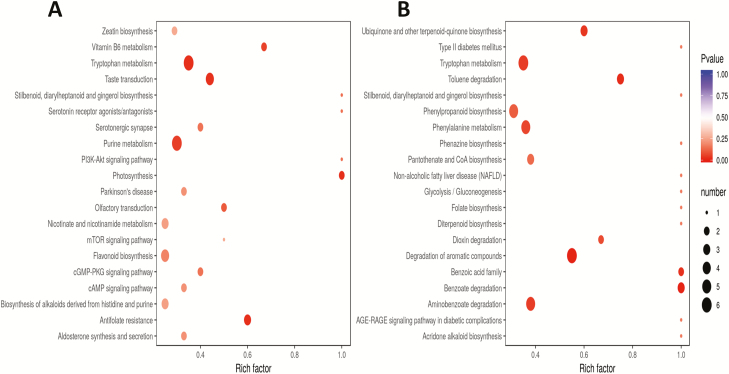
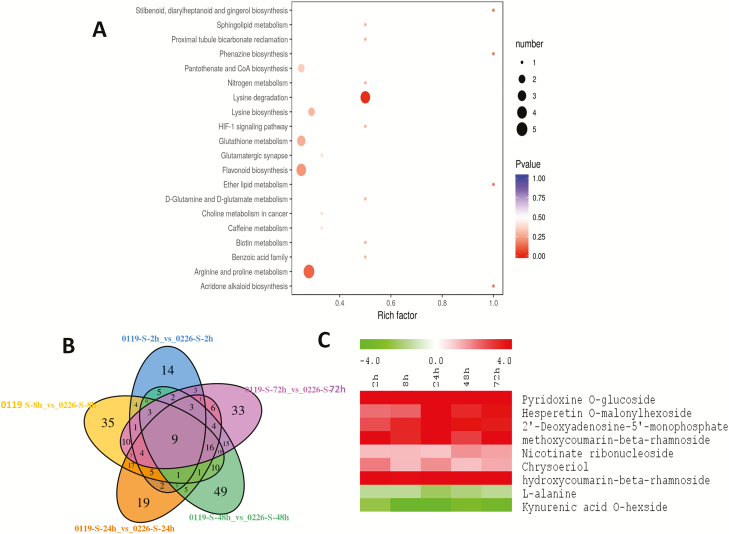
To get an insight into the important metabolites conferring salt tolerance in 0119, we analysed the DAMs between 0119 and 0226 under stress. In total, 295 metabolites were differentially accumulated between the two cultivars at least at one time point under stress [see Supporting Information—Table S7]. Kyoto Encyclopedia of Genes and Genomes enrichment analysis of the DAMs between the two cultivars at each time point under stress showed a dynamic metabolic adjustment over the stress durations (Figs 5–7). The most enriched biological pathways were purine metabolism, pyrimidine metabolism, arginine and proline metabolism, tryptophan metabolism, flavonoid biosynthesis, phenylpropanoid biosynthesis, glutathione metabolism and lysine degradation. We identified nine metabolites (pyridoxine O-glucoside, L-alanine, hesperetin O-malonylhexoside, kynurenic acid O-hexside, 2′-deoxyadenosine-5′-monophosphate, 4-hydroxy-7-methoxycoumarin-beta-rhamnoside, nicotinate ribonucleoside, chrysoeriol 6-C-hexoside 8-C-hexoside-O-hexoside and 7-hydroxycoumarin-beta-rhamnoside) constitutively and differentially accumulated in 0119 compared to 0226 during all the stress duration (Fig. 7B). These metabolites may represent major biomarkers for salt stress tolerance in hulless barley and interestingly, the majority of these compounds were increased in the tolerant accession, an indication of their positive functions to impart salt tolerance (Fig. 7C).
Figure 5.
(A) KEGG enrichment analysis of the DAMs between 0119 and 0226 at 2 h under salt stress; (B) KEGG enrichment analysis of the DAMs between 0119 and 0226 at 8 h under salt stress.
Figure 6.
