Abstract
Traditional qPCR assays for pneumococcal detection and serotype characterization require large sample volume, is expensive and labor intensive. We aimed to develop a quantitative nanofluidic Fluidigm assay to overcome some of these shortcomings. A quantitative Fluidigm assay was established to detect 11 bacterial pathogens, 55 pneumococcal serotypes and 6 serotypes of H. influenzae. The Fluidigm assay results were compared to conventional qPCR and culture. All reactions in the Fluidigm assay effectively amplified their respective targets with high sensitivity and specificity compared to qPCR. There was excellent concordance between qPCR and Fluidigm for detection of carriage prevalence (kappa > 0.75) and density (Rho > 0.95). Fluidigm identified an additional 7 (4.2%) serotypes over those detected by qPCR. There was a modest concordance between culture and Fluidigm for the majority of reactions detecting S. pneumoniae serotypes/serogroups (kappa > 0.6), with Fluidigm identifying an additional 113 (39.1%) serotypes. Discordant results between the three methods were associated with a low carriage density. The Fluidigm assay was able to detect common pneumococcal serotypes, H. influenzae serotypes, and other common nasopharyngeal bacterial organisms simultaneously. Deployment of this assay in epidemiological studies could provide better insight into the effect of PCV immunization on the nasopharyngeal microbiota in the community.
Subject terms: Infectious-disease diagnostics, Biological sciences
Introduction
The human nasopharynx is a common ecological niche for several respiratory pathogens including Streptococcus pneumoniae, Staphylococcus aureus, Moraxella catarrhalis, and Haemophilus influenzae1. There is a paucity of data and conflicting findings on the interactions between these bacteria in the human nasopharynx2–6. Furthermore, most nasopharyngeal colonization studies have used non-quantitate culture based methods, with a lack of quantitative data available to assist in understanding the dynamics of interaction of these bacteria in the nasopharynx7–9. Recently, molecular quantitative PCR-based methods have been developed, which has assisted our understanding of bacterial nasopharyngeal colonization and its relationship to disease9–12. Serotyping of pneumococci by qPCR, however, has several shortcomings including large sample volumes needed to distribute across all reactions and it being labor intensive10.
Fluidigm is a nanofluidic automated real-time PCR system that relies on microfluidic technology in which dynamic arrays of integrated fluidic circuits (IFCs) are used. These IFC’s contain thousands of controlled valves and interconnected channels in which molecules of biological samples and reagents can be automatically mixed in a variety of patterns. The instrument uses an array of non-fluidic chips called dynamic arrays for qPCR, in which a typical chip format allows for 9 216 PCR reactions (96.96 chip format; 96 samples × 96 assays) in a single qPCR run (www.fluidigm.co.za)13. Other advantages of Fluidigm over standard qPCR include a larger number of reactions per plate, making it more cost-effective and less time-consuming. Further, IFCs not only reduce the reaction volume from 10 µL–20 µL down to 10 nL scale, but the technology allows for validations as well as increased throughput of qPCR reactions13.
We aimed to develop a novel nanofluidic real-time PCR (Fluidigm) assay that simultaneously detected and quantified 11 bacterial pathogens in 96 different samples (92 samples and 4 controls) in a single run. The assay included serotyping for 55 pneumococcal serotypes (16 individual serotypes and 39 serotypes in 13 groups) and six H. influenzae (serotypes a-f) serotypes. We compared the results of the Fluidigm assay to that of traditional qPCR for detection of bacterial colonization and pneumococcal serotyping in a cohort of PCV-vaccinated African children. Our previous work had compared qPCR to standard culture methods for 46 pneumococcal serotypes (14 individual serotypes and 32 serotypes among 10 serogroups) and 6 bacterial pathogens in a different cohort of children10. In these analyses, we expand on the latter by comparing the results from the Fluidigm assay to culture and qPCR.
Results
From the initial 407 nasopharyngeal swab samples collected at the two study visits, 335 (82.3%) were available for subsequent molecular nanofluidic qPCR (Fluidigm) analysis (Supplementary Fig. 1). Of these available samples, 83.4% were Black-African and 49.1% were male. Data from the two study visits were combined for the main analysis as findings from both 9 and 16 months of age were similar.
Performance of the Fluidigm
All reactions were effective in amplifying their respective targets with high sensitivity and specificity in the Biomark HD system (Fluidigm), with the efficiency of the reactions ranging from 89% to 105% (Supplementary Table 1). Within the linear dynamic range, the correlation coefficients (r2) of the reactions were 0.99, except for reactions detecting H. influenzae E, IS481 (B. pertussis and B. holmesii) and S. pneumoniae serotype 19A, were r2 was 0.98 (Supplementary Table 1). The limit of detection (LLD) for most of the reactions were equivalent >10 copies, with exception to primer/probes that detected H. influenzae serotype E, IS481 (B. pertussis and B. holmesii) and pneumococcal serotypes/serogroups 5, 6A/B/C/D, 6C/D, 13 and 21, in which the LLD was 10 fold less (>100 copies). Further, no cross-reactivity occurred, with all primer and probes being specific for their respective target. Lastly, for all respective reactions, the intra- and inter-assay variation was <0.1, while the accuracy was within ±0.1 (Supplementary Table 1).
Detection of bacterial nasopharyngeal carriage by qPCR and Fluidigm
There was excellent concordance between qPCR and Fluidigm for the detection of S. pneumoniae (Kappa = 0.98), H. influenzae (Kappa = 0.97), M. catarrhalis (Kappa = 0.98), S. aureus (Kappa = 0.98) and S. pyogenes (Kappa = 0.96); and a high concordance between qPCR and Fluidigm for the detection of N. meningitides (Kappa = 0.75; Fig. 1, Table 1). All additional bacteria detected by either qPCR or Fluidigm had estimated copy numbers of <102 colony forming units (CFU)/ml per swab (Fig. 2) and thus discordance between the methods was strongly associated with the density of carriage. The sensitivity and specificity of the Fluidigm method compared to conventional qPCR were high with all reactions having a sensitivity >92% and a specificity of >99% (Table 1).
Figure 1.
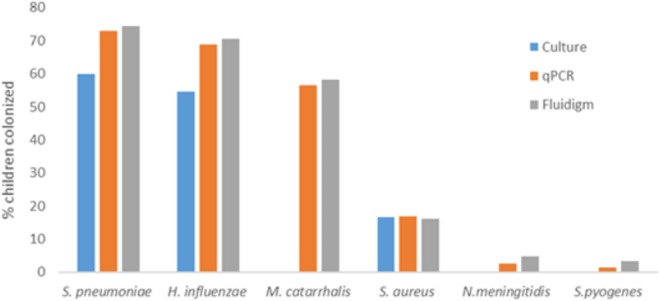
Prevalence of nasopharyngeal (NP) bacterial colonization in PCV-vaccinated, HIV-uninfected children as measured by culture, qPCR and Fluidigm. P-values of <0.05 was considered significant as determined by McNemar’s test.
Table 1.
