Abstract
Treatment of physeal fractures (15%–30% of all paediatric fractures) remains a challenge as in approximately 10% of the cases, significant growth disturbance may occur. Bioresorbable Magnesium-based implants represent a strategy to minimize damage (i.e., load support until bone healing without second surgery). Nevertheless, the absence of harmful effects of magnesium-implants and their degradation products on the growth plate should be confirmed. Here, the proteome of human mesenchymal stem cells undergoing chondrogenesis was evaluated when exposed to the products of various Magnesium-based materials degradation. The results of this study indicate that the materials induced regulation of proteins associated with cell chondrogenesis and cartilage formation, which should be beneficial for cartilage regeneration.
Keywords: Stem cells differentiation, Chondrogenesis, Proteomics, Magnesium biomaterial, Biodegradable implant
Graphical abstract
Highlights
-
•
Degradation products from Mg-based materials generated changes in protein expression.
-
•
Relevant proteins involved in cartilage formation were upregulated.
-
•
Potential application of especially Pure-Mg and Mg-10Gd for cartilage regeneration.
1. Introduction
Biodegradable magnesium (Mg)-based materials are promising candidates for substituting permanent implants for orthopaedic application as a second surgery and chronic inflammation will be avoided [1]. Furthermore, as an element, Mg is essential to the human body and, as metal, has mechanical properties close to the ones of bone. Special emphasis should be given to children, a sector of the population with a high incidence of bone damage [2]. Bone repair in children is a fast process, and the main requirement from an implant for appropriate healing is to reduce bone load bearing. Therefore, an additional and undesirable immobilisation of the patients will always be necessary to remove a permanent implant. Growing long bones have specific cartilaginous discs at both ends, growth plates, responsible for endochondral ossification and bone formation until adult stage. Any damage due to the implant application or removal, as well as due to the degradation products, could generate irreversible malformations [3,4].
Foetal bone development starts with stem cells condensation, chondrocyte differentiation, proliferation, maturation and ossification. Every step is characterised by changes in cell morphology, proliferation and extracellular matrix (ECM) production, and by a complex molecular regulation [5]. After stem cell condensation, most of the cells become chondrocytes with a rounded morphology and express specific genes such as SRY (sex determining region Y)-box9 (SOX9), aggrecan (ACAN; a proteoglycan) and collagen, type II COL2. The ECM is rich in COL2 and glycosaminoglycans (GAG). Then chondrocytes proliferate and synthesize more ECM, enlarging cartilage [[6], [7], [8], [9]]. Chondrocytes undergo hypertrophy (or maturation), showing a notably enlarged size, a high expression of collagen, type-X gene (COL10) [10,11], and regulating mineralisation of surrounding matrix by expressing other gene markers which are also bone markers, such as osteopontin (OPN) and collagen, type I alpha 1 (COL1A1) [5]. At the final stage of maturation, the chondrocytes can undergo apoptosis.
Previous proteomic studies of mesenchymal stem cell (MSC) chondrogenesis [[12], [13], [14], [15], [16], [17]] have shown that the majority of regulated proteins are related to cell metabolism (e.g., adenosine triphosphate (ATP) synthase subunits alpha (α) and beta (β), carbonyl reductase, aldose reductase, α-enolase, dihydropyrimidinase-like 2, glyceraldehyde-3-phosphate dehydrogenase, and glycogen phosphorylase), ECM and cytoskeleton (e.g., annexins, actin-related proteins, biglycan, chondroadherin, collagen α-2(VI) chain, collagen α-3(VI) chain, fibronectin, vimentin, gelsolin, procollagen-lysine, and transforming growth factor-beta-induced protein ig-h3 precursor), and response to stress (e.g., 78 kDa glucose-regulated protein, endoplasmin, peroxiredoxin-6, peptidyl-prolyl cis-trans isomerise A, superoxide dismutase, heat shock protein beta-1, and stress-induced phosphoprotein 1). Human umbilical cord perivascular (HUCPV) cells are mesenchymal stem cells (MSC), isolated from the vessels surface of umbilical cords, with a high proliferation rate and strong potential for differentiation into the skeletal lineages (both bone and cartilage) [[18], [19], [20]].
In order to prove the potential of Mg-based materials for application in cartlage treatment, a better understanding of the chondrogenic mechanisms influenced by Mg-based materials degradation is still necessary. This study aimed at (I) evaluating the differently expressed proteins during chondrogenesis under the influence of Mg-materials degradation products, contained in the degradation medium (extracts), and (II) determining which of those proteins are directly involved in the chondrogenic differentiation. For this purpose, differential proteomics via label-free quantification of HUCPV cells driven toward chondrogenesis under the influence of three materials (Pure-Mg, Mg-2Ag and Mg-10Gd) was performed. Silver (Ag) was selected as alloying elements due to its antibacterial properties and gadolinium (Gd) to improve material mechanical properties. Cyto- and biocompatibility of these two alloys have been earlier tested and demonstrated ( [21,22,23,24] for Ag and Gd, respectively).
2. Experimental procedures
2.1. Extract preparation and characterization
Extracts of Pure Mg (Pure-Mg, 99.95%), Mg with 2 wt% silver (Mg-2Ag) and Mg with 10 wt% gadolinium (Mg-10Gd) materials were prepared according to EN ISO standards 10993:5 [25] and 10993:12 [26]. Pure elutes were characterised (composition and pH) and diluted in differentiation medium to obtain a common concentration of Mg (i.e., 6.08 mM).
2.2. Induction of micropellets formation and chondrogenic differentiation
Ethical approval for the isolation of HUCPV was obtained from the Ethik-Kommission der Ärztekammer Hamburg. Umbilical cord samples were provided by Asklepios Klinik Altona immediately after caesarean sections of consenting donors. Cell micromasses were obtained from HUCPV cell (passage 2) pellets after 3 days. Chondrogenesis was then chemically induced for up to 11 days with or without Mg-extracts, followed by proteome analysis. For each group (i.e., control, Pure-Mg, Mg-2Ag, and Mg-10Gd) 3 biological replicates were established. Furthermore, 3 technical replicates (i.e, LC MSMS injections) were performed. Control group refers to micropellets driven toward chondrogenesis without any Mg extract (only differentiation medium). More details can be found in Supplemental experimental procedures.
2.3. Proteomic analysis
Proteins from the pellets were extracted using TissueLyzer II (QIAGEN), tryptic in-solution digested and then desalted.
For liquid chromatography–mass spectrometry (LC-MSMS) measurements, all the tryptic digested peptides were subjected to a nano-flow UPLC-column (DionexUltiMate 3000 RSLCnano, Thermo Scientific, Bremen, Germany) coupled via electrospray ionization (ESI) to an Orbitrap mass spectrometer (Orbitrap-Fusion, Thermo Fisher Scientific).
To compare the relative protein abundance, raw data files obtained from the LC-MSMS were processed by MaxQuant 1.5.2.8 [27]. These parameters were used for identification and label-free quantification: identification of the peptides against SwissProt database downloaded from UniProt in July 2015 (with internal contaminants database of MaxQuant); trypsin was used as an enzyme with one missed cleavage; carbamidomethylation on cysteine was set as fixed modification and oxidation of methionine as variable modifications; precursor mass of 20 ppm and fragment mass tolerance of 0.5 Da; and minimum peptide length of 6 amino acids for identification and match between runs.
Peptide spectrum match (PSM) and protein false discovery rate (FDR) were 0.01; and at least 2 ratio count for LFQ was used.
Perseus 1.5.2.6 [28] and Wolfram Mathematica 10.0 (Wolfram Research Europe Ltd., Oxfordshire, United Kingdom) were used for bioinformatics analysis.
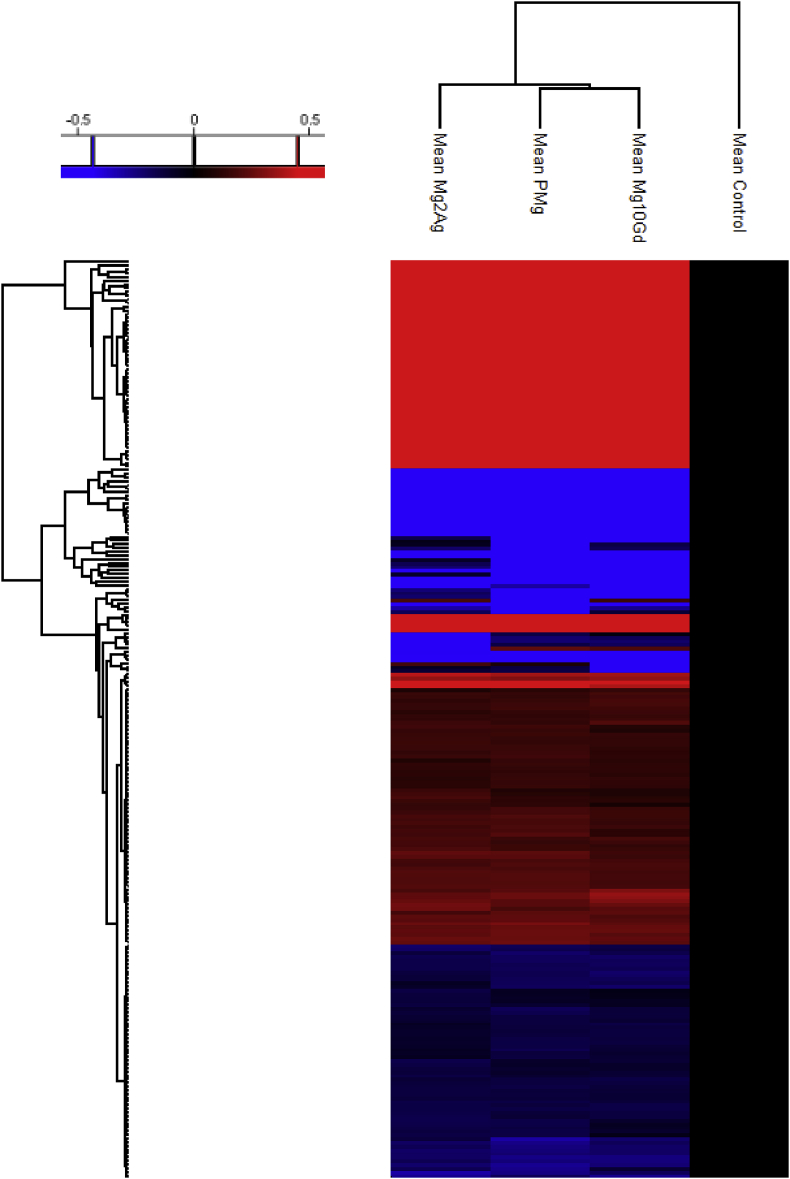
Heat maps (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5; Fig. A1), based on two-sided student's T test, prepared in Perseus, indicates the fold change and significance of each protein of HUCPV cells incubated for 11 days with Mg-alloys (Mg-10Gd, Mg-2Ag, and Pure-Mg) compared to control cells after 11 days incubation without Mg-alloys (permutation-based FDR of 0.01, s0 = 0.1).
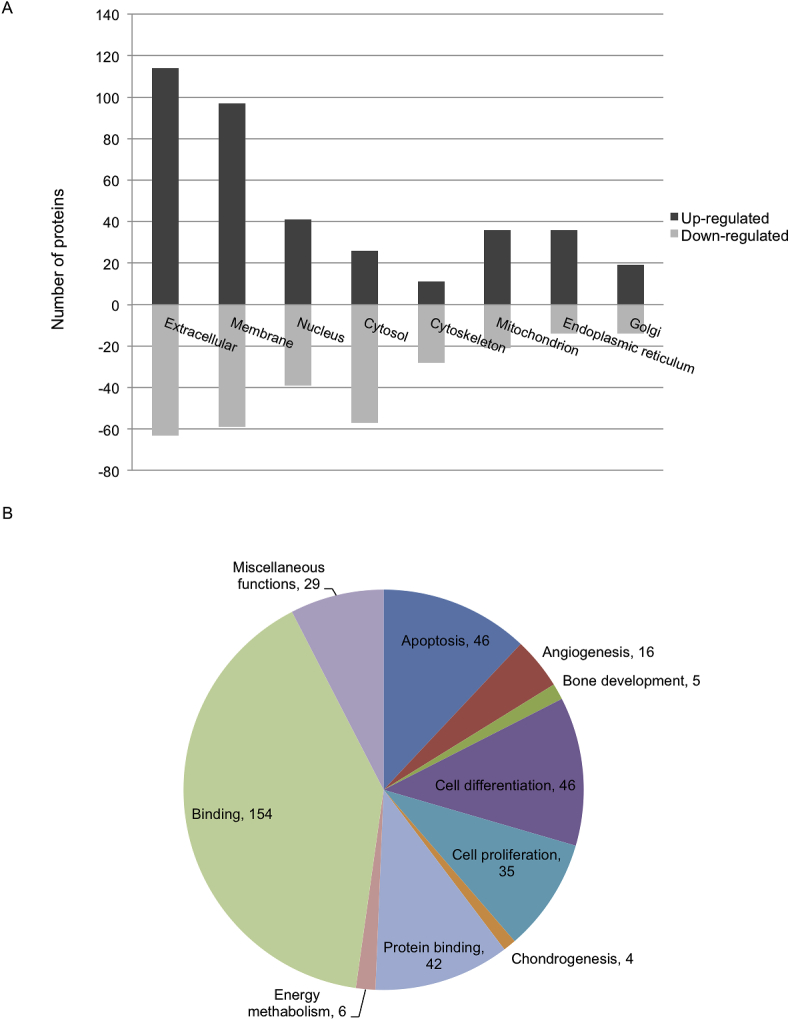
Fig. 1.
The number of regulated proteins with more than two-fold change in at least one of the Mg-alloys sorted according to (a) their location in the cells and (b) their involvement in physiological processes.
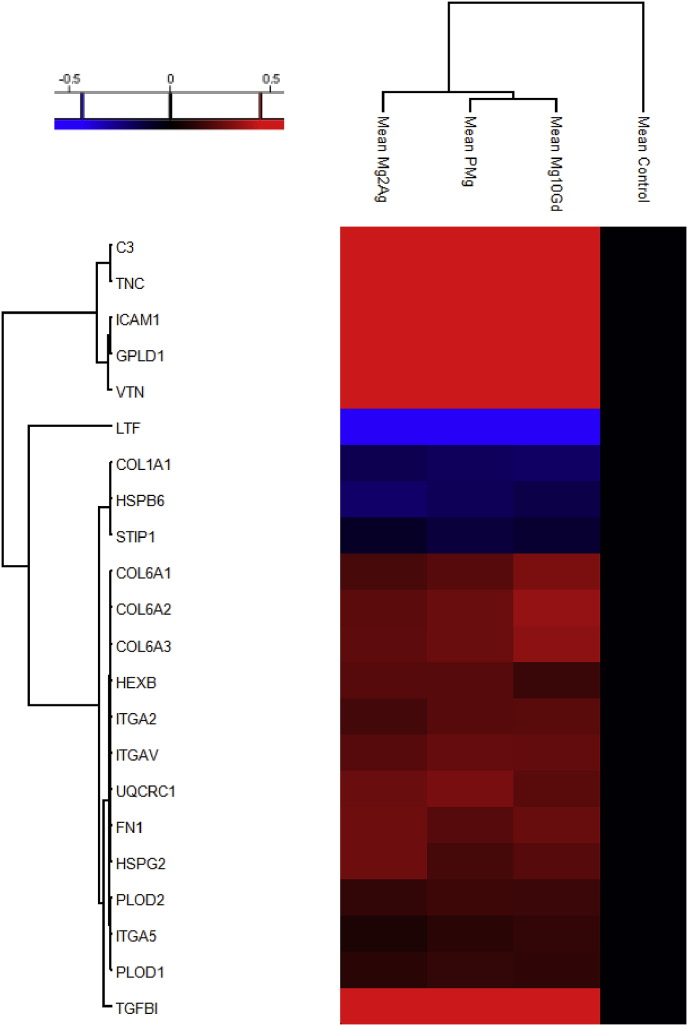
Fig. 2.
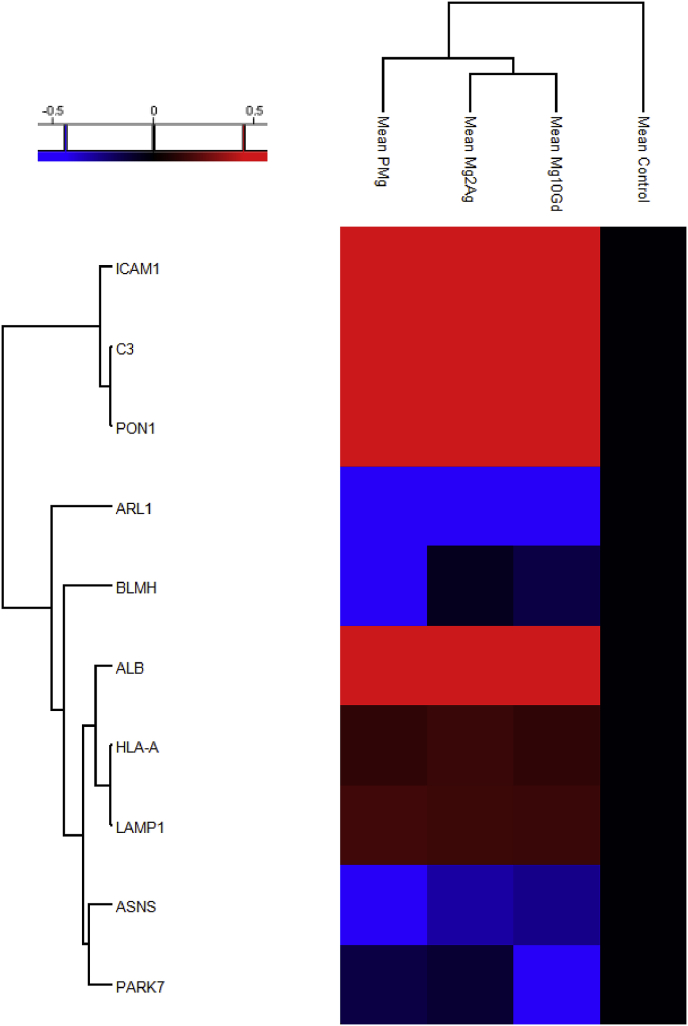
Heat-map and hierarchical clustering of the up- and down-regulated proteins involved in chondrogenesis and cartilage development (P-value = 0.05; min. fold-change of 2) in all Mg-alloys compared based on the mean values of the biological replicates (normalized to Control).
