Abstract
There have been several recent studies addressing the genetic architecture of depression. This review serves to take stock of what is known now about the genetics of depression, how it has increased our knowledge and understanding of its mechanisms and how the information and knowledge can be leveraged to improve the care of people affected. We identify four priorities for how the field of MD genetics research may move forward in future years, namely by increasing the sample sizes available for genome-wide association studies (GWAS), greater inclusion of diverse ancestries and low-income countries, the closer integration of psychiatric genetics with electronic medical records and developing the neuroscience toolkit for polygenic disorders.
1. Introduction
Major depressive disorder is a disabling syndrome characterised by persistent low mood and reduced enjoyment along with additional signs and symptoms including reduced concentration, energy, and self-esteem and altered appetite and sleep quality. Although many symptom combinations can lead to a diagnosis of major depressive disorder, the commonalities are its persistence, pervasiveness, and pathological extent. Depressed mood is a normal human emotion; in major depressive disorder, however, depressed mood becomes nearly unremitting, unshakable, and associated with other cognitive and physical symptoms. Thus, major depressive disorder is clinically heterogeneous, and individuals vary greatly in their symptom severity, treatment response, and outcome. Clinical heterogeneity may also reflect substantial causal heterogeneity, whereby individuals with different aetiologies are grouped under the same diagnosis.
In most single-sample genome-wide association studies (GWAS), structured diagnostic criteria of major depressive disorder have generally been used to define the trait of interest. The diagnostic criteria for major depressive disorder (Table 1) are provided in the American Psychiatric Association’s Diagnostic and Statistical Manual (DSM) (American Psychiatric Association, 2013). These criteria are generally preferred over the very similar criteria for moderate depressive disorder adopted by the 10th edition of the World Health Organisation’s International Classification of Diseases (World Health Organization, 1993). These two diagnostic systems are highly overlapping, and both require that the symptoms must be present most of the time for a two week period and are not better accounted for by another condition.
Table 1:
Diagnostic criteria for major depressive disorder and depressive episode
| DSM-5 Major Depressive Disorder | ICD-10 Moderate Depressive Episode |
|---|---|
|
Five or more symptoms, at least one of which must come from the ‘A’ criteria: ‘A’ criteria
|
Two or more symptoms from
|
‘B’ criteria
|
Three or more typical symptoms from
|
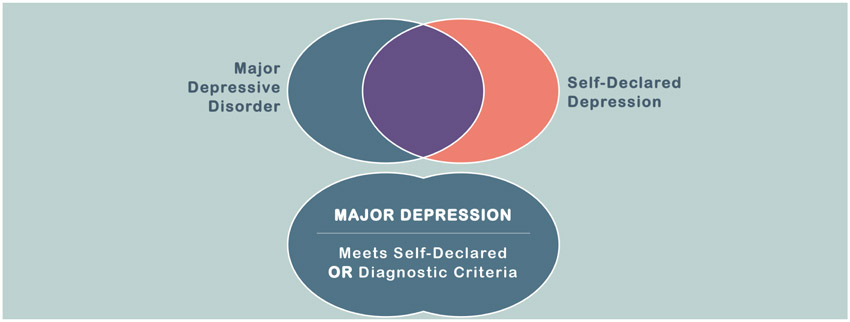
In recent years several large samples have become available that include ‘case’ definitions of depression that would not meet full DSM or ICD criteria. The company 23andMe recently provided a sample of more than a million individuals that self-reported the presence or absence of a depression diagnosis made by a healthcare professional (Howard et al., 2018a). A proportion of individuals reporting a health professional’s diagnosis of depression will not simultaneously meet DSM or ICD criteria and, to reflect this uncertainty, the Psychiatric Genomics Consortium (PGC) have adopted the term ‘Major Depression’ (MD) to include more minimally phenotyped samples. Genetic analyses of MD may therefore generate association findings that do not generalise to major depressive disorder, although this has not yet been demonstrated to the best of our knowledge. In addition, individuals endorsing 23andMe’s self-declared depression question would be expected to meet a threshold of clinical significance, whereas individuals meeting major depressive disorder criteria elicited through a reliable structured clinical interview (First et al., 2002; Kessler et al., 1998) may not have sought a health professional’s help for their symptoms of low mood. The effects of these different definitions on the results of genetic association studies are currently being explored and identified. Two recent studies have identified high genetic correlations between a major depressive disorder diagnosis made using full (Zeng et al., 2016) or limited (Howard et al., 2018b) DSM criteria and self-declared depression, whilst a recent study preprint suggests that the genetic correlations of a detailed DSM-based diagnosis versus minimally phenotyped MD-traits may be more distinct (Cai et al., 2018). MD is defined by the PGC for genetic research to encompass all individuals with major depressive disorder as well as participants endorsing self-rated and other more minimal-phenotyping criteria (Figure 1).
Figure 1:
Differences between major depressive disorder and major depression (MD)
MD is undeniably a leading global cause of disability, affecting at least 2-4% of the population at a given point in time, at least 16% over the course of a lifespan (Kessler et al., 2003) and accounting for more than 4% of all years lived with disability (Vos et al., 2017). MD affects countries irrespective of their gross domestic product, although the highest morbidity burden is borne by low- and middle-income countries (Patel, 2007). The burden of MD has increased worldwide since 1990, particularly in low and middle income countries, where the majority of the world’s population reside and healthcare services are generally less able to meet patient need. MD is associated with social disadvantage, a broad range of physical diseases, and shortened lifespan (Chesney et al., 2014). In contrast to many diseases that receive greater research funding, it has occupied a higher rank over time in the global burden of diseases (Woelbert et al., 2019) and its persistence is a substantial source of individual and family adversity.
The Psychiatric Genomics Consortium (PGC) Working Group for Major Depressive Disorder is an international consortium in more than 20 countries that was set up in response to the growing realisation that elucidating the genetic underpinnings of MD requires global cooperation (Levinson, 2006; Sullivan et al., 2018). Although twin and family studies have demonstrated a substantial contribution of genetic factors (Polderman et al., 2015), the lack of replicable molecular genetic associations, together with need for large sample sizes, and consistent approaches to quality control and analysis necessitated a global response from the psychiatric genetics community. The PGC Major Depressive Disorder Working Group has published three major meta-analyses of MD (Howard et al., 2018a; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al., 2013; Wray et al., 2018) and related traits and the results generated have been used in many downstream studies to characterise the genetics of depression and related traits. In addition, there have been relatively recent GWAS studies of depressive symptoms (Hek et al., 2013), self-declared depression (Howard et al., 2018b; Hyde et al., 2016) as well as both more broadly (Direk et al., 2017) and narrowly defined (Hall et al., 2018; Milaneschi et al., 2017) traits. This review serves to take stock of what is known now about the genetic architecture of MD, how it has affected our knowledge and understanding of depression and its mechanisms, and how the field of MD genetics research may move forward in future years.
2. Genetic approaches to MD
In the past 40 years, many twin and family studies of MD have established that liability to MD has a non-deterministic genetic component to its etiology as substantiated by a twin-heritability of 31-42% (Sullivan et al., 2000). This level of twin-heritability implies the need for large samples for reliable and reproducible gene identification (Levinson et al., 2014; Wray et al., 2012) but complex diseases with similar twin-heritabilities have had considerable success (e.g., type 2 diabetes). Twin-heritability provides support for the logic of genetic searches but does not elucidate the most critical feature, the genetic architecture (the number of loci underlying liability to a complex disorder along the effect sizes and frequencies of the loci) (Sullivan et al., 2012). For instance, a trait with a handful of loci each with strong effects (genotypic relative risks, GRR >2) would be more tractable to genetic analysis than a trait with hundreds of loci each with GRR 1.1-1.2.