(A) KEGG enrichment analysis of the DAMs between 0119 and 0226 at 24 h under salt stress; (B) KEGG enrichment analysis of the DAMs between 0119 and 0226 at 48 h under salt stress.
Figure 7.
(A) KEGG enrichment analysis of the DAMs between 0119 and 0226 at 72 h under salt stress; (B) Venn diagram depicting the shared and common DAMs between the time point series under salt stress in 0119 compared to 0226; (C) Log2 fold change of the constitutively detected DAMs between 0119 and 0226 during salt treatment.
Discussion
Extensive metabolite data for deciphering hulless barley responses and tolerance mechanisms to salinity stress
Metabolites are intermediates or end products of metabolism, playing significant physiological and biochemical roles in plants (Luo 2015). It is estimated that the total number of plant metabolites exceeds 200 000 (Pichersky et al. 2006) reflecting their multifaceted functions in plants’ life cycle. In hulless barley, the diversity of metabolites and their functional characterization have not yet been reported. Hulless barley is a resilient crop able to survive several environmental stresses (Li et al. 2016); therefore, investigating its metabolic responses to abiotic stress can provide important and novel insights into the tolerance mechanisms to abiotic stress in plants. Among the major abiotic stresses affecting the productivity of major food crops worldwide, salinity is of particular importance but limited progress has been made so far to enhance the crop’s salinity tolerance (Negrão et al. 2017). In the present investigation, a rich and diverse set of metabolites was detected for the first time in the leaves of two hulless barley cultivars. Under salt stress, the levels of numerous metabolites were altered, offering an excellent opportunity to identify novel salt-responsive compounds (Shen et al. 2016). The temporal metabolic profiling strategy adopted in this study coupled with the comparison of tolerant and sensitive cultivars highlighted that salinity stress-induced metabolic reprogramming is genotype-dependent and time-dependent in hulless barley. Our observations are in perfect agreement with the study of Zhao et al. (2014) who observed that metabolites were temporally and genotype-dependently regulated under salt stress in two contrasting rice genotypes. Similarly, Kim et al. (2007) revealed that a short-term salt stress affects the phenylpropanoid pathway for lignin and glycine betaine biosynthesis, while a long-term response tends to combine the induction of glycolysis and sucrose metabolism in A. thaliana.
Salinity stress principally disturbs the primary metabolism in hulless barley
Under abiotic stress, the plant metabolism is significantly perturbed (Sanchez et al. 2008). The main purpose of studying metabolic changes during stress response is to identify metabolites that allow for the reestablishment of homeostasis and normal metabolic fluxes. We observed that the salt-responsive metabolites in hulless barley were dominated by the primary metabolites. According to Slama et al. (2015), primary metabolites are the most important metabolites affected by stress, usually on account of the limited CO2 assimilation. In accordance with our findings, salt stress was reported to greatly influence the primary metabolism in various plant species such as maize (Mladenova 1990), rice (Wang et al. 2015), barley (Shelden et al. 2016), peanut (Cui et al. 2018), etc. Particularly, most of the detected salt-responsive primary metabolites belong to the classes of organic acids, nucleotide and its derivates, and amino acid derivatives. It is well-known that organic acid metabolism is of fundamental importance at the cellular level for various biochemical pathways, including energy production, the formation of precursors for amino acid biosynthesis and at the whole plant level in modulating adaptation to abiotic stresses (López-Bucio et al. 2000). As photosynthetic intermediates, they are essential to plants (Cockburn 1985) but they also have a potential role as metabolically active solutes for the osmotic adjustment and the balance of cation excess; therefore, their activity is crucial for plant responses to abiotic stresses such as salinity. The nucleotide metabolism is also of great importance to living organisms, as nucleotides are the building blocks of nucleic acids synthesis, energy sources, coenzymes for redox reactions and precursors for the synthesis of primary and secondary products (Moffatt and Ashihara 2002). Similarly, amino acids have various prominent functions in plants, particularly during protein biosynthesis. They represent the building blocks for several other biosynthesis pathways and participate in signalling processes during plant stress response (Hildebrandt et al. 2015).