Concordance between qPCR and Fluidigm for the detection of common nasopharyngeal pathogens in PCV-vaccinated, HIV-uninfected children.
| Bacterial Target | qPCR | Fluidigm | P-value | Concordance (kappa) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| S. pneumoniae | 244(72.8) | 246(73.4) | 0.16 | 0.98 | 98.8 | 100 |
| H. influenzae | 230(68.7) | 232(69.3) | 0.32 | 0.97 | 98.7 | 99 |
| M. catarrhalis | 189(56.4) | 192(57.3) | 0.08 | 0.98 | 98.4 | 100 |
| S. aureus | 57(17) | 55(16.4) | 0.16 | 0.98 | 100 | 99.3 |
| S. pyogenes | 11(3.3) | 12(3.6) | 0.32 | 0.96 | 91.7 | 100 |
| N. meningitidis | 3(0.9) | 5(1.5) | 0.16 | 0.75 | 91.7 | 100 |
Figure 2.
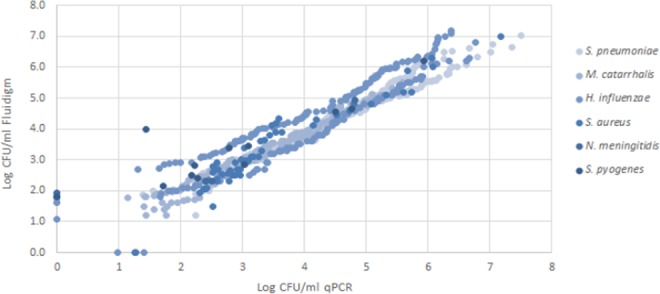
Density of colonization by common nasopharyngeal bacterial pathogens as determined by qPCR and Fluidigm.
B. bronchiseptica, B. parapertusis, B. pertussis, B. holmesii, and N. lactamica were not tested by conventional qPCR and thus could not be compared to Fluidigm; however, the overall detection prevalence by Fluidigm was 0.3%, 0%, 0%, 0%, and 4.2% respectively. Further H. influenzae was not serotyped by conventional qPCR and thus could not be compared to Fluidigm; however, of the H. influenzae positive samples, 3.9% were serotype a, 1.3% were serotype b, 2.6% were serotype c, 1.3% were serotype d, 1.7% were serotype e, 4.3% were serotype f, and 83.6% were non-typable (hinfNT).
Detection of bacterial nasopharyngeal carriage by culture and Fluidigm
There was excellent concordance between culture and Fluidigm assay for detection of S. aureus (Kappa = 0.86), and a modest concordance for detection of H. influenzae (Kappa = 0.63) and S. pneumoniae (Kappa = 0.59). The Fluidigm assay was more sensitive than culture in detecting S. pneumoniae (73.4% vs. 60%; p < 0.001) and H. influenzae (69.3% vs. 54.6%, p < 0.001; Fig. 1, Supplementary Table 2). Discordance for bacterial detection between culture and Fluidigm methods were associated with carriage density, with the majority of Fluidigm-positive but culture-negative samples being <104 CFU/ml per swab, and the majority of culture-positive but Fluidigm negative samples reported as “scant” growth (<5 colonies/plate) on culture.
Detection of pneumococcal serotype carriage by qPCR and Fluidigm
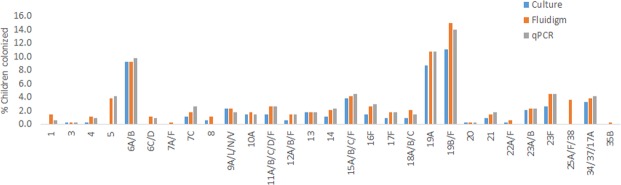
There was excellent concordance between qPCR and Fluidigm assays, with the majority of reactions detecting S. pneumoniae serotypes/serogroups (Kappa > 0.8) by both methods, with exception to serotype 1, for which the concordance was still high (Kappa = 0.75; Fig. 3, Table 2). Fluidigm was more sensitive in detecting serotypes/groups 1, 4, 10A, 18A/B/C, and 19B/F; while qPCR was more sensitive in detecting serotype 7C. In addition, the Fluidigm assay identified 7 (4.2%) additional serotypes compared to those identified by qPCR; and conversely, the qPCR method detected 2 (2.1%) additional serotypes not detected by Fluidigm. Discordant results between the methods were strongly associated with the density of carriage, with all additional pneumococcal serotypes/groups detected either by qPCR or Fluidigm having estimated copy numbers of <103 CFU/ml per swab (Fig. 4). The sensitivity and specificity of the Fluidigm compared to qPCR assay were high; i.e. sensitivity >80% and specificity >99.3%; Table 2.
Figure 3.
Prevalence of pneumococcal serotype colonization in PCV-vaccinated, HIV-uninfected children as measured by culture, qPCR and Fluidigm. P-values of <0.05 was considered significant as determined by McNemar’s test.
Table 2.
Concordance between qPCR and Fluidigm for the detection of pneumococcal serotypes in PCV-vaccinated, HIV-uninfected children.
| Serotype | qPCR | Fluidigm | P-value | Concordance (kappa) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| LytA | 244(72.8) | 246(73.4) | 0.16 | 0.98 | 98.8 | 100 |
| 1 | 3(0.9) | 5(1.5) | 0.16 | 0.75 | 95 | 100 |
| 3 | 1(0.3) | 1(0.3) | — | 1 | 100 | 100 |
| 4 | 3(0.9) | 4(1.2) | 0.32 | 0.86 | 80 | 100 |
| 5 | 13(3.9) | 13(3.9) | — | 1 | 100 | 100 |
| 6A/B | 31 (9.3) | 31 (9.3) | — | 1 | 100 | 100 |
| 6C/D | 2(0.6) | 2(0.6) | — | 1 | 100 | 100 |
| 7C | 8(2.4) | 6(1.8) | 0.16 | 0.85 | 100 | 99.3 |
| 9A/L/N/V | 8(2.4) | 8(2.4) | — | 1 | 100 | 100 |
| 10A | 5(1.5) | 6(1.8) | 0.32 | 0.91 | 83.3 | 100 |
| 11A/B/C/D/F | 9(2.7) | 9(2.7) | — | 1 | 100 | 100 |
| 12ABF/44/46 | 5(1.5) | 5(1.5) | — | 1 | 100 | 100 |
| 13 | 6(1.8) | 6(1.8) | — | 1 | 100 | 100 |
| 14 | 7(2.1) | 7(2.1) | — | 1 | 100 | 100 |
| 15A/B/C/F | 14(4.2) | 14(4.2) | — | 1 | 100 | 100 |
| 16F | 9(2.7) | 9(2.7) | — | 1 | 100 | 100 |
| 17F | 6(1.8) | 6(1.8) | — | 1 | 100 | 100 |
| 18A/B/C | 6(1.8) | 7(2.1) | 0.32 | 0.93 | 85.7 | 100 |
| 19A | 36(10.7) | 36(10.7) | — | 1 | 100 | 100 |
| 19B/F | 48(14.3) | 50(14.9) | 0.08 | 0.96 | 94 | 100 |
| 20 | 1(0.3) | 1(0.3) | — | 1 | 100 | 100 |
| 21 | 5(1.5) | 5(1.5) | — | 1 | 100 | 100 |
| 23A/B | 8(2.4) | 8(2.4) | — | 1 | 100 | 100 |
| 23F | 15(4.5) | 15(4.5) | — | 1 | 100 | 100 |
| 34/37/17F | 13(3.9) | 13(3.9) | — | 1 | 100 | 100 |
Figure 4.
