Abstract
The provision of model code is required for publication in CPT: Pharmacometrics & Systems Pharmacology, enabling quantitative systems pharmacology (QSP) model availability. A searchable repository of published QSP models would enhance model accessibility. We assess the feasibility of establishing such a resource based on 18 QSP models published in this journal. However, because of the diversity of software platforms (nine), file formats, and functionality, such a resource is premature. We evaluated 12 of the models (those coded in R, PK‐Sim/MoBi, and MATLAB) for functionality. Of the 12, only 4 were executable in that figures from the associated manuscript could be generated via a “run” script. Many researchers are aware of the challenges involved in repurposing published models. We offer some ideas to enable model sharing going forward, including annotation guidelines, standardized formats, and the inclusion of “run” scripts. If practitioners can agree to some minimum standards for the provision of model code, model reuse and extension would be accelerated.
Background
Reproducibility lies at the heart of scientific research. Yet a recent survey found that 70% of scientists have tried and failed to reproduce results from a published experiment.1 This is a critical problem for the pharmaceutical industry, where multimillion‐dollar drug development programs emanate from discoveries in academic laboratories. Scientists at Bayer have reported that only ~25% of published preclinical studies could be validated to the point at which projects could progress.2 Depressingly, Amgen scientists reported that only 6/53 (11%) “landmark” cancer studies could be reproduced in‐house.3 Although the specific publications were not disclosed, these same targets were likely pursued to no avail by many other academic and industrial groups.
One would presume that simulated results emanating from computational experiments would fare better. Anyone who has tried to recreate a model and replicate a simulation from an article may not be so sure. Indeed, the reproducibility of published computational research has been reported as similarly dismal (~25%).4 The barriers to computational reproducibility include a lack of standardization for building and representing models, a lack of documentation on usage, and a lack of transparency (sometimes intentional) by authors5 in addition to simple coding errors and typos.
Computational modeling and simulation is playing an increasingly critical role in drug research, development, and regulatory decision making. The US Food and Drug Administration has recently launched a number of initiatives to advance model‐informed drug development, outlined in the The Prescription Drug User Fee Act (PDUFA) VI.6 To fulfill this role, model reproducibility and methodological transparency are imperative. Clinical pharmacometrics (pharmacokinetic and exposure–response models) have largely achieved this out of necessity. Pharmacokinetic/pharmacodynamic model simulations are typically included in regulatory submissions, and reviewers will often attempt to independently replicate these results. Standardized software packages (i.e., NONMEM), annotation rules, and file formats smooth the process. Transparency and consistency can be worth millions if they mean expediting a drug approval.
Pharmacometrics as a discipline has, however, taken more than 40 years of evolution to reach this state. Quantitative systems pharmacology (QSP) lacks this degree of standardization as a result of both the newness of the discipline and fundamental differences from pharmacometrics the use of a wide diversity of data types, model formalisms, biological scales, and objectives beyond dose projection.7 Still, QSP results should be reproducible in that individuals should be able to replicate simulations from an article without contacting the authors. This is important if models are to be reused for new data or repurposed to new projects, as a lack of reproducibility hinders good models from “bubbling up” in the literature. The ease of reproducibility is also critical if QSP is to play a greater role in drug development and regulatory decisions.8
In “Ten Simple Rules for Reproducible Computational Research,” the last, but not least, important rule states that all data and model code should be publicly available and easily accessible.9 CPT: Pharmacometrics & Systems Pharmacology has a crucial role to play here. The provision of model code is required for publication, and as an open access journal, this ensures model availability. However, as files are buried in supplementary materials with no unique identifiers, structure, or standardized annotation, model accessibility remains a problem.
To help alleviate this problem, we set forth to establish a searchable repository of QSP models published in CPT: Pharmacometrics & Systems Pharmacology. Although QSP research is published in numerous journals (e.g., Journal of Pharmacokinetics and Pharmacodynamics, npj Systems Biology and Applications, and basic science and engineering journals), we limited the scope to assess feasibility of this American Society of Clinical Pharmacology and Therapeutics–directed project. We next describe the challenges faced in our (failed) attempt to achieve this goal.
Literature Search
A PubMed literature search was conducted (October 2018) to identify relevant publications and embedded models for inclusion in the repository. To do so, we queried PubMed with the following search terms:
“CPT: pharmacometrics & systems pharmacology”[Journal] AND (“Systems” [Title] OR “QSP” [Title]) AND Model* [Title].