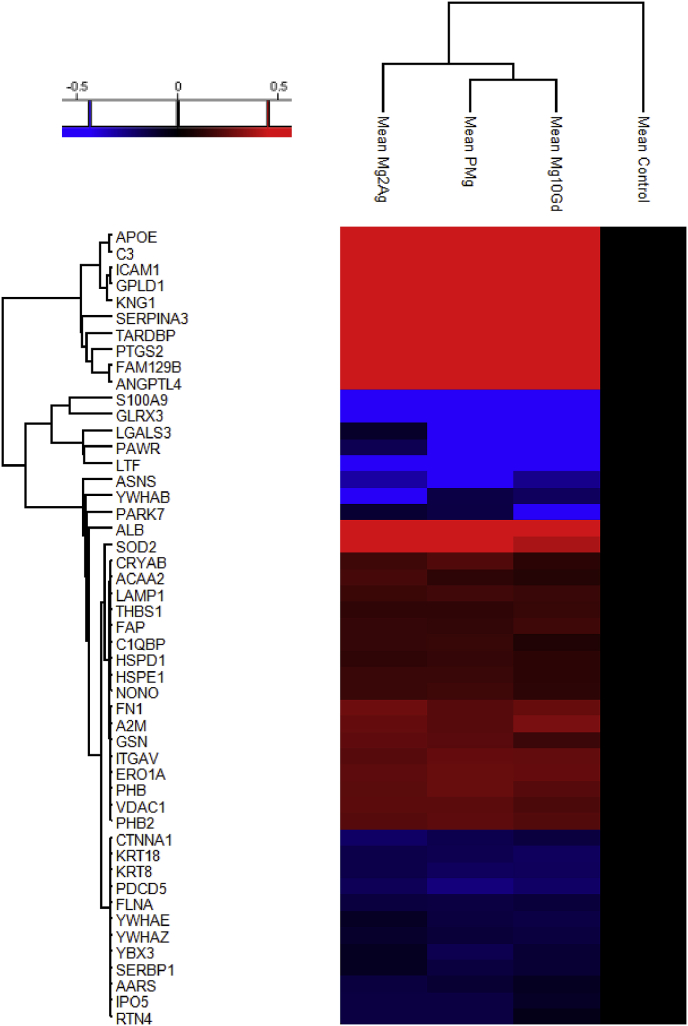
Fig. 3.
Heat-map and hierarchical clustering of the up- and down-regulated proteins involved in apoptosis (P-value = 0.05; min. fold-change of 2) in all Mg-alloys compared based on the mean values of the biological replicates (normalized to Control).
Fig. 4.
Heat-map and hierarchical clustering of the up- and down-regulated proteins involved in cellular response to toxicity (P-value = 0.05; min. fold-change of 2) in all Mg-alloys compared based on the mean values of the biological replicates (normalized to Control).
Fig. 5.
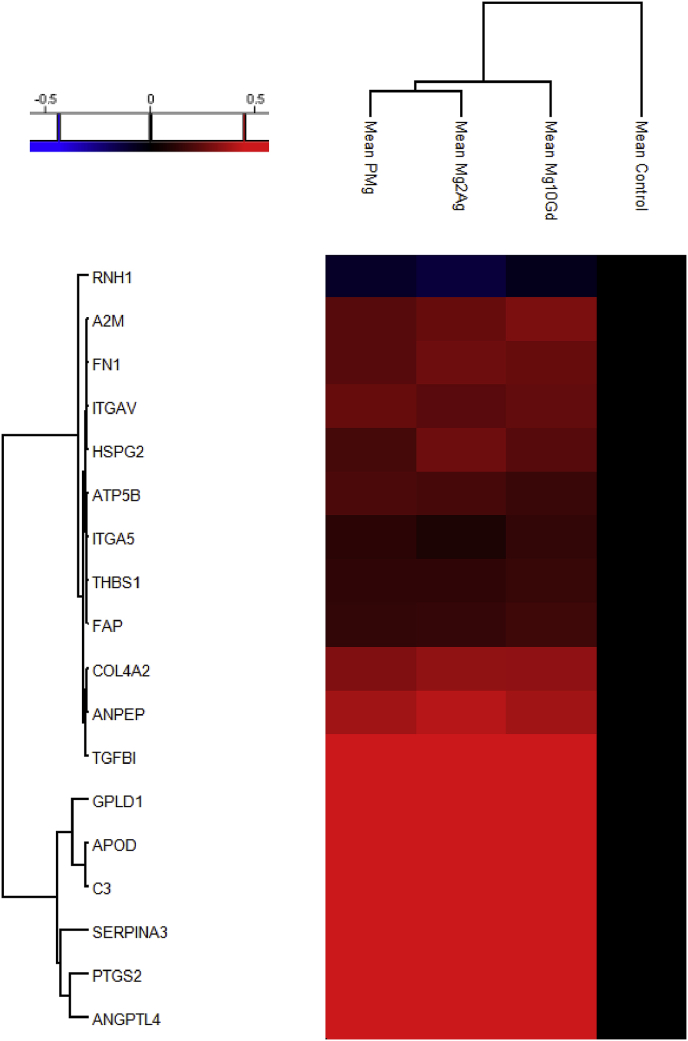
Heat-map and hierarchical clustering of the up- and down-regulated proteins involved in angiogenesis and bone formation (P-value = 0.05; min. fold-change of 2) in all Mg-alloys compared based on the mean values of the biological replicates (normalized to Control).
Other and more detailed experimental procedures are described in Supplemental experimental procedures.
3. Results
3.1. Composition of the extracts
As it can be observed in Table 1, Mg contents increased strongly in the extracts compared to the extraction media (α-MEM supplemented with 10% foetal bovine serum for mesenchymal stem cells (SC-FBS; Stem Cell Technologies, Vancouver, Canada) and 1% antibiotics Penicillin/Streptomycin (Pen strep; Invitrogen, Bremen, Germany)) while Ca and P ones decreased. To avoid osmotic choc and in order to study the effect of alloying element independently of Mg content, extracts were diluted with differentiation medium to obtain a common Mg concentration of about 6.08 mM.
Table 1.
Elemental characterisation of the extraction medium (growth medium) initial extracts (pure) and after dilution to a Mg concentration of 6.08 mM (diluted) measured via ICP-MS. All concentrations are in millimolar (mM).
| Mg (mM) | Ca (mM) | P (mM) | Gd (mM) | Ag (mM) | pH | |||
|---|---|---|---|---|---|---|---|---|
| Extract | Pure | Pure-Mg | 51.43 | 0.70 | 0.30 | n.d. | n.d. | 8.68 |
| Mg-10Gd | 80.64 | 0.32 | 0.32 | 2.16 x 10−3 | n.d. | 8.51 | ||
| Mg-2Ag | 50.60 | 0.54 | 0.54 | n.d. | 71 x 10−3 | 8.68 | ||
| Diluted | Pure-Mg | 6.08 | 1.79 | 1.26 | n.d. | n.d. | 8.15 | |
| Mg-10Gd | 6.08 | 1.77 | 1.26 | 1.63 x 10−4 | n.d. | 8.15 | ||
| Mg-2Ag | 6.08 | 1.72 | 1.16 | n.d. | 11 x 10−3 | 8.25 | ||
| Growth or expansion medium | 0.81 | 1.80 | 1.01 | n.d. | n.d. | 7.34 | ||
| Differentiation or chondrogenic medium | 0.82 | 1.89 | 1.37 | n.d. | n.d. | 7.40 | ||
3.2. Effects of the Mg-alloys degradation products on chondrogenic-differentiated HUCPV proteome
In order to determine the influence of Pure-Mg, Mg-10Gd, and Mg-2Ag extracts on HUCPV cells driven toward chondrogenesis, proteins from each condition were analysed with differential bottom-up proteomics using label-free quantification (LFQ).
246 significantly regulated proteins (Table A1) were found under the influence of the extracts and clustered in a heat map (Fig. A1). 136 proteins were upregulated in the presence of Mg-10Gd, Mg-2Ag and Pure-Mg (from which 134 proteins were common for the three extracts) while 110 proteins were downregulated. A Gene Ontology (GO) annotation downloaded from UniProt was performed for each protein of the list of regulated proteins. Clustering these proteins regarding their localisation in the cells (cellular compartment; Fig. 1a) indicated that the number of upregulated extracellular proteins and membrane proteins (involved not only in ECM composition) was considerably higher than downregulated ones in the presence of Mg alloys. Additionally, the number of regulated cytosol proteins was high. Cytosolic and cytoskeletal proteins were mostly downregulated. Fig. 1b shows the most affected biological processes. The highest number of regulated proteins were involved in cell binding, differentiation, apoptosis, and cell proliferation. To a lesser extent, proteins involved in angiogenesis, energy metabolism, bone development, and chondrogenesis were influenced by the extracts. Proteins involved in chondrogenesis and cartilage formation are depicted in Fig. 2. Heat maps in Fig. 3, Fig. 4, Fig. 5 illustrate the regulated proteins clustered according to their involvement in apoptosis, response to cell toxicity and angiogenesis, respectively.
In a second step, Mg-alloy specific proteins will be discussed.
3.2.1. Regulated proteins involved in chondrogenesis and cartilage formation
Fig. 2 illustrates the regulated chondrogenesis-related proteins. Four chondrogenesis-related proteins were upregulated by the extracts: glycosylphosphatidylinositol specific phospholipase D1 (GPLD1) was upregulated in the presence of Mg-alloys, while hexosaminidase B (beta polypeptide) (HEXB) and transforming growth factor, beta-induced, 68 kDa (TGFBI) were upregulated compared to the control. In addition, proteins involved in cartilage ECM formation and organisation (fibronectin 1 (FN1), collagen, type VI (COL6), tenascin C (TNC), intercellular adhesion molecule 1 (ICAM1), vitronectin (VTN), heparan sulfate proteoglycan 2 (HSPG2), procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 and 2 (PLOD1 (significantly in the presence of pure-Mg and Mg10Gd) and PLOD2) and integrins α-2 (ITGA2), α-5 (ITGA5)) were upregulated in the presence of Mg. COL1A1 was downregulated by all the extracts. ITGA5 was significantly upregulated in the presence of Mg-10Gd but not with the other extracts.
3.2.2. Regulated proteins involved in apoptosis
Regulated proteins involved in apoptosis were clustered in the heat map shown in Fig. 3. From the total 49 proteins identified, 29 were upregulated, while 20 of them were downregulated in the presence of Mg-alloys. S100 calcium binding protein A9 (S100A9) was completely absent in the presence of Mg-alloys. Galectin 3 (LGALS3) was downregulated in the presence of Mg-2Ag (in less than 2 fold) and absent in any biological replicates in the presence of the other Mg-alloys. On the other hand, 10 apoptotic-related regulated proteins in at least two biological replicates of each condition were only present after incubation of HUCPV with Mg-alloys. From those proteins, 6 were stimulators of apoptosis: C3, Kininogen-1 (KNG1), prostaglandin-endoperoxide synthase 2 (PTGS2), TAR DNA-binding protein (TARDBP), SERPINA3 and GPLD1 while 4 were inhibitors: apolipoprotein E (APOE), ICAM1, niban-like protein 1 (FAM129B) and angiopoietin-related protein 4 (ANGPTL4). Regarding the downregulated proteins in the presence of the extracts, programmed cell death protein 5 (PDCD5) and PRKC apoptosis WT1 regulator protein (PAWR) were positive regulators of apoptosis. The regulation of the other apoptotic-related proteins is significant in all Mg-alloys.
3.2.3. Regulated proteins involved in the cellular response to toxicity
The heat map in Fig. 4 indicates the regulated proteins involved in cellular response to toxic substances in the presence of Mg-alloys. 6 of those 10 proteins involved in the cellular response to toxicity were upregulated while 4 were downregulated. ICAM1, C3, and paraoxonase 1 (PON1) were only present in the HUCPV cells incubated with Mg-alloys.
3.2.4. Regulated proteins involved in angiogenesis
18 of angiogenesis-related proteins were regulated in the presence of Mg-alloys, 17 of those were significantly increased in the presence of at least one of the Mg-alloys. Apolipoprotein D (APOD), Complement Component 3 (C3), PTGS2, GPLD1, Alpha-1-antichymotrypsin (SERPINA3) and angiopoietin-like 4 (ANGPTL4) were present in at least two biological replicates of the HUCPV cells incubated with Mg-alloys, and not present in the control (Fig. 5). Ribonuclease/angiogenin inhibitor 1 (RNH1) was downregulated in the presence of all Mg-alloys. However, downregulation of this protein is significant only in the presence of Mg-2Ag. Moreover, ITGA5 was significantly upregulated in the presence of Mg-10Gd, while there was no significant change in the presence of the other Mg-alloys. Thrombospondin 1 (THBS1) was upregulated with all the extracts. The upregulation of the other angiogenesis-related proteins was significant in all Mg-alloys.
3.2.5. Regulated proteins in all biological replicates in the presence of Mg-10Gd & Mg-2Ag
The significantly regulated proteins in the presence of Mg-10Gd and Mg-2Ag (5 proteins) are listed in Table 2. Charged multivesicular body protein 4B (CHMP4B) was upregulated (while downregulated with Pure-Mg) and Asparagine synthetase (NARS) was downregulated by the three extracts (being also significantly decreased with Mg-10Gd and Mg-2Ag compared to Pure-Mg. Both proteins are involved in cell apoptosis and NARS also in response to toxic substrates. Thioredoxin reductase 1 (TXNRD1) was downregulated only with Mg-2Ag and Mg-10Gd. Keratin 19 (KRT19) was downregulated with the three extracts (but the downregulation was significantly lower than with Pure-Mg (Table A1). SERPINE2, an ECM protein, was present only in the incubated cells with the three Mg-alloys, exhibiting higher expression in Mg-10Gd and Mg-2Ag (in all of the biological replicates of Mg-10Gd and Mg-2Ag) than in Pure-Mg (Table A1).
Table 2.
Significantly regulated proteins (⇧ upregulation - ⇩ downregulation) under different conditions. Function based on UniProt database search. Protein names and symbol are according to Hugo Gene Nomenclature Committee – synonyms/previous names are italicised.
