Abstract
To reveal overwintering dormancy (diapause) mechanisms of Culex pipiens pallens (L.), global protein expression differences at three separate time points represent nondiapause, diapause preparation and overwintering diapause phases of Cx. pipiens pallens were compared using iTRAQ. Cx. pipiens pallens females accumulate more lipid droplets during diapause preparation and overwintering diapause maintenance than during the nondiapause phase. A total of 1030 proteins were identified, among which 1020 were quantified and compared. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG), Domain and Clusters of Orthologous Groups (COG) analyses revealed key groups of proteins, pathways and domains differentially regulated during diapause preparation and overwintering diapause maintenance phases in this mosquito, including major shifts in energy production and conversion, fatty acid metabolism, the citrate (TCA) cycle, and the cytoskeletal reorganization pathway. Our results provide novel insight into the molecular bases of diapause in mosquitoes and corroborate previously reported diapause-associated features in invertebrates. More interestingly, the phototransduction pathway exists in Cx. pipiens pallens, in particular, actin, rather than other proteins, appears to have substantial role in diapause regulation. In addition, the differential changes in calmodulin protein expression in each stage implicate its important regulatory role of the Cx. pipiens pallens biological clock. Finally, 24 proteins were selected for verification of differential expression using a parallel reaction monitoring strategy. The findings of this study provide a unique opportunity to explore the molecular modifications underlying diapause in mosquitoes and might therefore enable the future design and development of novel genetic tools for improving management strategies in mosquitoes.
Subject terms: Protein-protein interaction networks, Entomology
Introduction
Culex pipiens pallens is the primary vector of Wuchereria bancrofti filariasis, Japanese encephalitis virus, West Nile virus and Rickettsia felis in China1,2. Cx. pipiens pallens mosquitoes overwinter in a state of adulthood diapause3, and overwintering mosquitoes play significant roles in the population dynamics of these species and in the spread and prevalence of mosquito-borne diseases4. Therefore, clarifying overwintering diapause mechanisms is essential for improving mosquito management strategies.
The results of previous studies have increased our knowledge of the molecular regulatory mechanisms of diapause preparation, maintenance and termination, revealing several physiological themes that appear to be shared across the diapause response of multiple insect species, including shifts in metabolism5–8, increased lipid synthesis and storage9–11, upregulation of stress-response genes12,13, hormonal regulation during diapause induction12,14–20, and changes in insulin signalling8,19,21–23. Most previous studies on the molecular mechanisms of diapause regulation have been performed at the transcriptional level, leading to exciting progress with regard to the transcriptional basis of diapause in several insect taxa7,21,24–35. However, an overall relatively low correlation between the expression levels of mRNAs and the abundance of various proteins has been reported in a comparison of prediapause and non-diapause locust eggs31. As diapause has evolved independently multiple times during the evolutionary history of insects, it is unclear to what extent metabolic patterns are conserved across species that exhibit the same or different diapause syndromes. Overall, a single suite of mechanisms regulating diapause across all species and developmental stages is unlikely36, yet our understanding of the molecular mechanisms involved in generating, maintaining, and breaking diapause is still highly fragmentary14. This is especially true because proteomic information on diapause is limited and the mechanism by which organisms measure and interpret photoperiod remains completely unresolved and controversial. Studies on diapause-related changes in expression of proteins relevant to physiological processes can facilitate a better understanding of the specific mechanisms involved in diapause-driven physiological changes and their physiological responses to physical environmental cues as well as elusive seasonal adaption.
With the advent of highly advanced proteomic platforms based on isobaric tags for relative and absolute quantification (iTRAQ)37, proteomics allows for screening differentially expressed proteins at a large scale, which ultimately provides a direct measurement of protein expression levels and insight into the activity of relevant proteins. Changes in protein expression during diapause have mainly been explored using 2-dimensional gel electrophoresis (2-DE)38–43 and shotgun proteomics44 and using Itraq31,45 on other insects. In contrast, there are no reports of proteomic analysis of diapause in mosquitoes thus far.
In this study, iTRAQ was used to investigate the proteome of Cx. pipiens pallens collected in July 2016 (representing nondiapausing, SUM), November 2016 (diapause preparation, BW) and March 15–19, 2017 (the overwinter diapause maintenance stage of female mosquitoes, AW). The results reveal novel insight into the metabolic processes occurring in these phases and enhance our understanding of the mechanisms underlying overwintering diapause in mosquitoes. These findings may serve as a foundation and provide a platform for developing novel vector control methods based on genetic or chemical disruption of such key traits46.
Results
Ovarian development, lipid storage status, protein identification and expression of different protein (EDP) screening
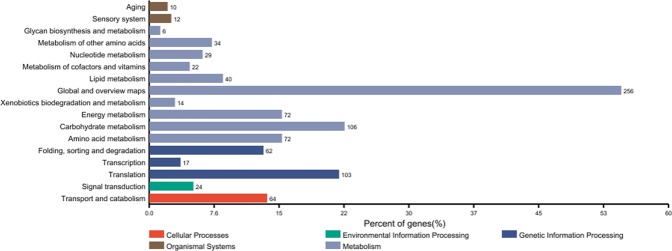
Ovarian development and lipid storage status were monitored (Table 1, Fig. 1). A total of 1,030 proteins were identified, among which quantitative information was obtained for 1,020. All the proteins were annotated by Gene Ontology, KEGG, COG, and Domain and Subcellular Location analyses (Supplementary Table 1). Interestingly, ageing (10 members) and sensory system (12 members) pathways were identified in KEGG pathway analysis (Fig. 2). There were 244 and 174 differentially expressed proteins identified between AW vs. BW and BW vs. SUM, respectively.
Table 1.
Dissection of Cx. pipiens pallens.
| Date | No. of dissections | Percent of fat bodies (%) | Percent of ovarian development (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | ++ | +++ | I | II | III | IV | V | |||
| 2016 | Nov. 1–8 | 105 | 8.6 | 43.8 | 47.6 | 98.1 | 1.9 | 0 | 0 | 0 |
| Nov. 12–20 | 88 | 5.7 | 30.7 | 63.7 | 97.7 | 0 | 0 | 2.3 | 0 | |
| Nov. 23–29 | 48 | 0 | 29.1 | 70.9 | 100 | 0 | 0 | 0 | 0 | |
| Dec. 3–7 | 79 | 0 | 11.4 | 88.6 | 100 | 0 | 0 | 0 | 0 | |
| Dec. 11–20 | 76 | 1.3 | 9.2 | 89.5 | 100 | 0 | 0 | 0 | 0 | |
| Dec. 25–30 | 135 | 0 | 10.4 | 89.6 | 100 | 0 | 0 | 0 | 0 | |
| 2017 | Jan. 4–9 | 120 | 0 | 5.8 | 94.2 | 100 | 0 | 0 | 0 | 0 |
| Jan. 11–19 | 146 | 0 | 5.5 | 94.5 | 100 | 0 | 0 | 0 | 0 | |
| Jan. 21–22 | 86 | 0 | 1.2 | 98.8 | 100 | 0 | 0 | 0 | 0 | |
| Feb. 8 | 5 | 0 | 0 | 100 | 100 | 0 | 0 | 0 | 0 | |
| Mar. 15–19 | 12 | 0 | 33.3 | 66.7 | 100 | 0 | 0 | 0 | 0 | |
| Mar. 23–28 | 12 | 41.7 | 50 | 8.3 | 0 | 100 | 0 | 0 | 0 | |
| Apr. 1–9 | 20 | 60 | 40 | 0 | 0 | 95 | 0 | 0 | 5 | |
| Apr. 11–16 | 6 | 100 | 0 | 0 | 16.7 | 0 | 0 | 0 | 83.3 | |
| Total | 938 | 4.1 | 16.1 | 79.8 | 95.6 | 3.5 | 0 | 0.2 | 0.7 | |
“+”: A few lipid droplets can be seen under the microscope.