The genetic study designs applied to MD have mirrored those used in other common, complex disorders. The three major genetic approaches, besides genome-wide association studies, are linkage analysis, candidate gene studies, and re-sequencing studies.
Linkage analysis evaluates pedigrees with many affected individuals to screen the genome to identify regions inherited from a common ancestor and present in affected individuals. This approach was sensible given findings for other biomedical diseases (albeit with simpler genetic architectures). At least 12 studies pursued this design from 1998-2010 (Camp and Cannon-Albright, 2005; Cloninger et al., 1998; Fullerton et al., 2003; Holmans et al., 2007; Kuo et al., 2007; McGuffin et al., 2005; Middeldorp et al., 2009; Nash et al., 2004; Neale et al., 2005; Nurnberger et al., 2001; Wray et al., 2008; Zubenko et al., 2003), and no replicable findings emerged. For this review, we reanalysed the reported linkage regions using partitioned LD score regression and found no significant overlap with current GWAS results for MD (in fact, the linkage regions tended to be depleted of MD SNP-heritability, enrichment=0.849, SE=0.078, P=0.055). Bioinformatic analysis of the linkage regions identified no biological themes. It is very likely that the assumptions of linkage analyses were not robust (with no rare genetic loci with large effect sizes existing) and that the sample sizes used were far too small.
Candidate gene association studies (circa 1995-2005) selected one or a few of the 21,000 genes in the human genome and compared allele frequencies in MD cases and controls. Candidate gene selection was based on prior knowledge, often derived from pharmacology (e.g., to study genetic variation in the serotonin transporter, the site of action of some antidepressants). The candidate gene approach has long been controversial (reviewed in Sullivan (Sullivan et al., 2001)) given its clear propensity to generate false positive findings (Sullivan, 2007, 2017). For instance, Farrell et al. (Farrell et al., 2015) evaluated 25 historical candidate genes for schizophrenia (e.g., COMT, DISC1, DTNBP1). These authors conducted a meta-analysis of the candidate gene literature, added common variant findings from the largest genomic study of schizophrenia available at the time, included unedited commentary from proponents of these genes or who introduced them into the literature, and included ratings from 24 schizophrenia geneticists. From empirical results, the historical candidate gene literature was essentially uninformative for the genetic basis of schizophrenia (in fact, the effect sizes reported by the initial studies could be excluded by subsequent studies with ~100% power). In many instances, authors who had studied these genes indicated that they no longer thought that they were involved. An independent study confirmed these general conclusions (Johnson et al., 2017).
But, what of candidate genes for MD? Recently, Border et al. (Border et al., 2019) evaluated 18 MDD candidate genes (e.g., SLC6A5, BDNF, COMT and HTR2A). In an extensive set of analyses of empirical data, they did not find much support for any candidate gene. We refer the reader to this paper for full details, but these authors concluded: “The study results do not support previous depression candidate gene findings, in which large genetic effects are frequently reported in samples orders of magnitude smaller than those examined here. Instead, the results suggest that early hypotheses about depression candidate genes were incorrect and that the large number of associations reported in the depression candidate gene literature are likely to be false positives.”
One candidate gene study deserves particular mention. One of the most highly cited papers in psychiatry (>4300 citations) was in Science in 2003 by Caspi et al. who reported a gene-environment association of SLC6A4/HTTLPR and early stress on risk for MD (Caspi et al., 2003). This paper remains contentious in some circles – many of the salient issues are discussed at length in a pre-specified public meta-analysis plan (Culverhouse et al., 2013) and in the response by Moffit and Caspi (Moffitt and Caspi, 2014). The meta-analysis (N=38,802) rather robustly did not support the claims in the original paper (N=837). For many readers of this literature, the paper by the late David Fergusson and colleagues tips the scale (Fergusson et al., 2011). This was an exceptionally similar study to Caspi et al.: both were longitudinal birth cohort studies in the south island of New Zealand (Christchurch and Dunedin) with dense prospective measurement and a focus on childhood development and risk for subsequent psychiatric disorders. Fergusson et al. state: “A series of 104 regression models were fitted to four mental health outcomes (depressive symptoms, major depression, anxiety disorder and suicidal ideation) observed at ages 18, 21, 25 and 30 using 13 measures of life-course stress that spanned childhood and adult stressors. No evidence was found that would support the hypothesis that 's' alleles of 5-HTTLPR are associated with increased responsivity to life stressors.” The lack of replication in a highly similar study is notable.
Whether one does or does not “believe” in the candidate gene approach, we now have better ways to secure replicable findings. This was a popular design but, in retrospect, the reproducible yield was negligible. Indeed, a high-level National Institute of Mental Health genomics panel recommended that “candidate gene studies of psychopathologic, cognitive, or behavioral phenotypes should be abandoned in favor of well powered, unbiased association studies” (NIMH, 2018) . The most enduring result of the candidate gene era may be the current unyielding commitment to statistical rigor and reproducibility.
There have been few whole exome or whole genome re-sequencing studies of MD, and none of notable size and genome coverage. A study using very low pass sequencing data suggest a role for genetic variation in mitochondrial DNA, but we are unaware of external replication (Cai et al., 2015). However, if experiences with schizophrenia and type 2 diabetes are relevant (i.e., meta-analyses of whole exome data for ~25,000 cases each found <10 confident associations), sequencing studies are unlikely to yield confident results until the sample sizes become extremely large (N >106) (Zuk et al., 2014).
Structural variation plays an important role in schizophrenia, but in MD the role of this class of genetic variation has been under-explored. Current studies suggest that rare copy number variants play a more minor role in MD than in schizophrenia and other neuropsychiatric disorders (Kendall et al., 2018), but are nonetheless more common in cases than controls. A recent meta-analysis of 4 cohorts (5780 cases, 6626 controls) also reported a greater burden of short (<100kb) deletions in MD that were enriched for likely enhancer elements (Zhang et al. 2019). These findings suggest that copy number variants play a role in the aetiology of MD through disruption of gene expression.