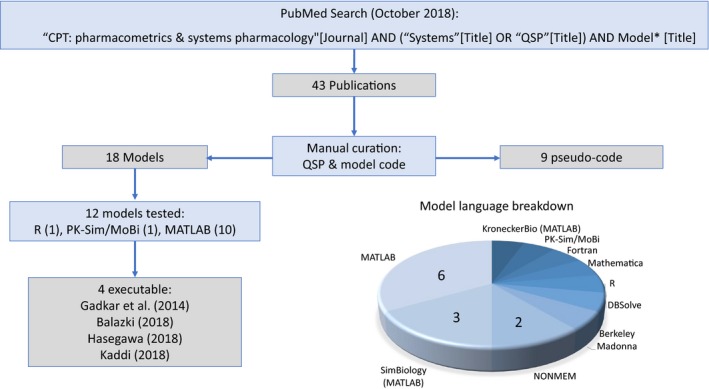
The search resulted in 43 publications. These publications were manually curated by reading abstracts to include only primary QSP research articles (i.e., excluding reviews and commentaries, model analyses, and methodology‐focused publications) that included intact model code in their supplemental information, resulting in a total of 18 (Table 1). The supplementary material packages were downloaded from each publication. An additional nine were found to include only pseudo‐code or references to other publications and were thus not considered further (Supplement 1 ). As a caveat, this excludes many relevant publications that do not include “systems” or “QSP” in their title10, 11 and highly impactful models published elsewhere.12 Although a more thorough analysis of the published literature would be valuable, this would require text‐mining methods, which is beyond the scope of this initial feasibility project. The results from our analyses of this limited set of models is nonetheless revealing.
Table 1.
Summary of 18 QSP models
| PMID | Title | Author, year | Language | Executable |
|---|---|---|---|---|
| 29637732 | Quantitative Systems Pharmacology Model of hUGT1A1‐modRNA Encoding for the UGT1A1 Enzyme to Treat Crigler‐Najjar Syndrome Type 1 | Apgar (2018) | KroneckerBio (MATLAB) | No |
| 28548387 | A Quantitative Systems Physiology Model of Renal Function and Blood Pressure Regulation: Model Description | Hallow (2017) | R | Error |
| 26312163 | Using a Systems Pharmacology Model of the Blood Coagulation Network to Predict the Effects of Various Therapies on Biomarkers | Nayak (2015) | SimBiology (MATLAB) | No |
| 26225228 | A Systems Pharmacology Model of Erythropoiesis in Mice Induced by Small Molecule Inhibitor of Prolyl Hydroxylase Enzymes | Singh (2015) | Fortran | Not tested |
| 28188981 | A Mechanistic Systems Pharmacology Model for Prediction of LDL Cholesterol Lowering by PCSK9 Antagonism in Human Dyslipidemic Populations | Gadkar (2014) | SimBiology (MATLAB) | Yes |
| 24918743 | Effects of IL‐1β–Blocking Therapies in Type 2 Diabetes Mellitus: A Quantitative Systems Pharmacology Modeling Approach to Explore Underlying Mechanisms | Palmér (2014) | Mathematica | Not tested |
| 23903463 | Quantitative Systems Pharmacology Model of NO Metabolome and Methemoglobin Following Long‐Term Infusion of Sodium Nitrite in Humans | Vega‐Villa (2013) | NONMEM | Not tested |
| 28941225 | Systems Pharmacology Model of Gastrointestinal Damage Predicts Species Differences and Optimizes Clinical Dosing Schedules | Shankaran (2017) | MATLAB | Error |
| 28571112 | A Translational Systems Pharmacology Model for Aβ Kinetics in Mouse, Monkey, and Human | Karelina (2017) | DBSolve | Not tested |
| 27537780 | A Systems Model for Ursodeoxycholic Acid Metabolism in Healthy and Patients With Primary Biliary Cirrhosis | Zuo (2016) | MATLAB | No |
| 27299938 | Systems Pharmacology Modeling of Prostate‐Specific Antigen in Patients With Prostate Cancer Treated With an Androgen Receptor Antagonist and Down‐Regulator | Mistry (2016) | MATLAB | No |
| 26783501 | A Systems Pharmacology Model for Predicting Effects of Factor Xa Inhibitors in Healthy Subjects: Assessment of Pharmacokinetics and Binding Kinetics | Zhou (2015) | SimBiology (MATLAB) | No |
| 26451331 | Application of a Systems Pharmacology‐Based Placebo Population Model to Analyze Long‐Term Data of Postmenopausal Osteoporosis | Berkhout (2015) | NONMEM | Not tested |
| 24402117 | Scale Reduction of a Systems Coagulation Model With an Application to Modeling Pharmacokinetic–Pharmacodynamic Data | Gulati (2014) | MATLAB | No |
| 23887363 | Integrated Pharmacometrics and Systems Pharmacology Model‐Based Analyses to Guide GnRH Receptor Modulator Development for Management of Endometriosis | Riggs (2012) | Berkeley Madonna | Not tested |