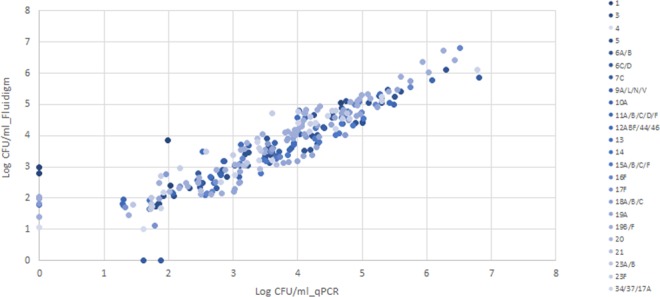
Density of colonization of pneumococcal serotypes as determined by qPCR and Fluidigm.
Serotypes/serogroups 7A/F, 8, 22A/F, 25AF/38 and 35B were only tested by Fluidigm, with an overall detection prevalence of 0.3%, 0.9%, 0.6%, 2.7% and 0.3%, respectively.
Detection of pneumococcal serotype carriage by culture and Fluidigm
Generally, there was high concordance between culture and the Fluidigm assay for detecting S. pneumoniae serotypes/serogroups. Culture identified 21 (10.9%) serotypes/serogroups not detected by the Fluidigm assay, and conversely the Fluidigm assay identifying as additional 113 (39.1%) serotypes not identified by culture methods (Fig. 3, Supplementary Table 3). This included higher detection prevalence by the Fluidigm assay for serotypes/serogroups 1 (1.5% vs. 1%; p = 0.03), 5 (8.9% vs. 0%; p = 0.003), 16 (2.7% vs. 1.5%, p = 0.05), 18A/B/C (1.8% vs. 0.9%, p = 0.05), 19B/F (14.9% vs. 11%, p = 0.002), 23F (4.5% vs. 2.7%, p = 0.03), and 25A/25F/38 (3.6% vs. 0%, p = 0.005). The additional serotypes detected only by Fluidigm mostly had densities <104 CFU/ml per swab. Furthermore, the majority of the additional serotypes detected by Fluidigm were carried concurrently with other pneumococcal serotypes, of 89% were a non-primary colonizing serotype (Supplementary Table 4). The sensitivity of the Fluidigm assay compared to culture were high for the majority of reactions; mainly having sensitivity >80%, with the exception of serotype 10A (60%) and serogroup 23A/B (71%). All Fluidigm reactions had high specificity (>95%) compared to culture (Supplementary Table 3).
Density of bacterial nasopharyngeal carriage as determined by qPCR and Fluidigm
There was an excellent correlation (Rho > 0.95) between qPCR and Fluidigm for measuring the density of colonization (Figs 2 and 4), with no significant differences observed between the two methods for all bacterial pathogens and pneumococcal serotypes (Table 3).
Table 3.
Density of bacterial nasopharyngeal carriage as determined by qPCR and Fluidigm.
| Bacteria | Density detected by qPCR | Density detected by Fluidigm | P-value |
|---|---|---|---|
| S. pneumoniae | 4.54(4.37–4.70) | 4.39(4.24–4.54) | 0.49 |
| H. influenzae | 4.38(4.21–4.55) | 4.56(4.39–4.73) | 0.18 |
| M. catarrhalis | 3.38(3.23–3.52) | 3.29(3.15–3.44) | 0.37 |
| S. aureus | 3.48(3.17–3.79) | 3.41(3.08–3.75) | 0.13 |
| S. pyogenes | 3.09(2.14–4.03) | 3.68(3.04–4.32) | 0.11 |
| N. meningitides | 3.24(−0.13–6.61) | 2.96(0.20–5.73) | 0.18 |
| Pneumococcal serotypes/serogroups | |||
| 1 | 2.90(−0.92–6.71) | 3.76(0.46–7.07) | 0.22 |
| 3 | 3.55 | 3.66 | — |
| 4 | 2.85(−0.27–5.97) | 2.30(−1.20–7.20) | 0.79 |
| 5 | 2.52(2.01–3.02) | 2.46(2.0–2.91) | 0.09 |
| 6A/B | 4.25(3.88–4.61) | 4.07(3.77–4.36) | 0.056 |
| 6C/D | 4.05(−1131–19.42) | 3.60(−5.11–12.30) | 0.54 |
| 7C | 3.49(1.73–5.25) | 3.27(1.89–4.67) | 0.33 |
| 9A/L/V/N | 3.31(2.52–4.10) | 3.23(2.52–3.95) | 0.16 |
| 10A | 3.23(2.47–3.98) | 3.62(2.64–4.1) | 0.17 |
| 11A/B/C/D/F | 3.29(2.03–4.56) | 3.21(2.16–4.27) | 0.51 |
| 12A/12B/12F/44/46 | 3.38(1.39–5.38) | 3.58(1.76–5.40) | 0.19 |
| 13 | 3.96(2.85–5.06) | 4.03(3.13–4.94) | 0.82 |
| 14 | 3.40(2.06–4.74) | 2.93(2.37–3.48) | 0.29 |
| 15A/B/C/F | 3.81(2.93–4.68) | 3.54(3.01–4.07) | 0.075 |
| 16F | 4.50(3.43–5.56) | 4.23(3.01–5.44) | 0.14 |
| 17F | 2.54(1.55–3.53) | 2.55(1.67–3.44) | 0.88 |
| 18A/B/C | 3.84(2.39–5.28) | 3.84(2.96–4.73) | 0.98 |
| 19A | 3.82(3.48–4.17) | 3.75(3.4–4.10) | 0.26 |
| 19B/F | 3.87(3.49–4.24) | 4.14(3.78–4.51) | 0.66 |
| 20 | ND | ND | |
| 21 | 3.38(2.23–4.53) | 3.13(2.58–3.68) | 0.55 |
| 23A/B | 2.92(−0.76–4.92) | 2.58(0.31–4.86) | 0.16 |
| 23F | 3.64(3.12–4.16) | 3.98(3.44–4.51) | 0.181 |
| 34/37/17A | 3.90(3.08–4.72) | 3.41(2.88–3.94) | 0.059 |
Numbers are geometric mean densities (95% Confidence intervals).
P-values of 0.05 were considered significant as determined by a paired student t-test.
ND, not done; too few variables to calculate.
Discussion
A reliable molecular nanofluidic real-time PCR (Fluidigm) system was established to simultaneously detect 11 bacterial pathogens, 55 pneumococcal serotypes (16 individual serotypes and 39 serotypes in 13 groups) and 6 serotypes of H. influenzae (serotypes a-f) in a single run14. There was high concordance between qPCR and Fluidigm, including for identifying bacterial colonization, density of bacterial targets and pneumococcal serotypes, with discordant results mainly associated with samples in which the carriage density was low (<102 CFU/ml per swab).