3. Insights from MD’s polygenic underpinnings
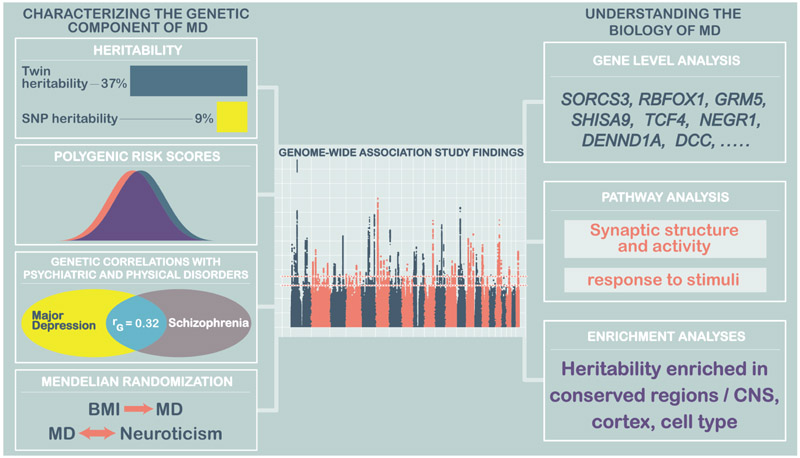
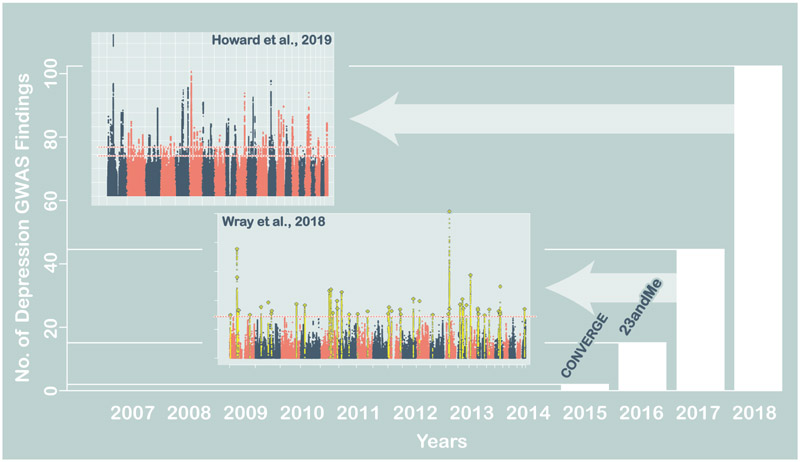
Genetic studies of MD show that the underlying liability to depression is polygenic. Extensive linkage, candidate gene and genome-wide association studies have confirmed that no loci of major effect exist, and imply that the heritable component of MD is due to thousands of loci each having a minor effect on liability to the disorder (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al., 2013). Genome-wide association studies, which test for single nucleotide polymorphism (SNP) associations with depression genome-wide, have now identified 102 common genetic variants associated with MD (Howard et al., 2019). These variants account for only a small proportion of genetic contribution to MD, but our recent progress indicates that, as in other disorders, expanding our sample sizes will continue to increase the number of associated variants. For polygenic disorders such as MD, follow-up studies focused on any specific variant are of limited value, because each variant has a modest effect on risk. However genome-wide methods assessing the impact of sets of variants can be highly informative, and extensive computational tool kits are now available to explore and exploit genome-wide results (Maier et al., 2018). Here, we describe how genetic results can generate insights to MD through estimating heritability, assessing individual-level risk, identifying genes or pathways critical for conferring liability to the disorder, highlighting links with treatment, and detecting pleiotropy, where genetic risk is shared with other disorders (Figure 3).
Figure 3:
Characterising the genetic component of MD and understanding its biology
The heritability of MD captured by genome-wide studies analysed in Howard et al. (2018a) is 8.9% (95% confidence interval 8.3% - 9.5%). This common genetic variation captures a substantial part of the twin-heritability of ~37%, and is expected to increase as genome-wide association studies of larger sample sizes with denser imputation uncover more of the genetic component of MD. Genome-wide association studies test the evidence for association with each genetic variant, combined across a set of MD cases and controls. The genome-wide results can be taken back to an individual-level risk measure, by calculating a polygenic risk score risk that captures genetic liability to MD. Here, effect sizes from genetic variants from a discovery GWAS are used to calculate polygenic risk scores in an set of individuals who were not part of the original GWAS. For a subset of variants, the PRS sums the number of risk alleles an individual carries, weighting each variant by its effect size (log(odds ratio)). Scores are calculated from a set of independent genetic variants that meet a pre-specified level of significance. The PRS score can be restricted to genome-wide significant loci, but experience shows us that reducing the p-value threshold increases prediction, and using an independent set of loci genome-wide (~100k variants) may maximise the level of prediction obtained, and forms a true ‘polygenic’ score. Polygenic risk scores have an approximately normal distribution, and when calculated in a set of MD cases and controls, scores are expected to be higher in MD cases than controls, reflecting the higher number of risk alleles carried by cases. Polygenic risk scores therefore summarise a large genotype matrix into a single variable per individual. They can be used to predict case-control status (and in MD they account for 2% of variance in case control status) or to assess genetic contribution to disease subtypes (MD polygenic risk scores are higher, for example in recurrent depression than single-episode depression, and in early onset cases). Further, they can be used to test for genetic overlap across disorders, so that polygenic risk scores for schizophrenia also predict MD case control status. Although these polygenic risk scores analyses attain statistical significance in research studies, the score only accounts for a small proportion of disease, or subtype risk. For example, a polygenic risk score in the top decile of scores confers a 2.5-fold increased risk of depression compared to a score in the lowest decile (Wray et al., 2018). The genetic component of depression captured by our existing studies is therefore most valuable for exploring disorder-level characteristics, with no current utility for determining the level of risk for an individual.
MD is frequently comorbid with disorders of physical health (Moussavi et al., 2007) and genetic studies allow us to identify the extent to which this is due to pleiotropy, where genetic variants contribute to both disorders. Such studies use summary statistics for genetic studies - a complete genome-wide listing of statistical significance and effect size for each variant tested. Making summary statistics open access for the scientific community has become standard research practice and enables a wide-range of follow-up studies to be performed. Two methodological tools, combined with the access to summary statistics, have further facilitated these studies: LD score regression (Bulik-Sullivan et al., 2015) to estimate the genetic correlation between disorders, and Mendelian randomization (Sterne et al., 2008) to explore causality.
MD shows strong genetic correlations (rg) with other psychiatric traits, with correlations of between 0.3-0.4 with schizophrenia, bipolar disorder, and ADHD, and lower values with anorexia nervosa (0.13) and Autism Spectrum Disorder (ASD, 0.13). These values indicate a common genetic predisposition across disorders in addition to genetic variants that are specific to a single diagnostic entity. Significant genetic correlations with social traits such as educational attainment and completing college (but not IQ) highlight the importance of environmental risk factors and the burden of depression, particularly in adolescence. Genetic correlations with age at menarche and menopause may point to novel physiological clues, perhaps implicating hormonal links that are also relevant to postpartum depression. Although strong phenotypic relationships exist between immune-mediated disorders and MD, the genetic correlations are modest, with significant results only detected with inflammatory bowel disease and Crohn’s disease. Further studies will be needed to assess whether this reflects non-genetic mechanisms or a lack of power to detect common genetic contributions.
Using the random assortment of alleles that occurs at meiosis, Mendelian Randomisation has been compared to a randomised controlled trial, allowing stronger causal inferences to be made between an exposure and an outcome. Mendelian randomisation studies (Burgess et al., 2013) determine the direction of causal effects between two traits, by comparing the effect sizes of two GWAS test statistics, using significant variants from the potentially causal factor. For example, using Mendelian Randomisation, studies have shown that the significant SNPs associated with years of education can be ranked in terms of their effect size. When the effect size for MD is estimated at the same variants, a significant negative association with MD risk was also detected, thus implying a causal effect of longer education on lower risk of MD. years of education and body mass index were both ‘causal’ for MD, but not vice versa (Tyrrell et al., 2018; Wray et al., 2018). Howard et al (2019), applying Mendelian Randomisation, also provided evidence that higher neuroticism is causally associated with greater liability to MD, and also that liability to MD may increase an individual’s tendency to smoke tobacco.
Characterisation of the genetic associations of MD using gene expression, functional annotation and pathway analysis indicate convergent signals for underlying biological mechanisms and identify potential routes for follow up studies. For example, MD is significantly associated with genes expressed in brain regions, specifically the cortex. This finding is confirmed by enrichment in neurons but not oligodendrocytes or astrocytes. Pathway analyses also implicate excitatory synapses and the modulation of synaptic neurotransmission and activity, providing further focus for translational studies on relevant aspects of human MD aetiology.