| 30270578 | A Quantitative Systems Pharmacology Kidney Model of Diabetes Associated Renal Hyperfiltration and the Effects of SGLT Inhibitors | Balazki (2018) | PK‐Sim/MoBi | Yes |
| 30043496 | Automated Scale Reduction of Nonlinear QSP Models With an Illustrative Application to a Bone Biology System | Hasegawa (2018) | MATLAB | Yes |
| 29920993 | Quantitative Systems Pharmacology Modeling of Acid Sphingomyelinase Deficiency and the Enzyme Replacement Therapy Olipudase Alfa Is an Innovative Tool for Linking Pathophysiology and Pharmacology | Kaddi (2018) | MATLAB | Yes |
QSP, quantitative systems pharmacology.
Model Analyses
Software diversity
The most striking although perhaps unsurprising finding was the diversity of software platforms employed (Figure 1 ). Although MATLAB (MathWorks, Natick, MA) and its dependents (SimBiology and KroneckerBio) predominated, NONMEM (ICON, Dublin, Ireland), R, PK‐Sim/MoBi (Bayer, Leverkusen, Germany), Mathematica (Wolfram Research, Champaign, IL, USA), DBSolve (InSysBio, Moskow, Russia), Berkeley Madonna (University of California, Berkeley, CA), and Fortran (IBM, Armonk, NY) were also represented. The file formats supplied in the supplements were also diverse, spanning .doc, .txt, .xlsx, .pdf, and .m files as well as the number of files per model from a single script to 23 individual files. Model annotation also varied extensively, from a single “run” script, which would automatically generate figures, to a set of ordinary differential equations (ODEs) requiring time, initial conditions, and parameter vectors as inputs to run.
Figure 1.
Workflow and results summary. QSP, quantitative systems pharmacology.
Model reproducibility
Lacking the technical proficiency or software to assess all 18 models, we settled on testing the subset written in R (1), PK‐Sim/MoBi (1), or MATLAB and its dependents (10). By “testing,” we evaluated how many cases contained scripts that would generate a simulation without extensive coding, i.e., executable.
Of the 10 MATLAB‐based models, the packages provided by Hasegawa and Duffull, 13 Kadidi et al.14 and Gadkar et al.15 all contained “readme” text files describing the code and a single “run” script that loaded multiple files, executed simulations, and generated pharmacokinetic/pharmacodynamic time course figures from the associated manuscripts (Figures S1–S3 ). The model of chemotherapy‐induced gastrointestinal damage by Shankaran et al.16 purported such in the code annotation, but the supplement did not contain a critical Excel file required to execute the simulation. The model of diabetes‐associated renal hyperfiltration by Balazki et al., 17 coded in PK‐Sim/MoBi, also contained both the MOBI model files as well as R‐based “run” scripts that generated figures from the manuscript (Figure S4 ). A model of renal physiology and control mechanisms developed by Hallow and Gebremichael18 contained well‐annotated R code and accompanying run scripts. However, we were unable to execute because of issues associated with the parameter file. In none of the other models tested were we able to generate a simulation using a similar run file. Thus, 4/12 models tested were deemed functionally executable. Note that nonexecutable does not mean that the code is erroneous or nonfunctional, nor say anything about the quality of the work, but only that it would take significant efforts to replicate a simulation from the associated manuscript using the files provided. Many models contained only the “base” model, i.e., the set of equations, with parameters often saved in a separate file, such that one would need to write custom code to load the model and execute simulations. This would rely on the text describing these simulations in sufficient detail to be able to reproduce the calculations.