| Protein name (gene name) synonym | Function | Fold change in Pure-Mg | Fold change in Mg-10Gd | Fold change in Mg-2Ag | |
|---|---|---|---|---|---|
| Significantly regulated proteins in Mg-10Gd and Mg-2Ag (not in Pure-Mg). | Charged multivesicular body protein 4B (CHMP4B) Chromatin modifying protein 4B | negative regulation of cell death | / | ⇧ 2.29 ±0.199 |
⇧ 2.57 ±0.068 |
| Eukaryotic translation initiation factor 3 subunit C (EIF3C) | translation initiation factor activity | / | ⇩ 2.04 ±0.0.97 |
⇩ 2.34 ±0.056 |
|
| Thioredoxin reductase 1 (TXNRD1) | cell proliferation, response to reactive oxygen species, thioredoxin-disulfide reductase activity | / | ⇩ 2.14 ±0.02 |
⇩ 2.24 ±0.018 |
|
| Keratin 19 (KRT19) | cell differentiation, involved in embryonic placenta development, structural constituent of cytoskeleton | / | ⇩ 16.22 ±0.319 |
⇩17.78 ±0.068 |
|
| Asparagine-tRNA ligase (NARS) | negative regulation of apoptotic process, response to toxic substance | / | ⇩ 4.17 ±0.083 |
⇩ 5.13 ±0.069 |
|
| Fold change in Pure-Mg | Fold change in Mg-10Gd | Fold change in Mg-2Ag | |||
| Significantly regulated proteins in Mg-10Gd and Pure-Mg (not in Mg-2Ag) | Transmembrane p24 trafficking protein 7 (TMED7) Transmembrane emp24 domain-containing protein 7 | protein transport | ⇧ 3.39 ±0.077 |
⇧ 2.82±0.0056 | / |
| Sideroflexin 3 (SFXN3) | transporter activity | Not present in control | Not present in control | / | |
| IKBKB interacting protein (IKBIP) Inhibitor of nuclear factor kappa-B kinase-interacting protein | response to X-ray | ⇧ 2.09 ±0.036 |
⇧ 2.29 ±0.072 |
/ | |
| Pyrophosphatase (inorganic) 1 (PPA1) PP | magnesium ion binding; phosphate-containing compound metabolic process | ⇩ 2.45 ±0.022 |
⇩ 2.00 ±0.060 |
/ | |
| Bleomycin hydrolase (BLMH) | aminopeptidase activity, proteolysis, response to toxic substance | Not present in Pure-Mg | ⇩ 2.19 ±0.056 |
/ | |
| Four and a half LIM domains protein 1 (FHL1) LIM protein SLIMMER (SLIM1) | cell differentiation, positive regulation of potassium ion transport, zinc ion binding | ⇩ 3.31 ±0.194 |
⇩ 2.69 ±0.065 |
/ | |
| Nexilin F-actin binding protein (NEXN) NELIN, nexilin | regulation of cytoskeleton organisation | Not present in Pure-Mg | Not present in Mg-10Gd | / | |
| AHNAK nucleoprotein (AHNAK) neuroblast differentiation-associated protein, desmoyokin | regulation of voltage-gated calcium channel activity | ⇩ 2.57 ±0.098 |
⇩ 2.69 ±0.24 |
/ | |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 (PLOD1) lysyl hydroxlase 1, LH1 | oxidation-reduction process, procollagen-lysine 5-dioxygenase activity | ⇧ 2.09 ±0.020 |
⇧ 2.00 ±0.133 |
/ | |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon (YWHAE) 14-3-3 protein epsilon | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process, positive regulation of protein insertion into mitochondrial membrane, involved in apoptotic signalling pathway | ⇩ 2.09 ±0.117 |
⇩ 2.24 ±0.178 |
/ | |
| Prostaglandin-endoperoxide synthase 2 (PTGS2) prostaglandin G/H synthase 2, cyclooxygenase 2, COX2 | positive regulation of apoptotic process, angiogenesis, involved in sprouting angiogenesis, bone mineralisation | Not present in control | Not present in control | / | |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) protein kinase C inhibitor protein 1 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signalling pathway | ⇩ 2.24 ±0.067 |
⇩ 2.75 ±0.078 |
/ | |
| ATP synthase, H+ transporting, mitochondrial Fo complex subunit B1(ATP5F1) | ATP biosynthetic process, transporter activity | ⇧ 2.19 ±0.061 |
⇧ 2.63 ±0.132 |
/ | |
| Galectin 3 (LGALS3) lectin, galactoside-binding, soluble, 3 | epithelial cell differentiation, regulation of extrinsic apoptotic signalling pathway via death domain receptors, regulation of T cell apoptotic process, regulation of T cell proliferation | Not present in Pure-Mg | Not present in Mg-10Gd | / | |
| Plastin 3 (PLS3) T-plastin | auditory receptor cell differentiation, bone development, calcium ion binding | ⇩ 2.04 ±0.071 |
⇩ 2.09 ±0.068 |
/ | |
| Cytochrome c oxidase subunit 4 isoform 1 (COX4I1) cytochrome c oxidase subunit IV, COX4 | generation of precursor metabolites and energy, response to nutrient | ⇧ 2.19 ±0.066 |
⇧ 2.04 ±0.041 |
/ | |
| Calpain 1 (CAPN1) calpain 1, (mu/I) large subunit | extracellular matrix disassembly, positive regulation of cell proliferation, proteolysis | ⇩ 3.47 ±0.083 |
⇩ 3.16 ±0.061 |
/ | |
| Lecithin-cholesterol acyltransferase (LCAT) phosphatidylcholine-sterol acyltransferase | lipoprotein biosynthetic process, response to copper ion | Not present in control | Not present in control | / | |
| H2A histone family, member (H2AFY) Core histone macro-H2A.1, MACROH2A1 | SH3/SH2 adaptor activity | ⇧ 2.04 ±0.099 |
⇧ 2.29 ±0.108 |
/ | |
| Actinin alpha 4 (ACTN4) | BAT3 complex binding, positive regulation of ER-associated ubiquitin-dependent protein catabolic process | ⇩ 2.09 ±0.035 |
⇩ 2.19 ±0.027 |
/ | |
| Orosomucoid 1 (ORM1) alpha-1-acid glycoprotein 1, OMD, ORM | metal ion binding, SMAD protein signal transduction, transport | Not present in Pure-Mg | Not present in Mg-10Gd | / | |
| Alpha fetoprotein (HPAFP) | oxygen transporter activity, heme binding | ⇩ 2.51 ±0.0137 |
⇩ 2.88 ±0.202 |
/ | |
| Fold change in Pure-Mg | Fold change in Mg-10Gd | Fold change in Mg-2Ag | |||
| Significantly regulated proteins in Mg-2Ag and Pure-Mg (not in Mg-10Gd) | REX2, RNA exonuclease 2 homolog (S. cerevisiae) (REXO2), Oligoribonuclease, mitochondrial precursor | 3′-5′ exonuclease activity, focal adhesion, nucleotide metabolic process | Not present in control | / | Not present in control |
| LIM domain and actin-binding protein 1 (LIMA1), Epithelial protein lost in neoplasm, EPLIN, FLJ38853 | focal adhesion, negative regulation of actin filament depolymerisation, stress fiber | ⇩ 4.07 ±0.036 |
/ | ⇩ 2.51 ±0.112 |
|
| Reticulon-4 (RTN4), ASY, Foocen, KIAA0886, My043, Nbla00271 | negative regulation of cell growth, of apoptotic process, regulation of cell migration | ⇩ 2.19 ±0.160 |
/ | ⇩ 2.14 ±0.14 |
|
| Histone cluster 1, H2bl (HIST1H2BL) Histone H2B type 1-L, H2BFC, Histone H2B.c | nucleosome assembly | ⇧ 2.24 ±0.044 |
/ | ⇧ 2.75 ±0.035 |
|
| Kinectin (KTN1), CG1, CG-1 antigen, kinesin receptor | microtubule-based movemen | Not present in Pure-Mg | / | Not present in Mg2Ag | |
| non-POU domain containing, octamer-binding (NONO), NonO protein, Non-POU domain-containing octamer-binding protein | DNA recombination, DNA repair, negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway [ | ⇧ 2.45 ±0.097 |
/ | ⇧ 2.24 ±0.040 |
|
| LIM and SH3 domain protein 1 (LASP1), Metastatic lymph node gene 50 protein,Lasp-1 | focal adhesion;ion transmembrane transporter activity, ion transport, zinc ion binding | ⇩ 4.07 ±0.089 |
/ | ⇩ 2.88 ±0.23 |
|
| Major vault protein (MVP), LRP, Lung resistance-related protein, Major vault protein | ERBB signaling pathway, protein transport | ⇧ 2.29 ±0.115 |
/ | ⇧ 2.04 ±0.006 |
|
| Caldesmon 1 (CALD1), CDM, L-caldesmon, Non-muscle caldesmon | movement of cell or subcellular component, muscle contraction | ⇩ 3.09 ±0.076 |
/ | ⇩ 2.04 ±0.090 |
|
| Nucleobindin 1 (NUCB1), CALNUC, DKFZp686A15286 | regulation of protein targeting | ⇧ 2.34 ±0.108 |
/ | ⇧ 2.40 ±0.008 |
|
| heat shock 10 kDa protein 1 (chaperonin 10), (HSPE1), 10 kDa heat shock protein, 10 kDa chaperonin, CPN10 | activation of cysteine-type endopeptidase activity involved in apoptotic process, osteoblast differentiation | ⇧ 2.24 ±0.107 |
/ | ⇧ 2.24 ±0.029 | |
| Branched-chain-amino-acid aminotransferase 1, cytosolic (BCAT1), BCAT(c), BCATC, BCT1, Branched-chain-amino-acid aminotransferase, cytosolic | aspartate biosynthetic process, cell proliferation | ⇩ 2.24 ±0.034 |
/ | ⇩ 2.14 ±0.103 |
|
| Tu translation elongation factor, mitochondrial (TUFM), Elongation factor Tu, mitochondrial, P43, COXPD4 | GTP binding, GTPase activity, mitochondrial translational elongation [ | ⇩ 2.29 ±0.041 |
/ | ⇩ 2.29 ±0.105 |
|
| Serum amyloid A4, constitutive (SAA4), Serum amyloid A-4 protein, CSAA, C-SAA | acute-phase response, cell chemotaxis, chemoattractant activity | Not present in control | / | Not present in control | |
| Phosphatidylethanolamine-binding protein 1 (PEBP1), HCNP, HCNPpp, Neuropolypeptide h3, PBP | ATP binding, serine-type endopeptidase inhibitor activity | ⇩ 3.89 ±0.110 |
/ | ⇩ 2.88 ±0.117 |
|
| Filamin-A, akoga (FLNA), ABP-280, ABPX, Actin-binding protein 280, Alpha-filamin | cell junction assembly, focal adhesion, negative regulation of apoptotic process, wound healing | ⇩ 2.14 ±0.062 |
/ | ⇩ 2.09 ±0.020 |
|
| Spermidine synthase (SRM), PAPT, Putrescine aminopropyltransferase, SPDSY | polyamine metabolic process, protein homodimerization activity, spermidine biosynthetic process | ⇩ 2.29 ±0.152 |
/ | ⇩ 2.40 ±0.067 |
|
| Ezrin (EZR), CVIL, CVL, Cytovillin, DKFZp762H157, Ezrin | focal adhesion, regulation of cell shape | ⇩2.19 ±0.116 |
/ | ⇩ 2.29 ±0.017 |
|
| Heat shock 60 kDa protein 1 (chaperonin) (HSP60), 60 kDa heat shock protein,Chaperonin 60, Chaperonin 60, CPN60, GROE | negative regulation of apoptotic process, positive regulation of interleukin-6,10,12 | ⇧ 2.14 ±0.099 |
/ | ⇧ 2.00 ±0.045 |
|
| l-lactate dehydrogenase B (LDHB), l-lactate dehydrogenase B chain, LHD heart subunit, LHD-B | l-lactate dehydrogenase activity, pyruvate metabolic process | ⇩ 2.04 ±0.034 |
/ | ⇩ 2.09 ±0.049 |
|
| Apolipoprotein A-II (APOA2), apoAII, ApoA-II, Apo-AII, Apolipoprotein A2 | acute inflammatory response, protein oxidation | Not present in control | / | Not present in control | |
| Crystallin alpha B (CRYAB), Alpha-crystallin B chain, Alpha (B)-crystalline, HspB5, HSPB5 | negative regulation of extrinsic apoptotic signaling pathway, positive regulation of cell aging, positive regulation of osteoblast differentiation, negative regulation of adipose tissue development, | ⇧ 3.02 ±0.084 |
/ | ⇧ 2.45 ±0.048 |
|
| Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (SERPINA1), A1A, A1AT, Alpha-1-antiproteinase, alpha-1-antitrypsin | acute-phase response, inflammatory response, serine-type endopeptidase inhibitor activity | Not present in control | / | Not present in control | |
| Serpin peptidase inhibitor, clade C (antithrombin), member 1 (SERPINC1) Antithrombin-III, AT3, ATIII, Serpin C1 | acute-phase response, serine-type endopeptidase inhibitor activity | Not present in control | / | Not present in control | |
| Haptoglobin-related protein (HPR), A-259H10.2, Haptoglobin-related protein, HP | extracellular matrix disassembly, negative regulation of cell proliferation, negative regulation of cell-cell adhesion mediated by cadherin, positive regulation of fibrinolysis, serine-type endopeptidase activity | Not present in control | / | Not present in control | |
| Glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) (GOT2), Aspartate aminotransferase, mitochondrial, FABP-1, FABPpm | canonical glycolysis,epithelial cell differentiation | ⇧ 2.63 ±0.088 |
/ | ⇧ 2.51 ±0.48 |
|
| Glutaredoxin 3 (GLRX3), Glutaredoxin-3, GRX3, GRX4, PICOT, PKC-interacting cousin of thioredoxin | osteoblast differentiation, regulation of apoptotic process | Not present in P-Mg | / | Not present in Mg-2Ag | |
| Programmed cell death protein 5 (PDCD5), Protein TFAR19, TF-1 cell apoptosis-related protein 19, TFAR19 | chloride transmembrane transport, lipid transport | ⇩ 3.63 ±0.067 |
/ | ⇩ 2.63 ±0.196 |
|
| Importin5 (IPO5), Imp5, Importin-5, Importin subunit beta-3 | positive regulation of apoptotic process, positive regulation of intrinsic apoptotic signaling pathway | ⇩2.14 ±0.039 |
/ | ⇩ 2.19 ±0.008 |
3.2.6. Regulated proteins in the presence of Mg-10Gd & Pure-Mg
Regulated proteins (22 proteins) in all of the biological replicates in HUCPV cells incubated with Mg-10Gd and Pure-Mg are listed in Table 2. Among them, 6 and 9 proteins expression was increased and decreased, respectively, compared to control cells. Three proteins were identified only in the presence of Mg-10Gd and Pure-Mg, whereas 3 proteins were absent in the presence of both extracts. Four proteins involved in the apoptotic process were regulated: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (YWHAE) and LGALS3 were downregulated, the latter being absent in cells incubated with those extracts. PTGS2 was remarkably upregulated with the three extracts, but significantly higher with Mg-10Gd and Pure-Mg than with Mg-2Ag. Among the proteins involved in transport, sideroflexin 3 (SFXN3) involved in iron homeostasis was absent in control cells and orosomucoid 2 (ORM2) was missing in the presence of Mg-alloys. Four downregulated proteins with a role in cell differentiation, four and a half LIM domains protein 1 (FHL1), PLS3, LGALS3, and calpain 1 (CAPN1) are listed in Table 2. Additionally, downregulation of calmodulin 1 (CALM1) was observed with the three extracts (although not significantly), being more notable with Mg-10Gd and Mg-Pure than with Mg-2Ag.
3.2.7. Regulated proteins in the presence of Mg-2Ag & Pure-Mg
In the presence of Mg-2Ag and pure-Mg, 29 proteins were significantly regulated in all of the biological replicates of these two conditions (Table 2). Eight of these proteins were up- and 13 of them were down-regulated. Six proteins were only present in all biological replicates of the incubated cells with Mg-2Ag and pure-Mg. Furthermore, there are two proteins which were presented only in the cells without Mg-alloys. Regulated proteins involved in apoptosis: Reticulon 4 (RTN4), Filamin-A (FLNA), Glutaredoxin-3 (GLRX3) and Importin-5 (IPO5) were down-regulated in the presence of Mg-2Ag and pure-Mg. 10 kDa heat shock protein (HSPE1) was up-regulated while 60 kDa heat shock protein (HSPE1) and Alpha-crystallin B chain (CRYAB) were up-regulated. Regarding proteins involved in cell differentiation, Aspartate aminotransferase (GOT2) was up-regulated and Glutaredoxin-3 (GLRX3) was down-regulated in cells cultured with Mg-2Ag and pure Mg. Moreover, the proteins involved in transportation such as LIM and SH3 domain protein 1 (LASP1), Major vault protein (MVP), Ezrin (EZR), Programmed cell death protein 5 (PDCD5), were down- or up-regulated in the presence of Mg-2Ag and pure-Mg. Among the proteins observed in the presence of Mg-2Ag and pure-Mg, which were absent in control cells, APOA2, Antithrombin-III (SERPINA3), and Alpha-1-antitrypsin (SERPINA1) are involved in the acute-phase response. Non-POU domain-containing octamer-binding protein (NONO) was upregulated in the presence of Mg-2Ag and pure-Mg.
4. Discussion
According to the overall results, it is obvious that the increased concentration of Mg2+ ions is responsible for the main effects observed in this study. In comparison to the effect of Mg2+ ions, Ag+-ions and Gd3+-ions have minor effects. Additionally, increased extracts pH probably have an influence on chondrogenesis. Indeed, lower pH (as observed in diabetes and aging) negatively influence bone homeostasis (altered bone structure and density). Furthermore, an alkaline pH (about 8) is optimal for alkaline phosphatase activity and hydroxyapatite precipitation while switching off osteoclast resorption [29,30]. Moreover, Moghadam et al. demonstrated that chondrogenesis was more efficient after short-term culture in alkaline medium [31]. In vitro and even in vivo magnesium-based material degradation is a complex mechanism accompanied by increased pH, ion released (increased osmolality) and other phenomenon. Therefore, the already observed positive effects of these biomaterials on bone healing are probably multifactorial and due to the synergistic effects of magnesium-based degradation. Furthermore, pH of the different extracts are similar thus, the proteomics variation measured between the different extracts are probably due to the material compositions themselves.
Mg2+ is an endogenous element in living organisms and its doubly charged ion involved in a multitude of physiological processes, in many cases enabling defined functions of proteins as their ligands. Living organisms are equipped with a fine-tuned system guaranteeing constant levels of Mg ions in the intra- and extracellular space. Thus, it is not surprising that the increase of Mg ions in the culture medium, will lead to an active cell reaction (e.g., regulation of 246 proteins). Extracellular proteins and cytosolic/cytoskeletal proteins were mostly upregulated and downregulated, respectively. Among the main cell functions of the proteins influenced by the extracts, cell attachment, growth, differentiation and survival or apoptosis were identified. Such functions are involved also in the interactions between chondrocytes and ECM and are important for cartilage homeostasis and cartilage repair [32]. Mg has a key role in cellular energy metabolism and Mg2+ ions are known to enhance the activity of adenosine triphosphate (ATP) synthase [33]. This enzyme consists of two main regions, F0 and F1 themselves composed of subunits. Here, accordingly, several subunits were upregulated (Fig. A1): from the F0 complex: ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 (ATP5F1) and from F1: ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 (ATP5A1), gamma polypeptide 1 (ATP5C1), beta polypeptide (ATP5B) and O subunit (ATP5O). Similarly, upregulation of voltage-dependent anion channel 1 (VDAC1) was induced by the extracts. This protein interacts with hexokinase and creatine kinase to convert newly generated ATP into high-energy storage molecules. Therefore the increased synthesis of VDAC1 is also associated with high metabolically active and energy-demanding cells [34]. A possible explanation may be that the increased Mg2+-concentration induces the increased synthesis of energy-rich (phosphate-rich) metabolites for binding free Mg2+- ions for maintaining Mg2+ homeostasis. Increased of energy-rich metabolites may increase biosynthesis by which the energy-rich metabolites are consumed [35].
Proteins involved in cholesterol metabolism were strongly upregulated by the extracts. Lecithin-cholesterol acyltransferase (LCAT) showed 2-fold higher expression with Mg-10Gd and Pure-Mg than with Mg-2Ag. This protein is the central enzyme involved in the extracellular metabolism of lipoproteins. Apolipoprotein A-I (APOA1) is the most potent phosphatidylcholine-sterol acyltransferase activator in plasma (although it can also be activated by APOE, APOC1 and APOA4). All those apolipoproteins involved in cholesterol efflux (as well as additional ones as APOD) were strongly upregulated with the three extracts (Appendix A). Both LCAT and APOAI lack or deficiency give rise to cartilage degeneration and the development of osteoarthritis (OA) [36,37]. Thus, the possible inhibitory effect of those extracts on OA (i.e., through the upregulation of the aforementioned proteins), is an interesting subject for future investigations.
The three extracts induced the upregulation of proteins involved in cartilage development (both ECM integrity and ECM-cell adhesion) (Appendix A). Among them, TNC is notably upregulated during cartilage development, and it is involved in ECM remodelling and cell differentiation [38]. HSPG2 is involved in the metabolism (synthesis and catabolism) of GAG, one of the main components of cartilage ECM. It is required for cartilage development, where it plays a role in ECM organization. ICAM1 (whose expression was not detected in control cells) has multiple functions, being relevant its role in cell adhesion (specifically integrin-mediated adhesion). Its expression in human chondrocytes can be induced by exogenous interleukin 1 α (IL1α), which was added to the culture medium in the study of Davies et al. in order to induce chondrogenesis [39]. Those results suggest a synergistic effect of IL1α in the presence of Mg-extracts or a direct effect of the Mg ions on ICAM1 expression.
Another group of proteins, upregulated by the three extracts, was the integrin family. Integrins are responsible for primary adhesion of cells to orthopaedic or dental implants, therefore addition of Mg ions to the surface of biomaterials enhance cell-material interaction, reducing the possibilities of implant rejection by the body [40]. Furthermore, the integrin family plays a major role in mediating cell-matrix interactions that are important in regulating cartilage development and repair. Integrins and cell-matrix interactions have been shown to be involved in chondrogenesis of MSC [38] and enhance MSC attachment to endochondral defects (enhancing its repair). Additionally, integrins are involved in the negative regulation of apoptosis. Upregulation of integrins in response to Mg extracts seems therefore beneficial, not only for enhancing chondrogenesis of HUCPV cells, but also for generating a good quality cartilaginous matrix. In native cartilage, chondrocytes express several members of the integrin family, which can serve as receptors for relevant proteins in the structure of the ECM (which also were upregulated under the influence of the extracts): ITGA5 is a receptor for FN1, ITGAV for VTN and ITGA2 for COL6. Since divalent cations, including Mg2+, are ligands for integrins and activate them, an increased cell adhesion of HUCPV cells under the influence of the three extracts is expected. The integrin-signalling proteins are important components of the cartilage ECM. VTN interacts with glycosaminoglycans and proteoglycans and serves as a cell-to-substrate adhesion molecule. Furthermore, it inhibits the membrane-damaging effect of the terminal cytolytic effect of the complement pathway. FN1 is involved in early chondrocyte differentiation events after birth. Its upregulation takes place during the condensation of stem cells [41]. COL4 is found in connective tissue. A notable upregulation of this protein has been reported during early stages of human MSC chondrogenesis (after 10 days), possibly due to the influence of this protein on Sox9 [42]. Two other chondrogenic-related proteins were upregulated by the three extracts: GPLD1, which stimulates chondrocyte differentiation and HEXB, which has a role in the catabolic process of chondroitin sulphate.
The presence of Mg-extracts on HUCPV cells during chondrogenesis also induced the upregulation of TGFBI, a protein involved in the cell-collagen interaction, and important for ECM remodelling during chondrocyte differentiation [43]. TGFBI overexpression positively enhances the proliferation and chondrogenic potential of human synovium-derived MSC [44]. Furthermore, TGFBI induces upregulation of integrins [45]. Therefore, Mg extracts may induce an enhancement of TGFBI, which will in turn upregulate integrin production, and subsequently, integrin-mediated cell adhesion and chondrogenesis. In addition, TGFBI induces expression of PLOD1 and PLOD2, proteins upregulated in regenerated cartilage in vivo (regarding the natural cartilage).