“+++”: More lipid droplets under the microscope.
“++++”: A stretch of lipid droplets under the microscope.
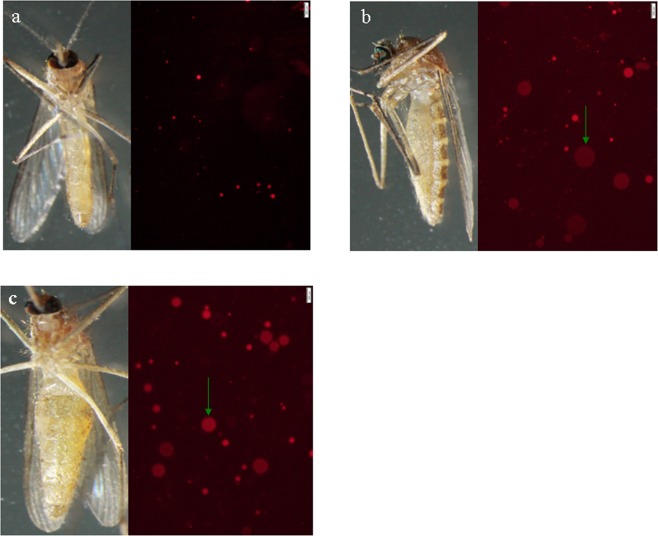
Figure 1.
(a) Slim females and Nile Red staining of lipid droplets in SUM adult females. (b,c) Fat females and Nile Red staining of lipid droplets in BW and AW adult females, respectively. Green arrow: larger lipid droplets.
Figure 2.
Statistics of differential proteins for KEGG annotation. The x-axis shows the percentage of proteins contained in different metabolic types; the y-axis shows the different metabolic types and number of proteins involved.
Protein expression patterns and function Clustering
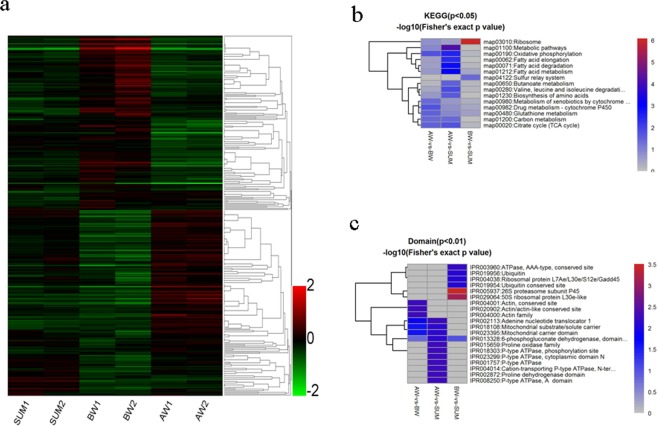
We employed hierarchical clustering to characterize changes in expression during the diapause preparation and overwintering diapause maintenance phases compared to a nondiapause phase control. All quantified proteins revealed clearly distinguishable increasing or decreasing expression during BW and AW (Fig. 3). Comparing the pattern of expression of different proteins between SUM, BW and AW, almost all of the identified proteins overlapped between stages, presenting a continuous dynamic of quantitative changes at different time points. Although the fold enrichment of each protein was quite small, almost all protein enrichment changes at the peak and the lowest valley were in BW, indicating that the abundance of various proteins changes in response to preparation for overwintering in Cx. pipiens pallens and that the diapause preparation process alters the normal developmental pathway into an alternative one. The individual ecological phenomenon of overwintering diapause was found to be very important in the long-term evolution of mosquitoes (Fig. 4). Interestingly, the phototransduction pathway exists, implicating an important regulatory role in this mosquito (Fig. 5). Furthermore, the differential changes in calmodulin protein expression in each group suggests a critical regulatory role associated with Cx. pipiens pallens’s biological clock. Of note is the differentially expressed protein actin in the phototransduction pathway. Actin-87E (B0WEY5), actin (B0FBP2) and actin (A0A173GY68) were downregulated in BW vs. SUM but upregulated in AW vs. BW; actin-2 (B0WEY6) was downregulated in AW vs. BW. These findings indicate that actin may play an important role in the regulation of this process in the phototransduction pathway.
Figure 3.
Clustering of protein expression patterns and functional enrichment. (a) Differential protein relative expression matrix heatmap of the three groups; the colour in the figure indicates the relative expression level of the protein in the sample. The red colour indicates that the protein has a higher expression level in the sample, and the green colour represents a lower expression level. Colour bars represent specific expression abundance. (b,c) Functional enrichment-based clustering for protein groups in KEGG and Domain. Clustering of the results of differential protein enrichment in different groups. The colour −log10 (Fisher exact test p value) represents the credibility of enrichment.
Figure 4.
Comparison of protein expression pattern clusters between SUM, BW and AW. For the expression trend line graph of each sub-cluster, the x-axis is the comparison sample group, and the y-axis is the relative expression level of the protein in the group of samples. Each line in the figure represents a protein, and the different colour representations show the relationship between relative expression and the mean value. Each graph shows one type of expression pattern, a trend that reflects changes in the expression of this group of proteins.
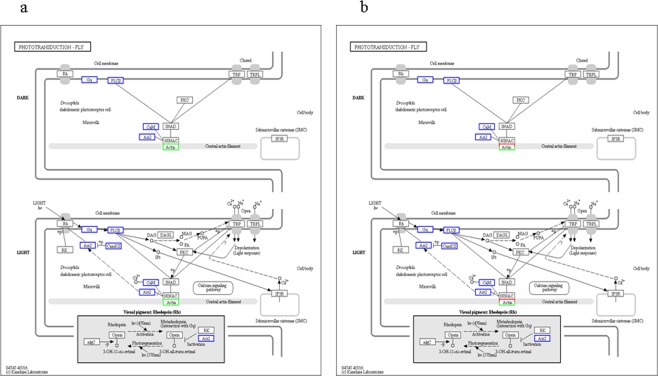
Figure 5.
(a) Representative KEGG phototransduction pathway in BW vs. SUM, map04745, and (b) phototransduction pathway in AW vs. BW, map04745; the rectangular nodes in the figure represent gene products. The blue border belongs to the background proteins, and the white colour indicates proteins not identified in this experiment. The red/green colours in the figure indicate to the differentially expressed proteins detected in this study, with red representing upregulated proteins and green downregulated proteins. Half red and half green indicates both upregulated and downregulated proteins for that gene product74–76.