Discordance between the methods included the qPCR method detecting an additional 3 bacterial species and 2 pneumococcal serotypes above those detected by Fluidigm, and conversely the Fluidigm assay detecting an additional 11 bacterial species and 7 pneumococcal serotypes above those detected by qPCR. Although most of these differences could not be confirmed by culture, due to the additional pneumococcal serotypes detected being non-primary (second or third colonizing serotypes; Supplementary Fig. 3) colonizers, one additional serogroup 19B/F isolate detected by Fluidigm and one additional serotype 7C isolate detected by qPCR were concordant with culture results. These small differences (<5%) in the detection of targets between qPCR and Fluidigm methods are, however, expected based on Poisson’s distribution that the assays had a 95% probability of detecting their respective targets with reasonable certainty14. This was further supported by the discordant readouts between the methods mainly being associated with samples where the density of carriage was very low (<102 CFU/ml). Furthermore, the higher yield on the Fluidigm assay could be due to a pre-amplification step that selected for targets in this assay (and not the qPCR assay).
While culture remains the referent standard for bacterial carriage and pneumococcal serotype characterization studies, these assays lack quantitative data which impose limitations on our understanding of the association between nasopharyngeal carriage and its relation to disease15–19. Recently, several molecular qPCR methods have been developed to overcome the above shortcoming of conventional culture assays. Nevertheless, most molecular methods are still limited by the large sample volume required to distribute across all reactions, cost (approximately $ 1.85 per sample per reaction in duplex) and processing multiple qPCR reactions being labor intensive and time-consuming10. Further, there is a limited number of quantitative studies that have concurrently described colonization of a diverse number of bacterial pathogens and pneumococcal serotypes.
Our Fluidigm assay addressed some of the challenges posed by qPCR assays, including analyzing a larger number of reactions per plate making the assay more cost-effective (approximately $ 0.37 per sample per reaction), less time-consuming and less labor intensive (9216 reactions per one Fluidigm plate versus 96 reactions per one qPCR plate). Further, IFCs not only reduces the reaction volume from 10 µL–20 µL down to 10 nL scale, but the technology allows for validations as well as increased parallelism and throughput of qPCR reactions13.
Notably, the Fluidigm assay was more sensitive in detecting serotype 5 (3.9% vs. 0%) and serogroup 25AF/38 (3.6% vs. 0%) compared to culture (Fig. 3). Half of the isolates typed as serogroup 22AF/38 were not identified on culture, while the other half were commonly found as non-primary colonizers (Supplementary Fig. 2), which might explain them not being identified by culture methods. The majority of serotype 5 isolates (12/13; 92%) were commonly isolated as non-primary serotypes, raising the possibility of false positive readouts from the Fluidigm assay. These results were consistent with findings when we analyzed using the qPCR compared to culture assay10. This is of particular importance for serotype 5 since the primers used also detect S. mitis in Silio. Nevertheless, we recommend that results from serotype 5 and serogroup 22AF/38 are interpreted with caution and further investigation is warranted.
The Biomark HD system in a new instrument that has only been used to serotype pneumococcus by one other group; however, their assay differed in that they used Evagreen (a dsDNA-binding dye) chemistry instead of TaqMan probes to quantify bacterial carriage. Further, the study by Dhoubhadel et al. was designed to only detect 50 pneumococcal serotypes (17 individual serotypes and 33 serotypes in 12 groups) and did not include any additional bacterial pathogens20,21. The performance of our assay was, however, comparable to that described by Dhoubhadel et al.
Limitations of our study include that Fluidigm assay was unable to discriminate between all pneumococcal serotypes within some serogroups. Also, although the Fluidigm assay included additional pneumococcal serotypes and other bacterial targets compared to those investigated for by qPCR10,11, we did not test for all known pneumococcal serotypes. This should be considered for future studies. Further, the LytA gene was chosen to detect overall pneumococcal colonization; however, recent studies reported other isolates within the S. mitis group also harbor LytA22. No serotype was discernible for a small percentage (7.9%) of LytA positive samples detected by Fluidigm in our study, suggesting they were either non-typable or belonged to one of the serotypes not included in our assay. Alternately, these could have been LytA positive non-pneumococcal species that were being identified. Future studies on pneumococcal colonization using qPCR or Fluidigm should consider including an alternative gene such as Xisco, which is purportedly only present in Streptococcus pneumoniae23. As an initial step for gene expression on the Biomark HD Fluidigm system manufacturers recommends Specific target amplification (STA) for each sample to be done by combining all primers in a pool. Due to the high similarity between some pneumococcal primers, non-specific binding of primers to each other resulted (results not shown) in cross-specificity between some of the pneumococcal serotyping targets. This was addressed by separating primers into two separate pools (Supplementary Table 5), and then combing the STA products from the two assays. Careful monitoring and optimization of the Fluidigm assay are thus needed in future studies, especially if the assay is to be expanded with additional serotypes.
In conclusion, we established a Fluidigm assay that was highly sensitive and specific. Using the Fluidigm assay enabled simultaneous detection of nasopharyngeal colonization by common pneumococcal serotypes, H. influenzae serotypes, and other common nasopharyngeal bacterial pathogens; and provided quantitative data in a single run.
Material and Methods
Study population
Archived nasopharyngeal swab samples collected from a PCV7-vaccinated cohort of HIV-uninfected infants in Soweto, South Africa were retrospectively analyzed. The study cohort has been previously described24,25. Briefly, infants enrolled between April 2005 and June 2006 were 6–12 weeks old at enrolment and included participants who were both HIV-exposed-uninfected (HEU) and HIV-unexposed. These infants were scheduled to receive three doses of PCV7 (i.e. Prevnar®; Wyeth Vaccines, NJ, USA) at 6, 10 and 14 weeks of age25,26. PCV immunization was introduced into the public EPI program in May 2009 and thus, during the course of the study, PCV immunization of children in Soweto (birth cohort 28,000 per annum) was limited mainly to study-participants (approximately 600)27. As described, standard culture methods were used to culture samples for S. pneumoniae and the Quellung method was undertaken for serotyping28.
Nasopharyngeal (NP) swabs were collected at several time intervals, including at 9 and 16 months of age. Swabs were stored in skim milk-tryptone-glucose-glycerol (STGG) transport media at the Respiratory and Meningeal Pathogen Research Unit (RMPRU) in South Africa29. Samples were screened previously for S. pneumoniae, H. influenzae, S. aureus, M. catarrhalis, S pyogenes, and N meningitides by qPCR11, and pneumococcal serotyping was done by qPCR on all samples that tested positive for S. pneumoniae as described30. In this study, we now developed and evaluated a Fluidigm assay which screened for an additional 9 serotypes/serogroups (7A/F, 8, 22A/F, 25AF/38 and 35B) not included in our earlier qPCR assay, as well as investigated for presence of additional bacteria (Bordetella bronchiseptica, Bordetella parapertusis, Bordetella pertussis, Bordetella holmesii, and Neisseria lactamica. The Fluidigm assay was also designed to serotype H. influenzae. The additional serotypes included in the Fluidigm assay were chosen based on the most frequently isolated non-vaccine serotypes associated with colonization in South Africa as detected by culture methods, at the time of study design.