Whilst there is an increasingly diverse set of tools for the downstream investigation of MD-associated genetic risk variants, including the layering of annotated genomic information from other databases, there remain significant gaps in understanding how findings relate to underlying mechanisms. Current neuroscientific methods are well developed for the investigation of single variants of large effect, but have been slow to respond to the realisation that all psychiatric disorders are polygenic. Addressing this issue is a priority for the neurosciences, if we are to fully capitalise on genetic advances.
4. Our changing concept of depression
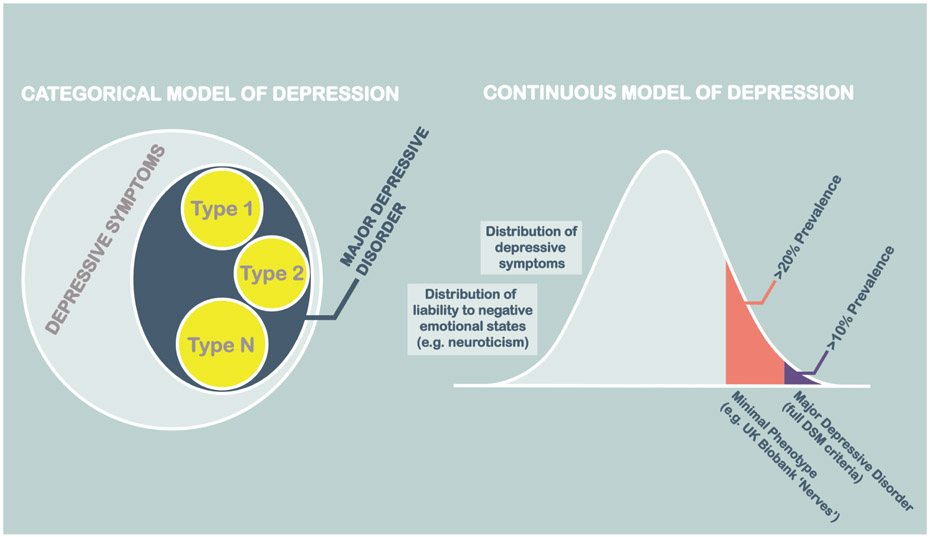
High pairwise genetic correlations between MD and other broad definitions of depression and related traits, whether ascertained through detailed clinical interview, self-report, or through quantitative measures of depressive symptoms has led some to question the validity of ‘binary’ or categorical models of MD, and its potential subtypes (Figure 4). Since these interrelated measures also vary markedly in cost and in the feasibility of their application to large datasets, the necessity of detailed clinical assessments for genetic research needs also to be examined and carefully justified.
Figure 4:
Our changing concept of depression
For GWAS, high genetic correlations of self-reported and interview-based depressive definitions suggest that these definitions may be almost exchangeable for common genetic variant discovery purposes (Howard et al., 2018b; Wray et al., 2018; Zeng et al., 2016). Very broadly defined depression-like traits, such as the that provided by the ‘Nerves’ measure in UK Biobank (UK Biobank Fields 2090, 2100), also have substantial genetic correlations with clinical interview ascertained and DSM defined major depressive disorder. Many other traits (e.g. antidepressant use, abbreviated scales and others; see Table 2) could also potentially be used to identify MD cases. The higher prevalence of more minimally-phenotyped depression traits is consistent with both traits lying on a continuum of shared genetic liability (Figure 4). Diagnoses based on full DSM based criteria may not be exchangeable with more minimally phenotyped traits for all purposes, however, as each trait may have a set of specific genetic or environmental risk factor associations. In a recent study, for example, the pedigree-based and environmental correlations of self-declared and DSM-based definitions were substantially less than 1 (Zeng et al., 2016). This was in marked contrast to the genetic correlation for common genetic variants, which was indistinguishable from 1 in the same sample. Broadly-defined measures may be expedient for SNP discovery, as they limit phenotyping to the most frugal, rapidly applied and scalable measures, but they may do so at the expense of future phenotypic stratification, clinical prediction, and translation. Whilst genetic studies using broader definitions, such as MD, may efficiently identify the many variants that are associated with both MD and more severe phenotypes, they may do so at the expense of failing to identify the variants specifically associated with more severe forms of illness. Future stratification of patients based on their genotype is likely to be most readily applied when the biological pathways impacted by these variants map onto clinical correlates and other individual characteristics. When these details are not provided by the measures used in GWAS discovery studies, identifying the clinical characteristics and other relevant correlated traits will require further samples in which there are more severely affected cases and more detailed clinical information, in which to test for their actionable associations. Clinical prediction may also be hampered, as the growing accuracy of MD prediction in independent studies may slow as the proportion of clinically-ascertained participants in GWAS study samples reduces.
Table 2:
Examples of different depths of depression phenotyping
| Source of information | |||
|---|---|---|---|
| Self-rated | Health record | Trained interview | |
| Diagnostic standard | CIDI touchscreen questionnaire (Davis et al., 2018a) | Diagnostic code E.g. DSM-5/ICD-10 (Davis et al., 2018b) | Structured diagnostic interview (Hall et al., 2018) |
| Multiple item (sub-diagnostic) | Probable depression (Smith et al., 2013) | NLP based text mining (Smoller, 2018) | PHQ-9 depression rating scale (Thombs et al., 2014) |
| Single item (sub-diagnostic) | Single question (Howard et al., 2018b) | Use of ‘depression’ or ‘antidepressant’ (Wigmore et al., 2019) | Evoked recollection of depression (Arroll et al., 2003) |
Our diagnostic concept of MD was originally based on attempts to classify distress and mental disorder into distinct categories that were anticipated to have specific aetiologies, treatments and outcomes (Kendell, 1987). Our genetic findings suggest that the discontinuity of major depressive disorder from more broadly defined pathological mental states (such as MD) may be unjustified for GWAS studies that seek to accelerate the discovery of common risk-associated genetic variants. Furthermore, continuous measures of non-pathological depressive symptoms and the personality trait of neuroticism, a tendency to experience negative emotions, also show substantial genetic correlations with MD (Luciano et al., 2018; Nagel et al., 2018). These findings suggest that non-pathological states also share a substantial proportion of their genetic architecture with MD and it may be possible to further leverage their findings for MD gene discovery in future.
Moving towards a more valid and predictive diagnostic structure is a major goal of psychiatric taxonomy and personalised medicine and stratification based on genetic factors aligns diagnosis and disease aetiology. Merely broadening the definition of depression is potentially associated with differences in genetic architecture and may also be associated with differences in environmental risk factors (Zeng et al., 2016). The first applications of data-driven machine learning algorithms to genetic data from other disorders provide an initial proof of concept that GWAS data may still be leveraged for stratification (Kim and Kim, 2018; Trakadis et al., 2018). Maintaining the development of new clinical research samples with detailed clinical and rich phenotyping remains a necessity for the validation of genetic findings from more broadly defined traits as well as for findings to be put to eventual clinical use.