Angiogenesis is a fundamental component of bone repair due to the development of blood vessels in the fracture callus [46] and a vital part of bone formation [46,47]. Hypertrophic cartilage produces angiogenic stimulators [[48], [49], [50]], unlike angiogenesis inhibitors, which are secreted by immature chondrocytes [51,52]. The 3 processes, chondrogenesis, angiogenesis and bone formation are closely related. Hence, some of the regulated proteins are involved in all of them (e.g., ITGA5 and COL1A1). Upregulation of 15 of the 16 regulated proteins involved in angiogenesis shows a hypertrophic stage of chondrocytes. Furthermore, THBS1 upregulation could be of interest since it has been shown that this protein inhibits vascular endothelial growth factor (VEGF)-induced migration in human microvascular cells [53].
COL1A1 was downregulated under the influence of the three extract. COL1A1 is involved in bone trabecula formation and final stage of cartilage development. Nevertheless, the downregulation of this protein versus the lack of effect on COL2 production is indicating a reduction in the ratio COL2/COL1 characteristic in cartilage tissue. PLS3 is involved in bone development and its downregulation in the presence of Mg-alloys may avoid cartilage mineralisation. Consequently, the downregulation of those two proteins is beneficial in order to keep the chondrogenic phenotype of the differentiated HUCPV cells. Furthermore, two proteins PTGS2 and GPLD1 were only detected in the presence of extracts. PTGS2 (or cyclooxygenase 2, COX 2) is responsible for production of inflammatory prostaglandins. Furthermore, PTGS2 is also associated with increased cell adhesion, phenotypic changes and resistance to apoptosis. PTGS2 is a target of nonsteroidal anti-inflammatory drugs (NSAID) including acetylsalicylic acid (“aspirin”) and isobutylphenylpropionic acid (“ibuprofen”). NSAID have been reported to have (controversial/negative) influence osteogenesis during bone fracture healing [54]. Welting et al. demonstrate the role of PTGS2 in chondrocyte maturation-hypertrophy [55]. GPLD1 hydrolyses the inositol phosphate linkage in proteins anchored by glycosylphosphatidylinositol (GPI) to the outer leaflet of the plasma membrane, thereby releasing the attached protein. Over 250 GPI-proteins are known, among them heparan sulfate proteoglycans (HSPG) [56], ephrin A ligands (for Eph receptors - the largest known subfamily of receptor protein-tyrosine kinases), putative adhesion/signalling molecules of the Ly6 family, and enzymes like alkaline phosphatase [57]. GPI-anchored proteins are believed to have a role in cell adhesion events involved in tissue patterning and cell signalling. Indeed, Ahrens et al. demonstrated that GPI-anchored proteins are necessary to the columnar tissue arrangement and the proper development of the growth plate [57].
Apoptosis is a tightly regulated process, inevitable and essential during development, particularly during formation of articular cartilage and endochondral ossification of growth plate [58]. Increased apoptosis in native cartilage is associated with matrix degradation. Induction of MSC chondrogenesis in vitro using micromasses formation models increases the possibility of apoptosis due to the severe hypoxic conditions that cells suffer in the centre of the spheres. Nevertheless, some differences in protein expression (involved in both positive and negative regulation of apoptosis) were found due to the action of the extracts (Fig. 4). On the one hand, 9 apoptosis-related proteins were found only in presence of extracts: 5 were stimulators of apoptosis (C3, KNG1, PTGS2, TARDBP, and GPLD1) and 4 inhibitors (APOE, ICAM1, FAM129B, and ANGPTL4). On the other hand, 2 proteins stimulating apoptosis were downregulated, S100A9 and LGALS3. S100A9 was completely absent in the presence of Mg-alloys. This protein also has a role in actin cytoskeleton reorganization, proinflammatory response and oxidant-scavenging. LGALS3 was downregulated in the presence of Mg-2Ag, and absent in the presence of Pure-Mg and Mg-10Gd, which may indicate a toxic action of Ag. Galectin-3 (LGALS3) is also found to be a potent inhibitor for osteoclastogenesis in vitro [59]. Those results show a clear influence on apoptosis and suggest a reduction of cell death by the extracts.
Since Mg-alloy degradation products might have undesired side effects like toxicity to the cells, regulated proteins involved in the response to toxic effect were evaluated. Six from the 10 regulated proteins known to be associated with stress response showed increased expression in the presence of extracts. From those six, four proteins were not found in the absence of the extracts, which might suggest a response of the cells toward toxicity; however, these proteins have also other roles in the cells. For instance, C3 is one of the stimulators for angiogenesis and ICAM1 is involved in cell migration and adhesion, therefore reinforcing cartilage repair.
Some proteins were regulated only by 2 extracts, or showed significant differences in the expression (always normalised to control) among the 3 extracts. In principle, proteins up- or downregulated only by Mg-2Ag or Mg-10Gd extracts should give information about the effects of the alloying elements (Ag and Gd) on HUCPV chondrogenesis, as Mg concentration was constant. CHMP4B, having a role apoptosis suppression, was upregulated with those extracts, could be beneficial for cell viability. However, NARS with the same function was downregulated. Regarding the other function of NARS in the cellular response to toxic substrate, its downregulation supports suggests a lack of toxic effect of Mg-2Ag and Mg-10Gd. Interestingly, serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2), was only expressed in the presence of the extracts, and its expression was significantly higher with Mg-10Gd and Mg-2Ag than with Pure-Mg. It has been shown that SERPINE2 expression in human chondrocytes might prevent cartilage catabolism by inhibiting the expression of matrix metallopeptidase 13 (MMP13), one of the most relevant collagenases, involved in cartilage breakdown in OA [60]. Therefore, alloying elements could have a positive effect on the maintenance of cartilage integrity. TXNRD1, protein involved in cell proliferation, was downregulated only with Mg-10Gd and Mg-2Ag. In correlation with this, a strong downregulation of the microtubule-associated protein 9 (MAP9) with Mg-10Gd extract was observed, while a slight upregulation was detected with Pure-Mg and Mg-2Ag. MAP9 is required for mitosis progression and cytokinesis (UniProt). Therefore, its downregulation could decrease cell cycle progression and cell division. The proteins commonly regulated only by Pure-Mg and Mg-2Ag as well as Pure-Mg and Mg-10Gd were mainly involved in apoptosis, indicating that Mg itself has a significant influence on this cellular process.
Mg-2Ag and Pure-Mg showed a beneficial effect on HUCPV viability by downregulating proteins positively involved in apoptosis (RTN4,FLNA, GLRX3, and IPO5) and upregulating two proteins involved in negative regulation of apoptosis (60 kDa heat shock protein and α-crystallin B chain). However, FLNA, having a role in protecting cells from apoptosis was also downregulated. An interesting upregulated protein in the presence of Mg-2Ag and Pure-Mg is non-POU domain containing, octamer-binding protein (NONO). This protein has a protective role in the regulation of oxidative stress-induced neuron intrinsic apoptotic signalling pathway, Furthermore, NONO promotes chondrogenesis by interacting with SOX9 thus allowing/promoting transcription of SOX9 target genes such COL2A1 [61]. Both functions suggest that NONO upregulation is beneficial for cartilage development and bone healing. Some proteins involved in acute-phase or response to inflammation (Apolipoprotein A-II2, ATIII and SERPINA1) were only observed under the influence of Mg-2Ag and Pure-Mg, making them interesting for further research.
Mg-10Gd and Pure-Mg showed beneficial effects on chondrogenesis and maintenance of cartilage integrity. First, LGALS3, which has a role in the regulation of extrinsic apoptotic signalling pathway via death domain receptors, was absent. PTGS2 (having roles previously described in apoptosis, angiogenesis and bone formation) was remarkably upregulated with the three extracts, but significantly lower with Pure-Mg and Mg-2Ag than with Mg-10Gd. In the second place, the downregulation of calpain 1 (CAPN1), supported by the decreased expression of calmodulin 1 (phosphorylase kinase, delta) (CALM1) (Appendix A), which expression has been reported to diminish during chondrogenic differentiation of stem cells [62], indicate that Mg-10d and Pure-Mg extracts could enhance cell chondrogenesis. This idea is also supported by the absence of CAPN1 protein in the presence of Mg-10Gd and Pure-Mg. The serum concentration of CAPN1 raises several folds during an acute phase response (the systemic answer to a local inflammatory stimulus). Therefore its absence suggests a lack or decrease of immunological reactions against the degradation products of the material [63].
To conclude, various regulated proteins were identified in response to Mg-alloy degradation products. Regulation of specific proteins indicate a positive effect on chondrogenesis (i.e., integrins, TGFBI, FN1, VTN, CALM1, NONO) and cell viability (apoptotic-related proteins), as well as possible influence on reducing or inhibiting OA (cholesterol metabolism-related proteins and SERPINE2) and acute-phase response (APOA2, ATIII and SERPINA1). These results show that the Mg-based materials have potential to stimulate cartilage in vitro. Further investigation in vivo will be pursuit to validate these results.
Author Contributions
A.M.S. and F.F. designed the study; A.M.S, M.O., M.W., M.M., and B.L. conducted cell cultures, processing, and data analysis; H.S., R.W.R. and B.L. provided intellectual contributions; A.M.S. and M.O. cowrote the paper.
Conflicts of interest
None.
Acknowledgements
This research was financially supported by the Helmholtz Virtual Institute VH-VI-523 (in vivo studies of biodegradable magnesium based implant materials). We thank Ute Kohlmeyer (Galab, Hamburg, Germany) for ICP-MS measurements and to Gabor Szakacs for his work in the fabrication of the materials (Magnesium Innovation Center - MagIC, Helmholtz Zentrum, Geesthacht, Germany). We would like to thank the Asklepios Klinik Altona (Hamburg, Germany) for providing the human umbilical cord samples.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Fig. A1.
Table A1.
| Protein name | Gene symbol | Log10 fold change compared to control |
||
|---|---|---|---|---|
| Mg-10Gd | Mg-2Ag | Pure-Mg | ||
| Apolipoprotein D | APOD | >1 | >1 | >1 |
| ApoD, Apo-D, Apolipoprotein D | ||||
| Plasminogen | PLG | >1 | >1 | >1 |
| Complement component 4 binding protein, alpha | C4BPA | >1 | >1 | >1 |
| C4b-binding protein alpha chain, C4bp, C4BP, Proline-rich protein, PRP | ||||
| Immunoglobulin heavy constant gamma 1 (G1m marker) | IGHG1 | >1 | >1 | >1 |
| Immunoglobulin heavy constant gamma 1 (G1m marker) | ||||
| Immunoglobulin heavy constant alpha 1 | IGHA1 | >1 | >1 | >1 |
| FLJ14473, FLJ35065, FLJ35500, FLJ36402, FLJ39698, FLJ40001, FLJ41548, FLJ41552, FLJ41789, FLJ43248, FLJ43594, FLJ44293, FLJ46028, FLJ46621, FLJ46724, FLJ46811, FLJ46824, FLJ90170, IgA1, Ig alpha-1 chain C region, MGC102857 | ||||
| Ceruloplasmin (Ferroxidase) | CP | >1 | >1 | >1 |
| Ceruloplasmin, ferroxidase | ||||
| Immunoglobulin kappa constant | IGKC | >1 | >1 | >1 |
| HCAK1, Ig kappa chain C region, Ig kappa chain V-I region HK101, Ig kappa chain V-I region Walker, Km, MGC111575, MGC62011, MGC72072, MGC88770, MGC88771, MGC88809 | ||||
| GC, vitamin D binding protein | GC | >1 | >1 | >1 |
| Vitamin D-binding protein (DBP) (VDB) (Gc protein-derived macrophage activating factor) (Gc-MAF) (GcMAF) (Gc-globulin) (Group-specific component) (Gc) (Vitamin D-binding protein-macrophage activating factor) (DBP-maf) | ||||
| Complement C3 | C3 | >1 | >1 | >1 |
| (C3 and PZP-like alpha-2-macroglobulin domain-containing protein 1) [Cleaved into: Complement C3 beta chain;C3-beta-c (C3bc);Complement C3 alpha chain; C3a anaphylatoxin; Acylation stimulating protein (ASP) (C3adesArg); Complement C3b alpha’ chain; Complement C3c alpha’ chain fragment 1; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment; Complement C3c alpha’ chain fragment 2], CPAMD1 | ||||
| immunoglobulin heavy constant mu | IGHM | >1 | >1 | >1 |
| AGM1, DKFZp686I15196, DKFZp686I15212, FLJ00385, Ig mu chain C region, MGC104996, MGC52291, MU, VH | ||||
| Paraoxonase 1 | PON1 | >1 | >1 | >1 |
| Serum paraoxonase/arylesterase 1 (PON 1) (EC 3.1.1.2) (EC 3.1.1.81) (EC 3.1.8.1) (Aromatic esterase 1) (A-esterase 1) (K-45) (Serum aryldialkylphosphatase 1), PON | ||||
| Apolipoprotein A4 | APOA4 | >1 | >1 | >1 |
| ApoA-IV, Apo-AIV, Apolipoprotein A4, Apolipoprotein A-IV, MGC142154, MGC142156 | ||||
| Tenascin C | TNC | >1 | >1 | >1 |
| 150-225, Cytotactin, Glioma-associated-extracellular matrix antigen, GMEM, GP, GP 150–225, Hexabrachion, HXB, JI, MGC167029, Myotendinous antigen, Neuronectin, Tenascin, Tenascin-C, TN, TN-C | ||||
| Apolipoprotein E | APOE | >1 | >1 | >1 |
| APOE | ||||
| serum amyloid A4, constitutive | SAA4 | >1 | >1 | >1 |
| Constitutively expressed serum amyloid A protein, CSAA, C-SAA, Serum amyloid A-4 protein | ||||
| Apolipoprotein M | APOM | >1 | >1 | >1 |
| Apolipoprotein M, ApoM, Apo-M, G3a, G3A, HSPC336, MGC22400, NG20, Protein G3a | ||||
| Transthyretin | TTR | >1 | >1 | >1 |
| ATTR, CTS, CTS1, HsT2651, PALB, Prealbumin, TBPA, Transthyretin | ||||
| Apolipoprotein A2 | APOA2 | >1 | >1 | >1 |
| apoAII, ApoA-II, Apo-AII, Apolipoprotein A2, Apolipoprotein A-II | ||||
| Hemopexin | HPX | >1 | >1 | >1 |
| Beta-1B-glycoprotein, FLJ56652, Hemopexin, HX | ||||
| Immunoglobulin heavy constant gamma 2 (G2m marker) | IGHG2 | >1 | >1 | >1 |
| Ig gamma-2 chain C region | ||||
| Haptoglobin-related protein | HPR | >1 | >1 | >1 |
| A-259H10.2, Haptoglobin-related protein, HP | ||||
| Apolipoprotein C3 | APOC3 | >1 | >1 | >1 |
| APOCIII, ApoC-III, Apo-CIII, Apolipoprotein C3, Apolipoprotein C-III, MGC150353 | ||||
| Nidogen 2 | NID2 | >1 | >1 | >1 |
| NID-2, Nidogen-2, Osteonidogen | ||||
| Alpha-1-B glycoprotein | A1BG | >1 | >1 | >1 |
| A1B, ABG, Alpha-1B-glycoprotein, Alpha-1-B glycoprotein, DKFZp686F0970, GAB, HYST2477 | ||||
| Inter-alpha-trypsin inhibitor heavy chain1 | ITIH1 | >1 | >1 | >1 |
| H1P, IATIH, IGHEP1, Inter-alpha-inhibitor heavy chain 1, Inter-alpha-trypsin inhibitor complex component III, Inter-alpha-trypsin inhibitor heavy chain H1, ITIH, ITI-HC1, ITI heavy chain H1, MGC126415, Serum-derived hyaluronan-associated protein, SHAP | ||||