Bacterial and pneumococcal reference isolates
Control strains for the pneumococcal serotypes (1, 2, 3, 4, 5, 6A, 6B, 6C, 6D, 7A, 7F, 7C, 8, 9A, 9L, 9N, 9V, 10A, 11A, 11B, 11C, 11D, 11F, 12A, 12B, 12F, 13, 14, 15A, 15B, 15C, 15F, 16A, 16F, 17A. 17F, 18A, 18B, 18C, 19A, 19B, 19F, 20, 21, 22A, 22F, 23A, 23B, 23F, 25A, 25F, 34, 35B and 37), H. influenzae serotypes (serotypes a-f), S. aureus, M. catarrhalis, N. meningitidis and S. pyogenes were obtained from the National Institute for Communicable Diseases (NICD). Additional isolates for B. bronchiseptica (ATCC® 4617), B. parapertusis (ATCC® 15311), B. pertussis (ATCC® 2397) and N. lactamica (ATCC® 23970) were purchased from Davies Diagnostics (South Africa), while an isolate for B. holmesii (ATCC® 51541) was purchased from LGC standards South Africa. DNA from these strains were used to optimize the Fluidigm reactions and as positive controls.
DNA extraction
Total nucleic acid was automatically extracted from nasopharyngeal swab samples stored in STGG, using the NucliSens® easyMAG® extraction system (BioMérieux, Marcy l′Etoile, France), according to manufactures instructions. Similarly, total nucleic acid was also extracted from pneumococcal reference strains (positive control strains) grown in Todd-Hewitt broth supplemented with 5% yeast. S. aureus, M. catarrhalis, S. pyogenes, N. meningitidis, and N. lactamica reference strains were grown in Todd-Hewitt broth alone, H. influenzae serotypes a-f were grown in Brain-Heart infusion (BHI) broth and Bordetella species were grown in potato broth. Extracted DNA from samples and reference strains were stored at −20 °C.
Real-time qPCR multiplex assay
Target DNA was previously pre-screened for S. pneumoniae, M. catarrhalis, H. influenzae, S. aureus, N. meningitidis and S. pyogenes as described10,11. All LytA positive samples (Cq < 35) were further molecularly serotyped for PCV7 serotypes/groups (4, 6A/B, 9A/L/N/V, 14, 18A/B/C, 19B/F and 23F) and non-vaccine serotypes/groups (1, 3, 4, 5, 6C/D, 10A, 11A/B/C/D/F, 12A/B/F, 13, 15A/B/C/F, 16F, 17F, 19A, 20, 21, 23A/B and 34/37/17A) as described10.
Fluidigm assay
ABI primer express software package, version 3.0 (Applied Biosystems, ABI, Foster City, USA) was used to design oligonucleotide primers and FAM dye-labelled MGB probes for additional targets not included in the multiplex qPCR assay. Primer and probe sequences used in the Fluidigm assay are described in Table 4. GAPDH and BexA were included to confirm the efficiency of the DNA extraction and to confirm the presence of non-typable H. influenzae, respectively. The Fluidigm method was unable to discriminate between some pneumococcal serotypes within a particular serogroup due to the genotypic similarities between the capsule loci of certain serotypes (including serotypes: 6A/B, 6C/D, 7A/F, 9A/L/N/V, 11A/B/C/D/F, 12A/B/F/44/46, 15A/B/C/F, 18A/B/C, 19B/F, 22A/F, 23A/B, 25A/25F/38 and 34/37/17A); however, the method was able to identify serotypes 1, 3, 4, 5, 7C, 8, 10A, 13, 14, 16F, 17F, 19A, 20, 21, 23F and 35B individually.
Table 4.
Oligonucleotide primers and probea sequences for quantitative molecular Fluidigm detection of respiratory pathogen.
| TargetName | Primer/Probename | Forward primer | Reverse primer | Probe | Reference |
|---|---|---|---|---|---|
| Streptococcus pneumoniae | lytA | TCTTACGCAATCTAGCAGATGAAGC | GTTGTTTGGTTGGTTATTCGTGC | TTTGCCGAAAACGCTTGATACAGGG | McAvinet al.31 |
| Haemophilus influenzae | IGA | CAAAATTGCCAAGATTAAATGCTT | TGCTCGCCATACTGCACA A | CCTGCGGTTAAACC | This study |
| Haemophilus influenzaetype A | HiA | GCAACCATCTTACAACTTAGCGAATAC | GGTCTGCGGTGTCCTGTGTT | AAGTGAAGCATGTCGCCATTCGTCCA | Maaroufi et al.32 |
| Haemophilus influenzaetype B | HiB | TGTTCGCCATAACTTCATCTTAGC | CTTACGCTTCTATCTCGGTGATTAATAA | CACAAAACTTCTCATTCTTCGAGCCTA | Maaroufi et al.32 |
| Haemophilus influenzaetype C | HiC | TCTGTGTAGATGATGGTTCAGTAG | TTAGGATATTTACGCTGCCATT | TGCAGCTAAGATTATT | Maaroufi et al.32 |
| Haemophilus influenzaetype D | HiD | TATTGATGACCGATACAACCTGTTTAAA | CCAGAAATTATTTCTCCGTTATGTTGA | AATGGTTGTAAAACTCTTCT | Maaroufi et al.32 |
| Haemophilus influenzaetype E | HiE | GTTGAAAACAAACCGCACTTT | ATCTTTAATTACCAGATCCCTTTCAT | AACGAATGTAGTGGTAGTTAGA | Maaroufi et al.32 |
| Haemophilus influenzaetype F | HiF | GGATAATCAAATACCACATTGGCTTA | GTAGATTAGCCTCAATAACATGTGAATTAA | TCATCGTGAGATCATTGATCACGAT | Maaroufi et al.32 |
| BexA | BexA | CTGAATTRGGYGATTATCTTTATGA | ACAATCAAAYTCAACHGAAAGHGA | AGGGATGAAAGCYCGRCTTGCAT | Maaroufi et al.32 |
| Staphylococcus aureus | SA | GCTCAGCAAATGCATCACAAA | CACTATATACTGTTGGATCTTCAGAACCA | AGATAACGGCGTAAATA | This Study |
| Moraxella catarrhalis | MCAT | CCGCTTTTACAACCACTGCTT | TGTATCGCCTGCCAAGACAA | CAGCTGTTAGCCAGCC | This Study |
| Streptococcus pyogenes | SPY | GCACTCGCTACTATTTCTTACCTCAA | GTCACAATGTCTTGGAAACCAGTAAT | CCGCAACTCATCAAGGATTTCTGTTACCA | CDC 2008 |
| Neisseria meningitidis | SodC | GCACACTTAGGTGATTTACCTGCAT | CCACCCGTGTGGATCATAATAGA | CATGATGGCACAGCAACAAATCCTGTTT | Thomas et al.33 |