PPI network analysis
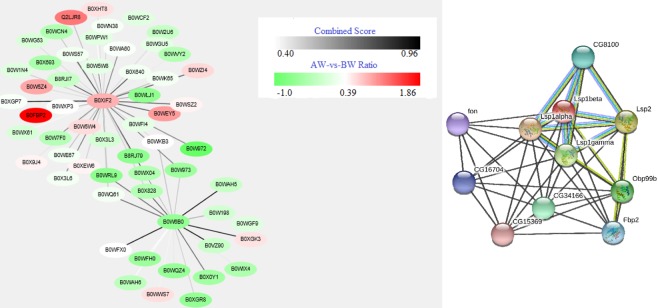
We then used STRING, which reveals functional associations between two groups, to further analyse the functional correlation of differentially expressed proteins between each group (Fig. 6). The mean combined score was 0.589; the maximum combined score between calmodulin (B0XIF2) and serine/threonine-protein phosphatase (B0XGP7) was 0.931, and the maximum combined score between glutathione S-transferase 1–6 (B0W6B0) and glutathione S-transferase (B0VZ90) was 0.952. The maximum and minimum ratio values for the AW and BW groups were 1.85 (CPIJ006534-PA, B0FBP2 (actin)) and −1.0 (CPIJ003588-PA, B0W972 (ubiquitin)), respectively.
Figure 6.
Calmodulin (B0XIF2), glutathione s-transferase (B0W6B0) and LSP-1 beta (B0XHC4) were revealed by Cytoscape. (a) Each circle in the figure represents a protein; the ratio value is the log2 (FC) value of the comparison group AW vs. BW, and the shade of the colour between the genes represents the combined score. (b) The interactions between expressed proteins are indicated by the connecting line, with the thickness of an edge indicating the confidence in that associations (thicker line indicates more evidence for interaction). Different line colours represent the types of evidence for the association: green = neighbourhood; red = gene fusion; pink = experiments; light green = text mining; blue = cooccurrence; dark blue = coexpression; purple = homology. Circle nodes indicate different proteins.
Verification and validation of differentially expressed proteins by a parallel reaction monitoring strategy
Using a parallel reaction monitoring (PRM)-quantifiable strategy, 24 differentially expressed proteins of Cx. pipiens pallens were confirmed. All proteins, peptides, and measured peptide peak areas are listed in Supplementary Table 2 and Fig. 7.
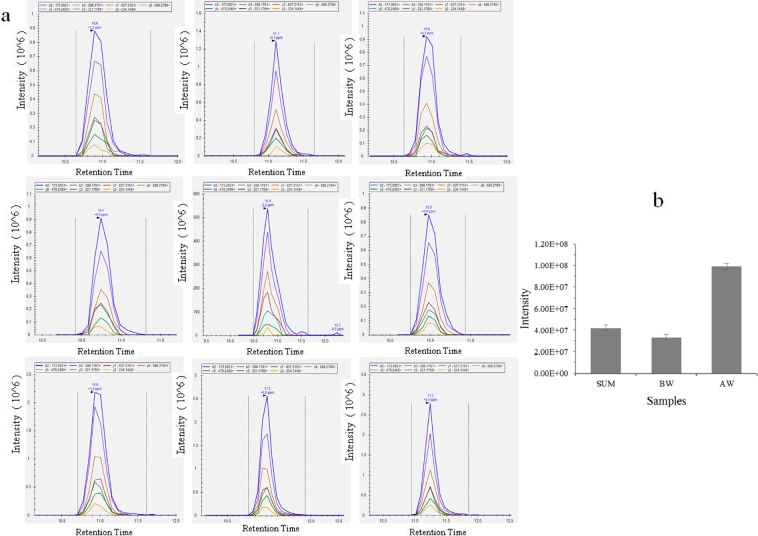
Figure 7.
(a) PRM for the abundance of glycerol-3-phosphate dehydrogenase was determined. Graphs displaying chromatograms of fragment ions extracted from the peptide TALVEADDFASGTSSK. The mass measurement error and retention time of the most intense transition are annotated above the peak. (b) Quantification performance of PRM analyses of the glycerol-3-phosphate dehydrogenase peptide. Highly consistent results were obtained, reflecting the excellent agreement between the intensities of peptides.
Discussion
Because most insects do not feed during the long period of diapause and increase lipid stores are often associated with hardiness against metabolic stress8,47–50, fat storage in diapause preparation and regulation of its utilization are important for successful overwintering. From November 2016, most Cx. pipiens pallens female mosquitoes entered a diapause preparation phase, accumulated more lipid droplets, arrested follicular development at stage I and maintained this stage until March 15–19, 2017. AW and BW female Cx. pipiens pallens mosquitoes were conspicuously larger and fatter than their SUM counterparts, and larger lipid droplets were observed in AW and BW. Indeed, no large fat lipid droplets were found in the SUM groups, revealing a dramatic increase in fat storage and the number of fat body cells in AW and BW of Cx. pipiens pallens (Table 1, Fig. 1), consistent with previous findings that Culex pipiens pupae and adults are larger and contain more fat than do their nondiapausing counterparts programmed for adult diapause51.
Because diapause is often accompanied by a decrease in metabolism, in accordance with reduced metabolism and energy production and an overall metabolic reduction, KEGG pathway enrichment analysis in BW vs. SUM specifically revealed downregulation of metabolic pathways, fatty acid biosynthesis, oxidative phosphorylation, starch and sucrose metabolism and phagosome pathways (Supplementary Figs 1–5). As reported previously in Cx. pipiens, fatty acid synthase is upregulated and fat metabolism genes, including multiple kinetic classes and genes involved in β-oxidation, an energy-generation step, are suppressed in early diapause21. Such findings are consistent with global suppression of the expression of proteins involved in fatty acid synthesis found in recent high-throughput genomic studies during other forms of hypometabolism, such as hibernation in ground squirrels52 and the mosquito Aedes albopictus27.
Redirection of energy production is among the defining characteristics of diapause53,54; however, a previous study associated upregulation of genes involved in fatty acid synthesis and sequestration at the molecular level in Cx. pipiens at the onset of diapause21. How can these two views be reconciled? It should be noted that the above study examined only a single time point during diapause; in contrast, the more comprehensive investigation of the functional roles of proteins in our study, along with our evaluation of the full course of diapause, would likely result in a clearer understanding of the intriguing diapause phenotype. Overall, this pattern suggests that overall metabolic and energy production rates are lower in BW, thus enabling the mosquito to economically utilize energy.