| Kininogen 1 | KNG1 | >1 | >1 | >1 |
| Alpha-2-thiol proteinase inhibitor, BDK, Fitzgerald factor, High molecular weight kininogen, HMWK, Kininogen-1, KNG, Williams-Fitzgerald-Flaujeac factor | ||||
| AE binding protein 1 | AEBP1 | >1 | >1 | >1 |
| ACLP, Adipocyte enhancer-binding protein 1, AE-binding protein 1, Aortic carboxypeptidase-like protein, FLJ33612 | ||||
| Perilipin 2 | PLIN2 | >1 | >1 | >1 |
| adipose differentiation-related protein, ADFP | ||||
| Apolipoprotein L1 | APOL1 | >1 | >1 | >1 |
| ApoL, APOL, Apo-L, APO-L, ApoL-I, APOL-I, Apolipoprotein L, Apolipoprotein L1, Apolipoprotein L-I, FSGS4 | ||||
| Ig kappa chain V-III region GOL (Rheumatoid factor) | >1 | >1 | >1 | |
| Tubulointerstitial nephritis antigen like 1 | TINAGL1 | >1 | >1 | >1 |
| ARG1, GIS5, Glucocorticoid-inducible protein 5, LCN7, LIECG3, OLRG2, OLRG-2, Oxidized LDL-responsive gene 2 protein, P3ECSL, PP6614, PSEC0088, TINAGL, TIN Ag-related protein, TINAGRP, TIN-Ag-RP, Tubulointerstitial nephritis antigen-like, Tubulointerstitial nephritis antigen-related protein, UNQ204/PRO230 | ||||
| Glutathione peroxidase 3 | GPX3 | >1 | >1 | >1 |
| Extracellular glutathione peroxidase, Glutathione peroxidase 3, GPx-3, GPXP, GPx-P, GSHPx-3, GSHPx-P, Plasma glutathione peroxidase | ||||
| Complement C1q C chain | C1QC | >1 | >1 | >1 |
| Complement component 1, q subcomponent, C chain, C1Q-C, C1QG, Complement C1q subcomponent subunit C, FLJ27103 | ||||
| Orosomucoid 2 | ORM2 | >1 | >1 | >1 |
| AGP2, AGP 2, AGP-B, AGP-B′, Alpha-1-acid glycoprotein 2, OMD 2, Orosomucoid-2 | ||||
| Apolipoprotein C-I | APOC1 | >1 | >1 | >1 |
| APOC1 | ||||
| Coagulation factor XII | F12 | >1 | >1 | >1 |
| Coagulation factor XII, HAE3, HAEX, HAF, Hageman factor | ||||
| serpin family C member 1 | SERPINC1 | >1 | >1 | >1 |
| Serpin peptidase inhibitor, clade C (antithrombin), member 1, Antithrombin-III, AT3, ATIII, MGC22579, PRO0309, Serpin C1 | ||||
| HtrA serine peptidase 1 | HTRA1 | >1 | >1 | >1 |
| ARMD7, HtrA, HTRA, IGFBP5-protease, L56, ORF480, PRSS11, Serine protease 11, Serine protease HTRA1 | ||||
| Amyloid P component, serum | APCS | >1 | >1 | >1 |
| 9.5S alpha-1-glycoprotein, MGC88159, PTX2, SAP, Serum amyloid P-component | ||||
| Intercellular adhesion molecule 1 | ICAM1 | >1 | >1 | >1 |
| BB2, CD54, ICAM-1, Intercellular adhesion molecule 1, Major group rhinovirus receptor, P3.58 | ||||
| Serpin family A member 3 | SERPINA3 | >1 | >1 | >1 |
| Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3, AACT, ACT, alpha-1-antichymotrypsin, Alpha-1-antichymotrypsin, Cell growth-inhibiting gene 24/25 protein, GIG24, GIG25, MGC88254, Serpin A3 | ||||
| Serpin family E member 2 | SERPINE2 | >1 | >1 | >1 |
| Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2, DKFZp686A13110, GDN, Glia-derived nexin, nexin, Peptidase inhibitor 7, PI7, PI-7, PN1, PN-1, PNI, Protease nexin 1, Protease nexin I, Serpin E2 | ||||
| RNA exonuclease 2 | REXO2 | >1 | >1 | >1 |
| REX2, RNA exonuclease 2 homolog (S. cerevisiae), CGI-114, DKFZp566E144, DKFZP566E144, MGC111570, Oligoribonuclease, mitochondrial, REX2, RFN, RNA exonuclease 2 homolog, SFN, Small fragment nuclease, SMFN | ||||
| Malectin | MLEC | >1 | >1 | >1 |
| Oligosaccharyltransferase complex subunit (non-catalytic), KIAA0152 | ||||
| Glycosylphosphatidylinositol specific phospholipase D1 | GPLD1 | >1 | >1 | >1 |
| Glycoprotein phospholipase D, Glycosyl-phosphatidylinositol-specific phospholipase D, GPIPLD, GPI-PLD, GPIPLDM, GPI-specific phospholipase D, MGC22590, Phosphatidylinositol-glycan-specific phospholipase D, PIGPLD, PI-G PLD, PIGPLD1 | ||||
| Progesterone receptor membrane component 1 | PGRMC1 | >1 | >1 | >1 |
| HPR6.6, Membrane-associated progesterone receptor component 1, mPR, MPR, PGRMC | ||||
| Vitronectin | VTN | >1 | >1 | >1 |
| Serum-spreading factor, S-protein, V75, Vitronectin, VN, VNT | ||||
| Serpin family A member 1 | SERPINA1 | >1 | >1 | >1 |
| Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, A1A, A1AT, AAT, Alpha-1-antiproteinase, alpha-1-antitrypsin, Alpha-1-antitrypsin, alpha1AT, Alpha-1 protease inhibitor, MGC23330, MGC9222, PI, PI1, PRO0684, PRO2209, PRO2275, Serpin A1 | ||||
| Family with sequence similarity 129, member B | FAM129B | >1 | >1 | >1 |
| bA356B19.6, C9orf88, DKFZP434H0820, FLJ13518, FLJ22151, FLJ22298, Meg-3, MEG-3, MINERVA, Niban-like protein 1, OC58, Protein FAM129B | ||||
| Angiopoietin- like 4 | ANGPTL4 | >1 | >1 | >1 |
| Angiopoietin-like protein 4, Angiopoietin-related protein 4, ANGPTL2, ARP4, FIAF, Hepatic fibrinogen/angiopoietin-related protein, HFARP, NL2, PGAR, pp1158, PP1158, PSEC0166, UNQ171/PRO197 | ||||
| TAR DNA-binding protein | TARDBP | >1 | >1 | >1 |
| ALS10, TAR DNA-binding protein 43, TDP43, TDP-43 | ||||
| Fibrillin1 | FBN1 | >1 | >1 | >1 |
| FBN, Fibrillin-1, MASS, MFS1, OCTD, SGS, SSKS, WMS | ||||
| Prostaglandin-endoperoxide synthase 2 | PTGS2 | >1 | >1 | >1 |
| COX2, COX-2, Cyclooxygenase-2, GRIPGHS, hCox-2, PGG/HS, PGHS-2, PGH synthase 2, PHS-2, PHS II, Prostaglandin-endoperoxide synthase 2, Prostaglandin G/H synthase 2, Prostaglandin H2 synthase 2 | ||||
| Lecithin-cholesterol acyltransferase | LCAT | >1 | >1 | >1 |
| Lecithin-cholesterol acyltransferase, Phosphatidylcholine-sterol acyltransferase, Phospholipid-cholesterol acyltransferase | ||||
| Apolipoprotein B | APOB | >1 | >1 | >1 |
| Apo B-100, Apolipoprotein B-100, FLDB, LDLCQ4 | ||||
| Glutaminyl-tRNA synthetase | QARS | >1 | >1 | >1 |
| GlnRS, GLNRS, Glutamine--tRNA ligase, Glutaminyl-tRNA synthetase, PRO2195 | ||||
| Sideroflexin 3 | SFXN3 | >1 | >1 | >1 |
| BA108L7.2, SFX3, Sideroflexin-3 | ||||
| Apolipoprotein A1 | APOA1 | 1.872 | 1.892 | 1.839 |
| ApoA-I, Apo-AI, Apolipoprotein A-I | ||||
| Albumin | ALB | 1.652 | 1.814 | 1.788 |
| DKFZp779N1935, GIG20, GIG42, PRO0883, PRO0903, PRO1341, PRO1708, PRO2044, PRO2619, PRO2675, Serum albumin, UNQ696/PRO1341 | ||||
| Haptoglobin | HP | 1.578 | 1.698 | 1.693 |
| BP, HP2ALPHA2, HPA1S, MGC111141 | ||||
| Transferrin | TF | 1.208 | 1.482 | 1.372 |
| Beta-1 metal-binding globulin, DKFZp781D0156, PRO1400, PRO1557, PRO2086, Serotransferrin, Siderophilin, Transferrin | ||||
| Transforming growth factor beta induced | TGFBI | 1.050 | 1.080 | 1.076 |
| Beta ig-h3, BIGH3, CDB1, CDG2, CDGG1, CSD, CSD1, CSD2, CSD3, EBMD, Kerato-epithelin, LCD1, RGD-CAP, RGD-containing collagen-associated protein, Transforming growth factor-beta-induced protein ig-h3 | ||||
| Superoxide dismutase 2 | SOD2 | 0.870 | 1.031 | 1.012 |
| IPOB, MNSOD, MVCD6 | ||||
| Alanyl aminopeptidase, membrane | ANPEP | 0.835 | 0.921 | 0.831 |
| Alanyl aminopeptidase, Aminopeptidase M, Aminopeptidase N, AP-M, APN, AP-N, CD13, gp150, GP150, hAPN, LAP1, Microsomal aminopeptidase, Myeloid plasma membrane glycoprotein CD13, p150, P150, PEPN | ||||
| Collagen type IV alpha 2 chain | COL4A2 | 0.764 | 0.770 | 0.691 |
| Collagen alpha-2(IV) chain, DKFZp686I14213, FLJ22259 | ||||
| Fibronectin 1 | FN1 | 0.577 | 0.616 | 0.503 |
| CIG, Cold-insoluble globulin, DKFZp686F10164, DKFZp686H0342, DKFZp686I1370, DKFZp686O13149, ED-B, Fibronectin, FINC, FN, FNZ, GFND, GFND2, LETS, MS | ||||
| Heparan sulfate proteoglycan 2 | HSPG2 | 0.503 | 0.613 | 0.415 |
| Basement membrane-specific heparan sulfate proteoglycan core protein, HSPG, perlecan, Perlecan, PLC, PRCAN, SJA, SJS, SJS1 | ||||
| Ubiquinol-cytochrome c reductase core protein I | UQCRC1 | 0.523 | 0.594 | 0.656 |
| Complex III subunit 1, Core protein I, Cytochrome b-c1 complex subunit 1, mitochondrial, D3S3191, QCR1, Ubiquinol-cytochrome-c reductase complex core protein 1, UQCR1 | ||||
| Ubiquinol-cytochrome c reductase core protein II | UQCRC2 | 0.540 | 0.581 | 0.584 |
| Complex III subunit 2, Core protein II, Cytochrome b-c1 complex subunit 2, mitochondrial, QCR2, Ubiquinol-cytochrome-c reductase complex core protein 2, UQCR2, | ||||
| Alpha-2-macroglobulin | A2M | 0.673 | 0.572 | 0.506 |
| Alpha-2-M, Alpha-2-macroglobulin, C3 and PZP-like alpha-2-macroglobulin domain-containing protein 5, CPAMD5, DKFZp779B086, FWP007, S863-7 | ||||
| Stomatin like 2 | STOML2 | 0.512 | 0.560 | 0.613 |
| Stomatin (EPB72)-like 2, EPB72-like protein 2, HSPC108, SLP2, SLP-2, Stomatin-like protein 2 | ||||
| Gelsolin | GSN | 0.375 | 0.548 | 0.514 |
| Actin-depolymerizing factor, ADF, AGEL, Brevin, DKFZp313L0718, Gelsolin | ||||
| Collagen type VI alpha 3 chain | COL6A3 | 0.741 | 0.543 | 0.594 |
| Collagen alpha-3(VI) chain, DKFZp686D23123, DKFZp686K04147, DKFZp686N0262, FLJ34702, FLJ98399 | ||||
| Endoplasmic reticulum oxidoreductase 1 alpha | ERO1A | 0.571 | 0.539 | 0.587 |
| Endoplasmic oxidoreductin-1-like protein, ERO1A, ERO1-alpha, ERO1-L, ERO1-L-alpha, ERO1-like protein alpha, Oxidoreductin-1-L-alpha, UNQ434/PRO865 | ||||
| Collagen type VI alpha 2 chain | COL6A2 | 0.778 | 0.530 | 0.590 |
| Collagen alpha-2(VI) chain, DKFZp586E1322, FLJ46862, PP3610 | ||||
| Voltage dependent anion channel 1 | VDAC1 | 0.454 | 0.529 | 0.530 |
| hVDAC1, MGC111064, Outer mitochondrial membrane protein porin 1, Plasmalemmal porin, PORIN, Porin 31HL, Porin 31HM, VDAC, VDAC-1, Voltage-dependent anion-selective channel protein 1 | ||||
| Prohibitin | PHB | 0.498 | 0.525 | 0.588 |
| PHB1 | ||||
| Integrin subunit alpha V | ITGAV | 0.562 | 0.512 | 0.573 |
| integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51, CD51, DKFZp686A08142, Integrin alpha-V, MSK8, Vitronectin receptor subunit alpha, VNRA | ||||
| Prohibitin 2 | PHB2 | 0.489 | 0.504 | 0.542 |
| BAP, Bap37, BCAP37, B-cell receptor-associated protein BAP37, D-prohibitin, MGC117268, p22, PNAS-141, Prohibitin-2, REA, Repressor of estrogen receptor activity | ||||
| Hexosaminidase subunit beta | HEXB | 0.348 | 0.503 | 0.504 |
| Beta-hexosaminidase subunit beta, Beta-N-acetylhexosaminidase subunit beta, Cervical cancer proto-oncogene 7 protein, HCC7, HCC-7, Hexosaminidase subunit B, N-acetyl-beta-glucosaminidase subunit beta | ||||
| DnaJ heat shock protein family (Hsp40) member B11 | DNAJB11 | 0.406 | 0.482 | 0.478 |
| ABBP2, ABBP-2, APOBEC1-binding protein 2, DJ9, DnaJ homolog subfamily B member 11, DnaJ protein homolog 9, EDJ, ER-associated DNAJ, ER-associated dnaJ protein 3, ER-associated Hsp40 co-chaperone, ERdj3, ERj3, ERJ3, ERj3p, hDj9, HDJ9, hDj-9, HEDJ, Human DnaJ protein 9, PRO1080, PSEC0121, PWP1-interacting protein 4, UNQ537, UNQ537/PRO1080 | ||||
| Fumarate hydratase | FH | 0.390 | 0.475 | 0.460 |
| Fumarase, Fumarase, Fumarate hydratase, mitochondrial, HLRCC, LRCC, MCL, MCUL1 | ||||
| ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | ATP5C1 | 0.405 | 0.468 | 0.485 |
| ATP5C, ATP5CL1, ATP synthase subunit gamma, mitochondrial, F-ATPase gamma subunit | ||||
| Enoyl CoA hydratase 1 | ECH1 | 0.397 | 0.456 | 0.468 |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial, HPXEL | ||||
| dihydrolipoamide S-succinyltransferase | DLST | 0.403 | 0.455 | 0.488 |
| 2-oxoglutarate dehydrogenase complex component E2, Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex, Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial, DLTS, E2K, OGDC-E2 | ||||
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | ATP5O | 0.418 | 0.453 | 0.435 |
| ATPO, ATP synthase subunit O, mitochondrial, Oligomycin sensitivity conferral protein, OSCP | ||||
| Histone cluster 1 H2B family member l | HIST1H2BL | 0.337 | 0.436 | 0.354 |
| dJ97D16.4, H2B/c, H2BFC, Histone H2B.c, Histone H2B type 1-L | ||||
| Collagen type VI alpha 1 chain | COL6A1 | 0.676 | 0.433 | 0.510 |
| Collagen alpha-1(VI) chain | ||||
| Acetyl-CoA acyltransferase 2 | ACAA2 | 0.243 | 0.428 | 0.295 |
| 3-ketoacyl-CoA thiolase, mitochondrial, Acetyl-CoA acyltransferase, Beta-ketothiolase, DSAEC, FLJ35992, FLJ95265, Mitochondrial 3-oxoacyl-CoA thiolase, T1 | ||||
| Malate dehydrogenase 2 | MDH2 | 0.332 | 0.426 | 0.422 |
| Malate dehydrogenase, mitochondrial, MDH, MGC:3559, M-MDH, MOR1, malate dehydrogenase 2, NAD (mitochondrial) | ||||
| ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | ATP5B | 0.360 | 0.425 | 0.456 |
| ATPMB, ATPSB, ATP synthase subunit beta, mitochondrial, MGC5231 | ||||
| Histone cluster 1 H4 family member a | HIST1H4A | 0.413 | 0.419 | 0.345 |
| Histone cluster 1, H4a, H4/a, H4FA, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, HIST2H4B, HIST4H4 | ||||
| Charged multivesicular body protein 4B | CHMP4B | 0.359 | 0.414 | < −1 |
| Chromatin modifying protein 4B, C20orf178, Charged multivesicular body protein 4b, CHMP4A, CHMP4b, Chromatin-modifying protein 4b, CTPP3, dJ553F4.4, hSnf7-2, hVps32-2, Shax1, SHAX1, SNF7, SNF7-2, SNF7 homolog associated with Alix 1, Vacuolar protein sorting-associated protein 32-2, Vps32-2, VPS32B | ||||
| Histone cluster 2 H2B family member e | HIST2H2BE | 0.366 | 0.407 | 0.315 |
| Histone cluster 2, H2be, GL105, H2B, H2B/q, H2B.1, H2BFQ, H2BGL105, H2BQ, Histone H2B.q, Histone H2B-GL105, Histone H2B type 2-E, MGC119802, MGC119804, MGC129733, MGC129734 | ||||
| Integrin subunit alpha 2 | ITGA2 | 0.520 | 0.404 | 0.511 |
| Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), BR, CD49 antigen-like family member B, CD49b, CD49B, Collagen receptor, GPIa, Integrin alpha-2, Platelet membrane glycoprotein Ia, VLA-2, VLA-2 subunit alpha, VLAA2 | ||||
| 5′-nucleotidase ecto | NT5E | 0.407 | 0.400 | 0.431 |
| 5′-NT, 5′-nucleotidase, CD73, E5NT, Ecto-5′-nucleotidase, eN, eNT, NT, NT5, NTE | ||||
| Brain abundant membrane attached signal protein 1 | BASP1 | 0.305 | 0.399 | 0.358 |
| 22 kDa neuronal tissue-enriched acidic protein, Brain acid soluble protein 1, CAP23, CAP-23, MGC8555, NAP22, NAP-22, Neuronal axonal membrane protein NAP-22 | ||||
| Glutamic-oxaloacetic transaminase 2 | GOT2 | 0.267 | 0.399 | 0.418 |
| Aspartate aminotransferase, mitochondrial, FABP-1, FABPpm, Fatty acid-binding protein, FLJ40994, Glutamate oxaloacetate transaminase 2, KAT4, KATIV, mAspAT, mitAAT, Plasma membrane-associated fatty acid-binding protein, Transaminase A | ||||
| Glutamate dehydrogenase 1 | GLUD1 | 0.338 | 0.397 | 0.420 |
| GDH, GDH1, GDH 1, GLUD, Glutamate dehydrogenase 1, mitochondrial, MGC132003 | ||||
| Keratin 9 | KRT9 | 0.196 | 0.393 | 0.241 |
| CK-9, Cytokeratin-9, EPPK, K9, Keratin, type I cytoskeletal 9, Keratin-9 | ||||
| Microtubule associated protein 9 | MAP9 | < −1 | 0.392 | 0.172 |
| ASAP, Aster-associated protein, FLJ21159, Microtubule-associated protein 9 | ||||
| Crystallin alpha B | CRYAB | 0.270 | 0.386 | 0.476 |
| Alpha(B)-crystallin, Alpha-crystallin B chain, CRYA2, CTPP2, Heat shock protein beta-5, HspB5, HSPB5, Renal carcinoma antigen NY-REN-27, Rosenthal fiber component | ||||
| Transmembrane p24 trafficking protein 9 | TMED9 | 0.432 | 0.385 | 0.455 |
| Glycoprotein 25L2, GP25L2, HSGP25L2G, Transmembrane emp24 domain-containing protein 9 | ||||
| Nucleobindin 1 | NUCB1 | 0.231 | 0.382 | 0.373 |
| CALNUC, DKFZp686A15286, FLJ40471, NUC, Nucleobindin-1 | ||||