5. How will genetics influence clinical practice?
Drug target identification
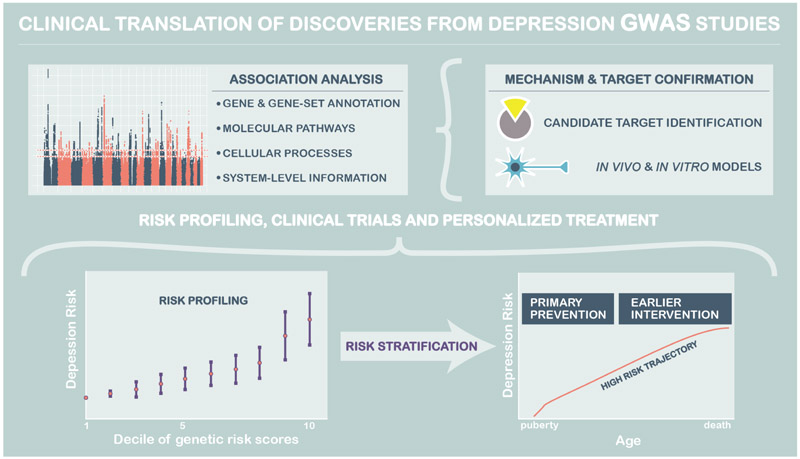
An early motivation of genetic studies was to identify ‘druggable targets’ through the identification of associated genes, subsequently identifying chemical ligands for these targets and then utilising these ligands in model systems and clinical trials (Figure 5). In theory, the same approach may also be used to repurpose existing medications with favourable side-effect and toxicity profiles, for new indications based on their receptor binding profile or other effects. Studies identifying risk associated variation in the D2 receptor pathway for schizophrenia (Ripke et al., 2014), and in the genetic targets of lipid-lowering therapy for cardiovascular disease, suggest this is a clinically valuable approach (Visscher et al., 2017). Wray et al. (2018) also identified enrichment of the targets of antidepressant treatment in the recent PGC MD meta-analysis. Basing drug development on the findings from MD case-control GWAS studies assumes that the mechanisms of effective treatment will be to reverse those leading to the case definitions adopted in the included studies. Studies of treatment response as a phenotype in its own right may also be a profitable approach to drug target discovery and to providing genomic risk scores that could be used to stratify individuals with depression by the treatments to which they are most likely to respond. These approaches are at an early stage, but are beginning to be applied by the PGC MDD and other investigators for both drug (Fabbri et al., 2019; Wigmore et al., 2019) and psychological therapies (Andersson et al., 2018).
Figure 5:
Clinical translation of genetic associations from GWAS studies
Risk stratification and early intervention has also been an explicit aim of genome wide association studies. Initially, this was based on the potentially mistaken belief that there would be variants of large effect in depression, and other psychiatric disorders. The polygenic architecture of depression has however enabled the use of polygenic risk profiling and has shown that individuals in the top decile of polygenic risk may have an approximately 2.5x odds increase in lifetime risk compared to those in the lowest decile. The predictive accuracy of polygenic risk scores should gradually improve as GWAS sample sizes increase and it is likely that, at least for those at the extremes of polygenic risk, algorithms to develop interventions and mitigate risk in those at highest liability of MD will be warranted.
Genetic risk scores for cardiovascular disease, several cancers, and type II diabetes can currently be used to stratify individuals recruited from the general population into categories with a more than 5-fold increase in risk (Khera et al., 2018). A strong case for assertively managing those at highest risk of these disorders in order to reduce their mortality has been made. Since these disorders are twice as common in people with MD than in the general population, individuals with MD may have much to gain from risk stratification along a spectrum of life-shortening comorbidities if that increases the likelihood of effective management. Risk score profiling may also be extended to more mechanistic studies, by first identifying individuals at high and low polygenic risk and using participant-derived cell lines and derived tissues to model MD in vitro. Polygenic scores can also be calculated using variants lying within specific biological pathways or gene sets to test mechanistic hypotheses using brain imaging and other physiological data. Early examples of this approach have used the NETRIN1-DCC gene pathway to examine the effect of risk-associated variation within this signalling pathway on brain white matter connectivity (Barbu et al., 2018).
Potentially causal relationships between MD and other quantitative traits identified using Mendelian Randomisation, such as the directional association between liability to increasing body mass index and risk of major depression in Wray et al (2018), also implicate potentially modifiable environmental exposures. These risk factors have important public health implications that may extend more broadly to individual behaviours and lifestyle interventions. Since modifying these exposures may reduce MD risk, these findings may directly impact the advice given by healthcare professionals in the clinic.
6. Moving forward
The recent success in genome-wide analysis of depression has confirmed that the disorder is tractable to standard genetic dissection tools, despite inherent heterogeneity in diagnosis. Depression now appears to be on a similar trajectory to other common, complex disorders, where GWAS in progressively increasing sample sizes has identified further numbers of significantly associated loci.
The need for larger samples of participants with major depressive disorder, MD and more severe forms of illness
The higher power acquired with large sample size identifies loci with lower effect sizes, which still contribute to improved prediction through polygenic risk scores. The most efficient strategy to detect increasing numbers of common risk-associated SNPs may be to increase sample sizes. The advantages of this strategy are perhaps most clearly shown by use of the lightly phenotyped 23andMe and UK Biobank cohorts, which have seen a doubling in the numbers of replicated risk-associated variants as well as increases in the accuracy of out-of-sample prediction of major depressive disorder cases from samples with more detailed diagnostic clinical phenotyping (Howard et al., 2018a; Wray et al., 2018). Given the high prevalence of depression, and its widespread assessment in population and health studies, this strategy should continue to increase the number of novel associated variants. Fewer richly-phenotyped clinical cohorts are available to dissect heterogeneity of depression, and our current understanding is limited to higher polygenic risk scores seen in early onset, recurrent and more severe depression cases.
Given the potential limitations of minimally-phenotyped samples as a sole means of identifying the genetic architecture of MD and major depressive disorder, samples with more detailed clinical and DSM-based phenotyping are clearly needed. Inclusion of cases from secondary psychiatric care, those requiring psychological therapies, drug treatments or electroconvulsive therapy will enable any variants identified in more minimally-phenotyped studies to be tested in clinically-relevant samples. This will help to ensure that any variants identified from minimally-phenotyped samples remain relevant to those at greatest clinical need. By comparing GWAS findings from minimally phenotyped, community-based, outpatient and inpatient samples, we may also be able to disambiguate the effects of genetic variation on the onset, severity and persistence of MD.
The need for greater geographical, ethnic and economic diversity
The substantial economic resources and growing genetic study sample sizes in countries of European ancestry has led to a marked global asymmetry in MD genetics. There is a severe lack, if not absence, of identified genetic risk factors for depression in almost all low-income countries, and in most non-European ancestries. Unaddressed, this will lead to a lack of information on the genetic architecture of depression in all countries. Genetic association information from different ancestries can be combined for greater power, fine mapping of functional polymorphisms. Studies using diverse ancestries may also enable triangulation of findings (e.g. confirming the findings of Mendelian Randomisation studies using different methods) in the presence of different macroeconomic and social environments. The CONVERGE consortium’s study of depression in Han Chinese women has so far been the only major published study of MD in a non-European ancestry population (CONVERGE consortium, 2015). There is now an urgent need to address MD in diverse populations in order to avoid the growing knowledge gap, enable risk stratification and to enable the broader clinical benefits of genetics to be realised in the countries at greatest need but with the lowest resources to meet those needs.
Mining rich clinical data and the electronic health record
The growing use of data acquired from non-clinical settings for genetic studies of depression has come with substantial benefits in terms of greater power, but may disadvantage phenotypic stratification, risk profiling and clinical translation. The goal of most research has been to obtain actionable insights for individuals affected by severe clinical disorder whereas many of the individuals in current GWAS studies have been included on the basis of traits that are neither necessary or sufficient for the diagnosis of major depressive disorder. Whether current variants, genes or pathways are associated with dimensions of clinical symptoms, or with specific treatment responses or outcomes, cannot be known without access to appropriately detailed longitudinal samples. These gaps in our understanding are unlikely to be fully bridged by adding genetic data to clinical trials, which are expensive and often conducted on small samples by genomic standards.