Energy metabolism is crucial for the survival of diapausing insects throughout winter. Consistent with these considerations, energy demands are fuelled by lipid stores, and Cx. pipiens pallens females begin to utilize triacylglyceride stores, convert stored fat to free fatty acids, and transport these fatty acids to mitochondria to meet energetic needs. We found that 43 proteins associated with energy production and conversion were upregulated during prolonged overwintering in female mosquitoes, indicating a general activation of lipid consumption. As shown in Supplementary Fig. 6, the six most enriched GO terms regarding biological processes of upregulated proteins are related to the oxidation-reduction process, generation of precursor metabolites and energy, energy derivation by oxidation of organic compounds, the single-organism metabolic process, the single-organism process, and cellular respiration, with high fold enrichment (log2) and a significant Fisher’s exact test p-value (-log10). KEGG pathway enrichment analysis provided additional evidence, specifically regarding the upregulation of metabolic pathways, oxidative phosphorylation, the citrate cycle (TCA cycle), carbon metabolism, fatty acid metabolism, starch and sucrose metabolism, fatty acid biosynthesis, glycolysis/gluconeogenesis and phototransduction (Supplementary Figs 7–15). In particular, differential gene expression analysis in diapausing Cx. pipien females provided evidence for increases in two separately upregulated glycogen-debranching enzymes involved in glycolysis and gluconeogenesis metabolism30. Moreover, examination of the enriched proteins in the over-represented energy production category and oxidative phosphorylation pathway showed changes at the protein level in overwintering diapause conditions and increased levels of several nicotinamide adenine dinucleotide dehydrogenase (NADH) subunits (complex), suggesting that overwintering diapause enhances energy production through the oxidative phosphorylation pathway.
In AW vs. BW, the components of the TCA pathway (Supplementary Fig. 10) upregulated in overwintering diapause are largely proteins that are involved directly in oxidative metabolism in the mitochondria and in alternative mitochondrial or cytosolic reactions that support glycolysis and gluconeogenesis. Conversely, the proteins acting in the initial steps in the TCA cycle involving the conversion of pyruvate from glycolysis to acetyl-CoA appear to be suppressed (pyruvate dehydrogenase E1 component subunit alpha, B0W2T1). This step is a key gateway for entry of carbon flux from the glycolytic pathway into the mitochondria and is responsible for catalysing the conversion of pyruvate and acyl-CoA to form acetyl-CoA. Isocitrate dehydrogenase [NADP] (B0VZM6), which was observed to be downregulated in AW vs. BW, is a key enzyme regulating the reaction between isocitric acid and NAD, a rate-limiting step of the TCA cycle. Downregulation of pyruvate dehydrogenase E1 component subunit alpha and isocitrate dehydrogenase [NADP] in overwintering diapause would enable mosquitoes to economically utilize energy reserves.
Our experiments also indicate that expression of certain cytoskeletal proteins is affected by overwintering diapause in Cx. pipiens pallens, as a relatively large number of cytoskeleton-associated proteins with a diversity of functions showed highly differential abundance in AW vs. BW and BW vs. SUM (Fig. 3c). For example, actin-87E (B0WEY5), actin (A0A173GY68, B0FBP2), myosin light chain 1 (B0X1Z6), tropomodulin (B0XBH8) and fimbrin/plastin (B0WA60) and tubulin beta chain (Q15G69) were downregulated in BW vs. SUM. Examination of component-enriched lists in the cytoskeleton category in AW and BW showed upregulation of spectrin alpha chain (B0X640), actin-87E (B0WEY5), tropomyosin invertebrate (B0X3L6), actin (A0A173GY68, B0FBP2), myosin light chain 1 (B0X1Z6), talin-1 (B0WXE5), and tubulin alpha chain (B0WK65). Actins constitute a large family of proteins with specific localization and regulation, and thus, levels of different actins might be elevated while levels of others might be decreased concomitantly during diapause55. These results correspond to observations in diapausing adult Cx. pipiens, whereby actin was found to be upregulated in early diapause but returned to low levels by late diapause56. According to data for wheat (Triticum aestivum), actin downregulation may contribute to the increased cold tolerance associated with dormancy57. In addition, a brain-specific actin was reported to be downregulated during pharate larval diapause of the gypsy moth Lymantria dispar58. Interestingly, in Drosophila americana, actinD1 expression levels have also been shown to reflect the observed differences in the life span of both diapausing and non-diapausing flies59. In our study, actin, conserved site, actin family, actin/actin-like conserved site were the most enriched domains for upregulated proteins in AW vs. BW (Supplementary Fig. 16). Taken together, our observations confirm the notion that cytoskeleton proteins show altered expression and cellular cytoskeletal rearrangement, suggesting fortification of structural components in response to overwinter diapausing and leading to increased cold tolerance and desiccation resistance in diapausing Cx. pipiens pallens.
Proline, a well-known energy source and cryoprotectant in some insects6, increases cold tolerance in insects; in fact upregulation of proline is commonly considered a potentially important source of metabolic fuel for overwintering60–62. In this study, pyrroline-5-carboxylate dehydrogenase (B0WKF4) and proline oxidase (B0WRS5) were upregulated in AW vs. BW, and upregulation of pyrroline-5-carboxylate reductase has also been reported for Culex pipiens30.
Calmodulin, which can bind the second messenger calcium and affect a number of target proteins, is an important regulatory protein that adjusts a wide range of cellular processes and development63. Chilling from 25 °C to 0 °C evoked a 40% increase in intracellular calcium concentrations in the tracheal cells of the freeze-tolerant Eurosta solidaginis and increased the activity of calcium/calmodulin-dependent protein kinase II (CaMKII)64. Additionally, calmodulin levels were decreased in pupal diapause induction in Helicoverpa armigera65, and calmodulin and calmodulin protein kinase II were decreased in larval diapause initiation40. Calcineurin A and FK506-binding protein, which are related to Ca2+ release, were uniquely identified in diapausing Bombyx mori eggs66. Moreover, the calcium ATPase gene was upregulated in 3-day-diapausing Tetranychus urticae67.
Interestingly, in the present study, the phototransduction signalling network that perceives day length and ultimately translates seasonal information into the developmental programme we recognize as diapause in Cx. pipiens pallens exhibited differential changes in calmodulin protein (B0XIF2) expression (Fig. 5) in each group, implicating a regulatory role for calmodulin in phototransduction pathways. Actin is a critical link in the regulation of proteins in the phototransduction pathway response to seasonal alteration in Cx. pipiens pallens, though details and insight into how calmodulin and actin affect diapause at both local and systemic levels and to what extent calmodulin signals are involved in diapauses of different developmental stages are lacking.
Diapause appears to have evolved multiple times in insect lineages, and photoperiodic signals are effective at evoking diapause only during the sensitive period of an insect’s life cycle, which is species specific and may occur well before the actual diapause stage14. Thus, we can anticipate species variation in how this calmodulin signalling pathway is exploited for regulating diapause in different species. The exciting prospect is that this is the first report of an association between calmodulin and actin with the mosquito biological clock, with connections to insect diapause at many levels, and these findings provide a first step towards understanding the protein regulatory network of phototransduction in diapausing mosquitoes. Thus, our results provide a unique opportunity to explore the molecular modifications underlying overwintering diapause in Cx. pipiens pallens, a major goal for deciphering the molecular mechanisms of diapause.