The sensitive and tolerant cultivars share a core metabolome fundamental to salinity stress responses
Despite their differential tolerance level, sensitive and tolerant genotypes share pools of metabolites which are fundamental to the stress responses. Here, by comparing 0226 and 0119, we uncovered several key metabolites commonly altered in both genotypes under stress. Most of these metabolites were reported to be involved in salt stress response in plants (Sanchez et al. 2008). Interestingly, these core metabolites were mainly increased under salt stress, indicating their positive roles in osmotic regulation and salinity endurance in hulless barley. Distinctly, high levels of ABA were constitutively observed in both cultivars throughout the salt stress application. Abscisic acid is described as a key hormone in plant adaptation to abiotic stress such as salt, osmotic and cold (Waśkiewicz et al. 2013). One major biochemical change in response to stress is the elevation of ABA levels, which in turn triggers the activation of stress-responsive genes (Wasilewska et al. 2008). The ABA signalling pathway can therefore be considered essential to salt stress response in hulless barley.
The efficient management of glycolysis and energy consumption are the key strategies for tolerating salt stress
In general, the metabolic changes observed in stressed plants varied in their significance according to the tolerant/sensitive phenotypes. Here, we observed that the sensitive cultivar experienced a more pronounced metabolic change as compared to the tolerant one. Similar to our findings, Widodo et al. (2009) and Shen et al. (2016) observed that within the cultivated and wild barley species, the metabolism under salt stress in the tolerant genotypes was less affected as compared to the sensitive ones. Recently, Fu et al. (2018) also demonstrated that rice, which is a salt sensitive species, experiences more serious osmotic stress and metabolic changes than barley, a salt tolerant species. In this study, further investigations into the temporal regulation of the salt-responsive metabolites indicated that the salt tolerance observed in the cultivar 0119 is due to its adaptive reaction at the different stress times. At the early salt stress stages, in contrast to 0226, the tolerant cultivar 0119 tended to save energy by limiting glycolysis activity, nucleotide metabolism and amino acid synthesis, while increasing antioxidant compounds such as flavonoids. And under a long-term exposure to salt stress, the tolerant cultivar 0119 highly supplied energy probably for Na+ exclusion from cells and strongly accumulated the metabolites serving as compatible solutes against the osmotic challenge. Recently, Wu et al. (2013) also observed that increasing glycolysis and energy consumption during the most stressful time under high salinity stress is a tolerance mechanism in the cultivated barley. In rice, a similar salt stress tolerance mechanism was reported (Zhao et al. 2014). Kim et al. (2007) also demonstrated that A. thaliana resists to a long-term salinity stress by inducing glycolysis and sucrose metabolism, indicating that it is a conserved salt-tolerance mechanism in plants (Sanchez et al. 2008).
Hulless barley provides novel salt-tolerance candidate biomarkers
Candidate metabolites as stress-tolerance biomarkers are useful in breeding programmes and for crop improvement strategies. By analysing the differential salt-affected metabolites in the highly tolerant cultivar 0119 compared to the sensitive one, we anticipated to uncover important salt-tolerance biomarkers. A total of 295 metabolites displayed differential accumulation between the two cultivars under stress, denoting that these compounds are the basis of their contrasting responses to salinity. The metabolites can be grouped into the classes of vitamins, amino acid derivatives, flavonoids, organic acids, nucleotide and its derivates and coumarins, which play essential functions in plant normal metabolism, development and responses to biotic and abiotic stresses (Buer et al. 2010; Dell’Aglio et al. 2017). Of these diverse metabolites, we pinpointed nine metabolites as the major salt-tolerance biomarkers in hulless barley and their up-accumulation in the tolerant cultivar further highlights their positive functions in salt tolerance. For example, pyridoxine glucoside accumulation was >5-fold in the tolerant cultivar than the sensitive one. Pyridoxine glucoside is a fraction of vitamin B6 in plant foods (Gregory et al. 1991). Colinas et al. (2016) reported that vitamin B6 is required for >200 catalytic reactions ranging from hormone biosynthesis to amino acid metabolism in