An alternative and complementary approach to conducting large studies and genetically enhanced RCTs in individuals with depression is to utilise the growing database of electronic health records (eHR) (McIntosh et al., 2016; Russ et al., 2018; Smoller, 2018). The eHR rarely contains genetic information, but can be augmented by adding low-cost genetic enhancements. DNA can be obtained by diverting samples obtained from dried heel-prick samples (obtained at birth for the purpose of diagnosing inborn errors of metabolism) or surplus blood samples obtained for routine laboratory investigation. DNA can also be obtained at scale and at low-cost by posting simple salivary DNA collection kits to people’s home address for returning to laboratory facilities. The eHR may also detail clinical symptoms assessed at interview, data on treatment response and the results of diagnostic tests and other investigations (Hafferty et al., 2018; Kerr et al., 2017) obtained blind to the patient’s genotype. These data may provide important clues to the phenome-wide longitudinal effects of genetic risk for depression and may enable in silico tests of treatment response stratified by genotype. Extracting this information from the eHR is challenging, but a growing number of methods for extracting coded and unstructured information are being developed and applied for more accurate case identification (Ford et al., 2016). These include the Clinical Record Information Search (CRIS) system applied to public healthcare provider data in the UK (Jackson et al., 2017) and billing records from private healthcare providers in the US (Pakhomov et al., 2007). Both of these examples employ natural language processing (NLP) as a means of extracting knowledge from the eHR in an unbiased way. The addition of information from unstructured text has also been shown to increase diagnostic accuracy when added to codified data from the eHR (Ford et al., 2016).
7. Summary
A major limitation in our current pipeline of mechanistic discoveries are the challenges faced in conducting experimental studies using variants identified by GWAS. This problem is surprisingly similar for most complex diseases including cardiovascular disease, type 2 diabetes, and common/non-syndromic cancers. The optimal path from robust genetic discoveries to a better mechanistic understanding of MD is currently being debated and discussed across a number of complex disorders, and the reader is directed to a number of excellent articles on this topic (Sestan and State, 2018; Visscher et al., 2017).
Progress is being made on several fronts. As described at length elsewhere (Sullivan and Geschwind, 2019) integrating functional genomic data can massively augment the interpretability of GWAS findings (Giusti-Rodriguez and Sullivan, 2018; Wang et al., 2018). However, perhaps the greatest explanatory power of GWAS findings for psychiatric disorders including MDD, is that the implicated genes in aggregate identify specific brain cell types (Bryois et al., 2019; Skene et al., 2018). Neuroscience has a range of tools for the manipulation of single risk genes for oligogenic disorders but application of these methods to hundreds of MDD risk genes may be impractical; however, the toolkit for manipulating specific brain cell types is rapidly improving and these approaches may be best for the post-GWAS era.
Figure 2:
Recent progress in genome-wide loci discovery from GWAS studies
Table 3:
Our top 4 priorities for genetic studies of MD
| Priority | Opportunity |
|---|---|
| Increase the sample sizes available for GWAS of MD, including cases meeting full DSM or ICD criteria for major depressive disorder | Improved knowledge of genetic architecture, more accurate genetic prediction, greater numbers of genetic instruments to discover modifiable environmental factors. Identification of genetic variants contributing to more severe and persistent clinically-defined definitions |
| Greater inclusion of diverse ancestries and low and middle-income countries | Representative inclusion of global ethnicities and cultures; improved fine-mapping of causal variants; stronger causal inferences based on consistently identified associations in different contexts |
| Integration with electronic medical records | Ability to examine longitudinal associations with clinical symptoms, treatment response and comorbid physical conditions; enables stratification of depression based on clinical factors; provides a platform for recruitment to clinical trials and observational studies |
| Developing the neurosciences of polygenic disorders | To identify the intermediate molecular, cellular, and systems biology of MD through simultaneous modelling of many low-penetrance risk alleles |
Highlights.
A review by McIntosh et al in this edition takes stock of recent rapid progress in the genetics of depression, how it has increased our mechanistic understanding and how this information could be used to improve patient care in future.
Acknowledgements
AMM is supported by a Wellcome Trust Strategic Award reference 104036/Z/14/Z and by UK Medical Research Council (MRC) support (grant references MC_PC_17209 and MC_PC_MR/R01910X/1). CML is part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. She is supported by MRC grant MR/N015746/1. PFS and the PGC has received major funding from the US National Institute of Mental Health and the US National Institute of Drug Abuse (U01 MH109528 and U01 MH1095320). PFS also gratefully acknowledges support from the Swedish Research Council (Vetenskapsrådet, award D0886501) and the Horizon 2020 Program of the European Union (COSYN, RIA grant agreement n°610307).
Footnotes
Declaration of interests
AMM currently receives research support from the Sackler Trust and has received past support from Eli Lilly and Janssen within the last 3 years. PFS reports the following potentially competing financial interests. Current: Lundbeck (advisory committee, grant recipient). Past three years: Pfizer (scientific advisory board), Element Genomics (consultation fee), and Roche (speaker reimbursement). CML receives research support from Eli Lilly and RGA Re-insurance.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders (DSM-5®) (American Psychiatric Pub). [Google Scholar]
- Andersson E, Crowley JJ, Lindefors N, Ljótsson B, Hedman-Lagerlöf E, Boberg J, El Alaoui S, Karlsson R, Lu Y, Mattheisen M, et al. (2018). Genetics of response to cognitive behavior therapy in adults with major depression: a preliminary report. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroll B, Khin N, and Kerse N (2003). Screening for depression in primary care with two verbally asked questions: cross sectional study. BMJ 327, 1144–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbu MC, Zeng Y, Shen X, Cox SR, Clarke T-K, Gibson J, Adams MJ, Johnstone M, Haley CS, Lawrie SM, et al. (2018). Association of Whole-Genome and NETRIN1 Signaling Pathway–Derived Polygenic Risk Scores for Major Depressive Disorder and White Matter Microstructure in the UK Biobank. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border R, Johnson EC, Evans LM, Smolen A, Berley N, Sullivan PF, and Keller MC (2019). No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples. Am. J. Psychiatry appiajp201818070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryois J, Skene NG, Hansen TF, and Kogelman LJA (2019). Genetic Identification of Cell Types Underlying Brain Complex Traits Yields Novel Insights Into the Etiology of Parkinson’s Disease. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM, Corvin A, et al. (2015). LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Butterworth A, and Thompson SG (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol 37, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N, Li Y, Chang S, Liang J, Lin C, Zhang X, Liang L, Hu J, Chan W, Kendler KS, et al. (2015). Genetic Control over mtDNA and Its Relationship to Major Depressive Disorder. Curr. Biol 25, 3170–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N, Kendler K, and Flint J (2018). Minimal phenotyping yields GWAS hits of low specificity for major depression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp NJ, and Cannon-Albright LA (2005). Dissecting the genetic etiology of major depressive disorder using linkage analysis. Trends Mol. Med 11, 138–144. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, et al. (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389. [DOI] [PubMed] [Google Scholar]