Additional interesting differences include larval serum proteins (Fig. 6), which are storage proteins that participate in metamorphosis by supplying energy and serving as a source of amino acids. Furthermore, protein LSP-1 beta (B0XHC4) was dramatically increased in BW, with a considerably lower abundance of protein LSP-1 beta in AW, in which its expression level was even lower than in SUM. Therefore, Cx. pipiens pallens likely synthesizes many serum haemolymph proteins not only to accelerate fat storage but also to strengthen its immune system sensitivity in the high-efficiency ingestion phage in preparation for overwinter. Although protein LSP-1 beta has been indicated as among the primary serum proteins in different physiological phages of Cx. pipiens pallens, including the larva, its function remains unknown. In addition, our results support the conclusion that increased expression of the LSP-1 beta protein in BW suggest that these proteins may serve as new targets for continued research towards understanding diapause in Cx. pipiens pallens and for developing genetic tools for improving mosquito management strategies.
Glutathione s-transferase (B0W6B0) (Supplementary Fig. 17), a stress response protein, was found to be highly expressed in the spruce budworm Choristoneura fumiferana throughout diapause, though it levels declined at the end of diapause68. Glutathione s-transferase is also involved in xenobiotic biodegradation and metabolism, and Cx. pipiens pallens virulence varies among SUM, BW and AW.
Conclusions
In summary, this study represents the first large-scale investigation of global protein expression differences during the diapause preparation and the overwintering diapause maintenance phases of the mosquito Cx. pipiens pallens. Using iTRAQ, we found major metabolic changes in several pathways and expression changes in important proteins involved in overwintering diapause regulation, revealing new insight into the physiology and regulation of overwintering diapause in Cx. pipiens pallens. These results provide comprehensive proteome insight and expand our understanding of the molecular mechanisms influencing overwintering diapause in Cx. pipiens pallens. This is the first study to identify the involvement of calmodulin, actin, and the phototransduction pathway throughout overwintering diapause in Cx. pipiens pallens, and LSP-1 beta, glutathione s-transferase may also have roles in this process. We anticipate that these results will prove useful in probing the molecular events that may be common to diapause in insects of different taxa and developmental stages.
Materials and Methods
Mosquito collection, dissection, and fat body staining with Nile red
Summer non-diapausing female Cx. pipiens pallens mosquitoes were collected in Weifang District in July 2016. Cx. pipiens pallens mosquitoes in diapause preparation and overwintering diapause maintenance stages were also collected from November 2016 until the 16th of April 2017 in Weifang District, Shandong Province until most females had terminated diapause. Collections were conducted during daylight hours from various resting sites, including animal sheds, barns, drains, large resting boxes, and other locations in and around people’s houses3; the samples were brought to the insectary after morphological identification. For each sampling occasion, some Cx. Pipiens pallens females were dissected under a dissecting microscope to determine their diapause status, and lipid storage status was monitored according to Zhao’s study69. The fat bodies were smashed using an anatomical needle and divided into 4 types: disperse, conglobation, small block, and large block. The number of lipid droplets was observed under a microscope with a 400× field of vision. Lipid droplets <50, 50–100, 100–200 and 200–400 were classified as minute, less, medium and more, respectively. Ovarian developmental stages were defined according to methods described by Spielman and Wong70 and Sim and Denlinger19. Fat content was monitored by Nile Red staining19. Both dissected and un-dissected mosquito samples were sorted into pools, frozen in liquid nitrogen and preserved for further analysis at −80 °C.
Protein extraction
For protein extraction, Cx. pipiens pallens mosquitoes in the diapause preparation (collected in November 2016) and overwintering diapause maintenance (collected in February 2017) stages (each from 4~5 adult females) were added to 1:50 (W/V) lysis buffer (8 M urea, 2 mM EDTA, 10 mM DTT and 1% protease inhibitor cocktail) and homogenized thoroughly with a tissue grinder. The samples were sonicated for 3 min and centrifuged at 13,000 × g (4 °C) for 10 min to remove debris, and the protein in the supernatant was precipitated with cold acetone for 3 h at −20 °C. After centrifugation at 4 °C and 12,000 × g for 10 min, the protein pellets was redissolved with urea buffer (8 M urea, 100 mM TEAB). The protein concentration was determined using a Modified Bradford Protein Assay Kit according to the manufacturer’s instructions.
Protein reduction, alkylation, isobaric labelling and sample cleanup
For digestion, 100 μg protein from each sample was first reduced with 10 mM DTT at 37 °C for 60 min and then alkylated with 25 mM iodoacetamide (IAM) at room temperature for 45 min in darkness. The protein pool of each sample was digested with Sequencing Grade Modified Trypsin with a ratio of protein:trypsin of 50:1 at 37 °C overnight and 100:1 for a second digestion for 4 h. After trypsin digestion, peptides were desalted using a Strata X SPE column, vacuum-dried, reconstituted in 25 μL 500 mM TEAB and processed according to the manufacturer’s protocol for the 8-plex iTRAQ kit (Applied Biosystems, Inc.)71. Briefly, one unit of iTRAQ reagent was added to the peptide solution after thawing and dissolving in 50 μL isopropanol. The peptide mixtures were incubated for 2 h at room temperature and then pooled and dried by vacuum centrifugation. The dried and labelled peptides were reconstituted with HPLC solution A (2% ACN, pH 10) and then fractionated by high-pH reverse-phase HPLC using a Waters Bridge Peptide BEH C18 (130 Å, 3.5 μm, 4.6 × 250 mm). Briefly, peptides were first separated by a gradient of 2% to 98% acetonitrile in pH 10 at a speed of 0.5 mL/min over 88 min into 60 fractions. The peptides were then combined into 20 fractions and dried by vacuum centrifugation. The peptide fractions were desalted using a Ziptip C18 according to the manufacturer’s instructions. The samples were dried under vacuum and stored at −20 °C until MS analyses were performed.
High-resolution LC-MS/MS analysis
NanoLC 1000 LC-MS/MS analysis was performed using a Proxeon EASY-nLC 1000 coupled to an LTQ-Orbitrap Elite. Trypsin digestion fractions were reconstituted in 0.1% FA and directly loaded onto a reversed-phase pre-column (Acclaim PepMap®100C18, 3 μm, 100 Å, 75 μm × 2 cm) at 5 μL/min in 100% solvent A (0.1 M acetic acid in water). Next, peptides eluted from the trap column were loaded onto a reversed-phase analytical column (Acclaim PepMap® RSLC C18, 2 μm, 100 Å, 50 μm × 15 cm). The gradient comprised an increase from 10% to 35% solvent B (0.1% FA in 98% ACN) over 60 min, 35% to 50% in 10 min and climbing to 100% in 5 min at a constant flow rate of 250 nL/min on an EASY-nLC 1000 system.
The eluent was sprayed via an NSI source at an electrospray voltage of 1.8 kV and then analysed by tandem mass spectrometry (MS/MS) using a LTQ-Orbitrap Elite. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS. Full-scan MS spectra (from m/z 350 to 1800) were acquired in the Orbitrap with a resolution of 60,000. Ion fragments were detected in the Orbitrap at a resolution of 15,000, and the 20 most intense precursors were selected for subsequent decision tree-based ion trap HCD fragmentation at a collision energy of 38% above a threshold ion count of 300 in the MS survey scan with 30.0-s dynamic exclusion. Full width at half maximum (FHMW) at 400 m/z using an AGC setting of 1e6 ions was used, and the fixed first mass was set as 100 m/z.