plants. Therefore, the key strategies for plant survival under stress conditions include mechanisms to maintain the homeostasis of B vitamins. Vitamin B6 was demonstrated to play a crucial role in protecting cells from oxidative stress because it exhibits antioxidant activity (Vanderschuren et al. 2013). Similarly, Huang et al. (2013) revealed that abiotic stresses increased the abundance of different B6 vitamers, particularly pyridoxine in tobacco plants. Therefore, we deduce that the high concentration of pyridoxine glucoside helps alleviate the osmotic stress caused by salinity stress in hulless barley. Two flavonoid compounds including hesperidin and chrysoeriol were also detected as important biomarkers for salt tolerance in hulless barley. Hesperidin is a plant flavanone (a subclass of flavonoids) naturally and abundantly found in plants such as citrus (Liu et al. 2013). The flavanone hesperetin (aglycone) is a glycoside form of hesperidin. Extensive works have shown that flavonoids are accumulated in plants under abiotic stresses and function as absorbing compounds or free radical scavengers to reduce the damages to plants (Pourcel et al. 2007; Buer et al. 2010). Likewise, chrysoeriol has been shown to possess a potent antioxidant activity (Mishra et al. 2003). However, so far, there was no report on the effect of salt stress on hesperitin and chrysoeriol accumulation in plants. Furthermore, their functions in stress tolerance in plants, especially salt stress tolerance are unknown. Besides, many of the salt-tolerance biomarkers detected in our study have not yet been described as associated with abiotic stress response in plants, implying that they may be novel compounds or exclusive salt-tolerance metabolites in hulless barley. Therefore, we propose that future studies may deeply investigate these newly identified compounds, as novel salt-tolerance candidate biomarkers in plants.
Conclusions
In this report, we generated for the first time extensive metabolic data under progressive salinity stress in two contrasting hulless barley cultivars. Salinity stress affects the metabolism of hulless barley genotypes and the stress-induced metabolic reprogramming is genotype-dependent and time-dependent. Furthermore, salinity stress mainly disturbs the primary metabolism in hulless barley, particularly, compounds from the classes of organic acids, nucleotide and its derivates, and amino acid derivatives. Several metabolites were commonly and differentially altered in both cultivars under salinity stress, representing the core metabolome fundamental to salinity stress responses in hulless barley. Our study highlighted the cardinal importance of ABA in salt stress response signalling in hulless barley. By comparing the metabolic responses triggered by the two contrasting cultivars under salinity stress, it was clear that the efficient management of glycolysis and energy consumption, especially during the most stressful time is the key strategy for tolerating salt stress. Finally, we detected nine salt-tolerance biomarkers, mostly unreported salt-tolerance metabolites, which may help improve salt stress tolerance in plants.
Availability of Data and Material
The data sets supporting the conclusion of this article are included within the article and the Supporting Information. The metabolite quantification data set will be provided upon request addressed to the corresponding author.
Sources of Funding
This work was supported by the Financial Special Fund (2017CZZX002, XZNKY-2018-C-021).
Contributions by the Authors
Conceptualization, Z.S., T.N.; methodology, Y.W., X.Z., Q.X.; formal analysis and data curation, X.M., H.Y., D.J.; writing-original draft preparation, Y.W.; writing-review and editing, Z.S., T.N. All authors discussed the results and contributed comments to the manuscript. All authors approved the final version of the manuscript.
Conflict of Interest
None declared.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. Full list of the metabolites detected in leaves of the two hulless barley cultivars using the widely targeted metabolomics approach.
Table S2. List of the differentially accumulated metabolites under salt stress in the tolerant cultivar 0119 and the sensitive cultivar 0226.
Table S3. List of the differentially accumulated metabolites at 2, 8, 24, 48 and 72 h under salt stress in the tolerant cultivar 0119.
Table S4. List of the differentially accumulated metabolites at 2, 8, 24, 48 and 72 h under salt stress in the sensitive cultivar 0226.
Table S5. List of the differentially accumulated metabolites constitutively detected at four and five time points under salt stress in the sensitive cultivar 0226.