| Signal sequence receptor subunit 4 | SSR4 | 0.362 | 0.378 | 0.437 |
| Signal sequence receptor, delta (translocon-associated protein delta), Signal sequence receptor subunit delta, SSR-delta, Translocon-associated protein subunit delta, TRAPD, TRAP-delta | ||||
| histone cluster 1 H2A family member j | HIST1H2AJ | 0.466 | 0.365 | 0.310 |
| Histone cluster 1, H2aj, dJ160A22.4, H2A/E, H2AFE, Histone H2A/e, Histone H2A type 1-J | ||||
| Lysosome associated membrane protein 1 | LAMP1 | 0.352 | 0.364 | 0.400 |
| CD107a, CD107 antigen-like family member A, LAMP-1, LAMPA, LGP120, Lysosome-associated membrane glycoprotein 1, Lysosome-associated membrane protein 1 | ||||
| Phosphoglycerate kinase 1 | PGK1 | 0.260 | 0.355 | 0.273 |
| Cell migration-inducing gene 10 protein, MGC117307, MGC142128, MGC8947, MIG10, OK/SW-cl.110, PGKA, Phosphoglycerate kinase 1, Primer recognition protein 2, PRP 2 | ||||
| Major histocompatibility complex, class I, A | HLA-A | 0.305 | 0.353 | 0.305 |
| HLA class I histocompatibility antigen, A-24 alpha chain (Aw-24) (HLA class I histocompatibility antigen, A-9 alpha chain) (MHC class I antigen A*24) | ||||
| Heat shock protein family E (Hsp10) member 1 | HSPE1 | 0.281 | 0.351 | 0.346 |
| 10 kDa chaperonin, 10 kDa heat shock protein, mitochondrial, Chaperonin 10, CPN10, Early-pregnancy factor, EPF, GROES, Hsp10, HSP10 | ||||
| Pyrroline-5-carboxylate reductase 1 | PYCR1 | 0.307 | 0.350 | 0.347 |
| ARCL2B, P5C, P5CR, P5CR 1, P5C reductase 1, PIG45, PP222, PRO3, PYCR, Pyrroline-5-carboxylate reductase 1, mitochondrial | ||||
| Proteasome 26S subunit, non-ATPase 14 | PSMD14 | 0.308 | 0.348 | 0.323 |
| Proteasome (prosome, macropain) 26S subunit, non-ATPase, 14, 26S proteasome-associated PAD1 homolog 1, 26S proteasome non-ATPase regulatory subunit 14, 26S proteasome regulatory subunit RPN11, pad1, PAD1, POH1, Rpn11, RPN11 | ||||
| Non-POU domain containing octamer binding | NONO | 0.287 | 0.347 | 0.386 |
| 54 kDa nuclear RNA-and DNA-binding protein, 55 kDa nuclear protein, DNA-binding p52/p100 complex, 52 kDa subunit, NMT55, NonO protein, Non-POU domain-containing octamer-binding protein, NRB54, P54, p54(nrb), p54nrb, P54NRB | ||||
| High mobility group AT-hook 1 | HMGA1 | 0.192 | 0.345 | 0.203 |
| High mobility group AT-hook protein 1, High mobility group protein A1, High mobility group protein HMG-I/HMG-Y, High mobility group protein R, HMGA1A, HMG-I(Y), HMGIY, HMG-R, MGC12816, MGC4242, MGC4854 | ||||
| Transmembrane p24 trafficking protein 10 | TMED10 | 0.410 | 0.342 | 0.329 |
| 21 kDa transmembrane-trafficking protein, p23, P24(DELTA), p24delta, S31I125, S31III125, TMP21, Tmp-21-I, Transmembrane emp24 domain-containing protein 10, Transmembrane protein Tmp21 | ||||
| Fibroblast activation protein alpha | FAP | 0.384 | 0.336 | 0.307 |
| 170 kDa melanoma membrane-bound gelatinase, DKFZp686G13158, DPPIV, FAPA, Fibroblast activation protein alpha, Integral membrane serine protease, Seprase | ||||
| Prolyl 4-hydroxylase subunit alpha 1 | P4HA1 | 0.329 | 0.334 | 0.352 |
| 4-PH alpha-1, C-P4Halpha(I), P4HA, Procollagen-proline,2-oxoglutarate-4-dioxygenase subunit alpha-1, Prolyl 4-hydroxylase subunit alpha-1, prolyl 4-hydroxylase, alpha polypeptide I | ||||
| Complement C1q binding protein | C1QBP | 0.201 | 0.331 | 0.337 |
| Complement component 1, q subcomponent binding protein, C1qBP, Complement component 1 Q subcomponent-binding protein, mitochondrial, GC1QBP, gC1qR, gC1Q-R, GC1q-R protein, Glycoprotein gC1qBP, HABP1, Hyaluronan-binding protein 1, Mitochondrial matrix protein p32, p32, p33, SF2p32, SF2P32 | ||||
| ER lipid raft associated 2 | ERLIN2 | 0.339 | 0.330 | 0.403 |
| C8orf2, Endoplasmic reticulum lipid raft-associated protein 2, Erlin-2, MGC87072, NET32, SPFH2, SPFH domain-containing protein 2, Stomatin-prohibitin-flotillin-HflC/K domain-containing protein 2, UNQ2441/PRO5003/PRO9924 | ||||
| Signal sequence receptor subunit 1 | SSR1 | 0.416 | 0.327 | 0.369 |
| Signal sequence receptor, alpha, DKFZp781N23103, FLJ14232, FLJ22100, FLJ23034, FLJ78242, FLJ93042, PSEC0262, Signal sequence receptor subunit alpha, SSR-alpha, Translocon-associated protein subunit alpha, TRAPA, TRAP-alpha | ||||
| Tropomodulin 3 | TMOD3 | 0.136 | 0.320 | 0.281 |
| Tropomodulin-3, Ubiquitous tropomodulin, UTMOD, U-Tmod | ||||
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | PLOD2 | 0.372 | 0.320 | 0.375 |
| LH2, Lysyl hydroxylase 2, Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2, TLH | ||||
| Major vault protein | MVP | 0.273 | 0.313 | 0.361 |
| LRP, Lung resistance-related protein, Major vault protein, VAULT1 | ||||
| Thrombospondin 1 | THBS1 | 0.342 | 0.305 | 0.302 |
| THBS, THBS-1, Thrombospondin-1, TSP, TSP1, TSP-1 | ||||
| Heat shock protein family D (Hsp60) member 1 | HSPD1 | 0.281 | 0.301 | 0.334 |
| 60 kDa chaperonin, 60 kDa heat shock protein, mitochondrial, Chaperonin 60, CPN60, GROEL, Heat shock protein 60, HLD4, Hsp60, HSP60, HSP-60, HSP65, HuCHA60, Mitochondrial matrix protein P1, P60 lymphocyte protein, SPG13 | ||||
| IKBKB interacting protein | IKBIP | 0.363 | 0.299 | 0.321 |
| Inhibitor of nuclear factor kappa-B kinase-interacting protein (I kappa-B kinase-interacting protein) (IKBKB-interacting protein) (IKK-interacting protein) | ||||
| ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | ATP5A1 | 0.290 | 0.290 | 0.312 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1, cardiac muscle, ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 2, non-cardiac muscle-like 2, ATP5AL2, ATPM | ||||
| ATP synthase, H+ transporting, mitochondrial Fo complex subunit B1 | ATP5F1 | 0.422 | 0.287 | 0.337 |
| ATP synthase F(0) complex subunit B1, mitochondrial (ATP synthase proton-transporting mitochondrial F(0) complex subunit B1) (ATP synthase subunit b) (ATPase subunit b) | ||||
| Atlastin GTPase 3 | ATL3 | 0.260 | 0.272 | 0.321 |
| Atlastin 3, DKFZP564J0863 | ||||
| Basigin (Ok blood group) | BSG | 0.286 | 0.266 | 0.302 |
| 5F7, Basigin, CD147, Collagenase stimulatory factor, EMMPRIN, Extracellular matrix metalloproteinase inducer, Leukocyte activation antigen M6, M6, OK, OK blood group antigen, TCSF, Tumor cell-derived collagenase stimulatory factor, UNQ6505/PRO21383 | ||||
| Adipocyte plasma membrane-associated protein | APMAP | 0.293 | 0.264 | 0.333 |
| Chromosome 20 open reading frame 3, C20orf3, BSCv, Protein BSCv, UNQ1869/PRO4305 | ||||
| H2A histone family member Y | H2AFY | 0.363 | 0.263 | 0.307 |
| Core histone macro-H2A.1, H2A/y, H2A.y, H2AF12M, H2AFJ, Histone H2A.y, Histone macroH2A1, MACROH2A1, MACROH2A1.1, macroH2A1.2, Medulloblastoma antigen MU-MB-50.205, mH2A1 | ||||
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | PLOD1 | 0.304 | 0.260 | 0.324 |
| FLJ42041, LH, LH1, LLH, Lysyl hydroxylase 1, PLOD, Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | ||||
| Cytochrome c oxidase subunit 4I1 | COX4I1 | 0.309 | 0.246 | 0.339 |
| COX4, COX4-1, COXIV, COX IV-1, Cytochrome c oxidase polypeptide IV, Cytochrome c oxidase subunit 4 isoform 1, mitochondrial, Cytochrome c oxidase subunit IV isoform 1, FLJ23483, MGC72016 | ||||
| Aspartyl-tRNA synthetase | DARS | 0.261 | 0.205 | 0.329 |
| Aspartate--tRNA ligase, Aspartyl-tRNA synthetase, cytoplasmic, AspRS, Cell proliferation-inducing gene 40 protein, DKFZp781B11202, MGC111579, PIG40 | ||||
| Integrin subunit alpha 5 | ITGA5 | 0.316 | 0.183 | 0.268 |
| CD49 antigen-like family member E, CD49e, Fibronectin receptor subunit alpha, FNRA, Integrin alpha-5, Integrin alpha-F, VLA-5, VLA5A | ||||
| Eukaryotic translation initiation factor 2 subunit beta | EIF2S2 | < −1 | −0.119 | < −1 |
| DKFZp686L18198, EIF2, EIF2B, EIF2beta, eIF-2-beta, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit beta, MGC8508 | ||||
| Bleomycin hydrolase | BLMH | −0.338 | −0.155 | < −1 |
| BH, Bleomycin hydrolase, BLM hydrolase, BMH | ||||
| FK506 binding protein 1A | FKBP1A | < −1 | −0.162 | −0.324 |
| 12 kDa FK506-binding protein, 12 kDa FKBP, FK506-binding protein 1A, FKBP1, FKBP12, FKBP-12, FKBP12C, FKBP-1A, Immunophilin FKBP12, Peptidyl-prolyl cis-trans isomerase FKBP1A, PKC12, PKCI2, PPIASE, PPIase FKBP1A, Rotamase | ||||
| SERPINE1 mRNA binding protein 1 | SERBP1 | −0.264 | −0.170 | −0.324 |
| CGI-55, CHD3IP, DKFZp564M2423, DKFZP564M2423, FLJ90489, HABP4L, PAI1 RNA-binding protein 1, PAIRBP1, PAI-RBP1, Plasminogen activator inhibitor 1 RNA-binding protein, SERPINE1 mRNA-binding protein 1 | ||||
| Y-box binding protein 3 | YBX3 | −0.264 | −0.174 | −0.385 |
| Cold-shock domain containing A1, CSDA1, dbpA, ZONAB | ||||
| A-kinase anchoring protein 12 | AKAP12 | −0.386 | −0.186 | −0.426 |
| A kinase (PRKA) anchor protein 12, AKAP-12, AKAP250, AKAP 250, A-kinase anchor protein 12, A-kinase anchor protein 250 kDa, DKFZp686M0430, DKFZp686O0331, FLJ20945, FLJ97621, Gravin, Myasthenia gravis autoantigen | ||||
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon | YWHAE | −0.346 | −0.191 | −0.321 |
| 14-3-3E, 14-3-3 protein epsilon, FLJ45465, FLJ53559, KCIP-1, MDCR, MDS | ||||
| Stress induced phosphoprotein 1 | STIP1 | −0.247 | −0.196 | −0.307 |
| Hop, HOP, Hsc70/Hsp90-organizing protein, IEF-SSP-3521, P60, Renal carcinoma antigen NY-REN-11, STI1, STI1L, Stress-induced-phosphoprotein 1, Transformation-sensitive protein IEF SSP 3521 | ||||
| Zyxin | ZYX | −0.285 | −0.206 | −0.339 |
| ESP-2, HED-2, Zyxin, Zyxin-2 | ||||
| Diazepam binding inhibitor, acyl-CoA binding protein | DBI | −0.483 | −0.212 | −0.445 |
| Diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein), ACBD1, ACBP, Acyl-CoA-binding protein, CCK-RP, Diazepam-binding inhibitor, Endozepine, EP, MGC70414 | ||||
| Myosin light chain 9 | MYL9 | −0.304 | −0.216 | −0.333 |
| 20 kDa myosin light chain, LC20, MGC3505, MLC2, MLC-2C, MRLC1, Myosin regulatory light chain 2, smooth muscle isoform, Myosin regulatory light chain 9, Myosin regulatory light chain MRLC1, Myosin regulatory light polypeptide 9, Myosin RLC, MYRL2 | ||||
| Keratin 7 | KRT7 | −0.305 | −0.217 | −0.307 |
| CK7, CK-7, Cytokeratin-7, K2C7, K7, Keratin, type II cytoskeletal 7, Keratin-7, MGC129731, MGC3625, Sarcolectin, SCL, Type-II keratin Kb7 | ||||
| Galectin 3 | LGALS3 | < −1 | −0.218 | < −1 |
| Lectin, galactoside-binding, soluble, 3, 35 kDa lectin, Carbohydrate-binding protein 35, CBP35, CBP 35, GAL3, Gal-3, Galactose-specific lectin 3, Galactoside-binding protein, GALBP, Galectin-3, GALIG, IgE-binding protein, L31, L-31, Laminin-binding protein, Lectin L-29, LGALS2, MAC2, MAC-2, Mac-2 antigen | ||||
| Calmodulin 1 | CALM1 | −0.321 | −0.223 | −0.342 |
| Calmodulin 1 (phosphorylase kinase, delta), CALM2, CALM3, CALML2, caM, CAMI, DD132, PHKD | ||||
| Myotrophin | MTPN | < −1 | −0.226 | < −1 |
| FLJ31098, FLJ99857, GCDP, Myotrophin, MYOTROPHIN, Protein V-1, V-1 | ||||
| Tropomyosin 4 | TPM4 | −0.329 | −0.232 | −0.312 |
| TM30p1, Tropomyosin-4, Tropomyosin alpha-4 chain | ||||
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | YWHAZ | −0.326 | −0.235 | −0.286 |
| 14-3-3 protein zeta/delta, 14-3-3-zeta, KCIP-1, MGC111427, MGC126532, MGC138156, Protein kinase C inhibitor protein 1, YWHAD | ||||
| Malate dehydrogenase1 | MDH1 | −0.258 | −0.244 | −0.320 |
| Malate dehydrogenase 1, NAD (soluble), Cytosolic malate dehydrogenase, Malate dehydrogenase, cytoplasmic, MDHA, MDH-s, MGC:1375, MOR2 | ||||
| Microtubule associated protein 4 | MAP4 | −0.290 | −0.247 | −0.418 |
| DKFZp779A1753, MAP-4, MGC8617, Microtubule-associated protein 4 | ||||
| Parkinsonism associated deglycase | PARK7 | < −1 | −0.260 | −0.339 |
| Parkinson disease (autosomal recessive, early onset) 7, DJ1, DJ-1, FLJ27376, FLJ34360, FLJ92274, Oncogene DJ1, Parkinson disease protein 7, Protein DJ-1 | ||||
| Cysteine and glycine rich protein 1 | CSRP1 | −0.297 | −0.263 | −0.319 |
| CRP, CRP1, CSRP, CYRP, Cysteine and glycine-rich protein 1, Cysteine-rich protein 1, D1S181E, DKFZp686M148 | ||||
| Actinin alpha 4 | ACTN4 | −0.344 | −0.280 | −0.321 |
| ACTININ-4, Alpha-actinin-4, DKFZp686K23158, F-actin cross-linking protein, FSGS, FSGS1, Non-muscle alpha-actinin 4 | ||||
| Pyrophosphatase (inorganic) 1 | PPA1 | −0.304 | −0.290 | −0.388 |
| Inorganic pyrophosphatase, IOPPP, MGC111556, PP, PP1, Ppase, PPase, Pyrophosphate phospho-hydrolase, SID6-8061 | ||||
| Alpha fetoprotein | AFP | −0.457 | −0.293 | −0.395 |
| Alpha-1-fetoprotein, Alpha-fetoglobulin, Alpha-fetoprotein, FETA, HPAFP | ||||
| AHNAK nucleoprotein | AHNAK | −0.426 | −0.299 | −0.411 |
| AHNAKRS, Desmoyokin, MGC5395, Neuroblast differentiation-associated protein AHNAK, PM227 | ||||
| Plastin 3 | PLS3 | −0.319 | −0.300 | −0.315 |
| Plastin-3, T-plastin | ||||
| Glycine-tRNA synthetase | GARS | −0.261 | −0.301 | −0.229 |
| AP-4-A synthetase, CMT2D, Diadenosine tetraphosphate synthetase, DSMAV, Glycine--tRNA ligase, Glycyl-tRNA synthetase, GlyRS, HMN5, SMAD1 | ||||
| Filamin C | FLNC | −0.269 | −0.301 | −0.298 |
| ABP-280, ABP280A, ABP-280-like protein, ABPA, ABPL, ABP-L, Actin-binding-like protein, Filamin-2, Filamin-C, FLJ10186, FLN2, FLNc, FLN-C, Gamma-filamin, filamin C, gamma | ||||
| Ribonuclease/angiogenin inhibitor 1 | RNH1 | −0.142 | −0.304 | −0.204 |
| MGC18200, MGC4569, MGC54054, Placental ribonuclease inhibitor, Placental RNase inhibitor, PRI, RAI, Ribonuclease/angiogenin inhibitor 1, Ribonuclease inhibitor, RNH | ||||
| Calpain 2 | CAPN2 | −0.183 | −0.304 | −0.183 |
| Calcium-activated neutral proteinase 2, Calpain-2 catalytic subunit, Calpain-2 large subunit, Calpain large polypeptide L2, Calpain M-type, CANP2, CANP 2, CANPL2, CANPml, FLJ39928, M-calpain, mCANP, Millimolar-calpain | ||||
| PDZ and LIM domain 3 | PDLIM3 | −0.497 | −0.305 | −0.712 |
| Actinin-associated LIM protein, ALP, Alpha-actinin-2-associated LIM protein, DKFZp686L0362, FLJ26096, PDZ and LIM domain protein 3 | ||||
| Ribosomal protein L10a | RPL10A | −0.224 | −0.307 | −0.247 |
| 60S ribosomal protein L10a, Csa-19, CSA-19, L10A, NEDD6, NEDD-6, Neural precursor cell expressed developmentally down-regulated protein 6 | ||||
| Solute carrier family 3 member 2 | SLC3A2 | −0.119 | −0.312 | −0.191 |
| Solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2, 4F2, 4F2 cell-surface antigen heavy chain, 4F2hc, 4F2HC, 4F2 heavy chain antigen, 4T2HC, CD98, CD98HC, Lymphocyte activation antigen 4F2 large subunit, MDU1, NACAE | ||||
| Caldesmon 1 | CALD1 | −0.375 | −0.313 | −0.488 |
| CAD, Caldesmon, CDM, HCAD, H-CAD, LCAD, L-CAD, MGC21352, NAG22 | ||||
| GDP dissociation inhibitor 2 | GDI2 | −0.486 | −0.315 | −0.396 |
| FLJ16452, FLJ37352, GDI-2, Guanosine diphosphate dissociation inhibitor 2, RABGDIB, Rab GDI beta, Rab GDP dissociation inhibitor beta | ||||
| Lactate dehydrogenase B | LDHB | −0.270 | −0.316 | −0.308 |
| LDH-B, LDH-H, LDH heart subunit, l-lactate dehydrogenase B chain, Renal carcinoma antigen NY-REN-46, TRG-5 | ||||
| Filamin A | FLNA | −0.300 | −0.318 | −0.333 |