Parallel reaction monitoring analysis
A label-free targeted parallel reaction monitoring (PRM) method72 was adopted to verify the reliability of our label-based proteomics. A total of 24 differentially expressed proteins, including one internal standard housekeeping protein, were chosen. For the SUM, BW, and AW groups, equal amounts of protein samples were analysed for semi-quantitative measurements, and each group underwent three replications. Peak areas were extracted from PRM mass spectrum data using Skyline software73.
Data analysis
The resulting MS/MS raw data were searched against the Culex Taxonomy database (Taxon identifier: 7174, include 21082 protein sequences) downloaded from Uniprot using Sequest software integration in Proteome Discoverer (version 1.3, Thermo Scientific). Trypsin was chosen as the enzyme, and two missed cleavages were allowed. Carbamidomethylation (C) was set as a fixed modification, and oxidation (M) and acetylation in N-Term were set as variable modifications. The searches were performed using a peptide mass tolerance of 20 ppm and a product ion tolerance of 0.02 Da, resulting in a 1% false discovery rate (FDR).
The Gene Ontology (GO) annotation proteome was derived from the UniProt-GOA database (www.http://ebi.ac.uk/GOA/). After obtaining the ID of a protein based on a protein search, the ID was converted into the ID of the UniProtKB database; GO annotation information for Culex quinquefasciatus was located based on the UniProKB ID (ftp://ftp.pir.georgetown.edu/databases/idmapping/uniprot_idmapping/idmapping_selected.tab.gz). Among the 1030 proteins identified, 841 were annotated. KEGG Pathway is part of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database74–76, which is a reference database for pathway mapping. KEGG Pathway analyses of identified proteins were extracted using the Search pathway tool in the KEGG Mapper platform (http://www.genome.jp/kegg/mapper.html); 711 proteins had corresponding KO numbers in the KEGG database with the species Culex quinquefasciatus, and 510 KO numbers were mapped to pathways. Proteins were identified by BLAST alignment and the COG protein database to predict function and conduct statistical analyses with a p value < 1e−5. Protein domain annotation was assessed using InterProScan77. GO, KEGG pathway and Domain enrichment of differential protein expression for all identified proteins was evaluated using the two-tailed Fisher’s exact test. Correction for multiple hypothesis testing was carried out under standard FDR control methods, and pathways with a corrected p value < 0.05 were considered the most significant. A heatmap was generated by the software Heml 1.0.378. The expression level of the differentially expressed proteins was taken as log2 for relative expression; the historical clustering method had an average linkage, and the similarity metric was the Pearson distance. The expression pattern of the protein was completed using the timeclust function in the R package ‘TCseq’. We also present the STRING network of some known/novel protein-protein interactions as an evidence view by using the String 9.0 (Search Tool for the Retrieval of In-teracting Genes/Proteins) database of physical and functional interactions (http://string-db.org/)79. Among 244 proteins, 199 were found in the STRING database for the species Culex quinquefasciatus. We selected the combined score >0.400 between nodes for PPI with Cytoscape (version 3.0), part of which is shown in the paper but only for important proteins.
Supplementary information
Acknowledgements
We thank Kai Yu, Xiaoli Huang, Yajun Guo of Shanghai MHelix BioTech Co., Ltd. for their assistance with experiments. We also thank Hongmei Liu and Haifang Wang (Shandong Institute of Parasitic Diseases) for their assistance with insect rearing and advice on the data analysis, respectively. The present study was supported by the National Natural Science Foundation of China (NSFC) (Grant No. 81672059, 81871685) and the Innovation Project of Shandong Academy of Medical Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Z.C.X. and G.M.Q. conceived and designed the experiments. M.F. contributed reagents/materials/analytic tools. W.D.D. and S.G.H. performed the experiments. H.X.L. collected and analysed the data. C.P., L.G.Z., G.X.X., L.L.J. W.H.W. analysed the data. Z.C.X. wrote the paper. M.F. helped review and edit the final paper. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chongxing Zhang, Email: chongxingzhang@aliyun.com.
Maoqing Gong, Email: gmq2005@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42961-w.
References
- 1.Turell MJ. Members of the Culex pipiens complex as vectors of viruses. The American Mosquito Control Association. 2012;28:123–126. doi: 10.2987/8756-971X-28.4.123. [DOI] [PubMed] [Google Scholar]
- 2.Zhang, J. et al. First report of Rickettsia felisin China. BMC Infectious Diseases14, 10.1186/s12879-014-0682-1 (2014).
- 3.Liu LJ, et al. Overwintering of Culex pipiens pallens (Diptera Culicidae) in Shandong, China. Journal of Entomological Science. 2016;51:314–320. doi: 10.18474/JES15-38.1. [DOI] [Google Scholar]
- 4.Denlinger DL, Armbruster PA. Mosquito Diapause. Annual Review of Entomology. 2014;59:73–93. doi: 10.1146/annurev-ento-011613-162023. [DOI] [PubMed] [Google Scholar]
- 5.Kukal O, Denlinger DL, Lee RE. Developmental and metabolic changes induced by anoxia in diapausing and nondiapausing flesh fly pupae. Journal of Comparative Physiology B-Biochemical Systemic and Environmental Physiology. 1991;160:683–689. doi: 10.1007/BF00571268. [DOI] [Google Scholar]
- 6.Michaud MR, Denlinger DL. Shifts in the carbohydrate, polyol, and amino acid pools during rapid cold-hardening and diapause-associated cold-hardening in flesh flies (Sarcophaga crassipalpis): a metabolomic comparison. Journal of Comparative Physiology B. 2007;177:753–763. doi: 10.1007/s00360-007-0172-5. [DOI] [PubMed] [Google Scholar]
- 7.Ragland GJ, Denlinger DL, Hahn DA. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proceedings of the National Academy of Sciences. 2010;107:14909–14914. doi: 10.1073/pnas.1007075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn DA, Denliner DL. Energetics of insect diapause. Annual Review of Entomology. 2011;56:103–121. doi: 10.1146/annurev-ento-112408-085436. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds JA, Hand SC. Embryonic diapause highlighted by differential expression of mRNAs for ecdysteroidogenesis, transcription and lipid sparing in the cricket Allonemobius socius. Journal of Experimental Biology. 2009;212:2074–2083. doi: 10.1242/jeb.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robich RM, Denlinger DL. Diapause in the mosquito Culex pipiens evokes a metabolic switch from blood feeding to sugar gluttony. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15912–15917. doi: 10.1073/pnas.0507958102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquitoCulex pipiens. Insect Molecular Biology. 2009;18:325–332. doi: 10.1111/j.1365-2583.2009.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denlinger, D. L., Yocum, G. D. & Rinehart, J. L. Hormonal control of diapause. Gilbert, L., Iatrou, K., Gill, S., editors. Elsevier Press, Amsterdam, The Netherlands, 615–650 (2005).