Table S6. List of the differentially accumulated metabolites constitutively detected at four and five time points under salt stress in the tolerant cultivar 0119.
Table S7. List of the differentially accumulated metabolites between the sensitive cultivar 0226 and the tolerant one 0119 at 2, 8, 24, 48 and 72 h under salt stress.
Acknowledgements
We thank Dr Komivi Dossa for his help in revising this manuscript.
Literature Cited
- Ashraf M, Athar HR, Harris PJC, Kwon TR. 2008. Some prospective strategies for improving crop salt tolerance. Advances in Agronomy 97:45–110. [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. 2010. Flavonoids: new roles for old molecules. Journal of Integrative Plant Biology 52:98–111. [DOI] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J. 2013. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Molecular Plant 6:1769–1780. [DOI] [PubMed] [Google Scholar]
- Cockburn W. 1985. Variation in photosynthetic acid metabolism in vascular plants: CAM and related phenomena. New Phytologist 101:3–24. [DOI] [PubMed] [Google Scholar]
- Colinas M, Eisenhut M, Tohge T, Pesquera M, Fernie AR, Weber AP, Fitzpatrick TB. 2016. Balancing of B6 vitamers is essential for plant development and metabolism in Arabidopsis. The Plant Cell 28:439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EA, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, Quilici D, Schlauch KA, Schooley DA, Cushman JC. 2007. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Functional & Integrative Genomics 7:111–134. [DOI] [PubMed] [Google Scholar]
- Cui F, Sui N, Duan G, Liu Y, Han Y, Liu S, Wan S, Li G. 2018. Identification of metabolites and transcripts involved in salt stress and recovery in peanut. Frontiers in Plant Science 9:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Nevo E, Wu DZ, Comadran J, Zhou MX, Qiu L, Chen ZH, Beiles A, Chen GX, Zhang GP. 2012. Tibet is one of the centers of domestication of cultivated barley. Proceedings of the National Academy of Sciences of the United States of America 109:16969–16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Aglio E, Boycheva S, Fitzpatrick TB. 2017. The pseudoenzyme PDX1.2 sustains vitamin B6 biosynthesis as a function of heat stress. Plant Physiology 174:2098–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Cuin TA, Svistunenko D, Smith SJ, Miller AJ, Shabala S, Sokolik A, Yurin V. 2010. Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. Journal of Cell Science 123:1468–1479. [DOI] [PubMed] [Google Scholar]
- Deng G, Liang J, Xu D, Long H, Pan Z, Yu M. 2013. The relationship between proline content, the expression level of P5CS (Δ1_Pyrroline_5_Carboxylate Synthetase), and drought tolerance in Tibetan hulless barley (Hordeum vulgare var. nudum). Russian Journal of Plant Physiology 60:693–700. [Google Scholar]
- Flowers TJ. 2004. Improving crop salt tolerance. Journal of Experimental Botany 55:307–319. [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. 2015. Plant salt tolerance: adaptations in halophytes. Annals of Botany 115:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga CG, Clowers BH, Moore RJ, Zink EM. 2010. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Analytical Chemistry 82:4165–4173. [DOI] [PubMed] [Google Scholar]
- Fu L, Shen Q, Kuang L, Yu J, Wu D, Zhang G. 2018. Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiology and Biochemistry 130:248–257. [DOI] [PubMed] [Google Scholar]
- Gengmao Z, Yu H, Xing S, Shihui L, Quanmei S, Changhai W. 2015. Salinity stress increases secondary metabolites and enzyme activity in safflower. Industrial Crops Production 64:175–181. [Google Scholar]
- Gregory JF, Trumbo PR, Bailey LB, Toth JP, Baumgartner TG, Cerda JJ. 1991. Bioavailability of pyridoxine-5’-beta-D-glucoside determined in humans by stable-isotopic methods. The Journal of Nutrition 121:177–186. [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun HP. 2015. Amino acid catabolism in plants. Molecular Plant 8:1563–1579. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. 1950. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular 347:1–32. [Google Scholar]