| Neisseria lactamica | LacZ | TTGCCCGAGAACCATTGTATC | GCGGTTCTTATCACGTTCTATATTTG | TATTGGAGCGGACTAAA | ThisStudy |
| Bordetella pertusis and Bordetella holmesii | IS481 | CAAGGCCGAACGCTTCAT | GAGTTCTGGTAGGTGTGAGCGTAA | CAGTCGGCCTTGCGTGAGTGGG | Tattiet al.34 |
| Bordetella holmesii | hIS1001 | GGCGACAGCGAGACAGAATC | GCCGCCTTGGCTCACTT | CGTGCAGATAGGCTTTTAGCTTGAGCGC | Tattiet al.34 |
| Bordetella parapertusis | pIS1001 | TCGAACGCGTGGAATGG | GGCCGTTGGCTTCAAATAGA | AGACCCAGGGCGCACGCTGTC | Tattiet al.34 |
| Bordetella pertusis, Bordetella parapertusis and Bordetella bronchiseptica | PtxS | CGCCAGCTCGTACTTC | GATACGGCCGGCATT | AATACGTCGACACTTATGGCGA | Tattiet al.34 |
| Streptococcus pneumoniae serotype 1 | 1 | CGTGCGGTAATTGAAGCTATGA | TGTGGCCCCAGCAACTCT | TGCTTGCCCTTGTATAGGGT | Azzariet al.35 |
| Streptococcus pneumoniae serotype 3 | 3 | GGTCAGCAGAAAGTATGCATTGG | TCGTTTATCCAGGGTCTGATGA | TATTGGATGTGGTTTATCGTGAAGA | Azzariet al.35 |
| Streptococcus pneumoniae serotype 4 | 4 | TGGGATGACATTTCTACGCACTA | CCGTCGCTGATGCTTTATCA | TCCTATTGGATGGTTAGTTGGTGA | Azzariet al.35 |
| Streptococcus pneumoniae serotype 5 | 5 | TTACGGGAGTATCTTATGTCTTTAATGG | CAGCATTCCAGTAGCCTAAAACTAGA | TTGTCTCAGCAACTCTATTTGGCTGTGGG | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 6A/B/C/D | 6A/B/C/D | AAGTTTGCACTAGAGTATGGGAAGGT | ACATTATGTCCRTGTCTTCGATACAAG | TGTTCTGCCCTGAGCAACTGG | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 6C/D | 6C/D | TTGGGATGATTGGTCGTATTAG | CTCTTCAATTAGTTCTTCAGTTCG | CCACGCAATTCGCCATC | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 7A/F | 7A/F | GATGGCATGTGGCAAACCA | TTTGCCCTCCTTAATCATTTCAC | TTGGCTATCGGCATGGTGGT | Azzariet al.35 |
| Streptococcus pneumoniae serotype 7C | 7C | CGTCAGGAATAGGTGCAATCTCT | TGAAATTCCAAGCGAAGCAA | TTC ATCTATTGGTTCTTATGGTGTT | ThisStudy |
| Streptococcus pneumoniae serotype 8 | 8 | CCACTCATCAGTTTCCCATATGTTT | TCAATAATTGAAGAAGCGAACGTT | TGATGGCAGATGGGTTGGGACGAG | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 9A/L/N/V | 9A/L/N/V | TGGAATGGGCAAAGGGTAGTA | TCGGTTCCCCAAGATTTTCTC | TTAATCATGCTAACGGCTCATCGA | Azzariet al.35 |
| Streptococcus pneumoniae serotype 10A | 10A | CCTCTCCTATCAACTATTACTCATTATACTACCT | AATAACCATAAGTCCCTAGATCATTCAAAG | TCATTACAACTCCCTATGTGACACGGGTCTTTT | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 11A/B/C/D/F | 11A/B/C/D/F | ACCGCATTTCTTATCGCACTATATT | TCTCCTTACCATCAAACATGTTAATCA | TGAATCAGTCTGACCGTTT | ThisStudy |
| Streptococcus pneumoniae serogroup 12ABF/44/46 | 12A/12B/12F/44/46 | GATTATTCGCTTGCCTCTTCATG | ATAGCCGAAATAAGCTTTCCAGAA | ATTTGTAAGCGGACGTGCGATT | Azzariet al.35 |
| Streptococcus pneumoniae serotype 13 | 13 | TCGGATTTAGTAGTAACCCCATTGA | TTCTTGATTGAGGATGCATTTCC | AGTAGTAAGAGATCATATTCAAG | ThisStudy |
| Streptococcus pneumoniae serotype 14 | 14 | CGACTGAAATGTCACTAGGAGAAGAT | AATACAGTCCATCAATTACTGCAATACTC | TGTCATTCGTTTGCCAATACTTGATGGTCTC | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 15A/B/C/F | 15A/B/C/F | TTGAATCAGGTAGATTGATTTCTGCTA | CTCTAGGAATCAAATACTGAGTCCTAATGA | CTCCGGCTTTTGTCTTCTCTGT | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 16F | 16F | GCAACTGGTATTTTTGATATTGGAGAA | CAAAGGAATGCCATGCCATA | AAAATGCTAACTTCGTTGGAGG | ThisStudy |
| Streptococcus pneumoniae serotype 17F | 17F | GTAAAGATTTCATGTCCTATAAGGGAGAA | AGGCGTCCCTGTTTATGAGAAG | TTGTACATGGTCTGGATTT | ThisStudy |
| Streptococcus pneumoniae serogroup 18A/B/C | 18A/B/C | CCTGTTGTTATTCACGCCTTACG | TTGCACTTCTCGAATAGCCTTACTC | AACCGTTGGCCCTTGTGGTGGA | Azzariet al.35 |
| Streptococcus pneumoniae serotype 19A | 19A | TTCGACGACGTATCAGCTTCA | TCATTGAGAGCCTTAACCTCTTCA | ACCCAAAACGGTTGACGCATTATACT | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 19B/F | 19B/F | GGTCATGCGAGATACGACAGAA | TCCTCATCAGTCCCAACCAATT | ACCTGAAGGAGTAGCTGCTGGAACGTTG | Azzariet al.35 |
| Streptococcus pneumoniae serotype 20 | 20 | AAAGATACTGGCTGAGGAGCTATCTATT | AGTCAAAAGTACTCAACCATTCTGATATATTC | AGGATAAGGTCTACTTTGTGGGAGTTC | Azzariet al.35 |
| Streptococcus pneumoniae serotype 21 | 21 | CCATTTGAAGGACCAGTTGTTG | AAAAAGCCACTATCAGGAATACCAA | AATGGCATTGCTTCGTAAA | ThisStudy |
| Streptococcus pneumoniae serotype 22A/F | 22A/F | TCTATTAAATAACCCATTGGAATTGAAACG | TCGCAATTGAAGACCACATAAACTG | TCCGTAAT”T”CGCTTATGGGCACATTCTCCA | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 23A/B/F | 23A/B/F | GGTGGACTTTCCGATGCAA | CACTGTCAACAAAAATGAGGTAATCTC | AAATGTCGGTATAGATAAAG | ThisStudy |
| Streptococcus pneumoniae serotype 23F | 23F | TGCTATTTGCGATCCTGTTCAT | AGAGCCTCCGTTGTTTCGTAAA | TTTCTCCGGCATCAAACGTTAAG | Azzariet al.35 |
| Streptococcus pneumoniae serogroup 25A/25F/38 | 25A/25F/38 | GTCTTACGTAGAACCTCTCTGGATGA | TGGTCCTACAAGCGACATGTG | TTGCCACAGATTTGGAATATTTTGGTCGG | ThisStudy |
| Streptococcus pneumoniae serogroup 34/37/17A | 34/37/17A | GGATACTATGTACGAACAGATGGACTTG | CTCACTAACTCGCCCGAATAAAC | CCGACTATACTCCATTTGA | ThisStudy |
| Streptococcus pneumoniae serotype 35B | 35B | GCATGGAGGTGGAGCATACA | TGTAAAGACTGCACAACTCGATATAAAA | CAATTTAAACAATATTAGTAAAGCGCAGGTCAAGCAAA | Azzariet al.35 |
Probea: Minor Grove Binding (MGB) FAM labelled TaqMan probes.