- 13.Rinehart JP, et al. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proceedings of the National Academy of Sciences. 2007;104:11130–11137. doi: 10.1073/pnas.0703538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denlinger DL. Regulation of diapause. Annual Review of Entomology. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- 15.Horie Y, Kanda T, Mochida Y. Sorbitol as an arrester of embryonic development in diapausing eggs of the silkworm, Bombyx mori. Journal of Insect Physiology. 2000;46:1009–1016. doi: 10.1016/S0022-1910(99)00212-7. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda M, et al. Induction of embryonic diapause and stimulation of ovary trehalase activity in the silkworm, Bombyx mori, by synthetic diapause hormone. Journal of Insect Physiology. 1993;39:889–895. doi: 10.1016/0022-1910(93)90122-8. [DOI] [Google Scholar]
- 17.Yamashita O. Diapause hormone of the silkworm, Bombyx mori: Structure, gene expression and function. Journal of Insect Physiology. 1996;42:669–679. doi: 10.1016/0022-1910(96)00003-0. [DOI] [Google Scholar]
- 18.Sim Cheolho, Denlinger David L. Juvenile hormone III suppresses forkhead of transcription factor in the fat body and reduces fat accumulation in the diapausing mosquito,Culex pipiens. Insect Molecular Biology. 2012;22(1):1–11. doi: 10.1111/j.1365-2583.2012.01166.x. [DOI] [PubMed] [Google Scholar]
- 19.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proceedings of the National Academy of Sciences. 2008;105:6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spielman A. Effect of synthetic juvenile hormone on ovarian diapause of Culex pipiens mosquitoes. Journal of Medical Entomology. 1974;11:223–225. doi: 10.1093/jmedent/11.2.223. [DOI] [PubMed] [Google Scholar]
- 21.Sim C, Denlinger DL. Transcription profiling and regulation of fat metabolism genes in diapausing adults of the mosquito Culex pipiens. Physiological Genomics. 2009;39:202–209. doi: 10.1152/physiolgenomics.00095.2009.-Culex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatar M, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 23.Williams KD, et al. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proceedings of the National Academy of Sciences. 2006;103:15911–15915. doi: 10.1073/pnas.0604592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz D, et al. Sympatric ecological speciation meets pyrosequencing: sampling the transcriptome of the apple maggot Rhagoletis pomonella. BMC Genomics. 2009;10:633. doi: 10.1186/1471-2164-10-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragland GJ, Egan SP, Feder JL, Berlocher SH, Hahn DA. Developmental trajectories of gene expression reveal candidates for diapause termination: a key life-history transition in the apple maggot fly Rhagoletis pomonella. Journal of Experimental Biology. 2011;214:3948–3960. doi: 10.1242/jeb.061085. [DOI] [PubMed] [Google Scholar]
- 26.Qi, X. et al. De novo transcriptome sequencing and analysis of Coccinella septempunctata L. in non-diapause, diapause and diapause-terminated states to identify diapause-associated genes. BMC Genomics16, 10.1186/s12864-015-2309-3 (2015). [DOI] [PMC free article] [PubMed]
- 27.Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. Deep sequencing reveals complex mechanisms of diapause preparation in the invasive mosquito, Aedes albopictus. Proceedings of the Royal Society B: Biological Sciences. 2013;280:2013.0143. doi: 10.1098/rspb.2013.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers P, et al. Divergence of the diapause transcriptome in apple maggot flies: winter regulation and post-winter transcriptional repression. The Journal of Experimental Biology. 2016;219:2613–2622. doi: 10.1242/jeb.140566. [DOI] [PubMed] [Google Scholar]
- 29.Kučerová, L. et al. Slowed aging during reproductive dormancy is reflected in genome-wide transcriptome changes in Drosophila melanogaster. BMC Genomics17, 10.1186/s12864-016-2383-1 (2016). [DOI] [PMC free article] [PubMed]
- 30.Kang DS, Cotton MA, Denlinger DL, Sim C. Comparative Transcriptomics Reveals Key Gene Expression Differences between Diapausing and Non-Diapausing Adults of Culex pipiens. PLOS ONE. 2016;11:e0154892. doi: 10.1371/journal.pone.0154892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu, X. et al. Transcriptomic and proteomic analysis of pre-diapause and non-diapause eggs of migratory locust, Locusta migratoria L. (Orthoptera: Acridoidea). Scientific Reports5, 10.1038/srep11402 (2015). [DOI] [PMC free article] [PubMed]
- 32.Emerson KJ, Bradshaw WE, Holzapfel CM. Microarrays Reveal Early Transcriptional Events during the Termination of Larval Diapause in Natural Populations of the Mosquito, Wyeomyia smithii. PLoS One. 2010;5:e9574. doi: 10.1371/journal.pone.0009574.g001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao, B. & Xu, W.-H. Identification of gene expression changes associated with the initiation of diapause in the brain of the cotton bollworm, Helicoverpa armigera. BMC Genomics12, 10.1186/1471-2164-12-224 (2011). [DOI] [PMC free article] [PubMed]
- 34.Etges WJ, et al. Transcriptional Differences between Diapausing and Non-Diapausing D. montana Females Reared under the Same Photoperiod and Temperature. Plos One. 2016;11:e0161852. doi: 10.1371/journal.pone.0161852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Poelchau MF, Armbruster PA. Global Transcriptional Dynamics of Diapause Induction in Non-Blood-Fed and Blood-Fed Aedes albopictus. PLOS Neglected Tropical Diseases. 2015;9:e0003724. doi: 10.1371/journal.pntd.0003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hand SC, Denlinger DL, Podrabsky JE, Roy R. Mechanisms of animal diapause: recent developments from nematodes, crustaceans, insects, and fish. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2016;310:R1193–R1211. doi: 10.1152/ajpregu.00250.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan C, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) Journal of Proteome Research. 2007;6:821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 38.Pavlides SC, Pavlides SA, Tammariello SP. Proteomic and phosphoproteomic profiling during diapause entrance in the flesh fly, Sarcophaga crassipalpis. Journal of Insect Physiology. 2011;57:635–644. doi: 10.1016/j.jinsphys.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Hedo M, Sánchez-López I, Eizaguirre M. Comparative analysis of hemolymph proteome maps in diapausing larvae and non-diapausing larvae of Sesamia nonagrioides. Proteome Sciences. 2012;10:58. doi: 10.1186/1477-5956-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Lu YX, Xu WH. Integrated proteomic and metabolomic analysis of larval brain associated with diapause induction and preparation in the cotton bollworm, Helicoverpa armigera. Journal of Proteome Research. 2012;11:1042–1053. doi: 10.1021/pr200796a. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Lu YX, Xu WH. Proteomic and metabolomic profiles of larval hemolymph associated with diapause in the cotton bollworm, Helicoverpa armigera. BMC Genomics. 2013;14:751. doi: 10.1186/1471-2164-14-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren XY ZL, et al. Proteomic research on diapause-related proteins in the female ladybird, Coccinella septempunctata L. Bulletin of Entomological Research. 2016;106:168–174. doi: 10.1017/S0007485315000954. [DOI] [PubMed] [Google Scholar]