Best Practice Recommendations for Publishing QSP Models
It would be both challenging and perhaps of little use to create a searchable database from such a jumble of files. We have provided the materials in a single folder for readers to access and assess for themselves (Supplement 2 ). As QSP models are coded in many different software platforms by practitioners from diverse backgrounds, there is no consistency in how models are annotated or executed, nor should there be at this stage. We seek here not to provide firm rules as to how model code should be provided but, rather, to start a discussion as to what can be done to make QSP model reproducbility and sharing easier than it exists today. The following sections provide some ideas.
Near term
The most immediately practical idea is the provision of a single “run” script that loads and simulates the model (perhaps calling multiple provided files) to generate at least one figure from the publication and an associated “readme” file that succinctly describes such. In our experience, the extra effort required to do so can save time and headache in the long run, minimizing email exchanges regarding questions about how to use the provided code.
Guidelines should be provided by journals as to how model code should be submitted and made accessible, rather than simply request one to “provide model code.” Although we do not intend to establish what this would be, annotation standards could start with those described in the minimum information requested in the annotation of biochemical models.19 Checklists are typically required for the description of experimental data and methods,20 and similar checklists could be created toward model code. Nature has in fact recently released guidelines for code and software submission.21
In addition to model annotation, sufficient details should be provided to describe individual simulations and analyses. Some existing tools provide this as part of their project files, for example, MATLAB SimBiology models are encoded as a single sbproj file, which can include experimental conditions (e.g., dosing protocols), and these can be exported to (open‐source) systems biology markup language (SBML). We are aware of at least one case of successful model sharing using such details. An SBML file and a single MATLAB “run” script22 was translated into R via the mgrsolve package and then used to create a R‐Shiny application without any input from the authors, with the entire process taking only of a few hours of work.23
Longer term
Authors could potentially be rewarded with a “badge” for articles that meet some threshold of model reproducibility during peer review. This would incentivize the provision of functional, well‐annotated code and “run” scripts.
This would of course place the burden of functional code on reviewers, who may lack the software or technical proficiency to do so in all cases. Alternatively, journals could establish an annually elected small group of professionals, postdocs, and/or doctorate candidates capable and willing to provide model evaluation tests. Similar to the model qualification procedure developed within the DDMoRe Consortium,24 this review group could ensure code functionality while granting young professionals valuable experience and insight into the review process. However, getting such a system established and integrated smoothly into the review process would be a significant undertaking.
As is the case for genomics data sets, journals could request authors to provide models in an established repository. Two such model repositories currently exist: BioModels25 geared toward cell‐ and molecular‐based systems models and DDMoRE26 for pharmacometrics. Consortiums have also been established to develop more comprehensive tissue‐ and disease‐focused models, such as DILIsym27 (drug‐induced liver injury), Certara's QSP Immunogenicity Consortium, or the Critical Path for Alzheimer's Disease; hence, these may serve in specific cases.
QSP models should ultimately be provided in an open‐source, standardized format. Although no single standard currently exists, there are a number of options, such as SBML,28 PharmML,29 and PK‐Sim/MoBi.30 However, given the extra effort required to do so and the fact that QSP work is rarely scripted in such platforms, such a requirement may simply impede publication rather than enhance sharing at this point. As an alternate “standard” format, an Excel or text file listing the full set of reactions, associated rate laws, and parameters could suffice. Such files can be converted into equations and executable code with relative ease by many software platforms.
A fundamental challenge with all the above is that results often require substantial computation on clusters or clouds and may depend on multiple software packages and data sets to generate. Furthermore, the results may depend on libraries for which the authors do not have permission to redistribute or complex chains of open‐source software configured in a particular way. Reproducibility in such cases is not simply a matter of providing access to the model and executable code but access to the computational environment used to generate the results.
Here we may look to the software industry for a potential solution, where the software‐as‐a‐service model has addressed this issue by bundling together software, data, and computation into a single offering. Particularly for cases for which a single “run” script does not suffice, authors could provide access not only to the model and code but also to the underling technical infrastructure. This removes the burden on the individual scientist to download software and replicate the computing environment. Consistent with this, Nature has recently launched a test trial with Code Ocean, a cloud‐based computing platform that enabes authors to share fully functional and executable code.31 Obviously, this raises new issues around how long such services would need to be maintained and who would bear that expense.
Given the diversity of opinions on how best to meet these challenges, workshops should be organized, perhaps through the American Society of Clinical Pharmacology and Therapeutics or the International Society of Pharmacometrics, to discuss practical strategies to move forward.