Pre-amplification of DNA for Fluidigm
Specific target amplification (STA) was done per manufactures recommendations as the initial step (pre-amplification of DNA) for the Biomark HD system (Fluidigm). Briefly, two separate pools containing 24X TaqMan assays in each pool as described in Supplementary Table 5, were prepared by mixing forward and reverse primers and diluted with 10 mM Tris/HCL and 0.1 mM EDTA (pH 8) to give a final concentration of 200 nM of each primer. PCR reactions were carried out for each STA pool in 5 µl volumes each containing 2.5 µL TaqMan PreAmp Master Mix (Fluidigm, CA, USA), 1.25 µL of pooled assay and 1.25 µL of DNA. Reactions were amplified with the T100 Thermal Cycler (Bio-Rad Laboratories, CA, USA) and cycling conditions included an initial activation at 95 °C for 10 minutes followed by 14 two-step cycles (denaturation at 95 °C for 15 seconds and annealing/extension at 60 °C for 4 minutes). STA products from the two PCR reactions were combined and diluted 1:5 with 10 mM Tris/HCL and 0.1 mM EDTA (pH 8).
Real-time qPCR using the Biomark HD System (Fluidigm)
Fluidigm was carried out with 96.96 dynamic arrays (Fluidigm Corporation, CA, USA) according to manufactures instructions. 10X assay mixture and sample pre-mix were prepared, with each assay being prepared in duplicate (making up a total of 96 assays). Briefly, each 10X assays contained 2.5 ul 20X TaqMan gene expression assay (Fluidigm), prepared by mixing forward primer, reverse primers and probe, and 2.5 ul 2X Assay Loading Reagent (Fluidigm). The final concentration (at 10X) for each primer was 9 μM and 2 μM for the probe. Sample pre-mix was made by combining 2.5 ul TaqMan Universal PCR Master Mix (2X (Life Technologies, 0.25 ul 20X GE Sample Loading Reagent (Fluidigm PN 100–7610) and 2.5 ul pre-amplified cDNA (diluted STA product containing a negligible concentration of carried-over STA primers) for each of the 96 sample inlets. IFC controller (Fluidigm) were used to prime 96.96 dynamic arrays IFC Chip (Fluidigm) with control line fluid. 5 µL of each assay and sample mix was then transferred into the appropriate inlets of the primed chip and loaded with the IFC controller. After loading, the chip was placed in the Biomark instrument for Fluidigm screening at 95 °C for 10 minutes followed by 40 cycles at 95 °C for 15 seconds and 60 °C for 1 minute. The data were analyzed with Real-Time PCR Analysis Software in the BIOMARK instrument (Fluidigm Corporation, CA, USA) using manually defined thresholds. Negative samples were defined as those with Cq values ≥ 35 for each bacterial species. A schematic diagram illustrating the workflow of the Fluidigm assay is shown in Supplementary Fig. 3.
All respective reactions included in the Fluidigm assay were optimized according to the MIQE guidelines. Briefly, Standard concentration curves, lower limits of detection (LLDs), correlation coefficient (r2), amplification efficiency, analytical specificity, intra-assay variation, accuracy and inter-assay variation for each reaction were calculated as described previously10.
Statistical analysis
Statistical analysis was performed with STATA Version 11.0 (Statacorp, Texas, USA). McNemar’s test was used to determine the sensitivities of culture/qPCR and Fluidigm. When analyzed for concordance, using kappa statistics, only common serogroups/types tested by both methods were included. Findings for non-typable (NT) pneumococcus were not compared between culture and qPCR/Fluidigm, as the qPCR and Fluidigm methods were not optimized to detect all pneumococcal serotypes and thus some untested serotypes might be misidentified as NT pneumococcus. Paired student t-tests were used to compare the mean bacterial concentrations detected by qPCR and Fluidigm, following log10 transformation of data. Results were considered significant when the p-value was ≤0.05.
Ethics approval
Ethical consent was obtained from the Medical Human Research Ethics Committee (HREC) of the University of Witwatersrand for the initial enrolment of the cohort [Vaccinated cohort: HREC: 040704, also registered under Clinical trials number NCT00099658] and all methods were performed in accordance with the relevant guidelines and regulations. The HREC also approved further testing of samples (M140907) in this study. At the time of the initial enrolment, written, informed consent had been obtained from the parents/guardians of all the study participants.
Supplementary information
Acknowledgements
This study was partially supported financially by grants from the Department for Science and Technology/National Research Foundation through the South African Research Chair Initiative and Medical Research Council: Respiratory and Meningeal Pathogens Research Unit.
Author Contributions
C.P.O., P.V.A. and S.A.M. conceived and designed the study. C.P.O. was responsible for data curation and analysis. Funding acquisition was undertaken by S.A.M. C.P.O wrote the first draft of the manuscript and all authors were involved in reviewing and editing the manuscript.
Data Availability
The data that support the findings of this study are available from the corresponding author upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Courtney P. Olwagen, Email: olwagenc@rmpru.co.za
Shabir A. Madhi, Email: madhis@rmpru.co.za
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42846-y.