- 43.Cheng W, et al. Proteomic analysis of pre-diapause, diapause and post-diapause larvae of the wheat blossom midge, Sitodiplosis mosellana (Diptera: Cecidomyiidae) European Journal of Entomology. 2009;106:29–35. doi: 10.14411/eje.2009.004. [DOI] [Google Scholar]
- 44.Wolschin F, Gadau J. Deciphering Proteomic Signatures of Early Diapause in Nasonia. PLOS ONE. 2009;4:e6394. doi: 10.1371/journal.pone.0006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan, Q.-Q. et al. Describing the Diapause-Preparatory Proteome of the Beetle Colaphellus bowringi and Identifying Candidates Affecting Lipid Accumulation Using Isobaric Tags for Mass Spectrometry-Based Proteome Quantification (iTRAQ). Frontiers in Physiology8, 10.3389/fphys.2017.00251 (2017). [DOI] [PMC free article] [PubMed]
- 46.Denlinger DL. Why study diapause? Entomological Research. 2008;38:1–9. doi: 10.1111/j.1748-5967.2008.00139.x. [DOI] [Google Scholar]
- 47.Adedokun TA, Denlinger DL. Metabolic reserves associated with pupal diapause in the flesh fly, Sarcophaga crassipalpis. Journal of Insect Physiology. 1985;31:229–233. doi: 10.1016/0022-1910(85)90124-6. [DOI] [Google Scholar]
- 48.Arrese EL, Soulages JL. Insect Fat Body: Energy, Metabolism, and Regulation. Annual Review of Entomology. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colinet H, Muratori F, Hance T. Cold-induced expression of diapause in Praon volucre: fitness cost and morpho-physiological characterization. Physiological Entomology. 2010;35:301–307. doi: 10.1111/j.1365-3032.2010.00743.x. [DOI] [Google Scholar]
- 50.Reynolds JA, Poelchau MF, Rahman Z, Armbruster PA, Denlinger DL. Transcript profiling reveals mechanisms for lipid conservation during diapause in the mosquito, Aedes albopictus. Journal of Insect Physiology. 2012;58:966–973. doi: 10.1016/j.jinsphys.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, Denlinger DL. Molecular structure of the prothoracicotropic hormone gene in the northern house mosquito, Culex pipiens, and its expression analysis in association with diapause and blood feeding. Insect Molecular Biology. 2011;20:201–213. doi: 10.1111/j.1365-2583.2010.01058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao CL, et al. Shotgun Proteomics Analysis of Hibernating Arctic Ground Squirrels. Molecular & Cellular Proteomics. 2010;9:313–326. doi: 10.1074/mcp.M900260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cellular and Molecular Life Sciences. 2010;67:2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hand SC, et al. Metabolic restructuring during energy-limited states: Insights from Artemia franciscana embryos and other animals. Journal of Insect Physiology. 2011;57:584–594. doi: 10.1016/j.jinsphys.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinehart JP, Robich RM, Denlinger DL. Isolation of diapause-regulated genes from the flesh fly, Sarcophagacrassipalpis by suppressive subtractive hybridization. Journal of Insect Physiology. 2010;56:603–609. doi: 10.1016/j.jinsphys.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 56.Kim, M., Robich, R. M., Rinehart, J. P. & Denlinger, D. L. Upregulation of two actin genes and redistribution of actin during diapause and cold stress in the northern house mosquito, Culex pipiens. Journal of Insect Physiology. (2006). [DOI] [PMC free article] [PubMed]
- 57.Ouellet F, Carpentier E, Cope MJTV, Monroy AF, Sarhan F. Regulation of a wheat actin-depolymerizing factor during cold acclimation. Plant Physiology. 2001;125:360–368. doi: 10.1104/pp.125.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee KY, Hiremath S, Denlinger DL. Expression of actin in the central nervous system is switched off during diapause in the gypsy moth, Lymantria dispar. Journal of Insect Physiology. 1998;44:221–226. doi: 10.1016/S0022-1910(97)00173-X. [DOI] [PubMed] [Google Scholar]
- 59.Reis M, Valer FB, Vieira CP, Vieira J. Drosophila americana diapausing females show features typical of young flies. PlosOne. 2015;10:e0138758. doi: 10.1371/journal.pone.0138758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimada, K. & Riihimaa, A. Cold-induced freezing tolerance in diapausing and non-diapausing larvae of Chymomyza costata (Diptera: Drosophilidae) with accumulation of trehalose and proline. CryoLetters. (1990).
- 61.Fields PG, et al. He effect of cold acclimation and deacclimation on cold tolerance, trehalose and free amino acid levels in Sitophilus granarius and Cryptolestes ferrugineus (Coleoptera) Journal of Insect Physiology. 1998;44:955–965. doi: 10.1016/S0022-1910(98)00055-9. [DOI] [PubMed] [Google Scholar]
- 62.Storey KB, Storey JM. Freeze tolerance in animals. Physiological Reviews. 1988;68:27–84. doi: 10.1152/physrev.1988.68.1.27. [DOI] [PubMed] [Google Scholar]
- 63.Cheung WY. Calmodulin plays a pivotal role in cellular regulation. Science. 1980;207:19–27. doi: 10.1126/science.6243188. [DOI] [PubMed] [Google Scholar]
- 64.Teets, N. M., Yi, S.X., Lee, R.E. Jr. & Denlinger, D. L. Calcium signaling mediates cold sensing in insect tissues. Proceedings of the National Academy of Sciences, 9154–9159 (2013). [DOI] [PMC free article] [PubMed]
- 65.Lu Y, Xu WH. Proteomic and Phosphoproteomic Analysis at Diapause Initiation in the Cotton Bollworm Helicoverpa armigera. Journal of Proteome Research. 2010;9:5053–5064. doi: 10.1021/pr100356t. [DOI] [PubMed] [Google Scholar]
- 66.Fan L, Lin J, Zhong Y, Liu J. Shotgun proteomic analysis on the diapause and non-diapause eggs of domesticated silkworm Bombyx mori. PLOS ONE. 2013;8:e60386. doi: 10.1371/journal.pone.0060386.g001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao JY, et al. Transcriptome and proteome analyses reveal complex mechanisms of reproductive diapause in the two-spotted spider mite,Tetranychus urticae. Insect Molecular Biology. 2017;26:215–232. doi: 10.1111/imb.12286. [DOI] [PubMed] [Google Scholar]
- 68.Feng QD, et al. Developmental expression and stress induction of glutathione S-transferase in the spruce budworm, Choristoneura fumiferana. Journal of Insect Physiology. 2001;47:1–10. doi: 10.1016/S0022-1910(00)00093-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhao TY. Research on Overwintering and Physiological Ecology of Anopheles sinensis in China. Medical Animal Control. 1989;5:64–72. [Google Scholar]
- 70.Spielman A, Wong J. Environmental control of ovarian diapause in Culex pipiens. Annals of the Entomological Society of America. 1973;66:905–907. doi: 10.1093/aesa/66.4.905. [DOI] [Google Scholar]
- 71.Noirell J. Methods in Quantitative Proteomics: Setting iTRAQ on the Right Track. Curr. Proteomics. 2011;8:17–30. [Google Scholar]
- 72.Wang WZ, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Analytical Chemistry. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 73.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Goto S. KEGG Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zdobnov E, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 78.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS One. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jensen LJ, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.