Going Forward
What should model sharing look like? Is providing the set of equations and parameters underlying a model sufficient? As with experimental protocols, the procedures involved in executing a simulation often contain many details and nuances not captured in a set of equations. At the other extreme, should one be able to generate every figure in an article from a single script? This is the bar set by bioinformatics,32 although probably an unreasonable expectation for QSP. Such a requirement may simply dissuade publishing, particularly for industry scientists who must balance the desire to publish with project timelines and the need to guard proprietary information. Given the diversity of QSP, it is unclear to what end of the spectrum should be required or is reasonable to request. The provision of a single “run” script, when feasible, is the most immediately tractable idea. Continued dialogue within the community via publications, workshops, or other meeting forums will be necessary to reach some consensus.
Many have had the experience of attempting to extract a model from an article and reproduce a result only to be met with errors and frustration, necessitating a back and forth with the authors, sometimes with success and sometimes to no avail. If we can find ways to lesson this hardship, the field will be better for it.
Funding
No funding was received for this work.
Conflict of Interests
The authors declared no competing interests for this work.
Supporting information
Figure S1. Pharmacokinetic/pharmacodynamic plots generated from Hasegawa et al. model.
Figure S2. Pharmacokinetic/pharmacodynamic plots generated from Kadidi et al.14 model.
Figure S3. Pharmacokinetic/pharmacodynamic plots generated from Gadkar et al. model.
Figure S4. Pharmacokinetic/pharmacodynamic plots generated from Balazki et al. model.
Supplementary Material S1. Results of Pubmed query.
Supplementary Material S2. Model files provided in supplementary materials of 18 publications.
Acknowledgments
We would like to thank Jeffery Kearns, Joshua Apgar, John Burke, Saroja Ramanujan, Gregory Ferl, Michael Block, Piet van der Graaf, and the ASCPT Quantitative Pharmacology Network leadership for providing ideas and feedback.
References
- 1. Baker, M. & Penny, D. Is there a reproducibility crisis? Nature 533, 452–454 (2016). [DOI] [PubMed] [Google Scholar]
- 2. Prinz, F. , Schlange, T. & Asadullah, K. Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 10, 712–713 (2011). [DOI] [PubMed] [Google Scholar]
- 3. Begley, C.G. & Ellis, L.M. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012). [DOI] [PubMed] [Google Scholar]
- 4. Stodden, V. , Seiler, J. & Ma, Z. An empirical analysis of journal policy effectiveness for computational reproducibility. Proc. Natl Acad. Sci. U S A 115, 2584–2589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waltemath, D. & Wolkenhauer, O. How modeling standards, software, and initiatives support reproducibility in systems biology and systems medicine. IEEE Trans. Biomed. Eng. 63, 1999–2006 (2016). [DOI] [PubMed] [Google Scholar]
- 6. US Food and Drug Administration . PDUFA Reauthorization Performance Goals and Procedures Fiscal Years 2018 Through 2022 (US Food and Drug Administration, Silver Spring, MD, 2018). [Google Scholar]
- 7. Gadkar, K. , Kirouac, D.C. , Mager, D.E. , Van Der Graaf, P.H. & Ramanujan, S. A six‐stage workflow for robust application of systems pharmacology. CPT Pharmacometrics Syst. Pharmacol. 5, 235–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peterson, M. & Riggs, M. FDA advisory meeting clinical pharmacology review utilizes a quantitative systems pharmacology (QSP) model: a watershed moment? CPT Pharmacometrics Syst. Pharmacol. 4, 189–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sandve, G.K. , Nekrutenko, A. , Taylor, J. & Hovig, E. Ten simple rules for reproducible computational research. PLoS Comput. Biol. 9, 1–4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirouac, D.C. et al Model‐based design of a decision tree for treating HER2 + cancers based on genetic and protein biomarkers. CPT Pharmacometrics Syst. Pharmacol. 4, 212–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen, X. , Hickling, T.P. & Vicini, P. A mechanistic, multiscale mathematical model of immunogenicity for therapeutic proteins: part 2‐model applications. CPT Pharmacometrics Syst. Pharmacol. 3, e134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peterson, M.C. & Riggs, M.M. A physiologically based mathematical model of integrated calcium homeostasis and bone remodeling. Bone 46, 49–63 (2010). [DOI] [PubMed] [Google Scholar]