- Chesney E, Goodwin GM, and Fazel S (2014). Risks of all-cause and suicide mortality in mental disorders: a meta-review. World Psychiatry 13, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Van Eerdewegh P, Goate A, Edenberg HJ, Blangero J, Hesselbrock V, Reich T, Nurnberger J Jr, Schuckit M, Porjesz B, et al. (1998). Anxiety proneness linked to epistatic loci in genome scan of human personality traits. Am. J. Med. Genet 81, 313–317. [DOI] [PubMed] [Google Scholar]
- CONVERGE consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse RC, Bowes L, Breslau N, Nurnberger JI Jr, Burmeister M, Fergusson DM, Munafò M, Saccone NL, Bierut LJ, and 5-HTTLPR, Stress, and Depression Consortium (2013). Protocol for a collaborative meta-analysis of 5-HTTLPR, stress, and depression. BMC Psychiatry 13, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, Dickens C, Fox E, Graham N, Holliday J, et al. (2018a). Mental health in UK Biobank: development, implementation and results from an online questionnaire completed by 157 366 participants. BJPsych Open 4, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Davis KAS, Bashford O, Jewell A, Shetty H, Stewart RJ, Sudlow CLM, and Hotopf MH (2018b). Using data linkage to electronic patient records to assess the validity of selected mental health diagnoses in English Hospital Episode Statistics (HES). PLoS One 13, e0195002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Direk N, Williams S, Smith JAJA, Ripke S, Air T, Amare ATAT, Amin N, Baune BTBTBT, Bennett DADA, Blackwood DHRDHR, et al. (2017). An Analysis of Two Genome-wide Association Meta-analyses Identifies a New Locus for Broad Depression Phenotype. Biol. Psychiatry 82, 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri C, Kasper S, Kautzky A, Bartova L, Dold M, Zohar J, Souery D, Montgomery S, Albani D, Raimondi I, et al. (2019). Genome-wide association study of treatment-resistance in depression and meta-analysis of three independent samples. Br. J. Psychiatry 214, 36–41. [DOI] [PubMed] [Google Scholar]
- Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’Donovan MC, Corvin A, Cichon S, and Sullivan PF (2015). Evaluating historical candidate genes for schizophrenia. Mol. Psychiatry 20, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergusson DM, Horwood LJ, Miller AL, and Kennedy MA (2011). Life stress, 5-HTTLPR and mental disorder: findings from a 30-year longitudinal study. Br. J. Psychiatry 198, 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Miriam G, and B.w. WJ (2002). Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. (SCID-I/P)(New York: Biometrics Research, New York State Psychiatric Institute). [Google Scholar]
- Ford E, Carroll JA, Smith HE, Scott D, and Cassell JA (2016). Extracting information from the text of electronic medical records to improve case detection: a systematic review. J. Am. Med. Inform. Assoc 23, 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton J, Cubin M, Tiwari H, Wang C, Bomhra A, Davidson S, Miller S, Fairburn C , Goodwin G, Neale MC, et al. (2003). Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am. J. Hum. Genet 72, 879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti-Rodriguez PM, and Sullivan PF (2018). Schizophrenia and a high-resolution map of the three-dimensional chromatin interactome of adult and fetal cortex. [Google Scholar]
- Hafferty JDJD, Campbell AIAI, Navrady LBLB, Adams MJMJ, MacIntyre D, Lawrie SMSM, Nicodemus K, Porteous DJDJ, and McIntosh AMAM (2018). Self-reported medication use validated through record linkage to national prescribing data. J. Clin. Epidemiol 94, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LSLSLS, Adams MJMJMJ, Arnau-Soler A, Clarke TKT-KT-KT-K, Howard DMDMDM, Zeng Y, Davies G, Hagenaars SPSPSP, Fernandez-Pujals AMAM, Gibson J, et al. (2018). Genome-wide meta-analyses of stratified depression in Generation Scotland and UK Biobank. Transl. Psychiatry 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Amin N, Bakshis E, Baumert J, Ding J, et al. (2013). A genome-wide association study of depressive symptoms. Biol. Psychiatry 73, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, Depaulo JR Jr, Knowles JA, Zubenko WN, Murphy-Eberenz K, Marta DH, et al. (2007). Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am. J. Psychiatry 164, 248–258. [DOI] [PubMed] [Google Scholar]
- Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Ward J, Wigmore EM, Alloza C, et al. (2018a). Genome-wide meta-analysis of depression in 807,553 individuals identifies 102 independent variants with replication in a further 1,507,153 individuals. [Google Scholar]
- Howard DMDM, Adams MJMJ, Shirali M, Clarke T-KTK, Marioni RERE, Davies G, Coleman JRIJRI, Alloza C, Shen X, Barbu MCMC, et al. (2018b). Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, Tung JY, Hinds DA, Perlis RH, and Winslow AR (2016). Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet 48, 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RG, Patel R, Jayatilleke N, Kolliakou A, Ball M, Gorrell G, Roberts A, Dobson RJ, and Stewart R (2017). Natural language processing to extract symptoms of severe mental illness from clinical text: the Clinical Record Interactive Search Comprehensive Data Extraction (CRIS-CODE) project. BMJ Open 7, e012012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, and Keller MC (2017). No Evidence That Schizophrenia Candidate Genes Are More Associated With Schizophrenia Than Noncandidate Genes. Biol. Psychiatry 82, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall KM, Rees E, Bracher-Smith M, Riglin L, Zammit S, O’Donovan MC, Owen MJ, Jones I, Kirov G, and Walters JTR (2018). The role of rare copy number variants in depression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendell RE (1987). Diagnosis and classification of functional psychoses. Br. Med. Bull 43, 499–513. [DOI] [PubMed] [Google Scholar]
- Kerr SMSM, Campbell A, Marten J, Vitart V, McIntosh AMAM, Porteous DJDJ, and Hayward C (2017). Electronic health record and genome-wide genetic data in Generation Scotland participants. Wellcome Open Research 2, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Andrews G, and Mroczek D The world health organization composite international diagnostic interview short-form (CIDI-SF). Int. J. Methods Psychiatr. Res, 7 (1998), pp. 171–185 [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, Rush AJ, Walters EE, Wang PS, and National Comorbidity Survey Replication (2003). The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 289, 3095–3105. [DOI] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-J, and Kim S-H (2018). Prediction of inherited genomic susceptibility to 20 common cancer types by a supervised machine-learning method. Proc. Natl. Acad. Sci. U. S. A 115, 1322–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P-H, Neale MC, Riley BP, Patterson DG, Walsh D, Prescott CA, and Kendler KS (2007). A genome-wide linkage analysis for the personality trait neuroticism in the Irish affected sib-pair study of alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet 144B, 463–468. [DOI] [PubMed] [Google Scholar]
- Levinson DF (2006). The Genetics of Depression: A Review. Biol. Psychiatry 60, 84–92. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR, and Sullivan PF (2014). Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol. Psychiatry 76, 510–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Hagenaars SPSP, Davies G, Hill WDD, Clarke TKT-K, Shirali M, Harris SESE, Marioni RERERE, Liewald DCDC, Fawns-Ritchie C, et al. (2018). Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat. Genet 50, 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Visscher PM, Robinson MR, and Wray NR (2018). Embracing polygenicity: a review of methods and tools for psychiatric genetics research. Psychol. Med 48, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DHR, Boomsma DI, et al. (2013). A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, Gill M, Korszun A, Maier W, Middleton L, et al. (2005). Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum. Mol. Genet 14, 3337–3345. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Stewart R, John A, Smith DJ, Davis K, Sudlow C, Corvin A, Nicodemus KK, Kingdon D, Hassan L, et al. (2016). Data science for mental health: a UK perspective on a global challenge. The Lancet Psychiatry 3, 993–998. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, Sullivan PF, Wray NR, Hottenga J-J, de Geus EJC, van den Berg M, Montgomery GW, Coventry WL, Statham DJ, Andrews G, et al. (2009). Suggestive linkage on chromosome 2, 8, and 17 for lifetime major depression. Am. J. Med. Genet. B Neuropsychiatr. Genet 150B, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milaneschi Y, Lamers F, Peyrot WJ, Baune BT, Breen G, Dehghan A, Forstner AJ, Grabe HJ, Homuth G, Kan C, et al. (2017). Genetic association of major depression with a typical features and obesity-related immunometabolic dysregulations. JAMA Psychiatry 74, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt TE, and Caspi A (2014). Bias in a protocol for a meta-analysis of 5-HTTLPR, stress, and depression. BMC Psychiatry 14, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi S, Chatterji S, Verdes E, Tandon A, Patel V, and Ustun B (2007). Depression, chronic diseases, and decrements in health: results from the World Health Surveys. Lancet 370, 851–858. [DOI] [PubMed] [Google Scholar]
- Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, Savage JE, Hammerschlag AR, Skene NG, Muñoz-Manchado AB, et al. (2018). Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet 50, 920–927. [DOI] [PubMed] [Google Scholar]
- Nash MW, Huezo-Diaz P, Williamson RJ, Sterne A, Purcell S, Hoda F, Cherny SS, Abecasis GR, Prince M, Gray JA, et al. (2004). Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum. Mol. Genet 13, 2173–2182. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sullivan PF, and Kendler KS (2005). A genome scan of neuroticism in nicotine dependent smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet 132B, 65–69. [DOI] [PubMed] [Google Scholar]
- NIMH (2018). Towards a Genomic Psychiatry: Recommendations of the Genomics Workgroup of the NAMHC. [Google Scholar]
- Nurnberger JI Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, et al. (2001). Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am. J. Psychiatry 158, 718–724. [DOI] [PubMed] [Google Scholar]
- Pakhomov SSV, Hemingway H, Weston SA, Jacobsen SJ, Rodeheffer R, and Roger VL (2007). Epidemiology of angina pectoris: role of natural language processing of the medical record. Am. Heart J 153, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V (2007). Mental health in low- and middle-income countries. British Medical Bulletin 81-82, 81–96. [DOI] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, and Posthuma D (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet 47, 702. [DOI] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, et al. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ TC, Woelbert E, Davis KAS, Hafferty JD, Ibrahim Z, Inkster B, John A, Lee W, Maxwell M, McIntosh AM, et al. (2018). How data science can advance mental health research. Nat Hum Behav 47, 114. [DOI] [PubMed] [Google Scholar]
- Sestan N, and State MW (2018). Lost in Translation: Traversing the Complex Path from Genomics to Therapeutics in Autism Spectrum Disorder. Neuron 100, 406–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Muñoz-Manchado AB, et al. (2018). Genetic identification of brain cell types underlying schizophrenia. Nat. Genet 50, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Nicholl BI, Cullen B, Martin D, Ul-Haq Z, Evans J, Gill JMR, Roberts B, Gallacher J, Mackay D, et al. (2013). Prevalence and characteristics of probable major depression and bipolar disorder within UK biobank: cross-sectional study of 172,751 participants. PLoS One 8, e75362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW (2018). The use of electronic health records for psychiatric phenotyping and genomics. Am. J. Med. Genet. B Neuropsychiatr. Genet 177, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne JAC, Timpson N, and Davey Smith G (2008). Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Interface [DOI] [PubMed] [Google Scholar]
- Sullivan PF (2007). Spurious genetic associations. Biol. Psychiatry 61, 1121–1126. [DOI] [PubMed] [Google Scholar]
- Sullivan PF (2017). How Good Were Candidate Gene Guesses in Schizophrenia Genetics? Biol. Psychiatry 82, 696–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, and Geschwind DH (2019). Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, and Kendler KS (2000). Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry 157, 1552–1562. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Eaves LJ, Kendler KS, and Neale MC (2001). Genetic case-control association studies in neuropsychiatry. Arch. Gen. Psychiatry 58, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, and O’Donovan M (2012). Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet 13, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, Cichon S, Edenberg HJ, Faraone SV, Gelernter J, et al. (2018). Psychiatric Genomics: An Update and an Agenda. Am. J. Psychiatry 175, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thombs BD, Benedetti A, Kloda LA, Levis B, Nicolau I, Cuijpers P, Gilbody S, loannidis JPA, McMillan D, Patten SB, et al. (2014). The diagnostic accuracy of the Patient Health Questionnaire-2 (PHQ-2), Patient Health Questionnaire-8 (PHQ-8), and Patient Health Questionnaire-9 (PHQ-9) for detecting major depression: protocol for a systematic review and individual patient data meta-analyses. Syst. Rev 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakadis YJ, Sardaar S, Chen A, Fulginiti V, and Krishnan A (2018). Machine learning in schizophrenia genomics, a case-control study using 5,090 exomes. Am. J. Med. Genet. B Neuropsychiatr. Genet [DOI] [PubMed] [Google Scholar]
- Tyrrell J, Mulugeta A, Wood AR, Zhou A, Beaumont RN, Tuke MA, Jones SE, Ruth KS, Yaghootkar H, Sharp S, et al. (2018). Using genetics to understand the causal influence of higher BMI on depression. Int. J. Epidemiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, and Yang J (2017). 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, and Others (2017). Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic .… Lancet 90, 1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, et al. (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigmore EM, Hafferty JD, Hall LS, Howard DM, Clarke T-K, Fabbri C, Lewis CM, Uher R, Navrady LB, Adams MJ, et al. (2019). Genome-wide association study of antidepressant treatment resistance in a population-based cohort using health service prescription data and meta-analysis with GENDEP. Pharmacogenomics J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelbert E, Kirtley A, Balmer N, and Dix S (2019). How much is spent on mental health research: developing a system for categorising grant funding in the UK. Lancet Psychiatry. [DOI] [PubMed] [Google Scholar]
- World Health Organization (1993). The ICD-10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research (World Health Organization). [Google Scholar]
- Wray NR, Middeldorp CM, Birley AJ, Gordon SD, Sullivan PF, Visscher PM, Nyholt DR, Willemsen G, de Geus EJC, Slagboom PE, et al. (2008). Genome-wide linkage analysis of multiple measures of neuroticism of 2 large cohorts from Australia and the Netherlands. Arch. Gen. Psychiatry 65, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DHR, Penninx BWJH, Gordon SD, Nyholt DR, Ripke S, MacIntyre DJ, McGhee KA, MacLean AW, et al. (2012). Genome-wide association study of major depressive disorder: New results, meta-analysis, and lessons learned. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Navarro P, Xia C, Amador C, Fernandez-Pujals AM, Thomson PA, Campbell A, Nagy R, Clarke T-K, Hafferty JD, et al. (2016). Shared Genetics and Couple-Associated Environment Are Major Contributors to the Risk of Both Clinical and Self-Declared Depression. EBioMedicine 14, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Abdellaoui A, Rucker J, de Jong S, Potash JB, Weissman MM, and Shi J Genome-wide burden of rare short deletions is enriched in Major Depressive Disorder in four cohorts. Biological Psychiatry (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko GS, Maher B, Hughes HB 3rd, Zubenko WN, Stiffler JS, Kaplan BB, and Marazita ML (2003). Genome-wide linkage survey for genetic loci that influence the development of depressive disorders in families with recurrent, early-onset, major depression. Am. J. Med. Genet. B Neuropsychiatr. Genet 123B, 1–18. [DOI] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, and Lander ES (2014). Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. U. S. A 111, E455–E464. [DOI] [PMC free article] [PubMed] [Google Scholar]