References
- 1.Garcia-Rodriguez JA, Fresnadillo Martinez MJ. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. The Journal of antimicrobial chemotherapy. 2002;50(Suppl S2):59–73. doi: 10.1093/jac/dkf506. [DOI] [PubMed] [Google Scholar]
- 2.Xu Q, Pichichero ME. Co-colonization by Haemophilus influenzae with Streptococcus pneumoniae enhances pneumococcal-specific antibody response in young children. Vaccine. 2014;32:706–711. doi: 10.1016/j.vaccine.2013.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Q, Almudervar A, Casey JR, Pichichero ME. Nasopharyngeal bacterial interactions in children. Emerging infectious diseases. 2012;18:1738–1745. doi: 10.3201/eid1811.111904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy TF, Bakaletz LO, Smeesters PR. Microbial interactions in the respiratory tract. The Pediatric infectious disease journal. 2009;28:S121–126. doi: 10.1097/INF.0b013e3181b6d7ec. [DOI] [PubMed] [Google Scholar]
- 5.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerging infectious diseases. 2008;14:1584–1591. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madhi SA, et al. Long-term effect of pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae–and associated interactions with Staphylococcus aureus and Haemophilus influenzae colonization–in HIV-Infected and HIV-uninfected children. J Infect Dis. 2007;196:1662–1666. doi: 10.1086/522164. [DOI] [PubMed] [Google Scholar]
- 7.Bogaert D, de Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. The Lancet Infectious Diseases. 2004;4:144–154. doi: 10.1016/s1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 8.Syrjänen RK, Kilpi TM, Kaijalainen TH, Herva EE, Takala AK. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. The Journal of infectious diseases. 2001;184:451–459. doi: 10.1086/322048. [DOI] [PubMed] [Google Scholar]
- 9.Satzke C, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine. 2013;32:165–179. doi: 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 10.Olwagen, C. P., Adrian, P. V. & Madhi, S. A. Comparison of traditional culture and molecular qPCR for detection of simultaneous carriage of multiple pneumococcal serotypes in African children. Scientific Reports7 (2017). [DOI] [PMC free article] [PubMed]
- 11.Olwagen CP, Adrian PV, Nunes MC, Madhi SA. Evaluation of the association of pneumococcal conjugate vaccine immunization and density of nasopharyngeal bacterial colonization using a multiplex quantitative polymerase chain reaction assay. Vaccine. 2018;36:3278–3285. doi: 10.1016/j.vaccine.2018.04.068. [DOI] [PubMed] [Google Scholar]
- 12.Rivera-Olivero IA, Blommaart M, Bogaert D, Hermans PW, de Waard JH. Multiplex PCR reveals a high rate of nasopharyngeal pneumococcal 7-valent conjugate vaccine serotypes co-colonizing indigenous Warao children in Venezuela. Journal of medical microbiology. 2009;58:584–587. doi: 10.1099/jmm.0.006726-0. [DOI] [PubMed] [Google Scholar]
- 13.Jang JS, et al. Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays. BMC genomics. 2011;12:144. doi: 10.1186/1471-2164-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 15.Gray BM, Converse GM, 3rd, Dillon HC., Jr. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980;142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 16.Zenni MK, et al. Streptococcus pneumoniae colonization in the young child: association with otitis media and resistance to penicillin. The Journal of pediatrics. 1995;127:533–537. doi: 10.1016/S0022-3476(95)70108-7. [DOI] [PubMed] [Google Scholar]
- 17.Faden H, et al. Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics. The Journal of infectious diseases. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 18.Mastro TD, et al. Antimicrobial resistance of pneumococci in children with acute lower respiratory tract infection in Pakistan. Lancet. 1991;337:156–159. doi: 10.1016/0140-6736(91)90813-5. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd-Evans N, et al. Nasopharyngeal carriage of pneumococci in Gambian children and in their families. The Pediatric infectious disease journal. 1996;15:866–871. doi: 10.1097/00006454-199610000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Dhoubhadel BG, et al. A novel high-throughput method for molecular serotyping and serotype-specific quantification of Streptococcus pneumoniae using a nanofluidic real-time PCR system. J Med Microbiol. 2014;63:528–539. doi: 10.1099/jmm.0.071464-0. [DOI] [PubMed] [Google Scholar]
- 21.Dhoubhadel BG, et al. Bacterial load of pneumococcal serotypes correlates with their prevalence and multiple serotypes is associated with acute respiratory infections among children less than 5 years of age. PloS one. 2014;9:e110777. doi: 10.1371/journal.pone.0110777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simões AS, et al. lytA-based identification methods can misidentify Streptococcus pneumoniae. Diagnostic microbiology and infectious disease. 2016;85:141–148. doi: 10.1016/j.diagmicrobio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Salvà-Serra F, Connolly G, Moore ER, Gonzales-Siles L. Detection of “Xisco” gene for identification of Streptococcus pneumoniae isolates. Diagnostic microbiology and infectious disease. 2018;90:248–250. doi: 10.1016/j.diagmicrobio.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Nunes MC, et al. Acquisition of Streptococcus pneumoniae in pneumococcal conjugate vaccine-naive South African children and their mothers. Pediatr Infect Dis J. 2013;32:e192–205. doi: 10.1097/INF.0b013e31828683a3. [DOI] [PubMed] [Google Scholar]
- 25.Madhi SA, et al. Effect of HIV infection status and anti-retroviral treatment on quantitative and qualitative antibody responses to pneumococcal conjugate vaccine in infants. J Infect Dis. 2010;202:355–361. doi: 10.1086/653704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madhi SA, et al. Immunogenicity following the first and second doses of 7-valent pneumococcal conjugate vaccine in HIV-infected and -uninfected infants. Vaccine. 2013;31:777–783. doi: 10.1016/j.vaccine.2012.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhi SA, Bamford L, Ngcobo N. Effectiveness of pneumococcal conjugate vaccine and rotavirus vaccine introduction into the South African public immunisation programme. S Afr Med J. 2014;104:228–234. doi: 10.7196/SAMJ.7597. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien KL, Nohynek H. Report from a WHO Working Group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr Infect Dis J. 2003;22:e1–11. doi: 10.1097/01.inf.0000049347.42983.77. [DOI] [PubMed] [Google Scholar]
- 29.O’Brien KL, et al. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. Journal of clinical microbiology. 2001;39:1021–1024. doi: 10.1128/JCM.39.3.1021-1024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olwagen CP, et al. Use of Multiplex Quantitative PCR To Evaluate the Impact of Pneumococcal Conjugate Vaccine on Nasopharyngeal Pneumococcal Colonization in African Children. mSphere. 2017;2:e00404–00417. doi: 10.1128/mSphere.00404-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAvin JC, et al. Sensitive and specific method for rapid identification of Streptococcus pneumoniae using real-time fluorescence PCR. J Clin Microbiol. 2001;39:3446–3451. doi: 10.1128/JCM.39.10.3446-3451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maaroufi Y, De Bruyne J-M, Heymans C, Crokaert F. Real-time PCR for determining capsular serotypes of Haemophilus influenzae. Journal of clinical microbiology. 2007;45:2305–2308. doi: 10.1128/JCM.00102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas JD, et al. sodC-based real-time PCR for detection of Neisseria meningitidis. PloS one. 2011;6:e19361. doi: 10.1371/journal.pone.0019361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatti, K. M., Sparks, K. N., Boney, K. O. & Tondella, M. L. A novel multi-target real-time PCR assay for the rapid diagnosis of Bordetella species in clinical specimens. Journal of clinical microbiology, JCM. 00601–00611 (2011). [DOI] [PMC free article] [PubMed]
- 35.Azzari C, et al. Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One. 2010;5:e9282. doi: 10.1371/journal.pone.0009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.