- 13. Hasegawa, C. & Duffull, S.B. Automated scale reduction of nonlinear QSP models with an illustrative application to a bone biology system. CPT Pharmacometrics Syst. Pharmacol. 7, 562–572 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaddi, C.D. et al Quantitative systems pharmacology modeling of acid sphingomyelinase deficiency and the enzyme replacement therapy olipudase alfa is an innovative tool for linking pathophysiology and pharmacology. CPT Pharmacometrics Syst. Pharmacol. 7, 442–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gadkar, K. et al A mechanistic systems pharmacology model for prediction of LDL cholesterol lowering by PCSK9 antagonism in human dyslipidemic populations. CPT Pharmacometrics Syst. Pharmacol. 3, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shankaran, H. et al Systems pharmacology model of gastrointestinal damage predicts species differences and optimizes clinical dosing schedules. CPT Pharmacometrics Syst. Pharmacol. 7, 26–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balazki, P. , Schaller, S. , Eissing, T. & Lehr, T. A quantitative systems pharmacology kidney model of diabetes associated renal hyperfiltration and the effects of SGLT inhibitors. CPT Pharmacometrics Syst. Pharmacol. 7, 788–797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hallow, K.M. & Gebremichael, Y. A quantitative systems physiology model of renal function and blood pressure regulation: model description. CPT Pharmacometrics Syst. Pharmacol. 6, 383–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Novère, N. et al Minimum information requested in the annotation of biochemical models (MIRIAM). Nat. Biotechnol. 23, 1509–1515 (2005). [DOI] [PubMed] [Google Scholar]
- 20. Kenall, A. et al Better reporting for better research: a checklist for reproducibility. Gigascience 4, 8–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. NatureResearch. Guidelines for authors submitting code & software <http://www.nature.com/documents/GuidelinesCodePublication.pdf>.
- 22. Kirouac, D.C. et al Clinical responses to ERK inhibition in BRAF V600E‐mutant colorectal cancer predicted using a computational model. NPJ Syst. Biol. Appl. 3, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Metrum Research Group. Clinical responses to ERK inhibition in BRAF{V600E}‐mutant colorectal cancer predicted using a computational model <https://metrumrg.shinyapps.io/cic-2017/>.
- 24. Harnisch, L. , Matthews, I. , Chard, J. & Karlsson, M.O. Drug and disease model resources: a consortium to create standards and tools to enhance model‐based drug development. CPT Pharmacometrics Syst. Pharmacol. 2, 1–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glont, M. et al BioModels: expanding horizons to include more modelling approaches and formats. Nucleic Acids Res. 46, D1248–D1253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith, M.K. et al Model description language (MDL): a standard for modeling and simulation. CPT Pharmacometrics Syst. Pharmacol. 6, 647–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shoda, L.K.M. , Woodhead, J.L. , Siler, S.Q. , Watkins, P.B. & Howell, B.A. Linking physiology to toxicity using DILIsym®, a mechanistic mathematical model of drug‐induced liver injury. Biopharm. Drug Dispos. 35, 33–49 (2014). [DOI] [PubMed] [Google Scholar]
- 28. Hucka, M. et al The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531 (2003). [DOI] [PubMed] [Google Scholar]
- 29. Bizzotto, R. et al PharmML in action: an interoperable language for modeling and simulation. CPT Pharmacometrics Syst. Pharmacol. 6, 651–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eissing, T. et al A computational systems biology software platform for multiscale modeling and simulation: integrating whole‐body physiology, disease biology, and molecular reaction networks. Front. Physiol. 2, 4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. [Editorial] Easing the burden of code review. Nat. Methods 15, 641 (2018). [DOI] [PubMed] [Google Scholar]
- 32. Lewis, J. , Breeze, C.E. , Charlesworth, J. , Maclaren, O.J. & Cooper, J. Where next for the reproducibility agenda in computational biology? BMC Syst. Biol. 10, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pharmacokinetic/pharmacodynamic plots generated from Hasegawa et al. model.
Figure S2. Pharmacokinetic/pharmacodynamic plots generated from Kadidi et al.14 model.
Figure S3. Pharmacokinetic/pharmacodynamic plots generated from Gadkar et al. model.
Figure S4. Pharmacokinetic/pharmacodynamic plots generated from Balazki et al. model.
Supplementary Material S1. Results of Pubmed query.
Supplementary Material S2. Model files provided in supplementary materials of 18 publications.