| ABP-280, ABPX, Actin-binding protein 280, Alpha-filamin, CVD1, DKFZp434P031, Endothelial actin-binding protein, Filamin-1, Filamin-A, FLN, FLN1, FLN-A, FMD, MNS, NHBP, Non-muscle filamin, OPD, OPD1, OPD2, XLVD, XMVD | ||||
| Branched chain amino acid transaminase 1 | BCAT1 | −0.232 | −0.325 | −0.350 |
| BCAT(c), BCATC, BCT1, Branched-chain-amino-acid aminotransferase, cytosolic, DKFZp686E12175, ECA39, MECA39, PNAS121, PP18, Protein ECA39 | ||||
| Alanine-tRNA synthetase | AARS | −0.173 | −0.326 | −0.255 |
| Alanine--tRNA ligase, Alanyl-tRNA synthetase, cytoplasmic, AlaRS, Renal carcinoma antigen NY-REN-42 | ||||
| Phosphoglucomutase 1 | PGM1 | −0.282 | −0.329 | −0.299 |
| Glucose phosphomutase 1, PGM 1, Phosphoglucomutase-1 | ||||
| Citrate synthase | CS | −0.099 | −0.334 | −0.181 |
| Citrate synthase, mitochondrial | ||||
| Reticulon 4 | RTN4 | −0.120 | −0.335 | −0.335 |
| ASY, Foocen, KIAA0886, My043, Nbla00271, Nbla10545, Neurite outgrowth inhibitor, Neuroendocrine-specific protein, Neuroendocrine-specific protein C homolog, NI220/250, NOGO, NOGO-A, Nogo-B, NOGOC, Nogo-C, Nogo protein, NSP, NSP-CL, Reticulon-4, Reticulon-5, RTN4-A, RTN4-B1, RTN4-B2, RTN4-C, RTN-x, RTN-X, SP1507 | ||||
| Eukaryotic translation initiation factor 3 subunit D | EIF3D | −0.221 | −0.337 | −0.218 |
| eIF3d, eIF3 p66, eIF3-p66, EIF3S7, eIF3-zeta, eIF-3-zeta, Eukaryotic translation initiation factor 3 subunit 7, Eukaryotic translation initiation factor 3 subunit D, MGC126526, MGC17258 | ||||
| Importin 5 | IPO5 | −0.199 | −0.337 | −0.332 |
| DKFZp686O1576, FLJ43041, IMB3, Imp5, Importin-5, Importin subunit beta-3, Karyopherin beta-3, KPNB3, MGC2068, Ran-binding protein 5, RanBP5, RANBP5 | ||||
| Fermitin family member 2 | FERMT2 | −0.278 | −0.338 | −0.253 |
| DKFZp686G11125, Fermitin family homolog 2, FLJ34213, FLJ44462, KIND2, Kindlin-2, MIG2, mig-2, MIG-2, Mitogen-inducible gene 2 protein, PH domain-containing family C member 1, Pleckstrin homology domain-containing family C member 1, PLEKHC1, UNC112, UNC112B | ||||
| Peroxiredoxin 6 | PRDX6 | −0.359 | −0.343 | −0.346 |
| 1-Cys, 1-Cys peroxiredoxin, 1-Cys PRX, 24 kDa protein, Acidic calcium-independent phospholipase A2, aiPLA2, Antioxidant protein 2, AOP2, KIAA0106, Liver 2D page spot 40, MGC46173, Non-selenium glutathione peroxidase, NSGPx, p29, Peroxiredoxin-6, PRX, Red blood cells page spot 12 | ||||
| Asparagine-tRNA synthetase | NARS | −0.353 | −0.343 | −0.309 |
| AsnRS, ASNRS, Asparagine--tRNA ligase, Asparaginyl-tRNA synthetase, cytoplasmic, NARS1 | ||||
| Thioredoxin reductase 1 | TXNRD1 | −0.329 | −0.355 | −0.288 |
| Gene associated with retinoic and IFN-induced mortality 12 protein, Gene associated with retinoic and interferon-induced mortality 12 protein, GRIM12, GRIM-12, KDRF, KM-102-derived reductase-like factor, MGC9145, Thioredoxin reductase 1, cytoplasmic, Thioredoxin reductase TR1, TR, TR1, Trxr1, TRXR1, TXNR | ||||
| Ezrin | EZR | −0.300 | −0.356 | −0.344 |
| CVIL, CVL, Cytovillin, DKFZp762H157, Ezrin, FLJ26216, MGC1584, p81, VIL2, Villin-2 | ||||
| Thioredoxin | TXN | −0.413 | −0.364 | −0.407 |
| ADF, ATL-derived factor, DKFZp686B1993, MGC61975, SASP, Surface-associated sulphydryl protein, Thioredoxin, TRDX, Trx, TRX, TRX1 | ||||
| Tu translation elongation factor, mitochondrial | TUFM | −0.181 | −0.364 | −0.357 |
| COXPD4, EFTu, EFTU, EF-Tu, EF-TuMT, Elongation factor Tu, mitochondrial, P43 | ||||
| Cell division cycle 37 | CDC37 | < −1 | −0.365 | < −1 |
| CDC37A, Hsp90 chaperone protein kinase-targeting subunit, Hsp90 co-chaperone Cdc37, p50Cdc37, P50CDC37 | ||||
| Keratin 8 | KRT8 | −0.434 | −0.365 | −0.444 |
| CARD2, CK8, CK-8, CYK8, Cytokeratin-8, K2C8, K8, Keratin, type II cytoskeletal 8, Keratin-8, KO, Type-II keratin Kb8 | ||||
| Keratin 18 | KRT18 | −0.437 | −0.367 | −0.399 |
| Cell proliferation-inducing gene 46 protein, CK-18, CYK18, Cytokeratin-18, K18, Keratin, type I cytoskeletal 18, Keratin-18, PIG46 | ||||
| Eukaryotic translation initiation factor 3 subunit C | EIF3C | −0.314 | −0.374 | −0.280 |
| eIF3c, EIF3CL, eIF3-p110, EIF3S8, FLJ53378, FLJ54400, FLJ54404, FLJ55450, FLJ55750, FLJ78287, MGC189737, MGC189744 | ||||
| Spermidine synthase | SRM | −0.236 | −0.375 | −0.356 |
| PAPT, Putrescine aminopropyltransferase, SPDSY, Spermidine synthase, SPS1, SRML1 | ||||
| Pentraxin 3 | PTX3 | −0.284 | −0.379 | −0.224 |
| Pentaxin-related protein PTX3, Pentraxin-related protein PTX3, TNFAIP5, TNF alpha-induced protein 5, TSG14, TSG-14, Tumor necrosis factor alpha-induced protein 5, Tumor necrosis factor-inducible gene 14 protein | ||||
| Collagen type I alpha 1 chain | COL1A1 | −0.468 | −0.387 | −0.441 |
| Alpha-1 type I collagen, Collagen alpha-1(I) chain, OI4, collagen, type I, alpha 1 | ||||
| Pro-apoptotic WT1 regulator | PAWR | < −1 | −0.400 | < −1 |
| PAR4, par-4, Par-4, PRKC apoptosis WT1 regulator protein, Prostate apoptosis response 4 protein | ||||
| LIM domain and actin binding 1 | LIMA1 | −0.266 | −0.400 | −0.610 |
| Epithelial protein lost in neoplasm, EPLIN, FLJ38853, LIM domain and actin-binding protein 1, MGC131726, PP624, SREBP3 | ||||
| Transgelin | TAGLN | −0.561 | −0.408 | −0.553 |
| 22 kDa actin-binding protein, DKFZp686B01212, DKFZp686P11128, Protein WS3-10, SM22, SM22-alpha, SMCC, Smooth muscle protein 22-alpha, TAGLN1, Transgelin, WS3-10 | ||||
| Programmed cell death 5 | PDCD5 | −0.479 | −0.415 | −0.564 |
| FLJ42784, MGC9294, Programmed cell death protein 5, Protein TFAR19, TF-1 cell apoptosis-related protein 19, TFAR19 | ||||
| EH domain containing 2 | EHD2 | −0.283 | −0.422 | < −1 |
| EH-domain-containing protein 2, FLJ96617, PAST2, PAST homolog 2 | ||||
| Histidyl-tRNA synthetase | HARS | < −1 | −0.444 | < −1 |
| FLJ20491, HisRS, Histidine--tRNA ligase, Histidyl-tRNA synthetase, cytoplasmic, HRS | ||||
| Phosphatidylethanolamine binding protein 1 | PEBP1 | −0.574 | −0.457 | −0.595 |
| HCNP, HCNPpp, Neuropolypeptide h3, PBP, PEBP, PEBP-1, Phosphatidylethanolamine-binding protein 1, Prostatic-binding protein, Raf kinase inhibitor protein, RKIP | ||||
| LIM and SH3 protein 1 | LASP1 | −0.435 | −0.460 | −0.606 |
| Lasp-1, LASP-1, LIM and SH3 domain protein 1, Metastatic lymph node gene 50 protein, MLN50, MLN 50 | ||||
| PDZ and LIM domain 5 | PDLIM5 | −0.481 | −0.460 | −0.520 |
| Enh, ENH, ENH1, Enigma homolog, Enigma-like PDZ and LIM domains protein, L9, LIM, PDZ and LIM domain protein 5 | ||||
| Catenin alpha 1 | CTNNA1 | −0.325 | −0.470 | −0.380 |
| Catenin (cadherin-associated protein), alpha 1, 102 kDa, Alpha E-catenin, Cadherin-associated protein, CAP102, Catenin alpha-1, FLJ36832, FLJ52416, Renal carcinoma antigen NY-REN-13 | ||||
| Tumor protein D54 like 2 | TPD52L2 | −0.469 | −0.481 | −0.542 |
| D54, DKFZp686A1765, hD54, Tumor protein D52-like 2, Tumor protein D54 | ||||
| Heat shock protein family B (small) member 6 | HSPB6 | −0.356 | −0.486 | −0.415 |
| Heat shock protein, alpha-crystallin-related, B6, FLJ32389, Heat shock 20 kDa-like protein p20, Heat shock protein beta-6, Hsp20, HspB6 | ||||
| Eukaryotic translation initiation factor 4 gamma 1 | EIF4G1 | −0.411 | −0.487 | < −1 |
| DKFZp686A1451, EIF4F, EIF4G, eIF-4G1, EIF-4G1, eIF-4G 1, eIF-4-gamma 1, EIF4GI, Eukaryotic translation initiation factor 4 gamma 1, p220, P220 | ||||
| LIM domain 7 | LMO7 | < −1 | −0.494 | < −1 |
| F-box only protein 20, FBX20, FBXO20, KIAA0858, LIM domain only protein 7, LMO-7, LOMP | ||||
| Ribosome binding protein 1 | RRBP1 | −0.532 | −0.498 | −0.646 |
| Ribosome binding protein 1 homolog 180 kDa (dog), 180 kDa ribosome receptor homolog, DKFZp586A1420, ES/130, ES/130-related protein, ES130, FLJ36146, hES, KIAA1398, MGC157720, MGC157721, Ribosome-binding protein 1, Ribosome receptor protein, RRp | ||||
| Thioredoxin domain containing 17 | TXNDC17 | < −1 | −0.521 | < −1 |
| 14 kDa thioredoxin-related protein, MGC14353, Protein 42-9-9, Thioredoxin domain-containing protein 17, Thioredoxin-like protein 5, TRP14, TXNL5 | ||||
| Tubulin folding cofactor A | TBCA | < −1 | −0.547 | < −1 |
| CFA, TCP1-chaperonin cofactor A, Tubulin-folding cofactor A, Tubulin-specific chaperone A | ||||
| Phosphoserine aminotransferase 1 | PSAT1 | −0.625 | −0.641 | −0.430 |
| EPIP, MGC1460, Phosphohydroxythreonine aminotransferase, Phosphoserine aminotransferase, PSA, PSAT | ||||
| Asparagine synthetase (glutamine-hydrolyzing) | ASNS | −0.622 | −0.707 | < −1 |
| Cell cycle control protein TS11, Glutamine-dependent asparagine synthetase, TS11 | ||||
| Glutaminase | GLS | −0.434 | −0.722 | −0.587 |
| AAD20, DKFZp686O15119, FLJ10358, GLS1, Glutaminase kidney isoform, mitochondrial, K-glutaminase, KIAA0838, l-glutamine amidohydrolase | ||||
| Calponin 1 | CNN1 | −1.106 | −0.982 | −1.022 |
| Calponin 1, basic, smooth muscle, Basic calponin, Calponin-1, Calponin H1, smooth muscle, Sm-Calp, SMCC | ||||
| Apolipoprotein A2 | APOA2 | −1.336 | −1.176 | −1.202 |
| ApoAII, ApoA-II, Apo-AII, Apolipoprotein A2, Apolipoprotein A-II | ||||
| Keratin 19 | KRT19 | −1.214 | −1.246 | < −1 |
| CK19, CK-19, Cytokeratin-19, K19, K1CS, Keratin, type I cytoskeletal 19, Keratin-19, MGC15366 | ||||
| Hemoglobin subunit alpha 1 | HBA1 | −1.225 | −1.325 | −1.284 |
| CD31, MGC126895, MGC126897 | ||||
| Transmembrane p24 trafficking protein 7 | TMED7 | 0.451 | < −1 | 0.533 |
| CGI-109, FLJ57776, FLJ90481, Transmembrane emp24 domain-containing protein 7 | ||||
| GDP dissociation inhibitor 1 | GDI1 | < −1 | < −1 | < −1 |
| FLJ41411, GDI-1, GDIL, Guanosine diphosphate dissociation inhibitor 1, MRX41, MRX48, Oligophrenin-2, OPHN2, Protein XAP-4, RABGD1A, RABGDIA, Rab GDI alpha, Rab GDP dissociation inhibitor alpha, XAP4, XAP-4 | ||||
| Aldehyde dehydrogenase 18 family member A1 | ALDH18A1 | −0.071 | < −1 | −0.350 |
| Aldehyde dehydrogenase family 18 member A1, Delta-1-pyrroline-5-carboxylate synthase, GSAS, MGC117316, P5CS, PYCS | ||||
| Cystatin B | CSTB | < −1 | < −1 | < −1 |
| CPI-B, CST6, Cystatin-B, EPM1, Liver thiol proteinase inhibitor, PME, Stefin-B, STFB | ||||
| Aldehyde dehydrogenase 1 family member L2 | ALDH1L2 | < −1 | < −1 | < −1 |
| Aldehyde dehydrogenase family 1 member L2, DKFZp686A16126, DKFZp686M064, DKFZp686P14145, FLJ36769, FLJ38508, MGC119536, MGC119537, mtFDH | ||||
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | YWHAB | −0.441 | < −1 | −0.346 |
| 14-3-3 protein beta/alpha, GW128, HS1, KCIP-1, Protein 1054, Protein kinase C inhibitor protein 1, YWHAA | ||||
| Lactotransferrin | LTF | < −1 | < −1 | < −1 |
| GIG12, HLF2, Lactoferrin, Lactotransferrin, LF, Talalactoferrin | ||||
| Calpain 1 | CAPN1 | −0.495 | < −1 | −0.537 |
| Calcium-activated neutral proteinase 1, Calpain-1 catalytic subunit, Calpain-1 large subunit, Calpain mu-type, CANP, CANP1, CANP 1, CANPL1, Cell proliferation-inducing gene 30 protein, Micromolar-calpain, muCANP, muCL, PIG30 | ||||
| Latexin | LXN | < −1 | < −1 | −0.706 |
| ECI, Endogenous carboxypeptidase inhibitor, Latexin, Protein MUM, TCI, Tissue carboxypeptidase inhibitor | ||||
| Four and a half LIM domains 1 | FHL1 | −0.432 | < −1 | −0.519 |
| bA535K18.1, FHL-1, FHL1A, FHL1B, FLH1A, Four and a half LIM domains protein 1, KYOT, KYO-T, MGC111107, Skeletal muscle LIM-protein 1, SLIM, SLIM1, SLIM-1, SLIMMER, XMPMA | ||||
| Nexilin F-actin binding protein | NEXN | < −1 | < −1 | < −1 |
| F-actin-binding protein, MGC104234, MGC138865, MGC138866, Nelin, NELIN, nexilin, Nexilin | ||||
| Orosomucoid 1 | ORM1 | < −1 | < −1 | < −1 |
| AGP1, AGP 1, AGP-A, Alpha-1-acid glycoprotein 1, OMD 1, ORM, Orosomucoid-1 | ||||
| Serine and arginine rich splicing factor 6 | SRSF6 | < −1 | < −1 | < −1 |
| Serine/arginine-rich splicing factor 6, SFRS6, splicing factor, arginine/serine-rich 6, B52, pre-mRNA splicing factor SRP55, SR splicing factor 6, SRP55 | ||||
| Kinectin 1 | KTN1 | < −1 | < −1 | < −1 |
| CG1, CG-1 antigen, KIAA0004, Kinectin, Kinesin receptor, KNT, MGC133337, MU-RMS-40.19 | ||||
| 6-phosphogluconolactonase | PGLS | < −1 | < −1 | < −1 |
| 6PGL | ||||
| ADP ribosylation factor like GTPase 1 | ARL1 | < −1 | < −1 | < −1 |
| ADP-ribosylation factor-like 1, ADP-ribosylation factor-like protein 1, ARFL1 | ||||
| Protein phosphatase 1 regulatory subunit 12A | PPP1R12A | < −1 | < −1 | < −1 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 12A, MBS, MGC133042, Myosin phosphatase-targeting subunit 1, Myosin phosphatase target subunit 1, MYPT1, Protein phosphatase 1 regulatory subunit 12A, Protein phosphatase myosin-binding subunit | ||||
| Synaptopodin 2 | SYNPO2 | < −1 | < −1 | < −1 |
| Genethonin-2, Myopodin, MYOPODIN, Synaptopodin-2 | ||||
| Glutaredoxin 3 | GLRX3 | < −1 | < −1 | < −1 |
| bA500G10.4, FLJ11864, GLRX4, Glutaredoxin-3, GRX3, GRX4, HUSSY-22, PICOT, PKC-interacting cousin of thioredoxin, PKCq-interacting protein, PKC-theta-interacting protein, Thioredoxin-like protein 2, TXNL2, TXNL3 | ||||
| NSFL1 cofactor | NSFL1C | < −1 | < −1 | < −1 |
| NSFL1 (p97) cofactor (p47), dJ776F14.1, FLJ46889, MGC3347, NSFL1 cofactor p47, p47, P47, p97 cofactor p47, UBX1, UBXD10, UBX domain-containing protein 2C, UBXN2C | ||||
| Cold inducible RNA binding protein | CIRBP | < −1 | < −1 | < −1 |
| A18HNRNP, A18 hnRNP, CIRP, Cold-inducible RNA-binding protein, Glycine-rich RNA-binding protein CIRP | ||||
| Heterogeneous nuclear ribonucleoprotein D like | HNRNPDL | < −1 | < −1 | < −1 |
| Heterogeneous nuclear ribonucleoprotein D-like (hnRNP D-like) (hnRNP DL) (AU-rich element RNA-binding factor) (JKT41-binding protein) (Protein laAUF1), HNRPDL, JKTBP | ||||
| Phosphoenolpyruvate carboxykinase 2, mitochondrial | PCK2 | < −1 | < −1 | < −1 |
| PEPCK, PEPCK2, PEPCK-M, Phosphoenolpyruvate carboxylase | ||||
| Desmin | DES | < −1 | < −1 | < −1 |
| CMD1I, CSM1, CSM2, FLJ12025, FLJ39719, FLJ41013, FLJ41793 | ||||
| Alpha fetoprotein | AFP | < −1 | < −1 | < −1 |
| Alpha-1-fetoprotein, Alpha-fetoglobulin, Alpha-fetoprotein, FETA, HPAFP | ||||
| Spectrin repeat containing nuclear envelope protein 1 | SYNE1 | < −1 | < −1 | < −1 |
| 8B, ARCA1, C6orf98, CPG2, dJ45H2.2, DKFZp781J13156, EDMD4, enaptin, Enaptin, FLJ30878, FLJ41140, KIAA0796, KIAA1262, KIAA1756, MYNE1, Myne-1, Myocyte nuclear envelope protein 1, nesprin-1, Nesprin-1, Nuclear envelope spectrin repeat protein 1, SCAR8, Synaptic nuclear envelope protein 1, Syne-1, SYNE-1B | ||||
| S100 calcium binding protein A9 | S100A9 | < −1 | < −1 | < −1 |
| 60B8AG, CAGB, Calgranulin-B, Calprotectin L1H subunit, CFAG, CGLB, L1AG, Leukocyte L1 complex heavy chain, LIAG, MAC387, MIF, Migration inhibitory factor-related protein 14, MRP14, MRP-14, NIF, p14, P14, Protein S100-A9, S100 calcium-binding protein A9 | ||||
| Ubiquitin conjugating enzyme E2 N | UBE2N | < −1 | < −1 | < −1 |
| Bendless-like ubiquitin-conjugating enzyme, BLU, MGC131857, MGC8489, Ubc13, UBC13, UbcH-ben, Ubiquitin carrier protein N, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-protein ligase N | ||||
Table B1.