- Huang S, Zhang J, Wang L, Huang L. 2013. Effect of abiotic stress on the abundance of different vitamin B6 vitamers in tobacco plants. Plant Physiology and Biochemistry 66:63–67. [DOI] [PubMed] [Google Scholar]
- Junker BH, Klukas C, Schreiber F. 2006. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ. 2001. Gene expression profiles during the initial phase of salt stress in rice. The Plant Cell 13:889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Bamba T, Harada K, Fukusaki E, Kobayashi A. 2007. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. Journal of Experimental Botany 58:415–424. [DOI] [PubMed] [Google Scholar]
- Kosmides AK, Kamisoglu K, Calvano SE, Corbett SA, Androulakis IP. 2013. Metabolomic fingerprinting: challenges and opportunities. Critical Reviews in Biomedical Engineering 41:205–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lai P, Li P, Zhao Y. 2016. Transformation of Cichorium intybus with the HvBADH1 gene enhanced the salinity tolerance of the transformants. South African Journal of Botany 102:110–119. [Google Scholar]
- Liang J, Chen X, Deng G, Pan Z, Zhang H, Li Q, Yang K, Long H, Yu M. 2017. Dehydration induced transcriptomic responses in two Tibetan hulless barley (Hordeum vulgare var. nudum) accessions distinguished by drought tolerance. BMC Genomics 18:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EH, Zhao P, Duan L, Zheng GD, Guo L, Yang H, Li P. 2013. Simultaneous determination of six bioactive flavonoids in citri reticulatae pericarpium by rapid resolution liquid chromatography coupled with triple quadrupole electrospray tandem mass spectrometry. Food Chemistry 141:3977–3983. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez V, Herrera-Estrella L. 2000. Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Science 160:1–13. [DOI] [PubMed] [Google Scholar]
- Luo J. 2015. Metabolite-based genome-wide association studies in plants. Current Opinion in Plant Biology 24:31–38. [DOI] [PubMed] [Google Scholar]
- Mishra B, Priyadarsini KI, Kumar MS, Unnikrishnan MK, Mohan H. 2003. Effect of O-glycosilation on the antioxidant activity and free radical reactions of a plant flavonoid, chrysoeriol. Bioorganic & Medicinal Chemistry 11:2677–2685. [DOI] [PubMed] [Google Scholar]
- Mladenova YI. 1990. Influence of salt stress on primary metabolism of Zea mays L. seedlings of model genotypes. Plant and Soil 123:217–222. [Google Scholar]
- Moffatt BA, Ashihara H. 2002. Purine and pyrimidine nucleotide synthesis and metabolism. In: Somerville CR, Meyerowitz EM eds. The arabidopsis book. Rockville, MD: American Society of Plant Biologists, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. 2002. Comparative physiology of salt and water stress. Plant, Cell & Environment 25:239–250. [DOI] [PubMed] [Google Scholar]
- Munns R, Gilliham M. 2015. Salinity tolerance of crops - what is the cost? The New Phytologist 208:668–673. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59:651–681. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Saito K. 2015. Integrated metabolomics for abiotic stress responses in plants. Current Opinion in Plant Biology 24:10–16. [DOI] [PubMed] [Google Scholar]
- Negrão S, Schmöckel S, Tester M. 2017. Evaluating physiological responses of plants to salinity stress. Annals of Botany 119:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N. 2006. Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311:808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I. 2007. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends in Plant Science 12:29–36. [DOI] [PubMed] [Google Scholar]
- Roy SJ, Negrão S, Tester M. 2014. Salt resistant crop plants. Current Opinion in Biotechnology 26:115–124. [DOI] [PubMed] [Google Scholar]
- Sanchez DH, Siahpoosh MR, Roessner U, Udvardi M, Kopka J. 2008. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiologia Plantarum 132:209–219. [DOI] [PubMed] [Google Scholar]
- Shabala S, Mackay A. 2011. Ion transport in halophytes. In: Turkan I, ed. Plant responses to drought and salinity stress: developments in a post-genomic era (advances in botanical research), Vol. 57 Cambridge, MA: Elsevier Academic Press, 151–199. [Google Scholar]