| Condition | Composition |
|---|---|
| Growth medium (expansion media) | MEM + 10% SC-FBS + 1% antibiotics penicillin/streptomycin |
| Differentiation medium (control) | αMEM + 10% SC-FBS + 1% antibiotics penicillin/streptomycin +0.28 nM l-Ascorbic acid 2-phosphate + 1 mM L-Cystein + 100 ng/mL IGF1 + 20 ng/mL TGFB1 + 10 ng/mL IL4 |
| Pure-Mg extract | Differentiation medium + pure extract |
| Mg-10Gd extract | Differentiation medium + pure extract |
| Mg-2Ag extract | Differentiation medium + pure extract |
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Brar H.S., Platt M.O., Sarntinoranont M., Martin P.I., Manuel M.V. Magnesium as a biodegradable and bioabsorbable material for medical implants. JOM. 2009;61(9):31–34. [Google Scholar]
- 2.Pichler K., Kraus T., Martinelli E., Sadoghi P., Musumeci G., Uggowitzer P.J., Weinberg A.M. Cellular reactions to biodegradable magnesium alloys on human growth plate chondrocytes and osteoblasts. Int. Orthop. 2014;38(4):881–889. doi: 10.1007/s00264-013-2163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pichler K., Schmidt B., Fischerauer E.E., Rinner B., Dohr G., Leithner A., Weinberg A.M. Behaviour of human physeal chondro-progenitorcells in early growth plate injury response in vitro. Int. Orthop. 2012;36(9):1961–1966. doi: 10.1007/s00264-012-1578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung R., Foster B.K., Xian C.J. Injury responses and repair mechanisms of the injured growth plate. Front Biosci (Schol Ed) 2011;3:117–125. doi: 10.2741/s137. [DOI] [PubMed] [Google Scholar]
- 5.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 6.Noonan K.J., Hunziker E.B., Nessler J., Buckwalter J.A. Changes in cell, matrix compartment, and fibrillar collagen volumes between growth-plate zones. J. Orthop. Res. 1998;16(4):500–508. doi: 10.1002/jor.1100160416. [DOI] [PubMed] [Google Scholar]
- 7.Temu T.M., Wu K.Y., Gruppuso P.A., Phornphutkul C. The mechanism of ascorbic acid-induced differentiation of ATDC5 chondrogenic cells. Am. J. Physiol. Endocrinol. Metab. 2010;299(2):E325–E334. doi: 10.1152/ajpendo.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancedda R., Descalzi Cancedda F., Castagnola P. Chondrocyte differentiation. Int. Rev. Cytol. 1995;159:265–358. doi: 10.1016/s0074-7696(08)62109-9. [DOI] [PubMed] [Google Scholar]
- 9.Stevens D.A., Williams G.R. Hormone regulation of chondrocyte differentiation and endochondral bone formation. Mol. Cell. Endocrinol. 1999;151(1–2):195–204. doi: 10.1016/s0303-7207(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 10.Nasu M., Takayama S., Umezawa A. Endochondral ossification model system: designed cell fate of human epiphyseal chondrocytes during long‐term implantation. J. Cell. Physiol. 2015;230(6):1376–1388. doi: 10.1002/jcp.24882. [DOI] [PubMed] [Google Scholar]
- 11.Bannister A.J., Oehler T., Wilhelm D., Angel P., Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11(12):2509–2514. [PubMed] [Google Scholar]
- 12.Ishihara T., Kakiya K., Takahashi K., Miwa H., Rokushima M., Yoshinaga T., Tanaka Y., Ito T., Togame H., Takemoto H., Amano M., Iwasaki N., Minami A., Nishimura S.-I. Discovery of novel differentiation markers in the early stage of chondrogenesis by glycoform-focused reverse proteomics and genomics. Biochim. Biophys. Acta Gen. Subj. 2014;1840(1):645–655. doi: 10.1016/j.bbagen.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 13.Rocha B., Calamia V., Mateos J., Fernández-Puente P., Blanco F.J., Ruiz-Romero C. Metabolic labeling of human bone marrow mesenchymal stem cells for the quantitative analysis of their chondrogenic differentiation. J. Proteome Res. 2012;11(11):5350–5361. doi: 10.1021/pr300572r. [DOI] [PubMed] [Google Scholar]
- 14.Çelebi B., Elçin A.E., Elçin Y.M. Proteome analysis of rat bone marrow mesenchymal stem cell differentiation. J. Proteome Res. 2010;9(10):5217–5227. doi: 10.1021/pr100506u. [DOI] [PubMed] [Google Scholar]
- 15.Wang D., Park J.S., Chu J.S., Krakowski A., Luo K., Chen D.J., Li S. Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor beta1 stimulation. J. Biol. Chem. 2004;279(42):43725–43734. doi: 10.1074/jbc.M407368200. [DOI] [PubMed] [Google Scholar]
- 16.Ji Y.H., Ji J.L., Sun F.Y., Zeng Y.Y., He X.H., Zhao J.X., Yu Y., Yu S.H., Wu W. Quantitative proteomics analysis of chondrogenic differentiation of C3H10T1/2 mesenchymal stem cells by iTRAQ labeling coupled with on-line two-dimensional LC/MS/MS. Mol. Cell. Proteomics. 2010;9(3):550–564. doi: 10.1074/mcp.M900243-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De la Fuente A., Mateos J., Lesende-Rodriguez I., Calamia V., Fuentes-Boquete I., de Toro F.J., Arufe M.C., Blanco F.J. Proteome analysis during chondrocyte differentiation in a new chondrogenesis model using human umbilical cord stroma mesenchymal stem cells. Mol. Cell. Proteomics. 2012;11(2) doi: 10.1074/mcp.M111.010496. M111 010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baksh D., Yao R., Tuan R.S. Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cell. 2007;25(6):1384–1392. doi: 10.1634/stemcells.2006-0709. [DOI] [PubMed] [Google Scholar]
- 19.Tsang W.P., Shu Y., Kwok P.L., Zhang F., Lee K.K.H., Tang M.K., Li G., Chan K.M., Chan W.-Y., Wan C. CD146+ human umbilical cord perivascular cells maintain stemness under hypoxia and as a cell source for skeletal regeneration. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0076153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarugaser R., Lickorish D., Baksh D., Hosseini M.M., Davies J.E. Human umbilical cord perivascular (HUCPV) cells: a source of mesenchymal progenitors. Stem Cell. 2005;23(2):220–229. doi: 10.1634/stemcells.2004-0166. [DOI] [PubMed] [Google Scholar]
- 21.Jahn K., Saito H., Taipaleenmaki H., Gasser A., Hort N., Feyerabend F., Schluter H., Rueger J.M., Lehmann W., Willumeit-Romer R., Hesse E. Intramedullary Mg2Ag nails augment callus formation during fracture healing in mice. Acta Biomater. 2016;36:350–360. doi: 10.1016/j.actbio.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z., Schade R., Luthringer B., Hort N., Rothe H., Muller S., Liefeith K., Willumeit-Romer R., Feyerabend F. Influence of the microstructure and silver content on degradation, cytocompatibility, and antibacterial properties of magnesium-silver alloys in vitro. Oxid Med Cell Longev. 2017;2017:8091265. doi: 10.1155/2017/8091265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad Agha N., Willumeit-Romer R., Laipple D., Luthringer B., Feyerabend F. The degradation interface of magnesium based alloys in direct contact with human primary osteoblast cells. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0157874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myrissa A., Agha N.A., Lu Y., Martinelli E., Eichler J., Szakács G., Kleinhans C., Willumeit-Römer R., Schäfer U., Weinberg A.-M. In vitro and in vivo comparison of binary Mg alloys and pure Mg. Mater. Sci. Eng. C. 2016;61:865–874. doi: 10.1016/j.msec.2015.12.064. [DOI] [PubMed] [Google Scholar]
- 25.I. 10993-5:2009 . 2009. Biological Evaluation of Medical Devices -- Part 5: Tests for in Vitro Cytotoxicity. [Google Scholar]
- 26.I. 10993-12:2012 . 2012. Biological Evaluation of Medical Devices -- Part 12: Sample Preparation and Reference Materials. [Google Scholar]
- 27.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Molecular & Cellular Proteomics; 2014. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 29.Massa A., Perut F., Chano T., Woloszyk A., Mitsiadis T.A., Avnet S., Baldini N. The effect of extracellular acidosis on the behaviour of mesenchymal stem cells in vitro. Eur. Cells Mater. 2017;33:252–267. doi: 10.22203/eCM.v033a19. [DOI] [PubMed] [Google Scholar]
- 30.Arnett T.R. Extracellular pH regulates bone cell function. J. Nutr. 2008;138(2):415S–418S. doi: 10.1093/jn/138.2.415S. [DOI] [PubMed] [Google Scholar]
- 31.Moghadam F.H., Tayebi T., Dehghan M., Eslami G., Nadri H., Moradi A., Vahedian-Ardakani H., Barzegar K. Differentiation of bone marrow mesenchymal stem cells into chondrocytes after short term culture in alkaline medium. Int. J. Hematol. Oncol. Stem Cell Res. 2014;8(4):12–19. [PMC free article] [PubMed] [Google Scholar]
- 32.Loeser R.F. Chondrocyte integrin expression and function. Biorheology. 2000;37(1–2):109–116. [PubMed] [Google Scholar]
- 33.Ko Y.H., Hong S., Pedersen P.L. Chemical mechanism of ATP synthase: magnesium plays a pivotal role in formation of the transition state where ATP is synthesized from ADP and inorganic phosphate. J. Biol. Chem. 1999;274(41):28853–28856. doi: 10.1074/jbc.274.41.28853. [DOI] [PubMed] [Google Scholar]
- 34.Shoshan-Barmatz V., Golan M. Mitochondrial VDAC1: function in cell life and death and a target for cancer therapy. Curr. Med. Chem. 2012;19(5):714–735. doi: 10.2174/092986712798992110. [DOI] [PubMed] [Google Scholar]
- 35.Huttemann M., Helling S., Sanderson T.H., Sinkler C., Samavati L., Mahapatra G., Varughese A., Lu G., Liu J., Ramzan R., Vogt S., Grossman L.I., Doan J.W., Marcus K., Lee I. Regulation of mitochondrial respiration and apoptosis through cell signaling: cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim. Biophys. Acta. 2012;1817(4):598–609. doi: 10.1016/j.bbabio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papachristou D.J., Blair H.C. Bone and high-density lipoprotein: the beginning of a beautiful friendship. World J. Orthoped. 2016;7(2):74–77. doi: 10.5312/wjo.v7.i2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.I.E. Triantaphyllidou, E. Kalyvioti, E. Karavia, I. Lilis, K.E. Kypreos, D.J. Papachristou, Perturbations in the HDL metabolic pathway predispose to the development of osteoarthritis in mice following long-term exposure to western-type diet, Osteoarthritis Cartilage 21(2) 322-330. [DOI] [PubMed]
- 38.Kozhemyakina E., Lassar A.B., Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142(5):817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies M.E., Dingle J.T., Pigott R., Power C., Sharma H. Expression of intercellular adhesion molecule 1 (Icam-1) on human articular cartilage chondrocytes. Connect. Tissue Res. 1991;26(3):207–216. doi: 10.3109/03008209109152439. [DOI] [PubMed] [Google Scholar]
- 40.Zreiqat H., Howlett C.R., Zannettino A., Evans P., Schulze-Tanzil G., Knabe C., Shakibaei M. Mechanisms of magnesium-stimulated adhesion of osteoblastic cells to commonly used orthopaedic implants. J. Biomed. Mater. Res. 2002;62(2):175–184. doi: 10.1002/jbm.10270. [DOI] [PubMed] [Google Scholar]
- 41.Kulyk W.M., Upholt W.B., Kosher R.A. Fibronectin gene expression during limb cartilage differentiation. Development. 1989;106(3):449–455. doi: 10.1242/dev.106.3.449. [DOI] [PubMed] [Google Scholar]
- 42.Cui J., Liu Y., Matumoto R., Uemura T. Highly expression and biological function of type VI collagen on the early events of chondrogenesis in human mesenchymal stem cells. J. Oral Tissue Eng. 2013;11(1):29–41. [Google Scholar]
- 43.Klamer S.E., Kuijk C.G.M., Hordijk P.L., van der Schoot C.E., von Lindern M., van Hennik P.B., Voermans C. BIGH3 modulates adhesion and migration of hematopoietic stem and progenitor cells. Cell Adhes. Migrat. 2013;7(5):434–449. doi: 10.4161/cam.26596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y.I., Ryu J.-S., Yeo J.E., Choi Y.J., Kim Y.S., Ko K., Koh Y.-G. Overexpression of TGF-β1 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cells. Biochem. Biophys. Res. Commun. 2014;450(4):1593–1599. doi: 10.1016/j.bbrc.2014.07.045. [DOI] [PubMed] [Google Scholar]
- 45.Imabuchi R., Ohmiya Y., Joon Kwon H., Onodera S., Kitamura N., Kurokawa T., Ping Gong J., Yasuda K. Gene expression profile of the cartilage tissue spontaneously regenerated in vivo by using a novel double-network gel: comparisons with the normal articular cartilage. BMC Muscoskelet. Disord. 2011;12 doi: 10.1186/1471-2474-12-213. 213-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hankenson K.D., Dishowitz M., Gray C., Schenker M. Angiogenesis in bone regeneration. Injury. 2011;42(6):556–561. doi: 10.1016/j.injury.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maes C., Carmeliet P., Moermans K., Stockmans I., Smets N., Collen D., Bouillon R., Carmeliet G. Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Mech. Dev. 2002;111(1–2):61–73. doi: 10.1016/s0925-4773(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 48.Alini M., Marriott A., Chen T., Abe S., Poole A.R. A novel angiogenic molecule produced at the time of chondrocyte hypertrophy during endochondral bone formation. Dev. Biol. 1996;176(1):124–132. doi: 10.1006/dbio.1996.9989. [DOI] [PubMed] [Google Scholar]
- 49.Carlevaro M.F., Albini A., Ribatti D., Gentili C., Benelli R., Cermelli S., Cancedda R., Cancedda F.D. Transferrin promotes endothelial cell migration and invasion: implication in cartilage neovascularization. J. Cell Biol. 1997;136(6):1375–1384. doi: 10.1083/jcb.136.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vu T.H., Shipley J.M., Bergers G., Berger J.E., Helms J.A., Hanahan D., Shapiro S.D., Senior R.M., Werb Z. MMP-9/Gelatinase B is a key regulator of growth plate Angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93(3):411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukunami C., Iyama K., Inoue H., Hiraki Y. Spatiotemporal pattern of the mouse chondromodulin-I gene expression and its regulatory role in vascular invasion into cartilage during endochondral bone formation. Int. J. Dev. Biol. 1999;43(1):39–49. [PubMed] [Google Scholar]
- 52.Moses M.A., Wiederschain D., Wu I., Fernandez C.A., Ghazizadeh V., Lane W.S., Flynn E., Sytkowski A., Tao T., Langer R. Troponin I is present in human cartilage and inhibits angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 1999;96(6):2645–2650. doi: 10.1073/pnas.96.6.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norbert S. Angiogenesis in osteoarthritis. Curr. Rheumatol. Rev. 2008;4(3):206–209. [Google Scholar]
- 54.Salari P., Abdollahi M. Controversial effects of non-steroidal anti-inflammatory drugs on bone: a review. Inflamm. Allergy - Drug Targets. 2009;8(3):169–175. doi: 10.2174/187152809788681065. [DOI] [PubMed] [Google Scholar]
- 55.Welting T.J., Caron M.M., Emans P.J., Janssen M.P., Sanen K., Coolsen M.M., Voss L., Surtel D.A., Cremers A., Voncken J.W., van Rhijn L.W. Inhibition of cyclooxygenase-2 impacts chondrocyte hypertrophic differentiation during endochondral ossification. Eur. Cells Mater. 2011;22:420–436. doi: 10.22203/ecm.v022a31. ; discussion 436-7. [DOI] [PubMed] [Google Scholar]
- 56.Bernfield M., Gotte M., Park P.W., Reizes O., Fitzgerald M.L., Lincecum J., Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 57.Ahrens M.J., Li Y., Jiang H., Dudley A.T. Convergent extension movements in growth plate chondrocytes require gpi-anchored cell surface proteins. Development (Camb.) 2009;136(20):3463–3474. doi: 10.1242/dev.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piróg K.A., Irman A., Young S., Halai P., Bell P.A., Boot-Handford R.P., Briggs M.D. Abnormal chondrocyte apoptosis in the cartilage growth plate is influenced by genetic background and deletion of CHOP in a targeted mouse model of pseudoachondroplasia. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0085145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y.-J., Kukita A., Teramachi J., Nagata K., Wu Z., Akamine A., Kukita T. A possible suppressive role of galectin-3 in upregulated osteoclastogenesis accompanying adjuvant-induced arthritis in rats. Lab. Invest. 2008;89(1):26–37. doi: 10.1038/labinvest.2008.111. [DOI] [PubMed] [Google Scholar]
- 60.Santoro A., Conde J., Scotece M., Abella V., Lois A., Lopez V., Pino J., Gomez R., Gomez-Reino J.J., Gualillo O. SERPINE2 inhibits IL-1α-induced MMP-13 expression in human chondrocytes: involvement of ERK/NF-κB/AP-1 pathways. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hata K., Nishimura R., Muramatsu S., Matsuda A., Matsubara T., Amano K., Ikeda F., Harley V.R., Yoneda T. Paraspeckle protein p54(nrb) links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J. Clin. Investig. 2008;118(9):3098–3108. doi: 10.1172/JCI31373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sekiya I., Vuoristo J.T., Larson B.L., Prockop D.J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc. Natl. Acad. Sci. U.S.A. 2002;99(7):4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hochepied T., Berger F.G., Baumann H., Libert C. Alpha(1)-Acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003;14(1):25–34. doi: 10.1016/s1359-6101(02)00054-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.