- Shelden MC, Dias DA, Jayasinghe NS, Bacic A, Roessner U. 2016. Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. Journal of Experimental Botany 67:3731–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Fu L, Dai F, Jiang L, Zhang G, Wu D. 2016. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genomics 17:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Cortes D, Miller G, Mittler R. 2008. Metabolomics for plant stress response. Physiologia Plantarum 132:199–208. [DOI] [PubMed] [Google Scholar]
- Slama I, Abdelly C, Bouchereau A, Flowers T, Savouré A. 2015. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Annals of Botany 115:433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Xu H, Sun N, Jiang L, Tian P, Yong Y, Yang W, Cai H, Cui G. 2017. Metabolomic analysis of alfalfa (Medicago sativa L.) root-symbiotic rhizobia responses under alkali stress. Frontiers in Plant Science 8:1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschuren H, Boycheva S, Li KT, Szydlowski N, Gruissem W, Fitzpatrick TB. 2013. Strategies for vitamin B6 biofortification of plants: a dual role as a micronutrient and a stress protectant. Frontiers in Plant Science 4:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ma R, Cao Q, Shan Z, Jiao Z. 2018. Enhanced tolerance to salt stress in highland barley seedlings (Hordeum vulgare ssp. vulgare) by gamma irradiation pretreatment. Acta Physiologiae Plantarum 40:174. [Google Scholar]
- Wang WS, Zhao XQ, Li M, Huang LY, Xu JL, Zhang F, Cui YR, Fu BY, Li ZK. 2016. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. Journal of Experimental Botany 67:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zheng D, Shen Y. 2008. Land use change and its driving forces on the Tibetan Plateau during 1990–2000. Catena 72:56–66. [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. 2008. An update on abscisic acid signaling in plants and more. Molecular Plant 1:198–217. [DOI] [PubMed] [Google Scholar]
- Waśkiewicz A, Beszterda M, Goliński P. 2013. ABA: role in plant signaling under salt stress. In: Ahmad P, Azooz MM, Prasad MNV eds. Salt stress in plants. New York, NY: Springer, 175–196. [Google Scholar]
- Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. 2009. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. Journal of Experimental Botany 60:4089–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Cai S, Chen M, Ye L, Chen Z, Zhang H, Dai F, Wu F, Zhang G. 2013. Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS One 8:e55431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu QJ, Wang YL, Wei ZX, Yuan HJ, Zengm XQ, Nyima T. 2017. Cloning and functional characterization of the HbSYR1 gene encoding a syntaxin-related protein in Tibetan hulless barley (Hordeum vulgare L. var. nudum HK f). Genetics and Molecular Research 16:gmr16038909. [DOI] [PubMed] [Google Scholar]
- Yong HY, Wang C, Bancroft I, Li F, Wu X, Kitashiba H, Nishio T. 2015. Identification of a gene controlling variation in the salt tolerance of rapeseed (Brassica napus L.). Planta 242:313–326. [DOI] [PubMed] [Google Scholar]
- Yuan H, Zeng X, Ling Z, Wei Z, Wang Y, Zhuang Z, Xu Q, Tang Y, Nyima T. 2017. Transcriptome profiles reveal cold acclimation and freezing tolerance of susceptible and tolerant hulless barley genotypes. Acta Physiologea Plantarum 39:275. [Google Scholar]
- Zeng X, Bai L, Wei Z, Yuan H, Wang Y, Xu Q, Tang Y, Nyima T. 2016. Transcriptome analysis revealed the drought-responsive genes in Tibetan hulless barley. BMC Genomics 17:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang W, Zhang F, Deng J, Li Z, Fu B. 2014. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS One 9:e108020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E, Koehl K, Kopka J. 2007. Comparative metabolome analysis of the salt response in breeding cultivars of rice. In: Jenks MA, Hasegawa PM, Jain SM, eds. Advances in molecular breeding toward drought and salt tolerant crops. Berlin, Heidelberg, New York: Springer-Verlag, 285–315. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusion of this article are included within the article and the Supporting Information. The metabolite quantification data set will be provided upon request addressed to the corresponding author.